Molecular insights into mechanisms regulating faithful chromosome separation in female meiosis
|
|
|
- Rodger Joseph
- 6 years ago
- Views:
Transcription
1 Cell Cycle ISSN: (Print) (Online) Journal homepage: Molecular insights into mechanisms regulating faithful chromosome separation in female meiosis Shen Yin, Xiao-Fang Sun, Heide Schatten & Qing-Yuan Sun To cite this article: Shen Yin, Xiao-Fang Sun, Heide Schatten & Qing-Yuan Sun (2008) Molecular insights into mechanisms regulating faithful chromosome separation in female meiosis, Cell Cycle, 7:19, , DOI: /cc To link to this article: Copyright 2008 Landes Bioscience Published online: 01 Oct Submit your article to this journal Article views: 122 View related articles Citing articles: 26 View citing articles Full Terms & Conditions of access and use can be found at Download by: [ ] Date: 28 November 2017, At: 03:51
2 [Cell Cycle 7:19, ; 1 October 2008]; 2008 Landes Bioscience Review Molecular insights into mechanisms regulating faithful chromosome separation in female meiosis Shen Yin, 1 Xiao-Fang Sun, 2 Heide Schatten 3 and Qing-Yuan Sun 1, * 1 State Key Laboratory of Reproductive Biology; Institute of Zoology; Chinese Academy of Sciences; Beijing, China; 2 Institute of Gynecology; The Third Affiliated Hospital of Guangzhou Medical College; Guangzhou, China; 3 Department of Veterinary Pathobiology; University of Missouri; Columbia, Missouri USA Abbreviations: APC/C, anaphase promoting complex/cyclosome; M, metaphase; A, anaphase Key words: chromosome separation, cohesin, meiosis, shugoshin, spindle checkpoint The faithful segregation of chromosomes into daughter cells in meiosis is crucial to produce healthy progeny. In gametogenesis, two consecutive rounds of chromosome separation occur with only one round of DNA replication, and the chromosome number is reduced to half to produce haploid gametes. Here, we discuss the molecular mechanisms underlying faithful chromosome separation in meiosis from three aspects: spindle checkpoint, two-step releases of cohesion, and the specific space-time protection of cohesin. Introduction During each cell cycle, accurate segregation of chromosomes must take place to ensure that each daughter cell receives exactly one copy of the genome in order to maintain genetic stability of species. In mitosis, sister chromatids separate during the metaphaseto-anaphase transition so that the two daughter cells receive the same chromosome numbers as the original mother cell. In meiosis, however, two consecutive rounds of chromosome separations take place with only one round of DNA replication, resulting in chromosome number reduction to exactly half to produce haploid gametes. Homologous chromosomes separate from each other during the first meiosis (meiosis I) while sister chromatids segregate during the second meiosis (meiosis II). 1,2 Failures in executing this process accurately results in aneuploidy, which is the cause for genetic disorders and aneuploid embryos In human meiosis, chromosome non-disjunction resulting in aneuploidy is the leading cause of trisomies. 11,12 Meiosis II resembles mitosis, while meiosis I is distinctly different and displays unique characteristics. First, in meiosis I, homologous chromosomes undergo pairing and recombination Single chromatid from each of the homologous chromosomes form synapses and chiasmata that allow and ensure recombination of genetic information. 13,16 Next, the kinetochores of sister chromatids are captured by microtubules that emanate from the same spindle pole (sister kinetochores co-orientation), which ensures that homologous chromosomes separate accurately to opposite poles of the spindle. Finally, a two-step releases of cohesin ensures that the sister chromatids are pulled together to the same pole of the meiosis I spindle; they only separate away from each other in meiosis II. 20,21 To avoid aneuploidy, meiotic cells have developed highly conserved mechanisms to ensure faithful segregation of homologous chromosomes in meiosis I and sister chromatids in meiosis II, respectively. Here, we discuss the underlying molecular mechanisms of faithful chromosome separation in meiosis from three aspects: spindle checkpoint, two-step releases of cohesion, and the specific space-time protection of cohesin. Spindle Checkpoint Kinetochores. In mitosis, sister chromatids are oriented toward opposite spindle poles, thus sister chromatids separate in anaphase to opposite directions. In contrast to mitosis, however, sister chromatids have to be oriented toward the same spindle pole in meiosis I, which ensures monopolar attachment and segregation of homologous chromosomes in meiosis I. The faithful separation of chromosomes requires accurate connections between chromosomes and microtubules, in which kinetochores play most significant roles, both in mitosis and meiosis. Kinetochores serve at least three functions: attaching chromosomes to the spindle, controlling chromosome movement, and maintaining the spindle checkpoint by sensing tension and/or attachment of spindle microtubules to kinetochores In mitosis, sister kinetochores attach to microtubules that emanate from opposite spindle poles prior to the onset of anaphase in a process termed sister kinetochore bi-orientation. In meiosis I, however, sister kinetochores from one homolog must attach to microtubules that extend from the same spindle pole, which is termed sister kinetochore co-orientation, that ensures homologous chromosomes separation in meiosis I, with sister chromatids being pulled to the same pole. Functional genomics has identified monopolin(mam1), a kinetochore protein required for the monopolar attachment and segregation of homologous chromosomes during meiosis I. 26 In monopolin *Correspondence to: Qing-Yuan Sun; State Key Laboratory of Reproductive Biology; Institute of Zoology; Chinese Academy of Sciences; #5 Datun Rd; Chaoyang, Beijing China; Tel./Fax: ; sunqy@ioz.ac.cn; sunqy1@ yahoo.com Submitted: 07/17/08; Accepted: 08/18/08 Previously published online as a Cell Cycle E-publication: Cell Cycle 2997
3 Figure 1. Spindle checkpoint pathway. Checkpoint is turned on when sister chromatids are not correctly attached to the spindle microtubules. All checkpoint proteins, Aurora B, Mps1, Bub1, CENP-E, BubR1, Bub3 Mad1 and Mad2 are sequentially binding to kinetochores. Mad2 directly combines Cdc20, thus inhibits the activity of APC/C. Inactivation of APC/C leads to stabilization of securin, which in turn inhibits separase. Plk1 kinase phosphorylates and removes cohesin along chromosome arms except for centromere. After all chromaids are correctly attached to the spindle, checkpoint is turned off. Mad2 detaches from kinetochores which releases the inhibition on Cdc20, and then Cdc20 activates APC/C, which ubiquitinates securin. Then ubiquitinated securin is degraded by proteasome and separase is activated, resulting in cleavage of centromeric cohesin. Thus, sister chromatids separate from each other in anaphase. mutants, kinetochores of sister chromatids fail to attach to microtubules emanating from the same spindle pole (co-orientation), but attach to opposite poles (bi-orientation) during meiosis I. A protein complex, Csm1/Lrs4, migrates to centromeres and combines with monopolin, which is essential for monopolar attachment of sister kinetochores in meiosis I. 27 When Polo-like kinase Cdc5 is lacking, localization of Lrs4 and monopolin proteins to kinetochores is defective, resulting in bipolar orientation of sister kinetochores in meiosis I. 28 In fission yeast, Aurora kinase Ark1 and Ipl1 is necessary for the faithful mono-orientation of sister chromatids in meiosis I. 29,30 Thus, it is implied that the monopolin complex regulates kinetochore-microtubule attachment by including Aurora B kinase to co-orient sister chromatids during meiosis I. In addition, the kinetochore proteins Pcs1, Mde4, casein kinase1 and heterochromatin are required for mono-polar attachment of sister kinetochores at meiosis I. 31,32 Spindle checkpoint pathway. If sister kinetochores are not correctly attached to spindle microtubules at anaphase onset, chromosome mis-segregation may occur, resulting in aneuploidy. Mitotic cells have developed a high-fidelity surveillance mechanism termed the spindle checkpoint to prevent chromosome missegregation by delaying onset of anaphase until all kinetochores of sister chromatids are correctly attached to the spindle The major components of this surveillance mechanism, originally identified in budding yeast, contain mitotic arrest-deficient (Mad) 1 3 proteins, budding uninhibited by benzimidazole (Bub) 1 3 proteins, and Mps Aside from these proteins, increasing evidence has implies Aurora B as a new important factor in the spindle checkpoint Spindle checkpoint is able to detect spindle damage or a single, unaligned chromosome in the spindle and to arrest the cell cycle at metaphase, which provides more time for all of the chromosomes to move to the spindle equator before chromosomes separate. Although it is not clear by which mechanisms the cells ensure that sister chromatids or homologous chromosomes attach to the microtubules and the spindle checkpoint proteins control anaphase onset, it appears that Aurora B, Mad1-3, Bub1, Bub3 and Mps1 are part of the same metaphase-to-anaphase-transition checkpoint pathway Aurora B may be the first checkpoint protein that binds to the kinetochores. 45,49 It is implicated that Aurora B play roles in metaphase chromosome alignment, 50,51 microtubule-kinetochore interactions 40 and kinetochore localization of Bub1, Mad2 and Mps1. 52 Then, Mps1 and/or Bub1 bind to kinetochores. As a protein kinase, Bub1 can bind and phosphorylate Bub3, 53 and be sensitive to tension across the centromere Bub1 accumulates on kinetochores in the absence of tension, 44,47,48,54,57 as a platform for other spindle checkpoint proteins. 58 Bub1 is also reported to control centromeric cohesin and loss of Bub1 function causes checkpoint impairment. 44,59-63 The results from depletion of Bub1 and Mps1 suggest that Bub1 and Mps1 bind to kinetochores at the same time and depend on each other, but they bind to kinetochores earlier than other checkpoint proteins ,49,64 CENP-E is a kinase-related microtubule motor protein 64,65 and participates in chromosome movement It also participates in spindle checkpoint by acting as a binding partner of BubR1, 69,70 or forming a link between microtubules and kinetochores. 49,71,72 The next proteins that bind to kinetochors are Bub3 and BubR1 (mammalian homolog of yeast Mad3). 73 As a platform, Bub3 associates with Mad2, Mad3 and Cdc20, which directly inhibit anaphase promoting complex/cyclosome (APC/C). 74,75 BubR1 is required for kinetochore loading of Mad1 and Mad2. 46 The last two checkpoint proteins binding to kinetochores are Mad1 and Mad It appears that Mad1 affects Mad2 binding to kinetochores. 79 As the last one in the checkpoint pathway, Mad2 has been proposed to be a marker for spindle checkpoint, since Mad2 appears to respond directly to microtubule 2998 Cell Cycle 2008; Vol. 7 Issue 19
4 Figure 2. Two-step releases of cohesin and specific space-time protection of centromeric cohesin during meiosis. In meiosis I, sister kinetochores of a homologous chromosome are arranged side-by-side, and they establish attachment to microtubules emanating from the same spindle pole (co-orientation). For synapsis and recombination, sister chromatids from one homologous chromosome are held together by chromosome arm cohesin and centromeric cohesin. Rec8, the target of separase, is the most important cohesin subunit in meiosis. The Plk1 kinase marks Rec8 by phosphorylation at chromatid arms for Rec8 degradation by separase. Shugoshin1 and PP2A are localized at centromeres by Bub1, which results in dephosphorylation of centromeric cohesin. After silencing of spindle checkpoint, at the transition of MI-AI, separase can only cleave phosphorylated arm cohesin along sister chromatids while centromeric cohesin remains unphosporylated by PP2A. Shugoshin1 collaborates with PP2A to protect cohesin by antagonizing phosphorylation from Plk1. In meiosis II, Shugoshin and PP2A disappear from kinetochores, thus centromeric cohesin is cleaved by separase to allow sister chromatid separation as is the case in mitosis. anchoring at the kinetochore plate 80 and directly binds to Cdc20, which is the activator of APC/C. 81 Studies in several model systems have proposed that the downstream target of the spindle checkpoint is Cdc20, an activator of APC/C, which is an 11-subunit complex containing ubiquitin ligase activity. 34 Cdc20 is a member of the cell cycle protein family including Cdc4 in S. cerevisiae, Fzy in Drosophila, Slp1 in S. pombe and p55cdc in human The association of APC/C with either Cdc20 or Cdh1 is essential for its activity. 83 The spindle checkpoint proteins transmit inhibitory signals to APC/C Cdc20 and thus prevents the metaphase-anaphase transition until all chromosomes have established a bipolar attachment to the spindle. 86 At the transition of metaphase to anaphase, APC/C Cdc20 activity triggers securin (separase inhibitor) degradation, thereby allowing separase to become active and dissolve the cohesin between sister chromatids. This cascade is highly conserved from yeast to human All these proteins work together and form a complicated checkpoint system. Once any spindle checkpoint protein is out of order, then the checkpoint system is turned off and faithful chromosome segregation is affected, resulting in aneuploidy. Spindle checkpoint proteins in mammalian meiosis I. Although the checkpoint pathway in mitosis has been widely studied, only a few spindle checkpoint proteins have been reported in meiosis. Mad1, 90 Mad2, and Bub1, 95 function during meiosis as checkpoint proteins, which ensures high fidelity meiotic chromosome segregation. 49,96 Mad1 is present in mouse oocytes and its localization is not exactly the same as in mitosis. 90,97 Mad1 functions by sensing attachment of chromosomes to microtubules rather than tension between microtubules and chromosomes, since Mad1 localization is not altered when tension is changed. 90,98 Adding anti-mad1 antibody to Xenopus egg extracts de-activates the checkpoint and prevents Mad2 from binding to unattached kinetochores. 79 Mad2 becomes activated and dissociated from Mad1 at kinetochores and is replenished by the pool of Mad1- free Mad2. 78 Thus, Mad1 participates in spindle checkpoint in meiosis by recruiting Mad2 binding to kinetochores. Mad2 is one of the best-studied spindle checkpoint proteins in mammalian meiosis. Similar Mad2 localization was observed in the pre-metaphse of meiosis I (Pre-MI) stage and differences in Mad2 localization were observed after reaching MI and past MI stages during meiosis in rat, 93 mouse 91 and pig. 99 The localization of Mad2 is similar to that of Mad1, but is clearly different from other checkpoint proteins, which still bind to kinetochores of metaphase stage cells. However, one common characteristic is that Mad2 binds to unattached kinetochores and is released from kinetochores when microtubules attach to kinetochores. 49 The function of Mad2 has been studied by Mad2 depletion, 100 antibody inhibition, overexpression, microtubule disruption and stabilization. Blocking Mad2 function induces premature anaphase in which chromosomes start to separate before they are correctly aligned at the metaphase plate in rat, 93 mouse 91,101 and pig oocytes. 99 On the other hand, overexpression of Mad2 results in metaphase arrest by activation of the spindle checkpoint in mammalian oocytes, 93,101 which indicates that excess of Mad2 prevents silencing of the spindle checkpoint. It appears that Mad2 senses the defect of microtubule attachment rather than tension. 54, We also reported the same results in oocyte meiosis, in which Mad2 can sense unattached kinetochores, but can not sense unaligned chromosomes. 93 It has been implicated that dynein, one spindle motor protein, also participates in the spindle checkpoint mechanism. 106,107 In mammalian oocytes, we reported that cytoplasmic dynein participates in meiotic checkpoint inactivation by transporting cytoplasmic Mad1 and Mad2 proteins from kinetochores to spindle poles. 98 Evidences for Bub1 as spindle checkpoint protein in meiosis came from mouse oocyte. 95,108 We reported that Bub1 could prevent precocious anaphase and chromosome misalignment, 95 which is consistent to the dual role for Bub1 in the spindle checkpoint and chromosome congression. 109 For the latter role, Bub1 may function through controlling Shugoshin (see below) localization according to results obtained from mitosis. 61,62 CENP-E function is essential for meiosis I in pig, 110 mouse, rat and frog, 111,112 since it is a binding partner of BubR1. The localization of MAP kinase kinase (MEK) and MAP kinase is similar to some spindle checkpoint proteins in oocytes. Immunodepletion of MAP kinase prevents checkpoint activation, which is rescued Cell Cycle 2999
5 by adding external MAP kinase. 119 Knockdown of MEK causes misalignment of chromosomes and error in chromosome separation. 118 Thus, MEK/MAP kinase may also participate in the spindle checkpoint pathway regulation in meiosis All these observations support the notion that spindle checkpoint is required for faithful segregation of chromosomes in oocyte meiosis I. Therefore, premature silencing of spindle checkpoint may be associated with an increase in aneuploidy of oocytes. Two-Step Releases of Cohesion When all homologous chromosomes are correctly attached to the spindle, the cohesion between chromosomes is cut off to initiate anaphase. It is known that cohesion of sister chromatids in eukaryotes is largely achieved by the cohesin complex, which is important for high fidelity chromosome segregation during anaphase. 124 In meiosis I, homologous chromosomes are connected by chiasmata. Then the kinetochores of sister chromatids are captured by the spindle fiber complex emanating from the same pole of spindle. When the homologous chromosomes are aligned correctly on the equator of spindle, anaphase I is initiated and sister chromatids are pulled to the same pole of the spindle. Importantly, only cohesins along the chromosome arms are removed and centromeric cohesins are maintained in meiosis I. Retention of centromeric cohesin is necessary for faithful segregation of sister chromatids in meiosis II. Thereby, the two-step releases of arm and centromeric cohesin accounts for segregation of homologous chromosomes in meiosis I and sister chromatids in meiosis II. The structure of cohesin. Cohesin is a multiprotein complex, conserved from yeast to humans, that mediates linkage of sister chromatids. It combines sister chromatids in the S stage and disappears from sister chromatids during the transition from metaphase to anaphase. 124 In mitosis of Saccharomyces cerevisiae, it includes at least four subunits, Smc1, Smc3 (Structural maintenance of chromosome, SMC), Scc1/Mcd1 and Scc3 (Sister chromaitd cohesion, SCC) Up to date, the paralogs of four subunits have been identified in Caenorhabditis elegans, Drosophila melanogaster, Xenopus laevis, Mus musculus, Homo sapiens. In human, three paralogs of Scc3 are found, namely SA1, SA2 and SA3, and SA3 is a meiosis-specific cohesin. 127 Two paralogs of Smc1 are found, namely Smc1α and Smc1β, the latter is meiosis-specific. 128 With the help of Scc3, Scc1 combines the Smc1-Smc3 heterodimer through its N and C motif, to form a large protein ring, which holds the sister chromatids together. 129,130 The ring-shaped structure holding sister chromatids tightly together contributes to the recombination of DNA and faithful chromosome segregation. 131 When the cell undergoes the transition from metaphase to anaphase, Scc1 cleavage by separase causes the protein ring to open and cohesins dissociate from centromeres, which initiates the segregation of sister chromatids. 129 Therefore, Scc1 is an important target controlling the transition from metaphase to anaphase. Except for the proteins described, one new cohesin protein, Pds5, has been identified in yeast and human, which is in physical contact with the cohesin complex. But their connection is weak, and thus Pds5 may be a cohesin-raleated protein. 132,133 In meiosis, Scc1/Mcd1 is replaced by Rec8, the specific meiotic protein, while the other three subunits, Scc3, Smc1 and Smc3 are consistent with those of mitosis. 134 Rec8 is the most important meiosis-specific cohesin subunit, which is degraded by separase and necessary for the segregation of chromosomes The paralog proteins have been identified in homo sapiens, Mus musculus, Caenorhabditis elegans, Drosophila melanogaster, Xenous laevis Arabidopsis thaliana. 1,127, These proteins are composed of a conserved protein family, the Rad21/Rec8 family. According to amino acid sequence analysis, Scc1 and Rec8 paralogs in higher eukaryotes contain highly conserved N and C-terminal motif. 140,141 Localization and degradation of cohesion. The combination of cohesin with chromosomes initiated at the S stage largely depends on the loading proteins, Scc2 and Scc Cohesin localizes strongly to the centromeres and randomly associates with multiple sites along the chromosome arms depending on the transcription convergent regions, 143 about every 5 10 kb. 144,145 The different localizations on chromosome arms and centromeres are the perquisite for the two-step release, which is necessary for segregation of homologous choromosomes in meiosis I and sister chromatids in meiosis II. In mitosis, during the transition from metaphase to anaphase, cohesin cleavage initiates the segregation of chromosomes. The specific endopeptidase, separase, is responsible for timely cohesin degradation The conserved His and Cys amino acids in the C terminal motif of separase are necessary for the enzyme activity, since mutation in either one of them causes the inactivation of separase. 149,150 Two different pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. 151 The first is prophase pathway. In prophase and pre-metaphase, the chromosome arm cohesin is first degraded, independent of separase. 137,152,153 Kinase phosphorylation plays important roles in this process. 154 The second is the anaphase pathway. The remaining centromeric cohesin is cleaved by separase at the transition from metaphase to anaphase. 144,151,155 The cleavage of arm cohesin and centromeric cohesin is triggered in two different ways, which may be the evolutionary predecessor of the two-step releases of cohesin in meiosis. In meiosis I, due to crossovers between homologous chromosome arms, cohesion along the arms must first be dissociated to allow the segregation of the homologous chromosomes. The arm Rec8 first dissociates from homologous chromosomes and sister chromatids, which introduces segregation of homologous chromosomes and the two arms of sister chromatids. 136, This is the first step release of cohesin from chromosomes. Notably, the removal of arm cohesin in meiosis I is a separase-dependent mechanism, 159 while in mitosis it is a separase-independent mechanism so-called prophase pathway. However, the centromeric Rec8 between sister chromatids persists until meiosis II, so that sister chromatids are held together and pulled towards the same pole of the spindle. 134,160 The second step release of cohesin occurs in meiosis II. As in mitosis, the cleavage of centromeric Rec8 by separase initiates the segregation of sister chromatids in mammalian meiosis II. 157 The two-step releases of arm cohesin and centromeric cohesin is the important molecular mechanism regulating two subsequent meiotic divisions. Activation of separase for cohesin release. It has been shown that separase is responsible for cleavage of cohesin subunit Rec8, the resolution of chiasmata and transition from metaphase to anaphase in meiosis. 159,161,162 For most of the cell cycle, separase combines with its inhibitory factor, securin, and loses its ability to dissociate cohesin. 148, Spindle checkpoint and APC/C control securin dissociation from 3000 Cell Cycle 2008; Vol. 7 Issue 19
6 separase. Receiving the inhibitory signals from checkpoint, stabilized securin with separase prevents cleavage of cohesin and metaphaseanaphase transition. 86, After all chromosomes have established correct attachments to the spindle fibers, the spindle checkpoint is inactivated and the APC/C Cdc20 ubiquitinates securin, and degrades securin by proteasomes, which in turn allows separase to become active and thus cohesion is eliminated. 162,172 In meiosis, mechanisms regulating separase are similar to those in mitosis. APC/C and securin are also key regulators in meiosis. Accumulation and degradation of securin is observed in both meiosis I and meiosis II, 173,174 which is the key for the timely activation of separase. At the onset of anaphase, APC/C-dependent cleavage of securin causes loss of its inhibition to separase, which in turn cleaves Rec8 and causes opening of the cohesin ring along chromosome arm. 162 Since in Xenopus oocytes APC/C is supposedly not required for meiosis I, the arm cohesin is removed by a separase-independent mechanism called prophase pathway. 175,176 Nevertheless, it is given serious credence by other results. 159,177 In mouse oocytes, conditional knockout of separase prevents removal of cohesin s Rec8 from chromosome arm and resolution of chiasma. mrna encoding wild-type but not catalytically inactive separase restores chiasma resolution. Thus, proteolytic activity of separase is essential for cohesin s removal from chromosome arms and chiasma resolution. 159 In conclusion, removal of cohesin along chromosome arms triggers segregation of homologs during meiosis I. However, cohesin localized around centromeres is not degraded during meiosis I, allowing sister chromatids to be moved to the same direction. 134,157,160,178 During second meiosis, the centromeric cohesin is removed, thus allowing the sister chromatids to separate. What are the mechanisms responsible for preventing centromeric cohesin from degradation in meiosis I? The following part will address this question. Specific Space-time Protection of Centromeric Cohesins During the First Meiosis Mei-S332/shugoshin. It is not clear how meiotic cells protect centromeric cohesins from separase cleavage in meiosis I but not in meiosis II. It had been suggested that there might be specific spacetime protecting mechanisms of centromeric cohesin in meiosis. Several factors had been proposed as candidates, such as Spo13, Bub1 and heterochromatin. In Saccharomyces cerevisiae, mutant Spo13 introduces defects of centromeric cohesin protection, and overexpression of Spo13 will block degradation of Rec It has been proposed that Spo13 protects meiotic cohesin at centromeres in meiosis I. 180 However, Spo13 is not a centromere-specific protein and no homologue has been found in other species, which leaves the possibility that it might indirectly play protective functions. Studies on Bub1 provided promising results. 181 When Bub1 gene is deleted in Schizosaccharomyces pombe, the cohesin Rec8 is never retained at centromeres at anaphase I and sister chromatids often move to opposite spindle poles during meiosis I. 63 But Bub1 is better known as a spindle checkpoint protein rather than a protector of Rec8. Similarly, pericentromeric heterochromatin is important in recruiting Rec8 in fission yeast, 156 but heterochromatin persists in both meiosis cycles and is not meiosis I-specific. The first break came from Mei-S332, which has been identified in Drosophila melanogaster. 182 The Mei-S332 gene encodes a 44 kda protein, and mutations in Mei-S332 cause premature separation of sister chromatids in late anaphase of meiosis I. 183 It contains two separable functional domains. The carboxy-terminal basic region is required for chromosomal localization and the amino-terminal coiled-coil domain may facilitate protein-protein interactions between MEI-S332 and male meiotic proteins. 184 Mei-S332 disappears from the chromosomes when sister chromatids separate at anaphase II. Therefore, the Mei-S332 protein was proposed to hold the centromere regions of sister chromatids together until anaphase II. 183 Mei-S332 localization is driven by the functional centromeric chromatin, and binding of Mei-S332 is regulated independent of kinetochore formation. 185 As a centromeric protein, Mei-S332 is the most likely candidate which is necessary for the protection of centromeric cohesin in meiosis I. However, the counterparts of Mei-S332 so far have not been identified in other species for over ten years, which obscures its significance. In 2004, the protector of centromeric cohesin Rec8, Shugoshin was first identified in yeast. 186,187 Shugoshin1 localizes at pericentromeric regions but dissociates at the onset of anaphase II. In Shugoshin1-deleted cells, centromeric Rec8 was lost during anaphase I. These data implied that Shugoshuin1 protects centromeric cohesion at anaphase I by safeguarding cohesin Rec8 from separase cleavage. 186,187 As of now, both Shugoshin 1 and 2 have been identified in Schizosaccharomyces pombe, A.thaliana, mouse and human, but only Shugoshin1 in Saccharomyces cerevisiae, C. elegans and N. crassa Notably, all Shugoshins share structural similarity to Mei-S332, for their similarity of the sequence architecture, including conserved N-terminal coiled-coil and C-terminal motif. 186,188,190 Therefore, these proteins define an orthologous family conserved in eukaryotic cells. The localization of Shuhoshin was controlled by Bub1 and Aurora B. 61,62,191,192 Shugoshin is degraded by the anaphasepromoting complex, allowing separation of sister centromeres in anaphase. 193 In meiosis I, the amount of meiotic cohesin subunit Rec8 retained at centromeres is reduced in Sgo1 mutant, but not in Sgo2 mutant cells, and Shugoshin1 appears to regulate cleavage of Rec8 by separase. 188 Depletion of Sgo1 in budding yeast not only causes frequent non-disjunction of homologous chromosomes at meiosis I but also random segregation of sister centromeres at meiosis II. 189 In maize, Shugoshin homolog ZMSgo1 is required for maintenance of centromeric cohesin during meiosis but it does not have mitotic functions. 194 Evidence of Shogoshin as a protector of centromeric cohesin was also reported in mammalian meiosis. In mouse oocytes, both Shogoshin proteins are centromeric proteins. Sister chromatids initiate precocious separation in Sgo2-RNAi oocytes while modest defect was observed in Sgo1-RNAi oocytes. 195 Thus, it is accepted that Shugoshin prevents precocious dissociation of cohesin from centromeres in meiosis I. The mechanism is conserved from yeast to mammals. These findings on Shugoshin protection of centromeric cohesin provide significant insights into the mechanism of faithful segregation of homologous chromosomes and sister chromatids in meiosis. In addition, Shugoshin may also be acting as a tension sensor for the spindle checkpoint. In fission yeast, centromeric Shugosin1 is required to sense lack of tension on mitotic chromosomes. 196 Vertebrate Shugoshin interacts with microtubules in vitro and it regulates kinetochore microtubule stability in vivo, consistent with direct microtubule interactions. 193 RNAi assays of Shugoshin1 Cell Cycle 3001
7 diminished Plk1 kinetochore binding and Shugoshin1 is phosphorylated by Plk1 in vitro. Thus, Shogoshin1 may play a role as tension sensor by regulating Plk1 kinetochore affinity. 197 A novel requirement for Shugoshin1 is to bias sister kinetochores toward bi-orientation in meiosis I. 198 Shugoshin2 is important for chromosome bi-orientation and it controls docking of the Passenger proteins on chromosomes in early mitotic cells. 199 In Schizosaccharomyces pombe, Shugoshin2 interacts with Bir1/Survivin and promotes Aurora kinase complex localization to the pericentromeric region, to correct erroneous attachment of kinetochores, thereby enabling tension-generating attachment. 200 Thus, Shugoshin2 might also be a component of the tension-sensing machinery. 201 The understanding of Shugoshin function gives new insights on spindle checkpoint and chromosome separation. Thus, except for protecting centrometic cohesin, Shugoshin plays a key role in ensuring faithful chromosome segregation in another way, i.e., sensing the absence of tension from sister kinetochores, which could explain Bub1 s dual roles in meiosis. 53,95 Shugoshin/PP2A: dephosphorylation antagonizes phosphorylation. Although it has been recognized that Shugoshin1 closely combines with Rec8 and protects it from separase in anaphase I, the underlying mechanisms are still elusive. The important role of protein phosphatase 2A (PP2A) and dephosphorylation of cohesin was found independently by two labs in ,203 It was demonstrated that Shgoshin1 recruits PP2A to centromeres and inactivation of PP2A causes loss of centromeric cohesin at anaphase I and random segregation of sister centromeres at meiosis II in both budding and fission yeast. 203 It was also shown that PP2A colocalizes with Shugoshin at centromeres and is required for the protection of centermeric cohesin in mitosis and in fission yeast meiosis. 202 Thus, phosphorylation of cohesin mediated by Plk1 kinase is required for degradation of cohesion by separase, while dephosphorylation of cohesin by PP2A is refractory to separase. It seems that the phosphorylation of cohesin is the trigger for separase to cleave cohesin. Shugoshin1 localizes to centromeres at anaphase I but not anaphase II, so PP2A only dephosphrylates cohesin at anaphase I and cohesin resists degradation by separase in anaphase I. Shugoshin1 and PP2A disappear from centromeres before anaphase II, not to antagonize phosphorylation of Plk1 on Rec8, which is then degraded by separase. Thus, sister chromatids can not segregate at anaphase I but only separate at anaphase II. PP2A and Shugoshin1 may complex and interact with each other, since PP2A is also required for centromeric localization of Shugoshin1 during mitosis of human cells. 204 Interestingly, mouse Shugoshin2 is solely responsible for the centromeric localization of PP2A and the protection of cohesin Rec PP2A comprises a group of protein family that is highly abundant and ubiquitously expressed serine/threonine phosphatases in eukaryotes. It is a large and complicated complex with multiple functions, composed of three distinct subunits (A, B and C subunits). The A subunit serves as a scaffold to accommodate the other two subunits as a structural subunit. The C subunit is the catalytic unit, which could combine with the A subunit and form the dimeric core enzyme. 205 In many cases, it is the B regulatory subunit that associates with the core catalytic subunits of PP2A to define the specific functions of PP2A. Multiple subunits allow generation of over 60 different heterotrimeric PP2A holoenzymes. PP2A accounts for the majority of the Ser/Thr phosphatase activity and has been implicated in the regulation of many signaling pathways, such as G 1 /S transition, mitotic entry and exit, cytokinesis, checkpoint, apoptosis. Recruiting PP2A to centromeres to antagonize phosphorylation by Polo kinase is the key protection of centromeric cohesin in anaphase I. Therefore, it seems that the balance of kinase and phosphatase activities is the key to the conserved mechanism that protects centromeric cohesin from separase-mediated cleavage during meiosis I. The Shugoshin1-PP2A model explains the long-standing puzzle, namely why the centromeric cohesin resists separase cleavage specifically during meiosis I, but not during meiosis II or mitosis. 206 Thus, PP2A s role in the control of sister chromatid cohesin, involving a direct interaction with Shugoshin, shows that PP2A is not merely a silent partner to kinases in regulating cell division. 207 Faithful chromosome segregation in meiosis is more complicated than in mitosis. Specific space-time cleavage of cohesins between sister chromatids are fundamental to this process, and any defect in this process will results in aneuploidy. The exact mechanisms require further in-depth exploration. Acknowledgment Q.Y.S. is currently supported by the National Basic Research program of China (2006CB944001, 2006CB504004); CAS program (KSCX2-YW-R-52) and NSFC project ( , ). References 1. Lee JY, Orr-Weaver TL. The molecular basis of sister-chromatid cohesion. Annu Rev Cell Dev Biol 2001; 17: Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 2003; 112: Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2001; 2: Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer 2001; 1: Matsuura S, Ito E, Tauchi H, Komatsu K, Ikeuchi T, Kajii T. Chromosomal instability syndrome of total premature chromatid separation with mosaic variegated aneuploidy is defective in mitotic-spindle checkpoint. Am J Hum Genet 2000; 67: Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy and cancer. Oncogene 2004; 23: Mailhes JB, Young D, London SN. Postovulatory ageing of mouse oocytes in vivo and premature centromere separation and aneuploidy. Biol Reprod 1998; 58: Zenzes MT, Casper RF. Cytogenetics of human oocytes, zygotes and embryos after in vitro fertilization. Hum Genet 1992; 88: Cowchock FS, Gibas Z, Jackson LG. Chromosome errors as a cause of spontaneous abortion: the relative importance of maternal age and obstetric history. Fertil Steril 1993; 59: Hassold T, Abruzzo M, Adkins K, Griffin D, Merrill M, Millie E, et al. Human aneuploidy: incidence, origin and etiology. Environ Mol Mutagen 1996; 28: Koehler KE, Hawley RS, Sherman S, Hassold T. Recombination and nondisjunction in humans and flies. Hum Mol Genet 1996; 5: Hunt PA, Hassold TJ. Sex matters in meiosis. Science 2002; 296: Garcia-Muse T, Boulton SJ. Meiotic recombination in Caenorhabditis elegans. Chromosome Res 2007; 15: Joyce EF, McKim KS. When specialized sites are important for synapsis and the distribution of crossovers. Bioessays 2007; 29: Wells JL, Pryce DW, McFarlane RJ. Homologous chromosome pairing in Schizosaccharomyces pombe. Yeast 2006; 23: Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 2006; 115: Vallente RU, Cheng EY, Hassold TJ. The synaptonemal complex and meiotic recombination in humans: new approaches to old questions. Chromosoma 2006; 115: McKim KS. When size does not matter: pairing sites during meiosis. Cell 2005; 123: Cohen PE, Pollack SE, Pollard JW. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr Rev 2006; 27: Watanabe Y, Kitajima TS. Shugoshin protects cohesin complexes at centromeres. Philos Trans R Soc Lond B Biol Sci 2005; 360: Gregan J, Rumpf C, Li Z, Cipak L. What makes centromeric cohesion resistant to separase cleavage during meiosis I but not during meiosis II? Cell Cycle 2008; 7: Fukagawa T. The kinetochore and spindle checkpoint in vertebrate cells. Front Biosci 2008; 13: Cell Cycle 2008; Vol. 7 Issue 19
8 23. Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 2008; 9: Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem 2007; 76: Gerton JL. Enhancing togetherness: kinetochores and cohesion. Genes Dev 2007; 21: Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell 2000; 103: Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, et al. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell 2003; 4: Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, et al. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol 2003; 5: Hauf S, Biswas A, Langegger M, Kawashima SA, Tsukahara T, Watanabe Y. Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-i. Embo J 2007; 26: Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell 2007; 128: Gregan J, Riedel CG, Pidoux AL, Katou Y, Rumpf C, Schleiffer A, et al. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr Biol 2007; 17: Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, et al. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell 2006; 126: Allshire RC. Centromeres, checkpoints and chromatid cohesion. Curr Opin Genet Dev 1997; 7: Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, et al. Budding yeast Cdc20: a target of the spindle checkpoint. Science 1998; 279: Musacchio A, Hardwick KG. The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol 2002; 3: Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell 1991; 66: Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 1991; 66: Listovsky T, Oren YS, Yudkovsky Y, Mahbubani HM, Weiss AM, Lebendiker M, et al. Mammalian Cdh1/Fzr mediates its own degradation. Embo J 2004; 23: Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J Cell Biol 2002; 158: Murata-Hori M, Wang YL. The kinase activity of aurora B is required for kinetochoremicrotubule interactions during mitosis. Curr Biol 2002; 12: Morrow CJ, Tighe A, Johnson VL, Scott MI, Ditchfield C, Taylor SS. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/C Cdc20. J Cell Sci 2005; 118: Jablonski SA, Chan GK, Cooke CA, Earnshaw WC, Yen TJ. The hbub1 and hbubr1 kinases sequentially assemble onto kinetochores during prophase with hbubr1 concentrating at the kinetochore plates in mitosis. Chromosoma 1998; 107: Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell 2001; 106: Sharp-Baker H, Chen RH. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3 and CENP-E, independently of its kinase activity. J Cell Biol 2001; 153: Vigneron S, Prieto S, Bernis C, Labbe JC, Castro A, Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell 2004; 15: Chen RH. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J Cell Biol 2002; 158: Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 1997; 89: Taylor SS, Hussein D, Wang Y, Elderkin S, Morrow CJ. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J Cell Sci 2001; 114: Wang WH, Sun QY. Meiotic spindle, spindle checkpoint and embryonic aneuploidy. Front Biosci 2006; 11: Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol 2001; 153: Kallio MJ, McCleland ML, Stukenberg PT, Gorbsky GJ. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol 2002; 12: Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2 and Cenp-E to kinetochores. J Cell Biol 2003; 161: Williams GL, Roberts TM, Gjoerup OV. Bub1: escapades in a cellular world. Cell Cycle 2007; 6: Skoufias DA, Andreassen PR, Lacroix FB, Wilson L, Margolis RL. Mammalian mad2 and bub1/bubr1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc Natl Acad Sci USA 2001; 98: Yu H, Tang Z. Bub1 multitasking in mitosis. Cell Cycle 2005; 4: Asakawa H, Kitamura K, Shimoda C. A novel Cdc20-related WD-repeat protein, Fzr1, is required for spore formation in Schizosaccharomyces pombe. Mol Genet Genomics 2001; 265: Logarinho E, Bousbaa H, Dias JM, Lopes C, Amorim I, Antunes-Martins A, et al. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J Cell Sci 2004; 117: Rischitor PE, May KM, Hardwick KG. Bub1 is a fission yeast kinetochore scaffold protein, and is sufficient to recruit other spindle checkpoint proteins to ectopic sites on chromosomes. PLoS ONE 2007; 2: Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci 2004; 117: Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol Cell 2004; 16: Tang Z, Sun Y, Harley SE, Zou H, Yu H. Human Bub1 protects centromeric sisterchromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci USA 2004; 101: Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol 2005; 15: Bernard P, Maure JF, Javerzat JP. Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat Cell Biol 2001; 3: Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell 2003; 14: Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, et al. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. Embo J 1991; 10: Cooke CA, Schaar B, Yen TJ, Earnshaw WC. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma 1997; 106: Wood KW, Sakowicz R, Goldstein LS, Cleveland DW. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell 1997; 91: Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol 1995; 128: Chan GK, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hbubr1. J Cell Biol 1998; 143: Chan GK, Jablonski SA, Sudakin V, Hittle JC, Yen TJ. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/ APC. J Cell Biol 1999; 146: Yao X, Anderson KL, Cleveland DW. The microtubule-dependent motor centromereassociated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J Cell Biol 1997; 139: Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol 2000; 2: Logarinho E, Resende T, Torres C, Bousbaa H. The human spindle assembly checkpoint protein bub3 is required for the establishment of efficient kinetochore-microtubule attachments. Mol Biol Cell 2008; 19: Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20 and MAD2. J Cell Biol 2001; 154: Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. Embo J 2001; 20: Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell 1999; 10: Sironi L, Mapelli M, Knapp S, De Antoni A, Jeang KT, Musacchio A. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a safety belt binding mechanism for the spindle checkpoint. Embo J 2002; 21: Chung E, Chen RH. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol Biol Cell 2002; 13: Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol 1998; 143: Waters JC, Chen RH, Murray AW, Gorbsky GJ, Salmon ED, Nicklas RB. Mad2 binding by phosphorylated kinetochores links error detection and checkpoint action in mitosis. Curr Biol 1999; 9: Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell 2001; 1: Matsumoto T. A fission yeast homolog of CDC20/p55CDC/Fizzy is required for recovery from DNA damage and genetically interacts with p34cdc2. Mol Cell Biol 1997; 17: Cell Cycle 3003
Lecture 10. G1/S Regulation and Cell Cycle Checkpoints. G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint
 Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Cytoskelet Prednáška 6 Mikrotubuly a mitóza
 Cytoskelet Prednáška 6 Mikrotubuly a mitóza Polarity of tubulin polymerization Nuclei Tubulin > C C Preferential addition of tubulin ar (+) ends Tubulin < C C Preferential loss of tubulin ar (+) ends Kinesin
Cytoskelet Prednáška 6 Mikrotubuly a mitóza Polarity of tubulin polymerization Nuclei Tubulin > C C Preferential addition of tubulin ar (+) ends Tubulin < C C Preferential loss of tubulin ar (+) ends Kinesin
Regulators of Cell Cycle Progression
 Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
基醫所. The Cell Cycle. Chi-Wu Chiang, Ph.D. IMM, NCKU
 基醫所 The Cell Cycle Chi-Wu Chiang, Ph.D. IMM, NCKU 1 1 Introduction to cell cycle and cell cycle checkpoints 2 2 Cell cycle A cell reproduces by performing an orderly sequence of events in which it duplicates
基醫所 The Cell Cycle Chi-Wu Chiang, Ph.D. IMM, NCKU 1 1 Introduction to cell cycle and cell cycle checkpoints 2 2 Cell cycle A cell reproduces by performing an orderly sequence of events in which it duplicates
Cell Cycle, Mitosis, and Microtubules. LS1A Final Exam Review Friday 1/12/07. Processes occurring during cell cycle
 Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
The Spindle Assembly Checkpoint and Its Defects in Human Cancer
 Kamla-Raj 2003 Int J Hum Genet, 3(2): 89-97 (2003) The Spindle Assembly Checkpoint and Its Defects in Human Cancer Gourish Mondal and Susanta Roychoudhury Human Genetics and Genomics Division, Indian Institute
Kamla-Raj 2003 Int J Hum Genet, 3(2): 89-97 (2003) The Spindle Assembly Checkpoint and Its Defects in Human Cancer Gourish Mondal and Susanta Roychoudhury Human Genetics and Genomics Division, Indian Institute
NOTHING TO DECLARE. Eugene Pergament, MD, PhD. FACMG. Northwestern Reproductive Genetics, Inc ORIGIN OF ANEUPLOIDY
 NOTHING TO DECLARE Eugene Pergament, MD, PhD. FACMG Northwestern Reproductive Genetics, Inc CHROMOSOME SEGREGATION ERRORS 1-2% 8-9% ~90% Origin of Aneuploidy 1. Cohesin complex 2.Kinetochore-Microtubule
NOTHING TO DECLARE Eugene Pergament, MD, PhD. FACMG Northwestern Reproductive Genetics, Inc CHROMOSOME SEGREGATION ERRORS 1-2% 8-9% ~90% Origin of Aneuploidy 1. Cohesin complex 2.Kinetochore-Microtubule
Cellular Reproduction, Part 2: Meiosis Lecture 10 Fall 2008
 Mitosis & 1 Cellular Reproduction, Part 2: Lecture 10 Fall 2008 Mitosis Form of cell division that leads to identical daughter cells with the full complement of DNA Occurs in somatic cells Cells of body
Mitosis & 1 Cellular Reproduction, Part 2: Lecture 10 Fall 2008 Mitosis Form of cell division that leads to identical daughter cells with the full complement of DNA Occurs in somatic cells Cells of body
Chapter 2. Mitosis and Meiosis
 Chapter 2. Mitosis and Meiosis Chromosome Theory of Heredity What structures within cells correspond to genes? The development of genetics took a major step forward by accepting the notion that the genes
Chapter 2. Mitosis and Meiosis Chromosome Theory of Heredity What structures within cells correspond to genes? The development of genetics took a major step forward by accepting the notion that the genes
Cell cycle and Apoptosis. Chalermchai Mitrpant
 Cell cycle and Apoptosis 2556 Chalermchai Mitrpant Overview of the cell cycle Outline Regulatory mechanisms controlling cell cycle Progression of the cell cycle Checkpoint of the cell cycle Phases of the
Cell cycle and Apoptosis 2556 Chalermchai Mitrpant Overview of the cell cycle Outline Regulatory mechanisms controlling cell cycle Progression of the cell cycle Checkpoint of the cell cycle Phases of the
Chapter 8: Cellular Reproduction
 Chapter 8: Cellular Reproduction 1. The Cell Cycle 2. Mitosis 3. Meiosis 2 Types of Cell Division 2n 1n Mitosis: occurs in somatic cells (almost all cells of the body) generates cells identical to original
Chapter 8: Cellular Reproduction 1. The Cell Cycle 2. Mitosis 3. Meiosis 2 Types of Cell Division 2n 1n Mitosis: occurs in somatic cells (almost all cells of the body) generates cells identical to original
To segregate sister chromatids evenly into daughter cells
 Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis Zhanyun Tang*, Yuxiao Sun, Sara E. Harley*, Hui Zou, and Hongtao Yu* Departments of *Pharmacology and Molecular
Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis Zhanyun Tang*, Yuxiao Sun, Sara E. Harley*, Hui Zou, and Hongtao Yu* Departments of *Pharmacology and Molecular
Regulation of APC Cdc20 by the spindle checkpoint Hongtao Yu
 706 Regulation of AP by the spindle checkpoint Hongtao Yu The spindle checkpoint ensures the fidelity of chromosome segregation in mitosis and meiosis. In response to defects in the mitotic apparatus,
706 Regulation of AP by the spindle checkpoint Hongtao Yu The spindle checkpoint ensures the fidelity of chromosome segregation in mitosis and meiosis. In response to defects in the mitotic apparatus,
How Cells Divide. Chapter 10
 How Cells Divide Chapter 10 Bacterial Cell Division Bacteria divide by binary fission. -the single, circular bacterial chromosome is replicated -replication begins at the origin of replication and proceeds
How Cells Divide Chapter 10 Bacterial Cell Division Bacteria divide by binary fission. -the single, circular bacterial chromosome is replicated -replication begins at the origin of replication and proceeds
Chapter 14. Cell Division
 Chapter 14 Cell Division 14.1. The Cell Cycle A eukaryotic cell cannot divide into two, the two into four, etc. unless two processes alternate: doubling of its genome (DNA) in S phase (synthesis phase)
Chapter 14 Cell Division 14.1. The Cell Cycle A eukaryotic cell cannot divide into two, the two into four, etc. unless two processes alternate: doubling of its genome (DNA) in S phase (synthesis phase)
BIOLOGY 4/6/2015. Cell Cycle - Mitosis. Outline. Overview: The Key Roles of Cell Division. identical daughter cells. I. Overview II.
 2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
Why do cells reproduce?
 Outline Cell Reproduction 1. Overview of Cell Reproduction 2. Cell Reproduction in Prokaryotes 3. Cell Reproduction in Eukaryotes 1. Chromosomes 2. Cell Cycle 3. Mitosis and Cytokinesis Examples of Cell
Outline Cell Reproduction 1. Overview of Cell Reproduction 2. Cell Reproduction in Prokaryotes 3. Cell Reproduction in Eukaryotes 1. Chromosomes 2. Cell Cycle 3. Mitosis and Cytokinesis Examples of Cell
Mitosis and the Cell Cycle
 Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
Mitosis & Meiosis. Diploid cells- (2n)- a cell that has 2 of each chromosome - 1 from mom, 1 from dad = 1 pair
 Mitosis & Meiosis Diploid cells- (2n)- a cell that has 2 of each chromosome - 1 from mom, 1 from dad = 1 pair The pair is called homologous chromosomes The homologous chromosomes contain the same gene
Mitosis & Meiosis Diploid cells- (2n)- a cell that has 2 of each chromosome - 1 from mom, 1 from dad = 1 pair The pair is called homologous chromosomes The homologous chromosomes contain the same gene
Ploidy and Human Cell Types. Cell Cycle and Mitosis. DNA and Chromosomes. Where It All Began 11/19/2014. Chapter 12 Pg
 Ploidy and Human Cell Types Cell Cycle and Mitosis Chapter 12 Pg. 228 245 Cell Types Somatic cells (body cells) have 46 chromosomes, which is the diploid chromosome number. A diploid cell is a cell with
Ploidy and Human Cell Types Cell Cycle and Mitosis Chapter 12 Pg. 228 245 Cell Types Somatic cells (body cells) have 46 chromosomes, which is the diploid chromosome number. A diploid cell is a cell with
Biology is the only subject in which multiplication is the same thing as division
 The Cell Cycle Biology is the only subject in which multiplication is the same thing as division Why do cells divide? For reproduction asexual reproduction For growth one-celled organisms from fertilized
The Cell Cycle Biology is the only subject in which multiplication is the same thing as division Why do cells divide? For reproduction asexual reproduction For growth one-celled organisms from fertilized
Genetics. Instructor: Dr. Jihad Abdallah Lecture 2 The cell cycle and Cell Division
 Genetics Instructor: Dr. Jihad Abdallah Lecture 2 The cell cycle and Cell Division 1 The cell cycle Living cells go through a series of stages known as the cell cycle. They undergo a continuous alternation
Genetics Instructor: Dr. Jihad Abdallah Lecture 2 The cell cycle and Cell Division 1 The cell cycle Living cells go through a series of stages known as the cell cycle. They undergo a continuous alternation
Chapter 8. The Cellular Basis of Reproduction and Inheritance. Lecture by Mary C. Colavito
 Chapter 8 The Cellular Basis of Reproduction and Inheritance PowerPoint Lectures for Biology: Concepts & Connections, Sixth Edition Campbell, Reece, Taylor, Simon, and Dickey Copyright 2009 Pearson Education,
Chapter 8 The Cellular Basis of Reproduction and Inheritance PowerPoint Lectures for Biology: Concepts & Connections, Sixth Edition Campbell, Reece, Taylor, Simon, and Dickey Copyright 2009 Pearson Education,
Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division
 Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division Question No. 1 of 10 1. Which of the following statements about mitosis is correct? Question #1 (A) Mitosis involves the dividing
Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division Question No. 1 of 10 1. Which of the following statements about mitosis is correct? Question #1 (A) Mitosis involves the dividing
Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle. 9.1 Multiple-Choice Questions
 Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle 9.1 Multiple-Choice Questions 1) Starting with a fertilized egg (zygote), a series of five cell divisions would produce an early embryo with how
Campbell Biology in Focus (Urry) Chapter 9 The Cell Cycle 9.1 Multiple-Choice Questions 1) Starting with a fertilized egg (zygote), a series of five cell divisions would produce an early embryo with how
Chapter 10 Chromosomes and Cell Reproduction
 Chapter 10 Chromosomes and Cell Reproduction Chromosomes Organisms grow by dividing of cells Binary Fission form of asexual reproduction that produces identical offspring (Bacteria) Eukaryotes have two
Chapter 10 Chromosomes and Cell Reproduction Chromosomes Organisms grow by dividing of cells Binary Fission form of asexual reproduction that produces identical offspring (Bacteria) Eukaryotes have two
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis
 Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Chromosomes Chromosomes were first observed by the German embryologist Walther Fleming in 1882. Chromosome number varies among organisms most
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Chromosomes Chromosomes were first observed by the German embryologist Walther Fleming in 1882. Chromosome number varies among organisms most
Creating Identical Body Cells
 Creating Identical Body Cells 5.A Students will describe the stages of the cell cycle, including DNA replication and mitosis, and the importance of the cell cycle to the growth of organisms 5.D Students
Creating Identical Body Cells 5.A Students will describe the stages of the cell cycle, including DNA replication and mitosis, and the importance of the cell cycle to the growth of organisms 5.D Students
Cellular Reproduction Chapter 8
 Cellular Reproduction Chapter 8 1. Importance of Cell Division 2. Eukaryotic Cell Cycle 3. Eukaryotic Chromosomes 4. Mitosis 5. Cytokinesis in animal and plant cells 6. Sexual Iife cycle 7. Meiosis 8.
Cellular Reproduction Chapter 8 1. Importance of Cell Division 2. Eukaryotic Cell Cycle 3. Eukaryotic Chromosomes 4. Mitosis 5. Cytokinesis in animal and plant cells 6. Sexual Iife cycle 7. Meiosis 8.
BIOLOGY. Cell Cycle - Mitosis. Outline. Overview: The Key Roles of Cell Division. identical daughter cells. I. Overview II.
 2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
2 Cell Cycle - Mitosis CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson Outline I. Overview II. Mitotic Phase I. Prophase II. III. Telophase IV. Cytokinesis III. Binary fission
Mitosis THE CELL CYCLE. In unicellular organisms, division of one cell reproduces the entire organism Multicellular organisms use cell division for..
 Mitosis THE CELL CYCLE In unicellular organisms, division of one cell reproduces the entire organism Multicellular organisms use cell division for.. Development from a fertilized cell Growth Repair Cell
Mitosis THE CELL CYCLE In unicellular organisms, division of one cell reproduces the entire organism Multicellular organisms use cell division for.. Development from a fertilized cell Growth Repair Cell
Origin of replication. Septum
 Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Chromatin Loop Solenoid Scaffold protein Scaffold protein Chromatin loop
Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Chromatin Loop Solenoid Scaffold protein Scaffold protein Chromatin loop
Bacterial cell. Origin of replication. Septum
 Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Scaffold protein Chromatin Loop Solenoid Scaffold protein Chromatin loop
Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Scaffold protein Chromatin Loop Solenoid Scaffold protein Chromatin loop
Cell Division and Mitosis
 Chromatin-Uncoiled DNA during interphase Cell Division and Mitosis Chromosomes-Tightly coiled DNA Chromatid-One half of a duplicated chromosome. Each is identical and called sister chromatids Centromere-The
Chromatin-Uncoiled DNA during interphase Cell Division and Mitosis Chromosomes-Tightly coiled DNA Chromatid-One half of a duplicated chromosome. Each is identical and called sister chromatids Centromere-The
Genes and Proteins. Key points: The DNA must be copied and then divided exactly so that each cell gets an identical copy.
 Mitosis Genes and Proteins Proteins do the work of the cell: growth, maintenance, response to the environment, reproduction, etc. Proteins are chains of amino acids. The sequence of amino acids in each
Mitosis Genes and Proteins Proteins do the work of the cell: growth, maintenance, response to the environment, reproduction, etc. Proteins are chains of amino acids. The sequence of amino acids in each
How deregulated cell cycles (might) trigger cancer. Dan Fisher. Friday 27th October Lucie Fisher, 3 Eddie Fisher, 6.
 How deregulated cell cycles (might) trigger cancer http://www.igmm.cnrs.fr Dan Fisher Friday 27th October 2017 Lucie Fisher, 3 Eddie Fisher, 6 1 transformed cells transformed cells Structure of this lecture
How deregulated cell cycles (might) trigger cancer http://www.igmm.cnrs.fr Dan Fisher Friday 27th October 2017 Lucie Fisher, 3 Eddie Fisher, 6 1 transformed cells transformed cells Structure of this lecture
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis
 Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Prokaryotes Have a Simpler Cell Cycle Cell division in prokaryotes takes place in two stages, which together make up a simple cell cycle 1. Copy
Breaking Up is Hard to Do (At Least in Eukaryotes) Mitosis Prokaryotes Have a Simpler Cell Cycle Cell division in prokaryotes takes place in two stages, which together make up a simple cell cycle 1. Copy
NOTES- CHAPTER 6 CHROMOSOMES AND CELL REPRODUCTION
 NOTES- CHAPTER 6 CHROMOSOMES AND CELL REPRODUCTION Section I Chromosomes Formation of New Cells by Cell Division New cells are formed when old cells divide. 1. Cell division is the same as cell reproduction.
NOTES- CHAPTER 6 CHROMOSOMES AND CELL REPRODUCTION Section I Chromosomes Formation of New Cells by Cell Division New cells are formed when old cells divide. 1. Cell division is the same as cell reproduction.
Chapter 12. The Cell Cycle
 Chapter 12 The Cell Cycle The Key Roles of Cell Division The ability of organisms to produce more of their own kind is the one characteristic that best distinguishes living things from nonliving things.
Chapter 12 The Cell Cycle The Key Roles of Cell Division The ability of organisms to produce more of their own kind is the one characteristic that best distinguishes living things from nonliving things.
Cell cycle and apoptosis
 Cell cycle and apoptosis Cell cycle Definition Stages and steps Cell cycle Interphase (G1/G0, S, and G2) Mitosis (prophase, metaphase, anaphase, telophase, karyokinesis, cytokinesis) Control checkpoints
Cell cycle and apoptosis Cell cycle Definition Stages and steps Cell cycle Interphase (G1/G0, S, and G2) Mitosis (prophase, metaphase, anaphase, telophase, karyokinesis, cytokinesis) Control checkpoints
meiosis asexual reproduction CHAPTER 9 & 10 The Cell Cycle, Meiosis & Sexual Life Cycles Sexual reproduction mitosis
 meiosis asexual reproduction CHAPTER 9 & 10 The Cell Cycle, Meiosis & Sexual Sexual reproduction Life Cycles mitosis Chromosomes Consists of a long DNA molecule (represents thousands of genes) Also consists
meiosis asexual reproduction CHAPTER 9 & 10 The Cell Cycle, Meiosis & Sexual Sexual reproduction Life Cycles mitosis Chromosomes Consists of a long DNA molecule (represents thousands of genes) Also consists
The Cell Cycle CHAPTER 12
 The Cell Cycle CHAPTER 12 The Key Roles of Cell Division cell division = reproduction of cells All cells come from pre-exisiting cells Omnis cellula e cellula Unicellular organisms division of 1 cell reproduces
The Cell Cycle CHAPTER 12 The Key Roles of Cell Division cell division = reproduction of cells All cells come from pre-exisiting cells Omnis cellula e cellula Unicellular organisms division of 1 cell reproduces
BIOLOGY - CLUTCH CH.12 - CELL DIVISION.
 !! www.clutchprep.com CONCEPT: CELL DIVISION Cell division is the process by which one cell splits into two or more daughter cells. Cell division generally requires that cells produce enough materials,
!! www.clutchprep.com CONCEPT: CELL DIVISION Cell division is the process by which one cell splits into two or more daughter cells. Cell division generally requires that cells produce enough materials,
Cell division functions in 1. reproduction, 2. growth, and 3. repair
 Cell division functions in 1. reproduction, 2. growth, and 3. repair What do you think you are looking at here??? Can something like you or I do this??? Fig. 12.1 How did you start out? How did you grow?
Cell division functions in 1. reproduction, 2. growth, and 3. repair What do you think you are looking at here??? Can something like you or I do this??? Fig. 12.1 How did you start out? How did you grow?
The spindle assembly checkpoint: Preventing chromosome mis-segregation during mitosis and meiosis
 FEBS Letters 580 (2006) 2888 2895 Minireview The spindle assembly checkpoint: Preventing chromosome mis-segregation during mitosis and meiosis Nicolas Malmanche a, André Maia a, Claudio E. Sunkel a,b,
FEBS Letters 580 (2006) 2888 2895 Minireview The spindle assembly checkpoint: Preventing chromosome mis-segregation during mitosis and meiosis Nicolas Malmanche a, André Maia a, Claudio E. Sunkel a,b,
Meiosis. Oh, and a little bit of mitosis
 Meiosis Oh, and a little bit of mitosis Haploid Cells- The sex cells (egg and sperm) only contain half of the genetic diversity that diploid cells do. For humans this would mean 23 single chromosomes.
Meiosis Oh, and a little bit of mitosis Haploid Cells- The sex cells (egg and sperm) only contain half of the genetic diversity that diploid cells do. For humans this would mean 23 single chromosomes.
The Cell Cycle and How Cells Divide
 The Cell Cycle and How Cells Divide 1 Phases of the Cell Cycle The cell cycle consists of Interphase normal cell activity The mitotic phase cell divsion INTERPHASE Growth G 1 (DNA synthesis) Growth G 2
The Cell Cycle and How Cells Divide 1 Phases of the Cell Cycle The cell cycle consists of Interphase normal cell activity The mitotic phase cell divsion INTERPHASE Growth G 1 (DNA synthesis) Growth G 2
Mitosis/Meiosis Simulation Activities
 Mitosis/Meiosis Simulation Activities In this simulation, you will demonstrate an understanding of mitosis, meiosis, segregation, independent assortment, and crossing over, all processes involved with
Mitosis/Meiosis Simulation Activities In this simulation, you will demonstrate an understanding of mitosis, meiosis, segregation, independent assortment, and crossing over, all processes involved with
Cell Cycle and Cell Division
 122 Cell Cycle and Cell Division 1. Meiosis I is reductional division. Meiosis II is equational division due to [1988] (a) pairing of homologous chromosomes (b) crossing over (c) separation of chromatids
122 Cell Cycle and Cell Division 1. Meiosis I is reductional division. Meiosis II is equational division due to [1988] (a) pairing of homologous chromosomes (b) crossing over (c) separation of chromatids
Chapter 12. living /non-living? growth repair renew. Reproduction. Reproduction. living /non-living. fertilized egg (zygote) next chapter
 Chapter 12 How cells divide Reproduction living /non-living? growth repair renew based on cell division first mitosis - distributes identical sets of chromosomes cell cycle (life) Cell Division in Bacteria
Chapter 12 How cells divide Reproduction living /non-living? growth repair renew based on cell division first mitosis - distributes identical sets of chromosomes cell cycle (life) Cell Division in Bacteria
Molecular Cell Biology. Prof. D. Karunagaran. Department of Biotechnology. Indian Institute of Technology Madras
 Molecular Cell Biology Prof. D. Karunagaran Department of Biotechnology Indian Institute of Technology Madras Module 6 Cell Division, Cell Cycle, Cell Growth and Differentiation, Programmed Cell Death
Molecular Cell Biology Prof. D. Karunagaran Department of Biotechnology Indian Institute of Technology Madras Module 6 Cell Division, Cell Cycle, Cell Growth and Differentiation, Programmed Cell Death
BIOLOGY. The Cell Cycle CAMPBELL. Reece Urry Cain Wasserman Minorsky Jackson. Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick
 CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 12 The Cell Cycle Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick The Key Roles of Cell Division The ability
CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 12 The Cell Cycle Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick The Key Roles of Cell Division The ability
CH 9: The Cell Cycle Overview. Cellular Organization of the Genetic Material. Distribution of Chromosomes During Eukaryotic Cell Division
 CH 9: The Cell Cycle Overview The ability of organisms to produce more of their own kind best distinguishes living things from nonliving matter The continuity of life is based on the reproduction of cells,
CH 9: The Cell Cycle Overview The ability of organisms to produce more of their own kind best distinguishes living things from nonliving matter The continuity of life is based on the reproduction of cells,
Outline Interphase Mitotic Stage Cell Cycle Control Apoptosis Mitosis Mitosis in Animal Cells Cytokinesis Cancer Prokaryotic Cell Division
 The Cell Cycle and Cellular Reproduction Chapter 9 Outline Interphase Mitotic Stage Cell Cycle Control Apoptosis Mitosis Mitosis in Animal Cells Cytokinesis Cancer Prokaryotic Cell Division 1 2 Interphase
The Cell Cycle and Cellular Reproduction Chapter 9 Outline Interphase Mitotic Stage Cell Cycle Control Apoptosis Mitosis Mitosis in Animal Cells Cytokinesis Cancer Prokaryotic Cell Division 1 2 Interphase
Chapt 15: Molecular Genetics of Cell Cycle and Cancer
 Chapt 15: Molecular Genetics of Cell Cycle and Cancer Student Learning Outcomes: Describe the cell cycle: steps taken by a cell to duplicate itself = cell division; Interphase (G1, S and G2), Mitosis.
Chapt 15: Molecular Genetics of Cell Cycle and Cancer Student Learning Outcomes: Describe the cell cycle: steps taken by a cell to duplicate itself = cell division; Interphase (G1, S and G2), Mitosis.
The questions below refer to the following terms. Each term may be used once, more than once, or not at all.
 The questions below refer to the following terms. Each term may be used once, more than once, or not at all. a) telophase b) anaphase c) prometaphase d) metaphase e) prophase 1) DNA begins to coil and
The questions below refer to the following terms. Each term may be used once, more than once, or not at all. a) telophase b) anaphase c) prometaphase d) metaphase e) prophase 1) DNA begins to coil and
The Cell Cycle CAMPBELL BIOLOGY IN FOCUS SECOND EDITION URRY CAIN WASSERMAN MINORSKY REECE
 CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 9 The Cell Cycle Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION Overview: The Key
CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 9 The Cell Cycle Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION Overview: The Key
2014 Pearson Education, Inc.
 2 The Cell Cycle CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson The Key Roles of Cell Division The ability of organisms to produce more of their own kind best distinguishes living
2 The Cell Cycle CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson The Key Roles of Cell Division The ability of organisms to produce more of their own kind best distinguishes living
Chapter 8 The Cell Cycle
 What molecule stores your genetic information or determines everything about you? DNA a nucleic acid How are DNA molecules arranged in the nucleus? As you can see DNA is: Chapter 8 The Cell Cycle 1. Arranged
What molecule stores your genetic information or determines everything about you? DNA a nucleic acid How are DNA molecules arranged in the nucleus? As you can see DNA is: Chapter 8 The Cell Cycle 1. Arranged
Why do cells divide? Cells divide in order to make more cells they multiply in order to create a larger surface to volume ratio!!!
 Why do cells divide? Cells divide in order to make more cells they multiply in order to create a larger surface to volume ratio!!! Chromosomes Are made of chromatin: a mass of genetic material composed
Why do cells divide? Cells divide in order to make more cells they multiply in order to create a larger surface to volume ratio!!! Chromosomes Are made of chromatin: a mass of genetic material composed
Cell cycle control (mammalian)
 Apr. 21, 2005 Cell cycle control (mammalian) Basic mechanisms & protein components Checkpoints Chap. 21, by Lodish et al., 5 th ed. 2004 Chap. 17, by Alberts et al., 4 th ed. 2002 鍾明怡 mychung@vghtpe.gov.tw
Apr. 21, 2005 Cell cycle control (mammalian) Basic mechanisms & protein components Checkpoints Chap. 21, by Lodish et al., 5 th ed. 2004 Chap. 17, by Alberts et al., 4 th ed. 2002 鍾明怡 mychung@vghtpe.gov.tw
The Cell Cycle. Chapter 12. Biology Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 12 The Cell Cycle PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
Chapter 12 The Cell Cycle PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
LECTURE PRESENTATIONS
 LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
Reproduction is a fundamental property of life. Cells are the fundamental unit of life. Reproduction occurs at the cellular level with one mother
 Cell Division ision Reproduction is a fundamental property of life. Cells are the fundamental unit of life. Reproduction occurs at the cellular level with one mother cell giving rise to two daughter cells.
Cell Division ision Reproduction is a fundamental property of life. Cells are the fundamental unit of life. Reproduction occurs at the cellular level with one mother cell giving rise to two daughter cells.
Cell Division. Chromosome structure. Made of chromatin (mix of DNA and protein) Only visible during cell division
 Chromosome structure Made of chromatin (mix of DNA and protein) Only visible during cell division Chromosome structure The DNA in a cell is packed into an elaborate, multilevel system of coiling and folding.
Chromosome structure Made of chromatin (mix of DNA and protein) Only visible during cell division Chromosome structure The DNA in a cell is packed into an elaborate, multilevel system of coiling and folding.
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
General Embryology. School of Medicine Department of Anatomy and Histology School of medicine The University of Jordan
 General Embryology 2019 School of Medicine Department of Anatomy and Histology School of medicine The University of Jordan https://www.facebook.com/dramjad-shatarat What is embryology? Is the science that
General Embryology 2019 School of Medicine Department of Anatomy and Histology School of medicine The University of Jordan https://www.facebook.com/dramjad-shatarat What is embryology? Is the science that
Mitosis. AND Cell DiVISION
 Mitosis AND Cell DiVISION Cell Division Characteristic of living things: ability to reproduce their own kind. Cell division purpose: When unicellular organisms such as amoeba divide to form offspring reproduction
Mitosis AND Cell DiVISION Cell Division Characteristic of living things: ability to reproduce their own kind. Cell division purpose: When unicellular organisms such as amoeba divide to form offspring reproduction
The Cell Cycle 4/10/12. Chapter 12. Overview: The Key Roles of Cell Division
 LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
Chapter 12 The Cell Cycle
 Chapter 12 The Cell Cycle Objectives Describe how cell reproduction contributes to repair and growth. Compare and contrast prokaryotic and eukaryotic cell division. Compare and contrast asexual and sexual
Chapter 12 The Cell Cycle Objectives Describe how cell reproduction contributes to repair and growth. Compare and contrast prokaryotic and eukaryotic cell division. Compare and contrast asexual and sexual
Genetics and Cellular Function
 Genetics and Cellular Function DNA replication and the cell cycle Mitosis Mitosis Mitosis: division of cells that results in daughter cells with the same the genetic information that the original cell
Genetics and Cellular Function DNA replication and the cell cycle Mitosis Mitosis Mitosis: division of cells that results in daughter cells with the same the genetic information that the original cell
The metaphase to anaphase transition
 Eur. J. Biochem. 263, 14±19 (1999) q FEBS 1999 MINIREVIEW The metaphase to anaphase transition A case of productive destruction Katie A. Farr and Orna Cohen-Fix The Laboratory of Molecular and Cellular
Eur. J. Biochem. 263, 14±19 (1999) q FEBS 1999 MINIREVIEW The metaphase to anaphase transition A case of productive destruction Katie A. Farr and Orna Cohen-Fix The Laboratory of Molecular and Cellular
University of Bristol - Explore Bristol Research
 Shandilya, J., Medler, K., & Roberts, S. G. E. (2016). Regulation of AURORA B function by mitotic checkpoint protein MAD2. Cell Cycle, 15(16), 2196-2201. https://doi.org/10.1080/15384101.2016.1200773 Peer
Shandilya, J., Medler, K., & Roberts, S. G. E. (2016). Regulation of AURORA B function by mitotic checkpoint protein MAD2. Cell Cycle, 15(16), 2196-2201. https://doi.org/10.1080/15384101.2016.1200773 Peer
The Cell Cycle. Dr. SARRAY Sameh, Ph.D
 The Cell Cycle Dr. SARRAY Sameh, Ph.D Overview When an organism requires additional cells (either for growth or replacement of lost cells), new cells are produced by cell division (mitosis) Somatic cells
The Cell Cycle Dr. SARRAY Sameh, Ph.D Overview When an organism requires additional cells (either for growth or replacement of lost cells), new cells are produced by cell division (mitosis) Somatic cells
-The cell s hereditary endowment of DNA -Usually packaged into chromosomes for manageability
 Binary Fission-Bacterial Cell Division -Asexual reproduction of prokaryotes -No mitosis -Circular DNA and organelles replicate, the copies migrate to opposite sides of the elongating cell, and the cell
Binary Fission-Bacterial Cell Division -Asexual reproduction of prokaryotes -No mitosis -Circular DNA and organelles replicate, the copies migrate to opposite sides of the elongating cell, and the cell
3/19/17. Chromosomes. Chromosome Structure. Chromosome Structure. Chromosome Structure. Chapter 10: Cell Growth & Division
 Chapter 10: Cell Growth & Division Section 2: The Process of Cell Division Chromosomes DNA (deoxyribonucleic acid) a molecule that stores genetic information, which controls the development and functioning
Chapter 10: Cell Growth & Division Section 2: The Process of Cell Division Chromosomes DNA (deoxyribonucleic acid) a molecule that stores genetic information, which controls the development and functioning
Major concepts: Notes: Cell Reproduction: From One Cell to Two. Why do Cells Reproduce?
 Grade 7 Standard: Life Science 1e Students know cells divide to increase their numbers through a process of mitosis, which results in two daughter cells with identical sets of chromosomes. Major concepts:
Grade 7 Standard: Life Science 1e Students know cells divide to increase their numbers through a process of mitosis, which results in two daughter cells with identical sets of chromosomes. Major concepts:
The Cell Cycle. Packet #9. Thursday, August 20, 2015
 1 The Cell Cycle Packet #9 2 Introduction Cell Cycle An ordered sequence of events in the life of a dividing eukaryotic cell and is a cellular asexual reproduction. The contents of the parent s cell nucleus
1 The Cell Cycle Packet #9 2 Introduction Cell Cycle An ordered sequence of events in the life of a dividing eukaryotic cell and is a cellular asexual reproduction. The contents of the parent s cell nucleus
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
The Cell Cycle. Chapter 12. PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece
 Chapter 12 The Cell Cycle PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Overview:
Chapter 12 The Cell Cycle PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Overview:
Organisms that reproduce Sexually are made up of two different types of cells.
 MEIOSIS Organisms that reproduce Sexually are made up of two different types of cells. 1. Somatic Cells are body cells and contain the normal number of chromosomes.called the Diploid number (the symbol
MEIOSIS Organisms that reproduce Sexually are made up of two different types of cells. 1. Somatic Cells are body cells and contain the normal number of chromosomes.called the Diploid number (the symbol
LECTURE PRESENTATIONS
 LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
MITOSIS AND THE CELL CYCLE PowerPoint Notes
 1 Name: Date: MITOSIS AND THE CELL CYCLE PowerPoint Notes THE FUNCTIONS OF CELL DIVISION 1. Cell division is vital for all. living organisms This is the only process that can create. new cells 2. Cell
1 Name: Date: MITOSIS AND THE CELL CYCLE PowerPoint Notes THE FUNCTIONS OF CELL DIVISION 1. Cell division is vital for all. living organisms This is the only process that can create. new cells 2. Cell
Why do cells divide? The Cell Cycle: Cell Growth, Cell Division. Making new cells. Getting the right stuff. Overview of mitosis 1/5/2015
 Why do cells divide? The Cell Cycle: Cell Growth, Cell Division For reproduction asexual reproduction one-celled organisms For growth from fertilized egg to multi-celled organism For repair & renewal replace
Why do cells divide? The Cell Cycle: Cell Growth, Cell Division For reproduction asexual reproduction one-celled organisms For growth from fertilized egg to multi-celled organism For repair & renewal replace
Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008
 Cell Theory 1 Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008 Cell theory: All organisms are made of cells All cells arise from preexisting cells How do new cells arise? Cell division the reproduction
Cell Theory 1 Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008 Cell theory: All organisms are made of cells All cells arise from preexisting cells How do new cells arise? Cell division the reproduction
Chapter 12 The Cell Cycle: Cell Growth, Cell Division
 Chapter 12 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a period at the end of a sentence And now look at you How did you get from there to
Chapter 12 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a period at the end of a sentence And now look at you How did you get from there to
The spindle checkpoint and chromosome segregation in meiosis
 REVIEW The spindle checkpoint and chromosome segregation in meiosis Gary J. Gorbsky Cell Cycle & Cancer Biology, Oklahoma Medical Research Foundation, OK, USA Keywords anaphase-promoting complex, aneuploidy,
REVIEW The spindle checkpoint and chromosome segregation in meiosis Gary J. Gorbsky Cell Cycle & Cancer Biology, Oklahoma Medical Research Foundation, OK, USA Keywords anaphase-promoting complex, aneuploidy,
LECTURE PRESENTATIONS
 LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
LECTURE PRESENTATIONS For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Chapter 12 The Cell Cycle Lectures by Erin
Chapter 10 How Cell Divide
 Chapter 10 How Cell Divide Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. **Important study hints** You get the idea by now!! http://www.hercampus.com/school/wisconsin/it-s-finals-time-5-ways-prepare-week-we-re-all-dreading
Chapter 10 How Cell Divide Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. **Important study hints** You get the idea by now!! http://www.hercampus.com/school/wisconsin/it-s-finals-time-5-ways-prepare-week-we-re-all-dreading
Molecular causes of aneuploidy in mammalian eggs
 PRIMER 3719 Development 140, 3719-3730 (2013) doi:10.1242/dev.090589 2013. Published by The Company of Biologists Ltd Molecular causes of aneuploidy in mammalian eggs Keith T. Jones* and Simon I. R. Lane
PRIMER 3719 Development 140, 3719-3730 (2013) doi:10.1242/dev.090589 2013. Published by The Company of Biologists Ltd Molecular causes of aneuploidy in mammalian eggs Keith T. Jones* and Simon I. R. Lane
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
Mitosis vs. Meiosis. The Somatic Cell Cycle (Mitosis) The somatic cell cycle consists of 3 phases: interphase, m phase, and cytokinesis.
 Mitosis vs. Meiosis In order for organisms to continue growing and/or replace cells that are dead or beyond repair, cells must replicate, or make identical copies of themselves. In order to do this and
Mitosis vs. Meiosis In order for organisms to continue growing and/or replace cells that are dead or beyond repair, cells must replicate, or make identical copies of themselves. In order to do this and
DAPI ASY1 DAPI/ASY1 DAPI RAD51 DAPI/RAD51. Supplementary Figure 1. Additional information on meiosis in R. pubera. a) The
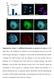 a % 10 Number of crossover per bivalent b 0 1 c DAPI/telomere 80 1 60 40 1 2 20 d 0 0 1 2 >=3 DAPI ASY1 DAPI/ASY1 e DAPI RAD51 DAPI/RAD51 Supplementary Figure 1. Additional information on meiosis in R.
a % 10 Number of crossover per bivalent b 0 1 c DAPI/telomere 80 1 60 40 1 2 20 d 0 0 1 2 >=3 DAPI ASY1 DAPI/ASY1 e DAPI RAD51 DAPI/RAD51 Supplementary Figure 1. Additional information on meiosis in R.
The Cell Cycle. Chapter 12. Biology. Edited by Shawn Lester. Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 12 The Cell Cycle Edited by Shawn Lester PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions
Chapter 12 The Cell Cycle Edited by Shawn Lester PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions
The Cell Cycle. Chapter 10
 The Cell Cycle Chapter 10 Why Do Cells Divide? Unicellular 1. Reproduction Multicellular 1. Grow 2. Repair 3. Development/reproduction Types of Division Prokaryotic cells Binary fission = asexual reproduction
The Cell Cycle Chapter 10 Why Do Cells Divide? Unicellular 1. Reproduction Multicellular 1. Grow 2. Repair 3. Development/reproduction Types of Division Prokaryotic cells Binary fission = asexual reproduction
Name. A.P. Biology Chapter 12 The Cell Cycle
 A.P. Biology Chapter 12 The Cell Cycle Name Living species MUST possess the ability to r if they are to flourish. The Cell Cycle follows the life of a cell from its o until its d. The Key Roles Of Cell
A.P. Biology Chapter 12 The Cell Cycle Name Living species MUST possess the ability to r if they are to flourish. The Cell Cycle follows the life of a cell from its o until its d. The Key Roles Of Cell
Chromosomes Can Congress To The Metaphase Plate Prior. To Bi-Orientation
 Chromosomes Can Congress To The Metaphase Plate Prior To Bi-Orientation Tarun M. Kapoor 1,2, Michael Lampson 1, Polla Hergert 3, Lisa Cameron 2,4, Daniela Cimini 4, E.D. Salmon 2,4, Bruce F. McEwen 3,
Chromosomes Can Congress To The Metaphase Plate Prior To Bi-Orientation Tarun M. Kapoor 1,2, Michael Lampson 1, Polla Hergert 3, Lisa Cameron 2,4, Daniela Cimini 4, E.D. Salmon 2,4, Bruce F. McEwen 3,
PowerPoint Image Slideshow
 COLLEGE BIOLOGY PHYSICS Chapter 10 # Cell Chapter Reproduction Title PowerPoint Image Slideshow CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 12 The Cell Cycle 2014 Pearson
COLLEGE BIOLOGY PHYSICS Chapter 10 # Cell Chapter Reproduction Title PowerPoint Image Slideshow CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 12 The Cell Cycle 2014 Pearson
The form of cell division by which gametes, with half the number of chromosomes, are produced. Chromosomes
 & Karyotypes The form of cell division by which gametes, with half the number of chromosomes, are produced. Homologous Chromosomes Pair of chromosomes (maternal and paternal) that are similar in shape,
& Karyotypes The form of cell division by which gametes, with half the number of chromosomes, are produced. Homologous Chromosomes Pair of chromosomes (maternal and paternal) that are similar in shape,
