Differential Expression of reg-i and reg-ii Genes During Aging in the Normal Mouse
|
|
|
- Cuthbert Tate
- 6 years ago
- Views:
Transcription
1 Journal of Gerontology: BIOLOGICAL SCIENCES 1996, Vol. 51A, No. 5, B308-B315 Copyright 1996 by The Gerontological Society of America Differential Expression of reg-i and reg-ii Genes During Aging in the Normal Mouse Riccardo Perfetti, 1 Josephine M. Egan, 1 Michael E. Zenilman, 2 and Alan R. Shuldiner 3 'Diabetes Unit, Laboratory of Clinical Physiology, National Institute on Aging, National Institutes of Health. department of Surgery, and 'Division of Geriatric Medicine and Gerontology, Johns Hopkins University School of Medicine, Baltimore. A cdna termed reg (for regenerating gene) has been isolated from a rat pancreatic DNA library. Reg expression has been shown to correlate with changes in /3 cell mass and function. This finding has been recently challenged by studies showing a non-fi-cell-dependent regulation of reg expression. All studies to date, however, have neglected the fact that two nonallelic reg genes (reg-i and reg-ii) exist in several species. In studying the regulation of each individual copy gene, we investigated reg-i and -II gene expression in a naturally occurring modification of fi-cellphysiology: normal aging. RNA was isolated from individual pancreata of1-, 3-, 9-, 20-, and 30-month-old C57BL/6J mice (n 2= 3 per group) and subjected to slot-blot analysis using homologous probes for reg-i, reg-ii, insulin, and elastase-i. A progressive age-dependent decrease in total reg mrna levels (reg-i and -II) was detected. At 30 months of age, total reg mrna levels were approximately 45% of the level detected in 1-month-old mice (p =.01). This paralleled the decrease in insulin mrna levels (p =.01), which fell below 50%; by contrast, mrna levels for elastase-i increased with age (p =.05). Analysis of RNA isolated from purified islets did not reveal any mrna for reg, suggesting that in the normal mouse, reg is primarily a product of the exocrine pancreas. Reg mrna were detectable in RNA extracts from stomach, duodenum, and small intestine. By hybridization of total pancreatic RNA with oligonucleotide probes which specifically recognize reg-i or reg-ii sequences, we show that reg-i mrna levels declined with age (p =.001) while reg-ii mrna levels remained unchanged. These data demonstrate that in mouse pancreas the two nonallelic reg genes are differentially expressed during aging and that the decrease in reg-i mrna levels parallels the decrease in insulin gene expression. Differential regulation of reg-i and reg-ii genes may explain the presence of conflicting data in the current literature. MATURE pancreatic islets of Langerhans are composed of cells that slowly undergo mitosis and that exhibit poor capability to regenerate after injury (Swenne, 1983; Hellerstrom et al., 1988). Administration of poly(adpribose) synthase inhibitors, such as nicotinamide, to rats subjected to 90% pancreatectomy induces regeneration of pancreatic islets and amelioration of diabetes (Okamoto, 1981; Yonemura et al., 1984, 1988). Terazono et al. (1988) isolated and characterized a cdna from a rat regenerating pancreatic-derived cdna library, designated reg (for regenerating gene). The 166-amino acid peptide has also been identified in mouse (Unno et al., 1993), bovine (Cai et al., 1990), and human (Watanabe et al., 1990) and is homologous to the C-type animal lectins, a family of proteins with known mitogenic properties (Drickamer, 1988). Two nonallelic reg genes have been identified in human (Watanabe et al., 1990), rat (Kamimura et al., 1992), and mouse (Unno et al., 1993) pancreatic cdna libraries. In several models, reg expression has been shown to mirror changes in (S-cell mass and/or P-cell function. Reg mrna levels increased markedly in islets induced to proliferate by pancreatectomy and nicotinamide treatment compared to normal islets (Terazono et al., 1988). The increase in reg expression was temporally correlated with the increase in size of the regenerating islets and the decrease in glycosuria. Increased reg gene expression has also been demonstrated in the hyperplastic islets of NON (non-obese) mice after treatment with aurothioglucose (Terazono et al., 1988). In another model of pancreatic regeneration in which the pancreatic duct of a rat is subjected to nonocclusive obstruction (wrapping), reg expression increased coordinately with the appearance of new islets (Zenilman et al., 1993). Further, subcutaneous implantation of a solid insulinoma tumor into rats caused a dramatic reduction in the volume of the endogenous islets (Chick et al., 1977). This phenomenon was associated with coordinate suppression of both reg and insulin gene expression (Miyaura et al., 1991). Removal of the tumor resulted in a rapid induction of reg gene expression, proliferation of endocrine P cells, and the restoration of endogenous insulin gene expression and secretion (Miyaura etal., 1991). Although these studies appear to link changes in reg gene expression with changes in (3-cell mass, Smith et al. (1994) have shown that reg may be induced even in the absence of proliferative changes. This finding discouraged many laboratories from performing further studies to explore the possible role of reg as a growth and maintenance factor for pancreatic 3 cells. We believe that a fundamental technical problem may have contributed to the generation of conflicting data. Both proponents and opponents of the view that reg may be important in (3-cell physiology have neglected previous reports demonstrating that in several animal species there are two independent, nonallelic reg genes located on different chromosomes (Watanabe et al., 1990; Kaminura et al., 1992; Unno et al., 1993). All studies to date, either in support of or against a role for reg in (3-cell physiology, have B308
2 PANCREATIC reg-i AND -II GENE EXPRESSION DURING AGING B309 been performed using cdna probes that were not able to distinguish between the two reg mrnas. We believe that this may have largely contributed to the presence of conflicting results in the literature. One of the aims of this work is to provide experimental evidence for the need to support a revision of those studies, taking into account the two nonallelic reg genes individually. In this study, we investigated the regulation of pancreatic reg-i and reg-ii gene expression during normal aging in the mouse. mrna Insulin Up Down Elastase-I Up Down Table 1. Sequence of Oligonucleotide Primers Used to Generate Mouse cdna Probes Primers Size (bp) 5-'GTCCCGCCGTGAAGTGGAGG-3' 171 5' -GTTGC AGTAGTTCTCC AG-3' 5'-ATGCTGCGCTTCCTGTTC-3' 393 5' - AG A ATACTGGCCGTTC ACCTT-3' MATERIALS AND METHODS Animals. One-, 3-, 9-, 20-, and 30-month-old C57BL/ 6J male mice (n ^ 3 per group) were housed at the Gerontology Research Center, National Institute on Aging. They were fed a stock diet and maintained in a controlled environment (12-h light/dark cycle; 22 C). Prior to sacrifice by decapitation, they had free access to water but were fasted overnight. Pancreata were quickly removed and used for subsequent experiments. All procedures were in accordance with National Institutes of Health guidelines and were approved by the Animal Resource Section of the National Institute on Aging. Oligonucleotide primers and probes. Oligonucleotides, synthesized on an Applied Biosystems (Foster City, CA) DNA synthesizer were cleaved from the resin with concentrated ammonium hydroxide at room temperature, deprotected by heating to 55 C overnight, and purified over Nensorb Prep columns (New England Nuclear, Boston, MA). Polymerase chain reaction (PCR) primers were designed according to the published mouse or rat sequences for reg, insulin, and elastase-i cdnas (MacDonald et al., 1982; Wentworth et al., 1986; Unno et al., 1993) (Table 1). Homologous cdna probes were generated by reverse transcription (RT)-PCR (see below). To measure total reg mrna levels, a 246-bp reg-ii cdna probe that hybridizes to mrnas for both reg-i and reg-ii was generated by RT- PCR. In addition, two oligonucleotides specific for reg-i or reg-ii mrna were used to detect the relative abundance of each gene individually (Table 1 and Figure 1). Oligonucleotide and cdna probes were radiolabeled using [7-32 P]ATP (polynucleotide kinase) and [a- 32 P]dCTP (random priming), respectively (Amersham, Arlington Heights, IL). Synthesis ofpancreatic cdna probes by RT-PCR. One microgram of mouse pancreatic RNA extracted from a 3- month-old male was used as template for RT which was performed at 37 C in a final volume of 20 (xl containing 50 mm KC1, 10 mm Tris-HCl (ph 8.3 at 25 C), 1.5 mm MgCl 2, 0.01 mg/ml gelatin, 0.2 mm each dntp, 40 units/ tube RNasin (Promega, Madison, WI), 0.5 \im downstream primer (Table 1), and 7 units/tube avian myeloblastosis virus reverse transcriptase (Promega) (Saiki et al., 1988; ShuldinerandPerfetti, 1993). The second DNA strand was synthesized during the first cycle of PCR in which 5 jxl of the RT reaction mixture was used in a final volume of 50 xl containing 50 mm KC1, 10 mm TrisHCl (ph 8.3 at 25 C), 1.5 mm MgCl mg/ml Reg-I Reg-II U,,S" GGCTCAGAACAATGTATACC 3' D 5' GTTGGCAGTGCTTGGAGCTC 3 1 Reg-II standard Reg-I standard Reg-II probe I). 5' GCTAGGAACGCCTACTTCATCC 3 Primers Tor Reg-II standard Primers for Reg I standard U S 1 GCCTCGCTGGTTAAGGAGAGT 3' I Primers for Reg-II prol D 5 1 GGCTCTGAACTTGCAGACAAAT 3 1 J Figure 1. Schematic representation of reg-i and reg-ii mrnas and the oligonucleotide primers and probe used. The coding region of reg is composed of six exons. Primers used to obtain reg-i and reg-ii standards are indicated as U, and D,, and U n and D n, respectively. The reg-i product was 395 bp long, and the reg-ii product was 418 bp long. Primers used to obtain the 246-bp reg-ii cdna probe are indicated as U and D; oligonucleotide probes that specifically recognize reg-i or reg-ii sequences are indicated as I* and II*. The oligonucleotide that was common to mrnas for both reg-i and reg-ii is indicated as C*. gelatin, 0.2 mm each dntp, 0.5 \xm upstream primer (Table 1), 0.5 (xm downstream primer (Table 1), 1 unit/tube Taq polymerase (Perkin-Elmer/Cetus, Norwalk, CT), and approximately 50 xl of paraffin oil. Thirty-nine PCR cycles were performed. Each cycle consisted of denaturation (1 min at 94 C), annealing (1 min at 55 C), and extension (1 min at 72 C), except the first cycle in which the denaturation time was increased to 5 min, and the last cycle in which the extension time was increased to 10 min. For RT-PCR of elastase-i, the conditions for the amplification were identical to those described above except that the annealing temperature was decreased to 50 C. RT-PCR products were subjected to electrophoresis on a composite gel consisting of 1% agarose (Bethesda Research Laboratories, Gaithersburg, I
3 B310 PERFETTIET AL. MD) and 2% Nusieve (FMC Byproducts, Rockland, ME). The gel was stained with ethidium bromide and photographed to verify that the amplified products corresponded to the predicted size. Synthesis ofreg-i and reg-ii cdna standards. A PCRbased cloning strategy was used to obtain partial cdna sequences of mouse reg-i and reg-ii using gene-specific oligonucleotide primers specific for each sequence (Figure 1). Mouse reg-i and reg-ii PCR products were ligated into pcr-h plasmid (Invitrogen, San Diego, CA). Plasmid DNA was purified using a Qiagen (Chatsworth, CA) kit. Purified plasmid DNA (1-3 (ig) was subjected to dideoxy sequence analysis (Sequenase, United States Biochemical, Cleveland, OH). Reg-I and reg-ii cdnas were used as standards to demonstrate that hybridization with gene-specific oligonucleotide probes was (7) specific, and (2) that the specific activity of the radiolabeled probes was similar. RNA extraction of solid tissue and Northern blot analysis. Total RNA was prepared from fresh tissue (pancreas, esophagus, stomach, duodenum, small intestine, colon, heart, lung, brain, kidney, spleen, and testis) by homogenization in guanidinium thiocyanate (Chirgwin et al., 1979). Pancreata were collected from all age groups described above, while the other tissues were obtained only from 3- month-old mice. RNA samples (10 fxg) were subjected to electrophoresis on a 1.5% agarose, 2.2 M formaldehyde gel. Ribosomal RNA was visualized by staining with ethidium bromide. RNA was transferred to nylon filters (Schleicher & Schuell, Keene, NH), fixed by baking for 2 h in a vacuum oven at 80 C, and hybridized with an oligonucleotide probe homologous to the mouse 18S ribosomal subunit (5'- ACGGTATCTGATCGTCTTCGAA-3'). Prehybridization and hybridization were performed at 42 C in 5 x Denhardt's solution containing 50 mm Tris-HCl (ph 7.5), 1 M NaCl, 0.1% sodium pyrophosphate, 1% sodium dodecyl sulfate (SDS), 10% dextran sulfate, and 100 n-g/ml denaturated salmon sperm DNA. Filters were washed twice under stringent conditions (2 x SSPE, 0.05% SDS; 45 C, 15 min) and then exposed to Kodak (Eastman Kodak, Rochester, NY) XAR-5 film with enhancing screens at -70 C for 24 h. RNA isolation from mouse islets of Langerhans. Total RNA from freshly isolated primary cultures of mouse pancreatic islets from 3-month-old mice was isolated using a modification of the guanidinium thiocyanate-phenol-chloroform method (Chomczynski and Sacchi, 1987). Briefly, cells were homogenized by passage through 18- or 22-gauge needles in the presence of denaturing solution [4 M guanidinium thiocyanate, 25 mm sodium citrate (ph 7), 0.5% N- lauroylsarcosine, 0.1 M 2-mercaptoethanol]. Then, 0.1 ml of 2 M sodium acetate (ph 4), 1 ml of phenol, and 0.2 ml of chloroformisoamyl alcohol mixture (49:1) were added to the homogenate. The suspension was shaken vigorously and cooled on ice for 15 min. Samples were centrifuged at 10,000 x g for 20 min at 4 C. After centrifugation, the aqueous phase (1 ml) was transferred to a new tube, mixed with 1 ml of isopropanol, and placed at -20 C for at least 1 h. RNA was pelleted by centrifugation at 20,000 x g for 20 min. The RNA pellet was dissolved in 0.3 ml of denaturing solution, transferred to a new tube, and precipitated with 1 volume of isopropanol at-20 C for 1 h. After centrifugation for 10 min at 4 C, the RNA pellet was resuspended in 75% ethanol, sedimented, vacuum dried, and dissolved in 50 xl 0.5% SDS by heating at 65 C for 10 min and vortex mixing. Slot-blot analysis. Fifteen micrograms of total RNA from pancreas and other tissues were diluted in sterile TE buffer [10 mm TrisHCl, 1 mm EDTA (ph 7.4)] to a final volume of 50 xl. Twenty microliters of 37% formaldehyde and 20 xl 10 x standard saline citrate (SSC) were added, and the solution was placed at 60 C for 15 min. The samples were then diluted with 1 ml of cold 10 x SSC, and 300 \i\ of the solution was applied in triplicate to a nylon membrane in a slot-blot manifold. The wells were washed three times with 400 xl cold 10 x SSC, air dried, and baked in a vacuum oven at 80 C for 2 h. The membranes were hybridized with X 10 6 cpm of 32 P-radiolabeled probe per ml of hybridization solution. Hybridization conditions were the same as those described for Northern blot analysis. The blots were washed twice in 2 x SSPE and 0.5% SDS for 15 min at 55 C when hybridized with cdna probes, or twice in 2x SSPE and 0.5% SDS for 15 min at 42 C when hybridized with oligonucleotide probes, then subjected to autoradiography. Quantification ofmrna levels. The relative levels of mrna for reg-i, reg-ii, insulin, and elastase-i between mice of different ages were quantified using a Betascope 703 blot analyzer (Betagen, Waltham, MA). After quantification, the blots were stripped by washing twice with 0.01 x SSPE and 0.1% SDS at 80 C. To correct for possible differences in loading of RNA, data were normalized by rehybridizing the same blots with 32 P-labeled oligo-dt 20. Statistical analyses. The data were expressed as mean ± SEM of three to five replicates and analyzed by one-way analysis of variance (ANOVA). Regression analysis was performed for all data presented. RESULTS Subcloning of mouse reg-i and reg-ii cdnas and validation of probe specificity. To generate cdna standards for reg-i and reg-ii, RT-PCR was performed using oligonucleotide primers specific for each sequence (U, and D, for reg-i, and U u and D u for reg-ii) (Figure 1). The identity of the two products was confirmed by their difference in size, as detected by agarose gel electrophoresis: reg-i and reg-ii PCR products were 395 and 418 bp long, respectively (data not shown). Both products were subcloned and sequenced to further confirm their identity (data not shown). Figure 2 shows oligonucleotide hybridization of serial dilutions (1:10) of reg-i and reg-ii cdna standards with genespecific oligonucleotide probes (reg-i probe, Left; reg-ii probe, Center) and with an oligonucleotide probe common to both sequences (probe C, Right). The two gene-specific probes recognized exclusively their respective sequences, and the common probe recognized both reg-i and reg-ii sequences (Figure 2).
4 PANCREATIC reg-i AND -II GENE EXPRESSION DURING AGING B311 Oligonucleotide probes Reg-I* Reg-II* C* (Reg-I & -II) t Reg-I and -II 1 2 REG STANDARDS II I* 5 1 CCCCTTGGCTGAAAAAGAC 3 1 II* 5* GCCTACAGCTCCTATTGTTAC 3' C* 5' CCAATGTCTGGACTGGACT 3' Insulin Figure 2. Gene-specific oligonucleotide hybridization. (Upper) Serial dilutions (1:10) of reg-i and reg-ii cdnas were subjected to slot-blot hybridization in triplicate with oligonucleotide probes that specifically recognized reg-i (Left), and reg-ii (Center), as well as probe C (Right) that recognized both sequences. (Lower) Sequences of oligonucleotide probes used for gene-specific hybridization. Elastase-1 The same membranes containing serial dilutions of reg-i and reg-ii cdna standards were hybridized with the 246-bp reg-ii cdna probe. With the hybridization and wash conditions employed, this probe recognized both reg-i and reg-ii sequences (data not shown). Northern blot analysis of pancreatic and extrapancreatic RNA. Northern blot analysis was performed for all samples to verify the integrity of the RNA by ethidium-bromide staining and visualization of the 28S and 18S ribosomal bands. In addition, hybridization of RNA samples with an oligonucleotide probe homologous to the mouse 18S ribosomal subunit revealed that no significant degradation occurred (data not shown). Reg mrna in whole pancreas and in islets oflangerhans. Northern blot analysis of total RNA extracted from 3- month-old mouse islets and from 3-month-old whole pancreas was performed with cdna probes for mouse insulin, elastase-i, and reg-ii (which recognized equally well both reg-i and reg-ii sequences). Reg mrna was detectable in the whole pancreas but not in the isolated islets (Figure 3). This indicated that in the normal mouse, reg is primarily, if not exclusively, a product of the exocrine pancreas. Insulin mrna was detectable in both samples, and elastase-i was present exclusively in whole pancreas (Figure 3). Reg gene expression in extrapancreatic tissues. Slotblot analysis of total RNA from several extrapancreatic Figure 3. Northern blot analysis of RNA from whole pancreas and isolated islets. Ten micrograms of total RNA isolated from 3-month-old mouse pancreas (lane 1) and from 3-month-old mouse islets (lane 2) were subjected to Northern blot analysis as described in Materials and Methods. The membrane was hybridized with the 246-bp reg-ii cdna probe, which recognizes equally reg-i and reg-ii mrnas (Top). After washing, the same membranes were hybridized with insulin (Middle) and elastase-i (Bottom) cdna probes. mouse tissues with the 246-bp reg-ii cdna probe that recognized both reg-i and reg-ii sequences showed that reg mrna was detectable in several segments of the gastrointestinal tract. Reg mrna was present in extracts from duodenum, small intestine, and colon (Figure 4). No reg mrna was detected in both esophagus and stomach. In addition, reg mrna was absent in brain, cerebellum, heart, lung, liver, spleen, kidney, and testis (Figure 4). Hybridization of the same membranes with the mouse insulin cdna probe was negative for all tissues with the exception of the pancreas, demonstrating that the extrapancreatic samples were not contaminated with pancreatic RNA. Finally, hybridization with an oligonucleotide probe homologous to the poly(a) tail of all mrnas demonstrated that an approximately equal amount of RNA was used for each individual sample (Figure 4).
5 B312 PERFETTIETAL. B Esophagus Stomach Duodenum Small intestine Colon Brain Reg Oligo dt 2 o T Cerebellum Heart Lung Liver Spleen Kidney Testis Pancreas.3 3 ft it : 60 : >«S 40 : 20 : 30 Figure 4. Detection of reg mrna in extrapancreatic tissues. Five micrograms of total RNA obtained from a 3-month-old mouse were subjected to slot-blot analysis and hybridized with the 246-bp reg-ii probe, which recognizes equally reg-i and reg-ii mrnas (B) and insulin-ii cdna probes (C). The data were normalized by hybridizing the same membrane with oligonucleotide dt^ (A). Pancreatic reg and insulin gene expression during normal aging. Total reg mrna levels (reg-i and reg-ii), as detected with the 246-bp reg-ii cdna probe, decreased progressively with age (Figure 5). At 30 months of age, reg mrna levels were approximately 46.8 ± 7.9% of the level detected in 1-month-old mice (p =.01). This decrease was parallel to the age-dependent decline in insulin mrna levels which fell by 43.3 ± 10.1% with age (p =.02) (Figure 6). Regression analysis of either reg or insulin mrna levels showed a constant decrease of mrna levels for both of them per each month of age (Figures 5 and 6). By contrast, elastase-i mrna levels showed a modest but significant increase with age. At 30 months of age (p =.05), elastase-i mrna levels were approximately 124 ± 8.1% of the level detected in 1-month-old mice (p =.05) (Figure 7). Differential expression of reg-i and reg-ii genes during normal aging. Hybridization with a reg cdna probe that recognized both reg-i and reg-ii sequences showed that total reg mrna levels decreased markedly with age (Figure 3). In order to investigate if there was any difference in the relative expression of reg-i versus reg-ii, we took advantage of a DNA segment (corresponding to a region encoding exon 2) that was substantially different between reg-i and reg-ii sequences to design oligonucleotide probes that specifically hybridized to mrna for either reg-i or reg-ii (Figures 1 and Figure 5. Quantitative slot-blot analysis for reg mrna. Five micrograms of pancreatic RNA from 1-, 3-, 9-, 20-, and 30-month-old mice were subjected to slot-blot analysis with the 246-bp mouse reg-ii probe, which recognizes equally reg-i and reg-ii mrnas. The data were normalized by hybridizing the same membranes with oligo-dt w and were expressed as a percentage of the mean value observed in the 1-month-old group. As evaluated by one-way ANOVA, there was a statistically significant decrease in reg (p =.01) mrna levels. Regression analysis of the data showed that reg mrna levels decreased.007 relative units per month of age (p <.0002). The two blots on the right represent two individual experiments, while the values shown on the graph indicate the mean of three different experiments. For graphic purposes, the age scale on the x axes was drawn using an equal arbitrary spacing between bars. 2). Total pancreatic RNA from 3-, 9-, and 30-month-old mice was then subjected to slot-blot analysis in duplicate and hybridized with reg-i and reg-ii-specific oligonucleotide probes. We found that with age, the two nonallelic mouse reg genes were differentially regulated. There was an agedependent decline in reg-i mrna levels (p =.001), while there was no significant variation in reg-ii mrna levels (Figure 8). At 9 and 30 months of age, reg-i mrna levels were, respectively, 59.2 ± 16.9% and 39.9 ± 28.1% of the levels detected in 3-month-old mice (p =.01 at 9 months, p =.003 at 30 months). Regression analysis of the data showed a statistically significant decrease in reg-i mrna levels per each month of age (Figure 8). DISCUSSION The reg gene was cloned from a regenerating pancreas following surgical subtotal pancreatectomy and treatment with nicotinamide (Terazono et al., 1988). Interventions that cause a reduction in (3-cell mass or suppression of (3-cell function (i.e., implantation of an insulinoma) (Miyamura et
6 PANCREATIC reg-i AND -II GENE EXPRESSION DURING AGING B Insulin Oligo dt 2 o Age (months) Figure 7. Quantitative slot-blot analysis for elastase-i mrna. Five micrograms of pancreatic RNA from 3-, 9-, and 30-month-old mice were subjected to slot-blot analysis and hybridized with the elastase-i cdna probe. The data were normalized by hybridizing the same membrane with oligo-dt^ and were expressed as a percentage of the value observed in the 3-month-old group. Regression analysis of the data showed that elastase mrna levels increased.16 relative units per month of age (p <.01). The graph shown represents the mean of three different experiments. For graphic purposes, the age scale on the x axes was drawn using an equal arbitrary spacing between bars. Figure 6. Quantitative slot-blot analysis for insulin mrna. Five micrograms of pancreatic RNA from 1-, 3-, 9-, 20-, and 30-month-old mice were subjected to slot-blot analysis with the insulin-ii cdna probe. The data were normalized by hybridizing the same membranes with digo-dt^) and were expressed as a percentage of the mean value observed in the 1-monthold group. As evaluated by one-way ANOVA, there was a statistically significant decrease in insulin mrna levels (p -.005). Regression analysis of the data showed that insulin mrna levels decreased.07 relative units per month of age (/; <.0001). The two blots on the right represent two individual experiments, while the values shown on the graph indicate the mean of three different experiments. For graphic purposes, the age scale on the x axes was drawn using an equal arbitrary spacing between bars. al., 1991) result in a decrease in reg mrna levels, while interventions that stimulate islet proliferation (i.e., removal of an insulinoma) (Miyamura et al., 1991) or surgical wrapping/partial occlusion of the pancreatic duct (Zenilman et al., 1993) are associated with marked increases in reg mrna levels. Watanabe et al. (1994) have shown that administration of recombinant reg to 90% depancreatized rats induces (3-cell proliferation. Furthermore, reg protein has been shown to induce DNA synthesis in isolated islets in culture (Watanabe et al., 1994) as well as in islet-derived cell lines (Zenilman et al., 1993). In this study, we demonstrate that the mouse reg genes are expressed in the exocrine pancreas but not in normal pancreatic islets of 3-month-old mice. We also found that the stomach, the duodenum, and the small intestine exhibit detectable levels of reg mrna. This is in contrast with previous observations which have shown extrapancreatic expression of reg only in the gall bladder (Terazono et al., 1988), a tissue that we did not analyze. Others have detected increased expression of reg (Satomura et al., 1993) and reg- 30 related genes (Lasserre et al., 1992), respectively, in the diseased pancreas and in neoplastic colonic tissue, further linking enhanced reg expression to the proliferative state. The common embryological origin of the exocrine pancreas and the gastrointestinal tract could represent the biological basis for the detection of reg mrna in the gut. The negative results presented by other groups may be related to a greater level of sensitivity achieved in our laboratory. We demonstrated that there was an age-dependent decrease in reg mrna levels that closely parallels decreases in insulin mrna levels. Since reg is mainly a product of the exocrine pancreas, we investigated the expression of elastase-i, which is one of the major exocrine gene products, to discern whether the decrease in mrna levels was a generic phenomenon associated with aging of the whole organ or with a more specific event. Elastase-I mrna levels showed a moderate but significant increase with age. We did not measure reg protein levels; therefore, we do not have the experimental evidence that changes in reg mrna are followed by analogous changes in protein levels. While our findings do not allow extrapolation concerning the protein(s) levels, others have extensively showed that changes in reg mrna levels are followed by parallel variations in the functional status of pancreatic (3 cells (i.e., increased insulin synthesis is associated with increased reg mrna levels, while suppression of insulin synthesis is paralleled by decreased reg mrna levels), implying that these phenomena are likely mediated by the counterpart protein(s) (Terazono etal., 1988;Miyauraetal., 1991). Interestingly, we found that the two nonallelic mouse reg genes were differentially regulated during aging. While reg- I mrna levels progressively declined with age, reg-ii mrna levels remained unchanged. This finding may help to further explain why different laboratories reported conflict-
7 B314 PERFETTIETAL. 120 Age (months) Figure 8. Differential expression of the two nonallelic reg genes during aging. Five micrograms of pancreatic RNA from 3-, 9-, and 30-month-old mice were subjected to slot-blot analysis and hybridized with reg-1 specific (Top) and reg-u specific (Bottom) oligonucleotide probes. The data were normalized by hybridizing the same membranes with oligo-dt M and were expressed as a percentage of the mean value observed in the 3-month-old group. There was a significant decrease in reg-i mrna levels with age (p =.0001, ANOVA), while reg-il mrna levels were unchanged. Regression analysis of reg-i showed a decrease of 7.5 relative units per month of age (p <.01). Regression analysis of reg-ii mrna levels did not show significant variation with age. The graph shown represents the mean of three different experiments. For graphic purposes, the age scale on the x axes was drawn using an equal arbitrary spacing between bars. ing results regarding reg expression and (3-cell physiology (Smith et al., 1994; Watanabe et al., 1994). All studies to date have neglected to consider each of the two nonallelic genes individually (Terazono et al., 1988; Miyamura et al., 1991; Smith et al., 1994; Watanabe et al., 1994). This may have led to inaccurate data interpretation. It is possible to speculate that if a difference in gene expression or biological activity among the two nonallelic genes exists, this difference may have been either greatly underestimated or artificially magnified, depending on the sensitivity and the specificity of the technique used. Over the last few years, it has been extensively reported that reg belongs to a large gene family that shares a significant degree of nucleotide homology (Drickamer, 1988; Kamimura et al., 1992; Lasserre et al., 1992; Zenilman et al., 1993; Watanabe et al., 1994); despite such high structural similarity, very different biological functions have been claimed for individual proteins that belong to the reg gene family. Therefore, in studying their regulation, it is necessary to prove that the technique used is gene-specific and that it eliminates any possible crossreactivity with other very similar genes. On the basis of our findings, it is likely that those studies postulating a crucial role of reg in fj-cell physiology and/or islet mass may have used assay conditions that measure mostly reg-i mrna levels. On the other hand, reports challenging this relationship may have mostly detected reg-ii mrna levels that, according to our study, do not parallel the expression of the insulin gene. The study of recently duplicated generic loci offers a unique opportunity to investigate how two genes, initially with identical patterns of expression and function, gain individual characteristics over time. We have previously shown that in Xenopus laeves the two nonallelic insulin genes (Shuldiner et al., 1991) and the two nonallelic insulinlike growth factor I (IGF-I) genes (Perfetti et al., 1994) are differentially expressed during development. It is hypothesized that duplication may offer a selective advantage to the organism since the duplicated allele may act as a "backup" for its partner (Soares et al., 1985; Graf and Kobel, 1991). Thus genetic alterations that may have been lethal at a nonduplicated locus are no longer lethal. Alternatively, duplicated loci may evolve differential functions and may be the basis for the early events leading to the evolution of gene families (Kobel and Du Pasquier, 1986). We hypothesize that early in the course of the evolution of gene families, progression to differential function first involves alterations in regulatory regions resulting in the differential expression (e.g., developmental and/or tissue-specific) of an otherwise identical or highly similar protein product. Later, further mutations in coding regions may have resulted in protein products with distinct (or overlapping) biological functions. On the basis of sequence analysis, Unno et al. (1993) demonstrated that mouse reg-i and rat reg-i genes are more closely related than mouse reg-i and reg-ii genes, suggesting that the ancestral gene for reg may have been duplicated prior to divergence between mouse and rat. On the basis of this hypothesis, it is conceivable that the differential expression of the two reg genes may be associated with a different biological function of the two gene products. In conclusion, we show that in the normal mouse (7) the two nonallelic reg genes are differentially expressed during aging, and (2) decreases in reg-i expression parallel decreases in insulin expression during aging. ACKNOWLEDGMENTS This work was supported in part by the National Institute on Aging, the Johns Hopkins University Department of Medicine, the American Foundation of Aging Research, and the Chesapeake Education and Research Trust. We would like to thank Drs. Whaseon Lee-Kwon and Yihong Wang for critical reading of the manuscript. We also extend our gratitude to Mr. David Brindley for his editorial assistance. The present address for Dr. Michael E. Zenilman is Albert Einstein Medical College, New York, NY Address correspondence to Dr. Riccardo Perfetti, Diabetes Unit, Laboratory of Clinical Physiology, National Institute on Aging, National Institutes of Health, Gerontology Research Center, Room 2B02, 4940 Eastern Avenue, Baltimore, MD REFERENCES Cai, L.; Harris, W.R.; Marshak, D.R.; Gross, J.; Crabb, J.W. Structural
8 PANCREATIC reg-i AND -II GENE EXPRESSION DURING AGING B315 analysis of bovine pancreatic thread protein. J. Protein Chem. 9: ;1990. Chick, W.L.; Warren, S.; Chute, R.N.; Like, A.A.; Lauris, V.; Kitchen, K.C. A transplantable insulinoma in the rat. Proc. Natl. Acad. Sci. USA 74: ; Chirgwin, J.M.; Przybyla, D.M.; Smart, J.E.; Burridge, K.; Thomas, G.P. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18: ; Chomczynski, P.; Sacchi, H. Single step method RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: ; Drickamer, K. Two distinct classes of carbohydrate-recognition domains in animal lectins. J. Biol. Chem. 263: ; Graf, J.D.; Kobel, H.R. Genetics of Xenopus laevis. Methods Cell Biol. 36:19-34; Hellerstrom, C; Swenne, I.; Anderson, A. Islet cell replication and diabetes. In: Lefebvre, P.J.; Pipeleers, D.G., eds. The pathology of the endocrine pancreas. Heidelberg, Germany: Springer-Verlag, 1988: Kamimura, T.; West, C; Beutler, E. Sequence of a cdna encoding a rat reg-2 protein. Gene 118: ; Kobel, H.R.; Du Pasquier, L. Genetics of polyploid Xenopus. Trends Genet. 2: ; Lasserre, C; Christa, L.; Simon, M.T.; Vernier, P.; Brechot, C. A novel gene (HIP) activated in human primary liver cancer. Cancer Res. 52: ; MacDonald, R.J.; Swift, G.H.; Quinto, C; Swain, W.; Pictet, R.L.; Nikovits, W.; Rutter, W.J. Primary structure of the two distinct rat pancreatic preproelastases determined by sequence analysis of the complete cloned messenger ribonucleic acid sequences. Biochemistry 21: ; Miyaura, C; Chen, L.; Appel, M.; Alam, T.; Inman, L.; Hughes, S.D.; Milburn, J.L.; Unger, R.H.; Newgard, C.B. Expression of Reg/PSP, a pancreatic exocrine gene: relationship to changes in islet B-cell mass. Mol. Endocrinol. 5: ; Okamoto, H. Regulation of proinsulin synthesis on pancreatic islets and a new aspect to insulin-dependent diabetes. Mol. Cell. Biochem. 37:43-46; Perfetti, R.; Scott, L.A.; Shuldiner, A.R. The two nonallelic insulin-like growth factor-i (IGF-I) genes in Xenopus laevis are differentially regulated during early development. Endocrinology 135: ; Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer directed enzymatic amplification of DNA with thermostable DNA polymerase. Science 239: ;1988. Satomura, Y.; Sawabu, N.; Ohta, H.; Watanabe, H.; Yamakawa, O.; Motoo, Y.; Okai, T.; Toya, D.; Makino, H.; Okamoto, H. The immunohistochemical evaluation of PSP/reg-protein in normal and diseased human pancreatic tissues. Int. J. Pancreatol. 13:59 67; Shuldiner, A.; Perfetti, R. The polymerase chain reaction. In: De Pablo, F.; Scanes, C; Weintraub, B.D., eds. Techniques in endocrine research. San Diego, CA: Academic Press, 1993: Shuldiner, A.R.; Phillips, S.; Robert, C.T., Jr.; LeRoith, D.; Roth, J. Two nonallelic insulin genes in Xenopus laevis are expressed differentially during neurulation in prepancreatic embryos. Proc. Natl. Acad. Sci. USA 88: ; Smith, F.E.; Bonner-Weir, S.; Leahy, J.L.; Laufgraben, M.J.; Ogawa, Y.; Rosen, K.M.; Villa-Komaroff, L. Pancreatic reg/pancreatic stone protein (PSP) gene expression does not correlate with beta-cell growth and regeneration in rats. Diabetologia 37: ; Soares, M.B.; Schon, E.; Henderson, A.; Karathanasis, S.K.; Cate, R.; Zeitlin, S.; Chirgwin, J.; Efstratiadis, A. RNA-mediated duplication: the rat preproinsulin-i gene is a functional retroposon. Mol. Cell. Biol. 5: ; Swenne, I. Effects of aging on the regenerative capacity of the pancreatic B- cell of the rat. Diabetes 32:14-19; Terazono, K.; Yamamoto, H.; Takasawa, S.; Shiga, K.; Yonekura, Y.; Tochino, Y.; Okamoto, H. A novel gene activated in regenerating islets. J. Biol. Chem. 263: ; Unno, M.; Yonekura, H.; Nakagawara, K.; Watanabe, T.; Miyashita, H.; Moriizumi, S.; Okamoto, H.; Itoh, T.; Teraoka, H. Structure, chromosomal localization, and expression of mouse reg genes, reg I and II: a novel type of reg gene, reg II, exists in the mouse genome. J. Biol. Chem. 268: ; Watanabe, T.; Yonekemura, H.; Terazono, K.; Yamamoto, H.; Okamoto, H. Complete nucleotide sequence of the human reg gene and its expression in normal and tumoral tissues. J. Biol. Chem. 265: ;1990. Watanabe, T.; Yonemura, Y.; Yonekura, H.; Suzuki, Y.; Myhashita, H.; Sugiyama, K.; Moriizumi, S.; Unno, M.; Tanara, O.; Kondo, H.; Bone, A.; Takasawa, S.; Okamoto, H. Pancreatic beta-cell replication and amelioration of surgical diabetes by reg protein. Proc. Natl. Acad. Sci. USA 91: ; Wentworth, B.M.; Schaefer, I.M.; Villa-Komaroff, L.; Chirgwin, J.M. Characterization of the two nonallelic genes encoding mouse preproinsulin. J. Mol. Evol. 23: ; Yonemura, Y.; Takashima, T.; Miwa, K.; Miyazaki, 1.; Yamamoto, H.; Okamoto, H. Amelioration of diabetes mellitus in partially depancreatized rats by poly(adp-ribose) synthase inhibitors: evidence of islet B- cell regeneration. Diabetes 33: ; Yonemura, Y.; Takashima, T.; Matsuda, Y.; Miwa, K.; Sudiyma, K.; Miyazaki, I.; Yamamoto, H.; Okamoto, H. Induction of islet B-cell regeneration in partially pancreatectomized rats by poly(adp-ribose) synthetase inhibitors. Int. J. Pancreatol. 3:73-82; Zenilman, M.E.; Perfetti, R.; Shuldiner, A.R. Pancreatic regenerating gene (reg) expression during islet growth in the rat. Surg. Forum 44: ;1993. Received November 22, 1995 Accepted March 20, 1996
a Regenerating (reg) Gene Pancreas Treated wi th
 Endocrine Journal 1995, 42(5), 649-653 Increased Expression of Protein in Neonatal Rat Streptozotocin a Regenerating (reg) Gene Pancreas Treated wi th ToMIO OHNO, CHIKARA ISHII, NoRIHIRO KATO, YOSHITO
Endocrine Journal 1995, 42(5), 649-653 Increased Expression of Protein in Neonatal Rat Streptozotocin a Regenerating (reg) Gene Pancreas Treated wi th ToMIO OHNO, CHIKARA ISHII, NoRIHIRO KATO, YOSHITO
Product Manual. Omni-Array Sense Strand mrna Amplification Kit, 2 ng to 100 ng Version Catalog No.: Reactions
 Genetic Tools and Reagents Universal mrna amplification, sense strand amplification, antisense amplification, cdna synthesis, micro arrays, gene expression, human, mouse, rat, guinea pig, cloning Omni-Array
Genetic Tools and Reagents Universal mrna amplification, sense strand amplification, antisense amplification, cdna synthesis, micro arrays, gene expression, human, mouse, rat, guinea pig, cloning Omni-Array
PRODUCT: RNAzol BD for Blood May 2014 Catalog No: RB 192 Storage: Store at room temperature
 PRODUCT: RNAzol BD for Blood May 2014 Catalog No: RB 192 Storage: Store at room temperature PRODUCT DESCRIPTION. RNAzol BD is a reagent for isolation of total RNA from whole blood, plasma or serum of human
PRODUCT: RNAzol BD for Blood May 2014 Catalog No: RB 192 Storage: Store at room temperature PRODUCT DESCRIPTION. RNAzol BD is a reagent for isolation of total RNA from whole blood, plasma or serum of human
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Doctor of Philosophy
 Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h)
 Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
CHAPTER 7: REAGENTS AND SOLUTIONS
 7.1. ANALYSIS OF MODULATION OF SOD ENZYME Acetic acid (cat. no. 11007, Glaxo Qualigen, India): Bovine Serum Albumin stock solution (BSA, 1mg/ml): 1 mg of standard bovine serum albumin (cat. no. A2153,
7.1. ANALYSIS OF MODULATION OF SOD ENZYME Acetic acid (cat. no. 11007, Glaxo Qualigen, India): Bovine Serum Albumin stock solution (BSA, 1mg/ml): 1 mg of standard bovine serum albumin (cat. no. A2153,
Generating Mouse Models of Pancreatic Cancer
 Generating Mouse Models of Pancreatic Cancer Aom Isbell http://www2.massgeneral.org/cancerresourceroom/types/gi/index.asp Spring/Summer 1, 2012 Alexandros Tzatsos, MD PhD Bardeesy Lab: Goals and Objectives
Generating Mouse Models of Pancreatic Cancer Aom Isbell http://www2.massgeneral.org/cancerresourceroom/types/gi/index.asp Spring/Summer 1, 2012 Alexandros Tzatsos, MD PhD Bardeesy Lab: Goals and Objectives
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer. Application Note
 Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab)
 Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab) Updated 12/3/02 Reagents: ChIP sonication Buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mm Tris 8.1, 150 mm NaCl, 5 mm EDTA): 10 ml 10 % Triton
Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab) Updated 12/3/02 Reagents: ChIP sonication Buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mm Tris 8.1, 150 mm NaCl, 5 mm EDTA): 10 ml 10 % Triton
Hepatitis B Virus Genemer
 Product Manual Hepatitis B Virus Genemer Primer Pair for amplification of HBV Viral Specific Fragment Catalog No.: 60-2007-10 Store at 20 o C For research use only. Not for use in diagnostic procedures
Product Manual Hepatitis B Virus Genemer Primer Pair for amplification of HBV Viral Specific Fragment Catalog No.: 60-2007-10 Store at 20 o C For research use only. Not for use in diagnostic procedures
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana
 Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
Annexure III SOLUTIONS AND REAGENTS
 Annexure III SOLUTIONS AND REAGENTS A. STOCK SOLUTIONS FOR DNA ISOLATION 0.5M Ethylene-diamine tetra acetic acid (EDTA) (ph=8.0) 1M Tris-Cl (ph=8.0) 5M NaCl solution Red cell lysis buffer (10X) White cell
Annexure III SOLUTIONS AND REAGENTS A. STOCK SOLUTIONS FOR DNA ISOLATION 0.5M Ethylene-diamine tetra acetic acid (EDTA) (ph=8.0) 1M Tris-Cl (ph=8.0) 5M NaCl solution Red cell lysis buffer (10X) White cell
Work-flow: protein sample preparation Precipitation methods Removal of interfering substances Specific examples:
 Dr. Sanjeeva Srivastava IIT Bombay Work-flow: protein sample preparation Precipitation methods Removal of interfering substances Specific examples: Sample preparation for serum proteome analysis Sample
Dr. Sanjeeva Srivastava IIT Bombay Work-flow: protein sample preparation Precipitation methods Removal of interfering substances Specific examples: Sample preparation for serum proteome analysis Sample
Protocol for Gene Transfection & Western Blotting
 The schedule and the manual of basic techniques for cell culture Advanced Protocol for Gene Transfection & Western Blotting Schedule Day 1 26/07/2008 Transfection Day 3 28/07/2008 Cell lysis Immunoprecipitation
The schedule and the manual of basic techniques for cell culture Advanced Protocol for Gene Transfection & Western Blotting Schedule Day 1 26/07/2008 Transfection Day 3 28/07/2008 Cell lysis Immunoprecipitation
CHAPTER 4 RESULTS. showed that all three replicates had similar growth trends (Figure 4.1) (p<0.05; p=0.0000)
 CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
The Schedule and the Manual of Basic Techniques for Cell Culture
 The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
Luminescent platforms for monitoring changes in the solubility of amylin and huntingtin in living cells
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Absence of Kaposi s Sarcoma-Associated Herpesvirus in Patients with Pulmonary
 Online Data Supplement Absence of Kaposi s Sarcoma-Associated Herpesvirus in Patients with Pulmonary Arterial Hypertension Cornelia Henke-Gendo, Michael Mengel, Marius M. Hoeper, Khaled Alkharsah, Thomas
Online Data Supplement Absence of Kaposi s Sarcoma-Associated Herpesvirus in Patients with Pulmonary Arterial Hypertension Cornelia Henke-Gendo, Michael Mengel, Marius M. Hoeper, Khaled Alkharsah, Thomas
Online Data Supplement. Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2
 Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format:
 Supplementary Protocol 1. Adaptor preparation: For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format: Per reaction X96 10X NEBuffer 2 10 µl 10 µl x 96 5 -GATC
Supplementary Protocol 1. Adaptor preparation: For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format: Per reaction X96 10X NEBuffer 2 10 µl 10 µl x 96 5 -GATC
Islet viability assay and Glucose Stimulated Insulin Secretion assay RT-PCR and Western Blot
 Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Phosphate buffered saline (PBS) for washing the cells TE buffer (nuclease-free) ph 7.5 for use with the PrimePCR Reverse Transcription Control Assay
 Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Materials and Methods , The two-hybrid principle.
 The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
Human Immunodeficiency Virus-1 (HIV-1) Genemer. Primer Pair for amplification of HIV-1 Specific DNA Fragment
 Product Manual Human Immunodeficiency Virus-1 (HIV-1) Genemer Primer Pair for amplification of HIV-1 Specific DNA Fragment Catalog No.: 60-2002-10 Store at 20 o C For research use only. Not for use in
Product Manual Human Immunodeficiency Virus-1 (HIV-1) Genemer Primer Pair for amplification of HIV-1 Specific DNA Fragment Catalog No.: 60-2002-10 Store at 20 o C For research use only. Not for use in
GENE Forward primer Reverse primer FABP4 CCTTTGTGGGGACCTGGAAA TGACCGGATGACGACCAAGT CD68 AATGTGTCCTTCCCACAAGC GGCAGCAAGAGAGATTGGTC
 Published in "" which should be cited to refer to this work. mrna extraction and RT-PCR Total RNA from 5 15 mg of crushed white adipose tissue was isolated using the technique described by Chomczynski
Published in "" which should be cited to refer to this work. mrna extraction and RT-PCR Total RNA from 5 15 mg of crushed white adipose tissue was isolated using the technique described by Chomczynski
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Chromatin IP (Isw2) Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles.
 Chromatin IP (Isw2) 7/01 Toshi last update: 06/15 Reagents Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles. 2.5 M glycine. TBS:
Chromatin IP (Isw2) 7/01 Toshi last update: 06/15 Reagents Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles. 2.5 M glycine. TBS:
DETECTION OF HEPATITIS C VIRUS cdna SEQUENCE BY THE POLYMERASE CHAIN REACTION IN HEPATOCELLULAR CARCINOMA TISSUES
 Jpn. J. Med. Sci. Biol., 43, 89-94, 1990. Short Communication DETECTION OF HEPATITIS C VIRUS cdna SEQUENCE BY THE POLYMERASE CHAIN REACTION IN HEPATOCELLULAR CARCINOMA TISSUES Tetsuo YONEYAMA, Kenji TAKEUCHI,
Jpn. J. Med. Sci. Biol., 43, 89-94, 1990. Short Communication DETECTION OF HEPATITIS C VIRUS cdna SEQUENCE BY THE POLYMERASE CHAIN REACTION IN HEPATOCELLULAR CARCINOMA TISSUES Tetsuo YONEYAMA, Kenji TAKEUCHI,
Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples
 DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
Role of Paired Box9 (PAX9) (rs ) and Muscle Segment Homeobox1 (MSX1) (581C>T) Gene Polymorphisms in Tooth Agenesis
 EC Dental Science Special Issue - 2017 Role of Paired Box9 (PAX9) (rs2073245) and Muscle Segment Homeobox1 (MSX1) (581C>T) Gene Polymorphisms in Tooth Agenesis Research Article Dr. Sonam Sethi 1, Dr. Anmol
EC Dental Science Special Issue - 2017 Role of Paired Box9 (PAX9) (rs2073245) and Muscle Segment Homeobox1 (MSX1) (581C>T) Gene Polymorphisms in Tooth Agenesis Research Article Dr. Sonam Sethi 1, Dr. Anmol
Norgen s HIV proviral DNA PCR Kit was developed and validated to be used with the following PCR instruments: Qiagen Rotor-Gene Q BioRad icycler
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com HIV Proviral DNA PCR Kit Product # 33840 Product Insert Background Information
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com HIV Proviral DNA PCR Kit Product # 33840 Product Insert Background Information
AIDS - Knowledge and Dogma. Conditions for the Emergence and Decline of Scientific Theories Congress, July 16/ , Vienna, Austria
 AIDS - Knowledge and Dogma Conditions for the Emergence and Decline of Scientific Theories Congress, July 16/17 2010, Vienna, Austria Reliability of PCR to detect genetic sequences from HIV Juan Manuel
AIDS - Knowledge and Dogma Conditions for the Emergence and Decline of Scientific Theories Congress, July 16/17 2010, Vienna, Austria Reliability of PCR to detect genetic sequences from HIV Juan Manuel
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Norgen s HIV Proviral DNA PCR Kit was developed and validated to be used with the following PCR instruments: Qiagen Rotor-Gene Q BioRad T1000 Cycler
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com HIV Proviral DNA PCR Kit Product# 33840 Product Insert Intended
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com HIV Proviral DNA PCR Kit Product# 33840 Product Insert Intended
Manual (Second edition)
 Reagent for RNA Extraction ISOGENⅡ Manual (Second edition) Code No. 311-07361 Code No. 317-07363 NIPPON GENE CO., LTD. Table of contents I Product description 1 II Product content 1 III Storage 1 IV Precautions
Reagent for RNA Extraction ISOGENⅡ Manual (Second edition) Code No. 311-07361 Code No. 317-07363 NIPPON GENE CO., LTD. Table of contents I Product description 1 II Product content 1 III Storage 1 IV Precautions
Supplemental Materials and Methods Plasmids and viruses Quantitative Reverse Transcription PCR Generation of molecular standard for quantitative PCR
 Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Gastric Carcinoma with Lymphoid Stroma: Association with Epstein Virus Genome demonstrated by PCR
 Gastric Carcinoma with Lymphoid Stroma: Association with Epstein Virus Genome demonstrated by PCR Pages with reference to book, From 305 To 307 Irshad N. Soomro,Samina Noorali,Syed Abdul Aziz,Suhail Muzaffar,Shahid
Gastric Carcinoma with Lymphoid Stroma: Association with Epstein Virus Genome demonstrated by PCR Pages with reference to book, From 305 To 307 Irshad N. Soomro,Samina Noorali,Syed Abdul Aziz,Suhail Muzaffar,Shahid
E.Z.N.A. SQ Blood DNA Kit II. Table of Contents
 E.Z.N.A. SQ Blood DNA Kit II Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Blood Storage and DNA Yield...4 Preparing Reagents...5 100-500 μl Whole Blood Protocol...6
E.Z.N.A. SQ Blood DNA Kit II Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Blood Storage and DNA Yield...4 Preparing Reagents...5 100-500 μl Whole Blood Protocol...6
Validation Report: VERSA Mini PCR Workstation Reverse Transcription of Avian Flu RNA and Amplification of cdna & Detection of H5N1
 I. Objectives Validation Report: VERSA Mini PCR Workstation Reverse Transcription of Avian Flu RNA and Amplification of cdna & Detection of H5N1 1. To ensure stability of RNA (highly thermolabile and degradatively
I. Objectives Validation Report: VERSA Mini PCR Workstation Reverse Transcription of Avian Flu RNA and Amplification of cdna & Detection of H5N1 1. To ensure stability of RNA (highly thermolabile and degradatively
MasterPure RNA Purification Kit
 Cat. No. MCR85102 The MasterPure RNA Purification Kit pro vides all of the reagents necessary to recover RNA from a wide variety of biological sources. This kit uses a rapid desalting process 1 to remove
Cat. No. MCR85102 The MasterPure RNA Purification Kit pro vides all of the reagents necessary to recover RNA from a wide variety of biological sources. This kit uses a rapid desalting process 1 to remove
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Human Rotavirus A. genesig Standard Kit. Non structural protein 5 (NSP5) 150 tests. Primerdesign Ltd. For general laboratory and research use only
 TM Primerdesign Ltd Human Rotavirus A Non structural protein 5 (NSP5) genesig Standard Kit 150 tests For general laboratory and research use only 1 Introduction to Human Rotavirus A Rotavirus is a genus
TM Primerdesign Ltd Human Rotavirus A Non structural protein 5 (NSP5) genesig Standard Kit 150 tests For general laboratory and research use only 1 Introduction to Human Rotavirus A Rotavirus is a genus
Life Sciences 1A Midterm Exam 2. November 13, 2006
 Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Human Rotavirus B. Non structural protein 5 (NSP5) 150 tests. Quantification of Human Rotavirus B genomes Advanced kit handbook HB10.01.
 PCR Max Ltd TM qpcr test Human Rotavirus B Non structural protein 5 (NSP5) 150 tests For general laboratory and research use only 1 Introduction to Human Rotavirus B Rotavirus is a genus of double-stranded
PCR Max Ltd TM qpcr test Human Rotavirus B Non structural protein 5 (NSP5) 150 tests For general laboratory and research use only 1 Introduction to Human Rotavirus B Rotavirus is a genus of double-stranded
SUPPLEMENTAL MATERIAL. Supplementary Methods
 SUPPLEMENTAL MATERIAL Supplementary Methods Culture of cardiomyocytes, fibroblasts and cardiac microvascular endothelial cells The isolation and culturing of neonatal rat ventricular cardiomyocytes was
SUPPLEMENTAL MATERIAL Supplementary Methods Culture of cardiomyocytes, fibroblasts and cardiac microvascular endothelial cells The isolation and culturing of neonatal rat ventricular cardiomyocytes was
Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator. of the Interaction with Macrophages
 Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator of the Interaction with Macrophages Yohei Sanada, Takafumi Yamamoto, Rika Satake, Akiko Yamashita, Sumire Kanai, Norihisa Kato, Fons AJ van
Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator of the Interaction with Macrophages Yohei Sanada, Takafumi Yamamoto, Rika Satake, Akiko Yamashita, Sumire Kanai, Norihisa Kato, Fons AJ van
RNA/DNA Stabilization Reagent for Blood/Bone Marrow
 For general laboratory use. Not for use in diagnostic procedures. FOR IN VITRO USE ONLY. RNA/DNA Stabilization Reagent for Blood/Bone Marrow For simultaneous cell lysis and stabilization of nucleic acids
For general laboratory use. Not for use in diagnostic procedures. FOR IN VITRO USE ONLY. RNA/DNA Stabilization Reagent for Blood/Bone Marrow For simultaneous cell lysis and stabilization of nucleic acids
Product Contents. 1 Specifications 1 Product Description. 2 Buffer Preparation... 3 Protocol. 3 Ordering Information 4 Related Products..
 INSTRUCTION MANUAL Quick-RNA MidiPrep Catalog No. R1056 Highlights 10 minute method for isolating RNA (up to 1 mg) from a wide range of cell types and tissue samples. Clean-Spin column technology allows
INSTRUCTION MANUAL Quick-RNA MidiPrep Catalog No. R1056 Highlights 10 minute method for isolating RNA (up to 1 mg) from a wide range of cell types and tissue samples. Clean-Spin column technology allows
Antibodies: LB1 buffer For 50 ml For 10ml For 30 ml Final 1 M HEPES, ph 2.5 ml 0.5 ml 1.5 ml 50mM. 5 M NaCl 1.4 ml 280 µl 0.
 Experiment: Date: Tissue: Purpose: ChIP-Seq Antibodies: 11x cross-link buffer: Regent Stock Solution Final Vol for 10 ml of 11xstock concentration 5 M NaCl 0.1M 0.2 ml 0.5 M EDTA 1 mm 20 ul 0.5 M EGTA,
Experiment: Date: Tissue: Purpose: ChIP-Seq Antibodies: 11x cross-link buffer: Regent Stock Solution Final Vol for 10 ml of 11xstock concentration 5 M NaCl 0.1M 0.2 ml 0.5 M EDTA 1 mm 20 ul 0.5 M EGTA,
Product # Kit Components
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com Pneumocystis jirovecii PCR Kit Product # 42820 Product Insert Background Information
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com Pneumocystis jirovecii PCR Kit Product # 42820 Product Insert Background Information
RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using
 Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
Aldehyde Dehydrogenase-2 Genotype Detection in Fingernails among Non-alcoholic Northeastern Thai Population and Derived Gene Frequency
 ScienceAsia 28 (2002) : 99-103 Aldehyde Dehydrogenase-2 Genotype Detection in Fingernails among Non-alcoholic Northeastern Thai Population and Derived Gene Frequency Paiboon Mongconthawornchai a, *, Somsong
ScienceAsia 28 (2002) : 99-103 Aldehyde Dehydrogenase-2 Genotype Detection in Fingernails among Non-alcoholic Northeastern Thai Population and Derived Gene Frequency Paiboon Mongconthawornchai a, *, Somsong
Differentiation-induced Changes of Mediterranean Fever Gene (MEFV) Expression in HL-60 Cell
 Differentiation-induced Changes of Mediterranean Fever Gene (MEFV) Expression in HL-60 Cell Wenxin Li Department of Biological Sciences Fordham University Abstract MEFV is a human gene that codes for an
Differentiation-induced Changes of Mediterranean Fever Gene (MEFV) Expression in HL-60 Cell Wenxin Li Department of Biological Sciences Fordham University Abstract MEFV is a human gene that codes for an
Mitochondrial DNA Isolation Kit
 Mitochondrial DNA Isolation Kit Catalog Number KA0895 50 assays Version: 01 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
Mitochondrial DNA Isolation Kit Catalog Number KA0895 50 assays Version: 01 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
MORIMOTO LAB BUFFER AND SOLUTION RECIPES
 MORIMOTO LAB BUFFER AND SOLUTION RECIPES 30% Acrylamide 100ml 29g 2X Acyrlamide 1g N,N -methylenebisacrylamide Add ~300ml ddh 2 O. Heat to 37 C to dissolve chemicals. Adjust final volume to 500ml with
MORIMOTO LAB BUFFER AND SOLUTION RECIPES 30% Acrylamide 100ml 29g 2X Acyrlamide 1g N,N -methylenebisacrylamide Add ~300ml ddh 2 O. Heat to 37 C to dissolve chemicals. Adjust final volume to 500ml with
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supporting Online Material Material and Methods References Supplemental Figures S1, S2, and S3
 Supporting Online Material Material and Methods References Supplemental Figures S1, S2, and S3 Sarbassov et al. 1 Material and Methods Materials Reagents were obtained from the following sources: protein
Supporting Online Material Material and Methods References Supplemental Figures S1, S2, and S3 Sarbassov et al. 1 Material and Methods Materials Reagents were obtained from the following sources: protein
For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin
 Materials and Methods Pair Feeding Experiment For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin (0.005%), which is average daily food intake of mice fed control HFD ad libitum at week
Materials and Methods Pair Feeding Experiment For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin (0.005%), which is average daily food intake of mice fed control HFD ad libitum at week
Genotype analysis by Southern blots of nine independent recombinated ES cell clones by
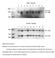 Supplemental Figure 1 Selected ES cell clones show a correctly recombined conditional Ngn3 allele Genotype analysis by Southern blots of nine independent recombinated ES cell clones by hybridization with
Supplemental Figure 1 Selected ES cell clones show a correctly recombined conditional Ngn3 allele Genotype analysis by Southern blots of nine independent recombinated ES cell clones by hybridization with
A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis
 APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
Item Catalog Number Manufacturer 1,4-Dithioerythritol (1 g) D9680 Sigma-Aldrich
 SOP: Nuclei isolation from fresh mouse tissues and DNaseI treatment Date modified: 01/12/2011 Modified by: E. Giste/ T. Canfield (UW) The following protocol describes the isolation of nuclei and subsequent
SOP: Nuclei isolation from fresh mouse tissues and DNaseI treatment Date modified: 01/12/2011 Modified by: E. Giste/ T. Canfield (UW) The following protocol describes the isolation of nuclei and subsequent
Post-meiotic transcription of phosphoglycerate-kinase 2 in mouse testes
 Bioscience Reports 5, 1087-1091 (i985) 1087 Printed in Great Britain Post-meiotic transcription of phosphoglycerate-kinase 2 in mouse testes Robert P. ERICKSON I, Alan M. MICHELSON 2, Michael P. ROSENBERG
Bioscience Reports 5, 1087-1091 (i985) 1087 Printed in Great Britain Post-meiotic transcription of phosphoglycerate-kinase 2 in mouse testes Robert P. ERICKSON I, Alan M. MICHELSON 2, Michael P. ROSENBERG
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
HIV-1 Genemer Detection Kit Ready to Use Amplification Kit for HIV-1 Specific DNA Fragment Analysis
 Product Manual HIV-1 Genemer Detection Kit Ready to Use Amplification Kit for HIV-1 Specific DNA Fragment Analysis For research use only. Not for use in diagnostic procedures for clinical purposes Catalog
Product Manual HIV-1 Genemer Detection Kit Ready to Use Amplification Kit for HIV-1 Specific DNA Fragment Analysis For research use only. Not for use in diagnostic procedures for clinical purposes Catalog
1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics
 1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics to gain mechanistic insight! 4. Return to step 2, as
1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics to gain mechanistic insight! 4. Return to step 2, as
WHO Prequalification of Diagnostics Programme PUBLIC REPORT. Product: VERSANT HIV-1 RNA 1.0 Assay (kpcr) Number: PQDx
 WHO Prequalification of Diagnostics Programme PUBLIC REPORT Product: VERSANT HIV-1 RNA 1.0 Assay (kpcr) Number: PQDx 0115-041-00 Abstract The VERSANT HIV-1 RNA 1.0 Assay (kpcr) with product codes 10375763,
WHO Prequalification of Diagnostics Programme PUBLIC REPORT Product: VERSANT HIV-1 RNA 1.0 Assay (kpcr) Number: PQDx 0115-041-00 Abstract The VERSANT HIV-1 RNA 1.0 Assay (kpcr) with product codes 10375763,
Midi Plant Genomic DNA Purification Kit
 Midi Plant Genomic DNA Purification Kit Cat #:DP022MD/ DP022MD-50 Size:10/50 reactions Store at RT For research use only 1 Description: The Midi Plant Genomic DNA Purification Kit provides a rapid, simple
Midi Plant Genomic DNA Purification Kit Cat #:DP022MD/ DP022MD-50 Size:10/50 reactions Store at RT For research use only 1 Description: The Midi Plant Genomic DNA Purification Kit provides a rapid, simple
EXO-DNA Circulating and EV-associated DNA extraction kit
 Datasheet EXO-DNA Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
Datasheet EXO-DNA Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
Pair-fed % inkt cells 0.5. EtOH 0.0
 MATERIALS AND METHODS Histopathological analysis Liver tissue was collected 9 h post-gavage, and the tissue samples were fixed in 1% formalin and paraffin-embedded following a standard procedure. The embedded
MATERIALS AND METHODS Histopathological analysis Liver tissue was collected 9 h post-gavage, and the tissue samples were fixed in 1% formalin and paraffin-embedded following a standard procedure. The embedded
Lack of PTC Gene (ret Proto-oncogene Rearrangement) in Human Thyroid Tumors
 Endocrinol japon 1991, 38 (6), 627-632 Lack of PTC Gene (ret Proto-oncogene Rearrangement) in Human Thyroid Tumors HIROYUKI NAMBA*, SHUNICHI YAMASHITA*, HAI-CHENG PEI, NAOFUMISHIKAWA**, MARIA C. VILLADOLID,
Endocrinol japon 1991, 38 (6), 627-632 Lack of PTC Gene (ret Proto-oncogene Rearrangement) in Human Thyroid Tumors HIROYUKI NAMBA*, SHUNICHI YAMASHITA*, HAI-CHENG PEI, NAOFUMISHIKAWA**, MARIA C. VILLADOLID,
Acute myocardial infarction (MI) on initial presentation was diagnosed if there was 20 minutes
 SUPPLEMENTAL MATERIAL Supplemental Methods Diagnosis for acute myocardial infarction Acute myocardial infarction (MI) on initial presentation was diagnosed if there was 20 minutes or more of chest pain
SUPPLEMENTAL MATERIAL Supplemental Methods Diagnosis for acute myocardial infarction Acute myocardial infarction (MI) on initial presentation was diagnosed if there was 20 minutes or more of chest pain
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr)
 Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Supplementary Material
 Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Product Contents. 1 Specifications 1 Product Description. 2 Buffer Preparation... 3 Protocol. 3 Ordering Information 4
 INSTRUCTION MANUAL Quick-RNA Midiprep Kit Catalog No. R1056 Highlights 10 minute method for isolating RNA (up to 1 mg) from a wide range of cell types and tissue samples. Clean-Spin column technology allows
INSTRUCTION MANUAL Quick-RNA Midiprep Kit Catalog No. R1056 Highlights 10 minute method for isolating RNA (up to 1 mg) from a wide range of cell types and tissue samples. Clean-Spin column technology allows
Geneaid DNA Isolation Kit
 Instruction Manual Ver. 02.21.17 For Research Use Only Geneaid DNA Isolation Kit GEB100, GEB01K, GEB01K+ GEC150, GEC1.5K, GEC1.5K+ GET150, GET1.5K, GET1.5K+ GEE150, GEE1.5K, GEE1.5K+ Advantages Sample:
Instruction Manual Ver. 02.21.17 For Research Use Only Geneaid DNA Isolation Kit GEB100, GEB01K, GEB01K+ GEC150, GEC1.5K, GEC1.5K+ GET150, GET1.5K, GET1.5K+ GEE150, GEE1.5K, GEE1.5K+ Advantages Sample:
Supplemental Experimental Procedures
 Cell Stem Cell, Volume 2 Supplemental Data A Temporal Switch from Notch to Wnt Signaling in Muscle Stem Cells Is Necessary for Normal Adult Myogenesis Andrew S. Brack, Irina M. Conboy, Michael J. Conboy,
Cell Stem Cell, Volume 2 Supplemental Data A Temporal Switch from Notch to Wnt Signaling in Muscle Stem Cells Is Necessary for Normal Adult Myogenesis Andrew S. Brack, Irina M. Conboy, Michael J. Conboy,
Molecular Detection of BCR/ABL1 for the Diagnosis and Monitoring of CML
 Molecular Detection of BCR/ABL1 for the Diagnosis and Monitoring of CML Imran Mirza, MD, MS, FRCPC Pathology & Laboratory Medicine Institute Sheikh Khalifa Medical City, Abu Dhabi, UAE. imirza@skmc.ae
Molecular Detection of BCR/ABL1 for the Diagnosis and Monitoring of CML Imran Mirza, MD, MS, FRCPC Pathology & Laboratory Medicine Institute Sheikh Khalifa Medical City, Abu Dhabi, UAE. imirza@skmc.ae
Week 3 The Pancreas: Pancreatic ph buffering:
 Week 3 The Pancreas: A gland with both endocrine (secretion of substances into the bloodstream) & exocrine (secretion of substances to the outside of the body or another surface within the body) functions
Week 3 The Pancreas: A gland with both endocrine (secretion of substances into the bloodstream) & exocrine (secretion of substances to the outside of the body or another surface within the body) functions
7.014 Problem Set 7 Solutions
 MIT Department of Biology 7.014 Introductory Biology, Spring 2005 7.014 Problem Set 7 Solutions Question 1 Part A Antigen binding site Antigen binding site Variable region Light chain Light chain Variable
MIT Department of Biology 7.014 Introductory Biology, Spring 2005 7.014 Problem Set 7 Solutions Question 1 Part A Antigen binding site Antigen binding site Variable region Light chain Light chain Variable
PNGase F Instruction Manual
 PNGase F Instruction Manual Catalog Number 170-6883 Bio-Rad Laboratories, 2000 Alfred Nobel Dr., Hercules, CA 94547 4006094 Rev A Table of Contents Section 1 Introduction...1 Section 2 Kit Components and
PNGase F Instruction Manual Catalog Number 170-6883 Bio-Rad Laboratories, 2000 Alfred Nobel Dr., Hercules, CA 94547 4006094 Rev A Table of Contents Section 1 Introduction...1 Section 2 Kit Components and
Avian Influenza A H5N8
 TM Primerdesign Ltd Avian Influenza A H5N8 Hemagglutinin (HA) gene & Neuraminidase (NA) gene genesig Standard Kit 150 tests For general laboratory and research use only 1 Introduction to Avian Influenza
TM Primerdesign Ltd Avian Influenza A H5N8 Hemagglutinin (HA) gene & Neuraminidase (NA) gene genesig Standard Kit 150 tests For general laboratory and research use only 1 Introduction to Avian Influenza
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A. Real-Time RT-PCR Detection.
 For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
Human Rotavirus A. genesig Advanced Kit. Non structural protein 5 (NSP5) 150 tests. Primerdesign Ltd. For general laboratory and research use only
 TM Primerdesign Ltd Human Rotavirus A Non structural protein 5 (NSP5) genesig Advanced Kit 150 tests For general laboratory and research use only 1 Introduction to Human Rotavirus A Rotavirus is a genus
TM Primerdesign Ltd Human Rotavirus A Non structural protein 5 (NSP5) genesig Advanced Kit 150 tests For general laboratory and research use only 1 Introduction to Human Rotavirus A Rotavirus is a genus
Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/-
 Supplemental Material Results. Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/- and Slc2a7 -/- mice. The expression of AE1 in the kidney was examined in Slc26a7 KO mice.
Supplemental Material Results. Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/- and Slc2a7 -/- mice. The expression of AE1 in the kidney was examined in Slc26a7 KO mice.
Kit Components Product # EP42720 (24 preps) MDx 2X PCR Master Mix 350 µl Cryptococcus neoformans Primer Mix 70 µl Cryptococcus neoformans Positive
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com Cryptococcus neoformans End-Point PCR Kit Product# EP42720 Product
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com Cryptococcus neoformans End-Point PCR Kit Product# EP42720 Product
Expression of Reg and cytokeratin 20 during ductal cell differentiation and proliferation in a mouse model of autoimmune diabetes
 European Journal of Endocrinology (1999) 141 644 652 ISSN 0804-4643 EXPERIMENTAL STUDY Expression of Reg and cytokeratin 20 during ductal cell differentiation and proliferation in a mouse model of autoimmune
European Journal of Endocrinology (1999) 141 644 652 ISSN 0804-4643 EXPERIMENTAL STUDY Expression of Reg and cytokeratin 20 during ductal cell differentiation and proliferation in a mouse model of autoimmune
APPENDIX-I. The compositions of media used for the growth and differentiation of Pseudomonas aeruginosa are as follows:
 APPENDIX-I The compositions of media used for the growth and differentiation of Pseudomonas aeruginosa are as follows: COMPOSITION OF DIFFERENT MEDIA STOCK CULTURE AGAR (AYERS AND JOHNSON AGAR) Gms/Litre
APPENDIX-I The compositions of media used for the growth and differentiation of Pseudomonas aeruginosa are as follows: COMPOSITION OF DIFFERENT MEDIA STOCK CULTURE AGAR (AYERS AND JOHNSON AGAR) Gms/Litre
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Histogram showing hybridization signals for chicken (left) and quail (right) genomic DNA analyzed by Chicken GeneChip (n=3). www.nature.com/nature 1 Supplementary Figure 2. Independent
Supplementary Figure 1. Histogram showing hybridization signals for chicken (left) and quail (right) genomic DNA analyzed by Chicken GeneChip (n=3). www.nature.com/nature 1 Supplementary Figure 2. Independent
Supporting Information
 Translation of DNA into Synthetic N-Acyloxazolidines Xiaoyu Li, Zev. J. Gartner, Brian N. Tse and David R. Liu* Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts
Translation of DNA into Synthetic N-Acyloxazolidines Xiaoyu Li, Zev. J. Gartner, Brian N. Tse and David R. Liu* Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts
AquaPreserve DNA/RNA/Protein Order # Preservation and Extraction Kit 8001MT, 8060MT
 AquaPreserve DNA/RNA/Protein Order # Preservation and Extraction Kit 8001MT, 8060MT MoBiTec GmbH 2014 Page 2 Contents 1. Description... 3 2. Kit Contents... 3 3. Terms & Conditions... 3 4. AquaPreserve
AquaPreserve DNA/RNA/Protein Order # Preservation and Extraction Kit 8001MT, 8060MT MoBiTec GmbH 2014 Page 2 Contents 1. Description... 3 2. Kit Contents... 3 3. Terms & Conditions... 3 4. AquaPreserve
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep
 SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
EXO-DNAc Circulating and EV-associated DNA extraction kit
 Datasheet EXO-DNAc Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
Datasheet EXO-DNAc Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
General Biology 1004 Chapter 11 Lecture Handout, Summer 2005 Dr. Frisby
 Slide 1 CHAPTER 11 Gene Regulation PowerPoint Lecture Slides for Essential Biology, Second Edition & Essential Biology with Physiology Presentation prepared by Chris C. Romero Neil Campbell, Jane Reece,
Slide 1 CHAPTER 11 Gene Regulation PowerPoint Lecture Slides for Essential Biology, Second Edition & Essential Biology with Physiology Presentation prepared by Chris C. Romero Neil Campbell, Jane Reece,
Characterization of Double-Stranded RNA Satellites Associated with the Trichomonas vaginalis Virus
 JOURNAL OF VIROLOGY, Nov. 1995, p. 6892 6897 Vol. 69, No. 11 0022-538X/95/$04.00 0 Copyright 1995, American Society for Microbiology Characterization of Double-Stranded RNA Satellites Associated with the
JOURNAL OF VIROLOGY, Nov. 1995, p. 6892 6897 Vol. 69, No. 11 0022-538X/95/$04.00 0 Copyright 1995, American Society for Microbiology Characterization of Double-Stranded RNA Satellites Associated with the
