Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme Biosynthesis in Methanoarchaea
|
|
|
- Scot Fleming
- 5 years ago
- Views:
Transcription
1 JOURNAL OF BACTERIOLOGY, July 2000, p Vol. 182, No /00/$ Copyright 2000, American Society for Microbiology. All Rights Reserved. Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme Biosynthesis in Methanoarchaea MARION GRAUPNER, HUIMIN XU, AND ROBERT H. WHITE* Department of Biochemistry, Virginia Polytechnic Institute and State University, Blacksburg, Virginia Received 22 February 2000/Accepted 14 April 2000 Two putative malate dehydrogenase genes, MJ1425 and MJ0490, from Methanococcus jannaschii and one from Methanothermus fervidus were cloned and overexpressed in Escherichia coli, and their gene products were tested for the ability to catalyze pyridine nucleotide-dependent oxidation and reduction reactions of the following -hydroxy -keto acid pairs: (S)-sulfolactic acid and sulfopyruvic acid; (S)- -hydroxyglutaric acid and -ketoglutaric acid; (S)-lactic acid and pyruvic acid; and 1-hydroxy-1,3,4,6-hexanetetracarboxylic acid and 1-oxo-1,3,4,6-hexanetetracarboxylic acid. Each of these reactions is involved in the formation of coenzyme M, methanopterin, coenzyme F 420, and methanofuran, respectively. Both the MJ1425-encoded enzyme and the MJ0490-encoded enzyme were found to function to different degrees as malate dehydrogenases, reducing oxalacetate to (S)-malate using either NADH or NADPH as a reductant. Both enzymes were found to use either NADH or NADPH to reduce sulfopyruvate to (S)-sulfolactate, but the /K m value for the reduction of sulfopyruvate by NADH using the MJ1425-encoded enzyme was 20 times greater than any other combination of enzymes and pyridine nucleotides. Both the M. fervidus and the MJ1425-encoded enzyme catalyzed the NAD -dependent oxidation of (S)-sulfolactate to sulfopyruvate. The MJ1425-encoded enzyme also catalyzed the NADH-dependent reduction of -ketoglutaric acid to (S)-hydroxyglutaric acid, a component of methanopterin. Neither of the enzymes reduced pyruvate to (S)-lactate, a component of coenzyme F 420. Only the MJ1425-encoded enzyme was found to reduce 1-oxo-1,3,4,6-hexanetetracarboxylic acid, and this reduction occurred only to a small extent and produced an isomer of 1-hydroxy-1,3,4,6-hexanetetracarboxylic acid that is not involved in the biosynthesis of methanofuran c. We conclude that the MJ1425-encoded enzyme is likely to be involved in the biosynthesis of both coenzyme M and methanopterin. The biosynthesis of the methanogenic cofactors coenzyme M (2-mercaptoethanesulfonic acid), methanopterin, coenzyme F 420, and methanofuran c (Fig. 1) requires the generation of an -hydroxy acid that either becomes a component in the final structure or serves as an intermediate in the formation of the coenzyme. In the case of coenzyme M, (S)-sulfolactate, formed from phosphoenolpyruvate (PEP) and bisulfite, is an intermediate in the biosynthesis (28 30). In the case of methanopterin (24, 25) and several related modified folates (33, 34, 36), (S)- hydroxyglutaric acid (23) is incorporated into the coenzyme during its biosynthesis (32, 35). For coenzyme F 420 (6) and its -polyglutamate derivatives (7, 8, 18), (S)-hydroxypropionic acid (S-lactic acid) becomes a part of the final structure. Finally, two (1R)-diastereomers of 1-hydroxy-1,3,4,6-hexanetetracarboxylic acid (HHTCA) serve as intermediates in the biosynthesis of the 1,3,4,6-hexanetetracarboxylic acid (HTCA) moiety of methanofuran (17; unpublished results), and another diastereomer of HHTCA [(1S)-HHTCA] is a component of methanofuran c (31). The HHTCA intermediates in HTCA biosynthesis have the same absolute stereochemistry at the -hydroxy acid portion of the molecule as the pantothenic acid moiety of coenzyme A, the only other cofactor containing an -hydroxy acid (10). At present, nothing is known about the enzymes required for the formation and the metabolism of these -hydroxy acids and * Corresponding author. Mailing address: Department of Biochemistry (0308), Virginia Polytechnic Institute and State University, Blacksburg, VA Phone: (540) Fax: (540) rhwhite@vt.edu. -keto acids in the methanoarchaea. Based on the S-stereochemistry of the (S)-hydroxyglutaric acid present in methanopterin and the (S)-lactic acid present in coenzyme F 420,itis likely that each is formed by the reduction of the corresponding -keto acid by a NAD(P)H-dependent dehydrogenase related to the lactate/malate dehydrogenase group of enzymes (1, 3, 9). Likewise, it could be argued, again based on stereochemical grounds, that the oxidation of (S)-sulfolactate to sulfopyruvate occurring during the biosynthesis of coenzyme M could also be carried out by an enzyme related to the lactate/ malate dehydrogenases. Two malate dehydrogenases, designated MdhI and MdhII, were recently isolated from Methanobacterium thermoautotrophicum strain Marburg (22). Via the N-terminal sequences, the genes encoding the malate dehydrogenases would correspond to those encoded from the M. thermoautotrophicum (strain H) genes MT1205 and MT0188, respectively (22). From genomic sequence data (21), the sequences of M. thermoautotrophicum genes MT1205 and MT0188 have 53.5 and 48.6% sequence identity, respectively, to the Methanococcus jannaschii genes MJ1425 and MJ0490 (2). In both of these organisms, these are the only two genes with any clear sequence homology to the lactate/malate family of dehydrogenases (2, 21). The M. jannaschii gene MJ0490, producing the MdhII enzyme, has 49% sequence identity to the (S)-lactate/malate dehydrogenases from bacteria and eukaryotes (1, 9), and it aligns well with the Archaeoglobus fulgidus AF0855-encoded malate dehydrogenase (16). From many sequence comparisons and sitedirected mutagensis of members of this family of dehydrogenases, it could be shown that the structure of the amino acid at 3688
2 VOL. 182, HYDROXY ACID DEHYDROGENASES 3689 Downloaded from FIG. 1. Methanogenic coenzymes that contain -hydroxy acids as components of their structures. (S)-Sulfolactate is an intermediate in the biosynthesis of coenzyme M, and (S)-malate is an intermediate in the partial citric acid cycle. one conserved position can determine the substrate specificity and the coenzyme-binding specificity (9). A sequence comparison of the MJ0490 gene with those lactate/malate dehydrogenases would predict that the MJ0490 enzyme should prefer (S)-malate over (S)-lactate and NADP over NAD. The MJ1425-encoded enzyme, on the contrary, does not fit as well into the family of lactate/malate dehydrogenases. The MJ1425- encoded enzyme has 44% sequence identity with the malate dehydrogenases from the methanoarchaea Methanothermus fervidus (12) and the MTH1205-encoded enzyme mentioned above from M. thermoautotrophicum. The MJ1425-encoded enzyme has only 12% sequence identity to a malate dehydrogenase from Bacillus subtilis. It is not clear why the M. jannaschii and M. thermoautotrophicum genomes contain two genes for malate dehydrogenases. Taking into account the lack of specificity of many of the known dehydrogenases (3, 15, 37), we considered it likely that one or more of these archaeal enzymes may be the enzyme(s) used for producing one or more of the (S)- -hydroxy acids required for the biosynthesis of methanoarchaeal coenzymes. To test these hypotheses, we have cloned and overexpressed the MJ0490 and MJ1425 genes from M. jannaschii and a malate dehydrogenase gene from M. fervidus in Escherichia coli and tested their protein products for the ability to reduce or oxidize desired coenzyme biosynthetic intermediates. MATERIALS AND METHODS Preparation of substrates. The S and R stereoisomers of sulfolactate were prepared by nitrous acid deamination of (S)-cysteic acid and (R)-cysteic acid (M. Graupner and R. H. White, unpublished results). Sulfopyruvate was prepared as previously described (29). 1-Oxo-1,3,4,6-hexanetetracarboxylic acid (KHTCA) was prepared by the condensation of the dimethylketal derivative of -ketoglutarate with the dimethyl ester of -bromoglutaric acid (Graupner and White, unpublished results). The compound used as substrate consisted of a racemic mixture of one part of erythro-khtca and two parts of threo-khtca. Oxalacetate, pyruvate, (S)-lactate, (S)-2-hydroxyglutaric acid, and (R)-2-hydroxyglutaric acid were obtained from Sigma Chemical Co. Identification, cloning, and high-level expression of the gene product. Expression of MJ1425 and MJ0490 genes in E. coli was accomplished by the following procedure. The MJ1425 and MJ0490 genes were amplified by PCR, using genomic DNA from M. jannaschii (David E. Graham, Urbana, Ill.) as the template. The primers, 5 CATGCATATGATTTTAAAACCAGAAAATGAA 3 and 5 GATCGGATCCTTATTCAATATAGTCCTCAAT 3 derived from the MJ1425 DNA sequence and primers 5 CATGCATATGAAAGTTACAATTA TAGGAGC 3 and 5 GATCGGATCCTTATAAGTTTTTAACTTCTTC 3 derived from MJ0490 DNA sequence were used. The PCR products, purified via absorption and desorption to a Qia quick spin column, were digested with NdeI and BamHI and were cloned into NdeI-BamHI-digested pt7-7 plasmid vector to obtain the constructs pt7-7-mj1425 and pt7-7-mj0490. The constructs were transformed to E. coli TB1 for plasmid preparation and to E. coli BL21(DE3) for on December 30, 2018 by guest
3 3690 GRAUPNER ET AL. J. BACTERIOL. TABLE 1. NADH- and NADPH-dependent enzymatic reductions of various -keto acids by the MJ1425-encoded enzyme, the MJ0490-encoded enzyme, and the MF-malate dehydrogenase Substrate Cosubstrate a MJ1425 (MdhI) MJ0490 (MdhII) MF (MdhIII) /K m /K m /K m Oxalacetate NADH Oxalacetate NADPH Sulfopyruvate NADH , ,700 Sulfopyruvate NADPH Ketoglutarate NADH NA b NA KHTCA NADH NA NA a The cosubstrate concentrations were 0.3 mm NADH and 0.3 mm NADPH. b NA, no activity detectable ( 0.1 U/mg) at substrate concentrations up to 10 mm. protein expression. The expression plasmid for the malate dehydrogenase from Methanothermus fervidus (12) was a generous gift from R. Hensel (University of Essen, Essen, Germany); it was treated in the same manner as the M. jannaschii gene containing plasmids. The cells of E. coli BL21(DE3) containing the pt7-7-mj1425 or pt7-7-mj0490 plasmid were incubated in Luria-Bertani (LB) medium supplemented with 100 mg of ampicillin per liter at 37 C to an absorbance at 600 nm of 1.0. Protein production was then induced by the addition of isopropylthio- -Dgalactoside (IPTG) to a final concentration of 0.1 mm, and the cells were grown for an additional 4 h at 37 C. The cells were harvested by centrifugation (4,000 g, 5 min) and were frozen at 20 C until used. High-level expression of the desired gene products was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% polyacrylamide) of the SDS-soluble cellular proteins. Preparation and analysis of cell extracts. Cell extracts were prepared by sonication of the cell pellets ( 300 mg [wet weight]) suspended in 3 ml of buffer [50 mm N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) (ph 7.0), 10 mm MgCl 2, 20 mm mercaptoethanol] followed by centrifugation (14,000 g, 10 min). SDS-PAGE analysis of the pellets and the cell extracts showed that most ( 90%) of the overexpressed proteins were present in a soluble form. Heating the extracts at 80 C for 30 min followed by centrifugation (14,000 g, 10 min) removed most of the E. coli proteins and left essentially pure solutions of the overexpressed proteins (95% pure). These solutions were used for the analyses reported here. The protein concentrations were determined with the Bio-Rad Protein Assay. Measurement of enzymatic activities. The activities of the enzymes were measured spectrophotometrically at 366 nm at 70 C in 1-ml quartz cuvettes (12). The 366-nm wavelength was used so that higher concentrations of reduced pyridine nucleotides could be used. NADH and NADPH had molar absorptivities of 1,800 M 1 cm 1 and 1,900 M 1 cm 1, respectively, at 70 C at 366 nm. For the determination of the K m and values of the various substrates, the 1.0-ml assay mixture contained 0.1 M potassium phosphate (ph 8.0), 0.3 mm NADH or NADPH, and 0 to 8 mm concentrations of the indicated substrates. Oxalacetate at 1 and 2 mm was used for the determination of the K m and of NADH and NADPH, respectively. The reaction was started by the addition of 10 l of a 1:10 dilution of the protein solution (2 to 5 mg/ml; Bio-Rad Protein Assay), and the time-dependent decrease in NADH/NADPH absorbance was monitored for 4 min. One unit of enzyme activity refers to 1 mol of NADH or NADPH oxidized per min. The oxidative activity was determined by following the reduction of NAD or NADP at 366 nm. The buffer used in this assay consisted of 0.4 M hydrazine and 1 M glycine, ph 9.5 (11). The concentrations of NAD and NADP were 2 mm; the reductant substrates were present at 0 to 8 mm. For the determination of K m and and their associated errors, the kinetic data were fitted with Lineweaver-Burke equation using KaleidaGraph for Macintosh (version 3.08d). Product identification. The products of the NADH/NADPH-dependent reductions of each of the substrates were identified by gas chromatography-mass spectrometry (GC-MS). The general procedure in each case was to incubate the enzyme with the substrates, isolate the products, convert the products into suitable methyl ester derivatives, and analyze them by GC-MS using a known sample as a reference. In the case of the identification of sulfolactate, 100 l of a cell extract containing the protein encoded from gene MJ1425 was mixed with 275 l of 50 mm TES (ph 8.0) and incubated for 1hat50 C in the presence of 13 mm NADH and 6.7 mm sulfopyruvate. After the addition of an equal volume of 95% ethanol, the sample was heated for 5 min at 100 C and centrifuged (10 min, 14,000 g) to produce a clear solution. After evaporation of the ethanol, the residue was dissolved in 0.5 ml of water, passed through a Dowex 50-8X (H ) column (0.5 by 1.5 cm), and evaporated to dryness. After the sample was dissolved in methanol (100 l), an ether solution of diazomethane (300 l) was added which generated a cloudy yellow color, and the sample was clarified by centrifugation (5,000 g, 5 min). The resulting separated clear solution was then evaporated to dryness, dissolved in methylene dichloride, and analyzed by GC-MS (29). The remaining compounds were assayed as their methyl esters, as previously described (13). The absolute stereochemistry of malate and -hydroxyglutaric acid was established by GC-MS of their methyl ester derivatives using a type G-TA Chiraldex column (0.25 mm by 40 m; Advanced Separation Technologies Inc., Whippany, N.J.) programmed from 95 to 180 C at 3 C per min. GC-MS was used in these analyses so that positive identification of the GC peaks could be established even in the complex mixtures. Both of these samples gave very well separated peaks that were easily assigned to the respective R and S isomers. RESULTS AND DISCUSSION Reactions catalyzed by the overproduced enzymes. The MJ1425-encoded enzyme, which we call MdhI, catalyzed the NADH-dependent reduction of oxalacetate, -ketoglutarate, sulfopyruvate, and to a much lower extent, 1-oxo-1,3,4,6-hexanetetracarboxylic acid (KHTCA) (Table 1). The /K m for reduction by NADPH was less than 2% of those observed with NADH. The MJ0490-encoded enzyme, which we call MdhII, catalyzed the reduction of only oxalacetate and sulfopyruvate. Either NADH or NADPH serves as reductant for these reactions, with NADPH being only marginally effective for the MdhI catalyzing reduction of oxalacetate and -ketoglutarate. Based on the measured /K m for these different substrates and coenzymes, it is clear that the NADH-dependent reduction of sulfopyruvate by the MJ1425-derived enzyme is the most efficient reaction measured. The M. fervidus MdhIII (MF- MdhIII) catalyzed only the reduction of oxalacetate and sulfopyruvate. Like the MJ1425-encoded enzyme, this reduction proceeds more efficiently with NADH than with NADPH. Although the amino acid sequence of the M. fervidus enzyme is homologous to the MJ1425-encoded protein, this enzyme did not catalyze the reduction of -ketoglutarate. Since the structure of the methanopterin C 1 carrier in M. fervidus is not known, this finding may indicate that these cells simply do not produce -hydroxyglutarate, since it is not needed for the biosynthesis of their C 1 carrier. Because of the differences in the specificity of this enzyme compared with both the MJ1425- and MJ0490-encoded enzymes, we are calling this enzyme MdhIII. A consistent observation among the MdhI and MdhIII enzymes is the much lower K m for the substrates with NADH compared to those with NADPH. We have not found any examples of such large differences in the K m of substrate changes by the different pyridine nucleotides. The results can, however, be rationalized by the decreased affinity of the anionic substrates for the enzyme that has bound the more anionic NADPH. The K m and for NADH with the MJ1425- derived enzyme were, respectively, mm and U/mg at 1 mm oxalacetate. The K m and for the MJ0490-derived enzyme were, respectively, mm and U/mg at 2 mm oxalacetate.
4 VOL. 182, HYDROXY ACID DEHYDROGENASES 3691 TABLE 2. NAD - and NADP -dependent enzymatic oxidations of various -hydroxy acids by the MJ1425-encoded enzyme, the MJ0490-encoded enzyme, and the MF-malate dehydrogenase Substrate Cosubstrate a MJ1425 (MdhI) MJ0490 (MdhII) MF (MdhIII) /K m /K m /K m (S)-Malate NAD (R)-Malate NAD ND b ND ND ND (S)-Malate NADP ND ND (S)-Sulfolactate NAD NA c (S)-Sulfolactate NADP ND ND (S)- -Hydroxyglutarate NAD NA ND ND a The cosubstrate concentrations were 1 mm NAD or 1 mm NADP. b ND, not determined. c NA, no activity detectable ( 0.1 U/mg) at substrate concentrations of up to 10 mm. The MJ1425-encoded enzyme catalyzed the oxidation of the S isomers of malate, -hydroxyglutarate, and sulfolactate (Table 2). No reaction could be detected for the (R)-sulfolactate and the (R)- -hydroxyglutaric acid, indicating that the MJ1425- encoded enzyme oxidizes only the S isomers. The MJ0490- encoded enzyme catalyzed the oxidation of (S)-malate and (S)-sulfolactate (observed only in the presence of NADP), whereas the MF-Mdh catalyzed these reactions only in the presence of NAD. Again, the MF-malate dehydrogenase as we expected from its relationship to the MJ1425-encoded enzyme did not accept (S)- -hydroxyglutarate as a substrate. (S)- and (R)-lactate were not oxidized by any of the enzymes with NAD or NADP. The K m and values for the MJ1425-encoded enzyme (Table 1) indicate that the enzyme at expected physiological concentrations of substrates would produce both (S)-hydroxyglutaric acid and (S)-malate. If the MJ1425-encoded enzyme were present at a concentration high enough to supply all of the malate needed by the cells, it would also produce far more (S)-hydroxyglutaric acid than would be needed for the biosynthesis of methanopterin, despite the fact the enzyme has a higher K m for -ketoglutarate than for oxalacetate. Thus, MdhI can clearly produce the (S)- -hydroxyglutaric acid required for the biosynthesis of methanopterin by the reduction of -ketoglutarate. Sulfopyruvate was found to be reduced to sulfolactate by all of the enzymes much more efficiently than oxalacetate (Table 1). Furthermore, the reaction kinetics of the MJ1425-encoded enzyme and the MF-malate dehydrogenase showed substrate inhibition at very low sulfopyruvate concentrations (0.1 mm for both enzymes), as has been reported with other malate dehydrogenases (14). This phenomenon has also been observed with the malate dehydrogenase from ox heart mitochondria but with oxalacetate used as the substrate (5). The malate dehydrogenase from pig heart mitochondria has been shown to use sulfopyruvate very poorly as substrate (26); the /K m value was 460 times less than that for oxalacetate. Sulfolactate was not a substrate for the chicken liver NADPdependent malate enzyme but was in fact an inhibitor (27). The MJ0490-encoded enzyme also prefers sulfopyruvate as a substrate, but the differences of the /K m values compared to those for oxalacetate are not as pronounced as with the MJ1425-encoded enzyme or the MF-malate dehydrogenase. In the biosynthesis-relevant direction that is, the oxidation of (S)-sulfolactate to sulfopyruvate (30) only the MJ1425-encoded enzyme and the MF-malate dehydrogenase were found to oxidize (S)-sulfolactate using NAD as the oxidant, whereas the MJ0490-encoded enzyme prefered to oxidize (S)-malate over (S)-sulfolactate using NADP (Table 2). None of the enzymes catalyzed the oxidation of (R)-sulfolactate. These results indicate the possible involvement of the MJ1425-encoded enzyme and the MF MdhIII in the coenzyme M biosynthetic pathway and are consistent with only the (S)-sulfolactate being an intermediate in coenzyme M biosynthesis, as previously described (30). The MJ1425-encoded enzyme could carry out the reduction of the KHTCA with NADH. Despite the fact that the high K m of 15 mm makes the reduction of questionable biochemical relevance, the GC-MS analysis of the produced isomer was shown to be xylo-hhtca, an isomer different from that involved in HTCA biosynthesis (31). Considering the stereospecificity of the MJ1425-encoded enzyme for the (S)-isomers (Table 2), the isomer produced by the MJ1425-encoded enzyme could be assigned to (S)-xylo-HHTCA. From these results, we conclude that the naturally occurring isomers of HHTCA must be produced by an enzyme that reduces the keto acid group to the hydroxy acid with R stereochemistry, as opposed to the S stereochemistry observed here. These results clearly demonstrate that neither of these enzymes is involved in the biosynthesis of the HHTCA intermediates used in HTCA biosynthesis. Pyruvate was found not to serve as a substrate for either enzyme with either of the reduced pyridine nucleotides. Thus, the source of the (S)-lactate present in coenzyme F 420 is still unknown. This finding could indicate that the lactate moiety of F 420 may arise by an alternate route, perhaps by the reduction of PEP. If the MJ1425-encoded enzyme is indeed involved in the biosynthesis of these coenzymes, we may expect that this gene could be in a cluster of other genes that are involved in the biosynthesis of the coenzymes. In M. jannaschii, the MJ1425 gene is located within a group of genes that has no clear relationship to coenzyme biosynthesis. However, the homologous gene in M. thermoautotrophicum, MHT1205, is next to the recently established sulfopyruvate decarboxylase genes MTH1206 and MTH1207 encoding another enzyme involved in coenzyme M biosynthesis. This finding thus establishes a genetic link of this gene to the biosynthesis of coenzyme M. In conclusion, the data presented here are consistent with the idea that the MdhI enzyme may participate in the biosynthesis of coenzyme M and methanopterin by catalyzing the oxidation of sulfolactate to sulfopyruvate in the biosynthetic pathway to coenzyme M and in supplying -hydroxyglutarate for the biosynthesis of methanopterin. This enzyme would thus function in these cells in a multiple capacity, not only functioning as part of a NADPH:NAD transhydrogenase (22) but also supplying metabolites for the biosynthesis of coenzymes. This enzyme thus joins an established but growing list of enzymes,
5 3692 GRAUPNER ET AL. J. BACTERIOL. such as hexokinase (4), transaminases (20), fatty acid synthases, acetohydroxy acid synthases (23a), nucleoside mono and diphosphate kinases (19), and the AksA enzyme involved in the biosynthesis of coenzyme B (13), that are able to catalyze more than one metabolically essential reaction. ACKNOWLEDGMENT This work was supported in part by National Science Foundation grant MCB REFERENCES 1. Birktoft, J. J., R. T. Fernley, R. A. Bradshaw, and L. J. Banaszak Amino acid sequence homology among the 2-hydroxy acid dehydrogenases: mitochondrial and cytoplasmic malate dehydrogenases from a homologous system with lactate dehydrogenase. Proc. Natl. Acad. Sci. USA 79: Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J.-F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, J. F. Weidman, J. L. Fuhrmann, D. Nguyen, T. R. Utterback, J. M. Kelley, J. D. Peterson, P. W. Sadow, M. C. Hanna, M. D. Cotton, K. M. Roberts, M. A. Hurst, B. P. Kaine, M. Borodovsky, H.-P. Klenk, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273: Bur, D., M. A. Lutaen, H. Wynn, L. R. Provencher, J. B. Jones, M. Gold, J. D. Friesen, A. R. Clark, and J. J. Holbrook An evaluation of the substrate specificity and asymmetric synthesis potential of the cloned L-lactate dehydrogenase from Bacillus stearothermophilus. Can. J. Chem. 67: Crane, R. K Hexokinases and pentokinases, p In P. D. Boyer, H. Lardy, and K. Myrback (ed.), The enzymes, vol. 6. Academic Press, New York, N.Y. 5. Davies, D., and E. Kun Isolation and properties of malic dehydrogenase from ox-heart mitochondria. Biochem. J. 1957: Eirich, L. D., G. D. Vogels, and R. S. Wolfe Proposed structure of coenzyme F 420 from Methanobacterium. Biochemistry 17: Gorris, L. G. M., C. van der Drift, and G. D. Vogels Separation and quantification of cofactors from methanogenic bacteria by high-performance liquid chromatography: optimum and routine analyses. J. Microbiol. Methods 8: Gorris, L. G. M., and C. van der Drift Cofactor contents of methanogenic bacteria reviewed. Biofactors 4: Goward, C. R., and D. J. Nicholls Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci. 3: Hill, R. K., and T. H. Chan The absolute configuration of pantothenic acid. Biochem. Biophys. Res. Commun. 38: Hohorst, J. J L-( )-Malate determination with malate dehydrogenase and DPN, p In H.-U. Bergmeyer (ed.), Methods of enzymatic analysis. Academic Press, New York, N.Y. 12. Honka, E., S. Fabry, T. Niermann, P. Palm, and R. Hensel Properties and primary structure of the L-malate dehydrogenases from the extremely thermophilic archaebacterium Methanothermus fervidus. Eur. J. Biochem. 188: Howell, D. M., K. Harich, H. Xu, and R. H. White The -keto acid chain elongation reactions involved in the biosynthesis of coenzyme B (7- mercaptoheptanoylthreonine phosphate) in methanogenic Archaea. Biochemistry 37: Kagawa, T., and P. L. Bruno NADP-malate dehydrogenase from leaves of Zea mays: purification and physical, chemical and kinetic properties. Arch. Biochem. Biophys. 260: Kim, M.-J., and G. M. Whitesides L-Lactate dehydrogenase: substrate specificity and use as a catalyst in the synthesis of homochiral 2-hydroxy acids. J. Am. Chem. Soc. 110: Langelandsvik, A. S., I. H. Steen, N. K. Birkeland, and T. Lien Properties and primary structure of a thermostable L-malate dehydrogenase from Archaeoglobus fulgidus. Arch. Microbiol. 168: Leigh, J. A., L. L. Reinhart, Jr., and R. S. Wolfe Structure of methanofuran, the carbon dioxide reduction factor of Methanobacterium thermoautotrophicum. J. Am. Chem. Soc. 106: Lin, X., and R. H. White Occurrence of coenzyme F 420 and its -monoglutamyl derivative in nonmethanogenic archaebacteria. J. Bacteriol. 168: Parks, R. E., Jr., and R. P. Agarwal Nucleoside diphosphokinases, p In P. D. Boyer (ed.), The enzymes, 3d ed., vol. VIII. Academic Press, New York, N.Y. 20. Pittard, A. J Biosynthesis of the aromatic amino acids, p In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C. 21. Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Potheir, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. M. Church, C. J. Daniels, J. Mao, P. Rice, J. Nölling, and J. N. Reeve Complete genome sequence of Methanobacterium thermoautrophicum. J. Bacteriol. 179: Thompson, H., A. Tersteegen, R. K. Thauer, and R. Hedderich Two malate dehydrogenases in Methanobacterium thermoautotrophicum. Arch. Microbiol. 170: Thurston-Solow, B., and R. H. White The absolute stereochemistry of 2-hydroxyglutaric acid present in methanopterin. Chirality 9: a.Umbarger, H. E Biosynthesis of branched-chain amino acids, p In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C. 24. Van Beelen, P., A. P. M. Strassen, J. W. G. Bosch, G. D. Vogels, W. Guijt, and C. A. G. Haasnoot Elucidation of the structure of methanopterin, a coenzyme from Methanobacterium thermoautotrophicum, using two-dimensional nuclear-magnetic-resonance techniques. Eur. J. Biochem. 138: Van Beelen, P., J. F. A. Labro, J. T. Labro, J. T. Keltjens, W. J. Geerts, G. D. Vogels, W. H. Laarhoven, and C. A. G. Haasnoot Derivatives of methanopterin, a coenzyme involved in methanogenesis. Eur. J. Biochem. 139: Weinstein, C. L., and O. W. Griffith Sulfopyruvate: chemical and enzymatic syntheses and enzymatic assay. Anal Biochem. 156: Weinstein, C. L., and O. W. Griffith Cysteinesulfonate and -sulfopyruvate metabolism: partitioning between decarboxylation, transamination, and reduction pathways. J. Biol. Chem. 263: White, R. H Biosynthesis of coenzyme M (2-mercaptoethanesulfonic acid). Biochemistry 24: White, R. H Intermediates in the biosynthesis of coenzyme M (2- mercaptoethanesulfonic acid). Biochemistry 25: White, R. H Characterization of the enzymatic conversion of sulfopyruvate and L-cysteine into coenzyme M (mercaptoethanesulfonic acid). Biochemistry 27: White, R. H Structural diversity in the methanofuran from different methanogenic bacteria. J. Bacteriol. 170: White, R. H Biosynthesis of methanopterin. Biochemistry 29: White, R. H Structure of the modified folates in the thermophilic archaebacteria Pyrococcus furiosus. Biochemistry 32: White, R. H Structures of the modified folates in the extremely thermophilic archaebacteria Thermococcus litoralis. J. Bacteriol. 175: White, R. H Biosynthesis of methanopterin. Biochemistry 35: White, R. H Structural characterization of modified folates in Archaea. Methods Enzymol. 281: Wilks, H. M., D. J. Halsall, T. Atkinson, W. N. Chia, A. R. Clarke, and J. J. Holbook Designs for a broad substrate specificity keto acid dehydrogenase. Biochemistry 29:
CHE 242 Exam 3 Practice Questions
 CHE 242 Exam 3 Practice Questions Glucose metabolism 1. Below is depicted glucose catabolism. Indicate on the pathways the following: A) which reaction(s) of glycolysis are irreversible B) where energy
CHE 242 Exam 3 Practice Questions Glucose metabolism 1. Below is depicted glucose catabolism. Indicate on the pathways the following: A) which reaction(s) of glycolysis are irreversible B) where energy
Respiration. Respiration. How Cells Harvest Energy. Chapter 7
 How Cells Harvest Energy Chapter 7 Respiration Organisms can be classified based on how they obtain energy: autotrophs: are able to produce their own organic molecules through photosynthesis heterotrophs:
How Cells Harvest Energy Chapter 7 Respiration Organisms can be classified based on how they obtain energy: autotrophs: are able to produce their own organic molecules through photosynthesis heterotrophs:
Ahmad Ulnar. Faisal Nimri ... Dr.Faisal
 24 Ahmad Ulnar Faisal Nimri... Dr.Faisal Fatty Acid Synthesis - Occurs mainly in the Liver (to store excess carbohydrates as triacylglycerols(fat)) and in lactating mammary glands (for the production of
24 Ahmad Ulnar Faisal Nimri... Dr.Faisal Fatty Acid Synthesis - Occurs mainly in the Liver (to store excess carbohydrates as triacylglycerols(fat)) and in lactating mammary glands (for the production of
Vocabulary. Chapter 19: The Citric Acid Cycle
 Vocabulary Amphibolic: able to be a part of both anabolism and catabolism Anaplerotic: referring to a reaction that ensures an adequate supply of an important metabolite Citrate Synthase: the enzyme that
Vocabulary Amphibolic: able to be a part of both anabolism and catabolism Anaplerotic: referring to a reaction that ensures an adequate supply of an important metabolite Citrate Synthase: the enzyme that
Review of Carbohydrate Digestion
 Review of Carbohydrate Digestion Glycolysis Glycolysis is a nine step biochemical pathway that oxidizes glucose into two molecules of pyruvic acid. During this process, energy is released and some of it
Review of Carbohydrate Digestion Glycolysis Glycolysis is a nine step biochemical pathway that oxidizes glucose into two molecules of pyruvic acid. During this process, energy is released and some of it
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
Respiration. Respiration. Respiration. How Cells Harvest Energy. Chapter 7
 How Cells Harvest Energy Chapter 7 Organisms can be classified based on how they obtain energy: autotrophs: are able to produce their own organic molecules through photosynthesis heterotrophs: live on
How Cells Harvest Energy Chapter 7 Organisms can be classified based on how they obtain energy: autotrophs: are able to produce their own organic molecules through photosynthesis heterotrophs: live on
How Cells Harvest Energy. Chapter 7. Respiration
 How Cells Harvest Energy Chapter 7 Respiration Organisms classified on how they obtain energy: autotrophs: produce their own organic molecules through photosynthesis heterotrophs: live on organic compounds
How Cells Harvest Energy Chapter 7 Respiration Organisms classified on how they obtain energy: autotrophs: produce their own organic molecules through photosynthesis heterotrophs: live on organic compounds
Methods of Enzyme Assay. By: Amal Alamri
 Methods of Enzyme Assay By: Amal Alamri Introduction: All enzyme assays measure either the consumption of substrate or production of product over time. Different enzymes require different estimation methods
Methods of Enzyme Assay By: Amal Alamri Introduction: All enzyme assays measure either the consumption of substrate or production of product over time. Different enzymes require different estimation methods
Metabolic engineering some basic considerations. Lecture 9
 Metabolic engineering some basic considerations Lecture 9 The 90ties: From fermentation to metabolic engineering Recruiting heterologous activities to perform directed genetic modifications of cell factories
Metabolic engineering some basic considerations Lecture 9 The 90ties: From fermentation to metabolic engineering Recruiting heterologous activities to perform directed genetic modifications of cell factories
Methods of Enzyme Assay
 Methods of Enzyme Assay Introduction All enzyme assays measure either the consumption of substrate or production of product over time. Different enzymes require different estimation methods dependingon
Methods of Enzyme Assay Introduction All enzyme assays measure either the consumption of substrate or production of product over time. Different enzymes require different estimation methods dependingon
BASIC ENZYMOLOGY 1.1
 BASIC ENZYMOLOGY 1.1 1.2 BASIC ENZYMOLOGY INTRODUCTION Enzymes are synthesized by all living organisms including man. These life essential substances accelerate the numerous metabolic reactions upon which
BASIC ENZYMOLOGY 1.1 1.2 BASIC ENZYMOLOGY INTRODUCTION Enzymes are synthesized by all living organisms including man. These life essential substances accelerate the numerous metabolic reactions upon which
Energy Production In A Cell (Chapter 25 Metabolism)
 Energy Production In A Cell (Chapter 25 Metabolism) Large food molecules contain a lot of potential energy in the form of chemical bonds but it requires a lot of work to liberate the energy. Cells need
Energy Production In A Cell (Chapter 25 Metabolism) Large food molecules contain a lot of potential energy in the form of chemical bonds but it requires a lot of work to liberate the energy. Cells need
MULTIPLE CHOICE QUESTIONS
 MULTIPLE CHOICE QUESTIONS 1. Which of the following statements concerning anabolic reactions is FALSE? A. They are generally endergonic. B. They usually require ATP. C. They are part of metabolism. D.
MULTIPLE CHOICE QUESTIONS 1. Which of the following statements concerning anabolic reactions is FALSE? A. They are generally endergonic. B. They usually require ATP. C. They are part of metabolism. D.
PAPER No. : 16 Bioorganic and biophysical chemistry MODULE No. : 25 Coenzyme-I Coenzyme A, TPP, B12 and biotin
 Subject Paper No and Title Module No and Title Module Tag 16, Bio organic and Bio physical chemistry 25, Coenzyme-I : Coenzyme A, TPP, B12 and CHE_P16_M25 TABLE OF CONTENTS 1. Learning Outcomes 2. Introduction
Subject Paper No and Title Module No and Title Module Tag 16, Bio organic and Bio physical chemistry 25, Coenzyme-I : Coenzyme A, TPP, B12 and CHE_P16_M25 TABLE OF CONTENTS 1. Learning Outcomes 2. Introduction
Citrate Cycle. Lecture 28. Key Concepts. The Citrate Cycle captures energy using redox reactions
 Citrate Cycle Lecture 28 Key Concepts The Citrate Cycle captures energy using redox reactions Eight reactions of the Citrate Cycle Key control points in the Citrate Cycle regulate metabolic flux What role
Citrate Cycle Lecture 28 Key Concepts The Citrate Cycle captures energy using redox reactions Eight reactions of the Citrate Cycle Key control points in the Citrate Cycle regulate metabolic flux What role
Chapter 9. Cellular Respiration and Fermentation
 Chapter 9 Cellular Respiration and Fermentation Energy flows into an ecosystem as sunlight and leaves as heat Photosynthesis generates O 2 and organic molecules, which are used in cellular respiration
Chapter 9 Cellular Respiration and Fermentation Energy flows into an ecosystem as sunlight and leaves as heat Photosynthesis generates O 2 and organic molecules, which are used in cellular respiration
Yield of energy from glucose
 Paper : Module : 05 Yield of Energy from Glucose Principal Investigator, Paper Coordinator and Content Writer Prof. Ramesh Kothari, Professor Dept. of Biosciences, Saurashtra University, Rajkot - 360005
Paper : Module : 05 Yield of Energy from Glucose Principal Investigator, Paper Coordinator and Content Writer Prof. Ramesh Kothari, Professor Dept. of Biosciences, Saurashtra University, Rajkot - 360005
True or False: 1. Reactions are called endergonic if they occur spontaneously and release free energy.
 True or False: 1. Reactions are called endergonic if they occur spontaneously and release free energy. 2. Enzymes catalyze chemical reactions by lowering the activation energy 3. Biochemical pathways are
True or False: 1. Reactions are called endergonic if they occur spontaneously and release free energy. 2. Enzymes catalyze chemical reactions by lowering the activation energy 3. Biochemical pathways are
Under aerobic conditions, pyruvate enters the mitochondria where it is converted into acetyl CoA.
 Under aerobic conditions, pyruvate enters the mitochondria where it is converted into acetyl CoA. Acetyl CoA is the fuel for the citric acid cycle, which processes the two carbon acetyl unit to two molecules
Under aerobic conditions, pyruvate enters the mitochondria where it is converted into acetyl CoA. Acetyl CoA is the fuel for the citric acid cycle, which processes the two carbon acetyl unit to two molecules
Module No. # 01 Lecture No. # 19 TCA Cycle
 Biochemical Engineering Prof. Dr. Rintu Banerjee Department of Agricultural and Food Engineering Asst. Prof. Dr. Saikat Chakraborty Department of Chemical Engineering Indian Institute of Technology, Kharagpur
Biochemical Engineering Prof. Dr. Rintu Banerjee Department of Agricultural and Food Engineering Asst. Prof. Dr. Saikat Chakraborty Department of Chemical Engineering Indian Institute of Technology, Kharagpur
Independent Study Guide Metabolism I. Principles of metabolism (section 6.1) a. Cells must: (figure 6.1) i. Synthesize new components
 Independent Study Guide Metabolism I. Principles of metabolism (section 6.1) a. Cells must: (figure 6.1) i. Synthesize new components (anabolism/biosynthesis) ii. Harvest energy and convert it to a usable
Independent Study Guide Metabolism I. Principles of metabolism (section 6.1) a. Cells must: (figure 6.1) i. Synthesize new components (anabolism/biosynthesis) ii. Harvest energy and convert it to a usable
Chapter 8 Mitochondria and Cellular Respiration
 Chapter 8 Mitochondria and Cellular Respiration Cellular respiration is the process of oxidizing food molecules, like glucose, to carbon dioxide and water. The energy released is trapped in the form of
Chapter 8 Mitochondria and Cellular Respiration Cellular respiration is the process of oxidizing food molecules, like glucose, to carbon dioxide and water. The energy released is trapped in the form of
BY: RASAQ NURUDEEN OLAJIDE
 BY: RASAQ NURUDEEN OLAJIDE LECTURE CONTENT INTRODUCTION CITRIC ACID CYCLE (T.C.A) PRODUCTION OF ACETYL CoA REACTIONS OF THE CITIRC ACID CYCLE THE AMPHIBOLIC NATURE OF THE T.C.A CYCLE THE GLYOXYLATE CYCLE
BY: RASAQ NURUDEEN OLAJIDE LECTURE CONTENT INTRODUCTION CITRIC ACID CYCLE (T.C.A) PRODUCTION OF ACETYL CoA REACTIONS OF THE CITIRC ACID CYCLE THE AMPHIBOLIC NATURE OF THE T.C.A CYCLE THE GLYOXYLATE CYCLE
Respiration. Organisms can be classified based on how they obtain energy: Autotrophs
 Respiration rganisms can be classified based on how they obtain energy: Autotrophs Able to produce their own organic molecules through photosynthesis Heterotrophs Live on organic compounds produced by
Respiration rganisms can be classified based on how they obtain energy: Autotrophs Able to produce their own organic molecules through photosynthesis Heterotrophs Live on organic compounds produced by
The Conservation of Homochirality and Prebiotic Synthesis of Amino Acids
 The Conservation of Homochirality and Prebiotic Synthesis of Amino Acids Harold J. Morowitz SFI WORKING PAPER: 2001-03-017 SFI Working Papers contain accounts of scientific work of the author(s) and do
The Conservation of Homochirality and Prebiotic Synthesis of Amino Acids Harold J. Morowitz SFI WORKING PAPER: 2001-03-017 SFI Working Papers contain accounts of scientific work of the author(s) and do
III. 6. Test. Respiració cel lular
 III. 6. Test. Respiració cel lular Chapter Questions 1) What is the term for metabolic pathways that release stored energy by breaking down complex molecules? A) anabolic pathways B) catabolic pathways
III. 6. Test. Respiració cel lular Chapter Questions 1) What is the term for metabolic pathways that release stored energy by breaking down complex molecules? A) anabolic pathways B) catabolic pathways
BIOLOGY - CLUTCH CH.9 - RESPIRATION.
 !! www.clutchprep.com CONCEPT: REDOX REACTIONS Redox reaction a chemical reaction that involves the transfer of electrons from one atom to another Oxidation loss of electrons Reduction gain of electrons
!! www.clutchprep.com CONCEPT: REDOX REACTIONS Redox reaction a chemical reaction that involves the transfer of electrons from one atom to another Oxidation loss of electrons Reduction gain of electrons
Marah Bitar. Faisal Nimri ... Nafeth Abu Tarboosh
 8 Marah Bitar Faisal Nimri... Nafeth Abu Tarboosh Summary of the 8 steps of citric acid cycle Step 1. Acetyl CoA joins with a four-carbon molecule, oxaloacetate, releasing the CoA group and forming a six-carbon
8 Marah Bitar Faisal Nimri... Nafeth Abu Tarboosh Summary of the 8 steps of citric acid cycle Step 1. Acetyl CoA joins with a four-carbon molecule, oxaloacetate, releasing the CoA group and forming a six-carbon
Prerequisites Protein purification techniques and protein analytical methods. Basic enzyme kinetics.
 Case 19 Purification of Rat Kidney Sphingosine Kinase Focus concept The purification and kinetic analysis of an enzyme that produces a product important in cell survival is the focus of this study. Prerequisites
Case 19 Purification of Rat Kidney Sphingosine Kinase Focus concept The purification and kinetic analysis of an enzyme that produces a product important in cell survival is the focus of this study. Prerequisites
Metabolism Energy Pathways Biosynthesis. Catabolism Anabolism Enzymes
 Topics Microbial Metabolism Metabolism Energy Pathways Biosynthesis 2 Metabolism Catabolism Catabolism Anabolism Enzymes Breakdown of complex organic molecules in order to extract energy and dform simpler
Topics Microbial Metabolism Metabolism Energy Pathways Biosynthesis 2 Metabolism Catabolism Catabolism Anabolism Enzymes Breakdown of complex organic molecules in order to extract energy and dform simpler
Citric Acid Cycle: Central Role in Catabolism. Entry of Pyruvate into the TCA cycle
 Citric Acid Cycle: Central Role in Catabolism Stage II of catabolism involves the conversion of carbohydrates, fats and aminoacids into acetylcoa In aerobic organisms, citric acid cycle makes up the final
Citric Acid Cycle: Central Role in Catabolism Stage II of catabolism involves the conversion of carbohydrates, fats and aminoacids into acetylcoa In aerobic organisms, citric acid cycle makes up the final
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site.
 Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
CITRIC ACID CYCLE ERT106 BIOCHEMISTRY SEM /19 BY: MOHAMAD FAHRURRAZI TOMPANG
 CITRIC ACID CYCLE ERT106 BIOCHEMISTRY SEM 1 2018/19 BY: MOHAMAD FAHRURRAZI TOMPANG Chapter Outline (19-1) The central role of the citric acid cycle in metabolism (19-2) The overall pathway of the citric
CITRIC ACID CYCLE ERT106 BIOCHEMISTRY SEM 1 2018/19 BY: MOHAMAD FAHRURRAZI TOMPANG Chapter Outline (19-1) The central role of the citric acid cycle in metabolism (19-2) The overall pathway of the citric
Syllabus for BASIC METABOLIC PRINCIPLES
 Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
Chapter 7 Cellular Respiration and Fermentation*
 Chapter 7 Cellular Respiration and Fermentation* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. Life Is Work
Chapter 7 Cellular Respiration and Fermentation* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. Life Is Work
4. Which step shows a split of one molecule into two smaller molecules? a. 2. d. 5
 1. Which of the following statements about NAD + is false? a. NAD + is reduced to NADH during both glycolysis and the citric acid cycle. b. NAD + has more chemical energy than NADH. c. NAD + is reduced
1. Which of the following statements about NAD + is false? a. NAD + is reduced to NADH during both glycolysis and the citric acid cycle. b. NAD + has more chemical energy than NADH. c. NAD + is reduced
The citric acid cycle Sitruunahappokierto Citronsyracykeln
 The citric acid cycle Sitruunahappokierto Citronsyracykeln Ove Eriksson BLL/Biokemia ove.eriksson@helsinki.fi Metabolome: The complete set of small-molecule metabolites to be found in a cell or an organism.
The citric acid cycle Sitruunahappokierto Citronsyracykeln Ove Eriksson BLL/Biokemia ove.eriksson@helsinki.fi Metabolome: The complete set of small-molecule metabolites to be found in a cell or an organism.
TCA CYCLE (Citric Acid Cycle)
 TCA CYCLE (Citric Acid Cycle) TCA CYCLE The Citric Acid Cycle is also known as: Kreb s cycle Sir Hans Krebs Nobel prize, 1953 TCA (tricarboxylic acid) cycle The citric acid cycle requires aerobic conditions!!!!
TCA CYCLE (Citric Acid Cycle) TCA CYCLE The Citric Acid Cycle is also known as: Kreb s cycle Sir Hans Krebs Nobel prize, 1953 TCA (tricarboxylic acid) cycle The citric acid cycle requires aerobic conditions!!!!
BIOL 158: BIOLOGICAL CHEMISTRY II
 BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
AP Bio Photosynthesis & Respiration
 AP Bio Photosynthesis & Respiration Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. What is the term used for the metabolic pathway in which
AP Bio Photosynthesis & Respiration Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. What is the term used for the metabolic pathway in which
Mock Exam All of the following are oxidizing agents EXCEPT a. NADP+ b. NADH c. FAD d. e. cytochromes
 Mock Exam 2 1. The Calvin cycle differs from the citric acid cycle in that it a. produces ATP b. directly requires light to run c. depends on the products of an electron transport chain d. occurs in a
Mock Exam 2 1. The Calvin cycle differs from the citric acid cycle in that it a. produces ATP b. directly requires light to run c. depends on the products of an electron transport chain d. occurs in a
BIOLOGY. Cellular Respiration and Fermentation CAMPBELL. Reece Urry Cain Wasserman Minorsky Jackson
 CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 9 Cellular Respiration and Fermentation Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick Figure 9.2 Light energy
CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 9 Cellular Respiration and Fermentation Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick Figure 9.2 Light energy
Cellular Respiration. 3. In the figure, which step of the citric acid cycle requires both NAD+ and ADP as reactants? a. Step 1. c. Step 3 b.
 Cellular Respiration 1. Enzymes are organic catalysts. How do they increase the rate of chemical reactions? a. By decreasing the free-energy change of the reaction b. By increasing the free-energy change
Cellular Respiration 1. Enzymes are organic catalysts. How do they increase the rate of chemical reactions? a. By decreasing the free-energy change of the reaction b. By increasing the free-energy change
Sheet #13. #Citric acid cycle made by zaid al-ghnaneem corrected by amer Al-salamat date 11/8/2016. Here we go.. Record #18
 1 Sheet #13 #Citric acid cycle made by zaid al-ghnaneem corrected by amer Al-salamat date 11/8/2016 Here we go.. Record #18 2 Three processes play central role in aerobic metabolism: 1) The citric acid
1 Sheet #13 #Citric acid cycle made by zaid al-ghnaneem corrected by amer Al-salamat date 11/8/2016 Here we go.. Record #18 2 Three processes play central role in aerobic metabolism: 1) The citric acid
Role of Sodium in Determining Alternate Pathways of Aerobic Citrate Catabolism in Aerobacter aerogenes
 JOURNAL OF BACTERIOLOGY, Aug. 1969, p. 389-394 Copyright 1969 American Society for Microbiology Vol. 99, No. 2 Printed in U.S.A. Role of Sodium in Determining Alternate Pathways of Aerobic Citrate Catabolism
JOURNAL OF BACTERIOLOGY, Aug. 1969, p. 389-394 Copyright 1969 American Society for Microbiology Vol. 99, No. 2 Printed in U.S.A. Role of Sodium in Determining Alternate Pathways of Aerobic Citrate Catabolism
Cellular Respiration: Harvesting Chemical Energy
 Chapter 9 Cellular Respiration: Harvesting Chemical Energy You should be able to: 1. Explain how redox reactions are involved in energy exchanges. Name and describe the three stages of cellular respiration;
Chapter 9 Cellular Respiration: Harvesting Chemical Energy You should be able to: 1. Explain how redox reactions are involved in energy exchanges. Name and describe the three stages of cellular respiration;
Lecture 11 - Biosynthesis of Amino Acids
 Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
BIOCHEMISTRY and MOLECULAR BIOLOGY INTERNATIONAL Pages 48]-486
![BIOCHEMISTRY and MOLECULAR BIOLOGY INTERNATIONAL Pages 48]-486 BIOCHEMISTRY and MOLECULAR BIOLOGY INTERNATIONAL Pages 48]-486](/thumbs/93/111729446.jpg) Vol. 41, No. 3, March 1997 BIOCHEMISTRY and MOLECULAR BIOLOGY INTERNATIONAL Pages 48]-486 INACTIVATION OF ACONITASE IN YEAST EXPOSED TO OXIDATIVE STRESS Keiko Murakami and Masataka Yoshino* Department
Vol. 41, No. 3, March 1997 BIOCHEMISTRY and MOLECULAR BIOLOGY INTERNATIONAL Pages 48]-486 INACTIVATION OF ACONITASE IN YEAST EXPOSED TO OXIDATIVE STRESS Keiko Murakami and Masataka Yoshino* Department
Harvesting energy: photosynthesis & cellular respiration
 Harvesting energy: photosynthesis & cellular respiration Learning Objectives Know the relationship between photosynthesis & cellular respiration Know the formulae of the chemical reactions for photosynthesis
Harvesting energy: photosynthesis & cellular respiration Learning Objectives Know the relationship between photosynthesis & cellular respiration Know the formulae of the chemical reactions for photosynthesis
Protein & Enzyme Lab (BBT 314)
 Protein & Enzyme Lab (BBT 314) Experiment 3 A: Determination of the enzyme ALT or SGPT activity in serum by enzymatic method using Bioanalyzer Background: Alanine aminotransferase (glutamate pyruvate transaminase)
Protein & Enzyme Lab (BBT 314) Experiment 3 A: Determination of the enzyme ALT or SGPT activity in serum by enzymatic method using Bioanalyzer Background: Alanine aminotransferase (glutamate pyruvate transaminase)
Biologic Oxidation BIOMEDICAL IMPORTAN
 Biologic Oxidation BIOMEDICAL IMPORTAN Chemically, oxidation is defined as the removal of electrons and reduction as the gain of electrons. Thus, oxidation is always accompanied by reduction of an electron
Biologic Oxidation BIOMEDICAL IMPORTAN Chemically, oxidation is defined as the removal of electrons and reduction as the gain of electrons. Thus, oxidation is always accompanied by reduction of an electron
ANSC/NUTR 618 Lipids & Lipid Metabolism
 I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
Chemistry 107 Exam 4 Study Guide
 Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Biological oxidation II. The Cytric acid cycle
 Biological oxidation II The Cytric acid cycle Outline The Cytric acid cycle (TCA tricarboxylic acid) Central role of Acetyl-CoA Regulation of the TCA cycle Anaplerotic reactions The Glyoxylate cycle Localization
Biological oxidation II The Cytric acid cycle Outline The Cytric acid cycle (TCA tricarboxylic acid) Central role of Acetyl-CoA Regulation of the TCA cycle Anaplerotic reactions The Glyoxylate cycle Localization
Energetics of carbohydrate and lipid metabolism
 Energetics of carbohydrate and lipid metabolism 1 Metabolism: The sum of all the chemical transformations taking place in a cell or organism, occurs through a series of enzymecatalyzed reactions that constitute
Energetics of carbohydrate and lipid metabolism 1 Metabolism: The sum of all the chemical transformations taking place in a cell or organism, occurs through a series of enzymecatalyzed reactions that constitute
Cellular Respiration and Fermentation
 CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
Supplemental data, Section 1:
 Supplemental data, Section 1: In the following section, we described the conflicting knowledge of some of the dead ends that are listed in Table S4 and our decision on how to resolve them. Thiamine Biosynthesis:
Supplemental data, Section 1: In the following section, we described the conflicting knowledge of some of the dead ends that are listed in Table S4 and our decision on how to resolve them. Thiamine Biosynthesis:
Chapter 9: Cellular Respiration
 Chapter 9: Cellular Respiration To perform their many tasks, living cells require energy from outside sources. Energy stored in food utimately comes from the sun. Photosynthesis makes the raw materials
Chapter 9: Cellular Respiration To perform their many tasks, living cells require energy from outside sources. Energy stored in food utimately comes from the sun. Photosynthesis makes the raw materials
Hind Abu Tawileh. Moh Tarek & Razi Kittaneh. Ma moun
 26 Hind Abu Tawileh Moh Tarek & Razi Kittaneh... Ma moun Cofactors are non-protein compounds, they are divided into 3 types: Protein-based. Metals: if they are bounded tightly (covalently) to the enzyme
26 Hind Abu Tawileh Moh Tarek & Razi Kittaneh... Ma moun Cofactors are non-protein compounds, they are divided into 3 types: Protein-based. Metals: if they are bounded tightly (covalently) to the enzyme
If you ate a clown, would it taste funny? Oh, wait, that s cannibalism . Anabolism
 If you ate a clown, would it taste funny? Oh, wait, that s cannibalism. Anabolism is about putting things together. Anabolism: The Use of Energy in Biosynthesis Anabolism energy from catabolism is used
If you ate a clown, would it taste funny? Oh, wait, that s cannibalism. Anabolism is about putting things together. Anabolism: The Use of Energy in Biosynthesis Anabolism energy from catabolism is used
CHAPTER 5 MICROBIAL METABOLISM
 CHAPTER 5 MICROBIAL METABOLISM I. Catabolic and Anabolic Reactions A. Metabolism - The sum of all chemical reactions within a living cell either releasing or requiring energy. (Overhead) Fig 5.1 1. Catabolism
CHAPTER 5 MICROBIAL METABOLISM I. Catabolic and Anabolic Reactions A. Metabolism - The sum of all chemical reactions within a living cell either releasing or requiring energy. (Overhead) Fig 5.1 1. Catabolism
enzymatic determinations of kecap Page 1 of 13
 enzymatic determinations of kecap Page 1 of 13 Contains enzymatic determinations of: - citrate - glycerol - glucose, fructose, starch - galactose - L-glutamic acid - formic acid - malate - ethanol - acetate
enzymatic determinations of kecap Page 1 of 13 Contains enzymatic determinations of: - citrate - glycerol - glucose, fructose, starch - galactose - L-glutamic acid - formic acid - malate - ethanol - acetate
Growth. Principles of Metabolism. Principles of Metabolism 1/18/2011. The role of ATP energy currency. Adenosine triphosphate
 Metabolism: Fueling Cell Growth Principles of Metabolism Cells (including your own) must: Synthesize new components (anabolism/biosynthesis) Harvest energy and convert it to a usable form (catabolism)
Metabolism: Fueling Cell Growth Principles of Metabolism Cells (including your own) must: Synthesize new components (anabolism/biosynthesis) Harvest energy and convert it to a usable form (catabolism)
Glycolysis Part 2. BCH 340 lecture 4
 Glycolysis Part 2 BCH 340 lecture 4 Regulation of Glycolysis There are three steps in glycolysis that have enzymes which regulate the flux of glycolysis These enzymes catalyzes irreversible reactions of
Glycolysis Part 2 BCH 340 lecture 4 Regulation of Glycolysis There are three steps in glycolysis that have enzymes which regulate the flux of glycolysis These enzymes catalyzes irreversible reactions of
Transport. Oxidation. Electron. which the en the ETC and. of NADH an. nd FADH 2 by ation. Both, Phosphorylation. Glycolysis Glucose.
 Electron Transport Chain and Oxidation Phosphorylation When one glucose molecule is oxidized to six CO 2 molecules by way of glycolysiss and TCA cycle, considerable amount of energy (ATP) is generated.
Electron Transport Chain and Oxidation Phosphorylation When one glucose molecule is oxidized to six CO 2 molecules by way of glycolysiss and TCA cycle, considerable amount of energy (ATP) is generated.
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 30 Amino Acid Degradation and the Urea Cycle 2013 W. H. Freeman and Company Chapter 30 Outline Amino acids are obtained from the
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 30 Amino Acid Degradation and the Urea Cycle 2013 W. H. Freeman and Company Chapter 30 Outline Amino acids are obtained from the
Chapter 24 Lecture Outline
 Chapter 24 Lecture Outline Carbohydrate Lipid and Protein! Metabolism! In the catabolism of carbohydrates, glycolysis converts glucose into pyruvate, which is then metabolized into acetyl CoA. Prepared
Chapter 24 Lecture Outline Carbohydrate Lipid and Protein! Metabolism! In the catabolism of carbohydrates, glycolysis converts glucose into pyruvate, which is then metabolized into acetyl CoA. Prepared
Objective: You will be able to construct an explanation for how each phase of respiration captures and stores free energy.
 Objective: You will be able to construct an explanation for how each phase of respiration captures and stores free energy. Do Now: Compare and contrast the three black equations below ADP + P + Energy
Objective: You will be able to construct an explanation for how each phase of respiration captures and stores free energy. Do Now: Compare and contrast the three black equations below ADP + P + Energy
Cellular Respiration and Fermentation
 CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
Integration Of Metabolism
 Integration Of Metabolism Metabolism Consist of Highly Interconnected Pathways The basic strategy of catabolic metabolism is to form ATP, NADPH, and building blocks for biosyntheses. 1. ATP is the universal
Integration Of Metabolism Metabolism Consist of Highly Interconnected Pathways The basic strategy of catabolic metabolism is to form ATP, NADPH, and building blocks for biosyntheses. 1. ATP is the universal
7 Cellular Respiration and Fermentation
 CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
Student Number: To form the polar phase when adsorption chromatography was used.
 Name: Student Number: April 14, 2001, 1:30 AM - 4:30 PM Page 1 (of 4) Biochemistry II Lab Section Final Examination Examiner: Dr. A. Scoot 1. Answer ALL questions in the space provided.. 2. The last page
Name: Student Number: April 14, 2001, 1:30 AM - 4:30 PM Page 1 (of 4) Biochemistry II Lab Section Final Examination Examiner: Dr. A. Scoot 1. Answer ALL questions in the space provided.. 2. The last page
Photosynthesis in chloroplasts. Cellular respiration in mitochondria ATP. ATP powers most cellular work
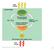 Light energy ECOSYSTEM CO + H O Photosynthesis in chloroplasts Cellular respiration in mitochondria Organic molecules + O powers most cellular work Heat energy 1 becomes oxidized (loses electron) becomes
Light energy ECOSYSTEM CO + H O Photosynthesis in chloroplasts Cellular respiration in mitochondria Organic molecules + O powers most cellular work Heat energy 1 becomes oxidized (loses electron) becomes
Citrate Cycle Supplemental Reading
 Citrate Cycle Supplemental Reading Key Concepts - The Citrate Cycle captures energy using redox reactions - Eight enzymatic reactions of the Citrate Cycle - Key control points in the citrate cycle regulate
Citrate Cycle Supplemental Reading Key Concepts - The Citrate Cycle captures energy using redox reactions - Eight enzymatic reactions of the Citrate Cycle - Key control points in the citrate cycle regulate
Cellular Pathways That Harvest Chemical Energy. Cellular Pathways That Harvest Chemical Energy. Cellular Pathways In General
 Cellular Pathways That Harvest Chemical Energy A. Obtaining Energy and Electrons from Glucose Lecture Series 12 Cellular Pathways That Harvest Chemical Energy B. An Overview: Releasing Energy from Glucose
Cellular Pathways That Harvest Chemical Energy A. Obtaining Energy and Electrons from Glucose Lecture Series 12 Cellular Pathways That Harvest Chemical Energy B. An Overview: Releasing Energy from Glucose
LAB 6 Fermentation & Cellular Respiration
 LAB 6 Fermentation & Cellular Respiration INTRODUCTION The cells of all living organisms require energy to keep themselves alive and fulfilling their roles. Where does this energy come from? The answer
LAB 6 Fermentation & Cellular Respiration INTRODUCTION The cells of all living organisms require energy to keep themselves alive and fulfilling their roles. Where does this energy come from? The answer
Background knowledge
 Background knowledge This is the required background knowledge: State three uses of energy in living things Give an example of an energy conversion in a living organism State that fats and oils contain
Background knowledge This is the required background knowledge: State three uses of energy in living things Give an example of an energy conversion in a living organism State that fats and oils contain
Photosynthesis in chloroplasts CO2 + H2O. Cellular respiration in mitochondria ATP. powers most cellular work. Heat energy
 Figure 9-01 LE 9-2 Light energy ECOSYSTEM Photosynthesis in chloroplasts CO2 + H2O Cellular respiration in mitochondria Organic + O molecules 2 powers most cellular work Heat energy LE 9-UN161a becomes
Figure 9-01 LE 9-2 Light energy ECOSYSTEM Photosynthesis in chloroplasts CO2 + H2O Cellular respiration in mitochondria Organic + O molecules 2 powers most cellular work Heat energy LE 9-UN161a becomes
Regulation of Citric Acid Cycle
 Paper : 04 Metabolism of carbohydrates Module : 30 Principal Investigator, Paper Coordinator and Content Writer Dr. Ramesh Kothari, Professor UGC-CAS Department of Biosciences Saurashtra University, Rajkot-5
Paper : 04 Metabolism of carbohydrates Module : 30 Principal Investigator, Paper Coordinator and Content Writer Dr. Ramesh Kothari, Professor UGC-CAS Department of Biosciences Saurashtra University, Rajkot-5
How Cells Harvest Chemical Energy
 Chapter 6 How Cells Harvest Chemical Energy INTRODUCTION TO CELLULAR RESIRATION hotosynthesis and cellular respiration provide energy for life Cellular respiration makes and consumes O During the oxidation
Chapter 6 How Cells Harvest Chemical Energy INTRODUCTION TO CELLULAR RESIRATION hotosynthesis and cellular respiration provide energy for life Cellular respiration makes and consumes O During the oxidation
FIRST BIOCHEMISTRY EXAM Tuesday 25/10/ MCQs. Location : 102, 105, 106, 301, 302
 FIRST BIOCHEMISTRY EXAM Tuesday 25/10/2016 10-11 40 MCQs. Location : 102, 105, 106, 301, 302 The Behavior of Proteins: Enzymes, Mechanisms, and Control General theory of enzyme action, by Leonor Michaelis
FIRST BIOCHEMISTRY EXAM Tuesday 25/10/2016 10-11 40 MCQs. Location : 102, 105, 106, 301, 302 The Behavior of Proteins: Enzymes, Mechanisms, and Control General theory of enzyme action, by Leonor Michaelis
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question.
 Respiration Practice Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) Which of the following statements describes NAD+? A) NAD+ can donate
Respiration Practice Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) Which of the following statements describes NAD+? A) NAD+ can donate
(de novo synthesis of glucose)
 Gluconeogenesis (de novo synthesis of glucose) Gluconeogenesis Gluconeogenesis is the biosynthesis of new glucose. The main purpose of gluconeogenesis is to maintain the constant blood Glc concentration.
Gluconeogenesis (de novo synthesis of glucose) Gluconeogenesis Gluconeogenesis is the biosynthesis of new glucose. The main purpose of gluconeogenesis is to maintain the constant blood Glc concentration.
Supporting information (protein purification, kinetic characterization, product isolation, and characterization by NMR and mass spectrometry):
 Supporting Information Mechanistic studies of a novel C-S lyase in ergothioneine biosynthesis: the involvement of a sulfenic acid intermediate Heng Song, 1 Wen Hu, 1,2 Nathchar Naowarojna, 1 Ampon Sae
Supporting Information Mechanistic studies of a novel C-S lyase in ergothioneine biosynthesis: the involvement of a sulfenic acid intermediate Heng Song, 1 Wen Hu, 1,2 Nathchar Naowarojna, 1 Ampon Sae
How Cells Harvest Chemical Energy
 How Cells Harvest Chemical Energy Global Athlete Outreach Program US CytoThesis Systems Medicine Center www.cytothesis.us US OncoTherapy Systems BioMedicine Group CytoThesis Bioengineering Research Group
How Cells Harvest Chemical Energy Global Athlete Outreach Program US CytoThesis Systems Medicine Center www.cytothesis.us US OncoTherapy Systems BioMedicine Group CytoThesis Bioengineering Research Group
CH 7: Cell Respiration and Fermentation Overview. Concept 7.1: Catabolic pathways yield energy by oxidizing organic fuels
 CH 7: Cell Respiration and Fermentation Overview Living cells require energy from outside sources Some animals obtain energy by eating plants, and some animals feed on other organisms Energy flows into
CH 7: Cell Respiration and Fermentation Overview Living cells require energy from outside sources Some animals obtain energy by eating plants, and some animals feed on other organisms Energy flows into
Chapter 9 Overview. Aerobic Metabolism I: The Citric Acid Cycle. Live processes - series of oxidation-reduction reactions. Aerobic metabolism I
 n n Chapter 9 Overview Aerobic Metabolism I: The Citric Acid Cycle Live processes - series of oxidation-reduction reactions Ingestion of proteins, carbohydrates, lipids Provide basic building blocks for
n n Chapter 9 Overview Aerobic Metabolism I: The Citric Acid Cycle Live processes - series of oxidation-reduction reactions Ingestion of proteins, carbohydrates, lipids Provide basic building blocks for
CIII Advances in Engineering Metabolism & Microbial Conversion
 CIII Advances in Engineering Metabolism & Microbial Conversion George Bennett, Ka-Yiu San, Rice University Manipulation and Balance of Reducing Equivalents to Enhance Productivity of Chemicals in E. coli
CIII Advances in Engineering Metabolism & Microbial Conversion George Bennett, Ka-Yiu San, Rice University Manipulation and Balance of Reducing Equivalents to Enhance Productivity of Chemicals in E. coli
Conversion of green note aldehydes into alcohols by yeast alcohol dehydrogenase
 Conversion of green note aldehydes into alcohols by yeast alcohol dehydrogenase M.-L. Fauconnier 1, A. Mpambara 1, J. Delcarte 1, P. Jacques 2, P. Thonart 2 & M. Marlier 1 1 Unité de Chimie Générale et
Conversion of green note aldehydes into alcohols by yeast alcohol dehydrogenase M.-L. Fauconnier 1, A. Mpambara 1, J. Delcarte 1, P. Jacques 2, P. Thonart 2 & M. Marlier 1 1 Unité de Chimie Générale et
Student Number: THE UNIVERSITY OF MANITOBA April 16, 2007, 9:00 AM -12:00 PM Page 1 (of 4) Biochemistry II Laboratory Section Final Examination
 Name: Student Number: THE UNIVERSITY OF MANITOBA April 16, 2007, 9:00 AM -12:00 PM Page 1 (of 4) Biochemistry II Laboratory Section Final Examination MBIO / CHEM.2370 Examiner: Dr. A. Scoot 1. Answer ALL
Name: Student Number: THE UNIVERSITY OF MANITOBA April 16, 2007, 9:00 AM -12:00 PM Page 1 (of 4) Biochemistry II Laboratory Section Final Examination MBIO / CHEM.2370 Examiner: Dr. A. Scoot 1. Answer ALL
BIL 256 Cell and Molecular Biology Lab Spring, Tissue-Specific Isoenzymes
 BIL 256 Cell and Molecular Biology Lab Spring, 2007 Background Information Tissue-Specific Isoenzymes A. BIOCHEMISTRY The basic pattern of glucose oxidation is outlined in Figure 3-1. Glucose is split
BIL 256 Cell and Molecular Biology Lab Spring, 2007 Background Information Tissue-Specific Isoenzymes A. BIOCHEMISTRY The basic pattern of glucose oxidation is outlined in Figure 3-1. Glucose is split
Chapter 7: How Cells Harvest Energy AP
 Chapter 7: How Cells Harvest Energy AP Essential Knowledge 1.B.1 distributed among organisms today. (7.1) 1.D.2 Organisms share many conserved core processes and features that evolved and are widely Scientific
Chapter 7: How Cells Harvest Energy AP Essential Knowledge 1.B.1 distributed among organisms today. (7.1) 1.D.2 Organisms share many conserved core processes and features that evolved and are widely Scientific
III. Metabolism The Citric Acid Cycle
 Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
7/5/2014. Microbial. Metabolism. Basic Chemical Reactions Underlying. Metabolism. Metabolism: Overview
 PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University Basic Chemical Reactions Underlying Metabolism Metabolism C H A P T E R 5 Microbial Metabolism Collection
PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University Basic Chemical Reactions Underlying Metabolism Metabolism C H A P T E R 5 Microbial Metabolism Collection
Enzymatic Assay of PYRUVATE KINASE (EC ) From Rabbit Liver
 Enzymatic Assay of PYRUVATE KINASE PRINCIPLE: Phospho(enol)pyruvate + ADP Pyruvate Kinase > Pyruvate + ATP Mg2 + Pyruvate + ß-NADH Lactic Dehydrogenase > Lactate + ß-NAD Abbreviations used: ADP = Adenosine
Enzymatic Assay of PYRUVATE KINASE PRINCIPLE: Phospho(enol)pyruvate + ADP Pyruvate Kinase > Pyruvate + ATP Mg2 + Pyruvate + ß-NADH Lactic Dehydrogenase > Lactate + ß-NAD Abbreviations used: ADP = Adenosine
Coenzymes, vitamins and trace elements 209. Petr Tůma Eva Samcová
 Coenzymes, vitamins and trace elements 209 Petr Tůma Eva Samcová History and nomenclature of enzymes 1810, Gay-Lussac made an experiment with yeats alter saccharide to ethanol and CO 2 Fermentation From
Coenzymes, vitamins and trace elements 209 Petr Tůma Eva Samcová History and nomenclature of enzymes 1810, Gay-Lussac made an experiment with yeats alter saccharide to ethanol and CO 2 Fermentation From
Dual nucleotide specificity of bovine glutamate dehydrogenase
 Biochem J. (1980) 191, 299-304 Printed in Great Britain 299 Dual nucleotide specificity of bovine glutamate dehydrogenase The role of negative co-operativity Stephen ALX and J. llis BLL Department ofbiochemistry,
Biochem J. (1980) 191, 299-304 Printed in Great Britain 299 Dual nucleotide specificity of bovine glutamate dehydrogenase The role of negative co-operativity Stephen ALX and J. llis BLL Department ofbiochemistry,
Cellular Respiration. Overview of Cellular Respiration. Lecture 8 Fall Overview of Cellular Respiration. Overview of Cellular Respiration
 Overview of Cellular Respiration 1 Cellular Respiration Lecture 8 Fall 2008 All organisms need ATP to do cellular work Cellular Respiration: The conversion of chemical energy of carbon compounds into another
Overview of Cellular Respiration 1 Cellular Respiration Lecture 8 Fall 2008 All organisms need ATP to do cellular work Cellular Respiration: The conversion of chemical energy of carbon compounds into another
