Exome sequencing of a patient with suspected mitochondrial disease reveals a likely multigenic etiology
|
|
|
- Frank Bates
- 5 years ago
- Views:
Transcription
1 Craigen et al. BMC Medical Genetics 2013, 14:83 RESEARCH ARTICLE Open Access Exome sequencing of a patient with suspected mitochondrial disease reveals a likely multigenic etiology William J Craigen 1,2, Brett H Graham 1, Lee-Jun Wong 1, Fernando Scaglia 1,4, Richard Alan Lewis 1,3 and Penelope E Bonnen 1,5* Abstract Background: The clinical features of mitochondrial disease are complex and highly variable, leading to challenges in establishing a specific diagnosis. Despite being one of the most commonly occurring inherited genetic diseases with an incidence of 1/5000, ~90% of these complex patients remain without a DNA-based diagnosis. We report our efforts to identify the pathogenetic cause for a patient with typical features of mitochondrial disease including infantile cataracts, CPEO, ptosis, progressive distal muscle weakness, and ataxia who carried a diagnosis of mitochondrial disease for over a decade. Methods: Whole exome sequencing and bioinformatic analysis of these data were conducted on the proband. Results: Exome sequencing studies showed a homozygous splice site mutation in SETX, which is known to cause Spinocerebellar Ataxia, Autosomal Recessive 1 (SCAR1). Additionally a missense mutation was identified in a highly conserved position of the OCRL gene, which causes Lowe Syndrome and Dent Disease 2. Conclusions: This patient s complex phenotype reflects a complex genetic etiology in which no single gene explained the complete clinical presentation. These genetic studies reveal that this patient does not have mitochondrial disease but rather a genocopy caused by more than one mutant locus. This study demonstrates the benefit of exome sequencing in providing molecular diagnosis to individuals with complex clinical presentations. Keywords: Encephalomyopathy, Lowe syndrome, OCRL, SETX, Diagnostics, Genocopy Background The clinical presentation of mitochondrial disease is highly complex and variable, resulting in diagnostic challenges. Numerous manifestations include subacute necrotizing encephalomyelopathy (Leigh disease), cardiomyopathy, skeletal myopathy, anemia, intestinal pseudo-obstruction, liver failure, sensorineural hearing loss, diabetes and other endocrinopathies, renal tubulopathy, and growth failure. Chronic progressive external ophthalmoplegia (CPEO) is among the most commonly observed features, reported in nearly two-thirds of patients with mitochondrial myopathy. Molecular diagnosis is often pursued but the * Correspondence: pbonnen@bcm.edu 1 Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA 5 Human Genome Sequencing Center, Baylor College of Medicine, Houston, Texas, USA Full list of author information is available at the end of the article majority of these complex patients remain without a DNA-based diagnosis. Pediatric-onset mitochondrial diseases are a diverse group and estimated to occur with an incidence of 1/5,000 [1]. Most cases (85 90%) are likely due to nuclear-encoded mutations, and autosomal recessive transmission is observed most commonly [1,2]. Despite their common occurrence, little is known about the molecular genetic basis of mitochondrial disorders. We applied whole exome sequencing to identify pathogenic mutations in a patient with suspected pediatric-onset mitochondrial disease. The patient s clinical presentation included classic hallmarks of mitochondrial disease: congenital cataracts, CPEO, early onset symmetric bilateral ptosis, progressive constitutional weakness, and ataxia. Exome sequencing revealed mutations in multiple genes 2013 Craigen et al.; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License ( which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
2 Craigen et al. BMC Medical Genetics 2013, 14:83 Page 2 of 7 that, when considered in aggregate, explain the patient s phenotype. Methods Sample DNA Informed consent was obtained from the subject and his parents for research and publication of clinical details according to a protocol approved by the Institutional Review Board for Baylor College of Medicine and Affiliated Hospitals. Genomic DNA was extracted from peripheral leukocytes and saliva according to standard protocols. Sequencing We performed sequencing with a custom capture design called VCROME developed at the BCM Human Genome Sequencing Center that enriches for approximately 20,000 genes comprising a total of 45 Mb of captured genomic sequence [3]. Coding exons in the Vega, Consensus Coding Sequence (CCDS) exons, and RefSeq annotation sets were targeted in addition to mirnas from mirbase (release 10). In addition, 8 MB of untranslated regions (UTR) were enriched in this capture design. Probe pools were manufactured by Roche NimbleGen (Madison, WI). Samples were sequenced in a paired-end strategy on an Illumina HiSeq 2000 instrument with one lane per sample. Targeted re-sequencing of both SETX and OCRL was completed for the propositus and each parent with primers SETXF1 TGACATTTCCTGTCAATGC CTC, SETXR1 GAATAACTCCCGGTGAGCCC, OCRLF1 GGACAGGAGGTAGCCAGAGAT, and OCRLR1 CGG CTTACTTTCTTATACTTGGC. The region was PCRamplified and sequenced by di-deoxy terminator sequencing on an ABI 3730XL. All chromatograms were inspected manually, and genotype calls were based on bi-directional sequence reads with phred quality score > 20. Sequence data analysis and bioinformatics Sequence data were aligned with BWA software to human reference sequence HG19 [4]. Duplicate reads were removed with the Picard program. Recalibration and realignment of the data were accomplished with GATK [5,6]. Single nucleotide variants (SNVs) were called with Samtools [7]. Small insertions and deletions (InDels) were determined by GATK. Quality control filtering of variants was based on coverage, strand bias, mapping quality, and base quality. Annotation of variants was conducted with internally developed Perl scripts. Prediction for potential functional consequences of variants was conducted with SIFT [8], PolyPhen2 [9], and PhyloP [10] using dbnsfp [11] and its scoring system which scales each of these metrics from 0 to 1 with 1 being most damaging. The evolutionary conservation of missense mutations was also determined by Genomic Evolutionary Rate Profiling (GERP) which approximates evolutionary constraint at a locus by maximum likelihood estimation [12,13]. The GERP++ score reported here is based on the alignment of 35 mammalian species and the maximum GERP score for this analysis is Clinical description The patient is a 33-year-old male, born to a then 20-year old mother following an uncomplicated pregnancy. His parents are first cousins of Persian descent. Infantile cataracts were noted at 3 months of age and the lenses removed shortly thereafter. Hypospadias (without cryptorchidism) was surgically repaired in infancy. His motor development was thought to be normal during early childhood, but CPEO, ptosis, and progressive distal muscle weakness developed in early adolescence, and ataxia and tremors became apparent as a young adult. A muscle biopsy was performed when the patient was an adolescent, but the results of the evaluation are not available; however, the family was told the results were inconclusive. Surgical correction of bilateral ptosis was not successful. Because of progressive ataxia, a magnetic resonance imaging (MRI) of the brain was performed when the patient was 25 years old and it demonstrated cerebellar atrophy. The family history was notable for a younger sibling who died at 8 months of age of congenital heart disease. His mother has cerulean cataracts that do not interfere with her vision and were identified when she was in her 20 s. The maternal grandmother had cataract surgery when she was in her 50s. The patient s mother had 3 siblings including a brother who underwent cataract surgery at 4 years of age and who died at 39 years of age from an accident. A second brother has no history of cataracts, while a sister also has cataracts. One of the sister s two children has cataracts that have not required surgery. In the father s family there is no history of cataracts. The physical examination of the patient was notable for bilateral ptosis, complete CPEO with a small angle esotropia and a normal retinal exam, neck weakness, extremity weakness that was more prominent in the lower extremities, upper extremity tremors, and symmetrically markedly reduced reflexes. Relevant clinical biochemical and molecular diagnostic testing performed at 30 years of age included plasma thymidine level, urine organic acids analysis, carbohydratedeficient transferrin analysis, a complete metabolic panel, and TSH, all of which were normal. The blood lactate was increased at 2.6 mm (normal range mm), as was creatine kinase at 279 U/L (normal range U/L). Genetic diagnostic testing included whole mitochondrial genome sequencing which detected one homoplasmic rare variant (m.3110c > T) in the 16S rrna
3 Craigen et al. BMC Medical Genetics 2013, 14:83 Page 3 of 7 gene. This variant has been reported in Mitomap as a polymorphism ( Additional diagnostic sequencing of three nuclear genes known to cause CPEO; POLG, C10ORF2, and ANT1, also found no deleterious mutations, and OPA1 sequencing was also normal. Results Exome sequencing of the proband resulted in 127X average coverage of targeted regions and 98% of targeted bases having at least 10 reads. Variant analysis revealed 17,328 total coding single nucleotide variants (SNVs) with a transition/transversion ratio of 3.3, and 188 total small insertions and deletions (InDels) (Table 1). Due to the patient s clinical diagnosis of suspected pediatriconset mitochondrial disease, exome sequence results were analyzed with an assumed recessive genetic model. Fifteen (15) homozygous or hemizygous SNVs and 4 homozygous InDels were not reported in dbsnp137. One of the SNVs is predicted to affect splicing. Four of the SNVs were non-synonymous missense mutations; of these two were predicted to damage protein function due to high conservation across species (Table 2). Analysis of results of exome sequencing showed that the patient is homozygous for a splice site mutation in senataxin (SETX). This mutation, SETX c g > A (GenBank Accession Number: NM_ ), is in the 1 position of exon 10 of the SETX transcript. SETX has a total of 24 exons encoding a protein of 2677 amino acids long and contains at its C terminus a seven-motif domain that places it in the superfamily 1 of helicases. Recessive mutations in SETX cause Spinocerebellar Ataxia, Autosomal Recessive 1 (SCAR1, OMIM ). Patients typically develop juvenile-onset progressive ataxia, and some but not all patients exhibit oculomotor apraxia, hence the condition has also been termed ataxia with oculomotor apraxia type 2 (AOA2). Other features include increases in serum alpha-fetoprotein and creatine kinase. The clinical phenotype previously reported for SCAR1 parallels the ataxia, tremor, cerebellar atrophy, and hyporeflexia observed in the patient described here, thus, the homozygous SETX splice site mutation likely accounts for the patient s ataxia. Heterozygous missense substitutions in SETX can give rise to dominant juvenile amyotrophic lateral sclerosis (ALS4, OMIM ). The patient also has several clinical features that have not been reported to occur in SCAR1. In particular, the patient does not have ocular apraxia but rather CPEO, ptosis, and a history of bilateral congenital cataracts, features not reported in a recent survey of 90 mutation positive cases of SCAR1/AOA2 [14]. Given this clinical picture, the possibility that more than one gene locus gave rise to the patient s phenotype was entertained, and the identified variants were re-analyzed with both recessive and dominant genetic models. Further review of the list of genes harboring variants not found in public databases and predicted to be damaging to protein function failed to identify any genes that have been annotated as part of the mitochondrial proteome, but did reveal an additional missense variant in the known disease gene, OCRL. Mutations in OCRL cause Lowe Syndrome as well as Dent2 disease. The oculocerebrorenal syndrome of Lowe includes variable involvement of the eyes, brain, and kidneys that could possibly explain the patient s historical congenital cataracts. The missense variant, OCRL c.850g > A; p.e284k (GenBank Accession Number: NM_ ), was identified in exon 10. Most persons with Lowe Syndrome have mutations in exons and many of these are missense [15]. Two different tests for evolutionary conservation, GERP and PhyloP, both showed that this base position is highly constrained. The GERP test for evolutionary sequence conservation yielded a score of 5.44, which is the maximum or most constrained score possible (Table 2). PhyloP analysis also showed significant evidence for conservation (score = 1). Software programs that predict whether a missense mutation is damaging to protein function scored this variant with the most damaging score (Sift = 1, PolyPhen2 = 1). Additionally, this OCRL variant is not present in either dbsnp or HGMD. There is substantial bioinformatics support that this mutation damages protein function and therefore would manifest as Lowe Syndrome in this male, however, we were unable to complete a full clinical assessment for Lowe Syndrome in the patient because the patient was not available for further clinical investigations. Likewise, Table 1 Summary of coding variants identified through exome sequencing A ALL KNOWN NOVEL Ti/Tv Total Het Hom Total Het Hom Total Het Hom B ALL KNOWN NOVEL INSERTION DELETION HET HOM NOVEL HOM A: Single nucleotide variants and B: small insertions and deletions. Variants are listed as novel if not present in dbsnp137.
4 Table 2 Summary of novel non-synonymous coding homozygous variants in patient CHROM POS REF ALT Gene Position Variant type Amino acid change GERP PhyloP SIFT Polyphen2 OMIM disorders C T SETX Splice Splice Ataxia-ocular apraxia-2, (3); Amyotrophic lateral sclerosis4, juvenile, (3) X G A OCRL Exon_CDS Missense E284K Lowe syndrome, (3); Dent disease 2, (3) C T ARHGAP24 Exon_CDS Missense R47C X G A ARSF Exon_CDS Missense G374S X G A TKTL1 Exon_CDS Missense R7K TAGCAGTGACAGCAGCAAC T DSPP Exon_CDS Deletion Dentinogenesis imperfecta, Shields type II/III, / (3); Deafness,autosomal dominant 36, with dentinogenesis, (3); TAA T C10orf68 Exon_CDS Deletion CA C PLA2G4E Exon_CDS Deletion TC T C14orf64 5 UTR Deletion Craigen et al. BMC Medical Genetics 2013, 14:83 Page 4 of 7
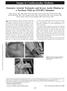
5 Craigen et al. BMC Medical Genetics 2013, 14:83 Page 5 of 7 due to a lack of tissue from the patient we were not able to confirm directly if the SETX c g > A results in the loss of exon 10 or otherwise affects this protein. DNA samples from each parent were sequenced. Each was heterozygous for the SETX splice site mutation, and the mother was found to be a carrier of the OCRL variant. The patient s mother did not display the characteristic cortical flecks in the lens typical of Lowe Syndrome carriers, but instead she has bilateral cerulean cataracts that were first discovered when she was in her 20 s. The mother s pedigree shows three other family members with cataracts, one who had an early age of onset necessitating surgery (Figure 1). The possibility of an independent locus segregating in this family that confers autosomal dominant cataract was evaluated. We queried the patient s exome variants present in genes known to cause all inherited forms of cataract, reviewed in Huang et al [16]. No variants that were predicted to damage protein function were identified in any known cataract gene. This includes four known cerulean cataract genes: MAF, CRYG, CRYBB2, and GJA3. Ascertainment of CRYG and CRYBB2 coding region was 100% in the exome dataset. However, part of the first exon of MAF and the last exon of GJA3 are both repeat rich and as a result was not targeted by exome capture. Attempts to sequence these gene regions by Sanger sequencing were unsuccessful. Discussion We describe a patient with a complex clinical presentation including congenital cataracts, CPEO, ptosis, progressive systemic neuromuscular weakness, cerebellar atrophy, ataxia, and tremors, and the results of sequencing his exome. Exome sequencing revealed potentially pathogenic variants in two genes: SETX and OCRL, each of which may contribute to this complex phenotype. Recently, senataxin was found to harbor RNA/DNA helicase activity important in mrna transcription, with an ability to unwind RNA/DNA hybrids formed behind the elongating RNA Polymerase II between the nascent RNA transcript and single-stranded DNA template. This activity is likely important in the termination of Figure 1 Pedigree of patient with ataxia and congenital cataracts. Four generations of the family of a proband with ataxia and congenital cataracts. Genotypes for the SETX mutation chr9:135,176,191, C > T and the OCRL mutation is chrx: , G > A are shown for the proband and his parents. Several members of the maternal lineage were noted to have early onset cataracts.
6 Craigen et al. BMC Medical Genetics 2013, 14:83 Page 6 of 7 transcription at the polya addition signal [17], and studies in yeast point to a similar 3 processing function in noncoding RNAs [18], suggesting that perturbations in global gene expression may underlie the pleiotropic features of the disease. This study represents the challenges in diagnosing patients with complex clinical phenotypes and the interpretation of exome sequence data, a technology that is moving rapidly into clinical diagnostics. This patient s initial diagnosis of mitochondrial disease was suggested by his findings and offered without the benefit of genome-wide sequence data. Exome sequencing revealed that his complex phenotype reflects a complex genetic etiology in which no single gene explained the complete clinical presentation. The mutation found in SETX partially explains his clinical diagnosis, while the pathogenicity of the OCRL missense variant remains undetermined. There is strong bioinformatics support for pathogenicity, however, the patient was not available for further clinical investigations and the patient s mother did not exhibit the classic ocular phenotype of heterozygotes of Lowe Syndrome mutations. We considered the possibility of an independent locus causing the congenital cataracts in this patient. Exome data for genes known to cause dominantly inherited cataracts were queried and no variants with predicted pathogenicity were found. In summary, exome sequencing improves the diagnostic reservoir, and some patients may require further clinical and/or functional studies to achieve a complete diagnosis. Conclusions Exome sequencing studies identified mutations in multiple independent loci in a patient with typical features of mitochondrial disease including infantile cataracts, CPEO, ptosis, progressive distal muscle weakness, and ataxia who carried a diagnosis of mitochondrial disease for over a decade. Sequencing revealed a homozygous splice site mutation in SETX, which is known to cause Spinocerebellar Ataxia, Autosomal Recessive 1 (SCAR1), as well as a missense mutation in a highly conserved position of the OCRL gene, which causes Lowe Syndrome and Dent Disease 2. This patient s complex phenotype reflects a complex genetic etiology in which no single gene explained the complete clinical presentation. These genetic studies reveal that this patient does not have mitochondrial disease but rather a genocopy caused by more than one mutant locus. This study demonstrates the benefit of exome sequencing in providing molecular diagnosis to individuals with complex clinical presentations. Competing interests The authors declare no competing interests. Authors contributions PEB participated in study design, drafting of the manuscript, collection and analyses of data. WC participated in collection of samples and data interpretation. WC, BG, RL, FS clinically evaluated the patient and family. LW conducted diagnostic screening. All authors read and approved the final manuscript. Acknowledgments We thank the family for participating in this study. This material is based in part upon work supported by the Texas Norman Hackerman Advanced Research Program under Grant No. [THECB] 02006; NHARP proposal number Dr. Lewis is a Senior Scientific Investigator of Research to Prevent Blindness, New York, N.Y., that provided some unrestricted funds for these studies. PEB is on the clinical advisory board for InVitae, Inc. Author details 1 Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA. 2 Department of Pediatrics, Baylor College of Medicine, Houston, Texas, USA. 3 Department of Ophthalmology, Baylor College of Medicine, Houston, Texas, USA. 4 Texas Children s Hospital, Clinical Care Center, Houston, Texas, USA. 5 Human Genome Sequencing Center, Baylor College of Medicine, Houston, Texas, USA. Received: 2 November 2012 Accepted: 6 August 2013 Published: 16 August 2013 References 1. Debray FG, Lambert M, Mitchell GA: Disorders of mitochondrial function. Curr Opin Pediatr 2008, 20(4): Thorburn DR: Mitochondrial disorders: prevalence, myths and advances. J Inherit Metab Dis 2004, 27(3): Bainbridge MN, Wang M, Wu Y, Newsham I, Muzny DM, Jefferies JL, Albert TJ, Burgess DL, Gibbs RA: Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol 2011, 12(7):R Li H, Durbin R: Fast and accurate short read alignment with burrowswheeler transform. Bioinformatics 2009, 25(14): DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al: A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011, 43(5): McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al: The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010, 20(9): Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP: The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25(16): Ng PC, Henikoff S: Predicting deleterious amino acid substitutions. Genome Res 2001, 11(5): Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR: A method and server for predicting damaging missense mutations. Nat Methods 2010, 7(4): Siepel A, Pollard KS, Haussler D: New methods for detecting lineage-specific selection, Proceedings of the 10th International Conference on Research in Computational Molecular Biology RECOMB. Berlin Heidelberg New York: Springer; 2006: ISBN Liu X, Jian X, Boerwinkle E: dbnsfp: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat 2011, 32(8): Cooper GM, Stone EA, Asimenos G, Program NCS, Green ED, Batzoglou S, Sidow A: Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 2005, 15(7): Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S: Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 2010, 6(12):e Anheim M, Monga B, Fleury M, Charles P, Barbot C, Salih M, Delaunoy JP, Fritsch M, Arning L, Synofzik M, et al: Ataxia with oculomotor apraxia type 2: clinical, biological and genotype/phenotype correlation study of a cohort of 90 patients. Brain 2009, 132(Pt 10):
7 Craigen et al. BMC Medical Genetics 2013, 14:83 Page 7 of Nussbaum RL, Suchy SF, Lin T: Database of the OCRL1 mutations causing lowe syndrome, National Human Genome Research Institute, National Institutes of Health. select_db=ocrl. 16. Huang B, He W: Molecular characteristics of inherited congenital cataracts. Eur J Med Genet 2010, 53(6): Baranski TJ, Faust PL, Kornfeld S: Generation of a lysosomal enzyme targeting signal in the secretory protein pepsinogen. Cell 1990, 63(2): Finkel JS, Chinchilla K, Ursic D, Culbertson MR: Sen1p performs two genetically separable functions in transcription and processing of U5 small nuclear RNA in Saccharomyces cerevisiae. Genetics 2010, 184(1): doi: / Cite this article as: Craigen et al.: Exome sequencing of a patient with suspected mitochondrial disease reveals a likely multigenic etiology. BMC Medical Genetics :83. Submit your next manuscript to BioMed Central and take full advantage of: Convenient online submission Thorough peer review No space constraints or color figure charges Immediate publication on acceptance Inclusion in PubMed, CAS, Scopus and Google Scholar Research which is freely available for redistribution Submit your manuscript at
Multiple gene sequencing for risk assessment in patients with early-onset or familial breast cancer
 www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Multiple gene sequencing for risk assessment in patients with early-onset or familial breast cancer Supplementary Materials Supplementary
www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Multiple gene sequencing for risk assessment in patients with early-onset or familial breast cancer Supplementary Materials Supplementary
CITATION FILE CONTENT/FORMAT
 CITATION For any resultant publications using please cite: Matthew A. Field, Vicky Cho, T. Daniel Andrews, and Chris C. Goodnow (2015). "Reliably detecting clinically important variants requires both combined
CITATION For any resultant publications using please cite: Matthew A. Field, Vicky Cho, T. Daniel Andrews, and Chris C. Goodnow (2015). "Reliably detecting clinically important variants requires both combined
CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE
 CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE is_clinical dbsnp MITO GENE chr1 13273 G C heterozygous - - -. - DDX11L1 chr1 949654 A G Homozygous 52 - - rs8997 - ISG15 chr1 1021346 A G heterozygous
CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE is_clinical dbsnp MITO GENE chr1 13273 G C heterozygous - - -. - DDX11L1 chr1 949654 A G Homozygous 52 - - rs8997 - ISG15 chr1 1021346 A G heterozygous
Variant Detection & Interpretation in a diagnostic context. Christian Gilissen
 Variant Detection & Interpretation in a diagnostic context Christian Gilissen c.gilissen@gen.umcn.nl 28-05-2013 So far Sequencing Johan den Dunnen Marja Jakobs Ewart de Bruijn Mapping Victor Guryev Variant
Variant Detection & Interpretation in a diagnostic context Christian Gilissen c.gilissen@gen.umcn.nl 28-05-2013 So far Sequencing Johan den Dunnen Marja Jakobs Ewart de Bruijn Mapping Victor Guryev Variant
Identifying Mutations Responsible for Rare Disorders Using New Technologies
 Identifying Mutations Responsible for Rare Disorders Using New Technologies Jacek Majewski, Department of Human Genetics, McGill University, Montreal, QC Canada Mendelian Diseases Clear mode of inheritance
Identifying Mutations Responsible for Rare Disorders Using New Technologies Jacek Majewski, Department of Human Genetics, McGill University, Montreal, QC Canada Mendelian Diseases Clear mode of inheritance
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research
 Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
TITLE: - Whole Genome Sequencing of High-Risk Families to Identify New Mutational Mechanisms of Breast Cancer Predisposition
 AD Award Number: W81XWH-13-1-0336 TITLE: - Whole Genome Sequencing of High-Risk Families to Identify New Mutational Mechanisms of Breast Cancer Predisposition PRINCIPAL INVESTIGATOR: Mary-Claire King,
AD Award Number: W81XWH-13-1-0336 TITLE: - Whole Genome Sequencing of High-Risk Families to Identify New Mutational Mechanisms of Breast Cancer Predisposition PRINCIPAL INVESTIGATOR: Mary-Claire King,
Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library
 Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library Marilou Wijdicks International Product Manager Research For Life Science Research Only. Not for Use in Diagnostic Procedures.
Advance Your Genomic Research Using Targeted Resequencing with SeqCap EZ Library Marilou Wijdicks International Product Manager Research For Life Science Research Only. Not for Use in Diagnostic Procedures.
NGS in neurodegenerative disorders - our experience
 Neurology Clinic, Clinical Center of Serbia Faculty of Medicine, University of Belgrade Belgrade, Serbia NGS in neurodegenerative disorders - our experience Marija Branković, MSc Belgrade, 2018 Next Generation
Neurology Clinic, Clinical Center of Serbia Faculty of Medicine, University of Belgrade Belgrade, Serbia NGS in neurodegenerative disorders - our experience Marija Branković, MSc Belgrade, 2018 Next Generation
MEDICAL GENOMICS LABORATORY. Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG)
 Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Single Gene (Monogenic) Disorders. Mendelian Inheritance: Definitions. Mendelian Inheritance: Definitions
 Single Gene (Monogenic) Disorders Mendelian Inheritance: Definitions A genetic locus is a specific position or location on a chromosome. Frequently, locus is used to refer to a specific gene. Alleles are
Single Gene (Monogenic) Disorders Mendelian Inheritance: Definitions A genetic locus is a specific position or location on a chromosome. Frequently, locus is used to refer to a specific gene. Alleles are
Novel Heterozygous Mutation in YAP1 in A Family with Isolated Ocular Colobomas
 Novel Heterozygous Mutation in YAP1 in A Family with Isolated Ocular Colobomas Julius T. Oatts 1, Sarah Hull 2, Michel Michaelides 2, Gavin Arno 2, Andrew R. Webster 2*, Anthony T. Moore 1,2* 1. Department
Novel Heterozygous Mutation in YAP1 in A Family with Isolated Ocular Colobomas Julius T. Oatts 1, Sarah Hull 2, Michel Michaelides 2, Gavin Arno 2, Andrew R. Webster 2*, Anthony T. Moore 1,2* 1. Department
Exploding Genetic Knowledge in Developmental Disabilities. Disclosures. The Genetic Principle
 Exploding Genetic Knowledge in Developmental Disabilities How to acquire the data and how to make use of it Elliott H. Sherr MD PhD Professor of Neurology & Pediatrics UCSF Disclosures InVitae: clinical
Exploding Genetic Knowledge in Developmental Disabilities How to acquire the data and how to make use of it Elliott H. Sherr MD PhD Professor of Neurology & Pediatrics UCSF Disclosures InVitae: clinical
6/12/2018. Disclosures. Clinical Genomics The CLIA Lab Perspective. Outline. COH HopeSeq Heme Panels
 Clinical Genomics The CLIA Lab Perspective Disclosures Raju K. Pillai, M.D. Hematopathologist / Molecular Pathologist Director, Pathology Bioinformatics City of Hope National Medical Center, Duarte, CA
Clinical Genomics The CLIA Lab Perspective Disclosures Raju K. Pillai, M.D. Hematopathologist / Molecular Pathologist Director, Pathology Bioinformatics City of Hope National Medical Center, Duarte, CA
Dr Rick Tearle Senior Applications Specialist, EMEA Complete Genomics Complete Genomics, Inc.
 Dr Rick Tearle Senior Applications Specialist, EMEA Complete Genomics Topics Overview of Data Processing Pipeline Overview of Data Files 2 DNA Nano-Ball (DNB) Read Structure Genome : acgtacatgcattcacacatgcttagctatctctcgccag
Dr Rick Tearle Senior Applications Specialist, EMEA Complete Genomics Topics Overview of Data Processing Pipeline Overview of Data Files 2 DNA Nano-Ball (DNB) Read Structure Genome : acgtacatgcattcacacatgcttagctatctctcgccag
variant led to a premature stop codon p.k316* which resulted in nonsense-mediated mrna decay. Although the exact function of the C19L1 is still
 157 Neurological disorders primarily affect and impair the functioning of the brain and/or neurological system. Structural, electrical or metabolic abnormalities in the brain or neurological system can
157 Neurological disorders primarily affect and impair the functioning of the brain and/or neurological system. Structural, electrical or metabolic abnormalities in the brain or neurological system can
CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY
 CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY More genetic information requires cutting-edge interpretation techniques Whole Exome Sequencing For certain patients the combination of symptoms does not allow
CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY More genetic information requires cutting-edge interpretation techniques Whole Exome Sequencing For certain patients the combination of symptoms does not allow
CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY
 CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY More genetic information requires cutting-edge interpretation techniques Whole Exome Sequencing For some patients, the combination of symptoms does not allow
CentoXome FUTURE'S KNOWLEDGE APPLIED TODAY More genetic information requires cutting-edge interpretation techniques Whole Exome Sequencing For some patients, the combination of symptoms does not allow
NGS panels in clinical diagnostics: Utrecht experience. Van Gijn ME PhD Genome Diagnostics UMCUtrecht
 NGS panels in clinical diagnostics: Utrecht experience Van Gijn ME PhD Genome Diagnostics UMCUtrecht 93 Gene panels UMC Utrecht Cardiovascular disease (CAR) (5 panels) Epilepsy (EPI) (11 panels) Hereditary
NGS panels in clinical diagnostics: Utrecht experience Van Gijn ME PhD Genome Diagnostics UMCUtrecht 93 Gene panels UMC Utrecht Cardiovascular disease (CAR) (5 panels) Epilepsy (EPI) (11 panels) Hereditary
Genetic diagnosis of limb girdle muscular dystrophy type 2A, A Case Report
 Genetic diagnosis of limb girdle muscular dystrophy type 2A, A Case Report Roshanak Jazayeri, MD, PhD Assistant Professor of Medical Genetics Faculty of Medicine, Alborz University of Medical Sciences
Genetic diagnosis of limb girdle muscular dystrophy type 2A, A Case Report Roshanak Jazayeri, MD, PhD Assistant Professor of Medical Genetics Faculty of Medicine, Alborz University of Medical Sciences
Welcome to the Genetic Code: An Overview of Basic Genetics. October 24, :00pm 3:00pm
 Welcome to the Genetic Code: An Overview of Basic Genetics October 24, 2016 12:00pm 3:00pm Course Schedule 12:00 pm 2:00 pm Principles of Mendelian Genetics Introduction to Genetics of Complex Disease
Welcome to the Genetic Code: An Overview of Basic Genetics October 24, 2016 12:00pm 3:00pm Course Schedule 12:00 pm 2:00 pm Principles of Mendelian Genetics Introduction to Genetics of Complex Disease
CHAPTER IV RESULTS Microcephaly General description
 47 CHAPTER IV RESULTS 4.1. Microcephaly 4.1.1. General description This study found that from a previous study of 527 individuals with MR, 48 (23 female and 25 male) unrelated individuals were identified
47 CHAPTER IV RESULTS 4.1. Microcephaly 4.1.1. General description This study found that from a previous study of 527 individuals with MR, 48 (23 female and 25 male) unrelated individuals were identified
Whole exome sequencing identifies a novel DFNA9 mutation, C162Y
 Clin Genet 2013: 83: 477 481 Printed in Singapore. All rights reserved Short Report 2012 John Wiley & Sons A/S. Published by Blackwell Publishing Ltd CLINICAL GENETICS doi: 10.1111/cge.12006 Whole exome
Clin Genet 2013: 83: 477 481 Printed in Singapore. All rights reserved Short Report 2012 John Wiley & Sons A/S. Published by Blackwell Publishing Ltd CLINICAL GENETICS doi: 10.1111/cge.12006 Whole exome
Identification of genomic alterations in cervical cancer biopsies by exome sequencing
 Chapter- 4 Identification of genomic alterations in cervical cancer biopsies by exome sequencing 105 4.1 INTRODUCTION Athough HPV has been identified as the prime etiological factor for cervical cancer,
Chapter- 4 Identification of genomic alterations in cervical cancer biopsies by exome sequencing 105 4.1 INTRODUCTION Athough HPV has been identified as the prime etiological factor for cervical cancer,
DOES THE BRCAX GENE EXIST? FUTURE OUTLOOK
 CHAPTER 6 DOES THE BRCAX GENE EXIST? FUTURE OUTLOOK Genetic research aimed at the identification of new breast cancer susceptibility genes is at an interesting crossroad. On the one hand, the existence
CHAPTER 6 DOES THE BRCAX GENE EXIST? FUTURE OUTLOOK Genetic research aimed at the identification of new breast cancer susceptibility genes is at an interesting crossroad. On the one hand, the existence
Concurrent Practical Session ACMG Classification
 Variant Effect Prediction Training Course 6-8 November 2017 Prague, Czech Republic Concurrent Practical Session ACMG Classification Andreas Laner / Anna Benet-Pagès 1 Content 1. Background... 3 2. Aim
Variant Effect Prediction Training Course 6-8 November 2017 Prague, Czech Republic Concurrent Practical Session ACMG Classification Andreas Laner / Anna Benet-Pagès 1 Content 1. Background... 3 2. Aim
Assessing Laboratory Performance for Next Generation Sequencing Based Detection of Germline Variants through Proficiency Testing
 Assessing Laboratory Performance for Next Generation Sequencing Based Detection of Germline Variants through Proficiency Testing Karl V. Voelkerding, MD Professor of Pathology University of Utah Medical
Assessing Laboratory Performance for Next Generation Sequencing Based Detection of Germline Variants through Proficiency Testing Karl V. Voelkerding, MD Professor of Pathology University of Utah Medical
Genomic structural variation
 Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
Mutational and phenotypical spectrum of phenylalanine hydroxylase deficiency in Denmark
 Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE. Dr. Bahar Naghavi
 2 CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE Dr. Bahar Naghavi Assistant professor of Basic Science Department, Shahid Beheshti University of Medical Sciences, Tehran,Iran 3 Introduction Over 4000
2 CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE Dr. Bahar Naghavi Assistant professor of Basic Science Department, Shahid Beheshti University of Medical Sciences, Tehran,Iran 3 Introduction Over 4000
Pedigree Construction Notes
 Name Date Pedigree Construction Notes GO TO à Mendelian Inheritance (http://www.uic.edu/classes/bms/bms655/lesson3.html) When human geneticists first began to publish family studies, they used a variety
Name Date Pedigree Construction Notes GO TO à Mendelian Inheritance (http://www.uic.edu/classes/bms/bms655/lesson3.html) When human geneticists first began to publish family studies, they used a variety
MutationTaster & RegulationSpotter
 MutationTaster & RegulationSpotter Pathogenicity Prediction of Sequence Variants: Past, Present and Future Dr. rer. nat. Jana Marie Schwarz Klinik für Pädiatrie m. S. Neurologie Exzellenzcluster NeuroCure
MutationTaster & RegulationSpotter Pathogenicity Prediction of Sequence Variants: Past, Present and Future Dr. rer. nat. Jana Marie Schwarz Klinik für Pädiatrie m. S. Neurologie Exzellenzcluster NeuroCure
Unifactorial or Single Gene Disorders. Hanan Hamamy Department of Genetic Medicine and Development Geneva University Hospital
 Unifactorial or Single Gene Disorders Hanan Hamamy Department of Genetic Medicine and Development Geneva University Hospital Training Course in Sexual and Reproductive Health Research Geneva 2011 Single
Unifactorial or Single Gene Disorders Hanan Hamamy Department of Genetic Medicine and Development Geneva University Hospital Training Course in Sexual and Reproductive Health Research Geneva 2011 Single
Supplementary Information
 1 Supplementary Information MATERIALS AND METHODS Study subjects Exome-sequenced HBOC patients Clinical characteristics of 24 exome-sequenced HBOC patients are presented in Table 1. Table 1. Clinical characteristics
1 Supplementary Information MATERIALS AND METHODS Study subjects Exome-sequenced HBOC patients Clinical characteristics of 24 exome-sequenced HBOC patients are presented in Table 1. Table 1. Clinical characteristics
CRISPR/Cas9 Enrichment and Long-read WGS for Structural Variant Discovery
 CRISPR/Cas9 Enrichment and Long-read WGS for Structural Variant Discovery PacBio CoLab Session October 20, 2017 For Research Use Only. Not for use in diagnostics procedures. Copyright 2017 by Pacific Biosciences
CRISPR/Cas9 Enrichment and Long-read WGS for Structural Variant Discovery PacBio CoLab Session October 20, 2017 For Research Use Only. Not for use in diagnostics procedures. Copyright 2017 by Pacific Biosciences
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier
 Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier Test Disease Population Triad Disease name Leber congenital amaurosis OMIM number for disease 204000 Disease alternative
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier Test Disease Population Triad Disease name Leber congenital amaurosis OMIM number for disease 204000 Disease alternative
Supplementary Figure 1
 Supplementary Figure 1 Sanger-sequencing chromatograms for the two families. Shown are the Sanger sequencing chromatograms for the two BACH2 mutations (A.II.1 left; B.II.1 and B.III.2 right) and unaffected
Supplementary Figure 1 Sanger-sequencing chromatograms for the two families. Shown are the Sanger sequencing chromatograms for the two BACH2 mutations (A.II.1 left; B.II.1 and B.III.2 right) and unaffected
Corporate Medical Policy
 Corporate Medical Policy Invasive Prenatal (Fetal) Diagnostic Testing File Name: Origination: Last CAP Review: Next CAP Review: Last Review: invasive_prenatal_(fetal)_diagnostic_testing 12/2014 3/2018
Corporate Medical Policy Invasive Prenatal (Fetal) Diagnostic Testing File Name: Origination: Last CAP Review: Next CAP Review: Last Review: invasive_prenatal_(fetal)_diagnostic_testing 12/2014 3/2018
Supplementary appendix
 Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Ellingford JM, Sergouniotis PI, Lennon R, et
Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Ellingford JM, Sergouniotis PI, Lennon R, et
Hands-On Ten The BRCA1 Gene and Protein
 Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Rare Variant Burden Tests. Biostatistics 666
 Rare Variant Burden Tests Biostatistics 666 Last Lecture Analysis of Short Read Sequence Data Low pass sequencing approaches Modeling haplotype sharing between individuals allows accurate variant calls
Rare Variant Burden Tests Biostatistics 666 Last Lecture Analysis of Short Read Sequence Data Low pass sequencing approaches Modeling haplotype sharing between individuals allows accurate variant calls
CANCER GENETICS PROVIDER SURVEY
 Dear Participant, Previously you agreed to participate in an evaluation of an education program we developed for primary care providers on the topic of cancer genetics. This is an IRB-approved, CDCfunded
Dear Participant, Previously you agreed to participate in an evaluation of an education program we developed for primary care providers on the topic of cancer genetics. This is an IRB-approved, CDCfunded
Supplemental Data. De Novo Truncating Mutations in WASF1. Cause Intellectual Disability with Seizures
 The American Journal of Human Genetics, Volume 13 Supplemental Data De Novo Truncating Mutations in WASF1 Cause Intellectual Disability with Seizures Yoko Ito, Keren J. Carss, Sofia T. Duarte, Taila Hartley,
The American Journal of Human Genetics, Volume 13 Supplemental Data De Novo Truncating Mutations in WASF1 Cause Intellectual Disability with Seizures Yoko Ito, Keren J. Carss, Sofia T. Duarte, Taila Hartley,
The Organism as a system
 The Organism as a system PATIENT 1: Seven-year old female with a history of normal development until age two. At this point she developed episodic vomiting, acidosis, epilepsy, general weakness, ataxia
The Organism as a system PATIENT 1: Seven-year old female with a history of normal development until age two. At this point she developed episodic vomiting, acidosis, epilepsy, general weakness, ataxia
Non-Mendelian inheritance
 Non-Mendelian inheritance Focus on Human Disorders Peter K. Rogan, Ph.D. Laboratory of Human Molecular Genetics Children s Mercy Hospital Schools of Medicine & Computer Science and Engineering University
Non-Mendelian inheritance Focus on Human Disorders Peter K. Rogan, Ph.D. Laboratory of Human Molecular Genetics Children s Mercy Hospital Schools of Medicine & Computer Science and Engineering University
Genetic Testing and Analysis. (858) MRN: Specimen: Saliva Received: 07/26/2016 GENETIC ANALYSIS REPORT
 GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11396 A total of 2078 samples from a large sequencing project at decode were used in this study, 219 samples from 78 trios with two grandchildren who were not also members of other trios,
doi:10.1038/nature11396 A total of 2078 samples from a large sequencing project at decode were used in this study, 219 samples from 78 trios with two grandchildren who were not also members of other trios,
Investigating rare diseases with Agilent NGS solutions
 Investigating rare diseases with Agilent NGS solutions Chitra Kotwaliwale, Ph.D. 1 Rare diseases affect 350 million people worldwide 7,000 rare diseases 80% are genetic 60 million affected in the US, Europe
Investigating rare diseases with Agilent NGS solutions Chitra Kotwaliwale, Ph.D. 1 Rare diseases affect 350 million people worldwide 7,000 rare diseases 80% are genetic 60 million affected in the US, Europe
Case Report Intermediate MCAD Deficiency Associated with a Novel Mutation of the ACADM Gene: c.1052c>t
 Case Reports in Genetics Volume 215, Article ID 5329, 4 pages http://dx.doi.org/1.1155/215/5329 Case Report Intermediate MCAD Deficiency Associated with a Novel Mutation of the ACADM Gene: c.152c>t Holli
Case Reports in Genetics Volume 215, Article ID 5329, 4 pages http://dx.doi.org/1.1155/215/5329 Case Report Intermediate MCAD Deficiency Associated with a Novel Mutation of the ACADM Gene: c.152c>t Holli
Computational Systems Biology: Biology X
 Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature13908 Supplementary Tables Supplementary Table 1: Families in this study (.xlsx) All families included in the study are listed. For each family, we show: the genders of the probands and
doi:10.1038/nature13908 Supplementary Tables Supplementary Table 1: Families in this study (.xlsx) All families included in the study are listed. For each family, we show: the genders of the probands and
DNA-seq Bioinformatics Analysis: Copy Number Variation
 DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
Chapter 1 : Genetics 101
 Chapter 1 : Genetics 101 Understanding the underlying concepts of human genetics and the role of genes, behavior, and the environment will be important to appropriately collecting and applying genetic
Chapter 1 : Genetics 101 Understanding the underlying concepts of human genetics and the role of genes, behavior, and the environment will be important to appropriately collecting and applying genetic
Images in Cardiovascular Medicine
 Images in Cardiovascular Medicine Extensive Arterial Tortuosity and Severe Aortic Dilation in a Newborn With an EFEMP2 Mutation Maria Iascone, BSc, PhD; Maria Elena Sana, PhD; Laura Pezzoli, BSc; Paolo
Images in Cardiovascular Medicine Extensive Arterial Tortuosity and Severe Aortic Dilation in a Newborn With an EFEMP2 Mutation Maria Iascone, BSc, PhD; Maria Elena Sana, PhD; Laura Pezzoli, BSc; Paolo
Merging single gene-level CNV with sequence variant interpretation following the ACMGG/AMP sequence variant guidelines
 Merging single gene-level CNV with sequence variant interpretation following the ACMGG/AMP sequence variant guidelines Tracy Brandt, Ph.D., FACMG Disclosure I am an employee of GeneDx, Inc., a wholly-owned
Merging single gene-level CNV with sequence variant interpretation following the ACMGG/AMP sequence variant guidelines Tracy Brandt, Ph.D., FACMG Disclosure I am an employee of GeneDx, Inc., a wholly-owned
Molecular Diagnostic Laboratory 18 Sequencing St, Gene Town, ZY Tel: Fax:
 Molecular Diagnostic Laboratory 18 Sequencing St, Gene Town, ZY 01234 Tel: 555-920-3333 Fax: 555-920-3334 www.moldxlaboratory.com Patient Name: Jane Doe Specimen type: Blood, peripheral DOB: 04/05/1990
Molecular Diagnostic Laboratory 18 Sequencing St, Gene Town, ZY 01234 Tel: 555-920-3333 Fax: 555-920-3334 www.moldxlaboratory.com Patient Name: Jane Doe Specimen type: Blood, peripheral DOB: 04/05/1990
AD (Leave blank) TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients
 AD (Leave blank) Award Number: W81XWH-12-1-0444 TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients PRINCIPAL INVESTIGATOR: Mark A. Watson, MD PhD CONTRACTING ORGANIZATION:
AD (Leave blank) Award Number: W81XWH-12-1-0444 TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients PRINCIPAL INVESTIGATOR: Mark A. Watson, MD PhD CONTRACTING ORGANIZATION:
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier/Additional Provider
 Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier/Additional Provider TEST DISORDER/CONDITION POPULATION TRIAD Submitting laboratory: Exeter RGC Approved: Sept 2013 1. Disorder/condition
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier/Additional Provider TEST DISORDER/CONDITION POPULATION TRIAD Submitting laboratory: Exeter RGC Approved: Sept 2013 1. Disorder/condition
Variant Classification. Author: Mike Thiesen, Golden Helix, Inc.
 Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Human Genetic Disorders
 Human Genetic Disorders HOMOLOGOUS CHROMOSOMES Human somatic cells have 23 pairs of homologous chromosomes 23 are inherited from the mother and 23 from the father HOMOLOGOUS CHROMOSOMES Autosomes o Are
Human Genetic Disorders HOMOLOGOUS CHROMOSOMES Human somatic cells have 23 pairs of homologous chromosomes 23 are inherited from the mother and 23 from the father HOMOLOGOUS CHROMOSOMES Autosomes o Are
Title:Exome sequencing helped the fine diagnosis of two siblings afflicted with atypical Timothy syndrome (TS2)
 Author's response to reviews Title:Exome sequencing helped the fine diagnosis of two siblings afflicted with atypical Timothy syndrome (TS2) Authors: Sebastian Fröhler (Sebastian.Froehler@mdc-berlin.de)
Author's response to reviews Title:Exome sequencing helped the fine diagnosis of two siblings afflicted with atypical Timothy syndrome (TS2) Authors: Sebastian Fröhler (Sebastian.Froehler@mdc-berlin.de)
Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
 Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Introduction to genetic variation. He Zhang Bioinformatics Core Facility 6/22/2016
 Introduction to genetic variation He Zhang Bioinformatics Core Facility 6/22/2016 Outline Basic concepts of genetic variation Genetic variation in human populations Variation and genetic disorders Databases
Introduction to genetic variation He Zhang Bioinformatics Core Facility 6/22/2016 Outline Basic concepts of genetic variation Genetic variation in human populations Variation and genetic disorders Databases
MEDICAL GENOMICS LABORATORY. Non-NF1 RASopathy panel by Next-Gen Sequencing and Deletion/Duplication Analysis of SPRED1 (NNP-NG)
 Non-NF1 RASopathy panel by Next-Gen Sequencing and Deletion/Duplication Analysis of SPRED1 (NNP-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with
Non-NF1 RASopathy panel by Next-Gen Sequencing and Deletion/Duplication Analysis of SPRED1 (NNP-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies
 Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
TITLE: - Whole Genome Sequencing of High-Risk Families to Identify New Mutational Mechanisms of Breast Cancer Predisposition
 AD Award Number: W81XWH-13-1-0336 TITLE: - Whole Genome Sequencing of High-Risk Families to Identify New Mutational Mechanisms of Breast Cancer Predisposition PRINCIPAL INVESTIGATOR: Mary-Claire King,
AD Award Number: W81XWH-13-1-0336 TITLE: - Whole Genome Sequencing of High-Risk Families to Identify New Mutational Mechanisms of Breast Cancer Predisposition PRINCIPAL INVESTIGATOR: Mary-Claire King,
Advances in genetic diagnosis of neurological disorders
 Acta Neurol Scand 2014: 129 (Suppl. 198): 20 25 DOI: 10.1111/ane.12232 2014 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd ACTA NEUROLOGICA SCANDINAVICA Review Article Advances in genetic diagnosis
Acta Neurol Scand 2014: 129 (Suppl. 198): 20 25 DOI: 10.1111/ane.12232 2014 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd ACTA NEUROLOGICA SCANDINAVICA Review Article Advances in genetic diagnosis
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier
 Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier Test Disease Population Triad Disease name Choroideremia OMIM number for disease 303100 Disease alternative names please
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier Test Disease Population Triad Disease name Choroideremia OMIM number for disease 303100 Disease alternative names please
NGS for Cancer Predisposition
 NGS for Cancer Predisposition Colin Pritchard MD, PhD University of Washington Dept. of Lab Medicine AMP Companion Society Meeting USCAP Boston March 22, 2015 Disclosures I am an employee of the University
NGS for Cancer Predisposition Colin Pritchard MD, PhD University of Washington Dept. of Lab Medicine AMP Companion Society Meeting USCAP Boston March 22, 2015 Disclosures I am an employee of the University
Germline Testing for Hereditary Cancer with Multigene Panel
 Germline Testing for Hereditary Cancer with Multigene Panel Po-Han Lin, MD Department of Medical Genetics National Taiwan University Hospital 2017-04-20 Disclosure No relevant financial relationships with
Germline Testing for Hereditary Cancer with Multigene Panel Po-Han Lin, MD Department of Medical Genetics National Taiwan University Hospital 2017-04-20 Disclosure No relevant financial relationships with
Personalis ACE Clinical Exome The First Test to Combine an Enhanced Clinical Exome with Genome- Scale Structural Variant Detection
 Personalis ACE Clinical Exome The First Test to Combine an Enhanced Clinical Exome with Genome- Scale Structural Variant Detection Personalis, Inc. 1350 Willow Road, Suite 202, Menlo Park, California 94025
Personalis ACE Clinical Exome The First Test to Combine an Enhanced Clinical Exome with Genome- Scale Structural Variant Detection Personalis, Inc. 1350 Willow Road, Suite 202, Menlo Park, California 94025
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits
 AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
Using large-scale human genetic variation to inform variant prioritization in neuropsychiatric disorders
 Using large-scale human genetic variation to inform variant prioritization in neuropsychiatric disorders Kaitlin E. Samocha Hurles lab, Wellcome Trust Sanger Institute ACGS Summer Scientific Meeting 27
Using large-scale human genetic variation to inform variant prioritization in neuropsychiatric disorders Kaitlin E. Samocha Hurles lab, Wellcome Trust Sanger Institute ACGS Summer Scientific Meeting 27
Reporting TP53 gene analysis results in CLL
 Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Molecular Diagnosis of Genetic Diseases: From 1 Gene to 1000s
 ROMA, 23 GIUGNO 2011 Molecular Diagnosis of Genetic Diseases: From 1 Gene to 1000s USSD Lab Genetica Medica Ospedali Riuniti Bergamo The problem What does the clinician/patient want to know?? Diagnosis,
ROMA, 23 GIUGNO 2011 Molecular Diagnosis of Genetic Diseases: From 1 Gene to 1000s USSD Lab Genetica Medica Ospedali Riuniti Bergamo The problem What does the clinician/patient want to know?? Diagnosis,
A Likelihood-Based Framework for Variant Calling and De Novo Mutation Detection in Families
 A Likelihood-Based Framework for Variant Calling and De Novo Mutation Detection in Families Bingshan Li 1 *, Wei Chen 2, Xiaowei Zhan 3, Fabio Busonero 3,4, Serena Sanna 4, Carlo Sidore 4, Francesco Cucca
A Likelihood-Based Framework for Variant Calling and De Novo Mutation Detection in Families Bingshan Li 1 *, Wei Chen 2, Xiaowei Zhan 3, Fabio Busonero 3,4, Serena Sanna 4, Carlo Sidore 4, Francesco Cucca
Nature Genetics: doi: /ng Supplementary Figure 1. Country distribution of GME samples and designation of geographical subregions.
 Supplementary Figure 1 Country distribution of GME samples and designation of geographical subregions. GME samples collected across 20 countries and territories from the GME. Pie size corresponds to the
Supplementary Figure 1 Country distribution of GME samples and designation of geographical subregions. GME samples collected across 20 countries and territories from the GME. Pie size corresponds to the
Molecular Characterization of the NF2 Gene in Korean Patients with Neurofibromatosis Type 2: A Report of Four Novel Mutations
 Korean J Lab Med 2010;30:190-4 DOI 10.3343/kjlm.2010.30.2.190 Original Article Diagnostic Genetics Molecular Characterization of the NF2 Gene in Korean Patients with Neurofibromatosis Type 2: A Report
Korean J Lab Med 2010;30:190-4 DOI 10.3343/kjlm.2010.30.2.190 Original Article Diagnostic Genetics Molecular Characterization of the NF2 Gene in Korean Patients with Neurofibromatosis Type 2: A Report
Benefits and pitfalls of new genetic tests
 Benefits and pitfalls of new genetic tests Amanda Krause Division of Human Genetics, NHLS and University of the Witwatersrand Definition of Genetic Testing the analysis of human DNA, RNA, chromosomes,
Benefits and pitfalls of new genetic tests Amanda Krause Division of Human Genetics, NHLS and University of the Witwatersrand Definition of Genetic Testing the analysis of human DNA, RNA, chromosomes,
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
BWA alignment to reference transcriptome and genome. Convert transcriptome mappings back to genome space
 Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Basic Definitions. Dr. Mohammed Hussein Assi MBChB MSc DCH (UK) MRCPCH
 Basic Definitions Chromosomes There are two types of chromosomes: autosomes (1-22) and sex chromosomes (X & Y). Humans are composed of two groups of cells: Gametes. Ova and sperm cells, which are haploid,
Basic Definitions Chromosomes There are two types of chromosomes: autosomes (1-22) and sex chromosomes (X & Y). Humans are composed of two groups of cells: Gametes. Ova and sperm cells, which are haploid,
Clinical Genetics. Functional validation in a diagnostic context. Robert Hofstra. Leading the way in genetic issues
 Clinical Genetics Leading the way in genetic issues Functional validation in a diagnostic context Robert Hofstra Clinical Genetics Leading the way in genetic issues Future application of exome sequencing
Clinical Genetics Leading the way in genetic issues Functional validation in a diagnostic context Robert Hofstra Clinical Genetics Leading the way in genetic issues Future application of exome sequencing
Illuminating the genetics of complex human diseases
 Illuminating the genetics of complex human diseases Michael Schatz Sept 27, 2012 Beyond the Genome @mike_schatz / #BTG2012 Outline 1. De novo mutations in human diseases 1. Autism Spectrum Disorder 2.
Illuminating the genetics of complex human diseases Michael Schatz Sept 27, 2012 Beyond the Genome @mike_schatz / #BTG2012 Outline 1. De novo mutations in human diseases 1. Autism Spectrum Disorder 2.
Corporate Medical Policy
 Corporate Medical Policy Genetic Testing for Duchenne and Becker Muscular Dystrophy File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_duchenne_and_becker_muscular_dystrophy
Corporate Medical Policy Genetic Testing for Duchenne and Becker Muscular Dystrophy File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_duchenne_and_becker_muscular_dystrophy
A. Incorrect! Cells contain the units of genetic they are not the unit of heredity.
 MCAT Biology Problem Drill PS07: Mendelian Genetics Question No. 1 of 10 Question 1. The smallest unit of heredity is. Question #01 (A) Cell (B) Gene (C) Chromosome (D) Allele Cells contain the units of
MCAT Biology Problem Drill PS07: Mendelian Genetics Question No. 1 of 10 Question 1. The smallest unit of heredity is. Question #01 (A) Cell (B) Gene (C) Chromosome (D) Allele Cells contain the units of
Variant Annotation and Functional Prediction
 Variant Annotation and Functional Prediction Copyrighted 2018 Isabelle Schrauwen and Suzanne M. Leal This exercise touches on several functionalities of the program ANNOVAR to annotate and interpret candidate
Variant Annotation and Functional Prediction Copyrighted 2018 Isabelle Schrauwen and Suzanne M. Leal This exercise touches on several functionalities of the program ANNOVAR to annotate and interpret candidate
VARIANT PRIORIZATION AND ANALYSIS INCORPORATING PROBLEMATIC REGIONS OF THE GENOME ANIL PATWARDHAN
 VARIANT PRIORIZATION AND ANALYSIS INCORPORATING PROBLEMATIC REGIONS OF THE GENOME ANIL PATWARDHAN Email: apatwardhan@personalis.com MICHAEL CLARK Email: michael.clark@personalis.com ALEX MORGAN Email:
VARIANT PRIORIZATION AND ANALYSIS INCORPORATING PROBLEMATIC REGIONS OF THE GENOME ANIL PATWARDHAN Email: apatwardhan@personalis.com MICHAEL CLARK Email: michael.clark@personalis.com ALEX MORGAN Email:
PROGRESS: Beginning to Understand the Genetic Predisposition to PSC
 PROGRESS: Beginning to Understand the Genetic Predisposition to PSC Konstantinos N. Lazaridis, MD Associate Professor of Medicine Division of Gastroenterology and Hepatology Associate Director Center for
PROGRESS: Beginning to Understand the Genetic Predisposition to PSC Konstantinos N. Lazaridis, MD Associate Professor of Medicine Division of Gastroenterology and Hepatology Associate Director Center for
TOWARDS ACCURATE GERMLINE AND SOMATIC INDEL DISCOVERY WITH MICRO-ASSEMBLY. Giuseppe Narzisi, PhD Bioinformatics Scientist
 TOWARDS ACCURATE GERMLINE AND SOMATIC INDEL DISCOVERY WITH MICRO-ASSEMBLY Giuseppe Narzisi, PhD Bioinformatics Scientist July 29, 2014 Micro-Assembly Approach to detect INDELs 2 Outline 1 Detecting INDELs:
TOWARDS ACCURATE GERMLINE AND SOMATIC INDEL DISCOVERY WITH MICRO-ASSEMBLY Giuseppe Narzisi, PhD Bioinformatics Scientist July 29, 2014 Micro-Assembly Approach to detect INDELs 2 Outline 1 Detecting INDELs:
Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep
 Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep Tom Walsh, PhD Division of Medical Genetics University of Washington Next generation sequencing Sanger sequencing gold
Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep Tom Walsh, PhD Division of Medical Genetics University of Washington Next generation sequencing Sanger sequencing gold
Supplemental Data: Detailed Characteristics of Patients with MKRN3. Patient 1 was born after an uneventful pregnancy. She presented in our
 1 2 Supplemental Data: Detailed Characteristics of Patients with MKRN3 Mutations 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 Patient 1 was born after an uneventful pregnancy. She presented
1 2 Supplemental Data: Detailed Characteristics of Patients with MKRN3 Mutations 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 Patient 1 was born after an uneventful pregnancy. She presented
No mutations were identified.
 Hereditary High Cholesterol Test ORDERING PHYSICIAN PRIMARY CONTACT SPECIMEN Report date: Aug 1, 2017 Dr. Jenny Jones Sample Medical Group 123 Main St. Sample, CA Kelly Peters Sample Medical Group 123
Hereditary High Cholesterol Test ORDERING PHYSICIAN PRIMARY CONTACT SPECIMEN Report date: Aug 1, 2017 Dr. Jenny Jones Sample Medical Group 123 Main St. Sample, CA Kelly Peters Sample Medical Group 123
MEDICAL GENOMICS LABORATORY. Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG)
 Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with receipt) Saliva (OGR-575 DNA Genotek;
Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with receipt) Saliva (OGR-575 DNA Genotek;
THIAMINE TRANSPORTER TYPE 2 DEFICIENCY
 THIAMINE TRANSPORTER TYPE 2 DEFICIENCY WHAT IS THE THIAMINE TRANSPORTER TYPE 2 DEFICIENCY (hthtr2)? The thiamine transporter type 2 deficiency (hthtr2) is a inborn error of thiamine metabolism caused by
THIAMINE TRANSPORTER TYPE 2 DEFICIENCY WHAT IS THE THIAMINE TRANSPORTER TYPE 2 DEFICIENCY (hthtr2)? The thiamine transporter type 2 deficiency (hthtr2) is a inborn error of thiamine metabolism caused by
Exam #2 BSC Fall. NAME_Key correct answers in BOLD FORM A
 Exam #2 BSC 2011 2004 Fall NAME_Key correct answers in BOLD FORM A Before you begin, please write your name and social security number on the computerized score sheet. Mark in the corresponding bubbles
Exam #2 BSC 2011 2004 Fall NAME_Key correct answers in BOLD FORM A Before you begin, please write your name and social security number on the computerized score sheet. Mark in the corresponding bubbles
LEIDEN, THE NETHERLANDS
 Full-length CYP2D6 diplotyping for better drug dosage and response management Henk Buermans, PhD Leiden University Medical Center Human Genetics, LGTC LEIDEN, THE NETHERLANDS CYP2D6 Function Metabolism
Full-length CYP2D6 diplotyping for better drug dosage and response management Henk Buermans, PhD Leiden University Medical Center Human Genetics, LGTC LEIDEN, THE NETHERLANDS CYP2D6 Function Metabolism
Lecture 17: Human Genetics. I. Types of Genetic Disorders. A. Single gene disorders
 Lecture 17: Human Genetics I. Types of Genetic Disorders A. Single gene disorders B. Multifactorial traits 1. Mutant alleles at several loci acting in concert C. Chromosomal abnormalities 1. Physical changes
Lecture 17: Human Genetics I. Types of Genetic Disorders A. Single gene disorders B. Multifactorial traits 1. Mutant alleles at several loci acting in concert C. Chromosomal abnormalities 1. Physical changes
Pedigree Analysis Why do Pedigrees? Goals of Pedigree Analysis Basic Symbols More Symbols Y-Linked Inheritance
 Pedigree Analysis Why do Pedigrees? Punnett squares and chi-square tests work well for organisms that have large numbers of offspring and controlled mating, but humans are quite different: Small families.
Pedigree Analysis Why do Pedigrees? Punnett squares and chi-square tests work well for organisms that have large numbers of offspring and controlled mating, but humans are quite different: Small families.
