ATTACHMENT OF RIBOSOMES TO MEMBRANES DURING POLYSOME FORMATION IN MOUSE SARCOMA 180 CELLS
|
|
|
- Gavin Bradford
- 5 years ago
- Views:
Transcription
1 Published Online: 1 June, 1971 Supp Info: Downloaded from jcb.rupress.org on November 2, 2018 ATTACHMENT OF RIBOSOMES TO MEMBRANES DURING POLYSOME FORMATION IN MOUSE SARCOMA 180 CELLS SE YONG LEE, VELIBOR KRSMANOVIC, and GEORGE BRAWERMAN From the Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut Dr. Se Yong Lee's and Dr. George Brawerman's present address is the Department of Biochemistry-Pharmacology, Tufts University School of Medicine, Boston, Massachusetts ABSTRACT Addition of nutrients to starved mouse S-180 cells leads to rapid conversion of ribosomal monomers to polysomes. During this process, a portion of the ribosomes originally found in the 17,000 g (10 min centrifugation) supernatant of cell lysates becomes firmly attached to structures sedimenting at 500 g (5 min centrifugation). Electron microscopy of sections of the intact cells showed the change from randomly distributed ribosomal particles to clusters. Association with membranes also became evident. The material sedimenting at 500 g comprised nuclei enclosed in an extensive endoplasmic reticulum (ER) network. This fraction prepared from recovering cells showed numerous ribosome clusters associated with the ER network. The appearance of many of these clusters indicated that the ribosomal particles were not directly bound to the membranes. RNase treatment released about 40% of the attached ribosomes as monomers, and ethylenediaminetetraacetic acid released 60% as subunits. It is suggested that during polysome formation a portion of the ribosomes becomes attached to the membranes through the intermediary of messenger RNA. INTRODUCTION In animal cells, a portion of the ribosomes has been shown to be attached to membranes of the endoplasmic reticulum, while the rest is free in the cytoplasm. The proportion of bound ribosomes is high in differentiated cells active in synthesis of proteins for secretion (7, 8, 10). Studies of the mode of attachment of liver ribosomes have indicated that it is the large ribosomal subunit that is bound directly to the membrane (4, 11). HeLa cells have also been reported to have a significant proportion of ribosomes bound to membranes in the same manner, although no obvious secretory function is evident for these cells (1). We present evidence for a form of polysome association with membranes in which the binding appears to be through the mrna.i In the course of a study of polysome formation initiated by restoration of nutrients to starved S-180 cells (6), it was observed that a portion of the ribosomes becomes rapidly attached to membranes. With the present method used for cell lysis, an extensive endoplasmic reticulum network remained attached to the nuclei, and could be washed free of 'Abbreviations : mrna, messenger RNA ; RNase, ribonuclease ; EDTA, ethylenediaminetetraacetic acid. THE JOURNAL OF CELL BIOLOGY VOLUME 49, pages
2 nonattached polysomes. This permitted the characterization of the bound polysome fraction both by biochemical studies and by electron microscopy. EXPERIMENTAL Tumor Cells Mouse sarcoma 180 cells, kindly supplied by Dr. Sartorelli (Department of Pharmacology, Yale University), were maintained by weekly transfers into the peritoneal cavity of albino mice. Incubations and Cell Fractionations The detailed procedures have been described elsewhere (6). The cells, harvested from the peritoneal cavity 5-7 days after inoculation, were washed several times in warm Krebs bicarbonate buffer. For the starvation treatment, the cells were incubated with gentle agitation at 37 C for 30 min in the same buffer, at a cell density of 1 X 10 7 /ml. Recovery was initiated by addition of glucose, bovine serum, and a complete mixture of amino acids. The cells were harvested after rapid cooling at 0 C, and washed once with ice-cold 1% NaCl. For lysis, the cells were swollen by washing twice with ice-cold 10 mss Tris-HCl (ph 7.6), 10 mm KC1, and 1 mm MgCls, then gently suspended in 2.5 vol of 50 mat Tris (ph 7.6), 130 mm KCI, 6.5 mss MgCl2, 6.5 mm ß-mercaptoethanol, 0.13% Triton X-100, and 13% sucrose. The resulting lysate was centrifuged at 500 g for 5 min. The pellet was used as nuclear fraction and the supernatant as cytoplasmic fraction. Nucleic Acid Determinations The samples were precipitated with ice-cold 0.2 N HC1O4 and the pellets were washed with 5 ml of the same solution. They were next suspended in 4 ml of 0.1 N KOH and kept at 100 C for 10 min to hydrolyze the RNA. After chilling in ice, 1 ml of 2.5 N HCIO4 was added to precipitate the DNA and proteins. The suspensions were centrifuged, the supernatants were used for RNA determinations, and the pellets were treated with 0.5 N HC1O4 at 70 C for 20 min to solubilize the DNA. After chilling, the suspensions were centrifuged and the supernatants were used for DNA determinations. The amounts of DNA and RNA were obtained by optical density measurements at 260 mµ. The average nucleotide molar extinction values at 260 mµ used in this study were 9200 for DNA and 11,900 for RNA. Electron Microscopy The preparation of the specimens, as well as the examination of the electron micrographs, were kindly performed by Dr. Samuel Dales (The Public Health Research Institute of The City of New York, Inc.). The nuclear pellets were washed by suspension in 10 ml of ice-cold 10% sucrose, 50 mm Tris (ph 7.6), and 3 m m CaC12, and by recentrifugation. The cell samples, washed in 1 % NaCl, were used as such. The pellets were prepared for thin sectioning and electron microscopy as described by Dales et al. (5). Sections A in thickness were mounted on naked grids and examined in a Philips EM 300 electron microscope fitted with an anticontamination device. The images were recorded on Kodak electron image plates at instrumental magnifications of ,000. RESULTS Changes in Ribosome Distribution in S-180 Cells Subjected to Starvation and Recovery Addition of nutrients to cells incubated in Krebs bicarbonate solution (see Experimental section) leads to rapid conversion of the ribosomal monomers accumulated during the starvation treatment to polysomes (Fig. 1). The disappearance of polysomes during starvation appears to be due to ribosome runoff coupled with a block in the initiation process, as indicated by the fact that the resulting ribosomal monomers are free of mrna and peptidyl-transfer trna (6). The mrna released from the polysomes appears to be fully functional, as evidenced by its rapid re-incorporation into polysomes after the addition of nutrients (6). The formation of polysomes in starved cells supplemented with nutrients is accompanied by the attachment of a portion of the ribosomes to structures sedimenting at 500 g, as indicated by determinations of RNA distribution in subcellular fractions (Tables I and II). The procedure used for cell lysis leaves the great bulk of the RNA distributed between the nuclear fraction (500 g pellet) and the 17,000 g supernatant fraction (Table I). The ribosomal material in the nuclear fraction appears to be firmly bound, since washing hardly reduces the RNA content of this fraction. The amount of RNA in the nuclear fraction of starved cells tended to vary. Values as low as 12% of the total cellular RNA were obtained in some experiments. The value of 25% shown in Table II, however, was more typical. The shift of ribosomes from cytoplasm to nuclear fraction was most rapid within the first 10 min, but it continued for about 1 hr (Table II) THE JOURNAL of CELL BIOLOGY VOLUME 49, 1971
3 t2.5 E 2.0 STARVED CELLS SOS NUTRIENTS 10 min NUTRIENTS 60 min.a Q o o S BOS FIGURE 1 Polysome formation in starved S-180 cells after administration of nutrients. Cells were subjected to starvation and recovery treatments as described in the Experimental section ml samples of cytoplasmic fractions (containing 40,ug RNA) were layered over 4.5 ml of linear 10-30% sucrose gradients in 50 mbt Tris (ph 7.6), 50 mm KCI, and M902, with a 0.5 ml cushion of 2 M sucrose at the bottom of the centrifuge tubes. Centrifugation was done at 45,000 rpm for 25 min in the SW 50 Spinco rotor. Contents of tubes were then pumped from the bottom through a UV detector and the absorbancy at 257 mµ was recorded. TABLE I Distribution of RNA in Subcellular Fractions of Recovered Cells Cell subjected to recovery treatment for 30 min. Subcellular fractions obtained by successive centrifugations of cell lysate at 500 g for 5 min and 17,000 g for 10 min ; 500 g pellet was washed by resuspending in lysing medium (see Experimental section) and by recentrifuging at 500 g for 5 min. Fraction RNA content Distribution of Ribosomes in Cytological Preparations of Whole Cells Examination of the recovering cells by electron microscopy showed the expected change from a random distribution of ribosomes in the cytoplasm of starved cells (Fig. 2) to the clustering typical for polysomal aggregates in the cells exposed to nutrients for 10 min (Fig. 4). Most of the ribosomes are free in the starved cells, but frequent association with the endoplasmic reticulum is evident in the recovering cells. After 60 min of recovery, the appearance of the cells is about the same as in the 10 min sample, although there appears to MS % of total 500 g pellet, crude g pellet, washed ,000 g pellet ,000 g supernatant TABLE II Changes in RNA Content of Nuclear Fraction During Polysome Formation Cell suspension preincubated in complete medium for 30 min, then washed and subjected to procedure for starvation and recovery. Nuclear fraction represents 500 g pellet washed with 10% sucrose, 50 mm Tris (ph 7.6), and 3 MM CaCI2. Values expressed as micrograms nucleic acid per sample. State of cells RNA$ Nuclear fraction DNA* RNA DNA Total RNA in ccll' % of total RNA in nuclear fraction Starved Recovering, 10 min ) min min * Variations in DNA and in total RNA can be attributed to sampling errors, since no consistent pattern of variations was observed in different experiments. $ Microscopic examination of cell lysates showed the virtual absence of intact cells. Thus, the changes in RNA content of nuclear fractions cannot be attributed to contamination by intact cells. be a greater proportion of membrane-bound polysomes (Fig. 6). The characteristic arrangement of ribosomes in rosette-like patterns is evident where the cisternae of the endoplasmic LEE, KRSMANOVIC, AND BRAWERMAN Membrane-Bound Polysomes 685
4 FIGURE 2 Portion of an intact cell from a suspension subjected to starvation treatment. The cytoplasm contains mitochondria, elements of the endoplasmic reticulum, and ribosomes. The "free" ribosomes appear to be randomly distributed throughout the cytoplasmic matrix. N, nucleus; M, mitochondria. X 44,500. FIGURE 3 Low power micrograph of section through a washed 500 g pellet from lysate of starved cells, showing nuclei (N) and remaining cytoplasmic components (C) of several cells. Numerous swollen mitochondria (M) and vesicles of the endoplasmic reticulum remain associated with nuclei, but few ribosomes remain (arrows). X 14,
5 reticulum were sectioned tangentially. These patterns have been described by Palade (9). Distribution of Ribosomes in Preparations from Nuclear Fractions The significance of ribosome attachment to structures in the nuclear fraction became apparent when the latter was examined by electron microscopy. The method of cell lysis used in this study leaves the nuclei enclosed by a swollen endoplasmic reticulum network. Numerous ribosome clusters are associated with this network in preparations from recovering cells (Figs. 5 and 7), while relatively few ribosomes are present in preprations derived from starved cells (Fig. 3). Fig. 7, with its higher magnification, reveals some details of the polysome-membrane association. Many of the clusters appear to overlie and hang away from the membranes. Nature of Attachment of Ribosomes to Endoplasmic Reticulum The conversion of ribosomes previously "free" in starved cells to polysomes attached to membranes could be the result of interaction of the ribosomal monomers with membrane-associated mrna. In this case the attachment should be through the intermediary of the mrna molecule. Direct attachment of ribosomes to membranes through the large subunit, as described by Sabatini et al. (11), could also take place. Ribosomes bound in the latter fashion are not released by RNase treatment (4), and EDTA treatment leads to preferential release of the small ribosomal subunit (11). As seen in Fig. 8, RNase treatment of the nuclear fracton of recovered cells releases ribosomal monomers, and EDTA leads to release of the two ribosomal subunits in apparently equal proportion. Sedimentation profiles of material released from the equivalent fraction from starved cells showed considerably less ribosomal material, but nearly equal amounts of the slowly sedimenting UV-absorbing material. The latter probably represents nonribosomal RNA, since the amounts released were not dependent on the ribosome content of the nuclear fraction. The same appears to be true of the acid-soluble material produced by the RNase treatment, since the absolute amounts produced from the two types of nuclear fractions were the same (see Table III). Moreover, the conditions for RNase digestion used here do not lead to ribosome destruction (unpublished experiments). The total amounts of RNA released from the nuclear fractions of starved and recovered cells are listed in Table III. The differences between the amounts released from the two types of nuclear fractions account for only a portion of the ribosomes shifted to the nuclear fraction during recovery (60% in the case of the EDTA treatment and 46% in the case of the RNase treatment). TALE III Release of Ribosomal Material from Nuclear Fraction of Recovering Cells Samples of washed nuclear fractions (1.2 mg DNA per sample), suspended in 10% sucrose, 50 mm Tris (ph 7.6), and 3 mm CaC12, were treated as described in Fig. 8, or left untreated. All samples were then centrifuged at 600 g for 10 min, and nucleic acid analyses were done on both pellets and supernatant fluids. Values are expressed as ratios of RNA to DNA in untreated nuclear fractions. Treatments of nuclear fraction Wash EDTA RNase Released Residue Released Residue Released Residue Acid solublet Starved , Recovering, 60 min Material shifted or released* 0, * Values express, in the case of "Wash," the amount of ribosomal material shifted to structures in the nuclear fraction during recovery, and in other cases, the portions of this material released by the RNase and EDTA treatments. $ Represents material hydrolyzed to acid-soluble components by enzyme treatment, as determined by difference between amounts of RNA originally in nuclear fraction and amounts remaining after treatment. LEE, KRSMANOVIC, AND BRAWERMAN Membrane-Bound Polysomes 68 7
6 FIGURE 4 Appearance of intact cell 10 min after addition of nutrients to suspension of starved cells. Many ribosomes, some in rosette configuration, are associated with endoplasmic reticulum cisternae (arrows). Clustering of ribosomes in the cytoplasmic matrix is also apparent. N, nucleus ; M, mitochondria. X 45,000. FIGURE 5 Section through a washed 500 g pellet of lysed cells from 10 min recovered culture. Numerous clusters of ribosomes apparently connected to membranous cell components are clearly evident (arrows). N, nucleus ; M, mitochondria ; L, lipid-filled vacuole. X 20, THE JOURNAL OF CELL BIOLOGY VOLUME 49, 1971
7 FIGURE 6 Appearance of intact cell 60 min after addition of nutrients to starved cell suspension. Clusters of ribosomes in the matrix and associated with the cisternae are evident (arrows). Where the membranes of tangentially sectioned cisternae are displayed en face, the rosette pattern of ribosomes is observed (upper arrow). N, nucleus ; M, mitochondria. X 35,000. FiGuRE 7 Section through a washed 500 g pellet of lysed cells from 60 min recovered culture. Clusters of membrane-related ribosomes (arrows) are readily apparent. In this sample, several A type virus-like particles (V) occur, possessing the morphology described for the leukemia-sarcoma murine agents (3). N, nucleus. X 55,
8 Treatment of the nuclear fraction of recovered cells with EDTA concentrations ranging from 10 to 30 mm yielded similar amounts of ribosomal subunits. The release of monomers by treatment with RNase was very inefficient at 0 C, and increased wtih the temperature of incubation. It is not known whether the temperature of 10 C selected for the experiments described in Fig. 8 and Table III represents the optimum, since higher temperatures were not tried. Incubation of the nuclear fraction at 10 C without enzyme did not lead to any release of ribosomes. Attempts to separate the membranous structures from the nuclei, in order to permit further characterization of the membrane-bound polysomes, proved unsuccessful. The structures in the nuclear fraction resisted vigorous mechanical shear, even in the presence of 2 M sucrose. Treatment with 16/0 Triton X-100 released very little, if any, ribosomal material. 0.2% sodium deoxycholate caused little release, and higher concentrations led to lysis of the nuclei. DISCUSSION This report describes the rapid attachment of ribosomes to membranes of the endoplasmic reticulum during polysome formation in mouse sarcoma cells recovering from starvation. The membrane-bound ribosomes are seen in the electron micrographs as clusters, an indication that they are attached in the form of polysomes. The appearance of the ribosomal particles in sections of whole cells, as well as in the nuclear fractions, indicates that two different types of ribosome binding may occur in the recovering cells. Ribosomes aligned in rows along the cisternae of the endoplasmic reticulum, as well as the rosette-like patterns, are clearly evident. These patterns are typical for ribosomes bound to membranes through the intermediary of the large ribosomal subunits, as described in microsomal fractions of guinea pig liver and of HeLa cells (1, 11). A second type of binding is suggested by the appearance of many of the ribosome clusters, which seem to hang away from the membranes (Figs. 5 and 7). The occurrence of two types of binding is also suggested by the effect of treatment with RNase, which causes the release of about half of the ribosomes as monomers. Ribosomes attached directly to the membranes have been shown not to be released by treatment with RNase (1, 4). The sensitivity to RNase in the present case indicated that the material is more susceptible to the enzyme than are the ribosomes. Polysomes attached through the intermediary of mrna could account for the RNase effect as well as for the appearance in the electron micrographs. The effect of EDTA, which dissociates ribosomes into subunits, is more difficult to understand in terms of the two types of binding postulated above. This treatment should release the small subunits preferentially from the ribosomes bound directly to the membranes (11) and both subunits equally from the ribosomes not attached in this manner. Zone centrifugation of the released particles (Fig. 8) should have shown an 0.6 u E N- N 0.4 A RNase B EDTA U Z a w ô 0.2 w a 80S 0 -ON FIGURE 8 Zone centrifugation patterns of material released by RNase and EDTA from washed nuclear fraction of cells subjected to recovery treatment for 30 min. 500 g pellet of cell lysate was washed once in 10% sucrose, 50 mm Tris (ph 7.6), and 3 mm CaC12, and resuspended in the same solution at a concentration equivalent to the one in the original lysate. Suspensions were incubated with 10 Ag/ml of pancreatic RNase at 10 C for 20 min (A) ; and with 20 mm EDTA in the cold (B). After treatments, suspensions were centrifuged at 600 g for 10 min, and 0.2 ml samples of supernatants (containing 0.5 A260 my units for A and 0.4, units for B) were centrifuged through 5-ml linear 10-30% sucrose gradients in 50 mm Tris (ph 7.6), 50 Dam KCl and 1 HIM MgC12 at 45,000 rpm in the SW Spinco rotor for 30 min (A) and 90 min (B). 690 THE JOURNAL OF CELL BIOLOGY. VOLUME 49, 1971
9 excess amount of the small subunits. It is possible that a portion of the released small subunits became degraded. Also, the amount of ribosomal material not released by EDTA (Table III) seems excessive. This suggests that a substantial portion of the ribosomes is in such a configuration that it resists dissociation into subunits. The occurrence of "stuck" monomers is also indicated in the study of Sabatini et al. (II). The present results suggest that S-180 cells may contain mrna associated with membranes and that this RNA may function as a template for protein synthesis in this state. The concept of mrna association with membranes is not a novel one (12), but convincing evidence has been lacking due to the difficulty in characterizing the mrna. Membrane-associated polysome structures similar to those described here have been found in poliovirus-infected HeLa cells (5) and in rat liver (2). Uninfected cells also appear to possess membrane-bound polysomes that can be released by RNase (Rosbach and Penman, personal communication). Additional evidence, however, will be required before this type of polysome structure can be established as a normal feature of animal cells. The release of ribosomes from membrane-bound mrna during starvation and their reattachment during recovery could be readily explained in terms of interruption and resumption of polypeptide chain initiation (6). The relation between attachment to membranes and initiation of protein synthesis, however, may not be a direct one, since ribosome attachment seems to proceed more slowly than polysome formation (compare Fig. 1 and Table II). Ribosomes bound directly to the membranes also appear to be released and reattached during the starvation-recovery treatment. The mechanism of attachment is now known in this latter case. It would appear from the present results that the ribosomes must be engaged in protein synthesis in order to remain bound to the membranes. The authors are very grateful to Dr. Samuel Dales for the preparation and interpretation of the electron micrographs, as well as for useful discussions. They also wish to thank Mrs. Mary Ann Krutz for her very competent technical assistance. This work was supported by a United States Public Health Service Research Career Development Award (GM-K3-3295) from the National Institute of General Medical Sciences to Dr. Brawerman and a research grant from the United States Public Health Service (GM-11527). Received for publication 24 August 1970, and in revised form 11 January REFERENCES 1. ATTARDI, B., B. CRAVIOTO, and G. ATTARDI J. Mol. Biol. 44 : BENEDETTI, E. L., W. S. BONT, and H. BLOEMEN- DAL Lab. Invest. 15 : BERNHARD, W., and M. Gu$RIN C. R. Acad. Sci. Ser. D. 248 : BLOBEL, G., and V. R. POTTER J. Mol. Biol. 26 : DALES, S., H. J. EGGERS, I. TAMM, and G. E. PALADE Virology. 26 : LEE, S. Y., V. KRSMANOVIC, and G. BRAWERMAN Biochemistry. 10 : PALADE, G. E J. Biophys. Biochem. Cyto l. 2 (4, Suppl.) : PALADE, G. E In Microsomal Particles and Protein Synthesis. R. B. Roberts, editor. Pergamon Press, New York PALADE, G. E Proc. Nat. Acad. Sci. U. S. A. 52 : Porter, K. R In The Cell. J. Brachet and A. Mirsky, editors. Academic Press, Inc., New York. 2 : SABATINI, D. D., Y. TAsHIRO, and G. E. PALADE J. Mol. Biol. 19 : SHAPOT, V., and H. C. PITOT Biochim. Biophys. Acta. 119 :37. LEE, KRSMANOVIC, AND BRAWERMAN Membrane-Bound Polysomes 69 1
10 mm KCl in a Ti-15 zonal rotor at 35,000 rpm for 16 hr at
 Proc. Nat. Acad. SCi. USA Vol. 68, No. 11, pp. 2752-2756, November 1971 Translation of Exogenous Messenger RNA for Hemoglobin on Reticulocyte and Liver Ribosomes (initiation factors/9s RNA/liver factors/reticulocyte
Proc. Nat. Acad. SCi. USA Vol. 68, No. 11, pp. 2752-2756, November 1971 Translation of Exogenous Messenger RNA for Hemoglobin on Reticulocyte and Liver Ribosomes (initiation factors/9s RNA/liver factors/reticulocyte
Chlamydomonas reinhardtii (chloramphenicol/puromycin reaction/electron microscopy/synchronous cultures)
 Proc. Nat. Acad. Sci. USA Vol. 70, No. 5, pp. 1554-1558, May 1973 Attachment of Chloroplast Polysomes to Thylakoid Membranes in Chlamydomonas reinhardtii (chloramphenicol/puromycin reaction/electron microscopy/synchronous
Proc. Nat. Acad. Sci. USA Vol. 70, No. 5, pp. 1554-1558, May 1973 Attachment of Chloroplast Polysomes to Thylakoid Membranes in Chlamydomonas reinhardtii (chloramphenicol/puromycin reaction/electron microscopy/synchronous
Single Essential Amino Acids (valine/histidine/methiotiine/high-temperature inhibition)
 Proc. Nat. Acad. Sci. USA Vol. 68, No. 9, pp. 2057-2061, September 1971 Regulation of Protein Synthesis Initiation in HeLa Cells Deprived of Single ssential Amino Acids (valine/histidine/methiotiine/high-temperature
Proc. Nat. Acad. Sci. USA Vol. 68, No. 9, pp. 2057-2061, September 1971 Regulation of Protein Synthesis Initiation in HeLa Cells Deprived of Single ssential Amino Acids (valine/histidine/methiotiine/high-temperature
Thursday, October 16 th
 Thursday, October 16 th Good morning. Those of you needing to take the Enzymes and Energy Quiz will start very soon. Students who took the quiz Wednesday: Please QUIETLY work on the chapter 6 reading guide.
Thursday, October 16 th Good morning. Those of you needing to take the Enzymes and Energy Quiz will start very soon. Students who took the quiz Wednesday: Please QUIETLY work on the chapter 6 reading guide.
PRODUCT: RNAzol BD for Blood May 2014 Catalog No: RB 192 Storage: Store at room temperature
 PRODUCT: RNAzol BD for Blood May 2014 Catalog No: RB 192 Storage: Store at room temperature PRODUCT DESCRIPTION. RNAzol BD is a reagent for isolation of total RNA from whole blood, plasma or serum of human
PRODUCT: RNAzol BD for Blood May 2014 Catalog No: RB 192 Storage: Store at room temperature PRODUCT DESCRIPTION. RNAzol BD is a reagent for isolation of total RNA from whole blood, plasma or serum of human
FOCUS SubCell. For the Enrichment of Subcellular Fractions. (Cat. # ) think proteins! think G-Biosciences
 169PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name FOCUS SubCell For the Enrichment of Subcellular Fractions (Cat. # 786 260) think
169PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name FOCUS SubCell For the Enrichment of Subcellular Fractions (Cat. # 786 260) think
Distribution of Ribosomes in Hypophysectomized Rat Liver. An Improved Method for the Determination of Free and Membrane-bound Ribosomes
 Agr. Biol. Chem., 37 (2), 219 `224, 1973 Distribution of Ribosomes in Hypophysectomized Rat Liver An Improved Method for the Determination of Free and Membrane-bound Ribosomes Masamichi TAKAGI, Tadashi
Agr. Biol. Chem., 37 (2), 219 `224, 1973 Distribution of Ribosomes in Hypophysectomized Rat Liver An Improved Method for the Determination of Free and Membrane-bound Ribosomes Masamichi TAKAGI, Tadashi
Biol110L-Cell Biology Lab Spring Quarter 2012 Module 1-4 Friday April 13, 2012 (Start promptly; work fast; the protocols take ~4 h)
 Biol110L-Cell Biology Lab Spring Quarter 2012 Module 1-4 Friday April 13, 2012 (Start promptly; work fast; the protocols take ~4 h) A. Microscopic Examination of the Plasma Membrane and Its Properties
Biol110L-Cell Biology Lab Spring Quarter 2012 Module 1-4 Friday April 13, 2012 (Start promptly; work fast; the protocols take ~4 h) A. Microscopic Examination of the Plasma Membrane and Its Properties
PREPARATION OF IF- ENRICHED CYTOSKELETAL PROTEINS
 TMM,5-2011 PREPARATION OF IF- ENRICHED CYTOSKELETAL PROTEINS Ice-cold means cooled in ice water. In order to prevent proteolysis, make sure to perform all steps on ice. Pre-cool glass homogenizers, buffers
TMM,5-2011 PREPARATION OF IF- ENRICHED CYTOSKELETAL PROTEINS Ice-cold means cooled in ice water. In order to prevent proteolysis, make sure to perform all steps on ice. Pre-cool glass homogenizers, buffers
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting
 Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
CELLS. Cells. Basic unit of life (except virus)
 Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
1. (a) (i) Ability to distinguish points (close together); 1 (ii) Electrons have a shorter wavelength; 1
 1. (a) (i) Ability to distinguish points (close together); 1 Electrons have a shorter wavelength; 1 (b) (i) Golgi / nucleus / mitochondrion / endoplasmic reticulum / chromosome / larger ribosomes; R Membrane
1. (a) (i) Ability to distinguish points (close together); 1 Electrons have a shorter wavelength; 1 (b) (i) Golgi / nucleus / mitochondrion / endoplasmic reticulum / chromosome / larger ribosomes; R Membrane
Work-flow: protein sample preparation Precipitation methods Removal of interfering substances Specific examples:
 Dr. Sanjeeva Srivastava IIT Bombay Work-flow: protein sample preparation Precipitation methods Removal of interfering substances Specific examples: Sample preparation for serum proteome analysis Sample
Dr. Sanjeeva Srivastava IIT Bombay Work-flow: protein sample preparation Precipitation methods Removal of interfering substances Specific examples: Sample preparation for serum proteome analysis Sample
The Fine Structure of the Epithelial Cells of the Mouse Prostate* II. Ventral Lobe Epithelium
 Published Online: 1 June, 1960 Supp Info: http://doi.org/10.1083/jcb.7.3.511 Downloaded from jcb.rupress.org on September 28, 2018 The Fine Structure of the Epithelial Cells of the Mouse Prostate* II.
Published Online: 1 June, 1960 Supp Info: http://doi.org/10.1083/jcb.7.3.511 Downloaded from jcb.rupress.org on September 28, 2018 The Fine Structure of the Epithelial Cells of the Mouse Prostate* II.
Biology 12 Cell Structure and Function. Typical Animal Cell
 Biology 12 Cell Structure and Function Typical Animal Cell Vacuoles: storage of materials and water Golgi body: a series of stacked disk shaped sacs. Repackaging centre stores, modifies, and packages proteins
Biology 12 Cell Structure and Function Typical Animal Cell Vacuoles: storage of materials and water Golgi body: a series of stacked disk shaped sacs. Repackaging centre stores, modifies, and packages proteins
AP Biology
 Tour of the Cell (1) 2007-2008 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Cell Size Why organelles? Specialized structures - specialized
Tour of the Cell (1) 2007-2008 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Cell Size Why organelles? Specialized structures - specialized
BY F. BROWN, B. CARTWRIGHT AND DOREEN L. STEWART Research Institute (Animal Virus Diseases), Pirbright, Surrey. (Received 22 August 1962) SUMMARY
 J. gen. Microbial. (1963), 31, 179186 Prinied in Great Britain 179 The Effect of Various Inactivating Agents on the Viral and Ribonucleic Acid Infectivities of FootandMouth Disease Virus and on its Attachment
J. gen. Microbial. (1963), 31, 179186 Prinied in Great Britain 179 The Effect of Various Inactivating Agents on the Viral and Ribonucleic Acid Infectivities of FootandMouth Disease Virus and on its Attachment
Explain that each trna molecule is recognised by a trna-activating enzyme that binds a specific amino acid to the trna, using ATP for energy
 7.4 - Translation 7.4.1 - Explain that each trna molecule is recognised by a trna-activating enzyme that binds a specific amino acid to the trna, using ATP for energy Each amino acid has a specific trna-activating
7.4 - Translation 7.4.1 - Explain that each trna molecule is recognised by a trna-activating enzyme that binds a specific amino acid to the trna, using ATP for energy Each amino acid has a specific trna-activating
Experiment 1. Isolation of Glycogen from rat Liver
 Experiment 1 Isolation of Glycogen from rat Liver Figure 35: FIG-2, Liver, PAS, 100x. Note the presence of a few scattered glycogen granules (GG). Objective To illustrate the method for isolating glycogen.
Experiment 1 Isolation of Glycogen from rat Liver Figure 35: FIG-2, Liver, PAS, 100x. Note the presence of a few scattered glycogen granules (GG). Objective To illustrate the method for isolating glycogen.
Blocking by Histones of Accessibility to DNA in Chromatin (DNase/RNA polymerase/dna polymerase)
 Proc. Nat. Acad. Sci. USA Vol. 69, No. 8, pp. 2115-2119, August 1972 Blocking by Histones of Accessibility to in Chromatin (/RNA polymerase/ polymerase) ALFRED E. MIRSKY AND BERT SILVERMAN The Rockefeller
Proc. Nat. Acad. Sci. USA Vol. 69, No. 8, pp. 2115-2119, August 1972 Blocking by Histones of Accessibility to in Chromatin (/RNA polymerase/ polymerase) ALFRED E. MIRSKY AND BERT SILVERMAN The Rockefeller
Wilmington, Delaware cells were harvested in the cold and pelleted. The cell. pellet was suspended in 2 ml of cold buffer consisting
 JOURNAL OF VIROLOGY, June 1969, p. 599-64 Vol. 3, No. 6 Copyright 1969 American Society for Microbiology Printed in U.S.A. Sindbis Virus-induced Viral Ribonucleic Acid Polymerasel T. SREEVALSAN' AND FAY
JOURNAL OF VIROLOGY, June 1969, p. 599-64 Vol. 3, No. 6 Copyright 1969 American Society for Microbiology Printed in U.S.A. Sindbis Virus-induced Viral Ribonucleic Acid Polymerasel T. SREEVALSAN' AND FAY
Problem-solving Test: The Mechanism of Protein Synthesis
 Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
The following protocol describes the isolation of nuclei from tissue. Item. Catalog No Manufacturer
 SOP: Nuclei isolation from tissue and DNaseI treatment Date modified: 090923 Modified by: P. Sabo. (UW) The following protocol describes the isolation of nuclei from tissue. Ordering Information Item.
SOP: Nuclei isolation from tissue and DNaseI treatment Date modified: 090923 Modified by: P. Sabo. (UW) The following protocol describes the isolation of nuclei from tissue. Ordering Information Item.
PRODUCT INFORMATION & MANUAL
 PRODUCT INFORMATION & MANUAL Mitochondrial Extraction Kit NBP2-29448 Research use only. Not for diagnostic or therapeutic procedures www.novusbio.com P: 303.760.1950 P: 888.506.6887 F: 303.730.1966 technical@novusbio.com
PRODUCT INFORMATION & MANUAL Mitochondrial Extraction Kit NBP2-29448 Research use only. Not for diagnostic or therapeutic procedures www.novusbio.com P: 303.760.1950 P: 888.506.6887 F: 303.730.1966 technical@novusbio.com
/searchlist/6850.html Tour of the Cell 1
 http://www.studiodaily.com/main /searchlist/6850.html Tour of the Cell 1 2011-2012 Cytology: science/study of cells To view cells: Light microscopy resolving power: measure of clarity Electron microscopy
http://www.studiodaily.com/main /searchlist/6850.html Tour of the Cell 1 2011-2012 Cytology: science/study of cells To view cells: Light microscopy resolving power: measure of clarity Electron microscopy
ELECTRON MICROSCOPIC STUDIES ON EQUINE ENCEPHALOSIS VIRUS
 Onderstepoort]. vet. Res. 40 (2), 53-58 (1973) ELECTRON MICROSCOPIC STUDIES ON EQUINE ENCEPHALOSIS VIRUS G. LECATSAS, B. J. ERASMUS and H. J. ELS, Veterinary Research Institute, Onderstepoort ABSTRACT
Onderstepoort]. vet. Res. 40 (2), 53-58 (1973) ELECTRON MICROSCOPIC STUDIES ON EQUINE ENCEPHALOSIS VIRUS G. LECATSAS, B. J. ERASMUS and H. J. ELS, Veterinary Research Institute, Onderstepoort ABSTRACT
Western Immunoblotting Preparation of Samples:
 Western Immunoblotting Preparation of Samples: Total Protein Extraction from Culture Cells: Take off the medium Wash culture with 1 x PBS 1 ml hot Cell-lysis Solution into T75 flask Scrap out the cells
Western Immunoblotting Preparation of Samples: Total Protein Extraction from Culture Cells: Take off the medium Wash culture with 1 x PBS 1 ml hot Cell-lysis Solution into T75 flask Scrap out the cells
RETICULUM MEMBRANES AFTER IN VIVO DISASSEMBLY OF
 RETENTION OF MRNA ON THE ENDOPLASMIC RETICULUM MEMBRANES AFTER IN VIVO DISASSEMBLY OF POLYSOMES BY AN INHIBITOR OF INITIATION MILTON ADESNIK, MAURICIO LANDE, TERENCE MARTIN, and DAVID D. SABATINI. From
RETENTION OF MRNA ON THE ENDOPLASMIC RETICULUM MEMBRANES AFTER IN VIVO DISASSEMBLY OF POLYSOMES BY AN INHIBITOR OF INITIATION MILTON ADESNIK, MAURICIO LANDE, TERENCE MARTIN, and DAVID D. SABATINI. From
Name: Date: Block: Biology 12
 Name: Date: Block: Biology 12 Provincial Exam Review: Cell Processes and Applications January 2003 Use the following diagram to answer questions 1 and 2. 1. Which labelled organelle produces most of the
Name: Date: Block: Biology 12 Provincial Exam Review: Cell Processes and Applications January 2003 Use the following diagram to answer questions 1 and 2. 1. Which labelled organelle produces most of the
Item Catalog Number Manufacturer 1,4-Dithioerythritol (1 g) D9680 Sigma-Aldrich
 SOP: Nuclei isolation from fresh mouse tissues and DNaseI treatment Date modified: 01/12/2011 Modified by: E. Giste/ T. Canfield (UW) The following protocol describes the isolation of nuclei and subsequent
SOP: Nuclei isolation from fresh mouse tissues and DNaseI treatment Date modified: 01/12/2011 Modified by: E. Giste/ T. Canfield (UW) The following protocol describes the isolation of nuclei and subsequent
Chapter 7. (7-1 and 7-2) A Tour of the Cell
 Chapter 7 (7-1 and 7-2) A Tour of the Cell Microscopes as Windows to the World of Cells Cells were first described in 1665 by Robert Hooke. By the mid-1800s, the accumulation of scientific evidence led
Chapter 7 (7-1 and 7-2) A Tour of the Cell Microscopes as Windows to the World of Cells Cells were first described in 1665 by Robert Hooke. By the mid-1800s, the accumulation of scientific evidence led
FIRST MIDTERM EXAMINATION
 FIRST MIDTERM EXAMINATION 1. True or false: because enzymes are produced by living organisms and because they allow chemical reactions to occur that would not otherwise occur, enzymes represent an exception
FIRST MIDTERM EXAMINATION 1. True or false: because enzymes are produced by living organisms and because they allow chemical reactions to occur that would not otherwise occur, enzymes represent an exception
Explain the reason for this difference in resolving power.
 1. (a) An electron microscope has a much greater resolving power than an optical microscope. (i) Explain the meaning of the term resolving power. Explain the reason for this difference in resolving power.
1. (a) An electron microscope has a much greater resolving power than an optical microscope. (i) Explain the meaning of the term resolving power. Explain the reason for this difference in resolving power.
CONTROLLED PROTEOLYSIS OF NASCENT POLYPEPTIDES IN RAT LIVER CELL FRACTIONS. I. Location of the Polypeptides within Ribosomes
 CONTROLLED PROTEOLYSIS OF NASCENT POLYPEPTIDES IN RAT LIVER CELL FRACTIONS I. Location of the Polypeptides within Ribosomes G. BLOBEL and D. D. SABATI NI From The Rockefeller University, New York 10021
CONTROLLED PROTEOLYSIS OF NASCENT POLYPEPTIDES IN RAT LIVER CELL FRACTIONS I. Location of the Polypeptides within Ribosomes G. BLOBEL and D. D. SABATI NI From The Rockefeller University, New York 10021
Human Saliva as a Convenient Source of Ribonuclease. By S. BRADBURY
 Human Saliva as a Convenient Source of Ribonuclease 323 By S. BRADBURY (From the Cytological Laboratory, Department of Zoology, University Museum, Oxford) SUMMARY Saliva, heated to 80 C for 10 minutes
Human Saliva as a Convenient Source of Ribonuclease 323 By S. BRADBURY (From the Cytological Laboratory, Department of Zoology, University Museum, Oxford) SUMMARY Saliva, heated to 80 C for 10 minutes
CELL PART OF THE DAY. Chapter 7: Cell Structure and Function
 CELL PART OF THE DAY Chapter 7: Cell Structure and Function Cell Membrane Cell membranes are composed of two phospholipid layers. Cell membrane is flexible, not rigid The cell membrane has two major functions.
CELL PART OF THE DAY Chapter 7: Cell Structure and Function Cell Membrane Cell membranes are composed of two phospholipid layers. Cell membrane is flexible, not rigid The cell membrane has two major functions.
Translation Activity Guide
 Translation Activity Guide Student Handout β-globin Translation Translation occurs in the cytoplasm of the cell and is defined as the synthesis of a protein (polypeptide) using information encoded in an
Translation Activity Guide Student Handout β-globin Translation Translation occurs in the cytoplasm of the cell and is defined as the synthesis of a protein (polypeptide) using information encoded in an
SACE Stage 2 Biology Notes - Cells
 SACE Biology Year 2016 Mark 20.00 Pages 26 Published Jan 4, 2017 SACE Stage 2 Biology Notes - Cells By Elizabeth (99.75 ATAR) Powered by TCPDF (www.tcpdf.org) Your notes author, Elizabeth. Elizabeth achieved
SACE Biology Year 2016 Mark 20.00 Pages 26 Published Jan 4, 2017 SACE Stage 2 Biology Notes - Cells By Elizabeth (99.75 ATAR) Powered by TCPDF (www.tcpdf.org) Your notes author, Elizabeth. Elizabeth achieved
Objectives: Prof.Dr. H.D.El-Yassin
 Protein Synthesis and drugs that inhibit protein synthesis Objectives: 1. To understand the steps involved in the translation process that leads to protein synthesis 2. To understand and know about all
Protein Synthesis and drugs that inhibit protein synthesis Objectives: 1. To understand the steps involved in the translation process that leads to protein synthesis 2. To understand and know about all
Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab)
 Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab) Updated 12/3/02 Reagents: ChIP sonication Buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mm Tris 8.1, 150 mm NaCl, 5 mm EDTA): 10 ml 10 % Triton
Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab) Updated 12/3/02 Reagents: ChIP sonication Buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mm Tris 8.1, 150 mm NaCl, 5 mm EDTA): 10 ml 10 % Triton
Cell Structure. Present in animal cell. Present in plant cell. Organelle. Function. strength, resist pressure created when water enters
 Cell Structure Though eukaryotic cells contain many organelles, it is important to know which are in plant cells, which are in animal cells and what their functions are. Organelle Present in plant cell
Cell Structure Though eukaryotic cells contain many organelles, it is important to know which are in plant cells, which are in animal cells and what their functions are. Organelle Present in plant cell
A Tour of the Cell. reference: Chapter 6. Reference: Chapter 2
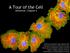 A Tour of the Cell reference: Chapter 6 Reference: Chapter 2 Monkey Fibroblast Cells stained with fluorescent dyes to show the nucleus (blue) and cytoskeleton (yellow and red fibers), image courtesy of
A Tour of the Cell reference: Chapter 6 Reference: Chapter 2 Monkey Fibroblast Cells stained with fluorescent dyes to show the nucleus (blue) and cytoskeleton (yellow and red fibers), image courtesy of
TRANSPORT OF AMINO ACIDS IN INTACT 3T3 AND SV3T3 CELLS. Binding Activity for Leucine in Membrane Preparations of Ehrlich Ascites Tumor Cells
 Journal of Supramolecular Structure 4:441 (401)-447 (407) (1976) TRANSPORT OF AMINO ACIDS IN INTACT 3T3 AND SV3T3 CELLS. Binding Activity for Leucine in Membrane Preparations of Ehrlich Ascites Tumor Cells
Journal of Supramolecular Structure 4:441 (401)-447 (407) (1976) TRANSPORT OF AMINO ACIDS IN INTACT 3T3 AND SV3T3 CELLS. Binding Activity for Leucine in Membrane Preparations of Ehrlich Ascites Tumor Cells
Trident Membrane Protein Extraction Kit
 Cat. No. Size Shelf life GTX16373 5/ 20 tests 12 months at the appropriate storage temperatures (see below) Contents Component Storage Amount for 5 tests Amount for 20 tests Buffer A -20 o C 2.5 ml 10
Cat. No. Size Shelf life GTX16373 5/ 20 tests 12 months at the appropriate storage temperatures (see below) Contents Component Storage Amount for 5 tests Amount for 20 tests Buffer A -20 o C 2.5 ml 10
SBI3U7 Cell Structure & Organelles. 2.2 Prokaryotic Cells 2.3 Eukaryotic Cells
 SBI3U7 Cell Structure & Organelles 2.2 Prokaryotic Cells 2.3 Eukaryotic Cells No nucleus Prokaryotic Cells No membrane bound organelles Has a nucleus Eukaryotic Cells Membrane bound organelles Unicellular
SBI3U7 Cell Structure & Organelles 2.2 Prokaryotic Cells 2.3 Eukaryotic Cells No nucleus Prokaryotic Cells No membrane bound organelles Has a nucleus Eukaryotic Cells Membrane bound organelles Unicellular
Introduction. Biochemistry: It is the chemistry of living things (matters).
 Introduction Biochemistry: It is the chemistry of living things (matters). Biochemistry provides fundamental understanding of the molecular basis for the function and malfunction of living things. Biochemistry
Introduction Biochemistry: It is the chemistry of living things (matters). Biochemistry provides fundamental understanding of the molecular basis for the function and malfunction of living things. Biochemistry
Cell Structure & Function. Source:
 Cell Structure & Function Source: http://koning.ecsu.ctstateu.edu/cell/cell.html Definition of Cell A cell is the smallest unit that is capable of performing life functions. http://web.jjay.cuny.edu/~acarpi/nsc/images/cell.gif
Cell Structure & Function Source: http://koning.ecsu.ctstateu.edu/cell/cell.html Definition of Cell A cell is the smallest unit that is capable of performing life functions. http://web.jjay.cuny.edu/~acarpi/nsc/images/cell.gif
Chapter 1 Plasma membranes
 1 of 5 TEXTBOOK ANSWERS Chapter 1 Plasma membranes Recap 1.1 1 The plasma membrane: keeps internal contents of the cell confined to one area keeps out foreign molecules that damage or destroy the cell
1 of 5 TEXTBOOK ANSWERS Chapter 1 Plasma membranes Recap 1.1 1 The plasma membrane: keeps internal contents of the cell confined to one area keeps out foreign molecules that damage or destroy the cell
(A) Cell membrane (B) Ribosome (C) DNA (D) Nucleus (E) Plasmids. A. Incorrect! Both prokaryotic and eukaryotic cells have cell membranes.
 High School Biology - Problem Drill 03: The Cell No. 1 of 10 1. Which of the following is NOT found in prokaryotic cells? #01 (A) Cell membrane (B) Ribosome (C) DNA (D) Nucleus (E) Plasmids Both prokaryotic
High School Biology - Problem Drill 03: The Cell No. 1 of 10 1. Which of the following is NOT found in prokaryotic cells? #01 (A) Cell membrane (B) Ribosome (C) DNA (D) Nucleus (E) Plasmids Both prokaryotic
Ultrastructure of Mycoplasmatales Virus laidlawii x
 J. gen. Virol. (1972), I6, 215-22I Printed in Great Britain 2I 5 Ultrastructure of Mycoplasmatales Virus laidlawii x By JUDY BRUCE, R. N. GOURLAY, AND D. J. GARWES R. HULL* Agricultural Research Council,
J. gen. Virol. (1972), I6, 215-22I Printed in Great Britain 2I 5 Ultrastructure of Mycoplasmatales Virus laidlawii x By JUDY BRUCE, R. N. GOURLAY, AND D. J. GARWES R. HULL* Agricultural Research Council,
Don t Freak Out. Test on cell organelle on Friday!
 Cell Structure 1 Don t Freak Out Test on cell organelle on Friday! This test should be a buffer test and help raise your overall test score. All information will come from this week! 2 Cells Provide Compartments
Cell Structure 1 Don t Freak Out Test on cell organelle on Friday! This test should be a buffer test and help raise your overall test score. All information will come from this week! 2 Cells Provide Compartments
Notes Chapter 7 Cell Structure and Function Hooke looked at cork under a simple microscope and found tiny chambers he named cells.
 Notes Chapter 7 Cell Structure and Function 7.1 Cell discovery and Theory 1665 Hooke looked at cork under a simple microscope and found tiny chambers he named cells. Cells are the basic structural and
Notes Chapter 7 Cell Structure and Function 7.1 Cell discovery and Theory 1665 Hooke looked at cork under a simple microscope and found tiny chambers he named cells. Cells are the basic structural and
Chapter 4. A Tour of the Cell. Lectures by Edward J. Zalisko
 Chapter 4 A Tour of the Cell PowerPoint Lectures for Campbell Essential Biology, Fifth Edition, and Campbell Essential Biology with Physiology, Fourth Edition Eric J. Simon, Jean L. Dickey, and Jane B.
Chapter 4 A Tour of the Cell PowerPoint Lectures for Campbell Essential Biology, Fifth Edition, and Campbell Essential Biology with Physiology, Fourth Edition Eric J. Simon, Jean L. Dickey, and Jane B.
Replication of Sindbis Virus V. Polyribosomes and mrna in Infected Cells
 JOURNAL OF VIROLOGY, Sept. 1974, p. 552-559 Vol. 14, No. 3 Copyright @ 1974 American Society for Microbiology Printed in U.S.A. Replication of Sindbis Virus V. Polyribosomes and mrna in Infected Cells
JOURNAL OF VIROLOGY, Sept. 1974, p. 552-559 Vol. 14, No. 3 Copyright @ 1974 American Society for Microbiology Printed in U.S.A. Replication of Sindbis Virus V. Polyribosomes and mrna in Infected Cells
ab CytoPainter Golgi/ER Staining Kit
 ab139485 CytoPainter Golgi/ER Staining Kit Instructions for Use Designed to detect Golgi bodies and endoplasmic reticulum by microscopy This product is for research use only and is not intended for diagnostic
ab139485 CytoPainter Golgi/ER Staining Kit Instructions for Use Designed to detect Golgi bodies and endoplasmic reticulum by microscopy This product is for research use only and is not intended for diagnostic
ab Membrane Fractionation Kit Instructions for Use For the rapid and simple separation of membrane, cytosolic and nuclear cellular fractions.
 ab139409 Membrane Fractionation Kit Instructions for Use For the rapid and simple separation of membrane, cytosolic and nuclear cellular fractions. This product is for research use only and is not intended
ab139409 Membrane Fractionation Kit Instructions for Use For the rapid and simple separation of membrane, cytosolic and nuclear cellular fractions. This product is for research use only and is not intended
Cell Theory. Passive Transport
 Cell Theory 4 basic concepts of cell theory are: Cells are the units of structure (building blocks) of all animals and plants. Cells are the smallest unit of function in all animals and plants. Cells originate
Cell Theory 4 basic concepts of cell theory are: Cells are the units of structure (building blocks) of all animals and plants. Cells are the smallest unit of function in all animals and plants. Cells originate
Cell Cell
 Go to cellsalive.com. Select Interactive Cell Models: Plant and Animal. Fill in the information on Plant and Animal Organelles, then Click on Start the Animation Select Plant or Animal Cell below the box.
Go to cellsalive.com. Select Interactive Cell Models: Plant and Animal. Fill in the information on Plant and Animal Organelles, then Click on Start the Animation Select Plant or Animal Cell below the box.
Starch grains - excess sugars
 (a) Membrane system - site of light reactions (photosynthesis) - chlorpophyll pigments - enzymes - electron carriers - flattened, fluid-filled sacs (called thylakoids which are stacked to form grana) -
(a) Membrane system - site of light reactions (photosynthesis) - chlorpophyll pigments - enzymes - electron carriers - flattened, fluid-filled sacs (called thylakoids which are stacked to form grana) -
Instructions for Use. APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests
 3URGXFW,QIRUPDWLRQ Sigma TACS Annexin V Apoptosis Detection Kits Instructions for Use APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests For Research Use Only. Not for use in diagnostic procedures.
3URGXFW,QIRUPDWLRQ Sigma TACS Annexin V Apoptosis Detection Kits Instructions for Use APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests For Research Use Only. Not for use in diagnostic procedures.
PMT. Contains ribosomes attached to the endoplasmic reticulum. Genetic material consists of linear chromosomes. Diameter of the cell is 1 µm
 1. (a) Complete each box in the table, which compares a prokaryotic and a eukaryotic cell, with a tick if the statement is correct or a cross if it is incorrect. Prokaryotic cell Eukaryotic cell Contains
1. (a) Complete each box in the table, which compares a prokaryotic and a eukaryotic cell, with a tick if the statement is correct or a cross if it is incorrect. Prokaryotic cell Eukaryotic cell Contains
TRANSFER OF PREMELANOSOMES INTO THE KERATINIZING CELLS OF ALBINO HAIR FOLLICLE
 TRANSFER OF PREMELANOSOMES INTO THE KERATINIZING CELLS OF ALBINO HAIR FOLLICLE PAUL F. PARAKKAL. From the Department of Dermatology, Boston University School of Medicine, Boston, Massachusetts 02118 INTRODUCTION
TRANSFER OF PREMELANOSOMES INTO THE KERATINIZING CELLS OF ALBINO HAIR FOLLICLE PAUL F. PARAKKAL. From the Department of Dermatology, Boston University School of Medicine, Boston, Massachusetts 02118 INTRODUCTION
ab Lipid Peroxidation (MDA) Assay kit (Colorimetric/ Fluorometric)
 Version 10b Last updated 19 December 2018 ab118970 Lipid Peroxidation (MDA) Assay kit (Colorimetric/ Fluorometric) For the measurement of Lipid Peroxidation in plasma, cell culture and tissue extracts.
Version 10b Last updated 19 December 2018 ab118970 Lipid Peroxidation (MDA) Assay kit (Colorimetric/ Fluorometric) For the measurement of Lipid Peroxidation in plasma, cell culture and tissue extracts.
Laboratory 8 Succinate Dehydrogenase Activity in Cauliflower Mitochondria
 BIO354: Cell Biology Laboratory 1 I. Introduction Laboratory 8 Succinate Dehydrogenase Activity in Cauliflower Mitochondria In eukaryotic cells, specific functions are localized to different types of organelles.
BIO354: Cell Biology Laboratory 1 I. Introduction Laboratory 8 Succinate Dehydrogenase Activity in Cauliflower Mitochondria In eukaryotic cells, specific functions are localized to different types of organelles.
Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
 Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Catalog Number KA1538 48 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use...
Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Catalog Number KA1538 48 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use...
3- Cell Structure and Function How do things move in and out of cells? A Quick Review Taft College Human Physiology
 3- Cell Structure and Function How do things move in and out of cells? A Quick Review Taft College Human Physiology How do things move in and out of cells? Things may move through cell membranes by Passive
3- Cell Structure and Function How do things move in and out of cells? A Quick Review Taft College Human Physiology How do things move in and out of cells? Things may move through cell membranes by Passive
10/13/11. Cell Theory. Cell Structure
 Cell Structure Grade 12 Biology Cell Theory All organisms are composed of one or more cells. Cells are the smallest living units of all living organisms. Cells arise only by division of a previously existing
Cell Structure Grade 12 Biology Cell Theory All organisms are composed of one or more cells. Cells are the smallest living units of all living organisms. Cells arise only by division of a previously existing
Published Online: 25 November, 1956 Supp Info: on November 16, 2018 jcb.rupress.org Downloaded from
 Published Online: 25 November, 1956 Supp Info: http://doi.org/10.1083/jcb.2.6.799 Downloaded from jcb.rupress.org on November 16, 2018 B~IEF NOrmS 799 Permanganate--A New Fixative for Electron Microscopy.*
Published Online: 25 November, 1956 Supp Info: http://doi.org/10.1083/jcb.2.6.799 Downloaded from jcb.rupress.org on November 16, 2018 B~IEF NOrmS 799 Permanganate--A New Fixative for Electron Microscopy.*
Electron Microscopy of Small Cells: Mycoplasma hominis
 JOURNAL of BAcTRiowOY, Dc. 1969, p. 1402-1408 Copyright 0 1969 American Society for Microbiology Vol. 100, No. 3 Printed In U.S.A. NOTES Electron Microscopy of Small Cells: Mycoplasma hominis JACK MANILOFF
JOURNAL of BAcTRiowOY, Dc. 1969, p. 1402-1408 Copyright 0 1969 American Society for Microbiology Vol. 100, No. 3 Printed In U.S.A. NOTES Electron Microscopy of Small Cells: Mycoplasma hominis JACK MANILOFF
Chromatin IP (Isw2) Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles.
 Chromatin IP (Isw2) 7/01 Toshi last update: 06/15 Reagents Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles. 2.5 M glycine. TBS:
Chromatin IP (Isw2) 7/01 Toshi last update: 06/15 Reagents Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles. 2.5 M glycine. TBS:
The Cell Organelles. Eukaryotic cell. The plasma membrane separates the cell from the environment. Plasma membrane: a cell s boundary
 Eukaryotic cell The Cell Organelles Enclosed by plasma membrane Subdivided into membrane bound compartments - organelles One of the organelles is membrane bound nucleus Cytoplasm contains supporting matrix
Eukaryotic cell The Cell Organelles Enclosed by plasma membrane Subdivided into membrane bound compartments - organelles One of the organelles is membrane bound nucleus Cytoplasm contains supporting matrix
The endoplasmic reticulum is a network of folded membranes that form channels through the cytoplasm and sacs called cisternae.
 Endoplasmic reticulum (ER) The endoplasmic reticulum is a network of folded membranes that form channels through the cytoplasm and sacs called cisternae. Cisternae serve as channels for the transport of
Endoplasmic reticulum (ER) The endoplasmic reticulum is a network of folded membranes that form channels through the cytoplasm and sacs called cisternae. Cisternae serve as channels for the transport of
BIL 256 Cell and Molecular Biology Lab Spring, Tissue-Specific Isoenzymes
 BIL 256 Cell and Molecular Biology Lab Spring, 2007 Background Information Tissue-Specific Isoenzymes A. BIOCHEMISTRY The basic pattern of glucose oxidation is outlined in Figure 3-1. Glucose is split
BIL 256 Cell and Molecular Biology Lab Spring, 2007 Background Information Tissue-Specific Isoenzymes A. BIOCHEMISTRY The basic pattern of glucose oxidation is outlined in Figure 3-1. Glucose is split
The Microscopic World of Cells. The Microscopic World of Cells. The Microscopic World of Cells 9/21/2012
 Organisms are either: Single-celled, such as most prokaryotes and protists or Multicelled, such as plants, animals, and most fungi How do we study cells? Light microscopes can be used to explore the structures
Organisms are either: Single-celled, such as most prokaryotes and protists or Multicelled, such as plants, animals, and most fungi How do we study cells? Light microscopes can be used to explore the structures
virus-i (RAV-1) or Rous associated virus-2 (RAV-2), do not transform but do produce
 ISOLATION OF NONINFECTIOUS PARTICLES CONTAINING ROUS SARCOMA VIRUS RNA FROM THE MEDIUM OF ROUS SARCOMA VIRUS-TRANSFORMED NONPRODUCER CELLS* BY HARRIET LATHAM ROBINSONt VIRUS LABORATORY, UNIVERSITY OF CALIFORNIA,
ISOLATION OF NONINFECTIOUS PARTICLES CONTAINING ROUS SARCOMA VIRUS RNA FROM THE MEDIUM OF ROUS SARCOMA VIRUS-TRANSFORMED NONPRODUCER CELLS* BY HARRIET LATHAM ROBINSONt VIRUS LABORATORY, UNIVERSITY OF CALIFORNIA,
E.Z.N.A. SQ Blood DNA Kit II. Table of Contents
 E.Z.N.A. SQ Blood DNA Kit II Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Blood Storage and DNA Yield...4 Preparing Reagents...5 100-500 μl Whole Blood Protocol...6
E.Z.N.A. SQ Blood DNA Kit II Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Blood Storage and DNA Yield...4 Preparing Reagents...5 100-500 μl Whole Blood Protocol...6
For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format:
 Supplementary Protocol 1. Adaptor preparation: For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format: Per reaction X96 10X NEBuffer 2 10 µl 10 µl x 96 5 -GATC
Supplementary Protocol 1. Adaptor preparation: For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format: Per reaction X96 10X NEBuffer 2 10 µl 10 µl x 96 5 -GATC
Midi Plant Genomic DNA Purification Kit
 Midi Plant Genomic DNA Purification Kit Cat #:DP022MD/ DP022MD-50 Size:10/50 reactions Store at RT For research use only 1 Description: The Midi Plant Genomic DNA Purification Kit provides a rapid, simple
Midi Plant Genomic DNA Purification Kit Cat #:DP022MD/ DP022MD-50 Size:10/50 reactions Store at RT For research use only 1 Description: The Midi Plant Genomic DNA Purification Kit provides a rapid, simple
Yara shwabkeh. Osama Alkhader. Heba Kalbouneh
 2 Yara shwabkeh Osama Alkhader Heba Kalbouneh CELL OVERVIEW -Note ; the important thing is to know how the organelles appear under the microscope - the stains we usually use in Histology are composed of
2 Yara shwabkeh Osama Alkhader Heba Kalbouneh CELL OVERVIEW -Note ; the important thing is to know how the organelles appear under the microscope - the stains we usually use in Histology are composed of
Supplementary Information
 Supplementary Information Structural aspects of messenger RNA maintenance by the ribosome Lasse B. Jenner 1,2, Natalia Demeshkina 1,3, Gulnara Yusupova 1,3* and Marat Yusupov 1,3*. 1 Institut de Génétique
Supplementary Information Structural aspects of messenger RNA maintenance by the ribosome Lasse B. Jenner 1,2, Natalia Demeshkina 1,3, Gulnara Yusupova 1,3* and Marat Yusupov 1,3*. 1 Institut de Génétique
Cells & Cell Organelles
 Cells & Cell Organelles The Building Blocks of Life AP Biology 2008-2009 Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal
Cells & Cell Organelles The Building Blocks of Life AP Biology 2008-2009 Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal
Study Guide for Biology Chapter 5
 Class: Date: Study Guide for Biology Chapter 5 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Which of the following led to the discovery of cells? a.
Class: Date: Study Guide for Biology Chapter 5 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Which of the following led to the discovery of cells? a.
Objectives. By the end of the lesson you should be able to: State the 2 types of cells Relate the structure to function for all the organelles
 Biology 11 THE Cell Objectives By the end of the lesson you should be able to: State the 2 types of cells Relate the structure to function for all the organelles Types of Cells There are two types of cells:
Biology 11 THE Cell Objectives By the end of the lesson you should be able to: State the 2 types of cells Relate the structure to function for all the organelles Types of Cells There are two types of cells:
Structures in Cells. Cytoplasm. Lecture 5, EH1008: Biology for Public Health, Biomolecules
 Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
EXPERIMENT 13: Isolation and Characterization of Erythrocyte
 EXPERIMENT 13: Isolation and Characterization of Erythrocyte Day 1: Isolation of Erythrocyte Steps 1 through 6 of the Switzer & Garrity protocol (pages 220-221) have been performed by the TA. We will be
EXPERIMENT 13: Isolation and Characterization of Erythrocyte Day 1: Isolation of Erythrocyte Steps 1 through 6 of the Switzer & Garrity protocol (pages 220-221) have been performed by the TA. We will be
Membranes of Saccharomyces cerevisiae
 JOURNAL OF BACTERIOLOGY, Aug. 1967, p. 475-481 Vol. 94, No. 2 Copyright 1967 American Society for Microbiology Printed in U.S.A. Membranes of Saccharomyces cerevisiae HAROLD P. KLEIN, CAROL VOLKMANN, AND
JOURNAL OF BACTERIOLOGY, Aug. 1967, p. 475-481 Vol. 94, No. 2 Copyright 1967 American Society for Microbiology Printed in U.S.A. Membranes of Saccharomyces cerevisiae HAROLD P. KLEIN, CAROL VOLKMANN, AND
Links amino acids / synthesises / makes protein; C Involved in modifying / packaging protein / forms glycoproteins / forms vesicles; 3
 1. (a) A Carries the (genetic) code / genetic instructions / DNA / makes mrna / transcription / makes ribosomes; B Links amino acids / synthesises / makes protein; C Involved in modifying / packaging protein
1. (a) A Carries the (genetic) code / genetic instructions / DNA / makes mrna / transcription / makes ribosomes; B Links amino acids / synthesises / makes protein; C Involved in modifying / packaging protein
ELECTRON MICROSCOPIC STUDY OF THE FORMATION OF BLUETONGUE VIRUS*
 Onderstepoort J. vet. Res. (1968), 35 (1), 139-150 Printed in the Repub. of S. Afr. by The Government Printer, Pretoria ELECTRON MICROSCOPIC STUDY OF THE FORMATION OF BLUETONGUE VIRUS* G. LECATSAS, Veterinary
Onderstepoort J. vet. Res. (1968), 35 (1), 139-150 Printed in the Repub. of S. Afr. by The Government Printer, Pretoria ELECTRON MICROSCOPIC STUDY OF THE FORMATION OF BLUETONGUE VIRUS* G. LECATSAS, Veterinary
In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question.
 CHAPTER 3 TEST Cell Structure Circle T if the statement is true or F if it is false. T F 1. Small cells can transport materials and information more quickly than larger cells can. T F 2. Newly made proteins
CHAPTER 3 TEST Cell Structure Circle T if the statement is true or F if it is false. T F 1. Small cells can transport materials and information more quickly than larger cells can. T F 2. Newly made proteins
EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
 EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
Intracellular Compartments and Protein Sorting
 Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
Cell Overview. Hanan Jafar BDS.MSc.PhD
 Cell Overview Hanan Jafar BDS.MSc.PhD THE CELL is made of: 1- Nucleus 2- Cell Membrane 3- Cytoplasm THE CELL Formed of: 1. Nuclear envelope 2. Chromatin 3. Nucleolus 4. Nucleoplasm (nuclear matrix) NUCLEUS
Cell Overview Hanan Jafar BDS.MSc.PhD THE CELL is made of: 1- Nucleus 2- Cell Membrane 3- Cytoplasm THE CELL Formed of: 1. Nuclear envelope 2. Chromatin 3. Nucleolus 4. Nucleoplasm (nuclear matrix) NUCLEUS
CELLS and TRANSPORT Student Packet SUMMARY CELL MEMBRANES ARE SELECTIVELY PERMEABLE DUE TO THEIR STRUCTURE Hydrophilic head
 CELLS and TRANSPORT Student Packet SUMMARY CELL MEMBRANES ARE SELECTIVELY PERMEABLE DUE TO THEIR STRUCTURE Hydrophilic head Hydrophobic tail Hydrophobic regions of protein Hydrophilic regions of protein
CELLS and TRANSPORT Student Packet SUMMARY CELL MEMBRANES ARE SELECTIVELY PERMEABLE DUE TO THEIR STRUCTURE Hydrophilic head Hydrophobic tail Hydrophobic regions of protein Hydrophilic regions of protein
OCR (A) Biology GCSE. Topic 1: Cell Level Systems
 OCR (A) Biology GCSE Topic 1: Cell Level Systems Notes (Content in bold is for higher tier only) Cell structures Microscopes (1.1a and c) Light (optical) microscopes The specimen is placed onto a slide,
OCR (A) Biology GCSE Topic 1: Cell Level Systems Notes (Content in bold is for higher tier only) Cell structures Microscopes (1.1a and c) Light (optical) microscopes The specimen is placed onto a slide,
Nucleic acids. Nucleic acids are information-rich polymers of nucleotides
 Nucleic acids Nucleic acids are information-rich polymers of nucleotides DNA and RNA Serve as the blueprints for proteins and thus control the life of a cell RNA and DNA are made up of very similar nucleotides.
Nucleic acids Nucleic acids are information-rich polymers of nucleotides DNA and RNA Serve as the blueprints for proteins and thus control the life of a cell RNA and DNA are made up of very similar nucleotides.
Extracting DNA from cheek cells: a classroom experiment for Year 7 upwards
 Extracting DNA from cheek cells: a classroom experiment for Year 7 upwards Dr Kathryn Scott Research Administrator, Zitzmann Group, Department of Biochemistry Lecturer in Biochemistry, Christ Church Extracting
Extracting DNA from cheek cells: a classroom experiment for Year 7 upwards Dr Kathryn Scott Research Administrator, Zitzmann Group, Department of Biochemistry Lecturer in Biochemistry, Christ Church Extracting
10 The Golgi Apparatus: The First 100 Years
 2 Structure With no cell compartment or organelle has morphology served such a pivotal role in its discovery and investigation as with the apparatus of Golgi. The original description of the apparato reticulo
2 Structure With no cell compartment or organelle has morphology served such a pivotal role in its discovery and investigation as with the apparatus of Golgi. The original description of the apparato reticulo
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
