Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs.
|
|
|
- Hugh Glenn
- 5 years ago
- Views:
Transcription
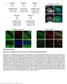
1 Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs. (a) CNA analysis of expression microarray data obtained from 15 tumors in the SV40Tag GEMM 52. (b) CNA analysis of acga data obtained from the same tumors. 26 of 27 aberrations detected by expression analysis were also identified by the acgh analysis (false detection rate of 0.037), demonstrating that expression-based karyotyping can accurately capture the landscape of aneuploidy and large CNAs in breast cancer GEMMs.
2 Supplementary Figure 2: Chromosomal aberrations are a late event in breast cancer tumorigenesis, and further aberrations are acquired during the derivation and propagation of cell lines. (a) Following chromosomal aberrations in another study with the SV40Tag mouse model (GSE50813) confirms that large CNAs occur, or are selected, only at the progression of non-malignant lesions to invasive carcinomas. Presented are moving average plots of gene expression profiles from various stages of tumor development. (b) Quantification of the prevalence of chromosomal aberrations in normal tissues, premalignant tissues/lesions and invasive carcinomas derived from all mouse models combined (Supplementary Data 4; only samples from studies with CNA-harboring tumors are included). *, p=1x10-5 and p=2.7x10-11 (chi-squared test), for the comparison of tumors to normal and to premalignant tissues, respectively. (c) Following chromosomal aberrations in tumors derived by injecting 4T1 cells into recipient mice (GSE54773) confirms that metastasis is not necessarily associated with increased burden of aneuploidy and large CNAs. Presented are moving average plots of gene expression profiles from primary tumors and metastases. Only one additional CNA was detected in one out of six brain and lung metastases. (d) Freshlyderived cell lines harbor many more CNAs than the primary tumors from which they are derived. Quantification of the prevalence of chromosomal aberrations in primary tumors and in the cell lines derived from them, in two genomically stable GEMMs, Her2/Neu and Wnt/βcat. *, p=2x10-4 and p<1x10-16 (Fisher s exact test) for Her2/Neu and Wnt/βcat, respectively.
3 Supplementary Figure 3: Breast cancer cell lines from the cancer cell line encyclopedia represent all molecular subtypes of the human disease and exhibit the common mutations identified in the tumors. (a) A pie chart describing the PAM50 molecular subtypes of the 57 breast cancer cell lines, as evaluated by expression signature analysis. (b) A bar chart showing the prevalence of common breast cancer mutations/alterations in the breast cancer CCLE cell lines.
4 Supplementary Figure 4: Schematics of the timeline of CNA acquisition/selection during GEMM breast cancer tumorigenesis. This schematic model describes the timeline of breast cancer tumorigenesis. The major wave of genomic instability (i.e., acquisition or selection of chromosomal aberrations) occurs during the progression of a pre-malignant tissue to an invasive carcinoma; additional waves of instability arise during cell line derivation and propagation.
5 Supplementary Figure 5: Representative results from CNA analysis of genomically stable and unstable GEMMs. Upper panel: moving average plots of global gene expression levels along the genome of 5 normal mammary samples (blue lines) and 5 tumor samples (orange lines) from two mouse models generated in the same study (GSE223938): the Her2/Neu model (left) and the p53 -/- model (right). Lower panel: piecewise constant fit (PCF) detection of amplifications (in red) and deletions (in blue) in the same samples. Note the significant DGI difference between the two GEMMs. The data from the p53 -/- model are the same as in Fig. 1a.
6
7 Supplementary Figure 6: Driver-specific degree of genomic instability in breast cancer GEMMs. (a) The status of p53 is a major determinant of genomic instability in breast cancer GEMMs. Presented is a comparison of genomic instability in models with data available for both p53 +/+ and p53 +/- background. *, p=2x10-4 (Fisher s exact test). (b) The degree of genomic instability, as estimated by autocorrelation between proximate genes, in three representative GEMMs. *,p=1.2x10-14, p=5.5x10-3 and p=2.2x10-16 (Mann-Whitney U test), for the PyMT/Etv6-Ntrk3, Etv6-Ntrk3/p53 and PyMT/p53 comparisons, respectively. (c) The degree of genomic instability, as estimated by the proportion of genes affected by CNAs, across the 11 most common GEMMs. (d) The prevalence of chromosomal aberrations in tumors from nine genetic models and one chemical model (DMBA) of breast cancer (GSE3165), showing that some genetic models are as stable as the chemical model. (e) The degree of genomic instability observed in the breast cancer GEMMs is not a result of the mouse strains used for their generation. Shown is the prevalence of chromosomal aberrations in 784 tumors from eight GEMMs generated on the FVB/N strain background. The observed DGI of each model is highly similar to that observed when all strains are analyzed together (compare to Fig. 3a). (f) Also shown is the prevalence of chromosomal aberrations in four GEMMs, each generated on more than one strain background. No significant differences in DGI are observed between different backgrounds within each model.
8 Supplementary Figure 7: Unique landscapes of aneuploidy and large CNAs in breast cancer GEMMs. Heat maps of the chromosomal landscapes of the 11 GEMMs analyzed. Gains are shown in red, losses in blue. Heat maps correspond to the frequency plots presented in Fig. 4a.
9 Supplementary Figure 8: Binomial distribution analysis of recurrent CNAs in breast cancer GEMMs. Binomial distribution test for recurrence of chromosomal aberrations in each of the 11 GEMMs analyzed. Red dots denote significantly gained regions, and blue dots denote significantly lost regions (Bonferronicorrected p < 0.05).
10 Supplementary Figure 9: Chromosomal aberrations are a late event in lymphoma and prostate cancer GEMMs. (a) Following chromosomal aberrations in the Eμ-Myc mouse model of lymphoma (GSE32239). Presented are moving average plots of gene expression profiles from wildtype B lymphocytes from control mice, premalignant B lymphocytes from transgenic mice, and malignant B lymphocytes from transgenic mice. (b) Following chromosomal aberrations in the SV40Tag mouse models of prostate cancer (GSE53202). Presented are moving average plots of gene expression profiles from wildtype prostate tissues, hyperplasias and/or prostatic intraepithelial neoplasias (PINs) from tumors in the Pb- TagAPT 121 (APT) model, and adenocarcinomas from the Pb-T/tag (TRAMP) model. (c) The prevalence of chromosomal aberrations in intrinsic to the model and not determined by tumor latency in the lymphoma Eμ-Myc GEMM.
11 Supplementary Figure 10: Comparative oncogenomics identifies BIRC5 as a general oncogene that promotes tumorigenesis across breast cancer subtypes. (a) A Venn diagram presenting the number of over-expressed genes in three GEMMs with recurrent amplification of 11qE1-E2: PyMT, Brca1 -/- and Met. In each model, expression levels were compared between tumors and normal mammary tissues 10. Only one gene, Birc5, was significantly over-expressed in all three models. (b) Alteration frequency of BIRC5 in three large-scale cancer genomic studies, revealing that BIRC5 is commonly amplified in human breast cancer. Data were obtained from cbioportal 53 ( (c) High expression of BIRC5 is associated with worse prognosis in human breast cancer patients. Presented are Kaplan-Meier plots of patients overall survival, based on a cohort of 1,117 patients of all molecular subtypes 46.
12 Supplementary Figure 11: Comparative oncogenomics identifies SFN as a putative co-driver gene that cooperates with HER2 during breast tumorigenesis. (a) Out of the 22 genes that reside within mouse chromosome 4 and the syntenic region on human chromosome 1p, and are downregulated in the Her2/Neu GEMM, only two genes were found to be connected to HER2 in a network analysis: SFN and EPS15. Of these, only SFN interacts directly with HER2 in a protein-protein interaction (PPI) analysis, shown here. (For reference, a PPI analysis of HER2 together with all human 1p genes identifies ~40 times as many interactions.) (b) SFN gene expression level is anti-correlated with the protein expression level of HER2 in human breast tumors. (c) High expression of SFN is associated with worse prognosis in basal subtype tumors, in line with previous findings 35. Presented are Kaplan-Meier plots of patients overall survival, based on a cohort of 204 basal subtype patients 46.
13 Supplementary Figure 12: Downregulation of SFN promotes in vitro tumorigenesis of human breast cancer cell lines of the HER2-enriched subtype. (a) Immunoblot analysis of SFN protein levels in human breast cancer cell lines, following shrnamediated knockdown or CRISPR/Cas9-mediated knockout of SFN. (b) A quantification of the reduction in protein levels. (c) Decreased migration of the basal cell line and increased migration of the HER2- enriched cell lines, following the knockdown/knockout of SFN, as evaluated by a transwell migration assay. *, p<0.05 (Student s t-test). (d). Decreased invasion of the basal cell line and increased invasion of the HER2-enriched cell lines, following the knockdown/knockout of SFN, as evaluated by a transwell invasion assay. *, p<0.05 (Student s t-test). (e) Decreased colony formation of the basal cell line and increased colony formation of the HER2-enriched cell lines, following the knockdown/knockout of SFN, as evaluated by a soft-agar assay. *, p<0.05 (Student s t-test).
14 Supplementary Figure 13: Comparative oncogenomics pipeline to identify candidate co-driver genes underlying the recurrence of driver-specific CNAs. Schematic outlining of the strategy that we applied to the driver-specific CNAs. The candidate genes identified by this strategy are listed in Supplementary Data 9. GEMMs, genetically-engineered mouse models; CNAs, copy number alterations; DEGs, differentially expressed genes; GE, gene expression.
15 Supplementary Figure 14: Uncropped western blots Uncropped scans of western blots displayed in the main Figures
16 Supplementary Table 1 Kruskal-Wallis rank sun test by altered genes: Chi-squared= , df=10, p< BRCA1-/- Etv6-Ntrk3 Her2/Neu Met Myc Pik3ca Pten-/- PyMT SV40 Tag Wnt/β-cat Etv6- Ntrk3 1 Her2/Neu < < Met 1 1 < Myc < Pik3ca < Pten-/ PyMT < < < < SV40 Tag < < < < Wnt/β-cat < < < p53-/ < < < < < < < Kruskal-Wallis rank sun test by CNA prevalence (# of events per sample): Chi-squared= , df=10, p< BRCA1-/- Etv6-Ntrk3 Her2/Neu Met Myc Pik3ca Pten-/- PyMT SV40 Tag Wnt/β-cat Etv6- Ntrk Her2/Neu < < Met 1 1 < Myc < Pik3ca < Pten-/ PyMT < < < < < SV40 Tag < < < < < Wnt/β-cat < < < < p53-/ < < < < < < Supplementary Table 1: Statistically significant DGI differences between breast cancer GEMMs The variation analysis between the 11 most common breast cancer GEMMs was performed by a one-way ANOVA on ranks (Kruskal-Wallis) test, followed by a post-hoc Dunn s test to compare each pair of GEMMs. Shown are the Bonferroni corrected p-values for the pair-wise analyses, based either on CNA prevalence (i.e., number of aberrations per sample) or on the proportion of altered genes per sample. Pairwise comparisons revealed statistically-significant differences between low, medium and high DGI models.
17 Mouse model Recurrent aberrations (>10% of sampels, regardless of statistical significance) Recurrent aberrations (Statistically significant by a binomial distribution test) Frequency of aberrations (regional minimum/maximum) Binomial test p-value for significance within model (maximum of region) GISTIC2.0 q-value for significance within model (maximum of region) Chi-squared test p-value for significance between models (maximum of region) Supplementary Table 2 PymT - Amp 11qE1-E / E E-44 N.S. Wnt/β-cat Pik3ca Her2/Neu - Del / E E E-03 Myc Amp 15 Amp E E E-04 Pten-/- Amp 4qB1-C6 Amp 4qB1-C / E E E-41 Amp 14qA1- A3 Amp 14qA / E E E-11 Del 17qB1-B3 Del 17qB1-B / E E E-36 Amp 3qG3-H N.S. 2.60E-03 N.S. Amp 17qC-E N.S. 2.60E-03 N.S. Etv6-Ntrk3 Amp N.S. 1.50E E-05 Met Amp /0.132 N.S. 6.62E-03 N.S. Amp 11qA N.S. N.S. 6.80E-04 Amp 11qE1-E N.S. 1.30E-02 N.S. Amp N.S. 2.28E E-17 Amp N.S. 6.62E-03 N.S. BRCA1-/- Amp 3qF2.1 Amp 3qF N.S. 2.57E-02 N.S. Amp 11qE2 Amp 11qE N.S. 2.58E-04 N.S. Del 17qB1-B3 Del 17qB1-B E E E-35 Del 6qA1-A N.S. 2.34E E-16 Del 10qD2-D N.S. 5.77E E-07 Del 12qA1.1- D N.S. 2.56E-03 N.S. SV40 Tag Amp 3 Amp E E E-09 Amp 18 Amp E E E-38 Amp X Amp X E E E-45 Amp N.S. 1.23E-02 N.S. Amp N.S. 1.72E E-10 Del 9qA5.1- A N.S. 3.11E E-16 p53-/- Amp 6qA1-C1 Amp 6qA1-B / E E E-12 Amp 6qG1-G3 Amp 6qG1-G E E E-06 Amp 8qA1.1- Amp 8qA1.1- A4 A / E E E-19 Del 8qB1.2-E2 Del 8qB1.1-E / E E E-28 Del 12 Del / E E E-43 Del 14 Del / E E E-18 Amp 3qF N.S. 2.11E E-05 Amp 15qA1-C N.S. 3.60E-02 N.S.
18 Supplementary Table 2: Recurrent CNAs in breast cancer mouse models A summary of the recurrent aberrations in the 11 most common breast cancer GEMMs. Presented are all aberrations present in>10% of tumor samples, and those identified as significant by a binomial distribution test. 34 of the 35 events were confirmed to be significant by a GISTIC2.0 analysis. Recurrent aberrations were subjected to a chi-squared test, to examine their model-specificity. Adjusted p-values are mentioned for the binomial and chi-squared tests, and q-values are mentioned for the GISTIC2.0 analysis. Significant model-specific recurrent events are highlighted in blue.
19 Mouse Model Recurrent transgene-specific aberrations (statistically significant in both tests) Human chromosomes containing synteny blocks Human synteny blocks Syntenic regions significantly altered in the same direction in human breast cancer with the activation of the same pathway Supplementary Table 3 Her2/Neu Del 4 1,6,8,9 Chr 1:0.9M-61.7M, Chr 6:87.1M-99.8M, Chr 8:55.7M-61.8M, Chr 8:86.0M-96.2M, Chr 1p: 0.9M-61.7M Chr 9:6.8M-38.5M, Chr 9:80.4M-83.6M, Chr 9:97.3M-120.7M Myc Amp 15 5,8,12,22 Chr 5:8.9M-42.9M, Chr 8:96.4M-144.6M, Chr 12:33.1M-34.1M, Chr 8q: 96.4M-144.6M Chr 12:38.2M-54.7M, Chr 22:35.6M-50.8M p53-/- Amp 6qA1-B1 7 Chr 7:93.1M-97.9M, Chr 7:7.1M-12.5M, Chr 7:112.5M-128.5M, - Chr 7:128.7M-149.9M Amp 6qG1-G3 12 Chr 12:9.7M-32.4M Chr 12p: 9.7M-32.4M Amp 8qA1.1-A4 8,13, 19 Chr 8:0.4M-18.1M, Chr 13:102.9M M, - Del 8qB1.1-E2 1,4,8,10,16,19,21,22 Del 12 2,7,9,14 Del 14 3,6,8,10,13,14 Chr 19:7.1M-8.1M Chr 1:229.2M-235.2M, Chr 4:140.3M-150.0M, Chr 4:162.6M-190.0M, Chr 8:18.1M-20.3M, Chr 10:32.8M-34.9M, Chr 19:12.6M-14.6M, Chr 19:16.1M-19.7M, Chr 22:33.3M-35.5M, Chr 16: M Chr 12:0.2M-17.8M, Chr 2:94.6M-94.7M, Chr 7:12.5M-22.5M, Chr 7:105.6M-112.5M, Chr 7:157.4M-159.1M, Chr 9:40.4M-40.5M, Chr 9:43.0M-43.1M, Chr 9:64.5M-66.0M, Chr 14:24.7M-51.8M, Chr 14:58.2M-105.9M Chr 3:15.2M-16.3M, Chr 3:23.1M-27.7M, Chr 3:52.3M-64.0M, Chr 6:39.1M-39.3M, Chr 8:9.8M-11.9M, Chr 8:20.3M-29.3M, Chr 10:45.9M-50.0M, Chr 10:73.1M-79.5M, Chr 10:80.3M-87.2M, Chr 13:19.6M-24.9M, Chr 4q:140.3M-150.0M, Chr 4q:162.6M-190.0M, Chr 8:18.1M-20.3M Chr 14q:24.7M-51.8M, Chr 14q:58.2M-105.9M Chr 8p:9.8M-11.9M, Chr 8p:20.3M-29.3M, Chr 14q:19.7M-24.7M, Chr 14q:52.2M-58.2M,
20 Chr 13:40.9M-102.4M, Chr 14:19.7M-24.7M, Chr 14:52.2M-58.2M, Pten-/- Amp 4qB1-C6 1,9 Chr 1:58.7M-67.1M, Chr 9:6.8M-38.5M, Chr 9:80.4M-83.6M, - Chr 9:97.3M-120.7M Amp 14qA1 3 Chr 3:58.0M-64.0M - Del 17qB1-B3 6,19,21 Chr 6:29.4M-33.3M, Chr 19:8.3M-8.7M, Chr 19:15.2M-15.7M, - Chr 21:42.1M-43.7M Brca1-/- Del 17qB1-B3 6,19,21 Chr 6:29.4M-33.3M, Chr 19:8.3M-8.7M, Chr 19:15.2M-15.7M, - Chr 21:42.1M-43.7M SV40 Tag Amp 3 NA NA - Amp 18 NA NA - Amp X NA NA - Supplementary Table 3: Syntenic aberrations in breast cancer GEMMs and in human breast tumors that activate the same pathway A summary of the 15 model-specific CNAs identified in breast cancer GEMMs, together with their syntenic human chromosomal regions. Highlighted are syntenic regions that are significantly altered in the same direction in human tumors that activate the same pathway, as judged by gene expression signatures 33.
ARTICLE RESEARCH. Macmillan Publishers Limited. All rights reserved
 Extended Data Figure 6 Annotation of drivers based on clinical characteristics and co-occurrence patterns. a, Putative drivers affecting greater than 10 patients were assessed for enrichment in IGHV mutated
Extended Data Figure 6 Annotation of drivers based on clinical characteristics and co-occurrence patterns. a, Putative drivers affecting greater than 10 patients were assessed for enrichment in IGHV mutated
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Computer Science, Biology, and Biomedical Informatics (CoSBBI) Outline. Molecular Biology of Cancer AND. Goals/Expectations. David Boone 7/1/2015
 Goals/Expectations Computer Science, Biology, and Biomedical (CoSBBI) We want to excite you about the world of computer science, biology, and biomedical informatics. Experience what it is like to be a
Goals/Expectations Computer Science, Biology, and Biomedical (CoSBBI) We want to excite you about the world of computer science, biology, and biomedical informatics. Experience what it is like to be a
Nature Genetics: doi: /ng Supplementary Figure 1. Somatic coding mutations identified by WES/WGS for 83 ATL cases.
 Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Genetic alterations of histone lysine methyltransferases and their significance in breast cancer
 Genetic alterations of histone lysine methyltransferases and their significance in breast cancer Supplementary Materials and Methods Phylogenetic tree of the HMT superfamily The phylogeny outlined in the
Genetic alterations of histone lysine methyltransferases and their significance in breast cancer Supplementary Materials and Methods Phylogenetic tree of the HMT superfamily The phylogeny outlined in the
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
Supplementary Tables. Supplementary Figures
 Supplementary Files for Zehir, Benayed et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients Supplementary Tables Supplementary Table 1: Sample
Supplementary Files for Zehir, Benayed et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients Supplementary Tables Supplementary Table 1: Sample
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Quantification of myelin fragments in the aging brain (a) Electron microscopy on corpus callosum is shown for a 18-month-old wild type mice. Myelin fragments (arrows) were detected
Supplementary Figure 1 Quantification of myelin fragments in the aging brain (a) Electron microscopy on corpus callosum is shown for a 18-month-old wild type mice. Myelin fragments (arrows) were detected
Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors
 Wang et al LEGENDS TO SUPPLEMENTARY INFORMATION Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors A. Induced expression of estrogen receptor α (ERα) in AME vs PDA
Wang et al LEGENDS TO SUPPLEMENTARY INFORMATION Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors A. Induced expression of estrogen receptor α (ERα) in AME vs PDA
Nature Genetics: doi: /ng Supplementary Figure 1. Phenotypic characterization of MES- and ADRN-type cells.
 Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma.
 Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Table S1. List of PTPRK-RSPO3 gene fusions in TCGA's colon cancer cohort. Chr. # of Gene 2. Chr. # of Gene 1
 Supplementary Tale S1. List of PTPRK-RSPO3 gene fusions in TCGA's colon cancer cohort TCGA Case ID Gene-1 Gene-2 Chr. # of Gene 1 Chr. # of Gene 2 Genomic coordiante of Gene 1 at fusion junction Genomic
Supplementary Tale S1. List of PTPRK-RSPO3 gene fusions in TCGA's colon cancer cohort TCGA Case ID Gene-1 Gene-2 Chr. # of Gene 1 Chr. # of Gene 2 Genomic coordiante of Gene 1 at fusion junction Genomic
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplemental Figure legends
 Supplemental Figure legends Supplemental Figure S1 Frequently mutated genes. Frequently mutated genes (mutated in at least four patients) with information about mutation frequency, RNA-expression and copy-number.
Supplemental Figure legends Supplemental Figure S1 Frequently mutated genes. Frequently mutated genes (mutated in at least four patients) with information about mutation frequency, RNA-expression and copy-number.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
ANGPTL2 increases bone metastasis of breast cancer cells through. Tetsuro Masuda, Motoyoshi Endo, Yutaka Yamamoto, Haruki Odagiri, Tsuyoshi
 Masuda et al. Supplementary information for ANGPTL2 increases bone metastasis of breast cancer cells through enhancing CXCR4 signaling Tetsuro Masuda, Motoyoshi Endo, Yutaka Yamamoto, Haruki Odagiri, Tsuyoshi
Masuda et al. Supplementary information for ANGPTL2 increases bone metastasis of breast cancer cells through enhancing CXCR4 signaling Tetsuro Masuda, Motoyoshi Endo, Yutaka Yamamoto, Haruki Odagiri, Tsuyoshi
Supplementary Figure S1. Generation of LSL-EZH2 conditional transgenic mice.
 Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
Supplementary Fig. 1: ATM is phosphorylated in HER2 breast cancer cell lines. (A) ATM is phosphorylated in SKBR3 cells depending on ATM and HER2
 Supplementary Fig. 1: ATM is phosphorylated in HER2 breast cancer cell lines. (A) ATM is phosphorylated in SKBR3 cells depending on ATM and HER2 activity. Upper panel: Representative histograms for FACS
Supplementary Fig. 1: ATM is phosphorylated in HER2 breast cancer cell lines. (A) ATM is phosphorylated in SKBR3 cells depending on ATM and HER2 activity. Upper panel: Representative histograms for FACS
Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the. mutated sequence.
 Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the mutated sequence. 1 Supplementary Figure 2. Expression of mir-182 and SMAD7 in various cell lines. (A) Basal levels of mir-182 expression
Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the mutated sequence. 1 Supplementary Figure 2. Expression of mir-182 and SMAD7 in various cell lines. (A) Basal levels of mir-182 expression
Only Estrogen receptor positive is not enough to predict the prognosis of breast cancer
 Young Investigator Award, Global Breast Cancer Conference 2018 Only Estrogen receptor positive is not enough to predict the prognosis of breast cancer ㅑ Running head: Revisiting estrogen positive tumors
Young Investigator Award, Global Breast Cancer Conference 2018 Only Estrogen receptor positive is not enough to predict the prognosis of breast cancer ㅑ Running head: Revisiting estrogen positive tumors
Expanded View Figures
 Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung
 Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier
 Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes.
 Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Atlas representations of the midcingulate (MCC) region targeted in this study compared against the anterior cingulate (ACC) region commonly reported. Coronal sections are shown on
Supplementary Figure 1 Atlas representations of the midcingulate (MCC) region targeted in this study compared against the anterior cingulate (ACC) region commonly reported. Coronal sections are shown on
Abstract. Optimization strategy of Copy Number Variant calling using Multiplicom solutions APPLICATION NOTE. Introduction
 Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
AP VP DLP H&E. p-akt DLP
 A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis
 The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/7/283/283ra54/dc1 Supplementary Materials for Clonal status of actionable driver events and the timing of mutational processes in cancer evolution
www.sciencetranslationalmedicine.org/cgi/content/full/7/283/283ra54/dc1 Supplementary Materials for Clonal status of actionable driver events and the timing of mutational processes in cancer evolution
underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
 Supplementary Figures Figure S1. Patient cohorts and study design. To define and interrogate the genetic alterations underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
Supplementary Figures Figure S1. Patient cohorts and study design. To define and interrogate the genetic alterations underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
Chapter 4 Cellular Oncogenes ~ 4.6 -
 Chapter 4 Cellular Oncogenes - 4.2 ~ 4.6 - Many retroviruses carrying oncogenes have been found in chickens and mice However, attempts undertaken during the 1970s to isolate viruses from most types of
Chapter 4 Cellular Oncogenes - 4.2 ~ 4.6 - Many retroviruses carrying oncogenes have been found in chickens and mice However, attempts undertaken during the 1970s to isolate viruses from most types of
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary. properties of. network types. randomly sampled. subsets (75%
 Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
Supplemental Information. Integrated Genomic Analysis of the Ubiquitin. Pathway across Cancer Types
 Cell Reports, Volume 23 Supplemental Information Integrated Genomic Analysis of the Ubiquitin Pathway across Zhongqi Ge, Jake S. Leighton, Yumeng Wang, Xinxin Peng, Zhongyuan Chen, Hu Chen, Yutong Sun,
Cell Reports, Volume 23 Supplemental Information Integrated Genomic Analysis of the Ubiquitin Pathway across Zhongqi Ge, Jake S. Leighton, Yumeng Wang, Xinxin Peng, Zhongyuan Chen, Hu Chen, Yutong Sun,
Award Number: W81XWH TITLE: Characterizing an EMT Signature in Breast Cancer. PRINCIPAL INVESTIGATOR: Melanie C.
 AD Award Number: W81XWH-08-1-0306 TITLE: Characterizing an EMT Signature in Breast Cancer PRINCIPAL INVESTIGATOR: Melanie C. Bocanegra CONTRACTING ORGANIZATION: Leland Stanford Junior University Stanford,
AD Award Number: W81XWH-08-1-0306 TITLE: Characterizing an EMT Signature in Breast Cancer PRINCIPAL INVESTIGATOR: Melanie C. Bocanegra CONTRACTING ORGANIZATION: Leland Stanford Junior University Stanford,
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis
 Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Materials
 Supplementary Materials Supplementary figure 1. Taxonomic representation summarized at genus level. Fecal microbiota from a separate set of Jackson and Harlan mice prior to irradiation. A taxon was included
Supplementary Materials Supplementary figure 1. Taxonomic representation summarized at genus level. Fecal microbiota from a separate set of Jackson and Harlan mice prior to irradiation. A taxon was included
Supplemental File. TRAF6 is an amplified oncogene bridging the Ras and nuclear factor-κb cascade in human lung cancer
 Supplemental File TRAF6 is an amplified oncogene bridging the Ras and nuclear factor-κb cascade in human lung cancer Daniel T. Starczynowski, William W. Lockwood, Sophie Delehouzee, Raj Chari, Joanna Wegrzyn,
Supplemental File TRAF6 is an amplified oncogene bridging the Ras and nuclear factor-κb cascade in human lung cancer Daniel T. Starczynowski, William W. Lockwood, Sophie Delehouzee, Raj Chari, Joanna Wegrzyn,
Discovery Dataset. PD Liver Luminal B/ Her-2+ Letrozole. PD Supraclavicular Lymph node. PD Supraclavicular Lymph node Luminal B.
 Discovery Dataset 11T pt1cpn2am1(liver) 2009 2010 Liver / Her-2+ 2011 Death Letrozole CHT pt1cpn0(sn)m0 Supraclavicular Lymph node Death 12T 2003 2006 2006 2008 Anastrozole Local RT+Examestane Fulvestrant
Discovery Dataset 11T pt1cpn2am1(liver) 2009 2010 Liver / Her-2+ 2011 Death Letrozole CHT pt1cpn0(sn)m0 Supraclavicular Lymph node Death 12T 2003 2006 2006 2008 Anastrozole Local RT+Examestane Fulvestrant
Cytogenetics 101: Clinical Research and Molecular Genetic Technologies
 Cytogenetics 101: Clinical Research and Molecular Genetic Technologies Topics for Today s Presentation 1 Classical vs Molecular Cytogenetics 2 What acgh? 3 What is FISH? 4 What is NGS? 5 How can these
Cytogenetics 101: Clinical Research and Molecular Genetic Technologies Topics for Today s Presentation 1 Classical vs Molecular Cytogenetics 2 What acgh? 3 What is FISH? 4 What is NGS? 5 How can these
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1. Galectin-3 is present within tumors. (A) mrna expression levels of Lgals3 (galectin-3) and Lgals8 (galectin-8) in the four classes of cell lines as determined
Supplementary Figures Supplementary Fig. 1. Galectin-3 is present within tumors. (A) mrna expression levels of Lgals3 (galectin-3) and Lgals8 (galectin-8) in the four classes of cell lines as determined
CONTRACTING ORGANIZATION: Rush University Medical Center Chicago, IL 60612
 AD Award Number: W81XWH-11-1-0302 TITLE: Yin and Yang of Heparanase in breast Tumor Initiation PRINCIPAL INVESTIGATOR: Xiulong Xu, Ph.D. CONTRACTING ORGANIZATION: Rush University Medical Center Chicago,
AD Award Number: W81XWH-11-1-0302 TITLE: Yin and Yang of Heparanase in breast Tumor Initiation PRINCIPAL INVESTIGATOR: Xiulong Xu, Ph.D. CONTRACTING ORGANIZATION: Rush University Medical Center Chicago,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
Microarray Analysis and Liver Diseases
 Microarray Analysis and Liver Diseases Snorri S. Thorgeirsson M.D., Ph.D. Laboratory of Experimental Carcinogenesis Center for Cancer Research, NCI, NIH Application of Microarrays to Cancer Research Identifying
Microarray Analysis and Liver Diseases Snorri S. Thorgeirsson M.D., Ph.D. Laboratory of Experimental Carcinogenesis Center for Cancer Research, NCI, NIH Application of Microarrays to Cancer Research Identifying
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed.
 Supplementary Note The potential association and implications of HBV integration at known and putative cancer genes of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed. Human telomerase
Supplementary Note The potential association and implications of HBV integration at known and putative cancer genes of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed. Human telomerase
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
La Jolla, CA Approved for Public Release; Distribution Unlimited
 AD Award Number: TITLE: Suppression of Breast Cancer Progression by Tissue Factor PRINCIPAL INVESTIGATOR: Wolfram Ruf, M.D. CONTRACTING ORGANIZATION: The Scripps Research Institute La Jolla, CA 92037 REPORT
AD Award Number: TITLE: Suppression of Breast Cancer Progression by Tissue Factor PRINCIPAL INVESTIGATOR: Wolfram Ruf, M.D. CONTRACTING ORGANIZATION: The Scripps Research Institute La Jolla, CA 92037 REPORT
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Session 4 Rebecca Poulos
 The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 28
The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 28
Characterisation of structural variation in breast. cancer genomes using paired-end sequencing on. the Illumina Genome Analyser
 Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
SHREE ET AL, SUPPLEMENTAL MATERIALS. (A) Workflow for tumor cell line derivation and orthotopic implantation.
 SHREE ET AL, SUPPLEMENTAL MATERIALS SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure 1. Derivation and characterization of TS1-TGL and TS2-TGL PyMT cell lines and development of an orthotopic
SHREE ET AL, SUPPLEMENTAL MATERIALS SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure 1. Derivation and characterization of TS1-TGL and TS2-TGL PyMT cell lines and development of an orthotopic
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
James C. Fleet, PhD Professor Dept of Nutrition Science Purdue University
 James C. Fleet, PhD Professor Dept of Nutrition Science Purdue University Overview What are we trying to model? Vitamin D biology Cancer Is there in vivo proof of principle for vitamin D/cancer relationship?
James C. Fleet, PhD Professor Dept of Nutrition Science Purdue University Overview What are we trying to model? Vitamin D biology Cancer Is there in vivo proof of principle for vitamin D/cancer relationship?
Supplementary Figure 1. Experimental paradigm. A combination of genome and exome sequencing coupled with array-comparative genome hybridization was
 Supplementary Figure 1. Experimental paradigm. A combination of genome and exome sequencing coupled with array-comparative genome hybridization was performed on a total of 85 SS patients. Data filtration
Supplementary Figure 1. Experimental paradigm. A combination of genome and exome sequencing coupled with array-comparative genome hybridization was performed on a total of 85 SS patients. Data filtration
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Bredel M, Scholtens DM, Yadav AK, et al. NFKBIA deletion in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Bredel M, Scholtens DM, Yadav AK, et al. NFKBIA deletion in
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines.
 Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
Supplemental Figure S1. RANK expression on human lung cancer cells.
 Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
Supplementary figures
 Supplementary figures Supplementary Figure 1. B cells stimulated with pokeweed mitogen display normal mitotic figures but not cells infected with B95-8. The figures show cells stimulated with pokeweed
Supplementary figures Supplementary Figure 1. B cells stimulated with pokeweed mitogen display normal mitotic figures but not cells infected with B95-8. The figures show cells stimulated with pokeweed
HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA LEO TUNKLE *
 CERNA SEARCH METHOD IDENTIFIED A MET-ACTIVATED SUBGROUP AMONG EGFR DNA AMPLIFIED LUNG ADENOCARCINOMA PATIENTS HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA Email:
CERNA SEARCH METHOD IDENTIFIED A MET-ACTIVATED SUBGROUP AMONG EGFR DNA AMPLIFIED LUNG ADENOCARCINOMA PATIENTS HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA Email:
Genomic tests to personalize therapy of metastatic breast cancers. Fabrice ANDRE Gustave Roussy Villejuif, France
 Genomic tests to personalize therapy of metastatic breast cancers Fabrice ANDRE Gustave Roussy Villejuif, France Future application of genomics: Understand the biology at the individual scale Patients
Genomic tests to personalize therapy of metastatic breast cancers Fabrice ANDRE Gustave Roussy Villejuif, France Future application of genomics: Understand the biology at the individual scale Patients
Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) 108 (b-c) (d) (e) (f) (g)
 Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) In four mice, cre-dependent expression of the hyperpolarizing opsin Arch in pyramidal cells
Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) In four mice, cre-dependent expression of the hyperpolarizing opsin Arch in pyramidal cells
A genomic analysis of mouse models of breast cancer reveals molecular features of mouse models and relationships to human breast cancer
 Hollern and Andrechek Breast Cancer Research 2014, 16:R59 RESEARCH ARTICLE Open Access A genomic analysis of mouse models of breast cancer reveals molecular features of mouse models and relationships to
Hollern and Andrechek Breast Cancer Research 2014, 16:R59 RESEARCH ARTICLE Open Access A genomic analysis of mouse models of breast cancer reveals molecular features of mouse models and relationships to
Nature Getetics: doi: /ng.3471
 Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
PAX8-PPARγ Fusion Protein in thyroid carcinoma
 Sidney H. Ingbar Lecture, ATA 10/17/2013: PAX8-PPARγ Fusion Protein in thyroid carcinoma Ronald J. Koenig, MD, PhD Division of Metabolism, Endocrinology & Diabetes University of Michigan Learning Objectives
Sidney H. Ingbar Lecture, ATA 10/17/2013: PAX8-PPARγ Fusion Protein in thyroid carcinoma Ronald J. Koenig, MD, PhD Division of Metabolism, Endocrinology & Diabetes University of Michigan Learning Objectives
Supplemental Table 1 Molecular Profile of the SCLC Cell Line Panel
 Supplemental Table 1 Molecular Profile of the SCLC Cell Line Panel p53 RB Myc Cell Line Mutation A Mutation A Amplification B COR-L103 C p.y234c p.d584e L-Myc NCI-H526 p.s33_splice None N-Myc NCI-H1048
Supplemental Table 1 Molecular Profile of the SCLC Cell Line Panel p53 RB Myc Cell Line Mutation A Mutation A Amplification B COR-L103 C p.y234c p.d584e L-Myc NCI-H526 p.s33_splice None N-Myc NCI-H1048
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma. Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute
 Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making
 Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making November 20, 2014 Capturing Value in Next Generation Sequencing Symposium Douglas Johnson MD, MSCI Vanderbilt-Ingram
Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making November 20, 2014 Capturing Value in Next Generation Sequencing Symposium Douglas Johnson MD, MSCI Vanderbilt-Ingram
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
doi:10.1038/nature15260 Supplementary Data 1: Gene expression in individual basal/stem, luminal, and luminal progenitor cells. Box plots show expression levels for each gene from the 49-gene differentiation
Supplementary Figure 1. Copy Number Alterations TP53 Mutation Type. C-class TP53 WT. TP53 mut. Nature Genetics: doi: /ng.
 Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
(A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC,
 Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
Identification of Causal Genetic Drivers of Human Disease through Systems-Level Analysis of Regulatory Networks
 Biologists are from Venus, Mathematicians are from Mars, They cosegregate on Earth, And conditionally associate to create a DIGGIT. Identification of Causal Genetic Drivers of Human Disease through Systems-Level
Biologists are from Venus, Mathematicians are from Mars, They cosegregate on Earth, And conditionally associate to create a DIGGIT. Identification of Causal Genetic Drivers of Human Disease through Systems-Level
Supplementary Information
 Supplementary Information Supplementary Figure 1. Effect of mir mimics and anti-mirs on DTPs a, Representative fluorescence microscopy images of GFP vector control or mir mimicexpressing parental and DTP
Supplementary Information Supplementary Figure 1. Effect of mir mimics and anti-mirs on DTPs a, Representative fluorescence microscopy images of GFP vector control or mir mimicexpressing parental and DTP
Nature Genetics: doi: /ng Supplementary Figure 1. Details of sequencing analysis.
 Supplementary Figure 1 Details of sequencing analysis. (a) Flow chart showing which patients fall into each category and were used for analysis. (b) Graph showing the average and median coverage for all
Supplementary Figure 1 Details of sequencing analysis. (a) Flow chart showing which patients fall into each category and were used for analysis. (b) Graph showing the average and median coverage for all
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Nature Medicine: doi: /nm.3967
 Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Nature Immunology: doi: /ni Supplementary Figure 1. RNA-Seq analysis of CD8 + TILs and N-TILs.
 Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Nature Neuroscience: doi: /nn Supplementary Figure 1. Trial structure for go/no-go behavior
 Supplementary Figure 1 Trial structure for go/no-go behavior a, Overall timeline of experiments. Day 1: A1 mapping, injection of AAV1-SYN-GCAMP6s, cranial window and headpost implantation. Water restriction
Supplementary Figure 1 Trial structure for go/no-go behavior a, Overall timeline of experiments. Day 1: A1 mapping, injection of AAV1-SYN-GCAMP6s, cranial window and headpost implantation. Water restriction
5 th July 2016 ACGS Dr Michelle Wood Laboratory Genetics, Cardiff
 5 th July 2016 ACGS Dr Michelle Wood Laboratory Genetics, Cardiff National molecular screening of patients with lung cancer for a national trial of multiple novel agents. 2000 NSCLC patients/year (late
5 th July 2016 ACGS Dr Michelle Wood Laboratory Genetics, Cardiff National molecular screening of patients with lung cancer for a national trial of multiple novel agents. 2000 NSCLC patients/year (late
User s Manual Version 1.0
 User s Manual Version 1.0 #639 Longmian Avenue, Jiangning District, Nanjing,211198,P.R.China. http://tcoa.cpu.edu.cn/ Contact us at xiaosheng.wang@cpu.edu.cn for technical issue and questions Catalogue
User s Manual Version 1.0 #639 Longmian Avenue, Jiangning District, Nanjing,211198,P.R.China. http://tcoa.cpu.edu.cn/ Contact us at xiaosheng.wang@cpu.edu.cn for technical issue and questions Catalogue
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Computational Investigation of Homologous Recombination DNA Repair Deficiency in Sporadic Breast Cancer
 University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 11-16-2017 Computational Investigation of Homologous Recombination DNA Repair
University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 11-16-2017 Computational Investigation of Homologous Recombination DNA Repair
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD
 Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Journal: Nature Methods
 Journal: Nature Methods Article Title: Network-based stratification of tumor mutations Corresponding Author: Trey Ideker Supplementary Item Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure
Journal: Nature Methods Article Title: Network-based stratification of tumor mutations Corresponding Author: Trey Ideker Supplementary Item Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure
Material and Methods. Flow Cytometry Analyses:
 Material and Methods Flow Cytometry Analyses: Immunostaining of breast cancer cells for HER2 was performed by incubating cells with anti- HER2/neu APC (Biosciences, Cat# 340554), anti-her2/neu PE (Biosciences,
Material and Methods Flow Cytometry Analyses: Immunostaining of breast cancer cells for HER2 was performed by incubating cells with anti- HER2/neu APC (Biosciences, Cat# 340554), anti-her2/neu PE (Biosciences,
Breast Cancer Statistics
 1 in 8 Breast Cancer Statistics Incidence Mortality Prevalence 2 Breast Cancer Incidence Breast Cancer Mortality Breast Cancer Prevalence ~$100,000 Female Breast Anatomy Breasts consist mainly of fatty
1 in 8 Breast Cancer Statistics Incidence Mortality Prevalence 2 Breast Cancer Incidence Breast Cancer Mortality Breast Cancer Prevalence ~$100,000 Female Breast Anatomy Breasts consist mainly of fatty
Session 4 Rebecca Poulos
 The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 20
The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 20
