Pde1 Phosphodiesterase Modulates Cyclic AMP Levels through a Protein Kinase A-Mediated Negative Feedback Loop in Cryptococcus neoformans
|
|
|
- Morgan Gibbs
- 5 years ago
- Views:
Transcription
1 EUKARYOTIC CELL, Dec. 2005, p Vol. 4, No /05/$ doi: /ec Copyright 2005, American Society for Microbiology. All Rights Reserved. Pde1 Phosphodiesterase Modulates Cyclic AMP Levels through a Protein Kinase A-Mediated Negative Feedback Loop in Cryptococcus neoformans Julie K. Hicks, 1 Yong-Sun Bahn, 1 and Joseph Heitman 1,2,3 * Departments of Molecular Genetics and Microbiology, 1 Medicine, 2 and Pharmacology and Cancer Biology, 3 Duke University Medical Center, Durham, North Carolina Received 25 July 2005/Accepted 20 September 2005 The virulence of the human pathogenic fungus Cryptococcus neoformans is regulated by a cyclic AMP (camp)-dependent protein kinase A (PKA) signaling cascade that promotes mating and the production of melanin and capsule. In this study, genes encoding homologs of the Saccharomyces cerevisiae low- and highaffinity phosphodiesterases, PDE1 and PDE2, respectively, were deleted in serotype A strains of C. neoformans. The resulting mutants exhibited moderately elevated levels of melanin and capsule production relative to the wild type. Epistasis experiments indicate that Pde1 functions downstream of the G subunit Gpa1, which initiates camp-dependent signaling in response to an extracellular signal. Previous work has shown that the PKA catalytic subunit Pka1 governs camp levels via a negative feedback loop. Here we show that a pde1 pka1 mutant strain exhibits camp levels that are dramatically increased ( 15-fold) relative to those in a pka1 single mutant strain and that a site-directed mutation in a consensus PKA phosphorylation site reduces Pde1 function. These data provide evidence that fluctuations in camp levels are modulated by both Pka1- dependent regulation of Pde1 and another target that comprise a robust negative feedback loop to tightly constrain intracellular camp levels. Signal transduction cascades are the primary means by which external cues are communicated to the nuclei of eukaryotic organisms. The resulting changes in gene transcription are manifested as an appropriate response to the given cue. The importance of signal transduction cascades is perhaps best reflected in the high degree of cascade component conservation that exists between animals and fungi, despite the fact that they diverged from a common ancestor over 800 million years ago (29). In fungi, mitogen-activated protein kinase and cyclic AMP (camp)-dependent signal transduction cascades are two major signal transduction conduits. The highly conserved components of the camp signaling cascade have been co-opted by many different fungi for a variety of purposes, including regulation of asexual and sexual spore production, mycotoxin production, pseudohyphal growth, mating, pathogenesis, and virulence (1, 16, 17, 20, 32, 36, 42, 48). The camp-dependent signaling pathway has been well characterized for the human-pathogenic basidiomycete fungus Cryptococcus neoformans. Both serotype A (C. neoformans var. grubii) and serotype D (C. neoformans var. neoformans) strains infect immunocompromised individuals and cause meningoencephalitis (6). The virulence of this fungus depends on several factors, including the ability to grow at 37 C and the production of melanin and a polysaccharide capsule (6). Additionally, the ability to mate and grow filamentously also likely has a role in survival and propagation in the environment and may generate infectious propagules (23). * Corresponding author. Mailing address: Department of Molecular Genetics and Microbiology, 322 CARL Bldg., Duke University Medical Center, Research Dr., Durham, NC Phone: (919) Fax: (919) heitm001@duke.edu. C. neoformans virulence factor production and mating ability are regulated by the camp-dependent signaling pathway (for reviews, see references 12 and 29). Components of this pathway that have been identified include a G subunit of a heterotrimeric G protein (Gpa1), adenylyl cyclase (Cac1), the PKA catalytic (Pka1 and Pka2) and regulatory (Pkr1) subunits, and an adenylyl cyclase-associated protein (Aca1) that works independently of Gpa1 to activate adenylyl cyclase (1 3, 13, 19) (Fig. 1). Deletion of the GPA1, CAC,orACA1 gene results in a loss of mating and capsule and melanin production and attenuates virulence. These defects can be rescued by the addition of exogenous camp (1 3). Deletion of the Pkr1 regulatory subunit results in increased capsule production and hypervirulence (13). The roles of Pka1 and Pka2 in serotype A and serotype D have diverged. In serotype A, Pka1 plays the primary role in the regulation of mating, capsule and melanin production, and virulence, while Pka2 plays only an ancillary role (3, 13). In serotype D, Pka2 contributes to the regulation of virulence trait production but is not required for virulence (13, 19). In Saccharomyces cerevisiae, intracellular camp levels are influenced by the activity of the low- and high-affinity phosphodiesterases (PDEases), Pde1 and Pde2, respectively. These phosphodiesterases are structurally distinct, sharing virtually no sequence similarity. Pde2 is a high-affinity PDEase that is represented in organisms ranging from yeast to mammals (7). In contrast, Pde1, the low-affinity PDEase, is less well characterized but is found in a range of organisms, including S. cerevisiae, Schizosaccharomyces pombe, Candida albicans, Dictyostelium discoideum, Vibrio fischeri, Leishmania mexicana, Trypanosoma brucei, and Trypanosoma cruzi (9, 11, 14, 22, 27, 28, 34, 39). Pde1 and Pde2 have distinct functions. Pde2 regulates basal 1971
2 1972 HICKS ET AL. EUKARYOT. CELL FIG. 1. Model for the role of Pde1 and Pde2 in the camp signaling cascade. Virulence factor production is regulated by the camp-dependent signaling pathway via the following components: an unidentified seven-transmembrane receptor(s); the G subunit (Gpa1); adenylyl cyclase (Cac1) and its associated protein, Aca1; PKA, composed of the Pkr1 regulatory subunit and the Pka1 and Pka2 catalytic subunits; and the PDEase Pde1. In serotype A, the Pka1 catalytic subunit plays a major role in regulating virulence factor production, while the Pka2 subunit plays only a minor role. Intracellular camp levels are modulated through a feedback loop controlled by Pka1. When active, Pka1 positively regulates Pde1 and may also negatively regulate Cac1, exerting control over both the production (Cac1) and degradation (Pde1) of camp. Although Pde2 has no apparent role in the regulation of intracellular camp levels, it is possible that Pde2 may be regulated by Pka2. Solid lines and arrows indicate experimentally proven portions of the pathway, and dashed lines and arrows indicate putative portions of the pathway for which there is, as yet, no experimental evidence. Gray arrows and lines indicate minor roles in the pathway. camp levels in both S. cerevisiae and C. albicans (5, 30). Because the regulation of basal camp levels is important in determining tolerance to stress, including temperature extremes and high salt and heavy metal concentrations, Pde2 has a pivotal role in stress tolerance (30, 37). In C. albicans, Pde2 is also involved in filamentation, nutrient sensing, entry into stationary phase, and cell wall and membrane integrity (5, 25). In contrast, S. cerevisiae Pde1 does not significantly affect the basal levels of camp and does not confer any obvious mutant phenotype. However, in both S. cerevisiae and S. pombe, Pde1 does regulate camp levels induced by the presence of glucose (21, 30). Furthermore, the camp degradation activity of Pde1 in S. cerevisiae is positively regulated by the PKA catalytic subunits (35). The regulation of Pde1 activity is also observed in S. pombe, but it is not clear if this regulation is effected directly via PKA catalytic or allosteric activation by camp, as occurs in D. discoideum (21, 31). In humans, it has been suggested that some PDEases may be regulated independently of PKA (8). Here we report the characterization of Pde1 and Pde2 from C. neoformans. We show that, unlike in S. cerevisiae and C. albicans, Pde2 has no apparent role in regulating intracellular camp levels, and deletion of the gene results in subtle mutant phenotypes. However, Pde1 clearly is part of the camp-dependent signaling pathway that regulates virulence attributes. Not only does Pde1 disruption rescue gpa1 mutant phenotypes, but we also provide evidence that Pde1 is activated by Pka1 and that it is an important part of the PKA-activated negative feedback loop that governs intracellular camp levels in this pathogenic fungus. MATERIALS AND METHODS C. neoformans strains and media. All strains used for this study are listed in Table 1. C. neoformans strains were grown on standard S. cerevisiae medium (41). Selective medium for biolistic transformation, Niger seed medium, V8 medium, and Dulbecco s modified Eagle s medium (DMEM) were prepared as described previously (1, 18, 44). For camp suppression experiments, camp was added at a concentration of 10 mm to the media. pka1 ::URA5 transformants were selected on synthetic medium lacking uracil and containing 1 M sorbitol. pde1 ::NAT and pde2 ::NAT transformants were selected on yeast extract-peptone-dextrose (YPD) medium containing 100 g/ml nourseothricin. pde1 ::NEO and pde2 ::NEO were selected on YPD medium containing 200 g/ml G418. Genotypes were confirmed by both Southern hybridization and expression analysis. Identification of C. neoformans PDE1 and PDE2 genes. The C. neoformans PDE1 and PDE2 genes were identified in serotype A and D strains by performing a tblastn search of the Duke Bioinformatics C. neoformans serotype A database ( and the Stanford Genome Technology Center serotype D database ( using the S. cerevisiae Pde1 and Pde2 protein sequences. Disruption of PKA1, PDE1, and PDE2 genes. The pka1, pde1, and pde2 null mutants were generated by overlap PCR as described previously (10). The primer sequences used for this study are available upon request. To construct the pka1 ::URA5 allele, in the first round of PCR the 5 end of the PKA1 gene was amplified with primers 9352 and 9354, the 3 end of the gene was amplified with primers 9355 and 9357, and the URA5 gene was amplified with primers 9352 and Primers 9352 and 9357 were then used in an overlap amplification with the first three products as templates to yield the 4.0-kb PCR product bearing the pka1 ::URA5 allele. The gel-extracted PKA1 disruption cassette was precipitated onto 0.6- g gold microcarrier beads (0.8- m; Bioworld Inc.) and transformed into the MAT ura5 serotype A strain F99 by biolistic transformation (44). The pka1 ::URA5 allele deletes the entire PKA1 open reading frame (ORF). The PDE1 and PDE2 gene ORFs were disrupted by biolistic transformation of the congenic C. neoformans serotype A strains H99 and KN99a with constructs generated by overlap PCR (10). The 5 and 3 regions of the PDE1 and PDE2 genes in serotype A strains were PCR amplified with the following primers: 10403/10404 and 10347/10348 for the 5 regions of the PDE1 and PDE2 genes, respectively, and 10405/10406 and 10349/10350 for the 3 regions of PDE1 and PDE2, respectively. M13F and M13R primers were used to generate Nat r dominant selectable markers from the template pnatstm#58 (for PDE1) or pnatstm#102 (for PDE2), with a unique signature tag (33), and Neo r dominant selectable markers from the template pjaf1 (15). The PDE1 disruption alleles were generated by overlap PCR using primers and The PDE2 disruption alleles were generated by overlap PCR using primers and Prototrophic wild-type strains were biolistically transformed with the gel-extracted PDE1 and PDE2 disruption alleles as described previously (10). pde1 and pde2 strains were screened by diagnostic PCR for the 5 and 3 junctions and by Southern blot analysis using specific probes generated by PCR with the primers 10423/10424 and 11206/11207 for the PDE1 and PDE2 genes, respectively (data not shown).
3 VOL. 4, 2005 Pde1 MODULATES camp LEVELS IN C. NEOFORMANS 1973 TABLE 1. Strains used for this study Strain Relevant genotype Parent strains Reference H99 MAT 38 F99 MAT ura5 (FOA r ) 47 CHM3 MAT lac1 ::NAT 19 JKH7 MAT pka1 ::URA5 ura5 This study JKH33 MAT pde2 ::NAT-STM#102 This study JKH38 MATa pde2 ::NAT-STM#102 JKH33 PPW196 This study JKH63 MAT pde1 ::NAT-STM#58 This study JKH70 MAT pde1 ::NAT-STM#58 JKH38 JKH63 This study pde2 ::NAT-STM#102 JKH73 MATa pde1 ::NAT-STM#58 JKH38 JKH63 This study JKH85 MATa hog1 ::NEO pka1 ::URA5 YSB81 YSB112 This study JKH94 MAT pde2 ::NAT-STM#102 JKH33 JKH85 This study pka1 ::URA5 JKH95 MATa pde1 ::NAT-STM#58 JKH70 JKH85 This study pka1 ::URA5 JKH101 MATa pde2 ::NAT-STM#102 JKH33 YSB85 This study gpa1 ::NEO JKH109 MAT pde1 ::NAT STM#58 JKH70 JKH85 This study pde2 ::NAT-STM#102 pka1 ::URA5 JKH111 MAT pde1 ::NAT-STM#58 JKH70 YSB85 This study gpa1 ::NEO JKH116 MATa pde1 ::NAT-STM#58 JKH70 YSB85 This study pde2 ::NAT-STM#102 gpa1 ::NEO JKH124 MATa pde1 ::NAT-STM#58 JKH70 JKH85 This study pde2 ::NAT-STM#102 pka1 ::URA5 hog1 ::NEO JKH173 MATa pde1 ::NAT-STM#58 JKH73 YSB83 This study gpa1 ::NAT-STM#5 JKH190 MATa pde1 ::NAT-STM#58 This study gpa1 ::NAT-STM#5 PDE1 JKH238 MATa pde1 ::NAT-STM#58 This study gpa1 ::NAT pde1 S226A JKH271 MATa pde1 ::NAT-STM#58 This study pka1 ::URA5 pde1 S226A PPW196 MATa ura5 crg1::ura5 3 YSB81 MAT ura5 hog1 ::NAT-STM#177 pka1 ::URA5 4 YSB83 MAT gpa1 ::NAT-STM#5 3 YSB85 MATa gpa1 ::NEO 3 YSB112 MATa hog1 ::NEO 4 YSB194 MAT pka2 ::NAT-STM#205 3 YSB198 MATa pka2 ::NEO 3 Complementation of the pde1 mutant. The serotype A PDE1 gene for complementation of the pde1 mutant was amplified via PCR using primers and 13459, with wild-type (H99) genomic DNA as the template. This 3.8-kb product included the promoter, ORF, and terminator regions of the PDE1 gene and was initially cloned into the pcr2.1-topo vector (Invitrogen), creating plasmid pjh106. pjh106 was digested with ApaI and NotI, and the fragment was ligated into pjaf12 (3), creating plasmid pjh108. Strain JKH173 was transformed with plasmid pjh108, using the biolistic apparatus (44). Site-directed mutagenesis. To mutate the putative PKA phosphorylation site in Pde1, we constructed plasmid pjh168, containing the pde1 S226A mutation, via overlap PCR (10). An 1.3-kb SalI fragment comprising the PDE1 sequence containing a mutation in the sole putative PKA phosphorylation site was created by first amplifying the regions upstream of the putative phosphorylation site (primers and 14509) and downstream of the site (primers and 14508). The pde1 S226A allele was generated by overlap PCR with primers and This fragment was cloned into the pcr2.1-topo vector (Invitrogen) to create plasmid pjh155 and sequenced. pjh108 was digested with SalI, and the wild-type 1.3-kb fragment was replaced with the pde1 S226A -containing 1.3-kb SalI fragment from pjh155 to create plasmid pjh168. Strains JKH238 and JKH271, containing the pde1 S226A mutation, were generated by biolistically transforming plasmid pjh168 into strains JKH173 and JKH95, respectively. The presence of the mutated allele in strains JKH238 and JKH271 was further confirmed by PCR amplification and sequencing of the relevant region. Expression analysis. Fungal strains were inoculated into 5 ml YPD medium and grown overnight at 30 C. Fifty milliliters of YPD medium in a 125-ml flask was inoculated with 500 l of overnight culture and grown at 250 rpm and 30 C for 5 h prior to harvesting. RNAs were isolated from the harvested cultures with Trizol (Gibco-BRL) according to the manufacturer s instructions. Fifteen micrograms (as indicated by spectrophotometric measurement) of RNA was separated in a denaturing gel and transferred to a nylon membrane. The resulting blots were probed with PDE1 and PDE2 gene-specific probes. Capsule assays. To assess capsule and cell sizes, each strain was inoculated onto a plate of DMEM and grown at either 30 C (see Fig. 3) or 37 C (see Fig. 4 and 6) for 2 days. Aliquots of all cultures were stained with India ink and viewed with a 100 oil objective. Twenty cells from each culture were chosen at random, and the cell and capsule sizes were measured using Axiovision 3.0 software (Carl Zeiss Microscopy). The relative capsule diameter was calculated with the following equation: (D w D c ) 100/D w, where D w and D c indicate the diameters of the whole-cell body (D w ; cell plus capsule) and the cell body only (D c ) (49). The Bonferroni multiple comparison test was performed with Prism 4.0 software (GraphPad Software) to compare each strain to every other strain (3, 49). Each experiment was done in triplicate (total n 60).
4 1974 HICKS ET AL. EUKARYOT. CELL FIG. 2. C. neoformans Pde1 and Pde2 exhibit signature sequences of low- and high-affinity camp PDEases. MacVector software (Accelrys) was used to perform Clustal W comparisons of Pde1 (A) and Pde2 (B). Both proteins contain the conserved signature sequences found in low- and high-affinity homologs, respectively, from S. cerevisiae, S. pombe, and C. albicans. The Pde1 signature sequence is H-x-H-L-D-H-(LIVM)-x-(GS)- (LIVMA)-(LIVM) 2 -x-s-(ap), and the Pde2 signature sequence is H-D-(LIVMFY)-x-H-x-(AG)-x 2 -(NQ)-x-(LIVMFY). Only the signature sequences are shown, and the numbers indicate the positions of the sequences in the respective proteins. Accession numbers for the proteins are as follows: CnPde1A, AY864841; CnPde1, AY864842; ScPde1, CAA64139; SpPde1, CAA20848; CaPde1, XP720545; CnPde2A, AY874131; CnPde2, AY874132; ScPde2, CAA99689; CaPde2, AAM Laccase assays. To quantitate melanin production, the wild-type (H99) and pde1 (JKH63), pde2 (JKH33), pde1 pde2 (JKH70), and lac1 (CHM3) mutant strains were grown in 25 ml of L-3,4-dihydroxyphenylalanine (L-DOPA) medium (0.1% L-asparagine, 0.1% glucose, 0.3% KH 2 PO 4, 0.01% L-DOPA, 0.025% MgSO 4 7H 2 O; ph 5.6) at 30 C and 250 rpm for 16 h. The cultures were then incubated at 25 C and 250 rpm for 24 h, at which time 1 ml of culture was centrifuged, and the supernatant was spectrophotometrically read for the optical density at a wavelength of 475 nm (OD 475 ). camp assays. Examinations of intracellular camp levels were performed by growing the wild-type (H99) and pka1 (JKH7), gpa1 (YSB83), pde1 (JKH63), pde2 (JKH33), pde1 pde2 (JKH70), pde1 pka1 (JKH95), pde2 pka1 (JKH94), pde1 pde2 pka1 (JKH109), pde1 gpa1 (JKH111), pde2 gpa1 (JKH101), and pde1 pde2 gpa1 (JKH116) mutant strains in 50 ml of YPD medium for 24 h. The cultures were centrifuged and washed twice with double-distilled water (ddh 2 O) and once with MES buffer (10 mm morpholineethanesulfonic acid [MES; ph 6.0], 0.5 mm EDTA) before being resuspended in MES buffer. Fifteen milliliters of MES buffer was then inoculated in order to give the cultures a final OD 600 of 2.0. Following2hofincubation at 30 C and 250 rpm, 1-ml samples of each culture were collected via filtration on m filters 0 min, 0.5 min, 1.0 min, and 3.0 min after the addition of 2% glucose. The cell-containing filters were incubated in a petri plate containing 1 ml of formic acid:butanol:ddh 2 O (3.7:76.3:20) for 1 h prior to transfer of the liquid to a microcentrifuge tube, centrifugation at 13,000 rpm for 10 min, and transfer of the supernatant to a new tube. The resulting liquid was lyophilized, at which time it was resuspended in the assay buffer supplied with the camp Biotrak enzyme immunoassay system (Amersham Biosciences). The assay was performed according to the manufacturer s instructions. Nucleotide sequence accession numbers. The PDE1 and PDE2 serotype A gene sequences have been deposited in the NCBI GenBank database and have the following accession numbers: AY (PDE1A) and AY (PDE2A). The PDE1 and PDE2 gene sequences from serotype D have also been deposited in the NCBI GenBank database and have the following accession numbers: AY (PDE1 ) and AY (PDE2 ). RESULTS C. neoformans contains homologs of low- and high-affinity PDEases, Pde1 and Pde2. S. cerevisiae expresses two PDEases, the low-affinity form, Pde1, and the high-affinity form, Pde2. Pde1 functions in response to agonist-induced increases in camp, while Pde2 regulates basal camp levels (30). Using the S. cerevisiae Pde1 and Pde2 protein sequences, we performed a tblastn search to identify C. neoformans serotype A and serotype D Pde1 and Pde2 homologs. One sequence for each protein was identified from each database and designated Pde1 and Pde2. Both Pde1 and Pde2 have the conserved signature sequences for the low- and high-affinity PDEases, respectively (Fig. 2). The serotype A and D Pde1 proteins share 94% identity, and the serotype A and D Pde2 proteins share 93% identity. In contrast, the Pde1 and Pde2 proteins share only 12 to 14% identity, similar to the Pde1 and Pde2 proteins of S. cerevisiae (13% identity). Deletion of PDE1 and PDE2 enhances melanin and capsule production. The entire ORFs of the PDE1 and PDE2 genes were deleted from a serotype A wild-type strain to examine their role in the production of virulence factors. Expression analysis confirmed that the entire ORF had been deleted in both cases (data not shown). The wild-type strain and the pde1, pde2, and pde1 pde2 mutant strains were inoculated on Niger seed agar (1) and incubated at 37 C for 2 days to examine melanin production. This qualitative assay did not reveal any obvious differences between the mutant strains and the wild type (Fig. 3A). Nevertheless, a quantitative analysis of laccase production in L-DOPA medium (19) showed that the laccase activities of the mutants were increased relative to that of the wild-type strain (Fig. 3B). When 10 mm exogenous camp was added, all strains exhibited laccase activities that were indistinguishable from each other (Fig. 3B). The lac1 control strain, in which the LAC1 laccase gene has been eliminated, exhibited essentially no laccase activity in the presence or absence of exogenous camp. To examine the capsule, the wild-type and mutant strains were inoculated onto DMEM (18) and incubated at 37 C for 2 days. By visual observation and quantitative analysis, capsule production by the mutant strains was modestly enhanced relative to that of the wild type (Fig. 3C and D). The mating ability of the pde1 and pde2 mutants was also examined, but no apparent mutant phenotype was observed (data not shown). The pde1 mutation restores virulence trait production in a gpa1 mutant. We next examined if, like Pde1 and Pde2 in S. cerevisiae, the C. neoformans Pde1 and/or Pde2 protein functions in the camp-dependent PKA signaling pathway (reviewed in reference 12). Gpa1 is the G subunit that functions early in this pathway, and deletion of GPA1 compromises mel-
5 VOL. 4, 2005 Pde1 MODULATES camp LEVELS IN C. NEOFORMANS 1975 anin and capsule production and reduces mating (1, 3). Additionally, gpa1 mutants have low basal camp levels relative to wild-type strains. We hypothesized that if Pde1 and/or Pde2 functions downstream of Gpa1, then in epistasis analysis gpa1 mutant strains also containing either the pde1 or pde2 mutation should have higher levels of camp than the gpa1 single mutant, and these elevated camp levels might suffice to suppress gpa1 mutant phenotypes. This hypothesis was based on the assumption that deletion of PDE1 and/or PDE2 would result in a loss of PDEase activity and thereby reduce the degradation of even the basal levels of camp present in the gpa1 strain. To perform epistasis analysis, pde1 gpa1, pde2 gpa1, and pde1 pde2 gpa1 mutant strains were generated via meiotic crosses in a serotype A background. These strains were then inoculated onto Niger seed agar and DMEM to examine melanin and capsule production, respectively. Additionally, each mutant strain was mated with a wild-type strain, and relative mating abilities were determined by visualizing filament production. Melanin production by the gpa1 mutant was fully rescued by the pde1 mutation, but not by the pde2 mutation (Fig. 4A), and mating ability was also partially restored by the pde1 but not the pde2 mutation (Fig. 4D). The capsule production of gpa1 mutant cells was clearly rescued by the pde1 mutant and may have been modestly rescued by the pde2 mutation (Fig. 4B and C). The wild-type PDE1 gene complemented the pde1 mutation when a pde1 gpa1 strain was transformed with the wild-type PDE1 gene. The resulting complemented strain (pde1 gpa1 PDE1) exhibited melanin, capsule, and mating defects similar to those of the gpa1 single mutant (Fig. 4A, C, and D). To determine if the observed phenotypes of the pde1 gpa1 and pde2 gpa1 double mutant strains and the pde1 pde2 gpa1 triple mutant strain are correlated with intracellular camp levels, camp assays were performed on samples collected after glucose induction. As previously reported (2, 3), wild-type camp levels increased rapidly following the addition of glucose to glucose-starved cells, and gpa1 mutants failed to mount a camp increase in response to glucose (Fig. 4E). A similar increase was also observed for the pde1 strain, and although the error in the measurements was large, there was a trend towards higher camp levels than those in the wild type. Consistent with the results of epistasis analysis, the pde1 mutation enhanced camp levels in the pde1 gpa1 mutant, and glucose clearly enhanced camp production in the pde1 pde2 gpa1 mutant strain (Fig. 4E). FIG. 3. Deletion of the PDE1 and PDE2 genes causes modest increases in melanin and capsule production. (A) Serotype A wild-type (H99) and pde1 (JKH63), pde2 (JKH33), and pde1 pde2 (JKH70) mutant strains were grown on Niger seed agar, with or without 10 mm camp, and incubated for 2 days at 37 C prior to being photographed. Melanin production was not obviously different in any of the mutant strains relative to the wild-type strain. (B) Melanin production in serotype A strains was quantitated by growing wild-type (H99) and pde1 (JKH63), pde2 (JKH33), pde1 pde2 (JKH70), and lac1 (CHM3) mutant strains at 250 rpm and 30 C for 16 h in L-DOPA medium, with or without 10 mm camp, prior to transferring cultures to 250 rpm and 25 C for 24 h. Spectrophotometric readings of the culture supernatants were taken to determine the OD 475. One unit of laccase is equal to an OD 475 of (50). Each bar represents an average of five independent replicates. Standard deviations are indicated by error bars. (C) Serotype A wild-type (H99) and pde1 (JKH63), pde2 (JKH33), and pde1 pde2 (JKH70) mutant strains were inoculated onto agar-based DMEM (18), with or without 10 mm camp, incubated at 30 C for 2 days, and examined by the exclusion of India ink (magnification, 1,000). (D) Quantitative measurements of relative capsule diameters (see Materials and Methods). A total of 60 cells (20 cells from three independent experiments) were measured for each strain, and error bars indicate standard deviations.
6 1976 HICKS ET AL. EUKARYOT. CELL
7 VOL. 4, 2005 Pde1 MODULATES camp LEVELS IN C. NEOFORMANS 1977 FIG. 5. pde1 mutation results in highly elevated intracellular camp levels in a pka1 background. Intracellular camp levels were measured in a serotype A wild-type (H99) strain and the pka1 (JKH7), pde1 pka1 (JKH95), pde2 pka1 (JKH94), pde1 pde2 pka1 (JKH109), pde1 (JKH63), pde2 (JKH33), and pde1 pde2 (JKH70) mutant strains. The pde1 pka1 and pde1 pde2 pka1 strains produced significantly higher levels of camp than any of the other strains, and the scale for the data shown in panel A was expanded to illustrate this (B). The pka1 and pde2 pka1 strains had levels higher than those of the other strains, and the scale for the data in panel B was expanded to show this (C). Intracellular camp levels are elevated in a pde1 background. In S. cerevisiae, the PKA catalytic subunits regulate camp levels through a negative feedback loop in which Pde1 is activated by PKA phosphorylation (30). Pka1, the C. neoformans PKA catalytic subunit, also negatively regulates camp levels, but the regulatory mechanism was not known (13). We tested whether this negative feedback loop involves a positive regulation of Pde1 by Pka1. Previous work has shown that pka1 mutants have elevated intracellular camp levels relative to a wild-type strain (13). One plausible model is that activated Pka1 normally activates Pde1, resulting in camp degradation, whereas the pka1 mutant is unable to activate Pde1. It is also possible that Pka1 has other targets in addition to Pde1 as part of the regulation it imposes on camp levels (35). Also, Pde1 may be subject to both Pka1-dependent and -independent regulation (8), allowing for a minimal degradation of camp even in the absence of a functional Pka1. In either scenario, pde1 pka1 mutants may have elevated camp levels relative to the pka1 mutant. To test this, intracellular camp levels were measured during glucose sensing (Fig. 5). The wild-type strain and the pde1, pde2, and pde1 pde2 mutant strains all showed similarly elevated camp levels in response to glucose (Fig. 5C). The pka1 mutant had camp levels that were significantly higher ( 50-fold) than those in the wild-type strain and the pde1, pde2, and pde1 pde2 mutant strains (Fig. 5B), as expected based on its established role in a negative feedback loop (13, 46). The pde1 pka1 mutant exhibited camp levels that were approximately 10 times higher than those found in the pka1 single mutant strain and 500-fold higher than those in the wild-type strain (Fig. 5A), indicating that Pde1 plays a significant role in constraining camp levels, even in the absence of Pka1. These data also suggest that Pka1 has additional targets besides Pde1, because the phenotype conferred by the pka1 mutation on camp levels is exerted even in the absence of Pde1 (compare the pde1 mutant and the pde1 pka1 mutant in Fig. 5). Also, these data provide further support that Pde2 plays only a minor role, if any, in regulating camp levels. Elimination of a PKA phosphorylation site causes a loss of Pde1 function. The S. cerevisiae Pde1 enzyme has one consensus PKA phosphorylation site [(K/R)-(K/R)-(X)-(S/T)] (26). Mutagenesis of the serine residue in this site (pde1 S252A )results in reduced Pde1 activity (30). To determine if Pka1 phos- FIG. 4. pde1 mutation restores mating and melanin and capsule production of gpa1 mutants. (A) Serotype A wild-type (H99), gpa1 (YSB83), pde1 gpa1 (JKH111), pde2 gpa1 (JKH101), pde1 pde2 gpa1 (JKH116), pde1 (JKH63), pde2 (JKH33), and pde1 pde2 (JKH70) mutant, and pde1 gpa1 PDE1 complemented strains were grown on Niger seed agar and incubated for 2 days at 37 C prior to being photographed. (B) The strains used for panel A were grown in YPD overnight prior to being plated onto agar-based DMEM, incubated for 2 days at 37 C, resuspended in ddh 2 O, stained with India ink, and photographed at a magnification of 1,000. (C) For each strain used for panels A and B, a total of 60 cells (20 cells from three independent experiments) were treated as described in panel B and then measured (see Materials and Methods). Error bars indicate standard deviations. (D) MAT and MATa strains were cocultured on V8 (ph 5.0) medium and incubated at room temperature for 14 days in the dark prior to being photographed (magnification, 100). These strains included a (H99 KN99a), gpa1 a (YSB83 KN99a), pde1 gpa1 a (JKH111 KN99a), pde2 gpa1 a (JKH101 H99), pde1 pde2 gpa1 a (JKH116 H99), and pde1 gpa1 PDE1 a (JKH190 H99). (E) Intracellular camp levels were measured in each of the strains used for panels A to C.
8 1978 HICKS ET AL. EUKARYOT. CELL phorylates and subsequently activates Pde1 in C. neoformans, we examined the Pde1 sequence for putative PKA phosphorylation sites, using the PROSCAN site/signature sequence recognition program, and found that the serotype A Pde1 protein contains one putative PKA phosphorylation site (RKKS). We predicted that if Pka1 activates Pde1 by phosphorylation of the serine residue in this site, then mutagenesis of the serine to an alanine would result in a Pde1 protein that was unable to be activated and would hence act as a pde1 allele. To test this hypothesis, we generated a PDE1 allele identical to the construct that was utilized to complement the pde1 mutation in a gpa1 background, except for an S226A mutation at the serine residue in the putative PKA phosphorylation site. Both the pde1 gpa1 strain and the pde1 pka1 strain were transformed with the pde1 S226A allele. Northern analysis indicated that the pde1 S226A transcript level was comparable to that of the wild-type PDE1 gene, although this does not exclude the possibility that the resulting phenotypes could be attributable to a loss of function or instability of the pde1 S226A protein. The pde1 gpa1 pde1 S226A transformant was examined for the ability to produce melanin and capsule. As predicted if the pde1 S226A allele acts as a pde1 null mutant, melanin and capsule production in the pde1 gpa1 pde1 S226A strain was indistinguishable from that in the pde1 gpa1 mutant (Fig. 6A and B). This is in contrast to the pde1 gpa1 PDE1-complemented strain, in which the melanin and capsule defects conferred by the gpa1 mutation were readily apparent. When the pde1 S226A allele was placed in a pde1 pka1 background, camp levels were elevated similarly to the levels found in a pde1 pka1 mutant (Fig. 6C), again suggesting that the pde1 S226A allele acts like a pde1 null mutant. DISCUSSION Cyclic AMP signaling plays multiple roles in regulating development in eukaryotic organisms as diverse as slime molds, fungi, and humans. Here we define the PDE1 and PDE2 genes in the basidiomycete fungus C. neoformans and their biological function in camp signaling. Although similar to proteins in the divergent fungal model S. cerevisiae in some aspects, both the Pde1 and Pde2 proteins also show functional distinctions from their S. cerevisiae counterparts. The PDE1 gene encodes a phosphodiesterase protein that has a significant role in regulating camp levels in the cell. For S. cerevisiae, it has been determined that Pde1 is responsible for regulating agonistinduced camp levels, based on camp assays showing that the initial, transient spike in camp levels that occurs after glucose addition was enhanced in a pde1 mutant (30). In the C. neoformans pde1 mutant, only a modest enhancement of camp levels was observed upon glucose induction. Pde1 may be regulated through other pathways in addition to the campdependent signaling pathway (Fig. 1) (8), a hypothesis supported by our observation that in a pde1 pka1 mutant, camp levels are much higher than in a pka1 single mutant, suggesting that Pka1-independent regulation of Pde1 may occur such that in a pka1 mutant there is still some activity of Pde1 allowing for camp degradation. Also, it is possible that the feedback inhibition of camp levels initiated by Pka1 (see below) is robust enough that camp levels do not escalate substantially when Pde1 is absent. Deletion of the PDE1 gene by itself conferred only modest increases in melanin and capsule production in otherwise wildtype cells. However, deletion of PDE1 in combination with mutation of the G subunit Gpa1 remediated the melanin, capsule, and mating defects caused by the gpa1 single mutation. camp assays showed significantly higher camp levels in a pde1 gpa1 double mutant than in a gpa1 single mutant, supporting a model in which the deletion of PDE1 in a gpa1 background results in camp levels that are sufficient to substantially activate the protein kinase catalytic subunit Pka1. The observation that the pde1 pde2 gpa1 mutant strain responded to glucose in the absence of a functional G subunit suggests that this response is independent of the G-proteincoupled receptor. As in S. cerevisiae, it is possible that C. neoformans is able to sense intracellular glucose levels, possibly as the metabolite glucose-6-phosphate (40), but that intracellular glucose levels stimulate camp to such a low extent that only in the absence of one or both phosphodiesterases is the increase in camp levels detectable. Previous studies have shown that in C. neoformans, activated Pka1 mediates a negative feedback loop that controls intracellular camp levels (13, 46). However, the mechanism for this regulation had not been elucidated. S. pombe also exhibits negative feedback regulation of camp levels, and it has been established that Pde1 is part of this regulatory network, although the relationship between Pde1 and the PKA catalytic subunit Pka1 is not clear (21). In contrast, for S. cerevisiae it has been definitively shown that the PKA catalytic subunits phosphorylate serine residue 252 in Pde1, causing the activation and subsequent degradation of camp. Site-directed mutagenesis of this residue to create a pde1 S252A allele resulted in decreased Pde1 activity in vitro (30). Our data suggest that a similar situation to that in S. cerevisiae exists in C. neoformans. A single putative PKA phosphorylation site was found in Pde1, at residues 223 to 226. A site-directed mutation (pde1 S226A )in the serine residue of this putative phosphorylation site reduced the protein activity of Pde1. Additionally, both the pde1 pka1 mutant and the pde1 pka1 pde1 S226A mutant had camp levels that were elevated relative to that in the pka1 single mutant. This suggests that Pde1 is activated independently of Pka1, in addition to Pka1-dependent regulation, and that Pka1 targets other proteins in addition to Pde1 as part of the negative feedback loop that modulates intracellular camp levels (Fig. 1). Additional candidate targets of Pka1 include proteins upstream of Pka1 in the camp-dependent signaling pathway, such as the Pkr1 protein kinase A regulatory subunit and the Cac1 adenylyl cyclase protein homologs from divergent C. neoformans serotypes, which both have putative conserved PKA phosphorylation sites. In S. cerevisiae, the Bcy1 PKA regulatory subunit is not thought to be a candidate for additional feedback regulation, while the Cyr1 adenylyl cyclase is (43). In S. pombe, camp levels are controlled by the regulation of both adenylyl cyclase and the Pde1 homolog, Cgs2 (21). Studies with S. pombe suggest that shortly after the glucose-stimulated activation of adenylyl cyclase, the Cgs2 phosphodiesterase is also activated, resulting in a modulation of camp levels (45). Sitedirected mutagenesis of the sites in the C. neoformans Cac1 and Pkr1 proteins should elucidate whether these candidate proteins are targets of Pka1 and if they have roles in the
9 VOL. 4, 2005 Pde1 MODULATES camp LEVELS IN C. NEOFORMANS 1979 Downloaded from FIG. 6. Elimination of a PKA phosphorylation site results in a loss of Pde1 activity and increased camp levels in a pka1 background. Site-directed mutagenesis was utilized to eliminate the single PKA phosphorylation site in Pde1 (pde1 S226A ). The mutant PDE1 alleles were introduced into the pde1 gpa1 background, and the resultant pde1 gpa1 pde1 S226A (JKH238) strain was compared with the wild-type strain (H99) and the gpa1 (YSB83), pde1 (JKH63), pde1 gpa1 (JKH173), and pde1 gpa1 PDE1 (JKH190) mutant strains. Melanin (A) and capsule (B) production were examined as described in Materials and Methods. Cultures for examinations of capsule production were incubated at 37 C for 2 days prior to being observed by India ink staining. Bar, 10 m. (C) camp levels were also examined as described in Materials and Methods. The pde1 pka1 and pde1 pka1 pde1 S226A strains produced significantly higher levels of camp than any of the other strains, and the pka1 strain also had levels higher than those of the wild type or the pde1 mutant; the scale for the data shown in the graph on the left was expanded to illustrate this. on September 9, 2018 by guest negative feedback loop that regulates camp levels in C. neoformans. The role of Pde2 in regulating camp (30) production is less obvious than that of Pde1. Pde2 contains the class I signature sequence found in numerous high-affinity PDEase proteins, and RNA analysis confirmed that a transcript was produced. However, deletion of the PDE2 gene has only modest effects on mating or melanin or capsule production, does not result in a significant change from the wild-type strain with regard to camp levels, and results in no change in heat shock or oxidative stress resistance. Pde2 regulates heat shock sensitivity in S. cerevisiae (30); that it does not in C. neoformans supports other evidence that the camp signaling pathway is not as involved in the response to environmental stresses as it is in other fungi (24). We noted that the pde2 mutation appears to weakly rescue capsule production in a gpa1 strain (Fig. 4A) and thus might function under some more limited physiological contexts, in contrast to Pde1. These data suggest that Pde2 either
10 1980 HICKS ET AL. EUKARYOT. CELL is not in the same signaling pathway as Gpa1, does not function similarly to Pde1, or is not regulated in the same manner as Pde1. It is probable that C. neoformans contains two related but distinct feedback regulation systems controlling the camp pathway, one involving camp degradation (via induction of the Pde1 phosphodiesterase) and another involving camp production (via the repression of adenylyl cyclase). This dual regulation suggests that precise regulation of the camp pathway may be critical to the maintenance of virulence attributes of the pathogen. The fact that several other fungi, including the nonpathogenic organisms S. cerevisiae and S. pombe, share this regulatory network supports the contention that the constraint of intracellular camp levels is critical for cellular function in many organisms. Therefore, future studies of C. neoformans will investigate the impact of this strict constraint on the camp pathway in fungal survival and virulence during the course of disease. ACKNOWLEDGMENTS We thank Xiaorong Lin, Jeffrey Batten, Chaoyang Xue, and Alex Idnurm for helpful suggestions in the preparation of the manuscript. This research was funded in part by an NIH interdisciplinary AIDS training grant (AI ) (J.K.H.) and by RO1 grant AI39115 and PO1 grant AI44975 (J.H.). REFERENCES 1. Alspaugh, J. A., J. R. Perfect, and J. Heitman Cryptococcus neoformans mating and virulence are regulated by the G-protein subunit GPA1 and camp. Genes Dev. 11: Alspaugh, J. A., R. Pukkila-Worley, T. Harashima, L. M. Cavallo, D. Funnell, G. M. Cox, J. R. Perfect, J. W. Kronstad, and J. Heitman Adenylyl cyclase functions downstream of the G protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1: Bahn, Y. S., J. K. Hicks, S. S. Giles, G. M. Cox, and J. Heitman Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot. Cell 3: Bahn, Y. S., K. Kojima, G. M. Cox, and J. Heitman Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16: Bahn, Y. S., J. Staab, and P. Sundstrom Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol. Microbiol. 50: Casadevall, A., and J. R. Perfect Cryptococcus neoformans. ASM Press, Washington, D.C. 7. Charbonneau, H., N. Beier, K. A. Walsh, and J. A. Beavo Identification of a conserved domain among cyclic nucleotide phosphodiesterases from diverse species. Proc. Natl. Acad. Sci. USA 83: Conti, M Phosphodiesterases and cyclic nucleotide signaling in endocrine cells. Mol. Endocrinol. 14: D Angelo, M. A., S. Sanguineti, J. M. Reece, L. Birnbaumer, H. N. Torres, and M. M. Flawia Identification, characterization and subcellular localization of TcPDE1, a novel camp-specific phosphodiesterase from Trypanosoma cruzi. Biochem. J. 378: Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. D. Berrios, C. M. Hull, C. D Souza, P. Wang, and J. Heitman A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148: De Voti, J., G. Seydoux, D. Beach, and M. McLeod Interaction between ran1 protein kinase and camp dependent protein kinase as negative regulators of fission yeast meiosis. EMBO J. 10: D Souza, C., and J. Heitman Conserved camp signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25: D Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21: Dunlap, P. V., and S. M. Callahan Characterization of a periplasmic 3 :5 -cyclic nucleotide phosphodiesterase gene, cdpp, from the marine symbiotic bacterium Vibrio fischeri. J. Bacteriol. 175: Fraser, J. A., R. L. Subaran, C. B. Nichols, and J. Heitman Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island. Eukaryot. Cell 2: Gao, S., and D. L. Nuss Distinct roles for two G protein subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc. Natl. Acad. Sci. USA 93: Gold, S., G. Duncan, K. Barrett, and J. Kronstad camp regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 8: Granger, D. L., J. R. Perfect, and D. T. Durack Virulence of Cryptococcus neoformans: regulation of capsule synthesis by carbon dioxide. J. Clin. Investig. 76: Hicks, J. K., C. A. D Souza, G. M. Cox, and J. Heitman Cyclic-AMP dependent protein kinase catalytic subunits have divergent roles in virulence factor production in two varieties of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 3: Hicks, J. K., J. Yu, N. P. Keller, and T. H. Adams Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G-protein-dependent signaling pathway. EMBO J. 16: Hoffman, C. S Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem. Soc. Trans. 33: Hoyer, L. L., L. B. Cieslinski, M. M. McLaughlin, T. J. Torphy, A. R. Shatzman, and G. P. Livi A Candida albicans cyclic nucleotide phosphodiesterase: cloning and expression in Saccharomyces cerevisiae and biochemical characterization of the recombinant enzyme. Microbiology 140: Hull, C. M., and J. Heitman Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36: Idnurm, A., Y.-S. Bahn, K. Nielsen, X. Lin, J. Fraser, and J. Heitman Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3: Jung, W. H., P. Warn, E. Ragni, L. Popolo, C. D. Nunn, M. P. Turner, and L. Stateva Deletion of PDE2, the gene encoding the high-affinity camp phosphodiesterase, results in changes of the cell wall and membrane in Candida albicans. Yeast 22: Kemp, B. E., and R. B. Pearson Protein kinase recognition sequence motifs. Trends Biochem. Sci. 15: Kunz, S., T. Kloeckner, L.-O. Essen, T. Seebeck, and M. Boshart TbPDE1, a novel class I phosphodiesterase of Trypanosoma brucei. Eur. J. Biochem. 271: Lacombe, M.-L., G. J. Podgorski, J. Franke, and R. H. Kessin Molecular cloning and developmental expression of the cyclic nucleotide phosphodiesterase gene of Dictyostelium discoideum. J. Biol. Chem. 261: Lengeler, K. B., R. C. Davidson, C. D Souza, T. Harashima, W.-C. Sheng, P. Wang, X. Pan, M. Waugh, and J. Heitman Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64: Ma, P., S. Wera, P. V. Dijck, and J. M. Thevelein The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced camp signaling. Mol. Biol. Cell 10: Meima, M. E., K. E. Weening, and P. Schaap Characterization of a camp-stimulated camp phosphodiesterase in Dictyostelium discoideum. J. Biol. Chem. 278: Mitchell, T. K., and R. A. Dean The camp-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea. Plant Cell 7: Nelson, R. T., J. Hua, B. Pryor, and J. K. Lodge Identification of virulence mutants of the fungal pathogen Cryptococcus neoformans using signature-tagged mutagenesis. Genetics 157: Nikawa, J., P. Sass, and M. Wigler Cloning and characterization of the low affinity camp phosphodiesterase gene of S. cerevisiae. Mol. Cell. Biol. 7: Nikawa, J.-I., S. Cameron, T. Toda, K. M. Ferguson, and M. Wigler Rigorous feedback control of camp levels in Saccharomyces cerevisiae. Genes Dev. 1: Pan, X., and J. Heitman Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: Park, J. I., C. M. Grant, and I. W. Dawes The high-affinity camp phosphodiesterase of Saccharomyces cerevisiae is the major determinant of camp levels in stationary phase: involvement of different branches of the Ras-cyclic AMP pathway in stress responses. Biochem. Biophys. Res. Commun. 327: Perfect, J. R., N. Ketabchi, G. M. Cox, C. W. Ingram, and C. L. Beiser Karyotyping of Cryptococcus neoformans as an epidemiological tool. J. Clin. Microbiol. 31: Rascon, A., M. E. Viloria, L. De-Chiara, and M. E. Dubra Characterization of cyclic AMP phosphodiesterases in Leishmania mexicana and purification of soluble form. Mol. Biochem. Parasitol. 106:
Received 16 July 2004/Accepted 21 August 2004
 EUKARYOTIC CELL, Dec. 2004, p. 1476 1491 Vol. 3, No. 6 1535-9778/04/$08.00 0 DOI: 10.1128/EC.3.6.1476 1491.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Adenylyl Cyclase-Associated
EUKARYOTIC CELL, Dec. 2004, p. 1476 1491 Vol. 3, No. 6 1535-9778/04/$08.00 0 DOI: 10.1128/EC.3.6.1476 1491.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Adenylyl Cyclase-Associated
Specialization of the HOG pathway and its impact on differentiation and virulence. of Cryptococcus neoformans
 Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans Yong-Sun Bahn, Kaihei Kojima, Gary M. Cox, and Joseph Heitman,,,, Departments of Molecular Genetics
Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans Yong-Sun Bahn, Kaihei Kojima, Gary M. Cox, and Joseph Heitman,,,, Departments of Molecular Genetics
Received 24 September 2007/Accepted 8 October 2007
 EUKARYOTIC CELL, Dec. 2007, p. 2278 2289 Vol. 6, No. 12 1535-9778/07/$08.00 0 doi:10.1128/ec.00349-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ssk2 Mitogen-Activated Protein
EUKARYOTIC CELL, Dec. 2007, p. 2278 2289 Vol. 6, No. 12 1535-9778/07/$08.00 0 doi:10.1128/ec.00349-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ssk2 Mitogen-Activated Protein
Adenylyl Cyclase Functions Downstream of the G Protein Gpa1 and Controls Mating and Pathogenicity of Cryptococcus neoformans
 EUKARYOTIC CELL, Feb. 2002, p. 75 84 Vol. 1, No. 1 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.1.75 84.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Adenylyl Cyclase Functions
EUKARYOTIC CELL, Feb. 2002, p. 75 84 Vol. 1, No. 1 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.1.75 84.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Adenylyl Cyclase Functions
Pheromones Stimulate Mating and Differentiation via Paracrine and Autocrine Signaling in Cryptococcus neoformans
 EUKARYOTIC CELL, June 2002, p. 366 377 Vol. 1, No. 3 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.3.366 377.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Pheromones Stimulate
EUKARYOTIC CELL, June 2002, p. 366 377 Vol. 1, No. 3 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.3.366 377.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Pheromones Stimulate
The RGS Protein Crg2 Regulates Pheromone and Cyclic AMP Signaling in Cryptococcus neoformans
 EUKARYOTIC CELL, Sept. 2008, p. 1540 1548 Vol. 7, No. 9 1535-9778/08/$08.00 0 doi:10.1128/ec.00154-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. The RGS Protein Crg2 Regulates
EUKARYOTIC CELL, Sept. 2008, p. 1540 1548 Vol. 7, No. 9 1535-9778/08/$08.00 0 doi:10.1128/ec.00154-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. The RGS Protein Crg2 Regulates
Identification of ENA1 as a Virulence Gene of the Human Pathogenic Fungus Cryptococcus neoformans through Signature-Tagged Insertional Mutagenesis
 EUKARYOTIC CELL, Mar. 2009, p. 315 326 Vol. 8, No. 3 1535-9778/09/$08.00 0 doi:10.1128/ec.00375-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Identification of ENA1 as a Virulence
EUKARYOTIC CELL, Mar. 2009, p. 315 326 Vol. 8, No. 3 1535-9778/09/$08.00 0 doi:10.1128/ec.00375-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Identification of ENA1 as a Virulence
Mating-Type-Specific and Nonspecific PAK Kinases Play Shared and Divergent Roles in Cryptococcus neoformans
 EUKARYOTIC CELL, Apr. 2002, p. 257 272 Vol. 1, No. 2 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.2.257 272.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Mating-Type-Specific
EUKARYOTIC CELL, Apr. 2002, p. 257 272 Vol. 1, No. 2 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.2.257 272.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Mating-Type-Specific
Received 11 August 2011/Accepted 9 September 2011
 EUKARYOTIC CELL, Nov. 2011, p. 1455 1464 Vol. 10, No. 11 1535-9778/11/$12.00 doi:10.1128/ec.05207-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. The Casein Kinase I Protein
EUKARYOTIC CELL, Nov. 2011, p. 1455 1464 Vol. 10, No. 11 1535-9778/11/$12.00 doi:10.1128/ec.05207-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. The Casein Kinase I Protein
Amt2 Permease Is Required To Induce Ammonium-Responsive Invasive Growth and Mating in Cryptococcus neoformans
 EUKARYOTIC CELL, Feb. 2008, p. 237 246 Vol. 7, No. 2 1535-9778/08/$08.00 0 doi:10.1128/ec.00079-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Amt2 Permease Is Required To Induce
EUKARYOTIC CELL, Feb. 2008, p. 237 246 Vol. 7, No. 2 1535-9778/08/$08.00 0 doi:10.1128/ec.00079-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Amt2 Permease Is Required To Induce
Pleiotropic roles of the Msi1-like protein, Msl1, in Cryptococcus neoformans
 EC Accepts, published online ahead of print on 5 October 2012 Eukaryotic Cell doi:10.1128/ec.00261-12 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11 12
EC Accepts, published online ahead of print on 5 October 2012 Eukaryotic Cell doi:10.1128/ec.00261-12 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11 12
The F-Box Protein Fbp1 Regulates Sexual Reproduction and Virulence in Cryptococcus neoformans
 EUKARYOTIC CELL, June 2011, p. 791 802 Vol. 10, No. 6 1535-9778/11/$12.00 doi:10.1128/ec.00004-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. The F-Box Protein Fbp1 Regulates
EUKARYOTIC CELL, June 2011, p. 791 802 Vol. 10, No. 6 1535-9778/11/$12.00 doi:10.1128/ec.00004-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. The F-Box Protein Fbp1 Regulates
Cryptococcal Titan Cell Formation Is Regulated by G-Protein Signaling in Response to Multiple Stimuli
 EUKARYOTIC CELL, Oct. 2011, p. 1306 1316 Vol. 10, No. 10 1535-9778/11/$12.00 doi:10.1128/ec.05179-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. Cryptococcal Titan Cell Formation
EUKARYOTIC CELL, Oct. 2011, p. 1306 1316 Vol. 10, No. 10 1535-9778/11/$12.00 doi:10.1128/ec.05179-11 Copyright 2011, American Society for Microbiology. All Rights Reserved. Cryptococcal Titan Cell Formation
Cryptococcus neoformans is an opportunistic fungal pathogen of
 A Ric8/Synembryn Homolog Promotes Gpa1 and Gpa2 Activation To Respectively Regulate Cyclic AMP and Pheromone Signaling in Cryptococcus neoformans Jinjun Gong, a Jacob D. Grodsky, b Zhengguang Zhang, c
A Ric8/Synembryn Homolog Promotes Gpa1 and Gpa2 Activation To Respectively Regulate Cyclic AMP and Pheromone Signaling in Cryptococcus neoformans Jinjun Gong, a Jacob D. Grodsky, b Zhengguang Zhang, c
Lecture 15. Signal Transduction Pathways - Introduction
 Lecture 15 Signal Transduction Pathways - Introduction So far.. Regulation of mrna synthesis Regulation of rrna synthesis Regulation of trna & 5S rrna synthesis Regulation of gene expression by signals
Lecture 15 Signal Transduction Pathways - Introduction So far.. Regulation of mrna synthesis Regulation of rrna synthesis Regulation of trna & 5S rrna synthesis Regulation of gene expression by signals
DNA Mutations Mediate Microevolution between Host- Adapted Forms of the Pathogenic Fungus Cryptococcus neoformans
 DNA Mutations Mediate Microevolution between Host- Adapted Forms of the Pathogenic Fungus Cryptococcus neoformans Denise A. Magditch 1, Tong-Bao Liu 2, Chaoyang Xue 2, Alexander Idnurm 1 * 1 Division of
DNA Mutations Mediate Microevolution between Host- Adapted Forms of the Pathogenic Fungus Cryptococcus neoformans Denise A. Magditch 1, Tong-Bao Liu 2, Chaoyang Xue 2, Alexander Idnurm 1 * 1 Division of
8 Suppression Analysis
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 8 Suppression Analysis OVERVIEW Suppression
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 8 Suppression Analysis OVERVIEW Suppression
Impact of Mating Type, Serotype, and Ploidy on the Virulence of Cryptococcus neoformans
 INFECTION AND IMMUNITY, July 2008, p. 2923 2938 Vol. 76, No. 7 0019-9567/08/$08.00 0 doi:10.1128/iai.00168-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. Impact of Mating Type,
INFECTION AND IMMUNITY, July 2008, p. 2923 2938 Vol. 76, No. 7 0019-9567/08/$08.00 0 doi:10.1128/iai.00168-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. Impact of Mating Type,
Laboratory of Clinical Investigation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892
 The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence Y. C. Chang, L. A. Penoyer, and K. J. Kwon-Chung* Laboratory of Clinical Investigation, National
The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence Y. C. Chang, L. A. Penoyer, and K. J. Kwon-Chung* Laboratory of Clinical Investigation, National
Chapter 9. Cellular Signaling
 Chapter 9 Cellular Signaling Cellular Messaging Page 215 Cells can signal to each other and interpret the signals they receive from other cells and the environment Signals are most often chemicals The
Chapter 9 Cellular Signaling Cellular Messaging Page 215 Cells can signal to each other and interpret the signals they receive from other cells and the environment Signals are most often chemicals The
Sex-Specific Homeodomain Proteins Sxi1 and Sxi2a Coordinately Regulate Sexual Development in Cryptococcus neoformans
 EUKARYOTIC CELL, Mar. 2005, p. 526 535 Vol. 4, No. 3 1535-9778/05/$08.00 0 doi:10.1128/ec.4.3.526 535.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Sex-Specific Homeodomain
EUKARYOTIC CELL, Mar. 2005, p. 526 535 Vol. 4, No. 3 1535-9778/05/$08.00 0 doi:10.1128/ec.4.3.526 535.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Sex-Specific Homeodomain
Cell Signaling part 2
 15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
Luminescent platforms for monitoring changes in the solubility of amylin and huntingtin in living cells
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Function and Regulation of Yeast Hexose Transporters
 MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 1999, p. 554 569 Vol. 63, No. 3 1092-2172/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Function and Regulation of
MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 1999, p. 554 569 Vol. 63, No. 3 1092-2172/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Function and Regulation of
Received 25 October 2009/Accepted 14 January 2010
 EUKARYOTIC CELL, Mar. 2010, p. 360 378 Vol. 9, No. 3 1535-9778/10/$12.00 doi:10.1128/ec.00309-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. Comparative Transcriptome Analysis
EUKARYOTIC CELL, Mar. 2010, p. 360 378 Vol. 9, No. 3 1535-9778/10/$12.00 doi:10.1128/ec.00309-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. Comparative Transcriptome Analysis
Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37 6C and in vivo
 Microbiology (2004), 150, 1547 1558 DOI 10.1099/mic.0.26928-0 Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37 6C and in vivo Joanne M. Kingsbury,
Microbiology (2004), 150, 1547 1558 DOI 10.1099/mic.0.26928-0 Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37 6C and in vivo Joanne M. Kingsbury,
Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION
 Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION 1. Feedback a. Negative feedback mechanisms maintain dynamic homeostasis for a particular condition (variable) by regulating physiological processes,
Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION 1. Feedback a. Negative feedback mechanisms maintain dynamic homeostasis for a particular condition (variable) by regulating physiological processes,
BIOLOGY. Cell Communication CAMPBELL. Reece Urry Cain Wasserman Minorsky Jackson. Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick
 CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 11 Cell Communication Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick Cellular Messaging Cells can signal to
CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 11 Cell Communication Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick Cellular Messaging Cells can signal to
Transcription Factor Nrg1 Mediates Capsule Formation, Stress Response, and
 Transcription Factor Nrg1 Mediates Capsule Formation, Stress Response, and Pathogenesis in Cryptococcus neoformans Kari L. Cramer, Quincy D. Gerrald, Connie B. Nichols, Michael S. Price and J. Andrew Alspaugh
Transcription Factor Nrg1 Mediates Capsule Formation, Stress Response, and Pathogenesis in Cryptococcus neoformans Kari L. Cramer, Quincy D. Gerrald, Connie B. Nichols, Michael S. Price and J. Andrew Alspaugh
School of Medicine and Public Health, University of Wisconsin-Madison, Madison, WI 53706
 EC Accepts, published online ahead of print on 7 July 2014 Eukaryotic Cell doi:10.1128/ec.00152-14 Copyright 2014, American Society for Microbiology. All Rights Reserved. 1 1 2 3 4 5 6 7 8 9 10 11 12 13
EC Accepts, published online ahead of print on 7 July 2014 Eukaryotic Cell doi:10.1128/ec.00152-14 Copyright 2014, American Society for Microbiology. All Rights Reserved. 1 1 2 3 4 5 6 7 8 9 10 11 12 13
Quorum Sensing-Mediated, Cell Density-Dependent Regulation of Growth and Virulence in Cryptococcus neoformans
 RESEARCH ARTICLE Quorum Sensing-Mediated, Cell Density-Dependent Regulation of Growth and Virulence in Cryptococcus neoformans Patrícia Albuquerque, a,b,c André M. Nicola, a,c Edward Nieves, d,e Hugo Costa
RESEARCH ARTICLE Quorum Sensing-Mediated, Cell Density-Dependent Regulation of Growth and Virulence in Cryptococcus neoformans Patrícia Albuquerque, a,b,c André M. Nicola, a,c Edward Nieves, d,e Hugo Costa
Materials and Methods , The two-hybrid principle.
 The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
Unisexual reproduction enhances fungal competitiveness by promoting. habitat exploration via hyphal growth and sporulation
 EC Accepts, published online ahead of print on 21 June 2013 Eukaryotic Cell doi:10.1128/ec.00147-13 Copyright 2013, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11 12 13
EC Accepts, published online ahead of print on 21 June 2013 Eukaryotic Cell doi:10.1128/ec.00147-13 Copyright 2013, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11 12 13
Cellular Signaling Pathways. Signaling Overview
 Cellular Signaling Pathways Signaling Overview Signaling steps Synthesis and release of signaling molecules (ligands) by the signaling cell. Transport of the signal to the target cell Detection of the
Cellular Signaling Pathways Signaling Overview Signaling steps Synthesis and release of signaling molecules (ligands) by the signaling cell. Transport of the signal to the target cell Detection of the
Supporting Information
 Supporting Information Dauvillée et al. 10.1073/pnas.0907424106 Fig. S1. Iodine screening of the C. cohnii mutant bank. Each single colony was grown on rich-medium agar plates then vaporized with iodine.
Supporting Information Dauvillée et al. 10.1073/pnas.0907424106 Fig. S1. Iodine screening of the C. cohnii mutant bank. Each single colony was grown on rich-medium agar plates then vaporized with iodine.
Received 27 June 2003/Accepted 26 July 2003
 EUKARYOTIC CELL, Oct. 2003, p. 1036 1045 Vol. 2, No. 5 1535-9778/03/$08.00 0 DOI: 10.1128/EC.2.5.1036 1045.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Recapitulation of
EUKARYOTIC CELL, Oct. 2003, p. 1036 1045 Vol. 2, No. 5 1535-9778/03/$08.00 0 DOI: 10.1128/EC.2.5.1036 1045.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Recapitulation of
Vets 111/Biov 111 Cell Signalling-2. Secondary messengers the cyclic AMP intracellular signalling system
 Vets 111/Biov 111 Cell Signalling-2 Secondary messengers the cyclic AMP intracellular signalling system The classical secondary messenger model of intracellular signalling A cell surface receptor binds
Vets 111/Biov 111 Cell Signalling-2 Secondary messengers the cyclic AMP intracellular signalling system The classical secondary messenger model of intracellular signalling A cell surface receptor binds
Unique roles of the unfolded protein response pathway in fungal. development and differentiation. Kwang Woo Jung, Yee Seul So, & Yong Sun Bahn *
 Supplementry Informtion Unique roles of the unfolded protein response pthwy in fungl development nd differentition Kwng Woo Jung, Yee Seul So, & Yong Sun Bhn * Contents Supplementry Figure S1 Supplementry
Supplementry Informtion Unique roles of the unfolded protein response pthwy in fungl development nd differentition Kwng Woo Jung, Yee Seul So, & Yong Sun Bhn * Contents Supplementry Figure S1 Supplementry
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene
 Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
TKB1 Competent Cells. Instruction Manual. Research Use Only. Not for Use in Diagnostic Procedures. Catalog # Revision B
 TKB1 Competent Cells Instruction Manual Catalog #200134 Revision B Research Use Only. Not for Use in Diagnostic Procedures. 200134-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement
TKB1 Competent Cells Instruction Manual Catalog #200134 Revision B Research Use Only. Not for Use in Diagnostic Procedures. 200134-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
 EC Accepts, published online ahead of print on 23 August 2013 Eukaryotic Cell doi:10.1128/ec.00169-13 Copyright 2013, American Society for Microbiology. All Rights Reserved. 1 TITLE: Nitrogen source-dependent
EC Accepts, published online ahead of print on 23 August 2013 Eukaryotic Cell doi:10.1128/ec.00169-13 Copyright 2013, American Society for Microbiology. All Rights Reserved. 1 TITLE: Nitrogen source-dependent
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
Tropentag 2012, Göttingen, Germany September 19-21, 2012
 Tropentag 2012, Göttingen, Germany September 19-21, 2012 Conference on International Research on Food Security, Natural Resource Management and Rural Development organised by: Georg-August Universität
Tropentag 2012, Göttingen, Germany September 19-21, 2012 Conference on International Research on Food Security, Natural Resource Management and Rural Development organised by: Georg-August Universität
Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana
 Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
Biochemistry: Signaling in the Nitrogen Assimilation Pathway of Arabidopsis Thaliana 38 CAMERON E. NIENABER ʻ04 Abstract Long recognized as essential plant nutrients and metabolites, inorganic and organic
Cell Communication. Chapter 11. Biology Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 11 Cell Communication PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp
Chapter 11 Cell Communication PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp
2017/18 PRODUCT CATALOGUE SCHIZOSACCHAROMYCES POMBE
 2017/18 PRODUCT CATALOGUE SCHIZOSACCHAROMYCES POMBE CONTENTS 3 Schizosaccharomyces Pombe 4 Complex Media 5 YE Broth 5 YE Agar 6 YES Broth 7 YES Agar 8 YSO Broth 9 YSO Agar 10 ME Broth 11 ME Agar 22 SP
2017/18 PRODUCT CATALOGUE SCHIZOSACCHAROMYCES POMBE CONTENTS 3 Schizosaccharomyces Pombe 4 Complex Media 5 YE Broth 5 YE Agar 6 YES Broth 7 YES Agar 8 YSO Broth 9 YSO Agar 10 ME Broth 11 ME Agar 22 SP
Interaction of Cryptococcus neoformans Rim101 and Protein Kinase A Regulates Capsule
 Interaction of Cryptococcus neoformans Rim101 and Protein Kinase A Regulates Capsule Teresa R. O Meara, Diana Norton, Michael S. Price, Christie Hay, Meredith F. Clements, Connie B. Nichols, J. Andrew
Interaction of Cryptococcus neoformans Rim101 and Protein Kinase A Regulates Capsule Teresa R. O Meara, Diana Norton, Michael S. Price, Christie Hay, Meredith F. Clements, Connie B. Nichols, J. Andrew
Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy
 Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
Course Title Form Hours subject
 Course Title Form Hours subject Types, and structure of chromosomes L 1 Histology Karyotyping and staining of human chromosomes L 2 Histology Chromosomal anomalies L 2 Histology Sex chromosomes L 1 Histology
Course Title Form Hours subject Types, and structure of chromosomes L 1 Histology Karyotyping and staining of human chromosomes L 2 Histology Chromosomal anomalies L 2 Histology Sex chromosomes L 1 Histology
Sexual Development in Cryptococcus neoformans Requires CLP1, a Target of the Homeodomain Transcription Factors Sxi1 and Sxi2a
 EUKARYOTIC CELL, Jan. 2008, p. 49 57 Vol. 7, No. 1 1535-9778/08/$08.00 0 doi:10.1128/ec.00377-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Sexual Development in Cryptococcus
EUKARYOTIC CELL, Jan. 2008, p. 49 57 Vol. 7, No. 1 1535-9778/08/$08.00 0 doi:10.1128/ec.00377-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Sexual Development in Cryptococcus
Interaction between genetic background and the mating type locus in Cryptococcus neoformans virulence potential
 Genetics: Published Articles Ahead of Print, published on June 18, 2005 as 10.1534/genetics.105.045039 Interaction between genetic background and the mating type locus in Cryptococcus neoformans virulence
Genetics: Published Articles Ahead of Print, published on June 18, 2005 as 10.1534/genetics.105.045039 Interaction between genetic background and the mating type locus in Cryptococcus neoformans virulence
Carbon dioxide production of wild type and PDC1 mutant Saccharomyces cerevisiae in D-glucose
 Carbon dioxide production of wild type and PDC1 mutant Saccharomyces cerevisiae in D-glucose Luke Gooding, Grace Lam, Simran Parmar, Jessica Sham Abstract To study the differences in respiration between
Carbon dioxide production of wild type and PDC1 mutant Saccharomyces cerevisiae in D-glucose Luke Gooding, Grace Lam, Simran Parmar, Jessica Sham Abstract To study the differences in respiration between
Transcriptional Network of Multiple Capsule and Melanin Genes Governed by the Cryptococcus neoformans Cyclic AMP Cascade
 EUKARYOTIC CELL, Jan. 2005, p. 190 201 Vol. 4, No. 1 1535-9778/05/$08.00 0 doi:10.1128/ec.4.1.190 201.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Transcriptional Network
EUKARYOTIC CELL, Jan. 2005, p. 190 201 Vol. 4, No. 1 1535-9778/05/$08.00 0 doi:10.1128/ec.4.1.190 201.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Transcriptional Network
Receptor mediated Signal Transduction
 Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
ß-Galactosidase Repression in Escherichia coli B23 Using Minimal Concentrations of Glucose and Sucrose
 ß-Galactosidase Repression in Escherichia coli B23 Using Minimal Concentrations of Glucose and Sucrose JILLIAN CLARK, JACQUIE HUDSON, ROBIN MAK, CHRISTA McPHERSON, AND CARMEN TSIN Department of Microbiology
ß-Galactosidase Repression in Escherichia coli B23 Using Minimal Concentrations of Glucose and Sucrose JILLIAN CLARK, JACQUIE HUDSON, ROBIN MAK, CHRISTA McPHERSON, AND CARMEN TSIN Department of Microbiology
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Amplification of Capsule-associated Genes from Cryptococcus neoformans
 Amplification of Capsule-associated Genes from Cryptococcus neoformans Randa Alarousy *1, Heidy Abo El Yazeed 2, Hosam Kotb 3, Khaled Abdalla 4, and Mohamed Refai 2 1 Department of Microbiology and Immunology,
Amplification of Capsule-associated Genes from Cryptococcus neoformans Randa Alarousy *1, Heidy Abo El Yazeed 2, Hosam Kotb 3, Khaled Abdalla 4, and Mohamed Refai 2 1 Department of Microbiology and Immunology,
Journal of Experimental Microbiology and Immunology (JEMI) Vol. 9:6-10 Copyright April 2006, M&I, UBC
 The Effect of Cyclic 3, 5 -Adenosine Monophosphate on the Effect of Glucose, Fructose, and Sucrose Supplements on the Induction of lacz in Escherichia coli B23 Grown in Minimal Medium with Glycerol JOSHUA
The Effect of Cyclic 3, 5 -Adenosine Monophosphate on the Effect of Glucose, Fructose, and Sucrose Supplements on the Induction of lacz in Escherichia coli B23 Grown in Minimal Medium with Glycerol JOSHUA
Cover Page. The handle holds various files of this Leiden University dissertation.
 Cover Page The handle http://hdl.handle.net/1887/20360 holds various files of this Leiden University dissertation. Author: Eijk, Patrick van Title: Nucleotide excision repair in yeast Date: 2012-12-13
Cover Page The handle http://hdl.handle.net/1887/20360 holds various files of this Leiden University dissertation. Author: Eijk, Patrick van Title: Nucleotide excision repair in yeast Date: 2012-12-13
Biol220 Cell Signalling Cyclic AMP the classical secondary messenger
 Biol220 Cell Signalling Cyclic AMP the classical secondary messenger The classical secondary messenger model of intracellular signalling A cell surface receptor binds the signal molecule (the primary
Biol220 Cell Signalling Cyclic AMP the classical secondary messenger The classical secondary messenger model of intracellular signalling A cell surface receptor binds the signal molecule (the primary
Supplementary Figure 1: Transcription factors required for temperature-dependent growth of C. neoformans. C. neoformans strains were grown overnight
 Continued Supplementary Figure 1: Transcription factors required for temperature-dependent growth of C. neoformans. C. neoformans strains were grown overnight in liquid YPD medium at 30 C, serially diluted
Continued Supplementary Figure 1: Transcription factors required for temperature-dependent growth of C. neoformans. C. neoformans strains were grown overnight in liquid YPD medium at 30 C, serially diluted
Signal Transduction: G-Protein Coupled Receptors
 Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
CHAPTER 4 RESULTS. showed that all three replicates had similar growth trends (Figure 4.1) (p<0.05; p=0.0000)
 CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER
 REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER RNA Splicing Lecture 3, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Splice
REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER RNA Splicing Lecture 3, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Splice
GENETICS OF CELL PROLIFERATION. S. Cameron K. O Neil M. Riggs L. Rodgers G. Waitches
 GENETICS OF CELL PROLIFERATION M W igler C. Birchmeier D. Broek T. Kataoka S. Powers T. Toda G. Bolger J. Colicelli K. Ferguson J. Field T. Michaeli J. Nikawa P Sass S. Sharma D. Young S. Cameron K. O
GENETICS OF CELL PROLIFERATION M W igler C. Birchmeier D. Broek T. Kataoka S. Powers T. Toda G. Bolger J. Colicelli K. Ferguson J. Field T. Michaeli J. Nikawa P Sass S. Sharma D. Young S. Cameron K. O
Saccharomyces cerevisiae?
 JOURNAL OF BACTERIOLOGY, Aug. 1983, p. 623-627 21-9193/83/8623-5$2.O/ Copyright 1983, American Society for Microbiology Vol. 155, No. 2 What Is the Function of Nitrogen Catabolite Repression in Saccharomyces
JOURNAL OF BACTERIOLOGY, Aug. 1983, p. 623-627 21-9193/83/8623-5$2.O/ Copyright 1983, American Society for Microbiology Vol. 155, No. 2 What Is the Function of Nitrogen Catabolite Repression in Saccharomyces
Part-4. Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death
 Part-4 Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death 95 1. Introduction The process of replicating DNA and dividing cells can be described as a series of coordinated
Part-4 Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death 95 1. Introduction The process of replicating DNA and dividing cells can be described as a series of coordinated
2013 W. H. Freeman and Company. 12 Signal Transduction
 2013 W. H. Freeman and Company 12 Signal Transduction CHAPTER 12 Signal Transduction Key topics: General features of signal transduction Structure and function of G protein coupled receptors Structure
2013 W. H. Freeman and Company 12 Signal Transduction CHAPTER 12 Signal Transduction Key topics: General features of signal transduction Structure and function of G protein coupled receptors Structure
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
The Ras GTPases are membrane-associated binary switches that
 Plasma Membrane Localization Is Required for RasA-Mediated Polarized Morphogenesis and Virulence of Aspergillus fumigatus Jarrod R. Fortwendel, f Praveen R. Juvvadi, a Luise E. Rogg, a Yohannes G. Asfaw,
Plasma Membrane Localization Is Required for RasA-Mediated Polarized Morphogenesis and Virulence of Aspergillus fumigatus Jarrod R. Fortwendel, f Praveen R. Juvvadi, a Luise E. Rogg, a Yohannes G. Asfaw,
Recipes for Media and Solution Preparation SC-ura/Glucose Agar Dishes (20mL/dish, enough for 8 clones)
 Protocol: 300 ml Yeast culture preparation Equipment and Reagents needed: Autoclaved toothpicks Shaker Incubator set at 30 C Incubator set at 30 C 60 mm 2 sterile petri dishes Autoclaved glass test tubes
Protocol: 300 ml Yeast culture preparation Equipment and Reagents needed: Autoclaved toothpicks Shaker Incubator set at 30 C Incubator set at 30 C 60 mm 2 sterile petri dishes Autoclaved glass test tubes
The Effects of Alcohol and Nicotine on Microbial Flora. Jeff Van Kooten Grade 11 Pittsburgh Central Catholic High School
 The Effects of Alcohol and Nicotine on Microbial Flora Jeff Van Kooten Grade 11 Pittsburgh Central Catholic High School Microbial Flora The internal and external flora has eukaryotic fungi, protists, and
The Effects of Alcohol and Nicotine on Microbial Flora Jeff Van Kooten Grade 11 Pittsburgh Central Catholic High School Microbial Flora The internal and external flora has eukaryotic fungi, protists, and
Cell Communication. Local and Long Distance Signaling
 Cell Communication Cell to cell communication is essential for multicellular organisms Some universal mechanisms of cellular regulation providing more evidence for the evolutionary relatedness of all life
Cell Communication Cell to cell communication is essential for multicellular organisms Some universal mechanisms of cellular regulation providing more evidence for the evolutionary relatedness of all life
Plasma membranes. Plasmodesmata between plant cells. Gap junctions between animal cells Cell junctions. Cell-cell recognition
 Cell Communication Cell Signaling Cell-to-cell communication is essential for multicellular organisms Communicate by chemical messengers Animal and plant cells have cell junctions that directly connect
Cell Communication Cell Signaling Cell-to-cell communication is essential for multicellular organisms Communicate by chemical messengers Animal and plant cells have cell junctions that directly connect
AP Biology Summer Assignment Chapter 3 Quiz
 AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
MRC-Holland MLPA. Description version 08; 30 March 2015
 SALSA MLPA probemix P351-C1 / P352-D1 PKD1-PKD2 P351-C1 lot C1-0914: as compared to the previous version B2 lot B2-0511 one target probe has been removed and three reference probes have been replaced.
SALSA MLPA probemix P351-C1 / P352-D1 PKD1-PKD2 P351-C1 lot C1-0914: as compared to the previous version B2 lot B2-0511 one target probe has been removed and three reference probes have been replaced.
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Factors influencing the Aspergillus fumigatus survival into the host mediated by the calcineurin pathway
 Factors influencing the Aspergillus fumigatus survival into the host mediated by the calcineurin pathway Gustavo H. Goldman Universidade de São Paulo, Brazil Calcineurin is a calmodulin/ca +2 dependent
Factors influencing the Aspergillus fumigatus survival into the host mediated by the calcineurin pathway Gustavo H. Goldman Universidade de São Paulo, Brazil Calcineurin is a calmodulin/ca +2 dependent
SACCHAROMYCES CEREVISIAE GENOMIC LIBRARY SCREENING IN SEARCH FOR THE GENE RESPONSIBLE FOR INDUCTIVE ACTIVE GLYCEROL
 SACCHAROMYCES CEREVISIAE GENOMIC LIBRARY SCREENING IN SEARCH FOR THE GENE RESPONSIBLE FOR INDUCTIVE ACTIVE GLYCEROL UPTAKE R.P. Oliveira, and C. Lucas Departamento de Biologia da Universidade do Minho.
SACCHAROMYCES CEREVISIAE GENOMIC LIBRARY SCREENING IN SEARCH FOR THE GENE RESPONSIBLE FOR INDUCTIVE ACTIVE GLYCEROL UPTAKE R.P. Oliveira, and C. Lucas Departamento de Biologia da Universidade do Minho.
Relationship of the Glyoxylate Pathway to the Pathogenesis of Cryptococcus neoformans
 INFECTION AND IMMUNITY, Oct. 2002, p. 5684 5694 Vol. 70, No. 10 0019-9567/02/$04.00 0 DOI: 10.1128/IAI.70.10.5684 5694.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Relationship
INFECTION AND IMMUNITY, Oct. 2002, p. 5684 5694 Vol. 70, No. 10 0019-9567/02/$04.00 0 DOI: 10.1128/IAI.70.10.5684 5694.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Relationship
Hands-On Ten The BRCA1 Gene and Protein
 Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Supplemental Data. Hiruma et al. Plant Cell. (2010) /tpc Col-0. pen2-1
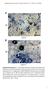 A Ch B Col-0 Cg pen2-1 Supplemental Figure 1. Trypan Blue Staining of Leaves Inoculated with Adapted and Nonadapted Colletotrichum Species.(A) Conidial suspensions of C. higginsianum MAFF305635 (Ch) were
A Ch B Col-0 Cg pen2-1 Supplemental Figure 1. Trypan Blue Staining of Leaves Inoculated with Adapted and Nonadapted Colletotrichum Species.(A) Conidial suspensions of C. higginsianum MAFF305635 (Ch) were
Discovery. Gerry Graham* and Rob Nibbs SUMMARY BACKGROUND
 D6 Gerry Graham* and Rob Nibbs Cancer Research Campaign Laboratories, The Beaton Institute for Cancer Research, Garscube Estate Switchback Road, Bearsdon, Glasgow G61 1BD, UK * corresponding author tel:
D6 Gerry Graham* and Rob Nibbs Cancer Research Campaign Laboratories, The Beaton Institute for Cancer Research, Garscube Estate Switchback Road, Bearsdon, Glasgow G61 1BD, UK * corresponding author tel:
SURVEILLANCE TECHNICAL
 CHAPTER 5 SURVEILLANCE TECHNICAL ASPECTS 55 Protect - detect - protect Polio eradication strategies can be summed up as protect and detect protect children against polio by vaccinating them, and detect
CHAPTER 5 SURVEILLANCE TECHNICAL ASPECTS 55 Protect - detect - protect Polio eradication strategies can be summed up as protect and detect protect children against polio by vaccinating them, and detect
SALSA MLPA probemix P315-B1 EGFR
 SALSA MLPA probemix P315-B1 EGFR Lot B1-0215 and B1-0112. As compared to the previous A1 version (lot 0208), two mutation-specific probes for the EGFR mutations L858R and T709M as well as one additional
SALSA MLPA probemix P315-B1 EGFR Lot B1-0215 and B1-0112. As compared to the previous A1 version (lot 0208), two mutation-specific probes for the EGFR mutations L858R and T709M as well as one additional
Muscular Dystrophy. Biol 405 Molecular Medicine
 Muscular Dystrophy Biol 405 Molecular Medicine Duchenne muscular dystrophy Duchenne muscular dystrophy is a neuromuscular disease that occurs in ~ 1/3,500 male births. The disease causes developmental
Muscular Dystrophy Biol 405 Molecular Medicine Duchenne muscular dystrophy Duchenne muscular dystrophy is a neuromuscular disease that occurs in ~ 1/3,500 male births. The disease causes developmental
The elements of G protein-coupled receptor systems
 The elements of G protein-coupled receptor systems Prostaglandines Sphingosine 1-phosphate a receptor that contains 7 membrane-spanning domains a coupled trimeric G protein which functions as a switch
The elements of G protein-coupled receptor systems Prostaglandines Sphingosine 1-phosphate a receptor that contains 7 membrane-spanning domains a coupled trimeric G protein which functions as a switch
Ch. 18 Regulation of Gene Expression
 Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Conservation of morphogenesis and virulence in Cryptococcus neoformans. Xiaorong Lin Texas A&M University
 Conservation of morphogenesis and virulence in Cryptococcus neoformans Xiaorong Lin Texas A&M University Cryptococcus infection Lymphatic and hematogenous dissemination Pulmonary infection Cryptococcal
Conservation of morphogenesis and virulence in Cryptococcus neoformans Xiaorong Lin Texas A&M University Cryptococcus infection Lymphatic and hematogenous dissemination Pulmonary infection Cryptococcal
FUNDAMENTALS OF BIOCHEMISTRY, CELL BIOLOGY AND BIOPHYSICS Vol. I - Biochemistry of Vitamins, Hormones and Other Messenger Molecules - Chris Whiteley
 BIOCHEMISTRY OF VITAMINS, HORMONES AND OTHER MESSENGER MOLECULES Chris Whiteley Department of Biochemistry and Microbiology, Rhodes University, Grahamstown, South Africa Keywords: phosphorylation, phosphorylase,
BIOCHEMISTRY OF VITAMINS, HORMONES AND OTHER MESSENGER MOLECULES Chris Whiteley Department of Biochemistry and Microbiology, Rhodes University, Grahamstown, South Africa Keywords: phosphorylation, phosphorylase,
Supplemental Data Macrophage Migration Inhibitory Factor MIF Interferes with the Rb-E2F Pathway
 Supplemental Data Macrophage Migration Inhibitory Factor MIF Interferes with the Rb-E2F Pathway S1 Oleksi Petrenko and Ute M. Moll Figure S1. MIF-Deficient Cells Have Reduced Transforming Ability (A) Soft
Supplemental Data Macrophage Migration Inhibitory Factor MIF Interferes with the Rb-E2F Pathway S1 Oleksi Petrenko and Ute M. Moll Figure S1. MIF-Deficient Cells Have Reduced Transforming Ability (A) Soft
Molecular Cell Biology - Problem Drill 19: Cell Signaling Pathways and Gene Expression
 Molecular Cell Biology - Problem Drill 19: Cell Signaling Pathways and Gene Expression Question No. 1 of 10 1. Which statement about cell signaling is correct? Question #1 (A) Cell signaling involves receiving
Molecular Cell Biology - Problem Drill 19: Cell Signaling Pathways and Gene Expression Question No. 1 of 10 1. Which statement about cell signaling is correct? Question #1 (A) Cell signaling involves receiving
DURING eukaryotic growth, developmental transitions
 INVESTIGATION Analysis of Cryptococcus neoformans Sexual Development Reveals Rewiring of the Pheromone-Response Network by a Change in Transcription Factor Identity Emilia K. Kruzel,* Steven S. Giles,*
INVESTIGATION Analysis of Cryptococcus neoformans Sexual Development Reveals Rewiring of the Pheromone-Response Network by a Change in Transcription Factor Identity Emilia K. Kruzel,* Steven S. Giles,*
Cch1 Mediates Calcium Entry in Cryptococcus neoformans and Is Essential in Low-Calcium Environments
 EUKARYOTIC CELL, Oct. 2006, p. 1788 1796 Vol. 5, No. 10 1535-9778/06/$08.00 0 doi:10.1128/ec.00158-06 Copyright 2006, American Society for Microbiology. All Rights Reserved. Cch1 Mediates Calcium Entry
EUKARYOTIC CELL, Oct. 2006, p. 1788 1796 Vol. 5, No. 10 1535-9778/06/$08.00 0 doi:10.1128/ec.00158-06 Copyright 2006, American Society for Microbiology. All Rights Reserved. Cch1 Mediates Calcium Entry
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
Gene expression regulation during cellular differentiation. Dom Helmlinger CRBM 24/10/2017
 Gene expression regulation during cellular differentiation Dom Helmlinger CRBM 24/10/2017 Outline 1. Gene expression regulation: why and how? 2. Yeast as model systems. 3. Control of sexual differentiation
Gene expression regulation during cellular differentiation Dom Helmlinger CRBM 24/10/2017 Outline 1. Gene expression regulation: why and how? 2. Yeast as model systems. 3. Control of sexual differentiation
