g membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation.
|
|
|
- Justin Higgins
- 5 years ago
- Views:
Transcription
1 g membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. Maarten G. Wolf, Martin Hoefling, Camilo Aponte-Santamaría, Helmut Grubmüller and Gerrit Groenhof Maarten G. Wolf Computational Biomolecular Chemistry Max-Planck-Institute for Biophysical Chemistry Am Faßberg 11 D Göttingen, Germany Martin Hoefling Department for Theoretical and Computational Biophysics Max-Planck-Institute for Biophysical Chemistry Am Faßberg 11 D Göttingen, Germany Camilo Aponte-Santamaría Department for Theoretical and Computational Biophysics Max-Planck-Institute for Biophysical Chemistry Am Faßberg 11 D Göttingen, Germany Helmut Grubmüller Department for Theoretical and Computational Biophysics Max-Planck-Institute for Biophysical Chemistry Am Faßberg 11 D Göttingen, Germany Gerrit Groenhof Computational Biomolecular Chemistry Max-Planck-Institute for Biophysical Chemistry Am Faßberg 11 D Göttingen, Germany 1
2 g membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. Maarten G. Wolf, Martin Hoefling, Camilo Aponte-Santamaría, Helmut Grubmüller and Gerrit Groenhof Abstract To efficiently insert a protein into an equilibrated and fully hydrated membrane with minimal membrane perturbation we present a computational tool, called g membed, which is part of the Gromacs suite of programs. The input consists of an equilibrated membrane system, either flat or curved, and a protein structure in the right position and orientation with respect to the lipid bilayer. g membed first decreases the width of the protein in the xy-plane and removes all molecules (generally lipids and waters) that overlap with the narrowed protein. Then the protein is grown back to its full size in a short molecular dynamics simulation (typically 1,000 steps), thereby pushing the lipids away to optimally accommodate the protein in the membrane. After embedding the protein in the membrane, both the lipid properties and the hydration layer are still close to equilibrium. Thus only a short equilibration run (less then 1 ns in the cases tested) is required to re-equilibrate the membrane. Its simplicity makes g membed very practical for use in scripting and high-throughput molecular dynamics simulations. Keywords : computer simulations, membrane, Gromacs, molecular dynamics, protein insertion 1 Introduction The number of computer simulations to probe the molecular details of membrane proteins is rapidly increasing [1]. Proteins embedded in or associated with membranes are of interest because they perform various crucial tasks, including (selective) transport, signal transduction and energy conversion. It is estimated that in most organisms 20-30% of the proteins encoded in the genome are membrane proteins [2] and that many drug targets are located at or near the cell surface (60% in 2006) [3]. Nevertheless, these percentages are not reflected in the ratio of studies between soluble and membrane proteins. Due to the requirement of a lipid bilayer, membrane proteins are more complicated to crystallize or study with NMR. Consequently less than 1% of the structurally resolved proteins are membrane proteins [4]. However, the number of membrane protein structures in the pdb-database is rising quickly (see 2
3 Proteins xtal.html for a maintained list) and thus their availability for computational studies. Also on the simulation side, setting up a membrane protein system requires additional efforts compared to soluble proteins. First, an appropriate membrane model has to be selected for which parameters are available in the desired force field. Setting up plain membrane simulations can now be considered standard procedure [1] and hence will not be further discussed here. Furthermore, many equilibrated membrane patches and the corresponding force field parameters can be downloaded from webservers (URLs given in [1]) maintained by research groups specialized in membrane simulations. Second, and crucially, once a suitable membrane is selected the protein has to be embedded in the membrane before the production runs can be performed. Several protocols have been suggested to insert a protein into a lipid bilayer, but up to now no standard has been established, mainly for reasons discussed below. The simplest way to insert a protein is to remove lipids and solvent molecules that overlap with the protein after combining the coordinate sets (X,Y,Z). However, the highly disordered nature of lipid tails results in an irregular and oversized hole and hence a poor lipid-protein packing. Long simulation runs are then needed to equilibrate the system. To obtain a good lipid-protein packing one can build a lipid bilayer around a protein from preequilibrated and prehydrated single lipids [5]. Unfortunately, assembling the bilayer from uncorrelated lipids will result in bad contacts (overlapping atoms), which require extensive optimisation by, for instance, rigid body translation and rotations [5]. A more sophisticated method uses repulsive forces to create a hole at the intended protein position before inserting the protein itself[6, 7]. This will result in a proper lipid-protein packing in an equilibrated membrane, but needs considerable tuning by the user to find the optimal hole generating parameters [8]. Recently, it has also been proposed to inflate the lipid bilayer, insert the protein and then, alternatingly deflate the lipid bilayer in small steps and perform an energy minimization, until the area per lipid reaches the reference equilibrium value [8]. However, this method cannot be applied with a solvated membrane and thus requires resolvation. Furthermore, it is not clear if the membrane remains in equilibrium during the deflation procedure. The lipid tails that were entangled in the equilibrated membrane are likely to clash when going from a very diluted (inflated) to a fully packed membrane, resulting in a different packing of the lipid tails. Finally, with coarse grained simulations it is possible to let the lipids self-assemble around the membrane protein [9]. Unfortunately, the level of detail of coarse grained simulation might not be sufficient to assess the relevant interactions in membrane proteins 3
4 adequately. Also the inverse mapping problem, i.e. obtaining the atom positions from the coarse grained coordinates, is highly non-trivial. To address the issue more systematically, we propose that an insertion method needs to fulfil four requirements: (1) It should be easy to use, without the requirement of parameter tuning or manual intervention during or after the embedding process. (2) It should yield a structure that is close to equilibrium, to reduce the equilibration simulation time before production runs. (3) It should be easily automated, to allow for large scale high-throughput simulations. (4) It should be distributed as part of a popular MD package, to maintain availability, functionality and retraceability. None of the aforementioned methods satisfies these four criteria and therefore we developed a new Gromacs [10] tool, g membed, that grows a protein into a lipid bilayer during a short md simulation (1,000 steps typically). By using an already hydrated and equilibrated lipid bilayer the output system (protein embedded in the solvated membrane) only requires a short equilibration run (maximal 1 ns in our test systems) to reequilibrate the membrane part of the system. Finally, by making g membed part of Gromacs availability, functionality and retraceability is guaranteed. 2 Method The basic idea is to slowly grow the protein into an already equilibrated membrane, thereby pushing away the lipids and waters and fit the protein nicely into the lipid bilayer. Starting point of the growth process is the narrowed protein, where the narrowing of the protein is performed within the membrane plane, but not in the direction of the membrane normal. In the limiting case all protein atoms start on a line perpendicular to the membrane plane. Shrinking the protein in 2D instead of 3D is crucial, as it avoids severe perturbation of the lipids, especially when approaching the limiting cases (a line instead of a point). The growth phase itself is a short md run. After each md step the size of the protein is slightly increased until the protein has reached its initial size (see Figure 1 for a typical example). During this process all protein-protein interactions, including bonded interactions, are switched off. The method, as implemented into the Gromacs tool g membed (embedding a protein within a membrane) requires a file with the protein structure overlapping the lipid bilayer spanning the xy-plane at the desired position and orientation as input (we assume the user already has this knowledge about the system). Optimal placement of the protein with respect to the membrane will avoid needlessly-long equilibration 4
5 runs after embedding. In the case of a curved bilayer (e.g. a vesicle), it is important that the protein is overlapping a membrane part that has its normal directed along the z-axis. g membed then performs the following steps to embed the protein (or any other desired group of atoms defined as a Gromacs index group) within a lipid bilayer. 1. Protein narrowing. The coordinates of the atoms in the protein are scaled with respect to the geometrical center of the transmembrane part of the protein by a user specified scaling factor of the original (input) coordinates in the xy-plane and, if applicable, in the z-direction. Normally the protein should not be scaled in the z-direction. However in special cases, such as a protein that has the same height as the bilayer, increasing the size of the protein in the z-direction prevents lipids to envelop the protein during the growth phase. 2. Remove overlapping molecules. Every molecule not part of the protein for which at least one atom is within a user defined radius of a protein atom will be removed. If the difference between the number of lipids removed from the lower (n lower ) and the upper (n upper ) membrane leaflet is not equal to a user defined number (n diff ), i.e. n lower n upper n diff, additional lipids will be removed, such that this equality will be obtained. This option is usefull when inserting an asymetrically shaped protein. 3. Growth phase. Iterate steps 3b and 3a nxy + nz times to grow the narrowed protein to its original size. (a) md step. Do a normal md step. (b) Protein resizing. Change the atom coordinates of the protein by linear interpolation between the coordinates of the narrowed protein (step 1) and those of the input configuration by s i = r i = r geom + s i (r 0 r geom ) (1) s i,x = s i,y = s 0,xy + i (1 s0,xy) n xy and s i,z = s 0,z i n xy s i,x = s i,y = 1 and s i,z = s 0,z + (i nxy) (1 s0,z) n z n xy < i n xy + n z (2) with r i the protein atom coordinates at step i, r 0 the input atom coordinates of the protein, r geom the coordinates of the geometrical center of the transmembrane region, s i the scaling factor at step i, s 0,xy and s 0,z the initial scaling factors and n xy and n z the number of embedding 5
6 steps in the xy-plane and the z-direction, respectively. This way the atom coordinates of the proteins reach the input configuration in n xy steps in the xy-plane and, if applicable, after the xy-dimension is completely expanded in n z steps in the z-dimension. The output will be a file containing the protein structure properly embedded in the membrane. All atom coordinates of the protein in the output file will be equal to the coordinates provided in the input (see the Appendix for a user manual). 3 Case studies As a test we have applied g membed to insert four proteins of various shape and size: Integrin αiibβ3 2K9J [11], β-barrel Platform 2JMM [12], Reaction center 1PRC [13] and Yeast Aquaporin 2W2E [14] (see Table i). We have embedded the first two into a united-atom (POPC [15, 16]) and the latter two into an all-atom (DOPC [17]) membrane. In addition we have embedded the β-barrel Platform within the lipid bilayer of a vesicle (POPE), see Figure 2. The crystal waters available in the Reaction Centers pdb entry were preserved and considered as part of the protein. Initially, the width of the protein in these tests was scaled to 10% of the full-size protein (s 0,xy = 0.1). All proteins listed in Table i were inserted successfully within the bilayer within 1,000 md steps. To assess the time required to reach equilibrium after embedding the protein within the membrane, a 10 ns standard md simulation was performed after the embedding (insertion within the vesicle not included). Figure 3 shows the area per lipid (A lip ) as a function of time, where A lip is obtained through A lip = 2 (A xy A prot )/n lip (3) with n lip the number of lipids in the membrane, A xy the area spanned by the x- and y-box vectors (membrane plane) and A prot the protein area estimated by projecting all the atoms of the protein that overlap the membrane slab on a two-dimensional square grid in the xy-plane and summing the area of all grid elements that contain a protein atom. From Figure 3 it is clear that already within 1 ns the system reaches the equilibrium area per lipid. For a pure popc and dopc membrane system we found an area per lipid of 0.59 and 0.62, respectively, which is slightly higher than the values we found for the protein-membrane system. This discrepancy can partly be explained by the fact that the protein area is 6
7 overestimated, resulting in an underestimation of the area per lipid. Furthermore, since the influence of proteins on lipid properties is still not completely understood, it is not clear whether the area per lipid should actually be unchanged after protein insertion [18, 19]. Reaching the equilibrium area per lipid does not necessarily imply that the membrane is in equilibrium. As a more rigorous test, we therefore also analyzed the deuterium order parameter, S CD, a measure for the amount of disorder in the lipid tails. Comparing the deuterium order parameter before and after embedding the protein into the membrane did not show any significant differences. In addition, we calculated the density profile of water normal to the membrane plane to compare the hydration of the membrane. Also here we found no significant difference before and after embedding the protein. Monitoring the deuterium order parameter and the water density profile normal to the membrane over time indicate that the membrane is still in equilibrium. To determine the time required to reach area equilibrium, we calculated the relaxation time τ associated with the area per lipid by fitting A lip =< A lip > A 0 exp( t/τ) (4) to the area per lipid as a function of time (Figure 3). In equation 4, < A lip > is the ensemble average of the area per lipid, where we averaged over the 2-10 ns time interval and A 0 the fitted difference with < A lip > at t=0. The resulting relaxation times are listed in Table ii, column 0.1. The average relaxation time within the scaling factor range (including data not listed in Table ii) is 180 ± 190 ps. From this value and the values listed in Table ii we expect that a 1 ns equilibration run suffices after embedding a protein in an equilibrated membrane using g membed. Nevertheless, as the number of cases studied here is limited, the required equilibration time might differ for some specific systems. Finally, we examined the influence of the protein scaling factor s 0,xy used in the first step of the embedding process. We expect that a larger initial scaling factor implies slower equilibration, since more lipids have to be moved to accomodate the protein. We embedded a Yeast Aquaporin tetramer and the β-barrel Platform starting with initial scaling factors ranging from 0.1 to 0.7, followed by a 10 ns standard simulation. The root mean square deviation (rmsd) of the Yeast Aquaporin tetramer with respect to the x-ray structure approaches the same value, 0.15 nm, for all initial scaling factors, whereas the rmsd for the β-barrel Platform ranges from 0.3 to 0.6 nm without any correlation to the initial scaling factor. 7
8 These findings indicate that the protein structure is unperturbed by the initial scaling factor. Table ii shows the relaxation times of the area per lipid for different initial scaling factors. These values show that indeed our hypothesis that a smaller scaling factor implies slower equilibration is true. When the scaling factor approaches a value, for which the removed lipid area is only slightly smaller than the estimated protein area (0.6 and 0.7 for the β-barrel Platform and Yeast Aquaporin, respectively) the perturbation to the membrane as measured by the area per lipid, deuterium order parameter and water density profile is minimal in the cases tested. The length of the simulation to equilibrate the system is then mainly determined by the equilibration of the protein. 4 Conclusions We have presented a program, g membed, for efficient insertion of a protein structure into a hydrated and equilibrated membrane. g membed can handle both flat and curved membranes, thereby for instance also allowing embedding of protein structures within vesicle bilayers. The program only minimally perturbs the properties and the hydration of the system during protein insertion. As a consequence short subsequent equilibration runs suffice, enabling drastically increased throughput. In addition, one set of parameters (set as default) can be aplied to insert proteins of various shape and size into an arbitrary lipid bilayer, suggesting this set to be useful for most proteins. We consider the following two setups most useful. (1) The protein is embedded within a membrane slab that is relatively small compared to the protein size, so that a small number of lipids should be removed. Consequently, the initial scaling factor s 0,xy needs to be set to a very small value (e.g. 0.1 of the full protein size). After embedding, a 1 ns equilibration is recommended to reequilibrate the membrane, but longer equilibration times may be required for the protein. This setup is the default for g membed. (2) Alternatively, if the protein is to be embedded within a membrane slab that is large with respect to the proteins size, the protein scaling factor s 0,xy can be set to a value, such that g membed removes a lipid portion that is slightly smaller than the estimated protein area (depending on the protein size a scaling factor between 0.5 and 0.7). In this case, the perturbation of the system is even smaller, and the length of the equilibration run is determined by the time it takes to equilibrate the protein structure. 8
9 5 Computational Details 5.1 g membed As g membed uses the molecular dynamics code implemented in Gromacs [10], all system options available to a standard md simulation also apply to g membed. The Integrin αiibβ3 and the β-barrel Platform were simulated using the OPLS-AA force field [20] and embedded in the popc lipid bilayer [15, 16] combined with simple point charge (SPC) water [21]. For the Reaction-Centre and Yeast Aquaporin we used the Amber03 parameter set [22] with the dopc membrane [17] and TIP3P water [23]. During embedding of the β-barrel Platform within the pope vesicle we applied the Gromos-87 force field [24] with SPC water. To insert a protein in the membrane by g membed, 1,000 time steps were performed. A Van der Waals cut-off of 1.4 nm was used and the electrostatic interactions were treated using PME [25] with a real space cut-off of 1.0 nm. All bonds were constraint using the LINCS algorithm [26], allowing a time step of 2 fs. The temperature and pressure were kept constant at 300 K and 1 bar using velocity rescaling [27] and Berendsen semi-isotropic pressure coupling [28], respectively. Protein-protein interactions were excluded and the atoms of the protein were set to the position specified in step 3b of the g membed protocol. 5.2 Equilibration runs After embedding the protein in the membrane a 10 ns equilibration run was performed. The same system parameters were used as during embedding, with the exception that the temperature of the popc simulations was set to 323 K to prevent ordering of the bilayer. Also, the protein-protein interactions are not excluded and the atom positions of the protein are no longer fixed. Acknowledgement We thank Ulrike Gerischer for carefully reading the manuscript and Bert de Groot for sharing the coordinate file and force field parameters of the vesicle. 9
10 Appendix A Manual The required input for g membed is a tpr-file containing the group to embed (further called protein) and the (solvated) membrane. The protein should be placed at the desired position and orientation overlapping the lipid bilayer by using for instance pymol [29]. If the position and orientation of the protein is satisfactory, merge the protein and (solvated) membrane structure files into one (e.g. merged.gro) and create a matching topology file. Download sample.mdp from our website ( ggroenh/membed.html) or set the following options in an mdp file. integrator energygrp freezegrps freezedim energygrp excl = md = Protein (or other group that you want to insert) = Protein = Y Y Y = Protein Protein Generate the input file for g membed, input.tpr, with the gromacs pre-processor. grompp -f sample.mdp -c merged.gro -p merged.top -o input.tpr Then run g membed -f input.tpr -p merged.top -xyinit 0.1 -xyend 1.0 -nxy 1000 or g membed -f input.tpr -p merged.top -xyinit 0.1 -xyend 1.0 -nxy zinit 1.1 -zend 1.0 -nz 100 and select the group that has to be inserted. The options of g membed are -f the required input tpr-file, -p to update the number of molecules in the topology file, -xyinit and -zinit the scaling factor for the width and height, respectively, of the protein in the membrane plane at the start of the embedding and -nxy and -nz the number of steps to reset -xyinit and -zinit to -xyend and -zend, respectively. If the protein is (very) asymmetric in shape use -ndiff to remove more lipids from the lower (negative integer) or upper (positive integer) leaflet. Finally, a 1 ns equilibration run should be run before performing any production runs. grompp -f equi.mdp -c membedded.gro -p merged.top -o equilibrate.tpr mdrun -deffnm equilibrate 10
11 When the group to embed is not a default group, such as a protein and its crystal water, an ndx file should also be provided to g membed. Make sure all the molecule types in the group to embed are unique, e.g. the molecule type of the crystal waters should be different from that of the solvent. Also the freeze and energy exclusion parameters in the mdp file should be changed to match the name of the group to embed. Note that the program will also issue a warning for most common mistakes we have encountered (will be updated). References [1] Biggin, P. and Bond, P., Methods Mol. Biol., 2008, 443, [2] Wallin, E. and von Heijne, G., Protein Sci., 1998, 7(4), [3] Overington, J.; Al-Lazikani, B. and Hopkins, A., Nat. Rev. Drug Discov., 2006, 5(12), [4] Carpenter, E.; Beis, K.; Cameron, A. and Iwata, S., Curr. Opin. Struct. Biol., 2008, 18, [5] Woolf, T. B. and Roux, B., Proteins, 1996, 24(1), [6] Faraldo-Gómez, J.; Smith, G. and Sansom, M., Eur. Biophys. J., 2002, 31(3), [7] Shen, L.; Bassolino, D. and Stouch, T., Biophys. J., 1997, 73(1), [8] Kandt, C.; Ash, W. and Tieleman, D., Methods, 2007, 41(4), [9] Scott, K.; Bond, P.; Ivetac, A.; Chetwynd, A.; Khalid, S. and Sansom, M., Structure, 2008, 16(4), [10] Hess, B.; Kutzner, C.; van der Spoel, D. and Lindahl, E., J. Chem. Theor. Comp., 2008, 4(3), [11] Lau, T.-L.; Kim, C.; Ginsberg, M. and Ulmer, T., EMBO J., 2009, 28(9), [12] Johansson, M.; Alioth, S.; Hu, K.; Walser, R.; Koebnik, R. and Pervushin, K., Biochemistry, 2007, 46(5), [13] Deisenhofer, J.; Epp, O.; Sinning, I. and Michel, H., J. Mol. Biol., 1995, 246(3), [14] Fischer, G.; Kosinska-Eriksson, U.; Aponte-Santamaría, C.; Palmgren, M.; Geijer, C.; Hedfalk, K.; Hohmann, S.; de Groot, B. L.; Neutze, R. and Lindkvist-Petersson, K., PLoS Biol., 2009, 7(6), e [15] Berger, O.; Edholm, O. and Jähnig, F., Biophys. J., 1997, 72(5), [16] Tieleman, P.; MacCallum, J.; Ash, W.; Kandt, C.; Xu, Z. and Monticelli, L., J. Phys.: Condens. Matter, 2006, 18, S1221 S1234. [17] Siu, S. W. I.; Vácha, R.; Jungwirth, P. and Böckmann, R. A., J. Chem. Phys., 2008, 128(12), [18] Periole, X.; Huber, T.; Marrink, S.-J. and Sakmar, T. P., J. Am. Chem. Soc., 2007, 129(33), [19] Ollila, O. H. S.; Risselada, H. J.; Louhivuori, M.; Lindahl, E.; Vattulainen, I. and Marrink, S. J., Phys. Rev. Lett., 2009, 102(7),
12 [20] Kaminski, G.; Friesner, R.; Tirado-Rives, J. and Jorgensen, W., J. Phys. Chem. B, 2001, 105, [21] Berendsen, H. J. C.; Postma, J. P. M.; van Gunsteren, W. F. and Hermans, J., In Pullman Editors Intermolecular Forces, D. Reidel Publishing Company, Dordrecht, 1981; pages [22] Sorin, E. J. and Pande, V. S., Biophys. J., 2005, 88, [23] Jorgensen, W.; Chandrasekhar, J.; Madura, J.; Impey, R. and Klein, M., J. Chem. Phys., 1983, 79, [24] van Gunsteren, W. F. and Berendsen, H. J. C., Gromos-87 manual Biomos BV Nijenborgh 4,1947 AG Groningen, The Netherlands, [25] Darden, T.; York, D. and Pedersen, L., J. Chem. Phys., 1993, 98, [26] Hess, B.; Bekker, H.; Berendsen, H. J. C. and Fraaije, J. G. E. M., J. Comput. Chem., 1997, 18, [27] Bussi, G.; Donadio, D. and Parrinello, M., J. Chem. Phys., 2007, 126, [28] Berendsen, H.; Postma, J.; DiNola, A. and Haak, J., J. Chem. Phys., 1984, 81, [29] DeLano, W., The pymol molecular graphics system. DeLano Scientific LLC, Palo Alto, CA, USA
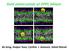
13 Figure 1: The embedding proces. Three snapshot of a protein (Reaaction Center, see table i) grown within the membrane using g membed are shown in top view (upper) and side view (lower). The protein, membrane and water are displayed as Van der Waals spheres and colored yellow, blue and red/white, respectively. The width of the protein in the membrane plane is 0.1, 0.55 and 1.0 for step 1, 500 and 1,000, respectively. Also the length of the box-vectors are shown. 13
14 Figure 2: A protein embedded in a vesicle bilayer. The β-barrel Platform (yellow) has been embedded within a POPE vesicle (blue) in water (red/white) with g membed. 14
15 Figure 3: Equilibrating a system after embedding a protein with g membed into a membrane starting with an initial scaling factor of 0.1. The area per lipid (A lip, eq 3) as a function of simulation time shows that within 1 ns the equilibrium area per lipid is reached. 15
16 pdb-code atoms (nr) A prot (nm 2 ) membrane Integrin αiibβ3 [11] 2K9J popc β-barrel Platform [12] 1 2JMM popc Reaction Center [13] 1PRC dopc Yeast Aquaporin [14] 2W2E dopc β-barrel Platform [12] 1 2JMM pope (vesicle) 1 The discrepancies are due to the use of different force fields. Table i: Systems used to test g membed. Protein (membrane) initial protein scaling factor s 0,xy Integrin αiibβ3 (popc) 2 β-barrel Platform (popc) Reaction Center (dopc) 306 Yeast Aquaporin (dopc) Table ii: Relaxation time τ(ps) of the area per lipid after embedding the protein in a membrane with different initial protein sizes. 16
g_membed: Efficient Insertion of a Membrane Protein into an Equilibrated Lipid Bilayer with Minimal Perturbation
 g_membed: Efficient Insertion of a Membrane Protein into an Equilibrated Lipid Bilayer with Minimal Perturbation MAARTEN G. WOLF, 1 MARTIN HOEFLING, 2 CAMILO APONTE-SANTAMARÍA, 2 HELMUT GRUBMÜLLER, 2 GERRIT
g_membed: Efficient Insertion of a Membrane Protein into an Equilibrated Lipid Bilayer with Minimal Perturbation MAARTEN G. WOLF, 1 MARTIN HOEFLING, 2 CAMILO APONTE-SANTAMARÍA, 2 HELMUT GRUBMÜLLER, 2 GERRIT
Supplementary information for Effects of Stretching Speed on. Mechanical Rupture of Phospholipid/Cholesterol Bilayers: Molecular
 Supplementary information for Effects of Stretching Speed on Mechanical Rupture of Phospholipid/Cholesterol Bilayers: Molecular Dynamics Simulation Taiki Shigematsu, Kenichiro Koshiyama*, and Shigeo Wada
Supplementary information for Effects of Stretching Speed on Mechanical Rupture of Phospholipid/Cholesterol Bilayers: Molecular Dynamics Simulation Taiki Shigematsu, Kenichiro Koshiyama*, and Shigeo Wada
Supplementary Information
 Coarse-Grained Molecular Dynamics Simulations of Photoswitchable Assembly and Disassembly Xiaoyan Zheng, Dong Wang,* Zhigang Shuai,* MOE Key Laboratory of Organic Optoelectronics and Molecular Engineering,
Coarse-Grained Molecular Dynamics Simulations of Photoswitchable Assembly and Disassembly Xiaoyan Zheng, Dong Wang,* Zhigang Shuai,* MOE Key Laboratory of Organic Optoelectronics and Molecular Engineering,
The effect of orientation dynamics in melittin as antimicrobial peptide in lipid bilayer calculated by free energy method
 Journal of Physics: Conference Series PAPER OPEN ACCESS The effect of orientation dynamics in melittin as antimicrobial peptide in lipid bilayer calculated by free energy method To cite this article: Sri
Journal of Physics: Conference Series PAPER OPEN ACCESS The effect of orientation dynamics in melittin as antimicrobial peptide in lipid bilayer calculated by free energy method To cite this article: Sri
We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field
 Parameterization of the fullerene coarse-grained model We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field for lipids 1 and proteins 2. In the MARTINI force
Parameterization of the fullerene coarse-grained model We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field for lipids 1 and proteins 2. In the MARTINI force
Supporting Information for Origin of 1/f noise in hydration dynamics on lipid membrane surfaces
 Supporting Informati for Origin of /f noise in hydrati dynamics lipid membrane surfaces Eiji Yamamoto, Takuma kimoto, Masato Yasui, 2 and Kenji Yasuoka Department of Mechanical Engineering, Keio University,
Supporting Informati for Origin of /f noise in hydrati dynamics lipid membrane surfaces Eiji Yamamoto, Takuma kimoto, Masato Yasui, 2 and Kenji Yasuoka Department of Mechanical Engineering, Keio University,
Ethanol Induces the Formation of Water-Permeable Defects in Model. Bilayers of Skin Lipids
 Electronic Supplementary Material (ESI) for Chemical Communications. This journal is The Royal Society of Chemistry 2015 Ethanol Induces the Formation of Water-Permeable Defects in Model Bilayers of Skin
Electronic Supplementary Material (ESI) for Chemical Communications. This journal is The Royal Society of Chemistry 2015 Ethanol Induces the Formation of Water-Permeable Defects in Model Bilayers of Skin
Phase Behavior of a Phospholipid/Fatty Acid/Water Mixture Studied in Atomic Detail
 Published on Web 01/19/2006 Phase Behavior of a Phospholipid/Fatty Acid/Water Mixture Studied in Atomic Detail Volker Knecht,*, Alan E. Mark,, and Siewert-Jan Marrink Contribution from the Max Planck Institute
Published on Web 01/19/2006 Phase Behavior of a Phospholipid/Fatty Acid/Water Mixture Studied in Atomic Detail Volker Knecht,*, Alan E. Mark,, and Siewert-Jan Marrink Contribution from the Max Planck Institute
Massive oxidation of phospholipid membranes. leads to pore creation and bilayer. disintegration
 Massive oxidation of phospholipid membranes leads to pore creation and bilayer disintegration Lukasz Cwiklik and Pavel Jungwirth Institute of Organic Chemistry and Biochemistry, Academy of Sciences of
Massive oxidation of phospholipid membranes leads to pore creation and bilayer disintegration Lukasz Cwiklik and Pavel Jungwirth Institute of Organic Chemistry and Biochemistry, Academy of Sciences of
Supporting Information: Revisiting partition in. hydrated bilayer systems
 Supporting Information: Revisiting partition in hydrated bilayer systems João T. S. Coimbra, Pedro A. Fernandes, Maria J. Ramos* UCIBIO, REQUIMTE, Departamento de Química e Bioquímica, Faculdade de Ciências,
Supporting Information: Revisiting partition in hydrated bilayer systems João T. S. Coimbra, Pedro A. Fernandes, Maria J. Ramos* UCIBIO, REQUIMTE, Departamento de Química e Bioquímica, Faculdade de Ciências,
Conformational Flexibility of the Peptide Hormone Ghrelin in Solution and Lipid Membrane Bound: A Molecular Dynamics Study
 Open Access Article The authors, the publisher, and the right holders grant the right to use, reproduce, and disseminate the work in digital form to all users. Journal of Biomolecular Structure & Dynamics,
Open Access Article The authors, the publisher, and the right holders grant the right to use, reproduce, and disseminate the work in digital form to all users. Journal of Biomolecular Structure & Dynamics,
Supplementary Information: A Critical. Comparison of Biomembrane Force Fields: Structure and Dynamics of Model DMPC, POPC, and POPE Bilayers
 Supplementary Information: A Critical Comparison of Biomembrane Force Fields: Structure and Dynamics of Model DMPC, POPC, and POPE Bilayers Kristyna Pluhackova,, Sonja A. Kirsch, Jing Han, Liping Sun,
Supplementary Information: A Critical Comparison of Biomembrane Force Fields: Structure and Dynamics of Model DMPC, POPC, and POPE Bilayers Kristyna Pluhackova,, Sonja A. Kirsch, Jing Han, Liping Sun,
Supporting information for: Hydration dynamics of a peripheral membrane. protein
 Supporting information for: Hydration dynamics of a peripheral membrane protein Olivier Fisette,, Christopher Päslack,,, Ryan Barnes, J. Mario Isas, Ralf Langen, Matthias Heyden, Songi Han, and Lars V.
Supporting information for: Hydration dynamics of a peripheral membrane protein Olivier Fisette,, Christopher Päslack,,, Ryan Barnes, J. Mario Isas, Ralf Langen, Matthias Heyden, Songi Han, and Lars V.
Membrane Simulations in GROMACS Lipid bilayers and membrane proteins
 Membrane Simulations in GROMACS Lipid bilayers and membrane proteins Justin A. Lemkul September 13, 2013 jalemkul@outerbanks.umaryland.edu Objectives Learn the fundamentals of Self-consistent force fields
Membrane Simulations in GROMACS Lipid bilayers and membrane proteins Justin A. Lemkul September 13, 2013 jalemkul@outerbanks.umaryland.edu Objectives Learn the fundamentals of Self-consistent force fields
Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes
 Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes Kevin Gasperich Advisor: Dmitry Kopelevich ABSTRACT The recent rapid development of applications for fullerenes has led to an increase
Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes Kevin Gasperich Advisor: Dmitry Kopelevich ABSTRACT The recent rapid development of applications for fullerenes has led to an increase
Supporting material. Membrane permeation induced by aggregates of human islet amyloid polypeptides
 Supporting material Membrane permeation induced by aggregates of human islet amyloid polypeptides Chetan Poojari Forschungszentrum Jülich GmbH, Institute of Complex Systems: Structural Biochemistry (ICS-6),
Supporting material Membrane permeation induced by aggregates of human islet amyloid polypeptides Chetan Poojari Forschungszentrum Jülich GmbH, Institute of Complex Systems: Structural Biochemistry (ICS-6),
Improving PME on distributed computer systems
 Improving PME on distributed computer systems Carsten Kutzner Max-Planck-Institut für Biophysikalische Chemie, Göttingen Theoretical and Computational Biophysics Department Outline how to get optimum performance
Improving PME on distributed computer systems Carsten Kutzner Max-Planck-Institut für Biophysikalische Chemie, Göttingen Theoretical and Computational Biophysics Department Outline how to get optimum performance
Coarse grained simulations of Lipid Bilayer Membranes
 Coarse grained simulations of Lipid Bilayer Membranes P. B. Sunil Kumar Department of Physics IIT Madras, Chennai 600036 sunil@iitm.ac.in Atomistic MD: time scales ~ 10 ns length scales ~100 nm 2 To study
Coarse grained simulations of Lipid Bilayer Membranes P. B. Sunil Kumar Department of Physics IIT Madras, Chennai 600036 sunil@iitm.ac.in Atomistic MD: time scales ~ 10 ns length scales ~100 nm 2 To study
Biological Membranes. Lipid Membranes. Bilayer Permeability. Common Features of Biological Membranes. A highly selective permeability barrier
 Biological Membranes Structure Function Composition Physicochemical properties Self-assembly Molecular models Lipid Membranes Receptors, detecting the signals from outside: Light Odorant Taste Chemicals
Biological Membranes Structure Function Composition Physicochemical properties Self-assembly Molecular models Lipid Membranes Receptors, detecting the signals from outside: Light Odorant Taste Chemicals
Supporting information for: Quantifying. lateral inhomogeneity of. cholesterol-containing membranes
 Supporting information for: Quantifying lateral inhomogeneity of cholesterol-containing membranes Celsa Díaz-Tejada, Igor Ariz-Extreme, Neha Awasthi, and Jochen S. Hub Georg-August-University Göttingen,
Supporting information for: Quantifying lateral inhomogeneity of cholesterol-containing membranes Celsa Díaz-Tejada, Igor Ariz-Extreme, Neha Awasthi, and Jochen S. Hub Georg-August-University Göttingen,
Coarse-Grained Molecular Dynamics for Copolymer- Vesicle Self-Assembly. Case Study: Sterically Stabilized Liposomes.
 Coarse-Grained Molecular Dynamics for Copolymer- Vesicle Self-Assembly. Case Study: Sterically Stabilized Liposomes. Alexander Kantardjiev 1 and Pavletta Shestakova 1 1 Institute of Organic Chemistry with
Coarse-Grained Molecular Dynamics for Copolymer- Vesicle Self-Assembly. Case Study: Sterically Stabilized Liposomes. Alexander Kantardjiev 1 and Pavletta Shestakova 1 1 Institute of Organic Chemistry with
Structural Characterization on the Gel to Liquid-Crystal Phase Transition of Fully Hydrated DSPC and DSPE Bilayers
 8114 J. Phys. Chem. B 2009, 113, 8114 8123 Structural Characterization on the Gel to Liquid-Crystal Phase Transition of Fully Hydrated DSPC and DSPE Bilayers Shan-Shan Qin and Zhi-Wu Yu* Key Lab of Bioorganic
8114 J. Phys. Chem. B 2009, 113, 8114 8123 Structural Characterization on the Gel to Liquid-Crystal Phase Transition of Fully Hydrated DSPC and DSPE Bilayers Shan-Shan Qin and Zhi-Wu Yu* Key Lab of Bioorganic
Water-phospholipid interactions at the interface of lipid. membranes: comparison of different force fields
 Water-phospholipid interactions at the interface of lipid membranes: comparison of different force fields Jianjun Jiang, 1,2 Weixin Li, 1,3 Liang Zhao, 1 Yuanyan Wu, 1 Peng Xiu, 4 Guoning Tang, 3 and Yusong
Water-phospholipid interactions at the interface of lipid membranes: comparison of different force fields Jianjun Jiang, 1,2 Weixin Li, 1,3 Liang Zhao, 1 Yuanyan Wu, 1 Peng Xiu, 4 Guoning Tang, 3 and Yusong
RELEASE OF CONTENT THROUGH MECHANO- SENSITIVE GATES IN PRESSURISED LIPOSOMES. Martti Louhivuori University of Groningen
 RELEASE OF CONTENT THROUGH MECHANO- SENSITIVE GATES IN PRESSURISED LIPOSOMES Martti Louhivuori University of Groningen www.cgmartini.nl MARTINI coarse-grained model water P4 butane Qo Na DPPC cholesterol
RELEASE OF CONTENT THROUGH MECHANO- SENSITIVE GATES IN PRESSURISED LIPOSOMES Martti Louhivuori University of Groningen www.cgmartini.nl MARTINI coarse-grained model water P4 butane Qo Na DPPC cholesterol
Alchembed: A Computational Method for Incorporating Multiple Proteins into Complex Lipid Geometries
 This is an open access article published under a Creative Commons Attribution (CC-BY) License, which permits unrestricted use, distribution and reproduction in any medium, provided the author and source
This is an open access article published under a Creative Commons Attribution (CC-BY) License, which permits unrestricted use, distribution and reproduction in any medium, provided the author and source
Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale
 Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale Rakesh Gupta and Beena Rai* TATA Research Development & Design Centre, TCS Innovation labs, Pune India,
Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale Rakesh Gupta and Beena Rai* TATA Research Development & Design Centre, TCS Innovation labs, Pune India,
Properties of water hydrating the galactolipid and phospholipid bilayers: a molecular dynamics simulation study*
 Regular paper Vol. 62, No 3/2015 475 481 http://dx.doi.org/10.18388/abp.2015_1077 Properties of water hydrating the galactolipid and phospholipid bilayers: a molecular dynamics simulation study* Michał
Regular paper Vol. 62, No 3/2015 475 481 http://dx.doi.org/10.18388/abp.2015_1077 Properties of water hydrating the galactolipid and phospholipid bilayers: a molecular dynamics simulation study* Michał
Supporting Information. to the article. Conical lipids in flat bilayers induce packing defects similar to that induced by positive curvature
 Supporting Information to the article Conical lipids in flat bilayers induce packing defects similar to that induced by positive curvature Lydie Vamparys 1,2,3, Romain Gautier 4, Stefano Vanni 4, W.F.
Supporting Information to the article Conical lipids in flat bilayers induce packing defects similar to that induced by positive curvature Lydie Vamparys 1,2,3, Romain Gautier 4, Stefano Vanni 4, W.F.
Physicochemical Properties of Nanoparticles Regulate Translocation across Pulmonary. Surfactant Monolayer and Formation of Lipoprotein Corona
 Physicochemical Properties of Nanoparticles Regulate Translocation across Pulmonary Surfactant Monolayer and Formation of Lipoprotein Corona Supplementary Information Guoqing Hu 1 *, Bao Jiao 1, Xinghua
Physicochemical Properties of Nanoparticles Regulate Translocation across Pulmonary Surfactant Monolayer and Formation of Lipoprotein Corona Supplementary Information Guoqing Hu 1 *, Bao Jiao 1, Xinghua
FIRM. Full Iterative Relaxation Matrix program
 FIRM Full Iterative Relaxation Matrix program FIRM is a flexible program for calculating NOEs and back-calculated distance constraints using the full relaxation matrix approach. FIRM is an interactive
FIRM Full Iterative Relaxation Matrix program FIRM is a flexible program for calculating NOEs and back-calculated distance constraints using the full relaxation matrix approach. FIRM is an interactive
Pacific Symposium on Biocomputing 12:40-50(2007)
 PREDICTING STRUCTURE AND DYNAMICS OF LOOSELY- ORDERED PROTEIN COMPLEXES: INFLUENZA HEMAGGLUTININ FUSION PEPTIDE PETER M. KASSON Medical Scientist Training Program Stanford University, Stanford CA 94305
PREDICTING STRUCTURE AND DYNAMICS OF LOOSELY- ORDERED PROTEIN COMPLEXES: INFLUENZA HEMAGGLUTININ FUSION PEPTIDE PETER M. KASSON Medical Scientist Training Program Stanford University, Stanford CA 94305
Supporting Information
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 018 Supporting Information Modulating Interactions between Ligand-Coated Nanoparticles and Phase-Separated
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 018 Supporting Information Modulating Interactions between Ligand-Coated Nanoparticles and Phase-Separated
Translocation of C 60 and Its Derivatives Across a Lipid Bilayer
 Translocation of C 60 and Its Derivatives Across a Lipid Bilayer Rui Qiao* NANO LETTERS 2007 Vol. 7, No. 3 614-619 Department of Mechanical Engineering, Clemson UniVersity, Clemson, South Carolina 29634
Translocation of C 60 and Its Derivatives Across a Lipid Bilayer Rui Qiao* NANO LETTERS 2007 Vol. 7, No. 3 614-619 Department of Mechanical Engineering, Clemson UniVersity, Clemson, South Carolina 29634
Modeling and Molecular Dynamics of Membrane Proteins
 Modeling and Molecular Dynamics of Membrane Proteins Emad Tajkhorshid Department of Biochemistry, Center for Biophysics and Computational Biology, and Beckman Institute University of Illinois at Urbana-Champaign
Modeling and Molecular Dynamics of Membrane Proteins Emad Tajkhorshid Department of Biochemistry, Center for Biophysics and Computational Biology, and Beckman Institute University of Illinois at Urbana-Champaign
Flip-Flop Induced Relaxation Of Bending Energy: Implications For Membrane Remodeling
 Biophysical Journal, Volume 97 Supporting Material Flip-Flop Induced Relaxation Of Bending Energy: Implications For Membrane Remodeling Raphael Jeremy Bruckner, Sheref S. Mansy, Alonso Ricardo, L. Mahadevan,
Biophysical Journal, Volume 97 Supporting Material Flip-Flop Induced Relaxation Of Bending Energy: Implications For Membrane Remodeling Raphael Jeremy Bruckner, Sheref S. Mansy, Alonso Ricardo, L. Mahadevan,
Interactions Between a Voltage Sensor and a Toxin via Multiscale Simulations
 1558 Biophysical Journal Volume 98 April 2010 1558 1565 Interactions Between a Voltage Sensor and a Toxin via Multiscale Simulations Chze Ling Wee, David Gavaghan, and Mark S. P. Sansom * Department of
1558 Biophysical Journal Volume 98 April 2010 1558 1565 Interactions Between a Voltage Sensor and a Toxin via Multiscale Simulations Chze Ling Wee, David Gavaghan, and Mark S. P. Sansom * Department of
Molecular dynamics simulation of lipid-protein ensembles
 International Journal of Bioelectromagnetism Vol. 6, No. 1, pp. xx - xx, 2004 www/ijbem.org Molecular dynamics simulation of lipid-protein ensembles Kenichi Yamanishi a, Lukas Pichl a Yuko Nitahara-Kasahara
International Journal of Bioelectromagnetism Vol. 6, No. 1, pp. xx - xx, 2004 www/ijbem.org Molecular dynamics simulation of lipid-protein ensembles Kenichi Yamanishi a, Lukas Pichl a Yuko Nitahara-Kasahara
University of Groningen. Lipids out of equilibrium Tieleman, D. Peter; Marrink, Siewert. Published in: Journal of the American Chemical Society
 University of Groningen Lipids out of equilibrium Tieleman, D. Peter; Marrink, Siewert Published in: Journal of the American Chemical Society DOI: 10.1021/ja0624321 IMPORTANT NOTE: You are advised to consult
University of Groningen Lipids out of equilibrium Tieleman, D. Peter; Marrink, Siewert Published in: Journal of the American Chemical Society DOI: 10.1021/ja0624321 IMPORTANT NOTE: You are advised to consult
Simulationen von Lipidmembranen
 Simulationen von Lipidmembranen Thomas Stockner thomas.stockner@meduniwien.ac.at Molecular biology Molecular modelling Membranes environment Many cellular functions occur in or around membranes: energy
Simulationen von Lipidmembranen Thomas Stockner thomas.stockner@meduniwien.ac.at Molecular biology Molecular modelling Membranes environment Many cellular functions occur in or around membranes: energy
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. (a) Uncropped version of Fig. 2a. RM indicates that the translation was done in the absence of rough mcirosomes. (b) LepB construct containing the GGPG-L6RL6-
Supplementary Figures Supplementary Figure 1. (a) Uncropped version of Fig. 2a. RM indicates that the translation was done in the absence of rough mcirosomes. (b) LepB construct containing the GGPG-L6RL6-
Topic 7b: Biological Membranes
 Topic 7b: Biological Membranes Overview: Why does life need a compartment? Nature of the packaging what is it made of? properties? New types of deformations in 2D Applications: Stretching membranes, forming
Topic 7b: Biological Membranes Overview: Why does life need a compartment? Nature of the packaging what is it made of? properties? New types of deformations in 2D Applications: Stretching membranes, forming
Application of Molecular Modelling and EPR Spectroscopy to Lipid Membranes a Combined Approach
 Armenian Journal of Physics, 2016, vol. 9, issue 2, pp. 159-166 Application of Molecular Modelling and EPR Spectroscopy to Lipid Membranes a Combined Approach Andrea Catte and Vasily S. Oganesyan * School
Armenian Journal of Physics, 2016, vol. 9, issue 2, pp. 159-166 Application of Molecular Modelling and EPR Spectroscopy to Lipid Membranes a Combined Approach Andrea Catte and Vasily S. Oganesyan * School
Molecular dynamics simulations of charged and neutral lipid bilayers: treatment of electrostatic interactions
 Vol. 50 No. 3/2003 789 798 QUARTERLY Molecular dynamics simulations of charged and neutral lipid bilayers: treatment of electrostatic interactions Tomasz Róg, Krzysztof Murzyn and Marta Pasenkiewicz-Gierula
Vol. 50 No. 3/2003 789 798 QUARTERLY Molecular dynamics simulations of charged and neutral lipid bilayers: treatment of electrostatic interactions Tomasz Róg, Krzysztof Murzyn and Marta Pasenkiewicz-Gierula
Supplementary Information
 Supplementary Information Effect of head group and lipid tail oxidation in the cell membrane revealed through integrated simulations and experiments M Yusupov 1, K Wende 2, S Kupsch, E C Neyts 1, S Reuter
Supplementary Information Effect of head group and lipid tail oxidation in the cell membrane revealed through integrated simulations and experiments M Yusupov 1, K Wende 2, S Kupsch, E C Neyts 1, S Reuter
Self-Assembly of Diamondoid Molecules and Derivatives
 [1] Self-Assembly of Diamondoid Molecules and Derivatives (MD Simulations and DFT Calculations) Yong Xue 1 and G.Ali Mansoori 2 University of Illinois at Chicago, (M/C 063) Chicago, IL 60607-7052, USA.
[1] Self-Assembly of Diamondoid Molecules and Derivatives (MD Simulations and DFT Calculations) Yong Xue 1 and G.Ali Mansoori 2 University of Illinois at Chicago, (M/C 063) Chicago, IL 60607-7052, USA.
Molecular Dynamics Simulations of Mixed Micelles Modeling Human Bile
 Biochemistry 2002, 41, 5375-5382 5375 Molecular Dynamics Simulations of Mixed Micelles Modeling Human Bile S. J. Marrink* and A. E. Mark Department of Biophysical Chemistry, UniVersity of Groningen, Nijenborgh
Biochemistry 2002, 41, 5375-5382 5375 Molecular Dynamics Simulations of Mixed Micelles Modeling Human Bile S. J. Marrink* and A. E. Mark Department of Biophysical Chemistry, UniVersity of Groningen, Nijenborgh
Molecular modeling of the pathways of vesicle membrane interaction. Tongtao Yue and Xianren Zhang
 Molecular modeling of the pathways of vesicle membrane interaction Tongtao Yue and Xianren Zhang I. ELECTRONIC SUPPLEMENTARY INFORMATION (ESI): METHODS Dissipative particle dynamics method The dissipative
Molecular modeling of the pathways of vesicle membrane interaction Tongtao Yue and Xianren Zhang I. ELECTRONIC SUPPLEMENTARY INFORMATION (ESI): METHODS Dissipative particle dynamics method The dissipative
Self-Assembly of Diamondoid Molecules and Derivatives (MD Simulations and DFT Calculations)
 Int. J. Mol. Sci. 2010, 11, 288-303; doi:10.3390/ijms11010288 Article OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Self-Assembly of Diamondoid Molecules
Int. J. Mol. Sci. 2010, 11, 288-303; doi:10.3390/ijms11010288 Article OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Self-Assembly of Diamondoid Molecules
Kinetics, Statistics, and Energetics of Lipid Membrane Electroporation Studied by Molecular Dynamics Simulations
 Biophysical Journal Volume 95 August 2008 1837 1850 1837 Kinetics, Statistics, and Energetics of Lipid Membrane Electroporation Studied by Molecular Dynamics Simulations Rainer A. Böckmann,* Bert L. de
Biophysical Journal Volume 95 August 2008 1837 1850 1837 Kinetics, Statistics, and Energetics of Lipid Membrane Electroporation Studied by Molecular Dynamics Simulations Rainer A. Böckmann,* Bert L. de
Interaction Between Amyloid-b (1 42) Peptide and Phospholipid Bilayers: A Molecular Dynamics Study
 Biophysical Journal Volume 96 February 2009 785 797 785 Interaction Between Amyloid-b (1 42) Peptide and Phospholipid Bilayers: A Molecular Dynamics Study Charles H. Davis and Max L. Berkowitz * Department
Biophysical Journal Volume 96 February 2009 785 797 785 Interaction Between Amyloid-b (1 42) Peptide and Phospholipid Bilayers: A Molecular Dynamics Study Charles H. Davis and Max L. Berkowitz * Department
Dynamics and energetics of solute permeation through the Plasmodium falciparum aquaglyceroporin
 PAPER www.rsc.org/pccp Physical Chemistry Chemical Physics Dynamics and energetics of solute permeation through the Plasmodium falciparum aquaglyceroporin Camilo Aponte-Santamaría, a Jochen S. Hub b and
PAPER www.rsc.org/pccp Physical Chemistry Chemical Physics Dynamics and energetics of solute permeation through the Plasmodium falciparum aquaglyceroporin Camilo Aponte-Santamaría, a Jochen S. Hub b and
MARTINI Coarse-Grained Model of Triton TX-100 in Pure DPPC. Monolayer and Bilayer Interfaces. Supporting Information
 MARTINI Coarse-Grained Model of Triton TX-100 in Pure DPPC Monolayer and Bilayer Interfaces. Antonio Pizzirusso a, Antonio De Nicola* a, Giuseppe Milano a a Dipartimento di Chimica e Biologia, Università
MARTINI Coarse-Grained Model of Triton TX-100 in Pure DPPC Monolayer and Bilayer Interfaces. Antonio Pizzirusso a, Antonio De Nicola* a, Giuseppe Milano a a Dipartimento di Chimica e Biologia, Università
H 2 O. Liquid, solid, and vapor coexist in the same environment
 Water H 2 O Liquid, solid, and vapor coexist in the same environment WATER MOLECULES FORM HYDROGEN BONDS Water is a fundamental requirement for life, so it is important to understand the structural and
Water H 2 O Liquid, solid, and vapor coexist in the same environment WATER MOLECULES FORM HYDROGEN BONDS Water is a fundamental requirement for life, so it is important to understand the structural and
Asymmetry of lipid bilayers induced by monovalent salt: Atomistic molecular-dynamics study
 THE JOURNAL OF CHEMICAL PHYSICS 122, 244902 2005 Asymmetry of lipid bilayers induced by monovalent salt: Atomistic molecular-dynamics study Andrey A. Gurtovenko a Laboratory of Physics and Helsinki Institute
THE JOURNAL OF CHEMICAL PHYSICS 122, 244902 2005 Asymmetry of lipid bilayers induced by monovalent salt: Atomistic molecular-dynamics study Andrey A. Gurtovenko a Laboratory of Physics and Helsinki Institute
The Interaction of Phospholipase A2 with a Phospholipid Bilayer: Coarse-Grained Molecular Dynamics Simulations
 Biophysical Journal Volume 95 August 2008 1649 1657 1649 The Interaction of Phospholipase A2 with a Phospholipid Bilayer: Coarse-Grained Molecular Dynamics Simulations Chze Ling Wee,* Kia Balali-Mood,*
Biophysical Journal Volume 95 August 2008 1649 1657 1649 The Interaction of Phospholipase A2 with a Phospholipid Bilayer: Coarse-Grained Molecular Dynamics Simulations Chze Ling Wee,* Kia Balali-Mood,*
Interactions of Polyethylenimines with Zwitterionic and. Anionic Lipid Membranes
 Interactions of Polyethylenimines with Zwitterionic and Anionic Lipid Membranes Urszula Kwolek, Dorota Jamróz, Małgorzata Janiczek, Maria Nowakowska, Paweł Wydro, Mariusz Kepczynski Faculty of Chemistry,
Interactions of Polyethylenimines with Zwitterionic and Anionic Lipid Membranes Urszula Kwolek, Dorota Jamróz, Małgorzata Janiczek, Maria Nowakowska, Paweł Wydro, Mariusz Kepczynski Faculty of Chemistry,
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/3/eaaq0762/dc1 Supplementary Materials for Structures of monomeric and oligomeric forms of the Toxoplasma gondii perforin-like protein 1 Tao Ni, Sophie I. Williams,
advances.sciencemag.org/cgi/content/full/4/3/eaaq0762/dc1 Supplementary Materials for Structures of monomeric and oligomeric forms of the Toxoplasma gondii perforin-like protein 1 Tao Ni, Sophie I. Williams,
Lipid Bilayers Are Excellent For Cell Membranes
 Lipid Bilayers Are Excellent For Cell Membranes ydrophobic interaction is the driving force Self-assembly in water Tendency to close on themselves Self-sealing (a hole is unfavorable) Extensive: up to
Lipid Bilayers Are Excellent For Cell Membranes ydrophobic interaction is the driving force Self-assembly in water Tendency to close on themselves Self-sealing (a hole is unfavorable) Extensive: up to
Supporting Information: Atomistic Model for Near-Quantitative. Simulations of Langmuir Monolayers
 Supporting Information: Atomistic Model for Near-Quantitative Simulations of Langmuir Monolayers Matti Javanainen,,, Antti Lamberg, Lukasz Cwiklik,, Ilpo Vattulainen,,, and Samuli Ollila,,# Laboratory
Supporting Information: Atomistic Model for Near-Quantitative Simulations of Langmuir Monolayers Matti Javanainen,,, Antti Lamberg, Lukasz Cwiklik,, Ilpo Vattulainen,,, and Samuli Ollila,,# Laboratory
Andrey A. Gurtovenko*, and Ilpo Vattulainen,,
 J. Phys. Chem. B 2008, 112, 1953-1962 1953 Effect of NaCl and KCl on Phosphatidylcholine and Phosphatidylethanolamine Lipid Membranes: Insight from Atomic-Scale Simulations for Understanding Salt-Induced
J. Phys. Chem. B 2008, 112, 1953-1962 1953 Effect of NaCl and KCl on Phosphatidylcholine and Phosphatidylethanolamine Lipid Membranes: Insight from Atomic-Scale Simulations for Understanding Salt-Induced
Gold nanocrystals at DPPC bilayer. Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad
 Gold nanocrystals at DPPC bilayer Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad Coarse-grained mapping strategy of a DPPC lipid molecule. The structure of gold nanocrystals (bare gold nanoparticles)
Gold nanocrystals at DPPC bilayer Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad Coarse-grained mapping strategy of a DPPC lipid molecule. The structure of gold nanocrystals (bare gold nanoparticles)
Role of Surface Ligands in Nanoparticle Permeation through a Model. Membrane: A Coarse grained Molecular Dynamics Simulations Study
 Role of Surface Ligands in Nanoparticle Permeation through a Model Membrane: A Coarse grained Molecular Dynamics Simulations Study Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad * Department of
Role of Surface Ligands in Nanoparticle Permeation through a Model Membrane: A Coarse grained Molecular Dynamics Simulations Study Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad * Department of
Bilayer Deformation, Pores & Micellation Induced by Oxidized Lipids
 Supporting Information Bilayer Deformation, Pores & Micellation Induced by Oxidized Lipids Phansiri Boonnoy 1, Viwan Jarerattanachat 1,2, Mikko Karttunen 3*, and Jirasak Wongekkabut 1* 1 Department of
Supporting Information Bilayer Deformation, Pores & Micellation Induced by Oxidized Lipids Phansiri Boonnoy 1, Viwan Jarerattanachat 1,2, Mikko Karttunen 3*, and Jirasak Wongekkabut 1* 1 Department of
Simulationen von Lipidmembranen
 Simulationen von Lipidmembranen Thomas Stockner Thomas.stockner@meduniwien.ac.at Summary Methods Force Field MD simulations Membrane simulations Application Oxidized lipids Anesthetics Molecular biology
Simulationen von Lipidmembranen Thomas Stockner Thomas.stockner@meduniwien.ac.at Summary Methods Force Field MD simulations Membrane simulations Application Oxidized lipids Anesthetics Molecular biology
Molecular Dynamics Simulations of Hydrophilic Pores in Lipid Bilayers Leontiadou, Hari; Mark, Alan E.; Marrink, Siewert
 University of Groningen Molecular Dynamics Simulations of Hydrophilic Pores in Lipid Bilayers Leontiadou, Hari; Mark, Alan E.; Marrink, Siewert Published in: Biophysical Journal DOI: 10.1016/S0006-3495(04)74275-7
University of Groningen Molecular Dynamics Simulations of Hydrophilic Pores in Lipid Bilayers Leontiadou, Hari; Mark, Alan E.; Marrink, Siewert Published in: Biophysical Journal DOI: 10.1016/S0006-3495(04)74275-7
Term Definition Example Amino Acids
 Name 1. What are some of the functions that proteins have in a living organism. 2. Define the following and list two amino acids that fit each description. Term Definition Example Amino Acids Hydrophobic
Name 1. What are some of the functions that proteins have in a living organism. 2. Define the following and list two amino acids that fit each description. Term Definition Example Amino Acids Hydrophobic
GridMAT-MD: A Grid-based Membrane Analysis Tool for use with Molecular Dynamics
 GridMAT-MD: A Grid-based Membrane Analysis Tool for use with Molecular Dynamics William J. Allen, Justin A. Lemkul, and David R. Bevan Department of Biochemistry, Virginia Tech User s Guide Version 1.0.2
GridMAT-MD: A Grid-based Membrane Analysis Tool for use with Molecular Dynamics William J. Allen, Justin A. Lemkul, and David R. Bevan Department of Biochemistry, Virginia Tech User s Guide Version 1.0.2
Model of an Asymmetric DPPC/DPPS Membrane: Effect of Asymmetry on the Lipid Properties. A Molecular Dynamics Simulation Study
 2358 J. Phys. Chem. B 2006, 110, 2358-2363 Model of an Asymmetric DPPC/DPPS Membrane: Effect of Asymmetry on the Lipid Properties. A Molecular Dynamics Simulation Study J. J. López Cascales,*, T. F. Otero,
2358 J. Phys. Chem. B 2006, 110, 2358-2363 Model of an Asymmetric DPPC/DPPS Membrane: Effect of Asymmetry on the Lipid Properties. A Molecular Dynamics Simulation Study J. J. López Cascales,*, T. F. Otero,
Membrane Potential and Electrostatics of Phospholipid Bilayers with Asymmetric Transmembrane Distribution of Anionic Lipids
 J. Phys. Chem. B 2008, 112, 4629-4634 4629 Membrane Potential and Electrostatics of Phospholipid Bilayers with Asymmetric Transmembrane Distribution of Anionic Lipids Andrey A. Gurtovenko*, and Ilpo Vattulainen,,
J. Phys. Chem. B 2008, 112, 4629-4634 4629 Membrane Potential and Electrostatics of Phospholipid Bilayers with Asymmetric Transmembrane Distribution of Anionic Lipids Andrey A. Gurtovenko*, and Ilpo Vattulainen,,
Lipid-Protein Interactions of Integral Membrane Proteins: A Comparative Simulation Study
 Biophysical Journal Volume 87 December 2004 3737 3749 3737 Lipid-Protein Interactions of Integral Membrane Proteins: A Comparative Simulation Study Sundeep S. Deol,* Peter J. Bond,* Carmen Domene,* y and
Biophysical Journal Volume 87 December 2004 3737 3749 3737 Lipid-Protein Interactions of Integral Membrane Proteins: A Comparative Simulation Study Sundeep S. Deol,* Peter J. Bond,* Carmen Domene,* y and
0.5 nm nm acyl tail region (hydrophobic) 1.5 nm. Hydrophobic repulsion organizes amphiphilic molecules: These scales are 5 10xk B T:
 Lecture 31: Biomembranes: The hydrophobic energy scale and membrane behavior 31.1 Reading for Lectures 30-32: PKT Chapter 11 (skip Ch. 10) Upshot of last lecture: Generic membrane lipid: Can be cylindrical
Lecture 31: Biomembranes: The hydrophobic energy scale and membrane behavior 31.1 Reading for Lectures 30-32: PKT Chapter 11 (skip Ch. 10) Upshot of last lecture: Generic membrane lipid: Can be cylindrical
Effects of Epicholesterol on the Phosphatidylcholine Bilayer: A Molecular Simulation Study
 1818 Biophysical Journal Volume 84 March 2003 1818 1826 Effects of Epicholesterol on the Phosphatidylcholine Bilayer: A Molecular Simulation Study Tomasz Róg and Marta Pasenkiewicz-Gierula Department of
1818 Biophysical Journal Volume 84 March 2003 1818 1826 Effects of Epicholesterol on the Phosphatidylcholine Bilayer: A Molecular Simulation Study Tomasz Róg and Marta Pasenkiewicz-Gierula Department of
Molecular Dynamics Simulation of Melittin in a Dimyristoylphosphatidylcholine Bilayer Membrane
 Biophysical Journal Volume 75 October 1998 1603 1618 1603 Molecular Dynamics Simulation of Melittin in a Dimyristoylphosphatidylcholine Bilayer Membrane Simon Bernèche, Mafalda Nina, and Benoît Roux Membrane
Biophysical Journal Volume 75 October 1998 1603 1618 1603 Molecular Dynamics Simulation of Melittin in a Dimyristoylphosphatidylcholine Bilayer Membrane Simon Bernèche, Mafalda Nina, and Benoît Roux Membrane
Molecular Dynamics Simulations of the Anchoring and Tilting of the Lung-Surfactant Peptide SP-B 1-25 in Palmitic Acid Monolayers
 Biophysical Journal Volume 89 December 2005 3807 3821 3807 Molecular Dynamics Simulations of the Anchoring and Tilting of the Lung-Surfactant Peptide SP-B 1-25 in Palmitic Acid Monolayers Hwankyu Lee,*
Biophysical Journal Volume 89 December 2005 3807 3821 3807 Molecular Dynamics Simulations of the Anchoring and Tilting of the Lung-Surfactant Peptide SP-B 1-25 in Palmitic Acid Monolayers Hwankyu Lee,*
Molecular dynamics investigation of myelin basic protein stability on lipid membranes
 Molecular dynamics investigation of myelin basic protein stability on lipid membranes Kyrylo Bessonov, George Harauz* *This study was conducted under the supervision of Professor George Harauz, Department
Molecular dynamics investigation of myelin basic protein stability on lipid membranes Kyrylo Bessonov, George Harauz* *This study was conducted under the supervision of Professor George Harauz, Department
Structural Change in Lipid Bilayers and Water Penetration Induced by Shock Waves: Molecular Dynamics Simulations
 2198 Biophysical Journal Volume 91 September 2006 2198 2205 Structural Change in Lipid Bilayers and Water Penetration Induced by Shock Waves: Molecular Dynamics Simulations Kenichiro Koshiyama,* Tetsuya
2198 Biophysical Journal Volume 91 September 2006 2198 2205 Structural Change in Lipid Bilayers and Water Penetration Induced by Shock Waves: Molecular Dynamics Simulations Kenichiro Koshiyama,* Tetsuya
obtained for the simulations of the E2 conformation of SERCA in a pure POPC lipid bilayer (blue) and in a
 Supplementary Figure S1. Distribution of atoms along the bilayer normal. Normalized density profiles obtained for the simulations of the E2 conformation of SERCA in a pure POPC lipid bilayer (blue) and
Supplementary Figure S1. Distribution of atoms along the bilayer normal. Normalized density profiles obtained for the simulations of the E2 conformation of SERCA in a pure POPC lipid bilayer (blue) and
Comparing Simulations of Lipid Bilayers to Scattering Data: The GROMOS 43A1-S3 Force Field
 pubs.acs.org/jpcb Comparing Simulations of Lipid Bilayers to Scattering Data: The GROMOS 43A1-S3 Force Field Anthony R. Braun, Jonathan N. Sachs, and John F. Nagle*, Department of Biomedical Engineering,
pubs.acs.org/jpcb Comparing Simulations of Lipid Bilayers to Scattering Data: The GROMOS 43A1-S3 Force Field Anthony R. Braun, Jonathan N. Sachs, and John F. Nagle*, Department of Biomedical Engineering,
Karen Doniza 1*, Hui Lin Ong 2, Al Rey Villagracia 1, Aristotle Ubando 5 Melanie David 1, Nelson Arboleda Jr. 1,3, Alvin Culaba 5, Hideaki Kasai 4
 A Molecular Dynamics Study on the Permeability of Oxygen, Carbon Dioxide and Xenon on dioleoyl-phosphatidylcholine (DOPC) and 1- palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) Double Lipid
A Molecular Dynamics Study on the Permeability of Oxygen, Carbon Dioxide and Xenon on dioleoyl-phosphatidylcholine (DOPC) and 1- palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) Double Lipid
Phospholipid Component Volumes: Determination and Application to Bilayer Structure Calculations
 734 Biophysical Journal Volume 75 August 1998 734 744 Phospholipid Component Volumes: Determination and Application to Bilayer Structure Calculations Roger S. Armen, Olivia D. Uitto, and Scott E. Feller
734 Biophysical Journal Volume 75 August 1998 734 744 Phospholipid Component Volumes: Determination and Application to Bilayer Structure Calculations Roger S. Armen, Olivia D. Uitto, and Scott E. Feller
Accelerating potential of mean force calculations for lipid membrane permeation: System size, reaction coordinate, solute-solute distance, and cutoffs
 Accelerating potential of mean force calculations for lipid membrane permeation: System size, reaction coordinate, solute-solute distance, and cutoffs Naomi Nitschke, Kalina Atkovska, and Jochen S. Hub
Accelerating potential of mean force calculations for lipid membrane permeation: System size, reaction coordinate, solute-solute distance, and cutoffs Naomi Nitschke, Kalina Atkovska, and Jochen S. Hub
Lipid Packing Drives the Segregation of Transmembrane Helices into Disordered Lipid Domains in Model Membranes
 Supporting Information Lipid Packing Drives the Segregation of Transmembrane Helices into Disordered Lipid Domains in Model Membranes Lars V. Schäfer, Djurre H. de Jong, Andrea Holt, Andrzej J. Rzepiela,
Supporting Information Lipid Packing Drives the Segregation of Transmembrane Helices into Disordered Lipid Domains in Model Membranes Lars V. Schäfer, Djurre H. de Jong, Andrea Holt, Andrzej J. Rzepiela,
The Influence of Ceramide Tail Length on the. Structure of Bilayers Composed of Stratum. Corneum Lipids
 The Influence of Ceramide Tail Length on the Structure of Bilayers Composed of Stratum Corneum Lipids T. C. Moore, R. Hartkamp, C. R. Iacovella, A. L. Bunge, and C. M c Cabe Condensed Title: Stratum Corneum
The Influence of Ceramide Tail Length on the Structure of Bilayers Composed of Stratum Corneum Lipids T. C. Moore, R. Hartkamp, C. R. Iacovella, A. L. Bunge, and C. M c Cabe Condensed Title: Stratum Corneum
Multi-compartment encapsulation of communicating droplets and droplet networks in hydrogel as a model for artificial cells
 Multi-compartment encapsulation of communicating droplets and droplet networks in hydrogel as a model for artificial cells Mariam Bayoumi 1, Hagan Bayley 2, Giovanni Maglia 1,3, K. Tanuj Sapra 4 1 Department
Multi-compartment encapsulation of communicating droplets and droplet networks in hydrogel as a model for artificial cells Mariam Bayoumi 1, Hagan Bayley 2, Giovanni Maglia 1,3, K. Tanuj Sapra 4 1 Department
Computational analysis of local membrane properties
 J Comput Aided Mol Des (2013) 27:845 858 DOI 10.1007/s10822-013-9684-0 Computational analysis of local membrane properties Vytautas Gapsys Bert L. de Groot Rodolfo Briones Received: 27 May 2013 / Accepted:
J Comput Aided Mol Des (2013) 27:845 858 DOI 10.1007/s10822-013-9684-0 Computational analysis of local membrane properties Vytautas Gapsys Bert L. de Groot Rodolfo Briones Received: 27 May 2013 / Accepted:
Free Volume Properties of Sphingomyelin, DMPC, DPPC, and PLPC Bilayers
 PUBLISHED IN: JOURNAL OF COMPUTATIONAL AND THEORETICAL NANOSCIENCE, VOL. 2, PP. 40-43 (2005). Free Volume Properties of Sphingomyelin, DMPC, DPPC, and PLPC Bilayers M. Kupiainen, E. Falck, S. Ollila, P.
PUBLISHED IN: JOURNAL OF COMPUTATIONAL AND THEORETICAL NANOSCIENCE, VOL. 2, PP. 40-43 (2005). Free Volume Properties of Sphingomyelin, DMPC, DPPC, and PLPC Bilayers M. Kupiainen, E. Falck, S. Ollila, P.
A proposed mechanism for tear film breakup: a molecular level view by employing in-silico approach
 Journal for Modeling in Ophthalmology 2018; 1:19-23 Meeting highlights article: ARVO 2017 A proposed mechanism for tear film breakup: a molecular level view by employing in-silico approach Lukasz Cwiklik
Journal for Modeling in Ophthalmology 2018; 1:19-23 Meeting highlights article: ARVO 2017 A proposed mechanism for tear film breakup: a molecular level view by employing in-silico approach Lukasz Cwiklik
University of Groningen. Ion transport across transmembrane pores Leontiadou, Hari; Mark, Alan E.; Marrink, Siewert. Published in: Biophysical Journal
 University of Groningen Ion transport across transmembrane pores Leontiadou, Hari; Mark, Alan E.; Marrink, Siewert Published in: Biophysical Journal DOI: 10.1529/biophysj.106.101295 IMPORTANT NOTE: You
University of Groningen Ion transport across transmembrane pores Leontiadou, Hari; Mark, Alan E.; Marrink, Siewert Published in: Biophysical Journal DOI: 10.1529/biophysj.106.101295 IMPORTANT NOTE: You
Defining the Membrane-Associated State of the PTEN Tumor Suppressor Protein
 Biophysical Journal Volume 104 February 2013 613 621 613 Defining the Membrane-Associated State of the PTEN Tumor Suppressor Protein Craig N. Lumb and Mark S. P. Sansom* Department of Biochemistry, University
Biophysical Journal Volume 104 February 2013 613 621 613 Defining the Membrane-Associated State of the PTEN Tumor Suppressor Protein Craig N. Lumb and Mark S. P. Sansom* Department of Biochemistry, University
Inter-Species Cross-Seeding: Stability and Assembly of Rat - Human Amylin Aggregates. Workalemahu M. Berhanu
 Inter-Species Cross-Seeding: Stability and Assembly of Rat - Human Amylin Aggregates Workalemahu M. Berhanu University of Oklahoma Department of Chemistry and Biochemistry STRUCTURE OF AMYLOIDS & ROLE
Inter-Species Cross-Seeding: Stability and Assembly of Rat - Human Amylin Aggregates Workalemahu M. Berhanu University of Oklahoma Department of Chemistry and Biochemistry STRUCTURE OF AMYLOIDS & ROLE
Life Sciences 1a. Practice Problems 4
 Life Sciences 1a Practice Problems 4 1. KcsA, a channel that allows K + ions to pass through the membrane, is a protein with four identical subunits that form a channel through the center of the tetramer.
Life Sciences 1a Practice Problems 4 1. KcsA, a channel that allows K + ions to pass through the membrane, is a protein with four identical subunits that form a channel through the center of the tetramer.
RSC Advances.
 This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after
This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after
University of Groningen. Molecular dynamics simulation of a lipid diamond cubic phase Marrink, Siewert; Tieleman, D.P
 University of Groningen Molecular dynamics simulation of a lipid diamond cubic phase Marrink, Siewert; Tieleman, D.P Published in: Journal of the American Chemical Society DOI: 10.1021/ja016012h IMPORTANT
University of Groningen Molecular dynamics simulation of a lipid diamond cubic phase Marrink, Siewert; Tieleman, D.P Published in: Journal of the American Chemical Society DOI: 10.1021/ja016012h IMPORTANT
Arginine-rich peptides destabilize the plasma membrane, consistent with a pore formation translocation mechanism of cell penetrating peptides
 Supplementary materials for: Arginine-rich peptides destabilize the plasma membrane, consistent with a pore formation translocation mechanism of cell penetrating peptides H. D. Herce 1,2, A. E. Garcia
Supplementary materials for: Arginine-rich peptides destabilize the plasma membrane, consistent with a pore formation translocation mechanism of cell penetrating peptides H. D. Herce 1,2, A. E. Garcia
Molecular Dynamics Simulation of a Dipalmitoylphosphatidylcholine Bilayer with NaCl
 Biophysical Journal Volume 84 June 2003 3743 3750 3743 Molecular Dynamics Simulation of a Dipalmitoylphosphatidylcholine Bilayer with NaCl Sagar A. Pandit,* David Bostick, y and Max L. Berkowitz* *Department
Biophysical Journal Volume 84 June 2003 3743 3750 3743 Molecular Dynamics Simulation of a Dipalmitoylphosphatidylcholine Bilayer with NaCl Sagar A. Pandit,* David Bostick, y and Max L. Berkowitz* *Department
PROCEEDINGS OF THE YEREVAN STATE UNIVERSITY
 PROCEEDINGS OF THE YEREVAN STATE UNIVERSITY Physical and Mathematical Sciences 2018, 52(3), p. 217 221 P h y s i c s STUDY OF THE SWELLING OF THE PHOSPHOLIPID BILAYER, DEPENDING ON THE ANGLE BETWEEN THE
PROCEEDINGS OF THE YEREVAN STATE UNIVERSITY Physical and Mathematical Sciences 2018, 52(3), p. 217 221 P h y s i c s STUDY OF THE SWELLING OF THE PHOSPHOLIPID BILAYER, DEPENDING ON THE ANGLE BETWEEN THE
Coarse-grained model for phospholipid/cholesterol bilayer employing inverse Monte Carlo with thermodynamic constraints
 THE JOURNAL OF CHEMICAL PHYSICS 126, 075101 2007 Coarse-grained model for phospholipid/cholesterol bilayer employing inverse Monte Carlo with thermodynamic constraints Teemu Murtola Laboratory of Physics,
THE JOURNAL OF CHEMICAL PHYSICS 126, 075101 2007 Coarse-grained model for phospholipid/cholesterol bilayer employing inverse Monte Carlo with thermodynamic constraints Teemu Murtola Laboratory of Physics,
Improved Coarse-Grained Modeling of Cholesterol-Containing Lipid Bilayers
 This is an open access article published under an ACS AuthorChoice License, which permits copying and redistribution of the article or any adaptations for non-commercial purposes. pubs.acs.org/jctc Improved
This is an open access article published under an ACS AuthorChoice License, which permits copying and redistribution of the article or any adaptations for non-commercial purposes. pubs.acs.org/jctc Improved
Journal of Biomechanical Science and Engineering
 013456789 Bulletin of the JSME Vol.11, No.1, 016 Journal of Biomechanical Science and Engineering Line tension of the pore edge in phospholipid/cholesterol bilayer from stretch molecular dynamics simulation
013456789 Bulletin of the JSME Vol.11, No.1, 016 Journal of Biomechanical Science and Engineering Line tension of the pore edge in phospholipid/cholesterol bilayer from stretch molecular dynamics simulation
