NEW PRODUCTS May 2013
|
|
|
- Edward Morrison
- 5 years ago
- Views:
Transcription
1 The World s Leading Producer of Ubiquitin-Related Research Products E3 Ligase Kits MuRF1/S5a Ubiquitination Kit K Kit MuRF1 (Muscle-specific RING-finger protein 1) is a RING-finger E3 ligase found in striated muscle (heart and skeletal) and iris tissues. The protein contains both RING and B-box zinc fingers, and a coiled-coil tripartite fold known as TRIM. MuRF1 ligase activity regulates the proteasomal degradation of cardiac troponin 1 and other sarcomeric-associated proteins. This kit is designed for in vitro MuRF1-mediated ubiquitination of user-supplied substrates such as cardiac troponin 1, MYH7, or muscle creatine kinase. Ubiquitinated proteins can be used in downstream applications, or analyzed by Western blot using antibodies specific for the target protein. A control substrate, S5a (SP-400) and detection antibody (AF5540) are included in the kit. MDM2/p53 Ubiquitination Kit, version B K-200B 1 Kit The RING-finger ubiquitin E3 ligase MDM2 (Murine Double Minute 2) is an important regulator of the tumor suppressor protein p53. MDM2 ligase activity is at least partially responsible for the ubiquitination and subsequent proteasomal degradation of p53. This kit is designed for in vitro ubiquitination of p53 (SP-450) by MDM2. The ubiquitinated protein can be detected by Western blot using the supplied α-p53 monoclonal antibody (MAB1355). This kit can be used as a positive control in studies that monitor the activities of other known or putative E3 ligase enzymes that ubiquitinate p53. Ubiquitinated p53 substrate can also be used to study E4 ligase activities of enzymes such as UBE4B, or the activity of deubiquitinating enzymes such as USP7 (E-518) MDM2/S5a Ubiquitination Kit K Kit The RING-finger ubiquitin E3 ligase MDM2 (Murine Double Minute 2) is an important regulator of the tumor suppressor protein p53. MDM2 ligase activity is at least partially responsible for the ubiquitination and subsequent proteasomal degradation of p53 and other proteins. This kit is designed for in vitro MDM2-mediated ubiquitination of user-supplied substrates such as p53 (SP-450), β-arrestin, and DYRK2. Ubiquitinated proteins can be used in downstream applications, or analyzed by Western blot using antibodies specific for the target protein. A control substrate, S5a (SP-400) and detection antibody (AF5540) are included in the kit. RNF4/di-SUMO3 Ubiquitination Kit K Kit RNF4 (SNURF) is a RING-finger ubiquitin E3 ligase that ubiquitinates and mediates the proteasomal destruction of PML/RARα, a fusion protein that is a hallmark of acute promyelocytic leukemia (APL). APL can sometimes be treated effectively with arsenic trioxide, which induces PML/RARα modification by small ubiquitin-like modifiers (SUMO). RNF4 subsequently binds to and ubiquitinates the poly-sumoylated PML/RARα, thereby targeting it to the proteasome. RNF4 contains four SUMO-interacting motifs (SIMs) that function to recruit this ligase to a variety of poly-sumoylated substrates. RNF4 will autoubiquitinate in vitro, and will also ubiquitinate poly-sumo chains. This kit is designed for in vitro RNF4-mediated ubiquitination of user-supplied, poly-sumoylated substrates such as PML, PEA3, CENP1, and PARP1. The resulting proteins can be used in downstream applications, or analyzed by Western blot using antibodies specific for the target protein. A control substrate, di-sumo3 (ULC-300) and detection antibody (MAB2959) are included in the kit. E6AP/S5a Ubiquitination Kit NEW PRODUCTS May 2013 K Kit E6AP (E6-Associated Protein) is a HECT domain ubiquitin E3 ligase that ubiquitinates and mediates the proteasomal destruction of substrate proteins. HECT domain ligases use an active site cysteine to accept charged ubiquitin from ubiquitin-e2 thioester complexes for subsequent transfer to substrate proteins; in this way HECT class ligases are distinct from most RING class ligases in that the latter facilitate transfer of ubiquitin from charged E2 s directly to substrate proteins without an E3-ubiquitin thioester intermediate. This kit is designed for in vitro E6AP-mediated ubiquitination of user-supplied, reported substrates such as β-arrestin, and DYRK2. Ubiquitinated proteins can be used in downstream applications, or analyzed by Western blot using antibodies specific for the target protein. A control substrate, S5a (SP-400) and detection antibody (AF5540) are included in the kit. E6AP/E6/p53 Ubiquitination Kit K Kit E6AP (E6-Associated Protein) (E3-230) is a HECT domain ubiquitin E3 ligase that ubiquitinates and mediates the proteasomal destruction of substrate proteins. HECT domain ligases use an active site cysteine to accept charged ubiquitin from ubiquitin-e2 thioester complexes for subsequent transfer to substrate proteins; in this way HECT class ligases are distinct from most RING class ligases in that the latter facilitate transfer of ubiquitin from charged E2 s directly to substrate proteins without an E3-ubiquitin thioester intermediate. E6 (Early protein 6) (AP-120) is a viral protein produced in cells infected with the Human Papillomavirus. E6 forms a complex with the host cell E6AP generating a ligase activity that polyubiquitinates tumor suppressors p53 and p73 and targets them to the 26S proteasome for degradation. As a result DNA damage and chromosomal instabilities increase, often leading to cell proliferation and cancer. The E6/E6AP complex also targets other substrates for ubiquitination, such as TERT, BAK1, FADD, and pro-casp8 none of which appear to be substrates for E6AP in the absence of E6. This kit is designed for in vitro E6AP/ E6-mediated ubiquitination of user-supplied substrates. Ubiquitinated proteins can be used in downstream applications, or analyzed by Western blot using antibodies specific for the target protein. A control substrate, His6-FLAG-p53 (SP-452) and detection antibody (MAB1355) are included in the kit. CHIP/Luciferase Ubiquitination Kit K Kit CHIP (Carboxy terminus of HSP70-Interacting Protein) is a U-Box ubiquitin E3 ligase that ubiquitinates and mediates the proteasomal destruction of misfolded chaperone substrates. CHIP functions in coordination with several chaperone complexes, including HSP40, HSP70, and HSP90. CHIP activity may be modulated by the deubuiquitinase Ataxin-3, which restricts the length of ubiquitin chains attached to CHIP substrates and prevents further chain extension. This kit is designed for in vitro ubiquitination of user-supplied substrates by CHIP or the CHIP/HSP70/HSP40 ternary complex. Ubiquitinated proteins can be used in downstream applications, or analyzed by Western blot using antibodies specific for the target protein. A luciferase control substrate and detection antibody are included in the kit. NOTE: Kit contains reagents sufficient for 10 x 30 μl reactions and 5 Western Blots (mini-gel format).
2 E3 Ligases Ubiquitin E3 Ligase Ring Finger 4 (RNF4), E µg RNF4 (small nuclear ring finger protein, SNURF) is a RING-finger ubiquitin E3 ligase that ubiquitinates and mediates the proteasomal destruction of targets such as PML, PEA3, CENP1, and PARP1. In addition to the RING domain, RNF4 contains four SUMO-interacting motifs (SIMs) that function to recruit this ligase to poly-sumoylated substrates. RNF4 will autoubiquitinate in vitro, and will also ubiquitinate poly-sumo chains. This recombinant protein is untagged. Purity: > 95% by SDS-PAGE MW: 21 kda Ubiquitin E3 Ligase CHIP (Stub1), E µg CHIP (Carboxy terminus of Hsp70-Interacting Protein) is a U-Box ubiquitin E3 ligase that ubiquitinates and mediates the proteasomal destruction of misfolded chaperone substrates. CHIP functions in coordination with several chaperone complexes, including Hsp40, Hsp70, and Hsp90. CHIP activity may be modulated by the deubuiquitinase Ataxin-3, which restricts the length of ubiquitin chains attached to CHIP substrates and prevents further chain extension. This protein contains a C-terminal 6-His tag. Purity: > 90% by SDS-PAGE MW: 36 kda Ubiquitin E3 Ligase E6AP (UBE3A), E µg E6AP (E6-Associated Protein) is a HECT domain ubiquitin E3 ligase that ubiquitinates and mediates the proteasomal destruction of substrate proteins. HECT domain ligases use an active site cysteine to accept charged ubiquitin from ubiquitin-e2 thioester complexes for subsequent transfer to substrate proteins; in this way HECT class ligases are distinct from most RING class ligases in that the latter facilitate transfer of ubiquitin from charged E2 s directly to substrate proteins without an E3-ubiquitin thioester intermediate. The viral HPV-E6 protein (present in cells infected with human papillomavirus) forms a complex with E6AP to generate a ligase activity that polyubiquitinates the tumor suppressors p53 and p73 and targets them to the 26S proteasome for degradation. This protein contains a C-terminal 6-His tag. Purity: > 95% by SDS-PAGE MW: 102 kda Non-hydrolyzable Chains Non-hydrolyzable di-ub (K11-linked) UCN μg Linkage specific, non-hydrolyzable di-ubiquitin is resistant to the activity of enzymes (DUB s) that cleave the isopeptide linkage between adjacent ubiquitin molecules. It can be used to investigate binding interactions between diubiquitin and proteins that contain elements such as ubiquitin-associated domains (UBAs) or ubiquitin-interacting motifs (UIMs). Biological roles of K11- linked polyubiquitin are less understood than K48- or K63-linked Ub chains, though recent evidence points to a role for K11 linkages in the degradation of the anaphase-promoting complex (APC/C), and in TNF -stimulated NF- B activation. Purity: > 95 % by SDS-PAGE MW: 17 kda Deubiquitinating Enzymes Ubiquitin Specific Protease 25 E μg USP25 (Ubiquitin Specific Protease 25) is a deubiquitinating enzyme of the C19 family that is expressed in many human tissue types. USP25 activity is modulated by a sumoylation event that is dependent on the presence of a SUMO-interaction domain (SIM) located in the amino-terminal region of all USP25 isoforms. Sumoylation occurs within one of two ubiquitin interaction motifs (UIMs) that are required for efficient in vitro hydrolysis of polyubiquitin chains (K48-linked or K63-linked) by USP25. While sumoylation impairs USP25 activity again polyubiquitin substrates, it does not affect its hydrolysis of ubiquitin-amc. This protein contains a C-terminal 6-His tag. Purity: >90% by SDS-PAGE MW: 123 kda STAM-binding protein AMSH, E µg AMSH (STAM binding protein) is a zinc metalloprotease belonging to the JAMM (JAB1/MPN/Mov34) family of deubiquitinating enzymes (DUBs). This enzyme functions at the endosome, where it is involved in the sorting of various cellsurface receptors to lysozomes. AMSH cleaves K63-linked but not K48-linked polyubiquitin chains, and the in vitro activity of AMSH is greatly increased in the presence of STAM protein. This recombinant protein is full-length. Purity: > 90% by SDS-PAGE MW: 48 kda Signal transducing adapter molecule 1, E µg STAM-1 (Signal Transducing Adapter Molecule 1) is involved in multiple signal transduction pathways through interactions mediated by ITAM, SH3, UIM, and VHS domains located within the protein. Roles in growth factor receptor downregulation, T-cell development, and induction of DNA synthesis following IL-2 stimulation have been identified. This full-length recombinant protein has no intrinsic deubiquitinase activity, but is useful in stimulating the in vitro activity of the JAMM-class deubiquitinase AMSH. Purity: > 90% by SDS-PAGE MW: 59 kda Deubiquitinase USP9x, isoform 2, His6 E µL USP9x (homologue of drosophila Protein Fat Facets) is a deubiquitinating enzyme of the C19 peptidase family. USP9x is an essential component of TGFß signaling, where it functions at least partially through deubiquitination of SMAD4. USP9x may also generate important signaling events in pancreatic ductal adenocarcinoma and chronic myelogenous leukemia. This recombinant Purity: > 85% by SDS-PAGE MW: 292 kda Deubiquitinase Otubain 2, His6 E µL Otubain-2 is a C65 type peptidase and a member of the ovarian tumor (OTU) protein super-family found in eukaryotes, viruses and pathogenic bacterium. Otubain-2 associates with the E3 ubiquitin ligases TRAF3 and TRAF6, and has been shown to negatively regulate virus-induced type I IFN induction and the antiviral responses mediated by these ligases. This recombinant protein contains a C-terminal 6-His tag. Purity: >90% by SDS-PAGE MW: 28 kda Deubiquitinase CYLD, His6 E µg RNF4 (small nuclear ring finger protein, SNURF) is a RING-finger ubiquitin E3 ligase that ubiquitinates and mediates the proteasomal destruction of targets such as PML, PEA3, CENP1, and PARP1. In addition to the RING domain, RNF4 contains four SUMO-interacting motifs (SIMs) that function to recruit this ligase to poly-sumoylated substrates. RNF4 will autoubiquitinate in vitro, and will also ubiquitinate poly-sumo chains. This recombinant protein is untagged. Purity: > 95% by SDS-PAGE MW: 21 kda Deubiquitinase FAM105B/OTULIN, E µg FAM105B ( OTULIN ) is a C65 type peptidase and a member of the ovarian tumor (OTU) protein super-family. OTULIN has exquisite specificity for linear (methionine-1-linked) ubiquitin linkages and has been shown to play a role in the regulation of NF-kB mediated signaling. This recombinant protein is untagged. Purity: > 95% by SDS-PAGE MW: 41 kda
3 Deubiquitinase ZRANB1/Trabid, E µg µ Zinc Finger, RAN-binding Domain Containing 1 (ZRANB1), also known as TRAFbinding Domain-containing Protein (Trabid), is a C64 type peptidase and a member of the ovarian tumor (OTU) protein super-family with a predicted molecular weight of 81 kda (1). The human protein shares 99% amino acid sequence identity with its mouse ortholog. ZRANB1 preferentially cleaves K29-, K33-, and K63-linked poly-ubiquitin chains (2). It has been shown to play a role in the regulation of Wnt signaling via deubiquitination of APC (3,4). This recombinant protein contains a C-terminal 6-His tag. Purity: > 95% by SDS-PAGE MW: 82 kda E2 Conjugating Enzymes Ubiquitin conjugating enzyme UBE2G1, E μg UBE2G1 (E2-17K) is a homologue of the yeast Ubc7 protein. This E2 forms polyubiquitin chains in vitro, and may serve as an E2 for the CRL4Cdt2 ligase complex. This Purity: >95% by SDS-PAGE MW: 20.3 kd Ubiquitin conjugating enzyme UBE2D4 E μg UBE2D (UbcH5) enzymes are human homologues of the yeast UBC4/5 family and play many important regulatory roles in inflammation and cancer. UBE2D4 (UbcH5D) has been reported to generate multiple ubiquitin chain linkage types in vitro, and may serve as the E2 carrier protein for numerous ubiquitin ligases. This protein contains N-terminal 6-His tag. Purity: >95% by SDS-PAGE MW: 17.5 kd Ubiquitin conjugating enzyme UBE2J2, isoform 1 E μg UBE2J2 is a transmembrane, endoplasmic reticulum-associated E2 that may be involved in targeting retrotranslocated, ER-associated proteins for proteasomal degradation (ERAD). This E2 is utilized by the γ-hv68 viral mk3 ligase in the polyubiquitination of substrates on serine, and/or threonine residues. The transmembrane and lumenal domains of this protein have been removed. This Purity: >95% by SDS-PAGE MW: 26.3 kd Ubiquitin conjugating enzyme UBE2R2, E μg UBE2R2 (CDC34B) is highly homologous to both UBE2R1, and the yeast CDC34 protein. UBE2R2 is phosphorylated by Casein Kinase 2 in vivo and in vitro, and may play a role in β-catenin stability. This E2 has been reported to form K48- isopeptide linkages in vitro. This Purity:>95% by SDS-PAGE MW: 28.0 kd Ubiquitin conjugating enzyme UBE2Q1, isoform 2, E µg UBE2Q1 is the E2 enzyme encoded by NICE-5, one of the genes of the human epidermal differentiation complex (EDC). The protein forms thiolester complexes with ubiquitin in vitro, and contains an N-terminal 6-His tag. Purity:> 90% by SDS-PAGE MW: 29.3 kd Ubiquitin conjugating enzyme UBE2W, isoform 2, E µg UBE2W (Ubc16) may play a role in Fanconi anemia tumor suppressor pathway by mono-ubiquitinating FANCD2 when in complex with the E3 ligase FANCL. This E2 has also been shown to catalyze the autoubiquitination of the BRCA1/ BARD1 E3 complex, and to form K11 linked polyubiquitin chains in vitro. This Purity: >90% by SDS-PAGE MW: 19.5 kd Ubiquitin conjugating enzyme UBE2V2 E µg UBE2V2 (MMS2) is not an active E2 in the absence of UBE2N. The UBE2V2/ UBE2N heterodimer catalyzes the synthesis of K63-linked polyubiquitin chains and may be an E2 for the CHFR E3 ligase. This protein forms polyubiquitin chains in vitro and contains an N-terminal 6-His tag. Purity: > 95% by SDS-PAGE MW: 17.2 kd Ubiquitin conjugating enzyme UBE2W, isoform 1 E µg UBE2W (Ubc16) may play a role in Fanconi anemia tumor suppressor pathway by mono-ubiquitinating FANCD2 when in complex with the E3 ligase FANCL. This E2 has also been shown to catalyze the autoubiquitination of the BRCA1/ BARD1 E3 complex, and to form K11 linked polyubiquitin chains in vitro. This Purity: > 90% by SDS-PAGE MW: 18.1 kd Ubiquitin conjugating enzyme UBE2Q2, isoform 1 E µg UBE2Q2 has implicated roles in cell-cycle regulation, apoptosis, and cancer, and has been reported to form K48-type linkages in some in vitro assays. This Purity: > 95% by SDS-PAGE MW: 43.6 kd Ubiquitin conjugating enzyme UBE2J1, E µg UBE2J1 is a human homologue of the yeast Ubc6 protein. In yeast, this tailanchored transmembrane E2 is localized to the cytoplasmic surface of the ER and participates in ER-associated degradation (ERAD) of misfolded proteins. UBE2J1 is reported to play a role in degradation of misfolded MHC class I molecules via a mechanism requiring the E3 ubiquitin ligase Hrd-1. This protein contains an N-terminal 6-His tag, and the C-terminal transmembrane and lumenal amino acid sequences have been removed. Purity: > 95% by SDS-PAGE MW: 32.0 kd Ubiquitin conjugating enzyme UBE2Q1, isoform 1, E µg UBE2Q1 is the E2 enzyme encoded by NICE-5, one of the genes of the human epidermal differentiation complex (EDC). The protein forms thioester complexes with ubiquitin in vitro, and contains an N-terminal 6-His tag. Purity: >90% by SDS-PAGE MW: 46.9 kd
4 Ubiquitin-like Modifiers Ubiquitin-like modifier FUBI/MNSFβ, UL µg The Ubiquitin-like protein FUBI/MNSFß (Monoclonal Nonspecific Suppressor Factor Beta) is encoded by the FAU gene, and is translated as a pro-form consisting of FUBI fused to the ribosomal S30 protein. S30 is cleaved in a posttranslational reaction, releasing the mature 74 amino acid FUBI/MNSFß protein. FUBI has a C-terminal gly-gly motif common to many ubiquitin-like proteins. FUBI/MNSFß conjugation to Bcl-G has been shown to regulate the ERK 1/2- MAPK cascade in macrophage cell lines, and may be implicated in TLR4- mediated signal transduction. Conjugation to endophilin II regulates dectin-1- mediated phagocytosis and inflammatory responses, and may be implicated in TLR2 signaling pathway. Purity: > 95% by SDS-PAGE MW: 7.8 kda Ubiquitin-related modifier 1 protein, UL µg URM1 (Ubiquitin-related modifier 1) is a protein that functions in the posttranslational modification of proteins via the Urmylation pathway. In humans, Urm1 is thiocarboxylated (-COSH) at its C-terminus by the MOCS3 protein in a two-step process that is highly reminiscent of ubiquitin activation by E1 enzymes. It s been suggested that Urm1 plays a role in oxidative-stress response in mammals, and that the protein may be an evolutionary link between ancient ubiquitin progenitors and the eukaryotic ubiquitin/ubiquitin-like modification systems. This protein is untagged. Purity: >99% by SDS-PAGE MW: 11 kda Ubiquitin Mutants Ubiquitin mutant L73P, UM-L73P 1 mg This ubiquitin (Ub) contains a mutation of leucine 73 to proline. Ub L73P can form an E1-catalyzed active thioester at the C-terminus allowing the molecule to be transferred to E2 carrier proteins, and possibly to E3 ligases and their substrates. Polyubiquitin chains generated using L73P are highly resistant to disassembly by deubiquitinases. Potential uses of this protein include generating ubiquitin conjugates that are more stable than their native counterparts in the presence of deubiquitinase enzymes. Purity: > 95% by SDS-PAGE MW: 8.5 kda DCA-linked Non-hydrolyzable Chains Di-Ubiquitin/Ub2, K6-DCA-linked UCD µg Dichloroacetone (DCA) linked forms of Ub2 are chemically linked via a DCA linker using cysteine introduced into the ubiquitin sequence at the appropriate residues to mimic a native isopeptide linkage. Reports have demonstrated that the DCA linkage is stable and cannot be processed by deubiquitinating enzymes. Additionally, the orientation of the two ubiquitins in DCA-linked Ub2 Di-Ubiquitin/Ub2, 6-DCA-linked, Agarose, UCD µl Di-Ubiquitin/Ub2, 11-DCA-linked, UCD µg Di-Ubiquitin/Ub2, 11-DCA-linked, Agarose, UCD µl demonstrated that the DCA linkage is stable and cannot be processed by deubiquitinases. Additionally, the orientation of the two ubiquitins in DCA-linked Ub2 Di-Ubiquitin/Ub2, K29-DCA-linked UCD µg Di-Ubiquitin/Ub2, 29-DCA-linked, Agarose, UCD µl Di-Ubiquitin/Ub2, 76-DCA-linked, UCD µg Dichloroacetone (DCA) linked forms of Ub2 are chemically linked via a DCA linker using cysteine introduced into the ubiquitin sequence at the appropriate residues to mimic a native isopeptide linkage. Reports have demonstrated that the DCA linkage is stable and cannot be processed by deubiquitinating enzymes. Additionally, the orientation of the two ubiquitins in DCA-linked Ub2 Although Ub is not a naturally occurring linkage, this di-ubiquitin may be a useful control in experiments using the other DCA-linked di-ubiquitins. demonstrated that the DCA linkage is stable and cannot be processed by deubiquitinases. Additionally, the orientation of the two ubiquitins in DCA-linked Ub2
5 Di-Ubiquitin/Ub2, 76-DCA-linked, Agarose, UCD µl Although Ub is not a naturally occurring linkage, this di-ubiquitin may be a useful control in experiments using the other DCA-linked di-ubiquitins. Di-Ubiquitin/Ub2, 33-DCA-linked, UCD µg Di-Ubiquitin/Ub2, 33-DCA-linked, Agarose, UCD µL Di-Ubiquitin/Ub2, 48-DCA-linked, UCD µg proteins in the absence of added deubiquitinase inhibitors Di-Ubiquitin/Ub2, 48-DCA-linked, Agarose, UCD µL Di-Ubiquitin/Ub2, 63-DCA-linked, UCD µg Dichloroacetone (DCA) linked forms of Ub2 are chemically linked via a DCA linker using cysteine introduced into the ubiquitin sequence at the appropriate residues to mimic a native isopeptide linkage. Reports have demonstrated that the DCA linkage is stable and cannot be processed by deubiquitinating enzymes. Additionally, the orientation of the two ubiquitins in DCA-linked Ub2 Di-Ubiquitin/Ub2, 63-DCA-linked, Agarose, UCD µL SUMO Chains Poly-SUMO2 Hydrolysis Resistant Chains (3-8) ULN µg Sentrin protease (SENP)-resistant poly-sumo2 chains can be used to investigate mechanisms of chain recognition by SUMO-targeted ubiquitin ligases (STUbl s) or other proteins that contain SUMO-interacting domains (SIM s). These K11- linked chains are formed enzymatically with recombinant SUMO2 containing a mutation of glutamine 90 to proline, which renders them approximately 500- fold more resistant to disassembly by SENP s than are wild-type chains. Monoand di-sumo2 have been removed from the chain mixture. Purity: >90% by SDS-PAGE Poly-SUMO3 Hydrolysis Resistant Chains (3-8), ULN µg Sentrin protease (SENP)-resistant poly-sumo3 chains can be used to investigate mechanisms of chain recognition by SUMO-targeted ubiquitin ligases (STUbl s) or other proteins that contain SUMO-interacting domains (SIM s). These K-11 linked chains are formed enzymatically with recombinant SUMO3 containing a mutation of glutamine 89 to proline, which renders them approximately 500- fold more resistant to disassembly by SENP s than are wild-type chains. Monoand di-sumo3 have been removed from the chain mixture. Purity: >90% by SDS-PAGE Inhibitors PS-341, 26S Proteasome Inhibitor Synthetic I mg The dipeptide-boronic acid analogue PS-341 is the active ingredient in Bortezomib (Velcade, Millennium Pharmaceuticals). This potent, cell permeable molecule inhibits primarily the chymotryptic activity of the proteasome without substantially altering tryptic or caspase-like activities. PS-341 inhibits proliferation of a number of tumor cell lines with nanomolar potency, and is approved by the USFDA for the treatment of multiple myeloma. Purity: >99% by HPLC and TLC FW: 384
6 Nedd8 E1 Activating Enzyme Inhibitor Synthetic I mg NAE Inhibitor contains the active component of MLN4924, a cell-permeable, small molecule inhibitor of Nedd8 activating enzyme (NAE). By inhibiting the Neddylation pathway this molecule disrupts cullin-ring ligase activity, leading to inhibition of proliferation or apoptotic death of various human tumor cells through the deregulation of S-phase DNA synthesis. Mutations in UBA3, a gene encoding one of the subunits of Nedd8 activating enzyme, have been reported to cause resistance to MLN4924 in some tumor cell lines. Purity: >99% by HPLC FW: 444 Buffers 10X E3 Ligase Reaction Buffer B-71 5 ml E3 Ligase Reaction Buffer is formulated to support the in vitro activity of E3 Ligase enzymes. This buffer is similar in composition to 10X Ubiquitin Conjugation Reaction Buffer (B-70), but contains a higher concentration of reductant for those enzymes that benefit from additional disulfide-reducing potential. Fully compatible with E1 and E2 enzyme activity, this buffer can be used to supplement the ubiquitin-protein conjugation kit (K-960), fraction II conjugation kits (K-930, K-935), and E3-ligase enzymes. 10X HSP Reaction Buffer B-72 5 ml HSP Reaction Buffer is formulated to support the in vitro enzymatic activity of Heat Shock Proteins. This buffer is similar in composition to 10X E3 Ligase Reaction Buffer (B-71), but contains potassium chloride that improves the catalytic function of HSP70 in vitro. Fully compatible with E1, E2, and E3 ubiquitin ligase enzyme activities, this buffer can be used for in vitro protein refolding experiments, or in ubiquitination reactions that require HSP activity. This buffer does not contain Mg2+-ATP. Associated Proteins E6, Human Papilloma Virus type 16, recombinant AP µg μ E6 (Early protein 6) is a viral protein produced in cells infected with the Human Papillomavirus. E6 forms a complex with the host cell ubiquitin ligase E6AP (E6-Associated-Protein) generating a ligase activity that polyubiquitinates tumor suppressors p53 and p73 and targets them to the 26S proteasome for degradation. As a result DNA damage and chromosomal instabilities increase, often leading to cell proliferation and cancer. The E6/E6AP complex also targets other substrates for ubiquitination, such as TERT, BAK1, FADD, and pro- CASP8 none of which appear to be substrates for E6AP in the absence of E6. This protein is untagged, and contains six cysteine-to-serine substitutions at positions 23, 58, 87, 104, 118, and 147 (HPV type 16 sequence numbering). Purity: > 90% by SDS-PAGE MW: 19 kda Substrate Proteins -FLAG-p53, SP µg Tumor suppressor protein p53, a nuclear transcription factor, plays an essential role in the regulation of the cell cycle and is frequently mutated or inactivated in many cancers. Numerous post-translational modifications modulate p53 activity including ubiquitination, phosphorylation, acetylation and methylation. The stability of p53 is regulated via the ubiquitin-proteasome pathway (UPP). MDM2 is an oncogenic ubiquitin E3 ligase that ubiquitinates p53, inhibits its transcriptional activity and promotes its degradation. Other E3 ligases that promote the proteasome-mediated degradation of p53 include Pirh2, COP1 and p300. USP7 (HAUSP) stabilizes p53 by deubiquitination and induces p53- dependent cell growth repression and apoptosis. Additional factors such as p14arf and MdmX also modulate p53 function via the UPP. This recombinant protein contains N-terminal 6-His and FLAG tags. Purity: >75% by SDS-PAGE MW: 46 kda Buffers HSP70/HSPA1A, AP μg Heat shock proteins (HSPs) are a family of highly conserved stress response proteins. Heat shock proteins function primarily as molecular chaperones by facilitating the folding of other cellular proteins, preventing protein aggregation or targeting improperly folded proteins to specific degradative pathways. HSPs are typically expressed at low levels under normal physiological conditions, but are dramatically up-regulated in response to cellular stress. HSP70 is a 72 kda member of the heat shock protein 70 family of proteins, and is also known as HSPA1A, HSP70-1, and HSP72. HSP70 and HSP40 are required to target some misfolded proteins to the Ubiquitin E3 ligase CHIP (Stub1) for subsequent ubiquitination. Purity: > 95% by SDS-PAGE MW: 70 kda E3 Ligase Kits Deconjugating Enzym HSP40/DNAJB1, AP µg Heat shock protein 40 (HSP40, also known as DNAJB1) is the human homologue of the bacterial DnaJ heat shock protein. Heat shock proteins (HSPs) are a highly conserved family of stress response proteins. HSPs function primarily as molecular chaperones, facilitating the folding of other cellular proteins, preventing protein aggregation, or targeting improperly folded proteins to specific degradative pathways. Heat Shock Proteins are ubiquitously expressed in all organisms, and they are induced in response to various types of environmental stresses like heat, cold, and oxygen deprivation. HSP40 is a stress inducible chaperone that co localizes with HSP70 and can bind unfolded proteins and prevent protein denaturation and aggregation. The conserved amino terminal J domain can interact with HSP70 and stimulate its ATPase activity. HSP40 and HSP70 are required to target some misfolded proteins to the Ubiquitin E3 ligase CHIP (Stub1) for subsequent ubiquitination. The protein contains a C-terminal 6-His tag. Purity: > 95% by SDS-PAGE MW: 39 kda
Protein methylation CH 3
 Protein methylation CH 3 methionine S-adenosylmethionine (SAM or adomet) Methyl group of the methionine is activated by the + charge of the adjacent sulfur atom. SAM S-adenosylhomocysteine homocysteine
Protein methylation CH 3 methionine S-adenosylmethionine (SAM or adomet) Methyl group of the methionine is activated by the + charge of the adjacent sulfur atom. SAM S-adenosylhomocysteine homocysteine
Proteasomes. When Death Comes a Knock n. Warren Gallagher Chem412, Spring 2001
 Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
PDF hosted at the Radboud Repository of the Radboud University Nijmegen
 PDF hosted at the Radboud Repository of the Radboud University Nijmegen The following full text is a publisher's version. For additional information about this publication click this link. http://hdl.handle.net/2066/122889
PDF hosted at the Radboud Repository of the Radboud University Nijmegen The following full text is a publisher's version. For additional information about this publication click this link. http://hdl.handle.net/2066/122889
The Nobel Prize in Chemistry 2004
 The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
E2Select Ubiquitin Conjugation Kit
 E2Select Ubiquitin Conjugation Kit Catalog Number K-982 This kit contains reagents sufficient to perform 2 Ubiquitination reactions for each E2 included in the kit. This package insert must be read in
E2Select Ubiquitin Conjugation Kit Catalog Number K-982 This kit contains reagents sufficient to perform 2 Ubiquitination reactions for each E2 included in the kit. This package insert must be read in
The Role of Ubiquitin and Ubiquitin-Like Modification Systems in Papillomavirus Biology
 Viruses 2014, 6, 3584-3611; doi:10.3390/v6093584 Review OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses The Role of Ubiquitin and Ubiquitin-Like Modification Systems in Papillomavirus Biology
Viruses 2014, 6, 3584-3611; doi:10.3390/v6093584 Review OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses The Role of Ubiquitin and Ubiquitin-Like Modification Systems in Papillomavirus Biology
Study of different types of ubiquitination
 Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Signal Transduction Cascades
 Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
KEY CONCEPT QUESTIONS IN SIGNAL TRANSDUCTION
 Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Post-translational modifications of proteins in gene regulation under hypoxic conditions
 203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
Practice Exam 2 MCBII
 1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
Innate Immunity & Inflammation
 Innate Immunity & Inflammation The innate immune system is an evolutionally conserved mechanism that provides an early and effective response against invading microbial pathogens. It relies on a limited
Innate Immunity & Inflammation The innate immune system is an evolutionally conserved mechanism that provides an early and effective response against invading microbial pathogens. It relies on a limited
Cellular functions of protein degradation
 Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Biochemistry 2 Recita0on Amino Acid Metabolism
 Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
p53 and Apoptosis: Master Guardian and Executioner Part 2
 p53 and Apoptosis: Master Guardian and Executioner Part 2 p14arf in human cells is a antagonist of Mdm2. The expression of ARF causes a rapid increase in p53 levels, so what would you suggest?.. The enemy
p53 and Apoptosis: Master Guardian and Executioner Part 2 p14arf in human cells is a antagonist of Mdm2. The expression of ARF causes a rapid increase in p53 levels, so what would you suggest?.. The enemy
Introduction to pathology lecture 5/ Cell injury apoptosis. Dr H Awad 2017/18
 Introduction to pathology lecture 5/ Cell injury apoptosis Dr H Awad 2017/18 Apoptosis = programmed cell death = cell suicide= individual cell death Apoptosis cell death induced by a tightly regulated
Introduction to pathology lecture 5/ Cell injury apoptosis Dr H Awad 2017/18 Apoptosis = programmed cell death = cell suicide= individual cell death Apoptosis cell death induced by a tightly regulated
The ubiquitin-proteasome system. in bone biology. University of Nottingham. ECTS PhD Training July 5 th Dr Rob Layfield
 The ubiquitin-proteasome system in bone biology Dr Rob Layfield University of Nottingham ECTS PhD Training July 5 th 2009 Images reproduced from: http://www.bostonbiochem.com/overview.php?prod=ubchains
The ubiquitin-proteasome system in bone biology Dr Rob Layfield University of Nottingham ECTS PhD Training July 5 th 2009 Images reproduced from: http://www.bostonbiochem.com/overview.php?prod=ubchains
ab E3 Ligase Auto- Ubiquitilylation Assay Kit
 ab139469 E3 Ligase Auto- Ubiquitilylation Assay Kit Instructions for Use For testing ubiquitin E3 ligase activity through assessment of their ability to undergo auto-ubiquitinylation This product is for
ab139469 E3 Ligase Auto- Ubiquitilylation Assay Kit Instructions for Use For testing ubiquitin E3 ligase activity through assessment of their ability to undergo auto-ubiquitinylation This product is for
C-Phycocyanin (C-PC) is a n«sjfc&c- waefc-jduble phycobiliprotein. pigment isolated from Spirulina platensis. This water- soluble protein pigment is
 ' ^Summary C-Phycocyanin (C-PC) is a n«sjfc&c- waefc-jduble phycobiliprotein pigment isolated from Spirulina platensis. This water- soluble protein pigment is of greater importance because of its various
' ^Summary C-Phycocyanin (C-PC) is a n«sjfc&c- waefc-jduble phycobiliprotein pigment isolated from Spirulina platensis. This water- soluble protein pigment is of greater importance because of its various
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
General information. Cell mediated immunity. 455 LSA, Tuesday 11 to noon. Anytime after class.
 General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
B F. Location of MHC class I pockets termed B and F that bind P2 and P9 amino acid side chains of the peptide
 Different MHC alleles confer different functional properties on the adaptive immune system by specifying molecules that have different peptide binding abilities Location of MHC class I pockets termed B
Different MHC alleles confer different functional properties on the adaptive immune system by specifying molecules that have different peptide binding abilities Location of MHC class I pockets termed B
E2 enzymes: more than just middle men
 REVIEW Cell Research (2016) 26:423-440. www.nature.com/cr npg E2 enzymes: more than just middle men Mikaela D Stewart 1, *, Tobias Ritterhoff 1, *, Rachel E Klevit 1, Peter S Brzovic 1 1 Department of
REVIEW Cell Research (2016) 26:423-440. www.nature.com/cr npg E2 enzymes: more than just middle men Mikaela D Stewart 1, *, Tobias Ritterhoff 1, *, Rachel E Klevit 1, Peter S Brzovic 1 1 Department of
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
SUPPLEMENTARY INFORMATION
 Figure S1. Silver staining and immunoblotting of the purified TAK1 kinase complex. The TAK1 kinase complex was purified through tandem affinity methods (Protein A and FLAG), and aliquots of the purified
Figure S1. Silver staining and immunoblotting of the purified TAK1 kinase complex. The TAK1 kinase complex was purified through tandem affinity methods (Protein A and FLAG), and aliquots of the purified
Cancer. Throughout the life of an individual, but particularly during development, every cell constantly faces decisions.
 Cancer Throughout the life of an individual, but particularly during development, every cell constantly faces decisions. Should it divide? Yes No--> Should it differentiate? Yes No-->Should it die? Yes-->Apoptosis
Cancer Throughout the life of an individual, but particularly during development, every cell constantly faces decisions. Should it divide? Yes No--> Should it differentiate? Yes No-->Should it die? Yes-->Apoptosis
Coenzyme Q 10. R. JÜRGEN DOHMEN Institute for Genetics, University of Cologne, Germany
 Coenzyme Q 10 Ubiquitin is a small protein of 76 residues that is conjugated posttranslationally to other proteins. It is ubiquitously present in all eukaryotes, hence its name, and is one of the most
Coenzyme Q 10 Ubiquitin is a small protein of 76 residues that is conjugated posttranslationally to other proteins. It is ubiquitously present in all eukaryotes, hence its name, and is one of the most
Molecular Graphics Perspective of Protein Structure and Function
 Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Key Concept B F. How do peptides get loaded onto the proper kind of MHC molecule?
 Location of MHC class I pockets termed B and F that bind P and P9 amino acid side chains of the peptide Different MHC alleles confer different functional properties on the adaptive immune system by specifying
Location of MHC class I pockets termed B and F that bind P and P9 amino acid side chains of the peptide Different MHC alleles confer different functional properties on the adaptive immune system by specifying
RAS Genes. The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes.
 ۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
Cell Signaling part 2
 15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
Chapter 6. Antigen Presentation to T lymphocytes
 Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Emerging Role of the Ubiquitin-proteasome System as Drug Targets
 Send Orders of Reprints at reprints@benthamscience.org Current Pharmaceutical Design, 2013, 19, 000-000 1 Emerging Role of the Ubiquitin-proteasome System as Drug Targets Gozde Kar 1, Ozlem Keskin 1, Franca
Send Orders of Reprints at reprints@benthamscience.org Current Pharmaceutical Design, 2013, 19, 000-000 1 Emerging Role of the Ubiquitin-proteasome System as Drug Targets Gozde Kar 1, Ozlem Keskin 1, Franca
UbcH (E2) Enzyme Kit. Catalog Number K-980B. This kit contains reagents for the qualitative analysis of UbcH Enzyme usage.
 UbcH (E2) Enzyme Kit Catalog Number K-980B This kit contains reagents for the qualitative analysis of UbcH Enzyme usage. This package insert must be read in its entirety before using this product. For
UbcH (E2) Enzyme Kit Catalog Number K-980B This kit contains reagents for the qualitative analysis of UbcH Enzyme usage. This package insert must be read in its entirety before using this product. For
2013 John Wiley & Sons, Inc. All rights reserved. PROTEIN SORTING. Lecture 10 BIOL 266/ Biology Department Concordia University. Dr. S.
 PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
Degrading the Barriers to Drug Discovery in Ubiquitin E3 Ligase Pathways
 Degrading the Barriers to Drug Discovery in iquitin E3 Ligase Pathways Blaine N. Armbruster, PhD. Sr. Manager - Discovery & Development Solutions EMD Millipore 1 Oct, 2012 Outline Introduction ti to EMD
Degrading the Barriers to Drug Discovery in iquitin E3 Ligase Pathways Blaine N. Armbruster, PhD. Sr. Manager - Discovery & Development Solutions EMD Millipore 1 Oct, 2012 Outline Introduction ti to EMD
TNFSF13B tumor necrosis factor (ligand) superfamily, member 13b NF-kB pathway cluster, Enrichment Score: 3.57
 Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Cell cycle, signaling to cell cycle, and molecular basis of oncogenesis
 Cell cycle, signaling to cell cycle, and molecular basis of oncogenesis MUDr. Jiří Vachtenheim, CSc. CELL CYCLE - SUMMARY Basic terminology: Cyclins conserved proteins with homologous regions; their cellular
Cell cycle, signaling to cell cycle, and molecular basis of oncogenesis MUDr. Jiří Vachtenheim, CSc. CELL CYCLE - SUMMARY Basic terminology: Cyclins conserved proteins with homologous regions; their cellular
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001
 BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
Protein Modification Overview DEFINITION The modification of selected residues in a protein and not as a component of synthesis
 Lecture Four: Protein Modification & Cleavage [Based on Chapters 2, 9, 10 & 11 Berg, Tymoczko & Stryer] (Figures in red are for the 7th Edition) (Figures in Blue are for the 8th Edition) Protein Modification
Lecture Four: Protein Modification & Cleavage [Based on Chapters 2, 9, 10 & 11 Berg, Tymoczko & Stryer] (Figures in red are for the 7th Edition) (Figures in Blue are for the 8th Edition) Protein Modification
Insulin mrna to Protein Kit
 Insulin mrna to Protein Kit A 3DMD Paper BioInformatics and Mini-Toober Folding Activity Student Handout www.3dmoleculardesigns.com Insulin mrna to Protein Kit Contents Becoming Familiar with the Data...
Insulin mrna to Protein Kit A 3DMD Paper BioInformatics and Mini-Toober Folding Activity Student Handout www.3dmoleculardesigns.com Insulin mrna to Protein Kit Contents Becoming Familiar with the Data...
The role of deubiquitinating enzymes in disease processes
 The role of deubiquitinating enzymes in disease processes Thesis submitted to Department of Pharmaceutical Biosciences, School of Pharmacy, Faculty of Mathematics and Natural Sciences, University of Oslo
The role of deubiquitinating enzymes in disease processes Thesis submitted to Department of Pharmaceutical Biosciences, School of Pharmacy, Faculty of Mathematics and Natural Sciences, University of Oslo
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
VII SYNOPSIS. Cancer is a disease, causes due to failure of control of growth and. cell proliferation mechanisms. There are genetic control systems,
 VII SYPSIS Cancer is a disease, causes due to failure of control of growth and cell proliferation mechanisms. There are genetic control systems, regulate the balance between cell birth and death in response
VII SYPSIS Cancer is a disease, causes due to failure of control of growth and cell proliferation mechanisms. There are genetic control systems, regulate the balance between cell birth and death in response
HIV-1 genome organization
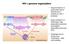 HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
T cell maturation. T-cell Maturation. What allows T cell maturation?
 T-cell Maturation What allows T cell maturation? Direct contact with thymic epithelial cells Influence of thymic hormones Growth factors (cytokines, CSF) T cell maturation T cell progenitor DN DP SP 2ry
T-cell Maturation What allows T cell maturation? Direct contact with thymic epithelial cells Influence of thymic hormones Growth factors (cytokines, CSF) T cell maturation T cell progenitor DN DP SP 2ry
Antibodies for Unfolded Protein Response
 Novus-lu-2945 Antibodies for Unfolded rotein Response Unfolded roteins ER lumen GR78 IRE-1 GR78 ERK Cytosol GR78 TRAF2 ASK1 JNK Activator Intron RIDD elf2α Degraded mrna XB1 mrna Translation XB1-S (p50)
Novus-lu-2945 Antibodies for Unfolded rotein Response Unfolded roteins ER lumen GR78 IRE-1 GR78 ERK Cytosol GR78 TRAF2 ASK1 JNK Activator Intron RIDD elf2α Degraded mrna XB1 mrna Translation XB1-S (p50)
SHORT LINEAR MOTIFS. Holger Dinkel EMBO Practical Course Computational analysis of protein-protein interactions From sequences to networks
 Holger Dinkel EMBO Practical Course Computational analysis of protein-protein interactions From sequences to networks IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19
Holger Dinkel EMBO Practical Course Computational analysis of protein-protein interactions From sequences to networks IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19
Ch. 18 Regulation of Gene Expression
 Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
All intracellular proteins and many extracellular proteins
 Review Protein Degradation by the Ubiquitin Proteasome Pathway in Normal and Disease States Stewart H. Lecker,* Alfred L. Goldberg, and William E. Mitch *Nephrology Division, Beth Israel Deaconess, and
Review Protein Degradation by the Ubiquitin Proteasome Pathway in Normal and Disease States Stewart H. Lecker,* Alfred L. Goldberg, and William E. Mitch *Nephrology Division, Beth Israel Deaconess, and
Lecture #27 Lecturer A. N. Koval
 Lecture #27 Lecturer A. N. Koval Hormones Transduce Signals to Affect Homeostatic Mechanisms Koval A. (C), 2011 2 Lipophilic hormones Classifying hormones into hydrophilic and lipophilic molecules indicates
Lecture #27 Lecturer A. N. Koval Hormones Transduce Signals to Affect Homeostatic Mechanisms Koval A. (C), 2011 2 Lipophilic hormones Classifying hormones into hydrophilic and lipophilic molecules indicates
Cell Biology from an Immune Perspective
 Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai Cell Biology from an Immune Perspective In this lecture we will very briefly
Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai Cell Biology from an Immune Perspective In this lecture we will very briefly
Index. Index 371. Aggresome, cystic fibrosis transmembrane conductance regulator,
 Index 371 Index Aggresome, cystic fibrosis transmembrane conductance regulator, centrosome-associated inclusions, characterization of soluble and insoluble fractions, 319 formation induction, 318, 320,
Index 371 Index Aggresome, cystic fibrosis transmembrane conductance regulator, centrosome-associated inclusions, characterization of soluble and insoluble fractions, 319 formation induction, 318, 320,
Protein sorting (endoplasmic reticulum) Dr. Diala Abu-Hsasan School of Medicine
 Protein sorting (endoplasmic reticulum) Dr. Diala Abu-Hsasan School of Medicine dr.abuhassand@gmail.com An overview of cellular components Endoplasmic reticulum (ER) It is a network of membrane-enclosed
Protein sorting (endoplasmic reticulum) Dr. Diala Abu-Hsasan School of Medicine dr.abuhassand@gmail.com An overview of cellular components Endoplasmic reticulum (ER) It is a network of membrane-enclosed
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS
 CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS Summary of the regulation of cyclin/cdk complexes during celll cycle Cell cycle phase Cyclin-cdk complex inhibitor activation Substrate(s) G1 Cyclin D/cdk 4,6
CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS Summary of the regulation of cyclin/cdk complexes during celll cycle Cell cycle phase Cyclin-cdk complex inhibitor activation Substrate(s) G1 Cyclin D/cdk 4,6
Biochemistry 673 Lecture 2 Jason Kahn, UMCP Introduction to steroid hormone receptor (nuclear receptor) signalling
 Biochemistry 673 Lecture 2 Jason Kahn, UMCP Introduction to steroid hormone receptor (nuclear receptor) signalling Resources: Latchman Lodish chapter 10, 20 Helmreich, chapter 11 http://www.nursa.org,
Biochemistry 673 Lecture 2 Jason Kahn, UMCP Introduction to steroid hormone receptor (nuclear receptor) signalling Resources: Latchman Lodish chapter 10, 20 Helmreich, chapter 11 http://www.nursa.org,
The ubiquitin proteasome system in cardiovascular disease Basic mechanisms
 ECS Congress 2010 Stockholm, Sweden 28 Aug 2010 01 Sep 2010 The ubiquitin proteasome system in cardiovascular disease Basic mechanisms Saskia Schlossarek Department of Experimental and Clinical Pharmacology
ECS Congress 2010 Stockholm, Sweden 28 Aug 2010 01 Sep 2010 The ubiquitin proteasome system in cardiovascular disease Basic mechanisms Saskia Schlossarek Department of Experimental and Clinical Pharmacology
Intracellular Compartments and Protein Sorting
 Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
Toll-like Receptor Signaling
 Toll-like Receptor Signaling 1 Professor of Medicine University of Massachusetts Medical School, Worcester, MA, USA Why do we need innate immunity? Pathogens multiply very fast We literally swim in viruses
Toll-like Receptor Signaling 1 Professor of Medicine University of Massachusetts Medical School, Worcester, MA, USA Why do we need innate immunity? Pathogens multiply very fast We literally swim in viruses
Chapter 31. Completing the Protein Life Cycle: Folding, Processing and Degradation. Biochemistry by Reginald Garrett and Charles Grisham
 Chapter 31 Completing the Protein Life Cycle: Folding, Processing and Degradation Biochemistry by Reginald Garrett and Charles Grisham Essential Question How are newly synthesized polypeptide chains transformed
Chapter 31 Completing the Protein Life Cycle: Folding, Processing and Degradation Biochemistry by Reginald Garrett and Charles Grisham Essential Question How are newly synthesized polypeptide chains transformed
Apoptosis Oncogenes. Srbová Martina
 Apoptosis Oncogenes Srbová Martina Cell Cycle Control point Cyclin B Cdk1 Cyclin D Cdk4 Cdk6 Cyclin A Cdk2 Cyclin E Cdk2 Cyclin-dependent kinase (Cdk) have to bind a cyclin to become active Regulation
Apoptosis Oncogenes Srbová Martina Cell Cycle Control point Cyclin B Cdk1 Cyclin D Cdk4 Cdk6 Cyclin A Cdk2 Cyclin E Cdk2 Cyclin-dependent kinase (Cdk) have to bind a cyclin to become active Regulation
Effects of Second Messengers
 Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
Intrinsic cellular defenses against virus infection
 Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Signaling Through Immune System Receptors (Ch. 7)
 Signaling Through Immune System Receptors (Ch. 7) 1. General principles of signal transduction and propagation. 2. Antigen receptor signaling and lymphocyte activation. 3. Other receptors and signaling
Signaling Through Immune System Receptors (Ch. 7) 1. General principles of signal transduction and propagation. 2. Antigen receptor signaling and lymphocyte activation. 3. Other receptors and signaling
Summary of Endomembrane-system
 Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Antigen Presentation to T lymphocytes
 Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antibodies and T cell receptors
Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antibodies and T cell receptors
Molecular biology :- Cancer genetics lecture 11
 Molecular biology :- Cancer genetics lecture 11 -We have talked about 2 group of genes that is involved in cellular transformation : proto-oncogenes and tumour suppressor genes, and it isn t enough to
Molecular biology :- Cancer genetics lecture 11 -We have talked about 2 group of genes that is involved in cellular transformation : proto-oncogenes and tumour suppressor genes, and it isn t enough to
Ub-CHOP-Reporter Deubiquitination Assay Kit Catalog Number PR1001
 - 1 - MANUAL Ub-CHOP-Reporter Deubiquitination Assay Kit Catalog Number PR1001 all products are for research use only not intended for human or animal diagnostic or therapeutic uses LifeSensors, Inc.,
- 1 - MANUAL Ub-CHOP-Reporter Deubiquitination Assay Kit Catalog Number PR1001 all products are for research use only not intended for human or animal diagnostic or therapeutic uses LifeSensors, Inc.,
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting
 Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled
 Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
This Review is part of a thematic series on Ubiquitination, which includes the following articles:
 This Review is part of a thematic series on Ubiquitination, which includes the following articles: Regulation of G Protein and Mitogen-Activated Protein Kinase Signaling by Ubiquitination: Insights From
This Review is part of a thematic series on Ubiquitination, which includes the following articles: Regulation of G Protein and Mitogen-Activated Protein Kinase Signaling by Ubiquitination: Insights From
three forms: c-cbl, CBLb and CBL-3) MDM2, MDMX and SCF Fbxo11
 Supplementary information S1 (table) Reported Neddylation Substrates Target protein Biological function of target Effect of neddylation E2 E3 Regulation of neddylation Cullins RING ubiquitin E3 ligases
Supplementary information S1 (table) Reported Neddylation Substrates Target protein Biological function of target Effect of neddylation E2 E3 Regulation of neddylation Cullins RING ubiquitin E3 ligases
Chapter 11. B cell generation, Activation, and Differentiation. Pro-B cells. - B cells mature in the bone marrow.
 Chapter B cell generation, Activation, and Differentiation - B cells mature in the bone marrow. - B cells proceed through a number of distinct maturational stages: ) Pro-B cell ) Pre-B cell ) Immature
Chapter B cell generation, Activation, and Differentiation - B cells mature in the bone marrow. - B cells proceed through a number of distinct maturational stages: ) Pro-B cell ) Pre-B cell ) Immature
Previous Class. Today. Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism. Protein Kinase Catalytic Properties
 Previous Class Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism Today Protein Kinase Catalytic Properties Protein Phosphorylation Phosphorylation: key protein modification
Previous Class Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism Today Protein Kinase Catalytic Properties Protein Phosphorylation Phosphorylation: key protein modification
Section 1 Proteins and Proteomics
 Section 1 Proteins and Proteomics Learning Objectives At the end of this assignment, you should be able to: 1. Draw the chemical structure of an amino acid and small peptide. 2. Describe the difference
Section 1 Proteins and Proteomics Learning Objectives At the end of this assignment, you should be able to: 1. Draw the chemical structure of an amino acid and small peptide. 2. Describe the difference
Structure and Function of Antigen Recognition Molecules
 MICR2209 Structure and Function of Antigen Recognition Molecules Dr Allison Imrie allison.imrie@uwa.edu.au 1 Synopsis: In this lecture we will examine the major receptors used by cells of the innate and
MICR2209 Structure and Function of Antigen Recognition Molecules Dr Allison Imrie allison.imrie@uwa.edu.au 1 Synopsis: In this lecture we will examine the major receptors used by cells of the innate and
G-Protein Signaling. Introduction to intracellular signaling. Dr. SARRAY Sameh, Ph.D
 G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
BIO360 Quiz #1. September 14, Name five of the six Hallmarks of Cancer (not emerging hallmarks or enabling characteristics ): (5 points)
 Name: BIO360 Quiz #1 September 14, 2012 1. Name five of the six Hallmarks of Cancer (not emerging hallmarks or enabling characteristics ): (5 points) 2. The controversial hypothesis that only a small subset
Name: BIO360 Quiz #1 September 14, 2012 1. Name five of the six Hallmarks of Cancer (not emerging hallmarks or enabling characteristics ): (5 points) 2. The controversial hypothesis that only a small subset
Chapter 9. Cellular Signaling
 Chapter 9 Cellular Signaling Cellular Messaging Page 215 Cells can signal to each other and interpret the signals they receive from other cells and the environment Signals are most often chemicals The
Chapter 9 Cellular Signaling Cellular Messaging Page 215 Cells can signal to each other and interpret the signals they receive from other cells and the environment Signals are most often chemicals The
There are approximately 30,000 proteasomes in a typical human cell Each proteasome is approximately 700 kda in size The proteasome is made up of 3
 Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
Chapt 15: Molecular Genetics of Cell Cycle and Cancer
 Chapt 15: Molecular Genetics of Cell Cycle and Cancer Student Learning Outcomes: Describe the cell cycle: steps taken by a cell to duplicate itself = cell division; Interphase (G1, S and G2), Mitosis.
Chapt 15: Molecular Genetics of Cell Cycle and Cancer Student Learning Outcomes: Describe the cell cycle: steps taken by a cell to duplicate itself = cell division; Interphase (G1, S and G2), Mitosis.
Cell cycle and Apoptosis. Chalermchai Mitrpant
 Cell cycle and Apoptosis 2556 Chalermchai Mitrpant Overview of the cell cycle Outline Regulatory mechanisms controlling cell cycle Progression of the cell cycle Checkpoint of the cell cycle Phases of the
Cell cycle and Apoptosis 2556 Chalermchai Mitrpant Overview of the cell cycle Outline Regulatory mechanisms controlling cell cycle Progression of the cell cycle Checkpoint of the cell cycle Phases of the
Transformation of Normal HMECs (Human Mammary Epithelial Cells) into Metastatic Breast Cancer Cells: Introduction - The Broad Picture:
 Transformation of Normal HMECs (Human Mammary Epithelial Cells) into Metastatic Breast Cancer Cells: Introduction - The Broad Picture: Spandana Baruah December, 2016 Cancer is defined as: «A disease caused
Transformation of Normal HMECs (Human Mammary Epithelial Cells) into Metastatic Breast Cancer Cells: Introduction - The Broad Picture: Spandana Baruah December, 2016 Cancer is defined as: «A disease caused
Antigen processing and presentation. Monika Raulf
 Antigen processing and presentation Monika Raulf Lecture 25.04.2018 What is Antigen presentation? AP is the display of peptide antigens (created via antigen processing) on the cell surface together with
Antigen processing and presentation Monika Raulf Lecture 25.04.2018 What is Antigen presentation? AP is the display of peptide antigens (created via antigen processing) on the cell surface together with
Amino acids. Side chain. -Carbon atom. Carboxyl group. Amino group
 PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
Antigen presenting cells
 Antigen recognition by T and B cells - T and B cells exhibit fundamental differences in antigen recognition - B cells recognize antigen free in solution (native antigen). - T cells recognize antigen after
Antigen recognition by T and B cells - T and B cells exhibit fundamental differences in antigen recognition - B cells recognize antigen free in solution (native antigen). - T cells recognize antigen after
BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001
 BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) In the
BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) In the
Molecular Cell Biology - Problem Drill 19: Cell Signaling Pathways and Gene Expression
 Molecular Cell Biology - Problem Drill 19: Cell Signaling Pathways and Gene Expression Question No. 1 of 10 1. Which statement about cell signaling is correct? Question #1 (A) Cell signaling involves receiving
Molecular Cell Biology - Problem Drill 19: Cell Signaling Pathways and Gene Expression Question No. 1 of 10 1. Which statement about cell signaling is correct? Question #1 (A) Cell signaling involves receiving
Cell Physiology Final Exam Fall 2008
 Cell Physiology Final Exam Fall 2008 Guys, The average on the test was 69.9. Before you start reading the right answers please do me a favor and remember till the end of your life that GLUCOSE TRANSPORT
Cell Physiology Final Exam Fall 2008 Guys, The average on the test was 69.9. Before you start reading the right answers please do me a favor and remember till the end of your life that GLUCOSE TRANSPORT
VIII Curso Internacional del PIRRECV. Some molecular mechanisms of cancer
 VIII Curso Internacional del PIRRECV Some molecular mechanisms of cancer Laboratorio de Comunicaciones Celulares, Centro FONDAP Estudios Moleculares de la Celula (CEMC), ICBM, Facultad de Medicina, Universidad
VIII Curso Internacional del PIRRECV Some molecular mechanisms of cancer Laboratorio de Comunicaciones Celulares, Centro FONDAP Estudios Moleculares de la Celula (CEMC), ICBM, Facultad de Medicina, Universidad
TRIM5 requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription
 Manuscript EMBO-2014-90361 TRIM5 requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription Adam J Fletcher, Devin E Christensen, Chad Nelson, Choon Ping Tan, Torsten Schaller,
Manuscript EMBO-2014-90361 TRIM5 requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription Adam J Fletcher, Devin E Christensen, Chad Nelson, Choon Ping Tan, Torsten Schaller,
Islamic University Faculty of Medicine
 Islamic University Faculty of Medicine 2012 2013 2 Proteins are found in all living systems, ranging from bacteria and archaea through the unicellular eukaryotes, to plants, fungi, and animals. In all
Islamic University Faculty of Medicine 2012 2013 2 Proteins are found in all living systems, ranging from bacteria and archaea through the unicellular eukaryotes, to plants, fungi, and animals. In all
Enzyme-coupled Receptors. Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors
 Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Significance of the MHC
 CHAPTER 8 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
CHAPTER 8 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
CELL BIOLOGY - CLUTCH CH THE IMMUNE SYSTEM.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF HOST DEFENSES The human body contains three lines of against infectious agents (pathogens) 1. Mechanical and chemical boundaries (part of the innate immune system)
!! www.clutchprep.com CONCEPT: OVERVIEW OF HOST DEFENSES The human body contains three lines of against infectious agents (pathogens) 1. Mechanical and chemical boundaries (part of the innate immune system)
Chapter 11. B cell generation, Activation, and Differentiation. Pro-B cells. - B cells mature in the bone marrow.
 Chapter B cell generation, Activation, and Differentiation - B cells mature in the bone marrow. - B cells proceed through a number of distinct maturational stages: ) Pro-B cell ) Pre-B cell ) Immature
Chapter B cell generation, Activation, and Differentiation - B cells mature in the bone marrow. - B cells proceed through a number of distinct maturational stages: ) Pro-B cell ) Pre-B cell ) Immature
Phospho-AKT Sampler Kit
 Phospho-AKT Sampler Kit E 0 5 1 0 0 3 Kits Includes Cat. Quantity Application Reactivity Source Akt (Ab-473) Antibody E021054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit Akt (Phospho-Ser473) Antibody
Phospho-AKT Sampler Kit E 0 5 1 0 0 3 Kits Includes Cat. Quantity Application Reactivity Source Akt (Ab-473) Antibody E021054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit Akt (Phospho-Ser473) Antibody
The post-translational modification, SUMOylation, and cancer (Review)
 INTERNATIONAL JOURNAL OF ONCOLOGY 52: 1081-1094, 2018 The post-translational modification, SUMOylation, and cancer (Review) Zhi-Jian Han 1, Yan-Hu Feng 1, Bao-Hong Gu 2, Yu-Min Li 2 and Hao Chen 2 1 Key
INTERNATIONAL JOURNAL OF ONCOLOGY 52: 1081-1094, 2018 The post-translational modification, SUMOylation, and cancer (Review) Zhi-Jian Han 1, Yan-Hu Feng 1, Bao-Hong Gu 2, Yu-Min Li 2 and Hao Chen 2 1 Key
