Supplementary Information
|
|
|
- Leonard Baldwin
- 6 years ago
- Views:
Transcription
1 Gureaso et al., Supplementary Information Supplementary Information Membrane-dependent Signal Integration by the Ras ctivator Son of Sevenless Jodi Gureaso, William J. Galush, Sean Boyevisch, Holger Sondermann, Dafna Bar-Sagi, Jay T. Groves and John Kuriyan
2 Gureaso et al., Supplementary Information Supplementary Figure Lipid attachment does not affect the intrinsic nucleotide release rates from Ras. To ensure that membrane anchorage of Ras does not alter the intrinsic rate of nucleotide release, we too advantage of an isolated dc5 domain construct of the SOS homolog Ras Guanine Nucleotide Releasing Factor (RasGRF), which is active in the absence of an allosteric activator. The rate of fluorescently labeled mant-dgdp release from Ras in solution and membrane-bound Ras (~, molecules µm ) in the presence of SOS cat and RasGRF dc5 is compared (mant-dgdp echanged for GDP). The bul volume concentration of Ras, RasGRF dc5, and SOS cat is µm (when present). Note that membrane attachment does not alter the intrinsic nucleotide release rate from Ras alone or the RasGRF dc5 -catalyzed rate.
3 Gureaso et al., Supplementary Information Supplementary Figure The enhanced affinity of Ras-GTP for the allosteric site of SOS underlies the positive feedbac activation of Ras by SOS. The rates for SOS cat - catalyzed nucleotide echange for Ras coupled to lipid vesicles when fluorescently labeled mant-dgdp is echanged for either unlabeled GDP or GTP are compared for a given surface density of Ras. lthough the rates are shown for three Ras surface densities, the bul volume concentrations of Ras and SOS are the same for each measurement ( µm). Note that the replacement of GDP for GTP on Ras results in a ~- fold increase in the rate of SOS cat -catalyzed nucleotide release for an equal surface density of membrane-bound Ras.
4 Gureaso et al., Supplementary Information Supplementary Figure 3 PH domain-dependent binding of SOS to PIP. The binding of SOS proteins to lipid vesicles composed of a miture of phosphatidylcholine (P), phosphatidylserine (PS) and the relevant phospholipid species at ratios of :: is assayed using a sucrose density gradient flotation assay (see Supplementary Methods). Binding of the PH domain containing SOS constructs, SOS DP and SOS HDP, to vesicles containing PIP is observed. No binding to vesicles containing PIP is observed for a mutant form of SOS DP, SOS DP(K5E R59E), in which the two basic residues that are critical for PIP binding are replaced by glutamate. The results shown are representative of five independent eperiments.
5 Gureaso et al., Supplementary Information Supplementary Figure Lateral mobility of Ras-coupled supported lipid bilayers. Membrane-lined Ras mobility is detected by fluorescence recovery after photobleaching (FRP) eperiments. High intensity illumination is used to bleach a small region of the membrane, bleaching most of the BODIPY-GDP fluorophores. fter 9 minutes the bleached fluorophores have laterally diffused out of the spot, as indicated by intensity traces along the yellow line. [White bar = µm]
6 Gureaso et al., Supplementary Information Supplementary Figure 5 Schematic of supported lipid bilayer eperimental setup. planar bilayer in a µl well presents mobile, lipid-lined Ras to the solution (left). small region of bilayer is removed by scratching with a needle (center, bar = µm). Upon addition of SOS, signal from the bilayer decays with time, while the scratched region controls for signal from fluorescent nucleotide in solution (right).
7 Gureaso et al., Supplementary Information Supplementary Table alculation of Ras molecules per vesicle and Ras surface density (molecules µm ) mol% Ras-coupled maleimide-lipids Number of Ras molecules per vesicle Surface density per vesicle (Ras µm ). 3 mol% Ras-coupled maleimide-lipids is calculated using final protein and lipid concentrations after Ras-conjugation (see Methods). The number of Ras molecules per vesicle and the surface density of Ras (Ras molecules per µm ) are calculated based on the following assumptions: Radius of vesicle 5 nm, surface area of vesicle = πr 3 nm (.3 µm ) Surface area of lipid. nm, number of surface lipids per vesicle 3.9 Number of Ras molecules per vesicle = (mol% Ras-coupled lipids/) (surface lipids/vesicle) (.) (3.9 ) 39 Ras molecules/vesicle Surface density of Ras molecules per µm = (Ras molecules/vesicle)/(surface area of vesicle) (39 Ras/vesicle)/(.3 µm /vesicle) 3 Ras/µm
8 Gureaso et al., Supplementary Information Supplementary Methods Liposome Binding ssays The binding of SOS proteins to phospholipid vesicles composed of a miture of phosphatidylcholine (P), phosphatidylserine (PS) and the relevant phospholipid species at ratios of :: was assayed using flotation through a sucrose density gradient to separate protein-associated vesicles from free protein. Phospholipid vesicles ( µg total lipid, corresponding to ~ µm lipid of interest) were incubated with SOS proteins ( ng) at room temperature for min. Binding reactions were mied with 5 µl of.5 M sucrose in binding buffer (5 mm HEPES-NaOH [ph.] and mm Nal) to mae a final concentration of. M sucrose. µl sample of this miture was layered onto the bottom of a polycarbonate ( mm) centrifuge tube (Becman) and overlaid with µl of.5 M sucrose in binding buffer followed by µl of binding buffer. entrifugation was carried out at, g for min at room temperature in a TL rotor (Becman). Phospholipid vesicles were recovered by flotation through the gradient and phospholipid-associated proteins were analyzed using SDS PGE followed by silver staining (SilverQuest, Invitrogen) after normalizing for lipid recovery.
9 Gureaso et al., Supplementary Information Supplementary Discussion Ras/SOS reaction networ model model was constructed to simulate the interaction of membrane-bound Ras with SOS, where SOS is able to bind two Ras molecules, but only catalyzes nucleotide echange at the catalytic site. We use a convention where Ras SOS indicates Ras bound in the allosteric site, SOS Ras indicates Ras bound in the catalytic site, and Ras SOS Ras indicates Ras in both allosteric and catalytic sites. The Ras-bound nucleotide is indicated by a * or, where Ras* is a protein loaded with fluorescent nucleotide, and Ras is a protein with non-fluorescent nucleotide. Our model contains the following reactions: Ras*SOS - Ras* SOS SOS Ras* - SOS Ras* Ras* SOS Ras* - Ras* SOS Ras* Ras*SOS Ras* - Ras* SOS Ras* Ras SOS - Ras SOS SOS Ras SOS Ras - Ras SOS Ras Ras SOS Ras - Ras SOS Ras Ras SOS Ras - Ras* SOS Ras Ras* SOS Ras - Ras SOS Ras* - Ras SOS Ras* Ras SOS Ras* - Ras SOS Ras*
10 Gureaso et al., Supplementary Information Ras*SOS Ras Ras* SOS Ras - fast Ras* SOS Ras* Ras* SOS Ras SOS Ras* slow SOS Ras fast Ras SOS Ras* Ras SOS Ras where the rate constants are: = binding to allosteric site (empty catalytic site) = unbinding from allosteric site (empty catalytic site) = binding to catalytic site (empty allosteric site) = unbinding from catalytic site (empty allosteric site) = binding to allosteric site (filled catalytic site) = unbinding from allosteric site (filled catalytic site) = binding to catalytic site (filled allosteric site) = unbinding from catalytic site (empty allosteric site) slow = nucleotide echange rate constant with no allosteric Ras bound fast = nucleotide echange rate constant with allosteric Ras bound and the or subscripts indicate the absence or presence, respectively, of Ras in the nonechanging, allosteric binding site. Thus, is the forward rate constant to bind Ras to the allosteric site of SOS, while is the forward rate constant for Ras binding to the allosteric site of SOS Ras (i.e. SOS with a Ras already in the catalytic site). ll rate constants have units of s (discussed below). Variables fast and slow are pseudo-first order constants for an ecess of unlabeled nucleotide. In order to have a unified scheme that deals with both solution and membrane-bound components, we consider all species and rates in terms of the absolute number of molecules rather than their volume or surface concentrations. Thus, for unimolecular reactions, rates are epressed in the form d i = i () l where i is number of molecules i and l is the rate constant for the reaction in units of s. For bimolecular interactions, the rate of reaction of species i is simply proportional to: i) the number i, ii) the probability of encountering a molecule of type j, and iii) a rate constant establishing the probability of the reaction occurring. This leads to bimolecular reaction rates of the form
11 Gureaso et al., Supplementary Information di = P () i j l where i is number of molecules i, P j is the probability of encountering a molecule of species j (given by j s volume or area fraction of the maimum close-paced density), and the rate constant l in units of s. It is important to note that this analysis avoids the need to consider rate constants of mied dimensionality that relate interactions between solution and surface species. dditionally, all rate constants are directly intercomparable, regardless of what chemical components are involved. s a matter of clarification, the forward bimolecular reaction i j Product is often epressed as d[ i ] = i j molar (3) where i is the usual molar concentration of i, and molar is the usual molar bimolecular rate constant. Note that the left-hand side of (3) can be re-epressed as d[ i ] = d i () N V = d i N V (5) where i is the number of molecules of i, N is vogadro s number and V is the volume (assumed to be liter). Our molecular-based formalism applied to the same bimolecular reaction given by equation () can be written as d i = i j j ma l () where jma is the maimum close paced number of j in liter and l is once again the forward rate constant in units of s. ombining equations (5) and (), d[ i ] so, = N V i j l = i j molar () j ma N V N V
12 Gureaso et al., Supplementary Information l N V = molar () j ma s an eample, converting the value of below into molar concentration terms, one finds that the dissociation constant for Ras binding to the allosteric site ( / ) is in the micromolar range. We chose rate constants for the simulation that qualitatively describe the rate phenomena present in both vesicle and supported bilayer eperiments. We have observed that BODIPY-GDP echanges several fold faster than mant-dgdp, thus we set the rate constant of nucleotide echange on supported bilayers to 3X the value used for vesicle simulations. We chose this ratio to best describe the behavior of both systems. The specific rate constants chosen were: = s = s = s = s = s = s = s = s slow = s fast = 5 s (vesicle); 5 s (supported bilayer) The above model leads to the following eleven differential equations, which were solved using Matlab (MathWors) for the real eperimental conditions for both the vesicle and supported bilayer systems. We analyzed the behavior of the sum of the fluorescent Ras species with respect to time to simulate the eperimental observable. The term P j (the probability of molecule i encountering molecule j) is the volume fraction of j, given by j divided by jma, where jma is the maimum close paced number of j in the volume or area. d d = = ma ma ma 3 ma ma 3 3ma 5 ma 3 ma ma ma 5 d3 = ma ma 3ma
13 Gureaso et al., Supplementary Information slow d ma 5 ma ma = fast d 5 5 ma 5 3ma 3 5 = ma 3ma 3 9 ma 9 ma ma ma d = ma 9 ma ma d = slow d ma 9 ma ma = fast d 9 ma 9 ma 9 = fast d 5 ma 3ma 3 = fast d ma ma = where = Ras* = SOS 3 = Ras* SOS = SOS Ras* 5 = Ras* SOS Ras* = Ras = Ras SOS = SOS Ras 9 = Ras SOS Ras* = Ras* SOS Ras = Ras SOS Ras
14 Gureaso et al., Supplementary Information References. Miller, E., ntonny, B., Hamamoto, S. & Scheman, R. argo selection into OPII vesicles is driven by the Secp subunit. Embo J, 5-3 ().. Freedman, T.S. et al. Ras-induced conformational switch in the Ras activator Son of sevenless. Proc Natl cad Sci U S 3, 9- ().
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Beck et al., http://www.jcb.org/cgi/content/full/jcb.201011027/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Membrane binding of His-tagged proteins to Ni-liposomes.
Supplemental material Beck et al., http://www.jcb.org/cgi/content/full/jcb.201011027/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Membrane binding of His-tagged proteins to Ni-liposomes.
Supplementary Materials. High affinity binding of phosphatidylinositol-4-phosphate. by Legionella pneumophila DrrA
 Supplementary Materials High affinity binding of phosphatidylinositol-4-phosphate by Legionella pneumophila DrrA Running title: Molecular basis of PtdIns(4)P-binding by DrrA Stefan Schoebel, Wulf Blankenfeldt,
Supplementary Materials High affinity binding of phosphatidylinositol-4-phosphate by Legionella pneumophila DrrA Running title: Molecular basis of PtdIns(4)P-binding by DrrA Stefan Schoebel, Wulf Blankenfeldt,
Supplementary Figure 1. Overview of steps in the construction of photosynthetic protocellular systems
 Supplementary Figure 1 Overview of steps in the construction of photosynthetic protocellular systems (a) The small unilamellar vesicles were made with phospholipids. (b) Three types of small proteoliposomes
Supplementary Figure 1 Overview of steps in the construction of photosynthetic protocellular systems (a) The small unilamellar vesicles were made with phospholipids. (b) Three types of small proteoliposomes
NANO 243/CENG 207 Course Use Only
 L9. Drug Permeation Through Biological Barriers May 3, 2018 Lipids Lipid Self-Assemblies 1. Lipid and Lipid Membrane Phospholipid: an amphiphilic molecule with a hydrophilic head and 1~2 hydrophobic tails.
L9. Drug Permeation Through Biological Barriers May 3, 2018 Lipids Lipid Self-Assemblies 1. Lipid and Lipid Membrane Phospholipid: an amphiphilic molecule with a hydrophilic head and 1~2 hydrophobic tails.
ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP
 ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP Peera Jaru-Ampornpan 1, Kuang Shen 1,3, Vinh Q. Lam 1,3, Mona Ali 2, Sebastian Doniach 2, Tony Z. Jia 1, Shu-ou Shan 1 Supplementary
ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP Peera Jaru-Ampornpan 1, Kuang Shen 1,3, Vinh Q. Lam 1,3, Mona Ali 2, Sebastian Doniach 2, Tony Z. Jia 1, Shu-ou Shan 1 Supplementary
The Annexin V Apoptosis Assay
 The Annexin V Apoptosis Assay Development of the Annexin V Apoptosis Assay: 1990 Andree at al. found that a protein, Vascular Anticoagulant α, bound to phospholipid bilayers in a calcium dependent manner.
The Annexin V Apoptosis Assay Development of the Annexin V Apoptosis Assay: 1990 Andree at al. found that a protein, Vascular Anticoagulant α, bound to phospholipid bilayers in a calcium dependent manner.
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
A Primer on G Protein Signaling. Elliott Ross UT-Southwestern Medical Center
 A Primer on G Protein Signaling Elliott Ross UT-Southwestern Medical Center Receptor G Effector The MODULE Rhodopsins Adrenergics Muscarinics Serotonin, Dopamine Histamine, GABA b, Glutamate Eiscosanoids
A Primer on G Protein Signaling Elliott Ross UT-Southwestern Medical Center Receptor G Effector The MODULE Rhodopsins Adrenergics Muscarinics Serotonin, Dopamine Histamine, GABA b, Glutamate Eiscosanoids
Transport and Concentration of Charged Molecules in a Lipid Membrane
 Transport and Concentration of Charged Molecules in a Lipid Membrane Johannes S. Roth 1, Matthew R. Cheetham 1,2, and Stephen D. Evans* 1 1 University of Leeds, 2 Current Address: University of Oxford
Transport and Concentration of Charged Molecules in a Lipid Membrane Johannes S. Roth 1, Matthew R. Cheetham 1,2, and Stephen D. Evans* 1 1 University of Leeds, 2 Current Address: University of Oxford
The MOLECULES of LIFE
 The MOLECULES of LIFE Physical and Chemical Principles Solutions Manual Prepared by James Fraser and Samuel Leachman Chapter 16 Principles of Enzyme Catalysis Problems True/False and Multiple Choice 1.
The MOLECULES of LIFE Physical and Chemical Principles Solutions Manual Prepared by James Fraser and Samuel Leachman Chapter 16 Principles of Enzyme Catalysis Problems True/False and Multiple Choice 1.
Lab Results: 1. Document the initial and final egg masses. 2. Calculate the percent change
 Lab Results: 1. Document the initial and final egg masses. 2. Calculate the percent change 3. Draw an arrow showing which way water traveled (in or out of the egg) on your post lab. CHI- SQUARE: What if
Lab Results: 1. Document the initial and final egg masses. 2. Calculate the percent change 3. Draw an arrow showing which way water traveled (in or out of the egg) on your post lab. CHI- SQUARE: What if
BILAYER CHANNEL RECONSTITUTION
 (1) 1% Agar Salt Bridge 1.0 g Agar 3.75g KCl in 100ml distilled water, store at 4 o C. BILAYER CHANNEL RECONSTITUTION (2) Cs solution: (Cesium Methanesulfonate) 1) 50 mm Cs + solution 0.209 MOPS, 10mM
(1) 1% Agar Salt Bridge 1.0 g Agar 3.75g KCl in 100ml distilled water, store at 4 o C. BILAYER CHANNEL RECONSTITUTION (2) Cs solution: (Cesium Methanesulfonate) 1) 50 mm Cs + solution 0.209 MOPS, 10mM
GPCR. 2. Briefly describe the steps in PKA activation by a GPCR signal. You are encouraged to include a sketch.
 Biochemical Signaling Many of the most critical biochemical signaling pathways originate with an extracellular signal being recognized by a GPCR or a RTK. In this activity, we will explore these two signaling
Biochemical Signaling Many of the most critical biochemical signaling pathways originate with an extracellular signal being recognized by a GPCR or a RTK. In this activity, we will explore these two signaling
Signal Transduction: G-Protein Coupled Receptors
 Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
Gas Exchange in the Tissues
 Gas Exchange in the Tissues As the systemic arterial blood enters capillaries throughout the body, it is separated from the interstitial fluid by only the thin capillary wall, which is highly permeable
Gas Exchange in the Tissues As the systemic arterial blood enters capillaries throughout the body, it is separated from the interstitial fluid by only the thin capillary wall, which is highly permeable
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
Biol220 Cell Signalling Cyclic AMP the classical secondary messenger
 Biol220 Cell Signalling Cyclic AMP the classical secondary messenger The classical secondary messenger model of intracellular signalling A cell surface receptor binds the signal molecule (the primary
Biol220 Cell Signalling Cyclic AMP the classical secondary messenger The classical secondary messenger model of intracellular signalling A cell surface receptor binds the signal molecule (the primary
Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants
 Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants Species UDP-2,3- diacylglucosamine hydrolase specific activity (nmol min -1 mg -1 ) Fold vectorcontrol specific
Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants Species UDP-2,3- diacylglucosamine hydrolase specific activity (nmol min -1 mg -1 ) Fold vectorcontrol specific
Aminoglycoside activity observed on single pre-translocation ribosome complexes
 correction notice Nat. Chem. Biol. 6, 54 62 (2010) Aminoglycoside activity observed on single pre-translocation ribosome complexes Michael B Feldman, Daniel S Terry, Roger B Altman & Scott C Blanchard
correction notice Nat. Chem. Biol. 6, 54 62 (2010) Aminoglycoside activity observed on single pre-translocation ribosome complexes Michael B Feldman, Daniel S Terry, Roger B Altman & Scott C Blanchard
Physical effects underlying the transition from primitive to modern cell membranes
 Physical effects underlying the transition from primitive to modern cell membranes Itay Budin and Jack W. Szostak* *To whom correspondence should be addressed. Email: szostak@molbio.mgh.harvard.edu This
Physical effects underlying the transition from primitive to modern cell membranes Itay Budin and Jack W. Szostak* *To whom correspondence should be addressed. Email: szostak@molbio.mgh.harvard.edu This
Signal Transduction Cascades
 Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
SelectScreen Biochemical Kinase Profiling Service
 Page 1 of 11 ASSAY THEORY 2 ADAPTA ASSAY CONDITIONS 3 ADAPTA ASSAY CONTROLS 4 ADAPTA DATA ANALYSIS 5 KINASE-SPECIFIC ASSAY CONDITIONS 6 CAMK1 (CaMK1) 6 CDK7/cyclin H/MNAT1 6 CDK9/cyclin T1 6 CHUK (IKK
Page 1 of 11 ASSAY THEORY 2 ADAPTA ASSAY CONDITIONS 3 ADAPTA ASSAY CONTROLS 4 ADAPTA DATA ANALYSIS 5 KINASE-SPECIFIC ASSAY CONDITIONS 6 CAMK1 (CaMK1) 6 CDK7/cyclin H/MNAT1 6 CDK9/cyclin T1 6 CHUK (IKK
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
ASSAY THEORY 2 ADAPTA ASSAY CONDITIONS 3 ADAPTA ASSAY CONTROLS 4 ADAPTA DATA ANALYSIS 5
 Page 1 of 11 ASSAY THEORY 2 ADAPTA ASSAY CONDITIONS 3 ADAPTA ASSAY CONTROLS 4 ADAPTA DATA ANALYSIS 5 KINASE-SPECIFIC ASSAY CONDITIONS 6 CAMK1 (CaMK1) 6 CDK4/cyclin D1 6 CDK4/cyclin D3 6 CDK6/cyclin D1
Page 1 of 11 ASSAY THEORY 2 ADAPTA ASSAY CONDITIONS 3 ADAPTA ASSAY CONTROLS 4 ADAPTA DATA ANALYSIS 5 KINASE-SPECIFIC ASSAY CONDITIONS 6 CAMK1 (CaMK1) 6 CDK4/cyclin D1 6 CDK4/cyclin D3 6 CDK6/cyclin D1
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
MCB II MCDB 3451 Exam 1 Spring, minutes, close everything and be concise!
 MCB II MCDB 3451 Exam 1 Spring, 2016 50 minutes, close everything and be concise! Name ID NOTE: QUESTIONS ARE NOT ALL WORTH THE SAME POINTS Total (100) Grade EXAM 1, 2016 MCBII Name 1. Which is UNIQUE
MCB II MCDB 3451 Exam 1 Spring, 2016 50 minutes, close everything and be concise! Name ID NOTE: QUESTIONS ARE NOT ALL WORTH THE SAME POINTS Total (100) Grade EXAM 1, 2016 MCBII Name 1. Which is UNIQUE
Chapter 12: Membranes. Voet & Voet: Pages
 Chapter 12: Membranes Voet & Voet: Pages 390-415 Slide 1 Membranes Essential components of all living cells (define boundry of cells) exclude toxic ions and compounds; accumulation of nutrients energy
Chapter 12: Membranes Voet & Voet: Pages 390-415 Slide 1 Membranes Essential components of all living cells (define boundry of cells) exclude toxic ions and compounds; accumulation of nutrients energy
Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow
 SUPPLEMENTARY DATA Supplementary Figure Legends Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow cytometry analysis of PMVs labelled with annexin-v-pe (Guava technologies)
SUPPLEMENTARY DATA Supplementary Figure Legends Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow cytometry analysis of PMVs labelled with annexin-v-pe (Guava technologies)
CHAPTER 4. Tryptophan fluorescence quenching by brominated lipids
 CHAPTER 4 Tryptophan fluorescence quenching by brominated lipids 102 4.1 INTRODUCTION The structure and dynamics of biological macromolecules have been widely studied with fluorescence quenching. The accessibility
CHAPTER 4 Tryptophan fluorescence quenching by brominated lipids 102 4.1 INTRODUCTION The structure and dynamics of biological macromolecules have been widely studied with fluorescence quenching. The accessibility
Monitoring the waiting time sequence of single Ras
 Monitoring the waiting time sequence of single Ras GTPase activation events using liposome functionalized zero-mode waveguides Supporting Information Sune M. Christensen, Meredith G. Triplet, Christopher
Monitoring the waiting time sequence of single Ras GTPase activation events using liposome functionalized zero-mode waveguides Supporting Information Sune M. Christensen, Meredith G. Triplet, Christopher
CHEM 527 SECOND EXAM FALL 2006
 CEM 527 SECD EXAM FALL 2006 YUR AME: TES: 1. Where appropriate please show work if in doubt show it anyway. 2. Pace yourself you may want to do the easier questions first. 3. Please note the point value
CEM 527 SECD EXAM FALL 2006 YUR AME: TES: 1. Where appropriate please show work if in doubt show it anyway. 2. Pace yourself you may want to do the easier questions first. 3. Please note the point value
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/6/e1700338/dc1 Supplementary Materials for HIV virions sense plasma membrane heterogeneity for cell entry Sung-Tae Yang, Alex J. B. Kreutzberger, Volker Kiessling,
advances.sciencemag.org/cgi/content/full/3/6/e1700338/dc1 Supplementary Materials for HIV virions sense plasma membrane heterogeneity for cell entry Sung-Tae Yang, Alex J. B. Kreutzberger, Volker Kiessling,
PHSI3009 Frontiers in Cellular Physiology 2017
 Overview of PHSI3009 L2 Cell membrane and Principles of cell communication L3 Signalling via G protein-coupled receptor L4 Calcium Signalling L5 Signalling via Growth Factors L6 Signalling via small G-protein
Overview of PHSI3009 L2 Cell membrane and Principles of cell communication L3 Signalling via G protein-coupled receptor L4 Calcium Signalling L5 Signalling via Growth Factors L6 Signalling via small G-protein
Supplementary Information
 Supplementary Information Supplementary Figure 1. Fabrication processes of bud-mimicking PDMS patterns based on a combination of colloidal, soft, and photo-lithography a-d, The polystyrene (PS) particles
Supplementary Information Supplementary Figure 1. Fabrication processes of bud-mimicking PDMS patterns based on a combination of colloidal, soft, and photo-lithography a-d, The polystyrene (PS) particles
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Nature Biotechnology: doi: /nbt.3828
 Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
Electronic Supporting Information
 Modulation of raft domains in a lipid bilayer by boundary-active curcumin Manami Tsukamoto a, Kenichi Kuroda* b, Ayyalusamy Ramamoorthy* c, Kazuma Yasuhara* a Electronic Supporting Information Contents
Modulation of raft domains in a lipid bilayer by boundary-active curcumin Manami Tsukamoto a, Kenichi Kuroda* b, Ayyalusamy Ramamoorthy* c, Kazuma Yasuhara* a Electronic Supporting Information Contents
Lipid surface concentration estimation, conversion of fluorescence
 Lipid surface concentration estimation, conversion of fluorescence intensity, and data analysis In order to be able to fit the catalysis data with the kinetic model we proposed, the fluorescence intensity
Lipid surface concentration estimation, conversion of fluorescence intensity, and data analysis In order to be able to fit the catalysis data with the kinetic model we proposed, the fluorescence intensity
BIOLOGY 103 Spring 2001 MIDTERM LAB SECTION
 BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
MEMBRANE STRUCTURE. Lecture 8. Biology Department Concordia University. Dr. S. Azam BIOL 266/
 1 MEMBRANE STRUCTURE Lecture 8 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Plasma Membrane 2 Plasma membrane: The outer boundary of the cell that separates it from the world
1 MEMBRANE STRUCTURE Lecture 8 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Plasma Membrane 2 Plasma membrane: The outer boundary of the cell that separates it from the world
Chapter 20. Cell - Cell Signaling: Hormones and Receptors. Three general types of extracellular signaling. endocrine signaling. paracrine signaling
 Chapter 20 Cell - Cell Signaling: Hormones and Receptors Three general types of extracellular signaling endocrine signaling paracrine signaling autocrine signaling Endocrine Signaling - signaling molecules
Chapter 20 Cell - Cell Signaling: Hormones and Receptors Three general types of extracellular signaling endocrine signaling paracrine signaling autocrine signaling Endocrine Signaling - signaling molecules
Biological Membranes. Lipid Membranes. Bilayer Permeability. Common Features of Biological Membranes. A highly selective permeability barrier
 Biological Membranes Structure Function Composition Physicochemical properties Self-assembly Molecular models Lipid Membranes Receptors, detecting the signals from outside: Light Odorant Taste Chemicals
Biological Membranes Structure Function Composition Physicochemical properties Self-assembly Molecular models Lipid Membranes Receptors, detecting the signals from outside: Light Odorant Taste Chemicals
Biology. Slide 1 / 74. Slide 2 / 74. Slide 3 / 74. Membranes. Vocabulary
 Slide 1 / 74 Slide 2 / 74 iology Membranes 2015-10-28 www.njctl.org Vocabulary Slide 3 / 74 active transport carrier protein channel protein concentration gradient diffusion enzymatic activity facilitated
Slide 1 / 74 Slide 2 / 74 iology Membranes 2015-10-28 www.njctl.org Vocabulary Slide 3 / 74 active transport carrier protein channel protein concentration gradient diffusion enzymatic activity facilitated
Biology 2180 Laboratory #3. Enzyme Kinetics and Quantitative Analysis
 Biology 2180 Laboratory #3 Name Introduction Enzyme Kinetics and Quantitative Analysis Catalysts are agents that speed up chemical processes and the catalysts produced by living cells are called enzymes.
Biology 2180 Laboratory #3 Name Introduction Enzyme Kinetics and Quantitative Analysis Catalysts are agents that speed up chemical processes and the catalysts produced by living cells are called enzymes.
Key Concepts - All Cells Use Energy Energy Conversions - Reactions Absorb or Release Energy Endergonic, Exergonic - ATP is Cellular Energy
 Key Concepts - All Cells Use Energy Energy Conversions - Reactions Absorb or Release Energy Endergonic, Exergonic - ATP is Cellular Energy ATP Cycle - Enzymes Speed Up Reactions Enzyme Function, Factors
Key Concepts - All Cells Use Energy Energy Conversions - Reactions Absorb or Release Energy Endergonic, Exergonic - ATP is Cellular Energy ATP Cycle - Enzymes Speed Up Reactions Enzyme Function, Factors
BIOCHEMISTRY 460 FIRST HOUR EXAMINATION FORM A (yellow) ANSWER KEY February 11, 2008
 WRITE YOUR AND I.D. NUMBER LEGIBLY ON EVERY PAGE PAGES WILL BE SEPARATED FOR GRADING! CHECK TO BE SURE YOU HAVE 6 PAGES, (print): ANSWERS INCLUDING COVER PAGE. I swear/affirm that I have neither given
WRITE YOUR AND I.D. NUMBER LEGIBLY ON EVERY PAGE PAGES WILL BE SEPARATED FOR GRADING! CHECK TO BE SURE YOU HAVE 6 PAGES, (print): ANSWERS INCLUDING COVER PAGE. I swear/affirm that I have neither given
Dynamic Partitioning of a GPI-Anchored Protein in Glycosphingolipid-Rich Microdomains Imaged by Single-Quantum Dot Tracking
 Additional data for Dynamic Partitioning of a GPI-Anchored Protein in Glycosphingolipid-Rich Microdomains Imaged by Single-Quantum Dot Tracking Fabien Pinaud 1,3, Xavier Michalet 1,3, Gopal Iyer 1, Emmanuel
Additional data for Dynamic Partitioning of a GPI-Anchored Protein in Glycosphingolipid-Rich Microdomains Imaged by Single-Quantum Dot Tracking Fabien Pinaud 1,3, Xavier Michalet 1,3, Gopal Iyer 1, Emmanuel
Motor coupling through lipid membranes enhances transport velocities for ensembles of myosin Va
 Motor coupling through lipid membranes enhances transport velocities for ensembles of myosin Va Shane R. Nelson, Kathleen M. Trybus, and David M. Warshaw Department of Molecular Physiology and iophysics,
Motor coupling through lipid membranes enhances transport velocities for ensembles of myosin Va Shane R. Nelson, Kathleen M. Trybus, and David M. Warshaw Department of Molecular Physiology and iophysics,
Enzymes Part III: regulation II. Dr. Mamoun Ahram Summer, 2017
 Enzymes Part III: regulation II Dr. Mamoun Ahram Summer, 2017 Advantage This is a major mechanism for rapid and transient regulation of enzyme activity. A most common mechanism is enzyme phosphorylation
Enzymes Part III: regulation II Dr. Mamoun Ahram Summer, 2017 Advantage This is a major mechanism for rapid and transient regulation of enzyme activity. A most common mechanism is enzyme phosphorylation
μ i = chemical potential of species i C i = concentration of species I
 BIOE 459/559: Cell Engineering Membrane Permeability eferences: Water Movement Through ipid Bilayers, Pores and Plasma Membranes. Theory and eality, Alan Finkelstein, 1987 Membrane Permeability, 100 Years
BIOE 459/559: Cell Engineering Membrane Permeability eferences: Water Movement Through ipid Bilayers, Pores and Plasma Membranes. Theory and eality, Alan Finkelstein, 1987 Membrane Permeability, 100 Years
3) How many different amino acids are proteogenic in eukaryotic cells? A) 12 B) 20 C) 25 D) 30 E) None of the above
 Suggesting questions for Biochemistry 1 and 2 and clinical biochemistry 1) Henderson Hasselbalch Equation shows: A) The relationship between ph and the concentration of an acid and its conjugate base B)
Suggesting questions for Biochemistry 1 and 2 and clinical biochemistry 1) Henderson Hasselbalch Equation shows: A) The relationship between ph and the concentration of an acid and its conjugate base B)
Cell Signaling (part 1)
 15 Cell Signaling (part 1) Introduction Bacteria and unicellular eukaryotes respond to environmental signals and to signaling molecules secreted by other cells for mating and other communication. In multicellular
15 Cell Signaling (part 1) Introduction Bacteria and unicellular eukaryotes respond to environmental signals and to signaling molecules secreted by other cells for mating and other communication. In multicellular
Comercial Rafer in Zaragoza
 Comercial Rafer in Zaragoza 06.04.2017 ~A novel affinity method to isolate and identify intact Exosomes~ Taku Funakoshi (Mr.) Wako Chemicals GmbH Laboratory Chemicals Division Manager Research fields of
Comercial Rafer in Zaragoza 06.04.2017 ~A novel affinity method to isolate and identify intact Exosomes~ Taku Funakoshi (Mr.) Wako Chemicals GmbH Laboratory Chemicals Division Manager Research fields of
Shape Transformations of Lipid Bilayers Following Rapid Cholesterol
 Biophysical Journal, Volume 111 Supplemental Information Shape Transformations of Lipid Bilayers Following Rapid Cholesterol Uptake Mohammad Rahimi, David Regan, Marino Arroyo, Anand Bala Subramaniam,
Biophysical Journal, Volume 111 Supplemental Information Shape Transformations of Lipid Bilayers Following Rapid Cholesterol Uptake Mohammad Rahimi, David Regan, Marino Arroyo, Anand Bala Subramaniam,
AMPK Phosphorylation Assay Kit
 AMPK Phosphorylation Assay Kit Catalog Number KA3789 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
AMPK Phosphorylation Assay Kit Catalog Number KA3789 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) (b) (c) (d) (e) (f) (g) .
 Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Biosignals, Chapter 8, rearranged, Part I
 Biosignals, Chapter 8, rearranged, Part I Nicotinic Acetylcholine Receptor: A Ligand-Binding Ion Channel Classes of Receptor Proteins in Eukaryotes, Heterotrimeric G Proteins Signaling View the Heterotrimeric
Biosignals, Chapter 8, rearranged, Part I Nicotinic Acetylcholine Receptor: A Ligand-Binding Ion Channel Classes of Receptor Proteins in Eukaryotes, Heterotrimeric G Proteins Signaling View the Heterotrimeric
MAE 545: Lecture 14 (11/10) Mechanics of cell membranes
 MAE 545: ecture 14 (11/10) Mechanics of cell membranes Cell membranes Eukaryotic cells E. Coli FIBROBAST 10 mm E. COI nuclear pore complex 1 mm inner membrane plasma membrane secretory complex ribosome
MAE 545: ecture 14 (11/10) Mechanics of cell membranes Cell membranes Eukaryotic cells E. Coli FIBROBAST 10 mm E. COI nuclear pore complex 1 mm inner membrane plasma membrane secretory complex ribosome
Identification of influential proteins in the classical retinoic acid signaling pathway
 Ghaffari and Petzold Theoretical Biology and Medical Modelling (2018) 15:16 https://doi.org/10.1186/s12976-018-0088-7 RESEARCH Open Access Identification of influential proteins in the classical retinoic
Ghaffari and Petzold Theoretical Biology and Medical Modelling (2018) 15:16 https://doi.org/10.1186/s12976-018-0088-7 RESEARCH Open Access Identification of influential proteins in the classical retinoic
Chapter 14 - Electron Transport and Oxidative Phosphorylation
 Chapter 14 - Electron Transport and Oxidative Phosphorylation The cheetah, whose capacity for aerobic metabolism makes it one of the fastest animals Prentice Hall c2002 Chapter 14 1 14.4 Oxidative Phosphorylation
Chapter 14 - Electron Transport and Oxidative Phosphorylation The cheetah, whose capacity for aerobic metabolism makes it one of the fastest animals Prentice Hall c2002 Chapter 14 1 14.4 Oxidative Phosphorylation
[3]
![[3] [3]](/thumbs/73/69261795.jpg) 1 (a) Describe the structure of a plasma (cell surface) membrane............................... [3] (b) A student investigated the movement of substances through the cell surface membrane of yeast cells
1 (a) Describe the structure of a plasma (cell surface) membrane............................... [3] (b) A student investigated the movement of substances through the cell surface membrane of yeast cells
Single patch chip for planar lipid bilayer assays: Ion channels characterization and screening
 RTN Mid-Term Activity Molecular basis of antibiotic translocation Single patch chip for planar lipid bilayer assays: Ion channels characterization and screening Mohamed Kreir April 2008 Overview Planar
RTN Mid-Term Activity Molecular basis of antibiotic translocation Single patch chip for planar lipid bilayer assays: Ion channels characterization and screening Mohamed Kreir April 2008 Overview Planar
Notes 2-4. Chemical Reactions and Enzymes
 Notes 2-4 Chemical Reactions and Enzymes Chemical Reaction: A process that changes one set of chemicals into another set of chemicals Reactants: Elements entered into the reaction Products: Elements or
Notes 2-4 Chemical Reactions and Enzymes Chemical Reaction: A process that changes one set of chemicals into another set of chemicals Reactants: Elements entered into the reaction Products: Elements or
Supplementary information. Membrane curvature enables N-Ras lipid anchor sorting to. liquid-ordered membrane phases
 Supplementary information Membrane curvature enables N-Ras lipid anchor sorting to liquid-ordered membrane phases Jannik Bruun Larsen 1,2,3, Martin Borch Jensen 1,2,3,6, Vikram K. Bhatia 1,2,3,7, Søren
Supplementary information Membrane curvature enables N-Ras lipid anchor sorting to liquid-ordered membrane phases Jannik Bruun Larsen 1,2,3, Martin Borch Jensen 1,2,3,6, Vikram K. Bhatia 1,2,3,7, Søren
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/12/e1601838/dc1 Supplementary Materials for General and programmable synthesis of hybrid liposome/metal nanoparticles Jin-Ho Lee, Yonghee Shin, Wooju Lee, Keumrai
advances.sciencemag.org/cgi/content/full/2/12/e1601838/dc1 Supplementary Materials for General and programmable synthesis of hybrid liposome/metal nanoparticles Jin-Ho Lee, Yonghee Shin, Wooju Lee, Keumrai
ELECTRONIC SUPPORTING INFORMATION
 ELECTRONIC SUPPORTING INFORMATION Calculation of the number of integrated peptide amphiphiles and encapsulated material per liposome The number of both membrane integrated liposome and encapsulated material
ELECTRONIC SUPPORTING INFORMATION Calculation of the number of integrated peptide amphiphiles and encapsulated material per liposome The number of both membrane integrated liposome and encapsulated material
Supplementary Information. Supplementary Figures
 Supplementary Information Supplementary Figures Supplementary Figure 1: Mutational analysis of the ADP-based coupled ATPase-AK activity. (a) Proposed model for the coupled ATPase/AK reaction upon addition
Supplementary Information Supplementary Figures Supplementary Figure 1: Mutational analysis of the ADP-based coupled ATPase-AK activity. (a) Proposed model for the coupled ATPase/AK reaction upon addition
SUPPLEMENTARY DATA. Materials and Methods
 SUPPLEMENTARY DATA Materials and Methods HPLC-UV of phospholipid classes and HETE isomer determination. Fractionation of platelet lipid classes was undertaken on a Spherisorb S5W 150 x 4.6 mm column (Waters
SUPPLEMENTARY DATA Materials and Methods HPLC-UV of phospholipid classes and HETE isomer determination. Fractionation of platelet lipid classes was undertaken on a Spherisorb S5W 150 x 4.6 mm column (Waters
Structure and Activation of the Visual Pigment Rhodopsin
 Seminar 02.12.2011 Structure and Activation of the Visual Pigment Rhodopsin by Steven O.Smith, 2010 Constantin Schneider FB Physik, FU Berlin GPCR: Overview Rhodopsin is a G protein-coupled receptor (GPCR)
Seminar 02.12.2011 Structure and Activation of the Visual Pigment Rhodopsin by Steven O.Smith, 2010 Constantin Schneider FB Physik, FU Berlin GPCR: Overview Rhodopsin is a G protein-coupled receptor (GPCR)
Biology Movement across the Cell Membrane
 Biology 160 - Movement across the Cell Membrane Prelab Information Movement is one of the characteristics of life. The ability to control the movement of material across the cell membrane is an incredibly
Biology 160 - Movement across the Cell Membrane Prelab Information Movement is one of the characteristics of life. The ability to control the movement of material across the cell membrane is an incredibly
Regulation of cell function by intracellular signaling
 Regulation of cell function by intracellular signaling Objectives: Regulation principle Allosteric and covalent mechanisms, Popular second messengers, Protein kinases, Kinase cascade and interaction. regulation
Regulation of cell function by intracellular signaling Objectives: Regulation principle Allosteric and covalent mechanisms, Popular second messengers, Protein kinases, Kinase cascade and interaction. regulation
ALLOSTERIC REGULATION OF GPCR ACTIVITY BY PHOSPHOLIPIDS
 Supplementary Information ALLOSTERIC REGULATION OF GPCR ACTIVITY BY PHOSPHOLIPIDS Rosie Dawaliby 1, Cataldo Trubbia 1, Cédric Delporte 3,4, Matthieu Masureel 2, Pierre Van Antwerpen 3,4, Brian K. Kobilka
Supplementary Information ALLOSTERIC REGULATION OF GPCR ACTIVITY BY PHOSPHOLIPIDS Rosie Dawaliby 1, Cataldo Trubbia 1, Cédric Delporte 3,4, Matthieu Masureel 2, Pierre Van Antwerpen 3,4, Brian K. Kobilka
Engineering a polarity-sensitive biosensor for time-lapse imaging of apoptotic processes and degeneration
 nature methods Engineering a polarity-sensitive biosensor for time-lapse imaging of apoptotic processes and degeneration Yujin E Kim, Jeannie Chen, Jonah R Chan & Ralf Langen Supplementary figures and
nature methods Engineering a polarity-sensitive biosensor for time-lapse imaging of apoptotic processes and degeneration Yujin E Kim, Jeannie Chen, Jonah R Chan & Ralf Langen Supplementary figures and
supplementary information
 Figure S1 Nucleotide binding status of RagA mutants. Wild type and mutant forms of MycRagA was transfected into HEK293 cells and the transfected cells were labeled with 32 Pphosphate. MycRagA was immunoprecipitated
Figure S1 Nucleotide binding status of RagA mutants. Wild type and mutant forms of MycRagA was transfected into HEK293 cells and the transfected cells were labeled with 32 Pphosphate. MycRagA was immunoprecipitated
Chapter 11: Enzyme Catalysis
 Chapter 11: Enzyme Catalysis Matching A) high B) deprotonated C) protonated D) least resistance E) motion F) rate-determining G) leaving group H) short peptides I) amino acid J) low K) coenzymes L) concerted
Chapter 11: Enzyme Catalysis Matching A) high B) deprotonated C) protonated D) least resistance E) motion F) rate-determining G) leaving group H) short peptides I) amino acid J) low K) coenzymes L) concerted
Zool 3200: Cell Biology Exam 4 Part II 2/3/15
 Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Receptor mediated Signal Transduction
 Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
1. For the following reaction, at equilibrium [S] = 5 mm, [P] = 0.5 mm, and k f = 10 s -1. k f
![1. For the following reaction, at equilibrium [S] = 5 mm, [P] = 0.5 mm, and k f = 10 s -1. k f 1. For the following reaction, at equilibrium [S] = 5 mm, [P] = 0.5 mm, and k f = 10 s -1. k f](/thumbs/77/75734355.jpg) 1. For the following reaction, at equilibrium [S] = 5 mm, [P] = 0.5 mm, and k f = 10 s -1. S k f k r P a) Calculate K eq (the equilibrium constant) and k r. b) A catalyst increases k f by a factor of 10
1. For the following reaction, at equilibrium [S] = 5 mm, [P] = 0.5 mm, and k f = 10 s -1. S k f k r P a) Calculate K eq (the equilibrium constant) and k r. b) A catalyst increases k f by a factor of 10
Methods and Materials
 a division of Halcyonics GmbH Anna-Vandenhoeck-Ring 5 37081 Göttingen, Germany Application Note Micostructured lipid bilayers ANDREAS JANSHOFF 1), MAJA GEDIG, AND SIMON FAISS Fig.1: Thickness map of microstructured
a division of Halcyonics GmbH Anna-Vandenhoeck-Ring 5 37081 Göttingen, Germany Application Note Micostructured lipid bilayers ANDREAS JANSHOFF 1), MAJA GEDIG, AND SIMON FAISS Fig.1: Thickness map of microstructured
2 A mechanism determining the stability of echinocytes
 Membrane shear elasticity and stability of spiculated red cells A. Iglic Faculty of Electrical and Computer Engineering, University of Ljublijana, Trzaska 25, 61000 Ljublijana, Slovenia 8291 Abstract In
Membrane shear elasticity and stability of spiculated red cells A. Iglic Faculty of Electrical and Computer Engineering, University of Ljublijana, Trzaska 25, 61000 Ljublijana, Slovenia 8291 Abstract In
MCB*4010 Midterm Exam / Winter 2008
 MCB*4010 Midterm Exam / Winter 2008 Name: ID: Instructions: Answer all 4 questions. The number of marks for each question indicates how many points you need to provide. Write your answers in point form,
MCB*4010 Midterm Exam / Winter 2008 Name: ID: Instructions: Answer all 4 questions. The number of marks for each question indicates how many points you need to provide. Write your answers in point form,
Chromatography on Immobilized Artificial Membrane
 Chromatography on Immobilized Artificial Membrane Principles of measuring compounds affinity to phospholipids using immobilized artificial membrane chromatography Chromatography on Immobilized Artificial
Chromatography on Immobilized Artificial Membrane Principles of measuring compounds affinity to phospholipids using immobilized artificial membrane chromatography Chromatography on Immobilized Artificial
A Homogeneous Phosphoinositide 3-Kinase Assay on Phospholipid FlashPlate Platforms. Busi Maswoswe, Hao Xie, Pat Kasila and Li-an Yeh
 A Homogeneous Phosphoinositide 3-Kinase Assay on Phospholipid FlashPlate Platforms Busi Maswoswe, Hao Xie, Pat Kasila and Li-an Yeh Abstract Phosphoinositide 3-kinases (PI 3-kinase) consist of a family
A Homogeneous Phosphoinositide 3-Kinase Assay on Phospholipid FlashPlate Platforms Busi Maswoswe, Hao Xie, Pat Kasila and Li-an Yeh Abstract Phosphoinositide 3-kinases (PI 3-kinase) consist of a family
Selective protection of an ARF1-GTP signaling axis by a bacterial scaffold induces bidirectional trafficking arrest.
 Selective protection of an ARF1-GTP signaling axis by a bacterial scaffold induces bidirectional trafficking arrest. Andrey S. Selyunin, L. Evan Reddick, Bethany A. Weigele, and Neal M. Alto Supplemental
Selective protection of an ARF1-GTP signaling axis by a bacterial scaffold induces bidirectional trafficking arrest. Andrey S. Selyunin, L. Evan Reddick, Bethany A. Weigele, and Neal M. Alto Supplemental
Name: Student Number
 UNIVERSITY OF GUELPH CHEM 454 ENZYMOLOGY Winter 2003 Quiz #1: February 13, 2003, 11:30 13:00 Instructor: Prof R. Merrill Instructions: Time allowed = 80 minutes. Total marks = 34. This quiz represents
UNIVERSITY OF GUELPH CHEM 454 ENZYMOLOGY Winter 2003 Quiz #1: February 13, 2003, 11:30 13:00 Instructor: Prof R. Merrill Instructions: Time allowed = 80 minutes. Total marks = 34. This quiz represents
Chapter 1 Membrane Structure and Function
 Chapter 1 Membrane Structure and Function Architecture of Membranes Subcellular fractionation techniques can partially separate and purify several important biological membranes, including the plasma and
Chapter 1 Membrane Structure and Function Architecture of Membranes Subcellular fractionation techniques can partially separate and purify several important biological membranes, including the plasma and
This week s topic will be: Evidence for the Fluid Mosaic Model. Developing theories, testing hypotheses and techniques for visualizing cells
 Tutorials, while not mandatory, will allow you to improve your final grade in this course. Thank you for your attendance to date. These notes are not a substitute for the discussions that we will have
Tutorials, while not mandatory, will allow you to improve your final grade in this course. Thank you for your attendance to date. These notes are not a substitute for the discussions that we will have
Supplemental Data Figure S1 Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells.
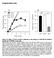 Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Instructions. Fuse-It-mRNA easy. Shipping and Storage. Overview. Kit Contents. Specifications. Note: Important Guidelines
 Membrane fusion is a highly efficient method for transfecting various molecules and particles into mammalian cells, even into sensitive and primary cells. The Fuse-It reagents are cargo-specific liposomal
Membrane fusion is a highly efficient method for transfecting various molecules and particles into mammalian cells, even into sensitive and primary cells. The Fuse-It reagents are cargo-specific liposomal
Annexin V-PE Apoptosis Detection Kit
 Annexin V-PE Apoptosis Detection Kit Catalog Number KA0716 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
Annexin V-PE Apoptosis Detection Kit Catalog Number KA0716 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
Data Sheet. PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002
![Data Sheet. PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002 Data Sheet. PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002](/thumbs/79/80181612.jpg) Data Sheet PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002 DESCRIPTION: The PCSK9[Biotinylated]-LDLR Binding Assay Kit is designed for screening and profiling purposes. PCSK9 is known to function
Data Sheet PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002 DESCRIPTION: The PCSK9[Biotinylated]-LDLR Binding Assay Kit is designed for screening and profiling purposes. PCSK9 is known to function
Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013
 Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Lipids: Membranes Testing Fluid Mosaic Model of Membrane Structure: Cellular Fusion
 Models for Membrane Structure NEW MODEL (1972) Fluid Mosaic Model proposed by Singer & Nicholson Lipids form a viscous, twodimensional solvent into which proteins are inserted and integrated more or less
Models for Membrane Structure NEW MODEL (1972) Fluid Mosaic Model proposed by Singer & Nicholson Lipids form a viscous, twodimensional solvent into which proteins are inserted and integrated more or less
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. Schematic of Ras biochemical coupled assay.
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. Schematic of Ras biochemical coupled assay.
Biology Movement Across the Cell Membrane
 Biology 160 - Movement Across the Cell Membrane Prelab Information Movement is one of the characteristics of life. The ability to control the movement of material across the cell membrane is an incredibly
Biology 160 - Movement Across the Cell Membrane Prelab Information Movement is one of the characteristics of life. The ability to control the movement of material across the cell membrane is an incredibly
Translating Molecular Detection into a Temperature Test Using. Target-Responsive Smart Thermometer
 Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2018 Supporting Information Translating Molecular Detection into a Temperature Test Using Target-Responsive
Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2018 Supporting Information Translating Molecular Detection into a Temperature Test Using Target-Responsive
Signaling. Dr. Sujata Persad Katz Group Centre for Pharmacy & Health research
 Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
Which DNA sequence is most likely to form a hairpin structure? x indicates any nucleotide.
 Which DNA sequence is most likely to form a hairpin structure? x indicates any nucleotide. A. xxxgtcagtxxxxtatgcgxxx B. xxxtcgtatxxxxgtccgaxxx C. xxxcactgtxxxxgtactgxxx D. xxxgtcagtxxxxcctagaxxx E. xxxgtcatcxxxxgatgacxxx
Which DNA sequence is most likely to form a hairpin structure? x indicates any nucleotide. A. xxxgtcagtxxxxtatgcgxxx B. xxxtcgtatxxxxgtccgaxxx C. xxxcactgtxxxxgtactgxxx D. xxxgtcagtxxxxcctagaxxx E. xxxgtcatcxxxxgatgacxxx
G-Protein Signaling. Introduction to intracellular signaling. Dr. SARRAY Sameh, Ph.D
 G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
