Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition
|
|
|
- Steven Chandler
- 6 years ago
- Views:
Transcription
1 In the format provided y the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition Chloé Béziat 1,2, Elke Barez 1,2, Mugurel I. Feraru 1, Doris Lucyshyn 1 and Jürgen Kleine-Vehn 1,2 * The phytohormone auxin induces or represses growth depending on its concentration and the underlying tissue type. However, it remains unknown how auxin signalling is modulated to allow tissues transiting etween repression and promotion of growth. Here we used apical hook development as a model for growth transitions in plants. A PIN-FORMED (PIN)-dependent intercellular auxin transport module defines an auxin maximum that is causal for growth repression during the formation of the apical hook. Our data illustrate that growth transition for apical hook opening is largely independent of this PIN module, ut requires the PIN-LIKES (PILS) putative auxin carriers at the endoplasmic reticulum. PILS proteins reduce nuclear auxin signalling in the apical hook, leading to the de-repression of growth and the onset of hook opening. We also show that the phytochrome (phy) B-reliant light-signalling pathway directly regulates PILS gene activity, therey enaling light perception to repress nuclear auxin signalling and to control growth. We propose a novel mechanism, in which PILS proteins allow external signals to alter tissue sensitivity to auxin, defining differential growth rates. T1 Department of Applied Genetics and Cell Biology, University of Natural Resources and Life Sciences (BOKU), Muthgasse 18, 119 Vienna, Austria. 2 Department of Plant Systems Biology, VIB and Department of Plant Biotechnology and Bioinformatics, Ghent University, Technologiepark 927, 952 Gent, Belgium. * juergen.kleine-vehn@oku.ac.at NATURE PLANTS DOI: 1.138/nplants Macmillan Pulishers Limited, part of Springer Nature. All rights reserved.
2 a 2 ns pin pin4-3 ns Hours after light exposure 2 1 ns pin3 pin Hours after light exposure Supplementary Figure 1. Intercellular auxin transport and hook opening. a,, Kinetics of apical hook opening during light exposure of pin3, pin4 single (a) and pin3 pin4 doule mutants () compared to seedlings (germinated in dark). Please note that pin3 pin4 doule mutants do not form closed apical hooks. For comparative reasons, the initial angle of opening was aligned (n 12). The opening kinetics (K) was statistically evaluated as descried in Supplementary Fig. 2. We did not detect statistically significant differences etween and pin mutants (ns).
3 a Y plateau K Y=(Y - Plateau)*exp(-K*X) + Plateau Y=(18-5)*exp(-K*X) Y plateau K pils2 pils Hours after light exposure constants: Y = 18 (angle during the maintenance phase) plateau= 5 (angle at full opening) variales: K=? (defines the speed of the opening) Y plateau X K Y= IF( X<X, Y, Plateau+(Y -Plateau)*exp(-K*(X-X ))) Y= IF( X<X, 18, 5+(18-5)*exp(-K*(X-X ))) 2 Y plateau 5 X 1 pils2 pils5 K 15 constants: Y = 18 (angle during the maintenance phase) plateau= 5 (angle at full opening) variales: X =? (defines the end of maintenance phase) K=? (defines the speed of opening) Supplementary Figure 2. Statistical analysis of the apical hook kinetics. a, Apical hook opening in light. Seedlings germinated in the dark were, in the early maintenance phase, exposed to light. The apical hook opening kinetics were monitored from the start of the light exposure onwards as shown in Fig. 4f and 3a as well as in Supplementary Fig.1a,;Fig. 8c and 11. Using non-linear regression, the data points were fitted to a model, representing a single phase exponential decay. The variales (K), defining the opening speed, of and pils2 pils5 seedlings have een statistically compared using an Extra sum of squares F test., Apical hook development in dark. Seedlings were kept in the dark. The apical hook developmental kinetics were monitored from 12 hours after germination onwards as shown in Fig. 2; 6a,,c as well as in Supplementary Fig. 5a,; 6d; 7; 8; 9a. Using non-linear regression, the data points were fitted to a model, representing a plateau phase, followed y a single phase exponential decay. The variales (X ) and (K), defining the end of the maintenance phase and opening speed respectively, of and pils2 pils5 seedlings have een statistically compared using an Extra sum of squares F test. Left panels depict schematic overviews of the used model and the preset constants and variales.right panels depict the apical hook kinetics of and pils2 pils5 seedlings in light (a) or in dark () along with the respective regression curves. Graphpad ( was used for all the analysis.
4 a 3 FORMATION 3 MAINTENANCE 3 OPENING Distance from the hook outer side (µm) 1 2 Distance from the hook outer side (µm) 1 2 Distance from the hook outer side (µm) Distance from the hook outer side (µm) 1 2 Distance from the hook outer side (µm) 1 2 Distance from the hook outer side (µm) Supplementary Figure 3. The activity of ppils2 and ppils5 is asymmetric during the 3 sequential phases of hook development. a, Quantification of GUS expression during formation, maintenance and opening phases in seedlings expressing ppils2::gus (a) or ppils5::gus (). The quantification was performed from the outer to the inner side of the hook along the arrow indicated in the pictures illustrating the staining in the different phases. Both lines showed increased expression towards the inner side of apical hooks. Arrow represents 15 µm.
5 DMSO NPA ppils2::gus DMSO NPA *** 2 1 *** S U pp IL S5 ::G S U S U ns pp IL S2 ::G ppils5::gus ppils3::gus 3 pp IL S3 ::G a Supplementary Figure 4. Intercellular auxin transport regulates PILS expression in the hook zone. a, Expressions patterns of 3 days dark grown seedlings expressing ppils2::gus, ppils3::gus, ppils5::gus grown on medium supplemented with NPA (5 µm) or DMSO control. Scale ar, 1 µm. The white rectangle represents the region of interest (ROI) where the quantification was performed., Quantification of mean grey values in the ROI of respective lines grown on NPA or DMSO (n=1). Two-way ANOVA followed y Bonferroni post-hoc test was performed comparing DMSO and NPA for each transgenic line ***p<.1. ns : non significant.
6 a pils2 ** pils5 ns pils2pils5 *** ** pils3 ns pils2 pils3 ns pils2 pils3 pils Supplementary Figure 5. PILS functional redundancy during apical hook opening. a,, Kinetics of apical hook development in, pils2, pils5, and pils2pils5 dark-grown seedlings (n 12) (a) and in, pils3, pils2pils3, pils2pils3pils5 dark-grown seedlings (n 12) (). Errors ars represent standard error of the mean. For the statistics X (end of maintenance phase) and K (speed of opening) were compared (see Supplementary Fig. 2) to (a) or pils2pils3pils5 (). Statistical significance is indicated y *p<,5 **p<.1 ***p<.1; ns: non significant.
7 a AT1G7652 ATG TGA 5 3 PILS3.5 k pils3-1 Promoter UTR Exon Intron c pils3-1 pils3-1 PILS3 Tuulin Expression fold change pils3-1 *** PILS3 expression d ppils3::pils3:gfp in pils ns Supplementary Figure 6. pils3 knockdown mutant characterization and complementation. a, PILS3 gene contains 8 exons. The analysed T-DNA insertion is depicted in the figure (arrow head). Scale ar, 5p., RT-PCR of PILS3 expression level in pils3-1 compared to (upper and), the expression level of the tuulin as a control gene is also presented in the same lane (lower and). Note that PILS3 expression is knocked-down in pils3-1 compared to. c, Quantitative RT-PCR of PILS3 expression in and pils3-1 normalized to EIF1A expression. Statistical significance was evaluated with the student T-test; the p value is indicated y ***p<.1. d, Kinetics of apical hook development in and ppils3::pils3:gfp in pils3 dark grown seedlings (n 12). For the statistics X (end of maintenance phase) and K (speed of opening) were compared to (see Supplementary Fig. 2) (p>,4). Errors ars represent standard error of the mean. Both parameters were statistically not significant (ns).
8 a 4h 6h PILS3OX *** pils3 *** 2 PILS3OX 15h pils h 4h 6h d pils3 4h 6h ** ** f PILS3OX 8 6 ** ** h 8 15 pdr5::rfp in PILS3OX e pdr5::gfp in pils3 1 6 c Supplementary Figure 7. PILS3 is required for apical hook development. a, Representative pictures of apical hooks in dark-grown wild type (), PILS3OX and pils3 seedlings during formation (15 hours), maintenance (4 hours) and early opening (6 hours). Scale ars, 3 µm., Kinetics of apical hook development in, PILS3OX and pils3 dark-grown seedlings (n 12). For the statistics X (end of maintenance phase) and K (speed of opening) were compared to (see Supplementary Fig. 2 ). Significant results for oth parameters are indicated ***p<.1. c, e, DR5 promoter activity in pils3 (c) as well as in PILS3OX (e) compared to dark-grown seedlings at 15h (formation), 4h (maintenance) and 6h (opening) time points after germination. Colour code (lack to green or red) depicts (low to high) pdr5rev::gfp/rfp1er signal intensity. Scale ars, 2 µm. d, f, Quantification of DR5 promoter activity in pils3 (d) as well as in PILS3OX (f) compared to dark-grown seedlings at 15, 4 and 6 hours after germination (n 5). The region of interest (ROI) was delimited to the inner side of the hook. Error ars represent standard error of the mean. Statistical significance was evaluated with two-way ANOVA followed y Bonferroni post-hoc test y comparing the 2 mutant lines at the different time point; the p value is indicated y **p<.1 PILS3OX: p35s::gfp:pils3
9 a PILS2 OX PILS3 OX PILS5 OX c PILS2 OX *** PILS2 OX *** Hours after light exposure Supplementary Figure 8. PILS2 overexpressor shows faster hook opening. a, Representative pictures of GFP signal in, PILS2 OX, PILS3 OX, PILS5 OX dark grown seedlings, Scale ar, 2 µm., Kinetic of apical hook development in and PILS2 OX dark grown seedlings (n 12). For the statistics X (end of maintenance phase) and K (speed of opening) (see Supplementary Fig. 2 ) were compared to. Statistical significance are indicated for oth parameters y ***p<.1. c, Kinetic of apical hook opening during light exposure in and PILS2 OX dark grown seedlings (n 12). For the statistics K (speed of opening) was compared to (see Supplementary Fig. 2a). Significant results are indicated ***p<.1. Error ars represent standard error of the mean. PILS2 OX : p35s::gfp:pils2; PILS3 OX : p35s::gfp:pils3; PILS5 OX : p35s::pils5:gfp
10 a 25 2 PILS5 OX *** ACC *** PILS5 OX ACC*** PILS3 OX PILS5 OX c *** *** ACC 2 1 *** ACC PILS3 OX PILS5 OX Supplementary Figure 9. Ethylene dependent hook exaggeration mediated y PIN is unchanged in PILS OX. a, Kinetics of apical hook development in and PILS5 OX grown on control medium or supplemented with ethylene precursor (ACC 5 µm). For the statistics X (end of maintenance phase) and K (speed of opening) (see Supplementary Fig. 2) were compared to. Statistical significance is indicated for oth parameters y ***p<.1., Representative pictures of, PILS3 OX, PILS5 OX 4 DAG on control medium or supplemented with ACC 5 µm. Scale ar, 15 µm. c, Apical hook angle measurement 4 DAG in the respective lines and treatments (n=25). Two-way ANOVA followed y Bonferroni post-hoc test was performed comparing the treatment and control of the respective lines. ***p<.1. PILS3 OX : p35s::gfp:pils3 PILS5 OX : p35s::pils5:gfp
11 a 1h 2h 1h 2h 3h VV VV V V ppils5::gus h h 3h ppils5::gus 3 1 ppils5::gus 2 a 1 15 c 5 Hours after light exposure Supplementary Figure 1. Light regulates PILS5 expression in a complex manner. a,, Expression patterns of ppils5::gus in apical hook maintenance phase of dark-grown seedlings or after 1, 2 or 3 hours of light exposure. Scale ars, 2 µm (a), 1 µm (). c, GUS quantification of ppils5 in dark or after 1, 2 or 3 hours of light exposure (n 8). The region of quantification was the apical hook region (). Error ars represent standard error of the mean. Statistical significance was evaluated y one way ANOVA followed y multiple comparisons Tukey s test p<,5. Distinct letters indicate statistically significant differences.
12 5h h a pils3 PILS3 OX 2 pils3*** Hours after light exposure 2 PILS3 OX *** Hours after light exposure Supplementary Figure 11. PILS3 defines growth transition in light. a, Representative pictures of apical hooks darkgrown or after eing exposed 5 hours to light in wild type (), pils3 and PILS 3O X seedlings. Scale ars, 2 µm., Kinetics of apical hook opening during light exposure in pils3 and PILS 3OX darkgrown seedlings compared to (n 12). For the statistics K (speed of opening) (see Supplementary Fig. 2a) were compared to. Statistical significance is indicated y ***p<.1. Error ars represent standard error of the mean. PILS3 OX : p35s::gfp:pils3.
13 a > ppils2 CCGAGATCAGAATTTTGGATCCGGTACCCGCCGATTCATGCTTTTTAATCTTCTTAACCAATTACGTACAGACACGAGTTA GTATGTCTGGTTTTGGGTTCCACGTGGATCTGTTTGGTTTGCGGCAGCTATTTTATTCCCACGTGCGATTACTGATCAAAT CTCAAGCCAAGCGTTCCCTTTTGTCGTCTTGTTCTGATTTAATATTAAACCGGAAATGACCGATCATTCAGCCGCTAGTGT TTGGTTTGTATATTTTGACGCATTAATGGGCTTATAATTGGGCTTCTTTTTTAGACTCGTGTTGTCGCTAGCCCAATGGAT AATCTTTGACAGTATCGTTCAACCACAGATTATCTTCTTCGCACGATGTGATAAATACAAAATTTTCAAATC > ppils3: TTCCAATAGTACAAATCAAGATTTTCAATATATGTCAAGTTTGTTATTTTCATTCACGGACATTAAAACGAGTGGAGCATT TCAATTTGACATATCTTTGTCCTTTTCTTTTTGTTACAGATATCTTTTGTTCACGTTCTTTGAATCATCATATTCATAGCT ATGAGATCATAGAAATATCAAATACTGTTAATTAACCTCTTTAAAAATACATATTCACAAGTCTCTTGTGTGAGTAGAAAT CCTATACAAACAAGTTTATTCATGCTCTAATAAGATAGATAAATAAGAAGATAGCTCATGACATGGCCTTCATTAGGCTTG TTGCATATACAATAAGCTTACATATAAGGAATTAAGGATGGTCTAATATCTCTTGCTTCACATGAGAAGAGTTATAGGATT CGAGTCACATAAGAGATTTTTATTGTAGTGTGTATAATATATCCCACTTCTACAATTATCCAGTCTATTGTGTATTTTTCA TTAATAAATCACTAAAATTTGCACTTTGACAATATATACTTAAATATTTGTATCTCGTATATATATATATACATATTTTTA GCACTGTATGATAATTCTAATTATTCGCTAATTGTTTTTTTTTGGTAAGAATTAGTCGACTATTTTGATATTGGGGTTGTT ACATTTATATCTTGCATTGAATTTTATGGACGGACACTGGTCGCAGCCTCACACACTAATTTGAATTTTCCGTTATCATTA ATATTCGATCGTTTCGTTGGTACGCTTCCATCACACAATTAATTTAAGCCACCACACTATTATCTTATTCTCATTCAGTTT TGTTTCCATTTCAAATTTCTTACGACGCATATGAAAATGTACATTTTTATAACAATTCCCCTTAAAAATAGATGACAAAAA GAAAAATGTTTTTTTTAAAAAAATATATAAAATAAAATAAATAATAAATCTTCAACAATCGATGAACACTTTGCAATATCT CCACACCTTGGGCAAGAACCAGAAAAAAACATGGGGCACGTTTCGTAGTCGATGTGACGAAAGGCATGACGACGGTTACCA TATAAATTCGTCTTCTCAGATAAAGCCTTATTATTCTCAATAGTGTTATACGACAATATACGAAACAAACAAAGACCCCGC TTGAGTTGAGTTTTCCAAAAATTGTGGAACTGTTTCCTCTTTCTTTTTGTACATAAACTAGGTTACAAAAAATTGGAAAAT ATACAAAATCAAAGGACATAAACTTACGAGATATATCATAACTCATAAGTTGTGAAAGTTGGAATTCAAAAAGGAGCTAAC TATGAATCTATGATACGTTATTTTGTTGCTTATACGGAACAAAACAATTCCAAAAACAAAGAAGATGTAAAAGTCTGACCC GGATCATGCGGACACCAAGTTGTACATACAAAATATGTAGGATGAAAATATGAAATTCTCTTCCCATATGCATAGAATAAT AATTTACCTTAGTTAAAAAGATTAGTTTCTAAGTTTGCCCCAAATAAAGAGATTAGTTTCTTGAAAAAAGCATTTT >ppils5: ACACCTACACGATGACACCTACACGATTACACTGTTGCAGGCTGCTCCCACTACAAGCATGCTTACTCTAGAT TATAGCTTAGTTTTCCAAGTAAACAAAAGCAACTAGATTCAAACCAACTTCTTTCACATGTAACTACTTGTTC TCTCTTTGACTTCTTCAGTATGGTTTGAAAGCAAAATAGGCATTATTCAACAGTCTGCTTTCTAATTAATCCT TTGAGATTGCAGTTTACGAAAATTATCCAAGGGATTCATAATTTTAGTTGAAGATATAAAGCAATTTCCTTAG CAGATTACAATTACCTTTAGCTGTTACTTGTCTGTTTAGCTTCGATTGGTCTAGTAAACAGTTTTGGTTTGGA TTAATCATATTTCTTATTCTCTGTTACCGTTTTGTTGGATCTCAAATTTCTTTGATGATTCTAGCAAGTGGTT GAGTTTAATTGATCTGTATTTTATCCATTAAGCTGTTTTGCAGTAGCAGGTGAAAAACAAACGAATTCACTGT GTTGAATTTCATTCACCGGAATAATCAGAGGCTACGACGGCGAAGGATGACGATGGAAATCTTTTGATTACTC ATCATACACGTGTTGATAACATGTGGCTTTGAAAGGAATTATCTGATCAAATCTTCTTCTTCAATATTGAGTT TGAGTTGGGATTTAGTTTGTTTTCGTAACTGGTTATGTTGTAGGGCTTTTTTAGCCCATGTTTGTAATGCTTA CCTCAATAAAATATGAACTTGTGAGAAAAAAAAAAAGATTTGGGTTTTAGTTAAAAGAGGAGAAAGAAATGTT TAAAATGATGAGCTAAATTTTAAATAAGTTGTAAGATTAAGCAACAAATGAGACTTGTAGAAATTGCAAAAGT TGGGACCTAAATCTTGAGAAGCCACTAGAATAAGGACCTTATTTTATTAATGTTGTGGCCCTTGAGAAAAATA ATATTGGGTAAGAAAATAAACAACTTATAGTTTTCAAAAACACAAAAACACAAAACCCTCAATAGTTGGAAGA GGGCTCCCCAAGACCGTATTCGCGTCCAGAATCGTAACATTTTCGATAAGATTAAAAAAACCTGCAAAATCTG GTCCGCACGTTTTTAGCAACACAACAAAGTATGACGAGAAGTTCCACGATTTTTGCAGGTATAGTACATTCGC TTTTCGCCACCTCATTGCCTTGGATTCATACACTGGATCGGTTTAATCTTCTTCTCTGTTTTTCCATGTCAAC TATTATATAGTCATTGTATTTGTATTTATTTTCTATATATTGCACCGAAAAAAGCAAGAGAGGGATATGGTTC ACATGAACTATGTAGGAGGTGAATAAATTAGACACATGTATTCTTAATTTCAGGATCACATGAATTGTGATAG TTCGGTGCTCAGTAGAACTTATAAAAAAGCACAACACTTATTCTTTTTGTCGGATATGATAATCCGCTTTTGA TTTTTTTTAAATATATGGAGTTTTCGATAGTCCAAAAAAAAGTTTCATGCTTAGGCAGACAAAAACTTTAATC TTTGTGTGTGGACGACATATTTGTTTTAACTTGCTAGTGAATTTTTCTTTAATCTACCTTTGTTTTGTATATA TATCTAACCATTATAATTATGTATATGTTAGCTGATTCCAAACGATATCATAAGTTCTAATGAGTAATGATTG GTCAATTTATCTTGTTGTAAATTGTTCAATATACGTGACGTGGTCCACTTCTATCCTCCAAGACTCCAGCTAC ACAAAGACACAGTAACACAAACGCATACGCATATGCATGTGTCGTGTCTTTGTGTGTGTATGTTGTATATATT AATATATTATCTAAACTATACCATTTTCATGAAATTTATAATATATTTTCTCAAATGATTTAAGAAGATAATG ATATGTTTACGAGAGTAAAAAAATTAATGATCTTCCAGTAATCAATAAATATAGTAAATTGATATCAGATCTT GTTGCCAACTATAGAATTGAACAAAATACAAAACTGTCATCGCCATGTTATATTATCGTAAAGC putative PIF5 inding sites predicted y Supplementary Figure 12. Predicted PIF5 inding motifs in PILS2, PILS3 and PILS5 promoters. a, G oxes predicted in silico positions are depicted in red letters in PILS2, PILS3 and PILS5 promoters sequences. The underlined sites were considered for ChIP. Proaility of inding was in silico defined for ppils2 (p>95%), as well as for ppils3 and ppils5 (p>85%).
14 Supplementary Tale 1. Primers used in this study. Genotyping primers pils2-2 FW: ATTGCTCAAGGTGAATCCAT pils2-2 RW: AGACCAATCACGGTTAAACA Salk_LB1-3:ATTTTGCCGATTTCGGAAC pils5-2 FW: CCCTTGTTTGGATCATGGTA pils5-2 RW: TCTTACTGCACCGAAAATGA; Salk_LB1-3:ATTTTGCCGATTTCGGAAC pils3-1 FW: AAAGGCATGACGACGGTTAC pils3-1 RW: AAGAGCGTCTCCAAAATTTGG; Salk_LB1-3: ATTTTGCCGATTTCGGAAC pif5 FW: CCATCCATTCAGAGGCTTC RW: ATCGTTCGTGGCTTCTCAG; Salk_LB1-3:ATTTTGCCGATTTCGGAAC RT PCR primers PILS3 FW: AAGCTTTTGGAGCTGTTCA PILS3 RW: CTAAGCTACAAGCCACATGA Tuulin FW: ACTCGTTGGGAGGAGGAACT Tuulin RW: ACACCAGACATAGTAGCAGAAATCAAG q PCR primers PILS3 FW: AGGCGACCATGCAAGTGTTG PILS3 RW: GTGGTACAGCTAGATGACAGTGAG EIF1A FW: CTGGAGGTTTTGAGGCTGGTAT EIF1A RW: CCAAGGGTGAAAGCAAGAAGA Primers used for Chromatin immunoprecipitation PILS2 G ox FW: GTTTGGTTTGCGGCAGCTAT RW: CTAGCGGCTGAATGATCGGT PILS3 G ox FW: AGTCTCTTGTGTGAGTAGAAATCCT RW: CAACAAGCCTAATGAAGGCCA WAG2 G ox FW: GGCCCACAGATATGTGATTAG RW: TCAAAATATTATGAGGGATTCGC FT 3 UTR FW: CATTTTATGATACGAGTAACGAACGGTG RW: CACTCTCATTTTCCTCCCCCTCTC Cloning primers
15 ppils1-attb4_fw: GGGGACAACTTTGTATAGAAAAGTTGCGGATTCAAGCTGATGCAATGCCTG ppils1-attb1_rw: GGGGACTGCTTTTTTGTACAAACTTGCCTTCGCTTTCAATCTGATACC ppils3-attb4_fw: GGGGACAACTTTGTATAGAAAAGTTGCGGAAGCTAATTCTCTGAGACATAGC ppils3-attb1_rw: GGGGACTGCTTTTTTGTACAAACTTGCCTTTTTACTATCAACGCGAGAATC ppils4-attb4_fw: GGGGACAACTTTGTATAGAAAAGTTGCGGTGAAGCTTTTGGAGCTGTTCAT ppils4_attb1_rw: GGGGACTGCTTTTTTGTACAAACTTGCCTCTACTTCCAAAAGTTAGAATC ppils6_attb4_fw: GGGGACAACTTTGTATAGAAAAGTTGCGCCATCGAGAGATTTAATTTGATC ppils6_attb1_rw: GGGGACTGCTTTTTTGTACAAACTTGCTCTGAGAATTTTGAGGGTGGAC ppils7_attb4_fw: GGGGACAACTTTGTATAGAAAAGTTGCGCTGCGGACAACAAAAAAGTGTTAG ppils7_attb1_rw: GGGGACTGCTTTTTTGTACAAACTTGCCCCTAATCTCTTCTTTTTCTTTTCT
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figures
 Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
A bts-1 (SALK_016526)
 Electronic Supplementary Material (ESI) for Metallomics. This journal is The Royal Society of Chemistry 217 A ts-1 (SALK_16526) BTS (At3g1829) ATG tsl1 (SALK_1554) 2 p BTSL1 (At1g7477) ATG tsl2 (SAIL_615_HO1)
Electronic Supplementary Material (ESI) for Metallomics. This journal is The Royal Society of Chemistry 217 A ts-1 (SALK_16526) BTS (At3g1829) ATG tsl1 (SALK_1554) 2 p BTSL1 (At1g7477) ATG tsl2 (SAIL_615_HO1)
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM.
 Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
IP: anti-gfp VPS29-GFP. IP: anti-vps26. IP: anti-gfp - + +
 FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
Neocortex Zbtb20 / NFIA / Sox9
 Neocortex / NFIA / Sox9 Supplementary Figure 1. Expression of, NFIA, and Sox9 in the mouse neocortex at. The lower panels are higher magnification views of the oxed area. Arrowheads indicate triple-positive
Neocortex / NFIA / Sox9 Supplementary Figure 1. Expression of, NFIA, and Sox9 in the mouse neocortex at. The lower panels are higher magnification views of the oxed area. Arrowheads indicate triple-positive
Supplemental Data. Müller-Xing et al. (2014). Plant Cell /tpc
 Supplemental Figure 1. Phenotypes of iclf (clf-28 swn-7 CLF pro :CLF-GR) plants. A, Late rescue of iclf plants by renewed DEX treatment; senescent inflorescence with elongated siliques (arrow; 90 DAG,
Supplemental Figure 1. Phenotypes of iclf (clf-28 swn-7 CLF pro :CLF-GR) plants. A, Late rescue of iclf plants by renewed DEX treatment; senescent inflorescence with elongated siliques (arrow; 90 DAG,
Supplemental Data. Wang et al. (2013). Plant Cell /tpc
 Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
EGFR shrna A: CCGGCGCAAGTGTAAGAAGTGCGAACTCGAGTTCGCACTTCTTACACTTGCG TTTTTG. EGFR shrna B: CCGGAGAATGTGGAATACCTAAGGCTCGAGCCTTAGGTATTCCACATTCTCTT TTTG
 Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Supplementary Information
 1 Supplementary Information A role for primary cilia in glutamatergic synaptic integration of adult-orn neurons Natsuko Kumamoto 1,4,5, Yan Gu 1,4, Jia Wang 1,4, Stephen Janoschka 1,2, Ken-Ichi Takemaru
1 Supplementary Information A role for primary cilia in glutamatergic synaptic integration of adult-orn neurons Natsuko Kumamoto 1,4,5, Yan Gu 1,4, Jia Wang 1,4, Stephen Janoschka 1,2, Ken-Ichi Takemaru
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
Supplemental Information. Figures. Figure S1
 Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
Supporting Information
 Supporting Information Lee et al. 10.1073/pnas.0910950106 Fig. S1. Fe (A), Zn (B), Cu (C), and Mn (D) concentrations in flag leaves from WT, osnas3-1, and OsNAS3-antisense (AN-2) plants. Each measurement
Supporting Information Lee et al. 10.1073/pnas.0910950106 Fig. S1. Fe (A), Zn (B), Cu (C), and Mn (D) concentrations in flag leaves from WT, osnas3-1, and OsNAS3-antisense (AN-2) plants. Each measurement
Supplementary Information. Cofilin Regulates Nuclear Architecture through a Myosin-II Dependent Mechanotransduction Module
 Supplementary Information Cofilin Regulates Nuclear Architecture through a Myosin-II Dependent Mechanotransduction Module O Neil Wiggan, Bryce Schroder, Diego Krapf, James R. Bamurg and Jennifer G. DeLuca
Supplementary Information Cofilin Regulates Nuclear Architecture through a Myosin-II Dependent Mechanotransduction Module O Neil Wiggan, Bryce Schroder, Diego Krapf, James R. Bamurg and Jennifer G. DeLuca
a 0,8 Figure S1 8 h 12 h y = 0,036x + 0,2115 y = 0,0366x + 0,206 Labeling index Labeling index ctrl shrna Time (h) Time (h) ctrl shrna S G2 M G1
 (GFP+ BrdU+)/GFP+ Labeling index Labeling index Figure S a, b, y =,x +, y =,x +,,,,,,,, Time (h) - - Time (h) c d S G M G h M G S G M G S G h Time of BrdU injection after electroporation (h) M G S G M
(GFP+ BrdU+)/GFP+ Labeling index Labeling index Figure S a, b, y =,x +, y =,x +,,,,,,,, Time (h) - - Time (h) c d S G M G h M G S G M G S G h Time of BrdU injection after electroporation (h) M G S G M
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
Supplementary Table 1. List of primers used in this study
 Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplemental Information. Spatial Auxin Signaling. Controls Leaf Flattening in Arabidopsis
 Current Biology, Volume 27 Supplemental Information Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis Chunmei Guan, Binbin Wu, Ting Yu, Qingqing Wang, Naden T. Krogan, Xigang Liu, and Yuling
Current Biology, Volume 27 Supplemental Information Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis Chunmei Guan, Binbin Wu, Ting Yu, Qingqing Wang, Naden T. Krogan, Xigang Liu, and Yuling
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
Supplementary information. The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic. Spindle Orientation
 Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplemental Data. Shin et al. Plant Cell. (2012) /tpc YFP N
 MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Data. Di Giorgio et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. Synteny analysis of NIP4;1 and NIP4;2. Examination of the NIP4;1-NIP4;2 region in rabidopsis thaliana and equivalent evolutionary regions in other dicot genomes are shown on the
Supplemental Figure 1. Synteny analysis of NIP4;1 and NIP4;2. Examination of the NIP4;1-NIP4;2 region in rabidopsis thaliana and equivalent evolutionary regions in other dicot genomes are shown on the
Supplementary Information
 Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
tom tom 24hpf tom tom 48hpf tom 60hpf tom tom 72hpf tom
 a 24hpf c 48hpf d e 60hpf f g 72hpf h i j k ISV ISV Figure 1. Vascular integrity defects and endothelial regression in mutant emryos. (a,c,e,g,i) Bright-field and (,d,f,h,j) corresponding fluorescent micrographs
a 24hpf c 48hpf d e 60hpf f g 72hpf h i j k ISV ISV Figure 1. Vascular integrity defects and endothelial regression in mutant emryos. (a,c,e,g,i) Bright-field and (,d,f,h,j) corresponding fluorescent micrographs
Supplemental Data. Deinlein et al. Plant Cell. (2012) /tpc
 µm Zn 2+ 15 µm Zn 2+ Growth (% of control) empty vector NS1 NS2 NS3 NS4 S. pombe zhfδ Supplemental Figure 1. Functional characterization of. halleri NS genes in Zn 2+ hypersensitive S. pombe Δzhf mutant
µm Zn 2+ 15 µm Zn 2+ Growth (% of control) empty vector NS1 NS2 NS3 NS4 S. pombe zhfδ Supplemental Figure 1. Functional characterization of. halleri NS genes in Zn 2+ hypersensitive S. pombe Δzhf mutant
Table S1. Total and mapped reads produced for each ChIP-seq sample
 Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Nature Immunology: doi: /ni eee Supplementary Figure 1
 eee Supplementary Figure 1 Hyphae induce NET release, but yeast do not. (a) NET release by human peripheral neutrophils stimulated with a hgc1 yeast-locked C. albicans mutant (yeast) or pre-formed WT C.
eee Supplementary Figure 1 Hyphae induce NET release, but yeast do not. (a) NET release by human peripheral neutrophils stimulated with a hgc1 yeast-locked C. albicans mutant (yeast) or pre-formed WT C.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples.
 Supplementary files Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples. Table S3. Specificity of AGO1- and AGO4-preferred 24-nt
Supplementary files Table S1. Relative abundance of AGO1/4 proteins in different organs. Table S2. Summary of smrna datasets from various samples. Table S3. Specificity of AGO1- and AGO4-preferred 24-nt
Table S1. Primer sequences used for qrt-pcr. CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT ACTB AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT LCOR
 Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis
 Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
BIOL2005 WORKSHEET 2008
 BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table.
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tale. Type of file: VI Title of file for HTML: Supplementary Movie 1 Description:
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tale. Type of file: VI Title of file for HTML: Supplementary Movie 1 Description:
Conditional and reversible disruption of essential herpesvirus protein functions
 nature methods Conditional and reversible disruption of essential herpesvirus protein functions Mandy Glaß, Andreas Busche, Karen Wagner, Martin Messerle & Eva Maria Borst Supplementary figures and text:
nature methods Conditional and reversible disruption of essential herpesvirus protein functions Mandy Glaß, Andreas Busche, Karen Wagner, Martin Messerle & Eva Maria Borst Supplementary figures and text:
Bone Marrow Pop. (% Total) Mature Pool (Absolute %) Immature Pool (Absolute %) A10 EC Control A10 EC Control A10 EC Control
 Bone Marrow Pop. (% Total) Mature Pool (Asolute %) Immature Pool (Asolute %) A10 EC A10 EC A10 EC Myeloid 50.7 57.5 37.5 46.2 13.2 11.3 Erythroid 38.3 23.2 33.3 16.8 9.3 6.3 Lymphocytes 13.8 19.0 - - -
Bone Marrow Pop. (% Total) Mature Pool (Asolute %) Immature Pool (Asolute %) A10 EC A10 EC A10 EC Myeloid 50.7 57.5 37.5 46.2 13.2 11.3 Erythroid 38.3 23.2 33.3 16.8 9.3 6.3 Lymphocytes 13.8 19.0 - - -
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplemental Figure S1. Sequence feature and phylogenetic analysis of GmZF351. (A) Amino acid sequence alignment of GmZF351, AtTZF1, SOMNUS, AtTZF5,
 Supplemental Figure S. Sequence feature and phylogenetic analysis of GmZF35. (A) Amino acid sequence alignment of GmZF35, AtTZF, SOMNUS, AtTZF5, and OsTZF. Characters with black background indicate conserved
Supplemental Figure S. Sequence feature and phylogenetic analysis of GmZF35. (A) Amino acid sequence alignment of GmZF35, AtTZF, SOMNUS, AtTZF5, and OsTZF. Characters with black background indicate conserved
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells
 Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplemental Data. Beck et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
Figure legends of supplementary figures
 Figure legends of supplementary figures Figure 1. Phenotypic analysis of rice early flowering1 () plants and enhanced expression of floral identity genes in.. Leaf emergence of,, and plants with complementary
Figure legends of supplementary figures Figure 1. Phenotypic analysis of rice early flowering1 () plants and enhanced expression of floral identity genes in.. Leaf emergence of,, and plants with complementary
293T cells were transfected with indicated expression vectors and the whole-cell extracts were subjected
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
Supplemental Data. Wu et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. FIM5 is preferentially expressed in stamen and mature pollen. The expression data of FIM5 was extracted from Arabidopsis efp browser (http://www.bar.utoronto.ca/efp/development/),
Supplemental Figure 1. FIM5 is preferentially expressed in stamen and mature pollen. The expression data of FIM5 was extracted from Arabidopsis efp browser (http://www.bar.utoronto.ca/efp/development/),
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Table I: PHT1 transporter family comparison at the amino acid level. BLAST program was used to obtain percentage of similarity and identity (in bold).
 Supplemental Tables Table I: PHT1 transporter family comparison at the amino acid level. BLAST program was used to obtain percentage of similarity and identity (in bold). PHT1;1 PHT1;2 PHT1;3 PHT1;4 PHT1;5
Supplemental Tables Table I: PHT1 transporter family comparison at the amino acid level. BLAST program was used to obtain percentage of similarity and identity (in bold). PHT1;1 PHT1;2 PHT1;3 PHT1;4 PHT1;5
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Bidirectional optogenetic modulation of the tonic activity of CEA PKCδ + neurons in vitro. a, Top, Cell-attached voltage recording illustrating the blue light-induced increase in
Supplementary Figure 1 Bidirectional optogenetic modulation of the tonic activity of CEA PKCδ + neurons in vitro. a, Top, Cell-attached voltage recording illustrating the blue light-induced increase in
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Quantification of myelin fragments in the aging brain (a) Electron microscopy on corpus callosum is shown for a 18-month-old wild type mice. Myelin fragments (arrows) were detected
Supplementary Figure 1 Quantification of myelin fragments in the aging brain (a) Electron microscopy on corpus callosum is shown for a 18-month-old wild type mice. Myelin fragments (arrows) were detected
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Behavioural effects of ketamine in non-stressed and stressed mice. Naive C57BL/6 adult male mice (n=10/group) were given a single dose of saline vehicle or ketamine (3.0 mg/kg,
Supplementary Figure 1. Behavioural effects of ketamine in non-stressed and stressed mice. Naive C57BL/6 adult male mice (n=10/group) were given a single dose of saline vehicle or ketamine (3.0 mg/kg,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
APOLs with low ph dependence can kill all African trypanosomes
 SUPPLEMENTARY INFORMATION Letters DOI: 1.138/s41564-17-34-1 In the format provided by the authors and unedited. APOLs with low ph dependence can kill all African trypanosomes Frédéric Fontaine 1, Laurence
SUPPLEMENTARY INFORMATION Letters DOI: 1.138/s41564-17-34-1 In the format provided by the authors and unedited. APOLs with low ph dependence can kill all African trypanosomes Frédéric Fontaine 1, Laurence
TGF-β Signaling Regulates Neuronal C1q Expression and Developmental Synaptic Refinement
 Supplementary Information Title: TGF-β Signaling Regulates Neuronal C1q Expression and Developmental Synaptic Refinement Authors: Allison R. Bialas and Beth Stevens Supplemental Figure 1. In vitro characterization
Supplementary Information Title: TGF-β Signaling Regulates Neuronal C1q Expression and Developmental Synaptic Refinement Authors: Allison R. Bialas and Beth Stevens Supplemental Figure 1. In vitro characterization
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained
 Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
Supplementary Figure 1
 Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
SUPPLEMENTARY RESULTS
 SUPPLEMENTARY RESULTS Supplementary Table 1. hfpr1- Flpln-CHO hfpr2-flpln-cho pec 50 E max (%) Log( /K A) Log( /K A) N pec 50 E max (%) Log( /K A) Log( /K A) n ERK1/2 phosphorylation fmlp 9.0±0.6 80±7
SUPPLEMENTARY RESULTS Supplementary Table 1. hfpr1- Flpln-CHO hfpr2-flpln-cho pec 50 E max (%) Log( /K A) Log( /K A) N pec 50 E max (%) Log( /K A) Log( /K A) n ERK1/2 phosphorylation fmlp 9.0±0.6 80±7
Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation.
 ` Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation. (a) Overexpression of hlrrk2 in HeLa cells enhances total protein synthesis in [35S] methionine/cysteine incorporation assays.
` Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation. (a) Overexpression of hlrrk2 in HeLa cells enhances total protein synthesis in [35S] methionine/cysteine incorporation assays.
Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 )
 770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
Supplementary Figure 1. Baf60c and baf180 are induced during cardiac regeneration in zebrafish. RNA in situ hybridization was performed on paraffin
 Supplementary Figure 1. Baf60c and baf180 are induced during cardiac regeneration in zebrafish. RNA in situ hybridization was performed on paraffin sections from sham-operated adult hearts (a and i) and
Supplementary Figure 1. Baf60c and baf180 are induced during cardiac regeneration in zebrafish. RNA in situ hybridization was performed on paraffin sections from sham-operated adult hearts (a and i) and
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
Supplementary Information
 Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Divergent effects of intrinsically active MEK variants on developmental Ras signaling Yogesh Goyal,2,3,4, Granton A. Jindal,2,3,4, José L. Pelliccia 3, Kei Yamaya 2,3, Eyan Yeung
SUPPLEMENTARY MATERIAL Divergent effects of intrinsically active MEK variants on developmental Ras signaling Yogesh Goyal,2,3,4, Granton A. Jindal,2,3,4, José L. Pelliccia 3, Kei Yamaya 2,3, Eyan Yeung
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Nature Biotechnology: doi: /nbt.3828
 Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
Nature Biotechnology: doi: /nbt Supplementary Figure 1. PL gene expression in tomato fruit.
 Supplementary Figure 1 PL gene expression in tomato fruit. Relative expression of five PL-coding genes measured in at least three fruit of each genotype (cv. Alisa Craig) at four stages of development,
Supplementary Figure 1 PL gene expression in tomato fruit. Relative expression of five PL-coding genes measured in at least three fruit of each genotype (cv. Alisa Craig) at four stages of development,
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity.
 Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
Metabolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients
 Metaolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients Franck Chiappini, Audrey Coilly, Hanane Kadar, Philippe Gual, Alert Tran, Christophe Desterke, Didier
Metaolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients Franck Chiappini, Audrey Coilly, Hanane Kadar, Philippe Gual, Alert Tran, Christophe Desterke, Didier
Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad
 Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad libitum conditions detecting PER2 protein in brain and
Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad libitum conditions detecting PER2 protein in brain and
Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function
 ONLINE DATA SUPPLEMENTS Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function Supplementary Figures Figure S1 Effect of Ad-p27-126TS on the expression
ONLINE DATA SUPPLEMENTS Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function Supplementary Figures Figure S1 Effect of Ad-p27-126TS on the expression
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Figure S1 Expression of AHL gene family members in diploid (Ler Col) and triploid (Ler
 Supplemental material Supplemental figure legends Figure S Expression of AHL gene family members in diploid (Ler ) and triploid (Ler osd) seeds. AHLs from clade B are labelled with (I), and AHLs from clade
Supplemental material Supplemental figure legends Figure S Expression of AHL gene family members in diploid (Ler ) and triploid (Ler osd) seeds. AHLs from clade B are labelled with (I), and AHLs from clade
Supporting Information Table of Contents
 Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
Supplemental Data. Candat et al. Plant Cell (2014) /tpc Cytosol. Nucleus. Mitochondria. Plastid. Peroxisome. Endomembrane system
 Cytosol Nucleus 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 PSORT MultiLoc YLoc SubLoc BaCelLo WoLF PSORT
Cytosol Nucleus 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 PSORT MultiLoc YLoc SubLoc BaCelLo WoLF PSORT
Nature Neuroscience: doi: /nn Supplementary Figure 1. Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites.
 Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were
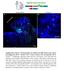 Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were electroporated with β- Catenin S33Y in PiggyBac expression
Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were electroporated with β- Catenin S33Y in PiggyBac expression
days days and gbt-i.cd Recipient 20
 gbt-i. GFP+ Resident memory cells: gbt-i.gfp+ Recruited memory cells: gbt-i.cd45.1+ 1 2-3 gbt-i. flu.gb sc. CD45.1+ Graft with gbt-i.gfp+ 1 Recipient 1 re- 3 36 Graft with gbt-i.gfp+ and gbt-i.cd45.1+
gbt-i. GFP+ Resident memory cells: gbt-i.gfp+ Recruited memory cells: gbt-i.cd45.1+ 1 2-3 gbt-i. flu.gb sc. CD45.1+ Graft with gbt-i.gfp+ 1 Recipient 1 re- 3 36 Graft with gbt-i.gfp+ and gbt-i.cd45.1+
mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons
 Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
MII. Supplement Figure 1. CapZ β2. Merge. 250ng. 500ng DIC. Merge. Journal of Cell Science Supplementary Material. GFP-CapZ β2 DNA
 A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
SUPPLEMENTARY INFORMATION
 Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Formation of the AA5x. a, Camera lucida drawing of embryo at 48 hours post fertilization (hpf, modified from Kimmel et al. Dev Dyn. 1995 203:253-310). b, Confocal microangiogram
Supplementary Figure 1. Formation of the AA5x. a, Camera lucida drawing of embryo at 48 hours post fertilization (hpf, modified from Kimmel et al. Dev Dyn. 1995 203:253-310). b, Confocal microangiogram
s -1 (A) and (B) 2 s 1 in WT, riq1, and the riq1 mutant complemented by
 A 250-2 s -1 B 250-2 s -1 Supplemental Figure 1. The NPQ Phenotype in riq Mutants Was Complemented by Introduction of RIQ Genes. (A) and (B) 2 s 1 in, riq1, and the riq1 mutant complemented by the RIQ1
A 250-2 s -1 B 250-2 s -1 Supplemental Figure 1. The NPQ Phenotype in riq Mutants Was Complemented by Introduction of RIQ Genes. (A) and (B) 2 s 1 in, riq1, and the riq1 mutant complemented by the RIQ1
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
GFP/Iba1/GFAP. Brain. Liver. Kidney. Lung. Hoechst/Iba1/TLR9!
 Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1
 Supplemental Data Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1 and kan1-11 kan2-5 double mutants. A, The numbers of hydathodes in different leaves of Col-0, as2-1 rev-1,
Supplemental Data Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1 and kan1-11 kan2-5 double mutants. A, The numbers of hydathodes in different leaves of Col-0, as2-1 rev-1,
Analysis of the peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) cistrome reveals novel co-regulatory role of ATF4
 Khozoie et al. BMC Genomics 2012, 13:665 RESEARCH ARTICLE Open Access Analysis of the peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) cistrome reveals novel co-regulatory role of ATF4 Combiz Khozoie
Khozoie et al. BMC Genomics 2012, 13:665 RESEARCH ARTICLE Open Access Analysis of the peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) cistrome reveals novel co-regulatory role of ATF4 Combiz Khozoie
* * A3027. A4623 e A3507 A3507 A3507
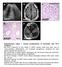 a c L A327 d e A37 A37 A37 Supplementary Figure 1. Clinical manifestations of individuals with mutations. (a) Renal ultrasound of right kidney in A327 reveals small renal cysts, loss of corticomedullary
a c L A327 d e A37 A37 A37 Supplementary Figure 1. Clinical manifestations of individuals with mutations. (a) Renal ultrasound of right kidney in A327 reveals small renal cysts, loss of corticomedullary
Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent Protein.
 prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Hippocampal sections from new-born Pten+/+ and PtenFV/FV pups were stained with haematoxylin and eosin (H&E) and were imaged at (a) low and (b) high
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Hippocampal sections from new-born Pten+/+ and PtenFV/FV pups were stained with haematoxylin and eosin (H&E) and were imaged at (a) low and (b) high
Behavioral generalization
 Supplementary Figure 1 Behavioral generalization. a. Behavioral generalization curves in four Individual sessions. Shown is the conditioned response (CR, mean ± SEM), as a function of absolute (main) or
Supplementary Figure 1 Behavioral generalization. a. Behavioral generalization curves in four Individual sessions. Shown is the conditioned response (CR, mean ± SEM), as a function of absolute (main) or
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2294 Figure S1 Localization and function of cell wall polysaccharides in root hair cells. (a) Spinning-disk confocal sections of seven day-old A. thaliana seedlings stained with 0.1% S4B
DOI: 10.1038/ncb2294 Figure S1 Localization and function of cell wall polysaccharides in root hair cells. (a) Spinning-disk confocal sections of seven day-old A. thaliana seedlings stained with 0.1% S4B
Supplemental Fig. 1. Relative mrna Expression. Relative mrna Expression WT KO WT KO RT 4 0 C
 Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative

 Supplementary Figure 1 a OD c. 1. 1... Time [min] 1 p
Supplementary Figure 1 a OD c. 1. 1... Time [min] 1 p