Title: Single Cell Dual Adherent-Suspension Co-Culture Micro-Environment for Studying Tumor- Stromal Interactions
|
|
|
- Helen Pitts
- 5 years ago
- Views:
Transcription
1 Electronic Supplementary Material (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2016 Title: Single Cell Dual Adherent-Suspension Co-Culture Micro-Environment for Studying Tumor- Stromal Interactions Authors and affiliations: Yu-Chih Chen a, c^, Zhixiong Zhang a^, Shamileh Fouladdel c, Yadwinder Deol c, Patrick N. Ingram b, Sean P. McDermott c, Ebrahim Azizi c, Max S. Wicha c, and Euisik Yoon* a, b a Department of Electrical Engineering and Computer Science, University of Michigan, 1301 Beal Avenue, Ann Arbor, MI ; b Department of Biomedical Engineering, University of Michigan, 2200 Bonisteel, Blvd. Ann Arbor, MI , USA c University of Michigan Comprehensive Cancer Center, 1500 E. Medical Center Drive, Ann Arbor, MI 48109, USA ^Authors made equal contributions *Corresponding author Yu-Chih Chen 1301 Beal Avenue, Ann Arbor, MI , USA Tel: ; yuchchen@umich.edu. Euisik Yoon 1301 Beal Avenue, Ann Arbor, MI , USA Tel: ; esyoon@umich.edu.
2 Supplementary Tables Supplementary Table 1. The 96-gene panel chosen to identify the oncogenic signature of breast cancer cells. ABCB1 CD146 EZH2 ITGB3 NFKB1 SOX2 ABCG2 CD20 FBXW7 JAG2 NOTCH1 STAP2 AKT1 CD24 GAPDH KRT18 NOTCH2 TAZ AKT3 CD3D GATA3 KRT19 NOTCH3 TGFb1 ALDH1a1 CD44 gp130 KRT5 NUMB TGFbR1 ALDH1a3 CD45 GSK3B KRT7 O ct4 TM4SF1 AMOTL2 CDH1 HER2 KRT8 p53 TMEM57 ANXA3 CDH2 HES1 LIN28A p63 TNKS1BP1 AR CDH3 HEY2 MCL1 PCNA TSPAN6 BAX CTNNB1 HPRT1 MET PGR Twist1 BCL2 CXCR1 ID1 MKI67 PI3K UXT BRCA1 CXCR4 ID2 MMP9 PTEN Vimentin CCND1 DLL1 IL6 MTOR RAB7A WNT2 CD11B EGFR IL6R MUC1 SLUG YAP1 CD133 EpCAM IL8 NANOG SNAI1 ZEB1 CD14 ESR1 ITGA6 NESTIN SOCS3 ZEB2
3 Supplementary Table 2. The significantly different genes between mono-cultured T47D and cocultured T47D cells. GeneID P-Value T47D_Mono.Average T47D_Co.Average KRT8 1.12E GATA3 1.35E STAP2 3.79E ANXA3 1.26E PI3K 5.45E RAB7A 6.68E NUMB 1.66E TNKS1BP1 1.60E KRT E HES1 6.28E HER2 8.57E MUC1 9.26E NOTCH3 1.60E CDH1 1.51E UXT 1.78E MTOR 3.19E CTNNB1 4.85E MKI E KRT SOCS CD MCL BAX PTEN DLL PGR JAG CXCR ITGA AMOTL TSPAN O ct HPRT CDH NOTCH
4 Supplementary Table 3. The significantly different genes between MKI67 high and MKI67 low cells in co-culture group. GeneID P-Value MKI67+.Average MKI67-.Average MKI E EZH2 3.08E JAG2 6.42E CCND PCNA BRCA ANXA NOTCH BAX HPRT ID
5 Supplementary Figures Supplemental Fig. 1. Fabrication process of the dual adherent-suspension co-culture microenvironment.
6 Supplementary Fig. 2: The presented dual culture platform. Cancer cells are loaded into a suspension chamber for clonal sphere formation while the stromal cells are cultured in the adherent chamber in close proximity for interaction through small connecting channels.
7 Supplemental Fig. 3. Simulations of the distribution of the concentration (mole/l) of the proteins secreted by fibroblast when 50 Pa was applied to the adherent port of the chamber (the flow condition right after media exchange) using COMSOL 5.1. We assume that one cell secretes 1000 proteins per second, 10 fibroblasts are captured in the outer chamber, and the protein diffusion coefficient is 1-10 m 2 /s. 1-4 (Scale bar: 100 µm)
8 Supplemental Fig. 4. Simulations of the distribution of the concentration (mole/l) of the proteins secreted by cancer cell when 50 Pa was applied to the adherent port of the chamber (the flow condition right after media exchange) using COMSOL 5.1. We assume that one cell secretes 1000 proteins per second, 1 cancer cell is captured in the inner chamber, and the protein diffusion coefficient is 1-10 m 2 /s. 1-4 (Scale bar: 100 µm)
9 Supplemental Fig. 5. Simulations of the distribution of the concentration (mole/l) of the proteins secreted by fibroblast when there is no flow using COMSOL 5.1. We assume that one cell secretes 1000 proteins per second, 10 fibroblasts are captured in the outer chamber, and the protein diffusion coefficient is 1-10 m 2 /s. 1-4 (Scale bar: 100 µm)
10 Supplemental Fig. 6. Simulations of the distribution of the concentration (mole/l) of the proteins secreted by cancer cell when there is no flow using COMSOL 5.1. We assume that one cell secretes 1000 proteins per second, 1 cancer cell is captured in the inner chamber, and the protein diffusion coefficient is 1-10 m 2 /s. 1-4 (Scale bar: 100 µm)
11 Supplemental Fig. 7. Principal component analysis (PCA) plot showing log2 gene expression data of single cells from T47D breast cancer cell line. T47D B1st (Red) and T47D BulkR (Blue) are the same single cells isolated by C1 and analyzed by BioMark HD in 2 separate RT-qPCR experiments as technical replicates. T47D B3rd (Green) and T47D B1st (Red) are biological replicates of 2 independent C1/BioMark HD experiments.
12 Supplemental Fig. 8. The plasma etching process to remove residual polyhema. (a) Before Etching, (b) Proper etching only removes the un-desired polyhema residues. (b) Over-etching can etch the polyhema in the indented well and expose the PDMS substrate.
13 Supplemental Fig. 9. The selectivity of the adherent-suspension co-culture micro-environment of (a) MDA-MB-231 and (b) C2C12 cells. For both cell lines, no cell adhere in the polyhemacoated indented well, but cells can adhere on the un-coated. (scale bar: 100 µm)
14 Supplemental Fig. 10. COMSOL simulations of the cell loading process: (a) pressure, (b) flow velocity in the XY-plane of 1.5 µm height (the half of the height of the interaction channel), and (c) the flow velocity in the XZ-plane. 50 Pa was applied to the adherent port of the chamber to simulate the flow condition right after media exchange. The results suggest that once a cell settles into the indented well, it is unlikely to be washed out by reversing flow. (Scale bar: 100 µm)
15 Supplemental Fig. 11. Representative images of single breast cancer cells isolated within the C1 chip. Cells of different sizes and morphology can be captured on-chip.
16 Supplementary Fig. 12. PCA clustering of single cell expression analysis for co-cultured (red, N=37) and mono-cultured (green, N=22) T47D cells compared. The results were not normalized.
17 Supplementary Fig. 13. Heatmap hierarchical clustering of single cell expression analysis for between co-cultured (red, N=37) and mono-cultured (green, N=22) T47D cells. The results were not normalized.
18 Supplementary Fig. 14. Violin plots of all 96 genes of co-cultured (red, N = 37) and mono-cultured (green, N = 22) cells. The results were not normalized.
19 Supplementary Fig. 15: PCA clustering of single cell expression analysis for co-cultured (red, N=37) and mono-cultured (green, N=22) T47D cells compared. The results were normalized by GAPDH.
20 Supplementary Fig. 16. Heatmap hierarchical clustering of single cell expression analysis for between co-cultured (red, N=37) and mono-cultured (green, N=22) T47D cells. The results were normalized by GAPDH.
21 Supplementary Fig. 17. Violin plots of all 96 genes of co-cultured (red, N = 37) and mono-cultured (green, N = 22) cells. The results were normalized by GAPDH.
22 Supplementary Fig. 18: PCA clustering of single cell expression analysis for MKI67 high (green, N=10) and MKI 67 low (red, N=10) cells in the co-cultured group. The results were normalized by GAPDH.
23 Supplementary Fig. 19. Heatmap hierarchical clustering of single cell expression analysis for MKI67 high (green, N=10) and MKI 67 low (red, N=10) cells in the co-cultured group. The results were normalized by GAPDH.
24 Supplementary Fig. 20. Violin plots of all 96 genes of MKI67 high (green, N=10) and MKI 67 low (red, N=10) cells in the co-cultured group. The results were normalized by GAPDH.
25 Supplementary Fig. 21: PCA clustering of single cell expression analysis for MKI67 high (green, N=1) and MKI 67 low (red, N=10) cells in the mono-cultured group. The results were normalized by GAPDH.
26 Supplementary Fig. 22. Heatmap hierarchical clustering of single cell expression analysis for MKI67 high (green, N=1) and MKI 67 low (red, N=10) cells in the mono-cultured group. The results were normalized by GAPDH.
27 Supplementary Fig. 23. Violin plots of all 96 genes of MKI67 high (green, N=1) and MKI 67 low (red, N=10) cells in the mono-cultured group. The results were normalized by GAPDH.
28 Reference: 1. Henn A. D.; Rebhahn J.; Brown M. A.; Murphy A. J.; Coca M. N.; Hyrien O.; Pellegrin T.; Mosmann T.; Zand M. S. J. Immunol. 2009, 183, Alexis J. Torres, Abby S. Hill, and J. Christopher Love, Anal Chem. 2014, 86(23), Love J. C. Ronan J. L. Grotenbreg G. M. van der Veen A. G. Ploegh H. L. Nat. Biotechnol. 2006, 24, Charlet M. Kromenaker S. J. Srienc F. Biotechnol. Bioeng. 1995, 47,
Supplemental Information. High-Throughput Microfluidic Labyrinth for the. Label-free Isolation of Circulating Tumor Cells
 Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Finite-element analysis of cell cluster dynamics in different cluster trap architectures. (a) Cluster-Chip (b) Filter (c) A structure identical to the Cluster-Chip except that one
Supplementary Figure 1 Finite-element analysis of cell cluster dynamics in different cluster trap architectures. (a) Cluster-Chip (b) Filter (c) A structure identical to the Cluster-Chip except that one
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
The Avatar System TM Yields Biologically Relevant Results
 Application Note The Avatar System TM Yields Biologically Relevant Results Liquid biopsies stand to revolutionize the cancer field, enabling early detection and noninvasive monitoring of tumors. In the
Application Note The Avatar System TM Yields Biologically Relevant Results Liquid biopsies stand to revolutionize the cancer field, enabling early detection and noninvasive monitoring of tumors. In the
Supplementary Information and Figure legends
 Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Table S1: Analysis of Notch gene rearrangements in triple negative breast cancer subtypes
 Supplemental Tables Table S1: Analysis of Notch gene rearrangements in triple negative breast cancer subtypes NOTCH1 or NOTCH2 Basal Immune Luminal AR Mesenchymal Stem Like WT 27 (87%) 24 (100%) 4 (66%)
Supplemental Tables Table S1: Analysis of Notch gene rearrangements in triple negative breast cancer subtypes NOTCH1 or NOTCH2 Basal Immune Luminal AR Mesenchymal Stem Like WT 27 (87%) 24 (100%) 4 (66%)
Controllable organization and high throughput production of recoverable 3D tumors using pneumatic microfluidics
 Electronic Supplementary Material (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information (ESI) for Lab on a Chip This journal is The Royal Society
Electronic Supplementary Material (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information (ESI) for Lab on a Chip This journal is The Royal Society
Oncology Biomarker Panel
 Oncology Biomarker Panel HTG EdgeSeq Oncology Biomarker Panel The HTG EdgeSeq Oncology Biomarker Panel profiles tumor samples in order to identify therapeutic targets and drug response markers using an
Oncology Biomarker Panel HTG EdgeSeq Oncology Biomarker Panel The HTG EdgeSeq Oncology Biomarker Panel profiles tumor samples in order to identify therapeutic targets and drug response markers using an
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study. February 2018
 Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
The mir-199a/brm/egr1 axis is a determinant of anchorage-independent growth in epithelial tumor cell lines
 Supplementary information Supplementary Figure -9 Supplementary Table -4 The mir-99a/brm/egr axis is a determinant of anchorage-independent growth in epithelial tumor cell lines Kazuyoshi Kobayashi, Kouhei
Supplementary information Supplementary Figure -9 Supplementary Table -4 The mir-99a/brm/egr axis is a determinant of anchorage-independent growth in epithelial tumor cell lines Kazuyoshi Kobayashi, Kouhei
Supplementary Figure (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were
 Supplementary Figure 1. Gd@C 82 (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were treated with PBS, Gd@C 82 (OH) 22, C 60 (OH) 22 or GdCl
Supplementary Figure 1. Gd@C 82 (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were treated with PBS, Gd@C 82 (OH) 22, C 60 (OH) 22 or GdCl
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier
 Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester
 Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry Please do 2017 not adjust margins Nanoscale Electronic Supplementary Information Mono-fullerenols modulating
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry Please do 2017 not adjust margins Nanoscale Electronic Supplementary Information Mono-fullerenols modulating
Fluxion Biosciences and Swift Biosciences Somatic variant detection from liquid biopsy samples using targeted NGS
 APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
Treating patients affordably: personalized medicine in India
 economic Treating patients affordably: personalized medicine in India biologic B Moni Abraham Kuriakose MB ChB, MD, FDSRCS, FFDRCS, FRCS Ed., FRCS, Dip AB Professor and Director, Department of Surgical
economic Treating patients affordably: personalized medicine in India biologic B Moni Abraham Kuriakose MB ChB, MD, FDSRCS, FFDRCS, FRCS Ed., FRCS, Dip AB Professor and Director, Department of Surgical
TP53 Mutation, Epithelial-Mesenchymal Transition, and Stemlike Features in Breast Cancer Subtypes
 Journal of Biomedicine and Biotechnology Volume 0, Article ID 0, pages doi:0./0/0 Research Article TP Mutation, Epithelial-Mesenchymal Transition, and Stemlike Features in Breast Cancer Subtypes Danila
Journal of Biomedicine and Biotechnology Volume 0, Article ID 0, pages doi:0./0/0 Research Article TP Mutation, Epithelial-Mesenchymal Transition, and Stemlike Features in Breast Cancer Subtypes Danila
Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or
 Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or ALDH-positive cell population by qpcr. Data represent
Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or ALDH-positive cell population by qpcr. Data represent
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
AdnaNews. News from the meeting on Advances in Circulating Tumor Cells (ACTC), Reythymnon, Crete, Greece October 8-11, 2014.
 News from the meeting on Advances in Circulating Tumor Cells (ACTC), Reythymnon, Crete, Greece October 8-11, 2014. The ACTC meeting (www.actc2014.org) is together with the ISMRC meeting one of the most
News from the meeting on Advances in Circulating Tumor Cells (ACTC), Reythymnon, Crete, Greece October 8-11, 2014. The ACTC meeting (www.actc2014.org) is together with the ISMRC meeting one of the most
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
Genomic Analyses across Six Cancer Types Identify Basal-like Breast Cancer as a Unique Molecular Entity
 Genomic Analyses across Six Cancer Types Identify Basal-like Breast Cancer as a Unique Molecular Entity Aleix Prat, Barbara Adamo, Cheng Fan, Vicente Peg, Maria Vidal, Patricia Galván, Ana Vivancos, Paolo
Genomic Analyses across Six Cancer Types Identify Basal-like Breast Cancer as a Unique Molecular Entity Aleix Prat, Barbara Adamo, Cheng Fan, Vicente Peg, Maria Vidal, Patricia Galván, Ana Vivancos, Paolo
A High-Throughput Photodynamic Therapy Screening Platform with On-Chip Control of Multiple Microenvironmental Factors
 A High-Throughput Photodynamic Therapy Screening Platform with On-Chip Control of Multiple Microenvironmental Factors Xia Lou a, Gwangseong Kim b, Hyung Ki Yoon b, Yong-Eun Koo Lee b, Raoul Kopelman b,
A High-Throughput Photodynamic Therapy Screening Platform with On-Chip Control of Multiple Microenvironmental Factors Xia Lou a, Gwangseong Kim b, Hyung Ki Yoon b, Yong-Eun Koo Lee b, Raoul Kopelman b,
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Correlation between LKB1 and YAP expression in human lung cancer samples. (a) Representative photos showing LKB1 and YAP immunohistochemical staining in human
Supplementary Figures Supplementary Figure 1 Correlation between LKB1 and YAP expression in human lung cancer samples. (a) Representative photos showing LKB1 and YAP immunohistochemical staining in human
1. Microfluidic device characteristics and cell compartmentalization
 Supplementary figures: 1. Microfluidic device characteristics and cell compartmentalization Microfluidic channels were molded in PDMS (Fig. S1 A and B) over silicon masters, which were structured with
Supplementary figures: 1. Microfluidic device characteristics and cell compartmentalization Microfluidic channels were molded in PDMS (Fig. S1 A and B) over silicon masters, which were structured with
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing)
 Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
(A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square),
 Supplementary Figures and Tables Figure S1. Validation of EMT-selective small molecules (A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square), and HMLE_Twist (black diamond) cells
Supplementary Figures and Tables Figure S1. Validation of EMT-selective small molecules (A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square), and HMLE_Twist (black diamond) cells
3D Tissue Models. Simple, Low Cost Fabrication. Simple, Robust Protocols
 3D Tissue Models SynVivo is a physiological, cell-based microfluidic platform that provides a morphologically and physiologically realistic microenvironment allowing real-time study of cellular behavior,
3D Tissue Models SynVivo is a physiological, cell-based microfluidic platform that provides a morphologically and physiologically realistic microenvironment allowing real-time study of cellular behavior,
Electronic Supplementary Information (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2012
 Electronic Supplementary Information (ESI) for Lab on a Chip Electronic Supplementary Information Construction of oxygen and chemical concentration gradients in a single microfluidic device for studying
Electronic Supplementary Information (ESI) for Lab on a Chip Electronic Supplementary Information Construction of oxygen and chemical concentration gradients in a single microfluidic device for studying
Supplementary Data Dll4-containing exosomes induce capillary sprout retraction ina 3D microenvironment
 Supplementary Data Dll4-containing exosomes induce capillary sprout retraction ina 3D microenvironment Soheila Sharghi-Namini 1, Evan Tan 1,2, Lee-Ling Sharon Ong 1, Ruowen Ge 2 * and H. Harry Asada 1,3
Supplementary Data Dll4-containing exosomes induce capillary sprout retraction ina 3D microenvironment Soheila Sharghi-Namini 1, Evan Tan 1,2, Lee-Ling Sharon Ong 1, Ruowen Ge 2 * and H. Harry Asada 1,3
CD Marker Antibodies. atlasantibodies.com
 CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Layered-IHC (L-IHC): A novel and robust approach to multiplexed immunohistochemistry So many markers and so little tissue
 Page 1 The need for multiplex detection of tissue biomarkers. There is a constant and growing demand for increased biomarker analysis in human tissue specimens. Analysis of tissue biomarkers is key to
Page 1 The need for multiplex detection of tissue biomarkers. There is a constant and growing demand for increased biomarker analysis in human tissue specimens. Analysis of tissue biomarkers is key to
Identifying genomic signatures in circulating breast tumour cells
 Identifying genomic signatures in circulating breast tumour cells 9 th ISMRC 2013, Paris, France September 25th, 2013 NISHA KANWAR PhD Candidate Department of Laboratory Medicine and Pathobiology University
Identifying genomic signatures in circulating breast tumour cells 9 th ISMRC 2013, Paris, France September 25th, 2013 NISHA KANWAR PhD Candidate Department of Laboratory Medicine and Pathobiology University
Valve-Based Microfluidic Compression Platform: Single Axon Injury and Regrowth
 Supplementary Material (ESI) for Lab on a Chip This Journal is The Royal Society of Chemistry 2011 Valve-Based Microfluidic Compression Platform: Single Axon Injury and Regrowth Suneil Hosmane a, Adam
Supplementary Material (ESI) for Lab on a Chip This Journal is The Royal Society of Chemistry 2011 Valve-Based Microfluidic Compression Platform: Single Axon Injury and Regrowth Suneil Hosmane a, Adam
ONLINE DATA SUPPLEMENT. Single Cell RNA-Sequencing Identifies Diverse Roles of Epithelial Cells in. Idiopathic Pulmonary Fibrosis
 ONLINE DATA SUPPLEMENT Single Cell RNA-Sequencing Identifies Diverse Roles of Epithelial Cells in Idiopathic Pulmonary Fibrosis Yan Xu Takako Mizuno Anusha Sridharan Yina Du Minzhe Guo Jie Tang Kathryn
ONLINE DATA SUPPLEMENT Single Cell RNA-Sequencing Identifies Diverse Roles of Epithelial Cells in Idiopathic Pulmonary Fibrosis Yan Xu Takako Mizuno Anusha Sridharan Yina Du Minzhe Guo Jie Tang Kathryn
Supplementary information. Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls
 Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
PKCζ Promotes Breast Cancer Invasion by Regulating Expression of E-cadherin and Zonula Occludens-1 (ZO-1) via NFκB-p65
 SUPPLEMENTARY INFORMATION TITLE: PKCζ Promotes Breast Cancer Invasion by Regulating Expression of E-cadherin and Zonula Occludens-1 (ZO-1) via NFκB-p65 RUNNING TITLE: PKCζ-NFκB Signaling in Breast Cancer
SUPPLEMENTARY INFORMATION TITLE: PKCζ Promotes Breast Cancer Invasion by Regulating Expression of E-cadherin and Zonula Occludens-1 (ZO-1) via NFκB-p65 RUNNING TITLE: PKCζ-NFκB Signaling in Breast Cancer
Supplementary Figure 1: Experimental design. DISCOVERY PHASE VALIDATION PHASE (N = 88) (N = 20) Healthy = 20. Healthy = 6. Endometriosis = 33
 DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
SureSelect Cancer All-In-One Custom and Catalog NGS Assays
 SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
Comparative study of multicellular tumor spheroid formation methods and implications for drug screening
 Supporting Information Comparative study of multicellular tumor spheroid formation methods and implications for drug screening Maria F. Gencoglu, Lauren E. Barney, Christopher L. Hall, Elizabeth A. Brooks,
Supporting Information Comparative study of multicellular tumor spheroid formation methods and implications for drug screening Maria F. Gencoglu, Lauren E. Barney, Christopher L. Hall, Elizabeth A. Brooks,
SUPPLEMENTAL INFORMATIONS
 1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
Supporting Information for
 Supporting Information for Rupture of Lipid Membranes Induced by Amphiphilic Janus Nanoparticles Kwahun Lee, Liuyang Zhang, Yi Yi, Xianqiao Wang, Yan Yu* Department of Chemistry, Indiana University, Bloomington,
Supporting Information for Rupture of Lipid Membranes Induced by Amphiphilic Janus Nanoparticles Kwahun Lee, Liuyang Zhang, Yi Yi, Xianqiao Wang, Yan Yu* Department of Chemistry, Indiana University, Bloomington,
Phosphorylation Site Company Cat #
 Supplemental Table 1. Antibodies used for RPPA analysis. Label Protein Phosphorylation Site Company Cat # Used on MDA_CLSS Used on MDA_Pilot 4EBP1 4EBP1 Cell Signaling 9452 No 4EBP1.pS65 4EBP1 S65 Cell
Supplemental Table 1. Antibodies used for RPPA analysis. Label Protein Phosphorylation Site Company Cat # Used on MDA_CLSS Used on MDA_Pilot 4EBP1 4EBP1 Cell Signaling 9452 No 4EBP1.pS65 4EBP1 S65 Cell
Supplementary Figures
 Supplementary Figures Figure S1. Validation of kinase regulators of ONC201 sensitivity. Validation and screen results for changes in cell viability associated with the combination of ONC201 treatment (1
Supplementary Figures Figure S1. Validation of kinase regulators of ONC201 sensitivity. Validation and screen results for changes in cell viability associated with the combination of ONC201 treatment (1
Supplemental Table S1
 Supplemental Table S. Tumorigenicity and metastatic potential of 44SQ cell subpopulations a Tumorigenicity b Average tumor volume (mm ) c Lung metastasis d CD high /4 8. 8/ CD low /4 6./ a Mice were injected
Supplemental Table S. Tumorigenicity and metastatic potential of 44SQ cell subpopulations a Tumorigenicity b Average tumor volume (mm ) c Lung metastasis d CD high /4 8. 8/ CD low /4 6./ a Mice were injected
Supplementary Figure 1. Basal level EGFR across a panel of ESCC lines. Immunoblots demonstrate the expression of phosphorylated and total EGFR as
 Supplementary Figure 1. Basal level EGFR across a panel of ESCC lines. Immunoblots demonstrate the expression of phosphorylated and total EGFR as well as their downstream effectors across a panel of ESCC
Supplementary Figure 1. Basal level EGFR across a panel of ESCC lines. Immunoblots demonstrate the expression of phosphorylated and total EGFR as well as their downstream effectors across a panel of ESCC
supplementary information
 DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
Dynamic cohesin-mediated chromatin architecture controls epithelial mesenchymal plasticity in cancer
 Article Dynamic cohesin-mediated chromatin architecture controls epithelial mesenchymal plasticity in cancer Jiyeon Yun,, Sang-Hyun Song, Hwang-Phill Kim, Sae-Won Han,, Eugene C Yi & Tae-You Kim,,, Abstract
Article Dynamic cohesin-mediated chromatin architecture controls epithelial mesenchymal plasticity in cancer Jiyeon Yun,, Sang-Hyun Song, Hwang-Phill Kim, Sae-Won Han,, Eugene C Yi & Tae-You Kim,,, Abstract
TITLE: A Novel Immune-Intact Mouse Model of Prostate Cancer Bone Metastasis: Mechanisms of Chemotaxis and Bone Colonization
 AWARD NUMBER: W81XWH-16-1-0174 TITLE: A Novel Immune-Intact Mouse Model of Prostate Cancer Bone Metastasis: Mechanisms of Chemotaxis and Bone Colonization PRINCIPAL INVESTIGATOR: Srinivas Nandana CONTRACTING
AWARD NUMBER: W81XWH-16-1-0174 TITLE: A Novel Immune-Intact Mouse Model of Prostate Cancer Bone Metastasis: Mechanisms of Chemotaxis and Bone Colonization PRINCIPAL INVESTIGATOR: Srinivas Nandana CONTRACTING
RNA preparation from extracted paraffin cores:
 Supplementary methods, Nielsen et al., A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor positive breast cancer.
Supplementary methods, Nielsen et al., A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor positive breast cancer.
Supplementary Material
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2018 Supplementary Material 1 Supplemental Figures Supplemental Figure S1. Mechanical properties
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2018 Supplementary Material 1 Supplemental Figures Supplemental Figure S1. Mechanical properties
Genetic heterogeneity and transcriptional plasticity drive resistance during immuno- and targeted therapy
 Genetic heterogeneity and transcriptional plasticity drive resistance during immuno- and targeted therapy Reinhard Dummer, Daniel Widmer, Daniela Mihic, Phil Cheng, Lars French, Verena Paulitzschke, Mitch
Genetic heterogeneity and transcriptional plasticity drive resistance during immuno- and targeted therapy Reinhard Dummer, Daniel Widmer, Daniela Mihic, Phil Cheng, Lars French, Verena Paulitzschke, Mitch
Bezzi et al., Supplementary Figure 1 *** Nature Medicine: doi: /nm Pten pc-/- ;Zbtb7a pc-/- Pten pc-/- ;Pml pc-/- Pten pc-/- ;Trp53 pc-/-
 Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes.
 Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Associating GWAS Information with the Notch Signaling Pathway Using Transcription Profiling
 Cancer Informatics Original Research Open Access Full open access to this and thousands of other papers at http://www.la-press.com. Associating GWAS Information with the Notch Signaling Pathway Using Transcription
Cancer Informatics Original Research Open Access Full open access to this and thousands of other papers at http://www.la-press.com. Associating GWAS Information with the Notch Signaling Pathway Using Transcription
Supplementary Figure 1 a
 Supplementary Figure 1 a b d c Supplementary Figure 1. Poor sensitivity for mutation detection using established single-cell RNA-sequencing methods. (a) Representative bioanalyzer trace showing expected
Supplementary Figure 1 a b d c Supplementary Figure 1. Poor sensitivity for mutation detection using established single-cell RNA-sequencing methods. (a) Representative bioanalyzer trace showing expected
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Frequency(%) KRAS G12 KRAS G13 KRAS A146 KRAS Q61 KRAS K117N PIK3CA H1047 PIK3CA E545 PIK3CA E542K PIK3CA Q546. EGFR exon19 NFS-indel EGFR L858R
 Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Analyses of Intravesicular Exosomal Proteins Using a Nano-Plasmonic System
 Supporting Information Analyses of Intravesicular Exosomal Proteins Using a Nano-Plasmonic System Jongmin Park 1, Hyungsoon Im 1.2, Seonki Hong 1, Cesar M. Castro 1,3, Ralph Weissleder 1,4, Hakho Lee 1,2
Supporting Information Analyses of Intravesicular Exosomal Proteins Using a Nano-Plasmonic System Jongmin Park 1, Hyungsoon Im 1.2, Seonki Hong 1, Cesar M. Castro 1,3, Ralph Weissleder 1,4, Hakho Lee 1,2
Mesenchymal Stem Cells Reshape and Provoke Proliferation of Articular. State Key Laboratory of Bioreactor Engineering, East China University of
 Mesenchymal Stem Cells Reshape and Provoke Proliferation of Articular Chondrocytes by Paracrine Secretion Lei Xu, Yuxi Wu, Zhimiao Xiong, Yan Zhou, Zhaoyang Ye *, Wen-Song Tan * State Key Laboratory of
Mesenchymal Stem Cells Reshape and Provoke Proliferation of Articular Chondrocytes by Paracrine Secretion Lei Xu, Yuxi Wu, Zhimiao Xiong, Yan Zhou, Zhaoyang Ye *, Wen-Song Tan * State Key Laboratory of
Surface-Enhanced Raman Scattering Active Gold Nanoparticles. with Enzyme-Mimicking Activities for Measuring Glucose and
 Surface-Enhanced Raman Scattering Active Gold Nanoparticles with Enzyme-Mimicking Activities for Measuring Glucose and Lactate in Living Tissues Yihui Hu, Hanjun Cheng, Xiaozhi Zhao, Jiangjiexing Wu, Faheem
Surface-Enhanced Raman Scattering Active Gold Nanoparticles with Enzyme-Mimicking Activities for Measuring Glucose and Lactate in Living Tissues Yihui Hu, Hanjun Cheng, Xiaozhi Zhao, Jiangjiexing Wu, Faheem
What is the status of the technologies of "precision medicine?
 Session 2: What is the status of the technologies of "precision medicine? Gideon Blumenthal, MD, Clinical Team Leader, Thoracic and Head/Neck Oncology, Center for Drug Evaluation and Research (CDER), U.S.
Session 2: What is the status of the technologies of "precision medicine? Gideon Blumenthal, MD, Clinical Team Leader, Thoracic and Head/Neck Oncology, Center for Drug Evaluation and Research (CDER), U.S.
Supplementary Materials and Methods
 DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
Peng Zhang 1, Michelle Yueqin Chen 1, Diana Caridha 1, William J. Smith 2 & Peter K. Chiang 1
 Peng Zhang 1, Michelle Yueqin Chen 1, Diana Caridha 1, William J. Smith 2 & Peter K. Chiang 1 1 Walter Reed Army Institute of Research, 2 United States Army Medical Research Institute of Chemical Defense
Peng Zhang 1, Michelle Yueqin Chen 1, Diana Caridha 1, William J. Smith 2 & Peter K. Chiang 1 1 Walter Reed Army Institute of Research, 2 United States Army Medical Research Institute of Chemical Defense
In situ drug-receptor binding kinetics in single cells: a quantitative label-free study of anti-tumor drug resistance
 Supplementary Information for In situ drug-receptor binding kinetics in single cells: a quantitative label-free study of anti-tumor drug resistance Wei Wang 1, Linliang Yin 2,3, Laura Gonzalez-Malerva
Supplementary Information for In situ drug-receptor binding kinetics in single cells: a quantitative label-free study of anti-tumor drug resistance Wei Wang 1, Linliang Yin 2,3, Laura Gonzalez-Malerva
SUPPLEMENTARY INFORMATION
 In the format provided by the authors and unedited. 2 3 4 DOI: 10.1038/NMAT4893 EGFR and HER2 activate rigidity sensing only on rigid matrices Mayur Saxena 1,*, Shuaimin Liu 2,*, Bo Yang 3, Cynthia Hajal
In the format provided by the authors and unedited. 2 3 4 DOI: 10.1038/NMAT4893 EGFR and HER2 activate rigidity sensing only on rigid matrices Mayur Saxena 1,*, Shuaimin Liu 2,*, Bo Yang 3, Cynthia Hajal
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Round Table: Tissue Biopsy versus Liquid Biopsy. César A. Rodríguez Hospital Universitario de Salamanca-IBSAL
 Round Table: Tissue Biopsy versus Liquid Biopsy César A. Rodríguez Hospital Universitario de Salamanca-IBSAL Introduction Classic Advantages of liquid biopsy collection over standard biopsy Standard biopsy
Round Table: Tissue Biopsy versus Liquid Biopsy César A. Rodríguez Hospital Universitario de Salamanca-IBSAL Introduction Classic Advantages of liquid biopsy collection over standard biopsy Standard biopsy
Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis
 Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis Yan Xu,, Barry R. Stripp, Jeffrey A. Whitsett JCI Insight. 2017;1(20):e90558. https://doi.org/10.1172/jci.insight.90558.
Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis Yan Xu,, Barry R. Stripp, Jeffrey A. Whitsett JCI Insight. 2017;1(20):e90558. https://doi.org/10.1172/jci.insight.90558.
Expanded View Figures
 EMO Molecular Medicine Proteomic map of squamous cell carcinomas Hanibal ohnenberger et al Expanded View Figures Figure EV1. Technical reproducibility. Pearson s correlation analysis of normalised SILC
EMO Molecular Medicine Proteomic map of squamous cell carcinomas Hanibal ohnenberger et al Expanded View Figures Figure EV1. Technical reproducibility. Pearson s correlation analysis of normalised SILC
(A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC,
 Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, MD
 AD Award Number: W81XWH-11-1-0126 TITLE: Chemical strategy to translate genetic/epigenetic mechanisms to breast cancer therapeutics PRINCIPAL INVESTIGATOR: Xiang-Dong Fu, PhD CONTRACTING ORGANIZATION:
AD Award Number: W81XWH-11-1-0126 TITLE: Chemical strategy to translate genetic/epigenetic mechanisms to breast cancer therapeutics PRINCIPAL INVESTIGATOR: Xiang-Dong Fu, PhD CONTRACTING ORGANIZATION:
Simulation of Chemotractant Gradients in Microfluidic Channels to Study Cell Migration Mechanism in silico
 Simulation of Chemotractant Gradients in Microfluidic Channels to Study Cell Migration Mechanism in silico P. Wallin 1*, E. Bernson 1, and J. Gold 1 1 Chalmers University of Technology, Applied Physics,
Simulation of Chemotractant Gradients in Microfluidic Channels to Study Cell Migration Mechanism in silico P. Wallin 1*, E. Bernson 1, and J. Gold 1 1 Chalmers University of Technology, Applied Physics,
stomach p63/krt5 CDH17 CEACAM5 CEACAM7 ANXA13 ANXA10 CLDN18 VSIG1 TFF2 VIL1 GDA
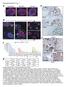 Supplementary Fig. 1 a esophagus Barrett s e FABP1 Ecad stomach TFF3 b p63/krt5 CDH17 REG4 CDH17 f c signal intensity p63/krt5 EsoSC 5000 4000 3000 2000 CDH17 1500 1000 Barrett s KRT14 KRT6A KRT13 KRT5
Supplementary Fig. 1 a esophagus Barrett s e FABP1 Ecad stomach TFF3 b p63/krt5 CDH17 REG4 CDH17 f c signal intensity p63/krt5 EsoSC 5000 4000 3000 2000 CDH17 1500 1000 Barrett s KRT14 KRT6A KRT13 KRT5
Case Study - Informatics
 bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
Validation & Assay Performance Summary
 Validation & Assay Performance Summary CellSensor DBE-bla MDA-MB-468 Cell Line Cat. no. K1814 Pathway Description The phosphatidylinositol-3-kinase (PI3K) signaling cascade is essential for cell growth
Validation & Assay Performance Summary CellSensor DBE-bla MDA-MB-468 Cell Line Cat. no. K1814 Pathway Description The phosphatidylinositol-3-kinase (PI3K) signaling cascade is essential for cell growth
Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were
 Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were separated via ultracentrifugation and lysed to analyze
Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were separated via ultracentrifugation and lysed to analyze
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Diagram of BBB and brain chips.
 Supplementary Figure 1 Diagram of BBB and brain chips. (a) Schematic of the BBB Chip demonstrates the 3 parts of the chip, Top PDMS channel, membrane and Bottom PDMS channel; (b) Image of 2 BBB Chips,
Supplementary Figure 1 Diagram of BBB and brain chips. (a) Schematic of the BBB Chip demonstrates the 3 parts of the chip, Top PDMS channel, membrane and Bottom PDMS channel; (b) Image of 2 BBB Chips,
Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63.
 Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Breast Cancer and Activated Signaling Pathways: Avenues for New Therapies
 Breast Cancer and Activated Signaling Pathways: Avenues for New Therapies Princess Margaret Cancer Center University Health Network Michael Reedijk MD PhD FRCSC Surgical Oncology No conflicts of interest
Breast Cancer and Activated Signaling Pathways: Avenues for New Therapies Princess Margaret Cancer Center University Health Network Michael Reedijk MD PhD FRCSC Surgical Oncology No conflicts of interest
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Joint Department of Biomedical Engineering
 Electronic Supplementary Material (ESI) for Biomaterials Science. This journal is The Royal Society of Chemistry 2018 Supplementary Data Systemic Delivery of CRISPR/Cas9 with PEG-PLGA Nanoparticles for
Electronic Supplementary Material (ESI) for Biomaterials Science. This journal is The Royal Society of Chemistry 2018 Supplementary Data Systemic Delivery of CRISPR/Cas9 with PEG-PLGA Nanoparticles for
Islet Oxygen Consumption Assay using the XF24 Islet Capture Microplate Shirihai Lab and Seahorse Bioscience Aug
 1 Islet Oxygen Consumption Assay using the XF24 Islet Capture Microplate Shirihai Lab and Seahorse Bioscience Aug 6 2009 XF Analyzers are most commonly used with an adherent monolayer of cells attached
1 Islet Oxygen Consumption Assay using the XF24 Islet Capture Microplate Shirihai Lab and Seahorse Bioscience Aug 6 2009 XF Analyzers are most commonly used with an adherent monolayer of cells attached
Supplementary Table 1: Motor-clutch gene mrna expression Myosin Motors
 Supplementary Table 1: Motor-clutch gene mrna expression Myosin Motors Symbol Description Expression Rank MYL6 myosin, light chain 6, alkali, smooth muscle and non-muscle 11903 79 MYH9 myosin, heavy chain
Supplementary Table 1: Motor-clutch gene mrna expression Myosin Motors Symbol Description Expression Rank MYL6 myosin, light chain 6, alkali, smooth muscle and non-muscle 11903 79 MYH9 myosin, heavy chain
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Welcome. Nanostring Immuno-Oncology Summit. September 21st, FOR RESEARCH USE ONLY. Not for use in diagnostic procedures.
 Welcome Nanostring Immuno-Oncology Summit September 21st, 2017 1 FOR RESEARCH USE ONLY. Not for use in diagnostic procedures. FOR RESEARCH USE ONLY. Not for use in diagnostic procedures. Agenda 4:00-4:30
Welcome Nanostring Immuno-Oncology Summit September 21st, 2017 1 FOR RESEARCH USE ONLY. Not for use in diagnostic procedures. FOR RESEARCH USE ONLY. Not for use in diagnostic procedures. Agenda 4:00-4:30
Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the. mutated sequence.
 Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the mutated sequence. 1 Supplementary Figure 2. Expression of mir-182 and SMAD7 in various cell lines. (A) Basal levels of mir-182 expression
Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the mutated sequence. 1 Supplementary Figure 2. Expression of mir-182 and SMAD7 in various cell lines. (A) Basal levels of mir-182 expression
Negative Regulation of c-myc Oncogenic Activity Through the Tumor Suppressor PP2A-B56α
 Negative Regulation of c-myc Oncogenic Activity Through the Tumor Suppressor PP2A-B56α Mahnaz Janghorban, PhD Dr. Rosalie Sears lab 2/8/2015 Zanjan University Content 1. Background (keywords: c-myc, PP2A,
Negative Regulation of c-myc Oncogenic Activity Through the Tumor Suppressor PP2A-B56α Mahnaz Janghorban, PhD Dr. Rosalie Sears lab 2/8/2015 Zanjan University Content 1. Background (keywords: c-myc, PP2A,
Supplementary Information
 Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Color Key PCA. mir- 15a let-7c 106b let-7b let-7a 16 10b 99a 26a 20b 374b 19b 135b 125b a-5p 199b-5p 93 92b MES PN.
 1123 83 528 816 84 2 17 718 Comp3 3 2 1 1 2 3 4 Comp2 a b Color Key MES PN PCA 3 2 1 1 2 3 4 Comp1 1 2 3 1 2 3 4-2 -1 1 2 mi- 15a let-7c 16b let-7b let-7a 16 1b 99a 26a 2b 374b 19b 135b 125b 9 34 125a-5p
1123 83 528 816 84 2 17 718 Comp3 3 2 1 1 2 3 4 Comp2 a b Color Key MES PN PCA 3 2 1 1 2 3 4 Comp1 1 2 3 1 2 3 4-2 -1 1 2 mi- 15a let-7c 16b let-7b let-7a 16 1b 99a 26a 2b 374b 19b 135b 125b 9 34 125a-5p
ras Multikinase Inhibitor Multikinase Inhibitor 0.1
 a ras ** * ** * ** ** ** ** un in et m lu Se ib SL G 32 W 7 So 50 ra 74 fe ni b LY W 294 or 0 tm 02 R an ap n a in Ev my er cin ol im BE us Z2 3 En P 5 za I13 st 0 au rin D as SP ati 60 nib C 012 is 5
a ras ** * ** * ** ** ** ** un in et m lu Se ib SL G 32 W 7 So 50 ra 74 fe ni b LY W 294 or 0 tm 02 R an ap n a in Ev my er cin ol im BE us Z2 3 En P 5 za I13 st 0 au rin D as SP ati 60 nib C 012 is 5
