Supplemental Information:
|
|
|
- Abraham Flynn
- 5 years ago
- Views:
Transcription
1 SupplementalInformation: Content: FigureS1.AccessibilityoftheL.interrogansproteomebyLC MSanalysis. FigureS2.Functionalannotationoftheidentifiedproteins. FigureS3.DistributionofMS1 featureintensities. FigureS4.ComparisonoftheDDA anddirected(inl)lc MS/MSstrategyemployed. FigureS5.PerformanceofthedirectedMS workflowusingdifferentlc MS/MSplatforms. FigureS6.Reproducibilityoflabel freequantificationofthel.interrogansproteome. FigureS7.Correlationoftheestimatedproteinabundances(incopies/cell)witharecentlypublished study. FigureS8.Determinationofcellularproteinconcentrations. FigureS9.Significantproteinchangesincontrolsamplesafter24and48hourswithoutanytreatment. Figure S10. Impact of relative (in fold) and absolute (in copies/cell) protein level changes on the L. interrogansproteome. FigureS11.ProteinsofthebacterialchemotaxispathwaycoveredinclusterS 5asassignedbyDAVID usingthekeggdatabase. Figure S12. Proteins of the TCA cycle covered in cluster S 5 as assigned by DAVID using the KEGG database. Figure S13. Proteins of the oxidative phosphorylation pathway covered in cluster S 5 as assigned by DAVIDusingtheKEGGdatabase. TableSI.ProteinabundancelevelsdeterminedbystableisotopedilutionLC MS. TableSII.Identifiedfeaturesduringdiscoveryphase(seexls file). TableSIII.Identifiedproteotypicpeptides(PTPs)duringdiscoveryphase(seexls file). TableSIV.Identifiedproteinsduringdiscoveryphase(seexls file). TableSV.Monitoredproteinsduringscoringphase(alltreatments,seexls file). 1
2 FigureS1: Figure S1. Accessibility of the L. interrogans proteome by LC MS analysis. Number of proteins with their correspondingnumberofproteotypicpeptidesavailableforlc MS/MSanalysisaftertrypticcleavage. 2
3 FigureS2: FigureS2.Functionalannotationoftheidentifiedproteins.DistributionoftheproteinsidentifiedbytheLConly(red)andOGE/LC(green)MSanalysisincomparisontoallpredictedgenes(blue)accordingtotheir functionalcategoriesasassignedbydavid(huangetal,2007). 3
4 FigureS3: Figure S3. Distribution of MS1 feature intensities. Binned extracted precursor ion intensities (log 10 ) of all 13,113features(blue)identifiedandtheselected4953proteotypicpeptides(green)forproteinquantification. 4
5 FigureS4: FigureS4.ComparisonoftheDDA anddirected(inl)lc MS/MSstrategyemployed.(A)Analysisofthesame peptidesample(control)withthesamenumberoflc MS/MSrunsusingeitherthedata dependentacquisition (DDA,blue)orthedirected(INL,red)LC MS/MSapproach.Thenumbersofpeptidesandproteinsidentifiedby eitherapproachareindicated.(b)distributionoftheproteinsidentifiedbythedifferentapproachesaccording totheirabundanceasdeterminedbycodonbias. 5
6 FigureS5: FigureS5.PerformanceofthedirectedMS workflowusingdifferentlc MS/MSplatforms.(A)Numberof timesaproteinwasidentifiedin6differentcontrolsamplesusingdirectedmassspectrometryontheltq FT ICRLC MS/MSplatform.(B)AnalysisofthesamesamplesonadifferentLC(ProxeonEasy LC)andMS(Orbitrap Velos)systemusingthesamepeptidemasslistsgeneratedinthediscoveryphase.(C)Venndiagramshowing thenumberofoverlappingandspecificproteinfractionsidentified. 6
7 FigureS6: FigureS6.Reproducibilityoflabel freequantificationofthel.interrogansproteome.(a C)Scatterplotsof proteinms intensities(log 2 )betweenvariousuntreatedreplicatecontrolsamplesasdeterminedbythe Progenesissoftware,includingtherespectivesquaredPearsoncorrelationR 2.(D F)Distributionoftheprotein ratios(log 2 )determinedfromthesamesamplesasindicatedina Cversusthesignificancevaluecalculated fromtheanovaanalysis.thecoefficientofvarianceaswellasthesignificantthresholdssetforp value (ANOVA,<0.05)andratio(>1.5 fold)indicatedasredlinesareshown. 7
8 FigureS7: FigureS7.Correlationoftheestimatedproteinabundances(incopies/cell)witharecentlypublishedstudy. Comparisonoftheproteinabundancesdeterminedwitharecentlypublisheddatasetusingasimilarapproach (Malmströmetal,2009). 8
9 FigureS8: FigureS8.Determinationofcellularproteinconcentrations.(A)Averageextractedprecursorionintensitiesof thethreemostabundantpeptidesperprotein(log 10 )plottedagainsttheirproteinconcentrationincopies/cell (log 10 )determinedusingthespikedinheavylabeledreferencepeptides.(b)distributionoffold errorrates calculatedbybootstrappingaccordingto(malmströmetal,2009). 9
10 FigureS9: Figure S9. Significant protein changes detected in control samples. Volcano plots showing the significant proteinabundancechangesdetectedincontrolsamplesthatwerenotsubjectedtoanytreatmentafter(a)24 and (B) 48 hours. Protein regulations considered as significant ((ANOVA, <0.05) and ratio (>1.5 fold)) are indicatedwithintheredlinestogetherwiththenumberofup anddown regulatedproteins. 10
11 FigureS10: Figure S10. Impact of relative (in fold) and absolute (in copies/cell) protein level changes on the L. interrogansproteome.(a)averagerelativeabundanceratios(log 10 )ofallproteinsgroupedinto6clustersof different concentration levels in the cell (control state). (B) Like A, using absolute protein concentration changes (log 10 ) instead fold changes. The standard deviations calculated are indicated as error bars. (C) Hierarchical clustering of relative protein abundance changes (log 10 ) to the corresponding untreated control samplesincopiespercell(log 10 )forall24treatments.thecolumndendrogramrepresentingtheclusteringof thedifferentiallyperturbedsamplesisdisplayedandtheclusters(1 6)obtainedareindicated. 11
12 Fig.S11: FigureS11.ProteinsofthebacterialchemotaxispathwaycoveredinclusterS 5asassignedbyDAVID(Huang etal,2007)usingthekeggdatabase(kanehisaetal,2010).proteinspresentintheclusterareindicatedwith aredstar.green(white)labeledgenesarepresent(notpresent)inthel.interrogansgenome. 12
13 Fig.S12: FigureS12.ProteinsoftheTCAcyclecoveredinclusterS 5asassignedbyDAVID(Huangetal,2007)using thekeggdatabase(kanehisaetal,2010).proteinspresentintheclusterareindicatedwitharedstar.green (white)labeledgenesarepresent(notpresent)inthel.interrogansgenome. 13
14 Fig.S13: FigureS13.ProteinsoftheoxidativephosphorylationpathwaycoveredinclusterS 5asassignedbyDAVID (Huangetal,2007)usingtheKEGGdatabase(Kanehisaetal,2010).Proteinspresentintheclusterare indicatedwitharedstar.green(white)labeledgenesarepresent(notpresent)inthel.interrogansgenome. 14
15 Table SI. Protein abundance levels determined by stable isotope dilution LC MS Protein Protein Name Peptide Sequence Peptide Copies/Cell 1 Protein Copies/Cell 2 gi ref YP_ fatty acid synthase subunit beta TEVITHANLVR gi ref YP_ DNA directed RNA polymerase beta subunit ITNLDYLPNLIQIQK gi ref YP_ DNA directed RNA polymerase beta subunit TFDLGEVGR 1219 gi ref YP_ DNA directed RNA polymerase beta' subunit FATSDLNDLYR gi ref YP_ flagellar hook protein ENIGGVNPQQVGLGSLIAAIDK gi ref YP_ flagellar hook protein VATAVFNNPAGLDK 437 gi ref YP_ ATP synthase subunit A ILEVPVGPELLGR 14,633 12,691 gi ref YP_ ATP synthase subunit A TSIALDTILNQK 10,749 gi ref YP_ ATP synthase subunit B FSQAGSEVSALLGR 10,042 10,042 gi ref YP_ MreB GIVLTGGGCLLR gi ref YP_ MreB TGGDEFDEAIIK 2921 gi ref YP_ GroEL AVTAAVESIQK 10,572 13,649 gi ref YP_ GroEL VEDALSATR 16,726 gi ref YP_ GroES ESDILAVVK 13,686 11,704 gi ref YP_ GroES VGDTVLYGK 9723 gi ref YP_ flagellar M ring protein GFTPDGPAGTEPNIAPGYK gi ref YP_ flagellar M ring protein IISDFEEDLEK 60 gi ref YP_ ATP dependent protease ATP binding subunit LLEEVSFEGPDLPESQR gi ref YP_ recombinase A IVEIYGPESSGK gi ref YP_ ATP dependent CLP protease like, proteolytic subunit IAEVFEELTGSK gi ref YP_ ATP dependent CLP protease like, proteolytic subunit IFLWGPVTDESSK 1401 gi ref YP_ ATP dependent CLP protease like, proteolytic subunit LNQILADACGHPISK 4015 gi ref YP_ ATP dependent protease AVDLIDEASSK gi ref YP_ ATP dependent protease IADIQLEGLR 437 gi ref YP_ S ribosomal protein S6 EFLINQNILR gi ref YP_ Hsp15 like protein ILELPTEVDSEK gi ref YP_ Hsp15 DVQVQLEK gi ref YP_ S ribosomal protein S5 FSFNALSVVGDQR gi ref YP_ fatty acid synthase subunit beta EFFDTSFK ) Endogenous peptide abundances determined by stable isotope dilution 2) If multiple peptides per protein were identified and quantified, the average abundance level is shown 15
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/398/rs12/dc1 Supplementary Materials for Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells Scott F. Rusin, Kate
www.sciencesignaling.org/cgi/content/full/8/398/rs12/dc1 Supplementary Materials for Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells Scott F. Rusin, Kate
Department of Chemistry, Université de Montréal, C.P. 6128, Succursale centre-ville, Montréal, Québec, H3C 3J7, Canada.
 Phosphoproteome dynamics of Saccharomyces cerevisiae under heat shock and cold stress Evgeny Kanshin 1,5, Peter Kubiniok 1,2,5, Yogitha Thattikota 1,3, Damien D Amours 1,3 and Pierre Thibault 1,2,4 * 1
Phosphoproteome dynamics of Saccharomyces cerevisiae under heat shock and cold stress Evgeny Kanshin 1,5, Peter Kubiniok 1,2,5, Yogitha Thattikota 1,3, Damien D Amours 1,3 and Pierre Thibault 1,2,4 * 1
Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triplenegative
 SUPPLEMENTARY INFORMATION Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triplenegative breast cancer Roman Camarda, Alicia Y. Zhou, Rebecca A. Kohnz, Sanjeev Balakrishnan, Celine
SUPPLEMENTARY INFORMATION Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triplenegative breast cancer Roman Camarda, Alicia Y. Zhou, Rebecca A. Kohnz, Sanjeev Balakrishnan, Celine
Digitizing the Proteomes From Big Tissue Biobanks
 Digitizing the Proteomes From Big Tissue Biobanks Analyzing 24 Proteomes Per Day by Microflow SWATH Acquisition and Spectronaut Pulsar Analysis Jan Muntel 1, Nick Morrice 2, Roland M. Bruderer 1, Lukas
Digitizing the Proteomes From Big Tissue Biobanks Analyzing 24 Proteomes Per Day by Microflow SWATH Acquisition and Spectronaut Pulsar Analysis Jan Muntel 1, Nick Morrice 2, Roland M. Bruderer 1, Lukas
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function
 Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
microrna PCR System (Exiqon), following the manufacturer s instructions. In brief, 10ng of
 SUPPLEMENTAL MATERIALS AND METHODS Quantitative RT-PCR Quantitative RT-PCR analysis was performed using the Universal mircury LNA TM microrna PCR System (Exiqon), following the manufacturer s instructions.
SUPPLEMENTAL MATERIALS AND METHODS Quantitative RT-PCR Quantitative RT-PCR analysis was performed using the Universal mircury LNA TM microrna PCR System (Exiqon), following the manufacturer s instructions.
Extended Mass Range Triple Quadrupole for Routine Analysis of High Mass-to-charge Peptide Ions
 Extended Mass Range Triple Quadrupole for Routine Analysis of High Mass-to-charge Peptide Ions Application Note Targeted Proteomics Authors Linfeng Wu, Christine A. Miller, Jordy Hsiao, Te-wei Chu, Behrooz
Extended Mass Range Triple Quadrupole for Routine Analysis of High Mass-to-charge Peptide Ions Application Note Targeted Proteomics Authors Linfeng Wu, Christine A. Miller, Jordy Hsiao, Te-wei Chu, Behrooz
Complexity DNA. Genome RNA. Transcriptome. Protein. Proteome. Metabolites. Metabolome
 DNA Genome Complexity RNA Transcriptome Systems Biology Linking all the components of a cell in a quantitative and temporal manner Protein Proteome Metabolites Metabolome Where are the functional elements?
DNA Genome Complexity RNA Transcriptome Systems Biology Linking all the components of a cell in a quantitative and temporal manner Protein Proteome Metabolites Metabolome Where are the functional elements?
Supporting Information. Lysine Propionylation to Boost Proteome Sequence. Coverage and Enable a Silent SILAC Strategy for
 Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
Expanded View Figures
 EMO Molecular Medicine Proteomic map of squamous cell carcinomas Hanibal ohnenberger et al Expanded View Figures Figure EV1. Technical reproducibility. Pearson s correlation analysis of normalised SILC
EMO Molecular Medicine Proteomic map of squamous cell carcinomas Hanibal ohnenberger et al Expanded View Figures Figure EV1. Technical reproducibility. Pearson s correlation analysis of normalised SILC
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
Automating Mass Spectrometry-Based Quantitative Glycomics using Tandem Mass Tag (TMT) Reagents with SimGlycan
 PREMIER Biosoft Automating Mass Spectrometry-Based Quantitative Glycomics using Tandem Mass Tag (TMT) Reagents with SimGlycan Ne uaca2-3galb1-4glc NAcb1 6 Gal NAca -Thr 3 Ne uaca2-3galb1 Ningombam Sanjib
PREMIER Biosoft Automating Mass Spectrometry-Based Quantitative Glycomics using Tandem Mass Tag (TMT) Reagents with SimGlycan Ne uaca2-3galb1-4glc NAcb1 6 Gal NAca -Thr 3 Ne uaca2-3galb1 Ningombam Sanjib
TECHNICAL NOTE. Accurate and fast proteomics analysis of human plasma with PlasmaDive and SpectroDive
 TECHNICAL NOTE Accurate and fast proteomics analysis of human plasma with PlasmaDive and SpectroDive In this technical note you will learn about: Step-by-step set-up of parallel reaction monitoring (PRM)
TECHNICAL NOTE Accurate and fast proteomics analysis of human plasma with PlasmaDive and SpectroDive In this technical note you will learn about: Step-by-step set-up of parallel reaction monitoring (PRM)
Human Neurology 3-Plex A
 Human Neurology 3-Plex A SUMMARY AND EXPLANATION OF THE TEST The Human N3PA assay is a digital immunoassay for the quantitative determination of total Tau, Aβ42, and Aβ40 in human plasma and CSF. Determination
Human Neurology 3-Plex A SUMMARY AND EXPLANATION OF THE TEST The Human N3PA assay is a digital immunoassay for the quantitative determination of total Tau, Aβ42, and Aβ40 in human plasma and CSF. Determination
Universal sample preparation method for proteome analysis
 nature methods Universal sample preparation method for proteome analysis Jacek R Wi niewski, Alexandre Zougman, Nagarjuna Nagaraj & Matthias Mann Supplementary figures and text: Supplementary Figure 1
nature methods Universal sample preparation method for proteome analysis Jacek R Wi niewski, Alexandre Zougman, Nagarjuna Nagaraj & Matthias Mann Supplementary figures and text: Supplementary Figure 1
APPLICATION NOTE. Highly reproducible and Comprehensive Proteome Profiling of Formalin-Fixed Paraffin-Embedded (FFPE) Tissues Slices
 APPLICATION NOTE Highly reproducible and Comprehensive Proteome Profiling of Formalin-Fixed Paraffin-Embedded (FFPE) Tissues Slices INTRODUCTION Preservation of tissue biopsies is a critical step to This
APPLICATION NOTE Highly reproducible and Comprehensive Proteome Profiling of Formalin-Fixed Paraffin-Embedded (FFPE) Tissues Slices INTRODUCTION Preservation of tissue biopsies is a critical step to This
MG4010 Course Information. Syllabus
 MG4010 Course Information Syllabus A one- semester course designed to introduce students to the basic tenets of biochemistry. The topics that will be discussed have been chosen to comply with the guidelines
MG4010 Course Information Syllabus A one- semester course designed to introduce students to the basic tenets of biochemistry. The topics that will be discussed have been chosen to comply with the guidelines
Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler
 Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler DR CALUX Boys Girls Database Systemic lupus erythematosus 4.4 0.0021 6.7
Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler DR CALUX Boys Girls Database Systemic lupus erythematosus 4.4 0.0021 6.7
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Student Number: THE UNIVERSITY OF MANITOBA April 10, 2006, 1:30 AM - 4:30 PM Page 1 (of 4) Biochemistry II Laboratory Section Final Examination
 Name: Student Number: April 10, 2006, 1:30 AM - 4:30 PM Page 1 (of 4) Biochemistry II Laboratory Section Final Examination Examiner: Dr. A. Scoot 1. Answer ALL questions in the space provided. 2. The back
Name: Student Number: April 10, 2006, 1:30 AM - 4:30 PM Page 1 (of 4) Biochemistry II Laboratory Section Final Examination Examiner: Dr. A. Scoot 1. Answer ALL questions in the space provided. 2. The back
Nature Structural & Molecular Biology: doi: /nsmb.3218
 Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
DNA codes for RNA, which guides protein synthesis.
 Section 3: DNA codes for RNA, which guides protein synthesis. K What I Know W What I Want to Find Out L What I Learned Vocabulary Review synthesis New RNA messenger RNA ribosomal RNA transfer RNA transcription
Section 3: DNA codes for RNA, which guides protein synthesis. K What I Know W What I Want to Find Out L What I Learned Vocabulary Review synthesis New RNA messenger RNA ribosomal RNA transfer RNA transcription
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
NIH Public Access Author Manuscript J Proteome Res. Author manuscript; available in PMC 2014 July 05.
 NIH Public Access Author Manuscript Published in final edited form as: J Proteome Res. 2013 July 5; 12(7): 3071 3086. doi:10.1021/pr3011588. Evaluation and Optimization of Mass Spectrometric Settings during
NIH Public Access Author Manuscript Published in final edited form as: J Proteome Res. 2013 July 5; 12(7): 3071 3086. doi:10.1021/pr3011588. Evaluation and Optimization of Mass Spectrometric Settings during
Activity: Biologically Important Molecules
 Activity: Biologically Important Molecules AP Biology Introduction We have already seen in our study of biochemistry that the molecules that comprise living things are carbon-based, and that they are thought
Activity: Biologically Important Molecules AP Biology Introduction We have already seen in our study of biochemistry that the molecules that comprise living things are carbon-based, and that they are thought
Explain that each trna molecule is recognised by a trna-activating enzyme that binds a specific amino acid to the trna, using ATP for energy
 7.4 - Translation 7.4.1 - Explain that each trna molecule is recognised by a trna-activating enzyme that binds a specific amino acid to the trna, using ATP for energy Each amino acid has a specific trna-activating
7.4 - Translation 7.4.1 - Explain that each trna molecule is recognised by a trna-activating enzyme that binds a specific amino acid to the trna, using ATP for energy Each amino acid has a specific trna-activating
chapter 1 - fig. 2 Mechanism of transcriptional control by ppar agonists.
 chapter 1 - fig. 1 The -omics subdisciplines. chapter 1 - fig. 2 Mechanism of transcriptional control by ppar agonists. 201 figures chapter 1 chapter 2 - fig. 1 Schematic overview of the different steps
chapter 1 - fig. 1 The -omics subdisciplines. chapter 1 - fig. 2 Mechanism of transcriptional control by ppar agonists. 201 figures chapter 1 chapter 2 - fig. 1 Schematic overview of the different steps
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified
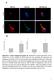 Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Supplementary Figure 1
 Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Electron Transport Chain and Oxidative phosphorylation
 Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Supplementary Figures and Notes for Digestion and depletion of abundant
 Supplementary Figures and Notes for Digestion and depletion of abundant proteins improves proteomic coverage Bryan R. Fonslow, Benjamin D. Stein, Kristofor J. Webb, Tao Xu, Jeong Choi, Sung Kyu Park, and
Supplementary Figures and Notes for Digestion and depletion of abundant proteins improves proteomic coverage Bryan R. Fonslow, Benjamin D. Stein, Kristofor J. Webb, Tao Xu, Jeong Choi, Sung Kyu Park, and
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Section 1 Proteins and Proteomics
 Section 1 Proteins and Proteomics Learning Objectives At the end of this assignment, you should be able to: 1. Draw the chemical structure of an amino acid and small peptide. 2. Describe the difference
Section 1 Proteins and Proteomics Learning Objectives At the end of this assignment, you should be able to: 1. Draw the chemical structure of an amino acid and small peptide. 2. Describe the difference
Improve Protein Analysis with the New, Mass Spectrometry- Compatible ProteasMAX Surfactant
 Improve Protein Analysis with the New, Mass Spectrometry- Compatible Surfactant ABSTRACT Incomplete solubilization and digestion and poor peptide recovery are frequent limitations in protein sample preparation
Improve Protein Analysis with the New, Mass Spectrometry- Compatible Surfactant ABSTRACT Incomplete solubilization and digestion and poor peptide recovery are frequent limitations in protein sample preparation
Enhancing Sequence Coverage in Proteomics Studies by Using a Combination of Proteolytic Enzymes
 Enhancing Sequence Coverage in Proteomics Studies by Using a Combination of Proteolytic Enzymes Dominic Baeumlisberger 2, Christopher Kurz 3, Tabiwang N. Arrey, Marion Rohmer 2, Carola Schiller 3, Thomas
Enhancing Sequence Coverage in Proteomics Studies by Using a Combination of Proteolytic Enzymes Dominic Baeumlisberger 2, Christopher Kurz 3, Tabiwang N. Arrey, Marion Rohmer 2, Carola Schiller 3, Thomas
Chapter 10. 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation)
 Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
Chapter 31. Completing the Protein Life Cycle: Folding, Processing and Degradation. Biochemistry by Reginald Garrett and Charles Grisham
 Chapter 31 Completing the Protein Life Cycle: Folding, Processing and Degradation Biochemistry by Reginald Garrett and Charles Grisham Essential Question How are newly synthesized polypeptide chains transformed
Chapter 31 Completing the Protein Life Cycle: Folding, Processing and Degradation Biochemistry by Reginald Garrett and Charles Grisham Essential Question How are newly synthesized polypeptide chains transformed
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Experimental design and workflow utilized to generate the WMG Protein Atlas.
 Supplementary Figure 1 Experimental design and workflow utilized to generate the WMG Protein Atlas. (a) Illustration of the plant organs and nodule infection time points analyzed. (b) Proteomic workflow
Supplementary Figure 1 Experimental design and workflow utilized to generate the WMG Protein Atlas. (a) Illustration of the plant organs and nodule infection time points analyzed. (b) Proteomic workflow
Unsupervised Identification of Isotope-Labeled Peptides
 Unsupervised Identification of Isotope-Labeled Peptides Joshua E Goldford 13 and Igor GL Libourel 124 1 Biotechnology institute, University of Minnesota, Saint Paul, MN 55108 2 Department of Plant Biology,
Unsupervised Identification of Isotope-Labeled Peptides Joshua E Goldford 13 and Igor GL Libourel 124 1 Biotechnology institute, University of Minnesota, Saint Paul, MN 55108 2 Department of Plant Biology,
Figure S1: Abundance of Fe related proteins in oceanic Synechococcus sp. strain WH8102 in. Ferredoxin. Ferredoxin. Fe ABC transporter
 0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
Loss of protein association causes cardiolipin degradation in Barth syndrome
 SUPPLEMENTARY INFORMATION Loss of protein association causes cardiolipin degradation in Barth syndrome Yang Xu 1, Colin K.L. Phoon 2, Bob Berno 5, Kenneth D Souza 6, Esthelle Hoedt 4, Guoan Zhang 4, Thomas
SUPPLEMENTARY INFORMATION Loss of protein association causes cardiolipin degradation in Barth syndrome Yang Xu 1, Colin K.L. Phoon 2, Bob Berno 5, Kenneth D Souza 6, Esthelle Hoedt 4, Guoan Zhang 4, Thomas
From Atoms to Cells: Fundamental Building Blocks. Models of atoms. A chemical connection
 From Atoms to Cells: A chemical connection Fundamental Building Blocks Matter - all materials that occupy space & have mass Matter is composed of atoms Atom simplest form of matter not divisible into simpler
From Atoms to Cells: A chemical connection Fundamental Building Blocks Matter - all materials that occupy space & have mass Matter is composed of atoms Atom simplest form of matter not divisible into simpler
Metabolomic and Proteomics Solutions for Integrated Biology. Christine Miller Omics Market Manager ASMS 2015
 Metabolomic and Proteomics Solutions for Integrated Biology Christine Miller Omics Market Manager ASMS 2015 Integrating Biological Analysis Using Pathways Protein A R HO R Protein B Protein X Identifies
Metabolomic and Proteomics Solutions for Integrated Biology Christine Miller Omics Market Manager ASMS 2015 Integrating Biological Analysis Using Pathways Protein A R HO R Protein B Protein X Identifies
COPD lungs show an attached stratified mucus layer that separate. bacteria from the epithelial cells resembling the protective colonic
 COPD lungs show an attached stratified mucus layer that separate bacteria from the epithelial cells resembling the protective colonic mucus SUPPLEMENTARY TABLES AND FIGURES Tables S1 S8, page 1 and separate
COPD lungs show an attached stratified mucus layer that separate bacteria from the epithelial cells resembling the protective colonic mucus SUPPLEMENTARY TABLES AND FIGURES Tables S1 S8, page 1 and separate
Mass Spectrometry and Proteomics - Lecture 4 - Matthias Trost Newcastle University
 Mass Spectrometry and Proteomics - Lecture 4 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk previously Peptide fragmentation Hybrid instruments 117 The Building Blocks of Life DNA RNA Proteins
Mass Spectrometry and Proteomics - Lecture 4 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk previously Peptide fragmentation Hybrid instruments 117 The Building Blocks of Life DNA RNA Proteins
Supplemental Information
 Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Supplementary Material. Contents include:
 Supplementary Material Contents include: 1. Supplementary Figures (p. 2-7) 2. Supplementary Figure Legends (p. 8-9) 3. Supplementary Tables (p. 10-12) 4. Supplementary Table Legends (p. 13) 1 Wellen_FigS1
Supplementary Material Contents include: 1. Supplementary Figures (p. 2-7) 2. Supplementary Figure Legends (p. 8-9) 3. Supplementary Tables (p. 10-12) 4. Supplementary Table Legends (p. 13) 1 Wellen_FigS1
Search settings MaxQuant
 Search settings MaxQuant Briefly, we used MaxQuant version 1.5.0.0 with the following settings. As variable modifications we allowed Acetyl (Protein N-terminus), methionine oxidation and glutamine to pyroglutamate
Search settings MaxQuant Briefly, we used MaxQuant version 1.5.0.0 with the following settings. As variable modifications we allowed Acetyl (Protein N-terminus), methionine oxidation and glutamine to pyroglutamate
NBCE Mock Board Questions Biochemistry
 1. Fluid mosaic describes. A. Tertiary structure of proteins B. Ribosomal subunits C. DNA structure D. Plasma membrane structure NBCE Mock Board Questions Biochemistry 2. Where in the cell does beta oxidation
1. Fluid mosaic describes. A. Tertiary structure of proteins B. Ribosomal subunits C. DNA structure D. Plasma membrane structure NBCE Mock Board Questions Biochemistry 2. Where in the cell does beta oxidation
Chapter 8 Microbial Metabolism: The Chemical Crossroads of Life
 Chapter 8 Microbial Metabolism: The Chemical Crossroads of Life / Building Your Knowledge 1) What are the two branches of metabolism? a. b. Which branch synthesizes large molecules from small subunits?
Chapter 8 Microbial Metabolism: The Chemical Crossroads of Life / Building Your Knowledge 1) What are the two branches of metabolism? a. b. Which branch synthesizes large molecules from small subunits?
SimGlycan. A high-throughput glycan and glycopeptide data analysis tool for LC-, MALDI-, ESI- Mass Spectrometry workflows.
 PREMIER Biosoft SimGlycan A high-throughput glycan and glycopeptide data analysis tool for LC-, MALDI-, ESI- Mass Spectrometry workflows SimGlycan software processes and interprets the MS/MS and higher
PREMIER Biosoft SimGlycan A high-throughput glycan and glycopeptide data analysis tool for LC-, MALDI-, ESI- Mass Spectrometry workflows SimGlycan software processes and interprets the MS/MS and higher
Quantification with Proteome Discoverer. Bernard Delanghe
 Quantification with Proteome Discoverer Bernard Delanghe Overview: Which approach to use? Proteome Discoverer Quantification Method What When to use Metabolic labeling SILAC Cell culture systems Small
Quantification with Proteome Discoverer Bernard Delanghe Overview: Which approach to use? Proteome Discoverer Quantification Method What When to use Metabolic labeling SILAC Cell culture systems Small
Limsoon Wong Global-COE Workshop on Engineering/Information Science for Integrated Life Science and Predictive Medicine 28 February 2012
 A Novel Contextualization Approach to Proteomic Profile Analysis Limsoon Wong Global-COE Workshop on Engineering/Information Science for Integrated Life Science and Predictive Medicine 28 February 2012
A Novel Contextualization Approach to Proteomic Profile Analysis Limsoon Wong Global-COE Workshop on Engineering/Information Science for Integrated Life Science and Predictive Medicine 28 February 2012
Supporting Information
 Supporting Information Phillips and Milo 10.1073/pnas.0907732106 Table S1. Database vital statistics Number of entries Number of distinct references Number of organisms Unique visitors per day Searches
Supporting Information Phillips and Milo 10.1073/pnas.0907732106 Table S1. Database vital statistics Number of entries Number of distinct references Number of organisms Unique visitors per day Searches
Week 3 The Pancreas: Pancreatic ph buffering:
 Week 3 The Pancreas: A gland with both endocrine (secretion of substances into the bloodstream) & exocrine (secretion of substances to the outside of the body or another surface within the body) functions
Week 3 The Pancreas: A gland with both endocrine (secretion of substances into the bloodstream) & exocrine (secretion of substances to the outside of the body or another surface within the body) functions
Insulin mrna to Protein Kit
 Insulin mrna to Protein Kit A 3DMD Paper BioInformatics and Mini-Toober Folding Activity Student Handout www.3dmoleculardesigns.com Insulin mrna to Protein Kit Contents Becoming Familiar with the Data...
Insulin mrna to Protein Kit A 3DMD Paper BioInformatics and Mini-Toober Folding Activity Student Handout www.3dmoleculardesigns.com Insulin mrna to Protein Kit Contents Becoming Familiar with the Data...
J. A. Mayfield et al. FIGURE S1. Methionine Salvage. Methylthioadenosine. Methionine. AdoMet. Folate Biosynthesis. Methylation SAH.
 FIGURE S1 Methionine Salvage Methionine Methylthioadenosine AdoMet Folate Biosynthesis Methylation SAH Homocysteine Homocystine CBS Cystathionine Cysteine Glutathione Figure S1 Biochemical pathway of relevant
FIGURE S1 Methionine Salvage Methionine Methylthioadenosine AdoMet Folate Biosynthesis Methylation SAH Homocysteine Homocystine CBS Cystathionine Cysteine Glutathione Figure S1 Biochemical pathway of relevant
Supplementary Figure 1:
 Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
DEPARTMENT OF SCIENCE
 DEPARTMENT OF SCIENCE COURSE OUTLINE Fall 2015 BC 2000 INTRODUCTORY BIOCHEMISTRY INSTRUCTOR: Philip Johnson PHONE: 780-539-2863 OFFICE: J224 E-MAIL: PJohnson@gprc.ab.ca OFFICE HOURS: Tuesdays 1000-1120
DEPARTMENT OF SCIENCE COURSE OUTLINE Fall 2015 BC 2000 INTRODUCTORY BIOCHEMISTRY INSTRUCTOR: Philip Johnson PHONE: 780-539-2863 OFFICE: J224 E-MAIL: PJohnson@gprc.ab.ca OFFICE HOURS: Tuesdays 1000-1120
Proteins are sometimes only produced in one cell type or cell compartment (brain has 15,000 expressed proteins, gut has 2,000).
 Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Amino acids. Side chain. -Carbon atom. Carboxyl group. Amino group
 PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
REGULATION OF ENZYME ACTIVITY. Medical Biochemistry, Lecture 25
 REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
Supplemental Information. Comprehensive Quantification. of the Modified Proteome Reveals Oxidative. Heart Damage in Mitochondrial Heteroplasmy
 Cell Reports, Volume 23 Supplemental Information Comprehensive Quantification of the Modified Proteome Reveals Oxidative Heart Damage in Mitochondrial Heteroplasmy Navratan Bagwan, Elena Bonzon-Kulichenko,
Cell Reports, Volume 23 Supplemental Information Comprehensive Quantification of the Modified Proteome Reveals Oxidative Heart Damage in Mitochondrial Heteroplasmy Navratan Bagwan, Elena Bonzon-Kulichenko,
Supplementary. properties of. network types. randomly sampled. subsets (75%
 Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
METABOLISM -Introduction- Serkan SAYINER, DVM PhD. Assist. Prof.
 METABOLISM -Introduction- Serkan SAYINER, DVM PhD. Assist. Prof. Near East University, Faculty of Veterinary Medicine, Department of Biochemistry serkan.sayiner@neu.edu.tr Overview Living organisms need
METABOLISM -Introduction- Serkan SAYINER, DVM PhD. Assist. Prof. Near East University, Faculty of Veterinary Medicine, Department of Biochemistry serkan.sayiner@neu.edu.tr Overview Living organisms need
1- Which of the following statements is TRUE in regards to eukaryotic and prokaryotic cells?
 Name: NetID: Exam 3 - Version 1 October 23, 2017 Dr. A. Pimentel Each question has a value of 4 points and there are a total of 160 points in the exam. However, the maximum score of this exam will be capped
Name: NetID: Exam 3 - Version 1 October 23, 2017 Dr. A. Pimentel Each question has a value of 4 points and there are a total of 160 points in the exam. However, the maximum score of this exam will be capped
Production Management: New Lessons from Biology. James E. Metherall, Ph.D., M.B.A. Associate Professor of Human Genetics University of Utah
 Production Management: New Lessons from Biology James E. Metherall, Ph.D., M.B.A. Associate Professor of Human Genetics University of Utah Major Points 1) Metabolic Regulation is an Extremely Complex Operations
Production Management: New Lessons from Biology James E. Metherall, Ph.D., M.B.A. Associate Professor of Human Genetics University of Utah Major Points 1) Metabolic Regulation is an Extremely Complex Operations
The Structure and Function of Biomolecules
 The Structure and Function of Biomolecules The student is expected to: 9A compare the structures and functions of different types of biomolecules, including carbohydrates, lipids, proteins, and nucleic
The Structure and Function of Biomolecules The student is expected to: 9A compare the structures and functions of different types of biomolecules, including carbohydrates, lipids, proteins, and nucleic
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
Characterization of the use of wastewater from hydrothermal liquefaction as a nitrogen source by the potential algal biofuel strain Picochlorum sp.
 Characterization of the use of wastewater from hydrothermal liquefaction as a nitrogen source by the potential algal biofuel strain Picochlorum sp. Shuyi Wang, Xinguo Shi, Fatima Foflonker, Debashish Bhattacharya,
Characterization of the use of wastewater from hydrothermal liquefaction as a nitrogen source by the potential algal biofuel strain Picochlorum sp. Shuyi Wang, Xinguo Shi, Fatima Foflonker, Debashish Bhattacharya,
ANSWERS Problem Set 8
 ANSWERS Problem Set 8 Problem 1. All oxidation steps in the pathway from glucose to CO 2 result in the production of NADH, except the succinate dehydrogenase (SDH) step in the TCA cycle, which yields FADH2.
ANSWERS Problem Set 8 Problem 1. All oxidation steps in the pathway from glucose to CO 2 result in the production of NADH, except the succinate dehydrogenase (SDH) step in the TCA cycle, which yields FADH2.
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 The timeline of the NGAG method for extraction of N-linked glycans and glycosite-containing peptides. The timeline can be changed based on the number of samples. Supplementary Figure
Supplementary Figure 1 The timeline of the NGAG method for extraction of N-linked glycans and glycosite-containing peptides. The timeline can be changed based on the number of samples. Supplementary Figure
Biol 219 Lec 7 Fall 2016
 Cellular Respiration: Harvesting Energy to form ATP Cellular Respiration and Metabolism Glucose ATP Pyruvate Lactate Acetyl CoA NAD + Introducing The Players primary substrate for cellular respiration
Cellular Respiration: Harvesting Energy to form ATP Cellular Respiration and Metabolism Glucose ATP Pyruvate Lactate Acetyl CoA NAD + Introducing The Players primary substrate for cellular respiration
Protein Homeostasis in Mitochondria: Chaperones and Proteases of the Mitochondrial Matrix Prof. Wolfgang Voos
 Protein Homeostasis in Mitochondria: Chaperones and Proteases of the Mitochondrial Matrix Institute for Biochemistry and Molecular Biology (IBMB) University of Bonn, Germany wolfgang.voos@uni-bonn.de 1
Protein Homeostasis in Mitochondria: Chaperones and Proteases of the Mitochondrial Matrix Institute for Biochemistry and Molecular Biology (IBMB) University of Bonn, Germany wolfgang.voos@uni-bonn.de 1
Bacterial Structures. Capsule or Glycocalyx TYPES OF FLAGELLA FLAGELLA. Average size: µm 2-8 µm Basic shapes:
 PROKARYOTIC One circular chromosome, not in a membrane No histones No organelles Peptidoglycan cell walls Binary fission EUKARYOTIC Paired chromosomes, in nuclear membrane Histones Organelles Polysaccharide
PROKARYOTIC One circular chromosome, not in a membrane No histones No organelles Peptidoglycan cell walls Binary fission EUKARYOTIC Paired chromosomes, in nuclear membrane Histones Organelles Polysaccharide
Aerobic Fate of Pyruvate. Chapter 16 Homework Assignment. Chapter 16 The Citric Acid Cycle
 Chapter 16 Homework Assignment The following problems will be due once we finish the chapter: 1, 3, 7, 10, 16, 19, 20 Additional Problem: Write out the eight reaction steps of the Citric Acid Cycle, using
Chapter 16 Homework Assignment The following problems will be due once we finish the chapter: 1, 3, 7, 10, 16, 19, 20 Additional Problem: Write out the eight reaction steps of the Citric Acid Cycle, using
Chapter 7 Cellular Respiration and Fermentation*
 Chapter 7 Cellular Respiration and Fermentation* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. Life Is Work
Chapter 7 Cellular Respiration and Fermentation* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. Life Is Work
Iron depletion enhances production of antimicrobials by Pseudomonas
 Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. Angela T. Nguyen 1, Jace W. Jones 1, Max A. Ruge 1, Maureen A. Kane 1, and Amanda G. Oglesby-Sherrouse 1,2 * University of
Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. Angela T. Nguyen 1, Jace W. Jones 1, Max A. Ruge 1, Maureen A. Kane 1, and Amanda G. Oglesby-Sherrouse 1,2 * University of
Application Note # LCMS-89 High quantification efficiency in plasma targeted proteomics with a full-capability discovery Q-TOF platform
 Application Note # LCMS-89 High quantification efficiency in plasma targeted proteomics with a full-capability discovery Q-TOF platform Abstract Targeted proteomics for biomarker verification/validation
Application Note # LCMS-89 High quantification efficiency in plasma targeted proteomics with a full-capability discovery Q-TOF platform Abstract Targeted proteomics for biomarker verification/validation
MITOCHONDRIA LECTURES OVERVIEW
 1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
MEMBRANE-BOUND ELECTRON TRANSFER AND ATP SYNTHESIS (taken from Chapter 18 of Stryer)
 MEMBRANE-BOUND ELECTRON TRANSFER AND ATP SYNTHESIS (taken from Chapter 18 of Stryer) FREE ENERGY MOST USEFUL THERMODYNAMIC CONCEPT IN BIOCHEMISTRY Living things require an input of free energy for 3 major
MEMBRANE-BOUND ELECTRON TRANSFER AND ATP SYNTHESIS (taken from Chapter 18 of Stryer) FREE ENERGY MOST USEFUL THERMODYNAMIC CONCEPT IN BIOCHEMISTRY Living things require an input of free energy for 3 major
Lipidomics Discovery Profiling and Targeted LC/MS Analysis in 3T3-L1 Differentiating Adipocytes
 Lipidomics Discovery Profiling and Targeted LC/MS Analysis in 3T3-L1 Differentiating Adipocytes Application Note Authors Theodore Sana and Steve Fischer Agilent Technologies, Inc. Santa Clara, CA, USA
Lipidomics Discovery Profiling and Targeted LC/MS Analysis in 3T3-L1 Differentiating Adipocytes Application Note Authors Theodore Sana and Steve Fischer Agilent Technologies, Inc. Santa Clara, CA, USA
The distribution of log 2 ratio (H/L) for quantified peptides. cleavage sites in each bin of log 2 ratio of quantified. peptides
 Journal: Nature Methods Article Title: Corresponding Author: Protein digestion priority is independent of their abundances Mingliang Ye and Hanfa Zou Supplementary Figure 1 Supplementary Figure 2 The distribution
Journal: Nature Methods Article Title: Corresponding Author: Protein digestion priority is independent of their abundances Mingliang Ye and Hanfa Zou Supplementary Figure 1 Supplementary Figure 2 The distribution
Details of Organic Chem! Date. Carbon & The Molecular Diversity of Life & The Structure & Function of Macromolecules
 Details of Organic Chem! Date Carbon & The Molecular Diversity of Life & The Structure & Function of Macromolecules Functional Groups, I Attachments that replace one or more of the hydrogens bonded to
Details of Organic Chem! Date Carbon & The Molecular Diversity of Life & The Structure & Function of Macromolecules Functional Groups, I Attachments that replace one or more of the hydrogens bonded to
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Chapter 14. Energy conversion: Energy & Behavior
 Chapter 14 Energy conversion: Energy & Behavior Why do you Eat and Breath? To generate ATP Foods, Oxygen, and Mitochodria Cells Obtain Energy by the Oxidation of Organic Molecules Food making ATP making
Chapter 14 Energy conversion: Energy & Behavior Why do you Eat and Breath? To generate ATP Foods, Oxygen, and Mitochodria Cells Obtain Energy by the Oxidation of Organic Molecules Food making ATP making
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12864 Supplementary Table 1 1 2 3 4 5 6 7 Peak Gene code Screen Function or Read analysis AMP reads camp annotation reads minor Tb927.2.1810 AMP ISWI Confirmed
SUPPLEMENTARY INFORMATION doi:10.1038/nature12864 Supplementary Table 1 1 2 3 4 5 6 7 Peak Gene code Screen Function or Read analysis AMP reads camp annotation reads minor Tb927.2.1810 AMP ISWI Confirmed
Week 2. Macromolecules
 Week 2 In living organisms, smaller molecules are often attached to each other to make larger molecules. These smaller molecules are sometimes called monomers, and the larger molecules made from these
Week 2 In living organisms, smaller molecules are often attached to each other to make larger molecules. These smaller molecules are sometimes called monomers, and the larger molecules made from these
Metabolomic Data Analysis with MetaboAnalyst
 Metabolomic Data Analysis with MetaboAnalyst User ID: guest6501 April 16, 2009 1 Data Processing and Normalization 1.1 Reading and Processing the Raw Data MetaboAnalyst accepts a variety of data types
Metabolomic Data Analysis with MetaboAnalyst User ID: guest6501 April 16, 2009 1 Data Processing and Normalization 1.1 Reading and Processing the Raw Data MetaboAnalyst accepts a variety of data types
LECTURE-15. itraq Clinical Applications HANDOUT. Isobaric Tagging for Relative and Absolute quantitation (itraq) is a quantitative MS
 LECTURE-15 itraq Clinical Applications HANDOUT PREAMBLE Isobaric Tagging for Relative and Absolute quantitation (itraq) is a quantitative MS based method for quantifying proteins subject to various different
LECTURE-15 itraq Clinical Applications HANDOUT PREAMBLE Isobaric Tagging for Relative and Absolute quantitation (itraq) is a quantitative MS based method for quantifying proteins subject to various different
GENERAL CHARACTERISTICS OF THE ENDOCRINE SYSTEM FIGURE 17.1
 GENERAL CHARACTERISTICS OF THE ENDOCRINE SYSTEM FIGURE 17.1 1. The endocrine system consists of glands that secrete chemical signals, called hormones, into the blood. In addition, other organs and cells
GENERAL CHARACTERISTICS OF THE ENDOCRINE SYSTEM FIGURE 17.1 1. The endocrine system consists of glands that secrete chemical signals, called hormones, into the blood. In addition, other organs and cells
A look at macromolecules (Text pages 38-54) What is the typical chemical composition of a cell? (Source of figures to right: Madigan et al.
 A look at macromolecules (Text pages 38-54) What is the typical chemical composition of a cell? (Source of figures to right: Madigan et al. 2002 Chemical Bonds Ionic Electron-negativity differences cause
A look at macromolecules (Text pages 38-54) What is the typical chemical composition of a cell? (Source of figures to right: Madigan et al. 2002 Chemical Bonds Ionic Electron-negativity differences cause
Cell Structure & Function. Source:
 Cell Structure & Function Source: http://koning.ecsu.ctstateu.edu/cell/cell.html Definition of Cell A cell is the smallest unit that is capable of performing life functions. http://web.jjay.cuny.edu/~acarpi/nsc/images/cell.gif
Cell Structure & Function Source: http://koning.ecsu.ctstateu.edu/cell/cell.html Definition of Cell A cell is the smallest unit that is capable of performing life functions. http://web.jjay.cuny.edu/~acarpi/nsc/images/cell.gif
How do we retain emphasis on function?
 Systems Biology Systems biology studies biological systems by systematically perturbing them (biologically, genetically, or chemically); monitoring the gene, protein, and informational pathway responses;
Systems Biology Systems biology studies biological systems by systematically perturbing them (biologically, genetically, or chemically); monitoring the gene, protein, and informational pathway responses;
Comprehensive Lipid Profiling of Human Liver Tissue Extracts of Non-Alcoholic Fatty Liver Disease
 Comprehensive Lipid Profiling of Human Liver Tissue Extracts of Non-Alcoholic Fatty Liver Disease Multiplexed Precursor Ion Scanning and LipidView Software Brigitte Simons 1 and Bianca Arendt 2 1 AB SCIEX,
Comprehensive Lipid Profiling of Human Liver Tissue Extracts of Non-Alcoholic Fatty Liver Disease Multiplexed Precursor Ion Scanning and LipidView Software Brigitte Simons 1 and Bianca Arendt 2 1 AB SCIEX,
Point total. Page # Exam Total (out of 90) The number next to each intermediate represents the total # of C-C and C-H bonds in that molecule.
 This exam is worth 90 points. Pages 2- have questions. Page 1 is for your reference only. Honor Code Agreement - Signature: Date: (You agree to not accept or provide assistance to anyone else during this
This exam is worth 90 points. Pages 2- have questions. Page 1 is for your reference only. Honor Code Agreement - Signature: Date: (You agree to not accept or provide assistance to anyone else during this
Proteins? Protein function. Protein folding. Protein folding diseases. Protein interactions. Macromolecular assemblies. The end product of Genes
 Proteins? Protein function Protein folding Protein folding diseases Protein interactions Macromolecular assemblies The end product of Genes Protein Unfolding DOD Acid Catalysis DOD HDOD + N H N D C N C
Proteins? Protein function Protein folding Protein folding diseases Protein interactions Macromolecular assemblies The end product of Genes Protein Unfolding DOD Acid Catalysis DOD HDOD + N H N D C N C
