against the ras protein inhibits insulin action in frog oocytes
|
|
|
- Dina Hamilton
- 5 years ago
- Views:
Transcription
1 Proc. Natl. Acad. Sci. USA Vol. 86, pp , October 1989 Biochemistry In vitro tyrosine phosphorylation studies on RAS proteins and calmodulin suggest that polylysine-like basic peptides or domains may be involved in interactions between insulin receptor kinase and its substrate (purified system/hras protein/kras protein) YOKO FUJITA-YAMAGUCHI*t, SATISH KATHURIA*, QIN-YU XU*, JAY M. MCDONALDt, HIROFUMI NAKANO, AND TOHRU KAMATA *Department of Molecular Genetics, Beckman Research Institute of the City of Hope, Duarte, CA 91010; *Departments of Pathology and Medicine, Washington University School of Medicine, Saint Louis, MO 63110; Tokyo Research Laboratories, Kyowa Hakko Kogyo Co., Machida-shi, Tokyo, Japan; and lprogram Resources, Inc., BCDP, National Cancer Institute-Frederick Cancer Research Facility, Frederick, MD Communicated by Rachmiel Levine, June 16, 1989 (received for review November 30, 1988) ABSTRACT We have investigated the in vitro tyrosine phosphorylation of the HRAS and KRAS proteins by human placental insulin receptor kinase. Purified HRAS proteins are not phosphorylated by purified insulin receptor kinase. Since the tyrosine phosphorylation of calmodulin by the insulin receptor kinase in vitro requires cofactors such as protamine and poly(l-lysine), we examined the possibility that poly(llysine) may also potentiate the interaction between RAS proteins and the insulin receptor. We found that purified HRAS proteins are indeed phosphorylated by purified insulin receptor kinase in the presence of poly(l-lysine). In contrast, the KRAS protein, which carries an extremely basic domain (residues , Lys-Asp-Glu-Lys6-Ser-Arg), is phosphorylated by the receptor kinase without the addition of basic proteins. We then determined whether the KRAS basic domain peptide plays a role similar to that of poly(l-lysine) and found that both the HRAS protein and calmodulin are phosphorylated by the receptor kinase in the presence of the KRAS basic domain peptide. Further examination of the role of poly(l-lysine) in potentiating tyrosine phosphorylation of the BRAS protein and calmodulin by purified insulin receptor kinase indicates that poly(l-lysine) affects the conformation of these protein substrates as well as that of the receptor kinase domain. These studies suggest that polylysine-like basic proteins or domains are required to establish the interaction between insulin receptor kinase and its substrate. The RAS genes, HRAS, NRAS, and KRAS, have been studied intensively since their transforming alleles were first identified in human tumors in They code for structurally related proteins of 21,000 daltons (p21) that bind guanine nucleotides, have GTPase activity, and are associated with the plasma membrane (1). RAS proteins show significant sequence homology with guanine nucleotide binding (G) proteins (2). This evidence suggests that RAS proteins may participate in the transduction of signals across cellular membranes. In an attempt to define the role of RAS proteins in transmembrane signaling systems, we have previously investigated a potential interaction between HRAS proteins and the insulin receptor and reported that insulin stimulates phosphorylation of the Harvey murine sarcoma virus transforming gene-encoded ras protein (v-ha-ras) in isolated membranes (3). Consistent with our observation, others have The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" in accordance with 18 U.S.C solely to indicate this fact. suggested that microinjection of monoclonal antibodies against the ras protein inhibits insulin action in frog oocytes (4, 5). However, we were not able to show a direct interaction between purified Ha-ras and insulin receptor proteins (3). O'Brien et al. (6) have also observed that the insulin receptor does not phosphorylate native Ha-ras protein. The phosphorylation of calmodulin by the insulin receptor in vitro has been found to require cofactors such as protamine and poly(l-lysine) (7) while in intact adipocytes calmodulin appears to be phosphorylated at tyrosine residues in an insulin-dependent manner (8). More recently, we have studied the role of poly(l-lysine) in phosphorylation of calmodulin and src-related peptide by purified insulin receptor kinase (9, 10). [A synthetic peptide resembling the tyrosine phosphorylation site of pp6src (Arg-Arg-Leu-Ile-Glu-Asp- Ala-Glu-Tyr-Ala-Ala-Arg-Gly) is routinely used to measure tyrosine-specific protein kinase activity (11).] We report here that poly(l-lysine) potentiates the tyrosine phosphorylation of purified HRAS proteins by the insulin receptor kinase in vitro. Based on these findings we predicted that, in contrast to the HRAS protein, the KRAS protein, which contains a poly(l-lysine)-like domain in its carboxyl terminal region, would interact directly with the insulin receptor. We found that the KRAS protein is indeed phosphorylated by the receptor kinase in vitro. We also found that a basic peptide from the KRAS protein potentiates the interaction between the receptor kinase and its substrate. We have further examined modalities of interaction between the receptor kinase, poly(l-lysine), and either substrate (HRAS or calmodulin), and we propose a model explaining our findings. MATERIALS AND METHODS Materials. Insulin receptor was purified 2400-fold from human placental membranes by sequential affinity chromatography on wheat germ agglutinin- and insulin-sepharose as described (12). The receptor was apparently pure and retained both insulin binding activity and tyrosine kinase activity (12-14). Human normal (wt) and oncogenic (T24) HRAS proteins were expressed in an Escherichia coli expression system (provided by R. Sweet, Smith Kline & French) (15) and were purified to homogeneity as described (3). Human v-kras protein expressed in E. coli was purified to Abbreviations: wt, wild-type HRAS protein; T24, T24 transforming (Gly -- Val at position 12) HRAS protein; BSA, bovine serum albumin. tto whom reprint requests should be addressed. 7306
2 Biochemistry: Fujita-Yamaguchi et al. homogeneity as described (16). A peptide corresponding to the last 14 amino acids in the carboxyl terminus of the human c-kras2 gene product (Lys6-Ser-Lys-Thr-Lys-Cys-Val- Leu-Met) was provided by K. C. Glenn (Monsanto). POly(Llysine) (Mr, 30,000-70,000), y-globulin (IgG), P3-galactodsidase, myosin, phosphorylase b, bovine serum albumin (BSA), and phosphoamino acids were obtained from Sigma. Purified porcine brain calmodulin was from Ocean Biologics (Edmonds, WA). Crystalline porcine insulin was supplied by Eli Lilly. Molecular weight markers and reagents for Na- DodSO4/PAGE were purchased from Bio-Rad. [y-32p]atp was from New England Nuclear. Phosphorylation of Protein Substrates by Purified Insulin Receptor Kinase. The standard condition was that the purified insulin receptor kinase ( tug) and the indicated amounts of protein substrates, such as purified RAS protein or calmodulin, were incubated for 60 min at 250C in the absence or presence of 0.1 pum insulin and/or 0.24 ttm poly(l-lysine) in 12.5,u of 50 mm Tris HCl buffer, ph 7.4/ 0.1% Triton X-100. The phosphorylation reaction was initiated by the addition of 2.5 1ul of 6 times-concentrated [y- 32P]ATP/metal ion mixtures that gave final concentrations of 40 /im ATP (-12,000 cpm/pmol), 2 mm MnCl2, and 15 mm MgCl2. The reaction was allowed to proceed for 20 min at 25 C and stopped by addition of 7.5,d of 3 timesconcentrated Laemmli sample buffer, followed by boiling for 5 min. NaDodSO4/PAGE was performed according to Laemmli (17) under reducing conditions. RESULTS Phosphorylation of HRAS Proteins by Purified Insulin Receptor Kinase in the Absence or Presence of Poly(L-Lysine). To examine the possibility that poly(l-lysine) may potentiate the interaction between HRAS proteins and the insulin receptor kinase, we used both normal (glycine at position 12; wt) and oncogenic (valine at position 12; T24) RAS proteins purified from E. coli cells that stably expressed one or the other RAS protein. As shown in Fig. 1 (lanes 3 and 5), the purified insulin receptor kinase did not phosphorylate either RAS protein. Phosphorylation of the RAS proteins was not observed even in the presence of insulin (data not shown). [We have previously shown that the receptor kinase does not phosphorylate calmodulin in the absence or presence of insulin (9).] However, the HRAS proteins were phosphorylated by the purified insulin receptor kinase in the presence of poly(llysine) (lanes 4 and 6). Incubation of RAS proteins with poly(l-lysine) and BSA (in place of the receptor kinase) did not cause phosphorylation of the RAS proteins, indicating Mr x ALa.- 'W AiAiL HaST24 H-at- -_ - BSA a :_i _ ras IR + + f PLL + _+-+ + _ FIG. 1. Phosphorylation of purified HRAS proteins (wt and T24) by purified insulin receptor kinase (IR) in the presence or absence of poly(l-lysine) (PLL). Insulin receptor kinase ("0.1,g) and HRAS protein (- 1,ug) were incubated in the presence or absence of 0.24,uM poly(l-lysine) at 25 C for 60 min in 8 IlI of 50 mm Tris HCl buffer, ph 7.4/0.1% Triton X-100. The phosphorylation reaction was performed as described in Materials and Methods. Samples were analyzed by NaDodSO4/10% PAGE and autoradiography. Proc. Natl. Acad. Sci. USA 86 (1989) 7307 that the poly(l-lysine)-induced phosphorylation of RAS proteins is not due to some other protein kinase contaminating the RAS protein preparations (lanes 7 and 8). As we have reported (10), poly(l-lysine) stimulates phosphorylation of the,3 subunit as well as the a and /31 subunits of the insulin receptor (Fig. 1, lane 2). [The,81 subunit, which is often found in purified insulin receptor preparations, is the N-terminal portion of the /8 subunit generated by proteolytic loss of the kinase domain (18, 19).] Of interest also is that phosphorylation of the receptor 8 subunit was seemingly enhanced in the presence of the RAS protein (lanes 3 and 5). Phosphoamino acid analysis has shown that both the HRAS proteins and the insulin receptor /3 subunit are phosphorylated exclusively on tyrosine residues (data not shown). Effect of Poly(L-Lysine) on Phosphorylation of Various Proteins by Purified Insulin Receptor Kinase. To test the possibility that poly(l-lysine) may be able to act on acidic proteins other than the RAS proteins and calmodulin, various proteins that are not known to be substrates for the kinase were examined. IgG,,3-galactosidase, myosin, and phosphorylase b (data not shown) as well as BSA (Fig. 2, lanes 8 and 10). The experiments showed that myosin, phosphorylase b, and BSA are not good substrates even in the presence of poly(l-lysine). /8-Galactosidase appeared to be phosphorylated by the insulin receptor kinase in the absence of poly(llysine) and this phosphorylation was enhanced when the receptor was activated in the presence of poly(l-lysine). The IgG heavy chain seemed to be slightly phosphorylated by the receptor kinase in the presence of poly(l-lysine), but the intensity of phosphorylation was much lower than that of receptor autophosphorylation or phosphorylation of the RAS proteins or calmodulin. Phosphorylation of the Purified KRAS Protein by Purified Insulin Receptor Kinase. Since the KRAS protein has a poly(l-lysine)-like domain in its carboxyl-terminal variable domain (residues , Lys-Asp-Glu-Lys6-Ser-Arg), we CaM H-ras BSA IR Insulin PLL ' + ( H-ros -CaM FIG. 2. Phosphorylation of the HRAS protein and calmodulin (CaM) by purified insulin receptor kinase (IR) under various preincubation conditions as indicated. Lanes 2, 5-10, and 12: the indicated proteins ((D) were incubated together at 25 C for 60 min prior to initiation of the phosphorylation reaction by addition of [y- 32P]ATP/metal ion mixtures. Lanes 1 and 11 and also lanes 3 and 13: the indicated proteins ((D) were similarly incubated and poly(l-lysine) (+) was addedjust prior to initiation of the phosphorylation reaction. Lanes 4 and 14: the indicated proteins (G)) were incubated together, and IR and insulin (ie) were incubated together, and the mixtures were combined just prior to initiation of the phosphorylation reaction. Assays were performed as described in Materials and Methods except that reaction mixtures for calmodulin contained 1 mm EGTA since the presence of Ca2+ inhibits the phosphorylation of calmodulin (9). Samples were analyzed by NaDodSO4/12.5% PAGE. The time course of 32p incorporation into the RAS proteins (or calmodulin) was linear under the conditions used.
3 7308 Biochemistry: Fujita-Yamaguchi et al Proc. Natl. Acad. Sci. USA 86 (1989) '_ K-ras-* -- H-ras A H-roef3-'-4 IR INS PLL + K-ras H-ras FIG. 3. Phosphorylation of the purified KRAS protein by purified insulin receptor kinase. KRAS protein (-2,ug) was incubated with insulin receptor kinase (IR; -0.1 ug) in the absence (lane 1) or presence (lane 2) of insulin (INS). Lanes 3 and 4: controls in which the HRAS protein (wt, -2,ug) was incubated with insulin receptor kinase (-0.1,ug) in the presence of insulin (lane 3) or 0.24 AM poly(l-lysine) (PLL) (lane 4). The phosphorylation reaction was performed as described in Materials and Methods. Samples were analyzed by NaDodSO4/1W0 PAGE and autoradiography. hypothesized that the KRAS protein may be phosphorylated by the insulin receptor kinase in the absence of cofactor basic proteins. Purified KRAS protein was phosphorylated by purified insulin receptor kinase (Fig. 3, lane 1) and this phosphorylation was enhanced in the presence of insulin (lane 2) whereas in control experiments phosphorylation of the HRAS protein by the receptor kinase was observed only in the presence of poly(l-lysine) (lanes 3 and 4). Phosphorylation of the HRAS Protein and Calmodulin by Purified Insulin Receptor Kinase in the Presence of the KRAS Basic Domain Peptide. To examine the ability of the poly(llysine)-like basic domain found in the KRAS protein to link insulin receptor kinase with its substrates, a peptide corresponding to the carboxyl-terminal domain of the c-kras2 protein (Lys6-Ser-Lys-Thr-Lys-Cys-Val-Leu-Met) (20) was used in place of poly(l-lysine). As shown in Fig. 4, both substrates were phosphorylated in the presence of the KRAS peptide. Higher concentrations of the peptide were required for the phosphorylation as compared to poly(l-lysine): ,M vs ,uM. In addition, the peptide was less effective than poly(l-lysine); phosphorylation in the presence of the KRAS peptide, although somewhat variable, averaged 2% and 9% of the effects achieved by poly(l-lysine) for calmodulin and HRAS protein, respectively. FIG. 4. B CaM-~ PLL M K-ras peptide p M Phosphorylation of the HRAS protein (A) and calmodulin (B) by purified insulin receptor kinase in the presence of the KRAS basic peptide. Insulin receptor kinase (-0.1 ikg) and HRAS protein (1,gg) or calmodulin (4,ug) were incubated in the presence of poly(l-lysine) (PLL) or the KRAS basic peptide and 0.1 um insulin at 25 C for 60 min. The phosphorylation reaction was performed as described in Materials and Methods except that the calmodulin reaction mixtures contained 1 mm EGTA since the presence of Ca2' inhibits the phosphorylation of calmodulin (9). Samples were analyzed by NaDodSO4/12.5% PAGE. The gels were exposed to Kodak XAR 5 film with one intensifying screen for 1 hr (B, lane 1), 5 hr (A, lane 1; B, lanes 2-7), or 15 hr (A, lanes 2-7). Phosphorylation of the HRAS Protein and Calmodulin by Purified Insulin Receptor Kinase After Various Preincubation Conditions. In our standard experiments, a substrate, the insulin receptor, insulin, and poly(l-lysine) were preincubated, and the phosphorylation reaction was initiated by addition of [y-32p]atp/metal ion mixtures. When the preincubation conditions were varied, 32p incorporation into the RAS protein or calmodulin was strikingly different from that obtained under the standard preincubation condition. A representative experiment is shown in Fig. 2, and a summary of these experiments is presented in Table 1. A scheme derived from these results is illustrated in Fig. 5. Lanes 1-4 and of Fig. 2 show the phosphorylation of calmodulin and the HRAS protein, respectively, under different conditions. Of the four preincubation conditions used, that shown in lanes 1 and 11 appears to be best for phosphorylation of either substrate by the receptor kinase. Under this condition, substrate, receptor, and insulin were preincubated first, then poly(l-lysine) was added just before the phosphorylation reaction was initiated. Hence, we used 32p incorporation into either substrate under this condition as a Table 1. Comparison of phosphorylation of the HRAS protein, calmodulin, and the insulin receptor 1 kinase under various preincubation conditions subunit by purified insulin receptor Phosphorylation of Phosphorylation of 18 subunit in presence of Preincubation conditions Addition just substrate, % substrate, % prior to reaction Fig. 2 lanes Calmodulin HRAS Form in Fig. 5 Calmodulin HRAS BSA 1. Substrate + IR + Ins* 0 0 VI Substrate + IR + PLL 1 and VII Ins 3. Substrate + IR + None 2 and ± ± Ins + PLL (3) (4) 4. IR + Ins + PLL Substrate 3 and ± ± 7.7 III (3) (4) 5. Substrate + PLL, None 4 and ± ± 3.3 VIII IR + Ins (3) (3) Experimental conditions are described in the legend to Fig. 2. Substrates were wt HRAS or calmodulin. Ins, insulin; IR, insulin receptor; PLL, poly(l-lysine). Phosphorylation was determined by excision of the appropriate band from the gel followed by quantitation of incorporated radioactivity by Cerenkov counting. Numbers in parentheses represent numbers of experiments. *Phosphorylation results extrapolated from many identical experiments as shown in Fig. 1 (lanes 3 and 5) and Fig. 2 (lane 9).
4 Biochemistry: Fujita-Yamaguchi et al. CaM, i10% VI H-ras, 0% CaM, 0% I' /IV ~~~~~~~~~~~-ras, Proc. Natl. Acad. Sci. USA 86 (1989) 7309 ~~~~~~~~CaM, 34% dd TPK Domain I,m,sm,3m I'M IV V. *H- 32Pincorporation into the src-peptide H-ras, 56% CaM, 33% I' V \ Acidic (19 a.a.) H-ras protein or CaM Carboxyl terminal [ KKNGR ( )] Poly L-lysine FIG. 5. Speculative schematic of our present and previous studies on the insulin receptor kinase. The insulin kinase.8 subunit is shown in the upper center (form I) with the kinase domain illustrated as a "horseshoe" shape with its catalytic site on the bottom, and four alternative conformations of the catalytic site, implying different degrees of kinase activity, are shown on the lower right. The boxes indicate the transmembrane domain (TM) and the acidic A-III domain residues ( ), which is the potential site for the poly(l-lysine) interaction previously proposed (10). The carboxyl-terminal region downstream from the A-III domain is comprised of 57 amino acid residues. This region contains a short stretch of basic domain (residues ; Lys2-Asn-Gly-Arg), which is indicated as a "knot." Insulin receptor kinase inactivation occurs as illustrated by the sequence I -* IV -+ V (21). Poly(L-lysine) activates both intact (form I) and partially inactivated (form IV) kinases to give form II (10). Neither the HRAS protein nor calmodulin can be phosphorylated by the purified insulin receptor kinase alone; I -- VI (3, 9). When poly(l-lysine) is added to the HRAS (or calmodulin)/receptor kinase mixture, the HRAS protein (or calmodulin) is phosphorylated by the receptor kinase; VI -* VII (this study and ref. 9). When substrates are added to the receptor kinase-poly(l-lysine) complex, phosphorylation of the HRAS protein or calmodulin is greatly reduced; II -- III. When the substrates and poly(l-lysine) are preincubated first and the receptor kinase is added later, the degree of phosphorylation ofthe substrates is decreased as compared to the controls; I -. VIII (this study). control (Table 1, condition 2; 100%) (form VII in Fig. 5). In the case of HRAS protein phosphorylation, the standard condition (lane 12) gave results approximately equal to those of the control condition (Table 1, condition 3; 89.6 ± 16.1%). In contrast, calmodulin phosphorylation was significantly reduced under this condition (Fig. 2, lane 2, and Table 1, condition 3; 48.8 ± 19.4%). When the receptor, insulin, and poly(l-lysine) were preincubated first, and the substrates added just before initiation of the phosphorylation reaction, HRAS protein phosphorylation was significantly reduced (Fig. 2, lane 13, and Table 1, condition 4; 14.4 ± 7.7%) and calmodulin phosphorylation was also lower than that obtained under the standard condition (Fig. 2, lane 3, and Table 1, condition 4; 34.2 ± 7.7%) (form III in Fig. 5). When the substrates and poly(l-lysine) were preincubated together and the receptor and insulin were preincubated together, and these mixtures were combined just before initiation of the phosphorylation reaction, the degree of phosphorylation of calmodulin and the HRAS protein was significantly lower than controls (Fig. 2, lane 4, and Table 1, condition 5; %; Fig. 2, lane 14, and Table 1, condition 5; 55.6 ± 3.3%, respectively) (form VIII in Fig. 5). Fig. 2 also shows the effects of poly(l-lysine) and substrates on the insulin receptor itself. Phosphorylation of the /3 subunit appeared to be enhanced in the presence of calmodulin or the RAS protein (Figs. 1 and 2). The degree of,/-subunit phosphorylation under different preincubation conditions (Fig. 2) is summarized in Table 1. These results indicate that interaction between the insulin receptor kinase and these substrates increase the kinase activity. DISCUSSION We have shown that purified HRAS proteins are phosphorylated by purified insulin receptor kinase in the presence of poly(l-lysine), whereas the purified KRAS protein is phosphorylated by the receptor kinase without the addition of basic proteins. The difference observed between the two RAS proteins can most likely be attributed to the amino acid sequence differences in their carboxyl-terminal domains (1). The human KRAS protein carries an extremely basic domain at residues (Lys-Asp-Glu-Lys6-Ser-Arg), whereas this domain in the human HRAS and NRAS protein consists of primarily neutral amino acid residues. Our studies using purified insulin receptor and RAS proteins suggested the possibility that RAS proteins may be phosphorylated at tyrosine residues in intact cells, although to our knowledge, this has not been reported. In this study, we have examined the molecular interactions between insulin receptor kinase and potential substrates in vitro. There always seems to be a critical gap between studies using intact cells and those using broken cell preparations. In other words, even if two protein components are shown to interact in an intact cell system, it is unlikely that an identical in vitro system can be established simply by mixing the purified protein components. We know that at least two insulin-sensitive phosphorylation systems, the HRAS pro-
5 7310 Biochemistry: Fujita-Yamaguchi et al. Proc. Natl. Acad. Sci. USA 86 (1989) teins and calmodulin, have this characteristic. In both cases, phosphorylation of the proteins has been observed in an insulin-dependent manner in intact cells, but when the purified proteins or calmodulin were incubated with purified insulin receptor kinase, neither were phosphorylated (3, 9). Whether or not calmodulin is directly phosphorylated by insulin receptor kinase in intact cells requires further investigation, since although Colca et al. (8) have reported that calmodulin is phosphorylated in an insulin-dependent manner in intact rat adipocytes, Blackshear and Haupt (22) have recently provided evidence that calmodulin is not phosphorylated in 3T3-L1 adipocytes. This in vitro study, as well as previous studies (7, 9) suggests that a poly(l-lysine)-like basic protein or domain is required for establishment of the direct interaction between the insulin-responsive protein and the insulin receptor in broken cells or purified systems. Polylysine-containing peptides have been shown to affect membrane-associated enzymes (20). Gatica et al. (20) have searched for protein sequences containing long stretches of lysine residues and found that the human KRAS2 gene has six consecutive lysine residues. A peptide corresponding to the last 14 amino acids in the carboxyl terminus of the human KRAS2 gene product was synthesized, and it also stimulated in vitro membrane protein phosphorylation. We hypothesized that this peptide might be a naturally occurring polylysine-like peptide, acting as a cofactor for phosphorylation of the HRAS protein and calmodulin. We found that the KRAS peptide, like poly(l-lysine), is effective in mediating phosphorylation of both of these substrates by insulin receptor kinase. These studies suggest that the basic domains of naturally occurring proteins may play a role in establishing interactions between the receptor kinase and its substrates. We have proposed a model for the interactions among poly(l-lysine), insulin receptor kinase, and either substrate (Fig. 5). This model represents one of the several possibilities which will require further experiments to verify. Although we cannot exclude a possible allosteric regulation on kinase activity by poly(l-lysine), the results of the various preincubation-condition experiments (Table 1) would most likely fit the hypothesis described here. Our previous studies suggest that the conformation of the carboxyl-terminal domain of the receptor kinase becomes relaxed (form IV) during puriflication and storage and that, after this conformational change occurs, the carboxyl-terminal -20-amino acid domain is readily proteolyzed, resulting in form V (21, 23). The kinase activity of the relaxed form (IV) is 410% of that of the native form (I), and this form (IV) can no longer be activated by insulin (21). Our recent studies (10) have suggested the following. (i) The native kinase (form I) and the relaxed kinase (form IV) may both be activated by incubation with poly(l-lysine). We have proposed that poly(l-lysine) activates the kinase by interaction with three acidic domains of the insulin receptor, including residues (an acidic A-III domain) (10), to give an activated kinase (form II) that is much more active than the native form (I; 10- to 60-fold). (ii) The form (V) that has lost the carboxyl-terminal -20 amino acids is an inactive kinase and the A-III domain may be such a conformation that poly(l-lysine) is no longer able to restore the kinase activity. We have also shown in the present and in a previous study (9) that the purified insulin receptor kinase (form I) is not able to phosphorylate calmodulin or the RAS proteins as illustrated in form VI. However, after addition of poly(l-lysine), the RAS protein (or calmodulin) can become phosphorylated by the kinase as illustrated in form VII. Although the molecular mechanism underlying this result is not yet clear, one possibility is that poly(l-lysine) binds to the A-Ill domain and consequently changes the conformation of the substrates (form VII). Our previous study also showed that poly(llysine) binds directly to both the insulin receptor and calmodulin and that calmodulin changes its conformation by interaction with poly(l-lysine) as determined by circular dichroism spectra (9). The poly(l-lysine)-activated kinase (form II) alone does not phosphorylate either the HRAS protein or calmodulin very effectively (form III). This indicates that poly(l-lysine) may bind very closely to the kinase domain and that the protein substrate is not able to fit into the catalytic domain, whereas the src-related peptide can easily fit and therefore be phosphorylated (10). When substrates and poly(l-lysine) are incubated first and then mixed with the kinase (form VIII), the degree of phosphorylation of the HRAS protein is between those observed under the other two conditions (forms VII and III) whereas calmodulin phosphorylation is at nearly the same level as that shown in form III. This possibly reflects the structural difference between the two substrates. These results suggest that the substrate-poly(l-lysine) complex cannot fit as well into the kinase domain (form VIII) as compared to form VII. These studies have focused on a potential role of basic protein cofactors in mediating the interaction between the insulin receptor kinase and insulin-responsive protein substrates. The studies provide an in vitro system useful for identification of naturally occurring cofactors, as we have shown for the basic HRAS peptide. Furthermore, functional changes in the activities of tyrosine-phosphorylated RAS proteins might best be investigated using an experimental system of this type. We thank Smith Kline & French Co. for providing E. coli cells which carry human RAS gene and Dr. K. C. Glenn (Monsanto Co., Saint Louis, MO) for the gift of c-ki-ras peptide. We also thank Faith Sorensen for typing this manuscript. This work was supported by National Institutes of Health Grant DK29770 and by a grant from the Diabetes Research and Education Foundation. 1. Barbacid, M. (1987) Annu. Rev. Biochem. 56, Hurly, J. B., Simon, M. I., Treplow, D. B., Robishaw, J. D. & Gilman, A. G. (1984) Science 226, Kamata, T., Kathuria, S. & Fujita-Yamaguchi, Y. (1987) Biochem. Biophys. Res. Commun. 144, Korn, L. J., Siebel, C. W., McCormick, F. & Roth, R. (1987) Science 236, Deshpande, A. K. & Kung, H.-F. (1987) Mol. Cell. Biol. 7, O'Brien, R. M., Siddle, K., Houslay, M. D. & Hall, A. (1987) FEBS Lett. 217, Sacks, D. B. & McDonald, J. M. (1988) J. Biol. Chem. 263, Colca, J. R., DeWald, D. B., Pearson, J. D., Palazak, B. J., Laurino, J. P. & McDonald J. M. (1987) J. Biol. Chem. 262, Sacks, D. B., Fujita-Yamaguchi, Y., Gale, R. D. & McDonald, J. M. (1989) Biochem. J., in press. 10. Fujita-Yamaguchi, Y., Sacks, D. B., McDonald, J. M., Sahal, D. & Kathuria, S. (1989) Biochem. J., in press. 11. Casnellie, J. E., Harrison, M. L., Pike, L. J., Hellstrom, E. & Krebs, E. G. (1982) Proc. Natl. Acad. Sci. USA 79, Fujita-Yamaguchi, Y., Choi, S., Sakamoto, Y. & Itakura, K. (1983) J. Biol. Chem. 258, Kasuga, M., Fujita-Yamaguchi, Y., Blithe, D. L., White, M. R. & Kahn, C. R. (1983) J. Biol. Chem. 258, Fujita-Yamaguchi, Y. & Kathuria, S. (1985) Proc. Nati. Acad. Sci. USA 82, Gross, M., Sweet, R. W., Sathe, G., Yokoyama, S., Fasano, 0. M. & Rosenberg, M. (1985) Mol. Cell. Biol. 5, Hara, M., Tamaoki, T. & Nakano, H. (1988) Oncogene Res. 2, Laemmli, U. K. (1970) Nature (London) 227, Fujita-Yamaguchi, Y. (1984) J. Biol. Chem. 259, Ebina, Y., Ellis, L., Jarnagin, K., Edery, M., Graf, L., Clauser, E., Ou, J.-H., Masiarz, F., Kan, Y. W., Goldfine, I. D., Roth, R. A. & Rutter, W. J. (1985) Cell 40, Gatica, M., Allende, C. C., Antonelli, M. & Allende, J. E. (1987) Proc. Nati. Acad. Sci. USA 84, Kathuria, S., Hartman, S., Grunfeld, C., Ramachandran, J. & Fujita- Yamaguchi, Y. (1986) Proc. Nati. Acad. Sci. USA 83, Blackshear, P. J. & Haupt, D. M. (1989) J. Biol. Chem. 264, Grunfeld, C., Shigenaga, J. K., Fujita-Yamaguchi, Y., McFarland, K. C., Burnier, J. & Ramachandran, J. (1987) Endocrinology 121,
Saccharomyces cerevisiae*
 THE JOURNAL OF BIOLOGICAL CHEMISTRY 1988 by The American Society for Biochemistry and Molecular Biology, Inc. Vol. 263, No. 29, Issue of October 15, pp. 14948-14955, 1988 Printed in U.S.A. Purification
THE JOURNAL OF BIOLOGICAL CHEMISTRY 1988 by The American Society for Biochemistry and Molecular Biology, Inc. Vol. 263, No. 29, Issue of October 15, pp. 14948-14955, 1988 Printed in U.S.A. Purification
Role of insulin receptor phosphorylation in the insulinomimetic effects of hydrogen peroxide
 Proc. Nadl. Acad. Sci. USA Vol. 84, pp. 8115-8119, November 1987 Medical Sciences Role of insulin receptor phosphorylation in the insulinomimetic effects of hydrogen peroxide (insulin action/hexose transport/tyrosine
Proc. Nadl. Acad. Sci. USA Vol. 84, pp. 8115-8119, November 1987 Medical Sciences Role of insulin receptor phosphorylation in the insulinomimetic effects of hydrogen peroxide (insulin action/hexose transport/tyrosine
Amino acids. Side chain. -Carbon atom. Carboxyl group. Amino group
 PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
Previous Class. Today. Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism. Protein Kinase Catalytic Properties
 Previous Class Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism Today Protein Kinase Catalytic Properties Protein Phosphorylation Phosphorylation: key protein modification
Previous Class Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism Today Protein Kinase Catalytic Properties Protein Phosphorylation Phosphorylation: key protein modification
CS612 - Algorithms in Bioinformatics
 Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular
 Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Copyright 2008 Pearson Education, Inc., publishing as Pearson Benjamin Cummings
 Concept 5.4: Proteins have many structures, resulting in a wide range of functions Proteins account for more than 50% of the dry mass of most cells Protein functions include structural support, storage,
Concept 5.4: Proteins have many structures, resulting in a wide range of functions Proteins account for more than 50% of the dry mass of most cells Protein functions include structural support, storage,
Page 8/6: The cell. Where to start: Proteins (control a cell) (start/end products)
 Page 8/6: The cell Where to start: Proteins (control a cell) (start/end products) Page 11/10: Structural hierarchy Proteins Phenotype of organism 3 Dimensional structure Function by interaction THE PROTEIN
Page 8/6: The cell Where to start: Proteins (control a cell) (start/end products) Page 11/10: Structural hierarchy Proteins Phenotype of organism 3 Dimensional structure Function by interaction THE PROTEIN
Chemistry 121 Winter 17
 Chemistry 121 Winter 17 Introduction to Organic Chemistry and Biochemistry Instructor Dr. Upali Siriwardane (Ph.D. Ohio State) E-mail: upali@latech.edu Office: 311 Carson Taylor Hall ; Phone: 318-257-4941;
Chemistry 121 Winter 17 Introduction to Organic Chemistry and Biochemistry Instructor Dr. Upali Siriwardane (Ph.D. Ohio State) E-mail: upali@latech.edu Office: 311 Carson Taylor Hall ; Phone: 318-257-4941;
Multiple-Choice Questions Answer ALL 20 multiple-choice questions on the Scantron Card in PENCIL
 Multiple-Choice Questions Answer ALL 20 multiple-choice questions on the Scantron Card in PENCIL For Questions 1-10 choose ONE INCORRECT answer. 1. Which ONE of the following statements concerning the
Multiple-Choice Questions Answer ALL 20 multiple-choice questions on the Scantron Card in PENCIL For Questions 1-10 choose ONE INCORRECT answer. 1. Which ONE of the following statements concerning the
AA s are the building blocks of proteins
 Chamras Chemistry 106 Lecture otes Chapter 24: Amino Acids, Peptides, and Proteins General Formula: () n (') α-amino Acids: (n = 1) Example: Amino Acids and Proteins: Glycine Alanine Valine AA s are the
Chamras Chemistry 106 Lecture otes Chapter 24: Amino Acids, Peptides, and Proteins General Formula: () n (') α-amino Acids: (n = 1) Example: Amino Acids and Proteins: Glycine Alanine Valine AA s are the
Lecture 4. Grouping Amino Acid 7/1/10. Proteins. Amino Acids. Where Are Proteins Located. Nonpolar Amino Acids
 Proteins Lecture 4 Proteins - Composition of Proteins (Amino Acids) Chapter 21 ection 1-6! Proteins are compounds of high molar mass consisting almost entirely of amino acid chain(s)! Molar masses range
Proteins Lecture 4 Proteins - Composition of Proteins (Amino Acids) Chapter 21 ection 1-6! Proteins are compounds of high molar mass consisting almost entirely of amino acid chain(s)! Molar masses range
AP Bio. Protiens Chapter 5 1
 Concept.4: Proteins have many structures, resulting in a wide range of functions Proteins account for more than 0% of the dry mass of most cells Protein functions include structural support, storage, transport,
Concept.4: Proteins have many structures, resulting in a wide range of functions Proteins account for more than 0% of the dry mass of most cells Protein functions include structural support, storage, transport,
LAB#23: Biochemical Evidence of Evolution Name: Period Date :
 LAB#23: Biochemical Evidence of Name: Period Date : Laboratory Experience #23 Bridge Worth 80 Lab Minutes If two organisms have similar portions of DNA (genes), these organisms will probably make similar
LAB#23: Biochemical Evidence of Name: Period Date : Laboratory Experience #23 Bridge Worth 80 Lab Minutes If two organisms have similar portions of DNA (genes), these organisms will probably make similar
BIOCHEMISTRY REVIEW. Overview of Biomolecules. Chapter 4 Protein Sequence
 BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 4 Protein Sequence 2 3 4 Are You Getting It?? A molecule of hemoglobin is compared with a molecule of lysozyme. Which characteristics do they share?
BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 4 Protein Sequence 2 3 4 Are You Getting It?? A molecule of hemoglobin is compared with a molecule of lysozyme. Which characteristics do they share?
Biomolecules: amino acids
 Biomolecules: amino acids Amino acids Amino acids are the building blocks of proteins They are also part of hormones, neurotransmitters and metabolic intermediates There are 20 different amino acids in
Biomolecules: amino acids Amino acids Amino acids are the building blocks of proteins They are also part of hormones, neurotransmitters and metabolic intermediates There are 20 different amino acids in
MEK1 Assay Kit 1 Catalog # Lot # 16875
 MEK1 Assay Kit 1 Kit Components Assay Dilution Buffer (ADB), Catalog # 20-108. Three vials, each containing 1.0ml of assay dilution buffer (20mM MOPS, ph 7.2, 25mM ß-glycerol phosphate, 5mM EGTA, 1mM sodium
MEK1 Assay Kit 1 Kit Components Assay Dilution Buffer (ADB), Catalog # 20-108. Three vials, each containing 1.0ml of assay dilution buffer (20mM MOPS, ph 7.2, 25mM ß-glycerol phosphate, 5mM EGTA, 1mM sodium
Effects of Second Messengers
 Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth 2 points.
 MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
CHAPTER 21: Amino Acids, Proteins, & Enzymes. General, Organic, & Biological Chemistry Janice Gorzynski Smith
 CHAPTER 21: Amino Acids, Proteins, & Enzymes General, Organic, & Biological Chemistry Janice Gorzynski Smith CHAPTER 21: Amino Acids, Proteins, Enzymes Learning Objectives: q The 20 common, naturally occurring
CHAPTER 21: Amino Acids, Proteins, & Enzymes General, Organic, & Biological Chemistry Janice Gorzynski Smith CHAPTER 21: Amino Acids, Proteins, Enzymes Learning Objectives: q The 20 common, naturally occurring
Proteins are sometimes only produced in one cell type or cell compartment (brain has 15,000 expressed proteins, gut has 2,000).
 Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Objective: You will be able to explain how the subcomponents of
 Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Electronic Supplementary Information. Table of Contents
 Electronic Supplementary Information Examination of native chemical ligation using peptidyl prolyl thioester Takahiro Nakamura, Akira Shigenaga, Kohei Sato, Yusuke Tsuda, Ken Sakamoto, and Akira Otaka*
Electronic Supplementary Information Examination of native chemical ligation using peptidyl prolyl thioester Takahiro Nakamura, Akira Shigenaga, Kohei Sato, Yusuke Tsuda, Ken Sakamoto, and Akira Otaka*
Introduction to Peptide Sequencing
 Introduction to Peptide equencing Quadrupole Ion Traps tructural Biophysics Course December 3, 2014 12/8/14 Introduction to Peptide equencing - athan Yates 1 Why are ion traps used to sequence peptides?
Introduction to Peptide equencing Quadrupole Ion Traps tructural Biophysics Course December 3, 2014 12/8/14 Introduction to Peptide equencing - athan Yates 1 Why are ion traps used to sequence peptides?
Amino Acids. Review I: Protein Structure. Amino Acids: Structures. Amino Acids (contd.) Rajan Munshi
 Review I: Protein Structure Rajan Munshi BBSI @ Pitt 2005 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2005 Amino Acids Building blocks of proteins 20 amino acids
Review I: Protein Structure Rajan Munshi BBSI @ Pitt 2005 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2005 Amino Acids Building blocks of proteins 20 amino acids
Molecular Biology. general transfer: occurs normally in cells. special transfer: occurs only in the laboratory in specific conditions.
 Chapter 9: Proteins Molecular Biology replication general transfer: occurs normally in cells transcription special transfer: occurs only in the laboratory in specific conditions translation unknown transfer:
Chapter 9: Proteins Molecular Biology replication general transfer: occurs normally in cells transcription special transfer: occurs only in the laboratory in specific conditions translation unknown transfer:
HPLC '88. Poster Presentation. Isolation of Thymosin B4 from Thymosin Fraction 5 by Reverse Phase HPLC
 Essentials in HPLC '88 Poster Presentation Isolation of Thymosin B4 from Thymosin Fraction 5 by Reverse Phase HPLC M. Badamchian, M.P. Strickler, M.J. Stone, A.L. Goldstein for Waters.bioresearchThe absolute,
Essentials in HPLC '88 Poster Presentation Isolation of Thymosin B4 from Thymosin Fraction 5 by Reverse Phase HPLC M. Badamchian, M.P. Strickler, M.J. Stone, A.L. Goldstein for Waters.bioresearchThe absolute,
Enzymes Part III: regulation II. Dr. Mamoun Ahram Summer, 2017
 Enzymes Part III: regulation II Dr. Mamoun Ahram Summer, 2017 Advantage This is a major mechanism for rapid and transient regulation of enzyme activity. A most common mechanism is enzyme phosphorylation
Enzymes Part III: regulation II Dr. Mamoun Ahram Summer, 2017 Advantage This is a major mechanism for rapid and transient regulation of enzyme activity. A most common mechanism is enzyme phosphorylation
Properties of amino acids in proteins
 Properties of amino acids in proteins one of the primary roles of DNA (but far from the only one!!!) is to code for proteins A typical bacterium builds thousands types of proteins, all from ~20 amino acids
Properties of amino acids in proteins one of the primary roles of DNA (but far from the only one!!!) is to code for proteins A typical bacterium builds thousands types of proteins, all from ~20 amino acids
2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry
 Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
The Structure and Function of Large Biological Molecules Part 4: Proteins Chapter 5
 Key Concepts: The Structure and Function of Large Biological Molecules Part 4: Proteins Chapter 5 Proteins include a diversity of structures, resulting in a wide range of functions Proteins Enzymatic s
Key Concepts: The Structure and Function of Large Biological Molecules Part 4: Proteins Chapter 5 Proteins include a diversity of structures, resulting in a wide range of functions Proteins Enzymatic s
Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A
 Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A Homework Watch the Bozeman video called, Biological Molecules Objective:
Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A Homework Watch the Bozeman video called, Biological Molecules Objective:
Short polymer. Dehydration removes a water molecule, forming a new bond. Longer polymer (a) Dehydration reaction in the synthesis of a polymer
 HO 1 2 3 H HO H Short polymer Dehydration removes a water molecule, forming a new bond Unlinked monomer H 2 O HO 1 2 3 4 H Longer polymer (a) Dehydration reaction in the synthesis of a polymer HO 1 2 3
HO 1 2 3 H HO H Short polymer Dehydration removes a water molecule, forming a new bond Unlinked monomer H 2 O HO 1 2 3 4 H Longer polymer (a) Dehydration reaction in the synthesis of a polymer HO 1 2 3
The rabbit femoral artery was prepared and each arterial ring was permeabilized
 Online Supplement Nakmura et al. cgmp-dependent relaxation of smooth muscle Materials and Methods Measurement of tension The rabbit femoral artery was prepared and each arterial ring was permeabilized
Online Supplement Nakmura et al. cgmp-dependent relaxation of smooth muscle Materials and Methods Measurement of tension The rabbit femoral artery was prepared and each arterial ring was permeabilized
BIO 311C Spring Lecture 15 Friday 26 Feb. 1
 BIO 311C Spring 2010 Lecture 15 Friday 26 Feb. 1 Illustration of a Polypeptide amino acids peptide bonds Review Polypeptide (chain) See textbook, Fig 5.21, p. 82 for a more clear illustration Folding and
BIO 311C Spring 2010 Lecture 15 Friday 26 Feb. 1 Illustration of a Polypeptide amino acids peptide bonds Review Polypeptide (chain) See textbook, Fig 5.21, p. 82 for a more clear illustration Folding and
2. Which of the following amino acids is most likely to be found on the outer surface of a properly folded protein?
 Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Self-association of α-chymotrypsin: Effect of amino acids
 J. Biosci., Vol. 13, Number 3, September 1988, pp. 215 222. Printed in India. Self-association of α-chymotrypsin: Effect of amino acids T. RAMAKRISHNA and M. W. PANDIT* Centre for Cellular and Molecular
J. Biosci., Vol. 13, Number 3, September 1988, pp. 215 222. Printed in India. Self-association of α-chymotrypsin: Effect of amino acids T. RAMAKRISHNA and M. W. PANDIT* Centre for Cellular and Molecular
Amino acids-incorporated nanoflowers with an
 Amino acids-incorporated nanoflowers with an intrinsic peroxidase-like activity Zhuo-Fu Wu 1,2,+, Zhi Wang 1,+, Ye Zhang 3, Ya-Li Ma 3, Cheng-Yan He 4, Heng Li 1, Lei Chen 1, Qi-Sheng Huo 3, Lei Wang 1,*
Amino acids-incorporated nanoflowers with an intrinsic peroxidase-like activity Zhuo-Fu Wu 1,2,+, Zhi Wang 1,+, Ye Zhang 3, Ya-Li Ma 3, Cheng-Yan He 4, Heng Li 1, Lei Chen 1, Qi-Sheng Huo 3, Lei Wang 1,*
Ionization of amino acids
 Amino Acids 20 common amino acids there are others found naturally but much less frequently Common structure for amino acid COOH, -NH 2, H and R functional groups all attached to the a carbon Ionization
Amino Acids 20 common amino acids there are others found naturally but much less frequently Common structure for amino acid COOH, -NH 2, H and R functional groups all attached to the a carbon Ionization
Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels. Supplementary Information
 Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels Craig T. Armstrong, Philip E. Mason, J. L. Ross Anderson and Christopher E. Dempsey
Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels Craig T. Armstrong, Philip E. Mason, J. L. Ross Anderson and Christopher E. Dempsey
Methionine (Met or M)
 Fig. 5-17 Nonpolar Fig. 5-17a Nonpolar Glycine (Gly or G) Alanine (Ala or A) Valine (Val or V) Leucine (Leu or L) Isoleucine (Ile or I) Methionine (Met or M) Phenylalanine (Phe or F) Polar Trypotphan (Trp
Fig. 5-17 Nonpolar Fig. 5-17a Nonpolar Glycine (Gly or G) Alanine (Ala or A) Valine (Val or V) Leucine (Leu or L) Isoleucine (Ile or I) Methionine (Met or M) Phenylalanine (Phe or F) Polar Trypotphan (Trp
1. Describe the relationship of dietary protein and the health of major body systems.
 Food Explorations Lab I: The Building Blocks STUDENT LAB INVESTIGATIONS Name: Lab Overview In this investigation, you will be constructing animal and plant proteins using beads to represent the amino acids.
Food Explorations Lab I: The Building Blocks STUDENT LAB INVESTIGATIONS Name: Lab Overview In this investigation, you will be constructing animal and plant proteins using beads to represent the amino acids.
Review II: The Molecules of Life
 Review II: The Molecules of Life Judy Wieber BBSI @ Pitt 2007 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2007 Outline Introduction Proteins Carbohydrates Lipids
Review II: The Molecules of Life Judy Wieber BBSI @ Pitt 2007 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2007 Outline Introduction Proteins Carbohydrates Lipids
Amino Acids. Amino Acids. Fundamentals. While their name implies that amino acids are compounds that contain an NH. 3 and CO NH 3
 Fundamentals While their name implies that amino acids are compounds that contain an 2 group and a 2 group, these groups are actually present as 3 and 2 respectively. They are classified as α, β, γ, etc..
Fundamentals While their name implies that amino acids are compounds that contain an 2 group and a 2 group, these groups are actually present as 3 and 2 respectively. They are classified as α, β, γ, etc..
Chemical Mechanism of Enzymes
 Chemical Mechanism of Enzymes Enzyme Engineering 5.2 Definition of the mechanism 1. The sequence from substrate(s) to product(s) : Reaction steps 2. The rates at which the complex are interconverted 3.
Chemical Mechanism of Enzymes Enzyme Engineering 5.2 Definition of the mechanism 1. The sequence from substrate(s) to product(s) : Reaction steps 2. The rates at which the complex are interconverted 3.
Chemical Nature of the Amino Acids. Table of a-amino Acids Found in Proteins
 Chemical Nature of the Amino Acids All peptides and polypeptides are polymers of alpha-amino acids. There are 20 a- amino acids that are relevant to the make-up of mammalian proteins (see below). Several
Chemical Nature of the Amino Acids All peptides and polypeptides are polymers of alpha-amino acids. There are 20 a- amino acids that are relevant to the make-up of mammalian proteins (see below). Several
Practice Problems 3. a. What is the name of the bond formed between two amino acids? Are these bonds free to rotate?
 Life Sciences 1a Practice Problems 3 1. Draw the oligopeptide for Ala-Phe-Gly-Thr-Asp. You do not need to indicate the stereochemistry of the sidechains. Denote with arrows the bonds formed between the
Life Sciences 1a Practice Problems 3 1. Draw the oligopeptide for Ala-Phe-Gly-Thr-Asp. You do not need to indicate the stereochemistry of the sidechains. Denote with arrows the bonds formed between the
BIO th September, 1997
 BIO 451 26th September, 1997 EXAM I This exam will be taken apart for grading. Please PRINT your name on each page. If you do not have sufficient room for your answer in the space provided, please continue
BIO 451 26th September, 1997 EXAM I This exam will be taken apart for grading. Please PRINT your name on each page. If you do not have sufficient room for your answer in the space provided, please continue
1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled
 Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Lipids: diverse group of hydrophobic molecules
 Lipids: diverse group of hydrophobic molecules Lipids only macromolecules that do not form polymers li3le or no affinity for water hydrophobic consist mostly of hydrocarbons nonpolar covalent bonds fats
Lipids: diverse group of hydrophobic molecules Lipids only macromolecules that do not form polymers li3le or no affinity for water hydrophobic consist mostly of hydrocarbons nonpolar covalent bonds fats
Four Classes of Biological Macromolecules. Biological Macromolecules. Lipids
 Biological Macromolecules Much larger than other par4cles found in cells Made up of smaller subunits Found in all cells Great diversity of func4ons Four Classes of Biological Macromolecules Lipids Polysaccharides
Biological Macromolecules Much larger than other par4cles found in cells Made up of smaller subunits Found in all cells Great diversity of func4ons Four Classes of Biological Macromolecules Lipids Polysaccharides
PROTEINS. Building blocks, structure and function. Aim: You will have a clear picture of protein construction and their general properties
 PROTEINS Building blocks, structure and function Aim: You will have a clear picture of protein construction and their general properties Reading materials: Compendium in Biochemistry, page 13-49. Microbiology,
PROTEINS Building blocks, structure and function Aim: You will have a clear picture of protein construction and their general properties Reading materials: Compendium in Biochemistry, page 13-49. Microbiology,
In this study an antiphosphotyrosine antibody prepared as. that become phosphorylated in an insulin-dependent way.
 Proc. Nati. Acad. Sci. USA Vol. 84, pp. 113-117, January 1987 Cell Biology Insulin rapidly stimulates phosphorylation of a 46-kDa membrane protein on tyrosine residues as well as phosphorylation of several
Proc. Nati. Acad. Sci. USA Vol. 84, pp. 113-117, January 1987 Cell Biology Insulin rapidly stimulates phosphorylation of a 46-kDa membrane protein on tyrosine residues as well as phosphorylation of several
Molecular Medicine: Gleevec and Chronic Myelogenous Leukemia
 Molecular Medicine: Gleevec and Chronic Myelogenous Leukemia Dec 14 & 19, 2006 Prof. Erin Shea Prof. Dan Kahne Cancer, Kinases and Gleevec: 1. What is CML? a. Blood cell maturation b. Philadelphia Chromosome
Molecular Medicine: Gleevec and Chronic Myelogenous Leukemia Dec 14 & 19, 2006 Prof. Erin Shea Prof. Dan Kahne Cancer, Kinases and Gleevec: 1. What is CML? a. Blood cell maturation b. Philadelphia Chromosome
9/16/15. Properties of Water. Benefits of Water. More properties of water
 Properties of Water Solid/Liquid Density Water is densest at 4⁰C Ice floats Allows life under the ice Hydrogen bond Ice Hydrogen bonds are stable Liquid water Hydrogen bonds break and re-form Benefits
Properties of Water Solid/Liquid Density Water is densest at 4⁰C Ice floats Allows life under the ice Hydrogen bond Ice Hydrogen bonds are stable Liquid water Hydrogen bonds break and re-form Benefits
Signal-Transduction Cascades - 2. The Phosphoinositide Cascade
 Signal-Transduction Cascades - 2 The Phosphoinositide Cascade Calcium ion as a second messenger Tyrosine kinase and receptor dimerization scribd.com Faisal Khatib JU The Phosphoinositide Cascade Used by
Signal-Transduction Cascades - 2 The Phosphoinositide Cascade Calcium ion as a second messenger Tyrosine kinase and receptor dimerization scribd.com Faisal Khatib JU The Phosphoinositide Cascade Used by
Macromolecules of Life -3 Amino Acids & Proteins
 Macromolecules of Life -3 Amino Acids & Proteins Shu-Ping Lin, Ph.D. Institute of Biomedical Engineering E-mail: splin@dragon.nchu.edu.tw Website: http://web.nchu.edu.tw/pweb/users/splin/ Amino Acids Proteins
Macromolecules of Life -3 Amino Acids & Proteins Shu-Ping Lin, Ph.D. Institute of Biomedical Engineering E-mail: splin@dragon.nchu.edu.tw Website: http://web.nchu.edu.tw/pweb/users/splin/ Amino Acids Proteins
Receptor mediated Signal Transduction
 Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Classification of amino acids: -
 Page 1 of 8 P roteinogenic amino acids, also known as standard, normal or primary amino acids are 20 amino acids that are incorporated in proteins and that are coded in the standard genetic code (subunit
Page 1 of 8 P roteinogenic amino acids, also known as standard, normal or primary amino acids are 20 amino acids that are incorporated in proteins and that are coded in the standard genetic code (subunit
Steps at which eukaryotic gene expression can be controlled. Cell 7.5
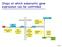 Steps at which eukaryotic gene expression can be controlled Cell 7.5 Protein Variability and Protein Activity Control Aminoacid sequence Three-dimensional shape (conformation) Function Protein processing
Steps at which eukaryotic gene expression can be controlled Cell 7.5 Protein Variability and Protein Activity Control Aminoacid sequence Three-dimensional shape (conformation) Function Protein processing
Chapter 23 Enzymes 1
 Chapter 23 Enzymes 1 Enzymes Ribbon diagram of cytochrome c oxidase, the enzyme that directly uses oxygen during respiration. 2 Enzyme Catalysis Enzyme: A biological catalyst. With the exception of some
Chapter 23 Enzymes 1 Enzymes Ribbon diagram of cytochrome c oxidase, the enzyme that directly uses oxygen during respiration. 2 Enzyme Catalysis Enzyme: A biological catalyst. With the exception of some
Judy Wieber. Department of Computational Biology. May 27, 2008
 Review II: The Molecules of Life Judy Wieber BBSI @ Pitt 2008 Department of Computational Biology University it of Pittsburgh School of Medicine i May 27, 2008 Outline Introduction Proteins Carbohydrates
Review II: The Molecules of Life Judy Wieber BBSI @ Pitt 2008 Department of Computational Biology University it of Pittsburgh School of Medicine i May 27, 2008 Outline Introduction Proteins Carbohydrates
ras proteins enhance the phosphorylation of a 38 kda protein (~38) in rat liver plasma membrane
 Volume 217, number 1, 74-80 FEB 04768 June 1987 ras proteins enhance the phosphorylation of a 38 kda protein (~38) in rat liver plasma membrane Ashok N. Hegde and M.R. Das Centre for Cellular & Molecular
Volume 217, number 1, 74-80 FEB 04768 June 1987 ras proteins enhance the phosphorylation of a 38 kda protein (~38) in rat liver plasma membrane Ashok N. Hegde and M.R. Das Centre for Cellular & Molecular
Four melanocyte-stimulating hormones have the following amino acid sequences:
 Assignment 14: Melanocyte-stimulating hormone belongs to a group called the melanocortins. This group includes ACTH, alpha-msh, beta-msh and gamma-msh; these peptides are all cleavage products of a large
Assignment 14: Melanocyte-stimulating hormone belongs to a group called the melanocortins. This group includes ACTH, alpha-msh, beta-msh and gamma-msh; these peptides are all cleavage products of a large
PROTEINS. Amino acids are the building blocks of proteins. Acid L-form * * Lecture 6 Macromolecules #2 O = N -C -C-O.
 Proteins: Linear polymers of amino acids workhorses of the cell tools, machines & scaffolds Lecture 6 Macromolecules #2 PRTEINS 1 Enzymes catalysts that mediate reactions, increase reaction rate Structural
Proteins: Linear polymers of amino acids workhorses of the cell tools, machines & scaffolds Lecture 6 Macromolecules #2 PRTEINS 1 Enzymes catalysts that mediate reactions, increase reaction rate Structural
G-Protein Signaling. Introduction to intracellular signaling. Dr. SARRAY Sameh, Ph.D
 G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
Point total. Page # Exam Total (out of 90) The number next to each intermediate represents the total # of C-C and C-H bonds in that molecule.
 This exam is worth 90 points. Pages 2- have questions. Page 1 is for your reference only. Honor Code Agreement - Signature: Date: (You agree to not accept or provide assistance to anyone else during this
This exam is worth 90 points. Pages 2- have questions. Page 1 is for your reference only. Honor Code Agreement - Signature: Date: (You agree to not accept or provide assistance to anyone else during this
The Basics: A general review of molecular biology:
 The Basics: A general review of molecular biology: DNA Transcription RNA Translation Proteins DNA (deoxy-ribonucleic acid) is the genetic material It is an informational super polymer -think of it as the
The Basics: A general review of molecular biology: DNA Transcription RNA Translation Proteins DNA (deoxy-ribonucleic acid) is the genetic material It is an informational super polymer -think of it as the
The elements of G protein-coupled receptor systems
 The elements of G protein-coupled receptor systems Prostaglandines Sphingosine 1-phosphate a receptor that contains 7 membrane-spanning domains a coupled trimeric G protein which functions as a switch
The elements of G protein-coupled receptor systems Prostaglandines Sphingosine 1-phosphate a receptor that contains 7 membrane-spanning domains a coupled trimeric G protein which functions as a switch
Amino acids & Protein Structure Chemwiki: Chapter , with most emphasis on 16.3, 16.4 and 16.6
 Amino acids & Protein Structure Chemwiki: Chapter 16. 16.1, 16.3-16.9 with most emphasis on 16.3, 16.4 and 16.6 1 1. Most jobs (except information storage) in cells are performed by proteins. 2. Proteins
Amino acids & Protein Structure Chemwiki: Chapter 16. 16.1, 16.3-16.9 with most emphasis on 16.3, 16.4 and 16.6 1 1. Most jobs (except information storage) in cells are performed by proteins. 2. Proteins
Biomolecules Amino Acids & Protein Chemistry
 Biochemistry Department Date: 17/9/ 2017 Biomolecules Amino Acids & Protein Chemistry Prof.Dr./ FAYDA Elazazy Professor of Biochemistry and Molecular Biology Intended Learning Outcomes ILOs By the end
Biochemistry Department Date: 17/9/ 2017 Biomolecules Amino Acids & Protein Chemistry Prof.Dr./ FAYDA Elazazy Professor of Biochemistry and Molecular Biology Intended Learning Outcomes ILOs By the end
9/6/2011. Amino Acids. C α. Nonpolar, aliphatic R groups
 Amino Acids Side chains (R groups) vary in: size shape charge hydrogen-bonding capacity hydrophobic character chemical reactivity C α Nonpolar, aliphatic R groups Glycine (Gly, G) Alanine (Ala, A) Valine
Amino Acids Side chains (R groups) vary in: size shape charge hydrogen-bonding capacity hydrophobic character chemical reactivity C α Nonpolar, aliphatic R groups Glycine (Gly, G) Alanine (Ala, A) Valine
Chemistry of Insulin. Based on Chemistry of Insulin by F. Sanger, Science 129, (1959)
 Chemistry of Insulin Based on Chemistry of Insulin by F. Sanger, Science 129, 1340-1344 (1959) Human Insulin The Sanger Method The DNP- amino acids are bright yellow substances and can be separated from
Chemistry of Insulin Based on Chemistry of Insulin by F. Sanger, Science 129, 1340-1344 (1959) Human Insulin The Sanger Method The DNP- amino acids are bright yellow substances and can be separated from
reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced to express
 Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
Quantitative LC-MS/MS Analysis of Glucagon. Veniamin Lapko, Ph.D June 21, 2011
 Quantitative LC-MS/MS Analysis of Glucagon Veniamin Lapko, Ph.D June 21, 2011 Contents Comparison with small molecule LC-MS/MS LC-MS/MS sensitivity of peptides detection Stability: neat vs. matrix solutions
Quantitative LC-MS/MS Analysis of Glucagon Veniamin Lapko, Ph.D June 21, 2011 Contents Comparison with small molecule LC-MS/MS LC-MS/MS sensitivity of peptides detection Stability: neat vs. matrix solutions
CHAPTER 10: REGULATORY STRATEGIES. Traffic signals control the flow of traffic
 CHAPTER 10: REGULATORY STRATEGIES Traffic signals control the flow of traffic INTRODUCTION CHAPTER 10 The activity of enzymes must often be regulated so that they function at the proper time and place.
CHAPTER 10: REGULATORY STRATEGIES Traffic signals control the flow of traffic INTRODUCTION CHAPTER 10 The activity of enzymes must often be regulated so that they function at the proper time and place.
Midterm 1 Last, First
 Midterm 1 BIS 105 Prof. T. Murphy April 23, 2014 There should be 6 pages in this exam. Exam instructions (1) Please write your name on the top of every page of the exam (2) Show all work for full credit
Midterm 1 BIS 105 Prof. T. Murphy April 23, 2014 There should be 6 pages in this exam. Exam instructions (1) Please write your name on the top of every page of the exam (2) Show all work for full credit
Lecture 19: Review of regulation
 Chem*3560 Lecture 19: Review of regulation What is meant by cooperative allosteric regulation? Positive cooperativity - characteristic is the sigmoidal binding/activity curve T-state has weaker affinity,
Chem*3560 Lecture 19: Review of regulation What is meant by cooperative allosteric regulation? Positive cooperativity - characteristic is the sigmoidal binding/activity curve T-state has weaker affinity,
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Cahn - Ingold - Prelog system. Proteins: Evolution, and Analysis Lecture 7 9/15/2009. The Fischer Convention (1) G (2) (3)
 Chapter 4 (1) G Proteins: Evolution, and Analysis Lecture 7 9/15/2009 A V L I M P F W Chapter 4 (2) S (3) T N Q Y C K R H D E The Fischer Convention Absolute configuration about an asymmetric carbon related
Chapter 4 (1) G Proteins: Evolution, and Analysis Lecture 7 9/15/2009 A V L I M P F W Chapter 4 (2) S (3) T N Q Y C K R H D E The Fischer Convention Absolute configuration about an asymmetric carbon related
Molecular Medicine: Gleevec and Chronic Myelogenous Leukemia. Dec 14 & 19, 2006 Prof. Erin O Shea Prof. Dan Kahne
 Molecular Medicine: Gleevec and Chronic Myelogenous Leukemia Dec 14 & 19, 2006 Prof. Erin Shea Prof. Dan Kahne 1 Cancer, Kinases and Gleevec: 1. What is CML? a. Blood cell maturation b. Philadelphia Chromosome
Molecular Medicine: Gleevec and Chronic Myelogenous Leukemia Dec 14 & 19, 2006 Prof. Erin Shea Prof. Dan Kahne 1 Cancer, Kinases and Gleevec: 1. What is CML? a. Blood cell maturation b. Philadelphia Chromosome
Chapter 3: Amino Acids and Peptides
 Chapter 3: Amino Acids and Peptides BINF 6101/8101, Spring 2018 Outline 1. Overall amino acid structure 2. Amino acid stereochemistry 3. Amino acid sidechain structure & classification 4. Non-standard
Chapter 3: Amino Acids and Peptides BINF 6101/8101, Spring 2018 Outline 1. Overall amino acid structure 2. Amino acid stereochemistry 3. Amino acid sidechain structure & classification 4. Non-standard
1. (38 pts.) 2. (25 pts.) 3. (15 pts.) 4. (12 pts.) 5. (10 pts.) Bonus (12 pts.) TOTAL (100 points)
 Moorpark College Chemistry 11 Spring 2010 Instructor: Professor Torres Examination #5: Section Five May 4, 2010 ame: (print) ame: (sign) Directions: Make sure your examination contains TWELVE total pages
Moorpark College Chemistry 11 Spring 2010 Instructor: Professor Torres Examination #5: Section Five May 4, 2010 ame: (print) ame: (sign) Directions: Make sure your examination contains TWELVE total pages
Towards a New Paradigm in Scientific Notation Patterns of Periodicity among Proteinogenic Amino Acids [Abridged Version]
![Towards a New Paradigm in Scientific Notation Patterns of Periodicity among Proteinogenic Amino Acids [Abridged Version] Towards a New Paradigm in Scientific Notation Patterns of Periodicity among Proteinogenic Amino Acids [Abridged Version]](/thumbs/84/90818314.jpg) Earth/matriX: SCIENCE TODAY Towards a New Paradigm in Scientific Notation Patterns of Periodicity among Proteinogenic Amino Acids [Abridged Version] By Charles William Johnson Earth/matriX Editions P.O.
Earth/matriX: SCIENCE TODAY Towards a New Paradigm in Scientific Notation Patterns of Periodicity among Proteinogenic Amino Acids [Abridged Version] By Charles William Johnson Earth/matriX Editions P.O.
Biochemistry 2 Recita0on Amino Acid Metabolism
 Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Chapter 3. Protein Structure and Function
 Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
Cells N5 Homework book
 1 Cells N5 Homework book 2 Homework 1 3 4 5 Homework2 Cell Ultrastructure and Membrane 1. Name and give the function of the numbered organelles in the cell below: A E B D C 2. Name 3 structures you might
1 Cells N5 Homework book 2 Homework 1 3 4 5 Homework2 Cell Ultrastructure and Membrane 1. Name and give the function of the numbered organelles in the cell below: A E B D C 2. Name 3 structures you might
Wheat Amino acids & Peptides for Hair Care. INCI Name EU/USA CAS # EINECS # Hydrolyzed wheat protein
 KELYAMIN Wheat Amino acids & Peptides for Hair Care Identification INCI Name EU/USA CAS # EINECS # Hydrolyzed wheat protein 70084-87-6 305-225-0 Composition % Liquid Powder Aqua Hydrolyzed wheat protein
KELYAMIN Wheat Amino acids & Peptides for Hair Care Identification INCI Name EU/USA CAS # EINECS # Hydrolyzed wheat protein 70084-87-6 305-225-0 Composition % Liquid Powder Aqua Hydrolyzed wheat protein
For questions 1-4, match the carbohydrate with its size/functional group name:
 Chemistry 11 Fall 2013 Examination #5 PRACTICE 1 For the first portion of this exam, select the best answer choice for the questions below and mark the answers on your scantron. Then answer the free response
Chemistry 11 Fall 2013 Examination #5 PRACTICE 1 For the first portion of this exam, select the best answer choice for the questions below and mark the answers on your scantron. Then answer the free response
SIMPLE BASIC METABOLISM
 SIMPLE BASIC METABOLISM When we eat food such as a tuna fish sandwich, the polysaccharides, lipids, and proteins are digested to smaller molecules that are absorbed into the cells of our body. As these
SIMPLE BASIC METABOLISM When we eat food such as a tuna fish sandwich, the polysaccharides, lipids, and proteins are digested to smaller molecules that are absorbed into the cells of our body. As these
ab Calcineurin Phosphatase Activity Assay Kit (Colorimetric)
 ab139461 Calcineurin Phosphatase Activity Assay Kit (Colorimetric) Instructions for Use For measuring Calcineurin phosphatase activity. This product is for research use only and is not intended for diagnostic
ab139461 Calcineurin Phosphatase Activity Assay Kit (Colorimetric) Instructions for Use For measuring Calcineurin phosphatase activity. This product is for research use only and is not intended for diagnostic
Insulin Resistance. Biol 405 Molecular Medicine
 Insulin Resistance Biol 405 Molecular Medicine Insulin resistance: a subnormal biological response to insulin. Defects of either insulin secretion or insulin action can cause diabetes mellitus. Insulin-dependent
Insulin Resistance Biol 405 Molecular Medicine Insulin resistance: a subnormal biological response to insulin. Defects of either insulin secretion or insulin action can cause diabetes mellitus. Insulin-dependent
1-To know what is protein 2-To identify Types of protein 3- To Know amino acids 4- To be differentiate between essential and nonessential amino acids
 Amino acids 1-To know what is protein 2-To identify Types of protein 3- To Know amino acids 4- To be differentiate between essential and nonessential amino acids 5-To understand amino acids synthesis Amino
Amino acids 1-To know what is protein 2-To identify Types of protein 3- To Know amino acids 4- To be differentiate between essential and nonessential amino acids 5-To understand amino acids synthesis Amino
Problem-solving Test: The Mechanism of Protein Synthesis
 Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
CDK9 Autophosphorylation Regulates High-Affinity Binding of the Human Immunodeficiency Virus Type 1 Tat P-TEFb Complex to TAR RNA
 MOLECULAR AND CELLULAR BIOLOGY, Sept. 2000, p. 6958 6969 Vol. 20, No. 18 0270-7306/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. CDK9 Autophosphorylation Regulates
MOLECULAR AND CELLULAR BIOLOGY, Sept. 2000, p. 6958 6969 Vol. 20, No. 18 0270-7306/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. CDK9 Autophosphorylation Regulates
Tala Saleh. Ahmad Attari. Mamoun Ahram
 23 Tala Saleh Ahmad Attari Minna Mushtaha Mamoun Ahram In the previous lecture, we discussed the mechanisms of regulating enzymes through inhibitors. Now, we will start this lecture by discussing regulation
23 Tala Saleh Ahmad Attari Minna Mushtaha Mamoun Ahram In the previous lecture, we discussed the mechanisms of regulating enzymes through inhibitors. Now, we will start this lecture by discussing regulation
Calcineurin Cellular Activity Assay Kit, Colorimetric Cat. No
 User Protocol 207007 Rev. 27-July-04 JSW Page 1 of 9 Calcineurin Cellular Activity Assay Kit, Colorimetric Cat. No. 207007 Introduction Calcineurin (CaN) is a neuronal form of the widely distributed Ca
User Protocol 207007 Rev. 27-July-04 JSW Page 1 of 9 Calcineurin Cellular Activity Assay Kit, Colorimetric Cat. No. 207007 Introduction Calcineurin (CaN) is a neuronal form of the widely distributed Ca
Reactions and amino acids structure & properties
 Lecture 2: Reactions and amino acids structure & properties Dr. Sameh Sarray Hlaoui Common Functional Groups Common Biochemical Reactions AH + B A + BH Oxidation-Reduction A-H + B-OH + energy ª A-B + H
Lecture 2: Reactions and amino acids structure & properties Dr. Sameh Sarray Hlaoui Common Functional Groups Common Biochemical Reactions AH + B A + BH Oxidation-Reduction A-H + B-OH + energy ª A-B + H
