Characterizing Cancer Subtypes Using Dual Analysis in Caleydo StratomeX
|
|
|
- Thomasine Randall
- 5 years ago
- Views:
Transcription
1 Visual Analytics for Biological Data Characterizing Cancer Subtypes Using Dual Analysis in Caleydo StratomeX Cagatay Turkay City University London Alexander Lex Harvard University Marc Streit Johannes Kepler University Hanspeter Pfister Harvard University Helwig Hauser University of Bergen A lthough cancers are colloquially referred to by the tissue from which they originate (for example, lung cancer), significant differences can exist between cancers from the same tissue. The differences are often characterized by various biomolecular properties. These different forms of cancer are called subtypes. Large-scale research projects In this approach, dual-analysis such as the Cancer Genome Atviews depict distributions las (TCGA; of genes or data samples nih.gov) elicit comprehensive within Caleydo. Significantgenomic and clinical datasets difference plots show the with the goals of characterizing elements of a cancer subtype the molecular alterations responsible for cancer and identithat differ significantly from fying and characterizing cancer other subtypes. Analysts subtypes. can characterize subtypes, Owing to next-generation seinvestigate how samples quencing and microarray techrelate to their subtype and nology, these projects can employ other groups, and create welllarge, heterogeneous datasets. defined subtypes based on However, deriving insight from statistical properties. these complex datasets remains a challenge. Current analysis relies largely on custom scripts to find interesting genes or clusters of patients (stratifications) in these datasets. To remedy this, we developed Caleydo Stra38 gtur.indd 38 March/April 014 tomex, an interactive visualization method to analyze and discover relationships in these datasets.1 Researchers can use StratomeX to evaluate overlaps and relationships of stratifications. However, StratomeX doesn t inherently enable analysts to identify the characteristic genes of candidate subtypes, nor does it communicate how patients relate to a given subtype. The former capability is important because the characteristic genes could also be causally involved in a subtype and thus might be a target for a therapeutic or diagnostic approach. With the latter capability, researchers can investigate how samples relate to a subtype to estimate the quality of candidate subtypes and build a deeper characterization of a subtype. To address these limitations, we integrated two techniques into StratomeX: dual analysis, a general high-dimensional data analysis methodology, and significant-difference plots, a novel visual representation of the differences between data subsets. With this approach, domain scientists can discover genes that are distinctive for specific subtypes. They can also observe the properties of a cluster s samples and compare how they behave in Published by the IEEE Computer Society /14/$ IEEE /7/14 7:00 PM
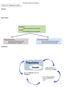
2 different datasets and clusters. These capabilities can provide a deeper understanding of stratifications. Moreover, scientists can employ the dualanalysis methodology to interactively generate stratifications. Biological Background and Analysis Tasks Subtype analysis is based on a variety of biomolecular datasets that capture different aspects of the process of life, ranging from the information stored in the genome to the functional products that trigger biochemical reactions in cells. Projects such as TCGA capture information on gene activity, factors influencing gene expression, and the genome s structure and sequence. An example of gene activity data is messenger RNA (mrna) data, which measures mrna s abundance in the cell. mrna is translated into proteins, which are the functional products. In addition, microrna (mirna) and DNA methylation influence gene expression and thus are important factors in many processes and diseases. All these factors play a role in the development of certain cancers, so a comprehensive analysis solution must take into account all these datasets, in addition to metadata such as clinical patient data. In this article, we demonstrate our approach by investigating mrna, mrna-seq (which relates to the same biological process as mrna but uses a different acquisition technique), mirna, and methylation data. However, a comprehensive analysis would also incorporate other datasets for instance, related to structural variations occurring on various scales in the genome. In previous research, we elicited subtype analysis tasks that dealt with finding and evaluating stratifications based on multiple datasets.1 We recently revisited those requirements in collaboration with domain scientists and found the need to supplement them with the following three tasks to further characterize stratifications. Find Distinctive Elements Identifying distinctive elements of clusters in a stratification provides a deeper understanding of why a particular cluster exists and how it relates to other clusters in the analysis. Distinctive elements are also good candidates to investigate as diagnostic markers or might even be causally involved in the disease. properties. Analysts can observe how strongly a cluster s members are related and explore whether they show similar properties in a dataset different from the one used for clustering. Create Clusters Analysts should be able to create clusters in an exploratory manner and interactively compare the intermediate results to metadata such as clinical data. Moreover, this manual clustering should enable analysts to merge observations of different datasets. The resulting clusters will be well defined in terms of statistical properties and richer in terms of the information sources included during construction. Methodological Building Blocks To enable the aforementioned tasks, our solution employs StratomeX and dual analysis. StratomeX Caleydo ( is an open-source visualization framework for biomolecular data analysis. It provides rich functionality for loading and handling multiple heterogeneous datasets as well as stratifications defined on the data. A core strength is its ability to slice datasets into meaningful subsets and flexibly combine multiple small visualizations of these subsets, using views such as histograms or heat maps, to create a fully integrated composite visualization.3 Other examples of visual methods that improve analysis of genomics data are the Hierarchical Cluster Explorer4 and Mayday.5 StratomeX, a Calyedo project, is a comparativevisualization technique that uses slicing. It lets analysts investigate the relationships between multiple stratifications, represented as columns. Each column consists of blocks, each corresponding to a group of patients. Ribbons of varying width visualize the overlap between neighboring stratifications, resulting in an overall appearance similar to Parallel Sets6 or Sankey Diagrams.7 Wide ribbons indicate a strong overlap between two groups; thin or absent ribbons correspond to only a few or no shared patients. Each block contains a visualization of the data for that group s patients. Analysts can switch between different types of visualizations. For numerical data, clustered heat maps are the default because they effectively communicate global trends and patterns. Compare Samples Investigating samples characteristics over several datasets and in comparison to other stratifications helps build a more complete picture of the samples Dual Analysis In this approach, the visual analysis occurs in parallel on both the data items and the dimensions. IEEE Computer Graphics and Applications gtur.indd /7/14 7:00 PM
3 Visual Analytics for Biological Data µ σ A single sample µ, σ A single gene µ vs. σ µ vs. σ Figure 1. Setting up dual-analysis views depicting the data as a D heat map. To construct the view depicting samples (the one with the yellow background), we computed the statistics for each sample (the mean, µ, and standard deviation, σ) using a row of the data. To construct the view of the genes (the one with the lightgreen background), we computed the statistics using a column of the data. We achieve this duality by using statistics computed over both the dataset rows and columns. For example, consider an mrna gene expression dataset given as a D data table with n rows and p columns, where each row corresponds to a single sample (patient) and each column to a single gene. The matrix cells contain the expression values. After normalizing the data appropriately, we calculate the central tendency (the mean, µ, or median) and the spread (the standard deviation, σ, or interquartile range, IQR), using each of the n samples and p genes separately. We calculate the robust counterparts of statistical moments to increase the statistics resistance to outlier values. Because experts are often accustomed to using nonrobust versions of the statistics (for example, μ or σ), our system incorporates such measures. This helps users quickly familiarize themselves with the information in the views, and at any point in an analysis, they can modify the set of statistics they re using. Figure 1 illustrates how we construct dualanalysis views. Visualizations of samples have a yellow background, with each point representing a sample; visualizations of genes have a light-green background, with each point depicting a gene. The computed statistics determine a point s location in a scatterplot. We can elaborate the analysis by using statistics other than the first two statistical moments. For the analyses in this article, we also computed skewness, which indicates a distribution s asymmetry (and the asymmetry s direction), and kurtosis, which characterizes its peakedness. Characterizing Subtypes To facilitate subtype characterization, we incorporate dual-analysis scatterplots and significantdifference plots as blocks. We also use these visualizations as separate linked views to enhance interactive visual exploration and achieve tasks such as manual cluster creation. Dual-Analysis Views Figure shows the embedded dual-analysis views. The sample scatterplots (yellow) display only those samples that are members of the represented cluster. The gene scatterplots (green) display the statistics for all the genes computed, using only the members of the represented cluster. We enhance interactive exploration by enabling a selection mechanism that s linked with all the views in StratomeX. Users can select both samples (see the second cluster in the second column of Figure ) and genes (see the second cluster in the third column in Figure ) at the same time. In Figure, the ribbons in StratomeX highlight the selection of the samples. 40 March/April 014
4 Figure. Dual-analysis views in StratomeX. The first column shows a four-cluster stratification for a microrna (mirna) dataset. The scatterplots show the median versus the interquartile range (IQR) for the cluster samples. The second column shows a three-cluster stratification for a messenger RNA (mrna) dataset, again showing samples. The third column uses the same three-cluster stratification for the same dataset but shows genes instead of samples. The sample scatterplots (yellow) depict the statistical characteristics of each cluster s members; the gene scatterplots (green) depict statistics computed for the genes using only the samples from the cluster represented by the block. The selection of samples is highlighted in the first two columns and in the ribbons. The selection of the genes enables investigation of the distribution of expression values for the genes for different clusters in a stratification. Significant-Difference Plots We previously used plots to effectively display the changes in statistical computations in response to a user s selection. 8 Here, we extend that approach with the determination and communication of the visualized differences significance. Figure 3 illustrates how we construct significantdifference plots (we call them just difference plots from now on). The user first selects (brushes) a subset of samples. In response, the system automatically calculates µ and σ for each gene using only the set of selected samples B (μ B and σ B ) and the rest of the samples R (μ R and σ R ) separately. We then compute the differences between the values with Δ µ = µ B μ R, Δ σ = σ B σ R, (1) where Δ μ and Δ σ are data vectors of size p, the number of genes. The difference plot then visualizes these values for all p. When no difference exists between a gene s expression values for B and R, we place that gene at the origin (0, 0). While building this view, we compare the selected set of samples to the rest instead of comparing against all the samples. This approach avoids overlap between the two compared sets. The difference plot on the right in Figure 3 displays the distribution of the differences in the statistic computations in response to the (sample) selection in the scatterplot (see the left of Figure 3). In this example, most genes have lower µ values for the selected items; that is, they re on the left of the y-axis. Communicating significance. An important consideration for analyzing differences between two subsets is statistical significance whether the difference is not likely due to chance. As in many other domains, researchers who analyze genomic data use statistical hypothesis tests to test for IEEE Computer Graphics and Applications 41
5 Visual Analytics for Biological Data µ R σ R Sample selection µ B σ B Differences for a single gene µ vs. σ µ vs. σ Figure 3. A significant-difference plot. The plot visualizes the differences between the selected samples (B) and unselected samples (R) for the genes. Δ μ and Δ σ are data vectors of size p, the number of genes. Genes that differ significantly are red; all others are blue. significance. 9 So, we enhance difference plots with integrated statistical hypothesis testing. As the hypothesis-testing procedure, we use the two-sample Welch s t-test. 10 This test doesn t assume that the two subsets have equal variance, which makes it suitable for our application. We perform the test on B and R and test against the (null) hypothesis that they have equal central tendencies. We compute t and the degrees of freedom, d.f.: µ B µ R t =, sb sr + NB NR ( sb NB + sr NR ) df..= s N ( N 1)+( s N ) ( N 1), ( ) B B B R R R where µ i is the sample mean, s i is the sample variance, and N i is the sample size of B and R. We then use these values together with the t- distribution and test the null hypothesis with a significance level of 0.05, employing a two-tail strategy. We perform this test for all p. For each gene, we store whether it shows a significant difference between B and R. We communicate this information by modifying the color of each gene in the difference plot. Genes with significant differences are red; the others are blue (see the right side of Figure 3). This enhancement lets analysts get immediate feedback on the differences significance. On the basis of this initial assessment, analysts can employ more-advanced routines to confirm the significance of the changes between the two subsets. Difference plots as blocks. While constructing these blocks, we again compute Δ μ and Δ σ for each gene, using Equation 1. Here, however, B corresponds to the samples that are members of the represented cluster, and R corresponds to the rest of the samples. We also compute the differences significance and color the visualization accordingly. The resulting blocks communicate which genes are more distinctive for each cluster. Moreover, the selection mechanism lets analysts compare these distinctive genes across different clusters. We show an example of this feature later. Case Studies We demonstrated our approach s effectiveness by analyzing a comprehensive invasive breast carcinoma (BRCA) dataset collected by the TCGA consortium. We used the mrna expression data, mirna sequencing data, and methylation data from more than 800 breast cancer patients. First, we loaded the BRCA data, which is available prepackaged for Caleydo. For comparison and evaluation, we used a recently published reference study that provided a stratification of samples March/April 014
6 (a) (b) (c) Figure 4. Using difference plots to find descriptive genes. (a) We marked descriptive genes for the HER-enriched subtype. A comparison to a reference study 11 showed the marked genes relevance. (b) We selected underexpressed genes for the Luminal-A subtype. They showed overexpression for the Basal-like subtype; that is, they constituted good features to discriminate these two subtypes. (c) The overexpressed genes for Luminal-A could also be considered good discriminators for this subtype but showed similar expression profiles for the Basal-like and HER-enriched subtypes. The case studies aimed to demonstrate how our approach lets analysts execute the three tasks we described earlier. Find Distinctive Elements We first compared the significantly distinctive genes suggested by our computations with those that the reference study identified. That study reported four subtypes: Luminal-A, Basal-like, Luminal-B, and HER-enriched (see Figure 4a). It used unsupervised clustering to identify a list of genes that are differentially expressed for the HER-enriched subtype (see supplementary Table 7 of the study 11 ). We selected the seven most significantly underexpressed genes (AGR3, ESR1, GFRA1, NPY1R, PGR, SER- PINA3, and SUSD3) and the 10 most significantly overexpressed genes (ABCA1, CALML5, CLCA, CRYM, DCD, GLYATL, MUCL1, NXPH1, PNMT, IEEE Computer Graphics and Applications 43
7 Visual Analytics for Biological Data (a) (b) Figure 5. (a) Investigating the sample profiles for the Basal-like subtype (column 1) over three datasets (mrna, mirna, and DNA methylation). The subtype contained samples with lower values and high variance for the mrna data and usually higher values for the mirna data. In the methylation data, however, we saw no dominant characteristic. (b) We selected core members of a cluster from an unsupervised stratification of mrna data (circled in the left column) and visualized them with a mirna stratification (the middle column) and the subtypes. The selected members corresponded to a subgroup in the Luminal-A subtype (circled in the right column). and SOX11). All seven underexpressed genes and six of the 10 overexpressed genes were identical to the ones in the reference study. This match demonstrates that our approach quickly yields relevant results in determining descriptive genes. Next, we explored the expression characteristics of the Luminal-A subtype s distinctive genes compared to the other subtypes. We first selected the significantly underexpressed genes for the Luminal- A subtype (see Figure 4b). These genes (AQP9, FAM83D, GGH, MCM10, and MMP1 being some of the lowest) were often overexpressed for the Basal-like subtype. We concluded that they re potentially good markers to distinguish the Luminal- A subtype from the Basal-like subtype. Similarly, when we selected the overexpressed genes for the Luminal-A subtype (see Figure 4c), we observed that they were underexpressed for the Basal-like subtype. However, unlike the previous set, they showed expression profiles similar to the HER-enriched subtype. Consequently, these genes carried less distinctive characteristics compared to the previous set. Compare Samples Here, we investigated how different datasets share certain properties of samples from a particular subtype for instance, outliers or trends. We first investigated the characteristics of samples from the Basal-like subtype by considering the mrna, mirna, and methylation datasets. We created a view with the subtypes from the reference study as the first column and unstratified versions of the three datasets as the following columns (see Figure 5a). After selecting the samples, we observed that they had lower expression values with a high variance in the mrna dataset and higher expression values in the mirna dataset. When looking at their methylation values, however, we saw no dominant characteristics. We used the same approach to determine the characteristics of a cluster computed as a result of an unsupervised clustering of the mrna dataset (see the left column in Figure 5b). We selected the second cluster s core members those with similar expression values and variance. These samples didn t show any dominant characteristics in an unsupervised clustering of the mirna data (see the middle column in Figure 5b). However, when considering the reference subtypes from the BRCA study (see the right column in Figure 5b), we observed that the selected samples constituted a subgroup of the Luminal-A subtype. We could also see that these samples were the overexpressed Luminal- A members with a lower variance. So, we conclude that we can use the second cluster from the mrna stratification to determine a Luminal-A subgroup. Create Clusters Sometimes in tumor subtype analysis, the stratification information isn t readily available. In these cases, we use dual analysis to manually create stratifications, instead of automated methods. This ability lets analysts discover structures 44 March/April 014
8 Kurtosis mrna σ Step 1 (a) Skewness (b) µ IQR mrna-seq σ Step (c) Median (d) µ σ Step 3 µ (e) (f) Figure 6. Manual clustering of an unstratified mrna dataset using dual-analysis views. (a) We selected negatively skewed genes through a skewness-versus-kurtosis visualization. (b) The difference plot for the samples updated automatically; we observed a group of samples with lower values and marked them as our first cluster. (c) We then switched to the mrna-seq dataset and selected genes that were more highly expressed with a large variety in the values. We identified two groups and marked them as (d) cluster and (e) cluster 3. (f) For validation, we compared our stratification with the subtypes from the reference study and observed a significant overlap of the subtypes. through different views of multiple datasets and represent these structures as a stratification. We performed the manual clustering process on the BRCA data. We used dual-analysis views as separate linked views rather than embedding them in StratomeX; that is, selections in any view were highlighted in the others. We brought up two linked views of the mrna dataset: a skewness-versus-kurtosis visualization of the genes (see Figure 6a) and a difference plot for the samples for Δ μ versus Δ σ (see Figure 6b). We added two other views of the mrnaseq dataset: median versus IQR visualization of the genes (see Figure 6c) and difference plots for the samples for Δ μ versus Δ σ (see Figures 6d and 6e). Then, we marked an unstratified mrna dataset as the target for the manual clustering (through a user interface not shown in the images). The clustering involved three steps. In step 1, we used skewness. High skewness indicated that a gene had nonuniform expression levels over the samples and thus was a good candidate to be a discriminator between subtypes. So, in this example, we selected the left-skewed genes (the ones with negative skew values) (see Figure 6a) and selected a group of samples that were visually separate from the rest (see the left of Figure 6b). At this point, we marked this subset of samples as a stratification of the mrna dataset (the first cluster in the first column in Figure 6f). In step, we switched to the mrna-seq dataset and selected the genes with higher expression values and higher variety (see Figure 6c). The difference plot updated automatically; we selected the samples with higher expression values and lower variance (see Figure 6d). From the difference plot, we selected the samples in the lower-right IEEE Computer Graphics and Applications 45
9 Visual Analytics for Biological Data quadrant that is, high values and variety. Instead of using the observed visual structures to guide selection (as in step 1), we used the visualization axes another strategy for making interesting selections. We made this selection because we expected to see higher variance and higher values for the samples in response to the selection of genes in Figure 6c. We finished this step by marking the sample selection as a second cluster. Our interactive approach enables analysts to merge interesting structures observed in several datasets using different perspectives on the data. In step 3, without updating the selection of genes, we selected the samples with higher variety but smaller mrna-seq values (see Figure 6e). This selection corresponded to the difference plot s upper-left quadrant that is, lower values and higher variety. We marked this selection as the third cluster. We left the rest of the samples as an unclustered set. We compared our custom stratification to the one from the reference study (see Figure 6f). The cluster from step 1, characterized by genes with negative skewness, almost completely overlapped with the Basal-like subtype. The cluster from step corresponded largely to a subgroup of the Luminal- A subtype. Finally, more than half of the samples from the cluster from step 3 belonged to the HER-enriched subtype. This overlap between the manually created clusters and the reference subtypes showed that manual clustering can produce relevant results. Our interactive approach enables analysts to merge interesting structures observed in several datasets using different perspectives on the data for example, using the skewness-versus-kurtosis view for the mrna dataset and the median-versus-iqr view for the mrna-seq dataset. This flexibility leads to outcomes that aren t so straightforward to generate through automated methods. Moreover, manual clustering provides a way to externalize analysis findings. Analysts can compare manually generated clusters with automatically computed results such as hierarchical clustering. Throughout these studies, we ve seen that our approach facilitates the characterization of cancer subtypes by enabling an investigation of them over both the samples and the genes. In the future, we aim to include advanced statistical tests and procedures, such as analysis of variance, Bonferroni correction, 9 and dimension reduction. We ll include these methods through integration of the R statistical-computing environment. 1 We also plan to extend difference plots to compare more than two groups. Furthermore, besides comparing one cluster to all the other elements, we plan to implement mechanisms to compare clusters with each other. Acknowledgments We thank Nils Gehlenborg, Samuel Gratzl, and Christian Partl for their input. Austrian Science Fund grant J 3437-N15 and US Air Force Research Laboratory and DARPA grant FA C-0300 supported this research. Cagatay Turkay carried out his research mostly at the University of Bergen, and his research is funded by the university s Department of Informatics. References 1. A. Lex et al., StratomeX: Visual Analysis of Large- Scale Heterogeneous Genomics Data for Cancer Subtype Characterization, Computer Graphics Forum, vol. 31, no. 3, 01, pp C. Turkay, P. Filzmoser, and H. Hauser, Brushing Dimensions a Dual Visual Analysis Model for High-Dimensional Data, IEEE Trans. Visualization and Computer Graphics, vol. 17, no. 1, 011, pp A. Lex et al., VisBricks: Multiform Visualization of Large, Inhomogeneous Data, IEEE Trans. Visualization and Computer Graphics, vol. 17, no. 1, 011, pp J. Seo and B. Shneiderman, Interactively Exploring Hierarchical Clustering Results, Computer, vol. 35, no. 7, 00, pp J. Dietzsch, N. Gehlenborg, and K. Nieselt, Mayday a Microarray Data Analysis Workbench, Bioinformatics, vol., no. 8, 006, pp R. Kosara, F. Bendix, and H. Hauser, Parallel Sets: Interactive Exploration and Visual Analysis of Categorical Data, IEEE Trans. Visualization and Computer Graphics, vol. 1, no. 4, 006, pp P. Riehmann, M. Hanfler, and B. Froehlich, Interactive Sankey Diagrams, Proc. 005 IEEE Symp. Information Visualization (InfoVis 05), 005, pp C. Turkay, J. Parulek, and H. Hauser, Dual Analysis of DNA Microarrays, Proc. 01 Conf. Knowledge Management and Knowledge Technologies (i-know 1), 01, article D.B. Allison et al., Microarray Data Analysis: From 46 March/April 014
10 Disarray to Consolidation and Consensus, Nature Reviews Genetics, vol. 7, no. 1, 006, pp G.D. Ruxton, The Unequal Variance t-test Is an Underused Alternative to Student s t-test and the Mann Whitney U Test, Behavioral Ecology, vol. 17, no. 4, 006, pp D. Koboldt et al., Comprehensive Molecular Portraits of Human Breast Tumours, Nature, vol. 490, no. 7418, 01, pp R Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, 013; fullrefman.pdf. Cagatay Turkay is a faculty member and lecturer in applied data science at the gicentre in the Department of Computer Science at City University London. His research focuses on the tight integration of interactive visualizations, data analysis techniques, and supporting exploratory knowledge and the capabilities of experts. Turkay received a PhD from the University of Bergen. Contact him at turkay.cagatay.1@city.ac.uk. Alexander Lex is a postdoctoral visualization researcher at the Visual Computing Group at the Harvard School of Engineering and Applied Sciences. His research interests are data visualization, especially for molecular biology, and human computer interaction. Lex received a PhD from the Graz University of Technology s Institute for Computer Graphics and Vision. Contact him at alex@seas.harvard.edu. Marc Streit is an assistant professor at the Institute of Computer Graphics at Johannes Kepler University Linz, where he leads the visualization group. His research interests include information visualization, visual analytics, and biological data visualization, particularly the integrated analysis of large heterogeneous data. Streit received his PhD in visualization from the Graz University of Technology. Contact him at marc.streit@jku.at. Hanspeter Pfister is the Gordon McKay Professor of Computer Science at Harvard University s School of Engineering and Applied Sciences. His research in visual computing lies at the intersection of visualization, computer graphics, and computer vision. Pfister received a PhD in computer science from the State University of New York at Stony Brook. He received the 010 IEEE Visualization Technical Achievement Award. He s coeditor of Point-Based Graphics (Elsevier, 007). Contact him at pfister@seas.harvard.edu. Helwig Hauser is a professor at the University of Bergen s Informatics Department, where he leads a visualization research group. His research interests include interactive visual analysis; flow visualization; illustrative visualization; and visualization for medicine, geosciences, biology, fluid dynamics, and so on. Hauser received a habilitation in visualization from the Vienna University of Technology. He received the 006 Heinz-Zemanek Award in computer science (for research on generalizing focus+context visualization) and the 013 Dirk Bartz Prize for visual computing in medicine (for pioneering high-quality visualization of sonographic medical data). He s on the Eurographics Conference on Visualization steering committee. Contact him at helwig.hauser@uib.no. GET HOT TOPIC INSIGHTS FROM INDUSTRY LEADERS Our bloggers keep you up on the latest Cloud, Big Data, Programming, Enterprise and Software strategies. Our multimedia, videos and articles give you technology solutions you can use. Our professional development information helps your career. Visit ComputingNow.computer.org. Your resource for technical development and leadership. Visit IEEE Computer Graphics and Applications gtur.indd /7/14 7:00 PM
StratomeX Visual Analysis of Large-Scale Heterogeneous Genomics Data for Cancer Subtype Characterization
 StratomeX Visual Analysis of Large-Scale Heterogeneous Genomics Data for Cancer Subtype Characterization Alexander Lex 1, Marc Streit 2, Hans-Jörg Schulz 3, Christian Partl 1, Dieter Schmalstieg 1, Peter
StratomeX Visual Analysis of Large-Scale Heterogeneous Genomics Data for Cancer Subtype Characterization Alexander Lex 1, Marc Streit 2, Hans-Jörg Schulz 3, Christian Partl 1, Dieter Schmalstieg 1, Peter
Supplementary Information
 Supplementary Information Guided Visual Exploration of Genomic Stratifications in Cancer Marc Streit 1,6, Alexander Lex 2,6, Samuel Gratzl¹, Christian Partl³, Dieter Schmalstieg³, Hanspeter Pfister², Peter
Supplementary Information Guided Visual Exploration of Genomic Stratifications in Cancer Marc Streit 1,6, Alexander Lex 2,6, Samuel Gratzl¹, Christian Partl³, Dieter Schmalstieg³, Hanspeter Pfister², Peter
Nature Methods: doi: /nmeth.3115
 Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
correspondence Guided visual exploration of genomic stratifications in cancer npg 2014 Nature America, Inc. All rights reserved.
 correspondence npg 2014 Nature America, Inc. All rights reserved. motor behaviors. The first was the forward optomotor response (OMR) 1,2, in which swimming is elicited by visual gratings moving in the
correspondence npg 2014 Nature America, Inc. All rights reserved. motor behaviors. The first was the forward optomotor response (OMR) 1,2, in which swimming is elicited by visual gratings moving in the
T. R. Golub, D. K. Slonim & Others 1999
 T. R. Golub, D. K. Slonim & Others 1999 Big Picture in 1999 The Need for Cancer Classification Cancer classification very important for advances in cancer treatment. Cancers of Identical grade can have
T. R. Golub, D. K. Slonim & Others 1999 Big Picture in 1999 The Need for Cancer Classification Cancer classification very important for advances in cancer treatment. Cancers of Identical grade can have
Identification of Tissue Independent Cancer Driver Genes
 Identification of Tissue Independent Cancer Driver Genes Alexandros Manolakos, Idoia Ochoa, Kartik Venkat Supervisor: Olivier Gevaert Abstract Identification of genomic patterns in tumors is an important
Identification of Tissue Independent Cancer Driver Genes Alexandros Manolakos, Idoia Ochoa, Kartik Venkat Supervisor: Olivier Gevaert Abstract Identification of genomic patterns in tumors is an important
CS Information Visualization Sep. 10, 2012 John Stasko. General representation techniques for multivariate (>3) variables per data case
 Topic Notes Multivariate Visual Representations 1 CS 7450 - Information Visualization Sep. 10, 2012 John Stasko Agenda General representation techniques for multivariate (>3) variables per data case But
Topic Notes Multivariate Visual Representations 1 CS 7450 - Information Visualization Sep. 10, 2012 John Stasko Agenda General representation techniques for multivariate (>3) variables per data case But
Introduction. Introduction
 Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
StratomeX: Visual Analysis of Large-Scale Heterogeneous Genomics Data for Cancer Subtype Characterization
 Eurographics Conference on Visualization (EuroVis) 2012 S. Bruckner, S. Miksch, and H. Pfister (Guest Editors) Volume 31 (2012), Number 3 StratomeX: Visual Analysis of Large-Scale Heterogeneous Genomics
Eurographics Conference on Visualization (EuroVis) 2012 S. Bruckner, S. Miksch, and H. Pfister (Guest Editors) Volume 31 (2012), Number 3 StratomeX: Visual Analysis of Large-Scale Heterogeneous Genomics
Unit 1 Exploring and Understanding Data
 Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
MBios 478: Systems Biology and Bayesian Networks, 27 [Dr. Wyrick] Slide #1. Lecture 27: Systems Biology and Bayesian Networks
![MBios 478: Systems Biology and Bayesian Networks, 27 [Dr. Wyrick] Slide #1. Lecture 27: Systems Biology and Bayesian Networks MBios 478: Systems Biology and Bayesian Networks, 27 [Dr. Wyrick] Slide #1. Lecture 27: Systems Biology and Bayesian Networks](/thumbs/80/82116384.jpg) MBios 478: Systems Biology and Bayesian Networks, 27 [Dr. Wyrick] Slide #1 Lecture 27: Systems Biology and Bayesian Networks Systems Biology and Regulatory Networks o Definitions o Network motifs o Examples
MBios 478: Systems Biology and Bayesian Networks, 27 [Dr. Wyrick] Slide #1 Lecture 27: Systems Biology and Bayesian Networks Systems Biology and Regulatory Networks o Definitions o Network motifs o Examples
Chapter 1. Introduction
 Chapter 1 Introduction 1.1 Motivation and Goals The increasing availability and decreasing cost of high-throughput (HT) technologies coupled with the availability of computational tools and data form a
Chapter 1 Introduction 1.1 Motivation and Goals The increasing availability and decreasing cost of high-throughput (HT) technologies coupled with the availability of computational tools and data form a
ANOVA in SPSS (Practical)
 ANOVA in SPSS (Practical) Analysis of Variance practical In this practical we will investigate how we model the influence of a categorical predictor on a continuous response. Centre for Multilevel Modelling
ANOVA in SPSS (Practical) Analysis of Variance practical In this practical we will investigate how we model the influence of a categorical predictor on a continuous response. Centre for Multilevel Modelling
Quantitative Methods in Computing Education Research (A brief overview tips and techniques)
 Quantitative Methods in Computing Education Research (A brief overview tips and techniques) Dr Judy Sheard Senior Lecturer Co-Director, Computing Education Research Group Monash University judy.sheard@monash.edu
Quantitative Methods in Computing Education Research (A brief overview tips and techniques) Dr Judy Sheard Senior Lecturer Co-Director, Computing Education Research Group Monash University judy.sheard@monash.edu
Stepwise method Modern Model Selection Methods Quantile-Quantile plot and tests for normality
 Week 9 Hour 3 Stepwise method Modern Model Selection Methods Quantile-Quantile plot and tests for normality Stat 302 Notes. Week 9, Hour 3, Page 1 / 39 Stepwise Now that we've introduced interactions,
Week 9 Hour 3 Stepwise method Modern Model Selection Methods Quantile-Quantile plot and tests for normality Stat 302 Notes. Week 9, Hour 3, Page 1 / 39 Stepwise Now that we've introduced interactions,
Biostatistics for Med Students. Lecture 1
 Biostatistics for Med Students Lecture 1 John J. Chen, Ph.D. Professor & Director of Biostatistics Core UH JABSOM JABSOM MD7 February 14, 2018 Lecture note: http://biostat.jabsom.hawaii.edu/education/training.html
Biostatistics for Med Students Lecture 1 John J. Chen, Ph.D. Professor & Director of Biostatistics Core UH JABSOM JABSOM MD7 February 14, 2018 Lecture note: http://biostat.jabsom.hawaii.edu/education/training.html
Summarizing Data. (Ch 1.1, 1.3, , 2.4.3, 2.5)
 1 Summarizing Data (Ch 1.1, 1.3, 1.10-1.13, 2.4.3, 2.5) Populations and Samples An investigation of some characteristic of a population of interest. Example: You want to study the average GPA of juniors
1 Summarizing Data (Ch 1.1, 1.3, 1.10-1.13, 2.4.3, 2.5) Populations and Samples An investigation of some characteristic of a population of interest. Example: You want to study the average GPA of juniors
V. Gathering and Exploring Data
 V. Gathering and Exploring Data With the language of probability in our vocabulary, we re now ready to talk about sampling and analyzing data. Data Analysis We can divide statistical methods into roughly
V. Gathering and Exploring Data With the language of probability in our vocabulary, we re now ready to talk about sampling and analyzing data. Data Analysis We can divide statistical methods into roughly
AP Statistics. Semester One Review Part 1 Chapters 1-5
 AP Statistics Semester One Review Part 1 Chapters 1-5 AP Statistics Topics Describing Data Producing Data Probability Statistical Inference Describing Data Ch 1: Describing Data: Graphically and Numerically
AP Statistics Semester One Review Part 1 Chapters 1-5 AP Statistics Topics Describing Data Producing Data Probability Statistical Inference Describing Data Ch 1: Describing Data: Graphically and Numerically
Here are the various choices. All of them are found in the Analyze menu in SPSS, under the sub-menu for Descriptive Statistics :
 Descriptive Statistics in SPSS When first looking at a dataset, it is wise to use descriptive statistics to get some idea of what your data look like. Here is a simple dataset, showing three different
Descriptive Statistics in SPSS When first looking at a dataset, it is wise to use descriptive statistics to get some idea of what your data look like. Here is a simple dataset, showing three different
MODEL-BASED CLUSTERING IN GENE EXPRESSION MICROARRAYS: AN APPLICATION TO BREAST CANCER DATA
 International Journal of Software Engineering and Knowledge Engineering Vol. 13, No. 6 (2003) 579 592 c World Scientific Publishing Company MODEL-BASED CLUSTERING IN GENE EXPRESSION MICROARRAYS: AN APPLICATION
International Journal of Software Engineering and Knowledge Engineering Vol. 13, No. 6 (2003) 579 592 c World Scientific Publishing Company MODEL-BASED CLUSTERING IN GENE EXPRESSION MICROARRAYS: AN APPLICATION
Visual Analytics of Cohort Study Data B. Preim (Visualization, Magdeburg)
 Visual Analytics of Cohort Study Data B. Preim (Visualization, Magdeburg) B. Preim Visual Analytics What is visual analytics? Integrates visualization techniques, data analysis and interaction techniques
Visual Analytics of Cohort Study Data B. Preim (Visualization, Magdeburg) B. Preim Visual Analytics What is visual analytics? Integrates visualization techniques, data analysis and interaction techniques
bivariate analysis: The statistical analysis of the relationship between two variables.
 bivariate analysis: The statistical analysis of the relationship between two variables. cell frequency: The number of cases in a cell of a cross-tabulation (contingency table). chi-square (χ 2 ) test for
bivariate analysis: The statistical analysis of the relationship between two variables. cell frequency: The number of cases in a cell of a cross-tabulation (contingency table). chi-square (χ 2 ) test for
Visualizing Temporal Patterns by Clustering Patients
 Visualizing Temporal Patterns by Clustering Patients Grace Shin, MS 1 ; Samuel McLean, MD 2 ; June Hu, MS 2 ; David Gotz, PhD 1 1 School of Information and Library Science; 2 Department of Anesthesiology
Visualizing Temporal Patterns by Clustering Patients Grace Shin, MS 1 ; Samuel McLean, MD 2 ; June Hu, MS 2 ; David Gotz, PhD 1 1 School of Information and Library Science; 2 Department of Anesthesiology
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE
 EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
Module 3: Pathway and Drug Development
 Module 3: Pathway and Drug Development Table of Contents 1.1 Getting Started... 6 1.2 Identifying a Dasatinib sensitive cancer signature... 7 1.2.1 Identifying and validating a Dasatinib Signature... 7
Module 3: Pathway and Drug Development Table of Contents 1.1 Getting Started... 6 1.2 Identifying a Dasatinib sensitive cancer signature... 7 1.2.1 Identifying and validating a Dasatinib Signature... 7
Statistical analysis DIANA SAPLACAN 2017 * SLIDES ADAPTED BASED ON LECTURE NOTES BY ALMA LEORA CULEN
 Statistical analysis DIANA SAPLACAN 2017 * SLIDES ADAPTED BASED ON LECTURE NOTES BY ALMA LEORA CULEN Vs. 2 Background 3 There are different types of research methods to study behaviour: Descriptive: observations,
Statistical analysis DIANA SAPLACAN 2017 * SLIDES ADAPTED BASED ON LECTURE NOTES BY ALMA LEORA CULEN Vs. 2 Background 3 There are different types of research methods to study behaviour: Descriptive: observations,
Nature Medicine: doi: /nm.3967
 Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Motivation: Fraud Detection
 Outlier Detection Motivation: Fraud Detection http://i.imgur.com/ckkoaop.gif Jian Pei: CMPT 741/459 Data Mining -- Outlier Detection (1) 2 Techniques: Fraud Detection Features Dissimilarity Groups and
Outlier Detection Motivation: Fraud Detection http://i.imgur.com/ckkoaop.gif Jian Pei: CMPT 741/459 Data Mining -- Outlier Detection (1) 2 Techniques: Fraud Detection Features Dissimilarity Groups and
Data and Statistics 101: Key Concepts in the Collection, Analysis, and Application of Child Welfare Data
 TECHNICAL REPORT Data and Statistics 101: Key Concepts in the Collection, Analysis, and Application of Child Welfare Data CONTENTS Executive Summary...1 Introduction...2 Overview of Data Analysis Concepts...2
TECHNICAL REPORT Data and Statistics 101: Key Concepts in the Collection, Analysis, and Application of Child Welfare Data CONTENTS Executive Summary...1 Introduction...2 Overview of Data Analysis Concepts...2
1) What is the independent variable? What is our Dependent Variable?
 1) What is the independent variable? What is our Dependent Variable? Independent Variable: Whether the font color and word name are the same or different. (Congruency) Dependent Variable: The amount of
1) What is the independent variable? What is our Dependent Variable? Independent Variable: Whether the font color and word name are the same or different. (Congruency) Dependent Variable: The amount of
Before we get started:
 Before we get started: http://arievaluation.org/projects-3/ AEA 2018 R-Commander 1 Antonio Olmos Kai Schramm Priyalathta Govindasamy Antonio.Olmos@du.edu AntonioOlmos@aumhc.org AEA 2018 R-Commander 2 Plan
Before we get started: http://arievaluation.org/projects-3/ AEA 2018 R-Commander 1 Antonio Olmos Kai Schramm Priyalathta Govindasamy Antonio.Olmos@du.edu AntonioOlmos@aumhc.org AEA 2018 R-Commander 2 Plan
VU Biostatistics and Experimental Design PLA.216
 VU Biostatistics and Experimental Design PLA.216 Julia Feichtinger Postdoctoral Researcher Institute of Computational Biotechnology Graz University of Technology Outline for Today About this course Background
VU Biostatistics and Experimental Design PLA.216 Julia Feichtinger Postdoctoral Researcher Institute of Computational Biotechnology Graz University of Technology Outline for Today About this course Background
Lecture Outline. Biost 517 Applied Biostatistics I. Purpose of Descriptive Statistics. Purpose of Descriptive Statistics
 Biost 517 Applied Biostatistics I Scott S. Emerson, M.D., Ph.D. Professor of Biostatistics University of Washington Lecture 3: Overview of Descriptive Statistics October 3, 2005 Lecture Outline Purpose
Biost 517 Applied Biostatistics I Scott S. Emerson, M.D., Ph.D. Professor of Biostatistics University of Washington Lecture 3: Overview of Descriptive Statistics October 3, 2005 Lecture Outline Purpose
EBCC Data Analysis Tool (EBCC DAT) Introduction
 Instructor: Paul Wolfgang Faculty sponsor: Yuan Shi, Ph.D. Andrey Mavrichev CIS 4339 Project in Computer Science May 7, 2009 Research work was completed in collaboration with Michael Tobia, Kevin L. Brown,
Instructor: Paul Wolfgang Faculty sponsor: Yuan Shi, Ph.D. Andrey Mavrichev CIS 4339 Project in Computer Science May 7, 2009 Research work was completed in collaboration with Michael Tobia, Kevin L. Brown,
Measuring the User Experience
 Measuring the User Experience Collecting, Analyzing, and Presenting Usability Metrics Chapter 2 Background Tom Tullis and Bill Albert Morgan Kaufmann, 2008 ISBN 978-0123735584 Introduction Purpose Provide
Measuring the User Experience Collecting, Analyzing, and Presenting Usability Metrics Chapter 2 Background Tom Tullis and Bill Albert Morgan Kaufmann, 2008 ISBN 978-0123735584 Introduction Purpose Provide
Kidane Tesfu Habtemariam, MASTAT, Principle of Stat Data Analysis Project work
 1 1. INTRODUCTION Food label tells the extent of calories contained in the food package. The number tells you the amount of energy in the food. People pay attention to calories because if you eat more
1 1. INTRODUCTION Food label tells the extent of calories contained in the food package. The number tells you the amount of energy in the food. People pay attention to calories because if you eat more
Cancer Informatics Lecture
 Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
WDHS Curriculum Map Probability and Statistics. What is Statistics and how does it relate to you?
 WDHS Curriculum Map Probability and Statistics Time Interval/ Unit 1: Introduction to Statistics 1.1-1.3 2 weeks S-IC-1: Understand statistics as a process for making inferences about population parameters
WDHS Curriculum Map Probability and Statistics Time Interval/ Unit 1: Introduction to Statistics 1.1-1.3 2 weeks S-IC-1: Understand statistics as a process for making inferences about population parameters
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Population. Sample. AP Statistics Notes for Chapter 1 Section 1.0 Making Sense of Data. Statistics: Data Analysis:
 Section 1.0 Making Sense of Data Statistics: Data Analysis: Individuals objects described by a set of data Variable any characteristic of an individual Categorical Variable places an individual into one
Section 1.0 Making Sense of Data Statistics: Data Analysis: Individuals objects described by a set of data Variable any characteristic of an individual Categorical Variable places an individual into one
National Academies Next Generation SAMPLE Researchers TITLE Initiative HERE
 National Academies Next Generation SAMPLE Researchers TITLE Initiative HERE Dennis A. Dean, II, PhD Sanofi Auditorium July 13, 2017 sevenbridges.com A little about me Research Experience Analytics and
National Academies Next Generation SAMPLE Researchers TITLE Initiative HERE Dennis A. Dean, II, PhD Sanofi Auditorium July 13, 2017 sevenbridges.com A little about me Research Experience Analytics and
Nature Immunology: doi: /ni Supplementary Figure 1. RNA-Seq analysis of CD8 + TILs and N-TILs.
 Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Identifying Thyroid Carcinoma Subtypes and Outcomes through Gene Expression Data Kun-Hsing Yu, Wei Wang, Chung-Yu Wang
 Identifying Thyroid Carcinoma Subtypes and Outcomes through Gene Expression Data Kun-Hsing Yu, Wei Wang, Chung-Yu Wang Abstract: Unlike most cancers, thyroid cancer has an everincreasing incidence rate
Identifying Thyroid Carcinoma Subtypes and Outcomes through Gene Expression Data Kun-Hsing Yu, Wei Wang, Chung-Yu Wang Abstract: Unlike most cancers, thyroid cancer has an everincreasing incidence rate
AN INFORMATION VISUALIZATION APPROACH TO CLASSIFICATION AND ASSESSMENT OF DIABETES RISK IN PRIMARY CARE
 Proceedings of the 3rd INFORMS Workshop on Data Mining and Health Informatics (DM-HI 2008) J. Li, D. Aleman, R. Sikora, eds. AN INFORMATION VISUALIZATION APPROACH TO CLASSIFICATION AND ASSESSMENT OF DIABETES
Proceedings of the 3rd INFORMS Workshop on Data Mining and Health Informatics (DM-HI 2008) J. Li, D. Aleman, R. Sikora, eds. AN INFORMATION VISUALIZATION APPROACH TO CLASSIFICATION AND ASSESSMENT OF DIABETES
HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA LEO TUNKLE *
 CERNA SEARCH METHOD IDENTIFIED A MET-ACTIVATED SUBGROUP AMONG EGFR DNA AMPLIFIED LUNG ADENOCARCINOMA PATIENTS HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA Email:
CERNA SEARCH METHOD IDENTIFIED A MET-ACTIVATED SUBGROUP AMONG EGFR DNA AMPLIFIED LUNG ADENOCARCINOMA PATIENTS HALLA KABAT * Outreach Program, mircore, 2929 Plymouth Rd. Ann Arbor, MI 48105, USA Email:
Observational studies; descriptive statistics
 Observational studies; descriptive statistics Patrick Breheny August 30 Patrick Breheny University of Iowa Biostatistical Methods I (BIOS 5710) 1 / 38 Observational studies Association versus causation
Observational studies; descriptive statistics Patrick Breheny August 30 Patrick Breheny University of Iowa Biostatistical Methods I (BIOS 5710) 1 / 38 Observational studies Association versus causation
Exercises: Differential Methylation
 Exercises: Differential Methylation Version 2018-04 Exercises: Differential Methylation 2 Licence This manual is 2014-18, Simon Andrews. This manual is distributed under the creative commons Attribution-Non-Commercial-Share
Exercises: Differential Methylation Version 2018-04 Exercises: Differential Methylation 2 Licence This manual is 2014-18, Simon Andrews. This manual is distributed under the creative commons Attribution-Non-Commercial-Share
COMPARATIVE STUDY ON FEATURE EXTRACTION METHOD FOR BREAST CANCER CLASSIFICATION
 COMPARATIVE STUDY ON FEATURE EXTRACTION METHOD FOR BREAST CANCER CLASSIFICATION 1 R.NITHYA, 2 B.SANTHI 1 Asstt Prof., School of Computing, SASTRA University, Thanjavur, Tamilnadu, India-613402 2 Prof.,
COMPARATIVE STUDY ON FEATURE EXTRACTION METHOD FOR BREAST CANCER CLASSIFICATION 1 R.NITHYA, 2 B.SANTHI 1 Asstt Prof., School of Computing, SASTRA University, Thanjavur, Tamilnadu, India-613402 2 Prof.,
Detection and Classification of Lung Cancer Using Artificial Neural Network
 Detection and Classification of Lung Cancer Using Artificial Neural Network Almas Pathan 1, Bairu.K.saptalkar 2 1,2 Department of Electronics and Communication Engineering, SDMCET, Dharwad, India 1 almaseng@yahoo.co.in,
Detection and Classification of Lung Cancer Using Artificial Neural Network Almas Pathan 1, Bairu.K.saptalkar 2 1,2 Department of Electronics and Communication Engineering, SDMCET, Dharwad, India 1 almaseng@yahoo.co.in,
Supplementary Materials Extracting a Cellular Hierarchy from High-dimensional Cytometry Data with SPADE
 Supplementary Materials Extracting a Cellular Hierarchy from High-dimensional Cytometry Data with SPADE Peng Qiu1,4, Erin F. Simonds2, Sean C. Bendall2, Kenneth D. Gibbs Jr.2, Robert V. Bruggner2, Michael
Supplementary Materials Extracting a Cellular Hierarchy from High-dimensional Cytometry Data with SPADE Peng Qiu1,4, Erin F. Simonds2, Sean C. Bendall2, Kenneth D. Gibbs Jr.2, Robert V. Bruggner2, Michael
Business Research Methods. Introduction to Data Analysis
 Business Research Methods Introduction to Data Analysis Data Analysis Process STAGES OF DATA ANALYSIS EDITING CODING DATA ENTRY ERROR CHECKING AND VERIFICATION DATA ANALYSIS Introduction Preparation of
Business Research Methods Introduction to Data Analysis Data Analysis Process STAGES OF DATA ANALYSIS EDITING CODING DATA ENTRY ERROR CHECKING AND VERIFICATION DATA ANALYSIS Introduction Preparation of
TWO HANDED SIGN LANGUAGE RECOGNITION SYSTEM USING IMAGE PROCESSING
 134 TWO HANDED SIGN LANGUAGE RECOGNITION SYSTEM USING IMAGE PROCESSING H.F.S.M.Fonseka 1, J.T.Jonathan 2, P.Sabeshan 3 and M.B.Dissanayaka 4 1 Department of Electrical And Electronic Engineering, Faculty
134 TWO HANDED SIGN LANGUAGE RECOGNITION SYSTEM USING IMAGE PROCESSING H.F.S.M.Fonseka 1, J.T.Jonathan 2, P.Sabeshan 3 and M.B.Dissanayaka 4 1 Department of Electrical And Electronic Engineering, Faculty
Chapter 1: Exploring Data
 Chapter 1: Exploring Data Key Vocabulary:! individual! variable! frequency table! relative frequency table! distribution! pie chart! bar graph! two-way table! marginal distributions! conditional distributions!
Chapter 1: Exploring Data Key Vocabulary:! individual! variable! frequency table! relative frequency table! distribution! pie chart! bar graph! two-way table! marginal distributions! conditional distributions!
SubLasso:a feature selection and classification R package with a. fixed feature subset
 SubLasso:a feature selection and classification R package with a fixed feature subset Youxi Luo,3,*, Qinghan Meng,2,*, Ruiquan Ge,2, Guoqin Mai, Jikui Liu, Fengfeng Zhou,#. Shenzhen Institutes of Advanced
SubLasso:a feature selection and classification R package with a fixed feature subset Youxi Luo,3,*, Qinghan Meng,2,*, Ruiquan Ge,2, Guoqin Mai, Jikui Liu, Fengfeng Zhou,#. Shenzhen Institutes of Advanced
Gene Selection for Tumor Classification Using Microarray Gene Expression Data
 Gene Selection for Tumor Classification Using Microarray Gene Expression Data K. Yendrapalli, R. Basnet, S. Mukkamala, A. H. Sung Department of Computer Science New Mexico Institute of Mining and Technology
Gene Selection for Tumor Classification Using Microarray Gene Expression Data K. Yendrapalli, R. Basnet, S. Mukkamala, A. H. Sung Department of Computer Science New Mexico Institute of Mining and Technology
Applied Statistical Analysis EDUC 6050 Week 4
 Applied Statistical Analysis EDUC 6050 Week 4 Finding clarity using data Today 1. Hypothesis Testing with Z Scores (continued) 2. Chapters 6 and 7 in Book 2 Review! = $ & '! = $ & ' * ) 1. Which formula
Applied Statistical Analysis EDUC 6050 Week 4 Finding clarity using data Today 1. Hypothesis Testing with Z Scores (continued) 2. Chapters 6 and 7 in Book 2 Review! = $ & '! = $ & ' * ) 1. Which formula
Pythia WEB ENABLED TIMED INFLUENCE NET MODELING TOOL SAL. Lee W. Wagenhals Alexander H. Levis
 Pythia WEB ENABLED TIMED INFLUENCE NET MODELING TOOL Lee W. Wagenhals Alexander H. Levis ,@gmu.edu Adversary Behavioral Modeling Maxwell AFB, Montgomery AL March 8-9, 2007 1 Outline Pythia
Pythia WEB ENABLED TIMED INFLUENCE NET MODELING TOOL Lee W. Wagenhals Alexander H. Levis ,@gmu.edu Adversary Behavioral Modeling Maxwell AFB, Montgomery AL March 8-9, 2007 1 Outline Pythia
Understandable Statistics
 Understandable Statistics correlated to the Advanced Placement Program Course Description for Statistics Prepared for Alabama CC2 6/2003 2003 Understandable Statistics 2003 correlated to the Advanced Placement
Understandable Statistics correlated to the Advanced Placement Program Course Description for Statistics Prepared for Alabama CC2 6/2003 2003 Understandable Statistics 2003 correlated to the Advanced Placement
EXTRACT THE BREAST CANCER IN MAMMOGRAM IMAGES
 International Journal of Civil Engineering and Technology (IJCIET) Volume 10, Issue 02, February 2019, pp. 96-105, Article ID: IJCIET_10_02_012 Available online at http://www.iaeme.com/ijciet/issues.asp?jtype=ijciet&vtype=10&itype=02
International Journal of Civil Engineering and Technology (IJCIET) Volume 10, Issue 02, February 2019, pp. 96-105, Article ID: IJCIET_10_02_012 Available online at http://www.iaeme.com/ijciet/issues.asp?jtype=ijciet&vtype=10&itype=02
Biostatistics. Donna Kritz-Silverstein, Ph.D. Professor Department of Family & Preventive Medicine University of California, San Diego
 Biostatistics Donna Kritz-Silverstein, Ph.D. Professor Department of Family & Preventive Medicine University of California, San Diego (858) 534-1818 dsilverstein@ucsd.edu Introduction Overview of statistical
Biostatistics Donna Kritz-Silverstein, Ph.D. Professor Department of Family & Preventive Medicine University of California, San Diego (858) 534-1818 dsilverstein@ucsd.edu Introduction Overview of statistical
Use Case 9: Coordinated Changes of Epigenomic Marks Across Tissue Types. Epigenome Informatics Workshop Bioinformatics Research Laboratory
 Use Case 9: Coordinated Changes of Epigenomic Marks Across Tissue Types Epigenome Informatics Workshop Bioinformatics Research Laboratory 1 Introduction Active or inactive states of transcription factor
Use Case 9: Coordinated Changes of Epigenomic Marks Across Tissue Types Epigenome Informatics Workshop Bioinformatics Research Laboratory 1 Introduction Active or inactive states of transcription factor
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Six Sigma Glossary Lean 6 Society
 Six Sigma Glossary Lean 6 Society ABSCISSA ACCEPTANCE REGION ALPHA RISK ALTERNATIVE HYPOTHESIS ASSIGNABLE CAUSE ASSIGNABLE VARIATIONS The horizontal axis of a graph The region of values for which the null
Six Sigma Glossary Lean 6 Society ABSCISSA ACCEPTANCE REGION ALPHA RISK ALTERNATIVE HYPOTHESIS ASSIGNABLE CAUSE ASSIGNABLE VARIATIONS The horizontal axis of a graph The region of values for which the null
Knowledge discovery tools 381
 Knowledge discovery tools 381 hours, and prime time is prime time precisely because more people tend to watch television at that time.. Compare histograms from di erent periods of time. Changes in histogram
Knowledge discovery tools 381 hours, and prime time is prime time precisely because more people tend to watch television at that time.. Compare histograms from di erent periods of time. Changes in histogram
Analysis and Interpretation of Data Part 1
 Analysis and Interpretation of Data Part 1 DATA ANALYSIS: PRELIMINARY STEPS 1. Editing Field Edit Completeness Legibility Comprehensibility Consistency Uniformity Central Office Edit 2. Coding Specifying
Analysis and Interpretation of Data Part 1 DATA ANALYSIS: PRELIMINARY STEPS 1. Editing Field Edit Completeness Legibility Comprehensibility Consistency Uniformity Central Office Edit 2. Coding Specifying
Brain Tumour Diagnostic Support Based on Medical Image Segmentation
 Brain Tumour Diagnostic Support Based on Medical Image Segmentation Z. Měřínský, E. Hošťálková, A. Procházka Institute of Chemical Technology, Prague Department of Computing and Control Engineering Abstract
Brain Tumour Diagnostic Support Based on Medical Image Segmentation Z. Měřínský, E. Hošťálková, A. Procházka Institute of Chemical Technology, Prague Department of Computing and Control Engineering Abstract
Seamless Audio Splicing for ISO/IEC Transport Streams
 Seamless Audio Splicing for ISO/IEC 13818 Transport Streams A New Framework for Audio Elementary Stream Tailoring and Modeling Seyfullah Halit Oguz, Ph.D. and Sorin Faibish EMC Corporation Media Solutions
Seamless Audio Splicing for ISO/IEC 13818 Transport Streams A New Framework for Audio Elementary Stream Tailoring and Modeling Seyfullah Halit Oguz, Ph.D. and Sorin Faibish EMC Corporation Media Solutions
Leveraging Standards for Effective Visualization of Early Efficacy in Clinical Trial Oncology Studies
 PharmaSUG 2018 - Paper DV-26 Leveraging Standards for Effective Visualization of Early Efficacy in Clinical Trial Oncology Studies Kelci Miclaus and Lili Li, JMP Life Sciences SAS Institute ABSTRACT Solid
PharmaSUG 2018 - Paper DV-26 Leveraging Standards for Effective Visualization of Early Efficacy in Clinical Trial Oncology Studies Kelci Miclaus and Lili Li, JMP Life Sciences SAS Institute ABSTRACT Solid
Expert-guided Visual Exploration (EVE) for patient stratification. Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland
 Expert-guided Visual Exploration (EVE) for patient stratification Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland Oncoscape.sttrcancer.org Paul Lisa Ken Jenny Desert Eric The challenge Given - patient clinical
Expert-guided Visual Exploration (EVE) for patient stratification Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland Oncoscape.sttrcancer.org Paul Lisa Ken Jenny Desert Eric The challenge Given - patient clinical
Introduction to Statistical Data Analysis I
 Introduction to Statistical Data Analysis I JULY 2011 Afsaneh Yazdani Preface What is Statistics? Preface What is Statistics? Science of: designing studies or experiments, collecting data Summarizing/modeling/analyzing
Introduction to Statistical Data Analysis I JULY 2011 Afsaneh Yazdani Preface What is Statistics? Preface What is Statistics? Science of: designing studies or experiments, collecting data Summarizing/modeling/analyzing
Representing Process Variation by Means of a Process Family
 Representing Process Variation by Means of a Process Family Borislava I. Simidchieva, Leon J. Osterweil, Lori A. Clarke Laboratory for Advanced Software Engineering Research University of Massachusetts,
Representing Process Variation by Means of a Process Family Borislava I. Simidchieva, Leon J. Osterweil, Lori A. Clarke Laboratory for Advanced Software Engineering Research University of Massachusetts,
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES
 ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
TERRORIST WATCHER: AN INTERACTIVE WEB- BASED VISUAL ANALYTICAL TOOL OF TERRORIST S PERSONAL CHARACTERISTICS
 TERRORIST WATCHER: AN INTERACTIVE WEB- BASED VISUAL ANALYTICAL TOOL OF TERRORIST S PERSONAL CHARACTERISTICS Samah Mansoour School of Computing and Information Systems, Grand Valley State University, Michigan,
TERRORIST WATCHER: AN INTERACTIVE WEB- BASED VISUAL ANALYTICAL TOOL OF TERRORIST S PERSONAL CHARACTERISTICS Samah Mansoour School of Computing and Information Systems, Grand Valley State University, Michigan,
RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays
 Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Explore. sexcntry Sex according to country. [DataSet1] D:\NORA\NORA Main File.sav
![Explore. sexcntry Sex according to country. [DataSet1] D:\NORA\NORA Main File.sav Explore. sexcntry Sex according to country. [DataSet1] D:\NORA\NORA Main File.sav](/thumbs/82/84761267.jpg) EXAMINE VARIABLES=nc228 BY sexcntry /PLOT BOXPLOT HISTOGRAM NPPLOT /COMPARE GROUPS /STATISTICS DESCRIPTIVES /CINTERVAL 95 /MISSING LISTWISE /NOTOTAL. Explore Notes Output Created Comments Input Missing
EXAMINE VARIABLES=nc228 BY sexcntry /PLOT BOXPLOT HISTOGRAM NPPLOT /COMPARE GROUPS /STATISTICS DESCRIPTIVES /CINTERVAL 95 /MISSING LISTWISE /NOTOTAL. Explore Notes Output Created Comments Input Missing
Using Analytical and Psychometric Tools in Medium- and High-Stakes Environments
 Using Analytical and Psychometric Tools in Medium- and High-Stakes Environments Greg Pope, Analytics and Psychometrics Manager 2008 Users Conference San Antonio Introduction and purpose of this session
Using Analytical and Psychometric Tools in Medium- and High-Stakes Environments Greg Pope, Analytics and Psychometrics Manager 2008 Users Conference San Antonio Introduction and purpose of this session
Visual Exploration of Classification Models for Risk Assessment
 Visual Exploration of Classification Models for Risk Assessment M.A. Migut M. Worring Intelligent Systems Lab Amsterdam, University of Amsterdam, Amsterdam, The Netherlands M.A.Migut@uva.nl M.Worring@uva.nl
Visual Exploration of Classification Models for Risk Assessment M.A. Migut M. Worring Intelligent Systems Lab Amsterdam, University of Amsterdam, Amsterdam, The Netherlands M.A.Migut@uva.nl M.Worring@uva.nl
SPRING GROVE AREA SCHOOL DISTRICT. Course Description. Instructional Strategies, Learning Practices, Activities, and Experiences.
 SPRING GROVE AREA SCHOOL DISTRICT PLANNED COURSE OVERVIEW Course Title: Basic Introductory Statistics Grade Level(s): 11-12 Units of Credit: 1 Classification: Elective Length of Course: 30 cycles Periods
SPRING GROVE AREA SCHOOL DISTRICT PLANNED COURSE OVERVIEW Course Title: Basic Introductory Statistics Grade Level(s): 11-12 Units of Credit: 1 Classification: Elective Length of Course: 30 cycles Periods
10/4/2007 MATH 171 Name: Dr. Lunsford Test Points Possible
 Pledge: 10/4/2007 MATH 171 Name: Dr. Lunsford Test 1 100 Points Possible I. Short Answer and Multiple Choice. (36 points total) 1. Circle all of the items below that are measures of center of a distribution:
Pledge: 10/4/2007 MATH 171 Name: Dr. Lunsford Test 1 100 Points Possible I. Short Answer and Multiple Choice. (36 points total) 1. Circle all of the items below that are measures of center of a distribution:
Statistics is a broad mathematical discipline dealing with
 Statistical Primer for Cardiovascular Research Descriptive Statistics and Graphical Displays Martin G. Larson, SD Statistics is a broad mathematical discipline dealing with techniques for the collection,
Statistical Primer for Cardiovascular Research Descriptive Statistics and Graphical Displays Martin G. Larson, SD Statistics is a broad mathematical discipline dealing with techniques for the collection,
Journal: Nature Methods
 Journal: Nature Methods Article Title: Network-based stratification of tumor mutations Corresponding Author: Trey Ideker Supplementary Item Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure
Journal: Nature Methods Article Title: Network-based stratification of tumor mutations Corresponding Author: Trey Ideker Supplementary Item Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure
Nature Getetics: doi: /ng.3471
 Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
A Comparison of Collaborative Filtering Methods for Medication Reconciliation
 A Comparison of Collaborative Filtering Methods for Medication Reconciliation Huanian Zheng, Rema Padman, Daniel B. Neill The H. John Heinz III College, Carnegie Mellon University, Pittsburgh, PA, 15213,
A Comparison of Collaborative Filtering Methods for Medication Reconciliation Huanian Zheng, Rema Padman, Daniel B. Neill The H. John Heinz III College, Carnegie Mellon University, Pittsburgh, PA, 15213,
Statistical Summaries. Kerala School of MathematicsCourse in Statistics for Scientists. Descriptive Statistics. Summary Statistics
 Kerala School of Mathematics Course in Statistics for Scientists Statistical Summaries Descriptive Statistics T.Krishnan Strand Life Sciences, Bangalore may be single numerical summaries of a batch, such
Kerala School of Mathematics Course in Statistics for Scientists Statistical Summaries Descriptive Statistics T.Krishnan Strand Life Sciences, Bangalore may be single numerical summaries of a batch, such
Expanded View Figures
 Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
SUPPLEMENTARY INFORMATION In format provided by Javier DeFelipe et al. (MARCH 2013)
 Supplementary Online Information S2 Analysis of raw data Forty-two out of the 48 experts finished the experiment, and only data from these 42 experts are considered in the remainder of the analysis. We
Supplementary Online Information S2 Analysis of raw data Forty-two out of the 48 experts finished the experiment, and only data from these 42 experts are considered in the remainder of the analysis. We
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Set the stage: Genomics technology. Jos Kleinjans Dept of Toxicogenomics Maastricht University, the Netherlands
 Set the stage: Genomics technology Jos Kleinjans Dept of Toxicogenomics Maastricht University, the Netherlands Amendment to the latest consolidated version of the REACH legislation REACH Regulation 1907/2006:
Set the stage: Genomics technology Jos Kleinjans Dept of Toxicogenomics Maastricht University, the Netherlands Amendment to the latest consolidated version of the REACH legislation REACH Regulation 1907/2006:
Extraction of Blood Vessels and Recognition of Bifurcation Points in Retinal Fundus Image
 International Journal of Research Studies in Science, Engineering and Technology Volume 1, Issue 5, August 2014, PP 1-7 ISSN 2349-4751 (Print) & ISSN 2349-476X (Online) Extraction of Blood Vessels and
International Journal of Research Studies in Science, Engineering and Technology Volume 1, Issue 5, August 2014, PP 1-7 ISSN 2349-4751 (Print) & ISSN 2349-476X (Online) Extraction of Blood Vessels and
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Experimental design and workflow utilized to generate the WMG Protein Atlas.
 Supplementary Figure 1 Experimental design and workflow utilized to generate the WMG Protein Atlas. (a) Illustration of the plant organs and nodule infection time points analyzed. (b) Proteomic workflow
Supplementary Figure 1 Experimental design and workflow utilized to generate the WMG Protein Atlas. (a) Illustration of the plant organs and nodule infection time points analyzed. (b) Proteomic workflow
Session 4 Rebecca Poulos
 The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 20
The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 20
What is Data? Part 2: Patterns & Associations. INFO-1301, Quantitative Reasoning 1 University of Colorado Boulder
 What is Data? Part 2: Patterns & Associations INFO-1301, Quantitative Reasoning 1 University of Colorado Boulder August 29, 2016 Prof. Michael Paul Prof. William Aspray Overview This lecture will look
What is Data? Part 2: Patterns & Associations INFO-1301, Quantitative Reasoning 1 University of Colorado Boulder August 29, 2016 Prof. Michael Paul Prof. William Aspray Overview This lecture will look
User Guide. Association analysis. Input
 User Guide TFEA.ChIP is a tool to estimate transcription factor enrichment in a set of differentially expressed genes using data from ChIP-Seq experiments performed in different tissues and conditions.
User Guide TFEA.ChIP is a tool to estimate transcription factor enrichment in a set of differentially expressed genes using data from ChIP-Seq experiments performed in different tissues and conditions.
Semiotics and Intelligent Control
 Semiotics and Intelligent Control Morten Lind 0rsted-DTU: Section of Automation, Technical University of Denmark, DK-2800 Kgs. Lyngby, Denmark. m/i@oersted.dtu.dk Abstract: Key words: The overall purpose
Semiotics and Intelligent Control Morten Lind 0rsted-DTU: Section of Automation, Technical University of Denmark, DK-2800 Kgs. Lyngby, Denmark. m/i@oersted.dtu.dk Abstract: Key words: The overall purpose
Computer Science, Biology, and Biomedical Informatics (CoSBBI) Outline. Molecular Biology of Cancer AND. Goals/Expectations. David Boone 7/1/2015
 Goals/Expectations Computer Science, Biology, and Biomedical (CoSBBI) We want to excite you about the world of computer science, biology, and biomedical informatics. Experience what it is like to be a
Goals/Expectations Computer Science, Biology, and Biomedical (CoSBBI) We want to excite you about the world of computer science, biology, and biomedical informatics. Experience what it is like to be a
Experimental Psychology
 Title Experimental Psychology Type Individual Document Map Authors Aristea Theodoropoulos, Patricia Sikorski Subject Social Studies Course None Selected Grade(s) 11, 12 Location Roxbury High School Curriculum
Title Experimental Psychology Type Individual Document Map Authors Aristea Theodoropoulos, Patricia Sikorski Subject Social Studies Course None Selected Grade(s) 11, 12 Location Roxbury High School Curriculum
Current State of Visualization of EHR data: What's needed? What's next? (Session No: S50)
 Current State of Visualization of EHR data: What's needed? What's next? (Session No: S50) Panel: Danny Wu, University of Michigan Vivian West, Duke University Dawn Dowding, Columbia University Hadi Kharrazi,
Current State of Visualization of EHR data: What's needed? What's next? (Session No: S50) Panel: Danny Wu, University of Michigan Vivian West, Duke University Dawn Dowding, Columbia University Hadi Kharrazi,
Using Bayesian Networks to Analyze Expression Data. Xu Siwei, s Muhammad Ali Faisal, s Tejal Joshi, s
 Using Bayesian Networks to Analyze Expression Data Xu Siwei, s0789023 Muhammad Ali Faisal, s0677834 Tejal Joshi, s0677858 Outline Introduction Bayesian Networks Equivalence Classes Applying to Expression
Using Bayesian Networks to Analyze Expression Data Xu Siwei, s0789023 Muhammad Ali Faisal, s0677834 Tejal Joshi, s0677858 Outline Introduction Bayesian Networks Equivalence Classes Applying to Expression
LifelogExplorer: A Tool for Visual Exploration of Ambulatory Skin Conductance Measurements in Context
 LifelogExplorer: A Tool for Visual Exploration of Ambulatory Skin Conductance Measurements in Context R.D. Kocielnik 1 1 Department of Mathematics & Computer Science, Eindhoven University of Technology,
LifelogExplorer: A Tool for Visual Exploration of Ambulatory Skin Conductance Measurements in Context R.D. Kocielnik 1 1 Department of Mathematics & Computer Science, Eindhoven University of Technology,
