Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion
|
|
|
- Morgan Sharp
- 5 years ago
- Views:
Transcription
1
2
3
4 Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol _s_at NM_ /// NM_ /// NM_ Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC FC ALK shrna FC ALK inhibitors _s_at NM_ growth arrest-specific 1 GAS _at NM_ serum/glucocorticoid regulated kinase SGK _at NM_ oncostatin M OSM _s_at NM_ growth arrest-specific 1 GAS _s_at NM_ interleukin 2 receptor, alpha IL2RA _s_at NM_ regulator of G-protein signalling 16 RGS _at NM_ inducible T-cell co-stimulator ICOS _s_at NR_ cytidine monophosphate-n-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate CMAH monooxygenase) _at NM_ chemokine (C-C motif) ligand 20 CCL _x_at NM_ hydroxysteroid (17-beta) dehydrogenase 7 HSD17B _s_at NM_ v-myc myelocytomatosis viral oncogene homolog (avian) MYC _at NM_ proliferation-associated 2G4, 38kDa PA2G _s_at NM_ proliferation-associated 2G4, 38kDa PA2G _at NM_ /// tumor necrosis factor receptor superfamily, NM_ member 8 TNFRSF _at NM_ chromosome 13 open reading frame 18 C13orf _at NM_ Down syndrome critical region gene 1-like 1 DSCR1L _at NM_ Jun dimerization protein p21snft SNFT _at NM_ NTF2-like export factor 1 NXT _s_at NM_ chromosome 1 open reading frame 33 C1orf _at NM_ BCL2-related protein A1 BCL2A _at NM_ /// NM_ /// NM_ NM_ /// NM_ guanine nucleotide binding protein-like 3 (nucleolar) GNL _at interleukin 1 receptor accessory protein IL1RAP _s_at NM_ KIAA1199 KIAA NM_ /// _s_at NM_ /// solute carrier family 43, member 3 SLC43A NM_ _s_at NM_ FK506 binding protein 11, 19 kda FKBP _at NM_ early growth response 2 (Krox-20 homolog, Drosophila) EGR _at NM_ prefoldin subunit 2 PFDN _at NM_ dyskeratosis congenita 1, dyskerin DKC _x_at NM_ splicing factor, arginine/serine-rich 7, 35kDa SFRS _at NM_ lectin, galactoside-binding, soluble, 1 (galectin 1) LGALS _s_at NM_ /// HIST2H2AA /// histone 2, H2aa /// similar to Histone H2A.o NM_ /// LOC /// (H2A/o) (H2A.2) (H2a-615) /// histone H2A/r XM_ H2A/R _at NM_ placenta-specific 8 PLAC _s_at NM_ spectrin, alpha, non-erythrocytic 1 (alphafodrin) SPTAN _s_at NM_ protein kinase C and casein kinase substrate in neurons 2 PACSIN _s_at NM_ solute carrier family 35, member E2 SLC35E _at NM_ /// NM_ /// NM_ TGFB1-induced anti-apoptotic factor 1 /// myosin XVIIIA TIAF1 /// MYO18A _at NM_ ras homolog gene family, member B RHOB _s_at NM_ tubulin, alpha 3 TUBA _s_at NM_ spectrin, alpha, non-erythrocytic 1 (alphafodrin) SPTAN _at NM_ /// NM_ AHNAK nucleoprotein (desmoyokin) AHNAK _s_at NM_ thioredoxin interacting protein TXNIP _at NM_ Ras and Rab interactor 2 RIN _s_at NM_ mitogen-activated protein kinase kinase kinase 12 MAP3K _s_at NM_ thioredoxin interacting protein TXNIP _s_at NM_ thioredoxin interacting protein TXNIP _at NM_ /// host cell factor C1 regulator 1 (XPO1 NM_ /// dependent) NM_ HCFC1R
5 Figure S1. Specificity of NPM-ALK knock-down by inducible shrna. (A) ALK transcripts are significantly down-modulated after shrna induction. ALK mrna expression levels of TS-TTA-A5, TS-TTA-A5M, and Su-DHL1 cells (21 samples) are shown as assessed by microarray analysis. (B) Eisen plot of the expression of the selected genes as assessed by microarray analysis. (C) OAS1 gene transcripts do not correlated with shrna induction. Gene expression quantification of the classic interferon target gene OAS1 as assessed by microarray analysis. (D) The genes specifically expressed in TS-TTA-A5M cells represent a minimum subset. Eisen plot of the expression of the genes uniquely modulated after DOX treatment of TTA-A5M cells (84 hours). Figure S2. Validation of the NPM-ALK signature in Su-DHL1 cells. (A) ALK signature in SuDHL-1 cells. Identification of genes specifically expressed in Su-DHL1-TTA-A5 cells untreated versus treated with DOX (72 hours) by supervised analysis (3 versus 3). (B) Eisen plot of the expression values of the 103 common transcripts (72 increased and 31 decreased genes), which overlap after the comparison of the gene lists identified by the supervised analysis related to either to ALK silencing in Su-DHL-1-TTA-A5 or TS-TTA-A5 cells. Figure S3. ALK signaling regulates the expression of C/EBPβ in lymphoid and non-lymphoid cells. (A) C/EBPβ expression is down-regulated after ALK silencing. In the histogram C/EBPβ mrna expression levels (21 samples) are represented as assessed by microarray analysis. (B) The expression of C/EBPβ among the CEBP family members correlates the most with that of ALK. The expression of C/EBP family genes was evaluated in all experimental conditions (21 samples) for absolute correlation with ALK. The CEBPB gene clustered within the branch including the ALK probe set (green rectangle). (C) ALK expression is upregulated after tetracycline induction in HEK-293T NPM-ALK Tet-On cells. In the histogram ALK mrna expression levels (6 samples) are represented as assessed by microarray analysis. (D) CEBPB expression is concomitantly upregulated after tetracycline
6 induction in HEK-293T NPM-ALK Tet-On cells. In the histogram CEBPB mrna expression levels (6 samples) are represented as assessed by microarray analysis. (E) CEBPG//B/D expressions are most closely related to ALK expression in non-lymphoid cells. Expression of CEBP family genes evaluated in different experimental conditions (6 samples) for absolute correlation with ALK. The C/EBPG/B/G genes clustered within the branch including the ALK probe set (green rectangle). Table S1. Common transcripts concordantly modulated across shrna- and ALK inhibitor-treated TS- TTA-A5 cells. FC: Fold Change
TNFSF13B tumor necrosis factor (ligand) superfamily, member 13b NF-kB pathway cluster, Enrichment Score: 3.57
 Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Table S1 Differentially expressed genes showing > 2 fold changes and p <0.01 for 0.01 mm NS-398, 0.1mM ibuprofen and COX-2 RNAi.
 Table S1 Differentially expressed genes showing > 2 fold changes and p
Table S1 Differentially expressed genes showing > 2 fold changes and p
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
U118MG. Supplementary Figure 1 U373MG U118MG 3.5 A CCF-SSTG
 A172 CCF-SSTG1 15 - - - 1 1 1 2 2 3 4 6 7 8 10101112131617192022-1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22 T98G U373MG - - - 1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22-1 1 1 2 2 3 4 6 7
A172 CCF-SSTG1 15 - - - 1 1 1 2 2 3 4 6 7 8 10101112131617192022-1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22 T98G U373MG - - - 1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22-1 1 1 2 2 3 4 6 7
* Kyoto Encyclopedia of Genes and Genomes.
 Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and Bcl2A1 as critical target genes
 Research article Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and Bcl2A1 as critical target genes Roberto Piva, 1 Elisa Pellegrino, 1 Michela Mattioli, 2 Luca Agnelli,
Research article Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and Bcl2A1 as critical target genes Roberto Piva, 1 Elisa Pellegrino, 1 Michela Mattioli, 2 Luca Agnelli,
RALYL Hypermethylation: A Potential Diagnostic Marker of Esophageal Squamous Cell Carcinoma (ESCC) Junwei Liu, MD
 RALYL Hypermethylation: A Potential Diagnostic Marker of Esophageal Squamous Cell Carcinoma (ESCC) Junwei Liu, MD Aurora Healthcare, Milwaukee, WI INTRODUCTION v Epigenetic aberration and genetic alteration
RALYL Hypermethylation: A Potential Diagnostic Marker of Esophageal Squamous Cell Carcinoma (ESCC) Junwei Liu, MD Aurora Healthcare, Milwaukee, WI INTRODUCTION v Epigenetic aberration and genetic alteration
Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection.
 Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection. Genes found to be significantly upregulated (FDR2) in Y strain
Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection. Genes found to be significantly upregulated (FDR2) in Y strain
Table S1. Differentially expressed transcripts between hepatic inkt cells. sorted form WT and plck-hcd1d tg mice. Differentially expressed
 Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
SUPPLEMENTARY DATA. Assessment of cell survival (MTT) Assessment of early apoptosis and cell death
 Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
SUPPLEMENTARY FIG. S2. b-galactosidase staining of
 SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
Epstein-Barr virus driven promoter hypermethylated genes in gastric cancer
 RESEARCH FUND FOR THE CONTROL OF INFECTIOUS DISEASES Epstein-Barr virus driven promoter hypermethylated genes in gastric cancer J Yu *, KF To, QY Liang K e y M e s s a g e s 1. Somatostatin receptor 1
RESEARCH FUND FOR THE CONTROL OF INFECTIOUS DISEASES Epstein-Barr virus driven promoter hypermethylated genes in gastric cancer J Yu *, KF To, QY Liang K e y M e s s a g e s 1. Somatostatin receptor 1
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION
 CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Information
 Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Supporting Information
 Supporting Information Gorgun et al. 10.1073/pnas.0901166106 SI Text Animals, Disease Detection and Infusion of CLL Cells. Mice were examined for lymphadenopathy and splenomegaly and at specific ages or
Supporting Information Gorgun et al. 10.1073/pnas.0901166106 SI Text Animals, Disease Detection and Infusion of CLL Cells. Mice were examined for lymphadenopathy and splenomegaly and at specific ages or
Fetal-maternal alignment of regulatory T cells correlates with Interleukin 10 mediated Bcl-
 Fetal-maternal alignment of regulatory T cells correlates with Interleukin 10 mediated Bcl- 2 upregulation in pregnancy B. Santner-Nanan, K. Straubinger, P. Hsu, G. Parnell, B. Tang, B. Xu, A. Makris,
Fetal-maternal alignment of regulatory T cells correlates with Interleukin 10 mediated Bcl- 2 upregulation in pregnancy B. Santner-Nanan, K. Straubinger, P. Hsu, G. Parnell, B. Tang, B. Xu, A. Makris,
Mouse Meda-4 : chromosome 5G bp. EST (547bp) _at. 5 -Meda4 inner race (~1.8Kb)
 Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Cover Page. The handle holds various files of this Leiden University dissertation.
 Cover Page The handle http://hdl.handle.net/1887/20177 holds various files of this Leiden University dissertation. Author: Kester, Maria Sophia van (Marloes) Title: Molecular aspects of cutaneous T-cell
Cover Page The handle http://hdl.handle.net/1887/20177 holds various files of this Leiden University dissertation. Author: Kester, Maria Sophia van (Marloes) Title: Molecular aspects of cutaneous T-cell
RAS Genes. The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes.
 ۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
Innate Immunity & Inflammation
 Innate Immunity & Inflammation The innate immune system is an evolutionally conserved mechanism that provides an early and effective response against invading microbial pathogens. It relies on a limited
Innate Immunity & Inflammation The innate immune system is an evolutionally conserved mechanism that provides an early and effective response against invading microbial pathogens. It relies on a limited
Table 1: NFkB Target Gene Sets 1.Genes included in the subset of 15k genes selected according to a MAD-based variation filter
 Supplementary Information NFkB target gene sets The NFkB target gene sets are listed below (Table1). In addition to the previously performed mapping of Affymetrix probesets and corresponding Lymphochip
Supplementary Information NFkB target gene sets The NFkB target gene sets are listed below (Table1). In addition to the previously performed mapping of Affymetrix probesets and corresponding Lymphochip
BIO360 Fall 2013 Quiz 1
 BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice
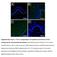 Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Numerous hypothesis tests were performed in this study. To reduce the false positive due to
 Two alternative data-splitting Numerous hypothesis tests were performed in this study. To reduce the false positive due to multiple testing, we are not only seeking the results with extremely small p values
Two alternative data-splitting Numerous hypothesis tests were performed in this study. To reduce the false positive due to multiple testing, we are not only seeking the results with extremely small p values
To compare the relative amount of of selected gene expression between sham and
 Supplementary Materials and Methods Gene Expression Analysis To compare the relative amount of of selected gene expression between sham and mice given renal ischemia-reperfusion injury (IRI), ncounter
Supplementary Materials and Methods Gene Expression Analysis To compare the relative amount of of selected gene expression between sham and mice given renal ischemia-reperfusion injury (IRI), ncounter
Regulating the Regulators for Cancer Immunotherapy: LAG-3 Finally Catches Up. Drew Pardoll Sidney Kimmel Cancer Center Johns Hopkins
 Regulating the Regulators for Cancer Immunotherapy: LAG-3 Finally Catches Up Drew Pardoll Sidney Kimmel Cancer Center Johns Hopkins The hostile immune microenvironment within a tumor Stat3 Stat3 NK Lytic
Regulating the Regulators for Cancer Immunotherapy: LAG-3 Finally Catches Up Drew Pardoll Sidney Kimmel Cancer Center Johns Hopkins The hostile immune microenvironment within a tumor Stat3 Stat3 NK Lytic
mirna Dr. S Hosseini-Asl
 mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
Megan S. Lim MD PhD. Translating Mass Spectrometry-Based Proteomics of Malignant Lymphoma into Clinical Application
 Translating Mass Spectrometry-Based Proteomics of Malignant Lymphoma into Clinical Application Megan S. Lim MD PhD FRCPC Department of Pathology, University of Michigan, Ann Arbor, MI Proteomics is a multi-faceted
Translating Mass Spectrometry-Based Proteomics of Malignant Lymphoma into Clinical Application Megan S. Lim MD PhD FRCPC Department of Pathology, University of Michigan, Ann Arbor, MI Proteomics is a multi-faceted
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at
 Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
Supporting Information. Evaluation of Toxicity and Gene Expression Changes Triggered by Oxide Nanoparticles
 Evaluation of Toxicity and Gene Expression Changes Triggered Bull. Korean Chem. Soc. 2011, Vol. 32, No. 6 1 DOI 10.5012/bkcs.2011.32.6. Supporting Information Evaluation of Toxicity and Gene Expression
Evaluation of Toxicity and Gene Expression Changes Triggered Bull. Korean Chem. Soc. 2011, Vol. 32, No. 6 1 DOI 10.5012/bkcs.2011.32.6. Supporting Information Evaluation of Toxicity and Gene Expression
An integrated transcriptomic and functional genomic approach identifies a. type I interferon signature as a host defense mechanism against Candida
 Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
Structure and Function of Fusion Gene Products in. Childhood Acute Leukemia
 Structure and Function of Fusion Gene Products in Childhood Acute Leukemia Chromosomal Translocations Chr. 12 Chr. 21 der(12) der(21) A.T. Look, Science 278 (1997) Distribution Childhood ALL TEL-AML1 t(12;21)
Structure and Function of Fusion Gene Products in Childhood Acute Leukemia Chromosomal Translocations Chr. 12 Chr. 21 der(12) der(21) A.T. Look, Science 278 (1997) Distribution Childhood ALL TEL-AML1 t(12;21)
Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream signals in in vitro Symbol Gene ID RefSeqID Clone ID
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream
Chapter 20. Cell - Cell Signaling: Hormones and Receptors. Three general types of extracellular signaling. endocrine signaling. paracrine signaling
 Chapter 20 Cell - Cell Signaling: Hormones and Receptors Three general types of extracellular signaling endocrine signaling paracrine signaling autocrine signaling Endocrine Signaling - signaling molecules
Chapter 20 Cell - Cell Signaling: Hormones and Receptors Three general types of extracellular signaling endocrine signaling paracrine signaling autocrine signaling Endocrine Signaling - signaling molecules
Supplementary Material. Table S1. Summary of mapping results
 Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
HIV-1 Tat second exon limits the extent of Tat-mediated modulation. of interferon-stimulated genes in antigen presenting cells.
 HIV-1 Tat second exon limits the extent of Tat-mediated modulation of interferon-stimulated genes in antigen presenting cells The Harvard community has made this article openly available. Please share
HIV-1 Tat second exon limits the extent of Tat-mediated modulation of interferon-stimulated genes in antigen presenting cells The Harvard community has made this article openly available. Please share
Chapter 6: Cancer Pathways. Other Pathways. Cancer Pathways
 Chapter 6: Cancer Pathways Limited number of pathways control proliferation and differentiation Transmit signals from growth factors, hormones, cell-to-cell communications/interactions Pathways turn into
Chapter 6: Cancer Pathways Limited number of pathways control proliferation and differentiation Transmit signals from growth factors, hormones, cell-to-cell communications/interactions Pathways turn into
Transduction of lentivirus to human primary CD4+ T cells
 Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Introduction to Cancer Biology
 Introduction to Cancer Biology Robin Hesketh Multiple choice questions (choose the one correct answer from the five choices) Which ONE of the following is a tumour suppressor? a. AKT b. APC c. BCL2 d.
Introduction to Cancer Biology Robin Hesketh Multiple choice questions (choose the one correct answer from the five choices) Which ONE of the following is a tumour suppressor? a. AKT b. APC c. BCL2 d.
SUPPLEMENTARY DATA Expression of complement factors is induced in DBA/2 mice following administration of C. albicans
 SUPPLEMENTARY DATA Expression of complement factors is induced in DBA/2 mice following administration of C. albicans water-soluble mannoprotein-beta-glucan complex (CAWS) Noriko Nagi-Miura 1,, Daisuke
SUPPLEMENTARY DATA Expression of complement factors is induced in DBA/2 mice following administration of C. albicans water-soluble mannoprotein-beta-glucan complex (CAWS) Noriko Nagi-Miura 1,, Daisuke
Oncolytic virus strategy
 Oncolytic viruses Oncolytic virus strategy normal tumor NO replication replication survival lysis Oncolytic virus strategy Mechanisms of tumor selectivity of several, some of them naturally, oncolytic
Oncolytic viruses Oncolytic virus strategy normal tumor NO replication replication survival lysis Oncolytic virus strategy Mechanisms of tumor selectivity of several, some of them naturally, oncolytic
CHAPTER INTRODUCTION
 CHAPTER 6 IDENTIFICATION OF RHEUMATOID ARTHRITIS SIGNIFICANT TARGET PROTEINS AND THEIR PATHWAYS THROUGH THE COMBINATORIAL APPROACH (RA-GIM, RA-DTP AND MICROARRAY DATANALYSIS OF RA) 6.1 INTRODUCTION The
CHAPTER 6 IDENTIFICATION OF RHEUMATOID ARTHRITIS SIGNIFICANT TARGET PROTEINS AND THEIR PATHWAYS THROUGH THE COMBINATORIAL APPROACH (RA-GIM, RA-DTP AND MICROARRAY DATANALYSIS OF RA) 6.1 INTRODUCTION The
5.1 Introduction The DSCR1 gene family
 Chapter 5 5.1 Introduction Several endogenous regulators of calcineurin signalling pathway are important for cellular homeostasis and a role of cyclophilin B emerged in the previous chapter. Yet another
Chapter 5 5.1 Introduction Several endogenous regulators of calcineurin signalling pathway are important for cellular homeostasis and a role of cyclophilin B emerged in the previous chapter. Yet another
CO 2 tolerance of Atlantic salmon post-smolts in recirculating aquaculture systems
 CO 2 tolerance of Atlantic salmon post-smolts in recirculating aquaculture systems Vasco Mota*, Tom Nilsen, Elizabeth Ytteborg, Grete Baeverfjord, Aleksei Krasnov, Jelena Kolarevic, Lars Ebbesson, Steven
CO 2 tolerance of Atlantic salmon post-smolts in recirculating aquaculture systems Vasco Mota*, Tom Nilsen, Elizabeth Ytteborg, Grete Baeverfjord, Aleksei Krasnov, Jelena Kolarevic, Lars Ebbesson, Steven
Applied Biosystems models 7500 (Fast block), 7900HT (Fast block), StepOnePlus, ViiA 7 (Fast block)
 RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human HIV Infection and Host Response Cat. no. 330231 PAHS-051YA For pathway expression analysis Format Format A Format C Format D Format
A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy se
 A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy serves as a defence mechanism that prevents or retards
A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy serves as a defence mechanism that prevents or retards
 Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
OVARIAN CANCER POSSIBLE TUMOR-SUPPRESSIVE ROLE OF BATF2 IN OVARIAN CANCER
 POSSIBLE TUMOR-SUPPRESSIVE ROLE OF BATF2 IN OVARIAN CANCER Rissa Fedora, OMS-III OVARIAN CANCER 5 th leading cause of death among American women Risk factors: Age Gender female Greater lifetime ovulations
POSSIBLE TUMOR-SUPPRESSIVE ROLE OF BATF2 IN OVARIAN CANCER Rissa Fedora, OMS-III OVARIAN CANCER 5 th leading cause of death among American women Risk factors: Age Gender female Greater lifetime ovulations
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
Subject Index. Bcl-2, apoptosis regulation Bone marrow, polymorphonuclear neutrophil release 24, 26
 Subject Index A1, apoptosis regulation 217, 218 Adaptive immunity, polymorphonuclear neutrophil role 31 33 Angiogenesis cancer 178 endometrium remodeling 172 HIV Tat induction mechanism 176 inflammatory
Subject Index A1, apoptosis regulation 217, 218 Adaptive immunity, polymorphonuclear neutrophil role 31 33 Angiogenesis cancer 178 endometrium remodeling 172 HIV Tat induction mechanism 176 inflammatory
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Follicular Lymphoma. ced3 APOPTOSIS. *In the nematode Caenorhabditis elegans 131 of the organism's 1031 cells die during development.
 Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai Follicular Lymphoma 1. Characterized by t(14:18) translocation 2. Ig heavy
Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai Follicular Lymphoma 1. Characterized by t(14:18) translocation 2. Ig heavy
Molecular measurements for radiation-induced PTC risk modelling
 Molecular measurements for radiation-induced PTC risk modelling DoReMi 3rd Periodic Meeting Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences
Molecular measurements for radiation-induced PTC risk modelling DoReMi 3rd Periodic Meeting Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences
Excessive fatty acid oxidation induces muscle atrophy in cancer cachexia
 SUPPLEMENTARY INFORMATION Excessive fatty acid oxidation induces muscle atrophy in cancer cachexia Tomoya Fukawa, Benjamin Chua Yan-Jiang, Jason Chua Min-Wen, Elwin Tan Jun-Hao, Dan Huang, Chao-Nan Qian,
SUPPLEMENTARY INFORMATION Excessive fatty acid oxidation induces muscle atrophy in cancer cachexia Tomoya Fukawa, Benjamin Chua Yan-Jiang, Jason Chua Min-Wen, Elwin Tan Jun-Hao, Dan Huang, Chao-Nan Qian,
SIRT6 histone deacetylase functions as a potential oncogene in human melanoma -
 SIRT6 histone deacetylase functions as a potential oncogene in human melanoma - Garcia-Peterson et al Supplementary Figure S:Tissue microarray (TMA) staining and organization. A) Diagram of the TMA indicating
SIRT6 histone deacetylase functions as a potential oncogene in human melanoma - Garcia-Peterson et al Supplementary Figure S:Tissue microarray (TMA) staining and organization. A) Diagram of the TMA indicating
Lecture #27 Lecturer A. N. Koval
 Lecture #27 Lecturer A. N. Koval Hormones Transduce Signals to Affect Homeostatic Mechanisms Koval A. (C), 2011 2 Lipophilic hormones Classifying hormones into hydrophilic and lipophilic molecules indicates
Lecture #27 Lecturer A. N. Koval Hormones Transduce Signals to Affect Homeostatic Mechanisms Koval A. (C), 2011 2 Lipophilic hormones Classifying hormones into hydrophilic and lipophilic molecules indicates
What would you observe if you fused a G1 cell with a S cell? A. Mitotic and pulverized chromosomes. B. Mitotic and compact G1 chromosomes.
 What would you observe if you fused a G1 cell with a S cell? A. Mitotic and pulverized chromosomes. B. Mitotic and compact G1 chromosomes. C. Mostly non-compact G1 chromosomes. D. Compact G1 and G2 chromosomes.
What would you observe if you fused a G1 cell with a S cell? A. Mitotic and pulverized chromosomes. B. Mitotic and compact G1 chromosomes. C. Mostly non-compact G1 chromosomes. D. Compact G1 and G2 chromosomes.
Characterization of Complete Response to IL-2 Using Gene Expression Analysis and Tissue Array Validation in Metastatic RCCa
 Characterization of Complete Response to IL-2 Using Gene Expression Analysis and Tissue Array Validation in Metastatic RCCa Allan J. Pantuck, M.D. UCLA Department of Urology Society of Biologic Therapy
Characterization of Complete Response to IL-2 Using Gene Expression Analysis and Tissue Array Validation in Metastatic RCCa Allan J. Pantuck, M.D. UCLA Department of Urology Society of Biologic Therapy
MAPK Pathway
 MAPK Pathway Mitogen-activated protein kinases (MAPK) are proteins that are serine/ threonine specific kinases which are activated by a wide range of stimuli including proinflammatory cytokines, growth
MAPK Pathway Mitogen-activated protein kinases (MAPK) are proteins that are serine/ threonine specific kinases which are activated by a wide range of stimuli including proinflammatory cytokines, growth
Cytokines, adhesion molecules and apoptosis markers. A comprehensive product line for human and veterinary ELISAs
 Cytokines, adhesion molecules and apoptosis markers A comprehensive product line for human and veterinary ELISAs IBL International s cytokine product line... is extremely comprehensive. The assays are
Cytokines, adhesion molecules and apoptosis markers A comprehensive product line for human and veterinary ELISAs IBL International s cytokine product line... is extremely comprehensive. The assays are
Determination Differentiation. determinated precursor specialized cell
 Biology of Cancer -Developmental Biology: Determination and Differentiation -Cell Cycle Regulation -Tumor genes: Proto-Oncogenes, Tumor supressor genes -Tumor-Progression -Example for Tumor-Progression:
Biology of Cancer -Developmental Biology: Determination and Differentiation -Cell Cycle Regulation -Tumor genes: Proto-Oncogenes, Tumor supressor genes -Tumor-Progression -Example for Tumor-Progression:
Supplementary information. Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls
 Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
BIO360 Fall 2013 Quiz 1
 BIO360 Fall 2013 Quiz 1 Name: Key 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function
BIO360 Fall 2013 Quiz 1 Name: Key 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function
KEY CONCEPT QUESTIONS IN SIGNAL TRANSDUCTION
 Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Developing Best in Class BET Inhibitors for Oncology & AI: from Discovery to the Clinic. Kevin G. McLure, PhD EpiCongress July 2014
 Developing Best in Class BET Inhibitors for Oncology & AI: from Discovery to the Clinic Kevin G. McLure, PhD EpiCongress July 2014 Zenith Epigenetics Overview Formed from Resverlogix as an independent
Developing Best in Class BET Inhibitors for Oncology & AI: from Discovery to the Clinic Kevin G. McLure, PhD EpiCongress July 2014 Zenith Epigenetics Overview Formed from Resverlogix as an independent
Supplemental Figure 1
 Supplemental Figure 1 A B Immune Tissues Non-Immune Tissues Immune Tissues Non-Immune Tissues thymus fetal liver spleen germinal center B cells spleen blood lymph node leukemia T cell (Jurkat) tonsil lymphoma
Supplemental Figure 1 A B Immune Tissues Non-Immune Tissues Immune Tissues Non-Immune Tissues thymus fetal liver spleen germinal center B cells spleen blood lymph node leukemia T cell (Jurkat) tonsil lymphoma
Validated Mouse Quantitative RT-PCR Genes Gene Gene Bank Accession Number Name
 5HT1a NM_008308.4 Serotonin Receptor 1a 5HT1b NM_010482.1 Serotonin Receptor 1b 5HT2a NM_172812.2 Serotonin Receptor 2a ACO-2 NM_080633.2 Aconitase 2 Adora2A NM_009630.02 Adenosine A2a Receptor Aif1 (IbaI)
5HT1a NM_008308.4 Serotonin Receptor 1a 5HT1b NM_010482.1 Serotonin Receptor 1b 5HT2a NM_172812.2 Serotonin Receptor 2a ACO-2 NM_080633.2 Aconitase 2 Adora2A NM_009630.02 Adenosine A2a Receptor Aif1 (IbaI)
MicroRNA dysregulation in cancer. Systems Plant Microbiology Hyun-Hee Lee
 MicroRNA dysregulation in cancer Systems Plant Microbiology Hyun-Hee Lee Contents 1 What is MicroRNA? 2 mirna dysregulation in cancer 3 Summary What is MicroRNA? What is MicroRNA? MicroRNAs (mirnas) -
MicroRNA dysregulation in cancer Systems Plant Microbiology Hyun-Hee Lee Contents 1 What is MicroRNA? 2 mirna dysregulation in cancer 3 Summary What is MicroRNA? What is MicroRNA? MicroRNAs (mirnas) -
Toll-like Receptor Signaling
 Toll-like Receptor Signaling 1 Professor of Medicine University of Massachusetts Medical School, Worcester, MA, USA Why do we need innate immunity? Pathogens multiply very fast We literally swim in viruses
Toll-like Receptor Signaling 1 Professor of Medicine University of Massachusetts Medical School, Worcester, MA, USA Why do we need innate immunity? Pathogens multiply very fast We literally swim in viruses
SUPPLEMENTARY INFORMATION. Divergent TLR7/9 signaling and type I interferon production distinguish
 SUPPLEMENTARY INFOATION Divergent TLR7/9 signaling and type I interferon production distinguish pathogenic and non-pathogenic AIDS-virus infections Judith N. Mandl, Ashley P. Barry, Thomas H. Vanderford,
SUPPLEMENTARY INFOATION Divergent TLR7/9 signaling and type I interferon production distinguish pathogenic and non-pathogenic AIDS-virus infections Judith N. Mandl, Ashley P. Barry, Thomas H. Vanderford,
Numerous hypothesis tests were performed in this study. To reduce the false positive due to
 Two alternative data-splitting Numerous hypothesis tests were performed in this study. To reduce the false positive due to multiple testing, we are not only seeking the results with extremely small p values
Two alternative data-splitting Numerous hypothesis tests were performed in this study. To reduce the false positive due to multiple testing, we are not only seeking the results with extremely small p values
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
Supplementary Table S1: Gene expression targets
 Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Computational Biology I LSM5191
 Computational Biology I LSM5191 Aylwin Ng, D.Phil Lecture 6 Notes: Control Systems in Gene Expression Pulling it all together: coordinated control of transcriptional regulatory molecules Simple Control:
Computational Biology I LSM5191 Aylwin Ng, D.Phil Lecture 6 Notes: Control Systems in Gene Expression Pulling it all together: coordinated control of transcriptional regulatory molecules Simple Control:
Identificación de vulnerabilidades en tumores sólidos: aportes hacia una medicina personalizada?
 Identificación de vulnerabilidades en tumores sólidos: aportes hacia una medicina personalizada? Alberto Ocana Albacete University Hospital Salamanca May 19th, 2016 WHAT IS PERSONALIZED MEDICINE? Molecular
Identificación de vulnerabilidades en tumores sólidos: aportes hacia una medicina personalizada? Alberto Ocana Albacete University Hospital Salamanca May 19th, 2016 WHAT IS PERSONALIZED MEDICINE? Molecular
T Cell Activation. Patricia Fitzgerald-Bocarsly March 18, 2009
 T Cell Activation Patricia Fitzgerald-Bocarsly March 18, 2009 Phases of Adaptive Immune Responses Phases of T cell responses IL-2 acts as an autocrine growth factor Fig. 11-11 Clonal Expansion of T cells
T Cell Activation Patricia Fitzgerald-Bocarsly March 18, 2009 Phases of Adaptive Immune Responses Phases of T cell responses IL-2 acts as an autocrine growth factor Fig. 11-11 Clonal Expansion of T cells
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr
 Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
Basis and Clinical Applications of Interferon
 Interferon Therapy Basis and Clinical Applications of Interferon JMAJ 47(1): 7 12, 2004 Jiro IMANISHI Professor, Kyoto Prefectural University of Medicine Abstract: Interferon (IFN) is an antiviral substance
Interferon Therapy Basis and Clinical Applications of Interferon JMAJ 47(1): 7 12, 2004 Jiro IMANISHI Professor, Kyoto Prefectural University of Medicine Abstract: Interferon (IFN) is an antiviral substance
Antibodies for Unfolded Protein Response
 Novus-lu-2945 Antibodies for Unfolded rotein Response Unfolded roteins ER lumen GR78 IRE-1 GR78 ERK Cytosol GR78 TRAF2 ASK1 JNK Activator Intron RIDD elf2α Degraded mrna XB1 mrna Translation XB1-S (p50)
Novus-lu-2945 Antibodies for Unfolded rotein Response Unfolded roteins ER lumen GR78 IRE-1 GR78 ERK Cytosol GR78 TRAF2 ASK1 JNK Activator Intron RIDD elf2α Degraded mrna XB1 mrna Translation XB1-S (p50)
Table 1A. Genes enriched as over-expressed in DPM treatment group
 Table 1A. s enriched as over-expressed in DPM treatment group Affy Probeset mrna Accession Description 10538247 Npy NM_023456 Mus musculus neuropeptide Y (Npy), mrna. 2.0024 10598062 --- NC_005089 gi 34538597
Table 1A. s enriched as over-expressed in DPM treatment group Affy Probeset mrna Accession Description 10538247 Npy NM_023456 Mus musculus neuropeptide Y (Npy), mrna. 2.0024 10598062 --- NC_005089 gi 34538597
Effects of Ole e 1 allergen on human bronchial epithelial cells cultured at air-liquid interface (No. JIACI-D )
 1 SUPPLEMENTAL TABLES Effects of Ole e 1 allergen on human bronchial epithelial cells cultured at air-liquid interface (No. JIACI-D-17-00149) Juan C. López-Rodríguez 1, Guillermo Solís-Fernández 1, Rodrigo
1 SUPPLEMENTAL TABLES Effects of Ole e 1 allergen on human bronchial epithelial cells cultured at air-liquid interface (No. JIACI-D-17-00149) Juan C. López-Rodríguez 1, Guillermo Solís-Fernández 1, Rodrigo
Supplemental Table 1:
 Supplemental Table 1: Gene up-regulated in remnant kidneys of the lesion-prone FVB/N mice as compared to the resistant B6D2F1 animals 2 months after 75% nephron reduction. Gene Symbol Gene Name F FDR Accession
Supplemental Table 1: Gene up-regulated in remnant kidneys of the lesion-prone FVB/N mice as compared to the resistant B6D2F1 animals 2 months after 75% nephron reduction. Gene Symbol Gene Name F FDR Accession
Venky Ramakrishna PhD Celldex Therapeutics, Hampton NJ, USA
 Antibody Targeting of Human CD27 with Varlilumab (CDX-1127) identifies genes and pathways related to inflammation Venky Ramakrishna PhD Celldex Therapeutics, Hampton NJ, USA www.celldextherapeutics.com
Antibody Targeting of Human CD27 with Varlilumab (CDX-1127) identifies genes and pathways related to inflammation Venky Ramakrishna PhD Celldex Therapeutics, Hampton NJ, USA www.celldextherapeutics.com
Sequence Coverage (%) Profilin-1 P UD 2
 Protein Name Accession Number (Swissprot) Sequence Coverage (%) No. of MS/MS Queries Mascot Score 1 Reference Cytoskeletal proteins Beta-actin P60709 37 14 298 Alpha-actin P68032 33 10 141 20 Beta-actin-like
Protein Name Accession Number (Swissprot) Sequence Coverage (%) No. of MS/MS Queries Mascot Score 1 Reference Cytoskeletal proteins Beta-actin P60709 37 14 298 Alpha-actin P68032 33 10 141 20 Beta-actin-like
Supporting Information
 Supporting Information M1 macrophage-derived nanovesicles potentiate the anticancer efficacy of immune checkpoint inhibitors Yeon Woong Choo, 1, Mikyung Kang, 2, Han Young Kim, 1 Jin Han, 1 Seokyung Kang,
Supporting Information M1 macrophage-derived nanovesicles potentiate the anticancer efficacy of immune checkpoint inhibitors Yeon Woong Choo, 1, Mikyung Kang, 2, Han Young Kim, 1 Jin Han, 1 Seokyung Kang,
Molecular Cell Biology. Prof. D. Karunagaran. Department of Biotechnology. Indian Institute of Technology Madras
 Molecular Cell Biology Prof. D. Karunagaran Department of Biotechnology Indian Institute of Technology Madras Module 9 Molecular Basis of Cancer, Oncogenes and Tumor Suppressor Genes Lecture 2 Genes Associated
Molecular Cell Biology Prof. D. Karunagaran Department of Biotechnology Indian Institute of Technology Madras Module 9 Molecular Basis of Cancer, Oncogenes and Tumor Suppressor Genes Lecture 2 Genes Associated
Supplementary Material 1
 Supplementary Material 1 Legend of Supplementary Figure 1. Heat-map generated using class comparison methods and clustered using analysis of variance test (ANOVA), as described in the Methods section.
Supplementary Material 1 Legend of Supplementary Figure 1. Heat-map generated using class comparison methods and clustered using analysis of variance test (ANOVA), as described in the Methods section.
Principles of cell signaling Lecture 4
 Principles of cell signaling Lecture 4 Johan Lennartsson Molecular Cell Biology (1BG320), 2014 Johan.Lennartsson@licr.uu.se 1 Receptor tyrosine kinase-induced signal transduction Erk MAP kinase pathway
Principles of cell signaling Lecture 4 Johan Lennartsson Molecular Cell Biology (1BG320), 2014 Johan.Lennartsson@licr.uu.se 1 Receptor tyrosine kinase-induced signal transduction Erk MAP kinase pathway
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004
 Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent Protein.
 prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
Lectins: selected topics 3/2/17
 Lectins: selected topics 3/2/17 Selected topics Regulation of T-cell receptor signaling Thymic selection of self vs. non-self T-cells Essentials of Glycobiology Second Edition Signaling pathways associated
Lectins: selected topics 3/2/17 Selected topics Regulation of T-cell receptor signaling Thymic selection of self vs. non-self T-cells Essentials of Glycobiology Second Edition Signaling pathways associated
T-cell activation T cells migrate to secondary lymphoid tissues where they interact with antigen, antigen-presenting cells, and other lymphocytes:
 Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
T-cell activation T cells migrate to secondary lymphoid tissues where they interact with antigen, antigen-presenting cells, and other lymphocytes:
 Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Cancer. Throughout the life of an individual, but particularly during development, every cell constantly faces decisions.
 Cancer Throughout the life of an individual, but particularly during development, every cell constantly faces decisions. Should it divide? Yes No--> Should it differentiate? Yes No-->Should it die? Yes-->Apoptosis
Cancer Throughout the life of an individual, but particularly during development, every cell constantly faces decisions. Should it divide? Yes No--> Should it differentiate? Yes No-->Should it die? Yes-->Apoptosis
Expanded View Figures
 Molecular Systems iology Immediate late genes decode ERK signal duration Florian Uhlitz et al Expanded View Figures Figure EV. cquired samples from HEK9ΔRF:ER cells. Microarrays - Untreated I +U6-5 6 7
Molecular Systems iology Immediate late genes decode ERK signal duration Florian Uhlitz et al Expanded View Figures Figure EV. cquired samples from HEK9ΔRF:ER cells. Microarrays - Untreated I +U6-5 6 7
3) How many different amino acids are proteogenic in eukaryotic cells? A) 12 B) 20 C) 25 D) 30 E) None of the above
 Suggesting questions for Biochemistry 1 and 2 and clinical biochemistry 1) Henderson Hasselbalch Equation shows: A) The relationship between ph and the concentration of an acid and its conjugate base B)
Suggesting questions for Biochemistry 1 and 2 and clinical biochemistry 1) Henderson Hasselbalch Equation shows: A) The relationship between ph and the concentration of an acid and its conjugate base B)
CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS
 CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS Summary of the regulation of cyclin/cdk complexes during celll cycle Cell cycle phase Cyclin-cdk complex inhibitor activation Substrate(s) G1 Cyclin D/cdk 4,6
CELL CYCLE MOLECULAR BASIS OF ONCOGENESIS Summary of the regulation of cyclin/cdk complexes during celll cycle Cell cycle phase Cyclin-cdk complex inhibitor activation Substrate(s) G1 Cyclin D/cdk 4,6
