GWAS Meets Microarray: Are the Results of Genome- Wide Association Studies and Gene-Expression Profiling Consistent? Prostate Cancer as an Example
|
|
|
- Kenneth Chandler
- 5 years ago
- Views:
Transcription
1 GWAS Meets Microarray: Are the Results of Genome- Wide Association Studies and Gene-Expression Profiling Consistent? Prostate Cancer as an Example Ivan P. Gorlov 1 *, Gary E. Gallick 1, Olga Y. Gorlova 2, Christopher Amos 2, Christopher J. Logothetis 1 1 Department of Genitourinary Medical Oncology, The University of Texas M. D. Anderson Cancer Center, Houston, Texas, United States of America, 2 Department of Epidemiology, The University of Texas M. D. Anderson Cancer Center, Houston, Texas, United States of America Abstract Background: Genome-wide association studies (GWASs) and global profiling of gene expression (microarrays) are two major technological breakthroughs that allow hypothesis-free identification of candidate genes associated with tumorigenesis. It is not obvious whether there is a consistency between the candidate genes identified by GWAS (GWAS genes) and those identified by profiling gene expression (microarray genes). Methodology/Principal Findings: We used the Cancer Genetic Markers Susceptibility database to retrieve single nucleotide polymorphisms from candidate genes for prostate cancer. In addition, we conducted a large meta-analysis of gene expression data in normal prostate and prostate tumor tissue. We identified 13,905 genes that were interrogated by both GWASs and microarrays. On the basis of P values from GWASs, we selected 1,649 most significantly associated genes for functional annotation by the Database for Annotation, Visualization and Integrated Discovery. We also conducted functional annotation analysis using same number of the top genes identified in the meta-analysis of the gene expression data. We found that genes involved in cell adhesion were overrepresented among both the GWAS and microarray genes. Conclusions/Significance: We conclude that the results of these analyses suggest that combining GWAS and microarray data would be a more effective approach than analyzing individual datasets and can help to refine the identification of candidate genes and functions associated with tumor development. Citation: Gorlov IP, Gallick GE, Gorlova OY, Amos C, Logothetis CJ (2009) GWAS Meets Microarray: Are the Results of Genome-Wide Association Studies and Gene- Expression Profiling Consistent? Prostate Cancer as an Example. PLoS ONE 4(8): e6511. doi: /journal.pone Editor: Eshel Ben-Jacob, Tel Aviv University, Israel Received May 11, 2009; Accepted June 29, 2009; Published August 4, 2009 Copyright: ß 2009 Gorlov et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Funding: This study was supported by the David Koch Center for Applied Research in Genitourinary Cancer. Partial support for this study has been provided by US National Institute of Health grants R01CA A2 to CA and AR subcontract to OG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Competing Interests: The authors have declared that no competing interests exist. * ipgorlov@mdanderson.org Introduction Microarray technology allows simultaneous assessment of the expression of virtually all genes in the genome. This approach has been widely used to identify candidate genes associated with cancer development and progression [1 3]. Genome-wide association studies (GWASs) have recently emerged as a powerful tool to identify genetic polymorphisms associated with cancer risk [4,5]. In a GWAS, hundreds of thousands of single nucleotide polymorphisms (SNPs) are genotyped in a large number of cases and controls. A difference in allelic or genotype frequencies between cases and controls suggests an association between cancer risk and the SNP and a linked gene or regulatory region. Whether these two approaches produce comparable results has not been examined. Recently Chen et al. [6] identified the genes that tend to be differentially expressed across various experiential conditions and states using gene-expression data from the Gene Expression Omnibus (GEO). They found that differentially expressed genes are more likely to be detected as disease variants in association studies. In this study, we undertook a more direct approach to link GWAS and microarray data. We performed functional annotations of the top genes identified in prostate cancer GWASs and the same number of the top candidate genes identified in a meta-analysis of the gene-expression data for normal prostate and prostate tumor. The results of our analyses indicate that these two approaches yield similar results at the functional level. Materials and Methods Several prostate cancer GWASs were recently conducted, and a number of candidate genes were identified (Table 1) [7 10]. Though only a few SNPs with the genome-wide level of significance, 10 27, were identified in these studies, a number of SNPs were significant at the level of individual tests but nonsignificant after correction for multiple testing. Such SNPs likely indicate enrichment with causal SNPs that do not reach the genome-wide significance level because of their small effect size or low allele frequency [11]. The GWAS data for this analysis were retrieved from the Cancer Genetic Markers Susceptibility (CGEMS) database, cancer.gov/about/executive_summary.asp. We used the Oncomine database to conduct a PLoS ONE 1 August 2009 Volume 4 Issue 8 e6511
2 Table 1. Candidate genes identified in prostate cancer GWAS. Gene GeneID Comparison PMID Microarray Ps CTBP nonaggressive vs. aggressive prostate cancer EHBP case control study SLC22A case control study LMTK case control study KLK case control study KLK3 354 case control study NUDT case control study DAB2IP nonaggressive vs. aggressive prostate cancer HNF1B 6928 nonaggressive vs. aggressive prostate cancer HPC1(RNASEL) 6041 controls vs. sporadic prostate cancer JAZF nonaggressive vs. aggressive prostate cancer MSMB 4477 nonaggressive vs. aggressive prostate cancer MYC 4609 controls vs. cases doi: /journal.pone t001 meta-analysis of the number of studies comparing gene expression in normal prostate tissue with that of localized prostate tumor tissue [12]. The complete list of the studies used in the meta-analysis can be found in the supplementary materials (Table S1). We used an extension of Stouffer s method [13] for the meta-analysis. This approach is based on estimating the standard normal deviation, Z, and is similar to the approach recently proposed by Ochsner et al. [14]. The meta-analysis identified a number of genes differentially expressed between normal prostate and prostate tumor. Results As an initial validation of our hypothesis that GWASs and microarrays tend to identify the same genes, we used a metaanalysis of the Oncomine gene-expression data to assess the expression of the GWAS-identified genes (Table 1). We found that all but three (HNF1B, EHBP1, and LMTK2) of the genes were differentially expressed between the normal and tumorous prostate. Therefore, 10 of 13 (77%) of the GWAS genes were differentially expressed in the transition from normal prostate to prostate cancer that is higher than one can expect to detect among randomly chosen 13 genes 21.1 (x 2 = 20.9, df = 1, P,0.0001). The prostate cancer GWAS data from CGEMS Phase 1A and Phase 1B, were used in the analysis. We limited our analysis to the gene-associated SNPs to make GWAS and microarray results comparable. We followed the CGEMS designation of the geneassociated SNPs. A total 63,831 gene-associated SNPs belonging to 16,550 unique genes were identified. For each gene, a SNP with the smallest P value was used to characterize an association. If a given SNP was associated with multiple genes, all those associations were included in our analysis. Because in many cases aliases rather than official gene names were used in GWAS, we linked various gene identifiers to the official gene names and EntrezIDs using the latest version of the NCBI gene database (accessed January 17, 2009). Overlapping of the unique GWAS and microarray genes demonstrated that 13,905 genes were assessed in both GWAS and microarray analyses. The list of the genes with corresponding GWAS and microarray P values is shown in Table S1. To assess whether the GWAS and microarray analyses tend to identify similar sets of genes we assess a correlation between log(p) values based on GWAS data and log(p) values based on the analysis of gene expression. We found a small but significant (because of the large sample size) positive correlation between GWAS and microarray log(p)s (Figure 1). The Database for Annotation, Visualization and Integrated Discovery (DAVID) [15] was used for the functional annotation of GWAS and microarray genes. We selected genes with GWAS P values #0.01. A total of 1,649 genes were identified. We used exactly the same number of the top genes identified in the metaanalysis of the gene-expression data. To control for possible biases in gene selection, we used the list of 13,905 genes as background. Functional annotation charts were used to retrieve an extended annotation coverage that included more than 40 annotation categories [15]. A functional chart for the top GWAS genes can be found in Table S2. Many cell adhesion related categories are among the top annotation categories. Clustering of the terms of functional annotations summarized all types of the functional description used by DAVID, identifying cell adhesion as the top cluster, followed by plasma membrane and fibronectin. Functional annotation of the top differentially expressed genes identified cytoskeleton, focal adhesion, extracellular matrix, and cell adhesion as the top annotation terms (Table S3). Clustering of the terms of functional annotation demonstrated cytoskeleton, actin cytoskeleton, extracellular matrix, and cell adhesion among the top identified clusters. Figure 2 shows the results of the clustering of functional terms by DAVID based on the analysis of the top GWAS and differentially expressed genes (see also Table S4). In both lists, most of the top functional clusters derived from GWAS and microarray data are directly or indirectly related to cell adhesion. We next looked for an overlap between the top 1,649 GWAS and the top 1,649 differentially expressed genes. We identified 248 appearing in both lists genes (see supplementary materials for the list of the genes). This number is higher than would be expected by chance. If we randomly sample 1,649 genes from among the 13,905 genes, the expected number of the genes found in two independent samples would be (1,649/13,905)ˆ2*1,649 = The functional annotation of these 248 genes identified cytoskeleton, focal adhesion, and actin binding as top functional categories. Functional clustering of the genes identified cell migration, cell motility, cytoskeleton, and cell adhesion as the top clusters. PLoS ONE 2 August 2009 Volume 4 Issue 8 e6511
3 Figure 1. The plot of GWAS and microarray log(p)s. Black line shows the linear regression curve, red line moving average computed using a sliding window of 100 points. Spearman s rank-order correlation coefficient: r = 0.043, N = 13905, P = doi: /journal.pone g001 Discussion GWAS and microarray analyses both allow unbiased identification of candidate genes and pathways associated with cancer development. These two approaches each have advantages and drawbacks. By combining data from multiple expression studies, analyses of gene expressions have the statistical power to detect even small differences in gene expression between normal and tumor tissues. On the other hand, because genes in the human genome are involved in multiple interactions, modulation of the expression of a single gene may cause a ripple effect on multiple downstream targets, making it difficult to separate causal and induced changes in gene expression. This is unlikely to be an issue in GWASs. GWASs, however, are often statistically underpowered to detect SNPs with small effect size. When we compared candidate genes for prostate cancer identified by GWAS with those identified by microarray, we noted a significant positive correlation between the GWAS and microarray log(p)s. The correlation was small, with the Pearson rank correlation coefficient being only 0.04, but positive correlation between two ranks is expected to be driven by a relatively small number of causal genes. Not all causal genes will be detected by GWAS. Even if the gene is mechanistically linked to prostate tumorigenesis, it can be detected by GWAS only if it carries genetic variants that modulate its function. On the other hand, genes identified by microarray analysis are expected to be a mix of causal genes and the genes that are differentially expressed because of the ripple effect of the causal genes. This suggests that only a fraction of the genes significant in both analyses are causal genes. We found that the top GWAS and differentially expressed candidates were enriched in cell adhesion genes. If we consider all known cell adhesion genes in the genome, only 74 genes or 10% of them were among the top differentially expressed genes. If the cell adhesion pathway is associated with prostate tumorigenesis, one can expect that other cell adhesion genes those that did not make it to the top 1,649 genes also will tend to be significantly positively associated. We found that the average GWAS-derived P value for the cell adhesion genes that failed to reach the top 1649 was lower than the average value for the GWAS genes (t test = 2.9, df = 13,902, P = 0.001). A similar result was obtained for the P values derived from the analysis of the gene expression: the absolute Z score was higher among cell adhesion genes (excluding those among the top 1649 genes) than was the average Z score (t test = 1.81, df = 17811, P = 0.07 on the two-tailed test and P = 0.03 on the one-tailed test). This suggests that cell adhesion function as a whole is associated with prostate tumorigenesis. Both GWAS and microarray genes form functional clusters related to different aspects of cell adhesion, including cell adhesion itself, cell junction, extracellular matrix glycoproteins, fibronectin, actin cytoskeleton, and cell motility. Several other clusters also show a mechanistic association with cell adhesion. For example, cadherin uptake from the cell surface by endocytosis regulates the level of the free cadherins on the cell surface and therefore cell adhesion [16]. Also, zinc finger proteins with the LIM domain are important for focal adhesion and cell adhesion to fibronectin [17,18]. The modulation of the cell adhesion function seems not to be limited to any specific adhesion type but includes cadherins, integrins, and selectins as well as adhesion molecules associated with tight junctions. The results of a number of studies suggested the involvement of the cell adhesion system in prostate cancer development. Cadherins play a role in regulating tumor cell proliferation through cyclins and cyclin-dependent kinases [19]. Integrins are involved in different aspects of prostate tumorigenesis, including cell proliferation, cell motility, and apoptosis [20 22]. Modulation of cell adhesion can play an important role in epithelial-tomesenchymal transition that is believed to be a key step in malignant transformation [23 25]. Also the results of a number of studies suggestd an involvement of cell adhesion in angiogenesis [26 28]. GWAS-identified genes are considered to be cancer susceptibility genes that are mainly associated with tumor initiation. We believe, however, that genes identified by GWAS are also likely to include genes important for tumor progression. Indeed, the detection of tumor is usually symptomatic: the tumor needs to reach a certain size to be detected. This suggests that genes involved in tumor progression will be among GWAS-detected candidate genes. Therefore, GWAS and gene expression analysis may target essentially the same set of genes, providing the theoretical basis for the joint analysis of GWAS and microarray data. PLoS ONE 3 August 2009 Volume 4 Issue 8 e6511
4 Figure 2. Clustering of the functional annotation terms based on GWAS- (upper panel) and microarray-derived genes (lower panel). Functional clusters related to cell adhesion are shown in blue. Detailed information on the composition of clusters can be found in Table S4. doi: /journal.pone g002 In summary, our analysis found a considerable overlap between prostate cancer genes identified by GWAS and those identified by global profiling of the gene expression. We identified cell adhesion as a biological function associated with prostate tumorigenesis. The results of this study also suggest that combining GWAS and microarray data might be a more effective approach than using just the analysis of the individual datasets, and can help to refine the identification of candidate genes and/or functions involved in tumor development. Supporting Information Table S1 Found at: doi: /journal.pone s001 (0.83 MB PDF) Table S2 Found at: doi: /journal.pone s002 (0.07 MB PLoS ONE 4 August 2009 Volume 4 Issue 8 e6511
5 Table S3 Found at: doi: /journal.pone s003 (0.33 MB Table S4 Found at: doi: /journal.pone s004 (0.33 MB References 1. Lacroix L, Commo F, Soria JC (2008) Gene expression profiling of non-smallcell lung cancer. Expert Rev Mol Diagn 8: Konstantinopoulos PA, Spentzos D, Cannistra SA (2008) Gene-expression profiling in epithelial ovarian cancer. Nat Clin Pract Oncol 5: Bao T, Davidson NE (2008) Gene expression profiling of breast cancer. Adv Surg 42: McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9: Browning SR (2008) Missing data imputation and haplotype phase inference for genome-wide association studies. Hum Genet 124: Chen R, Morgan AA, Dudley J, Deshpande T, Li L, et al. (2008) FitSNPs: highly differentially expressed genes are more likely to have variants associated with disease. Genome Biol 9: R Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, et al. (2007) Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst 99: Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, et al. (2007) Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet 39: Nam RK, Zhang WW, Loblaw DA, Klotz LH, Trachtenberg J, et al. (2008) A genome-wide association screen identifies regions on chromosomes 1q25 and 7p21 as risk loci for sporadic prostate cancer. Prostate Cancer Prostatic Dis 11: Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, et al. (2008) Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet 40: Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI (2008) Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am J Hum Genet 82: Gorlov IP, Gorlova OY, Efstathiou E, Logothetis CJ (2009) Candidate pathways and genes for prostate cancer: a meta-analysis of gene expression data. BMC Cancer, In press. 13. Rosenthal R (1979) The file drawer problem and tolerance for null results. Psychol Bull 86: Ochsner SA, Steffen DL, Hilsenbeck SG, Chen ES, Watkins C, et al. (2009) GEMS (Gene Expression MetaSignatures), a Web resource for querying metaanalysis of expression microarray datasets: 17beta-estradiol in MCF-7 cells. Cancer Res 69: Author Contributions Conceived and designed the experiments: IPG OYG CL. Performed the experiments: IPG CIA. Analyzed the data: IPG GG OYG. Wrote the paper: IPG GG CIA CL. 15. Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: Delva E, Kowalczyk AP (2009) Regulation of cadherin trafficking. Traffic 10: Brown MC, Perrotta JA, Turner CE (1998) Serine and threonine phosphorylation of the paxillin LIM domains regulates paxillin focal adhesion localization and cell adhesion to fibronectin. Mol Biol Cell 9: Hansen MD, Beckerle MC (2006) Opposing roles of zyxin/lpp ACTA repeats and the LIM domain region in cell-cell adhesion. J Biol Chem 281: Mason MD, Davies G, Jiang WG (2002) Cell adhesion molecules and adhesion abnormalities in prostate cancer. Crit Rev Oncol Hematol 41: Moschos SJ, Drogowski LM, Reppert SL, Kirkwood JM (2007) Integrins and cancer. Oncology (Williston Park) 21(9 Suppl 3): Fornaro M, Manes T, Languino LR (2001) Integrins and prostate cancer metastases. Cancer Metastasis Rev 20(3-4): Goel H, Li J, Kogan S, Languino L (2008) Integrins in prostate cancer progression. Endocr Relat Cancer 15: Ke XS, Qu Y, Goldfinger N, Rostad K, Hovland R, et al. (2008) Epithelial to mesenchymal transition of a primary prostate cell line with switches of cell adhesion modules but without malignant transformation. PLoS ONE 3(10): e Takkunen M, Ainola M, Vainionpaa N, Grenman R, Patarroyo M, et al. (2008) Epithelial-mesenchymal transition downregulates laminin alpha5 chain and upregulates laminin alpha4 chain in oral squamous carcinoma cells. Histochem Cell Biol 130: Etienne-Manneville S (2008) Polarity proteins in migration and invasion. Oncogene 27: Piao M, Mori D, Satoh T, Sugita Y, Tokunaga O (2006) Inhibition of endothelial cell proliferation, in vitro angiogenesis, and the down-regulation of cell adhesion-related genes by genistein.combined with a cdna microarray analysis. Endothelium 13: Bazas VM, Lukyanova NY, Demash DV, Galakhin KO, Myasoedov DV (2008) Relation between cell-to-cell adhesion and angiogenesis and clinico-morphological prognostic factors in patients with gastric cancer. Exp Oncol 30: Ramjaun AR, Hodivala-Dilke K (2009) The role of cell adhesion pathways in angiogenesis. Int J Biochem Cell Biol 41: PLoS ONE 5 August 2009 Volume 4 Issue 8 e6511
Supplementary webappendix
 Supplementary webappendix This webappendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Hartman M, Loy EY, Ku CS, Chia KS. Molecular
Supplementary webappendix This webappendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Hartman M, Loy EY, Ku CS, Chia KS. Molecular
Additional Disclosure
 Additional Disclosure The Genetics of Prostate Cancer: Clinical Implications William J. Catalona, MD Collaborator with decode genetics, Inc. Non-paid consultant with no financial interest or support Northwestern
Additional Disclosure The Genetics of Prostate Cancer: Clinical Implications William J. Catalona, MD Collaborator with decode genetics, Inc. Non-paid consultant with no financial interest or support Northwestern
Assessing Accuracy of Genotype Imputation in American Indians
 Assessing Accuracy of Genotype Imputation in American Indians Alka Malhotra*, Sayuko Kobes, Clifton Bogardus, William C. Knowler, Leslie J. Baier, Robert L. Hanson Phoenix Epidemiology and Clinical Research
Assessing Accuracy of Genotype Imputation in American Indians Alka Malhotra*, Sayuko Kobes, Clifton Bogardus, William C. Knowler, Leslie J. Baier, Robert L. Hanson Phoenix Epidemiology and Clinical Research
Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer
 Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer risk in stage 1 (red) and after removing any SNPs within
Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer risk in stage 1 (red) and after removing any SNPs within
Massoud Houshmand National Institute for Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran
 QUID 2017, pp. 669-673, Special Issue N 1- ISSN: 1692-343X, Medellín-Colombia NON-GENE REGION AND INFLUENCES TUMOR CHARACTERISTICS BY LOW-RISK ALLELES IN BREAST CANCER (Recibido el 21-06-2017. Aprobado
QUID 2017, pp. 669-673, Special Issue N 1- ISSN: 1692-343X, Medellín-Colombia NON-GENE REGION AND INFLUENCES TUMOR CHARACTERISTICS BY LOW-RISK ALLELES IN BREAST CANCER (Recibido el 21-06-2017. Aprobado
CS2220 Introduction to Computational Biology
 CS2220 Introduction to Computational Biology WEEK 8: GENOME-WIDE ASSOCIATION STUDIES (GWAS) 1 Dr. Mengling FENG Institute for Infocomm Research Massachusetts Institute of Technology mfeng@mit.edu PLANS
CS2220 Introduction to Computational Biology WEEK 8: GENOME-WIDE ASSOCIATION STUDIES (GWAS) 1 Dr. Mengling FENG Institute for Infocomm Research Massachusetts Institute of Technology mfeng@mit.edu PLANS
Mendelian Randomization
 Mendelian Randomization Drawback with observational studies Risk factor X Y Outcome Risk factor X? Y Outcome C (Unobserved) Confounders The power of genetics Intermediate phenotype (risk factor) Genetic
Mendelian Randomization Drawback with observational studies Risk factor X Y Outcome Risk factor X? Y Outcome C (Unobserved) Confounders The power of genetics Intermediate phenotype (risk factor) Genetic
BST227 Introduction to Statistical Genetics. Lecture 4: Introduction to linkage and association analysis
 BST227 Introduction to Statistical Genetics Lecture 4: Introduction to linkage and association analysis 1 Housekeeping Homework #1 due today Homework #2 posted (due Monday) Lab at 5:30PM today (FXB G13)
BST227 Introduction to Statistical Genetics Lecture 4: Introduction to linkage and association analysis 1 Housekeeping Homework #1 due today Homework #2 posted (due Monday) Lab at 5:30PM today (FXB G13)
Award Number: W81XWH TITLE: Characterizing an EMT Signature in Breast Cancer. PRINCIPAL INVESTIGATOR: Melanie C.
 AD Award Number: W81XWH-08-1-0306 TITLE: Characterizing an EMT Signature in Breast Cancer PRINCIPAL INVESTIGATOR: Melanie C. Bocanegra CONTRACTING ORGANIZATION: Leland Stanford Junior University Stanford,
AD Award Number: W81XWH-08-1-0306 TITLE: Characterizing an EMT Signature in Breast Cancer PRINCIPAL INVESTIGATOR: Melanie C. Bocanegra CONTRACTING ORGANIZATION: Leland Stanford Junior University Stanford,
Tumor microenvironment Interactions and Lung Cancer Invasiveness. Pulmonary Grand Rounds Philippe Montgrain, M.D.
 Tumor microenvironment Interactions and Lung Cancer Invasiveness Pulmonary Grand Rounds Philippe Montgrain, M.D. February 26, 2009 Objectives Review epithelial mesenchymal transition (EMT), and its implications
Tumor microenvironment Interactions and Lung Cancer Invasiveness Pulmonary Grand Rounds Philippe Montgrain, M.D. February 26, 2009 Objectives Review epithelial mesenchymal transition (EMT), and its implications
Cancer-Drug Associations: A Complex System
 Cancer-Drug Associations: A Complex System Ertugrul Dalkic 1,2,3, Xuewei Wang 1,4, Neil Wright 1,5, Christina Chan 1,2,3,4,6,7 * 1 Center for Systems Biology, Michigan State University, East Lansing, Michigan,
Cancer-Drug Associations: A Complex System Ertugrul Dalkic 1,2,3, Xuewei Wang 1,4, Neil Wright 1,5, Christina Chan 1,2,3,4,6,7 * 1 Center for Systems Biology, Michigan State University, East Lansing, Michigan,
Supplementary Figure S1A
 Supplementary Figure S1A-G. LocusZoom regional association plots for the seven new cross-cancer loci that were > 1 Mb from known index SNPs. Genes up to 500 kb on either side of each new index SNP are
Supplementary Figure S1A-G. LocusZoom regional association plots for the seven new cross-cancer loci that were > 1 Mb from known index SNPs. Genes up to 500 kb on either side of each new index SNP are
Deciphering the Role of micrornas in BRD4-NUT Fusion Gene Induced NUT Midline Carcinoma
 www.bioinformation.net Volume 13(6) Hypothesis Deciphering the Role of micrornas in BRD4-NUT Fusion Gene Induced NUT Midline Carcinoma Ekta Pathak 1, Bhavya 1, Divya Mishra 1, Neelam Atri 1, 2, Rajeev
www.bioinformation.net Volume 13(6) Hypothesis Deciphering the Role of micrornas in BRD4-NUT Fusion Gene Induced NUT Midline Carcinoma Ekta Pathak 1, Bhavya 1, Divya Mishra 1, Neelam Atri 1, 2, Rajeev
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes
 Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Large-scale identity-by-descent mapping discovers rare haplotypes of large effect. Suyash Shringarpure 23andMe, Inc. ASHG 2017
 Large-scale identity-by-descent mapping discovers rare haplotypes of large effect Suyash Shringarpure 23andMe, Inc. ASHG 2017 1 Why care about rare variants of large effect? Months from randomization 2
Large-scale identity-by-descent mapping discovers rare haplotypes of large effect Suyash Shringarpure 23andMe, Inc. ASHG 2017 1 Why care about rare variants of large effect? Months from randomization 2
The Angiopoietin Axis in Cancer
 Ang2 Ang1 The Angiopoietin Axis in Cancer Tie2 An Overview: The Angiopoietin Axis Plays an Essential Role in the Regulation of Tumor Angiogenesis Growth of a tumor beyond a limiting size is dependent upon
Ang2 Ang1 The Angiopoietin Axis in Cancer Tie2 An Overview: The Angiopoietin Axis Plays an Essential Role in the Regulation of Tumor Angiogenesis Growth of a tumor beyond a limiting size is dependent upon
of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed.
 Supplementary Note The potential association and implications of HBV integration at known and putative cancer genes of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed. Human telomerase
Supplementary Note The potential association and implications of HBV integration at known and putative cancer genes of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed. Human telomerase
Negative Regulation of c-myc Oncogenic Activity Through the Tumor Suppressor PP2A-B56α
 Negative Regulation of c-myc Oncogenic Activity Through the Tumor Suppressor PP2A-B56α Mahnaz Janghorban, PhD Dr. Rosalie Sears lab 2/8/2015 Zanjan University Content 1. Background (keywords: c-myc, PP2A,
Negative Regulation of c-myc Oncogenic Activity Through the Tumor Suppressor PP2A-B56α Mahnaz Janghorban, PhD Dr. Rosalie Sears lab 2/8/2015 Zanjan University Content 1. Background (keywords: c-myc, PP2A,
Crosstalk between Adiponectin and IGF-IR in breast cancer. Prof. Young Jin Suh Department of Surgery The Catholic University of Korea
 Crosstalk between Adiponectin and IGF-IR in breast cancer Prof. Young Jin Suh Department of Surgery The Catholic University of Korea Obesity Chronic, multifactorial disorder Hypertrophy and hyperplasia
Crosstalk between Adiponectin and IGF-IR in breast cancer Prof. Young Jin Suh Department of Surgery The Catholic University of Korea Obesity Chronic, multifactorial disorder Hypertrophy and hyperplasia
Single SNP/Gene Analysis. Typical Results of GWAS Analysis (Single SNP Approach) Typical Results of GWAS Analysis (Single SNP Approach)
 High-Throughput Sequencing Course Gene-Set Analysis Biostatistics and Bioinformatics Summer 28 Section Introduction What is Gene Set Analysis? Many names for gene set analysis: Pathway analysis Gene set
High-Throughput Sequencing Course Gene-Set Analysis Biostatistics and Bioinformatics Summer 28 Section Introduction What is Gene Set Analysis? Many names for gene set analysis: Pathway analysis Gene set
DOES THE BRCAX GENE EXIST? FUTURE OUTLOOK
 CHAPTER 6 DOES THE BRCAX GENE EXIST? FUTURE OUTLOOK Genetic research aimed at the identification of new breast cancer susceptibility genes is at an interesting crossroad. On the one hand, the existence
CHAPTER 6 DOES THE BRCAX GENE EXIST? FUTURE OUTLOOK Genetic research aimed at the identification of new breast cancer susceptibility genes is at an interesting crossroad. On the one hand, the existence
Novel Biomarkers (Kallikreins) for Prognosis and Therapy Response in Ovarian cancer
 Novel Biomarkers (Kallikreins) for Prognosis and Therapy Response in Ovarian cancer Eleftherios P. Diamandis, M.D., Ph.D., FRCP(C) EORTC-NCI-ASCO Meeting,November 16, 2007 Yousef GM, Diamandis EP. Endocr.
Novel Biomarkers (Kallikreins) for Prognosis and Therapy Response in Ovarian cancer Eleftherios P. Diamandis, M.D., Ph.D., FRCP(C) EORTC-NCI-ASCO Meeting,November 16, 2007 Yousef GM, Diamandis EP. Endocr.
MORE ON SNPS AND PCA. Terrence P McGarty White Paper No 119 November, 2014
 MORE ON SNPS AND PCA Terrence P McGarty White Paper No 119 November, 2014 There has been a continuing focus on SNPs and Prostate Cancer. This document reviews some of this recent works and adds to what
MORE ON SNPS AND PCA Terrence P McGarty White Paper No 119 November, 2014 There has been a continuing focus on SNPs and Prostate Cancer. This document reviews some of this recent works and adds to what
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded
 Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, Maryland
 AD Award Number: W81XWH-06-1-0181 TITLE: Analysis of Ethnic Admixture in Prostate Cancer PRINCIPAL INVESTIGATOR: Cathryn Bock, Ph.D. CONTRACTING ORGANIZATION: Wayne State University Detroit, MI 48202 REPORT
AD Award Number: W81XWH-06-1-0181 TITLE: Analysis of Ethnic Admixture in Prostate Cancer PRINCIPAL INVESTIGATOR: Cathryn Bock, Ph.D. CONTRACTING ORGANIZATION: Wayne State University Detroit, MI 48202 REPORT
Utility of Incorporating Genetic Variants for the Early Detection of Prostate Cancer
 Utility of Incorporating Genetic Variants for the Early Detection of Prostate Cancer Robert K. Nam, 1,9 William W. Zhang, 1 John Trachtenberg, 6 Arun Seth, 2 Laurence H. Klotz, 1 Aleksandra Stanimirovic,
Utility of Incorporating Genetic Variants for the Early Detection of Prostate Cancer Robert K. Nam, 1,9 William W. Zhang, 1 John Trachtenberg, 6 Arun Seth, 2 Laurence H. Klotz, 1 Aleksandra Stanimirovic,
Network-assisted data analysis
 Network-assisted data analysis Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Protein identification in shotgun proteomics Protein digestion LC-MS/MS Protein
Network-assisted data analysis Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Protein identification in shotgun proteomics Protein digestion LC-MS/MS Protein
CHAPTER VII CONCLUDING REMARKS AND FUTURE DIRECTION. Androgen deprivation therapy is the most used treatment of de novo or recurrent
 CHAPTER VII CONCLUDING REMARKS AND FUTURE DIRECTION Stathmin in Prostate Cancer Development and Progression Androgen deprivation therapy is the most used treatment of de novo or recurrent metastatic PCa.
CHAPTER VII CONCLUDING REMARKS AND FUTURE DIRECTION Stathmin in Prostate Cancer Development and Progression Androgen deprivation therapy is the most used treatment of de novo or recurrent metastatic PCa.
The Hallmarks of Cancer
 The Hallmarks of Cancer Theresa L. Hodin, Ph.D. Clinical Research Services Theresa.Hodin@RoswellPark.org Hippocrates Cancer surgery, circa 1689 Cancer Surgery Today 1971: Nixon declares War on Cancer
The Hallmarks of Cancer Theresa L. Hodin, Ph.D. Clinical Research Services Theresa.Hodin@RoswellPark.org Hippocrates Cancer surgery, circa 1689 Cancer Surgery Today 1971: Nixon declares War on Cancer
CCN1: A NOVEL TARGET FOR PANCREATIC CANCER. Andrew Leask.
 CCN1: A NOVEL TARGET FOR PANCREATIC CANCER Andrew Leask CIHR Group in Skeletal Development and Remodeling, Division of Oral Biology and Department of Physiology and Pharmacology, Schulich School of Medicine
CCN1: A NOVEL TARGET FOR PANCREATIC CANCER Andrew Leask CIHR Group in Skeletal Development and Remodeling, Division of Oral Biology and Department of Physiology and Pharmacology, Schulich School of Medicine
What can we contribute to cancer research and treatment from Computer Science or Mathematics? How do we adapt our expertise for them
 From Bioinformatics to Health Information Technology Outline What can we contribute to cancer research and treatment from Computer Science or Mathematics? How do we adapt our expertise for them Introduction
From Bioinformatics to Health Information Technology Outline What can we contribute to cancer research and treatment from Computer Science or Mathematics? How do we adapt our expertise for them Introduction
Epstein-Barr virus driven promoter hypermethylated genes in gastric cancer
 RESEARCH FUND FOR THE CONTROL OF INFECTIOUS DISEASES Epstein-Barr virus driven promoter hypermethylated genes in gastric cancer J Yu *, KF To, QY Liang K e y M e s s a g e s 1. Somatostatin receptor 1
RESEARCH FUND FOR THE CONTROL OF INFECTIOUS DISEASES Epstein-Barr virus driven promoter hypermethylated genes in gastric cancer J Yu *, KF To, QY Liang K e y M e s s a g e s 1. Somatostatin receptor 1
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
 Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
2) Cases and controls were genotyped on different platforms. The comparability of the platforms should be discussed.
 Reviewers' Comments: Reviewer #1 (Remarks to the Author) The manuscript titled 'Association of variations in HLA-class II and other loci with susceptibility to lung adenocarcinoma with EGFR mutation' evaluated
Reviewers' Comments: Reviewer #1 (Remarks to the Author) The manuscript titled 'Association of variations in HLA-class II and other loci with susceptibility to lung adenocarcinoma with EGFR mutation' evaluated
On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles
 On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles Ying-Wooi Wan 1,2,4, Claire M. Mach 2,3, Genevera I. Allen 1,7,8, Matthew L. Anderson 2,4,5 *, Zhandong Liu 1,5,6,7 * 1 Departments of Pediatrics
On the Reproducibility of TCGA Ovarian Cancer MicroRNA Profiles Ying-Wooi Wan 1,2,4, Claire M. Mach 2,3, Genevera I. Allen 1,7,8, Matthew L. Anderson 2,4,5 *, Zhandong Liu 1,5,6,7 * 1 Departments of Pediatrics
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Introduction to Genetics and Genomics
 2016 Introduction to enetics and enomics 3. ssociation Studies ggibson.gt@gmail.com http://www.cig.gatech.edu Outline eneral overview of association studies Sample results hree steps to WS: primary scan,
2016 Introduction to enetics and enomics 3. ssociation Studies ggibson.gt@gmail.com http://www.cig.gatech.edu Outline eneral overview of association studies Sample results hree steps to WS: primary scan,
MRC-Holland MLPA. Description version 06; 23 December 2016
 SALSA MLPA probemix P417-B2 BAP1 Lot B2-1216. As compared to version B1 (lot B1-0215), two reference probes have been added and two target probes have a minor change in length. The BAP1 (BRCA1 associated
SALSA MLPA probemix P417-B2 BAP1 Lot B2-1216. As compared to version B1 (lot B1-0215), two reference probes have been added and two target probes have a minor change in length. The BAP1 (BRCA1 associated
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies
 Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
Introduction to the Genetics of Complex Disease
 Introduction to the Genetics of Complex Disease Jeremiah M. Scharf, MD, PhD Departments of Neurology, Psychiatry and Center for Human Genetic Research Massachusetts General Hospital Breakthroughs in Genome
Introduction to the Genetics of Complex Disease Jeremiah M. Scharf, MD, PhD Departments of Neurology, Psychiatry and Center for Human Genetic Research Massachusetts General Hospital Breakthroughs in Genome
A bioinformatics driven cancer study
 The 11th China-Japan-Korea Bioinformatics Training Course & Symposium, June 16-18, Suzhou, China A bioinformatics driven cancer study Chao Li and Yixue Li yxli@sibs.ac.cn Shanghai Institutes for Bio-Medicine
The 11th China-Japan-Korea Bioinformatics Training Course & Symposium, June 16-18, Suzhou, China A bioinformatics driven cancer study Chao Li and Yixue Li yxli@sibs.ac.cn Shanghai Institutes for Bio-Medicine
UNIVERSITY OF CALIFORNIA, LOS ANGELES
 UNIVERSITY OF CALIFORNIA, LOS ANGELES BERKELEY DAVIS IRVINE LOS ANGELES MERCED RIVERSIDE SAN DIEGO SAN FRANCISCO UCLA SANTA BARBARA SANTA CRUZ DEPARTMENT OF EPIDEMIOLOGY SCHOOL OF PUBLIC HEALTH CAMPUS
UNIVERSITY OF CALIFORNIA, LOS ANGELES BERKELEY DAVIS IRVINE LOS ANGELES MERCED RIVERSIDE SAN DIEGO SAN FRANCISCO UCLA SANTA BARBARA SANTA CRUZ DEPARTMENT OF EPIDEMIOLOGY SCHOOL OF PUBLIC HEALTH CAMPUS
In vitro scratch assay: method for analysis of cell migration in vitro labeled fluorodeoxyglucose (FDG)
 In vitro scratch assay: method for analysis of cell migration in vitro labeled fluorodeoxyglucose (FDG) 1 Dr Saeb Aliwaini 13/11/2015 Migration in vivo Primary tumors are responsible for only about 10%
In vitro scratch assay: method for analysis of cell migration in vitro labeled fluorodeoxyglucose (FDG) 1 Dr Saeb Aliwaini 13/11/2015 Migration in vivo Primary tumors are responsible for only about 10%
Supplementary Tables. Supplementary Figures
 Supplementary Files for Zehir, Benayed et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients Supplementary Tables Supplementary Table 1: Sample
Supplementary Files for Zehir, Benayed et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients Supplementary Tables Supplementary Table 1: Sample
Conditions. Name : dummy Age/sex : xx Y /x. Lab No : xxxxxxxxx. Rep Centre : xxxxxxxxxxx Ref by : Dr. xxxxxxxxxx
 Name : dummy Age/sex : xx Y /x Lab No : xxxxxxxxx Rep Centre : xxxxxxxxxxx Ref by : Dr. xxxxxxxxxx Rec. Date : xx/xx/xx Rep Date : xx/xx/xx GENETIC MAPPING FOR ONCOLOGY Conditions Melanoma Prostate Cancer
Name : dummy Age/sex : xx Y /x Lab No : xxxxxxxxx Rep Centre : xxxxxxxxxxx Ref by : Dr. xxxxxxxxxx Rec. Date : xx/xx/xx Rep Date : xx/xx/xx GENETIC MAPPING FOR ONCOLOGY Conditions Melanoma Prostate Cancer
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Correlation between expression and significance of δ-catenin, CD31, and VEGF of non-small cell lung cancer
 Correlation between expression and significance of δ-catenin, CD31, and VEGF of non-small cell lung cancer X.L. Liu 1, L.D. Liu 2, S.G. Zhang 1, S.D. Dai 3, W.Y. Li 1 and L. Zhang 1 1 Thoracic Surgery,
Correlation between expression and significance of δ-catenin, CD31, and VEGF of non-small cell lung cancer X.L. Liu 1, L.D. Liu 2, S.G. Zhang 1, S.D. Dai 3, W.Y. Li 1 and L. Zhang 1 1 Thoracic Surgery,
Supplementary note: Comparison of deletion variants identified in this study and four earlier studies
 Supplementary note: Comparison of deletion variants identified in this study and four earlier studies Here we compare the results of this study to potentially overlapping results from four earlier studies
Supplementary note: Comparison of deletion variants identified in this study and four earlier studies Here we compare the results of this study to potentially overlapping results from four earlier studies
The lymphoma-associated NPM-ALK oncogene elicits a p16ink4a/prb-dependent tumor-suppressive pathway. Blood Jun 16;117(24):
 DNA Sequencing Publications Standard Sequencing 1 Carro MS et al. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006 Jun;5(11) 2 Lassandro L et al. The DNA sequence
DNA Sequencing Publications Standard Sequencing 1 Carro MS et al. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006 Jun;5(11) 2 Lassandro L et al. The DNA sequence
CELL BIOLOGY - CLUTCH CH CELL JUNCTIONS AND TISSUES.
 !! www.clutchprep.com CONCEPT: CELL-CELL ADHESION Cells must be able to bind and interact with nearby cells in order to have functional and strong tissues Cells can in two main ways - Homophilic interactions
!! www.clutchprep.com CONCEPT: CELL-CELL ADHESION Cells must be able to bind and interact with nearby cells in order to have functional and strong tissues Cells can in two main ways - Homophilic interactions
Supplementary Figure 1: Func8onal Network Analysis of Kinases Significantly Modulated by MERS CoV Infec8on and Conserved Across All Time Points
 A. B. 8 4 Supplementary Figure : Func8onal Network Analysis of Kinases Significantly Modulated by MERS CoV Infec8on and Conserved Across All Time Points Examined. A) Venn diagram analysis of kinases significantly
A. B. 8 4 Supplementary Figure : Func8onal Network Analysis of Kinases Significantly Modulated by MERS CoV Infec8on and Conserved Across All Time Points Examined. A) Venn diagram analysis of kinases significantly
Predictive Assays in Radiation Therapy
 Outline Predictive Assays in Radiation Therapy Radiation Biology Introduction Early predictive assays Recent trends in predictive assays Examples for specific tumors Summary Lecture 4-23-2014 Introduction
Outline Predictive Assays in Radiation Therapy Radiation Biology Introduction Early predictive assays Recent trends in predictive assays Examples for specific tumors Summary Lecture 4-23-2014 Introduction
Reduced expression of SOX7 in ovarian cancer: a novel tumor suppressor through the Wnt/β-catenin signaling pathway
 Reduced expression of SOX7 in ovarian cancer: a novel tumor suppressor through the Wnt/β-catenin signaling pathway Huidi Liu, M.D. & Ph.D Genomics Research Centre Harbin Medical University, China 15804606535@163.com
Reduced expression of SOX7 in ovarian cancer: a novel tumor suppressor through the Wnt/β-catenin signaling pathway Huidi Liu, M.D. & Ph.D Genomics Research Centre Harbin Medical University, China 15804606535@163.com
Cancer Genetics. What is Cancer? Cancer Classification. Medical Genetics. Uncontrolled growth of cells. Not all tumors are cancerous
 Session8 Medical Genetics Cancer Genetics J avad Jamshidi F a s a U n i v e r s i t y o f M e d i c a l S c i e n c e s, N o v e m b e r 2 0 1 7 What is Cancer? Uncontrolled growth of cells Not all tumors
Session8 Medical Genetics Cancer Genetics J avad Jamshidi F a s a U n i v e r s i t y o f M e d i c a l S c i e n c e s, N o v e m b e r 2 0 1 7 What is Cancer? Uncontrolled growth of cells Not all tumors
Supplementary Figure 1 Dosage correlation between imputed and genotyped alleles Imputed dosages (0 to 2) of 2-digit alleles (red) and 4-digit alleles
 Supplementary Figure 1 Dosage correlation between imputed and genotyped alleles Imputed dosages (0 to 2) of 2-digit alleles (red) and 4-digit alleles (green) of (A) HLA-A, HLA-B, (C) HLA-C, (D) HLA-DQA1,
Supplementary Figure 1 Dosage correlation between imputed and genotyped alleles Imputed dosages (0 to 2) of 2-digit alleles (red) and 4-digit alleles (green) of (A) HLA-A, HLA-B, (C) HLA-C, (D) HLA-DQA1,
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD
 Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Figure S4. 15 Mets Whole Exome. 5 Primary Tumors Cancer Panel and WES. Next Generation Sequencing
 Figure S4 Next Generation Sequencing 15 Mets Whole Exome 5 Primary Tumors Cancer Panel and WES Get coverage of all variant loci for all three Mets Variant Filtering Sequence Alignments Index and align
Figure S4 Next Generation Sequencing 15 Mets Whole Exome 5 Primary Tumors Cancer Panel and WES Get coverage of all variant loci for all three Mets Variant Filtering Sequence Alignments Index and align
White Paper Guidelines on Vetting Genetic Associations
 White Paper 23-03 Guidelines on Vetting Genetic Associations Authors: Andro Hsu Brian Naughton Shirley Wu Created: November 14, 2007 Revised: February 14, 2008 Revised: June 10, 2010 (see end of document
White Paper 23-03 Guidelines on Vetting Genetic Associations Authors: Andro Hsu Brian Naughton Shirley Wu Created: November 14, 2007 Revised: February 14, 2008 Revised: June 10, 2010 (see end of document
Host Genomics of HIV-1
 4 th International Workshop on HIV & Aging Host Genomics of HIV-1 Paul McLaren École Polytechnique Fédérale de Lausanne - EPFL Lausanne, Switzerland paul.mclaren@epfl.ch Complex trait genetics Phenotypic
4 th International Workshop on HIV & Aging Host Genomics of HIV-1 Paul McLaren École Polytechnique Fédérale de Lausanne - EPFL Lausanne, Switzerland paul.mclaren@epfl.ch Complex trait genetics Phenotypic
VIII Curso Internacional del PIRRECV. Some molecular mechanisms of cancer
 VIII Curso Internacional del PIRRECV Some molecular mechanisms of cancer Laboratorio de Comunicaciones Celulares, Centro FONDAP Estudios Moleculares de la Celula (CEMC), ICBM, Facultad de Medicina, Universidad
VIII Curso Internacional del PIRRECV Some molecular mechanisms of cancer Laboratorio de Comunicaciones Celulares, Centro FONDAP Estudios Moleculares de la Celula (CEMC), ICBM, Facultad de Medicina, Universidad
38 Int'l Conf. Bioinformatics and Computational Biology BIOCOMP'16
 38 Int'l Conf. Bioinformatics and Computational Biology BIOCOMP'16 PGAR: ASD Candidate Gene Prioritization System Using Expression Patterns Steven Cogill and Liangjiang Wang Department of Genetics and
38 Int'l Conf. Bioinformatics and Computational Biology BIOCOMP'16 PGAR: ASD Candidate Gene Prioritization System Using Expression Patterns Steven Cogill and Liangjiang Wang Department of Genetics and
CoINcIDE: A framework for discovery of patient subtypes across multiple datasets
 Planey and Gevaert Genome Medicine (2016) 8:27 DOI 10.1186/s13073-016-0281-4 METHOD CoINcIDE: A framework for discovery of patient subtypes across multiple datasets Catherine R. Planey and Olivier Gevaert
Planey and Gevaert Genome Medicine (2016) 8:27 DOI 10.1186/s13073-016-0281-4 METHOD CoINcIDE: A framework for discovery of patient subtypes across multiple datasets Catherine R. Planey and Olivier Gevaert
Intercellular indirect communication
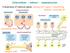 Intercellular indirect communication transmission of chemical signals: sending cell signal transmitting tissue hormone medium receiving cell hormone intercellular fluid blood neurocrine neurotransmitter
Intercellular indirect communication transmission of chemical signals: sending cell signal transmitting tissue hormone medium receiving cell hormone intercellular fluid blood neurocrine neurotransmitter
Genetic Predictors of Radiosensitivity.
 Genetic Predictors of Radiosensitivity. Richard G. Stock, MD Professor, Director of Genito-Urinary Oncology Department of Radiation Oncology Ichan School of Medicine at Mount Sinai New York, NY DISCLOSURE
Genetic Predictors of Radiosensitivity. Richard G. Stock, MD Professor, Director of Genito-Urinary Oncology Department of Radiation Oncology Ichan School of Medicine at Mount Sinai New York, NY DISCLOSURE
Fundamental research on breast cancer in Belgium. Rosita Winkler
 Fundamental research on breast cancer in Belgium Rosita Winkler Medline search for «breast cancer» and Belgium limits: english, posted in the last 5 years. Result: 484 papers - fundamental / clinical -
Fundamental research on breast cancer in Belgium Rosita Winkler Medline search for «breast cancer» and Belgium limits: english, posted in the last 5 years. Result: 484 papers - fundamental / clinical -
Human population sub-structure and genetic association studies
 Human population sub-structure and genetic association studies Stephanie A. Santorico, Ph.D. Department of Mathematical & Statistical Sciences Stephanie.Santorico@ucdenver.edu Global Similarity Map from
Human population sub-structure and genetic association studies Stephanie A. Santorico, Ph.D. Department of Mathematical & Statistical Sciences Stephanie.Santorico@ucdenver.edu Global Similarity Map from
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier
 Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes.
 Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
The diagnostic value of determination of serum GOLPH3 associated with CA125, CA19.9 in patients with ovarian cancer
 European Review for Medical and Pharmacological Sciences 2017; 21: 4039-4044 The diagnostic value of determination of serum GOLPH3 associated with CA125, CA19.9 in patients with ovarian cancer H.-Y. FAN
European Review for Medical and Pharmacological Sciences 2017; 21: 4039-4044 The diagnostic value of determination of serum GOLPH3 associated with CA125, CA19.9 in patients with ovarian cancer H.-Y. FAN
Quantitative Data Analysis Assignment Sample Newessays.co.uk
 Absorbance Quantitative Data Analysis Assignment Sample Newessays.co.uk Part A: Results of the Study Is there a difference of curve profile between the MTT assay and the cell number? What do the different
Absorbance Quantitative Data Analysis Assignment Sample Newessays.co.uk Part A: Results of the Study Is there a difference of curve profile between the MTT assay and the cell number? What do the different
SALSA MLPA probemix P315-B1 EGFR
 SALSA MLPA probemix P315-B1 EGFR Lot B1-0215 and B1-0112. As compared to the previous A1 version (lot 0208), two mutation-specific probes for the EGFR mutations L858R and T709M as well as one additional
SALSA MLPA probemix P315-B1 EGFR Lot B1-0215 and B1-0112. As compared to the previous A1 version (lot 0208), two mutation-specific probes for the EGFR mutations L858R and T709M as well as one additional
Diabetes Mellitus and Breast Cancer
 Masur K, Thévenod F, Zänker KS (eds): Diabetes and Cancer. Epidemiological Evidence and Molecular Links. Front Diabetes. Basel, Karger, 2008, vol 19, pp 97 113 Diabetes Mellitus and Breast Cancer Ido Wolf
Masur K, Thévenod F, Zänker KS (eds): Diabetes and Cancer. Epidemiological Evidence and Molecular Links. Front Diabetes. Basel, Karger, 2008, vol 19, pp 97 113 Diabetes Mellitus and Breast Cancer Ido Wolf
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis
 The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
Src-INACTIVE / Src-INACTIVE
 Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer
 Supplementary Information Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer Lars A. Forsberg, Chiara Rasi, Niklas Malmqvist, Hanna Davies, Saichand
Supplementary Information Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer Lars A. Forsberg, Chiara Rasi, Niklas Malmqvist, Hanna Davies, Saichand
Relationship between SPOP mutation and breast cancer in Chinese population
 Relationship between SPOP mutation and breast cancer in Chinese population M.A. Khan 1 *, L. Zhu 1 *, M. Tania 1, X.L. Xiao 2 and J.J. Fu 1 1 Key Laboratory of Epigenetics and Oncology, The Research Center
Relationship between SPOP mutation and breast cancer in Chinese population M.A. Khan 1 *, L. Zhu 1 *, M. Tania 1, X.L. Xiao 2 and J.J. Fu 1 1 Key Laboratory of Epigenetics and Oncology, The Research Center
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
For more information about how to cite these materials visit
 Author(s): Kerby Shedden, Ph.D., 2010 License: Unless otherwise noted, this material is made available under the terms of the Creative Commons Attribution Share Alike 3.0 License: http://creativecommons.org/licenses/by-sa/3.0/
Author(s): Kerby Shedden, Ph.D., 2010 License: Unless otherwise noted, this material is made available under the terms of the Creative Commons Attribution Share Alike 3.0 License: http://creativecommons.org/licenses/by-sa/3.0/
Lack of association of IL-2RA and IL-2RB polymorphisms with rheumatoid arthritis in a Han Chinese population
 Lack of association of IL-2RA and IL-2RB polymorphisms with rheumatoid arthritis in a Han Chinese population J. Zhu 1 *, F. He 2 *, D.D. Zhang 2 *, J.Y. Yang 2, J. Cheng 1, R. Wu 1, B. Gong 2, X.Q. Liu
Lack of association of IL-2RA and IL-2RB polymorphisms with rheumatoid arthritis in a Han Chinese population J. Zhu 1 *, F. He 2 *, D.D. Zhang 2 *, J.Y. Yang 2, J. Cheng 1, R. Wu 1, B. Gong 2, X.Q. Liu
Identifying Novel Targets for Non-Small Cell Lung Cancer Just How Novel Are They?
 Identifying Novel Targets for Non-Small Cell Lung Cancer Just How Novel Are They? Dubovenko Alexey Discovery Product Manager Sonia Novikova Solution Scientist September 2018 2 Non-Small Cell Lung Cancer
Identifying Novel Targets for Non-Small Cell Lung Cancer Just How Novel Are They? Dubovenko Alexey Discovery Product Manager Sonia Novikova Solution Scientist September 2018 2 Non-Small Cell Lung Cancer
The clinical relevance of circulating, cell-free and exosomal micrornas as biomarkers for gynecological tumors
 Department of Tumor Biology The clinical relevance of circulating, cell-free and exosomal micrornas as biomarkers for gynecological tumors cfdna Copenhagen April 6-7, 2017 Heidi Schwarzenbach, PhD Tumor
Department of Tumor Biology The clinical relevance of circulating, cell-free and exosomal micrornas as biomarkers for gynecological tumors cfdna Copenhagen April 6-7, 2017 Heidi Schwarzenbach, PhD Tumor
High AU content: a signature of upregulated mirna in cardiac diseases
 https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Identificación de vulnerabilidades en tumores sólidos: aportes hacia una medicina personalizada?
 Identificación de vulnerabilidades en tumores sólidos: aportes hacia una medicina personalizada? Alberto Ocana Albacete University Hospital Salamanca May 19th, 2016 WHAT IS PERSONALIZED MEDICINE? Molecular
Identificación de vulnerabilidades en tumores sólidos: aportes hacia una medicina personalizada? Alberto Ocana Albacete University Hospital Salamanca May 19th, 2016 WHAT IS PERSONALIZED MEDICINE? Molecular
APPLICATION NOTE. Highly reproducible and Comprehensive Proteome Profiling of Formalin-Fixed Paraffin-Embedded (FFPE) Tissues Slices
 APPLICATION NOTE Highly reproducible and Comprehensive Proteome Profiling of Formalin-Fixed Paraffin-Embedded (FFPE) Tissues Slices INTRODUCTION Preservation of tissue biopsies is a critical step to This
APPLICATION NOTE Highly reproducible and Comprehensive Proteome Profiling of Formalin-Fixed Paraffin-Embedded (FFPE) Tissues Slices INTRODUCTION Preservation of tissue biopsies is a critical step to This
Chromatin marks identify critical cell-types for fine-mapping complex trait variants
 Chromatin marks identify critical cell-types for fine-mapping complex trait variants Gosia Trynka 1-4 *, Cynthia Sandor 1-4 *, Buhm Han 1-4, Han Xu 5, Barbara E Stranger 1,4#, X Shirley Liu 5, and Soumya
Chromatin marks identify critical cell-types for fine-mapping complex trait variants Gosia Trynka 1-4 *, Cynthia Sandor 1-4 *, Buhm Han 1-4, Han Xu 5, Barbara E Stranger 1,4#, X Shirley Liu 5, and Soumya
Genetic variability of genes involved in DNA repair influence treatment outcome in osteosarcoma
 Genetic variability of genes involved in DNA repair influence treatment outcome in osteosarcoma M.J. Wang, Y. Zhu, X.J. Guo and Z.Z. Tian Department of Orthopaedics, Xinxiang Central Hospital, Xinxiang,
Genetic variability of genes involved in DNA repair influence treatment outcome in osteosarcoma M.J. Wang, Y. Zhu, X.J. Guo and Z.Z. Tian Department of Orthopaedics, Xinxiang Central Hospital, Xinxiang,
Cell Polarity and Cancer
 Cell Polarity and Cancer Pr Jean-Paul Borg Email: jean-paul.borg@inserm.fr Features of malignant cells Steps in Malignant Progression Cell polarity, cell adhesion, morphogenesis and tumorigenesis pathways
Cell Polarity and Cancer Pr Jean-Paul Borg Email: jean-paul.borg@inserm.fr Features of malignant cells Steps in Malignant Progression Cell polarity, cell adhesion, morphogenesis and tumorigenesis pathways
Nature Methods: doi: /nmeth.3115
 Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figures
 Supplementary Figures Supplementary Fig 1. Comparison of sub-samples on the first two principal components of genetic variation. TheBritishsampleisplottedwithredpoints.The sub-samples of the diverse sample
Supplementary Figures Supplementary Fig 1. Comparison of sub-samples on the first two principal components of genetic variation. TheBritishsampleisplottedwithredpoints.The sub-samples of the diverse sample
PATHOBIOLOGY OF NEOPLASIA
 PATHOBIOLOGY OF NEOPLASIA Department of Pathology Gadjah Mada University School of Medicine dr. Harijadi Blok Biomedis, 6 Maret 2009 [12] 3/17/2009 1 The pathobiology of neoplasia Normal cells Malignant
PATHOBIOLOGY OF NEOPLASIA Department of Pathology Gadjah Mada University School of Medicine dr. Harijadi Blok Biomedis, 6 Maret 2009 [12] 3/17/2009 1 The pathobiology of neoplasia Normal cells Malignant
Protein kinase C expression is deregulated in chronic lymphocytic leukemia
 Protein kinase C expression is deregulated in chronic lymphocytic leukemia Kabir, Nuzhat N.; Rönnstrand, Lars; Kazi, Julhash U. Published in: Leukemia & Lymphoma DOI: 10.3109/10428194.2013.769220 2013
Protein kinase C expression is deregulated in chronic lymphocytic leukemia Kabir, Nuzhat N.; Rönnstrand, Lars; Kazi, Julhash U. Published in: Leukemia & Lymphoma DOI: 10.3109/10428194.2013.769220 2013
number Done by Corrected by Doctor Maha Shomaf
 number 19 Done by Waseem Abo-Obeida Corrected by Abdullah Zreiqat Doctor Maha Shomaf Carcinogenesis: the molecular basis of cancer. Non-lethal genetic damage lies at the heart of carcinogenesis and leads
number 19 Done by Waseem Abo-Obeida Corrected by Abdullah Zreiqat Doctor Maha Shomaf Carcinogenesis: the molecular basis of cancer. Non-lethal genetic damage lies at the heart of carcinogenesis and leads
Gene-Environment Interactions
 Gene-Environment Interactions What is gene-environment interaction? A different effect of an environmental exposure on disease risk in persons with different genotypes," or, alternatively, "a different
Gene-Environment Interactions What is gene-environment interaction? A different effect of an environmental exposure on disease risk in persons with different genotypes," or, alternatively, "a different
Identifying Relevant micrornas in Bladder Cancer using Multi-Task Learning
 Identifying Relevant micrornas in Bladder Cancer using Multi-Task Learning Adriana Birlutiu (1),(2), Paul Bulzu (1), Irina Iereminciuc (1), and Alexandru Floares (1) (1) SAIA & OncoPredict, Cluj-Napoca,
Identifying Relevant micrornas in Bladder Cancer using Multi-Task Learning Adriana Birlutiu (1),(2), Paul Bulzu (1), Irina Iereminciuc (1), and Alexandru Floares (1) (1) SAIA & OncoPredict, Cluj-Napoca,
Supplementary Online Content
 Supplementary Online Content Hartwig FP, Borges MC, Lessa Horta B, Bowden J, Davey Smith G. Inflammatory biomarkers and risk of schizophrenia: a 2-sample mendelian randomization study. JAMA Psychiatry.
Supplementary Online Content Hartwig FP, Borges MC, Lessa Horta B, Bowden J, Davey Smith G. Inflammatory biomarkers and risk of schizophrenia: a 2-sample mendelian randomization study. JAMA Psychiatry.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
