Functional genomics identifies a requirement of pre-mrna splicing factors for sister chromatid cohesion
|
|
|
- Job Sullivan
- 5 years ago
- Views:
Transcription
1 The EMBO Journal Peer Review Process File - EMBO Manuscript EMBO Functional genomics identifies a requirement of pre-mrna splicing factors for sister chromatid cohesion Sriramkumar Sundaramoorthy, María Dolores Vázquez-Novelle, Sergey Lekomtsev, Michael Howell and Mark Petronczki Corresponding author: Mark Petronczki, Cancer Research UK London Research Institute Review timeline: Submission date: 14 February 2014 Editorial Decision: 01 April 2014 Revision received: 03 August 2014 Accepted: 12 August 2014 Transaction Report: (Note: With the exception of the correction of typographical or spelling errors that could be a source of ambiguity, letters and reports are not edited. The original formatting of letters and referee reports may not be reflected in this compilation.) Editor: Anne Nielsen 1st Editorial Decision 01 April 2014 Thank you for submitting your manuscript for consideration by The EMBO Journal and my apologies for the unusual duration of the review period in this case. Your study has now been seen by three referees whose comments are shown below. As you will see from the reports, all referees express interest in the findings reported in your manuscript and emphasize the high technical quality of the experimental data. However, while ref #1 and ref #2 consequently favour publication following minor revisions, ref #3 is more critical and questions the overall novelty and specificity of findings. In light of these rather divergent recommendations, we had an additional round of consultation with all three refs to specifically address the conceptual concerns raised. The outcome of these discussions is that while splicing in general may be expected to impact on numerous biological pathways (as pointed out by ref #3), the main finding of your work is the surprisingly specific effect on sororin - as acknowledged by all three referees and strongly supported by refs #1 and #2 in these additional discussions. We would therefore be happy to pursue publication of an adequately revised version of your manuscript. We realize that some of the suggestions made by ref #3 may be outside the scope of the present work, but we would ask you to focus your efforts on the following points: -> Please include controls for knockdown efficiency (ref#1) -> Please comment on the general correlation between observed phenotypes and sororin expression EMBO 1
2 The EMBO Journal Peer Review Process File - EMBO levels (refs #1 and #2) -> Please provide further experimental data to address the mechanistic basis for the high sororin sensitivity to altered splicing (refs #2 and #3) by -> assessing mrna transcription and turnover rates for sororin and controls -> testing if splicing sensitivity is correlated with specific sequence features -> addressing splicing sensitivity for other cohesin factors -> In addition, please discuss/address all minor issues raised by all three referees Given the referees' positive recommendations, we offer you the opportunity to submit a revised version of the manuscript, addressing the comments of all three reviewers. I should add that it is EMBO Journal policy to allow only a single round of revision, and acceptance or rejection of your manuscript will therefore depend on the completeness of your responses in this revised version. Please do not hesitate to contact me if you have questions related to the review process and the requests made by the referees. When preparing your letter of response to the referees' comments, please bear in mind that this will form part of the Review Process File, and will therefore be available online to the community. For more details on our Transparent Editorial Process, please visit our website: We generally allow three months as standard revision time. As a matter of policy, competing manuscripts published during this period will not negatively impact on our assessment of the conceptual advance presented by your study. However, we request that you contact the editor as soon as possible upon publication of any related work, to discuss how to proceed. Should you foresee a problem in meeting this three-month deadline, please let us know in advance and we may be able to grant an extension. Thank you for the opportunity to consider your work for publication. I look forward to your revision. REFEREE REPORTS: Referee #1: Based on RNAi screening, the authors found that the depletion of MFAP1 causes abnormal nuclear morphology, a hallmark of the defect in mitotic progression. They confirmed that mitotic progression is defective due to a cohesion defect in MFAP1-depleted cells. Other splicing factors such as SART1 also cause a similar phenotype. Moreover, the authors show that the cohesion defect is detected in interphase, and that the stability of the cohesin-chromatin interaction becomes less stable in MFAP1-depleted cells. They examined the expression of sororin, which maintains the stable association of cohesin with chromatin until mitosis. The sororin protein levels are decreased in MFAP1-depleted cells due to a defect in the splicing of the sororin pre-mrna. Cells expressing an intronless version of sororin rescue the cohesion defect seen in MFAP1-depleted cells. Furthermore, the depletion of Wapl, a counteracter of sororin, also rescues the phenotype. Thus, the authors conclude that splicing factors are required for the maintenance of cohesion and faithful mitotic progression, and that the splicing of the sororin mrna is the critical target for this pathway. Overall, the work in the paper is of high quality, and for the most part supports the important conclusions reported. Comments; 1) Depletion of splicing factors and Wapl should be examined by immunoblot. (Fig.3 and Fig. 8). 2) In general the phenotype caused by RNAi looks very strong (~100%). Please explain this measurement in detail. EMBO 2
3 The EMBO Journal Peer Review Process File - EMBO Referee #2: The manuscript by Sundaarmoorthy et al. describes the relevance of multiple pre-mrna splicing factors for sister chromatid cohesion. Although there were previous reported evidences for mitotic defects induced by defective splicing, the molecular reason for these observations was not clear. The manuscripts by Petronczki's and Peters' groups make a strong link between the splicing machinery and the protein levels of sororin as a consequence of aberrant splicing. The study by Sundaarmoorthy et al therefore addresses a very important issue and the results are exciting, novel and of high quality. In general, the manuscript is scientifically sound and technically very solid. The analyses are meticulously performed and the take-home message is very significant for our understanding on the links between gene transcription, splicing, chromosome segregation and, perhaps, cancer biology. Hence, I strongly recommend publication. I only have some minor comments that the authors may consider to improve the manuscript. How the phenotypes observed in Fig. 1B compare to downregulation of sororin? It seems from this panel that the cell fate after MFAP1 sirna is very diverse. Did cells die also in interphase? It is difficult to see single chromatids in Fig. 3B (/b etc.) and it will be even worst in the reduced published figure so adding insets may help to understand the phenotype. Some insets sin other panels (e.g. 4A) could also be enlarged. Statistics are missing in some histograms (Fig. 6). Why sororin? Is it any clue from the analysis about which specific sequences make the sororin gene susceptible to the lack of splicing factors? Again, these are minor points and do not diminish my enthusiasm for this study. Referee #3: In this study the authors aimed at identifying new factors involved in mitosis. Based on published datasets, the authors established a list of 718 potential candidates whose involvement in mitosis were further tested by a systematic sirna-based screen. From 718 candidates the list shrunk to 50 genes whose sirna appeared to have a real impact on nuclear morphology. This list was further reduced to 7 confirmed genes whose depletion had a consistent effect on nuclear shape. From this list, the depletion of the splicing factor MFAP1 resulted in particularly severe nuclear fragmentation. By using imaging techniques, the authors determined that in cells lacking MFAP1, chromosomes could not be aligned at the metaphase plate due to a loss of sister chromatid cohesion, suggesting that MFAP1 could have a function in sister chromatid cohesion in addition to its well characterized role as core pre-mrna splicing factor. However, by performing sirna-mediated depletions, the authors identified 18 other splicing factors that had a similar effect on chromatid cohesion as that observed upon MFAP1 depletion. Thus, the authors convincingly established a general link between pre-mrna splicing and chromatid cohesion, but apparently no specific or direct effect of MFAP1 on mitosis. Further experiments excluded a potential defect in cohesin loading onto chromatin, but demonstrated that loss of spliceosome components resulted in a specific and dramatic decrease in the essential chromatid cohesion factor sororin. Consistent with a requirement for pre-mrna splicing, complementation experiments in which sororin was expressed from an intronless transgene restored sister chromatid cohesion in cells depleted of splicing factors. This study demonstrates that the integrity of the spliceosome is required to maintain an appropriate level of spliced sororin mrna and its encoded protein sororin, which plays a role in chromatid cohesion. This study is well written and understandable to a large audience. The experimental procedures are well described and controlled. However, spliceosomal factors are essential for virtually all cellular processes, as splicing is an upstream event in gene expression and literally controls the expression of nearly all genes in mammalian cells. Therefore, the question addressed in this study does not represent a significant step forward in our understanding of the chromatid cohesion process, in addition to failing to identify new factors directly involved in mitosis. Thus, this study is not suitable for publication in EMBO J. in its present form. EMBO 3
4 The EMBO Journal Peer Review Process File - EMBO An interesting finding in this study is that the protein levels of the essential chromatid cohesion factors SMC1, SMC3, SCC1 and SA2 are not altered upon sirna depletion of the essential splicing factors NHP2L1, MFAP1, SART1 and CDC5L. Only the level of the sororin protein appears to be reduced. One obvious explanation would be that sororin has a high rate of transcription and protein turnover, which would explain its acute sensitivity to a reduction in splicing activity. However this point is not investigated and only briefly mentioned in the discussion. This manuscript would greatly benefit if further experiments providing insight into how stalling a general process like splicing appears to affect only one chromatid cohesion factor, i.e. sororin. Therefore I would greatly encourage the authors to determine sororin's half-life and its transcription rate, and determine whether this provides a satisfying explanation for sororin's exceptional sensitivity towards a general reduction in splicing. In addition the authors could compare the effect of depletion of spliceosome components on sororin to other known short lived proteins. Moreover, the authors should investigate whether only the splicing of sororin is blocked upon NHP2L1, MFAP1, SART1 and CDC5L depletion or whether the splicing of the other cohesion factors SMC1, SMC3, SCC1 and SA2 is also affected. In summary, any experiment that would help us understand the specific effect observed on sororin would greatly improve the significance and general interest of this manuscript. 2. Page 5: For 7 genes, two or more out of four sirna duplexes recapitulated the phenotype observed in the primary screen (Supplementary Table 2). This suggests that many of 50 hits selected from the primary screen were caused by off-target depletion effects. These are not necessarily off-target effects. A single RNAi duplex does not always inhibit efficiently gene expression. 3. Page 9: Like MFAP1, NHP2L1 and SART1 are components of the U5 and U4/U6 tri-snrnp complex MFAP1 is not a component of tri-snrnp but rather a non-snrnp, protein factor that joins the spliceosome at the spliceosomal B complex stage. 4. Page9:...Prp19 complex that promotes the catalytic activation of the spliceosome for cleavage at the 5' splicing site This should be rephrased to "...Prp19 complex required for the activation of the spliceosome before the first step of the splicing reaction" 5. Fig. 1B vs Fig. S1: could the authors explain the difference in penetrance observed. In Fig. 1B only 16 out of 50 cells successfully passed cell division while in Fig. S1 the defects were observed in only ~25% cells. 6. Fig. 4C: a loading control is missing 7. Fig. 6A: it seems that sirna-mediated depletion of sororin leads to co-depletion of SCC1. Does sororin affect the level of SCC1? Why don't CDC5L cells, which have a dramatically reduced level of sororin, show any reduction in SCC1? 8. Fig. 6C: in contrast to MFAP1 and NHPL2L1, SART1 and CDC5L cells show a two-fold reduction in the level of Exon2 of sororin (pre)-mrna. Is this a significant reproducible change? If so, do these cells have a transcription related defect (in the case of the sororin gene)? What is the steady state ratio of the sororin pre-mrna to its mrna? What is the half-life of sororin mrna? 9. Fig. 7 shows much milder effects of splicing factor knockdown as compared to Fig. 3 and Fig. S2, even in the case of CDC5L for which RNAi treatment was performed for nearly the same time. Could the authors comment on this. 10. Fig. 8B: the cartoon is confusing. "Premature loss of chromatid cohesion" should be next to free cohesion, right? Please indicate in the legend the subunits of the cohesin "triangles". 11. Page 12 (and Page 16 of Discussion): Expression of the intronless sororin transgene did not however restore mitotic progression in cells transfected with sirnas targeting spliceosome subunits... It is not clear based on which data this conclusion has been made. 12. Page 17: in the last paragraph of the Discussion the authors describe the potential biomedical implications of their findings, in particular to chronic lymphocytic leukaemia. Defects in the splicing factors described in this manuscript lead to mitotic arrest or cell death rather than uncontrolled cell proliferation characteristic for chronic lymphocytic leukaemia. Please clarify this point. 1st Revision - authors' response 03 August 2014 EMBO 4
5 Response to referees comments EMBOJ We would like to thank all reviewers for their comments, suggestions and criticism. To address the comments, we have added new experiments and Figures to the revised version of our manuscript and we have made changes to the manuscript text. Also we include an additional figure at the end of this document. Please find below a point-bypoint response to the reviewers comments. Referee #1: Based on RNAi screening, the authors found that the depletion of MFAP1 causes abnormal nuclear morphology, a hallmark of the defect in mitotic progression. They confirmed that mitotic progression is defective due to a cohesion defect in MFAP1- depleted cells. Other splicing factors such as SART1 also cause a similar phenotype. Moreover, the authors show that the cohesion defect is detected in interphase, and that the stability of the cohesin-chromatin interaction becomes less stable in MFAP1- depleted cells. They examined the expression of sororin, which maintains the stable association of cohesin with chromatin until mitosis. The sororin protein levels are decreased in MFAP1-depleted cells due to a defect in the splicing of the sororin premrna. Cells expressing an intronless version of sororin rescue the cohesion defect seen in MFAP1-depleted cells. Furthermore, the depletion of Wapl, a counteracter of sororin, also rescues the phenotype. Thus, the authors conclude that splicing factors are required for the maintenance of cohesion and faithful mitotic progression, and that the splicing of the sororin mrna is the critical target for this pathway. Overall, the work in the paper is of high quality, and for the most part supports the important conclusions reported. Comments; 1) Depletion of splicing factors and Wapl should be examined by immunoblot. (Fig.3 and Fig. 8). In the revised version of our manuscript we have added immunoblots probing the depletion of WAPL and that of splicing factors, against which antibodies were available. These new data are now included as Expanded View Figure E3A and E8B. Furthermore, the WAPL + splicing factor co-depletion experiment aimed at testing suppression of cohesion defects was repeated. Figure 8A now displays the mean values for cohesion status from the 2 repeats. 2) In general the phenotype caused by RNAi looks very strong (~100%). Please explain this measurement in detail. The depletion of several factors (incl. sororin, SGOL1, SCC1, CDC5L, ) causes highly penetrant defects in sister chromatid cohesion as detected by chromosome spread assays. In our experience, highly penetrant cell division phenotypes can be achieved by reverse transfection of sirna duplexes into human cells depending on the function and protein level threshold requirement of the particular factor targeted (e.g.: Lekomtsev et al., Naure 2012; Su et al., DevCell 2011). Chromosome spreading from mitotic cells is likely to exacerbate cohesion defects to a small degree as even in control cells around 10% of spread karyotypes can display a loss of cohesion. To address, this we conducted FISH experiments in intact mitotic cells following MFAP1 depletion (Fig. 1B). These
6 experiments demonstrated that centromeres of sister chromatids were separated in mitotic cells even without the procedure of chromosome spreading. In our manuscript we have followed the conventional classification of sister chromatid cohesion status into three categories of increasing phenotypic severity: normal (X-shaped), parallel chromatids without connection at central construction, and fully split (no alignment of chromatids observed). For each experiment or experimental repeat, 100 spread karyotypes (derived from 100 cells) were assessed (as detailed in Figure legends) and classified according to the aforementioned categories. Within the karyotype/chromosome spreads of a single cell, the vast majority of chromatids or chromosomes typically conforms to one of the categories and the spread was therefore classified accordingly. A value of e.g. 90% split sister chromatids in graphs is the result of 100 observed karyotypes displaying no association between sister chromatids. For percentages of split sister chromatids specified in micrographs, the categories of karyotypes with parallel and fully split chromatids were combined. We agree that this could be confusing and thank the reviewer for pointing this out. To clarify this point we have added a short description to the material and methods section of the revised manuscript. Also we have changed the wording for in the % associated with micrographs to % spreads with separated chromatids. Referee #2: The manuscript by Sundaarmoorthy et al. describes the relevance of multiple premrna splicing factors for sister chromatid cohesion. Although there were previous reported evidences for mitotic defects induced by defective splicing, the molecular reason for these observations was not clear. The manuscripts by Petronczki's and Peters' groups make a strong link between the splicing machinery and the protein levels of sororin as a consequence of aberrant splicing. The study by Sundaarmoorthy et al therefore addresses a very important issue and the results are exciting, novel and of high quality. In general, the manuscript is scientifically sound and technically very solid. The analyses are meticulously performed and the take-home message is very significant for our understanding on the links between gene transcription, splicing, chromosome segregation and, perhaps, cancer biology. Hence, I strongly recommend publication. I only have some minor comments that the authors may consider to improve the manuscript. How the phenotypes observed in Fig. 1B compare to downregulation of sororin? It seems from this panel that the cell fate after MFAP1 sirna is very diverse. Did cells die also in interphase? To compare the effect of loss of sororin to the effect of loss of MFAP1, we have analyzed the fate of cells and their mitotic phenotype by live-cell imaging following transfection of sororin sirna. The cell fate data and mean mitotic duration following sororin depletion are presented in a Figure for Referees that has been attached at the end of this letter. As shown in the Figure, loss of sororin causes a fully penetrant mitotic arrest with 70% of cells undergoing cell death during the arrest and the remaining cells exiting mitosis in an aberrant fashion. The mean time of sororin-depleted cells spent in mitosis was 678 min compared to 510 min for loss of MFAP1 (control 46 min) (Fig. 1B). Thus, the loss of sororin leads to a slightly more uniform and penetrant mitotic defect than
7 MFAP1 loss. The effect of MFAP1 loss is more pleiotropic due to the protein s role in splicing. Nevertheless, loss of MFAP1 (and other splicing factors) causes a mitotic phenotype consistent with a reduction in sororin levels, with the majority of cells dying during a mitotic arrest or exiting mitosis in an aberrant fashion following a protracted arrest. The terminal phenotype of loss of MFAP1 (and other splicing factors) is cell death. This is presumably caused by combination of defective mitosis and gene expression failure. A minor fraction (10%) of MFAP1-depleted cells died prior to entry into mitosis. For mitotic duration analysis only cells entering mitosis were scored. In summary, our data suggest that loss of MFAP1 causes a more pleiotropic defect than depletion of sororin finally resulting in cell death. However, prior to cell death the vast majority of MFAP1-depleted cells arrest in mitosis with split sister chromatids in a mitotic checkpoint-dependent manner (Figure 1). It is difficult to see single chromatids in Fig. 3B (/b etc.) and it will be even worst in the reduced published figure so adding insets may help to understand the phenotype. Some insets sin other panels (e.g. 4A) could also be enlarged. We thank the reviewer for pointing this out. In the revised version of the manuscript we have included enlarged insets of chromosomes/chromatids for Figure panels 2A, 2C, 3B and 8A. Furthermore, we have enlarged the insets of FISH signals shown in Figure 4A. Statistics are missing in some histograms (Fig. 6). For the revised version of the manuscript, we have repeated the immunoblot analysis shown in Fig. 6A and we have conducted a new qrt-pcr analysis (three biological repeats) of intron retention for sororin and core cohesin subunit RNAs following depletion of splicing factors. Figure 6 now shows the mean of the data obtained from the biological repeats and contains error for standard deviation. Why sororin? Is it any clue from the analysis about which specific sequences make the sororin gene susceptible to the lack of splicing factors? We have explored this interesting question. Analysis of 5 and 3 splice site sequences (MaxEntScan splice site score algorithm; and the presumptive position of the branch site with regard to the 3 splice site in of the first intron of sororin did not reveal particular features that would explain a pronounced sensitivity of sororin pre-mrna splicing to the depletion of spliceosomes. In the future, global analysis of spliceosome assembly and branch site usage may be able to address this question if correlated with aberrant intron retention upon loss of specific spliceosome components. Based on the transcriptome-wide analysis of intron retention in the accompanying manuscript by the Peters lab, it is clear that depletion of the splicing factor SNW1 affects intron removal in a number of genes thus suggesting that this effect is not unique to sororin. However, amongst the factors essential for cohesion, depletion of spliceosome subunits had the strongest impact on intron retention in sororin RNA (Fig. 6B). In the revised version of the manuscript we show that both sororin protein and mrna have a higher turnover than other cohesin subunit (Fig 6C and E7). In G1 cells sororin protein has an extraordinary short half-life of less than 2 hours (Fig. 6C and E7B), consistent with it having been identified as an APC/C substrate (Rankin et al., 2005). This short protein half-life sets sororin apart from other cohesin proteins. The high turnover of
8 sororin RNA and protein gene products may synergize with a potentially stringent requirement for intact spliceosomes for intron removal to cause a drastic and selective reduction of sororin protein following depletion of splicing factors. We have amended the discussion section of our revised manuscript to include the new data and to better reflect the aforementioned points. Again, these are minor points and do not diminish my enthusiasm for this study. Referee #3: In this study the authors aimed at identifying new factors involved in mitosis. Based on published datasets, the authors established a list of 718 potential candidates whose involvement in mitosis were further tested by a systematic sirna-based screen. From 718 candidates the list shrunk to 50 genes whose sirna appeared to have a real impact on nuclear morphology. This list was further reduced to 7 confirmed genes whose depletion had a consistent effect on nuclear shape. From this list, the depletion of the splicing factor MFAP1 resulted in particularly severe nuclear fragmentation. By using imaging techniques, the authors determined that in cells lacking MFAP1, chromosomes could not be aligned at the metaphase plate due to a loss of sister chromatid cohesion, suggesting that MFAP1 could have a function in sister chromatid cohesion in addition to its well characterized role as core pre-mrna splicing factor. However, by performing sirna-mediated depletions, the authors identified 18 other splicing factors that had a similar effect on chromatid cohesion as that observed upon MFAP1 depletion. Thus, the authors convincingly established a general link between pre-mrna splicing and chromatid cohesion, but apparently no specific or direct effect of MFAP1 on mitosis. Further experiments excluded a potential defect in cohesin loading onto chromatin, but demonstrated that loss of spliceosome components resulted in a specific and dramatic decrease in the essential chromatid cohesion factor sororin. Consistent with a requirement for pre-mrna splicing, complementation experiments in which sororin was expressed from an intronless transgene restored sister chromatid cohesion in cells depleted of splicing factors. This study demonstrates that the integrity of the spliceosome is required to maintain an appropriate level of spliced sororin mrna and its encoded protein sororin, which plays a role in chromatid cohesion. This study is well written and understandable to a large audience. The experimental procedures are well described and controlled. However, spliceosomal factors are essential for virtually all cellular processes, as splicing is an upstream event in gene expression and literally controls the expression of nearly all genes in mammalian cells. Therefore, the question addressed in this study does not represent a significant step forward in our understanding of the chromatid cohesion process, in addition to failing to identify new factors directly involved in mitosis. Thus, this study is not suitable for publication in EMBO J. in its present form. We thank the reviewer for his/her comments and criticism. Our work demonstrates that one of the acute consequences of compromised splicing in human cells is a failure of cohesin to stably associate with chromatin and to hold sister chromatids together. Furthermore, we provide the mechanistic basis for this effect. Cohesin and sister chromatid cohesion are essential for several pivotal chromosomal processes including chromosome segregation, DNA damage response and repair, and gene regulation. Our work highlights an early cellular consequence of derailing a major gene expression system in human cells. Furthermore, it emphasizes the delicate balance that cells have to strike between the WAPL-dependent cohesin release activity and the sororin-
9 dependent mechanism that counteracts WAPL and promotes the establishment of long-lived connections between sister chromatids. It also provides one explanation for the reported involvement of spliceosome subunits in mitosis. One of the splicing factors that we identify to be important for sister chromatid cohesion, SF3B1, has recently emerged as one of the major recurrently mutated driver genes in hyperproliferative haematopoietic disorders, such as chronic lymphocytic leukaemia (CLL) and myelodysplastic syndrome (MDS). Heterozygous somatic mutations in SF3B1 are detected in CLL cells as frequently as mutations in the classic tumor suppressor p53 (Wang et al., NEJM 2011; Rossi et al., Blood 2011; Queseda et al., Nature Genetics 2011; Landau et al., Cell 2013). SF3B1 mutations occur in 65% of patients suffering from a subtype of MDS (MDS with ring sideroblasts) (PMIDs: Papaemmanuil et al., NEJM 2011; Yoshida et al., Nature 2011). Our findings connecting SF3B1 to sister chromatid cohesion and cohesin s association with chromatin may therefore have clinical implications and could inform about aspects of CLL and MDS pathology. Testing and modelling the effects of patient mutations in SF3B1 on cohesin dynamics is a goal worth pursuing in the future. Based on the aforementioned points, we believe that reporting on the connection between splicing and cohesin dynamics/cohesion that we have uncovered here is important and relevant. 1. An interesting finding in this study is that the protein levels of the essential chromatid cohesion factors SMC1, SMC3, SCC1 and SA2 are not altered upon sirna depletion of the essential splicing factors NHP2L1, MFAP1, SART1 and CDC5L. Only the level of the sororin protein appears to be reduced. One obvious explanation would be that sororin has a high rate of transcription and protein turnover, which would explain its acute sensitivity to a reduction in splicing activity. However this point is not investigated and only briefly mentioned in the discussion. This manuscript would greatly benefit if further experiments providing insight into how stalling a general process like splicing appears to affect only one chromatid cohesion factor, i.e. sororin. Therefore I would greatly encourage the authors to determine sororin's half-life and its transcription rate, and determine whether this provides a satisfying explanation for sororin's exceptional sensitivity towards a general reduction in splicing. In addition the authors could compare the effect of depletion of spliceosome components on sororin to other known short-lived proteins. Moreover, the authors should investigate whether only the splicing of sororin is blocked upon NHP2L1, MFAP1, SART1 and CDC5L depletion or whether the splicing of the other cohesion factors SMC1, SMC3, SCC1 and SA2 is also affected. In summary, any experiment that would help us understand the specific effect observed on sororin would greatly improve the significance and general interest of this manuscript. We have addressed several of these points and included new experiments and dataset in the revised version of our manuscript. Analysis of 5 and 3 splice site sequences (MaxEntScan splice site score algorithm; and the position of the presumptive branch site (TAACT motif present) with regard to the 3 splice site in of the first intron of sororin did not reveal particular features that would explain a pronounced sensitivity of sororin pre-mrna splicing to the depletion of spliceosomes. Based on the transcriptome-wide analysis of intron retention in the accompanying manuscript by the Peters lab, it is clear that depletion of the splicing factor SNW1 affects intron removal in a number of genes. This suggests that the effect is not unique to sororin. However, amongst the factors essential for cohesion, depletion of spliceosome subunits had the strongest impact on intron retention in sororin RNA (Fig. 6B). In a new set of experiments (triplicate biological repeat), we used intron-exon junction specific
10 primer pairs to track intron retention. We now demonstrate that enhanced intron retention in sororin RNA but not in RNAs encoding the core cohesin subunits SCC1, SA2, SMC3 or SMC1 (Fig. 6B). Thus, our results support a selective effect on sororin splicing. While the molecular basis for this selectivity remains to be determined, we found that both sororin protein and mrna have a higher turnover than other cohesin subunit in the revised version of the manuscript (Fig 6C and E7). For these experiments, cells were treated with cycloheximide or actinomycin D. In G1 cells sororin protein has an extraordinarily short half-life of less than 2 hours (Fig. 6C and E7B). This short protein half-life sets sororin apart from other cohesin proteins. Our targeted analysis is supported by a yet unpublished global analysis of protein and RNA half-life by the Selbach lab (Berlin) that utilizes metabolic pulse labeling (as developed in Schwannhäuser et al., Nature 2011) (Henrik Zauber and Matthias Selbach, personal communication). The high turnover of sororin RNA and in particular sororin protein may synergize with a potentially stringent requirement for intact spliceosomes for intron removal to cause a drastic and selective reduction of sororin protein following depletion of splicing factors. We have amended the discussion section of our revised manuscript to include the new data and to better reflect the aforementioned points. In summary, our newly included data indicate the selective retention of introns in sororin RNA following splicing factor depletion compared to the RNAs of other cohesin subunits. Furthermore, we provide data demonstrating higher turnover of sororin gene products compared to the products of core cohesin subunits. The mechanistic basis for a selective or stringent requirement of intact spliceosomes for the splicing of sororin RNA (and others) remains unknown at this point. It will be facilitated by the global analyses of spliceosome assembly on RNA and branch site usage if correlated with aberrant intron retention upon loss of specific spliceosome components. In our opinion, deciphering why sororin RNA splicing is particularly sensitive to the depletion of spliceosome components, although clearly an interesting question, is beyond the scope of this manuscript. 2. Page 5: For 7 genes, two or more out of four sirna duplexes recapitulated the phenotype observed in the primary screen (Supplementary Table 2). This suggests that many of 50 hits selected from the primary screen were caused by off-target depletion effects. These are not necessarily off-target effects. A single RNAi duplex does not always inhibit efficiently gene expression. In the revised version of the manuscript, we have removed the sentence suggesting that the remaining 43 primary screen hits were caused by off-target effects. 3. Page 9: Like MFAP1, NHP2L1 and SART1 are components of the U5 and U4/U6 trisnrnp complex MFAP1 is not a component of tri-snrnp but rather a non-snrnp, protein factor that joins the spliceosome at the spliceosomal B complex stage. We thank the reviewer for pointing out this mistake. We have removed Like MFAP1, from the above sentence. 4. Page9:...Prp19 complex that promotes the catalytic activation of the spliceosome for cleavage at the 5' splicing site This should be rephrased to "...Prp19 complex required for the activation of the spliceosome before the first step of the splicing reaction" The sentence has been rephrased based on the reviewer s suggestion.
11 5. Fig. 1B vs Fig. S1: could the authors explain the difference in penetrance observed. In Fig. 1B only 16 out of 50 cells successfully passed cell division while in Fig. S1 the defects were observed in only ~25% cells. The apparent discrepancy in phenotypic penetrance between Fig. 1B and Fig S1 (now E1) stems from the different methods of analysis. For Figure 1B, the fate of cells was tracked over a long period using time-lapse microscopy (from 32 hrs post transfection onwards). For Figure S1/E1, the analysis was performed by fixing cells at a particular timepoint following sirna transfection (at 52 hrs post transfection). The latter approach presents a terminal and static picture of cellular phenotypes. Many MFAP1 depleted cells underwent mitotic arrest and cell death (in mitosis) before fixation. These cells will be lost and not captured using a fixed cell analysis by immunofluorescence. However, time-lapse tracking allows us to analyze and score the fate of all cells over time. Thus, the phenotypic penetrance is lower in Figure S1/E1 as strongly depleted cells undergo mitotic arrest/cell death (as shown in Fig. 1B) before the time point of analysis. In our experience, time-lapse recording present a more reliable view of the mitotic aberrations and phenotypic penetrance. 6. Fig. 4C: a loading control is missing To provide a loading control, we have probed the samples with an antibody against α-tubulin (see Figure 4C). Please note that the level of SMC3 acetylation was calculated using the ratios of AcSMC3 signal intensity to SMC3 signal intensity for each condition (using detection of fluorescently labeled secondary antibodies). 7. Fig. 6A: it seems that sirna-mediated depletion of sororin leads to co-depletion of SCC1. Does sororin affect the level of SCC1? Why don't ΔCDC5L cells, which have a dramatically reduced level of sororin, show any reduction in SCC1? We have conducted the quantitative immunoblot analysis of sororin protein levels shown in Figure 6A twice. In both experiments, sororin sirna transfection caused a reduction in SCC1 protein levels to around 50%. The analysis was performed in G2 synchronized cells. Two possible interpretations could account for this effect. (1) complete depletion of sororin interferes with the stability of SCC1 protein by e.g. increasing the degradation of SCC1 in the soluble pool. (2) sororin sirna could affect SCC1 expression through off-target effects. If the latter is correct, the reduction of SCC1 does not measurably contribute to cohesion defects in sororin sirna transfected cells as of expression of an sirna-resistant sororin transgene completely restores cohesion after depletion of the endogenous counterpart (Fig. 7A). If the former hypothesis is correct, strong but incomplete reduction in sororin through depletion of splicing factors is not sufficient to destabilize SCC1. At this point, the basis of this reduction in SCC1 levels following sororin depletion is unclear. However, in our opinion, this does not affect the main conclusions of our work. 8. Fig. 6C: in contrast to ΔMFAP1 and ΔNHPL2L1,ΔSART1 and ΔCDC5L cells show a two-fold reduction in the level of Exon2 of sororin (pre)-mrna. Is this a significant reproducible change? If so, do these cells have a transcription related defect (in the case of the sororin gene)? What is the steady state ratio of the sororin pre-mrna to its mrna? What is the half-life of sororin mrna?
12 In the revised version of our manuscript we have replaced the intron retention analysis with a new set of data (Fig. 6B). In this analysis we detected selective strong retention (6 to 16 fold) of sororin introns 1 and 2 following depletion of all tested splicing factors. In contrast, we do only detect minor changes in the retention of tested introns for SCC1, SA2, SMC3, and SMC1. The analysis used an exon only amplicon for normalization of intron-exon levels for all genes. This revealed that depletion of splicing factors reduced the combined levels of unprocessed + processed RNAs of all genes tested (sororin, SCC1, SA2, SMC3, and SMC1) around 1.5 to 3 fold depending on the splicing factor depleted. This suggests that interfering with splicing decreases the cellular transcript levels of the genes tested. However, sororin transcript levels were not reduced stronger than the levels of other genes tested. Only sororin RNA showed a strong increase in intron retention after normalization of intron-exon amplicon to the exon only amplicon for each gene (Fig 6B). 9. Fig. 7 shows much milder effects of splicing factor knockdown as compared to Fig. 3 and Fig. S2, even in the case of CDC5L for which RNAi treatment was performed for nearly the same time. Could the authors comment on this. In the case of CDC5L, the difference in phenotypic severity between Fig. 3 and Fig. 7 is explained by the fact that pools of 4 different sirna duplexes were used for depletion experiment in Figure 3 whereas a single sirna duplex (oligo 4) was used for the experiment in Fig. 7. As shown in the deconvolution dataset in Figure S2 (now E2), the CDC5L oligo4 caused a cohesion defect in 70% of spreads, which is slightly less than the 81% defect shown in Figure 7. Thus, for CDC5L, the phenotype obtained in Fig.7 is not milder than the one obtained in Fig. S2/E2 but actually slightly stronger. The use and identity of single sirna duplexes versus sirna pools duplexes is specified in the method section of the manuscript. The differences in phenotypic severity between Fig.3 and Fig. 7 for the other splicing factors (MFAP1, NHP2L1 and SART1) can be explained by the fact that cells were collected for chromosome spread analysis earlier than for the experiments in Fig. 3 and S2/E2. For Fig. 3 and S2/E2, cells were collected 56 hr post transfection (including 4 hr nocodazole treatment). In contrast, for the experiment in Fig 7 cells were collected after 40/34/40 hr for depletion of MFAP1/NHP2L1/SART1, respectively (all time intervals include 4 hr of nocodazole treatment). The timings are specified in the figure legends of the paper. The reason for a reduced exposure to sirnas for Fig7 was to reduce the impact of cohesion fatigue which can make a contribution to the loss of sister chromatid cohesion as mentioned in the discussion section of our manuscript and as elaborated on in more detail in the accompanying manuscript by the Peters lab. 10. Fig. 8B: the cartoon is confusing. "Premature loss of chromatid cohesion" should be next to free cohesion, right? Please indicate in the legend the subunits of the cohesin "triangles". We agree that the position of the text in the figure was not appropriate. In the revised version we have changed the position of the text box accordingly and placed the text box between the two separating chromatids. Furthermore, we have labeled the subunits of the cohesin complex in the model. 11. Page 12 (and Page 16 of Discussion): Expression of the intronless sororin transgene did not however restore mitotic progression in cells transfected with sirnas targeting spliceosome subunits... It is not clear based on which data this conclusion has been made.
13 To address this point, we have used time-lapse microscopy to measure the mitotic duration of cells expressing either an AcFL tag or an intronless and RNAi-resistant AcFL-tagged version of sororin (Figure E8B). Both stable transgenic cell lines were transfected with control sirna duplexes or duplexes targeting Sororin, MFAP1, NHP2L1, SART1 or CDC5L. Expression of the transgenic and sirna-resistant copy of sororin rescued the mitotic arrest following sororin sirna transfection but not following transfection with sirna duplexes targeting splicing factors. These data are now presented in Figure E8A of the revised manuscript and support the above statement in the manuscript. 12. Page 17: in the last paragraph of the Discussion the authors describe the potential biomedical implications of their findings, in particular to chronic lymphocytic leukaemia. Defects in the splicing factors described in this manuscript lead to mitotic arrest or cell death rather than uncontrolled cell proliferation characteristic for chronic lymphocytic leukaemia. Please clarify this point. Complete loss of splicing function as well as a penetrant defect in sister chromatid cohesion is likely to be cell lethal. This condition would clearly be incompatible with neoplastic growth in hyperproliferative disorders such CLL and MDS. However, the genetic changes detected in CLL patients are heterozygous somatic mutations. The most frequent mutation detected in SF3B1 in CLL patients is the somatic heterozygous mutation K700E. Our finding that depletion of SF3B1 results in dramatic loss of sister chromatid cohesion (Fig. 3A and B) raises the possibility that the heterozygous patient mutations such as K700E could alter the dynamics of cohesin-chromatin association and thereby change the fidelity of chromosome disjunction, DNA repair and gene regulation, all hallmarks of cancer, in a manner that is compatible with cell proliferation. Testing this hypothesis will require modeling the patient mutation K700E in a heterozygous and homozygous configuration in experimentally accessible cell models. This is an important goal for future research. Therefore, studies based on the findings presented in this manuscript could contribute to our understanding of the molecular basis of CLL and MDS. Given the prevalence of SF3B1 mutations in CLL and MDS, deciphering the impact of these mutations on cell physiology is of significant biomedical importance. To clarify the fact that disease-associated mutations of SF3B1 are not expected to be complete loss of-function situations we have added the term heterozygous to the discussion section of the revised version of our manuscript.
14
15 The EMBO Journal Peer Review Process File - EMBO nd Editorial Decision 12 August 2014 Thank you for submitting your revised manuscript to The EMBO Journal. Your study has now been seen by two of the original referees (comments included below) and as you will see they both find that all criticisms have been adequately addressed. I am therefore happy to inform you that your manuscript has been accepted for publication with us. REFEREE REPORTS: Referee #1: I think that the authors properly addressed my concerns. The manuscript should be published immediately. Referee #2: The authors have properly addressed all my questions. In particular, I appreciate the effort to explain the specific requirement for the spliceosome machinery in maintaining sororin levels given the high turnover of this protein. I therefore recommend publication. EMBO 5
NFIL3/E4BP4 controls Type 2 T helper cell cytokine expression
 Manuscript EMBO-2010-75657 NFIL3/E4BP4 controls Type 2 T helper cell cytokine expression Masaki Kashiwada, Suzanne L. Cassel, John D. Colgan and Paul B. Rothman Corresponding author: Paul Rothman, University
Manuscript EMBO-2010-75657 NFIL3/E4BP4 controls Type 2 T helper cell cytokine expression Masaki Kashiwada, Suzanne L. Cassel, John D. Colgan and Paul B. Rothman Corresponding author: Paul Rothman, University
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication
 Manuscript EMBO-2010-74756 Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication Tsai-Ling Liao, Chung-Yi Wu, Wen-Chi Su, King-Song Jeng and Michael Lai Corresponding
Manuscript EMBO-2010-74756 Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication Tsai-Ling Liao, Chung-Yi Wu, Wen-Chi Su, King-Song Jeng and Michael Lai Corresponding
Regulators of Cell Cycle Progression
 Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Myelin suppresses axon regeneration by PIR-B/SHPmediated inhibition of Trk activity
 Manuscript EMBO-2010-76298 Myelin suppresses axon regeneration by PIR-B/SHPmediated inhibition of Trk activity Yuki Fujita, Shota Endo, Toshiyuki Takai and Toshihide Yamashita Corresponding author: Toshihide
Manuscript EMBO-2010-76298 Myelin suppresses axon regeneration by PIR-B/SHPmediated inhibition of Trk activity Yuki Fujita, Shota Endo, Toshiyuki Takai and Toshihide Yamashita Corresponding author: Toshihide
GW182 proteins cause PABP dissociation from silenced mirna targets in the absence of deadenylation
 Manuscript EMBO-2012-83275 GW182 proteins cause PABP dissociation from silenced mirna targets in the absence of deadenylation Latifa Zekri, Duygu Kuzuoğlu-Öztürk and Elisa Izaurralde Corresponding author:
Manuscript EMBO-2012-83275 GW182 proteins cause PABP dissociation from silenced mirna targets in the absence of deadenylation Latifa Zekri, Duygu Kuzuoğlu-Öztürk and Elisa Izaurralde Corresponding author:
Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly
 Manuscript EMBO-2015-91176 Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly Camino De Juan Romero, Carl Bruder, Ugo Tomasello, Jose Miguel Sanz-Anquela
Manuscript EMBO-2015-91176 Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly Camino De Juan Romero, Carl Bruder, Ugo Tomasello, Jose Miguel Sanz-Anquela
Foxm1 Transcription Factor is Required for Lung Fibrosis and Epithelial to Mesenchymal Transition.
 Manuscript EMBO-2012-82682 Foxm1 Transcription Factor is Required for Lung Fibrosis and Epithelial to Mesenchymal Transition. David Balli, Vladimir Ustiyan, Yufang Zhang, I-Ching Wang, Alex J. Masino,
Manuscript EMBO-2012-82682 Foxm1 Transcription Factor is Required for Lung Fibrosis and Epithelial to Mesenchymal Transition. David Balli, Vladimir Ustiyan, Yufang Zhang, I-Ching Wang, Alex J. Masino,
The unfolded protein response is shaped by the NMD pathway
 Manuscript EMBOR-2014-39696 The unfolded protein response is shaped by the NMD pathway Rachid Karam, Chih-Hong Lou, Heike Kroeger, Lulu Huang, Jonathan H. Lin and Miles F. Wilkinson Corresponding author:
Manuscript EMBOR-2014-39696 The unfolded protein response is shaped by the NMD pathway Rachid Karam, Chih-Hong Lou, Heike Kroeger, Lulu Huang, Jonathan H. Lin and Miles F. Wilkinson Corresponding author:
1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics
 1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics to gain mechanistic insight! 4. Return to step 2, as
1. Identify and characterize interesting phenomena! 2. Characterization should stimulate some questions/models! 3. Combine biochemistry and genetics to gain mechanistic insight! 4. Return to step 2, as
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes
 Manuscript EMBO-2009-71812 Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes Thomas S. Jacques, Alexander Swales, Monika J. Brzozowski, Nico
Manuscript EMBO-2009-71812 Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes Thomas S. Jacques, Alexander Swales, Monika J. Brzozowski, Nico
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis
 Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Chromosomes Chromosomes were first observed by the German embryologist Walther Fleming in 1882. Chromosome number varies among organisms most
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Chromosomes Chromosomes were first observed by the German embryologist Walther Fleming in 1882. Chromosome number varies among organisms most
Novel RNAs along the Pathway of Gene Expression. (or, The Expanding Universe of Small RNAs)
 Novel RNAs along the Pathway of Gene Expression (or, The Expanding Universe of Small RNAs) Central Dogma DNA RNA Protein replication transcription translation Central Dogma DNA RNA Spliced RNA Protein
Novel RNAs along the Pathway of Gene Expression (or, The Expanding Universe of Small RNAs) Central Dogma DNA RNA Protein replication transcription translation Central Dogma DNA RNA Spliced RNA Protein
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
The Biology and Genetics of Cells and Organisms The Biology of Cancer
 The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
Cell Cycle. Trends in Cell Biology
 Cell Cycle Trends in Cell Biology Cell Cycle The orderly sequence of events by which a cell duplicates its contents and divides into two Daughter Cells Activities of a cell from one cell division to the
Cell Cycle Trends in Cell Biology Cell Cycle The orderly sequence of events by which a cell duplicates its contents and divides into two Daughter Cells Activities of a cell from one cell division to the
Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle. 9.1 Multiple-Choice Questions
 Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle 9.1 Multiple-Choice Questions 1) Starting with a fertilized egg (zygote), a series of five cell divisions would produce an early embryo with how
Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle 9.1 Multiple-Choice Questions 1) Starting with a fertilized egg (zygote), a series of five cell divisions would produce an early embryo with how
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis
 Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Prokaryotes Have a Simpler Cell Cycle Cell division in prokaryotes takes place in two stages, which together make up a simple cell cycle 1. Copy
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Prokaryotes Have a Simpler Cell Cycle Cell division in prokaryotes takes place in two stages, which together make up a simple cell cycle 1. Copy
CELL CYCLE REGULATION AND CANCER. Cellular Reproduction II
 CELL CYCLE REGULATION AND CANCER Cellular Reproduction II THE CELL CYCLE Interphase G1- gap phase 1- cell grows and develops S- DNA synthesis phase- cell replicates each chromosome G2- gap phase 2- cell
CELL CYCLE REGULATION AND CANCER Cellular Reproduction II THE CELL CYCLE Interphase G1- gap phase 1- cell grows and develops S- DNA synthesis phase- cell replicates each chromosome G2- gap phase 2- cell
Cell Cycle, Mitosis, and Microtubules. LS1A Final Exam Review Friday 1/12/07. Processes occurring during cell cycle
 Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Growth and Division *
 OpenStax-CNX module: m46034 1 Cell Growth and Division * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you will
OpenStax-CNX module: m46034 1 Cell Growth and Division * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you will
EGFR kinase activity is required for TLR4 signaling and the septic shock response
 Manuscript EMBOR-2015-40337 EGFR kinase activity is required for TLR4 signaling and the septic shock response Saurabh Chattopadhyay, Manoj Veleeparambil, Darshana Poddar, Samar Abdulkhalek, Sudip K. Bandyopadhyay,
Manuscript EMBOR-2015-40337 EGFR kinase activity is required for TLR4 signaling and the septic shock response Saurabh Chattopadhyay, Manoj Veleeparambil, Darshana Poddar, Samar Abdulkhalek, Sudip K. Bandyopadhyay,
The RNA binding protein Arrest (Bruno) regulates alternative splicing to enable myofibril maturation in Drosophila flight muscle
 Manuscript EMBOR-2014-39791 The RNA binding protein Arrest (Bruno) regulates alternative splicing to enable myofibril maturation in Drosophila flight muscle Maria L. Spletter, Christiane Barz, Assa Yeroslaviz,
Manuscript EMBOR-2014-39791 The RNA binding protein Arrest (Bruno) regulates alternative splicing to enable myofibril maturation in Drosophila flight muscle Maria L. Spletter, Christiane Barz, Assa Yeroslaviz,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
A genome-scale screen reveals context-dependent ovarian cancer sensitivity to mirna overexpression
 A genome-scale screen reveals context-dependent ovarian cancer sensitivity to mirna overexpression Benjamin B. Shields, Chad V. Pecot, Hua Gao, Elizabeth McMillan, Malia Potts, Christa Nagel, Scott Purinton,
A genome-scale screen reveals context-dependent ovarian cancer sensitivity to mirna overexpression Benjamin B. Shields, Chad V. Pecot, Hua Gao, Elizabeth McMillan, Malia Potts, Christa Nagel, Scott Purinton,
A novel fragile X syndrome mutation reveals a conserved role for the carboxy-terminus in FMRP localization and function
 A novel fragile X syndrome mutation reveals a conserved role for the carboxy-terminus in FMRP localization and function Zeynep Okray, Celine E.F. de Esch, Hilde Van Esch, Koen Devriendt, Annelies Claeys,
A novel fragile X syndrome mutation reveals a conserved role for the carboxy-terminus in FMRP localization and function Zeynep Okray, Celine E.F. de Esch, Hilde Van Esch, Koen Devriendt, Annelies Claeys,
Cell-to-cell variability of alternative RNA splicing
 Cell-to-cell variability of alternative RNA splicing Zeev Waks, Allon Klein, Pamela Silver Corresponding author: Pamela Silver, Harvard Medical School Review timeline: Submission date: 10 March 2011 Editorial
Cell-to-cell variability of alternative RNA splicing Zeev Waks, Allon Klein, Pamela Silver Corresponding author: Pamela Silver, Harvard Medical School Review timeline: Submission date: 10 March 2011 Editorial
REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER
 REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER RNA Splicing Lecture 3, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Splice
REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER RNA Splicing Lecture 3, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Splice
Minor point: In Table S1 there seems to be a typo - multiplicity of the data for the Sm3+ structure 134.8?
 PEER REVIEW FILE Reviewers' comments: Reviewer #1 (Remarks to the Author): Finogenova and colleagues investigated aspects of the structural organization of the Nuclear Exosome Targeting (NEXT) complex,
PEER REVIEW FILE Reviewers' comments: Reviewer #1 (Remarks to the Author): Finogenova and colleagues investigated aspects of the structural organization of the Nuclear Exosome Targeting (NEXT) complex,
RECAP (1)! In eukaryotes, large primary transcripts are processed to smaller, mature mrnas.! What was first evidence for this precursorproduct
 RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
Fig. 1. Simple columnar epithelial cells lining the small intestine.
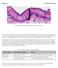 Mitosis Prelab Reading Fig. 1. Simple columnar epithelial cells lining the small intestine. The tall cells pictured in Fig. 1 form the lining of the small intestine in humans and other animals. These cells
Mitosis Prelab Reading Fig. 1. Simple columnar epithelial cells lining the small intestine. The tall cells pictured in Fig. 1 form the lining of the small intestine in humans and other animals. These cells
UNDERSTANDING THE ROLE OF ATP HYDROLYSIS IN THE SPLICEOSOME
 UNDERSTANDING THE ROLE OF ATP HYDROLYSIS IN THE SPLICEOSOME RNA Splicing Lecture 2, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Eight essential ATPases
UNDERSTANDING THE ROLE OF ATP HYDROLYSIS IN THE SPLICEOSOME RNA Splicing Lecture 2, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Eight essential ATPases
BIOLOGY - CLUTCH CH.12 - CELL DIVISION.
 !! www.clutchprep.com CONCEPT: CELL DIVISION Cell division is the process by which one cell splits into two or more daughter cells. Cell division generally requires that cells produce enough materials,
!! www.clutchprep.com CONCEPT: CELL DIVISION Cell division is the process by which one cell splits into two or more daughter cells. Cell division generally requires that cells produce enough materials,
TRIM5 requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription
 Manuscript EMBO-2014-90361 TRIM5 requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription Adam J Fletcher, Devin E Christensen, Chad Nelson, Choon Ping Tan, Torsten Schaller,
Manuscript EMBO-2014-90361 TRIM5 requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription Adam J Fletcher, Devin E Christensen, Chad Nelson, Choon Ping Tan, Torsten Schaller,
GENETIC ANALYSIS OF RAS SIGNALING PATHWAYS IN CELL PROLIFERATION, MIGRATION AND SURVIVAL
 Manuscript EMBO-2009-72496 GENETIC ANALYSIS OF RAS SIGNALING PATHWAYS IN CELL PROLIFERATION, MIGRATION AND SURVIVAL Matthias Drosten, Alma Dhawahir, Eleanor Sum, Jelena Urosevic, Carmen Lechuga, Luis Esteban,
Manuscript EMBO-2009-72496 GENETIC ANALYSIS OF RAS SIGNALING PATHWAYS IN CELL PROLIFERATION, MIGRATION AND SURVIVAL Matthias Drosten, Alma Dhawahir, Eleanor Sum, Jelena Urosevic, Carmen Lechuga, Luis Esteban,
Unit 9: The Cell Cycle
 Unit 9: The Cell Cycle Name: Period: Test Date: 1 Table of Contents Title of Page Page Number Teacher Stamp Unit 9 Warm-Ups 3-4 Cell Cycle/Interphase Notes 5-6 DNA Replication Notes 7-8 DNA replication
Unit 9: The Cell Cycle Name: Period: Test Date: 1 Table of Contents Title of Page Page Number Teacher Stamp Unit 9 Warm-Ups 3-4 Cell Cycle/Interphase Notes 5-6 DNA Replication Notes 7-8 DNA replication
Ploidy and Human Cell Types. Cell Cycle and Mitosis. DNA and Chromosomes. Where It All Began 11/19/2014. Chapter 12 Pg
 Ploidy and Human Cell Types Cell Cycle and Mitosis Chapter 12 Pg. 228 245 Cell Types Somatic cells (body cells) have 46 chromosomes, which is the diploid chromosome number. A diploid cell is a cell with
Ploidy and Human Cell Types Cell Cycle and Mitosis Chapter 12 Pg. 228 245 Cell Types Somatic cells (body cells) have 46 chromosomes, which is the diploid chromosome number. A diploid cell is a cell with
Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old mdx mice
 Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old mdx mice Mozzetta C., Consalvi S., Saccone V., Tierney M., Diamantini
Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old mdx mice Mozzetta C., Consalvi S., Saccone V., Tierney M., Diamantini
Mitosis and the Cell Cycle
 Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
The Cell Cycle. Dr. SARRAY Sameh, Ph.D
 The Cell Cycle Dr. SARRAY Sameh, Ph.D Overview When an organism requires additional cells (either for growth or replacement of lost cells), new cells are produced by cell division (mitosis) Somatic cells
The Cell Cycle Dr. SARRAY Sameh, Ph.D Overview When an organism requires additional cells (either for growth or replacement of lost cells), new cells are produced by cell division (mitosis) Somatic cells
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
(a) Reproduction. (b) Growth and development. (c) Tissue renewal
 100 µm 200 µm 20 µm (a) Reproduction (b) Growth and development (c) Tissue renewal 1 20 µm 2 0.5 µm Chromosomes DNA molecules Chromosome arm Centromere Chromosome duplication (including DNA synthesis)
100 µm 200 µm 20 µm (a) Reproduction (b) Growth and development (c) Tissue renewal 1 20 µm 2 0.5 µm Chromosomes DNA molecules Chromosome arm Centromere Chromosome duplication (including DNA synthesis)
RECAP (1)! In eukaryotes, large primary transcripts are processed to smaller, mature mrnas.! What was first evidence for this precursorproduct
 RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins.
 Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
FGF22 signaling regulates synapse formation during postinjury remodeling of the spinal cord
 Manuscript EMBO-2014-90578 FGF22 signaling regulates synapse formation during postinjury remodeling of the spinal cord Anne Jacobi, Kristina Loy, Anja M Schmalz, Mikael Hellsten, Hisashi Umemori, Martin
Manuscript EMBO-2014-90578 FGF22 signaling regulates synapse formation during postinjury remodeling of the spinal cord Anne Jacobi, Kristina Loy, Anja M Schmalz, Mikael Hellsten, Hisashi Umemori, Martin
Lecture 10. G1/S Regulation and Cell Cycle Checkpoints. G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint
 Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Antithetical NFATc1-Sox2 and p53-mir200 signaling networks govern pancreatic cancer cell plasticity
 The EMBO Journal Peer Review Process File - EMBO-2014-89574 Manuscript EMBO-2014-89574 Antithetical NFATc1-Sox2 and p53-mir200 signaling networks govern pancreatic cancer cell plasticity Shiv K. Singh,
The EMBO Journal Peer Review Process File - EMBO-2014-89574 Manuscript EMBO-2014-89574 Antithetical NFATc1-Sox2 and p53-mir200 signaling networks govern pancreatic cancer cell plasticity Shiv K. Singh,
An Introduction to Genetics. 9.1 An Introduction to Genetics. An Introduction to Genetics. An Introduction to Genetics. DNA Deoxyribonucleic acid
 An Introduction to Genetics 9.1 An Introduction to Genetics DNA Deoxyribonucleic acid Information blueprint for life Reproduction, development, and everyday functioning of living things Only 2% coding
An Introduction to Genetics 9.1 An Introduction to Genetics DNA Deoxyribonucleic acid Information blueprint for life Reproduction, development, and everyday functioning of living things Only 2% coding
BIOLOGY 4/6/2015. Cell Cycle - Mitosis. Outline. Overview: The Key Roles of Cell Division. identical daughter cells. I. Overview II.
 2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
Targeted proteomics reveals strain-specific changes in the mouse insulin and central metabolic pathways after sustained high-fat diet
 Molecular Systems Biology Peer Review Process File Targeted proteomics reveals strain-specific changes in the mouse insulin and central metabolic pathways after sustained high-fat diet Eduard Sabidó, Yibo
Molecular Systems Biology Peer Review Process File Targeted proteomics reveals strain-specific changes in the mouse insulin and central metabolic pathways after sustained high-fat diet Eduard Sabidó, Yibo
Cell Division. Chromosome structure. Made of chromatin (mix of DNA and protein) Only visible during cell division
 Chromosome structure Made of chromatin (mix of DNA and protein) Only visible during cell division Chromosome structure The DNA in a cell is packed into an elaborate, multilevel system of coiling and folding.
Chromosome structure Made of chromatin (mix of DNA and protein) Only visible during cell division Chromosome structure The DNA in a cell is packed into an elaborate, multilevel system of coiling and folding.
Chapter 12. living /non-living? growth repair renew. Reproduction. Reproduction. living /non-living. fertilized egg (zygote) next chapter
 Chapter 12 How cells divide Reproduction living /non-living? growth repair renew based on cell division first mitosis - distributes identical sets of chromosomes cell cycle (life) Cell Division in Bacteria
Chapter 12 How cells divide Reproduction living /non-living? growth repair renew based on cell division first mitosis - distributes identical sets of chromosomes cell cycle (life) Cell Division in Bacteria
Why do cells reproduce?
 Outline Cell Reproduction 1. Overview of Cell Reproduction 2. Cell Reproduction in Prokaryotes 3. Cell Reproduction in Eukaryotes 1. Chromosomes 2. Cell Cycle 3. Mitosis and Cytokinesis Examples of Cell
Outline Cell Reproduction 1. Overview of Cell Reproduction 2. Cell Reproduction in Prokaryotes 3. Cell Reproduction in Eukaryotes 1. Chromosomes 2. Cell Cycle 3. Mitosis and Cytokinesis Examples of Cell
A. Incorrect! All the cells have the same set of genes. (D)Because different types of cells have different types of transcriptional factors.
 Genetics - Problem Drill 21: Cytogenetics and Chromosomal Mutation No. 1 of 10 1. Why do some cells express one set of genes while other cells express a different set of genes during development? (A) Because
Genetics - Problem Drill 21: Cytogenetics and Chromosomal Mutation No. 1 of 10 1. Why do some cells express one set of genes while other cells express a different set of genes during development? (A) Because
5/25/2015. Replication fork. Replication fork. Replication fork. Replication fork
 Mutations Chapter 5 Cellular Functions Lecture 3: and Cell Division Most DNA mutations alter the protein product May Make it function better (rarely) Change its function Reduce its function Make it non-functional
Mutations Chapter 5 Cellular Functions Lecture 3: and Cell Division Most DNA mutations alter the protein product May Make it function better (rarely) Change its function Reduce its function Make it non-functional
Where Splicing Joins Chromatin And Transcription. 9/11/2012 Dario Balestra
 Where Splicing Joins Chromatin And Transcription 9/11/2012 Dario Balestra Splicing process overview Splicing process overview Sequence context RNA secondary structure Tissue-specific Proteins Development
Where Splicing Joins Chromatin And Transcription 9/11/2012 Dario Balestra Splicing process overview Splicing process overview Sequence context RNA secondary structure Tissue-specific Proteins Development
Mitosis. An Introduction to Genetics. An Introduction to Cell Division
 Mitosis An Introduction to Genetics An Introduction to Cell Division DNA is Packaged in Chromosomes Cell Cycle Mitosis and Cytokinesis Variations in Cell Division Cell Division and Cancer An Introduction
Mitosis An Introduction to Genetics An Introduction to Cell Division DNA is Packaged in Chromosomes Cell Cycle Mitosis and Cytokinesis Variations in Cell Division Cell Division and Cancer An Introduction
Omnis cellula e cellula
 Chapter 12 The Cell Cycle Omnis cellula e cellula 1855- Rudolf Virchow German scientist all cells arise from a previous cell Every cell from a cell In order for this to be true, cells must have the ability
Chapter 12 The Cell Cycle Omnis cellula e cellula 1855- Rudolf Virchow German scientist all cells arise from a previous cell Every cell from a cell In order for this to be true, cells must have the ability
Almost every cell in the human body has an identical set of 46 chromosomes, produced through the process of mitosis.
 M I T O S I S Mitosis Mitosis is the type of cell division that occurs for growth (adding new cells) and repair (replacing old or damaged cells). It results in two daughter cells that have identical chromosomes
M I T O S I S Mitosis Mitosis is the type of cell division that occurs for growth (adding new cells) and repair (replacing old or damaged cells). It results in two daughter cells that have identical chromosomes
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa
 SUPPLEMENTARY FIGURES Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa tet-on cells that stably express HP1α-CFP, HP1β-CFP, or HP1γ-CFP were monitored with livecell
SUPPLEMENTARY FIGURES Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa tet-on cells that stably express HP1α-CFP, HP1β-CFP, or HP1γ-CFP were monitored with livecell
Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel meca homologue mecc
 Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel meca homologue mecc Ewan M. Harrison, Gavin K. Paterson, Matthew T.G. Holden, Jesper Larsen, Marc Stegger, Anders
Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel meca homologue mecc Ewan M. Harrison, Gavin K. Paterson, Matthew T.G. Holden, Jesper Larsen, Marc Stegger, Anders
Chapter 2. Mitosis and Meiosis
 Chapter 2. Mitosis and Meiosis Chromosome Theory of Heredity What structures within cells correspond to genes? The development of genetics took a major step forward by accepting the notion that the genes
Chapter 2. Mitosis and Meiosis Chromosome Theory of Heredity What structures within cells correspond to genes? The development of genetics took a major step forward by accepting the notion that the genes
Editorial Note: this manuscript has been previously reviewed at another journal that is not operating a transparent peer review scheme.
 Editorial Note: this manuscript has been previously reviewed at another journal that is not operating a transparent peer review scheme. This document only contains reviewer comments and rebuttal letters
Editorial Note: this manuscript has been previously reviewed at another journal that is not operating a transparent peer review scheme. This document only contains reviewer comments and rebuttal letters
LECTURE PRESENTATIONS
 LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
Cell division functions in 1. reproduction, 2. growth, and 3. repair
 Cell division functions in 1. reproduction, 2. growth, and 3. repair What do you think you are looking at here??? Can something like you or I do this??? Fig. 12.1 How did you start out? How did you grow?
Cell division functions in 1. reproduction, 2. growth, and 3. repair What do you think you are looking at here??? Can something like you or I do this??? Fig. 12.1 How did you start out? How did you grow?
Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy
 Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
Tankyrase 1 regulates centrosome function by controlling CPAP stability
 Manuscript EMBOR-2011-35655 Tankyrase 1 regulates centrosome function by controlling CPAP stability Mi Kyung Kim, Charles Dudognon and Susan Smith Corresponding author: Susan Smith, The Skirball Institute
Manuscript EMBOR-2011-35655 Tankyrase 1 regulates centrosome function by controlling CPAP stability Mi Kyung Kim, Charles Dudognon and Susan Smith Corresponding author: Susan Smith, The Skirball Institute
Mitosis THE CELL CYCLE. In unicellular organisms, division of one cell reproduces the entire organism Multicellular organisms use cell division for..
 Mitosis THE CELL CYCLE In unicellular organisms, division of one cell reproduces the entire organism Multicellular organisms use cell division for.. Development from a fertilized cell Growth Repair Cell
Mitosis THE CELL CYCLE In unicellular organisms, division of one cell reproduces the entire organism Multicellular organisms use cell division for.. Development from a fertilized cell Growth Repair Cell
Bacterial cell. Origin of replication. Septum
 Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Scaffold protein Chromatin Loop Solenoid Scaffold protein Chromatin loop
Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Scaffold protein Chromatin Loop Solenoid Scaffold protein Chromatin loop
Origin of replication. Septum
 Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Chromatin Loop Solenoid Scaffold protein Scaffold protein Chromatin loop
Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Chromatin Loop Solenoid Scaffold protein Scaffold protein Chromatin loop
BIOLOGY. The Cell Cycle CAMPBELL. Reece Urry Cain Wasserman Minorsky Jackson. Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick
 CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 12 The Cell Cycle Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick The Key Roles of Cell Division The ability
CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 12 The Cell Cycle Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick The Key Roles of Cell Division The ability
CELL CYCLE INTRODUCTION PART I ANIMAL CELL CYCLE INTERPHASE EVOLUTION/HEREDITY UNIT. Activity #3
 AP BIOLOGY EVOLUTION/HEREDITY UNIT Unit 1 Part 3 Chapter 12 Activity #3 INTRODUCTION CELL CYCLE NAME DATE PERIOD The nuclei in cells of eukaryotic organisms contain chromosomes with clusters of genes,
AP BIOLOGY EVOLUTION/HEREDITY UNIT Unit 1 Part 3 Chapter 12 Activity #3 INTRODUCTION CELL CYCLE NAME DATE PERIOD The nuclei in cells of eukaryotic organisms contain chromosomes with clusters of genes,
Mitosis. AND Cell DiVISION
 Mitosis AND Cell DiVISION Cell Division Characteristic of living things: ability to reproduce their own kind. Cell division purpose: When unicellular organisms such as amoeba divide to form offspring reproduction
Mitosis AND Cell DiVISION Cell Division Characteristic of living things: ability to reproduce their own kind. Cell division purpose: When unicellular organisms such as amoeba divide to form offspring reproduction
Chapter 12. The Cell Cycle
 Chapter 12 The Cell Cycle The Key Roles of Cell Division The ability of organisms to produce more of their own kind is the one characteristic that best distinguishes living things from nonliving things.
Chapter 12 The Cell Cycle The Key Roles of Cell Division The ability of organisms to produce more of their own kind is the one characteristic that best distinguishes living things from nonliving things.
Chapter 10. Cell Growth and Division
 Chapter 10 Cell Growth and Division Cell Growth A. Limits to Cell Growth 1. Two main reasons why cells divide: a. Demands on DNA as the cell get too large Cell Growth b. Moving nutrients and waste across
Chapter 10 Cell Growth and Division Cell Growth A. Limits to Cell Growth 1. Two main reasons why cells divide: a. Demands on DNA as the cell get too large Cell Growth b. Moving nutrients and waste across
The Cell Cycle CAMPBELL BIOLOGY IN FOCUS SECOND EDITION URRY CAIN WASSERMAN MINORSKY REECE
 CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 9 The Cell Cycle Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION Overview: The Key
CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 9 The Cell Cycle Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION Overview: The Key
Cellular Reproduction, Part 2: Meiosis Lecture 10 Fall 2008
 Mitosis & 1 Cellular Reproduction, Part 2: Lecture 10 Fall 2008 Mitosis Form of cell division that leads to identical daughter cells with the full complement of DNA Occurs in somatic cells Cells of body
Mitosis & 1 Cellular Reproduction, Part 2: Lecture 10 Fall 2008 Mitosis Form of cell division that leads to identical daughter cells with the full complement of DNA Occurs in somatic cells Cells of body
Cell Cycle. Interphase, Mitosis, Cytokinesis, and Cancer
 Cell Cycle Interphase, Mitosis, Cytokinesis, and Cancer Cell Division One cell divides into 2 new identical daughter cells. Chromosomes carry the genetic information (traits) of the cell How many Chromosomes
Cell Cycle Interphase, Mitosis, Cytokinesis, and Cancer Cell Division One cell divides into 2 new identical daughter cells. Chromosomes carry the genetic information (traits) of the cell How many Chromosomes
Stages of Mitosis. Introduction
 Name: Due: Stages of Mitosis Introduction Mitosis, also called karyokinesis, is division of the nucleus and its chromosomes. It is followed by division of the cytoplasm known as cytokinesis. Both mitosis
Name: Due: Stages of Mitosis Introduction Mitosis, also called karyokinesis, is division of the nucleus and its chromosomes. It is followed by division of the cytoplasm known as cytokinesis. Both mitosis
Supplementary information. The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic. Spindle Orientation
 Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Src-dependent autophagic degradation of Ret in FAKsignaling defective cancer cells
 Manuscript EMBOR-2011-35678 Src-dependent autophagic degradation of Ret in FAKsignaling defective cancer cells Emma Sandilands, Bryan Serrels, Simon Wilkinson and Margaret C Frame Corresponding author:
Manuscript EMBOR-2011-35678 Src-dependent autophagic degradation of Ret in FAKsignaling defective cancer cells Emma Sandilands, Bryan Serrels, Simon Wilkinson and Margaret C Frame Corresponding author:
Name: Cell division and cancer review
 Name: Cell division and cancer review 1. What type of cell undergoes meiosis? Gamete cells or Somatic cells 2. Define homologous chromosomes. 2 chromosomes with similar structure 3. For each of the following
Name: Cell division and cancer review 1. What type of cell undergoes meiosis? Gamete cells or Somatic cells 2. Define homologous chromosomes. 2 chromosomes with similar structure 3. For each of the following
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
BIOLOGY. Cell Cycle - Mitosis. Outline. Overview: The Key Roles of Cell Division. identical daughter cells. I. Overview II.
 2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
Wnt mediated protein stabilization ensures proper mitotic microtubule assembly and chromosome segregation
 Manuscript EMBOR-2014-39410 Wnt mediated protein stabilization ensures proper mitotic microtubule assembly and chromosome segregation Ailine Stolz, Kim Neufeld, Norman Ertych and Holger Bastians Corresponding
Manuscript EMBOR-2014-39410 Wnt mediated protein stabilization ensures proper mitotic microtubule assembly and chromosome segregation Ailine Stolz, Kim Neufeld, Norman Ertych and Holger Bastians Corresponding
A DNA/HDAC dual-targeting drug CY with significantly enhanced anticancer potency
 A DNA/HDAC dual-targeting drug CY190602 with significantly enhanced anticancer potency Chuan Liu, Hongyu Ding, Xiaoxi Li, Christian P. Pallasch, Liya Hong, Dianwu Guo, Yi Chen, Difei Wang, Wei Wang, Yajie
A DNA/HDAC dual-targeting drug CY190602 with significantly enhanced anticancer potency Chuan Liu, Hongyu Ding, Xiaoxi Li, Christian P. Pallasch, Liya Hong, Dianwu Guo, Yi Chen, Difei Wang, Wei Wang, Yajie
Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008
 Cell Theory 1 Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008 Cell theory: All organisms are made of cells All cells arise from preexisting cells How do new cells arise? Cell division the reproduction
Cell Theory 1 Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008 Cell theory: All organisms are made of cells All cells arise from preexisting cells How do new cells arise? Cell division the reproduction
Chapter 4 Genetics and Cellular Function. The Nucleic Acids (medical history) Chromosome loci. Organization of the Chromatin. Nucleotide Structure
 Chapter 4 Genetics and Cellular Function The Nucleic Acids (medical history) Nucleus and nucleic acids Protein synthesis and secretion DNA replication and the cell cycle Chromosomes and heredity Organization
Chapter 4 Genetics and Cellular Function The Nucleic Acids (medical history) Nucleus and nucleic acids Protein synthesis and secretion DNA replication and the cell cycle Chromosomes and heredity Organization
Inhibitors of mitochondrial Kv1.3 channels induce Bax/Bakindependent death of cancer cells
 Manuscript EMM-2011-01039 Inhibitors of mitochondrial Kv1.3 channels induce Bax/Bakindependent death of cancer cells Luigi Leanza, Brian Henry, Nicola Sassi, Mario Zoratti, K. George Chandy, Erich Gulbins
Manuscript EMM-2011-01039 Inhibitors of mitochondrial Kv1.3 channels induce Bax/Bakindependent death of cancer cells Luigi Leanza, Brian Henry, Nicola Sassi, Mario Zoratti, K. George Chandy, Erich Gulbins
Cell Division Mitosis Notes
 Cell Division Mitosis Notes Cell Division process by which a cell divides into 2 new cells Why do cells need to divide? 1.Living things grow by producing more cells, NOT because each cell increases in
Cell Division Mitosis Notes Cell Division process by which a cell divides into 2 new cells Why do cells need to divide? 1.Living things grow by producing more cells, NOT because each cell increases in
General Biology. Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells, or cell division
 General Biology Course No: BNG2003" Credits: 3.00 " " " 8. The Cell Cycle Prof. Dr. Klaus Heese Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells,
General Biology Course No: BNG2003" Credits: 3.00 " " " 8. The Cell Cycle Prof. Dr. Klaus Heese Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells,
TITLE: The Role Of Alternative Splicing In Breast Cancer Progression
 AD Award Number: W81XWH-06-1-0598 TITLE: The Role Of Alternative Splicing In Breast Cancer Progression PRINCIPAL INVESTIGATOR: Klemens J. Hertel, Ph.D. CONTRACTING ORGANIZATION: University of California,
AD Award Number: W81XWH-06-1-0598 TITLE: The Role Of Alternative Splicing In Breast Cancer Progression PRINCIPAL INVESTIGATOR: Klemens J. Hertel, Ph.D. CONTRACTING ORGANIZATION: University of California,
Physiological release of endogenous tau is stimulated by neuronal activity
 Manuscript EMBOR-2013-37101 Physiological release of endogenous tau is stimulated by neuronal activity Emma C Phillips, Dawn HW Lau, Wendy Noble, Diane P Hanger and Amy M Pooler Corresponding author: Amy
Manuscript EMBOR-2013-37101 Physiological release of endogenous tau is stimulated by neuronal activity Emma C Phillips, Dawn HW Lau, Wendy Noble, Diane P Hanger and Amy M Pooler Corresponding author: Amy
Cell Division Mitosis Notes
 Cell Division Mitosis Notes Cell Division process by which a cell divides into 2 new cells Why do cells need to divide? 1.Living things grow by producing more cells, NOT because each cell increases in
Cell Division Mitosis Notes Cell Division process by which a cell divides into 2 new cells Why do cells need to divide? 1.Living things grow by producing more cells, NOT because each cell increases in
How Cells Divide. Chapter 10
 How Cells Divide Chapter 10 Bacterial Cell Division Bacteria divide by binary fission. -the single, circular bacterial chromosome is replicated -replication begins at the origin of replication and proceeds
How Cells Divide Chapter 10 Bacterial Cell Division Bacteria divide by binary fission. -the single, circular bacterial chromosome is replicated -replication begins at the origin of replication and proceeds
CELL CYCLE INTRODUCTION PART I ANIMAL CELL CYCLE INTERPHASE
 CELL CYCLE INTRODUCTION The nuclei in cells of eukaryotic organisms contain chromosomes with clusters of genes, discrete units of hereditary information consisting of double-stranded DNA. Structural proteins
CELL CYCLE INTRODUCTION The nuclei in cells of eukaryotic organisms contain chromosomes with clusters of genes, discrete units of hereditary information consisting of double-stranded DNA. Structural proteins
CKIP-1 couples Smurf1 ubiquitin ligase with Rpt6 subunit of proteasome to promote substrate degradation
 Manuscript EMBO-2012-36312 CKIP-1 couples Smurf1 ubiquitin ligase with Rpt6 subunit of proteasome to promote substrate degradation Yifang Wang, Jing Nie, Yiwu Wang, Luo Zhang, Guichun Xing, Ping Xie, Fuchu
Manuscript EMBO-2012-36312 CKIP-1 couples Smurf1 ubiquitin ligase with Rpt6 subunit of proteasome to promote substrate degradation Yifang Wang, Jing Nie, Yiwu Wang, Luo Zhang, Guichun Xing, Ping Xie, Fuchu
