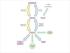
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the
|
|
|
- Caroline Ward
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
2 Supplementary Figure 2. Comparison of the alp, lom, and flu gene clusters. The three gene clusters are responsible for the biosynthesis of kinamycin, lomaiviticin and fluostatin, respectively. The DNA region labeled as region 1 is extended in this study. The genes in black are indicated as mini-pks, in green as the conserved six-gene subregion responsible for the diazo assembly.
3 Supplementary Figure 3. Phylogenetic analysis of the oxygenases in the alp, lom and flu clusters. The tree was constructed by Neighbor-Joining method and the bootstrap (500 replicates) values for the main clades are shown. Scale bar represents 10% dissimilarity. The oxygenases labeled in black or blue are the candidates discussed in this study for A-ring transformation in the kinamycin, fluostatin, and lomaiviticin biosynthesis.
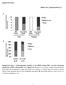
4 Supplementary Figure 4. PCR confirmation of alp1u double-deletion mutant. Lane 1, DNA ladder; lane 2, alp1u giving a band 631 bp. lane 3, the starting strain giving a band corresponding to a fragment of 1,204 bp in length. Both detected bands were correct in size. Supplementary Figure 5. SDS-PAGE analysis of the purified Alp1U. Lane 1, protein marker; lane 2, Alp1U giving a band corresponding to a protein of 37 kda. The detected band was correct in size.
5 Supplementary Figure 6. LC-MS spectrum of kinamycin E (5) under ESI negative mode. Supplementary Figure 7. HPLC analysis of kinamycin E (5) dissolved in (CD 3 ) 2 SO. (A) before NMR measurement; (B) after NMR measurement. Supplementary Figure 8. SDS-PAGE analysis of the purified Lom6. Lane 1, protein marker; lane 2, Lom6 giving a band corresponding to a protein of 30 kda. The detected band was correct in size.
6 Supplementary Figure 9. LC-MS spectrum of kinamycin F (2) under ESI negative mode. Supplementary Figure 10. HPLC profiles of Lom6 reaction with longer incubation time. (A) standard epoxykinamycin (1); (B) 10 min; (C) 20 min; (D) 50 min
7 Epoxykinamycin FL-120B (4) 1 H NMR (500 MHz, in (CD 3 ) 2 SO) Supplementary Figure H NMR spectrum for epoxykinamycin FL-120B (4).
8 Epoxykinamycin FL-120B (4) 13 C NMR (125 MHz, in (CD 3 ) 2 SO) Supplementary Figure C NMR spectrum for epoxykinamycin FL-120B (4).
9 Epoxykinamycin FL-120B (4) COSY (500 MHz, in (CD 3 ) 2 SO) Supplementary Figure 13. COSY spectrum for epoxykinamycin FL-120B (4).
10 Epoxykinamycin FL-120B (4) HSQC (500 MHz, in (CD 3 ) 2 SO) Supplementary Figure 14. HSQC spectrum for epoxykinamycin FL-120B (4).
11 Epoxykinamycin FL-120B (4) HMBC (500 MHz, in (CD 3 ) 2 SO) Supplementary Figure 15. HMBC spectrum for epoxykinamycin FL-120B (4).
12 Epoxykinamycin FL-120B (4) NOESY (500 MHz, in (CD 3 ) 2 SO) Supplementary Figure 16. NOESY spectrum for epoxykinamycin FL-120B (4).
13 Kinamycin E (5) 1 H NMR (500 MHz, in (CD 3 ) 2 SO) Supplementary Figure H NMR spectrum for kinamycin E (5).
14 Kinamycin E (5) 13 C NMR (125 MHz, in (CD 3 ) 2 SO) Supplementary Figure C NMR spectrum for kinamycin E (5).
15 Kinamycin E (5) HMBC (500 MHz, in (CD 3 ) 2 SO) Supplementary Figure 19. HMBC spectrum for kinamycin E (5).
16 Kinamycin E (5) NOESY (500 MHz, in (CD 3 ) 2 SO) Supplementary Figure 20. NOESY spectrum for kinamycin E (5).
17 Supplementary Table 1. Predicted functions of the gene products in the alp cluster. gene predicted function note alpz alpy alpx alpw alpv alpu alpt alps alpr alpq alpp2 alpp alpo alpn alpm alpl2 alpl alpk alpj alpi alpa alpb alpc alpd alpe alpf alpg alph gamma-butyrolactone binding protein hypothetical protein putative carboxyl transferase TetR-family transcriptional regulator SARP family pathway specific regulator SARP family pathway specific regulator SARP family pathway specific regulator putative thioesterase putative polyketide synthase putative chain length factor protein hypothetical protein putative cyclase putative acyl-coa dehydrogenase putative phosphopantetheinyl transferase peptide synthase condensation domain conserved hypothetical protein JadX-like protein oxygenase oxidase polyketide cyclase ketoacyl synthase chain length determinant acyl carrier protein polyketide ketoreductase cyclase/dehydratase oxygenase oxygenase putative O-methyltransferase the previously proposed alp cluster
18 Continued gene predicted function note alp1a conserved hypothetical protein alp1b putative myo-inositol phosphate synthase alp1c putative transferase alp1d alp1e putative abasic site repairing enzyme conserved hypothetical protein detoxification and DNA damage-repair alp1f putative metal-dependent hydrolase alp1g conserved hypothetical protein alp1h putative type I phosphodiesterase alp1i putative ferredoxin alp1j hypothetical protein alp1k putative amidase diazo assembly alp1l putative glutamine synthetase machinery alp1m putative adenylosuccinate lyase alp1n putative acetyltransferase alp1o two-component system regulator alp1p putative efflux protein alp1q conserved hypothetical protein export and regulation alp1r putative antibiotic antiporter alp1s putative oxygenase alp1t NDP-sugar epimerase alp1u epoxy hydrolase A-ring transformation alp1v putative oxygenase alp1w putative glutamine amidotransferase diazo assembly alp1x putative transcriptional regulator machinery region 1
19 Supplementary Table 2. The NMR data of epoxykinamycin FL-120B (4) Position 13 C-NMR a 1 H-NMR b HMBC b NOESY (s, 1H) C-4a H-2, H-4, H OH 5.69 (s, 1H) C-1, C (t, 1H, J = 6.0 Hz) C-1, C-3, C-11b, C-12 H (s, 1H) C-3, C-5, C-11b, C-12 H a a a OH (s, 1H) C-6a, C-7, C (d, 1H, J = 7.5 Hz) C-6a, C-7, C (t, 1H, J = 7.5 Hz) C-7, C-10a (d, 1H, J = 7.5 Hz) C-6a, C-8 10a a b (s, 3H) C-2, C-3, C (s, 3H) C-13 a Recorded at 125 MHz in (CD 3 ) 2 SO. b Recorded at 500 MHz in (CD 3 ) 2 SO.
20 Supplementary Table 3. The NMR data of kinamycin E (5) Position 13 C-NMR a 1 H-NMR b HMBC b NOESY (dd, 1H, J = 3.5, 6.0 Hz) C-3, C-4a H-2, H-12 1-OH 5.18 (d, 1H, J = 3.5 Hz) (t, 1H, J = 6.0 Hz) C-1, C-3, C-11b, C-12 H-1 2-OH 5.26 (d, 1H, J = 6.0 Hz) OH 5.10 (s, 1H) (s, 1H) C-2, C-5, C-11b, C-12 H-12 4a a a OH (s, 1H) C-6a, C-7, C (d, 1H, J = 8.5 Hz) C-6a, C-7, C (t, 1H, J = 8.5 Hz) C-7, C-10a (d, 1H, J = 8.5 Hz) C-6a, C-8 10a a b (s, 3H) C-2, C-3, C (s, 3H) C-13 a Recorded at 125 MHz in (CD 3 ) 2 SO. b Recorded at 500 MHz in (CD 3 ) 2 SO.
21 Supplementary Table 4. List of primers used in this study. Primers Restriction sites Sequences 1U-upF HindIII 5' CCCAAGCTTGTGCAGATCCCCGACGCC 3' 1U-upR XbaI 5' GCTCTAGAGGCCAGTTCCCGGTCCGA 3' 1U-dnF XbaI 5' GCTCTAGATACGCCAACGCCTACAACAGC 3' 1U-dnR EcoRI 5' GGAATTCCCCATGTCGTAGCGGATGTC 3' CK-1U-F 5' CGAGGACATGGGTAGATGACGC 3' CK-1U-R 5' GGCCGGGCAGTTTGTCGTAGA 3' 1U-F NdeI 5' GGAATTCCATATGCATGGGTAGATGACGCTGAAC 3' 1U-R EcoRI 5' GGAATTCGAGGTGGCGTCAGCCGA 3' Ealp1U-F NdeI 5' GGAATTCCATATGCTCGCCCTGGGCACGG 3' Ealp1U-R HindIII 5' CCCAAGCTTGGAGGTGGCGTCAGCCGAAGAAG 3'
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function
 Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted. residues are colored red. Numbers indicate the position of each residue.
 Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
Supplementary Information
 Supplementary Information Assembly and Clustering of Natural Antibiotics Guides Target Identification Chad W. Johnston 1,2, Michael A. Skinnider 1,2, Chris A. Dejong 1,2, Philip N. Rees 1,2, Gregory M.
Supplementary Information Assembly and Clustering of Natural Antibiotics Guides Target Identification Chad W. Johnston 1,2, Michael A. Skinnider 1,2, Chris A. Dejong 1,2, Philip N. Rees 1,2, Gregory M.
Lomaiviticin Biosynthesis Employs a New Strategy for Starter Unit Generation
 Lomaiviticin Biosynthesis Employs a New Strategy for Starter Unit Generation The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters
Lomaiviticin Biosynthesis Employs a New Strategy for Starter Unit Generation The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters
Supplementary Figure 1. HGL-DTG levels in uninduced and herbivory induced leaves. Total HGL-
 Supplementary Figure 1. HGL-DTG levels in uninduced and herbivory induced leaves. Total HGL- DTG concentrations [F 1,4 = 36.88, P 0.0037; significant differences (threshold: P 0.05) between means (± SE)
Supplementary Figure 1. HGL-DTG levels in uninduced and herbivory induced leaves. Total HGL- DTG concentrations [F 1,4 = 36.88, P 0.0037; significant differences (threshold: P 0.05) between means (± SE)
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
Supporting Information
 Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
Figure S3 Differentially expressed fungal and plant genes organised by catalytic activity ontology Organisation of E. festucae (A) and L.
 sak WT Number of branches 3 WT sak Figure S1 Vasculature of plants infected with the E. festucae sak mutant. Light micrographs of perennial ryegrass blade tissue showing branching between the vasculature
sak WT Number of branches 3 WT sak Figure S1 Vasculature of plants infected with the E. festucae sak mutant. Light micrographs of perennial ryegrass blade tissue showing branching between the vasculature
Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66
 SUPPORTING INFORMATION belonging to the manuscript: Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66 by Changsheng Wu 1, 2, Gilles P. van Wezel 1, *, and Young Hae
SUPPORTING INFORMATION belonging to the manuscript: Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66 by Changsheng Wu 1, 2, Gilles P. van Wezel 1, *, and Young Hae
Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis
 Supporting information Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis Martin F. Kreutzer and Markus Nett * Table of contents SI-2, SI-3 SI-4 SI-5 SI-6
Supporting information Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis Martin F. Kreutzer and Markus Nett * Table of contents SI-2, SI-3 SI-4 SI-5 SI-6
Investigations of Pactamycin Biosynthesis Andrew Osborn
 Investigations of Pactamycin Biosynthesis Andrew Osborn Research Mentors: Dr. Taifo Mahmud, Dr. Kerry McPhail A Bioresource Research Thesis Seminar Presentation Streptomyces pactum produces the antitumor
Investigations of Pactamycin Biosynthesis Andrew Osborn Research Mentors: Dr. Taifo Mahmud, Dr. Kerry McPhail A Bioresource Research Thesis Seminar Presentation Streptomyces pactum produces the antitumor
Figure S1: Abundance of Fe related proteins in oceanic Synechococcus sp. strain WH8102 in. Ferredoxin. Ferredoxin. Fe ABC transporter
 0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus
 Supplemental Information Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus Torben Hofeditz, 1 Claudia Eva-Maria Unsin 2, Jutta
Supplemental Information Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus Torben Hofeditz, 1 Claudia Eva-Maria Unsin 2, Jutta
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol
 Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
Supplementary Information
 Supplementary Information New Highly Oxygenated Germacranolides from Carpesium divaricatum and their Cytotoxic Activity Tao Zhang, 1 Jin-Guang Si, 1, 2 Qiu-Bo Zhang, 1 Gang Ding, 1 and Zhong-Mei Zou 1*
Supplementary Information New Highly Oxygenated Germacranolides from Carpesium divaricatum and their Cytotoxic Activity Tao Zhang, 1 Jin-Guang Si, 1, 2 Qiu-Bo Zhang, 1 Gang Ding, 1 and Zhong-Mei Zou 1*
Supplemental Data Functional characterization of core components of the Bacillus subtilis c- di-
 Supplemental Data Functional characterization of core components of the Bacillus subtilis c- di- GMP signaling pathway Xiaohui Gao 1,3, Sampriti Mukherjee 2, Paige M. Matthews 1, Loubna A. Hammad 1, Daniel
Supplemental Data Functional characterization of core components of the Bacillus subtilis c- di- GMP signaling pathway Xiaohui Gao 1,3, Sampriti Mukherjee 2, Paige M. Matthews 1, Loubna A. Hammad 1, Daniel
Bacterial cellulose synthesis mechanism of facultative
 1 2 3 4 Bacterial cellulose synthesis mechanism of facultative anaerobe Kaihua Ji 1 +, Wei Wang 2, 3 +, Bing Zeng 1, Sibin Chen 1, Qianqian Zhao 4, Yueqing Chen 1, Guoqiang Li 1* and Ting Ma 1* 5 6 7 Figure
1 2 3 4 Bacterial cellulose synthesis mechanism of facultative anaerobe Kaihua Ji 1 +, Wei Wang 2, 3 +, Bing Zeng 1, Sibin Chen 1, Qianqian Zhao 4, Yueqing Chen 1, Guoqiang Li 1* and Ting Ma 1* 5 6 7 Figure
Supplementary Information
 Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and Gerard D. Wright.
 Supplementary Data for TetX is a Flavin-Dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and
Supplementary Data for TetX is a Flavin-Dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and
Supplementary Information. A new strategy for aromatic ring alkylation in cylindrocyclophane biosynthesis
 Supplementary Information A new strategy for aromatic ring alkylation in cylindrocyclophane biosynthesis Hitomi Nakamura, Erica E. Schultz, and Emily P. Balskus* Corresponding author: balskus@chemistry.harvard.edu
Supplementary Information A new strategy for aromatic ring alkylation in cylindrocyclophane biosynthesis Hitomi Nakamura, Erica E. Schultz, and Emily P. Balskus* Corresponding author: balskus@chemistry.harvard.edu
Minjia Tan, Ph.D Shanghai Institute of Materia Medica, Chinese Academy of Sciences
 Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5 Minjia Tan, Ph.D Shanghai Institute of Materia Medica, Chinese Academy of Sciences Lysine: the most frequently modified
Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5 Minjia Tan, Ph.D Shanghai Institute of Materia Medica, Chinese Academy of Sciences Lysine: the most frequently modified
Supplementary Material (ESI) for Chemical Communications This journal is (c) The Royal Society of Chemistry 2008
 Experimental Details Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich Chemical Company and were used as received. 2-DOS and neamine were kindly provided by Dr. F. Huang. Paromamine
Experimental Details Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich Chemical Company and were used as received. 2-DOS and neamine were kindly provided by Dr. F. Huang. Paromamine
SDR families. SDR10E Fatty acyl-coa reductase FACR1_HUMAN 544 x x SDR11E 3 beta-hydroxysteroid dehydrogenase 3BHS1_HUMAN 254 x x
 SDR1E UDP-glucose 4-epimerase GALE_HUMAN 5305 x x x x SDR2E dtdp-d-glucose 4,6-dehydratase TGDS_HUMAN 4194 x x x x SDR3E GDP-mannose 4,6 dehydratase GMDS_HUMAN 2314 x x x x SDR4E GDP-L-fucose synthetase
SDR1E UDP-glucose 4-epimerase GALE_HUMAN 5305 x x x x SDR2E dtdp-d-glucose 4,6-dehydratase TGDS_HUMAN 4194 x x x x SDR3E GDP-mannose 4,6 dehydratase GMDS_HUMAN 2314 x x x x SDR4E GDP-L-fucose synthetase
Marine Streptomyces sp. derived antimycin analogues. suppress HeLa cells via depletion HPV E6/E7 mediated by
 Marine Streptomyces sp. derived antimycin analogues suppress HeLa cells via depletion HPV E6/E7 mediated by ROS-dependent ubiquitin proteasome system Weiyi Zhang 1, +, Qian Che 1, 2, +, Hongsheng Tan 1,
Marine Streptomyces sp. derived antimycin analogues suppress HeLa cells via depletion HPV E6/E7 mediated by ROS-dependent ubiquitin proteasome system Weiyi Zhang 1, +, Qian Che 1, 2, +, Hongsheng Tan 1,
Supporting Information for: New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans
 Supporting Information for: New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans Matthew T. Henke a, Alexandra A. Soukup b, Anthony W. Goering a, Ryan A.
Supporting Information for: New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans Matthew T. Henke a, Alexandra A. Soukup b, Anthony W. Goering a, Ryan A.
Supporting information
 Supporting information Rhabdopeptide/Xenortide-like Peptides from Xenorhabdus innexi with Terminal Amines Showing Potent Anti-protozoal Activity Lei Zhao,, Marcel Kaiser,, Helge B. Bode *,, Molekulare
Supporting information Rhabdopeptide/Xenortide-like Peptides from Xenorhabdus innexi with Terminal Amines Showing Potent Anti-protozoal Activity Lei Zhao,, Marcel Kaiser,, Helge B. Bode *,, Molekulare
number Done by Corrected by Doctor F. Al-Khateeb
 number 23 Done by A. Rawajbeh Corrected by Doctor F. Al-Khateeb Ketone bodies Ketone bodies are used by the peripheral tissues like the skeletal and cardiac muscles, where they are the preferred source
number 23 Done by A. Rawajbeh Corrected by Doctor F. Al-Khateeb Ketone bodies Ketone bodies are used by the peripheral tissues like the skeletal and cardiac muscles, where they are the preferred source
GtfA and GtfB are both required for protein O-glycosylation in Lactobacillus plantarum
 Supplemental information for: GtfA and GtfB are both required for protein O-glycosylation in Lactobacillus plantarum I-Chiao Lee, Iris I. van Swam, Satoru Tomita, Pierre Morsomme, Thomas Rolain, Pascal
Supplemental information for: GtfA and GtfB are both required for protein O-glycosylation in Lactobacillus plantarum I-Chiao Lee, Iris I. van Swam, Satoru Tomita, Pierre Morsomme, Thomas Rolain, Pascal
WT siz1-2 siz1-3. WT siz1-2 siz1-3. -Pi, 0.05 M IAA. -Pi, 2.5 M NPA
 A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
Mouse Meda-4 : chromosome 5G bp. EST (547bp) _at. 5 -Meda4 inner race (~1.8Kb)
 Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
USA. Supplemental Information
 Cloning and sequencing of the kedarcidin biosynthetic gene cluster from Streptoalloteichus sp. ATCC 53650 revealing new insights into biosynthesis of the enediyne family of antitumor antibiotics Jeremy
Cloning and sequencing of the kedarcidin biosynthetic gene cluster from Streptoalloteichus sp. ATCC 53650 revealing new insights into biosynthesis of the enediyne family of antitumor antibiotics Jeremy
Ahmad Ulnar. Faisal Nimri ... Dr.Faisal
 24 Ahmad Ulnar Faisal Nimri... Dr.Faisal Fatty Acid Synthesis - Occurs mainly in the Liver (to store excess carbohydrates as triacylglycerols(fat)) and in lactating mammary glands (for the production of
24 Ahmad Ulnar Faisal Nimri... Dr.Faisal Fatty Acid Synthesis - Occurs mainly in the Liver (to store excess carbohydrates as triacylglycerols(fat)) and in lactating mammary glands (for the production of
Figure 1 Original Advantages of biological reactions being catalyzed by enzymes:
 Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
Supplementary Information
 Supplementary Information J. Braz. Chem. Soc., Vol. 24, No. 12, S1-S21, 2013. Printed in Brazil - 2013 Sociedade Brasileira de Química 0103-5053 $6.00+0.00 SI Rui He, a,b,c Bochu Wang,*,b Toshiyuki Wakimoto,
Supplementary Information J. Braz. Chem. Soc., Vol. 24, No. 12, S1-S21, 2013. Printed in Brazil - 2013 Sociedade Brasileira de Química 0103-5053 $6.00+0.00 SI Rui He, a,b,c Bochu Wang,*,b Toshiyuki Wakimoto,
Chapter 4. Erythronolide B and the Erythromycins
 124 Chapter 4 Erythronolide B and the Erythromycins Isolation and tructure Isolated in 1952 from the soil bacteria actinomycetes, 1 the erythromycins are a distinguished family of natural products by virtue
124 Chapter 4 Erythronolide B and the Erythromycins Isolation and tructure Isolated in 1952 from the soil bacteria actinomycetes, 1 the erythromycins are a distinguished family of natural products by virtue
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS
 SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
Biosynthesis of Fatty Acids
 Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study.
 Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study. In this study, one fluorophosphonate (FP, 1), three nitrophenol phosphonate probes
Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study. In this study, one fluorophosphonate (FP, 1), three nitrophenol phosphonate probes
MITOCW watch?v=ddt1kusdoog
 MITOCW watch?v=ddt1kusdoog The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
MITOCW watch?v=ddt1kusdoog The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein
![2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein](/thumbs/86/94743397.jpg) Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
Very-Long Chain Fatty Acid Biosynthesis
 Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Supplementary figure legends
 Supplementary figure legends Fig. S1. Lineweaver-Burk plot of putrescine uptake by YeeF. An overnight culture of SK629 was inoculated in 100-mL LBG medium in 500-mL Erlenmeyer flasks. The medium was supplemented
Supplementary figure legends Fig. S1. Lineweaver-Burk plot of putrescine uptake by YeeF. An overnight culture of SK629 was inoculated in 100-mL LBG medium in 500-mL Erlenmeyer flasks. The medium was supplemented
Synthesis of Fatty Acids and Triacylglycerol
 Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
Identification of Mutation(s) in. Associated with Neutralization Resistance. Miah Blomquist
 Identification of Mutation(s) in the HIV 1 gp41 Subunit Associated with Neutralization Resistance Miah Blomquist What is HIV 1? HIV-1 is an epidemic that affects over 34 million people worldwide. HIV-1
Identification of Mutation(s) in the HIV 1 gp41 Subunit Associated with Neutralization Resistance Miah Blomquist What is HIV 1? HIV-1 is an epidemic that affects over 34 million people worldwide. HIV-1
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
Acaulins A and B, Trimeric Macrodiolides from Acaulium. Table of Contents
 Supporting Information Acaulins A and B, Trimeric Macrodiolides from Acaulium sp. H-JQSF Ting Ting Wang, Ying Jie Wei, Hui Ming Ge, Rui Hua Jiao, Ren Xiang Tan* * Corresponding author. E-mail: rxtan@nju.edu.cn
Supporting Information Acaulins A and B, Trimeric Macrodiolides from Acaulium sp. H-JQSF Ting Ting Wang, Ying Jie Wei, Hui Ming Ge, Rui Hua Jiao, Ren Xiang Tan* * Corresponding author. E-mail: rxtan@nju.edu.cn
Fatty acid synthesis. Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai - 116
 Fatty acid synthesis Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai 116 Harper s biochemistry 24 th ed, Pg 218 Fatty acid Synthesis Known as
Fatty acid synthesis Dr. Nalini Ganesan M.Sc., Ph.D Associate Professor Department of Biochemistry SRMC & RI (DU) Porur, Chennai 116 Harper s biochemistry 24 th ed, Pg 218 Fatty acid Synthesis Known as
LIPID METABOLISM
 LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
MILK BIOSYNTHESIS PART 3: FAT
 MILK BIOSYNTHESIS PART 3: FAT KEY ENZYMES (FROM ALL BIOSYNTHESIS LECTURES) FDPase = fructose diphosphatase Citrate lyase Isocitrate dehydrogenase Fatty acid synthetase Acetyl CoA carboxylase Fatty acyl
MILK BIOSYNTHESIS PART 3: FAT KEY ENZYMES (FROM ALL BIOSYNTHESIS LECTURES) FDPase = fructose diphosphatase Citrate lyase Isocitrate dehydrogenase Fatty acid synthetase Acetyl CoA carboxylase Fatty acyl
Protein Class/Name. Pathways. bat00250, bat00280, bat00410, bat00640, bat bat00270, bat00330, bat00410, bat00480
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 1. Proteins Increased in Either Soil or Laboratory media Table 1A. Proteins Increased
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 1. Proteins Increased in Either Soil or Laboratory media Table 1A. Proteins Increased
Characterizing fatty acids with advanced multinuclear NMR methods
 Characterizing fatty acids with advanced multinuclear NMR methods Fatty acids consist of long carbon chains ending with a carboxylic acid on one side and a methyl group on the other. Most naturally occurring
Characterizing fatty acids with advanced multinuclear NMR methods Fatty acids consist of long carbon chains ending with a carboxylic acid on one side and a methyl group on the other. Most naturally occurring
Supplementary Materials
 Supplementary Materials Relative enthalpies (H) and Gibbs free energy (G) in kcal/mol for the cyclization process. The relative energies are calculated taking IIb as the reference. The solvent values are
Supplementary Materials Relative enthalpies (H) and Gibbs free energy (G) in kcal/mol for the cyclization process. The relative energies are calculated taking IIb as the reference. The solvent values are
Chapter 23 Enzymes 1
 Chapter 23 Enzymes 1 Enzymes Ribbon diagram of cytochrome c oxidase, the enzyme that directly uses oxygen during respiration. 2 Enzyme Catalysis Enzyme: A biological catalyst. With the exception of some
Chapter 23 Enzymes 1 Enzymes Ribbon diagram of cytochrome c oxidase, the enzyme that directly uses oxygen during respiration. 2 Enzyme Catalysis Enzyme: A biological catalyst. With the exception of some
Oxidation of Long Chain Fatty Acids
 Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
Stabilization and enhanced reactivity of actinorhodin polyketide synthase minimal complex in polymer/nucleotide coacervate droplets
 Stabilization and enhanced reactivity of actinorhodin polyketide synthase minimal complex in polymer/nucleotide coacervate droplets John Crosby,* Tom Treadwell, Michelle Hammerton, Konstantinos Vasilakis,
Stabilization and enhanced reactivity of actinorhodin polyketide synthase minimal complex in polymer/nucleotide coacervate droplets John Crosby,* Tom Treadwell, Michelle Hammerton, Konstantinos Vasilakis,
Chapter 10. 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation)
 Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
Supporting Information
 Supporting Information Bar-Even et al. 10.1073/pnas.0907176107 Fig. S1. A scheme of the reductive pentose phosphate cycle, as inspired from (81). Dashed line corresponds to RUBISCO's oxygenase reaction
Supporting Information Bar-Even et al. 10.1073/pnas.0907176107 Fig. S1. A scheme of the reductive pentose phosphate cycle, as inspired from (81). Dashed line corresponds to RUBISCO's oxygenase reaction
Very-Long Chain Fatty Acid Biosynthesis
 Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Supporting information
 Supporting information for Pentaketide Ansamycin Microansamycins A I from Micromonospora sp. Reveal Diverse Post- PKS Modifications Jianxiong Wang,, Wen Li,, Haoxin Wang, and Chunhua Lu *, Key Laboratory
Supporting information for Pentaketide Ansamycin Microansamycins A I from Micromonospora sp. Reveal Diverse Post- PKS Modifications Jianxiong Wang,, Wen Li,, Haoxin Wang, and Chunhua Lu *, Key Laboratory
Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding
 SUPPLEMENTARY INFORMATION: Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding Gong Zhang 1,2, Magdalena Hubalewska 1 & Zoya Ignatova 1,2 1 Department of Cellular
SUPPLEMENTARY INFORMATION: Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding Gong Zhang 1,2, Magdalena Hubalewska 1 & Zoya Ignatova 1,2 1 Department of Cellular
6. How Are Fatty Acids Produced? 7. How Are Acylglycerols and Compound Lipids Produced? 8. How Is Cholesterol Produced?
 Lipid Metabolism Learning bjectives 1 How Are Lipids Involved in the Generationand Storage of Energy? 2 How Are Lipids Catabolized? 3 What Is the Energy Yield from the xidation of Fatty Acids? 4 How Are
Lipid Metabolism Learning bjectives 1 How Are Lipids Involved in the Generationand Storage of Energy? 2 How Are Lipids Catabolized? 3 What Is the Energy Yield from the xidation of Fatty Acids? 4 How Are
BCM 221 LECTURES OJEMEKELE O.
 BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
Genome-wide association studies for understanding pathogen evolution. Samuel K. Sheppard
 Genome-wide association studies for understanding pathogen evolution Samuel K. Sheppard Pathogenic bacteria Population genomics and evolution Sheppard et al. Science 2008; 11: 237-239. Sheppard et al.
Genome-wide association studies for understanding pathogen evolution Samuel K. Sheppard Pathogenic bacteria Population genomics and evolution Sheppard et al. Science 2008; 11: 237-239. Sheppard et al.
Supplementary Figure 1. High-affinity methane oxidation (HAMO) dynamics of soils with added methane at ppmv for 1 time and 10 times.
 Supplementary Figure 1. High-affinity methane oxidation () dynamics of soils with added methane at 10000 ppmv for 1 time and 10 times. After the complete consumption of 10000 ppmv methane, the measurement
Supplementary Figure 1. High-affinity methane oxidation () dynamics of soils with added methane at 10000 ppmv for 1 time and 10 times. After the complete consumption of 10000 ppmv methane, the measurement
Overview of the Expressway Cell-Free Expression Systems. Expressway Mini Cell-Free Expression System
 Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
CHAPTER 6 METABOLIC PATHWAY RECONSTRUCTION
 66 CHAPTER 6 METABOLIC PATHWAY RECONSTRUCTION 6.1 GENERAL A metabolic pathway is a series of biochemical reactions that occur within a cell and these chemical reactions are catalyzed by certain enzymes.
66 CHAPTER 6 METABOLIC PATHWAY RECONSTRUCTION 6.1 GENERAL A metabolic pathway is a series of biochemical reactions that occur within a cell and these chemical reactions are catalyzed by certain enzymes.
Applying Molecular Networking for the Detection of
 Supporting Information Applying Molecular Networking for the Detection of Natural Sources and Analogues of the Selective Gq Protein Inhibitor FR900359 Raphael Reher, Markus Kuschak, Nina Heycke, Suvi Annala,
Supporting Information Applying Molecular Networking for the Detection of Natural Sources and Analogues of the Selective Gq Protein Inhibitor FR900359 Raphael Reher, Markus Kuschak, Nina Heycke, Suvi Annala,
Supplementary Fig. A
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2015 10 17 33 41 42 99 Pseudomonas_alcaligenes_strain_IAM12411 NR_043419.1 Pseudomonas_oryzihabitans_strain_L-1
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2015 10 17 33 41 42 99 Pseudomonas_alcaligenes_strain_IAM12411 NR_043419.1 Pseudomonas_oryzihabitans_strain_L-1
MDSC 1102/VM1102 Cardiovascular and Renal. Purine nucleotide metabolism
 MDSC 1102/VM1102 Cardiovascular and Renal Purine nucleotide metabolism Dr. J. Foster Biochemistry Unit, Dept. Preclinical Sciences Faculty of Medical Sciences, U.W.I. Learning Objectives Discuss purineand
MDSC 1102/VM1102 Cardiovascular and Renal Purine nucleotide metabolism Dr. J. Foster Biochemistry Unit, Dept. Preclinical Sciences Faculty of Medical Sciences, U.W.I. Learning Objectives Discuss purineand
COMMUNICATION Acyl chain elongation drives ketosynthase substrate selectivity in trans-acyl transferase polyketide synthases
 Acyl chain elongation drives ketosynthase substrate selectivity in trans-acyl transferase polyketide synthases Matthew Jenner [a], José Pedro Afonso [a], Hannah R. Bailey [a], Sarah Frank [b], Annette
Acyl chain elongation drives ketosynthase substrate selectivity in trans-acyl transferase polyketide synthases Matthew Jenner [a], José Pedro Afonso [a], Hannah R. Bailey [a], Sarah Frank [b], Annette
Synthesis, evaluation of anti-hiv-1 and anti-hcv activity of novel 2,3 -dideoxy- 2,2 -difluoro-4 -azanucleosides
 Synthesis, evaluation of anti-hiv-1 and anti-hcv activity of novel 2,3 -dideoxy- 2,2 -difluoro-4 -azanucleosides Saúl Martínez-Montero, a,b Susana Fernández, a Yogesh S. Sanghvi, c Emmanuel A. Theodorakis,*
Synthesis, evaluation of anti-hiv-1 and anti-hcv activity of novel 2,3 -dideoxy- 2,2 -difluoro-4 -azanucleosides Saúl Martínez-Montero, a,b Susana Fernández, a Yogesh S. Sanghvi, c Emmanuel A. Theodorakis,*
GENERAL FEATURES OF FATTY ACIDS BIOSYNTHESIS
 1 GENERAL FEATURES OF FATTY ACIDS BIOSYNTHESIS 1. Fatty acids may be synthesized from dietary glucose via pyruvate. 2. Fatty acids are the preferred fuel source for the heart and the primary form in which
1 GENERAL FEATURES OF FATTY ACIDS BIOSYNTHESIS 1. Fatty acids may be synthesized from dietary glucose via pyruvate. 2. Fatty acids are the preferred fuel source for the heart and the primary form in which
Very-Long Chain Fatty Acid Biosynthesis
 Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Very-Long Chain Fatty Acid Biosynthesis Objectives: 1. Review information on the isolation of mutants deficient in VLCFA biosynthesis 2. Generate hypotheses to explain the absence of mutants with lesions
Avermectin. 2. Physical data (Avermectin B1a) White powder. C 48 H 72 O 14 ; mol wt Sol. in CHCl 3, acetone, MeOH. Insol. in H 2 O.
 Chapter Chapter 1 Avermectin 1. Discovery, producing organism and structures 1,) The discovery of avermectins was the result of a collaboration with Merck harp & Dohm Research Laboratories. Natural products
Chapter Chapter 1 Avermectin 1. Discovery, producing organism and structures 1,) The discovery of avermectins was the result of a collaboration with Merck harp & Dohm Research Laboratories. Natural products
SUPPLEMENTARY INFORMATION
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 2 ARTICLE NUMBER: 17084 Metabolic anticipation in Mycobacterium tuberculosis Hyungjin Eoh, Zhe Wang, Emilie Layre,
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 2 ARTICLE NUMBER: 17084 Metabolic anticipation in Mycobacterium tuberculosis Hyungjin Eoh, Zhe Wang, Emilie Layre,
Supplemental Figures
 Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
Department of Chemistry and Biochemistry, University of California Santa Cruz, 1156 High Street, Santa Cruz, CA 95064, USA
 Supplemental Information: Integration of high-content phenotypic screening and untargeted metabolomics analysis for the comprehensive functional annotation of complex natural products libraries. Kenji
Supplemental Information: Integration of high-content phenotypic screening and untargeted metabolomics analysis for the comprehensive functional annotation of complex natural products libraries. Kenji
gliomas. Fetal brain expected who each low-
 Supplementary Figure S1. Grade-specificity aberrant expression of HOXA genes in gliomas. (A) Representative RT-PCR analyses of HOXA gene expression in human astrocytomas. Exemplified glioma samples include
Supplementary Figure S1. Grade-specificity aberrant expression of HOXA genes in gliomas. (A) Representative RT-PCR analyses of HOXA gene expression in human astrocytomas. Exemplified glioma samples include
Supporting Information
 Supporting Information Harries et al. 1.173/pnas.9923916 A Fig. S1. Disruption of microfilaments within epidermal cells after treatment with 5 M Lat. Images of N. benthamiana cells are from plants expressing
Supporting Information Harries et al. 1.173/pnas.9923916 A Fig. S1. Disruption of microfilaments within epidermal cells after treatment with 5 M Lat. Images of N. benthamiana cells are from plants expressing
[U- 13 C5] glutamine. Glutamate. Acetyl-coA. Citrate. Citrate. Malate. Malate. Isocitrate OXIDATIVE METABOLISM. Oxaloacetate CO2.
![[U- 13 C5] glutamine. Glutamate. Acetyl-coA. Citrate. Citrate. Malate. Malate. Isocitrate OXIDATIVE METABOLISM. Oxaloacetate CO2. [U- 13 C5] glutamine. Glutamate. Acetyl-coA. Citrate. Citrate. Malate. Malate. Isocitrate OXIDATIVE METABOLISM. Oxaloacetate CO2.](/thumbs/76/74112154.jpg) Supplementary Figures a. Relative mrna levels Supplementary Figure 1 (Christofk) 3.0 2.5 2.0 1.5 1.0 0.5 0.0 LAT1 Fumarate Succinate Palmitate Acetyl-coA Oxaloacetate OXIDATIVE METABOLISM α-ketoglutarate
Supplementary Figures a. Relative mrna levels Supplementary Figure 1 (Christofk) 3.0 2.5 2.0 1.5 1.0 0.5 0.0 LAT1 Fumarate Succinate Palmitate Acetyl-coA Oxaloacetate OXIDATIVE METABOLISM α-ketoglutarate
Supplemental Information
 Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
University of Guelph Department of Chemistry and Biochemistry Structure and Function In Biochemistry
 University of Guelph Department of Chemistry and Biochemistry 19-356 Structure and Function In Biochemistry Final Exam, April 21, 1997. Time allowed, 120 min. Answer questions 1-30 on the computer scoring
University of Guelph Department of Chemistry and Biochemistry 19-356 Structure and Function In Biochemistry Final Exam, April 21, 1997. Time allowed, 120 min. Answer questions 1-30 on the computer scoring
Expression constructs
 Gene expressed in bebe3 ZmBEa Expression constructs 35S ZmBEa Pnos:Hygromycin r 35S Pnos:Hygromycin r 35S ctp YFP Pnos:Hygromycin r B -1 Chl YFP- Merge Supplemental Figure S1: Constructs Used for the Expression
Gene expressed in bebe3 ZmBEa Expression constructs 35S ZmBEa Pnos:Hygromycin r 35S Pnos:Hygromycin r 35S ctp YFP Pnos:Hygromycin r B -1 Chl YFP- Merge Supplemental Figure S1: Constructs Used for the Expression
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
Biosynthesis of Fatty Acids. By Dr.QUTAIBA A. QASIM
 Biosynthesis of Fatty Acids By Dr.QUTAIBA A. QASIM Fatty Acids Definition Fatty acids are comprised of hydrocarbon chains terminating with carboxylic acid groups. Fatty acids and their associated derivatives
Biosynthesis of Fatty Acids By Dr.QUTAIBA A. QASIM Fatty Acids Definition Fatty acids are comprised of hydrocarbon chains terminating with carboxylic acid groups. Fatty acids and their associated derivatives
Supplementary Figures and Tables
 Supplementary Figures and Tables Supplementary Figure 1. Study design and sample collection. S.japonicum were harvested from C57 mice at 8 time points after infection. Total number of samples for RNA-Seq:
Supplementary Figures and Tables Supplementary Figure 1. Study design and sample collection. S.japonicum were harvested from C57 mice at 8 time points after infection. Total number of samples for RNA-Seq:
Department of Chemistry, University of California, Davis, One Shields Avenue, Davis, CA 95616, USA
 Journal of Industrial Microbiology & Biotechnology (2001) 27, 378 385 D 2001 Nature Publishing Group 1367-5435/01 $17.00 www.nature.com/jim The biosynthetic gene cluster for the anticancer drug bleomycin
Journal of Industrial Microbiology & Biotechnology (2001) 27, 378 385 D 2001 Nature Publishing Group 1367-5435/01 $17.00 www.nature.com/jim The biosynthetic gene cluster for the anticancer drug bleomycin
Aflatoxin biosynthesis and regulation
 Aflatoxin biosynthesis and regulation Kenneth Ehrlich Southern Regional Research Center New rleans Introduction AF/ST biosynthesis involves expression of at least 25 genes Genes are organized in a cluster
Aflatoxin biosynthesis and regulation Kenneth Ehrlich Southern Regional Research Center New rleans Introduction AF/ST biosynthesis involves expression of at least 25 genes Genes are organized in a cluster
Supplemental data. Uppalapati et al. (2012) Plant Cell /tpc
 PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
Coenzymes. Coenzymes 9/11/2018. BCMB 3100 Introduction to Coenzymes & Vitamins
 BCMB 3100 Introduction to Coenzymes & Vitamins Cofactors Essential ions Coenzymes Cosubstrates Prosthetic groups Coenzymes structure/function/active group Vitamins 1 Coenzymes Some enzymes require for
BCMB 3100 Introduction to Coenzymes & Vitamins Cofactors Essential ions Coenzymes Cosubstrates Prosthetic groups Coenzymes structure/function/active group Vitamins 1 Coenzymes Some enzymes require for
Electron Transport Chain and Oxidative phosphorylation
 Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Protein Class/Name KEGG Pathways
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 4. Proteins Increased in Either Soil medium at 37 o C and 25 o C Table 4A. Proteins
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 4. Proteins Increased in Either Soil medium at 37 o C and 25 o C Table 4A. Proteins
Stereospecificity of Enoylreductase Domains from Modular Polyketide
 Stereospecificity of Enoylreductase Domains from Modular Polyketide Synthases Luyun Zhang, a,c Junjie Ji, b,c Meijuan Yuan, a Yuanyuan Feng, a Lei Wang, a Zixin Deng, a Linquan Bai, a Jianting Zheng* a
Stereospecificity of Enoylreductase Domains from Modular Polyketide Synthases Luyun Zhang, a,c Junjie Ji, b,c Meijuan Yuan, a Yuanyuan Feng, a Lei Wang, a Zixin Deng, a Linquan Bai, a Jianting Zheng* a
igem2013 Microbiology BMB SDU
 igem2013 Microbiology BMB SDU Project type: Creation of Biobrick with Prenyltransferase Project title: Biobrick_Prenyltransferase Sub project: Creation date: 13.08.09 Written by: SIS Performed by: PRA,
igem2013 Microbiology BMB SDU Project type: Creation of Biobrick with Prenyltransferase Project title: Biobrick_Prenyltransferase Sub project: Creation date: 13.08.09 Written by: SIS Performed by: PRA,
Supplementary Information
 Supplementary Information Figure S1 Sequencing quality. Sample screenshot of re-sequencing raw file quality generated by FastQC. For both the graphs, scores were based on the Illumina 1.5 quality score.
Supplementary Information Figure S1 Sequencing quality. Sample screenshot of re-sequencing raw file quality generated by FastQC. For both the graphs, scores were based on the Illumina 1.5 quality score.
Influenza hemagglutinin protein stability correlates with fitness and seasonal evolutionary dynamics
 Influenza hemagglutinin protein stability correlates with fitness and seasonal evolutionary dynamics Eili Y. Klein, Ph.D. Assistant Professor, Department of Emergency Medicine & Fellow, Center for Disease
Influenza hemagglutinin protein stability correlates with fitness and seasonal evolutionary dynamics Eili Y. Klein, Ph.D. Assistant Professor, Department of Emergency Medicine & Fellow, Center for Disease
Analysis of Uncomplexed and Copper-complexed Methanobactin with UV/Visible Spectrophotometry, Mass Spectrometry and NMR Spectrometry
 Analysis of Uncomplexed and Copper-complexed Methanobactin with UV/Visible Spectrophotometry, Mass Spectrometry and NMR Spectrometry Lee Behling, Alan DiSpirito, Scott Hartsel, Larry Masterson, Gianluigi
Analysis of Uncomplexed and Copper-complexed Methanobactin with UV/Visible Spectrophotometry, Mass Spectrometry and NMR Spectrometry Lee Behling, Alan DiSpirito, Scott Hartsel, Larry Masterson, Gianluigi
The biosynthetic gene cluster for the anticancer
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Liangcheng Du Publications Published Research - Department of Chemistry December 2001 The biosynthetic gene cluster for
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Liangcheng Du Publications Published Research - Department of Chemistry December 2001 The biosynthetic gene cluster for
Table removed due to copyright reasons.
 atural Product Biosynthesis Relevance of natural products in the pharmaceutical industry (see Table 2 in Butler J. at. Prod. 2004, 67, 2141-2153.)) Table removed due to copyright reasons. 2 * a phosphopantetheinyl
atural Product Biosynthesis Relevance of natural products in the pharmaceutical industry (see Table 2 in Butler J. at. Prod. 2004, 67, 2141-2153.)) Table removed due to copyright reasons. 2 * a phosphopantetheinyl
