1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created.
|
|
|
- Barnard Simpson
- 5 years ago
- Views:
Transcription
1 Figure S1 Schematic of the in silico TCRminer and MiXCR validation. 1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created. Then, 100,000 simulated 80 bp RNA-Seq reads were generated using R package Polyester(50). Number of reads recovered by TCRminer and MiXCR are shown Figure S2 TCR gene segment recovery from in silico-generated simulated NGS TCR reads by TCRminer and MiXCR. V and J sequences were extracted from simulated RNA-Seq datasets using MiXCR or TCRMiner. The abundance of the recovered TCR sequences
2 is plotted along the Y-axis. The abundance of the simulated TCR sequences is plotted on the X-axis Figure S3 TCR gene correlations with T cell surface markers and transcription factors. For all graphs, discovery RNA-Seq dataset was used and the Spearman correlation coefficients and p-values are displayed on each plot. P-values for Spearman's test were calculated using the asymptotic t approximation. Significant correlations are outlined in blue. Psoriasis RNA-Seq datasets are depicted with red dots and healthy control RNA- Seq datasets are depicted with blue dots. (A) Scatter plots of TRA versus TRB and TRG versus gene expression correlation levels in psoriatic plaque samples are shown. (B) Expression of CCR6, a psoriasis-associated T cell marker, correlated with both TRB and. (C) CD4 and CD8A correlations with TRB and in healthy controls. (D) TRB and correlation with transcription factors in psoriatic plaque samples.(e) Correlation of transcription factors with psoriasis-associated cytokines in psoriatic plaque samples Figure S4 Meta-analysis of TRGV5, TRAJ39 and TRBV3-1 correlations with psoriasis-relevant cytokines. Differences in gene expressions were calculated (normalized gene expression of non-lesional samples were subtracted from their corresponding normalized gene expression within the matching lesional sample). Spearman correlation 41
3 coefficients for differences in gene expressions were then calculated. Forest plots of correlation coefficients are shown. Each black box is representative of the study weight for each data set and horizontal lines are 95% CI. Diamonds represent Standardized Mean Differences for results of all studies combined. Extremes of diamonds give 95% CI
4 TRVB MiXCR TCRMiner Percent (MiXCR) Percent (TCRMiner) Percent in model Percent in model TRAJ Percent (MiXCR) Percent (TCRMiner) Percent in model Percent in model
5
6 T B RORC FOXP3 BX21 (T-bet) GATA3 GATA3 CD4 CD8 TR CCR6 A Psoriasis B Psoriasis E Transcription Factor Correlations C D TRA TRB TRG TRB Healthy Control TRB Psoriasis #$%&'($'$ TNF CCL27 IL12RB1 CXCL9 CXCL10 IL17B IL17A TBX21 TBX21 TBX21 TBX21 RORC RORC IL34 IL37 IL18 IL17F IL17C GATA3 GATA3 GATA3 GATA3 RORC RORC IFNG IL12RB2 IL36A CXCL9 CXCL10 TRB TRB STAT1 STAT1 STAT1 STAT1 STAT1 CCL22 CTLA4 CCL19 IDO1 LAG3 TRB TRB FOXP3 FOXP3 FOXP3 FOXP3 FOXP3
7 TRGV5 - IL17C TRAJ39 - IL17A TRBV3-1 - IL17A TRAJ39 - IL36A TRBV3-1 - IL36A
8 Supplementary Tables Table S1. Benchmarking TCR extraction efficiency by rna-seq of cultured skin cells with no TCR Extraction TCR rearranged clones with MIXCR - Human Cell type Sample Total reads TRA TRB TRG Fibroblast ENCFF065GSV Fibroblast ENCFF509HFH Fibroblast ENCFF627NPY Fibroblast ENCFF737TKD Keratinocyte ENCFF000IDC Keratinocyte ENCFF000IDF Keratinocyte SRR Keratinocyte SRR Keratinocyte SRR Total per millon RNA-Seq reads <0.001 <0.001 <0.001 <0.001 Extraction TCR reads with TCRminer - Human Fibroblast ENCFF065GSV Fibroblast ENCFF509HFH Fibroblast ENCFF627NPY Fibroblast ENCFF737TKD Keratinocyte ENCFF000IDC Keratinocyte ENCFF000IDF Keratinocyte SRR Keratinocyte SRR Keratinocyte SRR Total per millon RNA-Seq reads <0.002 Table S2. Benchmarking TCR extraction efficiency using simulated data MIXCR test with simulated TCR transcriptome TRA TRB TRG Simulated clones Found clones Not uniquely or incorrect identified V-regions Not uniquely or incorrect identified J-regions CDR3 errors TCRminer test with simulated TCR transcriptome Simulated clones Found TCR chains Found V-regions Found J-regions Not uniquely or incorrect identified V-regions Not uniquely or incorrect identified J-regions Table S3. TCR recovery rate using simulated data Chain Simulated RNA-Seq MiXCR TCR miner MiXCR TCR miner reads recovery rate recovery rate recovery percent recovery percent TRA TRB TRG
9 Table S4. TCR V/J gene usage in the setting of psoriasis vs healthy controls in Tsoi et al dataset. TCR Segment Normal Psoriasis Segment Type Normal Psoriasis Fold Change p-value FDR Bonferroni TRAJ e e e e e-02 TRAJ e e e e e-27 TRAJ e e e e-248 TRAJ43 7.5e e e e e-02 TRAJ e e e e e-04 TRB20/OR e e e e e-02 TRBV26/OR e e e e e-19 TRBV e e e e e-06 TRBV e e e e TRBV e e e e TRGV5 9.01e e e e-13 TRGVA 1.88e e e e e-08
10 Table S5. Unpaired analysis of TCR V/J gene usage for lesional versus non-lesional samples in Tsoi et al dataset. TCR Segment Normal Psoriasis Segment Type Sample Number Normal Psoriasis Normal Psoriasis Fold Change p-value FDR Bonferroni TRAJ e e e-01 TRAJ e e e-11 TRAJ e e e-01 TRAJ e e e+00 TRAJ e e e-67 TRAJ e e e-01 TRAJ e e e-02 TRBV26/OR e e e-06 TRBV29/OR e e e-02 TRBV e e e-05 TRBV e e e-03 TRBV e e e-02 TRGV e e e-04 TRGVA e e e-01
11 Table S6. TCR V/J gene usage in the setting of psoriasis vs healthy controls in Weidinger dataset TCR Segment Normal Psoriasis Segment Type Sample Number Fold Normal Psoriasis Normal Psoriasis Change p-value FDR Bonferroni TRAJ e e e-03 TRAJ e e e-17 TRAJ e e e-51 TRAJ e e e-01 TRAV e e e-04 TRBV e e e-02 TRBV20/OR e e e-01 TRBV26/OR e e e-02 TRBV e e e-09 TRBV e e e-04 TRBV e e e-01 TRBV e e e-07 V e e e-02 TRGV e e e-03 TRGVA e e e+00
12 Table S7. Association between HLA-C*06 and TCR V- J- gene usage in psoriasis in Tsoi et al dataset. TCR Segment Other HLA HLA-C06 Sample Number Other HLA HLA-C06 Fold change p-value FDR Bonferroni TRAJ e e e e e-03 TRBV26/ OR e e e e e-01 Table S8. Association between HLA-C*06 and TCR V- J- gene usage in healthy individuals in Tsoi et al dataset. TCR Segment Other HLA HLA-C06 Sample Number Other HLA HLA-C06 Fold change p-value FDR Bonferroni TRAV e e e e e-03 TRBV e e e e e-03 TRAJ3 5.36e e e e e-02 TRAJ e e e e e-02 TRBV e e e e e-02 TRBV e e e e e-01 TRBV e e e e e-01
13 Table S9. T cell clones that were discovered in multiple psoriatic plaques or control biopsies Chain AA. Seq. CDR3 Normal skin samples TRA CAVNDYKLSF 1 4 TRA CAVMDSSYKLIF 0 3 TRA CAAMDSSYKLIF 0 2 TRA CALDAGNNRKLIW 0 2 TRA CALDSGGYQKVTF 0 2 TRA CALNTDKLIF 1 1 TRA CALYSGGGADGLTF 0 2 TRA CAVNGNARLMF 0 2 TRA CAVNRDDKIIF 0 2 TRA CAVQGSQGNLIF 2 0 TRA CAVRDGDYKLSF 1 1 TRA CAVRRMDSSYKLIF 0 2 TRA CAVSGTGRRALTF 0 2 TRA CAVSNQAGTALIF 0 2 TRA CAVYSGAGSYQLTF 0 2 TRA CLVGYNTDKLIF 1 1 TRB CASSDSSGSYNEQFF 2 0 TRB CASSTINSPLHF 1 1 TRB CATSRDSNTGVGNQPQHF 0 2 TRG CATWD_DKKLF 1 1 Psoriasis lesional skin samples Table S10. The Shannon Diversity Index for healthy and psoriatic CDR3 sequences Chain Healthy Control Psoriasis p-value TRA e-07 TRB e-08 Table S11. The Shannon Diversity Index for healthy and psoriatic V/J sequences Chain Healthy Control Psoriasis p-value TRA e-14 TRB e-08 TRG e e-02
14 Table S12. TCR gene segments correlation with T cell markers TCRminer Gene TRA TRA p-val TRB TRB p-val TRG TRG p-val p-val CD e <2e e e-06 CD8A 0.64 <2e <2e e GATA TBX e e e RORC MIXCR Gene TRA TRA clones TRB TRB clones TRG TRG clones clones clones p-val clones p-val clones p-val clones p-val CD e e NA NA CD8A e NA NA GATA NA NA TBX e NA NA RORC NA NA
15 Table S13. TCRminer correlation with psoriasis-related genes Gene TRA TRA p-val TRB TRB p-val TRG TRG p-val p-val IFNG IL1A IL1B IL IL6R IL IL12A IL12B IL17A IL17B IL17C IL17D IL17F IL17RA e IL17RC IL IL IL22RA IL22RA IL23A IL23R CXCL CXCL CCR e e e CCL TNF 0.63 <2e <2e e TNFSF e e TGFB e <2e TGFB TGFB e-07 RORgammaT
16 Table S14. TCR gene segments correlation with IL-17 TRGV Gene IL17A P-val TRGV TRGV TRGV TRGV TRGV TRGV TRGV5P TRGV TRGV V Gene IL17A P-val V V V TRBV Gene IL17A P-val TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV26/OR TRBV TRBV TRBV TRBV29/OR TRBV TRBV TRBV
17 TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRBV TRAV Gene IL17A P-val TRAV TRAV TRAV TRAV TRAV TRAV TRAV TRAV TRAV14/DV TRAV TRAV TRAV TRAV TRAV TRAV TRAV TRAV TRAV23/DV TRAV TRAV TRAV TRAV TRAV TRAV29/DV TRAV TRAV TRAV TRAV
18 TRAV36/DV TRAV TRAV38-2/DV TRAV TRAV TRAV TRAV TRAV TRAV TRAV TRAV TRAV TRAV TRAV Table S15. TRGV5 correlation with IL-36 genes Gene Correlation p-val FDR IL36A e-06 4e-05 IL36B IL36RN IL36G
qpcr-array Analysis Service
 qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
Supplemental Information. Lung gd T Cells Mediate Protective Responses. during Neonatal Influenza Infection. that Are Associated with Type 2 Immunity
 Immunity, Volume 9 Supplemental Information Lung gd T Cells Mediate Protective Responses during Neonatal Influenza Infection that re ssociated with Type Immunity Xi-zhi J. Guo, Pradyot Dash, Jeremy Chase
Immunity, Volume 9 Supplemental Information Lung gd T Cells Mediate Protective Responses during Neonatal Influenza Infection that re ssociated with Type Immunity Xi-zhi J. Guo, Pradyot Dash, Jeremy Chase
Eosinophils! 40! 30! 20! 10! 0! NS!
 A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
Nature Genetics: doi: /ng Supplementary Figure 1. Workflow of CDR3 sequence assembly from RNA-seq data.
 Supplementary Figure 1 Workflow of CDR3 sequence assembly from RNA-seq data. Paired-end short-read RNA-seq data were mapped to human reference genome hg19, and unmapped reads in the TCR regions were extracted
Supplementary Figure 1 Workflow of CDR3 sequence assembly from RNA-seq data. Paired-end short-read RNA-seq data were mapped to human reference genome hg19, and unmapped reads in the TCR regions were extracted
Transduction of lentivirus to human primary CD4+ T cells
 Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
To compare the relative amount of of selected gene expression between sham and
 Supplementary Materials and Methods Gene Expression Analysis To compare the relative amount of of selected gene expression between sham and mice given renal ischemia-reperfusion injury (IRI), ncounter
Supplementary Materials and Methods Gene Expression Analysis To compare the relative amount of of selected gene expression between sham and mice given renal ischemia-reperfusion injury (IRI), ncounter
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
Supplementary Table 1. Functional avidities of survivin-specific T-cell clones against LML-peptide
 Supplementary Table 1. Functional avidities of survivin-specific T-cell clones against LML-peptide pulsed T2 cells. clone avidity by 4-hour 51 Cr-release assay 50% lysis at E:T 10:1 [LML peptide, M] #24
Supplementary Table 1. Functional avidities of survivin-specific T-cell clones against LML-peptide pulsed T2 cells. clone avidity by 4-hour 51 Cr-release assay 50% lysis at E:T 10:1 [LML peptide, M] #24
Supplementary Table 1 Clinicopathological characteristics of 35 patients with CRCs
 Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Supplementary Figure 1. IL-12 serum levels and frequency of subsets in FL patients. (A) IL-12
 1 Supplementary Data Figure legends Supplementary Figure 1. IL-12 serum levels and frequency of subsets in FL patients. (A) IL-12 serum levels measured by multiplex ELISA (Luminex) in FL patients before
1 Supplementary Data Figure legends Supplementary Figure 1. IL-12 serum levels and frequency of subsets in FL patients. (A) IL-12 serum levels measured by multiplex ELISA (Luminex) in FL patients before
CD44
 MR1-5-OP-RU CD24 CD24 CD44 MAIT cells 2.78 11.2 WT RORγt- GFP reporter 1 5 1 4 1 3 2.28 1 5 1 4 1 3 4.8 1.6 8.1 1 5 1 4 1 3 1 5 1 4 1 3 3.7 3.21 8.5 61.7 1 2 1 3 1 4 1 5 TCRβ 2 1 1 3 1 4 1 5 CD44 1 2 GFP
MR1-5-OP-RU CD24 CD24 CD44 MAIT cells 2.78 11.2 WT RORγt- GFP reporter 1 5 1 4 1 3 2.28 1 5 1 4 1 3 4.8 1.6 8.1 1 5 1 4 1 3 1 5 1 4 1 3 3.7 3.21 8.5 61.7 1 2 1 3 1 4 1 5 TCRβ 2 1 1 3 1 4 1 5 CD44 1 2 GFP
Supplementary information CD4 T cells are required for both development and maintenance of disease in a new model of reversible colitis
 Supplementary information CD4 T cells are required for both development and maintenance of disease in a new model of reversible colitis rasseit and Steiner et al. .. Supplementary Figure 1 % of initial
Supplementary information CD4 T cells are required for both development and maintenance of disease in a new model of reversible colitis rasseit and Steiner et al. .. Supplementary Figure 1 % of initial
Supplementary Figure 1. Flow cytometry panels used for BD Canto (A) and BD Fortessa (B).
 Intra Immune nalysis Surface Supplementary Figure 1. Flow cytometry panels used for D Canto () and D Fortessa (). Name Fluorochrome ID F488 PE PerCp-Cy5.5 PC Paclue PE-Cy7 PC-H7 Lympho* 1 CD56 CD8 CD16
Intra Immune nalysis Surface Supplementary Figure 1. Flow cytometry panels used for D Canto () and D Fortessa (). Name Fluorochrome ID F488 PE PerCp-Cy5.5 PC Paclue PE-Cy7 PC-H7 Lympho* 1 CD56 CD8 CD16
Supplementary. presence of the. (c) mrna expression. Error. in naive or
 Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
CD4+ T Helper T Cells, and their Cytokines in Immune Defense and Disease
 CD4+ T Helper T Cells, and their Cytokines in Immune Defense and Disease Andrew Lichtman M.D., Ph.D. Brigham and Women s Hospital Harvard Medical School Lecture outline Intro to T cell mediated immunity
CD4+ T Helper T Cells, and their Cytokines in Immune Defense and Disease Andrew Lichtman M.D., Ph.D. Brigham and Women s Hospital Harvard Medical School Lecture outline Intro to T cell mediated immunity
Supplementary appendix
 Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kennedy GA, Varelias A, Vuckovic S, et al.
Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kennedy GA, Varelias A, Vuckovic S, et al.
Supplementary Appendix
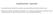 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Johnson DB, Balko JM, Compton ML, et al. Fulminant myocarditis
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Johnson DB, Balko JM, Compton ML, et al. Fulminant myocarditis
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 A β-strand positions consistently places the residues at CDR3β P6 and P7 within human and mouse TCR-peptide-MHC interfaces. (a) E8 TCR containing V β 13*06 carrying with an 11mer
Supplementary Figure 1 A β-strand positions consistently places the residues at CDR3β P6 and P7 within human and mouse TCR-peptide-MHC interfaces. (a) E8 TCR containing V β 13*06 carrying with an 11mer
SUPPLEMENTAL INFORMATIONS
 1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
Nature Immunology: doi: /ni Supplementary Figure 1. Cytokine pattern in skin in response to urushiol.
 Supplementary Figure 1 Cytokine pattern in skin in response to urushiol. Wild-type (WT) and CD1a-tg mice (n = 3 per group) were sensitized and challenged with urushiol (uru) or vehicle (veh). Quantitative
Supplementary Figure 1 Cytokine pattern in skin in response to urushiol. Wild-type (WT) and CD1a-tg mice (n = 3 per group) were sensitized and challenged with urushiol (uru) or vehicle (veh). Quantitative
The Genetic Epidemiology of Rheumatoid Arthritis. Lindsey A. Criswell AURA meeting, 2016
 The Genetic Epidemiology of Rheumatoid Arthritis Lindsey A. Criswell AURA meeting, 2016 Overview Recent successes in gene identification genome wide association studies (GWAS) clues to etiologic pathways
The Genetic Epidemiology of Rheumatoid Arthritis Lindsey A. Criswell AURA meeting, 2016 Overview Recent successes in gene identification genome wide association studies (GWAS) clues to etiologic pathways
Molecular Profiling of Tumor Microenvironment Alex Chenchik, Ph.D. Cellecta, Inc.
 Molecular Profiling of Tumor Microenvironment Alex Chenchik, Ph.D. Cellecta, Inc. Cellecta, Inc. Founded: April 2006 Headquarters: Mountain View, CA 12 SBIR Grants Custom Service Provider for Functional
Molecular Profiling of Tumor Microenvironment Alex Chenchik, Ph.D. Cellecta, Inc. Cellecta, Inc. Founded: April 2006 Headquarters: Mountain View, CA 12 SBIR Grants Custom Service Provider for Functional
Incomplete immune response to Coxsackie B viruses. associates with early autoimmunity against insulin
 Incomplete immune response to Coxsackie B viruses associates with early autoimmunity against insulin Michelle P. Ashton 1, Anne Eugster 1, Denise Walther 1, Natalie Daehling 1, Stephanie Riethausen 2,
Incomplete immune response to Coxsackie B viruses associates with early autoimmunity against insulin Michelle P. Ashton 1, Anne Eugster 1, Denise Walther 1, Natalie Daehling 1, Stephanie Riethausen 2,
Bezzi et al., Supplementary Figure 1 *** Nature Medicine: doi: /nm Pten pc-/- ;Zbtb7a pc-/- Pten pc-/- ;Pml pc-/- Pten pc-/- ;Trp53 pc-/-
 Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
T cell Receptor. Chapter 9. Comparison of TCR αβ T cells
 Chapter 9 The αβ TCR is similar in size and structure to an antibody Fab fragment T cell Receptor Kuby Figure 9-3 The αβ T cell receptor - Two chains - α and β - Two domains per chain - constant (C) domain
Chapter 9 The αβ TCR is similar in size and structure to an antibody Fab fragment T cell Receptor Kuby Figure 9-3 The αβ T cell receptor - Two chains - α and β - Two domains per chain - constant (C) domain
Deconvolution of Heterogeneous Wound Tissue Samples into Relative Macrophage Phenotype Composition via Models based on Gene Expression
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Nature Immunology: doi: /ni Supplementary Figure 1. RNA-Seq analysis of CD8 + TILs and N-TILs.
 Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Supplementary Figure 1. Example of gating strategy
 Supplementary Figure 1. Example of gating strategy Legend Supplementary Figure 1: First, gating is performed to include only single cells (singlets) (A) and CD3+ cells (B). After gating on the lymphocyte
Supplementary Figure 1. Example of gating strategy Legend Supplementary Figure 1: First, gating is performed to include only single cells (singlets) (A) and CD3+ cells (B). After gating on the lymphocyte
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Hasan et al analyse the transcriptional program used by pathogenic Th17 cells raised in the presence of IL-23 as compared to
Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Hasan et al analyse the transcriptional program used by pathogenic Th17 cells raised in the presence of IL-23 as compared to
Supplementary Information
 Supplementary Information Vedolizumab is associated with changes in innate rather than adaptive immunity in patients with inflammatory bowel disease Sebastian Zeissig, Elisa Rosati, C. Marie Dowds, Konrad
Supplementary Information Vedolizumab is associated with changes in innate rather than adaptive immunity in patients with inflammatory bowel disease Sebastian Zeissig, Elisa Rosati, C. Marie Dowds, Konrad
Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer
 Supplementary Information Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer Grit S. Herter-Sprie, Shohei Koyama, Houari Korideck, Josephine Hai, Jiehui Deng, Yvonne Y. Li, Kevin A. Buczkowski,
Supplementary Information Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer Grit S. Herter-Sprie, Shohei Koyama, Houari Korideck, Josephine Hai, Jiehui Deng, Yvonne Y. Li, Kevin A. Buczkowski,
D CD8 T cell number (x10 6 )
 IFNγ Supplemental Figure 1. CD T cell number (x1 6 ) 18 15 1 9 6 3 CD CD T cells CD6L C CD5 CD T cells CD6L D CD8 T cell number (x1 6 ) 1 8 6 E CD CD8 T cells CD6L F Log(1)CFU/g Feces 1 8 6 p
IFNγ Supplemental Figure 1. CD T cell number (x1 6 ) 18 15 1 9 6 3 CD CD T cells CD6L C CD5 CD T cells CD6L D CD8 T cell number (x1 6 ) 1 8 6 E CD CD8 T cells CD6L F Log(1)CFU/g Feces 1 8 6 p
A fresh approach to Immuno-oncology: Ex vivo analysis of drug efficacy in fresh patient tumortissue
 A fresh approach to Immuno-oncology: Ex vivo analysis of drug efficacy in fresh patient tumortissue Soner Altiok, MD, PhD Chief Scientific Officer Nilogen Oncosystems Tampa, Florida - Nilogen Oncosystems,
A fresh approach to Immuno-oncology: Ex vivo analysis of drug efficacy in fresh patient tumortissue Soner Altiok, MD, PhD Chief Scientific Officer Nilogen Oncosystems Tampa, Florida - Nilogen Oncosystems,
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplemental Information. Regulatory T Cells Exhibit Distinct. Features in Human Breast Cancer
 Immunity, Volume 45 Supplemental Information Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer George Plitas, Catherine Konopacki, Kenmin Wu, Paula D. Bos, Monica Morrow, Ekaterina V.
Immunity, Volume 45 Supplemental Information Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer George Plitas, Catherine Konopacki, Kenmin Wu, Paula D. Bos, Monica Morrow, Ekaterina V.
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Effector T Cells and
 1 Effector T Cells and Cytokines Andrew Lichtman, MD PhD Brigham and Women's Hospital Harvard Medical School 2 Lecture outline Cytokines Subsets of CD4+ T cells: definitions, functions, development New
1 Effector T Cells and Cytokines Andrew Lichtman, MD PhD Brigham and Women's Hospital Harvard Medical School 2 Lecture outline Cytokines Subsets of CD4+ T cells: definitions, functions, development New
Nature Genetics: doi: /ng Supplementary Figure 1
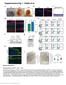 Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was
 Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Table 1. T-cell receptor sequences of HERV-K(HML-2)-specific CD8 + T cell clone.
 Supplementary Table 1. T-cell receptor sequences of HERV-K(HML-2)-specific CD8 + T cell clone. alpha beta ATGCTCCTGCTGCTCGTCCCAGTGCTCGAGGTGATTTTTACTCTGGGAGGAACCAGAGCC CAGTCGGTGACCCAGCTTGACAGCCACGTCTCTGTCTCTGAAGGAACCCCGGTGCTGCTG
Supplementary Table 1. T-cell receptor sequences of HERV-K(HML-2)-specific CD8 + T cell clone. alpha beta ATGCTCCTGCTGCTCGTCCCAGTGCTCGAGGTGATTTTTACTCTGGGAGGAACCAGAGCC CAGTCGGTGACCCAGCTTGACAGCCACGTCTCTGTCTCTGAAGGAACCCCGGTGCTGCTG
4. Th1-related gene expression in infected versus mock-infected controls from Fig. 2 with gene annotation.
 List of supplemental information 1. Graph of mouse weight loss during course of infection- Line graphs showing mouse weight data during course of infection days 1 to 10 post-infections (p.i.). 2. Graph
List of supplemental information 1. Graph of mouse weight loss during course of infection- Line graphs showing mouse weight data during course of infection days 1 to 10 post-infections (p.i.). 2. Graph
An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action
 An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action Supplementary Figure : Expression levels of toll-like receptor 4 (Tlr4) in muse lung does not change throughout the
An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action Supplementary Figure : Expression levels of toll-like receptor 4 (Tlr4) in muse lung does not change throughout the
NMED-A65251A. Supplementary Figures.
 NMED-A65251A Supplementary Figures. Sup. Fig. 1. ILC3 cells are the main source of in obese mice a. We gated on T cells (upper panels) or T cells (lower panels), and examined production. b. CD45 + - IL-13
NMED-A65251A Supplementary Figures. Sup. Fig. 1. ILC3 cells are the main source of in obese mice a. We gated on T cells (upper panels) or T cells (lower panels), and examined production. b. CD45 + - IL-13
AbSeq on the BD Rhapsody system: Exploration of single-cell gene regulation by simultaneous digital mrna and protein quantification
 BD AbSeq on the BD Rhapsody system: Exploration of single-cell gene regulation by simultaneous digital mrna and protein quantification Overview of BD AbSeq antibody-oligonucleotide conjugates. High-throughput
BD AbSeq on the BD Rhapsody system: Exploration of single-cell gene regulation by simultaneous digital mrna and protein quantification Overview of BD AbSeq antibody-oligonucleotide conjugates. High-throughput
Cancer, inflammation, and immunity crosstalk
 Contact Technical Support BRCsupport@QIAGEN.COM 1-800-362-7737 Cancer, inflammation, and immunity crosstalk Jesse Liang, Ph.D. Legal disclaimer QIAGEN products shown here are intended for molecular biology
Contact Technical Support BRCsupport@QIAGEN.COM 1-800-362-7737 Cancer, inflammation, and immunity crosstalk Jesse Liang, Ph.D. Legal disclaimer QIAGEN products shown here are intended for molecular biology
An innovative multi-dimensional NGS approach to understanding the tumor microenvironment and evolution
 An innovative multi-dimensional NGS approach to understanding the tumor microenvironment and evolution James H. Godsey, Ph.D. Vice President, Research & Development Clinical Sequencing Division (CSD) Life
An innovative multi-dimensional NGS approach to understanding the tumor microenvironment and evolution James H. Godsey, Ph.D. Vice President, Research & Development Clinical Sequencing Division (CSD) Life
Immune reprogramming via PD-1 inhibition enhances early-stage lung cancer survival
 Immune reprogramming via PD-1 inhibition enhances early-stage lung cancer survival Geoffrey J. Markowitz, 1,2,3 Lauren S. Havel, 1,2,3 Michael J.P. Crowley, 3,4 Yi Ban, 1,2,3 Sharrell B. Lee, 1,3 Jennifer
Immune reprogramming via PD-1 inhibition enhances early-stage lung cancer survival Geoffrey J. Markowitz, 1,2,3 Lauren S. Havel, 1,2,3 Michael J.P. Crowley, 3,4 Yi Ban, 1,2,3 Sharrell B. Lee, 1,3 Jennifer
ILC1 and ILC3 isolation and culture Following cell sorting, we confirmed that the recovered cells belonged to the ILC1, ILC2 and
 Supplementary Methods and isolation and culture Following cell sorting, we confirmed that the recovered cells belonged to the, ILC2 and subsets. For this purpose we performed intracellular flow cytometry
Supplementary Methods and isolation and culture Following cell sorting, we confirmed that the recovered cells belonged to the, ILC2 and subsets. For this purpose we performed intracellular flow cytometry
Transcript-indexed ATAC-seq for immune profiling
 Transcript-indexed ATAC-seq for immune profiling Technical Journal Club 22 nd of May 2018 Christina Müller Nature Methods, Vol.10 No.12, 2013 Nature Biotechnology, Vol.32 No.7, 2014 Nature Medicine, Vol.24,
Transcript-indexed ATAC-seq for immune profiling Technical Journal Club 22 nd of May 2018 Christina Müller Nature Methods, Vol.10 No.12, 2013 Nature Biotechnology, Vol.32 No.7, 2014 Nature Medicine, Vol.24,
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer
 Resource Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer Graphical Abstract Authors George Plitas, Catherine Konopacki, Kenmin Wu,..., Ekaterina V. Putintseva, Dmitriy M. Chudakov,
Resource Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer Graphical Abstract Authors George Plitas, Catherine Konopacki, Kenmin Wu,..., Ekaterina V. Putintseva, Dmitriy M. Chudakov,
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
A genome-wide association study identifies vitiligo
 A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
CT-2A * * * * * * NRRP (particles per cell)
 CT-2A Viability (%) 8 6 4 2 TNF-α: AT-46 GDC-917 LCL161 Monomer AZD-5582 Birinapant Dimer Supplementary Figure 1. s potently synergize with TNF-α to induce the death of glioblastoma cells. Viability of
CT-2A Viability (%) 8 6 4 2 TNF-α: AT-46 GDC-917 LCL161 Monomer AZD-5582 Birinapant Dimer Supplementary Figure 1. s potently synergize with TNF-α to induce the death of glioblastoma cells. Viability of
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1. ETBF activate Stat3 in B6 and Min mice colons
 Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Evidence for antigen-driven TCRβ chain convergence in the tumor infiltrating T cell repertoire
 Evidence for antigen-driven TCRβ chain convergence in the tumor infiltrating T cell repertoire Geoffrey M. Lowman, PhD Senior Staff Scientist EACR - 02 July 2018 For Research Use Only. Not for use in diagnostic
Evidence for antigen-driven TCRβ chain convergence in the tumor infiltrating T cell repertoire Geoffrey M. Lowman, PhD Senior Staff Scientist EACR - 02 July 2018 For Research Use Only. Not for use in diagnostic
An integrated transcriptomic and functional genomic approach identifies a. type I interferon signature as a host defense mechanism against Candida
 Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
Profiling immunoglobulin repertoires across multiple human tissues by RNA sequencing
 Profiling immunoglobulin repertoires across multiple human tissues by RNA sequencing SergheiMangul, Igor Mandric, Harry Taegyun Yang, NicolasStrauli, Dennis Montoya, Jeremy Rotman, Will Van Der Wey, Jiem
Profiling immunoglobulin repertoires across multiple human tissues by RNA sequencing SergheiMangul, Igor Mandric, Harry Taegyun Yang, NicolasStrauli, Dennis Montoya, Jeremy Rotman, Will Van Der Wey, Jiem
Tissue distribution and clonal diversity of the T and B cell repertoire in type 1 diabetes
 Tissue distribution and clonal diversity of the T and B cell repertoire in type 1 diabetes Howard R. Seay, 1 Erik Yusko, 2 Stephanie J. Rothweiler, 1 Lin Zhang, 1 Amanda L. Posgai, 1 Martha Campbell-Thompson,
Tissue distribution and clonal diversity of the T and B cell repertoire in type 1 diabetes Howard R. Seay, 1 Erik Yusko, 2 Stephanie J. Rothweiler, 1 Lin Zhang, 1 Amanda L. Posgai, 1 Martha Campbell-Thompson,
SUPPLEMENTARY APPENDIX
 SUPPLEMENTARY APPENDIX 1) Supplemental Figure 1. Histopathologic Characteristics of the Tumors in the Discovery Cohort 2) Supplemental Figure 2. Incorporation of Normal Epidermal Melanocytic Signature
SUPPLEMENTARY APPENDIX 1) Supplemental Figure 1. Histopathologic Characteristics of the Tumors in the Discovery Cohort 2) Supplemental Figure 2. Incorporation of Normal Epidermal Melanocytic Signature
Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL).
 Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL). (b) Depiction of a MTZ array generated by NAFL. (c-e) IgG production
Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL). (b) Depiction of a MTZ array generated by NAFL. (c-e) IgG production
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Introduction. Introduction
 Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
Current practice, needs and future directions in immuno-oncology research testing
 Current practice, needs and future directions in immuno-oncology research testing Jose Carlos Machado IPATIMUP - Porto, Portugal ESMO 2017- THERMO FISHER SCIENTIFIC SYMPOSIUM Immune Therapies are Revolutionizing
Current practice, needs and future directions in immuno-oncology research testing Jose Carlos Machado IPATIMUP - Porto, Portugal ESMO 2017- THERMO FISHER SCIENTIFIC SYMPOSIUM Immune Therapies are Revolutionizing
High-Throughput Sequencing Course
 High-Throughput Sequencing Course Introduction Biostatistics and Bioinformatics Summer 2017 From Raw Unaligned Reads To Aligned Reads To Counts Differential Expression Differential Expression 3 2 1 0 1
High-Throughput Sequencing Course Introduction Biostatistics and Bioinformatics Summer 2017 From Raw Unaligned Reads To Aligned Reads To Counts Differential Expression Differential Expression 3 2 1 0 1
Supplementary Figure 1
 Supplementary Figure 1 Identification of IFN-γ-producing CD8 + and CD4 + T cells with naive phenotype by alternative gating and sample-processing strategies. a. Contour 5% probability plots show definition
Supplementary Figure 1 Identification of IFN-γ-producing CD8 + and CD4 + T cells with naive phenotype by alternative gating and sample-processing strategies. a. Contour 5% probability plots show definition
Supplementary Figure 1
 d f a IL7 b IL GATA RORγt h HDM IL IL7 PBS Ilra R7 PBS HDM Ilra R7 HDM Foxp Foxp Ilra R7 HDM HDM Ilra R7 HDM. 9..79. CD + FOXP + T reg cell CD + FOXP T conv cell PBS Ilra R7 PBS HDM Ilra R7 HDM CD + FOXP
d f a IL7 b IL GATA RORγt h HDM IL IL7 PBS Ilra R7 PBS HDM Ilra R7 HDM Foxp Foxp Ilra R7 HDM HDM Ilra R7 HDM. 9..79. CD + FOXP + T reg cell CD + FOXP T conv cell PBS Ilra R7 PBS HDM Ilra R7 HDM CD + FOXP
and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the
 Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols.
 Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
SUPPLEMENTAL DATA. Table S1. Genes in each pathway selected for analysis.
 SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
Effect of Cytomegalovirus Infection on Immune Responsiveness. Rene van Lier Sanquin Blood Supply Foundation
 Effect of Cytomegalovirus Infection on Immune Responsiveness Rene van Lier Sanquin Blood Supply Foundation OUTLINE CMV infection and vaccination responses Effects of CMV infection on CD8 + T cell numbers
Effect of Cytomegalovirus Infection on Immune Responsiveness Rene van Lier Sanquin Blood Supply Foundation OUTLINE CMV infection and vaccination responses Effects of CMV infection on CD8 + T cell numbers
CPM (x 10-3 ) Tregs +Teffs. Tregs alone ICOS CLTA-4
 A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
January 25, 2017 Scientific Research Process Name of Journal: ESPS Manuscript NO: Manuscript Type: Title: Authors: Correspondence to
 January 25, 2017 Scientific Research Process Name of Journal: World Journal of Gastroenterology ESPS Manuscript NO: 31928 Manuscript Type: ORIGINAL ARTICLE Title: Thiopurine use associated with reduced
January 25, 2017 Scientific Research Process Name of Journal: World Journal of Gastroenterology ESPS Manuscript NO: 31928 Manuscript Type: ORIGINAL ARTICLE Title: Thiopurine use associated with reduced
SUPPLEMENTARY FIG. S2. b-galactosidase staining of
 SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
The quest for the identification of the genetic determinants of cancer immune responsiveness
 2015 Sidra Medical and Research Center The quest for the identification of the genetic determinants of cancer immune responsiveness Francesco M Marincola - Chief Research Officer Sidra Medical and Research
2015 Sidra Medical and Research Center The quest for the identification of the genetic determinants of cancer immune responsiveness Francesco M Marincola - Chief Research Officer Sidra Medical and Research
Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III
 Supplementary Materials: Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III patient samples. Genomic DNA samples extracted from punch biopsies from either FFPE or frozen tumor
Supplementary Materials: Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III patient samples. Genomic DNA samples extracted from punch biopsies from either FFPE or frozen tumor
HTG EdgeSeq Immuno-Oncology Assay Gene List
 A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
Effector mechanisms of cell-mediated immunity: Properties of effector, memory and regulatory T cells
 ICI Basic Immunology course Effector mechanisms of cell-mediated immunity: Properties of effector, memory and regulatory T cells Abul K. Abbas, MD UCSF Stages in the development of T cell responses: induction
ICI Basic Immunology course Effector mechanisms of cell-mediated immunity: Properties of effector, memory and regulatory T cells Abul K. Abbas, MD UCSF Stages in the development of T cell responses: induction
Figure S1. A Non-lesional Lesional
 Figure S1 Non-lesional Lesional GEN1 Non-lesional GEN1 Lesional C Normal Negative control Figure S1. Dermal Populations of + cells are found in human skin. Immunohistochemistry for on fixed, frozen skin
Figure S1 Non-lesional Lesional GEN1 Non-lesional GEN1 Lesional C Normal Negative control Figure S1. Dermal Populations of + cells are found in human skin. Immunohistochemistry for on fixed, frozen skin
Kazutaka Kitaura 1, Tadasu Shini 2,3, Takaji Matsutani 1* and Ryuji Suzuki 1,2
 Kitaura et al. BMC Immunology (2016) 17:38 DOI 10.1186/s12865-016-0177-5 RESEARCH ARTICLE A new high-throughput sequencing method for determining diversity and similarity of T cell receptor (TCR) α and
Kitaura et al. BMC Immunology (2016) 17:38 DOI 10.1186/s12865-016-0177-5 RESEARCH ARTICLE A new high-throughput sequencing method for determining diversity and similarity of T cell receptor (TCR) α and
Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated
 1 Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated (U) EGFR TL mice (n=7), Kras mice (n=7), PD-1 blockade
1 Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated (U) EGFR TL mice (n=7), Kras mice (n=7), PD-1 blockade
Supplementary Information
 Supplementary Information An orally available, small-molecule interferon inhibits viral replication Hideyuki Konishi 1, Koichi Okamoto 1, Yusuke Ohmori 1, Hitoshi Yoshino 2, Hiroshi Ohmori 1, Motooki Ashihara
Supplementary Information An orally available, small-molecule interferon inhibits viral replication Hideyuki Konishi 1, Koichi Okamoto 1, Yusuke Ohmori 1, Hitoshi Yoshino 2, Hiroshi Ohmori 1, Motooki Ashihara
HARNESS THE POWER OF THE IMMUNE SYSTEM TO FIGHT CANCER
 OncoPept TM HARNESS THE POWER OF THE IMMUNE SYSTEM TO FIGHT CANCER OncoPept identifies and delivers priortized T-cell neo-epitopes from the patient's tumor mutanome 2 What is OncoPept? OncoPept is an integrated
OncoPept TM HARNESS THE POWER OF THE IMMUNE SYSTEM TO FIGHT CANCER OncoPept identifies and delivers priortized T-cell neo-epitopes from the patient's tumor mutanome 2 What is OncoPept? OncoPept is an integrated
Supplementary Information
 Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded
 Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Table S1. Viral load and CD4 count of HIV-infected patient population
 Table S1. Viral load and CD4 count of HIV-infected patient population Subject ID Viral load (No. of copies per ml of plasma) CD4 count (No. of cells/µl of blood) 28 7, 14 29 7, 23 21 361,99 94 217 7, 11
Table S1. Viral load and CD4 count of HIV-infected patient population Subject ID Viral load (No. of copies per ml of plasma) CD4 count (No. of cells/µl of blood) 28 7, 14 29 7, 23 21 361,99 94 217 7, 11
Interferon-gamma pathway is activated in a chronically HBV infected chimpanzee that controls HBV following ARC-520 RNAi treatment
 Interferon-gamma pathway is activated in a chronically HBV infected chimpanzee that controls HBV following ARC-520 RNAi treatment Zhao Xu, Deborah Chavez, Jason E. Goetzmann, Robert E. Lanford and Christine
Interferon-gamma pathway is activated in a chronically HBV infected chimpanzee that controls HBV following ARC-520 RNAi treatment Zhao Xu, Deborah Chavez, Jason E. Goetzmann, Robert E. Lanford and Christine
Supplementary Figure 1. Localization of face patches (a) Sagittal slice showing the location of fmri-identified face patches in one monkey targeted
 Supplementary Figure 1. Localization of face patches (a) Sagittal slice showing the location of fmri-identified face patches in one monkey targeted for recording; dark black line indicates electrode. Stereotactic
Supplementary Figure 1. Localization of face patches (a) Sagittal slice showing the location of fmri-identified face patches in one monkey targeted for recording; dark black line indicates electrode. Stereotactic
Supplementary information. The proton-sensing G protein-coupled receptor T-cell death-associated gene 8
 1 Supplementary information 2 3 The proton-sensing G protein-coupled receptor T-cell death-associated gene 8 4 (TDAG8) shows cardioprotective effects against myocardial infarction 5 Akiomi Nagasaka 1+,
1 Supplementary information 2 3 The proton-sensing G protein-coupled receptor T-cell death-associated gene 8 4 (TDAG8) shows cardioprotective effects against myocardial infarction 5 Akiomi Nagasaka 1+,
A Quick-Start Guide for rseqdiff
 A Quick-Start Guide for rseqdiff Yang Shi (email: shyboy@umich.edu) and Hui Jiang (email: jianghui@umich.edu) 09/05/2013 Introduction rseqdiff is an R package that can detect differential gene and isoform
A Quick-Start Guide for rseqdiff Yang Shi (email: shyboy@umich.edu) and Hui Jiang (email: jianghui@umich.edu) 09/05/2013 Introduction rseqdiff is an R package that can detect differential gene and isoform
Supplemental Figure 1. Protein L
 Supplemental Figure 1 Protein L m19delta T m1928z T Suppl. Fig 1. Expression of CAR: B6-derived T cells were transduced with m19delta (left) and m1928z (right) to generate CAR T cells and transduction
Supplemental Figure 1 Protein L m19delta T m1928z T Suppl. Fig 1. Expression of CAR: B6-derived T cells were transduced with m19delta (left) and m1928z (right) to generate CAR T cells and transduction
Andrea s Final Exam Review PCB 3233 Spring Practice Final Exam
 NOTE: Practice Final Exam Although I am posting the answer key for this practice exam, I want you to use this practice to gauge your knowledge, and try to figure out the right answer by yourself before
NOTE: Practice Final Exam Although I am posting the answer key for this practice exam, I want you to use this practice to gauge your knowledge, and try to figure out the right answer by yourself before
As outlined under External contributions (see appendix 7.1), the group of Prof. Gröne at the
 3 RESULTS As outlined under External contributions (see appendix 7.1), the group of Prof. Gröne at the DKFZ in Heidelberg (Dept. of Cellular and Molecular pathology) contributed to this work by performing
3 RESULTS As outlined under External contributions (see appendix 7.1), the group of Prof. Gröne at the DKFZ in Heidelberg (Dept. of Cellular and Molecular pathology) contributed to this work by performing
An atlas of mouse CD4 + T cell transcriptomes
 Stubbington et al. Biology Direct (2015) 10:14 DOI 10.1186/s13062-015-0045-x RESEARCH An atlas of mouse CD4 + T cell transcriptomes Michael JT Stubbington 1, Bidesh Mahata 1,2, Valentine Svensson 1, Andrew
Stubbington et al. Biology Direct (2015) 10:14 DOI 10.1186/s13062-015-0045-x RESEARCH An atlas of mouse CD4 + T cell transcriptomes Michael JT Stubbington 1, Bidesh Mahata 1,2, Valentine Svensson 1, Andrew
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Information
 Supplementary Information Supplementary Figure 1. Short-term coreceptor and costimulation blockade induces tolerance to pluripotent human ESCs. (A) Schematic diagram showing experimental approaches and
Supplementary Information Supplementary Figure 1. Short-term coreceptor and costimulation blockade induces tolerance to pluripotent human ESCs. (A) Schematic diagram showing experimental approaches and
RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB
 RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB CSF-NGS January 22, 214 Contents 1 Introduction 1 2 Experimental Details 1 3 Results And Discussion 1 3.1 ERCC spike ins............................................
RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB CSF-NGS January 22, 214 Contents 1 Introduction 1 2 Experimental Details 1 3 Results And Discussion 1 3.1 ERCC spike ins............................................
