a-dS. Short title: Create 1 new species (Ampivirus A) in a new genus (Ampivirus) (e.g. 6 new species in the genus Zetavirus) Modules attached
|
|
|
- Baldric Houston
- 5 years ago
- Views:
Transcription
1 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the separate document Help with completing a taxonomic proposal Please try to keep related proposals within a single document; you can copy the modules to create more than one genus within a new family, for example. MODULE : TITLE, AUTHORS, etc Code assigned: a-dS (to be completed by ICTV officers) Short title: Create new species (Ampivirus A) in a new genus (Ampivirus) (e.g. 6 new species in the genus Zetavirus) Modules attached (modules and are required) Author(s): Roland Zell, Eric Delwart, Alexander E. Gorbalenya, Tapani Hovi, Andrew M.Q. King, Nick J. Knowles, A. Michael Lindberg, Mark A. Pallansch, Ann C. Palmenberg, Gabor Reuter, Peter Simmonds, Tim Skern, Glyn Stanway and Teruo Yamashita Corresponding author with address: Roland Zell (roland.zell@med.uni-jena.de) List the ICTV study group(s) that have seen this proposal: A list of study groups and contacts is provided at If in doubt, contact the appropriate subcommittee chair (fungal, invertebrate, plant, prokaryote or vertebrate viruses) Picornaviridae Study Group ICTV Study Group comments (if any) and response of the proposer: Date first submitted to ICTV: 5/06/206 Date of this revision (if different to above): ICTV-EC comments and response of the proposer: Page of 0
2 MODULE 2: NEW SPECIES creating and naming one or more new species. If more than one, they should be a group of related species belonging to the same genus. All new species must be placed in a higher taxon. This is usually a genus although it is also permissible for species to be unassigned within a subfamily or family. Wherever possible, provide sequence accession number(s) for one isolate of each new species proposed. Code aS (assigned by ICTV officers) To create new species within: Genus: Ampivirus (new) Subfamily: Family: Picornaviridae Order: Picornavirales Name of new species: Representative isolate: (only per species please) Fill in all that apply. If the higher taxon has yet to be created (in a later module, below) write (new) after its proposed name. If no genus is specified, enter unassigned in the genus box. Ampivirus A newt picornavirus [newt/203/hun] KP77040 GenBank sequence accession number(s) Reasons to justify the creation and assignment of the new species: Explain how the proposed species differ(s) from all existing species. o If species demarcation criteria (see module 3) have previously been defined for the genus, explain how the new species meet these criteria. o If criteria for demarcating species need to be defined (because there will now be more than one species in the genus), please state the proposed criteria. Further material in support of this proposal may be presented in the Appendix, Module A novel picornavirus was detected in faecal samples of the smooth newt (Lissotriton vulgaris) in Hungary. Virus isolates are not available. The sequence exhibits significant similarity to picornaviruses and a picornavirus genome layout (compare Figure ): VPg+5'UTR[AB-C-D/2A-2B-2C Hel /3A-3B VPg -3C Prot -3D Pol ]3'UTR-poly(A) Ampivirus proteins P, 2C and 3CD show only very low amino acid similarities with the orthologous proteins of other picornaviruses. Alignments reveal amino acid identities ranging from: P:.-6.0%, 2C Hel : %, 3D Pol : %. Further typical picornavirus features of the ampivirus polyprotein sequence are: (i) two rhv-like domains (Pfam database) corresponding to VP0, VP3 (ii) NTP-binding motif of 2C Hel (G447DSQCGKT, Walker A motif, and a D499DLLQ motif) (iii) putative 3C proteinase catalytic triad (H96, D2025, D2089ICG) (iv) RNA-dependent RNA polymerase motifs (K235DELR, G2494LPSG, L2539GDD, F2598LKS) Phylogenetic trees comprising reference sequences of the order Picornavirales indicate that ampivirus clusters at the root of the picornavirus branch (compare Figure 2; Appendix). Figures 3, 4 (Appendix) show phylogenetic trees of the picornavirus P and 3CD proteins. Page 2 of 0
3 MODULE 3: NEW GENUS creating a new genus Ideally, a genus should be placed within a higher taxon. Code bS (assigned by ICTV officers) To create a new genus within: Subfamily: Family: Picornaviridae Order: Picornavirales Fill in all that apply. If the higher taxon has yet to be created (in a later module, below) write (new) after its proposed name. If no family is specified, enter unassigned in the family box naming a new genus Code cS (assigned by ICTV officers) To name the new genus: Ampivirus Assigning the type species and other species to a new genus Code dS (assigned by ICTV officers) To designate the following as the type species of the new genus Ampivirus A Every genus must have a type species. This should be a well characterized species although not necessarily the first to be discovered The new genus will also contain any other new species created and assigned to it (Module 2) and any that are being moved from elsewhere (Module 7b). Please enter here the TOTAL number of species (including the type species) that the genus will contain: Reasons to justify the creation of a new genus: Additional material in support of this proposal may be presented in the Appendix, Module Ampivirus is the most divergent picornavirus presently known (Figures 2, 4; Appendix). It has only little sequence similarity to the other picornaviruses. It is the first picornavirus with a capsid protein (VP) that has similarity to the CRPV capsid superfamily (Pfam database) of the dicistroviruses, iflaviruses, marnaviruses and some diatom viruses (labyrnavirus, bacillarnavirus) (Figure ; Appendix). Other unique features of Ampivirus are rather large 3C Pro and 3D Pol proteins. Origin of the new genus name: Ampivirus: from amphibian picornavirus Reasons to justify the choice of type species: only a single species Page 3 of 0
4 Species demarcation criteria in the new genus: If there will be more than one species in the new genus, list the criteria being used for species demarcation and explain how the proposed members meet these criteria. only a single species Page 4 of 0
5 MODULE : APPENDIX: supporting material additional material in support of this proposal References: Reuter G, Boros A, Toth Z, Phan TG, Delwart E, Pankovics P A highly divergent picornavirus in an amphibian, the smooth newt (Lissotriton vulgaris). J Gen Virol 96: Annex: Include as much information as necessary to support the proposal, including diagrams comparing the old and new taxonomic orders. The use of Figures and Tables is strongly recommended but direct pasting of content from publications will require permission from the copyright holder together with appropriate acknowledgement as this proposal will be placed on a public web site. For phylogenetic analysis, try to provide a tree where branch length is related to genetic distance. Genome organization: Fig. Ampivirus A (KP77040) 5'-NTR 970 nt nt 97 M AB rhv 337 aa Q 884 A 885 Q 337 G 338 E 606 S 607?? Q 298 T 299 Q 674 F 675 Q 920 T 92 Q 25 M 252 C rhv 269 aa D CRPV 2A 2B 2C Hel 3A 377 aa 20 aa 3B 36 aa 3C Pro 23 aa 3D Pol 562 aa nt 92 N 273 3'-NTR 34nt Figure : Schematic depiction of the Ampivirus genome organization. The open reading frames are indicated by boxes. Positions of putative aa cleavage sites and the lengths of the deduced proteins are shown as proposed by Reuter et al. (205). Arrows ( ) indicate the putative 3C pro processing sites, whereas question marks (?) hint at two unknown cleavage sites. The sizes of the respective proteins (shaded gray) is unclear. Rhv and CRPV domains proposed by the Pfam database are also indicated Page 5 of 0
6 00 AB050782, Fabavirus, Patchouli mild mosaic virus 99 AB084450, Fabavirus, Broad bean wilt virus 00 NC_003549, Comovirus, Cowpea mosaic virus 59 D0095, Nepovirus, Grapevine fanleaf virus 93 NC_003785, Sadwavirus, Satsuma dwarf virus DQ388879, Torradovirus, Tomato torrado virus AJ62357, Cheravirus, Cherry rasp leaf virus Secoviridae 00 NC_003628, Sequivirus, Parsnip yellow fleck virus EU980442, Sequivirus, Carrot necrotic dieback virus [Anthriscus] 76 NC_00632, Waikavirus, Rice tungro spherical virus 00 U67839, Waikavirus, Maize chlorotic dwarf waikavirus [Tennessee] AY365064, Ectropis obliqua picorna-like virus Iflaviridae AB000906, Iflavirus, Infectious flacherie virus AF092924, Sacbrood virus Iflaviridae 58 NC_004830, Deformed wing virus Iflaviridae Iflaviridae 99 NC_0437, Slow bee paralysis virus (Iflaviridae) KF360262, unassigned, Aphis glycines virus [ApGlV] 00 KM05260, unassigned, Soybean-associated bicistronic virus [North Dakota] 00 KJ420969, unassigned, Arivirus [A09777/676/200] KJ420970, unassigned, Arivirus 2 [A09777/676/200] KM07736, unassigned, Fesavirus KM259869, unassigned, Antarctic picorna-like virus 00 KJ802403, unassigned, Cripavirus [NB-/20/HUN] AB07037, Cripavirus, Himetobi P virus AF022937, Cripavirus, Rhopalosiphum padi virus EU282007, Cripavirus, Rhopalosiphum padi virus [RhPV6] NC_003924, Cripavirus, Cricket paralysis virus NC_00834, Cripavirus, Drosophila C virus 64 NC_006559, Aparavirus, Solenopsis invicta virus- 00 AF50629, Aparavirus, Acute bee paralysis virus NC_00528, Marnavirus, Heterosigma akashiwo RNA virus [SOG263] AB93726, Labyrnavirus, Aurantiochytrium ssrna virus KM25987, unassigned, Antarctic picorna-like virus 3 JN6660, unassigned, Marine RNA virus SF KF478836, unassigned, Marine RNA virus SF-3 KF4290, unassigned, Marine RNA virus SF-2 EF98242, unassigned, Marine RNA virus JP-B KM259872, unassigned, Antarctic picorna-like virus 4 NC_024489, unassigned, Asterionellopsis glacialis RNA virus NC_0222, Bacillarnavirus, Chaetoceros socialis f. radians RNA virus NC_0863, Bacillarnavirus, Rhizosolenia setigera RNA virus-0 AB375474, Bacillarnavirus, Chaetoceros tenuissimus RNA virus KM259870, unassigned, Antarctic picorna-like virus 2 NC_025889, unassigned, Chaetoceros tenuissimus RNA virus type-ii [SS0-6V] NC_009757, unassigned, Marine RNA virus JP-A AB639040, unassigned, Chaetoceros sp. RNA virus 2 [Csp02RNAV0] KT25902, unassigned, Husavirus [ACS78] NC_023637, unassigned, Posavirus NC_023638, unassigned, Posavirus 2 KU75459, unassigned, Pow Burn virus 00 KM434233, unassigned, Fisavirus [HAL] NC_0648, unassigned, Halastavi arva RNA virus KP77040, Ampivirus, Ampivirus A [NEWT/203/HUN] 00 AF40683, Sapelovirus, Sapelovirus A [PEV8-V3] 60 V049, Enterovirus, Enterovirus C poliovirus [Mahoney] M886, Cardiovirus, Cardiovirus A [R] 76 KC935379, Kunsagivirus, Kunsagivirus A 56 JQ36470, Pasivirus, Pasivirus A [swine/france/20] 98 L0297, Parechovirus, Parechovirus A [Harris] 70 AB937989, unassigned, Crohivirus [shrew/zm54/zambia/202] 0,5 Dicistroviridae Marnaviridae Picornaviridae Figure 2: Phylogenetic analyses of the 3CD protein of representative members of the Picornavirales using maximum likelihood tree inference (MEGA 5.2). 60 sequences were retrieved from GenBank. Presented are GenBank accession numbers, genus names, species names and types. If available, designations of isolates [in square brackets] are given. Yet unassigned viruses are printed in blue. Proposed Ampivirus is printed in red and indicated by a dot ( ). Numbers at nodes indicate bootstrap values obtained after 200 replications. The optimal substitution model (GTR+G+I) was determined with MEGA 5.2. The scale indicates substitutions/site. Page 6 of 0
7 P HQ595340, unassigned, bat picornavirus [NC6A] HQ59534, unassigned, bat picornavirus [LMH22A] HQ595342, unassigned, bat picornavirus 2 [MH9F] HQ595343, unassigned, bat picornavirus 2 [SK7F] 0.84 JN83356, unassigned, canine picornavirus [dog/hong Kong/325F/2008] KJ64694, unassigned, bat picornavirus [BtRs-PicoV/YN200] JQ84852, unassigned, Ia io picornavirus HQ595344, unassigned, bat picornavirus 3 [TLC5F] HQ595345, unassigned, bat picornavirus 3 [TLC2F] KJ64693, unassigned, bat picornavirus [BtRh-PicoV/SC203] 0.88 JN5727, unassigned, feline picornavirus [356F] JN5729, unassigned, feline picornavirus [02F] JN5725, unassigned, feline picornavirus [cat/hong Kong/073F/2007] JN5726, unassigned, feline picornavirus [27F] 0.87 JN5728, unassigned, feline picornavirus [66F] 0.54 KJ64687, unassigned, bat picornavirus [BtMf-PicoV/FJ202] KJ64690, unassigned, bat picornavirus [BtMf-PicoV-/GD202] KJ64699, unassigned, bat picornavirus [BtMf-PicoV-2/SAX20] 0.85 LC03658, unassigned, bovine picornavirus [Bo-2-/2009/JPN] LC036582, unassigned, bovine picornavirus [Bo-2-38/2009/JPN] LC036579, unassigned, bovine picornavirus [Bo-2-3/2009/JPN] LC036580, unassigned, bovine picornavirus [Bo-2-7/2009/JPN] LC00697, unassigned, bovine picornavirus [Bo--39/2009/JPN] KP345888, unassigned, dromedary camel enterovirus 2 [20CC] KP345887, unassigned, dromedary camel enterovirus [9CC] 0.52 DQ092770, Enterovirus, EV-F BEV-2 [BEV-26-RM2] DQ092769, Enterovirus, EV-E BEV- [LC-R4] AF363453, Enterovirus, EV-G [UKG-40-73] U2252, Enterovirus, EV-A7 [BrCr] AF326766, Enterovirus, EV-J [SV6-63] D00820, Enterovirus, EV-D70 [J670-7] AF20894, Enterovirus, EV-H [A-2 plaque virus] 0.9 K022, Enterovirus, RV-B4 [959] 0.52 EF86077, Enterovirus, RV-C3 [HRV-QPM] L2497, Enterovirus, RV-A6 [757] 0.52 V049, Enterovirus, EV-C PV [Mahoney] M33854, Enterovirus, EV-B CVB3 [Nancy] KP233897, unassigned, rabovirus [Berlin/Jan20/0572] KJ950883, unassigned, rat picornavirus [RPV/NYC-B0] AF40683, Sapelovirus, Sapelovirus A, porcine sapelovirus [V3] AY064708, Sapelovirus, Sapelovirus B, simian sapelovirus [SV2-2383] KJ64696, unassigned, bat picornavirus [BtVs-PicoV/SC203] 0.9 KJ64689, unassigned, bat picornavirus [BtMa-PicoV/FJ202] JN420368, unassigned, Californian sealion sapelovirus [62] JN674502, unassigned, quail picornavirus KT880670, unassigned, phacovirus [Pf-CHK/PhV] FR72744, unassigned, pigeon picornavirus B [pigeon/norway/03/64/2003] KC56080, unassigned, pigeon picornavirus B [GAL-7/200/Hungary] AY563023, Sapelovirus, Avian sapelovirus [TW90A] 0.84 KJ64688, unassigned, bat picornavirus [BtRlep-PicoV/FJ202] KJ64692, unassigned, bat picornavirus [BtRa-PicoV/JS203] KJ64685, unassigned, bat picornavirus [BtRf-PicoV-/YN202] KJ64697, unassigned, bat picornavirus [BtNv-PicoV/SC203] KM87363, unassigned, tortoise picornavirus [9-05] KM87364, unassigned, tortoise picornavirus [44-0] KM87365, unassigned, tortoise picornavirus [203-T4] KM8736, unassigned, tortoise picornavirus [4-04] KM87362, unassigned, tortoise picornavirus [5-04] KM87367, unassigned, tortoise picornavirus [5-03] KM87366, unassigned, tortoise picornavirus [24-4-0] JF973687, Mosavirus, Mosavirus A [M-7] KF95846, Mosavirus, Mosavirus A2 [SZAL6-MoV/20/HUN] KM396707, unassigned, lesavirus [Mis0308/202] KM396708, unassigned, lesavirus 2 [Nai0805/202] JQ94880, Hunnivirus, Hunnivirus A [BHUV/2008/HUN] 0.9 AF23769, Teschovirus, Teschovirus A [Talfan] 0.65 EU236594, Aphthovirus, Bovine rhinitis B virus [EC] JN936206, Aphthovirus, Bovine rhinitis A virus 2 [H-] X0087, Aphthovirus, Foot-and-mouth disease virus O [Kaufbeuren] X96870, Aphthovirus, Equine rhinitis A virus [PERV] X9687, Erbovirus, Equine rhinitis B virus [P436/7] KM589358, unassigned, bovine picornavirus [TCH6] KP054277, unassigned, bat picornavirus [BatPV/V3/3/Hun] KP054276, unassigned, bat picornavirus [BatPV/V2/3/Hun] KP054274, unassigned, bat picornavirus [BatPV/V8/3/Hun] KP054275, unassigned, bat picornavirus [BatPV/V/3/Hun] KP054273, unassigned, bat picornavirus [BatPV/V/3/Hun] KP054278, unassigned, bat picornavirus [BatPV/V4/3/Hun] JQ8485, Mischivirus, Mischivirus A [China/200] KP00644, unassigned, African bat icavirus [TNo3] FJ438902, Cosavirus, Cosavirus A [0553] JN867758, unassigned, human cosavirus F [PK5006] FJ438907, unassigned, human cosavirus B [2263] FJ555055, unassigned, human cosavirus E [Australia/6] FJ438908, unassigned, human cosavirus D [5004] M886, Cardiovirus, Cardiovirus A, encephalomyocarditis virus [R] M20562, Cardiovirus, Cardiovirus B, Theiler's murine encephalomyelitis virus [GDVII] JQ864242, Cardiovirus, Cardiovirus C, boone cardiovirus [rat/usa/200] JX683808, Cardiovirus, Cardiovirus C2, boone cardiovirus 2 [rat/usa/202] DQ64257, Senecavirus, Senecavirus A, Seneca Valley virus [SVV-00] KJ690629, unassigned, chicken proventriculitis virus [CPV/Korea/03] KF9687, unassigned, chicken megrivirus [chicken/chk-iv-chv/203/hun] KF9686, unassigned, chicken megrivirus [chicken/b2-chv/202/hun] KF979336, unassigned, chicken picornavirus 5 [27C] 0.97 HM7599, Megrivirus, Melegrivirus A, turkey hepatitis virus [2993D] KF979335, unassigned, chicken picornavirus 4 [5C] KC663628, unassigned, duck megrivirus [LY] 0.74 KC876003, unassigned, mesivirus [HK2] KC8837, unassigned, mesivirus 2 [pigeon/galii5-pimev/20/hun] JN89202, Dicipivirus, Cadicivirus A, canine picodicistrovirus [209] JF973686, Rosavirus, Rosavirus A, murine rosavirus [M-7] KJ934637, Kobuvirus, Aichivirus A5, roller kobuvirus [SZAL6-KoV/20/HUN] JQ898342, Kobuvirus, Aichivirus A6, Kathmandu sewage kobuvirus [KoV-SewKTM] 0.89 JF755427, Kobuvirus, Aichivirus A3, murine kobuvirus [M-5/USA/200] AB0045, Kobuvirus, Aichivirus A [A846-88] KJ958930, Kobuvirus, Aichivirus A4, feline kobuvirus [2D240] KF83027, Kobuvirus, Aichivirus A4, feline kobuvirus [FK-3] KM09960, Kobuvirus, Aichivirus A4, feline kobuvirus [FeKoV/TE/52/IT/3] JN38733, Kobuvirus, Aichivirus A2, canine kobuvirus [dog-an2d-usa-2009] 0.8 GU245693, Kobuvirus, Aichivirus B3, ovine kobuvirus [sheep/tb3/hun/2009] KF006985, Kobuvirus, Aichivirus B2, ferret kobuvirus [MpKoV38] AB084788, Kobuvirus, Aichivirus B, bovine kobuvirus [U-] EU , Kobuvirus, Aichivirus C, porcine kobuvirus [swine/s-/hun/2007/hungary] KF793927, Kobuvirus, Aichivirus C2, caprine kobuvirus [2Q08] LC055960, unassigned, kagovirus 2 [cattle/kagoshima-2-24-kov/205/jpn] LC05596, unassigned, kagovirus [cattle/kagoshima--22-kov/204/jpn] KJ6469, unassigned, bat picornavirus [BtMf-PicoV-2/GD202] KJ64686, unassigned, bat picornavirus [BtMr-PicoV/JX200] KT325852, unassigned, rabbit picornavirus [Rabbit0/203/HUN] GQ79640, Salivirus, Salivirus A [NG-J] 0.97 KF74227, Sicinivirus, Sicinivirus A [UCC00] 0.82 KF97933, Sicinivirus, Sicinivirus A2, chicken picornavirus [55C] 0.94 JQ6963, Gallivirus, Gallivirus A [turkey/m76/20/hun] GU82406, Passerivirus, Passerivirus A, turdivirus [thrush/hong Kong/00356/2007] 0.6 GU82408, Oscivirus, Oscivirus A, turdivirus 2 [robin/hong Kong/077/2006] 0.72 GU8240, Oscivirus, Oscivirus A2, turdivirus 3 [thrush/hong Kong/0878/2006] KF38772, Sakobuvirus, Sakobuvirus A, feline sakobuvirus [FFUP/Portugal/202] KJ4577, unassigned, tortoise rafivirus [UF4] KF979334, unassigned, chicken picornavirus 3 [45C] 0.97 KT880669, unassigned, chicken picornavirus [Pf-CHK/AsV] KF979333, unassigned, chicken picornavirus 2 [44C] KC465954, Avisivirus, Avisivirus A [turkey/m76-tuasv/20/hun] KC64703, Avisivirus, Avisivirus A [USA-IN] KM203656, unassigned, orivirus [chicken/pf-chk/203/hun] KT880667, unassigned, orivirus 2 [Pf-CHK] KJ000696, unassigned, aalivirus (duck picornavirus) [GL/2] DQ249299, Avihepatovirus, Avihepatovirus A, duck hepatitis A virus [03D] JQ36470, Pasivirus, Pasivirus A, swine pasivirus [swine/france/20] AB937989, unassigned, crohivirus [shew/zm54] 0.57 AF327920, Parechovirus, Parechovirus B, Ljungan virus [87-02] HF677705, unassigned, sebokele virus [An/B/227/d] L0297, Parechovirus, Parechovirus A, human parechovirus [Harris] 0.72 KF006989, unassigned, ferret parechovirus [MpPeV] KJ64698, unassigned, bat picornavirus [BtMf-PicoV-/SAX20] JQ84853, unassigned, Rhinolophus affinis picornavirus KC935379, Kunsagivirus, Kunsagivirus A, roller kunsagivirus [roller/szal6-kuv/20/hun] unassigned, Bat Kunsagivirus [bat/cameroon/203] 0.92 EU42040, Aquamavirus, Aquamavirus A, seal picornavirus [HO-02-2] KC843627, Potamipivirus, Potamipivirus A, eel picornavirus [F5/05] 0.9 KF8395, Limnipivirus, Limnipivirus C, fathead minnow picornavirus [fhm//mn/usa/200] KF8396, Limnipivirus, Limnipivirus C, fathead minnow picornavirus [fhm/2/mn/usa/200] KF874490, Limnipivirus, Limnipivirus C, fathead minnow picornavirus [fhm/20/il/usa/200] KC465953, Limnipivirus, Limnipivirus C, fathead minnow picornavirus [09-283] KF306267, Limnipivirus, Limnipivirus B, carp picornavirus [F37/06] JX34222, Limnipivirus, Limnipivirus A, bluegill picornavirus [04-032] KT452695, unassigned, hedgehog hepatovirus [Igel75Erieur204] KT452698, unassigned, hedgehog hepatovirus [Igel68Erieur204] KT45269, unassigned, hedgehog hepatovirus [Igel8Erieur204] KT45274, unassigned, bat hepatovirus [M32Eidhel200] KT87758, unassigned, tupaia hepatovirus [TN] KT452730, unassigned, bat hepatovirus [BUO2BF86Colafr200] KT452729, unassigned, bat hepatovirus [FOAF48Rhilan200] KT452742, unassigned, bat hepatovirus [SMG8520Minmav204] 0.72 KT452735, unassigned, rodent hepatovirus [CIV459Lopsik2004] KR703607, unassigned, phopivirus [NewEngland/USA/20] KT452637, unassigned, rodent hepatovirus [RMU0637Micarv200] KT45264, unassigned, rodent hepatovirus [KS50Myogla20] KT452644, unassigned, rodent hepatovirus [KS230Crimig20] M4707, Hepatovirus, Hepatovirus A, hepatitis A virus [HM-75] D00924, Hepatovirus, Hepatovirus A, simian hepatitis A virus [AGM-27] KT22962, unassigned, woodchuck hepatovirus [3ID] KT2296, unassigned, woodchuck hepatovirus [2ID] KT452685, unassigned, rodent hepatovirus [KEF2Sigmas202] KT452658, unassigned, shrew hepatovirus [KS2232Sorara202] KT45266, unassigned, shrew hepatovirus [KS2289Sorara202] AJ22573, Tremovirus, Tremovirus A, avian encephalomyelitis virus [Calnek vaccine strain] 0.87 KP77040, Ampivirus, Ampivirus A [NEWT/203/HUN] KP230449, unassigned, falcovirus [kestrel/vove0622/203/hun] 0.5 Page 7 of 0
8 Figure 3 (previous page): Phylogenetic analyses of picornavirus P using Bayesian tree inference (MrBayes 3.2). 78 picornavirus sequences were retrieved from GenBank. Presented are GenBank accession numbers, genus names, species names and types. If available, common names and designations of isolates [in square brackets] are given. Yet unassigned viruses are printed in blue. Proposed name is printed in red and indicated by a dot ( ). Numbers at nodes indicate posterior probabilities obtained after 6,000,000 generations. The optimal substitution model (GTR+G+I) was determined with MEGA 5. The scale indicates substitutions/site. Page 8 of 0
9 3CD KJ958930, Kobuvirus, Aichivirus A4, feline kobuvirus [2D240] KM09960, Kobuvirus, Aichivirus A4, feline kobuvirus [FeKoV/TE/52/IT/3] KF83027, Kobuvirus, Aichivirus A4, feline kobuvirus [FK-3] JN38733, Kobuvirus, Aichivirus A2, canine kobuvirus [dog-an2d-usa-2009] JF755427, Kobuvirus, Aichivirus A3, murine kobuvirus [M-5-USA-200] AB0045, Kobuvirus, Aichivirus A [A846-88] JQ898342, Kobuvirus, Aichivirus A6, Kathmandu sewage kobuvirus [KoV-SewKTM] KJ934637, Kobuvirus, Aichivirus A5, roller kobuvirus [SZAL6-KoV/20/HUN] AB084788, Kobuvirus, Aichivirus B, bovine kobuvirus [U-] GU245693, Kobuvirus, Aichivirus B3, ovine kobuvirus [sheep/tb3/hun/2009] KF006985, Kobuvirus, Aichivirus B2, ferret kobuvirus [ferret/mpkov38/nl/200] EU787450, Kobuvirus, Aichivirus C, porcine kobuvirus [sw/s-/hun/2007] KF793927, Kobuvirus, Aichivirus C2, caprine kobuvirus [2Q08] LC055960, unassigned, kagovirus 2 [cattle/kagoshima-2-24-kov/205/jpn] LC05596, unassigned, kagovirus [cattle/kagoshima--22-kov/204/jpn] GQ79640, Salivirus, Salivirus A [hu/ng-j/nigeria/2007] JQ6963, Gallivirus, Gallivirus A [turkey/hun/m76/20] GU82406, Passerivirus, Passerivirus A, turdivirus [thrush/hong Kong/00356/2007] KF74227, Sicinivirus, Sicinivirus A [chicken/ucc00/eire] KF97933, Sicinivirus, Sicinivirus A [ChPV-/chicken/55C/Hong Kong/2008] KF38772, Sakobuvirus, Sakobuvirus A [FFUP/Portugal/202] KJ64686, unassigned, bat picornavirus [BtMr-PicoV/JX200] KJ6469, unassigned, bat picornavirus [BtMf-PicoV-2/GD202] KT325852, unassigned, rabbit picornavirus [Rabbit0/203/HUN] GU82408, Oscivirus, Oscivirus A, turdivirus 2 [robin/hong Kong/077/2006) GU8240, Oscivirus, Oscivirus A2, turdivirus 3 [robin/hong Kong/ ] KJ4577, unassigned, tortoise rafivirus [UF4] KF9686, unassigned, chicken megrivirus [chicken/b2-chv/202/hun] KF9687, unassigned, chicken megrivirus [chicken/chk-iv-chv/203/hun] KJ690629, unassigned, chicken proventriculitis virus [CPV/Korea/03] 0.5 HM7599, Megrivirus, Melegrivirus A, turkey hepatitis virus [2993D] 0.55 KF979336, unassigned, chicken picornavirus 5 [27C] KF979335, unassigned, chicken picornavirus 4 [5C] KC663628, unassigned, duck megrivirus [LY] KC8837, unassigned, pigeon mesivirus 2 [pigeon/galii5-pimev/20/hun] KC876003, unassigned, mesivirus [HK2] JN89202, Dicipivirus, Cadicivirus A, canine picodicistrovirus [209] JF973686, Rosavirus, Rosavirus A [mouse/usa/m-7/200] KP054278, unassigned, bat picornavirus [BatPV/V4/3/Hun] KP054276, unassigned, bat picornavirus [BatPV/V2/3/Hun] KP054275, unassigned, bat picornavirus [BatPV/V/3/Hun] 0.9 KP054277, unassigned, bat picornavirus [BatPV/V3/3/Hun] KP054274, unassigned, bat picornavirus [BatPV/V8/3/Hun] KP054273, unassigned, bat picornavirus [BatPV/V/3/Hun] JQ8485, Mischivirus, Mischivirus A [bat/china/200] KP00644, unassigned, African bat icavirus [TNo3] M20562, Cardiovirus, Cardiovirus B, Theiler's murine encephalomyelitis virus [GDVII] M886, Cardiovirus, Cardiovirus A, encephalomyocarditis virus [R] JQ864242, Cardiovirus, Cardiovirus C, Boone cardiovirus [rat/usa/200] JX683808, Cardiovirus, Cardiovirus C2, Boone cardiovirus 2 [rat/usa/202] DQ64257, Senecavirus, Senecavirus A, Seneca Valley virus [cc/us/svv-00] FJ438908, unassigned, human cosavirus D [5004] FJ555055, unassigned, human cosavirus E [Australia/6] FJ438907, unassigned, human cosavirus B [2263] FJ438902, Cosavirus, Cosavirus A [0553] JN867758, unassigned, human cosavirus F [PK5006] EU236594, Aphthovirus, Bovine rhinitis B virus [EC] JN936206, Aphthovirus, Bovine rhinitis A virus 2 [H-] X0087, Aphthovirus, Foot-and-mouth disease virus O [Kaufbeuren] X96870, Aphthovirus, Equine rhinitis A virus [PERV] KM396707, unassigned, lesavirus [Mis0308/202] KM396708, unassigned, lesavirus 2 [Nai0805/202] JQ94880, Hunnivirus, Hunnivirus A [cattle/hun/2008] AF23769, Teschovirus, Teschovirus A, porcine teschovirus [Talfan] X9687, Erbovirus, Erbovirus A, equine rhinitis B virus [P436/7] KM589358, unassigned, bovine erbo-like picornavirus [TCH6] KM87363, unassigned, tortoise picornavirus [9-05] JN5729, unassigned, feline picornavirus [02F] JN5725, unassigned, feline picornavirus [cat/hong Kong/073F/20079 JN5726, unassigned, feline picornavirus [27F] JN5728, unassigned, feline picornavirus [66F] KJ64687, unassigned, bat picornavirus [BtMf-PicoV/FJ202] 0.69 KJ64699, unassigned, bat picornavirus [BtMf-PicoV-2/SAX20] KJ64690, unassigned, bat picornavirus [BtMf-PicoV-/GD202] HQ595344, unassigned, bat picornavirus [TLC5F] HQ595345, unassigned, bat picornavirus [TLC2F] KJ64693, unassigned, bat picornavirus [BtRh-PicoV/SC203] HQ595340, unassigned, bat picornavirus [NC6A] HQ59534, unassigned, bat picornavirus [LMH22A] HQ595342, unassigned, bat picornavirus [MH9F] HQ595343, unassigned, bat picornavirus [SK7F] JQ84852, unassigned, Ia io picornavirus KJ64694, unassigned, bat picornavirus [BtRs-PicoV/YN200] JN83356, unassigned, canine picornavirus [dog/hong Kong/325F/2008] LC036579, unassigned, bovine picornavirus [Bo-2-3/2009/JPN] LC03658, unassigned, bovine picornavirus [Bo-2-/2009/JPN] LC036580, unassigned, bovine picornavirus [Bo-2-7/2009/JPN] LC036582, unassigned, bovine picornavirus [Bo-2-38/2009/JPN] LC00697, unassigned, Bovine picornavirus [Bo--39/2009/JPN] AF40683, Sapelovirus, Sapelovirus A, porcine sapelovirus [V3] AY064708, Sapelovirus, Sapelovirus B, simian sapelovirus [SV2-2383] KJ64696, unassigned, bat picornavirus [BtVs-PicoV/SC203] KJ64689, unassigned, bat picornavirus [BtMa-PicoV/FJ202] AY563023, Sapelovirus, Avian sapelovirus [TW90A] JN420368, unassigned, Californian sealion sapelovirus [62] KJ64688, unassigned, bat picornavirus [BtRlep-PicoV/FJ202] KJ64692, unassigned, bat picornavirus [BtRa-PicoV/JS203] KJ64685, unassigned, bat picornavirus [BtRf-PicoV-/YN202] KJ64697, unassigned, bat picornavirus [BtNv-PicoV/SC203] M33854, Enterovirus, EV-B CV-B3 [Nancy] AF326766, Enterovirus, EV-J [SV6-63] D00820, Enterovirus, EV-D70 [J670-7] V049, Enterovirus, EV-C PV [Mahoney] U2252, Enterovirus, EV-A7 [BrCr] KP345888, unassigned, Dromedary camel enterovirus 2 [20CC] KP345887, unassigned, Dromedary camel enterovirus [9CC] DQ092770, Enterovirus, EV-F BEV-2 [BEV-26 RM2] DQ092769, Enterovirus, EV-E BEV- [LC-R4] AF363453, Enterovirus, EV-G [UKG/40/73] AF20894, Enterovirus, EV-H [A-2 plaque virus] L2497, Enterovirus, RV-A6 [757] EF86077, Enterovirus, RV-C3 [HRV-QPM] K022, Enterovirus, RV-B4 [959] KP233897, unassigned, rabovirus [Berlin/Jan20/0572] KJ950883, unassigned, rat picornavirus [rat/nyc-b0/usa/200] FR72744, unassigned, pigeon picornavirus B [pigeon/norway/03/64/2003] KC56080, unassigned, pigeon picornavirus B [GAL-7/200/Hungary] FR72745, unassigned, pigeon picornavirus A [pigeon/norway/03/603-7/2003] JN674502, unassigned, quail picornavirus [quail/hun/200] KT880670, unassigned, phacovirus [Pf-CHK/PhV] KP230449, unassigned, falcovirus [kestrel/vove0622/203/hun] KJ64684, unassigned, bat picornavirus [BtRf-PicoV-2/YN202] KT452637, unassigned, rodent hepatovirus [RMU0637Micarv200] KT45264, unassigned, rodent hepatovirus [KS50Myogla20] KT452644, unassigned, rodent hepatovirus []KS230Crimig20] KT22962, unassigned, woodchuck hepatovirus [3ID] KT2296, unassigned, woodchuck hepatovirus [2ID] KT452685, unassigned, rodent hepatovirus [KEF2Sigmas202] M4707, Hepatovirus, Hepatovirus A, hepatitis A virus [HM-75] D00924, Hepatovirus, Hepatovirus A, simian hepatitis A virus [AGM-27] KT452658, unassigned, shrew hepatovirus [KS2232Sorara202] KT45266, unassigned, shrew hepatovirus [KS2289Sorara202] KT452695, unassigned, hedgehog hepatovirus [Igel75Erieur204] KT452698, unassigned, hedgehog hepatovirus [Igel68Erieur204] KT45269, unassigned, hedgehog hepatovirus [Igel8Erieur204] KT45274, unassigned, bat hepatovirus [M32Eidhel200] KT87758, unassigned, tupaia hepatovirus [TN] KT452730, unassigned, bat hepatovirus [BUO2BF86Colafr200] KT452729, unassigned, bat hepatovirus [FOAF48Rhilan200] KR703607, unassigned, phopivirus [NewEngland/USA/20] KT452742, unassigned, bat hepatovirus [SMG8520Minmav204] KT452735, unassigned, rodent hepatovirus [CIV459Lopsik2004] AJ22573, Tremovirus, Tremovirus A, avian encephalomyelitis virus [Calnek vaccine strain] 0.95KF8395, Limnipivirus, Limnipivirus C, fathead minnow picornavirus [fhm//mn/usa/200] KF8396, Limnipivirus, Limnipivirus C, fathead minnow picornavirus [fhm/2/mn/usa/200] KF874490, Limnipivirus, Limnipivirus C, fathead minnow picornavirus [fhm/20/il/usa/200] KF306267, Limnipivirus, Limnipivirus B, carp picornavirus [F37/06] JX34222, Limnipivirus, Limnipivirus A, bluegill picornavirus [04-032] KC843627, Potamipivirus, Potamipivirus A, eel picornavirus [F5/05] KJ64698, unassigned, bat picornavirus [BtMf-PicoV-/SAX20] AF327920, Parechovirus, Parechovirus B, Ljungan virus [87-02] HF677705, unassigned, sebokele virus [An/B/227/d] L0297, Parechovirus, Parechovirus A, human parechovirus [Harris] KF006989, unassigned, ferret parechovirus [ferret/mppev/nl] JQ36470, Pasivirus, Pasivirus A [swine/france/20] AB937989, unassigned, crohivirus [shrew/zm54/zambia/202] KC465954, Avisivirus, Avisivirus A [turkey/m76-tuasv/20/hun] KC64703, Avisivirus, Avisivirus A [turkey/usa/in/200] KF979334, unassigned, chicken picornavirus 3 [45C] KT880669, unassigned,chicken picornavirus [Pf-CHK/AsV] KF979333, unassigned, chicken picornavirus 2 [44C] KJ000696, unassigned, aalivirus [duck/gl/2/china/202] KM203656, unassigned, orivirus [chicken/pf-chk/203/hun] KT880667, unassigned, orivirus 2 [Pf-CHK] DQ249299, Avihepatovirus, Avihepatovirus A, duck hepatitis A virus [03D] KC935379, Kunsagivirus, Kunsagivirus A [roller/szal6-kuv/20/hun] unassigned, bat kunsagivirus EU42040, Aquamavirus, Aquamavirus A, seal picornavirus [HO-02-2] KP77040, Ampivirus, Ampivirus A [NEWT/203/HUN] KM87364, unassigned, tortoise picornavirus [44-0] KM87365, unassigned, tortoise picornavirus [203-T4] KM87362, unassigned, tortoise picornavirus [5-04] KM8736, unassigned, tortoise picornavirus [4-04] KM87366, unassigned, tortoise picornavirus [24-4-0] KM87367, unassigned, tortoise picornavirus [5-03] JF973687, Mosavirus, Mosavirus A [mouse/m-7/usa/200] KF95846, Mosavirus,Mosavirus A2 [SZAL6-MoV/20/HUN] JN5727, unassigned, feline picornavirus [356F] 0.77 Page 9 of 0
10 Figure 4 (previous page): Phylogenetic analyses of picornavirus 3CD gene regions using Bayesian tree inference (MrBayes 3.2). 78 sequences were retrieved from GenBank. Presented are GenBank accession numbers, genus names, species names and types. If available, common names and designations of isolates [in square brackets] are given. Yet unassigned viruses are printed in blue. Proposed names are printed in red and indicated by a dot ( ). Numbers at nodes indicate posterior probabilities obtained after 4,750,000 generations. The optimal substitution model (GTR+G+I) was determined with MEGA 5. The scale indicates substitutions/site. Page 0 of 0
a-dS (modules 1 and 11 are required)
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
a-dS. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
a-dV. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
aV. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
Identification and characterization of novel picornaviruses by molecular biological methods. Ph.D. Thesis. Péter Pankovics
 Identification and characterization of novel picornaviruses by molecular biological methods Ph.D. Thesis Péter Pankovics Supervisor: 1 Gábor Reuter, M.D., Ph.D., Med. Habil. Program leader: 2 Levente Emődy,
Identification and characterization of novel picornaviruses by molecular biological methods Ph.D. Thesis Péter Pankovics Supervisor: 1 Gábor Reuter, M.D., Ph.D., Med. Habil. Program leader: 2 Levente Emődy,
aM. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
a-hV. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
aV (modules 1 and 9 are required)
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
aD (modules 1 and 10 are required)
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
Virus isolate from carp: genetic characterization reveals a novel picornavirus with two aphthovirus 2A-like sequences
 Journal of General Virology (2014), 95, 80 90 DOI 10.1099/vir.0.058172-0 Virus isolate from carp: genetic characterization reveals a novel picornavirus with two aphthovirus 2A-like sequences Jeannette
Journal of General Virology (2014), 95, 80 90 DOI 10.1099/vir.0.058172-0 Virus isolate from carp: genetic characterization reveals a novel picornavirus with two aphthovirus 2A-like sequences Jeannette
aP. Code assigned: Short title: Remove (abolish) the species Narcissus symptomless virus in the genus Carlavirus, family Betaflexiviridae
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
The Origin, Evolution and Diagnosis of Seneca Valley Virus, a New Vesicular Disease-Causing Picornavirus of Pigs
 The Origin, Evolution and Diagnosis of Seneca Valley Virus, a New Vesicular Disease-Causing Picornavirus of Pigs Nick J. Knowles, Katarzyna Bachanek-Bankowska, Jemma Wadsworth, Veronica L. Fowler & Donald
The Origin, Evolution and Diagnosis of Seneca Valley Virus, a New Vesicular Disease-Causing Picornavirus of Pigs Nick J. Knowles, Katarzyna Bachanek-Bankowska, Jemma Wadsworth, Veronica L. Fowler & Donald
aM (modules 1 and 10 are required)
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
a,bD. Code assigned: Short title: Two new species in the genus Orthohepadnavirus (e.g. 6 new species in the genus Zetavirus) Modules attached
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
ÁNTSZ Regional Institute of State Public Health Service, Pécs, Hungary
 Journal of eneral Virology (2013), 94, 2029 2035 DOI 10.1099/vir.0.054676-0 Short ommunication orrespondence ábor Reuter reuter.gabor@ddr.antsz.hu enetic characterization of a novel picornavirus distantly
Journal of eneral Virology (2013), 94, 2029 2035 DOI 10.1099/vir.0.054676-0 Short ommunication orrespondence ábor Reuter reuter.gabor@ddr.antsz.hu enetic characterization of a novel picornavirus distantly
Highly diverse population of Picornaviridae and other members of the Picornavirales, in Cameroonian fruit bats
 Yinda et al. BMC Genomics (2017) 18:249 DOI 10.1186/s12864-017-3632-7 RESEARCH ARTICLE Open Access Highly diverse population of Picornaviridae and other members of the Picornavirales, in Cameroonian fruit
Yinda et al. BMC Genomics (2017) 18:249 DOI 10.1186/s12864-017-3632-7 RESEARCH ARTICLE Open Access Highly diverse population of Picornaviridae and other members of the Picornavirales, in Cameroonian fruit
aP (modules 1 and 9 are required)
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
ICTVdB Virus Descriptions
 [Home] [Index of Viruses ] [Descriptions] [ Character List ] [ Picture Gallery ] [ Interactive Key ] [ Data Entry] [ 2002 ICTV] ICTVdB Virus Descriptions Descriptions are generated automatically from the
[Home] [Index of Viruses ] [Descriptions] [ Character List ] [ Picture Gallery ] [ Interactive Key ] [ Data Entry] [ 2002 ICTV] ICTVdB Virus Descriptions Descriptions are generated automatically from the
aV. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
aP (modules 1 and 9 are required) Caulimoviridae
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
aV (modules 1 and 9 are required)
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
aV. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
Novel Positive-Sense, Single-Stranded RNA ( plus ssrna) Virus with Di-Cistronic Genome from Intestinal Content of Freshwater Carp (Cyprinus carpio)
 Edinburgh Research Explorer Novel Positive-Sense, Single-Stranded RNA ( plus ssrna) Virus with Di-Cistronic Genome from Intestinal Content of Freshwater Carp (Cyprinus carpio) Citation for published version:
Edinburgh Research Explorer Novel Positive-Sense, Single-Stranded RNA ( plus ssrna) Virus with Di-Cistronic Genome from Intestinal Content of Freshwater Carp (Cyprinus carpio) Citation for published version:
Analysis of the interaction between the 2C protein of parechoviruses and lipid droplets. Ali Aidarous Khrid
 Analysis of the interaction between the 2C protein of parechoviruses and lipid droplets Ali Aidarous Khrid A thesis submitted for the degree of Doctor of Philosophy School of Biological Sciences University
Analysis of the interaction between the 2C protein of parechoviruses and lipid droplets Ali Aidarous Khrid A thesis submitted for the degree of Doctor of Philosophy School of Biological Sciences University
Aichi Virus 1: Environmental Occurrence and Behavior
 Pathogens 2015, 4, 256-268; doi:10.3390/pathogens4020256 Review OPEN ACCESS pathogens ISSN 2076-0817 www.mdpi.com/journal/pathogens Aichi Virus 1: Environmental Occurrence and Behavior Masaaki Kitajima,
Pathogens 2015, 4, 256-268; doi:10.3390/pathogens4020256 Review OPEN ACCESS pathogens ISSN 2076-0817 www.mdpi.com/journal/pathogens Aichi Virus 1: Environmental Occurrence and Behavior Masaaki Kitajima,
Sequence analysis for VP4 of enterovirus 71 isolated in Beijing during 2007 to 2008
 16 2009 3 4 1 Journal of Microbes and Infection, March 2009, Vol. 4, No. 1 2007 2008 71 VP4 1, 2, 2, 2, 1, 2, 2, 2, 1, 2 1., 100730; 2., 100020 : 2007 2008 71 ( EV71), 2007 3 EV71( 1, 2 ) 2008 5 EV71(
16 2009 3 4 1 Journal of Microbes and Infection, March 2009, Vol. 4, No. 1 2007 2008 71 VP4 1, 2, 2, 2, 1, 2, 2, 2, 1, 2 1., 100730; 2., 100020 : 2007 2008 71 ( EV71), 2007 3 EV71( 1, 2 ) 2008 5 EV71(
aP. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
aD. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
Complete Genome Analysis of Three Novel Picornaviruses from Diverse Bat Species
 JOURNAL OF VIROLOGY, Sept. 2011, p. 8819 8828 Vol. 85, No. 17 0022-538X/11/$12.00 doi:10.1128/jvi.02364-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. Complete Genome Analysis
JOURNAL OF VIROLOGY, Sept. 2011, p. 8819 8828 Vol. 85, No. 17 0022-538X/11/$12.00 doi:10.1128/jvi.02364-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. Complete Genome Analysis
aP. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
Vincent Racaniello
 Vincent Racaniello vrr1@columbia.edu www.virology.ws Poliomyelitis Polio (grey), myelon (marrow) = Greek itis (inflammation of) = Latin A common, acute viral disease characterized clinically by a brief
Vincent Racaniello vrr1@columbia.edu www.virology.ws Poliomyelitis Polio (grey), myelon (marrow) = Greek itis (inflammation of) = Latin A common, acute viral disease characterized clinically by a brief
Supplementary Fig. S1: E. helvum cytochrome b haplotype Bayesian phylogeny with E. dupreanum used as an outgroup. Three main clades are identified:
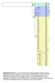 Supplementary Fig. S1: E. helvum cytochrome b haplotype Bayesian phylogeny with E. dupreanum used as an outgroup. Three main clades are identified: A, B and C. Posterior probabilities are shown on the
Supplementary Fig. S1: E. helvum cytochrome b haplotype Bayesian phylogeny with E. dupreanum used as an outgroup. Three main clades are identified: A, B and C. Posterior probabilities are shown on the
Chinese Journal of Biochemistry and Molecular Biology ; (untranslated region,utr) (internal ribosome entry site, IRES). ,.
 ISSN 100727626 CN 1123870ΠQ 2007 7 Chinese Journal of Biochemistry and Molecular Biology 23 (7) :513 518 RNA ( IRES),,,, (, 430072 ;, 430072) 3 RNA 5 (untranslated region,utr) (internal ribosome entry
ISSN 100727626 CN 1123870ΠQ 2007 7 Chinese Journal of Biochemistry and Molecular Biology 23 (7) :513 518 RNA ( IRES),,,, (, 430072 ;, 430072) 3 RNA 5 (untranslated region,utr) (internal ribosome entry
NOTES. Characterization of a Canine Homolog of Human Aichivirus. Received 6 June 2011/Accepted 22 August 2011
 JOURNAL OF VIROLOGY, Nov. 2011, p. 11520 11525 Vol. 85, No. 21 0022-538X/11/$12.00 doi:10.1128/jvi.05317-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. NOTES Characterization
JOURNAL OF VIROLOGY, Nov. 2011, p. 11520 11525 Vol. 85, No. 21 0022-538X/11/$12.00 doi:10.1128/jvi.05317-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. NOTES Characterization
The vast majority of emerging infectious diseases (EIDs) in humans
 RESEARCH ARTICLE crossmark Discovery of a Novel Hepatovirus (Phopivirus of Seals) Related to Human Hepatitis A Virus S. J. Anthony, a,b,c J. A. St. Leger, d E. Liang, a,c A. L. Hicks, a M. D. Sanchez-Leon,
RESEARCH ARTICLE crossmark Discovery of a Novel Hepatovirus (Phopivirus of Seals) Related to Human Hepatitis A Virus S. J. Anthony, a,b,c J. A. St. Leger, d E. Liang, a,c A. L. Hicks, a M. D. Sanchez-Leon,
Taxonomic changes and additions for human and animal viruses,
 JCM Accepted Manuscript Posted Online 19 October 2016 J. Clin. Microbiol. doi:10.1128/jcm.01525-16 Copyright 2016, American Society for Microbiology. All Rights Reserved. 1 2 3 Taxonomic changes and additions
JCM Accepted Manuscript Posted Online 19 October 2016 J. Clin. Microbiol. doi:10.1128/jcm.01525-16 Copyright 2016, American Society for Microbiology. All Rights Reserved. 1 2 3 Taxonomic changes and additions
Picornaviruses (family Picornaviridae), with small, nonenveloped
 omparative omplete enome nalysis of hicken and Turkey Megriviruses (Family Picornaviridae): Long 3= ntranslated Regions with a Potential Second Open Reading Frame and Evidence for Possible Recombination
omparative omplete enome nalysis of hicken and Turkey Megriviruses (Family Picornaviridae): Long 3= ntranslated Regions with a Potential Second Open Reading Frame and Evidence for Possible Recombination
CELL SURFACE INTERACTIONS OF COXSACKIEVIRUS A9 AND HUMAN PARECHOVIRUS 1. Pirjo Merilahti
 CELL SURFACE INTERACTIONS OF COXSACKIEVIRUS A9 AND HUMAN PARECHOVIRUS 1 Pirjo Merilahti TURUN YLIOPISTON JULKAISUJA ANNALES UNIVERSITATIS TURKUENSIS Sarja - ser. D osa - tom. 1283 Medica - Odontologica
CELL SURFACE INTERACTIONS OF COXSACKIEVIRUS A9 AND HUMAN PARECHOVIRUS 1 Pirjo Merilahti TURUN YLIOPISTON JULKAISUJA ANNALES UNIVERSITATIS TURKUENSIS Sarja - ser. D osa - tom. 1283 Medica - Odontologica
Picornaviruses. Virion. Genome. Genes and proteins. Viruses and hosts. Diseases. Distinctive characteristics
 Picornaviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Naked icosahedral capsid (T=1) Diameter of 30 nm Genome Linear single-stranded RNA, positive
Picornaviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Naked icosahedral capsid (T=1) Diameter of 30 nm Genome Linear single-stranded RNA, positive
Identification and characterisation of tomato torrado virus, a new plant picorna-like virus from tomato
 Arch Virol (2007) 152: 881 890 DOI 10.1007/s00705-006-0917-6 Printed in the Netherlands Identification and characterisation of tomato torrado virus, a new plant picorna-like virus from tomato M. Verbeek
Arch Virol (2007) 152: 881 890 DOI 10.1007/s00705-006-0917-6 Printed in the Netherlands Identification and characterisation of tomato torrado virus, a new plant picorna-like virus from tomato M. Verbeek
bV. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
Isolation and Molecular Characterization of a Novel Picornavirus from Baitfish in the USA
 Isolation and Molecular Characterization of a Novel Picornavirus from Baitfish in the USA Nicholas B. D. Phelps 1,2 *, Sunil K. Mor 1, Anibal G. Armien 1,2, William Batts 3, Andrew E. Goodwin 4, Lacey
Isolation and Molecular Characterization of a Novel Picornavirus from Baitfish in the USA Nicholas B. D. Phelps 1,2 *, Sunil K. Mor 1, Anibal G. Armien 1,2, William Batts 3, Andrew E. Goodwin 4, Lacey
Recombination and Selection in the Evolution of Picornaviruses and Other Mammalian Positive-Stranded RNA Viruses
 JOURNAL OF VIROLOGY, Nov. 2006, p. 11124 11140 Vol. 80, No. 22 0022-538X/06/$08.00 0 doi:10.1128/jvi.01076-06 Copyright 2006, American Society for Microbiology. All Rights Reserved. Recombination and Selection
JOURNAL OF VIROLOGY, Nov. 2006, p. 11124 11140 Vol. 80, No. 22 0022-538X/06/$08.00 0 doi:10.1128/jvi.01076-06 Copyright 2006, American Society for Microbiology. All Rights Reserved. Recombination and Selection
Yip, CY; Lo, KL; Que, TL; Lee, RA; Chan, KH; Yuen, KY; Woo, PCY; Lau, SKP
 Title Author(s) Epidemiology of human parechovirus, Aichi virus and salivirus in fecal samples from hospitalized children with gastroenteritis in Hong Kong Yip, CY; Lo, KL; Que, TL; Lee, RA; Chan, KH;
Title Author(s) Epidemiology of human parechovirus, Aichi virus and salivirus in fecal samples from hospitalized children with gastroenteritis in Hong Kong Yip, CY; Lo, KL; Que, TL; Lee, RA; Chan, KH;
The Bunyaviridae Study Group supports this proposal. RM Elliott, 19/06/2014
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
A highly divergent picornavirus in a marine mammal. ACCEPTED
 JVI Accepts, published online ahead of print on 17 October 2007 J. Virol. doi:10.1128/jvi.01240-07 Copyright 2007, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights Reserved.
JVI Accepts, published online ahead of print on 17 October 2007 J. Virol. doi:10.1128/jvi.01240-07 Copyright 2007, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights Reserved.
Enterovirus D68-US Outbreak 2014 Laboratory Perspective
 Enterovirus D68-US Outbreak 2014 Laboratory Perspective Allan Nix Team Lead Picornavirus Laboratory Polio and Picornavirus Laboratory Branch PAHO / SARInet Webinar November 6, 2014 National Center for
Enterovirus D68-US Outbreak 2014 Laboratory Perspective Allan Nix Team Lead Picornavirus Laboratory Polio and Picornavirus Laboratory Branch PAHO / SARInet Webinar November 6, 2014 National Center for
a-cP. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
Sequencing of Porcine Enterovirus Groups II and III Reveals Unique Features of Both Virus Groups
 REFERENCES CONTENT ALERTS Sequencing of Porcine Enterovirus Groups II and III Reveals Unique Features of Both Virus Groups Andi Krumbholz, Malte Dauber, Andreas Henke, Eckhard Birch-Hirschfeld, Nick J.
REFERENCES CONTENT ALERTS Sequencing of Porcine Enterovirus Groups II and III Reveals Unique Features of Both Virus Groups Andi Krumbholz, Malte Dauber, Andreas Henke, Eckhard Birch-Hirschfeld, Nick J.
An Integrated Bioinformatics and Computational Biophysics Approach to Enterovirus Surveillance and Research.
 An Integrated Bioinformatics and Computational Biophysics Approach to Enterovirus Surveillance and Research. A thesis submitted in fulfilment of the requirements of the degree of Doctor of Philosophy Jason
An Integrated Bioinformatics and Computational Biophysics Approach to Enterovirus Surveillance and Research. A thesis submitted in fulfilment of the requirements of the degree of Doctor of Philosophy Jason
Molecular analysis of duck hepatitis virus type 1 reveals a novel lineage close to the genus Parechovirus in the family Picornaviridae
 Journal of General Virology (2006), 87, 3307 3316 DOI 10.1099/vir.0.81804-0 Molecular analysis of duck hepatitis virus type 1 reveals a novel lineage close to the genus Parechovirus in the family Picornaviridae
Journal of General Virology (2006), 87, 3307 3316 DOI 10.1099/vir.0.81804-0 Molecular analysis of duck hepatitis virus type 1 reveals a novel lineage close to the genus Parechovirus in the family Picornaviridae
Complete genome sequence analysis of Seneca Valley virus-001, a novel oncolytic picornavirus
 Journal of General Virology (2008), 89, 1265 1275 DOI 10.1099/vir.0.83570-0 Complete genome sequence analysis of Seneca Valley virus-001, a novel oncolytic picornavirus Laura M. Hales, 1 3 Nick J. Knowles,
Journal of General Virology (2008), 89, 1265 1275 DOI 10.1099/vir.0.83570-0 Complete genome sequence analysis of Seneca Valley virus-001, a novel oncolytic picornavirus Laura M. Hales, 1 3 Nick J. Knowles,
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Detection of HEV in Estonia. Anna Ivanova, Irina Golovljova, Valentina Tefanova
 Detection of HEV in Estonia Anna Ivanova, Irina Golovljova, Valentina Tefanova National Institute for Health Development Department of Virology Zoonotic pathogens Tick-borne pathogens: TBEV, Borrelia sp.,
Detection of HEV in Estonia Anna Ivanova, Irina Golovljova, Valentina Tefanova National Institute for Health Development Department of Virology Zoonotic pathogens Tick-borne pathogens: TBEV, Borrelia sp.,
Framework for the evaluation of. and scientific impacts of plant viruses. technologies.
 Framework for the evaluation of biosecurity, commercial, regulatory and scientific impacts of plant viruses and viroids identified by NGS technologies. Sebastien Massart, Thierry Candresse, José Gil, Christophe
Framework for the evaluation of biosecurity, commercial, regulatory and scientific impacts of plant viruses and viroids identified by NGS technologies. Sebastien Massart, Thierry Candresse, José Gil, Christophe
First report of human salivirus/klassevirus in respiratory specimens of a child with fatal adenovirus infection
 Virus Genes (2016) 52:620 624 DOI 10.1007/s11262-016-1361-7 First report of human salivirus/klassevirus in respiratory specimens of a child with fatal adenovirus infection Na Pei 1,3 Jiaosheng Zhang 2
Virus Genes (2016) 52:620 624 DOI 10.1007/s11262-016-1361-7 First report of human salivirus/klassevirus in respiratory specimens of a child with fatal adenovirus infection Na Pei 1,3 Jiaosheng Zhang 2
Evaluation of Seneca Valley Virus Levels Mouse to Man
 Evaluation of Seneca Valley Virus Levels Mouse to Man EMEA/IHC Meeting on Virus Shedding Rotterdam, The Netherlands October 30, 2007 Paul Hallenbeck Neotropix, Inc. 351 Phoenixville Pike Malvern, PA 19380
Evaluation of Seneca Valley Virus Levels Mouse to Man EMEA/IHC Meeting on Virus Shedding Rotterdam, The Netherlands October 30, 2007 Paul Hallenbeck Neotropix, Inc. 351 Phoenixville Pike Malvern, PA 19380
aV. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
Mapping of the influenza A hemagglutinin serotypes evolution by the ISSCOR method
 Regular paper Supplementary Materials pp 441 452 Vol. 61, 2014 on-line at: www.actabp.pl Mapping of the influenza A hemagglutinin serotypes evolution by the ISSCOR method Jan P. Radomski 1,4 *, Piotr P.
Regular paper Supplementary Materials pp 441 452 Vol. 61, 2014 on-line at: www.actabp.pl Mapping of the influenza A hemagglutinin serotypes evolution by the ISSCOR method Jan P. Radomski 1,4 *, Piotr P.
High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage.
 JVI Accepts, published online ahead of print on 29 August 2012 J. Virol. doi:10.1128/jvi.00869-12 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 High variety of known
JVI Accepts, published online ahead of print on 29 August 2012 J. Virol. doi:10.1128/jvi.00869-12 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 High variety of known
Recombinaison expérimentale intra-et interespèce chez les Rhinovirus et Quantification d'arn de Rhinovirus par RT-PCR en temps réel.
 Thesis Recombinaison expérimentale intra-et interespèce chez les Rhinovirus et Quantification d'arn de Rhinovirus par RT-PCR en temps réel SCHIBLER, Manuel Abstract Les rhinovirus sont des petits virus
Thesis Recombinaison expérimentale intra-et interespèce chez les Rhinovirus et Quantification d'arn de Rhinovirus par RT-PCR en temps réel SCHIBLER, Manuel Abstract Les rhinovirus sont des petits virus
Virology Journal. Open Access. Abstract
 Virology Journal BioMed Central Research The use of RNA-dependent RNA polymerase for the taxonomic assignment of Picorna-like viruses (order Picornavirales) infecting Apis mellifera L. populations Andrea
Virology Journal BioMed Central Research The use of RNA-dependent RNA polymerase for the taxonomic assignment of Picorna-like viruses (order Picornavirales) infecting Apis mellifera L. populations Andrea
Ch10 Classification and Nomenclature of Viruses 國立台灣海洋大學海洋生物研究所陳歷歷
 Ch10 Classification and Nomenclature of Viruses 國立台灣海洋大學海洋生物研究所陳歷歷 At a glance 10.1 History of virus classification and nomenclature Some of characteristics can be used for the purposes of classification:
Ch10 Classification and Nomenclature of Viruses 國立台灣海洋大學海洋生物研究所陳歷歷 At a glance 10.1 History of virus classification and nomenclature Some of characteristics can be used for the purposes of classification:
University of Canberra Research Publication Collection
 - University of Canberra Research Publication Collection Faculty of Health 2012 - Peer-Reviewed Journal Article for Dr. Reena Ghildyal (Associate Professor, University of Canberra) This is the peer reviewed
- University of Canberra Research Publication Collection Faculty of Health 2012 - Peer-Reviewed Journal Article for Dr. Reena Ghildyal (Associate Professor, University of Canberra) This is the peer reviewed
An automated global clade classification tool for H1 hemagglutinin genes from swine influenza A viruses
 An automated global clade classification tool for H1 hemagglutinin genes from swine influenza A viruses Yun Zhang J. Craig Venter Institute June 25, 2017 Outline A web-based, automated H1 clade classification
An automated global clade classification tool for H1 hemagglutinin genes from swine influenza A viruses Yun Zhang J. Craig Venter Institute June 25, 2017 Outline A web-based, automated H1 clade classification
Envelope e_ :------, Envelope glycoproteins e_ ~ Single-stranded RNA ----, Nucleocapsid
 Virus Antib(tdies Your Expertise, Our Antibodies, Accelerated Discovery. Envelope e_---------:------, Envelope glycoproteins e_-------1~ Single-stranded RNA ----, Nucleocapsid First identified in 1989,
Virus Antib(tdies Your Expertise, Our Antibodies, Accelerated Discovery. Envelope e_---------:------, Envelope glycoproteins e_-------1~ Single-stranded RNA ----, Nucleocapsid First identified in 1989,
Introduction: RNA viruses
 SECTION I Introduction: RNA viruses Carol Shoshkes Reiss Viruses that infect the central nervous system may cause acute, chronic, or latent infections. In some cases, the diseases manifested are attributable
SECTION I Introduction: RNA viruses Carol Shoshkes Reiss Viruses that infect the central nervous system may cause acute, chronic, or latent infections. In some cases, the diseases manifested are attributable
Characterization of a canine homolog of human Aichivirus.
 JVI Accepts, published online ahead of print on 31 August 2011 J. Virol. doi:10.1128/jvi.05317-11 Copyright 2011, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights Reserved.
JVI Accepts, published online ahead of print on 31 August 2011 J. Virol. doi:10.1128/jvi.05317-11 Copyright 2011, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights Reserved.
Supplementary Online Content
 Supplementary Online Content Rodger AJ, Cambiano V, Bruun T, et al; PARTNER Study Group. Sexual activity without condoms and risk of HIV transmission in serodifferent couples when the HIVpositive partner
Supplementary Online Content Rodger AJ, Cambiano V, Bruun T, et al; PARTNER Study Group. Sexual activity without condoms and risk of HIV transmission in serodifferent couples when the HIVpositive partner
Susanne Johansson, 1 Bo Niklasson, 2 Jacob Maizel, 3 Alexander E. Gorbalenya, 4 * and A. Michael Lindberg 1 *
 JOURNAL OF VIROLOGY, Sept. 2002, p. 8920 8930 Vol. 76, No. 17 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.17.8920 8930.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Molecular
JOURNAL OF VIROLOGY, Sept. 2002, p. 8920 8930 Vol. 76, No. 17 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.17.8920 8930.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Molecular
Chih-Yu Wu,* Chu-Fang Lo, Chang-Jen Huang, Hon-Tsen Yu, and Chung-Hsiung Wang*,1
 Virology 294, 312 323 (2002) doi:10.1006/viro.2001.1344, available online at http://www.idealibrary.com on The Complete Genome Sequence of Perina nuda Picorna-like Virus, An Insect-Infecting RNA Virus
Virology 294, 312 323 (2002) doi:10.1006/viro.2001.1344, available online at http://www.idealibrary.com on The Complete Genome Sequence of Perina nuda Picorna-like Virus, An Insect-Infecting RNA Virus
Change of Major Genotype of Enterovirus 71 in Outbreaks of Hand-Foot-and-Mouth Disease in Taiwan between 1998 and 2000
 JOURNAL OF CLINICAL MICROBIOLOGY, Jan. 2002, p. 10 15 Vol. 40, No. 1 0095-1137/02/$04.00 0 DOI: 10.1128/JCM.40.1.10 15.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Change
JOURNAL OF CLINICAL MICROBIOLOGY, Jan. 2002, p. 10 15 Vol. 40, No. 1 0095-1137/02/$04.00 0 DOI: 10.1128/JCM.40.1.10 15.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Change
Novel Picornavirus in Turkey Poults with Hepatitis, California, USA
 RESEARCH Novel Picornavirus in Turkey Poults with Hepatitis, California, USA Kirsi S. Honkavuori, H. L. Shivaprasad, Thomas Briese, Craig Street, David L. Hirschberg, Stephen K. Hutchison, and W. Ian Lipkin
RESEARCH Novel Picornavirus in Turkey Poults with Hepatitis, California, USA Kirsi S. Honkavuori, H. L. Shivaprasad, Thomas Briese, Craig Street, David L. Hirschberg, Stephen K. Hutchison, and W. Ian Lipkin
First Identification and Characterization of Porcine Enterovirus G in the United States
 First Identification and Characterization of Porcine Enterovirus G in the United States Srivishnupriya Anbalagan 1, Richard A. Hesse 2, Ben M. Hause 1 * 1 Newport Laboratories, Inc., Worthington, Minnesota,
First Identification and Characterization of Porcine Enterovirus G in the United States Srivishnupriya Anbalagan 1, Richard A. Hesse 2, Ben M. Hause 1 * 1 Newport Laboratories, Inc., Worthington, Minnesota,
Rajesh Kannangai Phone: ; Fax: ; *Corresponding author
 Amino acid sequence divergence of Tat protein (exon1) of subtype B and C HIV-1 strains: Does it have implications for vaccine development? Abraham Joseph Kandathil 1, Rajesh Kannangai 1, *, Oriapadickal
Amino acid sequence divergence of Tat protein (exon1) of subtype B and C HIV-1 strains: Does it have implications for vaccine development? Abraham Joseph Kandathil 1, Rajesh Kannangai 1, *, Oriapadickal
MIRTRIYKPTYTTTTPPTCHTPIKMEEDPREKMHPQSMWRLVRLRAQRLLSYSESTDLSTREFLEDVSKS I...R...M...D.A...I.G.
 A. FDLV iiiiiiiiiiiiiii1 AnaPV(1110RN043)iiii1 AnaPV(1110RN047)iiii1 AnaPV(1201RN001)I ii1 AnaPV(1201RN003) iii1 AnaPV(1203RN009) iii1 Biti-CA98 iiiiiiiiii1 Dasy-GER00 iiiiiiiii1 Xeno-USA99 iiiiiiiii1
A. FDLV iiiiiiiiiiiiiii1 AnaPV(1110RN043)iiii1 AnaPV(1110RN047)iiii1 AnaPV(1201RN001)I ii1 AnaPV(1201RN003) iii1 AnaPV(1203RN009) iii1 Biti-CA98 iiiiiiiiii1 Dasy-GER00 iiiiiiiii1 Xeno-USA99 iiiiiiiii1
Evidence for enteroviral persistence in humans
 Journal of General Virology (1997), 78, 307 312. Printed in Great Britain...... SHORT COMMUNICATION Evidence for enteroviral persistence in humans Daniel N. Galbraith, Carron Nairn and Geoffrey B. Clements
Journal of General Virology (1997), 78, 307 312. Printed in Great Britain...... SHORT COMMUNICATION Evidence for enteroviral persistence in humans Daniel N. Galbraith, Carron Nairn and Geoffrey B. Clements
Nucleotide sequences and taxonomy of satsuma dwarf virus
 Journal of General Virology (1999), 80, 793 797. Printed in Great Britain... SHORT COMMUNICATION Nucleotide sequences and taxonomy of satsuma dwarf virus Toru Iwanami, 1 Yoshiko Kondo 1 and Alexander V.
Journal of General Virology (1999), 80, 793 797. Printed in Great Britain... SHORT COMMUNICATION Nucleotide sequences and taxonomy of satsuma dwarf virus Toru Iwanami, 1 Yoshiko Kondo 1 and Alexander V.
Translation. Host Cell Shutoff 1) Initiation of eukaryotic translation involves many initiation factors
 Translation Questions? 1) How does poliovirus shutoff eukaryotic translation? 2) If eukaryotic messages are not translated how can poliovirus get its message translated? Host Cell Shutoff 1) Initiation
Translation Questions? 1) How does poliovirus shutoff eukaryotic translation? 2) If eukaryotic messages are not translated how can poliovirus get its message translated? Host Cell Shutoff 1) Initiation
The 5 0 untranslated region of Perina nuda virus (PnV) possesses a strong internal translation activity in baculovirus-infected insect cells
 FEBS Letters 581 (2007) 3120 3126 The 5 0 untranslated region of Perina nuda virus (PnV) possesses a strong internal translation activity in baculovirus-infected insect cells Tzong-Yuan Wu a,b,1, Chih-Yu
FEBS Letters 581 (2007) 3120 3126 The 5 0 untranslated region of Perina nuda virus (PnV) possesses a strong internal translation activity in baculovirus-infected insect cells Tzong-Yuan Wu a,b,1, Chih-Yu
Coronaviruses. Virion. Genome. Genes and proteins. Viruses and hosts. Diseases. Distinctive characteristics
 Coronaviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Spherical enveloped particles studded with clubbed spikes Diameter 120-160 nm Coiled helical
Coronaviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Spherical enveloped particles studded with clubbed spikes Diameter 120-160 nm Coiled helical
NOMENCLATURE & CLASSIFICATION OF PLANT VIRUSES. P.N. Sharma Department of Plant Pathology, CSK HPKV, Palampur (H.P.)
 NOMENCLATURE & CLASSIFICATION OF PLANT VIRUSES P.N. Sharma Department of Plant Pathology, CSK HPKV, Palampur (H.P.) What is the purpose of classification? To make structural arrangement comprehension for
NOMENCLATURE & CLASSIFICATION OF PLANT VIRUSES P.N. Sharma Department of Plant Pathology, CSK HPKV, Palampur (H.P.) What is the purpose of classification? To make structural arrangement comprehension for
HBV. Next Generation Sequencing, data analysis and reporting. Presenter Leen-Jan van Doorn
 HBV Next Generation Sequencing, data analysis and reporting Presenter Leen-Jan van Doorn HBV Forum 3 October 24 th, 2017 Marriott Marquis, Washington DC www.forumresearch.org HBV Biomarkers HBV biomarkers:
HBV Next Generation Sequencing, data analysis and reporting Presenter Leen-Jan van Doorn HBV Forum 3 October 24 th, 2017 Marriott Marquis, Washington DC www.forumresearch.org HBV Biomarkers HBV biomarkers:
Received 18 August 2000/Accepted 31 July 2001
 JOURNAL OF VIROLOGY, Nov. 2001, p. 10244 10249 Vol. 75, No. 21 0022-538X/01/$04.00 0 DOI: 10.1128/JVI.75.21.10244 10249.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved. The
JOURNAL OF VIROLOGY, Nov. 2001, p. 10244 10249 Vol. 75, No. 21 0022-538X/01/$04.00 0 DOI: 10.1128/JVI.75.21.10244 10249.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved. The
... Department of Biological Sciences, John Tabor Laboratories, University of Essex, Colchester CO4 3SQ, UK
 Journal of General Virology (2000), 81, 201 207. Printed in Great Britain... The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in
Journal of General Virology (2000), 81, 201 207. Printed in Great Britain... The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in
Report concerning identification of the Northern European isolate of BTV (26 th August 2006)
 Report concerning identification of the Northern European isolate of BTV (26 th August 2006) Peter Mertens, Carrie Batten, Simon Anthony, Lydia Kgosana, Karin Darpel, Kasia Bankowska, Eva Veronesi, Abid
Report concerning identification of the Northern European isolate of BTV (26 th August 2006) Peter Mertens, Carrie Batten, Simon Anthony, Lydia Kgosana, Karin Darpel, Kasia Bankowska, Eva Veronesi, Abid
Genome Sequencing and Analysis of a Porcine Delta Coronavirus from Eastern China
 ResearchArticle imedpub Journals http://www.imedpub.com/ DOI: 10.21767/2248-9215.100025 European Journal of Experimental Biology Genome Sequencing and Analysis of a Porcine Delta Coronavirus from Eastern
ResearchArticle imedpub Journals http://www.imedpub.com/ DOI: 10.21767/2248-9215.100025 European Journal of Experimental Biology Genome Sequencing and Analysis of a Porcine Delta Coronavirus from Eastern
Identifying Hosts of Families of Viruses: A Machine Learning Approach
 Identifying Hosts of Families of Viruses: A Machine Learning Approach Anil Raj 1 *, Michael Dewar 2, Gustavo Palacios 3, Raul Rabadan 4, Christopher H. Wiggins 5 1 Department of Applied Physics and Applied
Identifying Hosts of Families of Viruses: A Machine Learning Approach Anil Raj 1 *, Michael Dewar 2, Gustavo Palacios 3, Raul Rabadan 4, Christopher H. Wiggins 5 1 Department of Applied Physics and Applied
Viral Taxonomic Classification
 Viruses Part I Viral Taxonomic Classification Order>> -virales Family>> - viridae Subfamily>> -virinae Genus>> -virus Species Order>> Picornavirales Family>> Picornaviridae Subfamily>> Picornavirinae Genus>>
Viruses Part I Viral Taxonomic Classification Order>> -virales Family>> - viridae Subfamily>> -virinae Genus>> -virus Species Order>> Picornavirales Family>> Picornaviridae Subfamily>> Picornavirinae Genus>>
CONSTRUCTION OF PHYLOGENETIC TREE USING NEIGHBOR JOINING ALGORITHMS TO IDENTIFY THE HOST AND THE SPREADING OF SARS EPIDEMIC
 CONSTRUCTION OF PHYLOGENETIC TREE USING NEIGHBOR JOINING ALGORITHMS TO IDENTIFY THE HOST AND THE SPREADING OF SARS EPIDEMIC 1 MOHAMMAD ISA IRAWAN, 2 SITI AMIROCH 1 Institut Teknologi Sepuluh Nopember (ITS)
CONSTRUCTION OF PHYLOGENETIC TREE USING NEIGHBOR JOINING ALGORITHMS TO IDENTIFY THE HOST AND THE SPREADING OF SARS EPIDEMIC 1 MOHAMMAD ISA IRAWAN, 2 SITI AMIROCH 1 Institut Teknologi Sepuluh Nopember (ITS)
HAV HBV HCV HDV HEV HGV
 Viral Hepatitis HAV HBV HCV HDV HEV HGV Additional well-characterized viruses that can cause sporadic hepatitis, such as yellow fever virus, cytomegalovirus, Epstein-Barr virus, herpes simplex virus, rubella
Viral Hepatitis HAV HBV HCV HDV HEV HGV Additional well-characterized viruses that can cause sporadic hepatitis, such as yellow fever virus, cytomegalovirus, Epstein-Barr virus, herpes simplex virus, rubella
(ii) The effective population size may be lower than expected due to variability between individuals in infectiousness.
 Supplementary methods Details of timepoints Caió sequences were derived from: HIV-2 gag (n = 86) 16 sequences from 1996, 10 from 2003, 45 from 2006, 13 from 2007 and two from 2008. HIV-2 env (n = 70) 21
Supplementary methods Details of timepoints Caió sequences were derived from: HIV-2 gag (n = 86) 16 sequences from 1996, 10 from 2003, 45 from 2006, 13 from 2007 and two from 2008. HIV-2 env (n = 70) 21
Using a fast clustering method for viral segment lineage determination, applied to the H9 influenza hemagglutinin.
 Using a fast clustering method for viral segment lineage determination, applied to the H9 influenza hemagglutinin. Andrew R. Dalby Department of Biomedical Sciences, University of Westminster, 5 New Cavendish
Using a fast clustering method for viral segment lineage determination, applied to the H9 influenza hemagglutinin. Andrew R. Dalby Department of Biomedical Sciences, University of Westminster, 5 New Cavendish
Suspected zoonotic transmission of rotavirus group A in Danish adults
 Downloaded from orbit.dtu.dk on: Jul 04, 2018 Suspected zoonotic transmission of rotavirus group A in Danish adults Midgley, S. E.; Hjulsager, Charlotte Kristiane; Larsen, Lars Erik; Falkenhorst, G.; Böttiger,
Downloaded from orbit.dtu.dk on: Jul 04, 2018 Suspected zoonotic transmission of rotavirus group A in Danish adults Midgley, S. E.; Hjulsager, Charlotte Kristiane; Larsen, Lars Erik; Falkenhorst, G.; Böttiger,
Diversity of picornaviruses in rural Bolivia
 Journal of General Virology (2013), 94, 2017 2028 DOI 10.10/vir.3827-0 Diversity of picornaviruses in rural Bolivia W. Allan Nix, 1 Nino Khetsuriani, 1 Silvia Peñaranda, 1 Kaija Maher, 1 Linda Venczel,
Journal of General Virology (2013), 94, 2017 2028 DOI 10.10/vir.3827-0 Diversity of picornaviruses in rural Bolivia W. Allan Nix, 1 Nino Khetsuriani, 1 Silvia Peñaranda, 1 Kaija Maher, 1 Linda Venczel,
The Fecal Viral Flora of Wild Rodents
 The Fecal Viral Flora of Wild Rodents Tung G. Phan 1,2, Beatrix Kapusinszky 1,2,3, Chunlin Wang 4, Robert K. Rose 5, Howard L. Lipton 6, Eric L. Delwart 1,2 * 1 Blood Systems Research Institute, San Francisco,
The Fecal Viral Flora of Wild Rodents Tung G. Phan 1,2, Beatrix Kapusinszky 1,2,3, Chunlin Wang 4, Robert K. Rose 5, Howard L. Lipton 6, Eric L. Delwart 1,2 * 1 Blood Systems Research Institute, San Francisco,
Sequence analysis and genomic organization of Aphid lethal paralysis virus: a new member of the family Dicistroviridae
 Journal of General Virology (2002), 83, 3131 3138. Printed in Great Britain... Sequence analysis and genomic organization of Aphid lethal paralysis virus: a new member of the family Dicistroviridae M.
Journal of General Virology (2002), 83, 3131 3138. Printed in Great Britain... Sequence analysis and genomic organization of Aphid lethal paralysis virus: a new member of the family Dicistroviridae M.
Sequencing-Based Identification of a Novel Coronavirus in Ferrets with Epizootic Catarrhal Enteritis and Development of Molecular Diagnostic Tests
 Sequencing-Based Identification of a Novel Coronavirus in Ferrets with Epizootic Catarrhal Enteritis and Development of Molecular Diagnostic Tests A. Wise, Matti Kiupel,, C. Isenhour, R. Maes Coronaviruses
Sequencing-Based Identification of a Novel Coronavirus in Ferrets with Epizootic Catarrhal Enteritis and Development of Molecular Diagnostic Tests A. Wise, Matti Kiupel,, C. Isenhour, R. Maes Coronaviruses
