Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted. residues are colored red. Numbers indicate the position of each residue.
|
|
|
- Brittany Alannah Woods
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue.
2 Supplementary Figure 2. SDS-PAGE analysis of purified recombinant proteins. 12.5% acrylamide gel was used for GodD, GodE, GodF, and LazF. 15% acrylamide gel was used for GodH. BLUE Star Prestained Protein-Ladder (NIPPON Genetics Co, Ltd.) was used as a molecular weight marker.
3 Supplementary Figure 3. The biosynthetic gene cluster for lactazoles.
4 Supplementary Figure 4. LC-MS analysis of flavin co-factor bound to recombinant GodE and LazF. a, chromatograms extracted at m/z 457 [M+H] + were shown. Chromatograms from denatured GodE and LazF were compared with that of authentic flavin mononucleotide (FMN). b, MS 2 spectra of FMN (m/z 457) detected in each sample.
5 Supplementary Figure 5. LC-MS analysis of co-factor bound to recombinant GodH. The methanol extract of GodH (a) was compared with authentic acetyl-coa (b), Chromatograms were extracted at m/z 808 [M-H] -. MS 2 spectra derived from m/z 808
6 precursor ions were shown in boxes.
7 Supplementary Figure 6. MALDI-TOF-MS analysis of the reaction product of GodF reaction with GodA*. Red line is from enzymatic reaction and black line from control. Calculated and observed m/z values in MALDI-TOF-MS analyses are summarized in Supplementary Data.
8 Supplementary Figure 7. MALDI-TOF-MS analysis of the reaction catalyzed by GodD and GodE with GodA*1 2 and GodA*2 3. Red lines are from enzymatic reactions and black lines from control. Calculated and observed m/z values in MALDI- TOF-MS analyses are summarized in Supplementary Data.
9
10 Supplementary Figure 8. MALDI-TOF-MS analysis of the reaction catalyzed by GodD and GodE with GodA*1 9-T2A/T5A/C8A, GodA*1 6-T2A/T5A, and GodA*1 3-T2A. Red lines are from enzymatic reactions and black lines from control. Calculated and observed m/z values in MALDI-TOF-MS analyses are summarized in Supplementary Data.
11 Supplementary Figure 9. MALDI-TOF-MS analysis of the reaction catalyzed by GodD and GodE with truncated mutants. GodA*1 8 (a), GodA*2 9 (b), GodA*1 5 (c), and GodA*2 6 (d) were used as substrates. Red lines are from enzymatic reactions and black lines from control. Calculated and observed m/z values in MALDI-TOF-MS analyses are summarized in Supplementary Data.
12 Supplementary Figure 10. MALDI-TOF-MS analysis of the reaction catalyzed by GodD and GodE with GodA*1-3 mutants whose X1/X2 residues were substituted with S/T/C residues. GodA*1 3-A1C/T2A (a), GodA*1 3-A1T/T2A (b), GodA*1 3- A1S/T2A (c), GodA*1 3-T2A/V3C (d), GodA*1 3-T2A/V3T (e), and GodA*1 3- T2A/V3S (f) were used as substrates. Red lines are from enzymatic reactions and black lines from control. Calculated and observed m/z values in MALDI-TOF-MS analyses are summarized in Supplementary Data.
13 Supplementary Figure 11. MALDI-TOF-MS analysis of the reaction catalyzed by GodD and GodE with GodA*1 9-A1T/T2A (a), GodA*1 6-A1T/T2A (b), GodA*1 6-T5I/I6T (c), GodA*1 9-T2V/V3T (d), GodA*1 6-T2V/V3T (e), and GodA*1 9- L7C/C8L (f). Red lines are from enzymatic reactions and black lines from control. Calculated and observed m/z values in MALDI-TOF-MS analyses are summarized in Supplementary Data.
14 Supplementary Figure 12. MALDI-TOF-MS analysis of the reaction catalyzed by GodD and GodE with GodA*1 9--1A1 (a), GodA*1 9-3A4 (b), and GodA*1 9-6A7 (c). Red lines are from enzymatic reactions and black lines from control. Calculated and observed m/z values in MALDI-TOF-MS analyses are summarized in Supplementary Data.
15 Supplementary Figure 13. MALDI-TOF-MS analysis of the reaction catalyzed by GodD and GodE with GodA*1 3-T2S. Red line is from enzymatic reaction and black line from control. Calculated and observed m/z values in MALDI-TOF-MS analyses are summarized in Supplementary Data.
16 Supplementary Figure 14. MALDI-TOF-MS analysis of the reaction catalyzed by GodD and GodE with GodA*1-3 mutants which harbor the mutation at X1/X2 residues. GodA*1 3-A1E (a), GodA*1 3-A1R (b), GodA*1 3-A1Y (c), GodA*1 3- A1W (d), GodA*1 3-V3R (e), GodA*1 3-V3W (f), and GodA*1 3-V3E (g) were used as substrates. Red lines are from enzymatic reactions and black lines from control. Calculated and observed m/z values in MALDI-TOF-MS analyses are summarized in Supplementary Data.
17
18 Supplementary Figure 15. MALDI-TOF-MS analysis of the reaction catalyzed by GodD, GodE, GodF, and LazF with GodA* (a), GodA*1 16 (b), GodA*1 13 (c), GodA*1-9 (d) and GodA*1 6 (e). Red lines are from enzymatic reactions and black lines from control. Calculated and observed m/z values in MALDI-TOF-MS analyses are summarized in Supplementary Data.
19 Supplementary Figure 16. MALDI-TOF-MS analysis of the reaction catalyzed by GodD, GodE, GodF, and LazF with GodA*-T5A and GodA*-S15A. Red lines are from enzymatic reactions and black lines from control. Calculated and observed m/z values in MALDI-TOF-MS analyses are summarized in Supplementary Data.
20 Supplementary Figure 17. MALDI-TOF-MS analysis of the reaction catalyzed by GodD, GodE, GodF, and LazF with GodA*LP(-25) (-1) and GodA*LP(-6) (-1) Red lines are from enzymatic reactions and black lines from control. Calculated and observed m/z values in MALDI-TOF-MS analyses are summarized in Supplementary Data.
21 Supplementary Figure 18. LC-MS analyses of the metabolites produced by Streptomyces sp. TP-A0584 goda harboring pgodr and goda mutants. Red
22 diamondss indicate the major GS derivatives. Gray diamond indicates the minor derivative mentioned in the text. Chromatograms were extracted at 254 nm.
23 Supplementary Figure 19. MS fragmentation analysis of A1S. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn. The fragment, i (y16, b15) was observed as a divalent ion.
24 Supplementary Figure 20. MS fragmentation analysis of V3S. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicat the internal peptide fragment derived from fragments ym and a/bn. The fragment, i (y16, b15) was observed as a divalent ion.
25 Supplementary Figure 21. MS fragmentation analysis of I6S. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn. The fragment, i (y16, b15) was observed as a divalent ion.
26 Supplementary Figure 22. MS fragmentation analysis of the major product of L7S. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn. The fragment, i (y16, b15) was observed as a divalent ion.
27 Supplementary Figure 23. MS fragmentation analysis of G10S. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn. The fragment, i (y16, b15) was observed as a divalent ion.
28 Supplementary Figure 24. MS fragmentation analysis of G11S. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn. The fragment, i (y16, b15) was observed as a divalent ion.
29 Supplementary Figure 25. MS fragmentation analysis of L13S. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn.
30 Supplementary Figure 26. MS fragmentation analysis of A16S. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn. The fragment, i (y13, b15) was observed as a divalent ion.
31 Supplementary Figure 27. MS fragmentation analysis of G17S. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn. The fragment, i (y16, b15) was observed as a divalent ion.
32 Supplementary Figure 28. MS fragmentation analysis of V19S. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn. The fragment, i (y16, b15) was observed as a divalent ion.
33 Supplementary Figure 29. MS fragmentation analysis of the minor product of L7S. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn.
34 Supplementary Figure 30. MS fragmentation analysis of I6Y. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn. The fragment, i (y16, b15) was observed as a divalent ion.
35 Supplementary Figure 31. MS fragmentation analysis of V19Y. MS E spectrum and assignment for fragmentation are shown. The labels, i (ym, a/bn) indicate the internal peptide fragment derived from fragments ym and a/bn. The fragment, i (y16, b15) was observed as a divalent ion.
36 Supplementary Figure H NMR spectrum of GS-10SA11 (DMSO-d6, 600 MHz)
37 Supplementary Figure C NMR spectrum of GS-10SA11 (DMSO-d6, 150 MHz). Asterisk indicates the signal derived from impurity.
38 Supplementary Figure 34. COSY spectrum of GS-10SA11 (DMSO-d6)
39 Supplementary Figure 35. HSQC spectrum of GS-10SA11 (DMSO-d6)
40 Supplementary Figure 36. CT-HMBC-1 1 spectrum of GS-10SA11 (DMSO-d6)
41 Supplementary Figure 37. TOCSY spectrum of GS-10SA11 (DMSO-d6)
42 Supplementary Figure 38. Correlations observed in 2D NMR analyses of GS-10SA11. Bold lines indicate correlations in COSY and TOCSY analyses. Arrows indicate correlations in CT-HMBC-1.
43 Supplementary Table 1. Summary of NMR analysis of GS-10SA11 (DMSO-d6). No. residue δ C δ H (m, J in Hz) 1 AcAla NH (d,7.8) Ca (quint, 7.8) Cb (d, 7.2) Ac-CO Ac-Me (s) 2 Mz Me (s)** C=O Val NH (d, 9.0) C=O Ca (dd, 6.6, 9.0) Cb (m) Cg (d, 6.6) Cg (m) 4 Dha NH (s) Ca Cb (s) 5.69 (s) 5 Mz Me (s) C=O Ile NH (d, 9.6) C=O Ca (m) Cb (m) Cg (m) 1.06 (m) Cg (m)
44 Cδ (t, 7.2) 7 Leu NH (d, 8.4) Ca (q, 7.5) Cb (t, 7.2) Cg (m) Cd (m) Cd (d, 7.2) 8 Tz (s) C=O Ser NH (d, 7.8) C=O Ca (m) Cb (m) 3.68 (m) -OH (brt. 4.8) 10 Gly NH (t, 6.0) Ca (dd, 5.4, 16.8) 4.5 (m) 11 Oz * (s) C=O Ala NH (d, 8.4) C=O Ca (m) Cβ (d, 7.2) 13 Gly NH (t, 5.4) Ca (d, 6.0) 14 Mz Me (s) C=O
45 15 Leu NH (d, 9.0) C=O Ca (td, 4.2, 9.0) Cβ (m) 1.59 (m) Cg (m) Cd (m) Cd (m) 16 Dha NH (s) Ca Cβ (s) 5.70 (s) 17 Oz * (s) C=O Ala NH (d, 7.2) C=O Ca (m) Cβ (d, 7.2) 19 Gly NH (t, 6.0) Ca (dd, 6.0, 16.8) 4.58 (dd, 7.2, 16.2) 20 Tz (s) C=O Val NH (d, 9.0) C=O Ca (dd, 5.4, 9.0) Cβ (m) Cg (m) Cg (m) *: These signals are exchangeable. Assignments were based on chemical shifts in the
46 literature because no correlations were observed in 2D NMR spectra. **: This signal was overlapped by the signal derived from residual DMSO.
47 Supplementary Table 2. Observed and calculated m/z of designer GS analogs produced in vivo, and estimated chemical formula of monoisotopic ions. Name Observed Calculated Chemical formula GS-10SA [M+H] [M+H] + C78 H104 N21 O22 S2 GS-tandem [M+2H] [M+2H] 2+ C142 H192 N38 O38 S4
48 Supplementary Table 3. Sequences of godd and goda-tandem. gene Sequence (shown from 5 to 3 ) godd ATGGCACCGGTGTTACACTTTCGCCGCAGCTTTAAGGTGGACCTG CTGCAGGGTGACGGTGTTTACCTGACCAGTGATCGTGGTGAGAGC ACAGTGCTGCGTGGTCAGCTGGTTGAGAGCTTAAGTCCGCTGCTG ATGGCAGGCCGCACAGAAGATGATCTGGTTACCGCCCTGGCAGGT ATGTTCCCTGCACAGCGCATCTTAAGCGCACTGCGCCAACTGGAA AAAGCAGGCTACGTTCGCCGTGCCGAGGCAGCAACCGGTGCAGA GACCTTTGCCGCAGGTTTCTGGGAGAGCAGTGGTGTGGACGGCA GTACCGCACTGGCCCGTCTGGCAAGCCGTACCGTTCGTATTACAG GTGTTGGCCATGCCGGCAGCGAAGGTGATGTGGTTCGTCGTGTGC GCAAAGCAGGTAGCAGTCTGGGCCTGCATCTGACCGACGGCAGC GCAGACCTGACAGTGGTTGTGACCGTGGACTATCTGGAGCCGGAA CTGGCCCGCATCAACGCAGAGGCCTTAGCCGATGGCCGCCCGTG GATGCTGGCAAAACCTGGCGGTAGCGTTGCCTGGTTTGGTCCGTT CTTCCGCCCGGGCGAAAGCGCATGCTGGGCCTGTCTGGCCCATC GTCTGAGCGGCAACCGCATGATCGAAACCTATGTGGCCAAAGCAC CGACACGCAGCGCAAGCTATAGCCCGACCGTGAACCTGCCGGCC ACACAAACCGCAGCAGAGGAGTTAACCGCCCTGCACGCAGCCAAA TGGTTAGCAGGCGTTTATGCCGCCCAGGCCAGTGAACACAGTGCA CGCCCGCAGGGTCTGAATCCGGATCTGCTGCACACATTCGATGCA GTTACCTTAGCCGCACAGGAGCACCTGGTGTTTCGTCGTCCGCAG TGCCCTGAGTGTGGTGATGGCGAGTTAATGGCCCGCCAGCATCAG AGTCCGGTTCGTCTGCGCAGCATGCCGAAGGTTACACTGGTGGAC GGTGGTCATCGTAGCAAAGACCCGCAGCGTATGCTGGACCTGCAT GGCCACCTGATTAGCAGCGCCCTGGGTCCGGTTACCGGCTTACAG AAGGTTCCGAGCGTTTGGCCTGGCTTCCATGCCTATACCGCCGGT CAAAATTTCGCCATCCCGATGAGCCGTCCTGGTGATTTACGTGTGG GCCTGCGTAGTCAGAGCTGCGGCAAAGGTATGAGCGATTTACAGG CCCGCGCAAGTGCACTGGGTGAAGCCCTGGAGCGTTACAGTGGT GTTTATCAGGGCGACGAAGCACGCATTACCGCAAGTTACGACGATC TGGGCGATCGCGCAATCGCCCCGAACGATTTAGCCCTGTATAGTGC CCGCCAGTTCGACGAACGCGAAGAGTGGAACAACCGCGACGTTC ATTTCCACCGTGTGCTGGCCCCTTTCGACACCGCAGCACCGATTG ACTGGACCCCGGTGTGGAGTCTGACCATGCAGCGCCATCGCTATG TTCCTACCGCCAGCCTGTTCTACGGTTACCCTCTGGATCGCGACCA
49 goda-tandem TCAGTATGCAGCCGCCGATAGCAATGGCAGCGCAGCCGGTACCAG CATCGAGGACGCAGTGCTGCAAGGTTTTATGGAGCTGGTTGAGCG TGACAGTGTGGCCCTGTGGTGGTATAACCGTGTGCAACGTCCTGA AGTGGATCTGCAGAGTTTCGGCGAGCCGTATTTTCTGGAGTGGCT GGCCCAATATCGCAGTCTGAATCGTGAGGCATGGGTGCTGGATCT GACCAGCGATTTCGGTATCCCGGTGATGGCAGCCATTAGCCGTCG TATCGACAAGCCGGCCGAAGACATCCTGATCGCATTCGGTGCACA CTTTGACGCCCGTATTGCCGTTGGTCGTGCCCTGACAGAGATGAA CCAATTCCTGCCGGCCGTTGTTCACGCCAAACCGGAAGGTGGTGG TTATACATACCCGGATCCGGCCCAGCAGCATTGGTGGCAAACCGCC ACACTGGCCAACCAACCTTATCTGCGCCCGCTGAGTGCACCGCGT CGTACCGCAGGTGACTTCCCTGTGCACGAAAGTCTGGATCTGCTG GATGATCTGCATCGCGCACAAGCAACCGTGGAAGAGCACGGCATG GAGCTGCTGGTGATCAATCAGACCCGCCCGGACGTGGGTCTGCC GGTTGTTAAGGTGATTGTGCCTGGCATGCGTCACTTCTGGCCGCG TTTCGCCCCGGGTCGCTTATACGATGTGCCGGTGAAACTGGGCTG GGTTAGCCAGCAGACCCGCGAAGAGGACCTGAATCCGATTGGCGT GTTTATTTGA ATGGAGAACGTCCAGACCCTGGCGATCGACGACATCGAGAACATC GACGCTGAGGTGACCATCGAGGAGCTTTCCTCGACCAACGGCGC CGCCACCGTCAGCACCATCCTGTGCAGCGGCGGCACCCTCAGCT CGGCCGGCTGCGTCGCCACCGTCAGCACCATCCTGTGCAGCGGC GGCACCCTCAGCTCGGCCGGCTGCGTCTGA
50 Supplementary Table 4. Primers used for gene cloning. Restriction sites are underlined and corresponding restriction enzymes are shown in parentheses. Name pgoddopt-f-ndei pgoddopt-r-xhoi gode-n-nde gode-cter-hind godf-n-nde godf-c-xho lazf-ndei lazf-r-xhoi godh-n-nde godh-c-xho Sequence (restriction sites were underlined) GGGGGGCATATGGCACCGGTGTTAC (NdeI) GGGCTCGAGTTAAATAAACACGCCAATCGG (XhoI) GGTGTTCATCATATGATTGTCTCCGACACG (NdeI) CGGGCGAAGCTTTTGGGACTCGTCCTCCAG (HindIII) GGACGAGTCCATATGACCTCGCCCGCCGCA (NdeI) GATTCGCTCGAGTCAGACAAATGCCACCAG (XhoI) GGGGGGCATATGACCACCCACGCG (NdeI) GGGCTCGAGCTCCATGATCACCG (XhoI) GAAGGATTCCATATGAATGAGATCACCTGG (NdeI) TGCGTTCTCGAGTTATGACTGAAGTGAAAC (XhoI)
51 Supplementary Table 5. Primers used for the syntheses of DNA templates for in vitro translation. No. Name Sequence (restriction site) 1 goda*-f1 GGCGTAATACGACTCACTATAGGGTTAACTTTAACAAGGAGAAAAACAT 2 goda*-f2 GGCGTAATACGACTCACTATAG 3 goda*-r1 GATATTCTCAATATCGTCAATTGCCAGAGTTTGCACGTTTTCCATGTTTTTCTCCTTGTT 4 goda*-r2 ACCATTAGTAGAGCTCAGTTCCTCAATCGTAACCTCAGCGTCGATATTCTCAATATCGTC 5 goda*-r3 CCCCCGCTACACAAGATCGTGCTCACGGTAGCTGCACCATTAGTAGAGCTCAG 6 goda*-r4 CGAAGCTTAAACGCAACCTGCAGAACTAAGAGTACCCCCGCTACACAAG 7 goda*truncate_r2 CACCATTGGTAGAGCTCAGTTCCTCAATGGTAACCTCAGCGTCGATATTCTCAATATCGTC 8 goda*1 9_R3 CGAAGCTTATGAACATAAGATGGTAGACACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 9 goda*1 6-R3 CGAAGCTTAGATGGTAGACACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 10 goda*1 3-R3 CGAAGCTTACACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 11 goda*1 2-R3 CGAAGCTTAGGTAGCTTCTGCACCATTAGTAGAGCTCAG 12 goda*2 3-R3 CGAAGCTTACACGGTTTCTGCACCATTAGTAGAGCTCAG 13 goda*1 9T2A/T5A/C8A-R3 CGAAGCTTATGAAGCTAAGATAGCAGACACTGCAGCTTCTGCACCATTAGTAGAGCTCAG 14 goda*1 6-T2A/T5A-R3 CGAAGCTTAGATAGCAGACACTGCAGCTTCTGCACCATTAGTAGAGCTCAG 15 goda*1 3-T2A -R3 CGAAGCTTACACTGCAGCTTCTGCACCATTAGTAGAGCTCAG 16 goda*1 8-R3 CGAAGCTTAACATAAGATGGTAGACACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 17 goda*2 9-R3 CGAAGCTTATGAACATAAGATGGTAGACACGGTTTCTGCACCATTAGTAGAGCTCAG 18 goda*1 5-R3 CGAAGCTTAGGTAGACACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 19 goda*2 6-R3 CGAAGCTTAGATGGTAGACACGGTTTCTGCACCATTAGTAGAGCTCAG 20 goda*1 3-A1C/T2A-R3 CGAAGCTTACACAGCACATTCTGCACCATTAGTAGAGCTCAG
52 21 goda*1 3-A1T/T2A-R3 CGAAGCTTACACAGCGGTTTCTGCACCATTAGTAGAGCTCAG 22 goda*1 3-A1S/T2A-R3 CGAAGCTTACACAGCAGATTCTGCACCATTAGTAGAGCTCAG 23 goda*1 3-T2A/V3C-R3 CGAAGCTTAACATGCAGCTTCTGCACCATTAGTAGAGCTCAG 24 goda*1 3-T2A/V3T-R3 CGAAGCTTAGGTTGCAGCTTCTGCACCATTAGTAGAGCTCAG 25 goda*1 3-T2A/V3S-R3 CGAAGCTTAAGATGCAGCTTCTGCACCATTAGTAGAGCTCAG 26 goda*1 9-A1T/T2A-R3 CGAAGCTTATGAACATAAGATGGTAGACACAGCGGTTTCTGCACCATTAGTAGAGCTCAG 27 goda*1 6-A1T/T2A -R3 CGAAGCTTAGATGGTAGACACAGCGGTTTCTGCACCATTAGTAGAGCTCAG 28 goda*1 6-T5I/I6T-R3 CGAAGCTTAGGTGATAGACACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 29 goda*1 9-T2V/V3T-R3 CGAAGCTTATGAACATAAGATGGTAGAGGTCACAGCTTCTGCACCATTAGTAGAGCTCAG 30 goda*1 6-T2V/V3T -R3 CGAAGCTTAGATGGTAGAGGTCACAGCTTCTGCACCATTAGTAGAGCTCAG 31 goda*1 9-L7C/C8L-R3 CGAAGCTTATGATAAACAGATGGTAGACACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 32 goda* A 1-R3 CGAAGCTTATGAACATAAGATGGTAGACACGGTAGCTGCTTCTGCACCATTAGTAGAGCTCAG 33 goda*1 9-3A 4-R3 CGAAGCTTATGAACATAAGATGGTAGATGCCACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 34 goda*1 9-6A7 -R3 CGAAGCTTATGAACATAATGCGATGGTAGACACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 35 goda*1 3-T2C-R3 CGAAGCTTACACACAAGCTTCTGCACCATTAGTAGAGCTCAG 36 goda*1 3-T2S-R3 CGAAGCTTACACTGAAGCTTCTGCACCATTAGTAGAGCTCAG 37 goda*1 3-A1E-R3 CGAAGCTTACACGGTTTCTTCTGCACCATTAGTAGAGCTCAG 38 goda*1 3-A1R-R3 CGAAGCTTACACGGTACGTTCTGCACCATTAGTAGAGCTCAG 39 goda*1 3-A1Y-R3 CGAAGCTTACACGGTATATTCTGCACCATTAGTAGAGCTCAG 40 goda*1 3-A1W-R3 CGAAGCTTACACGGTCCATTCTGCACCATTAGTAGAGCTCAG 41 goda*1-3-v3r-r3 CGAAGCTTAACGGGTAGCTTCTGCACCATTAGTAGAGCTCAG 42 goda*1 3-V3Y-R3 CGAAGCTTAATAGGTAGCTTCTGCACCATTAGTAGAGCTCAG
53 43 goda*1 3-V3W-R3 CGAAGCTTACCAGGTAGCTTCTGCACCATTAGTAGAGCTCAG 44 goda*1 3-V3E-R3 CGAAGCTTATTCGGTAGCTTCTGCACCATTAGTAGAGCTCAG 45 goda*1 16-R3 CGAAGCTTATGCAGAACTAAGAGTACCCCCGCTACACAAG 46 goda*1 13-R3 CGAAGCTTAAAGAGTACCCCCGCTACACAAG 47 goda*t5a-r3 CCCCCGCTACACAAGATTGCGCTCACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 48 goda*t15a-r4 CGAAGCTTAAACGCAACCTGCTGCACTAAGAGTACCCCCGCTACACAAG 49 goda*s4a-r3 CCCCCGCTACACAAGATCGTAGCCACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 50 goda*s14a-r4 CGAAGCTTAAACGCAACCTGCAGATGCAAGAGTACCCCCGCTACACAAG 51 goda* LP(-25) (-1)-R1 GATATTCTCAATATCGTCAATTGCCAGCTTTTTCTTCATGTTTTTCTCCTTGTT 52 goda* LP(-20) (-1)-R1 GCGTCGATATTCTCAATCTTTTTCTTCATGTTTTTCTCCTTGTT 53 goda* LP(-20) (-1)-R2 CACCATTAGTAGAGCTCAGTTCCTCAATCGTAACCTCAGCGTCGATATTCTCAAT 54 goda* LP(-15) (-1)-R1 GTAGAGCTCAGTTCCTCAATCGTAACCTCAGCCTTTTTCTTCATGTTTTTCTCCTTGTT 55 goda* LP(-15) (-1)-R2 CCCCCGCTACACAAGATCGTGCTCACGGTAGCTTCTGCACCATTAGTAGAGCTCAGTTCCTCAAT 56 goda* LP(-10) (-1)-R1 CCATTAGTAGAGCTCAGTTCCTCCTTTTTCTTCATGTTTTTCTCCTTGTT 57 goda* LP(-10) (-1)-R2 CCCCCGCTACACAAGATCGTGCTCACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 58 goda* LP(-6) (-1)-R1 GCTTCTGCACCATTAGTAGACTTTTTCTTCATGTTTTTCTCCTTGTT 59 goda* LP(-6) (-1)-R2 GAGTACCCCCGCTACACAAGATCGTGCTCACGGTAGCTTCTGCACCATTAGT 60 goda* LP(-6) (-1)-R3 CGAAGCTTAAACGCAACCTGCAGAACTAAGAGTACCCCCGCTACA 61 goda*-10sa11-r3 GACCCAGAACACAAGATCGTTGACACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 62 goda*-10sa11-r4 CGAAGCTTAAACGCAACCTGCAGATGAAAGAGTACCAGCTGACCCAGAACACAAG 63 goda*-tandem-r3 GTACCCCCGCTACATAAGATCGTGCTCACGGTAGCTTCTGCACCATTAGTAGAGCTCAG 64 goda*-tandem-r4 CAGAATGGTAGAAACAGTTGCAACGCAACCTGCAGAACTAAGGGTACCCCCGCTACAT
54 Supplementary Table 6. List of primers used for the synthesis of DNA templated for GodA derivatives. Primers are indicated by the numbers in Supplementary Table 5. extension 1st PCR 2nd PCR 3rd PCR 4th PCR Name Forward primer Reverse primer Forward primer Reverse primer Forward primer Reverse primer Forward primer Reverse primer Forward primer Reverse primer GodA* N/A GodA* N/A N/A GodA* N/A N/A GodA* N/A N/A GodA* N/A N/A GodA* N/A N/A GodA*1 9/T2A/T5A/C8A N/A N/A GodA*1 6-T2A/T5A N/A N/A GodA*1 3-T2A N/A N/A GodA* N/A N/A GodA* N/A N/A GodA* N/A N/A GodA* N/A N/A GodA*1 3-A1C/T2A N/A N/A GodA*1 3-A1T/T2A N/A N/A
55 GodA*1 3- A1S/T2A N/A N/A GodA*1 3-T2A/V3C N/A N/A GodA*1 3-T2A/V3T N/A N/A GodA*1 3-T2A/V3S N/A N/A GodA*1 9-A1T/T2A N/A N/A GodA*1 6-A1T/T2A N/A N/A GodA*1 6-T5I/I6T N/A N/A GodA*1 9-T2V/V3T N/A N/A GodA*1 6-T2V/V3T N/A N/A GodA*1 9-L7C/C8L N/A N/A GodA*1 9--1A N/A N/A GodA*1 9-3A N/A N/A GodA*1 9-6A N/A N/A GodA*1 3-T2C N/A N/A GodA*1 3-T2S N/A N/A GodA*1 3-A1E N/A N/A GodA*1 3-A1R N/A N/A GodA*1 3-A1Y N/A N/A GodA*1 3-A1W N/A N/A GodA*1-3-V3R N/A N/A GodA*1 3-V3Y N/A N/A GodA*1 3-V3W N/A N/A
56 GodA*1 3-V3E N/A N/A GodA* N/A N/A GodA* N/A N/A GodA*-T5A N/A GodA*-S15A N/A GodA*-S4A N/A GodA*-S14A N/A GodA*LP(-25) (-1) N/A GodA*LP(-20) (-1) N/A GodA*LP(-15) (-1) N/A N/A GodA*LP(-10) (-1) N/A N/A GodA*LP(-6) (-1) N/A N/A GodA*-10SA N/A GodA*-tandem
57 Supplementary Table 7. Primers for site-directed mutagenesis of goda. # Name Sequence 1 A1S_sense TCGACCAACGGCGCCTCCACCGTCAGCACC 2 A1S_anti GGTGCTGACGGTGGAGGCGCCGTTGGTCGA 3 V3S_sense AACGGCGCCGCCACCAGCAGCACCATCCTG 4 V3S_anti CAGGATGGTGCTGCTGGTGGCGGCGCCGTT 5 I6S_sense GCCACCGTCAGCACCAGCCTGTGCAGCGGC 6 I6S_anti GCCGCTGCACAGGCTGGTGCTGACGGTGGC 7 I6Y_sense GCCACCGTCAGCACCTACCTGTGCAGCGGC 8 I6Y_anti GCCGCTGCACAGGTAGGTGCTGACGGTGGC 9 L7S_sense ACCGTCAGCACCATCTCGTGCAGCGGCGGC 10 L7S_anti GCCGCCGCTGCACGAGATGGTGCTGACGGT 11 G10S_sense ACCATCCTGTGCAGCAGCGGCACCCTCAGC 12 G10S_anti GCTGAGGGTGCCGCTGCTGCACAGGATGGT 13 G11S_sense ATCCTGTGCAGCGGCAGCACCCTCAGCTCG 14 G11S_anti CGAGCTGAGGGTGCTGCCGCTGCACAGGAT 15 L13S_sense TGCAGCGGCGGCACCTCCAGCTCGGCCGGC 16 L13S_anti GCCGGCCGAGCTGGAGGTGCCGCCGCTGCA 17 A16S_sense GGCACCCTCAGCTCGTCCGGCTGCGTCTGA 18 A16S_anti TCAGACGCAGCCGGACGAGCTGAGGGTGCC 19 G17S_sense ACCCTCAGCTCGGCCAGCTGCGTCTGATCG 20 G17S_anti CGATCAGACGCAGCTGGCCGAGCTGAGGGT 21 V19S_sense AGCTCGGCCGGCTGCAGCTGATCGGTCGTC 22 V19S_anti GACGACCGATCAGCTGCAGCCGGCCGAGCT 23 V19Y_sense AGCTCGGCCGGCTGCTACTGATCGGTCGTC 24 V19Y_anti GACGACCGATCAGTAGCAGCCGGCCGAGCT 25 10SA11_sense ATCCTGTGCAGCGGCAGCGCCGGCACCCTC 26 10SA11_anti GAGGGTGCCGGCGCTGCCGCTGCACAGGAT
58 Supplementary References 1 Furihata, K. & Seto, H. Constant time HMBC (CT-HMBC), a new HMBC technique useful for improving separation of cross peaks. Tetrahedron Letters 39, , doi: /s (98) (1998).
Supporting Information
 Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
Supplementary Figure 1
 Supplementary Figure 1 Isolation of mt-trnas and RNA-MS analysis of mt-trna Asn from M. nudus (a)m. nudus mt-trnas were isolated by RCC and resolved by 10% denaturing PAGE. The gel was stained with SYBR
Supplementary Figure 1 Isolation of mt-trnas and RNA-MS analysis of mt-trna Asn from M. nudus (a)m. nudus mt-trnas were isolated by RCC and resolved by 10% denaturing PAGE. The gel was stained with SYBR
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol
 Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced to express
 Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the
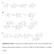 Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis
 Supporting information Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis Martin F. Kreutzer and Markus Nett * Table of contents SI-2, SI-3 SI-4 SI-5 SI-6
Supporting information Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis Martin F. Kreutzer and Markus Nett * Table of contents SI-2, SI-3 SI-4 SI-5 SI-6
Supporting Information
 Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany A Convergent Synthesis of N-Linked Glycopeptides Clyde M. Kaneshiro, Katja Michael* 1. LC-MS analysis of crude glycopeptide 16. 2. ESI-TOF
Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany A Convergent Synthesis of N-Linked Glycopeptides Clyde M. Kaneshiro, Katja Michael* 1. LC-MS analysis of crude glycopeptide 16. 2. ESI-TOF
Supplementary Materials: An NMR Guided Screening Method for Selective Fragment Docking and Synthesis of a Warhead Inhibitor
 Molecules 216, 21, 846; doi:1.339/molecules217846 S1 of S14 Supplementary Materials: An NMR Guided Screening Method for Selective Fragment Docking and Synthesis of a Warhead Inhibitor Ram B. Khattri, Daniel
Molecules 216, 21, 846; doi:1.339/molecules217846 S1 of S14 Supplementary Materials: An NMR Guided Screening Method for Selective Fragment Docking and Synthesis of a Warhead Inhibitor Ram B. Khattri, Daniel
Identification, characterization of a flavin-containing monooxygenase MoA and its function in a
 1 Applied Microbiology and Biotechnology 2 Supplementary Material 3 4 Identification, characterization of a flavin-containing monooxygenase MoA and its function in a specific sophorolipid molecule metabolism
1 Applied Microbiology and Biotechnology 2 Supplementary Material 3 4 Identification, characterization of a flavin-containing monooxygenase MoA and its function in a specific sophorolipid molecule metabolism
Analysis of Uncomplexed and Copper-complexed Methanobactin with UV/Visible Spectrophotometry, Mass Spectrometry and NMR Spectrometry
 Analysis of Uncomplexed and Copper-complexed Methanobactin with UV/Visible Spectrophotometry, Mass Spectrometry and NMR Spectrometry Lee Behling, Alan DiSpirito, Scott Hartsel, Larry Masterson, Gianluigi
Analysis of Uncomplexed and Copper-complexed Methanobactin with UV/Visible Spectrophotometry, Mass Spectrometry and NMR Spectrometry Lee Behling, Alan DiSpirito, Scott Hartsel, Larry Masterson, Gianluigi
Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66
 SUPPORTING INFORMATION belonging to the manuscript: Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66 by Changsheng Wu 1, 2, Gilles P. van Wezel 1, *, and Young Hae
SUPPORTING INFORMATION belonging to the manuscript: Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66 by Changsheng Wu 1, 2, Gilles P. van Wezel 1, *, and Young Hae
Supporting Information. Copyright Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, 2008
 Supporting Information Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 9 Weinheim, 008 Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 9 Weinheim, 008 Supporting Information for Streptomyces Phospholipase D Mutants
Supporting Information Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 9 Weinheim, 008 Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 9 Weinheim, 008 Supporting Information for Streptomyces Phospholipase D Mutants
L-Carnosine-Derived Fmoc-Tripeptides Forming ph- Sensitive and Proteolytically Stable Supramolecular
 Supporting Information: L-Carnosine-Derived Fmoc-Tripeptides Forming ph- Sensitive and Proteolytically Stable Supramolecular Hydrogels Rita Das Mahapatra, a Joykrishna Dey* a, and Richard G. Weiss b a
Supporting Information: L-Carnosine-Derived Fmoc-Tripeptides Forming ph- Sensitive and Proteolytically Stable Supramolecular Hydrogels Rita Das Mahapatra, a Joykrishna Dey* a, and Richard G. Weiss b a
Supporting Information
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2014 Supporting Information 1. General Information...S2 a. Materials b. HPLC
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2014 Supporting Information 1. General Information...S2 a. Materials b. HPLC
Structural Characterization of Prion-like Conformational Changes of the Neuronal Isoform of Aplysia CPEB
 Structural Characterization of Prion-like Conformational Changes of the Neuronal Isoform of Aplysia CPEB Bindu L. Raveendra, 1,5 Ansgar B. Siemer, 2,6 Sathyanarayanan V. Puthanveettil, 1,3,7 Wayne A. Hendrickson,
Structural Characterization of Prion-like Conformational Changes of the Neuronal Isoform of Aplysia CPEB Bindu L. Raveendra, 1,5 Ansgar B. Siemer, 2,6 Sathyanarayanan V. Puthanveettil, 1,3,7 Wayne A. Hendrickson,
Supplementary Information
 Supplementary Information New Highly Oxygenated Germacranolides from Carpesium divaricatum and their Cytotoxic Activity Tao Zhang, 1 Jin-Guang Si, 1, 2 Qiu-Bo Zhang, 1 Gang Ding, 1 and Zhong-Mei Zou 1*
Supplementary Information New Highly Oxygenated Germacranolides from Carpesium divaricatum and their Cytotoxic Activity Tao Zhang, 1 Jin-Guang Si, 1, 2 Qiu-Bo Zhang, 1 Gang Ding, 1 and Zhong-Mei Zou 1*
Supporting information
 Supporting information Rhabdopeptide/Xenortide-like Peptides from Xenorhabdus innexi with Terminal Amines Showing Potent Anti-protozoal Activity Lei Zhao,, Marcel Kaiser,, Helge B. Bode *,, Molekulare
Supporting information Rhabdopeptide/Xenortide-like Peptides from Xenorhabdus innexi with Terminal Amines Showing Potent Anti-protozoal Activity Lei Zhao,, Marcel Kaiser,, Helge B. Bode *,, Molekulare
Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus
 Supplemental Information Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus Torben Hofeditz, 1 Claudia Eva-Maria Unsin 2, Jutta
Supplemental Information Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus Torben Hofeditz, 1 Claudia Eva-Maria Unsin 2, Jutta
VOL. 56 NO. 2, FEB THE JOURNAL OF ANTIBIOTICS pp YM and YM , Novel Thiopeptide Antibiotics Produced
 VOL. 56 NO. 2, FEB. 2003 THE JOURNAL OF ANTIBIOTICS pp. 129-134 YM-266183 and YM-266184, Novel Thiopeptide Antibiotics Produced by Bacillus cereus Isolated from a Marine Sponge II. Structure Elucidation
VOL. 56 NO. 2, FEB. 2003 THE JOURNAL OF ANTIBIOTICS pp. 129-134 YM-266183 and YM-266184, Novel Thiopeptide Antibiotics Produced by Bacillus cereus Isolated from a Marine Sponge II. Structure Elucidation
2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry
 Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
Preparation, isolation and characterization of N α -Fmoc-peptide isocyanates: Solution synthesis of oligo-α-peptidyl ureas
 SUPPORTING INFORMATION Preparation, isolation and characterization of N α -Fmoc-peptide isocyanates: Solution synthesis of oligo-α-peptidyl ureas Vommina V. Suresh Babu*, Basanagoud S. Patil, and Rao Venkataramanarao
SUPPORTING INFORMATION Preparation, isolation and characterization of N α -Fmoc-peptide isocyanates: Solution synthesis of oligo-α-peptidyl ureas Vommina V. Suresh Babu*, Basanagoud S. Patil, and Rao Venkataramanarao
Supplementary Information
 Supplementary Information J. Braz. Chem. Soc., Vol. 24, No. 12, S1-S21, 2013. Printed in Brazil - 2013 Sociedade Brasileira de Química 0103-5053 $6.00+0.00 SI Rui He, a,b,c Bochu Wang,*,b Toshiyuki Wakimoto,
Supplementary Information J. Braz. Chem. Soc., Vol. 24, No. 12, S1-S21, 2013. Printed in Brazil - 2013 Sociedade Brasileira de Química 0103-5053 $6.00+0.00 SI Rui He, a,b,c Bochu Wang,*,b Toshiyuki Wakimoto,
Predictive PP1Ca binding region in BIG3 : 1,228 1,232aa (-KAVSF-) HEK293T cells *** *** *** KPL-3C cells - E E2 treatment time (h)
 Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 The timeline of the NGAG method for extraction of N-linked glycans and glycosite-containing peptides. The timeline can be changed based on the number of samples. Supplementary Figure
Supplementary Figure 1 The timeline of the NGAG method for extraction of N-linked glycans and glycosite-containing peptides. The timeline can be changed based on the number of samples. Supplementary Figure
Supplementary Information
 Supplementary Information Precursors of trnas are stabilized by methylguanosine cap structures Takayuki Ohira and Tsutomu Suzuki Department of Chemistry and Biotechnology, Graduate School of Engineering,
Supplementary Information Precursors of trnas are stabilized by methylguanosine cap structures Takayuki Ohira and Tsutomu Suzuki Department of Chemistry and Biotechnology, Graduate School of Engineering,
Cytochalasins from an Australian marine sediment-derived Phomopsis sp. (CMB-M0042F): Acid-mediated intra-molecular cycloadditions enhance
 SUPPORTING INFORMATION Cytochalasins from an Australian marine sediment-derived Phomopsis sp. (CMB-M42F): Acid-mediated intra-molecular cycloadditions enhance chemical diversity Zhuo Shang, Ritesh Raju,
SUPPORTING INFORMATION Cytochalasins from an Australian marine sediment-derived Phomopsis sp. (CMB-M42F): Acid-mediated intra-molecular cycloadditions enhance chemical diversity Zhuo Shang, Ritesh Raju,
Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis
 Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis Hongcheng Liu*, Gomathinayagam Ponniah, Alyssa Neil, Rekha Patel, Bruce Andrien Protein Characterization,
Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis Hongcheng Liu*, Gomathinayagam Ponniah, Alyssa Neil, Rekha Patel, Bruce Andrien Protein Characterization,
Applying Molecular Networking for the Detection of
 Supporting Information Applying Molecular Networking for the Detection of Natural Sources and Analogues of the Selective Gq Protein Inhibitor FR900359 Raphael Reher, Markus Kuschak, Nina Heycke, Suvi Annala,
Supporting Information Applying Molecular Networking for the Detection of Natural Sources and Analogues of the Selective Gq Protein Inhibitor FR900359 Raphael Reher, Markus Kuschak, Nina Heycke, Suvi Annala,
Overview of the Expressway Cell-Free Expression Systems. Expressway Mini Cell-Free Expression System
 Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Supplementary Information
 Supplementary Information Archaeal Elp3 catalyzes trna wobble uridine modification at C5 via a radical mechanism Kiruthika Selvadurai, Pei Wang, Joseph Seimetz & Raven H Huang* Department of Biochemistry,
Supplementary Information Archaeal Elp3 catalyzes trna wobble uridine modification at C5 via a radical mechanism Kiruthika Selvadurai, Pei Wang, Joseph Seimetz & Raven H Huang* Department of Biochemistry,
Characterizing fatty acids with advanced multinuclear NMR methods
 Characterizing fatty acids with advanced multinuclear NMR methods Fatty acids consist of long carbon chains ending with a carboxylic acid on one side and a methyl group on the other. Most naturally occurring
Characterizing fatty acids with advanced multinuclear NMR methods Fatty acids consist of long carbon chains ending with a carboxylic acid on one side and a methyl group on the other. Most naturally occurring
Supplementary Figure 1. HGL-DTG levels in uninduced and herbivory induced leaves. Total HGL-
 Supplementary Figure 1. HGL-DTG levels in uninduced and herbivory induced leaves. Total HGL- DTG concentrations [F 1,4 = 36.88, P 0.0037; significant differences (threshold: P 0.05) between means (± SE)
Supplementary Figure 1. HGL-DTG levels in uninduced and herbivory induced leaves. Total HGL- DTG concentrations [F 1,4 = 36.88, P 0.0037; significant differences (threshold: P 0.05) between means (± SE)
Study of On-Resin Convergent Synthesis of N-Linked. Glycopeptides Containing a Large High Mannose N- Linked Oligosaccharide
 Supporting Information Study of On-Resin Convergent Synthesis of N-Linked Glycopeptides Containing a Large High Mannose N- Linked Oligosaccharide Rui Chen and Thomas J. Tolbert* Department of Chemistry,
Supporting Information Study of On-Resin Convergent Synthesis of N-Linked Glycopeptides Containing a Large High Mannose N- Linked Oligosaccharide Rui Chen and Thomas J. Tolbert* Department of Chemistry,
Electronic supplementary information Poly(vinyl)chloride supported palladium nanoparticles: Catalyst for rapid hydrogenation reactions
 Electronic supplementary information Poly(vinyl)chloride supported palladium nanoparticles: Catalyst for rapid hydrogenation reactions Hosahalli P. Hemantha and Vommina V. Sureshbabu* Peptide Research
Electronic supplementary information Poly(vinyl)chloride supported palladium nanoparticles: Catalyst for rapid hydrogenation reactions Hosahalli P. Hemantha and Vommina V. Sureshbabu* Peptide Research
Supplementary Information. Amino group carrier protein-mediated secondary metabolite biosynthesis in Streptomyces
 Supplementary Information Amino group carrier protein-mediated secondary metabolite biosynthesis in Streptomyces Fumihito Hasebe 1, Kenichi Matsuda 1, Taro Shiraishi 1, Yushi Futamura 2, Takeshi Nakano
Supplementary Information Amino group carrier protein-mediated secondary metabolite biosynthesis in Streptomyces Fumihito Hasebe 1, Kenichi Matsuda 1, Taro Shiraishi 1, Yushi Futamura 2, Takeshi Nakano
Your Name: Question 1. Spectrum Prediction I: Ethyl Acetoacetate. (15 points) ppm ppm ppm ppm. J(A,D) = 8 Hz = 0.
 Question 1. Spectrum Prediction I: Ethyl Acetoacetate. (15 points) A B C D 4.202 ppm 3.451 ppm 2.273 ppm 1.288 ppm J(A,D) = 8 Hz = 0.08 ppm (in CDCl 3 ) Draw the 100 MHz H-NMR spectrum to scale. Draw splitting
Question 1. Spectrum Prediction I: Ethyl Acetoacetate. (15 points) A B C D 4.202 ppm 3.451 ppm 2.273 ppm 1.288 ppm J(A,D) = 8 Hz = 0.08 ppm (in CDCl 3 ) Draw the 100 MHz H-NMR spectrum to scale. Draw splitting
Marine Streptomyces sp. derived antimycin analogues. suppress HeLa cells via depletion HPV E6/E7 mediated by
 Marine Streptomyces sp. derived antimycin analogues suppress HeLa cells via depletion HPV E6/E7 mediated by ROS-dependent ubiquitin proteasome system Weiyi Zhang 1, +, Qian Che 1, 2, +, Hongsheng Tan 1,
Marine Streptomyces sp. derived antimycin analogues suppress HeLa cells via depletion HPV E6/E7 mediated by ROS-dependent ubiquitin proteasome system Weiyi Zhang 1, +, Qian Che 1, 2, +, Hongsheng Tan 1,
Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random
 S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
Double charge of 33kD peak A1 A2 B1 B2 M2+ M/z. ABRF Proteomics Research Group - Qualitative Proteomics Study Identifier Number 14146
 Abstract The 2008 ABRF Proteomics Research Group Study offers participants the chance to participate in an anonymous study to identify qualitative differences between two protein preparations. We used
Abstract The 2008 ABRF Proteomics Research Group Study offers participants the chance to participate in an anonymous study to identify qualitative differences between two protein preparations. We used
Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and Gerard D. Wright.
 Supplementary Data for TetX is a Flavin-Dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and
Supplementary Data for TetX is a Flavin-Dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and
Beta Amyloid Peptides
 Beta Amyloid Peptides The Most Comprehensive Collection for Alzheimer s Disease Academic Services Pharmaceutical Services Beta Amyloid Peptides The Most Comprehensive Collection for Alzheimer s Disease
Beta Amyloid Peptides The Most Comprehensive Collection for Alzheimer s Disease Academic Services Pharmaceutical Services Beta Amyloid Peptides The Most Comprehensive Collection for Alzheimer s Disease
2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein
![2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein](/thumbs/86/94743397.jpg) Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis
 Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary Information
 Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
THE JOURNAL OF ANTIBIOTICS. Polyketomycin, a New Antibiotic from Streptomyces sp. MK277-AF1. II. Structure Determination
 THE JOURNAL OF ANTIBIOTICS Polyketomycin, a New Antibiotic from Streptomyces sp. MK277-AF1 II. Structure Determination ISAO MOMOSE, WEI CHEN, HIKARU NAKAMURA, HIROSHI NAGANAWA, HIRONOBU IINUMA and TOMIO
THE JOURNAL OF ANTIBIOTICS Polyketomycin, a New Antibiotic from Streptomyces sp. MK277-AF1 II. Structure Determination ISAO MOMOSE, WEI CHEN, HIKARU NAKAMURA, HIROSHI NAGANAWA, HIRONOBU IINUMA and TOMIO
Figure S7: ATR FTIR spectra of enantiomeric UP-PLA-UPy and sc-upy-pla-upy. Figure S9: SEM microphotographs sc-upy-pla-oh in 1,4-dioxane and chloroform
 Supporting Information for Macromolecules, DOI: Supramolecular Polylactides by the Cooperative Interaction of the End Groups and Stereocomplexation By M. Brzeziński*, T. Biela Table of Contents Figure
Supporting Information for Macromolecules, DOI: Supramolecular Polylactides by the Cooperative Interaction of the End Groups and Stereocomplexation By M. Brzeziński*, T. Biela Table of Contents Figure
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular
 Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Supplementary Information
 Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
SUPPLEMENTARY INFORMATION. An orthogonal ribosome-trnas pair by the engineering of
 SUPPLEMENTARY INFORMATION An orthogonal ribosome-trnas pair by the engineering of peptidyl transferase center Naohiro Terasaka 1 *, Gosuke Hayashi 1 *, Takayuki Katoh 1, and Hiroaki Suga 1,2 1 Department
SUPPLEMENTARY INFORMATION An orthogonal ribosome-trnas pair by the engineering of peptidyl transferase center Naohiro Terasaka 1 *, Gosuke Hayashi 1 *, Takayuki Katoh 1, and Hiroaki Suga 1,2 1 Department
Scheme S1. Synthesis of glycose-amino ligand.
 Scheme S1. Synthesis of glycose-amino ligand. 5-Chloro-1-pentyl-2,3,4,6-tetra-O-acetyl-ß-D-glucopyranoside S2 To a solution of penta-o-acetyl-ß-d-glucopyranoside S1 (3.0 g, 7.69 mmol) and 5-chloropentan-1-ol
Scheme S1. Synthesis of glycose-amino ligand. 5-Chloro-1-pentyl-2,3,4,6-tetra-O-acetyl-ß-D-glucopyranoside S2 To a solution of penta-o-acetyl-ß-d-glucopyranoside S1 (3.0 g, 7.69 mmol) and 5-chloropentan-1-ol
The N-terminal loop of IRAK-4 death domain regulates ordered assembly of the Myddosome signalling scaffold
 The N-terminal loop of IRAK-4 death domain regulates ordered assembly of the Myddosome signalling scaffold Anthony C.G. Dossang * 1, Precious G. Motshwene * 1, Yang Yang* 1, Martyn F. Symmons*, Clare E.
The N-terminal loop of IRAK-4 death domain regulates ordered assembly of the Myddosome signalling scaffold Anthony C.G. Dossang * 1, Precious G. Motshwene * 1, Yang Yang* 1, Martyn F. Symmons*, Clare E.
Facile Cu(II) mediated conjugation of thioesters and thioacids to peptides and proteins under mild conditions
 Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Facile Cu(II) mediated conjugation of thioesters and thioacids to peptides
Electronic Supplementary Material (ESI) for Organic & Biomolecular Chemistry. This journal is The Royal Society of Chemistry 2018 Facile Cu(II) mediated conjugation of thioesters and thioacids to peptides
Naoya Takahashi, Keiya Hirota and Yoshitaka Saga* Supplementary material
 Supplementary material Facile transformation of the five-membered exocyclic E-ring in 13 2 -demethoxycarbonyl chlorophyll derivatives by molecular oxygen with titanium oxide in the dark Naoya Takahashi,
Supplementary material Facile transformation of the five-membered exocyclic E-ring in 13 2 -demethoxycarbonyl chlorophyll derivatives by molecular oxygen with titanium oxide in the dark Naoya Takahashi,
ESI and MALDI Mass Spectrometry. S. Sankararaman Department of Chemistry Indian Institute of Technology Madras Chennai
 ESI and MALDI Mass Spectrometry S. Sankararaman Department of Chemistry Indian Institute of Technology Madras Chennai 600036 sanka@iitm.ac.in MODULE 22 ESI-Q MASS SPECTROMETRY - examples Mol. Wt = 1140
ESI and MALDI Mass Spectrometry S. Sankararaman Department of Chemistry Indian Institute of Technology Madras Chennai 600036 sanka@iitm.ac.in MODULE 22 ESI-Q MASS SPECTROMETRY - examples Mol. Wt = 1140
Microwave heating in peptide side chain modification via sulfhydryl reaction
 Microwave heating in peptide side chain modification via sulfhydryl reaction E. Calce and S. De Luca* Institute of Biostructures and Bioimaging, National Research Council, 80134 Naples, Italy stefania.deluca@cnr.it
Microwave heating in peptide side chain modification via sulfhydryl reaction E. Calce and S. De Luca* Institute of Biostructures and Bioimaging, National Research Council, 80134 Naples, Italy stefania.deluca@cnr.it
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
Isolation, Purification and Molecular Weight Determination of Antihypertensive Peptides Derived from
 2011, Vol. 32, No. 02 213 1,2 1 1, * 1 1 (1. 361012 2. 350002) AS.1398 (Porphyra haitanesis)ace Sephadex G-15 (RP-HPLC) (MALDI-TOF-MS) 2000D ACE IC50 0.73mg/mL Sephadex G-15 6 E IC50 0.67mg/mL E RP-HPLC
2011, Vol. 32, No. 02 213 1,2 1 1, * 1 1 (1. 361012 2. 350002) AS.1398 (Porphyra haitanesis)ace Sephadex G-15 (RP-HPLC) (MALDI-TOF-MS) 2000D ACE IC50 0.73mg/mL Sephadex G-15 6 E IC50 0.67mg/mL E RP-HPLC
In vitro conversion of chanoclavine-i aldehyde to the stereoisomers festuclavine and pyroclavine was controlled by the second reduction step
 1 Electronic Supporting Information to: In vitro conversion of chanoclavinei aldehyde to the stereoisomers festuclavine and pyroclavine was controlled by the second reduction step Marco Matuschek a, Christiane
1 Electronic Supporting Information to: In vitro conversion of chanoclavinei aldehyde to the stereoisomers festuclavine and pyroclavine was controlled by the second reduction step Marco Matuschek a, Christiane
The Impact of a Transposon Insertion in phzf2 on the Specialized Metabolite. Production and Interkingdom Interactions of Pseudomonas aeruginosa
 1 2 3 Supplemental Material The Impact of a Transposon Insertion in phzf2 on the Specialized Metabolite Production and Interkingdom Interactions of Pseudomonas aeruginosa 4 5 6 7 Vanessa V. Phelan a, Wilna
1 2 3 Supplemental Material The Impact of a Transposon Insertion in phzf2 on the Specialized Metabolite Production and Interkingdom Interactions of Pseudomonas aeruginosa 4 5 6 7 Vanessa V. Phelan a, Wilna
Supporting Information
 Investigation of self-immolative linkers in the design of hydrogen peroxide metalloprotein inhibitors Jody L. Major Jourden, Kevin B. Daniel, and Seth M. Cohen* Department of Chemistry and Biochemistry,
Investigation of self-immolative linkers in the design of hydrogen peroxide metalloprotein inhibitors Jody L. Major Jourden, Kevin B. Daniel, and Seth M. Cohen* Department of Chemistry and Biochemistry,
Supporting Information for: New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans
 Supporting Information for: New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans Matthew T. Henke a, Alexandra A. Soukup b, Anthony W. Goering a, Ryan A.
Supporting Information for: New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans Matthew T. Henke a, Alexandra A. Soukup b, Anthony W. Goering a, Ryan A.
Supporting Information. A Two-In-One Fluorescent Sensor With Dual Channels to. Discriminate Zn 2+ and Cd 2+
 Electronic Supplementary Material (ESI) for RS Advances Supporting Information A Two-In-One Fluorescent Sensor With Dual hannels to Discriminate Zn 2 and d 2 Li-Kun Zhang, a Guang-Fu Wu, a Ying Zhang,
Electronic Supplementary Material (ESI) for RS Advances Supporting Information A Two-In-One Fluorescent Sensor With Dual hannels to Discriminate Zn 2 and d 2 Li-Kun Zhang, a Guang-Fu Wu, a Ying Zhang,
PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System
 Application Note LC/MS PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System Purpose This application note describes an automated workflow
Application Note LC/MS PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System Purpose This application note describes an automated workflow
Supporting information to Amino-functional polyester dendrimers based on bis-mpa as nonviral vectors for sirna delivery
 Supporting information to Amino-functional polyester dendrimers based on bis-mpa as nonviral vectors for sirna delivery P. Stenström, D. Manzanares, Y. Zhang, V. Ceña and M. Malkoch* * To whom correspondence
Supporting information to Amino-functional polyester dendrimers based on bis-mpa as nonviral vectors for sirna delivery P. Stenström, D. Manzanares, Y. Zhang, V. Ceña and M. Malkoch* * To whom correspondence
Characterization of Disulfide Linkages in Proteins by 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry. Supporting Information
 Characterization of Disulfide Linkages in Proteins by 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry M. Montana Quick, Christopher M. Crittenden, Jake A. Rosenberg, and Jennifer S. Brodbelt
Characterization of Disulfide Linkages in Proteins by 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry M. Montana Quick, Christopher M. Crittenden, Jake A. Rosenberg, and Jennifer S. Brodbelt
Nature Methods: doi: /nmeth.3177
 Supplementary Figure 1 Characterization of LysargiNase, trypsin and LysN missed cleavages. (a) Proportion of peptides identified in LysargiNase and trypsin digests of MDA-MB-231 cell lysates carrying 0,
Supplementary Figure 1 Characterization of LysargiNase, trypsin and LysN missed cleavages. (a) Proportion of peptides identified in LysargiNase and trypsin digests of MDA-MB-231 cell lysates carrying 0,
Supplementary Figure 1. ESI/MS/MS analyses of native and de-acetylated S2A Supplementary Figure 2. Partial 1D 1H NMR spectrum of S2A
 Supplementary Figure 1. ESI/MS/MS analyses of native and de-acetylated S2A. Panel A, Positive ESI mass spectra of native (N) and de-acetylated (DA) non-active S2A were obtained, and demonstrated a shift
Supplementary Figure 1. ESI/MS/MS analyses of native and de-acetylated S2A. Panel A, Positive ESI mass spectra of native (N) and de-acetylated (DA) non-active S2A were obtained, and demonstrated a shift
Supplementary Information
 Supplementary Information Efficient use of the Dmab Protecting Group: Applications for the Solid-Phase Synthesis of N- Linked Glycopeptides Trent Conroy, Katrina A. Jolliffe and Richard J. Payne* School
Supplementary Information Efficient use of the Dmab Protecting Group: Applications for the Solid-Phase Synthesis of N- Linked Glycopeptides Trent Conroy, Katrina A. Jolliffe and Richard J. Payne* School
Supplementary Material (ESI) for Chemical Communications This journal is (c) The Royal Society of Chemistry 2008
 Experimental Details Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich Chemical Company and were used as received. 2-DOS and neamine were kindly provided by Dr. F. Huang. Paromamine
Experimental Details Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich Chemical Company and were used as received. 2-DOS and neamine were kindly provided by Dr. F. Huang. Paromamine
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Chemical constituents from Agrimonia pilosa Ledeb. and their chemotaxonomic significance Wei-jie Liu, Xue-qian Hou, Hao Chen, Jing-yu Liang*, Jian-bo Sun** Department of Natural
SUPPLEMENTARY MATERIAL Chemical constituents from Agrimonia pilosa Ledeb. and their chemotaxonomic significance Wei-jie Liu, Xue-qian Hou, Hao Chen, Jing-yu Liang*, Jian-bo Sun** Department of Natural
Lecture 3. Tandem MS & Protein Sequencing
 Lecture 3 Tandem MS & Protein Sequencing Nancy Allbritton, M.D., Ph.D. Department of Physiology & Biophysics 824-9137 (office) nlallbri@uci.edu Office- Rm D349 Medical Science D Bldg. Tandem MS Steps:
Lecture 3 Tandem MS & Protein Sequencing Nancy Allbritton, M.D., Ph.D. Department of Physiology & Biophysics 824-9137 (office) nlallbri@uci.edu Office- Rm D349 Medical Science D Bldg. Tandem MS Steps:
Supporting Information
 Supporting Information A new series of cytotoxic pyrazoline derivatives as potential anticancer agents induces cell cycle arrest and apoptosis Hong Wang 1,, Jinhong Zheng 1,, Weijie Xu 1, Cheng Chen 1,
Supporting Information A new series of cytotoxic pyrazoline derivatives as potential anticancer agents induces cell cycle arrest and apoptosis Hong Wang 1,, Jinhong Zheng 1,, Weijie Xu 1, Cheng Chen 1,
Trans-stereospecific polymerization of butadiene and random copolymerization with styrene using borohydrido rare earths / magnesium dialkyl catalysts
 Trans-stereospecific polymerization of butadiene and random copolymerization with styrene using borohydrido rare earths / magnesium dialkyl catalysts A. Ventura a, T. Chenal b,c,d,e, *, M. Bria b,f, F.
Trans-stereospecific polymerization of butadiene and random copolymerization with styrene using borohydrido rare earths / magnesium dialkyl catalysts A. Ventura a, T. Chenal b,c,d,e, *, M. Bria b,f, F.
Synthesis of an Alleged Byproduct Precursor in Iodixanol Preparation
 SUPPORTING INFORMATION Synthesis of an Alleged Byproduct Precursor in Iodixanol Preparation Marianne Bore Haarr, Emil Lindbäck, Torfinn Håland, * and Magne O. Sydnes * Department of Mathematics and Natural
SUPPORTING INFORMATION Synthesis of an Alleged Byproduct Precursor in Iodixanol Preparation Marianne Bore Haarr, Emil Lindbäck, Torfinn Håland, * and Magne O. Sydnes * Department of Mathematics and Natural
Synthesis of Dichotomin A: Use of a Penicillamine Derived Pseudoproline to Furnish Native Valine Residues
 Supplementary Material Synthesis of Dichotomin A: Use of a Penicillamine Derived Pseudoproline to Furnish ative Valine Residues Michelle S. Y. Wong A, Deni Taleski A, Katrina A. Jolliffe A, B A School
Supplementary Material Synthesis of Dichotomin A: Use of a Penicillamine Derived Pseudoproline to Furnish ative Valine Residues Michelle S. Y. Wong A, Deni Taleski A, Katrina A. Jolliffe A, B A School
Genetic information flows from mrna to protein through the process of translation
 Genetic information flows from mrn to protein through the process of translation TYPES OF RN (RIBONUCLEIC CID) RN s job - protein synthesis (assembly of amino acids into proteins) Three main types: 1.
Genetic information flows from mrn to protein through the process of translation TYPES OF RN (RIBONUCLEIC CID) RN s job - protein synthesis (assembly of amino acids into proteins) Three main types: 1.
Manja Henze, Dorothee Merker and Lothar Elling. 1. Characteristics of the Recombinant β-glycosidase from Pyrococcus
 S1 of S17 Supplementary Materials: Microwave-Assisted Synthesis of Glycoconjugates by Transgalactosylation with Recombinant Thermostable β-glycosidase from Pyrococcus Manja Henze, Dorothee Merker and Lothar
S1 of S17 Supplementary Materials: Microwave-Assisted Synthesis of Glycoconjugates by Transgalactosylation with Recombinant Thermostable β-glycosidase from Pyrococcus Manja Henze, Dorothee Merker and Lothar
Acaulins A and B, Trimeric Macrodiolides from Acaulium. Table of Contents
 Supporting Information Acaulins A and B, Trimeric Macrodiolides from Acaulium sp. H-JQSF Ting Ting Wang, Ying Jie Wei, Hui Ming Ge, Rui Hua Jiao, Ren Xiang Tan* * Corresponding author. E-mail: rxtan@nju.edu.cn
Supporting Information Acaulins A and B, Trimeric Macrodiolides from Acaulium sp. H-JQSF Ting Ting Wang, Ying Jie Wei, Hui Ming Ge, Rui Hua Jiao, Ren Xiang Tan* * Corresponding author. E-mail: rxtan@nju.edu.cn
Analysis of fatty acid metabolism using Click-Chemistry and HPLC-MS
 Analysis of fatty acid metabolism using Click-Chemistry and HPLC-MS Alexander J. Pérez and Helge B. Bode -Supporting Information- Contents Experimental section Supplementary figures NMR spectra Page S2
Analysis of fatty acid metabolism using Click-Chemistry and HPLC-MS Alexander J. Pérez and Helge B. Bode -Supporting Information- Contents Experimental section Supplementary figures NMR spectra Page S2
CHAPTER-6 IDENTIFICATION, AND CHARACTERISATION OF DEGRADATION IMPURITY IN VALSARTAN TABLETS
 129 CHAPTER-6 IDENTIFICATION, AND CHARACTERISATION OF DEGRADATION IMPURITY IN VALSARTAN TABLETS 130 6.1. Introduction Valsartan is an orally active specific angiotensin II blocker effective in lowering
129 CHAPTER-6 IDENTIFICATION, AND CHARACTERISATION OF DEGRADATION IMPURITY IN VALSARTAN TABLETS 130 6.1. Introduction Valsartan is an orally active specific angiotensin II blocker effective in lowering
Supporting Information
 Supporting Information Wiley-VCH 2008 69451 Weinheim, Germany Chemoselective Peptide Cyclization via Induced Traceless Staudinger Ligation Rolf Kleineweischede, Christian P.R. Hackenberger* Institute for
Supporting Information Wiley-VCH 2008 69451 Weinheim, Germany Chemoselective Peptide Cyclization via Induced Traceless Staudinger Ligation Rolf Kleineweischede, Christian P.R. Hackenberger* Institute for
Christophe Lincheneau, Bernard Jean-Denis and Thorfinnur Gunnlaugsson* Electronic Supplementary Information
 Self-assembly formation of mechanically interlocked [2]- and [3]catenanes using lanthanide ion [Eu(III)] templation and ring closing metathesis reactions Christophe Lincheneau, Bernard Jean-Denis and Thorfinnur
Self-assembly formation of mechanically interlocked [2]- and [3]catenanes using lanthanide ion [Eu(III)] templation and ring closing metathesis reactions Christophe Lincheneau, Bernard Jean-Denis and Thorfinnur
Supporting Information
 Supporting Information Zheng, Y. et al, Lipoxygenases mediate the effect of essential fatty acid in skin barrier formation: A proposed role in releasing omega-hydroxyceramide for construction of the corneocyte
Supporting Information Zheng, Y. et al, Lipoxygenases mediate the effect of essential fatty acid in skin barrier formation: A proposed role in releasing omega-hydroxyceramide for construction of the corneocyte
This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth 2 points.
 MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
Supporting Information. Development of N, S-doped carbon dots as a novel matrix for the. analysis of small molecules by negative ion MALDI-TOF MS
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 16 Supporting Information Development of N, S-doped carbon dots as a novel matrix for the analysis
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 16 Supporting Information Development of N, S-doped carbon dots as a novel matrix for the analysis
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004
 Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Thiol-Ene Photoimmobilization of Chymotrypsin on Polysiloxane Gels for Enzymatic Peptide Synthesis
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2018 Supporting information for: Thiol-Ene Photoimmobilization of Chymotrypsin on Polysiloxane Gels
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2018 Supporting information for: Thiol-Ene Photoimmobilization of Chymotrypsin on Polysiloxane Gels
Biochemistry 2 Recita0on Amino Acid Metabolism
 Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Supporting Information. Chemoenzymatic Synthesis of Galectin Binding. Glycopolymers
 Supporting Information Chemoenzymatic Synthesis of Galectin Binding Glycopolymers Jessica H. Ennist, Henry R. Termuehlen, Samuel P. Bernhard, Mackenzie S. Fricke, and Mary J. Cloninger* Department of Chemistry
Supporting Information Chemoenzymatic Synthesis of Galectin Binding Glycopolymers Jessica H. Ennist, Henry R. Termuehlen, Samuel P. Bernhard, Mackenzie S. Fricke, and Mary J. Cloninger* Department of Chemistry
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
TWO NEW ELLAGIC ACID GLYCOSIDES FROM LEAVES OF DIPLOPANAX STACHYANTHUS
 Journal of Asian Natural Products Research, December 2004, Vol. 6 (4), pp. 271 276 TWO NEW ELLAGIC ACID GLYCOSIDES FROM LEAVES OF DIPLOPANAX STACHYANTHUS XIAO-HONG YAN and YUE-WEI GUO* State Key Laboratory
Journal of Asian Natural Products Research, December 2004, Vol. 6 (4), pp. 271 276 TWO NEW ELLAGIC ACID GLYCOSIDES FROM LEAVES OF DIPLOPANAX STACHYANTHUS XIAO-HONG YAN and YUE-WEI GUO* State Key Laboratory
Cahn - Ingold - Prelog system. Proteins: Evolution, and Analysis Lecture 7 9/15/2009. The Fischer Convention (1) G (2) (3)
 Chapter 4 (1) G Proteins: Evolution, and Analysis Lecture 7 9/15/2009 A V L I M P F W Chapter 4 (2) S (3) T N Q Y C K R H D E The Fischer Convention Absolute configuration about an asymmetric carbon related
Chapter 4 (1) G Proteins: Evolution, and Analysis Lecture 7 9/15/2009 A V L I M P F W Chapter 4 (2) S (3) T N Q Y C K R H D E The Fischer Convention Absolute configuration about an asymmetric carbon related
Phosphorylated glycosphingolipids essential for cholesterol mobilization in C. elegans
 SUPPLEMENTARY INFORMATION Phosphorylated glycosphingolipids essential for cholesterol mobilization in C. elegans Sebastian Boland, Ulrike Schmidt, Vyacheslav Zagoriy, Julio L. Sampaio, Raphael Fritsche,
SUPPLEMENTARY INFORMATION Phosphorylated glycosphingolipids essential for cholesterol mobilization in C. elegans Sebastian Boland, Ulrike Schmidt, Vyacheslav Zagoriy, Julio L. Sampaio, Raphael Fritsche,
Supporting Information. Lysine Propionylation to Boost Proteome Sequence. Coverage and Enable a Silent SILAC Strategy for
 Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
Protein Synthesis. What are proteins and what are they composed of? What are some of their functions? What determines the final function of a protein?
 Protein Synthesis What are proteins and what are they composed of? What are some of their functions? What determines the final function of a protein? What is needed to make specific proteins and what is
Protein Synthesis What are proteins and what are they composed of? What are some of their functions? What determines the final function of a protein? What is needed to make specific proteins and what is
High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. R. J. Rose, E. Damoc, E. Denisov, A. Makarov, A. J. R.
 High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies R. J. Rose, E. Damoc, E. Denisov, A. Makarov, A. J. R. Heck SUPPLEMENTARY INFORMATION HCD multipole C -trap Transport octapole
High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies R. J. Rose, E. Damoc, E. Denisov, A. Makarov, A. J. R. Heck SUPPLEMENTARY INFORMATION HCD multipole C -trap Transport octapole
Colloidal metal oxide nanocrystal catalysis by sustained chemically driven ligand displacement
 Colloidal metal oxide nanocrystal catalysis by sustained chemically driven ligand displacement Jonathan De Roo 1,2,3 *, Isabel Van Driessche 1, José C. Martins 2 and Zeger Hens 3,4 * 1 Sol-gel Center for
Colloidal metal oxide nanocrystal catalysis by sustained chemically driven ligand displacement Jonathan De Roo 1,2,3 *, Isabel Van Driessche 1, José C. Martins 2 and Zeger Hens 3,4 * 1 Sol-gel Center for
Supporting Information. Nitrodibenzofuran: a One- and Two-Photon Sensitive Protecting Group that is Superior to
 Supporting Information Nitrodibenzofuran: a One- and Two-Photon Sensitive Protecting Group that is Superior to Brominated Hydroxycoumarin for Thiol Caging in Peptides M. Mohsen Mahmoodi, Daniel Abate-Pella,
Supporting Information Nitrodibenzofuran: a One- and Two-Photon Sensitive Protecting Group that is Superior to Brominated Hydroxycoumarin for Thiol Caging in Peptides M. Mohsen Mahmoodi, Daniel Abate-Pella,
