Comprehensive isolation, identification, and nucleic acid analysis of single breast cancer cells: CTC-isoTECH
|
|
|
- Ashley Carroll
- 5 years ago
- Views:
Transcription
1 Comprehensive isolation, identification, and nucleic acid analysis of single breast cancer cells: CTC-isoTECH Lori M. Millner 1,2*, Lindsay N. Strotman 1,2, Kevin S. Goudy 1,2, Mark W. Linder 1,2, Roland Valdes, Jr. 1,2 1 University of Louisville, School of Medicine, Department of Pathology and Laboratory Medicine; 511 South Floyd Street, MDR 221, Louisville KY PGXL Technologies, 201 E. Floyd St., Suite 306, Louisville KY *Corresponding author: Lori Millner. lmmill10@louisville.edu Citation: Lori Millner, et al. Comprehensive isolation, identification, and nucleic acid analysis of single breast cancer cells: CTC-isoTECH. Cancer Research Frontiers May; 2(2): doi: / Lori Millner, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Competing Interests: The authors declare no competing financial interests. Received Jan 8, 2016; Revised Mar 18, 2016; Accepted Mar 31, Published May 27, 2016 Abstract The ability to isolate, phenotypically characterize, and profile the gene signature of single circulating tumor cells (CTCs) will provide deeper insight into cancer metastasis and will lead to improved diagnosis and treatment of cancer patients. Using current methods based on positive selection of epithelial markers, up to 40% of patients with highly aggressive metastatic breast cancer have no CTCs detected. This may be due to CTCs undergoing the process of epithelial to mesenchymal transition (EMT). This process causes epithelial characteristics to be down regulated as more mesenchymal features develop allowing the cells to have higher motility and increased ability to evade immune detection. Methods to enable capture and characterization of heterogeneous CTCs including those with non-epithelial phenotypes are needed. Additionally, having the ability to isolate pure, single CTCs allows for individual cell analysis providing information that is masked by bulk analysis. Here we provide details on an integrated method for isolating and characterizing heterogeneous single breast cancer cells based on a multiantigen, negative depletion strategy followed by sorting with dielectrophoresis (DEPArray). This negative depletion method removes unwanted leukocytes and allows heterogeneous, epithelial and non-epithelial cells to be retained. We were able to visually verify the isolation of single tumor cells with 100% purity. Following sorting, next generation sequencing and real-time PCR of single cells is described. This method allows single, heterogeneous CTCs to be analyzed which will have implications for prognosis and treatment options. Keywords: Circulating tumor cells (CTCs), breast cancer, negative enrichment, dielectrophoresis, next generation sequencing Introduction:
2 A circulating tumor cell (CTC) is a tumor cell that has been shed from a tumor and is found in systemic circulation. It is thought that these cells are shed from either primary or metastatic tumors, intravasate into the systemic circulation and have the potential to form metastatic lesions. The clinical significance of CTCs is very high because the major cause of morbidity in breast cancer patients is metastatic cancer (1). These cells must have the ability to withstand the shear forces encountered in the systemic circulation and the ability to evade immune detection (2). In order to create a secondary metastatic lesion, CTCs must extravasate into microvasculature and initiate neoangiogenesis. Because of these unique and demanding requirements, CTCs likely possess traits that enable them to carry out these tasks resulting in exploitable markers for identification and characterization. Various method to enrich, isolate, and detect CTCs have been reported and are reviewed elsewhere (3). The only Food and Drug Administration (FDA) cleared method (CellSearch by Janssen) for identifying and enumerating epithelial CTCs uses peripheral blood mononuclear cells (PBMCs) enriched for cells expressing epithelial-cell adhesion molecule (EpCAM) and positively identifies CTCs based on expression of cytokeratins (CK) 8, 18, or 19. Since these markers are expressed in epithelial cells, this method identifies and enumerates only CTCs with an epithelial phenotype. Several studies have reported that overall and progression free survival is well correlated with the number of epithelial CTCs in metastatic breast, prostate, and colon cancers (4, 5) However, CTCs are not detectable in close to 40% of metastatic breast cancer patients (6-8). Additionally, CTC heterogeneity has been reported from a single blood draw of a single patient (9, 10). The process of epithelial to mesenchymal transition (EMT) is thought to be partly responsible for CTCs that are not detectable using epithelial markers. EMT is a biologic process that allows an epithelial cell to assume a mesenchymal phenotype including enhanced migratory capacity, invasiveness, elevated resistance to apoptosis and greatly increased production of ECM components (11, 12). EMT is known to occur early in development, wound healing, as well as in metastasis and has been reviewed elsewhere (13-17). A definition inclusive of CTCs that have experienced EMT needs to be established. In this study, we describe a method independent of epithelial phenotype status to enrich and isolate 2 distinct breast cancer cell line subtypes. A heterogeneous model is important because currently, only CTCs that express EpCAM are captured. We have chosen to include a breast cancer cell line that has undetectable or very low levels of EpCAM expression (MDA-MB-231, basal B) and a cell line that has moderate and varying EpCAM expression SK-Br-3 (luminal/ HER2). We used a combination of EpCAM, HER2, and CD44 for positive identification. EpCAM was chosen because it is the most commonly used extracellular marker for epithelial CTC detection. To detect non-epithelial cell types, HER2 and CD44 were chosen because they are likely to be expressed on more aggressive or non-epithelial CTCs. HER2 expression has been detected on CTCs in early and metastatic cancer and is not dependent on the primary tumor HER2 status (18, 19). In several studies, HER2 expression was found more frequently on CTCs than the primary tumor, and expression on CTCs has been associated with poor clinical outcome of early breast cancer (19-21). CD44 is involved in tumorigenesis, and it s presence on CTCs has been associated with lower overall survival than those with no CD44 expression (22). CD44 has been described as a marker of tumor-initiating cells and high expression has been observed on a subset of metastasis-initiating CTCs (23, 24). Studies have been conducted to characterize patients in which
3 no CTCs are detected by examining epithelial markers including EpCAM and cytokeratin. In metastatic (7) and non-metastatic (25) cancer, patients with the most aggressive diseases were found to have no detectable CTCs using epithelial markers. In metastatic breast cancer, it was observed that a higher proportion of patients with poor prognosis (high grade, hormone receptor and HER-2 negative disease, IBC patients, and patients with brain metastasis) had no detectable CTCs (7). Patients younger than 50 years old with early breast cancer possessing HER2-amplified and G1-G2 tumors had a higher possibility of having no detectable CTCs (25). It is not known if these patients have no CTCs or if they have CTCs that are simply not detected by epithelial markers. However, in a recent study, 64% of clinical samples with negative or undetermined EpCAM positive CTCs revealed at least one putative CTC (EpCAM neg CK pos /CD45 neg event) (8). Because cancer patients with no detectable epithelial CTCs may be at increased risk for having more aggressive disease, it is critical to develop methods to isolate and characterize these potential CTCs. The goal of this study was to demonstrate feasibility to identify, isolate, and characterize single cells taken from a heterogeneous cell mixture including epithelial and non-epithelial type cells. To accomplish this goal, we describe a method based on dielectrophoresis to identify, capture, and characterize 2 subtypes of single tumor cells. This method using the DEPArray (Silicon BioSystems, Italy) is based on a microchip containing microelectrodes capable of creating dielectrophoretic cages that can house and manipulate cells to allow sorting (26). Fluorescently labeled cells can be visualized during the entire process to ensure 100% purity and absolute assurance of cell capture. Because this method is gentle on the cell, single cells can then be further characterized by methods including NGS and RT-PCR. Sequencing a single CTC allows unique genetic profiles to be revealed that are masked by bulk analysis. A few previous studies have been conducted using the DEPArray to isolate single cells in order to perform single cell analysis by sequencing (27-30). Only a single paper has been previously published describing NGS on single tumor cells isolated by DEPArray. Small cell lung cancer cells enriched by RosetteSep Human Circulating Epithelial Tumor Cell Cocktail and isolated by DEPArray were analyzed by NGS whole genome sequencing (31). Here we describe NGS sequencing of DEPArray isolated single cells of both epithelial and mesenchymal phenotype using a targeted cancer panel. Methods & Materials Cell Culture Subtypes were chosen based on molecular classification, invasion potential using modified Boyden chamber assays, and expression of EMT markers (32, 33). The breast cancer cell lines SK- BR-3 and MDA-MB-231, purchased from ATCC were cultured in DMEM Medium (Lonza, Basal, Switzerland) containing 10 % fetal bovine serum (FBS) (Life Technologies, Carlsbad, CA) and 1 % mycozap (Lonza). All cells were cultured at 37 C and maintained under 5 % CO2 in polystyrene flasks until confluent (70-80%). Cells were released using a 0.25% trypsin/edta solution and collected via centrifugation. Blood Processing Blood samples from healthy donors (Biospecialty, USA) were collected into K3EDTA (BD Biosciences, USA) tubes and shipped within 24 hours. Breast cancer cell lines, a surrogate for CTCs, were spiked in before performing a 1-step RBC lysis and fixation (ebiosciences) step on 5 ml whole blood for 30 minutes at room temperature (RT). Next, the sample was centrifuged at 500 RCF for 5 minutes and washed with 50 ml 1xPBS. Before
4 the second centrifugation, a count of the cells was obtained using a Milipore Sceptor (Becton Dickinson, Franklin Lakes, NJ). Positive Enrichment and Negative Depletion For the negative depletion, each sample was resuspended in 80 µl automacs buffer (Miltenyi, Germany) for every 10 7 cells previously enumerated. Next, a cocktail of 20 µl of CD45 and 10 µl CD66 magnetic beads (Miltenyi) for every 10 7 cells was mixed with each sample. The sample was then incubated for 30 minutes at 4 C. Next, 3 ml of automacs buffer was added to the sample, which was centrifuged at 200 RCF for 10 minutes. The sample was resuspended in 1 ml automacs buffer and passed over a LS column (Miltenyi) primed with 3 ml automacs buffer contained in a high gradient magnetic field. Three wash steps with 3 ml of automacs buffer were performed to allow the CTCs to pass through the flow through representing the positive fraction. Finally, the column was removed from the high gradient magnetic field and 5 ml automacs buffer was added to plunge the remaining depleted leukocytes (negative fraction). For positive enrichment, the same method was performed, however 300 µl automacs buffer and 100 µl EpCAM magnetic beads (Miltenyi) were added to the sample. For the positive enrichment, the flow through containing leukocytes represents the negative fraction and the plunged fraction containing CTCs represents the positive fraction. Cell Imaging For quantification experiments, cells were incubated for fifteen minutes with 2 mm calcein AM (Life Technologies, USA) in 1xPBS at 37 C. The cells were centrifuged and washed once in 1xPBS, then counted using a hemocytometer. 20 µl of a 5,000 cells/ml dilution was added per 5 ml whole blood or to 80 µl 1xPBS in a 96 well plate, which served as a control to determine total cell count (n=8). Cells were captured and processed as described above. Positive and negative fractions were added to a 96 well plate and imaged using a Nikon Eclipse fluorescent microscope. Cells were enumerated using ImageJ and divided by the average total cell count to determine percent capture efficiency. Cell Staining For cell staining heterogeneity studies, cells were stained at a concentration of 5x10^5 cells/100 µl. Cells were stained with Hoechst (Life Technologies) at a concentration of 1 ug/ul, CD44 antibody conjugated with APC (1:100, Biolegend ), EpCAM antibody conjugated with PE (1:100, Biolegend ) and HER2 antibody conjugated with FITC (1:100, Biolegend ) for 15 minutes at 4 C. Cells were then fixed with 1% paraformaldehyde for 5 minutes at room temperature. To stain cells spiked into whole blood that have undergone negative depletion, 100 µl automacs buffer containing 1 % Tween-20 (Sigma) was added to the sample, which were then stained overnight at 4 C with Hoechst (Life Tech) at a concentration of 1 ug/ul, cytokeratin conjugated to FITC (1:25, Miltenyi), EpCAM conjugated to PE (1:25, Miltenyi) and CD45 conjugated to APC (1:25, Biolegend). DEPArray single cell sorting Cell sorting by the DEPArray was carried out by manufacturer s instructions. Briefly, 14 µl (13,000-40,000 cells) of sample was injected onto the DEPArray cartridge. Next, 830 µl of SB115 buffer (Silicon Biosystems, Bologna, Italy) was also injected onto the cartridge for later cell recovery. Following cartridge loading, 9.26 µl was automatically injected by the system onto the cartridge microchamber where non-uniform electric gradient was applied causing cells to become trapped in individual dielectrophoretic cages. Images were taken covering the entire surface area of the microchamber using 4 different fluorescent channels including PE, FITC, APC, and DAPI/Hoechst as well as brightfield images. Cells were automatically detected by the
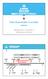
5 Figure 1. CTC-isoTECH platform description. Stage 1 includes whole blood processing, a negative immunomagnetic depletion of unwanted white blood cells followed by staining of the enriched cells. Stage 2 consists of single cell sorting by the DEPArray and, lastly, Stage 3 provides for molecular analysis of the single cells including gene expression analysis and next generation sequencing. DEPArray based on Hoechst intensity. Captured images of the cells were visually confirmed by the user and cells of interest were moved to the parking chamber. Once in the parking chamber, cells were visually confirmed and then moved individually to the sorting chamber and sorted into individual 0.2 ml microfuge tubes. 100 µl of buffer was added and cells were centrifuged to remove supernatant. Single Cell Transcriptional Analysis Transcriptional analysis was performed using the Single Cell-to-Ct kit (Ambion) per the manufacturer s instructions on single cells isolated with the above described method. Briefly, 10 µl of cell lysis buffer was added to each single cell sorted by the DEPArray and allowed to incubate for 5 minutes at room temperature followed by the addition of 1 µl of stop solution. 4.5 µl of reverse transcription mix was added followed by reverse transcription reaction and
6 Figure 2. Negative depletion followed by positive identification (EpCAM, HER2, CD44) of breast cancer cell lines (a) SKBR (epithelial) and (b) MDA (mesenchymal) spiked into whole blood. % Positive and % Negative refer to the the positive (expected) fraction and the negative (unexpected) fraction. cycles of targeted pre-amplification was performed by adding Pre-Amp mix and diluted TaqMan Gene Expression Assays. Finally, preamplified cdna products were subjected to realtime PCR for 4 marker genes and 4 housekeeping genes using an ABI The genes included were the epithelial marker EpCAM, the stem cell marker CD44, the marker for proliferation HER2, and the adhesion marker CDH1 as well as the reference genes ACTB, GAPDH, RPL13A, and RPL37A. Florigenic PCR reactions were performed at 40 cycles using TaqMan Gene Expression Assays and TaqMan Universal PCR Master Mix. A minus RT sample was included for each gene as a negative control. Because only single cells were used, all Ct values less than or equal to 35 were considered positive, while those that were greater than 35 were considered negative (34, 35). Due to the large cell-to-cell variation of any one gene when examining single cells, no normalization was used when comparing Ct values as a qualitative comparison (36). Single Cell Whole Genome Amplification Prior to sequencing, whole genome amplification of single cells was conducted by cell lysis, Mse I restriction digest, adaptor ligation and PCR amplification using the Ampli1 whole genome amplification (WGA) kit (Silicon Biosystems) per the manufacturer s instructions. Briefly, 2 µl of lysis mix was added to single cells sorted by the DEPArray and incubated for 15 hours. The DNA is then digested by adding 2 µl of digestion mix and incubated for 3 hours followed by addition of 5 µl of a ligation mix and an overnight ligation reaction. The primary PCR was carried out by adding 40 µl of the primary PCR mix and a 44 cycle PCR reaction (Eppendorf). A quality control end point PCR was carried out (Ampli1 QC kit, Silicon Biosystems) for 4 amplicons of 91, , 299, and 614 base pairs. Samples that had at least 3 out of 4 amplicons present were considered successfully amplified and were used for sequencing. Single Cell Sequencing Genomic DNA was extracted from SKBR and MDA cells using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer s directions. Bulk genomic and single cell WGA DNA from SKBR and MDA cells were then quantified using the GeneRead DNA QuantiMIZE kit (Qiagen) according to manufacturer s directions. Whole exonic regions of 24 genes were amplified from
7 Figure 3. DEPArray images of single breast cancer cells. (a) DEPArray images of SKBR-3 cells that were spiked into whole blood followed by negative depletion. The remaining cells were stained with EpCAM, HER2, CD45, and Hoechst combined and analyzed by DEPArray. (b) SKBR and MDA-231 cells were combined, stained with EpCAM, HER2, CD44, and Hoechst in a single tube, and analyzed by DEPArray. (c) A single breast cell line, SKBR-3 was stained with EpCAM, HER2, CD44, and Hoechst and analyzed by DEPArray. SKBR- 3 cells typically express high levels of EpCAM and HER2 and low or no expression of CD44 (stem cell marker). ng of DNA per sample that was split into 4 pools, which then underwent multiplex PCR using the GeneRead DNAseq Clinically Relevant Tumor Panel (Qiagen, Version 2). Next, PCR amplicons of each sample were pooled and purified using the Agencourt AMPure XP Beads (Beckman Coulter, Brea, CA, USA). To construct a barcoded Illumina DNA library, the GeneRead DNA Library I Core Kit (Qiagen) and GeneRead Adapter I Set A 12-plex (Qiagen) kits were used according to manufacturer s directions. A size selection of the fragments was then performed using the Agencourt AMPure XP Beads. A final PCR was performed to amplify adapter-carrying fragments
8 Table 1. Breast cancer subtypes used to model patient CTC heterogeneity Breast Cancer Cell Line Molecular classification HER2 EpCAM CD44 SK-BR-3 (epithelial) Luminal/HER2 + +/- MDA-MB-231 (non-epithelial) Basal B + and amplicons, which was again purified using the Agencourt AMPure XP Beads. Finally, the concentration of the amplicon library was determined by the GeneRead DNA seq Library Quant Assay (Qiagen) according to the manufacturer s protocol by qpcr. To ensure quality of the correct amplicon size it was analyzed using the QIAxcel DNA high resolution cartridge in a QIAxcel, a gel capillary electrophoresis instrument. Afterwards, samples were diluted to a final concentration of 2 nm and pooled according to manufacturer s instructions (Illumina) before being added to a MiSeq flow cell v2 (Illumina) to undergo sequencing on a MiSeq instrument, which generated fastq files. NGS Analysis Paired de-multiplexed fastq files from a MiSeq Illumina sequencer using Qiagen s Clinically Relevant tumor panel were imported into the CLC Biomedical Workbench (CLC Bio, version 2.5.1). Adapter sequences and bases with low quality scores (less than 20) were trimmed and reads were mapped to the reference genome, HG19. Variants were detected with the CLC variant caller using a threshold of 10% allele frequency and 450 minimum coverage. Results: This paper describes the CTC-isoTECH platform, which is an integrated method that starts with the enrichment of heterogeneous CTCs from whole blood followed by their identification, isolation, and downstream characterization (Figure 1). To enrich CTCs from whole blood, this method employs a negative depletion technique of CTCs from whole blood by removing CD45 and CD66b expressing cells from whole blood. When performing this negative depletion method with 50 SKBR-3 cells spiked into 5 ml of whole blood, we achieved an average of 84.2±8.2% recovery of our SKBR-3 cells in the positive (expected) fraction (Figure 2a). For the more mesenchymal cell line MDA-MB-231, we achieved an average recovery of 88.9±5.4% in the positive (expected) fraction (Figure 2b). This method resulted in a 402 fold depletion of leukocytes by reducing an average of approximately 13 million leukocytes prior to depletion down to 32,000 cells after depletion. This stringent depletion allows the cells to be sorted into single cell populations by the DEPArray which has a cartridge limitation of less than 80,000 cells. To demonstrate our ability to distinguish cancer cells from peripheral blood monocyte cells (PBMCs), we performed an integrated experiment by spiking SKBR cells into whole blood, followed by negative depletion of CD45 and CD66b expressing cells and DEPArray analysis. We were able to distinguish SKBR-3 (EpCAM+, HER+, CD45- ) cells from PBMCs (CD45+) (Figure 3a). To demonstrate our ability to identify and sort single heterogeneous cancer cells, a mixture of SKBR
9 Table 2. Single cell gene expression. CD44 CDH1 EPCAM ERBB2 GAPDH RPL13A RPL37A ACTB SKBR SKBR SKBR SKBR 4 + SKBR MDA MDA MDA MDA MDA Pos Con Neg Con RT-PCR was conducted on single cells sorted by the DEPArray. The top section examines single SKBR cells while the lower section examines single MDA cells. Expression is given as positive (+) or negative, with positive expression being any Ct value less than 35. (epithelial phenotype) and MDA-MB-231 (basal/mesenchymal phenotype) cells were stained with EpCAM, HER2, CD44, and Hoechst and analyzed by DEPArray (Figure 3b). Two distinct phenotypes were observed. The putative SKBR-3 cells were found to be EpCAM and HER2 positive while the putative MDA-MB-231 cells were found to be singly positive for the stem cell marker, CD44. In an experiment to examine heterogeneity on the single cell level within a single cell line, we stained SKBR-3 cells and analyzed single cell expression by DEPArray (Figure 3c). The majority of cells were found to express the expected phenotype of EpCAM +, HER2 +, and CD44 -. Two subsets of cells with unexpected phenotypes were identified; 27% of cells expressed low or no EpCAM with high HER2 expression and 0.5% of cells expressed only the stem cell marker, CD44. This demonstrates the ability of the DEPArray to detect heterogeneous and rare cell populations. Following DEPArray sorting, SKBR-3 (epithelial phenotype) and MDA-MB-231 (mesenchymal phenotype) cells were further characterized by single cell real-time quantitative PCR (RT-PCR). Four target genes including EpCAM, ERBB2, CD44, and CDH1 and four housekeeping genes including GAPDH, ACTB, RPL13A and RPL37A were interrogated in 5 single SKBR cells (Table 2). Using a CT of 35 to discriminate between positivity and negativity, 3/5 cells were positive for at least one target gene and one housekeeping gene. Two cells (SKBR 1 and 4) were found to express no target genes and one housekeeping genes, ACTB or GAPDH. Expression varied in all target genes, including EpCAM, ERBB2, CD44, and CDH1. Less
10 Table 3. Single cell NGS examining a single (a) SKBR-3 cell replicates or single (b) MDA replicates. Column 1 refers to the chromosome, column 2 is the position on the reference genome, column 3 refers to the type of variant, column 4 is the variant allele, column 5 is the gene name, and column 6 is the dbsnp ID, and columns 7-9 refer to the presence (X) or absence (blank) of the variants in each of 3 triplicates. Table 3a. SKBR-3 single cell replicates of NGS Chromosome Region Type Variant gene dbsnp ID SKBR Rep 1 SKBR Rep 2 SKBRRep SNV G PGDFRA X X X ^ Insertion A PGDFRA X X X SNV A BRAF X X X SNV T TP X X X SNV C TP X X X SNV G LY X X X SNV C DDR X X X SNV A DDR X X X SNV A PIK3CA X X X SNV A HIVEP1 X X X SNV T ERBB X X X Table 3b. MDA-MB-231 single cell replicates of NGS Chromosome Region Type Variant gene dbsnp ID MDA Rep 1 MDA Rep 2 MDA Rep Insertion T PIK3CA X X X Insertion TT PIK3CA X X X Deletion - PIK3CA X X X Deletion - PIK3CA X X X SNV T FIP1L X X X SNV G TP53 X X X Deletion - TP X X X SNV C ERBB2 X X X SNV T ERBB2 X X X MNV TA ERBB2 X X X heterogeneity was observed in the 5 single MDA- MB-231 cells that were examined (Table 2). All MDA-231 cells expressed CD44 and the housekeeping gene, ACTB. The only target gene with heterogeneity in expression was ERBB2. These experiments demonstrate our ability to characterize single cells by gene expression following DEPArray sorting to the single cell level
11 In addition to single cell RT-PCR, cells sorted by DEPArray were also further characterized using next generation sequencing (NGS) by Illumina s massively parallel sequencing technology. Following DEPArray sorting, single SKBR-3 and MDA-231 cells were subjected to whole genome amplification and library preparation for NGS. A targeted 24 gene panel was analyzed for each single cell and single nucleotide variants (SNVs) were reported for 3 replicates of each single cell. We found 100% concordance in both the single SKBR-3 cell and in the single MDA-231 cell (Table 3). This experiment demonstrates feasibility to perform NGS on single cells sorted by the DEPArray. Discussion: The majority of CTC capture methods are based on enrichment and/or identification of epithelial markers. Studies have shown that through the process of epithelial to mesenchymal transition (EMT) known to occur in cancer, epithelial markers are down-regulated in favor of mesenchymal markers that allow for increased motility and ability to evade immune detection (2). Therefore, using epithelial markers as the basis for enrichment and identification will not detect those cells that express only mesenchymal markers. To combat this limitation, we describe here a method of negative depletion following identification using both epithelial and mesenchymal targets. Circulating tumor cells have demonstrated the potential to be key players in metastasis and therefore in cancer morbidity (37). CTCs have been well associated with overall survival and progression free survival (4) and have great potential to serve as biomarkers and therapeutic monitors. Because they can be obtained serially in a non-invasive manner, the use of CTCs has been touted as a non invasive liquid biopsy. CTCs may possess potential druggable targets and studies have been conducted to enumerate CTCs before, during, and after treatment. Although there is great interest in studying these cells, technical challenges have not been fully addressed to allow for robust and reproducible methods to study these cells. The identification, capture, and characterization of single CTCs remains a challenge and much work is underway to address these limitations (38). Single cell analysis is growing in popularity for studying disease and has been recently reviewed (39). Research is growing on single cell sequencing (40-42), single cell transcriptomics, and single cell metabolomics. Because circulating tumor cells have been shown to be heterogeneous even from the same blood draw (43), it is crucial to be able to capture and characterize single CTCs. This will ultimately allow researchers to develop methods that will eradicate those cells from cancer patients and prevent metastasis. Our study demonstrates the ability to examine single, heterogeneous CTCs regardless of EMT status. Two heterogeneous subtypes that have varying expression of epithelial markers were sorted from a mixed population. We were able to identify extracellular protein phenotypes using mean fluorescent intensity and then sort the cells into single cell populations. These single cells were further characterized by gene expression and NGS. The ability to perform genetic analysis on single CTCs from patient samples will provide physicians with the most comprehensive information to determine the best course of treatment for each patient. The ability to study single, heterogeneous CTCs will also allow researchers to discover new drug targets and methods of preventing metastasis from occurring. Because it is likely that only a small subset of a patient s CTCs are capable of causing metastases, it is crucial to characterize a comprehensive collection of a patient s CTCs including epithelial and non-epithelial subtypes as we have demonstrated here
12 Abbreviations: CK, cytokeratins; CTCs, circulating tumor cells; EpCAM, epithelial cell adhesion molecule; EMT, epithelial to mesenchymal transition; FDA, Food and Drug Administration; NGS, next-generation sequencing; PBMC, peripheral blood mononuclear cells; SNVs, single nucleotide variants Funding Sources: NCI Contract #HHSN C References 1. Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer Jun;3(6): DOI: /nrc Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity Aug;21(2): DOI: /j.immuni Millner LM, Linder MW, Valdes R, Jr. Circulating tumor cells: a review of present methods and the need to identify heterogeneous phenotypes. Ann Clin Lab Sci Summer;43(3): Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med Aug 19;351(8): DOI: /NEJMoa Gazzaniga P, Raimondi C, Gradilone A, Biondi Zoccai G, Nicolazzo C, Gandini O, et al. Circulating tumor cells in metastatic colorectal cancer: do we need an alternative cutoff? J Cancer Res Clin Oncol Aug;139(8): DOI: /s Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res Feb 1;13(3): DOI: / CCR Mego M, De Giorgi U, Dawood S, Wang X, Valero V, Andreopoulou E, et al. Characterization of metastatic breast cancer patients with nondetectable circulating tumor cells. Int J Cancer Jul 15;129(2): DOI: /ijc Schneck H, Gierke B, Uppenkamp F, Behrens B, Niederacher D, Stoecklein NH, et al. EpCAM-Independent Enrichment of Circulating Tumor Cells in Metastatic Breast Cancer. PLoS One. 2015;10(12):e DOI: /journal.pone Negin BP, Cohen SJ. Circulating tumor cells in colorectal cancer: past, present, and future challenges. Curr Treat Options Oncol Jun;11(1-2):1-13. DOI: /s Resel Folkersma L, Olivier Gomez C, San Jose Manso L, Veganzones de Castro S, Galante Romo I, Vidaurreta Lazaro M, et al. Immunomagnetic quantification of circulating tumoral cells in patients with prostate cancer: clinical and pathological correlation. Arch Esp Urol Jan-Feb;63(1): Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest Jun;119(6): DOI: /JCI
13 12. Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest Dec;112(12): DOI: /JCI Roxanis I. Occurrence and significance of epithelial-mesenchymal transition in breast cancer. J Clin Pathol Jun;66(6): DOI: /jclinpath Drasin DJ, Robin TP, Ford HL. Breast cancer epithelial-to-mesenchymal transition: examining the functional consequences of plasticity. Breast Cancer Res. 2011;13(6):226. DOI: /bcr Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27: DOI: /annurev-cellbio Takebe N, Warren RQ, Ivy SP. Breast cancer growth and metastasis: interplay between cancer stem cells, embryonic signaling pathways and epithelial-to-mesenchymal transition. Breast Cancer Res. 2011;13(3):211. DOI: /bcr Krasnapolski MA, Todaro LB, de Kier Joffe EB. Is the epithelial-to-mesenchymal transition clinically relevant for the cancer patient? Curr Pharm Biotechnol Nov;12(11): Kallergi G, Agelaki S, Papadaki MA, Nasias D, Matikas A, Mavroudis D, et al. Expression of truncated human epidermal growth factor receptor 2 on circulating tumor cells of breast cancer patients. Breast Cancer Res. 2015;17:113. DOI: /s x. 19. Fehm T, Muller V, Aktas B, Janni W, Schneeweiss A, Stickeler E, et al. HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial. Breast Cancer Res Treat Nov;124(2): DOI: /s x. 20. Wulfing P, Borchard J, Buerger H, Heidl S, Zanker KS, Kiesel L, et al. HER2-positive circulating tumor cells indicate poor clinical outcome in stage I to III breast cancer patients. Clin Cancer Res Mar 15;12(6): DOI: / CCR Ignatiadis M, Kallergi G, Ntoulia M, Perraki M, Apostolaki S, Kafousi M, et al. Prognostic value of the molecular detection of circulating tumor cells using a multimarker reverse transcription-pcr assay for cytokeratin 19, mammaglobin A, and HER2 in early breast cancer. Clin Cancer Res May 1;14(9): DOI: / CCR Ascolani G, Occhipinti A, Lio P. Modelling circulating tumour cells for personalised survival prediction in metastatic breast cancer. PLoS Comput Biol May;11(5):e DOI: /journal.pcbi Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med Apr 10;5(180):180ra48. DOI: /scitranslmed Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol Jun;31(6): DOI: /nbt Nadal R, Fernandez A, Sanchez-Rovira P, Salido M, Rodriguez M, Garcia-Puche JL, et al. Biomarkers characterization of circulating tumour cells in breast cancer patients. Breast Cancer Res. 2012;14(3):R71. DOI: /bcr Fuchs AB, Romani A, Freida D, Medoro G, Abonnenc M, Altomare L, et al. Electronic sorting and recovery of single live cells from microlitre sized samples. Lab Chip Jan;6(1): DOI: /b505884h. 27. Fabbri F, Carloni S, Zoli W, Ulivi P, Gallerani G, Fici P, et al. Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRAS mutation status in pure CTCs. Cancer Lett Jul 10;335(1): DOI: /j.canlet Carpenter EL, Rader J, Ruden J, Rappaport EF, Hunter KN, Hallberg PL, et al. Dielectrophoretic capture and genetic analysis of single neuroblastoma tumor cells. Front Oncol. 2014;4:201. DOI: /fonc Pestrin M, Salvianti F, Galardi F, De Luca F, Turner N, Malorni L, et al. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol Oncol Apr;9(4): DOI: /j.molonc
14 30. Polzer B, Medoro G, Pasch S, Fontana F, Zorzino L, Pestka A, et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol Med Nov;6(11): DOI: /emmm Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med Aug;20(8): DOI: /nm Nagaraja GM, Othman M, Fox BP, Alsaber R, Pellegrino CM, Zeng Y, et al. Gene expression signatures and biomarkers of noninvasive and invasive breast cancer cells: comprehensive profiles by representational difference analysis, microarrays and proteomics. Oncogene Apr 13;25(16): DOI: /sj.onc Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell Dec;10(6): DOI: /j.ccr Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol Dec;29(12): DOI: /nbt Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell Apr 20;18(4): DOI: /j.devcel Livak KJ, Wills QF, Tipping AJ, Datta K, Mittal R, Goldson AJ, et al. Methods for qpcr gene expression profiling applied to 1440 lymphoblastoid single cells. Methods Jan;59(1):71-9. DOI: /j.ymeth Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nature reviews Cancer Jun;3(6): DOI: /nrc Millner LM, Linder, M. W., Valdes, R. Jr.,. Advances in Cell Separation Technologies: Isolation of Circulating Tumor Cells. Association of Clinical Scientists, Mobile, Al. May 23-27, Eberwine J, Sul JY, Bartfai T, Kim J. The promise of single-cell sequencing. Nat Methods Jan;11(1): Ren SC, Qu M, Sun YH. Investigating intratumour heterogeneity by single-cell sequencing. Asian J Androl Nov;15(6): DOI: /aja Lasken RS. Single-cell sequencing in its prime. Nat Biotechnol Mar;31(3): DOI: /nbt The biology of genomes. Single-cell sequencing tackles basic and biomedical questions. Science May 25;336(6084): DOI: /science Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7(5):e DOI: /journal.pone
Cover Letter. Reviewer 1:
 Cover Letter Michael Yang, M.D., Ph.D. Managing Editor of Cancer Research Frontiers 1188 Willis Ave, #109, Albertson, NY 11507, USA Phone: +1-917-426-1571 http://cancer-research-frontiers.org/ Dear Dr.
Cover Letter Michael Yang, M.D., Ph.D. Managing Editor of Cancer Research Frontiers 1188 Willis Ave, #109, Albertson, NY 11507, USA Phone: +1-917-426-1571 http://cancer-research-frontiers.org/ Dear Dr.
A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis
 APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
Fluxion Biosciences and Swift Biosciences Somatic variant detection from liquid biopsy samples using targeted NGS
 APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
A Novel CTC-Detecting Technique Using TelomeScan and Its Clinical Applications
 A Novel CTC-Detecting Technique Using TelomeScan and Its Clinical Applications Yasuo Urata CEO and President Oncolys BioPharma Inc. February 16, 2013 Telomere Length is a Limiting Factor for Cell Replication
A Novel CTC-Detecting Technique Using TelomeScan and Its Clinical Applications Yasuo Urata CEO and President Oncolys BioPharma Inc. February 16, 2013 Telomere Length is a Limiting Factor for Cell Replication
NGS in tissue and liquid biopsy
 NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
Performance Characteristics BRCA MASTR Plus Dx
 Performance Characteristics BRCA MASTR Plus Dx with drmid Dx for Illumina NGS systems Manufacturer Multiplicom N.V. Galileïlaan 18 2845 Niel Belgium Table of Contents 1. Workflow... 4 2. Performance Characteristics
Performance Characteristics BRCA MASTR Plus Dx with drmid Dx for Illumina NGS systems Manufacturer Multiplicom N.V. Galileïlaan 18 2845 Niel Belgium Table of Contents 1. Workflow... 4 2. Performance Characteristics
The Presence and Persistence of Resistant and Stem Cell- Like Tumor Cells as a Major Problem in Ovarian Cancer
 Welcome! The Presence and Persistence of Resistant and Stem Cell- Like Tumor Cells as a Major Problem in Ovarian Cancer Prof. Sabine Kasimir-Bauer Department of Gynecology and Obstetrics University Hospital
Welcome! The Presence and Persistence of Resistant and Stem Cell- Like Tumor Cells as a Major Problem in Ovarian Cancer Prof. Sabine Kasimir-Bauer Department of Gynecology and Obstetrics University Hospital
Disclosure. Summary. Circulating DNA and NGS technology 3/27/2017. Disclosure of Relevant Financial Relationships. JS Reis-Filho, MD, PhD, FRCPath
 Circulating DNA and NGS technology JS Reis-Filho, MD, PhD, FRCPath Director of Experimental Pathology, Department of Pathology Affiliate Member, Human Oncology and Pathogenesis Program Disclosure of Relevant
Circulating DNA and NGS technology JS Reis-Filho, MD, PhD, FRCPath Director of Experimental Pathology, Department of Pathology Affiliate Member, Human Oncology and Pathogenesis Program Disclosure of Relevant
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester
 Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
CTC in clinical studies: Latest reports on GI cancers
 CTC in clinical studies: Latest reports on GI cancers François-Clément Bidard, MD PhD GI cancers are characterized by Multimodal treatment strategies Treatments are adapted to tumor burden & prognosis
CTC in clinical studies: Latest reports on GI cancers François-Clément Bidard, MD PhD GI cancers are characterized by Multimodal treatment strategies Treatments are adapted to tumor burden & prognosis
Exosome DNA Extraction Kits
 Exosome DNA Extraction Kits Summary Section 5 Introduction 40 EXO-DNAc 41 EXO-DNA 43 Introduction Genomic DNA Extractiom Kits Ordering informations Products can be purchased directly in our on-line shop:
Exosome DNA Extraction Kits Summary Section 5 Introduction 40 EXO-DNAc 41 EXO-DNA 43 Introduction Genomic DNA Extractiom Kits Ordering informations Products can be purchased directly in our on-line shop:
Challenges for use of CTCs as a Diagnostic. Farideh Z. Bischoff, Ph.D. Interim CSO Sr. Director, Translational Clinical Development Biocept, Inc.
 Challenges for use of CTCs as a Diagnostic Farideh Z. ischoff, Ph.D. Interim CSO Sr. Director, Translational Clinical Development iocept, Inc. Current Technology for CTC Testing Existing CTC testing platform
Challenges for use of CTCs as a Diagnostic Farideh Z. ischoff, Ph.D. Interim CSO Sr. Director, Translational Clinical Development iocept, Inc. Current Technology for CTC Testing Existing CTC testing platform
High Sensitivity Immunomagnetic CTC Isolation as Compared to Alternative Isolation Methods
 High Sensitivity Immunomagnetic CTC Isolation as Compared to Alternative Isolation Methods 1. Introduction: An overview of CTC isolation methods 2. Challenges for direct comparisons of CTC recovery 3.
High Sensitivity Immunomagnetic CTC Isolation as Compared to Alternative Isolation Methods 1. Introduction: An overview of CTC isolation methods 2. Challenges for direct comparisons of CTC recovery 3.
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
AVENIO ctdna Analysis Kits The complete NGS liquid biopsy solution EMPOWER YOUR LAB
 Analysis Kits The complete NGS liquid biopsy solution EMPOWER YOUR LAB Analysis Kits Next-generation performance in liquid biopsies 2 Accelerating clinical research From liquid biopsy to next-generation
Analysis Kits The complete NGS liquid biopsy solution EMPOWER YOUR LAB Analysis Kits Next-generation performance in liquid biopsies 2 Accelerating clinical research From liquid biopsy to next-generation
Detecting Oncogenic Mutations in Whole Blood
 WHITE PAPER Detecting Oncogenic Mutations in Whole Blood Analytical validation of Cynvenio Biosystems LiquidBiopsy circulating tumor cell (CTC) capture and next-generation sequencing (NGS) September 2013
WHITE PAPER Detecting Oncogenic Mutations in Whole Blood Analytical validation of Cynvenio Biosystems LiquidBiopsy circulating tumor cell (CTC) capture and next-generation sequencing (NGS) September 2013
Supplementary Online Content
 Supplementary Online Content Fumagalli D, Venet D, Ignatiadis M, et al. RNA Sequencing to predict response to neoadjuvant anti-her2 therapy: a secondary analysis of the NeoALTTO randomized clinical trial.
Supplementary Online Content Fumagalli D, Venet D, Ignatiadis M, et al. RNA Sequencing to predict response to neoadjuvant anti-her2 therapy: a secondary analysis of the NeoALTTO randomized clinical trial.
Characterisation of structural variation in breast. cancer genomes using paired-end sequencing on. the Illumina Genome Analyser
 Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Circulating Tumor Cells (CTC) Technologies
 Table of Contents MIR036 I. SCOPE AND METHODOLOGY Scope of the Study Analytics and data presented in this report pertain to several parameters such as - Research Methodology This report is uniquely researched
Table of Contents MIR036 I. SCOPE AND METHODOLOGY Scope of the Study Analytics and data presented in this report pertain to several parameters such as - Research Methodology This report is uniquely researched
Cellecta Overview. Started Operations in 2007 Headquarters: Mountain View, CA
 Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
PG-Seq NGS Kit for Preimplantation Genetic Screening
 Application Note: PG-Seq Validation Study PG-Seq NGS Kit for Preimplantation Genetic Screening Validation using Multi (5-10) Cells and Single Cells from euploid and aneuploid cell lines Introduction Advances
Application Note: PG-Seq Validation Study PG-Seq NGS Kit for Preimplantation Genetic Screening Validation using Multi (5-10) Cells and Single Cells from euploid and aneuploid cell lines Introduction Advances
Medical Coverage Policy Circulating Tumor DNA and. Circulating Tumor Cells for Cancer Management (Liquid Biopsy)
 Medical Coverage Policy Circulating Tumor DNA and Circulating Tumor Cells for Cancer Management (Liquid Biopsy) EFFECTIVE DATE: 12 01 2016 POLICY LAST UPDATED: 07 17 2018 OVERVIEW Circulating tumor DNA
Medical Coverage Policy Circulating Tumor DNA and Circulating Tumor Cells for Cancer Management (Liquid Biopsy) EFFECTIVE DATE: 12 01 2016 POLICY LAST UPDATED: 07 17 2018 OVERVIEW Circulating tumor DNA
Extracellular Vesicle RNA isolation Kits
 Extracellular Vesicle RNA isolation Kits Summary Section 4 Introduction 42 Exo-TotalRNA and TumorExo-TotalRNA isolation kits 43 Extracellular Vesicle RNA extraction kits Ordering information Products can
Extracellular Vesicle RNA isolation Kits Summary Section 4 Introduction 42 Exo-TotalRNA and TumorExo-TotalRNA isolation kits 43 Extracellular Vesicle RNA extraction kits Ordering information Products can
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Finite-element analysis of cell cluster dynamics in different cluster trap architectures. (a) Cluster-Chip (b) Filter (c) A structure identical to the Cluster-Chip except that one
Supplementary Figure 1 Finite-element analysis of cell cluster dynamics in different cluster trap architectures. (a) Cluster-Chip (b) Filter (c) A structure identical to the Cluster-Chip except that one
Abstract. Optimization strategy of Copy Number Variant calling using Multiplicom solutions APPLICATION NOTE. Introduction
 Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples
 DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
Circulating tumor cells as biomarker for hormonal treatment in breast and prostate cancer. Michal Mego
 National Cancer Institute, Slovakia Translational Research Unit Circulating tumor cells as biomarker for hormonal treatment in breast and prostate cancer Michal Mego 2 nd Department of Oncology, Faculty
National Cancer Institute, Slovakia Translational Research Unit Circulating tumor cells as biomarker for hormonal treatment in breast and prostate cancer Michal Mego 2 nd Department of Oncology, Faculty
EXO-DNAc Circulating and EV-associated DNA extraction kit
 Datasheet EXO-DNAc Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
Datasheet EXO-DNAc Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits
 AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
AdnaNews. News from the meeting on Advances in Circulating Tumor Cells (ACTC), Reythymnon, Crete, Greece October 8-11, 2014.
 News from the meeting on Advances in Circulating Tumor Cells (ACTC), Reythymnon, Crete, Greece October 8-11, 2014. The ACTC meeting (www.actc2014.org) is together with the ISMRC meeting one of the most
News from the meeting on Advances in Circulating Tumor Cells (ACTC), Reythymnon, Crete, Greece October 8-11, 2014. The ACTC meeting (www.actc2014.org) is together with the ISMRC meeting one of the most
Primary Adult Naïve CD4+ CD45RA+ Cells. Prepared by: David Randolph at University of Alabama, Birmingham
 Primary Adult Naïve CD4+ CD45RA+ Cells Prepared by: David Randolph (drdrdr@uab.edu) at University of Alabama, Birmingham Goal: To obtain large numbers of highly pure primary CD4+ CD45RO- CD25- cells from
Primary Adult Naïve CD4+ CD45RA+ Cells Prepared by: David Randolph (drdrdr@uab.edu) at University of Alabama, Birmingham Goal: To obtain large numbers of highly pure primary CD4+ CD45RO- CD25- cells from
La biopsia liquida. Aldo Scarpa. Anatomia Patologica e ARC-NET Centro di Ricerca Applicata sul Cancro
 La biopsia liquida Aldo Scarpa Anatomia Patologica e ARC-NET Centro di Ricerca Applicata sul Cancro Azienda Ospedaliera Universitaria Integrata di Verona Obstacles to precision oncology Genomic heterogeneity
La biopsia liquida Aldo Scarpa Anatomia Patologica e ARC-NET Centro di Ricerca Applicata sul Cancro Azienda Ospedaliera Universitaria Integrata di Verona Obstacles to precision oncology Genomic heterogeneity
DNA Methylation of Tumor Suppressor and Metastasis Suppressor Genes in Circulating Tumor Cells and corresponding Circulating Tumor DNA
 DNA Methylation of Tumor Suppressor and Metastasis Suppressor Genes in Circulating Tumor Cells and corresponding Circulating Tumor DNA Maria Chimonidou 1, Areti Strati 1, Nikos Malamos 2, Vasilis Georgoulias
DNA Methylation of Tumor Suppressor and Metastasis Suppressor Genes in Circulating Tumor Cells and corresponding Circulating Tumor DNA Maria Chimonidou 1, Areti Strati 1, Nikos Malamos 2, Vasilis Georgoulias
Integrated platform for liquid biopsy-based personalized cancer medicine
 Integrated platform for liquid biopsy-based personalized cancer medicine Dr. Bernhard Polzer Fraunhofer ITEM-Regensburg Personalized Tumor Therapy Personalized cancer therapy Primary tumor single tumor
Integrated platform for liquid biopsy-based personalized cancer medicine Dr. Bernhard Polzer Fraunhofer ITEM-Regensburg Personalized Tumor Therapy Personalized cancer therapy Primary tumor single tumor
QIAGEN Complete Solutions for Liquid Biopsy Molecular Testing
 QIAGEN Complete Solutions for Liquid Biopsy Molecular Testing Christopher Swagell, PhD Market Development Manager, Advanced Molecular Pathology QIAGEN 1 Agenda QIAGEN Solid Tumor Testing and Liquid Biopsy
QIAGEN Complete Solutions for Liquid Biopsy Molecular Testing Christopher Swagell, PhD Market Development Manager, Advanced Molecular Pathology QIAGEN 1 Agenda QIAGEN Solid Tumor Testing and Liquid Biopsy
Molecular profiling of single circulating tumor cells with diagnostic intention
 Research Article Molecular profiling of single circulating tumor cells with diagnostic intention Bernhard Polzer 1,, Gianni Medoro 2,, Sophie Pasch 3, Francesca Fontana 2, Laura Zorzino 4, Aurelia Pestka
Research Article Molecular profiling of single circulating tumor cells with diagnostic intention Bernhard Polzer 1,, Gianni Medoro 2,, Sophie Pasch 3, Francesca Fontana 2, Laura Zorzino 4, Aurelia Pestka
Survey Results Q1. How would you best describe your organization?
 Survey Results Q1. How would you best describe your organization? Q2. How high would you rate the priority of Circulating Tumor Cells in you organization? Q3. What do you think is the biggest challenge
Survey Results Q1. How would you best describe your organization? Q2. How high would you rate the priority of Circulating Tumor Cells in you organization? Q3. What do you think is the biggest challenge
The Avatar System TM Yields Biologically Relevant Results
 Application Note The Avatar System TM Yields Biologically Relevant Results Liquid biopsies stand to revolutionize the cancer field, enabling early detection and noninvasive monitoring of tumors. In the
Application Note The Avatar System TM Yields Biologically Relevant Results Liquid biopsies stand to revolutionize the cancer field, enabling early detection and noninvasive monitoring of tumors. In the
Profiles of gene expression & diagnosis/prognosis of cancer. MCs in Advanced Genetics Ainoa Planas Riverola
 Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, MD
 AD Award Number: W81XWH-11-1-0126 TITLE: Chemical strategy to translate genetic/epigenetic mechanisms to breast cancer therapeutics PRINCIPAL INVESTIGATOR: Xiang-Dong Fu, PhD CONTRACTING ORGANIZATION:
AD Award Number: W81XWH-11-1-0126 TITLE: Chemical strategy to translate genetic/epigenetic mechanisms to breast cancer therapeutics PRINCIPAL INVESTIGATOR: Xiang-Dong Fu, PhD CONTRACTING ORGANIZATION:
EXO-DNA Circulating and EV-associated DNA extraction kit
 Datasheet EXO-DNA Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
Datasheet EXO-DNA Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
Cytogenetics 101: Clinical Research and Molecular Genetic Technologies
 Cytogenetics 101: Clinical Research and Molecular Genetic Technologies Topics for Today s Presentation 1 Classical vs Molecular Cytogenetics 2 What acgh? 3 What is FISH? 4 What is NGS? 5 How can these
Cytogenetics 101: Clinical Research and Molecular Genetic Technologies Topics for Today s Presentation 1 Classical vs Molecular Cytogenetics 2 What acgh? 3 What is FISH? 4 What is NGS? 5 How can these
Introduction of an NGS gene panel into the Haemato-Oncology MPN service
 Introduction of an NGS gene panel into the Haemato-Oncology MPN service Dr. Anna Skowronska, Dr Jane Bryon, Dr Samuel Clokie, Dr Yvonne Wallis and Professor Mike Griffiths West Midlands Regional Genetics
Introduction of an NGS gene panel into the Haemato-Oncology MPN service Dr. Anna Skowronska, Dr Jane Bryon, Dr Samuel Clokie, Dr Yvonne Wallis and Professor Mike Griffiths West Midlands Regional Genetics
Products for cfdna and mirna isolation. Subhead Circulating Cover nucleic acids from plasma
 MACHEREY-NAGEL Products for cfdna and mirna isolation Bioanalysis Subhead Circulating Cover nucleic acids from plasma n Flexible solutions for small and large blood plasma volumes n Highly efficient recovery
MACHEREY-NAGEL Products for cfdna and mirna isolation Bioanalysis Subhead Circulating Cover nucleic acids from plasma n Flexible solutions for small and large blood plasma volumes n Highly efficient recovery
Focus Application. Compound-Induced Cytotoxicity
 xcelligence System Real-Time Cell Analyzer Focus Application Compound-Induced Cytotoxicity Featured Study: Using the Time Resolving Function of the xcelligence System to Optimize Endpoint Viability and
xcelligence System Real-Time Cell Analyzer Focus Application Compound-Induced Cytotoxicity Featured Study: Using the Time Resolving Function of the xcelligence System to Optimize Endpoint Viability and
CTC molecular characterization: Are we ready to move forward with clinical testing?
 CTC molecular characterization: Are we ready to move forward with clinical testing? Michail Ignatiadis MD, PhD Jules Bordet Institute, Université Libre de Bruxelles Brussels, Belgium Breast cancer: Diagnostics
CTC molecular characterization: Are we ready to move forward with clinical testing? Michail Ignatiadis MD, PhD Jules Bordet Institute, Université Libre de Bruxelles Brussels, Belgium Breast cancer: Diagnostics
For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format:
 Supplementary Protocol 1. Adaptor preparation: For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format: Per reaction X96 10X NEBuffer 2 10 µl 10 µl x 96 5 -GATC
Supplementary Protocol 1. Adaptor preparation: For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format: Per reaction X96 10X NEBuffer 2 10 µl 10 µl x 96 5 -GATC
Consensus statement between CM-Path, CRUK and the PHG Foundation following on from the Liquid Biopsy workshop on the 8th March 2018
 Consensus statement between CM-Path, CRUK and the PHG Foundation following on from the Liquid Biopsy workshop on the 8th March 2018 Summary: This document follows on from the findings of the CM-Path The
Consensus statement between CM-Path, CRUK and the PHG Foundation following on from the Liquid Biopsy workshop on the 8th March 2018 Summary: This document follows on from the findings of the CM-Path The
Liquid Biopsy: Implications for Cancer Staging & Therapy
 Prof. Klaus Pantel, MD, PhD Institut für Tumorbiologie Liquid Biopsy: Implications for Cancer Staging & Therapy Tumor cell dissemination and cancer dormancy Primary tumor Local relapse Cancer cells disseminate
Prof. Klaus Pantel, MD, PhD Institut für Tumorbiologie Liquid Biopsy: Implications for Cancer Staging & Therapy Tumor cell dissemination and cancer dormancy Primary tumor Local relapse Cancer cells disseminate
AbSeq on the BD Rhapsody system: Exploration of single-cell gene regulation by simultaneous digital mrna and protein quantification
 BD AbSeq on the BD Rhapsody system: Exploration of single-cell gene regulation by simultaneous digital mrna and protein quantification Overview of BD AbSeq antibody-oligonucleotide conjugates. High-throughput
BD AbSeq on the BD Rhapsody system: Exploration of single-cell gene regulation by simultaneous digital mrna and protein quantification Overview of BD AbSeq antibody-oligonucleotide conjugates. High-throughput
ExoQuick Exosome Isolation and RNA Purification Kits
 ExoQuick Exosome Isolation and RNA Purification Kits Cat # EQ806A-1, EQ806TC-1, EQ808A-1 User Manual Storage: Please see individual components Version 2 8/14/2018 A limited-use label license covers this
ExoQuick Exosome Isolation and RNA Purification Kits Cat # EQ806A-1, EQ806TC-1, EQ808A-1 User Manual Storage: Please see individual components Version 2 8/14/2018 A limited-use label license covers this
Next generation diagnostics Bringing high-throughput sequencing into clinical application
 Next generation diagnostics Bringing high-throughput sequencing into clinical application Leonardo A. Meza-Zepeda, PhD Translational Genomics Group Institute for Cancer Research Leonardo.Meza-Zepeda@rr-research.no
Next generation diagnostics Bringing high-throughput sequencing into clinical application Leonardo A. Meza-Zepeda, PhD Translational Genomics Group Institute for Cancer Research Leonardo.Meza-Zepeda@rr-research.no
EXAMPLE. - Potentially responsive to PI3K/mTOR and MEK combination therapy or mtor/mek and PKC combination therapy. ratio (%)
 Dr Kate Goodhealth Goodhealth Medical Clinic 123 Address Road SUBURBTOWN NSW 2000 Melanie Citizen Referring Doctor Your ref Address Dr John Medico 123 Main Street, SUBURBTOWN NSW 2000 Phone 02 9999 9999
Dr Kate Goodhealth Goodhealth Medical Clinic 123 Address Road SUBURBTOWN NSW 2000 Melanie Citizen Referring Doctor Your ref Address Dr John Medico 123 Main Street, SUBURBTOWN NSW 2000 Phone 02 9999 9999
Youngnam Cho. National Cancer Center Biomarker Branch
 Youngnam Cho National Cancer Center Biomarker Branch Contents 1. Liquid Biopsy 2. Circulating Tumor Cells from Blood 3. Cell-free DNA from Blood 1. Liquid biopsy Cancer Diagnosis IMAGING TISSUE BIOPSY
Youngnam Cho National Cancer Center Biomarker Branch Contents 1. Liquid Biopsy 2. Circulating Tumor Cells from Blood 3. Cell-free DNA from Blood 1. Liquid biopsy Cancer Diagnosis IMAGING TISSUE BIOPSY
SALSA MLPA probemix P315-B1 EGFR
 SALSA MLPA probemix P315-B1 EGFR Lot B1-0215 and B1-0112. As compared to the previous A1 version (lot 0208), two mutation-specific probes for the EGFR mutations L858R and T709M as well as one additional
SALSA MLPA probemix P315-B1 EGFR Lot B1-0215 and B1-0112. As compared to the previous A1 version (lot 0208), two mutation-specific probes for the EGFR mutations L858R and T709M as well as one additional
LIQUID BIOPSY
 www.idisantiago.es LIQUID BIOPSY Miguel Abal Investigador I3SNS Oncoloxía Médica Traslacional Instituto de Investigación Sanitaria de Santiago (IDIS) Complexo Hospitalario Universitario de Santiago/SERGAS
www.idisantiago.es LIQUID BIOPSY Miguel Abal Investigador I3SNS Oncoloxía Médica Traslacional Instituto de Investigación Sanitaria de Santiago (IDIS) Complexo Hospitalario Universitario de Santiago/SERGAS
Early dissemination in prostate cancer
 Early dissemination in prostate cancer Miodrag Guzvic, University of Regensburg, Germany Adjuvant Palliative M0 Initiation Diagnosis Surgery Metastasis Death Intervention window to delay or prevent metastasis
Early dissemination in prostate cancer Miodrag Guzvic, University of Regensburg, Germany Adjuvant Palliative M0 Initiation Diagnosis Surgery Metastasis Death Intervention window to delay or prevent metastasis
Focus Application. Compound-Induced Cytotoxicity
 xcelligence System Real-Time Cell Analyzer Focus Application Compound-Induced Cytotoxicity For life science research only. Not for use in diagnostic procedures. Featured Study: Using the Time Resolving
xcelligence System Real-Time Cell Analyzer Focus Application Compound-Induced Cytotoxicity For life science research only. Not for use in diagnostic procedures. Featured Study: Using the Time Resolving
Phosphate buffered saline (PBS) for washing the cells TE buffer (nuclease-free) ph 7.5 for use with the PrimePCR Reverse Transcription Control Assay
 Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Accel-Amplicon Panels
 Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
 MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research
 Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Lukas Bubendorf Pathologie. Liquid biopsies
 Lukas Bubendorf Pathologie Liquid biopsies Liquid biopsies 1. Circulating cell-free tumor-dna (ctdna) 2. Circulating tumor cells (CTC) Source: Sysmex CTCs ctdna ctrna exosomes Quantification Protein RNA
Lukas Bubendorf Pathologie Liquid biopsies Liquid biopsies 1. Circulating cell-free tumor-dna (ctdna) 2. Circulating tumor cells (CTC) Source: Sysmex CTCs ctdna ctrna exosomes Quantification Protein RNA
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer. Application Note
 Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Supplementary Data 1. Alanine substitutions and position variants of APNCYGNIPL. Applied in
 Supplementary Data 1. Alanine substitutions and position variants of APNCYGNIPL. Applied in Supplementary Fig. 2 Substitution Sequence Position variant Sequence original APNCYGNIPL original APNCYGNIPL
Supplementary Data 1. Alanine substitutions and position variants of APNCYGNIPL. Applied in Supplementary Fig. 2 Substitution Sequence Position variant Sequence original APNCYGNIPL original APNCYGNIPL
Circulating Cell-Free DNA Pre-analytics: Importance of ccfdna Stabilization and Extraction for Liquid Biopsy Applications
 Circulating Cell-Free DNA Pre-analytics: Importance of ccfdna Stabilization and Extraction for Liquid Biopsy Applications Martin Schlumpberger, Ass.Director R&D, QIAGEN GmbH IQNpath 2017 - Circulating
Circulating Cell-Free DNA Pre-analytics: Importance of ccfdna Stabilization and Extraction for Liquid Biopsy Applications Martin Schlumpberger, Ass.Director R&D, QIAGEN GmbH IQNpath 2017 - Circulating
Bulfoni et al. Breast Cancer Research (2016) 18:30 DOI /s
 Bulfoni et al. Breast Cancer Research (2016) 18:30 DOI 10.1186/s13058-016-0687-3 RESEARCH ARTICLE In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial-to-mesenchymal
Bulfoni et al. Breast Cancer Research (2016) 18:30 DOI 10.1186/s13058-016-0687-3 RESEARCH ARTICLE In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial-to-mesenchymal
Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library
 Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library Marilou Wijdicks International Product Manager Research For Life Science Research Only. Not for Use in Diagnostic Procedures.
Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library Marilou Wijdicks International Product Manager Research For Life Science Research Only. Not for Use in Diagnostic Procedures.
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr)
 Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Supplementary Information
 Supplementary Information - chimeric fusion transcript in human gastric cancer promotes tumorigenesis through activation of PI3K/AKT signaling Sun Mi Yun, Kwiyeom Yoon, Sunghoon Lee, Eunjeong Kim, Seong-Ho
Supplementary Information - chimeric fusion transcript in human gastric cancer promotes tumorigenesis through activation of PI3K/AKT signaling Sun Mi Yun, Kwiyeom Yoon, Sunghoon Lee, Eunjeong Kim, Seong-Ho
PRECISION INSIGHTS. Liquid GPS. Blood-based tumor profiling and quantitative monitoring. Reveal more with cfdna + cfrna.
 PRECISION INSIGHTS Liquid GPS Blood-based tumor profiling and quantitative monitoring Reveal more with cfdna + cfrna www.nanthealth.com Why Blood-Based Tumor Profiling? Although tissue-based molecular
PRECISION INSIGHTS Liquid GPS Blood-based tumor profiling and quantitative monitoring Reveal more with cfdna + cfrna www.nanthealth.com Why Blood-Based Tumor Profiling? Although tissue-based molecular
Supplemental Experimental Procedures
 Cell Stem Cell, Volume 2 Supplemental Data A Temporal Switch from Notch to Wnt Signaling in Muscle Stem Cells Is Necessary for Normal Adult Myogenesis Andrew S. Brack, Irina M. Conboy, Michael J. Conboy,
Cell Stem Cell, Volume 2 Supplemental Data A Temporal Switch from Notch to Wnt Signaling in Muscle Stem Cells Is Necessary for Normal Adult Myogenesis Andrew S. Brack, Irina M. Conboy, Michael J. Conboy,
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
ccfdna Webinar Series: The Basics and Beyond
 ccfdna Webinar Series: The Basics and Beyond Part I Maxwell RSC ccfdna Plasma Kit and Maxwell RSC instrument are For Research Use Only. Not for Use in Diagnostic Procedures. Introduction to Circulating,
ccfdna Webinar Series: The Basics and Beyond Part I Maxwell RSC ccfdna Plasma Kit and Maxwell RSC instrument are For Research Use Only. Not for Use in Diagnostic Procedures. Introduction to Circulating,
CytoSelect Tumor- Endothelium Adhesion Assay
 Product Manual CytoSelect Tumor- Endothelium Adhesion Assay Catalog Number CBA- 215 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cancer metastasis comprises several
Product Manual CytoSelect Tumor- Endothelium Adhesion Assay Catalog Number CBA- 215 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cancer metastasis comprises several
MRC-Holland MLPA. Description version 29;
 SALSA MLPA KIT P003-B1 MLH1/MSH2 Lot 1209, 0109. As compared to the previous lots 0307 and 1006, one MLH1 probe (exon 19) and four MSH2 probes have been replaced. In addition, one extra MSH2 exon 1 probe,
SALSA MLPA KIT P003-B1 MLH1/MSH2 Lot 1209, 0109. As compared to the previous lots 0307 and 1006, one MLH1 probe (exon 19) and four MSH2 probes have been replaced. In addition, one extra MSH2 exon 1 probe,
RNA preparation from extracted paraffin cores:
 Supplementary methods, Nielsen et al., A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor positive breast cancer.
Supplementary methods, Nielsen et al., A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor positive breast cancer.
Instructions for Use. RealStar Influenza Screen & Type RT-PCR Kit /2017 EN
 Instructions for Use RealStar Influenza Screen & Type RT-PCR Kit 4.0 05/2017 EN RealStar Influenza Screen & Type RT-PCR Kit 4.0 For research use only! (RUO) 164003 INS-164000-EN-S01 96 05 2017 altona
Instructions for Use RealStar Influenza Screen & Type RT-PCR Kit 4.0 05/2017 EN RealStar Influenza Screen & Type RT-PCR Kit 4.0 For research use only! (RUO) 164003 INS-164000-EN-S01 96 05 2017 altona
Incorporating pharmacodynamic, response and patient selection biomarkers. Paul Elvin PhD Chief Translational Science Officer Aptus Clinical
 Incorporating pharmacodynamic, response and patient selection biomarkers Paul Elvin PhD Chief Translational Science Officer Aptus Clinical 22 Oncology drug development Biomarkers key for: Strong hypothesis
Incorporating pharmacodynamic, response and patient selection biomarkers Paul Elvin PhD Chief Translational Science Officer Aptus Clinical 22 Oncology drug development Biomarkers key for: Strong hypothesis
MATERIALS AND METHODS. Neutralizing antibodies specific to mouse Dll1, Dll4, J1 and J2 were prepared as described. 1,2 All
 MATERIALS AND METHODS Antibodies (Abs), flow cytometry analysis and cell lines Neutralizing antibodies specific to mouse Dll1, Dll4, J1 and J2 were prepared as described. 1,2 All other antibodies used
MATERIALS AND METHODS Antibodies (Abs), flow cytometry analysis and cell lines Neutralizing antibodies specific to mouse Dll1, Dll4, J1 and J2 were prepared as described. 1,2 All other antibodies used
In vitro human regulatory T cell expansion
 - 1 - Human CD4 + CD25 + regulatory T cell isolation, Workflow in vitro expansion and analysis In vitro human regulatory T cell expansion Introduction Regulatory T (Treg) cells are a subpopulation of T
- 1 - Human CD4 + CD25 + regulatory T cell isolation, Workflow in vitro expansion and analysis In vitro human regulatory T cell expansion Introduction Regulatory T (Treg) cells are a subpopulation of T
AD (Leave blank) TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients
 AD (Leave blank) Award Number: W81XWH-12-1-0444 TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients PRINCIPAL INVESTIGATOR: Mark A. Watson, MD PhD CONTRACTING ORGANIZATION:
AD (Leave blank) Award Number: W81XWH-12-1-0444 TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients PRINCIPAL INVESTIGATOR: Mark A. Watson, MD PhD CONTRACTING ORGANIZATION:
National Surgical Adjuvant Breast and Bowel Project (NSABP) Foundation Annual Progress Report: 2011 Formula Grant
 National Surgical Adjuvant Breast and Bowel Project (NSABP) Foundation Annual Progress Report: 2011 Formula Grant Reporting Period July 1, 2012 June 30, 2013 Formula Grant Overview The NSABP Foundation
National Surgical Adjuvant Breast and Bowel Project (NSABP) Foundation Annual Progress Report: 2011 Formula Grant Reporting Period July 1, 2012 June 30, 2013 Formula Grant Overview The NSABP Foundation
Instructions for Use. RealStar Influenza S&T RT-PCR Kit /2017 EN
 Instructions for Use RealStar Influenza S&T RT-PCR Kit 3.0 01/2017 EN RealStar Influenza S&T RT-PCR Kit 3.0 For research use only! (RUO) 163003 INS-163000-EN-S02 96 01 2017 altona Diagnostics GmbH Mörkenstr.
Instructions for Use RealStar Influenza S&T RT-PCR Kit 3.0 01/2017 EN RealStar Influenza S&T RT-PCR Kit 3.0 For research use only! (RUO) 163003 INS-163000-EN-S02 96 01 2017 altona Diagnostics GmbH Mörkenstr.
In vitro human regulatory T cell expansion
 - 1 - Human CD4 + CD25 + CD127 dim/- regulatory T cell Workflow isolation, in vitro expansion and analysis In vitro human regulatory T cell expansion Introduction Regulatory T (Treg) cells are a subpopulation
- 1 - Human CD4 + CD25 + CD127 dim/- regulatory T cell Workflow isolation, in vitro expansion and analysis In vitro human regulatory T cell expansion Introduction Regulatory T (Treg) cells are a subpopulation
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Cryopreservation alters CD62L expression by CD4 T cells. Freshly isolated (left) or cryopreserved PBMCs (right) were stained with the mix of antibodies described
Supplementary Figure 1 Supplementary Figure 1: Cryopreservation alters CD62L expression by CD4 T cells. Freshly isolated (left) or cryopreserved PBMCs (right) were stained with the mix of antibodies described
Rapid antigen-specific T cell enrichment (Rapid ARTE)
 Direct ex vivo characterization of human antigen-specific CD154+CD4+ T cell Rapid antigen-specific T cell enrichment (Rapid ARTE) Introduction Workflow Antigen (ag)-specific T cells play a central role
Direct ex vivo characterization of human antigen-specific CD154+CD4+ T cell Rapid antigen-specific T cell enrichment (Rapid ARTE) Introduction Workflow Antigen (ag)-specific T cells play a central role
Commercially available HLA Class II tetramers (Beckman Coulter) conjugated to
 Class II tetramer staining Commercially available HLA Class II tetramers (Beckman Coulter) conjugated to PE were combined with dominant HIV epitopes (DRB1*0101-DRFYKTLRAEQASQEV, DRB1*0301- PEKEVLVWKFDSRLAFHH,
Class II tetramer staining Commercially available HLA Class II tetramers (Beckman Coulter) conjugated to PE were combined with dominant HIV epitopes (DRB1*0101-DRFYKTLRAEQASQEV, DRB1*0301- PEKEVLVWKFDSRLAFHH,
Direct ex vivo characterization of human antigen-specific CD154 + CD4 + T cells Rapid antigen-reactive T cell enrichment (Rapid ARTE)
 Direct ex vivo characterization of human antigen-specific CD154 + CD4 + T cells Rapid antigen-reactive T cell enrichment (Rapid ARTE) Introduction Workflow Antigen (ag)-specific T cells play a central
Direct ex vivo characterization of human antigen-specific CD154 + CD4 + T cells Rapid antigen-reactive T cell enrichment (Rapid ARTE) Introduction Workflow Antigen (ag)-specific T cells play a central
Enabling Personalized
 Molecular Enabling Personalized Diagnostics Medicine- Targeted Sequencing: NGS-based solutions Silvia Dorn Roel Reinders- Andreas Diplas Friday, 19.06.2015 Company Overview Founded in April 2011 Development
Molecular Enabling Personalized Diagnostics Medicine- Targeted Sequencing: NGS-based solutions Silvia Dorn Roel Reinders- Andreas Diplas Friday, 19.06.2015 Company Overview Founded in April 2011 Development
SUPPLEMENTARY INFORMATION. Involvement of IL-21 in the epidermal hyperplasia of psoriasis
 SUPPLEMENTARY INFORMATION Involvement of IL-21 in the epidermal hyperplasia of psoriasis Roberta Caruso 1, Elisabetta Botti 2, Massimiliano Sarra 1, Maria Esposito 2, Carmine Stolfi 1, Laura Diluvio 2,
SUPPLEMENTARY INFORMATION Involvement of IL-21 in the epidermal hyperplasia of psoriasis Roberta Caruso 1, Elisabetta Botti 2, Massimiliano Sarra 1, Maria Esposito 2, Carmine Stolfi 1, Laura Diluvio 2,
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Liquid biopsy in lung cancer: The EGFR paradigm
 Liquid biopsy in lung cancer: The EGFR paradigm Lynette M. Sholl, M.D. Brigham and Women s Hospital Dana Farber Cancer Institute Department of Pathology Boston, MA Disclosure of Relevant Financial Relationships
Liquid biopsy in lung cancer: The EGFR paradigm Lynette M. Sholl, M.D. Brigham and Women s Hospital Dana Farber Cancer Institute Department of Pathology Boston, MA Disclosure of Relevant Financial Relationships
ExpressArt FFPE Clear RNAready kit
 Features and Example Results General problems with FFPE samples Formalin-fixation of tissues results in severe RNA fragmentation, as well as in RNA RNA, RNA-DNA and RNA protein cross-linking, which impairs
Features and Example Results General problems with FFPE samples Formalin-fixation of tissues results in severe RNA fragmentation, as well as in RNA RNA, RNA-DNA and RNA protein cross-linking, which impairs
Cell Migration and Invasion Assays INCUCYTE LIVE-CELL ANALYSIS SYSTEM. Real-time automated measurements of cell motility inside your incubator
 Cell Migration and Invasion Assays INCUCYTE LIVE-CELL ANALYSIS SYSTEM Real-time automated measurements of cell motility inside your incubator See the whole story Real-time cell motility visualization and
Cell Migration and Invasion Assays INCUCYTE LIVE-CELL ANALYSIS SYSTEM Real-time automated measurements of cell motility inside your incubator See the whole story Real-time cell motility visualization and
EXOSOMES & MICROVESICLES
 The Sample Preparation Experts EXOSOMES & MICROVESICLES The New Standard in Exosome Purification and RNA Isolation Best-in-Class, Pure & Simple Exosome Purification, Fractionation of Exosomal Free & Circulating
The Sample Preparation Experts EXOSOMES & MICROVESICLES The New Standard in Exosome Purification and RNA Isolation Best-in-Class, Pure & Simple Exosome Purification, Fractionation of Exosomal Free & Circulating
Evaluation of MIA FORA NGS HLA test and software. Lisa Creary, PhD Department of Pathology Stanford Blood Center Research & Development Group
 Evaluation of MIA FORA NGS HLA test and software Lisa Creary, PhD Department of Pathology Stanford Blood Center Research & Development Group Disclosure Alpha and Beta Studies Sirona Genomics Reagents,
Evaluation of MIA FORA NGS HLA test and software Lisa Creary, PhD Department of Pathology Stanford Blood Center Research & Development Group Disclosure Alpha and Beta Studies Sirona Genomics Reagents,
2/10/2016. Evaluation of MIA FORA NGS HLA test and software. Disclosure. NGS-HLA typing requirements for the Stanford Blood Center
 Evaluation of MIA FORA NGS HLA test and software Lisa Creary, PhD Department of Pathology Stanford Blood Center Research & Development Group Disclosure Alpha and Beta Studies Sirona Genomics Reagents,
Evaluation of MIA FORA NGS HLA test and software Lisa Creary, PhD Department of Pathology Stanford Blood Center Research & Development Group Disclosure Alpha and Beta Studies Sirona Genomics Reagents,
Molecular Profiling of Tumor Microenvironment Alex Chenchik, Ph.D. Cellecta, Inc.
 Molecular Profiling of Tumor Microenvironment Alex Chenchik, Ph.D. Cellecta, Inc. Cellecta, Inc. Founded: April 2006 Headquarters: Mountain View, CA 12 SBIR Grants Custom Service Provider for Functional
Molecular Profiling of Tumor Microenvironment Alex Chenchik, Ph.D. Cellecta, Inc. Cellecta, Inc. Founded: April 2006 Headquarters: Mountain View, CA 12 SBIR Grants Custom Service Provider for Functional
Circulating Tumor DNA and Circulating Tumor Cells for Management (Liquid Biopsy) of Solid Tumor Cancers
 NOTE: This policy is not effective until December 1, 2018. To view the current policy, click here. Medical Policy Manual Laboratory, Policy No. 46 Circulating Tumor DNA and Circulating Tumor Cells for
NOTE: This policy is not effective until December 1, 2018. To view the current policy, click here. Medical Policy Manual Laboratory, Policy No. 46 Circulating Tumor DNA and Circulating Tumor Cells for
