Definition of MHC Supertypes Through Clustering of MHC Peptide-Binding Repertoires
|
|
|
- Rodger Burke
- 5 years ago
- Views:
Transcription
1 11 Definition of MHC Supertypes Through Clustering of MHC Peptide-Binding Repertoires Pedro A. Reche and Ellis L. Reinherz Summary Identification of peptides that can bind to major histocompatibility complex (MHC) molecules is important for anticipation of T-cell epitopes and for the design of epitope-based vaccines. Population coverage of epitope vaccines is, however, compromised by the extreme polymorphism of MHC molecules, which is in fact the basis for their differential peptide binding. Therefore, grouping of MHC molecules into supertypes according to peptide-binding specificity is relevant for optimizing the composition of epitope-based vaccines. Despite the fact that the peptidebinding specificity of MHC molecules is linked to their specific amino acid sequences, it is unclear how amino sequence differences correlate with peptide-binding specificities. In this chapter, we detail a method for defining MHC supertypes based on the analysis and subsequent clustering of their peptide-binding repertoires Key Words: MHC; supertypes; clustering; peptide-binding repertoire 1. Introduction Major histocompatibility complex (MHC) molecules play a key role in the immune system by capturing peptide antigens for display on cell surfaces. Subsequently, these peptide MHC (pmhc) complexes are recognized by T cells through their T-cell receptors (TCRs). MHC molecules fall into two Address for correspondence: Department of Immunology, Faculated de Medicina, Universidad Complutense de Madrid, Ave Complutense S/N Madrid, 28040, SPAIN. TL: ; FX: ; parecheg@med.ucm.es From: Methods in Molecular Biology, vol. 409: Immunoinformatics: Predicting Immunogenicity In Silico Edited by: D. R. Flower Humana Press Inc., Totowa, NJ 163
2 164 Reche and Reinherz major classes, MHC class I (MHCI) and MHC class II (MHCII). Antigens presented by MHCI and MHCII are recognized by two distinct sets of T cells, CD8 + T and CD4 + T cells, respectively (1). Because T-cell recognition is limited to those peptides presented by MHC molecules, prediction of peptides that can bind to MHC molecules is important for anticipating T-cell epitopes and designing epitope-based vaccines (2 4). Furthermore, the availability of computational methods that can readily identify potential epitopes from primary protein sequences has fueled a new epitope discovery-driven paradigm in vaccine development. A major complication to the development of epitope-based vaccines is the extreme polymorphism of the MHC molecules. In the human, MHC molecules are known as human leukocyte antigens (HLAs), and there are hundreds of allelic variants of class I (HLA I) and class II (HLA II) molecules. These HLA allelic variants bind distinct sets of peptides (5) and are expressed at vastly variable frequencies in different ethnic groups (6). Consequently, the potential population coverage of epitope-based vaccines is greatly compromised. Interestingly, it has been noted that some HLA molecules can bind largely overlapping sets of peptides (7,8). Therefore, grouping of MHC molecules into supertypes according to peptide-binding specificity is of relevance for the formulation of epitope vaccines providing a wide population coverage. The first supertypes were defined by Sidney, Sette, and co-workers (7,8) (hereafter Sidney Sette et al.) upon inspection of the reported peptide-binding motifs of individual HLA alleles. However, the relationships between peptidebinding specificities of HLA molecules may be too subtle to be defined by visual inspection of these peptide-binding motifs. Furthermore, such sequence patterns have proven to be too simple to describe the binding ability of a peptide to a given MHC molecule (9,10). In view of these limitations, we developed an alternative method to define MHC supertypes by clustering the peptide-binding repertoire of MHC molecules. The core of the method consists of the generation of a distance matrix whose coefficients are inversely proportional to the peptide binders shared by any two MHC molecules. Subsequently, this distance matrix is fed to a phylogenic clustering algorithm to establish the kinship among the distinct MHC peptide-binding repertoires. The peptide-binding repertoire of any given MHC molecule is unknown, and thereby, defining supertypes through this method requires the estimation of the peptide-binding repertoire of MHC molecules. In this chapter, we will use position-specific scoring matrices (PSSMs) as the predictor of peptide MHC binding (11,12) and describe in detail the generation of supertypes using, for example, a selection of HLA class I (HLA I) molecules for which PSSMs are readily available.
3 Definition of MHC Supertypes Materials 2.1. Prediction of Peptide MHC Binding Repertoires We consider the peptide-binding repertoire of any MHC molecule as the subset of peptides predicted to bind from a reference set consisting of a random protein of 1,000 amino acids. A selection of public online resources that can be used for the prediction of peptide MHC binding is summarized in Table 1. In our study PSSMs derived from aligned MHC ligands as the predictors of peptide MHC binding (11,12). In this approach, the binding potential of any peptide sequence (query) to the MHC molecule is determined by its similarity to a set of known peptide MHC binders and can be obtained by comparing the query to the PSSM. Prediction of peptide MHC binding is threshold-dependent, and here we use the same threshold for all MHC molecules. Thus, the size of the peptide-binding repertoire of all MHC molecules is considered to be same (same number of peptides) Supertype Construction MHC supertypes are derived following the general scheme illustrated in Fig. 1. First, the overlap between the predicted peptide-binding repertoires (see Section 2.1) of any two MHC molecules, pmhc i and pmhc j, is computed as the number of peptide binders shared by the two molecules. Let that number be n ij. Subsequently, a distance coefficient (d ij ) is defined as follows: d ij = N n ij (1) where N is the size of the peptide-binding repertoire of the MHC molecule. Thus, if the peptide-binding repertoire between two MHC molecules is identical, then d ij = 0. Alternatively, if they share no peptides in common, d ij will match the size of the binding repertoire, N. Through the repetition of this process over all distinct pairs of MHC molecules, a quadratic distance matrix is derived containing the d ij coefficients for all distinct pairs of MHC molecules. Once the distance matrix is obtained, we use the Phylogeny Inference Package (PHYLIP; evolution.genetics.washington.edu/phylip.html) (13) to generate a phylogenic tree where the MHC molecules appear clustered according to their peptide-binding specificity. Specifically, within the PHYLIP package one must use applications such as kitsch and neighbor that take distance matrices as input. The kitsch application uses a Fitch Margoliash criterion and assumes an evolutionary clock (14). On the other hand, the neighbor application uses the popular neighbor-joining method to derive an unrooted tree without the assumption of a
4 Table 1 Public online resources for the prediction of peptide major histocompatibility complex (MHC) binding Name URL Method Class Reference HLABIND QM I (22) MHCpred QM I and II (23) PROPRED QM II (24) SVMHC SVM I (25) SYFPEITHI EpitopePrediction.htm Motif matrix I and II (26) RANKPEP Motif PSSM I and II (11) NetMHC ANN I (27) PREDEP Structure based I (28) ANN, Artificial Neural Network; PSSM, position-specific scoring matrix; QM, quantitative matrix.
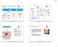
5 Definition of MHC Supertypes 167 Fig. 1. Strategy to define major histocompatibility complex (MHC) supertypes. MHC supertypes are identified as follows: (1) estimate number of common peptides, n ij, between the binding repertoires of any two MHC molecules, pmhc i and pmhc j ; (2) obtain a distance matrix whose coefficients, d ij, are inversely proportional to the peptide-binding overlap between any pair of MHC molecules; and (3) derive a dendrogram using a phylogenic clustering algorithm to visualize MHC supertypes (groups of MHC molecules with similar peptide-binding specificity). N is the size of the peptide-binding repertoire of the MHC molecule. clock (15). For instance, to generate a tree using the neighbor-joining algorithm method one can use the command: echo Y neighbor > /dev/null This command will generate a tree from a distance matrix that must be named as infile using the default options of the neighbor application. Likewise, one may use similar commands to generate trees using other applications. In any case, these applications will generate two files, one named outfile displaying the tree and another named treefile describing the tree in NEWICK format, which can be used to visualize and manipulate the tree using third party applications such as TREEVIEW ( taxonomy.zoology.gla.ac.uk/rod/treeview.html). 3. Methods 3.1. HLA I Supertypes Definition of MHC supertypes using the method described here requires the estimation of the peptide-binding repertoire of the MHC molecules using predictors of peptide MHC binding. The prediction of peptide MHCII binding is generally less reliable than that of peptide MHCI binding (12). Therefore, to illustrate the definition of MHC supertypes, we focused on 55 HLA I molecules
6 168 Reche and Reinherz (human MHCI) for which we can readily predict their peptide-binding repertoires using PSSMs (see Section 2). Given that MHCI ligands are usually nine residues in length, we selected PSSMs for the prediction of binders of that same size (nine residues). In previous studies we have shown that depending on the specific MHCI molecule, the accuracy of peptide MHCI binding predictions is optimal by considering as binders the top 2 5% scoring peptides (2 5% threshold) within a protein query (12). Here we have estimated the peptidebinding repertoire of the selected HLA I molecules using a 2% threshold. Thus, following the method described above with a Fitch and Margoliash clustering algorithm (14) (Section 2.2; kitsch application), we generated the phylogenic tree, which is shown in Fig. 2. In this tree, HLA I molecules with similar peptide-binding specificity (large overlap in their peptide-binding repertoires) branch together in groups or supertypes. The relationship between the peptidebinding specificities of HLA I molecules is extensive, and although affinities are mostly confined to alleles belonging to the same gene, they also reach to alleles belonging to different genes (Fig. 2, B15 cluster; B 4002 and A 2902; and A 2402 and B 3801). We clearly identified the classic A2, A3, B7, B27, and B44 supertypes previously defined by Sidney Sette et al., as well as three new potential supertypes, BX, AB, and B57 (Fig. 2). Furthermore, this analysis indicates that classic HLA I supertypes may be larger than that previously thought. For instance, the A2 supertype would also include the A0207, A0209, and A0214, and the A3 supertype will also include A Combined Phenotypic Frequency of HLA I Supertypes HLA I-restricted peptides are the targets of CD8 + cytotoxic T lymphocytes (CTLs). The population protection coverage (PPC) of a vaccine composed of CTL epitopes is given by the combined phenotypic frequency (CPF) of the HLA I molecules restricting the epitopes, and it can be computed from the gene and haplotype frequencies (16). Using the allelic and haplotype frequencies reported by Cao et al. (17) corresponding to five major American ethnic groups (Black, Caucasian, Hispanic, Native American, and Asian), we have computed the CPF for the HLA I supertypes defined in the previous section (Section 3.1), and the values are tabulated in Table 2. Targeting HLA I supertypes for the prediction of promiscuous peptide binders allows to minimize the total number of predicted epitopes without compromising the population coverage required in the design of multi-epitope vaccines. However, including many distantly related HLA I molecules in the supertypes may result in too few or no epitopes predicted to bind to all the alleles included in the supertype. Therefore, for the CPF calculations, we have limited the composition of HLA I supertypes to
7 Definition of MHC Supertypes 169 Fig. 2. Human leukocyte antigen (HLA) I supertypes. This figure shows an unroot dendrogram reflecting the relationships between the peptide-binding specificities of HLA I molecules. The closer the HLA I alleles branch, the larger the overlap between their peptide-binding repertoires. Groups of HLA I alleles with similar peptide-binding specificities branch together defining supertypes (shaded groups). include only those HLA I alleles with 20% peptide-binding overlap (pairwise between any pair of alleles). The A2, A3, and B7 supertypes have the largest CPF in the five studied ethnic groups, providing a CPF close to 90%, regardless of ethnicity. To increase the CPF to 95% in all ethnicities, it is necessary to include at least two more supertypes. Specifically, the supertypes A2, A3, B7, B15, and A24 or B44 represent the minimal supertypic combination providing a CPF 95%. These results indicate that as few as five epitopes restricted by the mentioned HLA I supertypes may be enough to develop a vaccine eliciting CTL responses in the whole population, regardless of ethnicity.
8 Table 2 Cumulative phenotype frequency of defined supertypes Supertype Alleles Blacks (%) Caucasians (%) Hispanics (%) N.A. Natives (%) Asians (%) A2 A , A A3 A 0301, A 1101, A 3101, A 3301, A 6801, A 6601 B7 B 0702, B 3501, B , B 5301, B 5401 B15 A 0101, B 1501_B62, B1502 A24 A 2402, B B44 B 4402, B B57 B , B5801, B 1503 ABX A 2902, B B27 B , B 2709, B 3909 BX B 1509, B 1510, B AB A 2902, B N.A., North American.
9 Definition of MHC Supertypes Conclusions HLA molecules are represented by hundreds of allelic variants displaying distinct peptide-binding specificities, and grouping them into supertypes is relevant for developing epitope-based vaccines with a wide PPC. The peptidebinding specificity of HLA molecules stems from the specific amino acids lining their binding groove, and consequently, supertypes may be defined from structural analysis (18 20). However, it is not always clear how amino acid sequence differences among HLA molecules translate into distinct peptide-binding specificities. Indeed, structure-based methods for the prediction of peptide MHC binding are still in their infancy. Therefore, in thischapter, we described a method for defining HLA supertypes based on the analysis and subsequent clustering of their predicted peptide-binding repertoires. Furthermore, we have shown that the method can identify experimentally defined HLA I supertypes, suggesting in addition new potential relationships between the peptide-binding specificity of HLA I molecules. When the predictor of peptide MHC binding is a specificity matrix such as a PSSM, clustering of the HLA molecules according to peptide-binding specificity may alternatively be achieved by comparison of the matrix coefficients (21). However, it is important to stress that the clustering method described here to derive supertypes can be applied in combination with any predictor of peptide MHC binding. Although, not indicated in this chapter, minor differences in the defined supertypes appear depending on the phylogenic algorithm used to cluster the HLA I molecules. There are also two other limitations to the method described here. First, the method is limited by both the quality and availability of the peptide MHC binding predictors. Thus, we do not discard the possibility that the fine structure of the supertypes may suffer some changes as new and better predictors of peptide MHC binding develop. The second limitation is that we have considered the size of the peptide-binding repertoire of all MHC molecules to be the same. However, that might not always be the case. Indeed, it has been noted that, for instance, the A 0201 appears to be quite promiscuous, binding larger sets of peptides than the other HLA I molecules (Azouz, Reinhold, and Reinherz, unpublished results). References 1. Margulies, D.H Interactions of TCRs with MHC-peptide complexes: a quantitative basis for mechanistic models. Curr Opin Immunol 9: Yu, K., Petrovsky, N., Schonbach, C., Koh, J.Y., and Brusic, V Methods for prediction of peptide binding to MHC molecules: a comparative study. Mol Med 8:
10 172 Reche and Reinherz 3. Flower, D Towards in silico prediction of immunogenic epitopes. Trends Immunol 24: Flower, D., and Doytchinova, I.A Immunoinformatics and the prediction of immunogenicity. Appl Bioinformatics 1: Reche, P.A., and Reinherz, E.L Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J Mol Biol 331: David W. Gjertson, and Paul I. Terasaki, E. (Eds) HLA American Society for Histocompatibility and Immunogenetics, Lenexa. 7. Sette, A., and Sidney, J Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50: Sette, A., and Sidney, J HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr Opin Immunol 10: Bouvier, M., and Wiley, D.C Importance of peptide amino acid and carboxyl termini to the stability of MHC class I molecules. Science 265: Ruppert, J., Sidney, J., Celis, E., Kubo, T., Grey, H.M., and Sette, A Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell 74: Reche, P.A., Glutting, J.-P., and Reinherz, E.L Prediction of MHC class I binding peptides using profile motifs. Hum Immunol 63: Reche, P.A., Glutting, J.-P, Zhang, H., and Reinherz, E.L Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics 56: Retief, J.D Phylogenetic analysis using PHYLIP. Methods Mol Biol 132: Fitch, W.M., and Margoliash, E Construction of phylogenetic trees. Science 155: Saitou, N., and Nei, M The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: Dawson, D.V., Ozgur, M., Sari, K., Ghanayem, M., and Kostyu, D.D Ramifications of HLA class I polymorphism and population genetics for vaccine development. Genet Epidemiol 20: Cao, K., Hollenbach, J., Shi, X., Shi, W., Chopek, M., and Fernandez-Vina, M.A Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol 62: Doytchinova, I.A., Guan, P., and Flower, D.R Quantitative structure-activity relationships and the prediction of MHC supermotifs. Methods 34: Doytchinova, I.A., and Flower, D.R In silico identification of supertypes for class II MHCs. J Immunol 174:
11 Definition of MHC Supertypes Doytchinova, I.A., Guan, P., and Flower, D.R Identifying human MHC supertypes using bioinformatic methods. J Immunol 172: Lund, O., Nielsen, M., Kesmir, C., Petersen, A.G., Lundegaard, C., Worning, P., Sylvester-Hvid, C., Lamberth, K., Roder, G., Justesen, S., Buus, S., and Brunak, S Definition of supertypes for HLA molecules using clustering of specificitymatrices. Immunogenetics 55: Parker, K.C., Bednarek, M.A., and Coligan, J.E Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side chains. J Immunol 152: Guan, P., Doytchinova, I.A., Zygouri, C., and Flower, D MHCPred: a server for quantitative prediction of peptide-mhc binding. Nucleic Acids Res 31: Singh, H., and Raghava, G.P ProPred: prediction of HLA-DR binding sites. Bioinformatics 17: Donnes, P., and Elofsson, A Prediction of MHC class I binding peptides, using SVMHC. BMC Bioinformatics 3: Rammensee, H.G., Bachmann, J., Emmerich, N.P.N., Bacho, O.A., and Stevanovic, S SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50: Buus, S., Lauemoller, S.L., Worning, P., Kesmir, C., Frimurer, T., Corbet, S., Fomsgaard, A., Hilden, J., Holm, A., and Brunak, S Sensitive quantitative predictions of peptide-mhc binding by a Query by Committee artificial neural network approach. Tissue Antigens 62: Altuvia, Y., Sette, A., Sidney, J., Southwood, S., and Margalit, H A structure based algorithm to predict potential binding peptides to MHC molecules with hydrophobic binding pockets. Hum Immunol 58:1 11.
Definition of MHC supertypes through clustering of MHC peptide binding repertoires
 Definition of MHC supertypes through clustering of MHC peptide binding repertoires Pedro A. Reche and Ellis L. Reinherz Laboratory of Immunobiology and Department of Medical Oncology, Dana-Farber Cancer
Definition of MHC supertypes through clustering of MHC peptide binding repertoires Pedro A. Reche and Ellis L. Reinherz Laboratory of Immunobiology and Department of Medical Oncology, Dana-Farber Cancer
PEPVAC: a web server for multi-epitope vaccine development based on the prediction of supertypic MHC ligands
 W138 W142 doi:10.1093/nar/gki357 PEPVAC: a web server for multi-epitope vaccine development based on the prediction of supertypic MHC ligands Pedro A. Reche 1,2, * and Ellis L. Reinherz 1,2 1 Laboratory
W138 W142 doi:10.1093/nar/gki357 PEPVAC: a web server for multi-epitope vaccine development based on the prediction of supertypic MHC ligands Pedro A. Reche 1,2, * and Ellis L. Reinherz 1,2 1 Laboratory
NetMHCpan, a Method for Quantitative Predictions of Peptide Binding to Any HLA-A and -B Locus Protein of Known Sequence
 NetMHCpan, a Method for Quantitative Predictions of Peptide Binding to Any HLA-A and -B Locus Protein of Known Sequence Morten Nielsen 1 *, Claus Lundegaard 1, Thomas Blicher 1, Kasper Lamberth 2, Mikkel
NetMHCpan, a Method for Quantitative Predictions of Peptide Binding to Any HLA-A and -B Locus Protein of Known Sequence Morten Nielsen 1 *, Claus Lundegaard 1, Thomas Blicher 1, Kasper Lamberth 2, Mikkel
Eur. J. Immunol : Antigen processing 2295
 Eur. J. Immunol. 2005. 35: 2295 2303 Antigen processing 2295 Antigen processing An integrative approach to CTL epitope prediction: A combined algorithm integrating MHC class I binding, TAP transport efficiency,
Eur. J. Immunol. 2005. 35: 2295 2303 Antigen processing 2295 Antigen processing An integrative approach to CTL epitope prediction: A combined algorithm integrating MHC class I binding, TAP transport efficiency,
A HLA-DRB supertype chart with potential overlapping peptide binding function
 A HLA-DRB supertype chart with potential overlapping peptide binding function Arumugam Mohanapriya 1,2, Satish Nandagond 1, Paul Shapshak 3, Uma Kangueane 1, Pandjassarame Kangueane 1, 2 * 1 Biomedical
A HLA-DRB supertype chart with potential overlapping peptide binding function Arumugam Mohanapriya 1,2, Satish Nandagond 1, Paul Shapshak 3, Uma Kangueane 1, Pandjassarame Kangueane 1, 2 * 1 Biomedical
CS229 Final Project Report. Predicting Epitopes for MHC Molecules
 CS229 Final Project Report Predicting Epitopes for MHC Molecules Xueheng Zhao, Shanshan Tuo Biomedical informatics program Stanford University Abstract Major Histocompatibility Complex (MHC) plays a key
CS229 Final Project Report Predicting Epitopes for MHC Molecules Xueheng Zhao, Shanshan Tuo Biomedical informatics program Stanford University Abstract Major Histocompatibility Complex (MHC) plays a key
NIH Public Access Author Manuscript Immunogenetics. Author manuscript; available in PMC 2014 September 01.
 NIH Public Access Author Manuscript Published in final edited form as: Immunogenetics. 2013 September ; 65(9): 655 665. doi:10.1007/s00251-013-0714-9. MHCcluster, a method for functional clustering of
NIH Public Access Author Manuscript Published in final edited form as: Immunogenetics. 2013 September ; 65(9): 655 665. doi:10.1007/s00251-013-0714-9. MHCcluster, a method for functional clustering of
Predicting Protein-Peptide Binding Affinity by Learning Peptide-Peptide Distance Functions
 Predicting Protein-Peptide Binding Affinity by Learning Peptide-Peptide Distance Functions Chen Yanover and Tomer Hertz,2 School of Computer Science and Engineering 2 The Center for Neural Computation,
Predicting Protein-Peptide Binding Affinity by Learning Peptide-Peptide Distance Functions Chen Yanover and Tomer Hertz,2 School of Computer Science and Engineering 2 The Center for Neural Computation,
The Immune Epitope Database Analysis Resource: MHC class I peptide binding predictions. Edita Karosiene, Ph.D.
 The Immune Epitope Database Analysis Resource: MHC class I peptide binding predictions Edita Karosiene, Ph.D. edita@liai.org IEDB Workshop October 29, 2015 Outline Introduction MHC-I peptide binding prediction
The Immune Epitope Database Analysis Resource: MHC class I peptide binding predictions Edita Karosiene, Ph.D. edita@liai.org IEDB Workshop October 29, 2015 Outline Introduction MHC-I peptide binding prediction
Use of BONSAI decision trees for the identification of potential MHC Class I peptide epitope motifs.
 Use of BONSAI decision trees for the identification of potential MHC Class I peptide epitope motifs. C.J. SAVOIE, N. KAMIKAWAJI, T. SASAZUKI Dept. of Genetics, Medical Institute of Bioregulation, Kyushu
Use of BONSAI decision trees for the identification of potential MHC Class I peptide epitope motifs. C.J. SAVOIE, N. KAMIKAWAJI, T. SASAZUKI Dept. of Genetics, Medical Institute of Bioregulation, Kyushu
Prediction of MHC-Peptide Binding: A Systematic and Comprehensive Overview
 Current Pharmaceutical Design, 2009, 15, 3209-3220 3209 Prediction of MHC-Peptide Binding: A Systematic and Comprehensive Overview Esther M. Lafuente and Pedro A. Reche * Laboratory of ImmuneMedicine,
Current Pharmaceutical Design, 2009, 15, 3209-3220 3209 Prediction of MHC-Peptide Binding: A Systematic and Comprehensive Overview Esther M. Lafuente and Pedro A. Reche * Laboratory of ImmuneMedicine,
Profiling HLA motifs by large scale peptide sequencing Agilent Innovators Tour David K. Crockett ARUP Laboratories February 10, 2009
 Profiling HLA motifs by large scale peptide sequencing 2009 Agilent Innovators Tour David K. Crockett ARUP Laboratories February 10, 2009 HLA Background The human leukocyte antigen system (HLA) is the
Profiling HLA motifs by large scale peptide sequencing 2009 Agilent Innovators Tour David K. Crockett ARUP Laboratories February 10, 2009 HLA Background The human leukocyte antigen system (HLA) is the
ProPred1: prediction of promiscuous MHC Class-I binding sites. Harpreet Singh and G.P.S. Raghava
 BIOINFORMATICS Vol. 19 no. 8 2003, pages 1009 1014 DOI: 10.1093/bioinformatics/btg108 ProPred1: prediction of promiscuous MHC Class-I binding sites Harpreet Singh and G.P.S. Raghava Institute of Microbial
BIOINFORMATICS Vol. 19 no. 8 2003, pages 1009 1014 DOI: 10.1093/bioinformatics/btg108 ProPred1: prediction of promiscuous MHC Class-I binding sites Harpreet Singh and G.P.S. Raghava Institute of Microbial
Selection of epitope-based vaccine targets of HCV genotype 1 of Asian origin: a systematic in silico approach
 www.bioinformation.net Hypothesis Volume 8(20) Selection of epitope-based vaccine targets of HCV genotype 1 of Asian origin: a systematic in silico approach Abida Shehzadi 1 *, Shahid Ur Rehman 1, 2 &
www.bioinformation.net Hypothesis Volume 8(20) Selection of epitope-based vaccine targets of HCV genotype 1 of Asian origin: a systematic in silico approach Abida Shehzadi 1 *, Shahid Ur Rehman 1, 2 &
Vaccine Design: A Statisticans Overview
 GoBack : A Statisticans Overview. Surajit Ray sray@samsi.info Surajit Ray Samsi PostDoc Seminar: Nov 2: 2004 - slide #1 The Chinese are credited with making the observation that deliberately infecting
GoBack : A Statisticans Overview. Surajit Ray sray@samsi.info Surajit Ray Samsi PostDoc Seminar: Nov 2: 2004 - slide #1 The Chinese are credited with making the observation that deliberately infecting
IMMUNOINFORMATICS: Bioinformatics Challenges in Immunology
 Bioinformatics 1 -- Lecture 22 IMMUNOINFORMATICS: Bioinformatics Challenges in Immunology Most slides courtesy of Julia Ponomarenko, San Diego Supercomputer Center or Oliver Kohlbacher, WSI/ZBIT, Eberhard-Karls-
Bioinformatics 1 -- Lecture 22 IMMUNOINFORMATICS: Bioinformatics Challenges in Immunology Most slides courtesy of Julia Ponomarenko, San Diego Supercomputer Center or Oliver Kohlbacher, WSI/ZBIT, Eberhard-Karls-
BIOINFORMATICS. June Immunological Bioinformatics
 IMMUNOLOGICAL BIOINFORMATICS MEHTEIDHWLEFSATKLSSCDSFTSTINELNHCLSLRT YLVGNSLSLADLCVWATLKGNAAWQEQLKQKKAPVH VKRWFGFLEAQQAFQSVGTKWDVSTTKARVAPEKKQ DVGKFVELPGAEMGKVTVRFPPEASGYLHIGHAKAA LLNQHYQVNFKGKLIMRFDDTNPEKEKEDFEKVILE
IMMUNOLOGICAL BIOINFORMATICS MEHTEIDHWLEFSATKLSSCDSFTSTINELNHCLSLRT YLVGNSLSLADLCVWATLKGNAAWQEQLKQKKAPVH VKRWFGFLEAQQAFQSVGTKWDVSTTKARVAPEKKQ DVGKFVELPGAEMGKVTVRFPPEASGYLHIGHAKAA LLNQHYQVNFKGKLIMRFDDTNPEKEKEDFEKVILE
Antigen Presentation to T lymphocytes
 Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antigen processing: How are
Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antigen processing: How are
MetaMHC: a meta approach to predict peptides binding to MHC molecules
 W474 W479 Nucleic Acids Research, 2010, Vol. 38, Web Server issue Published online 18 May 2010 doi:10.1093/nar/gkq407 MetaMHC: a meta approach to predict peptides binding to MHC molecules Xihao Hu 1, Wenjian
W474 W479 Nucleic Acids Research, 2010, Vol. 38, Web Server issue Published online 18 May 2010 doi:10.1093/nar/gkq407 MetaMHC: a meta approach to predict peptides binding to MHC molecules Xihao Hu 1, Wenjian
1 Introduction Epitope prediction Prediction methods used for MHC class I... 4
 Contents 1 Introduction... 1 2 Epitope prediction... 1 2.1 Prediction methods used for MHC class I... 4 3 Algorithms used for machine learning... 5 4 References... 10 1 Introduction This document supplements
Contents 1 Introduction... 1 2 Epitope prediction... 1 2.1 Prediction methods used for MHC class I... 4 3 Algorithms used for machine learning... 5 4 References... 10 1 Introduction This document supplements
Antigen Recognition by T cells
 Antigen Recognition by T cells TCR only recognize foreign Ags displayed on cell surface These Ags can derive from pathogens, which replicate within cells or from pathogens or their products that cells
Antigen Recognition by T cells TCR only recognize foreign Ags displayed on cell surface These Ags can derive from pathogens, which replicate within cells or from pathogens or their products that cells
Methods and protocols for prediction of immunogenic epitopes Joo ChuanTong, Tin Wee Tan and Shoba Ranganathan
 BRIEFINGS IN BIOINFORMATICS. VOL 8. NO 2. 96^108 Advance Access publication October 31, 2006 doi:10.1093/bib/bbl038 Methods and protocols for prediction of immunogenic epitopes Joo ChuanTong, Tin Wee Tan
BRIEFINGS IN BIOINFORMATICS. VOL 8. NO 2. 96^108 Advance Access publication October 31, 2006 doi:10.1093/bib/bbl038 Methods and protocols for prediction of immunogenic epitopes Joo ChuanTong, Tin Wee Tan
Major histocompatibility complex class I binding predictions as a tool in epitope discovery
 IMMUNOLOGY REVIEW ARTICLE Major histocompatibility complex class I binding predictions as a tool in epitope discovery Claus Lundegaard, Ole Lund, Søren Buus and Morten Nielsen Department of Systems Biology,
IMMUNOLOGY REVIEW ARTICLE Major histocompatibility complex class I binding predictions as a tool in epitope discovery Claus Lundegaard, Ole Lund, Søren Buus and Morten Nielsen Department of Systems Biology,
Short peptide epitope design from hantaviruses causing HFRS
 www.bioinformation.net Volume 13(7) Hypothesis Short peptide epitope design from hantaviruses causing HFRS Sathish Sankar*, Mageshbabu Ramamurthy, Balaji Nandagopal, Gopalan Sridharan Sri Sakthi Amma Institute
www.bioinformation.net Volume 13(7) Hypothesis Short peptide epitope design from hantaviruses causing HFRS Sathish Sankar*, Mageshbabu Ramamurthy, Balaji Nandagopal, Gopalan Sridharan Sri Sakthi Amma Institute
International Journal of Pharma and Bio Sciences ANTIGEN PROTEIN FROM SCHISTOSOMA MANSONI: NEW PARADIGM OF SYNTHETIC VACCINE DEVELOPMENT
 International Journal of Pharma and Bio Sciences ANTIGEN PROTEIN FROM SCHISTOSOMA MANSONI: NEW PARADIGM OF SYNTHETIC VACCINE DEVELOPMENT GOMASE V.S.* and CHITLANGE N.R. School of Technology, S.R.T.M. University,
International Journal of Pharma and Bio Sciences ANTIGEN PROTEIN FROM SCHISTOSOMA MANSONI: NEW PARADIGM OF SYNTHETIC VACCINE DEVELOPMENT GOMASE V.S.* and CHITLANGE N.R. School of Technology, S.R.T.M. University,
Machine Intelligence Approach for Development of MHC Binders and Fragment Based Peptide Vaccines from Tetrahymena pyriformis
 Global Journal of Biotechnology & Biochemistry 5 (3): 161-165, 2010 ISSN 2078-466X IDOSI Publications, 2010 Machine Intelligence Approach for Development of MHC Binders and Fragment Based Peptide Vaccines
Global Journal of Biotechnology & Biochemistry 5 (3): 161-165, 2010 ISSN 2078-466X IDOSI Publications, 2010 Machine Intelligence Approach for Development of MHC Binders and Fragment Based Peptide Vaccines
SMPD 287 Spring 2015 Bioinformatics in Medical Product Development. Final Examination
 Final Examination You have a choice between A, B, or C. Please email your solutions, as a pdf attachment, by May 13, 2015. In the subject of the email, please use the following format: firstname_lastname_x
Final Examination You have a choice between A, B, or C. Please email your solutions, as a pdf attachment, by May 13, 2015. In the subject of the email, please use the following format: firstname_lastname_x
Workshop presentation: Development and application of Bioinformatics methods
 Workshop presentation: Development and application of Bioinformatics methods Immune system Innate fast Addaptive remembers Cellular Cytotoxic T lymphocytes (CTL) Helper T lymphocytes (HTL) Humoral B lymphocytes
Workshop presentation: Development and application of Bioinformatics methods Immune system Innate fast Addaptive remembers Cellular Cytotoxic T lymphocytes (CTL) Helper T lymphocytes (HTL) Humoral B lymphocytes
BIOINF 3360 Computational Immunomics
 BIOINF 3360 Computational Immunomics Oliver Kohlbacher Summer 2011 11. Summary TYOTK Things you ought to know! Caveat: These are just examples, not the full list of potential ti exam questions! TYOTK:
BIOINF 3360 Computational Immunomics Oliver Kohlbacher Summer 2011 11. Summary TYOTK Things you ought to know! Caveat: These are just examples, not the full list of potential ti exam questions! TYOTK:
BIOINF 3360 Computational Immunomics
 BIOINF 3360 Computational Immunomics Oliver Kohlbacher Summer 2011 11. Summary TYOTK Things you ought to know! Caveat: These are just examples, not the full list of potential exam questions! TYOTK: Immunology
BIOINF 3360 Computational Immunomics Oliver Kohlbacher Summer 2011 11. Summary TYOTK Things you ought to know! Caveat: These are just examples, not the full list of potential exam questions! TYOTK: Immunology
Degenerate T-cell Recognition of Peptides on MHC Molecules Creates Large Holes in the T-cell Repertoire
 Degenerate T-cell Recognition of Peptides on MHC Molecules Creates Large Holes in the T-cell Repertoire Jorg J. A. Calis*, Rob J. de Boer, Can Keşmir Theoretical Biology & Bioinformatics, Utrecht University,
Degenerate T-cell Recognition of Peptides on MHC Molecules Creates Large Holes in the T-cell Repertoire Jorg J. A. Calis*, Rob J. de Boer, Can Keşmir Theoretical Biology & Bioinformatics, Utrecht University,
A community resource benchmarking predictions of peptide binding to MHC-I molecules
 Downloaded from orbit.dtu.dk on: Dec 17, 2017 A community resource benchmarking predictions of peptide binding to MHC-I molecules Peters, B; Bui, HH; Pletscher-Frankild, Sune; Nielsen, Morten; Lundegaard,
Downloaded from orbit.dtu.dk on: Dec 17, 2017 A community resource benchmarking predictions of peptide binding to MHC-I molecules Peters, B; Bui, HH; Pletscher-Frankild, Sune; Nielsen, Morten; Lundegaard,
HLA Amino Acid Polymorphisms and Kidney Allograft Survival. Supplemental Digital Content
 HLA Amino Acid Polymorphisms and Kidney Allograft Survival Supplemental Digital Content Malek Kamoun, MD, PhD a, Keith P. McCullough, MS b, Martin Maiers, MS c, Marcelo A. Fernandez Vina, MD, PhD d, Hongzhe
HLA Amino Acid Polymorphisms and Kidney Allograft Survival Supplemental Digital Content Malek Kamoun, MD, PhD a, Keith P. McCullough, MS b, Martin Maiers, MS c, Marcelo A. Fernandez Vina, MD, PhD d, Hongzhe
Significance of the MHC
 CHAPTER 7 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
CHAPTER 7 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
Epitope discovery and Rational vaccine design Morten Nielsen
 Epitope discovery and Rational vaccine design Morten Nielsen mniel@cbs.dtu.dk Bioinformatics in a nutshell List of peptides that have a given biological feature Mathematical model (neural network, hidden
Epitope discovery and Rational vaccine design Morten Nielsen mniel@cbs.dtu.dk Bioinformatics in a nutshell List of peptides that have a given biological feature Mathematical model (neural network, hidden
In silico methods for rational design of vaccine and immunotherapeutics
 In silico methods for rational design of vaccine and immunotherapeutics Morten Nielsen, CBS, Department of Systems Biology, DTU and Instituto de Investigaciones Biotecnológicas, Universidad de San Martín,
In silico methods for rational design of vaccine and immunotherapeutics Morten Nielsen, CBS, Department of Systems Biology, DTU and Instituto de Investigaciones Biotecnológicas, Universidad de San Martín,
Historical definition of Antigen. An antigen is a foreign substance that elicits the production of antibodies that specifically binds to the antigen.
 Historical definition of Antigen An antigen is a foreign substance that elicits the production of antibodies that specifically binds to the antigen. Historical definition of Antigen An antigen is a foreign
Historical definition of Antigen An antigen is a foreign substance that elicits the production of antibodies that specifically binds to the antigen. Historical definition of Antigen An antigen is a foreign
Major Histocompatibility Complex Class II Prediction
 American Journal of Bioinformatics Research 2012, 2(1): 14-20 DOI: 10.5923/j.bioinformatics.20120201.03 Major Histocompatibility Complex Class II Prediction Zeinab Abd El Halim *, Amr Badr, Khaled Tawfik,
American Journal of Bioinformatics Research 2012, 2(1): 14-20 DOI: 10.5923/j.bioinformatics.20120201.03 Major Histocompatibility Complex Class II Prediction Zeinab Abd El Halim *, Amr Badr, Khaled Tawfik,
The BLAST search on NCBI ( and GISAID
 Supplemental materials and methods The BLAST search on NCBI (http:// www.ncbi.nlm.nih.gov) and GISAID (http://www.platform.gisaid.org) showed that hemagglutinin (HA) gene of North American H5N1, H5N2 and
Supplemental materials and methods The BLAST search on NCBI (http:// www.ncbi.nlm.nih.gov) and GISAID (http://www.platform.gisaid.org) showed that hemagglutinin (HA) gene of North American H5N1, H5N2 and
Identification and characterization of merozoite surface protein 1 epitope
 Identification and characterization of merozoite surface protein epitope Satarudra Prakash Singh,2, Bhartendu Nath Mishra 2* Amity Institute of Biotechnology, Amity University, Uttar Pradesh, Gomti Nagar,
Identification and characterization of merozoite surface protein epitope Satarudra Prakash Singh,2, Bhartendu Nath Mishra 2* Amity Institute of Biotechnology, Amity University, Uttar Pradesh, Gomti Nagar,
New technologies for studying human immunity. Lisa Wagar Postdoctoral fellow, Mark Davis lab Stanford University School of Medicine
 New technologies for studying human immunity Lisa Wagar Postdoctoral fellow, Mark Davis lab Stanford University School of Medicine New strategies: Human immunology is ideal for a systems approach We have
New technologies for studying human immunity Lisa Wagar Postdoctoral fellow, Mark Davis lab Stanford University School of Medicine New strategies: Human immunology is ideal for a systems approach We have
Immunogens and Antigens *
 Immunogens and Antigens * Jeffrey K. Actor, Ph.D. Pathology and Laboratory Medicine The University of Texas-Houston Medical School * Special thanks to Dr. L. Scott Rodkey, Ph.D. Immunogen vs. Antigen Immunogen-Agent
Immunogens and Antigens * Jeffrey K. Actor, Ph.D. Pathology and Laboratory Medicine The University of Texas-Houston Medical School * Special thanks to Dr. L. Scott Rodkey, Ph.D. Immunogen vs. Antigen Immunogen-Agent
Contents. Just Classifier? Rules. Rules: example. Classification Rule Generation for Bioinformatics. Rule Extraction from a trained network
 Contents Classification Rule Generation for Bioinformatics Hyeoncheol Kim Rule Extraction from Neural Networks Algorithm Ex] Promoter Domain Hybrid Model of Knowledge and Learning Knowledge refinement
Contents Classification Rule Generation for Bioinformatics Hyeoncheol Kim Rule Extraction from Neural Networks Algorithm Ex] Promoter Domain Hybrid Model of Knowledge and Learning Knowledge refinement
Leukemia (2007) 21, & 2007 Nature Publishing Group All rights reserved /07 $
 REVIEW (2007) 21, 1859 1874 & 2007 Nature Publishing Group All rights reserved 0887-6924/07 $30.00 www.nature.com/leu Department of Immunohematology and Blood Transfusion, Leiden University Medical Center,
REVIEW (2007) 21, 1859 1874 & 2007 Nature Publishing Group All rights reserved 0887-6924/07 $30.00 www.nature.com/leu Department of Immunohematology and Blood Transfusion, Leiden University Medical Center,
MHC Tetramers and Monomers for Immuno-Oncology and Autoimmunity Drug Discovery
 MHC Tetramers and Monomers for Immuno-Oncology and Autoimmunity Drug Discovery Your Partner in Drug Discovery and Research MHC Tetramer Background T-Cell Receptors recognize and bind to complexes composed
MHC Tetramers and Monomers for Immuno-Oncology and Autoimmunity Drug Discovery Your Partner in Drug Discovery and Research MHC Tetramer Background T-Cell Receptors recognize and bind to complexes composed
The Major Histocompatibility Complex (MHC)
 The Major Histocompatibility Complex (MHC) An introduction to adaptive immune system before we discuss MHC B cells The main cells of adaptive immune system are: -B cells -T cells B cells: Recognize antigens
The Major Histocompatibility Complex (MHC) An introduction to adaptive immune system before we discuss MHC B cells The main cells of adaptive immune system are: -B cells -T cells B cells: Recognize antigens
Integrated modeling of the major events in the MHC class I antigen processing pathway
 Integrated modeling of the major events in the MHC class I antigen processing pathway PIERRE DÖNNES AND OLIVER KOHLBACHER Department for Simulation of Biological Systems, WSI/ZBIT, Eberhard Karls University
Integrated modeling of the major events in the MHC class I antigen processing pathway PIERRE DÖNNES AND OLIVER KOHLBACHER Department for Simulation of Biological Systems, WSI/ZBIT, Eberhard Karls University
Cover Page. The handle holds various files of this Leiden University dissertation.
 Cover Page The handle http://hdl.handle.net/1887/20898 holds various files of this Leiden University dissertation. Author: Jöris, Monique Maria Title: Challenges in unrelated hematopoietic stem cell transplantation.
Cover Page The handle http://hdl.handle.net/1887/20898 holds various files of this Leiden University dissertation. Author: Jöris, Monique Maria Title: Challenges in unrelated hematopoietic stem cell transplantation.
Immuno-Oncology Therapies and Precision Medicine: Personal Tumor-Specific Neoantigen Prediction by Machine Learning
 Immuno-Oncology Therapies and Precision Medicine: Personal Tumor-Specific Neoantigen Prediction by Machine Learning Yi-Hsiang Hsu, MD, SCD Sep 16, 2017 yihsianghsu@hsl.harvard.edu Director & Associate
Immuno-Oncology Therapies and Precision Medicine: Personal Tumor-Specific Neoantigen Prediction by Machine Learning Yi-Hsiang Hsu, MD, SCD Sep 16, 2017 yihsianghsu@hsl.harvard.edu Director & Associate
Evaluating Classifiers for Disease Gene Discovery
 Evaluating Classifiers for Disease Gene Discovery Kino Coursey Lon Turnbull khc0021@unt.edu lt0013@unt.edu Abstract Identification of genes involved in human hereditary disease is an important bioinfomatics
Evaluating Classifiers for Disease Gene Discovery Kino Coursey Lon Turnbull khc0021@unt.edu lt0013@unt.edu Abstract Identification of genes involved in human hereditary disease is an important bioinfomatics
MHC-based vaccination approaches: progress and perspectives
 MHC-based vaccination approaches: progress and perspectives Narinder K. Mehra and Gurvinder Kaur The major histocompatibility complex (MHC) harbours genes whose primary function in regulating immune responsiveness
MHC-based vaccination approaches: progress and perspectives Narinder K. Mehra and Gurvinder Kaur The major histocompatibility complex (MHC) harbours genes whose primary function in regulating immune responsiveness
CONVENTIONAL VACCINE DEVELOPMENT
 CONVENTIONAL VACCINE DEVELOPMENT PROBLEM Lethal germ Dead mouse LIVE VACCINES Related but harmless germ gives protection against lethal pathogen. Examples are the original pox vaccine and some TB vaccines
CONVENTIONAL VACCINE DEVELOPMENT PROBLEM Lethal germ Dead mouse LIVE VACCINES Related but harmless germ gives protection against lethal pathogen. Examples are the original pox vaccine and some TB vaccines
2017 EFI/DGI Meeting Teaching Session I
 2017 EFI/DGI Meeting Teaching Session I Mannheim, Wednesday, 31.05.2017 10:30 12:00 Role of non-classical HLA in Stem Cell Transplantation 10:30 10:40 Non Classical HLA-Molecules J. Mytilineos, Ulm 10:40
2017 EFI/DGI Meeting Teaching Session I Mannheim, Wednesday, 31.05.2017 10:30 12:00 Role of non-classical HLA in Stem Cell Transplantation 10:30 10:40 Non Classical HLA-Molecules J. Mytilineos, Ulm 10:40
Major Histocompatibility Complex (MHC) and T Cell Receptors
 Major Histocompatibility Complex (MHC) and T Cell Receptors Historical Background Genes in the MHC were first identified as being important genes in rejection of transplanted tissues Genes within the MHC
Major Histocompatibility Complex (MHC) and T Cell Receptors Historical Background Genes in the MHC were first identified as being important genes in rejection of transplanted tissues Genes within the MHC
Host Genomics of HIV-1
 4 th International Workshop on HIV & Aging Host Genomics of HIV-1 Paul McLaren École Polytechnique Fédérale de Lausanne - EPFL Lausanne, Switzerland paul.mclaren@epfl.ch Complex trait genetics Phenotypic
4 th International Workshop on HIV & Aging Host Genomics of HIV-1 Paul McLaren École Polytechnique Fédérale de Lausanne - EPFL Lausanne, Switzerland paul.mclaren@epfl.ch Complex trait genetics Phenotypic
Indian Journal of Nephrology Indian J Nephrol 2001;11: 88-97
 88 Indian Journal of Nephrology Indian J Nephrol 2001;11: 88-97 ARTICLE HLA gene and haplotype frequency in renal transplant recipients and donors of Uttar Pradesh (North India) S Agrawal, AK Singh, RK
88 Indian Journal of Nephrology Indian J Nephrol 2001;11: 88-97 ARTICLE HLA gene and haplotype frequency in renal transplant recipients and donors of Uttar Pradesh (North India) S Agrawal, AK Singh, RK
6/7/17. Immune cells. Co-evolution of innate and adaptive immunity. Importance of NK cells. Cells of innate(?) immune response
 Immune cells Co-evolution of innate and adaptive immunity 1 2 Importance of NK cells Cells of innate(?) immune response Patients with NK cell deficiency may lead to fatal infections and have an increased
Immune cells Co-evolution of innate and adaptive immunity 1 2 Importance of NK cells Cells of innate(?) immune response Patients with NK cell deficiency may lead to fatal infections and have an increased
Alessandra Franco MD PhD UCSD School of Medicine Department of Pediatrics Division of Allergy Immunology and Rheumatology
 Immunodominant peptides derived from the heavy constant region of IgG1 stimulate natural regulatory T cells: identification of pan- HLA binders for clinical translation Alessandra Franco MD PhD UCSD School
Immunodominant peptides derived from the heavy constant region of IgG1 stimulate natural regulatory T cells: identification of pan- HLA binders for clinical translation Alessandra Franco MD PhD UCSD School
HLA and antigen presentation. Department of Immunology Charles University, 2nd Medical School University Hospital Motol
 HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
The major histocompatibility complex (MHC) is a group of genes that governs tumor and tissue transplantation between individuals of a species.
 Immunology Dr. John J. Haddad Chapter 7 Major Histocompatibility Complex The major histocompatibility complex (MHC) is a group of genes that governs tumor and tissue transplantation between individuals
Immunology Dr. John J. Haddad Chapter 7 Major Histocompatibility Complex The major histocompatibility complex (MHC) is a group of genes that governs tumor and tissue transplantation between individuals
Immuno-Oncology Therapies and Precision Medicine: Personal Tumor-Specific Neoantigen Prediction by Machine Learning
 Immuno-Oncology Therapies and Precision Medicine: Personal Tumor-Specific Neoantigen Prediction by Machine Learning Yi-Hsiang Hsu, MD, SCD Sep 16, 2017 yihsianghsu@hsl.harvard.edu HSL GeneticEpi Center,
Immuno-Oncology Therapies and Precision Medicine: Personal Tumor-Specific Neoantigen Prediction by Machine Learning Yi-Hsiang Hsu, MD, SCD Sep 16, 2017 yihsianghsu@hsl.harvard.edu HSL GeneticEpi Center,
Phase of immune response
 Antigen and antigen recognition by lymphocytes Antigen presentation to T lymphocytes Sanipa Suradhat Department of Veterinary Microbiology Faculty of Veterinary Science Phase of immune response 1 Phase
Antigen and antigen recognition by lymphocytes Antigen presentation to T lymphocytes Sanipa Suradhat Department of Veterinary Microbiology Faculty of Veterinary Science Phase of immune response 1 Phase
SUPPLEMENTARY INFORMATION
 Supplementary Notes 1: accuracy of prediction algorithms for peptide binding affinities to HLA and Mamu alleles For each HLA and Mamu allele we have analyzed the accuracy of four predictive algorithms
Supplementary Notes 1: accuracy of prediction algorithms for peptide binding affinities to HLA and Mamu alleles For each HLA and Mamu allele we have analyzed the accuracy of four predictive algorithms
Convolutional and LSTM Neural Networks
 Convolutional and LSTM Neural Networks Vanessa Jurtz January 11, 2017 Contents Neural networks and GPUs Lasagne Peptide binding to MHC class II molecules Convolutional Neural Networks (CNN) Recurrent and
Convolutional and LSTM Neural Networks Vanessa Jurtz January 11, 2017 Contents Neural networks and GPUs Lasagne Peptide binding to MHC class II molecules Convolutional Neural Networks (CNN) Recurrent and
Key Concept B F. How do peptides get loaded onto the proper kind of MHC molecule?
 Location of MHC class I pockets termed B and F that bind P and P9 amino acid side chains of the peptide Different MHC alleles confer different functional properties on the adaptive immune system by specifying
Location of MHC class I pockets termed B and F that bind P and P9 amino acid side chains of the peptide Different MHC alleles confer different functional properties on the adaptive immune system by specifying
B F. Location of MHC class I pockets termed B and F that bind P2 and P9 amino acid side chains of the peptide
 Different MHC alleles confer different functional properties on the adaptive immune system by specifying molecules that have different peptide binding abilities Location of MHC class I pockets termed B
Different MHC alleles confer different functional properties on the adaptive immune system by specifying molecules that have different peptide binding abilities Location of MHC class I pockets termed B
CD3 TCR V V CDR3 CDR1,2 CDR1,2 MHC-I TCR V 1- CDR2 TCR V CDR1 CHO CDR3 CDR1 CDR3 CDR3 N CDR1 CDR3 CDR1 CDR2 CDR2 TCR V MHC-II MHC-I
 1 A CD3 T C C TCR V V CDR3 CDR1,2 1 CDR1,2 2 B TCR V MHC-I C 1- CDR3 N CDR1 CDR2 CDR1 CDR3 C CHO 1- CDR2 CDR3 N CDR1 CDR1 CDR3 TCR V C TCR V CDR2 2- CDR2 1- MHC-I TCR V MHC-II 2 3 TCR / CD3 TCR MHC 65
1 A CD3 T C C TCR V V CDR3 CDR1,2 1 CDR1,2 2 B TCR V MHC-I C 1- CDR3 N CDR1 CDR2 CDR1 CDR3 C CHO 1- CDR2 CDR3 N CDR1 CDR1 CDR3 TCR V C TCR V CDR2 2- CDR2 1- MHC-I TCR V MHC-II 2 3 TCR / CD3 TCR MHC 65
Received 25 April 2002/Accepted 21 May 2002
 JOURNAL OF VIROLOGY, Sept. 2002, p. 8757 8768 Vol. 76, No. 17 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.17.8757 8768.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Clustering
JOURNAL OF VIROLOGY, Sept. 2002, p. 8757 8768 Vol. 76, No. 17 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.17.8757 8768.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Clustering
Designing of interferon-gamma inducing MHC class-ii binders
 Dhanda et al. Biology Direct 2013, 8:30 http://www.biologydirect.com/content/8/1/30 RESEARCH Open Access Designing of interferon-gamma inducing MHC class-ii binders Sandeep Kumar Dhanda, Pooja Vir and
Dhanda et al. Biology Direct 2013, 8:30 http://www.biologydirect.com/content/8/1/30 RESEARCH Open Access Designing of interferon-gamma inducing MHC class-ii binders Sandeep Kumar Dhanda, Pooja Vir and
Basic Immunology. Lecture 5 th and 6 th Recognition by MHC. Antigen presentation and MHC restriction
 Basic Immunology Lecture 5 th and 6 th Recognition by MHC. Antigen presentation and MHC restriction Molecular structure of MHC, subclasses, genetics, functions. Antigen presentation and MHC restriction.
Basic Immunology Lecture 5 th and 6 th Recognition by MHC. Antigen presentation and MHC restriction Molecular structure of MHC, subclasses, genetics, functions. Antigen presentation and MHC restriction.
Supplementary Table 1. Data collection and refinement statistics (molecular replacement).
 Supplementary Table 1. Data collection and refinement statistics (molecular replacement). Data set statistics HLA A*0201- ALWGPDPAAA PPI TCR PPI TCR/A2- ALWGPDPAAA PPI TCR/A2- ALWGPDPAAA Space Group P2
Supplementary Table 1. Data collection and refinement statistics (molecular replacement). Data set statistics HLA A*0201- ALWGPDPAAA PPI TCR PPI TCR/A2- ALWGPDPAAA PPI TCR/A2- ALWGPDPAAA Space Group P2
Rajesh Kannangai Phone: ; Fax: ; *Corresponding author
 Amino acid sequence divergence of Tat protein (exon1) of subtype B and C HIV-1 strains: Does it have implications for vaccine development? Abraham Joseph Kandathil 1, Rajesh Kannangai 1, *, Oriapadickal
Amino acid sequence divergence of Tat protein (exon1) of subtype B and C HIV-1 strains: Does it have implications for vaccine development? Abraham Joseph Kandathil 1, Rajesh Kannangai 1, *, Oriapadickal
Determinants of Immunogenicity and Tolerance. Abul K. Abbas, MD Department of Pathology University of California San Francisco
 Determinants of Immunogenicity and Tolerance Abul K. Abbas, MD Department of Pathology University of California San Francisco EIP Symposium Feb 2016 Why do some people respond to therapeutic proteins?
Determinants of Immunogenicity and Tolerance Abul K. Abbas, MD Department of Pathology University of California San Francisco EIP Symposium Feb 2016 Why do some people respond to therapeutic proteins?
E.J. Glass. AFRC Institute of Animal Physiology and Genetics Research, Edinburgh Research Station, Roslin, Midlothian, EH25 9PS, U.K.
 THE ROLE OF MHC CLASS II IN IMMUNE RESPONSES IN CATTLE E.J. Glass AFRC Institute of Animal Physiology and Genetics Research, Edinburgh Research Station, Roslin, Midlothian, EH25 9PS, U.K. 388 SUMMARY Polymorphism
THE ROLE OF MHC CLASS II IN IMMUNE RESPONSES IN CATTLE E.J. Glass AFRC Institute of Animal Physiology and Genetics Research, Edinburgh Research Station, Roslin, Midlothian, EH25 9PS, U.K. 388 SUMMARY Polymorphism
MHC class I MHC class II Structure of MHC antigens:
 MHC class I MHC class II Structure of MHC antigens: MHC class I antigens consist of a transmembrane heavy chain (α chain) that is non-covalently associated with β2- microglobulin. Membrane proximal domain
MHC class I MHC class II Structure of MHC antigens: MHC class I antigens consist of a transmembrane heavy chain (α chain) that is non-covalently associated with β2- microglobulin. Membrane proximal domain
DIRECT IDENTIFICATION OF NEO-EPITOPES IN TUMOR TISSUE
 DIRECT IDENTIFICATION OF NEO-EPITOPES IN TUMOR TISSUE Eustache Paramithiotis PhD Vice President, Biomarker Discovery & Diagnostics 17 March 2016 PEPTIDE PRESENTATION BY MHC MHC I Antigen presentation by
DIRECT IDENTIFICATION OF NEO-EPITOPES IN TUMOR TISSUE Eustache Paramithiotis PhD Vice President, Biomarker Discovery & Diagnostics 17 March 2016 PEPTIDE PRESENTATION BY MHC MHC I Antigen presentation by
Vaccine Design: A Statisticans Overview
 GoBack : A Statisticans Overview. Surajit Ray sray@samsi.info Surajit Ray Samsi PostDoc Seminar: Nov 2: 2004 - slide #1 The Chinese are credited with making the observation that deliberately infecting
GoBack : A Statisticans Overview. Surajit Ray sray@samsi.info Surajit Ray Samsi PostDoc Seminar: Nov 2: 2004 - slide #1 The Chinese are credited with making the observation that deliberately infecting
Significance of the MHC
 CHAPTER 8 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
CHAPTER 8 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
Newsletter 2018 vol
 August 30, 2018 Newsletter 2018 vol.2 23-32 TARGETING TOOLS FOR A DNA VACCINE CARRIER, A DESIGN IN PREVENTION OF CANCER Sharif Mohammad Shaheen* and Mustafezur Rahman Faculty of Allied Health Sciences,
August 30, 2018 Newsletter 2018 vol.2 23-32 TARGETING TOOLS FOR A DNA VACCINE CARRIER, A DESIGN IN PREVENTION OF CANCER Sharif Mohammad Shaheen* and Mustafezur Rahman Faculty of Allied Health Sciences,
A general overview of Immune system and. Shuyan Li 2/4/2009
 A general overview of Immune system and Immunology Shuyan Li 2/4/2009 Definition Immune system a collection of biological processes within an organism that protects against disease by identifying and killing
A general overview of Immune system and Immunology Shuyan Li 2/4/2009 Definition Immune system a collection of biological processes within an organism that protects against disease by identifying and killing
Immunology - Lecture 2 Adaptive Immune System 1
 Immunology - Lecture 2 Adaptive Immune System 1 Book chapters: Molecules of the Adaptive Immunity 6 Adaptive Cells and Organs 7 Generation of Immune Diversity Lymphocyte Antigen Receptors - 8 CD markers
Immunology - Lecture 2 Adaptive Immune System 1 Book chapters: Molecules of the Adaptive Immunity 6 Adaptive Cells and Organs 7 Generation of Immune Diversity Lymphocyte Antigen Receptors - 8 CD markers
Tumor neoantigens: building a framework for personalized cancer immunotherapy
 Tumor neoantigens: building a framework for personalized cancer immunotherapy Matthew M. Gubin, 1,2 Maxim N. Artyomov, 1,2 Elaine R. Mardis, 3,4 and Robert D. Schreiber 1,2 Series Editor: Yiping Yang 1
Tumor neoantigens: building a framework for personalized cancer immunotherapy Matthew M. Gubin, 1,2 Maxim N. Artyomov, 1,2 Elaine R. Mardis, 3,4 and Robert D. Schreiber 1,2 Series Editor: Yiping Yang 1
The Major Histocompatibility Complex of Genes
 The Major Histocompatibility Complex of Genes Topic 4 The Major Histocompatibility Complex Outline of Lectures The immunological reasons for transplant rejection How the MHC was discovered using inbred
The Major Histocompatibility Complex of Genes Topic 4 The Major Histocompatibility Complex Outline of Lectures The immunological reasons for transplant rejection How the MHC was discovered using inbred
Adaptive Immune Response Day 2. The Adaptive Immune Response
 Adaptive Immune Response Day 2 Chapter 16 The Adaptive Immune Response 1 The B cell receptor vs. the T cell receptor. The B cell receptor vs. the T cell receptor. 2 Which T cells have CD4 and which have
Adaptive Immune Response Day 2 Chapter 16 The Adaptive Immune Response 1 The B cell receptor vs. the T cell receptor. The B cell receptor vs. the T cell receptor. 2 Which T cells have CD4 and which have
LIPOPREDICT: Bacterial lipoprotein prediction server
 www.bioinformation.net Server Volume 8(8) LIPOPREDICT: Bacterial lipoprotein prediction server S Ramya Kumari, Kiran Kadam, Ritesh Badwaik & Valadi K Jayaraman* Centre for Development of Advanced Computing
www.bioinformation.net Server Volume 8(8) LIPOPREDICT: Bacterial lipoprotein prediction server S Ramya Kumari, Kiran Kadam, Ritesh Badwaik & Valadi K Jayaraman* Centre for Development of Advanced Computing
PREDIVAC: CD4+ T-cell epitope prediction for vaccine design that covers 95% of HLA class II DR protein diversity
 Oyarzún et al. BMC Bioinformatics 2013, 14:52 RESEARCH ARTICLE Open Access PREDIVAC: CD4+ T-cell epitope prediction for vaccine design that covers 95% of HLA class II DR protein diversity Patricio Oyarzún
Oyarzún et al. BMC Bioinformatics 2013, 14:52 RESEARCH ARTICLE Open Access PREDIVAC: CD4+ T-cell epitope prediction for vaccine design that covers 95% of HLA class II DR protein diversity Patricio Oyarzún
Exploring HIV Evolution: An Opportunity for Research Sam Donovan and Anton E. Weisstein
 Microbes Count! 137 Video IV: Reading the Code of Life Human Immunodeficiency Virus (HIV), like other retroviruses, has a much higher mutation rate than is typically found in organisms that do not go through
Microbes Count! 137 Video IV: Reading the Code of Life Human Immunodeficiency Virus (HIV), like other retroviruses, has a much higher mutation rate than is typically found in organisms that do not go through
Computational prediction of immunodominant antigenic regions & potential protective epitopes for dengue vaccination
 Indian J Med Res 144, October 2016, pp 587-591 DOI: 10.4103/0971-5916.200894 Quick Response Code: Computational prediction of immunodominant antigenic regions & potential protective epitopes for dengue
Indian J Med Res 144, October 2016, pp 587-591 DOI: 10.4103/0971-5916.200894 Quick Response Code: Computational prediction of immunodominant antigenic regions & potential protective epitopes for dengue
New trends in donor selection in Europe: "best match" versus haploidentical. Prof Jakob R Passweg
 New trends in donor selection in Europe: "best match" versus haploidentical Prof Jakob R Passweg HSCT change in donor type: 1990-2015 9000 H S C T 8000 7000 6000 5000 4000 HLA identical sibling/twin Haplo-identical
New trends in donor selection in Europe: "best match" versus haploidentical Prof Jakob R Passweg HSCT change in donor type: 1990-2015 9000 H S C T 8000 7000 6000 5000 4000 HLA identical sibling/twin Haplo-identical
Potential cross reactions between HIV 1 specific T cells and the microbiome. Andrew McMichael Suzanne Campion
 Potential cross reactions between HIV 1 specific T cells and the microbiome Andrew McMichael Suzanne Campion Role of the Microbiome? T cell (and B cell) immune responses to HIV and Vaccines are influenced
Potential cross reactions between HIV 1 specific T cells and the microbiome Andrew McMichael Suzanne Campion Role of the Microbiome? T cell (and B cell) immune responses to HIV and Vaccines are influenced
Machine Learning For Personalized Cancer Vaccines. Alex Rubinsteyn February 9th, 2018 Data Science Salon Miami
 Machine Learning For Personalized Cancer Vaccines Alex Rubinsteyn February 9th, 2018 Data Science Salon Miami OpenVax @ Mount Sinai Focus: personalized cancer vaccines Machine learning for immunology Cancer
Machine Learning For Personalized Cancer Vaccines Alex Rubinsteyn February 9th, 2018 Data Science Salon Miami OpenVax @ Mount Sinai Focus: personalized cancer vaccines Machine learning for immunology Cancer
Immune Epitope Database NEWSLETTER Volume 6, Issue 2 July 2009
 Immune Epitope Database NEWSLETTER Volume 6, Issue 2 http://www.iedb.org July 2009 In response to concerns of a swine flu epidemic, the IEDB Curation curation team updated the database s collection of
Immune Epitope Database NEWSLETTER Volume 6, Issue 2 http://www.iedb.org July 2009 In response to concerns of a swine flu epidemic, the IEDB Curation curation team updated the database s collection of
Principles of phylogenetic analysis
 Principles of phylogenetic analysis Arne Holst-Jensen, NVI, Norway. Fusarium course, Ås, Norway, June 22 nd 2008 Distance based methods Compare C OTUs and characters X A + D = Pairwise: A and B; X characters
Principles of phylogenetic analysis Arne Holst-Jensen, NVI, Norway. Fusarium course, Ås, Norway, June 22 nd 2008 Distance based methods Compare C OTUs and characters X A + D = Pairwise: A and B; X characters
The Major Histocompatibility Complex
 The Major Histocompatibility Complex Today we will discuss the MHC The study of MHC is necessary to understand how an immune response is generated. And these are the extra notes with respect to slides
The Major Histocompatibility Complex Today we will discuss the MHC The study of MHC is necessary to understand how an immune response is generated. And these are the extra notes with respect to slides
HLA and antigen presentation. Department of Immunology Charles University, 2nd Medical School University Hospital Motol
 HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
TCR-p-MHC 10/28/2013. Disclosures. Rheumatoid Arthritis, Psoriatic Arthritis and Autoimmunity: good genes, elegant mechanisms, bad results
 Rheumatoid Arthritis, Psoriatic Arthritis and Autoimmunity: good genes, elegant mechanisms, bad results The meaning and significance of HLA associations Disclosures No relevant commercial relationships
Rheumatoid Arthritis, Psoriatic Arthritis and Autoimmunity: good genes, elegant mechanisms, bad results The meaning and significance of HLA associations Disclosures No relevant commercial relationships
Cellular Pathology of immunological disorders
 Cellular Pathology of immunological disorders SCBM344 Cellular and Molecular Pathology Witchuda Payuhakrit, Ph.D (Pathobiology) witchuda.pay@mahidol.ac.th Objectives Describe the etiology of immunological
Cellular Pathology of immunological disorders SCBM344 Cellular and Molecular Pathology Witchuda Payuhakrit, Ph.D (Pathobiology) witchuda.pay@mahidol.ac.th Objectives Describe the etiology of immunological
Neoantigen Identification using ATLAS Across Multiple Tumor Types Highlights Limitations of Prediction Algorithms
 Neoantigen Identification using ATLAS Across Multiple Tumor Types Highlights Limitations of Prediction Algorithms Wendy Broom, Ph.D. Keystone: Translational Systems Immunology, Snowbird February 1st 2018
Neoantigen Identification using ATLAS Across Multiple Tumor Types Highlights Limitations of Prediction Algorithms Wendy Broom, Ph.D. Keystone: Translational Systems Immunology, Snowbird February 1st 2018
CONSTRUCTION OF PHYLOGENETIC TREE USING NEIGHBOR JOINING ALGORITHMS TO IDENTIFY THE HOST AND THE SPREADING OF SARS EPIDEMIC
 CONSTRUCTION OF PHYLOGENETIC TREE USING NEIGHBOR JOINING ALGORITHMS TO IDENTIFY THE HOST AND THE SPREADING OF SARS EPIDEMIC 1 MOHAMMAD ISA IRAWAN, 2 SITI AMIROCH 1 Institut Teknologi Sepuluh Nopember (ITS)
CONSTRUCTION OF PHYLOGENETIC TREE USING NEIGHBOR JOINING ALGORITHMS TO IDENTIFY THE HOST AND THE SPREADING OF SARS EPIDEMIC 1 MOHAMMAD ISA IRAWAN, 2 SITI AMIROCH 1 Institut Teknologi Sepuluh Nopember (ITS)
Advantage of rare HLA supertype in HIV disease progression
 Advantage of rare HLA supertype in HIV disease progression Elizabeth Trachtenberg 1,7,Bette Korber 2,3,7,Cristina Sollars 1, Thomas B Kepler 3,4,Peter T Hraber 3, Elizabeth Hayes 1,Robert Funkhouser 2,3,Michael
Advantage of rare HLA supertype in HIV disease progression Elizabeth Trachtenberg 1,7,Bette Korber 2,3,7,Cristina Sollars 1, Thomas B Kepler 3,4,Peter T Hraber 3, Elizabeth Hayes 1,Robert Funkhouser 2,3,Michael
