|
|
|
- Jennifer Greer
- 5 years ago
- Views:
Transcription
1 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Scope The Atlas of Genetics and Cytogenetics in Oncology and Haematology is a peer reviewed on-line journal in open access, devoted to genes, cytogenetics, and clinical entities in cancer, and cancer-prone diseases. It presents structured review articles ( cards ) on genes, leukaemias, solid tumours, cancer-prone diseases, and also more traditional review articles ( deep insights ) on the above subjects and on surrounding topics. It also present case reports in hematology and educational items in the various related topics for students in Medicine and in Sciences. Editorial correspondance Jean-Loup Huret Genetics, Department of Medical Information, University Hospital F Poitiers, France tel or jlhuret@atlasgeneticsoncology.org or Editorial@AtlasGeneticsOncology.org The Atlas of Genetics and Cytogenetics in Oncology and Haematology is published 4 times a year by ARMGHM, a non profit organisation. Philippe Dessen is the Database Director, and Alain Bernheim the Chairman of the on-line version (Gustave Roussy Institute Villejuif France). ATLAS - ISSN Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 95
2 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Scope The Atlas of Genetics and Cytogenetics in Oncology and Haematology is a peer reviewed on-line journal in open access, devoted to genes, cytogenetics, and clinical entities in cancer, and cancer-prone diseases. It presents structured review articles ( cards ) on genes, leukaemias, solid tumours, cancer-prone diseases, and also more traditional review articles ( deep insights ) on the above subjects and on surrounding topics. It also present case reports in hematology and educational items in the various related topics for students in Medicine and in Sciences. Editorial correspondance Jean-Loup Huret Genetics, Department of Medical Information, University Hospital F Poitiers, France tel or jlhuret@atlasgeneticsoncology.org or Editorial@AtlasGeneticsOncology.org The Atlas of Genetics and Cytogenetics in Oncology and Haematology is published 4 times a year by ARMGHM, a non profit organisation. Philippe Dessen is the Database Director, and Alain Bernheim the Chairman of the on-line version (Gustave Roussy Institute Villejuif France). ATLAS - ISSN The PDF version of the Atlas of Genetics and Cytogenetics in Oncology and Haematology is a reissue of the original articles published in collaboration with the Institute for Scientific and Technical Information (INstitut de l Information Scientifique et Technique - INIST) of the French National Center for Scientific Research (CNRS) on its electronic publishing platform I-Revues. Online and PDF versions of the Atlas of Genetics and Cytogenetics in Oncology and Haematology are hosted by INIST-CNRS.
3 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Editor Jean-Loup Huret (Poitiers, France) Volume 4, Number 3, July - September 2000 Table of contents Gene Section GAS7 (growth arrest-specific 7) 95 Jean-Loup Huret KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog) 96 Lidia Larizza, Alessandro Beghini KITLG (KIT ligand) 99 Lidia Larizza, Alessandro Beghini CBP (CREB-binding protein) 101 Jean-Loup Huret CIITA (MHC class II transactivator) 103 Jean-Loup Huret EIF4A2 (eukaryotic translation initiation factor 4A, isoform 2) 105 Jean-Loup Huret IGH (Immunoglobulin Heavy) 107 Marie-Paule Lefranc IGK (Immunoglobulin Kappa) 111 Marie-Paule Lefranc IGL (Immunoglobulin Lambda) 114 Marie-Paule Lefranc TRA (T cell Receptor Alpha) 117 Marie-Paule Lefranc TRB (T cell Receptor Beta) 119 Marie-Paule Lefranc TRD (T cell Receptor Delta) 121 Marie-Paule Lefranc TRG (T cell Receptor Gamma) 123 Marie-Paule Lefranc Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3)
4 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Leukaemia Section Systemic mast cell disease (SMCD) 125 Lidia Larizza, Alessandro Beghini t(11;17)(q23;p13) 127 Jean-Loup Huret t(11;17)(q23;q12) MLL/RARa 128 Jean-Loup Huret +4 or trisomy Alessandro Beghini 3q27 rearrangements in non Hodgkin lymphoma, t(3;var)(q27;var) in non Hodgkin lymphoma 131 Antonio Cuneo, Jean-Loup Huret Solid Tumour Section Soft tissue tumors: Lipoma / benign lipomatous tumors 135 Nils Mandahl Soft tissue tumors: Liposarcoma / malignant lipomatous tumors 138 Nils Mandahl Uterus: Carcinoma of the cervix 142 Niels B Atkin Sot tissue tumors: Extraskeletal myxoid chondrosarcoma 145 Jérome Couturier Nervous system: Medulloblastoma 147 Anne Marie Capodano Nervous system: Meningioma 149 Anne Marie Capodano Cancer Prone Disease Section Cowden disease 153 Michel Longy Familial /sporadic gastrointestinal stromal tumors (GISTs) 155 Lidia Larizza, Alessandro Beghini Piebaldism 157 Lidia Larizza, Alessandro Beghini Educational Items Section Chromosomes, Chromosome Anomalies 159 Jean-Loup Huret, Claude Leonard, John RK Savage Malignant blood diseases 174 Jean-Loup Huret Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3)
5 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Short Communication GAS7 (growth arrest-specific 7) Jean-Loup Huret Genetics, Dept Medical Information, University of Poitiers, CHU Poitiers Hospital, F Poitiers, France (JLH) Published in Atlas Database: June 2000 Online updated version : DOI: /2042/37631 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Other names: KIAA0394 HGNC (Hugo): GAS7 Location: 17p13 Probe(s) - Courtesy Mariano Rocchi, Resources for Molecular Cytogenetics. DNA/RNA Description Genomic sequence 167 kb; 14 exons. Transcription 8 kb mrna; various splicings in the mouse. Protein Description 412 aa. Expression In quiescent/terminally differenciated cells, like other GAS (herien the name Growth Arrest-Specific); selective expression in growth-arrested fibroblasts; GAS7 is predominantly expressed in the brain (in particular, in the mouse, in Purkinje cells of the cerebellum, at lower level in other tissues such as the heart and the testes); gas7 expression is associated with neurite formation (and overproduction of gas7 in neuroblastoma cells promotes neurite-like outgrowth) which suggests that GAS7 may have a role in promoting, and possibly maintaining, maturation and morphological differentiation of cerebellar neurons. Implicated in Treatment-related acute non lymphocytic leukemia with t(11;17)(q23;p13) Disease Only one case: a 13-year-old boy, with a M4 ANLL and a karyotype with an apparent del(11)(q23) (finally proved to be in reality a cryptic t(11;17)(q23;p13)); four months later the patient died; other cases of t(11;17)(q23;p13) have been described but the involvement of GAS7 has not been proved. Hybrid/Mutated gene 5' MLL-3' GAS7; breakpoint in MLL intron 8, in MLL bcr; GAS7 breakpoint upstream of exon 1; fusion of MLL exon 7 or exon 8 to GAS7 exon 2. References Ju YT, Chang AC, She BR, Tsaur ML, Hwang HM, Chao CC, Cohen SN, Lin-Chao S. gas7: A gene expressed preferentially in growth-arrested fibroblasts and terminally differentiated Purkinje neurons affects neurite formation. Proc Natl Acad Sci U S A Sep 15;95(19): Megonigal MD, Cheung NK, Rappaport EF, Nowell PC, Wilson RB, Jones DH, Addya K, Leonard DG, Kushner BH, Williams TM, Lange BJ, Felix CA. Detection of leukemia-associated MLL-GAS7 translocation early during chemotherapy with DNA topoisomerase II inhibitors. Proc Natl Acad Sci U S A Mar 14;97(6): This article should be referenced as such: Huret JL. GAS7 (growth arrest-specific 7). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3):95. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 95
6 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog) Lidia Larizza, Alessandro Beghini Department of Biology and Genetics for Medical Sciences, Medical Faculty, University of Milan, Via Viotti 3/5, Milan, Italy (LL, AB) Published in Atlas Database: June 2000 Online updated version : DOI: /2042/37632 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Other names: SCFR (Stem Cell Factor Receptor); CD117 HGNC (Hugo): KIT Location: 4q12 Local order: centromere-pdgfra-kit-kdrtelomere. KIT (4q12) - Courtesy Mariano Rocchi, Resources for Molecular Cytogenetics. DNA/RNA Description Spans 89 kb; 21 exons; size of intron 1: 37,4 kb. Transcription 5,23 kb mrna; alternative splicing of exon 9 gives rise to two isoforms, KitA and Kit, that differ by the presence or absence of four aminoacids. Protein Description 976 aa; 145 kda; type III receptor tyrosine kinase; contains an extracellular domains with 5 Ig-like loops, a highly hydrophobic transmenbrane domain (23 aa), and an intracellular domain with tyrosine kinase activity split by a kinase insert (KI) in an ATP-binding region and in the phosphotransferase domain. Expression Hematopoietic stem cells, mast cells, melanocytes, germ-cell lineages and ICCs (Interstitial cells of Cajal). Localisation Plasma membrane. Function SCF/MGF receptor with tyrosine kinase activity; binding of ligand (SCF) induces receptor dimerization, autophosphorylation and signal transduction via molecules containing SH2- domains. Homology With CSF-1R, PDGFRb, PDGFRa, and FLT3. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 96
7 KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog) Larizza L, Beghini A Mutations Note: See diagrams: Loss-of-function mutations, and Gain-of-function mutations. Germinal In piebaldism, and in familial gastrointestinal stromal tumours (see below). Somatic In aggressive mastocytosis, mast cell leukemia, ANLL with/without mast cell involvement, myeloproliferative disorders, colon carcinoma and gastrointestinal stromal tumours and germ cell tumors (GCCs). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 97
8 KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog) Larizza L, Beghini A Implicated in Piebaldism Disease Autosomal dominant disorder of pigmentation; loss of function abnormalities of the c-kit gene have been demonstrated in 59% of the typical patients. Familial gastrointestinal stromal tumours and sporadic gastrointestinal stromal tumours (GISTs) Disease GISTs are the most common mesenchymal tumors in the human digestive tract; they originate from kitexpressing cells (ICCs), and often have activating c-kit mutations clustered in the juxtamembrane domain. Systemic mast cell disease (SMCD) Disease Mast cell hyperplasia in the bone marrow, liver, spleen, lymph nodes, gastrointestinal tract and skin; gain of function mutations are detected in most patients. Prognosis Depending on the four clinical entities recognized: indolent form, form associated with hematologic disorder, aggressive SMCD and mast cell leukemia; leukemic transformation with mast cell involvement is characterized by rapid progression of disease with a survival time less than 1 year. Oncogenesis Clinical features of malignant hematopoietic cell growth are influenced by the time, the location of c-kit mutative events, and the number of associated lesions. Core binding factor leukemias (ANLL- M2 with t(8;21) (link), (ANLL-M4Eo with inv(16)) Disease Characterized by disruption and loss of CBFa2/AML1 - CBFb/PEBP2b function. Myelomonoblastic leukemia cells are marked by combined positivity for the stem cell antigens CD34, CD117 and high frequency of c-kit mutations (see Figure on CBF leukemia and KIT mutations). To be noted Note Loss of expression of c-kit appears to be associated with progression of some tumors (melanoma) and autocrine/paracrine stimulation of the c-kit/scf system may participate in human solid tumors such as lung, breast, testicular and gynecological malignancies. References Vandenbark GR, decastro CM, Taylor H, Dew-Knight S, Kaufman RE. Cloning and structural analysis of the human c- kit gene. Oncogene Jul;7(7): Ezoe K, Holmes SA, Ho L, Bennett CP, Bolognia JL, Brueton L, Burn J, Falabella R, Gatto EM, Ishii N. Novel mutations and deletions of the KIT (steel factor receptor) gene in human piebaldism. Am J Hum Genet Jan;56(1):58-66 Longley BJ, Tyrrell L, Lu SZ, Ma YS, Langley K, Ding TG, Duffy T, Jacobs P, Tang LH, Modlin I. Somatic c-kit activating mutation in urticaria pigmentosa and aggressive mastocytosis: establishment of clonality in a human mast cell neoplasm. Nat Genet Mar;12(3):312-4 Andre C, Hampe A, Lachaume P, Martin E, Wang XP, Manus V, Hu WX, Galibert F. Sequence analysis of two genomic regions containing the KIT and the FMS receptor tyrosine kinase genes. Genomics Jan 15;39(2): Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Muhammad Tunio G, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science Jan 23;279(5350): Gari M, Goodeve A, Wilson G, Winship P, Langabeer S, Linch D, Vandenberghe E, Peake I, Reilly J. c-kit proto-oncogene exon 8 in-frame deletion plus insertion mutations in acute myeloid leukaemia. Br J Haematol Jun;105(4): Longley BJ Jr, Metcalfe DD, Tharp M, Wang X, Tyrrell L, Lu SZ, Heitjan D, Ma Y. Activating and dominant inactivating c- KIT catalytic domain mutations in distinct clinical forms of human mastocytosis. Proc Natl Acad Sci U S A Feb 16;96(4): Sakurai S, Fukasawa T, Chong JM, Tanaka A, Fukayama M. C-kit gene abnormalities in gastrointestinal stromal tumors (tumors of interstitial cells of Cajal. Jpn J Cancer Res Dec;90(12): Tian Q, Frierson HF Jr, Krystal GW, Moskaluk CA. Activating c-kit gene mutations in human germ cell tumors. Am J Pathol Jun;154(6): Beghini A, Peterlongo P, Ripamonti CB, Larizza L, Cairoli R, Morra E, Mecucci C. C-kit mutations in core binding factor leukemias. Blood Jan 15;95(2):726-7 Lux ML, Rubin BP, Biase TL, Chen CJ, Maclure T, Demetri G, Xiao S, Singer S, Fletcher CD, Fletcher JA. KIT extracellular and kinase domain mutations in gastrointestinal stromal tumors. Am J Pathol Mar;156(3):791-5 This article should be referenced as such: Larizza L, Beghini A. KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 98
9 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review KITLG (KIT ligand) Lidia Larizza, Alessandro Beghini Department of Biology and Genetics for Medical Sciences, Medical Faculty, University of Milan, Via Viotti 3/5, Milan, Italy (LL, AB) Published in Atlas Database: June 2000 Online updated version : DOI: /2042/37633 This article is an update of : Beghini A, Larizza L. MGF (Mast cell growth factor). Atlas Genet Cytogenet Oncol Haematol 1999;3(1):6-7. This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Other names: MGF (Mast Cell Growth Factor); SCF (Stem Cell factor) HGNC (Hugo): KITLG Location: 12q22 DNA/RNA Description Genomic sequence not known; 9 exons. Transcription 1,4 Kb mrna; alternative splicing gives rise to different transcripts, mainly represented by those for a membrane and a soluble form. Protein Description The membrane bound form is a surface molecule of 248 aa, that includes 23 aa of the highly hydrophobic transmembrane domain; the second form corresponds to a soluble protein constituted by the first 165 aa of the extracellular domain released by a posttranslational processing, consisting in a proteolytic cleavage of the mature SCF in the extracellular juxtamembrane region; the full length transcripts encode for a transmembrane precursor of the soluble protein; an alternative splicing that involves the region corresponding to exon 6 of the SCF cdna, which contains the proteolytic cleavage site, encodes for a surface molecule. Expression SCF transcripts have been found in the cells surrounding kit-positive cells, such as granulosa and Sertoli cells, bone marrow stromal cells and in fibroblasts, keratinocytes and mature granulocytes; SCF expression of peripheral lymphocytes and monocytes is still controversial. Localisation Plasma membrane or interstitial space. Function SCF/MGF binding of receptor KIT, with tyrosine kinase activity, induces receptor dimerization, autophosphorylation and signal transduction via molecules containing SH2-domains; the soluble and the transmembrane protein have a different biological activity; the soluble form mainly stimulates cellular proliferation; the membrane-bound isoform induces an activation of the receptor more prolonged than the soluble one. Homology With PDGFRb, PDGFRa, and CSF-1. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 99
10 KITLG (KIT ligand) Larizza L, Beghini A Mutations Germinal Human mutations are yet unknown in human MGF/SCF gene; mouse mutations at the murine steel (Sl) locus that encodes MGF are known and give rise to deficiencies in pigment cells, germ cells, and blood cells; in particular the steel-dickie (Sld) mouse has a 4.0-kb intragenic deletion that truncates the Sl coding sequence; Sld mice are only capable of encoding a soluble truncated growth factor that lacks both transmembrane and cytoplasmic domains. Implicated in Mastocytosis Disease In skin from patients with mastocytosis, MGF was found prevalently free in the dermis and in extracellular spaces between keratinocytes suggesting the presence of a soluble form of the protein; altered distribution of mast cell growth factor in the skin of patients with cutaneous mastocytosis is consistent with abnormal production of the soluble form of the factor, resulting by an increased cleavage of SCF with excessive release of a soluble form from the normally membrane bound form; no sequence abnormalities were detected in MGF mrna. Gynecological tumors Disease Findings obtained on three cervical carcinomas (ovarian serous adenocarcinoma, small cell carcinoma and ovarian immature teratoma) and two gynecological cancer cell lines (ME180 and HGCM) demonstrate coexpression of c-kit receptor and SCF; these observations are consistent with the possibility that an autocrine activation of SCF/KIT system might be involved in gynecological malignancies. Small-cell lung cancer Disease SCF is expressed in small-cell lung cancer (SCLC); abundant expression of SCF and c-kit mrna was seen in 32% of SCLC cell lines and 66% of SCLC tumors; an autocrine mechanism in the pathogenesis of SCLC is strongly suggested. References ANTONOV IP, SIZONENKO TP. [Some data on the state of the hematoencephalic barrier in cerebral cysticercosis]. Zh Nevropatol Psikhiatr Im S S Korsakova. 1963;63:213-6 Martin FH, Suggs SV, Langley KE, Lu HS, Ting J, Okino KH, Morris CF, McNiece IK, Jacobsen FW, Mendiaz EA. Primary structure and functional expression of rat and human stem cell factor DNAs. Cell Oct 5;63(1): Zsebo KM, Williams DA, Geissler EN, Broudy VC, Martin FH, Atkins HL, Hsu RY, Birkett NC, Okino KH, Murdock DC. Stem cell factor is encoded at the Sl locus of the mouse and is the ligand for the c-kit tyrosine kinase receptor. Cell Oct 5;63(1): Hibi K, Takahashi T, Sekido Y, Ueda R, Hida T, Ariyoshi Y, Takagi H, Takahashi T. Coexpression of the stem cell factor and the c-kit genes in small-cell lung cancer. Oncogene Dec;6(12): Ferrari S, Grande A, Manfredini R, Tagliafico E, Zucchini P, Torelli G, Torelli U. Expression of interleukins 1, 3, 6, stem cell factor and their receptors in acute leukemia blast cells and in normal peripheral lymphocytes and monocytes. Eur J Haematol Mar;50(3):141-8 Longley BJ Jr, Morganroth GS, Tyrrell L, Ding TG, Anderson DM, Williams DE, Halaban R. Altered metabolism of mast-cell growth factor (c-kit ligand) in cutaneous mastocytosis. N Engl J Med May 6;328(18): Inoue M, Kyo S, Fujita M, Enomoto T, Kondoh G. Coexpression of the c-kit receptor and the stem cell factor in gynecological tumors. Cancer Res Jun 1;54(11): Ramenghi U, Ruggieri L, Dianzani I, Rosso C, Brizzi MF, Camaschella C, Pietsch T, Saglio G. Human peripheral blood granulocytes and myeloid leukemic cell lines express both transcripts encoding for stem cell factor. Stem Cells Sep;12(5):521-6 This article should be referenced as such: Larizza L, Beghini A. KITLG (KIT ligand). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 100
11 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review CBP (CREB-binding protein) Jean-Loup Huret Genetics, Dept Medical Information, University of Poitiers, CHU Poitiers Hospital, F Poitiers, France (JLH) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37634 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Other names: CREBBP (CREB binding protein (Rubinstein-Taybi syndrome)); RTS (Rubinstein- Taybi syndrome); RSTS HGNC (Hugo): CREBBP Location: 16p13.3 CBP (16p22) - Courtesy Mariano Rocchi, Resources for Molecular Cytogenetics. DNA/RNA Description The gene spans about 190 kb; transcription from centromere to telomere. Transcription 8.7 kb mrna, with a 7.3 kb coding sequence. Protein Description 2442 amino acids; 265 kda; from NH2-term, is made of a CREB-Binding domain, a bromodomain. Cystidine/Histidine-rich domains, and Glutamine-rich domains in COOH-term. Expression Wide expression; expression in the whole embryo as well. Localisation Nucleus. Function Binds specifically to the DNA-binding protein CREB and connects it to the basal transcriptional machinery: transcription coactivator, with P300; has histone acetyltransferase activity; essential role in embryogenesis, cell differentiation, apoptosis, and proliferation; involved in the regulation of cell cycle during G1/S transition. Homology P300. Implicated in t(8;16)(p11;p13)/m4 ANLL --> MOZ/CBP Disease Acute non lymphocytic leukemia (ANLL) and treatment related ANLL (t-anll). Prognosis Poor: remission is obtained in half cases; survival is often less than 1 year. Cytogenetics +8 as an additional anomalies in half cases. Hybrid/Mutated gene 5' MOZ - 3' CBP. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 101
12 CBP (CREB-binding protein) Huret JL Abnormal protein N-term finger motifs and acetyl transferase from MOZ fused to most of CBP, with a breakpoint in 5' of the CREB binding domain of CBP. t(11;16)(q23;p13)/t-anll --> MLL/CBP Disease Treatment related ANLL (t-anll); should be very close to the t(11;22)(q23;q13). Prognosis Likely to be poor. Hybrid/Mutated gene 5' MLL - 3' CBP. Abnormal protein N-term AT hook and DNA methyltransferase from MLL fused to most of CBP; variable breakpoint in CBP: either in 5' of the CREB binding domain (like in the t(8;16)), or just upstream of the bromodomain. Rubinstein-taybi syndrome Note Due to CBP haploinsufficiency. Disease Autosomal dominant disorder with mental retardation, facial dysmorphia, broad thumbs/halluces, cardiac anomalies, and an increased risk of medulloblastoma, meningioma, and neuroblastoma. References Borrow J, Stanton VP Jr, Andresen JM, Becher R, Behm FG, Chaganti RS, Civin CI, Disteche C, Dubé I, Frischauf AM, Horsman D, Mitelman F, Volinia S, Watmore AE, Housman DE. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREBbinding protein. Nat Genet Sep;14(1):33-41 Eckner R. p300 and CBP as transcriptional regulators and targets of oncogenic events. Biol Chem Nov;377(11):685-8 Giles RH, Petrij F, Dauwerse HG, den Hollander AI, Lushnikova T, van Ommen GJ, Goodman RH, Deaven LL, Doggett NA, Peters DJ, Breuning MH. Construction of a 1.2- Mb contig surrounding, and molecular analysis of, the human CREB-binding protein (CBP/CREBBP) gene on chromosome 16p13.3. Genomics May 15;42(1): Goldman PS, Tran VK, Goodman RH. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog Horm Res. 1997;52:103-19; discussion Sobulo OM, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett NA, Rowley JD, Zeleznik-Le NJ. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3). Proc Natl Acad Sci U S A Aug 5;94(16): Taki T, Sako M, Tsuchida M, Hayashi Y. The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood Jun 1;89(11): Giordano A, Avantaggiati ML. p300 and CBP: partners for life and death. J Cell Physiol Nov;181(2): Partanen A, Motoyama J, Hui CC. Developmentally regulated expression of the transcriptional cofactors/histone acetyltransferases CBP and p300 during mouse embryogenesis. Int J Dev Biol Sep;43(6): Yuan ZM, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Shioya H, Utsugisawa Y, Shi Y, Weichselbaum R, Kufe D. Function for p300 and not CBP in the apoptotic response to DNA damage. Oncogene Oct 7;18(41): Ait-Si-Ali S, Polesskaya A, Filleur S, Ferreira R, Duquet A, Robin P, Vervish A, Trouche D, Cabon F, Harel-Bellan A. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene May 11;19(20): This article should be referenced as such: Huret JL. CBP (CREB-binding protein). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 102
13 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review CIITA (MHC class II transactivator) Jean-Loup Huret Genetics, Dept Medical Information, University of Poitiers, CHU Poitiers Hospital, F Poitiers, France (JLH) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37639 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Other names: MHC2TA (MHC class II transactivator); C2TA HGNC (Hugo): CIITA Location: 16p13 DNA/RNA Transcription 6.7 kb mrna; coding sequence: 3392 bp. Protein Description 1130 amino acids. Function Non-DNA-binding transcriptional coactivator; highly regulated; IFN-gamma-inducible; transactivator of the major histocompatibility complex (MHC) class II genes, herein the name; DM and Ii (molecules associated to MHC class II) are also regulated by MHC2TA; MHC2TA also transactivates MHC class I; lymphoid cells of MHC2TA-null mice do not express MHC class II. Implicated in t(3;16)(q27;p13)/ BCL6 Disease Non-Hodgkin lymphoma. Prognosis Yet unknown (only 1 case available with certainty). Hybrid/Mutated gene 5' MHC2TA - 3' BCL6; MHC2TA fuses with the second exon of BCL6. Abnormal protein No fusion protein, but promoter exchange. Oncogenesis BCL6 is a transcription repressor; it is supposed that substitution of the promoter of BCL6 may be responsible for BCL6 deregulation. Bare lymphocyte syndrome (BLS) Note At least five complementation groups. Disease Severe immunodeficiency due to failure to express MHC class II genes. References Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell Oct 8;75(1): Wright KL, Chin KC, Linhoff M, Skinner C, Brown JA, Boss JM, Stark GR, Ting JP. CIITA stimulation of transcription factor binding to major histocompatibility complex class II and associated promoters in vivo. Proc Natl Acad Sci U S A May 26;95(11): Fontes JD, Kanazawa S, Nekrep N, Peterlin BM. The class II transactivator CIITA is a transcriptional integrator. Microbes Infect Sep;1(11):863-9 Yoshida S, Kaneita Y, Aoki Y, Seto M, Mori S, Moriyama M. Identification of heterologous translocation partner genes fused to the BCL6 gene in diffuse large B-cell lymphomas: 5'-RACE and LA - PCR analyses of biopsy samples. Oncogene Dec 23;18(56): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 103
14 CIITA (MHC class II transactivator) Huret JL Girdlestone J. Synergistic induction of HLA class I expression by RelA and CIITA. Blood Jun 15;95(12): Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev May 1;14(9): This article should be referenced as such: Huret JL. CIITA (MHC class II transactivator). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 104
15 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Short Communication EIF4A2 (eukaryotic translation initiation factor 4A, isoform 2) Jean-Loup Huret Genetics, Dept Medical Information, University of Poitiers, CHU Poitiers Hospital, F Poitiers, France (JLH) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37635 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Other names: DDX2B HGNC (Hugo): EIF4A2 Location: 3q28 Probe(s) - Courtesy Mariano Rocchi, Resources for Molecular Cytogenetics. DNA/RNA Transcription 1.9 kb mrna; 1223 bp coding sequence. Protein Description 407 amino acids; 4.6 kda; NH2-term, ATP binding site, DEAD box, COOH-term. Expression Wide; also expressed in the embryo. Function ADP/ATP binding; RNA helicase; binds mrna to the ribosome: role in initiation of protein synthesis; also - binds single strand DNA. Homology DEAD box proteins, in particular other EIF4A. Implicated in t(3;18)(q27;p11.2) --> EIF4A2/ BCL6 Disease Non-Hodgkin lymphoma. Prognosis Yet unknown (only 1 case available). Hybrid/Mutated gene 5' EIF4A2-3' BCL6; EIF4A2 fuses with the second exon of BCL6. Abnormal protein No fusion protein, but promoter exchange. Oncogenesis BCL6 is a transcription repressor; it is supposed that substitution of the promoter of BCL6 may be responsible for BCL6 deregulation. References Sudo K, Takahashi E, Nakamura Y. Isolation and mapping of the human EIF4A2 gene homologous to the murine protein synthesis initiation factor 4A-II gene Eif4a2. Cytogenet Cell Genet. 1995;71(4):385-8 Lorsch JR, Herschlag D. The DEAD box protein eif4a. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry Feb 24;37(8): Li Q, Imataka H, Morino S, Rogers GW Jr, Richter-Cook NJ, Merrick WC, Sonenberg N. Eukaryotic translation initiation factor 4AIII (eif4aiii) is functionally distinct from eif4ai and eif4aii. Mol Cell Biol Nov;19(11): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 105
16 EIF4A2 (eukaryotic translation initiation factor 4A, isoform 2) Huret JL Yoshida S, Kaneita Y, Aoki Y, Seto M, Mori S, Moriyama M. Identification of heterologous translocation partner genes fused to the BCL6 gene in diffuse large B-cell lymphomas: 5'-RACE and LA - PCR analyses of biopsy samples. Oncogene Dec 23;18(56): This article should be referenced as such: Huret JL. EIF4A2 (eukaryotic translation initiation factor 4A, isoform 2). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 106
17 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review IGH (Immunoglobulin Heavy) Marie-Paule Lefranc IMGT, LIGM, IGH, UPR CNRS 1142, 141 rue de la Cardonille, Montpellier Cedex 5, France (MPL) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37636 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity HGNC (Hugo): IGH@ Location: 14q32.33 For complete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Note The human IGH locus is located on the chromosome 14 at band 14q32.33, at the telomeric extremity of the long arm; the orientation of the locus has been determined by the analysis of translocations, involving the IGH locus, in leukemia and lymphoma. DNA/RNA Description The human IGH locus at 14q32.33 spans 1250 kilobases (kb). It consists of 123 to 129 IGHV genes, depending from the haplotypes, 27 IGHD segments belonging to 7 subgroups, 9 IGHJ segments, and 11 IGHC genes. Eighty-two to 88 IGHV genes belong to 7 subgroups, whereas 41 pseudogenes, which are too divergent to be assigned to subgroups, have been assigned to 4 clans. Seven non-mapped IGHV genes have been described as insertion/deletion polymorphism but have not yet been precisely located. The most 5' IGHV genes occupy a position very close to the chromosome 14q telomere whereas the IGHC genes are in a more centromeric position. The potentiel genomic IGH repertoire is more limited since it comprises functional IGHV genes belonging to 6 or 7 subgroups depending from the haplotypes 23 IGHD, 6 IGHJ, and 9 IGHC genes. Thirty-five IGH genes have been found outside the main locus in other chromosomal localizations. These genes designated as orphons cannot contribute to the synthesis of the immunoglobulin chains, even if they have an Open Reading Frame (ORF). 9 IGHV orphons and 10 IGHD orphons have been described on chromosome 15 (15q11.2), and 16 IGHV orphons on chromosome 16 (16p11.2). In addition, one IGHC processed gene, IGHEP2 is localised on chromosome 9 (9p24.2-p24.1) This is so far the only processed Ig gene described. The total number of human IGH genes per haploid genome is 170 to 176 (206 to 212 genes, if the orphons and the processed gene are included) of which 77 to 84 genes are functional. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 107
18 IGH (Immunoglobulin Heavy) Lefranc MP IGH V-GENE: Green box: Functional; Yellow box: Open reading frame; Red: Pseudogene; Grey triangle: Not sequenced, not found. D-GENE: Blue: Functional; Blue open box: Open reading frame. J-GENE: Grey: Functional. C-GENE: Blue: Functional; Blue dashed box: Open reading frame; Blue open box: Pseudogene. GENES NOT RELATED: Purple open box: Pseudogene. For compete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Protein Description Proteins encoded by the IGH locus are the immunoglobulin heavy chains. They result from the recombination (or rearrangement), at the DNA level, of three genes: IGHV, IGHD and IGHJ, with deletion of the intermediary DNA to create a rearranged IGHV-D- J gene. The rearranged IGHV-D-J gene is transcribed with the IGHM gene and translated into an immunoglobulin mu chain. The gamma, alpha or epsilon heavy chains, result from a new recombination (or switch), again at the DNA level, between sequences designated as "Switch" and localized upstream of the IGHM and of Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 108
19 IGH (Immunoglobulin Heavy) Lefranc MP each of the functional IGHG, IGHA and IGHE constant genes. This recombination, accompanied by the deletion of the intermediary DNA, allows the IGHV-D-J initially transcribed with the IGHM, to be now transcribed with a IGHG, IGHA or IGHE gene, and translated into a gamma, alpha or epsilon chain. Translation of the variable germline genes involved in the IGHV-D-J rearrangements are available at IMGT Repertoire Compared to the germline genes, the rearranged variable genes will acquire somatic mutations during the B cell differentiation in the lymph nodes, which will considerably increase their diversity. These somatic mutations can be analysed using IMGT/V-QUEST tool. Mutations Note Mutations which correspond to allelic polymorphisms of the functional germline IGHV, IGHD, IGHJ and IGHC genes are described in the IMGT database. Implicated in Translocations which frequently result from errors of the recombinase enzyme complexe (RAG1, RAG2, etc.), responsable of the Immunoglobulin and T cell receptor V-J and V-D- J rearrangements, or from errors of the switch enzyme. IGHV, IGHD or IGHJ recombination signals or isolated heptamer (first case) or switch sequences (second case) are observed at the breakpoints. for human immunoglobulin heavy chains. Proc Natl Acad Sci U S A Jul;76(7): Ellison J, Buxbaum J, Hood L. Nucleotide sequence of a human immunoglobulin C gamma 4 gene. DNA. 1981;1(1):11-8 Rabbitts TH, Forster A, Milstein CP. Human immunoglobulin heavy chain genes: evolutionary comparisons of C mu, C delta and C gamma genes and associated switch sequences. Nucleic Acids Res Sep 25;9(18): Ravetch JV, Siebenlist U, Korsmeyer S, Waldmann T, Leder P. Structure of the human immunoglobulin mu locus: characterization of embryonic and rearranged J and D genes. Cell Dec;27(3 Pt 2): Battey J, Max EE, McBride WO, Swan D, Leder P. A processed human immunoglobulin epsilon gene has moved to chromosome 9. Proc Natl Acad Sci U S A Oct;79(19): Ellison J, Hood L. Linkage and sequence homology of two human immunoglobulin gamma heavy chain constant region genes. Proc Natl Acad Sci U S A Mar;79(6): Ellison JW, Berson BJ, Hood LE. The nucleotide sequence of a human immunoglobulin C gamma1 gene. Nucleic Acids Res Jul 10;10(13): Flanagan JG, Rabbitts TH. Arrangement of human immunoglobulin heavy chain constant region genes implies evolutionary duplication of a segment containing gamma, epsilon and alpha genes. Nature Dec 23;300(5894): Flanagan JG, Rabbitts TH. The sequence of a human immunoglobulin epsilon heavy chain constant region gene, and evidence for three non-allelic genes. EMBO J. 1982;1(5): Kirsch IR, Morton CC, Nakahara K, Leder P. Human immunoglobulin heavy chain genes map to a region of translocations in malignant B lymphocytes. Science Apr 16;216(4543):301-3 Max EE, Battey J, Ney R, Kirsch IR, Leder P. Duplication and deletion in the human immunoglobulin epsilon genes. Cell Jun;29(2):691-9 c-immunoglobulin genes IgH at 14q32.33, in normal cells: PAC 998D24 - Courtesy Mariano Rocchi. t(3;14)(q27;q32) t(4;14)(p16;q32) t(5;14)(q31;q32) t(8;14)(q11;q32) t(8;14)(q24;q32) t(10;14)(q24;q32) t(11;14)(q13;q32) t(14;18)(q32;q21) t(14;19)(q32;q13.1) References Croce CM, Shander M, Martinis J, Cicurel L, D'Ancona GG, Dolby TW, Koprowski H. Chromosomal location of the genes McBride OW, Battey J, Hollis GF, Swan DC, Siebenlist U, Leder P. Localization of human variable and constant region immunoglobulin heavy chain genes on subtelomeric band q32 of chromosome 14. Nucleic Acids Res Dec 20;10(24): Lefranc MP, Lefranc G, de Lange G, Out TA, van den Broek PJ, van Nieuwkoop J, Radl J, Helal AN, Chaabani H, van Loghem E. Instability of the human immunoglobulin heavy chain constant region locus indicated by different inherited chromosomal deletions. Mol Biol Med Sep;1(2): Lefranc MP, Lefranc G, de Lange G, Out TA, van den Broek PJ, van Nieuwkoop J, Radl J, Helal AN, Chaabani H, van Loghem E. Instability of the human immunoglobulin heavy chain constant region locus indicated by different inherited chromosomal deletions. Mol Biol Med Sep;1(2): White MB, Shen AL, Word CJ, Tucker PW, Blattner FR. Human immunoglobulin D: genomic sequence of the delta heavy chain. Science May 10;228(4700):733-7 Huck S, Fort P, Crawford DH, Lefranc MP, Lefranc G. Sequence of a human immunoglobulin gamma 3 heavy chain constant region gene: comparison with the other human C gamma genes. Nucleic Acids Res Feb 25;14(4): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 109
20 IGH (Immunoglobulin Heavy) Lefranc MP Bensmana M, Huck S, Lefranc G, Lefranc MP. The human immunoglobulin pseudo-gamma IGHGP gene shows no major structural defect. Nucleic Acids Res Apr 11;16(7):3108 Buluwela L, Albertson DG, Sherrington P, Rabbitts PH, Spurr N, Rabbitts TH. The use of chromosomal translocations to study human immunoglobulin gene organization: mapping DH segments within 35 kb of the C mu gene and identification of a new DH locus. EMBO J Jul;7(7): Ichihara Y, Matsuoka H, Kurosawa Y. Organization of human immunoglobulin heavy chain diversity gene loci. EMBO J Dec 20;7(13): Huck S, Lefranc G, Lefranc MP. A human immunoglobulin IGHG3 allele (Gmb0,b1,c3,c5,u) with an IGHG4 converted region and three hinge exons. Immunogenetics. 1989;30(4):250-7 Bensmana M, Chuchana P, Lefranc G, Lefranc MP. Sequence of the CH1 and hinge-ch2 exons of the human immunoglobulin IGHA2 A2m(2) allele: comparison with the nonallelic and allelic IGHA genes. Cytogenet Cell Genet. 1991;56(2):128 Shin EK, Matsuda F, Nagaoka H, Fukita Y, Imai T, Yokoyama K, Soeda E, Honjo T. Physical map of the 3' region of the human immunoglobulin heavy chain locus: clustering of autoantibody-related variable segments in one haplotype. EMBO J Dec;10(12): Cook GP, Tomlinson IM, Walter G, Riethman H, Carter NP, Buluwela L, Winter G, Rabbitts TH. A map of the human immunoglobulin VH locus completed by analysis of the telomeric region of chromosome 14q. Nat Genet Jun;7(2):162-8 Cook GP, Tomlinson IM. The human immunoglobulin VH repertoire. Immunol Today May;16(5): Corbett SJ, Tomlinson IM, Sonnhammer EL, Buck D, Winter G. Sequence of the human immunoglobulin diversity (D) segment locus: a systematic analysis provides no evidence for the use of DIR segments, inverted D segments, "minor" D segments or D-D recombination. J Mol Biol Jul 25;270(4): Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med Dec 7;188(11): Pallarès N, Lefebvre S, Contet V, Matsuda F, Lefranc MP. The human immunoglobulin heavy variable genes. Exp Clin Immunogenet. 1999;16(1):36-60 Ruiz M, Pallarès N, Contet V, Barbi V, Lefranc MP. The human immunoglobulin heavy diversity (IGHD) and joining (IGHJ) segments. Exp Clin Immunogenet. 1999;16(3): Scaviner D, Barbié V, Ruiz M, Lefranc MP. Protein displays of the human immunoglobulin heavy, kappa and lambda variable and joining regions. Exp Clin Immunogenet. 1999;16(4): Lefranc M-P.. Locus Map and Genomic repertoire of the Human Immunoglobulin Genes (Review) The immunologist. 2000; 8: Lefranc M-P.. Nomenclature of the human immunoglobulin genes (Review) Current Protocols in Immunology. 2000, Wiley, J. and Sons, New York, Supplement 40, A.1P.1-A.1P.37. Lefranc MP. Nomenclature of the human immunoglobulin heavy (IGH) genes. Exp Clin Immunogenet. 2001;18(2): Lefranc M-P. and Lefranc G.. The Immunoglobulin FactsBook (Review) Academic Press, London, UK. 2001, ISBN: X. This article should be referenced as such: Lefranc MP. IGH (Immunoglobulin Heavy). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 110
21 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review IGK (Immunoglobulin Kappa) Marie-Paule Lefranc IMGT, LIGM, IGH, UPR CNRS 1142, 141 rue de la Cardonille, Montpellier Cedex 5, France (MPL) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37637 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity HGNC (Hugo): IGK@ Location: 2p12 For complete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Note The human IGK locus is located on chromosome 2 on the short arm, at band 2p12. The orientation of the locus has been determined by the analysis of translocations, involving the IGK locus, in leukemia and lymphoma. DNA/RNA Description The human IGK locus at 2p12 spans 1820 kb. It consists of 76 IGKV genes belonging to 7 subgroups, 5 IGKJ segments, and a unique IGKC gene. The 76 IGKV genes are organized in two clusters separated by 800 kb. The IGKV distal cluster (the most 5' from IGKC and in the most centromeric position) spans 400 kb and comprises 36 genes. The IGKV proximal cluster (in 3' of the locus, closer to IGKC, and in the most telomeric position) spans 600 kb and comprises 40 genes. The potential genomic IGK repertoire comprises 31 to 35 functional IGKV genes belonging to 5 subgroups, the 5 IGKJ segments, and the unique IGKC gene. One rare IGKV haplotype has been described which contains only the proximal cluster. This haplotype comprises the 40 proximal IGKV genes belonging to 7 subgroups, of which 17 to 19 are functional and belong to 5 subgroups. Twenty-eight IGKV orphons have been identified and sequenced: 3 on the short arm of chromosome 2 but outside of the main IGK locus, 13 on the long arm of chromosome 2, 6 on chromosome 22, one on chromosome 1, one on chromosome 15, and 4 outside of chromosome 2. If both the proximal and distal IGKV clusters are present, the total number of human IGK genes per haploid genome is 82 (110 genes, if the orphons are included) of which are functional. If only the proximal IGKV cluster is present, the total number of genes per haploid genome is 46 (74 genes, if the orphons are included) of which genes are functional. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 111
22 IGK (Immunoglobulin Kappa) Lefranc MP IGK V-GENE: Green box: Functional; Yellow box: Open reading frame; Red: Pseudogene. J-GENE: Grey: Functional. C-GENE: Blue: Functional. For compete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Protein Description Proteins encoded by the IGK locus are the immunoglobulin kappa chains. They result from the recombination (or rearrangement), at the DNA level, of two genes: IGKV and IGKJ, with deletion of the intermediary DNA to create a rearranged IGKV-J gene. The rearranged IGKV-J gene is transcribed with the IGKC gene and translated into an immunoglobulin kappa chain. Translation of the variable germline genes involved in the IGKV-J rearrangements are available at IMGT Repertoire. Compared to the germline genes, the rearranged variable genes will acquire somatic mutations during the B cell differentiation in the lymph nodes, which will considerably increase their diversity. These somatic mutations can be analysed using IMGT/V-QUEST tool. Mutations Note Mutations which correspond to allelic polymorphisms of the functional germline IGKV, IGKJ and IGKC genes are described in the IMGT database. Implicated in Translocations which frequently result from errors of the recombinase enzyme complexe (RAG1, RAG2, etc.), which is responsable of the Immunoglobulin and T cell receptor V-J and V-D- J rearrangements. IGKV or IGKJ recombination signals or isolated heptamer are observed at the breakpoints. c-immunoglobulin genes IgK at 2p12, in normal cells: PAC 1117G4 - Courtesy Mariano Rocchi. t(2;3)(p12;q27) t(2;8)(p12;q24) References Hieter PA, Max EE, Seidman JG, Maizel JV Jr, Leder P. Cloned human and mouse kappa immunoglobulin constant and J region genes conserve homology in functional segments. Cell Nov;22(1 Pt 1): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 112
23 IGK (Immunoglobulin Kappa) Lefranc MP Hieter PA, Maizel JV Jr, Leder P. Evolution of human immunoglobulin kappa J region genes. J Biol Chem Feb 10;257(3): Malcolm S, Barton P, Murphy C, Ferguson-Smith MA, Bentley DL, Rabbitts TH. Localization of human immunoglobulin kappa light chain variable region genes to the short arm of chromosome 2 by in situ hybridization. Proc Natl Acad Sci U S A Aug;79(16): Huber C, Schäble KF, Huber E, Klein R, Meindl A, Thiebe R, Lamm R, Zachau HG. The V kappa genes of the L regions and the repertoire of V kappa gene sequences in the human germ line. Eur J Immunol Nov;23(11): Zachau HG. The immunoglobulin kappa locus-or-what has been learned from looking closely at one-tenth of a percent of the human genome. Gene Dec 15;135(1-2): Cox JP, Tomlinson IM, Winter G. A directory of human germline V kappa segments reveals a strong bias in their usage. Eur J Immunol Apr;24(4): Schäble K, Thiebe R, Flügel A, Meindl A, Zachau HG. The human immunoglobulin kappa locus: pseudogenes, unique and repetitive sequences. Biol Chem Hoppe Seyler Mar;375(3): Barbié V, Lefranc MP. The human immunoglobulin kappa variable (IGKV) genes and joining (IGKJ) segments. Exp Clin Immunogenet. 1998;15(3): Scaviner D, Barbié V, Ruiz M, Lefranc MP. Protein displays of the human immunoglobulin heavy, kappa and lambda variable and joining regions. Exp Clin Immunogenet. 1999;16(4): Brashear A, Watts MW, Marchetti A, Magar R, Lau H, Wang L. Duration of effect of botulinum toxin type A in adult patients with cervical dystonia: a retrospective chart review. Clin Ther Dec;22(12): Brashear A, Watts MW, Marchetti A, Magar R, Lau H, Wang L. Duration of effect of botulinum toxin type A in adult patients with cervical dystonia: a retrospective chart review. Clin Ther Dec;22(12): Lefranc M-P. Locus Map and Genomic repertoire of the Human Immunoglobulin Genes (Review) The immunologist. 2000; 8: Lefranc M-P. Nomenclature of the human immunoglobulin genes (Review) Current Protocols in Immunology. 2000, Wiley, J. and Sons, New York, Supplement 40, A.1P.1-A.1P.37. Lefranc MP. Nomenclature of the human immunoglobulin kappa (IGK) genes. Exp Clin Immunogenet. 2001;18(3): Lefranc M-P. and Lefranc G. The Immunoglobulin FactsBook (Review) Academic Press, London, UK. 2001, ISBN: X. This article should be referenced as such: Lefranc MP. IGK (Immunoglobulin Kappa). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 113
24 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review IGL (Immunoglobulin Lambda) Marie-Paule Lefranc IMGT, LIGM, IGH, UPR CNRS 1142, 141 rue de la Cardonille, Montpellier Cedex 5, France (MPL) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37638 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity HGNC (Hugo): IGL@ Location: 22q11.2 For complete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Note The human IGL locus is located on chromosome 22 on the long arm, at band 22q11.2. The orientation of the locus has been determined by the analysis of translocations, involving the IGL locus, in leukemia and lymphoma. Sequencing of the long arm of chromosome 22 showed that it encompasses about 35 megabases of DNA and that the IGL locus is localized at 6 megabases from the centromere. Although the correlation between DNA sequences and chromosomal bands has not yet been made, the localization of the IGL locus can be refined to 22q11.2. DNA/RNA Description The human IGL locus at 22q11.2 spans 1050 kb. It consists of 70 to 71 IGLV genes, localized on 900 kb, 7 to 11 IGLJ and 7 to 11 IGLC genes depending on the haplotypes, each IGLC gene being preceded by one IGLJ segment. Fifty-six to 57 genes belong to 11 subgroups, whereas 14 pseudogenes which are too divergent to be assigned to subgroups, have been assigned to 3 clans. The most 5' IGLV genes occupy the more centromeric position, whereas the IGLC genes, in 3' of the locus, are the most telomeric genes in the IGL locus. The potential genomic IGL repertoire comprises 29 to 32 functional IGLV genes belonging to 10 subgroups, 4 to 5 IGLJ, and 4 to 5 IGLC functional genes in the 7- IGLC gene haplotype. One, 2, 3 or 4 additional IGLC genes, each one probably preceded by one IGLJ, have been shown to characterize IGLC haplotypes with 8, 9, 10 or 11 genes, but these genes have not yet been sequenced. Two IGLV orphons have been identified on chromosome 8 at 8q11.2 and one of them belonging to subgroup 8 has been sequenced. The recent sequencing of the chromosome 22q showed that the IGL locus is localized at 6 megabases from the centromere. Two IGLC orphons and two IGLV orphons have also been characterized on 22q outside of the major IGL locus (See also IMGT Repertoire). The total number of human IGL genes per haploid genome is (90-99 genes, if the orphons are included) of which genes are functional. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 114
25 IGL (Immunoglobulin Lambda) Lefranc MP Protein IGL V-GENE: Green box: Functional; Yellow box: Open reading frame; Red: Pseudogene. J-GENE: Grey: Functional. C-GENE: Blue: Functional; Blue open box: Pseudogene; Blue triangle: Not sequenced. GENES NOT RELATED: Purple open box: Pseudogene. For compete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Description Proteins encoded by the IGL locus are the immunoglobulin lambda chains. They result from the recombination (or rearrangement), at the DNA level, of two genes: IGLV and IGLJ, with deletion of the intermediary DNA to create a rearranged IGLV-J gene. The rearranged IGLV-J gene is transcribed with one of the IGLC genes and translated into an immunoglobulin lambda chain. Translation of the variable germline genes involved in the IGLV-J rearrangements are available at IMGT Repertoire. Compared to the germline genes, the rearranged variable genes will acquire somatic mutations during the B cell differentiation in the lymph nodes, which will considerably increase their diversity. These somatic mutations can be analysed using the IMGT/V- QUEST. Mutations Note Mutations which correspond to allelic polymorphisms of the functional germline IGLV, IGLJ and IGLC genes are described in the IMGT database. Implicated in Translocations which frequently result from errors of the recombinase enzyme complexe (RAG1, RAG2, etc.), responsable of the Immunoglobulin and T cell receptor V-J and V-D- J rearrangements. IGLV or IGLJ recombination signals or isolated heptamer are observed at the breakpoints. c-immunoglobulin gene IgL at 22q11, in normal cells: PAC 1019H10 and PAC 869I1 - Courtesy Mariano Rocchi. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 115
26 IGL (Immunoglobulin Lambda) Lefranc MP t(3;22)(q27;q11) t(8;22)(q24;q11) References Emanuel BS, Cannizzaro LA, Magrath I, Tsujimoto Y, Nowell PC, Croce CM. Chromosomal orientation of the lambda light chain locus: V lambda is proximal to C lambda in 22q11. Nucleic Acids Res Jan 25;13(2):381-7 Dariavach P, Lefranc G, Lefranc MP. Human immunoglobulin C lambda 6 gene encodes the Kern+Oz-lambda chain and C lambda 4 and C lambda 5 are pseudogenes. Proc Natl Acad Sci U S A Dec;84(24): Ghanem N, Dariavach P, Bensmana M, Chibani J, Lefranc G, Lefranc MP. Polymorphism of immunoglobulin lambda constant region genes in populations from France, Lebanon and Tunisia. Exp Clin Immunogenet. 1988;5(4): Vasicek TJ, Leder P. Structure and expression of the human immunoglobulin lambda genes. J Exp Med Aug 1;172(2): Frippiat JP, Williams SC, Tomlinson IM, Cook GP, Cherif D, Le Paslier D, Collins JE, Dunham I, Winter G, Lefranc MP. Organization of the human immunoglobulin lambda light-chain locus on chromosome 22q11.2. Hum Mol Genet Jun;4(6): Kawasaki K, Minoshima S, Schooler K, Kudoh J, Asakawa S, de Jong PJ, Shimizu N. The organization of the human immunoglobulin lambda gene locus. Genome Res Sep;5(2): Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits JL, Wang J, Shimizu N. One-megabase sequence analysis of the human immunoglobulin lambda gene locus. Genome Res Mar;7(3): Lefranc MP, Pallarès N, Frippiat JP. Allelic polymorphisms and RFLP in the human immunoglobulin lambda light chain locus. Hum Genet May;104(5):361-9 Lefranc MP, Pallarès N, Frippiat JP. Allelic polymorphisms and RFLP in the human immunoglobulin lambda light chain locus. Hum Genet May;104(5):361-9 Scaviner D, Barbié V, Ruiz M, Lefranc MP. Protein displays of the human immunoglobulin heavy, kappa and lambda variable and joining regions. Exp Clin Immunogenet. 1999;16(4): Lefranc M-P.. Nomenclature of the human immunoglobulin genes (Review) Current Protocols in Immunology. 2000, Wiley, J. and Sons, New York, Supplement 40, A.1P.1-A.1P.37. Lefranc M-P.. Locus Map and Genomic repertoire of the Human Immunoglobulin Genes (Review) The immunologist. 2000; 8: Lefranc MP. Nomenclature of the human immunoglobulin lambda (IGL) genes. Exp Clin Immunogenet. 2001;18(4): Lefranc M-P. and Lefranc G.. The Immunoglobulin FactsBook (Review) Academic Press, London, UK. 2001, ISBN: X. This article should be referenced as such: Lefranc MP. IGL (Immunoglobulin Lambda). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 116
27 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review TRA (T cell Receptor Alpha) Marie-Paule Lefranc IMGT, LIGM, IGH, UPR CNRS 1142, 141 rue de la Cardonille, Montpellier Cedex 5, France (MPL) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37640 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity HGNC (Hugo): TRA@ Location: 14q11.2 For complete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Note The human TRA locus is located on the chromosome 14 on the long arm at band 14q11.2. The orientation of the locus has been determined by the analysis of translocations, involving the TRA and TRD loci, in leukemia and lymphoma. DNA/RNA Description The human TRA locus at 14q11.2 spans 1000 kilobases (kb). It consists of 54 TRAV genes belonging to 41 subgroups, 61 TRAJ segments localized on 71 kb, and a unique TRAC gene. The most 5' TRAV genes occupy the most centromeric position, whereas the TRAC genes, 3' of the locus, is the most telomeric gene in the TRA locus. The organization of the TRAJ segments on a large area is quite unusual and has not been observed in the other immunoglobulin or T cell receptor loci. Moreover the TRD locus is nestled in the TRA locus between the TRAV and TRAJ segments. V-Jrearrangements in the TRA locus therefore result in deletion of the TRD genes localized on the same chromosome. That deletion occurs in two steps, that is a deletion of the TRD genes, involving specific sequences located upstream from TRDC (sequence pseudo J alpha) would take place before the TRAV-J rearrangement. The potentiel genomic TRA repertoire comprises functional TRAV genes belonging to subgroups, 50 functional TRAJ segments, and the unique TRAC gene. Among the variable genes are included five genes designated as TRAV/DV which belong to five different subgroups and which have been found rearranged either to TRAJ or to TRDD segments and can therefore be used in the synthesis of alpha or delta chains. The total number of human TRA genes per haploid genome is 116 of which 96 to 98 genes are functional. Enhancer sequences have been characterized 4.5 kb 3' from TRAC. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 117
28 TRA (T cell Receptor Alpha) Lefranc MP TRA/TRD V-GENE: Green box: Functional; Red: Pseudogene. D-GENE: Blue: Functional. J-GENE: Yellow: Functional; Pale yellow: Open reading frame; Red: pseudogene. C-GENE: Blue: Functional. For compete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Protein Description Proteins encoded by the TRA locus are the T cell receptor alpha chains. They result from the recombination (or rearrangement), at the DNA level, of two genes: TRAV and TRAJ, with deletion of the intermediary DNA to create a rearranged TRAV-J gene. The rearranged TRAV-J gene is transcribed with the TRAC gene and translated into an T cell receptor alpha chain. Translation of the variable germline genes involved in the TRAV-J rearrangements are available at IMGT Repertoire. TRA V-J rearrangements can be analysed using the IMGT/V-QUEST tool. Mutations Note Mutations which correspond to allelic polymorphisms of the functional germline TRAV, TRAJ and TRAC genes are described in the IMGT database. Implicated in Translocations which frequently result from errors of the recombinase enzyme complex (RAG1, RAG2, etc.), which is responsable of the Immunoglobulin and T cell receptor V-J and V-D- J rearrangements. TRAV or TRAJ recombination signals or isolated heptamer are observed at the breakpoints. t(1;14)(p32;q11) t(8;14)(q24;q11) t(10;14)(q24;q11) t(11;14)(p13;q11) t(14;14)(q11;q32) References Lefranc M-P. Nomenclature of the human T cell Receptor genes (Review). Current Protocols in Immunology 2000, Wiley, J. and Sons, New York, Suppl 40, A.1O.1-A.1O.23 Lefranc M-P. and Lefranc G.. The T cell Receptor FactsBook (Review) Academic Press London UK 2001 Lefranc M-P. Locus Map and Genomic repertoire of the Human Immunoglobulin Genes (Review). The immunologist, 2000, 8,72-79 This article should be referenced as such: Lefranc MP. TRA (T cell Receptor Alpha). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 118
29 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review TRB (T cell Receptor Beta) Marie-Paule Lefranc IMGT, LIGM, IGH, UPR CNRS 1142, 141 rue de la Cardonille, Montpellier Cedex 5, France (MPL) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37641 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity HGNC (Hugo): TRB@ Location: 7q35 For complete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Note The human TRB locus is located on chromosome 7 on the long arm, at band 7q35. The orientation of the locus has been determined by the analysis of translocations, involving the TRB locus, in leukemia and lymphoma. DNA/RNA Description The human TRB locus at 7q35 spans 620 kb. It consists of TRBV genes belonging to 32 subgroups. Except for TRBV30, localized downstream of the TRBC2 gene, in inverted orientation of transcription, all the other TRBV genes are located upstream of a duplicated D-J-C-cluster, which comprises, for the first part one TRBD, six TRBJ, and the TRBC1 gene, and for the second part, one TRBD, eight TRBJ, and the TRBC2 gene. The most 5' TRBV genes occupy the most centromeric position, whereas the TRBV30 gene, 3' of the locus, is the most telomeric gene in the TRB locus. The potentiel repertoire consists of functional TRBV genes belonging to subgroups, the two TRBD, thirteen TRBJ (6 from the first cluster and 7 from the second cluster), and the two TRBC genes. Six TRBV orphons have been localized on chromosome 9 at 9p21. Enhancer sequences have been characterized 5.5 kb 3' from TRBC2. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 119
30 TRB (T cell Receptor Beta) Lefranc MP TRB V-GENE: Green box: Functional; Yellow box: Open reading frame; Red: Pseudogene. D-GENE: Blue: Functional. J-GENE: Grey: Functional. C-GENE: Blue: Functional. GENES NOT RELATED: Purple: Functional; Purple open box: Pseudogene. For compete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Protein Description Proteins encoded by the TRB locus are the T cell receptor beta chains. They result from the recombination (or rearrangement), at the DNA level, of three genes: TRBV, TRBD and TRBJ, with deletion of the intermediary DNA to create a rearranged TRBV-D- J gene. The rearranged TRBV-D-J gene is transcribed with one of the two TRBC genes and translated into a T cell receptor beta chain. Translation of the variable germline genes involved in the TRBV-D-J rearrangements are available at IMGT Repertoire Protein displays. TRB V-D-J rearrangements can be analysed using the IMGT/V- QUEST tool. Mutations Note Mutations which correspond to allelic polymorphisms of the functional germline TRBV, TRBD, TRBJ and TRBC genes are described in the IMGT database. Implicated in Translocations which frequently result from errors of the recombinase enzyme complexe (RAG1, RAG2, etc.), responsable of the Immunoglobulin and T cell receptor V-J and V-D- J rearrangements. TRBV, TRBD or TRBJ recombination signals or isolated heptamer are frequently observed at the breakpoints. t(7;9)(q35;q34) t(7;10)(q35;q24) t(7;11)(q35;p13) t(7;19)(q35;p13) References Lefranc M-P. Nomenclature of the human T cell Receptor genes (Review) Current Protocols in Immunology 2000, Wiley, J. and Sons, New York, Suppl 40, A.1O.1-A.1O.23 Lefranc M-P. and Lefranc G. The T cell Receptor FactsBook (Review) Academic Press London UK 2001 Lefranc M-P. Locus Map and Genomic repertoire of the Human Immunoglobulin Genes (Review). The immunologist, 2000, 8,72-79 This article should be referenced as such: Lefranc MP. TRB (T cell Receptor Beta). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 120
31 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review TRD (T cell Receptor Delta) Marie-Paule Lefranc IMGT, LIGM, IGH, UPR CNRS 1142, 141 rue de la Cardonille, Montpellier Cedex 5, France (MPL) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37642 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity HGNC (Hugo): TRD@ Location: 14q11.2 For complete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Note The TRD locus is embedded in the TRA locus, between the TRAV and TRAJ genes. The orientation of the locus has been determined by the analysis of translocations, involving the TRD locus, in leukemia and lymphoma. DNA/RNA Description The human TRD locus at 14q11.2 comprises a cluster of one TRDV gene (TRDV2), three TRDD segments, and four TRDJ segments, upstream of the unique TRDC gene; another TRDV gene (TRDV3) is localized downstream of the TRDC gene, in inverted orientation of transcription. This cluster spans 60 kb and is localized inside the TRA locus, between the TRAV genes and the TRAJ segments. One TRDV gene (TRDV1) is localized at 360 kb upstream of the TRDC gene, among the TRAV genes. Five variable genes have been found rearranged to both (D)J segments of the TRD locus and TRAJ segments, and can therefore be used for the synthesis of both delta and alpha chains. These genes are described as TRAV/DV. The TRDV genes are unique members of different subgroups. All the TRD genes are functional, with the exception of one TRAV/DV, which has been found either functional or as a pseudogene. Enhancer sequences have been described between the TRDJ3 and the TRDC gene. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 121
32 TRD (T cell Receptor Delta) Lefranc MP TRA/TRD V-GENE: Green box: Functional; Red: Pseudogene. D-GENE: Blue: Functional. J-GENE: Yellow: Functional; Pale yellow: Open reading frame; Red: Pseudogene. C-GENE: Blue: Functional. For compete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Protein Description Proteins encoded by the TRD locus are the T cell receptor delta chains. They result from the recombination (or rearrangement), at the DNA level, of three genes: TRDV, TRDD and TRDJ, with deletion of the intermediary DNA to create a rearranged TRDV-D- J gene. The rearranged TRDV-D-J gene is transcribed with the TRDC gene and translated into a T cell receptor delta chain. Translation of the variable germline genes involved in the TRDV-D-J rearrangements are available at IMGT Repertoire Protein displays. TRD V-D-J rearrangements can be analysed using the IMGT/V- QUEST tool. Mutations Note Mutations which correspond to allelic polymorphisms of the functional germline TRDV, TRDD, TRDJ and TRDC genes are described in the IMGT database. Implicated in Translocations which frequently result from errors of the recombinase enzyme complexe (RAG1, RAG2, etc.), responsable of the Immunoglobulin and T cell receptor V-J and V-D- J rearrangements. TRDV, TRDD or TRDJ recombination signals or isolated heptamer are frequently observed at the breakpoints. t(1;14)(p32;q11) t(10;14)(q24;q11) t(11;14)(p13;q11) t(11;14)(p15;q11) References Lefranc M-P. Nomenclature of the human T cell Receptor genes (Review). Current Protocols in Immunology 2000, Wiley, J. and Sons, New York, Suppl 40, A.1O.1-A.1O.23. Lefranc M-P. and Lefranc G. The T cell Receptor FactsBook (Review) Academic Press London UK Lefranc M-P. Locus Map and Genomic repertoire of the Human Immunoglobulin Genes (Review). The immunologist, 2000, 8, This article should be referenced as such: Lefranc MP. TRD (T cell Receptor Delta). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 122
33 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review TRG (T cell Receptor Gamma) Marie-Paule Lefranc IMGT, LIGM, IGH, UPR CNRS 1142, 141 rue de la Cardonille, Montpellier Cedex 5, France (MPL) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37643 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity HGNC (Hugo): TRG@ Location: 7p15-p14 For complete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Note The human TRG locus is located on chromosome 7, at band 7p15-p14 The orientation of the locus has been determined by the analysis of chromosome 7 inversions inv(7)(p15-q35), involving the TRG and TRB loci in ataxia telangiectasia patients, and in leukaemia. DNA/RNA Description The human TRG locus at 7p15-p14 spans 160 kb. It consists of TRGV genes belonging to 6 subgroups, upstream of a duplicated J-C-cluster, which comprises, for the first part, three TRGJ and the TRGC1 gene, and for the second part, two TRGJ and the TRGC2 gene. The most 5' TRGV genes occupy the most centromeric position, whereas the TRGC2 gene, 3' of the locus, is the most telomeric in the TRG locus. The potentiel repertoire consists of 4-6 functional TRGV genes belonging to two subgroups, the 5 TRGJ and the 2 TRGC genes. Polymorphisms in the number of TRGV genes and in the exon number of the TRGC2 gene have been described in different populations. Enhancer and silencer sequences have been characterized downstream of the TRGC2 gene. TRG V-GENE: Green box: Functional; Yellow box: Open reading frame; Red: Pseudogene; Triangle: Not sequenced. J-GENE: Grey: Functional. C-GENE: Blue: Functional. For compete Figure, see the international ImMunoGeneTics information system; Copyright IMGT. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 123
34 TRG (T cell Receptor Gamma) Lefranc MP Protein Description Proteins encoded by the TRG locus are the T cell receptor gamma chains. They result from the recombination (or rearrangement), at the DNA level, of two genes: TRGV and TRGJ, with deletion of the intermediary DNA to create a rearranged TRGV-J gene. The rearranged TRGV-J gene is transcribed with one of the two TRGC genes and translated into an T cell receptor gamma chain. Translation of the variable germline genes involved in the TRGV-J rearrangements are available at IMGT Repertoire Protein displays. TRG V-J rearrangements can be analysed using the IMGT/V-QUEST tool. Mutations Note Mutations which correspond to allelic polymorphisms of the functional germline TRGV, TRGJ and TRGC genes are described in the IMGT database. Implicated in Inversions which result from errors of the recombinase enzyme complexe (RAG1, RAG2, etc.), responsable of the Immunoglobulin and T cell receptor V-J and V-D-J rearrangements. TRGV or TRGJ recombination signals or isolated heptamer are observed at the breakpoints. References Lefranc M-P. Nomenclature of the human T cell Receptor genes (Review). Current Protocols in Immunology 2000, Wiley, J. and Sons, New York, Suppl 40, A.1O.1-A.1O.23. Lefranc M-P. and Lefranc G. The T cell Receptor FactsBook (Review) Academic Press London UK Lefranc M-P. Locus Map and Genomic repertoire of the Human Immunoglobulin Genes (Review). The immunologist, 2000, 8, This article should be referenced as such: Lefranc MP. TRG (T cell Receptor Gamma). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 124
35 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Leukaemia Section Mini Review Systemic mast cell disease (SMCD) Lidia Larizza, Alessandro Beghini Department of Biology and Genetics for Medical Sciences, Medical Faculty, University of Milan, Via Viotti 3/5, Milan, Italy (LL, AB) Published in Atlas Database: June 2000 Online updated version : DOI: /2042/37644 This article is an update of : Larizza L, Beghini A. Systemic mast cell disease (SMCD). Atlas Genet Cytogenet Oncol Haematol 1999;3(1): This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Note Mastocytosis is a heterogeneous clinical entity which is classified into four categories: 1- indolent mastocytosis (the most common form), 2- mastocytosis with an associated hematologic disorder, 3- mast cell leukemia and 4- aggressive mastocytosis. Clinics and pathology Phenotype/cell stem origin Mast cell Etiology Involvement of KIT/SCF has been demonstrated in a few cases, but the diversity of the clinical pattern has not yet been elucidated;increased soluble SCF has been reported in the skin of patient with indolent mastocytosis; c-kit mutations have been identified in patients with all forms of sporadic mastocytosis. Clinics Indolent mastocytosis involves the skin, bone marrow and gastrointestinal tract; clinical features range from a single cutaneous nodule to multiple pigmented macules resulting from increased epidermal melanin and papules (urticaria pigmentosa) or diffuse cutaneous involvement; bullae, vescicles and abnormal telangiectasia may be seen; gastrointestinal involvement leads to symptoms such as nausea, vomiting and abdominal pain. In mastocytosis with an associated hematological disorder the urticaria pigmentosa symptoms are accompanied by a variety of haematological findings due to mast cell infiltrates to bone marrow, spleen, liver and lymph nodes. Mast cell leukemia is characterized by proliferation and infiltration of immature mast cells in bone marrow, peripheral blood and various extramedullary tissues. Aggressive mastocytosis is characterized by aggressive involvement of several haematopoietic organs. Pathology Accumulation of mast cells in various organs and release of mast cell mediators which are responsible for the different clinical signs. Prognosis Highly dependent on the form being severe, often fatal, in all types with the exception of the indolent form. Genes involved and proteins KIT Location 4q12 DNA/RNA 21 exons Protein Transmembrane SCF/MGF receptor with tyrosine kinase activity; binding of ligand (SCF) induces receptor dimerization, autophosphorylation and signal transduction via molecules containing SH2- domains. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 125
36 Systemic mast cell disease (SMCD) Larizza L, Beghini A Somatic mutations Gly560Val, Asp816Val, Asp816Tyr, Asp820Gly. Asp816Val in peripheral blood lymphocytes (mastocytosis with an associated hematological disorder: AHD). Asp816Val in skin and spleen mast cells from patients with aggressive mastocytosis. Asp816Tyr in blasts from a patient with ANLL-M2 with mast cell involvement. Asp820 Gly in blasts from a patient with aggressive SMCD. Asp816Val and Gly560Val have been found in a human mast cell leukemia cell line (HMC1). Note All mutations with the exception of Gly560Val cluster to c-kit exon 17. Direct or indirect evidence has been provided that mutations affecting codon 816 promote ligand-independent autophosphorylation of the mutant receptor. SCF/MGF Location 12q22 DNA/RNA 9 exons Protein Soluble SCF: 248 aminoacids containing a proteolytic cleavage site encoded by exon 6 sequences, which is processed, giving rise to an active form (soluble) of 165 aminoacids; membrane-bound SCF: 220 aminoacids, results from alternative splicing of exon 6. Note: increased soluble SCF has been detected in the skin of patients with indolent mastocytosis; SCFspecific transcripts are detected by in situ RT-PCR in mast cell infiltrates in papulae from mastocytosis patients. References Furitsu T, Tsujimura T, Tono T, Ikeda H, Kitayama H, Koshimizu U, Sugahara H, Butterfield JH, Ashman LK, Kanayama Y. Identification of mutations in the coding sequence of the proto-oncogene c-kit in a human mast cell leukemia cell line causing ligand-independent activation of c-kit product. J Clin Invest Oct;92(4): Longley BJ Jr, Morganroth GS, Tyrrell L, Ding TG, Anderson DM, Williams DE, Halaban R. Altered metabolism of mast-cell growth factor (c-kit ligand) in cutaneous mastocytosis. N Engl J Med May 6;328(18): Nagata H, Worobec AS, Oh CK, Chowdhury BA, Tannenbaum S, Suzuki Y, Metcalfe DD. Identification of a point mutation in the catalytic domain of the protooncogene c-kit in peripheral blood mononuclear cells of patients who have mastocytosis with an associated hematologic disorder. Proc Natl Acad Sci U S A Nov 7;92(23): Longley BJ, Tyrrell L, Lu SZ, Ma YS, Langley K, Ding TG, Duffy T, Jacobs P, Tang LH, Modlin I. Somatic c-kit activating mutation in urticaria pigmentosa and aggressive mastocytosis: establishment of clonality in a human mast cell neoplasm. Nat Genet Mar;12(3):312-4 Pignon JM, Giraudier S, Duquesnoy P, Jouault H, Imbert M, Vainchenker W, Vernant JP, Tulliez M. A new c-kit mutation in a case of aggressive mast cell disease. Br J Haematol Feb;96(2):374-6 Beghini A, Cairoli R, Morra E, Larizza L. In vivo differentiation of mast cells from acute myeloid leukemia blasts carrying a novel activating ligand-independent C-kit mutation. Blood Cells Mol Dis Jun;24(2): This article should be referenced as such: Larizza L, Beghini A. Systemic mast cell disease (SMCD). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 126
37 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Leukaemia Section Short Communication t(11;17)(q23;p13) Jean-Loup Huret Genetics, Dept Medical Information, University of Poitiers, CHU Poitiers Hospital, F Poitiers, France (JLH) Published in Atlas Database: June 2000 Online updated version : DOI: /2042/37645 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Note The translocation may be hidden, and can present as a del(11)(q23). Clinics and pathology Disease Acute non lymphocytic leukemia (ANLL), acute lymphocytic leukemia (ALL) Clinics Poorly known: a few cases, but without ascertainement of GAS7 involvement (a gene in 17p13); they are cases of MDS in transformation, ANLL, bi-phenotypic leukemia, and ALL; one the other hand, one case of t(11;17) with MLL and GAS7 involvementshas been described: a 13-year-old boy, with a M4 treatmentrelated ANLL and a karyotype with an apparent del(11)(q23) (a cryptic t(11;17)(q23;p13)); four months later the patient died. Genes involved and proteins MLL Location 11q23 DNA/RNA 21 exons, spanning over 100 kb; kb mrna. Protein 431 kda; contains two DNA binding motifs (a AT hook, and Zinc fingers), a DNA methyl transferase motif, a bromodomain; transcriptional regulatory factor; nuclear localisation. GAS7 Location 17p13 DNA/RNA Genomic sequence 167 kb; 14 exons. Protein Expression in quiescent/terminally differenciated cells, especially in the brain. Result of the chromosomal anomaly Hybrid gene Description 5' MLL-3' GAS7; breakpoint in MLL intron 8, in MLL bcr; GAS7 breakpoint upstream of exon 1; fusion of MLL exon 7 or exon 8 to GAS7 exon 2. References Harrison CJ, Cuneo A, Clark R, Johansson B, Lafage- Pochitaloff M, Mugneret F, Moorman AV, Secker-Walker LM. Ten novel 11q23 chromosomal partner sites. European 11q23 Workshop participants. Leukemia May;12(5): Megonigal MD, Cheung NK, Rappaport EF, Nowell PC, Wilson RB, Jones DH, Addya K, Leonard DG, Kushner BH, Williams TM, Lange BJ, Felix CA. Detection of leukemia-associated MLL-GAS7 translocation early during chemotherapy with DNA topoisomerase II inhibitors. Proc Natl Acad Sci U S A Mar 14;97(6): This article should be referenced as such: Huret JL. t(11;17)(q23;p13). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3):127. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 127
38 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Leukaemia Section Short Communication t(11;17)(q23;q12) MLL/RARa Jean-Loup Huret Genetics, Dept Medical Information, University of Poitiers, CHU Poitiers Hospital, F Poitiers, France (JLH) Published in Atlas Database: June 2000 Online updated version : DOI: /2042/37646 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Clinics and pathology Disease M5 acute non lymphocytic leukemia (ANLL). Clinics Poorly known: one case, a 39 year old man with 47,XY,+5,t(11;17)(q23;q12). Genes involved and proteins MLL Location 11q23 DNA/RNA 21 exons, spanning over 100 kb; kb mrna. Protein 431 kda; contains two DNA binding motifs (a AT hook, and Zinc fingers), a DNA methyl transferase motif, a bromodomain; transcriptional regulatory factor; nuclear localisation. RARa Location 17q12-21 Protein Wide expression; nuclear receptor; binds specific DNA sequences: HRE (hormone response elements); ligand and dimerization domain; role in growth and differentiation. References Shekhter-Levin S, Gollin SM, Kaplan SS, Redner RL. Involvement of the MLL and RARalpha genes in a patient with acute monocytic leukemia with t(11;17)(q23;q12) Leukemia Mar;14(3):520-2 This article should be referenced as such: Huret JL. t(11;17)(q23;q12) MLL/RARa. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3):128. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 128
39 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Leukaemia Section Short Communication +4 or trisomy 4 Alessandro Beghini Department of Biology and Genetics for Medical Sciences, Medical Faculty, University of Milan, Via Viotti 3/5, Milan, Italy (AB) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37648 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Clinics and pathology Disease Acute non lymphocytic leukaemia (ANLL) Epidemiology +4 as the sole anomaly is a rare chromosomal abnormality associated with a specific subtype of primary ANLL and secondary (treatment related) ANLL with myelomonocytic morphology; it has been found with the same frequence in the M1-M2 and M4 French-American-British (FAB) phenotypes. Association of +4 with double minute chromosomes has been described in ten cases; five with AML-M2, two with AML-M4, one with refractory anemia with excess of blasts in transformation (RAEB-T), one with chronic myelomonocytic leukemia (CMMoL) and one with unclassified preleukemia. The coincidence of +4 with t(8;21) or its variant t(6;21;8), observed in at least two cases of ANLL (M1 and M2), is therefore recurrent. Prognosis Apparently +4 has no prognostic significance in ANLL; with the exception of the cases bearing c-kit mutations who are associated with a rapid disease progression. Disease Acute lymphocytic leukaemia (ALL) Epidemiology +4 has been described in two cases of T-cell acute lymphoblastic leukemia as the sole chromosomal anomaly. Combined trisomies of chromosomes 4 and 10 are found in children with B-progenitor cell acute lymphoblastic leukemia with a favourable prognostic association. Prognosis Patients with chromosomes 4 and 10 trisomies have an extremely favourable 4-year event-free survival (EFS) after antimetabolite-based chemotherapy. Disease Thecoma of the ovary Epidemiology +4 associated with +12 has been described in two cases of fibrothecoma and in one case of thecoma; it has been suggested that acquisition of trisomy 4 may constitute a second cytogenetic step in tumor progression of ovarian thecoma/fibrothecoma. Prognosis No prognostic significance. Disease +4 has been found as the sole aberration only in one case of uterine leiomyoma. Genetics Note Trisomy 4 was proven to lead to duplication of Asp816Tyr mutation of c-kit gene (that maps to 4q12) in a case of M2-ANLL with mast cell involvement. References Mecucci C, Van Orshoven A, Tricot G, Michaux JL, Delannoy A, Van den Berghe H. Trisomy 4 identifies a subset of acute nonlymphocytic leukemias. Blood May;67(5): Harris MB, Shuster JJ, Carroll A, Look AT, Borowitz MJ, Crist WM, Nitschke R, Pullen J, Steuber CP, Land VJ. Trisomy of leukemic cell chromosomes 4 and 10 identifies children with B- progenitor cell acute lymphoblastic leukemia with a very low risk of treatment failure: a Pediatric Oncology Group study. Blood Jun 15;79(12): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 129
40 +4 or trisomy 4 Beghini A Mrózek K, Limon J, Debniak J, Emerich J. Trisomy 12 and 4 in a thecoma of the ovary. Gynecol Oncol Apr;45(1):66-8 Kwong YL, Ha SY, Liu HW, Chan LC. Trisomy 4 may occur in a broad range of hematologic malignancies. Cancer Genet Cytogenet Sep;69(2): Feuring-Buske M, Haase D, Könemann S, Troff C, Grove D, Hiddemann W, Wörmann B. Trisomy 4 in 'stem cell-like' leukemic cells of a patient with AML. Leukemia Aug;9(8): O'Malley F, Rayeroux K, Cole-Sinclair M, Tong M, Campbell LJ. MYC amplification in two further cases of acute myeloid leukemia with trisomy 4 and double minute chromosomes. Cancer Genet Cytogenet Mar;109(2):123-5 Beghini A, Ripamonti CB, Castorina P, Pezzetti L, Doneda L, Cairoli R, Morra E, Larizza L. Trisomy 4 leading to duplication of a mutated KIT allele in acute myeloid leukemia with mast cell involvement. Cancer Genet Cytogenet May;119(1):26-31 This article should be referenced as such: Beghini A. +4 or trisomy 4. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 130
41 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Leukaemia Section Mini Review 3q27 rearrangements in non Hodgkin lymphoma, t(3;var)(q27;var) in non Hodgkin lymphoma Antonio Cuneo, Jean-Loup Huret Hematology Section, Department of Biomedical Sciences, University of Ferrara, Corso Giovecca 203, Ferrara, Italy (AC), Genetics, Dept Medical Information, University of Poitiers, CHU Poitiers Hospital, F Poitiers, France (JLH) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37647 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Note 3q27 rearrangements occur in distinct clinicopathological entities of B-cell non Hodgkin lymphoma (NHL), including diffuse large cell lymphoma (DLCL), follicle centre cell lymphoma (FCCL) and marginal zone B-cell lymphoma (MZBCL) in the REAL classification; very rare cases were also reported with mantle cell lymphoma and chronic lymphocytic leukemia. 3q27 breaks are usually, but not invariably, associated with rearrangements of the BCL6 gene located at the 3q27 chromosome band; likewise rearrangements of this gene may occur without detectable 3q27 breaks. t(3;22)(q27;q11) - Courtesy Diane H. Norback, Eric B. Johnson, Sara Morrison-Delap Cytogenetics at thewaisman Center. Clinics and pathology Disease Diffuse large cell lymphoma (DLCL) Note This biologically heterogeneous group of lymphomas in the REAL proposal accounts for as many as 40% of NHL in western countries and includes the entities of centroblastic lymphoma, immunoblastic lymphoma and B-cell anaplastic lymphoma recognized by the Kiel classification. Phenotype/cell stem origin The cell of origin is probably a large transformed B- cell, frequently deriving from the follicle centre, harbouring somatic hypermutation of the Ig genes and ongoing mutations (antigen driven stimulation). The phenotype is usually CD19+, CD22+, CD10-/+, SIg+. Epidemiology 10-20% of DLCL carry 3q27 translocations detectable at banding analysis, appoximately 50% of which may be expected to be associated with BCL6 rearrangement; molecular genetic methods proved very efficient in demonstrating this genetic lesion and studies using southern blotting detecting BCL6 breaks in the 4.0 kb major breakpoint region showed 20-30% of unselected DLCL to be rearranged. Pathology There is no distinctive histological features in DLCL with 3q27/BCL6 rearrangement as compared with other Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 131
42 3q27 rearrangements in non Hodgkin lymphoma Cuneo A, Huret JL DLCL; the proliferation consists of a diffuse infiltrate of large cells with vescicular nuclei and prominent nucleoli with basophilic cytoplasm; criteria for distinguishing those cases with a predominance of immunoblasts or of anaplastic B-cells were put forward but were felt not to be enough reproducible as to allow for proper categorization of distinct pathological entities; 3q27 abnormalities were seen in similar frequency in the immunoblastic variant and in the centroblastic variant of DLCL in a study. Prognosis A predominance of extra-nodal forms and a relatively favourable outcome was observed in BCL6-rearranged DLCL but BCL6 failed as a prognostic indicator when compared to other molecular genetic lesions; thus, the assessment of the prognostic significance of 3q27 or BCL6 breaks in DLCL needs further investigation in prospective studies. Disease Follicle centre cell lymphoma (FCCL) Note FCCL accounts for approximately 30-40% of all NHL in western countries. Phenotype/cell stem origin The neoplasia derives from centrocytes / centroblasts unable to progress through the germinal centre, carrying somatic hypermutation of the IgV genes and a pan-b+, CD10+/-, CD5-, sig+ phenotype. Epidemiology 3q27 translocations involving the chromosome regions where Ig genes are located (2p11: IgK, 14q32: IgH, 22q11: IgL) were detected in 6.5% of FCCL; a 16% incidence for any 3q27 break was reported; the association of 3q27/BCL6 involvement with the classical t(14;18) was described; molecular genetic studies found a 6-14% incidence for BCL6 rearrangement in FCCL. Prognosis No specific correlation was established between 3q27 breaks and specific clinicopathological features of FCCL. Disease Marginal zone B-cell lymphoma (MZBCL) Note 7-8% of NHL show the clinicopathological features of MZBCL. Phenotype/cell stem origin The transformed cells derive form marginal zone lymphocytes harbouring hypermutated IgV genes with the following phenotype: pan-b+, CD5-/+, CD10-, CD23-, CD11c+/-, cyig +(40% of the cells), sigm+ bright, sigd-. Epidemiology A minority of MZBCL may carry a 3q27/BCL6 translocation, mostly t(3;14)(q27;q32). Clinics There is no distinctive clinicopathological feature in this cytogenetic subset of MZBCL, but a predominance of extra-nodal forms over splenic and nodal types and an excess of large blast-like cells were noted. Genetics Note Below are listed translocations involving -or likely to involve- BCL6 in 3q27, and a partner gene in the other breakpoint. Cytogenetics Cytogenetics morphological t(2;3)(p12;q27): the gene in 2p12 is IgK. t(3;3)(q27;q29): the gene in 3q29 is TFRC, the transferrin receptor. t(3;4)(q27;p13): the gene in 4p13 is RHOH, a GTPase of the Ras superfamily; role in signal transduction. t(3;6)(q27;p22): the gene in 6p22 is histone H1F1, an architectural protein with a role in chromatin condensation and in gene regulation. t(3;6)(q27;p21.2): the gene in 6p21.2 is PIM-1, a protein kinase. t(3;7)(q27;p12): the gene in 7p12 is ZNFN1A1/Ikaros, a Zn finger protein involved in cell differentiation. t(3;8)(q27;q24) t(3;11)(q27;q23): the gene in 11q23 is OBF1, a B-cell specific transcriptional coactivator. t(3;13)(q27;q14): the gene in 13q14 is LCP1/L-plastin, a gene which belongs to an actin-binding protein family. t(3;14)(q27;q32): the gene in 14q32 is IgH. t(3;15)(q27;q22). t(3;16)(q27;p13): the gene in 16p13 is MHC2TA/CIITA, a Class II histocompatibility complex transactivator. t(3;17)(q27;q11). t(3;18)(q27;p11.2): the gene in 18p11.2 is EIF4A2, a DEAD box helicase. t(3;22)(q27;q11): the gene in 22q11 is IgL. t(3;?)(q27;?): the gene is HSP89A, a member of the HSP90 sub-family of the heat-shock protein (HSP) family. Finally, breakpoints in 1p34, 1p32, 2q21, 3p14, 6q23, 12p13, 14q11, 16p11.2,and 16p13 have also been described. However, cases of apparently simple translocations involving 3q27 -but not 14q32- (e.g. t(1;3)(q21;q37), or t((3;6)(q27;p25)) have disclosed insertion of IgH sequences within the 3q27 breakpoint. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 132
43 3q27 rearrangements in non Hodgkin lymphoma Cuneo A, Huret JL Moreover, in a substantial percentage of cases, a breakpoint in 3q27 in NHL is accompanied with germline BCL6: another gene is likely to be implicated in these cases (or else, the rearranged sequence, although distant, still disregulates BCL6). Cytogenetics molecular 3q27 anomalies are often associated with well known primary anomalies such as t(8;14)(q24;q32), t(11;14)(q13;q32), t(14,18)(q32;q21). Genes involved and proteins BCL6 Location 3q27 Note BCL6 mutations are regarded as a genetic marker of B- cell transition through the germinal center. DNA/RNA 10 exons; alternative splicing of exons 1 (1a and 1b), without modification of the open reading frame. Protein Transcription factor; belongs to the Krüppel family, with a N-term BTB/POZ domain and 6 zinc fingers; transcription repressor. Result of the chromosomal anomaly Hybrid gene Note The translocation partners of BCL6 are not confined to the immunoglobulin superfamily, contrarily to the situation found with c-myc, BCL1, or BCL2. Description Breakpoint in the first non-coding exon (containing the 2 promoters) or the first intron of BCL6; the partner gene therefore fuses with the second exon of BCL6, resulting in a 5' partner - 3' BCL6 fusion transcript; it is supposed that substitution of the promoter of BCL6 may be responsible for BCL6 regulation, or that a break in the breakpoint cluster region of BCL6 may inhibit a sequence involved in BCL6 regulation; partners other than immunoglobulin lack homology with switch regions, VDJ sequences, or Chi sequences. Fusion protein Description No fusion protein; the 5' regulatory region of BCL6 is replaced by the 5' regulatory region of the partner gene. References Baron BW, Nucifora G, McCabe N, Espinosa R 3rd, Le Beau MM, McKeithan TW. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc Natl Acad Sci U S A Jun 1;90(11): Bastard C, Deweindt C, Kerckaert JP, Lenormand B, Rossi A, Pezzella F, Fruchart C, Duval C, Monconduit M, Tilly H. LAZ3 rearrangements in non-hodgkin's lymphoma: correlation with histology, immunophenotype, karyotype, and clinical outcome in 217 patients. Blood May 1;83(9): Offit K, Lo Coco F, Louie DC, Parsa NZ, Leung D, Portlock C, Ye BH, Lista F, Filippa DA, Rosenbaum A. Rearrangement of the bcl-6 gene as a prognostic marker in diffuse large-cell lymphoma. N Engl J Med Jul 14;331(2):74-80 Tilly H, Rossi A, Stamatoullas A, Lenormand B, Bigorgne C, Kunlin A, Monconduit M, Bastard C. Prognostic value of chromosomal abnormalities in follicular lymphoma. Blood Aug 15;84(4): Dallery E, Galiègue-Zouitina S, Collyn-d'Hooghe M, Quief S, Denis C, Hildebrand MP, Lantoine D, Deweindt C, Tilly H, Bastard C. TTF, a gene encoding a novel small G protein, fuses to the lymphoma-associated LAZ3 gene by t(3;4) chromosomal translocation. Oncogene Jun 1;10(11): Wlodarska I, Mecucci C, Stul M, Michaux L, Pittaluga S, Hernandez JM, Cassiman JJ, De Wolf-Peeters C, Van den Berghe H. Fluorescence in situ hybridization identifies new chromosomal changes involving 3q27 in non-hodgkin's lymphomas with BCL6/LAZ3 rearrangement. Genes Chromosomes Cancer Sep;14(1):1-7 Miura I, Ohshima A, Takahashi N, Hashimoto K, Nimura T, Utsumi S, Saito M, Miki T, Hirosawa S, Miura AB. A new nonrandom chromosomal translocation t(3;6)(q27;p21.3) associated with BCL6 rearrangement in two patients with non- Hodgkin's lymphoma. Int J Hematol Oct;64(3-4): Akasaka T, Miura I, Takahashi N, Akasaka H, Yonetani N, Ohno H, Fukuhara S, Okuma M. A recurring translocation, t(3;6)(q27;p21), in non-hodgkin's lymphoma results in replacement of the 5' regulatory region of BCL6 with a novel H4 histone gene. Cancer Res Jan 1;57(1):7-12 Dierlamm J, Pittaluga S, Stul M, Wlodarska I, Michaux L, Thomas J, Verhoef G, Verhest A, Depardieu C, Cassiman JJ, Hagemeijer A, De Wolf-Peeters C, Van den Berghe H. BCL6 gene rearrangements also occur in marginal zone B-cell lymphoma. Br J Haematol Sep;98(3): Maes M, Depardieu C, Dargent JL, Hermans M, Verhaeghe JL, Delabie J, Pittaluga S, Troufléau P, Verhest A, De Wolf- Peeters C. Primary low-grade B-cell lymphoma of MALT-type occurring in the liver: a study of two cases. J Hepatol Nov;27(5):922-7 Ohno H, Fukuhara S. Significance of rearrangement of the BCL6 gene in B-cell lymphoid neoplasms. Leuk Lymphoma Sep;27(1-2):53-63 Chaganti SR, Chen W, Parsa N, Offit K, Louie DC, Dalla- Favera R, Chaganti RS. Involvement of BCL6 in chromosomal aberrations affecting band 3q27 in B-cell non-hodgkin lymphoma. Genes Chromosomes Cancer Dec;23(4):323-7 Chaganti SR, Rao PH, Chen W, Dyomin V, Jhanwar SC, Parsa NZ, Dalla-Favera R, Chaganti RS. Deregulation of BCL6 in non-hodgkin lymphoma by insertion of IGH sequences in complex translocations involving band 3q27. Genes Chromosomes Cancer Dec;23(4): Kramer MH, Hermans J, Wijburg E, Philippo K, Geelen E, van Krieken JH, de Jong D, Maartense E, Schuuring E, Kluin PM. Clinical relevance of BCL2, BCL6, and MYC rearrangements in diffuse large B-cell lymphoma. Blood Nov 1;92(9): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 133
44 3q27 rearrangements in non Hodgkin lymphoma Cuneo A, Huret JL Daudignon A, Bisiau H, Le Baron F, Laï JL, Wetterwald M, Galiègue-Zouitina S, Morel P, Duthilleul P. Four cases of follicular lymphoma with t(14;18)(q32;q21) and t(3;4)(q27;p13) with LAZ3 (BCL6) rearrangement. Cancer Genet Cytogenet Jun;111(2): Galiègue-Zouitina S, Quief S, Hildebrand MP, Denis C, Detourmignies L, Laï JL, Kerckaert JP. Nonrandom fusion of L- plastin(lcp1) and LAZ3(BCL6) genes by t(3;13)(q27;q14) chromosome translocation in two cases of B-cell non-hodgkin lymphoma. Genes Chromosomes Cancer Oct;26(2): Richardson MA. Research of complementary/alternative medicine therapies in oncology: promising but challenging. J Clin Oncol Nov;17(11 Suppl):38-43 Schlegelberger B, Zwingers T, Harder L, Nowotny H, Siebert R, Vesely M, Bartels H, Sonnen R, Hopfinger G, Nader A, Ott G, Müller-Hermelink K, Feller A, Heinz R. Clinicopathogenetic significance of chromosomal abnormalities in patients with blastic peripheral B-cell lymphoma. Kiel-Wien-Lymphoma Study Group. Blood Nov 1;94(9): Yoshida S, Kaneita Y, Aoki Y, Seto M, Mori S, Moriyama M. Identification of heterologous translocation partner genes fused to the BCL6 gene in diffuse large B-cell lymphomas: 5'-RACE and LA - PCR analyses of biopsy samples. Oncogene Dec 23;18(56): Hosokawa Y, Maeda Y, Ichinohasama R, Miura I, Taniwaki M, Seto M. The Ikaros gene, a central regulator of lymphoid differentiation, fuses to the BCL6 gene as a result of t(3;7)(q27;p12) translocation in a patient with diffuse large B- cell lymphoma. Blood Apr 15;95(8): Xu WS, Liang RH, Srivastava G. Identification and characterization of BCL6 translocation partner genes in primary gastric high-grade B-cell lymphoma: heat shock protein 89 alpha is a novel fusion partner gene of BCL6. Genes Chromosomes Cancer Jan;27(1):69-75 This article should be referenced as such: Cuneo A, Huret JL. 3q27 rearrangements in non Hodgkin lymphoma, t(3;var)(q27;var) in non Hodgkin lymphoma. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 134
45 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Solid Tumour Section Mini Review Soft tissue tumors: Lipoma / benign lipomatous tumors Nils Mandahl Department of Clinical Genetics, Lund University Hospital, Lund, Sweden (NM) Published in Atlas Database: May 2000 Online updated version : DOI: /2042/37650 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Note Lipomas are benign adipose tissue tumors with many subtypes, constituting one-third of all soft tissue tumors. Classification Ordinary lipoma: the solitary, ordinary lipomas represent the most common soft tissue tumors, with subcutaneous tumors being much more common than the deep-seated ones. Epidemiology: the incidence of lipomas is about one in 1000 inhabitants per year, but is probably underestimated since many lesions cause few problems; they occur most frequently between 30 and 70 years of age, with a peak incidence between 40 and 60 years. Clinics: solitary lipomas are slow-growing masses, most frequently located in the upper back, neck, shoulder, abdomen, and the proximal portions of the extremities. Evolution: surgery is required primarily when the tumors reach large size and cause cosmetic problems or complications due to their anatomical site; there is no risk of progression to malignancy, and recurrences are rare after shelling-out. Angiolipoma: angiolipomas show characteristic histological features and occur primarily as subcutaneous, painful nodules; multiple lesions are much more common than solitary ones; they are usually smaller than solitary, ordinary lipomas and present at an earlier age, with the dominating site being the forearm; an increased familial incidence has been noted. Spindle cell/pleomorphic lipoma: the characteristic feature of spindle cell lipoma is the replacement of mature fat by collagen-forming spindle cells; these rare tumors typically occur subcutaneously in the neck and shoulders of men aged 45 to 65 years. Pleomorphic lipoma probably represents a highly pleomorphic variant of spindle cell lipoma and show similar clinical features; both of these types of lesions may be confused with liposarcoma. Lipoblastoma: lipoblastoma is a tumor of the infancy; most tumors occur before three years of age and may occasionally be seen already at birth; the site is primarily the upper and lower extremities; there may be a close resemblance to myxoid liposarcoma. Hibernoma: hibernoma is chiefly a tumor of adults, although in average occurring at lower ages than solitary, ordinary lipomas; the most common sites are the scapular and interscapular regions, mediastinum and upper thorax. Angiomyo-lipoma: angiomyolipoma is a hamartomatous lesion in the kidneys of adult patients, with a preponderance of women. Chondroid lipoma: chondroid lipoma is a rare tumor occurring in the subcutis or muscle of adults; it may be confused with liposarcoma and chondrosarcoma, and shows microscopic features of both lipoma and hibernoma. Cytogenetics Cytogenetics Morphological Ordinary lipoma: The majority of tumors show fairly simple, structural chromosome aberrations; numerical changes are rare; seemingly balanced aberrations dominate over unbalanced changes, as also indicated by the finding of no copy number changes in smaller series of lipomas investigated by CGH. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 135
46 Soft tissue tumors: Lipoma / benign lipomatous tumors Mandahl N In more than two-thirds of cases with karyotypic changes, there is an involvement of chromosome segment 12q13-15, which may recombine with a large variety of other chromosomal segments; so far, more than 80 of the 320 chromosome bands in the standard karyotype have been involved in these rearrangements. The most frequent aberration is t(3;12)(q27-28;q14-15), found in one-fifth of cases with 12q13-15 changes. Other recurrently involved chromosome segments include 1p36, 1p32-34, 2p22-24, 2q35-37, 5q33, 10q22, 11q13, 12p11, 12q24, 13q12-14, and 21q21-22; all recurrent changes combined represent 75% of cases with 12q13-15 changes and 45-50% of all karyotypically abnormal ordinary lipomas among cases without 12q13-15 changes, one subset of tumors display translocations involving 6p21-23 and another subset have loss of 13q material with breakpoints in 13q12-14 and/or 13q22; many of the latter aberrations are seen as del(13)(q12q22); these 6p and 13q changes are found at similar frequencies, each occuring in 6-8% of ordinary lipomas with cytogenetic aberrations. Some 15-20% of the aberrant tumors have rearrangements involving neither 6p21-23, 12q13-15, 13q12-14 nor 13q22. Cytogenetics Molecular The rearrangements frequently affect the high mobility group protein gene HMGIC in 12q15, with most breakpoints occuring in the large intron 3; the outcome of the 3;12-translocations is the formation of a chimeric gene involving HMGIC and LPP. LPP is a member of the LIM protein gene family, containing a leucin-zipper motif in its amino-terminal region and three LIM domains in its carboxy-terminal region. Result of the chromosomal anomaly: the HMGIC/LPP transcripts frequently contain coding sequences for the three DNA-binding domains of HMGIC and two or three LIM domains from LPP; HMGIC/LPP is not specific for lipomas, but has also been identified in a series of pulmonary chondroid hamartomas with t(3;12), and HMGIC is rearranged in a variety of benign tumors. Another fusion has been with LHFP in a lipoma with t(12;13)(q13-15;q12); the expressed HMGIC/LHFP fusion transcript encoded the three DNA binding domains of HMGIC and 69 amino acids from frameshifted LHFP sequences. A candidate gene for rearrangements affecting 6p21 in lipomas is HMGIY. Cytogenetics Morphological Angiolipoma: subcutaneous angiolipomas have a normal karyotype as shown by chromosome banding. Spindle cell/pleomorphic lipoma: few cases have been investigated cytogenetically; the characteristic features are losses of chromosome 13 and 16 sequences, in particular involving the segments 13q12 and 16q13-qter, respectively. Lipoblastoma: few cases have been investigated cytogenetically; they have all had rearrangements of 8q11-13 in common; this segment has been found to recombine with a variety of other chromosome bands; preliminary data have indicated that the PLAG1 gene in 8q12 is affected, resulting in a similar promoter swapping as has been described in pleomorphic adenomas of the salivary gland. Hibernoma: the characteristic feature is rearrangements of 11q13, which may recombine with a variety of other chromosome bands; in contrast to ordinary lipomas, no translocations involving both 11q13 and 12q13-15 have been found in hibernomas. FISH analyses have demonstrated that both homologs of chromosome 11, including the seemingly normal one, have deletions encompassing the multiple endocrine neoplasia type I, MEN1, locus and a second region about 3 Mb distal to MEN1. Angiomyo-lipoma: few cases have been investigated cytogenetically; the only recurrent change identified has been trisomy 7; however, interphase FISH analysis did not reveal any significant fraction of cells with +7 in the angiomyolipomas investigated. Chondroid lipoma: a cytogenetically identical rearrangement, t(11;16)(q13;p13), found in the only two cases reported indicate that this is a recurrent aberration. References Peulve P, Thomine E, Hemet J. Cytogenetic analysis of a rare case of pediatric myxolipoma. Ann Genet. 1990;33(4):222-4 Sreekantaiah C, Leong SP, Karakousis CP, McGee DL, Rappaport WD, Villar HV, Neal D, Fleming S, Wankel A, Herrington PN. Cytogenetic profile of 109 lipomas. Cancer Res Jan 1;51(1): Debiec-Rychter M, Saryusz-Wolska H, Salagierski M. Cytogenetic analysis of renal angiomyolipoma. Genes Chromosomes Cancer Jan;4(1):101-3 Fletcher JA, Kozakewich HP, Schoenberg ML, Morton CC. Cytogenetic findings in pediatric adipose tumors: consistent rearrangement of chromosome 8 in lipoblastoma. Genes Chromosomes Cancer Jan;6(1):24-9 Mrózek K, Karakousis CP, Bloomfield CD. Chromosome 12 breakpoints are cytogenetically different in benign and malignant lipogenic tumors: localization of breakpoints in lipoma to 12q15 and in myxoid liposarcoma to 12q13.3. Cancer Res Apr 1;53(7): Tayyeb MT, Neff JR, Bridge JA. A case report of fibrolipoma with t(12;16)(q13;q24). Cancer Genet Cytogenet Jun;67(2):145-6 Dal Cin P, Sciot R, De Wever I, Van Damme B, Van den Berghe H. New discriminative chromosomal marker in adipose tissue tumors. The chromosome 8q11-q13 region in lipoblastoma. Cancer Genet Cytogenet Dec;78(2):232-5 Mandahl N, Höglund M, Mertens F, Rydholm A, Willén H, Brosjö O, Mitelman F. Cytogenetic aberrations in 188 benign and borderline adipose tissue tumors. Genes Chromosomes Cancer Mar;9(3): Mandahl N, Mertens F, Willén H, Rydholm A, Brosjö O, Mitelman F. A new cytogenetic subgroup in lipomas: loss of Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 136
47 Soft tissue tumors: Lipoma / benign lipomatous tumors Mandahl N chromosome 16 material in spindle cell and pleomorphic lipomas. J Cancer Res Clin Oncol. 1994;120(12): Meloni AM, Spanier SS, Bush CH, Stone JF, Sandberg AA. Involvement of 10q22 and 11q13 in hibernoma. Cancer Genet Cytogenet Jan;72(1):59-64 Mertens F, Rydholm A, Brosjö O, Willén H, Mitelman F, Mandahl N. Hibernomas are characterized by rearrangements of chromosome bands 11q Int J Cancer Aug 15;58(4):503-5 Ashar HR, Fejzo MS, Tkachenko A, Zhou X, Fletcher JA, Weremowicz S, Morton CC, Chada K. Disruption of the architectural factor HMGI-C: DNA-binding AT hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell Jul 14;82(1):57-65 Bridge JA, DeBoer J, Walker CW, Neff JR. Translocation t(3;12)(q28;q14) in parosteal lipoma. Genes Chromosomes Cancer Jan;12(1):70-2 Enzinger FM, Weiss SW. Lipoma In: Soft tissue tumors. 3rd ed. Mosby. St. Louis Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet Aug;10(4): Ashar HR, Cherath L, Przybysz KM, Chada K. Genomic characterization of human HMGIC, a member of the accessory transcription factor family found at translocation breakpoints in lipomas. Genomics Jan 15;31(2): Fletcher CD, Akerman M, Dal Cin P, de Wever I, Mandahl N, Mertens F, Mitelman F, Rosai J, et al. Correlation between clinicopathological features and karyotype in lipomatous tumors. A report of 178 cases from the Chromosomes and Morphology (CHAMP) Collaborative Study Group. Am J Pathol Feb;148(2): Mandahl N. Cytogenetics and molecular genetics of bone and soft tissue tumors. Adv Cancer Res. 1996;69:63-99 Petit MM, Mols R, Schoenmakers EF, Mandahl N, Van de Ven WJ. LPP, the preferred fusion partner gene of HMGIC in lipomas, is a novel member of the LIM protein gene family. Genomics Aug 15;36(1): Dal Cin P, Sciot R, Van Poppel H, Baert L, Van Damme B, Van den Berghe H. Chromosome analysis in angiomyolipoma. Cancer Genet Cytogenet Dec;99(2):132-4 Kanazawa C, Mitsui T, Shimizu Y, Saitoh E, et al. Chromosomal aberration in lipoblastoma: a case with 46,XX,ins(8;6)(q11.2;q13q27). Cancer Genet Cytogenet Jun;95(2):163-5 Merscher S, Marondel I, Pedeutour F, Gaudray P, Kucherlapati R, Turc-Carel C. Identification of new translocation breakpoints at 12q13 in lipomas. Genomics Nov 15;46(1):70-7 Sciot R, Akerman M, Dal Cin P, De Wever I, Fletcher CD, Mandahl N, Mertens F, Mitelman F, Rosai J, Rydholm A, Tallini G, Van den Berghe H, Vanni R, Willen H. Cytogenetic analysis of subcutaneous angiolipoma: further evidence supporting its difference from ordinary pure lipomas: a report of the CHAMP Study Group. Am J Surg Pathol Apr;21(4):441-4 Szymanska J, Virolainen M, Tarkkanen M, Wiklund T, et al. Overrepresentation of 1q21-23 and 12q13-21 in lipoma-like liposarcomas but not in benign lipomas: a comparative genomic hybridization study. Cancer Genet Cytogenet Nov;99(1):14-8 Tallini G, Dal Cin P, Rhoden KJ, Chiapetta G, Manfioletti G, Giancotti V, Fusco A, Van den Berghe H, Sciot R. Expression of HMGI-C and HMGI(Y) in ordinary lipoma and atypical lipomatous tumors: immunohistochemical reactivity correlates with karyotypic alterations. Am J Pathol Jul;151(1):37-43 Nanjangud G, Naresh KN, Nair CN, Parikh B, Dixit PH, Advani SH, Amare PS. Translocation (11;14)(q13;q32) and overexpression of cyclin D1 protein in a CD23-positive lowgrade B-cell neoplasm. Cancer Genet Cytogenet Oct 1;106(1):37-43 Petit MM, Swarts S, Bridge JA, Van de Ven WJ. Expression of reciprocal fusion transcripts of the HMGIC and LPP genes in parosteal lipoma. Cancer Genet Cytogenet Oct 1;106(1):18-23 Rogalla P, Kazmierczak B, Meyer-Bolte K, Tran KH, Bullerdiek J. The t(3;12)(q27;q14-q15) with underlying HMGIC-LPP fusion is not determining an adipocytic phenotype. Genes Chromosomes Cancer Jun;22(2):100-4 Sreekantaiah C. The cytogenetic and molecular characterization of benign and malignant soft tissue tumors. Cytogenet Cell Genet. 1998;82(1-2):13-29 Willén H, Akerman M, Dal Cin P, De Wever I, Fletcher CD, Mandahl N, Mertens F, Mitelman F, Rosai J, Rydholm A, Sciot R, Tallini G, Van den Berghe H, Vanni R. Comparison of chromosomal patterns with clinical features in 165 lipomas: a report of the CHAMP study group. Cancer Genet Cytogenet Apr 1;102(1):46-9 Aman P. Fusion genes in solid tumors. Semin Cancer Biol Aug;9(4): Gisselsson D, Domanski HA, Höglund M, Carlén B, Mertens F, Willén H, Mandahl N. Unique cytological features and chromosome aberrations in chondroid lipoma: a case report based on fine-needle aspiration cytology, histopathology, electron microscopy, chromosome banding, and molecular cytogenetics. Am J Surg Pathol Oct;23(10): Gisselsson D, Höglund M, Mertens F, Dal Cin P, Mandahl N. Hibernomas are characterized by homozygous deletions in the multiple endocrine neoplasia type I region. Metaphase fluorescence in situ hybridization reveals complex rearrangements not detected by conventional cytogenetics. Am J Pathol Jul;155(1):61-6 Petit MM, Schoenmakers EF, Huysmans C, Geurts JM, Mandahl N, Van de Ven WJ. LHFP, a novel translocation partner gene of HMGIC in a lipoma, is a member of a new family of LHFP-like genes. Genomics May 1;57(3): Thomson TA, Horsman D, Bainbridge TC. Cytogenetic and cytologic features of chondroid lipoma of soft tissue. Mod Pathol Jan;12(1):88-91 This article should be referenced as such: Mandahl N. Soft tissue tumors: Lipoma / benign lipomatous tumors. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 137
48 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Solid Tumour Section Mini Review Soft tissue tumors: Liposarcoma / malignant lipomatous tumors Nils Mandahl Department of Clinical Genetics, Lund University Hospital, Lund, Sweden (NM) Published in Atlas Database: May 2000 Online updated version : DOI: /2042/37651 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Note Liposarcomas are adipose tissue tumors, including lowmalignant to highly malignant subtypes, constituting 10-15% of soft tissue sarcomas. Classification Well-differentiated liposarcoma: the well-differentiated liposarcomas are tumors of low grade malignancy that may recur locally but not metastasize; the terminolgy of subtypes is not straightforward; three related sybtypes have been distinguished: lipoma-like (the most common form), inflammatory, and sclerosing; other terms that have been suggested to describe at least subsets of these tumors are atypical lipoma and atypical lipomatous tumor. Myxoid liposarcoma/round cell liposarcoma: myxoid liposarcoma is the most common form of liposarcomas, constituting about half of the cases, with a relatively favorable prognosis; the much less common, and more aggressive round cell liposarcoma is regarded as a poorly differentiated variant of myxoid liposarcoma; pure round cell liposarcomas are very rare, and more often the tumors represent mixed liposarcomas with both myxoid and round cell components at different proportions; in recurrences the round cell component may increase. Pleomorphic liposarcoma: the pleomorphic liposarcomas are highly malignant tumors showing a disorderly growth pattern and extensive cellular pleomorphism. Clinics and pathology Epidemiology The reported annual incidence of liposarcoma is in the range of 2.5 per million; liposarcomas are tumors of adult life with a median age of years; patients younger than 15 years are rare; men are slightly more often affected than women. Clinics The major sites are the lower extremities and the retroperitoneum; most tumors range from 5 to 10 cm in diameter, but much larger tumors are not rarely seen. Evolution The risk of distant metastases relate to the type and degree of histological differentiation; welldifferentiated liposarcomas may occasionally dedifferentiate to highly malignant tumors that may metastasize. Prognosis The survival rates are primarily dependent on the histological type, and patients with well-differentiated and myxoid liposarcomas, on the one hand, fare much better than round cell and pleomorphic liposarcomas on the other hand. Cytogenetics Cytogenetics Morphological Well-differentiated liposarcoma / atypical lipomatous tumor Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 138
49 Soft tissue tumors: Liposarcoma / malignant lipomatous tumors Mandahl N The vast majority of this subset of tumors, irrespective of whether classified as atypical lipomatous tumor, atypical lipoma, or well-differentiated liposarcoma, is characterized by the presence of one or more supernumerary ring chromosome or giant marker chromosome. Frequently, these show an extensive intratumor variability in size and number; this has been attributed to mitotic irregularities due to breakage-fusion-bridge cycles, which are also associated with the observed nuclear atypia. In about one-third of the cases, there are, in addition to rings and markers, a few other numerical and/or structural aberrations; these changes do not show any obvious non-random pattern, with the exception of loss of 13q material. Another characteristic feature, seen in the majority of these tumors, is the high frequency of telomeric associations, showing a non-random pattern with a preferential involvement of the 11p telomere. some tumors with minimal atypia have been reported to show gain of 12q15-q24 sequences rather than rings and markers or balanced translocations of 12q13-15, which is a typical feature of benign, ordinary lipomas. Cytogenetics Molecular The ring and giant marker chromosomes have been shown to contain regularly material from 12q and, occasionally, material from another chromosome that may vary from case to case, except for the frequent occurrence of amplified sequences from 1q21 in inter- /intramuscular tumors. Several tumor-associated genes, localized to 12q13- q21, are amplified; these include in particular MDM2, but also SAS, CDK4, and HMGIC; the size of the amplicon vary, and two or more of these genes as well as other sequences may be co-amplified, although frequently at different levels; with few exceptions, MDM2 is not only amplified but also overexpressed. Most rings are negative for chromosome specific centromere probes by FISH, but have centromeric activity as indicated by the positivity for anti CENP-C antibodies. Cytogenetics Morphological Myxoid liposarcoma / round cell liposarcoma: The specific rearrangement t(12;16)(q13;p11), or variants involving one or more additional chromosomes in complex translocations, is found in about 90% of myxoid liposarcomas, including tumors with a mixture of myxoid and round cell components; in one-third of the cases this translocation is the sole cytogenetic anomaly; the most common secondary aberration, seen in 6-7% of the cases, is trisomy 8. The alternative t(12;22)(q13;q12), most often seen in seemingly unbalanced or in complex rearrangements, has been identified in about 5% of the cases; it should be noted that a cytogenetically, but not molecularly, indistinguishable 12;22-translocation has been identified as the characteristic aberration inclear cell sarcoma of the tendons and aponeuroses. among the few cases reported as pure round cell liposarcoma that have been investigated cytogenetically, t(12;16) is rare and the majority of cases have had fairly complex, unspecific aberrations. Cytogenetics Molecular The molecular genetic consequences of the t(12;16) is the formation of a fusion gene, involving FUS in 16p11 and CHOP in 12q13, encoding a chimeric protein; different fusion transcripts have been identified, containing the 5 promotor part of FUS, most often with exons 1-5 or alternatively either exons 1-7 or 1-8, and the entire coding region of CHOP, i.e., exons 1-4 or 2-4; this gene fusion may be identified by RT-PCR as well as by genomic PCR; the CHOP protein belongs to the C/EBP family of basic leucin zipper group of transcription factors. The rearrangements involving 12q13 and 22q12 result in a related gene fusion, affecting the EWS and CHOP genes; thus, the two closely related genes FUS and EWS seem to be interchangable when fused to CHOP; both FUS and EWS carry a central RNA-binding RNP- 1 motif, and possibly these proteins can also bind to DNA. Cytogenetics Morphological Pleomorphic liposarcoma Few cases have been cytogenetically characterized; they invariably show complex karyotypic changes, with no characteristic changes identified, and an extensive intratumor heterogeneity. References Aman P, Ron D, Mandahl N, Fioretos T, Heim S, Arheden K, Willén H, Rydholm A, Mitelman F. Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11). Genes Chromosomes Cancer Nov;5(4): Sreekantaiah C, Karakousis CP, Leong SP, Sandberg AA. Cytogenetic findings in liposarcoma correlate with histopathologic subtypes. Cancer May 15;69(10): Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature Jun 17;363(6430):640-4 Dal Cin P, Kools P, Sciot R, De Wever I, Van Damme B, Van de Ven W, Van den Berghe H. Cytogenetic and fluorescence in situ hybridization investigation of ring chromosomes characterizing a specific pathologic subgroup of adipose tissue tumors. Cancer Genet Cytogenet Jul 15;68(2):85-90 Leach FS, Tokino T, Meltzer P, Burrell M, Oliner JD, Smith S, Hill DE, Sidransky D, Kinzler KW, Vogelstein B. p53 Mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res May 15;53(10 Suppl): Rabbitts TH, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet Jun;4(2): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 139
50 Soft tissue tumors: Liposarcoma / malignant lipomatous tumors Mandahl N Barone MV, Crozat A, Tabaee A, Philipson L, Ron D. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G1/S arrest. Genes Dev Feb 15;8(4): Mandahl N, Höglund M, Mertens F, Rydholm A, Willén H, Brosjö O, Mitelman F. Cytogenetic aberrations in 188 benign and borderline adipose tissue tumors. Genes Chromosomes Cancer Mar;9(3): Mandahl N, Mertens F, Åman P, Rydholm A, Brosjö O, Willén H, Mitelman F.. Nonrandom secondary chromosome aberrations in liposarcomas with t(12;16). Int J Oncol. 1994;4: Panagopoulos I, Mandahl N, Ron D, Höglund M, Nilbert M, Mertens F, Mitelman F, Aman P. Characterization of the CHOP breakpoints and fusion transcripts in myxoid liposarcomas with the 12;16 translocation. Cancer Res Dec 15;54(24): Pedeutour F, Suijkerbuijk RF, Forus A, Van Gaal J, Van de Klundert W, Coindre JM, Nicolo G, Collin F, Van Haelst U, Huffermann K. Complex composition and co-amplification of SAS and MDM2 in ring and giant rod marker chromosomes in well-differentiated liposarcoma. Genes Chromosomes Cancer Jun;10(2):85-94 BuesoRamos CE, Yang Y, Manshouri T, Feltz L, Ayala A, Glassman AB, Albitar M. Molecular abnormalities of MDM-2 in human sarcomas. Int J Oncol. 1995;7: de Boer AG, Wijker W, Bartelsman JF, de Haes HC. Inflammatory Bowel Disease Questionnaire: cross-cultural adaptation and further validation. Eur J Gastroenterol Hepatol Nov;7(11): Forus A, Weghuis DO, Smeets D, Fodstad O, Myklebost O, van Kessel AG. Comparative genomic hybridization analysis of human sarcomas: I. Occurrence of genomic imbalances and identification of a novel major amplicon at 1q21-q22 in soft tissue sarcomas. Genes Chromosomes Cancer Sep;14(1):8-14 Knight JC, Renwick PJ, Dal Cin P, Van den Berghe H, Fletcher CD. Translocation t(12;16)(q13;p11) in myxoid liposarcoma and round cell liposarcoma: molecular and cytogenetic analysis. Cancer Res Jan 1;55(1):24-7 Nakayama T, Toguchida J, Wadayama B, Kanoe H, Kotoura Y, Sasaki MS. MDM2 gene amplification in bone and soft-tissue tumors: Association with tumor progression in differentiated adipose-tissue tumors. Int J Cancer Oct20;64(5): Aman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, Höglund M, Forster A, Rabbitts TH, Ron D, Mandahl N, Mitelman F. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics Oct 1;37(1):1-8 Fletcher CD, Akerman M, Dal Cin P, de Wever I, Mandahl N, Mertens F, Mitelman F, Rosai J, Rydholm A, Sciot R, Tallini G, van den Berghe H, van de Ven W, Vanni R, Willen H. Correlation between clinicopathological features and karyotype in lipomatous tumors. A report of 178 cases from the Chromosomes and Morphology (CHAMP) Collaborative Study Group. Am J Pathol Feb;148(2): Panagopoulos I, Aman P, Mertens F, Mandahl N, Rydholm A, Bauer HF, Mitelman F. Genomic PCR detects tumor cells in peripheral blood from patients with myxoid liposarcoma. Genes Chromosomes Cancer Oct;17(2):102-7 Panagopoulos I, Höglund M, Mertens F, Mandahl N, Mitelman F, Aman P. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene Feb 1;12(3): Rosai J, Akerman M, Dal Cin P, DeWever I, Fletcher CD, Mandahl N, Mertens F, Mitelman F, Rydholm A, Sciot R, Tallini G, Van den Berghe H, Van de Ven W, Vanni R, Willen H. Combined morphologic and karyotypic study of 59 atypical lipomatous tumors. Evaluation of their relationship and differential diagnosis with other adipose tissue tumors (a report of the CHAMP Study Group). Am J Surg Pathol Oct;20(10): Szymanska J, Tarkkanen M, Wiklund T, Virolainen M, Blomqvist C, Asko-Seljavaara S, Tukiainen E, Elomaa I, Knuutila S. Gains and losses of DNA sequences in liposarcomas evaluated by comparative genomic hybridization. Genes Chromosomes Cancer Feb;15(2):89-94 Tallini G, Akerman M, Dal Cin P, De Wever I, Fletcher CD, Mandahl N, Mertens F, Mitelman F, Rosai J, Rydholm A, Sciot R, Van den Berghe H, Van den Ven W, Vanni R, Willen H. Combined morphologic and karyotypic study of 28 myxoid liposarcomas. Implications for a revised morphologic typing, (a report from the CHAMP Group). Am J Surg Pathol Sep;20(9): Aoki T, Hisaoka M, Kouho H, Hashimoto H, Nakata H. Interphase cytogenetic analysis of myxoid soft tissue tumors by fluorescence in situ hybridization and DNA flow cytometry using paraffin-embedded tissue. Cancer Jan 15;79(2): Berner JM, Meza-Zepeda LA, Kools PF, Forus A, Schoenmakers EF, Van de Ven WJ, Fodstad O, Myklebost O. HMGIC, the gene for an architectural transcription factor, is amplified and rearranged in a subset of human sarcomas. Oncogene Jun 19;14(24): Panagopoulos I, Lassen C, Isaksson M, Mitelman F, Mandahl N, Aman P. Characteristic sequence motifs at the breakpoints of the hybrid genes FUS/CHOP, EWS/CHOP and FUS/ERG in myxoid liposarcoma and acute myeloid leukemia. Oncogene Sep;15(11): Pilotti S, Della Torre G, Lavarino C, Di Palma S, Sozzi G, Minoletti F, Rao S, Pasquini G, Azzarelli A, Rilke F, Pierotti MA. Distinct mdm2/p53 expression patterns in liposarcoma subgroups: implications for different pathogenetic mechanisms. J Pathol Jan;181(1):14-24 Wolf M, Aaltonen LA, Szymanska J, Tarkkanen M, Blomqvist C, Berner JM, Myklebost O, Knuutila S. Complexity of 12q13-22 amplicon in liposarcoma: microsatellite repeat analysis. Genes Chromosomes Cancer Jan;18(1):66-70 Gisselsson D, Höglund M, Mertens F, Mitelman F, Mandahl N. Chromosomal organization of amplified chromosome 12 sequences in mesenchymal tumors detected by fluorescence in situ hybridization. Genes Chromosomes Cancer Nov;23(3): Mandahl N, Mertens F, Willén H, Rydholm A, Kreicbergs A, Mitelman F. Nonrandom pattern of telomeric associations in atypical lipomatous tumors with ring and giant marker chromosomes. Cancer Genet Cytogenet May;103(1):25-34 Mertens F, Fletcher CD, Dal Cin P, De Wever I, Mandahl N, Mitelman F, Rosai J, Rydholm A, Sciot R, Tallini G, Van den Berghe H, Vanni R, Willén H. Cytogenetic analysis of 46 pleomorphic soft tissue sarcomas and correlation with morphologic and clinical features: a report of the CHAMP Study Group. Chromosomes and MorPhology. Genes Chromosomes Cancer May;22(1):16-25 Aman P. Fusion genes in solid tumors. Semin Cancer Biol Aug;9(4): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 140
51 Soft tissue tumors: Liposarcoma / malignant lipomatous tumors Mandahl N Gisselsson D, Höglund M, Mertens F, Johansson B, Dal Cin P, Van den Berghe H, Earnshaw WC, Mitelman F, Mandahl N. The structure and dynamics of ring chromosomes in human neoplastic and non-neoplastic cells. Hum Genet Apr;104(4): Kanoe H, Nakayama T, Hosaka T, Murakami H, Yamamoto H, Nakashima Y, Tsuboyama T, Nakamura T, Ron D, Sasaki MS, Toguchida J. Characteristics of genomic breakpoints in TLS- CHOP translocations in liposarcomas suggest the involvement of Translin and topoisomerase II in the process of translocation. Oncogene Jan 21;18(3):721-9 Pedeutour F, Forus A, Coindre JM, Berner JM, Nicolo G, Michiels JF, Terrier P, Ranchere-Vince D, Collin F, Myklebost O, Turc-Carel C. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer Jan;24(1):30-41 Stein U, Eder C, Karsten U, Haensch W, Walther W, Schlag PM. GLI gene expression in bone and soft tissue sarcomas of adult patients correlates with tumor grade. Cancer Res Apr 15;59(8): Thelin-Järnum S, Lassen C, Panagopoulos I, Mandahl N, Aman P. Identification of genes differentially expressed in TLS- CHOP carrying myxoid liposarcomas. Int J Cancer Sep 24;83(1):30-3 This article should be referenced as such: Mandahl N. Soft tissue tumors: Liposarcoma / malignant lipomatous tumors. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 141
52 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Solid Tumour Section Mini Review Uterus: Carcinoma of the cervix Niels B Atkin Department of Cancer Research, Mount Vernon Hospital, Northwood, Middlesex, UK (NBA) Published in Atlas Database: May 2000 Online updated version : DOI: /2042/37649 This article is an update of : Atkin NB. Carcinoma of the Cervix Uteri. Atlas Genet Cytogenet Oncol Haematol 1999;3(3): This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Classification Note - Squamous cell carcinoma (80%); - Adenocarcinoma (10%); - Adenoacanthoma (10%). Clinics and pathology Disease Carcinoma of the cervix uteri; usually arises in the transitional zone between squamous and columnar cell epithelium. Etiology Infection with high-risk forms of the human papillomavirus (HPV) is established as the major factor: a secondary factor is cigarette smoking; recent evidence suggests that a polymorphic variant of the tumour suppressor P53 (p53arg) may represent a risk factor for cervical carcinogenesis. Epidemiology Over 470,000 new cases are diagnosed annually worldwide. Clinics Haematuria. Cytology Cervical smears confirm the diagnosis of carcinoma or may reveal the presence of the disease in its preinvasive (preclinical) stage. Pathology Three grades of preinvasive carcinoma-in-situ (CIN) are recognised: I (which usually undergoes spontaneous resolution), II and III; Carcinomas are staged as follows: IA: early invasive, not grossly visible; IB: usually grossly visible, but confined to the cervix; IIA: spread to the upper two thirds of the vagina only; IIB: lateral extension into the parametrium; IIIA: involvement of the lowest third of the vagina; IIIB: involvement of the pelvic side wall or hydronephrosis; IVA: bladder or rectal involvement; IVB: distant metastasis. Treatment Radiotherapy and/or surgery; late stages: radiotherapy supplemented by chemotherapy (e.g. cisplatin). Evolution Preinvasive stage, detectable by cervical cytology, shows a peak incidence between 25 and 40 years; that of invasive cancer is years, thus indicating that the preinvasive usually progresses to the invasive stage over a very prolonged period. Prognosis Preinvasive lesions are curable by local removal; stage I and early IIA cases may expect 80-90% five year survival; later cases show survival rates of 65-20% or less. Cytogenetics Note Polyploidisation, with modes in the triploid region or above, is common, particularly in the preinvasive phase where it may be linked to the frequent spindle anomalies that result, for instance, in the "three group" metaphases seen in histological sections and chromosome preparations; structural changes are commonest in chromosomes 1, 3, 5, 11 and 17 where, Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 142
53 Uterus: Carcinoma of the cervix Atkin NB except in chromosome 5, they most often result in short-arm deletions. Cytogenetics Morphological Chromosome 1: changes may also result in the acquisition of additional long-arm material (as is common in other types of carcinoma), e.g. in the form of a 1q isochromosome. Chromosome 3: additional material on 3q has been shown by comparative genomic hybridization (CGH) in 90% of carcinomas and this gain may occur at the point of transition from severe dysplasia to invasive carcinoma; recent studies suggest involvement of the htr gene which encodes the RNA component of telomerase; loss of heterozygosity (LOH) studies indicate that there are two regions on 3p where tumour suppressor genes may be situated: at 3pl4.2 (FHIT gene) and at 3q21, gene not yet identified. Chromosome 4: LOH studies suggest that at least two genes are important, at 4p16 and 4q Chromosome 5: an i(5p), often in two or more copies, is a frequent finding in cervical carcinomas, and this is consistent with CGH studies which show amplification of 5p, particularly in advanced stages. Chromosome 6: LOH studies show a high frequency of loss in the region 6p21.3-p25. Chromosome 11: possible gene loss on both chromosome arms are suggested by LOH studies, at 11p15 and 11q23; identities of the genes have yet to be determined. Chromosome 17: G-banding and LOH studies have shown the nonrandom loss of 17q, where the P53 gene is situated (at 17p13.3); mutations or loss of this gene are, however, relatively infrequent compared with other types of tumour, perhaps because there is instead interaction between p53 protein and the HPV E6 viral gene in most carcinomas of the cervix; indeed, p53 appears to be more frequently mutated in HPVnegative tumours. Role of HPV: types 16 and 18 are associated with about 70% of cervical carcinomas (other high-risk types include 31, 33, 35, 39, 51, 52, and 56); these high-risk types are often demonstrable in the moderate and severe stages of preinvasive malignancy (CIN II and III); in these lesions they are commonly situated extrachromosomally while in carcinomas they are integrated into chromosomes at random locations, where they undergo disruption of the HPV E2 viral transcriptional regulatory protein; integration may thereby provide a selective advantage resulting in uncontrolled cellular proliferation leading to aneuploidy; it has recently been shown that a single finding of HPV DNA in a Pap smear from healthy women confers an increased risk of future invasive carcinoma that is positive for the same type of virus. Another recent study suggests that integration of highrisk HPV DNA in cervical swabs or tissue removed from patients with CIN II or III strongly suggests that progression to carcinoma will occur. References Howley PM. Role of the human papillomaviruses in human cancer. Cancer Res Sep 15;51(18 Suppl):5019s-5022s Cannistra SA, Niloff JM. Cancer of the uterine cervix. N Engl J Med Apr 18;334(16): Heselmeyer K, Schröck E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci U S A Jan 9;93(1): Mullokandov MR, Kholodilov NG, Atkin NB, Burk RD, Johnson AB, Klinger HP. Genomic alterations in cervical carcinoma: losses of chromosome heterozygosity and human papilloma virus tumor status. Cancer Res Jan 1;56(1): Atkin NB. Cytogenetics of carcinoma of the cervix uteri: a review. Cancer Genet Cytogenet May;95(1):33-9 Kersemaekers AM, Hermans J, Fleuren GJ, van de Vijver MJ. Loss of heterozygosity for defined regions on chromosomes 3, 11 and 17 in carcinomas of the uterine cervix. Br J Cancer. 1998;77(2): Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh IM, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature May 21;393(6682): Dellas A, Torhorst J, Jiang F, Proffitt J, Schultheiss E, Holzgreve W, Sauter G, Mihatsch MJ, Moch H. Prognostic value of genomic alterations in invasive cervical squamous cell carcinoma of clinical stage IB detected by comparative genomic hybridization. Cancer Res Jul 15;59(14): Kirchhoff M, Rose H, Petersen BL, Maahr J, Gerdes T, Lundsteen C, Bryndorf T, Kryger-Baggesen N, Christensen L, Engelholm SA, Philip J. Comparative genomic hybridization reveals a recurrent pattern of chromosomal aberrations in severe dysplasia/carcinoma in situ of the cervix and in advanced-stage cervical carcinoma. Genes Chromosomes Cancer Feb;24(2): Klaes R, Woerner SM, Ridder R, Wentzensen N, Duerst M, Schneider A, Lotz B, Melsheimer P, von Knebel Doeberitz M. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res Dec 15;59(24): Lazo PA. The molecular genetics of cervical carcinoma. Br J Cancer Aug;80(12): Mitra AB. Genetic deletion and human papillomavirus infection in cervical cancer: loss of heterozygosity sites at 3p and 5p are important genetic events. Int J Cancer Jul 30;82(3):322-4 Thomas M, Pim D, Banks L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene Dec 13;18(53): Wallin KL, Wiklund F, Angström T, Bergman F, Stendahl U, Wadell G, Hallmans G, Dillner J. Type-specific persistence of human papillomavirus DNA before the development of invasive cervical cancer. N Engl J Med Nov 25;341(22): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 143
54 Uterus: Carcinoma of the cervix Atkin NB Sugita M, Tanaka N, Davidson S, Sekiya S, Varella-Garcia M, West J, Drabkin HA, Gemmill RM. Molecular definition of a small amplification domain within 3q26 in tumors of cervix, ovary, and lung. Cancer Genet Cytogenet Feb;117(1):9-18 This article should be referenced as such: Atkin NB. Uterus: Carcinoma of the cervix. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 144
55 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Solid Tumour Section Mini Review Soft tissue tumors: Extraskeletal myxoid chondrosarcoma Jérome Couturier Department of Pathology, Institut Curie, Paris, France (JC) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37652 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Clinics and pathology Disease Malignant tumour of soft tissue origin, distinct from the primary skeletal chondrosarcoma with myxoid alteration. Epidemiology It is a rare tumour: 2.3% of soft tissue sarcomas in a Japanese series; mean ages reported in various series range from 46 to 57 years, this tumour being exceptional in children and adolescents; males are affected about twice as often as females. Clinics Location: deep soft tissues of the lower extremities in about 75% of the cases, especially the thigh, the popliteal fossa, and the buttock; occasionally, a bone involvement may exist, as a minor component. Pathology Macroscopic findings: the tumour presents as lobulated or multinodular mass, generally well circumscribed by a distinct fibrous capsule. The size of the tumour at the time of diagnosis may vary from 1 to about 20 cm (mean size: about 7 cm). Histology: typically, tumour nodules are composed of round or slightly elongated cells, with features of chondroblasts, separated by mucoid substance; differentiated cartilage cells are rare; histological diagnosis may be very difficult, especially in highly cellular forms devoid of myxoid matrix. Tumour cells generally show positivity for vimentin, S- 100 protein, occasionally for EMA, and negativity for cytokeratin. Treatment Treatment: surgical excision, with adjuvant chemotherapy in case of lymph nodes or metastasis. Cytogenetics Cytogenetics Morphological Cytogenetic studies have demonstrated the presence of a recurrent translocation t(9;22)(q22;q12); it results in the fusion of the EWSR1 gene on chromosome 22 with TEC (or CHN) gene on chromosome 9. Recently, a variant translocation t(9;17)(q22;q11) has been identified, fusing the gene TEC to gene TAF2N (TAFII68, or RBP56). Genes involved and proteins TEC Location 9q22 DNA / RNA Transcripts: 2.6 kb and 3.7 kb. Protein Signaling mediator; activate the c-fos promoter; role in growth and differentiation processes of hematopoietic tissues. EWSR1 Location 22q12 DNA / RNA 17 exons; 2.4 kb mrna. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 145
56 Soft tissue tumors: Extraskeletal myxoid chondrosarcoma Couturier J Protein RNA-binding protein; transcription repressor. TAF2N Location 17q11.1-q11.2 DNA / RNA 16 exons; alternative splicing; 2.2 bp mrna. Protein RNA-binding protein; part of thetfiid and RNA polymerase II complex. Result of the chromosomal anomaly Fusion Protein Description The EWS/TEC(CHN) gene fusion encodes a fusion protein in which the C-terminal RNA-binding domain of EWS is replaced by the entire TEC protein. TEC is a member of the steroid/thyroid receptor gene superfamily; the EWS/TEC fusion protein is a potent transcriptional activator. The TAF2N/TEC fusion, in which exon 6 of TAF2N(TAFII68, or RBP56) is fused to the entire coding region of TEC, is structurally and functionally very similar to the EWS/TEC fusion. References Hinrichs SH, Jaramillo MA, Gumerlock PH, Gardner MB, Lewis JP, Freeman AE. Myxoid chondrosarcoma with a translocation involving chromosomes 9 and 22. Cancer Genet Cytogenet Jan 15;14(3-4): Clark J, Benjamin H, Gill S, Sidhar S, Goodwin G, Crew J, Gusterson BA, Shipley J, Cooper CS. Fusion of the EWS gene to CHN, a member of the steroid/thyroid receptor gene superfamily, in a human myxoid chondrosarcoma. Oncogene Jan 18;12(2): Antonescu CR, Argani P, Erlandson RA, Healey JH, Ladanyi M, Huvos AG. Skeletal and extraskeletal myxoid chondrosarcoma: a comparative clinicopathologic, ultrastructural, and molecular study. Cancer Oct 15;83(8): Attwooll C, Tariq M, Harris M, Coyne JD, Telford N, Varley JM. Identification of a novel fusion gene involving htafii68 and CHN from a t(9;17)(q22;q11.2) translocation in an extraskeletal myxoid chondrosarcoma. Oncogene Dec 9;18(52): Bjerkehagen B, Dietrich C, Reed W, Micci F, Saeter G, Berner A, Nesland JM, Heim S. Extraskeletal myxoid chondrosarcoma: multimodal diagnosis and identification of a new cytogenetic subgroup characterized by t(9;17)(q22;q11). Virchows Arch Nov;435(5): Labelle Y, Bussières J, Courjal F, Goldring MB. The EWS/TEC fusion protein encoded by the t(9;22) chromosomal translocation in human chondrosarcomas is a highly potent transcriptional activator. Oncogene May 27;18(21): Panagopoulos I, Mencinger M, Dietrich CU, Bjerkehagen B, Saeter G, Mertens F, Mandahl N, Heim S. Fusion of the RBP56 and CHN genes in extraskeletal myxoid chondrosarcomas with translocation t(9;17)(q22;q11). Oncogene Dec 9;18(52): Rubin BP, Fletcher JA. Skeletal and extraskeletal myxoid chondrosarcoma: related or distinct tumors? Adv Anat Pathol Jul;6(4): Sjögren H, Meis-Kindblom J, Kindblom LG, Aman P, Stenman G. Fusion of the EWS-related gene TAF2N to TEC in extraskeletal myxoid chondrosarcoma. Cancer Res Oct 15;59(20): This article should be referenced as such: Couturier J. Soft tissue tumors Extraskeletal myxoid chondrosarcoma. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 146
57 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Solid Tumour Section Mini Review Nervous system: Medulloblastoma Anne Marie Capodano Laboratoire de Cytogénétique Oncologique, Hôpital de la Timone, 264 rue Saint Pierre, Marseille, France (AMC) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37653 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Clinics and pathology Disease Medulloblastomas are malignant invasive embryonal tumours of the cerebellum with a tendency to metastasize in the central nervous system (CNS). This tumor is more frequently found in children. Epidemiology It represents 10 at 20 % of brain tumours and 30 % of tumours localized in posterior fossa; annual incidence is 0,5 per children; peak of occurrence at 7 years. Pathology Belongs to the primitive neurectodermal tumours (PNET): highly malignant embryonal tumours of the CNS with predominant neuronal differentiation. Several variants medulloblastoma are recognized in the OMS classification: Classic medulloblastoma composed of densely jacked round-cells with round to oval hyperchromatic nuclei. Desmoplastic medulloblastoma represents a variant with abundant reticulin and collagen. Large cell medulloblastoma is a rare variant composed of cells with large round nuclei. Histological features of a typical medulloblastoma: Homer-Wright rosettes - Anne Marie Capodano. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 147
58 Nervous system: Medulloblastoma Capodano AM Immuno histo chemistry: Classic medulloblastoma is strongly immuno-reactive for Vimentin. Some tumours are immunoreactive for NSE, Synaptophysine and GSAP. Treatment The treatment associates total surgical resection and radiotherapy or, according to the age, chemotherapy. Prognosis Survival without recurrence is 50 at 70 %; depends on the quality of surgical resection and on the presence of metastases at the time of diagnosis. Cytogenetics i(17q) - R-banding. Cytogenetics Morphological The most common specific abnormality in medulloblastomas, which is present in approximately 50 % of cases, is isochromosome 17q [i(17q)]. The breakpoint is in the proximal portion of p-arm at 17p11.2, so that the resultant structure is dicentric. In a few cases, partial or complete loss of 17p occurs through interstitial deletion, unbalanced translocation or monosomy 17. Chromosome 1 is also involved in medulloblastomas. The most frequent abnormalities are unbalanced translocations, deletions and duplications. Rearrangements of chromosome 1 often result in trisomy 1q without loss of the p-arm. Others less common chromosomal changes are: deletions of 6q, 9q, 10q, 11q, 11p and 16q, monosomy 22 and in rare cases double minutes. Cytogenetics Molecular Isochromosome 17q has been observed in interphase nuclei using fluorescence in situ hybridization. This technique is used in particular when only a few metaphases are obtained or when only normal diploid cells are obtained in culture. Genes involved and proteins Note Studies on loss of heterozygosity (LOH) have confirmed loss of portions of 17p in % of cases. Some studies showed a correlation between LOH for 17p and a poor response to therapy and shortened survival. Mutations of p53 gene located on 17p13 have been found in only 5-10 % of these tumors. Expression of PAX5 and PAX6 mrna was shown in 70 % of medulloblastomas. The precise mechanism by which these genes are involved remains unknown. Inactivation of PTCH tumor suppressor gene occurs in a subset of medulloblastomas. References Burger PC, Grahmann FC, Bliestle A, Kleihues P. Differentiation in the medulloblastoma. A histological and immunohistochemical study. Acta Neuropathol. 1987;73(2): Bigner SH, Mark J, Friedman HS, Biegel JA, Bigner DD. Structural chromosomal abnormalities in human medulloblastoma. Cancer Genet Cytogenet Jan;30(1): Biegel JA, Rorke LB, Packer RJ, Sutton LN, Schut L, Bonner K, Emanuel BS. Isochromosome 17q in primitive neuroectodermal tumors of the central nervous system. Genes Chromosomes Cancer Nov;1(2): Bigner SH, Vogelstein B. Cytogenetics and molecular genetics of malignant gliomas and medulloblastoma. Brain Pathol Sep;1(1):12-8 Cogen PH, Daneshvar L, Metzger AK, Edwards MS. Deletion mapping of the medulloblastoma locus on chromosome 17p. Genomics Oct;8(2): Giangaspero F, Chieco P, Ceccarelli C, Lisignoli G, Pozzuoli R, Gambacorta M, Rossi G, Burger PC. "Desmoplastic" versus "classic" medulloblastoma: comparison of DNA content, histopathology and differentiation. Virchows Arch A Pathol Anat Histopathol. 1991;418(3): Nakagawa H, Inazawa J, Misawa S, Tanaka S, Takashima T, Taniwaki M, Abe T, Kashima K. Detection of an i(17q) chromosome by fluorescent in situ hybridization with a chromosome 17 alpha satellite DNA probe. Cancer Genet Cytogenet Sep;62(2):140-3 Vagner-Capodano AM, Zattara-Cannoni H, et al. Detection of i(17q) chromosome by fluorescent in situ hybridization (FISH) with interphase nuclei in medulloblastoma. Cancer Genet Cytogenet Nov;78(1):1-6 Reardon DA, Michalkiewicz E, Boyett JM, Sublett JE, Entrekin RE, Ragsdale ST, Valentine MB, Behm FG, Li H, Heideman RL, Kun LE, Shapiro DN, Look AT. Extensive genomic abnormalities in childhood medulloblastoma by comparative genomic hybridization. Cancer Res Sep 15;57(18): Lescop S, Lellouch-Tubiana A, Vassal G, Besnard-Guerin C. Molecular genetic studies of chromosome 11 and chromosome 22q DNA sequences in pediatric medulloblastomas. J Neurooncol Sep;44(2): McLendon RE, Friedman HS, Fuchs HE, Kun LE, Bigner SH. Diagnostic markers in paediatric medulloblastoma: a Paediatric Oncology Group Study. Histopathology Feb;34(2): This article should be referenced as such: Capodano AM. Nervous system: Medulloblastoma. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 148
59 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Solid Tumour Section Mini Review Nervous system: Meningioma Anne Marie Capodano Laboratoire de Cytogénétique Oncologique, Hôpital de la Timone, 264 rue Saint Pierre, Marseille, France (AMC) Published in Atlas Database: July 2000 Online updated version : DOI: /2042/37654 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Classification Macroscopic aspect of a parassagital meningioma - Anne Marie Capodano. Clinics and pathology Disease Meningiomas are tumors arising from cells of the meningeal covering of the brain and spinal cord. These tumors are generally slow growing masses. Neurological signs and symptoms appear by compression of adjacent structures. Etiology Meningiomas are known to be induced by radiation with an average time interval to tumor appearence of years. The majority of patients with radioinduced meningiomas have received irradiation for tinea capitis or for primary brain tumor. Epidemiology Meningiomas account for 15-25% of primary intracranial and intraspinal neoplasms, with an annual incidence of approximatively 6 per individuals. Meningiomas are often multiple in patients with neurofibromatosis type 2 (NF2). Sporadic meningiomas may also be multiple. Meningiomas are most common during the sixth and seventh decade of life, but can occur in both childrens and in the elderly. There is a marked high frequency in females. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 149
60 Nervous system: Meningioma Capodano AM Histological features of a fibroblastic meningioma - Anne Marie Capodano. Clinics The vast majority of meningiomas arise within the intracranial, orbital, and intravertebral cavities. Pathology According to the World Health Classification (WHO 1993), the tumors are defined as Meningiothelial meningioma, Fibroblastic meningioma, Transitional meningioma, Psamommatous meningioma, Angiomatous meningioma, Chordoid meningioma, and are classified according to increased degrees of anaplasia in grades I, II and III. 90% of meningiomas are slowly growing benign tumors that histologically correspond to grade I according WHO classification. 6-8% of meningiomas are designated as atypical meningiomas: WHO grade II. These tumors show a tendancy for local recurrence even after complete resection. 2-3% of meningiomas exibit histological signs of malignancy: these tumors are classified as anaplastic malignant meningiomas of WHO grade III. They have a high risk for local recurrence and metastasis. Treatment The treatment consists of total surgical resection of tumor. Prognosis The major evolution is recurrence. The tumor grade provides the most useful predictor of recurrence. Benign meningiomas have a recurrence rate of about 7-20%. Atypical meningiomas recur in 29-38% of cases, and anaplastic meningiomas in 50-78% of cases. So, proliferation indices have been used to predict recurrence and survival. Cytogenetics Cytogenetics Morphological Meningiomas were among the first solid tumors recognized as having cytogenetic alterations. The most consistent change reported in benign meningiomas is partial (del(22)(q12)) or total deletion of chromosome 22. Loss of chromosome 22 more often occurs in meningiomas grade I. Other karyotypic abnormalities, associated or not with monosomy 22, are seen in grade II (atypical meningiomas), and grade III (anaplastic meningiomas); The most frequent abnormalities changes are deletion of the short arm of chromosome 1, partial or complete loss of chromosome 10, and loss of chromosome 14. Unstable chromosome alterations including rings, dicentrics and telomeric associations, have been observed. A statistical correlation between fibroblastic type and some chromosome abnormalities (monosomy 22 and telomeric associations), was reported. Studies support a postulated role of chromosome 22 as the primary event in the developpement of the majority of the meningiomas. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 150
61 Nervous system: Meningioma Capodano AM Top: del(22q) (G-banding) - Courtesy G. Reza Hafez, Eric B. Johnson, Sara Morrison-Delap Cytogenetics at the Waisman Center; bottom: partial karyotype of a fibroblatic meningioma cell; there was hypoploidy (39,XX), a monosommy 22 (arrow), and tas (arrowheads) - Courtesy Anne Marie Capodano. Genes involved and proteins Note Allelic losses: Molecular genetic findings using polymorphic DNA markers, confirmed that half of meningiomas have allelic loss of band q12 on chromosome 22. Atypical and anaplastic meningiomas often show allelic losses of chromosomal arms 1p, 9q, 10q, 14q, and 17p. LOH of chromosome 14 was the most frequent abnormality in atypical meningiomas: for this reason it is considered to be a step of malignant progression. NF2 Location 22q12 DNA / RNA Tumor suppressor gene. Protein Called merlin or schwannomin. Germinal mutations In neurofibromatosis type 2 patients. Somatic mutations Mutations in the NF2 gene are detected in aproximatively in 60% of sporadic meningiomas. The majority of mutations are small insertions, deletions or non-sens mutations that affect splice sites. The common effect of such mutations is a truncated merlin protein. The frequency of NF2 gene mutations varies according to the meningiomas types. Few mutations of NF2 gene were observed in meningothelial meningiomas : only 25% of cases % of fibroblastic and transitional meningiomas carry NF2 gene mutations. Mutations and allelic loss events are also found in other tumors (schwannomas) and in neurofibromatosis type 2 tumours. Note LOH studies on chromosome 22 have also detected losses of genetic material outside the NF2 region. NF2 is likely to be the major tumor suppressor gene of meningiomas, but other genes localized in other loci on chromosome 22 are probably involved. Another candidate gene on chromosome 22 is MN1 wich has been implicated in a case of translocation in a meningioma. PTEN mutations, a gene localized in 10q23, were described in anaplastic meningiomas. Rare mutations were reported on the CDKN2A gene. References Mark J, Levan G, Mitelman F. Identification by fluorescence of the G chromosome lost in human meningomas. Hereditas. 1972;71(1):163-8 Zulch KJ. International histological classification of tumors. Histological types of tumors of the central nervous system. Genova, World Health Organization,1979; Yamada K, Kondo T, Yoschioka M, Oami H. Cytogenetic studies in 20 human brain tumors : association of n22 chromosome abnormalities with tumors of the brain. Cancer Genet Cytogenet. 1980;2: Zankl H, Zang KD. Correlation between clinical and cytogenetical data in 180 meningiomas. Cancer Genet Cytogenet. 1980;2: Tedeschi F, Fragnito C, Brizzi R, Lechi A, Trabattoni G, Pietrini V. On the pathology of meningiomas. A study of 412 cases. Acta Neuropathol Suppl. 1981;7: Rouleau GA, Wertelecki W, Haines JL, Hobbs WJ, Trofatter JA, Seizinger BR, Martuza RL, Superneau DW, Conneally PM, Gusella JF. Genetic linkage of bilateral acoustic neurofibromatosis to a DNA marker on chromosome 22. Nature Sep 17-23;329(6136):246-8 Seizinger BR, de la Monte S, Atkins L, Gusella JF, Martuza RL. Molecular genetic approach to human meningioma: loss of genes on chromosome 22. Proc Natl Acad Sci U S A Aug;84(15): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 151
62 Nervous system: Meningioma Capodano AM Russell DS, Rubinstein LJ. - Pathology of Tumors of the Nervous System, 1989; 5th edition, London. Dumanski JP, Rouleau GA, Nordenskjöld M, Collins VP. Molecular genetic analysis of chromosome 22 in 81 cases of meningioma. Cancer Res Sep 15;50(18): Maier H, Ofner D, Hittmair A, Kitz K, Budka H. Classic, atypical, and anaplastic meningioma: three histopathological subtypes of clinical relevance. J Neurosurg Oct;77(4): Vagner-Capodano AM, Grisoli F, Gambarelli D, Figarella D, Pellissier JF. Telomeric association of chromosomes in human meningiomas. Ann Genet. 1992;35(2):69-74 Rey JA, Bello MJ, de Campos JM, Vaquero J, Kusak ME, Sarasa JL, Pestaña A. Abnormalities of chromosome 22 in human brain tumors determined by combined cytogenetic and molecular genetic approaches. Cancer Genet Cytogenet Mar;66(1):1-10 Lekanne Deprez RH, Bianchi AB, Groen NA, Seizinger BR, Hagemeijer A, van Drunen E, Bootsma D, Koper JW, Avezaat CJ, Kley N. Frequent NF2 gene transcript mutations in sporadic meningiomas and vestibular schwannomas. Am J Hum Genet Jun;54(6): Lindblom A, Ruttledge M, Collins VP, Nordenskjöld M, Dumanski JP. Chromosomal deletions in anaplastic meningiomas suggest multiple regions outside chromosome 22 as important in tumor progression. Int J Cancer Feb 1;56(3):354-7 Lekanne Deprez RH, Riegman PH, Groen NA, Warringa UL, van Biezen NA, Molijn AC, Bootsma D, de Jong PJ, Menon AG, Kley NA. Cloning and characterization of MN1, a gene from chromosome 22q11, which is disrupted by a balanced translocation in a meningioma. Oncogene Apr 20;10(8): Lekanne Deprez RH, Riegman PH, van Drunen E, Warringa UL, Groen NA, Stefanko SZ, Koper JW, Avezaat CJ, Mulder PG, Zwarthoff EC. Cytogenetic, molecular genetic and pathological analyses in 126 meningiomas. J Neuropathol Exp Neurol Mar;54(2): Louis DN, Ramesh V, Gusella JF. Neuropathology and molecular genetics of neurofibromatosis 2 and related tumors. Brain Pathol Apr;5(2): Papi L, De Vitis LR, Vitelli F, Ammannati F, Mennonna P, Montali E, Bigozzi U. Somatic mutations in the neurofibromatosis type 2 gene in sporadic meningiomas. Hum Genet Mar;95(3): Wellenreuther R, Kraus JA, Lenartz D, Menon AG, Schramm J, Louis DN, Ramesh V, Gusella JF, Wiestler OD, von Deimling A. Analysis of the neurofibromatosis 2 gene reveals molecular variants of meningioma. Am J Pathol Apr;146(4): Ono Y, Ueki K, Joseph JT, Louis DN. Homozygous deletions of the CDKN2/p16 gene in dural hemangiopericytomas. Acta Neuropathol. 1996;91(3):221-5 Kleihues P, Cavenee WK. Tumours of the Nervous System. Pathology and Genetics Weber RG, Boström J, Wolter M, Baudis M, Collins VP, Reifenberger G, Lichter P. Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci U S A Dec 23;94(26): Zattara-Cannoni H, Gambarelli D, Dufour H, Figarella D, Vollot F, Grisoli F, Vagner-Capodano AM. Contribution of cytogenetics and FISH in the diagnosis of meningiomas. A study of 189 tumors. Ann Genet. 1998;41(3): This article should be referenced as such: Capodano AM. Nervous system: Meningioma. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 152
63 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Cancer Prone Disease Section Mini Review Cowden disease Michel Longy Unite de Genetique Oncologique, Institut Bergonie, 180, rue de Saint-Genes, Bordeaux, France (ML) Published in Atlas Database: June 2000 Online updated version : DOI: /2042/37655 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Alias: Multiple hamartoma syndrome Inheritance: Autosomal dominant; high penetrance (close to 100% by the age of 30 yrs); highly variable expressivity (between and within families). Clinics Phenotype and clinics Clinical manifestations usually occur during the 2nd and 3rd decade; they are dystrophic, hamartomatous or tumoral lesions including the following to variable extend: - mucocutaneous papillomatous lesions (facial papules, sometimes related to trichilemmoma; oral papillomatosis with cobblestone gingiva; acral keratoses); - both dystrophic and adenomatous multinodular goiter; - intestinal tract polyps with variable histologies; - adenosis and fibrocystic disease of the breast; - macrocephaly; - lipomas; - genito-urinary abnormalities. Overlapping syndromes Bannayan-Riley-Ruvalcaba syndrome including precocious stigmata of Cowden disease (macrocephaly, lipomas, genital pigmented macules, hamartomatous intestinal tract polyps) is considered as a pediatric form of Cowden disease. Lhermitte Duclos syndrome or dysplastic gangliocytoma of the cerebelum is a rare and complex hamartomatous condition of the cerebellum which can occur alone but also in association with Cowden disease. Juvenile polyposis and Peutz Jeghers syndrome: Cowden disease, by its intestinal tract lesions can be linked to the scope of hereditary hamartomatous polyposis; molecular diagnosis can be useful in distinguishing juvenile polyposis, Peutz Jeghers syndrome or Cowden disease/bannayan. Neoplastic risk The main neoplastic risks arethyroid carcinoma (follicular type) and breast carcinoma of various histological types which are reported in respectively 15% of the patients and 30% of the affected women. Other tumor types occur rarely but more frequently than expected in the general population: renal cell carcinoma, neuroendocrine cell carcinoma, germ cell tumor, malignant melanoma, endometrial carcinoma. Genes involved and proteins PTEN (or MMAC1 or TEP1) Location 10q23 Protein Expression: 403 amino-acids, phosphatase with tumor suppressive effects, negative regulator of the PI3K/Akt signal cell pathway by dephosphorylating PIP3. Mutations Germinal: To date, at least 110 mutations have been described; they are observed along the various exons of the gene except the 9th (never described) and the 1st (very few reports); a mutational hot spot is observed in exon 5 in relation with the catalytic core motif; in the great majority of cases, inactivating mutations are observed, either by protein truncation, or by misense mutation within the phosphatase domain. Somatic: A lot of somatic mutations (more than 300) have been described in several tumor types but mainly in glioblastoma and in endometrial carcinoma; they lead to a biallelic inactivation of the gene more often by a combination of point mutation and large deletion of the second allele. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 153
64 Cowden disease Longy M References LLOYD KM 2nd, DENNIS M. Cowden's disease. A possible new symptom complex with multiple system involvement. Ann Intern Med Jan;58: Brownstein MH, Wolf M, Bikowski JB. Cowden's disease: a cutaneous marker of breast cancer. Cancer Jun;41(6): Salem OS, Steck WD. Cowden's disease (multiple hamartoma and neoplasia syndrome). A case report and review of the English literature. J Am Acad Dermatol May;8(5): Starink TM, van der Veen JP, Arwert F, de Waal LP, de Lange GG, Gille JJ, Eriksson AW. The Cowden syndrome: a clinical and genetic study in 21 patients. Clin Genet Mar;29(3): Case records of the Massachusetts General Hospital. Weekly clinicopathological exercises. Case A 56-year-old man with a substernal goiter, multiple cutaneous and mucosal lesions, and a positive stool test for occult blood. N Engl J Med Jun 11;316(24): Padberg GW, Schot JD, Vielvoye GJ, Bots GT, de Beer FC. Lhermitte-Duclos disease and Cowden disease: a single phakomatosis. Ann Neurol May;29(5): Longy M, Lacombe D. Cowden disease. Report of a family and review. Ann Genet. 1996;39(1):35-42 Nelen MR, Padberg GW, Peeters EA, Lin AY, van den Helm B, Frants RR, Coulon V, Goldstein AM, van Reen MM, Easton DF, Eeles RA, Hodgsen S, Mulvihill JJ, Murday VA, Tucker MA, Mariman EC, Starink TM, Ponder BA, Ropers HH, Kremer H, Longy M, Eng C. Localization of the gene for Cowden disease to chromosome 10q Nat Genet May;13(1):114-6 Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, Eng C, Parsons R. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet May;16(1):64-7 Marsh DJ, Coulon V, Lunetta KL, Rocca-Serra P, Dahia PL, Zheng Z, Liaw D, Caron S, Duboué B, Lin AY, Richardson AL, Bonnetblanc JM, Bressieux JM, Cabarrot-Moreau A, Chompret A, Demange L, Eeles RA, Yahanda AM, Fearon ER, Fricker JP, Gorlin RJ, Hodgson SV, Huson S, Lacombe D, Eng C. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet Mar;7(3): This article should be referenced as such: Longy M. Cowden disease. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 154
65 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Cancer Prone Disease Section Mini Review Familial / sporadic gastrointestinal stromal tumors (GISTs) Lidia Larizza, Alessandro Beghini Department of Biology and Genetics for Medical Sciences, Medical Faculty, University of Milan, Via Viotti 3/5, Milan, Italy (LL, AB) Published in Atlas Database: June 2000 Online updated version : DOI: /2042/37656 This article is an update of : Larizza L, Beghini A. Familial gastrointestinal stromal tumors (GISTs). Atlas Genet Cytogenet Oncol Haematol 1999;3(1):43 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Note A recently described familial cancer syndrome characterized by development of multiple GISTs in different family members. Inheritance: Autosomal dominant. Clinics Phenotype and clinics Symptoms are attributable to development of benign and malignant GISTs. Hyperpigmentation and mast-cell disease may be associated. Etiology: GISTs originate from the CD34+/KIT+ interstitial cells of Cajal (ICCs) which development depends on the SCF/KIT interaction; germline/somatic KIT mutations in familial/solitary GISTs. Pathology: mesenchymal tumours developped in the gastrointestinal wall mainly characterized by positivity for both KIT and CD34; precursor tumour cells are likely ICCs that are located in and near the circular muscle layer of the stomach, small intestine and large intestine. Genes involved and proteins KIT Location 4q12 DNA/RNA Description: 21 exons. Protein Description: Transmembrane SCF/MGF receptor with tyrosine kinase activity; binding of ligand (SCF) induces receptor dimerization, autophosphorylation and signal transduction via molecules containing SH2- domains. Mutations Germinal: Small deletion of one of two consecutive valine residues (codon 559 or 560, GTTGTT). Somatic: Simple in frame deletions, point mutations, deletion and point mutations are mainly clustered in exon 11 (from codon 550 to 584), but a few have been also identified in exon 9 and exon 13; all mutations are predicted to lead to constitutive phosphorylation and kinase activation. The percentage of GISTs positive for c-kit mutations in exon 11 has been estimated to be 57%. Use of c-kit mutation as unfavourable prognostic marker is under debate. References Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Muhammad Tunio G, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science Jan 23;279(5350): Nishida T, Hirota S, Taniguchi M, Hashimoto K, Isozaki K, Nakamura H, Kanakura Y, Tanaka T, Takabayashi A, Matsuda H, Kitamura Y. Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat Genet Aug;19(4):323-4 Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 155
66 Familial /sporadic gastrointestinal stromal tumors (GISTs) Larizza L, Beghini A Sakurai S, Fukasawa T, Chong JM, Tanaka A, Fukayama M. C-kit gene abnormalities in gastrointestinal stromal tumors (tumors of interstitial cells of Cajal. Jpn J Cancer Res Dec;90(12): Taniguchi M, Nishida T, Hirota S, Isozaki K, Ito T, Nomura T, Matsuda H, Kitamura Y. Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res Sep 1;59(17): Lux ML, Rubin BP, Biase TL, Chen CJ, Maclure T, Demetri G, Xiao S, Singer S, Fletcher CD, Fletcher JA. KIT extracellular and kinase domain mutations in gastrointestinal stromal tumors. Am J Pathol Mar;156(3):791-5 This article should be referenced as such: Larizza L, Beghini A. Familial /sporadic gastrointestinal stromal tumors (GISTs). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 156
67 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Cancer Prone Disease Section Mini Review Piebaldism Lidia Larizza, Alessandro Beghini Department of Biology and Genetics for Medical Sciences, Medical Faculty, University of Milan, Via Viotti 3/5, Milan, Italy (LL, AB) Published in Atlas Database: June 2000 Online updated version : DOI: /2042/37657 This article is an update of : Larizza L, Beghini A. Piebaldism. Atlas Genet Cytogenet Oncol Haematol 1999;3(1):44-45 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Note Defect in melanocyte development; one of the first genetic disorders for which a pedigree was presented in Inheritance Autosomal dominant; frequency is about 2.5/10 5 newborns. Clinics Phenotype and clinics Congenital patches of white skin and white hair, principally located on the scalp, forehead, chest and abdomen and on the limbs; several patients report lifelong severe constipation; a hierarchical correlation has been elaborated between severe or mild phenotypic traits and the associated KIT mutations; in a few patients with interstitial deletions mental retardation and congenital anomalies have been also described. Etiology: defective melanoblasts proliferation, survival and migration from the neural crest during development and defective migration of enteric-plexus ganglion cells from the neural crest to the gut. Pathology: white spotting in human piebaldism results from the absence of melanocytes from the nonpigmented patches of skin and from hairbulbs in the white patches of hair; occasionally, individuals lack ganglion cells of the intestinal enteric neural plexus, which like melanoblasts, are derived from the neural crest. Neoplastic risk An increased risk of epithelioma has been reported. Prognosis In contrast to vitiligo, piebaldism is both congenital and non-progressive. Cytogenetics Inborn conditions A few patients with interstitial deletions of chromosome 4q12-q21.1 have been identified; they are charaterized by multiple congenital anomalies, short stature and mental retardation. Genes involved and proteins KIT Location In 4q12. DNA/RNA Description: 21 exons. Protein Description: Transmembrane SCF/MGF receptor with tyrosine kinase activity; binding of ligand (SCF) induces receptor dimerization, autophosphorylation and signal transduction via molecules containing SH2- domains. Mutations Germinal: Loss of function mutations resulting in haploinsufficiency of the receptor; different kinds of point mutations have been identified (diagram): Missense substitutions (Glu583Lys; Phe584Leu; Ala621Thr; His650Pro; Gly664Arg; Gly791Arg; Arg796Gly; Val812Gly; Glu861Ala) and small deletions (641del2; 892 del12) in the intracellular tyrosine kinase domain; correlate with severe piebald Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 157
68 Piebaldism Larizza L, Beghini A phenotypes, because of dominant-negative inhibition of the KIT receptor via formation of impaired receptor heterodimers between a normal and a mutant KIT monomer, and a 75% decrease of KIT- dependent signal transduction. Proximal frameshifts (84del1; 249del4); Trp557Term; and missense mutations (Cys136Arg; Ala178Thr; Met318Gly) associated with a mild piebald phenotype, the result of pure haploinsufficiency due to a 50% decrease of KIT-dependent signal transduction. Distal frameshifts: 630insA; and splice junction mutations (IVS1+4G-A; IVS12+1G- A), located near the intracellular TK domain associated with variable phenotypes, as the truncated polypeptides via incorporation into nonfunctional receptor heterodimers would decrease KIT-dependent signal transduction by 50-75%, depending on their stability. Complete deletions of the entire KIT gene ("null" mutations) result in a mild- intermediate phenotype. PDGFRA Location 4q12 Note Is also deleted in patients with interstitial cytogenetic deletions (contiguous gene syndrome). SCF/MGF Location 12q22 Note No alteration of this gene has been so far identified in typical patients; at difference with the mouse system, where "steel" mice bearing SCF mutations show the "white spotting" phenotype likewise W mice bearing kit mutations; however, as mutations of KIT could not be detected in a consistent fraction of these patients, involvement of SCF is still an open question. References Ezoe K, Holmes SA, Ho L, Bennett CP, Bolognia JL, Brueton L, Burn J, Falabella R, Gatto EM, Ishii N. Novel mutations and deletions of the KIT (steel factor receptor) gene in human piebaldism. Am J Hum Genet Jan;56(1):58-66 Riva P, Milani N, Gandolfi P, Larizza L. A 12-bp deletion (7818del12) in the c-kit protooncogene in a large Italian kindred with piebaldism. Hum Mutat. 1995;6(4):343-5 Fleischman RA, Gallardo T, Mi X. Mutations in the ligandbinding domain of the kit receptor: an uncommon site in human piebaldism. J Invest Dermatol Nov;107(5):703-6 Spritz RA, Beighton P. Piebaldism with deafness: molecular evidence for an expanded syndrome. Am J Med Genet Jan 6;75(1):101-3 This article should be referenced as such: Larizza L, Beghini A. Piebaldism. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 158
69 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Educational Items Section Chromosomes, Chromosome Anomalies Jean-Loup Huret, Claude Léonard, John RK Savage Genetics, Dept Medical Information, University of Poitiers, CHU Poitiers Hospital, F Poitiers, France (JLH); Service de cytogénétique, Hôpital du Kremlin-Bicêtre, 78 rue du Général-Leclerc, Le Kremlin- Bicêtre, France (CL); MRC Radiation and Genome Stability Unit, Harwell, Didcot, OX11 0RD, UK (JRKS) Published in Atlas Database: May 2000 Online updated version : DOI: /2042/37658 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology CHROMOSOMES Introduction Chromosomes carry most of the genetic material and therefore they: carry inherited traits carry the organisation of the cell life heredity: each pair of homologues consists of one paternal and one maternal chromosome diploidy (2 lots). The intact set is passed to each daughter cell at every mitosis. cell life: will be perturbed if regular segregation fails - during embryogenesis (constitutional anomalies). - in a cancer (acquired anomalies). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 159
70 Chromosomes, Chromosome Anomalies Huret JL et al. Classification The normal human karyotype is made of 46 chromosomes: 22 pairs of autosomes, numbered from 1 to 22 by order of decreasing length 1 pair of gonosomes, or sex chromosomes: XX in the female, XY in the male. Each chromosome has a centromere (CEN), region which contains the kinetochore, a micro-tubule organising centre (MTOC) responsible for attachment of the chromosome to the spindle apparatus at mitosis. The 2 sister-chromatids are principally held together at the para-centric heterochromatin at opposite ends of the centromeric region. CEN divides the chromosome into two arms: the short arm (p arm) and the long arm (q arm). Convention places the p-arm at the top in diagrammatic representations. Each arm terminates (pter, qter) in a telomere, a highly conserved repetitive gene sequence which inhibits endend fusion, and which is important for attachments of chromosome ends to the nuclear envelope, particularly during meiosis. It is thought by many that diminution of telomere size is associated with cell ageing. When the short arm is nearly as long as the long arm, the chromosome is said metacentric; if it is shorter, the chromosome is said sub-metacentric; when it is very short, but still visible, the chromosome is said to be sub-telocentric; when extremely short, virtually invisible, the chromosome is said acrocentric. In the human karyotype, chromosome pairs 13, 14, 15, 21, 22 are acrocentric, and Y is sub-telocentric. In mammalian cells, the p-arm of many acrocentric chromosomes carry nucleolar organising regions (NORs) which contain genes coding for ribosomal RNA. This is true for all five pairs of acrocentrics in human cells. Certain staining techniques cause the chromosomes to take on a banded appearance, each arm presenting a sequence of dark and light bands of varying intensities. Patterns are specific and repeatable for each chromosome, allowing unambiguous identification and longitudinal mapping for locating gene positions and characterising structural changes. The number of bands observed is not fixed but is related dynamically to the state of chromosome contraction. Thus, prophase chromosomes have many more bands than metaphase ones. Patterns, and the nomenclature for defining positional mapping have been standardised to allow cytogeneticists to communicate and archive information for medical purposes. Numbering begins from the centromere and continues outward to the end of each arm. Conventionally, the arms are divided into a number of regions by means of easily recognisable "land-mark" bands, and bands numbered sequentially within each. Sub-bands are catered for by using a decimal system. e.g. the place of the star in the Figure 2 is: 21q22.3. CHROMOSOME ANOMALIES CONSTITUTIONAL versus ACQUIRED HOMOGENEOUS versus MOSAIC NUMERICAL versus STRUCTURAL A chromosome anomaly can be: CONSTITUTIONAL: All the tissues ("the whole patient") hold the same anomaly (Figure). The chromosome error was already present in the embryo. It could have occurred before fertilisation, being present in one of the 2 gametes, or possibly in the fertilised zygote. If the anomaly is unbalanced (i.e. if some genes are not present in 2 copies, but in 1 or in 3), the patient is likely to present with 1- dysmorphy and/or 2- visceral malformations, and/or a 3- developmental/psychomotor delay (triad). "Constitutional anomalies" herein refers to the chromosome inborn syndromes, such as trisomy 21, Turner syndromes, and others. ACQUIRED: Only one organ is involved, the other tissues being normal. The patient has a cancer of the affected organ. "Acquired anomalies" herein refers to malignancies. Note: many of the descriptions in this paper, particularly the references to behaviour at Meiosis, cover the general field of structural changes. It is important to realise that relatively few aberrations that occur lead directly to cancer, although some of them will introduce conditions within the cell that may trigger other events that can cause malignant transformation. The terms "constitutional" and "acquired" are really quite general terms, and can be applied to any persistent change encountered in clinical practice. Within the context of this paper, the term Acquired anomalies will apply exclusively to malignant situations. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 160
71 Chromosomes, Chromosome Anomalies Huret JL et al. A chromosome anomaly can be: HOMOGENEOUS: When all the cells (studied) carry the anomaly. e.g. 1: a constitutional anomaly having occurred in a parental gamete (e.g. + 21) will be found in each of the cells of the resulting child (homogeneous trisomy 21). e.g. 2: an acquired anomaly in a leukaemia found in all the bone marrow cells studied (when growth of the normal cells is inhibited by the malignant process and cannot divide in culture (e.g. t(9;22) in chronic myelocytic leukaemia (CML)). Note: In practice, when an acquired anomaly is said homogeneous, it only means that no normal cell was karyotyped within the scored sample. MOSAIC: When only some cells carry the anomaly whilst others are normal (or carry another anomaly). Thus one may find clones of cells carrying a particular change, obviously all derived from the original cell where the anomaly first arose. e.g. 1: A non-disjunction (e.g. + 21) having occurred in the zygote after a few cell divisions: Only some of the embryo cells (and later, of the child s cells) will carry the anomaly (46, XY/47, XY, +21). e.g. 2: Very common in leukaemia and other cancers subject to continuous chromosome change. Only a percentage of mitoses carry the anomalies, while other cells are normal. An example from an acute lymphoblastic leukaemia with one normal clone, one clone with a specific change, and a third with additional changes (46, XY / 46, XY, t(4;11) / 46, XY, t(4;11), i(7q) ). A chromosome anomaly can be: NUMERICAL: If there is one (or more) chromosome(s) in excess (trisomy) (e.g. +21) or missing (monosomy) (e.g. XO if a gonosome is lost, or -5). Note that the karyotype is always unbalanced in case of a numerical anomaly. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 161
72 Chromosomes, Chromosome Anomalies Huret JL et al. STRUCTURAL: If structural changes occur within the chromosomes themselves, not necessarily accompanied by any numerical change. The change is balanced, if there is no loss or gain of genetic material. Unbalanced, if there is deletion and/or duplication of chromosome segment(s). CHROMOSOME ANOMALIES MECHANISMS AND NOMENCLATURE I - NUMERICAL ANOMALIES A - HOMOGENEOUS 1 - Homogeneous due to meiotic non-disjunction 1.1. Autosomes - Non disjunction in first meiotic division produces 4 unbalanced gametes. - Non disjunction in second division produces 2 unbalanced and 2 normal gametes. Gametes with an extra autosome produce trisomic zygotes. The majority of trisomies are non-viable (e.g. trisomy 16) and a miscarriage occurs, sometimes so early that nothing is noticed. A few trisomies are more or less compatible with life, e.g. trisomies 21, 13, 18, and 8. Nullosomic gametes (missing one chromosome) produce monosomies. Monosomies are more deleterious than trisomies and almost all lead to early miscarriage. The only autosomal monosomy in humans which might be compatible with life is monosomy 21, but this is still a debatable situation. Non-disjunction can affect each pair of chromosomes and rarely more than one pair may be involved in the same meiotic cell (multiple non-disjunction most often involves a gonosome). Non-disjunction is not a rare event, but its occurrence is generally underestimated due to the early spontaneous elimination of most unbalanced conceptuses. With reference to the drawings, remember that, in the male, all 4 types of gametes will be effectively present in the sperm. In the female, Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 162
73 Chromosomes, Chromosome Anomalies Huret JL et al. only 1 of the types (presumably randomly selected on each occasion?) will normally participate in fertilisation, the other three being eliminated in the polar bodies) Gonosomes Gonosome unbalance is much less deleterious, and various trisomies can occur, as well as monosomy X (There must always be at least one X for viability). 2 - Homogenous due to a fertilisation anomaly Produce polyploidy. Triploidies are the most frequent, 3N = 69 chromosomes: e.g. 69, XXX, or 69, XXY, or 69, XYY. These are found in 20 % of spontaneous miscarriages. A live birth can occur, but the baby dies shortly afterwards. Mechanisms of formation of triploidies: Digyny: non-expulsion of the 2nd polar body. Diandry: fertilisation of 1 oocyte I by 2 spermatozoa. Diandry is 4 times more frequent than digyny. Tetraploidy, 4N = 92 chromosomes. Found in 6 % of spontaneous miscarriages. Literature records very few live births, but with death soon after. Hydatidiform moles are usually polyploid. Thus, these fertilisation errors are frequent, comprising about 2 to 3 % of fertilised eggs. B - MOSAICISM A mosaic individual is made of 2 (or more) cell populations characterised by difference(s) in the chromosomes. These cell populations, however, come from 1, and only 1, zygote (When recording, a mosaic is denoted by a slash between the various clones observed, e.g. trisomy 21 presenting as a mosaic: 46, XY / 47, XY, +21). Numerical anomaly is usually due to a mitotic nondisjunction: 1 daughter cell will get both chromatids of one of the homologues, the other none; so the former will be trisomic, the latter monosomic. Note: Viability of the two daughter cells may differ. In the above-mentioned trisomy 21 example, the clone monosomic for 21 is non-viable and has disappeared. The phenotype of surviving individuals is more or less affected, according to the proportion of the various clones. Variability of clone proportions is affected by various factors: The precocity of the event e.g. (45, X / 47, XXX) with no cells having 46 chromosomes: zygotic event; (45, X / 46, XX / 47, XXX): post-zygotic event. If 46, XX cells are the most numerous, the anomaly must have occurred late in development; if it occurred at the 32-cell, or 64-cell stage, all, none or part of the embryo could be affected, since by this stage, the cells destined for the primitive streak, and hence the embryo proper have been segregated, and the aberration might be confined to the membranes or placenta. - The distribution of the cell populations during embryogenesis. In this case, the proportions of the various clones will vary from one organ to another. - in vivo selection pressure on the different clones (this may occur in vitro as well, when the cell populations have different kinetics). A mosaic must not be confused with a chimaera. In a chimaera, the cells originate from two (or more) zygotes. They are produced by: Mixture, or exchange of cells, from different zygotes (e.g. early fusion of 2 embryos). Cross blood circulation between dizygotic twins with grafting of blood progenitors. Syngamy: abnormal fertilisation (e.g. 2 spermatozoa, the oocyte, plus the 2nd polar body, making one individual XX/XY with so-called "2 fathers and 2 mothers"). In chimaeras, the 2 cell populations may or may not have different karyotypes, the genes, however, are different. Note: Mosaicism is frequent in malignancies, either because normal cells can still be karyotyped, or because the malignant clone produces sub-clones with additional anomalies (clonal evolution). gametes O Y X XY YY XX XYY XXY XXYY O X XY* XX* XYY XXY XXYY X X XY XX XXY XYY XXX XXYY XXXY XX XX* XXY XXX XXXY XXYY XXXX XXXXY XXX XXX XXXY XXXX XXXXY XXXXX XXXX XXXX XXXXY XXXXX Table: Zygotes produced for each type gamete: Empty boxes indicate a non-viable conceptus. Boxes XX and XY with are normal zygotes from normal gametes. Boxes with * are normal zygotes from unbalanced gametes. II - STRUCTURAL ANOMALIES A - Introduction Visually, chromosomes can appear to break, and broken ends can rejoin in various ways: Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 163
74 Chromosomes, Chromosome Anomalies Huret JL et al. either as they were: restitution; or, in case of 2 (or more) breaks, with interactive rejoining to make a structural aberration (exchanges). Initial breaks are thought to be at the level of the DNA, and are probably frequent events. DNA repair then occurs. For various reasons, DNA repair is insufficient in chromosome instability syndromes. Most often, the break occurs in a non-coding sequence, and does not result in a mutation. Initial breaks can occur anywhere, short arms of acrocentics included. Ultimately, what is important for the individual, is to retain 2 (normal) copies of each gene, no more, no less. This is particularly true for the embryo, where a full balanced genetic complement is vital for normal development. Embryos with unbalanced constitutional anomalies have 1 or 3 copies of a whole set of genes, and abnormal development results. Note: a full balanced complement is not absolutely necessary for the functioning of many differentiated tissue cells, particularly if they are not called upon to divide. Nevertheless, relatively small imbalances can have dire consequences, even in somatic cells. A good example is the case of the Rb gene, implicated in the formation of retinoblastoma. Normal individuals carry 2 functional copies, but one of these can be inactivated by mutation or removal (loss of heterozygosity) and the cell continues normal function through the normal allele (which is now acting as a tumour suppressor gene). Loss of the second allele by removal (or mutation) leads to the formation of the tumour." It is important that these 2 gene copies are normal: Should the break occur within the domain of a gene, wrong re-joining can in activate it, switch on, or off its activity at the wrong time, or produce a hybrid gene with bits of an oncogene, encoding for a fusion protein with oncogenic properties (see Malignant blood diseases). Note: Many of the structural aberrations formed are cell lethal, and are soon eliminated from the cell population. Of those that survive and are transmitted, the most frequent are translocations, small inversions and deletions. Note: Rearranged chromosomes that are transmitted are called derivative chromosomes (der) and they are numbered according to the centromere they carry. Thus a reciprocal translocation between chromosome 7 and chromosome 14 will result in a der(7) and a der(14). B - Main structural anomalies 1 - Reciprocal translocation A mutual exchange between terminal segments from the arms of 2 chromosomes. Provided that there is no loss or alteration at the points of exchange, the new arrangement is genetically balanced, and called a: Balanced rearrangement. Recorded as t, followed by a bracket with the numerals of the 2 chromosomes, and a second bracket indicating the presumptive breakpoints (e.g. t(9;22)(q34;q11)). Transmission to descendants (constitutional anomalies) At meiosis, where there is pairing of homologous chromosome segments (normal chromosomes form a bivalent), followed by crossing-over, translocations may form a quadrivalent (tetravalent, in Greek) and this leads to segregation problems. At meiosis anaphase I, chromosomes separate without centromere separation; this separation occurs at anaphase 2. Segregation of chromatids in the case of a quadrivalent (Figure) can be according the following: alternate type, which produces normal gametes, or gametes with the parental balanced translocation. The baby will have a normal phenotype (unless cryptic imbalance is present). adjacent 1 type, (this is frequent): Associates a normal chromosome (e.g. chromosome a) with the rearranged (or derivative) from the other pair (der(b)). It gives rise to "duplication-deficiency": there is an excess of some bits and a lack of other bits. adjacent 2 type, (this is very rare): Associates a normal chromosome with the derivative from the same pair (e.g. a + der(a)). 3:1 type, (this is rare): - Either a derivative chromosome and the 2 normal homologues (e.g. a, b, der(b)) segregate to one daughter cell, and the other derivative (der(a)) to the other, - Or a normal homologue with the 2 derivative chromosomes (b, der(a), der(b)) to one cell, and the normal chromosome (a) to the other. In either case, this will result in zygotes with 47 or 45 chromosomes. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 164
75 Chromosomes, Chromosome Anomalies Huret JL et al. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 165
76 Chromosomes, Chromosome Anomalies Huret JL et al. Characteristics: Reciprocal translocations are, in most cases, balanced rearrangements and the carrier has a normal phenotype. At meiosis, they enhance malsegregations (especially when an acrocentric is involved in the translocation): Adjacent 1, adjacent 2, or 3:1 types lead to miscarriages, or to the birth of a malformed child. The more unbalanced a zygote is, the less the probability that the child will reach birth. Crossing over during meiosis has no consequence on the structure/morphology of the chromosomes (which is not the case in inversions or in some other rearrangements). Breakpoints can occur at the centromeres, leading to whole arm exchanges. Complex translocations: Three, or more breaks and more than two chromosomes can participate in exchange, leading to some very complicated rearrangements. The surviving, balanced forms are seen usually as cyclical translocations. The recent introduction of FISH-painting indicates that such complex translocations are much more frequent than we have realised. Note There will be no mechanical transmission problems at mitosis. Note: Reciprocal and Complex translocations can also occur in somatic cells at any time after birth; they are particularly frequent in cancer processes. 2 - Robertsonian translocation Fusion of 2 acrocentrics very close to the centromeres, most often in the p arms, giving rise to a dicentric chromosome (having 2 centromeres). The rearranged chromosome includes the long arms of the 2 acrocentrics, while most of the short arm material is lost. Almost always, one of the centromeres is inactivated, so that the translocation behaves as a monocentric giving no segregation problems. The karyotype of a Robertsonian carrier has therefore 45 chromosomes. However, it is said to be balanced, as the loss of the short arm has no phenotypic effect. Recorded as t, with the numerals of each of the 2 chromosomes followed by q in brackets (e.g. t(14q21q)). Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 166
77 Chromosomes, Chromosome Anomalies Huret JL et al. Characteristics: Centric fusions represent the most common chromosome anomaly; they have played an important role in speciation. The role of the acrocentrics in nucleolar organisation favours Robertsonian translocations. Those NORs that are active in a cell form functional nucleoli. Frequently, two, or more, of these nucleoli fuse, thus bringing the parent p-arms into very close proximity within the nucleus, and this will favour interchange formation between them. A dicentric-forming event close to the centromeres will delete the terminal regions of the acrocentric short arms, leaving a dicentric Robertsonian translocation. However, in certain cases, the presence of a nucleolus can act as a physical barrier, precluding close proximity and reducing the probability of interchange. They can occur de novo, or be transmitted through several generations. They are prone to malsegregations (Figure); Robertsonian translocations involving chromosomes 13 and/or 21 produce viable embryos with trisomies 13 or 21. The proportion of associations between the various acrocentrics in human cells is variable, the association being the most frequent. Robertsonian translocations between homologues always lead to unbalanced gametes. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 167
78 Chromosomes, Chromosome Anomalies Huret JL et al. 3 - Deletion Loss of a segment, either interstitial or terminal, from a chromosome (Figure). Invariably, but not always, results in the loss of important genetic material. This loss is sometimes called "partial monosomy". Deletion is therefore an unbalanced rearrangement. Recorded as del, followed by a bracket with the number of the chromosome, and a second bracket indicating the breakpoint(s) and the deleted region (e.g. del(5)(q14q34)); 2 breakpoints are recorded when the deletion is clearly interstitial; only 1 breakpoint is recorded when the deletion seems terminal. A true terminal deletion would leave the surviving chromosome without a telomere. For a long time, cytogeneticists have believed that these telomeres have a special structure, and are functional necessities for the integrity of the chromosome. If this were so, apparently terminal deletions must actually be interstitial, being capped by a telomere. FISH-painting using telomerespecific probes has shown this supposition to be correct Constitutional deletion Deletion in an autosome: Has major phenotypic repercussions (e.g. del(18p); del(18q); del(4p): Wolf- Hirschhorn syndrome; del(5p): cri du chat syndrome; (see chromosome inborn syndromes); therefore these heavily handicapped persons cannot transmit the anomaly to any descendant. The rearrangement most often occurs de novo (only 10 to 15 % of deletion cases come from the malsegregation of a parental rearrangement. The deletion may be accompanied by partial trisomy of another chromosome (duplication/deficiency): See section on reciprocal translocations). Special case: Microdeletions; may be transmitted (e.g. del(13)(q1400q1409): retinoblastoma). Deletion in a gonosome: Causes sexual differentiation and gametogenesis impairments (except distal Yq deletions) (e.g.: del(xp): Turner syndrome) Acquired deletion An example would be the loss of a tumour suppressor gene (e.g.: del(13)(q1400q1409): retinoblastoma). 4 - Ring Can be a centric or acentric event. Persistent (transmitted) rings are always centric. A centric ring involves the deletion (often small) of the ends of both arms (including the telomeres) and rejoining of the median segment to itself in a circular structure. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 168
79 Chromosomes, Chromosome Anomalies Huret JL et al. Is an unbalanced rearrangement, for although the terminal segments lost may not involve vital genetic material, duplication anomalies which occur in ring structures often lead to mechanical problems at mitosis, accompanied by continuous changes in ring size and composition. If sister-chromatid exchanges follow chromosome replication (Figure, right), the ring can form a dicentric ring, or a pair of interlocked rings which will lead to bridge breakage and loss at anaphase of mitosis. The ensuing fusion-fission cycle leads to variable ring sizes and additional duplications and losses of genetic material. Multiple and inter-locked rings can also be produced. Recorded as r, followed by a bracket with the number of the chromosome, and a second bracket indicating the breakpoints, if they are identifiable (e.g. r(13)(p12q33)). Bearing in mind the instability of ring composition mentioned above, break-point designations may not be accurate, or represent the initial change. Arise most often de novo, and are rarely transmitted to descendants (because a ring is unstable, cell divisions lead to impaired gametogenesis; see Figure, left part). frequency of mosaicisms. The repercussions on the phenotype are therefore variable, with signs of trisomy or of deletion. In humans, the most frequent ring in constitutional anomalies concerns chromosome Inversion Inversion occurs when a segment of chromosome breaks, and rejoining within the chromosome effectively inverts it. Recorded as inv, followed by a bracket with the number of the chromosome, and a second bracket indicating the breakpoints, where these can be determined (e.g. inv(9)(p11q13)). Only large inversions are normally detected. Inversions are, in the main, balanced rearrangements, and the carrier has a normal phenotype. (Note: If one break-point is in the middle of a gene imbalance for this particular gene will result). 5a - Paracentric inversion An inversion is termed paracentric when the segment involved lies wholly within one chromosome arm. Are rare (or more likely rarely detected since the majority probably involve very small segments). The most frequent paracentric inversions in constitutional anomalies involve chromosomes 3, 7, and 14. Carriers are often fertile (males as much as females), and about half of descendants have a normal karyotype, and the other half have the balanced rearrangement (like the parent). There are only a few offsprings with unbalanced forms, since unbalanced forms are often too deleterious to give rise to a viable conceptus. There are a few recorded cases of malformed descendants having an apparent balanced constitution. At meiosis, there is pairing of homologous segments, which results in the formation of an inversion loop. Crossing over within the loop (see top of the Figure) produces an acentric fragment (lost) and a chromosome bridge linking the 2 centromeres at anaphase. The bridge either: 1. Disrupts, and, according to where it breaks, there will be duplication/deficiency of certain segments in the daughter cells, or 2. prevents cell separation producing only 1 daughter cell with double the amount of genetic material, or 3. the dicentric will be excluded from both daughter cells, and will form a micronucleus, or 4. the dicentric is included entire into one daughter cell. In the last case, there will be, at telophase of second division: one normal cell, one cell with the balanced inversion, one cell devoid of this chromatid, and one cell with the dicentric. This dicentric will either: i) enter the fission-fusion cycle (leading to complex and numerous rearrangements), or ii) prevent diakinesis (leading to a tetraploidy), or iii) inactivate 1 of its 2 centromeres, which would stabilise the rearrangement. Other crossing over are possible, some would lead to 100 % of unbalanced products (see bottom of the Figure). In practice, a high selection pressure favours normal daughter cells, or those carrying the balanced inversion (as often in genetics, we have the paradox that a very heavy anomaly will have less consequence on the offspring, as the sex cells/eggs/embryos carrying it will be efficiently eliminated: the observable sample is highly biased, a general rule in biology). 5b - Pericentric inversion An inversion is said pericentric when the two breakpoints involved are sited on opposite sides of the centromere, and rejoining effectively inverts the central centromere-bearing segment. Some pericentric inversions are very frequent, and are called chromosome variants: inv(9)(p11q13): found in 1/400 individuals (with large geographic variations). Offsprings with unbalanced forms have not been regularly found (crossing over in heterochromatin is very exceptional). inv(y): found in 1 to 2/1000 male individuals. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 169
80 Chromosomes, Chromosome Anomalies Huret JL et al. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 170
81 Chromosomes, Chromosome Anomalies Huret JL et al. A pericentric inversion can provoke miscarriages, sterility (more often in the male), and lead to unbalanced products at meiosis. During meiosis, crossing over in the inversion loop will produce recombinant chromosomes (rec) with duplication of one segment and deficiency of another (a duplication p - deficiency q will be recorded as rec dup(p)). Note: Duplicated-deficient segments are those outside of the inversion loop (see Figure, bottom right). If the inversion is large, the probability of crossing over in the inversion loop will be higher, and duplicateddeficient segments (outside the loop) smaller. However, the risk will then be greater, since the probability that the conceptus is viable is higher. Conversely, a small inversion has a lower probability of crossing over in the small inversion loop. However, if it occurs, the very large duplicated-deficient segments will have a strong negative selection pressure effect, and the risk of a malformed offspring will be lower. Notes on paracentric and pericentric inversions: Crossing over outside the inverted segments (out of the loop) are without consequence. Wherever the crossing over occurs in the loop, the consequence will be the same. 2 (or an even number of) chiasmata within the loop cancel each other. 6 - Isochromosome Loss of a complete arm, "replaced" by the duplication of the other arm (equivalent to a monosomy for one arm and trisomy for the other). This is an unbalanced rearrangement. Recorded as i, followed by a bracket with the number of the chromosome and the arm (e.g. i(17q) or i(17)(q10): duplication of the q arm and loss of the p arm). This rearrangement is frequent on X chromosome (Turner syndrome with i(xq)). It is also frequent as an acquired anomaly in cancers (e.g. i(17q), secondary anomaly in chronic myelocytic leukaemia). Mechanisms of formation of an isochromosome are varied (Figure). If it arises in the first meiotic division, the duplicated material will be heterozygous. In somatic cells, the most likely origin is from an isochromatid deletion, with sister union, occurring within the centromeric region. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 171
82 Chromosomes, Chromosome Anomalies Huret JL et al. 7 - Insertion An interstitial segment of a chromosome is deleted and transferred to a new position in some other chromosome, or occasionally, into its homologue, or even somewhere else within the same chromosome. The inserted segment may be positioned with its original orientation (with respect to the centromere) or inverted. This is usually a balanced rearrangement. Recorded as ins, followed by a bracket with the number of the chromosome which receives the segment preceding the number of the chromosome which donates the segment (if different). A second bracket indicates the one breakpoint where it inserts, followed by the 2 breakpoints which define the ends of the deleted segment. e.g. ins(2)(p13q31q34) and ins(5;2)(p12;q31q34): the segment q31q34 from a chromosome 2 is inserted respectively in p13 of this chromosome 2, and in p12 of a chromosome 5. An insertion can be direct (dir ins) if the segment keeps its orientation in relation to the centromere (the most proximal band remaining the closest to the centromere; In the example above, band q31 precedes band q34). An insertion can be inverted (inv ins) if the most proximal band becomes the farthest from the centromere e.g. ins(2)(p13q34q31) and ins(5;2)(p12;q34q31), the distal band number preceding the proximal one. This aberration can be balanced and stable in somatic cells, and be transmitted for many cell generations. However, it is pretty devastating at meiosis. In many cases the inserted segment will not be large enough to cause the formation of a quadrivalent. Even so, random segregation at Meiosis 1 means that half of the gametes will be imbalanced. If the segment is large enough the permit occasional quadrivalent formation, then, as Figure shows, 25% of gametes will be normal if the insertion is direct, but none if the insertion is inverted (where we have the added complication of a dicentric bridge and acentric fragment to complicate further the situation). (Note: it is difficult to imagine pairing without a "loop", but this remains speculation. What it would actually look like in reality is an interesting thought. Someone, studying synaptonemal complexes must have seen a pachytene/diplotene spread of such a configuration - if not in man, certainly in mouse or hamster). Interesting exercise for students: work out all the gametic constitutions which result. 8 - Duplication Direct: A segment of chromosome is repeated, once or several times, the duplicated segment keeping the same orientation with respect to the centromere ("tandem duplication"). Inverted: The duplicated segment takes the opposite orientation. Is an unbalanced rearrangement. Recorded as dup, followed by a bracket with the number of the chromosome, and a second bracket indicating the breakpoint(s) and the duplicated region. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 172
83 Chromosomes, Chromosome Anomalies Huret JL et al. 9 - Dicentric A chromosome with 2 centromeres: Simplistically, it is the alternative rejoining mode of the reciprocal translocation, but it can originate by several other mechanisms. It is an unbalanced rearrangement, leading to mechanical separation problems at anaphase ("bridges"). Recorded as dic, or psu dic (pseudo dicentric), when one of the centromeres inactivates, precluding anaphase bridge formation. Inactivation seems principally to be a function of the intercalary distance between the centromeres. Persistent dicentrics are frequent in the case of Robertsonian translocations, but very rare as a constitutional anomaly, unless the short arm of an acrocentric is involved. Rare as an acquired anomaly. Dicentrics (other than Robertsonian translocations) are highly unstable unless: One of the centromeres inactivates. The inter-centromeric distance is very short so that the 2 centromeres can act as one. The only proofs of the presence of 2 active centromeres are: The presence of bridges at anaphase. The presence of non-disjunctions. The presence of isochromosomes from each of the 2 chromosomes, resulting from breaks in the bridge with lateral fusion (Sister Union fusion, between chromatids). Proof that inactivation of 1 centromere occurred is obvious when the 2 chromatids are separated instead of being tightly attached at the centromere location (Premature separation of a centromeric region may also result from several other causes - reduction in paracentric heterochromatin for one, so this "proof" is not absolute) Complex Rearrangements Involving more than 2 chromosomes and/or more than 3 breakpoints. As pointed out above, such aberrations now appear to be much more frequent than we have realised. Many changes seen in cancer cells are of this type. Frequency of malformations in apparently balanced carriers (genetic counselling) Marker A non-recognisable, persistent chromosome, recorded as mar. Either small supernumerary element in the constitutional karyotype, with or without phenotypic repercussions. Issue in prenatal diagnosis. Or a variable sized, often big, element in a cancer process. Since FISH-painting techniques have been developed, many markers have been shown to be highly rearranged chromosomes, involving many participants and many breakpoints Double minute; Homogeneously staining region Double minute: recorded as DM. Appear as very small, usually paired dots. Quite often numerous, but because they are acentric, segregation is irregular and numbers very variable. In the simplest case they represent interstitial deletions, and would normally be rapidly lost from a cell population. Multiple copies indicates a much more complex situation. There is evidence from some mouse cell lines that supernumerary chromosomes and multiple double minutes are interchangeable aberrations. Homogeneously staining region: recorded as HSR. Variable sized, often important, material multiply duplicated in (a) chromosome(s). Experimentally, HSR regions can be produced in response to chronic exposure to certain toxins. DM and HSR may indicate (onco)gene amplification when found in malignant processes, particularly in solid tumours. This article should be referenced as such: Huret JL, Leonard C, Savage JRK. Chromosomes, Chromosome Anomalies. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3): Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 173
84 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Educational Items Section Malignant blood diseases Jean-Loup Huret Genetics, Dept Medical Information, University of Poitiers, CHU Poitiers Hospital, F Poitiers France (JLH) Published in Atlas Database: June 2000 Online updated version : DOI: /2042/37659 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence Atlas of Genetics and Cytogenetics in Oncology and Haematology I-Introduction II- Myeloproliferative syndromes II-1. Chronic myeloid leukemia (CML) II- 2. Other myeloproliferative syndromes III- Myelodysplastic syndromes (MDS) IV- Acute non lymphoblastic leucemias (ANLL) V- Secondary acute leukemias VI- Acute lymphoblastic leukemias (ALL) VII- Non hodgkin's lymphomias VII- 1. Chronic lymphoid leukemia (CLL) VII- 2. Non Hodgkin's lymphomas (NHL) VIII- Main chromosome anomalies in malignant blood diseases IX- Domino game I- Introduction Malignant blood diseases may be classified: According to the clinical course: chronic leukemias acute leukemias According to the lineage: lymphoid lineage: B or T myeloid lineage: o myeloproliferative syndromes: quantitative anomalies o myelodysplastic syndromes: o qualitative anomalies acute myeloid leukemias (or acute non lymphoblastic leukemias ) According to the primary site: leukemia: originates in the bone marrow; flows into the peripheral blood lymphoma: originates in the lymph nodes; invades bone marrow and blood The cell Morphology (according to the FAB (French- American-British) classification of leukemias), the Immunophenotype and the Cytogenetic findings (MIC) allow a specific classification. II- Myeloproliferative syndromes Myeloproliferations: quantitatives anomalies of the myeloid lineage. II-1. Chronic myeloid leukemia (CML) malignant monoclonal process involving a pluripotent hematopoietic progenitor (therefore, most of the lineages are implicated) splenomegaly, high leukocyte count, basophilia, immature cells in the peripheral blood, low leucocyte alkaline phosphatase, bone marrow expansion with increased neutrophil lineage prognosis: chronic phase, followed by blast crises, ending in an acute transformation; median survival used to be of 4 yrs before the new treatments Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 174
85 Malignant blood diseases Huret JL Chromosome anomalies: t(9;22)(q34;q11) chromosome 22 appears shorter and was called Philadelphia chromosome (noted Ph) translocates (part of) an oncogene, ABL, sitting usually in 9q34, next to (part of) another oncogene, BCR (breakpoint cluster region), in 22q11 --> production of a hybrid gene 5' BCR-3'ABL the normal ABL is transcribed into a m-rna of 6 to 7 kbases, which produces a protein (tyrosine kinase) of 145 kdalton the hybrid gene BCR-ABL, result of the translocation t(9;22), is transcribed into a m- RNA of 8.5 kb, which produces a protein of 210 kda with: 1) an increased protein kinase activity 2) an increased half-life, as compared to normal ABL In a percentage of cases, there is a variant translocation, also implicating a third chromosome (e.g. t(1;9;22)); the implication of chromosome 9 or chromosome 22 may even be hidden (e.g. t(12;22); at times, finally, the karyotype seems normal ("Ph-CML"); however, the gene hybride BCR-ABL is always present (otherwise, it is NOT a CML!) therefore the translocation t(9;22) is the specific anomaly found in CML however, this anomaly is not pathognomonic, as it may also be found in ALL or in ANLL additionnal anomalies : most often found at the time of the blast crisis, they may also be present at diagnosis; mainly: +Ph, and/or +8, and/or (17q), and/or +19, and/or -7; clonal evolution II- 2. Other myeloproliferative syndromes Polycytemia vera (PV) : red cell lineage mainly; median survival: 10 to 15 yrs Idiopathic myelofibrosis (or agnogenic myeloid metaplasia) : splenic metaplasia with progressive myelofibrosis ; survival is very variable (3 to 15 yrs) Chromosome anomalies: rare at diagnosis: del(20q), or +8, or +9, or del (13q), or partial trisomy for 1q frequent during acute transformation: anomalies are the one found in usual ANLL or in secondary leukemias (see below) Essential thrombocythemia (ET): megakaryocytic lineage mainly; survival = 10 yrs; chromosome anomalies are rare III- Myelodysplastic syndromes (MDS) Dysmyelopoiesis: qualitative anomalies of the myeloid lineage Classified according to the FAB: refractory anemia without excess of blasts (RA) refractory anemia with excess of blasts (RAEB) refractory anemia with ringed sideroblasts (RARS) chronic myelomonocytic leukemia (CMML) Aside : secondary myelodysplasias (see secondary acute leukemias) Chromosome anomalies : del(5q) (or -5, of identical signification) del(7q) (or -7, equivalent) +8 various structural rearrangements of: 11q, 12p, or chromosome 3 IV- Acute non lymphoblastic leucemias (ANLL) or acute myeloid leukemias (AML), the term myeloid being a bit confusing massive proliferation of myeloid precursors; the chromosome anomaly bears a prognostic value Classified according to the FAB: M1 : myeloblastic without maturation M2 : myeloblastic with maturation M3 : promyelocytic M4 : myelomonocytic M5 : monocytic M6 : erythroleukemia M7 : megakaryoblastic Chromosome anomalies, main entities: t(8;21)(q22;q22) : mainly in M2-ANLL; genes ETO and AML1 t(15;17)(q25;q21) : (quasi) pathognomonic of M3- ANLL; genes PML and RARA fair prognosis if DIC is prevented and with the new treatments (differenciation therapy) (and also as compared with other ANLL) inv(16)(p13q22): pathognomonic of M4-ANLL with eosinophilia; genes MYH11 and CBFb good prognosis: median survival = 5 yrs t(9;22)(q34;q11): rare in ANLL; most often in M1 or M2 ANLL; BCR-ABL as in CML in half cases (protein bcr-abl of 210 kda, called P210), break at a different Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 175
86 Malignant blood diseases Huret JL locus in the other half cases with a m-rna of 7 to 7.5 kb, and production of a bcr-abl protein of 190 kda (named P190) with even a higher transforming ability than P210); very poor prognosis t(6;9(p23;q34) : low specificity; often associated with basophilia; genes DEK and CAN; poor prognosis 3q21 rearrangements : associated with thrombocytosis; very poor prognosis 11q23 rearrangements (M4, M5, biphenotypic acute leukemias) of which is the t(9;11)(p22;q23) Other: del (20q), +8, del (5q), del (7q), 12p rearrangements. V- Secondary acute leukemias induced leukemias: treatment related (or "therapy related") leukemia (after chemo and/or radiotherapy for a prior cancer), or leukemia after professional exposure to carcinogenetic (genotoxic) chemicals or physical agents very poor prognosis Chromosome anomalies : frequent, often complex: multiple monosomies (hypoploidy) del(5q) or -5 del(7q) or -7 rearrangements 6p, 12p, 17p, 11q23... VI- Acute lymphoblastic leukemias (ALL) heavy proliferation of B or T lymphoid precursors, the immunophenotyping (CD, Ig) allows the recognition of the lineage involved in the malignant process, and the degree of maturation of the malignant cell the morphology differenciates ALL1 and 2 on one hand, and ALL3 with large Burkitt-type cells on the other hand --> MIC classification (Morphology, Immunophenotype, Cytogenetics) allows to define entities with given prognoses ALL often occur in childhood Chromosome anomalies, main entities: t(4;11)(q21;q23) : immature (CD19+) B-cell; occurs often in childhood, especially very early (congenital leukemia, before 1 yr); very poor prognosis (median survival below 1 yr), the treatment being a bone marrow graft; genes MLL in 11q23 and AF4 in 4q21 other 11q23 ; MLL and a shared clinical profile t(9;22)(q34;q11) : B-cell; very poor prognosis; at the molecular level: ABL and BCR ; P210 in half cases, P190 in the other half, as is in ANLL with t(9;22) t(12;21)(p12;q22) : CD10+ B ALL in childhood; genes ETV6 and AML1 t(8;14)(q24;q32) and variants t(2;8)(p12;q24) and t(8;22)(q24;q11): t(8;14) being the most frequent; quasi pathognomonic of L3-ALL and Burkitt lymphoma (mature B malignant cell); the prognosis was poor until recently, where new treatments are accompanied with better outcome; MYC in 8q24; immunoglobulin heavychains (IgH) in 14q32, or light-chains K (Ig K) in 2p12 and L (IgL) in 22q11; these translocations set the oncogene under the regulation of immunoglobulin transcription stimulating sequences (active in the B- lineage), leading to overexpression t(11;14)(p13;q11), t(8;14)(q24;q11) and t(10;14)(q24;q11) : T-cell leukemia; T-cell receptor (TCR D et A) belonging to the immunoglobulin superfamilly in 14q11; RBTN2 in 11p13, HOX11 in 10q24, and, obviously, MYC in 8q24; comparable to the above, with here an oncogene under the regulation of the T-cell receptor transcription stimulating sequences (active in the T-lineage), leading to overexpression del(6q), 9p rearrangements, 12p rearrangements, quasi-haploidy, hyperploidy (hyperploidy _ 50 ; hyperploidy > 50, they are of good prognosis), are not rare ALL VII- Non hodgkin's lymphomas classified into numerous categories (see non Hodgkin lymphomas classification according to the cell and tissue morphology, and correlated with the prognosis (low to high grades) chronic lymphoid leukemia is considered as a leukemia by the haematologists and as a low grade lymphoma by the pathologists Chronic lymphoid leukemia (CLL): often a very slow process (10-15 yrs), at times very fast Chromosome anomalies: +12, 14q32 rearrangements, del(6q), 13q rearrangements, del(11q), +3, +18, non identifiable markers; often as associated anomalies Non Hodgkin's lymphomas (NHL) chromosome anomalies: t(14;18)(q32;q21) : typically, found in small cleaved B-cell lymphomas; BCL2 (B cell lymphoma 2) in 18q21, a gene of the BCL2/BAX familly, implicated in the abrogation/induction of apoptosis ("programmed cell death"), immunoglobulin heavy-chain (IgH) in 14q32; BCL2 (protein of the inner membrane of the mitochondria), in case of a translocation t(14;18), is set under the regulation of immunoglobulin transcription stimulating sequences (active in the B-lineage), and overexpressed (as above) other 14q32 rearrangements: of which is the t(11;14)(q13;q32) often seen in mantle cell lymphomas 14q11 rearrangements: T-cell lymphomas; TCR A et D (T-cell receptor) in 14q11, and, at the breakpoint on the partner chromosome, an 176
87 Malignant blood diseases Huret JL oncogene, overexpressed when put under the regulation of the T-cell receptor transcription stimulating sequences (active in the T-lineage) various rearrangements, unrecognizable markers, multiple and complex anomalies are not rares in NHL VIII- Main chromosome anomalies in malignant blood diseases 1 chrom 1 rearrangements various 2 t(2;8)(p12;q24) L3-ALL and Burkitt 4 t(4;11)(q21;q23) ALL 5 del(5q) or -5 MDS, ANLL, Second Leuk. 6 del(6q) ALL, CLL, NHL 7 del(7q) or -7 MDS, ANLL, Second Leuk. 8 t(2;8) see chromosome 2 t(8;14)(q24;q32) (8;14)(q24;q11) t(8;21)(q22;q22) L3-ALL and Burkitt T-ALL M2-ANLL +8 various, myeloid 9 t(9;22)(q34;q11) CML, ANLL, ALL del(9p) ALL +9 various 11 t(4;11) see chromosome 4 t(11;14)(p13;q11) T-ALL t(11;14)(q13;q32) del(11q) NHL MDS, ANLL, CLL CLL, NHL t(12;21)(p12;q22) ALL 13 del(13q) various 14 t(8;14) see chromosome 8 (11;14) see chromosome 11 t(14;18)(q32;q21) inv(14)(q11q32) NHL T-lymphocyte 15 t(15;17)(q22;q12) M3-ANLL 16 16q22 rearrangement M4-ANLL 17 t(15;17) see chromosome 15 i(17q) CML 18 t(14;18) see chromosome del(20q) myeloid 21 t(8;21) see chromosome 8 t(12;21) see chromosome t(8;22) see chromosome 8 t(9;22) see chromosome 9 Other hypoploidy hyperploidy marker Second Leuk., ALL Second Leuk., ALL, NHL Second Leuk., CLL, NHL Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3) 177
88 Malignant blood diseases Huret JL IX- Domino game This article should be referenced as such: Huret JL. Malignant blood diseases. Atlas Genet Cytogenet Oncol Haematol. 2000; 4(3):
89 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Instructions to Authors Manuscripts submitted to the Atlas must be submitted solely to the Atlas. Iconography is most welcome: there is no space restriction. The Atlas publishes "cards", "deep insights", "case reports", and "educational items". Cards are structured review articles. Detailed instructions for these structured reviews can be found at: for reviews on genes, for reviews on leukaemias, for reviews on solid tumours, for reviews on cancer-prone diseases. According to the length of the paper, cards are divided, into "reviews" (texts exceeding 2000 words), "mini reviews" (between), and "short communications" (texts below 400 words). The latter category may not be accepted for indexing by bibliographic databases. Deep Insights are written as traditional papers, made of paragraphs with headings, at the author's convenience. No length restriction. Case Reports in haematological malignancies are dedicated to recurrent -but rare- chromosomes abnormalities in leukaemias/lymphomas. Cases of interest shall be: 1- recurrent (i.e. the chromosome anomaly has already been described in at least 1 case), 2- rare (previously described in less than 20 cases), 3- with well documented clinics and laboratory findings, and 4- with iconography of chromosomes. It is mandatory to use the specific "Submission form for Case reports": see Educational Items must be didactic, give full information and be accompanied with iconography. Translations into French, German, Italian, and Spanish are welcome. Subscription: The Atlas is FREE! Corporate patronage, sponsorship and advertising Enquiries should be addressed to Editorial@AtlasGeneticsOncology.org. Rules, Copyright Notice and Disclaimer Conflicts of Interest: Authors must state explicitly whether potential conflicts do or do not exist. Reviewers must disclose to editors any conflicts of interest that could bias their opinions of the manuscript. The editor and the editorial board members must disclose any potential conflict. Privacy and Confidentiality Iconography: Patients have a right to privacy. Identifying details should be omitted. If complete anonymity is difficult to achieve, informed consent should be obtained. Property: As "cards" are to evolve with further improvements and updates from various contributors, the property of the cards belongs to the editor, and modifications will be made without authorization from the previous contributor (who may, nonetheless, be asked for refereeing); contributors are listed in an edit history manner. Authors keep the rights to use further the content of their papers published in the Atlas, provided that the source is cited. Copyright: The information in the Atlas of Genetics and Cytogenetics in Oncology and Haematology is issued for general distribution. All rights are reserved. The information presented is protected under international conventions and under national laws on copyright and neighbouring rights. Commercial use is totally forbidden. Information extracted from the Atlas may be reviewed, reproduced or translated for research or private study but not for sale or for use in conjunction with commercial purposes. Any use of information from the Atlas should be accompanied by an acknowledgment of the Atlas as the source, citing the uniform resource locator (URL) of the article and/or the article reference, according to the Vancouver convention. Reference to any specific commercial products, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or favouring. The views and opinions of contributors and authors expressed herein do not necessarily state or reflect those of the Atlas editorial staff or of the web site holder, and shall not be used for advertising or product endorsement purposes. The Atlas does not make any warranty, express or implied, including the warranties of merchantability and fitness for a particular purpose, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, and shall not be liable whatsoever for any damages incurred as a result of its use. In particular, information presented in the Atlas is only for research purpose, and shall not be used for diagnosis or treatment purposes. No responsibility is assumed for any injury and/or damage to persons or property for any use or operation of any methods products, instructions or ideas contained in the material herein. See also: "Uniform Requirements for Manuscripts Submitted to Biomedical Journals: Writing and Editing for Biomedical Publication - Updated October 2004": ATLAS - ISSN
90
Gene Section Mini Review
 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review CREBBP (CREB binding protein) Cristina Gervasini Division of Medical Genetics,
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review CREBBP (CREB binding protein) Cristina Gervasini Division of Medical Genetics,
The Human T Cell Receptor Alpha Variable (TRAV) Genes
 IMGT Locus in Focus Exp Clin Immunogenet 2000;17:83 96 Received: September 23, 1999 The Human T Cell Receptor Alpha Variable (TRAV) Genes Dominique Scaviner Marie-Paule Lefranc Laboratoire d ImmunoGénétique
IMGT Locus in Focus Exp Clin Immunogenet 2000;17:83 96 Received: September 23, 1999 The Human T Cell Receptor Alpha Variable (TRAV) Genes Dominique Scaviner Marie-Paule Lefranc Laboratoire d ImmunoGénétique
The Human T cell Receptor Beta Variable (TRBV) Genes
 IMGT Locus in Focus Exp Clin Immunogenet 2000;17:42 54 Received: July 3, 1999 The Human T cell Receptor Beta Variable (TRBV) Genes Géraldine Folch Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire,
IMGT Locus in Focus Exp Clin Immunogenet 2000;17:42 54 Received: July 3, 1999 The Human T cell Receptor Beta Variable (TRBV) Genes Géraldine Folch Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire,
The Human T Cell Receptor Alpha Joining (TRAJ) Genes
 IMGT Locus in Focus Exp Clin Immunogenet 2000;17:97 106 Received: October 15, 1999 The Human T Cell Receptor Alpha Joining (TRAJ) Genes Dominique Scaviner Marie-Paule Lefranc Laboratoire d ImmunoGénétique
IMGT Locus in Focus Exp Clin Immunogenet 2000;17:97 106 Received: October 15, 1999 The Human T Cell Receptor Alpha Joining (TRAJ) Genes Dominique Scaviner Marie-Paule Lefranc Laboratoire d ImmunoGénétique
The Human T Cell Receptor Beta Diversity (TRBD) and Beta Joining (TRBJ) Genes
 IMGT Locus in Focus Exp Clin Immunogenet 2000;17:107 114 Received: October 15, 1999 The Human T Cell Receptor Beta Diversity (TRBD) and Beta Joining (TRBJ) Genes Géraldine Folch Marie-Paule Lefranc Laboratoire
IMGT Locus in Focus Exp Clin Immunogenet 2000;17:107 114 Received: October 15, 1999 The Human T Cell Receptor Beta Diversity (TRBD) and Beta Joining (TRBJ) Genes Géraldine Folch Marie-Paule Lefranc Laboratoire
Nomenclature of the Human Immunoglobulin Heavy (IGH) Genes
 Locus in Focus Exp Clin Immunogenet 2001;18:100 116 Received: August 31, 2000 Nomenclature of the Human Immunoglobulin Heavy (IGH) Genes Marie-Paule Lefranc Chairperson, Nomenclature Committee, CNRS, Université
Locus in Focus Exp Clin Immunogenet 2001;18:100 116 Received: August 31, 2000 Nomenclature of the Human Immunoglobulin Heavy (IGH) Genes Marie-Paule Lefranc Chairperson, Nomenclature Committee, CNRS, Université
The Human Immunoglobulin Heavy Variable Genes
 IMGT Locus on Focus Exp Clin Immunogenet 1999;16:36 60 Received: September 25, 1998 The Human Immunoglobulin Heavy Variable Genes Nathalie Pallarès a Sophie Lefebvre a Valérie Contet a Fumihiko Matsuda
IMGT Locus on Focus Exp Clin Immunogenet 1999;16:36 60 Received: September 25, 1998 The Human Immunoglobulin Heavy Variable Genes Nathalie Pallarès a Sophie Lefebvre a Valérie Contet a Fumihiko Matsuda
The Teleostei Immunoglobulin Heavy IGH Genes
 IMGT Locus in Focus Exp Clin Immunogenet 2000;17:148 161 Received: November 19, 1999 The Teleostei Immunoglobulin Heavy IGH Genes Sylvaine Artéro Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire,
IMGT Locus in Focus Exp Clin Immunogenet 2000;17:148 161 Received: November 19, 1999 The Teleostei Immunoglobulin Heavy IGH Genes Sylvaine Artéro Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire,
Gene Section Mini Review
 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review IL22RA1 (interleukin 22 receptor, alpha 1) Pascal Gelebart, Raymond Lai Department
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review IL22RA1 (interleukin 22 receptor, alpha 1) Pascal Gelebart, Raymond Lai Department
Molecular Hematopathology Leukemias I. January 14, 2005
 Molecular Hematopathology Leukemias I January 14, 2005 Chronic Myelogenous Leukemia Diagnosis requires presence of Philadelphia chromosome t(9;22)(q34;q11) translocation BCR-ABL is the result BCR on chr
Molecular Hematopathology Leukemias I January 14, 2005 Chronic Myelogenous Leukemia Diagnosis requires presence of Philadelphia chromosome t(9;22)(q34;q11) translocation BCR-ABL is the result BCR on chr
Haematology Probes for Multiple Myeloma
 Haematology Probes for Multiple Myeloma MULTIPLE MYELOMA Multiple myeloma (MM) is a plasma cell neoplasm, characterised by the accumulation of clonal plasma cells in the bone marrow and by very complex
Haematology Probes for Multiple Myeloma MULTIPLE MYELOMA Multiple myeloma (MM) is a plasma cell neoplasm, characterised by the accumulation of clonal plasma cells in the bone marrow and by very complex
Molecular Diagnosis. Nucleic acid based testing in Oncology
 Molecular Diagnosis Nucleic acid based testing in Oncology Objectives Describe uses of NAT in Oncology Diagnosis, Prediction, monitoring. Genetics Screening, presymptomatic testing, diagnostic testing,
Molecular Diagnosis Nucleic acid based testing in Oncology Objectives Describe uses of NAT in Oncology Diagnosis, Prediction, monitoring. Genetics Screening, presymptomatic testing, diagnostic testing,
Nucleic Acid Testing - Oncology. Molecular Diagnosis. Gain/Loss of Nucleic Acid. Objectives. MYCN and Neuroblastoma. Molecular Diagnosis
 Nucleic Acid Testing - Oncology Molecular Diagnosis Nucleic acid based testing in Oncology Gross alterations in DNA content of tumors (ploidy) Gain/Loss of nucleic acids Markers of Clonality Oncogene/Tumor
Nucleic Acid Testing - Oncology Molecular Diagnosis Nucleic acid based testing in Oncology Gross alterations in DNA content of tumors (ploidy) Gain/Loss of nucleic acids Markers of Clonality Oncogene/Tumor
The Human Immunoglobulin Kappa Variable (IGKV) Genes and Joining (IGKJ) Segments
 IMGT Locus on Focus Exp Clin Immunogenet 1998;15:171 183 Received: June 13, 1998 Valérie Barbié Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire, CNRS, Université Montpellier II, Montpellier,
IMGT Locus on Focus Exp Clin Immunogenet 1998;15:171 183 Received: June 13, 1998 Valérie Barbié Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire, CNRS, Université Montpellier II, Montpellier,
Development of B and T lymphocytes
 Development of B and T lymphocytes What will we discuss today? B-cell development T-cell development B- cell development overview Stem cell In periphery Pro-B cell Pre-B cell Immature B cell Mature B cell
Development of B and T lymphocytes What will we discuss today? B-cell development T-cell development B- cell development overview Stem cell In periphery Pro-B cell Pre-B cell Immature B cell Mature B cell
Case 089: Therapy Related t(8;21)(q22;q22) AML and
 Case 089: Therapy Related t(8;21)(q22;q22) AML and SM-AHNMD ST Pullarkat 1, V Pullarkat 2, SH Kroft 3, CS Wilson 4, M Thein 5, WW Grody 1, and RK Brynes 5 1 UCLA, 2 City of Hope National Med Ctr, 3 Med
Case 089: Therapy Related t(8;21)(q22;q22) AML and SM-AHNMD ST Pullarkat 1, V Pullarkat 2, SH Kroft 3, CS Wilson 4, M Thein 5, WW Grody 1, and RK Brynes 5 1 UCLA, 2 City of Hope National Med Ctr, 3 Med
The Development of Lymphocytes: B Cell Development in the Bone Marrow & Peripheral Lymphoid Tissue Deborah A. Lebman, Ph.D.
 The Development of Lymphocytes: B Cell Development in the Bone Marrow & Peripheral Lymphoid Tissue Deborah A. Lebman, Ph.D. OBJECTIVES 1. To understand how ordered Ig gene rearrangements lead to the development
The Development of Lymphocytes: B Cell Development in the Bone Marrow & Peripheral Lymphoid Tissue Deborah A. Lebman, Ph.D. OBJECTIVES 1. To understand how ordered Ig gene rearrangements lead to the development
The development of clonality testing for lymphomas in the Bristol Genetics Laboratory. Dr Paula Waits Bristol Genetics Laboratory
 The development of clonality testing for lymphomas in the Bristol Genetics Laboratory Dr Paula Waits Bristol Genetics Laboratory Introduction The majority of lymphoid malignancies belong to the B cell
The development of clonality testing for lymphomas in the Bristol Genetics Laboratory Dr Paula Waits Bristol Genetics Laboratory Introduction The majority of lymphoid malignancies belong to the B cell
Role of FISH in Hematological Cancers
 Role of FISH in Hematological Cancers Thomas S.K. Wan PhD,FRCPath,FFSc(RCPA) Honorary Professor, Department of Pathology & Clinical Biochemistry, Queen Mary Hospital, University of Hong Kong. e-mail: wantsk@hku.hk
Role of FISH in Hematological Cancers Thomas S.K. Wan PhD,FRCPath,FFSc(RCPA) Honorary Professor, Department of Pathology & Clinical Biochemistry, Queen Mary Hospital, University of Hong Kong. e-mail: wantsk@hku.hk
oncogenes-and- tumour-suppressor-genes)
 Special topics in tumor biochemistry oncogenes-and- tumour-suppressor-genes) Speaker: Prof. Jiunn-Jye Chuu E-Mail: jjchuu@mail.stust.edu.tw Genetic Basis of Cancer Cancer-causing mutations Disease of aging
Special topics in tumor biochemistry oncogenes-and- tumour-suppressor-genes) Speaker: Prof. Jiunn-Jye Chuu E-Mail: jjchuu@mail.stust.edu.tw Genetic Basis of Cancer Cancer-causing mutations Disease of aging
Test Name Results Units Bio. Ref. Interval. Positive
 LL - LL-ROHINI (NATIONAL REFERENCE 135091534 Age 36 Years Gender Female 1/9/2017 120000AM 1/9/2017 105316AM 2/9/2017 104147AM Ref By Final LEUKEMIA GENETIC ROFILE ANY SIX MARKERS, CR QUALITATIVE AML ETO
LL - LL-ROHINI (NATIONAL REFERENCE 135091534 Age 36 Years Gender Female 1/9/2017 120000AM 1/9/2017 105316AM 2/9/2017 104147AM Ref By Final LEUKEMIA GENETIC ROFILE ANY SIX MARKERS, CR QUALITATIVE AML ETO
The lymphoma-associated NPM-ALK oncogene elicits a p16ink4a/prb-dependent tumor-suppressive pathway. Blood Jun 16;117(24):
 DNA Sequencing Publications Standard Sequencing 1 Carro MS et al. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006 Jun;5(11) 2 Lassandro L et al. The DNA sequence
DNA Sequencing Publications Standard Sequencing 1 Carro MS et al. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006 Jun;5(11) 2 Lassandro L et al. The DNA sequence
MPL W515L K mutation
 MPL W515L K mutation BCR-ABL genotyping The exact chromosomal defect in Philadelphia chromosome is a translocation. Parts of two chromosomes, 9 and 22, switch places. The result is a fusion gene, created
MPL W515L K mutation BCR-ABL genotyping The exact chromosomal defect in Philadelphia chromosome is a translocation. Parts of two chromosomes, 9 and 22, switch places. The result is a fusion gene, created
 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Scope The Atlas of Genetics and Cytogenetics in Oncology and Haematology is a peer reviewed on-line journal
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Scope The Atlas of Genetics and Cytogenetics in Oncology and Haematology is a peer reviewed on-line journal
Nomenclature of the Human Immunoglobulin Lambda (IGL) Genes
 Locus in Focus Exp Clin Immunogenet 2001;18:242 254 Received: March 17, 2001 Nomenclature of the Human Immunoglobulin Lambda (IGL) Genes Marie-Paule Lefranc Nomenclature Committee, CNRS, Université Montpellier
Locus in Focus Exp Clin Immunogenet 2001;18:242 254 Received: March 17, 2001 Nomenclature of the Human Immunoglobulin Lambda (IGL) Genes Marie-Paule Lefranc Nomenclature Committee, CNRS, Université Montpellier
Molecular Pathology of Lymphoma (Part 1) Rex K.H. Au-Yeung Department of Pathology, HKU
 Molecular Pathology of Lymphoma (Part 1) Rex K.H. Au-Yeung Department of Pathology, HKU Lecture outline Time 10:00 11:00 11:15 12:10 12:20 13:15 Content Introduction to lymphoma Review of lymphocyte biology
Molecular Pathology of Lymphoma (Part 1) Rex K.H. Au-Yeung Department of Pathology, HKU Lecture outline Time 10:00 11:00 11:15 12:10 12:20 13:15 Content Introduction to lymphoma Review of lymphocyte biology
Molecular Markers in Acute Leukemia. Dr Muhd Zanapiah Zakaria Hospital Ampang
 Molecular Markers in Acute Leukemia Dr Muhd Zanapiah Zakaria Hospital Ampang Molecular Markers Useful at diagnosis Classify groups and prognosis Development of more specific therapies Application of risk-adjusted
Molecular Markers in Acute Leukemia Dr Muhd Zanapiah Zakaria Hospital Ampang Molecular Markers Useful at diagnosis Classify groups and prognosis Development of more specific therapies Application of risk-adjusted
Educational Items Section
 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Educational Items Section Hemoglobin genes; Sickle-cell anemia - Thalassemias Jean-Loup Huret, Xavier Troussard
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Educational Items Section Hemoglobin genes; Sickle-cell anemia - Thalassemias Jean-Loup Huret, Xavier Troussard
IMGT Collier de Perles: the New Look for IgSF and MhcSF in IMGT
 IMGT Collier de Perles: the New Look for IgSF and MhcSF in IMGT Quentin Kaas and Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire Université Montpellier, UPR CNRS 1142, IGH Institut Universitaire
IMGT Collier de Perles: the New Look for IgSF and MhcSF in IMGT Quentin Kaas and Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire Université Montpellier, UPR CNRS 1142, IGH Institut Universitaire
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Introduction to Cancer Biology
 Introduction to Cancer Biology Robin Hesketh Multiple choice questions (choose the one correct answer from the five choices) Which ONE of the following is a tumour suppressor? a. AKT b. APC c. BCL2 d.
Introduction to Cancer Biology Robin Hesketh Multiple choice questions (choose the one correct answer from the five choices) Which ONE of the following is a tumour suppressor? a. AKT b. APC c. BCL2 d.
Molecular Oncology, oncology parameters see each test
 Molecular Oncology, oncology parameters see each test DPD deficiency Dihydropyrimidine dehydrogenase deficiency (DPD deficiency) is an autosomal recessive metabolic disorder in which there is absent or
Molecular Oncology, oncology parameters see each test DPD deficiency Dihydropyrimidine dehydrogenase deficiency (DPD deficiency) is an autosomal recessive metabolic disorder in which there is absent or
Ig light chain rearrangement: Rescue pathway
 B Cell Development Ig light chain rearrangement: Rescue pathway There is only a 1:3 chance of the join between the V and J region being in frame Vk Jk Ck Non-productive Rearrangement Light chain has a
B Cell Development Ig light chain rearrangement: Rescue pathway There is only a 1:3 chance of the join between the V and J region being in frame Vk Jk Ck Non-productive Rearrangement Light chain has a
Leukaemia Section Review
 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL INIST-CNRS Leukaemia Section Review t(15;17)(q24;q21) PML/RARA Pino J. Poddighe, Daniel Olde Weghuis Department of Clinical
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL INIST-CNRS Leukaemia Section Review t(15;17)(q24;q21) PML/RARA Pino J. Poddighe, Daniel Olde Weghuis Department of Clinical
Determination Differentiation. determinated precursor specialized cell
 Biology of Cancer -Developmental Biology: Determination and Differentiation -Cell Cycle Regulation -Tumor genes: Proto-Oncogenes, Tumor supressor genes -Tumor-Progression -Example for Tumor-Progression:
Biology of Cancer -Developmental Biology: Determination and Differentiation -Cell Cycle Regulation -Tumor genes: Proto-Oncogenes, Tumor supressor genes -Tumor-Progression -Example for Tumor-Progression:
JAK2 V617F analysis. Indication: monitoring of therapy
 JAK2 V617F analysis BCR-ABL genotyping The exact chromosomal defect in Philadelphia chromosome is a translocation. Parts of two chromosomes, 9 and 22, switch places. The result is a fusion gene, created
JAK2 V617F analysis BCR-ABL genotyping The exact chromosomal defect in Philadelphia chromosome is a translocation. Parts of two chromosomes, 9 and 22, switch places. The result is a fusion gene, created
Chapter 4 Cellular Oncogenes ~ 4.6 -
 Chapter 4 Cellular Oncogenes - 4.2 ~ 4.6 - Many retroviruses carrying oncogenes have been found in chickens and mice However, attempts undertaken during the 1970s to isolate viruses from most types of
Chapter 4 Cellular Oncogenes - 4.2 ~ 4.6 - Many retroviruses carrying oncogenes have been found in chickens and mice However, attempts undertaken during the 1970s to isolate viruses from most types of
Submitted to Leukemia as a Letter to the Editor, May Male preponderance in chronic lymphocytic leukemia utilizing IGHV 1-69.
 Submitted to Leukemia as a Letter to the Editor, May 2007 To the Editor, Leukemia :- Male preponderance in chronic lymphocytic leukemia utilizing IGHV 1-69. Gender plays an important role in the incidence,
Submitted to Leukemia as a Letter to the Editor, May 2007 To the Editor, Leukemia :- Male preponderance in chronic lymphocytic leukemia utilizing IGHV 1-69. Gender plays an important role in the incidence,
Probable involvement of immunoglobulin superfamily genes in most recurrent chromosomal rearrangements from ataxia telangiectasia
 Hum Genet (1986) 72 : 210-214 Springer-Verlag 1986 Probable involvement of immunoglobulin superfamily genes in most recurrent chromosomal rearrangements from ataxia telangiectasia A. Aurias and B. Dutrillaux
Hum Genet (1986) 72 : 210-214 Springer-Verlag 1986 Probable involvement of immunoglobulin superfamily genes in most recurrent chromosomal rearrangements from ataxia telangiectasia A. Aurias and B. Dutrillaux
Cancer. The fundamental defect is. unregulated cell division. Properties of Cancerous Cells. Causes of Cancer. Altered growth and proliferation
 Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer. The fundamental defect is. unregulated cell division. Properties of Cancerous Cells. Causes of Cancer. Altered growth and proliferation
 Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
The Adaptive Immune Response. T-cells
 The Adaptive Immune Response T-cells T Lymphocytes T lymphocytes develop from precursors in the thymus. Mature T cells are found in the blood, where they constitute 60% to 70% of lymphocytes, and in T-cell
The Adaptive Immune Response T-cells T Lymphocytes T lymphocytes develop from precursors in the thymus. Mature T cells are found in the blood, where they constitute 60% to 70% of lymphocytes, and in T-cell
Prepared by: Dr.Mansour Al-Yazji
 C L L CLL Prepared by: Abd El-Hakeem Abd El-Rahman Abu Naser Ahmed Khamis Abu Warda Ahmed Mohammed Abu Ghaben Bassel Ziad Abu Warda Nedal Mostafa El-Nahhal Dr.Mansour Al-Yazji LEUKEMIA Leukemia is a form
C L L CLL Prepared by: Abd El-Hakeem Abd El-Rahman Abu Naser Ahmed Khamis Abu Warda Ahmed Mohammed Abu Ghaben Bassel Ziad Abu Warda Nedal Mostafa El-Nahhal Dr.Mansour Al-Yazji LEUKEMIA Leukemia is a form
Generation of antibody diversity October 18, Ram Savan
 Generation of antibody diversity October 18, 2016 Ram Savan savanram@uw.edu 441 Lecture #10 Slide 1 of 30 Three lectures on antigen receptors Part 1 : Structural features of the BCR and TCR Janeway Chapter
Generation of antibody diversity October 18, 2016 Ram Savan savanram@uw.edu 441 Lecture #10 Slide 1 of 30 Three lectures on antigen receptors Part 1 : Structural features of the BCR and TCR Janeway Chapter
Peking University People's Hospital, Peking University Institute of Hematology
 Qian Jiang, M.D. Peking University People's Hospital, Peking University Institute of Hematology No. 11 Xizhimen South Street, Beijing, 100044, China. Phone number: 86-10-66583802 Mobile: 86-13611115100
Qian Jiang, M.D. Peking University People's Hospital, Peking University Institute of Hematology No. 11 Xizhimen South Street, Beijing, 100044, China. Phone number: 86-10-66583802 Mobile: 86-13611115100
Initial Diagnosis and Treatment 81 Male
 Case SH2017-0359 Shiraz Fidai 1, Sandeep Gurbuxani 1, Girish Venkataraman 1, Gordana Raca 2, Madina Sukhanova 3, Michelle M Le Beau 3, Y. Lynn Wang 4, Mir Alikhan 4, Megan M.McNerney 4, Yuri Kobzev 4,
Case SH2017-0359 Shiraz Fidai 1, Sandeep Gurbuxani 1, Girish Venkataraman 1, Gordana Raca 2, Madina Sukhanova 3, Michelle M Le Beau 3, Y. Lynn Wang 4, Mir Alikhan 4, Megan M.McNerney 4, Yuri Kobzev 4,
Test Name Results Units Bio. Ref. Interval. Positive
 LL - LL-ROHINI (NATIONAL REFERENCE 135091533 Age 28 Years Gender Male 1/9/2017 120000AM 1/9/2017 105415AM 4/9/2017 23858M Ref By Final LEUKEMIA DIAGNOSTIC COMREHENSIVE ROFILE, ANY 6 MARKERS t (1;19) (q23
LL - LL-ROHINI (NATIONAL REFERENCE 135091533 Age 28 Years Gender Male 1/9/2017 120000AM 1/9/2017 105415AM 4/9/2017 23858M Ref By Final LEUKEMIA DIAGNOSTIC COMREHENSIVE ROFILE, ANY 6 MARKERS t (1;19) (q23
Case #16: Diagnosis. T-Lymphoblastic lymphoma. But wait, there s more... A few weeks later the cytogenetics came back...
 Case #16: Diagnosis T-Lymphoblastic lymphoma But wait, there s more... A few weeks later the cytogenetics came back... 46,XY t(8;13)(p12;q12)[12] Image courtesy of Dr. Xinyan Lu Further Studies RT-PCR
Case #16: Diagnosis T-Lymphoblastic lymphoma But wait, there s more... A few weeks later the cytogenetics came back... 46,XY t(8;13)(p12;q12)[12] Image courtesy of Dr. Xinyan Lu Further Studies RT-PCR
Classification of Hematologic Malignancies. Patricia Aoun MD MPH
 Classification of Hematologic Malignancies Patricia Aoun MD MPH Objectives Know the basic principles of the current classification system for hematopoietic and lymphoid malignancies Understand the differences
Classification of Hematologic Malignancies Patricia Aoun MD MPH Objectives Know the basic principles of the current classification system for hematopoietic and lymphoid malignancies Understand the differences
HLA and antigen presentation. Department of Immunology Charles University, 2nd Medical School University Hospital Motol
 HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
membrane form secreted form 13 aa 26 aa K K V V K K 3aa
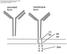 Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai secreted form membrane form 13 aa 26 aa K K V V K K 3aa Hapten monosaccharide
Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai secreted form membrane form 13 aa 26 aa K K V V K K 3aa Hapten monosaccharide
Andrea s SI Session PCB Practice Test Test 3
 Practice Test Test 3 READ BEFORE STARTING PRACTICE TEST: Remember to please use this practice test as a tool to measure your knowledge, and DO NOT use it as your only tool to study for the test, since
Practice Test Test 3 READ BEFORE STARTING PRACTICE TEST: Remember to please use this practice test as a tool to measure your knowledge, and DO NOT use it as your only tool to study for the test, since
of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed.
 Supplementary Note The potential association and implications of HBV integration at known and putative cancer genes of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed. Human telomerase
Supplementary Note The potential association and implications of HBV integration at known and putative cancer genes of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed. Human telomerase
RAS Genes. The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes.
 ۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
Genome of Hepatitis B Virus. VIRAL ONCOGENE Dr. Yahwardiah Siregar, PhD Dr. Sry Suryani Widjaja, Mkes Biochemistry Department
 Genome of Hepatitis B Virus VIRAL ONCOGENE Dr. Yahwardiah Siregar, PhD Dr. Sry Suryani Widjaja, Mkes Biochemistry Department Proto Oncogen and Oncogen Oncogen Proteins that possess the ability to cause
Genome of Hepatitis B Virus VIRAL ONCOGENE Dr. Yahwardiah Siregar, PhD Dr. Sry Suryani Widjaja, Mkes Biochemistry Department Proto Oncogen and Oncogen Oncogen Proteins that possess the ability to cause
Methylation status of SOCS1 and SOCS3 in BCR-ABL negative and. JAK2V617F negative chronic myeloproliferative disorders.
 Methylation status of SOCS1 and SOCS3 in BCR-ABL negative and JAK2V617F negative chronic myeloproliferative disorders. To the Editor BCR-ABL negative Chronic Myeloproliferative Disorders (s) are a heterogeneous
Methylation status of SOCS1 and SOCS3 in BCR-ABL negative and JAK2V617F negative chronic myeloproliferative disorders. To the Editor BCR-ABL negative Chronic Myeloproliferative Disorders (s) are a heterogeneous
Objectives. Morphology and IHC. Flow and Cyto FISH. Testing for Heme Malignancies 3/20/2013
 Molecular Markers in Hematologic Malignancy: Ways to locate the needle in the haystack. Objectives Review the types of testing for hematologic malignancies Understand rationale for molecular testing Marcie
Molecular Markers in Hematologic Malignancy: Ways to locate the needle in the haystack. Objectives Review the types of testing for hematologic malignancies Understand rationale for molecular testing Marcie
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10495 WWW.NATURE.COM/NATURE 1 2 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 3 4 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 5 6 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 7 8 WWW.NATURE.COM/NATURE
doi:10.1038/nature10495 WWW.NATURE.COM/NATURE 1 2 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 3 4 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 5 6 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 7 8 WWW.NATURE.COM/NATURE
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION
 CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
Introduction. Introduction. Lymphocyte development (maturation)
 Introduction Abbas Chapter 8: Lymphocyte Development and the Rearrangement and Expression of Antigen Receptor Genes Christina Ciaccio, MD Children s Mercy Hospital January 5, 2009 Lymphocyte development
Introduction Abbas Chapter 8: Lymphocyte Development and the Rearrangement and Expression of Antigen Receptor Genes Christina Ciaccio, MD Children s Mercy Hospital January 5, 2009 Lymphocyte development
Cancer Genetics. What is Cancer? Cancer Classification. Medical Genetics. Uncontrolled growth of cells. Not all tumors are cancerous
 Session8 Medical Genetics Cancer Genetics J avad Jamshidi F a s a U n i v e r s i t y o f M e d i c a l S c i e n c e s, N o v e m b e r 2 0 1 7 What is Cancer? Uncontrolled growth of cells Not all tumors
Session8 Medical Genetics Cancer Genetics J avad Jamshidi F a s a U n i v e r s i t y o f M e d i c a l S c i e n c e s, N o v e m b e r 2 0 1 7 What is Cancer? Uncontrolled growth of cells Not all tumors
HLA and antigen presentation. Department of Immunology Charles University, 2nd Medical School University Hospital Motol
 HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
Mixed Phenotype Acute Leukemias
 Mixed Phenotype Acute Leukemias CHEN GAO; AMY M. SANDS; JIANLAN SUN NORTH AMERICAN JOURNAL OF MEDICINE AND SCIENCE APR 2012 VOL 5 NO.2 INTRODUCTION Most cases of acute leukemia can be classified based
Mixed Phenotype Acute Leukemias CHEN GAO; AMY M. SANDS; JIANLAN SUN NORTH AMERICAN JOURNAL OF MEDICINE AND SCIENCE APR 2012 VOL 5 NO.2 INTRODUCTION Most cases of acute leukemia can be classified based
Myeloproliferative Disorders - D Savage - 9 Jan 2002
 Disease Usual phenotype acute leukemia precursor chronic leukemia low grade lymphoma myeloma differentiated Total WBC > 60 leukemoid reaction acute leukemia Blast Pro Myel Meta Band Seg Lymph 0 0 0 2
Disease Usual phenotype acute leukemia precursor chronic leukemia low grade lymphoma myeloma differentiated Total WBC > 60 leukemoid reaction acute leukemia Blast Pro Myel Meta Band Seg Lymph 0 0 0 2
Supplementary webappendix
 Supplementary webappendix This webappendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Hartman M, Loy EY, Ku CS, Chia KS. Molecular
Supplementary webappendix This webappendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Hartman M, Loy EY, Ku CS, Chia KS. Molecular
Antibodies and T Cell Receptor Genetics Generation of Antigen Receptor Diversity
 Antibodies and T Cell Receptor Genetics 2008 Peter Burrows 4-6529 peterb@uab.edu Generation of Antigen Receptor Diversity Survival requires B and T cell receptor diversity to respond to the diversity of
Antibodies and T Cell Receptor Genetics 2008 Peter Burrows 4-6529 peterb@uab.edu Generation of Antigen Receptor Diversity Survival requires B and T cell receptor diversity to respond to the diversity of
Katrina L. Lancaster-Shorts, Joanna Chaffin, Natasha M. Savage. Department of Pathology, Augusta University, Augusta, GA, USA;
 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Leukaemia Section Review Katrina L. Lancaster-Shorts, Joanna Chaffin, Natasha M. Savage Department of Pathology,
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Leukaemia Section Review Katrina L. Lancaster-Shorts, Joanna Chaffin, Natasha M. Savage Department of Pathology,
Characterisation of structural variation in breast. cancer genomes using paired-end sequencing on. the Illumina Genome Analyser
 Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Molecular Markers. Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC
 Molecular Markers Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC Overview Testing methods Rationale for molecular testing
Molecular Markers Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC Overview Testing methods Rationale for molecular testing
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
Introduction retroposon
 17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
Combinations of morphology codes of haematological malignancies (HM) referring to the same tumour or to a potential transformation
 Major subgroups according to the World Health Organisation (WHO) Classification Myeloproliferative neoplasms (MPN) Myeloid and lymphoid neoplasms with eosinophilia and abnormalities of PDGFRA, PDGFRB or
Major subgroups according to the World Health Organisation (WHO) Classification Myeloproliferative neoplasms (MPN) Myeloid and lymphoid neoplasms with eosinophilia and abnormalities of PDGFRA, PDGFRB or
Chapter 5. Generation of lymphocyte antigen receptors
 Chapter 5 Generation of lymphocyte antigen receptors Structural variation in Ig constant regions Isotype: different class of Ig Heavy-chain C regions are encoded in separate genes Initially, only two of
Chapter 5 Generation of lymphocyte antigen receptors Structural variation in Ig constant regions Isotype: different class of Ig Heavy-chain C regions are encoded in separate genes Initially, only two of
Immunology - Lecture 2 Adaptive Immune System 1
 Immunology - Lecture 2 Adaptive Immune System 1 Book chapters: Molecules of the Adaptive Immunity 6 Adaptive Cells and Organs 7 Generation of Immune Diversity Lymphocyte Antigen Receptors - 8 CD markers
Immunology - Lecture 2 Adaptive Immune System 1 Book chapters: Molecules of the Adaptive Immunity 6 Adaptive Cells and Organs 7 Generation of Immune Diversity Lymphocyte Antigen Receptors - 8 CD markers
Richter s Syndrome: Risk, Predictors and Treatment
 Richter s Syndrome: Risk, Predictors and Treatment 10/23/2015 John N. Allan MD Assistant Professor of Medicine Division of Hematology and Medical Oncology CLL Research Center Weill Cornell Medicine Agenda
Richter s Syndrome: Risk, Predictors and Treatment 10/23/2015 John N. Allan MD Assistant Professor of Medicine Division of Hematology and Medical Oncology CLL Research Center Weill Cornell Medicine Agenda
SH/EAHP WORKSHOP 2017 CASE 210 PRESENTATION
 SH/EAHP WORKSHOP 2017 CASE 210 PRESENTATION Jonathon H Gralewski DO, MS, Ginell R Post MD, PhD, Youzhong Yuan MD September 9, 2017 Clinical History 60 year old male with history of c-maf high-risk IgG
SH/EAHP WORKSHOP 2017 CASE 210 PRESENTATION Jonathon H Gralewski DO, MS, Ginell R Post MD, PhD, Youzhong Yuan MD September 9, 2017 Clinical History 60 year old male with history of c-maf high-risk IgG
Intronic BCL-6 mutations are preferentially targeted to the translocated allele in t(3;14)(q27;q32) non-hodgkin B-cell lymphoma
 NEOPLASIA Brief report Intronic BCL-6 mutations are preferentially targeted to the translocated allele in t(3;14)(q27;q32) non-hodgkin B-cell lymphoma Fabrice Jardin, Christian Bastard, Nathalie Contentin,
NEOPLASIA Brief report Intronic BCL-6 mutations are preferentially targeted to the translocated allele in t(3;14)(q27;q32) non-hodgkin B-cell lymphoma Fabrice Jardin, Christian Bastard, Nathalie Contentin,
Non-Hodgkin lymphomas (NHLs) Hodgkin lymphoma )HL)
 Non-Hodgkin lymphomas (NHLs) Hodgkin lymphoma )HL) Lymphoid Neoplasms: 1- non-hodgkin lymphomas (NHLs) 2- Hodgkin lymphoma 3- plasma cell neoplasms Non-Hodgkin lymphomas (NHLs) Acute Lymphoblastic Leukemia/Lymphoma
Non-Hodgkin lymphomas (NHLs) Hodgkin lymphoma )HL) Lymphoid Neoplasms: 1- non-hodgkin lymphomas (NHLs) 2- Hodgkin lymphoma 3- plasma cell neoplasms Non-Hodgkin lymphomas (NHLs) Acute Lymphoblastic Leukemia/Lymphoma
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Principles of Adaptive Immunity
 Principles of Adaptive Immunity Chapter 3 Parham Hans de Haard 17 th of May 2010 Agenda Recognition molecules of adaptive immune system Features adaptive immune system Immunoglobulins and T-cell receptors
Principles of Adaptive Immunity Chapter 3 Parham Hans de Haard 17 th of May 2010 Agenda Recognition molecules of adaptive immune system Features adaptive immune system Immunoglobulins and T-cell receptors
ONE STEP MULTIPLEX RT-PCR FOR BCRlABL GENE IN MALAY PATIENTS DIAGNOSED AS LEUKAEMIA
 ONE STEP MULTIPLEX RT-PCR FOR BCRlABL GENE IN MALAY PATIENTS DIAGNOSED AS LEUKAEMIA 1Rosline H, 1Majdan R, 1Wan Zaidah A, 1Rapiaah M, 1Selamah G, 2A A Baba, 3D M Donald 1Department of Haematology, 2Department
ONE STEP MULTIPLEX RT-PCR FOR BCRlABL GENE IN MALAY PATIENTS DIAGNOSED AS LEUKAEMIA 1Rosline H, 1Majdan R, 1Wan Zaidah A, 1Rapiaah M, 1Selamah G, 2A A Baba, 3D M Donald 1Department of Haematology, 2Department
Template for Reporting Results of Monitoring Tests for Patients With Chronic Myelogenous Leukemia (BCR-ABL1+)
 Template for Reporting Results of Monitoring Tests for Patients With Chronic Myelogenous Leukemia (BCR-ABL1+) Version: CMLBiomarkers 1.0.0.2 Protocol Posting Date: June 2017 This biomarker template is
Template for Reporting Results of Monitoring Tests for Patients With Chronic Myelogenous Leukemia (BCR-ABL1+) Version: CMLBiomarkers 1.0.0.2 Protocol Posting Date: June 2017 This biomarker template is
Tumor suppressor genes D R. S H O S S E I N I - A S L
 Tumor suppressor genes 1 D R. S H O S S E I N I - A S L What is a Tumor Suppressor Gene? 2 A tumor suppressor gene is a type of cancer gene that is created by loss-of function mutations. In contrast to
Tumor suppressor genes 1 D R. S H O S S E I N I - A S L What is a Tumor Suppressor Gene? 2 A tumor suppressor gene is a type of cancer gene that is created by loss-of function mutations. In contrast to
Template for Reporting Results of Biomarker Testing of Specimens From Patients With Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma
 Template for Reporting Results of Biomarker Testing of Specimens From Patients With Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma Version: CLLBiomarkers 1.0.0.2 Protocol Posting Date: June 2017
Template for Reporting Results of Biomarker Testing of Specimens From Patients With Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma Version: CLLBiomarkers 1.0.0.2 Protocol Posting Date: June 2017
Introduction to Genetics
 Introduction to Genetics Table of contents Chromosome DNA Protein synthesis Mutation Genetic disorder Relationship between genes and cancer Genetic testing Technical concern 2 All living organisms consist
Introduction to Genetics Table of contents Chromosome DNA Protein synthesis Mutation Genetic disorder Relationship between genes and cancer Genetic testing Technical concern 2 All living organisms consist
Corrigenda. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (revised 4th edition): corrections made in second print run
 Corrigenda WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (revised 4th edition): corrections made in second print run In addition to corrections of minor typographical errors, corrections
Corrigenda WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (revised 4th edition): corrections made in second print run In addition to corrections of minor typographical errors, corrections
Lymphoma: What You Need to Know. Richard van der Jagt MD, FRCPC
 Lymphoma: What You Need to Know Richard van der Jagt MD, FRCPC Overview Concepts, classification, biology Epidemiology Clinical presentation Diagnosis Staging Three important types of lymphoma Conceptualizing
Lymphoma: What You Need to Know Richard van der Jagt MD, FRCPC Overview Concepts, classification, biology Epidemiology Clinical presentation Diagnosis Staging Three important types of lymphoma Conceptualizing
T cell Receptor. Chapter 9. Comparison of TCR αβ T cells
 Chapter 9 The αβ TCR is similar in size and structure to an antibody Fab fragment T cell Receptor Kuby Figure 9-3 The αβ T cell receptor - Two chains - α and β - Two domains per chain - constant (C) domain
Chapter 9 The αβ TCR is similar in size and structure to an antibody Fab fragment T cell Receptor Kuby Figure 9-3 The αβ T cell receptor - Two chains - α and β - Two domains per chain - constant (C) domain
Structure and Function of Fusion Gene Products in. Childhood Acute Leukemia
 Structure and Function of Fusion Gene Products in Childhood Acute Leukemia Chromosomal Translocations Chr. 12 Chr. 21 der(12) der(21) A.T. Look, Science 278 (1997) Distribution Childhood ALL TEL-AML1 t(12;21)
Structure and Function of Fusion Gene Products in Childhood Acute Leukemia Chromosomal Translocations Chr. 12 Chr. 21 der(12) der(21) A.T. Look, Science 278 (1997) Distribution Childhood ALL TEL-AML1 t(12;21)
Innate immunity (rapid response) Dendritic cell. Macrophage. Natural killer cell. Complement protein. Neutrophil
 1 The immune system The immune response The immune system comprises two arms functioning cooperatively to provide a comprehensive protective response: the innate and the adaptive immune system. The innate
1 The immune system The immune response The immune system comprises two arms functioning cooperatively to provide a comprehensive protective response: the innate and the adaptive immune system. The innate
ADx Bone Marrow Report. Patient Information Referring Physician Specimen Information
 ADx Bone Marrow Report Patient Information Referring Physician Specimen Information Patient Name: Specimen: Bone Marrow Site: Left iliac Physician: Accession #: ID#: Reported: 08/19/2014 - CHRONIC MYELOGENOUS
ADx Bone Marrow Report Patient Information Referring Physician Specimen Information Patient Name: Specimen: Bone Marrow Site: Left iliac Physician: Accession #: ID#: Reported: 08/19/2014 - CHRONIC MYELOGENOUS
Chronic Myeloproliferative Disorders
 Chronic Myeloproliferative Disorders Chronic Myeloproliferative Disorders Cytogenetic and Molecular Genetic Abnormalities Editor Barbara J. Bain, London 15 figures, 4 in color, and 18 tables, 2003 ABC
Chronic Myeloproliferative Disorders Chronic Myeloproliferative Disorders Cytogenetic and Molecular Genetic Abnormalities Editor Barbara J. Bain, London 15 figures, 4 in color, and 18 tables, 2003 ABC
Ch. 18 Regulation of Gene Expression
 Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
mirna Dr. S Hosseini-Asl
 mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
CANCER. Inherited Cancer Syndromes. Affects 25% of US population. Kills 19% of US population (2nd largest killer after heart disease)
 CANCER Affects 25% of US population Kills 19% of US population (2nd largest killer after heart disease) NOT one disease but 200-300 different defects Etiologic Factors In Cancer: Relative contributions
CANCER Affects 25% of US population Kills 19% of US population (2nd largest killer after heart disease) NOT one disease but 200-300 different defects Etiologic Factors In Cancer: Relative contributions
B Lymphocyte Development and Activation
 Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai 09/26/05; 9 AM Shiv Pillai B Lymphocyte Development and Activation Recommended
Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai 09/26/05; 9 AM Shiv Pillai B Lymphocyte Development and Activation Recommended
BIT 120. Copy of Cancer/HIV Lecture
 BIT 120 Copy of Cancer/HIV Lecture Cancer DEFINITION Any abnormal growth of cells that has malignant potential i.e.. Leukemia Uncontrolled mitosis in WBC Genetic disease caused by an accumulation of mutations
BIT 120 Copy of Cancer/HIV Lecture Cancer DEFINITION Any abnormal growth of cells that has malignant potential i.e.. Leukemia Uncontrolled mitosis in WBC Genetic disease caused by an accumulation of mutations
Helminth worm, Schistosomiasis Trypanosomes, sleeping sickness Pneumocystis carinii. Ringworm fungus HIV Influenza
 Helminth worm, Schistosomiasis Trypanosomes, sleeping sickness Pneumocystis carinii Ringworm fungus HIV Influenza Candida Staph aureus Mycobacterium tuberculosis Listeria Salmonella Streptococcus Levels
Helminth worm, Schistosomiasis Trypanosomes, sleeping sickness Pneumocystis carinii Ringworm fungus HIV Influenza Candida Staph aureus Mycobacterium tuberculosis Listeria Salmonella Streptococcus Levels
