Supporting Information for: New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans
|
|
|
- Barbra Franklin
- 6 years ago
- Views:
Transcription
1 Supporting Information for: New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans Matthew T. Henke a, Alexandra A. Soukup b, Anthony W. Goering a, Ryan A. McClure c, Regan J. Thomson c, Nancy P. Keller b,d,e and Neil L. Kelleher a,c,f a Department of Molecular Biosciences, c Department of Chemistry and f Feinberg School of Medicine, Northwestern University, Evanston, IL 60208, USA b Department of Genetics, d Department of Bacteriology, and e Department of Medical Microbiology and Immunology University of Wisconsin, Madison, Wisconsin, 53706, USA Corresponding author: Neil L. Kelleher; (847) ; 2170 Campus Dr. Evanston, IL 60208; n-kelleher@northwestern.edu Table S1. Summary of NMR data for aspercryptins A1 and A2 Figure S1. Stereochemical determination of aspercryptins A1 and A2 Figure S2. Example of stable isotopes labeling with aspercryptin A1 Figure S3. Annotated MS/MS spectrum of aspercryptin A1 Figure S4. Annotated MS/MS spectrum of aspercryptin A2 Table S2. Summary of NMR data for aspercryptins A1 and A2 Figure S5. Quantitation of all aspercryptins in wildtype and rpda KD Figure S6. Putative structures for 13 additional aspercryptins Table S3. ORF analysis of genes in the aspercryptin biosynthetic gene cluster Figure S7. Transcript levels of aspercryptin gene cluster in rpda KD and overexpression of transcription factor atnn Figure S8. Conservation of the aspercryptin biosynthetic gene cluster across Aspergilli Figure S9. A proposal for the biosynthesis of the aspercryptins. Figure S10. Interrupted organization of fatty acid genes in the aspercryptin gene cluster Figure S11. Structures of members from other natural products that contain an α-amino fatty acid Table S4. Adenylation domain predictions for AN7884/AtnA Table S5. Strains used in this study NMR data for the aspercryptins and of synthetic fatty amino acid standards References 1. Chiang, Y. M., Ahuja, M., Oakley, C. E., Entwistle, R., Asokan, A., Zutz, C., Wang, C. C., and Oakley, B. R. (2016) Development of Genetic Dereplication Strains in Aspergillus nidulans Results in the Discovery of Aspercryptin, Angewandte Chemie 55, Rottig, M., Medema, M. H., Blin, K., Weber, T., Rausch, C., and Kohlbacher, O. (2011) NRPSpredictor2--a web server for predicting NRPS adenylation domain specificity, Nucleic acids research 39, W
2 Table S1. 1 H (600 MHz) and 13 C (150 MHz) NMR data for aspercryptin A1 and A2 in DMSOd 6. Full NMR data can be found below. position δ H (J, Hz) δ C position δ H (J, Hz) δ C serine 2-amino-octanoic acid (2aoa) (dd, 6.2, 4.3) (dd) (dd, 11, 4.7); 3.42 (dd, 10.6, 6.6) ; alanine a ; b threonine asparagine (broad) 25 8 (t) (d) (m) ; (d) (s, broad); 6.85 (s, broad) isoleucine (d) 2-amino-dodecanol (2adol) (t) (m, broad) (dd, 11,5.4); 3.2 (dd, 5.4, 11) ; ; ; b c a These values are for the alanine residue in aspercryptin A2 instead of the initial serine. b These positions are overlapping and cannot be disambiguated. c These values are based on synthetic standard of 2-amino-dodecanol. 2
3 Figure S1. Stereochemical determination of the aspercryptins following derivatization with Marfey s reagent and comparison with amino acid standards by HPLC-MS. (A) D- and L-serine standards (Selected Ion Chromatogram, or SIC: m/z ±10 ppm) confirm the serine of aspercryptin A1 is the D-enantiomer (B) D-/L-alanine standard (SIC: m/z ±10 ppm) confirms the alanine of aspercryptin A2 is actually a mixture of the D- and L-enantiomers. The epimer with L-alanine is epi-aspercryptin A2. (C) Standards of all four stereoisomers of threonine (SIC: m/z ) confirm that D-allo-threonine is present in the aspercryptins. (D) D-/L- isoleucine standard (SIC: m/z ±10 ppm) confirms the aspercryptins contain L- isoleucine. (E) Synthetic D-/L-2-amino-octanoic acid standard (SIC: m/z ±10 ppm) confirms the aspercryptins contain L-2-amino-octanoic acid. (F) D-/L-aspartate standard (SIC: m/z ±10 ppm) confirms that the aspercryptins contain L-asparagine. (G) Synthetic D-/L- 2-amino-dodecanol standard (SIC: m/z ±10 ppm) confirms the aspercryptins contain the L-enantiomer. 3
4 Figure S2. Metabolic labeling of aspercryptin A1 with three of its amino acid precursors containing stable isotopes. Shown are the mass spectra for aspercryptin A (A) with no label, (B) with 13 C 6 15 N 1 -isoleucine, (C) 13 C 4 15 N 1 -threonine, and (D) d 3 -serine. These results support the structural assignments made for the aspercryptins by NMR and MS. Note that the isotopic pattern in panel C does not correspond to the incorporation of more than one threonine: it is complicated due to the partial conversion of the labeled-threonine to isoleucine, and subsequent incorporation of both a heavy isoleucine and a heavy threonine into aspercryptin A1. The isotope pattern of panel D shows a +2 Da incorporation, indicative of an exchange of one deuterium for a proton; this is consistent with epimerization of the Ser residue in aspercryptin A1. 4
5 Figure S3. Annotation of the MS fragmentation spectrum of aspercryptin A1 (m/z ). For clarity b-ions are in blue and y-ions are in red. Unless noted otherwise, observed mass errors for all fragment ions were <1 part-per-million (ppm). Immonium ions for the isoleucine and 2- amino-octanoic acid residues are displayed on the spectrum (low m/z ions labelled with black text at the far left). Loss of ammonia is a common neutral loss from peptides containing asparagine. 5
6 Figure S4. Annotation of the MS fragmentation spectrum of aspercryptin A2 (m/z ). For clarity b-ions are in blue and y-ions are in red. The b-ions are shifted down by exactly the mass of an oxygen, while the y-ions have their same values as they do for aspercryptin A1. This localizes the oxygen to the N-terminus, and thus represents a serine-for-alanine substitution. Unless noted otherwise, observed mass errors for all fragment ions were <1 part-per-million (ppm). Immonium ions for the isoleucine and 2-amino-octanoic acid residues are displayed on the spectrum. Loss of ammonia is a common neutral loss from peptides containing asparagine. 6
7 Table S2. The formulae and expression ratios for the 18 aspercryptins from Figure 2, including the five aspercryptins elucidated by NMR (bold) and the 13 additional aspercryptins whose putative structures are supported by comparing MS/MS spectra. a NMR structures described in this work; b NMR structures described by Chiang et al. 1 ; c canonical sequence from which derivatives have been identified by shifts in characteristic fragment ions (in blue) Name m/z Formula aspercryptin A1 a,c C 37 H 71 N 7 O 9 [rpda KD ] [wildtype] RT Structure with diagnostic ions aspercryptin A2 a (and epi-form) aspercryptin A3 aspercryptin A C 37 H 71 N 7 O C 36 H 69 N 7 O C 35 H 67 N 7 O aspercryptin A5 aspercryptin A6 aspercryptin A C 36 H 69 N 7 O C 35 H 67 N 7 O C 34 H 66 N 6 O 7 aspercryptin B1 b C 47 H 79 N 7 O 12 aspercryptin B3 b C 46 H 77 N 7 O aspercryptin B2 aspercryptin B4 aspercryptin C1 aspercryptin C3 aspercryptin C2 aspercryptin C4 aspercryptin C6 aspercryptin D C 47 H 79 N 7 O C 45 H 75 N 7 O C 39 H 73 N 7 O C 38 H 71 N 7 O C 39 H 73 N 7 O C 37 H 69 N 7 O C 37 H 69 N 7 O C 40 H 75 N 7 O
8 Figure S5. Quantitation of 17 aspercryptins described here (aspercryptin A2 and epi-aspercryptin A2 are not distinguished here). Error bars are 1 standard deviation from 3 biological replicates. 8
9 Figure S6. Putative structures for the 13 aspercryptins based on analysis of MS/MS spectral differences. Heavily annotated MS/MS spectra of aspercryptins solved by NMR (blue asterisk) were used as the basis for comparison to other metabolites (see Supporting Information Figures S3 and S4). Note that stereochemical assignment has not been made for the MS-based structures; though, they are likely match the stereocenters of the experimentally determined aspercryptins. This chart is arranged hierarchically where the letter across the top describes the N-terminus, and the number along the side described the amino acid sequence. Note that epi-aspercryptin A2 is boxed to show that it does not slot into the chart as shown. 9
10 Table S3. Description of ORFs in the aspercryptin biosynthetic gene cluster from atna (An7884) to atnn (AN7872). Gene name AspGD designation protein size (aa) Description largely provided by AspGD atna AN NRPS; A 1 T-CA 2 TE-CA 3 T-CA 4 T-CA 5 T-CA 6 T-Red 6 modules; reductase domain atnb AN YCII-related domain atnc AN Predicted transmembrane transporter 9 TM helices atnd AN NAD(P)-binding; involved in oxidoreductase activity atne AN Cytochrome P450 heme binding; N-terminus (7-29) is likely transmembrane atnf AN Fatty acid synthase, subunit α (ACP-KR-KS) without PPT domain atng AN ABC-transporter; probably involved in export of mature compound atnh AN Aminotransferase predicted for branched chain amino acid metabolism; PLP binding site atni AN RTA-like protein; may bind to toxic compounds to prevent toxicity; integral membrane protein 6 TM helices atnj AN Aminotransferase predicted for branched chain amino acid metabolism; PLP binding site atnk AN Unknown atnl AN hydroxybenzoyl-CoA thioesterase. Hits to bacterial thioesterase domains atnm AN Fatty acid synthase, subunit β (AT-ER-DH-MPT) AN Unknown atnn AN Transcription factor 10
11 Figure S7. Transcript levels experimentally establish the borders of the AN7872-AN7884 gene cluster. (A) Northern blots show the up-regulation of the AN7872-AN7884 (aspercryptin) gene cluster in rpda KD when grown under repressive (-xylose) conditions, which increases global histone acetylation through lower levels of the histone deacetylase, RpdA. (B) Northern blots show the up-regulation of the gene cluster due to overexpression of the transcription factor AN7872 (alca::an7872) under inducing conditions. 11
12 Figure S8. Conservation of the aspercryptin biosynthetic gene cluster across various Aspergillus spp. Conservation between A. nidulans and A. versicolor appears to be greatest and shows the cluster boundaries quite clearly to be atna (AN7884) to atnn (AN7872). 12
13 Figure S9. A proposed biosynthesis for aspercryptin A1. Biosynthesis of the α-amino fatty acids likely occurs in 2 phases: 1) assembly of the fatty acids octanoic and dodecanoic acids by the FAS subunits AtnF and AtnM and 2) conversion to the 2-amino fatty acids through hydroxylation by AtnE, oxidation by AtnD and transamination by AtnH or AtnJ. Unusually for lipopeptide NPs, both fatty acid monomers are loaded as α-amino fatty acids through activation by dedicated adenylation domains. 13
14 Figure S10. Depiction of the chromosomal organization of the four copies of the genetic cassette encoding Fatty Acid Synthases (FASs) in the A. nidulans genome. It appears that six atn genes interrupt the FAS elements only in the case of the aspercryptin gene cluster (top); shown in green are the aminotransferases that are putatively required for conversion of the short chain fatty acids into their α-amino acids analogs for subsequent incorporation by NRPS to produce the aspercryptins. 14
15 Figure S11. Structures of known classes of lipopeptides where lipid portion is thought to be incorporated as an α-amino acid by an NRPS. All these compounds are cyclic, unlike the aspercryptins, which represent the first backward lipopeptide with an α-amino fatty acid. Table S4. Substrate predictions for the adenylation domains of AtnA (AN7884) by NRPSredictor2 2, which typically does not provide high-confidence predictions for substrates of fungal A domains. Module Stachelhaus code 1 D L G L S G M I L K 15 Nearest neighbor prediction 1 2-amino adipic acid Score Proposed substrate 50% serine/alanine 2 2 D V G F T G C V Y K cysteine 60% threonine 3 D A L F M G G V Y K leucine 60% isoleucine 4 D A M M V G V M I K 3,5-dihydroxyphenyl-glycine 50% 2-aminooctanoic acid 5 D V A F T G S I W K valine 60% asparagine 2-aminododecanoic acid 1 The nearest neighbor prediction is based on the most similar sequence to the extracted Stachelhaus code within a 6 D V L F I G G V A K cysteine 70% database of annotated A-domains. 2 In the case of aspercryptin A2 biosynthesis, alanine would be activated by the A- domain. Table S5. Strains used in this study
16 Strain Name Genotype RJMP1.1 TAAS393.2 TAAS394.2 TAAS176.3 TAAS395.1 TAAS217.1 pyrg89; pyroa4; ribob2; nkua::argb; vea1 AN7884(p)::A. parasticus pyrg::alca(p)::an7884); pyrg89; pyroa4; ribob2; nkua::argb; vea1 AN7872(p)::A. parasiticus pyrg::alca(p)::an7872); pyrg89; pyroa4; ribob2; nkua::argb; vea1 AN7884:: A. fumigatus ribob; pyrg89; pyroa4; ribob2; nkua::argb; vea1 AN7872(p)::A. parasiticus pyrg::alca(p)::an7872; AN7884:: A. fumigatus ribob; pyrg89; pyroa4; ribob2; nkua::argb; vea1 AN7872(p)::A. parasiticus pyrg::alca(p)::an7872; AN7880:: A. fumigatus ribob; pyrg89; pyroa4; ribob2; nkua::argb; vea1 Parent Strain(s) RJMP1.1 RJMP1.1 RJMP1.1 TAAS393.2 TAAS393.2 Source (Palmer et al., 2013) This study This study This study This study This study 16
17 1) 1 H-NMR of aspercryptins A1 and A2 in DMSO-d 6 NMR data for the aspercryptins A1 and A2 17
18 2) 13 C-NMR of aspercryptins A1 and A2 in DMSO-d 6 18
19 3) HSQC of aspercryptins A1 and A2 in DMSO-d 6 19
20 4) band-selective HSQC of aspercryptins A1 and A2 in DMSO-d 6 20
21 5) HMBC of aspercryptins A1 and A2 in DMSO-d 6 21
22 6) COSY of aspercryptins A1 and A2 in DMSO-d 6 22
23 7) TOCSY of aspercryptins A1 and A2 in DMSO-d 6 23
24 8) 1D-TOCSYs of aspercryptins A1 and A2 in DMSO-d 6 24
25 NMR data for fatty amino acid standards used to determine stereochemistry 1 H-NMR of synthetic 2-amino-dodecanol in DMSO-d 6 25
26 13 C-NMR of synthetic 2-amino-dodecanol in DMSO-d 6 + TFA 26
27 1 H-NMR of synthetic 2-amino-octanoic acid in DMSO-d 6 + TFA 27
28 13 C-NMR of synthetic 2-amino-octanoic acid in DMSO-d 6 + TFA 28
Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis
 Supporting information Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis Martin F. Kreutzer and Markus Nett * Table of contents SI-2, SI-3 SI-4 SI-5 SI-6
Supporting information Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis Martin F. Kreutzer and Markus Nett * Table of contents SI-2, SI-3 SI-4 SI-5 SI-6
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted. residues are colored red. Numbers indicate the position of each residue.
 Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
AMINO ACIDS STRUCTURE, CLASSIFICATION, PROPERTIES. PRIMARY STRUCTURE OF PROTEINS
 AMINO ACIDS STRUCTURE, CLASSIFICATION, PROPERTIES. PRIMARY STRUCTURE OF PROTEINS Elena Rivneac PhD, Associate Professor Department of Biochemistry and Clinical Biochemistry State University of Medicine
AMINO ACIDS STRUCTURE, CLASSIFICATION, PROPERTIES. PRIMARY STRUCTURE OF PROTEINS Elena Rivneac PhD, Associate Professor Department of Biochemistry and Clinical Biochemistry State University of Medicine
Supporting Information. Lysine Propionylation to Boost Proteome Sequence. Coverage and Enable a Silent SILAC Strategy for
 Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
Nitrogen Metabolism. Overview
 Nitrogen Metabolism Pratt and Cornely Chapter 18 Overview Nitrogen assimilation Amino acid biosynthesis Nonessential aa Essential aa Nucleotide biosynthesis Amino Acid Catabolism Urea Cycle Juicy Steak
Nitrogen Metabolism Pratt and Cornely Chapter 18 Overview Nitrogen assimilation Amino acid biosynthesis Nonessential aa Essential aa Nucleotide biosynthesis Amino Acid Catabolism Urea Cycle Juicy Steak
Introduction to Peptide Sequencing
 Introduction to Peptide equencing Quadrupole Ion Traps tructural Biophysics Course December 3, 2014 12/8/14 Introduction to Peptide equencing - athan Yates 1 Why are ion traps used to sequence peptides?
Introduction to Peptide equencing Quadrupole Ion Traps tructural Biophysics Course December 3, 2014 12/8/14 Introduction to Peptide equencing - athan Yates 1 Why are ion traps used to sequence peptides?
Level and activity of D-amino acids in mouse brain tissue and blood
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 SUPPLEMENTARY INFORMATION Level and activity of D-amino acids in mouse brain tissue and blood Choyce A. Weatherly 1, Siqi Du 1, Curran Parpia 1, Polan T. Santos 2, Adam
1 2 3 4 5 6 7 8 9 10 11 12 13 14 SUPPLEMENTARY INFORMATION Level and activity of D-amino acids in mouse brain tissue and blood Choyce A. Weatherly 1, Siqi Du 1, Curran Parpia 1, Polan T. Santos 2, Adam
These are example problems, which are similar to those you may see on the final exam.
 MCB102 / Metabolism Problem Set #3 Spring 2008 These are example problems, which are similar to those you may see on the final exam. QUESTION 1: /. Circle the correct answer, but if the answer is provide
MCB102 / Metabolism Problem Set #3 Spring 2008 These are example problems, which are similar to those you may see on the final exam. QUESTION 1: /. Circle the correct answer, but if the answer is provide
For questions 1-4, match the carbohydrate with its size/functional group name:
 Chemistry 11 Fall 2013 Examination #5 PRACTICE 1 ANSWERS For the first portion of this exam, select the best answer choice for the questions below and mark the answers on your scantron. Then answer the
Chemistry 11 Fall 2013 Examination #5 PRACTICE 1 ANSWERS For the first portion of this exam, select the best answer choice for the questions below and mark the answers on your scantron. Then answer the
PHAR3316 Pharmacy biochemistry Exam #2 Fall 2010 KEY
 1. How many protons is(are) lost when the amino acid Asparagine is titrated from its fully protonated state to a fully deprotonated state? A. 0 B. 1 * C. 2 D. 3 E. none Correct Answer: C (this question
1. How many protons is(are) lost when the amino acid Asparagine is titrated from its fully protonated state to a fully deprotonated state? A. 0 B. 1 * C. 2 D. 3 E. none Correct Answer: C (this question
Table removed due to copyright reasons.
 atural Product Biosynthesis Relevance of natural products in the pharmaceutical industry (see Table 2 in Butler J. at. Prod. 2004, 67, 2141-2153.)) Table removed due to copyright reasons. 2 * a phosphopantetheinyl
atural Product Biosynthesis Relevance of natural products in the pharmaceutical industry (see Table 2 in Butler J. at. Prod. 2004, 67, 2141-2153.)) Table removed due to copyright reasons. 2 * a phosphopantetheinyl
2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry
 Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
Introduction to proteins and protein structure
 Introduction to proteins and protein structure The questions and answers below constitute an introduction to the fundamental principles of protein structure. They are all available at [link]. What are
Introduction to proteins and protein structure The questions and answers below constitute an introduction to the fundamental principles of protein structure. They are all available at [link]. What are
BIOLOGY 103 Spring 2001 MIDTERM LAB SECTION
 BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
Amino Acid Metabolism
 Amino Acid Metabolism Last Week Most of the Animal Kingdom = Lazy - Most higher organisms in the animal kindom don t bother to make all of the amino acids. - Instead, we eat things that make the essential
Amino Acid Metabolism Last Week Most of the Animal Kingdom = Lazy - Most higher organisms in the animal kindom don t bother to make all of the amino acids. - Instead, we eat things that make the essential
Nitrogen Metabolism. Overview
 Nitrogen Metabolism Pratt and Cornely Chapter 18 Overview Nitrogen assimilation Amino acid biosynthesis Nonessential aa Essential aa Nucleotide biosynthesis Amino Acid Catabolism Urea Cycle Juicy Steak
Nitrogen Metabolism Pratt and Cornely Chapter 18 Overview Nitrogen assimilation Amino acid biosynthesis Nonessential aa Essential aa Nucleotide biosynthesis Amino Acid Catabolism Urea Cycle Juicy Steak
For questions 1-4, match the carbohydrate with its size/functional group name:
 Chemistry 11 Fall 2013 Examination #5 PRACTICE 1 For the first portion of this exam, select the best answer choice for the questions below and mark the answers on your scantron. Then answer the free response
Chemistry 11 Fall 2013 Examination #5 PRACTICE 1 For the first portion of this exam, select the best answer choice for the questions below and mark the answers on your scantron. Then answer the free response
Biomolecular Mass Spectrometry
 Lipids ot different than other organic small molecules Carbohydrates Polymers of monosaccharides linked via glycosidic bonds (acetals/ ketals) many different combinationsvery interesting no time ucleic
Lipids ot different than other organic small molecules Carbohydrates Polymers of monosaccharides linked via glycosidic bonds (acetals/ ketals) many different combinationsvery interesting no time ucleic
Biochemistry - I. Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture 1 Amino Acids I
 Biochemistry - I Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture 1 Amino Acids I Hello, welcome to the course Biochemistry 1 conducted by me Dr. S Dasgupta,
Biochemistry - I Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture 1 Amino Acids I Hello, welcome to the course Biochemistry 1 conducted by me Dr. S Dasgupta,
Proteins consist in whole or large part of amino acids. Simple proteins consist only of amino acids.
 Today we begin our discussion of the structure and properties of proteins. Proteins consist in whole or large part of amino acids. Simple proteins consist only of amino acids. Conjugated proteins contain
Today we begin our discussion of the structure and properties of proteins. Proteins consist in whole or large part of amino acids. Simple proteins consist only of amino acids. Conjugated proteins contain
Lecture 11 - Biosynthesis of Amino Acids
 Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
Section 1 Proteins and Proteomics
 Section 1 Proteins and Proteomics Learning Objectives At the end of this assignment, you should be able to: 1. Draw the chemical structure of an amino acid and small peptide. 2. Describe the difference
Section 1 Proteins and Proteomics Learning Objectives At the end of this assignment, you should be able to: 1. Draw the chemical structure of an amino acid and small peptide. 2. Describe the difference
Nitrogen Metabolism. Pratt and Cornely Chapter 18
 Nitrogen Metabolism Pratt and Cornely Chapter 18 Overview Nitrogen assimilation Amino acid biosynthesis Nonessential aa Essential aa Nucleotide biosynthesis Amino Acid Catabolism Urea Cycle Juicy Steak
Nitrogen Metabolism Pratt and Cornely Chapter 18 Overview Nitrogen assimilation Amino acid biosynthesis Nonessential aa Essential aa Nucleotide biosynthesis Amino Acid Catabolism Urea Cycle Juicy Steak
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol
 Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
Molecular Biology. general transfer: occurs normally in cells. special transfer: occurs only in the laboratory in specific conditions.
 Chapter 9: Proteins Molecular Biology replication general transfer: occurs normally in cells transcription special transfer: occurs only in the laboratory in specific conditions translation unknown transfer:
Chapter 9: Proteins Molecular Biology replication general transfer: occurs normally in cells transcription special transfer: occurs only in the laboratory in specific conditions translation unknown transfer:
Supporting information
 Supporting information Rhabdopeptide/Xenortide-like Peptides from Xenorhabdus innexi with Terminal Amines Showing Potent Anti-protozoal Activity Lei Zhao,, Marcel Kaiser,, Helge B. Bode *,, Molekulare
Supporting information Rhabdopeptide/Xenortide-like Peptides from Xenorhabdus innexi with Terminal Amines Showing Potent Anti-protozoal Activity Lei Zhao,, Marcel Kaiser,, Helge B. Bode *,, Molekulare
9/6/2011. Amino Acids. C α. Nonpolar, aliphatic R groups
 Amino Acids Side chains (R groups) vary in: size shape charge hydrogen-bonding capacity hydrophobic character chemical reactivity C α Nonpolar, aliphatic R groups Glycine (Gly, G) Alanine (Ala, A) Valine
Amino Acids Side chains (R groups) vary in: size shape charge hydrogen-bonding capacity hydrophobic character chemical reactivity C α Nonpolar, aliphatic R groups Glycine (Gly, G) Alanine (Ala, A) Valine
H 2 C H 2 N C CH O N C CH 3 CH 2 H O. aspartame
 1 The addition of sucrose, table sugar, to food and drink has been linked to the increased risk of obesity and insulin resistance. Aspartame is used as an alternative to sugar. The structure of aspartame
1 The addition of sucrose, table sugar, to food and drink has been linked to the increased risk of obesity and insulin resistance. Aspartame is used as an alternative to sugar. The structure of aspartame
SUPPLEMENTARY DATA Supplementary Figure 1. Body weight and fat mass of AdicerKO mice.
 SUPPLEMENTARY DATA Supplementary Figure 1. Body weight and fat mass of AdicerKO mice. Twelve week old mice were subjected to ad libitum (AL) or dietary restriction (DR) regimens for three months. (A) Body
SUPPLEMENTARY DATA Supplementary Figure 1. Body weight and fat mass of AdicerKO mice. Twelve week old mice were subjected to ad libitum (AL) or dietary restriction (DR) regimens for three months. (A) Body
Biomimetic Chirality Sensing with Pyridoxal-5 -phosphate
 Supporting Information Biomimetic Chirality Sensing with Pyridoxal-5 -phosphate Samantha L. Pilicer, Pegah R. Bakhshi, Keith W. Bentley, Christian Wolf Department of Chemistry, Georgetown University, Washington,
Supporting Information Biomimetic Chirality Sensing with Pyridoxal-5 -phosphate Samantha L. Pilicer, Pegah R. Bakhshi, Keith W. Bentley, Christian Wolf Department of Chemistry, Georgetown University, Washington,
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the
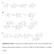 Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supplementary Information
 Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
Biochemistry 2 Recita0on Amino Acid Metabolism
 Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Student name ID # 2. (4 pts) What is the terminal electron acceptor in respiration? In photosynthesis?
 1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? 2. (4 pts) What is the terminal electron acceptor
1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? 2. (4 pts) What is the terminal electron acceptor
Figure S1: Abundance of Fe related proteins in oceanic Synechococcus sp. strain WH8102 in. Ferredoxin. Ferredoxin. Fe ABC transporter
 0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
Applying Molecular Networking for the Detection of
 Supporting Information Applying Molecular Networking for the Detection of Natural Sources and Analogues of the Selective Gq Protein Inhibitor FR900359 Raphael Reher, Markus Kuschak, Nina Heycke, Suvi Annala,
Supporting Information Applying Molecular Networking for the Detection of Natural Sources and Analogues of the Selective Gq Protein Inhibitor FR900359 Raphael Reher, Markus Kuschak, Nina Heycke, Suvi Annala,
Supplementary information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supplementary information Genomics-Driven Discovery of a Linear Lipopeptide
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Supplementary information Genomics-Driven Discovery of a Linear Lipopeptide
Glycolysis. Cellular Respiration
 Glucose is the preferred carbohydrate of cells. In solution, it can change from a linear chain to a ring. Energy is stored in the bonds of the carbohydrates. Breaking these bonds releases that energy.
Glucose is the preferred carbohydrate of cells. In solution, it can change from a linear chain to a ring. Energy is stored in the bonds of the carbohydrates. Breaking these bonds releases that energy.
For questions 1-4, match the carbohydrate with its size/functional group name:
 Chemistry 11 Fall 2009 Examination #5 ANSWER KEY For the first portion of this exam, select the best answer choice for the questions below and mark the answers on your scantron. Then answer the free response
Chemistry 11 Fall 2009 Examination #5 ANSWER KEY For the first portion of this exam, select the best answer choice for the questions below and mark the answers on your scantron. Then answer the free response
Iron depletion enhances production of antimicrobials by Pseudomonas
 Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. Angela T. Nguyen 1, Jace W. Jones 1, Max A. Ruge 1, Maureen A. Kane 1, and Amanda G. Oglesby-Sherrouse 1,2 * University of
Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. Angela T. Nguyen 1, Jace W. Jones 1, Max A. Ruge 1, Maureen A. Kane 1, and Amanda G. Oglesby-Sherrouse 1,2 * University of
I) Choose the best answer: 1- All of the following amino acids are neutral except: a) glycine. b) threonine. c) lysine. d) proline. e) leucine.
 1- All of the following amino acids are neutral except: a) glycine. b) threonine. c) lysine. d) proline. e) leucine. 2- The egg white protein, ovalbumin, is denatured in a hard-boiled egg. Which of the
1- All of the following amino acids are neutral except: a) glycine. b) threonine. c) lysine. d) proline. e) leucine. 2- The egg white protein, ovalbumin, is denatured in a hard-boiled egg. Which of the
Reactions and amino acids structure & properties
 Lecture 2: Reactions and amino acids structure & properties Dr. Sameh Sarray Hlaoui Common Functional Groups Common Biochemical Reactions AH + B A + BH Oxidation-Reduction A-H + B-OH + energy ª A-B + H
Lecture 2: Reactions and amino acids structure & properties Dr. Sameh Sarray Hlaoui Common Functional Groups Common Biochemical Reactions AH + B A + BH Oxidation-Reduction A-H + B-OH + energy ª A-B + H
Practice Problems 3. a. What is the name of the bond formed between two amino acids? Are these bonds free to rotate?
 Life Sciences 1a Practice Problems 3 1. Draw the oligopeptide for Ala-Phe-Gly-Thr-Asp. You do not need to indicate the stereochemistry of the sidechains. Denote with arrows the bonds formed between the
Life Sciences 1a Practice Problems 3 1. Draw the oligopeptide for Ala-Phe-Gly-Thr-Asp. You do not need to indicate the stereochemistry of the sidechains. Denote with arrows the bonds formed between the
Biomolecules: amino acids
 Biomolecules: amino acids Amino acids Amino acids are the building blocks of proteins They are also part of hormones, neurotransmitters and metabolic intermediates There are 20 different amino acids in
Biomolecules: amino acids Amino acids Amino acids are the building blocks of proteins They are also part of hormones, neurotransmitters and metabolic intermediates There are 20 different amino acids in
Biology. Lectures winter term st year of Pharmacy study
 Biology Lectures winter term 2008 1 st year of Pharmacy study 3 rd Lecture Chemical composition of living matter chemical basis of life. Atoms, molecules, organic compounds carbohydrates, lipids, proteins,
Biology Lectures winter term 2008 1 st year of Pharmacy study 3 rd Lecture Chemical composition of living matter chemical basis of life. Atoms, molecules, organic compounds carbohydrates, lipids, proteins,
Fundamentals of Organic Chemistry CHEM 109 For Students of Health Colleges
 Fundamentals of Organic Chemistry CHEM 109 For Students of Health Colleges Credit hrs.: (2+1) King Saud University College of Science, Chemistry Department CHEM 109 CHAPTER 9. AMINO ACIDS, PEPTIDES AND
Fundamentals of Organic Chemistry CHEM 109 For Students of Health Colleges Credit hrs.: (2+1) King Saud University College of Science, Chemistry Department CHEM 109 CHAPTER 9. AMINO ACIDS, PEPTIDES AND
MS/MS Scan Modes. Eötvös University, Budapest April 16, MS/MS Scan Modes. Árpád Somogyi. Product Ion Scan Select. Scan. Precursor Ion Scan Scan
 MS/MS Modes Árpád Somogyi Eötvös University, Budapest April 16, 2012 MS/MS Modes Product Ion Precursor Ion Neutral Loss Δ ed Reaction Monitoring (SRM) 1 modes in a triple quadrupole (QqQ) (one quadrupole
MS/MS Modes Árpád Somogyi Eötvös University, Budapest April 16, 2012 MS/MS Modes Product Ion Precursor Ion Neutral Loss Δ ed Reaction Monitoring (SRM) 1 modes in a triple quadrupole (QqQ) (one quadrupole
Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus
 Supplemental Information Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus Torben Hofeditz, 1 Claudia Eva-Maria Unsin 2, Jutta
Supplemental Information Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus Torben Hofeditz, 1 Claudia Eva-Maria Unsin 2, Jutta
Amino Acid Oxidation and the Urea Cycle
 Amino Acid Oxidation and the Urea Cycle Amino Acids: Final class of biomolecules whose oxidation contributes significantly to the generation of energy Undergo oxidation in three metabolic circumstances
Amino Acid Oxidation and the Urea Cycle Amino Acids: Final class of biomolecules whose oxidation contributes significantly to the generation of energy Undergo oxidation in three metabolic circumstances
Classification of amino acids: -
 Page 1 of 8 P roteinogenic amino acids, also known as standard, normal or primary amino acids are 20 amino acids that are incorporated in proteins and that are coded in the standard genetic code (subunit
Page 1 of 8 P roteinogenic amino acids, also known as standard, normal or primary amino acids are 20 amino acids that are incorporated in proteins and that are coded in the standard genetic code (subunit
Proteins are sometimes only produced in one cell type or cell compartment (brain has 15,000 expressed proteins, gut has 2,000).
 Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Lecture 10 - Protein Turnover and Amino Acid Catabolism
 Lecture 10 - Protein Turnover and Amino Acid Catabolism Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction 2 Proteins are degraded into amino acids. Protein
Lecture 10 - Protein Turnover and Amino Acid Catabolism Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction 2 Proteins are degraded into amino acids. Protein
Lecture 3. Tandem MS & Protein Sequencing
 Lecture 3 Tandem MS & Protein Sequencing Nancy Allbritton, M.D., Ph.D. Department of Physiology & Biophysics 824-9137 (office) nlallbri@uci.edu Office- Rm D349 Medical Science D Bldg. Tandem MS Steps:
Lecture 3 Tandem MS & Protein Sequencing Nancy Allbritton, M.D., Ph.D. Department of Physiology & Biophysics 824-9137 (office) nlallbri@uci.edu Office- Rm D349 Medical Science D Bldg. Tandem MS Steps:
Org/Biochem Final Lec Form, Spring 2012 Page 1 of 6
 Page 1 of 6 Missing Complete Protein and Question #45 Key Terms: Fill in the blank in the following 25 statements with one of the key terms in the table. Each key term may only be used once. Print legibly.
Page 1 of 6 Missing Complete Protein and Question #45 Key Terms: Fill in the blank in the following 25 statements with one of the key terms in the table. Each key term may only be used once. Print legibly.
CS612 - Algorithms in Bioinformatics
 Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
1. (38 pts.) 2. (25 pts.) 3. (15 pts.) 4. (12 pts.) 5. (10 pts.) Bonus (12 pts.) TOTAL (100 points)
 Moorpark College Chemistry 11 Spring 2010 Instructor: Professor Torres Examination #5: Section Five May 4, 2010 ame: (print) ame: (sign) Directions: Make sure your examination contains TWELVE total pages
Moorpark College Chemistry 11 Spring 2010 Instructor: Professor Torres Examination #5: Section Five May 4, 2010 ame: (print) ame: (sign) Directions: Make sure your examination contains TWELVE total pages
Moorpark College Chemistry 11 Fall Instructor: Professor Gopal. Examination # 5: Section Five May 7, Name: (print)
 Moorpark College Chemistry 11 Fall 2013 Instructor: Professor Gopal Examination # 5: Section Five May 7, 2013 Name: (print) Directions: Make sure your examination contains TEN total pages (including this
Moorpark College Chemistry 11 Fall 2013 Instructor: Professor Gopal Examination # 5: Section Five May 7, 2013 Name: (print) Directions: Make sure your examination contains TEN total pages (including this
AMINO ACID METABOLISM. Sri Widia A Jusman Dept. of Biochemistry & Molecular Biology FMUI
 AMINO ACID METABOLISM Sri Widia A Jusman Dept. of Biochemistry & Molecular Biology FMUI Amino acids derived from dietary protein absorbed from intestine through blood taken up by tissues used for biosynthesis
AMINO ACID METABOLISM Sri Widia A Jusman Dept. of Biochemistry & Molecular Biology FMUI Amino acids derived from dietary protein absorbed from intestine through blood taken up by tissues used for biosynthesis
Phosphorylated glycosphingolipids essential for cholesterol mobilization in C. elegans
 SUPPLEMENTARY INFORMATION Phosphorylated glycosphingolipids essential for cholesterol mobilization in C. elegans Sebastian Boland, Ulrike Schmidt, Vyacheslav Zagoriy, Julio L. Sampaio, Raphael Fritsche,
SUPPLEMENTARY INFORMATION Phosphorylated glycosphingolipids essential for cholesterol mobilization in C. elegans Sebastian Boland, Ulrike Schmidt, Vyacheslav Zagoriy, Julio L. Sampaio, Raphael Fritsche,
Objective: You will be able to explain how the subcomponents of
 Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Lecture 3: 8/24. CHAPTER 3 Amino Acids
 Lecture 3: 8/24 CHAPTER 3 Amino Acids 1 Chapter 3 Outline 2 Amino Acid Are Biomolecules and their Atoms Can Be Visualized by Two Different Ways 1) Fischer projections: Two dimensional representation of
Lecture 3: 8/24 CHAPTER 3 Amino Acids 1 Chapter 3 Outline 2 Amino Acid Are Biomolecules and their Atoms Can Be Visualized by Two Different Ways 1) Fischer projections: Two dimensional representation of
Properties of amino acids in proteins
 Properties of amino acids in proteins one of the primary roles of DNA (but far from the only one!!!) is to code for proteins A typical bacterium builds thousands types of proteins, all from ~20 amino acids
Properties of amino acids in proteins one of the primary roles of DNA (but far from the only one!!!) is to code for proteins A typical bacterium builds thousands types of proteins, all from ~20 amino acids
Post-translational modifications of proteins in gene regulation under hypoxic conditions
 203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
Biomolecules Amino Acids & Protein Chemistry
 Biochemistry Department Date: 17/9/ 2017 Biomolecules Amino Acids & Protein Chemistry Prof.Dr./ FAYDA Elazazy Professor of Biochemistry and Molecular Biology Intended Learning Outcomes ILOs By the end
Biochemistry Department Date: 17/9/ 2017 Biomolecules Amino Acids & Protein Chemistry Prof.Dr./ FAYDA Elazazy Professor of Biochemistry and Molecular Biology Intended Learning Outcomes ILOs By the end
So where were we? But what does the order mean? OK, so what's a protein? 4/1/11
 So where were we? We know that DNA is responsible for heredity Chromosomes are long pieces of DNA DNA turned out to be the transforming principle We know that DNA is shaped like a long double helix, with
So where were we? We know that DNA is responsible for heredity Chromosomes are long pieces of DNA DNA turned out to be the transforming principle We know that DNA is shaped like a long double helix, with
Metabolite identification in metabolomics: Database and interpretation of MSMS spectra
 Metabolite identification in metabolomics: Database and interpretation of MSMS spectra Jeevan K. Prasain, PhD Department of Pharmacology and Toxicology, UAB jprasain@uab.edu utline Introduction Putative
Metabolite identification in metabolomics: Database and interpretation of MSMS spectra Jeevan K. Prasain, PhD Department of Pharmacology and Toxicology, UAB jprasain@uab.edu utline Introduction Putative
AMINO ACIDS NON-ESSENTIAL ESSENTIAL
 Edith Frederika Introduction A major component of food is PROTEIN The protein ingested as part of our diet are not the same protein required by the body Only 40 to 50 gr of protein is required by a normal
Edith Frederika Introduction A major component of food is PROTEIN The protein ingested as part of our diet are not the same protein required by the body Only 40 to 50 gr of protein is required by a normal
Gentilucci, Amino Acids, Peptides, and Proteins. Peptides and proteins are polymers of amino acids linked together by amide bonds CH 3
 Amino Acids Peptides and proteins are polymers of amino acids linked together by amide bonds Aliphatic Side-Chain Amino Acids - - H CH glycine alanine 3 proline valine CH CH 3 - leucine - isoleucine CH
Amino Acids Peptides and proteins are polymers of amino acids linked together by amide bonds Aliphatic Side-Chain Amino Acids - - H CH glycine alanine 3 proline valine CH CH 3 - leucine - isoleucine CH
E.coli Core Model: Metabolic Core
 1 E.coli Core Model: Metabolic Core 2 LEARNING OBJECTIVES Each student should be able to: Describe the glycolysis pathway in the core model. Describe the TCA cycle in the core model. Explain gluconeogenesis.
1 E.coli Core Model: Metabolic Core 2 LEARNING OBJECTIVES Each student should be able to: Describe the glycolysis pathway in the core model. Describe the TCA cycle in the core model. Explain gluconeogenesis.
بسم هللا الرحمن الرحيم
 بسم هللا الرحمن الرحيم Biochemistry Lec #1 Dr. Nafith AbuTarboush- (30.6.2014) Amino Acids 1 Campbell and Farrell s Biochemistry, Chapter 3 (pp.66-76) Introduction: Biochemistry is two courses; one is
بسم هللا الرحمن الرحيم Biochemistry Lec #1 Dr. Nafith AbuTarboush- (30.6.2014) Amino Acids 1 Campbell and Farrell s Biochemistry, Chapter 3 (pp.66-76) Introduction: Biochemistry is two courses; one is
Introduction to Biochemistry Midterm exam )ومن أحياها(
 Introduction to Biochemistry Midterm exam 2016-2017 )ومن أحياها( 1. Which of the following amino (in a peptide chain) would probably be found at a beta bend or turn? a. lysine * b. Gly c. arg d. asn 2.
Introduction to Biochemistry Midterm exam 2016-2017 )ومن أحياها( 1. Which of the following amino (in a peptide chain) would probably be found at a beta bend or turn? a. lysine * b. Gly c. arg d. asn 2.
The N-terminal loop of IRAK-4 death domain regulates ordered assembly of the Myddosome signalling scaffold
 The N-terminal loop of IRAK-4 death domain regulates ordered assembly of the Myddosome signalling scaffold Anthony C.G. Dossang * 1, Precious G. Motshwene * 1, Yang Yang* 1, Martyn F. Symmons*, Clare E.
The N-terminal loop of IRAK-4 death domain regulates ordered assembly of the Myddosome signalling scaffold Anthony C.G. Dossang * 1, Precious G. Motshwene * 1, Yang Yang* 1, Martyn F. Symmons*, Clare E.
Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66
 SUPPORTING INFORMATION belonging to the manuscript: Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66 by Changsheng Wu 1, 2, Gilles P. van Wezel 1, *, and Young Hae
SUPPORTING INFORMATION belonging to the manuscript: Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66 by Changsheng Wu 1, 2, Gilles P. van Wezel 1, *, and Young Hae
Application of a new capillary HPLC- ICP-MS interface to the identification of selenium-containing proteins in selenized yeast
 Application of a new capillary HPLC- ICP-MS interface to the identification of selenium-containing proteins in selenized yeast Application note Food supplements Authors Juliusz Bianga and Joanna Szpunar
Application of a new capillary HPLC- ICP-MS interface to the identification of selenium-containing proteins in selenized yeast Application note Food supplements Authors Juliusz Bianga and Joanna Szpunar
Chapter 3: Amino Acids and Peptides
 Chapter 3: Amino Acids and Peptides BINF 6101/8101, Spring 2018 Outline 1. Overall amino acid structure 2. Amino acid stereochemistry 3. Amino acid sidechain structure & classification 4. Non-standard
Chapter 3: Amino Acids and Peptides BINF 6101/8101, Spring 2018 Outline 1. Overall amino acid structure 2. Amino acid stereochemistry 3. Amino acid sidechain structure & classification 4. Non-standard
Macromolecules of Life -3 Amino Acids & Proteins
 Macromolecules of Life -3 Amino Acids & Proteins Shu-Ping Lin, Ph.D. Institute of Biomedical Engineering E-mail: splin@dragon.nchu.edu.tw Website: http://web.nchu.edu.tw/pweb/users/splin/ Amino Acids Proteins
Macromolecules of Life -3 Amino Acids & Proteins Shu-Ping Lin, Ph.D. Institute of Biomedical Engineering E-mail: splin@dragon.nchu.edu.tw Website: http://web.nchu.edu.tw/pweb/users/splin/ Amino Acids Proteins
Supporting Information
 Supporting Information Mechanism of inactivation of -aminobutyric acid aminotransferase by (1S,3S)-3-amino-4-difluoromethylenyl-1- cyclopentanoic acid (CPP-115) Hyunbeom Lee, 1, Emma H. Doud, 1,2 Rui Wu,
Supporting Information Mechanism of inactivation of -aminobutyric acid aminotransferase by (1S,3S)-3-amino-4-difluoromethylenyl-1- cyclopentanoic acid (CPP-115) Hyunbeom Lee, 1, Emma H. Doud, 1,2 Rui Wu,
The Structure and Function of Macromolecules
 The Structure and Function of Macromolecules Macromolecules are polymers Polymer long molecule consisting of many similar building blocks. Monomer the small building block molecules. Carbohydrates, proteins
The Structure and Function of Macromolecules Macromolecules are polymers Polymer long molecule consisting of many similar building blocks. Monomer the small building block molecules. Carbohydrates, proteins
(30 pts.) 16. (24 pts.) 17. (20 pts.) 18. (16 pts.) 19. (5 pts.) 20. (5 pts.) TOTAL (100 points)
 Moorpark College Chemistry 11 Spring 2009 Instructor: Professor Torres Examination # 5: Section Five April 30, 2009 ame: (print) ame: (sign) Directions: Make sure your examination contains TWELVE total
Moorpark College Chemistry 11 Spring 2009 Instructor: Professor Torres Examination # 5: Section Five April 30, 2009 ame: (print) ame: (sign) Directions: Make sure your examination contains TWELVE total
Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A
 Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A Homework Watch the Bozeman video called, Biological Molecules Objective:
Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A Homework Watch the Bozeman video called, Biological Molecules Objective:
Bioactivity Based Molecular Networking for the Discovery of Drug Lead in Natural Product Bioassay-Guided Fractionation
 Bioactivity Based Molecular Networking for the Discovery of Drug Lead in Natural Product Bioassay-Guided Fractionation Louis-Félix Nothias,,, Mélissa Nothias-Esposito,, # Ricardo da Silva,, Mingxun Wang,,,
Bioactivity Based Molecular Networking for the Discovery of Drug Lead in Natural Product Bioassay-Guided Fractionation Louis-Félix Nothias,,, Mélissa Nothias-Esposito,, # Ricardo da Silva,, Mingxun Wang,,,
Amino acid Catabolism
 Enzymatic digestion of dietary proteins in gastrointestinal-tract. Amino acid Catabolism Amino acids: 1. There are 20 different amino acid, they are monomeric constituents of proteins 2. They act as precursors
Enzymatic digestion of dietary proteins in gastrointestinal-tract. Amino acid Catabolism Amino acids: 1. There are 20 different amino acid, they are monomeric constituents of proteins 2. They act as precursors
Your Name: Question 1. Spectrum Prediction I: Ethyl Acetoacetate. (15 points) ppm ppm ppm ppm. J(A,D) = 8 Hz = 0.
 Question 1. Spectrum Prediction I: Ethyl Acetoacetate. (15 points) A B C D 4.202 ppm 3.451 ppm 2.273 ppm 1.288 ppm J(A,D) = 8 Hz = 0.08 ppm (in CDCl 3 ) Draw the 100 MHz H-NMR spectrum to scale. Draw splitting
Question 1. Spectrum Prediction I: Ethyl Acetoacetate. (15 points) A B C D 4.202 ppm 3.451 ppm 2.273 ppm 1.288 ppm J(A,D) = 8 Hz = 0.08 ppm (in CDCl 3 ) Draw the 100 MHz H-NMR spectrum to scale. Draw splitting
Amino acids. (Foundation Block) Dr. Essa Sabi
 Amino acids (Foundation Block) Dr. Essa Sabi Learning outcomes What are the amino acids? General structure. Classification of amino acids. Optical properties. Amino acid configuration. Non-standard amino
Amino acids (Foundation Block) Dr. Essa Sabi Learning outcomes What are the amino acids? General structure. Classification of amino acids. Optical properties. Amino acid configuration. Non-standard amino
M1 - Renal, Fall 2007
 University of Michigan Deep Blue deepblue.lib.umich.edu 2007-09 M1 - Renal, Fall 2007 Lyons, R.; Burney, R. Lyons, R., Burney, R. (2008, August 07). Renal. Retrieved from Open.Michigan - Educational Resources
University of Michigan Deep Blue deepblue.lib.umich.edu 2007-09 M1 - Renal, Fall 2007 Lyons, R.; Burney, R. Lyons, R., Burney, R. (2008, August 07). Renal. Retrieved from Open.Michigan - Educational Resources
Activities for the α-helix / β-sheet Construction Kit
 Activities for the α-helix / β-sheet Construction Kit The primary sequence of a protein, composed of amino acids, determines the organization of the sequence into the secondary structure. There are two
Activities for the α-helix / β-sheet Construction Kit The primary sequence of a protein, composed of amino acids, determines the organization of the sequence into the secondary structure. There are two
If you like us, please share us on social media. The latest UCD Hyperlibrary newsletter is now complete, check it out.
 Sign In Forgot Password Register username username password password Sign In If you like us, please share us on social media. The latest UCD Hyperlibrary newsletter is now complete, check it out. ChemWiki
Sign In Forgot Password Register username username password password Sign In If you like us, please share us on social media. The latest UCD Hyperlibrary newsletter is now complete, check it out. ChemWiki
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 30 Amino Acid Degradation and the Urea Cycle 2013 W. H. Freeman and Company Chapter 30 Outline Amino acids are obtained from the
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 30 Amino Acid Degradation and the Urea Cycle 2013 W. H. Freeman and Company Chapter 30 Outline Amino acids are obtained from the
Solutions to Quiz I
 Solutions to 7.014 Quiz I lass Average = 75 Median = 75 Grade Range % A 87-100 18 B 74-86 39 62-73 31 D 48-61 8 F 0-47 4 Question 1 (26 points) The protein complex, LETMEGTHRUIN (LMGT for short), acts
Solutions to 7.014 Quiz I lass Average = 75 Median = 75 Grade Range % A 87-100 18 B 74-86 39 62-73 31 D 48-61 8 F 0-47 4 Question 1 (26 points) The protein complex, LETMEGTHRUIN (LMGT for short), acts
NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA. Citrus responses to huanglongbing
 NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA Citrus responses to huanglongbing HLB is Vector-borne Disease! Vector psyllids Pathogen Ca. Liberibacter asiaticus. Host Citrus spp.
NABIL KILLINY ASSISTANT PROFESSOR PLANT PATHOLOGY UNIVERSITY OF FLORIDA Citrus responses to huanglongbing HLB is Vector-borne Disease! Vector psyllids Pathogen Ca. Liberibacter asiaticus. Host Citrus spp.
Copyright 2008 Pearson Education, Inc., publishing as Pearson Benjamin Cummings
 Concept 5.4: Proteins have many structures, resulting in a wide range of functions Proteins account for more than 50% of the dry mass of most cells Protein functions include structural support, storage,
Concept 5.4: Proteins have many structures, resulting in a wide range of functions Proteins account for more than 50% of the dry mass of most cells Protein functions include structural support, storage,
Figure S6. A-J) Annotated UVPD mass spectra for top ten peptides found among the peptides identified by Byonic but not SEQUEST + Percolator.
 Extending Proteome Coverage by Combining MS/MS Methods and a Modified Bioinformatics Platform adapted for Database Searching of Positive and Negative Polarity 193 nm Ultraviolet Photodissociation Mass
Extending Proteome Coverage by Combining MS/MS Methods and a Modified Bioinformatics Platform adapted for Database Searching of Positive and Negative Polarity 193 nm Ultraviolet Photodissociation Mass
Unsupervised Identification of Isotope-Labeled Peptides
 Unsupervised Identification of Isotope-Labeled Peptides Joshua E Goldford 13 and Igor GL Libourel 124 1 Biotechnology institute, University of Minnesota, Saint Paul, MN 55108 2 Department of Plant Biology,
Unsupervised Identification of Isotope-Labeled Peptides Joshua E Goldford 13 and Igor GL Libourel 124 1 Biotechnology institute, University of Minnesota, Saint Paul, MN 55108 2 Department of Plant Biology,
Supplementary Figure S1. Appearacne of new acetyl groups in acetylated lysines using 2,3-13 C 6 pyruvate as a tracer instead of labeled glucose.
 Supplementary Figure S1. Appearacne of new acetyl groups in acetylated lysines using 2,3-13 C 6 pyruvate as a tracer instead of labeled glucose. (a) Relative levels of both new acetylation and all acetylation
Supplementary Figure S1. Appearacne of new acetyl groups in acetylated lysines using 2,3-13 C 6 pyruvate as a tracer instead of labeled glucose. (a) Relative levels of both new acetylation and all acetylation
PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System
 Application Note LC/MS PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System Purpose This application note describes an automated workflow
Application Note LC/MS PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System Purpose This application note describes an automated workflow
From sugar unit A From sugar unit B From sugar unit C
 ame 215 F12-Exam o. Page 2 I. (26 points) Raffinose is a trisaccharide occurring in cottonseed meal. Answer the following questions as directed in the boxes. (1) ( points) Label each of the glycosidic
ame 215 F12-Exam o. Page 2 I. (26 points) Raffinose is a trisaccharide occurring in cottonseed meal. Answer the following questions as directed in the boxes. (1) ( points) Label each of the glycosidic
Page 8/6: The cell. Where to start: Proteins (control a cell) (start/end products)
 Page 8/6: The cell Where to start: Proteins (control a cell) (start/end products) Page 11/10: Structural hierarchy Proteins Phenotype of organism 3 Dimensional structure Function by interaction THE PROTEIN
Page 8/6: The cell Where to start: Proteins (control a cell) (start/end products) Page 11/10: Structural hierarchy Proteins Phenotype of organism 3 Dimensional structure Function by interaction THE PROTEIN
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
Algal Biofuels Research: Using basic science to maximize fuel output. Jacob Dums, PhD candidate, Heike Sederoff Lab March 9, 2015
 Algal Biofuels Research: Using basic science to maximize fuel output Jacob Dums, PhD candidate, jtdums@ncsu.edu Heike Sederoff Lab March 9, 2015 Outline Research Approach Dunaliella Increase Oil Content
Algal Biofuels Research: Using basic science to maximize fuel output Jacob Dums, PhD candidate, jtdums@ncsu.edu Heike Sederoff Lab March 9, 2015 Outline Research Approach Dunaliella Increase Oil Content
