Activation of a PML/TP53 axis underlies acute promyelocytic. leukemia cure
|
|
|
- Amanda Quinn
- 5 years ago
- Views:
Transcription
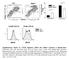
1 Activation of a PML/TP53 ais underlies acute promyelocytic leukemia cure Julien Ablain, Kim Rice, Hassane Soilihi, Aurélien de Reynies, Saverio Minucci & Hugues de Thé a PLZF-RARA vs PLZF-RARA pathway name RA low & int & high RA int & high not low RA int & high source h 1h h 1h h 1h h 1h h 1h GO:37 immune system process 7.E%15 1.3E%17 1.E%.E% *** *** * *** * ** GO:955 immune response.5e% 7.1E%1.7E%.1 * ** * *** * * GO:95 defense response 1.3E%7.E%11.E%.1E% * ** NS *** NS * GO:1 membrane 1.E%7 1.1E%9 1.E%.9 ** NS NS ** NS NS GO:95 response to eternal stimulus 1.3E%5 1.E%9 1.7E%3 3.1E% NS ** NS *** NS NS GO:59 response to stimulus 5.E%5 1.5E%9.9E%.E% NS *** NS *** NS *** GO:95 inflammatory response 1.E%.E%9.1E% 1.E% NS ** NS *** NS * GO:5177 response to other organism 1.E%5.1E%.. * * NS * NS NS GO:911 response to wounding.e% 5.7E%.1E%3 3.5E% NS * NS *** NS NS GO:5 plasma membrane 9.7E%.E%..93 * NS NS *** NS NS GO:917 response to bacterium 3.E%.7E%7 1. * * NS NS NS NS GO:31 intrinsic to membrane 3.E%.3E%7 5.1E%.95 ** NS NS * NS NS GO:7 receptor activity.9e%7.1e%5.7e%.9 * NS NS * NS NS GO:399 myeloid cell differentiation 7.E%7 1.E%.5 9.E% ** NS NS NS NS NS GO:5135 interphase E%7 NS NS NS NS NS NS GO:517 regulation of cell cycle..5.e%.e%7 NS * NS NS NS NS GO:59 chromosome E%7 NS NS NS NS NS *** GO: cell cycle process e%7 NS * NS NS NS *** GO:7 DNA replication initiation E%7 NS NS NS NS NS ** GO:75 cell cycle checkpoint.71.1.e% 1.7E%7 NS * NS NS NS ** GO:95 response to stress 3.3E%.E%7 5.E%.3E% NS *** NS ** NS *** GO:1 DNA-dependent DNA replication E%9 NS * NS NS NS *** GO:79 cell cycle e%9 NS ** NS NS NS *** GO:97 response to DNA damage stimulus E%9 NS ** NS NS NS *** GO:59 DNA metabolic process E%1 NS ** NS NS * *** GO: DNA replication e%1 NS ** NS NS * *** NS:P>1 %5 *:1 % <P<1 %5 **:1 %11 <P<1 % ***:P<1 %11 b Untreated RA low RA high h % of cells 7 h Nature Medicine: doi:1.13/nm.31
2 c % of GFP positive cells RA: Ø low high d Fold induction ( h) Serpine1 Dusp5 Egr RA: low int high int high RA: low int high int high RA: low int high int high Fold induction ( h) Fold induction ( h) PLZF-RARA PLZF-RARA PLZF-RARA e % Annein-V positive cells Untreat. Do RA int RA high f Relative Nucleus Area (piels/mm)," 3,5" 5,",3" 15," 1,1" 5 P =. P = 3.1 " RA: untreat"ra"low"ra"high" Ø Figure S1: High-dose RA triggers cell cycle arrest with features of senescence, not apoptosis in mouse APLs. (a) GO terms associated with genes differentially regulated in the different eperimental settings. The p values are indicated. (b) Cell-cycle analysis of GFP-positive bone marrow cells from APL mice treated with low (1.5) or high (5) RA doses for h or 7 h. DNA content was assessed by propidium iodide staining. Percentages of cells in each cell-cycle phase are shown in a representative histogram. (c) Analysis of GFP-marked leukemia cells in RA-treated APL mice. Mouse APLs were stably transduced with the MLS.1SHC-GFP retroviral vector and GFP-positive APL bone marrow cells were sorted and re-transplanted into recipient mice. APL mice were treated with low (1.5) or high (5) RA doses for 7 h and GFP-positive cell percentages were assessed by flow cytometry. (d) Epression levels of Serpine1, Dusp5 and Egr1 in bone marrow cells of h RA-treated APL mice relative to untreated (mean of three independent mice, Nature Medicine: doi:1.13/nm.31
3 measured by Q-PCR). (e) Proportion of Annein-V-positive bone marrow cells in APL mice treated for 1 h as indicated. Doorubicin (Do) was used as a positive control. (f) Quantification of nucleus size in GFP-marked leukemia cells following treatment of APL mice with RA low (1.5) and RA high (5) after 7 h (n = 1). a 1. Trp53 Fold induction (1 h) Untreat. RA int RA high b Trp53 ctrl#1 ctrl# sh#1 sh# c Relative colony numbers 1 1 P =.5 P =. Untreated RA 1 7 M ctrl sh Trp53 #1 sh Trp53 # d 7 d 1 1 d 3 P =. n = P = 5.1 n Time to death (d) Time to death of secondary recipients (d) ctrl #1 sh Trp53 #1 ctrl #1 + RA int sh Trp53 #1 + RA int treatment f Survival benefit (RA int vs untreated) P =. Trp53 +/+ Trp53 /! e 7 d d P =.5 n = P =.1 n Time to death (d) Time to death of secondary recipients (d) ctrl # sh Trp53 # ctrl # + RA int sh Trp53 # + RA int treatment g RA 1 M: Trp53 +/+ Trp53 /! + + Figure S: A key role for Trp53 activation in RA-induced loss of APL-initiating cell self-renewal in vivo. (a) Epression levels of Trp53 in bone marrow cells from APL mice treated with RA int (1) or RA high (1) for 1 h relative to untreated (untreated: n =, RA int: n =, RA high: n =, measured by epression arrays). (b) Nature Medicine: doi:1.13/nm.31
4 Western blot analysis of Trp53 in -transformed mouse hematopoietic progenitors after retroviral transduction of vectors epressing scrambled shrnas (ctrl #1 and ctrl #) or shrnas directed against Trp53 (sh #1 and sh #). (c) Relative numbers of colonies (sh #1 or #/ctrl colony ratios) formed by transformed mouse hematopoietic progenitors after retroviral transduction of vectors epressing scrambled shrna (ctrl) or shrnas against Trp53 (sh #1 and sh #), and grown in methylcellulose for 7 d in the presence or absence of 1 7 M RA. Data represent the mean ± SD of three independent eperiments. (d) & (e) Left panels: survival of mice inoculated with APL cells transduced with the vectors described in (b), and treated for 7 d with RA int (1). Right panels: survival of secondary recipients of bone marrow cells from the mice described above but that had been treated for 3 d with RA int (1). (f) Quantification of the survival benefit (treated/control survival ratios) of 7 d RA int (1) treatment of mice injected with Trp53 +/+ or Trp53 / APL cells (n=). (g) Western blot analysis of in Trp53 +/+ or Trp53 / APL cells treated e vivo by 1 M RA for 1 d. Nature Medicine: doi:1.13/nm.31
5 a Pml d Survival benefit (RA int vs untreated) ctrl sh#1 sh# P = Pml +/+ Pml /! b P = P =. Relative colony numbers 1 1 Untreated ctrl sh Pml #1 sh Pml # e Pml +/+ RA 1 RA 7 M Pml /! RA 1 M: + + c Bone marrow of APL mice Pml /! Pml +/+ CD11b Pml /! Pml+/+ Gr1 Untreated RA int d 3 RA int d Figure S3: Loss of Pml impedes RA-triggered APL clearance. (a) Western blot analysis of Pml in -transformed mouse hematopoietic progenitors after retroviral transduction of vectors epressing scrambled shrna (ctrl) or shrnas directed against Pml (sh #1 and sh #). (b) Relative numbers of colonies (sh #1 or #/ctrl colony ratios) formed by transformed mouse hematopoietic progenitors after retroviral transduction of vectors epressing scrambled shrna (ctrl) or shrnas against Pml (sh #1 and sh #), and grown in methylcellulose for 7 d in the presence or absence of 1 7 M RA. Data represent the mean ± SD of three independent eperiments. (c) MGG staining and FACS analysis of bone marrow cells of mice injected with Pml +/+ or Pml / APL cells and treated with RA int (1) for 3 or d. (d) Quantification of the survival benefit (treated/control survival ratios) of 7 d RA int (1) treatment of mice injected with Pml +/+ or Pml / APL cells (n=). (e) Western blot analysis of in Pml +/+ or Pml / APL cells treated e vivo by 1 M RA for 1 d. Nature Medicine: doi:1.13/nm.31
6 a Pathway'name GO:313()(regulation(of(tissue(remodeling GO:517()(positive(regulation(of(secretion GO:91()(regulation(of(apoptosis GO:191()(regulation(of(cell(death GO:571()(positive(regulation(of(protein(secretion GO:19()(cell(death GO:15()(death GO:715()(signal(transduction GO:99()(regulation(of(signal(transduction GO:57()(regulation(of(protein(secretion GO:1()(regulation(of(cell(communication GO:915()(apoptosis GO:151()(programmed(cell(death GO:19()(positive(regulation(of(cell(death GO:771()(tissue(remodeling GO:93()(secretion GO:51()(positive(regulation(of(protein(transport GO:51()(regulation(of(secretion GO:71()(cell(surface(receptor(linked(signaling(pathway GO:93()(protein(secretion GO:53()(negative(regulation(of(cellular(process GO:775()(multicellular(organismal(development GO:117()(regulation(of(cytokine(production GO:99()(negative(regulation(of(signal(transduction GO:71()(multicellular(organismal(homeostasis GO:()(induction(of(apoptosis(by(etracellular(signals GO:11()(cytokine(production GO:3373()(response(to(vitamin GO:715()(cell(communication GO:59()(response(to(stimulus GO:3()(negative(regulation(of(apoptosis GO:39()(negative(regulation(of(programmed(cell(death GO:5()(negative(regulation(of(cell(proliferation GO:95()(response(to(eternal(stimulus GO:5()(negative(regulation(of(cell(death GO:97()(endocytosis GO:35()(positive(regulation(of(apoptosis GO:737()(regulation(of(ERK1(and(ERK(cascade GO:73()(cellular(ion(homeostasis GO:3()(positive(regulation(of(programmed(cell(death GO:397()(response(to(cytokine(stimulus GO:917()(induction(of(apoptosis GO:35()(developmental(process GO:17()(regulation(of(cell(proliferation GO:15()(induction(of(programmed(cell(death GO:553()(bone(resorption GO:59()(homeostatic(process GO:5171()(cellular(response(to(stimulus GO:513()(regulation(of(protein(transport GO:5715()(positive(regulation(of(cytokine(secretion p53(response p,value 5.7E)7 1.E) 1.E).E).5E) 7.3E) 7.5E) 1.1E)5 1.E)5 1.E)5 1.E)5 1.E)5 1.7E)5.E)5.1E)5 3.1E)5 3.E)5 3.3E)5 3.E)5 3.5E)5 3.E)5 3.7E)5 3.7E)5 3.E)5.1E)5 5.E)5.5E)5.7E)5 7.E)5.E)5.E)5.7E)5 9.E)5 1.E) 1.1E) 1.3E) 1.E) 1.E) 1.E) 1.5E) 1.5E) 1.5E) 1.5E) 1.E) 1.E) 1.E) 1.7E) 1.7E) 1.7E) 1.7E).E)3 b RA: Nra1 Vdr Gs Tgm Pr7 Bcll15 Cd3 Lif Intu Serpine1 Ddit3 Klk1b1 Ndrg Gm191 Lrp1 Mmp5 Il1a Asb Ccl Dgke Validation h - 1 h Pml /! Pml +/+ Pml /! Trp53 I H I H I H I H target Pml +/+ Log induction Figure S: Pml-dependent transcriptional changes. (a) GO terms associated with genes deregulated in bone marrow cells of RA-treated Pml +/+ APL mice but unaffected in those of their Pml / counterparts. Genes differentially regulated between Pml +/+ and Pml / APL cells in untreated mice were ecluded. (b) Heatmap showing epression levels of the genes described in Figure 5b in bone marrow cells of RA int- (I: 1) and RA high (H: 1)-treated Pml +/+ or Pml / APL mice relative to untreated mice. Genes reported to be transcriptionally regulated by Trp53 or demonstrating Trp53 binding in non-apl cell lines (L. Attardi, personal communication) are indicated. Nature Medicine: doi:1.13/nm.31
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplement Material. Spleen weight (mg) LN cells (X106) Acat1-/- Acat1-/- Mouse weight (g)
 Supplement Material A Spleen weight (mg) C Mouse weight (g) 1 5 1 2 9 6 3 2 5 2 1 5 Male LN cells (X16) 4 ** ** Female B 3 2 1 Supplemental Figure I. Spleen weight (A), Inguinal lymph node (LN) cell number
Supplement Material A Spleen weight (mg) C Mouse weight (g) 1 5 1 2 9 6 3 2 5 2 1 5 Male LN cells (X16) 4 ** ** Female B 3 2 1 Supplemental Figure I. Spleen weight (A), Inguinal lymph node (LN) cell number
Supplemental Information. Granulocyte-Monocyte Progenitors and. Monocyte-Dendritic Cell Progenitors Independently
 Immunity, Volume 47 Supplemental Information Granulocyte-Monocyte Progenitors and Monocyte-endritic ell Progenitors Independently Produce Functionally istinct Monocytes lberto Yáñez, Simon G. oetzee, ndre
Immunity, Volume 47 Supplemental Information Granulocyte-Monocyte Progenitors and Monocyte-endritic ell Progenitors Independently Produce Functionally istinct Monocytes lberto Yáñez, Simon G. oetzee, ndre
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 MSI2 interactors are associated with the riboproteome and are functionally relevant. (a) Coomassie blue staining of FLAG-MSI2 immunoprecipitated complexes. (b) GO analysis of MSI2-interacting
Supplementary Figure 1 MSI2 interactors are associated with the riboproteome and are functionally relevant. (a) Coomassie blue staining of FLAG-MSI2 immunoprecipitated complexes. (b) GO analysis of MSI2-interacting
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
Prise en Charge des LAM-3. Hervé Dombret Hôpital Saint-Louis Institut Universitaire d Hématologie Université Paris Diderot
 Prise en Charge des LAM-3 Hervé Dombret Hôpital Saint-Louis Institut Universitaire d Hématologie Université Paris Diderot Basal transcription therapy by ATRA h Transcrip onal repression PML RA PML Blast
Prise en Charge des LAM-3 Hervé Dombret Hôpital Saint-Louis Institut Universitaire d Hématologie Université Paris Diderot Basal transcription therapy by ATRA h Transcrip onal repression PML RA PML Blast
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
SUPPLEMENTARY INFORMATION
 a. Smo+/+ b. Smo+/+ 5.63 5.48 c. Lin- d. e. 6 5 4 3 Ter119 Mac B T Sca1 Smo+/+ 25 15 2 o BMT 2 1 5 * Supplementary Figure 1: Deletion of Smoothened does not alter the frequency of hematopoietic lineages
a. Smo+/+ b. Smo+/+ 5.63 5.48 c. Lin- d. e. 6 5 4 3 Ter119 Mac B T Sca1 Smo+/+ 25 15 2 o BMT 2 1 5 * Supplementary Figure 1: Deletion of Smoothened does not alter the frequency of hematopoietic lineages
X P. Supplementary Figure 1. Nature Medicine: doi: /nm Nilotinib LSK LT-HSC. Cytoplasm. Cytoplasm. Nucleus. Nucleus
 a b c Supplementary Figure 1 c-kit-apc-eflu780 Lin-FITC Flt3-Linc-Kit-APC-eflu780 LSK Sca-1-PE-Cy7 d e f CD48-APC LT-HSC CD150-PerCP-cy5.5 g h i j Cytoplasm RCC1 X Exp 5 mir 126 SPRED1 SPRED1 RAN P SPRED1
a b c Supplementary Figure 1 c-kit-apc-eflu780 Lin-FITC Flt3-Linc-Kit-APC-eflu780 LSK Sca-1-PE-Cy7 d e f CD48-APC LT-HSC CD150-PerCP-cy5.5 g h i j Cytoplasm RCC1 X Exp 5 mir 126 SPRED1 SPRED1 RAN P SPRED1
Supplementary Figure 1. Successful excision of genes from WBM lysates and
 Supplementary Information: Supplementary Figure 1. Successful excision of genes from WBM lysates and survival of mice with different genotypes. (a) The proper excision of Pten, p110α, p110α and p110δ was
Supplementary Information: Supplementary Figure 1. Successful excision of genes from WBM lysates and survival of mice with different genotypes. (a) The proper excision of Pten, p110α, p110α and p110δ was
Control shrna#9 shrna#12. shrna#12 CD14-PE CD14-PE
 a Control shrna#9 shrna#12 c Control shrna#9 shrna#12 e Control shrna#9 shrna#12 h 14 12 CFU-E BFU-E GEMM GM b Colony number 7 6 5 4 3 2 1 6 pm A pa pc CFU-E BFU-E GEMM GM pu pgm A p pg B d f CD11b-APC
a Control shrna#9 shrna#12 c Control shrna#9 shrna#12 e Control shrna#9 shrna#12 h 14 12 CFU-E BFU-E GEMM GM b Colony number 7 6 5 4 3 2 1 6 pm A pa pc CFU-E BFU-E GEMM GM pu pgm A p pg B d f CD11b-APC
Chapt 15: Molecular Genetics of Cell Cycle and Cancer
 Chapt 15: Molecular Genetics of Cell Cycle and Cancer Student Learning Outcomes: Describe the cell cycle: steps taken by a cell to duplicate itself = cell division; Interphase (G1, S and G2), Mitosis.
Chapt 15: Molecular Genetics of Cell Cycle and Cancer Student Learning Outcomes: Describe the cell cycle: steps taken by a cell to duplicate itself = cell division; Interphase (G1, S and G2), Mitosis.
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Bezzi et al., Supplementary Figure 1 *** Nature Medicine: doi: /nm Pten pc-/- ;Zbtb7a pc-/- Pten pc-/- ;Pml pc-/- Pten pc-/- ;Trp53 pc-/-
 Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Review and Public RAC Discussion of Protocol #
 Review and Public RAC Discussion of Protocol #0508 725 A phase I pilot study of safety and feasibility of stem cell therapy for AIDS lymphoma using stem cells treated with a lentivirus vector encoding
Review and Public RAC Discussion of Protocol #0508 725 A phase I pilot study of safety and feasibility of stem cell therapy for AIDS lymphoma using stem cells treated with a lentivirus vector encoding
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr
 Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Tbk1-TKO! DN cells (%)! 15! 10!
 a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
Table S1. New colony formation 7 days after stimulation with doxo and VCR in JURKAT cells
 Table S1. New colony formation 7 days after stimulation with and in JURKAT cells drug co + number of colonies 7±14 4±7 48±11 JURKAT cells were stimulated and analyzed as in Table 1. Drug concentrations
Table S1. New colony formation 7 days after stimulation with and in JURKAT cells drug co + number of colonies 7±14 4±7 48±11 JURKAT cells were stimulated and analyzed as in Table 1. Drug concentrations
sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5
 sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
Nature Immunology: doi: /ni.3412
 Supplementary Figure 1 Gata1 expression in heamatopoietic stem and progenitor populations. (a) Unsupervised clustering according to 100 top variable genes across single pre-gm cells. The two main cell
Supplementary Figure 1 Gata1 expression in heamatopoietic stem and progenitor populations. (a) Unsupervised clustering according to 100 top variable genes across single pre-gm cells. The two main cell
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/2/1/ra81/dc1 Supplementary Materials for Delivery of MicroRNA-126 by Apoptotic Bodies Induces CXCL12- Dependent Vascular Protection Alma Zernecke,* Kiril Bidzhekov,
www.sciencesignaling.org/cgi/content/full/2/1/ra81/dc1 Supplementary Materials for Delivery of MicroRNA-126 by Apoptotic Bodies Induces CXCL12- Dependent Vascular Protection Alma Zernecke,* Kiril Bidzhekov,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 EGFR inhibition activates signaling pathways (a-b) EGFR inhibition activates signaling pathways (a) U251EGFR cells were treated with erlotinib (1µM) for the indicated times followed
Supplementary Figure 1 EGFR inhibition activates signaling pathways (a-b) EGFR inhibition activates signaling pathways (a) U251EGFR cells were treated with erlotinib (1µM) for the indicated times followed
Recombinant Protein Expression Retroviral system
 Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Stem cells: units of development and regeneration. Fernando D. Camargo Ph.D. Whitehead Fellow Whitehead Institute for Biomedical Research.
 Stem cells: units of development and regeneration Fernando D. Camargo Ph.D. Whitehead Fellow Whitehead Institute for Biomedical Research Concepts 1. Embryonic vs. adult stem cells 2. Hematopoietic stem
Stem cells: units of development and regeneration Fernando D. Camargo Ph.D. Whitehead Fellow Whitehead Institute for Biomedical Research Concepts 1. Embryonic vs. adult stem cells 2. Hematopoietic stem
(A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and a
 Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplemental Figure 1. Protein L
 Supplemental Figure 1 Protein L m19delta T m1928z T Suppl. Fig 1. Expression of CAR: B6-derived T cells were transduced with m19delta (left) and m1928z (right) to generate CAR T cells and transduction
Supplemental Figure 1 Protein L m19delta T m1928z T Suppl. Fig 1. Expression of CAR: B6-derived T cells were transduced with m19delta (left) and m1928z (right) to generate CAR T cells and transduction
BCR-ABL - LSK BCR-ABL + LKS - (%)
 Marker Clone BCR-ABL + LSK (%) BCR-ABL + LKS - (%) BCR-ABL - LSK (%) P value vs. BCR-ABL + LKS - P value vs. BCR-ABL - LSK CD2 RM2-5 12.9 ± 3.6 36.7 ± 6.5 19.3 ± 2.4 0.01 0.10 CD5 53-7.3 13.9 ± 3.2 20.8
Marker Clone BCR-ABL + LSK (%) BCR-ABL + LKS - (%) BCR-ABL - LSK (%) P value vs. BCR-ABL + LKS - P value vs. BCR-ABL - LSK CD2 RM2-5 12.9 ± 3.6 36.7 ± 6.5 19.3 ± 2.4 0.01 0.10 CD5 53-7.3 13.9 ± 3.2 20.8
Imtiyaz et al., Fig. S1
 . Imtiyaz et al., Fig. S1 1. 1.1 1% O.1.5 Lin/Sca-1/IL-7Rα GMPs.17 MPs.3 Days 3% O.1 MEPs.35 D3 Days 1, N, N, H, H 1 1 Days Supplemental Figure S1. Macrophage maturation, proliferation and survival are
. Imtiyaz et al., Fig. S1 1. 1.1 1% O.1.5 Lin/Sca-1/IL-7Rα GMPs.17 MPs.3 Days 3% O.1 MEPs.35 D3 Days 1, N, N, H, H 1 1 Days Supplemental Figure S1. Macrophage maturation, proliferation and survival are
ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1
 ZH, Li et al, page 1 ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1 Zhenhu Li 1,4,Yuan Zhang 1,4, Zhiduo Liu 1, Xiaodong Wu 1, Yuhan Zheng 1, Zhiyun Tao 1, Kairui Mao 1,
ZH, Li et al, page 1 ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1 Zhenhu Li 1,4,Yuan Zhang 1,4, Zhiduo Liu 1, Xiaodong Wu 1, Yuhan Zheng 1, Zhiyun Tao 1, Kairui Mao 1,
W/T Itgam -/- F4/80 CD115. F4/80 hi CD115 + F4/80 + CD115 +
 F4/8 % in the peritoneal lavage 6 4 2 p=.15 n.s p=.76 CD115 F4/8 hi CD115 + F4/8 + CD115 + F4/8 hi CD115 + F4/8 + CD115 + MHCII MHCII Supplementary Figure S1. CD11b deficiency affects the cellular responses
F4/8 % in the peritoneal lavage 6 4 2 p=.15 n.s p=.76 CD115 F4/8 hi CD115 + F4/8 + CD115 + F4/8 hi CD115 + F4/8 + CD115 + MHCII MHCII Supplementary Figure S1. CD11b deficiency affects the cellular responses
Suppl Video: Tumor cells (green) and monocytes (white) are seeded on a confluent endothelial
 Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, Maryland
 AD Award Number: W81XWH-13-1-0057 TITLE: Role of TIRAP in Myelodysplastic Syndromes PRINCIPAL INVESTIGATOR: Linda Ya-ting Chang CONTRACTING ORGANIZATION: British Columbia Cancer Agency Branch Vancouver,
AD Award Number: W81XWH-13-1-0057 TITLE: Role of TIRAP in Myelodysplastic Syndromes PRINCIPAL INVESTIGATOR: Linda Ya-ting Chang CONTRACTING ORGANIZATION: British Columbia Cancer Agency Branch Vancouver,
Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts
 Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts (TIG-3 cells) were rendered senescent by either serial passage
Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts (TIG-3 cells) were rendered senescent by either serial passage
Impaired DNA replication within progenitor cell pools promotes leukemogenesis
 Impaired DNA replication within progenitor cell pools promotes leukemogenesis Ganna Bilousova, University of Colorado Andriy Marusyk, University of Colorado Christopher Porter, Emory University Robert
Impaired DNA replication within progenitor cell pools promotes leukemogenesis Ganna Bilousova, University of Colorado Andriy Marusyk, University of Colorado Christopher Porter, Emory University Robert
MicroRNAs Modulate the Noncanonical NF- B Pathway by Regulating IKK Expression During Macrophage Differentiation
 MicroRNAs Modulate the Noncanonical NF- B Pathway by Regulating IKK Expression During Macrophage Differentiation Tao Li 1 *, Michael J. Morgan 1 *, Swati Choksi 1, Yan Zhang 1, You-Sun Kim 2#, Zheng-gang
MicroRNAs Modulate the Noncanonical NF- B Pathway by Regulating IKK Expression During Macrophage Differentiation Tao Li 1 *, Michael J. Morgan 1 *, Swati Choksi 1, Yan Zhang 1, You-Sun Kim 2#, Zheng-gang
Supplemental Figure 1
 Supplemental Figure 1 A S100A4: SFLGKRTDEAAFQKLMSNLDSNRDNEVDFQEYCVFLSCIAMMCNEFFEGFPDK Overlap: SF G DE KLM LD N D VDFQEY VFL I M N FF G PD S100A2: SFVGEKVDEEGLKKLMGSLDENSDQQVDFQEYAVFLALITVMCNDFFQGCPDR
Supplemental Figure 1 A S100A4: SFLGKRTDEAAFQKLMSNLDSNRDNEVDFQEYCVFLSCIAMMCNEFFEGFPDK Overlap: SF G DE KLM LD N D VDFQEY VFL I M N FF G PD S100A2: SFVGEKVDEEGLKKLMGSLDENSDQQVDFQEYAVFLALITVMCNDFFQGCPDR
Supplementary Figure 1. Dynamic Response of WT and mir-21 -/- mice to caerulein. (a) Representative histological sections of mouse pancreas stained
 Supplementary Figure 1. Dynamic Response of WT and mir-21 -/- mice to caerulein. (a) Representative histological sections of mouse pancreas stained with hematoxylin from caerulein-treated WT and mir-21
Supplementary Figure 1. Dynamic Response of WT and mir-21 -/- mice to caerulein. (a) Representative histological sections of mouse pancreas stained with hematoxylin from caerulein-treated WT and mir-21
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
IKK-dependent activation of NF-κB contributes to myeloid and lymphoid leukemogenesis by BCR-ABL1
 Supplemental Figures BLOOD/2014/547943 IKK-dependent activation of NF-κB contributes to myeloid and lymphoid leukemogenesis by BCR-ABL1 Hsieh M-Y and Van Etten RA Supplemental Figure S1. Titers of retroviral
Supplemental Figures BLOOD/2014/547943 IKK-dependent activation of NF-κB contributes to myeloid and lymphoid leukemogenesis by BCR-ABL1 Hsieh M-Y and Van Etten RA Supplemental Figure S1. Titers of retroviral
SUPPLEMENTARY INFORMATION
 Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa
 SUPPLEMENTARY FIGURES Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa tet-on cells that stably express HP1α-CFP, HP1β-CFP, or HP1γ-CFP were monitored with livecell
SUPPLEMENTARY FIGURES Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa tet-on cells that stably express HP1α-CFP, HP1β-CFP, or HP1γ-CFP were monitored with livecell
Acute Myeloid Leukemia with Recurrent Cytogenetic Abnormalities
 Acute Myeloid Leukemia with Recurrent Cytogenetic Abnormalities Acute Myeloid Leukemia with recurrent cytogenetic Abnormalities -t(8;21)(q22;q22)(aml/eto) -inv(16) or t(16;16) -t(15;17) -11q23 Acute Myeloid
Acute Myeloid Leukemia with Recurrent Cytogenetic Abnormalities Acute Myeloid Leukemia with recurrent cytogenetic Abnormalities -t(8;21)(q22;q22)(aml/eto) -inv(16) or t(16;16) -t(15;17) -11q23 Acute Myeloid
Supplementary Figure 1
 Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
SUPPLEMENTARY METHODS
 SUPPLEMENTARY METHODS Histological analysis. Colonic tissues were collected from 5 parts of the middle colon on day 7 after the start of DSS treatment, and then were cut into segments, fixed with 4% paraformaldehyde,
SUPPLEMENTARY METHODS Histological analysis. Colonic tissues were collected from 5 parts of the middle colon on day 7 after the start of DSS treatment, and then were cut into segments, fixed with 4% paraformaldehyde,
Hematopoiesis. - Process of generation of mature blood cells. - Daily turnover of blood cells (70 kg human)
 Hematopoiesis - Process of generation of mature blood cells - Daily turnover of blood cells (70 kg human) 1,000,000,000,000 total cells 200,000,000,000 red blood cells 70,000,000,000 neutrophils Hematopoiesis
Hematopoiesis - Process of generation of mature blood cells - Daily turnover of blood cells (70 kg human) 1,000,000,000,000 total cells 200,000,000,000 red blood cells 70,000,000,000 neutrophils Hematopoiesis
AHI-1 interacts with BCR-ABL and modulates BCR-ABL transforming activity and imatinib response of CML stem/progenitor cells
 AHI-1 interacts with BCR-ABL and modulates BCR-ABL transforming activity and imatinib response of CML stem/progenitor cells Liang L. Zhou, 1 Yun Zhao, 1 Ashley Ringrose, 1 Donna DeGeer, 1 Erin Kennah,
AHI-1 interacts with BCR-ABL and modulates BCR-ABL transforming activity and imatinib response of CML stem/progenitor cells Liang L. Zhou, 1 Yun Zhao, 1 Ashley Ringrose, 1 Donna DeGeer, 1 Erin Kennah,
SUPPLEMENTARY INFORMATION doi: /nature12026
 doi:1.138/nature1226 a 4 35 3 MCSF level (pg/ml) 25 2 15 1 5 1h3 3h 5h 7h 15h 24h b MPP (CD135 KSL) HSC (CD34 CD15 KSLF) c % 4 ** LPS 3 GFP pos cells 2 PU.1 GFP LPS 1 FSCA Ctl NI 24h LPS Sup.Fig.1 Effect
doi:1.138/nature1226 a 4 35 3 MCSF level (pg/ml) 25 2 15 1 5 1h3 3h 5h 7h 15h 24h b MPP (CD135 KSL) HSC (CD34 CD15 KSLF) c % 4 ** LPS 3 GFP pos cells 2 PU.1 GFP LPS 1 FSCA Ctl NI 24h LPS Sup.Fig.1 Effect
SHREE ET AL, SUPPLEMENTAL MATERIALS. (A) Workflow for tumor cell line derivation and orthotopic implantation.
 SHREE ET AL, SUPPLEMENTAL MATERIALS SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure 1. Derivation and characterization of TS1-TGL and TS2-TGL PyMT cell lines and development of an orthotopic
SHREE ET AL, SUPPLEMENTAL MATERIALS SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure 1. Derivation and characterization of TS1-TGL and TS2-TGL PyMT cell lines and development of an orthotopic
Therapy-related MDS/AML with KMT2A (MLL) Rearrangement Following Therapy for APL Case 0328
 Therapy-related MDS/AML with KMT2A (MLL) Rearrangement Following Therapy for APL Case 0328 Kenneth N. Holder, Leslie J. Greebon, Gopalrao Velagaleti, Hongxin Fan, Russell A. Higgins Initial Case: Clinical
Therapy-related MDS/AML with KMT2A (MLL) Rearrangement Following Therapy for APL Case 0328 Kenneth N. Holder, Leslie J. Greebon, Gopalrao Velagaleti, Hongxin Fan, Russell A. Higgins Initial Case: Clinical
FIG S1 Examination of eif4b expression after virus infection. (A) A549 cells
 Supplementary Figure Legends FIG S1 Examination of expression after virus infection. () 549 cells were infected with herpes simplex virus (HSV) (MOI = 1), and harvested at the indicated times, followed
Supplementary Figure Legends FIG S1 Examination of expression after virus infection. () 549 cells were infected with herpes simplex virus (HSV) (MOI = 1), and harvested at the indicated times, followed
EML Erythroid and Neutrophil Differentiation Protocols Cristina Pina 1*, Cristina Fugazza 2 and Tariq Enver 3
 EML Erythroid and Neutrophil Differentiation Protocols Cristina Pina 1*, Cristina Fugazza 2 and Tariq Enver 3 1 Department of Haematology, University of Cambridge, Cambridge, UK; 2 Dipartimento de Biotecnologie
EML Erythroid and Neutrophil Differentiation Protocols Cristina Pina 1*, Cristina Fugazza 2 and Tariq Enver 3 1 Department of Haematology, University of Cambridge, Cambridge, UK; 2 Dipartimento de Biotecnologie
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10134 Supplementary Figure 1. Anti-inflammatory activity of sfc. a, Autoantibody immune complexes crosslink activating Fc receptors, promoting activation of macrophages, and WWW.NATURE.COM/NATURE
doi:10.1038/nature10134 Supplementary Figure 1. Anti-inflammatory activity of sfc. a, Autoantibody immune complexes crosslink activating Fc receptors, promoting activation of macrophages, and WWW.NATURE.COM/NATURE
Intracellular MHC class II molecules promote TLR-triggered innate. immune responses by maintaining Btk activation
 Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Stewart et al. CD36 ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer
 NFκB (fold induction) Stewart et al. ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer a. mrna (fold induction) 5 4 3 2 1 LDL oxldl Gro1a MIP-2 RANTES mrna (fold induction)
NFκB (fold induction) Stewart et al. ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer a. mrna (fold induction) 5 4 3 2 1 LDL oxldl Gro1a MIP-2 RANTES mrna (fold induction)
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
% of live splenocytes. STAT5 deletion. (open shapes) % ROSA + % floxed
 Supp. Figure 1. a 14 1 1 8 6 spleen cells (x1 6 ) 16 % of live splenocytes 5 4 3 1 % of live splenocytes 8 6 4 b 1 1 c % of CD11c + splenocytes (closed shapes) 8 6 4 8 6 4 % ROSA + (open shapes) % floxed
Supp. Figure 1. a 14 1 1 8 6 spleen cells (x1 6 ) 16 % of live splenocytes 5 4 3 1 % of live splenocytes 8 6 4 b 1 1 c % of CD11c + splenocytes (closed shapes) 8 6 4 8 6 4 % ROSA + (open shapes) % floxed
Effective Targeting of Quiescent Chronic Myelogenous
 Cancer Cell, Volume 7 Supplemental Information Effective Targeting of Quiescent Chronic Myelogenous Leukemia Stem Cells by Histone Deacetylase Inhibitors in Combination with Imatinib Mesylate Bin Zhang,
Cancer Cell, Volume 7 Supplemental Information Effective Targeting of Quiescent Chronic Myelogenous Leukemia Stem Cells by Histone Deacetylase Inhibitors in Combination with Imatinib Mesylate Bin Zhang,
Fluorescence in-situ Hybridization (FISH) ETO(RUNX1T1)/AML1(RUNX1) or t(8;21)(q21.3;q22)
 PML/RARA t(15;17) Translocation Assay Result : nuc ish(pml 2)(RARA 2)[200] : 200/200(100%) interphase nuclei show normal 2O 2G signals for PML/RARA : is Negative for t(15;17)(q22;q21.1) 2 Orange 2 Green
PML/RARA t(15;17) Translocation Assay Result : nuc ish(pml 2)(RARA 2)[200] : 200/200(100%) interphase nuclei show normal 2O 2G signals for PML/RARA : is Negative for t(15;17)(q22;q21.1) 2 Orange 2 Green
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured
 Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Prolonged mitotic arrest induces a caspase-dependent DNA damage
 SUPPLEMENTARY INFORMATION Prolonged mitotic arrest induces a caspase-dependent DNA damage response at telomeres that determines cell survival Karolina O. Hain, Didier J. Colin, Shubhra Rastogi, Lindsey
SUPPLEMENTARY INFORMATION Prolonged mitotic arrest induces a caspase-dependent DNA damage response at telomeres that determines cell survival Karolina O. Hain, Didier J. Colin, Shubhra Rastogi, Lindsey
Pathologic Stage. Lymph node Stage
 ASC ASC a c Patient ID BMI Age Gleason score Non-obese PBMC 1 22.1 81 6 (3+3) PBMC 2 21.9 6 6 (3+3) PBMC 3 22 84 8 (4+4) PBMC 4 24.6 68 7 (3+4) PBMC 24. 6 (3+3) PBMC 6 24.7 73 7 (3+4) PBMC 7 23. 67 7 (3+4)
ASC ASC a c Patient ID BMI Age Gleason score Non-obese PBMC 1 22.1 81 6 (3+3) PBMC 2 21.9 6 6 (3+3) PBMC 3 22 84 8 (4+4) PBMC 4 24.6 68 7 (3+4) PBMC 24. 6 (3+3) PBMC 6 24.7 73 7 (3+4) PBMC 7 23. 67 7 (3+4)
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1. Galectin-3 is present within tumors. (A) mrna expression levels of Lgals3 (galectin-3) and Lgals8 (galectin-8) in the four classes of cell lines as determined
Supplementary Figures Supplementary Fig. 1. Galectin-3 is present within tumors. (A) mrna expression levels of Lgals3 (galectin-3) and Lgals8 (galectin-8) in the four classes of cell lines as determined
Highly Significant Antiviral Activity of HIV-1 LTR-Specific Tre-
 Supporting Information Hauber et al. page 1 Text S1 - Supporting Information Highly Significant Antiviral Activity of HIV-1 LTR-Specific Tre- Recombinase in Humanized Mice Ilona Hauber 1, Helga Hofmann-Sieber
Supporting Information Hauber et al. page 1 Text S1 - Supporting Information Highly Significant Antiviral Activity of HIV-1 LTR-Specific Tre- Recombinase in Humanized Mice Ilona Hauber 1, Helga Hofmann-Sieber
Done By : WESSEN ADNAN BUTHAINAH AL-MASAEED
 Done By : WESSEN ADNAN BUTHAINAH AL-MASAEED Acute Myeloid Leukemia Firstly we ll start with this introduction then enter the title of the lecture, so be ready and let s begin by the name of Allah : We
Done By : WESSEN ADNAN BUTHAINAH AL-MASAEED Acute Myeloid Leukemia Firstly we ll start with this introduction then enter the title of the lecture, so be ready and let s begin by the name of Allah : We
Targeting tumour associated macrophages in anti-cancer therapies. Annamaria Gal Seminar Series on Drug Discovery Budapest 5 January 2018
 Targeting tumour associated macrophages in anti-cancer therapies Annamaria Gal Seminar Series on Drug Discovery Budapest 5 January 2018 Macrophages: Professional phagocytes of the myeloid lineage APC,
Targeting tumour associated macrophages in anti-cancer therapies Annamaria Gal Seminar Series on Drug Discovery Budapest 5 January 2018 Macrophages: Professional phagocytes of the myeloid lineage APC,
Supplemental Table S1
 Supplemental Table S. Tumorigenicity and metastatic potential of 44SQ cell subpopulations a Tumorigenicity b Average tumor volume (mm ) c Lung metastasis d CD high /4 8. 8/ CD low /4 6./ a Mice were injected
Supplemental Table S. Tumorigenicity and metastatic potential of 44SQ cell subpopulations a Tumorigenicity b Average tumor volume (mm ) c Lung metastasis d CD high /4 8. 8/ CD low /4 6./ a Mice were injected
Supplementary Figure 1. Deletion of Smad3 prevents B16F10 melanoma invasion and metastasis in a mouse s.c. tumor model.
 A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung
 Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
SALL1 functions as a tumor suppressor in breast cancer by regulating cancer cell senescence and metastasis through the NuRD complex
 Ma et al. Molecular Cancer (2018) 17:78 https://doi.org/10.1186/s12943-018-0824-y RESEARCH SALL1 functions as a tumor suppressor in breast cancer by regulating cancer cell senescence and metastasis through
Ma et al. Molecular Cancer (2018) 17:78 https://doi.org/10.1186/s12943-018-0824-y RESEARCH SALL1 functions as a tumor suppressor in breast cancer by regulating cancer cell senescence and metastasis through
Evaluation of STAT3 Signaling in Macrophages Using a Lentiviral Reporter System
 Evaluation of STAT3 Signaling in Macrophages Using a Lentiviral Reporter System Schwertfeger Laboratory Emily Hartsough Breast Cancer Prevalence Adapted from Siegel et. al Cancer Statistics. 2016 Tumor
Evaluation of STAT3 Signaling in Macrophages Using a Lentiviral Reporter System Schwertfeger Laboratory Emily Hartsough Breast Cancer Prevalence Adapted from Siegel et. al Cancer Statistics. 2016 Tumor
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature775 4 O.D. (595-655) 3 1 -ζ no antibody isotype ctrl Plated Soluble 1F6 397 7H11 Supplementary Figure 1 Soluble and plated anti- Abs induce -! signalling. B3Z cells stably expressing!
doi: 1.138/nature775 4 O.D. (595-655) 3 1 -ζ no antibody isotype ctrl Plated Soluble 1F6 397 7H11 Supplementary Figure 1 Soluble and plated anti- Abs induce -! signalling. B3Z cells stably expressing!
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure S1: Fibroblast-induced elongation of cancer cells requires direct contact with living fibroblasts. A. Representative images of HT29-GFP cultured in the presence
SUPPLEMENTARY FIGURES Supplementary Figure S1: Fibroblast-induced elongation of cancer cells requires direct contact with living fibroblasts. A. Representative images of HT29-GFP cultured in the presence
Nature Medicine: doi: /nm.2109
 HIV 1 Infects Multipotent Progenitor Cells Causing Cell Death and Establishing Latent Cellular Reservoirs Christoph C. Carter, Adewunmi Onafuwa Nuga, Lucy A. M c Namara, James Riddell IV, Dale Bixby, Michael
HIV 1 Infects Multipotent Progenitor Cells Causing Cell Death and Establishing Latent Cellular Reservoirs Christoph C. Carter, Adewunmi Onafuwa Nuga, Lucy A. M c Namara, James Riddell IV, Dale Bixby, Michael
Transduction of lentivirus to human primary CD4+ T cells
 Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Sirt1 Hmg20b Gm (0.17) 24 (17.3) 877 (857)
 3 (0.17) 24 (17.3) Sirt1 Hmg20 Gm4763 877 (857) c d Suppl. Figure 1. Screen validation for top candidate antagonists of Dot1L (a) Numer of genes with one (gray), two (cyan) or three (red) shrna scored
3 (0.17) 24 (17.3) Sirt1 Hmg20 Gm4763 877 (857) c d Suppl. Figure 1. Screen validation for top candidate antagonists of Dot1L (a) Numer of genes with one (gray), two (cyan) or three (red) shrna scored
SUPPLEMENTARY INFORMATION. Supp. Fig. 1. Autoimmunity. Tolerance APC APC. T cell. T cell. doi: /nature06253 ICOS ICOS TCR CD28 TCR CD28
 Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic
 Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Supplementary Material
 Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4
 Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4 (n=3 independent differentiation experiments for each
Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4 (n=3 independent differentiation experiments for each
Supplementary Figure 1
 d f a IL7 b IL GATA RORγt h HDM IL IL7 PBS Ilra R7 PBS HDM Ilra R7 HDM Foxp Foxp Ilra R7 HDM HDM Ilra R7 HDM. 9..79. CD + FOXP + T reg cell CD + FOXP T conv cell PBS Ilra R7 PBS HDM Ilra R7 HDM CD + FOXP
d f a IL7 b IL GATA RORγt h HDM IL IL7 PBS Ilra R7 PBS HDM Ilra R7 HDM Foxp Foxp Ilra R7 HDM HDM Ilra R7 HDM. 9..79. CD + FOXP + T reg cell CD + FOXP T conv cell PBS Ilra R7 PBS HDM Ilra R7 HDM CD + FOXP
PTEN opposes negative selection and enables oncogenic transformation of pre-b cells
 Supplementary information PTEN opposes negative selection and enables oncogenic transformation of pre-b cells Seyedmehdi Shojaee, Lai N. Chan, Maike Buchner, Valeria Cazzaniga, Kadriye Nehir Cosgun, Huimin
Supplementary information PTEN opposes negative selection and enables oncogenic transformation of pre-b cells Seyedmehdi Shojaee, Lai N. Chan, Maike Buchner, Valeria Cazzaniga, Kadriye Nehir Cosgun, Huimin
Supplemental Figure 1. Activated splenocytes upregulate Serpina3g and Serpina3f expression.
 Relative Serpin expression 25 2 15 1 5 Serpina3f 1 2 3 4 5 6 8 6 4 2 Serpina3g 1 2 3 4 5 6 C57BL/6 DBA/2 Supplemental Figure 1. Activated splenocytes upregulate Serpina3g and Serpina3f expression. Splenocytes
Relative Serpin expression 25 2 15 1 5 Serpina3f 1 2 3 4 5 6 8 6 4 2 Serpina3g 1 2 3 4 5 6 C57BL/6 DBA/2 Supplemental Figure 1. Activated splenocytes upregulate Serpina3g and Serpina3f expression. Splenocytes
Supplementary Figure S1 Supplementary Figure S2
 Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x
 Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x magnification). B: Second HC skin stained for TSLP receptor in brown
Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x magnification). B: Second HC skin stained for TSLP receptor in brown
Irf1 fold changes (D) 24h 48h. p-p65. t-p65. p-irf3. t-irf3. β-actin SKO TKO 100% 80% 60% 40% 20%
 Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
EGFR shrna A: CCGGCGCAAGTGTAAGAAGTGCGAACTCGAGTTCGCACTTCTTACACTTGCG TTTTTG. EGFR shrna B: CCGGAGAATGTGGAATACCTAAGGCTCGAGCCTTAGGTATTCCACATTCTCTT TTTG
 Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Supplementary Figure 1. Expression of CUGBP1 in non-parenchymal liver cells treated with TGF-β
 Supplementary Figures Supplementary Figure 1. Expression of CUGBP1 in non-parenchymal liver cells treated with TGF-β and LPS. Non-parenchymal liver cells were isolated and treated with or without TGF-β
Supplementary Figures Supplementary Figure 1. Expression of CUGBP1 in non-parenchymal liver cells treated with TGF-β and LPS. Non-parenchymal liver cells were isolated and treated with or without TGF-β
TITLE: Properties of Leukemia Stem Cells in a Novel Model of CML Progression to Lymphoid Blast Crisis
 AD Award Number: W81XWH-05-1-0608 TITLE: Properties of Leukemia Stem Cells in a Novel Model of CML Progression to Lymphoid Blast Crisis PRINCIPAL INVESTIGATOR: Craig T. Jordan, Ph.D. CONTRACTING ORGANIZATION:
AD Award Number: W81XWH-05-1-0608 TITLE: Properties of Leukemia Stem Cells in a Novel Model of CML Progression to Lymphoid Blast Crisis PRINCIPAL INVESTIGATOR: Craig T. Jordan, Ph.D. CONTRACTING ORGANIZATION:
SUPPORTING INFORMATIONS
 SUPPORTING INFORMATIONS Mice MT/ret RetCD3ε KO α-cd25 treated MT/ret Age 1 month 3 mnths 6 months 1 month 3 months 6 months 1 month 3 months 6 months 2/87 Survival 87/87 incidence of 17/87 1 ary tumor
SUPPORTING INFORMATIONS Mice MT/ret RetCD3ε KO α-cd25 treated MT/ret Age 1 month 3 mnths 6 months 1 month 3 months 6 months 1 month 3 months 6 months 2/87 Survival 87/87 incidence of 17/87 1 ary tumor
TITLE: Effects of Hematopoietic Stem Cell Age on CML Disease Progression
 AD Award Number: W81XWH-04-1-0795 TITLE: Effects of Hematopoietic Stem Cell Age on CML Disease Progression PRINCIPAL INVESTIGATOR: Kenneth Dorshkind, Ph.D. CONTRACTING ORGANIZATION: University of California,
AD Award Number: W81XWH-04-1-0795 TITLE: Effects of Hematopoietic Stem Cell Age on CML Disease Progression PRINCIPAL INVESTIGATOR: Kenneth Dorshkind, Ph.D. CONTRACTING ORGANIZATION: University of California,
