FUNCTIONAL INTERACTIONS BETWEEN ERYTHROID KRUPPEL-LIKE FACTOR (EKLF/KLF1) AND PROTEIN PHOSPHATASE PPM1B/PP2Cß
|
|
|
- Silvester Simmons
- 5 years ago
- Views:
Transcription
1 SUPPLEMENTAL INFORMATION FOR FUNCTIONAL INTERACTIONS BETWEEN ERYTHROID KRUPPEL-LIKE FACTOR (EKLF/KLF1) AND PROTEIN PHOSPHATASE PPM1B/PP2Cß Yvette Y. Yien and James J. Bieker FIGURE S1. Induction of EKLF and M2-agarose pulldown of EKLF-interacting proteins. MEL stable cell lines expressing Zn-inducible FL-EKLF were treated with 100 µm Zn for 18 hours to express FL-EKLF. Cell lysates from untreated (-Zn) or treated (+Zn) samples were immunoprecipitated with anti- FLAG M2 agarose. Bound proteins were eluted with FLAG peptide and concentrated using a centricon unit with a 10kDa cutoff. Top: After SDS-PAGE, the gel was fixed with 10% methanol and stained with colloidal blue. Six bands, labelled 1 to 6 as shown, were sliced out of the gel and subjected to mass spectrometric analysis. Molecular weight markers are on the left, and major proteins in each band on the right (see Table S1). Bottom: 5% of cell lysate were subjected to SDS-PAGE electrophoresis and immunolotting with M2-HRP (Sigma) to verify that FL-EKLF was induced. FIGURE S2. EKLF likely interacts with multiple isoforms of Ppm1b. Amino acid alignment of the four Ppm1b isoforms are shown. Of the 10 Ppm1b peptides identified from the gel in Fig. S1 (and tabulated in Table S2), 9 peptide sequences are present in all 4 isoforms; one is unique to the largest isoform (isoform 1). TABLE S1. Proteins identified by mass spectrometry in FL-EKLF pulldown experiment shown in Fig. S1. Accession numbers are given for protein identity in the International Protein Index, TABLE S2. Peptides corresponding to non-keratin proteins that were identified by mass spectrometry from the gel in Fig. S1. TABLE S3. Nucleotide sequences for sippm1b that were used for nucleofection in Fig. 2.
2
3
4 Sheet1 B01 Identified Proteins (4) Accession Number Molecular Weight Otud4 protein (Otud 4) IPI kda Keratin type I cytoskeletal 19 IPI (+3) 45 kda B02 Identified Proteins (5) Accession Number Molecular Weight Isoform 1 of SCY1-like protein 2 (Scyl 2) IPI kda Keratin, type II cytoskeletal 79 IPI kda Krt19 Keratin, type I cytoskeletal 19 IPI (+3) 45 kda B03 Identified Proteins (6) Accession Number Molecular Weight Zyxin IPI (+1) 61 kda Heat shock cognate 71 kda protein (Hspa 8) IPI (+2) 71 kda Isoform 1 of SCY1-like protein 2 (Scyl 2) IPI kda Keratin 15, isoform CRA_a IPI (+1) 49 kda Keratin, type II cytoskeletal 2 epidermal IPI (+1) 71 kda B04 Identified Proteins (4) Accession Number Molecular Weight Ppm1b, Protein phosphatase 1B IPI kda Keratin, type II cytoskeletal 5 IPI kda B05 Identified Proteins (4) Accession Number Molecular Weight Ppm1b, Protein phosphatase 1B IPI (+5) 52 kda Keratin, type I cytoskeletal 42 IPI kda Kruppel-like factor 1 (EKLF) IPI kda B06 Identified Proteins (6) Accession Number Molecular Weight Ppm1b, Isoform Beta-1 of Protein phosphatase 1B IPI kda Actin, cytoplasmic 1 IPI kda Tropomodulin-3 IPI kda Keratin, type II cytoskeletal 5 IPI kda Keratin 15, isoform CRA_a IPI kda Klf1 Kruppel-like factor 1 (EKLF) IPI kda Page 1
5 Sheet1 Protein name Protein accession numbers No. unique peptides Peptide sequence Best Peptide identification probability Peptide start index Peptide stop index IPI:IPI Tax_Id=10090 Gene_Symbol=Ppm1b Putative uncharacterized protein IPI ALGDYDYK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Ppm1b Putative uncharacterized protein IPI DSELDKHLESR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Ppm1b Putative uncharacterized protein IPI GPTEQLVSPEPEVYEIVR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Ppm1b Putative uncharacterized protein IPI HVIEAVYSR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Ppm1b Putative uncharacterized protein IPI ILSAENIPNLPPGGGLAGK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Ppm1b Putative uncharacterized protein IPI IQNAGGSVMIQR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Ppm1b Putative uncharacterized protein IPI QLLEEMLTSYR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Ppm1b Putative uncharacterized protein IPI SGEEGMPDLAHVMR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Ppm1b Putative uncharacterized protein IPI VNGSLAVSR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Ppm1b Putative uncharacterized protein IPI YGLSSMQGWR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Zyx zyxin IPI ,IPI FAPVAPK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Zyx zyxin IPI ,IPI FAPVVAPKPK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Zyx zyxin IPI ,IPI FSPGAPSGPGPQPNQK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Zyx zyxin IPI ,IPI GPLSQAPTPAPK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Zyx zyxin IPI ,IPI SPGGPGPLTLK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Zyx zyxin IPI ,IPI VNPFRPGDSEPPVAAGAQR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Scyl2 Isoform 1 of SCY1-like protein 2 IPI LGSSSLTSIPEEVR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Scyl2 Isoform 1 of SCY1-like protein 2 IPI LILPELGPVFK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Scyl2 Isoform 1 of SCY1-like protein 2 IPI QEVAVFVFDKK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Scyl2 Isoform 1 of SCY1-like protein 2 IPI VTADVTSAVMGNPVTR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Scyl2 Isoform 1 of SCY1-like protein 2 IPI YGLLQVSEGLSFLHSSVK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Actb Actin, cytoplasmic 1 IPI AGFAGDDAPR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Actb Actin, cytoplasmic 1 IPI EITALAPSTMK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Actb Actin, cytoplasmic 1 IPI GYSFTTTAER 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Actb Actin, cytoplasmic 1 IPI VAPEEHPVLLTEAPLNPK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Tmod3 Tropomodulin-3 IPI MLEENTNILK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Tmod3 Tropomodulin-3 IPI QQLGTSVELEMAK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Tmod3 Tropomodulin-3 IPI SNDPVAVAFADMLK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Otud4 Otud4 protein IPI AVAEQVLHSQSR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Otud4 Otud4 protein IPI KADIDLASGSSAVEGK 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Klf1 Kruppel-like factor 1 IPI AGSVGGFFPR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Klf1 Kruppel-like factor 1 IPI SEETQDLGPGPPNPTGPSLH 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Hspa8 Heat shock cognate 71 kda protein IPI ,IPI ,IPI0 2 DAGTIAGLNVLR 95.00% IPI:IPI Tax_Id=10090 Gene_Symbol=Hspa8 Heat shock cognate 71 kda protein IPI ,IPI ,IPI0 2 TTPSYVAFTDTER 95.00% Page 1
6 Catalog no. Target sequence si1 SI TAGCCTAACTACACACATCAA si2 SI CGACATAACATGAGTATTGTA si3 SI ATCACAAACCTTGCAATCCAA
Supplementary Figure S1
 Supplementary Figure S1 Supplementary Figure S1. PARP localization patterns using GFP-PARP and PARP-specific antibody libraries GFP-PARP localization in non-fixed (A) and formaldehyde fixed (B) GFP-PARPx
Supplementary Figure S1 Supplementary Figure S1. PARP localization patterns using GFP-PARP and PARP-specific antibody libraries GFP-PARP localization in non-fixed (A) and formaldehyde fixed (B) GFP-PARPx
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
5 Identification of Binding Partners of the Annexin A2 / P11 Complex by Chemical Cross-Linking
 5 Identification of Binding Partners of the Annexin A2 / P11 Complex by Chemical Cross-Linking In the quest of the omics sciences for holistic schemes, the identification of binding partners of proteins
5 Identification of Binding Partners of the Annexin A2 / P11 Complex by Chemical Cross-Linking In the quest of the omics sciences for holistic schemes, the identification of binding partners of proteins
Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth.
 Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth. a. Immunoblot for Usp9x protein in NRAS mutant melanoma cells
Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth. a. Immunoblot for Usp9x protein in NRAS mutant melanoma cells
Supplementary Information
 Supplementary Information Targeted Disruption of the EZH2/EED Complex Inhibits EZH2- dependent Cancer Woojin Kim 1,2,3, Gregory H. Bird 2,3,4, Tobias Neff 5, Guoji Guo 1,2,3, Marc A. Kerenyi 1,2,3, Loren
Supplementary Information Targeted Disruption of the EZH2/EED Complex Inhibits EZH2- dependent Cancer Woojin Kim 1,2,3, Gregory H. Bird 2,3,4, Tobias Neff 5, Guoji Guo 1,2,3, Marc A. Kerenyi 1,2,3, Loren
Supplementary Fig. 1. Identification of acetylation of K68 of SOD2
 Supplementary Fig. 1. Identification of acetylation of K68 of SOD2 A B H. sapiens 54 KHHAAYVNNLNVTEEKYQEALAK 75 M. musculus 54 KHHAAYVNNLNATEEKYHEALAK 75 X. laevis 55 KHHATYVNNLNITEEKYAEALAK 77 D. rerio
Supplementary Fig. 1. Identification of acetylation of K68 of SOD2 A B H. sapiens 54 KHHAAYVNNLNVTEEKYQEALAK 75 M. musculus 54 KHHAAYVNNLNATEEKYHEALAK 75 X. laevis 55 KHHATYVNNLNITEEKYAEALAK 77 D. rerio
Effect of Taurine on Acinar Cell Apoptosis and Pancreatic Fibrosis in Dibutyltin Dichloride-induced Chronic Pancreatitis
 212 66 4 329334 Effect of Taurine on Acinar Cell Apoptosis and Pancreatic Fibrosis in Dibutyltin Dichloride-induced Chronic Pancreatitis a,c a* b b a a b a a b c 33 66 4 ʼ 6 6 6 28 6 5 5 5 28 45 ʼ ʼ ʼ
212 66 4 329334 Effect of Taurine on Acinar Cell Apoptosis and Pancreatic Fibrosis in Dibutyltin Dichloride-induced Chronic Pancreatitis a,c a* b b a a b a a b c 33 66 4 ʼ 6 6 6 28 6 5 5 5 28 45 ʼ ʼ ʼ
a b G75 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server.
 a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR-2B cells untreated (-) or stimulated (+) for 45 min
 Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
Structural Characterization of Prion-like Conformational Changes of the Neuronal Isoform of Aplysia CPEB
 Structural Characterization of Prion-like Conformational Changes of the Neuronal Isoform of Aplysia CPEB Bindu L. Raveendra, 1,5 Ansgar B. Siemer, 2,6 Sathyanarayanan V. Puthanveettil, 1,3,7 Wayne A. Hendrickson,
Structural Characterization of Prion-like Conformational Changes of the Neuronal Isoform of Aplysia CPEB Bindu L. Raveendra, 1,5 Ansgar B. Siemer, 2,6 Sathyanarayanan V. Puthanveettil, 1,3,7 Wayne A. Hendrickson,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Posch et al., http://www.jcb.org/cgi/content/full/jcb.200912046/dc1 Figure S1. Biochemical characterization of the interaction between
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Posch et al., http://www.jcb.org/cgi/content/full/jcb.200912046/dc1 Figure S1. Biochemical characterization of the interaction between
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR
 Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
N α -Acetylation of yeast ribosomal proteins and its effect on protein synthesis
 JOURNAL OF PROTEOMICS 74 (2011) 431 441 available at www.sciencedirect.com www.elsevier.com/locate/jprot N α -Acetylation of yeast ribosomal proteins and its effect on protein synthesis Masahiro Kamita
JOURNAL OF PROTEOMICS 74 (2011) 431 441 available at www.sciencedirect.com www.elsevier.com/locate/jprot N α -Acetylation of yeast ribosomal proteins and its effect on protein synthesis Masahiro Kamita
Section 1 Proteins and Proteomics
 Section 1 Proteins and Proteomics Learning Objectives At the end of this assignment, you should be able to: 1. Draw the chemical structure of an amino acid and small peptide. 2. Describe the difference
Section 1 Proteins and Proteomics Learning Objectives At the end of this assignment, you should be able to: 1. Draw the chemical structure of an amino acid and small peptide. 2. Describe the difference
Protocol for Western Blo
 Protocol for Western Blo ng SDS-PAGE separa on 1. Make appropriate percentage of separa on gel according to the MW of target proteins. Related recommenda ons and rou ne recipes of separa on/stacking gels
Protocol for Western Blo ng SDS-PAGE separa on 1. Make appropriate percentage of separa on gel according to the MW of target proteins. Related recommenda ons and rou ne recipes of separa on/stacking gels
TSH Receptor Monoclonal Antibody (49) Catalog Number MA3-218 Product data sheet
 Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 TSH Receptor Monoclonal Antibody (49) Catalog Number MA3-218 Product data sheet Details
Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 TSH Receptor Monoclonal Antibody (49) Catalog Number MA3-218 Product data sheet Details
Schwarz et al. Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis
 Schwarz et al Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis and Sorting Pathway Supplemental Data Supplemental Fie 1: AMPARs undergo activity-mediated ubiquitination
Schwarz et al Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis and Sorting Pathway Supplemental Data Supplemental Fie 1: AMPARs undergo activity-mediated ubiquitination
A. List of selected proteins with high SILAC (H/L) ratios identified in mass
 Supplementary material Figure S1. Interaction between UBL5 and FANCI A. List of selected proteins with high SILAC (H/L) ratios identified in mass spectrometry (MS)-based analysis of UBL5-interacting proteins,
Supplementary material Figure S1. Interaction between UBL5 and FANCI A. List of selected proteins with high SILAC (H/L) ratios identified in mass spectrometry (MS)-based analysis of UBL5-interacting proteins,
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Improve Protein Analysis with the New, Mass Spectrometry- Compatible ProteasMAX Surfactant
 Improve Protein Analysis with the New, Mass Spectrometry- Compatible Surfactant ABSTRACT Incomplete solubilization and digestion and poor peptide recovery are frequent limitations in protein sample preparation
Improve Protein Analysis with the New, Mass Spectrometry- Compatible Surfactant ABSTRACT Incomplete solubilization and digestion and poor peptide recovery are frequent limitations in protein sample preparation
33VASTVNGATSANNHGEPPS51PADARPR58
 Pro-rich region Trans-membrane region 214 246 359 381 UL50 1 397 211SSRTAS216PPPPPR222 NLS CR1 CR2 CR3 CR4 UL53 1 376 11RERRS15ALRS19LLRKRRR25 33VASTVNGATSANNHGEPPS51PADARPR58 FIG S1. UL97 phosphorylation
Pro-rich region Trans-membrane region 214 246 359 381 UL50 1 397 211SSRTAS216PPPPPR222 NLS CR1 CR2 CR3 CR4 UL53 1 376 11RERRS15ALRS19LLRKRRR25 33VASTVNGATSANNHGEPPS51PADARPR58 FIG S1. UL97 phosphorylation
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Helen Kim, Ph.D. and John Cutts. Dept of Pharmacology & Toxicology University of Alabama at Birmingham
 Understanding the actions of a dietary anti-oxidant at the protein and small molecule level using top-down proteomics, enzyme assays and mass spectrometry elen Kim, Ph.D. and John Cutts Mar 9, 2012 UAB
Understanding the actions of a dietary anti-oxidant at the protein and small molecule level using top-down proteomics, enzyme assays and mass spectrometry elen Kim, Ph.D. and John Cutts Mar 9, 2012 UAB
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Structural vs. nonstructural proteins
 Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Depleting Lipoproteins from Serum
 Depleting Lipoproteins from Serum Kathy K. Foxx Kalen Biomedical LLC President For decades, fetal bovine serum (FBS) has been used as a supplement for cell-culture media, providing the growth factors that
Depleting Lipoproteins from Serum Kathy K. Foxx Kalen Biomedical LLC President For decades, fetal bovine serum (FBS) has been used as a supplement for cell-culture media, providing the growth factors that
Name Animal source Vendor Cat # Dilutions
 Supplementary data Table S1. Primary and Secondary antibody sources Devi et al, TXNIP in mitophagy A. Primary Antibodies Name Animal source Vendor Cat # Dilutions 1. TXNIP mouse MBL KO205-2 1:2000 (WB)
Supplementary data Table S1. Primary and Secondary antibody sources Devi et al, TXNIP in mitophagy A. Primary Antibodies Name Animal source Vendor Cat # Dilutions 1. TXNIP mouse MBL KO205-2 1:2000 (WB)
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
Supplementary Figure 1. Method development.
 Supplementary Figure 1 Method development. Titration experiments to determine standard antibody:lysate concentration. Lysates (~2 mg of total proteins) were prepared from cells expressing FLAG- tagged
Supplementary Figure 1 Method development. Titration experiments to determine standard antibody:lysate concentration. Lysates (~2 mg of total proteins) were prepared from cells expressing FLAG- tagged
STUDIES ON MUSTARD-STIMULATED PROTEASES AND INHIBITORS IN HUMAN EPIDERMAL KERATINOCYTES (HEK): DEVELOPMENT OF ANTIVESICANT DRUGS
 STUDIES ON MUSTARD-STIMULATED PROTEASES AND INHIBITORS IN HUMAN EPIDERMAL KERATINOCYTES (HEK): DEVELOPMENT OF ANTIVESICANT DRUGS Xiannu Jin 1, Radharaman Ray 2, Guang Xu 1 and Prabhati Ray 1 1 Department
STUDIES ON MUSTARD-STIMULATED PROTEASES AND INHIBITORS IN HUMAN EPIDERMAL KERATINOCYTES (HEK): DEVELOPMENT OF ANTIVESICANT DRUGS Xiannu Jin 1, Radharaman Ray 2, Guang Xu 1 and Prabhati Ray 1 1 Department
Strep-tag HRP Detection Kit (Horse Radish Peroxidase)
 Strep-tag HRP Detection Kit (Horse Radish Peroxidase) General protocol for the detection of Strep-tag fusion proteins Last date of revision April 2013 Version PR16-0006 www.strep-tag.com For research use
Strep-tag HRP Detection Kit (Horse Radish Peroxidase) General protocol for the detection of Strep-tag fusion proteins Last date of revision April 2013 Version PR16-0006 www.strep-tag.com For research use
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System
 Application Note LC/MS PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System Purpose This application note describes an automated workflow
Application Note LC/MS PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System Purpose This application note describes an automated workflow
Protein MultiColor Stable, Low Range
 Product Name: DynaMarker Protein MultiColor Stable, Low Range Code No: DM670L Lot No: ******* Size: 200 μl x 3 (DM670 x 3) (120 mini-gel lanes) Storage: 4 C Stability: 12 months at 4 C Storage Buffer:
Product Name: DynaMarker Protein MultiColor Stable, Low Range Code No: DM670L Lot No: ******* Size: 200 μl x 3 (DM670 x 3) (120 mini-gel lanes) Storage: 4 C Stability: 12 months at 4 C Storage Buffer:
Supplementary Figure S1 Supplementary Figure S2
 Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This
 SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
- 1 - Cell types Monocytes THP-1 cells Macrophages. LPS Treatment time (Hour) IL-6 level (pg/ml)
 Supplementary Table ST1: The dynamic effect of LPS on IL-6 production in monocytes and THP-1 cells after GdA treatment. Monocytes, THP-1 cells and macrophages (5x10 5 ) were incubated with 10 μg/ml of
Supplementary Table ST1: The dynamic effect of LPS on IL-6 production in monocytes and THP-1 cells after GdA treatment. Monocytes, THP-1 cells and macrophages (5x10 5 ) were incubated with 10 μg/ml of
Mammalian Tissue Protein Extraction Reagent
 Mammalian Tissue Protein Extraction Reagent Catalog number: AR0101 Boster s Mammalian Tissue Protein Extraction Reagent is a ready-to-use Western blot related reagent solution used for efficient extraction
Mammalian Tissue Protein Extraction Reagent Catalog number: AR0101 Boster s Mammalian Tissue Protein Extraction Reagent is a ready-to-use Western blot related reagent solution used for efficient extraction
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the
 Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Global profiling of dynamic protein palmitoylation
 Nature Methods Global profiling of dynamic protein palmitoylation Brent R Martin, Chu Wang, Alexander Adibekian, Sarah E Tully & Benjamin F Cravatt Supplementary Figure 1 Supplementary Figure 2 Supplementary
Nature Methods Global profiling of dynamic protein palmitoylation Brent R Martin, Chu Wang, Alexander Adibekian, Sarah E Tully & Benjamin F Cravatt Supplementary Figure 1 Supplementary Figure 2 Supplementary
!"#$%!&%##&'()*+,-.&/01*-0)*,(&%,2345()
 !"#$%!&%##&'()*+,-.&/01*-0)*,(&%,2345() Date of Submission Name: Email: Lab Antibody Name: Target: Company/ Source: Catalog Number, database ID, laboratory Lot Number Antibody Description: Target Description:
!"#$%!&%##&'()*+,-.&/01*-0)*,(&%,2345() Date of Submission Name: Email: Lab Antibody Name: Target: Company/ Source: Catalog Number, database ID, laboratory Lot Number Antibody Description: Target Description:
Supplementary Figure 1. Supernatants electrophoresis from CD14+ and dendritic cells. Supernatants were resolved by SDS-PAGE and stained with
 Supplementary Figure 1. Supernatants electrophoresis from CD14+ and dendritic cells. Supernatants were resolved by SDS-PAGE and stained with Coomassie brilliant blue. One µg/ml recombinant human (rh) apo-e
Supplementary Figure 1. Supernatants electrophoresis from CD14+ and dendritic cells. Supernatants were resolved by SDS-PAGE and stained with Coomassie brilliant blue. One µg/ml recombinant human (rh) apo-e
The time has come: SINGLE CELL western blotting. Parrinello Natalia TJC
 The time has come: SINGLE CELL western blotting Parrinello Natalia TJC 08.07.2014 To fully understand diverse and often rare behaviors in complex cell populations, researchers need analytical tools that:
The time has come: SINGLE CELL western blotting Parrinello Natalia TJC 08.07.2014 To fully understand diverse and often rare behaviors in complex cell populations, researchers need analytical tools that:
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
Supplemental information contains 7 movies and 4 supplemental Figures
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
Anticancer pyrroloquinazoline LBL1 targets nuclear lamins
 Supporting Information Anticancer pyrroloquinazoline LBL1 targets nuclear lamins Bingbing X. Li, #,* Jingjin Chen, # Bo Chao, # Larry L. David, & Xiangshu Xiao #,* # Program in Chemical Biology, Department
Supporting Information Anticancer pyrroloquinazoline LBL1 targets nuclear lamins Bingbing X. Li, #,* Jingjin Chen, # Bo Chao, # Larry L. David, & Xiangshu Xiao #,* # Program in Chemical Biology, Department
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Separation of Main Proteins in Plasma and Serum
 BCH 471 Experiment (2) Separation of Main Proteins in Plasma and Serum PLASMA PROTEINS Mw The main plasma proteins are: þ Albumin (36-50 g/l), Mw 66.241kDa. þ Globulins (18-32 g/l), Mw of globulins Cover
BCH 471 Experiment (2) Separation of Main Proteins in Plasma and Serum PLASMA PROTEINS Mw The main plasma proteins are: þ Albumin (36-50 g/l), Mw 66.241kDa. þ Globulins (18-32 g/l), Mw of globulins Cover
For the rapid, sensitive and accurate quantification of Ras in various samples
 ab128504 Ras Assay Kit Instructions for Use For the rapid, sensitive and accurate quantification of Ras in various samples This product is for research use only and is not intended for diagnostic use.
ab128504 Ras Assay Kit Instructions for Use For the rapid, sensitive and accurate quantification of Ras in various samples This product is for research use only and is not intended for diagnostic use.
Supplementary Information. Preferential associations between co-regulated genes reveal a. transcriptional interactome in erythroid cells
 Supplementary Information Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells Stefan Schoenfelder, * Tom Sexton, * Lyubomira Chakalova, * Nathan
Supplementary Information Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells Stefan Schoenfelder, * Tom Sexton, * Lyubomira Chakalova, * Nathan
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
Table S1: Summary of seminal characteristics of ejaculates collected by manual massage from Australian saltwater crocodiles.
 Electronic supplementary material Table S1: Summary of seminal characteristics of ejaculates collected by manual massage from Australian saltwater crocodiles. Seminal characteristic Mean ± SEM Range Volume
Electronic supplementary material Table S1: Summary of seminal characteristics of ejaculates collected by manual massage from Australian saltwater crocodiles. Seminal characteristic Mean ± SEM Range Volume
Title: Vectorization of biomacromolecules into cells using extracellular vesicles with enhanced internalization induced by macropinocytosis
 Scientific Reports Supplementary information Title: Vectorization of biomacromolecules into cells using extracellular vesicles with enhanced internalization induced by macropinocytosis Authors: Ikuhiko
Scientific Reports Supplementary information Title: Vectorization of biomacromolecules into cells using extracellular vesicles with enhanced internalization induced by macropinocytosis Authors: Ikuhiko
TECHNICAL BULLETIN. R 2 GlcNAcβ1 4GlcNAcβ1 Asn
 GlycoProfile II Enzymatic In-Solution N-Deglycosylation Kit Product Code PP0201 Storage Temperature 2 8 C TECHNICAL BULLETIN Product Description Glycosylation is one of the most common posttranslational
GlycoProfile II Enzymatic In-Solution N-Deglycosylation Kit Product Code PP0201 Storage Temperature 2 8 C TECHNICAL BULLETIN Product Description Glycosylation is one of the most common posttranslational
Cell Lysis Buffer. Catalog number: AR0103
 Cell Lysis Buffer Catalog number: AR0103 Boster s Cell Lysis Buffer is a ready-to-use Western blot related reagent solution used for efficient extraction of total soluble protein in nondenatured state
Cell Lysis Buffer Catalog number: AR0103 Boster s Cell Lysis Buffer is a ready-to-use Western blot related reagent solution used for efficient extraction of total soluble protein in nondenatured state
HiPer Western Blotting Teaching Kit
 HiPer Western Blotting Teaching Kit Product Code: HTI009 Number of experiments that can be performed: 5/20 Duration of Experiment: ~ 2 days Day 1: 6-8 hours (SDS- PAGE and Electroblotting) Day 2: 3 hours
HiPer Western Blotting Teaching Kit Product Code: HTI009 Number of experiments that can be performed: 5/20 Duration of Experiment: ~ 2 days Day 1: 6-8 hours (SDS- PAGE and Electroblotting) Day 2: 3 hours
So where were we? But what does the order mean? OK, so what's a protein? 4/1/11
 So where were we? We know that DNA is responsible for heredity Chromosomes are long pieces of DNA DNA turned out to be the transforming principle We know that DNA is shaped like a long double helix, with
So where were we? We know that DNA is responsible for heredity Chromosomes are long pieces of DNA DNA turned out to be the transforming principle We know that DNA is shaped like a long double helix, with
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
supplementary information
 DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
The Schedule and the Manual of Basic Techniques for Cell Culture
 The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
Supplementary Figure 1: Co-localization of reconstituted L-PTC and dendritic cells
 a CD11c Na + K + ATPase Na + K + ATPase CD11c x-y CD11c Na + K + ATPase Na + K + ATPase CD11c x-z c b x-y view BoNT NAPs CD11c BoNT CD11c NAPs BoNT NAPs CD11c 90 x-z view Apical Basolateral Supplementary
a CD11c Na + K + ATPase Na + K + ATPase CD11c x-y CD11c Na + K + ATPase Na + K + ATPase CD11c x-z c b x-y view BoNT NAPs CD11c BoNT CD11c NAPs BoNT NAPs CD11c 90 x-z view Apical Basolateral Supplementary
BIL 256 Cell and Molecular Biology Lab Spring, Tissue-Specific Isoenzymes
 BIL 256 Cell and Molecular Biology Lab Spring, 2007 Background Information Tissue-Specific Isoenzymes A. BIOCHEMISTRY The basic pattern of glucose oxidation is outlined in Figure 3-1. Glucose is split
BIL 256 Cell and Molecular Biology Lab Spring, 2007 Background Information Tissue-Specific Isoenzymes A. BIOCHEMISTRY The basic pattern of glucose oxidation is outlined in Figure 3-1. Glucose is split
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
Supplementary Materials
 Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
Supplementary figure 1
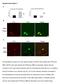 Supplementary figure 1 (A) Quantitative analysis of F-actin signal intensity in NIH3T3 cells treated with PTD4-myc- RBD. NIH3T3 cells were treated with PTD4-myc-RBD as described. Please note the increase
Supplementary figure 1 (A) Quantitative analysis of F-actin signal intensity in NIH3T3 cells treated with PTD4-myc- RBD. NIH3T3 cells were treated with PTD4-myc-RBD as described. Please note the increase
Tel: ; Fax: ;
 Tel.: +98 216 696 9291; Fax: +98 216 696 9291; E-mail: mrasadeghi@pasteur.ac.ir Tel: +98 916 113 7679; Fax: +98 613 333 6380; E-mail: abakhshi_e@ajums.ac.ir A Soluble Chromatin-bound MOI 0 1 5 0 1 5 HDAC2
Tel.: +98 216 696 9291; Fax: +98 216 696 9291; E-mail: mrasadeghi@pasteur.ac.ir Tel: +98 916 113 7679; Fax: +98 613 333 6380; E-mail: abakhshi_e@ajums.ac.ir A Soluble Chromatin-bound MOI 0 1 5 0 1 5 HDAC2
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Cell sphingolipids and S1P bound to endogenous TRAF2. Sphingolipid Cell pmol/mg TRAF2 immunoprecipitate pmol/mg Sphingomyelin 4200 ± 250 Not detected Monohexosylceramide 311 ± 18
Supplementary Table 1. Cell sphingolipids and S1P bound to endogenous TRAF2. Sphingolipid Cell pmol/mg TRAF2 immunoprecipitate pmol/mg Sphingomyelin 4200 ± 250 Not detected Monohexosylceramide 311 ± 18
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death
 www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death Supplementary
www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death Supplementary
Chemical Chaperones Mitigate Experimental Asthma By Attenuating Endoplasmic
 Chemical Chaperones Mitigate Experimental Asthma By Attenuating Endoplasmic Reticulum Stress Lokesh Makhija, BE, Veda Krishnan, MSc, Rakhshinda Rehman, MTech, Samarpana Chakraborty, MSc, Shuvadeep Maity,
Chemical Chaperones Mitigate Experimental Asthma By Attenuating Endoplasmic Reticulum Stress Lokesh Makhija, BE, Veda Krishnan, MSc, Rakhshinda Rehman, MTech, Samarpana Chakraborty, MSc, Shuvadeep Maity,
Predictive PP1Ca binding region in BIG3 : 1,228 1,232aa (-KAVSF-) HEK293T cells *** *** *** KPL-3C cells - E E2 treatment time (h)
 Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Expanded View Figures
 Sarah Kit Leng Lui et al USP26 stabilizes SM7 MO reports xpanded View igures igure V1. USP26 enhances SM2 phosphorylation and T-b-mediated transcription. raph representing relative luciferase values obtained
Sarah Kit Leng Lui et al USP26 stabilizes SM7 MO reports xpanded View igures igure V1. USP26 enhances SM2 phosphorylation and T-b-mediated transcription. raph representing relative luciferase values obtained
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
Nature Immunology: doi: /ni.3866
 Nature Immunology: doi:10.1038/ni.3866 Supplementary Figure 1 The effect of TIPE2 on chemotaxis. a, The expression of TIPE2 in dhl-60c, dhl-60t, TIPE2-expressing and 15/16Q-expressing dhl-60t neutrophils
Nature Immunology: doi:10.1038/ni.3866 Supplementary Figure 1 The effect of TIPE2 on chemotaxis. a, The expression of TIPE2 in dhl-60c, dhl-60t, TIPE2-expressing and 15/16Q-expressing dhl-60t neutrophils
Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis
 Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Materials
 Supplementary Materials Figure S1. MTT Cell viability assay. To measure the cytotoxic potential of the oxidative treatment, the MTT [3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl tetrazolium bromide] assay
Supplementary Materials Figure S1. MTT Cell viability assay. To measure the cytotoxic potential of the oxidative treatment, the MTT [3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl tetrazolium bromide] assay
SUPPLEMENTARY INFORMATION
 a c e doi:10.1038/nature10407 b d f Supplementary Figure 1. SERCA2a complex analysis. (a) Two-dimensional SDS-PAGE gels of SERCA2a complexes. A silver-stained SDSPAGE gel is shown, which reveals a 12 kda
a c e doi:10.1038/nature10407 b d f Supplementary Figure 1. SERCA2a complex analysis. (a) Two-dimensional SDS-PAGE gels of SERCA2a complexes. A silver-stained SDSPAGE gel is shown, which reveals a 12 kda
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
Supplemental information
 Supplemental information PI(3)K p11δ controls the sucellular compartmentalization of TLR4 signaling and protects from endotoxic shock Ezra Aksoy, Salma Taoui, David Torres, Sandrine Delauve, Aderrahman
Supplemental information PI(3)K p11δ controls the sucellular compartmentalization of TLR4 signaling and protects from endotoxic shock Ezra Aksoy, Salma Taoui, David Torres, Sandrine Delauve, Aderrahman
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Appendix. Table of Contents
 Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Table S1. Primers and fluorescent probes used for qrt-pcr analysis of relative expression levels of PPP family phosphatases. gene name forward primer, 5-3 probe, 5-3 reverse primer,
SUPPLEMENTARY MATERIAL Table S1. Primers and fluorescent probes used for qrt-pcr analysis of relative expression levels of PPP family phosphatases. gene name forward primer, 5-3 probe, 5-3 reverse primer,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/308/ra4/dc1 Supplementary Materials for Antipsychotics Activate mtorc1-dependent Translation to Enhance Neuronal Morphological Complexity Heather Bowling, Guoan
www.sciencesignaling.org/cgi/content/full/7/308/ra4/dc1 Supplementary Materials for Antipsychotics Activate mtorc1-dependent Translation to Enhance Neuronal Morphological Complexity Heather Bowling, Guoan
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Evaluating Variability of Allergens in Commodity Crops (Peanuts)
 Evaluating Variability of Allergens in Commodity Crops (Peanuts) Peggy Ozias-Akins Laura Ramos Paola Faustinelli Ye Chu University of Georgia Manel Jordana Katherine Arias McMaster University Soheila Maleki
Evaluating Variability of Allergens in Commodity Crops (Peanuts) Peggy Ozias-Akins Laura Ramos Paola Faustinelli Ye Chu University of Georgia Manel Jordana Katherine Arias McMaster University Soheila Maleki
Supplemental Material:
 Supplemental Material: MATERIALS AND METHODS RNA interference Mouse CHOP sirna (ON-TARGETplus SMARTpool Cat# L-062068-00) and control sirna (ON-TARGETplus Control) were purchased from Dharmacon. Transfection
Supplemental Material: MATERIALS AND METHODS RNA interference Mouse CHOP sirna (ON-TARGETplus SMARTpool Cat# L-062068-00) and control sirna (ON-TARGETplus Control) were purchased from Dharmacon. Transfection
Supplementary material: Materials and suppliers
 Supplementary material: Materials and suppliers Electrophoresis consumables including tris-glycine, acrylamide, SDS buffer and Coomassie Brilliant Blue G-2 dye (CBB) were purchased from Ameresco (Solon,
Supplementary material: Materials and suppliers Electrophoresis consumables including tris-glycine, acrylamide, SDS buffer and Coomassie Brilliant Blue G-2 dye (CBB) were purchased from Ameresco (Solon,
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
EhLimA, a Novel LIM Protein, Localizes to the Plasma Membrane in Entamoeba histolytica
 EUKARYOTIC CELL, Sept. 2007, p. 1646 1655 Vol. 6, No. 9 1535-9778/07/$08.00 0 doi:10.1128/ec.00177-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. EhLimA, a Novel LIM Protein,
EUKARYOTIC CELL, Sept. 2007, p. 1646 1655 Vol. 6, No. 9 1535-9778/07/$08.00 0 doi:10.1128/ec.00177-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. EhLimA, a Novel LIM Protein,
% of live splenocytes. STAT5 deletion. (open shapes) % ROSA + % floxed
 Supp. Figure 1. a 14 1 1 8 6 spleen cells (x1 6 ) 16 % of live splenocytes 5 4 3 1 % of live splenocytes 8 6 4 b 1 1 c % of CD11c + splenocytes (closed shapes) 8 6 4 8 6 4 % ROSA + (open shapes) % floxed
Supp. Figure 1. a 14 1 1 8 6 spleen cells (x1 6 ) 16 % of live splenocytes 5 4 3 1 % of live splenocytes 8 6 4 b 1 1 c % of CD11c + splenocytes (closed shapes) 8 6 4 8 6 4 % ROSA + (open shapes) % floxed
SD-1 SD-1: Cathepsin B levels in TNF treated hch
 SD-1 SD-1: Cathepsin B levels in TNF treated hch. A. RNA and B. protein extracts from TNF treated and untreated human chondrocytes (hch) were analyzed via qpcr (left) and immunoblot analyses (right) for
SD-1 SD-1: Cathepsin B levels in TNF treated hch. A. RNA and B. protein extracts from TNF treated and untreated human chondrocytes (hch) were analyzed via qpcr (left) and immunoblot analyses (right) for
Supplemental Information. Inhibition of the Proteasome b2 Site Sensitizes. Triple-Negative Breast Cancer Cells
 Cell Chemical Biology, Volume 24 Supplemental Information Inhibition of the Proteasome b2 Site Sensitizes Triple-Negative Breast Cancer Cells to b5 Inhibitors and Suppresses Nrf1 Activation Emily S. Weyburne,
Cell Chemical Biology, Volume 24 Supplemental Information Inhibition of the Proteasome b2 Site Sensitizes Triple-Negative Breast Cancer Cells to b5 Inhibitors and Suppresses Nrf1 Activation Emily S. Weyburne,
