1 Supplementary Table Description
|
|
|
- Phillip Thornton
- 5 years ago
- Views:
Transcription
1 1 Supplementary Table Description We created a table that contains information for human genes, including structural annotations and numerical summaries from our single-cell analysis and GTEx validation. The first ten columns contain indicators for whether the gene was classified as a member of each of our structures; these names are denoted as the full name of the structure followed by indicator. The following two columns contain indicators of membership of the gene sets published by Eisenberg and Levanon [2003] and by Lin et al. [2017] and denoted with the author names. Summary statistics, including the mean, standard deviation, and correlation, for each single cell dataset are calculated for the genes expressed in all six scrna-seq datasets. The mean and standard deviation of the log + 1 transformed expressions for each of the genes is calculated for each of the datasets; these variables are denoted with the five digit GEO accession number (GSE#####) followed by either a mean or sd suffix to denote which summary. The correlation with the unadjusted cell total is also included for each dataset, as well as the mean unadjusted cell total across the six datasets. Our analysis in the main text primarily looks at correlations with adjusted cell totals; we provide the mean correlation across the six datasets for each gene with each of the adjusted cell totals. These variables have the name sc cormean less followed by the name of the structure removed during the cell total calculation. Prior to analysis, the GTEx data is normalized according to an adjusted RPKM transformation. The total number of reads for a given sample is calculated and divided by 1,000,000. Each individual entry is then transformed with log (entry + 1/ total samples per million). Then, the mean and standard deviation of each gene are calculated for each of the tissue types contained within the data. F-statistics and their respective degrees of freedom are calculated for each of the tissue types that contain subtissue types; these F-statistics are a measure of how differentially expressed the gene is between the subtissue types. Note that genes that are stably expressed will have low F-statistics. These variables are denoted with GTEx tissue type statistic, where the tissue type and statistic are replaced with the appropriate designation; the options for the statistic are mean, standard deviation, F statistic, or degrees of freedom. Note that we have removed the Transformed fibroblasts from the Skin tissue type, as these were prepared differently than the other samples. 2 Data Sources 2.1 Experimental Data Data were retrieved from the Gene Expression Omnibus [Edgar et al., 2002]. Selection criteria for data included being conducted with the Fluidigm C1 platform for human cells and including ERCC spike-ins. The accession numbers are: GSE77288 [Tung et al., 2017], GSE84686 [Das et al., 2017], GSE79130, 1
2 Table 1: Data Source Information GEO sample Tissue type Number cells Number genes Stem cells 2,592 20, T cells 96 25, Kidney 47 18, Kidney 76 17, Nasal epithelium 96 16, Nasal epithelium 96 15,670 GSE89235, GSE89236, and GSE89237 [Arguel et al., 2017]. The last four data sources compose data from one experiment collected by the same lab. All except for GSE84686 also used unique molecular identifiers (UMIs) [Kivioja et al., 2012]. General information on these data sources can be found in Table 1. Genes in GSE77288 were reported by Ensembl identifiers, while genes in the other five datasets were reported by gene symbol. We transformed the Ensembl identifiers in GSE77288 to gene symbols using getgenes from mygene [Wu et al., 2013, Xin et al., 2016]. The Ensembl identifiers for which a gene symbol could not be identified were removed from the dataset, and multiple Ensembl identifiers that matched with the same gene symbol were summed. One cell was removed from each of GSE89236 and GSE These cells had two and zero UMIs measured, respectively. 2.2 Structural Annotation Dictionary Our structural annotation dictionary combines a list of human genes from Ensembl s Biomart with the Gene Ontology Consortium s (GO) information on genes gathered with mygene in R [Wu et al., 2013, Xin et al., 2016]. Gene names and not Ensembl identifiers were used. The query through mygene returns the top matches from GO for a given gene symbol. We retained only the top match. Any genes not identified from GO were removed. The biological process, molecular function, and cellular component were returned from GO. Each of these fields could have no values or multiple values. We considered the 1,629 unique cellular components for mapping the genes into ten broad cell structure (or location) categories. The cellular categories that were mapped to the cell structures were identified with a combination of text matching and review. First terms associated with the structure of interest were identified with the text term in the second column of Table 2. The cellular components that matched that text string were reviewed for appropriate fit. Cellular component categories that did not seem to fit were removed with the text string(s) specified in the third column of Table 2. The cell structures we considered are: nucleus, endoplasmic reticulum, Golgi apparatus, cytoplasm, membrane, mitochondria, ribosome, mitochondrial ribosome, ribonucleoprotein 2
3 Table 2: Structural Annotation Dictionary Criteria Structure Matching Term Removal Term(s) Nucleus nucl mitochond, nucleotide, ribonucleoprotein, male, endonuclease, ribonuclease Endoplasmic reticulum reticu sarcoplasmic, cortical Golgi bodies Golgi vesicle Cytoplasm cytop side of, axon, nuclear pore mitochond, Golgi, nucl, endoplasmic, organ, secretory, lysosom, endosome, ruffle, Membrane photoreceptor, vacuol, platelet, membrane, synaptic, peroxisomal, sperm, cytoplasmic vesicle vesic, acrosomal, granule, membrane, basement, melanosome, coat, azurophil granule neuron, phago, sarcoplasmic, membrane muscle, omegasome, tubular, spine, attack, junctional, transport, ER, chain, bounded, succinate, lamellar Ribosome ribosom mitochond Mitochondria mitochond ribosom Mitochondria ribosome mitochond, ribosom Ribonucleoprotein complex nucl, ribonucleoprotein Cytosolic ribosome ribosom, cytosolic complex, and cytosolic ribosome. The mapping was not one-to-one. In fact, each cellular component could be mapped to no structure, one structure, or multiple structures. There was nesting within the ribosomal structures with the cytosolic ribosome being a subset of the ribosome. In addition, each gene could have zero to 41 cellular components as annotated by GO, with 75% of genes having four or fewer cellular components. 2.3 Cytosolic Ribosomal Genes The 101 cytosolic ribosome genes from the scrna-seq datasets are: COA1, DDX3X, EIF2A, EIF2AK4, EIF2D, EIF4G1, FXR2, GEMIN5, LARP 4, MRPL1, MRPL4, MRPS18A, MRPS18C, MARPS5, NAA10, NHP2, NU- FIP1, RPL10, RPL10A, RPL11, RPL12, RPL13 RPL13A, RPL14, RPL23A, RPL24, RPL26, RPL26L1, RPL27, RPL27A, RPL28, RPL29, RPL3, RPL30, 3
4 Table 3: Overlap of Gene Sets Structure All Genes E & L Lin et al. Top 100 Number of Genes 8, Eisenberg & Levanon (2003) Lin et al. (2017) Top 100 Correlations RPL31, RPL32, RPL34, RPL35, RPL35A, RPL36, RPL36A, RPL36AL, RPL37, RPL37A, RPL38, RPL39, RPL39L, RPL4, RPL41, RPL5, RPL6, RPL7, RPL7A, RPL7L1, RPL8, RPL9, RPLP0, RPLP1, RPLP2, RPS10, RPS11, RPS12, RPS14, RPS15, RPS15A, RPS16, RPS18, RPS19, RPS2, RPS20, RPS21, RPS23, RPS24, RPS25, RPS26, RPS27, RPS27A, RPS27L, RPS28, RPS29, RPS3, RPS3A, RPS4X, RPS5, RPS6, RPS7, RPS9, RPSA, RSL1D1, RSL24D1, SURF6, ZNF622 The 89 cytosolic ribosome genes from the GTEx dataset are: APOD, DDX3X, EIF2A, EIF2AK4, FXR2, GEMIN5, HBA1, HBA2, MCTS1, MRPL4, MRPS18A, MRPS18C, MRPS5, NAA11, NHP2, NUFIP1, PPARGC1A, RPL10A, RPL10L, RPL11, RPL12, RPL13, RPL13AP3, RPL14, RPL17, RPL18, RPL18A, RPL19, RPL21, RPL22, RPL23, RPL23A, RPL24, RPL26, RPL26L1, RPL27, RPL27A, RPL28, RPL29, RPL30, RPL31, RPL32, RPL36, RPL36A, RPL36AL, RPL37, RPL37A, RPL38, RPL39, RPL39L, RPL39P5, RPL3L, RPL41, RPL7, RPLP1, RPLP2, RPS10, RPS10P5, RPS11, RPS12, RPS13, RPS14, RPS15, RPS16, RPS17, RPS18, RPS19, RPS20, RPS23, RPS25, RPS27, RPS27A, RPS27L, RPS28, RPS29, RPS3A, RPS4X, RPS5, RPS6, RPS7, RPS8, RPS9, RPSA, RSL1D1, RSL24D1, SNU13, SURF6, UBA52, ZNF622 3 Stability Measures Let i represent the gene, j a dataset, k a cell, G some set of genes, and c i,j,k the expression recorded for gene i in cell k in dataset j. Let s G,j be a vector representing the sum of the set of genes G; in other words, s G,j,k = i G c i,j,k is the kth entry in the vector s G,j. Conversely, let s G,j be a vector representing the sum of the genes not contained in the set of genes G, or s G,j,k = i / G c i,j,k is the kth entry of the vector s G,j. The correlations that serve as a measure of absolute stability for a given gene i and a given dataset j are calculated by r ERCC,i,j = cor(c i,j, s E,j ) where E is the set of ERCC spike-ins. The overall measure of absolute stability for gene i is r ERCC,i = j=1 r ERCC,i,j. The correlations that serve as a measure of proportional stability for a given gene i, a given dataset j, and a given cell structure with gene set S are defined as r S,i,j = cor(c i,j, s S,j ). The overall measure of stability for gene i is r S,i = j=1 r S,i,j. 4
5 We calculated an additional measure of proportional stability that was comparable across all genes. This uses an unadjusted cell total, unlike the measures of proportional stability defined above that uses a cell total adjusting for a given structure. s j is a vector where s j,k = I i=1 c i,j,k; that is, we sum over all of the genes for a given cell. The measure for a given dataset is r i,j = cor(c i,j, s j ) and is summarized over the 6 datasets by r i = j=1 r i,j. 4 Additional Figures The following figures provide extensions or additional information to those given in the paper. Figure 2 to Figure 8 in particular provide additional information that relate to many figures directly contained within the paper. 4.1 Correlation Histograms for Each Dataset Supplementary Figures 9 to 20 show the histograms of ERCC correlations found in Supplementary Figures 2 and 3 for each of the distinct datasets. Supplementary Figures 21 to 32 show the histograms of correlations found in Figure 3 and Supplementary Figure 4 for each of the distinct datasets. 4.2 Singular Value Decomposition of Single GTEx Tissue Types Supplementary Figures 33 to 38 show the first two left singular values from a singular value decomposition from a single tissue. These are colored by detailed tissue subtypes. In each of these figures, the top left panel shows the singular value decomposition using all genes as features. The top right panel shows the singular value decomposition using the bulk stably expressed genes from Eisenberg and Levanon [2003] as features. The bottom left panel shows the singular value decomposition from the single cell stably expressed genes from Lin et al. [2017] as features. The bottom right panel shows the singular value decomposition using the 89 cytosolic ribosomal genes found in the GTEx data as features. Note that we removed the Transformed fibroblasts subtype from the skin samples, due to technical differences in their preparation prior to experimental procedures. 4.3 Singular Value Decomposition of Two GTEx Tissue Types Supplementary Figures 39 to 80 show the singular value decomposition using the first two left singular values. These figures are colored by the two tissue types. The top left panel shows the singular value decomposition using all genes as features. The top right panel shows the singular value decomposition using the bulk genes presented in Eisenberg and Levanon [2003] as features. The bottom left uses the single cell stably expressed genes from Lin et al. [2017] 5
6 as features. The bottom right uses the cytosolic ribosomal genes as features. The first figure for each of these tissue types are colored by the general tissue type. The second figure for each of these tissue types are colored by the detailed sub-tissue type with shapes representing the two different tissue types. Genes that are more stably expressed across tissue types result in clusters that are less easily distinguishable. In these figures, the singular value decomposition from the cytosolic ribosomal genes result in the most overlap between clusters, indicating that the cytosolic ribosomal genes are more stably expressed across both general tissue types and across the more detailed tissue subtypes. 4.4 Singular Value Decomposition With Equal Number of Genes from GTEx The four different singular value decompositions shown in Supplementary Figures 33 to 80 have different numbers of genes used as features; we also provide the analogous figures that have been randomly subsetted to 89 genes. Supplementary Figures 81 to 106 show that the number of features does not explain the differences in separation of tissue clusters. 6
7 Figure 1: Scatterplots of the mean and standard deviation of ERCC spike-ins and the endogenous genes on the log scale for each of the six datasets. From these graphs, it appears that the variance exhibited by the spike-ins is smaller than those for endogenous genes, even after accounting for the mean measured expression. 7
8 Figure 2: Histograms of the average correlations of each gene with the ERCC total over the six datasets considered, with comparisons between the gene set of interest and the remaining genes. Note that the x-axis ranges from to
9 Figure 3: Histograms of the average correlations of each gene with the ERCC total over the six datasets considered, with comparisons between the gene set of interest and the remaining genes. Note that the x-axis ranges from to
10 Figure 4: Histograms of the average correlations of each gene over the six datasets considered, with comparisons between the set of genes of interest and the remaining genes. 10
11 Figure 5: Histogram of the average correlation of cytosolic ribosomal genes compared to the remaining genes when correlated with the adjusted cell total with respect to the ribosomal genes. Because the cytosolic ribosomal genes are a subset of the ribosomal genes, we consider the possibility that the correlations seen in Figure 3 could be inflated due to the correlation between cytosolic ribosomal genes and the remaining ribosomal genes. Using the adjusted cell total removing the ribosomal genes should reduce the artificial inflation. The figure here displays a similar distribution to the one in Figure 3, indicating that any inflation was minimal. 11
12 Figure 6: The average correlations of Beta-actin (ACTB) and GAPDH with the cell total over the six datasets. Note that, unlike in Figure 3, the correlations here are with the unadjusted cell total rather than an adjusted cell total. This allows correlations between ACTB, GAPDH, and the cytosolic ribosomal genes to be comparable. 12
13 Figure 7: Summary characteristics of the expression of cytosolic ribosomal genes across cells and across datasets. The boxplot on the top shows all of the expression measurements by dataset for the cytosolic ribosomal genes, transformed with the log + 1. For comparison, the proportion of zeros in each of these datasets is 80%, 41%, 42%, 63%, 76%, and 82%, respectively and means (on the log + 1 scale) of 1.23, 1.26, 1.37, 0.71, 0.45, and 0.30, respectively. The boxplot on the bottom looks at the expression (log + 1) of each of the cytosolic ribosomal genes within GSE Analogous boxplots for each of the datasets can be found in Supplementary Figure 8. 13
14 Figure 8: The boxplots of the distributions of each cytosolic ribosomal gene in each dataset. This expands the bottom panel of Supplementary Figure 7 to all datasets. We can observe the variation and relative stability in the expression of cytosolic ribosomal genes. 14
15 Figure 9: Histograms of the correlations of each gene with the ERCC total for each dataset, with comparisons between the ribosomal genes and the remaining genes. 15
16 Figure 10: Histogram of the correlations of each gene with the ERCC total for each dataset, with comparisons between the cytosolic ribosomal genes and the remaining genes. 16
17 Figure 11: Histogram of the correlations of each gene with the ERCC total for each dataset, with comparisons between the mitochondrial genes and the remaining genes. 17
18 Figure 12: Histogram of the correlations of each gene with the ERCC total for each dataset, with comparisons between the nuclear genes and the remaining genes. 18
19 Figure 13: Histogram of the correlations of each gene with the ERCC total for each dataset, with comparisons between the endoplasmic reticulum genes and the remaining genes. 19
20 Figure 14: Histogram of the correlations of each gene with the ERCC total for each dataset, with comparisons between the Golgi body genes and the remaining genes. 20
21 Figure 15: Histogram of the correlations of each gene with the ERCC total for each dataset, with comparisons between the cytoplasmic genes and the remaining genes. 21
22 Figure 16: Histogram of the correlations of each gene with the ERCC total for each dataset, with comparisons between the mitochondrial ribosomal genes and the remaining genes. 22
23 Figure 17: Histogram of the correlations of each gene with the ERCC total for each dataset, with comparisons between the ribonucleoprotein genes and the remaining genes. 23
24 Figure 18: Histogram of the correlations of each gene with the ERCC total for each dataset, with comparisons between the membrane genes and the remaining genes. 24
25 Figure 19: Histogram of the correlations of each gene with the ERCC total for each dataset, with comparisons between the stably expressed genes from Eisenberg and Levanon [2003] and the remaining genes. 25
26 Figure 20: Histogram of the correlations of each gene with the ERCC total for each dataset, with comparisons between the single cell stably expressed genes from Lin et al. [2017] and the remaining genes. 26
27 Figure 21: Histograms of the correlations of each gene with the adjusted cell total for ribosomal genes for each dataset, with comparisons between the ribosomal genes and the remaining genes. 27
28 Figure 22: Histogram of the correlations of each gene with the adjusted cell total for cytosolic ribosomal genes for each dataset,, with comparisons between the cytosolic ribosomal genes and the remaining genes. 28
29 Figure 23: Histogram of the correlations of each gene with the adjusted cell total for mitochondrial genes for each dataset, with comparisons between the mitochondrial genes and the remaining genes. 29
30 Figure 24: Histogram of the correlations of each gene with the adjusted cell total for nuclear genes for each dataset, with comparisons between the nuclear genes and the remaining genes. 30
31 Figure 25: Histogram of the correlations of each gene with the adjusted cell total for endoplasmic reticulum genes for each dataset, with comparisons between the endoplasmic reticulum genes and the remaining genes. 31
32 Figure 26: Histogram of the correlations of each gene with the adjusted cell total for Golgi body genes for each dataset, with comparisons between the Golgi body genes and the remaining genes. 32
33 Figure 27: Histogram of the correlations of each gene with the adjusted cell total for cytoplasmic genes for each dataset, with comparisons between the cytoplasmic genes and the remaining genes. 33
34 Figure 28: Histogram of the correlations of each gene with the adjusted cell total for mitochondrial ribosomal genes for each dataset, with comparisons between the mitochondrial ribosomal genes and the remaining genes. 34
35 Figure 29: Histogram of the correlations of each gene with the adjusted cell total for ribonucleoprotein genes for each dataset, with comparisons between the ribonucleoprotein genes and the remaining genes. 35
36 Figure 30: Histogram of the correlations of each gene with the adjusted cell total for membrane genes for each dataset, with comparisons between the membrane genes and the remaining genes. 36
37 Figure 31: Histogram of the correlations of each gene with the adjusted cell total for stably expressed genes from Eisenberg and Levanon [2013] for each dataset, with comparisons between the stably expressed genes and the remaining genes. 37
38 Figure 32: Histogram of the correlations of each gene with the adjusted cell total for single cell stably expressed genes from Lin et al. [2017] for each dataset, with comparisons between the single cell stably expressed genes and the remaining genes. 38
39 Figure 33: The first and second singular vectors of the GTEx data from the brain. 39
40 Figure 34: The first and second singular vectors of the GTEx data from the adipose tissue. 40
41 Figure 35: The first and second singular vectors of the GTEx data from the blood vessels. 41
42 Figure 36: The first and second singular vectors of the GTEx data from the esophagus. 42
43 Figure 37: The first and second singular vectors of the GTEx data from the heart. 43
44 Figure 38: The first and second singular vectors of the GTEx data from the skin. 44
45 Figure 39: The first and second singular vectors of the GTEx data from the adipose tissue and the heart. 45
46 Figure 40: The first and second singular vectors of the GTEx data from the adipose tissue and the heart. 46
47 Figure 41: The first and second singular vectors of the GTEx data from the adipose tissue and the muscle. 47
48 Figure 42: The first and second singular vectors of the GTEx data from the adipose tissue and the muscle. 48
49 Figure 43: The first and second singular vectors of the GTEx data from the brain and the adipose tissue. 49
50 Figure 44: The first and second singular vectors of the GTEx data from the brain and the adipose tissue. 50
51 Figure 45: The first and second singular vectors of the GTEx data from the brain and the blood vessel. 51
52 Figure 46: The first and second singular vectors of the GTEx data from the brain and the blood vessel. 52
53 Figure 47: The first and second singular vectors of the GTEx data from the brain and the esophagus. 53
54 Figure 48: The first and second singular vectors of the GTEx data from the brain and the esophagus. 54
55 Figure 49: The first and second singular vectors of the GTEx data from the brain and the heart. 55
56 Figure 50: The first and second singular vectors of the GTEx data from the brain and the heart. 56
57 Figure 51: The first and second singular vectors of the GTEx data from the brain and the muscle. 57
58 Figure 52: The first and second singular vectors of the GTEx data from the brain and the muscle. 58
59 Figure 53: The first and second singular vectors of the GTEx data from the brain and the skin. 59
60 Figure 54: The first and second singular vectors of the GTEx data from the brain and the skin. 60
61 Figure 55: The first and second singular vectors of the GTEx data from the blood vessel and the adipose tissue. 61
62 Figure 56: The first and second singular vectors of the GTEx data from the blood vessel and the adipose tissue. 62
63 Figure 57: The first and second singular vectors of the GTEx data from the blood vessel and the heart. 63
64 Figure 58: The first and second singular vectors of the GTEx data from the blood vessel and the heart. 64
65 Figure 59: The first and second singular vectors of the GTEx data from the blood vessel and the muscle. 65
66 Figure 60: The first and second singular vectors of the GTEx data from the blood vessel and the muscle. 66
67 Figure 61: The first and second singular vectors of the GTEx data from the esophagus and the adipose tissue. 67
68 Figure 62: The first and second singular vectors of the GTEx data from the esophagus and the adipose tissue. 68
69 Figure 63: The first and second singular vectors of the GTEx data from the esophagus and the blood vessel. 69
70 Figure 64: The first and second singular vectors of the GTEx data from the esophagus and the blood vessel. 70
71 Figure 65: The first and second singular vectors of the GTEx data from the esophagus and the heart. 71
72 Figure 66: The first and second singular vectors of the GTEx data from the esophagus and the heart. 72
73 Figure 67: The first and second singular vectors of the GTEx data from the esophagus and the muscle. 73
74 Figure 68: The first and second singular vectors of the GTEx data from the esophagus and the muscle. 74
75 Figure 69: The first and second singular vectors of the GTEx data from the heart and the muscle. 75
76 Figure 70: The first and second singular vectors of the GTEx data from the heart and the muscle. 76
77 Figure 71: The first and second singular vectors of the GTEx data from the skin and the adipose tissue. 77
78 Figure 72: The first and second singular vectors of the GTEx data from the skin and the adipose tissue. 78
79 Figure 73: The first and second singular vectors of the GTEx data from the skin and the blood vessel. 79
80 Figure 74: The first and second singular vectors of the GTEx data from the skin and the blood vessel. 80
81 Figure 75: The first and second singular vectors of the GTEx data from the skin and the esophagus. 81
82 Figure 76: The first and second singular vectors of the GTEx data from the skin and the esophagus. 82
83 Figure 77: The first and second singular vectors of the GTEx data from the skin and the heart. 83
84 Figure 78: The first and second singular vectors of the GTEx data from the skin and the heart. 84
85 Figure 79: The first and second singular vectors of the GTEx data from the skin and the muscle. 85
86 Figure 80: The first and second singular vectors of the GTEx data from the skin and the muscle. 86
87 Figure 81: The first and second singular vectors of the GTEx data from the brain. 87
88 Figure 82: The first and second singular vectors of the GTEx data from the adipose tissue. 88
89 Figure 83: The first and second singular vectors of the GTEx data from the blood vessels. 89
90 Figure 84: The first and second singular vectors of the GTEx data from the esophagus. 90
91 Figure 85: The first and second singular vectors of the GTEx data from the heart. 91
92 Figure 86: The first and second singular vectors of the GTEx data from the skin. 92
93 Figure 87: The first and second singular vectors of the GTEx data from the adipose tissue and the heart. 93
94 Figure 88: The first and second singular vectors of the GTEx data from the adipose tissue and the muscle. 94
95 Figure 89: The first and second singular vectors of the GTEx data from the brain and the adipose tissue. 95
96 Figure 90: The first and second singular vectors of the GTEx data from the brain and the blood vessel. 96
97 Figure 91: The first and second singular vectors of the GTEx data from the brain and the esophagus. 97
98 Figure 92: The first and second singular vectors of the GTEx data from the brain and the heart. 98
99 Figure 93: The first and second singular vectors of the GTEx data from the brain and the muscle. 99
100 Figure 94: The first and second singular vectors of the GTEx data from the blood vessel and the adipose tissue. 100
101 Figure 95: The first and second singular vectors of the GTEx data from the blood vessel and the heart. 101
102 Figure 96: The first and second singular vectors of the GTEx data from the blood vessel and the muscle. 102
103 Figure 97: The first and second singular vectors of the GTEx data from the esophagus and the adipose tissue. 103
104 Figure 98: The first and second singular vectors of the GTEx data from the esophagus and the blood vessel. 104
105 Figure 99: The first and second singular vectors of the GTEx data from the esophagus and the heart. 105
106 Figure 100: The first and second singular vectors of the GTEx data from the esophagus and the muscle. 106
107 Figure 101: The first and second singular vectors of the GTEx data from the heart and the muscle. 107
108 Figure 102: The first and second singular vectors of the GTEx data from the skin and the adipose tissue. 108
109 Figure 103: The first and second singular vectors of the GTEx data from the skin and the blood vessel. 109
110 Figure 104: The first and second singular vectors of the GTEx data from the skin and the esophagus. 110
111 Figure 105: The first and second singular vectors of the GTEx data from the skin and the heart. 111
112 Figure 106: The first and second singular vectors of the GTEx data from the skin and the muscle. 112
113 References Arguel, M.-J., LeBrigand, K., Paquet, A., RuizGarcía, S., Zaragosi, L.-E., Barbry, P., and Waldmann, R. (2017). A cost effective 5 selective single cell transcriptome profiling approach with improved UMI design. Nucleic Acids Research, 45(7), e48 e48. Das, A., Rouault-Pierre, K., Kamdar, S., Gomez-Tourino, I., Wood, K., Donaldson, I., Mein, C. A., Bonnet, D., Hayday, A. C., and Gibbons, D. L. (2017). Adaptive from Innate: Human IFN-γ + CD4 + T Cells Can Arise Directly from CXCL8-Producing Recent Thymic Emigrants in Babies and Adults. The Journal of Immunology, 199(5), Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research, 30(1), Eisenberg, E. and Levanon, E. Y. (2003). Human housekeeping genes are compact. Trends in Genetics, 19(7), Eisenberg, E. and Levanon, E. Y. (2013). Human housekeeping genes, revisited. Trends in Genetics, 29(10), Kivioja, T., Vähärautio, A., Karlsson, K., Bonke, M., Enge, M., Linnarsson, S., and Taipale, J. (2012). Counting absolute numbers of molecules using unique molecular identifiers. Nature Methods, 9(1), Lin, Y., Ghazanfar, S., Strbenac, D., Wang, A., Patrick, E., Speed, T., Yang, J., and Yang, P. (2017). Housekeeping genes, revisited at the single-cell level. biorxiv. Tung, P.-Y., Blischak, J. D., Hsiao, C. J., Knowles, D. A., Burnett, J. E., Pritchard, J. K., and Gilad, Y. (2017). Batch effects and the effective design of single-cell gene expression studies. Scientific reports, 7, Wu, C., MacLeod, I., and Su, A. I. (2013). BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Research, 41(D1), D561 D565. Xin, J., Mark, A., Afrasiabi, C., Tsueng, G., Juchler, M., Gopal, N., Stupp, G. S., Putman, T. E., Ainscough, B. J., Griffith, O. L., Torkamani, A., Whetzel, P. L., Mungall, C. J., Mooney, S. D., Su, A. I., and Wu, C. (2016). High-performance web services for querying gene and variant annotation. Genome Biology, 17(1),
By: Brooke Sheppard
 By: Brooke Sheppard What is a Cell? Cells are the basic structure of life for all organisms. Cells are microscopic, which means we can only view cells under a microscope. There are animal cells and plant
By: Brooke Sheppard What is a Cell? Cells are the basic structure of life for all organisms. Cells are microscopic, which means we can only view cells under a microscope. There are animal cells and plant
Structures in Cells. Cytoplasm. Lecture 5, EH1008: Biology for Public Health, Biomolecules
 Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells. Lecture 5, EH1008: Biology for Public Health, Biomolecules.
 Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Essentials of Anatomy and Physiology, 9e (Marieb) Chapter 3 Cells and Tissues. Short Answer. Figure 3.1
 Essentials of Anatomy and Physiology, 9e (Marieb) Chapter 3 Cells and Tissues Short Answer Figure 3.1 Using Figure 3.1, match the following: 1) The illustration of simple cuboidal epithelium is. Answer:
Essentials of Anatomy and Physiology, 9e (Marieb) Chapter 3 Cells and Tissues Short Answer Figure 3.1 Using Figure 3.1, match the following: 1) The illustration of simple cuboidal epithelium is. Answer:
Homeostatic Control Systems
 Homeostatic Control Systems In order to maintain homeostasis, control system must be able to Detect deviations from normal in the internal environment that need to be held within narrow limits Integrate
Homeostatic Control Systems In order to maintain homeostasis, control system must be able to Detect deviations from normal in the internal environment that need to be held within narrow limits Integrate
Chapters 2 and 3. Pages and Pages Prayer Attendance Homework
 Chapters 2 and 3 Pages 44-45 and Pages 59-62 Prayer Attendance Homework The Cell The cell is the basic unit of life on Earth, separated from its environment by a membrane and sometimes an outer wall. Prokaryotic
Chapters 2 and 3 Pages 44-45 and Pages 59-62 Prayer Attendance Homework The Cell The cell is the basic unit of life on Earth, separated from its environment by a membrane and sometimes an outer wall. Prokaryotic
Cells. 1. Smallest living structures. 2. Basic structural and functional units of the body. 3. Derived from pre-existing cells. 4. Homeostasis.
 Cells The Cell The human body has about 75 trillion cells All tissues and organs are made up of cells Smallest functional unit of life Cytology Histology Cytology Epithelial cells Fibroblasts Erythrocytes
Cells The Cell The human body has about 75 trillion cells All tissues and organs are made up of cells Smallest functional unit of life Cytology Histology Cytology Epithelial cells Fibroblasts Erythrocytes
Cell Structure and Organelles SBI4U 2016/10/14
 Cell Structure and Organelles SBI4U 2016/10/14 Inside the cell These are generalizations, not rules! Everything inside the cell membrane besides the nucleus is called the cytoplasm; The liquid is known
Cell Structure and Organelles SBI4U 2016/10/14 Inside the cell These are generalizations, not rules! Everything inside the cell membrane besides the nucleus is called the cytoplasm; The liquid is known
Name: Per/row: Cell Structure and Function Practice: Use Ch 4 in Mader Biology
 Cell Structure and Function Practice: Use Ch 4 in Mader Biology Name: Per/row: 1. Write the name of the cell part in the box next to its description/function. Cell membrane Centrioles Chloroplast Chromatin
Cell Structure and Function Practice: Use Ch 4 in Mader Biology Name: Per/row: 1. Write the name of the cell part in the box next to its description/function. Cell membrane Centrioles Chloroplast Chromatin
Plasma Membrane. comprised of a phospholipid bilayer and embedded proteins separates the cells s contents from its surroundings
 Cell Organelles Plasma Membrane comprised of a phospholipid bilayer and embedded proteins separates the cells s contents from its surroundings Cytosol the fluid Cytoplasm cell interior, everything outside
Cell Organelles Plasma Membrane comprised of a phospholipid bilayer and embedded proteins separates the cells s contents from its surroundings Cytosol the fluid Cytoplasm cell interior, everything outside
The Microscopic World of Cells. The Microscopic World of Cells. The Microscopic World of Cells 9/21/2012
 Organisms are either: Single-celled, such as most prokaryotes and protists or Multicelled, such as plants, animals, and most fungi How do we study cells? Light microscopes can be used to explore the structures
Organisms are either: Single-celled, such as most prokaryotes and protists or Multicelled, such as plants, animals, and most fungi How do we study cells? Light microscopes can be used to explore the structures
Organelles of the Cell & How They Work Together. Packet #5
 Organelles of the Cell & How They Work Together Packet #5 Developed by Mr. Barrow 2018 1 Introduction Organization of cells is basically similar in all cells. Additionally, most cells are tiny Ranging
Organelles of the Cell & How They Work Together Packet #5 Developed by Mr. Barrow 2018 1 Introduction Organization of cells is basically similar in all cells. Additionally, most cells are tiny Ranging
Organelles of the Cell & How They Work Together. Packet #7
 Organelles of the Cell & How They Work Together Packet #7 Introduction Introduction Organization of cells is basically similar in all cells. Additionally, most cells are tiny Ranging from 1 1000 cubic
Organelles of the Cell & How They Work Together Packet #7 Introduction Introduction Organization of cells is basically similar in all cells. Additionally, most cells are tiny Ranging from 1 1000 cubic
Organelles of the Cell & How They Work Together. Packet #5
 Organelles of the Cell & How They Work Together Packet #5 Developed by Mr. Barrow 2018 1 Introduction Organization of cells is basically similar in all cells. Additionally, most cells are tiny Ranging
Organelles of the Cell & How They Work Together Packet #5 Developed by Mr. Barrow 2018 1 Introduction Organization of cells is basically similar in all cells. Additionally, most cells are tiny Ranging
Human height. Length of some nerve and muscle cells. Chicken egg. Frog egg. Most plant and animal cells Nucleus Most bacteria Mitochondrion
 10 m 1 m 0.1 m 1 cm Human height Length of some nerve and muscle cells Chicken egg Unaided eye 1 mm Frog egg 100 µm 10 µm 1 µm 100 nm 10 nm Most plant and animal cells Nucleus Most bacteria Mitochondrion
10 m 1 m 0.1 m 1 cm Human height Length of some nerve and muscle cells Chicken egg Unaided eye 1 mm Frog egg 100 µm 10 µm 1 µm 100 nm 10 nm Most plant and animal cells Nucleus Most bacteria Mitochondrion
The endoplasmic reticulum is a network of folded membranes that form channels through the cytoplasm and sacs called cisternae.
 Endoplasmic reticulum (ER) The endoplasmic reticulum is a network of folded membranes that form channels through the cytoplasm and sacs called cisternae. Cisternae serve as channels for the transport of
Endoplasmic reticulum (ER) The endoplasmic reticulum is a network of folded membranes that form channels through the cytoplasm and sacs called cisternae. Cisternae serve as channels for the transport of
5/12/2015. Cell Size. Relative Rate of Reaction
 Cell Makeup Chapter 4 The Cell: The Fundamental Unit of Life We previously talked about the cell membrane The cytoplasm is everything inside the membrane, except the nucleus Includes Cytosol = liquid portion
Cell Makeup Chapter 4 The Cell: The Fundamental Unit of Life We previously talked about the cell membrane The cytoplasm is everything inside the membrane, except the nucleus Includes Cytosol = liquid portion
basic unit structure and function
 Chapter 3 Cells Introduction The cell is the basic unit of structure and function in living things. Cells vary in their shape, size, and arrangements, but all cells have similar components with a particular
Chapter 3 Cells Introduction The cell is the basic unit of structure and function in living things. Cells vary in their shape, size, and arrangements, but all cells have similar components with a particular
4/12/17. Cells. Cell Structure. Ch. 2 Cell Structure and Func.on. Range of Cell Sizes BIOL 100
 Ch. 2 Cell Structure and Func.on BIOL 100 Cells Fundamental units of life Cell theory All living things are composed of one or more cells. The cell is the most basic unit of life. All cells come from pre-existing
Ch. 2 Cell Structure and Func.on BIOL 100 Cells Fundamental units of life Cell theory All living things are composed of one or more cells. The cell is the most basic unit of life. All cells come from pre-existing
CELL PART OF THE DAY. Chapter 7: Cell Structure and Function
 CELL PART OF THE DAY Chapter 7: Cell Structure and Function Cell Membrane Cell membranes are composed of two phospholipid layers. Cell membrane is flexible, not rigid The cell membrane has two major functions.
CELL PART OF THE DAY Chapter 7: Cell Structure and Function Cell Membrane Cell membranes are composed of two phospholipid layers. Cell membrane is flexible, not rigid The cell membrane has two major functions.
A Tour of the Cell Lecture 2, Part 1 Fall 2008
 Cell Theory 1 A Tour of the Cell Lecture 2, Part 1 Fall 2008 Cells are the basic unit of structure and function The lowest level of structure that can perform all activities required for life Reproduction
Cell Theory 1 A Tour of the Cell Lecture 2, Part 1 Fall 2008 Cells are the basic unit of structure and function The lowest level of structure that can perform all activities required for life Reproduction
The Cell Organelles. Eukaryotic cell. The plasma membrane separates the cell from the environment. Plasma membrane: a cell s boundary
 Eukaryotic cell The Cell Organelles Enclosed by plasma membrane Subdivided into membrane bound compartments - organelles One of the organelles is membrane bound nucleus Cytoplasm contains supporting matrix
Eukaryotic cell The Cell Organelles Enclosed by plasma membrane Subdivided into membrane bound compartments - organelles One of the organelles is membrane bound nucleus Cytoplasm contains supporting matrix
CHAPTER 4 - CELLS. All living things are made up of one or more cells. A cell is the smallest unit that can carry on all of the processes of life.
 CHAPTER 4 - CELLS Objectives Name the scientists who first observed living and nonliving cells. Summarize the research that led to the development of the cell theory. State the three principles of the
CHAPTER 4 - CELLS Objectives Name the scientists who first observed living and nonliving cells. Summarize the research that led to the development of the cell theory. State the three principles of the
October 26, Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
BIOSC 041. v Today s lecture. v Today s lab. v Note- Monday is a holiday good time to do some reading!
 BIOSC 041 v Today s lecture Review questions Chapter 6, Cells More review questions v Today s lab Quick review of lab safety The Scientific Method start thinking about which environments you might want
BIOSC 041 v Today s lecture Review questions Chapter 6, Cells More review questions v Today s lab Quick review of lab safety The Scientific Method start thinking about which environments you might want
CELL PARTS TYPICAL ANIMAL CELL
 AP BIOLOGY CText Reference, Campbell v.8, Chapter 6 ACTIVITY1.12 NAME DATE HOUR CELL PARTS TYPICAL ANIMAL CELL ENDOMEMBRANE SYSTEM TYPICAL PLANT CELL QUESTIONS: 1. Write the name of the cell part in the
AP BIOLOGY CText Reference, Campbell v.8, Chapter 6 ACTIVITY1.12 NAME DATE HOUR CELL PARTS TYPICAL ANIMAL CELL ENDOMEMBRANE SYSTEM TYPICAL PLANT CELL QUESTIONS: 1. Write the name of the cell part in the
Looking Inside Cells
 Looking Inside Cells Inner Life of a Cell http://www.bing.com/videos/search?q=inside +cell+animation&form=hdrsc3#view=detail &mid=4ba834420ea307a061374ba834420ea 307A06137 Cell Defined Cells-Basic unit
Looking Inside Cells Inner Life of a Cell http://www.bing.com/videos/search?q=inside +cell+animation&form=hdrsc3#view=detail &mid=4ba834420ea307a061374ba834420ea 307A06137 Cell Defined Cells-Basic unit
10/5/2015. Cell Size. Relative Rate of Reaction
 The Cell Biology 102 Fundamental unit of life Smallest unit that displays all the basic elements of life Lecture 5: Cells Cell Theory 1. All living things are made of one or more cells Cell Theory 2. The
The Cell Biology 102 Fundamental unit of life Smallest unit that displays all the basic elements of life Lecture 5: Cells Cell Theory 1. All living things are made of one or more cells Cell Theory 2. The
A Tour of the Cell. reference: Chapter 6. Reference: Chapter 2
 A Tour of the Cell reference: Chapter 6 Reference: Chapter 2 Monkey Fibroblast Cells stained with fluorescent dyes to show the nucleus (blue) and cytoskeleton (yellow and red fibers), image courtesy of
A Tour of the Cell reference: Chapter 6 Reference: Chapter 2 Monkey Fibroblast Cells stained with fluorescent dyes to show the nucleus (blue) and cytoskeleton (yellow and red fibers), image courtesy of
Smart cell lesson (1).notebook February 02, 2015
 Cells 1 TEKS Related to the Lesson (12) Organisms and environments. The student knows that living systems at all levels of organization demonstrate the complementary nature of structure and function. The
Cells 1 TEKS Related to the Lesson (12) Organisms and environments. The student knows that living systems at all levels of organization demonstrate the complementary nature of structure and function. The
Cell Theory All living matter is composed of one or more The cell is the structural and functional unit of life All cells come from pre-existing cell
 Cell Theory All living matter is composed of one or more The cell is the structural and functional unit of life All cells come from pre-existing cell Prokeryotic Bacteria or archaea Cell wall, small circular
Cell Theory All living matter is composed of one or more The cell is the structural and functional unit of life All cells come from pre-existing cell Prokeryotic Bacteria or archaea Cell wall, small circular
(a) TEM of a plasma. Fimbriae. Nucleoid. Ribosomes. Plasma membrane. Cell wall Capsule. Bacterial chromosome
 0 m m 0. m cm mm 00 µm 0 µm 00 nm 0 nm Human height Length of some nerve and muscle cells Chicken egg Frog egg Most plant and animal cells Most bacteria Smallest bacteria Viruses Proteins Unaided eye Light
0 m m 0. m cm mm 00 µm 0 µm 00 nm 0 nm Human height Length of some nerve and muscle cells Chicken egg Frog egg Most plant and animal cells Most bacteria Smallest bacteria Viruses Proteins Unaided eye Light
6. What surrounds the nucleus? How many membranes does it have?
 Biology-R track Study Guide: 7.2 Cell Structure Cell Organization 1. What are the 2 major parts that you can divide the eukaryotic cell into? 2. What part is the fluid portion of the cell outside the nucleus?
Biology-R track Study Guide: 7.2 Cell Structure Cell Organization 1. What are the 2 major parts that you can divide the eukaryotic cell into? 2. What part is the fluid portion of the cell outside the nucleus?
What Are Cell Membranes?
 What Are Cell Membranes? Chapter 5, Lesson 1 24 Directions Match each term in Column A with its meaning in Column B. Write the letter on the line. Column A 1. cytoplasm 2. cytosol 3. extracellular matrix
What Are Cell Membranes? Chapter 5, Lesson 1 24 Directions Match each term in Column A with its meaning in Column B. Write the letter on the line. Column A 1. cytoplasm 2. cytosol 3. extracellular matrix
A Tour of the Cell. reference: Chapter 6. Reference: Chapter 2
 A Tour of the Cell reference: Chapter 6 Reference: Chapter 2 Monkey Fibroblast Cells stained with fluorescent dyes to show the nucleus (blue) and cytoskeleton (yellow and red fibers), image courtesy of
A Tour of the Cell reference: Chapter 6 Reference: Chapter 2 Monkey Fibroblast Cells stained with fluorescent dyes to show the nucleus (blue) and cytoskeleton (yellow and red fibers), image courtesy of
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question.
 Exam Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) All of the following are synthesized along various sites of the endoplasmic reticulum
Exam Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) All of the following are synthesized along various sites of the endoplasmic reticulum
Cell Cell
 Go to cellsalive.com. Select Interactive Cell Models: Plant and Animal. Fill in the information on Plant and Animal Organelles, then Click on Start the Animation Select Plant or Animal Cell below the box.
Go to cellsalive.com. Select Interactive Cell Models: Plant and Animal. Fill in the information on Plant and Animal Organelles, then Click on Start the Animation Select Plant or Animal Cell below the box.
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting
 Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
The Cell. Copyright 2003 Pearson Education, Inc. publishing as Benjamin Cummings
 The Cell Cell Theory The cell is the basic structural and functional unit of life The organism activity depends on individual and collective activity of cells Biochemical activities of cells are dictated
The Cell Cell Theory The cell is the basic structural and functional unit of life The organism activity depends on individual and collective activity of cells Biochemical activities of cells are dictated
1) All organisms are made up of one or more cells and the products of those cells.
 CELL ORGANELLES - NOTES CELL THEORY Cells are the basic unit of life. The Cell Theory states that: 1) All organisms are made up of one or more cells and the products of those cells. 2) All cells carry
CELL ORGANELLES - NOTES CELL THEORY Cells are the basic unit of life. The Cell Theory states that: 1) All organisms are made up of one or more cells and the products of those cells. 2) All cells carry
Cells and Tissues 3PART A. PowerPoint Lecture Slide Presentation by Patty Bostwick-Taylor, Florence-Darlington Technical College
 PowerPoint Lecture Slide Presentation by Patty Bostwick-Taylor, Florence-Darlington Technical College Cells and Tissues 3PART A Cells and Tissues Carry out all chemical activities needed to sustain life
PowerPoint Lecture Slide Presentation by Patty Bostwick-Taylor, Florence-Darlington Technical College Cells and Tissues 3PART A Cells and Tissues Carry out all chemical activities needed to sustain life
Parts (organelles) of the Cell
 Lesson 07 Cell Organelles.notebook Parts (organelles) of the Cell Parts that provide structure and support Cell Membrane 1) Is a barrier for the cell. It controls what goes in and out of the cell. 2) It
Lesson 07 Cell Organelles.notebook Parts (organelles) of the Cell Parts that provide structure and support Cell Membrane 1) Is a barrier for the cell. It controls what goes in and out of the cell. 2) It
Cells Alive- Internet Lesson
 Name Date Period Cells Alive Webquest Cells Alive- Internet Lesson URL: www.cellsalive.com Objective: You will look at computer models of cells, learn the functions and the descriptions of the cells and
Name Date Period Cells Alive Webquest Cells Alive- Internet Lesson URL: www.cellsalive.com Objective: You will look at computer models of cells, learn the functions and the descriptions of the cells and
A Tour of the Cell. Ch. 7
 A Tour of the Cell Ch. 7 Cell Theory O All organisms are composed of one or more cells. O The cell is the basic unit of structure and organization of organisms. O All cells come from preexisting cells.
A Tour of the Cell Ch. 7 Cell Theory O All organisms are composed of one or more cells. O The cell is the basic unit of structure and organization of organisms. O All cells come from preexisting cells.
Nucleic acids. Nucleic acids are information-rich polymers of nucleotides
 Nucleic acids Nucleic acids are information-rich polymers of nucleotides DNA and RNA Serve as the blueprints for proteins and thus control the life of a cell RNA and DNA are made up of very similar nucleotides.
Nucleic acids Nucleic acids are information-rich polymers of nucleotides DNA and RNA Serve as the blueprints for proteins and thus control the life of a cell RNA and DNA are made up of very similar nucleotides.
Ch 2: The Cell. Goals: Anatomy of a typical cell Cell Membrane Discussion of internal structure of a cell with emphasis on the various organelles
 Ch 2: The Cell Goals: Anatomy of a typical cell Cell Membrane Discussion of internal structure of a cell with emphasis on the various organelles Developed by John Gallagher, MS, DVM Some Terminology: 1.
Ch 2: The Cell Goals: Anatomy of a typical cell Cell Membrane Discussion of internal structure of a cell with emphasis on the various organelles Developed by John Gallagher, MS, DVM Some Terminology: 1.
Lysosomes. Gr: lysis solution, soma body. Membrane bounded vesicles. Usually round ovoid or irregular electron dense bodies m.
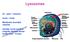 Lysosomes Gr: lysis solution, soma body Membrane bounded vesicles Usually round ovoid or irregular electron dense bodies 0.05 0.5 m. Lysosomes No. varies from a few to several hundred per cell, in different
Lysosomes Gr: lysis solution, soma body Membrane bounded vesicles Usually round ovoid or irregular electron dense bodies 0.05 0.5 m. Lysosomes No. varies from a few to several hundred per cell, in different
The Study of Cells The diversity of the cells of the body The following figure shows the proportion of cell size of the variety of cells in the body
 Adapted from Martini Human Anatomy 7th ed. Chapter 2 Foundations: The Cell Introduction There are trillions of cells in the body Cells are the structural building blocks of all plants and animals Cells
Adapted from Martini Human Anatomy 7th ed. Chapter 2 Foundations: The Cell Introduction There are trillions of cells in the body Cells are the structural building blocks of all plants and animals Cells
Zool 3200: Cell Biology Exam 4 Part I 2/3/15
 Name: Key Trask Zool 3200: Cell Biology Exam 4 Part I 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Name: Key Trask Zool 3200: Cell Biology Exam 4 Part I 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
CHAPTER 3: Cells. All Organisms Are Composed of Cells 3/24/2014. A cell is the smallest unit of life that can function independently.
 CHAPTER 3: Cells BIO 121 All Organisms Are Composed of Cells A cell is the smallest unit of life that can function independently. Section 3.1 Amoeba Wim van Egmond/Visuals Unlimited All Organisms Are Composed
CHAPTER 3: Cells BIO 121 All Organisms Are Composed of Cells A cell is the smallest unit of life that can function independently. Section 3.1 Amoeba Wim van Egmond/Visuals Unlimited All Organisms Are Composed
Cell Anatomy Anatomy = the study of the structures and components of an organism
 Cell Anatomy Anatomy = the study of the structures and components of an organism -Types of Cells: 1) Prokaryotic = simple, primitive = no membrane bound nucleus, only a dense, nuclear area = single-celled
Cell Anatomy Anatomy = the study of the structures and components of an organism -Types of Cells: 1) Prokaryotic = simple, primitive = no membrane bound nucleus, only a dense, nuclear area = single-celled
Renata Schipp Medical Biology Department
 Renata Schipp Medical Biology Department Deffinition of cell The cell is the smallest structural and functional unit of all known living organisms The cell was discovered by Robert Hooke in 1665 and also
Renata Schipp Medical Biology Department Deffinition of cell The cell is the smallest structural and functional unit of all known living organisms The cell was discovered by Robert Hooke in 1665 and also
Cells & Cell Organelles
 Cells & Cell Organelles The Building Blocks of Life AP Biology 2008-2009 Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal
Cells & Cell Organelles The Building Blocks of Life AP Biology 2008-2009 Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal
Supplement to SCnorm: robust normalization of single-cell RNA-seq data
 Supplement to SCnorm: robust normalization of single-cell RNA-seq data Supplementary Note 1: SCnorm does not require spike-ins, since we find that the performance of spike-ins in scrna-seq is often compromised,
Supplement to SCnorm: robust normalization of single-cell RNA-seq data Supplementary Note 1: SCnorm does not require spike-ins, since we find that the performance of spike-ins in scrna-seq is often compromised,
Cell morphology. Cell organelles structure and function. Chapter 1: UNIT 1. Dr. Charushila Rukadikar
 UNIT 1 Cell morphology Cell organelles structure and function Chapter 1: Dr. Charushila Rukadikar Assistant Professor Department Of Physiology ZMCH, Dahod Physiology The science that is concerned with
UNIT 1 Cell morphology Cell organelles structure and function Chapter 1: Dr. Charushila Rukadikar Assistant Professor Department Of Physiology ZMCH, Dahod Physiology The science that is concerned with
THE CELL Cells: Part 1
 THE CELL Cells: Part 1 OBJECTIVES By the end of the lesson you should be able to: State the 2 types of cells Relate the structure to function for all the organelles TYPES OF CELLS There are two types of
THE CELL Cells: Part 1 OBJECTIVES By the end of the lesson you should be able to: State the 2 types of cells Relate the structure to function for all the organelles TYPES OF CELLS There are two types of
Cells & Cell Organelles
 Cells & Cell Organelles Doing Life s Work 2009 2010 1 Types of cells bacteria cells Prokaryote no organelles animal cells Eukaryotes organelles plant cells 2 Cell size comparison Animal cell Bacterial
Cells & Cell Organelles Doing Life s Work 2009 2010 1 Types of cells bacteria cells Prokaryote no organelles animal cells Eukaryotes organelles plant cells 2 Cell size comparison Animal cell Bacterial
Ch. 6 A Tour of the Cell BIOL 222
 Ch. 6 A Tour of the Cell BIOL 222 Overview: The Fundamental Units of Life All organisms are made of cells The cell is the simplest collec=on of ma>er that can live Cell structure is correlated to cellular
Ch. 6 A Tour of the Cell BIOL 222 Overview: The Fundamental Units of Life All organisms are made of cells The cell is the simplest collec=on of ma>er that can live Cell structure is correlated to cellular
Plant Weight Week g 1.3 g 1.5 g 1 1 g 1.7 g 2 g 2 1 g 1.6 g 1.8 g 3 1 g 1.3 g 1.5 g 4 1 g 1 g 1 g. Plant Weight Week 1. Plant Weight Week 2
 9/11-9/12 Sci 7 AGENDA 1. Lecture 2. Organelle Flip Page 3. Processing Task 4. Fix your notebooks LEVEL ZERO VOICE CATALYST (10 minutes, individual work): 1. List all of the characteristics of life and
9/11-9/12 Sci 7 AGENDA 1. Lecture 2. Organelle Flip Page 3. Processing Task 4. Fix your notebooks LEVEL ZERO VOICE CATALYST (10 minutes, individual work): 1. List all of the characteristics of life and
DEPARTMENT OF PHYSICAL EDUCATION
 DEPARTMENT OF PHYSICAL EDUCATION Subject:- Anatomy, Physiology and Health Education Class:- B.P. Ed. Semester- I Presented By:- Dr. Mahesh Singh Dhapola Cell Theory All living things are made up of cells.
DEPARTMENT OF PHYSICAL EDUCATION Subject:- Anatomy, Physiology and Health Education Class:- B.P. Ed. Semester- I Presented By:- Dr. Mahesh Singh Dhapola Cell Theory All living things are made up of cells.
Cell Structure and Function. Biology 12 Unit 1 Cell Structure and Function Inquiry into Life pages and 68-69
 Cell Structure and Function Biology 12 Unit 1 Cell Structure and Function Inquiry into Life pages 45 59 and 68-69 Assignments for this Unit Pick up the notes/worksheet for this unit and the project There
Cell Structure and Function Biology 12 Unit 1 Cell Structure and Function Inquiry into Life pages 45 59 and 68-69 Assignments for this Unit Pick up the notes/worksheet for this unit and the project There
Cytosol the fluid Cytoplasm cell interior, everything outside the nucleus but within the cell membrane, includes the organelles, cytosol, and
 Cell Organelles Plasma Membrane comprised of a phospholipid bilayer and embedded proteins Outer surface has oligosaccharides separates the cells s contents from its surroundings Cytosol the fluid Cytoplasm
Cell Organelles Plasma Membrane comprised of a phospholipid bilayer and embedded proteins Outer surface has oligosaccharides separates the cells s contents from its surroundings Cytosol the fluid Cytoplasm
Early scientists who observed cells made detailed sketches of what they saw.
 Early scientists who observed cells made detailed sketches of what they saw. Early scientists who observed cells made detailed sketches of what they saw. CORK Early scientists who observed cells made detailed
Early scientists who observed cells made detailed sketches of what they saw. Early scientists who observed cells made detailed sketches of what they saw. CORK Early scientists who observed cells made detailed
Cell Structure & Function. Source:
 Cell Structure & Function Source: http://koning.ecsu.ctstateu.edu/cell/cell.html Definition of Cell A cell is the smallest unit that is capable of performing life functions. http://web.jjay.cuny.edu/~acarpi/nsc/images/cell.gif
Cell Structure & Function Source: http://koning.ecsu.ctstateu.edu/cell/cell.html Definition of Cell A cell is the smallest unit that is capable of performing life functions. http://web.jjay.cuny.edu/~acarpi/nsc/images/cell.gif
Title: Sep 10 7:59 PM (1 of 36) Ch 3 Cell Organelles and Transport
 Title: Sep 10 7:59 PM (1 of 36) Ch 3 Cell Organelles and Transport Title: Sep 10 8:02 PM (2 of 36) Cell organelles Nucleus: contains DNA Title: Sep 10 8:03 PM (3 of 36) Nuclear envelope double membrane
Title: Sep 10 7:59 PM (1 of 36) Ch 3 Cell Organelles and Transport Title: Sep 10 8:02 PM (2 of 36) Cell organelles Nucleus: contains DNA Title: Sep 10 8:03 PM (3 of 36) Nuclear envelope double membrane
SBI3U7 Cell Structure & Organelles. 2.2 Prokaryotic Cells 2.3 Eukaryotic Cells
 SBI3U7 Cell Structure & Organelles 2.2 Prokaryotic Cells 2.3 Eukaryotic Cells No nucleus Prokaryotic Cells No membrane bound organelles Has a nucleus Eukaryotic Cells Membrane bound organelles Unicellular
SBI3U7 Cell Structure & Organelles 2.2 Prokaryotic Cells 2.3 Eukaryotic Cells No nucleus Prokaryotic Cells No membrane bound organelles Has a nucleus Eukaryotic Cells Membrane bound organelles Unicellular
Cancer Informatics Lecture
 Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
Chapter 2 Cell. Zhou Li Prof. Dept. of Histology and Embryology
 Chapter 2 Cell Zhou Li Prof. Dept. of Histology and Embryology The inner life of the cell Ⅰ. Plasma membrane (Plasmalemma) 1.1 The structure Unit membrane: inner layer 3-layered structure outer layer mediat
Chapter 2 Cell Zhou Li Prof. Dept. of Histology and Embryology The inner life of the cell Ⅰ. Plasma membrane (Plasmalemma) 1.1 The structure Unit membrane: inner layer 3-layered structure outer layer mediat
CELL STRUCTURE AND CELL ORGANISATION
 CELL STRUCTURE AND CELL ORGANISATION CHAPTER 2 https://wickedbiology.wordpress.com Cellular components of animal & plant cells https://wickedbiology.wordpress.com Plant Cells Cell wall Plasma membrane
CELL STRUCTURE AND CELL ORGANISATION CHAPTER 2 https://wickedbiology.wordpress.com Cellular components of animal & plant cells https://wickedbiology.wordpress.com Plant Cells Cell wall Plasma membrane
Study Guide for Biology Chapter 5
 Class: Date: Study Guide for Biology Chapter 5 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Which of the following led to the discovery of cells? a.
Class: Date: Study Guide for Biology Chapter 5 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Which of the following led to the discovery of cells? a.
Cell Structure and Function
 Cell Structure and Function Many Scientists Contributed to the Cell Theory! Hooke discovered cells while looking at cork under the microscope! Leewenhoek was the first to observe bacteria! Schleiden discovered
Cell Structure and Function Many Scientists Contributed to the Cell Theory! Hooke discovered cells while looking at cork under the microscope! Leewenhoek was the first to observe bacteria! Schleiden discovered
LECTURE 3 CELL STRUCTURE
 LECTURE 3 CELL STRUCTURE HISTORY The cell was first discovered by Robert Hooke in 1665 examining very thin slices of cork and saw a multitude of tiny pores that remarked looked like the walled compartments
LECTURE 3 CELL STRUCTURE HISTORY The cell was first discovered by Robert Hooke in 1665 examining very thin slices of cork and saw a multitude of tiny pores that remarked looked like the walled compartments
Practice Exam 2 MCBII
 1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
PROTEIN TRAFFICKING. Dr. SARRAY Sameh, Ph.D
 PROTEIN TRAFFICKING Dr. SARRAY Sameh, Ph.D Overview Proteins are synthesized either on free ribosomes or on ribosomes bound to endoplasmic reticulum (RER). The synthesis of nuclear, mitochondrial and peroxisomal
PROTEIN TRAFFICKING Dr. SARRAY Sameh, Ph.D Overview Proteins are synthesized either on free ribosomes or on ribosomes bound to endoplasmic reticulum (RER). The synthesis of nuclear, mitochondrial and peroxisomal
Bio10 Cell Structure SRJC
 3.) Cell Structure and Function Structure of Cell Membranes Fluid mosaic model Mixed composition: Phospholipid bilayer Glycolipids Sterols Proteins Fluid Mosaic Model Phospholipids are not packed tightly
3.) Cell Structure and Function Structure of Cell Membranes Fluid mosaic model Mixed composition: Phospholipid bilayer Glycolipids Sterols Proteins Fluid Mosaic Model Phospholipids are not packed tightly
Unit 2: More on Matter & Energy in Ecosystems. Macromolecules to Organelles to Cells
 IN: Unit 2: More on Matter & Energy in Ecosystems Macromolecules to Organelles to Cells Where are cells on the biological scale? Sub-Atomic Particles Atoms Molecules Macromolecules (proteins, lipids, nucleic
IN: Unit 2: More on Matter & Energy in Ecosystems Macromolecules to Organelles to Cells Where are cells on the biological scale? Sub-Atomic Particles Atoms Molecules Macromolecules (proteins, lipids, nucleic
In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question.
 CHAPTER 3 TEST Cell Structure Circle T if the statement is true or F if it is false. T F 1. Small cells can transport materials and information more quickly than larger cells can. T F 2. Newly made proteins
CHAPTER 3 TEST Cell Structure Circle T if the statement is true or F if it is false. T F 1. Small cells can transport materials and information more quickly than larger cells can. T F 2. Newly made proteins
3. Which cell has the greater ratio of surface area to volume?
 Chapter 4 Worksheet A Tour of the Cell Exercise 1 Metric System Review/Size and Scale of Our World (4.1) Use the information in the two modules and the chart in Module 4.2 to complete the following table
Chapter 4 Worksheet A Tour of the Cell Exercise 1 Metric System Review/Size and Scale of Our World (4.1) Use the information in the two modules and the chart in Module 4.2 to complete the following table
Molecular Cell Biology - Problem Drill 17: Intracellular Vesicular Traffic
 Molecular Cell Biology - Problem Drill 17: Intracellular Vesicular Traffic Question No. 1 of 10 1. Which of the following statements about clathrin-coated vesicles is correct? Question #1 (A) There are
Molecular Cell Biology - Problem Drill 17: Intracellular Vesicular Traffic Question No. 1 of 10 1. Which of the following statements about clathrin-coated vesicles is correct? Question #1 (A) There are
Eukaryotic Cell Structure
 Eukaryotic Cell Structure Vocabulary listed for Chapter 7.3: cell wall, chromatin, nucleolus, ribosome, cytoplasm, endoplasmic reticulum, Golgi apparatus, vacuole, lysosome, chloroplast, plastid, chlorophyll,
Eukaryotic Cell Structure Vocabulary listed for Chapter 7.3: cell wall, chromatin, nucleolus, ribosome, cytoplasm, endoplasmic reticulum, Golgi apparatus, vacuole, lysosome, chloroplast, plastid, chlorophyll,
Chapter 4: Cell Structure and Function
 Chapter 4: Cell Structure and Function Robert Hooke Fig. 4-2, p.51 The Cell Smallest unit of life Can survive on its own or has potential to do so Is highly organized for metabolism Senses and responds
Chapter 4: Cell Structure and Function Robert Hooke Fig. 4-2, p.51 The Cell Smallest unit of life Can survive on its own or has potential to do so Is highly organized for metabolism Senses and responds
Lab 3: Cellular Structure and Function
 Lab 3: Cellular Structure and Function What is the basic unit of life? The simplest form of life is the cell! All living things are either: unicellular (only one cell) multicellular (many cells make one
Lab 3: Cellular Structure and Function What is the basic unit of life? The simplest form of life is the cell! All living things are either: unicellular (only one cell) multicellular (many cells make one
Cellular compartments
 Cellular compartments 1. Cellular compartments and their function 2. Evolution of cellular compartments 3. How to make a 3D model of cellular compartment 4. Cell organelles in the fluorescent microscope
Cellular compartments 1. Cellular compartments and their function 2. Evolution of cellular compartments 3. How to make a 3D model of cellular compartment 4. Cell organelles in the fluorescent microscope
Eukaryotic cells are essentially two envelope systems. Nuclear materials are separated from cytoplasm by nuclear membrane. Complex structure Also
 Dr. Gugale Pritesh Ramanlal M.Sc., Ph.D., B.Ed., D.M.L.T. Email id - pritesh.gugale09@gmail.com Contact numbernumber- 8446475310 Eukaryotic cells are essentially two envelope systems. Nuclear materials
Dr. Gugale Pritesh Ramanlal M.Sc., Ph.D., B.Ed., D.M.L.T. Email id - pritesh.gugale09@gmail.com Contact numbernumber- 8446475310 Eukaryotic cells are essentially two envelope systems. Nuclear materials
Cytology II Study of Cells
 Cytology II Study of Cells Biology 20 Cellular Basis of Life 1. Basic unit of Life 2. Composed of one or more cells 3. Arises from pre-existing cells Asexual (Mitosis)/Sexual (Meiosis) 4. Surrounded by
Cytology II Study of Cells Biology 20 Cellular Basis of Life 1. Basic unit of Life 2. Composed of one or more cells 3. Arises from pre-existing cells Asexual (Mitosis)/Sexual (Meiosis) 4. Surrounded by
Chapter 3. Chapter 3. Bellringer. Objectives. Chapter 3. Chapter 3. Vocabulary. Cell Wall. Identify the different parts of a eukaryotic cell.
 Bellringer Objectives Identify the different parts of a eukaryotic cell. Explain the function of each part of a eukaryotic cell. Plant cells and animal cells have many cell parts in common. But they also
Bellringer Objectives Identify the different parts of a eukaryotic cell. Explain the function of each part of a eukaryotic cell. Plant cells and animal cells have many cell parts in common. But they also
Cells & Cell Organelles. Doing Life s Work
 Cells & Cell Organelles Doing Life s Work AP Biology 2009-2010 Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal cell
Cells & Cell Organelles Doing Life s Work AP Biology 2009-2010 Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal cell
10 m Human height 1 m Length of some nerve and muscle cells eye 100 mm (10 cm) Chicken egg aid n 10 mm
 Biology 112 Unit Three Chapter Four 1 Cell Sizes Smallest Bacteria Largest Bird egg Longest Giraffe s Nerve Cell Most Cells Diameter of 0.7µm to 105 µm 2 10 m 1 m 100 mm (10 cm) 10 mm (1 cm) Human height
Biology 112 Unit Three Chapter Four 1 Cell Sizes Smallest Bacteria Largest Bird egg Longest Giraffe s Nerve Cell Most Cells Diameter of 0.7µm to 105 µm 2 10 m 1 m 100 mm (10 cm) 10 mm (1 cm) Human height
Structure & Function of Cells
 Anatomy & Physiology 101-805 Unit 4 Structure & Function of Cells Paul Anderson 2011 Anatomy of a Generalised Cell Attached or bound ribosomes Cilia Cytosol Centriole Mitochondrion Rough endoplasmic reticulum
Anatomy & Physiology 101-805 Unit 4 Structure & Function of Cells Paul Anderson 2011 Anatomy of a Generalised Cell Attached or bound ribosomes Cilia Cytosol Centriole Mitochondrion Rough endoplasmic reticulum
Structure and Function of Cells
 Structure and Function of Cells Learning Outcomes Explain the cell theory Explain why cell size is usually very small Describe the Fluid Mosaic Model of membranes Describe similarities and differences
Structure and Function of Cells Learning Outcomes Explain the cell theory Explain why cell size is usually very small Describe the Fluid Mosaic Model of membranes Describe similarities and differences
lysosomes Ingested materials Defective cell components Degrades macromolecules of all types:
 lysosomes Digests Ingested materials Defective cell components Degrades macromolecules of all types: Proteins Nucleic acids Carbohydrates Lipids Single membrane bound vesicle, contains up to 50 digestive
lysosomes Digests Ingested materials Defective cell components Degrades macromolecules of all types: Proteins Nucleic acids Carbohydrates Lipids Single membrane bound vesicle, contains up to 50 digestive
Cells. Variation and Function of Cells
 Cells Variation and Function of Cells Cell Theory states that: 1. All living things are made of cells 2. Cells are the basic unit of structure and function in living things 3. New cells are produced from
Cells Variation and Function of Cells Cell Theory states that: 1. All living things are made of cells 2. Cells are the basic unit of structure and function in living things 3. New cells are produced from
Chapter 7 Cell Structure and Function. Section Objectives: Relate advances in microscope technology to discoveries about cells and cell structure.
 Chapter 7 Cell Structure and Function Section Objectives: Relate advances in microscope technology to discoveries about cells and cell structure. Compare the operation of a microscope with that of an electron
Chapter 7 Cell Structure and Function Section Objectives: Relate advances in microscope technology to discoveries about cells and cell structure. Compare the operation of a microscope with that of an electron
Data mining with Ensembl Biomart. Stéphanie Le Gras
 Data mining with Ensembl Biomart Stéphanie Le Gras (slegras@igbmc.fr) Guidelines Genome data Genome browsers Getting access to genomic data: Ensembl/BioMart 2 Genome Sequencing Example: Human genome 2000:
Data mining with Ensembl Biomart Stéphanie Le Gras (slegras@igbmc.fr) Guidelines Genome data Genome browsers Getting access to genomic data: Ensembl/BioMart 2 Genome Sequencing Example: Human genome 2000:
Basic Structure of a Cell. copyright cmassengale
 Basic Structure of a Cell 1 Prokaryotes The first Cells Cells that lack a nucleus or membrane-bound organelles Includes bacteria Simplest type of cell Single, circular chromosome 2 Prokaryotes Nucleoid
Basic Structure of a Cell 1 Prokaryotes The first Cells Cells that lack a nucleus or membrane-bound organelles Includes bacteria Simplest type of cell Single, circular chromosome 2 Prokaryotes Nucleoid
A Tour of the Cell. Chapter 6. Biology Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 6 A Tour of the Cell PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp
Chapter 6 A Tour of the Cell PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp
A Tour of the Cell. Chapter 4. Most cells are microscopic. Cells vary in size and shape
 Chapter 4 A Tour of the Cell Most cells are microscopic Cells vary in size and shape 10 m Human height 1 m Length of some nerve and muscle cells 100 mm (10 cm) 10 mm (1 cm) Chicken egg Unaided eye 1 mm
Chapter 4 A Tour of the Cell Most cells are microscopic Cells vary in size and shape 10 m Human height 1 m Length of some nerve and muscle cells 100 mm (10 cm) 10 mm (1 cm) Chicken egg Unaided eye 1 mm
Renáta Schipp Gergely Berta Department of Medical Biology
 The cell III. Renáta Schipp Gergely Berta Department of Medical Biology Size and Biology Biology is a visually rich subject many of the biological events and structures are smaller than the unaided human
The cell III. Renáta Schipp Gergely Berta Department of Medical Biology Size and Biology Biology is a visually rich subject many of the biological events and structures are smaller than the unaided human
The Cell. BIOLOGY OF HUMANS Concepts, Applications, and Issues. Judith Goodenough Betty McGuire
 BIOLOGY OF HUMANS Concepts, Applications, and Issues Fifth Edition Judith Goodenough Betty McGuire 3 The Cell Lecture Presentation Anne Gasc Hawaii Pacific University and University of Hawaii Honolulu
BIOLOGY OF HUMANS Concepts, Applications, and Issues Fifth Edition Judith Goodenough Betty McGuire 3 The Cell Lecture Presentation Anne Gasc Hawaii Pacific University and University of Hawaii Honolulu
Human Epithelial Cells
 The Cell Human Epithelial Cells Plant Cells Cells have an internal structure Eukaryotic cells are organized Protective membrane around them that communicates with other cells Organelles have specific jobs
The Cell Human Epithelial Cells Plant Cells Cells have an internal structure Eukaryotic cells are organized Protective membrane around them that communicates with other cells Organelles have specific jobs
