SUPPLEMENTARY INFORMATION
|
|
|
- Adam Owen Mason
- 5 years ago
- Views:
Transcription
1 DOI: /ncb2419 Figure S1 NiGFP localization in Dl mutant dividing SOPs. a-c) time-lapse analysis of NiGFP (green) in Dl mutant SOPs (H2B-RFP, red; clones were identified by the loss of nlsgfp) showing three dividing mutant SOPs, colorcoded with dots at t=0 (a) and t=10 mn (b-c ). NiGFP specifically accumulated at the apical interface between daughter cells (surface view in c). NiGFP was not detected in the nuclei of Dl SOP progeny cells (white dots in c ). Genotype: N 55e11 w 1118 P[Ubx-flp] P[neur-Histone2B-RFP]619 / Y ; M[3xP3-RFP.attP. w+.nigfp]51d / + ; FRT82B P[ubi-nlsGFP] / FRT82B Dl Rev
2 Figure S2 Cell fate and nuclear NiGFP in numb RNAi and spdo RNAi pupae. a-e) the levels of nuclear NiGFP (green) were examined in SOPs (marked by H2B-RFP, red) in living pupae of the following genotypes: wild-type (i.e. N 55e11 NiGFP; a-a ), numb RNAi (N 55e11 NiGFP numb RNAi ; b-b,d) and spdo RNAi (N 55e11 NiGFP spdo RNAi ; c-c, e) pupae. Panels a-c show projected views of 3 confocal sections (with a Dz of 0.24mm; NiGFP channels are shown in a, b and c ). The corresponding intensity profile are shown in a, b and c (H2B-RFP profile in red and NiGFP profile in green). Nuclear NiGFP levels appeared to be higher in piia than in piib in wild-type controls (a ). Similarly, nuclear NiGFP levels appeared to be higher in numb RNAi piia and piia-like cells (b ) than in spdo RNAi piib and piib-like cells (c ) relative to their epidermal neighbours. Nuclear NiGFP was monitored over time as in Fig. 2e: no difference in nuclear NiGFP was detected between daughter cells upon silencing of numb (d; red line) and spdo (e; red line). The wild-type control (green line) shown in Fig. 2e is shown here as a reference. f-i) Analysis of cell fate transformations upon the silencing of spdo and/or numb. The fate of the sensory cells was examined in wild-type (f), numb RNAi (g), spdo RNAi (h) and numb RNAi spdo RNAi (i) pupae at hr APF using Cut (a nuclear sensory organ cell marker, red), HRP (a neuron marker, blue) and Su(H) (a socket cell marker, green). Silencing of numb phenocopied the numb mutant phenotype and led to piib-topiia transformations, as revealed by the loss of HRP-positive neurons and the gain of Su(H)-positive socket cells. The silencing of spdo also phenocopied the spdo mutant phenotype and resulted in opposite piia-topiib transformations, as revealed by the loss of Su(H)-positive socket cells and the gain of HRP-positive neurons. Double silencing of numb and spdo had the same effect as the silencing of spdo alone, consistent with spdo being epistatic over numb. Genotypes: a-a : N 55e11 w 1118 P[neur-Histone2B-RFP]619 / Y ; M[3xP3-RFP.attP. w+.nigfp]51d / + b-b, d: N 55e11 w 1118 P[neur-Histone2B-RFP]619 / Y ; M[3xP3-RFP.attP. w+.nigfp]51d / + ; pnr-gal4 P[UAS-dsRNA numb]-3779r-3 / + c-c, e: N 55e11 w 1118 P[neur-Histone2B-RFP]619 / Y ; M[3xP3-RFP.attP. w+.nigfp]51d / P[UAS-dsRNA spdo] ; pnr-gal4 / + f: w 1118 g: w 1118 ; ; pnr-gal4 P[UAS-dsRNA numb]-3779r-3 / + h: w 1118 P[UAS-dsRNA spdo] / + ; pnr-gal4 / + i: w 1118 P[UAS-dsRNA spdo] / + ; pnr-gal4 P[UAS-dsRNA numb]- 3779R-3 / + 2
3 Figure S3 Live analysis of NiGFP localization upon conditional inhibition of endocytosis. NiGFP (green) localisation in shi ts pupae. At permissive temperature (22 C; t=-1), NiGFP localized in relatively uniform manner at the cell cortex of epidermal cells (top panels: surface views at z=0mm) and in small apical endosomes (middle panels, z=-1 mm) in shi ts pupae. Temperature was shifted at restrictive temperature (32 C) at t=0 (telophase was monitored in SOPs using H2B-RFP, red). NiGFP progressively accumulated into distinct foci at the apical cortex (surface views) and NiGFP containing endosomes disappeared (z=-1 mm) and NiGFP accumulated in dots along the piia/piib interface 3-5 mm below the apical surface (bottom panels). These changes were not observed in shi ts pupae at 22 C, nor in wild-type pupae at 32 C (not shown; see Fig. 4d-d ). Genotype: shi ts / Y ; M[3xP3-RFP.attP.w+.NiGFP]51D / + ; P[neur-Histone2B-RFP] / + 3
4 Figure S4 numb-independent inhibition of Notch by Spdo. a-b ) Ectopic expression of Spdo in MARCM clones (GFP, green) resulted in increased SOP density (Senseless, red) in developing nota at 16 hr APF. This increased density is indicative of reduced Notch signalling. This inhibitory effect of Spdo on Notch activity was seen in both wild-type cells (a,a ) and numb mutant cells (b,b ), indicating that Spdo overexpression can inhibit the activity of Notch even in the absence of Numb. c) Analysis of cell fate decisions along clone borders in developing nota at 16 hr APF (as shown in a-b ). The bars illustrate the percentage of GFP-negative (red; outside the clone) and GFP-positive (yellow; inside the clone) SOPs that are located along clone borders (% values, given on the y-axis, are indicated above each bar; n is the total number of SOPs scored along clone borders). The genotypes scored in this clone border analysis are indicated above each pair of red/ yellow bars. GFP positive cells are either mutant and/or expressing spdo. Cells expressing Spdo prior to SOP selection were found to be more likely to become SOPs than their wild-type neighbors. This indicates that cells with high Spdo activity have low Notch receptor activity 1. This inhibitory effect of Spdo overexpression on Notch was also observed in numb mutant clones, showing that Spdo can down-regulate Notch in a Numb-independent manner. Since spdo cells mutant have the same probability to become SOPs than their wild-type neighbors, we conclude that endogenous Spdo has no significant role in the process of SOP selection. Control wild-type clones were generated using the parental FRT40A and FRT82B chromosomes. Genotypes: a, c: w 1118 hs-flp ptub-gal4 UAS-nlsGFP ; FRT40A / FRT40A ptub-gal80 ; P[UAS-Spdo]G1 / + b, c: w 1118 hs-flp ptub-gal4 UAS-nlsGFP ; FRT40A numb 15 / FRT40A ptub-gal80 ; P[UAS-Spdo]G1 / + c: w 1118 hs-flp ptub-gal4 UAS-nlsGFP ; FRT40A / FRT40A ptub-gal80 c: w 1118 hs-flp ptub-gal4 UAS-nlsGFP ; FRT40A numb 15 / FRT40A ptub- GAL80 c: w 1118 hs-flp ptub-gal4 UAS-nlsGFP ; FRT82B spdo G104 / FRT82B ptub- GAL80 c: w 1118 hs-flp ptub-gal4 UAS-nlsGFP ; FRT82B / FRT82B ptub-gal80 4
5 Figure S5 endocytosis of Notch by Spdo did not strictly require the NPAF motif of Spdo. Expression of GFP-Spdo (green in a-c ) and GFP-Spdo NP>AA (green in d-f ; the NPAF motif of Spdo that mediates interaction with Numb is deleted in GFP-Spdo NP>AA 2 ) in wing imaginal cells (ectopic expression of Spdo was directed in dorsal cells using ap-gal4) resulted in reduced levels of Notch at the apical cortex (NECD, red). Note that GFP-Spdo NP>AA localized baso-laterally whereas GFP-Spdo localized more apically. Panels b, b, e and e show high magnification views of a and d, respectively. Cross section views are shown in panels c, c, f and f. We conclude that the Spdo- Numb interaction is not strictly required for the down-regulation of Notch by Spdo. Genotypes: a-c : w 1118 ; ap-gal4 / UAS-GFP-Spdo d-f : w 1118 ; ap-gal4 / + ; UAS-GFP-Spdo NP>AA / + 5
6 Figure S6 Numb enhanced the down-regulation of Notch by Spdo. Analysis of the density of SOPs (marked by Senseless) in 16 hr APF pupae was used to investigate the effect of Numb and Spdo on the activity of Notch. Mild overexpression of Numb (b) and Spdo (c) had a weak inhibitory effect on Notch (compare with the negative control in a), as revealed by a mild increase in SOP number. Concomitant overexpression of Numb and Spdo resulted in a significant increase in SOP density (d), indicating that Numb enhanced the inhibitory effect of Spdo on the activity of Notch. Genotypes: a: w 1118 ; ; eq-gal4 / + b: w 1118 ; ; eq-gal4 / P[UAS-Numb] c: w 1118 ; ; eq-gal4 / P[UAS-Spdo]G1 d: w 1118 ; ; eq-gal4 / P[UAS-Numb] P[UAS-Spdo]G1 6
7 Supplemental Information - Movies Movie 1: live imaging of Notch in a wild-type SOP Spinning disk confocal sections corresponding to surface (z=0), subapical (z=-1mm) and basal (z=-5mm) views of NiGFP (green/white) are shown together with H2B-RFP (magenta; maximal projection of several confocal sections). Genotype: N 55e11 w 1118 P[neur-Histone2B-RFP]619 / Y ; M[3xP3-RFP.attP.w+.NiGFP]51D / + Movie 2: live imaging of cytokinesis in a wild-type SOP Actin (sgmca, white) and Histone2B (H2B-RFP, red) were imaged using spinning disk confocal microscopy. Confocal sections corresponding to surface (z=0), subapical (z=-3mm) and basal (z=-5mm) views are shown. A maximal projection of H2B-RFP channel is shown. Cytokinetic furrow initially forms basally. Furrow ingression is concomitant to chromatin decondensation. Note that apical actin accumulates in the newly born SOP daughter cells. This accumulation has been described as the Actin Rich Structure (ARS) 3. Genotype: w 1118 P[pNeur-H2B-RFP]619 / Y; P[pSqh-MoeABD-GFP] (sgmca ) /+ Movie 3: live imaging of Notch in a numb RNAi SOP Spinning disk confocal sections corresponding to surface (z=0) and basal (z=-5mm) views of NiGFP (green/white) are shown together with H2B-RFP (magenta; maximal projection of several confocal sections). A transient basal accumulation of NiGFP is indicated by an arrow. Genotype: N 55e11 w 1118 P[neur-Histone2B-RFP]619; M[3xP3-RFP.attP.w+.NiGFP]51D / + ; pnr-gal4 P[UAS-dsRNA numb]-3779r-3 / + Movie 4: live imaging of Notch in a spdo RNAi SOP Spinning disk confocal sections corresponding to surface (z=0) and subapical (z=-1mm) views of NiGFP (green/white) are shown together with H2B-RFP (magenta; maximal projection of several confocal sections). A transient apical accumulation of NiGFP is indicated by an arrow. Genotype: N 55e11 w 1118 P[neur-Histone2B-RFP]619; M[3xP3-RFP.attP.w+.NiGFP]51D / P[UAS-dsRNA spdo] ; pnr-gal4 / + 7
8 GENOTYPES PANELS w 1118 shi ts1 N 55e11 w 1118 / Y ; M[3xP3-RFP.attP.w+.NiGFP]51D / + N 55e11 w 1118 P[neur-Histone2B-RFP]619 / Y ; M[3xP3- RFP.attP.w+.NiGFP]51D / + N 55e11 w 1118 P[neur-PH-mRFP1]-965 / Y ; P[neur-PHmRFP1]-967 M[3xP3-RFP.attP.w+.NiGFP]51D / + d-d : N 55e11 w 1118 P[Ubx-flp] P[neur-Histone2B- RFP]619 / Y ; M[3xP3-RFP.attP.w+.NiGFP]51D / + ; FRT82B P[ubi-nlsGFP] / FRT82B neur IF65 N 55e11 w 1118 P[neur-PH-mRFP1]-965 / Y ; P[neur-PHmRFP1]-967 M[3xP3-RFP.attP.w+.NiGFP]51D / + N 55e11 w 1118 P[neur-Histone2B-RFP]619 / Y ; M[3xP3- RFP.attP.w+.NiGFP]51D / + ; pnr-gal4 P[UAS-dsRNA numb]-3779r-3 / + N 55e11 w 1118 P[neur-Histone2B-RFP]619 / Y ; M[3xP3- RFP.attP.w+.NiGFP]51D / P[UAS-dsRNA spdo] ; pnr-gal4 / + N 55e11 w 1118 P[neur-Histone2B-RFP]619 / Y ; M[3xP3- RFP.attP.w+.NiGFP]51D / P[UAS-dsRNA spdo] ; pnr-gal4 P[UAS-dsRNA numb]-3779r-3 / + N 55e11 w 1118 P[Ubx-flp] P[neur-Histone2B-RFP]619 / Y ; numb 15 FRT40A M[3xP3-RFP.attP.w+.NiGFP]51D / P[ubi-nlsGFP] FRT40A N 55e11 w 1118 P[Ubx-flp] P[neur-Histone2B-RFP]619 / Y ; M[3xP3-RFP.attP.w+.NiGFP]51D / + ; FRT82B P[ubinlsGFP] / FRT82B spdo G104 Fig. 2b,b ; Fig. 4b,b ; Fig. 4f; Fig. 6d-d Fig. 4c,c Fig. 2a,a Fig. 1c-c ; Fig. 1e-g ; Fig. 2c,c ; Fig. 2e; Fig. 3a; Fig. 4d-d ; Fig. 5a Fig. 2d Fig. 1d-d Fig. 1i,i' Fig. 3b; Fig. 4g Fig. 3c; Fig. 4h; Fig. 5b Fig. 3d Fig. 3e Fig. 3f
9 N 55e11 w 1118 P[neur-PH-mRFP1]-965 / Y ; M[3xP3- RFP.attP.w+.NiGFP]51D / + ; pnr-gal4 P[UAS-dsRNA numb]-3779r-3 / + Fig. 4a-a shi ts1 / Y ; M[3xP3-RFP.attP.w+.NiGFP]51D / + ; P[neur- Histone2B-RFP] / + Fig. 4e-e N 55e11 w 1118 P[Ubx-flp] P[neur-Histone2B-RFP]619 / Y ; M[3xP3-RFP.attP.w+.NiGFP]51D / + ; FRT82B P[ubinlsGFP] / FRT82B Dl Rev10 Fig. 5c,c N 55e11 w 1118 P[Ubx-flp] P[neur-Histone2B-RFP]619 / Y ; M[3xP3-RFP.attP.w+.NiGFP]51D / + ; FRT82B P[ubinlsGFP] / FRT82B Dl Rev10 spdo G104 Fig. 5d-d w 1118 ; ap-gal4 / + P[UAS-Spdo]G1 / + w 1118 ; neurp-gal4 / P[UAS-SpdoL2-Cherry] Fig. 5e-f Fig. 6a-c
Endocytosis by Numb breaks Notch symmetry at cytokinesis
 A R T I C L E S Endocytosis by Numb breaks Notch symmetry at cytokinesis Lydie Couturier 1,2, Nicolas Vodovar 1,2,3 and François Schweisguth 1,2,4 Cell-fate diversity can be generated by the unequal segregation
A R T I C L E S Endocytosis by Numb breaks Notch symmetry at cytokinesis Lydie Couturier 1,2, Nicolas Vodovar 1,2,3 and François Schweisguth 1,2,4 Cell-fate diversity can be generated by the unequal segregation
Supporting Information
 Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
Supplementary Figure S1: TIPF reporter validation in the wing disc.
 Supplementary Figure S1: TIPF reporter validation in the wing disc. a,b, Test of put RNAi. a, In wildtype discs the Dpp target gene Sal (red) is expressed in a broad stripe in the centre of the ventral
Supplementary Figure S1: TIPF reporter validation in the wing disc. a,b, Test of put RNAi. a, In wildtype discs the Dpp target gene Sal (red) is expressed in a broad stripe in the centre of the ventral
Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed
 Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed in the testes. The testes were immunostained with GFP
Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed in the testes. The testes were immunostained with GFP
glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal
 glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal Development in the Visual System of Drosophila Carole Chotard, Wendy Leung and Iris Salecker Supplemental Data Supplemental
glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal Development in the Visual System of Drosophila Carole Chotard, Wendy Leung and Iris Salecker Supplemental Data Supplemental
effects on organ development. a-f, Eye and wing discs with clones of ε j2b10 show no
 Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells.
 Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
Supplementary Figure 1. Nature Neuroscience: doi: /nn.4547
 Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
Unduplicated. Chromosomes. Telophase
 10-2 Cell Division The Cell Cycle Interphase Mitosis Prophase Cytokinesis G 1 S G 2 Chromatin in Parent Nucleus & Daughter Cells Chromatin Daughter Nuclei Telophase Mitotic Anaphase Metaphase Use what
10-2 Cell Division The Cell Cycle Interphase Mitosis Prophase Cytokinesis G 1 S G 2 Chromatin in Parent Nucleus & Daughter Cells Chromatin Daughter Nuclei Telophase Mitotic Anaphase Metaphase Use what
Supplementary Figure 1
 Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2294 Figure S1 Localization and function of cell wall polysaccharides in root hair cells. (a) Spinning-disk confocal sections of seven day-old A. thaliana seedlings stained with 0.1% S4B
DOI: 10.1038/ncb2294 Figure S1 Localization and function of cell wall polysaccharides in root hair cells. (a) Spinning-disk confocal sections of seven day-old A. thaliana seedlings stained with 0.1% S4B
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Expression of escargot (esg) and genetic approach for achieving IPC-specific knockdown. (a) esg MH766 -Gal4 UAS-cd8GFP (green) and esg-lacz B7-2-22 (red) show similar expression
Supplementary Figure 1 Expression of escargot (esg) and genetic approach for achieving IPC-specific knockdown. (a) esg MH766 -Gal4 UAS-cd8GFP (green) and esg-lacz B7-2-22 (red) show similar expression
Supplementary Figure 1. Flies form water-reward memory only in the thirsty state
 1 2 3 4 5 6 7 Supplementary Figure 1. Flies form water-reward memory only in the thirsty state Thirsty but not sated wild-type flies form robust 3 min memory. For the thirsty group, the flies were water-deprived
1 2 3 4 5 6 7 Supplementary Figure 1. Flies form water-reward memory only in the thirsty state Thirsty but not sated wild-type flies form robust 3 min memory. For the thirsty group, the flies were water-deprived
supplementary information
 DOI: 10.1038/ncb2153 Figure S1 Ectopic expression of HAUSP up-regulates REST protein. (a) Immunoblotting showed that ectopic expression of HAUSP increased REST protein levels in ENStemA NPCs. (b) Immunofluorescent
DOI: 10.1038/ncb2153 Figure S1 Ectopic expression of HAUSP up-regulates REST protein. (a) Immunoblotting showed that ectopic expression of HAUSP increased REST protein levels in ENStemA NPCs. (b) Immunofluorescent
Dominant-negative mutation in the 2 and 6 proteasome subunit genes affect alternative cell fate decisions in the Drosophila sense organ lineage
 Proc. Natl. Acad. Sci. USA Vol. 96, pp. 11382 11386, September 1999 Developmental Biology Dominant-negative mutation in the 2 and 6 proteasome subunit genes affect alternative cell fate decisions in the
Proc. Natl. Acad. Sci. USA Vol. 96, pp. 11382 11386, September 1999 Developmental Biology Dominant-negative mutation in the 2 and 6 proteasome subunit genes affect alternative cell fate decisions in the
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3443 In the format provided by the authors and unedited. Supplementary Figure 1 TC and SC behaviour during ISV sprouting. (a) Predicted outcome of TC division and competitive Dll4-Notch-mediated
DOI: 10.1038/ncb3443 In the format provided by the authors and unedited. Supplementary Figure 1 TC and SC behaviour during ISV sprouting. (a) Predicted outcome of TC division and competitive Dll4-Notch-mediated
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Brooks and Wallingford, http://www.jcb.org/cgi/content/full/jcb.201204072/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Quantification of ciliary compartments in control
Supplemental material Brooks and Wallingford, http://www.jcb.org/cgi/content/full/jcb.201204072/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Quantification of ciliary compartments in control
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. Procedures to independently control fly hunger and thirst states.
 Supplementary Figure 1 Procedures to independently control fly hunger and thirst states. (a) Protocol to produce exclusively hungry or thirsty flies for 6 h water memory retrieval. (b) Consumption assays
Supplementary Figure 1 Procedures to independently control fly hunger and thirst states. (a) Protocol to produce exclusively hungry or thirsty flies for 6 h water memory retrieval. (b) Consumption assays
ns ns hp761(lf); daf-28(lf) daf-28(gf) Figure S1
 A ns ns B 100 100 80 80 60 60 40 40 20 20 0 0% 0 0% hp761(lf); daf-28(lf) daf-28(gf) Figure S1 A 100 80 15 o C 22 o C 25 o C % Dauers 60 40 20 0 0% 0% * 0% 0% 0% Class I alleles Class II alleles daf-2(lf;ts)
A ns ns B 100 100 80 80 60 60 40 40 20 20 0 0% 0 0% hp761(lf); daf-28(lf) daf-28(gf) Figure S1 A 100 80 15 o C 22 o C 25 o C % Dauers 60 40 20 0 0% 0% * 0% 0% 0% Class I alleles Class II alleles daf-2(lf;ts)
Supplemental Information. Competition for Space Is Controlled. by Apoptosis-Induced Change. of Local Epithelial Topology
 Current Biology, Volume 28 Supplemental Information Competition for Space Is Controlled by Apoptosis-Induced Change of Local Epithelial Topology Alice Tsuboi, Shizue Ohsawa, Daiki Umetsu, Yukari Sando,
Current Biology, Volume 28 Supplemental Information Competition for Space Is Controlled by Apoptosis-Induced Change of Local Epithelial Topology Alice Tsuboi, Shizue Ohsawa, Daiki Umetsu, Yukari Sando,
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
Karen L.P. McNally, Amy S. Fabritius, Marina L. Ellefson, Jonathan R. Flynn, Jennifer A. Milan, and Francis J. McNally
 Developmental Cell, Volume 22 Supplemental Information Kinesin-1 Prevents Capture of the Oocyte Meiotic Spindle by the Sperm Aster Karen L.P. McNally, Amy S. Fabritius, Marina L. Ellefson, Jonathan R.
Developmental Cell, Volume 22 Supplemental Information Kinesin-1 Prevents Capture of the Oocyte Meiotic Spindle by the Sperm Aster Karen L.P. McNally, Amy S. Fabritius, Marina L. Ellefson, Jonathan R.
Ahtiainen et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB Ahtiainen et al., http ://www.jcb.org /cgi /content /full /jcb.201512074 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Distinct distribution of different cell cycle phases in the
Supplemental material JCB Ahtiainen et al., http ://www.jcb.org /cgi /content /full /jcb.201512074 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Distinct distribution of different cell cycle phases in the
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human
 Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
genome edited transient transfection, CMV promoter
 Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
100 mm Sucrose. +Berberine +Quinine
 8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
 Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cédric Maurange, Louise Cheng and Alex P. Gould Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cédric Maurange, Louise Cheng and Alex P. Gould Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
Developmental Biology
 Developmental Biology 327 (2009) 288 300 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology The HLH protein Extramacrochaetae is required
Developmental Biology 327 (2009) 288 300 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology The HLH protein Extramacrochaetae is required
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang and Page-McCaw, http://www.jcb.org/cgi/content/full/jcb.201403084/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Extracellular anti-wg staining is specific. Note
Supplemental material Wang and Page-McCaw, http://www.jcb.org/cgi/content/full/jcb.201403084/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Extracellular anti-wg staining is specific. Note
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
Supplementary Figures
 J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
Supplemental Information. Proprioceptive Opsin Functions. in Drosophila Larval Locomotion
 Neuron, Volume 98 Supplemental Information Proprioceptive Opsin Functions in Drosophila Larval Locomotion Damiano Zanini, Diego Giraldo, Ben Warren, Radoslaw Katana, Marta Andrés, Suneel Reddy, Stephanie
Neuron, Volume 98 Supplemental Information Proprioceptive Opsin Functions in Drosophila Larval Locomotion Damiano Zanini, Diego Giraldo, Ben Warren, Radoslaw Katana, Marta Andrés, Suneel Reddy, Stephanie
Supplementary Figure 1. Chimeric analysis of inner ears. (A-H) Chimeric inner ears with fluorescent ES cells and (I,J) Rainbow inner ears.
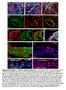 Supplementary Figure 1. himeric analysis of inner ears. (A-H) himeric inner ears with fluorescent ES cells and (I,J) Rainbow inner ears. (A,B) omposite images showing three colors in different vestibular
Supplementary Figure 1. himeric analysis of inner ears. (A-H) himeric inner ears with fluorescent ES cells and (I,J) Rainbow inner ears. (A,B) omposite images showing three colors in different vestibular
MII. Supplement Figure 1. CapZ β2. Merge. 250ng. 500ng DIC. Merge. Journal of Cell Science Supplementary Material. GFP-CapZ β2 DNA
 A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
The questions below refer to the following terms. Each term may be used once, more than once, or not at all.
 The questions below refer to the following terms. Each term may be used once, more than once, or not at all. a) telophase b) anaphase c) prometaphase d) metaphase e) prophase 1) DNA begins to coil and
The questions below refer to the following terms. Each term may be used once, more than once, or not at all. a) telophase b) anaphase c) prometaphase d) metaphase e) prophase 1) DNA begins to coil and
Tanimoto et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB Tanimoto et al., http ://www.jcb.org /cgi /content /full /jcb.201510064 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Method for aster 3D tracking, extended characterization of
Supplemental material JCB Tanimoto et al., http ://www.jcb.org /cgi /content /full /jcb.201510064 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Method for aster 3D tracking, extended characterization of
supplementary information
 DOI: 10.1038/ncb2157 Figure S1 Immobilization of histone pre-mrna to chromatin leads to formation of histone locus body with associated Cajal body. Endogenous histone H2b(e) pre-mrna is processed with
DOI: 10.1038/ncb2157 Figure S1 Immobilization of histone pre-mrna to chromatin leads to formation of histone locus body with associated Cajal body. Endogenous histone H2b(e) pre-mrna is processed with
Nature Neuroscience: doi: /nn Supplementary Figure 1. MADM labeling of thalamic clones.
 Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
SUPPLEMENTARY INFORMATION
 Figure S1. Loss of Ena/VASP proteins inhibits filopodia and neuritogenesis. (a) Bar graph of filopodia number per stage 1 control and mmvvee (Mena/ VASP/EVL-null) neurons at 40hrs in culture. Loss of all
Figure S1. Loss of Ena/VASP proteins inhibits filopodia and neuritogenesis. (a) Bar graph of filopodia number per stage 1 control and mmvvee (Mena/ VASP/EVL-null) neurons at 40hrs in culture. Loss of all
SUPPLEMENTARY INFORMATION
 SUPPLEENTRY INFORTION DOI: 1.138/ncb2577 Early Telophase Late Telophase B icrotubules within the ICB (percent of total cells in telophase) D G ultinucleate cells (% total) 8 6 4 2 2 15 1 5 T without gaps
SUPPLEENTRY INFORTION DOI: 1.138/ncb2577 Early Telophase Late Telophase B icrotubules within the ICB (percent of total cells in telophase) D G ultinucleate cells (% total) 8 6 4 2 2 15 1 5 T without gaps
Capu and Spire Assemble a Cytoplasmic Actin Mesh
 Developmental Cell 13 Supplemental Data Capu and Spire Assemble a Cytoplasmic Actin Mesh that Maintains Microtubule Organization in the Drosophila Oocyte Katja Dahlgaard, Alexandre A.S.F. Raposo, Teresa
Developmental Cell 13 Supplemental Data Capu and Spire Assemble a Cytoplasmic Actin Mesh that Maintains Microtubule Organization in the Drosophila Oocyte Katja Dahlgaard, Alexandre A.S.F. Raposo, Teresa
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
IP: anti-gfp VPS29-GFP. IP: anti-vps26. IP: anti-gfp - + +
 FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
Supplementary information. The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic. Spindle Orientation
 Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Supplementary Information
 Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Chapter 2. Mitosis and Meiosis
 Chapter 2. Mitosis and Meiosis Chromosome Theory of Heredity What structures within cells correspond to genes? The development of genetics took a major step forward by accepting the notion that the genes
Chapter 2. Mitosis and Meiosis Chromosome Theory of Heredity What structures within cells correspond to genes? The development of genetics took a major step forward by accepting the notion that the genes
Supplemental Data. Beck et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
sfigure 1: Detection of L-fucose in normal mouse renal cortex using the plant lectin LTL
 sfigure 1: Detection of L-fucose in normal mouse renal cortex using the plant lectin LTL LTL staining Negative control Fluorescence microscopy of normal (CL-11 +/+ ) mouse renal tissue after staining with
sfigure 1: Detection of L-fucose in normal mouse renal cortex using the plant lectin LTL LTL staining Negative control Fluorescence microscopy of normal (CL-11 +/+ ) mouse renal tissue after staining with
Supplemental Figure 1. Quantification of proliferation in thyroid of WT, Ctns -/- and grafted
 Supplemental Figure 1. Quantification of proliferation in thyroid of WT, Ctns -/- and grafted Ctns -/- mice. Cells immunolabeled for the proliferation marker (Ki-67) were counted in sections (n=3 WT, n=4
Supplemental Figure 1. Quantification of proliferation in thyroid of WT, Ctns -/- and grafted Ctns -/- mice. Cells immunolabeled for the proliferation marker (Ki-67) were counted in sections (n=3 WT, n=4
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
Supplementary Figure 1
 Supplementary Figure 1 14 12 SEM4C PLXN2 8 SEM4C C 3 Cancer Cell Non Cancer Cell Expression 1 8 6 6 4 log2 ratio Expression 2 1 4 2 2 p value.1 D Supplementary Figure 1. Expression of Sema4C and Plexin2
Supplementary Figure 1 14 12 SEM4C PLXN2 8 SEM4C C 3 Cancer Cell Non Cancer Cell Expression 1 8 6 6 4 log2 ratio Expression 2 1 4 2 2 p value.1 D Supplementary Figure 1. Expression of Sema4C and Plexin2
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila
 Article The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila Sarah K. Bowman, 1 Vivien Rolland, 1 Joerg Betschinger, 1,3 Kaolin A. Kinsey, 2 Gregory Emery,
Article The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila Sarah K. Bowman, 1 Vivien Rolland, 1 Joerg Betschinger, 1,3 Kaolin A. Kinsey, 2 Gregory Emery,
Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were
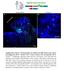 Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were electroporated with β- Catenin S33Y in PiggyBac expression
Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were electroporated with β- Catenin S33Y in PiggyBac expression
The Cell Cycle. Dr. SARRAY Sameh, Ph.D
 The Cell Cycle Dr. SARRAY Sameh, Ph.D Overview When an organism requires additional cells (either for growth or replacement of lost cells), new cells are produced by cell division (mitosis) Somatic cells
The Cell Cycle Dr. SARRAY Sameh, Ph.D Overview When an organism requires additional cells (either for growth or replacement of lost cells), new cells are produced by cell division (mitosis) Somatic cells
50mM D-Glucose. Percentage PI. L-Glucose
 Monica Dus, Minrong Ai, Greg S.B Suh. Taste-independent nutrient selection is mediated by a brain-specific Na+/solute cotransporter in Drosophila. Control + Phlorizin mm D-Glucose 1 2mM 1 L-Glucose Gr5a;Gr64a
Monica Dus, Minrong Ai, Greg S.B Suh. Taste-independent nutrient selection is mediated by a brain-specific Na+/solute cotransporter in Drosophila. Control + Phlorizin mm D-Glucose 1 2mM 1 L-Glucose Gr5a;Gr64a
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Information. Cofilin Regulates Nuclear Architecture through a Myosin-II Dependent Mechanotransduction Module
 Supplementary Information Cofilin Regulates Nuclear Architecture through a Myosin-II Dependent Mechanotransduction Module O Neil Wiggan, Bryce Schroder, Diego Krapf, James R. Bamurg and Jennifer G. DeLuca
Supplementary Information Cofilin Regulates Nuclear Architecture through a Myosin-II Dependent Mechanotransduction Module O Neil Wiggan, Bryce Schroder, Diego Krapf, James R. Bamurg and Jennifer G. DeLuca
Supplementary Information
 Supplementary Information Title Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis Authors Shin H. Kang, Ying Li, Masahiro Fukaya, Ileana Lorenzini,
Supplementary Information Title Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis Authors Shin H. Kang, Ying Li, Masahiro Fukaya, Ileana Lorenzini,
Cell Division Mitosis Notes
 Cell Division Mitosis Notes Cell Division process by which a cell divides into 2 new cells Why do cells need to divide? 1.Living things grow by producing more cells, NOT because each cell increases in
Cell Division Mitosis Notes Cell Division process by which a cell divides into 2 new cells Why do cells need to divide? 1.Living things grow by producing more cells, NOT because each cell increases in
Cell Cycle. Interphase, Mitosis, Cytokinesis, and Cancer
 Cell Cycle Interphase, Mitosis, Cytokinesis, and Cancer Cell Division One cell divides into 2 new identical daughter cells. Chromosomes carry the genetic information (traits) of the cell How many Chromosomes
Cell Cycle Interphase, Mitosis, Cytokinesis, and Cancer Cell Division One cell divides into 2 new identical daughter cells. Chromosomes carry the genetic information (traits) of the cell How many Chromosomes
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
supplementary information
 DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2697 Figure S1 Cytokeratin 5 is a specific marker for basal and intermediate cells in all mouse prostate lobes. (a) Immunofluorescence staining showing co-localization of YFP with p63 in
DOI: 10.1038/ncb2697 Figure S1 Cytokeratin 5 is a specific marker for basal and intermediate cells in all mouse prostate lobes. (a) Immunofluorescence staining showing co-localization of YFP with p63 in
Supplementary Figure S1 (a) (b)
 Supplementary Figure S1: IC87114 does not affect basal Ca 2+ level nor nicotineinduced Ca 2+ influx. (a) Bovine chromaffin cells were loaded with Fluo-4AM (1 μm) in buffer A containing 0.02% of pluronic
Supplementary Figure S1: IC87114 does not affect basal Ca 2+ level nor nicotineinduced Ca 2+ influx. (a) Bovine chromaffin cells were loaded with Fluo-4AM (1 μm) in buffer A containing 0.02% of pluronic
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supplemental Materials Molecular Biology of the Cell
 Supplemental Materials Molecular Biology of the Cell Garcia-Alvarez et al. Supplementary Figure Legends Figure S1.Expression and RNAi-mediated silencing of STIM1 in hippocampal neurons (DIV, days in vitro).
Supplemental Materials Molecular Biology of the Cell Garcia-Alvarez et al. Supplementary Figure Legends Figure S1.Expression and RNAi-mediated silencing of STIM1 in hippocampal neurons (DIV, days in vitro).
ROCK/Cdc42-mediated microglial motility and gliapse formation lead to phagocytosis of degenerating dopaminergic neurons in vivo
 Supplementary Information ROCK/Cdc42-mediated microglial motility and gliapse formation lead to phagocytosis of degenerating dopaminergic neurons in vivo Carlos Barcia* 1,2, Carmen M Ros 1,2, Valentina
Supplementary Information ROCK/Cdc42-mediated microglial motility and gliapse formation lead to phagocytosis of degenerating dopaminergic neurons in vivo Carlos Barcia* 1,2, Carmen M Ros 1,2, Valentina
Unit 6: Study Guide Cell Division. diploid gene allele interphase (G1, S, G2) prophase metaphase anaphase
 Unit 6: Study Guide Cell Division 1. Define: chromatin chromosome chromatid pair (sister chromatid) centromere spindle fibers haploid diploid gene allele interphase (G1, S, G2) prophase metaphase anaphase
Unit 6: Study Guide Cell Division 1. Define: chromatin chromosome chromatid pair (sister chromatid) centromere spindle fibers haploid diploid gene allele interphase (G1, S, G2) prophase metaphase anaphase
F-actin VWF Vinculin. F-actin. Vinculin VWF
 a F-actin VWF Vinculin b F-actin VWF Vinculin Supplementary Fig. 1. WPBs in HUVECs are located along stress fibers and at focal adhesions. (a) Immunofluorescence images of f-actin (cyan), VWF (yellow),
a F-actin VWF Vinculin b F-actin VWF Vinculin Supplementary Fig. 1. WPBs in HUVECs are located along stress fibers and at focal adhesions. (a) Immunofluorescence images of f-actin (cyan), VWF (yellow),
Mitosis/Meiosis Simulation Activities
 Mitosis/Meiosis Simulation Activities In this simulation, you will demonstrate an understanding of mitosis, meiosis, segregation, independent assortment, and crossing over, all processes involved with
Mitosis/Meiosis Simulation Activities In this simulation, you will demonstrate an understanding of mitosis, meiosis, segregation, independent assortment, and crossing over, all processes involved with
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5
 Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
Unit 6: CELL DIVISION PACKET
 Unit 6: CELL DIVISION PACKET This packet is designed to help you understand several concepts about Cell Division. As you practice the exercises on each handout, you will be able to: Use a model to illustrate
Unit 6: CELL DIVISION PACKET This packet is designed to help you understand several concepts about Cell Division. As you practice the exercises on each handout, you will be able to: Use a model to illustrate
Cell Division Mitosis Notes
 Cell Division Mitosis Notes Cell Division process by which a cell divides into 2 new cells Why do cells need to divide? 1.Living things grow by producing more cells, NOT because each cell increases in
Cell Division Mitosis Notes Cell Division process by which a cell divides into 2 new cells Why do cells need to divide? 1.Living things grow by producing more cells, NOT because each cell increases in
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
Nature Neuroscience: doi: /nn Supplementary Figure 1. Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites.
 Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Meigo governs dendrite targeting specificity by modulating Ephrin level and N-glycosylation
 Meigo governs dendrite targeting specificity by modulating Ephrin level and N-glycosylation Sayaka U. Sekine, Shuka Haraguchi, Kinhong Chao, Tomoko Kato, Liqun Luo, Masayuki Miura, and Takahiro Chihara
Meigo governs dendrite targeting specificity by modulating Ephrin level and N-glycosylation Sayaka U. Sekine, Shuka Haraguchi, Kinhong Chao, Tomoko Kato, Liqun Luo, Masayuki Miura, and Takahiro Chihara
Supplemental information contains 7 movies and 4 supplemental Figures
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
Border of Notch activity establishes a boundary between the two dorsal appendage tube cell types
 Developmental Biology 297 (2006) 461 470 www.elsevier.com/locate/ydbio Border of Notch activity establishes a boundary between the two dorsal appendage tube cell types Ellen J. Ward a,c, Xiaofeng Zhou
Developmental Biology 297 (2006) 461 470 www.elsevier.com/locate/ydbio Border of Notch activity establishes a boundary between the two dorsal appendage tube cell types Ellen J. Ward a,c, Xiaofeng Zhou
Supplementary Figure 1. Gene schematics of hyls-1, gasr-8 and k10g6.4, and TEM analysis of TFs in WT and hyls-1 cilia. (a) Gene structure of hyls-1,
 Supplementary Figure 1. Gene schematics of hyls-1, gasr-8 and k10g6.4, and TEM analysis of TFs in WT and hyls-1 cilia. (a) Gene structure of hyls-1, gasr-8 and k10g6.4 based on WormBase (http://wormbase.org),
Supplementary Figure 1. Gene schematics of hyls-1, gasr-8 and k10g6.4, and TEM analysis of TFs in WT and hyls-1 cilia. (a) Gene structure of hyls-1, gasr-8 and k10g6.4 based on WormBase (http://wormbase.org),
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Nature Neuroscience: doi: /nn Supplementary Figure 1. Trial structure for go/no-go behavior
 Supplementary Figure 1 Trial structure for go/no-go behavior a, Overall timeline of experiments. Day 1: A1 mapping, injection of AAV1-SYN-GCAMP6s, cranial window and headpost implantation. Water restriction
Supplementary Figure 1 Trial structure for go/no-go behavior a, Overall timeline of experiments. Day 1: A1 mapping, injection of AAV1-SYN-GCAMP6s, cranial window and headpost implantation. Water restriction
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
Supplementary Figure 1: GFAP positive nerves in patients with adenocarcinoma of
 SUPPLEMENTARY FIGURES AND MOVIE LEGENDS Supplementary Figure 1: GFAP positive nerves in patients with adenocarcinoma of the pancreas. (A) Images of nerves stained for GFAP (green), S100 (red) and DAPI
SUPPLEMENTARY FIGURES AND MOVIE LEGENDS Supplementary Figure 1: GFAP positive nerves in patients with adenocarcinoma of the pancreas. (A) Images of nerves stained for GFAP (green), S100 (red) and DAPI
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Fig. 1. Simple columnar epithelial cells lining the small intestine.
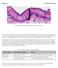 Mitosis Prelab Reading Fig. 1. Simple columnar epithelial cells lining the small intestine. The tall cells pictured in Fig. 1 form the lining of the small intestine in humans and other animals. These cells
Mitosis Prelab Reading Fig. 1. Simple columnar epithelial cells lining the small intestine. The tall cells pictured in Fig. 1 form the lining of the small intestine in humans and other animals. These cells
Mitosis and Cellular Division. EQ: How do the cells in our body divide?
 Mitosis and Cellular Division EQ: How do the cells in our body divide? Cell division is the process by which cellular material is divided between two new daughter cells. 1 Mother Cell 2 Daughter cells.
Mitosis and Cellular Division EQ: How do the cells in our body divide? Cell division is the process by which cellular material is divided between two new daughter cells. 1 Mother Cell 2 Daughter cells.
Cell Birth and Death. Chapter Three
 Cell Birth and Death Chapter Three Neurogenesis All neurons and glial cells begin in the neural tube Differentiated into neurons rather than ectoderm based on factors we have already discussed If these
Cell Birth and Death Chapter Three Neurogenesis All neurons and glial cells begin in the neural tube Differentiated into neurons rather than ectoderm based on factors we have already discussed If these
Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses
 Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses using an anti-cre antibody; testes at 1 week (left panel),
Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses using an anti-cre antibody; testes at 1 week (left panel),
An O-fucose site in the ligand binding domain inhibits Notch activation
 Research article 6411 An O-fucose site in the ligand binding domain inhibits Notch activation Liang Lei, Aiguo Xu, Vladislav M. Panin* and Kenneth D. Irvine Howard Hughes Medical Institute, Waksman Institute
Research article 6411 An O-fucose site in the ligand binding domain inhibits Notch activation Liang Lei, Aiguo Xu, Vladislav M. Panin* and Kenneth D. Irvine Howard Hughes Medical Institute, Waksman Institute
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Divergent effects of intrinsically active MEK variants on developmental Ras signaling Yogesh Goyal,2,3,4, Granton A. Jindal,2,3,4, José L. Pelliccia 3, Kei Yamaya 2,3, Eyan Yeung
SUPPLEMENTARY MATERIAL Divergent effects of intrinsically active MEK variants on developmental Ras signaling Yogesh Goyal,2,3,4, Granton A. Jindal,2,3,4, José L. Pelliccia 3, Kei Yamaya 2,3, Eyan Yeung
Outline Interphase Mitotic Stage Cell Cycle Control Apoptosis Mitosis Mitosis in Animal Cells Cytokinesis Cancer Prokaryotic Cell Division
 The Cell Cycle and Cellular Reproduction Chapter 9 Outline Interphase Mitotic Stage Cell Cycle Control Apoptosis Mitosis Mitosis in Animal Cells Cytokinesis Cancer Prokaryotic Cell Division 1 2 Interphase
The Cell Cycle and Cellular Reproduction Chapter 9 Outline Interphase Mitotic Stage Cell Cycle Control Apoptosis Mitosis Mitosis in Animal Cells Cytokinesis Cancer Prokaryotic Cell Division 1 2 Interphase
Why do cells divide? Cells divide in order to make more cells they multiply in order to create a larger surface to volume ratio!!!
 Why do cells divide? Cells divide in order to make more cells they multiply in order to create a larger surface to volume ratio!!! Chromosomes Are made of chromatin: a mass of genetic material composed
Why do cells divide? Cells divide in order to make more cells they multiply in order to create a larger surface to volume ratio!!! Chromosomes Are made of chromatin: a mass of genetic material composed
