Supplementary Appendix
|
|
|
- Justin Little
- 5 years ago
- Views:
Transcription
1 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med 2017;376: DOI: /NEJMoa
2 Table of Contents Supplementary methods... 3 Supplementary Figure S1 Distribution of mutations identified at low-frequency (< 20 patients) in the cohort... 5 Supplementary Figure S2 Distribution of number of driver mutations per patient... 6 Supplementary Figure S3 Variant allele fraction distribution by gene... 7 Supplementary Figure S4 Number of TP53 mutations per patient and effect on overall survival... 9 Supplementary Figure S5 TP53 mutation VAF distribution and and effect on overall survival Supplementary Figure S6 - TP53 mutation type and effect on overall survival Supplementary Figure S7 Clinical and genetic characteristics of TP53 mutations based on age Supplementary Figure S8 - TP53 mutations and pre-hct IPSS Supplementary Figure S9 Effect of complex karyotype on overall survival in patients with TP53 mutations Supplementary Figure S9 Distribution of co-mutations in patients with TP53 mutations Supplementary Figure S10 Mutation profile in patients 40 years of age with no TP53 mutation Supplementary Figure S11 Mutation profile in patients < 40 years of age with no TP53 mutation Supplementary Figure S12 Overall survival in 6 MDS subgroups identified by recursive partitioning analysis Supplementary Figure S13 Cumulative incidence of relapse and transplant-related mortality in all patients according to conditioning intensity Supplementary Figure S14 Cumulative incidence of relapse and NRM based on terminal nodestp53 mutations, based on JAK2 mutation status and conditioning intensity Supplementary Figure S15 Cumulative incidence of relapse and NRM in patients < 40 years of age without TP53 mutations, based on presence or absence of high risk features and according to conditioning intensity Supplementary Figure S16 Clinical and genetic characteristics of patients with biallelic SBDS mutations Supplementary Figure S17 Spectrum of driver mutations in therapy-related MDS Supplementary Table S1 List of participating transplant centers Supplementary Table S2 - Complete patient-related characteristics of the cohort Supplementary Table S3 - Complete transplantation-related characteristics of the cohort Supplementary Table S4 Complete disease-related clinical characteristics of the cohort Supplementary Table S5 - List of sequenced genes Supplementary Table S6- Criteria for identification of candidate driver mutations, which were subject to subsequent manual curation Supplementary Table S7 - Patient- and variant-level data in the entire cohort Supplementary Table S8 Frequency and distribution of mutations Supplementary Table S9 - Univariate associations of mutations with overall survival Supplementary Table S10 - Univariate associations of patient characteristics with overall survival Supplementary Table S11 - Univariate associations of transplant characteristics with overall survival Supplementary Table S12 - Univariate associations of disease characteristics with overall survival Supplementary Table S13 - Multivariable Cox model for OS Supplementary Table S14 - Multivariable Fine-Gray model for Relapse Supplementary Table S15 - Multivariable Fine-Gray model for NRM Supplementary Table S16 Association between gene mutations and primary or therapy-related MDS Supplementary Table S17 Association between gene mutations and older age ( 40 years) Supplementary Table S18 Competing risks regression models for relapse and non-relapse mortality according to conditioning intensity for terminal nodes of recursive partitioning model Supplementary Table S19 Competing risks regression models for relapse and non-relapse mortality after reduced-intensity conditioning (RIC) HCT according to gene mutation Supplementary Table S20 Competing risks regression models for relapse and transplant-related mortality after myeloablative conditioning (MAC) HCT according to gene mutation Supplementary Table S21 Clinical characteristics of patients with JAK2 mutated MDS... 95
3 Supplementary Table S22 Association between patient characteristics and overall survival in JAK2 mutated patients Supplementary Table S23 Cause of death according to terminal nodes of recursive partitioning model References
4 Supplementary methods Samples: Patient peripheral blood samples were collected prior to initiation of the preparative conditioning regimen. Samples were collected in Anticoagulant Citrate Dextrose Solution, Solution A (ACD-A) tubes and shipped for overnight delivery at ambient temperature. Blood samples were aliquotted into 1 milliliter cryovials on the day of receipt and maintained in -80 degrees Celsius frozen storage until the time of DNA extraction. Germline samples were uniformly unavailable. Library construction: Native genomic DNA was sheared and library constructed per manufacturer protocol (Agilent). Libraries were then quantified and pooled up to 24 samples per lane in equimolar amounts totaling 500ng of DNA. Each pool was then hybridized to Agilent Custom SureSelect In Solution Hybrid Capture RNA baits, consisting of probes, spanning kbp. Each capture reaction was washed, amplified, and sequenced on two lanes of an Illumina HiSeq bp paired end run. Variant Calling and annotation Fastq files were aligned to hg19 version of the human genome with BWA Single nucleotide and small insertion and deletion calling was performed with samtools mpileup and Varscan Pindel ,2 was used for FLT3-ITD calling at the specific genomic locus located at chromosome 13:28,608,000-28,608,600. Variants were annotated to include information about cdna and amino acid changes, sequence depth, number and percentage of reads supporting the variant allele, population allele frequency in 1000 Genomes release 2.2.2, 3 the Exome Sequencing Project, 4 Exome Aggregation Consortium, 5 and presence in Catalogue of Somatic Mutations in Cancer (COSMIC), version Variants were excluded if they had fewer than 15 total reads at the position, had fewer than 5 alternate reads, had variant allele fraction < 2%, fell outside of the target coordinates, had excessive read strand bias, had excessive number of calls in the local region, caused synonymous changes, or were recurrent small insertions/deletions at low variant allele fraction adjacent to homopolymer repeat regions. No germline tissue was available for evaluation of somatic status of mutations. As such, individual single nucleotide substitutions and small insertions or deletions were evaluated as candidate drivers of MDS or bone marrow failure based on gene-specific characteristics, as outlined in Table S4, then curated manually and classified as MDS driver mutations or pathogenic bone marrow failure mutations based on genetic criteria and literature review. 7,8 Variant level details are available in Table S5. All interpretation of variants was blinded to clinical characteristics and thus agnostic to variables including age, sex, treatment status, and clinical outcomes; the genetic analysis was completed and locked prior to merging with any clinical data. RAS pathway definition We used literature review of functional, genomic, and clinical evidence to define RAS pathway mutations prior to analysis and without reference to our sequencing results: NRAS, KRAS, PTPN11, CBL, NF1, RIT1, FLT3, KIT. Although the precise molecular consequence of individual mutations on intracellular signaling can be distinct, these mutations as a whole share a common role in the biology of myeloid malignancies. Specifically, mutations that cause aberrant activation of RAS/MAPK signaling are late events in disease progression that drive leukemic transformation of MDS We analyzed the impact of each gene individually and the impact of all pathway members together without post-hoc exclusions. Curation of DDX41 variants DDX41 has been reported to be mutated in the germline in the context of familial predisposition to MDS and somatically during disease intitiation or progression. 11,12 Because several of the reported disease-associated alleles are common in population-based exome studies and the effect size may be small or modified by genetic context, we uses variant allele fraction, population allele frequecy, and literature reports to assign DDX41 variants to candidate germline or somatic status for subsequent analyses. No germline material was available for definitive assessment. Statistics 3
5 Overall survival (OS) was defined as time from transplant until death from any cause. Subjects not confirmed dead were censored at the time last known to be alive. Univariate and multivariable analyses of OS were performed using Cox regression. Hazard ratios (HR) with 95% confidence intervals (CI) and Wald p-values were reported for covariates in multivariable Cox models. OS was estimated using the method of Kaplan and Meier (KM), and reported with 95% CIs based on Greenwood s formula. Differences in survival curves were assessed using log-rank tests. Median follow-up was calculated using the reverse KM method. Transplant-related mortality (NRM) was defined as death without relapse. Deaths after relapse were defined as competing risks (CR). Cumulative incidence between groups was assessed using Gray s test, and estimates were reported with 95% CIs. Cusing the method of Fine and Gray. Each mutation was assessed in the full data set individually with or without adjustment for those clinical and modifiable features identified as significantly associated with the outcome of interest through stepwise forward and backward procedures. OS used a Cox proportional hazards model and was adjusted for patient age in decade, Karnofsky performance status, IPSS-R risk score, therapy-related MDS, year of transplant, donor/recipient sex matching, and donor category. Time to NRM used a Fine and Gray competing risks regression model, and was adjusted for patient age in decade, donor category, IPSS-R risk score and Karnofsky performance status. Time to relapse used a Fine and Gray competing risks regression model, and was adjusted for IPSS-R risk score, conditioning intensity, bone marrow blasts at diagnosis, the use of in vivo T-cell depletion, TBI use, therapy-related MDS, monosomal karyotype and Karnofsky performance status. Recursive partitioning was used to generate a hierarchical model for OS integrating clinical and genetic characteristics. The variables considered included all mutations and non-modifiable clinical variables of age and sex; number of prior lines of therapy and prior diagnosis; therapy-related or de-novo MDS; Karnofsky performance score; pre-disposing conditions, monosomal karyotype, and cytogenetic risk factors; CMV infection; and blast, hemoglobin, platelet, and absolute neutrophil counts. Chi-square and Fisher s exact test was used to test for association between pairs of categorical variables. Odds ratios (OR) and 95% CIs were calculated for binary outcomes from contingency tables or logistic regression. The Wilcoxon rank-sum test was used to assess a location shift in the distribution of continuous variables between two groups. Descriptive statistics (proportions, medians, etc.) were reported with 95% CI or range. All p-values were two-sided, and adjustments for multiple hypothesis testing was performed using the method of Benjamini and Hochberg. Case definitions Cases were defined according to WHO criteria or published transplant literature. Therapy-related MDS was defined based on prior exposure to chemotherapy and/or radiation. 13 Transplant conditioning regimens were classified based on chemotherapy regimen and/or total body irradiation dose as myeloablative, reducedintensity, or non-myeloablative. 14 Availability of data All clinical and genetic data are available for request through CIBMTR: 4
6 Supplementary Figure S1 Distribution of mutations identified at low-frequency (< 20 patients) in the cohort. Shown is the frequency of driver mutations, limited to genes mutated in< 20 patients (<1.3%). Due to their low frequency in the study cohort, these mutations were not evaluated for their association with clinical outcomes. # mutated cases ETNK1 ATM BRCC3 CEBPA RAD21 PIGA BCORL1 GNB1 RIT1 GNAS MPL STAT3 EP300 LUC7L2 FLT3 SDS BRAF CALR CSNK1A1 CREBBP TERT SMC3 SH2B3 NOTCH1 TERC IKZF1 PDS5B KIT CTCF SMC1A SETD2 XRCC2 CBLB STAT5B RPS Frequency 5
7 Supplementary Figure S2 Distribution of number of driver mutations per patient Shown is number of driver mutations per patient in the study cohort (Median = 2, Mean = 2.8). 6
8 Supplementary Figure S3 Variant allele fraction distribution by gene Shown are variant allele fractions according to individual genes. Horizontal lines within boxes indicate median VAF. Box limits indicate the 25th and 75th percentiles, whiskers extend to the 10th and 90th percentiles, and outliers are represented by dots. 7
9 Supplementary Figure S4 Distribution of per-patient maximum somatic VAF Shown is the maximum variant allele fraction of putative somatic mutations in myeloid drivers. To infer the most abundant, clonal mutations in patients with multiple mutations, the mutation with higher VAF was used. Mutations in genes associated only with germline bone marrow failure syndromes are not included. Dotted line shows the median maximum somatic VAF (23%) and the limits of the grey shading indicate the 25th and 75th percentiles 8
10 Supplementary Figure S4 Number of TP53 mutations per patient and effect on overall survival In patients with TP53 mutations, 35% had more than one detectable TP53 mutation. Panel A shows the number of patients with TP53 mutated MDS based on number of distinct TP53 mutations. Panel B shows Kaplan Meier curves for OS in patients with TP53 mutations based on the presence of one or more than one independent TP53 mutation. Surival of patients with more than one TP53 mutation was not significant different than those with one TP53 mutation (HR 1.20, , p=0.18). 9
11 Supplementary Figure S5 TP53 mutation VAF distribution and and effect on overall survival Panel A shows the distribution of TP53 variant allele fractions, where box limits indicate the 25th and 75th percentiles, whiskers extend to the 1st and 99th percentiles, and outliers are represented by dots. The median TP53 VAF was 10% (range 2-85%). Panel B shows Kaplan Meier curves for OS in patients with TP53 mutations according to VAF above or below median. Surival of patients with TP53 VAF 10% was not significant different than those with TP53 VAF < 10% (HR 1.28, , p=0.07). 10
12 Supplementary Figure S6 - TP53 mutation type and effect on overall survival Panel A shows the localization of TP53 mutations based on position within the coding sequence. Missense mutations are shown in green, truncating mutations (frameshift, nonsense, splice site) are shown in red. Panel B shows the proportion of patients with TP53 mutated MDS with missense mutations only (71%, n=205), truncating mutations only (17%, n=48), or both truncating and missense mutations (13%, n=36). Panel C shows Kaplan Meier curves for OS in patients with TP53 mutations according to mutation type, as indicated. Patients with truncating mutations had poor survival compared to those with missense only (HR 1.61, , p = 0.006) or missense plus truncating mutations (HR 1.70, , p = 0.03). Panel C shows the distribution of all TP53 mutations based on amino acid position and mutation type where green are missense and red are truncating (frameshift indels, nonsense, splice site). 11
13 Supplementary Figure S7 Clinical and genetic characteristics of TP53 mutations based on age Panel A shows Kaplan Meier curves for OS in patients 18 according to TP53 mutation status. Panel B shows the distribution of TP53 variant allele fraction in patients according to the indicated age group. No significant differences were identified. Panel C shows the frequency of t-mds in patients according to TP53 mutation status in the indicated age groups. 12
14 Supplementary Figure S8 - TP53 mutations and pre-hct IPSS Panel A shows Kaplan Meier curves for OS in all patients according to IPSS score prior to transplantation. mutation status. Patients are grouped as lower (Low or Intermediate-1) or higher (Intermediate-2 or high) risk groups. Panel B shows the frequency of TP53 mutations in lower or higher risk groups prior to transplantation. Panel C shows Kaplan Meier curves for OS in all patients according to IPSS lower or higher risk groups and TP53 mutation status. 13
15 Supplementary Figure S9 Effect of complex karyotype on overall survival in patients with TP53 mutations. Shown is a Kaplan Meier curves for OS in patients TP53 mutations according to presence or absence of complex karyotype. 14
16 Supplementary Figure S9 Distribution of co-mutations in patients with TP53 mutations. A co-mutation plot shows nonsynonymous mutations in individual genes as labeled on the left. Mutations are depicted by colored bars and each column represents 1 of the 289 patients with TP53 mutations. TP53 PPM1D DNMT3A TET2 ASXL1 RUNX1 U2AF1 SF3B1 SRSF2 ZRSR2 BCOR BCORL1 SBDS EZH2 NRAS KRAS PTPN11 CBL NF1 RIT1 FLT3 WT1 IDH1 IDH2 PRPF8 ETV6 CEBPA GATA2 CUX1 STAG2 RAD21 BRAF CSF3R GNB1 CBLB SETBP1 PHF6 NPM1 JAK2 CALR PIGA STAT3 EP300 LUC7L2 DDX41 ATM NOTCH1 TERT RPS17 15
17 Supplementary Figure S10 Mutation profile in patients 40 years of age with no TP53 mutation A co-mutation plot shows nonsynonymous mutations in individual genes as labeled on the left. Mutations are depicted by colored bars and each column represents 1 of the 1011 patients 40 years of age without TP53 mutation. Colors reflect the three subgroups identified by recursive partitioning: RAS pathway mutation (blue), JAK2 mutation (gold), and all other patients (grey). TP53 NRAS KRAS PTPN11 CBL NF1 RIT1 FLT3 KIT BRAF JAK2 CALR MPL ASXL1 BCOR BCORL1 EZH2 DNMT3A TET2 WT1 IDH1 IDH2 SF3B1 SRSF2 U2AF1 ZRSR2 RUNX1 ETV6 CEBPA GATA2 SETBP1 CUX1 STAG2 PPM1D PHF6 NPM1 Other RAS pathway mutated JAK2 mutated RAS pathway and JAK2 unmutated 16
18 Supplementary Figure S11 Mutation profile in patients < 40 years of age with no TP53 mutation A co-mutation plot shows nonsynonymous mutations in individual genes as labeled on the left. Mutations are depicted by grey bars and each column represents 1 of the 214 patients under 40 years of age without TP53 mutation. Colored bars indicate the two subgroups identified by recursive partitioning: those with at least one high-risk feature (t-mds, platelets < 30 x 10 9 /L at HCT, or bone marrow blasts at diagnosis 15%) or with no high-risk features. Low Risk High Risk ASXL1 RUNX1 GATA2 SETBP1 BCOR STAG2 PIGA WT1 ETV6 NRAS KRAS PTPN11 CBL NF1 BCORL1 EZH2 TET2 DNMT3A U2AF1 SF3B1 SRSF2 ZRSR2 PRPF8 CEBPA IKZF1 CUX1 RAD21 SMC3 IDH1 IDH2 PDS5B FLT3 KIT BRAF CSF3R GNAS PHF6 NPM1 JAK2 CALR SH2B3 STAT3 STAT5B CSNK1A1 ETNK1 CREBBP EP300 LUC7L2 ATM BRCC3 XRCC2 17
19 Supplementary Figure S12 Overall survival in 6 MDS subgroups identified by recursive partitioning analysis. Panel A shows Kaplan Meier curves for OS in all patients in the cohort according to TP53 mutation status and age group. Panel B shows Kaplan Meier curves for OS in patients < 40 years old without TP53 mutations, according to the presence (High Risk) or absence (Low Risk) of at least one of the following clinical features: (t-mds, platelets < 30 x 10 9 /L at HCT, or bone marrow blasts at diagnosis 15%). 18
20 Supplementary Figure S13 Cumulative incidence of relapse and transplant-related mortality in all patients according to conditioning intensity. Panel A shows cumulative incidence of relapse and transplant-related mortality (NRM) in the entire cohort (n = 1514). Panels B and C show cumulative incidence of relapse and NRM in patients receiving myeloablative and reduced-intensity conditioning. 19
21 Supplementary Figure S14 Cumulative incidence of relapse and NRM based on terminal nodestp53 mutations, based on JAK2 mutation status and conditioning intensity. Shown are cumulative incidence of relapse and transplant-related mortality (NRM) patients in patients receiving myeloablative (MAC, solid lines) or reduced-intensity (RIC, dashed lines) conditioning. Panel A shows the total cohort according to presence (red) or absence (black) of TP53 mutations. Panel B shows patients 40 years of age without TP53 mutations according to presence (red) or absence (black) of RAS pathway mutations. Panel C shows patients 40 years of age without TP53 or RAS pathway mutations according to presence (red) or absence (black) of JAK2 mutations. P values are found in Table S18. 20
22 Supplementary Figure S15 Cumulative incidence of relapse and NRM in patients < 40 years of age without TP53 mutations, based on presence or absence of high risk features and according to conditioning intensity. Shown are cumulative incidence of relapse and transplant-related mortality (NRM) in patients < 40 years of age without TP53 mutations, based on presence or absence of high risk features in all patients (Panel A). Panels B and C show cumulative incidence of relapse and non-relapse mortality based on presence or absence of high risk features in patients according to treatment with myeloablative (solid lines) or reduced-intensity (dashed) conditioning. 21
23 Supplementary Figure S16 Clinical and genetic characteristics of patients with biallelic SBDS mutations. Panel A shows the age at MDS diagnosis of patients with two SBDS mutations (n = 7), one SBDS mutation (n = 11), or zero SBDS mutations (n = 1496). Box limits indicate the 25th and 75th percentiles, whiskers extend to the 1st and 99th percentiles, and outliers are represented by dots. Panel B shows sex-specific percentile height of patients with two or one SBDS mutations. Red circles depict the 2 patients known clinically to have Shwachman-Diamond Syndrome. Panel C shows the frequency of somatic TP53 mutations in patients with two SBDS mutations (100%, 7/7), one SBDS mutation (27%, 3/11), or zero SBDS mutations (19%, 279/1496). 22
24 Supplementary Figure S17 Spectrum of driver mutations in therapy-related MDS. The co-mutation plot shows the spectrum of driver mutations in t-mds, where mutations are depicted as colored bars and each column represents one of the 311 t-mds patients in the cohort. 23
25 Supplementary Table S1 List of participating transplant centers. Center name Helen DeVos Children's Hospital UCSF Benioff Children's Hospital - Oakland Stanford Health Care The Children's Hospital of Denver Louisiana State University Children's Hospital Steven and Alexandra Cohen Children's Medical Center of New York Montefiore Medical Center Rady Children's Hospital San Diego Children's Hospital of Wisconsin Washington University/St Louis Children's Hospital University of Tennessee Christiana Care Cook Children's Medical Center Cancer Transplant Institute at Virginia G. Piper Cancer Center Children's National Medical Center Roger Williams Medical Center Stony Brook University Hospital Pediatric BMT Program, Doernbecher Children's Hospital (OHSU) Scripps Blood & Marrow Transplant Program Johns Hopkins All Children's Hospital Children's Medical Center - Dallas Sarah Cannon BMT Center at Centennial Medical Center Cancer Institute of New Jersey Avera McKennan Transplant Institute Nationwide Children's Hospital SSM Health Saint Louis University Hospital The Children's Mercy Hospitals and Clinics Ann & Robert H. Lurie Children's Hospital of Chicago Banner Blood and Marrow Transplant Program 4564 University of Louisville Hospital/James Brown Cancer Center Dartmouth-Hitchcock Medical Center Children's Healthcare of Atlanta at Egleston Memorial Sloan Kettering Cancer Center - Peds Memorial Sloan-Kettering Cancer Center Sutter Cancer Center Georgia Cancer Center at Augusta University Health Beth Israel Deaconess Medical Center UT Southwestern Medical Center - BMT Program Duke University Medical Center; Pediatric BMT The Coleman Foundation Blood and Marrow Transplant Center, Rush Univer University of California-Davis Cancer Center St Jude Children's Research Hospital Blood & Marrow Transplant Center, Florida Hospital Medical Group Fox Chase Temple University Hospital Bone Marrow Transplant Program UMass Memorial Medical Center 24
26 University of Mississippi Medical Center - Jackson Philadelphia Children's Hospital Banner University Medical Center - Tucson Tufts New England Medical Center Cincinnati Children's Hospital Medical Center Mayo Clinic Florida - Jacksonville Indiana Blood & Marrow Transplantation Seidman Cancer Center - University Hospitals Case Medical Center Henry Ford Hospital Bone Marrow Transplant Program University of Kentucky Medical Center North Shore University Hospital University of Colorado Hospital Cedars-Sinai Medical Center University of Miami - Adults University of Pittsburgh Medical Center Osborn Hematopoietic Malignancy & Transplantation Program Oklahoma University Medical Center Dana Farber Cancer Institute & Boston Children's Hospital Yale New Haven Hospital Jewish Hospital Blood and Marrow Transplant Center University of Maryland School of Medicine Utah Blood and Marrow Transplant Program- Adults Latter Day Saints Hospital Thomas Jefferson University Medical City Dallas Hospital University of California, San Diego Medical Center Wake Forest Baptist Health Baylor College of Medicine Medical University of South Carolina West Penn Hospital Shands HealthCare & University of Florida University of North Carolina Hospitals - Chapel Hill Northwestern Memorial Hospital Mount Sinai Medical Center - New York UCLA Hematologic Malignancy/Stem Cell Transplantation Program University of Iowa Hospital & Clinics University of Nebraska Medical Center University of Rochester Medical Center University of Alabama Birmingham University of California - San Francisco - Adults Penn State Hershey Medical Center University of Wisconsin Hospital and Clinics Froedtert & Medical College of Wisconsin Virginia Commonwealth University Massey Cancer Center BMT Program Roswell Park Cancer Institute University of Chicago Medical Center Loyola University Medical Center Indiana University Hospital/Riley Hospital for Children 25
27 Texas Transplant Institute The Blood and Marrow Transplant Program at Northside Hospital New York Presbyterian Hospital University of Kansas Massachusetts General Hospital Colorado Blood Cancer Institute Abramson Cancer Center University - Pennsylvania Medical Center The Ohio State University Medical Center Emory University Mayo Clinic Arizona and Phoenix Children's Hospital Vanderbilt University Oregon Health and Science University Duke University - Adults Cleveland Clinic The University of Michigan Mayo Clinic Rochester Karmanos Cancer Institute Baylor University Medical Center The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins Masonic Cancer Center University of Minnesota Memorial Sloan Kettering Cancer Center - Adults Hackensack University Medical Center Washington University School of Medicine H Lee Moffitt Cancer Center City of Hope National Medical Center Fred Hutchinson Cancer Center Dana Farber Cancer Institute at Brigham and Women's Hospital - Adults MD Anderson Cancer Center Hospital De Clinicas Curitiba University Hospital, Rigshospitalet University Hospital Charite - Virchow University Hospital Charite Kinderklinik - Virchow University Leipzig, BMT Center University Hospital Mainz Deutsche Klinik fur Diagnostik - Wiesbaden UKE Hamburg, Klinik und Poliklinik fur Stammzelltransplantation Leiden University Medical Center 26
28 Supplementary Table S2 - Complete patient-related characteristics of the cohort. Karnofsky performance score No. of patients (%) (54) < (28) Missing 278 (18) Recipient age at transplant (2) (4) (4) (5) (11) (26) 60 and older 719 (47) Recipient sex Male 912 (60) Female 602 (40) Recipient race Caucasian 1397 (92) African American 57 (4) Asian 37 (2) Pacific Islander 3 (0) Native American 2 (0) Other 18 (1) Recipient ethnicity Caucasian, non-hispanic 1324 (87) African American, non-hispanic 54 (4) Asian race, non-hispanic 37 (2) Pacific Islander race, non-hispanic 3 (0) Native American race, non-hispanic 2 (0) Hispanic, Caucasian race 73 (5) Hispanic, African American race 3 (0) Missing 18 (1) 27
29 Supplementary Table S3 - Complete transplantation-related characteristics of the cohort. No. of patients (%) Conditioning regimen (1) Myeloablative 789 (52) Reduced intensity 582 (38) Non-myeloablative 130 (9) Missing 13 (1) Conditioning regimen (2) Myeloablative- TBI-based 116 (8) Myeloablative Chemo-based 673 (44) Reduced Intensity TBI-based 54 (4) Reduced Intensity Chemo-based 528 (35) Non-myeloablative TBI-based 85 (6) Non-myeloablative Chemo-based 45 (3) Missing 13 (1) Conditioning regimen (3) MAC-Cy/TBI/others 7 (0) MAC-CY/TBI 101 (7) MAC-TBI/others 8 (1) MAC-Bu/Flu 173 (11) MAC-Thiotepa based 8 (1) MAC-Bu/Cy/others 3 (0) MAC-Bu/Cy 216 (14) MAC-Bu/Flu + others 42 (3) MAC-Bu + others 188 (12) MAC-Flu/Melphalan 11 (1) MAC-Treosulfan-based 16 (1) MAC-others 16 (1) RIC-TBI+others 54 (4) RIC-Bu/Flu/others 3 (0) RIC-Bu/Flu 270 (18) RIC-Flu/Melphalan 210 (14) RIC-Flu/Melphalan/others 24 (2) RIC-Bu/others 2 (0) RIC-others 19 (1) NST-TBI/Cy/Flu 59 (4) NST-TBI/Flu 26 (2) NST-Flu/Cy 15 (1) NST-others 30 (2) Missing 13 (1) TBI use > 500 single dose or > 800 fractionated dose 116 (8) < 500 single dose or < 800 fractionated dose but other agents delivered at MAC dose 9 (1) single dose or fractionated dose 55 (4) TBI 200 cgy 134 (9) TBI, dosage missing 83 (5) Non-TBI regimen 1117 (74) 28
30 GVHD prophylaxis Ex vivo T-cell depletion 21 (1) CD34 selection 32 (2) Cyclophosphamide-based 19 (1) Tacrolimus-based 1134 (75) CSA-based 242 (16) None reported 24 (2) Missing 42 (3) in vivo T-cell depletion No 843 (56) Yes 605 (40) Missing 66 (4) Donor sex Male 972 (64) Female 454 (30) Missing 88 (6) Sex match (donor/recipient) M/M 610 (40) M/F 362 (24) F/M 250 (17) F/F 204 (13) Missing 88 (6) Graft type Bone marrow 221 (15) PBSC 1114 (74) Cord Blood 168 (11) Missing 11 (1) Donor type Unrelated 1165 (77) Related 181 (12) Cord blood 168 (11) Donor group/graft source 8/8 URD, PBSC 709 (47) HLA-identical siblings, BM 12 (1) HLA-identical siblings, PBSC 169 (11) 8/8 URD, BM 154 (10) 7/8 URD, BM 34 (2) 7/8 URD, PBSC 126 (8) 6/8 URD, BM 21 (1) 6/8 URD, PBSC 115 (8) UCBT 174 (11) Recipient CMV Negative 619 (41) Positive 877 (58) Missing 18 (1) CMV match (donor/recipient) Neg/Neg 439 (29) Neg/Pos 459 (30) 29
31 Pos/Neg 162 (11) Pos/Pos 379 (25) Missing 75 (5) Donor ethnicity Caucasian, non-hispanic 1086 (72) African American, non-hispanic 40 (3) Asian race, non-hispanic 27 (2) Native American race, non-hispanic 8 (1) Hispanic, Caucasian race 51 (3) Hispanic, African American race 1 (0) Hispanic, Native American race 1 (0) Hispanic, Unknown race 16 (1) Missing 284 (19) Donor race Caucasian 1130 (75) African American 41 (3) Asian 27 (2) Pacific Islander 7 (0) Native American 9 (1) Other 54 (4) Missing 246 (16) Donor age (11) (3) (35) (20) (15) (8) Missing 122 (8) Year of transplant (grouped) (4) (16) (21) (22) (33) (4) 30
32 Supplementary Table S4 Complete disease-related clinical characteristics of the cohort. No. of patients (%) Therapy-related MDS Primary MDS 1203 (79) t-mds 311 (21) Prior diagnosis for t-mds Predisposing condition 15 (1) HL 13 (1) NHL 74 (5) Breast cancer 56 (4) ALL 28 (2) CLL 37 (2) Other hem 39 (3) Missing 1252 (83) Predisposing conditions None reported 1394 (92) Yes 120 (8) Type of predisposing condition Inherited bone marrow failure syndrome 7 (0) Aplastic anemia 45 (3) Idiopathic thrombocytopenic purpura 9 (1) Paroxysmal nocturnal hemoglobinuria 3 (0) Other hematologic condition 46 (3) Other non-hematologic condition 10 (1) None reported 1394 (92) Bone marrow blasts at HCT (%) (25) (18) (16) (19) Missing 341 (23) Absolute neutrophil count at HCT (x 109/L) (59) < (32) Missing 137 (9) Platelets at HCT (x 10 9 /L ) (36) (23) < (36) Missing 85 (6) Hemoglobin (g/dl) (39) (35) <8 307 (20) Missing 81 (5) 31
33 Cytogenetic risk group Very good 10 (1) Good 569 (38) Intermediate 269 (18) Poor 287 (19) Very poor 125 (8) Missing 254 (17) Monosomal karyotype No 1090 (72) Yes 210 (14) Missing 214 (14) Prior MDS therapy None 509 (34) HMA only 761 (50) Chemo only 60 (4) HMA and Chemo 64 (4) Missing 120 (8) IPSS-R prior to HCT Very low 119 (8) Low 287 (19) Intermediate 340 (22) High 223 (15) Very high 171 (11) Missing 374 (25) IPSS-R prior to HCT Low 149 (10) Intermediate (29) Intermediate (24) High 61 (4) Missing 346 (24) MDS Classification at diagnosis MDS-NOS 226 (15) Refractory anemia 163 (11) Refractory anemia with excess blasts 136 (9) Refractory anemia with ring sideroblasts 81 (5) RAEB (17) RAEB (19) RCMD 201 (13) RCMD/RS 29 (2) 5q-syndrome 16 (1) Missing 114 (8) 32
34 Supplementary Table S5 - List of sequenced genes ANKRD26 CSF1R FANCC IDH2 NOTCH2 RPL11 SETBP1 TINF2 ASXL1 CSF3R FANCD2 IKZF1 NPM1 RPL23 SETD2 TP53 ATM CSNK1A1 FANCE JAK2 NRAS RPL26 SF1 U2AF1 ATRX CTC1 FANCF JAK3 PALB2 RPL35A SF3A1 U2AF2 B2M CTCF FANCG KIT PDS5B RPL36 SF3B1 VPS45 BCOR CUX1 FANCI KRAS PHF6 RPL5 SH2B3 WAS BCORL1 DDX41 FANCL LUC7L2 PIGA RPS10 SLX4 WRAP53 BRAF DKC1 FANCM MIR-142 PIGT RPS15 SMC1A WT1 BRCA1 DNMT3A FLT3 MPL PPM1D RPS17 SMC3 XRCC2 BRCA2 ELA2 G6PC3 MRE11A PRPF40B RPS19 SRSF2 ZRSR2 BRCC3 EP300 GATA1 MYD88 PRPF8 RPS24 STAG1 BRIP1 ERCC4 GATA2 MYH9 PTEN RPS26 STAG2 CALR ETNK1 GFI1 NBN PTPN11 RPS27A STAT3 CBL ETV6 GNAS NF1 RAD21 RPS29 STAT5B CBLB EZH2 GNB1 NHP2 RAD51C RPS7 TERC CEBPA FANCA HAX1 NOP10 RIT1 RUNX1 TERT CREBBP FANCB IDH1 NOTCH1 RMRP SBDS TET2 33
35 Supplementary Table S6- Criteria for identification of candidate driver mutations, which were subject to subsequent manual curation. Gene Criterion Gene Criterion FANCL type:frameshift KIT aa_range:810,830 and ExAC_Freq<0.01 aa_range:540,600 and ExAC_Freq<0.01 type:splice_site type:nonframeshift and ExAC_Freq<0.01 type:frameshift FANCM type:frameshift KRAS type:missense and ExAC_Freq<0.01 type:nonframeshift and ExAC_Freq<0.01 type:splice_site LUC7L2 type:frameshift FLT3 aa_match:d835 aa_match:n841 type:splice_site aa_match:y842 MPL aa_match:515 type:nonframeshift and aa_range:800,850 and aa_match:505 aa_match:n676 type:nonframeshift and aa_range:500,520 aa_match:n663 MRE11A type:frameshift aa_match:f691 aa_match:v592 type:splice_site aa_match:v579 MYD88 aa_match:l265 type:missense and aa_range:569,700 and MYH9 type:frameshift type:nonframeshift and aa_range:569,700 and type:frameshift and aa_range:569,648 type:splice_site G6PC3 type:frameshift type:missense and ExAC_Freq<0.01 NBN type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 type:splice_site GATA1 type:frameshift type:missense and ExAC_Freq<0.01 NF1 type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 type:splice_site GATA2 type:frameshift NHP2 type:frameshift type:splice_site type:splice_site type:missense and aa_range:200,398 type:missense and ExAC_Freq<0.01 position: , NOP10 type:frameshift GFI1 type:missense and ExAC_Freq<0.01 GNAS aa_match:r201 type:splice_site GNB1 aa_match:k57 type:missense and ExAC_Freq<0.01 aa_match:i80 NOTCH1 type:missense and aa_range:617,738 andexac_freq<0.01 aa_match:k89 type:missense and aa_range:500,616 andexac_freq<0.01 #aa_match:d76 type:frameshift and aa_range:2061,2555 #aa_match:k78 and aa_range:2061,2555 #aa_match:n88 NOTCH2 type:frameshift and aa_range:2010,2471 HAX1 type:frameshift and aa_range:2010,2471 NPM1 type:frameshift type:splice_site NRAS type:missense and ExAC_Freq<0.01 type:missense and ExAC_Freq<0.01 type:nonframeshift and ExAC_Freq<0.01 IDH1 aa_match:r132 #aa_match:g12 type:nonframeshift and aa_range:126,138 #aa_match:g13 IDH2 aa_match:r172 #aa_match:v14 aa_match:r140 #aa_match:i24 type:nonframeshift and aa_range:134,146 #aa_match:g60 type:nonframeshift and aa_range:164,180 #aa_match:q61 IKZF1 type:frameshift #aa_match:t74p #aa_match:a146 type:splice_site PALB2 type:frameshift JAK2 aa_match:v617f type:missense and aa_range:505,547 type:splice_site aa_match:v683 PDS5B type:frameshift aa_match:r867 aa_match:d873 type:splice_site aa_match:p933 JAK3 type:missense and ExAC_Freq<0.01 type:nonframeshift and ExAC_Freq<
36 Gene Criterion Gene Criterion ANKRD26 type:5utr and ExAC_Freq<0.01 CUX1 type:frameshift ASXL1 type:frameshift and aa_range:400,1540 and aa_range:400,1540 type:splice_site type:splice_site and ExAC_Freq<0.01 DDX41 type:missense and ExAC_Freq<0.01 ATM type:frameshift type:frameshift type:splice_site type:splice_site ATRX type:frameshift type:missense and aa_range:360,430 DKC1 type:frameshift type:splice_site B2M type:frameshift type:missense and ExAC_Freq<0.01 DNMT3A aa_match:r882 type:splice_site type:frameshift BCOR type:frameshift type:splice_site type:splice_site type:missense and aa_range:626,910 and BCORL1 type:frameshift type:missense and aa_range:290,374 and ELANE type:missense and ExAC_Freq<0.01 type:splice_site EP300 type:frameshift BRAF type:missense and aa_range:590,615 BRCA1 type:frameshift type:splice_site ERCC4 type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 type:splice_site BRCA2 type:frameshift ETNK1 aa_match:h243 aa_match:n244 type:splice_site ETV6 type:frameshift type:missense and ExAC_Freq<0.01 BRCC3 type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 type:splice_site type:missense and aa_range:338,424 and BRIP1 type:frameshift #ETS domain type:missense and aa_range:56,123 and type:splice_site #PNT domain CALR type:frameshift and aa_range:352,418 EZH2 type:frameshift CBL type:missense and aa_range:360,430 type:frameshift and aa_range:360,430 type:splice_site type:nonframeshift and aa_range:360,430 type:missense and aa_range:617,738 and type:splice_site type:missense and aa_range:500,616 and CBLB type:missense and aa_range:360,430 FANCA type:frameshift CEBPA type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 FANCB type:frameshift CREBBP type:frameshift type:splice_site type:splice_site FANCC type:frameshift CSF1R aa_match:y969 type:missense and aa_range:543,972 and type:splice_site type:frameshift FANCD2 type:frameshift CSF3R aa_match:t615 type:splice_site aa_match:t618 FANCE type:frameshift type:frameshift and aa_range:618,840 and aa_range:618,840 type:splice_site CSNK1A1 aa_match:e98 FANCF type:frameshift aa_match:d140 type:missense and ExAC_Freq<0.01 type:splice_site CTC1 type:frameshift FANCG type:frameshift type:splice_site type:splice_site CTCF type:frameshift FANCI type:frameshift type:splice_site type:splice_site 35
37 Gene Criterion Gene Criterion PHF6 type:frameshift RPS10 type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 type:missense and aa_range:239,330 and RPS15 type:frameshift type:missense and aa_range:42,132 and PIGA type:missense and ExAC_Freq<0.01 type:missense and ExAC_Freq<0.01 type:frameshift RPS17 type:frameshift PIGT type:frameshift type:missense and ExAC_Freq<0.01 RPS19 type:frameshift PPM1D type:frameshift and aa_range:421,605 and aa_range:421,605 type:missense and ExAC_Freq<0.01 PRPF40B type:frameshift RPS24 type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 PRPF8 type:frameshift RPS26 type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 PTEN type:missense and ExAC_Freq<0.01 RPS27A type:frameshift type:frameshift type:missense and ExAC_Freq<0.01 PTPN11 type:missense and ExAC_Freq<0.01 RPS29 type:frameshift RAD21 type:frameshift type:missense and ExAC_Freq<0.01 type:splice_site RPS7 type:frameshift RAD51C type:frameshift type:missense and ExAC_Freq<0.01 type:splice_site RUNX1 type:frameshift RIT1 aa_match:s35 aa_match:a57 type:splice_site aa_match:f82 type:missense and ExAC_Freq<0.01 aa_match:g95 SBDS type:missense and ExAC_Freq<0.01 aa_match:a77 type:nonframeshift and ExAC_Freq<0.01 aa_match:e81 type:splice_site aa_match:t83 type:frameshift aa_match:y89 aa_match:m90 SETBP1 type:missense and aa_range:855,880 RMRP type:missense and ExAC_Freq<0.01 SETD2 type:frameshift type:nonframeshift and ExAC_Freq<0.01 type:frameshift type:splice_site SF1 type:frameshift RPL11 type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 SF3A1 type:frameshift RPL23 type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 SF3B1 type:missense and aa_range:550,800 and RPL26 type:frameshift SH2B3 type:frameshift type:missense and ExAC_Freq<0.01 type:missense and aa_range:364,441 and RPL35A type:frameshift type:missense and aa_range:195,307 and SLX4 type:frameshift type:missense and ExAC_Freq<0.01 RPL36 type:frameshift type:splice_site SMC1A type:frameshift type:missense and ExAC_Freq<0.01 RPL5 type:frameshift type:splice_site type:missense and ExAC_Freq<
38 Gene Criterion Gene Criterion SMC3 type:frameshift TINF2 type:frameshift type:splice_site type:splice_site SRSF2 aa_match:p95 type:missense and aa_range:201,352 and type:nonframeshift and aa_range:85,100 TP53 type:frameshift type:frameshift and aa_range:85,100 STAG1 type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 type:splice_site U2AF1 aa_match:s34 STAG2 type:frameshift aa_match:q157 type:nonframeshift and aa_range:30-38 type:splice_site type:nonframeshift and aa_range: STAT3 type:missense and aa_range:584,674 U2AF2 type:nonframeshift and ExAC_Freq<0.01 type:nonframeshift and aa_range:584,674 type:frameshift STAT5B type:missense and aa_range:593,670 type:nonframeshift and aa_range:593,670 VPS45 type:missense and ExAC_Freq<0.01 TERC ExAC_Freq<0.01 type:nonframeshift and ExAC_Freq<0.01 TERT type:missense and ExAC_Freq<0.01 type:frameshift type:nonframeshift and ExAC_Freq<0.01 type:frameshift WAS type:missense and ExAC_Freq<0.01 type:nonframeshift and ExAC_Freq<0.01 TET2 type:frameshift type:frameshift type:splice_site WRAP53 type:frameshift type:missense and aa_range:1104,1481 and type:missense and aa_range:1843,2002 and type:splice_site type:nonframeshift and aa_range:1104,1481 and type:missense and ExAC_Freq<0.01 type:nonframeshift and aa_range:1843,2002 and WT1 type:frameshift type:splice_site XRCC2 type:frameshift type:splice_site type:missense and ExAC_Freq<0.01 ZRSR2 type:frameshift type:splice_site MIR142 ExAC_Freq<
39 Supplementary Table S7 - Patient- and variant-level data in the entire cohort. UPN Chr Position_Start Ref Alt Gene cdna AA Variant_Type VAF Depth C T ASXL1 c.c1249t p.r417x Nonsense C T ASXL1 c.c1249t p.r417x Nonsense A ASXL1 c insa p.y425fs fs-ins C T ASXL1 c.c1282t p.q428x Nonsense G ASXL1 c insg p.v435fs fs-ins C T ASXL1 c.c1549t p.q517x Nonsense G T ASXL1 c.g1696t p.e566x Nonsense A G ASXL1 c a>g Splice_Site G T ASXL1 c g>t Splice_Site G A ASXL1 c g>a Splice_Site C G ASXL1 c.c1730g p.s577x Nonsense C T ASXL1 c.c1762t p.q588x Nonsense C T ASXL1 c.c1762t p.q588x Nonsense C T ASXL1 c.c1762t p.q588x Nonsense A ASXL1 c insa p.y591-q592delinsx Nonsense A ASXL1 c insa p.y591-q592delinsx Nonsense A ASXL1 c insa p.y591-q592delinsx Nonsense A ASXL1 c insa p.y591-q592delinsx Nonsense C G ASXL1 c.c1773g p.y591x Nonsense C A ASXL1 c.c1773a p.y591x Nonsense C A ASXL1 c.c1773a p.y591x Nonsense C G ASXL1 c.c1773g p.y591x Nonsense C A ASXL1 c.c1773a p.y591x Nonsense C A ASXL1 c.c1773a p.y591x Nonsense C T ASXL1 c.c1774t p.q592x Nonsense C T ASXL1 c.c1774t p.q592x Nonsense C T ASXL1 c.c1774t p.q592x Nonsense G T ASXL1 c.g1804t p.e602x Nonsense TA ASXL1 c insta p.d616fs fs-ins TAAT ASXL1 c instaat p.d616-i617delinsdx Nonsense T ASXL1 c inst p.i617fs fs-ins A T ASXL1 c.a1852t p.k618x Nonsense C ASXL1 c insc p.a619fs fs-ins ASXL1 c del p del fs-del C T ASXL1 c.c1867t p.q623x Nonsense ASXL1 c del p del fs-del G ASXL1 c insg p.r625fs fs-ins ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del ASXL1 c del p del fs-del A ASXL1 c insa p.h633fs fs-ins A T ASXL1 c.a1900t p.r634x Nonsense G T ASXL1 c.g1903t p.e635x Nonsense ASXL1 c del p del fs-del ASXL1 c del p del fs-del A - ASXL1 c.1926dela p.g642fs fs-del G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins G ASXL1 c insg p.g642fs fs-ins
7/12/2016 TESTING. Objectives. New Directions in Aplastic Anemia: What's on the Horizon? Better way to evaluate clonal evolution?
 New Directions in Aplastic Anemia: What's on the Horizon? Amy E. DeZern, MD; MHS Assistant Professor Hematologic Malignancies Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins Baltimore, MD Objectives
New Directions in Aplastic Anemia: What's on the Horizon? Amy E. DeZern, MD; MHS Assistant Professor Hematologic Malignancies Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins Baltimore, MD Objectives
Supplementary Information
 Supplementary Information Table of Contents Supplementary methods... 2 Figure S1 - Variable DNA yield proportional to bone marrow aspirate cellularity.... 3 Figure S2 - Mutations by clinical ontogeny group....
Supplementary Information Table of Contents Supplementary methods... 2 Figure S1 - Variable DNA yield proportional to bone marrow aspirate cellularity.... 3 Figure S2 - Mutations by clinical ontogeny group....
Next Generation Sequencing in Haematological Malignancy: A European Perspective. Wolfgang Kern, Munich Leukemia Laboratory
 Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ
 Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ G E NETIC T E CHNOLOGIST MEDICAL G E NETICS, CARDIFF To Cover Background to the project Choice of panel Validation process Genes on panel, Protocol
Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ G E NETIC T E CHNOLOGIST MEDICAL G E NETICS, CARDIFF To Cover Background to the project Choice of panel Validation process Genes on panel, Protocol
The Center for PERSONALIZED DIAGNOSTICS
 The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
Laboratory Service Report
 Client C7028846-DLP Rochester Rochester, N 55901 Specimen Type Peripheral blood CR PDF Report available at: https://test.mmlaccess.com/reports/c7028846-zwselwql7p.ashx Indication for Test DS CR Pathogenic
Client C7028846-DLP Rochester Rochester, N 55901 Specimen Type Peripheral blood CR PDF Report available at: https://test.mmlaccess.com/reports/c7028846-zwselwql7p.ashx Indication for Test DS CR Pathogenic
No Other Company Discloses Higher Transplant Survival Rate. Infusions For Emerging Treatments. Date of Use. Recipient Age (yrs)
 Units Used In Transplants/Infusions No Other Company Discloses Higher Transplant Survival Rate Family Banking Provides Exclusive Access To Emerging Treatments With Your Own Cells 175 85% Type 1 Diabetes
Units Used In Transplants/Infusions No Other Company Discloses Higher Transplant Survival Rate Family Banking Provides Exclusive Access To Emerging Treatments With Your Own Cells 175 85% Type 1 Diabetes
New Directions in Aplastic Anemia
 11//15 New Directions in Aplastic Anemia ANDREW C. DI ETZ, MD, MSCR PEDIATRIC BLOOD AND MARROW TRANSPLANTATION CHILDREN S HOSPITAL LOS ANGELES, UNIVERSITY OF SOUTHERN CALIFORNIA Outline Ø Changes in Immunosuppressive
11//15 New Directions in Aplastic Anemia ANDREW C. DI ETZ, MD, MSCR PEDIATRIC BLOOD AND MARROW TRANSPLANTATION CHILDREN S HOSPITAL LOS ANGELES, UNIVERSITY OF SOUTHERN CALIFORNIA Outline Ø Changes in Immunosuppressive
The Evolving Role of Transplantation for MPN
 The Evolving Role of Transplantation for MPN (PMF, PV, ET) H.Joachim Deeg MD Fred Hutchinson Cancer Research Center, University of Washington, Seattle WA, SCCA jdeeg@fhcrc.org 10 th J. Niblack Conference
The Evolving Role of Transplantation for MPN (PMF, PV, ET) H.Joachim Deeg MD Fred Hutchinson Cancer Research Center, University of Washington, Seattle WA, SCCA jdeeg@fhcrc.org 10 th J. Niblack Conference
NeoTYPE Cancer Profiles
 NeoTYPE Cancer Profiles Multimethod Analysis of 25+ Hematologic Diseases and Solid Tumors Anatomic Pathology FISH Molecular The next generation of diagnostic, prognostic, and therapeutic assessment NeoTYPE
NeoTYPE Cancer Profiles Multimethod Analysis of 25+ Hematologic Diseases and Solid Tumors Anatomic Pathology FISH Molecular The next generation of diagnostic, prognostic, and therapeutic assessment NeoTYPE
Laboratory Service Report
 Specimen Type Peripheral blood CR PDF Report available at: https://test.mmlaccess.com/reports/c7028846-ih2xuglwpq.ashx Indication for Test DS CR Pathogenic utations Detected CR 1. JAK2: c.1849g>t;p.val617phe
Specimen Type Peripheral blood CR PDF Report available at: https://test.mmlaccess.com/reports/c7028846-ih2xuglwpq.ashx Indication for Test DS CR Pathogenic utations Detected CR 1. JAK2: c.1849g>t;p.val617phe
Out-Patient Billing CPT Codes
 Out-Patient Billing CPT Codes Updated Date: August 3, 08 Client Billed Molecular Tests HPV DNA Tissue Testing 8764 No Medicare Billed - Molecular Tests NeoARRAY NeoARRAY SNP/Cytogenetic No 89 NeoLAB NeoLAB
Out-Patient Billing CPT Codes Updated Date: August 3, 08 Client Billed Molecular Tests HPV DNA Tissue Testing 8764 No Medicare Billed - Molecular Tests NeoARRAY NeoARRAY SNP/Cytogenetic No 89 NeoLAB NeoLAB
Next generation sequencing analysis - A UK perspective. Nicholas Lea
 Next generation sequencing analysis - A UK perspective Nicholas Lea King s HMDC LMH is part of an integrated pathology service at King s Haematological Malignancy Diagnostic Centre (HMDC) HMDC serves population
Next generation sequencing analysis - A UK perspective Nicholas Lea King s HMDC LMH is part of an integrated pathology service at King s Haematological Malignancy Diagnostic Centre (HMDC) HMDC serves population
NeoTYPE Cancer Profiles
 NeoTYPE Cancer Profiles 30+ Multimethod Assays for Hematologic Diseases and Solid Tumors Molecular FISH Anatomic Pathology The next generation of diagnostic, prognostic, and therapeutic assessment What
NeoTYPE Cancer Profiles 30+ Multimethod Assays for Hematologic Diseases and Solid Tumors Molecular FISH Anatomic Pathology The next generation of diagnostic, prognostic, and therapeutic assessment What
Supplementary Figure 1
 Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
The preclinical efficacy of a novel telomerase inhibitor, imetelstat, in AML: A randomized trial in patient-derived xenografts
 The preclinical efficacy of a novel telomerase inhibitor, imetelstat, in AML: A randomized trial in patient-derived xenografts Claudia Bruedigam, Ph.D Gordon and Jessie Gilmour Leukaemia Research Laboratory
The preclinical efficacy of a novel telomerase inhibitor, imetelstat, in AML: A randomized trial in patient-derived xenografts Claudia Bruedigam, Ph.D Gordon and Jessie Gilmour Leukaemia Research Laboratory
Please Silence Your Cell Phones. Thank You
 Please Silence Your Cell Phones Thank You Utility of NGS and Comprehensive Genomic Profiling in Hematopathology Practice Maria E. Arcila M.D. Memorial Sloan Kettering Cancer Center New York, NY Disclosure
Please Silence Your Cell Phones Thank You Utility of NGS and Comprehensive Genomic Profiling in Hematopathology Practice Maria E. Arcila M.D. Memorial Sloan Kettering Cancer Center New York, NY Disclosure
Introduction of an NGS gene panel into the Haemato-Oncology MPN service
 Introduction of an NGS gene panel into the Haemato-Oncology MPN service Dr. Anna Skowronska, Dr Jane Bryon, Dr Samuel Clokie, Dr Yvonne Wallis and Professor Mike Griffiths West Midlands Regional Genetics
Introduction of an NGS gene panel into the Haemato-Oncology MPN service Dr. Anna Skowronska, Dr Jane Bryon, Dr Samuel Clokie, Dr Yvonne Wallis and Professor Mike Griffiths West Midlands Regional Genetics
Shall young patients with severe aplastic anemia without donors receive BMT from alternative source of HCT? Elias Hallack Atta, MD, PhD
 Shall young patients with severe aplastic anemia without donors receive BMT from alternative source of HCT? Elias Hallack Atta, MD, PhD Declaração de Conflito de Interesse Declaro que possuo conflito de
Shall young patients with severe aplastic anemia without donors receive BMT from alternative source of HCT? Elias Hallack Atta, MD, PhD Declaração de Conflito de Interesse Declaro que possuo conflito de
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance
Examining Genetics and Genomics of Acute Myeloid Leukemia in 2017
 Examining Genetics and Genomics of Acute Myeloid Leukemia in 2017 Elli Papaemmanuil, PhD Memorial Sloan Kettering Cancer Center New York, New York, United States Today s Talk Cancer genome introduction
Examining Genetics and Genomics of Acute Myeloid Leukemia in 2017 Elli Papaemmanuil, PhD Memorial Sloan Kettering Cancer Center New York, New York, United States Today s Talk Cancer genome introduction
Management of Myelodysplastic Syndromes
 Management of Myelodysplastic Syndromes Peter L. Greenberg, MD Stanford Cancer Institute Myelodysplastic Syndromes: Clinical & Molecular Advances for Disease Classification and Prognostication MDSs: A
Management of Myelodysplastic Syndromes Peter L. Greenberg, MD Stanford Cancer Institute Myelodysplastic Syndromes: Clinical & Molecular Advances for Disease Classification and Prognostication MDSs: A
ADVANCES IN THE MANAGEMENT OF MYELODYSPLASTIC SYNDROMES
 ADVANCES IN THE MANAGEMENT OF MYELODYSPLASTIC SYNDROMES Corey Cutler, MD MPH FRCPC Associate Professor of Medicine, Harvard Medical School Dana-Farber Cancer Institute, Boston, MA HCT Outcomes - MDS 2001-2011
ADVANCES IN THE MANAGEMENT OF MYELODYSPLASTIC SYNDROMES Corey Cutler, MD MPH FRCPC Associate Professor of Medicine, Harvard Medical School Dana-Farber Cancer Institute, Boston, MA HCT Outcomes - MDS 2001-2011
Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH )
 Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH2017-0314) Habibe Kurt, Joseph D. Khoury, Carlos E. Bueso-Ramos, Jeffrey L. Jorgensen, Guilin Tang, L. Jeffrey Medeiros, and
Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH2017-0314) Habibe Kurt, Joseph D. Khoury, Carlos E. Bueso-Ramos, Jeffrey L. Jorgensen, Guilin Tang, L. Jeffrey Medeiros, and
ASBMT MDS/MPN Update Sunil Abhyankar, MD
 ASBMT MDS/MPN Update Sunil Abhyankar, MD Professor of Medicine Medical Director, Pheresis and Cell Processing Division of Hematologic Malignancies and Cellular Therapeutics Department of Internal Medicine
ASBMT MDS/MPN Update Sunil Abhyankar, MD Professor of Medicine Medical Director, Pheresis and Cell Processing Division of Hematologic Malignancies and Cellular Therapeutics Department of Internal Medicine
Acute leukemia and myelodysplastic syndromes
 11/01/2012 Post-ASH meeting 1 Acute leukemia and myelodysplastic syndromes Peter Vandenberghe Centrum Menselijke Erfelijkheid & Afdeling Hematologie, UZ Leuven 11/01/2012 Post-ASH meeting 2 1. Acute myeloid
11/01/2012 Post-ASH meeting 1 Acute leukemia and myelodysplastic syndromes Peter Vandenberghe Centrum Menselijke Erfelijkheid & Afdeling Hematologie, UZ Leuven 11/01/2012 Post-ASH meeting 2 1. Acute myeloid
August 17, Dear Valued Client:
 August 7, 08 Re: CMS Announces 6-Month Period of Enforcement Discretion for Laboratory Date of Service Exception Policy Under the Medicare Clinical Laboratory Fee Schedule (the 4 Day Rule ) Dear Valued
August 7, 08 Re: CMS Announces 6-Month Period of Enforcement Discretion for Laboratory Date of Service Exception Policy Under the Medicare Clinical Laboratory Fee Schedule (the 4 Day Rule ) Dear Valued
Concomitant WT1 mutations predicted poor prognosis in CEBPA double-mutated acute myeloid leukemia
 Concomitant WT1 mutations predicted poor prognosis in CEBPA double-mutated acute myeloid leukemia Feng-Ming Tien, Hsin-An Hou, Jih-Luh Tang, Yuan-Yeh Kuo, Chien-Yuan Chen, Cheng-Hong Tsai, Ming Yao, Chi-Cheng
Concomitant WT1 mutations predicted poor prognosis in CEBPA double-mutated acute myeloid leukemia Feng-Ming Tien, Hsin-An Hou, Jih-Luh Tang, Yuan-Yeh Kuo, Chien-Yuan Chen, Cheng-Hong Tsai, Ming Yao, Chi-Cheng
SUPPLEMENTAL APPENDIX METZELER ET AL.: SPECTRUM AND PROGNOSTIC RELEVANCE OF DRIVER GENE MUTATIONS IN ACUTE MYELOID LEUKEMIA
 SUPPLEMENTAL APPENDIX TO METZELER ET AL.: SPECTRUM AND PROGNOSTIC RELEVANCE OF DRIVER GENE MUTATIONS IN ACUTE MYELOID LEUKEMIA SUPPLEMENTAL METHODS Treatment protocols In the AMLCG-1999 trial 1-3 (clinicaltrials.gov
SUPPLEMENTAL APPENDIX TO METZELER ET AL.: SPECTRUM AND PROGNOSTIC RELEVANCE OF DRIVER GENE MUTATIONS IN ACUTE MYELOID LEUKEMIA SUPPLEMENTAL METHODS Treatment protocols In the AMLCG-1999 trial 1-3 (clinicaltrials.gov
Published Ahead of Print on April 14, 2016, as doi: /haematol Copyright 2016 Ferrata Storti Foundation.
 Published Ahead of Print on April 14, 2016, as doi:10.3324/haematol.2016.143214. Copyright 2016 Ferrata Storti Foundation. Immunohistochemical pattern of p53 is a measure of TP53 mutation burden and adverse
Published Ahead of Print on April 14, 2016, as doi:10.3324/haematol.2016.143214. Copyright 2016 Ferrata Storti Foundation. Immunohistochemical pattern of p53 is a measure of TP53 mutation burden and adverse
Mutational Impact on Diagnostic and Prognostic Evaluation of MDS
 Mutational Impact on Diagnostic and Prognostic Evaluation of MDS Elsa Bernard, PhD Papaemmanuil Lab, Computational Oncology, MSKCC MDS Foundation ASH 2018 Symposium Disclosure Research funds provided by
Mutational Impact on Diagnostic and Prognostic Evaluation of MDS Elsa Bernard, PhD Papaemmanuil Lab, Computational Oncology, MSKCC MDS Foundation ASH 2018 Symposium Disclosure Research funds provided by
Samples Available for Recipient Only. Samples Available for Recipient and Donor
 Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
HCT for Myelofibrosis
 Allogeneic HSCT for MDS and Myelofibrosis Sunil Abhyankar, MD Professor Medicine, Medical Director, Pheresis and Cell Processing University of Kansas Hospital BMT Program April 27 th, 213 HCT for Myelofibrosis
Allogeneic HSCT for MDS and Myelofibrosis Sunil Abhyankar, MD Professor Medicine, Medical Director, Pheresis and Cell Processing University of Kansas Hospital BMT Program April 27 th, 213 HCT for Myelofibrosis
Chapter Two Incidence & prevalence
 Chapter Two Incidence & prevalence Science is the observation of things possible, whether present or past. Prescience is the knowledge of things which may come to pass, though but slowly. LEONARDO da Vinci
Chapter Two Incidence & prevalence Science is the observation of things possible, whether present or past. Prescience is the knowledge of things which may come to pass, though but slowly. LEONARDO da Vinci
Samples Available for Recipient and Donor
 Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
Samples Available for Recipient Only. Samples Available for Recipient and Donor
 Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
Molecular. Oncology & Pathology. Diagnostic, Prognostic, Therapeutic, and Predisposition Tests in Precision Medicine. Liquid Biopsy.
 Molecular Oncology & Pathology Hereditary Cancer Somatic Cancer Liquid Biopsy Next-Gen Sequencing qpcr Sanger Sequencing Diagnostic, Prognostic, Therapeutic, and Predisposition Tests in Precision Medicine
Molecular Oncology & Pathology Hereditary Cancer Somatic Cancer Liquid Biopsy Next-Gen Sequencing qpcr Sanger Sequencing Diagnostic, Prognostic, Therapeutic, and Predisposition Tests in Precision Medicine
Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation
 The new england journal of medicine Original Article Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation R.C. Lindsley, W. Saber, B.G. Mar, R. Redd, T. Wang, M.D. Haagenson,
The new england journal of medicine Original Article Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation R.C. Lindsley, W. Saber, B.G. Mar, R. Redd, T. Wang, M.D. Haagenson,
Disclosures. Dr. Hall is a paid consultant to the American College of Surgeons (ACS) as Associate Director of ACS-NSQIP
 Does Routine Drainage of the Operative Bed following Elective Distal Pancreatectomy reduce Complications? An Analysis of the ACS-NSQIP Pancreatectomy Demonstration Project Stephen W. Behrman, MD 1, Ben
Does Routine Drainage of the Operative Bed following Elective Distal Pancreatectomy reduce Complications? An Analysis of the ACS-NSQIP Pancreatectomy Demonstration Project Stephen W. Behrman, MD 1, Ben
Nature Genetics: doi: /ng Supplementary Figure 1. Bone marrow morphology in III-2 of family A.
 Supplementary Figure 1 Bone marrow morphology in III-2 of family A. (a) Pre-transplant peripheral blood smears from patient III-2 illustrating hypogranulated neutrophils (left) and hypolobated neutrophils
Supplementary Figure 1 Bone marrow morphology in III-2 of family A. (a) Pre-transplant peripheral blood smears from patient III-2 illustrating hypogranulated neutrophils (left) and hypolobated neutrophils
Supplemental Material. The new provisional WHO entity RUNX1 mutated AML shows specific genetics without prognostic influence of dysplasia
 Supplemental Material The new provisional WHO entity RUNX1 mutated AML shows specific genetics without prognostic influence of dysplasia Torsten Haferlach, 1 Anna Stengel, 1 Sandra Eckstein, 1 Karolína
Supplemental Material The new provisional WHO entity RUNX1 mutated AML shows specific genetics without prognostic influence of dysplasia Torsten Haferlach, 1 Anna Stengel, 1 Sandra Eckstein, 1 Karolína
February 9, The Vice President Old Executive Office Building Washington, DC Dear Mr. Vice President:
 February 9, 2016 The Vice President Old Executive Office Building Washington, DC 20501 Dear Mr. Vice President: As the nation s leading organizations focused on promoting cancer research and prevention,
February 9, 2016 The Vice President Old Executive Office Building Washington, DC 20501 Dear Mr. Vice President: As the nation s leading organizations focused on promoting cancer research and prevention,
SESSION 1 Reactive cytopenia and dysplasia
 SESSION 1 Reactive cytopenia and dysplasia Falko Fend, Tübingen & Alexandar Tzankov, Basel 1 Disclosure of speaker s interests (Potential) conflict of interest none Potentially relevant company relationships
SESSION 1 Reactive cytopenia and dysplasia Falko Fend, Tübingen & Alexandar Tzankov, Basel 1 Disclosure of speaker s interests (Potential) conflict of interest none Potentially relevant company relationships
Mayo alliance prognostic system for mastocytosis: clinical and hybrid clinical-molecular models
 REGULAR ARTICLE Mayo alliance prognostic system for mastocytosis: clinical and hybrid clinical-molecular models Animesh Pardanani, 1 Sahrish Shah, 1 Francesco Mannelli, 2 Yoseph C. Elala, 1 Paola Guglielmelli,
REGULAR ARTICLE Mayo alliance prognostic system for mastocytosis: clinical and hybrid clinical-molecular models Animesh Pardanani, 1 Sahrish Shah, 1 Francesco Mannelli, 2 Yoseph C. Elala, 1 Paola Guglielmelli,
DISCLOSURE Luca Malcovati, MD. No financial relationships to disclose
 ICUS, CCUS and CHIP Luca Malcovati, MD Department of Molecular Medicine, University of Pavia Medical School, & Department of Hematology Oncology, IRCCS Policlinico S. Matteo Foundation, Pavia, Italy DISCLOSURE
ICUS, CCUS and CHIP Luca Malcovati, MD Department of Molecular Medicine, University of Pavia Medical School, & Department of Hematology Oncology, IRCCS Policlinico S. Matteo Foundation, Pavia, Italy DISCLOSURE
2012 Medicaid and Partnership Chart
 2012 Medicaid and Chart or Alabama $525,000.00 $4,800.00 Minimum: 25,000.00 Alaska $525,000.00 Depends on area of state; Minimum: $113,640 $10,000 in Anchorage $1,656 Minimum:$1838.75 Maximum:$2,841 Minimum:
2012 Medicaid and Chart or Alabama $525,000.00 $4,800.00 Minimum: 25,000.00 Alaska $525,000.00 Depends on area of state; Minimum: $113,640 $10,000 in Anchorage $1,656 Minimum:$1838.75 Maximum:$2,841 Minimum:
Haploidentical Transplantation: The Answer to our Donor Problems? Mary M. Horowitz, MD, MS CIBMTR, Medical College of Wisconsin January 2017
 Haploidentical Transplantation: The Answer to our Donor Problems? Mary M. Horowitz, MD, MS CIBMTR, Medical College of Wisconsin January 2017 Allogeneic Transplant Recipients in the US, by Donor Type 9000
Haploidentical Transplantation: The Answer to our Donor Problems? Mary M. Horowitz, MD, MS CIBMTR, Medical College of Wisconsin January 2017 Allogeneic Transplant Recipients in the US, by Donor Type 9000
Frequency(%) KRAS G12 KRAS G13 KRAS A146 KRAS Q61 KRAS K117N PIK3CA H1047 PIK3CA E545 PIK3CA E542K PIK3CA Q546. EGFR exon19 NFS-indel EGFR L858R
 Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Cirrhosis and Liver Cancer Mortality in the United States : An Observational Study Supplementary Material
 Cirrhosis and Liver Cancer Mortality in the United States 1999-2016: An Observational Study Supplementary Material Elliot B. Tapper MD (1,2) and Neehar D Parikh MD MS (1,2) 1. Division of Gastroenterology
Cirrhosis and Liver Cancer Mortality in the United States 1999-2016: An Observational Study Supplementary Material Elliot B. Tapper MD (1,2) and Neehar D Parikh MD MS (1,2) 1. Division of Gastroenterology
IN ALLOGENEIC HEMATOPOIETIC CELL TRANSPLANT PATIENTS WITH BRINCIDOFOVIR: FINAL 36 WEEK RESULTS FROM THE ADVISE TRIAL
 TREATMENT OF ADENOVIRUS (AdV) INFECTION IN ALLOGENEIC HEMATOPOIETIC CELL TRANSPLANT PATIENTS WITH BRINCIDOFOVIR: FINAL 36 WEEK RESULTS FROM THE ADVISE TRIAL Vinod K. Prasad, MD, FRCP 1, Genovefa A. Papanicolaou,
TREATMENT OF ADENOVIRUS (AdV) INFECTION IN ALLOGENEIC HEMATOPOIETIC CELL TRANSPLANT PATIENTS WITH BRINCIDOFOVIR: FINAL 36 WEEK RESULTS FROM THE ADVISE TRIAL Vinod K. Prasad, MD, FRCP 1, Genovefa A. Papanicolaou,
Acute Myeloid Leukemia Progress at last
 Acute Myeloid Leukemia Progress at last Bruno C. Medeiros, MD September 9, 217 Introduction Mechanisms of leukemogenesis Emerging therapies in AML Previously untreated AML Relapsed and refractory patients
Acute Myeloid Leukemia Progress at last Bruno C. Medeiros, MD September 9, 217 Introduction Mechanisms of leukemogenesis Emerging therapies in AML Previously untreated AML Relapsed and refractory patients
ADRL Advanced Diagnostics Research Laboratory
 ADRL Advanced Diagnostics Research Laboratory John DeCoteau, MD FRCP Department of Pathology, Division of Hematopathology University of Saskatchewan Saskatchewan Cancer Agency ADRL Project Objectives New
ADRL Advanced Diagnostics Research Laboratory John DeCoteau, MD FRCP Department of Pathology, Division of Hematopathology University of Saskatchewan Saskatchewan Cancer Agency ADRL Project Objectives New
Myeloablative and Reduced Intensity Conditioning for HSCT Annalisa Ruggeri, MD, Hôpital Saint Antoine Eurocord- Hôpital Saint Louis, Paris
 Myeloablative and Reduced Intensity Conditioning for HSCT Annalisa Ruggeri, MD, Hôpital Saint Antoine Eurocord- Hôpital Saint Louis, Paris 18th ESH - EBMT Training Course on HSCT 8-10 May 2014, Vienna,
Myeloablative and Reduced Intensity Conditioning for HSCT Annalisa Ruggeri, MD, Hôpital Saint Antoine Eurocord- Hôpital Saint Louis, Paris 18th ESH - EBMT Training Course on HSCT 8-10 May 2014, Vienna,
Acute Myeloid Leukemia with RUNX1 and Several Co-mutations
 Case SH2017-0281 Acute Myeloid Leukemia with RUNX1 and Several Co-mutations James Bauer, MD, PhD David Yang, MD Erik Ranheim, MD, PhD Catherine Leith, MB, Bchir Clinical History Chief Complaint: 72 year
Case SH2017-0281 Acute Myeloid Leukemia with RUNX1 and Several Co-mutations James Bauer, MD, PhD David Yang, MD Erik Ranheim, MD, PhD Catherine Leith, MB, Bchir Clinical History Chief Complaint: 72 year
Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope
 Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope National Medical Center Disclosures I have no disclosures
Patricia Aoun MD, MPH Professor and Vice-Chair for Clinical Affairs Medical Director, Clinical Laboratories Department of Pathology City of Hope National Medical Center Disclosures I have no disclosures
West Midlands Regional Genetics Laboratory
 West Midlands Regional Genetics Laboratory Haemato-oncology service update letter October 2017 Dear colleagues, We are writing to outline the latest developments to our service, aiming to support the management
West Midlands Regional Genetics Laboratory Haemato-oncology service update letter October 2017 Dear colleagues, We are writing to outline the latest developments to our service, aiming to support the management
Fluxion Biosciences and Swift Biosciences Somatic variant detection from liquid biopsy samples using targeted NGS
 APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
Molecular profiling in confirming the diagnosis of early myelodysplastic syndrome
 Molecular profiling of early MDS Hematopathology - March 2016 Article Molecular profiling in confirming the diagnosis of early myelodysplastic syndrome Maya Thangavelu 1,*, Ryan Olson 2, Li Li 2, Wanlong
Molecular profiling of early MDS Hematopathology - March 2016 Article Molecular profiling in confirming the diagnosis of early myelodysplastic syndrome Maya Thangavelu 1,*, Ryan Olson 2, Li Li 2, Wanlong
Bone Marrow Transplantation in Myelodysplastic Syndromes. An overview for the Myelodysplasia Support Group of Ottawa
 Bone Marrow Transplantation in Myelodysplastic Syndromes An overview for the Myelodysplasia Support Group of Ottawa Objectives Provide brief review of marrow failure Re emphasize the importance of predictions
Bone Marrow Transplantation in Myelodysplastic Syndromes An overview for the Myelodysplasia Support Group of Ottawa Objectives Provide brief review of marrow failure Re emphasize the importance of predictions
CGC myeloid malignancy working group updates. Xinjie Xu & Rashmi Kanagal-Shamanna
 CGC myeloid malignancy working group updates Xinjie Xu & Rashmi Kanagal-Shamanna 8-9-2016 Group members Gordana Raca Children's Hospital Los Angeles Xinjie Xu University of Utah ARUP Laboratories Rashmi
CGC myeloid malignancy working group updates Xinjie Xu & Rashmi Kanagal-Shamanna 8-9-2016 Group members Gordana Raca Children's Hospital Los Angeles Xinjie Xu University of Utah ARUP Laboratories Rashmi
ASBMT MDS/MPN UPDATE
 ASBMT MDS/MPN UPDATE Sunil Abhyankar, MD Professor of Medicine Medical Director, Pheresis and Cell Processing Division of Hematologic Malignancies and Cellular Therapeutics Department of Internal Medicine
ASBMT MDS/MPN UPDATE Sunil Abhyankar, MD Professor of Medicine Medical Director, Pheresis and Cell Processing Division of Hematologic Malignancies and Cellular Therapeutics Department of Internal Medicine
Acute myeloid leukemia: prognosis and treatment. Dimitri A. Breems, MD, PhD Internist-Hematoloog Ziekenhuis Netwerk Antwerpen Campus Stuivenberg
 Acute myeloid leukemia: prognosis and treatment Dimitri A. Breems, MD, PhD Internist-Hematoloog Ziekenhuis Netwerk Antwerpen Campus Stuivenberg Patient Female, 39 years History: hypothyroidism Present:
Acute myeloid leukemia: prognosis and treatment Dimitri A. Breems, MD, PhD Internist-Hematoloog Ziekenhuis Netwerk Antwerpen Campus Stuivenberg Patient Female, 39 years History: hypothyroidism Present:
Volunteering in Oklahoma City, OK
 6/17/2010 Oklahoma City Profile - Volunteering in information on volunteering and civic engagement Volunteering in Oklahoma City, OK Statistics for this area were collected within the Oklahoma City Metropolitan
6/17/2010 Oklahoma City Profile - Volunteering in information on volunteering and civic engagement Volunteering in Oklahoma City, OK Statistics for this area were collected within the Oklahoma City Metropolitan
Genomic Medicine: What every pathologist needs to know
 Genomic Medicine: What every pathologist needs to know Stephen P. Ethier, Ph.D. Professor, Department of Pathology and Laboratory Medicine, MUSC Director, MUSC Center for Genomic Medicine Genomics and
Genomic Medicine: What every pathologist needs to know Stephen P. Ethier, Ph.D. Professor, Department of Pathology and Laboratory Medicine, MUSC Director, MUSC Center for Genomic Medicine Genomics and
The National Cancer Institute Cancer Centers Program. The Cancer Centers Program of the National Cancer Institute (NCI) supports major
 CANCER FACTS N a t i o n a l C a n c e r I n s t i t u t e N a t i o n a l I n s t i t u t e s o f H e a l t h D e p a r t m e n t o f H e a l t h a n d H u m a n S e r v i c e s The National Cancer Institute
CANCER FACTS N a t i o n a l C a n c e r I n s t i t u t e N a t i o n a l I n s t i t u t e s o f H e a l t h D e p a r t m e n t o f H e a l t h a n d H u m a n S e r v i c e s The National Cancer Institute
SUPPLEMENTARY FIG. S3. Kaplan Meier survival analysis followed with log-rank test of de novo acute myeloid leukemia patients selected by age <60, IA
 Supplementary Data Supplementary Appendix A: Treatment Protocols Treatment protocols of 123 cases patients were treated with the protocols as follows: 110 patients received standard DA (daunorubicin 45
Supplementary Data Supplementary Appendix A: Treatment Protocols Treatment protocols of 123 cases patients were treated with the protocols as follows: 110 patients received standard DA (daunorubicin 45
What s new in Blood and Marrow Transplant? Saar Gill, MD PhD Jan 22, 2016
 What s new in Blood and Marrow Transplant? Saar Gill, MD PhD Jan 22, 2016 Division of Hematology-Oncology University of Pennsylvania Perelman School of Medicine 1 Who should be transplanted and how? Updates
What s new in Blood and Marrow Transplant? Saar Gill, MD PhD Jan 22, 2016 Division of Hematology-Oncology University of Pennsylvania Perelman School of Medicine 1 Who should be transplanted and how? Updates
Myelodysplastic syndromes Impact of Biology. Lionel Adès Hopital Saint Louis Groupe Francophone des SMD. Épidémiologie
 Myelodysplastic syndromes Impact of Biology Lionel Adès Hopital Saint Louis Groupe Francophone des SMD Épidémiologie Incidence : 3 à 6 / 100 000 hab. / An Prédomine chez les sujets âgés Augmentation de
Myelodysplastic syndromes Impact of Biology Lionel Adès Hopital Saint Louis Groupe Francophone des SMD Épidémiologie Incidence : 3 à 6 / 100 000 hab. / An Prédomine chez les sujets âgés Augmentation de
Nature Medicine: doi: /nm.4439
 Figure S1. Overview of the variant calling and verification process. This figure expands on Fig. 1c with details of verified variants identification in 547 additional validation samples. Somatic variants
Figure S1. Overview of the variant calling and verification process. This figure expands on Fig. 1c with details of verified variants identification in 547 additional validation samples. Somatic variants
Ivosidenib (IVO; AG-120) in mutant IDH1 relapsed/refractory acute myeloid leukemia (R/R AML): Results of a phase 1 study
 7000 Ivosidenib (IVO; AG-120) in mutant IDH1 relapsed/refractory acute myeloid leukemia (R/R AML): Results of a phase 1 study Daniel A Pollyea 1, Courtney D DiNardo 2, Stéphane de Botton 3, Eytan M Stein
7000 Ivosidenib (IVO; AG-120) in mutant IDH1 relapsed/refractory acute myeloid leukemia (R/R AML): Results of a phase 1 study Daniel A Pollyea 1, Courtney D DiNardo 2, Stéphane de Botton 3, Eytan M Stein
Precision Medicine and Molecular Testing.
 Precision Medicine and Molecular Testing. David A. Sallman, MD Assistant Member Department of Malignant Hematology Moffitt Cancer Center david.sallman@moffitt.org Disclosures Research funding for Celgene
Precision Medicine and Molecular Testing. David A. Sallman, MD Assistant Member Department of Malignant Hematology Moffitt Cancer Center david.sallman@moffitt.org Disclosures Research funding for Celgene
PDX Tumor Biology Pla0orms for Drug Advancement Neal Goodwin, Ph.D. Vice President Corporate Research & Development
 PDX Tumor Biology Pla0orms for Drug Advancement Neal Goodwin, Ph.D. Vice President Corporate Research & Development ngoodwin@championsoncology.com +1-530-392-2741 Champions Oncology: Global provider of
PDX Tumor Biology Pla0orms for Drug Advancement Neal Goodwin, Ph.D. Vice President Corporate Research & Development ngoodwin@championsoncology.com +1-530-392-2741 Champions Oncology: Global provider of
Myelodysplastic syndromes and the new WHO 2016 classification
 Myelodysplastic syndromes and the new WHO 2016 classification 32nd General Annual Meeting of the Belgian Hematology Society 10-11 February 2017 Gregor Verhoef, Departement of Hematology, University Hospital
Myelodysplastic syndromes and the new WHO 2016 classification 32nd General Annual Meeting of the Belgian Hematology Society 10-11 February 2017 Gregor Verhoef, Departement of Hematology, University Hospital
Allogeneic Hematopoietic Cell Transplantation for Adult T Cell Acute Lymphoblastic Leukemia
 Allogeneic Hematopoietic Cell Transplantation for Adult T Cell Acute Lymphoblastic Leukemia Betty Ky Hamilton, Cleveland Clinic Lisa Rybicki, Cleveland Clinic Donna Abounader, Cleveland Clinic Kehinde
Allogeneic Hematopoietic Cell Transplantation for Adult T Cell Acute Lymphoblastic Leukemia Betty Ky Hamilton, Cleveland Clinic Lisa Rybicki, Cleveland Clinic Donna Abounader, Cleveland Clinic Kehinde
AAll s well that ends well; still the fine s the crown; Whate er the course, the end is the renown. WILLIAM SHAKESPEARE, All s Well That Ends Well
 AAll s well that ends well; still the fine s the crown; Whate er the course, the end is the renown. WILLIAM SHAKESPEARE, All s Well That Ends Well mthree TrEATMENT MODALITIES 7 ž 21 ATLAS OF ESRD IN THE
AAll s well that ends well; still the fine s the crown; Whate er the course, the end is the renown. WILLIAM SHAKESPEARE, All s Well That Ends Well mthree TrEATMENT MODALITIES 7 ž 21 ATLAS OF ESRD IN THE
Overview. Methods 9/11/2017. Next Generation Sequencing and Precision Medicine in Hematological Malignancies. Genotyping in hematology
 Overview Next Generation Sequencing and Precision Medicine in Hematological Malignancies Sharathkumar Bhagavathi, MD University of Iowa Carver College of Medicine NGS as a genotyping platform in hematopathology
Overview Next Generation Sequencing and Precision Medicine in Hematological Malignancies Sharathkumar Bhagavathi, MD University of Iowa Carver College of Medicine NGS as a genotyping platform in hematopathology
Bone Marrow Transplantation and the Potential Role of Iomab-B
 Bone Marrow Transplantation and the Potential Role of Iomab-B Hillard M. Lazarus, MD, FACP Professor of Medicine, Director of Novel Cell Therapy Case Western Reserve University 1 Hematopoietic Cell Transplantation
Bone Marrow Transplantation and the Potential Role of Iomab-B Hillard M. Lazarus, MD, FACP Professor of Medicine, Director of Novel Cell Therapy Case Western Reserve University 1 Hematopoietic Cell Transplantation
Accel-Amplicon Panels
 Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
CONSIDERATIONS IN DESIGNING ACUTE GVHD PREVENTION TRIALS: Patient Selection, Concomitant Treatments, Selecting and Assessing Endpoints
 CONSIDERATIONS IN DESIGNING ACUTE GVHD PREVENTION TRIALS: Patient Selection, Concomitant Treatments, Selecting and Assessing Endpoints CENTER FOR INTERNATIONAL BLOOD AND MARROW TRANSPLANT RESEARCH Potential
CONSIDERATIONS IN DESIGNING ACUTE GVHD PREVENTION TRIALS: Patient Selection, Concomitant Treatments, Selecting and Assessing Endpoints CENTER FOR INTERNATIONAL BLOOD AND MARROW TRANSPLANT RESEARCH Potential
Prevalence of Self-Reported Obesity Among U.S. Adults by State and Territory. Definitions Obesity: Body Mass Index (BMI) of 30 or higher.
 Prevalence of Self-Reported Obesity Among U.S. Adults by State and Territory Definitions Obesity: Body Mass Index (BMI) of 30 or higher. Body Mass Index (BMI): A measure of an adult s weight in relation
Prevalence of Self-Reported Obesity Among U.S. Adults by State and Territory Definitions Obesity: Body Mass Index (BMI) of 30 or higher. Body Mass Index (BMI): A measure of an adult s weight in relation
Deep Learning Analytics for Predicting Prognosis of Acute Myeloid Leukemia with Cytogenetics, Age, and Mutations
 Deep Learning Analytics for Predicting Prognosis of Acute Myeloid Leukemia with Cytogenetics, Age, and Mutations Andy Nguyen, M.D., M.S. Medical Director, Hematopathology, Hematology and Coagulation Laboratory,
Deep Learning Analytics for Predicting Prognosis of Acute Myeloid Leukemia with Cytogenetics, Age, and Mutations Andy Nguyen, M.D., M.S. Medical Director, Hematopathology, Hematology and Coagulation Laboratory,
Blast transformation in chronic myelomonocytic leukemia: Risk factors, genetic features, survival, and treatment outcome
 RESEARCH ARTICLE Blast transformation in chronic myelomonocytic leukemia: Risk factors, genetic features, survival, and treatment outcome AJH Mrinal M. Patnaik, 1 Emnet A. Wassie, 1 Terra L. Lasho, 2 Curtis
RESEARCH ARTICLE Blast transformation in chronic myelomonocytic leukemia: Risk factors, genetic features, survival, and treatment outcome AJH Mrinal M. Patnaik, 1 Emnet A. Wassie, 1 Terra L. Lasho, 2 Curtis
Donating your baby s cord to a public cord blood bank can save lives globally.
 Donating your baby s cord to a public cord blood bank can save lives globally. You can play an important role in saving lives by donating the blood contained in your baby s umbilical cord to LifeCord,
Donating your baby s cord to a public cord blood bank can save lives globally. You can play an important role in saving lives by donating the blood contained in your baby s umbilical cord to LifeCord,
HIV/AIDS and other Sexually Transmitted Diseases (STDs) in the Southern Region of the United States: Epidemiological Overview
 HIV/AIDS and other Sexually Transmitted Diseases (STDs) in the Southern Region of the United States: Epidemiological Overview Prepared by The Henry J. Kaiser Family Foundation for Southern States Summit
HIV/AIDS and other Sexually Transmitted Diseases (STDs) in the Southern Region of the United States: Epidemiological Overview Prepared by The Henry J. Kaiser Family Foundation for Southern States Summit
Identification and clinical detection of genetic alterations of pre-neoplastic lesions Time for the PML ome? David Sidransky MD Johns Hopkins
 Identification and clinical detection of genetic alterations of pre-neoplastic lesions Time for the PML ome? David Sidransky MD Johns Hopkins February 3-5, 2016 Lansdowne Resort, Leesburg, VA Molecular
Identification and clinical detection of genetic alterations of pre-neoplastic lesions Time for the PML ome? David Sidransky MD Johns Hopkins February 3-5, 2016 Lansdowne Resort, Leesburg, VA Molecular
Disclosers Updates: Management of Aplastic Anemia and Congenital Marrow Failure 5/9/2017
 2017 Updates: Management of Aplastic Anemia and Congenital Marrow Failure Sachit Patel, MD Department of Pediatrics Division of Hematology-Oncology Blood and Marrow Transplantation Disclosers None 1 Objectives:
2017 Updates: Management of Aplastic Anemia and Congenital Marrow Failure Sachit Patel, MD Department of Pediatrics Division of Hematology-Oncology Blood and Marrow Transplantation Disclosers None 1 Objectives:
GENETIC TESTING FOR FANCONI ANEMIA
 GENETIC TESTING FOR FANCONI ANEMIA Non-Discrimination Statement and Multi-Language Interpreter Services information are located at the end of this document. Coverage for services, procedures, medical devices
GENETIC TESTING FOR FANCONI ANEMIA Non-Discrimination Statement and Multi-Language Interpreter Services information are located at the end of this document. Coverage for services, procedures, medical devices
Quarterly Statistical Report
 01/04/2008 Page 1 of 26 INTERMACS Interagency Registry for Mechanically Assisted Circulatory Support Quarterly Statistical Report Implant dates: March 1, 2006 November 30, 2007 January 4, 2008 Prepared
01/04/2008 Page 1 of 26 INTERMACS Interagency Registry for Mechanically Assisted Circulatory Support Quarterly Statistical Report Implant dates: March 1, 2006 November 30, 2007 January 4, 2008 Prepared
New and Emerging Strategies in the Treatment of Patients with Higher risk Myelodysplastic Syndromes (MDS)
 Welcome to Managing Myelodysplastic Syndromes. My name is David Steensma. I am an Associate Professor of Medicine at Harvard Medical School and a faculty member in the Adult Leukemia Program at Dana Farber
Welcome to Managing Myelodysplastic Syndromes. My name is David Steensma. I am an Associate Professor of Medicine at Harvard Medical School and a faculty member in the Adult Leukemia Program at Dana Farber
Kevin Kelly, MD, Phd Acute Myeloid and Lymphoid Leukemias
 Kevin Kelly, MD, Phd Acute Myeloid and Lymphoid Leukemias Relevant financial relationships in the past twelve months by presenter or spouse/partner. Speakers bureau: Novartis, Janssen, Gilead, Bayer The
Kevin Kelly, MD, Phd Acute Myeloid and Lymphoid Leukemias Relevant financial relationships in the past twelve months by presenter or spouse/partner. Speakers bureau: Novartis, Janssen, Gilead, Bayer The
Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases.
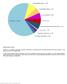 Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each
 Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Myelodysplastic syndrome is a highly heterogeneous hematopoietic
 SHORT COMMUNICATION Clinical Characteristics and Prognosis of 48 Patients with Mutations in Myelodysplastic Syndrome Yulu Tian #, Ruijuan Zhang #, Linhua Yang* Yang L. Clinical Characteristics and Prognosis
SHORT COMMUNICATION Clinical Characteristics and Prognosis of 48 Patients with Mutations in Myelodysplastic Syndrome Yulu Tian #, Ruijuan Zhang #, Linhua Yang* Yang L. Clinical Characteristics and Prognosis
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma. Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute
 Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Allogeneic Hematopoietic Stem-Cell Transplantation for Myelodysplastic Syndromes and Myeloproliferative Neoplasms. Policy Specific Section:
 Medical Policy Allogeneic Hematopoietic Stem-Cell Transplantation for Myelodysplastic Syndromes and Myeloproliferative Type: Medical Necessity and Investigational / Experimental Policy Specific Section:
Medical Policy Allogeneic Hematopoietic Stem-Cell Transplantation for Myelodysplastic Syndromes and Myeloproliferative Type: Medical Necessity and Investigational / Experimental Policy Specific Section:
Myelodysplasia/Myeloproliferative Neoplasms (MDS/MPN) Post-HCT Data
 Instructions for Myelodysplasia/Myeloproliferative Neoplasms (MDS/MPN) Post-HCT Data (Form 2114) This section of the CIBMTR Forms Instruction Manual is intended to be a resource for completing the Myelodysplasia/Myeloproliferative
Instructions for Myelodysplasia/Myeloproliferative Neoplasms (MDS/MPN) Post-HCT Data (Form 2114) This section of the CIBMTR Forms Instruction Manual is intended to be a resource for completing the Myelodysplasia/Myeloproliferative
Should Mutational Status in Primary Myelofibrosis (PMF) Guide Therapy..YES!!!
 Should Mutational Status in Primary Myelofibrosis (PMF) Guide Therapy..YES!!! Lindsay Anne Magura Rein, MD Division of Hematologic Malignancies and Cellular Therapy/BMT A Little Bit of History.. 1665 Advanced
Should Mutational Status in Primary Myelofibrosis (PMF) Guide Therapy..YES!!! Lindsay Anne Magura Rein, MD Division of Hematologic Malignancies and Cellular Therapy/BMT A Little Bit of History.. 1665 Advanced
Jennifer Hauenstein Oncology Cytogenetics Emory University Hospital Atlanta, GA
 Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Molecular Markers. Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC
 Molecular Markers Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC Overview Testing methods Rationale for molecular testing
Molecular Markers Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC Overview Testing methods Rationale for molecular testing
Myelodyspastic Syndromes
 Myelodyspastic Syndromes SUPPLEMENTARY APPENDIX Complex or monosomal karyotype and not blast percentage is associated with poor survival in acute myeloid leukemia and myelodysplastic syndrome patients
Myelodyspastic Syndromes SUPPLEMENTARY APPENDIX Complex or monosomal karyotype and not blast percentage is associated with poor survival in acute myeloid leukemia and myelodysplastic syndrome patients
