Membrane Simulations in GROMACS Lipid bilayers and membrane proteins
|
|
|
- Grant Walton
- 5 years ago
- Views:
Transcription
1 Membrane Simulations in GROMACS Lipid bilayers and membrane proteins Justin A. Lemkul September 13, 2013
2 Objectives Learn the fundamentals of Self-consistent force fields Membrane and membrane protein simulations Perform common analysis routines for lipids
3 Background References Theory Kandt, C.L., Ash, W.L., and Tieleman, D.P. (2007) Methods 41: Relevant to today s example Kandasamy, S.K. and Larson, R.G. (2006) Biophys. J. 90: Allen, W.J., Lemkul, J.A., and Bevan, D.R. (2009) J. Comput. Chem. 30: 1952.
4 References Theory Biomolecular force fields often built in stages Proteins, nucleic acids, lipids, other stuff Important to use self-consistent parameters Nowadays most force fields include support for all major macromolecule types
5 References Theory Lipid parameters (NOT exhaustive!) CHARMM36 Klauda, J.B. et al. (2010) J. Phys. Chem. B 114: Pastor, R.W. and MacKerell Jr., A.D. (2011) J. Phys. Chem. Lett. 2: OPLS-AA Ulmschneider, J.P. and Ulmschneider, M.B. (2009) J. Chem. Theory Comput. 5: GROMOS96 Kukol, A. (2009) J. Chem. Theory Comput. 5: 615. AMBER Siu, S.W.I. et al. (2008) J. Chem. Phys. 128:
6 Tutorial files provided membrane_tutorial/conf Sample configurations /data Sample data /gromos53a6_lipid.ff Force field /mdp parameters /scripts Accessory scripts /top Topologies /tpr input files Protocol online
7 Challenge #1: selecting a force field Challenge #2: building the starting configuration InflateGRO Kandt, C.L. et al. (2007) Methods 41: 475. Schmidt, T.H. and Kandt, C.L. (2012) J. Chem. Inf. Model. 52: g_membed Wolf, M.G. et al. (2010) J. Comput. Chem. 31: 2169.
8 Placing a protein: editconf center Protein COM coincident with membrane COM Protein COM above membrane COM
9 Placing a protein: editconf center editconf center x/2 y/2 z/2 box x y z Protein COM coincident with membrane COM Protein COM above membrane COM
10 Placing a protein: editconf center editconf center x/2 y/2 z/2 box x y z g_traj n p8.ndx ox nox noy z com editconf center x/2 y/2 z* -box x y z Protein COM coincident with membrane COM Protein COM above membrane COM
11 Topology generation: different approaches With CHARMM, simply use pdb2gmx Check contents of lipid.rtp With others, introduce lipid-specific terms lipid.itp (Berger parameters) Approach used here to combine Berger lipids with GROMOS96 More complicated procedure for combining allatom proteins with united-atom lipids
12 ffnonbonded.itp [ defaults ] [ atomtypes ] [ nonbond_params] [ pairtypes ] lipid.itp [ defaults ] [ atomtypes ] [ nonbond_params] Lipid-Lipid terms Lipid-GROMOS terms [ pairtypes ] ffbonded.itp [ bondtypes ] [ angletypes ] [ dihedraltypes ] [ dihedraltypes ]
13 Membrane-specific run settings comm_grps tc_grps ref_t for tens or hundreds of ns
14 Notes on cutoffs Berger: plain 1.0-nm cutoffs GROMOS96: 0.9/1.4-nm twin-range Here we use 1.2-nm (single-range) APL and membrane properties unaffected Peptide secondary structure
15 Analysis techniques to be covered Lateral diffusion/diffusion constant Membrane thickness Deuterium order parameters Area per lipid
16 Index groups Lateral diffusion: P8 atoms Membrane thickness: P8 atoms in each leaflet Deuterium order parameters: each acyl C atom
17 All P8 atoms Use make_ndx or g_select In make_ndx: a P8 In g_select: name P8; Top and bottom leaflet P8 atoms Use g_select (file p8_selection.dat) top_p8 = name P8 and (z > 3.2); bot_p8 = name P8 and (z < 3.2); top_p8; bot_p8; About middle of box
18 Acyl chain C atoms use make_ndx sn-1 and sn-2 chains separately a C34 a C36 a C37 a C50 a C15 a C17 a C18 a C31
19 Lateral diffusion echo 0 g_msd s md.tpr f traj.xtc n all_p8.ndx lateral z o msd_p8.xvg May have to deal with PBC separately for each leaflet Membrane thickness echo 0 1 g_dist s md.tpr f traj.xtc n top_bot_p8.ndx Only z-distance important (x/y should be 0) May not tell the whole story
20 Deuterium order parameters g_order s md.tpr f traj.xtc n sn1.ndx od deuter_sn1.xvg d z g_order s md.tpr f traj.xtc n sn2.ndx od deuter_sn2.xvg d z Important to consider equilibration time, usually need b Unsaturated lipids require unsat option separately for double bond atoms C n-1 C n =C n+1 C n+2
21 Area per lipid Trivial for pure membranes (Box-X * Box-Y)/(# of lipids per leaflet) Use g_energy to extract Box-X and Box-Y g_energy f ener.edr o box.xvg perl scripts/area.pl box.xvg 64
22 Area per lipid (and membrane thickness) Complicated problem if a protein is embedded Shameless self-promotion: GridMAT-MD Allen, W.J., Lemkul, J.A., and Bevan, D.R. (2009) J. Comput. Chem. 30: 1952.
Supplementary Information: A Critical. Comparison of Biomembrane Force Fields: Structure and Dynamics of Model DMPC, POPC, and POPE Bilayers
 Supplementary Information: A Critical Comparison of Biomembrane Force Fields: Structure and Dynamics of Model DMPC, POPC, and POPE Bilayers Kristyna Pluhackova,, Sonja A. Kirsch, Jing Han, Liping Sun,
Supplementary Information: A Critical Comparison of Biomembrane Force Fields: Structure and Dynamics of Model DMPC, POPC, and POPE Bilayers Kristyna Pluhackova,, Sonja A. Kirsch, Jing Han, Liping Sun,
Supporting Information
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 018 Supporting Information Modulating Interactions between Ligand-Coated Nanoparticles and Phase-Separated
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 018 Supporting Information Modulating Interactions between Ligand-Coated Nanoparticles and Phase-Separated
Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale
 Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale Rakesh Gupta and Beena Rai* TATA Research Development & Design Centre, TCS Innovation labs, Pune India,
Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale Rakesh Gupta and Beena Rai* TATA Research Development & Design Centre, TCS Innovation labs, Pune India,
Bilayer Deformation, Pores & Micellation Induced by Oxidized Lipids
 Supporting Information Bilayer Deformation, Pores & Micellation Induced by Oxidized Lipids Phansiri Boonnoy 1, Viwan Jarerattanachat 1,2, Mikko Karttunen 3*, and Jirasak Wongekkabut 1* 1 Department of
Supporting Information Bilayer Deformation, Pores & Micellation Induced by Oxidized Lipids Phansiri Boonnoy 1, Viwan Jarerattanachat 1,2, Mikko Karttunen 3*, and Jirasak Wongekkabut 1* 1 Department of
GridMAT-MD: A Grid-based Membrane Analysis Tool for use with Molecular Dynamics
 GridMAT-MD: A Grid-based Membrane Analysis Tool for use with Molecular Dynamics William J. Allen, Justin A. Lemkul, and David R. Bevan Department of Biochemistry, Virginia Tech User s Guide Version 1.0.2
GridMAT-MD: A Grid-based Membrane Analysis Tool for use with Molecular Dynamics William J. Allen, Justin A. Lemkul, and David R. Bevan Department of Biochemistry, Virginia Tech User s Guide Version 1.0.2
Conformational Flexibility of the Peptide Hormone Ghrelin in Solution and Lipid Membrane Bound: A Molecular Dynamics Study
 Open Access Article The authors, the publisher, and the right holders grant the right to use, reproduce, and disseminate the work in digital form to all users. Journal of Biomolecular Structure & Dynamics,
Open Access Article The authors, the publisher, and the right holders grant the right to use, reproduce, and disseminate the work in digital form to all users. Journal of Biomolecular Structure & Dynamics,
We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field
 Parameterization of the fullerene coarse-grained model We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field for lipids 1 and proteins 2. In the MARTINI force
Parameterization of the fullerene coarse-grained model We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field for lipids 1 and proteins 2. In the MARTINI force
Simulationen von Lipidmembranen
 Simulationen von Lipidmembranen Thomas Stockner Thomas.stockner@meduniwien.ac.at Summary Methods Force Field MD simulations Membrane simulations Application Oxidized lipids Anesthetics Molecular biology
Simulationen von Lipidmembranen Thomas Stockner Thomas.stockner@meduniwien.ac.at Summary Methods Force Field MD simulations Membrane simulations Application Oxidized lipids Anesthetics Molecular biology
Massive oxidation of phospholipid membranes. leads to pore creation and bilayer. disintegration
 Massive oxidation of phospholipid membranes leads to pore creation and bilayer disintegration Lukasz Cwiklik and Pavel Jungwirth Institute of Organic Chemistry and Biochemistry, Academy of Sciences of
Massive oxidation of phospholipid membranes leads to pore creation and bilayer disintegration Lukasz Cwiklik and Pavel Jungwirth Institute of Organic Chemistry and Biochemistry, Academy of Sciences of
Supporting material. Membrane permeation induced by aggregates of human islet amyloid polypeptides
 Supporting material Membrane permeation induced by aggregates of human islet amyloid polypeptides Chetan Poojari Forschungszentrum Jülich GmbH, Institute of Complex Systems: Structural Biochemistry (ICS-6),
Supporting material Membrane permeation induced by aggregates of human islet amyloid polypeptides Chetan Poojari Forschungszentrum Jülich GmbH, Institute of Complex Systems: Structural Biochemistry (ICS-6),
Molecular dynamics investigation of myelin basic protein stability on lipid membranes
 Molecular dynamics investigation of myelin basic protein stability on lipid membranes Kyrylo Bessonov, George Harauz* *This study was conducted under the supervision of Professor George Harauz, Department
Molecular dynamics investigation of myelin basic protein stability on lipid membranes Kyrylo Bessonov, George Harauz* *This study was conducted under the supervision of Professor George Harauz, Department
Simulationen von Lipidmembranen
 Simulationen von Lipidmembranen Thomas Stockner thomas.stockner@meduniwien.ac.at Molecular biology Molecular modelling Membranes environment Many cellular functions occur in or around membranes: energy
Simulationen von Lipidmembranen Thomas Stockner thomas.stockner@meduniwien.ac.at Molecular biology Molecular modelling Membranes environment Many cellular functions occur in or around membranes: energy
g_membed: Efficient Insertion of a Membrane Protein into an Equilibrated Lipid Bilayer with Minimal Perturbation
 g_membed: Efficient Insertion of a Membrane Protein into an Equilibrated Lipid Bilayer with Minimal Perturbation MAARTEN G. WOLF, 1 MARTIN HOEFLING, 2 CAMILO APONTE-SANTAMARÍA, 2 HELMUT GRUBMÜLLER, 2 GERRIT
g_membed: Efficient Insertion of a Membrane Protein into an Equilibrated Lipid Bilayer with Minimal Perturbation MAARTEN G. WOLF, 1 MARTIN HOEFLING, 2 CAMILO APONTE-SANTAMARÍA, 2 HELMUT GRUBMÜLLER, 2 GERRIT
The effect of orientation dynamics in melittin as antimicrobial peptide in lipid bilayer calculated by free energy method
 Journal of Physics: Conference Series PAPER OPEN ACCESS The effect of orientation dynamics in melittin as antimicrobial peptide in lipid bilayer calculated by free energy method To cite this article: Sri
Journal of Physics: Conference Series PAPER OPEN ACCESS The effect of orientation dynamics in melittin as antimicrobial peptide in lipid bilayer calculated by free energy method To cite this article: Sri
Supporting Information for Origin of 1/f noise in hydration dynamics on lipid membrane surfaces
 Supporting Informati for Origin of /f noise in hydrati dynamics lipid membrane surfaces Eiji Yamamoto, Takuma kimoto, Masato Yasui, 2 and Kenji Yasuoka Department of Mechanical Engineering, Keio University,
Supporting Informati for Origin of /f noise in hydrati dynamics lipid membrane surfaces Eiji Yamamoto, Takuma kimoto, Masato Yasui, 2 and Kenji Yasuoka Department of Mechanical Engineering, Keio University,
Interactions of Polyethylenimines with Zwitterionic and. Anionic Lipid Membranes
 Interactions of Polyethylenimines with Zwitterionic and Anionic Lipid Membranes Urszula Kwolek, Dorota Jamróz, Małgorzata Janiczek, Maria Nowakowska, Paweł Wydro, Mariusz Kepczynski Faculty of Chemistry,
Interactions of Polyethylenimines with Zwitterionic and Anionic Lipid Membranes Urszula Kwolek, Dorota Jamróz, Małgorzata Janiczek, Maria Nowakowska, Paweł Wydro, Mariusz Kepczynski Faculty of Chemistry,
Water-phospholipid interactions at the interface of lipid. membranes: comparison of different force fields
 Water-phospholipid interactions at the interface of lipid membranes: comparison of different force fields Jianjun Jiang, 1,2 Weixin Li, 1,3 Liang Zhao, 1 Yuanyan Wu, 1 Peng Xiu, 4 Guoning Tang, 3 and Yusong
Water-phospholipid interactions at the interface of lipid membranes: comparison of different force fields Jianjun Jiang, 1,2 Weixin Li, 1,3 Liang Zhao, 1 Yuanyan Wu, 1 Peng Xiu, 4 Guoning Tang, 3 and Yusong
Supplementary information for Effects of Stretching Speed on. Mechanical Rupture of Phospholipid/Cholesterol Bilayers: Molecular
 Supplementary information for Effects of Stretching Speed on Mechanical Rupture of Phospholipid/Cholesterol Bilayers: Molecular Dynamics Simulation Taiki Shigematsu, Kenichiro Koshiyama*, and Shigeo Wada
Supplementary information for Effects of Stretching Speed on Mechanical Rupture of Phospholipid/Cholesterol Bilayers: Molecular Dynamics Simulation Taiki Shigematsu, Kenichiro Koshiyama*, and Shigeo Wada
g membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation.
 g membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. Maarten G. Wolf, Martin Hoefling, Camilo Aponte-Santamaría, Helmut Grubmüller and Gerrit
g membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. Maarten G. Wolf, Martin Hoefling, Camilo Aponte-Santamaría, Helmut Grubmüller and Gerrit
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. (a) Uncropped version of Fig. 2a. RM indicates that the translation was done in the absence of rough mcirosomes. (b) LepB construct containing the GGPG-L6RL6-
Supplementary Figures Supplementary Figure 1. (a) Uncropped version of Fig. 2a. RM indicates that the translation was done in the absence of rough mcirosomes. (b) LepB construct containing the GGPG-L6RL6-
Physiologically-relevant levels of sphingomyelin, but not GM1, induces a beta-sheet-rich structure in the amyloid-beta(1-42) monomer
 https://helda.helsinki.fi Physiologically-relevant levels of sphingomyelin, but not GM1, induces a beta-sheet-rich structure in the amyloid-beta(1-42) monomer Owen, Michael C. 2018-09 Owen, M C, Kulig,
https://helda.helsinki.fi Physiologically-relevant levels of sphingomyelin, but not GM1, induces a beta-sheet-rich structure in the amyloid-beta(1-42) monomer Owen, Michael C. 2018-09 Owen, M C, Kulig,
Physicochemical Properties of Nanoparticles Regulate Translocation across Pulmonary. Surfactant Monolayer and Formation of Lipoprotein Corona
 Physicochemical Properties of Nanoparticles Regulate Translocation across Pulmonary Surfactant Monolayer and Formation of Lipoprotein Corona Supplementary Information Guoqing Hu 1 *, Bao Jiao 1, Xinghua
Physicochemical Properties of Nanoparticles Regulate Translocation across Pulmonary Surfactant Monolayer and Formation of Lipoprotein Corona Supplementary Information Guoqing Hu 1 *, Bao Jiao 1, Xinghua
Ethanol Induces the Formation of Water-Permeable Defects in Model. Bilayers of Skin Lipids
 Electronic Supplementary Material (ESI) for Chemical Communications. This journal is The Royal Society of Chemistry 2015 Ethanol Induces the Formation of Water-Permeable Defects in Model Bilayers of Skin
Electronic Supplementary Material (ESI) for Chemical Communications. This journal is The Royal Society of Chemistry 2015 Ethanol Induces the Formation of Water-Permeable Defects in Model Bilayers of Skin
Gold nanocrystals at DPPC bilayer. Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad
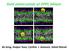 Gold nanocrystals at DPPC bilayer Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad Coarse-grained mapping strategy of a DPPC lipid molecule. The structure of gold nanocrystals (bare gold nanoparticles)
Gold nanocrystals at DPPC bilayer Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad Coarse-grained mapping strategy of a DPPC lipid molecule. The structure of gold nanocrystals (bare gold nanoparticles)
Supplementary Information
 Coarse-Grained Molecular Dynamics Simulations of Photoswitchable Assembly and Disassembly Xiaoyan Zheng, Dong Wang,* Zhigang Shuai,* MOE Key Laboratory of Organic Optoelectronics and Molecular Engineering,
Coarse-Grained Molecular Dynamics Simulations of Photoswitchable Assembly and Disassembly Xiaoyan Zheng, Dong Wang,* Zhigang Shuai,* MOE Key Laboratory of Organic Optoelectronics and Molecular Engineering,
Coarse-Grained Molecular Dynamics Study of Cyclic Peptide Nanotube Insertion into a Lipid Bilayer
 4780 J. Phys. Chem. A 2009, 113, 4780 4787 Coarse-Grained Molecular Dynamics Study of Cyclic Peptide Nanotube Insertion into a Lipid Bilayer Hyonseok Hwang* Department of Chemistry and Institute for Molecular
4780 J. Phys. Chem. A 2009, 113, 4780 4787 Coarse-Grained Molecular Dynamics Study of Cyclic Peptide Nanotube Insertion into a Lipid Bilayer Hyonseok Hwang* Department of Chemistry and Institute for Molecular
Break Desired Compound into 3 Smaller Ones
 Break Desired Compound into 3 Smaller nes A B C Indole ydrazine Phenol When creating a covalent link between model compounds move the charge on the deleted into the carbon to maintain integer charge (i.e.
Break Desired Compound into 3 Smaller nes A B C Indole ydrazine Phenol When creating a covalent link between model compounds move the charge on the deleted into the carbon to maintain integer charge (i.e.
Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes
 Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes Kevin Gasperich Advisor: Dmitry Kopelevich ABSTRACT The recent rapid development of applications for fullerenes has led to an increase
Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes Kevin Gasperich Advisor: Dmitry Kopelevich ABSTRACT The recent rapid development of applications for fullerenes has led to an increase
Computational analysis of local membrane properties
 J Comput Aided Mol Des (2013) 27:845 858 DOI 10.1007/s10822-013-9684-0 Computational analysis of local membrane properties Vytautas Gapsys Bert L. de Groot Rodolfo Briones Received: 27 May 2013 / Accepted:
J Comput Aided Mol Des (2013) 27:845 858 DOI 10.1007/s10822-013-9684-0 Computational analysis of local membrane properties Vytautas Gapsys Bert L. de Groot Rodolfo Briones Received: 27 May 2013 / Accepted:
John M. Leveritt III, Almudena Pino-Angeles, Themis Lazaridis*.
 The structure of a melittin-stabilized pore John M. Leveritt III, Almudena Pino-Angeles, Themis Lazaridis*. Department of Chemistry, The City College of New York, 160 Convent Ave, New York, NY, 10031,
The structure of a melittin-stabilized pore John M. Leveritt III, Almudena Pino-Angeles, Themis Lazaridis*. Department of Chemistry, The City College of New York, 160 Convent Ave, New York, NY, 10031,
Comparing Simulations of Lipid Bilayers to Scattering Data: The GROMOS 43A1-S3 Force Field
 pubs.acs.org/jpcb Comparing Simulations of Lipid Bilayers to Scattering Data: The GROMOS 43A1-S3 Force Field Anthony R. Braun, Jonathan N. Sachs, and John F. Nagle*, Department of Biomedical Engineering,
pubs.acs.org/jpcb Comparing Simulations of Lipid Bilayers to Scattering Data: The GROMOS 43A1-S3 Force Field Anthony R. Braun, Jonathan N. Sachs, and John F. Nagle*, Department of Biomedical Engineering,
Coarse-Grained Molecular Dynamics for Copolymer- Vesicle Self-Assembly. Case Study: Sterically Stabilized Liposomes.
 Coarse-Grained Molecular Dynamics for Copolymer- Vesicle Self-Assembly. Case Study: Sterically Stabilized Liposomes. Alexander Kantardjiev 1 and Pavletta Shestakova 1 1 Institute of Organic Chemistry with
Coarse-Grained Molecular Dynamics for Copolymer- Vesicle Self-Assembly. Case Study: Sterically Stabilized Liposomes. Alexander Kantardjiev 1 and Pavletta Shestakova 1 1 Institute of Organic Chemistry with
MARTINI Coarse-Grained Model of Triton TX-100 in Pure DPPC. Monolayer and Bilayer Interfaces. Supporting Information
 MARTINI Coarse-Grained Model of Triton TX-100 in Pure DPPC Monolayer and Bilayer Interfaces. Antonio Pizzirusso a, Antonio De Nicola* a, Giuseppe Milano a a Dipartimento di Chimica e Biologia, Università
MARTINI Coarse-Grained Model of Triton TX-100 in Pure DPPC Monolayer and Bilayer Interfaces. Antonio Pizzirusso a, Antonio De Nicola* a, Giuseppe Milano a a Dipartimento di Chimica e Biologia, Università
CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field
 CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field The Harvard community has made this article openly available. Please share
CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field The Harvard community has made this article openly available. Please share
Supplementary Information
 Supplementary Information Effect of head group and lipid tail oxidation in the cell membrane revealed through integrated simulations and experiments M Yusupov 1, K Wende 2, S Kupsch, E C Neyts 1, S Reuter
Supplementary Information Effect of head group and lipid tail oxidation in the cell membrane revealed through integrated simulations and experiments M Yusupov 1, K Wende 2, S Kupsch, E C Neyts 1, S Reuter
Carbon Compounds. Lesson Overview. Lesson Overview. 2.3 Carbon Compounds
 Lesson Overview Carbon Compounds Lesson Overview 2.3 THINK ABOUT IT In the early 1800s, many chemists called the compounds created by organisms organic, believing they were fundamentally different from
Lesson Overview Carbon Compounds Lesson Overview 2.3 THINK ABOUT IT In the early 1800s, many chemists called the compounds created by organisms organic, believing they were fundamentally different from
obtained for the simulations of the E2 conformation of SERCA in a pure POPC lipid bilayer (blue) and in a
 Supplementary Figure S1. Distribution of atoms along the bilayer normal. Normalized density profiles obtained for the simulations of the E2 conformation of SERCA in a pure POPC lipid bilayer (blue) and
Supplementary Figure S1. Distribution of atoms along the bilayer normal. Normalized density profiles obtained for the simulations of the E2 conformation of SERCA in a pure POPC lipid bilayer (blue) and
A: All atom molecular simulation systems
 Cholesterol level affects surface charge of lipid membranes in physiological environment Aniket Magarkar a, Vivek Dhawan b, Paraskevi Kallinteri a, Tapani Viitala c, Mohammed Elmowafy c, Tomasz Róg d,
Cholesterol level affects surface charge of lipid membranes in physiological environment Aniket Magarkar a, Vivek Dhawan b, Paraskevi Kallinteri a, Tapani Viitala c, Mohammed Elmowafy c, Tomasz Róg d,
Supporting Information: Revisiting partition in. hydrated bilayer systems
 Supporting Information: Revisiting partition in hydrated bilayer systems João T. S. Coimbra, Pedro A. Fernandes, Maria J. Ramos* UCIBIO, REQUIMTE, Departamento de Química e Bioquímica, Faculdade de Ciências,
Supporting Information: Revisiting partition in hydrated bilayer systems João T. S. Coimbra, Pedro A. Fernandes, Maria J. Ramos* UCIBIO, REQUIMTE, Departamento de Química e Bioquímica, Faculdade de Ciências,
Membranes & Membrane Proteins
 School on Biomolecular Simulations Membranes & Membrane Proteins Vani Vemparala The Institute of Mathematical Sciences Chennai November 13 2007 JNCASR, Bangalore Cellular Environment Plasma membrane extracellular
School on Biomolecular Simulations Membranes & Membrane Proteins Vani Vemparala The Institute of Mathematical Sciences Chennai November 13 2007 JNCASR, Bangalore Cellular Environment Plasma membrane extracellular
Structural Characterization on the Gel to Liquid-Crystal Phase Transition of Fully Hydrated DSPC and DSPE Bilayers
 8114 J. Phys. Chem. B 2009, 113, 8114 8123 Structural Characterization on the Gel to Liquid-Crystal Phase Transition of Fully Hydrated DSPC and DSPE Bilayers Shan-Shan Qin and Zhi-Wu Yu* Key Lab of Bioorganic
8114 J. Phys. Chem. B 2009, 113, 8114 8123 Structural Characterization on the Gel to Liquid-Crystal Phase Transition of Fully Hydrated DSPC and DSPE Bilayers Shan-Shan Qin and Zhi-Wu Yu* Key Lab of Bioorganic
Lesson Overview. Carbon Compounds. Lesson Overview. 2.3 Carbon Compounds
 Lesson Overview 2.3 THINK ABOUT IT In the early 1800s, many chemists called the compounds created by organisms organic, believing they were fundamentally different from compounds in nonliving things. We
Lesson Overview 2.3 THINK ABOUT IT In the early 1800s, many chemists called the compounds created by organisms organic, believing they were fundamentally different from compounds in nonliving things. We
Improved Coarse-Grained Modeling of Cholesterol-Containing Lipid Bilayers
 This is an open access article published under an ACS AuthorChoice License, which permits copying and redistribution of the article or any adaptations for non-commercial purposes. pubs.acs.org/jctc Improved
This is an open access article published under an ACS AuthorChoice License, which permits copying and redistribution of the article or any adaptations for non-commercial purposes. pubs.acs.org/jctc Improved
Biological Membranes. Lipid Membranes. Bilayer Permeability. Common Features of Biological Membranes. A highly selective permeability barrier
 Biological Membranes Structure Function Composition Physicochemical properties Self-assembly Molecular models Lipid Membranes Receptors, detecting the signals from outside: Light Odorant Taste Chemicals
Biological Membranes Structure Function Composition Physicochemical properties Self-assembly Molecular models Lipid Membranes Receptors, detecting the signals from outside: Light Odorant Taste Chemicals
Biochemistry. 2. Besides carbon, name 3 other elements that make up most organic compounds.
 Biochemistry Carbon compounds Section 3-1 1. What is an organic compound? 2. Besides carbon, name 3 other elements that make up most organic compounds. 3. Carbon dioxide, CO 2, is NOT an organic compound.
Biochemistry Carbon compounds Section 3-1 1. What is an organic compound? 2. Besides carbon, name 3 other elements that make up most organic compounds. 3. Carbon dioxide, CO 2, is NOT an organic compound.
INVESTIGATING THE VOLUME PERTURBATION OF NEURAL LIPID-MEMBRANE BY PROTEIN IN THE INITIAL STAGES OF ALZHEIMER S DISEASE
 INVESTIGATING THE VOLUME PERTURBATION OF NEURAL LIPID-MEMBRANE BY PROTEIN IN THE INITIAL STAGES OF ALZHEIMER S DISEASE SANTONA TULI WITH PROJECT COLLABORATORS GARNER COCHRAN AND AUSTIN WORKMAN Date: December
INVESTIGATING THE VOLUME PERTURBATION OF NEURAL LIPID-MEMBRANE BY PROTEIN IN THE INITIAL STAGES OF ALZHEIMER S DISEASE SANTONA TULI WITH PROJECT COLLABORATORS GARNER COCHRAN AND AUSTIN WORKMAN Date: December
in-silico Design of Nanoparticles for Transdermal Drug Delivery Application
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2018 in-silico Design of Nanoparticles for Transdermal Drug Delivery Application Rakesh Gupta and Beena
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2018 in-silico Design of Nanoparticles for Transdermal Drug Delivery Application Rakesh Gupta and Beena
Translocation of C 60 and Its Derivatives Across a Lipid Bilayer
 Translocation of C 60 and Its Derivatives Across a Lipid Bilayer Rui Qiao* NANO LETTERS 2007 Vol. 7, No. 3 614-619 Department of Mechanical Engineering, Clemson UniVersity, Clemson, South Carolina 29634
Translocation of C 60 and Its Derivatives Across a Lipid Bilayer Rui Qiao* NANO LETTERS 2007 Vol. 7, No. 3 614-619 Department of Mechanical Engineering, Clemson UniVersity, Clemson, South Carolina 29634
The. Crash Course. Basically, almost all living things are made up of these 4 Elements: - Carbon (C) - Nitrogen (N) - Hydrogen (H) - Oxygen (O)
 The Biochemistry Crash Course Basically, almost all living things are made up of these 4 Elements: - Carbon (C) - Nitrogen (N) - Hydrogen (H) - Oxygen (O) This exercise is designed to familiarize you with
The Biochemistry Crash Course Basically, almost all living things are made up of these 4 Elements: - Carbon (C) - Nitrogen (N) - Hydrogen (H) - Oxygen (O) This exercise is designed to familiarize you with
Accelerating potential of mean force calculations for lipid membrane permeation: System size, reaction coordinate, solute-solute distance, and cutoffs
 Accelerating potential of mean force calculations for lipid membrane permeation: System size, reaction coordinate, solute-solute distance, and cutoffs Naomi Nitschke, Kalina Atkovska, and Jochen S. Hub
Accelerating potential of mean force calculations for lipid membrane permeation: System size, reaction coordinate, solute-solute distance, and cutoffs Naomi Nitschke, Kalina Atkovska, and Jochen S. Hub
Alchembed: A Computational Method for Incorporating Multiple Proteins into Complex Lipid Geometries
 This is an open access article published under a Creative Commons Attribution (CC-BY) License, which permits unrestricted use, distribution and reproduction in any medium, provided the author and source
This is an open access article published under a Creative Commons Attribution (CC-BY) License, which permits unrestricted use, distribution and reproduction in any medium, provided the author and source
Simulation Study of the Structure and Phase Behavior of Ceramide Bilayers and the Role of Lipid Headgroup Chemistry
 pubs.acs.org/jctc Simulation Study of the Structure and Phase Behavior of Ceramide Bilayers and the Role of Lipid Headgroup Chemistry Shan Guo, Timothy C. Moore, Christopher R. Iacovella, L. Anderson Strickland,
pubs.acs.org/jctc Simulation Study of the Structure and Phase Behavior of Ceramide Bilayers and the Role of Lipid Headgroup Chemistry Shan Guo, Timothy C. Moore, Christopher R. Iacovella, L. Anderson Strickland,
Molecular Structure and Permeability at the Interface between Phase-Separated Membrane Domains
 SUPPORTING INFORMATION Molecular Structure and Permeability at the Interface between Phase-Separated Membrane Domains Rodrigo M. Cordeiro Universidade Federal do ABC, Avenida dos Estados 5001, CEP 09210-580,
SUPPORTING INFORMATION Molecular Structure and Permeability at the Interface between Phase-Separated Membrane Domains Rodrigo M. Cordeiro Universidade Federal do ABC, Avenida dos Estados 5001, CEP 09210-580,
Lessons of Slicing Membranes: Interplay of Packing, Free Area, and Lateral Diffusion in Phospholipid/Cholesterol Bilayers
 1076 Biophysical Journal Volume 87 August 2004 1076 1091 Lessons of Slicing Membranes: Interplay of Packing, Free Area, and Lateral Diffusion in Phospholipid/Cholesterol Bilayers Emma Falck,* Michael Patra,
1076 Biophysical Journal Volume 87 August 2004 1076 1091 Lessons of Slicing Membranes: Interplay of Packing, Free Area, and Lateral Diffusion in Phospholipid/Cholesterol Bilayers Emma Falck,* Michael Patra,
Phase Behavior of a Phospholipid/Fatty Acid/Water Mixture Studied in Atomic Detail
 Published on Web 01/19/2006 Phase Behavior of a Phospholipid/Fatty Acid/Water Mixture Studied in Atomic Detail Volker Knecht,*, Alan E. Mark,, and Siewert-Jan Marrink Contribution from the Max Planck Institute
Published on Web 01/19/2006 Phase Behavior of a Phospholipid/Fatty Acid/Water Mixture Studied in Atomic Detail Volker Knecht,*, Alan E. Mark,, and Siewert-Jan Marrink Contribution from the Max Planck Institute
Introduction. Conference paper
 DOI 1.1515/pac-214-523 Pure Appl. Chem. 214; 86(2): 215 222 Conference paper Wataru Shinoda* and Michael L. Klein Effective interaction between small unilamellar vesicles as probed by coarse-grained molecular
DOI 1.1515/pac-214-523 Pure Appl. Chem. 214; 86(2): 215 222 Conference paper Wataru Shinoda* and Michael L. Klein Effective interaction between small unilamellar vesicles as probed by coarse-grained molecular
Macromolecules Carbohydrates A COMPLEX COLORING EXPERIENCE
 Macromolecules Carbohydrates A COMPLEX COLORING EXPERIENCE Name: Per: Date: All plants, animals and microorganisms use carbohydrates as sources of energy. Carbohydrates are also used as structural building
Macromolecules Carbohydrates A COMPLEX COLORING EXPERIENCE Name: Per: Date: All plants, animals and microorganisms use carbohydrates as sources of energy. Carbohydrates are also used as structural building
Introduction to Macromolecules. If you were to look at the nutrition label of whole milk, what main items stick out?
 Introduction to Macromolecules Macromolecules are a set of molecules that are found in living organisms. Macromolecules essentially mean big molecules as the word macro means large. The functions of these
Introduction to Macromolecules Macromolecules are a set of molecules that are found in living organisms. Macromolecules essentially mean big molecules as the word macro means large. The functions of these
Lesson Overview. Carbon Compounds. Lesson Overview. 2.3 Carbon Compounds
 Lesson Overview 2.3 The Chemistry of Carbon What elements does carbon bond with to make up life s molecules? Carbon can bond with many elements, including Hydrogen, Oxygen, Phosphorus, Sulfur, and Nitrogen
Lesson Overview 2.3 The Chemistry of Carbon What elements does carbon bond with to make up life s molecules? Carbon can bond with many elements, including Hydrogen, Oxygen, Phosphorus, Sulfur, and Nitrogen
Computer Simulation of T-Cell Activation
 ISIM1 Computational Immunology Computer Simulation of T-Cell Activation W. Schreiner, M. Cibena, U. Omasits, M. Berger, R. Kobler, M. Neumann, O. Steinhauser Section for Biomedical Computersimulation and
ISIM1 Computational Immunology Computer Simulation of T-Cell Activation W. Schreiner, M. Cibena, U. Omasits, M. Berger, R. Kobler, M. Neumann, O. Steinhauser Section for Biomedical Computersimulation and
Molecular Dynamics Simulation of Melittin in a Dimyristoylphosphatidylcholine (DMPC) Bilayer Membrane
 Molecular Dynamics Simulation of Melittin in a Dimyristoylphosphatidylcholine (DMPC) Bilayer Membrane XIE ZHIYAO (Andy) Contact: zyxie5-c@my.cityu.edu.hk JIANG HUANBO (Bobby) Contact: huanjiang-c@my.cityu.edu.hk
Molecular Dynamics Simulation of Melittin in a Dimyristoylphosphatidylcholine (DMPC) Bilayer Membrane XIE ZHIYAO (Andy) Contact: zyxie5-c@my.cityu.edu.hk JIANG HUANBO (Bobby) Contact: huanjiang-c@my.cityu.edu.hk
Lipid14: The Amber Lipid Force Field
 pubs.acs.org/jctc Lipid14: The Amber Lipid Force Field Callum J. Dickson,, Benjamin D. Madej,,±, Åge A. Skjevik,, Robin M. Betz, Knut Teigen, Ian R. Gould,*, and Ross C. Walker*,,± Department of Chemistry
pubs.acs.org/jctc Lipid14: The Amber Lipid Force Field Callum J. Dickson,, Benjamin D. Madej,,±, Åge A. Skjevik,, Robin M. Betz, Knut Teigen, Ian R. Gould,*, and Ross C. Walker*,,± Department of Chemistry
How Alcohol Chain-Length and Concentration Modulate Hydrogen Bond Formation in a Lipid Bilayer
 2366 Biophysical Journal Volume 92 April 2007 2366 2376 How Alcohol Chain-Length and Concentration Modulate Hydrogen Bond Formation in a Lipid Bilayer Allison N. Dickey and Roland Faller* *Department of
2366 Biophysical Journal Volume 92 April 2007 2366 2376 How Alcohol Chain-Length and Concentration Modulate Hydrogen Bond Formation in a Lipid Bilayer Allison N. Dickey and Roland Faller* *Department of
Supporting Information: Atomistic Model for Near-Quantitative. Simulations of Langmuir Monolayers
 Supporting Information: Atomistic Model for Near-Quantitative Simulations of Langmuir Monolayers Matti Javanainen,,, Antti Lamberg, Lukasz Cwiklik,, Ilpo Vattulainen,,, and Samuli Ollila,,# Laboratory
Supporting Information: Atomistic Model for Near-Quantitative Simulations of Langmuir Monolayers Matti Javanainen,,, Antti Lamberg, Lukasz Cwiklik,, Ilpo Vattulainen,,, and Samuli Ollila,,# Laboratory
Sialic acids and diabetes : impact of desialylation on insulin receptor s functionality
 Journée scientifique du Centre de Calcul et de la Maison de la Simulation Sialic acids and diabetes : impact of desialylation on insulin receptor s functionality GUILLOT Alexandre Supervisors: DURLACH
Journée scientifique du Centre de Calcul et de la Maison de la Simulation Sialic acids and diabetes : impact of desialylation on insulin receptor s functionality GUILLOT Alexandre Supervisors: DURLACH
Interactions Between a Voltage Sensor and a Toxin via Multiscale Simulations
 1558 Biophysical Journal Volume 98 April 2010 1558 1565 Interactions Between a Voltage Sensor and a Toxin via Multiscale Simulations Chze Ling Wee, David Gavaghan, and Mark S. P. Sansom * Department of
1558 Biophysical Journal Volume 98 April 2010 1558 1565 Interactions Between a Voltage Sensor and a Toxin via Multiscale Simulations Chze Ling Wee, David Gavaghan, and Mark S. P. Sansom * Department of
Molecular dynamics simulations of charged and neutral lipid bilayers: treatment of electrostatic interactions
 Vol. 50 No. 3/2003 789 798 QUARTERLY Molecular dynamics simulations of charged and neutral lipid bilayers: treatment of electrostatic interactions Tomasz Róg, Krzysztof Murzyn and Marta Pasenkiewicz-Gierula
Vol. 50 No. 3/2003 789 798 QUARTERLY Molecular dynamics simulations of charged and neutral lipid bilayers: treatment of electrostatic interactions Tomasz Róg, Krzysztof Murzyn and Marta Pasenkiewicz-Gierula
Modification of the CHARMM Force Field for DMPC Lipid Bilayer
 Modification of the CHARMM Force Field for DMPC Lipid Bilayer CARL-JOHAN HÖGBERG, 1 ALEXEI M. NIKITIN, 1,2 ALEXANDER P. LYUBARTSEV 1 1 Division of Physical Chemistry, Arrhenius Laboratory, Stockholm University,
Modification of the CHARMM Force Field for DMPC Lipid Bilayer CARL-JOHAN HÖGBERG, 1 ALEXEI M. NIKITIN, 1,2 ALEXANDER P. LYUBARTSEV 1 1 Division of Physical Chemistry, Arrhenius Laboratory, Stockholm University,
Supplementary Information. A lithium-ion-active aerolysin nanopore for effectively trapping long. single-stranded DNA.
 Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2018 Supplementary Information A lithium-ion-active aerolysin nanopore for effectively trapping
Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2018 Supplementary Information A lithium-ion-active aerolysin nanopore for effectively trapping
Coarse grained simulations of Lipid Bilayer Membranes
 Coarse grained simulations of Lipid Bilayer Membranes P. B. Sunil Kumar Department of Physics IIT Madras, Chennai 600036 sunil@iitm.ac.in Atomistic MD: time scales ~ 10 ns length scales ~100 nm 2 To study
Coarse grained simulations of Lipid Bilayer Membranes P. B. Sunil Kumar Department of Physics IIT Madras, Chennai 600036 sunil@iitm.ac.in Atomistic MD: time scales ~ 10 ns length scales ~100 nm 2 To study
Structure and Dynamics of Sphingomyelin Bilayer: Insight Gained through Systematic Comparison to Phosphatidylcholine
 2976 Biophysical Journal Volume 87 November 2004 2976 2989 Structure and Dynamics of Sphingomyelin Bilayer: Insight Gained through Systematic Comparison to Phosphatidylcholine Perttu Niemelä,* Marja T.
2976 Biophysical Journal Volume 87 November 2004 2976 2989 Structure and Dynamics of Sphingomyelin Bilayer: Insight Gained through Systematic Comparison to Phosphatidylcholine Perttu Niemelä,* Marja T.
Methods and Materials
 a division of Halcyonics GmbH Anna-Vandenhoeck-Ring 5 37081 Göttingen, Germany Application Note Micostructured lipid bilayers ANDREAS JANSHOFF 1), MAJA GEDIG, AND SIMON FAISS Fig.1: Thickness map of microstructured
a division of Halcyonics GmbH Anna-Vandenhoeck-Ring 5 37081 Göttingen, Germany Application Note Micostructured lipid bilayers ANDREAS JANSHOFF 1), MAJA GEDIG, AND SIMON FAISS Fig.1: Thickness map of microstructured
Membrainy: a smart, unified membrane analysis tool
 Carr and MacPhee Source Code for Biology and Medicine (2015) 10:3 DOI 10.1186/s13029-015-0033-7 SOFTWARE Open Access Membrainy: a smart, unified membrane analysis tool Matthew Carr and Cait E MacPhee *
Carr and MacPhee Source Code for Biology and Medicine (2015) 10:3 DOI 10.1186/s13029-015-0033-7 SOFTWARE Open Access Membrainy: a smart, unified membrane analysis tool Matthew Carr and Cait E MacPhee *
Organic Compounds. (Carbon Compounds) Carbohydrates Lipids Proteins Nucleic Acids
 Organic Compounds (Carbon Compounds) Carbohydrates Lipids Proteins Nucleic Acids Carbon s Bonding Behavior Outer shell of carbon has 4 electrons; can hold 8 Each carbon atom can form covalent bonds with
Organic Compounds (Carbon Compounds) Carbohydrates Lipids Proteins Nucleic Acids Carbon s Bonding Behavior Outer shell of carbon has 4 electrons; can hold 8 Each carbon atom can form covalent bonds with
Interaction Between Amyloid-b (1 42) Peptide and Phospholipid Bilayers: A Molecular Dynamics Study
 Biophysical Journal Volume 96 February 2009 785 797 785 Interaction Between Amyloid-b (1 42) Peptide and Phospholipid Bilayers: A Molecular Dynamics Study Charles H. Davis and Max L. Berkowitz * Department
Biophysical Journal Volume 96 February 2009 785 797 785 Interaction Between Amyloid-b (1 42) Peptide and Phospholipid Bilayers: A Molecular Dynamics Study Charles H. Davis and Max L. Berkowitz * Department
Macromolecules Cut & Paste
 Macromolecules Cut & Paste Adapted from http://mrswords.weebly.com/uploads/1/5/2/4/15244382/ch_6-3_life_molecules_cut-out_lab.pdf INTRODUCTION Many of the molecules in living cells are so large that they
Macromolecules Cut & Paste Adapted from http://mrswords.weebly.com/uploads/1/5/2/4/15244382/ch_6-3_life_molecules_cut-out_lab.pdf INTRODUCTION Many of the molecules in living cells are so large that they
Diffusion, Osmosis and Active Transport
 Diffusion, Osmosis and Active Transport Particles like atoms, molecules and ions are always moving Movement increases with temperature (affects phases of matter - solid, liquid, gas) Solids - atoms, molecules
Diffusion, Osmosis and Active Transport Particles like atoms, molecules and ions are always moving Movement increases with temperature (affects phases of matter - solid, liquid, gas) Solids - atoms, molecules
Under the Influence of Alcohol: The Effect of Ethanol and Methanol on Lipid Bilayers
 Biophysical Journal Volume 90 February 2006 1121 1135 1121 Under the Influence of Alcohol: The Effect of Ethanol and Methanol on Lipid Bilayers Michael Patra,* y Emppu Salonen, z Emma Terama, z Ilpo Vattulainen,
Biophysical Journal Volume 90 February 2006 1121 1135 1121 Under the Influence of Alcohol: The Effect of Ethanol and Methanol on Lipid Bilayers Michael Patra,* y Emppu Salonen, z Emma Terama, z Ilpo Vattulainen,
Carbon Compounds (2.3) (Part 1 - Carbohydrates)
 Carbon Compounds (2.3) (Part 1 - Carbohydrates) The Chemistry of Carbon (Organic Chemistry) Organic Chemistry: The study of compounds that contain bonds between carbon atoms. Carbon can bond with many
Carbon Compounds (2.3) (Part 1 - Carbohydrates) The Chemistry of Carbon (Organic Chemistry) Organic Chemistry: The study of compounds that contain bonds between carbon atoms. Carbon can bond with many
Distribution of Amino Acids in a Lipid Bilayer from Computer Simulations
 Biophysical Journal Volume 94 May 2008 3393 3404 3393 Distribution of Amino Acids in a Lipid Bilayer from Computer Simulations Justin L. MacCallum, W. F. Drew Bennett, and D. Peter Tieleman Department
Biophysical Journal Volume 94 May 2008 3393 3404 3393 Distribution of Amino Acids in a Lipid Bilayer from Computer Simulations Justin L. MacCallum, W. F. Drew Bennett, and D. Peter Tieleman Department
The main biological functions of the many varied types of lipids include: energy storage protection insulation regulation of physiological processes
 Big Idea In the biological sciences, a dehydration synthesis (condensation reaction) is typically defined as a chemical reaction that involves the loss of water from the reacting molecules. This reaction
Big Idea In the biological sciences, a dehydration synthesis (condensation reaction) is typically defined as a chemical reaction that involves the loss of water from the reacting molecules. This reaction
Chp 2 (cont.) Organic Molecules. Spider s web and close up of capture strand - spider silk protein
 Chp 2 (cont.) Organic Molecules Spider s web and close up of capture strand - spider silk protein 1! Molecular Diversity is Based on Carbon An organic molecule contains both carbon and hydrogen. Ex: Methane
Chp 2 (cont.) Organic Molecules Spider s web and close up of capture strand - spider silk protein 1! Molecular Diversity is Based on Carbon An organic molecule contains both carbon and hydrogen. Ex: Methane
Chemotherapy efficiency increase via shock wave interaction with biological membranes: a molecular dynamics study
 Chemotherapy efficiency increase via shock wave interaction with biological membranes: a molecular dynamics study The MIT Faculty has made this article openly available. Please share how this access benefits
Chemotherapy efficiency increase via shock wave interaction with biological membranes: a molecular dynamics study The MIT Faculty has made this article openly available. Please share how this access benefits
Revisiting Hydrophobic Mismatch with Free Energy Simulation Studies of Transmembrane Helix Tilt and Rotation
 Biophysical Journal Volume 99 July 2010 175 183 175 Revisiting Hydrophobic Mismatch with Free Energy Simulation Studies of Transmembrane Helix Tilt and Rotation Taehoon Kim and Wonpil Im* Department of
Biophysical Journal Volume 99 July 2010 175 183 175 Revisiting Hydrophobic Mismatch with Free Energy Simulation Studies of Transmembrane Helix Tilt and Rotation Taehoon Kim and Wonpil Im* Department of
Application of Molecular Modelling and EPR Spectroscopy to Lipid Membranes a Combined Approach
 Armenian Journal of Physics, 2016, vol. 9, issue 2, pp. 159-166 Application of Molecular Modelling and EPR Spectroscopy to Lipid Membranes a Combined Approach Andrea Catte and Vasily S. Oganesyan * School
Armenian Journal of Physics, 2016, vol. 9, issue 2, pp. 159-166 Application of Molecular Modelling and EPR Spectroscopy to Lipid Membranes a Combined Approach Andrea Catte and Vasily S. Oganesyan * School
PHARMACOPHORE MODELING
 CHAPTER 9 PHARMACOPHORE MODELING 9.1 PHARMACOPHORES AS VIRTUAL SCREENING TOOLS The pharmacophore concept has been widely used over the past decades for rational drug design approaches and has now been
CHAPTER 9 PHARMACOPHORE MODELING 9.1 PHARMACOPHORES AS VIRTUAL SCREENING TOOLS The pharmacophore concept has been widely used over the past decades for rational drug design approaches and has now been
Lipids and Membranes
 Lipids and Membranes Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry I Philadelphia University Faculty of pharmacy Biological membranes are composed of lipid bilayers
Lipids and Membranes Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry I Philadelphia University Faculty of pharmacy Biological membranes are composed of lipid bilayers
Good Afternoon! 11/30/18
 Good Afternoon! 11/30/18 1. The term polar refers to a molecule that. A. Is cold B. Has two of the same charges C. Has two opposing charges D. Contains a hydrogen bond 2. Electrons on a water molecule
Good Afternoon! 11/30/18 1. The term polar refers to a molecule that. A. Is cold B. Has two of the same charges C. Has two opposing charges D. Contains a hydrogen bond 2. Electrons on a water molecule
Coarse-grained simulation studies of mesoscopic membrane phenomena
 Coarse-grained simulation studies of mesoscopic membrane phenomena Markus Deserno Department of Physics Carnegie Mellon University with: Ira R. Cooke, Gregoria Illya, Benedict J. Reynwar, Vagelis A. Harmandaris,
Coarse-grained simulation studies of mesoscopic membrane phenomena Markus Deserno Department of Physics Carnegie Mellon University with: Ira R. Cooke, Gregoria Illya, Benedict J. Reynwar, Vagelis A. Harmandaris,
Modeling and Molecular Dynamics of Membrane Proteins
 Modeling and Molecular Dynamics of Membrane Proteins Emad Tajkhorshid Department of Biochemistry, Center for Biophysics and Computational Biology, and Beckman Institute University of Illinois at Urbana-Champaign
Modeling and Molecular Dynamics of Membrane Proteins Emad Tajkhorshid Department of Biochemistry, Center for Biophysics and Computational Biology, and Beckman Institute University of Illinois at Urbana-Champaign
Proteins. Biomolecules. Nucleic Acids. The Building Blocks of Life
 Proteins Biomolecules Nucleic Acids The Building Blocks of Life Carbohydrates Lipids Biomolecules are 1. Organic molecules that are (at least 1 Carbon molecule and often chains of Carbon) They all contain.
Proteins Biomolecules Nucleic Acids The Building Blocks of Life Carbohydrates Lipids Biomolecules are 1. Organic molecules that are (at least 1 Carbon molecule and often chains of Carbon) They all contain.
Molecular modeling of the pathways of vesicle membrane interaction. Tongtao Yue and Xianren Zhang
 Molecular modeling of the pathways of vesicle membrane interaction Tongtao Yue and Xianren Zhang I. ELECTRONIC SUPPLEMENTARY INFORMATION (ESI): METHODS Dissipative particle dynamics method The dissipative
Molecular modeling of the pathways of vesicle membrane interaction Tongtao Yue and Xianren Zhang I. ELECTRONIC SUPPLEMENTARY INFORMATION (ESI): METHODS Dissipative particle dynamics method The dissipative
Molecular Models of Nanodiscs
 This is an open access article published under an ACS AuthorChoice License, which permits copying and redistribution of the article or any adaptations for non-commercial purposes. pubs.acs.org/jctc Molecular
This is an open access article published under an ACS AuthorChoice License, which permits copying and redistribution of the article or any adaptations for non-commercial purposes. pubs.acs.org/jctc Molecular
Physics of Cellular Materials: Biomembranes
 Physics of Cellular Materials: Biomembranes Tom Chou 1 1 Dept. of Biomathematics, UCL, Los ngeles, C 90095-1766 (Dated: December 6, 2002) Here I will review the mathematics and statistical physics associated
Physics of Cellular Materials: Biomembranes Tom Chou 1 1 Dept. of Biomathematics, UCL, Los ngeles, C 90095-1766 (Dated: December 6, 2002) Here I will review the mathematics and statistical physics associated
Free Volume Properties of Sphingomyelin, DMPC, DPPC, and PLPC Bilayers
 PUBLISHED IN: JOURNAL OF COMPUTATIONAL AND THEORETICAL NANOSCIENCE, VOL. 2, PP. 40-43 (2005). Free Volume Properties of Sphingomyelin, DMPC, DPPC, and PLPC Bilayers M. Kupiainen, E. Falck, S. Ollila, P.
PUBLISHED IN: JOURNAL OF COMPUTATIONAL AND THEORETICAL NANOSCIENCE, VOL. 2, PP. 40-43 (2005). Free Volume Properties of Sphingomyelin, DMPC, DPPC, and PLPC Bilayers M. Kupiainen, E. Falck, S. Ollila, P.
Name: Period: Date: Testing for Biological Macromolecules Lab
 Testing for Biological Macromolecules Lab Introduction: All living organisms are composed of various types of organic molecules, such as carbohydrates, starches, proteins, lipids and nucleic acids. These
Testing for Biological Macromolecules Lab Introduction: All living organisms are composed of various types of organic molecules, such as carbohydrates, starches, proteins, lipids and nucleic acids. These
Optimum Body and Leg Extension Selection in PLS CADD
 610 N. Whitney Way, Suite 160 Madison, WI 53705 Phone: 608.238.2171 Fax: 608.238.9241 Email:info@powline.com URL: http://www.powline.com Optimum Body and Leg Extension Selection in PLS CADD Introduction
610 N. Whitney Way, Suite 160 Madison, WI 53705 Phone: 608.238.2171 Fax: 608.238.9241 Email:info@powline.com URL: http://www.powline.com Optimum Body and Leg Extension Selection in PLS CADD Introduction
Structural Change in Lipid Bilayers and Water Penetration Induced by Shock Waves: Molecular Dynamics Simulations
 2198 Biophysical Journal Volume 91 September 2006 2198 2205 Structural Change in Lipid Bilayers and Water Penetration Induced by Shock Waves: Molecular Dynamics Simulations Kenichiro Koshiyama,* Tetsuya
2198 Biophysical Journal Volume 91 September 2006 2198 2205 Structural Change in Lipid Bilayers and Water Penetration Induced by Shock Waves: Molecular Dynamics Simulations Kenichiro Koshiyama,* Tetsuya
