Identification of Genetic Susceptibility Factors for Fibromyalgia
|
|
|
- Flora Page
- 5 years ago
- Views:
Transcription
1 Identification of Genetic Susceptibility Factors for Fibromyalgia Identificació de factors de susceptibilitat genètica per a fibromiàlgia Elisa Docampo Martínez ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents condicions d'ús: La difusió d aquesta tesi per mitjà del servei TDX ( i a través del Dipòsit Digital de la UB (diposit.ub.edu) ha estat autoritzada pels titulars dels drets de propietat intel lectual únicament per a usos privats emmarcats en activitats d investigació i docència. No s autoritza la seva reproducció amb finalitats de lucre ni la seva difusió i posada a disposició des d un lloc aliè al servei TDX ni al Dipòsit Digital de la UB. No s autoritza la presentació del seu contingut en una finestra o marc aliè a TDX o al Dipòsit Digital de la UB (framing). Aquesta reserva de drets afecta tant al resum de presentació de la tesi com als seus continguts. En la utilització o cita de parts de la tesi és obligat indicar el nom de la persona autora. ADVERTENCIA. La consulta de esta tesis queda condicionada a la aceptación de las siguientes condiciones de uso: La difusión de esta tesis por medio del servicio TDR ( y a través del Repositorio Digital de la UB (diposit.ub.edu) ha sido autorizada por los titulares de los derechos de propiedad intelectual únicamente para usos privados enmarcados en actividades de investigación y docencia. No se autoriza su reproducción con finalidades de lucro ni su difusión y puesta a disposición desde un sitio ajeno al servicio TDR o al Repositorio Digital de la UB. No se autoriza la presentación de su contenido en una ventana o marco ajeno a TDR o al Repositorio Digital de la UB (framing). Esta reserva de derechos afecta tanto al resumen de presentación de la tesis como a sus contenidos. En la utilización o cita de partes de la tesis es obligado indicar el nombre de la persona autora. WARNING. On having consulted this thesis you re accepting the following use conditions: Spreading this thesis by the TDX ( service and by the UB Digital Repository (diposit.ub.edu) has been authorized by the titular of the intellectual property rights only for private uses placed in investigation and teaching activities. Reproduction with lucrative aims is not authorized nor its spreading and availability from a site foreign to the TDX service or to the UB Digital Repository. Introducing its content in a window or frame foreign to the TDX service or to the UB Digital Repository is not authorized (framing). Those rights affect to the presentation summary of the thesis as well as to its contents. In the using or citation of parts of the thesis it s obliged to indicate the name of the author.
2
3 UNIVERSITATDEBARCELONA FACULTATDEBIOLOGIA DEPARTAMENTDEGENÈTICA GENÈTICAMOLECULARHUMANA BIENNI IDENTIFICATIONOFGENETICSUSCEPTIBILITYFACTORSFOR FIBROMYALGIA Identificaciódefactorsdesusceptibilitatgenèticaperafibromiàlgia MemòriapresentadaperElisaDocampoMartínezperaoptaraltítoldeDoctora perlauniversitatdebarcelona XavierEstivilliPallejà RaquelRabionetJanssen (Director) (Directora) DanielRaúlGrinbergVaisman (Tutor) TesirealitzadaalprogramaGensiMalaltiadelCentredeRegulacióGenòmica(CRG)
4
5 ABSTRACT Fibromyalgia(FM)isahighlydisablingsyndromedefinedbyalowpainthresholdandapermanentstateof pain.widespreadpainisaccompaniedbyaconstellationofsymptomssuchasfatigue,sleepdisturbances and cognitive impairment, among others. The mechanisms explaining this chronic pain remain unclear. Nowadays, the most established hypothesis underlying FM ethiopathogenesis is the existence of a dysfunction in pain processing, as supported by alterations in neuroimaging and neurotransmitters levels. TheetiologyofFMinvolvestheinteractionofenvironmentalandgeneticsusceptibilityfactors.Thegenetic contributiontofmhasbeenprovenbythepresenceofahigherconcordanceinmonozygoticthandizygotic twinsaswellasfamilyaggregation.however,theindividualgeneticandenvironmentalfactorsinvolvedhave notbeenidentified.theaimofthisthesiswastoelucidategeneticsusceptibilityfactorsforfibromyalgia.we assessed this objective through three main approaches: the identification of FM clinically homogeneous subgroups with a two step cluster analysis, a genomewide association study in order to evaluate the possible contribution of single nucleotide polymorphisms with Illumina 1 million duo array, and array comparative genomic hybridization experiments to identify regions varying in copy number that could be involved in FM susceptibility. In the cluster study, 48 variables were evaluated in 1,446 Spanish FM cases fulfilling1990acrfmcriteria.apartitioninganalysiswasperformedtofindgroupsofvariablessimilarto eachother.variablesclusteredintothreeindependentdimensions: symptomatology, comorbidities and clinical scales. Only the two first dimensions were considered for the construction of FM subgroups, classifying FM samples into three subgroups: low symptomatology and comorbidities (Cluster 1), high symptomatologyandcomorbidities(cluster2),andhighsymptomatologybutlowcomorbidities(cluster3). These subgroups showed differences in measures of disease severity and were further implemented in geneticanalysis.genomewideassociationstudywasperformedin300fmcasesand203controls.nosnp reachedgwasassociationthreshold,but21ofthemostassociatedsnpswerechosenforreplicationinover 900casesand900painfreecontrols.FourofthestrongestassociatedSNPsselectedforreplicationshoweda nominalassociationinthejointanalysis.inparticular,rs (myt1l)wasfoundtobeassociatedtofm withlowcomorbidities.arraycomparativegenomichybridizationdetected5differentiallyhybridizedregions, whichwerefollowedupbydirectgenotyping.followupexperimentsvalidatedassociationforoneofthese regions.anintronicdeletioninnrxn3wasassociatedtofemalecasesoffmandinparticularthosewithlow levels of comorbidities. This enhances the importance of gender in FM ethiopathogenesis and could be pointingtotheexistenceofadifferentgeneticbackgroundforfminmalesandfemales.italsohighlightsthe importanceofidentifyingfmhomogeneoussubgroupsforthedetectionoffmgeneticsusceptibilityfactors. If the proposed FM candidate genes are further validated in replication studies, this would constitute a change in the FM etiological concept, as several of our proposed candidates are known neuropsychiatric diseaseassociatedgenes(autism,addiction,mentaldisability).thiswouldhighlightanovelneurocognitive involvement in this disorder, currently considered as a musculoskeletal and affective disease.
6
7 CONTENTS INTRODUCTION...3 PAIN:AHOMEOSTATICEMOTIONESSENTIALFORTHESURVIVALOFSPECIES(CharlesDarwin)...5 Paindefinitionandclassification...5 Paincircuitryandstructures:howitworks...5 Visualizingpain...7 Mechanismsofpathologicalpain...8 THESYNAPSE...11 Definitionandstructure...11 Neurexinsandneuroligins...12 FIBROMYALGIA...14 Definitionandclinicalfeatures...14 Epidemiology...16 Diagnosticcriteria...16 Diseaseevolutionandprognosis:Fibromyalgiacomorbidities...18 Differentialdiagnoses...19 Clinicalimpactoffibromyalgia...21 Diseaseactivitymeasurement:clinicalscales...22 Symptomstreatment...22 Evidencesforabiologicalbasis...25 Fibromyalgiagenetics...30 GenomicsourcesofHUMANvariation...35 Singlenucleotidepolymorphisms...35 CopyNumberVariants...42 GENETICSUNDERLYINGCOMPLEXDISORDERSSUSCEPTIBLITY...52 Wherearewe?...52 Largecohortsandpopulationadmixture...52 Powercalculationandcorrectionformultipletesting...53 Dealingwithclinicaldata:Clusteranalysis...53 OBJECTIVES...55 MATERIALSANDMETHODS...59 SAMPLES:THEFIBROMYALGIABIOBANK...61 FMCLUSTERS:IDENTIFYINGFIBROMYALGIASUBGROUPS...61 Statisticalanalysis...62 GENOMEWIDEASSOCIATIONSTUDY...64 Qualitycontrol...65
8 MergingfilteredFMandfilteredcontrols...69 Associationanalysis...70 CheckingSNPsclusters...70 Imputation...71 SNPsannotation...71 Replication...71 AssessingSNPfunctioninsilico...72 CNVassessment:PennCNV...73 CNVmosaicismassessment...75 Arraycomparativegenomichybridization...75 Experimentalprotocol...75 Analysisofarraysresults...76 Validationofarrayresults...77 RESULTS...89 CLUSTERANALYSIS...91 GENOMEWIDEASSOCIATIONSTUDY...94 SNPgenotypinganalysis...94 CNVAssessment:PENNCNV AssessmentofCNVsinmosaicstate ARRAYCOMPARATIVEGENOMICHYBRIDIZATION Karray acgh1marray EvaluationofthepossiblefunctionalconsequencesofNRXN3_DEL DISCUSSION CONCLUSIONS RESUMEN REFERENCES ABBREVIATIONS ANNEXES SUPPLEMENTARYINFORMATION GLOSSARY SCALES PUBLICATIONS ACKNOWLEDGEMENTS...211
9 INTRODUCTION Pain,likelove,isallconsuming:whenyouhaveit,notmuchelsematters,andthereisnothingyoucandoaboutit. CliffordJ.Woolf
10
11 INTRODUCTION PAIN:AHOMEOSTATICEMOTIONESSENTIALFORTHESURVIVALOFSPECIES(CHARLESDARWIN) Paindefinitionandclassification Painistheabilitytodetectnoxiousstimuli.Itisaphysiologicalprotectivesystemessentialtoanorganism s survival: individuals who are incapable to detect painful stimuli do not engage in protective behaviours againstlifethreateningconditions(1). Based on its function and on the elapsed time from the painful stimulus, three kinds of pain can be considered: nociceptive, inflammatory and pathological pain (2). Nociceptive pain is the earlywarning physiological protective system, essential to detect and minimize contact with damaging stimuli. Acute nociceptivepainoccurswhenpowerfulornoxiousstimuliactivatenociceptors,specializedsensoryneurons characterized by high activation thresholds (3). This type of pain is not a clinical problem, except in the specificcontextofsurgeryandotherclinicalproceduresthatnecessarilyinvolvenoxiousstimuli,whereit mustbesurpassedbylocalandgeneralanaesthetics(4).aftertissuedamage,inflammatorypain,whichis causedbytheactivationoftheimmunesystem,assistsinthehealingoftheinjuredbodypart.bycreating hypersensitivity,ortenderness,itreducesfurtherriskofdamageandpromotesrecovery.inflammatorypain isadaptativebutitstillhastobereducedinpatientswithsevereand/orextensiveinjury.finally,thereisthe painthatisnotprotectivebutmaladaptative,resultingfromanabnormalfunctioningofthenervoussystem. Pathologicalpainisadiseasestateofthenervoussystem.Itcanoccurafternervousdamage(neuropathic pain),butalsoinconditionsinwhichthereisnosuchdamage(dysfunctionalpain).itistheconsequenceof amplified sensory signals in the central nervous system (CNS). Conditions that evoke dysfunctional pain includefibromyalgia(fm)andirritablebowelsyndromeamongmanyotherconditions. Paincircuitryandstructures:howitworks Noxiousstimuliaredetectedbynociceptors,whosecellbodiesarelocatedinthedorsalrootganglia(body sensibility) and the trigeminal ganglion (face sensibility). They have both a peripheral and central axonal branchthatinnervatetheirtargetorganandthespinalcord,respectively(5).thisspecializedsetofnerve fibersincludesunmyelinatedcfibersandthinly myelinateda fibers,whicharedistinct frommyelinated tactilesensors(afibers)andproprioceptors *1.Theperipheralterminalofthenociceptorwillonlyrespond toenvironmentalstimuli(painfulhot,coldormechanicalstimulation),whenthestimulusintensitiesreach thenoxiousrange.thephysicochemicalpropertiesofnoxiousstimuli,suchasheat,extremecold,pressure andchemicals,areconvertedtoelectricalactivitybytransientreceptorpotentialgeneratingchannels,and 1 TermsmarkedwithanasteriskaredefinedintheGlossarysection(Annexes). 5
12 INTRODUCTION this electrical activity is amplified by sodium channels to produce action potentials. Nociceptive afferents carrying these peripheral inputs form glutamatergic (excitatory) synapses onto secondorder neurons, mostlyinthesuperficialdorsalhorn.sensoryinputsareintegratedandprocessed,andthenetoutputfrom the spinal networks is carried by several pathways to distinct projections in the brain. These include the lateral spinothalamic tract, which projects to the lateral thalamus and has been implicated in sensory discriminative aspects of the pain experience (such as discriminating where the stimulus is? and, how intense is it?), and the medial aspect of the spinothalamic tract and the spinoparabrachial tract, which projecttothemedialthalamusandlimbicstructuresandarebelievedtomediateemotionalandaversive componentsofpain(6).finally,fromthesebrainstemandthalamicloci,theexperienceofpainisperceived inthecortex,whereseveralstructures,someofthembeingmoreassociatedwiththesensorydiscriminative properties(suchassomatosensorycortex)andotherswithemotionalaspects(suchasinsularcortex) are activated,andinformationissenttothespinalcordtoenablewithdrawalfromthenoxiousstimuli(5). Thepainascendingnetworkisregulatedbymechanismsofdescendinginhibitionanddescendingfacilitation (7). Thesedescendingpathwaysoriginateinthehypothalamus, thecortex,therostroventralmedullaand otherbrainstemnuclei,andcaninteractwithseveralneuronalelementsinthedorsalhorn:theterminalsof theprimaryafferentfibers,projectionneurons,intrinsicinhibitoryandexcitatoryinterneuronsandterminals ofotherdescendingpathways(figure1).multipleneurotransmittersareinvolved,andtheircolocalization, aswellasthepresenceofmultiplereceptorsperneurotransmitter,allowtheregulationofthenociception signalthroughdescendingfacilitatoryandinhibitorypathways(7).therefore,precisecontrolsmustexistto maintain a balance between activation and inhibition (8). Several theories point out that psychological factors are known to modulate pain perception. Variables such as attentional state, emotional context, hypnotic suggestions, attitudes, expectations or anaesthesiainduced changes in consciousness now have beenshowntoalterbothpainperceptionandforebrainpaintransmissioninhumans(9).thispsychological painmodulationisexertedbydescendingprojectionsfromfrontalbrainareas.ithasbeenproposedthatin a negative emotional state, activity in the frontal cortex is increased, enhancing the nociceptive signal transmittedfromthespinalcordtothebrainbyengagingdescendingfacilitatorypathways(10). Asasummary,inordertoensureaverypreciseregulationofthemessage,nociceptivetransmissionisvery complexandinvolvesdifferentpathwaysthatconnectdifferentcnsstructures. 6
13 INTRODUCTION Figure1:Paintransmissionandstructuresinvolved(basedonMillanetal(7).DP,Terminalsofdescendingpain;PAF, afferent fibres; IN, interneurons; MN, motoneurons; PN, projection neurons; and ETL, spinothalamic lateral tract. Synapsesthatarenotmarkedwithaplusoraminussymbolscanbebothactivatoryandinhibitory. Visualizingpain Formanyyears,thestudyofnociceptivepainwasrestrictedtotheanalysisofsensoryneuronsandcircuits inthespinalcord.onemainreasonwasthedifficultytoexaminehowthebrainprocessedpainsignalsin anaesthetized animals, when the standard definition of adequate anaesthesia is the loss of pain related behaviour. However, functional neuroimaging in human volunteers and patients allowed for definition of thosebrainareasactivatedbynociceptiveinputsasreviewedinshweinhardtetal(11).theprimarymodes of functional imaging that have been used in FM include functional magnetic resonance imaging (fmri), singlephoton emission computed tomography (SPECT), and positron emission tomography (PET). More detailsonthesetechniquesareincludedinbox1. 7
14 INTRODUCTION Box1:Imagingandfunctionalneuroimaging Neuroimaging methods infer brain activity from regional cerebral blood flow, glucose metabolism and neurochemicalconcentrations,andchangesinbrainfunctionfromstructuralmeasuresofwatermotilityand brainvolume. MRIproduceshighresolutionstructuralimagesbyplacingthepatientinastrongmagneticfieldthataligns all the body atomic nuclei. This alignment is altered by radio frequency fields, which produce a rotating magneticfieldinthenuclei.thesearedetectablebyascannerthatconstructsanimageofthescannedarea ofthebody.macroscopicchangesinbrainstructurecanbeevaluatedbyvoxelbasedmorphometry,which usesmriimagestoassessthevolumeofspecificbrainregions. FMRI measures changes in neural activity by measuring vascularization changes. Increased neural activity causesanincreaseddemandforoxygen,whichisensuredbythevascularsystembyincreasingtheamount of oxygenated haemoglobin. Because deoxygenated haemoglobin attenuates the MR signal, the vascular responseleadstoasignalincreasethatisrelatedtotheneuralactivity. Magnetic resonance spectroscopy (MRS) is used to measure the levels of different metabolites in body tissues.magneticresonance(mr)signalvariesamongthedifferentmolecules,andmrsevaluatesthesignal ofagivensourceandexpressesitsmagnitudeinrelationtoacontrolstandardmolecule(oftencreatinine). Thus,therelativeconcentrationofmoleculessuchasneurotransmitterscanbeinferred. SPECTevaluateschangesincerebralbloodflowwiththeinfusionofaradioactivetracer.Thistracerproduces gamma radiation that is detected by a camera (gamma camera) to acquire multiple 2D images, from multipleangles.acomputeristhenusedtoapplyatomographicreconstructionalgorithmtocreateathree dimensiondataset. InPET,whentheradioactivetracerdecays,apositronisdeliveredand,incombinationwithatissueelectron, itformsapositronium,whichisunstableandproducestwogammarayphotonsafteranaveragelifetimeof 125 picoseconds. The photons are detected by a camera, creating threedimensional images. Image resolutionofpetisbetterthanthatofspect,asphotonsproducemoreradiation. Mechanismsofpathologicalpain Althoughphysiologicalpainhasanimportantprotectivefunction,paincantakeonadiseasecharacterin pathological chronic states such as inflammation, neuropathy, cancer, viral infections chemotherapy and diabetes. This state is manifested as hyperalgesia (increased sensitivity to painful stimuli). Furthermore, individualswithchronicpainoftenshowdiseaseinduced,therapyresistantdeviationsfromnormaltactile sensations,suchasparesthesiasanddysestesias.finally,themostcommoncomplaintfromindividualswith chronicpainisspontaneouspain(6). 8
15 INTRODUCTION There are evidences, both at the CNS and at the peripheral nervous system level, indicating that these pathological pain states can be explained by an alteration in pain processing. In physiological conditions, thereisanenhancednetdescendinginhibitionafterinflammationinsitesofprimaryhyperalgesia.however, in models of neuropathic pain, the tactile allodynia (when a nonpainful stimulus becomes painful) after nerve injury is dependent upon a tonic activation of net descending facilitation. Thus, descending modulationofpersistentpaininvolvesbothinhibitionandfacilitation,inordertoimprovenociceptionatthe spinal level (12). This involves prolonged functional changes in the nervous system, evidenced by the developmentofdorsalhornhyperexcitability.animpairmentofthisendogeneouspainmodulationdueto animbalanceoftheexcitatoryandinhibitorytracts,knownaspaindisinhibition,canexplainthealteration inpainprocessingoccurringinindividualswithchronicpain(13). Permanentstimulationofthenociceptivepathwaysthatarepresentinchronicpainstatescanalsocausean increase in the magnitude of responses to a defined sensory stimulus, an increase in the level of spontaneous activity, or after discharges, which represent continued activity after the termination of a nociceptivestimulus.thisleadstoacentralamplificationofpainalsoknownascentralsensitization(higher excitabilityandspontaneousactivity)(6).fibresresponsibleofthetransmissionofnonpainfulstimuli(a fibers) get C fiber (nociceptive) qualities. As a result, touch becomes painful (Figure 2). This pathological enhancement of pain transmission is mediated by functional and structural changes. Functional plasticity includesmolecularchanges;forexample,persistentinflammationwithhigherandcontinuousamountsof inflammationmediatorscancauseaca 2+ ionchannelsensitization,leadingtoasynapticpotentiationthat finally causes central sensitization. Also, at the structural level, persistent nociceptive activity leads to an increase in the number and size of synaptic spines, causing changes in neuronal connectivity and cell proliferation(6). At the peripheral nervous system, continuous stimulation increases the excitability of nociceptors, in a processknownasperipheralsensitization.thisleadstoareductioninthepainthresholdand,therefore,to astateofhyperalgesia. 9
16 INTRODUCTION Figure2:Normalandcentralsensitization.Normalsensitization(topimages):primarysensoryneuronsthatencodelow intensity stimuli only activate central pathways leading to innocuous sensations, while high intensity stimuli that activate nociceptors only activate the central pathways that lead to pain. The induction of central sensitization in somatosensorypathways(bottomimages)isaccompaniedbyincreasesinsynapticefficacyandreductionsininhibition, leading to a central amplification. Pain response to noxious stimuli is enhanced, in amplitude, duration and spatial extent (hyperalgesia), while the strengthening of normally ineffective synapses recruits subliminal low threshold sensoryinputsthatactivatethepaincircuit(allodynia).thetwosensorypathwaysconverge(fromwolfetal(14)). 10
17 INTRODUCTION THESYNAPSE Definitionandstructure Neuralinformationispropagated,alonganeuron,asanelectricalsignal.Thistransmissionisnotcontinuous. The connection from one neuron to the other takes place through the synapses, whose existence was a matter of discussion during the end of the XIX th century (Box 2). Synapses are specialized intercellular junctions whose function is the transfer of information from a neuron to a target cell, usually another neuron. Synapses can be electrical or chemical, depending upon whether transmission occurs via direct propagation of the electrical stimulus in the presynaptic cell or via chemical intermediates. Electrical synapses are gap junctions between neurons; they can be either excitatory or inhibitory (15), allowing bidirectionalpropagationofthesignalandplayingaroleinsynchronizingneuronalactivity.atthechemical synapses, the presynaptic electrical signal is converted into neurotransmitters that bind to specific postsynapticreceptors,leadingtotheproductionofanewelectricalsignalinthepostsynapticneuron.we willfocusonchemicalsynapses,andwewillrefertothemhenceforwardassynapses(16,17). Synapses share many properties with other intercellular junctions, but they present differential features: theyareasymmetrical,transmitinformationbyanextremelyfastandtightlyregulatedmechanism,andare highly plastic (16, 17). In spite of their great morphological variability, all synapses share some common structuralfeatures.onthepresynapticcell,neurotransmittercontainingsynapticvesiclesaccumulateinthe active zone, where the neurotransmitter release occurs. These vesicles associate with the presynaptic plasmamembranethroughanetworkofscaffoldingproteinsknownasthecytomatrixofactivezones.the postsynaptic membrane includes an accumulation of neurotransmitter receptors, a thickening of the membrane,thepostsynapticdensityandasubmembranouselectrondensescaffold(16,18).forsynapsesto functionproperly,allthesecomponentsmustberecruitedandpreciselyalignedacrossthesynapticcleft,a 20nmwideextracellularspacethatseparatestwoneuronsatsynapticjunctions(18).Theproperreleaseof neurotransmittersintothesynapticcleftandtheirbindingtothereceptorsisfacilitatedbytheexistenceofa complexmatrixofadhesionmoleculeslocatedinboththepresynapticandpostsynapticneurons. Manyfamiliesofneuraladhesionmoleculeshavebeendescribed.Thesetransmembranemoleculesbindto eachotherextracellularlytopromoteadhesionbetweenthepresynapticandpostsynapticneurons.during development they enable the formation and specification of functional synapses, as they ensure stable contacts between neurons (8, 17, 18). Diversification of adhesion molecules through alternative splicing, amongothermechanisms,providesanimportantrepertoireofmoleculestoenablesynapsesfunctioninthe complexityandspecificityofthecns(19). 11
18 INTRODUCTION Box2:Continuityversuscontiguity:theXIXXX th debate The nature of the contact between neural cells was a controversial issue that dominated the thinking of neuroanatomistsandphysiologistsbetween1870and1920,withtwomaintheories: theneurontheory:neuronsasindependentcellularunits thereticulartheory:centralnervoussystemasacomplexsyncitium:anetworkof fibersthatareindirectcytoplasmiccontinuity Tosettlethequestionofcontinuityversuscontiguityitwasnecessarytodemonstratethefinalramifications ofthenervefibers.thiswasaccomplishedbysantiagoramónycajal.hisworkderivedinlargepartfromhis applicationofthechromesilverimpregnationmethodor reazionenera thathadbeenintroducedbygolgi in1873.thismethodofferedtwoadvantages.first,themethodstained,inanapparentlyrandommanner, onlyabout1%ofthecellsinanyparticularregionofthebrainorspinalcord.thismadeitpossibletostudy themorphologyofindividualnervecellsinisolationfromtheirneighbours.thesecondadvantagewasthat theneuronsthatwerestainedwereoftenimpregnatedthroughouttheirentireextent,sothatonecould clearlyvisualizecellbodies,axons,axoncollaterals,thefulldendriticarbourand,indevelopingbrains,axonal anddendriticgrowthcones.byexaminingindetailnervecellsandtheircontactsinhistologicalsectionsof almosteverybrainregion,cajalwasabletodescribenotonlydifferencesbetweenvarioustypesofnerve cellsbutalsothegreatvarietyofaxonalendingsfoundinthecentralnervoussystem.thisledhiminexorably toconcludethat,atthesitesofinteraction,theyarenotcontinuouswiththeircellulartargets,andtherefore notpartofadiffusenetwork.cajal swork,publishedinspanish,wasnotwidelyknownorappreciateduntil 1889,whenheattendedameetingoftheGermanAnatomicalSocietyinBerlinandattractedtheattention of Kölliker, who encouraged him to have his work translated into French or German. But it was a physiologist,charlessherrington,whocoinedthetermsynapsein1897,fromthegreek"syn"("together") and"haptein"("toclasp"). Oncethemorphologicalissueofhowthenervecellsinteracthadbeenresolved,attentionnaturally turned toward understanding the mechanism of synaptic transmission: was it electrical or was it chemical ExtractedfromSynapses,Chapter1Abriefhistoryofsynapsesandsynaptictransmission.2003CowanW.M.,SüdhofT.C.andStevensC.F. Neurexinsandneuroligins Neurexins are among the most widely studied adhesion and scaffolding molecules involved in synapse stability and function, and were discovered thanks to spider venom (Box 3). They are transmembrane proteins located in the presynaptic neuron that have three extracellular binding partners: neuroligins, dytroglycanandneurexophilins(20).inparticular,thebindingwithneuroliginshasshowntobeessentialfor thedevelopmentandfunctionofgabaergicandglutamatergicsynapses(17,21).theyaremainlyexpressed inthecns,andtheirdistributionacrossthedifferentbrainregionsisheterogeneous(22). In humans, there are three neurexin proteins encoded by three genes. Each gene has two independent promotersandgeneratestwoclassesoftranscripts,whichgiverisetotwoclassesofproteinswithdifferent length and domain composition: and neurexins. and neurexins share the transmembrane and 12
19 INTRODUCTION intracellularregions.theextracellularsequencesofthelongerisoformscontainsixlamininlikedomains (LNS)withthreeintercalatedepidermalgrowthfactor(EGF)likedomains,whereasisoformsonlycontain thelastlnsfollowingashortnterminalspecificsequence(18,23).neurexinsareessentialforsurvival, butredundant:knockout(ko)miceforthethreegeneswerealiveatbirthbutdiedthefirstday,doubleko mice died within the first week, and single KO mice exhibited impaired survival (24). neurexins are required for normal neurotransmitter release: their deletion impairs the function of Ca 2+ channels, in excitatoryandinhibitorysynapses (24).neurexinsalsoaffecttransmissionattheneuromuscularjunction (25).Synapseinducingactivityhasalsobeendemonstratedforneurexins. Neurexins bind to a family of postsynaptic transmembrane proteins, the neuroligins. The Neurexin/neuroligin celladhesion complex can promote the formation of de novo synapses and the differentiation of postsynaptic receptors, at least in vitro. The binding of neurexins to neuroligins is controlled by alternative splicing (18). There are five neuroligins (14 and Y) in humans. Neuroligin1 was identifiedasaresultofitsabilitytobindcertainisoformsofallthreeneurexins(26)(figure3).neurexins have five canonical sites of alternative splicing, and there is evidence suggesting that the different splice sitesareusedindependently,potentiallygeneratingmorethan1000differentisoforms.thesixneurexin( andofneurexin1,2,and3)isoformsareexpressedinallbrainregions,buttherelativeabundanceofeach isoformisinsomeextenttissuespecific(22).twodifferentneurexinscanbeexpressedinthesamecell(27). Figure 3: Neurexins and neuroligins in the synaptic cleft (taken from Sudhof et al (17)). The left part of the figure represents the presynaptic neuron, where and neurexins (NRXN) bind to postsynaptic neuroligins (NLGN). Their bindingisregulatedbyalternativesplicing(as):nrxnshavefive(2forshorterforms)(15)andnlgntwo(aandb) canonicalsitesofas.(l,lnsdomains;e,egflikedomains;andcho,carbohydrateattachmentsequence). RecentstudieshaveidentifiedmutationsinthegenesencodingNRXNsandneuroligins(NLGN)asacauseof schizophrenia(28),autism(29),andtourette ssyndrome(30).thesesynapticmoleculesparticipateinkey circuits influencing addictive behaviors (22, 31). Two members of the neurexin gene family have been associated with nicotine dependence, Neurexin 1 and Neurexin 3 (5, 32). Moreover, Neurexin 3 has also 13
20 INTRODUCTION been associated with smoking behaviour in schizophrenic patients (33), impulsivity and substance abuse (34),illegalsubstanceabuse(35),alcoholdependence(36)andobesity(37).Inadditiontosinglenucleotide variants(snps),largeandrarestructuralvariantsinvolvingneurexingeneshavebeendescribedinautism spectrumdisorders(3842)andschizophreniapatients(43). Box3:Neurexinsandspiders(27) Latrotoxin,acomponentofblackwidowspidervenom,triggersmassiveneurotransmitterreleasefromthe presynaptic nerve terminals of vertebrates, binding to specific receptors in the presynaptic plasma membrane by a mechanism that was unknown. Extensive peptide sequences from the purified bovine latrotoxin receptor were used to synthesize degenerate oligonucleotides, which were then used in polymerasechainreaction(pcr)experimentstoclonethecdnasequencesencodingthesepeptides.the PCRproductsandoligonucleotideswerethenusedasprobestoscreenratbraincDNAlibraries.Twosetsof overlapping cdnas that encoded homologous but distinct proteins were isolated, only one of which containedthepeptidesequencesobtainedfromtheisolatedlatrotoxinreceptor.eachsetofcdnaclones was highly polymorphic. The proteins encoded by these transcripts were referred as neurexins I and II becausetheyconstitutedneuronspecificcellsurfaceproteins. FIBROMYALGIA Definitionandclinicalfeatures Fibromyalgia(FM)isahighlydisablingsyndromedefinedbyalowpainthresholdandapermanentstateof pain.widespreadpainisaccompaniedbyaconstellationofsymptomssuchasfatigue;sleepdisturbances and cognitive impairment, among others. The mechanisms explaining this chronic pain remain unclear. Onsetofsymptomsisusuallygradual,butinsomecasesasuddenonsetfollowinganidentifiableeventhas beendescribed(44).fmisoftendebilitatingandfrustratingforphysiciansandpatients:thechronicnature ofthesymptoms,themultiplicityofpossibleaetiologiesandthelackofeffectivetreatments,makedifficult FMmanagement(45). Pain throughout the body is the pivotal symptom of FM (46). It is a cause of considerable suffering and functionalimpairment.painiscontinuous,althoughitisworseinthemorning,improvesduringthedayand worsensagainbynight.painisworsenedbystaticpositions,weatherchangesandphysicalburdens(47).it localizesinmusclesandjoints,mainlyshouldersandhips,mimickinginflammatoryjointdiseases.itpresents also neuropathic and visceral pain characteristics (48). Some patients also present pain with a superficial burning quality, increased sensitivity to painful stimuli (hyperalgesia) and features of allodynia (pain followinganinnocuousstimulus)(44). 14
21 INTRODUCTION The clinical understanding of FM has evolved over the last two decades, emphasizing the importance of symptoms other than pain that contribute to global suffering. More than 70% of the patients present fatigue.thiscanbecontinuousorappearasexhaustionperiodsof12days.sleepdisturbancesisthethird most frequent symptom and it is correlated with FM severity. Many components of sleep have been measured as abnormal in FM patients, including sleep quality, latency, duration and disturbances, and impaired daytime functioning (49). In particular, FM patients present alterations in the deepest stage of sleep,thedeltawavesleep(50).furthermore,poorqualityanddurationofsleephasbeenshowntohavea negativeimpactuponfatigueandaffect(51).thisclassicalfmtriad(pain,fatigueandsleepdisturbances)is accompanied by a wide spectrum of symptoms of different systems, since FM is a very heterogeneous disorder.theseadditionalsymptomsincludesensitivesymptoms,motorsymptoms,vegetativesymptoms, cognitivesymptoms,andaffectivesymptoms(figure4). Figure4:FMisaheterogeneousdisorder. FMpatientsfrequentlycomplainofcognitivesymptoms,popularlyreferredas fibrofog (52).Theseinclude difficulty with reading and calculation, memory impairment, forgetfulness, and even anomic aphasia (recallingnames)episodes.cognitivedeficitsaremoreprevalentinfmpatientsthaninpainfreeindividuals but they are reported by other chronic pain patients (osteoarthritis and rheumatoid arthritis patients). A studyshowedthat,regardlessofdiseasestatus,chronicpainpatientsdemonstratedcognitiveimpairment whenperformingeverydayattentionaltasksincomparisonwithmatchedpainfreecontrols(53).however, another study showed that these symptoms are more prevalent in FM than in other rheumatic diseases, suggesting the relevance to FM (52). Whether they are more prevalent in FM than in other chronic pain statesmaybeunclear,butwhatissureisthattheyconstituteamajorconcernforfmpatients.asaproofof concept,cognitivesymptomshavebeenincludedinthenewfmclassificationcriteria(14). Affective symptoms, mainly anxiety and depressive symptoms are also common FM features, as well as neurovegetativesymptoms,includingdizziness,sweatingorpalpitations. 15
22 INTRODUCTION FMpatients,whoarecharacterizedbyalowpainthreshold, presenttenderpoints,whicharepointsthatare painful under pressure. Physical examination findings also include other musculoskeletal unspecific alterations,suchasposturalalterations,muscularhypertonia,muscularcontractures,andpainfulstretching ofaffectedregions. Epidemiology FMisestimatedtoaffectmorethan5millionAmericans(2%5%oftheadultpopulation)(54) ).Theoverall prevalenceoffminthegeneralpopulationoffiveeuropeancountries( France,Germany,Italy,Portugaland Spain)is2.9%(55).Inparticular,accordingtotheEPISERstudy,FMaffects2.4%oftheSpanishpopulation over20yearsold(56).fmpatientsaremainlywomeninthefourthdecadeoflife,withafemale:maleratio of21:1(48).. Only a small number of FM epidemiological studies have been performed in nondeveloped countries, showingafrequencysimilartothatofdevelopedcountries: :2.5%inBraziland3.2%inBangladesh(57).This suggeststhatfmisnotadisorderspecificofdevelopedcountries.infact,astudyperformedintheamish population, which is an isolated society where modern life influence is absent, showed an increased prevalenceoffm(7.3%) )(58). Diagnosticcriteria Intheabsenceofsuitablediagnostic tests,fmdiagnosisisestablishedbyahistoryofsymptomsandthe exclusionofsomaticdiseasesexplainingthesesymptoms(14,59).inthebeginningofthenineties,withthe development of the American College of Rheumatology fibromyalgia criteria, FM was defined as the presenceofwidespreadpainwithdurationofatleast3months.thispainshouldbebilateral,,bothabove andbelowthewaist,anditshouldincludeaxialskeletalpainincombinationwithtendernessat11ormore of18specificpointsitesonthebody,calledtenderpoints(60)(figure5) ). Figure 5: 1990 American College of Rheumatology (ACR) FM classification criteria based on the presence of 11/18 tenderpoints.eachtenderpointislocatedonbothsidesofthebodyandpainfulunderpalpationwithapproximately4 kgofpressure. 16
23 INTRODUCTION Aseriesofobjectionstotheseclassificationcriteriadevelopedovertime(14).First,itbecameincreasingly clearthatthetenderpointcountwasrarelyperformedinprimarycare,wheremostfmarediagnosed,and when performed, was performed incorrectly. Consequently, in practice, FM diagnosis has often been a symptombaseddiagnosis.furthermore,thespecificityofthesetenderpointsbecameamatterofdiscussion becauseitwasknownasahallmarkoffmandwasusedbymalingererstosimulatethedisease.second,the importance of symptoms that had not been considered by the American College of Rheumatology (ACR) becameincreasinglyknownaskeyfmfeatures,asforexample,fatigue,cognitivesymptomsandsomatic symptoms. Finally, there was still another important problem with FM diagnosis: patients who improved failedtosatisfytheacr1990classificationdefinition.forallthesereasons,aneedtodevelopabroadbased severity scale able to detect clinical changes emerged. Based on a multicenter study of patients with diagnosisoffmandacontrolgroupofrheumaticdiseasepatientswithnoninflammatorydisorders,newfm diagnosticcriteriaweredeveloped(14). These2010FMcriteriawherenotmeanttoreplace1990ACRcriteria.TheyproposedanewdefinitionofFM with increasing recognition of the importance of cognitive problems and somatic symptoms. They were basedonawidespreadpainindex(wpi),withscoresbetween0and19,forthenumberofareasinwhich the patient had had pain over the previous week; a Symptom Severity (SS) score, where fatigue, sleep disturbancesandcognitivesymptomsseveritywerescoredfrom0to3,plustheextentofsomaticsymptoms in general. FM 2010 classification criteria were simplified a few months after their publication, by substitutingsomaticsymptomsingeneralbyheadaches,paininabdomen,anddepression(figure6).a031 FM(FS)scalewascreatedasthesumoftheWPIandtheSSscore,enablingtheuseofthese2010modified FMclassificationcriteriainstudieswithouttheneedofanexaminer. 17
24 INTRODUCTION Figure6:FM2010modifiedclassificationcriteria(fromWolfeetal2011)(61). Diseaseevolutionandprognosis:Fibromyalgiacomorbidities FMisachronicdisease.Usuallypainremainsatthesamelevel,withepisodesofenhancedclinicalactivity. Bettereconomicalandeducationallevels,aswellastheabsenceofpsychiatriccomorbidity,arelinkedwitha betterprognosis(62).adanishstudyshowed,however,thatdespitenooverallincreaseinmortality,fm patients have an elevated risk of suicide (10fold increase in mortality risk), liver cirrhosis/biliary tract disease(sixfoldincreaseinmortalityrisk)andcerebrovasculardisease(threefoldincreaseinrisk)(63).itis controversialwhetherfmpatientshaveahigherriskofcancer(64,65). Inadditiontothemultiplesymptomsaccompanyingchronicpain,comorbidconditionsareoftenpresentin FM;inparticularsomatoformdisorders * (Figure7).FMhaseventheoreticallybeendefinedasanoverlapof syndromesandsymptomsratherthanasadiscreteentity(66).infact,somephysiciansevenclaimthatfmis morearagbagthanadiseasebyitself.itisusualtofindinmanyscientificpublicationsthetermfibromyalgia syndrome(fms)insteadoffm.thechronicfatiguesyndrome(cfs),presentin2170%offmcases,isthe mostcommonoftheseoverlappingdisorders(box4)(45).psychiatriccomorbiditiesarealsofrequent,but the role they play in FM onset and development has not been well defined yet. Major depression, panic attacks;posttraumaticstressdisorderandpersonalitydisordersarethemostcommonlyassociatedwithfm. 18
25 INTRODUCTION Figure7:Fibromyalgiaoverlapswithothersomatoformsdisorders. Box4:Fibromyalgiaorchronicfatiguesyndrome? Numerous physicians underscore the difficulty in the diagnosis and classification of patients with somatic complaintsofdiffusepainandconstantfatigue,bygivingthemtwodiagnoses:fmandcfs.cfsisdefinedby unexplained,persistent,orrelapsingfatiguewithadefiniteonset.itisquitecontroversialwhethertheseare twodistinguishableentitiesordifferentmanifestationsofthesamedisorder.infact,2070%offmfulfills CFS diagnostic criteria. In the Spanish health care system, one curious think is that FM patients are controlledbyrheumatologistsandcfsbyinternists.infact,sometheoriesclaimthatthereisacontinuumin FMandCFSsymptomatologyandthatthosepatientsfulfillingbothFMandCFScriteriaaretheoneswitha moreseveredisease. Differentialdiagnoses ForadiagnosisofFM,firstalldisorderswherepainandfatiguearemajorsymptomshavetobediscarded (Box5).Mostofthesedisorderscanbediscardedthroughabloodtestincludingacompletebloodcount, kidneyandliverfunctiontests,creatinekinase,fastingserumcalciumandphosphorus,creactiveprotein/ erythrocytesedimentationrate,thyroidfunctiontests,andrheumatologicserologies.anyabnormalitiesin anyoftheseshouldbefollowedupwithmorefocusedandspecifictests(e.g.:thepresenceofanaemiaand high erythrocyte sedimentation is consistent with multiple myeloma; it should be discarded through a proteinelectrophoresisinbloodserum).furthermore,endocrinedisordersandinfectiousdiseasescouldbe mimicking FM because they are causing muscle pain and/or fatigue. Psychiatric disorders constitute a differentialdiagnosisforfm,asfmpatientsshowveryoftenaffectivesymptoms.whenpresentalongwith symptomsoffm,thephysicianmustcometoadecisionaboutwhichcauseswhat(45).thepresenceofpain andfatigueasmajorsymptomsconcentratingthepatientconcernwillpointouttoanfmdiagnosis.more 19
26 INTRODUCTION detailsabouttheotherdisorderslistedinbox5canbefoundintheglossarysection(annexes).itmustbe takenintoconsiderationthat,inmostofthesechronicdiseases,thediagnosisoffmisnotexcludedbythe presenceofotherdiseases,asitmaybesecondarytothese. Box5:Fibromyalgiadifferentialdiagnosis Autoimmunedisorders Celiacsprue Polymialgiarheumatica * Polymiositis * Seronegativespondyloarthropathy * Systemiclupuserythematosus * Endocrinedisorders Hyperpartiroidism Hypophosphatemia Hypothyroidism Infectiousdiseases HepatitisC HIVacuteinfection Nonviralmeningoencepahlitis Postviralencephalitisandmeningitis Malignancies Neoplasia:primarytumor(multiplemyeloma)andbonemetastasis Paraneoplasticdisorders * Musculoskeletaldisorders Costochondritis Lumbernerverootcompression Reflexsympatheticdystrophy * Spinalstenosis Neuromusculardisorders Miasteniagravis Multiplesclerosis Neuropathies Psychiatricdisorders Dysthimia Seasonalaffectivedisorder Melancholicmajordepression Sleepdisorders Obstructivesleepapnea Malingerers Finally,sinceFMisdiagnosedbasedonsymptomsandphysicalexamination,withoutanobjectivediagnostic test,ithasbecomeanappealingdiseaseformalingerers.thehighratesofdisabilityclaimsforfmpatients, despiteverylittleobjectivedata,explainthisphenomenon.thiscomplicatesevenmorethediagnosisoffm. 20
27 INTRODUCTION Clinicalimpactoffibromyalgia FMisoneoftherheumaticdiseaseswithahigherimpactonlife.Patientsreferbigconsequencesontheir lives in terms of physical activity, intellectual disability, emotional state, mental health and employment. Patientsthathaveapaidemploymentshowabetterprognosis(67). Despitethelackoforganicpathology,patientswithFMoftensufferdisruptionoftheirsocialstructureand thispotentiatestheirsymptoms(figure8).forexample,anexecutiveinacorporationmay,secondaryto severestress,haveanemotionalbreakdownthatevolvesintoachronicpainsyndrome.thismay,inturn, leadtoalossofemployment,disability,separationordivorce,andmaladaptiveillnessbehaviour(45). Figure8:Thefibromyalgiacycle.FMsymptomshaveabigimpactinpatients life,andthisaggravatessymptoms. Notonlyisthediseasebyitselfhighlydisabling.Therearestillmanyphysiciansandpeopleinpatient ssocial environmentthatarereluctanttoconsiderfmasadisease,andthinkthatfmpatientsarehypochondriacs ormalingerers(box6).thislackofcomprehensionwearsoutthepatient. FMcanhaveadevastatingeffectonpeople slivesbutalsotheirrelationshipwithfamily,friendsandwork colleagues(68).parents,siblingsandpartnersoffmpatientshaveaworselifequalitythanfamilymembers ofnonaffectedsubjects(67). Furthermore, FM constitutes a significant economic burden for society. FM subjects are found to have substantial costs, over 75% of which are driven by indirect costs from lost productivity, and these costs increase as FM severity increases (59). In addition to that, FM patients use more health resources than general population and incur similar costs as other rheumatic diseases such as rheumatoid arthritis (69). Given the prevalence of the disorder it has become a major concern as an economical issue. In order to optimize FM management and minimize its costs by reducing the number of consultations to general practitioners, in 2010, the Catalan Institute of Health (ICS) created FMCFS units. These units consist of multidisciplinaryteams,includingarheumatologist,apsychiatrist,apsychologistandanurse,toensurethe 21
28 INTRODUCTION bestmanagementoffmpatients.itremainstobeseenwhetherthisinitiativeleadstoareductionoffm costs.furthermore,evaluationofworkdisabilityinfmpatientsisacontroversialissue(47).disabilityclaims aremoreandmorefrequent.forthephysiciansitisdifficulttoevaluateapatientbasedonnonobjective arguments. In Catalonia, the final decision is taken by ICAMS (Institut Català d'avaluacions Mèdiques i Sanitàries).Lastyear,thefirstpatientwasdismissedofworkbecauseofFM. ( Finally, we should not forget that the economical impact is also at the personal level, since non pharmacologicaltreatmentsarenot coveredby thehealthservicesystem.fmpatienthastoassume the costofpsychotherapies,andalternativemedicinetreatments. Diseaseactivitymeasurement:clinicalscales FMdiagnosisisbasedonthepresenceofclinicalsymptoms.Thereisnotesttoconfirmandtomonitorize disease evolution or response to treatment. For this reason, in everyday clinics, clinical scales are widely usedforfmmanagement.someofthesescaleshavebeenspecificallydesignedforfm,suchasfibromyalgia Impact Questionnaire (FIQ), while most of them are used to measure FM symptoms (mainly psychiatric comorbidities) and impact on everyday life (life quality scales such as SF36). FIQ is easy to use and is sensitivetoclinicalevolution.thesescaleshavebeentranslatedandadaptedtodifferentlanguages.(see Scalesannex). Symptomstreatment Asstatedbefore,FMischaracterizedbypainplusalargeconstellationofsymptoms.Asthereisnocausefor thesesymptoms,treatmentisonlysymptomatic,withnoimpactondiseaseprognosisorevolution.infact, thereisnoeffectivetreatment.responseispoor,andthemodestbenefitsoffmtreatmentareacauseof frustrationforbothpatientsandphysicians(45).practitionershavetoemphasizethatitisachronicdisease and to fight against the stigma attached to this invisible illness (Box 6). They also have to take into consideration that most patients experience years of suffering and many doctors visits. Patients need validationoftheirsymptomsandneedtoknowthattheirdiseaseisreal:physician sempathyiscrucial(70). ThemajorgoalsofFMtreatmentaredecreasingpain,improvingsleepandestablishingaregularexercise program. Moreover, as each patient has a different constellation of symptoms, treatments have to be tailored (70), treatment programs have to be defined with care. Sometimes, due to the difficulty of the patient in coping with symptoms that don t improve in spite of the medication, there is a tendency to consultdifferentphysiciansatthesametime(publicsystem,privatesystem,andgeneralpractitioners). 22
29 INTRODUCTION Thiscanleadtoadangerouspharmacologicalcocktailthan,farfrombebeneficialtothepatient,mayresult intheaccumulationofsideeffectsratherthaninatherapeuticoutcome.inaddition,manyfmpatientsare unusuallysensitivetotheadverseeffectsofmedication(70).forthisreason,treatmentsshouldbestarted atlowdosesandincreasedgradually. Evidencebased guidelines suggest that FM has to be managed with multidisciplinary therapies involving medication and nonpharmacological interventions. This multidisciplinary treatment has been successfully implementedinfmmultidisciplinaryunits(71),whichhavealsohelpedinpatientscopingwithsymptoms andavoidingmulticonsultation. The relative role of role of each of the treatments varies among the different guidelines. Whereas the American Pain Society strongly recommends a pharmacological intervention (amytriptiline) plus non pharmacologicaltreatments(62),theeuropeanleagueagainstrheumatism(eular)stronglyrecommends multiplepharmacologicaltreatmentsonly(72).arecentsystematicreviewofrandomisedtrialshastriedto clarify these contradictory guidelines (59). From this study, a combination of pregabalin or serotonin noradrenaline reuptake inhibitors (SNRIs), as pharmacological interventions, and aerobic exercise and cognitivebehaviouraltherapy(cbt),asnonpharmacologicalinterventions,appearasthebestoptionsfor FMmanagement. As mentioned before, the treatment is individualized, mainly according to treatment response and medication side effects. A summary of the most common treatments, both pharmacological and non pharmacologicalfminterventions,ispresentedbelow. Pharmacologicaltreatments Paracetamolandmildopioids(tramadol) ParacetamolcanbeconsideredforFMtreatment,althoughthereislittleevidenceofresponse.Accordingto EULAR,mildopioidssuchastramadolarealsorecommendedforthemanagementofFM(72). Nonsteroidalantiinflammatorydrugs(NSAIDs)andglucocorticoids Accordingtodifferenttrials,NSAIDsandglucocorticoidsareineffectiveandarethereforenotrecommended inthetreatmentoffm(72).however,mostfmpatientsarecurrentusersofnsaids.theiravailabilityand commonusebythegeneralpopulationexplaintheiruseinfmtreatment. 23
30 INTRODUCTION Antidepressants Tricyclic antidepressants, amytriptiline in particular, are among the beststudied and most effective pharmaceutical interventions in FM. Serotonin reuptake inhibitors (SSRIs) have mostly been less effective than tricyclic medications, but newer studies demonstrate some efficacy of SSRIs. A randomized, double blindcrossovertrialoffluoxetine(andssris),amytriptilineandplaceboinfmpatientsshowedthatboth were effective in decreasing FM symptoms, and that the two drugs given simultaneously were more effectivethaneitherdrugalone.finally,ametaanalysisoffmtreatmentwithantidepressantsshowedthat antidepressantstendedtoimprovesleep,fatigue,pain,andwellbeing,butnottriggerpoints(45). Anticonvulsants Gabapentinandpregabalin,drugsthatwereinitiallysynthesizedtotreatepilepsy,haveshownefficacyinthe management of painful diabetic neuropathy and postherpetic neuralgia. Their mechanism of action likely consistsofbindingtothevoltagedependentcalciumchannelinthecns,blockingtheinfluxofcalciuminto the neuron and thereby reducing excitatory neurotransmitter release. Gabapentin showed significant improvement in pain, FIQ score and sleep, and it was generally well tolerated. Pregabalin significantly decreasedpain,and,forhigherdoses,leadtosignificantimprovementinsleepquality. Dopamineagonists FM patients may benefit of dopamine agonists if they also have restless leg syndrome. Pramipexole improvedpain,fatigue,functionandglobalstatus. Corticosteroidsinfiltrations For common tendinitis such as epicondilitis or trocanteritis, that do not improve with oral treatments, infiltrationswithcorticosteroidswillimprovesymptoms. Sympatheticblockade Regional sympathetic blockade through stellate ganglion blockade, with local bupivacaine injection, has showntodecreasetriggerpointsandrestingpain,butitdoesnotofferapracticaltherapeuticoptionforfm (45). 24
31 INTRODUCTION NonPharmacologicaltreatments Cognitivebehaviouraltherapy Cognitive behavioural therapy is a process that examines a patient s way of reacting to experiences and attempts to restructure maladaptive coping habits into effective coping skills. CBT has been shown to be effective in reducing disability in most of FM studies (45). Emotional and cognitive factors (thoughts, memories,beliefs,expectationsandfears)interactwithhowapatienthandlessensoryinput,includingpain perception(70). Balneotherapy Heatedpooltreatmentorbalneotherapywasreportedtobeeffectiveinimprovingpainandfunction(72). Aerobicexercise Individuallytailoredexerciseprogrammes,includingaerobicexerciseandstrengthtrainingcanbebeneficial tosomepatientswithfm(72).aquagymnandpilatesarethemostrecommendedexerciseprogrammesfor FMpatients,astheyaremildanddon timplyanoverchargethatcouldworsenfatigue. RelaxationandTaichi TaichihasshowntoreducediseaseactivityasassessedbyFIQandqualityoflife(SF36)inarandomized controlledtrial(73). Lifestylechanges Improvingsleephygieneandeatingahealthydiethavealsoshowntobebeneficial. Alternativemedicine InspiteofthemanytreatmentsofferedintheInternet,thereislittleresearchonalternativemedicineand FM.Acupunctureeffectsarenotclear(70).DesperateFMpatientsmaysuffertheabuseofcheaterswith bizarretreatmentsnotbasedonmedicalevidencesuchasanalozone. Evidencesforabiologicalbasis In the eighties and nineties there were several hypotheses pointing at almost any possible system alteration/dysregulation,asbeingresponsibleforfmsymptomatology:frommuscle,totheadrenalsystem, theimmunesystem,asleepdisorderandthenervoussystem. 25
32 INTRODUCTION Nowadays, the most established hypothesis underlying FM etiopathogenesis is the existence of a dysfunction in pain processing. Since pain and tenderness are the defining features of FM, it is currently attributedtoanincreaseincentralpainprocessing.however,therearestillotherhypothesesthatclaimthat FM could be an endocrine or an immune mediated disorder, or even due to the dysfunction of the autonomic nervous system. We will focus on the evidences supporting central and peripheral nervous systemalterations,astheynowappearasthemoreplausibleandmoreevidencebasedones,butwewill alsosummarizetheexistingevidencessupportingtheotherhypotheses. Box6:Fibromyalgia:alittlebitofhistoryandcontroversy AlthoughitmayseemthatFMisamoderndisorder,itwasinfactalreadydescribedinthebeginningofthe twentiethcentury.thefirstreferencestopatientswhohavechronicpain,an aching,stiffness,areadiness tofeelmuscularfatigue,interferencewithfreemuscularmovement,andveryoftenawantofenergyand vigour, wasmadebyralphstockman,anedinburghpathologistin1904.justafewyearsearlier,william Osler, in the third edition of his text The Principles and Practice of Medicine, defined myalgia in similar terms.becauseofwidevariabilityinsymptomsinpatientswithpainwithnoapparentreason,therewas littleenthusiasmamongclinicalinvestigatorstoexaminethesepatientsmorecloselyuntil1939,whenlewis andkellgrenenlistedvolunteersandsubjectedthemtoinjectionsofmusclesandligamentswithhypertonic saline(whichisverypainfulintheinjectionregion).thesedeepinjectionscauseddiscomfortinamyotomal, orreferredpattern,asopposedtoadermatomalorradicularpattern.aresurgenceofinterestinfmwas introduced in the 1980s when several groups of investigators published data showing that patients with fibrositisorfibromyalgiahadparticulartendernessinatleast11of18defined tenderpoints notsharedby asymptomaticcontrolpatients.duringthe1990showever,ithasbecomeapparentthathealthypeoplehave tender points,thatfm patientsareoftentenderallovertheir bodiesandcouldbestbecharacterizedas having a low pain threshold, and that FM, the chronic fatigue syndrome, exposure syndromes and somatoformdisordershaveconsiderableoverlapintheirclinicalmanifestations. Centralnervoussystempainprocessingdysfunction Extensive research suggests that FM s chronic widespread pain has a neurogenic origin. First of all, FM patientspresentstructuraldifferencesinthe brain. Twostudies havedetectedadecreaseingreymatter volume,whichwasgreaterthanincontrolsandtheequivalentto9.5timesthenormallosswithage(74). Thishasbeenpostulatedtoberelatedtostress,deficitsincognitivefunctionandimpairedendogenouspain inhibition, as many of the grey matter loss occur in regions related to stress and pain processing. Other studieshavedemonstratedchangesinvolumeofdifferentbrainstructuressuchastheamygdala,cingulated cortexandhippocampus,asreviewedingracelyetal(75). Furthermore,thereareseveralevidencesofcentralsensitizationatvariouslevelsinthenervoussystem(44). Neuroimagingstudiessupportthis,showingthatFMisassociatedwithaberrantprocessingofpainfulstimuli 26
33 INTRODUCTION inthecns.fmristudiesofthebraindemonstratethatinpatientswithfmapainresponsecanbeproduced usingamuchlowerpainstimulusthanincontrols(68).underthesamestimulusintensity,severalareasof the brain (secondary somatosensory cortex, insula, and the anterior cingulated cortex) consistently show greateractivationinfmpatientsthanincontrolindividualswithfmri.spectimaginghasshownreduced blood flow in the right thalamus of FM patients (75, 76). FM patients present a widespread reduction in thermal and mechanical pain thresholds and greater cerebral laser evoked potentials after mechanical stimuli(4).heatpainthresholdsandcerebraleventrelatedpotentialsfollowingpainfulco 2 laserstimulation are also altered in subjects with fibromyalgia syndrome (77). Differential central pain processing has also beenobservedfollowingrepetitiveintramuscularproton/prostaglandine(2)injectionsinfemalefmpatients andhealthycontrols(78). Thepainseemstoresultfromneurochemicalimbalancesinthecentralnervoussystemleadingtoa central amplification ofpainperception.pethasshownareductionoflevodopamineuptakewithinthebrainstem, thalamus, and multiple elements of the limbic cortex (79), and a reduction of opioid μ receptors binding potentialinstructuresplayingaroleinnociception,suchasamygdala,cingulatedandnucleusaccumbens, (80).ThiscouldexplainthelackofresponsetoopioidsinFMpatients.Inaddition,MRShasdemonstrated differences in concentrations of glutamate and combined glutamate/glutamine within the insula and posteriorgyrus(81)infmpatientsascomparedtocontrols.furthermore,thesechangesinglutamatelevels havebeenassociatedtochangesinpainperception(80). Moreover,widespreadpaininfibromyalgiaisrelatedtoadeficitofendogenouspaininhibition,duetoan imbalance in descending pathways as proven by altered levels of neurotransmitters in the CNS (82). Noradrenaline (NE) and 5hydroxytryptamine (serotonin) (5HT) are key neurotransmitters in descending inhibitory pain pathways and they have a significant modulatory effect on peripheral and central pain processing.levelsofprimarymetabolitesofne,5htanddopamine(da)arereducedinpatientswithfm (83, 84). Another study found correlation between the levels of glutamine and aspargine (glutamate and aspartatemetabolitesrespectively)andthenumberoftenderpoints(85).fourdifferentstudieshavealso foundanelevationofsubstancep(aneuropeptide releasedinspinalfluidwhenaxonsarestimulated)in cerebrospinal Fluid (CSF) of FM patients as reviewed in (76). Nerve growth factor also has shown to be elevatedinthecsfoffmpatients(86). Finally, enhanced central pain processing of FM patients is maintained by muscle afferent input: a randomized,doubleblind,placebocontrolledstudyshowedtheimportantroleofperipheralimpulseinput inmaintainingcentralsensitizationinfm(87). 27
34 INTRODUCTION Disorderedstressresponseandendocrineorhormonalfactors Thehypothalamicpituitaryadrenal(HPA)axisalongwiththelocuscoeruleussympatheticnervoussystemis theprincipalsystemofstressresponseinthebody.thehpaaxishasbeenfoundtobedysregulatedinfm patients. Several studies using different physiological stress generators showed enhanced or normal response of adrenocorticotrophic hormone (ACTH) with elevated levels of cortisol in the evening as reviewedindifranco(84).becausethe5htsysteminfluencesthehpaaxis,someofthesefindingscould becausedbythereducedlevelsof5htinplasma.thesealterationsarecommonlydetectedinpatientswith adefinedcauseofchronicpain,suggestingthatthesechangescouldbeaconsequenceoffmsymptoms, ratherthantheircause(45).inanycase,fmstudiessuggestanimpairedstressabilitytoactivatethesystem, ratherthananoverallincreasedfunctionofthehpasystem. Autonomicnervoussystemdysfunction The balance between sympathetic and parasympathetic systems is essential to preserve homeostasis. Studies show that FM patients have increased sympathetic and decreased parasympathetic tones, as assessedby24hourselectrocardiogramregistration(reviewedinmartínezlavin(88)).thisdysautonomiais characterized by a persistently hyperactive sympathetic nervous system that is hyporeactive to stress, as provenbydifferentstudiesusingorthostatismandtilttabletestamongotherstressors.thesealterations explainmanyofthefmsymptoms,suchasintestinaldysfunction,orheartpalpitations,andsuggestthatfm couldbeasympathicallymaintainedpainsyndrome.however,asinthecaseofthehpaaxisdysfunction, thevastmajorityofthescientificcommunityconsidersthesealterationsaconsequenceoffm,ratherthana cause. Sleepdisorder ThedeltasleepalterationsfoundinFMhaveshowntointerferewithsleepfunction,causingnonrestorative sleep, fatigue and musculoskeletal pain (89). In fact, experimental studies with normal control patients undergoing delta sleep disruption showed FM like symptomatology (90). The question is whether these alterationsarecauseorcontributingfactorstofm.inthissense,ithasbeenproposedthattheimpaired slowwavesleepcouldbecausingthedecreaseingrowthhormonesecretionthatispresentinfmpatients (91).Also,sleepdisturbancescouldbeaffectingalterationsin5HT,NA,DAandmelatoninlevelsinFM. Muscle MicroscopicalexaminationofFMmuscleshowedmusclefibreswereconnectedbyanetworkofreticularor elastic fibres, which are not present in muscle from healthy individuals (92). Also, other studies have detectedchangesinintramuscularmicrocirculationandinmuscleenergymetabolismasreviewedin(13).as 28
35 INTRODUCTION mentionedbefore,somehypothesespointoutthatperipheralactivation,whichcouldtakeplacethrough these muscular changes, is necessary for the development of central sensitization and pain disinhibition mechanisms. Inflammatory/autoimmunedisorder Chronicconditions,suchascancerorautoimmunedisorders,oftenhaveassociatedmooddisorders.Ithas beenshownthatproinflammatorycytokines,suchasinterleukine(il)1andtumoralnecrosisfactor(tnf), couldbeinvolvedinthedevelopmentofsicknessbehaviour,includingfatigueandincreasedpainsensitivity (reviewed in Dantzer et al (93)). This, and the fact that synergistic neuroimmune interactions promote sensitization to pain and the development of chronic pain (3), point out at a possible role of an immune dysregulationinfmpathogenesis.antinuclearantibodies(ana)havebeenshownin11%to30%ofallfm patients,buttherisktodevelopconnectivetissuediseasesdoesnotseemtoincreaseinfm,anddetection ofanadoesnothaveapredictivevalue.variousstudieshaveevaluatedinterleukinlevelsinfmpatients, with contradictory results. Some of them showed increased levels, notably IL8, while others show decreased levels (reviewed in Dadabhoy et al and Di Franco et al (76, 84)). A recent metaanalysis (94) concluded that only IL6 plasma levels were higher in FM patients compared to controls. The different methodologiesusedtomeasuredifferentcytokinesindifferenttissuesmaterialsexplaininpartthislackof reproducibilityacrossstudies.aposteriorworkhasshownincreasedcsfandserumlevelsofil8,proving thattherelationshipbetweenilandfmisstillacontroversialnonresolvedissue. Finally,theroleoftheimmunesysteminFMpathogenesisisalsosupportedbysomegeneticstudiesthat linkhumanleukocyteantigen(hla)locuswithfmfamilialcases(46). Environmentalfactors SeveraltriggerfactorsforthedevelopmentandonsetofFM,suchassurgery,stressfullifesituations(Box7), infections,traumatisms,andbloodtransfusionhavebeendescribed.upto40%ofpatientsreportthatthe onsetofsymptomswasprecededbysometriggeringevent,whichmightbeeitherpsychologicalorphysical. FMdevelopmentwouldbefavouredbyavulnerablepsychosocialsetting.Experiencesofphysicalandsexual assaultinadulthoodshowedastrongassociationwithfm(95).inaretrospectivecasecontrolstudy,fmwas significantly associated to physical trauma (a fracture, surgery, miscarriage or childbirth, and a traffic or othertypeofaccident)sixmonthspriortofmonset(96).otherfactorsdescribedinfmriskarelowlevelsof vitamin D in women (97), certain infections (98), lower levels of education, unemployment, divorce and obesity. Finally,severalinfectiousagentshavebeenlinkedtothedevelopmentofFM.Duetothesimilaritybetween theviralinfectionsandfmsymptomatology,severalviralagents,inparticularhepatitiscandbandhuman 29
36 INTRODUCTION immunodeficiencyvirus(hiv),havebeenexploredinfmepidemiologicalstudieswithinconclusiveresults,as reviewed in (99). The most plausible explanation for these findings is that after these chronic infections, patientsmaydevelopsecondaryfm. Box7:Painandwar Duringtimesofwar,chronicpainandfatiguewithnormalphysicalexaminationshasapproachedepidemic proportions.comrow,inhistextarthritisandalliedconditions,notedin1944that alargepercentageof oursoldiersenteringthemedicalserviceofanarmygeneralhospitalwithsymptomssimulatingfibrositis havedevelopedtheseonapsychogenicbasisandthatthesesymptomscannotberelievedbyheat,massage andexercise,butareabolishedbydischargefromthearmy.thepersiangulfsyndromemaybeanother exampleofthis,because45%ofdeployedveterans,comparedto15%ofthosenotdeployedtothefirst PersianGulfWar,developedaconstellationofsymptomsincludingmuscleandjointpain,fatigue,memory problems,headaches,andgastrointestinalcomplaints(45).aspecificmechanismthatmaylinkvaccination againstbiologicalwarfareagentsandlaterfmsymptomshasbeensuggested(100).hotopfetalconcluded that among veterans of Gulf war there was a specific relation between multiple vaccinations during deploymentandlaterillhealth(101). Fibromyalgiagenetics Theresponsetopainfulstimulihasageneticcomponent,asheritabilityisestimatedbetween22%and55% (102).However,theexplorationofgeneticcontributiontopainresponseandchronicpainstatesissofar scarce.copynumbervariants(cnv)havenotbeenexploredinfmorinotherchronicpainconditions(103). Regardingsinglenucleotidepolymorphisms(SNP),onlyonepaingenomewideassociationstudyhasbeen performed,evaluatingpainlevelsafterthirdmolarextraction.thisonlyfoundassociationatonesnp,which was in linkage disequilibrium with a gene encoding a zinc finger protein, but this was not evaluated in a replicationcohort(104).regardingchronicpain,astudyevaluating3295snpsrelatedtopainresearchin 348 cases of chronic temporomandibular disorders and 1612 controls failed to detect a statistically significantassociationaftercorrectingformultipletesting(105).asimilarsituationoccursinregardwithfm: littleisknownaboutgeneticfactorsunderlyingthischronicpainstate. 30
37 INTRODUCTION FMhasageneticcomponent:familyandtwinstudies FM has a genetic component, as there are evidences of family aggregation. Several studies have tried to assesstheoccurrenceoffmamongfamilymembersoffmpatients.first,astudyin50parentsandsiblings offmpatientsshowedthat52%oftheindividualspresentedfmsymptoms(106).anothershowedahigher prevalence of FM among offspring of FM mothers (28%) (107). Furthermore, this increased prevalence amongfamilymembersoffmpatientswasshowntobehigherinbloodfamilymembersthaninhusbandsof FMpatients(108),supportingthatgeneticfactorsmustprevailoverenvironmentalfactorsindetermining FMsusceptibility.Finally,geneticcontributiontoFMwasalsosupportedinastudy,showingthatFMfirst degreerelativeshadarelativeriskof8.5todevelopfm,ascomparedtorheumatoidarthritisfirstdegree relatives(109). SeveraltwinstudieshavealsobeenperformedinordertodissectoutFMgeneticfactors.First,astudyin11 yearoldfinnishtwinswithwidespreadpainshowedthatgeneticfactorsdidnotaccountfortwinsimilarity for widespread pain (110). However, pain was not chronic in these individuals, and therefore FM criteria were not fulfilled. Later, a twin study in the Swedish population showed that chronic widespread pain concordancewashigherinmonozygoticthanindizygotictwins(0.29versus0.16)andestimatedthatgenetic factors accounted for 48% to 54% of the total variance (111). Finally, a twin study in idiopathic chronic fatigueshowedalsoahigherconcordanceinmonozygotic(55%)thanindizygotictwins(19%).inconclusion, studiesperformedsofarsupporttheimportanceofgeneticfactorsinsusceptibilitytofmandchronicpain states. Fibromyalgiageneticstudies MostgeneticstudiesperformedsofarinFMhavebeencandidategenestudies.Thesestudieshavenotbeen abletoestablishacleargeneticassociation,astheypresentseverallimitations:mostofthemhave been performed in small cohorts; associations are not FM specific as they are shared with other disorders, in particular FM psychiatric comorbidities; they often present borderline significance without correction for multiple testing; and attempts to replicate them have shown contradictory results. So far, studies have focusedongenesrelatedtohlaandneurotransmitters.inastudy,where18fmpatientsand23controls were typed for HLA alleles, 67% of FM patients presented HLADR4 versus 30% of controls. Given the reducednumberofsamplesincludedinthestudy,fmhladr4frequencywasalsocomparedtoalarger cohort of controls (n=1676) and the difference remained statistical significant (112). However, not unexpectedly,asimilarstudy,performedin60fmcasesand159controls,failedtoreplicatethesefindings (113).Anotherstudy,investigatingtheHLAregion,showedgeneticlinkagetothisregioninananalysisof fortycaucasianfamilieswithfm,inwhichhlawastypedfora,banddrb1alleles(46).nevertheless,none 31
38 INTRODUCTION ofthesehlaassociationshavebeenreplicatedinlargerfmcohorts.ontheotherhand,variousstudieshave examinedthepossibleroleofneurotransmittersintheserotoninergic,dopaminergicandcatecholamiregic systemsinfm.theanalysesofserotonintransporterandcatecholomethyltransferasevariantsinfmare goodexamplesthatillustratethelimitationsoffmgeneticstudiesperformedsofar. ApossibleassociationofFMwithapolymorphismintheregulatoryregionoftheserotonintransportergene hasbeensuggested.thereisacommonvariantleadingtoashorterformofthepromoter,whichhasbeen associated to several FM psychiatric comorbidities, including affective and anxiety disorders. This variant showedahigherfrequencyinfmindividualsinrespectwithcontrols(62fmversus110controls,31%of shorthomozygousinfm,and16%incontrolsp=0.046),althoughitdidnotreachstatisticalsignificanceat thealleliclevel(114). Itwasaborderlineassociationthatwasreplicatedin anotherstudyconfirmingthe association(115),butnotinathirdonewhenanalysingfmpatientswithoutpsychiatriccomorbidities(116) (Table1).OtherserotonintransportervariantshavebeenfurtherexploredinFM,findingassociationofa SNP(rs6313)toFMbothattheallelicandgenotypiclevels,butagaininareducedsamplesetof168FM versus 115 controls and without replication. This variant had also been previously associated with schizophreniaandmigraine(117).itsassociationtofmwasnotreplicatedinasecondstudy(118)(table2). Acommonvariantaffectingcatecholamine smetabolismonthemodulationofresponsestosustainedpain in humans has also been explored in FM cases. Individuals homozygous for the Met158 allele of the catecholomethyltransferase(comt)polymorphism(val158met)showdiminishedregionalμopioidsystem responses to pain compared with heterozygotes (119). Several studies have evaluated this variant in FM cases with contradictory results, the one including more samples (115), only being able to detect an associationbetweenfmandthenumberoftenderpoints(table3).ametaanalysisshowednoassociation whenconsideringthree(vargasetal(120),tanderetal(121)&cohenetal(115))fmstudiesperformedon thisvariantuntilmarch2010(122).althoughanulteriorstudyshowedthatthefrequencyofcomtvariants associatedwithlowenzymeactivitywassignificantlyhigherin113fmpatientsversus65controls(123),the reducednumberofsamplesincludedandthemultiplecontradictoryresultsprovethatthisassociationstill hastobeestablished. Inaddition,geneticvariantsinthedopaminergicandadrenergicsystemshavealsobeenexploredinFMwith thesamelackofconsistencyintheresults,asreviewedinleeetal(122).oneworkperformedinmexican casesandcontrolswasabletofindanassociationtoa2adrenergicreceptorvariant,andreplicateditinan independentspanishdataset(124).however,theassociationdidnotpasscorrectionformultipletesting(as severalvariantsweretested),andthesizeofthestudycohortswassmallfortheexplorationofacommon variantinacomplexdisorder. 32
39 INTRODUCTION Finally,sofaronlyonestudyhasattemptedtoexplorethegeneticcontributiontoFMinagenomewide manner. Over 3200 SNPs in 350 genes implicated in pain transmission, inflammatory responses, and in influencingmoodandaffectivestatesassociatedwithchronicpainconditions,weregenotypedin496fm casesand348controls.however,thestrongestassociations(gabrb3(gammaaminobutyricacidareceptor, beta3)andtaar1(traceamineassociatedreceptor1)snps)didnotreplicateinindependentcohorts(105). Inconclusion,themanygeneticstudiesperformedsofarinFMhavenotbeenabletoidentifyitsgenetic component.asummaryincludingmostofthegeneticvariantsthathavebeenexploredinfmispresentedin Table4. Table1:Evaluationofapolymorphisminthepromoterregionoftheserotonintransporter (5HTTPRLR)geneinFMcasesshowcontradictoryresultsindifferentstudies. 5HTTPLR Short/longallele FM CONTROLS pvalue (Offenbaecher(114)) (Cohen2002(115)) * * (Gursoy2002(116)) NS (Potvin2010(125)) NS NS,nonsignificant.*jointanalysis * Table2:Evaluationofrs63135HT2A(serotonin2A)receptorgeneinFMcasesversuscontrols. 5HT2Areceptor rs6313 FM CONTROLS pvalue (Bondy1999(117)) (Gursoy2001(118)) NS (Tander2008(121)) NS NS,nonsignificant. Table3:EvaluationofcatecholOmethyltransferase(COMT)Val158Metpolymorphisminseveral FMcohorts. COMT rs4680(val158met) FM CONTROLS pvalue (Gursoy2003(126)) (Vargas2007(120)) (Tander2008(121)) NS NS (Cohen2002(115)) <0.05tenderpoints (Lee2012(122)) (Martinez2012(123)) <0.05 NS,nonsignificant 33
40 INTRODUCTION Gene Variant Table4:SummaryofgeneticvariantstestedinFMcohorts Number Individuals FMControls pvalue AAT E342K NS (127) ADRA1A (rs574584,rs138914,rs573542) (124) ADRB2 (rs ,rs ) (replicate) ADRB3 rs NS (124) COMT (rs6269,rs4633,rs4818,rs4680(val158met) rs (replicate) (replicate) NS NS Ref (124) (120) (120) (123) DAT VNTR NS (127) DRD3 Ser9Gly 3736 NS (125) DRD4 Exon3VNTR (128) enos Glue298Asp 9679 NS (129) GABR3 rs (105) GBP1 rs (105) HLA HLADR HTR2A rs NS (121) HTR3A HTR3B Exon142C/T Exon297G/A Exon3IVS3 Exon6576G/A Exon91377G/A Exon delAAGExon4IVS4 Exon5386A/C Exon6IVS6+72A/G NS NS NS NS NS NS NS NS NS (112) (130) (130) (130) (130) (130) (130) (130) (130) (130) IL4 Intron3VNTR NS (131) MAOA PromoterVNTR 941G/T NS (132) (131) MAOB Intron13G/A NS (132) SNC9 14SNPstested(rs ) (133) TAAR1 rs (105) TACR1 1354G/C NS (127) AAT, alpha1 antitrypsin; ADRA1A, adrenergic receptor alpha1a; ADRB2, adrenergic receptor beta2; ADRB3, adrenergic receptor beta3; COMT, catecholomethyltransferase; DAT, dopamine transporter; DRD3, DopamineD 3 receptor;drd4,dopamined 4 receptor;enos,endothelialnitricoxidesynthase;gabrb3,gammaaminobutyricacida receptor, beta 3; HTR2A, serotonin 2A receptor; HTR3A, serotonin 3A receptor; HTR3B, serotonin 3B receptor; IL4, interleukin4;maoa,monoamineoxidasea;maob,monoamineoxidaseb;taar1,traceamineassociatedreceptor 1);TACR1,substancePreceptor;VNTR,variablenumbertandemrepeat.NS,nonsignificant. 34
41 INTRODUCTION GENOMICSOURCESOFHUMANVARIATION Thehumangenomesequencewascompletedin2001(134,135).Thiswasthestartofanewerainhuman genetics,astheavailabilityofareferencesequenceconstitutedanincredibletoolforresearchersinorderto evaluate genetic variants and their contribution to disease susceptibility. It was claimed that genetic differencesbetweenindividualsaccountedforlessthan0.1%ofthe3millionnucleotidesinthehumandna sequence, and 1.4 million single nucleotide polymorphisms (SNPs) were detected. In the last years many effortshavebeenundertakeninordertoidentifyandinterprettheconsequencesofthevariantspresentin the human genome. These variants refer to several sources of variation, including SNPs, copy number variants, small polymorphic indels (one or two nucleotides), VNTRs, among others. Nowadays, the most commonly studied variants in relation with susceptibility to complex diseases are SNPs, and, to a lesser extent,copynumbervariants(cnv),whicharedescribedbelow. Singlenucleotidepolymorphisms ASNPisavariationinthehumangenomeaffectingasinglebasepairthatispresentinatleast5%ofagiven population.snpscanbeclassifiedinexonic,splicingandintronic,andexonicsnpsarefurtherdividedinto nonsynonymous and synonymous depending on whether they modify or not the amino acid sequence. MostSNPsarebiallelic.Nowadays,newsequencingtechnologieshaveallowedtheidentificationofover15 millionsnpsinthehumangenome(136). TheHapMapprojectwaspioneerintothedetectionofthiskindofvariants(Box8).Itwasanessentialtoolto develop many association studies, since maps of linkage disequilibrium helped in the selection of the variantstogenotype.beforethedevelopmentofnextgenerationsequencingtechnologies,itconstituteda keyresourceforresearcherstofindgeneticvariantsaffectinghealth,diseaseandresponsestodrugsand environmentalfactors. SNPsgenotyping Inthelastdecade,severaltechniqueshavebeendevelopedinordertoevaluateSNPsatdifferentscales: fromasinglevariant(kaspartaqmantechnology)todozensandhundredsofvariants(snplex,massarray, and Veracode technology) and even millions of variants with array based platforms for genome wide association(gwa)studies(table5).recently,somearraysareincludingvariantswithafrequencybelowthe polymorphic 5%level:variouscompanieshavedevelopedtheexomearrays,includingthousandsofrare codingvariantsidentifiedthroughnextgenerationsequencingapproaches.alsospecificarrayssuchasthe immunochip,whichincludesalltheassociationsdetectedingenesrelatedtotheimmunesystem(137)or Axiom GenomeWideHumanOrigins1Arraywhichisusedinpopulationgenetics.Finallyitisalsopossible todesigncustomarraysevaluatingthousandsofsnps. 35
42 INTRODUCTION Box8:TheHapMapProject TheinternationalHapMapProject( ordertobuildahaplotypemap(hapmap)ofthehumangenomethroughthestudyofsnps.itwasfocused originallyin270individualscomingfromthreepopulations:90samples(30triosoftwoparentsandachild) fromausutahpopulationwithnorthernandwesterneuropeanancestry(samplescollectedin1980bythe Centre d Etude du Polymorphisme Humain (CEPH)), 90 samples (30 trios of two parents and a child) collected from Yoruba people in Ibadan, Nigeria, 45 unrelated individuals from Tokyo, Japan and 45 unrelated individuals from Beijing, China. Currently, three phases have been completed including the genotypingofmorepopulationsandsnps. Four versions of a given chromosome are mostly identical and they only differ at a few bases, the SNPs, whichare biallelic (a). Haplotypes (b) are defined by the particular combination of alleles at adjacent locations (loci) on the chromosomethataretransmittedtogether.thegenotypingoftagsnps(c),whichareinhighlinkagedesequilibrium with the nearby SNPs, allows the identification of each of the four haplotypes without having to genotype all the variantsintheregion.(takenfromtheinternationalhapmapproject(138)). 36
43 INTRODUCTION Table5:SummaryofsomeofthemostcommonlyusedtechnologiestoevaluateSNPs. Name Variantsexplored AmountofDNA Technology Taqman 1 10to20ng SNPGenotypingAssaysincludetwoallelespecificprobesand apcrprimerpairtodetectspecificsnptargets.probes containdistinctfluorescentdyesandincludeanonfluorescent quencherthateliminatesbackgroundfluorescence.detection isachievedwithproven5 exonucleasechemistrybymeansof exonucleasecleavageofthe5 allelespecificdyewhich generatesthepermanentassaysignal(figure9).following PCRamplificationanendpointreadisperformedona thermocycler. SNPlex Upto48 37ng Allelespecificoligonucleotideprobes(ASO)andlocusspecific probeshybridizetothegenomictargetsequence.linkerswith universalpcrprimer bindingsequencesareligatedtothe probes.auniquezipcodesequenceisattachedatthe5endof thegenomicequivalentsequencewithineachaso.thisis followedbyuniversalpcrreactiontoamplifyligation products.biotinlabeledpcrproductsarelateroncapturedin streptavidincoatedmicrotiterplatesandfinallydetectedby capillaryelectrophoresis(139). Massarray Upto15 Upto36withSBE iplexgold 150ng(30μlat5ng/μl) 300ng(30μlat10ng/μl) Differentsizesproductsdependingontheallelearegenerated andsubsequentlydetectedwithmassspectrometry (MaldiTof). analysis/applications/snpgenotyping Veracode (array based) 48to ng(15μlat50ng/μl) Glassmicrobeads(240micronsinlengthby28micronsin diameter),withuniversaloligonucleotidesareused.whena laserpassesthroughthebead,itisdefractedbythe holographicelementscreatingacodeimagedetectedbyaccd camera. ( mn) GWAarray (array based) to2.5millions 500ngto1μg AffymetrixtechnologyDNAisdigested,amplifiedbyaPCR, fragmented,endlabelledandhybridizedagainstthearray containingtheprobes(oneforeachallele)totestsnps. Illuminatechnology(Infiniumtechnology).AnunlabeledDNA fragmentishybridizedandsubsequentenzymaticsinglebase extensionwithalabellednucleotideisperformed.dualcolor fluorescentstainingallowsthelabelednucleotidetobe detectedbyillumina siscanimagingsystem,whichidentifies bothcolorandsignalintensity. 37
44 INTRODUCTION Figure9:TaqmanSNPgenotypingsystem (fromhttp://www3.appliedbiosystems.com/cms/groups/mcb_marketing/documents/generaldocuments/) Genomewideassociationstudies Agenomewideassociationstudy(GWAS)isanexaminationofmanycommongeneticvariantsindifferent individuals to test if any variant is associated with a given trait. GWAS typically focus on associations betweensnps andtraitsordiseases.incontrasttomethods,whichspecificallytestoneorafewgenetic regions,gwasinvestigatetheentiregenome.theapproachisthereforesaidtobenoncandidatedriven,in contrasttogenespecific,candidatedrivenstudies.gwasidentifysnpsthatareassociatedwithadisease, but cannot, on their own, specify which genes are causal. SNPs showing the strongest associations are genotypedinareplicationcohortinordertoconfirmtheassociation.thisisperformedwiththedifferent genotypingplatformsexisting. The first strikingly successful GWAS was published in 2005 and investigated agerelated macular degeneration. It found two SNPs which had a significantly altered when comparing with healthy controls (140).Asof2013,GWASincludehundredsorthousandsofindividuals;morethan1,400humanGWAShave examinedover200diseasesandtraits;andover8,000snpassociationshavebeenfound(figure10).several GWAShavereceivedcriticismforomittingimportantqualitycontrolsteps,renderingthefindingsinvalid,but modernpublicationsaddresstheseissues.sincehundredsandthousandsofsamplesareused,ensuringthe homogeneityandthesamepopulationaloriginforbothcasesandcontrolsisamatterofconcern.andthis crucialaspectofthesekindsofstudiesisbecomingmoreandmoreimportantsinceitisnecessarytomerge differentstudiesthroughmetaanalysesinordertogetenoughpowertodetectvariantswithsmalleffect. Severalmethodshavebeenproposedtominimizethecostsofgenomewideassociationstudies:usingpools ofcasesandcontrols,sharingcontrolsacrosspopulations.ontheotherhandanaccurateconsiderationof 38
45 INTRODUCTION epidemiologicalcharacteristicsofthestudypopulationandtheselectionofthemosthomogenoussamples possiblecanincreasethepowerorreducecostsunderavarietyofconditions(141).theparadigmofthisis the success in the macular degeneration GWAS mentioned above (140), in which association to the complementfactorhgenewasdetectedwiththeinclusionofonly96casesand50controls.thiswasdueto the accuracy in the diagnosis, the homogeneity of the cases, and the fact that the association was very strongwithalargeeffect(oddsratioof7.4). The Welcome trust Case Control Consortium (WTCCC) was the first to release large scale GWAS where thousands of SNPs were genotyped in complex disorders (Box 9). It was the start of the Genome Wide Association(GWA)studiesera,withtheuseofverylargecohortsofcasesandcontrols.Itconstitutedagreat tool for researchers around the world, not only in terms of genotype data produced but also in the developmentofalgorithmsandstatisticalanalysismethodsforgwastudies. Figure10:Chromosomaldistributionofthehitsofthe1617GWASpublishedthroughJuly
46 INTRODUCTION Imputation Genotype imputation is the process of predicting genotypes at positions that are not directly typed in a dataset(142).thisisdonebasedonlinkagedisequilibrium(ld)valuesbetweenthegenotypedsnpsandthe SNPs identified in a reference panel. The most commonly used reference panels are HapMap and 1000genomes. The genotypes are imputed with uncertainty and a probability distribution over all three possiblegenotypesisproduced(figure11).thisuncertaintyistakenintoaccountinassociationanalysisof theimputeddata.imputationcanbeusedforfinemappingofsignalsdetectedfromdirectgenotypingorto capture new associations. The development of imputation methods has enabled the capture of more variants without directly genotyping them. Around 10 million SNPs can be imputed by using reference panels,sothat,thenumberofmarkersconsideredforassociationisconsiderablyincreased.ifthisshould modifystatisticalsignificancethresholdisstillamatterofconcern.finally,manyfactorssuchassamplesize should be considered as potential interfering factors in the imputation process, based on probabilistic genotypes. Figure11:Overviewiftheimputationprocess(figuremodifiedfromMarchinietal)(142). 40
47 INTRODUCTION Box9:TheWelcomeTrustCaseControlConsortium TheWellcomeTrustCaseControlConsortium(WTCCC)( groups across the UK which was established in The WTCCC aims were to exploit progress in understanding of patterns of human genome sequence variation along with advances in highthroughput genotypingtechnologies,andtoexploretheutility,designandanalysesofgenomewideassociation(gwa) studies. The WTCCC has substantially increased the number of genes known to play a role in the development of some of our most common diseases and has, to date, identified approximately 90 new variantsacrossallofthediseasesanalysed.in2007,thewtcccpresentedagwasof14000casesofseven common diseases and 3000 controls (143). WTCCC2 performed genomewide association studies in 13 disease conditions analysing over 60,000 samples: Ankylosing spondylitis, Barrett's oesophagus and oesophagealadenocarcinoma,glaucoma,ischaemicstroke,multiplesclerosis,parkinson'sdisease,psychosis endophenotypes,psoriasis,schizophrenia,ulcerativecolitisandvisceralleishmaniasis. WTCCC3isperforminggenomewideassociationstudiesinthefollowing4diseaseconditions:primarybiliary cirrhosis,anorexianervosa,preeclampsiainuksubjects,andtheinteractionsbetweendonorandrecipient DNArelatedtoearlyandlaterenaltransplantdysfunction.TheWTCCC3willalsocarryoutapilotinastudy ofthegeneticsofhostcontrolofhiv1infection.over40,000samplesarebeinganalysedusingtheillumina 660Kchip.TheWTCCC3usesthe6,000controlgenotypesgeneratedbytheWTCCC2. FromSNPsgenotypingtosequencing Intherecentyears,nextgenerationsequencingtechnologieshavedeterminedthebeginningofanewerain humangenetics.theincreaseinsequencingefficiencyandproportionaldecreaseincostsisenablingtheuse of massive sequencing in research projects, and therefore changing the scenario in the evaluation of genomicvariants(mostlysinglebasesnpsorrarevariants)(figure12).sequencingisreplacinggenotyping as the first step of the evaluation of variants linked to disease, since it allows not only the detection of common known SNPs but also the identification of novel mutations. Especially since the development of exomesequencing,manygeneticstudiesarenowbeingperformedbasedonthesetechniques,mostlyfor the study of Mendelian disorders, as the identification of causative genetic variation in these types of disordersismucheasier.thesetechnologiesarealsobeingusedforcomplexdiseases,buttheirapplication intocomplexdisorderswillneedthedevelopmentofstatisticaltestsapplicabletorarevariants.the1000 genomesproject(box10)hasconstitutedthelargestprojectsofartoapplynextgenerationsequencing.it can be defined as the equivalent of the HapMap project with the use of next generation sequencing. It combines both whole genome sequencing at different depths of coverage and exome sequencing in individualsfromdifferentpopulations.allthedataproducedbythisprojectcanknowbeusedasareference for researchers around the world. For instance, data from the 1000 genomes project is being used as a 41
48 INTRODUCTION referencepanelforgwasimputation,andalsototakeintoaccountvariantfrequenciesinperformingdata analysis for exome studies in several disorders. In a decade, the reference genome has moved into 1000 references.thisperfectlyillustratestherevolutionthatislivinghumangenetics. Figure12:EvolutionoftheDNAsequencingtechnologiesoverthepast30yearsand subsequentimprovementintherateofsequencing(takenfromstrattonetal(144)). Box10:The1000genomesproject Thegoalofthe1000GenomesProject( frequencies of at least 1% in the populations studied. To this purpose, low coverage whole genome sequencing of 179 individuals from four populations, highcoverage sequencing of two trios and exome sequencing of 697 individuals of seven populations have been performed in the pilot phase (The 1000 Genomes Project Consortium). These three experimental approaches are complementary as they provide differentinformation.thetriodesignallowsanaccuratediscoveryofmultiplevariantsacrossthegenome withmendeliantransmissionhelpingingenotypeestimation,inferenceofhaplotypesandqualitycontrol. The lowcoverage project identifies shared variants on common haplotypes. Finally, the exome design enablesaccuratediscoveryofcommonandrarevariantsbutonlyintheexome.atthetimeofwriting,2500 sampleshadbeensequenced. CopyNumberVariants In the last years, several publications have analyzed genomic structural variation, including insertions, deletions,translocationsanddeletionsofgeneticmaterial,asa new majorformofpolymorphism(145). Although the existence of structural variation has been known for a long time, this type of variationwas consideredtoconsistmostlyofrareindividualevents,andtheirrealimportanceremainedunknownuntil 42
49 INTRODUCTION the last few years. One type of structural variation are copy number variants (duplications, insertions, deletions):thesewereinitiallydefinedasdnasegmentslargerthan1kbanduptoseveralmegabases(mb) thatcanbefoundinavariablenumberofcopiesinthegenome.thissizerestrictionisnolongerconsidered in CNV definition, as thehigher resolution of the techniques has allowed the detection of smaller events (146). CNVs can be simple or complex, where the fragments duplicated vary in size between individuals, involvingseveralsubregions(figure13).over15%ofthehumangenomemaybeaffectedbycopynumber variation(147,148).accordingtothedatabaseofgenomicvariants( November2012,291,801CNVshadbeendescribedinthehumangenome(Figure14).Severalmechanisms areinvolvedinthegenerationofcnvs.inmostcases,copynumbervariantsaregeneratedbynonallelic homologous recombination (NAHR), but other mechanisms involve nonholomogous endjoining, fork stalling and template switiching (FoSTeS)/ microhomologymediated rearrangements (MMBIR) (149) and Line1mediatedretrotranspositionasreviewedinMalhotraetal(150)(Figures15and16). Figure13:Typesofstructuralvariants.CNVsincludedeletions,insertions,tandem duplicationsandunbalancedtranslocations(withlossorgainofgeneticmaterial). 43
50 INTRODUCTION Figure 14: Chromosomal distribution of CNVs identified by November Blue bars indicatereportedcnvs;redbarsindicatereportedinversionbreakpoints;greenbarsto theleftindicatesegmentalduplications(takenfromhttp://projects.tcag.ca/variation/). Figure 15: Mechanisms underlying CNV formation 1 (figure taken from Malhotra et al (150)).A)Nonallelichomologuousrecombination(NAHR)occursoverflankingsegmental duplicationsresultingindeletionandduplication.b)innonhomologousendjoining(nehj) doublestrandbreakarerepairedthoughseveralroundsofenzymaticactivitythatcanlead toanaccuraterepair,deletionsofdifferentsizesand,toalessextent,insertions. 44
51 INTRODUCTION Figure 16: Mechanisms underlying CNV formation 2 (Figure taken from Malhotra et al (150)).A)FoSTeS/MMBIR:whenareplicationforkencountersanick(shadowarrow),one arm of the fork breaks. The 3 singlestrand end of laggingstrand invades the sister leadingstrandguidedbyregionsofmicrohomology(mh)forminganewreplicationfork. Whether the template switch occurs in front of or behind the position of the original collapse determines a deletion or duplication. B) LINE1 retrotransposition. L1 is transcribedandtranslatedleadingtoal1ribonucleoprotein(rnp).l1rnpistransported to the nucleus and retroransposition occurs by target site primed reverse transcription (TPRT).DuringTPRTL1endonucleaseactivitynicksgenomicDNAexposingafree3 that servesasaprimerforreversetranscriptionofthel1rna.thisresultsintheinsertionofa 5 truncatedl1copyflankedbytargetsiteduplications.orf,openreadingframe. DespitemostCNVsarethoughttobebenignpolymorphisms,severalstudieshavealreadyprovenCNVsmay haveapathogenicroleintheaetiologyofdisorderssuchascharcotmarietoothneuropathytypei(151) and they have also been shown to contribute to complex diseases susceptibility such as HIV, Crohn s Disease,SystemicLupusErythematosusorpsoriasis(152155).However,wehavetotakeintoconsideration that the largest attempt to evaluate CNV role in complex disorders, in which acgh from 16,000 cases of eightcommondisordersand3,000controlswereanalyzed,failedtodetectanyassociation(156).themost obviousexplanationfortheeffectofthiskindofvariantscanbethealterationofgenedosage,buttheyalso maydisruptgenes,uncoverdeleteriousalleles,interrupttransvectioneffectsorhavepositionaleffectson theinternalorsurroundinggenes,suchasdistancingagenefromitsregulatoryregion(157). 45
52 INTRODUCTION Severalapproachesareusedinordertostudythiskindofpolymorphismsatagenomewidescale.Themost direct analysis consists of array comparative genomic hybridization (acgh), which allows for detection of gains or losses of genetic material in comparison to a reference sample. Also, given the amount of information generated by SNP arrays for whole genome association scans, several algorithms have been developedtoidentifycnvsbasedonintensityevaluation.finally,newmethods,suchaspairedendmapping (158) combine highthroughput sequencing and computational analysis in order to detect structural variation.inanycase,theresultsobtainedbythesemethodshavetobevalidatedwithquantitativemethods at individual or multiplex loci, such as quantitative realtime PCR, multiplex ligationdependent probe amplificationorfish. WholegenomemethodstodetectCNVs acgh InaCGH,DNAfromatestsampleandnormalreferencesamplearelabelledwithdifferentfluorophores,and hybridizedtoanarraycontainingseveralthousandprobesacrossthegenome.thefluorescenceintensityof thetestandofthereferencednaisthenmeasured,tocalculatetheratiobetweenthemandsubsequently the copy number changesforaparticularlocation inthe genome (Figure17).Adyeswapexperiment,in which the fluorophores are exchanged between the test and the control sample, is used in order to eliminatethefluorophorebias. Initially,aCGHwasperformedwithbacterialartificialchromosomes(BAC)thathadaresolutionofabout46 kb (159). Currently, the arrays used are more dense: for example, Agilent 2X400K microarray has about 420,00060bplongprobeswithamedianprobespacingof1,1KBandanaverageprobespatialresolutionof 6.6kb,coveringtheentiregenome,with84.8%ofCNVs<10kbbeingcoveredbyatleastoneprobe.Thishas allowedthedetectionofsmallerevents. 46
53 INTRODUCTION Figure17:aCGHexperiment.ThesameamountofcaseDNAandcontrolDNA are used, labelled with different fluorfores and hybridized against an array. RegionsshowingdifferentratiosoftheintensitiesareregionsvaryinginCNV. Astheresultsobtainedarerelativetothecontrolsample,theselectionofthecontrolsampleisimportant.In most studies of human CNVs, DNA from a male of European ancestry, NA10851, has been used as a reference for acgh (160). For this reason, Ju et al performed wholegenome sequencing and acgh of NA10851 in order to characterize its CNVs (160). A strategy used to minimize reference bias,istopool samples.byhybridizingapoolofcasesagainstapoolofcontrols,interindividualdifferencesmaybediluted anddifferencesincopynumberduetodiseasemaybeenhanced.thisapproachwassuccessfulindetecting anewcnvassociatedwithpsoriasissusceptibility(155).althoughpooleddnamayresolvereferencebiases it may also decrease the power of CNV detection in highly polymorphic regions (160) and lead to false positiveresults. SNParraysalgorithms:PennCNV CNVscanalsobeinferredfromSNPbasedarrays.Severalalgorithmshavebeendeveloped,usingBiallele frequency(baf)andlogrratio(lrr)todetect,inagivensample,regionsthatshowanincreaseordecrease ingenomicmaterialinrespecttotheothers.logrratioistheratioofintensityofthetwoallelesofeach SNP compared to the background total fluorescent intensity, and the BAF is the relative ratio of the fluorescent signals between the two probes/alleles at each SNP position. PennCNV (161) is possibly the widestusedalgorithmforcnvidentificationfromsnparraydata.itusesahiddenmarkovmodelalgorithm thatintegratesbothlrr andbaf toinfercnvsleadingtothe identificationofsixdifferentcopynumber 47
54 INTRODUCTION states(figure18).thisofferstheadvantagethatinformationisproducedfromalreadygenerateddata,but itsprincipallimitationisthattheresolutionandreliabilityofthecnvgenotypeislessthanthoseofferedby acgh. Figure 18: PennCNV allows the identification of six copy number statesbasedonlrrandbaf(figurestakenfromwangetal(161)). TheoverlapbetweenthedifferentexistingsoftwaretodetectCNVsfromSNParraydataispoor(Table6) (162).Forthisreason,inordertoidentifypossibleCNVeventsrelatedtodisease,manyauthorsproposeto implementseveralalgorithmsandonlyconsiderforvalidationtheoverlappingcnvs(163). Table6:ComparisonofSNParraybasedsoftwareforCNVprediction(takenformWinchesteretal(162) 48
55 INTRODUCTION Highthroughputsequencing:pairedendandexomealgorithms The development of Next Generation Sequencing (NGS) technologies, has lead to the implementation of different algorithms in order to evaluate CNVs (164) from NGS data. In particular, pairend sequencing is basedonthesequencingofadaptersligatedtobothendsofinsertsofdifferentsizes.adapterssequences areafterwardsmappedtothereferencegenome,allowingthedetection,withhighresolutionofinsertions, deletions and inversions (158). Also algorithms that combine the use of depth of coverage and distance between adapters sequence data are used to define regions varying in copy number, both in exome and wholegenomesequencingdata.whencompletelydevelopedandvalidated,thesemethodswillofferthe highestresolutionintermsofbreakpointsdefinitionandcopynumberstate. Validationofwholegenomemethods Various validation strategies have been applied to subsets of putative variants in each of the discovery reports.theseincludedfishofmetaphase,interphaseorfiberchromosomesusingvariousclonesorpcr amplified molecules; PCR or quantitative PCR (qpcr) for allele loss or quantitative variation; multiple ascertainment, whereby considerable weight was given to whether or not a putative variant was seen in more than one individual or had been reported in previous studies; array CGH to validate computational screening results, or for finest resolution of BACscreening results by oligonucleotide arrays; sequence analysisoffosmidinsertstoconfirmcallsandtoassesssomediscordantones;allelespecificfluorescence intensitiesandfamilialclustering(165). MultiplexPCR This validation approach for acgh results can only implemented in the cases of insertiondeletion polymorphismswithonlythreepossiblestates:0(homozygousdeleted),1(heterozygous)or2(homozygous non deleted) copies (Figure 19). First, in order to identify the breakpoints of a CNV region detected by a screeningmethod,alongrangepcrcanbeperformed.thedevelopmentofnewtechnologieswithhigher resolution has facilitated this analysis, by performing a computational analysis using the coordinates resultingfromtheacghorsequencingresultsandcheckingforknowncnvsintheavailablecataloguesof structural variants (Database of genomic variants among others). PCR primers are designed outside the deletedregion,whichwillonlyamplifyinadeletedsamplewhenthesizeofthecnvislargerthan34kb,or forsmallerinsertion/deletionswillgiveasmallerthanexpectedfragmentforthedeletedallele.sequencing thispcrproductandblastingittothereferencegenomewilldetectthebreakpoints.inparticular,inthe caseofthe2x400karray,sincethedesignofthisarrayisbasedonthecommoncnvsdetectedbyconradet al(146),thepcrprimerdesignandbreakpointdetectioniseveneasier,byusingthesupplementarydata fromthepaper,especiallyifthecnvhasbeenexperimentallyvalidated.oncethebreakpointsaredefined,it 49
56 INTRODUCTION ispossibletodesignamultiplexassay,usingtwosetsofprimers.apairofprimerslocatedinsidethedeleted region, which will only amplify in the nondeleted allele, and another set of primers located outside the deleted region, which will only amplify in the deleted allele. This will allow the genotyping of cases and controlsinareplicationcohortinordertoconfirmifthedifferenceincnvfrequenciesdetectedwiththe acghisassociatedtothestudiedphenotype. Figure19:FromaCGHresultstogenotypingofareplicationcohortwithamultiplexPCR. RealtimequantitativePCR Inthe casesofcomplexcnvs,inordertoquantify thenumber ofcopiesbothincasesandcontrols,itis necessarytoperformquantitativepcr.thistypeofvalidationrefersthenumberofcopiesinanyindividual toareferencesample.themostcommonlyusedtechniquetoquantifyistaqman (AppliedBiosystems).In this a pair of specific primers plus a specific probe with a fluorophore are used. Fluorescence is only producedwhentheprobeiscleavedwiththepcrextensionandthequencherisdetached(figure20).in additiontotheregionthatwewanttotest,aregionthatdoesnotvaryincopynumberisusedinorderto normalizethednaamount.roche Universalprobelibraryusesthesametechnology. 50
57 INTRODUCTION Figure20:Taqman technology ( technologies/realtimepcr/taqmanandsybrgreenchemistries.html) Othervalidationtechniques WhentheCNVeventsarelargeenough,visualizationthroughfluorescenceinsituhybridization(FISH)canbe usedinordertovalidateacghresults.itisbasedontheuseoffluorescentprobesthatbindspecificallyto complementaryregionsindnaofmetaphasecellsandaretargetedbyantibodiesfortheirdetection.the resolutionofthetechniquesis>3mb.forahigherresolution,fiberfishallowsthevisualizationofsmaller events,1400kb(166),bystretchingoutinterphasechromosomesinastraightline,byapplyingmechanical shear along the length of the slide. However, these visualization techniques of CNV events have the limitationthattheycannotbeperformedinahighthroughputmannerandthatcellsareneededinorderto perform the experiments. Their use will be more centered in family studies or merely as a secondary validationofthecnv. MLPA (multiple ligationdependent probe amplification) is also a widely used technique. It allows the detectionofinsertionsanddeletionsinamultiplexway(50locicanbetestedinthesameexperiment)and usingalowamountofdna(20ng)(167).however,thereproducibilityofthetechnique,whenusingcustom assays,isnothighandreducesthecostbenefitratioofthetechnique. 51
58 INTRODUCTION GENETICSUNDERLYINGCOMPLEXDISORDERSSUSCEPTIBLITY Wherearewe? The genetic model underlying susceptibility to complex disorders is still a matter of discussion. Different hypothesistrytoexplainhowgeneticfactorsplayaroleinthedevelopmentofthesedisorders.themost acceptedexplanationthesedaysincludesbothrarevariantswithalargeeffectandcommonvariantswitha smaller effect. The exploration of these kinds of variants presents different difficulties. In the case of commonvariantswithasmalleffect,itisessentialtoworkwithverylargecohortsinordertohaveenough powertodetect differencesamongcasesandcontrols.thisissomehowagreaterdealinheterogeneous disorders such as FM. The selection of the study subject s study is crucial, and another critical issue is phenotype accuracy. On the other hand, rare variants will probably be a new focus of analysis, as next generationsequencingtechnologies,suchasexomesequencing,havefacilitatedtheirdetection.oneofthe limitationintheanalysisofthesevariantsisdeterminedbyprivatevariants:howtoidentifyvariantslinked todiseasefromprivatevariantsthatcanbesomehowcommonbychancetoagroupofindividuals? Theapproachesusedinhumangeneticsforthestudyofgeneticsusceptibilitytodiseasesareseveraland have incredibly evolved over the past years. And this evolution has been in concordance with the development of new technologies allowing a more efficient evaluation of the genetic variants. Linkage studiesinfamilieshavebeenwidelyused.firstusingmicrosatellites,thensinglenucleotidepolymorphisms genotypedonarraysand,inthelastyears,byanalyzingexomedata.incomplexdiseases,themostcommon approach,abovefamilybasedtransmissiondisequilibrium,hasbeencasecontrolassociationstudies.initially itwasfocusedonthestudyofvariantsincandidategenes,thenongwasand,morerecently,byperforming exomesequencing,althoughtheresultsofthelatterhavenotbeenpublishedyet,astheanalysisiscomplex. Andthisisgoingtogetmoreandmorecomplexasthesenewtechniquesandthealgorithmsthatarebeing developedinordertoanalyseandinterprettheresultsarenowfocusingtotheanalysisofothervariants such as copy number variants. What is more, the possibility to explore epigenetic factors such as DNA methylationandhistonemodificationsopensanewfieldofresearchaddinganewcomplexityleveltothe studyofcomplexdisordersgeneticsusceptibility.finally,theencodeprojecthasenhancedtheimportance ofothernoncodingsequencesinhumangenome. Largecohortsandpopulationadmixture In most GWAS one of the principal limitations is population heterogeneity. By increasing the number of individuals included in a study, the population of study get more heterogeneous and then ancestry may becomeaconfounder.thissituationgetsdramaticinpopulationssuchasthatoftheunitedstatesinwhich 52
59 INTRODUCTION thereisextensivepopulationadmixture.itisessentialthattheconfoundingeffectcausedbythispopulation admixture be considered when performing the association analysis. STRUCTURE analysis using Ancestry Informative Markers (AIMs) scores would be most preferable for this kind of study compared to self reportedethnicity.aimsscoresshouldbeusedascovariatesinassociationanalyses. Populationadmixtureisamajorissueingeneticassociationstudies.However,theIberianPeninsulashowsa substantialgenetichomogeneity(168),onlyshowingquestioneddifferencesamongthebasque(169)and Canary island inhabitants (170). A recent paper by Julia et al (171), in which a whole genome scan was performed in Spanish controls and rheumatoid arthritis samples, only showed a west to east trend, but correction for ancestry informative markers in that study did not show a significant difference from the uncorrecteddata,notsupportinganeffectofpopulationstratification. Powercalculationandcorrectionformultipletesting InGWASoneoftheprincipallimitationsispower.Forcomplexdisorders,geneticfactorsareexpectedto have a limited effect. In fact, not considering the effect of the major histocompatibility complex in autoimmunedisorders,mostgwasresultsshowassociationswithoddratioslowerthan1.5.moreover,in ordertodetectthesemildassociationsthousandsofsamplesareneeded.severalsoftwarecanbeusedto performastatisticalpowercalculation,asforexamplequanto( samplesizenecessarytoreachstatisticalsignificanceforloweffectsvariants. An important issue in the human genetic studies nowadays is the correction for multiple testing. When different outputs are maesures/evaluated, finding an association with a pvalue 0.05 can be obtained by chance.forthisreasonandespeciallywhenperformingwholegenomeapproachesacorrectionformultiple testing is performed. The most astringent one is the Bonferroni in which the level of significance is established as 0.05 is divided by the number of tests performs. As in some case some of the markers evaluatedcanbelinked,alessastringentcorrectionmaybeperformed.forwholegenomeassociationscans thelevelofsignificancehasbeenestablishedat10e8. Dealingwithclinicaldata:Clusteranalysis Cluster analysis is a multivariate statistical technique, which evaluates the degree of similarity among heterogeneous variables to try to identify related groups of variables based on their similarities. This procedureattemptstoreducedata dimensionality tofewerlatentvariables. Ithasbeenappliedtoother disordersinanefforttounderstandtherelationshipsamongclinicalfeaturesandoutcomevariables(172).it wasfirstappliedintheoncologyfieldtogroupthesecondaryeffectsofchemotherapythatapparentlywere notrelatedatallsincetheyweremanifestationsofdifferentorgans(forexample:vomiting,aenemiaand 53
60 INTRODUCTION fever).ithasbeenlaterimplementedinordertolinkrelatedsymptomsortoidentifygroupsofhomogenous patientsofaparticulardisease. Many investigators have used cluster analysis to group FM symptoms, in order to define FM subgroups. These previous studies had different results, depending on sample sizes, variables studied and methods used.inmostcases,clusteranalysiswasusedtocategorizefmpatients,basedonsomaticorpsychological symptoms (173), quantitative sensory testing (174) or pressurepain thresholds and psychological factors (175). A recent study tried to discern clinically relevant subgroups across psychological and biomedical domains(176),whileanotherattemptedtoidentifyclustersofclinicalfeaturesmeaningfultofmpatients thatcorrespondedtotheirtreatmentprioritygoalsinthecontextofdesiredimprovement(177).finally,a morerecentwork(178)indicatedtheexistenceoftwolatentdimensionsunderlyingfmsymptomatology: FMcoresymptomsanddistress.Mostofthesestudieshavebeenperformedinsmallcohortstakinginto accountfewclinicalvariables,andusingclusteringtogroupeithervariablesorpatients.onlytwostudies were performed in large cohorts (179, 180). In these studies, patients were assessed through web based methodandtheirpurposewas,inonecase,toevaluatepatientsperceptionofsymptomsmanagement(179) and,intheothercase,toexaminedifferencesamongfmsubgroupsinhealthcareutilization(180).sofar,in spiteinthedifferentimplementationofclusteringproceduresintofmpatients,noclearfmsubgroupshave beenidentified. 54
61 OBJECTIVES
62
63 OBJECTIVES The aim of this thesis was to elucidate genetic susceptibility factors for fibromyalgia, a common, heterogeneousdisorderwhosegeneticcontributionhadnotbeenexploredinacomprehensivemanner.we assessedthisobjectivethroughthreemainapproaches: 1. TheidentificationofFMclinicallyhomogeneoussubgroupswithatwostepclusteranalyses: a) Constructionofdimensionsfromclinicalvariables,and b) IdentificationofFMsubphenotypes. 2. ToperformagenomewideassociationstudyofFMinordertoevaluatethepossiblecontributionof singlenucleotidepolymorphismsandalsotoinferregionsvaryingincopynumberandtheirpotential presenceinmosaicism.thisincludesthefollowingsubobjectives: a) AssociationanalysisofGWASdata, b) IdentificationofCNVs, c) ReplicationofstrongestassociatedSNPs,and d) ImplementationofclusteranalysisintotheGWASstudy. 3. Toperformarraycomparativegenomichybridizationexperimentstoidentifyregionsvaryingincopy numberthatcouldbeinvolvedinfmsusceptibility: a) acghexperimentsanalysisandvalidation, b) ImplementationofclusteranalysisintoaCGHanalysis,and c) EvaluationofpossiblefunctionalconsequencesofassociatedCNVs. To perform these analyses we had access to a large and very well characterized cohort of FM patients. 57
64
65 MATERIALSANDMETHODS
66
67 MATERIALSANDMETHODS SAMPLES:THEFIBROMYALGIABIOBANK We studied unrelated FM samples of a multicentric study, whose first purpose was the creation of the SpanishGeneticandClinicalDataBankofFibromyalgiaandChronicFatigueSyndrome,(FFSGCDB).FiveFM Units of five Spanish Hospitals (Hospital del Mar, Barcelona, Jordi Carbonell; Hospital Clínic i Provincial, Barcelona, Antonio Collado; Hospital de la Vall d Hebrón, Barcelona, Jose Alegre; Instituto General de Rehabilitaciónde Madrid,Madrid,JavierRivera;andHospitalGeneralde Guadalajara,Guadalajara,Javier Vidal)participatedinthecollectionofsamples.Atotalof1,510FMpatients,fulfilling1990ACRcriteria,were selected by rheumatologists. They were then evaluated by another set of physicians trained in the evaluationoffmpatients.allsampleswerespanishofcaucasianoriginandhadsignedinformedconsent beforeenrolment.theethicscommitteeatallrecruitmentcentersapprovedtheproject.theyallpassedthe samequestionnairesandphysicalexamination. Datacollectionfollowedastandardprotocolofquestionnairesandphysicalexaminationthatwererecorded by principal investigators of the FFSGCDB. It included collection of demographic variables (age, marital status, educational level, and occupational status), family and personal history of diseases (in particular, history of chronic fatigue, chronic pain, connective disorders, spine degenerative disease and psychopathology), time of disease evolution, presence of representative symptoms covering the wide spectrum of symptomatology in FM (musculoskeletal, neurological, autonomic and somatic), and treatments.coremeasuresoffmseveritywereassessedbydifferentspanishvalidatedscales:theintensity ofpainandfatiguewitha11pointsvisualanaloguescale(vas)(where0representednopainorfatigue,and 10themaximumpainorfatigue);thenumberoftenderpointswithstandardmanualexamination(181);the levelofanxietyordepressionwiththehospitalanxietyanddepressionscale(had)(182,183);thesleep qualitybythepittsburghsleepqualityindex(psqi)(184);andthegeneralhealthstatusbythefibromyalgia ImpactQuestionnaire(FIQ)(185),theFatigueImpactSeverityscale(FIS)(186,187)andtheQualityofLife survey(sf36)(188,189).finally,patientswereevaluatedforfukuda scfscriteria(190). 1,000controlsamplescamefromtheNationalDNABankofSalamanca).Theyhadlowlevelsofpainand fatigueasassessedbyaquestionnaire.wehadaccessto5μgofdnaoffmcasesand1μgofdnaofthe controlsamples. FMCLUSTERS:IDENTIFYINGFIBROMYALGIASUBGROUPS A twostep clustering procedure was performed in 1,446 FM patients in order to identify FM subgroups (Figure21).Afterexclusionoftreatmentandsociodemographicvariables,48variableswereselectedforthe clusteringanalysis.giventhemixednatureofthevariables,theseweretransformedintobinarytypes(0= mild;1=severe).forsymptoms(dichotomicvariables),thepresenceofthesymptomwascodifiedas1and 61
68 MATERIALSANDMETHODS theabsenceas0.forcontinuousvariables,themedianwasconsideredasacutoffvalue.forscales,values belowthemedianwerecodifiedas0andvaluesabovethemedianas1,whileforageofonset,asayounger ageofonset(38years)isconsideredmoresevere,thecodificationwasreversed.variablesincludedinthe clusteranalysis,aswellastheirmedians(interquartilerange),aresummarizedintable7. Figure21:IdentificationofFMsubgroupsthroughatwostepsclusteringprocedure.First,variableswere classifiedintodifferentdimensions,basedonvariables similarities.inasecondstep,variables dimensions wereusedtoidentifyfmsubgroups. Statisticalanalysis Buildingvariable sdimensions The underlying dimensions of FM were evaluated using partitioning cluster analysis.thisisamethodto partition data (clinical features) into meaningful subgroups when the number of subgroups and other informationabouttheircompositionmaybeunknown(191).inthissense,theprocedureattemptstoreduce datadimensionalitytofewerlatentvariables,which,inourstudy,aretermedfmdimensions. 62
69 MATERIALSANDMETHODS Table7:Variablesincludedintheclusteranalysis.Theyarelistedinalphabeticalorder. VARIABLES Adjustmentdisorder Ageofonset(38;p 25 :30;p 75 :45) Blackouts Concentrationproblems Connectivedisorder Dizziness ExcessivePerspiration Facialoedema Familyhistorychronicpain Familyhistoryofautoimmunedisorders Familyhistoryofchronicfatiguesyndrome Familyhistoryoffibromyalgia FatigueImpactScale(FIS)(66;p 25 :56.50;p 75 :75.00) Fatiguelevel(VAS110cm)(8;p 25 :6.4;p 75 :9) FibromyalgiaImpactQuestionnaire(FIQ)(74.66;p 25 :63.05;p 75 :84.25) Forgetfulness HADanxietysubscale(12;p 25 :8;p 75 :15) HADdepressionsubscale(10;p 25 :7;p 75 :14) Headache Impairedurination Intestinaldysfunction LifequalitySF36mentalsubscale(35;p 25 :25;p 75 :48) LifequalitySF36physicalsubscale(27;p 25 :22;p 75 :32) Majordepression Memorycomplaints Migratoryjointpain Monthsofpain(96;p 25 :48;p 75 :156) Morningstiffness Muscleweakness Muscularcontractures Onsettype Painlevel(VAS110cm)(7.5;p 25 :6.5;p 75 :8.5) Painsubtlemovementsimpairment Palpitations Panicattacks Personalhistoryofchronicpain Personalitydisorders PittsburghSleepQualityIndex(PSQI)(14;p 25 :11;p 75 :17) Postexercisefatigue Posttraumaticstressdisorder PreviousPersonalhistorypsychopathology SleepDisturbances Spineosteoarthritis Trembling TriggerPresence Visualaccommodationimpairment Widespreadpain 63
70 MATERIALSANDMETHODS The basic procedure behind partitioning cluster analysis is to construct subgroups with homogeneous objects, which in our study are variables corresponding to clinical features, based on a welldefined proximity measure. Given the noncontinuous nature of our variables, we used the Gower's general similaritymeasure(192).themostcommonpartitioningclusteringmethodisthekmeansalgorithm(193); however, we used a more robust approach called position around medoids (PAM), which is similar to K means,butgroupmembershipdependsonproximitytoanactualobservation(medoid)insteadofproximity toanaverageobject(centroid).thenumberofclustersorsubgroupswasdeterminedusingsilhouetteplots. Thenamesthatwereeventuallygiventoeachofthesubgroupsweredeterminedposthocinanattemptto characterizethenatureofeachfmdimension.theclusteringwasconstructedinaninitialsampleof559fm patients,andacrossvalidationofthetoolwasperformedinasecondsampleof887patients. Constructionofsamplessubgroups TheclusteringtooltodefinetheunderlyingFMdimensionswasusedtofurtherconstructpatientsynthetic indexes based on the clinical features composition of each FM dimension. The values of the synthetic indexeswerecalculatedfromlinearfunctions(oneperdimension)thatweightthedichotomousvariables constituting the specific dimension. The weighting factor is based on the silhouette value of the specific variableintheclustertowhichitisassigned,whichcanbeunderstoodasameasureofthecontributionof thatvariabletothedimension.thesilhouettevaluestake meaningfulvaluesfrom1(wellassigned)to1 (closertovariablesassignedtoneighbouringclusters),where0indicatesnoclearassignment.theresulting values of the synthetic indexes result into continuous measures for each FM dimension. The synthetic indexedvaluescalculatedpereachsampleanddimensionwherethenusedtofindpatientsubgroups.given nowthecontinuousnatureofthemeasuresweusedthekmeansclusteringproceduretogrouppatients withsimilarfmprofiles.allanalyseswereperformedintherenvironmentusingtheclusterpackage. GENOMEWIDEASSOCIATIONSTUDY 321FMcases,selectedbyFFSGCDBprincipalinvestigatorsforhavinglowlevelsofpsychiatriccomorbidities andbeingthebestfittingthefmdiagnosis,weregenotypedwithillumina1mduochip,whichinterrogates nearly 1.2 million loci per sample, containing tag SNPs, SNPs in genes, and SNPs and nonpolymorphic markersinknownandnovelcopynumbervariation(cnv)regions.themedianspacingbetweenmarkersis 1.5 kb (a mean of 2.4 kb), with the 90 th percentile for the gap size between SNPs being 6 kb. The array captures 95% of CEU HapMap population variation (HapMap 1) and also includes CNV targeted markers.genotypingwasperformedincegen(barcelonanode),followingthemanufacturer sprotocol.200 ngofdnapersamplewasused.abeadarrayreaderextractedimagesandreadfluorescenceintensities, andalldatawasuploadedintobeadstudiosoftwareforqualitycontrolandprocessing. 64
71 MATERIALSANDMETHODS Datafrom425Spanishcontrolsaged2044yearsold(generalSpanishpopulationfromGABRIELconsortium genotyped with Illumina 610quad chip was available. Genotyping hadbeenperformedincentrenationaldegénotypage( (Évry,France).Sincebothcases andcontrolshadbeengenotypedseparatelywithdifferentplatforms,inordertominimizeplatformbias, qualitycontrol(qc)procedureswereperformedseparatelyandqcedcasesandcontrolsweresubsequently mergedtogetherforassociationanalysis(figure22). Qualitycontrol Quality control was performed with PLINK software ( (194). TheseQCstepswereexecutedseparatelyintheFMandcontroldatasets,onlytakingintoaccounttheSNPs thatoverlappedbetweenthetwodatasets(582,892snps).theqcwasfirstperformedatsamplelevel. Figure22:GWASanalysispipeline:fromgenotypingtoreplicationofassociationresults 1. SampleQC 1.1. Origin The first step consisted of identifying and eliminating samples that were not of European origin. For this purpose,weranthegenomefunctionineachofourdatasets(fmandcontrols).first,weextractedsnps included in the 610quad array from FM, CEU, Yoruba and ChineseJapanese HapMap datasets (from HapMap2phase).HapMap610quaddatawerethenmergedwitheachofourdatasets(controlsandFM 65
72 MATERIALSANDMETHODS 610quaddata)intwoseparatefiles.Acrucialstepinthisprocedurewastogetallthegenotypedatainthe samestrandincontrols,fmandhapmapdata(whichallsnpallelesareinthepositivestrand).thiswas achieved with the PLINK flip strand function. Then, the Genome function (which calculates genomewide identity by descent (IBD) given identity by state (IBS) information to define pairwise similarities between samples) was used to compare samples pairwise (FM samples with HapMap samples and controls with HapMapsamples).Aninbreedingcoefficient(Pi_HAT)wasthencalculated: PI_HATP(IBD=2)+0.5*P(IBD=1)(proportionIBD) Thentheoutputofthisanalysiswasrunintotheidentitybystatefunction(clusterfunction)andsamples wereclustereddependingonhowmanygenotypesandallelestheirshared(identitybystate:ibs): (IBS2+0.5IBS1)N IBS2:numberofmarkerswherepairofindividualsshare2alleles IBS1:numberofmarkerswherepairofindividualsshare1allele N,numberofSNPs The two principal components that best classified the samples into these clusters were then plotted in a multidimensional scaling plot with R. Those outliers (FM samples or controls) notclustering with CEU sampleswereexcluded(figure23). Figure23:PCAplotsforcontrols(topimage)andFMsamples.SamplesthatdidnotclusterwithHapMapCEU sampleswhereexcludedfromtheanalysis.twocontrolsamplesandsixfmwereexcluded(redcircles). 66
73 MATERIALSANDMETHODS 1.2 Callrate PLINK scallratefunctionwasusedtoobtainsamplecallratesforthefmandcontroldatasets.thesecall rates were plotted in histograms with R software. The cut off value was defined by eliminating outliers (Figure24). Figure24:HistogramsrepresentingthegenotypingratesofFM(left)andcontrolsamples(right).NoFMsampleswere excluded;inthecontroldataset,thethreshold(markedwitharedline)wasestablishedat96%and3individualswere excluded. 1.3 Heterozygosity Alowlevelofheterozygositycouldbeindicatinginbreeding,andahighlevel,samples contamination.an algorithm developed by the group of Dr Eleftheria Zeggini (Sanger institute) was used to calculate heterozigosity levels in FM samples and in controls were calculated. The heterozygosity values of the sampleswerethenplottedinhistograms(figure25).thecutoffvaluewasdefinedbyeliminatingoutliers. For FM samples, cut off value was established at 30%: three samples were excluded. For controls, the thresholdwas29%andtwosampleswereexcluded. 1.4 Gendercheck Gender was imputed using genotype information from the X chromosome. Those samples discordant between phenotypic /database sex and imputed sex were excluded. Two FM samples and three controls wereexcludedforthisreason. 1.5 Relatedness Samplesthatpresentedadegreeofrelatednesshigherthanexpectedinarandomsamplewereexcluded. For this purpose we used the PI_HAT inbreeding coefficient values: samples presenting a PI_ HAT value higherorequalto0.05wereexcluded.fivefmsamplesandninecontrolswereexcluded. 67
74 MATERIALSANDMETHODS Afterallthesamplebasedfilteringprocedures,13sampleswereremovedfromFMdatasetand30inthe controldataset.theresultingfiltereddatasetsincluded582892snpsand308individualsinthefmdataset and582892snpsand395individualsinthecontroldataset.thesewerethenfilteredatthesnplevel. Figure25:HistogramsrepresentingpercentageofheterozygosityofFM(left)andcontrol(right)samples.A redlineindicatescutoffvalues. 2. SNPsQC 2.1 Genotypingrate TheGenotypingratesinbothdatasetswereobtainedwithPLINKandplottedinhistogramswithR(Figure 26).BothforFMsamplesandforcontrolsthecutoffvaluewasestablishedat96%:4846SNPswere excludedinthefmdatasetand5716snpsincontrols. Figure26:HistogramsrepresentingSNPscallrateinFM(left)andcontrols(right)datasets. Aredlineindicatedcutoffvalue. 68
75 MATERIALSANDMETHODS 2.2 HardyWeinbergEquilibrium ThethresholdforHardyWeinbergEquilibrium(HWE)inGWASisnotclearlyestablished.Manyinvestigators use a threshold of 10.e 8 claiming that they use the same multiple testing correction of association significance.sinceweusedareducedsamplenumber,wedecidedtobemorestringent,anduseda10.e 4 threshold.thisleadtotheexclusionof653snpsinthefmsamples,and883snpsinthecontroldataset. 2.3 MinimumAlleleFrequency SNPswithaminimumallelefrequency(MAF)<5%wereexcluded:64,610inFMand65,203incontrols.After QCatthesampleandSNPlevels,theFMdatasetincluded308cases(8malesand300females)and513,897 SNPsandthecontroldataset395controls(192malesand203females)and512,615SNPs. MergingfilteredFMandfilteredcontrols AsFMcasesandcontrolshadbeengenotypedwithdifferentplatforms,whenmergingbothdatasetswehad to face the problem of flipped strands. In the first step of QC, when checking for the samples European origin,bothfmandcontrolsdatasetswereflippedinordertohaveallthesnpsinthepositivestrandasitis inthehapmapdatasets.however,althoughplinkdetectsflippedstrands,itisnotabletodosointhecase ofa/tandc/gsnps.tosolvethisproblem,weusedtheplinkreferenceallelefunction(manuallyspecifies majoralleleandminoralleles)ontheoverlappingsnpsbetweenbothfiltereddatasets(505,454snps). ThenFMandcontrolsdatasetsweremerged.Weranthegenomeandclusterfunctionsand,again,thetwo principalcomponentsthatbestclassifiedthesamplesintotheseclusterswereplottedinamultidimensional scalingplotwithr.thiswasperformedonlywithcasesandcontrols,thenaddingceuhapmapdataand finallywithallthreehapmappopulations(figure27). Figure27:PCAplotsofFMandcontroldatasetswithHapMapCEU,CHB_JPTandYRIpopulations(left),and withhapmapceu(right).bothfmandcontroldatasetsclusterwithceuhapmapindividuals. 69
76 MATERIALSANDMETHODS Thegenderissue Since97%oftheFMsamplesarefemale(andover90%ofFMcasesinclinicalpractice)wedecidedtouse onlyfemalecontrolstohaveagendermatchedcontrolsetandtobeabletoevaluatethexchromosome (Figure 28). So in the end, 505,454 SNPs in 300 FM cases and 203 controls were considered for the associationanalysis. Associationanalysis Figure28:PCAplotofthemergedFMandcontrolsonlyconsidering females.therewasnoevidenceofpopulationstratification:the genomicinflationinthefemalesstudywasfinally AllelicassociationanalysiswasperformedwithPLINK(95%CI).QQplotsandannotationoftopSNPswere performedwiththewgasoftware( (195)andManhattanplotwithHaploviewsoftware(196). CheckingSNPsclusters SNPclusteringforSNPSshowingstrongestassociationwasonlycheckedinFMcases,sincewehadnoaccess tocontrolsrawdata.innormalconditions,threeclustersareexpected:oneforeachpossiblesnpgenotype (homozygousforaallele,heterozygousandhomozygousforballele).thepurposeofthisstepistodiscard spuriousassociationscausedbyasecondarysnporacnvoverlappingthesnp,whichwouldleadtoabad clusteringwithmorethanthreeclusters. 70
77 MATERIALSANDMETHODS Imputation Imputation was performed for fine mapping of the GWAS results following the workflow summarized in Figure 29. In brief, PLINK files were divided into the different chromosomes. Then file formats had to be modified in order to run the imputation software. This was done with GTOOL ( Then imputation was performed with Impute( kb and using as reference panels 1000Genomes and Hapmap3 CEU data. Association analysis of imputed data was performed with SNPTEST ( usinga frequentist test with method_score option that takes into account genotype uncertainty. Infoscore measures the genotype uncertainty: 0.8 value would correspond to an uncertainty of 20%. Usually, infoscore> 0.8 (and for some groupseven0.5)isusedasfilteringcriteriatoselectsnpswithagoodimputationquality. SNPsannotation Figure 29: Workflow of the imputation analysis. At each step of the procedure, the programsusedandfinallytheqcsteparelistedontheleftside,whiletheformatofthe files,thereferencepanelsusedandthefiltersapplied,arementionedontherightside. SNPs showing strongest association were annotated with WGA viewer software. Annotation included identifyingthenearestgeneandgeneticpositionofthesnp(coding,intronic,intergenic).thesnpandits genomicregionsrelationtodiseasewasevaluatedwithdecipher( Replication InordertovalidatetheGWASresults,21SNPsshowingstrongestassociationvaluesweregenotypedina replication set from our cohort, consisting in 982 additional FM cases and 971 controls coming from the BancoNacionaldeDNAdeSalamanca.Controlswereselectedforhavinglowlevelsofpainandfatigueas assessedbyaquestionnaire.genotypingwasperformedbykbiosciences,usingtheirpcrsnpgenotyping 71
78 MATERIALSANDMETHODS system (KASPar : Kbioscience, UK), which uses a competitive allelespecific PCR. As quality control, duplicates(2hapmapsamplesineachplate)andnegativecontrolswhereincluded.snpsnotfulfillinghwe, withamaf<5%oralowgenotypingrate(<95%),aswellassampleswithlowgenotypingrate(<95%),were excludedfromtheanalysis.associationanalysiswasperformedwithhmisc,nnetandsnpassocrpackages (genotypicandmultinomialanalyses).multinomialanalysiswasperformedinordertoevaluatethepossible effectoffmclustersinsnpassociation. Theprioritizationofthe21SNPstobegenotypedinthereplicationcohortwasbasedonseveralcriteria: Pvalue Signals:clusteringofSNPswithstrongassociationsinagenomicregion Function:SNPslocatedingenesorgeneregions(asassessedbyUCSCgenomebrowserandWGA viewersoftware) Imputationdatawasconsideredforfinemapping.ForGWASregionsshowingpositivesignals,understoodas severalsnpsshowingastrongassociationina50kbregion,imputationresultswereevaluated.snpsina window span of 100kb were considered, and those showing a stronger association than the directly genotypedsnpwherealsoincludedforreplication. AssessingSNPfunctioninsilico InordertoevaluatethefunctionofassociatedSNPsweuseddifferenttools.Firstofall,weusedPuppasuite ( relevantsnpswithinagene,basedondifferentcharacteristicsofthesnpitself,suchasvalidationstatus, type, frequency/population data and putative functional properties (pathological SNPs, SNPs disrupting potentialtranscriptionfactorbindingsites,snpslocatedinintron/exonboundaries). We also studied the SNPs possible cis effects in gene expression levels with Genevar ( This platform of database and web services is designedforintegrativeanalysisandvisualizationofsnpgeneassociationsineqtl(expressionquantitative traitloci)studies.itevaluatesthepossiblecorrelationbetweensnpgenotypesandgeneexpressionlevelsin differentdatasets:lymphoblastoidcelllinesfrom726hapmap3individuals(197),skin,adiposetissueand lymphoblastoidcelllinesfromfemaletwins(198),andinprimaryfibroblasts,lymphoblastoidcelllinesandt cells from umbilical cords of individuals of Western European origin (199). Briefly, data from expression arrays(illuminahumanht12andilluminawg6v3expressionarrays)andfromsnpsarrays(illumina1m duoandillumina550k)areusedtotestforassociationbetweenthesnp sgenotypeandmrnalevels.this is performed in a twostep process. First, SNPgene associations surrounding esnps/lead SNPs among tissues/populations are investigated with the SNP centric analysis (ciseqtlsnp). Then SNPprobe 72
79 MATERIALSANDMETHODS associationsacrosscelltypes/populationsareplotted(snpgeneassociationanalysis(eqtlsnpgene)).we tookintoaccountallprobeswithina1mbwindowcentredinthetranscriptionstartsiteofthegeneand performedassociationanalysisusingspearmanrankcorrelationandsettingthepvaluelimitfilterat Finally, after this year s publication of Encode results, we used the Regulome database ( RegulomeDB (200) is a database that annotates SNPs with known and predicted regulatory elements in the intergenic regions of the human genome. Known and predicted regulatorydnaelementsincluderegionsofdnaasehypersensitivity,transcriptionfactorsbindingsites,and promoterregionsthathavebeenbiochemicallycharacterizedtoregulatetranscriptionindifferentcelltypes (lymphocytes,glioblastomacelllines,osteoblastsandstemcells,amongothers). PathwayanalysisofGWASandreplicationresults SNPsshowingstrongestassociationwereanalyzedwithIngenuitySystemsPathwayanalysis(IPA)software ( and GeneSet analysis Toolkit v2 ( Briefly, for SNPs showing strongest allelic association in GWAS, the nearest gene Entrez Gene IDs were enteredintotheipadatabaseforacoreanalysisandintogenesetanalysistoolkitforgeneontology(go) biological processes, molecular function and cellular components analysis, and Kegg pathways analysis againsthumangenome,applyingbenjaminiandhochbergcorrection.keggpathwaymappingistheprocess tomapmoleculardatasets(inthepresentstudygenomics)tothekeggpathwaymapswhicharemanually drawn pathways representing our knowledge on the molecular interaction and reaction networks for metabolism,cellularprocess. CNVassessment:PennCNV Illuminaarrays,inadditiontoprovidinggenotypecalls,allowtheinferenceoftheproportionofhybridized sample that carries the B allele, which is called B allele frequency (BAF) ( In a normal sample, discrete BAFs of 0, 0.5 and 1.0 are expected for each locus, representing each of the possible genotypes (AA,AB and BB). Deviations from this expectation are indicative of aberrant copy number. Furthermore,theloggedratioofobservedprobeintensitytoexpectedintensity(LRR)isalsomeasured.Any deviations from zero in this metric are suggestive of a region varying in copy number as well. Several algorithms handle these signal intensity data coming from SNP arrays to infer CNV calls. Of these, we decidedtousepenncnvsoftware,asitisoneofthemostacceptedtools. PennCNV implements a hidden Markov model (HMM) that integrates multiple sources of information to infercnvcallsforindividualgenotypedsamples(161).itdiffersformsegmentationbasedalgorithmsinthat 73
80 MATERIALSANDMETHODS itconsiderssnpallelicratiodistributionandotherfactors,inadditiontosignalintensityalone.inaddition, PennCNVcanoptionally utilizefamilyinformation togeneratefamilybased CNVcallsby severaldifferent algorithms. Furthermore, PennCNV can generate CNV calls given a specific set of candidate CNV regions, throughavalidationcallingalgorithm.itallowssixstatesdefinitionofcnvevents(table8). Table8:CNVStatesdetectedbyPennCNV(takenfromWangetal) ). In our PennCNV based study, we selected regions includingatleastthree markers showingalterations in intensitydata.outofthese,weonlyselectedthoseaberrationspresentinatleast5%ofthesamples.we further selected those regions locatedinagene (+/50 kb). Then, CNV events having been previously reported as very frequent polymorphic events present in the general population and/or related to populationdifferenceswerealsodiscarded.finally,outofthecnvregionsthathadpassedalltheselection filters,thevalidationofpenncnvresultsstartedbythoseregionsthathadbeenalsodetectedbyacghin oursamples.furthermore,weperformedacomplementaryfilteringprocedureinordertodetectrareand largeevents,whichavoidedplateorbatcheffectsandwerenotpresentinthegeneralpopulation.basedon Zhangetal s(201)work,,wefollowedthepipelinesummarizedinfigure 30. Inclusion ofsamples that passed QCfor SNPanalysis Restrict analysis to autosomes Generation ofcnvcalls with PennCNVsoftware QCPennCNV output.remove samples not fulfilling any ofthe following critheria: LogRSD<0.3 Ballele Frequency drift <0.01 wavefactor<0.05or >0.05 < <239calls QCPennCNV output.removee CNVs with any ofthe following: CNVcalls with confidence score<10 CNVcallswith<10SNPs Low SNPdensity (< <1SNP/50kb onaverage) <100kb Exclude CNVs that overlap at50%or moreofits lenght with Ig,centromeric or telomeric regions Remove CNVs that overlap by >50% %oftheir lenght with commoncnvs(database ofgenomic variants) Lookfor recurrent events that targetthe same geneor region Check if the detected events arepresent incontrols Figure30:DetectionofrareCNVsalgorithmfromPennCNVoutput. 74
81 MATERIALSANDMETHODS CNVmosaicismassessment We used the Mosaic Alteration DetectionMAD algorithm developed by Gonzalez et al (202) for the detectionofcnvsinmosaicstatebyusingbafandlogrratio.briefly,whenthereisacnv(insertionor deletion) there are changes in LogRatio and BAF that overlap.an altered BAF that doesn t overlap with significativechangesinlogr(<0.2)isindicativeofacnveventinmosaicism.anabonormalaveragebaffor heterozygoussnps(notcenteredat0.5)withanormalaveragelogrvaluearound0isindicatingaprobable neutralcopynumberchangewithallelicimbalancesuggestiveofauniparentaldisomy(upd)inmosaicism; an abnormal average BAF accompanied by an altered logr ratio not reaching the chosen threshold for heterozygousdeletionsorduplicationscouldbeindicatingagainorlossinmosaicstate.bafvaluesandbaf standarddeviation(bdev)canbeusedtoinferethepercentatgeofcellscarryingthemosaicevent: L(proportionofcellswithaloss)=2Bdev/(0.5Bdev) G(proportionofcellswithagain)=2Bdev/(0.5+Bdev) U(proportionofcellswithcopynumberneutralchangeUPD)=2Bdev This algorithm allows the detection of four possible states: loss, gain, trisomy and lost of heterozygosity (LOH)duetoidentitybystate. ARRAYCOMPARATIVEGENOMICHYBRIDIZATION acgh experiments were performed with two sets of complementary platforms, Agilent 400K and Agilent 1M using a pooling strategy. This strategy aims to dilute rare CNV polymorphisms due to inter individual variability and highlight those common variants with a different frequency between cases and controls.threepoolsoffmsampleswheredesigned:fmwithoutfatigue(20samples),fmwithfatigue(20 samples)andearlyonsetfm(30samples).allthesamplesincludedinthepoolshadafamilyhistoryoffm. FM pools were hibridized against one pool of controls (50 samples). Samples technical requirements includeddnaintegrity(largednafragments,280260ratioaround1.8and260230ratioaround2)anda precise quantification ensuring that the same amount of each of the samples was included in the pool (concentrationrange50150ng/μl). Experimentalprotocol InordertoassessDNAqualityofthesamplesforinclusioninthepool,sampleswereruninadenaturinggel, DNAconcentrationwasquantifiedbyPicogreenandNanodrop,andthe280260and260230ratioswere calculated.samplesnotfulfillingqualitycontrolcriteriawereexcludedfromthepool.equalamountsofeach DNAweremixedintothepooledDNAusedforthehybridization.Areferencepool,generatedinthesame way,wascreated. 75
82 MATERIALSANDMETHODS EachFMpoolwashybridizedagainstthecontrolpoolontheAgilent400karray(directanddyeswap)and theagilent1marray(onlydirectastheywerecomplementing400kresults)(figure31).1μgofthereference and test pools was differentially labelled and both reference and onetest pools were competitively hybridized toamicroarray.then,inordertoavoidbiasdue to labelling, thetestandcontrolpools were labelledwiththeoppositedyeandhybridizedontoanewarray(dyeswap).hybridizationswereperformed accordingtoagilentoligonucleotidearraybasedcghforgenomicdnaanalysis(directmethod)protocol (Version4.0,June2006).Datawerefilteredtoexcludebadspotsandtoadjusttoolowortoohighintensities to more reasonable values, and then normalized to correct systematic errors due to technical reasons insteadofbiologicalvariability,sothatthemodalratioforthegenomewassettoastandardvalue0.0ona logarithmicscale. Figure31:ArrayCGHusingapoolingstrategy.ThreepoolsofFMsampleswithfamilyhistoryofFM weredesignedandeachwasdyedwithafluorophoreandhybridizedagainstapoolofpainfree controls on the 400k and 1M Agilent arrays. For the 400 K array the experiment was repeated exchangingthefluorophorebetweencasesandcontrolsinordertoavoidfluorophorebias. Analysisofarraysresults TheresultingdatawasanalyzedwithAgilent sgenomicworkbenchsoftware,usingtheadm2algorithm. Sincethestandarddeviationofthearrayswaslowerthanorequalto0.15(Table9),thecutoffvaluefor selecting probes showing differential hybridization was established at 0.3 (2 standard deviations). We selected regions with at least 3 aberrant probes and a mean/smoothed log2ratio>0.3. ADM2 algorithm identifies all intervals with consistently aberrant low or high log2 ratios based on a statistical score. It automaticallycalculatestheoptimalsizeofanaberrantregionandtestsregionsinwhichthestatisticalscore passauserdefinedthreshold.thisstatisticalscoreisbasedonlog2ratiosandnumberofprobes.thereis notafixedwindow:itsamplesadjacentprobestoarrivetoarobustestimationoftheboundariesofthe aberrantregion.furthermore,itincorporatesqualitydataofeachlogratio(probelogratioerror).forthis reason,itismorerobusttonoiseinthedata(presenceofnoisyprobes),especiallyiftheuserisinterestedin detectingsmallaberrantregions.finally,weselectedonlythoseregionsfulfillingthesecriteriainbothdirect anddyeswaphybridizations. 76
83 MATERIALSANDMETHODS Table9: Standarddeviationof400karrayhybridizations. Validationofarrayresults Hybridization FMcontrol FMcontrolDS FM_FCcontrol FM_FCcontrol DS FM_earlycontrol FM_earlycontrolDS MostCNVsdetectedintheaCGHexperimentswereshowntobeinsertionsdeletions(INDEL).Inthesecases, wefirstattemptedtodetectthebreakpointsofthecnv,andthendesignedamultiplexpcrtogenotypeitin ourentirecohortofcasesandcontrols. 1.BREAKPOINTSDETECTION We had to identify the breakpoints of the following CNVs: ACACA (detected both in 400K array and PennCNV)andWWOX(400Karray). Thepipelinesummarizedinfigure32wasfollowedup.IftheinitialPCR amplifiedseveralproducts,weperformedgelextractionofthebandcorrespondingtothedeletedallele.if gelextractionyieldwasinsufficientforasubsequentsequencingpcr,weperformedpcrsubcloning.this also was performed when the presence of repetitive sequences flanking the products didn t allow the obtainingofacleansequence.pcrconditionsandthedifferentexperimentsaredescribedrightafterwards. Figure32:Pipelineforbreakpointsdetection.Ifthestandardprotocol(bluearrow)didn tallow breakpointsdetectionadditionalexperiments(violetandredboxes)wereperformed. 77
84 MATERIALSANDMETHODS 1.1 PCRconditions For breakpoints detection we used the same mix components for the two CNVs evaluated. PCR program differedintheannealingtemperature and extension temperature and length, according to the expected size of the PCR products and the primers melting temperature (Table 10). Primers were designed with Primer3softwaretakingintoaccountaCGHpositiveprobescoordinatesandoverlappingCNVsdescribedin thedatabaseofgenomicvariants.primerssequencesaresummarizedintable11. We took advantage of data available in Conrad et al work and included, as positive controls, HapMap sampleswithknowngenotypesforthecnvs(table12). Table10:PCRconditionsforACACAandWWOXCNVsbreakpointsdetection. ACACA WWOX PCRprogram (30cicles) Mixcomponents 2 94ºC 30 94ºC 30 60ºC 1 72ºC 7 72ºC 2 94ºC 30 94ºC 30 60ºC 45 68ºC 7 68ºC 50ngDNA 10xRoche PCRreactionbuffer+Mg mMdNTPs 0.4pM/leachprimer 0.1U/lTaqPolymerase H 2 Otoreachafinalvolumeof25l Table11:Primersusedforbreakpointsdetection ACACA WWOX CNV Primers(5 3 ) ACACA_DelF:GGCCTCCTCTTCTAGCTGTTG ACACA_DelR:AACAGGTGCCCAATAAATGC WWOX_F1:TGGGTAGGAATCCTGCAGAC WWOX_R1:TGCCTAAAAGCACACACTGC WWOX_R2:GGGCATCCCAGTTTTCTACC WWOX_R3:CCTGCTTCCTGAACATTCCT Productsize(bp) 1200 Depending on primers combinations 78
85 MATERIALSANDMETHODS Table12:HapMapsampleswithdifferentgenotypesforWWOXandACACACNVsConradetal.. CNV Twocopies Onecopy Zerocopies ACACA NA10854;NA10855;NA10860 NA10847;NA10852 NA07055;NA07029;NA WWOX NA07019;NA06994 NA12056;NA12145;NA12864 NA06991;NA RemovalofPCRprimersandreagents 5lPCRproductswerecleanedupwith2lofUSB ExoSAPIT followinganincubationof15 at37ºplus 15 at80º. 1.3 SequencingPCR 1lofPCRproductafterExosap,wasaddedtoamixof1lBigDyeTerminator v3.1(appliedbiosystems), 1.5l5XBuffer,0.5lofeitherreverseorforwardprimer(10μM)and6lH 2 O,andasequencingPCR reactionwasperformedfollowingthepcrprogrambelow(30cicles): 30 95º 30 50º 3 60º ForMiniprepsproducts,thesequencingPCRwas performedwith400ngof DNAandoneofthevector s primers(table13). Table13:pGEMeasyT7andSP6universalprimerssequence. Primers(5 3 ) T7:TAATACGACTCACTATAGGG SP6:ATTTAGGTGACACTATAG 1.4 PurificationsequencingPCR Sequencing PCR products were purified with sepharose (Sephadex G50) columns. Briefly, 800 μl of sepharose were pipetted into a column and centrifuged at 1000 g for one minute for sepharose compactation. Flowthrough was discarded and 10 μl of water were added to the sepharose column and centrifuged for one minute again at 1000g. Finally, the column was introduced in a new eppendorf, the sequencingpcrloadedintothecolumnandcentrifugedfor1minuteat1000g.thepurifiedpcrwasrunina capillarysequenced(3730xlappliedbiosystems). 79
86 MATERIALSANDMETHODS 1.5 Sequenceanalysisandblasttohumangenome SequencingresultswereanalyzedwithCLCworkbenchwithstandardsettings.Onlycleansequenceswere selectedforblastanalysisinucscgenomebrowser. 1.6 PCRproductgelextraction PCRproductswererunona1.3%lowmeltingagarosegel,thebandscorrespondingtothedeletedalleles werecutusingauvtransilluminatorandextractedwithqiaquick GelExtractionKitfromQiagen,usinga microcentrifuge,accordingtoproductspecifications. 1.7 SubcloningPCRproducts TheamountofPCRproduct(insert)tobeincludedintheligationstepwascalculatedaccordingtheformula: ngofvector kbsizeofinsert insert:vectormolarratio=ngofinsert kbsizeofvector Wherethesizeofthevectoris3kb,vectorconcentration50ng/μlandinsert:vectorratioisrecommendedtobe1 Ligation of the extracted PCR product was performed with New England Biolabs Ligase and pgem easy vectoraccordingtomanualinstructions,withanovernightincubationina16 Cbath.Theligatedproduct wasthentransformedinjm109highefficientcompetentcellsthatwereplatedinlb/ampicilin/iptg/xgal platesovernightat37 C.Positivecolonieswerepickedthedayafterandincubatedfor1hourat37 Cin100 μloflysogenybroth(lb)medium(withampicilin),and1μloftheincubationwasusedtoperformthepcrto checkfortheinserts. The100μlofLBmedium(withampicilin)positivefortheinsert,wereincubatedin5mlofLBmedium(with ampicilin)overnightinashakerat37 Cat220revolutionsperminute(RPM).PlasmidDNAwaspurifiedthe day after with Qiagen Miniprep kit according to the manufacturers protocol and then quantified with Nanodrop. 2. CNVsGENOTYPING 2.1 MultiplexPCR Each genotyping reaction included both cases and controls in order to avoid possible bias, and negative (H 2 O)and,whenavailable,positivecontrols(HapMapsampleswithvalidatedgenotypesforeachCNV(146)). PCRconditionsareandprimersaresummarizedinTables14and15. 80
87 MATERIALSANDMETHODS Table14:PCRconditionsforCNVgenotyping.Greycellscorrespondtosharedcomponentsinbetweenreactions. ACACA GALNTL6 WWOX MYO5B PTPRD NRXN3 PCRprogram 2 94ºC 2 94ºC 2 94ºC 2 94ºC 2 94ºC 2 94ºC (30cicles) 30 94º 30 94º 30 94º 30 94º 30 94º 30 94º 30 60º 30 63º 30 61º 30 62º 30 60º 30 60º 1 72º 30 72º 30 72º 30 72º 30 72º 30 72º 7 72º 25 72º 7 72º 7 72º 7 72º 7 72º Mixcomponents 50ngDNA 75ng 10xRoche PCRreaction buffer+mg +2 dntps(mm) DelPrimers(pM) NonDelprimers(pM) TaqPolymerase(U) H 2 Otovolumeof25l 15l 2.2 PCRproductsdetection ACACAandPTPRDPCRproductswereloadedina2%AgarosegelandvisualizedwithaUVtransilluminator (GelDoc (BioRad)). WWOXandMYO5BPCRproductswereloadedina3%AgarosegelandvisualizedwithaUVtransilluminator (GelDoc (BioRad)). GALNTL6 CNV and NRXN3_DEL were genotyped by multiplex PCR with 5 FAM modification, followed by capillary electrophoresis in a 3730XL automatic sequencer and analysis with the Gene Mapper package (Applied Biosystems, Foster City, CA). Analysis was performed with the Gene Mapper package (Applied Biosystems). Samples showing peak intensities below 1000 fluorescent units or ratios of deleted allele to nondeleted allele<0.2or>5were notconsideredforanalysis.forcapillarydetection,nrxn3_del PCR reactionsweredilutedat1:15,and1μlofpcrdilutionwasthenaddedto9μlofaformamide/roxmixture (950μl+20μlper100samples),andsampleswereloadedinto3730XL.GALNTL6PCRproductswerenot diluted:1μlofthepcrwasaddedtotheformamide/roxmixture. 81
88 MATERIALSANDMETHODS Table15:PrimersforCNVgenotypingPCRreactions. Allele Primers(5 3 ) Productsize(bp) ACACDel ACACANonDel GALNTL6Del GALNTL6NonDel WWOXDel WWOXNonDel MYO5BDel MYO5BNonDel PTPRDDel PTPRDNonDel NRXN3Del NRXN3NonDel ACACA_DelF:GGCCTCCTCTTCTAGCTGTTG ACACA_DelR:AACAGGTGCCCAATAAATGC ACACA_F:GAGCCCATTAATCCAGAAAGG ACACA_R:TGACTTAGTGCCCATTCAAGG FF_Mi_A:[6FAM]GCAAGTAATGCCCAAGGAAA RF_Mi_del2:AGAGCATAAACCTCACAGGAC FF_Mi_wt:[6FAM]TGGTAATGAGCAGAGGAAAGG RF_Mi_wt5:TGAGCACTTACCCTGTCTGC WWOX_DelF:ATCTGGCCATGTCCTCATTT WWOX_DelR:TGTGACCTGATAACCGCTGA WWOX_F:AATGGGAATCTTTGCCTGTG WWOX_R:ATGGCAACTGACTTGGGAAG MYO5B_DelF:AACAGGCTGTCTCTTCCATGA MYO5B_DelR:CAGGGGTGGTTAGAATGAGG MYO5B_F:GAATGCATTTTGTCCAGCAGT MYO5B_R:CTCATAGAGGCGGTGTTCTTG PTPRD_DelF:GGGTGGTGGAAGGTGGTTAT PTPRD_DelR:GGTCTGGCATTTTGACATGA PTPRD_F:GCCAATTTCAGATCCTCAGC PTPRD_R:TTAGTGGCGTTCACACATGG NRXN3_FDel:CAGTCTTGACTGCTGGGTGAAC NRXN3_R:[6FAM]GTGACTGCTGATGAGCCACGC NRXN3_FNodel:GTGAGCACTCGATCCAGCATAA NRXN3_R:[6FAM]GTGACTGCTGATGAGCCACGC Del 980NonDel Veracodeassay NRXN3_del was also assessed with 3 SNPs included in a Veracode assay. Two of the SNPs were located withinthedeletedregion(rs andrs ),andwedesignedathirdsnpassay(nrxn3del)with eachofitsextensionprobesflankingthebreakpointsofthecnv.acombinationoftheresultsforthesesnps was used to assess the genotype. A sample was considered as homozygous deleted when failing in both SNPsincludedinthedeletedregionandamplyfinginthebreakpointsSNP;anheterozygoussampleforthe CNV was defined by presenting genotype for the three SNPs (the two inside the deletion having to be mandatorilyhomozygous);thehomozygousnondeletedsampleswerecharacterizedbythefailureofthe breakpointssnpandpresentinggenotypeatthesnpsinsidethecnvregion(beingeitherhomozygousor heterozygous). 82
89 MATERIALSANDMETHODS 2.4 Statisticalanalyses AllelicandgenotypicassociationanalyseswereperformedwithRSNpassocpackage.Associationanalysisof imputed data was performed with SNPTEST software. Quality control and association analyses of the VeracodeassaywereperformedwithPLINKsoftware.InteractionbetweenCNVsandNRXN3_DELandGWAS resultswastestedwithrsoftwareusingalikelihoodratiotest(lrt),whichcomparedthefullinteraction logisticmodel,amodelthatincludesaninteractiontermbetweensnps,tothesimpleradditivemodel,a modelthatincludesjustthesnpsasadditiveeffects. 3.EVALUATIONOFCNVFUNCTIONALCONSEQUENCES:NRXN3_DEL 3.1 GenomiccharacterizationofNRXN3_del:Veracodeassay We selected 45 SNPs covering the genomic region of NRXN3 using the Haploview software. These SNPs capture 42% of alleles in NRXN3 (1,460 Kb) and 66% in NRXN3 at r 2 0.8, based on the CEU HapMap genotypedsnps(version2,release21).fortheselection,potentialfunctionalvariants,variantslocatednear splicesitesandsnpsthathadbeenpreviouslyassociatedwithdisease(203)wereforcedtobeincludedas TagSNPs.Atotalof45SNPs(41TagSNPsand4singletonSNPs)and882FMsamplesand889controlswere included for genotyping. SNPs were genotyped using Illumina s VeraCode technology (Illumina), at the CeGen s (CEGEN) Barcelona node genotyping facility, following the manufacturer s protocol. The assays, developed for the VeraCode beads, were detected by Illumina s BeadXpress Reader System, and data analysiswasperformedusingillumina sbeadstudiosoftware(illumina,inc.,sandiego,ca,usa).asaquality control, 5% of the genotyped samples were duplicated, and notemplate controls and six HapMap trios (NA10840NA12286NA12287, NA12766NA12775NA12776, NA12818NA12829NA12830, NA12832 NA12842NA12843,NA12865NA12874NA12875,NA12877NA12889NA12890)wereincluded. 3.2 EvaluationofmRNAconsequencesofNRXN3_DEL SinceNRXN3isagenemainlyexpressedintheCNS,inordertoevaluateNRXN3_delconsequencesatthe mrna level, neuronal cell lines available in the laboratory were tested for NRXN3_del genotype: two neuroblastomacelllines(shsy5yandsknsh),aneuronalstemcellcelllineandtwoglioblastomacelllines (T98GandU87).NeuroblastomaandneuronalstemcellswereheterozygousforNRXN3_del,whileU87was homozygous deleted and T98G homozygous nondeleted. Therefore, U87 and T98G were selected to evaluatenrxn3_delpossiblefunctionalconsequences. 83
90 MATERIALSANDMETHODS T98GandU87CULTUREandDNAandRNAEXTRACTION T98GandU87glioblastomacelllineswereplatedandgrowninDulebecco smodifiedeaglemedium(dmem) supplementedwith10%ofheatinactivated(45minat56ºc)fetalbovineserumand1%ofpenicillinand streptomycin.mediumwasreplacedtwiceaweek.cellswerepassedwhenreachingconfluency(1:5every 48hoursapproximately).Aftertwopassescellswherecheckedforthepresenceofmycoplasmainfection withvenor GemMycoplasmadetectionkit (Minerva biolabs). Briefly,2μlofthe mediumwere collected andusedasatemplateinamycoplasmaspecificpcr.thus,mycoplasmainfectionwasdiscardedinbothcell lines.then,cellsweretripsinizedandpelletsforrnaanddnaextractionwerecollected. DNA extraction was performed with Wizard genomic DNA purification kit (Promega) according to the manufacturer sprotocol.rnaextractionwasperformedwithqiagenrnasepluskitaccordingtoproduct s protocolandincludingadnasepurificationstep.bothdnaandrnawerethenquantifiedwithnanodrop. AsbothU87andT98GDNAandRNApelletswereobtainedfromadifferentcellaliquot,withadifferentpass fromtheonesfromwhichdnahadbeenextractedandnrxn3_delhadbeenevaluated,nrxn3_delwas evaluatedagainconfirmingu87andt98ggenotypesforthisvariant. NRXN3TRANSCRIPTSINHUMANBRAINSAMPLES,ANDGLIOBLASTOMAANDHAPMAPCELLLINES Reversetranscriptase(RT)reactionwasperformedwith1μgofRNAwithsuperscriptIII(SSTIII)firststrand fromqiagen,includinganegativecontrol(withoutsst)followingmanufacturer sinstruction.rtstartedwith 5 at65ºc,followedby1 onice,then60 at50ºc,followedby15 at70ºcandafinalincubationof15 at 70ºC.1μlofcDNAwasusedinaPCRwithACTINprimersandanotherreactionwiththeforwardprimerin NRXN3exon18andthereverseinexon21inordertocapturethetwopossibletranscripts(withorwithout exon20)resultingfromalternativesplicinginsplicesite4(figure33). ACTINhousekeepingPCRreactionstartedwitha5 denaturationstepat95ºc,followedby30cyclesof30 at95ºc,30 at60ºc,and1 at72ºc,andafinalextensionstepof7 at72ºc.amplificationreactionswere performedwith1μlofcdnatemplate,10xroche PCRreactionBuffer+Mg,0.15mMdNTPs,0.4pMofeach oftheprimers(table16),0.1u/ltaqpolymerase,andh 2 Otoreachatotalvolumeof25l. 84
91 MATERIALSANDMETHODS Table16:Primersforexpressionhousekeepinggene(ACTIN)forcheckingtheperformanceoftheRTreactionand NRXN3primersfortranscriptsevaluation(takenfromOcchietal.work(204)) Primers(5 3 ) Productsize(bp) ACTIN_F:CTGGAACGGTGAAGGTGACA ACTIN_R:GGGAGAGGACTGGGCCATT NRXN3_F18:TCTTTGGGAAAAGTGGTGGG NRXN3_R21:ACCAAGATGCCATCCTTCAC NRXN3mRNAPCRreactionstartedwitha3 denaturationstepat94ºc,followedby37cyclesof30 at94ºc, 1 at65ºc,and1 at72ºc,andafinalextensionstepof7 at72ºc.amplificationreactionswereperformed with1μlofcdnatemplate,10xroche PCRreactionBuffer+Mg,0.16mMdNTPs,0.4pMofeachofthe primers (Table 17), 0.2 U/l Taq Polymerase, and H 2 O to reach a total volume of 25 l. 5 l of the PCR productwasloadedona2%agarosegelinordertocheckrtreaction(actin)andnrxn3transcripts.. Figure33:Neurexin3schemeincludingexonsandthetwocanonicalsitesofalternativesplicing.AsNRXN3_del(pink line)islocatedinintron1819,wewantedtoassesswhetheritmaybeinfluencingalternativesplicingatsplicingsite4, which may generate two different isoforms (with or without exon 20). SS4: splicing site 4; SS5: splicing site 5. Red arrowsrepresentpcrprimers.exonswithbluedashedlinesmaybediscardedthroughalternativesplicing.notethat both the numbering of exons and splicing sites don t start at number one because they also belong to the larger Neurexin3isoformandarethereforenumberedconsecutivelyafterNeurexin3exons116. CLONINGANDSEQUENCINGOFNRXN3TRANSCRIPTS Inordertocheckthatthetwotranscriptsobtainedcorrespondedtotheisoformswithandwithoutexon20, wesequencedbothproductsfromnrxn3mrnapcrreaction.forthiswegelextractedthecorrectband, purifieditandsubclonedit,andthenperformedsequencingpcrreactions. 85
92 MATERIALSANDMETHODS RELATIVEQUANTIFICATIONOFRTqPCRNRXN3TRANSCRIPTSinU87andT98GCELLLINES QuantificationofNRXN3transcriptsinthetwoglioblastomacelllineswasperformedwithTaqmanspecific geneexpressionassays,purchasedfromappliedbiosystemswebpage,followingmanufacturer sinstructions. InadditiontoaspecificassayforeachoftheNRXN3transcriptsresultingfromSS4#(Figure34),aNRXN3 assaymeasuringatranscriptnotaffectedbyalternativesplicingwaspurchasedinordertoevaluatethegene expressioninthecelllines.first,areversetranscription(rt)reactionwasperformedwithsuperscript III FirstStrand Synthesis SuperMix for qrtpcr (Applied Biosystems) according to the manufacturer instructions.rtreactionstartedby10 at25ºc,followedby30 at55ºcandicechilling.finally,rnasehwas added and the reaction was incubated 20 at 37ºC. Serial dilutions of the resulting cdna were then performedtochecktheefficiencyofthedifferentassaysinthetwoglioblastomacelllines(figure35).once theefficiencywasconfirmedasbeing2,reactionsincludingtwohousekeepinggenes(hprt1andtbp)and both NRXN3 assays in both cell lines were included in a single 384well plate experiment, in order to minimizebiasduetodifferentreactionsandplates.thiswasreplicatedinasecondexperimentwithanew RTreaction.qPCRreactionconsistedof40cyclesof15 at95ºand1 at60ºc.the10μlreactionconsistedof 5μlofTaqManuniversalPCRmastermixnoamperaseUNG,3.5μlofH 2 O,0.5μlofTaqmanGeneexpression assayand1μlofrtproduct.twoindependentrtreactionswereperformedforeachsample,andthen,for each RT, samples were run in quadruplicate. Reactions were run in an Applied Biosystems 7900HT Fast RealTimethermocycler.ResultswereanalysedwithSDSV2.1andRQmanager(AppliedBiosystems). Figure34:GenomiclocationofNRXN3Taqmangeneexpressionassays (NCBIB37;imagetakenfromTaqmanassayswebpageAppliedBiosystems). 86
93 MATERIALSANDMETHODS Figure35:AmplificationcurvesinU87cellline.a)HPRT1geneexpressionassay;(b)TBPgeneexpression assay;c)nrxn3geneexpressionassay.imagestakenfromrqmanagersoftware(appliedbiosystems). Relative quantification was calculated with the 2 Ct method to compare the expression values of both NRXN3transcriptsnormalizedtotwohousekeepinggenesHPRT1andTBP,usingthefollowintformula(205): RATIO=(Etest) Ct(ControlSample) /(Ereference) Ct(ControlSample) Then,wequantifiedNRXN3transcriptsinbothglioblastomacelllinesandcomparedtheratioofeachNRXN3 transcriptineachcellline. INSILICOEVALUATIONOFPTBPBINDINGSITESINNRXN3_DEL ThecandidateintronicregulatorysequencesweregatheredfromLaddetal(206)andwerealignedagainst thenrxn3_delgenomicsequenceobtainedfromucscgenomebrowser( clustalw2 ( Conservation of regulatory sequences present in NRXN3_delsequencewascheckedwithUCSCgenomebrowserconservationtrack. EVALUATIONOFPTBP2EXPRESSIONINT98GandU87CELLLINESANDMUSCLE WedesignedprimerstocheckPTPBP2expressioninT98GandU87celllines(Table17).Forwardprimerwas designedinexon2andreverseinexon4withagenomicspanof7921bp.ptbp2mrnapcrreactionstarted witha3 denaturationstepat94ºc,followedby30cyclesof30 at94ºc,30 at60ºc,and30 at72ºc,and a final extension step of 7 at 72ºC. Amplification reactions were performed with 1 μl of cdna template 87
94 MATERIALSANDMETHODS (T98G,U87,SHSYandmuscle),10XRoche PCRreactionBuffer+Mg,0.125mMdNTPs,0,4pMofeachofthe primers,0.1u/ltaqpolymerase,andh 2 Otoreachatotalvolumeof25l. Table17:PTBPprimerssequenceforcheckingtheexpressionofthegeneinU87andT98Gcelllines. Primers(5 3 ) PTBP2_F:GATGGTGCTCCTTCTCGTGTA PTBP2_R:TGCACTCTCGCTAACTGTGG Productsize(bp) 297bp 5 l of the PCR product were loaded on a 2% AgaroseGel in order to check the presence of PTBP2 PCR product.pcrproductwasthenpurifiedandsequenced. 88
95 RESULTS
96
97 RESULTS CLUSTERANALYSIS InordertoreduceFMheterogeneityweclassifiedclinicaldata(48variables)intosimplifieddimensionsthat definedfmsubgroups.1,446fmsamplesofcaucasianoriginwereincludedinthestudy.ofthese,97.2% werewomenwithameanageof49±10years;morethantwothirds(70.5%)weremarried;morethana third (37.5%) had a high school degree and an additional 19,3% had gone to university; and 80% of the patientshadapaidemployment. Aninitialsetof559unrelatedFMcaseswasconsideredfortheanalysis;thesameclusteranalysiswascross validatedinasecondsetof887cases.inthefirstdatasetof559unrelatedfmcases,theselected48clinical variablesclusteredintothreeindependentdimensions:fmsymptomsandtheircharacteristics(dimension1: symptomatology ),familialandpersonalcomorbidities(dimension2: comorbidities )andfmcoreclinical scales(dimension3)(figure36,table18).thecompositionoftheresultingdimensionswashomogeneous: only four variables (trembling, personal history of chronic pain, and SF36 mental and physical subscales) clustered in apparently unrelated dimensions. Since their weights within the respective dimension were among the lowest, their effect on the subsequent FM classification was reduced. Principal component analysisindicatedthepresenceoftwocorrelatedfactorsthatexplained31.59%oftotalvariability. Thisclusteringofthevariableswasreplicatedinthesecondcohortof887patients.Giventhisconfirmation,a globalanalysiswasperformedusingthewholecohorttoobtainaglobalweightforeachvariableinorderto performpatientclassification. Onlytwodimensions( symptomatology and comorbidities )wereconsideredfortheconstructionoffm subgroups.foreachfmsample,acompositeindexwascalculatedforthesetwodimensions.theresulting indexeswereusedtoclassify1,398outofthe1,446fmsamples,duetomissingdata. On one hand, these dimensions were considered as more reliable as they included more variables with higherweights.ontheotherhand,coreclinicalscaleswerenotincludedinthefmsubgroupingbecause theyweresubsequentlyusedfortheassessmentoftheresultingsubgroupsanddiseaseseverity.usingthe scales in this way allowed us to maximize the information from the scales instead of using them as dichotomizedvariables. 91
98 RESULTS Table18:Summaryofthethreedifferentdimensionsthatemergedafterclusteranalysis. VARIABLE VALUES DIMENSION WEIGHT NEIGHBOUR DIMENSION1:Symptomatology Widespreadpain 0=No1=yes Muscleweakness 0=No1=yes Postexercisefatigue 0=No1=yes Morningstiffness 0=No1=yes Muscularcontractures 0=No1=yes Concentrationproblems 0=No1=yes Memorycomplaints 0=No1=yes Onset 0=Progressive SleepDisturbances 0=No1=yes Forgetfulness 0=No1=yes Migratoryjointpain 0=No1=yes Headache 0=No1=yes Painsubtlemovementsimpairment 0=No1=yes Intestinaldysfunction 0=No1=yes Visualaccommodationimpairment 0=No1=yes Trigger 0=No1=yes Dizziness 0=No1=yes ExcessivePerspiration 0=No1=yes Monthsofpain(96;p 25 :48;p 75 :156) Personalhistoryofchronicpain 0=No1=yes Palpitations 0=No1=yes Ageofonset(38;p 25 :30;p 75 :45) DIMENSION2:Personalandfamily Posttraumaticstressdisorder 0=No1=yes Personalitydisorders 0=No1=yes Familyhistoryofautoimmunedisorders 0=No1=yes Familyhistoryofchronicfatiguesyndrome 0=No1=yes Panicattacks 0=No1=yes Familyhistoryoffibromyalgia 0=No1=yes Blackouts 0=No1=yes Facialoedema 0=No1=yes Connectivedisorder 0=FM Adjustmentdisorder 0=No1=yes PreviousPersonalhistorypsychopathology 0=No1=yes Majordepression 0=No1=yes Familyhistoryofchronicpain 0=No1=yes Impairedurination 0=No1=yes Spineosteoarthritis 0=No1=yes LifequalitySF36physicalsubscale(27;p 25 :22;p 75 :32) LifequalitySF36mentalsubscale(35;p 25 :25;p 75 :48) DIMENSION3:Scales HADdepressionsubscale(10;p 25 :7;p 75 :14) FibromyalgiaImpactQuestionnaire(FIQ)(74.66;p 25 :63.05; FatigueImpactScale(FIS)(66;p 25 :56.50;p 75 :75.00) Painlevel(VAS110cm)(7.5;p 25 :6.5;p 75 :8.5) PittsburghSleepQualityIndex(PSQI)(14;p 25 :11;p 75 :17) Fatiguelevel(VAS110cm)(8;p 25 :6.4;p 75 :9) HADanxietysubscale(12;p 25 :8;p 75 :15) NumberofTenderPoints(13;p 25 :13;p 75 :18) Trembling 0=No1=yes Thevariablesincludedineachdimensionarelistedandsortedbytheirweightedcontribution.Forcontinuousvariables, themedianwasusedascutoffvalueforbinarycodification.weightrepresentstherelationshipbetweenthesimilarity ofthevariabletotheothervariablesofthedimensionanditssimilaritytotheremainingvariables.neighbourrefersto theclosestdimension,excludingtheoneinwhichthevariablewasincluded. 92
99 RESULTS Component Component 1 These two components explain % of the point variability. Figure36:Clusteringofvariablesintothreedimensions.Crosses,triangles andcirclesrepresentvariablesassignedtoeachofthedimensions. Cases were classified into three subgroups: low symptomatology and low levels of familial and personal comorbidities (Cluster 1; 283 cases, 20.2%), high symptomatology and high comorbidities (Cluster 2; 357 cases,25.6%),andhighsymptomatologybutlowcomorbidities(cluster3;758cases,54.2%)(figure37). The resulting FM subgroups presented no differences in terms of gender, age, marital status, or employment. However, patients having higher levels of education (high school degree and above) were morerepresentedincluster1thantheoneshavinglowerlevels(65%vs.35%;p=0.015chisquaretest).this symptombasedclassificationcorrelatedwiththedatafromthescalesmeasuringpain,fatigue,psychiatric symptoms, and their impact in life, since individuals belonging to the low symptomatology and low comorbiditiesgroupalsohadlowermediansforthescales(table20). Figure37:Subgroupingoffibromyalgia(FM)samplesbasedonscoresin comorbidities (Yaxis)and symptoms (X axis).circlesrepresentfmpatientsandarecoloredbyclusterbasedontheclassificationbykmeans.blackcirclesare samplesofcluster1(lowlevelsofsymptomsandlowlevelsofcomorbidities),greencirclescluster2(highlevelsof symptomsandhighlevelsofcomorbidities),andredcirclescluster3(highsymptomsandlowcomorbidities). 93
100 RESULTS WhentherelationshipbetweenFMsubgroupsandcoremeasuresofseveritywasanalyzed,Cluster1was markedlydifferentfromcluster2andcluster3inallscales,identifyingthelessaffectedgroup.cluster2was moreaffectedthancluster3,butdifferenceswerenotassignificantaswhencomparingcluster1withthe othertwogroups(table19). TheFMsubgroupwithhighpainandcomorbiditieswasalsotheonewiththehighestleveloffatigue(fatigue VASwashigherthanpainVAS)and,infact,20%ofthepatientsinthisclusterfulfilledalsochronicfatigue syndromecriteria(fm+cfs)whereasintheothertwoclustersonly8%(cluster1)and11%(cluster3)ofthe patientswerefulfillingcfscriteria.althoughpainvaswashigherinthehighpainandcomorbiditiescluster, thenumberoftenderpointswasnotsignificantlydifferentbetweenbothhighpainclusters. Table19:Mediansofdifferentpain,psychiatricandqualityoflifescoresineachofthefibromyalgiaclinicalsubgroups. pvalueofmultinomialanalysis. VARIABLE FMCluster1 (N=283) FMCluster2 (N=357) FMCluster3 (N=758) Pvalue FibromyalgiaImpactQuestionnaire(FIQ) 59.46± ± ±14.38 <2.2E16 FatigueImpactScale(FIS) 65.10± ± ±16.46 <2.2E16 Painlevel(VAS110cm) 6.30± ± ±1.65 <2.2E16 Fatiguelevel(VAS110cm) 6.33± ± ±1.80 <2.2E16 NumberofTenderPoints 13.54± ± ± E15 LifequalitySF36physicalsubscale 30.7± ± ± E12 Lifequality(SF36)mental 42.12± ± ±13.57 <2.2E16 HADanxietysubscale 9.54± ± ±4.33 <2.2E16 HADdepressionsubscale 7.48± ± ±4.53 <2.2E16 PittsburghSleepQualityIndex(PSQI) 11.28± ± ±3.89 <2.2E16 Yearsofdiseaseevolution 11.14± ± ± GENOMEWIDEASSOCIATIONSTUDY SNPgenotypinganalysis 1. Populationstructure Therewasnoevidenceofpopulationstratificationinthesamplesconsideredforassociationanalysis,after removalofpcaoutliers,asillustratedbytheqqplot(figure38)andthegenomicinflationvalue(=1.013). 94
101 RESULTS Figure38:QQplotofFMGWAS.Observedpvaluesareplottedagainstexpectedpvaluesinanassociationstudyof500k SNPs.Thealmostperfectcorrelationbetweenobservedandexpectedvalueswasindicativeofabsenceofpopulation stratificationasprovenbyagenomicinflation()valueof1.013.figureobtainedwithwgaviewersoftware. 2. Genomewideassociationstudyfindings After QC procedures, 505,454 SNPs were considered for the anaysis in 300 FM and 203 controls. We performedallelicassociationanalysestofindlociassociatedwithfm.nosnpreachedgwassignificance.8 SNPsshowedapvalue<1X10 5 and69hadapvalue<1x10 4.SNPswithbetterpvaluesaresummarizedin Table20.Manhattanplot(Figure39)showedpossiblesignalsatchromosome3andX. Table20:SNPsshowingthestrongestallelicassociations(pvalue<104). SNP Pvalue Chromosome Coordinate(Hg18) Gene/Region rs ,14X106 X MCF2 rs ,20X STEAP1/STEAP2 rs ,52X ZBBX rs ,61X106 X ARMCX6 rs ,63X RP11518I13.1 rs ,62X PLCE1 rs ,06X NRXN1 rs ,94X TBX5 rs ,11X RP11478K15.2 rs ,12X RP1151O6.1 rs ,53X ZBBX rs ,61X ZBBX rs ,75X AP rs ,00X ZBBX rs ,21X SHISA6 rs ,35X105 X ARMCX4 rs ,35X105 X ARMCX4 rs ,48X AL rs ,50X AC
102 RESULTS rs ,60X MYT1L rs ,64X105 X ARMCX2 rs ,76X IFT140 rs ,81X105 X OTTHUMG rs ,82X OTTHUMG rs ,00X105 X LRG_128 rs ,05X CACNA2D1 rs ,10X NRXN1 rs ,23X RP11162K11.4 rs ,46X ZBBX rs ,79X105 X ARMCX4 rs ,91X AL rs ,05X ZNF438 rs ,06X ANK3 rs ,12X UNC5C rs ,39X SPNS3 rs ,43X TBX5 rs ,53X OTTHUMG rs ,61X AC rs ,68X SPNS3 rs ,72X OTTHUMG rs ,73X OTTHUMG rs ,75X NDUFV2 rs ,00X MACROD2 rs ,49X INPP4B rs ,68X AL rs ,99X105 X ARMCX4 rs ,19X SHISA6 rs ,27X CTSL3 rs ,42X OTTHUMG rs ,43X AC rs ,43X105 X ARMCX4 rs ,52X UNC5C rs ,61X CTSL3 rs ,62X OTTHUMG rs ,97X OTTHUMG rs ,15X STIM1 rs ,15X STIM1 rs ,15X STIM1 rs ,19X MYT1L rs ,24X AL rs ,28X RP4737E23.4 rs ,29X AL rs ,39X RP11738G5.1 rs ,53X CDK5RAP1 rs ,55X TBX5 rs ,84X RP1144H4.1 rs ,94X AKAP6 rs ,16X RP1144H4.1 rs ,24X105 X ARMCX4 rs ,36X ROR1 rs ,80X HMGCL rs ,18X CNPY1 rs ,25X EBF1 rs ,42X AL rs ,42X AC rs ,68X RP1144H4.1 rs ,73X105 X ARMCX4 SNPsarelistedondescendingorderbasedontheirpvalue.SNPsselectedforreplicationappearin bold. 96
103 RESULTS Figure39:SummaryofFMgenomewideassociationscanresults.NegativeLOG 10 pvaluesacrossthe genomeandbychromosomeareshown. 3.PathwayanalysisoftopassociatedSNPs 3.1 IngenuityPathwayAnalysis Thetop77SNPs(pvalue<1X10 4 )listedintable21,wereselectedtoperformpathwayanalysis.outofthe77 gene IDs introduced, only 53 were mapped. IPA pathway analysis identified two top networks that were overrepresentedinourgeneset:reproductivesystemdisease(16genes) )andneurologicaldisease(12genes) (Figure40). The analysis of canonical pathways showed overrepresentation ( p<0.05) of leucine degradation I, ketogenesis,, p7036k signaling, P13K Signaling in Blymphocytes and Dmyoinositol (1,4,5)trisphospate biosynthesis.thetopmolecularandcellularfunctionwascellulardevelopmentwith8molecules(p=1.24e E02). Figure40:NetworksidentifiedbyIPAsoftwareformGWAStopassociatedSNPs(imagetakenfromIPA softwareoutput).genesincludedinourgenesetappearinbold. 97
104 RESULTS 3.2 Geneset Only31ofthe77geneIDsweremappedinGenesetanalysisToolkittoperformGOanalysis.Thisanalysis identifiedtwomainmolecularfunctions:proteinbindingandionbinding(figure41),with16and14genes, respectively; in these categories, the analysis also identified metal ion binding as the only statistically significantoverrepresentedmolecularfunction(padjusted<0.05,benjaminicorrection)(figure42).mostof the mapped genes (18/31) encoded membrane or membrane related proteins, thus GO identified the membrane as the top cellular component (Figure 41), and a statistically significant overrepresentation of genes in the calcium channel complex. No biological process showed a statistically significant overrepresentationinthelistofgenesusedfortheanalysis.themostrelevant(althoughnonsignificant) biologicalprocessesrelatedtoourgenelistincludedcalciummediatedsignallingandcalciumiontransport, cardiacmuscledevelopmentand(figure42). Finally, Kegg pathways analysis (with N mapped gene Ids) indicated enrichment in inositol phosphate metabolism,phosphatidylinositolsignallingandmetabolicpathways. Figure41:Barchartofmolecularfunctioncategories(leftimage).Numberofgenesisplottedagainsteach category.barchartofcellularcomponentcategories(rightimage).numberofgenesisplottedagainst eachcategory.plotstakenfromgenesetgooutput. 98
105 RESULTS Figure42:GenesetoutputfromGOanalysis.Mostrepresentedbiologicalprocesses,molecular functionsandcellularcomponentsarerepresented.browncategoriesaretoptengocategories; statisticallysignificantenrichedgocategories(padjusted<0.05,benjaminicorrection)appearinred. 99
106 RESULTS 4. Imputation SNPsconsideredinGWASforassociationanalysiswereusedforimputation,and SNPswere imputed. Of these, SNPs were selected for having a good imputation quality (frequentist info score>0.8)andfinally forhavingaminimumallelefrequencyhigherthan0.05.sincethenumberof casesandcontrolswassmall,imputationdatawereonlyusedforfinemappingofsignalsdetectedindirect genotyping.threeimputedsnps,showingstrongerassociationthandirectlygenotypedsnp,wereincluded forreplication. 5. GWASreplication AfterQCprocedures,20SNPsin968casesand937controlswereconsideredfortheassociationanalysis (rs was discarded for not fulfilling HWE, p<0.05). We performed allelic association in the whole replication set and afterwards only in female samples. None of the selected SNPs was significant in the replicationanalysis,sincethecorrectionformultipletestingestablishedthesignificancethresholdat (Bonferronicorrection)(Table20). rs (p=0.025 in replication cohort), rs (p=0.039 in the replication cohort), rs (p=0.01inthefemalesreplicationcohort)andrs265015(p=0.016inthefemalesreplicationcohort)showed nominally significant associations in the replication study. However,the association for these SNPs in the replicationcohortwas for the oppositeallele of the one associated in the GWAS analysis. In fact, when addingthegwasdatatothereplicationdata,theseassociationswerelost. Wethenselectedthe4SNPsshowingthestrongestcombinedpvaluesandperformedanassociationtestin thesubsetsidentifiedbytheaforementionedclusteranalysis.wefirstperformedamultinomialanalysisin ordertotestforapossibleassociationbetweeneachsnpandthefmclustersgeneratedusingtheentirefm cohort(gwassamplesandsamplesincludedinthereplicationset)(supplementarytables14).thisshowed thatrs genotypesincluster3weredifferentfromthoseincluster2,althoughthisresultwasnot statisticallysignificant.nevertheless,wedecidedtoperformassociationanalysisforrs separately ineachfmcluster,bothinthegwassamples,inthereplicationcohortandwithafinaljointanalysis(table 21).Inthisanalysis,rs showedastrongerassociationinfemaleFMcasesbelongingtocluster3 than those belonging to cluster2, although the observed differences were not statistically significant (althoughtheorpointestimateofcluster2associationwasn tincludedinthecluster3or95%confidence intervalor95%confidenceintervalsoverlappedbetweengroups). 100
107 RESULTS Table21:SNPSselectedforreplication SNP Rank Type Gene/region PGWAS Prepli CombinedP Preplifem CombinedPfem rs intron_variant MCF2 2,14X X104 rs intergenic_variant STEAP1/STEAP2 3,20X rs intron_variant ZBBX 3,52X rs KB_downstream ARMCX6 3,61X rs intergenic_variant RP11518I13.1 3,63X rs intron_variant PLCE1 7,62X rs IM Intron_variant PLCE1 5,11X107 rs intron_variant NRXN1 8,06X rs intergenic_variant TBX5 8,94X rs IM intergenic_variant TBX5 1,49X rs intron_variant SHISA6 2,21X rs intergenic_variant 2,48X X104 rs intron_variant MYT1L 2,60X X104 rs intergenic_variant ZNF438 4,05X rs intron_variant ANK3 4,06X X104 rs intron_variant UNC5C 4,12X rs intron_variant LMO7 4,53X rs intron_variant MACROD2 5,00X rs IM intron_variant MACROD2 9,92X X X104 rs intron_variant LRG_164 7,15X rs intron_variant AKAP6 7,94X SNPsarelistedinascendingorderaccordingtoallelicassociationGWASpvalue(withtheexceptionofthethreeimputedSNPs).Type indicatestherelativepositionofthesnpwithrespecttothenearestgene,andgeneprovidesthegeneinwhichthesnpislocatedor thenearestgeneina500kbwindow.im:imputedsnp.type:snplocationuponthegeneaccordingtowgaviewerclassification; forrs andrs ,whicharelocatedonthexchromosome,allelicassociationwasonlyperformedinfemales subset. rs wasnotinhwenorinthewholecontrolset(pvalue=0.01)neitherinfemalescontrols(pvalue=0.005). In fact, both in the GWAS and replication datasets, the frequency of the protective allele A in FM cases belongingtocluster3wasslightlylowerthaninallthecases.asaconsequence,whenperformingthejoint analysisincludingallfemalecases(gwasandreplication)belongingtocluster3,theoverallsignificancewas increased (p=6.2x105). Association analysis results in the different FM subsets for rs are summarizedintable22. Table22:rs allelicassociationinthedifferentFMclustersinGWAS,replicationandjointcohorts rs F_FM F_CONTROLS Pvalue OR(95%CI) GWAS(300FMvs203C) X ( ) Replicationfem(940FMvs592C) ( ) GWAS+replicationfem(1240FMvs795C) X ( ) GWAScl3(196FMvs203C) X ( ) Replicationcl3(450FM vs592c) ( ) GWAScl3+replicationcl3(646 FMvs795C) X ( ) GWAScl1+replicationcl1(240FMvs795C) ( ) GWAScl13+replicationcl13(886FMvs795C) X ( ) GWAScl2+replicationcl2(304FMvs795C) ( ) F,frequencyoftheeffectallele(minorallele;AorTinrs );FM,femaleindividuals;C,controls. 101
108 RESULTS Finally,weevaluatedpossibleSNPsinLDwiththese4SNPsshowingthestrongestpvalue.Onlyrs and rs showed strong LD (r 2 >0.8) with other markers, as assessed by Haploview software (Supplementaryfigures1&2).rs wasincludedinablockanddefinedanhaplotypewithrs andrs ,taggingrs andrs FunctionalanalysisofSNPsselectedforreplication WeevaluatedinsilicothepossiblefunctionalconsequencesoftheSNPsselectedforreplicationwiththree differenttools:puppasuite,genevarandregulome.ofthe24snps(21plusthreeinld),puppasuiteonly foundfunctionalrelevancefortwosnps,rs981524andrs ,whicharehighlyconserved(figure43). Genvar was used to perform ciseqtlsnp analysis, finding a significant association of rs with STEAP2transcriptslevelsasassessedbytwoexpressionprobesintheciseQTLSNPanalysis(Figure44).SNP probeassociation splotwasnotavailableforthisdataset. rs (in almost perfect LD with rs , r 2 =0.95) showed a significant association with FAM13C transcriptslevels(oneexpressionprobe)intheciseqtlsnpanalysis(figure44).snpprobeassociationplot intheskinoffemaletwinsshowedacorrelationbetweenthesnpandfam13cgeneexpression(figure45). Figure43:SummaryofSNPpropertiesevaluatedbyPupasuiteinSNPsshowinganominalassociation inthereplicationcorhort.onlytwosnpsappearedtobeinconservedregions(asmarkedwithastar). 102
109 a) b) Figure44:a arrayprobes FAM13Cexpr areplotted a)rs inlymphobla ressionlevels dagainstchro Figure45:S FAM13C 6showedasta stoidcellline (oneprobe)i mosomalpos SNPprobeass Cgeneinfema atisticalsignif soffemaletw intheskinoff sition(1mbar significan sociationplot aletwin sskin RESULTS 103 icantassociat wins.b)rs790 femaletwins. roundthesnp tassociation forrs n.expressionl tionwithstea 06905showed Log10ofpva P).Dashedlin (p<0.001). 5andILMN_2 levelsareplot AP2expressio dastatistical lueoftheass erepresents expre ttedagainsts nlevelsintwo significantass sociationbetw thresholdfor essionprobef NPgenotypes oexpression sociationwith weenthesnps statistically from s. h s
110 RESULTS rs showed no significant association with any expression probe. However, both this SNP and rs (in almost perfect LD with rs , r 2 =0.90) presented a no statistically significant correlation(p>0.001),atoneexpressionprobe,withstng2geneexpressioninlymphocytes(seeannexes). Regulome analysis was only performed for 10 of the 24 SNPs, as the others did not have available information.theidentifiedfunctionalmarksweretfbindingsitesordnasepeak/histonemodificationswith minimalbindingevidence(table23). Table23:Resultsfromregulomedbanalysis. SNPS rs rs rs rs rs rs rs rs rs rs Functionalmark TFbindingorDNasepeak TFbindingorDNasepeak TFbindingorDNasepeak TFbindingorDNasepeak TFbindingorHistonemodifications TFbindingorHistonemodifications TFbindingorHistonemodifications TFbindingorHistonemodifications TFbindingorHistonemodifications TFbindingorHistonemodifications CNVAssessment:PENNCNV 1. Rawdataanalysis Wedetectedatotalof18648CNVeventsin317FMsamples,withamediansizeof20kbandincludinga meannumberof16snps(table24).thedistributionofthesecnvsamongthedifferentchromosomeswas, inglobal,inaccordancewiththechromosomesize(figure46),althoughtherewasahighernumberforcnv eventsinchromosome6,probablyduetothehlaregion. Table24:PennCNVresultsdescriptionofCNVsdetected. Min 1stQuartile Median Mean 3rdQuartile Max NºCNVs/sample Size(bp) NºSNPs Copynumber
111 RESULTS Chr21 Chr19 Chr17 Chr15 Chr13 Chr11 Chr9 CNV Size(Mb) Chr7 Chr5 Chr3 Chr Figure46:ChromosomaldistributionofCNVsdetectedbyPennCNV(red)andchromosesizes(datafromUCSC2003). OutofthedetectedCNVs,weselected94thatwerepresentinatleast5%ofthesamples(Table25)and thenfollowedaprioritisationpipelineforcnvvalidation(figure47).weendedupwithtwocnvsthatwere alsoidentifiedintheacghanalysis,locatedingng1andacaca. A3KbregionwithinGNG1genomicregionwasdetectedinaCGHasagain(Supplementarytable5),whereas PennCNV detected 59 samples with only one copy. Furthermore, although the aberration detected overlappedwithpreviouslydescribedcnvregions,thisoverlapwasonlypartial,despitethefactthatboth the genotyping array and the acgh design included probes fully overlapping the published CNVs. These circumstancesledustoprioritizethevalidationoftheothercnvvariant. 18,648CNVevents 94present inatleast 5%ofthe samples 43ingenesor <50kbgene 24inCNVs noninterindividualpolymorphisms 2present inacgh Figure47:PipelinefortheselectionofCNVstobevalidated.Weconsideredinterindividualpolymorphismsas thosecnvspreviouslydescribedaspopulationdependent,mainlyaffectinggenesinvolvedingeneenvironment interaction(gstt,adam,lce3,btln,citp450,amylasemetabolism,hla,ab,antigenrecognition). 105
112 RESULTS Table25:CNVregionsdetectedinatleast5%ofthesamples. Chromosome Start End NSNPs Length(bp) State Samples Gene chr state5,cn=3 47 TNFRSF18 chr state5,cn=3 24 TNFRSF18/TNFRSF4 chr state5,cn=3 21 SCNN1D/ACAP3 chr state5,cn=3 26 SCNN1D/ACAP3 chr state1,cn=0 31 FBLIM150kbupstream chr state2,cn= kbupstreamCD53 chr state1,cn=0 9 LCE3C chr state1,cn=0 59 NME7 chr state2,cn=1 58 RABGAP1L chr state2,cn=1 48 OR2T10 chr state2,cn=1 78 Intergenic chr state2,cn=1 197 Intergenic chr state5,cn=3 30 GALM chr state2,cn=1 31 Intergenic chr state1,cn=0 33 Intergenic chr state5,cn=3 33 RMND5Aclonesimilartoanaphase chr state5,cn=3 81 abparts chr state1,cn=0 38 Intergenic chr state2,cn=1 25 ZNF385B chr state1,cn=0 60 Intergenic chr state2,cn=1 21 CTDSPL chr state1,cn=0 42 SFMBT1 chr state2,cn=1 27 Intergenic chr state2,cn=1 40 EPHA3 chr state1,cn=0 21 Intergenic chr state2,cn=1 28 ZBTB20 chr state5,cn=3 21 Intergenic chr state2,cn=1 31 Intergenic chr state1,cn=0 37 BC chr state2,cn=1 70 CCDC50 chr state1,cn=0 32 Intergenic chr state1,cn=0 26 Intergenic chr state2,cn=1 129 Intergenic chr state2,cn=1 58 UGT2B17 chr state2,cn=1 116 Intergenic chr state1,cn=0 74 Intergenic chr state1,cn=0 30 Intergenic chr state1,cn=0 26 Intergenic chr state6,cn=4 32 Intergenic chr state2,cn=1 26 AK chr state2,cn=1 83 SGCD chr state1,cn=0 29 BTNL8 chr state2,cn=1 36 Intergenic chr state2,cn=1 40 HLAB chr state1,cn=0 249 DRB5 chr state5,cn=3 44 Intergenic chr state2,cn=1 49 Intergenic chr state1,cn=0 82 Intergenic chr state2,cn=1 122 Intergenic chr state5,cn=3 28 Intergenic chr state2,cn=1 31 Intergenic chr state6,cn=4 51 Intergenic chr state2,cn=1 59 GNG1 chr state1,cn=0 48 Intergenic chr state1,cn=0 49 CAV2 chr state2,cn=1 32 MGAM chr state1,cn=0 70 ERICH1 chr state5,cn=3 33 Intergenic 106
113 RESULTS chr state2,cn=1 193 ADAM chr state2,cn=1 43 Intergenic chr state1,cn=0 29 Intergenic chr state1,cn=0 75 Intergenic chr state2,cn=1 35 DQ594366/FAM27E3 chr state5,cn=3 29 IL15RA chr state2,cn=1 65 GENEDESERT chr state2,cn=1 23 SPLICEDest chr state1,cn=0 47 genedesert chr state2,cn=1 30 MGPRX1 chr state5,cn=3 28 Intergenic chr state2,cn=1 89 OR4C11 chr state2,cn=1 52 Intergenic chr state1,cn=0 98 Intergenic chr state5,cn=3 47 Intergenic chr state6,cn=4 43 Intergenic chr state2,cn=1 71 Intergenic chr state2,cn=1 55 STARD13 chr state2,cn=1 71 intergenic chr state5,cn=3 242 Abparts chr state5,cn=3 111 Abparts chr state5,cn=3 39 OR4N4 chr state5,cn=3 33 Intergenic chr state2,cn=1 34 Intergenic chr state2,cn=1 42 Intergenic chr state1,cn=0 32 Intergenic chr state5,cn=3 83 Intergenic chr state1,cn=0 41 ACACA chr state2,cn=1 24 DOK6 chr state2,cn=1 36 ZNF826 chr state2,cn=1 50 CYP2A7 chr state5,cn=3 53 Intergenic chr state2,cn=1 32 Intergenic chr state2,cn=1 25 BCAS1 chr state5,cn=3 42 abparts chr state5,cn=3 89 GSTT Coordinatesforbreakpoints,numberofSNPsincludedintheCNV,CNVsize,state(copies),numberofsamplespresenting theaberrationandgene/genomicpositionareprovided. ACACA PennCNVidentifieda1kbdeletedregion(0copies)in41FMsamples.ThiswasincludedinalargerCNV region detected by Conrad et al. (146), which had acgh support in our datasets. By acgh, this region appearedtohaveahigherfrequencyofthedeletedalleleincasesthanincontrols(table26). Table26:LRRof400KarrayhybridizationsinACACAPennCNVregion. PROBE CHR START END GENE FMvsC FMvsC_DS FM_FCvsC FM_FCvsC_DS FM_EARLYvsC FM_EARLYvsC_DS A_16_P chr ACACA A_16_P chr ACACA A_16_P chr ACACA A_16_P chr ACACA A_16_P chr ACACA TwoFMpools(withandwithoutfatigue)presentedadeletionbothindirectanddyeswaphybridizations. 107
114 RESULTS First,weattemptedtodeterminethebreakpointsofthisCNVregion.WedesignedPCRprimerstakinginto accountacghresultsratherthanpenncnvcoordinates,asacghresultsoverlappedwithcnvspreviously described (146, 207) and the smaller PennCNV region was included in this CNV region and couldn t be overlappingitcompletelyastherewerenoillumina sprobesintheremainingregion(figure48).weused CEUHapMapsamplesforwhichACACACNVgenotypeshadbeendeterminedinthepublicationbyConradet al.(146).onlyhomozygousdeletedandheterozygoussamplesamplifieda1200bpproduct(figure49)which wassequenced.astheresultingsequencesdidn tallowbreakpointsdetection,weperformedsubcloningof the deleted PCR products and sequenced the purified plasmid. We mapped the CNV s breakpoints, characterizingitasa3. 1kbdeletion(chr17: )(Figure48),locatedinanintronicregion withintheacacagene..wethendevelopedamultiplexpcrassayforgenotypingandusedittogenotype ourcohort. Figure48:GenomiclocationoftheCNVwithdefinedbreakpointsandthevariantpredictedbyPennCNV. PennCNVonlydetectedpartofthevariationbecauseoftheabsenceofIlluminaprobesintheupstream regionofthecnv(greenbox)imagetakenfromucscgenomebrowser). Figure49:HomozygousdeletedandheterozygousHapMapsamplesfor theacacacnvamplifieda1200bpproduct(2%agarosegel). 108
115 RESULTS Aftergenotyping200FMcasesandcontrols,nodifferenceswereobservedbetweenthetwogroups(Table 27;genotypicassociation,Fishertestp=0.2).Whatismore,theresultsfromthese400individualswerein contradictionwiththeexpectedresultsfromacgh,asthedeletionappearedmoreoftenincontrolsamples (Del: 27.9% FM, 31.5% controls). Given these discouraging results, the analysis of this region was not pursuedfurther. Table27:ACACACNVgenotypingresults. ACACA_CNV GENOTYPES CONTROLS (N=211) FM (N=209) Del/Del 23 (10.9%) 24 (11.4%) Del/NoDel 87 (41.2%) 69 (33.4%) NoDel/NoDel 101 (47.8%) 116 (55.5%) 2.Analysisofrarelargeevents PennCNVdetectedtheoccurrenceof25rare,largeevents(Supplementarytable6).Onlytwoofthesewere identifedinatleasttwosamples,andbothwerealsodetectedinasetofcontrolsofeuropeanorigin(data comingfromanotherstudythatwasongoinginthelaboratory).therefore,norarecnvwasselectedfor followup. AssessmentofCNVsinmosaicstate TheMADalgorithmidentified25nonLOHaberrationsweredetectedbythealgorithm.Outofthese,9were recurrent(table28) Table28:RecurrentnonLOHaberrationsinmosaicstate. Chromosome Coordinates(Hg18) Gene Aberration Samples chr kbupFMRD1 Gain 10 chr Gain 7 chr Gain 11 chr ANTXRL Gain 29 chr OVOS2 Gain 14 chr SLC2A14 Gain 11 chr HRNBP3 Gain 8 chr Gain 47 chr LRP5L Gain 9 Fiveofthemwerelocatedingenes.ThemostrecurrentwaslocatedinANTXRL(anthraxtoxinreceptorlike), whichwasageneofunknownfunction.theaberrationwithinovos2(figure50)waspresentin14samples (in8casesitincludedthewholegeneandin6samplesitinvolvedaregion13kbupstreamofthegene). 109
116 RESULTS Between21and48%ofthecells(dependingonthesample)werecarryingthegain.Theregionoverlapped withpreviouslyreportedcnvs(figure51).11samplespresenteda48kbgainincludingthetwofirstintrons and exons of SLC2A14. This gene encodes a ubiquitously expressed glucose transporter. Between 22 and 43%ofthecells(dependingonthesample)werecarryingthegain.Ninesamplespresentedagaininchr22: ,includingpartofLRP5L(lowdensityreceptorrelatedprotein) and8samplescarrieda CNVinchr17: ,locatedintheintronicregionofHRNBP3(hexaribonucleotidebinding protein3). Figure50:AberrationdetectedinOVOS2.Theregion(markedbyayellowoval)isidentifiedasit presentschangesinbafwithoutsignificantchangesinlogr.intheimage,thebaf(reddots)andlogr (blackdots)areplottedagainstchromosomalposition. Figure51:OVOS2mosaiceventoverlappedwithpreviouslydescribedCNVs.ImagetakenfromUCSCgenomebrowser. 110
117 RESULTS ARRAYCOMPARATIVEGENOMICHYBRIDIZATION 400Karray 1. Results UsingtheADM2algorithmprovidedbyAgilent ssoftware,wedetectedsevenregionsshowingdifferential hybridizationinfmsamplesincomparisontocontrolsbothindirectanddyeswaphybridizations(table29). Three regions (WDR60, DOCK5 and SIRPB1), were not considered for replication since they persistently appearedinalltheacghexperimentsperformedinthelaboratory(datanotshown). Table29:400kCNVaCGHresults.GenomiclocationisbasedonbuildHg18.DS:Dyeswap. Chromosome Cytoband Gene Start End Probes Log2ratio_Direct Log2ratio_DS Pools chr4 q34.1 GALNTL , , FMearly chr7 q36.3 WDR , , FMearly chr8 p21.2 DOCK , , FM_fatigue chr9 p23 PTPRD , ,828 FM chr14 q31.1 NRXN , , FM_fatigue FMearly chr16 q23.1 WWOX , , FMearly chr20 p13 SIRPB , , FM_fatigue 2. ValidationofaCGHresults Four regions coming from ADM2 analysis were selected for validation. An additional region, MYO5B, detected by the less restrictive ADM1 analysis was selected for validation because it constituted a good candidateforfm.thebreakpointsforfourofthemhadbeendefinedpreviouslyandbreakpointscoordinates were available in public databases (dbsnp and UCSC genome browser; hg18): GALNTL6:rs (chr4: ); PTPRD: rs (chr9: ); MYO5B: rs (chr18: )andnrxn3:ss (chr14: ).fortheremainingregion, WWOX, we had to identifie the indel s breakpoints. Then, for all regions, we designed multiplex PCR experiments to genotype them, first in a subset of 300 FM samples and 300 controls and, if there was validationofacghfindings,inourentirecasecontrolcohort(figure52). 111
118 RESULTS Figure52:FiveregionsfromaCGHresultswereselectedforreplication.Sizeofthevariantsisindicated(bp). 2.1 GALNTL6 Thefirstregionselectedforvalidationwasa5kbdeletiondetectedintheFM_earlypoolthatoverlapped with a previously described CNV (chr4: (hg18)). Although the overlap wasn t total, thiscouldbeexplainedbyalowdensityofarrayprobesinthecnvflankingregions(supplementarytable7). Taking advantage of a multiplex PCR assay designed in the laboratory (Figure 53), we genotyped a initial subsetof417controlsand460fmsamples,andfoundnominalassociationatthegenotypicandalleliclevels (p=0.04or(95%ci):0.78( )genotypicassociation,logadditivemodel;p=0.04or(95%ci)0.77( ) allelic association (Fisher test)). This wasn t confirmed when we increased the sample size to 893 controls and 1095 FM cases (p=0.55, genotypic association, logadditive model; p=0.56 allelic association (Fishertest)).SincethisregionwasdetectedonlyintheFM_EARLYpool(FMpatientswithageofonsetof thediseasebefore20yearsold),weconsideredthepossibilitythattheassociationwasspecifictotheearly onset cases. We performed a second analysis considering only early onset cases (age of onset<20) and, although we found a higher proportion of homozygous deleted samples in this subset, this was still not statisticallysignificant(p=0.29,genotypicassociation,logadditivemodel;p=0.33allelicassociation(fisher Test))(Table30). Figure53:GenemapperimageofaheterozygoussampleforGALNTL6CNVgenotyping. PeaksintensitiesareplottedagainstthePCRproductssizes(inbp). 112
119 RESULTS Table30:GenotypicdistributionofGALNTL6CNVina)Theinitialsubsetofcasesandcontrolsb)controls,FMcasesand FMofearlyonset. a) b) 2.2 WWOX GALNTL6CNV GENOTYPES CONTROLS (N=417) FM (N=460) Del/Del 273(65.5%) 744(70.4%) Del/NoDel 121(29.0%) 317(26.7%) NoDel/NoDel 23(5.5%) 34(2.8%) GALNTL6CNV GENOTYPES CONTROLS (N=893) FM (N=1095) FMearly (N=101) Del/Del 597(66.9%) 744(67.9%) 74(73.3%) Del/NoDel 265(29.7%) 317(28.9%) 23(22.8%) NoDel/NoDel 31(3.5%) 34(3.1%) 4(4.0%) In the 400K_CNV acgh experiments, analysis by the ADM2 algorithm detected an aberrant region at chr16q23.1 spanning over 11 probes. This region was shown as deleted in the FM_Early pool versus the controlsbothindirectanddyeswaphybridizations.theregion,ofalmost14kb,overlappedwithaknown CNV(146).TakingintoaccountthepositionsofaCGHprobes(Supplementarytable8)andthechromosomal positionsforthepublishedcnv,wedesignedapcrexperimentinordertodetectthecnv sbreakpoints.as thefm_earlypoolpresentedaloss,wegenotypedthesamplesincludedinthispool,expectingtodetectthe deleted allele in homozygous or heterozygous states. Three combinations of primers were tested. Single bandproductsweredirectlysequenced,whileforunspecificpcrreactions,thepcrproductwasloadedina lowmeltingagarosegel,forbandextractionandpurification,andtheresultingpurifiedproductwasthen sequenced(figure54). 113
120 RESULTS Figure54:WWOXCNVbreakpointsdetectionsPCRsloadedin2%agarosegel.a)F1R3combinationof primersresultedinaproductofapproximately1000bp.b)f1r1combinationdidnotamplifyandf1r2 resultedinaproductofapproximately1000bpbutwiththepresenceofunspecificbands(reddashed boxes).c)twoofthesamplesoff1r2reactionwereloadedina1.3%lowmeltingagarosegelforband isolation. WedetectedthebreakpointsfortheCNV,characterizingitasa13Kbdeletionlocatedintheintronicregion ofwwox(chr16: (hg18),andwedesignedamultiplexpcrassaytogenotypeit(figure 55).Wegenotyped619FMsamplesand691controlsandfoundnostatitiscallysignificantdifferences.In fact,therewasaslight,nonsignificant,increaseofthenondeletedalleleandthehomozygousnondeleted genotypeincases(allelicassociationfishertestp=0.10,or(95%ci)=1.13( );genotypicassociation log additive model p=0.10, OR (95%CI)=0.88 ( ); table 31) which was contradictory to the acgh results. 114
121 RESULTS Figure55: :WWOXmultiplexPCRloadedina3%agarosegel. Table31:GenotypicdistributionofWWOX_INDELincasesandcontrolsgenotypedwithamultiplexPCR. WWOX GENOTYPES CONTROLS (N=691) FM (N=619) NoDel/NoDel Del/NoDel Del/Del 172(24.9% ) 169(27.3%) 340(49.2% ) 314(50.7%) 179(25.9% ) 136(22.0%) 2.3 PTPRD TheCNVinPTPRDwasa500bpdeletioncomparedtothereferencegenome.Thisdeletionwasgenotyped byamultiplexpcr.sincethedeletionwassmall,theassayforthedeletedallele(450bp)generateda980bp productforthenondeletedallele.forthisreason,wedesignedaspecificassayforthenondeletedallele, generating a smaller product (219 bp) that ensured multiplex PCR with a balanced amplification of both alleles (Figure 56). PTPRD_INDEL genotyping of 283 controls and 303 cases didn t support acgh findings (Supplementarytable9).Wedidn tfindanyhomozygousnondeletedsamplesandthedistributionofthe othertwogenotypesdidnotdifferbetweencasesandcontrols(table32;p=0.7genotypicassociationlog additivemodel,allelicassociationfishertestp=0.08). Figure56:PTPRD_INDELmultiplexPCRloadedina2%Agarosegel.Two productscorrespondtothenondeletedalleleandonetothedeletedallele. 115
122 RESULTS Table32:GenotypicdistributionofPTPRD_INDELamongcasesandcontrols. PTPRDGENOTYPES Del/Del Del/Nodel CONTROLS (N=283) 200 (70.7%) 83 (29.3%) FM (N=303) 218(71.9%) 85 (28.1%) 2.4 MYO5B A3kbgaininMYO5BintronicregionwasdetectedinFMpool(bothindyrectanddyeswaphybridizations) withadm1algorithm(supplementarytable10).asitoverlappedwithapreviouslydescribedindel,weused thebreakpointscoordinatestodesignamultiplexpcr(figure57).wegenotypeditin350fmsamplesand 306controlsandfoundnostatitiscallysignificantdifferences(allelicassociationFishertestp=0.11;genotypic associationlogadditivemodelp=0.9)(table33).. Figure57:MYO5B_INDELmultiplexPCRproductloadedina3%agarosegel. Table33:GenotypicdistributionofMYOB_INDELincasesandcontrols. 2.5NRXN3 MYO5B_INDELGENOTYPES NoDel/NoDel Del/NoDel Del/Del CONTROLS (n=306) 142 (46.4%) 1333 (43.5%) 31 (10.1%) FM (n= =350) 161(46%) 153(43.7%) 36(10.3%) AdeletioninanintronicregionofNRXN3wastheonly400KarrayresultthatwaspresentintwoFMpools (Supplementary table 11). In order to validate this enrichment of NRXN3_DEL in FM samples, we first verified the published breakpoints (chr14: (hg18)) by sequencing a HapMap homozygousdeletedsampleandfoundaslightlysmallerevent(chr14: (hg18))(figure 58).ThenwedesignedamultiplexPCRandgenotypedtheCNV(Figure59)inaninitialsubsetof359FM cases and 378 controls, confirming the association of the deleted allele with FM (genotypic association, 116
123 RESULTS recessive model (homozygous deleted as risk genotype) p=0.0037, OR (95%CI) = 1.74( ); allelic associationp=0.12fishertest)(table34).wecompletedthegenotypingofourentirecohortwithboththe multiplexpcrassayandasnpbasedassayincludedinaveracodeexperiment,findingatrendtoassociation (genotypic association, dominant model p=0.064 OR 1.18 ( , allelic association Fisher Test p value=0.07,or(95%ci)=1.12( )).since97%offmcaseswerefemales,weperformedthesame analysis only considering female cases and controls and the assiociation became statistically significant (genotypic association, recessive model (homozygous deleted as risk genotype) p=0.021, OR 1.46 ( ),logadditivemodelp=0.014,OR1.22( ),allelicassociationFisherTestp=0.015OR(95%CI)= 1.22( )(Table35). WeperformedassociationanalysisonthesubsetsgeneratedbytheclusteranalysisthatidentifiedthreeFM subgroups. We found that there were differences among clusters, with an enrichment of the deletion in clusters 1 and (in a minor extent) 3 (interaction pvalue=0.046, supplementary table 12). We performed association analyses for each of the FM clusters against the controls and found that association was statisticalsignificantinfmsampleswithlowlevelsofcomorbidities(clusters1and3)(genotypicassociation, dominantmodel(homozygousdeletedandheterozygousasriskgenotypes)p=0.009,or(95%ci)=1.28( );allelicassociationFishertestp=0.019,OR(95%CI)=1.17( ))andtheassociationwasstronger ifconsideringonlyfemales(genotypicassociation,recessivemodel(homozygousdeletedasriskgenotype) p=0.019, OR (95%CI) = 1.49 ( ), log additive model p=0.004 OR (95%CI)=1.28 ( ), allelic associationfishertestp=0.004or(95%ci)=1.27( )(tables36) Figure58:NRXN3_DELbreakpointsdetection.NRXN3deletedallelesequencewasblastedagainstthe humanreferencegenome(hg18).a8870bpdeletion(chr14: )wasidentified. 117
124 Figure fromho Table34:a)G a) GENOTYPE Del/Del Del/NoDe NoDel/NoD Tables35:a) samples.c)g a) GENOTYPE Del/Del Del/NoDe NoDel/NoD c) GENOTYPE Del/Del Del/NoDe NoDel/NoD e59:nrxn3_ omozygousde Genotypicdis ES CO N 55 el 169 Del 154 Genotypicdis Genotypicdist ES CO N 118 el 386 Del 362 ES CONT N 48 el 207 Del 190 DELgenotypin eleted,hetero tributionofn NTROLS N=378 (14.6%) 9(44.7%) 4(40.7%) stributionofn tributionofnr NTROLS N=862 8(13.7%) 6(44.3%) 2(42.0%) TROLSfem N=445 8(10.8%) 7(46.5%) 0(42.7%) ng.multiplex ozygousandh NRXN3_DELam CNVinthe FM N=359 82(22.8%) 130(36.2%) 147(40.9%) NRXN3_DELa RXN3_DELam allth FM N= (14.9%) 657(47.0%) 532(38.1%) FMfem N= (15.0%) 640(47.1%) 514(37.8%) RESULTS 118 PCRproducts homozygousn mongcasesan samesubset b ) ) mongcasesa mongfemalec hefemalesam b) ) ) ) d) ) ) ) A N D srunina2%a ondeletedge ndcontrolsin ofsamples. b) ndcontrols.b casesandcont mples. ALLELES Nodel Del ALLELES NoD De ALLEL ES NoDel Del agarosegel(to enotypesforn itialdataset.b b)allelicdistri trols.d)allelic CONTROLS 477 (63.1%) 279 (36.9%) C Del 11 el 6 S CO op).genemap NRXN3_DELC b)allelicdistr ibutionofthe cdistribution FM CONTROLS 106(64.1%) 618(35.9%) ONTROLSfem 7(65.9%) 3(34.1%) pperimages CNV(bottom). ibutionofthe CNVinallthe ofthecnvin M 4(59%) 94(41%) FM 1721(61.7% 1073(38.3% FMfem 1668( (38.6. e e n %) %) %) %)
125 RESULTS Tables36:NRXN3_DELassociationacrossthedifferentclusters.a)Genotypic distributionamongthedifferentfmclusters.b)allelicdistributionclusters1and3. a) GENOTYPES CONTROLS N=862 FMCl1 N=277 FMCl2 N=341 FMCl3 N=730 FMCl1+Cl3 N=1007 FM N=1397 FMfemCl1+Cl3 N=980 Del/Del 118(13.7%) 50(18.1%) 49(14.4%) 104(14.2%) 154(15.3%) 208(14.9%) 150(15.3%) Del/NoDel 386(44.3%) 132(47.7%) 149(43.7%) 357(48.9%) 489(48.6%) 657(47.0%) 476(48.6%) NoDel/NoDel 362(42.0%) 95(34.3%) 143(41.9%) 269(36.8%) 364(36.1%) 532(38.1%) 354(36.1%) b) ALLELES CONTROLS FM(cl1+cl3) CONTROLSfem FM(cl1+cl3)fem NoDel 1106(64.1%) 1217(60.4%) 587(65.9%) 1184(60.4%) Del 618(35.9%) 797(39.6%) 303(34.1%) 776(39.6%) 3. CNVsinteractionevaluation WeevaluatedapossibleinteractionbetweenNRXN3_DELandGALNTL6CNVandWWOXCNVandfound thattherewasnoevidenceofinteraction(figure60). Figure 60: Plot representing interaction of NRXN3, GALNTL6 and WWOX CNVs. There was no evidence of statistical significantinteraction. Multiplecomparisonresultsare presentedintheplotsconsideringthecodominantmodelof geneticactionforbothtestedpaircnvcombinations.statisticalsignificancesarerepresentedbycolourscale;dark greenindicatesgreaterstatisticalsignificanceandyellowwhiteindicateslessstatisticalsignificance.theuppertriangle inthematrixcontainsthepvaluesfortheinteraction.thediagonalcontainspvaluesfromalikelihoodratiotest(lrt) forthecrudeeffectofeachcnv.thelowertrianglecontainspvaluesfromalrt,comparingthetwocnvadditive likelihoodtothebestofthesinglesnp. 119
126 RESULTS 4.NRXN3_DELGWASSNPsinteraction WeevaluatedapossibleinteractionbetweenNRXN3_DELand thefourgwassnpspresentinganominal associationinthejointanalysis.wethenperformedthesameanalysisconsideringonlyfemales,inorderto include the X chromosome variant rs There was no evidence of interaction; females there ws evidenceofanadditiveeffectofrs andrs (p=0.04)(figure61). Figure61:PlotsrepresentinginteractionofNRXN3_DELandrs ,rs ,rs inthewholecohort (leftplot)andinthefemalessubset(codominantmodel). 120
127 RESULTS acgh1marray Fortheanalysisofthe1MaCGHarrayweusedthesameselectioncriteriaasfortheanalysisofthe400K array,exceptthatweonlyhaddatafromdirect(insteadofdirectanddyeswap)experiments.fiveregions weredetectedaspotentiallydeletedorgainedbetweencasesandcontrols(table37)inoneormorepools. The CNV located near RXRA was discarded since FM and FM_FC presented a gain whereas FM_early presentedaloss;acarefulexaminationoftheacghresultsshowedthatthealteredprobesforscaperand SLC12A7 CNVs were actually the same probe that was duplicated. Finally SHANK3 and SCLA9A3 regions, were also discarded because not all probes in the regions were actually. For all these reason, we didn t undertakethevalidationof1marrayacghresults. Table38:1MaCGHresults.RegionsdetectedbyADM2algorithm,forregionsincludingatleast3probesand withalogratio>0.3. Chromosome Cytoband Gene Start End Probes Log2ratioDirect Pools chr9 q34.2 RXRA FMearly FM_fatigue FM chr15 q24.3 SCAPER FMearly FM chr5 P15.33 SLC9A FMearly FM chr5 p15.33 SLC12A FMearly chr22 q13.33 SHANK FMearly 121
128 RESULTS EvaluationofthepossiblefunctionalconsequencesofNRXN3_DEL 1.Veracodeassay AveracodeassayincludingseveralfunctionalandtaggingSNPsintheNRXN3genewasdesignedtotestfor association with other NRXN3 polymorphisms that could have been tagged by the deletion. After quality control,34snpsin869fmsamplesand843controlswereincludedforanalysis,as4snpsfailedhwe,7 SNPspresentedmissingness>0.05and12aMAF<5%,while59individualswereremovedforhavingalow genotypingrate.aftercorrectingformultipletesting(34markers,p< ,bonferronicorrection)nosnp showed significant association (supplementary table 13). The three assays included to genotype the NRXN3_DEL presenteda 100%concordancewith theresultsfromthe multiplexpcrassay.noothersnp appearedtobeaproxyofthedeletion.wethenappliedclusteranalysisandfoundnointeractionwithsnps associationandthedifferentclusters. 2.mRNAexperiments We performed a NRXN3 PCR with cdna obtained from two glioblastoma cell lines with a different homozygous genotypeforthedeletion,u87(del/del)andt98g(nondel/nondel),,andincludingaswella neuroblastomacellline(shsy)thatwasheterozygousforthedeletion.twoproductswereexpected,andin threeindependentexperiments(fromthreedifferentrtreactions)wefoundthattherewasadifferentratio ofthetwoproductsbetweencelllines(figure62). Figure62:mNRXN3PCRinthreeindependentRT.T:T98Gcellline;U:U987cellline; 11:SHSYcellline:11sinSST:RTreactionblank(withoutsuperscript); :PCRblank. 122
129 RESULTS Wesubclonedandsequencedeachoftheproductsandconfirmedthattheycorrespondedtotheexpected products:nrxn3withandwithoutexon20(figure63). Figure63:GenomicpositionofblatresultsofmRNANRXN3sequencedproducts.Top imagecorrespondstothesmallerpcrproductnotincludingexon20andbottomimage tothelargerfragmentincludingexon20.imagestakenfromucscgenomebrowser To quantify these differences we performed quantitative RTqPCR using commercial Taqman expression assays.sincet98gandu87celllinescamefromindividualsofdifferentgender,weusedahousekeeping gene notlocatedon the Xchromosome:TBP.Weperformed thesameexperimentusingcdnafromtwo differentrtexperimentsandconfirmedthedifferentratiobetweenthenrxn3isoformswithandwithout exon20(figure64)(table38).whereasbothisoformswherehighlyexpressedinu87cellline,thetranscript notincludingexon20 (exon1921)was 3.63timesmoreexpressedthantheoneincluding exon20. These results showed an enhanced exon 20 skipping in U87, carrying the NRXN3_DEL in homozygosity. We 123
130 RESULTS thereforeexploredapossiblemechanismthatcouldlinknrxn3_deltoalternativesplicingatsplicingsite4 (responsibleforexon20skipping).wefoundintheliteraturealinkbetweenalternativesplicingatsplicing site4andptbp2(polypirimidinebindingprotein2)molecules(alternativesplicinginhibitors)(208),through thespecificbindingofthesefactorsinintronsflankingexon20.asthepaperidentifiedptbp2bindingsitesin introns21and19weevaluatedinsilicothepresenceofthesemotifsinthedeletedregionandfoundsome of these motifs. In fact, they corresponded with some of the regions with high conservation rate inside NRXN3_DEL(Figure65). Assay T98GCt U87Ct TBP Exon Exon Figure64:U87(left)andT98G(right) )amplificationscurvesfortbp,eonx1921andexon21 assays.ctforeachassayineachofthetwocelllinesareindicatedinthetablebelow. Table38: :RatioofExon20andExon1921expression(referedtoTBPhousekeepinggene) inu87celllinerespect coparedtot98gcellline. Exon20/TBP RatioU87/T98G Exon1921/TBP Exo1921/Exon RATIO EXO20 =( EfficiencyExo on20) Ct(T98GU RATIO EXO1921 = =(EfficiencyExon1921) Ct(T U87) /(Efficienc T98GU87) /(Effic cytbp) Ct(T98GU87) ; ciencytbp) Ct (T98GU87) 124
131 RESULTS 1 9 CATGGTAGAGTGGCAATTTTGTTTTTCTGGCTCTTATATTGCAATGAATATTTTTTATTT GCAATTCT-CTTTTCTGTCT ****** * ******* ** Figure65:PTBP2silencerbindingsitesequenceinNRXN3_Del.Topofthefigure,blatresultofGCAATTCTCTTTTCTGTCT sequenceinamongnrxn3_del;below,thisnrxn3_delsequence(redcircle) hadhighlevelsofconservationamong species(imagetakenfromucscgenomebrowerhg18). Inordertoconfirmwhetherexon20skippingwasreallylinkedtothepresenceofNRXN3_DEL,weperformed thesameexperimentincdnacomingfromhapmapcelllineswithknowngenotypesforthedeletion.we didnolongerobservearelationbetweentheamountofeachofthetranscriptsandthegenotypesforthe NRXN3_DEL (Figure 66) ). To completely discard the possible effect of NRXN3_DEL in exon20 skipping in neural tissue (since the possible splicing inhibitors being involved in alternative splicing PTBP2 could be tissuespecific)wegenotyped25humanbrainsamplesfornrnx3_delandperformedthesamepcrincdna ofthreeofthesamplespresentingthethreepossiblenrxn3_delgenotypes.thisconfirmedtheabsenceof adirectcorrelationbetweennrxn3_delandexon20skipping(figure66). 125
132 RESULTS Figure66:NRXN3mRNAPCRinHapMapcelllinesandthreehumanbrain sampleswithdifferentgenotypesfornrxn3_del. 126
133 DISCUSSION
134
135 DISCUSSION Inthisthesiswehavetriedtoevaluate,withtwowholegenomeapproaches,thegeneticcontributiontoFM susceptibility. This constitutes the first attempt to dissect the possible role of SNPs and CNVs in FM ethiopathogenesisinacomprehensiveway.sincefmisaveryheterogeneousdisorder,wefirstattempted toidentifyhomogeneoussubgroupswithatwostepclusteringprocedure. Identificationoffibromyalgiasubgroupsthroughclusteranalysisofclinicaldata WehaveperformedaclusteranalysisofclinicaldataandsubsequentFMsubgroupinginalargecohortof FMpatients.Thesefindingswerereplicatedinasecondlargercohort,thusconferringastrongerrobustness toourresults.toourknowledgethisisthefirststudytoperformatwostepclusteringprocesstodefine variables dimensionsandsubsequentlyidentifyfmclinicalsubgroups.theinclusionofpersonalandfamily history of comorbidities and the collection of data through direct physician examination constitute also novelcontributionstofmclusteranalysis. Basedonourdata,thevariablesweregroupedinthreedimensions:FMsymptomsandtheircharacteristics (Dimension1),familialandpersonalcomorbidities(Dimension2),andscales(Dimension3).Thisclusteringof FM clinical variables into different dimensions is in agreement with previous studies (179). In fact, the resultingdimensionsarenotcompletelyunexpected,assomeoftheobservedclusteringcanbeattributed tothevariablesreferringtothesamesymptomororgan(i.e.muscularsymptomsindimension1). A novelty of our findings is that pain symptoms were grouped into the same dimension as cognitive symptoms.sincesymptomswithinaclustermayshareacommonidentifiableetiology(172),thisclustering couldbehighlightingthecnsimplicationinthephysiopathologyoffm(6).nevertheless,previousstudies did not show the clustering of FM core symptoms and cognitive symptoms. In some cases, cognitive symptoms were not considered (175177), or physical and psychological symptoms were considered separately (180) but in the study by Rutledge et al., where both were considered, pain and cognitive symptomsdidnotclustertogether.apossibleexplanationforthesecontradictoryresultsisthatrutledgeet al. evaluated in fact the patients management of the symptoms and which ones they wanted to be improved,andnotonlythepresenceofthesymptom.anotherpossibleexplanationcouldbethattheirdata werecollectedinadifferentway,throughonlinequestionnairesinsteadofthroughaphysician sinterview. Theclusteringoffamilyhistoryandpersonalcomorbiditiesintoaseconddimensionisalsoconsistentwith the fact that a family history of FM is linked with a more severe disease with more comorbidities (209). Personalhistoryofchronicpainbeforeextensivepain,however,clusteredinthefirstdimension,although 129
136 DISCUSSION presentingalowweightinthedimension.thefactthatthehistoryofchronicpaindidnotclusterwithother comorbiditiescouldbeindicatingthatitmaybeconsideredasasupportingfmcoresymptom.infact,itis difficulttoidentifytherealonsetoffibromyalgia,andwhetherthepreviousregionalchronicpainbelongsto thediseaseitself. Finally,theclusterofscaletypevariablesinthethirddimensioncouldbeduetothenatureofthevariables themselvesratherthantheirclinicalvalue.thismightbeindicatingalimitationoftheclusteringanalysis. However,wehavetotakeintoaccountthatscaleshavebeenreducedtoabinomialvariable,whichcould havereducedtheirclinicalvalue.inanycase,thislastdimensionwastheonewiththefewervariablesand thelowestweights,makingthedimensionlessreliablethantheothertwo.sincethemainpurposeofclinical scales is to measure severity (constituting also a screening method), they were finally used to evaluate differencesamongtheresultingfmsubgroups. Oncetheclusteringofthesampleswasperformedbasedontheirindexesinthefirstandseconddimension only,thesamplesformedthreegroups:lowsymptomatologyandlowfamiliarandpersonalcomorbidities (Cluster 1), high symptomatology and high familiar and personal comorbidities (Cluster 2) and high symptomatologybutlowfamilialandpersonalcomorbidities(cluster3). Wedidnotobservedifferencesinageorgenderamongthesubsets.Ithastobetakenintoaccountthatdue tothereducednumberofmalesincludedinthestudy(49individuals),wewereunderpoweredtoexcludea possible difference in the distribution of males and females among subgroups. We found no differences betweengroupsinage,neitherinageatthetimeofrecruitmentnorintheageofonset.thisisinagreement withpreviousstudies(173,180). PatientsincludedinCluster1showedmarkedlylowervaluesforallscales.Thiscouldbepointingtoamilder FMform,ortoalessevolveddisease.However,weevaluatedtimeofevolutionofthediseaseanddidnot observestatisticaldifferencesbetweenthethreeclusters,althoughpatientsbelongingtocluster2seemed to have a longer disease evolution. This would indicate that the differences among the clusters are not relatedtodiseasestaging,andcouldpointtotheidentificationofdifferentclinicalsubsets.aprospective studywouldbeusefultoelucidateifthelessaffectedcluster1couldhaveabetterprognosis. FMpatientsbelongingtoCluster2(highsymptomatologyandhighcomorbidities)werealsotheoneswith the highest levels of pain. This should not be unexpected, as most comorbidities are psychiatric, and individualswithpainhavebeenshowntobemorepronetodepression,becauseofpain sadverseeffectson mood and physical function (172). Furthermore, the number of depressive symptoms is a factor that has beenassociatedwiththedevelopmentofchronicwidespreadpain(210),andpreviousclusteranalysesin breastcancershowedthatdepression,fatigueandpainwereallsignificantlycorrelatedtoeachother(and 130
137 DISCUSSION tototalhealthstatus)(211).comorbidmedicalconditionscouldalsoberesponsibleforagreaterseverityof FMsymptomsinCluster2.However,differencesbetweenClusters2and3inFMcoremeasuresobservedin ourstudy,althoughstatisticallysignificant,werelimited.thiscouldbeindicativeofalimitedinfluenceof personal or family history of chronic pain and psychopathology in disease severity, showing that the presenceofhighnumberofsymptomsisthemainmarkerofdiseaseseverity. TheFMsubgroupsthatarisefromourstudyaresimilartotheonesdescribedinpreviousstudies(175,180, 212). Giesecke et al., for instance, also identified three subsets of patients, mainly based on pain and psychopathology:1)moderatemoodratings,moderatelevelsofcatastrophizingandperceivedcontrolover painandlowlevelsoftenderness;2)elevatedmoodratings,highlevelsofcatastrophizingandlowlevelof perceived control over pain and high levels of tenderness; and 3) normal mood ratings, low levels of catastrophizingandhighlevelofperceivedcontroloverpainandextremetenderness.theirgroupsshow similaritieswithourfindings,despiteanalyzingdifferentvariables.nevertheless,ourresultsandgiesecke s resultsarenotdirectlycomparable,astheyusedmeasurementsofexperimentalpainandsomevariables notincludedinourstudy.itisalsodifficulttocompareourresultswiththoseofthestudybyrehmetal.,as theyusedspecificpaincharacteristicsthatwerenotevaluatedinourwork.finally,inthestudybywilsonet al., they conducted a cluster analysis on the physical and psychological symptoms to identify subgroups. Subgroups I and IV of Wilson s classification seem to correspond to groups 2 and 1 of our classification, respectively.itwouldbeinterestingtotestthedegreeofsimilaritybetweentheresultinggroupsofeach study.intheirstudy,asinthepresentstudy,thegroupofpatientswithlowerlevelsofsymptomatologyhad highereducationlevels.differencesinthestudydesign(useofwebbasedsurveys,andexclusionofpersonal andfamilycomorbidities)couldexplaintheresultingfourgroupsinsteadofthree.however,boththestudy bywilsonetal.andthepresentstudyhighlighttheimportanceofsymptomsinfmpatientclassification. This is, indeed, in agreement with the new diagnostic and classification criteria of fibromyalgia (14). Nevertheless, comparison of classifications of different studies cannot be properly performed without actuallyapplyingalltheanalysistothesamesetofpatientsanddirectlycomparingthecompositionofeach of the resulting subgroups. In fact, it would be of interest to find a consensus regarding criteria for subgrouping of fibromyalgia patients. Performing the various types of classification in the same set of patientscouldhelpinidentifyingthemostrelevantcriteria.thesesubgroupscouldthenbereproducedin differentpatientpopulationsandusedforresponseanalysisinfuturetrials. We propose a definition of FM subgroups based on two clinical dimensions resulting from a clustering analysis,althoughitpresentsseverallimitations.wecannotdiscriminatetherelativeweightofpersonaland familyhistoryofcomorbidities,sincetheyclustertogether.also,wedonotknowifsomegroupofsymptoms (characteristics of pain, cognitive or autonomic symptoms) were more relevant than others. Finally, the 131
138 DISCUSSION transformationofclinicalscalesintobinomialvariablescouldhavelimitedtheirvalue.itwouldbepossibleto circumvent this last issue by implementing another statistical methodology allowing their inclusion in the clusteranalysisascontinuousvariables. Inspiteofthedescribedlimitations,theresultingsubgroupshavebeenusedinthereplicationofGWASdata and in the association study of CNV regions detected in acgh, observing that the associated variants detectedbehaveddifferentlyamongthedifferentfmsubgroups. EvaluationofSNPscontributiontofibromaylgiasusceptibility:agenomewideassociationstudy Wehaveperformedagenomewideassociationstudyconsideringover500000SNPsin300FMcasesand 203controls.Toourknowledge,thisisthefirstGWASperformedinFM.WehavenotidentifiedanySNP reachinggwasassociationthreshold,althoughtmanhattanplotshowedpossiblesignalsonchromosomes3 and X coming from SNPs with pvalues <10 4. These signals were not replicated but four of the 21 most significantly associated SNPs chosen for replication showed nominal association in the replication cohort. OutofthesefourSNPs,threewerelocatedinintronicregions:rs (MCF2gene),rs (ANK3 gene)andrs (myt1lgene),andrs wasintergenic. The protooncogen MCF2 is a member of a large family of GDPGTP exchange factors that modulate the activity of small GTPases of the Rho family. It is ubiquitously expressed and is involved, among other functions,inovogenesis,apoptoticprocessanddendriteproliferationinthecns.mcf2variantshavebeen associatedwithsusceptibilitytoautismandschizophrenia(213). ANK3(ankyrin3)belongstoafamilyofproteinsthatarebelievedtolinktheintegralmembraneproteinsto the underlying spectrinactin cytoskeleton. Ankyrins play key roles in activities such as cell motility, activation,proliferation,andmaintenanceofspecializedmembranedomains.ankyrin3isascaffoldprotein that has many essential functions in the brain, including organizational roles for subcellular domains in neuronssuchastheaxoninitialsegmentandnodesofranvier.itorchestratesthelocalizationofkeyion channels and GABAergic presynaptic terminals, and it is also involved in creating a diffusion barrier that limits transport into the axon and helps define axodendritic polarity. It is postulated that ANK3 similar structural and organizational roles at synaptic terminals. Variants in ANK3 have been associated to schizophrenia,bipolardisorder(214)andautism(215). rs showedanominalassociationinthewholecohort,and,whenconsideringonlyfemaleswith low levels of comorbidities (clusters 1 and 3), association got stronger both in GWAS and replication 132
139 DISCUSSION samples,andthejointpvaluewasimproved.rs (chr2: ;hg18)isinlocatedinthe thirdintronofmyt1l(myelintranscriptorfactor1like).myt1lencodesforapostmitoticneuronalspecific zincfingerproteinandbelongstothemyelintranscriptionfactor1(myt1)genefamily(myt1,myt1land NZF3). It spans over 500 kb, includes 25 exons and is involved in neuronal differentiation by recruiting hystonedeacetylase(216).studiesinratshaveshownthatitismainlyexpressedduringdevelopment(217), anditisstilldetectedintheadultbrainatlowlevels.itcaninducedifferentiationintoneuronsinhuman embryonicandpostnatalfibroblastsandinpluripotentcells,throughthemicrornamachinery(218)andin combination with other transcription factors (219, 220). MYT1L variants have been associated with neuropsychiatric disorders: rare CNVs and SNPs are involved in schizophrenia (221, 222), a duplication in autism(223),andvarioussnpsinchinesehancasesofmajordepression(224).noneofthesepreviously associatedsnpswereinldwithrs and,infact,thisvariantwasmorestronglyassociatedwithfm caseswithlowlevelsofpsychiatriccomorbidities.thiscouldbeindicatingthattheidentifiedassociationis reallyrelatedtofmandnottofmpsychiatriccomorbidities,suchasmajordepression.this,inadditionto theroleofmyt1lroleinneuronaldifferentiation,makesrs agoodcandidatesnpforfm,whichis characterizedbyacnsdysfunction(87).thepotentialfunctionaleffectofthisintronicvariantwasevaluated byexploringldintheregion.nofunctionalvariantsinmyt1lwereinldwithrs ,asitwasonly linkedwithintronicvariantsrs andrs Regarding possible regulatory effects in other genes, Genvar analysis revealed a possible correlation with SNTG2 (syntrophin gamma 2) expression in lymphocytes for both rs and rs This gene encodes a protein belonging to the syntrophin family. Syntrophins are cytoplasmic peripheral membrane proteinsthatbindtocomponentsofmechanosensitivesodiumchannelsandtothecarboxyterminaldomain of dystrophin and dystrophinrelated proteins. In particular, the PDZ domain of SNTG2 interacts with a proteiccomponentofamechanosensitivesodiumchannelthataffectschannelgating.accordingtogenvar findings, rs AA genotype (A being the risk allele for FM) is linked to lower expression levels of SNTG2. SNTG2 variants have been also associated to autism (225) and to suicide attempts in major depression(226).thus,rs implicationinfmsusceptibilitycouldalsobeduetochangesinsntg2 expressionlevels.aninterestingapproachtovalidatethiswouldbetoevaluatewhethertherearedifferent SNTG2 CNS expression levels in FM patients compared to controls (by measuring its levels in the cerebrospinalfluid).thiscouldalsobefurtherrelatedtothespecificgenotypesofthissnpinthesesamples. ThispotentialrolefortheCNSinFMgeneticssusceptibilitywasnotonlyhighlightedbythesethreeSNPs:IPA analysisofthe77gwassnpswithmostsignificantpvaluesrevealedneurologicaldiseaseasoneofthetwo topnetworks.furthermore,genesetgoanalysisresultsshowedionbinding(andcalciuminparticular)as oneofthetwomainmolecularfunctions,calciumchannelcomplexasthetopmolecularcomponent,and 133
140 DISCUSSION regulationofcalciummediatedsignallingandcalciumiontransportandneurogenesis,amongthetopten biological functions. Since calcium channels are essential for neurotransmitter release, the overrepresentationofthissystemconstitutesanadditionalargumentsupportingaroleforcnsinfm. AlthoughalltheresultsreportedabovesuggestFMassociationwithgenesexpressedintheCNS,wehaveto take these results with care since this GWAS presents several limitations.the first issue that we need to addressisthereducednumberofsamplesthatwewereconsidering.infact,toachieve80%powertodetect loweffectvariants(whicharetheonesexpectedincomplexdisordersuchasfm)itisnecessarytotakeinto accountthousandsofsamples.nevertheless,ithasbeenpossibletodetectassociationsthroughgwasin smaller sample sizes, as long as the cohorts of cases were very homogeneous, and, in these cases, the detectedvariantsshowedahighereffect.(227).inanycase,inourstudywewouldhavebeenabletodetect strongassociations,suchastheonesthathavebeenreportedinautoimmunedisordersinthehlalocus,and we have not seen any evidence pointing to such an association. This is an important finding since many attemptshavebeenundertakeninordertofindhlaassociationswithfm,inparticularwiththosealleles previously associated with rheumatic disorders (particularly, with the rheumatoid arthritis HLADR4 allele (112)).HLADR4associationtorheumatoidarthritispresentedanORrangingfrom2.36to4.1(143).Inour study,with300fmsamplesand203controls,formaf0.15wehadover90%powertodetectassociations with OR2.0, as calculated with Quanto software ( ). Thus, we should have detected an association at the level of HLA_DR4. Nonetheless, association to the HLA locus can not be completelydiscarded,sincehlaassociationfordisordersnotclassicallyconsideredasautoimmune(suchas schizophrenia),thatactuallyhavehlariskalleles,haveamuchloweror,of (228). Sinceweonlyhadaccesstothissamplecohort,inordertomaximizeourpower,wedecidedtouseavery wellcharacterizedsetofpatients,specificallyselectedforhavinglowlevelsofpsychiatriccomorbiditiesand a clear FM diagnosis. This was, in fact, confirmed by the enrichment of cluster3 individuals (high symptomatologyandlowcomorbidities)ingwassamples(supplementarytable13).inretrospect,itmight have been better to have the clustering results before starting the genetic study, in order to center the GWASoncluster3,butthiswasnotpossiblesincethecollectionofsampleswasdoneinparallelwiththe developmentoftheproject,andthus,theclusteringanalysiscouldnothavebeendonewiththeinitialsetof samples.thereducednumberofsampleswithgwasdatacouldexplainwhywewerenotabletodetectan associationreachinggwasthreshold.itcouldalsoresultinthedetectionoffalsepositivesamongthemost associatedvariants,andthiswouldleadtoalackofreplicationofthesevariants.inourcase,noneofthe variantswasstrictlyreplicatedwhenconsideringthewholecohort,asthejointanalysisofthegwasand replication cohorts did not result in an improved pvalue. Nevertheless, four of the SNPS selected for replicationpresentedanominalassociationinthereplicationcohortthatwasinthesamedirectionthatin 134
141 DISCUSSION thegwas.whatismore,thestratifiedanalysisofrs ,consideringfmsampleclusters,showeda replicationofthegwasresults.thishighlightstheimportanceofaprecisephenotypingofthesamplesin extremelycomplexdisorderslikefm. AninterestingfindingisthatassociationofreplicatedSNPswasstrongerwhenonlyconsideringfemales.In fact,manhattanplotdetectedasignalonxchromosome.interpretationofthisisdifficult:fmisadisorder thataffectswomeninover90%ofthecasesandthereiscontroversyastowhetherpainsensibilitydiffers among FM women and men. Small studies have been undertaken in order to evaluate this potential difference,buttheyshowcontradictoryresults(229,230).genderdifferencesinpainsensitivityareinfacta matter of discussion. Chronic pain is more frequent in females than in males, but studies addressing differentpainsensitivityamonggendersarenotconclusive.therehavebeenreported,womenfactorssuch asthehormonalcycle,thatmayplayaroleinpainresponseandthereforecomplicatethecomparisonwith men.however,someresearchershaveexploredpossibleunderlyingmechanismstoexplainsexdifferences in pain response and neurochemical differences have been identified (NMDA receptors, cytokine expression ).Infact,geneswithasexdependenteffectonpainhavebeenreported.Themelaconocortin1 receptor(mc1r)gene,forinstance,hasbeenreportedtomediatefemalespecificmechanismsofanalgesia, whereas toll like receptor 4 (TLR4) gene has been involved in spine pain processing only in males, as reviewed in Mogil et al. (231). Thus, our GWAS results would be in agreement with these reported sex differencesingenesinvolvedinpainperception. Inordertocompletelyconfirmourfindingsandovercomethereducedsamplesize,itwouldbenecessaryto replicatethedetectedassociationsinanothercohort.thisischallenginginfmsince,toourknowledge,this cohortisthelargestexistingcollectionoffibromyalgiadnasamples.weonlyknowofanotherfmcohort similar in size, and we are working with them in performing a reciprocal replication study, once an appropriate agreement is reached, since it involves several pharmaceutical companies. Other statistical methodssuchascommonpolygenicvariation(228)couldalsohelpindetectingfmassociatedvariantsof loweffect,althoughwedon tknowwhetherthisstatisticalmethodcouldbeimplementedinacohortofour samplesize. Evaluation of the contribution of copy number variants to fibromyalgia using genomewide association studydata In addition to evaluate genetic association through SNPs, the GWAS genotyping data was used for the identification of CNVs using the PennCNV algorithm. The analsysis of CNVs through SNP arrays presents severallimitations,whichwehaveobservedinourstudy.firstofall,availablealgorithmsusedtoinfercnvs fromsnpsarraysneedtheuseofmorethanonemethodtominimizetheerror.furthermore,theiraccuracy 135
142 DISCUSSION topredictcopynumberstateislimited,andtheresolutionfordetectingsmallvariantsisverylow.inour case,sinceweevaluatedaphenotypewithoutbroadclinicalcharacteristics,suchasschizophrenia,autismor mentalretardation,wedonotexpectittobeassociatedtoverylargecausativeevents,anditiswellknown thatsmalleventsaremissedbythistechnology.inparticular,theplatformthatwehaveused(illumina1m duo)isnotparticularlyenrichedincnvsprobes;ithasasmallernumberofcnvsprobescomparedtonewer arrays such as the Illumina Omni 1. After the filtering procedure we have only followed up one of the detectedcnvs,locatedintheacacagene.theanalysisofthiscnvwithadirectgenotypingmethoddidnot replicatethehighfrequencyofthehomozygousdeletedsamplesobservedinfmgwasdata.infact,the presenceandgenotypeofthecnvinthegwassamplesinwhichithadbeendetectedwasnotconfirmed by direct genotyping. Therefore, there were no differences between cases and controls when direct genotypingofthecnvwasperformed.thisillustratesthelimitationsofsnparrays(ormaybethisparticular algorithm) in copy number prediction. From 10 samples that appeared as homozygous deleted from PennCNVresults,onlyonepresentedthisgenotypebydirectmultiplexPCRgenotyping. Finally, we evaluated the possible presence of CNV events in mosaic state. CNV in mosaic state could constitute a good pathophysiological mechanism for FM. Changes could be due to interaction with environmental/socialfactorsandcouldappearacrosstheindividual slifetime.however,themainlimitation thatwehadtofacewasthatthedetectionofmosaicismshouldbeideallyperformedindifferenttissues, involvingtargettissuesifpossible:cnsandmuscleandcomparisonacrossthedifferenttissuesandacross time.wedidnothaveaccesstothistypeofdata,andweperformedthisanalysisbasedonsnpgenotype data.wefinallyconsideredinterestingtwooftheputativemosaicaberrations,bothlocatedonchromosome 12. One was located in SLC2A14, a glucose carrier gene located 30 kb away from SNPs that have been associatedwithalzheimer sdiseaseinnonapoe4allelecarriers(232,233).furthermore,glucosecarrier s genes have been associated with neuropsychiatric disorders such as adult attentiondeficit hyperactivity disorder (234). SLC2A14, a gene involved in CNS related disorders could therefore constitute a good candidateforfm,characterizedbyadysfunctioninpainprocessing.theothercnvregioninmosaicstate includedthefullcodingsequenceoftheovos2gene.ovos2(ovostatin2)encodesaproteinabletoinhibit all four classes of proteinases by a unique 'trapping' mechanism. It is ubiquitously expressed and it has shown to be overexpressed in ageing muscle and nucleus pulposus cells (235). Changes in OVOS2 copy numbercouldbeimplicatedinfmetiopathogenesisasacomplementaryfactor(initiatingorperpetuating) to an altered pain transmission. Nevertheless, we did not perform further experiments such as MLPA or quantitativepcrinordertovalidatetheseresults.althoughimplicationofbothgenesinfmsusceptibility seemsreasonable,wedidnotfollowthemupbecausetherewerenotadditionalevidences(snpsingwas orcnvsinacgh)supportingthemasfmcandidategenes,andadditionaltissuesofthepatientswerenot 136
143 DISCUSSION available.thisexploratoryanalysisshouldbepursuedinmuscleandnervoustissueinordertocompletea successfulvalidation. Contributionofcopynumbervariantstofibromyalgia:arraycomparativegenomichybridization WehavealsoexploredthepossiblecontributionofCNVtoFMgeneticsusceptibilityusingarraycomparative genomichybridization.tothispurpose,wehaveperformedacghwithtwotypesofarrays(agilent400kand 1M) using a pooling strategy. We detected seven regions that appeared to have different frequencies in casesandcontrols,andfiveofthemwereselectedforfollowup.ofthese,onlyonewasactuallyvalidatedas a susceptibility factor for FM. The deletion in the NRXN3 intronic region showed an association with susceptibilityforfminfemales,beingstrongerincaseswithlowlevelsofcomorbidities. GALNTL6emergedasdifferentialhybridizedintheearlyonsetpool.WeinitiallyvalidatedthisaCGHfinding inasubsetof300casesand300controlsbutassociationwaslostwhenenlargingthereplicationcohort.we werenotabletoreplicatethisfindingwhenonlyconsideringtheearlyonsetsamples.sincethenumberof earlyonsetcaseswassmall(100whenconsideringthosewithanageofonsetbefore20yearsoldasthe ones included in the acgh pool), we also explored GALNTL6 variant in the samples with an age of onset lower than 30 (30 representing the cohort p 25, the median age of onset being 38) with no evidence of associationeither.finally,weevaluatedthisvariantinthedifferentfmclustersbuttherewasnointeraction thatcouldbeexplainingtheinitialreplicationthatwasnotconfirmedwithfurthergenotyping. WealsointendedtoreplicateWWOX,PTPRDandMYO5BaCGHresults.WWOXencodesforaproteinwith anoxidoreductaseputativefunctionanditsexpressionisupregulatedinearlystagesofdevelopmentofthe peripheralandcentralnervoussysteminmouseembryos.thisproteinregulatesawidevarietyofmetabolic processes such as steroid metabolism, apoptosis, and tumour growth suppressor. It has been associated withdifferenttypesofsolidandbloodtumors(236),ithasbeenshowntobedownregulatedinapopulation ofhippocampalneuronsofindividualsaffectedwithalzheimer sdisease(237)andupregulatedafternerve injury(238). PTPRDencodesforamemberoftheproteintyrosinphosphatasefamily,whichmaybeimplicatedinneurite growthandneuronsaxonsguidance,assuggestedinstudiesofitsorthologsinchickenandfly.ithasbeen associated with several tumors and neuropsychiatric disorders, and has been shown to play a role in the developmentofinhibitorysynapses(239). MyosinVbisexpressedinseveralneuronalpopulationsandassociateswiththealphaamino3hydroxy5 methyl4isoxazolepropionatetypeglutamatereceptorsubunitglur1,mediatingitstransport(240).myo5b 137
144 DISCUSSION variantshavebeenassociatedwithbipolardisorder(241),andanothermemberofthegenefamily,myo18b hasshowntobeassociatedwithschizophrenia(228). Wecouldnotreplicatethesethreevariants.Sincetherewasnoevidenceofassociationaftergenotypinga subsetof300casesand300controlswedidnotcontinuethefollowupexperiments.wecannotcompletely excludetheparticipationofthesecnvsinthedisease,astheycouldbevariantswithaloweffectondisease whichwouldneedlargercohortsforvalidation.consideringalltheresultsemergingfromsnpsandcnvs analysisinfm,thesecouldconstitutegoodfmcandidates,astheyhavebeenshowntoplayaroleinthe CNS. Inordertoexploreapossibleadditiveeffectofthesevariants,explainingacausativerolenotdetectedby association analysis, we evaluated interaction between these CNVs. We did not detect nor an additive neitheranepistaticeffect.finally,themostplausibleexplanationisthatthesenotreplicatedcnvswerein factfalsepositivescausedbyabiasinthefrequenciesofthecnvsinthepooledsamples. Theuseofpoolsofsamples,althoughpresentingmanyadvantagesintermsofreductionofdataanalyzed andmoneyspent,canleadtothedetectionoffalsepositives.forcommonvariants,arandomdistributionof thedifferentgenotypescouldleadtoenrichmentincasesorcontrolsofaparticularvariantandthustofalse changesinhybridizationsignals.forrarervariants,aslightmistakeinthepreparationofthepoolleadingto thepresenceofalargeramountofoneormoresamplescarryingtheraregenotypecouldalsoleadtoafalse hybridizationdifferencebetweencasesandcontrols.theuseofpoolsmayalsoleadtoadecreasedpowerto detectvariantsinhighlypolymorphicregionsofthegenome.ontheseregions,thepoolwillrepresentan intermediatebetweenthepolymorphicandnonpolymorphicstates,resultinginsmallerrelativedifference inintensitythananonpolymorphicsinglereferencewouldyield(165). Anoptionthatmayovercometheselimitationsistoincreasethenumberofsamplesofthepool.Another optionwouldbetheuseofvariouscontrolpools,whichwillhelpingettingridoffalsepositivescausedby random bias in the selection of samples. Furthermore, by performing several hybridizations of the same controlpoolagainstdifferentpoolsandeliminatingrecurrentcnvsfromtheanalysisitshouldbepossibleto reduceerrorsduetorandombias.inourcase,sincewewereincludingfmcaseswithfamilyhistoryoffm and,atthetimeofpoolsdesign,weonlyhad300samplesavailable,wecouldnotincreasethenumberof casesincludedineachfmpool.forcontrols,weusedonepoolbutwetookintoaccountresultscoming fromthehybridizationofthesamepoolofcontrolsagainstpoolsofcasesfromotherdiseases,andthose variantsthatrecurrentlyappearedindifferenthybridizations(suchassirpb1)performedinourlaboratory were not considered for followup studies. Finally, interexperimental replication involved the same 138
145 DISCUSSION conditionsand/oranexperimentalalternate,suchas dyeswap ofthetwofluorochromelabelsbetween thetestandthereferencesamples. Finally,inretrospectandbasedonourGWASandaCGHfindings,weshouldhaveusedonlyfemalesforpools design.althoughtheproportionofmalesandfemaleswassimilarinfmandthecontrolspools,bothwitha majorproportionoffemales,itwouldhavebeenmoreinterestingtoonlyincludefemales.this,notonlyfor the different behavior of the variants in each gender, but also in order to explore the X chromosome structuralvariants,inparticularthoseinvolvingxlinkedneuroligin(nlgn4)(neurexinspartner). ForthedetectionofCNVregionsshowingdifferentialhybridizationbetweencasesandcontrols,weusedthe ADM2algorithm.Thisalgorithmposesdifferentadvantagesincomparisonwithothermethods.Forinstance, itusesavariablewindowsize,incontrasttothezscorealgorithm,thatuseswindowsoffixedsize,andit takes into account the quality score of each probe, not included in ADM1. However it presents some limitations:sinceitcalculatesthemedianofthelrrofthedetectedregion,theregionsappearinginthe resultshavetobecarefullychecked,sinceinsomecasespositiveprobesarenotconsecutiveandtherefore theregionasawholecannotbeimmediatelyconsideredasdifferentiallyhybridized.differentscenariosare thenpossible:alargecnvregionincludinganegativeprobethatpersistentlyfailsindifferenthybridizations would be considered as differentially hybridized, whereas a three probe region where one of the probes doesnotfulfilselectioncriteriawouldbelessreliableanddiscardedforfollowup.anotherlimitation,that weobservedmostprominentlyinthecaseof1marray,isthatitdoesnottakeintoaccountthoseprobes thatarerepeated,andsometimesaregioninwhichonlytwononrepeatedprobesfulfiltheselectioncriteria appearsasincludingamuchlargernumberofprobes. Finally,wehavebeenabletoreplicateaCGHresultsforNRXN3_DEL.Thegenotypingoftheinitialsubsetof samples(over300fmcasesand300controls)showedastatisticallysignificantassociationbetweenfmand thedeletion.whenwecompletedthegenotypingoftheentirecohort,theassociationremainedstatistically significantinfemales.thestrongerassociationintheinitialreplicationcohortcouldbeexplainedbythefact thatmostofthesamplesincludedinthissubsetweretheonesthathadbeenincludedinthegwas.they werethereforesamplesselectedbythecliniciansforhavingaclearandconfirmedfmphenotypeandlow levelsofcomorbidities.forthisreason,weexploredtheassociationofthecnvinthedifferentclustersand foundthat,whenconsideringsampleswithlowlevelsofcomorbidities(clusters1and3),theassociationwas replicated, and it was stronger if only considering females. NRXN3 is a good candidate for FM as it is essential for neuronal development and for signal transmission. As we have mentioned before, several variants in NRXN3 (both rare CNVs and SNPs) have been associated with different phenotypes, mainly neuropsychiatric disorders, including addictive behavior. Changes in NRXN3 and therefore in signal transmissioncouldexplainthecentralnervouspaindysfunctionthatisunderlyingthedisease. 139
146 DISCUSSION EvaluationofpossiblefunctionalconsequencesofNRXN3_DEL Sincethedetectedvariantwaslocatedinanintronicregion,weintendedtoevaluateitspossiblefunctional consequences.first,weassessedifthecnvcouldbeinldwithafunctionalsnp,orifitcouldbetaggedbya SNP, facilitating its detection. This was done by a Veracode genotyping assay. The Veracode experiment included 45 SNPs capturing the main functional SNPs in the gene and SNPs previously associated with disease, but it did not detect a proxy for the deletion. It could be useful to further saturate the region surrounding NRXN3_DEL, since the Veracode experiment, designed to capture most of the entire gene variability,onlyincluded15snpsina200kbwindowfromnrxn3_del.noneofthepreviouslyassociated SNPswithotherdisordersshowedassociationwithFMeither. In addition to assessing its possible link to other functional variants, we also evaluated possible consequences of NRXN3_DEL at the mrna level. We identified two neuronal cell lines that were homozygousdeletedandhomozygousnondeletedforthevariant.performingrtpcrexperimentstotest for mrna isoforms, we observed a different ratio of the two possible NRNX3 isoforms when amplifying cdna from exon18 to exon21 (with or without exon20). This was confirmed and quantified by a specific TaqmanRTqPCR.Furthermore,inaccordancewiththishypothesis,wefoundPTBP2(SS4#inhibitoryfactors) bindingsitesinthecnvregion,andthesecorrespondedtohighconservedregions.thepossiblecorrelation between exon20 skipping and NRXN3_DEL would have been of great interest, since exon 20 skipping in NRXN1isessentialforitslinkwithneuroligins:onlyneurexinsisoformswithoutexon20bindtoneuroligins (242). Wethenextendedthisanalysistoalargernumberofsamples,aswehadaccesstocelllinesfromHapMap sampleswithdifferentnrxn3_delgenotypes,butwedidnotseethiscorrelation.apossibleexplanationfor thiscouldhavebeenthatthebehaviorofthevariantcouldbedifferentdependingonthetissue,ashapmap cell lines are derived from lymphocytes, and the cell lines in which we observed the differences were neuronal.therefore,wethenperformedrtqpcronrnafromthreehumanbrainsamplescarryingeachof the possible genotypes for the deletion, where again we did not see the effect. Thus, the differences observed in the two glioblastoma cell lines could be due to many other factors, such as the tumor stage (T98G cell line comes from a grade III whereas U87 comes from a grade IV glioblastoma multiforme) or gender(t98gcamefromamaleandu87fromafemale). WecannotexcludeapossibleregulatoryeffectoftheCNVregionatotherlevels.Infact,wefoundbinding sitesforothersplicingregulators(supplementaryfigure4)amongthecnvregion.itcouldbepossiblethat 140
147 DISCUSSION the regulatory effect could affect alternative splicing at other sites: at ss#5 or in noncanonical sites of alternative splicing. In order to further explore these consequences, it would be of interest to clone the deletion and transfect it into a neuronal cell model and explore expression variants (by means of an expression array or RNAseq). However, the size of the deletion (8.9 kb) makes this difficult. Another possibility would be to transfect a minigene involving a few exons before and after the deletion (ranging fromnrxn3exon18toexon21)butwefindthesameissueregardingthesizeoftheminigene,whichwould belargerthan500kb,ifallintronicregionsareincluded. Apossiblesolutioninordertoexploreinacomprehensivemannertheputativefunctionalconsequencesof NRNX3_DEL would consist in evaluating the different changes, at the mrna level, in different samples (preferablyneuronal)carryingthedifferentnrxn3_delgenotypes.forthis,itwouldbenecessarytohavea minimalnumberofsamples(atleast10pergenotype)andtoevaluatethetranscriptswithnextgeneration sequencingtechniques(rnaseq).thelimitationofthiswouldbethepowertodetectdifferencesrelatedto thecnvandnottotheinterindividualvariabilityorvariabilityduetoavariablecelltypedistributioninthe sample.inordertominimizethis,itwouldbenecessarytoensurethesametissueforthernaextraction, and,ifpossible,toextendittosinglecellextraction(asthedifferentisoformscouldbelinked,notonlytothe brainarea,butalsotocelltype). Paintransmissionandgeneticsusceptibilitytofibromyalgia Insummary,bothGWASandaCGHresultspointatarolefortheCNSinFMgeneticsusceptibility.Infact, variantsdetectedbybothstudiesarelinked:calciumtransportappearsasoneofthemaingwasmolecular functions and the neurexinsneuroligins complex formation is dependent in calcium (24), and SNTG2 interactswithneuroligins3and4,whicharepartnersoftheneurexins(225).inthe14 th worldcongresson pain(internationalassociationforthestudyofpain(iasp))august2012,milan; evidencesforspecificneurophysiologicalalterationsinfmpatientswerereported.ofparticularinterestto thisstudy,functionalandmorphologicalimpairmentofsmallfibershavebeenreportedinfmcases(243). Individuals presenting those electrophysiological changes would be the ideal models to explore the association and the functionality of our detected genetic variants (by using, for example, tissue from peripheralnerve)inordertotrytoestablishacorrelationwithclinicalseverity,outcomeandresponseto treatment. In this meeting, another study evaluating gene expression of FM individuals also detected changesingenesimplicatedinpaintransmission,whichsupportsourfindings(244). InspiteofthedifficultiesencounteredinthestudyofgeneticfactorsofFM(clinicalheterogeneity,lackof previouslyvalidatedgeneticfactors,reducedavailabilityofreplicationcohortsandnonavailabilityoftarget tissue) we have been able to detect variants that can shed a light on genetic factors determining FM 141
148 DISCUSSION susceptibility.toourknowledge,onlyneurotransmitterrelatedgenes(includingreceptors,transportersand enzymesimplicatedinneurotransmittersmetabolism)hadbeentestedasfmsusceptibilitycandidates.the possible role of synaptic structural molecules such as ANK3 or NRXN3 and molecules implicated in CNS developmentandfunctioning,suchasmytl1,openanewwidefieldofresearchintermsofaetiologyand drugtargets.oneconsiderationthatwehavetotakeintoaccountisthatallofthesemoleculeshavebeen previouslyassociatedwithneuropsychiatricdisorders.ifthesesynapsegenesassociationsareconfirmedin other FM cohorts, FM could be considered as neuropsychiatric disorder more than a rheumatological disease. Someissuesremainunsolvedthough:althoughtheCNSappearsasthebestcandidatetargettissueforFM, themusclecouldalsohaveanactiveroleindiseasedevelopment.furthermore,itisnotclearwhetherthere isonlyonetypeoffmorthediseaseisactuallyamixtureofphenotypes,whichissomehowsuggestedby our cluster analysis. Finally, it seems that some of the detected variants have stronger association when consideringonlyfemales,whichwouldindicatethatitwouldbeimportanttoperformstudiesinmalesand femalesseparately,asthegeneticfactorscouldbegenderspecific. Inadditiontothereplicationinothercohortsandtheevaluationoffunctionalconsequencesofthedetected variants,othercomplementaryapproachescouldhelpintheunderstandingofthedisease.nextgeneration sequencing technologies could be implemented for the study of FM genetics. Considering our findings, a good approach could be targeted resequencing of genes encoding for synapse molecules. Furthermore, sincechronicpathologicalpainandinparticularfibromyalgiaisinfluencedbyenvironmentalsocialfactors, epigenetic changes may play an important role in FM susceptibility and development. The study of methylation changes and other regulatory marks, their evolution in time and their correlation with FM clinicalcourse,couldbeofgreathelpinordertounderstandthepathophysiology.andnaturalhistoryoffm Inthissense,thepossibleroleofchangesinthechromosomeX,assuggestedbyinitialgenomewidestudy results,couldfittogetherastheinactivatingmethylationofoneofthexcopiesinfemalescouldperhaps playarelevantroleinfmsusceptibility. 142
149 CONCLUSIONS
150
151 1. Fibromialgiaisanextremelycomplexclinicaldisorder.Theadditionofmultiplesmall effect genetic factors seems to be underlying FM susceptibility. It is therefore necessary to study large cohorts including thousands of samples in order to detect theseriskvariants. 2. ThedefinitionofclinicallyhomogeneousFMsubgroupsconstitutesakeystepforthe identificationoffmgeneticsusceptibilityfactorsleadingtoabetterunderstandingof itsbiologicalbasis.inthissense,ourworkindicatesthattheevaluationofpersonaland familyhistoryoffmcomorbiditiescanaddimportantinformationtofmclassification basedonsomaticsymptoms. 3. Our GWAS results point towards a possible contribution of CNS genes to FM susceptibility:pathwayanalysisoftopassociatedsnpsidentifiedneurologicaldisease andcalciumchannelpathwayasoverrepresented. 4. AnucleotidechangeintheMYT1LgeneshowedstatisticalsignificantassociationinFM sampleswithhighlevelsofsymptomatologyandlowlevelsofdiseasecomorbidities. This confirms the key role of nervous transmission in FM etiopathogenesis and highlightstheimportanceofidentifyingfmhomogeneoussubgroupsforthedetection offmgeneticsusceptibilityfactors. 5. Replicationanalysisshowedastrongerassociationwhenconsideringonlyfemalecases and controls. This enhances the importance of gender in FM ethiopathogenesis and couldbepointingtotheexistenceofadifferentgeneticbackgroundforfminmales andfemales. 145
152 6. The inference of CNVs in mosaic state also supported a role for the CNS, with the detectionofpossiblemosaiceventsinslc2a14.mosaicismanalysisalsoidentifieda CNV in mosaic state in a gene implicated in muscle degeneration, suggesting thus a roleformusculoskeletalsysteminfmethiopathogenesis. 7. Anintronicinsertion/deletionpolymorphismintheNRXN3gene(NRXN3_DEL)showed associationtofmfemalecaseswithlowlevelsofcomorbidities.sincethismoleculeis essentialinthedevelopment,maintenanceandfunctioningofthesynapse,thisresult constitutesanadditionalargumentsupportingadysfunctioninneuraltransmissionin FM. 8. APossiblefunctionalconsequenceofNRXN3_DELmaybeaffectingalternativesplicing, sincebindingsitesforsplicingregulatoryfactorsweredetectedintheregion.although apossibleeffectatsplicingsite4leadingtoaswitchintheexpressionoftwodifferent isoforms was discarded, the deletion could be affecting other sites of alternative splicing(canonicalornot). 9. IftheproposedFMcandidategeneswerefurthervalidatedinreplicationstudies,this wouldconstituteachangeinthefmetiologyconcept,asseveralofthesecandidates are known neuropsychiatric disease associated genes (autism, addiction and mental disability). This would highlight a novel neurocognitive involvement in this disorder, currentlyconsideredtoinvolvethemusculoskeletalandaffectivesystemsandcircuits. 146
153 RESUMEN
154
155 RESUMEN Eldoloresunmecanismofisiológicodedefensaanteagentesexternos.Existendiferentestiposdedolor:el doloragudo,eldolorinflamatorioyeldolorpatológico.tantoeldoloragudocomoelinflamatoriotienenuna funciónprotectora:elprimeroencondicionesbasalesyelsegundoparaprotegerregioneslesionadas.sin embargo,eldolorpatológicoesunaalteraciónenlatransmisióndeldolor.eldesarrollotécnológicodelas técnicasdeimagen,hapermitidoelestudioenprofundidaddeldolorpatológico.principalmenteelusodela resonanciamagnéticafuncionaljuntoaotrastécnicasdemedicinanuclear,hanpermitidovisualizareldolor. Alteraciones tanto a nivel del sistema nervioso periférico como del sistema nervioso central se han postuladocomocausantesdeestetipodedisfunciónenlatransmisióndeldolor.unadelaspatologíasmás frecuentescaracterizadaspordolorpatológicoeslafibromialgia. La fibromialgia (FM) es una enfermedad de etiología desconocida que se caracteriza por dolor crónico generalizado,juntoaunaampliaconstelacióndesíntomasacompañantes.enunporcentajemuyelevadode casos, la fatiga crónica (entidad en si misma) es, junto al dolor, uno de los síntomas predominantes. Los pacientes pueden presentar, además, alteraciones del sueño, depresión, ansiedad, rigidez articular, parestesias en extremidades, cefalea, así como una hipersensibilidad al dolor, que se manifiesta por la aparición de sensación dolorosa a la presión en múltiples lugares de inserción osteotendinosa. Según el estudio EPISER, la fibromialgia definida según los criterios del American College of Rheumatology, es una patología frecuente en España, con una prevalencia del 2,4% de la población mayor de 20 años, lo que suponeunos pacientesdefmenelestadoespañol.afectadeformapredominanteamujeres,con unarelaciónmujer:hombrede21:1. LaFMesunaenfermedadaltamenteincapacitante.Su impactoafectatantoalpacienteanivelfamiliary profesionalcomoasuentornolaboralypersonal.ademásconllevaungranimpactoeconómico,entérminos degastosanitarioydebajaslaborales.teniendoencuentalaelevadaprevalenciadelapatología,constituye actualmente un problema sanitario importante. Por este motivo, se han desarrollado unidades multiciplinariasparaelseguimientoyeltratamientoespecíficodelospacientesconelprincipalobjetivode mejorarlacalidadasistencialdedichospacientes. DadoquenoexistenpruebasobjetivasquepermitanevaluarlaFM,eldiagnósticosebasaenunahistoriade síntomasylaexclusióndeenfermedadessomáticasqueexpliquendichossíntomas.en1990,elamerican College of Rheumatology, estableció unos criterios diagnósticos que definían la FM como la presencia de dolor generalizado de más de tres meses de evolución. Dicho dolor debía ser bilateral, afectar al tronco superior e inferior e incuir dolor axial junto a la presencia de dolor a la presión en 11 de 18 puntos específicosdelcuerpo,denominadospuntosdolorosos. 149
156 RESUMEN Posteriormente,hanidoapareciendoobjecionesaestoscriteriosdiagnósticos.Labajatasadeexploración de los puntos dolorosos en la práctica clínica habitual, ha puesto en evidencia que el diagnóstico de FM acababa basándose más en la presencia de síntomas que en la exploración física. Por ello en 2010 sedesarrollaronunosnuevoscriteriosdiagnósticosqueincluíanunmayorreconocimientodelosproblemas cognitivosylossíntomassomáticos.estanuevadefinicióndefmsebasaenlaconstrucióndeuníndiceel WidespreadPainIndex(WPI)constituidoapartirdelapresenciaoausenciadediferentessíntomas. Dentro del diagnóstico diferencial de FM entran todas aquellas patologías que puedan causar dolor generalizado:desdeenfermedadestumoralesainfecciónvirales,pasandoporenfermedadesinflamatoriaso autoinmunes. Para descartar la mayoría de estas patologías, una analítica general es normalmente suficiente.ademásnohayqueolvidarlaexistenciaderentistasosimuladoresquebuscaneneldiagnóstico defmelbeneficiodefmentérminosdebajaslaboralesopensionesporinvalidez. Ademásdel usodecriteriosdiagnósticosparaelseguimientodelaenfermedadesmuy comúnel usode escalasdeactividad.dichasescalassontantoespecíficasdefm(fiq)comousadasenmúltiplespatologías para la monitorización de la calidad de vida del paciente (SF36) o la evaluación de sintomatología acompañante. Dadoquesetrata de unaentidaddeetiologíadesconocida,noexiste, hastael momentountratamiento curativo.únicamenteexistentratamientossintomáticosdelimitadoefectoterapéutico.estostratamientos combinanlaterapiafarmacológicaconlafísicaysuindicaciónnoestáclaramenteestablecida.losfármacos utilizados en FM son básicamente analgésicos así como antiepilépticos utilizados para el tratamiento del dolorneurpático.finalmente,dadoquelosenfermosdefmtienenunamayortasadeefectossecundarios, elestablecerunapautaterapéuticaadecuadarepresentaunretoparalosfacultativosypasaporrealizarun tratamientopersonalizado,prácticamenteamedida. Laausenciadesignosobjetivosquepermitanlaconfirmacióndeldiagnósticoasícomoelseguimientodela enfermedadhacenqueseauntemacontrovertido.inclusoalgunosfacultativosponenendudalaexistencia de dicha patología. Esta incomprehensión, tanto por parte del colectivo médico como de la sociedad dificultan el diagnóstico y el tratamiento de la enfermedad con el consecuente sufrimiento por parte del paciente. Pero además conlleva más inconvenientes. En muchos casos es considerada como un cajón desastredondevanaparar,deformaincorrecta,otrasentidadesmaldiagnosticadas. Labaseetiopatogénicaqueexplicaesteestadopermanentededoloresaúndesconocida.Sehanpostulado diferentes hipótesis (inmunológica, neurovegetativa, etiología vírica ) que expliquen las causa de la FM. Hastalafechalateoríamásplausibleeslaexistenciadeunadisfunciónenlatransmisióndeldolorconun desequilibrio entre las vías inhibitorias y activadoras del impulso doloroso. Alteraciones en los niveles de 150
157 RESUMEN diferentes neurotransmisores, así como en pruebas de resonancia magnética funcional o neurofisiología apoyandichahipótesis.lahipótesismásplausibleesquelafibromialgiaseaunprocesocomplejocausado porlaconfluenciadefactoresgenéticosyambientales. Los factores genéticos pueden explicar en gran medida la variabilidad en la percepción del dolor. Los estudios familiares han mostrado una considerable agregación familiar en fibromialgia y cuadros relacionados,sugiriendolaimportanciadelosfactoresgenéticoseneldesarrollodeestoscuadroslafm tieneuncomponentegenéticocomomuestralaexistenciadeagregaciónfamiliar.diferentesestudioshan evaluado la prevalencia de FM en familiares de afectos estimando una mayor recurrencia que en la poblacióngeneeral.dichamayorprevalenciahademostrado,además,sermayorenfamiliaresdesangreque enespososdepacientesdefm,loquevaafavordeunamayorimportanciadelosfactoresgenéticosquelos factoresambientalesendeterminarlasusceptibilidadadesarrollarlaenfermedad.numerososestudiosen gemelossehandesarrolladoenestesentidoy,aunqueenalgunoscasoslosresultadosseancontradictorios, en general muestran una mayor concordancia en gemelos monozigóticos que dizigóticos. Es, pues, un argumentomásafavordelaexistenciadefactoresdesusceptibilidadgenéticaparafm. Conelfindeevaluardichocomponentegenetico,sehanllevadoacabonumerososestudios.Losestudios realizados hasta la fecha han sido básicamente estudios de genes candidato y no han sido capaces de establecer asociaciones genéticas. Esto se debe a que presentaban numerosas limitaciones: la mayoría fueronrealizadosencohortespequeñas,losmarcadoresevluadoseranenlamayoríadeloscasosfacotres de susceptibilidad genética para comorbilidades psiquiátricas de la enfermedad, no presentabans asociaciones estadísticamente significativas tras corregir por multiple testing e intentos de replicar las asociacionesencontradasmostrabanresultadoscontradictorios.enparticular,polimorfismosdebaseúnica degenesimplicadosenlasvíasserotoninérgicas,dopaminérgicasocatecolaminérgicas,perohastalafecha ningúnestudiohasidocapazdeestablecerunmarcadorgenéticoparafm. Es por todo ello que la identificación de las bases biológicas de la fibromialgia precisa de un diseño de investigación que integre aproximaciones epidemiológicas y genómicas en un gran número de muestras biencaracterizadasclínicamente. La búsqueda de los factores de susceptibilidad genética a las enfermedades complejas se centra en la identificación de variantes del genoma que se encuentren con mayor frecuencia entre la población de enfermosencomparaciónconlapoblacióngeneral,conlaesperanzadequeellasmismasseanresponsables delaenfermedadobienqueseanmarcadores(debidoaldesequilibriodeligamiento)paralosverdaderos genes responsables. Los SNPs (polimorfismos de nucleótido único) representan la mayor fuente de variabilidad del genoma humano y constituyen una herramienta fundamental para realizar estudios de 151
158 RESUMEN asociación que permitan detectar los genes implicados en las enfermedades comunes que afectan a la población. Tanto en Estados Unidos como en el Reino Unido existen grandes iniciativas financiadas por el National InstituteofHealthyelWelcomeTrust,respectivamente,enelsenodelascualesseestángenotipandomás de snpsenmillaresdemuestrasdepacientesdedistintasenfermedades.losprimerosresultados delwelcometrustcasecontrolconsortium(wtccc)publicadosenlosúltimosaños,presentabandatos para14.000pacientesafectosdesieteenfermedadesdistintas(trastornobipolar,diabetesmellitustipo1, diabetes mellitus tipo 2, artritis reumatoide, enfermedad de Crohn, hipertensión y enfermedad cardiovascular)1,ademásdeotrosestudiosencáncer,asmayotrasenfermedadesinflamatorias.delmismo modo,yhaciendousodelasúltimastecnologíasdesecuenciación(nextgenerationsequencing),elconsorcio 1000genomes está produciendo datos de exoma y whole genome sequencing que vienen a completar el mapa de variablilidad del genoma humano.estos trabajos demuestran de forma clara que es posible identificar factores genéticos implicados en enfermedades comunes, abriendo nuevas vías biológicas implicadasenestosprocesos. La reciente identificación de variantes de número de copia (CNVs) en el genoma humano ha abierto un nuevo campo de estudio en la búsqueda de bases genéticas de las enfermedades. En algunas patologías complejasyasehaidentificadounarelaciónentrelasusceptibilidadapadecerlaenfermedadyvariantesde númerodecopiadeciertosgenes,comoporejemploenlupuseritematososistémico(genesfcgr3b32y C4A), psoriasis (LCE3C ylce3b) o susceptibilidad a la infección por HIV (gen CCL3L1)4,5. Por ello es importante tener en cuenta este tipo de variabilidad al investigar las causas genéticas de otras enfermedadescomplejas. Tanto para evluar SNPs como CNVs existen diferentes tecnologías. En el caso de los SNPs, además de la metodología implementada, el número de marcardores que se quiere evaluar determina el tipo de tecnologíaausar:desdeelgenotipadodeunúnicosnpmediantetaqmanhastalarealizacióndeunestudio degenomacompletoque,enlaactualidadpuedeevaluarmásde2.5millonesdesnps.enlosúlitmosaños, además de las tecnologías de genotipado, el desarrollo de la secuenciación de última generación ha permitido su uso más extendido. La progresiva rapidez así como el abaratamiento de costes, hace que problablemente,enlospróximosaños,lasecuenciaciónpaseasustituir,enlamayorpartedecasos,ala genotipación. Para realizar un barrido a nivel de todo el genoma en variantes en el número de copia, una de las tecnologías más utilizada es la hibridación genómica comparada basada en arrays, en la que la muestra problemaylamuestracontrolsonhibridadasdeformacompetitivaenelmismoarray.enlaimplementación de dicha tecnología se pueden usar muestras únicas o pooles de muestras. El uso de pooles tiene como 152
159 RESUMEN objetivo el diluir la variabilidad interindividual y resaltar las diferencias debidas a enfermedad. Asimismo, conlleva una reducción en el número de datos generados, así como en los costes.tras la detección de regionespresentandohibridaciñondiferencialentrecasosycontroles,dichasregionesdebenservalidadas mediantediferentestécnicascomopcr,fishopcrcuantitativaentiemporeal. Larealizacióndeestudiosdegenomacompleto,tantoaniveldeCNVscomodeSNPsnecesitadelusodeuna gran cantidad de muestras (del orden de millares) para tener el suficiente poder estadístico que permita detectarvariantesdebajoefecto.enelcasodelasenfermedadescomplejassehanpostuladodiferentes hipótesisqueexpliquenelcomponentegenético.secreequelasumaeinteraccióndeungrannúmerode variables de bajo riesgo serían las responsables de determinar susceptibilidad a desarrollar dicho tipo de patologías. Para alcanzar un tamaño muestral adecuado en muchos casos es necesario la integración de diferentes cohortes. Esto supone una limitación añadida: la heterogeniedad interpoblacional que puede añadir sesgos en el análisis y la disminución en la homogeneidad del fenotipo y en la exactitud en la valoración clínica. Todos estos factores deberán tenerse en cuenta a la hora de realizar un análisis de factoresdesusceptibilidadgenéticaparaunaenfermedadcomlafibromialgia Objetivosdelatesis Dado que la fibromialgia es una enfermedad de origen desconocido, compleja, frecuente y altamente incapacitante, con la presente tesis se ha pretendido estudiar e identificar variantes del genoma (polimorifsmodebaseúnicasinglenucleotidepolymorphisms SNPsyvariantesenelnúmerodecopia CNVs)asociadasaFM,conelobjetivodeprofundizarenlaetiologíadelaenfermedad.Paraello,sehan llevadoacabotresgrandesaproximaciones: 1. Identificacióndesubgruposclínicoshomogéneosdefibromialgiamedianteunanálisisdeclusters: a)construccióndedimensionesdevariablesclínicasparalaposterior b)identificacióndesubfenotiposdefm 2. Realización de un estudio de genoma completo (genome wide association study (GWAS)) para el análisisdirectodesnpsasícomoladetecciónderegionesvariablesennúmerodecopiaasociadasa enfermedad, y su potencial presencia en mosaicismo. Este objetivo incluye los siguientes subobjetivos: e) EstudiodeasociacióndelosdatosdelGWAS, f) IdentificacióndeCNVs, 153
160 RESUMEN g) ReplicaciondelosSNPsconunmayorniveldeasociación h) AplicacióndelanálisisdeclusterenlosresultadosdelGWAS 3. Realizacióndeexperimentosdehibridacióngenómicacomparadamediantearrays(aCGH)conelfin deidentificarregionesvariablesenelnúmerodecopiaasociadasafibromialgia: d) AnálsisyvalidacióndeexperimentosdeaCGH e) AplicacióndelanálisisdeclusterenlosresultadosdelaCGH f) Evaluación de las posibles consecuencias funcionales de las variatnes en número de copiaasociadas Parallevaracabodichosestudios,hemosdispuestodeunagrancohortedecasosdefibromialgia,muybien caracterizadosclínicamente. Seincluyeronenelstudio1510casosindependientesdeFMquecumplíanloscriteriosdiagnósticosparaFM delamericancollegeofrheumatologyde1990.estoscasosfueronrecogidosenunestudiomulticéntrico cuyoprincipalobjetivofuelacreacióndelbancoespañoldedatosclínicosyadndefibromialgiaysíndrome defatigacrónica.cincounidadesdefmdecincohospitalesespañoles(hospitaldelmar,barcelona,jordi Carbonell;HospitalClíniciProvincial,Barcelona,AntonioCollado;HospitaldelaValld Hebrón,Barcelona, JoseAlegre;InstitutoGeneraldeRehabilitacióndeMadrid,Madrid,JavierRivera;andHospitalGeneralde Guadalajara,Guadalajara,JavierVidal)participaronenlarecogidademuestras.Untotalde1,510pacientes defmfueronseleccionadosporreumatólogosparaserposteriormenteevluadosporotrogrupodemédicos entrenadosparalaevaluacióndeestetipodepacientes. Larecogidadedatosserealizómedianteunprotocoloestánadarddecuestionariosyexploraciónfísicaque incluía: datos sociodemográficos, historia personal y familiar de enfermedad, tiempo de evolución de la enfermedad,presenciadesíntomasdelamplioespectrodeafectacióndelafmytratamientos.lasmedidas centrales de actividad de la enfermedad fueron evaluadas mediante escalas validadas al español. 1,000 controlsamplescamefromthenationaldnabankofsalamanca). Asimismoseincluyeron1000muestrasdeindividuosespañolesconbajosnivelestantodedolorcomode fatigasegúnlasrespuestasauncuestionario. 154
161 RESUMEN Identificacióndesubgruposdefibromialgiamedianteunanálisisdeclusters Dada la heterogeneidad de la patología, se ha realizado, en primer lugar, un análisis de clusters. En un primer tiempo, se han construido dimensiones de variables (en base a su similitud) que en un segundo tiempohanservidoparaidentificarsubgruposdepacientes.elanálisissehallevadoacabo,inicialmente,en unsubsetde559casosdefmparaserreproducidoposteriormenteenunsegundogrupode887casos. Elestudiode48variablesclínicas(sintomatología,antecedentespersonalesdepsicopatologíaypatología osteoarticular, antecedentes familiares y escalas de medición de severidad de la enfermedad) en 1446 individuosnorelacionadosdiagnosticadosdefm,cumpliendoloscriteriosacrde1990yseleccionadosenel marco de la creación del banco de ADN de fibromialgia, ha dado lugar a la construcción de tres grandes dimensionesdevariables:sintomatología,antecedentes(personalesyfamiliares)yescalasdemedicióndela actividaddelaenfermedad.únicamentelasdosprimeras,porsermásrobustasalincluirunmayornúmero devariablesydemayorpeso,hansidoenunsegundotiempoutilizadasparalaidentificacióndesubgrupos de FM. Tres grupos de pacientes han sido identificados: FM con baja sintomatología y niveles bajos de comorbilidad, FM con elevada sintomatología y elevados niveles de comorbilidad y FM elevada sintomatología pero niveles bajos de comorbilidad. Las escalas clínicas que no han sido utilizadas para la construccióndedichossubgruposhanpermitidoevaluarquelossubgruposdetectadoseranclínicamente diferentes. En particular, los niveles de actividad de la enfermedad han sido diferentes de forma estadísticamente significativa en el grupo de FM con bajos niveles tanto de sintomatología como de comorbilidades. Estudiodeasociacióndegenomacompleto ParaelanálisisdelosSNPs,seharealizadounestudiodegenomacompleto(GWAS)medianteunarraycon más de 1 millón de marcadores, el Illumina 1M duo. Se han hibridado un total de 321 muestras de fibromialgia.pararealizarelestudiodeasociación,sehanutilizadolosdatosresultantesdelahibridaciónde 200muestrascontrolconelarray610QuaddeIllumina.Dadoquecasosycontrolessehangenotipadocon plataformasdiferentes,sehanrealizadocontrolesdecalidad(qualitycontrolqc)porseparadoencada cohorte, a nivel de muestras y de SNPs, con el software PLINK. Para las muestras se ha comprobado: el origen,elsexo,elparentesco,laheterogeneidadyelgenotypingcalling.aniveldelossnpssehaevaluado elgenotypingcalling,elequilibriodehardyweinbergylaminimumallelefrequency.unavezfiltradoslos 155
162 RESUMEN datosdecasosycontroles,seharealizadoelestudiodeasociación,medianteelsoftwareplink,de SNPsen300casosy203controles. Nosehandetectadodiferenciaspoblacionalessignificativasentrecasosycontroles,comomuestraelvalor del genomic inflation (=1.013). La asociación alélica no ha identificado SNPs con niveles de asociación inferiores al umbral establecido para un estudio de genoma completo, aunque 77 SNPs presentaron asociaciónconpvaloresinferioresa104. Se ha procedido entonces a la evaluación de dichos SNPs mediante los programas Ingenuity Systems Pathway analysis (IPA) y GeneSet analysis Toolkit v2. Se han identificado como principales pathways enfermedades reproductivas y enfermedades neurológicas. El transporte del calcio también ha aparecido comounprocesobiológicodestacado. Conelfinderealizarfinemapping,esdecir,obtenerunavisióndealtaresolucióndeunaregiónasociada paraaumentarlaposibilidaddequeunsnpcausalseaidentificadodirectamente,seharealizadoimputación con el software Imputev2, usando como referencia tanto el panel de CEU de Hapmap3 como datos de individuosceudel1000genomesproject.paraimplementardichosoftwarehasidonecesariotransformar losdatosalformatoplinkmedianteelsoftwaregtool.másde800000desnpshansidoimputadosdelos cualesmásdecuatromilloneshansidoconsideradosporsualtacalidadenlaimputaciónyporpresentar unamínimumallelefrequencysuperiora0.05.finalmente,sehaprocedidoalestudiodeasociacióndelos datosimputadosmedianteelsoftwaresnptest,usandoelmétododeasociaciónfrequentistquetieneen cuentalaincertezadelosgenotipos. Delos77SNPsmostrandomayoresnivelesdeasociaciónjuntoalosdatosdeimputacióndelasregiones cercanasadichossnps,hansidoseleccionados21snpsparasugenotipaciónenelrestodelacohorte.se han detectado diferentes SNPs asociados. Tras el QC, 20 SNPs en 968 FM y 937 controles han sido consideradosparaestudiodeasociación.laasociacióndetectadaenelgwasnohasidoreplicadadeforma estadísticamentesignificativa(p<0.0023trascorreccióndebonferroni).4snpshanpresentadoasociación nominalotendenciaalaasociaciónquehadadolugarunaasociacióndelordende10 4 alconsiderarlas muestrasgenotipadasenelgwasyenlaréplicadeformaconjunta:rs (mcf2),rs (ank3) andrs (myt1l)yrs (intergénico). DadoqueloscuatroSNPsconasociaciónnominalenelanálsisiconjunto,seencontrabanenregionesno codificantes, se han evaluado las posibles consecuencias funcionales de dichos SNPs. Mediante el uso de diferentessoftwares,sehandetectadoelevadosnivelesdeconservaciónparadosdeestossnps.asimismo, el análisis mediante el software Genvar, ha identificado una posible correlación de los genotipos de rs yrs (snpquestáendesequilibriodeligamientoconrs )conlaexpresiónen 156
163 RESUMEN linfocitosdelgensntg2.estudiosprevios,hanmostradovariantesdedichogenabasociadasconautismoe intentosdeautolisisendepresiónmayor. Posteriormente,sehallevadoacabolaevaluacióndelcomportamientodeestos4polimorfismosenlostres subgrupos de FM detectados mediante el análisis de clusters. Únicamente rs ha mostrado un comportamiento diferente, de forma que, al considerar únicamente el cluster 3 de pacientes (elevada sintomatologíaconbajosnivelesdecomorbilidad)laasociacióndetectadaenelgwassereplicaenelresto de la cohorte, con una mayor significación estadística de la asociación en el estudio conjunto (asociación alélicap=4.28x10 5,OR0.58( ). Elpolimorfismors esunavarianteintrónicadelgenMYT1L,quecodificaunaproteínazincfinger postmitóticaimplicadaenladiferenciaciónneuronal.variantesendichogenhansidoasociadasadiferentes trastornosneuropsiquiátricos.sehaevaluado,asimismo,laposibleexistenciadevariantesfuncionalesen desequilibrio de ligamiento con rs , identificándose únicamente como ligadas las variantes intrónicasrs yrs TambiénsehaprocedidoaladeteccióndeCNVsmedianteslosdatosgeneradosporelarraydeSNPs,en concretomedianteelanálisisdelabiallelefrequency(baf)yellogaritmodelaratioentreintensidadesde cada uno de los alelos posibles para cada SNP (log ratio). Para ello se ha utilizado el software PennCNV, detectándosemásde18000snps.sehaprocedidoentoncesalaseleccióndeloscnvsparavalidarteniendo encuentalossiguientescriterios:cnvspresentesenmásdel5%delasmuestras,localizadosengenesyno siendo elementos previamente relacionados con intervariabilidad poblacional. Dos CNVs han cumplido dichoscriterios. Se ha procedido al seguimiento de uno de ellos, localizado en el gen ACACA. Tras la identificación de los puntos de rotura mediante experimentos de PCR y posterior PCR secuenciación, previo subclonaje del producto de PCR, se ha diseñado una PCR multiplex. Con dicho diseño se han genotipado 209 FM y 211 controlessinidentificarunaasociaciónestadísticamentesignificativaquevalidaseloshallazgosdelanálisis mediantepenncnv. TambiénsehaprocedidoalfiltradodelasregionesdetectadasporPennCNVconelobjetivodeidentificar CNVs grandes y poco frecuentes asociados a FM. No se ha detectado ninguna región no presente en poblacióngeneralqueaparecieradeformarecurrenteenlasmuestrasdefm. Delmismomodo,sehaevaluadolaexistenciadeCNVenmosaicismoquepudieranestarasociadosaFM. Mediante el software Mosaic Alteration DetectionMAD se han identificado dos CNVs no presentes en población control. Una ganancia en mosaicismo en el gen OVOS2 y otra en el gen SLC2A14.Noseha 157
164 RESUMEN realizadolavalidaciónylareplicacióndeestoscnvenposiblemosaicismoporquenosedisponíadeotros tejidos para poder evaluar cambios en el número de copias entre los diferentes tejidos de un mismo individuo. EstudiodelaimplicacióndevariantesenelnúmerodecopiamedianteaCGH Para el estudio de los variantes en número de copia (CNV), se han llevado a cabo experimentos de hibridación genómica comparada por array(acgh), mediante el array 400K de Agilent, hibridando tres pooles de muestras de pacientes afectos de fibromialgia (FM) con un pool de controles. Los pooles de pacientes incluían casos de FM con antecedentes familiares para la enfermedad y las siguientes características:poolfm(individuossinfatiga),poolfmconfatigaypoolfmdeinicioprecoz(edaddeinicio de la enfermedad inferior o igual a 20 años).mediante el análisis informático de los resultados de dichas hibridaciones, se han identificado diferentes regiones mostrando diferencias de intensidad entre casos y controles.paradichoanálisissehautilizadoelalgoritmoadm2delsoftwaregenomicworkbenchdeagilent, estableciendocomocriteriosdeselecciónlapresenciadealmenostressondasconunlogratiosuperiora 0.3oinferiora0.3tantoenlashibridacionesdirectascomoenlashibridacionesdyeswap. Sehanrealizadotambiénhibridacionesconelarray1MilliondeAgilent,deformacomplementaria.Estavez únicamentesellevaronacabohibridacionesdirectas(nohubodyeswap). Enlashibridacionesdelarray400K,sehandetectado7regionesquemostrabanhibridacióndiferencialentre casos y controles. Dos de ellas han sido descartadas por aparecer de forma sistemática en todas las hibridacionesrealizadasenotrosestudiosquesellevabanacaboenellaboratorio.ademássehaincluidoen elseguimientounaregión,deformacomplementaria,detectadaporelalgortimoadm1(menosastringente), enelgenmyo5b,porconstituirunbuencandidatoparafibromialgia.enlosresultadosdelashibridaciones conelarray1million,nosehaseleccionadoningunaregiónparaelseguimientoporserlaregionespoco consistentes(presenciadesondasrepetidas,gananciasopérdidassegúnelpooldefm ). Así pues se ha realizado el seguimiento de las regiones localizadas en los genes GALTNL6, WWOX,MYO5B,PTPRDyNRXN3.Los puntos de rotura de cinco de las seis regiones seleccionadas para validación,estaban disponibles en las base datos públicas por lo que se han podido diseñar directamente experimentosdegenotipaciónmediantepcrmultiplex.paralavalidacióndelcnvlocalizadoenunaregión 158
165 RESUMEN intrónicadewwox,hasidonecesarioidentificarlospuntosderoturamedianteunapcryposteriorpcrde secuenciación. ParalavalidaciónyreplicacióndelasregionesdetectadasmedianteaCGH,sehanrealizadoexperimentosde PCRmultiplex.Sehangenotipadounapartedelacohorte(300casosy300controles)yenfuncióndelos resultadossehaporseguidoconelgenotipadodemásmuestrasosehadescartadolaregión.finalmente,en cinco de ellas no se ha encontrado asociación con FM, mientras que en la otra, NRXN3_DEL, se ha confirmadoelresultadodelexperimentodeacgh. NRXN3_DEL es una indel de 8.8 kb situada en el segundo intrón del gen neurexina 3 (NRXN3). NRXN3 codifica a un proteína transmembrana sitúada en las neuronas presinápticas siendo esencial para el desarrollo y funcionamiento de la sinapsis. Las neurexinas se unen a las neuroliginas (proteínas trans membrana potsinápticas) y el complejo neurexinaneuoligina puede promover la fomración de novode sinapsis y la diferenciación de receptores postsinápticos Dicha unión está estrechamente regulada por mecanismosdealternativesplicingenambasfamiliasdemoléculas.tantosnpscomocnvsenlosdiferentes genes neurexina (en seres humanos esxiten tres) y neuroligina se han asociado a trastornos neuropsiquiátricosyaadicciónadiferentessubstancias. Elgenotipadode359casosdeFMy378controleshamostradounaasociaciónestadísticamentesignificativa de FM con la deleción 359 (asociación genotípica, modelo recesivo (genotipo riesgo: homozigoto delecionado)p=0.0037,or(95%ci)=1.74( );asociaciónalélicap=0.12testdeconvirtiéndoseen una tendencia (asociación genotípica, modelo dominante, (genotipos de riesgo: homozigoto delecionado) p=0.064 OR 1.18 ( , asociación alélica, Test de Fisher p=0.07, OR (95%CI) = 1.12 ( )). Al considerar únicamente mujeres tanto en casos como en controles, la asociación sí que ha sido estadísticamente significativa (asociación genotípica, modelo recesivo (genotipo riesgo: homozigoto delecionado)p=0.021,or1.46( ),asociaciónalélicatestdefisherp=0.015or(95%ci)=1.22( )) Asimismo,alaplicarelanálisisdeclusters,sehadetectadounaasociaciónestadísticamentesignificativaal considerar los clusters 1 y 3 (FM con bajos niveles de comorbilidad) (asociación genotípica, modelo dominante p=0.009, OR (95%CI)=1.28( );asociación alélica test de Fisher p=0.019, OR(95%CI)=1.17( )). SehaevaluadolaposibleexistenciadeinteracciónentreNRXN3_DELylosotrosCNVsevaluados,asícomo los4snpsmostrandounaasociaciónnominalenelgwas,sinevidenciarseinteracciónalguna. 159
166 RESUMEN AltratarsedeunCNVsituadoenunaregiónintrónica,paraevaluarlasposiblesconsecuenciasfuncionales dedichavariante,sehadiseñadounexperimentoveracodeconelfindeanalizar48polimorfismosdebase única (SNPs) en la región y así poder establecer el desequilibrio de ligamiento existente. Se han seleccionadountotalde45tagsnps,snpsfuncionalesysnpssituadosensitiosdesplicingalternativo,a losquesehanañadido3snpsespecialmentediseñadosparagenotiparelcnv.ningúnsnphamostrado asociaciónconfmnihademostradoestarendesequilibriodeligamientoconelcnvcomoparapoderser su proxy. Dicho ensayo sí que ha demostrado su utilidad para genotipar el CNV ya que la concordancia entrelapcrmultiplexyelveracodehasidodel100%conlosehapodidogenotipareltotaldelacohorte deunaformamásrápidayprecisa,medianteelanálisisdelossnps. Porotraparte,sehanrealizadoexperimentosconlíneascelularesconelfindeevaluarlaposiblecorrelación entre el genotipo para NRXN3_DEL y los tránscritos resultantes. Se han seleccionado las líneas de glioblastomau87yt98gporprovenirdetejidoneuronalypresentardiferentesgenotiposparanrxn3_del (U87homozigotadelecionadayT98Ghomozigotanodelecionada)ysehaprocedidoaexperimentosdePCR amplificandocdna(provineintedelaextraccióndearndeambaslíneascelulares)delosexones19a21. Hanamplificadolasdosisoformasposiblesadichonivel(conysinelexon20)ypresentandounadiferente proporción de cada una en cada línea celular. Dichas diferencias de expresión han sido identificadas mediante PCR cuantitativa a tiempo real (ensayos Taqman) confirmando una mayor proporción de la isoformasinelexón20enlalíneacelularu87. Alconstatarenlabibliografíaqueexisteunaposiblerelaciónentreelsplicingalternativoenelsitiocanónico 4 y la presencia de secuencias de unión para inhibidores del splicing, hemos evaluado in silico la posible presencia de dichos motivos en la región NRXN3_DEL, confirmando la presencia de sitios de unión de splicing. Se han detectado secuencias para la unión de dichos factores en la región NRXN3_DEL. Además muchasdeestassecuenciaspresentabanunaelevadatasadeconservaciónentreespecies. Finalmente, para confirmar la relación entre el CNV y cambios en el splicing alternativo, evaluamos los tránscritosenlíneasdehapmapquepresentabandifenretesgenotiposparaladeleción.noencontramos relaciónyparadescartarqueelefectonofueradependientedetejido,tambiénhemosevaluadomuestras decerebrohumano(vermix)portadorasdediferentesgenotiposdenrnx3_del,descartandounarelación directa. 160
167 RESUMEN En la presente tesis, se ha llevado a cabo un estudio exhaustivo de los posibles factores genéticos implicadosenlasusceptibilidadadesarrollarfibromialgia.dadalaheterogeneidaddedichapatología,en un primer tiempo hemos realizado un análisis de clusters de variables clínicas que nos ha permitido identificartressubgruposdefm. ElanálisisdeclustersdevariablesclínicasdeFMharesultadoenlaidentificacióndetressubgruposdeFM: FM con bajos niveles de sintomatología y de comorbilidades, FM con elevados niveles tanto de sintomatologíacomodecomorbilidadesyfmconelevadosnivelesdesintomatologíaperobajosnivelesde comorbilidades. Los subgrupos identificados han resultado ser,además, diferentes a nivel de direntes escalasdemedicióndeactividadclínica.elanálisisdecluster,ademásdediferenciargruposclínicamente homogéneos de FM, ha mostrado la importancia de los antecedentes personales y familiares, para la correctaclasificacióndelospacientesencadaunodelosgrupos.estehechoapoyalanuevaclasificación de FM en la que se subraya la importancia de la sintomatología a la hora de evaluar la enfermedad. Finalmente,dichoestudiohasidoaplicadoalanálisisdevariantesgenéticas. Se ha realizado un estudio de genoma completo para el análisis de la posible contribución de SNPs en susceptiblidadadesarrollarfm.apesardequeelestudioinicialnohadetectadoningúnpolimorfismocon unniveldeasociaciónestadísticamentesignificativosegúnelumbralestablecidoparaestetipodeestudios, el análisis in silico de los variantes con una mayor asociación, ha mostrado una sobrerepresentación de enfermedades neurológicas. Dicho hallazgo iría en concordancia con las últimas hipótesis en las que se postula que la FM es debida a una disfunción en el sistema nervioso central y concretamente, en la transmisióndelimpulsodoloroso.además,elestudiodereplicaciónenelque3delos4snpsconmayor asociación (aunque nominal) están localizados en genes implicados en el sistema nervioso (MCF2,ANK3,MYT1L)loquevieneareforzarestaidea.Finalmente,laasociaciónders concasosde FM con baja comorbilidad y elevada sintomatología viene a confirmar la posible implicación del sistema nerviosoeneldesarrollodeestapatología. El proto oncogen MCF2 se expresa en diferentes tejidos y está implicado, entre otras funciones, en la ovogénesis,enlaproliferacióndendríticayenlaapoptosisenelsistemanerviosocentral.variantesenel genmcf2sehanasociadoconautismoyesquizofrenia. ANK3(ankyrina3)perteneceaunafamiliadeproteínasquejueganunpapelfundamentalenfuncionestales comomotilidadcellular,activaciónyproliferacióndedominiosespecializadosdemembrana.ank3tienesus principales funciones en el cerebro, implicada en el transporte a lo largo del axon y en la polaridad dendrítica.variantesenank3sehanasociadoconesquizofrenia,enfermedadbipolaryautismo. 161
168 RESUMEN rs ha mostrado asociación nominal en el estudio conjunto de toda la cohorte. Al considerar únicamentemujeresconnivelesbajosdecomorbilidad(clusters1y3),laasociaciónhasidosermayortanto enelgwascomoenlacohortedereplicaylasignificaciónestadísticadelaasociacióndelanálsisconjunto haresultadosuperior.rs (chr2: ;hg18)pertenecealtercerintróndelgenmyt1l (myelintranscriptorfactor1like).variantesenelgnemyt1l,tantocnvscomosnps,sehanasociadocon esquizofrenia,depresiónmayoryautismo. El estudio de variantes en el número de copia mediante acgh también va en la misma dirección, detectandoasociaciónenlasmuestrasconbajosnivelesdecomorbilidaddeunavarianteintrónicaenel genneurexina3,quecodificaporunamoléculatransmembranapresinápticaesencialparaeldesarrolloy la función de la sinapsis. Si bien parece que el ratio entre isoformas con y sin exón 20 no parece estar relacionada con la deleción, la presencia de motivos de unión de factores reguladores del splicing,en la región afectada por el CNV, podría indicar que NRXN3_DEL puede asociarse a otras alteraciones en el splicing.loscambiosenelratiodelasdosisoformasenu87yt98gpudieradeberseaotrosfacotrescomo el estadio de la enfermedad (del glioblastoma de origen) y el que provienen de individuos de sexo diferente.parapoderevaluardeformacorrectaposiblesconsecuenciasdenrxn3_delaniveldesplicing, seríanecesariorealizarexperimentosdernaseqendiferentesmuestrasdetejidocerebralprovinientesde individuoscongenotiposdiferentesparaladeleción. El estudio que hemos realizado presenta varias limitaciones. A nivel del GWAs el número reducido de muestras del que disponíamos, hace que no tuviéramos suficiente poder estadístico para detectar vairantesdebajoefecto.elanálisisdecnvsapartirdelosdatosdelgwastambiénhamostradoserpoco eficienteparadetectarcnvsdepequeñotamaño.yparalavalidacióndelosposiblescnvsenmosaico hubierasidonecesariodisponerdediferentestejidosdeunmismoindividuo.finalmente,elusodepooles enlosexperimentosdehibridacióngenómicacomparadapuedeexplicarelquenosehayanpodidovalidar cincodeloscnvsdetectados.cualquierpequeñoerroralcuantificarlasmuestrasparaprepararlospooles podríahaberdadolugaralasobrerepresentacióndeunamuestraenconcreto.tambiénhayqueteneren cuentaque,paravariantesfrecuentes,yconlaeleccióndelasmuestrasqueconformarelpool,sepuede introducirunsesgoaleatorioquedélugaraunfalsopositivo. Enestatesis,tantolosSNPscomelCNVdetectadoscomoposiblesasociacionesgenéticasaFM,pertenecen agenesquenosólosecaracterizanportenersuprincipalfunciónenelsistemanerviosocentral.además, dichosgeneshansidopreviamenteasociadosaenfermedadesneuropsiquiátricas.siseconfirmanestas asociacionesenotrascohortes,elconceptodefibromialgiacomoenfermedadreumatológicadaríaungiro 162
169 RESUMEN parapasarasituarsejuntoaenfermedadesneurocognitivas.asimismo,tantolossnpsquehanpresentado una asociación nominal en el estudio conjunto (datos GWAS junto a réplica), uno de ellos presentando asociación en fibromialgia con bajos niveles de comorbilidad, como la deleción del gen neurixina han presentado una mayor asociación al condirerar unícamente mujeres. Dicho hallazgo puede apuntar a la existenciadediferentesfactoresdesusceptibilidadgenéticaparafmsegúnelsexo. Enresumen,elpresenteestudiosugierelaimplicacióndevariantes,tantoSNPscomoCNVsimplicadosen elsistemanerviosoenlaetiopatogeniadefm.tambiénponedemanifiestolaimportanciaenidentificar subgrupos homogéneos de la enfermedad para el desarrollo exitoso de estudios genéticos. Dada la complejidad de la patología serán necesarias un mayor número de cohortes de mayor tamaño pero siempreycuandolaclasificaciónfenotípicaseaexhaustivaypermitaidentificardiferentessubgruposque puedantenerundiferentebackgroundgenético. Lasprincipalesconclusionesalasquesehallegadoenlapresentetesisson: 1 Lafibromialgiaesunaentidadextremadamentecompleja.Lasusceptibilidadgenéticaapadecerla enfermedadpareceresultardelasumadediferentesfactoresgenéticosdebajoefecto.esporello indispensable el uso de grandes cohortes que incluyan miles de muestras con el fin de poder detectardichasvariantes. 2 La identificación de subgrupos clínicamente homogéneos de la enfermedad constituye un paso indispensableparalaidentificacióndefactoresdesusceptibilidadgenéticaparafibromialgia.eneste sentido, la presente tesis apunta a que la inclusión de comorbilidades tanto personales como familiarespuedeaportarinformacióncomplementariadegranutilidadparalaclasificacióndelafm basadaensíntomassomáticos. 3 Los resultados del GWAS indican un posible contribución del sistema nervioso central en el desarrollodefibromialgia:lasenfermedadesneurológicasaparecencomosobrerepresentadasenel estudiodepathwaysrealizadoenlossnpsquepresentabanmayorasociación. 163
170 RESUMEN 4 UnSNPenelgenMYT1Lhapresentadoasociaciónestadísticamentesignificativaconfibromialgia connivelesbajosdecomorbilidad,poniendodemanifiestotantolaposibleimplicacióndelsistema nervioso central en la enfermedad, como la importancia de identificar subgrupos clínicamente homogéneosparaladeteccióndefactoresdesusceptibilidadgenéticaparafm. 5 El estudio de replicación del GWAS ha mostrado mayor asociación al considerar únicamente mujeres. Este hecho subraya la importancia del género en la etiopatogenia de la fibromialgia y sugierelaposibleexistenciadediferentesfactoresdesusceptibilidadgenéticaparacadaunodelos géneros. 6 LaposibleexistenciadelCNVSLC2A14.enmosaicismoenmuestrasdefibromialgiaconstituiríauna nuevaevidenciadelaposibleimplicacióndelsistemanerviosoenlaenfermedad.elotroposiblecnv enmosaicodetectado,implicadoendegeneraciónmuscular,apuntaríaalroldeotrosmecanismosa nivelosteomuscular. 7 Un insercióndeleción intrónica en el gen neurexina 3 ha mostrado asociación en mujeres con fibromialgia,yenparticular,enaquellasconbajosnivelesdecomorbilidad.dadoquelamoleculaes esencial para el desarrollo, mantenimiento y funcionamiento de la sinapsis, sería un nuevo argumentoafavordelaimplicacióndelsistemanerviosoenlaenfermedad. 8 Laconfirmacióndelasvariantesdetectadasennuevascohortesdefibromialgiasupondríaungiro conceptual de la enfermedad hacia una visión más neurocognitiva que osteomuscular. 164
171 REFERENCES
172
173 BasbaumAI,BautistaDM,ScherrerG,JuliusD.Cellularandmolecularmechanismsofpain.Cell.2009 Oct16;139(2): WoolfCJ.Whatisthisthingcalledpain?JClinInvest.2010Nov;120(11): GuillotX,SemeranoL,DeckerP,FalgaroneG,BoissierMC.Painandimmunity.JointBoneSpine. 2011May;79(3): WoolfCJ.Centralsensitization:implicationsforthediagnosisandtreatmentofpain.Pain.2010 Mar;152(3Suppl):S NussbaumJ,XuQ,PayneTJ,MaJZ,HuangW,GelernterJ,etal.Significantassociationofthe neurexin1gene(nrxn1)withnicotinedependenceineuropeanandafricanamericansmokers.hummol Genet.2008Jun1;17(11): KunerR.Centralmechanismsofpathologicalpain.NatMed.2010Nov;16(11): MillanMJ.Descendingcontrolofpain.ProgNeurobiol.2002Apr;66(6): LevinsonJN,ElHusseiniA.Newplayerstipthescalesinthebalancebetweenexcitatoryand inhibitorysynapses.molpain.2005;1: VillemureC,BushnellMC.Cognitivemodulationofpain:howdoattentionandemotioninfluence painprocessing?pain.2002feb;95(3): VillemureC,SchweinhardtP.Supraspinalpainprocessing:distinctrolesofemotionandattention. Neuroscientist.2010Jun;16(3): SchweinhardtP,BushnellMC.Painimaginginhealthanddiseasehowfarhavewecome?JClin Invest.2010Nov;120(11): RenK,DubnerR.Descendingmodulationinpersistentpain:anupdate.Pain.2002Nov;100(12): NielsenLA,HenrikssonKG.Pathophysiologicalmechanismsinchronicmusculoskeletalpain (fibromyalgia):theroleofcentralandperipheralsensitizationandpaindisinhibition.bestpractresclin Rheumatol.2007Jun;21(3): WolfeF,ClauwDJ,FitzcharlesMA,GoldenbergDL,KatzRS,MeaseP,etal.TheAmericanCollegeof Rheumatologypreliminarydiagnosticcriteriaforfibromyalgiaandmeasurementofsymptomseverity. ArthritisCareRes(Hoboken).2010May;62(5): RozentalR,GiaumeC,SprayDC.Gapjunctionsinthenervoussystem.BrainResBrainResRev.2000 Apr;32(1): CowanWM,Kandel,E.R..Abriefhistoryofsynapsesandsynaptictransmission.In:CowanWM, Südhof,T.C&Stevens,C.F.,editor.Synapses.Baltimore:TheJonhsHopkinsUniversityPress; SudhofTC.Neuroliginsandneurexinslinksynapticfunctiontocognitivedisease.Nature.2008Oct 16;455(7215): DeanC,DresbachT.Neuroliginsandneurexins:linkingcelladhesion,synapseformationand cognitivefunction.trendsneurosci.2006jan;29(1): ShapiroL,LoveJ,ColmanDR.Adhesionmoleculesinthenervoussystem:structuralinsightsinto functionanddiversity.annurevneurosci.2007;30: CraigAM,KangY.Neurexinneuroliginsignalinginsynapsedevelopment.CurrOpinNeurobiol.2007 Feb;17(1): CraigAM,GrafER,LinhoffMW.Howtobuildacentralsynapse:cluesfromcellculture.Trends Neurosci.2006Jan;29(1): UllrichB,UshkaryovYA,SudhofTC.Cartographyofneurexins:morethan1000isoformsgenerated byalternativesplicingandexpressedindistinctsubsetsofneurons.neuron.1995mar;14(3): MisslerM,FernandezChaconR,SudhofTC.Themakingofneurexins.JNeurochem.1998 Oct;71(4): MisslerM,ZhangW,RohlmannA,KattenstrothG,HammerRE,GottmannK,etal.Alphaneurexins coupleca2+channelstosynapticvesicleexocytosis.nature.2003jun26;423(6943): SonsMS,BuscheN,StrenzkeN,MoserT,ErnsbergerU,MoorenFC,etal.alphaNeurexinsare requiredforefficienttransmitterreleaseandsynaptichomeostasisatthemouseneuromuscularjunction. Neuroscience.2006;138(2):43346.
174 IchtchenkoK,HataY,NguyenT,UllrichB,MisslerM,MoomawC,etal.Neuroligin1:asplicesite specificligandforbetaneurexins.cell.1995may5;81(3): UshkaryovYA,PetrenkoAG,GeppertM,SudhofTC.Neurexins:synapticcellsurfaceproteinsrelated tothealphalatrotoxinreceptorandlaminin.science.1992jul3;257(5066): WalshT,McClellanJM,McCarthySE,AddingtonAM,PierceSB,CooperGM,etal.Rarestructural variantsdisruptmultiplegenesinneurodevelopmentalpathwaysinschizophrenia.science.2008apr 25;320(5875): JamainS,QuachH,BetancurC,RastamM,ColineauxC,GillbergIC,etal.MutationsoftheXlinked genesencodingneuroliginsnlgn3andnlgn4areassociatedwithautism.natgenet.2003may;34(1): LawsonYuenA,SaldivarJS,SommerS,PickerJ.FamilialdeletionwithinNLGN4associatedwith autismandtourettesyndrome.eurjhumgenet.2008may;16(5): LeinES,HawrylyczMJ,AoN,AyresM,BensingerA,BernardA,etal.Genomewideatlasofgene expressionintheadultmousebrain.nature.2007jan11;445(7124): BierutLJ,MaddenPA,BreslauN,JohnsonEO,HatsukamiD,PomerleauOF,etal.Novelgenes identifiedinahighdensitygenomewideassociationstudyfornicotinedependence.hummolgenet.2007 Jan1;16(1): NovakG,BoukhadraJ,ShaikhSA,KennedyJL,LeFollB.AssociationofapolymorphismintheNRXN3 genewiththedegreeofsmokinginschizophrenia:apreliminarystudy.worldjbiolpsychiatry.2009;10(4pt 3): StoltenbergSF,LehmannMK,ChristCC,HersrudSL,DaviesGE.Associationsamongtypesof impulsivity,substanceuseproblemsandneurexin3polymorphisms.drugalcoholdepend.2011jun LachmanHM,FannCS,BartzisM,EvgrafovOV,RosenthalRN,NunesEV,etal.Genomewide suggestivelinkageofopioiddependencetochromosome14q.hummolgenet.2007jun1;16(11): HishimotoA,LiuQR,DrgonT,PletnikovaO,WaltherD,ZhuXG,etal.Neurexin3polymorphismsare associatedwithalcoholdependenceandalteredexpressionofspecificisoforms.hummolgenet.2007dec 1;16(23): HeardCostaNL,ZillikensMC,MondaKL,JohanssonA,HarrisTB,FuM,etal.NRXN3isanovellocus forwaistcircumference:agenomewideassociationstudyfromthechargeconsortium.plosgenet.2009 Jun;5(6):e ChingMS,ShenY,TanWH,JesteSS,MorrowEM,ChenX,etal.DeletionsofNRXN1(neurexin1) predisposetoawidespectrumofdevelopmentaldisorders.amjmedgenetbneuropsychiatrgenet.2010 Jun5;153B(4): GauthierJ,SiddiquiTJ,HuashanP,YokomakuD,HamdanFF,ChampagneN,etal.Truncating mutationsinnrxn2andnrxn1inautismspectrumdisordersandschizophrenia.humgenet.2011 Oct;130(4): KimHG,KishikawaS,HigginsAW,SeongIS,DonovanDJ,ShenY,etal.Disruptionofneurexin1 associatedwithautismspectrumdisorder.amjhumgenet.2008jan;82(1): VaagsAK,LionelAC,SatoD,GoodenbergerM,SteinQP,CurranS,etal.RareDeletionsatthe Neurexin3LocusinAutismSpectrumDisorder.AmJHumGenet.2011Dec WisniowieckaKowalnikB,NesterukM,PetersSU,XiaZ,CooperML,SavageS,etal.Intragenic rearrangementsinnrxn1inthreefamilieswithautismspectrumdisorder,developmentaldelay,andspeech delay.amjmedgenetbneuropsychiatrgenet.2010jul;153b(5): KirovG,GumusD,ChenW,NortonN,GeorgievaL,SariM,etal.Comparativegenomehybridization suggestsarolefornrxn1andapba2inschizophrenia.hummolgenet.2008feb1;17(3): FitzcharlesMA,YunusMB.Theclinicalconceptoffibromyalgiaasachangingparadigminthepast20 years.painrestreat.2012;2012: BurkhamJ,HarrisE.D.Fibromyalgia:AchronicPainSyndrome.In:HarrisED,BuddR.,Genovese M.C.,FiresteinG.S.,sargentJ.S.,SledgeC.B.,editor.Kelley'stextbookofRheumatology.7ed.Pennsylvania: Elsevier;2005.p YunusMB,KhanMA,RawlingsKK,GreenJR,OlsonJM,ShahS.Geneticlinkageanalysisofmulticase familieswithfibromyalgiasyndrome.jrheumatol.1999feb;26(2):40812.
175 BallinaF.Fibromialgia.In:ReumatologíaSEd,editor.ManualSERdelasEnfermedadesReumáticas.4 ed.madrid:editorialmedicapanamericana;2004.p RiveraJAC,BallinaFJ,CarbonellJ,CarmonaL,CastelB,ColladoA,EsteveJJ,MartínezFG,TorneroJ, VallejoMA,VidalJ.ConsensusdocumentonfibromyalgiaoftheSpanishSocietyofRheumatology ReumatologíaClínica.2006;2S1: OsorioCD,GallinaroAL,LorenziFilhoG,LageLV.Sleepqualityinpatientswithfibromyalgiausingthe PittsburghSleepQualityIndex.JRheumatol.2006Sep;33(9): RoizenblattS,MoldofskyH,BeneditoSilvaAA,TufikS.Alphasleepcharacteristicsinfibromyalgia. ArthritisRheum.2001Jan;44(1): HamiltonNA,AffleckG,TennenH,KarlsonC,LuxtonD,PreacherKJ,etal.Fibromyalgia:theroleof sleepinaffectandinnegativeeventreactivityandrecovery.healthpsychol.2008jul;27(4): KatzRS,HeardAR,MillsM,LeavittF.Theprevalenceandclinicalimpactofreportedcognitive difficulties(fibrofog)inpatientswithrheumaticdiseasewithandwithoutfibromyalgia.jclinrheumatol. 2004Apr;10(2): DickB,EcclestonC,CrombezG.Attentionalfunctioninginfibromyalgia,rheumatoidarthritis,and musculoskeletalpainpatients.arthritisrheum.2002dec15;47(6): LawrenceRC,FelsonDT,HelmickCG,ArnoldLM,ChoiH,DeyoRA,etal.Estimatesoftheprevalence ofarthritisandotherrheumaticconditionsintheunitedstates.partii.arthritisrheum.2008jan;58(1): BrancoJC,BannwarthB,FaildeI,AbelloCarbonellJ,BlotmanF,SpaethM,etal.Prevalenceof fibromyalgia:asurveyinfiveeuropeancountries.seminarthritisrheum.2010jun;39(6): ReumatologíaSEd.EstudioEPISER.Prevalenciaeimpactodelasenfermedadesreumáticasenla poblaciónadultaespañola.in:reumatologíased,editor.madrid; MouraoAF,BlythFM,BrancoJC.Generalisedmusculoskeletalpainsyndromes.BestPractResClin Rheumatol.2010Dec;24(6): WhiteKP,ThompsonJ.FibromyalgiasyndromeinanAmishcommunity:acontrolledstudyto determinediseaseandsymptomprevalence.jrheumatol.2003aug;30(8): NueschE,HauserW,BernardyK,BarthJ,JuniP.Comparativeefficacyofpharmacologicalandnon pharmacologicalinterventionsinfibromyalgiasyndrome:networkmetaanalysis.annrheumdis.2012jun WolfeF,SmytheHA,YunusMB,BennettRM,BombardierC,GoldenbergDL,etal.TheAmerican CollegeofRheumatology1990CriteriafortheClassificationofFibromyalgia.ReportoftheMulticenter CriteriaCommittee.ArthritisRheum.1990Feb;33(2): WolfeF,ClauwDJ,FitzcharlesMA,GoldenbergDL,HauserW,KatzRS,etal.Fibromyalgiacriteriaand severityscalesforclinicalandepidemiologicalstudies:amodificationoftheacrpreliminarydiagnostic CriteriaforFibromyalgia.JRheumatol.2011Jun;38(6): GoldenbergDL,BurckhardtC,CroffordL.Managementoffibromyalgiasyndrome.JAMA.2004Nov 17;292(19): DreyerL,KendallS,DanneskioldSamsoeB,BartelsEM,BliddalH.MortalityinacohortofDanish patientswithfibromyalgia:increasedfrequencyofsuicide.arthritisrheum.2010oct;62(10): MacfarlaneGJ,JonesGT,KnektP,AromaaA,McBethJ,MikkelssonM,etal.Isthereportof widespreadbodypainassociatedwithlongtermincreasedmortality?datafromtheminifinlandhealth Survey.Rheumatology(Oxford).2007May;46(5): McBethJ,SymmonsDP,SilmanAJ,AllisonT,WebbR,BrammahT,etal.Musculoskeletalpainis associatedwithalongtermincreasedriskofcancerandcardiovascularrelatedmortality.rheumatology (Oxford).2009Jan;48(1): BennettRM,JonesJ,TurkDC,RussellIJ,MatallanaL.Aninternetsurveyof2,596peoplewith fibromyalgia.bmcmusculoskeletdisord.2007;8: BernardAL,PrinceA,EdsallP.Qualityoflifeissuesforfibromyalgiapatients.ArthritisCareRes.2000 Feb;13(1):4250.
176 ClauwDJ,ArnoldLM,McCarbergBH.Thescienceoffibromyalgia.MayoClinProc.2011 Sep;86(9): SilvermanS,DukesEM,JohnstonSS,BrandenburgNA,SadoskyA,HuseDM.Theeconomicburdenof fibromyalgia:comparativeanalysiswithrheumatoidarthritis.currmedresopin.2009apr;25(4): SumptonJE,MoulinDE.Fibromyalgia:presentationandmanagementwithafocuson pharmacologicaltreatment.painresmanag.2008novdec;13(6): CastelA,FontovaR,MontullS,PerinanR,PovedaMJ,MirallesI,etal.Theefficacyofa multidisciplinaryfibromyalgiatreatmentadaptedforwomenwithloweducationallevels:arandomised, controlledtrial.arthritiscareres(hoboken).2012aug CarvilleSF,ArendtNielsenS,BliddalH,BlotmanF,BrancoJC,BuskilaD,etal.EULARevidencebased recommendationsforthemanagementoffibromyalgiasyndrome.annrheumdis.2008apr;67(4): ZhouM,ZhouD,HeL.Arandomizedtrialoftaichiforfibromyalgia.NEnglJMed.2010Dec 2;363(23):2265;authorreply KuchinadA,SchweinhardtP,SeminowiczDA,WoodPB,ChizhBA,BushnellMC.Acceleratedbrain graymatterlossinfibromyalgiapatients:prematureagingofthebrain?jneurosci.2007apr 11;27(15): GracelyRH,AmbroseKR.Neuroimagingoffibromyalgia.BestPractResClinRheumatol.2011 Apr;25(2): DadabhoyD,CroffordLJ,SpaethM,RussellIJ,ClauwDJ.Biologyandtherapyoffibromyalgia. Evidencebasedbiomarkersforfibromyalgiasyndrome.ArthritisResTher.2008;10(4): GibsonSJ,LittlejohnGO,GormanMM,HelmeRD,GrangesG.Alteredheatpainthresholdsand cerebraleventrelatedpotentialsfollowingpainfulco2laserstimulationinsubjectswithfibromyalgia syndrome.pain.1994aug;58(2): DiersM,SchleyMT,RanceM,YilmazP,LauerL,RukwiedR,etal.Differentialcentralpainprocessing followingrepetitiveintramuscularproton/prostaglandine(2)injectionsinfemalefibromyalgiapatientsand healthycontrols.eurjpain.2011aug;15(7): WoodPB,PattersonJC,2nd,SunderlandJJ,TainterKH,GlabusMF,LilienDL.Reducedpresynaptic dopamineactivityinfibromyalgiasyndromedemonstratedwithpositronemissiontomography:apilot study.jpain.2007jan;8(1): HarrisRE,ClauwDJ,ScottDJ,McLeanSA,GracelyRH,ZubietaJK.Decreasedcentralmuopioid receptoravailabilityinfibromyalgia.jneurosci.2007sep12;27(37): FayedN,GarciaCampayoJ,MagallonR,AndresBergarecheH,LucianoJV,AndresE,etal.Localized 1HNMRspectroscopyinpatientswithfibromyalgia:acontrolledstudyofchangesincerebral glutamate/glutamine,inositol,choline,andnacetylaspartate.arthritisresther.2010;12(4):r JulienN,GoffauxP,ArsenaultP,MarchandS.Widespreadpaininfibromyalgiaisrelatedtoadeficit ofendogenouspaininhibition.pain.2005mar;114(12): LegangneuxE,MoraJJ,SpreuxVaroquauxO,ThorinI,HerrouM,AlvadoG,etal.Cerebrospinalfluid biogenicaminemetabolites,plasmarichplateletserotoninand[3h]imipraminereuptakeintheprimary fibromyalgiasyndrome.rheumatology(oxford).2001mar;40(3): DiFrancoM,IannuccelliC,ValesiniG.Neuroendocrineimmunologyoffibromyalgia.AnnNYAcad Sci.2010Apr;1193: LarsonAA,GiovengoSL,RussellIJ,MichalekJE.Changesintheconcentrationsofaminoacidsinthe cerebrospinalfluidthatcorrelatewithpaininpatientswithfibromyalgia:implicationsfornitricoxide pathways.pain.2000aug;87(2): SeidelMF,HerguijuelaM,ForkertR,OttenU.Nervegrowthfactorinrheumaticdiseases.Semin ArthritisRheum.2010Oct;40(2): StaudR,NagelS,RobinsonME,PriceDD.Enhancedcentralpainprocessingoffibromyalgiapatients ismaintainedbymuscleafferentinput:arandomized,doubleblind,placebocontrolledstudy.pain.2009 Sep;145(12): MartinezLavinM.Biologyandtherapyoffibromyalgia.Stress,thestressresponsesystem,and fibromyalgia.arthritisresther.2007;9(4):216.
177 MoldofskyH,LueFA.TherelationshipofalphaanddeltaEEGfrequenciestopainandmoodin 'fibrositis'patientstreatedwithchlorpromazineandltryptophan.electroencephalogrclinneurophysiol. 1980Oct;50(12): LentzMJ,LandisCA,RothermelJ,ShaverJL.Effectsofselectiveslowwavesleepdisruptionon musculoskeletalpainandfatigueinmiddleagedwomen.jrheumatol.1999jul;26(7): BennettRM,ClarkSR,CampbellSM,BurckhardtCS.LowlevelsofsomatomedinCinpatientswith thefibromyalgiasyndrome.apossiblelinkbetweensleepandmusclepain.arthritisrheum.1992 Oct;35(10): BartelsEM,DanneskioldSamsoeB.Histologicalabnormalitiesinmusclefrompatientswithcertain typesoffibrositis.lancet.1986apr5;1(8484): DantzerR,O'ConnorJC,FreundGG,JohnsonRW,KelleyKW.Frominflammationtosicknessand depression:whentheimmunesystemsubjugatesthebrain.natrevneurosci.2008jan;9(1): UceylerN,HauserW,SommerC.Systematicreviewwithmetaanalysis:cytokinesinfibromyalgia syndrome.bmcmusculoskeletdisord.2011;12: WalkerEA,KeeganD,GardnerG,SullivanM,BernsteinD,KatonWJ.Psychosocialfactorsin fibromyalgiacomparedwithrheumatoidarthritis:ii.sexual,physical,andemotionalabuseandneglect. PsychosomMed.1997NovDec;59(6): AlAllafAW,DunbarKL,HallumNS,NosratzadehB,TempletonKD,PullarT.Acasecontrolstudy examiningtheroleofphysicaltraumaintheonsetoffibromyalgiasyndrome.rheumatology(oxford).2002 Apr;41(4): AthertonK,BerryDJ,ParsonsT,MacfarlaneGJ,PowerC,HypponenE.VitaminDandchronic widespreadpaininawhitemiddleagedbritishpopulation:evidencefromacrosssectionalpopulation survey.annrheumdis.2009jun;68(6): BuskilaD,AtzeniF,SarziPuttiniP.Etiologyoffibromyalgia:thepossibleroleofinfectionand vaccination.autoimmunrev.2008oct;8(1): AblinJ,NeumannL,BuskilaD.Pathogenesisoffibromyalgiaareview.JointBoneSpine.2008 May;75(3): UnwinC,BlatchleyN,CokerW,FerryS,HotopfM,HullL,etal.HealthofUKservicemenwhoserved inpersiangulfwar.lancet.1999jan16;353(9148): HotopfM,DavidA,HullL,IsmailK,UnwinC,WesselyS.Roleofvaccinationsasriskfactorsforill healthinveteransofthegulfwar:crosssectionalstudy.bmj.2000may20;320(7246): NorburyTA,MacGregorAJ,UrwinJ,SpectorTD,McMahonSB.Heritabilityofresponsestopainful stimuliinwomen:aclassicaltwinstudy.brain.2007nov;130(pt11): MogilJS.Paingenetics:past,presentandfuture.TrendsGenet.2012Jun;28(6): KimH,RamsayE,LeeH,WahlS,DionneRA.Genomewideassociationstudyofacutepostsurgical paininhumans.pharmacogenomics.2009feb;10(2): SmithSB,MaixnerDW,FillingimRB,SladeG,GracelyRH,AmbroseK,etal.Largecandidategene associationstudyrevealsgeneticriskfactorsandtherapeutictargetsforfibromyalgia.arthritisrheum.2012 Feb;64(2): PellegrinoMJ,WaylonisGW,SommerA.Familialoccurrenceofprimaryfibromyalgia.ArchPhysMed Rehabil.1989Jan;70(1): BuskilaD,NeumannL,HazanovI,CarmiR.Familialaggregationinthefibromyalgiasyndrome.Semin ArthritisRheum.1996Dec;26(3): BuskilaD,NeumannL.Fibromyalgiasyndrome(FM)andnonarticulartendernessinrelativesof patientswithfm.jrheumatol.1997may;24(5): ArnoldLM,HudsonJI,HessEV,WareAE,FritzDA,AuchenbachMB,etal.Familystudyof fibromyalgia.arthritisrheum.2004mar;50(3): MikkelssonM,KaprioJ,SalminenJJ,PulkkinenL,RoseRJ.Widespreadpainamong11yearold Finnishtwinpairs.ArthritisRheum.2001Feb;44(2): KatoK,SullivanPF,EvengardB,PedersenNL.Importanceofgeneticinfluencesonchronic widespreadpain.arthritisrheum.2006may;54(5):16826.
178 BurdaCD,CoxFR,OsborneP.Histocompatabilityantigensinthefibrositis(fibromyalgia)syndrome. ClinExpRheumatol.1986OctDec;4(4): HorvenS,StilesTC,HolstA,MoenT.HLAantigensinprimaryfibromyalgiasyndrome.JRheumatol. 1992Aug;19(8): OffenbaecherM,BondyB,deJongeS,GlatzederK,KrugerM,SchoepsP,etal.Possibleassociation offibromyalgiawithapolymorphismintheserotonintransportergeneregulatoryregion.arthritisrheum. 1999Nov;42(11): CohenH,BuskilaD,NeumannL,EbsteinRP.Confirmationofanassociationbetweenfibromyalgia andserotonintransporterpromoterregion(5httlpr)polymorphism,andrelationshiptoanxietyrelated personalitytraits.arthritisrheum.2002mar;46(3): GursoyS.Absenceofassociationoftheserotonintransportergenepolymorphismwiththementally healthysubsetoffibromyalgiapatients.clinrheumatol.2002jun;21(3): BondyB,SpaethM,OffenbaecherM,GlatzederK,StratzT,SchwarzM,etal.TheT102C polymorphismofthe5ht2areceptorgeneinfibromyalgia.neurobioldis.1999oct;6(5): GursoyS,ErdalE,HerkenH,MadenciE,AlasehirliB.AssociationofT102Cpolymorphismofthe5 HT2Areceptorgenewithpsychiatricstatusinfibromyalgiasyndrome.RheumatolInt.2001Oct;21(2): ZubietaJK,HeitzegMM,SmithYR,BuellerJA,XuK,XuY,etal.COMTval158metgenotypeaffects muopioidneurotransmitterresponsestoapainstressor.science.2003feb21;299(5610): VargasAlarconG,FragosoJM,CruzRoblesD,VargasA,LaoVilladonigaJI,GarciaFructuosoF,etal. CatecholOmethyltransferasegenehaplotypesinMexicanandSpanishpatientswithfibromyalgia.Arthritis ResTher.2007;9(5):R TanderB,GunesS,BokeO,AlayliG,KaraN,BagciH,etal.Polymorphismsoftheserotonin2A receptorandcatecholomethyltransferasegenes:astudyonfibromyalgiasusceptibility.rheumatolint. 2008May;28(7): LeeYH,ChoiSJ,JiJD,SongGG.Candidategenestudiesoffibromyalgia:asystematicreviewand metaanalysis.rheumatolint.2012feb;32(2): MartinezJauandM,SitgesC,RodriguezV,PicornellA,RamonM,BuskilaD,etal.Painsensitivityin fibromyalgiaisassociatedwithcatecholomethyltransferase(comt)gene.eurjpain.2012apr VargasAlarconG,FragosoJM,CruzRoblesD,VargasA,MartinezA,LaoVilladonigaJI,etal. Associationofadrenergicreceptorgenepolymorphismswithdifferentfibromyalgiasyndromedomains. ArthritisRheum.2009Jul;60(7): PotvinS,LaroucheA,NormandE,deSouzaJB,GaumondI,MarchandS,etal.Norelationship betweentheinsdelpolymorphismoftheserotonintransporterpromoterandpainperceptionin fibromyalgiapatientsandhealthycontrols.eurjpain.2010aug;14(7): GursoyS,ErdalE,HerkenH,MadenciE,AlasehirliB,ErdalN.SignificanceofcatecholO methyltransferasegenepolymorphisminfibromyalgiasyndrome.rheumatolint.2003may;23(3): AblinJN,BarShiraA,YaronM,OrrUrtregerA.Candidategeneapproachinfibromyalgiasyndrome: associationanalysisofthegenesencodingsubstancepreceptor,dopaminetransporterandalpha1 antitrypsin.clinexprheumatol.2009sepoct;27(5suppl56):s BuskilaD,CohenH,NeumannL,EbsteinRP.Anassociationbetweenfibromyalgiaandthedopamine D4receptorexonIIIrepeatpolymorphismandrelationshiptonoveltyseekingpersonalitytraits.Mol Psychiatry.2004Aug;9(8): AlasehirliB,DemiryurekS,AricaE,GursoyS,DemiryurekAT.Noevidenceforanassociationbetween theglu298asppolymorphismoftheendothelialnitricoxidesynthasegeneandfibromyalgiasyndrome. RheumatolInt.2007Jan;27(3): FrankB,NieslerB,BondyB,SpathM,PongratzDE,AckenheilM,etal.Mutationalanalysisof serotoninreceptorgenes:htr3aandhtr3binfibromyalgiapatients.clinrheumatol.2004aug;23(4): SuSY,ChenJJ,LaiCC,ChenCM,TsaiFJ.Theassociationbetweenfibromyalgiaandpolymorphismof monoamineoxidaseaandinterleukin4.clinrheumatol.2007jan;26(1):126.
179 GursoyS,ErdalE,SezginM,BarlasIO,AydenizA,AlasehirliB,etal.WhichgenotypeofMAOgene thatthepatientshavearelikelytobemostsusceptibletothesymptomsoffibromyalgia?rheumatolint. 2008Feb;28(4): VargasAlarconG,AlvarezLeonE,FragosoJM,VargasA,MartinezA,VallejoM,etal.ASCN9Agene encodeddorsalrootgangliasodiumchannelpolymorphismassociatedwithseverefibromyalgia.bmc MusculoskeletDisord.2012;13: LanderES,LintonLM,BirrenB,NusbaumC,ZodyMC,BaldwinJ,etal.Initialsequencingandanalysis ofthehumangenome.nature.2001feb15;409(6822): VenterJC,AdamsMD,MyersEW,LiPW,MuralRJ,SuttonGG,etal.Thesequenceofthehuman genome.science.2001feb16;291(5507): ConsortiumTGP.Amapofhumangenomevariationfrompopulationscalesequencing.Nature.2010 Oct28;467(7319): CortesA,BrownMA.PromiseandpitfallsoftheImmunochip.ArthritisResTher.2011;13(1): Hapmap.TheInternationalHapMapProject.Nature.2003Dec18;426(6968): ToblerAR,ShortS,AndersenMR,PanerTM,BriggsJC,LambertSM,etal.TheSNPlexgenotyping system:aflexibleandscalableplatformforsnpgenotyping.jbiomoltech.2005dec;16(4): KleinRJ,ZeissC,ChewEY,TsaiJY,SacklerRS,HaynesC,etal.ComplementfactorHpolymorphismin agerelatedmaculardegeneration.science.2005apr15;308(5720): AmosCI.Successfuldesignandconductofgenomewideassociationstudies.HumMolGenet.2007 Oct15;16SpecNo.2:R MarchiniJ,HowieB.Genotypeimputationforgenomewideassociationstudies.NatRevGenet. 2010Jul;11(7): ConsortiumTWTCC.Genomewideassociationstudyof14,000casesofsevencommondiseasesand 3,000sharedcontrols.Nature.2007Jun7;447(7145): StrattonMR,CampbellPJ,FutrealPA.Thecancergenome.Nature.2009Apr9;458(7239): TuzunE,SharpAJ,BaileyJA,KaulR,MorrisonVA,PertzLM,etal.Finescalestructuralvariationof thehumangenome.natgenet.2005jul;37(7): ConradDF,PintoD,RedonR,FeukL,GokcumenO,ZhangY,etal.Originsandfunctionalimpactof copynumbervariationinthehumangenome.nature.2010apr1;464(7289): RedonR,IshikawaS,FitchKR,FeukL,PerryGH,AndrewsTD,etal.Globalvariationincopynumber inthehumangenome.nature.2006nov23;444(7118): EstivillX,ArmengolL.Copynumbervariantsandcommondisorders:fillingthegapsandexploring complexityingenomewideassociationstudies.plosgenet.2007oct;3(10): ZhangF,KhajaviM,ConnollyAM,TowneCF,BatishSD,LupskiJR.TheDNAreplication FoSTeS/MMBIRmechanismcangenerategenomic,genicandexoniccomplexrearrangementsinhumans. NatGenet.2009Jul;41(7): MalhotraD,SebatJ.CNVs:harbingersofararevariantrevolutioninpsychiatricgenetics.Cell.2012 Mar16;148(6): LupskiJR,deOcaLunaRM,SlaugenhauptS,PentaoL,GuzzettaV,TraskBJ,etal.DNAduplication associatedwithcharcotmarietoothdiseasetype1a.cell.1991jul26;66(2): GonzalezE,KulkarniH,BolivarH,ManganoA,SanchezR,CatanoG,etal.TheinfluenceofCCL3L1 genecontainingsegmentalduplicationsonhiv1/aidssusceptibility.science.2005mar4;307(5714): FellermannK,StangeDE,SchaeffelerE,SchmalzlH,WehkampJ,BevinsCL,etal.Achromosome8 geneclusterpolymorphismwithlowhumanbetadefensin2genecopynumberpredisposestocrohn diseaseofthecolon.amjhumgenet.2006sep;79(3): YangY,ChungEK,WuYL,SavelliSL,NagarajaHN,ZhouB,etal.Genecopynumbervariationand associatedpolymorphismsofcomplementcomponentc4inhumansystemiclupuserythematosus(sle):low copynumberisariskfactorforandhighcopynumberisaprotectivefactoragainstslesusceptibilityin EuropeanAmericans.AmJHumGenet.2007Jun;80(6):
180 decidr,riveiramunoze,zeeuwenpl,robargej,liaow,dannhauseren,etal.deletionofthelate cornifiedenvelopelce3bandlce3cgenesasasusceptibilityfactorforpsoriasis.natgenet.2009 Feb;41(2): CraddockN,HurlesME,CardinN,PearsonRD,PlagnolV,RobsonS,etal.Genomewideassociation studyofcnvsin16,000casesofeightcommondiseasesand3,000sharedcontrols.nature.2010apr 1;464(7289): LupskiJR,StankiewiczP.Genomicdisorders:molecularmechanismsforrearrangementsand conveyedphenotypes.plosgenet.2005dec;1(6):e KorbelJO,UrbanAE,AffourtitJP,GodwinB,GrubertF,SimonsJF,etal.Pairedendmappingreveals extensivestructuralvariationinthehumangenome.science.2007oct19;318(5849): IafrateAJ,FeukL,RiveraMN,ListewnikML,DonahoePK,QiY,etal.Detectionoflargescale variationinthehumangenome.natgenet.2004sep;36(9): JuYS,HongD,KimS,ParkSS,LeeS,ParkH,etal.Referenceunbiasedcopynumbervariantanalysis usingcghmicroarrays.nucleicacidsres.2010nov;38(20):e WangK,LiM,HadleyD,LiuR,GlessnerJ,GrantSF,etal.PennCNV:anintegratedhiddenMarkov modeldesignedforhighresolutioncopynumbervariationdetectioninwholegenomesnpgenotypingdata. GenomeRes.2007Nov;17(11): WinchesterL,YauC,RagoussisJ.ComparingCNVdetectionmethodsforSNParrays.BriefFunct GenomicProteomic.2009Sep;8(5): BerkelS,MarshallCR,WeissB,HoweJ,RoethR,MoogU,etal.MutationsintheSHANK2synaptic scaffoldinggeneinautismspectrumdisorderandmentalretardation.natgenet.2010jun;42(6): KoboldtDC,LarsonDE,ChenK,DingL,WilsonRK.Massivelyparallelsequencingapproachesfor characterizationofstructuralvariation.methodsmolbiol.2012;838: SchererSW,LeeC,BirneyE,AltshulerDM,EichlerEE,CarterNP,etal.Challengesandstandardsin integratingsurveysofstructuralvariation.natgenet.2007jul;39(7suppl):s FlorijnRJ,BondenLA,VrolijkH,WiegantJ,VaandragerJW,BaasF,etal.HighresolutionDNAFiber FISHforgenomicDNAmappingandcolourbarcodingoflargegenes.HumMolGenet.1995May;4(5): SchoutenJP,McElgunnCJ,WaaijerR,ZwijnenburgD,DiepvensF,PalsG.Relativequantificationof40 nucleicacidsequencesbymultiplexligationdependentprobeamplification.nucleicacidsres.2002jun 15;30(12):e FloresC,MacaMeyerN,GonzalezAM,OefnerPJ,ShenP,PerezJA,etal.Reducedgeneticstructure oftheiberianpeninsularevealedbyychromosomeanalysis:implicationsforpopulationdemography.eurj HumGenet.2004Oct;12(10): LaayouniH,CalafellF,BertranpetitJ.Agenomewidesurveydoesnotshowthegenetic distinctivenessofbasques.humgenet.2010apr;127(4): PinoYanesM,CorralesA,BasalduaS,HernandezA,GuerraL,VillarJ,etal.NorthAfricaninfluences andpotentialbiasincasecontrolassociationstudiesinthespanishpopulation.plosone.2011;6(3):e JuliaA,BallinaJ,CaneteJD,BalsaA,TorneroMolinaJ,NaranjoA,etal.Genomewideassociation studyofrheumatoidarthritisinthespanishpopulation:klf12asarisklocusforrheumatoidarthritis susceptibility.arthritisrheum.2008aug;58(8): AktasA,WalshD,RybickiL.Symptomclusters:mythorreality?PalliatMed.2010Jun;24(4): desouzajb,goffauxp,julienn,potvins,charestj,marchands.fibromyalgiasubgroups:profiling distinctsubgroupsusingthefibromyalgiaimpactquestionnaire.apreliminarystudy.rheumatolint.2009 Mar;29(5): HurtigIM,RaakRI,KendallSA,GerdleB,WahrenLK.Quantitativesensorytestinginfibromyalgia patientsandinhealthysubjects:identificationofsubgroups.clinjpain.2001dec;17(4): GieseckeT,WilliamsDA,HarrisRE,CuppsTR,TianX,TianTX,etal.Subgroupingoffibromyalgia patientsonthebasisofpressurepainthresholdsandpsychologicalfactors.arthritisrheum.2003 Oct;48(10): LoevingerBL,ShirtcliffEA,MullerD,AlonsoC,CoeCL.Delineatingpsychologicalandbiomedical profilesinaheterogeneousfibromyalgiapopulationusingclusteranalysis.clinrheumatol.2012dec27.
181 BennettRM,RussellJ,CappelleriJC,BushmakinAG,ZlatevaG,SadoskyA.Identificationofsymptom andfunctionaldomainsthatfibromyalgiapatientswouldliketoseeimproved:aclusteranalysis.bmc MusculoskeletDisord.2010;11: CuestaVargasA,LucianoJV,PenarrubiaMariaMT,GarciaCampayoJ,FernandezVergelR,Arroyo MoralesM,etal.Clinicaldimensionsoffibromyalgiasymptomsanddevelopmentofacombinedindexof severity:thecodiindex.qualliferes.2012feb RutledgeDN,MouttapaM,WoodPB.Symptomclustersinfibromyalgia:potentialutilityinpatient assessmentandtreatmentevaluation.nursres.2009sepoct;58(5): WilsonHD,RobinsonJP,TurkDC.Towardtheidentificationofsymptompatternsinpeoplewith fibromyalgia.arthritisrheum.2009apr15;61(4): OkifujiA,TurkDC,SinclairJD,StarzTW,MarcusDA.Astandardizedmanualtenderpointsurvey.I. Developmentanddeterminationofathresholdpointfortheidentificationofpositivetenderpointsin fibromyalgiasyndrome.jrheumatol.1997feb;24(2): ZigmondAS,SnaithRP.Thehospitalanxietyanddepressionscale.ActaPsychiatrScand.1983 Jun;67(6): HerreroMJ,BlanchJ,PeriJM,DePabloJ,PintorL,BulbenaA.Avalidationstudyofthehospital anxietyanddepressionscale(hads)inaspanishpopulation.genhosppsychiatry.2003julaug;25(4): BuysseDJ,ReynoldsCF,3rd,MonkTH,BermanSR,KupferDJ.ThePittsburghSleepQualityIndex:a newinstrumentforpsychiatricpracticeandresearch.psychiatryres.1989may;28(2): BurckhardtCS,ClarkSR,BennettRM.Thefibromyalgiaimpactquestionnaire:developmentand validation.jrheumatol.1991may;18(5): KruppLB,LaRoccaNG,MuirNashJ,SteinbergAD.Thefatigueseverityscale.Applicationtopatients withmultiplesclerosisandsystemiclupuserythematosus.archneurol.1989oct;46(10): BenitoLeonJ,MartinezMartinP,FradesB,MartinezGinesML,deAndresC,MecaLallanaJE,etal. Impactoffatigueinmultiplesclerosis:theFatigueImpactScaleforDailyUse(DFIS).MultScler.2007 Jun;13(5): WareJE,Jr.,SherbourneCD.TheMOS36itemshortformhealthsurvey(SF36).I.Conceptual frameworkanditemselection.medcare.1992jun;30(6): AlonsoJ,PrietoL,AntoJM.[TheSpanishversionoftheSF36HealthSurvey(theSF36health questionnaire):aninstrumentformeasuringclinicalresults].medclin(barc).1995may27;104(20): FukudaK,StrausSE,HickieI,SharpeMC,DobbinsJG,KomaroffA.Thechronicfatiguesyndrome:a comprehensiveapproachtoitsdefinitionandstudy.internationalchronicfatiguesyndromestudygroup. AnnInternMed.1994Dec15;121(12): FraleyCR,A.E.Howmanyclusters?Whichclusteringmethod?.TheComputerJournal. 1998;41(8): Gower.Ageneralcoefficientofsimilarityandsomeofitsproperties.Biometrics.1971;27: HartiganJAWMA.AlgorithmAS136:aKmeansclusteringalgorithm.ApplStat.1978;28: PurcellS,NealeB,ToddBrownK,ThomasL,FerreiraMA,BenderD,etal.PLINK:atoolsetforwhole genomeassociationandpopulationbasedlinkageanalyses.amjhumgenet.2007sep;81(3): GeD,ZhangK,NeedAC,MartinO,FellayJ,UrbanTJ,etal.WGAViewer:softwareforgenomic annotationofwholegenomeassociationstudies.genomeres.2008apr;18(4): BarrettJC,FryB,MallerJ,DalyMJ.Haploview:analysisandvisualizationofLDandhaplotypemaps. Bioinformatics.2005Jan15;21(2): StrangerBE,MontgomerySB,DimasAS,PartsL,StegleO,IngleCE,etal.Patternsofcisregulatory variationindiversehumanpopulations.plosgenet.2012;8(4):e NicaAC,PartsL,GlassD,NisbetJ,BarrettA,SekowskaM,etal.Thearchitectureofgeneregulatory variationacrossmultiplehumantissues:themutherstudy.plosgenet.2011;7(2):e DimasAS,DeutschS,StrangerBE,MontgomerySB,BorelC,AttarCohenH,etal.Commonregulatory variationimpactsgeneexpressioninacelltypedependentmanner.science.2009sep4;325(5945):
182 BoyleAP,HongEL,HariharanM,ChengY,SchaubMA,KasowskiM,etal.Annotationoffunctional variationinpersonalgenomesusingregulomedb.genomeres.2012sep;22(9): ZhangD,QianY,AkulaN,AllieyRodriguezN,TangJ,GershonES,etal.AccuracyofCNVDetection fromgwasdata.plosone.2011;6(1):e GonzalezJR,RodriguezSantiagoB,CaceresA,PiqueRegiR,RothmanN,ChanockSJ,etal.Afastand accuratemethodtodetectallelicgenomicimbalancesunderlyingmosaicrearrangementsusingsnparray data.bmcbioinformatics.2011;12: BierutLJ.Geneticvulnerabilityandsusceptibilitytosubstancedependence.Neuron.2011Feb 24;69(4): OcchiG,RampazzoA,BeffagnaG,AntonioDanieliG.Identificationandcharacterizationofheart specificsplicingofhumanneurexin3mrna(nrxn3).biochembiophysrescommun.2002oct 18;298(1): PfafflMW.AnewmathematicalmodelforrelativequantificationinrealtimeRTPCR.NucleicAcids Res.2001May1;29(9):e LaddAN,CooperTA.Findingsignalsthatregulatealternativesplicinginthepostgenomicera. GenomeBiol.2002Oct23;3(11):reviews ParkH,KimJI,JuYS,GokcumenO,MillsRE,KimS,etal.DiscoveryofcommonAsiancopynumber variantsusingintegratedhighresolutionarraycghandmassivelyparalleldnasequencing.natgenet.2010 May;42(5): ResnickM,SegallA,GGR,LupowitzZ,ZisapelN.Alternativesplicingofneurexins:aroleforneuronal polypyrimidinetractbindingprotein.neuroscilett.2008jul18;439(3): KashikarZuckS,LynchAM,SlaterS,GrahamTB,SwainNF,NollRB.Familyfactors,emotional functioning,andfunctionalimpairmentinjuvenilefibromyalgiasyndrome.arthritisrheum.2008oct 15;59(10): KindlerLL,JonesKD,PerrinN,BennettRM.Riskfactorspredictingthedevelopmentofwidespread painfromchronicbackorneckpain.jpain.2010dec;11(12): GastonJohanssonF,FallDicksonJM,BakosAB,KennedyMJ.Fatigue,pain,anddepressioninpre autotransplantbreastcancerpatients.cancerpract.1999sepoct;7(5): RehmSE,KoroschetzJ,GockelU,BroszM,FreynhagenR,TolleTR,etal.Acrosssectionalsurveyof 3035patientswithfibromyalgia:subgroupsofpatientswithtypicalcomorbiditiesandsensorysymptom profiles.rheumatology(oxford).2010jun;49(6): PitonA,GauthierJ,HamdanFF,LafreniereRG,YangY,HenrionE,etal.Systematicresequencingof Xchromosomesynapticgenesinautismspectrumdisorderandschizophrenia.MolPsychiatry.2011 Aug;16(8): SchulzeTG,DeteraWadleighSD,AkulaN,GuptaA,KassemL,SteeleJ,etal.TwovariantsinAnkyrin 3(ANK3)areindependentgeneticriskfactorsforbipolardisorder.MolPsychiatry.2009May;14(5): BiC,WuJ,JiangT,LiuQ,CaiW,YuP,etal.MutationsofANK3identifiedbyexomesequencingare associatedwithautismsusceptibility.hummutat.2012dec;33(12): RommE,NielsenJA,KimJG,HudsonLD.Myt1familyrecruitshistonedeacetylasetoregulateneural transcription.jneurochem.2005jun;93(6): KimJG,ArmstrongRC,vAgostonD,RobinskyA,WieseC,NagleJ,etal.Myelintranscriptionfactor1 (Myt1)oftheoligodendrocytelineage,alongwithacloselyrelatedCCHCzincfinger,isexpressedin developingneuronsinthemammaliancentralnervoussystem.jneuroscires.1997oct15;50(2): YooAS,SunAX,LiL,ShcheglovitovA,PortmannT,LiY,etal.MicroRNAmediatedconversionof humanfibroblaststoneurons.nature.2011aug11;476(7359): VierbuchenT,OstermeierA,PangZP,KokubuY,SudhofTC,WernigM.Directconversionof fibroblaststofunctionalneuronsbydefinedfactors.nature.2010feb25;463(7284): PfistererU,KirkebyA,TorperO,WoodJ,NelanderJ,DufourA,etal.Directconversionofhuman fibroblaststodopaminergicneurons.procnatlacadsciusa.2011jun21;108(25):
183 VrijenhoekT,BuizerVoskampJE,vanderSteltI,StrengmanE,SabattiC,GeurtsvanKesselA,etal. RecurrentCNVsdisruptthreecandidategenesinschizophreniapatients.AmJHumGenet.2008 Oct;83(4): LiW,WangX,ZhaoJ,LinJ,SongXQ,YangY,etal.Associationstudyofmyelintranscriptionfactor1 likepolymorphismswithschizophreniainhanchinesepopulation.genesbrainbehav.2012feb;11(1): MeyerKJ,AxelsenMS,SheffieldVC,PatilSR,WassinkTH.Germlinemosaictransmissionofanovel duplicationofpxdnandmyt1ltotwomalehalfsiblingswithautism.psychiatrgenet.2012jun;22(3): WangT,ZengZ,LiT,LiuJ,LiJ,LiY,etal.CommonSNPsinmyelintranscriptionfactor1like(MYT1L): associationwithmajordepressivedisorderinthechinesehanpopulation.plosone.2010;5(10):e YamakawaH,OyamaS,MitsuhashiH,SasagawaN,UchinoS,KohsakaS,etal.Neuroligins3and4X interactwithsyntrophingamma2,andtheinteractionsareaffectedbyautismrelatedmutations.biochem BiophysResCommun.2007Mar30;355(1): PerlisRH,RuderferD,MaussionG,ChambertK,GallagherP,TureckiG,etal.BipolarDisorderanda HistoryofSuicideAttemptsWithaDuplicationin5HTR1A.AmJPsychiatry.2012Nov1;169(11): HainesJL,HauserMA,SchmidtS,ScottWK,OlsonLM,GallinsP,etal.ComplementfactorHvariant increasestheriskofagerelatedmaculardegeneration.science.2005apr15;308(5720): PurcellSM,WrayNR,StoneJL,VisscherPM,O'DonovanMC,SullivanPF,etal.Commonpolygenic variationcontributestoriskofschizophreniaandbipolardisorder.nature.2009aug6;460(7256): AparicioVA,OrtegaFB,CarbonellBaezaA,FemiaP,TercedorP,RuizJR,etal.Aretheregender differencesinqualityoflifeandsymptomatologybetweenfibromyalgiapatients?amjmenshealth.2012 Jul;6(4): CastroSanchezAM,MataranPenarrochaGA,LopezRodriguezMM,LaraPalomoIC,ArendtNielsen L,FernandezdeLasPenasC.GenderDifferencesinPainSeverity,Disability,Depression,andWidespread PressurePainSensitivityinPatientswithFibromyalgiaSyndromeWithoutComorbidConditions.PainMed. 2012Nov MogilJS.Sexdifferencesinpainandpaininhibition:multipleexplanationsofacontroversial phenomenon.natrevneurosci.2012nov;13(12): ShulmanJM,ChipendoP,ChibnikLB,AubinC,TranD,KeenanBT,etal.Functionalscreeningof AlzheimerpathologygenomewideassociationsignalsinDrosophila.AmJHumGenet.2011Feb 11;88(2): WangW,YuJT,ZhangW,CuiWZ,WuZC,ZhangQ,etal.GeneticassociationofSLC2A14 polymorphismwithalzheimer'sdiseaseinahanchinesepopulation.jmolneurosci.2012jul;47(3): FrankeB,VasquezAA,JohanssonS,HoogmanM,RomanosJ,BoreattiHummerA,etal.Multicenter analysisoftheslc6a3/dat1vntrhaplotypeinpersistentadhdsuggestsdifferentialinvolvementofthe geneinchildhoodandpersistentadhd.neuropsychopharmacology.2010feb;35(3): MinogueBM,RichardsonSM,ZeefLA,FreemontAJ,HoylandJA.Characterizationofthehuman nucleuspulposuscellphenotypeandevaluationofnovelmarkergeneexpressiontodefineadultstemcell differentiation.arthritisrheum.2010dec;62(12): LewandowskaU,ZelazowskiM,SetaK,ByczewskaM,PluciennikE,BednarekAK.WWOX,thetumour suppressorgeneaffectedinmultiplecancers.jphysiolpharmacol.2009may;60suppl1: SzeCI,SuM,PugazhenthiS,JambalP,HsuLJ,HeathJ,etal.DownregulationofWWdomain containingoxidoreductaseinducestauphosphorylationinvitro.apotentialroleinalzheimer'sdisease.j BiolChem.2004Jul16;279(29): LiMY,LaiFJ,HsuLJ,LoCP,ChengCL,LinSR,etal.DramaticcoactivationofWWOX/WOX1with CREBandNFkappaBindelayedlossofsmalldorsalrootganglionneuronsuponsciaticnervetransectionin rats.plosone.2009;4(11):e TakahashiH,KatayamaK,SohyaK,MiyamotoH,PrasadT,MatsumotoY,etal.Selectivecontrolof inhibitorysynapsedevelopmentbyslitrk3ptpdeltatranssynapticinteraction.natneurosci.2012 Mar;15(3):38998,S12.
184 240. LiseMF,WongTP,TrinhA,HinesRM,LiuL,KangR,etal.InvolvementofmyosinVbinglutamate receptortrafficking.jbiolchem.2006feb10;281(6): SklarP,SmollerJW,FanJ,FerreiraMA,PerlisRH,ChambertK,etal.Wholegenomeassociation studyofbipolardisorder.molpsychiatry.2008jun;13(6): BoucardAA,ChubykinAA,ComolettiD,TaylorP,SudhofTC.Asplicecodefortranssynapticcell adhesionmediatedbybindingofneuroligin1toalphaandbetaneurexins.neuron.2005oct20;48(2): ÜçeylerN,AnnKathrinKahn,DanielZeller,SusanneKewenig,SarahKittelSchneider,Department ofpsychiatry,universityofkarlheinzreiners,claudiasommer.,editor.functionalandmorphological impairmentofsmallnervefibersinfibromaylgiasyndrome The14thinternationalassociationforthestudyofpain(IASP)worldcongressonpain;2012;Milan DepartmentofNeurology,UniversityofWürzburg SaliganL,MajorsB.,LukkahataiN.,HyungSukK.,Walitt,B.,editor.Geneexpressiondifferences betweenfibromyalgiaandpainfreecontrols 14thinternationalassociationforthestudyofpain(IASP)worldcongressonpain;2012;Milan. 178
185 ABBREVIATIONS
186
187 5HT 5hydroxytryptamine(serotonin) acgh Arraycomparativegenomichybridization ACR AmericanCollegeofRheumatology ACTH Adrenocorticotrophichormone ANA Antinuclearantibodies BAC Bacterialartificialchromosome BAF BalleleFrequency CBT CognitiveBehavioralTherapy CI ConfidenceInterval CFS ChronicFatigueSyndrome CNS CentralNervousSystem CNV CopyNumberVariant CSF CerebrospinalFluid CWP ChronicWidespreadPain DA Dopamine EULAR EuropeanLeagueAgainstRheumatism FIQ FibromyalgiaImpactQuestionnaire eqtl Expressionquantitativetraitloci FISH Fluorescenceinsituhybridization fmri FunctionalMagneticResonanceImaging FM Fibromyalgia FoSTeS GABA Forkstallingandtemplateswitching aminobutyricacid GO GeneOntology GWAS GenomeWideAssociationStudy HPA HypothalamicPituitaryAdrenalaxis HWE HardyWeinbergEquilibrium 181
188 IBD Identitybydescent IBS Identitybystate IL Interleukin INDEL InsertionDeletion IPA IngenuitySystemsPathwayanalysis Kb Kilobase L1 LINE1 LD Linkagedisequilibrium LB Lysogenybroth LNS LamininLikedomain LOH Lossofheterozygosity LRR LogRratio:loggedratioofobservedprobeintensitytoexpectedintensity LRT LikelihoodRatioTest MAD MosaicAlterationAlgorithm MAF MinimumAlleleFrequency MLPA MMBIR MultiplexLigationProbeAmplification Microhomologymediatedbreakinducedreplication MRS MagneticResonanceSpectroscopy NAHR Nonallelichomologuousrecombination NE Noradrenalin Ng Nanogram NHEJ Nonhomologousendjoining NGS NextGenerationSequencing NMDA NRXN NSAIDs NMethylDaspartate Neurexin Nonsteroidalantiinflammatorydrugs ORF Openreadingframe 182
189 PCR PolymeraseChainreaction PET PositronEmissionTomography PI PrincipalInvestigator PTBP2 Polypirimidinetractbindingprotein2 QC Qualitycontrol RPM Revolutionsperminute RT ReverseTranscription RTqPCR RealTimeQuantitativePolymeraseChainReaction SNP SingleNucleotidePolymorphism SNRIs SPECT SerotoninNoradrenalineReuptakeInhibitors SinglePhotonEmissionComputedTomography SSRIs SerotoninReuptakeInhibitors TCA TriciclicAntidepressants TF Transcriptionfactor TNF Tumoralnecrosisfactor UPD UniparentalDisomy WTCCC WelcometrustCaseControlConsortium 183
190
191 ANNEXES
192
193 SUPPLEMENTARYINFORMATION Supplementarytable1:Multinomialanalysisofrs acrossFMclusters.Interactionpvalue=0.40 Cl3 Cl2 OR(95%CI) Cl1 OR(95%CI) (N%) (N%) (N%) AA AG ( ) ( ) GG ( ) ( ) Trend ( ) ( ) PvalueTrend Supplementarytable2:Multinomialanalysisofrs acrossFMclusters.Interactionpvalue=0.39 Cl3 Cl2 OR(95%CI) Cl1 OR(95%CI) (N%) (N%) (N%) AA AG ( ) ( ) GG ( ) ( ) Trend ( ) ( ) PvalueTrend Supplementarytable3:Multinomialanalysisofrs acrossFMclusters.Interactionpvalue=0.193 Cl3 Cl2 OR(95%CI) Cl1 OR(95%CI) (N%) (N%) (N%) CC CT ( ) ( ) TT ( ) ( ) Trend ( ) (0.51.1) PvalueTrend Supplementarytable4:Multinomialanalysisofrs acrossFMclusters.Interactionpvalue=0.43 Cl3 Cl2 OR(95%CI) Cl1 OR(95%CI) (N%) (N%) (N%) AA AG ( ) (0.13.8) GG ( ) ( ) Trend ( ) ( ) PvalueTrend
194 Supplementarytable5:aCGHresultscorrespondingtotheGNG1regiondetectedbyPennCNValgorithm PROBE CHR START END GENE FMvsC FMvsC_DS FM_FCvsC FM_FCvsC_DS FM_EARLYvsC FM_EARLYvsC_DS A_16_P chr GNG A_16_P chr GNG A_16_P chr GNG A_16_P chr GNG Supplementarytable6:RarelargeCNVeventsdetectedbyPennCNVinFMsamples Chr SNP_CNV_posini SNP_CNV_posend nsnp Size Sample chr sample.0034_01 chr sample.0064_04 chr sample.0027_02 chr sample.0122_01 chr sample.0029_04 chr sample.0062_05 chr sample.0122_01 chr sample.0005_04 chr sample.0515_01 chr sample.0509_01 chr sample.0118_01 chr sample.0515_01 chr sample.0398_01 chr sample.0051_04 chr sample.0509_01 chr sample.0518_01 chr sample.0003_04 chr sample.0398_01 chr sample.0082_02 chr sample.0082_02 chr sample.0067_05 chr sample.0051_04 chr sample.0476_01 chr sample.0008_04 chr sample.0118_01 chr sample.0046_01 188
195 Supplementarytables711:Singleprobe400kCNVaCGHresults,asdetectedbyADM2algorithm(except MYO5BdetectedbyADM1).GenomiclocationisbasedonbuildHg18.DS:Dyeswap Supplementarytable7:Singleprobe400kCNVaCGHresultsinGALNTL6region PROBE CHR START END GENE FMvsC FMvsC_DS FM_FCvsC FM_FCvsC_DS FM_EARLYvsC FM_EARLYvsC_DS A_16_P chr GALNTL A_16_P chr GALNTL A_16_P chr GALNTL A_16_P chr GALNTL A_16_P chr GALNTL A_16_P chr GALNTL A_16_P chr GALNTL A_16_P chr GALNTL A_16_P chr GALNTL A_16_P chr GALNTL A_16_P chr GALNTL Supplementarytable8:Singleprobe400KaCGHresultsinWWOX_INDELregion. PROBE CHR START END GENE FMvsC FMvsC_DS FM_FCvsC FM_FCvs FM_EARLYvs FM_EARLY C_DS C C_DS A_16_P chr WWOX A_16_P chr WWOX A_16_P chr WWOX A_16_P chr WWOX A_16_P chr WWOX A_16_P chr WWOX A_16_P chr WWOX A_18_P chr WWOX A_18_P chr WWOX A_16_P chr WWOX A_16_P chr WWOX A_16_P chr WWOX A_16_P chr WWOX Supplementarytable9:aCGH400KarrayresultsinPTPRD_INDELregion. PROBE CHR START END GENE FMVsC FMvsC_DS FM_FCvsC FM_FCvsC_DS FM_EARLYvsC FM_EARLYvsC_DS A_18_P chr PTPRD A_16_P chr PTPRD A_18_P chr PTPRD A_16_P chr PTPRD A_16_P chr PTPRD
196 Supplementarytable10:aCGH400KarrayresultsinMYO5B_INDELregion. PROBE CHR START END GENE FMVsC FMvsC_DS FM_FCvsC FM_FCvsC_DS FM_EARLYvsC FM_EARLYvsC_DS A_16_P chr MYO5B A_16_P chr MYO5B A_16_P chr MYO5B A_16_P chr MYO5B A_18_P chr MYO5B A_18_P chr MYO5B A_16_P chr MYO5B Supplementarytable11:aCGH400KarrayresultsinNRXN3_INDELregion. PROBE START END GENE FMvsC FMvsC_DS FM_FCvsC FM_FCvsC_DS FM_EARLYvs FM_EARLY C vsc_ds A_16_P NRXN3 0, , , , , , A_18_P NRXN3 0, , , , , , A_16_P NRXN3 0, , , , , , A_16_P NRXN3 0, , , , , , A_16_P NRXN3 0, , , , , , A_18_P NRXN3 0, , , , , , A_16_P NRXN3 0, , , , , , A_18_P NRXN3 0, , , , , , A_16_P NRXN3 0, , , , , , A_16_P NRXN3 0, , , , , , A_16_P NRXN3 0, , , , , , A_18_P NRXN3 0, , , , , , A_16_P NRXN3 0, , , , , , A_16_P NRXN3 0, , , , , , A_18_P NRXN3 0, , , , , , A_16_P NRXN3 0, , , , , , A_18_P NRXN3 0, , , , , , A_18_P NRXN3 0, , , , , , acghresultsinnrxn3_indelgenomicregion Supplementarytable12:MultinomialanalysisofNRXN3_DELacrossFMclusters.Interactionpvalue=0.046 Cl3 Cl2 OR(95%CI) Cl1 OR(95%CI) (N%) (N%) (N%) DelDel DelNodel ( ) ( ) NodelNodel ( ) ( ) Trend ( ) ( ) PvalueTrend
197 Supplementarytable13:NRXN3Veracodeassociationanalysisresults CHR SNP POSITION A1 F_A F_U A2 CHISQ P OR SE L95 U95 14 rs G A rs A G rs G A rs A G rs G A rs G A rs A C rs A G rs G A rs A G rs G A rs A G rs A G rs A G rs A G rs C A rs T A rs C A rs A C rs A T rs T A rs G A rs A G rs A G rs A G rs C G rs G A rs A C rs G A rs G A rs A C rs G C rs G A rs A G Supplementarytable13:FMClustersproportionsamongGWASandreplicationsubsets Cluster1 Cluster2 Cluster3 GWAS(291) 66(22.7%) 29(10%) 196(67.3%) Replication(1135) 217(19.1%) 328(28.9%) 590(52%) Joint(1398) 283(20.2%) 357(25.6%) 758(54.2) 191
198 Suppementaryfigure1:Linkagedesequilibriumplotofrs D LDscoreisshowninsidetheboxes. 192
199 Supplementaryfigure2:Linkagedesequilibriumplotofrs D LDscoreisshowninsidetheboxes. 193
200 a) rs genotypesandsntg2expression(asassessedbyoneprobe)intwins lymphoblastyoidcelllines b) rs genotypesandsntg2expression(asassessedbyoneprobe)intwins lymphoblastyoidcelllines Supplementaryfigure3:SNPprobeassociationplotsforrs andrs Expressionlevelsforone expressionarrayprobeareplottedagainstsnpgenotypes. 194
201 ATGTTTSplicingenhancer 1 CGGCATTGATTAATCCTACTGTTTATTGGCTAAGTGTACATTTCCTATTGGTATGTTTCTT ATGTTT--- 6 ****** TTTSplicingsilencer 1 AAGAAAACCTGTGCTTTCCTTAATTTTTCTTGAATCCTGTGGCCTTCTCAGTCTATCCTTT TTT *** Supplementaryfigure4:S plicingfactorsbindingsitesdetectedinnrxn3_del.matchgingsequenceappearsinyellow. RedcirclerepresentstheconsensussequenceinUCSCgenomebrowserimageincludinglevelofconservationamong species. 195
Caracterització clínica, analítica i per imatge de la insuficiència cardíaca amb fracció d'ejecció preservada en pacients ambulatoris
 Caracterització clínica, analítica i per imatge de la insuficiència cardíaca amb fracció d'ejecció preservada en pacients ambulatoris Rut Andrea Riba ADVERTIMENT. La consulta d aquesta tesi queda condicionada
Caracterització clínica, analítica i per imatge de la insuficiència cardíaca amb fracció d'ejecció preservada en pacients ambulatoris Rut Andrea Riba ADVERTIMENT. La consulta d aquesta tesi queda condicionada
Intrauterine Growth Restriction (IUGR) and imprinted gene expression in the placenta: Role of PLAGL1 and analysis of the 6q24.
 Intrauterine Growth Restriction (IUGR) and imprinted gene expression in the placenta: Role of PLAGL1 and analysis of the 6q24.2 region Isabel Iglesias Platas ADVERTIMENT. La consulta d aquesta tesi queda
Intrauterine Growth Restriction (IUGR) and imprinted gene expression in the placenta: Role of PLAGL1 and analysis of the 6q24.2 region Isabel Iglesias Platas ADVERTIMENT. La consulta d aquesta tesi queda
ADVERTIMENT ADVERTENCIA WARNING
 ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents condicions d'ús: La difusió d aquesta tesi per mitjà del servei TDX (www.tesisenxarxa.net) ha estat autoritzada
ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents condicions d'ús: La difusió d aquesta tesi per mitjà del servei TDX (www.tesisenxarxa.net) ha estat autoritzada
biological psychology, p. 40 The study of the nervous system, especially the brain. neuroscience, p. 40
 biological psychology, p. 40 The specialized branch of psychology that studies the relationship between behavior and bodily processes and system; also called biopsychology or psychobiology. neuroscience,
biological psychology, p. 40 The specialized branch of psychology that studies the relationship between behavior and bodily processes and system; also called biopsychology or psychobiology. neuroscience,
Cardiac dysfunction by tissue Doppler in early- and late-onset fetal growth restriction
 Cardiac dysfunction by tissue Doppler in early- and late-onset fetal growth restriction Montserrat Comas Rovira ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents
Cardiac dysfunction by tissue Doppler in early- and late-onset fetal growth restriction Montserrat Comas Rovira ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents
Tratamiento e historia natural de la hepatitis crónica C en pacientes coinfectados por VIH-1
 Tratamiento e historia natural de la hepatitis crónica C en pacientes coinfectados por VIH-1 Javier Murillas Angoiti ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents
Tratamiento e historia natural de la hepatitis crónica C en pacientes coinfectados por VIH-1 Javier Murillas Angoiti ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents
Pharmacist interventions in depressed patients
 Pharmacist interventions in depressed patients Maria Rubio Valera ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents condicions d'ús: La difusió d aquesta tesi per
Pharmacist interventions in depressed patients Maria Rubio Valera ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents condicions d'ús: La difusió d aquesta tesi per
Prevalence, clinical correlates and factors associated with course and outcome of anxiety disorders in youth with bipolar disorder
 Prevalence, clinical correlates and factors associated with course and outcome of anxiety disorders in youth with bipolar disorder Regina Sala Cassola ADVERTIMENT. La consulta d aquesta tesi queda condicionada
Prevalence, clinical correlates and factors associated with course and outcome of anxiety disorders in youth with bipolar disorder Regina Sala Cassola ADVERTIMENT. La consulta d aquesta tesi queda condicionada
Universitat Internacional de Catalunya. Luca Gobbato
 Patients morbidity and root coverage outcomes by means of coronally advanced flap and the application of sub-eptithelial connective tissue graft with different surgical procedures. Luca Gobbato ADVERTIMENT.
Patients morbidity and root coverage outcomes by means of coronally advanced flap and the application of sub-eptithelial connective tissue graft with different surgical procedures. Luca Gobbato ADVERTIMENT.
Acetylcholine (ACh) Action potential. Agonists. Drugs that enhance the actions of neurotransmitters.
 Acetylcholine (ACh) The neurotransmitter responsible for motor control at the junction between nerves and muscles; also involved in mental processes such as learning, memory, sleeping, and dreaming. (See
Acetylcholine (ACh) The neurotransmitter responsible for motor control at the junction between nerves and muscles; also involved in mental processes such as learning, memory, sleeping, and dreaming. (See
The contribution of the hippocampus to language processing
 The contribution of the hippocampus to language processing Saleh Alamri ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents condicions d'ús: La difusió d aquesta tesi
The contribution of the hippocampus to language processing Saleh Alamri ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents condicions d'ús: La difusió d aquesta tesi
Pain Pathways. Dr Sameer Gupta Consultant in Anaesthesia and Pain Management, NGH
 Pain Pathways Dr Sameer Gupta Consultant in Anaesthesia and Pain Management, NGH Objective To give you a simplistic and basic concepts of pain pathways to help understand the complex issue of pain Pain
Pain Pathways Dr Sameer Gupta Consultant in Anaesthesia and Pain Management, NGH Objective To give you a simplistic and basic concepts of pain pathways to help understand the complex issue of pain Pain
Brain and behaviour (Wk 6 + 7)
 Brain and behaviour (Wk 6 + 7) What is a neuron? What is the cell body? What is the axon? The basic building block of the nervous system, the individual nerve cell that receives, processes and transmits
Brain and behaviour (Wk 6 + 7) What is a neuron? What is the cell body? What is the axon? The basic building block of the nervous system, the individual nerve cell that receives, processes and transmits
Chapter 11 Introduction to the Nervous System and Nervous Tissue Chapter Outline
 Chapter 11 Introduction to the Nervous System and Nervous Tissue Chapter Outline Module 11.1 Overview of the Nervous System (Figures 11.1-11.3) A. The nervous system controls our perception and experience
Chapter 11 Introduction to the Nervous System and Nervous Tissue Chapter Outline Module 11.1 Overview of the Nervous System (Figures 11.1-11.3) A. The nervous system controls our perception and experience
Receptors and Neurotransmitters: It Sounds Greek to Me. Agenda. What We Know About Pain 9/7/2012
 Receptors and Neurotransmitters: It Sounds Greek to Me Cathy Carlson, PhD, RN Northern Illinois University Agenda We will be going through this lecture on basic pain physiology using analogies, mnemonics,
Receptors and Neurotransmitters: It Sounds Greek to Me Cathy Carlson, PhD, RN Northern Illinois University Agenda We will be going through this lecture on basic pain physiology using analogies, mnemonics,
Lesson 14. The Nervous System. Introduction to Life Processes - SCI 102 1
 Lesson 14 The Nervous System Introduction to Life Processes - SCI 102 1 Structures and Functions of Nerve Cells The nervous system has two principal cell types: Neurons (nerve cells) Glia The functions
Lesson 14 The Nervous System Introduction to Life Processes - SCI 102 1 Structures and Functions of Nerve Cells The nervous system has two principal cell types: Neurons (nerve cells) Glia The functions
Unit 3: The Biological Bases of Behaviour
 Unit 3: The Biological Bases of Behaviour Section 1: Communication in the Nervous System Section 2: Organization in the Nervous System Section 3: Researching the Brain Section 4: The Brain Section 5: Cerebral
Unit 3: The Biological Bases of Behaviour Section 1: Communication in the Nervous System Section 2: Organization in the Nervous System Section 3: Researching the Brain Section 4: The Brain Section 5: Cerebral
Chapter 17. Nervous System Nervous systems receive sensory input, interpret it, and send out appropriate commands. !
 Chapter 17 Sensory receptor Sensory input Integration Nervous System Motor output Brain and spinal cord Effector cells Peripheral nervous system (PNS) Central nervous system (CNS) 28.1 Nervous systems
Chapter 17 Sensory receptor Sensory input Integration Nervous System Motor output Brain and spinal cord Effector cells Peripheral nervous system (PNS) Central nervous system (CNS) 28.1 Nervous systems
PSYC& 100: Biological Psychology (Lilienfeld Chap 3) 1
 PSYC& 100: Biological Psychology (Lilienfeld Chap 3) 1 1 What is a neuron? 2 Name and describe the functions of the three main parts of the neuron. 3 What do glial cells do? 4 Describe the three basic
PSYC& 100: Biological Psychology (Lilienfeld Chap 3) 1 1 What is a neuron? 2 Name and describe the functions of the three main parts of the neuron. 3 What do glial cells do? 4 Describe the three basic
Introduction to Neurobiology
 Biology 240 General Zoology Introduction to Neurobiology Nervous System functions: communication of information via nerve signals integration and processing of information control of physiological and
Biology 240 General Zoology Introduction to Neurobiology Nervous System functions: communication of information via nerve signals integration and processing of information control of physiological and
Nervous System. Master controlling and communicating system of the body. Secrete chemicals called neurotransmitters
 Nervous System Master controlling and communicating system of the body Interacts with the endocrine system to control and coordinate the body s responses to changes in its environment, as well as growth,
Nervous System Master controlling and communicating system of the body Interacts with the endocrine system to control and coordinate the body s responses to changes in its environment, as well as growth,
Neurons, Synapses, and Signaling
 Neurons, Synapses, and Signaling The Neuron is the functional unit of the nervous system. Neurons are composed of a cell body, which contains the nucleus and organelles; Dendrites which are extensions
Neurons, Synapses, and Signaling The Neuron is the functional unit of the nervous system. Neurons are composed of a cell body, which contains the nucleus and organelles; Dendrites which are extensions
Function of the Nervous System
 Nervous System Function of the Nervous System Receive sensory information, interpret it, and send out appropriate commands to form a response Composed of neurons (functional unit of the nervous system)
Nervous System Function of the Nervous System Receive sensory information, interpret it, and send out appropriate commands to form a response Composed of neurons (functional unit of the nervous system)
SYNAPTIC COMMUNICATION
 BASICS OF NEUROBIOLOGY SYNAPTIC COMMUNICATION ZSOLT LIPOSITS 1 NERVE ENDINGS II. Interneuronal communication 2 INTERNEURONAL COMMUNICATION I. ELECTRONIC SYNAPSE GAP JUNCTION II. CHEMICAL SYNAPSE SYNAPSES
BASICS OF NEUROBIOLOGY SYNAPTIC COMMUNICATION ZSOLT LIPOSITS 1 NERVE ENDINGS II. Interneuronal communication 2 INTERNEURONAL COMMUNICATION I. ELECTRONIC SYNAPSE GAP JUNCTION II. CHEMICAL SYNAPSE SYNAPSES
PAIN MANAGEMENT in the CANINE PATIENT
 PAIN MANAGEMENT in the CANINE PATIENT Laurie Edge-Hughes, BScPT, MAnimSt (Animal Physio), CAFCI, CCRT Part 1: Laurie Edge-Hughes, BScPT, MAnimSt (Animal Physio), CAFCI, CCRT 1 Pain is the most common reason
PAIN MANAGEMENT in the CANINE PATIENT Laurie Edge-Hughes, BScPT, MAnimSt (Animal Physio), CAFCI, CCRT Part 1: Laurie Edge-Hughes, BScPT, MAnimSt (Animal Physio), CAFCI, CCRT 1 Pain is the most common reason
The Brain & Homeostasis. The Brain & Technology. CAT, PET, and MRI Scans
 The Brain & Homeostasis Today, scientists have a lot of information about what happens in the different parts of the brain; however they are still trying to understand how the brain functions. We know
The Brain & Homeostasis Today, scientists have a lot of information about what happens in the different parts of the brain; however they are still trying to understand how the brain functions. We know
Impacte de la Síndrome d'apnea-hipoapnea Obstructiva del Son en l'obesitat greu
 Impacte de la Síndrome d'apnea-hipoapnea Obstructiva del Son en l'obesitat greu Impact of Obstructive Sleep Apnea in Severe Obesity Mercè Gasa Galmes ADVERTIMENT. La consulta d aquesta tesi queda condicionada
Impacte de la Síndrome d'apnea-hipoapnea Obstructiva del Son en l'obesitat greu Impact of Obstructive Sleep Apnea in Severe Obesity Mercè Gasa Galmes ADVERTIMENT. La consulta d aquesta tesi queda condicionada
NEURONS COMMUNICATE WITH OTHER CELLS AT SYNAPSES 34.3
 NEURONS COMMUNICATE WITH OTHER CELLS AT SYNAPSES 34.3 NEURONS COMMUNICATE WITH OTHER CELLS AT SYNAPSES Neurons communicate with other neurons or target cells at synapses. Chemical synapse: a very narrow
NEURONS COMMUNICATE WITH OTHER CELLS AT SYNAPSES 34.3 NEURONS COMMUNICATE WITH OTHER CELLS AT SYNAPSES Neurons communicate with other neurons or target cells at synapses. Chemical synapse: a very narrow
Spinal Cord Injury Pain. Michael Massey, DO CentraCare Health St Cloud, MN 11/07/2018
 Spinal Cord Injury Pain Michael Massey, DO CentraCare Health St Cloud, MN 11/07/2018 Objectives At the conclusion of this session, participants should be able to: 1. Understand the difference between nociceptive
Spinal Cord Injury Pain Michael Massey, DO CentraCare Health St Cloud, MN 11/07/2018 Objectives At the conclusion of this session, participants should be able to: 1. Understand the difference between nociceptive
NERVOUS SYSTEM 1 CHAPTER 10 BIO 211: ANATOMY & PHYSIOLOGY I
 BIO 211: ANATOMY & PHYSIOLOGY I 1 Ch 10 A Ch 10 B This set CHAPTER 10 NERVOUS SYSTEM 1 BASIC STRUCTURE and FUNCTION Dr. Lawrence G. Altman www.lawrencegaltman.com Some illustrations are courtesy of McGraw-Hill.
BIO 211: ANATOMY & PHYSIOLOGY I 1 Ch 10 A Ch 10 B This set CHAPTER 10 NERVOUS SYSTEM 1 BASIC STRUCTURE and FUNCTION Dr. Lawrence G. Altman www.lawrencegaltman.com Some illustrations are courtesy of McGraw-Hill.
Chronobiological related factors and weight loss evolution in severe obese patients undergoing bariatric surgery
 Chronobiological related factors and weight loss evolution in severe obese patients undergoing bariatric surgery Tania Pamela Ruiz Lozano ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l
Chronobiological related factors and weight loss evolution in severe obese patients undergoing bariatric surgery Tania Pamela Ruiz Lozano ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l
STRUCTURAL ORGANIZATION OF THE NERVOUS SYSTEM
 STRUCTURAL ORGANIZATION OF THE NERVOUS SYSTEM STRUCTURAL ORGANIZATION OF THE BRAIN The central nervous system (CNS), consisting of the brain and spinal cord, receives input from sensory neurons and directs
STRUCTURAL ORGANIZATION OF THE NERVOUS SYSTEM STRUCTURAL ORGANIZATION OF THE BRAIN The central nervous system (CNS), consisting of the brain and spinal cord, receives input from sensory neurons and directs
BIPN100 F15 Human Physiology 1 Lecture 3. Synaptic Transmission p. 1
 BIPN100 F15 Human Physiology 1 Lecture 3. Synaptic Transmission p. 1 Terms you should know: synapse, neuromuscular junction (NMJ), pre-synaptic, post-synaptic, synaptic cleft, acetylcholine (ACh), acetylcholine
BIPN100 F15 Human Physiology 1 Lecture 3. Synaptic Transmission p. 1 Terms you should know: synapse, neuromuscular junction (NMJ), pre-synaptic, post-synaptic, synaptic cleft, acetylcholine (ACh), acetylcholine
Tratamiento e historia natural de la hepatitis crónica C en pacientes coinfectados por VIH-1
 Tratamiento e historia natural de la hepatitis crónica C en pacientes coinfectados por VIH-1 Javier Murillas Angoiti ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents
Tratamiento e historia natural de la hepatitis crónica C en pacientes coinfectados por VIH-1 Javier Murillas Angoiti ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents
ANATOMY AND PHYSIOLOGY OF NEURONS. AP Biology Chapter 48
 ANATOMY AND PHYSIOLOGY OF NEURONS AP Biology Chapter 48 Objectives Describe the different types of neurons Describe the structure and function of dendrites, axons, a synapse, types of ion channels, and
ANATOMY AND PHYSIOLOGY OF NEURONS AP Biology Chapter 48 Objectives Describe the different types of neurons Describe the structure and function of dendrites, axons, a synapse, types of ion channels, and
Chapter 11: Nervous System and Nervous Tissue
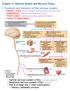 Chapter 11: Nervous System and Nervous Tissue I. Functions and divisions of the nervous system A. Sensory input: monitor changes in internal and external environment B. Integrations: make decisions about
Chapter 11: Nervous System and Nervous Tissue I. Functions and divisions of the nervous system A. Sensory input: monitor changes in internal and external environment B. Integrations: make decisions about
Chapter 11: Functional Organization of Nervous Tissue
 Chapter 11: Functional Organization of Nervous Tissue I. Functions of the Nervous System A. List and describe the five major nervous system functions: 1. 2. 3. 4. 5. II. Divisions of the Nervous System
Chapter 11: Functional Organization of Nervous Tissue I. Functions of the Nervous System A. List and describe the five major nervous system functions: 1. 2. 3. 4. 5. II. Divisions of the Nervous System
Introduction. Visual Perception Aditi Majumder, UCI. Perception is taken for granted!
 Introduction Visual Perception Perception is taken for granted! Slide 2 1 Perception is very complex Perceive Locate Identify/Recognize Different objects Their relationship with each other Qualitative
Introduction Visual Perception Perception is taken for granted! Slide 2 1 Perception is very complex Perceive Locate Identify/Recognize Different objects Their relationship with each other Qualitative
Fundamentals of the Nervous System and Nervous Tissue: Part C
 PowerPoint Lecture Slides prepared by Janice Meeking, Mount Royal College C H A P T E R 11 Fundamentals of the Nervous System and Nervous Tissue: Part C Warm Up What is a neurotransmitter? What is the
PowerPoint Lecture Slides prepared by Janice Meeking, Mount Royal College C H A P T E R 11 Fundamentals of the Nervous System and Nervous Tissue: Part C Warm Up What is a neurotransmitter? What is the
What is Pain? An unpleasant sensory and emotional experience associated with actual or potential tissue damage. Pain is always subjective
 Pain & Acupuncture What is Pain? An unpleasant sensory and emotional experience associated with actual or potential tissue damage. NOCICEPTION( the neural processes of encoding and processing noxious stimuli.)
Pain & Acupuncture What is Pain? An unpleasant sensory and emotional experience associated with actual or potential tissue damage. NOCICEPTION( the neural processes of encoding and processing noxious stimuli.)
Understanding pain and mental illness Impact on management principles
 Understanding pain and mental illness Impact on management principles Chris Alderman Consultant Psychopharmacologist Pain and mental illness - context PAIN MENTAL ILLNESS OTHER FACTORS (personality, history.
Understanding pain and mental illness Impact on management principles Chris Alderman Consultant Psychopharmacologist Pain and mental illness - context PAIN MENTAL ILLNESS OTHER FACTORS (personality, history.
Portions from Chapter 6 CHAPTER 7. The Nervous System: Neurons and Synapses. Chapter 7 Outline. and Supporting Cells
 CHAPTER 7 The Nervous System: Neurons and Synapses Chapter 7 Outline Neurons and Supporting Cells Activity in Axons The Synapse Acetylcholine as a Neurotransmitter Monoamines as Neurotransmitters Other
CHAPTER 7 The Nervous System: Neurons and Synapses Chapter 7 Outline Neurons and Supporting Cells Activity in Axons The Synapse Acetylcholine as a Neurotransmitter Monoamines as Neurotransmitters Other
Unit III. Biological Bases of Behavior
 Unit III Biological Bases of Behavior Module 9: Biological Psychology and Neurotransmission Module 10: The Nervous and Endocrine Systems Module 11: Studying the Brain, and Other Structures Module 12: The
Unit III Biological Bases of Behavior Module 9: Biological Psychology and Neurotransmission Module 10: The Nervous and Endocrine Systems Module 11: Studying the Brain, and Other Structures Module 12: The
Outline. Animals: Nervous system. Neuron and connection of neurons. Key Concepts:
 Animals: Nervous system Neuron and connection of neurons Outline 1. Key concepts 2. An Overview and Evolution 3. Human Nervous System 4. The Neurons 5. The Electrical Signals 6. Communication between Neurons
Animals: Nervous system Neuron and connection of neurons Outline 1. Key concepts 2. An Overview and Evolution 3. Human Nervous System 4. The Neurons 5. The Electrical Signals 6. Communication between Neurons
Chapter 12 Nervous Tissue. Copyright 2009 John Wiley & Sons, Inc. 1
 Chapter 12 Nervous Tissue Copyright 2009 John Wiley & Sons, Inc. 1 Terms to Know CNS PNS Afferent division Efferent division Somatic nervous system Autonomic nervous system Sympathetic nervous system Parasympathetic
Chapter 12 Nervous Tissue Copyright 2009 John Wiley & Sons, Inc. 1 Terms to Know CNS PNS Afferent division Efferent division Somatic nervous system Autonomic nervous system Sympathetic nervous system Parasympathetic
Name: Period: Chapter 2 Reading Guide The Biology of Mind
 Name: Period: Chapter 2 Reading Guide The Biology of Mind The Nervous System (pp. 55-58) 1. What are nerves? 2. Complete the diagram below with definitions of each part of the nervous system. Nervous System
Name: Period: Chapter 2 Reading Guide The Biology of Mind The Nervous System (pp. 55-58) 1. What are nerves? 2. Complete the diagram below with definitions of each part of the nervous system. Nervous System
Neurons, Synapses and Signaling. Chapter 48
 Neurons, Synapses and Signaling Chapter 48 Warm Up Exercise What types of cells can receive a nerve signal? Nervous Organization Neurons- nerve cells. Brain- organized into clusters of neurons, called
Neurons, Synapses and Signaling Chapter 48 Warm Up Exercise What types of cells can receive a nerve signal? Nervous Organization Neurons- nerve cells. Brain- organized into clusters of neurons, called
Body control systems. Nervous system. Organization of Nervous Systems. The Nervous System. Two types of cells. Organization of Nervous System
 Body control systems Nervous system Nervous system Quick Sends message directly to target organ Endocrine system Sends a hormone as a messenger to the target organ Slower acting Longer lasting response
Body control systems Nervous system Nervous system Quick Sends message directly to target organ Endocrine system Sends a hormone as a messenger to the target organ Slower acting Longer lasting response
Seizure: the clinical manifestation of an abnormal and excessive excitation and synchronization of a population of cortical
 Are There Sharing Mechanisms of Epilepsy, Migraine and Neuropathic Pain? Chin-Wei Huang, MD, PhD Department of Neurology, NCKUH Basic mechanisms underlying seizures and epilepsy Seizure: the clinical manifestation
Are There Sharing Mechanisms of Epilepsy, Migraine and Neuropathic Pain? Chin-Wei Huang, MD, PhD Department of Neurology, NCKUH Basic mechanisms underlying seizures and epilepsy Seizure: the clinical manifestation
LESSON 3.2 WORKBOOK How do our neurons communicate with each other?
 LESSON 3.2 WORKBOOK How do our neurons communicate with each other? This lesson introduces you to how one neuron communicates with another neuron during the process of synaptic transmission. In this lesson
LESSON 3.2 WORKBOOK How do our neurons communicate with each other? This lesson introduces you to how one neuron communicates with another neuron during the process of synaptic transmission. In this lesson
All questions below pertain to mandatory material: all slides, and mandatory homework (if any).
 ECOL 182 Spring 2008 Dr. Ferriere s lectures Lecture 6: Nervous system and brain Quiz Book reference: LIFE-The Science of Biology, 8 th Edition. http://bcs.whfreeman.com/thelifewire8e/ All questions below
ECOL 182 Spring 2008 Dr. Ferriere s lectures Lecture 6: Nervous system and brain Quiz Book reference: LIFE-The Science of Biology, 8 th Edition. http://bcs.whfreeman.com/thelifewire8e/ All questions below
The Nervous System. Biological School. Neuroanatomy. How does a Neuron fire? Acetylcholine (ACH) TYPES OF NEUROTRANSMITTERS
 Biological School The Nervous System It is all about the body!!!! It starts with an individual nerve cell called a NEURON. Synapse Neuroanatomy Neurotransmitters (chemicals held in terminal buttons that
Biological School The Nervous System It is all about the body!!!! It starts with an individual nerve cell called a NEURON. Synapse Neuroanatomy Neurotransmitters (chemicals held in terminal buttons that
Pain. Pain. Pain: One definition. Pain: One definition. Pain: One definition. Pain: One definition. Psyc 2906: Sensation--Introduction 9/27/2006
 Pain Pain Pain: One Definition Classic Paths A new Theory Pain and Drugs According to the international Association for the Study (Merskey & Bogduk, 1994), Pain is an unpleasant sensory and emotional experience
Pain Pain Pain: One Definition Classic Paths A new Theory Pain and Drugs According to the international Association for the Study (Merskey & Bogduk, 1994), Pain is an unpleasant sensory and emotional experience
Sensory coding and somatosensory system
 Sensory coding and somatosensory system Sensation and perception Perception is the internal construction of sensation. Perception depends on the individual experience. Three common steps in all senses
Sensory coding and somatosensory system Sensation and perception Perception is the internal construction of sensation. Perception depends on the individual experience. Three common steps in all senses
Chapter 9. Nervous System
 Chapter 9 Nervous System Central Nervous System (CNS) vs. Peripheral Nervous System(PNS) CNS Brain Spinal cord PNS Peripheral nerves connecting CNS to the body Cranial nerves Spinal nerves Neurons transmit
Chapter 9 Nervous System Central Nervous System (CNS) vs. Peripheral Nervous System(PNS) CNS Brain Spinal cord PNS Peripheral nerves connecting CNS to the body Cranial nerves Spinal nerves Neurons transmit
10.1: Introduction. Cell types in neural tissue: Neurons Neuroglial cells (also known as neuroglia, glia, and glial cells) Dendrites.
 10.1: Introduction Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Cell types in neural tissue: Neurons Neuroglial cells (also known as neuroglia, glia, and glial
10.1: Introduction Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Cell types in neural tissue: Neurons Neuroglial cells (also known as neuroglia, glia, and glial
The Nervous System. Chapter 4. Neuron 3/9/ Components of the Nervous System
 Chapter 4 The Nervous System 1. Components of the Nervous System a. Nerve cells (neurons) Analyze and transmit information Over 100 billion neurons in system Four defined regions Cell body Dendrites Axon
Chapter 4 The Nervous System 1. Components of the Nervous System a. Nerve cells (neurons) Analyze and transmit information Over 100 billion neurons in system Four defined regions Cell body Dendrites Axon
LECTURE STRUCTURE ASC171 NERVOUS SYSTEM PART 1: BACKGROUND 26/07/2015. Module 5
 LECTURE STRUCTURE PART 1: Background / Introduction PART 2: Structure of the NS, how it operates PART 3: CNS PART 4: PNS Why did the action potential cross the synaptic junction? To get to the other side
LECTURE STRUCTURE PART 1: Background / Introduction PART 2: Structure of the NS, how it operates PART 3: CNS PART 4: PNS Why did the action potential cross the synaptic junction? To get to the other side
Acute Pain NETP: SEPTEMBER 2013 COHORT
 Acute Pain NETP: SEPTEMBER 2013 COHORT Pain & Suffering an unpleasant sensory & emotional experience associated with actual or potential tissue damage, or described in terms of such damage International
Acute Pain NETP: SEPTEMBER 2013 COHORT Pain & Suffering an unpleasant sensory & emotional experience associated with actual or potential tissue damage, or described in terms of such damage International
Cardiovascular risk in patients infected by the human immunodeficiency virus compared with that of uninfected patients and general population
 Cardiovascular risk in patients infected by the human immunodeficiency virus compared with that of uninfected patients and general population Estudio del riesgo cardiovascular en pacientes infectados por
Cardiovascular risk in patients infected by the human immunodeficiency virus compared with that of uninfected patients and general population Estudio del riesgo cardiovascular en pacientes infectados por
CHAPTER 4 PAIN AND ITS MANAGEMENT
 CHAPTER 4 PAIN AND ITS MANAGEMENT Pain Definition: An unpleasant sensory and emotional experience associated with actual or potential tissue damage, or described in terms of such damage. Types of Pain
CHAPTER 4 PAIN AND ITS MANAGEMENT Pain Definition: An unpleasant sensory and emotional experience associated with actual or potential tissue damage, or described in terms of such damage. Types of Pain
CHAPTER 44: Neurons and Nervous Systems
 CHAPTER 44: Neurons and Nervous Systems 1. What are the three different types of neurons and what are their functions? a. b. c. 2. Label and list the function of each part of the neuron. 3. How does the
CHAPTER 44: Neurons and Nervous Systems 1. What are the three different types of neurons and what are their functions? a. b. c. 2. Label and list the function of each part of the neuron. 3. How does the
 Patrón de expresión de tres genes como marcador de predicción pronóstica en neuroblastoma. Mecanismos de regulación transcripcional y función de los genes seleccionados Gemma Mayol Ricart ADVERTIMENT.
Patrón de expresión de tres genes como marcador de predicción pronóstica en neuroblastoma. Mecanismos de regulación transcripcional y función de los genes seleccionados Gemma Mayol Ricart ADVERTIMENT.
Chapter 16. Sense of Pain
 Chapter 16 Sense of Pain Pain Discomfort caused by tissue injury or noxious stimulation, and typically leading to evasive action important /// helps to protect us lost of pain in diabetes mellitus = diabetic
Chapter 16 Sense of Pain Pain Discomfort caused by tissue injury or noxious stimulation, and typically leading to evasive action important /// helps to protect us lost of pain in diabetes mellitus = diabetic
2. When a neuron receives signals, an abrupt, temporary the inside becomes more positive in the polarity is generated (an action potential).
 Chapter 34 Integration and Control: Nervous Systems I. Neurons The Communication Specialists A. Functional Zones of a Neuron 1. The contains the nucleus and metabolic machinery for protein synthesis. 2.
Chapter 34 Integration and Control: Nervous Systems I. Neurons The Communication Specialists A. Functional Zones of a Neuron 1. The contains the nucleus and metabolic machinery for protein synthesis. 2.
MOLECULAR AND CELLULAR NEUROSCIENCE
 MOLECULAR AND CELLULAR NEUROSCIENCE BMP-218 November 4, 2014 DIVISIONS OF THE NERVOUS SYSTEM The nervous system is composed of two primary divisions: 1. CNS - Central Nervous System (Brain + Spinal Cord)
MOLECULAR AND CELLULAR NEUROSCIENCE BMP-218 November 4, 2014 DIVISIONS OF THE NERVOUS SYSTEM The nervous system is composed of two primary divisions: 1. CNS - Central Nervous System (Brain + Spinal Cord)
Pathophysiology of Pain
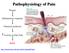 Pathophysiology of Pain Wound Inflammatory response Chemical mediators Activity in Pain Path PAIN http://neuroscience.uth.tmc.edu/s2/chapter08.html Chris Cohan, Ph.D. Dept. of Pathology/Anat Sci University
Pathophysiology of Pain Wound Inflammatory response Chemical mediators Activity in Pain Path PAIN http://neuroscience.uth.tmc.edu/s2/chapter08.html Chris Cohan, Ph.D. Dept. of Pathology/Anat Sci University
DO NOW: ANSWER ON PG 73
 DO NOW: ANSWER ON PG 73 1. Name 1 neurotransmitter that we have learned about. 2. Draw a basic graph of a neuron action potential. Label resting potential, threshold, depolarization, and repolarization
DO NOW: ANSWER ON PG 73 1. Name 1 neurotransmitter that we have learned about. 2. Draw a basic graph of a neuron action potential. Label resting potential, threshold, depolarization, and repolarization
PAIN IS A SUBJECTIVE EXPERIENCE: It is not a stimulus. MAJOR FEATURES OF THE PAIN EXPERIENCE: Sensory discriminative Affective (emotional) Cognitive
 PAIN PAIN IS A SUBJECTIVE EXPERIENCE: It is not a stimulus MAJOR FEATURES OF THE PAIN EXPERIENCE: Sensory discriminative Affective (emotional) Cognitive MEASUREMENT OF PAIN: A BIG PROBLEM Worst pain ever
PAIN PAIN IS A SUBJECTIVE EXPERIENCE: It is not a stimulus MAJOR FEATURES OF THE PAIN EXPERIENCE: Sensory discriminative Affective (emotional) Cognitive MEASUREMENT OF PAIN: A BIG PROBLEM Worst pain ever
Organization of the nervous system. [See Fig. 48.1]
![Organization of the nervous system. [See Fig. 48.1] Organization of the nervous system. [See Fig. 48.1]](/thumbs/90/103926552.jpg) Nervous System [Note: This is the text version of this lecture file. To make the lecture notes downloadable over a slow connection (e.g. modem) the figures have been replaced with figure numbers as found
Nervous System [Note: This is the text version of this lecture file. To make the lecture notes downloadable over a slow connection (e.g. modem) the figures have been replaced with figure numbers as found
BIOLOGY 12 NERVOUS SYSTEM PRACTICE
 1 Name: BIOLOGY 12 NERVOUS SYSTEM PRACTICE Date: 1) Identify structures X, Y and Z and give one function of each. 2) Which processes are involved in the movement of molecule Y from point X to point Z?
1 Name: BIOLOGY 12 NERVOUS SYSTEM PRACTICE Date: 1) Identify structures X, Y and Z and give one function of each. 2) Which processes are involved in the movement of molecule Y from point X to point Z?
The Nervous System Mark Stanford, Ph.D.
 The Nervous System Functional Neuroanatomy and How Neurons Communicate Mark Stanford, Ph.D. Santa Clara Valley Health & Hospital System Addiction Medicine and Therapy Services The Nervous System In response
The Nervous System Functional Neuroanatomy and How Neurons Communicate Mark Stanford, Ph.D. Santa Clara Valley Health & Hospital System Addiction Medicine and Therapy Services The Nervous System In response
Nervous system. The main regulation mechanism of organism's functions
 Nervous system The main regulation mechanism of organism's functions Questions Neuron The reflex arc The nervous centers Properties of the nervous centers The general principles of coordination Inhibition
Nervous system The main regulation mechanism of organism's functions Questions Neuron The reflex arc The nervous centers Properties of the nervous centers The general principles of coordination Inhibition
The Nervous System. Nervous System Functions 1. gather sensory input 2. integration- process and interpret sensory input 3. cause motor output
 The Nervous System Nervous System Functions 1. gather sensory input 2. integration- process and interpret sensory input 3. cause motor output The Nervous System 2 Parts of the Nervous System 1. central
The Nervous System Nervous System Functions 1. gather sensory input 2. integration- process and interpret sensory input 3. cause motor output The Nervous System 2 Parts of the Nervous System 1. central
San Francisco Chronicle, June 2001
 PAIN San Francisco Chronicle, June 2001 CONGENITAL INSENSITIVITY TO PAIN PAIN IS A SUBJECTIVE EXPERIENCE: It is not a stimulus MAJOR FEATURES OF THE PAIN EXPERIENCE: Sensory discriminative Affective (emotional)
PAIN San Francisco Chronicle, June 2001 CONGENITAL INSENSITIVITY TO PAIN PAIN IS A SUBJECTIVE EXPERIENCE: It is not a stimulus MAJOR FEATURES OF THE PAIN EXPERIENCE: Sensory discriminative Affective (emotional)
Physiology of synapses and receptors
 Physiology of synapses and receptors Dr Syed Shahid Habib Professor & Consultant Clinical Neurophysiology Dept. of Physiology College of Medicine & KKUH King Saud University REMEMBER These handouts will
Physiology of synapses and receptors Dr Syed Shahid Habib Professor & Consultant Clinical Neurophysiology Dept. of Physiology College of Medicine & KKUH King Saud University REMEMBER These handouts will
Neural Basis of Motor Control
 Neural Basis of Motor Control Central Nervous System Skeletal muscles are controlled by the CNS which consists of the brain and spinal cord. Determines which muscles will contract When How fast To what
Neural Basis of Motor Control Central Nervous System Skeletal muscles are controlled by the CNS which consists of the brain and spinal cord. Determines which muscles will contract When How fast To what
CHAPTER 4 PAIN AND ITS MANAGEMENT
 CHAPTER 4 PAIN AND ITS MANAGEMENT Pain Definition: An unpleasant sensory and emotional experience associated with actual or potential tissue damage, or described in terms of such damage. Types of Pain
CHAPTER 4 PAIN AND ITS MANAGEMENT Pain Definition: An unpleasant sensory and emotional experience associated with actual or potential tissue damage, or described in terms of such damage. Types of Pain
Anatomy of a Neuron. Copyright 2000 by BSCS and Videodiscovery, Inc. Permission granted for classroom use. Master 2.1
 Anatomy of a Neuron Master 2.1 Neurons Interact With Other Neurons Through Synapses Master 2.2 How Do Neurons Communicate? 1 2 3 4 5 6 Master 2.3 Neurons Communicate by Neurotransmission Neurons communicate
Anatomy of a Neuron Master 2.1 Neurons Interact With Other Neurons Through Synapses Master 2.2 How Do Neurons Communicate? 1 2 3 4 5 6 Master 2.3 Neurons Communicate by Neurotransmission Neurons communicate
Biology 218 Human Anatomy
 Chapter 17 Adapted form Tortora 10 th ed. LECTURE OUTLINE A. Overview of the Nervous System (p. 537) 1. The nervous system and the endocrine system are the body s major control and integrating centers.
Chapter 17 Adapted form Tortora 10 th ed. LECTURE OUTLINE A. Overview of the Nervous System (p. 537) 1. The nervous system and the endocrine system are the body s major control and integrating centers.
Okami Study Guide: Chapter 2 1
 Okami Study Guide: Chapter 2 1 Chapter Test 1. A cell that receives information and transmits it to other cells via an electrochemical process is called a(n) a. neuron b. hormone c. glia d. endorphin Answer:
Okami Study Guide: Chapter 2 1 Chapter Test 1. A cell that receives information and transmits it to other cells via an electrochemical process is called a(n) a. neuron b. hormone c. glia d. endorphin Answer:
Introduction to Physiological Psychology
 Introduction to Physiological Psychology Review Kim Sweeney ksweeney@cogsci.ucsd.edu www.cogsci.ucsd.edu/~ksweeney/psy260.html Today n Discuss Final Paper Proposal (due 3/10) n General Review 1 The article
Introduction to Physiological Psychology Review Kim Sweeney ksweeney@cogsci.ucsd.edu www.cogsci.ucsd.edu/~ksweeney/psy260.html Today n Discuss Final Paper Proposal (due 3/10) n General Review 1 The article
The Nervous System. Anatomy of a Neuron
 The Nervous System Chapter 38.1-38.5 Anatomy of a Neuron I. Dendrites II. Cell Body III. Axon Synaptic terminal 1 Neuron Connections dendrites cell body terminal cell body cell body terminals dendrites
The Nervous System Chapter 38.1-38.5 Anatomy of a Neuron I. Dendrites II. Cell Body III. Axon Synaptic terminal 1 Neuron Connections dendrites cell body terminal cell body cell body terminals dendrites
Fig Copyright 2002 Pearson Education, Inc., publishing as Benjamin Cummings
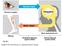 Fig. 48.1 Fig. 48.2 Axon endings are called synaptic terminals. They contain neurotransmitters which conduct a signal across a synapse. A synapse is the junction between a presynaptic and postsynaptic
Fig. 48.1 Fig. 48.2 Axon endings are called synaptic terminals. They contain neurotransmitters which conduct a signal across a synapse. A synapse is the junction between a presynaptic and postsynaptic
Chapter 45: Synapses Transmission of Nerve Impulses Between Neurons. Chad Smurthwaite & Jordan Shellmire
 Chapter 45: Synapses Transmission of Nerve Impulses Between Neurons Chad Smurthwaite & Jordan Shellmire The Chemical Synapse The most common type of synapse used for signal transmission in the central
Chapter 45: Synapses Transmission of Nerve Impulses Between Neurons Chad Smurthwaite & Jordan Shellmire The Chemical Synapse The most common type of synapse used for signal transmission in the central
Chapter 7. Objectives
 Chapter 7 The Nervous System: Structure and Control of Movement Objectives Discuss the general organization of the nervous system Describe the structure & function of a nerve Draw and label the pathways
Chapter 7 The Nervous System: Structure and Control of Movement Objectives Discuss the general organization of the nervous system Describe the structure & function of a nerve Draw and label the pathways
THE NERVOUS SYSTEM. Homeostasis Strand
 THE NERVOUS SYSTEM Homeostasis Strand Introduction In general, a nervous system has three overlapping functions : 1. Sensory input conduction of signals from sensory receptors to integration centres 2.
THE NERVOUS SYSTEM Homeostasis Strand Introduction In general, a nervous system has three overlapping functions : 1. Sensory input conduction of signals from sensory receptors to integration centres 2.
D) around, bypassing B) toward
 Nervous System Practice Questions 1. Which of the following are the parts of neurons? A) brain, spinal cord, and vertebral column B) dendrite, axon, and cell body C) sensory and motor D) cortex, medulla
Nervous System Practice Questions 1. Which of the following are the parts of neurons? A) brain, spinal cord, and vertebral column B) dendrite, axon, and cell body C) sensory and motor D) cortex, medulla
Primary Functions. Monitor changes. Integrate input. Initiate a response. External / internal. Process, interpret, make decisions, store information
 NERVOUS SYSTEM Monitor changes External / internal Integrate input Primary Functions Process, interpret, make decisions, store information Initiate a response E.g., movement, hormone release, stimulate/inhibit
NERVOUS SYSTEM Monitor changes External / internal Integrate input Primary Functions Process, interpret, make decisions, store information Initiate a response E.g., movement, hormone release, stimulate/inhibit
The Nervous System: Neural Tissue Pearson Education, Inc.
 13 The Nervous System: Neural Tissue Introduction Nervous System Characteristics Controls and adjust the activity of the body Provides swift but brief responses The nervous system includes: Central Nervous
13 The Nervous System: Neural Tissue Introduction Nervous System Characteristics Controls and adjust the activity of the body Provides swift but brief responses The nervous system includes: Central Nervous
Chapter 2. The Cellular and Molecular Basis of Cognition Cognitive Neuroscience: The Biology of the Mind, 2 nd Ed.,
 Chapter 2. The Cellular and Molecular Basis of Cognition Cognitive Neuroscience: The Biology of the Mind, 2 nd Ed., M. S. Gazzaniga, R. B. Ivry, and G. R. Mangun, Norton, 2002. Summarized by B.-W. Ku,
Chapter 2. The Cellular and Molecular Basis of Cognition Cognitive Neuroscience: The Biology of the Mind, 2 nd Ed., M. S. Gazzaniga, R. B. Ivry, and G. R. Mangun, Norton, 2002. Summarized by B.-W. Ku,
Lecture 22: A little Neurobiology
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 22: A little Neurobiology http://compbio.uchsc.edu/hunter/bio5099 Larry.Hunter@uchsc.edu Nervous system development Part of the ectoderm
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 22: A little Neurobiology http://compbio.uchsc.edu/hunter/bio5099 Larry.Hunter@uchsc.edu Nervous system development Part of the ectoderm
Neuroinflammation in first episodes of psychosis
 Neuroinflammation in first episodes of psychosis Miquel Bioque Alcázar ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents condicions d'ús: La difusió d aquesta tesi
Neuroinflammation in first episodes of psychosis Miquel Bioque Alcázar ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents condicions d'ús: La difusió d aquesta tesi
211MDS Pain theories
 211MDS Pain theories Definition In 1986, the International Association for the Study of Pain (IASP) defined pain as a sensory and emotional experience associated with real or potential injuries, or described
211MDS Pain theories Definition In 1986, the International Association for the Study of Pain (IASP) defined pain as a sensory and emotional experience associated with real or potential injuries, or described
PSYCH 260 Exam 2. March 2, Answer the questions using the Scantron form. Name:
 PSYCH 260 Exam 2 March 2, 2017 Answer the questions using the Scantron form. Name: 1 1 Main Please put in their proper order the steps that lead to synaptic communication between neurons. Begin with the
PSYCH 260 Exam 2 March 2, 2017 Answer the questions using the Scantron form. Name: 1 1 Main Please put in their proper order the steps that lead to synaptic communication between neurons. Begin with the
Outline. Neuron Structure. Week 4 - Nervous System. The Nervous System: Neurons and Synapses
 Outline Week 4 - The Nervous System: Neurons and Synapses Neurons Neuron structures Types of neurons Electrical activity of neurons Depolarization, repolarization, hyperpolarization Synapses Release of
Outline Week 4 - The Nervous System: Neurons and Synapses Neurons Neuron structures Types of neurons Electrical activity of neurons Depolarization, repolarization, hyperpolarization Synapses Release of
PHENOTYPIC AND TRANSCRIPTIONAL BIOMARKERS IN ORGAN TRANSPLANTATION. Isabel Puig-Pey Comas
 PHENOTYPIC AND TRANSCRIPTIONAL BIOMARKERS IN ORGAN TRANSPLANTATION Thesis presented by Isabel Puig-Pey Comas ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents condicions
PHENOTYPIC AND TRANSCRIPTIONAL BIOMARKERS IN ORGAN TRANSPLANTATION Thesis presented by Isabel Puig-Pey Comas ADVERTIMENT. La consulta d aquesta tesi queda condicionada a l acceptació de les següents condicions
Page 1. Neurons Transmit Signal via Action Potentials: neuron At rest, neurons maintain an electrical difference across
 Chapter 33: The Nervous System and the Senses Neurons: Specialized excitable cells that allow for communication throughout the body via electrical impulses Neuron Anatomy / Function: 1) Dendrites: Receive
Chapter 33: The Nervous System and the Senses Neurons: Specialized excitable cells that allow for communication throughout the body via electrical impulses Neuron Anatomy / Function: 1) Dendrites: Receive
Cellular Bioelectricity
 ELEC ENG 3BB3: Cellular Bioelectricity Notes for Lecture 24 Thursday, March 6, 2014 8. NEURAL ELECTROPHYSIOLOGY We will look at: Structure of the nervous system Sensory transducers and neurons Neural coding
ELEC ENG 3BB3: Cellular Bioelectricity Notes for Lecture 24 Thursday, March 6, 2014 8. NEURAL ELECTROPHYSIOLOGY We will look at: Structure of the nervous system Sensory transducers and neurons Neural coding
Brain and Behavior Lecture 13
 Brain and Behavior Lecture 13 Technology has improved our ability to know how the brain works. Case Study (Phineas Gage) Gage was a railroad construction foreman. An 1848 explosion forced a steel rod through
Brain and Behavior Lecture 13 Technology has improved our ability to know how the brain works. Case Study (Phineas Gage) Gage was a railroad construction foreman. An 1848 explosion forced a steel rod through
