Received 27 June 2003/Accepted 26 July 2003
|
|
|
- Elwin Lawrence
- 6 years ago
- Views:
Transcription
1 EUKARYOTIC CELL, Oct. 2003, p Vol. 2, No /03/$ DOI: /EC Copyright 2003, American Society for Microbiology. All Rights Reserved. Recapitulation of the Sexual Cycle of the Primary Fungal Pathogen Cryptococcus neoformans var. gattii: Implications for an Outbreak on Vancouver Island, Canada James A. Fraser, 1,2 Ryan L. Subaran, 1 Connie B. Nichols, 1 and Joseph Heitman 1,2,3,4 * Departments of Molecular Genetics and Microbiology, 1 Medicine, 3 and Pharmacology and Cancer Biology 4 and Howard Hughes Medical Institute, 2 Duke University Medical Center, Durham, North Carolina Received 27 June 2003/Accepted 26 July 2003 Cryptococcus neoformans is a human fungal pathogen that exists as three distinct varieties or sibling species: the predominantly opportunistic pathogens C. neoformans var. neoformans (serotype D) and C. neoformans var. grubii (serotype A) and the primary pathogen C. neoformans var. gattii (serotypes B and C). While serotypes A and D are cosmopolitan, serotypes B and C are typically restricted to tropical regions. However, serotype B isolates of C. neoformans var. gattii have recently caused an outbreak on Vancouver Island in Canada, highlighting the threat of this fungus and its capacity to infect immunocompetent individuals. Here we report a large-scale analysis of the mating abilities of serotype B and C isolates from diverse sources and identify unusual strains that mate robustly and are suitable for further genetic analysis. Unlike most isolates, which are of both the a and mating types but are predominantly sterile, the majority of the Vancouver outbreak strains are exclusively of the mating type and the majority are fertile. In an effort to enhance mating of these isolates, we identified and disrupted the CRG1 gene encoding the GTPase-activating protein involved in attenuating pheromone response. crg1 mutations dramatically increased mating efficiency and enabled mating with otherwise sterile isolates. Our studies provide a genetic and molecular foundation for further studies of this primary pathogen and reveal that the Vancouver Island outbreak may be attributable to a recent recombination event. Mating in the microbial world reassorts genomes, increases genetic diversity, and produces strains that can colonize new environmental niches. In several microorganisms, mating is also linked to pathogenicity. In the parasite Toxoplasma gondii, meiosis produces more-pathogenic isolates by recombining the genomes of two less-virulent strains (13, 42). In the plant fungal pathogen Ustilago maydis, mating is required for infection of maize because it is the filamentous dikaryon form that is pathogenic (for a review, see reference 32). In the human fungal pathogen Cryptococcus neoformans, the mating type has been linked to virulence (25) and sexual recombination produces spores, the suspected infectious propagule (43; J. Cohen, J. R. Perfect, and D. T. Durack, Letter, Lancet i:1301, 1982). C. neoformans is a haploid, encapsulated yeast that is the most common cause of fungal meningitis, which is fatal if untreated (17, 22, 29). C. neoformans strains can be classified into three divergent varieties and four serotypes based on capsular antigens: C. neoformans var. neoformans (serotype D), C. neoformans var. grubii (serotype A), and C. neoformans var. gattii (serotypes B and C). Serotypes A (the most clinically prevalent) and D are opportunistic pathogens that infect immunocompromised individuals throughout the world, whereas the divergent B and C serotypes are primary pathogens that * Corresponding author. Mailing address: Department of Molecular Genetics and Microbiology, 322 CARL Building, Research Dr., Duke University Medical Center, Durham, NC Phone: (919) Fax: (919) heitm001@duke.edu. The supplemental material for this article may be found at http: //ec.asm.org/. infect predominantly immunocompetent hosts (40). In contrast to serotypes A and D, which are cosmopolitan and most frequently associated with pigeon guano, the serotype B and C C. neoformans var. gattii is usually restricted to tropical and subtropical regions of the world, where it is found in association with woody debris of eucalyptus trees (10). Mating among serotype D strains (the least virulent variety) has been well characterized, and a congenic pair has been created to facilitate genetic analysis (16, 25). In contrast, the first accounts of the characterization of mating among serotype A C. neoformans var. grubii strains have only recently been reported (18, 35), an advance that was made possible by the discovery of a isolates long thought to be extinct (30, 45). The sexual system is bipolar (involving a and cell types) and is dependent on an unusually large mating type locus (28). Serotype A (C. neoformans var. grubii) strains exist predominantly as clonal isolates of the mating type throughout the world (12, 23, 47). In contrast, mixed populations of a and C. neoformans var. gattii strains have been identified colonizing hollows of eucalyptus trees in the Australian environment, where there is a relatively high incidence of cryptococcosis in native animals and indigenous human populations (4, 11, 14, 15). Despite the presence of both mating types, no meiotic recombination has been detected, suggesting that this ecological niche may support only clonal propagation of yeast cells and that sexual development occurs elsewhere if at all (15). Interest in the primary pathogenic form of C. neoformans has increased due to an outbreak of C. neoformans var. gattii infection on Vancouver Island in Canada. Prior to 1999, an average of two or three cases of cryptococcosis occurred annually in British Columbia and were caused by infections with 1036
2 VOL. 2, 2003 SEXUAL CYCLE OF C. NEOFORMANS VAR. GATTII 1037 serotype D. Since 1999, more than 66 human cases in otherwise healthy individuals, with at least four fatalities, have been reported and are attributable to infection with serotype B (M. Fyfe, W. Black, M. Romney, P. Kibsey, L. MacDougall, M. Starr, M. Pearce, C. Stephen, L. Stein, S. Mak, B. Emerson, J. Isaac-Renton, and D. Patrick, 5th Int. Conf. Cryptococcus Cryptococcosis, abstr. P1.1, 2002). There has also been an equally large increase in the rate of infection of animals, including dogs, cats, and porpoises (41). On Vancouver Island, C. neoformans var. gattii has been found in association with a number of native tree species (Douglas fir, alder, maple, and Garry oak) rather than eucalyptus trees, a previously identified environmental source of this variety. This expansion of the serotype B habitat to include nontropical areas increases the human risk of acquiring cryptococcosis and makes it paramount to understand the mechanisms by which C. neoformans var. gattii reproduces sexually. Our studies address the role of sexual reproduction in the life cycle and virulence of C. neoformans. The majority of C. neoformans var. gattii strains are sterile under most laboratory conditions. Although the perfect state of C. neoformans var. gattii was first reported over 25 years ago (21), these findings have proven difficult to reproduce (14, 26, 31, 34). Here we have recapitulated and extensively analyzed the sexual cycle of C. neoformans var. gattii. First, we describe conditions that support mating of a number of serotype B and C isolates from diverse sources. Second, we show that the vast majority of isolates from the Vancouver Island outbreak are fertile and that all are of the mating type. Third, we identify a pair of a and strains that mate robustly to produce viable meiotic progeny, providing a platform for genetic studies. Finally, we perform targeted gene disruptions in this variety and define a conserved role for the regulator of G protein signaling (RGS) protein Crg1 in pheromone sensing by demonstrating that crg1 strains mate more vigorously than wild-type strains. Taken together, our studies provide a genetic and molecular foundation for further studies of this primary pathogen and reveal that the Vancouver Island outbreak may be attributable to a recent recombination event in an unusual, fertile clade of C. neoformans. MATERIALS AND METHODS Strains, media, and mating assays. Reference strains used in this study were the congenic serotype D strains JEC20 (a) and JEC21 ( ) (16, 25). Strains were grown on yeast extract-peptone-dextrose (YEPD) medium. Mating assays were carried out on V8 medium (24) (5% [vol/vol] V8 juice, 3 mm KH 2 PO 4,4% [wt/vol] agar) at ph 5 or 7, carrot medium (5% [vol/vol] carrot juice, 3 mm KH 2 PO 4, 4% agar), or synthetic low ammonium dextrose (SLAD) medium. The ability to mate was tested against the serotype D tester strains JEC20 and JEC21, the serotype C strains NIH312 ( ) and B4546 (a), and crg1 mutant derivatives JF101 ( ) and JF109 (a). Strains were pregrown on YEPD agar for 2 days, and a small amount of cells was removed and patched onto solid mating medium either alone or mixed with a reference strain. Plates were incubated at room temperature in the dark for 9 days and assessed by light microscopy for filament and basidiospore formation. Molecular techniques. Standard methods were performed as described by Sambrook et al. (37). Serotyping was done using the Crypto Check serotyping kit from Iatron Laboratories (Tokyo). C. neoformans genomic DNA for Southern blot analysis was prepared as described by Pitkin et al. (36). C. neoformans biolistic transformation was performed using the technique for serotype D as described by Davidson et al. (7). Primers. Sequences of primers are as follows: STE12 U, CAATCTCAAAGCG GGGACAG; STE12 L, CTTTGTTTCGGTCCTAATACAGCC; JOHE6013, GCAACCAATCACTATGAA; JOHE6014, TGTGATCTGGTTCATGTA; JOHE8733, GCTGCGAGGATGTGAGCTGGA; JOHE8734, TCCATCTTG TTCAATCATAGACATGTTGGGCGAGTT; JOHE8735, AACTCGCCCA ACATGTCTATGATTGAACAAGATGGA; JOHE8736, ACCCCTTACCG CCTTCACTCAGAAGAACTCGTCAAG; JOHE8737, CTTGACGAGTTC TTCTGAGTGAAGGCGGTAAGGGGT; JOHE8738, GGTTTATCTGTAT TAACACGG; JOHE8896, CCCCTTCAACACAGCTCTTTA; JOHE8897, CAAGCCACCGGTCCTTATCCC; JOHE8987, AAGTTCGGACTGATTT ATTCA; JOHE8988, CGGAGGATAGAAGCTGTAAGCCAAAGTAAGG GGTAAGTTGGCAGA; JOHE8989, CCGTGTTAATACAGATAAACCGT CTTCAGAATACGAAAAGGACGA; JOHE8990, AAGCAAGACGGGAC TTATTGT; JOHE9365, CAGAAACGGTAACGGGAGCAC; JOHE9366, GCCCCTTCTTTCCTTATTCCT; JOHE9421, ATCCGCCCTCGAGTC AAA; JOHE9422, TGGCGACCGACTGTAGAT; JOHE9779, CCTTACTCCAT AACCCGCTAC; JOHE9780, AATTTCATAACGCTTTGGGAA; JOHE9787, ACACCGCCTGTTACAATGGAC; JOHE9788, CAGCGTTTGAAGATGGAC TTT; JOHE9574, ATTCGCCACCGGTTTATCTGTATTAAC; JOHE9575, ATT CGCCCTTTAAACGAGGATGTGAGC; JOHE9592, GAGTTCAGGCTTTTTC ATAGACATGTTGGGCGAGTT; JOHE9593, AACTCGCCCAACATGTCTAT GAAAAAGCCTGAACTC; JOHE9594, ACCCCTTACCGCCTTCACCTATTC CTTTGCCCTCGG; JOHE9595, CCGAGGGCAAAGGAATAGGTGAAGGCG GTAAGGGGT. Determination of mating type. Mating type was established by PCR using genomic DNA prepared with the Camgen yeast genomic DNA extraction kit (Whatman BioSciences Ltd., Cambridge, United Kingdom) with primers specific to the STE20a (JOHE9421-JOHE9422), STE12 (STE12 U-STE12 L), MFa (JOHE9787-JOHE9788), and MF (JOHE9779-JOHE9780) genes. PCR was repeated twice. Plasmids. The Neo r marker cassette clone pjaf1 was generated by combining the serotype A strain H99 ACT1 promoter (JOHE8733-JOHE8734), the transposon Tn5 Neo r gene (JOHE8735-JOHE8736), and the H99 TRP1 terminator (JOHE8737-JOHE8738) via overlap PCR using primer pair JOHE8733- JOHE8738 and cloned into plasmid pcr-topo2.1. The Hyg r marker cassette clone pjaf15 was created in an equivalent fashion by combining the ACT1 promoter (JOHE8733-JOHE9592), the Hyg r gene (JOHE9593-JOHE9594), and the TRP1 terminator (JOHE9595-JOHE8738) via overlap PCR using primer pair JOHE8733-JOHE8738 and cloned into plasmid pcr-topo2.1. Bacterial artificial chromosomes (BACs) containing the serotype B CRG1 gene (Australian isolates WM276 BAC 2B19 and E566 BAC 2B13) were identified by hybridization with a 1.2-kb CRG1 PCR fragment (JOHE6013-JOHE6014) from the serotype A strain H99. Based on results of Southern blot analysis, the CRG1 gene was subcloned as a 6.5-kb XhoI fragment into pbluescript SK SalI to yield plasmids pjaf2 (WM276) and pjaf3 (E566). Sequencing of the 6.5-kb subclones revealed only one single nucleotide polymorphism between the two isolates. The CRG1 reconstitution plasmid was created by amplifying the pch233 Nat r cassette (JOHE9574-JOHE9575), digesting it with AgeI/DraI, and using it to replace the NgoMIV/BsaA1 f1 origin fragment of pbluescript SK to give the Nat r cloning plasmid pjaf13. The WM276 CRG1 gene was then subcloned from pjaf2 into pjaf13 as a 6.6-kb ApaI/PstI fragment, yielding pjaf14. Disruption of the CRG1 gene. crg1 deletion alleles for the Australian isolates were generated by replacing the 3-kb Eco47III/SpeI fragment of the CRG1 gene in plasmid pjaf2 with the Nat r cassette from plasmid pch233 and the Neo r cassette from plasmid pjaf1 as an EcoRV/XbaI fragment to yield plasmids pjaf4 and pjaf6, respectively. With the use of the CRG1 gene sequence from strain WM276, overlap primers were designed to allow the construction of deletion alleles in strains NIH312, B4546, and R265 without the need to subclone and sequence the CRG1 gene. Building upon the technique of deletion allele generation by overlap PCR (6), we found that the process could be simplified by avoiding the use of chimeric primers for the marker segment, reducing the number of gene-specific primers required from six to four (Fig. 1). Furthermore, the design of the Neo r and Hyg r cassettes allows the same primer sets to be used for either a Neo r, Nat r,orhyg r disruption cassette. PCR was used to generate CRG1 5 (JOHE8987- JOHE9094), 3 (JOHE8989-JOHE8990), and Neo r (JOHE8733-JOHE8738) products, which were combined in a PCR overlap reaction (JOHE8987- JOHE8990) to yield strain-specific CRG1 deletion alleles. We used biolistic transformation for each strain of interest, introducing the appropriate circular plasmid (in strains WM276 and E566) or overlap PCR product (in strains B4546, NIH312, and R265) and selecting for resistance to G418 (200 g/ml) or nourseothricin (100 g/ml). crg1 transformants were identified by Southern blotting using the 6.5-kb WM276 CRG1 gene fragment as a probe (data not shown). Homologous integration frequencies were higher with overlap PCR products (7 of 60 for B4546, 6 of 70 for NIH312, and 15 of 32 for R265) than with circular
3 1038 FRASER ET AL. EUKARYOT. CELL A GSP1 B C GSP1 YFG1 5' GSP2 CRG1 JOHE8733 NAT/NEO/HYG GSP3 NAT/NEO/HYG JOHE8738 YFG1 3' YFG1 5' NAT/NEO/HYG YFG1 3' GSP4 GSP4 FIG. 1. Simplification of the creation of knockout constructs by using overlap PCR. We found that only four gene-specific primers (GSP), as opposed to six, were required to create knockout constructs. The flanking regions of the gene to be deleted were amplified (A) using one normal (GSP1-GSP4) and one chimeric (GSP2-GSP3) primer for each side. The marker cassette was amplified using the oligonucleotides JOHE8733 and JOHE8738, which work with the Nat r (NAT), Neo r (NEO), and Hyg r (HYG) cassettes. Like the products of normal overlap PCR, the three products were combined with the primers GSP1 and GSP4 (B) and amplified to yield the final knockout construct (C). For disruption of CRG1, the primers JOHE8987 (GSP1), JOHE9094 (GSP2), JOHE8989 (GSP3), and JOHE8990 (GSP4) were used. plasmid-borne crg1 deletion alleles (2 of 86 and 2 of 65 for the E566 Nat r and Neo r alleles, respectively, and 2 of 17 for the WM276 Neo r allele). Restriction fragment length polymorphism (RFLP) analysis. Fragments of the PLB1 (27) and SOD1 (3) genes were amplified from strains NIH312 and B4546 by PCR using the JOHE8896-JOHE8897 and JOHE9365-JOHE9366 primers pairs, respectively, and sequenced. Comparison revealed unique restriction sites in each gene a BglI restriction site present only in the PLB1 gene of strain NIH312 and a BsaJI restriction site present only in the SOD1 gene of strain B4546. For analysis of meiotic progeny and diploids, gene fragments were PCR amplified as described above and DNA was incubated with 1 U of the appropriate restriction enzyme and buffer for 2 h and separated on a 1.5% agarose gel. Slide matings. Glass microscope slides were sterilized, placed at a 20 angle in a plastic petri dish by using a sterile microfuge tube, and coated with a thin layer of molten V8 agar, with 5 ml of liquid V8 medium added to the bottom of each. The strains to be mated were resuspended in distilled H 2 O, and 10 l was pipetted at the apex of the slide and allowed to drain down the slide, creating a mating patch 3 to 4 cm in length and 0.5 cm wide. Slides were incubated at room temperature for 3 to 5 days in the dark. Microscopy and staining. For filament staining, slides were transferred to a fresh petri dish, washed three times with 10 ml of phosphate-buffered saline (PBS), fixed in 10 ml of 70% ethanol for 20 min, washed three times with PBS, permeabilized with 1% Triton X-100 in PBS for 10 min, and washed three times with PBS prior to staining. To visualize septa and DNA, filaments from matings were incubated in a 10-ml preparation of calcofluor white (fluorescent brightener 28 F-3397; Sigma) at 20 g/ml and a 5,000 dilution ofa5mmsolution of Sytox Green (Molecular Probes) in PBS for 20 min. Agar sections containing filaments were mounted on fresh microscope slides with 5 l of antifade (1 mg of p- phenylenediamine/ml in 50% glycerol). Bright field, differential interference microscopy (DIC), and fluorescent images were captured with a Zeiss Axioskop 2 Plus fluorescent microscope equipped with an AxioCam MRM digital camera and corresponding software. Environmental scanning electron microscopy (ESEM) was performed using a Philips XL30 ESEM TMP (FEI Company, Portland, Oreg.). Mating reactions were carried out on V8 plates, fixed by exposure to osmium tetroxide vapor for 30 min, and excised blocks of agar were adhered to prechilled stubs by using double-sided tape. The samples were viewed with a secondary electron detector at pressures of 5.2 to 5.5 torr and temperatures of 2.5 to 4.4 C and with a beam of 15 to 18 kv. Working distance ranged from 8.7 to 11 mm. Nucleotide sequence accession numbers. The sequence for the CRG1 genes of strains WM276 and E566 has been deposited in GenBank under accession number AY The accession numbers for the partial sequences of the SOD1 and PLB1 genes are AY and AY for NIH312 and AY and AY for B4546, respectively. RESULTS Intervarietal mating of C. neoformans var. gattii strains. Previous analyses of C. neoformans var. gattii mating involved intervarietal mating between serotype D tester strains and serotype B or C strains of the opposite mating type (21). We employed a similar approach and systematically analyzed the ability of a collection of 145 serotype B and C strains to mate with congenic serotype D a (JEC20) and (JEC21) strains on V8 ph 7 medium (see Table S1 in the supplemental material). Our collection consisted of three groups: laboratory serotype B (20) and C (14) isolates from diverse sources, Vancouver Island outbreak serotype B isolates (83), and Australian serotype B isolates (28). Analysis of the lab isolates revealed that 18 of 34 (53%) were sterile when mated with the appropriate serotype D tester strain. Of the 47% that did mate, mating was exclusively with partners of a single mating type (a or ). Accordingly, PCR analysis of these isolates with primers for -specific and a-specific genes revealed that each isolate was of a single mating type (see Table S1 in the supplemental material). The Australian isolates exhibited a different pattern. Of the clinical isolates, three of six (50%) mated with the tester strains. In contrast, 100% (22 of 22) of the environmental isolates were sterile. Overall, only 10% of isolates obtained from Australia were fertile. Importantly, the majority of the Vancouver Island outbreak isolates were fertile (70%, or 58 of 83). In contrast to isolates from Australia and elsewhere, which were both a and, all of the Vancouver isolates were of the mating type. Additionally, the mating abilities of the Vancouver Island outbreak isolates were not correlated with their clinical or environmental origins. A C. neoformans var. gattii pair can mate robustly. Most previous attempts to recapitulate mating of C. neoformans var. gattii used the type cultures NIH444 (serotype B and mating type ) and NIH191 (serotype C and mating type a). We found that both of these isolates mated with the appropriate serotype D tester strains, but mating to each other was much more difficult to detect and produced only a very weak mating reaction that was restricted to V8 ph 5 or carrot medium and required incubation for 6 weeks (data not shown). Further analysis of our collection revealed an alternative pair of serotype C isolates (NIH312,, and B4546, a) (Fig. 2) that mated with serotype D tester strains and robustly with each other. These isolates also mated with the type strains
4 VOL. 2, 2003 SEXUAL CYCLE OF C. NEOFORMANS VAR. GATTII 1039 A B C D E F FIG. 2. Morphology of mating C. neoformans var. gattii pairs. C. neoformans isolates were mated on V8 ph 7 medium on glass slides for 3 days, and morphology was observed. DIC microscopy allowed comparison of the mating ability and morphology of the congenic serotype D pair (JEC20 [serotype D and mating type a] JEC21 [serotype D and mating type ] [crosses are hereafter indicated in the form JEC20 Da JEC21 D ]) (A) with those of the robustly mating serotype C pair (B4546 Ca NIH312 C ) (B) and a serotype B-serotype C pair including one of the Vancouver isolates (B4546 Ca R265 B ) (C). All three of these C. neoformans var. gattii isolates underwent intervarietal mating with the serotype D congenic pair. (D) B4546 Ca JEC21 D. (E) JEC20 Da NIH312 C. (F) JEC20 Da R265 B. A comparison of morphologies of mating pairs (scale bar, 200 m) and basidial spore chain morphologies (inset) (scale bar, 20 m) is given. Downloaded from NIH444 and NIH191. We retested our collection of 146 isolates with this new mating pair. All of the strains that were mating competent with the serotype D testers also mated with these serotype C tester strains (see Table S1 in the supplemental material; Fig. 2). Additionally, we detected mating with 18 additional isolates from Vancouver Island (nine clinical or veterinary and nine environmental) that failed to mate with the serotype D strains, raising the percentage of mating-competent outbreak isolates from 70 to 91%. Morphology of mating serotype C strains. Based on examination by light microscopy and ESEM, filaments produced by mating of the serotype C a and strains B4546 and NIH312 exhibit morphological features characteristic of those produced by mating of basidiomycetes, including segregation of two haploid nuclei per filament cell and the production of fused clamp connections that enable faithful segregation of these nuclei. In contrast to serotype D crosses, which produce long peripheral hyphae, the intervarietal and serotype C intravarietal crosses produce abundant, shorter filaments primarily above the mating reaction. Calcofluor white staining revealed the presence of fused clamp connections (Fig. 3). Also unlike mating serotype D strains, mating serotype C strains form a protrusion from the subapical cell to which the clamp fuses, reminiscent of the peg structures observed in Coprinus cinereus (Fig. 3A) (20). This structure was not observed in serotype D in intervarietal crosses with serotype C (data not shown). Sytox Green staining revealed that each filament cell was dikaryotic and contained paired, comigrating nuclei (Fig. 3). ESEM was used to more closely examine the filaments produced during mating. In both serotype D and serotype C mating reactions, abundant filaments form, which in turn develop immature basidia. Four evenly spaced buds form simultaneously on the surface of the basidium and develop into basipetal chains of spores through consecutive rounds of budding (Fig. 4B). The serotype C rod-shaped spores are approximately twice as long as their lemon-shaped elliptical serotype D counterparts (Fig. 4B). The morphology of the mating Vancouver isolates differed from that of the serotype C isolates. While the serotype C mating pair produced intermediate-length aerial filaments over the vegetative mass, the Vancouver isolates produced basidia on stunted filaments close to the surface of the colony. However, the morphologies of the basidia and spores were the same as those of serotype C. Dominant selectable markers in C. neoformans var. gattii. To facilitate molecular studies of spore formation in C. neoformans var. gattii, we investigated the efficacy of several dominant markers in serotypes B and C. We confirmed that biolistic transformation of strains NIH312, B4546, R265, WM276, and E566 with the Nat r cassette (33) plasmid pch233 conferred resistance to nourseothricin (100 g/ml). Firstly, this verified that we could use biolistic transformation to transform a vari- on May 2, 2018 by guest
5 1040 FRASER ET AL. EUKARYOT. CELL A B C D Da x Dα Ca x Cα Ca x Dα ety of serotype B and C strains, and secondly it demonstrated that the serotype A strain H99 ACT1 promoter that drives the cassette can function in all four serotypes. Building upon the design of the Nat r cassette, we generated two derivatives by overlap PCR, replacing the Nat r coding region with the Tn5 Neo r gene (pjaf1) or the Escherichia coli Hyg r gene (pjaf15). Biolistic transformation of serotype A strain H99, serotype B strains R265, WM276, and E566, serotype C strains NIH312 and B4546, and the serotype D strain JEC21 with these plasmids confirmed that pjaf1 and pjaf15 were sufficient to confer resistance to G418 (200 g/ml) and hygromycin B (200 g/ml), respectively, in all four serotypes of C. neoformans. Previous marker cassettes designed to confer resistance to hygromycin B had not functioned in serotypes B and C (5). Differentially marked serotype C strains mate to produce recombinant spores. The morphology of C. neoformans var. gattii is distinct from those of C. neoformans var. neoformans and C. neoformans var. grubii, as C. neoformans var. gattii forms both round and elongated cells during infection (44) and elongated spores during mating (21). Elongated vegetative cells also form during growth on V8 medium irrespective of the mating partner and are difficult to distinguish from basidiospores during microdissection. Therefore, to select meiotic progeny, we differentially marked the serotype C mating pair NIH312 and B4546 by biolistic transformation with the Nat r and Neo r cassettes. These marked strains (MAT Nat r [JF65] and MATa G418 r ura5 [JF66]) were mated for 6 days and plated onto YEPD medium containing nourseothricin and G418. Similar to results with serotype D, stable Nat r G418 r diploids were obtained at 37 C that were thermally dimorphic, growing as yeast at 37 C but self-filamentously on V8 medium at 25 C (38). By selecting at 30 C, we could identify Nat r G418 r recombinant isolates that were not thermally dimorphic. To validate that these were bona fide meiotic progeny, we analyzed the segregation of four markers in a subset of the nonfilamentous isolates and observed recombinant progeny (Fig. 5). The markers included ura5 (from the a parent), RFLPs in the PLB1 and SOD1 genes, and the MAT locus (a and ). This analysis demonstrates that a random spore analysis approach can be used to isolate mating-competent meiotic progeny of serotype C. Mutation of the RGS protein Crg1. In Saccharomyces cerevisiae, mating is controlled by the pheromone response pathway, which is conserved in C. neoformans. Detection of pher- Da x Cα E Ca crg1 x Cα crg1 FIG. 3. Serotype C strains form dikaryon filaments and fused clamps during mating. Staining filaments from V8 ph 7 medium slide matings with calcofluor white and Sytox Green revealed that, as with serotype D (JEC20 Da JEC21 D [Da D ]) (A), both serotype C (B4546 Ca NIH312 C [Ca C ]) (B) and intervarietal serotype C-serotype D (B4546 Ca JEC21 D [Ca D ] and JEC20 Da NIH312 C [Da C ]) (C and D, respectively) crosses form dikaryotic filaments with paired nuclei (white arrows) and fused clamp connections (grey arrows). These same structures could also be observed in a crg1 bilateral cross (JF109 Ca JF101 C [Ca crg1 C crg1]) (E), supporting the observation that these strains undergo enhanced mating, as opposed to haploid fruiting which produces unfused clamps and uninucleate filaments. Scale bar, 10 m.
6 VOL. 2, 2003 SEXUAL CYCLE OF C. NEOFORMANS VAR. GATTII 1041 A D x D C x C B C x C C x C omones by a heterotrimeric G protein-coupled receptor initiates a conserved mitogen-activated protein kinase cascade to activate transcriptional regulators (for a review, see reference 9). In yeast, this pathway is negatively regulated by the RGS protein Sst2, which accelerates the intrinsic GTPase activity of the G subunit to silence signaling (1, 8). The C. neoformans SST2 homologue is the CRG1 gene, and crg1 mutations enhance mating of serotype A (35; P. Wang, personal communication). Here we show that a CRG1-targeted gene disruption enhances mating of serotypes B and C. The first targeted gene disruption in C. neoformans var. gattii, that of the serotype B SOD1 gene with the use of the URA5 gene, was recently reported (34). We disrupted the CRG1 gene in the serotype B strain WM276 by using dominant selectable markers that confer resistance to nourseothricin or G418. We also isolated crg1::neo r mutations in the serotype C mating pair NIH312 ( ) and B4546 (a) and the Vancouver Island clinical isolate R265 ( ). When cocultured with a strain of the opposite mating type, the crg1 mutant derivatives of NIH312, B4546, and R265 underwent much more rapid and proliferative mating reactions than the wild types, producing more hyphae and basidia (Fig. C x C D x D FIG. 4. Clamp cell formation and ESEM of serotype C basidia. (A) Staining with calcofluor white to analyze clamp cell formation revealed that the morphologies of mating pairs differ in serotype D and serotype C matings. In a serotype D cross (JEC20 Da JEC21 D [D D]) (top row, left to right), the clamp cell (large arrows) forms and fuses to the subapical filament cell. In a serotype C cross (B4546 Ca NIH312 C [C C]) (bottom row, left to right), clamp cell fusion involves the formation of a peglike protrusion (small arrows) from the filament subapical to the clamp cell (large arrows), to which the clamp fuses. Scale bar, 5 m. (B) ESEM of mating serotype C strains (B4546 Ca NIH312 C [C C]) shows the formation of four evenly spaced buds on the immature basidia, which develop into chains of smooth, rod-shaped spores. In contrast, mating serotype D strains (JEC20 Da JEC21 D [D D]) form more-spherical, rough basidiospore chains. Scale bar, 10 m. 6). This was also observed in intervarietal matings with serotype D strains. The enhanced mating of the crg1 mutant strains was restored to the wild-type level when the wild-type CRG1 gene was reintroduced (data not shown). Comparison of unilateral mutant crosses to bilateral mutant crosses revealed that the effect of the crg1 mutation is additive. Deletion of CRG1 was sufficient to allow mating on YEPD, a medium that usually strongly represses mating, in the case of a unilateral or bilateral cross (data not shown), indicating that this mutation is sufficient to bypass the nutritional limitation and desiccation signals normally required for C. neoformans mating. Mutation of CRG1 enables mating with a subset of previously sterile strains. Unlike the crg1 serotype C mating pair and Vancouver isolates, neither of the Australian crg1 strains exhibited any detectable mutant phenotype, nor could the CRG1 wild-type Australian isolates WM276 (serotype B and mating type ) or E566 (serotype B and mating type a) mate with crg1 strains. Also, attempts to identify fusion of the B4546 crg1::neo r strain JF109 and the E566 crg1::nat r strain JF9 by selecting for diploids at 37 C on YEPD medium containing nourseothricin and G418 failed to identify Nat r G418 r strains. The failure of the Australian isolate E566 to produce
7 1042 FRASER ET AL. EUKARYOT. CELL PLB1 SOD1 STE12α STE20a MFα MFa NIH 312 CDC B4546 JF65 JF66 Progeny 1 Progeny 2 Progeny 3 Progeny 4 Diploid URA MAT α a α a a a α α a/α FIG. 5. Analysis of meiotic progeny of a serotype C cross. The mating mixture of two marked strains MAT Nat r (JF65) and MATa G418 r ura5 (JF66) was plated onto YEPD medium containing both nourseothricin and G418 to select for recombinants. The parental strains and four progeny, in addition to diploid controls, were analyzed by PCR analysis for the segregation of RFLPs in the PLB1 and SOD1 genes and the mating type loci by using primers for the STE12, STE20a, MF, and MFa genes. Segregation of the ability to grow in the absence of uracil supplementation (URA5) and the ability to mate with the parental strains B4546 (serotype C and mating type a) and NIH312 (serotype C and mating type ) was also tested. mating structures is therefore due to a defect prior to fusion. This suggests that mutation of the CRG1 gene increases existing mating compatibility, leading to the production of more mating filaments. To test this hypothesis, we repeated our mating analysis of the original collection of 145 isolates by using NIH312 and B4546 crg1 mutants as tester strains. In all instances in which strains previously mated with NIH312 or B4546, this reaction was enhanced in a crg1 unilateral cross (see Table S1 in the supplemental material). In addition, one more serotype C strain (a total of 10 of 14, or 71%) and three more Vancouver Island environmental isolates (a total of 56 of 59, or 95%) mated. The crg1 strains are therefore useful tools for analyzing mating of C. neoformans var. gattii, allowing mating to be more readily observed and in some cases enabling otherwise sterile strains to mate. DISCUSSION Recapitulation of mating of C. neoformans var. gattii. The role of a sexual cycle in virulence of the human fungal pathogen C. neoformans is of great interest, as its importance in virulence is twofold. First, the vast majority of clinical cases are caused by strains of the mating type, and in serotype D, strains have been shown to be more pathogenic than a strains in a murine model (25). Second, the basidiospore is thought to be the infectious propagule. The sexual cycle and the components of the mating signaling cascade of C. neoformans var. neoformans have been studied intensely over the last decade, and recently this research has been extended to include serotype A C. neoformans var. grubii, in which a isolates and a complete sexual cycle have only recently been identified (18, 30, 35, 45). In contrast, the sexual cycle of C. neoformans var. gattii was first reported almost 30 years ago (21). Here we have recapitulated the original data on mating of C. neoformans var. gattii and extended these original studies by identifying strains that mate robustly with one another and with a large number of clinical and environmental isolates. Independently, Tscharke and Kwon-Chung have also identified serotype B strains capable of mating (R. L. Tscharke and K. J. Kwon- Chung, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. F-063, 2003). The recapitulation of data on mating and the discovery of robustly mating strains provide a platform for the generation of congenic serotype C strains and serve as an important foundation for genetic studies of this variety. As we have previously observed for serotype A (35), some C. neoformans var. gattii isolates are sterile, some are fertile, and others are capable of mating only with certain tester strains. This preference for mating partners can potentially be explained by three possible mechanisms. First, cryptic speciation events may be occurring that restrict mating between some members of the population, as has occurred in several other fungi, such as Coccidioides immitis and Coccidioides posadasii (19). Second, a spontaneous loss of mating ability may be occurring in nature or during laboratory passage, as has recently been reported for C. neoformans serotype D strains (46). Third, alterations in the mating type locus may lead to a loss or restriction of mating capacity, and we are currently investigating this possibility by sequencing the a and alleles of the MAT locus from strains WM276 and E566. Targeted gene disruption using dominant selectable markers in serotypes B and C. In addition to the availability of genetic analysis, the ability to perform targeted disruptions is an essential attribute with regard to a model organism. Our studies presented here demonstrate that the Nat r, Hyg r, and Neo r cassettes can be used as dominant selectable markers in C. neoformans var. gattii to readily disrupt genes by biolistic transformation and homologous recombination, providing valuable tools for facilitating molecular manipulation of the genome. The ability to transform a range of clinical and environmental isolates by using plasmids that can function in other varieties not only provides a tool for performing targeted disruptions in C. neoformans var. gattii but also allows the option
8 VOL. 2, 2003 SEXUAL CYCLE OF C. NEOFORMANS VAR. GATTII 1043 A B C D Ca x Cα Ca crg1 x Cα Ca x Cα crg1 Ca crg1 x Cα crg1 E F G H Ca x Bα Ca crg1 x Bα Ca x Bα crg1 Ca crg1 x Bα crg1 FIG. 6. The crg1 mutation accelerates mating. Mating C. neoformans isolates and crg1 mutant strains were incubated on V8 ph 7 medium on glass slides for 3 days, and their morphology was observed by using DIC microscopy. A comparison of the effects of the crg1 mutation on mating ability was performed on V8 ph 7 medium. Compared to the serotype C wild-type crosses (B4546 Ca NIH312 C [Ca C ] and B4546 Ca R265 B [Ca B ]) (A and E, respectively), introducing a crg1 mutant in unilateral crosses (JF109 crg1 Ca NIH312 C [Ca crg1 C ], B4546 Ca JF101 crg1 C [Ca C crg1 ], JF109 crg1 Ca R265 B [Ca crg1 B ], and B4546 Ca JF117 crg1 B [Ca B crg1 ]) (B, C, F, and G, respectively) increased the amount of filamentation and basidia production. This effect was even greater in bilateral crosses (JF109 crg1 Ca JF101 crg1 C [Ca crg1 C crg1 ] and JF109 crg1 Ca JF117 crg1 B [Ca crg1 B crg1 ]) (D and H, respectively). Scale bar, 200 m. Downloaded from of utilizing marker constructs and gene regulatory sequences designed for use in serotype A or D. The Vancouver Island outbreak is attributable to an unusually fertile set of isolates. Attention has recently focused on the primary pathogenic form of C. neoformans due to an outbreak on Vancouver Island, Canada. The presence of C. neoformans var. gattii on Vancouver Island indicates that this primary pathogen is not restricted to tropical regions. Our study reveals that, unlike isolates originating from elsewhere in the world, the strains from the Vancouver Island outbreak are exclusively of the mating type. Furthermore, the vast majority of the outbreak isolates are fertile. This observation gives rise to several possible models. In the first, mating and meiotic recombination in a fertile clade may have given rise to an isolate with increased virulence, expanded host range, or an altered environmental niche. In the second, the fertility of the isolates may increase the production of the infectious propagule, increasing the incidence of infection. Third, the outbreak may be attributable to a so far unobserved ability of these isolates to undergo haploid fruiting, which perhaps explains the presence of exclusively isolates. Finally, the relationship between the outbreak and the fertility of these isolates may be coincidental. Australian environmental isolates are sterile. In contrast to the fertility of isolates from Vancouver Island and elsewhere in the world is the infertility of the Australian environmental isolates, as we were unable to identify mating of any of these isolates. This finding supports previous reports that failed to identify recombining populations in association with eucalyptus trees (15). In contrast, 50% (three of six) of clinical isolates were fertile. These isolates may have originated elsewhere or may be due to the production of spores from rare environmental isolates. It is still possible that the conditions for mating of this subset of isolates differ from those for mating of the fertile strains identified thus far and that given appropriate conditions or mating partners these strains may be capable of undergoing sexual recombination. This represents an important avenue of study, as the C. neoformans var. gattii genome project that is currently under way uses a sterile Australian environmental isolate (WM276) (J. Kronstad, personal communication). One possibility is that these geographically isolated strains no longer have functional mating type loci. To investigate this prospect, we are currently sequencing the mating type loci of both (WM276) and a (E566) isolates. Molecular analysis of the mating type loci of these environmental isolates may reveal the reason for this infertility, providing not only the means to render the platform sequence reference strain WM276 fertile but also insight into the fertility of strains from elsewhere in the world. Disruption of the RGS protein-encoding gene CRG1 enhances mating of C. neoformans var. gattii. In C. neoformans var. grubii, loss of the RGS protein Crg1 required for pheromone desensitization enhances mating. Disruption of the CRG1 gene greatly enhanced mating of both serotype C isolates and an isolate from the Vancouver Island outbreak and on May 2, 2018 by guest
9 1044 FRASER ET AL. EUKARYOT. CELL conferred the ability to mate with a subset of partners that appeared to be sterile when crossed with a wild-type strain. Furthermore, the crg1 mutation overrides the nutritional and desiccation signals normally required for mating. In direct contrast to strains from Vancouver Island, environmental isolates originating from Australia had no identifiable sexual cycle despite deletion of the CRG1 gene in either or both strains in crosses. It is striking that the RGS protein mutations exert related phenotypes in the two divergent organisms S. cerevisiae and C. neoformans, enhancing pheromone responsiveness in both and enhancing mating of C. neoformans. This said, there are also interesting distinctions between the phenotypes of an S. cerevisiae sst2 mutant and the crg1 mutant of C. neoformans. In contrast to the alteration in cellular morphology and the reduced mating ability of an sst2 mutant (9, 39), crg1 strains show no alteration in growth morphology in the absence of a partner and undergo increased mating. Comparison of fungal RGS proteins reveals that all of them contain two N-terminal DEP (Dishevelled, Egl-10, Pleckstrin) domains, except the C. neoformans Crg1 protein, which appears to contain only one. In S. cerevisiae, these domains have been implicated in signaling the stress response pathway (2), and this function may have been lost in C. neoformans, perhaps explaining why crg1 mutant strains do not display any detrimental phenotypes. crg1 mutant strains therefore represent a valuable tool in the further analysis of mating and subsequent production of the infectious propagule by this primary human pathogen. In summary, these studies provide insight into a factor that may have contributed to the outbreak on Vancouver Island and a starting point for the generation of congenic pairs of this variety. Additionally, comparison of serotype B and C, Vancouver, and Australian isolates to the opportunistic serotype A and D pathogens provides a model to address the central question of how an opportunistic organism escapes immune control and evolves into a primary pathogen. ACKNOWLEDGMENTS We thank Jim Kronstad, Wiley Schell, June Kwon-Chung, and Wieland Meyer for strains and Leslie Eibest for valuable assistance in performing ESEM (NSF award no. DBI ). The Nat r cassette subclone pch233 was generously provided by Christina Hull. We thank Christina Hull, Ping Wang, Tom Mitchell, John Perfect, and Jim Kronstad for advice and comments on the manuscript. This work was supported in part by NIAID R01 grant AI50113 to Joseph Heitman and NIAID PO1 program project grant AI44975 to the Duke University Mycology Research Unit. Joseph Heitman is a Burroughs Wellcome Scholar in Molecular Pathogenic Mycology and an associate investigator of the Howard Hughes Medical Institute. REFERENCES 1. Apanovitch, D. M., K. C. Slep, P. B. Sigler, and H. G. Dohlman Sst2 is a GTPase-activating protein for Gpa1: purification and characterization of a cognate RGS-G protein pair in yeast. Biochemistry 37: Burchett, S. A., P. Flanary, C. Aston, L. Jiang, K. H. Young, P. Uetz, S. Fields, and H. G. Dohlman Regulation of stress response signaling by the N-terminal dishevelled/egl-10/pleckstrin domain of Sst2, a regulator of G protein signaling in Saccharomyces cerevisiae. J. Biol. Chem. 277: Chaturvedi, S., A. J. Hamilton, P. Hobby, G. Zhu, C. V. Lowry, and V. Chaturvedi Molecular cloning, phylogenetic analysis and three-dimensional modeling of Cu, Zn superoxide dismutase (CnSOD1) from three varieties of Cryptococcus neoformans. Gene 268: Chen, S., T. Sorrell, G. Nimmo, B. Speed, B. Currie, D. Ellis, D. Marriott, T. Pfeiffer, D. Parr, K. Byth, et al Epidemiology and host- and varietydependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Clin. Infect. Dis. 31: Cox, G. M., D. L. Toffaletti, and J. R. Perfect Dominant selection system for use in Cryptococcus neoformans. J. Med. Vet. Mycol. 34: Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D Souza, P. Wang, and J. Heitman A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148: Davidson, R. C., M. C. Cruz, R. A. Sia, B. Allen, J. A. Alspaugh, and J. Heitman Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 29: Dohlman, H. G., D. Apaniesk, Y. Chen, J. Song, and D. Nusskern Inhibition of G-protein signaling by dominant gain-of-function mutations in Sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: Dohlman, H. G., and J. W. Thorner Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70: Ellis, D. H., and T. J. Pfeiffer Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28: Fisher, D., J. Burrow, D. Lo, and B. Currie Cryptococcus neoformans in tropical northern Australia: predominantly variant gattii with good outcomes. Aust. N. Z. J. Med. 23: Franzot, S. P., J. S. Hamdan, B. P. Currie, and A. Casadevall Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J. Clin. Microbiol. 35: Grigg, M. E., S. Bonnefoy, A. B. Hehl, Y. Suzuki, and J. C. Boothroyd Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science 294: Halliday, C. L., T. Bui, M. Krockenberger, R. Malik, D. H. Ellis, and D. A. Carter Presence of and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J. Clin. Microbiol. 37: Halliday, C. L., and D. A. Carter Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J. Clin. Microbiol. 41: Heitman, J., B. Allen, J. A. Alspaugh, and K. J. Kwon-Chung On the origins of congenic MAT and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet. Biol. 28: Hull, C. M., and J. Heitman Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36: Keller, S. M., M. A. Viviani, M. C. Esposto, M. Cogliati, and B. L. Wickes Molecular and genetic characterization of a serotype A MATa Cryptococcus neoformans isolate. Microbiology 149: Koufopanou, V., A. Burt, and J. W. Taylor Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 94: Kues, U Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64: Kwon-Chung, K. J A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68: Kwon-Chung, K. J., and J. E. Bennett Cryptococcosis, p In K. J. Kwon-Chung and J. E. Bennett (ed.), Medical mycology. Lea & Febiger, Philadelphia, Pa. 23. Kwon-Chung, K. J., and J. E. Bennett Distribution of alpha and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108: Kwon-Chung, K. J., J. E. Bennett, and J. C. Rhodes Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Leeuwenhoek 48: Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60: Kwon-Chung, K. J., B. L. Wickes, L. Stockman, G. D. Roberts, D. Ellis, and D. H. Howard Virulence, serotype, and molecular characteristics of environmental strains of Cryptococcus neoformans var. gattii. Infect. Immun. 60: Latouche, G. N., T. C. Sorrell, and W. Meyer Isolation and characterisation of the phospholipase B gene of Cryptococcus neoformans var. gattii. FEMS Yeast Res. 2: Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester, F. S. Dietrich, and J. Heitman Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1: Lengeler, K. B., and J. Heitman Cryptococcus neoformans as a model fungal pathogen, p In H. D. Osiewacz (ed.), Molecular biology of fungal development. Marcel Dekker, Inc., New York, N.Y. 30. Lengeler, K. B., P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proc. Natl. Acad. Sci. USA 97: Madrenys, N., C. De Vroey, C. Raes-Wuytack, and J. M. Torres-Rodriguez.
Pheromones Stimulate Mating and Differentiation via Paracrine and Autocrine Signaling in Cryptococcus neoformans
 EUKARYOTIC CELL, June 2002, p. 366 377 Vol. 1, No. 3 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.3.366 377.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Pheromones Stimulate
EUKARYOTIC CELL, June 2002, p. 366 377 Vol. 1, No. 3 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.3.366 377.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Pheromones Stimulate
Sexual Cycle of Cryptococcus neoformans var. grubii and Virulence of Congenic a and Isolates
 INFECTION AND IMMUNITY, Sept. 2003, p. 4831 4841 Vol. 71, No. 9 0019-9567/03/$08.00 0 DOI: 10.1128/IAI.71.9.4831 4841.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Sexual
INFECTION AND IMMUNITY, Sept. 2003, p. 4831 4841 Vol. 71, No. 9 0019-9567/03/$08.00 0 DOI: 10.1128/IAI.71.9.4831 4841.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Sexual
Serotype AD Strains of Cryptococcus neoformans Are Diploid or Aneuploid and Are Heterozygous at the Mating-Type Locus
 INFECTION AND IMMUNITY, Jan. 2001, p. 115 122 Vol. 69, No. 1 0019-9567/01/$04.00 0 DOI: 10.1128/IAI.69.1.115 122.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved. Serotype AD
INFECTION AND IMMUNITY, Jan. 2001, p. 115 122 Vol. 69, No. 1 0019-9567/01/$04.00 0 DOI: 10.1128/IAI.69.1.115 122.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved. Serotype AD
Sex-Specific Homeodomain Proteins Sxi1 and Sxi2a Coordinately Regulate Sexual Development in Cryptococcus neoformans
 EUKARYOTIC CELL, Mar. 2005, p. 526 535 Vol. 4, No. 3 1535-9778/05/$08.00 0 doi:10.1128/ec.4.3.526 535.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Sex-Specific Homeodomain
EUKARYOTIC CELL, Mar. 2005, p. 526 535 Vol. 4, No. 3 1535-9778/05/$08.00 0 doi:10.1128/ec.4.3.526 535.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Sex-Specific Homeodomain
Interaction between genetic background and the mating type locus in Cryptococcus neoformans virulence potential
 Genetics: Published Articles Ahead of Print, published on June 18, 2005 as 10.1534/genetics.105.045039 Interaction between genetic background and the mating type locus in Cryptococcus neoformans virulence
Genetics: Published Articles Ahead of Print, published on June 18, 2005 as 10.1534/genetics.105.045039 Interaction between genetic background and the mating type locus in Cryptococcus neoformans virulence
Richard Malik Centre for Veterinary Education The University of Sydney
 Richard Malik Centre for Veterinary Education The University of Sydney 1 Pathology Update 3/8/2019 Pathology Update 3/8/2019 2 Pathology Update 3/8/2019 3 Pathology Update 3/8/2019 4 Pathology Update 3/8/2019
Richard Malik Centre for Veterinary Education The University of Sydney 1 Pathology Update 3/8/2019 Pathology Update 3/8/2019 2 Pathology Update 3/8/2019 3 Pathology Update 3/8/2019 4 Pathology Update 3/8/2019
Laboratory of Clinical Investigation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892
 The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence Y. C. Chang, L. A. Penoyer, and K. J. Kwon-Chung* Laboratory of Clinical Investigation, National
The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence Y. C. Chang, L. A. Penoyer, and K. J. Kwon-Chung* Laboratory of Clinical Investigation, National
develop capsules when released from the basidia. Budding may begin after encapsulation, thereby completing the life cycle.
 INTRODUCTION Cryptococcosis is an invasive fungal infection due to Cryptococcus neoformans which has become increasingly prevalent in immunocompromised patients. The microbiology and epidemiology of Cryptococcus
INTRODUCTION Cryptococcosis is an invasive fungal infection due to Cryptococcus neoformans which has become increasingly prevalent in immunocompromised patients. The microbiology and epidemiology of Cryptococcus
Sexual Development in Cryptococcus neoformans Requires CLP1, a Target of the Homeodomain Transcription Factors Sxi1 and Sxi2a
 EUKARYOTIC CELL, Jan. 2008, p. 49 57 Vol. 7, No. 1 1535-9778/08/$08.00 0 doi:10.1128/ec.00377-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Sexual Development in Cryptococcus
EUKARYOTIC CELL, Jan. 2008, p. 49 57 Vol. 7, No. 1 1535-9778/08/$08.00 0 doi:10.1128/ec.00377-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Sexual Development in Cryptococcus
Mating-Type-Specific and Nonspecific PAK Kinases Play Shared and Divergent Roles in Cryptococcus neoformans
 EUKARYOTIC CELL, Apr. 2002, p. 257 272 Vol. 1, No. 2 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.2.257 272.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Mating-Type-Specific
EUKARYOTIC CELL, Apr. 2002, p. 257 272 Vol. 1, No. 2 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.2.257 272.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Mating-Type-Specific
Discovery of a Modified Tetrapolar Sexual Cycle in Cryptococcus amylolentus and the Evolution of MAT in the Cryptococcus Species Complex
 Discovery of a Modified Tetrapolar Sexual Cycle in Cryptococcus amylolentus and the Evolution of MAT in the Cryptococcus Species Complex Keisha Findley 1,2., Sheng Sun 2., James A. Fraser 3, Yen-Ping Hsueh
Discovery of a Modified Tetrapolar Sexual Cycle in Cryptococcus amylolentus and the Evolution of MAT in the Cryptococcus Species Complex Keisha Findley 1,2., Sheng Sun 2., James A. Fraser 3, Yen-Ping Hsueh
Spores as Infectious Propagules of Cryptococcus neoformans
 INFECTION AND IMMUNITY, Oct. 2009, p. 4345 4355 Vol. 77, No. 10 0019-9567/09/$08.00 0 doi:10.1128/iai.00542-09 Copyright 2009, American Society for Microbiology. All Rights Reserved. Spores as Infectious
INFECTION AND IMMUNITY, Oct. 2009, p. 4345 4355 Vol. 77, No. 10 0019-9567/09/$08.00 0 doi:10.1128/iai.00542-09 Copyright 2009, American Society for Microbiology. All Rights Reserved. Spores as Infectious
CRYPTOCOCCUS neoformans is an encapsulated
 Copyright Ó 2007 by the Genetics Society of America DOI: 10.1534/genetics.107.078923 Genetic Analyses of a Hybrid Cross Between Serotypes A and D Strains of the Human Pathogenic Fungus Cryptococcus neoformans
Copyright Ó 2007 by the Genetics Society of America DOI: 10.1534/genetics.107.078923 Genetic Analyses of a Hybrid Cross Between Serotypes A and D Strains of the Human Pathogenic Fungus Cryptococcus neoformans
Molecular and genetic characterization of a serotype A MATa Cryptococcus neoformans isolate
 Microbiology (2003), 149, 131 142 DOI 10.1099/mic.0.25921-0 Molecular and genetic characterization of a serotype A MATa Cryptococcus neoformans isolate S. M. Keller, 1 M. A. Viviani, 2 M. C. Esposto, 2
Microbiology (2003), 149, 131 142 DOI 10.1099/mic.0.25921-0 Molecular and genetic characterization of a serotype A MATa Cryptococcus neoformans isolate S. M. Keller, 1 M. A. Viviani, 2 M. C. Esposto, 2
Cryptococcosis*,** *usually considered to be an opportunistic mycosis that often becomes systemic.
 Cryptococcosis*,** Definition - a chronic, subacute or rarely acute pulmonary, systemic or meningitic infection caused by Cryptococcus neoformans - the encapsulated Blastomycetes anamorph of Filobasidiella
Cryptococcosis*,** Definition - a chronic, subacute or rarely acute pulmonary, systemic or meningitic infection caused by Cryptococcus neoformans - the encapsulated Blastomycetes anamorph of Filobasidiella
Received 16 July 2004/Accepted 21 August 2004
 EUKARYOTIC CELL, Dec. 2004, p. 1476 1491 Vol. 3, No. 6 1535-9778/04/$08.00 0 DOI: 10.1128/EC.3.6.1476 1491.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Adenylyl Cyclase-Associated
EUKARYOTIC CELL, Dec. 2004, p. 1476 1491 Vol. 3, No. 6 1535-9778/04/$08.00 0 DOI: 10.1128/EC.3.6.1476 1491.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Adenylyl Cyclase-Associated
Epidemiology and Laboratory Diagnosis of Fungal Diseases
 Medical Mycology (BIOL 4849) Summer 2007 Dr. Cooper Epidemiology of Mycoses Epidemiology and Laboratory Diagnosis of Fungal Diseases Mycosis (pl., mycoses) - an infection caused by a fungus Two broad categories
Medical Mycology (BIOL 4849) Summer 2007 Dr. Cooper Epidemiology of Mycoses Epidemiology and Laboratory Diagnosis of Fungal Diseases Mycosis (pl., mycoses) - an infection caused by a fungus Two broad categories
Isolates of Cryptococcus neoformans from Infected Animals Reveal Genetic Exchange in Unisexual, Mating Type Populations
 EUKARYOTIC CELL, Oct. 2008, p. 1771 1780 Vol. 7, No. 10 1535-9778/08/$08.00 0 doi:10.1128/ec.00097-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. Isolates of Cryptococcus neoformans
EUKARYOTIC CELL, Oct. 2008, p. 1771 1780 Vol. 7, No. 10 1535-9778/08/$08.00 0 doi:10.1128/ec.00097-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. Isolates of Cryptococcus neoformans
Impact of Mating Type, Serotype, and Ploidy on the Virulence of Cryptococcus neoformans
 INFECTION AND IMMUNITY, July 2008, p. 2923 2938 Vol. 76, No. 7 0019-9567/08/$08.00 0 doi:10.1128/iai.00168-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. Impact of Mating Type,
INFECTION AND IMMUNITY, July 2008, p. 2923 2938 Vol. 76, No. 7 0019-9567/08/$08.00 0 doi:10.1128/iai.00168-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. Impact of Mating Type,
ACCEPTED. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange. in unisexual, α mating type populations.
 EC Accepts, published online ahead of print on 13 June 2008 Eukaryotic Cell doi:10.1128/ec.00097-08 Copyright 2008, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
EC Accepts, published online ahead of print on 13 June 2008 Eukaryotic Cell doi:10.1128/ec.00097-08 Copyright 2008, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
Transcription Factors Mat2 and Znf2 Operate Cellular Circuits Orchestrating Opposite- and Same-Sex Mating in Cryptococcus neoformans
 Transcription Factors Mat2 and Znf2 Operate Cellular Circuits Orchestrating Opposite- and Same-Sex Mating in Cryptococcus neoformans Xiaorong Lin 1,2 *, Jennifer C. Jackson 1, Marianna Feretzaki 2, Chaoyang
Transcription Factors Mat2 and Znf2 Operate Cellular Circuits Orchestrating Opposite- and Same-Sex Mating in Cryptococcus neoformans Xiaorong Lin 1,2 *, Jennifer C. Jackson 1, Marianna Feretzaki 2, Chaoyang
Conservation of morphogenesis and virulence in Cryptococcus neoformans. Xiaorong Lin Texas A&M University
 Conservation of morphogenesis and virulence in Cryptococcus neoformans Xiaorong Lin Texas A&M University Cryptococcus infection Lymphatic and hematogenous dissemination Pulmonary infection Cryptococcal
Conservation of morphogenesis and virulence in Cryptococcus neoformans Xiaorong Lin Texas A&M University Cryptococcus infection Lymphatic and hematogenous dissemination Pulmonary infection Cryptococcal
Cryptococcal Cell Morphology Affects Host Cell Interactions and Pathogenicity
 Cryptococcal Cell Morphology Affects Host Cell Interactions and Pathogenicity Laura H. Okagaki 1., Anna K. Strain 1., Judith N. Nielsen 2, Caroline Charlier 3, Nicholas J. Baltes 1, Fabrice Chrétien 3,4,
Cryptococcal Cell Morphology Affects Host Cell Interactions and Pathogenicity Laura H. Okagaki 1., Anna K. Strain 1., Judith N. Nielsen 2, Caroline Charlier 3, Nicholas J. Baltes 1, Fabrice Chrétien 3,4,
Amt2 Permease Is Required To Induce Ammonium-Responsive Invasive Growth and Mating in Cryptococcus neoformans
 EUKARYOTIC CELL, Feb. 2008, p. 237 246 Vol. 7, No. 2 1535-9778/08/$08.00 0 doi:10.1128/ec.00079-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Amt2 Permease Is Required To Induce
EUKARYOTIC CELL, Feb. 2008, p. 237 246 Vol. 7, No. 2 1535-9778/08/$08.00 0 doi:10.1128/ec.00079-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Amt2 Permease Is Required To Induce
Specialization of the HOG pathway and its impact on differentiation and virulence. of Cryptococcus neoformans
 Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans Yong-Sun Bahn, Kaihei Kojima, Gary M. Cox, and Joseph Heitman,,,, Departments of Molecular Genetics
Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans Yong-Sun Bahn, Kaihei Kojima, Gary M. Cox, and Joseph Heitman,,,, Departments of Molecular Genetics
Evidence of Sexual Recombination among Cryptococcus neoformans Serotype A Isolates in Sub-Saharan Africa
 EUKARYOTIC CELL, Dec. 2003, p. 1162 1168 Vol. 2, No. 6 1535-9778/03/$08.00 0 DOI: 10.1128/EC.2.6.1162 1168.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Evidence of Sexual
EUKARYOTIC CELL, Dec. 2003, p. 1162 1168 Vol. 2, No. 6 1535-9778/03/$08.00 0 DOI: 10.1128/EC.2.6.1162 1168.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Evidence of Sexual
The RGS Protein Crg2 Regulates Pheromone and Cyclic AMP Signaling in Cryptococcus neoformans
 EUKARYOTIC CELL, Sept. 2008, p. 1540 1548 Vol. 7, No. 9 1535-9778/08/$08.00 0 doi:10.1128/ec.00154-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. The RGS Protein Crg2 Regulates
EUKARYOTIC CELL, Sept. 2008, p. 1540 1548 Vol. 7, No. 9 1535-9778/08/$08.00 0 doi:10.1128/ec.00154-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. The RGS Protein Crg2 Regulates
Identification of ENA1 as a Virulence Gene of the Human Pathogenic Fungus Cryptococcus neoformans through Signature-Tagged Insertional Mutagenesis
 EUKARYOTIC CELL, Mar. 2009, p. 315 326 Vol. 8, No. 3 1535-9778/09/$08.00 0 doi:10.1128/ec.00375-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Identification of ENA1 as a Virulence
EUKARYOTIC CELL, Mar. 2009, p. 315 326 Vol. 8, No. 3 1535-9778/09/$08.00 0 doi:10.1128/ec.00375-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Identification of ENA1 as a Virulence
HYBRIDIZATION IN CRYPTOCOCCUS NEOFORMANS
 HYBRIDIZATION IN CRYPTOCOCCUS NEOFORMANS DEVELOPING A GENETIC LINKAGE MAP FROM AN INTERVARIETAL CROSS OF SEROTYPES A AND D AND THE ANALYSIS OF THE COSTS AND BENEFITS OF HYBRIDIZATION IN CRYPTCOCCUS NEOFORMANS
HYBRIDIZATION IN CRYPTOCOCCUS NEOFORMANS DEVELOPING A GENETIC LINKAGE MAP FROM AN INTERVARIETAL CROSS OF SEROTYPES A AND D AND THE ANALYSIS OF THE COSTS AND BENEFITS OF HYBRIDIZATION IN CRYPTCOCCUS NEOFORMANS
Unisexual reproduction enhances fungal competitiveness by promoting. habitat exploration via hyphal growth and sporulation
 EC Accepts, published online ahead of print on 21 June 2013 Eukaryotic Cell doi:10.1128/ec.00147-13 Copyright 2013, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11 12 13
EC Accepts, published online ahead of print on 21 June 2013 Eukaryotic Cell doi:10.1128/ec.00147-13 Copyright 2013, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11 12 13
Epidemiology of Gray Leaf Spot of Perennial Ryegrass Philip Harmon and Richard Latin. Objective
 Epidemiology of Gray Leaf Spot of Perennial Ryegrass Philip Harmon and Richard Latin Objective Rationale The continuing objective of this research is to investigate the environmental factors that influence
Epidemiology of Gray Leaf Spot of Perennial Ryegrass Philip Harmon and Richard Latin Objective Rationale The continuing objective of this research is to investigate the environmental factors that influence
DURING eukaryotic growth, developmental transitions
 INVESTIGATION Analysis of Cryptococcus neoformans Sexual Development Reveals Rewiring of the Pheromone-Response Network by a Change in Transcription Factor Identity Emilia K. Kruzel,* Steven S. Giles,*
INVESTIGATION Analysis of Cryptococcus neoformans Sexual Development Reveals Rewiring of the Pheromone-Response Network by a Change in Transcription Factor Identity Emilia K. Kruzel,* Steven S. Giles,*
Pde1 Phosphodiesterase Modulates Cyclic AMP Levels through a Protein Kinase A-Mediated Negative Feedback Loop in Cryptococcus neoformans
 EUKARYOTIC CELL, Dec. 2005, p. 1971 1981 Vol. 4, No. 12 1535-9778/05/$08.00 0 doi:10.1128/ec.4.12.1971 1981.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Pde1 Phosphodiesterase
EUKARYOTIC CELL, Dec. 2005, p. 1971 1981 Vol. 4, No. 12 1535-9778/05/$08.00 0 doi:10.1128/ec.4.12.1971 1981.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Pde1 Phosphodiesterase
Epidemiology of Fungal Diseases
 Lecture 2 Epidemiology of Fungal Diseases Disclaimer: This lecture slide presentation is intended solely for educational purposes. Many of the images contained herein are the property of the original owner,
Lecture 2 Epidemiology of Fungal Diseases Disclaimer: This lecture slide presentation is intended solely for educational purposes. Many of the images contained herein are the property of the original owner,
Adenylyl Cyclase Functions Downstream of the G Protein Gpa1 and Controls Mating and Pathogenicity of Cryptococcus neoformans
 EUKARYOTIC CELL, Feb. 2002, p. 75 84 Vol. 1, No. 1 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.1.75 84.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Adenylyl Cyclase Functions
EUKARYOTIC CELL, Feb. 2002, p. 75 84 Vol. 1, No. 1 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.1.75 84.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Adenylyl Cyclase Functions
Course Title Form Hours subject
 Course Title Form Hours subject Types, and structure of chromosomes L 1 Histology Karyotyping and staining of human chromosomes L 2 Histology Chromosomal anomalies L 2 Histology Sex chromosomes L 1 Histology
Course Title Form Hours subject Types, and structure of chromosomes L 1 Histology Karyotyping and staining of human chromosomes L 2 Histology Chromosomal anomalies L 2 Histology Sex chromosomes L 1 Histology
Host-parasite interactions: Evolutionary genetics of the House Finch- Mycoplasma epizootic
 Host-parasite interactions: Evolutionary genetics of the House Finch- Mycoplasma epizootic Scott V. Edwards Department of Organismic and Evolutionary Biology Harvard University Cambridge, MA USA http://www.oeb.harvard.edu/faculty/edwards
Host-parasite interactions: Evolutionary genetics of the House Finch- Mycoplasma epizootic Scott V. Edwards Department of Organismic and Evolutionary Biology Harvard University Cambridge, MA USA http://www.oeb.harvard.edu/faculty/edwards
Ch. 24 Speciation BIOL 221
 Ch. 24 Speciation BIOL 221 Speciation Speciation Origin of new, is at the focal point of evolutionary theory Microevolution consists of adaptations that evolve within a population confined to one gene
Ch. 24 Speciation BIOL 221 Speciation Speciation Origin of new, is at the focal point of evolutionary theory Microevolution consists of adaptations that evolve within a population confined to one gene
PRESENTER: DENNIS NYACHAE MOSE KENYATTA UNIVERSITY
 18/8/2016 SOURCES OF MICROBIAL CONTAMINANTS IN BIOSAFETY LABORATORIES IN KENYA PRESENTER: DENNIS NYACHAE MOSE KENYATTA UNIVERSITY 1 INTRODUCTION Contamination occurs through avoidable procedural errors
18/8/2016 SOURCES OF MICROBIAL CONTAMINANTS IN BIOSAFETY LABORATORIES IN KENYA PRESENTER: DENNIS NYACHAE MOSE KENYATTA UNIVERSITY 1 INTRODUCTION Contamination occurs through avoidable procedural errors
Chapter 19: The Genetics of Viruses and Bacteria
 Chapter 19: The Genetics of Viruses and Bacteria What is Microbiology? Microbiology is the science that studies microorganisms = living things that are too small to be seen with the naked eye Microorganisms
Chapter 19: The Genetics of Viruses and Bacteria What is Microbiology? Microbiology is the science that studies microorganisms = living things that are too small to be seen with the naked eye Microorganisms
The Human Fungal Pathogen Cryptococcus Can Complete Its Sexual Cycle during a Pathogenic Association with Plants
 Article The Human Fungal Pathogen Cryptococcus Can Complete Its Sexual Cycle during a Pathogenic Association with Plants Chaoyang Xue, 1 Yasuomi Tada, 2 Xinnian Dong, 2 and Joseph Heitman 1, * 1 Department
Article The Human Fungal Pathogen Cryptococcus Can Complete Its Sexual Cycle during a Pathogenic Association with Plants Chaoyang Xue, 1 Yasuomi Tada, 2 Xinnian Dong, 2 and Joseph Heitman 1, * 1 Department
DNA Mutations Mediate Microevolution between Host- Adapted Forms of the Pathogenic Fungus Cryptococcus neoformans
 DNA Mutations Mediate Microevolution between Host- Adapted Forms of the Pathogenic Fungus Cryptococcus neoformans Denise A. Magditch 1, Tong-Bao Liu 2, Chaoyang Xue 2, Alexander Idnurm 1 * 1 Division of
DNA Mutations Mediate Microevolution between Host- Adapted Forms of the Pathogenic Fungus Cryptococcus neoformans Denise A. Magditch 1, Tong-Bao Liu 2, Chaoyang Xue 2, Alexander Idnurm 1 * 1 Division of
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
Soo Chan Lee, Sujal Phadke, Sheng Sun, and Joseph Heitman*
 EC Accepts, published online ahead of print on 21 September 2012 Eukaryotic Cell doi:10.1128/ec.00242-12 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11
EC Accepts, published online ahead of print on 21 September 2012 Eukaryotic Cell doi:10.1128/ec.00242-12 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11
Received 3 August 1998/Returned for modification 10 September 1998/Accepted 15 October 1998
 JOURNAL OF CLINICAL MICROBIOLOGY, Feb. 1999, p. 315 320 Vol. 37, No. 2 0095-1137/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. New PCR Primer Pairs Specific for Cryptococcus
JOURNAL OF CLINICAL MICROBIOLOGY, Feb. 1999, p. 315 320 Vol. 37, No. 2 0095-1137/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. New PCR Primer Pairs Specific for Cryptococcus
Viral Genetics. BIT 220 Chapter 16
 Viral Genetics BIT 220 Chapter 16 Details of the Virus Classified According to a. DNA or RNA b. Enveloped or Non-Enveloped c. Single-stranded or double-stranded Viruses contain only a few genes Reverse
Viral Genetics BIT 220 Chapter 16 Details of the Virus Classified According to a. DNA or RNA b. Enveloped or Non-Enveloped c. Single-stranded or double-stranded Viruses contain only a few genes Reverse
Cryptococcus neoformans Mates on Pigeon Guano: Implications for the Realized Ecological Niche and Globalization
 EUKARYOTIC CELL, June 2007, p. 949 959 Vol. 6, No. 6 1535-9778/07/$08.00 0 doi:10.1128/ec.00097-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Cryptococcus neoformans Mates
EUKARYOTIC CELL, June 2007, p. 949 959 Vol. 6, No. 6 1535-9778/07/$08.00 0 doi:10.1128/ec.00097-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Cryptococcus neoformans Mates
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
Received 24 September 2007/Accepted 8 October 2007
 EUKARYOTIC CELL, Dec. 2007, p. 2278 2289 Vol. 6, No. 12 1535-9778/07/$08.00 0 doi:10.1128/ec.00349-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ssk2 Mitogen-Activated Protein
EUKARYOTIC CELL, Dec. 2007, p. 2278 2289 Vol. 6, No. 12 1535-9778/07/$08.00 0 doi:10.1128/ec.00349-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ssk2 Mitogen-Activated Protein
Kit Components Product # EP42720 (24 preps) MDx 2X PCR Master Mix 350 µl Cryptococcus neoformans Primer Mix 70 µl Cryptococcus neoformans Positive
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com Cryptococcus neoformans End-Point PCR Kit Product# EP42720 Product
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com Cryptococcus neoformans End-Point PCR Kit Product# EP42720 Product
Instructions for Use. APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests
 3URGXFW,QIRUPDWLRQ Sigma TACS Annexin V Apoptosis Detection Kits Instructions for Use APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests For Research Use Only. Not for use in diagnostic procedures.
3URGXFW,QIRUPDWLRQ Sigma TACS Annexin V Apoptosis Detection Kits Instructions for Use APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests For Research Use Only. Not for use in diagnostic procedures.
Knowing where to look: environmental sources of cryptococcal disease in the Pacific Northwest
 Knowing where to look: environmental sources of cryptococcal disease in the Pacific Northwest Karen H. Bartlett, PhD Associate Professor kbartlet@interchange.ubc.ca School of Environmental Health University
Knowing where to look: environmental sources of cryptococcal disease in the Pacific Northwest Karen H. Bartlett, PhD Associate Professor kbartlet@interchange.ubc.ca School of Environmental Health University
Phospholipid-Binding Protein Cts1 Controls Septation and Functions Coordinately with Calcineurin in Cryptococcus neoformans
 EUKARYOTIC CELL, Oct. 2003, p. 1025 1035 Vol. 2, No. 5 1535-9778/03/$08.00 0 DOI: 10.1128/EC.2.5.1025 1035.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Phospholipid-Binding
EUKARYOTIC CELL, Oct. 2003, p. 1025 1035 Vol. 2, No. 5 1535-9778/03/$08.00 0 DOI: 10.1128/EC.2.5.1025 1035.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Phospholipid-Binding
Cryptococcus gattii Fungal Meningitis. By Cassy Hedberg BI 234
 Cryptococcus gattii Fungal Meningitis By Cassy Hedberg BI 234 This disease s deadliest weapon is the fact you ve never heard of it -Kausik Datta, Post-doctoral Fellow at Johns Hopkins Medicine A deadly
Cryptococcus gattii Fungal Meningitis By Cassy Hedberg BI 234 This disease s deadliest weapon is the fact you ve never heard of it -Kausik Datta, Post-doctoral Fellow at Johns Hopkins Medicine A deadly
Cryptococcus neoformans Strains Preferentially Disseminate to the Central Nervous System during Coinfection
 INFECTION AND IMMUNITY, Aug. 2005, p. 4922 4933 Vol. 73, No. 8 0019-9567/05/$08.00 0 doi:10.1128/iai.73.8.4922 4933.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Cryptococcus
INFECTION AND IMMUNITY, Aug. 2005, p. 4922 4933 Vol. 73, No. 8 0019-9567/05/$08.00 0 doi:10.1128/iai.73.8.4922 4933.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Cryptococcus
Abstract. Introduction
 Cryptococcus gattii VGIII Isolates Causing Infections in HIV/AIDS Patients in Southern California: Identification of the Local Environmental Source as Arboreal Deborah J. Springer 1 *, R. Blake Billmyre
Cryptococcus gattii VGIII Isolates Causing Infections in HIV/AIDS Patients in Southern California: Identification of the Local Environmental Source as Arboreal Deborah J. Springer 1 *, R. Blake Billmyre
Chapter 14. In Vitro Measurement of Phagocytosis and Killing of Cryptococcus neoformans by Macrophages. André Moraes Nicola and Arturo Casadevall
 Chapter 14 In Vitro Measurement of Phagocytosis and Killing of Cryptococcus neoformans by Macrophages André Moraes Nicola and Arturo Casadevall Abstract Macrophages are pivotal cells in immunity against
Chapter 14 In Vitro Measurement of Phagocytosis and Killing of Cryptococcus neoformans by Macrophages André Moraes Nicola and Arturo Casadevall Abstract Macrophages are pivotal cells in immunity against
2046: Fungal Infection Pre-Infusion Data
 2046: Fungal Infection Pre-Infusion Data Fungal infections are significant opportunistic infections affecting transplant patients. Because these infections are quite serious, it is important to collect
2046: Fungal Infection Pre-Infusion Data Fungal infections are significant opportunistic infections affecting transplant patients. Because these infections are quite serious, it is important to collect
EOG Practice:,Evolution & Genetics [126663]
![EOG Practice:,Evolution & Genetics [126663] EOG Practice:,Evolution & Genetics [126663]](/thumbs/87/96059079.jpg) EOG Practice:,Evolution & Genetics [126663] Student Class Date 1. A particular peach tree produces peaches that are more resistant to disease than other peaches. What method would reproduce these EXACT
EOG Practice:,Evolution & Genetics [126663] Student Class Date 1. A particular peach tree produces peaches that are more resistant to disease than other peaches. What method would reproduce these EXACT
Luminescent platforms for monitoring changes in the solubility of amylin and huntingtin in living cells
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
CRYPTOCOCCUS neoformans is an important human tance (Matsushita 2000) and new infections (Piot et
 Copyright 2002 by the Genetics Society of America Isolation and Characterization of the Cryptococcus neoformans MATa Pheromone Gene Carol M. McClelland, Jianmin Fu, Gay L. Woodlee, Tara S. Seymour and
Copyright 2002 by the Genetics Society of America Isolation and Characterization of the Cryptococcus neoformans MATa Pheromone Gene Carol M. McClelland, Jianmin Fu, Gay L. Woodlee, Tara S. Seymour and
Transcription Factor Nrg1 Mediates Capsule Formation, Stress Response, and
 Transcription Factor Nrg1 Mediates Capsule Formation, Stress Response, and Pathogenesis in Cryptococcus neoformans Kari L. Cramer, Quincy D. Gerrald, Connie B. Nichols, Michael S. Price and J. Andrew Alspaugh
Transcription Factor Nrg1 Mediates Capsule Formation, Stress Response, and Pathogenesis in Cryptococcus neoformans Kari L. Cramer, Quincy D. Gerrald, Connie B. Nichols, Michael S. Price and J. Andrew Alspaugh
School of Medicine and Public Health, University of Wisconsin-Madison, Madison, WI 53706
 EC Accepts, published online ahead of print on 7 July 2014 Eukaryotic Cell doi:10.1128/ec.00152-14 Copyright 2014, American Society for Microbiology. All Rights Reserved. 1 1 2 3 4 5 6 7 8 9 10 11 12 13
EC Accepts, published online ahead of print on 7 July 2014 Eukaryotic Cell doi:10.1128/ec.00152-14 Copyright 2014, American Society for Microbiology. All Rights Reserved. 1 1 2 3 4 5 6 7 8 9 10 11 12 13
The F-Box Protein Fbp1 Regulates Sexual Reproduction and Virulence in Cryptococcus neoformans
 EUKARYOTIC CELL, June 2011, p. 791 802 Vol. 10, No. 6 1535-9778/11/$12.00 doi:10.1128/ec.00004-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. The F-Box Protein Fbp1 Regulates
EUKARYOTIC CELL, June 2011, p. 791 802 Vol. 10, No. 6 1535-9778/11/$12.00 doi:10.1128/ec.00004-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. The F-Box Protein Fbp1 Regulates
Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University
 Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
7-001b: Malt agar method for the detection of Alternaria dauci on Daucus carota (carrot)
 International Rules for Seed Testing Annexe to Chapter 7: Seed Health Testing Methods 7-001b: Malt agar method for the detection of Alternaria dauci on Daucus carota (carrot) Published by: International
International Rules for Seed Testing Annexe to Chapter 7: Seed Health Testing Methods 7-001b: Malt agar method for the detection of Alternaria dauci on Daucus carota (carrot) Published by: International
Relationship of the Glyoxylate Pathway to the Pathogenesis of Cryptococcus neoformans
 INFECTION AND IMMUNITY, Oct. 2002, p. 5684 5694 Vol. 70, No. 10 0019-9567/02/$04.00 0 DOI: 10.1128/IAI.70.10.5684 5694.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Relationship
INFECTION AND IMMUNITY, Oct. 2002, p. 5684 5694 Vol. 70, No. 10 0019-9567/02/$04.00 0 DOI: 10.1128/IAI.70.10.5684 5694.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Relationship
Calcium- and Calcineurin-Independent Roles for Calmodulin in Cryptococcus neoformans Morphogenesis and High-Temperature Growth
 EUKARYOTIC CELL, June 2005, p. 1079 1087 Vol. 4, No. 6 1535-9778/05/$08.00 0 doi:10.1128/ec.4.6.1079 1087.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Calcium- and Calcineurin-Independent
EUKARYOTIC CELL, June 2005, p. 1079 1087 Vol. 4, No. 6 1535-9778/05/$08.00 0 doi:10.1128/ec.4.6.1079 1087.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Calcium- and Calcineurin-Independent
Cryptococcus species identification by multiplex PCR
 Medical Mycology June 2008, 46, 377383 Cryptococcus species identification by multiplex PCR ANA LUSIA LEAL*, JOSIANE FAGANELLO*, MARIA CRISTINA BASSANESI $ & MARILENE H. VAINSTEIN* *Centro de Biotecnologia,
Medical Mycology June 2008, 46, 377383 Cryptococcus species identification by multiplex PCR ANA LUSIA LEAL*, JOSIANE FAGANELLO*, MARIA CRISTINA BASSANESI $ & MARILENE H. VAINSTEIN* *Centro de Biotecnologia,
Evolution of Sex. No area of evolutionary biology. mixture of strange phenomena than the evolution of sex. (Stearns, 1987)
 Evolution of Sex No area of evolutionary biology offers a more fascinating mixture of strange phenomena than the evolution of sex (Stearns, 1987) Sex Refers to union (SYNGAMY( SYNGAMY) ) of two genomes
Evolution of Sex No area of evolutionary biology offers a more fascinating mixture of strange phenomena than the evolution of sex (Stearns, 1987) Sex Refers to union (SYNGAMY( SYNGAMY) ) of two genomes
Non-Mendelian Inheritance of DNA-Induced Inositol Independence
 Proc. Nat. Acad. Sci. USA Vol. 70, No. 12, Part II, pp. 3875-3879, December 1973 Non-Mendelian Inheritance of DNA-Induced Inositol Independence in Neurospora (exosome/integration/meiosis/transformation)
Proc. Nat. Acad. Sci. USA Vol. 70, No. 12, Part II, pp. 3875-3879, December 1973 Non-Mendelian Inheritance of DNA-Induced Inositol Independence in Neurospora (exosome/integration/meiosis/transformation)
Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37 6C and in vivo
 Microbiology (2004), 150, 1547 1558 DOI 10.1099/mic.0.26928-0 Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37 6C and in vivo Joanne M. Kingsbury,
Microbiology (2004), 150, 1547 1558 DOI 10.1099/mic.0.26928-0 Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37 6C and in vivo Joanne M. Kingsbury,
Cryptococcal Titan Cell Formation Is Regulated by G-Protein Signaling in Response to Multiple Stimuli
 EUKARYOTIC CELL, Oct. 2011, p. 1306 1316 Vol. 10, No. 10 1535-9778/11/$12.00 doi:10.1128/ec.05179-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. Cryptococcal Titan Cell Formation
EUKARYOTIC CELL, Oct. 2011, p. 1306 1316 Vol. 10, No. 10 1535-9778/11/$12.00 doi:10.1128/ec.05179-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. Cryptococcal Titan Cell Formation
Growth and Mating of Cryptococcus neoformans var. grubii on Woody Debris
 Microb Ecol (2009) 57:757 765 DOI 10.1007/s00248-008-9452-1 FUNGAL MICROBIOLOGY Growth and Mating of Cryptococcus neoformans var. grubii on Woody Debris A. Botes & T. Boekhout & F. Hagen & H. Vismer &
Microb Ecol (2009) 57:757 765 DOI 10.1007/s00248-008-9452-1 FUNGAL MICROBIOLOGY Growth and Mating of Cryptococcus neoformans var. grubii on Woody Debris A. Botes & T. Boekhout & F. Hagen & H. Vismer &
Supplemental Material for. Figure S1. Identification of TetR responsive promoters in F. novicida and E. coli.
 Supplemental Material for Synthetic promoters functional in Francisella novicida and Escherichia coli Ralph L. McWhinnie and Francis E. Nano Department of Biochemistry and Microbiology, University of Victoria,
Supplemental Material for Synthetic promoters functional in Francisella novicida and Escherichia coli Ralph L. McWhinnie and Francis E. Nano Department of Biochemistry and Microbiology, University of Victoria,
Tropentag 2012, Göttingen, Germany September 19-21, 2012
 Tropentag 2012, Göttingen, Germany September 19-21, 2012 Conference on International Research on Food Security, Natural Resource Management and Rural Development organised by: Georg-August Universität
Tropentag 2012, Göttingen, Germany September 19-21, 2012 Conference on International Research on Food Security, Natural Resource Management and Rural Development organised by: Georg-August Universität
TEST NAME:review TEST ID: GRADE:07 Seventh Grade SUBJECT:Life and Physical Sciences TEST CATEGORY: My Classroom
 TEST NAME:review TEST ID:1070005 GRADE:07 Seventh Grade SUBJECT:Life and Physical Sciences TEST CATEGORY: My Classroom review Page 1 of 18 Student: Class: Date: 1. There are four blood types: A, B, AB,
TEST NAME:review TEST ID:1070005 GRADE:07 Seventh Grade SUBJECT:Life and Physical Sciences TEST CATEGORY: My Classroom review Page 1 of 18 Student: Class: Date: 1. There are four blood types: A, B, AB,
number Done by Corrected by Doctor د.حامد الزعبي
 number Fungi#1 Done by نرجس الس ماك Corrected by مهدي الشعراوي Doctor د.حامد الزعبي Introduction to Mycology -Terms: -Medical Mycology: The study of mycosis and their etiological agents -Mycosis: Disease
number Fungi#1 Done by نرجس الس ماك Corrected by مهدي الشعراوي Doctor د.حامد الزعبي Introduction to Mycology -Terms: -Medical Mycology: The study of mycosis and their etiological agents -Mycosis: Disease
(44) is microconidiating, fluffy, inositolless, isoleucineless, and valineless.
 THE EFFECT OF PHOTOREACTIVATION ON MUTATION FREQUENCY IN NEUROSPORA' JEANETTE SNYDER BROWN Stanford University, Stanford, California Received for publication April 1, 1951 Kelner (1949a) first reported
THE EFFECT OF PHOTOREACTIVATION ON MUTATION FREQUENCY IN NEUROSPORA' JEANETTE SNYDER BROWN Stanford University, Stanford, California Received for publication April 1, 1951 Kelner (1949a) first reported
Development and validation of benomyl birdseed agar for the isolation of Cryptococcus neoformans and Cryptococcus gattii from environmental samples
 Medical Mycology, 2014, 52, 417 421 doi: 10.1093/mmy/myt028 Advance Access Publication Date: 28 April 2014 Short Communication Short Communication Development and validation of benomyl birdseed agar for
Medical Mycology, 2014, 52, 417 421 doi: 10.1093/mmy/myt028 Advance Access Publication Date: 28 April 2014 Short Communication Short Communication Development and validation of benomyl birdseed agar for
Fungi. Eucaryotic Rigid cell wall(chitin, glucan) Cell membrane ergosterol Unicellular, multicellular Classic fungus taxonomy:
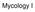 MYCOLOGY Mycology I Fungi Eucaryotic Rigid cell wall(chitin, glucan) Cell membrane ergosterol Unicellular, multicellular Classic fungus taxonomy: Morphology Spore formation FFungi Yeast Mold Yeastlike
MYCOLOGY Mycology I Fungi Eucaryotic Rigid cell wall(chitin, glucan) Cell membrane ergosterol Unicellular, multicellular Classic fungus taxonomy: Morphology Spore formation FFungi Yeast Mold Yeastlike
7-002a: Blotter method for the detection of Alternaria radicina on Daucus carota (carrot)
 International Rules for Seed Testing Annexe to Chapter 7: Seed Health Testing Methods 7-002a: Blotter method for the detection of Alternaria radicina on Daucus carota (carrot) Published by: International
International Rules for Seed Testing Annexe to Chapter 7: Seed Health Testing Methods 7-002a: Blotter method for the detection of Alternaria radicina on Daucus carota (carrot) Published by: International
Molecular Epidemiology of Clinical Cryptococcus neoformans Isolates in Seoul, Korea
 Mycobiology Research Article Molecular Epidemiology of Clinical Cryptococcus neoformans Isolates in Seoul, Korea So Hae Park 1, Mina Kim 2, Sei Ick Joo 3 and Soo Myung Hwang 1, * Department of Clinical
Mycobiology Research Article Molecular Epidemiology of Clinical Cryptococcus neoformans Isolates in Seoul, Korea So Hae Park 1, Mina Kim 2, Sei Ick Joo 3 and Soo Myung Hwang 1, * Department of Clinical
Cryptococcus neoformans var. grubii May Have Evolved
 Evidence that the Human Pathogenic Fungus Cryptococcus neoformans var. grubii May Have Evolved in Africa Anastasia P. Litvintseva 1 *, Ignazio Carbone 2, Jenny Rossouw 3, Rameshwari Thakur 4, Nelesh P.
Evidence that the Human Pathogenic Fungus Cryptococcus neoformans var. grubii May Have Evolved in Africa Anastasia P. Litvintseva 1 *, Ignazio Carbone 2, Jenny Rossouw 3, Rameshwari Thakur 4, Nelesh P.
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
7-001a: Blotter method for the detection of Alternaria dauci on Daucus carota (carrot)
 International Rules for Seed Testing Annexe to Chapter 7: Seed Health Testing Methods 7-001a: Blotter method for the detection of Alternaria dauci on Daucus carota (carrot) Published by: International
International Rules for Seed Testing Annexe to Chapter 7: Seed Health Testing Methods 7-001a: Blotter method for the detection of Alternaria dauci on Daucus carota (carrot) Published by: International
Cryptococcus neoformans is an opportunistic fungal pathogen of
 A Ric8/Synembryn Homolog Promotes Gpa1 and Gpa2 Activation To Respectively Regulate Cyclic AMP and Pheromone Signaling in Cryptococcus neoformans Jinjun Gong, a Jacob D. Grodsky, b Zhengguang Zhang, c
A Ric8/Synembryn Homolog Promotes Gpa1 and Gpa2 Activation To Respectively Regulate Cyclic AMP and Pheromone Signaling in Cryptococcus neoformans Jinjun Gong, a Jacob D. Grodsky, b Zhengguang Zhang, c
GENETIC EQUILIBRIUM. Chapter 16
 GENETIC EQUILIBRIUM Chapter 16 16-1 Population Genetics Population= number of organisms of the same species in a particular place at a point in time Gene pool= total genetic information of a population
GENETIC EQUILIBRIUM Chapter 16 16-1 Population Genetics Population= number of organisms of the same species in a particular place at a point in time Gene pool= total genetic information of a population
Ch. 23 The Evolution of Populations
 Ch. 23 The Evolution of Populations 1 Essential question: Do populations evolve? 2 Mutation and Sexual reproduction produce genetic variation that makes evolution possible What is the smallest unit of
Ch. 23 The Evolution of Populations 1 Essential question: Do populations evolve? 2 Mutation and Sexual reproduction produce genetic variation that makes evolution possible What is the smallest unit of
Received 10 May 2005/Accepted 1 August 2005
 EUKARYOTIC CELL, Oct. 2005, p. 1629 1638 Vol. 4, No. 10 1535-9778/05/$08.00 0 doi:10.1128/ec.4.10.1629 1638.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Comparative Gene
EUKARYOTIC CELL, Oct. 2005, p. 1629 1638 Vol. 4, No. 10 1535-9778/05/$08.00 0 doi:10.1128/ec.4.10.1629 1638.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Comparative Gene
Promiscuous mitochondria in Cryptococcus gattii
 RESEARCH ARTICLE Promiscuous mitochondria in Cryptococcus gattii Marjan Bovers 1, Ferry Hagen 1,2, Eiko E. Kuramae 1 & Teun Boekhout 1,2 1 CBS Fungal Biodiversity Centre, Uppsalalaan, Utrecht, The Netherlands;
RESEARCH ARTICLE Promiscuous mitochondria in Cryptococcus gattii Marjan Bovers 1, Ferry Hagen 1,2, Eiko E. Kuramae 1 & Teun Boekhout 1,2 1 CBS Fungal Biodiversity Centre, Uppsalalaan, Utrecht, The Netherlands;
SEX. Genetic Variation: The genetic substrate for natural selection. Sex: Sources of Genotypic Variation. Genetic Variation
 Genetic Variation: The genetic substrate for natural selection Sex: Sources of Genotypic Variation Dr. Carol E. Lee, University of Wisconsin Genetic Variation If there is no genetic variation, neither
Genetic Variation: The genetic substrate for natural selection Sex: Sources of Genotypic Variation Dr. Carol E. Lee, University of Wisconsin Genetic Variation If there is no genetic variation, neither
The Determination of the Genetic Order and Genetic Map for the Eye Color, Wing Size, and Bristle Morphology in Drosophila melanogaster
 Kudlac 1 Kaitie Kudlac March 24, 2015 Professor Ma Genetics 356 The Determination of the Genetic Order and Genetic Map for the Eye Color, Wing Size, and Bristle Morphology in Drosophila melanogaster Abstract:
Kudlac 1 Kaitie Kudlac March 24, 2015 Professor Ma Genetics 356 The Determination of the Genetic Order and Genetic Map for the Eye Color, Wing Size, and Bristle Morphology in Drosophila melanogaster Abstract:
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
