Nomenclature of the Human Immunoglobulin Lambda (IGL) Genes
|
|
|
- Melvyn Lawson
- 6 years ago
- Views:
Transcription
1 Locus in Focus Exp Clin Immunogenet 2001;18: Received: March 17, 2001 Nomenclature of the Human Immunoglobulin Lambda (IGL) Genes Marie-Paule Lefranc Nomenclature Committee, CNRS, Université Montpellier II, Montpellier, France Key Words Human W W Immunoglobulin W Lambda chain genes W Orphons Abstract Nomenclature of the Human Immunoglobulin Lambda (IGL) Genes, the 18th report of the Locus in Focus section, provides the first complete list of all the human IGL genes. The total number of human IGL genes per haploid genome is ( if the orphons are included), of which genes are functional. /Human Genome Organization (HUGO) gene names and definitions of the human IGL genes on chromosome 22q11.2 and IGL orphons on chromosomes 8 and 22 are provided with the gene functionality and the number of alleles, according to the rules of the Scientific chart, with the accession numbers of the reference sequences and with the of the Genome Database GDB and NCBI LocusLink databases, in which all the human IGL genes have been entered. The tables are available at the Marie-Paule page of, the international ImMunoGeneTics database ( created by Marie-Paule Lefranc, Université Montpellier II, CNRS, France. Copyright 2001 S. Karger AG, Basel Introduction Nomenclature of the Human Immunoglobulin Lambda (IGL) Genes is the 18th report of the Locus in Focus section launched in the April 1998 issue of Experimental and Clinical Immunogenetics [1 18]. This report comprises three tables and three figures entitled, respectively: (1) Complete list of the human IGL genes on chromosome 22 at 22q11.2; (2) Human IGL orphons on chromosomes 8 and 22; Correspondence between the human IGLV gene nomenclatures; (4) Chromosomal localization of the human IGL locus at 22q11.2; (5) Representation of the ABC Fax karger@karger.ch S. Karger AG, Basel /01/ $17.50/0 Accessible online at: Prof. Marie-Paule Lefranc, Laboratoire d ImmunoGénétique Moléculaire, LIGM UPR CNRS 1142 IGH, 141 rue de la Cardonille, F Montpellier Cedex 5 (France) Tel , Fax lefranc@ligm.igh.cnrs.fr, :
2 human IGL locus at 22q11.2, and (6) The CLASSIFICATION concept of the - ONTOLOGY, exemplified for the IGLV genes. The tables provide the first complete list of all the human IGL genes. The total number of human IGL genes per haploid genome is depending on the haplotypes ( if the orphons are included), of which genes are functional. /Human Genome Organization (HUGO) gene names and definitions of the human IGL genes on chromosome 22q11.2 and IGL orphons on chromosomes 8 and 22 are provided with the gene functionality and the number of alleles, according to the rules of the Scientific chart, with the accession numbers of the reference sequences and with the accession ID of the Genome Database GDB and NCBI LocusLink databases, in which all the human IGL genes have been entered. Detailed references for individual IGLV, IGLJ and IGLC genes are available in other reports [2, 7, 19]. These tables and figures are available at the Marie-Paule page of, the international ImMunoGeneTics database ( created by Marie-Paule Lefranc, Université Montpellier II, CNRS, Montpellier, France [20 23]. Fig. 1. Chromosomal localization of the human IGL locus at 22q11.2. The vertical line indicates the localization of the IGL locus at 22q11.2. The arrow indicates the orientation 5) 3) of the locus, and the gene group order in the locus. The arrow is proportional to the size of the locus, indicated in kilobases (kb). The total number of genes in the locus is shown between parentheses. Depending on the haplotype, there are 7 11 IGLC genes. In the 7-IGLC gene haplotype, each IGLC gene is preceded by one IGLJ gene in 5). Although the additional IGLC genes, in the 8-, 9-, 10- and 11-IGLC gene haplotypes have not yet been sequenced, they are probably preceded by one IGLJ gene. The number of functional genes defines the potential IGL repertoire, which comprises in the 7- IGLC gene haplotype, genes (29 33 IGLV, 4 5 IGLJ and 4 5 IGLC) per haploid genome. Human IGL Locus at 22q11.2 The human IGL locus is located at band 22q11.2 on the long arm of chromosome 22 [24, 25] (fig. 1). The orientation of the locus has been determined by the analysis of translocations involving the IGL locus in leukemia and lymphoma. Sequencing of the long arm of chromosome 22 showed that it encompasses about 35 megabases of DNA and that the IGL locus is localized at 6 megabases from the centromere [26]. The human IGL locus at 22q11.2 spans 1,050 kb (fig. 2). It consists of IGLV genes [2, 7, 27 30], localized on 900 kb, 7 11 IGLJ and 7 11 IGLC genes depending on the haplotypes, each IGLC gene being preceded by one IGLJ gene [31 34] (table 1). Fifty-six to 57 IGLV genes belong to 11 subgroups, whereas 17 pseudogenes which are too divergent to be assigned to subgroups, have been assigned to the clans (see header of table 1). The 5)-most IGLV genes occupy the more centromeric position, whereas the IGLC genes, in 3) of the locus, are the most telomeric genes in the IGL locus. The potential genomic IGL repertoire comprises func- Human IGL Gene Nomenclature Exp Clin Immunogenet 2001;18:
3 Fig. 2. Representation of the human IGL locus at 22q11.2. The boxes representing the genes are not to scale. Exons are not shown. A, B, C refer to three distinct V-CLUSTERs based on the IGLV gene subgroup content [29]. IGLV gene names are designated by a number for the subgroup [27, 29] followed by a hyphen and a number for the localization from 3) to 5) in the locus. Pseudogenes which could not be assigned to subgroups with functional genes are designated by a Roman numeral between parentheses, corresponding to the clans, followed by a hyphen and a number for the localization from 3) to 5) in the locus. tional IGLV genes belonging to 10 subgroups, 4 5 IGLJ, and 4 5 IGLC functional genes in the 7-IGLC gene haplotype. One, 2, 3 or 4 additional IGLC genes, each one probably preceded by 1 IGLJ, have been shown to characterize IGLC haplotypes with 8, 9, 10 or 11 genes [32, 35 37], but these genes have not yet been sequenced. The number of human IGL genes at 22q11.2 is of which genes are functional [19]. 244 Exp Clin Immunogenet 2001;18: Lefranc
4 Table 1. Complete list of the human IGL genes on chromosome 22 at 22q11.2 IGLV gene nomenclature: IGLV genes are designated by a number for the subgroup, followed by a hyphen and a number for the localization from 3) to 5) in the locus [19, 27, 29]. In the IGLV gene name column, the IGLV genes are listed, for each subgroup, according to their position from 3) to 5) in the locus. Pseudogenes which could not be assigned to subgroups with functional genes are designated by a Roman numeral between parentheses, corresponding to the clans, followed by a hyphen and a number for the localization from 3) to 5) in the locus. Clans comprise, respectively: clan I: IGLV1, IGLV2, IGLV6 and IGLV10 subgroup genes, and pseudogenes IGLV(I)-20, -38, -42, -56, -63, -68 and -70 clan II: IGLV3 subgroup genes clan III : IGLV7 and IGLV8 subgroup genes clan IV : IGLV5 and IGLV11 subgroup genes, and pseudogenes IGLV(IV)-53, -59, -64, -65 and IGLV(IV)-66-1 has been identified in D87004 by curators (G. Folch, V. Giudicelli and M.-P. Lefranc) clan V : IGLV4 and IGLV9 subgroup genes, and pseudogenes IGLV(V)-58 and -66 clan VI : pseudogenes IGLV(VI)-22-1 and IGLV(VI)-25-1 has been identified in D86994 by curators (N. Bosc and M.-P. Lefranc) clan VII : pseudogene IGLV(VII) An asterisk (*)indicates an allelic polymorphism by insertion/deletion which concerns the IGLV5-39 gene. One, two, three or four additional IGLC genes, each one probably preceded by one IGLJ, have been shown to characterize IGLC haplotypes with 8, 9, 10 or 11 genes [32, 35, 36], but these genes have not yet been sequenced and are not shown in this table. gene group gene name (1) functionality reference sequence accession numbers Number of alleles gene definition (2) GDB LocusLink IGLC IGLC1 F J Immunoglobulin lambda constant 1 GDB: IGLC2 F J Immunoglobulin lambda constant 2 GDB: IGLC3 F J Immunoglobulin lambda constant 3 GDB: IGLC4 P J Immunoglobulin lambda constant 4 GDB: IGLC5 P J Immunoglobulin lambda constant 5 GDB: IGLC6 F, P J Immunoglobulin lambda constant 6 GDB: IGLC7 F X Immunoglobulin lambda constant 7 GDB: IGLJ IGLJ1 F X Immunoglobulin lambda joining 1 GDB: IGLJ2 F M Immunoglobulin lambda joining 2 GDB: IGLJ3 F M Immunoglobulin lambda joining 3 GDB: IGLJ4 ORF X Immunoglobulin lambda joining 4 GDB: IGLJ5 ORF X Immunoglobulin lambda joining 5 GDB: IGLJ6 ORF M Immunoglobulin lambda joining 6 GDB: IGLJ7 F X Immunoglobulin lambda joining 7 GDB: IGLV IGLV1-36 F Z Immunoglobulin lambda variable 1-36 GDB: IGLV1-40 F M Immunoglobulin lambda variable 1-40 GDB: IGLV1-41 ORF, P M Immunoglobulin lambda variable 1-41 GDB: IGLV1-44 F Z Immunoglobulin lambda variable 1-44 GDB: IGLV1-47 F Z Immunoglobulin lambda variable 1-47 GDB: IGLV1-50 ORF M Immunoglobulin lambda variable 1-50 GDB: IGLV1-51 F Z Immunoglobulin lambda variable 1-51 GDB: Human IGL Gene Nomenclature Exp Clin Immunogenet 2001;18:
5 Table 1 (continued) gene group gene name (1) functionality reference sequence accession numbers Number of alleles gene definition (2) GDB LocusLink IGLV1-62 P D87022 Immunoglobulin lambda variable 1-62 GDB: IGLV2-5 P Z73641 Immunoglobulin lambda variable 2-5 GDB: IGLV2-8 F X Immunoglobulin lambda variable 2-8 GDB: IGLV2-11 F Z Immunoglobulin lambda variable 2-11 GDB: IGLV2-14 F Z Immunoglobulin lambda variable 2-14 GDB: IGLV2-18 F Z Immunoglobulin lambda variable 2-18 GDB: IGLV2-23 F X Immunoglobulin lambda variable 2-23 GDB: IGLV2-28 P X97466 Immunoglobulin lambda variable 2-28 GDB: IGLV2-33 ORF Z Immunoglobulin lambda variable 2-33 GDB: IGLV2-34 P D87013 Immunoglobulin lambda variable 2-34 GDB: IGLV3-1 F X Immunoglobulin lambda variable 3-1 GDB: IGLV3-2 P X97468 Immunoglobulin lambda variable 3-2 GDB: IGLV3-4 P D87024 Immunoglobulin lambda variable 3-4 GDB: IGLV3-6 P X97465 Immunoglobulin lambda variable 3-6 GDB: IGLV3-7 P X97470 Immunoglobulin lambda variable 3-7 GDB: IGLV3-9 F, P X Immunoglobulin lambda variable 3-9 GDB: IGLV3-10 F X Immunoglobulin lambda variable 3-10 GDB: IGLV3-12 F Z Immunoglobulin lambda variable 3-12 GDB: IGLV3-13 P X97463 Immunoglobulin lambda variable 3-13 GDB: IGLV3-15 P D87015 Immunoglobulin lambda variable 3-15 GDB: IGLV3-16 F X Immunoglobulin lambda variable 3-16 GDB: IGLV3-17 P X97472 Immunoglobulin lambda variable 3-17 GDB: IGLV3-19 F X Immunoglobulin lambda variable 3-19 GDB: IGLV3-21 F X Immunoglobulin lambda variable 3-21 GDB: IGLV3-22 F, P Z Immunoglobulin lambda variable 3-22 GDB: IGLV3-24 P X71968 Immunoglobulin lambda variable 3-24 GDB: IGLV3-25 F X Immunoglobulin lambda variable 3-25 GDB: IGLV3-26 P X97467 Immunoglobulin lambda variable 3-26 GDB: IGLV3-27 F D Immunoglobulin lambda variable 3-27 GDB: IGLV3-29 P Z73644 Immunoglobulin lambda variable 3-29 GDB: IGLV3-30 P Z73646 Immunoglobulin lambda variable 3-30 GDB: IGLV3-31 P X97469 Immunoglobulin lambda variable 3-31 GDB: IGLV3-32 ORF Z Immunoglobulin lambda variable 3-32 GDB: IGLV4-3 F X Immunoglobulin lambda variable 4-3 GDB: IGLV4-60 F Z Immunoglobulin lambda variable 4-60 GDB: IGLV4-69 F Z Immunoglobulin lambda variable 4-69 GDB: IGLV5-37 F Z Immunoglobulin lambda variable 5-37 GDB: (*)IGLV5-39 F Z Immunoglobulin lambda variable 5-39 GDB: Exp Clin Immunogenet 2001;18: Lefranc
6 IGLV5-45 F Z Immunoglobulin lambda variable 5-45 GDB: IGLV5-48 ORF Z Immunoglobulin lambda variable 5-48 GDB: IGLV5-52 F Z Immunoglobulin lambda variable 5-52 GDB: IGLV6-57 F Z Immunoglobulin lambda variable 6-57 GDB: IGLV7-35 P Z73660 Immunoglobulin lambda variable 7-35 GDB: IGLV7-43 F X Immunoglobulin lambda variable 7-43 GDB: IGLV7-46 F, P Z Immunoglobulin lambda variable 7-46 GDB: IGLV8-61 F Z Immunoglobulin lambda variable 8-61 GDB: IGLV9-49 F Z Immunoglobulin lambda variable 9-49 GDB: IGLV10-54 F Z Immunoglobulin lambda variable GDB: IGLV10-67 P Z73651 Immunoglobulin lambda variable GDB: IGLV11-55 ORF D Immunoglobulin lambda variable GDB: IGLV(I)-20 P D87007 Immunoglobulin lambda variable (I)-20 GDB: IGLV(I)-38 P D87009 Immunoglobulin lambda variable (I)-38 GDB: IGLV(I)-42 P X14613 Immunoglobulin lambda variable (I)-42 GDB: IGLV(I)-56 P D86996 Immunoglobulin lambda variable (I)-56 GDB: IGLV(I)-63 P D87022 Immunoglobulin lambda variable (I)-63 GDB: IGLV(I)-68 P D86993 Immunoglobulin lambda variable (I)-68 GDB: IGLV(I)-70 P D86993 Immunoglobulin lambda variable (I)-70 GDB: IGLV(IV)-53 P D86996 Immunoglobulin lambda variable (IV)-53 GDB: IGLV(IV)-59 P D87000 Immunoglobulin lambda variable (IV)-59 GDB: IGLV(IV)-64 P D87022 Immunoglobulin lambda variable (IV)-64 GDB: IGLV(IV)-65 P D87022 Immunoglobulin lambda variable (IV)-65 GDB: IGLV(IV)-66-1 P D87004 Immunoglobulin lambda variable (IV)-66-1 GDB: IGLV(V)-58 P D87000 Immunoglobulin lambda variable (V)-58 GDB: IGLV(V)-66 P D87004 Immunoglobulin lambda variable (V)-66 GDB: IGLV(VI)-22-1 P X71351 Immunoglobulin lambda variable (VI) IGLV(VI)-25-1 P D86994 Immunoglobulin lambda variable (VI) IGLV(VII)-41-1 P X99568 Immunoglobulin lambda variable (VII) (1) gene names have been approved by the Human Genome Organization (HUGO) Nomenclature Committee in Note that, in the HUGO symbols, parentheses in the names of pseudogenes assigned to clans are omitted. Otherwise all the gene names (gene symbols) are identical in and HUGO nomenclatures. (2) Gene definitions (full names) are identical (including parentheses) in and HUGO nomenclatures. Other entries concerning the IGL locus or groups, in the OMIM, GDB, and LocusLink genome databases, and in HUGO: designation definition a OMIM GDB LocusLink HUGO IGL locus Immunoglobulin lambda locus GDB: IGL@ IGLC group Immunoglobulin lambda constant group GDB: IGLC@ GLJ group Immunoglobulin lambda joining group GDB: IGLJ@ a Entry definitions are identical in, GDB, LocusLink and HUGO. Human IGL Gene Nomenclature Exp Clin Immunogenet 2001;18:
7 Table 2. Human IGL orphons on chromosomes 8 and 22 a On chromosome 8 at 8q11.2 gene group gene name (1) functionality reference sequence accession numbers Number of alleles gene definition (2) Chromosomal localization GDB LocusLink IGLV IGLV8/OR8-1 ORF, P Y08831 Immunoglobulin lambda variable 8/OR8-1 8q11.2 GDB: IGLV/OR8-2 Immunoglobulin lambda variable /OR8-2 8q11.2 (provisional) b On chromosome 22 gene group gene name (1) functionality reference sequence accession numbers Number of alleles gene definition (2) Chromosomal localization GDB LocusLink IGLC IGLC/OR22-1 AL Immunoglobulin lambda constant /OR q q12.3 GDB: IGLC/OR22-2 AL Immunoglobulin lambda constant /OR q q12.3 GDB: IGLV(IV)/OR22-1 AL Immunoglobulin lambda variable (IV)/OR q q12.1 GDB: IGLV(IV)/OR22-2 AL Immunoglobulin lambda variable (IV)/OR q q12.3 GDB: c Not localized Another orphon has been identified but not yet sequenced gene group gene name (1) functionality reference sequence accession numbers Number of alleles gene definition (2) Chromosomal localization GDB LocusLink IGLV IGLV8/OR2 Immunoglobulin lambda variable 8/OR2 (provisional) unknown (1) Note that in the HUGO symbols, slashes of the orphon names and parentheses are omitted. Otherwise the gene names (gene symbols) are identical in and HUGO nomenclatures. (2) Gene definitions (full names) are identical (including slashes and parentheses) in and HUGO nomenclatures. Other entries concerning the IGL orphons in the GDB and LocusLink genome databases. 248 Exp Clin Immunogenet 2001;18: Lefranc
8 Orphons Six IGL genes have been found outside the main locus in other chromosomal localizations (table 2). These genes, designated as orphons, cannot contribute to the synthesis of the immunoglobulin lambda chains, even if they have an open reading frame (ORF). Two IGLV orphons have been identified on chromosome 8 at 8q11.2 and one of them belonging to subgroup 8 has been sequenced [38]. Two IGLC orphons and two IGLV orphons have been characterized on 22q outside of the major IGL locus [26] (table 2). IGL Gene Nomenclature and Scientific Chart Gene Names Gene names (table 1, 2) are designated according to the gene name nomenclature for Ig and T cell receptors of all vertebrates based on the CLASSIFICATION concept of -ONTOLOGY [39] (Appendix 1) and according to the rules of the Scientific chart [20, 22] available at imgt.cines.fr. gene names and gene definitions for the human IG [40] and TR genes [41] were approved by the HUGO Nomenclature Committee in Note that in the HUGO symbols ( slashes and parentheses are omitted and capital letters replace the lowercase letters found in some provisional gene names. Otherwise the gene symbols and all the full names (including slashes and parentheses) are identical in and HUGO nomenclatures. Functionality Criteria of functionality (F = functional, P = pseudogene, ORF = open reading frame) (table 1, 2) have been described in the Scientific chart [1]. The definition of functionality is based on the sequence analysis. As examples, the instances functional (for germline V, D, J, and for C sequences) mean that the coding regions have an ORF without a stop codon, and that there is no described defect in the splicing sites and/or recombination signals and/or regulatory elements. According to the gravity of the identified defects, the functionality can be defined as ORF, pseudogene or vestigial (for germline V, D, J and for C genes) [1]. Complete definitions are available in the Scientific chart at the Marie-Paule page. Information on gene rearrangement, DNA transcription into mrna and RNA translation into a polypeptide chain is provided in the Germline gene tables in the Repertoire (columns designated as R, T and Pr, respectively), and has been published in a previous Locus in Focus report [2]. This information is extracted from the literature and through an /LIGM-DB sequence database search [21 23]. The /V-QUEST tool, available at the Home page at imgt.cines.fr, allows the identification of the germline IGLV and IGLJ genes from IGLV-J genomic rearrangements and transcripts, and provides translation and two-dimensional representation (Collier de Perles) of the variable regions [21, 22, 42]. Reference Sequences For each gene, an reference sequence accession number is given (table 1, 2). For the functional or ORF genes, the reference sequence accession number is that corresponding to the allele*01. Note that the number *01 does not necessarily mean that other alleles are already known: it signifies that any new polymorphic sequence will be described by comparison to that allele *01. Although the accession numbers are the same as those from the EMBL/GenBank/ Human IGL Gene Nomenclature Exp Clin Immunogenet 2001;18:
9 Table 3. Correspondence between the human IGLV nomenclatures. IGLV genes are listed from 3) in the IGL locus (top of the table) to 5) (bottom of the table), IGLV gene name [2, 19] Frippiat et al. [27] Williams, Frippiat et al. [29] Kawasaki et al. [30], IGLV gene name [2, 19] Frippiat et al. [27] Williams, Frippiat et al. [29] Kawasaki et al. [30] 3-1 3r a q 2-2P e c 5-1 (I) P P a 2-5 2a1 1-1P e a2 2-4P d 1-14P 3-7 3n 2-5P (VII)-41-1 lambdavgl 2-8 2c 1-2 (I)-42 V lambda A 1-15P 3-9 3j a p c e c i b f 2-9P g a d P a a f g 2-12P b d b l 2-13 (IV) P (I) P a h e 2-15 (I) P (VI)-22-1 lambdavg a 1-22P b2 1-7 (V) P d 2-16P (IV) P m a 5-4 (VI)-25-1 lambdavg a b 2-18P P (I) P b1 1-8P (IV) P c 2-20P (IV) P o 2-21P (V) P k 2-22P (IV) i1 2-23P b 1-25P f 1-9 (I) P P b c 3-1P (I) P DDBJ generalist databases, the content of the /LIGM-DB flat files differs in terms of the expert annotations added by. Alleles The number of alleles of the human IGLV, IGLJ and IGLC genes (table 1, 2) is determined according to Tables of alleles and Alignments of alleles, in the Repertoire, at A dash ( ) indicates that allele polymorphism of the pseudogenes has not been studied. Alignments of all known germline functional and ORF sequences assigned to the different alleles, by comparison to the allele *01, are displayed in another source [19]. Human IGL entries in 250 Exp Clin Immunogenet 2001;18: Lefranc
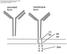
10 this source include 53 genes and 108 alleles, with a total of 213 sequences [19]. Genome Database Accession Numbers All /HUGO human IGL gene symbols, full names and reference sequence accession numbers have been entered into the Genome Database GDB, Toronto, Canada ( and into LocusLink at NCBI (National Center for Biotechnology Information), Bethesda, Md., USA ( Accession ID to these genome databases are provided in table 1 and 2. Links to OMIM (Online Mendelian Inheritance in Man, MIM) ( are cited when there are existing entries in OMIM. Links to the individual, GDB and LocusLink gene entries are available from imgt.cines.fr from Repertoire 1 Lists of IG and TR. Correspondences between Nomenclatures and Numberings Correspondence between the human IGLV gene nomenclatures is reported in table 3. In order to be able to easily compare sequences of immunoglobulins and T cell receptors, a unique numbering has been defined for the variable regions [42, 43]. Correspondence between the unique numbering and other numberings for the human IGLV genes is available from the Scientific chart and a previous report [42]. The unique numbering relies on the high conservation of the structure of the variable region. This numbering takes into account and combines the definition of the framework (FR) and complementarity determining regions (CDR) [44], structural data from X-ray diffraction studies [45], and the characterization of the hypervariable loops [46]. The unique numbering has allowed the redefinition of the limits of the FR and CDR [42]. The FR- and CDR- lengths themselves become crucial information characterizing the variable regions belonging to a group, a subgroup, and/or a gene. For example, for a germline gene of the human IGLV2 subgroup, the lengths of the 3 CDR-, expressed as the number of amino acids, are designated as [9.3.9] ( Repertoire12D and 3D structures) [7, 42]. The unique numbering is used as the output of the / V-QUEST alignment tool, and in the Alignments of alleles ( Repertoire1Proteins and alleles) [19]. Acknowledgments I thank Gérard Lefranc, Ruth Lovering, Donna Maglott, Chris Porter, Sue Povey, Conover Talbot and all my immunologist colleagues for their contribution to the Nomenclature Committee. I am grateful to Valérie Contet for the editorial work. is funded by the European Union s 5th PCRDT (QLG ) program, the Centre National de la Recherche Scientifique, the Ministère de la Recherche and the Ministère de l Education Nationale. Subventions have been received from Association pour la Recherche sur le Cancer and the Région Languedoc- Roussillon. Appendix 1 The CLASSIFICATION Concept of -ONTOLOGY The CLASSIFICATION concept of -ON- TOLOGY (fig. 3) organizes the immunogenetic knowledge that is useful for the naming and classification of the immunoglobulin genes [39]. Locus : A locus is a group of immunoglobulin genes that are ordered and localized in the same chromosomal location in a given species. The IGL locus (22q11.2) is one of the three main immunoglobulin loci in the human genome. Immunoglobulin genes have also been identified in other chromosomal locations outside the main loci, which represent new Human IGL Gene Nomenclature Exp Clin Immunogenet 2001;18:
11 Fig. 3. The CLASSIFICATION concept of -ONTOLOGY, exemplified for the IGLV genes. instances of the locus concept. However, the genes they contain, designated as orphons, are not functional. Group : A group is a set of genes which share the same gene type (V, D, J or C) and potentially participate in the synthesis of a polypeptide of the same chain type. By extension, a group includes the related pseudogenes and orphons. A 4-letter root designates the group, for example, IGLV, IGLJ, and IGLC for the immunoglobulin lambda genes. Subgroup : A subgroup is a set of genes which belong to the same group, in a given species, and which share at least 75% identity at the nucleotide level (in the germline configuration for V, D, and J). For example, the IGLV genes belong to 11 subgroups. Gene : A gene is defined as a DNA sequence that can be potentially transcribed and/or translated (this definition includes the regulatory elements in 5) and 3), and the introns, if present). Instances of the gene concept are gene names. By extension, orphons and pseudogenes are also instances of the gene concept. For each gene, has defined a reference sequence [20]. For the V, D, and J genes, the reference sequence corresponds to a germline entity. The rules for the choice of the reference sequences are described at in the Scientific chart. Allele : An allele is a polymorphic variant of a gene. Alleles are described, exhaustively and in a standardized way, for the four core coding regions, that is for the germline V-REGIONs, D-REGIONs and J-REGIONs and for the C-REGIONs of immunoglobulin genes. These alleles refer to sequence polymorphisms, with mutations described at the sequence level [1]. Their sequences are compared to the reference sequence designated as *01 (see Scientific chart at for description of mutations and allele nomenclature for sequence polymorphisms). 252 Exp Clin Immunogenet 2001;18: Lefranc
12 References 1 Lefranc MP: (ImMunoGeneTics) Locus on Focus. A new section of Experimental and Clinical Immunogenetics. Exp Clin Immunogenet 1998;15: Pallarès N, Frippiat JP, Giudicelli V, Lefranc MP: The human immunoglobulin lambda variable (IGLV) genes and joining (IGLJ) segments. Exp Clin Immunogenet 1998;15: Barbié V, Lefranc MP: The human immunoglobulin kappa variable (IGKV) genes and joining (IGKJ) segments. Exp Clin Immunogenet 1998;15: Martinez C, Lefranc MP: The mouse (Mus musculus) immunoglobulin kappa variable (IGKV) genes and joining (IGKJ) segments. Exp Clin Immunogenet 1998;15: Pallarès N, Lefebvre S, Contet V, Matsuda F, Lefranc MP: The human immunoglobulin heavy variable (IGHV) genes. Exp Clin Immunogenet 1999;16: Ruiz M, Pallarès N, Contet V, Barbié V, Lefranc MP: The human immunoglobulin heavy diversity (IGHD) and joining (IGHJ) segments. Exp Clin Immunogenet 1999;16: Scaviner D, Barbié V, Ruiz M, Lefranc MP: Protein displays of the human immunoglobulin heavy, kappa and lambda variable and joining regions. Exp Clin Immunogenet 1999;16: Folch G, Lefranc MP: The human T cell receptor beta variable (TRBV) genes. Exp Clin Immunogenet 2000; 17: Scaviner D, Lefranc MP: The human T cell receptor alpha variable (TRAV) genes. Exp Clin Immunogenet 2000;17: Scaviner D, Lefranc MP: The human T cell receptor alpha joining (TRAJ) genes. Exp Clin Immunogenet 2000;17: Folch G, Lefranc MP: The human T cell receptor beta diversity (TRBD) and beta joining (TRBJ) genes. Exp Clin Immunogenet 2000;17: Artero S, Lefranc MP: The Teleostei immunoglobulin heavy IGH genes. Exp Clin Immunogenet 2000;17: Artero S, Lefranc MP: The Teleostei immunoglobulin light IGL1 and IGL2 V, J and C genes. Exp Clin Immunogenet 2000;17: Folch G, Scaviner D, Contet V, Lefranc MP: Protein displays of the human T cell receptor alpha, beta, gamma and delta variable and joining regions. Exp Clin Immunogenet 2000;17: Bosc N, Lefranc MP: The mouse (Mus musculus) T cell receptor beta variable (TRBV), diversity (TRBD), and joining (TRBJ) genes. Exp Clin Immunogenet 2000;17: Bosc N, Contet V, Lefranc MP: The mouse (Mus musculus) T cell receptor delta variable (TRDV), diversity (TRDD), and joining (TRDJ) genes. Exp Clin Immunogenet 2001;18: Lefranc MP: Nomenclature of the human immunoglobulin heavy (IGH) genes. Exp Clin Immunogenet 2001;18: Lefranc MP: Nomenclature of the human immunoglobulin kappa (IGK) genes. Exp Clin Immunogenet 2001;18: Lefranc MP, Lefranc, G: The Immunoglobulin Facts Book. London, Academic Press, 2001, pp Lefranc MP, Giudicelli V, Ginestoux C, Bodmer J, Müller W, Bontrop R, Lemaître M, Malik A, Barbié V, Chaume D:, the international ImMunoGeneTics database. Nucleic Acids Res 1999;27: Ruiz M, Giudicelli V, Ginestoux C, Stoehr P, Robinson J, Bödmer J, Marsh S, Bontrop R, Lemaître M, Lefranc G, Chaume D, Lefranc MP:, the international ImMuno- GeneTics database. Nucleic Acids Res 2000;28: Lefranc MP: ImMunoGeneTics database. International BIOforum 2000;4: Lefranc MP:, the international ImMunoGeneTics database. Nucleic Acids Res 2001;29: Erikson J, Martinis J, Croce CM: Assignment of the genes for human lambda immunoglobulin chains to chromosome 22. Nature 1981;294: Emanuel BS, Cannizzaro LA, Magrath I, Tsujimoto Y, Nowell PC, Croce CM: Chromosomal orientation of the lambda light chain locus: V lambda proximal to C lambda in 22q11. Nucleic Acids Res 1985;13: Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, Ainscough R, Almeida JP, Babbage A, Bagguley C, Bailey J, Barlow K, Bates KN, Beasley O, Bird CP, Blakey S, Bridgeman AM, Buck D, Burgess J, Burrill WD, O Brien KP, et al: The DNA sequence of human chromosome 22. Nature 1999;402: Frippiat JP, Williams SC, Tomlinson IM, Cook GP, Cherif D, Le Paslier D, Collins JE, Dunham I, Winter G, Lefranc MP: Organization of the human immunoglobulin lambda light-chain locus on chromosome 22q11.2. Hum Mol Genet 1995;4: Kawasaki K, Minoshima S, Schooler K, Kudoh J, Asakaw S, de Jong PJ, Shimizu N: The organization of the human immunoglobulin lambda gene locus. Genome Res 1995;5: Williams SC, Frippiat JP, Tomlinson IM, Ignatovich O, Lefranc MP, Winter G: Sequence and evolution of the human germline V lambda repertoire. J Mol Biol 1996;264: Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits JL, Wang J, Shimizu N: One-megabase sequence analysis of the human immunoglobulin lambda gene locus. Genome Res 1997;7: Hieter PA, Hollis GF, Korsmeyer SJ, Waldmann TA, Leder P: Clustered arrangement of immunoglobulin lambda constant region genes in man. Nature 1981;294: Human IGL Gene Nomenclature Exp Clin Immunogenet 2001;18:
13 32 Taub RA, Hollis GF, Hieter PA, Korsmeyer SJ, Waldmann TA, Leder P: Variable amplification of immunoglobulin lambda light-chain genes in human populations. Nature 1983;304: Dariavach P, Lefranc G, Lefranc MP: Human immunoglobulin C lambda 6 gene encodes the Kern+Oz-lambda chain and C lambda 4 and C lambda 5 are pseudogenes. Proc Natl Acad Sci USA 1987;84: Vasicek TJ, Leder P: Structure and expression of the human immunoglobulin lambda genes. J Exp Med 1990;172: Ghanem N, Dariavach P, Bensmana M, Chibani J, Lefranc G, Lefranc MP: Polymorphism of immunoglobulin lambda constant region genes in populations from France, Lebanon and Tunisia. Exp Clin Immunogenet 1988;5: Kay PH, Moriuchi J, Ma PJ, Saueracher E: An unusual allelic from of the immunoglobulin lambda constant region genes in the Japanese. Immunogenetics 1992;35: Lefranc MP, Pallarès N, Frippiat JP: Allelic polymorphisms and RFLP in the human immunoglobulin lambda light chain locus. Hum Genet 1999;104: Frippiat JP, Dard P, Marsh S, Winter G, Lefranc MP: Immunoglobulin lambda light chain orphons on human chromosome 8q11.2. Eur J Immunol 1997;27: Giudicelli V, Lefranc MP: Ontology for immunogenetics: -ON- TOLOGY. Bioinformatics 1999;12: Lefranc MP: Nomenclature of the human immunoglobulin genes. Curr Protocols Immunol 2000;A.1P.1- A.1P Lefranc MP: Nomenclature of the human T cell Receptor genes. Curr Protocols Immunol 2000;A.1O.1- A.1O Lefranc MP: The unique numbering for Immunoglobulins, T cell receptors and Ig-like domains. Immunologist 1999;7: Lefranc MP: Unique database numbering system for immunogenetic analysis. Immunol Today 1997;8: Kabat EA, Wu TT, Reid-Miller M, Perry HM, Gottesman KS: Sequences of Proteins of Immunological Interest, ed 4. Washington, Public Health Service Satow Y, Cohen GH, Padlan EA, Davies DR: Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A. J Mol Biol 1986;190: Chothia C, Lesk AM: Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol 1987;196: Exp Clin Immunogenet 2001;18: Lefranc
The Human T Cell Receptor Alpha Joining (TRAJ) Genes
 IMGT Locus in Focus Exp Clin Immunogenet 2000;17:97 106 Received: October 15, 1999 The Human T Cell Receptor Alpha Joining (TRAJ) Genes Dominique Scaviner Marie-Paule Lefranc Laboratoire d ImmunoGénétique
IMGT Locus in Focus Exp Clin Immunogenet 2000;17:97 106 Received: October 15, 1999 The Human T Cell Receptor Alpha Joining (TRAJ) Genes Dominique Scaviner Marie-Paule Lefranc Laboratoire d ImmunoGénétique
The Human T Cell Receptor Beta Diversity (TRBD) and Beta Joining (TRBJ) Genes
 IMGT Locus in Focus Exp Clin Immunogenet 2000;17:107 114 Received: October 15, 1999 The Human T Cell Receptor Beta Diversity (TRBD) and Beta Joining (TRBJ) Genes Géraldine Folch Marie-Paule Lefranc Laboratoire
IMGT Locus in Focus Exp Clin Immunogenet 2000;17:107 114 Received: October 15, 1999 The Human T Cell Receptor Beta Diversity (TRBD) and Beta Joining (TRBJ) Genes Géraldine Folch Marie-Paule Lefranc Laboratoire
The Human T Cell Receptor Alpha Variable (TRAV) Genes
 IMGT Locus in Focus Exp Clin Immunogenet 2000;17:83 96 Received: September 23, 1999 The Human T Cell Receptor Alpha Variable (TRAV) Genes Dominique Scaviner Marie-Paule Lefranc Laboratoire d ImmunoGénétique
IMGT Locus in Focus Exp Clin Immunogenet 2000;17:83 96 Received: September 23, 1999 The Human T Cell Receptor Alpha Variable (TRAV) Genes Dominique Scaviner Marie-Paule Lefranc Laboratoire d ImmunoGénétique
The Human T cell Receptor Beta Variable (TRBV) Genes
 IMGT Locus in Focus Exp Clin Immunogenet 2000;17:42 54 Received: July 3, 1999 The Human T cell Receptor Beta Variable (TRBV) Genes Géraldine Folch Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire,
IMGT Locus in Focus Exp Clin Immunogenet 2000;17:42 54 Received: July 3, 1999 The Human T cell Receptor Beta Variable (TRBV) Genes Géraldine Folch Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire,
The Teleostei Immunoglobulin Heavy IGH Genes
 IMGT Locus in Focus Exp Clin Immunogenet 2000;17:148 161 Received: November 19, 1999 The Teleostei Immunoglobulin Heavy IGH Genes Sylvaine Artéro Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire,
IMGT Locus in Focus Exp Clin Immunogenet 2000;17:148 161 Received: November 19, 1999 The Teleostei Immunoglobulin Heavy IGH Genes Sylvaine Artéro Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire,
The Human Immunoglobulin Heavy Variable Genes
 IMGT Locus on Focus Exp Clin Immunogenet 1999;16:36 60 Received: September 25, 1998 The Human Immunoglobulin Heavy Variable Genes Nathalie Pallarès a Sophie Lefebvre a Valérie Contet a Fumihiko Matsuda
IMGT Locus on Focus Exp Clin Immunogenet 1999;16:36 60 Received: September 25, 1998 The Human Immunoglobulin Heavy Variable Genes Nathalie Pallarès a Sophie Lefebvre a Valérie Contet a Fumihiko Matsuda
The Human Immunoglobulin Kappa Variable (IGKV) Genes and Joining (IGKJ) Segments
 IMGT Locus on Focus Exp Clin Immunogenet 1998;15:171 183 Received: June 13, 1998 Valérie Barbié Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire, CNRS, Université Montpellier II, Montpellier,
IMGT Locus on Focus Exp Clin Immunogenet 1998;15:171 183 Received: June 13, 1998 Valérie Barbié Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire, CNRS, Université Montpellier II, Montpellier,
Nomenclature of the Human Immunoglobulin Heavy (IGH) Genes
 Locus in Focus Exp Clin Immunogenet 2001;18:100 116 Received: August 31, 2000 Nomenclature of the Human Immunoglobulin Heavy (IGH) Genes Marie-Paule Lefranc Chairperson, Nomenclature Committee, CNRS, Université
Locus in Focus Exp Clin Immunogenet 2001;18:100 116 Received: August 31, 2000 Nomenclature of the Human Immunoglobulin Heavy (IGH) Genes Marie-Paule Lefranc Chairperson, Nomenclature Committee, CNRS, Université
IMGT Collier de Perles: the New Look for IgSF and MhcSF in IMGT
 IMGT Collier de Perles: the New Look for IgSF and MhcSF in IMGT Quentin Kaas and Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire Université Montpellier, UPR CNRS 1142, IGH Institut Universitaire
IMGT Collier de Perles: the New Look for IgSF and MhcSF in IMGT Quentin Kaas and Marie-Paule Lefranc Laboratoire d ImmunoGénétique Moléculaire Université Montpellier, UPR CNRS 1142, IGH Institut Universitaire
Literature databases OMIM
 Literature databases OMIM Online Mendelian Inheritance in Man OMIM OMIM is a database that catalogues all the known diseases with a genetic component, and when possible links them to the relevant genes
Literature databases OMIM Online Mendelian Inheritance in Man OMIM OMIM is a database that catalogues all the known diseases with a genetic component, and when possible links them to the relevant genes
Annotation of Chimp Chunk 2-10 Jerome M Molleston 5/4/2009
 Annotation of Chimp Chunk 2-10 Jerome M Molleston 5/4/2009 1 Abstract A stretch of chimpanzee DNA was annotated using tools including BLAST, BLAT, and Genscan. Analysis of Genscan predicted genes revealed
Annotation of Chimp Chunk 2-10 Jerome M Molleston 5/4/2009 1 Abstract A stretch of chimpanzee DNA was annotated using tools including BLAST, BLAT, and Genscan. Analysis of Genscan predicted genes revealed
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10495 WWW.NATURE.COM/NATURE 1 2 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 3 4 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 5 6 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 7 8 WWW.NATURE.COM/NATURE
doi:10.1038/nature10495 WWW.NATURE.COM/NATURE 1 2 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 3 4 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 5 6 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 7 8 WWW.NATURE.COM/NATURE
Hands-On Ten The BRCA1 Gene and Protein
 Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Bioinformatics Laboratory Exercise
 Bioinformatics Laboratory Exercise Biology is in the midst of the genomics revolution, the application of robotic technology to generate huge amounts of molecular biology data. Genomics has led to an explosion
Bioinformatics Laboratory Exercise Biology is in the midst of the genomics revolution, the application of robotic technology to generate huge amounts of molecular biology data. Genomics has led to an explosion
SALSA MLPA KIT P050-B2 CAH
 SALSA MLPA KIT P050-B2 CAH Lot 0510, 0909, 0408: Compared to lot 0107, extra control fragments have been added at 88, 96, 100 and 105 nt. The 274 nt probe gives a higher signal in lot 0510 compared to
SALSA MLPA KIT P050-B2 CAH Lot 0510, 0909, 0408: Compared to lot 0107, extra control fragments have been added at 88, 96, 100 and 105 nt. The 274 nt probe gives a higher signal in lot 0510 compared to
Immunoglobulin light chain repertoire in chronic lymphocytic leukemia
 NEOPLASIA Immunoglobulin light chain repertoire in chronic lymphocytic leukemia Kostas Stamatopoulos, Chrysoula Belessi, Anastasia Hadzidimitriou, Tatjana Smilevska, Evangelia Kalagiakou, Katerina Hatzi,
NEOPLASIA Immunoglobulin light chain repertoire in chronic lymphocytic leukemia Kostas Stamatopoulos, Chrysoula Belessi, Anastasia Hadzidimitriou, Tatjana Smilevska, Evangelia Kalagiakou, Katerina Hatzi,
Antibodies and T Cell Receptor Genetics Generation of Antigen Receptor Diversity
 Antibodies and T Cell Receptor Genetics 2008 Peter Burrows 4-6529 peterb@uab.edu Generation of Antigen Receptor Diversity Survival requires B and T cell receptor diversity to respond to the diversity of
Antibodies and T Cell Receptor Genetics 2008 Peter Burrows 4-6529 peterb@uab.edu Generation of Antigen Receptor Diversity Survival requires B and T cell receptor diversity to respond to the diversity of
George R. Honig Junius G. Adams III. Human Hemoglobin. Genetics. Springer-Verlag Wien New York
 George R. Honig Junius G. Adams III Human Hemoglobin Genetics Springer-Verlag Wien New York George R. Honig, M.D., Ph.D. Professor and Head Department of Pediatrics, College of Medicine University of Illinois
George R. Honig Junius G. Adams III Human Hemoglobin Genetics Springer-Verlag Wien New York George R. Honig, M.D., Ph.D. Professor and Head Department of Pediatrics, College of Medicine University of Illinois
Antigen Receptor Structures October 14, Ram Savan
 Antigen Receptor Structures October 14, 2016 Ram Savan savanram@uw.edu 441 Lecture #8 Slide 1 of 28 Three lectures on antigen receptors Part 1 (Today): Structural features of the BCR and TCR Janeway Chapter
Antigen Receptor Structures October 14, 2016 Ram Savan savanram@uw.edu 441 Lecture #8 Slide 1 of 28 Three lectures on antigen receptors Part 1 (Today): Structural features of the BCR and TCR Janeway Chapter
Submitted to Leukemia as a Letter to the Editor, May Male preponderance in chronic lymphocytic leukemia utilizing IGHV 1-69.
 Submitted to Leukemia as a Letter to the Editor, May 2007 To the Editor, Leukemia :- Male preponderance in chronic lymphocytic leukemia utilizing IGHV 1-69. Gender plays an important role in the incidence,
Submitted to Leukemia as a Letter to the Editor, May 2007 To the Editor, Leukemia :- Male preponderance in chronic lymphocytic leukemia utilizing IGHV 1-69. Gender plays an important role in the incidence,
T cell Receptor. Chapter 9. Comparison of TCR αβ T cells
 Chapter 9 The αβ TCR is similar in size and structure to an antibody Fab fragment T cell Receptor Kuby Figure 9-3 The αβ T cell receptor - Two chains - α and β - Two domains per chain - constant (C) domain
Chapter 9 The αβ TCR is similar in size and structure to an antibody Fab fragment T cell Receptor Kuby Figure 9-3 The αβ T cell receptor - Two chains - α and β - Two domains per chain - constant (C) domain
38 Int'l Conf. Bioinformatics and Computational Biology BIOCOMP'16
 38 Int'l Conf. Bioinformatics and Computational Biology BIOCOMP'16 PGAR: ASD Candidate Gene Prioritization System Using Expression Patterns Steven Cogill and Liangjiang Wang Department of Genetics and
38 Int'l Conf. Bioinformatics and Computational Biology BIOCOMP'16 PGAR: ASD Candidate Gene Prioritization System Using Expression Patterns Steven Cogill and Liangjiang Wang Department of Genetics and
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins.
 Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
Immunoglobulin diversity gene usage predicts unfavorable outcome in a subset of chronic lymphocytic leukemia patients
 Research article Immunoglobulin diversity gene usage predicts unfavorable outcome in a subset of chronic lymphocytic leukemia patients Renee C. Tschumper, 1 Susan M. Geyer, 2 Megan E. Campbell, 2 Neil
Research article Immunoglobulin diversity gene usage predicts unfavorable outcome in a subset of chronic lymphocytic leukemia patients Renee C. Tschumper, 1 Susan M. Geyer, 2 Megan E. Campbell, 2 Neil
Introduction to Genetics
 Introduction to Genetics Table of contents Chromosome DNA Protein synthesis Mutation Genetic disorder Relationship between genes and cancer Genetic testing Technical concern 2 All living organisms consist
Introduction to Genetics Table of contents Chromosome DNA Protein synthesis Mutation Genetic disorder Relationship between genes and cancer Genetic testing Technical concern 2 All living organisms consist
Bio 111 Study Guide Chapter 17 From Gene to Protein
 Bio 111 Study Guide Chapter 17 From Gene to Protein BEFORE CLASS: Reading: Read the introduction on p. 333, skip the beginning of Concept 17.1 from p. 334 to the bottom of the first column on p. 336, and
Bio 111 Study Guide Chapter 17 From Gene to Protein BEFORE CLASS: Reading: Read the introduction on p. 333, skip the beginning of Concept 17.1 from p. 334 to the bottom of the first column on p. 336, and
DNA codes for RNA, which guides protein synthesis.
 Section 3: DNA codes for RNA, which guides protein synthesis. K What I Know W What I Want to Find Out L What I Learned Vocabulary Review synthesis New RNA messenger RNA ribosomal RNA transfer RNA transcription
Section 3: DNA codes for RNA, which guides protein synthesis. K What I Know W What I Want to Find Out L What I Learned Vocabulary Review synthesis New RNA messenger RNA ribosomal RNA transfer RNA transcription
LTA Analysis of HapMap Genotype Data
 LTA Analysis of HapMap Genotype Data Introduction. This supplement to Global variation in copy number in the human genome, by Redon et al., describes the details of the LTA analysis used to screen HapMap
LTA Analysis of HapMap Genotype Data Introduction. This supplement to Global variation in copy number in the human genome, by Redon et al., describes the details of the LTA analysis used to screen HapMap
IgCLL group activities in 2009 and beyond. Kostas Stamatopoulos Hematology Department and HCT Unit George Papanicolaou Hospital, Thessaloniki, Greece
 IgCLL group activities in 2009 and beyond Kostas Stamatopoulos Hematology Department and HCT Unit George Papanicolaou Hospital, Thessaloniki, Greece Troubleshooting Standardization Standardization and
IgCLL group activities in 2009 and beyond Kostas Stamatopoulos Hematology Department and HCT Unit George Papanicolaou Hospital, Thessaloniki, Greece Troubleshooting Standardization Standardization and
The development of clonality testing for lymphomas in the Bristol Genetics Laboratory. Dr Paula Waits Bristol Genetics Laboratory
 The development of clonality testing for lymphomas in the Bristol Genetics Laboratory Dr Paula Waits Bristol Genetics Laboratory Introduction The majority of lymphoid malignancies belong to the B cell
The development of clonality testing for lymphomas in the Bristol Genetics Laboratory Dr Paula Waits Bristol Genetics Laboratory Introduction The majority of lymphoid malignancies belong to the B cell
1 By Drs. Ingrid Waldron and. Jennifer Doherty, Department of Biology, University of Pennsylvania, These Teacher
 Teacher Preparation Notes for "From Gene to Protein via Transcription and Translation" 1 In this analysis and discussion activity, students learn (1) how genes provide the instructions for making a protein
Teacher Preparation Notes for "From Gene to Protein via Transcription and Translation" 1 In this analysis and discussion activity, students learn (1) how genes provide the instructions for making a protein
Data mining with Ensembl Biomart. Stéphanie Le Gras
 Data mining with Ensembl Biomart Stéphanie Le Gras (slegras@igbmc.fr) Guidelines Genome data Genome browsers Getting access to genomic data: Ensembl/BioMart 2 Genome Sequencing Example: Human genome 2000:
Data mining with Ensembl Biomart Stéphanie Le Gras (slegras@igbmc.fr) Guidelines Genome data Genome browsers Getting access to genomic data: Ensembl/BioMart 2 Genome Sequencing Example: Human genome 2000:
Identification and characterization of multiple splice variants of Cdc2-like kinase 4 (Clk4)
 Identification and characterization of multiple splice variants of Cdc2-like kinase 4 (Clk4) Vahagn Stepanyan Department of Biological Sciences, Fordham University Abstract: Alternative splicing is an
Identification and characterization of multiple splice variants of Cdc2-like kinase 4 (Clk4) Vahagn Stepanyan Department of Biological Sciences, Fordham University Abstract: Alternative splicing is an
HLA and antigen presentation. Department of Immunology Charles University, 2nd Medical School University Hospital Motol
 HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
Profiling immunoglobulin repertoires across multiple human tissues by RNA sequencing
 Profiling immunoglobulin repertoires across multiple human tissues by RNA sequencing SergheiMangul, Igor Mandric, Harry Taegyun Yang, NicolasStrauli, Dennis Montoya, Jeremy Rotman, Will Van Der Wey, Jiem
Profiling immunoglobulin repertoires across multiple human tissues by RNA sequencing SergheiMangul, Igor Mandric, Harry Taegyun Yang, NicolasStrauli, Dennis Montoya, Jeremy Rotman, Will Van Der Wey, Jiem
Sections 12.3, 13.1, 13.2
 Sections 12.3, 13.1, 13.2 Now that the DNA has been copied, it needs to send its genetic message to the ribosomes so proteins can be made Transcription: synthesis (making of) an RNA molecule from a DNA
Sections 12.3, 13.1, 13.2 Now that the DNA has been copied, it needs to send its genetic message to the ribosomes so proteins can be made Transcription: synthesis (making of) an RNA molecule from a DNA
MRC-Holland MLPA. Description version 08; 30 March 2015
 SALSA MLPA probemix P351-C1 / P352-D1 PKD1-PKD2 P351-C1 lot C1-0914: as compared to the previous version B2 lot B2-0511 one target probe has been removed and three reference probes have been replaced.
SALSA MLPA probemix P351-C1 / P352-D1 PKD1-PKD2 P351-C1 lot C1-0914: as compared to the previous version B2 lot B2-0511 one target probe has been removed and three reference probes have been replaced.
Introduction. 8 These authors contributed equally to this work
 ARTICLE Complete Haplotype Sequence of the Human Immunoglobulin Heavy-Chain Variable, Diversity, and Joining Genes and Characterization of Allelic and Copy-Number Variation Corey T. Watson, 1,7 Karyn M.
ARTICLE Complete Haplotype Sequence of the Human Immunoglobulin Heavy-Chain Variable, Diversity, and Joining Genes and Characterization of Allelic and Copy-Number Variation Corey T. Watson, 1,7 Karyn M.
Variant Classification. Author: Mike Thiesen, Golden Helix, Inc.
 Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Development of B and T lymphocytes
 Development of B and T lymphocytes What will we discuss today? B-cell development T-cell development B- cell development overview Stem cell In periphery Pro-B cell Pre-B cell Immature B cell Mature B cell
Development of B and T lymphocytes What will we discuss today? B-cell development T-cell development B- cell development overview Stem cell In periphery Pro-B cell Pre-B cell Immature B cell Mature B cell
MRC-Holland MLPA. Description version 14; 28 September 2016
 SALSA MLPA probemix P279-B3 CACNA1A Lot B3-0816. As compared to version B2 (lot B2-1012), one reference probe has been replaced and the length of several probes has been adjusted. Voltage-dependent calcium
SALSA MLPA probemix P279-B3 CACNA1A Lot B3-0816. As compared to version B2 (lot B2-1012), one reference probe has been replaced and the length of several probes has been adjusted. Voltage-dependent calcium
Computational Systems Biology: Biology X
 Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
Introduction to genetic variation. He Zhang Bioinformatics Core Facility 6/22/2016
 Introduction to genetic variation He Zhang Bioinformatics Core Facility 6/22/2016 Outline Basic concepts of genetic variation Genetic variation in human populations Variation and genetic disorders Databases
Introduction to genetic variation He Zhang Bioinformatics Core Facility 6/22/2016 Outline Basic concepts of genetic variation Genetic variation in human populations Variation and genetic disorders Databases
Introduction retroposon
 17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
Two categories of immune response. immune response. infection. (adaptive) Later immune response. immune response
 Ivana FELLNEROVÁ E-mail: fellneri@hotmail.com, mob. 732154801 Basic immunogenetic terminology innate and adaptive immunity specificity and polymorphism immunoglobuline gene superfamily immunogenetics MHC
Ivana FELLNEROVÁ E-mail: fellneri@hotmail.com, mob. 732154801 Basic immunogenetic terminology innate and adaptive immunity specificity and polymorphism immunoglobuline gene superfamily immunogenetics MHC
Significance of the MHC
 CHAPTER 7 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
CHAPTER 7 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
The lymphoma-associated NPM-ALK oncogene elicits a p16ink4a/prb-dependent tumor-suppressive pathway. Blood Jun 16;117(24):
 DNA Sequencing Publications Standard Sequencing 1 Carro MS et al. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006 Jun;5(11) 2 Lassandro L et al. The DNA sequence
DNA Sequencing Publications Standard Sequencing 1 Carro MS et al. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006 Jun;5(11) 2 Lassandro L et al. The DNA sequence
membrane form secreted form 13 aa 26 aa K K V V K K 3aa
 Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai secreted form membrane form 13 aa 26 aa K K V V K K 3aa Hapten monosaccharide
Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai secreted form membrane form 13 aa 26 aa K K V V K K 3aa Hapten monosaccharide
IVF Michigan, Rochester Hills, Michigan, and Reproductive Genetics Institute, Chicago, Illinois
 FERTILITY AND STERILITY VOL. 80, NO. 4, OCTOBER 2003 Copyright 2003 American Society for Reproductive Medicine Published by Elsevier Inc. Printed on acid-free paper in U.S.A. CASE REPORTS Preimplantation
FERTILITY AND STERILITY VOL. 80, NO. 4, OCTOBER 2003 Copyright 2003 American Society for Reproductive Medicine Published by Elsevier Inc. Printed on acid-free paper in U.S.A. CASE REPORTS Preimplantation
Chapter 5. Generation of lymphocyte antigen receptors
 Chapter 5 Generation of lymphocyte antigen receptors Structural variation in Ig constant regions Isotype: different class of Ig Heavy-chain C regions are encoded in separate genes Initially, only two of
Chapter 5 Generation of lymphocyte antigen receptors Structural variation in Ig constant regions Isotype: different class of Ig Heavy-chain C regions are encoded in separate genes Initially, only two of
Reporting TP53 gene analysis results in CLL
 Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
The Development of Lymphocytes: B Cell Development in the Bone Marrow & Peripheral Lymphoid Tissue Deborah A. Lebman, Ph.D.
 The Development of Lymphocytes: B Cell Development in the Bone Marrow & Peripheral Lymphoid Tissue Deborah A. Lebman, Ph.D. OBJECTIVES 1. To understand how ordered Ig gene rearrangements lead to the development
The Development of Lymphocytes: B Cell Development in the Bone Marrow & Peripheral Lymphoid Tissue Deborah A. Lebman, Ph.D. OBJECTIVES 1. To understand how ordered Ig gene rearrangements lead to the development
Supplementary Information. Preferential associations between co-regulated genes reveal a. transcriptional interactome in erythroid cells
 Supplementary Information Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells Stefan Schoenfelder, * Tom Sexton, * Lyubomira Chakalova, * Nathan
Supplementary Information Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells Stefan Schoenfelder, * Tom Sexton, * Lyubomira Chakalova, * Nathan
The Meaning of Genetic Variation
 Activity 2 The Meaning of Genetic Variation Focus: Students investigate variation in the beta globin gene by identifying base changes that do and do not alter function, and by using several CD-ROM-based
Activity 2 The Meaning of Genetic Variation Focus: Students investigate variation in the beta globin gene by identifying base changes that do and do not alter function, and by using several CD-ROM-based
SpliceDB: database of canonical and non-canonical mammalian splice sites
 2001 Oxford University Press Nucleic Acids Research, 2001, Vol. 29, No. 1 255 259 SpliceDB: database of canonical and non-canonical mammalian splice sites M.Burset,I.A.Seledtsov 1 and V. V. Solovyev* The
2001 Oxford University Press Nucleic Acids Research, 2001, Vol. 29, No. 1 255 259 SpliceDB: database of canonical and non-canonical mammalian splice sites M.Burset,I.A.Seledtsov 1 and V. V. Solovyev* The
HBV. Next Generation Sequencing, data analysis and reporting. Presenter Leen-Jan van Doorn
 HBV Next Generation Sequencing, data analysis and reporting Presenter Leen-Jan van Doorn HBV Forum 3 October 24 th, 2017 Marriott Marquis, Washington DC www.forumresearch.org HBV Biomarkers HBV biomarkers:
HBV Next Generation Sequencing, data analysis and reporting Presenter Leen-Jan van Doorn HBV Forum 3 October 24 th, 2017 Marriott Marquis, Washington DC www.forumresearch.org HBV Biomarkers HBV biomarkers:
HLA and antigen presentation. Department of Immunology Charles University, 2nd Medical School University Hospital Motol
 HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
Major Histocompatibility Complex (MHC) and T Cell Receptors
 Major Histocompatibility Complex (MHC) and T Cell Receptors Historical Background Genes in the MHC were first identified as being important genes in rejection of transplanted tissues Genes within the MHC
Major Histocompatibility Complex (MHC) and T Cell Receptors Historical Background Genes in the MHC were first identified as being important genes in rejection of transplanted tissues Genes within the MHC
PAH Mutation Analysis Consortium Database: a database for disease-producing and other allelic variation at the human PAH locus
 1996 Oxford University Press Nucleic Acids Research, 1996, Vol. 24, No. 1 127 131 PAH Mutation Analysis Consortium Database: a database for disease-producing and other allelic variation at the human PAH
1996 Oxford University Press Nucleic Acids Research, 1996, Vol. 24, No. 1 127 131 PAH Mutation Analysis Consortium Database: a database for disease-producing and other allelic variation at the human PAH
Variations in Chromosome Structure & Function. Ch. 8
 Variations in Chromosome Structure & Function Ch. 8 1 INTRODUCTION! Genetic variation refers to differences between members of the same species or those of different species Allelic variations are due
Variations in Chromosome Structure & Function Ch. 8 1 INTRODUCTION! Genetic variation refers to differences between members of the same species or those of different species Allelic variations are due
PALB2 c g>c is. VARIANT OF UNCERTAIN SIGNIFICANCE (VUS) CGI s summary of the available evidence is in Appendices A-C.
 Consultation sponsor (may not be the patient): First LastName [Patient identity withheld] Date received by CGI: 2 Sept 2017 Variant Fact Checker Report ID: 0000001.5 Date Variant Fact Checker issued: 12
Consultation sponsor (may not be the patient): First LastName [Patient identity withheld] Date received by CGI: 2 Sept 2017 Variant Fact Checker Report ID: 0000001.5 Date Variant Fact Checker issued: 12
General Biology 1004 Chapter 11 Lecture Handout, Summer 2005 Dr. Frisby
 Slide 1 CHAPTER 11 Gene Regulation PowerPoint Lecture Slides for Essential Biology, Second Edition & Essential Biology with Physiology Presentation prepared by Chris C. Romero Neil Campbell, Jane Reece,
Slide 1 CHAPTER 11 Gene Regulation PowerPoint Lecture Slides for Essential Biology, Second Edition & Essential Biology with Physiology Presentation prepared by Chris C. Romero Neil Campbell, Jane Reece,
Welcome to the Genetic Code: An Overview of Basic Genetics. October 24, :00pm 3:00pm
 Welcome to the Genetic Code: An Overview of Basic Genetics October 24, 2016 12:00pm 3:00pm Course Schedule 12:00 pm 2:00 pm Principles of Mendelian Genetics Introduction to Genetics of Complex Disease
Welcome to the Genetic Code: An Overview of Basic Genetics October 24, 2016 12:00pm 3:00pm Course Schedule 12:00 pm 2:00 pm Principles of Mendelian Genetics Introduction to Genetics of Complex Disease
Antibody-Cytokine- Autoimmune
 Antibody-Cytokine- Autoimmune Surasak Wongratanacheewin, Ph.D Dean, Graduate School, KKU Microbiology, Faculty of Medicine, KKU sura_wng@kku.ac.th การอบรมหล กส ตรประกาศน ยบ ตรการข นทะเบ ยนช วว ตถ ว นท
Antibody-Cytokine- Autoimmune Surasak Wongratanacheewin, Ph.D Dean, Graduate School, KKU Microbiology, Faculty of Medicine, KKU sura_wng@kku.ac.th การอบรมหล กส ตรประกาศน ยบ ตรการข นทะเบ ยนช วว ตถ ว นท
Handling Immunogenetic Data Managing and Validating HLA Data
 Handling Immunogenetic Data Managing and Validating HLA Data Steven J. Mack PhD Children s Hospital Oakland Research Institute 16 th IHIW & Joint Conference Sunday 3 June, 2012 Overview 1. Master Analytical
Handling Immunogenetic Data Managing and Validating HLA Data Steven J. Mack PhD Children s Hospital Oakland Research Institute 16 th IHIW & Joint Conference Sunday 3 June, 2012 Overview 1. Master Analytical
Biomarkers in Chronic Lymphocytic Leukemia: the art of synthesis. CLL Immunogenetics. Overview. Anastasia Chatzidimitriou
 Biomarkers in Chronic Lymphocytic Leukemia: the art of synthesis CLL Immunogenetics Overview Anastasia Chatzidimitriou Belgrade March 17, 2018 B cells: multiple receptors B cell receptor IG unique signature
Biomarkers in Chronic Lymphocytic Leukemia: the art of synthesis CLL Immunogenetics Overview Anastasia Chatzidimitriou Belgrade March 17, 2018 B cells: multiple receptors B cell receptor IG unique signature
Pre-mRNA has introns The splicing complex recognizes semiconserved sequences
 Adding a 5 cap Lecture 4 mrna splicing and protein synthesis Another day in the life of a gene. Pre-mRNA has introns The splicing complex recognizes semiconserved sequences Introns are removed by a process
Adding a 5 cap Lecture 4 mrna splicing and protein synthesis Another day in the life of a gene. Pre-mRNA has introns The splicing complex recognizes semiconserved sequences Introns are removed by a process
Generation of antibody diversity October 18, Ram Savan
 Generation of antibody diversity October 18, 2016 Ram Savan savanram@uw.edu 441 Lecture #10 Slide 1 of 30 Three lectures on antigen receptors Part 1 : Structural features of the BCR and TCR Janeway Chapter
Generation of antibody diversity October 18, 2016 Ram Savan savanram@uw.edu 441 Lecture #10 Slide 1 of 30 Three lectures on antigen receptors Part 1 : Structural features of the BCR and TCR Janeway Chapter
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Studying Alternative Splicing
 Studying Alternative Splicing Meelis Kull PhD student in the University of Tartu supervisor: Jaak Vilo CS Theory Days Rõuge 27 Overview Alternative splicing Its biological function Studying splicing Technology
Studying Alternative Splicing Meelis Kull PhD student in the University of Tartu supervisor: Jaak Vilo CS Theory Days Rõuge 27 Overview Alternative splicing Its biological function Studying splicing Technology
Phenotype analysis in humans using OMIM
 Outline: 1) Introduction to OMIM 2) Phenotype similarity map 3) Exercises Phenotype analysis in humans using OMIM Rosario M. Piro Molecular Biotechnology Center University of Torino, Italy 1 MBC, Torino
Outline: 1) Introduction to OMIM 2) Phenotype similarity map 3) Exercises Phenotype analysis in humans using OMIM Rosario M. Piro Molecular Biotechnology Center University of Torino, Italy 1 MBC, Torino
MRC-Holland MLPA. Description version 07; 26 November 2015
 SALSA MLPA probemix P266-B1 CLCNKB Lot B1-0415, B1-0911. As compared to version A1 (lot A1-0908), one target probe for CLCNKB (exon 11) has been replaced. In addition, one reference probe has been replaced
SALSA MLPA probemix P266-B1 CLCNKB Lot B1-0415, B1-0911. As compared to version A1 (lot A1-0908), one target probe for CLCNKB (exon 11) has been replaced. In addition, one reference probe has been replaced
Partial versus Productive Immunoglobulin Heavy Locus Rearrangements in Chronic Lymphocytic Leukemia: Implications for B-Cell Receptor Stereotypy
 Partial versus Productive Immunoglobulin Heavy Locus Rearrangements in Chronic Lymphocytic Leukemia: Implications for B-Cell Receptor Stereotypy Eugenia Tsakou, 1,2 Andreas Agathagelidis, 3,4 Myriam Boudjoghra,
Partial versus Productive Immunoglobulin Heavy Locus Rearrangements in Chronic Lymphocytic Leukemia: Implications for B-Cell Receptor Stereotypy Eugenia Tsakou, 1,2 Andreas Agathagelidis, 3,4 Myriam Boudjoghra,
3. What law of heredity explains that traits, like texture and color, are inherited independently of each other?
 Section 2: Genetics Chapter 11 pg. 308-329 Part 1: Refer to the table of pea plant traits on the right. Then complete the table on the left by filling in the missing information for each cross. 6. What
Section 2: Genetics Chapter 11 pg. 308-329 Part 1: Refer to the table of pea plant traits on the right. Then complete the table on the left by filling in the missing information for each cross. 6. What
Breast cancer. Risk factors you cannot change include: Treatment Plan Selection. Inferring Transcriptional Module from Breast Cancer Profile Data
 Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
PROTEIN SYNTHESIS. It is known today that GENES direct the production of the proteins that determine the phonotypical characteristics of organisms.
 PROTEIN SYNTHESIS It is known today that GENES direct the production of the proteins that determine the phonotypical characteristics of organisms.» GENES = a sequence of nucleotides in DNA that performs
PROTEIN SYNTHESIS It is known today that GENES direct the production of the proteins that determine the phonotypical characteristics of organisms.» GENES = a sequence of nucleotides in DNA that performs
SEX. Genetic Variation: The genetic substrate for natural selection. Sex: Sources of Genotypic Variation. Genetic Variation
 Genetic Variation: The genetic substrate for natural selection Sex: Sources of Genotypic Variation Dr. Carol E. Lee, University of Wisconsin Genetic Variation If there is no genetic variation, neither
Genetic Variation: The genetic substrate for natural selection Sex: Sources of Genotypic Variation Dr. Carol E. Lee, University of Wisconsin Genetic Variation If there is no genetic variation, neither
For all of the following, you will have to use this website to determine the answers:
 For all of the following, you will have to use this website to determine the answers: http://blast.ncbi.nlm.nih.gov/blast.cgi We are going to be using the programs under this heading: Answer the following
For all of the following, you will have to use this website to determine the answers: http://blast.ncbi.nlm.nih.gov/blast.cgi We are going to be using the programs under this heading: Answer the following
Stereotyped B Cell Receptors in Chronic Lymphocytic Leukaemia
 Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Medicine 405 Stereotyped B Cell Receptors in Chronic Lymphocytic Leukaemia Implications for Antigen Selection in Leukemogenesis
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Medicine 405 Stereotyped B Cell Receptors in Chronic Lymphocytic Leukaemia Implications for Antigen Selection in Leukemogenesis
Comprehensive translocation and clonality detection in lymphoproliferative disorders by next generation sequencing
 Comprehensive translocation and clonality detection in lymphoproliferative disorders by next generation sequencing Wren, D., Walker, B. A., Bruggemann, M., Catherwood, M. A., Pott, C., Stamatopoulos, K.,...
Comprehensive translocation and clonality detection in lymphoproliferative disorders by next generation sequencing Wren, D., Walker, B. A., Bruggemann, M., Catherwood, M. A., Pott, C., Stamatopoulos, K.,...
DEFINITIONS OF HISTOCOMPATIBILITY TYPING TERMS
 DEFINITIONS OF HISTOCOMPATIBILITY TYPING TERMS The definitions below are intended as general concepts. There will be exceptions to these general definitions. These definitions do not imply any specific
DEFINITIONS OF HISTOCOMPATIBILITY TYPING TERMS The definitions below are intended as general concepts. There will be exceptions to these general definitions. These definitions do not imply any specific
Translocations and clonality detection in lymphoproliferative disorders by capturebased Next-generation sequencing
 Translocations and clonality detection in lymphoproliferative disorders by capturebased Next-generation sequencing Dörte Wren MSc MPhil on behalf of the EuroClonality-NGS consortium Molecular Diagnostics
Translocations and clonality detection in lymphoproliferative disorders by capturebased Next-generation sequencing Dörte Wren MSc MPhil on behalf of the EuroClonality-NGS consortium Molecular Diagnostics
CHROMOSOMAL MICROARRAY (CGH+SNP)
 Chromosome imbalances are a significant cause of developmental delay, mental retardation, autism spectrum disorders, dysmorphic features and/or birth defects. The imbalance of genetic material may be due
Chromosome imbalances are a significant cause of developmental delay, mental retardation, autism spectrum disorders, dysmorphic features and/or birth defects. The imbalance of genetic material may be due
High AU content: a signature of upregulated mirna in cardiac diseases
 https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
Protein Synthesis
 Protein Synthesis 10.6-10.16 Objectives - To explain the central dogma - To understand the steps of transcription and translation in order to explain how our genes create proteins necessary for survival.
Protein Synthesis 10.6-10.16 Objectives - To explain the central dogma - To understand the steps of transcription and translation in order to explain how our genes create proteins necessary for survival.
MODULE NO.14: Y-Chromosome Testing
 SUBJECT Paper No. and Title Module No. and Title Module Tag FORENSIC SIENCE PAPER No.13: DNA Forensics MODULE No.21: Y-Chromosome Testing FSC_P13_M21 TABLE OF CONTENTS 1. Learning Outcome 2. Introduction:
SUBJECT Paper No. and Title Module No. and Title Module Tag FORENSIC SIENCE PAPER No.13: DNA Forensics MODULE No.21: Y-Chromosome Testing FSC_P13_M21 TABLE OF CONTENTS 1. Learning Outcome 2. Introduction:
Probable involvement of immunoglobulin superfamily genes in most recurrent chromosomal rearrangements from ataxia telangiectasia
 Hum Genet (1986) 72 : 210-214 Springer-Verlag 1986 Probable involvement of immunoglobulin superfamily genes in most recurrent chromosomal rearrangements from ataxia telangiectasia A. Aurias and B. Dutrillaux
Hum Genet (1986) 72 : 210-214 Springer-Verlag 1986 Probable involvement of immunoglobulin superfamily genes in most recurrent chromosomal rearrangements from ataxia telangiectasia A. Aurias and B. Dutrillaux
A HLA-DRB supertype chart with potential overlapping peptide binding function
 A HLA-DRB supertype chart with potential overlapping peptide binding function Arumugam Mohanapriya 1,2, Satish Nandagond 1, Paul Shapshak 3, Uma Kangueane 1, Pandjassarame Kangueane 1, 2 * 1 Biomedical
A HLA-DRB supertype chart with potential overlapping peptide binding function Arumugam Mohanapriya 1,2, Satish Nandagond 1, Paul Shapshak 3, Uma Kangueane 1, Pandjassarame Kangueane 1, 2 * 1 Biomedical
MRC-Holland MLPA. Description version 30; 06 June 2017
 SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0517, C1-0114. As compared to the previous B2 version (lot B2-0813, B2-0912), 11 target probes are replaced or added, and 10 new reference probes are
SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0517, C1-0114. As compared to the previous B2 version (lot B2-0813, B2-0912), 11 target probes are replaced or added, and 10 new reference probes are
Genomic structural variation
 Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
A Self-Propelled Biological Process Plk1-Dependent Product- Activated, Feed-Forward Mechanism
 A Self-Propelled Biological Process Plk1-Dependent Product- Activated, Feed-Forward Mechanism The Harvard community has made this article openly available. Please share how this access benefits you. Your
A Self-Propelled Biological Process Plk1-Dependent Product- Activated, Feed-Forward Mechanism The Harvard community has made this article openly available. Please share how this access benefits you. Your
MRC-Holland MLPA. Description version 29; 31 July 2015
 SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0114. As compared to the previous B2 version (lot 0813 and 0912), 11 target probes are replaced or added, and 10 new reference probes are included. P082
SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0114. As compared to the previous B2 version (lot 0813 and 0912), 11 target probes are replaced or added, and 10 new reference probes are included. P082
Human Genome Complexity, Viruses & Genetic Variability
 Human Genome Complexity, Viruses & Genetic Variability (Learning Objectives) Learn the types of DNA sequences present in the Human Genome other than genes coding for functional proteins. Review what you
Human Genome Complexity, Viruses & Genetic Variability (Learning Objectives) Learn the types of DNA sequences present in the Human Genome other than genes coding for functional proteins. Review what you
SALSA MLPA probemix P241-D2 MODY mix 1 Lot D As compared to version D1 (lot D1-0911), one reference probe has been replaced.
 mix P241-D2 MODY mix 1 Lot D2-0413. As compared to version D1 (lot D1-0911), one reference has been replaced. Maturity-Onset Diabetes of the Young (MODY) is a distinct form of non insulin-dependent diabetes
mix P241-D2 MODY mix 1 Lot D2-0413. As compared to version D1 (lot D1-0911), one reference has been replaced. Maturity-Onset Diabetes of the Young (MODY) is a distinct form of non insulin-dependent diabetes
Chapter 4 Cellular Oncogenes ~ 4.6 -
 Chapter 4 Cellular Oncogenes - 4.2 ~ 4.6 - Many retroviruses carrying oncogenes have been found in chickens and mice However, attempts undertaken during the 1970s to isolate viruses from most types of
Chapter 4 Cellular Oncogenes - 4.2 ~ 4.6 - Many retroviruses carrying oncogenes have been found in chickens and mice However, attempts undertaken during the 1970s to isolate viruses from most types of
Finding subtle mutations with the Shannon human mrna splicing pipeline
 Finding subtle mutations with the Shannon human mrna splicing pipeline Presentation at the CLC bio Medical Genomics Workshop American Society of Human Genetics Annual Meeting November 9, 2012 Peter K Rogan
Finding subtle mutations with the Shannon human mrna splicing pipeline Presentation at the CLC bio Medical Genomics Workshop American Society of Human Genetics Annual Meeting November 9, 2012 Peter K Rogan
7.014 Problem Set 7 Solutions
 MIT Department of Biology 7.014 Introductory Biology, Spring 2005 7.014 Problem Set 7 Solutions Question 1 Part A Antigen binding site Antigen binding site Variable region Light chain Light chain Variable
MIT Department of Biology 7.014 Introductory Biology, Spring 2005 7.014 Problem Set 7 Solutions Question 1 Part A Antigen binding site Antigen binding site Variable region Light chain Light chain Variable
Genetics 1. Down s syndrome is caused by an extra copy of cshromosome no 21. What percentage of
 Genetics 1. Down s syndrome is caused by an extra copy of cshromosome no 21. What percentage of offspring produced by an affected mother and a normal father would be affected by this disorder? (2003) 1)
Genetics 1. Down s syndrome is caused by an extra copy of cshromosome no 21. What percentage of offspring produced by an affected mother and a normal father would be affected by this disorder? (2003) 1)
RNA Processing in Eukaryotes *
 OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
 Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Scope The Atlas of Genetics and Cytogenetics in Oncology and Haematology is a peer reviewed on-line journal
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Scope The Atlas of Genetics and Cytogenetics in Oncology and Haematology is a peer reviewed on-line journal
