Supplementary Information: A Critical. Comparison of Biomembrane Force Fields: Structure and Dynamics of Model DMPC, POPC, and POPE Bilayers
|
|
|
- Garry Porter
- 5 years ago
- Views:
Transcription
1 Supplementary Information: A Critical Comparison of Biomembrane Force Fields: Structure and Dynamics of Model DMPC, POPC, and POPE Bilayers Kristyna Pluhackova,, Sonja A. Kirsch, Jing Han, Liping Sun, Zhenyan Jiang, Tobias Unruh, and Rainer A. Böckmann, Computational Biology, Department of Biology, Friedrich-Alexander-University of Erlangen-Nürnberg, Staudtstr. 5, Erlangen Lehrstuhl für Kristallografie und Strukturphysik, Department Physik, Friedrich-Alexander-University of Erlangen-Nürnberg, Staudtstr. 3, Erlangen kristyna.pluhackova@fau.de; rainer.boeckmann@fau.de 1
2 Table S1: Volumes of individual water molecule (in nm 3 ) simulated using force field specific parameter file (mdp files in GROMACS) calculated out of 4 ns simulations containing water molecules. Force field water model 320 K 300 K 308 K CHARMM36 TIPS3P Slipids TIP3P Lipid14 TIP3P GROMOS54a7 SPC
3 Figure S1: POPE simulation systems after 200 ns simulation time at 300 K. Only GROMOS54a7 which is partially in a gel phase doesn t exhibit a proper fluid phase. Hydrogen atoms were omitted for clarity. Water is shown as blue surface, DMPC as atom-color-coded sticks (carbon in green, oxygen in red, nitrogen in blue, and phosphorus in orange). 3
4 Figure S2: Evolution of the area per lipid over time. 4
5 Figure S3: Diffusion coefficients calculated in different ways. In each simulation the first 60 ns were excluded for equilibration. The center of mass (COM) motion of the whole bilayer (A) or separately of each monolayer (B-G) was removed. The mean square displacement (MSD) was calculated for lipid atoms (AE) or for the lipid center of masses (FG). In green columns (A, B, F), the diffusion was calculated by splitting the simulations in 10 ns windows (linear fit on 2-5 ns). In blue columns (C), the simulations were split in 70 ns time windows (linear fitting from 10 to 40 ns). In red (D, G) and yellow (E) columns, the diffusion was calculated from the whole ns simulations with linear fitting on ns and ns, respectively. The diffusion coefficient was calculated separately for each monolayer and averaged over all atoms/molecules and over time. Experimental findings were taken from (a) Almeida et al. (1992), 4 (b) Filippov et al. (2003), 5 (c) Ko chy et al. (1993), 6 and (d) Jin et al. (1999; 78 mol % PE). 7 The difference between the diffusion coefficients, calculated with the aid of the lipid atoms, and a removal of the bilayer (A) and monolayer (B) COM motion, is rather small. The same holds true for the diffusion coefficients obtained by lipid atoms (D) and lipid COM (G) for the whole 140 ns interval. Overall, the diffusion coefficients calculated over larger time windows exhibit smaller values compared to time windows of small duration. 5
6 6
7 Table S2: Detailed results of lipid chain protrusion analysis. Analysis performed on ns of simulations. Protrusions recorded at a single frame (10 ps) were excluded from the analysis. The value in paranthesis by median time denotes the interquartile range. Protrusion efficiency is a product of protrusion probability and the median time of a protrusion event. Simulated layer number of probability median time maximal protrusion System protrusions per lipid (IQR) time efficiency [1/µs] [ns] [ns] CHARMM36 DMPC lower (1.68) K upper (3.23) DMPC lower (0.50) K upper (0.37) POPC lower (1.19) K upper (0.83) POPC lower (0.43) K upper (0.41) POPE lower (0.99) K upper (0.32) Slipids DMPC lower (0.69) K upper (0.90) DMPC lower (0.55) K upper (0.55) POPC lower (0.83) K upper (1.11) POPC lower (0.60) K upper (0.37) POPE lower (0.82) K upper (0.75) Lipid14 DMPC lower (1.25) K upper (1.30) DMPC lower (0.51) K upper (0.55) POPC lower (0.63) K upper (0.70) POPC lower (0.45) K upper (0.49) POPE lower (0.78) K upper (0.72) GROMOS54a7 DMPC lower (1.74) K upper (1.51) DMPC lower (1.28) K upper (1.53) POPC lower (1.79) K upper (1.70) POPC lower (1.15) K upper (1.36) POPE lower (0.00) K upper (0.00)
8 Figure S4: Box plots of lipid protrusion times for systems under study. CHARMM36 is colored in purple, GROMOS54a7 in red, Lipid14 in blue, and Slipids in green. Horizontal top and bottom lines of each box define the first and third quartiles, the thick line inside the box is the median. The whiskers (horizontal lines outside the box) determine the last data point that is no more away than 1.5 times the interquartile range (box length) from the box. Box plots were generated with RStudio. 8 8
9 Figure S5: Distribution of PN vectors relative to the membrane normal for the POPE gel phase obtained by the GROMOS54a7 force field. The population of lipids with the PN vector smaller than 75 (an example lipid shown on the left) corresponds to 30%, the population of lipids with the PN vector larger than 75 (an example lipid shown on the right) makes up 70%. 9
10 Calculation of order parameters of unsaturated carbons Using the GROMACS s tool g order to calculate the order parameters around unsaturated carbons succeeds by following following workflow. The test case is a lipid containing one saturated chain (palmitoyl carbons named C2A - C2P) and one monounsaturated (oleoyl, carbons C1A-C1R) chain. Step 1. Calculation of standard saturated order parameters for all carbons. First index files have to be created for each chain individually containing only carbons for which order parameters should be calculated. Please note, that using g order no order parameter value can be calculated for the first and the last carbon in the chain. make ndx -f backmapped.gro -o sn1.ndx << EOF del 0-20 a C2A a C2B a C2C a C2D a C2E a C2F a C2G a C2H a C2I a C2J a C2K a C2L a C2M a C2N a C2O a C2P q EOF make ndx -f backmapped.gro -o sn2.ndx << EOF del 0-20 a C1A a C1B a C1C a C1D a C1E a C1F a C1G a C1H 10
11 a C1I a C1J a C1K a C1L a C1M a C1N a C1O a C1P a C1Q a C1R q EOF Next g order is used to calculate the order parameters for each chain. g order -s topol.tpr -f traj.xtc -n sn1.ndx -d z -od deuter sn1.xvg g order -s topol.tpr -f traj.xtc -n sn2.ndx -d z -od deuter sn2.xvg Step 2. Calculation of order parameters for unsaturated carbons The double bond is found between atoms C1I and C1J. Calculation of the order parameter for atom C1J ( - C1I = C1J C1K - ) succeeds following make ndx -f backmapped.gro -o sn2-c1j.ndx << EOF del 0-20 a C1I a C1J a C1K q EOF g order -unsat -f traj.xtc -s topol.tpr -n sn2-c1j.ndx -d z -od deuter C1J.xvg The order parameter for atom C1I is calculated following the opposite direction ( - C1J = C1I C1H - ). make ndx -f backmapped.gro -o sn2-c1i.ndx << EOF del 0-20 a C1J a C1I a C1H q EOF 11
12 g order -unsat -f traj.xtc -s topol.tpr -n sn2-c1i.ndx -d z -od deuter C1I.xvg Step 3. Combining results In the last step the order parameters in the file deuter sn2.xvg of atoms C1J and C1I are replaced by results obtained by calculation using the option -unsat and proper atom order. Figure S6: Deuterium order parameters of the unsaturated sn2 chain of POPC simulated by CHARMM36 at 320 K calculated based on C-H orientation (black) or C-C orientation (grey). The latter method is applied in GROMACS tool g order. The spread of the experimental data (crosses and triangles) is larger than the difference between the two theoretical methods. References (1) MacKerell, A. D.; Bashford, D.; Bellott,; Dunbrack, R. L.; Evanseck, J. D.; Field, M. J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, (2) Sun, Y.; Kollman, P. A. Hydrophobic solvation of methane and nonbond parameters of the TIP3P water model. J. Comput. Chem. 1995, 16, (3) Berendsen, H. J. C.; Postma, J. P. M.; van Gunsteren, W. F.; Hermans, J. Interaction models for water in relation to protein hydration. Intermolecular Forces 1981, (4) Almeida, P. F.; Vaz, W. L.; Thompson, T. Lateral diffusion in the liquid phases of dimyristoylphosphatidylcholine/cholesterol lipid bilayers: a free volume analysis. Biochemistry 1992, 31,
13 (5) Filippov, A.; Orädd, G.; Lindblom, G. Influence of cholesterol and water content on phospholipid lateral diffusion in bilayers. Langmuir 2003, 19, (6) Köchy, T.; Bayerl, T. M. Lateral diffusion coefficients of phospholipids in spherical bilayers on a solid support measured by 2 H-nuclear-magnetic-resonance relaxation. Phys. Rev. E 1993, 47, (7) Jin, A. J.; Edidin, M.; Nossal, R.; Gershfeld, N. A singular state of membrane lipids at cell growth temperatures. Biochemistry 1999, 38, (8) RStudio Team, RStudio: Integrated Development Environment for R. RStudio, Inc.: Boston, MA,
Interactions of Polyethylenimines with Zwitterionic and. Anionic Lipid Membranes
 Interactions of Polyethylenimines with Zwitterionic and Anionic Lipid Membranes Urszula Kwolek, Dorota Jamróz, Małgorzata Janiczek, Maria Nowakowska, Paweł Wydro, Mariusz Kepczynski Faculty of Chemistry,
Interactions of Polyethylenimines with Zwitterionic and Anionic Lipid Membranes Urszula Kwolek, Dorota Jamróz, Małgorzata Janiczek, Maria Nowakowska, Paweł Wydro, Mariusz Kepczynski Faculty of Chemistry,
Supporting Information: Revisiting partition in. hydrated bilayer systems
 Supporting Information: Revisiting partition in hydrated bilayer systems João T. S. Coimbra, Pedro A. Fernandes, Maria J. Ramos* UCIBIO, REQUIMTE, Departamento de Química e Bioquímica, Faculdade de Ciências,
Supporting Information: Revisiting partition in hydrated bilayer systems João T. S. Coimbra, Pedro A. Fernandes, Maria J. Ramos* UCIBIO, REQUIMTE, Departamento de Química e Bioquímica, Faculdade de Ciências,
Massive oxidation of phospholipid membranes. leads to pore creation and bilayer. disintegration
 Massive oxidation of phospholipid membranes leads to pore creation and bilayer disintegration Lukasz Cwiklik and Pavel Jungwirth Institute of Organic Chemistry and Biochemistry, Academy of Sciences of
Massive oxidation of phospholipid membranes leads to pore creation and bilayer disintegration Lukasz Cwiklik and Pavel Jungwirth Institute of Organic Chemistry and Biochemistry, Academy of Sciences of
Membrane Simulations in GROMACS Lipid bilayers and membrane proteins
 Membrane Simulations in GROMACS Lipid bilayers and membrane proteins Justin A. Lemkul September 13, 2013 jalemkul@outerbanks.umaryland.edu Objectives Learn the fundamentals of Self-consistent force fields
Membrane Simulations in GROMACS Lipid bilayers and membrane proteins Justin A. Lemkul September 13, 2013 jalemkul@outerbanks.umaryland.edu Objectives Learn the fundamentals of Self-consistent force fields
Bilayer Deformation, Pores & Micellation Induced by Oxidized Lipids
 Supporting Information Bilayer Deformation, Pores & Micellation Induced by Oxidized Lipids Phansiri Boonnoy 1, Viwan Jarerattanachat 1,2, Mikko Karttunen 3*, and Jirasak Wongekkabut 1* 1 Department of
Supporting Information Bilayer Deformation, Pores & Micellation Induced by Oxidized Lipids Phansiri Boonnoy 1, Viwan Jarerattanachat 1,2, Mikko Karttunen 3*, and Jirasak Wongekkabut 1* 1 Department of
Supplementary Information
 Coarse-Grained Molecular Dynamics Simulations of Photoswitchable Assembly and Disassembly Xiaoyan Zheng, Dong Wang,* Zhigang Shuai,* MOE Key Laboratory of Organic Optoelectronics and Molecular Engineering,
Coarse-Grained Molecular Dynamics Simulations of Photoswitchable Assembly and Disassembly Xiaoyan Zheng, Dong Wang,* Zhigang Shuai,* MOE Key Laboratory of Organic Optoelectronics and Molecular Engineering,
Supporting material. Membrane permeation induced by aggregates of human islet amyloid polypeptides
 Supporting material Membrane permeation induced by aggregates of human islet amyloid polypeptides Chetan Poojari Forschungszentrum Jülich GmbH, Institute of Complex Systems: Structural Biochemistry (ICS-6),
Supporting material Membrane permeation induced by aggregates of human islet amyloid polypeptides Chetan Poojari Forschungszentrum Jülich GmbH, Institute of Complex Systems: Structural Biochemistry (ICS-6),
The main biological functions of the many varied types of lipids include: energy storage protection insulation regulation of physiological processes
 Big Idea In the biological sciences, a dehydration synthesis (condensation reaction) is typically defined as a chemical reaction that involves the loss of water from the reacting molecules. This reaction
Big Idea In the biological sciences, a dehydration synthesis (condensation reaction) is typically defined as a chemical reaction that involves the loss of water from the reacting molecules. This reaction
Supplementary information for Effects of Stretching Speed on. Mechanical Rupture of Phospholipid/Cholesterol Bilayers: Molecular
 Supplementary information for Effects of Stretching Speed on Mechanical Rupture of Phospholipid/Cholesterol Bilayers: Molecular Dynamics Simulation Taiki Shigematsu, Kenichiro Koshiyama*, and Shigeo Wada
Supplementary information for Effects of Stretching Speed on Mechanical Rupture of Phospholipid/Cholesterol Bilayers: Molecular Dynamics Simulation Taiki Shigematsu, Kenichiro Koshiyama*, and Shigeo Wada
Biological Membranes. Lipid Membranes. Bilayer Permeability. Common Features of Biological Membranes. A highly selective permeability barrier
 Biological Membranes Structure Function Composition Physicochemical properties Self-assembly Molecular models Lipid Membranes Receptors, detecting the signals from outside: Light Odorant Taste Chemicals
Biological Membranes Structure Function Composition Physicochemical properties Self-assembly Molecular models Lipid Membranes Receptors, detecting the signals from outside: Light Odorant Taste Chemicals
We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field
 Parameterization of the fullerene coarse-grained model We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field for lipids 1 and proteins 2. In the MARTINI force
Parameterization of the fullerene coarse-grained model We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field for lipids 1 and proteins 2. In the MARTINI force
Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale
 Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale Rakesh Gupta and Beena Rai* TATA Research Development & Design Centre, TCS Innovation labs, Pune India,
Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale Rakesh Gupta and Beena Rai* TATA Research Development & Design Centre, TCS Innovation labs, Pune India,
Supporting Information
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 018 Supporting Information Modulating Interactions between Ligand-Coated Nanoparticles and Phase-Separated
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 018 Supporting Information Modulating Interactions between Ligand-Coated Nanoparticles and Phase-Separated
Supporting Information for Origin of 1/f noise in hydration dynamics on lipid membrane surfaces
 Supporting Informati for Origin of /f noise in hydrati dynamics lipid membrane surfaces Eiji Yamamoto, Takuma kimoto, Masato Yasui, 2 and Kenji Yasuoka Department of Mechanical Engineering, Keio University,
Supporting Informati for Origin of /f noise in hydrati dynamics lipid membrane surfaces Eiji Yamamoto, Takuma kimoto, Masato Yasui, 2 and Kenji Yasuoka Department of Mechanical Engineering, Keio University,
Structure of Dipalmitoylphosphatidylcholine/Cholesterol Bilayer at Low and High Cholesterol Concentrations: Molecular Dynamics Simulation
 Biophysical Journal Volume 77 October 1999 2075 2089 2075 Structure of Dipalmitoylphosphatidylcholine/Cholesterol Bilayer at Low and High Cholesterol Concentrations: Molecular Dynamics Simulation Alexander
Biophysical Journal Volume 77 October 1999 2075 2089 2075 Structure of Dipalmitoylphosphatidylcholine/Cholesterol Bilayer at Low and High Cholesterol Concentrations: Molecular Dynamics Simulation Alexander
obtained for the simulations of the E2 conformation of SERCA in a pure POPC lipid bilayer (blue) and in a
 Supplementary Figure S1. Distribution of atoms along the bilayer normal. Normalized density profiles obtained for the simulations of the E2 conformation of SERCA in a pure POPC lipid bilayer (blue) and
Supplementary Figure S1. Distribution of atoms along the bilayer normal. Normalized density profiles obtained for the simulations of the E2 conformation of SERCA in a pure POPC lipid bilayer (blue) and
Fluid Mozaic Model of Membranes
 Replacement for the 1935 Davson Danielli model Provided explanation for Gortner-Grendel lack of lipid and permitted the unit membrane model. Trans membrane protein by labelling Fry & Edidin showed that
Replacement for the 1935 Davson Danielli model Provided explanation for Gortner-Grendel lack of lipid and permitted the unit membrane model. Trans membrane protein by labelling Fry & Edidin showed that
Supporting Information: Atomistic Model for Near-Quantitative. Simulations of Langmuir Monolayers
 Supporting Information: Atomistic Model for Near-Quantitative Simulations of Langmuir Monolayers Matti Javanainen,,, Antti Lamberg, Lukasz Cwiklik,, Ilpo Vattulainen,,, and Samuli Ollila,,# Laboratory
Supporting Information: Atomistic Model for Near-Quantitative Simulations of Langmuir Monolayers Matti Javanainen,,, Antti Lamberg, Lukasz Cwiklik,, Ilpo Vattulainen,,, and Samuli Ollila,,# Laboratory
Supplementary Information A Hydrophobic Barrier Deep Within the Inner Pore of the TWIK-1 K2P Potassium Channel Aryal et al.
 Supplementary Information A Hydrophobic Barrier Deep Within the Inner Pore of the TWIK-1 K2P Potassium Channel Aryal et al. Supplementary Figure 1 TWIK-1 stability during MD simulations in a phospholipid
Supplementary Information A Hydrophobic Barrier Deep Within the Inner Pore of the TWIK-1 K2P Potassium Channel Aryal et al. Supplementary Figure 1 TWIK-1 stability during MD simulations in a phospholipid
Gold nanocrystals at DPPC bilayer. Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad
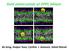 Gold nanocrystals at DPPC bilayer Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad Coarse-grained mapping strategy of a DPPC lipid molecule. The structure of gold nanocrystals (bare gold nanoparticles)
Gold nanocrystals at DPPC bilayer Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad Coarse-grained mapping strategy of a DPPC lipid molecule. The structure of gold nanocrystals (bare gold nanoparticles)
Elements & Macromolecules in Organisms
 Name: Period: Date: Elements & Macromolecules in Organisms Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body weight.
Name: Period: Date: Elements & Macromolecules in Organisms Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body weight.
Life Sciences 1a. Practice Problems 4
 Life Sciences 1a Practice Problems 4 1. KcsA, a channel that allows K + ions to pass through the membrane, is a protein with four identical subunits that form a channel through the center of the tetramer.
Life Sciences 1a Practice Problems 4 1. KcsA, a channel that allows K + ions to pass through the membrane, is a protein with four identical subunits that form a channel through the center of the tetramer.
B i o c h e m i s t r y N o t e s
 14 P a g e Carbon Hydrogen Nitrogen Oxygen Phosphorus Sulfur ~Major ~Found in all ~Found in most ~Found in all component of all organic organic molecules. molecules. ~Major structural atom in all organic
14 P a g e Carbon Hydrogen Nitrogen Oxygen Phosphorus Sulfur ~Major ~Found in all ~Found in most ~Found in all component of all organic organic molecules. molecules. ~Major structural atom in all organic
The Influence of Ceramide Tail Length on the. Structure of Bilayers Composed of Stratum. Corneum Lipids
 The Influence of Ceramide Tail Length on the Structure of Bilayers Composed of Stratum Corneum Lipids T. C. Moore, R. Hartkamp, C. R. Iacovella, A. L. Bunge, and C. M c Cabe Condensed Title: Stratum Corneum
The Influence of Ceramide Tail Length on the Structure of Bilayers Composed of Stratum Corneum Lipids T. C. Moore, R. Hartkamp, C. R. Iacovella, A. L. Bunge, and C. M c Cabe Condensed Title: Stratum Corneum
OCR A GCSE Chemistry. Topic 6: Global challenges. Organic chemistry. Notes.
 OCR A GCSE Chemistry Topic 6: Global challenges Organic chemistry Notes C6.2a recognise functional groups and identify members of the same homologous series Prefixes (beginning of the name) o remember
OCR A GCSE Chemistry Topic 6: Global challenges Organic chemistry Notes C6.2a recognise functional groups and identify members of the same homologous series Prefixes (beginning of the name) o remember
Biology Chapter 2 Review
 Biology Chapter 2 Review Vocabulary: Define the following words on a separate piece of paper. Element Compound Ion Ionic Bond Covalent Bond Molecule Hydrogen Bon Cohesion Adhesion Solution Solute Solvent
Biology Chapter 2 Review Vocabulary: Define the following words on a separate piece of paper. Element Compound Ion Ionic Bond Covalent Bond Molecule Hydrogen Bon Cohesion Adhesion Solution Solute Solvent
A: All atom molecular simulation systems
 Cholesterol level affects surface charge of lipid membranes in physiological environment Aniket Magarkar a, Vivek Dhawan b, Paraskevi Kallinteri a, Tapani Viitala c, Mohammed Elmowafy c, Tomasz Róg d,
Cholesterol level affects surface charge of lipid membranes in physiological environment Aniket Magarkar a, Vivek Dhawan b, Paraskevi Kallinteri a, Tapani Viitala c, Mohammed Elmowafy c, Tomasz Róg d,
Inorganic compounds: Usually do not contain carbon H 2 O Ca 3 (PO 4 ) 2 NaCl Carbon containing molecules not considered organic: CO 2
 Organic Chemistry The study of carbon-containing compounds and their properties. Biochemistry: Made by living things All contain the elements carbon and hydrogen Inorganic: Inorganic compounds: All other
Organic Chemistry The study of carbon-containing compounds and their properties. Biochemistry: Made by living things All contain the elements carbon and hydrogen Inorganic: Inorganic compounds: All other
Supplementary Information: Liquid-liquid phase coexistence in lipid membranes observed by natural abundance 1 H 13 C solid-state NMR
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the wner Societies 28 Supplementary Information: Liquid-liquid phase coexistence in lipid membranes observed
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the wner Societies 28 Supplementary Information: Liquid-liquid phase coexistence in lipid membranes observed
Lipids: Fats, Oils & Waxes: AP Biology
 Lipids: Fats, Oils & Waxes: Lipids long term energy storage concentrated energy *9 Cal/gram Lipids: Triglycerides Lipids are composed of C, H, O u long hydrocarbon chains (H-C) Family groups u fats u phospholipids
Lipids: Fats, Oils & Waxes: Lipids long term energy storage concentrated energy *9 Cal/gram Lipids: Triglycerides Lipids are composed of C, H, O u long hydrocarbon chains (H-C) Family groups u fats u phospholipids
WHAT IS A LIPID? OBJECTIVE The objective of this worksheet is to understand the structure and function of lipids
 WHAT IS A LIPID? OBJECTIVE The objective of this worksheet is to understand the structure and function of lipids PART A: Understanding Lipids Lipids are more commonly known as fats and include triglycerides,
WHAT IS A LIPID? OBJECTIVE The objective of this worksheet is to understand the structure and function of lipids PART A: Understanding Lipids Lipids are more commonly known as fats and include triglycerides,
Break Desired Compound into 3 Smaller Ones
 Break Desired Compound into 3 Smaller nes A B C Indole ydrazine Phenol When creating a covalent link between model compounds move the charge on the deleted into the carbon to maintain integer charge (i.e.
Break Desired Compound into 3 Smaller nes A B C Indole ydrazine Phenol When creating a covalent link between model compounds move the charge on the deleted into the carbon to maintain integer charge (i.e.
Chapter 3. Table of Contents. Section 1 Carbon Compounds. Section 2 Molecules of Life. Biochemistry
 Biochemistry Table of Contents Section 1 Carbon Compounds Section 2 Molecules of Life Section 1 Carbon Compounds Objectives Distinguish between organic and inorganic compounds. Explain the importance of
Biochemistry Table of Contents Section 1 Carbon Compounds Section 2 Molecules of Life Section 1 Carbon Compounds Objectives Distinguish between organic and inorganic compounds. Explain the importance of
Asymmetry of lipid bilayers induced by monovalent salt: Atomistic molecular-dynamics study
 THE JOURNAL OF CHEMICAL PHYSICS 122, 244902 2005 Asymmetry of lipid bilayers induced by monovalent salt: Atomistic molecular-dynamics study Andrey A. Gurtovenko a Laboratory of Physics and Helsinki Institute
THE JOURNAL OF CHEMICAL PHYSICS 122, 244902 2005 Asymmetry of lipid bilayers induced by monovalent salt: Atomistic molecular-dynamics study Andrey A. Gurtovenko a Laboratory of Physics and Helsinki Institute
Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random
 S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
Biochemistry 1 Recitation1 Cell & Water
 Biochemistry 1 Recitation1 Cell & Water There are several important themes that transcends the chemistry and bring the importance of understanding the cell biological differences between eukaryotes and
Biochemistry 1 Recitation1 Cell & Water There are several important themes that transcends the chemistry and bring the importance of understanding the cell biological differences between eukaryotes and
Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes
 Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes Kevin Gasperich Advisor: Dmitry Kopelevich ABSTRACT The recent rapid development of applications for fullerenes has led to an increase
Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes Kevin Gasperich Advisor: Dmitry Kopelevich ABSTRACT The recent rapid development of applications for fullerenes has led to an increase
Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation
 Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation Hang Yu, 1, 2, a) Wei Han, 1, 3, b) Wen Ma, 1, 2 1, 2, 3, c) and Klaus Schulten 1) Beckman
Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation Hang Yu, 1, 2, a) Wei Han, 1, 3, b) Wen Ma, 1, 2 1, 2, 3, c) and Klaus Schulten 1) Beckman
Biological Molecules
 Chemical Building Blocks of Life Chapter 3 Biological Molecules Biological molecules consist primarily of -carbon bonded to carbon, or -carbon bonded to other molecules. Carbon can form up to 4 covalent
Chemical Building Blocks of Life Chapter 3 Biological Molecules Biological molecules consist primarily of -carbon bonded to carbon, or -carbon bonded to other molecules. Carbon can form up to 4 covalent
3.1.3 Lipids. Source: AQA Spec
 alevelbiology.co.uk SPECIFICATION Triglycerides and phospholipids are two groups of lipid. Triglycerides are formed by the condensation of one molecule of glycerol and three molecules of fatty acid. A
alevelbiology.co.uk SPECIFICATION Triglycerides and phospholipids are two groups of lipid. Triglycerides are formed by the condensation of one molecule of glycerol and three molecules of fatty acid. A
Free Volume Properties of Sphingomyelin, DMPC, DPPC, and PLPC Bilayers
 PUBLISHED IN: JOURNAL OF COMPUTATIONAL AND THEORETICAL NANOSCIENCE, VOL. 2, PP. 40-43 (2005). Free Volume Properties of Sphingomyelin, DMPC, DPPC, and PLPC Bilayers M. Kupiainen, E. Falck, S. Ollila, P.
PUBLISHED IN: JOURNAL OF COMPUTATIONAL AND THEORETICAL NANOSCIENCE, VOL. 2, PP. 40-43 (2005). Free Volume Properties of Sphingomyelin, DMPC, DPPC, and PLPC Bilayers M. Kupiainen, E. Falck, S. Ollila, P.
Chemical Basis For Life Open Ended Questions:
 Chemical Basis For Life Open Ended Questions: Answer the following questions to the best of your ability: Make sure you read each question carefully and provide answers to all of the parts of the question.
Chemical Basis For Life Open Ended Questions: Answer the following questions to the best of your ability: Make sure you read each question carefully and provide answers to all of the parts of the question.
The effect of orientation dynamics in melittin as antimicrobial peptide in lipid bilayer calculated by free energy method
 Journal of Physics: Conference Series PAPER OPEN ACCESS The effect of orientation dynamics in melittin as antimicrobial peptide in lipid bilayer calculated by free energy method To cite this article: Sri
Journal of Physics: Conference Series PAPER OPEN ACCESS The effect of orientation dynamics in melittin as antimicrobial peptide in lipid bilayer calculated by free energy method To cite this article: Sri
Lecture-3. Water and Phospholipid
 Lecture-3 Water and Phospholipid Life on earth began in water and evolved there for three billion years before spreading onto land. Although most of the water in liquid form, it is also in solid form and
Lecture-3 Water and Phospholipid Life on earth began in water and evolved there for three billion years before spreading onto land. Although most of the water in liquid form, it is also in solid form and
Coarse grained simulations of Lipid Bilayer Membranes
 Coarse grained simulations of Lipid Bilayer Membranes P. B. Sunil Kumar Department of Physics IIT Madras, Chennai 600036 sunil@iitm.ac.in Atomistic MD: time scales ~ 10 ns length scales ~100 nm 2 To study
Coarse grained simulations of Lipid Bilayer Membranes P. B. Sunil Kumar Department of Physics IIT Madras, Chennai 600036 sunil@iitm.ac.in Atomistic MD: time scales ~ 10 ns length scales ~100 nm 2 To study
Andrey A. Gurtovenko*, and Ilpo Vattulainen,,
 J. Phys. Chem. B 2008, 112, 1953-1962 1953 Effect of NaCl and KCl on Phosphatidylcholine and Phosphatidylethanolamine Lipid Membranes: Insight from Atomic-Scale Simulations for Understanding Salt-Induced
J. Phys. Chem. B 2008, 112, 1953-1962 1953 Effect of NaCl and KCl on Phosphatidylcholine and Phosphatidylethanolamine Lipid Membranes: Insight from Atomic-Scale Simulations for Understanding Salt-Induced
Main Functions maintain homeostasis
 The Cell Membrane Main Functions The main goal is to maintain homeostasis. Regulates materials moving in and out of the cell. Provides a large surface area on which specific chemical reactions can occur.
The Cell Membrane Main Functions The main goal is to maintain homeostasis. Regulates materials moving in and out of the cell. Provides a large surface area on which specific chemical reactions can occur.
Neutron reflectivity in biology and medicine. Jayne Lawrence
 Neutron reflectivity in biology and medicine Jayne Lawrence Why neutron reflectivity studies? build up a detailed picture of the structure of a surface in the z direction n e u tro n s in n e u tro n s
Neutron reflectivity in biology and medicine Jayne Lawrence Why neutron reflectivity studies? build up a detailed picture of the structure of a surface in the z direction n e u tro n s in n e u tro n s
MOLECULAR DYNAMICS SIMULATION OF MIXED LIPID BILAYER WITH DPPC AND MPPC: EFFECT OF CONFIGURATIONS IN GEL-PHASE
 MOLECULAR DYNAMICS SIMULATION OF MIXED LIPID BILAYER WITH DPPC AND MPPC: EFFECT OF CONFIGURATIONS IN GEL-PHASE A Thesis Presented to The Academic Faculty by Young Kyoung Kim In Partial Fulfillment of the
MOLECULAR DYNAMICS SIMULATION OF MIXED LIPID BILAYER WITH DPPC AND MPPC: EFFECT OF CONFIGURATIONS IN GEL-PHASE A Thesis Presented to The Academic Faculty by Young Kyoung Kim In Partial Fulfillment of the
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/3/eaaq0762/dc1 Supplementary Materials for Structures of monomeric and oligomeric forms of the Toxoplasma gondii perforin-like protein 1 Tao Ni, Sophie I. Williams,
advances.sciencemag.org/cgi/content/full/4/3/eaaq0762/dc1 Supplementary Materials for Structures of monomeric and oligomeric forms of the Toxoplasma gondii perforin-like protein 1 Tao Ni, Sophie I. Williams,
Biochimica et Biophysica Acta
 iochimica et iophysica cta 1828 (2013) 1259 1270 Contents lists available at SciVerse ScienceDirect iochimica et iophysica cta journal homepage: www.elsevier.com/locate/bbamem Molecular dynamics study
iochimica et iophysica cta 1828 (2013) 1259 1270 Contents lists available at SciVerse ScienceDirect iochimica et iophysica cta journal homepage: www.elsevier.com/locate/bbamem Molecular dynamics study
Chapter 2 The Chemistry of Life Part 2
 Chapter 2 The Chemistry of Life Part 2 Carbohydrates are Polymers of Monosaccharides Three different ways to represent a monosaccharide Carbohydrates Carbohydrates are sugars and starches and provide
Chapter 2 The Chemistry of Life Part 2 Carbohydrates are Polymers of Monosaccharides Three different ways to represent a monosaccharide Carbohydrates Carbohydrates are sugars and starches and provide
BIOB111_CHBIO - Tutorial activity for Session 12
 BIOB111_CHBIO - Tutorial activity for Session 12 General topic for week 6 Session 12 Lipids Useful Links: 1. Animations on Cholesterol (its synthesis, lifestyle factors, LDL) http://www.wiley.com/college/boyer/0470003790/animations/cholesterol/cholesterol.htm
BIOB111_CHBIO - Tutorial activity for Session 12 General topic for week 6 Session 12 Lipids Useful Links: 1. Animations on Cholesterol (its synthesis, lifestyle factors, LDL) http://www.wiley.com/college/boyer/0470003790/animations/cholesterol/cholesterol.htm
Carbon. Isomers. The Chemical Building Blocks of Life
 The Chemical Building Blocks of Life Carbon Chapter 3 Framework of biological molecules consists primarily of carbon bonded to Carbon O, N, S, P or H Can form up to 4 covalent bonds Hydrocarbons molecule
The Chemical Building Blocks of Life Carbon Chapter 3 Framework of biological molecules consists primarily of carbon bonded to Carbon O, N, S, P or H Can form up to 4 covalent bonds Hydrocarbons molecule
Biology Unit 2 Elements & Macromolecules in Organisms Date/Hour
 Biology Unit 2 Name Elements & Macromolecules in rganisms Date/our Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body
Biology Unit 2 Name Elements & Macromolecules in rganisms Date/our Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body
Supplementary Information
 Supplementary Information Effect of head group and lipid tail oxidation in the cell membrane revealed through integrated simulations and experiments M Yusupov 1, K Wende 2, S Kupsch, E C Neyts 1, S Reuter
Supplementary Information Effect of head group and lipid tail oxidation in the cell membrane revealed through integrated simulations and experiments M Yusupov 1, K Wende 2, S Kupsch, E C Neyts 1, S Reuter
Diffusion and spectroscopy of water and lipids in fully hydrated dimyristoylphosphatidylcholine bilayer membranes
 Diffusion and spectroscopy of water and lipids in fully hydrated dimyristoylphosphatidylcholine bilayer membranes J. Yang, C. Calero, and J. Martí Citation: The Journal of Chemical Physics 14, 1491 (214);
Diffusion and spectroscopy of water and lipids in fully hydrated dimyristoylphosphatidylcholine bilayer membranes J. Yang, C. Calero, and J. Martí Citation: The Journal of Chemical Physics 14, 1491 (214);
Nutrition & Wellness for Life 2012 Chapter 6: Fats: A Concentrated Energy Source
 Tools: Printer 8.5 x 11 paper Scissors Directions: 1. Print 2. Fold paper in half vertically 3. Cut along dashed lines Copyright Goodheart-Willcox Co., Inc. All rights reserved. Tissue in which the body
Tools: Printer 8.5 x 11 paper Scissors Directions: 1. Print 2. Fold paper in half vertically 3. Cut along dashed lines Copyright Goodheart-Willcox Co., Inc. All rights reserved. Tissue in which the body
Molecular dynamics simulations of charged and neutral lipid bilayers: treatment of electrostatic interactions
 Vol. 50 No. 3/2003 789 798 QUARTERLY Molecular dynamics simulations of charged and neutral lipid bilayers: treatment of electrostatic interactions Tomasz Róg, Krzysztof Murzyn and Marta Pasenkiewicz-Gierula
Vol. 50 No. 3/2003 789 798 QUARTERLY Molecular dynamics simulations of charged and neutral lipid bilayers: treatment of electrostatic interactions Tomasz Róg, Krzysztof Murzyn and Marta Pasenkiewicz-Gierula
Simulationen von Lipidmembranen
 Simulationen von Lipidmembranen Thomas Stockner thomas.stockner@meduniwien.ac.at Molecular biology Molecular modelling Membranes environment Many cellular functions occur in or around membranes: energy
Simulationen von Lipidmembranen Thomas Stockner thomas.stockner@meduniwien.ac.at Molecular biology Molecular modelling Membranes environment Many cellular functions occur in or around membranes: energy
The Structure and Function of Biomolecules
 The Structure and Function of Biomolecules The student is expected to: 9A compare the structures and functions of different types of biomolecules, including carbohydrates, lipids, proteins, and nucleic
The Structure and Function of Biomolecules The student is expected to: 9A compare the structures and functions of different types of biomolecules, including carbohydrates, lipids, proteins, and nucleic
Experiment 12 Lipids. Structures of Common Fatty Acids Name Number of carbons
 Experiment 12 Lipids Lipids are a class of biological molecules that are insoluble in water and soluble in nonpolar solvents. There are many different categories of lipids and each category has different
Experiment 12 Lipids Lipids are a class of biological molecules that are insoluble in water and soluble in nonpolar solvents. There are many different categories of lipids and each category has different
Chemical Level Of Organization
 Chemical Level Of Organization List the Four Chemical Elements that Make Up Most of the Body s Mass Oxygen Carbon Hydrogen Nitrogen Distinguish Between Organic and Inorganic Compounds Organic Compounds:
Chemical Level Of Organization List the Four Chemical Elements that Make Up Most of the Body s Mass Oxygen Carbon Hydrogen Nitrogen Distinguish Between Organic and Inorganic Compounds Organic Compounds:
Water-phospholipid interactions at the interface of lipid. membranes: comparison of different force fields
 Water-phospholipid interactions at the interface of lipid membranes: comparison of different force fields Jianjun Jiang, 1,2 Weixin Li, 1,3 Liang Zhao, 1 Yuanyan Wu, 1 Peng Xiu, 4 Guoning Tang, 3 and Yusong
Water-phospholipid interactions at the interface of lipid membranes: comparison of different force fields Jianjun Jiang, 1,2 Weixin Li, 1,3 Liang Zhao, 1 Yuanyan Wu, 1 Peng Xiu, 4 Guoning Tang, 3 and Yusong
Activity Handout for Macromolecules - Station 1. Developed by Dr. Greg Perrier ACTIVITY MACROMOLECULES. Introduction
 Activity Handout for Macromolecules - Station 1 Developed by Dr. Greg Perrier ACTIVITY MACROMOLECULES Introduction In this exercise you will learn about the different functional groups that are added onto
Activity Handout for Macromolecules - Station 1 Developed by Dr. Greg Perrier ACTIVITY MACROMOLECULES Introduction In this exercise you will learn about the different functional groups that are added onto
Elements & Macromolecules in Organisms
 Elements & Macromolecules in rganisms Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body weight. All compounds can be
Elements & Macromolecules in rganisms Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body weight. All compounds can be
Structure and Dynamics of Sphingomyelin Bilayer: Insight Gained through Systematic Comparison to Phosphatidylcholine
 2976 Biophysical Journal Volume 87 November 2004 2976 2989 Structure and Dynamics of Sphingomyelin Bilayer: Insight Gained through Systematic Comparison to Phosphatidylcholine Perttu Niemelä,* Marja T.
2976 Biophysical Journal Volume 87 November 2004 2976 2989 Structure and Dynamics of Sphingomyelin Bilayer: Insight Gained through Systematic Comparison to Phosphatidylcholine Perttu Niemelä,* Marja T.
The Chemical Building Blocks of Life. Chapter 3
 The Chemical Building Blocks of Life Chapter 3 Biological Molecules Biological molecules consist primarily of -carbon bonded to carbon, or -carbon bonded to other molecules. Carbon can form up to 4 covalent
The Chemical Building Blocks of Life Chapter 3 Biological Molecules Biological molecules consist primarily of -carbon bonded to carbon, or -carbon bonded to other molecules. Carbon can form up to 4 covalent
Biological Molecules
 The Chemical Building Blocks of Life Chapter 3 Biological molecules consist primarily of -carbon bonded to carbon, or -carbon bonded to other molecules. Carbon can form up to 4 covalent bonds. Carbon may
The Chemical Building Blocks of Life Chapter 3 Biological molecules consist primarily of -carbon bonded to carbon, or -carbon bonded to other molecules. Carbon can form up to 4 covalent bonds. Carbon may
The Plasma Membrane - Gateway to the Cell
 The Plasma Membrane - Gateway to the Cell 1 Photograph of a Cell Membrane 2 Cell Membrane The cell membrane is flexible and allows a unicellular organism to move 3 Homeostasis Balanced internal condition
The Plasma Membrane - Gateway to the Cell 1 Photograph of a Cell Membrane 2 Cell Membrane The cell membrane is flexible and allows a unicellular organism to move 3 Homeostasis Balanced internal condition
Warm Up #8. What is a carbohydrate? What is a protein?
 Macromolecules Warm Up #8 What is a carbohydrate? What is a protein? Read Macromolecules As you read the article, complete the accompanying Biomolecule Chart This chart MUST be glued into your Notebook!
Macromolecules Warm Up #8 What is a carbohydrate? What is a protein? Read Macromolecules As you read the article, complete the accompanying Biomolecule Chart This chart MUST be glued into your Notebook!
Unit #2: Biochemistry
 Unit #2: Biochemistry STRUCTURE & FUNCTION OF FOUR MACROMOLECULES What are the four main biomolecules? How is each biomolecule structured? What are their roles in life? Where do we find them in our body?
Unit #2: Biochemistry STRUCTURE & FUNCTION OF FOUR MACROMOLECULES What are the four main biomolecules? How is each biomolecule structured? What are their roles in life? Where do we find them in our body?
Nutrition, Food, and Fitness. Chapter 6 Fats: A Concentrated Energy Source
 Nutrition, Food, and Fitness Chapter 6 Fats: A Concentrated Energy Source Tools: Printer (color optional) 4 sheets of 8.5 x 11 paper Scissors Directions: 1. Print 2. Fold paper in half vertically 3. Cut
Nutrition, Food, and Fitness Chapter 6 Fats: A Concentrated Energy Source Tools: Printer (color optional) 4 sheets of 8.5 x 11 paper Scissors Directions: 1. Print 2. Fold paper in half vertically 3. Cut
Gateway to the Cell 11/1/2012. The cell membrane is flexible and allows a unicellular organism to move FLUID MOSAIC MODEL
 Gateway to the Cell The cell membrane is flexible and allows a unicellular organism to move Isolates the cell, yet allows communication with its surroundings fluid mosaics = proteins (and everything else)
Gateway to the Cell The cell membrane is flexible and allows a unicellular organism to move Isolates the cell, yet allows communication with its surroundings fluid mosaics = proteins (and everything else)
Biochemistry. 2. Besides carbon, name 3 other elements that make up most organic compounds.
 Biochemistry Carbon compounds Section 3-1 1. What is an organic compound? 2. Besides carbon, name 3 other elements that make up most organic compounds. 3. Carbon dioxide, CO 2, is NOT an organic compound.
Biochemistry Carbon compounds Section 3-1 1. What is an organic compound? 2. Besides carbon, name 3 other elements that make up most organic compounds. 3. Carbon dioxide, CO 2, is NOT an organic compound.
Supporting Information Identification of Amino Acids with Sensitive Nanoporous MoS 2 : Towards Machine Learning-Based Prediction
 Supporting Information Identification of Amino Acids with Sensitive Nanoporous MoS 2 : Towards Machine Learning-Based Prediction Amir Barati Farimani, Mohammad Heiranian, Narayana R. Aluru 1 Department
Supporting Information Identification of Amino Acids with Sensitive Nanoporous MoS 2 : Towards Machine Learning-Based Prediction Amir Barati Farimani, Mohammad Heiranian, Narayana R. Aluru 1 Department
on the membrane interior. There are many difficulties that
 Biophysical Journal Volume 73 December 1997 297-2923 297 Structure and Dynamic Properties of Diunsaturated 1 -Palmitoyl-2-Linoleoyl-sn-Glycero-3-Phosphatidylcholine Lipid Bilayer from Molecular Dynamics
Biophysical Journal Volume 73 December 1997 297-2923 297 Structure and Dynamic Properties of Diunsaturated 1 -Palmitoyl-2-Linoleoyl-sn-Glycero-3-Phosphatidylcholine Lipid Bilayer from Molecular Dynamics
Elements & Macromolecules in Organisms
 Elements & Macromolecules in rganisms Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body weight. All compounds can be
Elements & Macromolecules in rganisms Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body weight. All compounds can be
SDS-Assisted Protein Transport Through Solid-State Nanopores
 Supplementary Information for: SDS-Assisted Protein Transport Through Solid-State Nanopores Laura Restrepo-Pérez 1, Shalini John 2, Aleksei Aksimentiev 2 *, Chirlmin Joo 1 *, Cees Dekker 1 * 1 Department
Supplementary Information for: SDS-Assisted Protein Transport Through Solid-State Nanopores Laura Restrepo-Pérez 1, Shalini John 2, Aleksei Aksimentiev 2 *, Chirlmin Joo 1 *, Cees Dekker 1 * 1 Department
Macromolecules Cut & Paste
 Macromolecules Cut & Paste Adapted from http://mrswords.weebly.com/uploads/1/5/2/4/15244382/ch_6-3_life_molecules_cut-out_lab.pdf INTRODUCTION Many of the molecules in living cells are so large that they
Macromolecules Cut & Paste Adapted from http://mrswords.weebly.com/uploads/1/5/2/4/15244382/ch_6-3_life_molecules_cut-out_lab.pdf INTRODUCTION Many of the molecules in living cells are so large that they
CHAPTER 2- BIOCHEMISTRY I. WATER (VERY IMPORTANT TO LIVING ORGANISMS) A. POLAR COMPOUND- 10/4/ H O KENNEDY BIOLOGY 1AB
 CHAPTER 2- BIOCHEMISTRY KENNEDY BIOLOGY 1AB I. WATER (VERY IMPORTANT TO LIVING ORGANISMS) WATER S UNIQUE PROPERTIES MAKE IT ESSENTIAL FOR ALL LIFE FUNCTIONS IT IS POLAR, AND HAS BOTH ADHESIVE AND COHESIVE
CHAPTER 2- BIOCHEMISTRY KENNEDY BIOLOGY 1AB I. WATER (VERY IMPORTANT TO LIVING ORGANISMS) WATER S UNIQUE PROPERTIES MAKE IT ESSENTIAL FOR ALL LIFE FUNCTIONS IT IS POLAR, AND HAS BOTH ADHESIVE AND COHESIVE
BIOLOGY 111. CHAPTER 3: Life's Components: Biological Molecules
 BIOLOGY 111 CHAPTER 3: Life's Components: Biological Molecules Life s Components: Biological Molecules 3.1 Carbon's Place in the Living World 3.2 Functional Groups 3.3 Carbohydrates 3.4 Lipids 3.5 Proteins
BIOLOGY 111 CHAPTER 3: Life's Components: Biological Molecules Life s Components: Biological Molecules 3.1 Carbon's Place in the Living World 3.2 Functional Groups 3.3 Carbohydrates 3.4 Lipids 3.5 Proteins
Ethanol Induces the Formation of Water-Permeable Defects in Model. Bilayers of Skin Lipids
 Electronic Supplementary Material (ESI) for Chemical Communications. This journal is The Royal Society of Chemistry 2015 Ethanol Induces the Formation of Water-Permeable Defects in Model Bilayers of Skin
Electronic Supplementary Material (ESI) for Chemical Communications. This journal is The Royal Society of Chemistry 2015 Ethanol Induces the Formation of Water-Permeable Defects in Model Bilayers of Skin
Carbohydrates, Lipids, Proteins, and Nucleic Acids
 Carbohydrates, Lipids, Proteins, and Nucleic Acids Is it made of carbohydrates? Organic compounds composed of carbon, hydrogen, and oxygen in a 1:2:1 ratio. A carbohydrate with 6 carbon atoms would have
Carbohydrates, Lipids, Proteins, and Nucleic Acids Is it made of carbohydrates? Organic compounds composed of carbon, hydrogen, and oxygen in a 1:2:1 ratio. A carbohydrate with 6 carbon atoms would have
Unit 6 Quiz This Friday
 Biology Warm Up Tuesday, November 9 1. Complete group evaluation form. Place completed form in blue bin by the end of class. Unit 6 Quiz This Friday Behavioral Consequences 1. Verbal Warning 2. Detention
Biology Warm Up Tuesday, November 9 1. Complete group evaluation form. Place completed form in blue bin by the end of class. Unit 6 Quiz This Friday Behavioral Consequences 1. Verbal Warning 2. Detention
Supporting information for: Quantifying. lateral inhomogeneity of. cholesterol-containing membranes
 Supporting information for: Quantifying lateral inhomogeneity of cholesterol-containing membranes Celsa Díaz-Tejada, Igor Ariz-Extreme, Neha Awasthi, and Jochen S. Hub Georg-August-University Göttingen,
Supporting information for: Quantifying lateral inhomogeneity of cholesterol-containing membranes Celsa Díaz-Tejada, Igor Ariz-Extreme, Neha Awasthi, and Jochen S. Hub Georg-August-University Göttingen,
Simulationen von Lipidmembranen
 Simulationen von Lipidmembranen Thomas Stockner Thomas.stockner@meduniwien.ac.at Summary Methods Force Field MD simulations Membrane simulations Application Oxidized lipids Anesthetics Molecular biology
Simulationen von Lipidmembranen Thomas Stockner Thomas.stockner@meduniwien.ac.at Summary Methods Force Field MD simulations Membrane simulations Application Oxidized lipids Anesthetics Molecular biology
Supplementary Figure 1. Overview of steps in the construction of photosynthetic protocellular systems
 Supplementary Figure 1 Overview of steps in the construction of photosynthetic protocellular systems (a) The small unilamellar vesicles were made with phospholipids. (b) Three types of small proteoliposomes
Supplementary Figure 1 Overview of steps in the construction of photosynthetic protocellular systems (a) The small unilamellar vesicles were made with phospholipids. (b) Three types of small proteoliposomes
Molecules Dancing in Membranes
 The Open-Access Journal for the Basic Principles of Diffusion Theory, Experiment and Application Molecules Dancing in Membranes Ilpo Vattulainen Laboratory of Physics & Helsinki Institute of Physics, Helsinki
The Open-Access Journal for the Basic Principles of Diffusion Theory, Experiment and Application Molecules Dancing in Membranes Ilpo Vattulainen Laboratory of Physics & Helsinki Institute of Physics, Helsinki
You Are What You Eat
 An Investigation of Macromolecules Student Materials Introduction....2 Pre-Lab Questions.5 Lab Protocol..6 Post-Lab Questions and Analysis 9 Last updated: September 26 th, 2017 1 Introduction When deciding
An Investigation of Macromolecules Student Materials Introduction....2 Pre-Lab Questions.5 Lab Protocol..6 Post-Lab Questions and Analysis 9 Last updated: September 26 th, 2017 1 Introduction When deciding
Changes in a Phospholipid Bilayer Induced by the Hydrolysis of a Phospholipase A 2 Enzyme: A Molecular Dynamics Simulation Study
 Biophysical Journal Volume 80 February 2001 565 578 565 Changes in a Phospholipid Bilayer Induced by the Hydrolysis of a Phospholipase A 2 Enzyme: A Molecular Dynamics Simulation Study Marja T. Hyvönen,*
Biophysical Journal Volume 80 February 2001 565 578 565 Changes in a Phospholipid Bilayer Induced by the Hydrolysis of a Phospholipase A 2 Enzyme: A Molecular Dynamics Simulation Study Marja T. Hyvönen,*
Fats & Fatty Acids. Answer part 2: 810 Cal 9 Cal/g = 90 g of fat (see above: each gram of fat provies 9 Cal)
 Fats & Fatty Acids Function of Fats Store energy (typically stored in the form of triglyceride fat molecules, shown on next page) Burn for energy (energy content is 9 Cal/g) Fatty acids are components
Fats & Fatty Acids Function of Fats Store energy (typically stored in the form of triglyceride fat molecules, shown on next page) Burn for energy (energy content is 9 Cal/g) Fatty acids are components
in-silico Design of Nanoparticles for Transdermal Drug Delivery Application
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2018 in-silico Design of Nanoparticles for Transdermal Drug Delivery Application Rakesh Gupta and Beena
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2018 in-silico Design of Nanoparticles for Transdermal Drug Delivery Application Rakesh Gupta and Beena
The Plasma Membrane - Gateway to the Cell
 The Plasma Membrane - Gateway to the Cell 1 Photograph of a Cell Membrane 2 Cell Membrane The cell membrane is flexible and allows a unicellular organism to move 3 Homeostasis Balanced internal condition
The Plasma Membrane - Gateway to the Cell 1 Photograph of a Cell Membrane 2 Cell Membrane The cell membrane is flexible and allows a unicellular organism to move 3 Homeostasis Balanced internal condition
Effects of Epicholesterol on the Phosphatidylcholine Bilayer: A Molecular Simulation Study
 1818 Biophysical Journal Volume 84 March 2003 1818 1826 Effects of Epicholesterol on the Phosphatidylcholine Bilayer: A Molecular Simulation Study Tomasz Róg and Marta Pasenkiewicz-Gierula Department of
1818 Biophysical Journal Volume 84 March 2003 1818 1826 Effects of Epicholesterol on the Phosphatidylcholine Bilayer: A Molecular Simulation Study Tomasz Róg and Marta Pasenkiewicz-Gierula Department of
The Atoms of Life. What are other elements would you expect to be on this list? Carbon Hydrogen Nitrogen Oxygen Phosphorous Sulfur (sometimes)
 Macromolecules The Atoms of Life The most frequently found atoms in the body are Carbon Hydrogen Nitrogen Oxygen Phosphorous Sulfur (sometimes) What are other elements would you expect to be on this list?
Macromolecules The Atoms of Life The most frequently found atoms in the body are Carbon Hydrogen Nitrogen Oxygen Phosphorous Sulfur (sometimes) What are other elements would you expect to be on this list?
Chem 60 Takehome Test 2 Student Section
 Multiple choice: 1 point each. Mark only one answer for each question. 1. are composed primarily of carbon and hydrogen, but may also include oxygen, nitrogen, sulfur, phosphorus, and a few other elements.
Multiple choice: 1 point each. Mark only one answer for each question. 1. are composed primarily of carbon and hydrogen, but may also include oxygen, nitrogen, sulfur, phosphorus, and a few other elements.
AFM In Liquid: A High Sensitivity Study On Biological Membranes
 University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2006 AFM In Liquid: A High Sensitivity Study On Biological Membranes Michael J. Higgins
University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2006 AFM In Liquid: A High Sensitivity Study On Biological Membranes Michael J. Higgins
Chapter 5 THE STRUCTURE AND FUNCTION OF LARGE BIOLOGICAL MOLECULES
 Chapter 5 THE STRUCTURE AND FUNCTION OF LARGE BIOLOGICAL MOLECULES You Must Know The role of dehydration synthesis in the formation of organic compounds and hydrolysis in the digestion of organic compounds.
Chapter 5 THE STRUCTURE AND FUNCTION OF LARGE BIOLOGICAL MOLECULES You Must Know The role of dehydration synthesis in the formation of organic compounds and hydrolysis in the digestion of organic compounds.
CH 7.2 & 7.4 Biology
 CH 7.2 & 7.4 Biology LABEL THE MEMBRANE Phospholipids Cholesterol Peripheral proteins Integral proteins Cytoskeleton Cytoplasm Extracellular fluid Most of the membrane A phospholipid bi-layer makes up
CH 7.2 & 7.4 Biology LABEL THE MEMBRANE Phospholipids Cholesterol Peripheral proteins Integral proteins Cytoskeleton Cytoplasm Extracellular fluid Most of the membrane A phospholipid bi-layer makes up
