The increasing complexity of the ubiquitin code
|
|
|
- Megan Peters
- 6 years ago
- Views:
Transcription
1 The increasing complexity of the ubiquitin code Richard Yau,2 and Michael Rape,2 * iquitylation is essential for signal transduction as well as cell division and differentiation in all eukaryotes. modifications range from a single ubiquitin molecule to complex polymeric chains, with different types of ubiquitylation often eliciting distinct outcomes. The recent identification of novel chain topologies has improved our understanding of how ubiquitylation establishes precise communication within cells. Here, we discuss how the increasing complexity of ubiquitylation is employed to ensure robust and faithful signal transduction in eukaryotic cells. Communication allows human societies to thrive. The basis for such interactions is our ability to articulate thoughts into words and sentences that can trigger particular responses from other individuals. Similarly, precise communication between and within cells is needed to support the development or homeostasis of a complex metazoan organism. The failure of cells to translate cues from their environment into proper action, be it division, differentiation or survival, can cause many diseases a dramatic example is cancer resulting from unabated growth factor signalling. Post-translational modification with the highly conserved 76-residue protein ubiquitin provides cells with an essential mechanism to establish precise communication. iquitylation is brought about by a cascade of three enzymes : E ubiquitin activating enzyme, E2 ubiquitin conjugating enzyme, and E3 ubiquitin protein ligase 4. In the simplest version of the process, monoubiquitylation, a single ubiquitin molecule is covalently attached to the ε-amino group of a lysine residue in a target protein. This reaction can also occur on multiple lysine residues of the substrate to yield a modification referred to as multi-monoubiquitylation (Fig. a). The very efficient monoubiquitylation of histone H2A, which is mediated by the Polycomb repressive complex and can account for up to 0% of the total histone H2A population, allowed the discovery of the first substrate of the ubiquitin pathway 5. Adding single ubiquitin subunits typically alters intra- or intermolecular interactions that in turn affect localization, complex formation or activity of the modified protein 6 (Fig. b). For example, monoubiquitylation mediates an interaction of the DNA polymerase processivity factor PCNA with translesion synthesis DNA polymerases 7,8, whereas the same modification on the SMAD4 transcription factor prevents it from binding its signalling partner SMAD2 (ref. 9). iquitin contains seven lysine residues that, together with its amino terminus, provide eight attachment sites for further ubiquitin molecules, thereby allowing the formation of polymeric chains (Fig. 2a). In homotypic ubiquitin conjugates, all building blocks of the chain are connected through the same lysine or methionine residue (Fig. 2b,c). These conjugates adopt distinct but often very dynamic conformational ensembles that can be recognized by effector proteins with linkagespecific ubiquitin-binding domains (UBDs) 0,. Recognition by UBDs couples the modification of a substrate to a downstream event, such as protein turnover, recruitment to a specific location or assembly of a signalling complex. Thus, similarly to words or sentences that elicit actions, s can encode information that is deciphered by UBDs to trigger specific biological outcomes. Providing the classic example for signalling through homotypic ubiquitin conjugates, K-linked chains are able to target proteins to the 26S proteasome for degradation (Fig. 2b). K linkages are the most abundant connection between ubiquitin molecules in cells, and their levels rapidly increase following proteasome inhibition 2. K-linked chains were originally discovered to promote the proteasomal turnover of short-lived model substrates 3, and they are now known to be the product of several E3 ligases associated with degradation, including the cell-cycle-associated E3 ligase SCF (ref. 4), the ER-associated degradation (ERAD) E3 gp78 (ref. 5), or E6-AP, an E3 that targets the tumour suppressor p53 for degradation following viral infection 6. Conversely, removal of K-linked chains from substrates by deubiquitylases (DUBs) often prevents degradation 7,8. Consistent with these observations in cells, K-linked chains efficiently trigger proteasomal degradation in purified systems 9,20. However, even for this paradigmatic type, the substrate can sometimes dictate an alternative fate: for example, attachment of K-linked chains to the yeast M4 protein inhibits its function as a transcription factor, but does not deliver the protein for proteasomal degradation 2. Following early work on K-linked chains, mutation of an additional residue, K, was found to impede formation of polymeric ubiquitin conjugates 22. Rather than triggering degradation, K-linked chains serve as molecular glue that allows for rapid and reversible formation of pivotal signalling complexes (Fig. 2c). K linkages are now known to regulate activation of the NF-κB transcription factor 23 25, DNA repair 7,26,27, innate immune responses 28, clearance of damaged Howard Hughes Medical Institute, University of California, Berkeley, California 94720, USA. 2 Department of Molecular and Cell Biology, University of California, Berkeley, California 94720, USA. * mrape@berkeley.edu NATURE CELL BIOLOGY VOLUME 8 NUMBER 6 JUNE
2 b a Monoubiquitylation mitochondria 29,30, and protein sorting 3,32 and they can also guide assembly of large protein complexes that drive mrna splicing 33 or translation 34,35. Reminiscent of second messengers in signalling cascades, K-linked chains are able to perform their functions even if they are not attached to substrate proteins 36,37. In agreement with these observations, E2s, E3s and UBDs involved in signal transduction, rather than protein degradation, produce and read K-linked conjugates,38,39, whereas K-specific DUBs terminate signalling events brought about by this chain topology 40,4. A series of proteomic and immunological approaches revealed that s composed of linkages involving the remaining five lysine residues as well as the α-amino group of the first methionine (M) of ubiquitin also exist in cells 2, Depending on the cellular context, several of these atypical chains are able to rival in abundance the prototypical K or K linkages 2,44,45. Accordingly, enzymes that can produce or disassemble such topologies, frequently with astonishing specificity, have been discovered 7,29,46,47. Recent progress in understanding how atypical chains are assembled and recognized has revealed a ubiquitin code that is much more complex than originally anticipated. We have learned new words in the ubiquitin signalling vocabulary, how they can be combined and how ubiquitin signals can be modified to provoke particular responses in the cell. Rather than being a simple code that assigns one specific output to each signal, ubiquitylation has emerged as a cellular language in which information transfer depends both on the nature of the signal and on the context in which it is sent. In this Review, we highlight these developments that have refined our appreciation of ubiquitin-dependent signalling as an essential communication module in eukaryotic cells. New words in the ubiquitin vocabulary The first atypical conjugates with assigned roles in signalling were K- and M-linked s (also referred to as linear chains). K- linked chains are abundant in asynchronously growing yeast, whereas in human cells they are preferentially produced during mitosis and early G 25,45,. Conversely, M-linked conjugates are rapidly synthesized Multimonoubiquitylation Figure Signalling by monoubiquitylation. (a) A schematic representation of monoubiquitylation and multi-monoubiquitylation. (b) Monoubiquitylation can recruit effector proteins to elicit a biological outcome (top), as illustrated with PCNA 7,8. Monoubiquitylation can also prevent protein interactions, requiring removal of the ubiquitin tag for signalling to occur (bottom). An example is provided by SMAD4, which depends on deubiquitylation to engage its signalling partner SMAD2 9. following activation of inflammatory signalling cascades 46,49 5. M linkages are also observed on pathogenic bacteria that have escaped vacuolar compartments and face ubiquitin-dependent autophagy in the cytoplasm 52,53. Both chain types are produced by linkage-specific enzymes: K-linked chains depend on an essential E3, the anaphasepromoting complex (APC/C) and its specific chain-elongating E2, e2s 47,54 58, whereas M-linked chains are the product of LUBAC, a heterotrimeric RBR-family E3 ligase that is composed of HOIP, HOIL-L and Sharpin 46,49 5,59. Each chain type can also be disassembled by OTUfamily DUBs with strong linkage preference: the K-specific Cezanne and the M-specific OTULIN 7,60,6. Interestingly, LUBAC interacts with its antagonist OTULIN, underscoring the dynamic nature and tight regulation of ubiquitin-dependent signaling Consistent with the production of K-linked chains during cell division, these conjugates target cell cycle regulators for degradation 20,47,65,66 (Fig. 2b). K linkages are recognized by the proteasome,67, and they increase in abundance in response to proteasome inhibition 2,42. In biophysical studies using proteasomes stripped of substrate delivery factors, single K-linked chains trigger proteasome binding and substrate degradation almost as efficiently as K-linked chains 20. K linkages are also found in conjugates that mediate cell-cycle-independent functions, including regulation of the Hifα transcription factor 68, ERAD in yeast 2, or the innate anti-viral immune response 69. As K-specific ubiquitin-binding domains have not yet been discovered, the molecular function of these chains in cell-cycle-independent reactions remains poorly understood. M-linked conjugates are recognized by linkage-specific UBDs, such as the UBAN domain 70,7, which couple modification of a substrate to integration into larger protein assemblies (Fig. 2c). Adding to a growing list of cellular functions, M-linked chains play pivotal roles in immune signalling and NF-κB activation 49 5,72, regulation of interferon production 73, and the control of Wnt signalling during blood vessel formation 62. It is interesting to note that M- and K-linked chains seem to encode information similar to canonical counterparts: whereas both M- and K-linked conjugates orchestrate the assembly of protein complexes, K- and K-linked chains can drive proteasomal degradation (Fig. 2b,c). How distinct chain topologies with similar functions are employed to fine-tune the consequences of ubiquitylation will be discussed below. More recently, additional linkages have been assigned potential functions in ubiquitin-dependent signalling. K6 linkages were observed during removal of damaged mitochondria from cells 29, K27 linkages have been implicated in regulating DNA repair and autoimmunity 74,75, K29 linkages have been attributed roles in proteasomal degradation 76, and K33-linked chains were proposed to regulate trafficking through the trans-golgi network 77. In contrast to M-, K-, K- and K-linked chains, for which linkage-specific ubiquitylation enzymes are known, these alternative linkages seem to be assembled by enzymes with mixed specificity. K6 linkages mediate functions of the E3 ligase Parkin, but this E3 also connects s through K, K and K 29,78,79. The bacterial E3 NleL produces both K6 and K linkages 80. UBR5 (a tumour suppressor E3 associated with the degradation of RORγt transcription factors 8, inhibition of telomeric RNF68 (ref. 82) and restriction to DNA damage signalling 83 ) assembles K and K29 linkages, whereas a poorly studied E3, KIAA0, connects ubiquitin molecules through both K29 and K 84. Moreover, 580 NATURE CELL BIOLOGY VOLUME 8 NUMBER 6 JUNE 206
3 the HECT-family E3 APEL produces K33 linkages as well as K and K linkages 85, and CUL3, which was proposed to produce K33-linked chains 77, also often monoubiquitylates substrates 86,87. This broader specificity of E3s is reflected by their opposing DUBs, which target multiple atypical linkages with similar efficiency. TRABID, for example, preferentially cleaves both K29 and K33 linkages 85,88 90 ; OTUD3, a DUB that has been implicated in stabilizing the tumour suppressor PTEN, takes on K6 and K linkages 7,9 ; and OTUD2, which binds the ubiquitin-selective segregase p97 (also known as Cdc or VCP, valosin-containing protein) and probably acts in ERAD 92, targets K, K27, K29 and K33 linkages 7. These observations indicate that atypical linkages between ubiquitin molecules might frequently be incorporated into heterotypic (rather than homotypic) conjugates, a notion that was recently supported by experimental evidence: both K6 and K linkages contribute to Parkin-dependent removal of damaged mitochondria 29,30,93,94, and both K and K linkages are involved in the efficient endocytosis of MHC class I molecules 95. Moreover, K29 linkages are incorporated into chains that also contain ubiquitin molecules connected through K 90. Combining signals with mixed and branched s In contrast to homotypic chains, where ubiquitin subunits are connected by a single linkage type, heterotypic conjugates contain multiple linkages and typically come in two types: mixed and branched. Mixed chains are composed of different linkages, but each ubiquitin is modified with only one other ubiquitin molecule (Fig. 3a). If, instead, a single ubiquitin subunit within a chain is modified with two or more ubiquitin molecules at a time, branched chain structures are generated (Fig. 3b). A cellular function for heterotypic chains was already suggested in studies of the yeast ubiquitin-fusion degradation (UFD) pathway, a system that has revealed many features of ubiquitin-dependent signalling 76. In model substrates of the UFD pathway, a ubiquitin molecule fused to the amino terminus of various proteins serves as the site of chain formation by two enzymes, the HECT E3 UFD4 and the U-box-containing UFD2 76,96. Whereas UFD4 preferentially assembles K29-linked chains 97, UFD2 connects ubiquitin molecules through K 98. Interestingly, mutation of either K29 or K in the fused ubiquitin of the model substrate interfered with formation and prevented proteasomal degradation, suggesting that this chain moiety is modified on both K29 and K 96. UFD2 can also cooperate with the K-specific HECT E3 RSP5 to promote degradation of the yeast transcription factor SPT23 99, whereas UFD4 collaborates with the E3 UBR to decorate a DNA repair protein with conjugates that contain both K29 and K linkages 00. These studies, therefore, suggest that UFD2 and UFD4 can assemble heterotypic conjugates, but the exact structure of these chains and the precise signalling function of each linkage remain poorly understood. Reminiscent of the synthesis of heterotypic conjugates by cooperating E3 ligases, the APC/C uses different E2 enzymes to assemble ubiquitin chains that drive the proteasomal degradation of cell cycle regulators. In human cells, chain formation is mostly initiated by the E2 e2c (also known as ch0), which binds the APC/C through a winged-helix domain in APC2 and a canonical E2-binding surface on the RINGdomain of APC Chain elongation proceeds through a different E2, e2s 54 56, which uses a unique C-terminal peptide to bind the APC/C at the same time as e2c 04. These APC/C-specific E2s have distinct linkage specificity: whereas e2c assembles mixed chains that b a K K33 K-linked c or -attached K-linked or K27/29 M K-linked or Free K-linked contain K, K and K linkages, e2s only produces K-linked chains 54,56,04. As e2s can attach K-linked chains to internal moieties of conjugates assembled by e2c, the combination of e2c and e2s activity leads to the formation of K/K-branched ubiquitin chains,05. These conjugates efficiently promote substrate recognition by the proteasome and the p97 segregase, allowing them to drive degradation of proteins that are engaged in stable interactions. Interestingly, p97 also recognizes K and K linkages for extraction of proteins from the endoplasmic reticulum and outer mitochondrial membranes 06,07, and heterotypic conjugates also seem to be involved in proteasomal processing of the Cubitus interruptus (Ci) transcriptional regulator 08. Thus, whereas rampant formation of randomly branched s can prevent proteasomal degradation of certain proteins 09, formation of K/K-branched conjugates seems to constitute a strong proteolytic signal. Modification of substrates with heterotypic s also plays important roles in non-proteolytic signalling. Binding of interleukin- (IL-) to its cognate receptor triggers a signalling cascade dependent on TRAF6, an E3 ligase that assembles K-linked chains 23,0, and the M-linkage-specific E3 LUBAC 46. Whereas K-linked chains recruit the TAK kinase to ligand-engaged IL- receptor (IL-R) complexes 24,25,36, M-linked chains are recognized by NEMO, a subunit of the IKK kinase that is activated by TAK 7. As shown by digestion with linkage-specific DUBs an approach referred to as icrest 7 the large majority of M-linked chains produced after IL- stimulation were attached to K K Degradation M-linked K6 26S proteasome Complex formation Figure 2 Signalling by homotypic s. (a) The structure of ubiquitin, with the eight potential attachment sites for chain formation shown in blue. (b) Homotypic K- and K-linked chains can drive proteasomal degradation. (c) Homotypic K- or M-linked s mediate complex assembly. NATURE CELL BIOLOGY VOLUME 8 NUMBER 6 JUNE
4 a b c f 26S proteasome Mixed Branched Signal coordination High local ubiquitin concentration d e DUB DUB Signal protection Signal editing Figure 3 Signalling by heterotypic s. (a) A schematic representation of mixed s. The different linkages were chosen arbitrarily. (b) A schematic representation of K/K-branched ubiquitin chains. (c) Signal coordination as a potential function of mixed or branched s, which could recruit multiple UBDs with distinct linkage specificity to bring components of signal transduction pathways into proximity. K-linked conjugates. By combining linkages that can be recognized by distinct UBDs, these M/K-linked chains might recruit both TAK and IKK to the same receptors (Fig. 3c), which could accelerate activation of IKK by TAK. Alternatively, decoration of K-linked chains with M-linked conjugates could prevent deubiquitylation by linkage-specific DUBs (Fig. 3d), thereby extending the time that the signalling pathway is active 2. It should be noted that the concept of heterotypic conjugates is not limited to s with multiple linkages, but could also include conjugates that are composed of distinct ubiquitin-like modifiers. The SUMO-targeted ubiquitin ligases (STUBLs) add s to proteins that are already decorated with the ubiquitin-like SUMO 3 5. At least in vitro, the STUBLs can directly ubiquitylate the previously attached SUMO tag 6. Appending ubiquitin to SUMOylated proteins tremendously alters the properties of the modified substrate: whereas SUMO usually signals through mediating protein interactions, addition of a by STUBLs frequently triggers complex disassembly or proteasomal degradation. The change in fate brought about by decorating SUMOylated proteins with proteolytic s is thus reminiscent to chain editing, in which a non-proteolytic ubiquitin chain type is replaced by a K-linked chain 7. STUBLs have now been implicated in several signalling pathways, including the cellular responses to arsenic poisoning and DNA damage, when SUMOylated proteins involved in handling chromatin need to be eliminated at various stages of the process Akin to sentences crafted from multiple words, the results described above have ascribed biological meaning to combinations of different (d) Mixed or branched chains could be protected from disassembly by linkagespecific DUBs. (e) Branching distinct s off an initial conjugate could alter signalling, for example if proteolytic K-linked chains would be branched off non-proteolytic K-linked conjugates. (f) Branching could provide a means to increase the local concentration of ubiquitin on substrates, a feature that should help recruit 26S proteasomes for degradation 20. topologies. As work in this area is still in its infancy, the mechanisms underlying assembly, recognition and function of heterotypic conjugates are not yet fully understood. Biophysical studies have implied that the distinct connections of a mixed or branched conjugate can signal independently of each other 23, and so branching K-linked chains off a K-linked conjugate might provide a means to terminate signalling events without the need to remove the activating signal (Fig. 3e). Moreover, branching allows E3 ligases to add multiple copies of a chain type to the same lysine residue in a substrate (Fig. 3f), and we hypothesize that the ensuing increase in the local ubiquitin concentration could explain the strong proteolytic signal derived from K/K-branched chains 20,. It is also possible that UBDs (or specific combinations of them) preferentially recognize branched conjugates, and that these conjugates might provoke unique reactions. So far, the analysis of heterotypic chains has been limited to indirect methods, such as digestion of conjugates in cell lysates with different combinations of linkage-specific DUBs or expression of ubiquitin variants that allowed for detection of branch signatures after treatment with sitespecific proteases 7,,90,. To understand the physiological relevance of the increasingly complex ubiquitin code, it will be important to develop new technologies that allow detection of mixed or branched ubiquitin chains in cells without expressing mutant forms of ubiquitin (Box ). Post-translational modification of ubiquitin Further expanding the signalling repertoire of ubiquitylation, ubiquitin has recently been found to be subject to post-translational modifications, including phosphorylation (Fig. 4a). As seen in large-scale proteomic 582 NATURE CELL BIOLOGY VOLUME 8 NUMBER 6 JUNE 206
5 studies, ubiquitin can be phosphorylated on most of its serine, threonine and tyrosine residues Under normal growth conditions, the overall concentration of phosphorylated ubiquitin seems to be low; in yeast cells, less than 0.5% of ubiquitin is phosphorylated at S65, and only % of the ubiquitin molecules in human cells carry a phosphate group at this site 78,27. This situation can change drastically during signalling, and ubiquitin molecules phosphorylated at S65 can make up ~20% of the ubiquitin pool attached to damaged mitochondria that are destined for removal by mitophagy 78. Our understanding of the consequences of ubiquitin phosphorylation for signal transduction greatly increased with the identification of the ubiquitin kinase encoded by the Parkinson s disease gene PINK. In healthy cells, PINK is imported into mitochondria, a reaction that results in the proteolytic release and cytoplasmic degradation of its kinase domain 28. If, however, a mitochondrion is damaged, import of PINK is prevented and the kinase accumulates on the outer membrane of the damaged organelle 29,30, where it phosphorylates S65 of ubiquitin as well as its counterpart in the ubiquitin-like domain of the RBR-family E3 ligase Parkin 78,79,3 34. The phosphorylated ubiquitin attached to mitochondrial membrane proteins helps to recruit and activate Parkin, and allows it to ubiquitylate more proteins on the damaged organelle that in turn are substrates for PINK. Phosphorylated ubiquitin also establishes a robust landing platform for the autophagy adaptors OPTN and NDP52 35,36, which, when phosphorylated by TBK, seem to recognize s of multiple topologies Thus, phosphorylation of ubiquitin sets in motion intertwined feedback loops that robustly decorate damaged mitochondria with s to elicit their selective removal from cells. Phosphorylation of ubiquitin can affect its structure as well as its interactions with conjugating enzymes, DUBs or ubiquitin-binding domains 27,34,35,38. As illustrated with Parkin, phosphorylation of ubiquitin can be a double-edged sword: whereas phospho-ubiquitin is specifically recognized by an allosteric site in Parkin to evoke striking structural rearrangements and catalytic activation of the E3 (Fig. 4b) 38, the catalytic domains of Parkin use phospho-ubiquitin less efficiently than unmodified ubiquitin and are unable to build ubiquitin chains that are composed entirely of phospho-ubiquitin 29,34. Phosphoubiquitin also impairs recognition by USP8 and USP30, two DUBs that have been implicated in preventing mitophagy 30,94,34, suggesting a mechanism that maintains a signal that triggers removal of the damaged organelle (Fig. 4c). How other post-translational modifications of ubiquitin impinge on cellular signalling events is less understood. iquitin can be phosphorylated at other residues, with S57 being the most prominently modified site 24,26, yet the responsible kinases remain unknown. Both yeast cells that naturally lack a PINK homologue and human cells that were engineered to live without PINK contain ubiquitin phosphorylated at S65 78,27, suggesting that additional kinases can target this residue. iquitin can also be acetylated at K6 and K, which in vitro can affect the build-up of K-, K- or K-linked chains 39. However, less than 0.03% of ubiquitin molecules are modified in this way, and the responsible acetylation enzyme is not known. Finally, effector proteins of pathogenic bacteria can convert Gln40 of ubiquitin into a glutamate residue, which can profoundly inhibit synthesis 40. Whether similar modifications can occur on ubiquitin conjugates and alter recognition by cellular UBDs is not yet known. BOX New technologies to decipher a complex ubiquitin code Understanding the connectivity between substrate-linked ubiquitin molecules in cells is critical to our ability to decipher the ubiquitin code. Current strategies that avoid expression of mutant ubiquitin variants are based on mass spectrometry 2,78,24, linkage-specific antibodies that can detect M, K, K or K linkages 44,45,49, and ubiquitin sensors that couple a linkage-specific ubiquitinbinding domain to a fluorescent reporter 52. These approaches are able to recognize particular linkages within a substrate-attached conjugate or at a particular cellular location, yet they cannot report on the presence of multiple linkages within the same chain or on the same ubiquitin molecule. Providing a step towards sequencing s, combinatorial treatment of ubiquitin conjugates with linkage-specific DUBs a powerful approach referred to as icrest 7 allows for analysis of more complex chain topologies. To fully understand the wide range of different heterotypic s, researchers will need to develop additional technologies. Such strategies could include: icrest approaches that employ DUBs with specificity for branched conjugates or even proteases with different target sites on ubiquitin; linkage-specific antibodies that either recognize branched structures or act as coincidence detectors for the presence of multiple linkages within one conjugate; or middle-down mass spectrometry approaches that avoid complete digestion of ubiquitin to retain information about multiple attachment sites on a single chain building block 50. When complemented with synthetic biology approaches to re-engineer the linkage specificity of ubiquitin ligases in cells, such a toolkit would put researchers in a position to unravel the remaining mysteries of ubiquitin-dependent signal transfer. Dense information in a ubiquitin cloud? The attachment of branched chains to cell cycle regulators or conjugation of heterotypic chains to outer mitochondrial membrane proteins results in a high local concentration of ubiquitin on the modified substrate or organelle. In the case of SUMOylation, clustered modifications on multiple subunits of protein complexes have been described as a SUMO cloud that provides a high-avidity signal for SUMO-binding proteins 4. Is it possible that some ubiquitin signals similarly depend on the local concentration, rather than the connectivity of ubiquitin molecules? In a surprising twist to the story of ubiquitin-dependent signalling, it was the prototypical function of linkage-specific ubiquitylation, proteasomal degradation, that provided evidence for a role of the local ubiquitin concentration in signal transduction. Ground-breaking work had established that 3 4 molecules linked through K of ubiquitin drive efficient recognition by the 26S proteasome 3,9, yet it has become difficult to rationalize this observation with recent structures of the 26S proteasome bound to a substrate 42. The large distance between ubiquitin receptors and the dynamic interactions of substrate delivery factors with the proteasome raised the possibility that multiple chain topologies could mediate degradation. Indeed, alternative chain types have been shown to mediate proteasomal turnover, including K-linked, K-linked and heterotypic conjugates 47,43,44. In some cases, short polypeptides or proteins with stretches of low structural complexity NATURE CELL BIOLOGY VOLUME 8 NUMBER 6 JUNE
6 a K Q40 Y59 K6 S57 Phosphorylation Acetylation Deamidation S65 PO 4 T66 T4 T2 c b can be delivered to the proteasome by monoubiquitylation or multimonoubiquitylation 45,46. In line with these observations, single-molecule studies recently showed that multiple K-linked di-ubiquitin conjugates provide a stronger degradation signal than the canonical tetra- 20. Initially, two di-ubiquitin entities cooperatively engage the proteasomal ubiquitin receptors Rpn0 and Rpn3, before addition of more di-ubiquitin signals accelerates turnover by increasing the local concentration of ubiquitin on the substrate. These findings are consistent with the notion that a large number of ubiquitylated proteins contain more than one attachment site for ubiquitin 42,43,24, and that increasing the number of lysine residues available for ubiquitylation can accelerate the turnover of model substrates 47. The proteasome itself seems to regulate the local ubiquitin concentration, as the proteasome-associated E3 HUL5 adds additional K-linked chains to proteins that are already bound to the degradative machine. Thus, reminiscent of a ubiquitin cloud, the local concentration of substrateattached ubiquitin molecules, rather than the linkage between them, might be an important determinant of proteasomal delivery. Conclusion Many historic examples illustrate the importance of precise communication, but few phrase it as pointedly as a line from a poem by Henry Wadsworth Longfellow about the ride of Paul Revere at the beginning of the American Revolutionary War: One if by land, and two if by sea. The number of lanterns hung from the tower of the Old North Church in Boston s North End informed Revere about the British approach on Boston and allowed him to mount an effective response. Although we are beginning to realize that ubiquitylation establishes a much more complex code than originally thought, the principles of information transfer are the same: distinct types of modification can trigger specific cellular responses that are at the heart of essential reactions, such as cell division, differentiation and survival. By deepening our understanding of the ubiquitin code in all its complexity the temporal and structural dynamics, the messages derived from mixed or branched chains, and the modification of its central player, ubiquitin we stand to learn much about the fundamental organization of a cell. Given the central role of E3 DUB P P P P E3 DUB Figure 4 Signalling by modified ubiquitin. (a) Sites for ubiquitin phosphorylation, acetylation and deamidation that were detected by mass spectrometry are mapped onto the structure of ubiquitin phosphorylated at S (b) Specific recognition of phosphorylated ubiquitin. As illustrated by the E3 ligase Parkin, binding of phospho-ubiquitin to an allosteric site can induce conformational changes that ultimately activate the E3 ligase 38. (c) Inhibition of ubiquitin recognition by phosphorylation can prevent DUBs from terminating signal transduction 34. ubiquitylation in cellular communication pathways relevant for human development and disease, such knowledge should ultimately result in innovative and effective therapeutic approaches. ACKNOWLEDGEMENTS We apologize to all colleagues whose work we were unable to include due to space constraints. We thank J. Schaletzky, A. Manford and A. Werner for reading this manuscript, and we are grateful to all members of our lab for many inspired discussions about the roles of ubiquitin in cell signalling. We also thank J. Campbell for discussions on the principles of communication. Work in our lab is funded by grants from the National Institutes of Health. M.R. is the K. Peter Hirth Chair of Cancer Biology at UC Berkeley and an Investigator of the Howard Hughes Medical Institute. ADDITIONAL INFORMATION Reprints and permissions information is available online at reprints. Correspondence should be addressed to M.R. COMPETING FINANCIAL INTERESTS M.R. is co-founder and consultant to Nurix, a biotechnology company that operates in the ubiquitin space.. Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 8, (202). 2. Deshaies, R. J. & Joazeiro, C. A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, (2009). 3. Ye, Y. & Rape, M. Building s: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 0, (2009). 4. Schulman, B. A. & Harper, J. W. iquitin-like protein activation by E enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 0, (2009). 5. Goldknopf, I. L., French, M. F., Musso, R. & Busch, H. Presence of protein A24 in rat liver nucleosomes. Proc. Natl Acad. Sci. USA 74, (977). 6. Husnjak, K. & Dikic, I. iquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 8, (202). 7. Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. RAD6- dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 49, 35 4 (2002). 8. Bienko, M. et al. iquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 30, (2005). 9. Dupont, S. et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell 36, (2009). 0. Ye, Y. et al. iquitin chain conformation regulates recognition and activity of interacting proteins. Nature 492, (202).. Liu, Z. et al. Lys-linked adopts multiple conformational states for specific target recognition. elife 4, e05767 (205). 2. Xu, P. et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 37, (2009). 3. Chau, V. et al. A multi is confined to specific lysine in a targeted short-lived protein. Science 243, (989). 4. Petroski, M. D. & Deshaies, R. J. Mechanism of lysine -linked ubiquitin-chain synthesis by the cullin-ring ubiquitin-ligase complex SCF-Cdc34. Cell 23, (2005). 5. Chen, B. et al. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl Acad. Sci. USA 03, (2006). 6. Scheffner, M., Huibregtse, J. M., Vierstra, R. D. & Howley, P. M. The HPV-6 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75, (993). 7. Mevissen, T. E. et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable restriction analysis. Cell 54, (203). 8. Schwickart, M. et al. Deubiquitinase USP9X stabilizes MCL and promotes tumour cell survival. Nature 4, (200). 9. Thrower, J. S., Hoffman, L., Rechsteiner, M. & Pickart, C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 9, (2000). 20. Lu, Y., Lee, B. H., King, R. W., Finley, D. & Kirschner, M. W. degradation by the proteasome: a single-molecule kinetic analysis. Science 3, (205). 2. Flick, K., Raasi, S., Zhang, H., Yen, J. L. & Kaiser, P. A ubiquitin-interacting motif protects polyubiquitinated Met4 from degradation by the 26S proteasome. Nat. Cell Biol. 8, (2006). 22. Spence, J., Sadis, S., Haas, A. L. & Finley, D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell Biol. 5, (995). 23. Deng, L. et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique poly. Cell 03, (2000). 24. Wang, C. et al. TAK is a ubiquitin-dependent kinase of MKK and IKK. Nature 42, (200). 25. Xu, M., Skaug, B., Zeng, W. & Chen, Z. J. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFα and IL-β. Mol Cell 36, (2009). 584 NATURE CELL BIOLOGY VOLUME 8 NUMBER 6 JUNE 206
7 26. Doil, C. et al. RNF68 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 36, (2009). 27. Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 36, (2009). 28. Gack, M. U. et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-Imediated antiviral activity. Nature 446, (2007). 29. Ordureau, A. et al. Defining roles of PARKIN and ubiquitin phosphorylation by PINK in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl Acad. Sci. USA 2, (205). 30. Cunningham, C. N. et al. USP30 and parkin homeostatically regulate atypical s on mitochondria. Nat. Cell Biol. 7, (205). 3. Lauwers, E., Jacob, C. & Andre, B. K-linked s as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 85, (2009). 32. Huang, F. et al. Lysine -linked polyubiquitination is required for EGF receptor degradation. Proc. Natl Acad. Sci. USA 0, (203). 33. Song, E. J. et al. The Prp9 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 24, (200). 34. Silva, G. M., Finley, D. & Vogel, C. K polyubiquitination is a new modulator of the oxidative stress response. Nat. Struct. Mol. Biol. 22, 6 23 (205). 35. Spence, J. et al. Cell cycle-regulated modification of the ribosome by a variant multi. Cell 02, (2000). 36. Xia, Z. P. et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 46, 4 9 (2009). 37. Zeng, W. et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polys in innate immunity. Cell 4, (200). 38. VanDemark, A. P., Hofmann, R. M., Tsui, C., Pickart, C. M. & Wolberger, C. Molecular insights into poly assembly: crystal structure of the Mms2/c3 heterodimer. Cell 05, (200). 39. Maspero, E. et al. Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat. Struct. Mol. Biol. 20, (203). 40. McCullough, J., Clague, M. J. & Urbe, S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol 66, (2004). 4. Cooper, E. M. et al. K-specific deubiquitination by two JAMM/MPN + complexes: BRISC-associated Brcc36 and proteasomal Poh. EMBO J. 28, 62 (2009). 42. Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44, (20). 43. Emanuele, M. J. et al. Global identification of modular cullin-ring ligase substrates. Cell 47, (20). 44. Matsumoto, M. L. et al. Engineering and structural characterization of a linear polyubiquitin-specific antibody. J. Mol. Biol., (202). 45. Matsumoto, M. L. et al. K-linked polyubiquitination in cell cycle control revealed by a K linkage-specific antibody. Mol. Cell 39, (200). 46. Tokunaga, F. et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol., (2009). 47. Jin, L., Williamson, A., Banerjee, S., Philipp, I. & Rape, M. Mechanism of ubiquitinchain formation by the human anaphase-promoting complex. Cell 33, (2008).. Meyer, H. J. & Rape, M. Enhanced protein degradation by branched s. Cell 57, (204). 49. Ikeda, F. et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 47, 7 64 (20). 50. Tokunaga, F., Nakagawa, T., Nakahara, M., Saeki, Y., Taniguchi, M., Sakata, S., Tanaka, K., Nakano, H., Iwai, K. SHARPIN is a component of the NF-κB activating linear assembly complex. Nature 47, 3 6 (20). 5. Gerlach, B. et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 47, (20). 52. van Wijk, S. J. et al. Fluorescence-based sensors to monitor localization and functions of linear and K-linked s in cells. Mol. Cell 47, (202). 53. Wild, P. et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, (20). 54. Williamson, A. et al. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl Acad. Sci. USA 06, (2009). 55. Garnett, M. J. et al. UBE2S elongates s on APC/C substrates to promote mitotic exit. Nat. Cell Biol., (2009). 56. Wu, T. et al. UBE2S drives elongation of K-linked s by the anaphase-promoting complex. Proc. Natl Acad. Sci. USA 07, (200). 57. Min, M., Mevissen, T. E., De Luca, M., Komander, D. & Lindon, C. Efficient APC/C substrate degradation in cells undergoing mitotic exit depends on K ubiquitin linkages. Mol. Biol. Cell 26, (205). 58. Baboshina, O. V. & Haas, A. L. Novel multi linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J. Biol. Chem. 27, (996). 59. Kirisako, T. et al. A ubiquitin ligase complex assembles linear polys. EMBO J. 25, (2006). 60. Bremm, A., Freund, S. M. & Komander, D. Lys-linked s adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat. Struct. Mol. Biol. 7, (200). 6. Keusekotten, K. et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met-linked polyubiquitin. Cell 53, (203). 62. Rivkin, E. et al. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature 498, (203).. Elliott, P. R. et al. Molecular basis and regulation of OTULIN-LUBAC interaction. Mol. Cell 54, (204). 64. Schaeffer, V. et al. Binding of OTULIN to the PUB domain of HOIP controls NF-κB signaling. Mol. Cell 54, (204). 65. Song, L., Craney, A. & Rape, M. Microtubule-dependent regulation of mitotic protein degradation. Mol. Cell 53, (204). 66. Song, L. & Rape, M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol. Cell 38, (200). 67. Castaneda, C. A., Kashyap, T. R., Nakasone, M. A., Krueger, S. & Fushman, D. Unique structural, dynamical, and functional properties of k-linked polys. Structure 2, 68 8 (203). 68. Bremm, A., Moniz, S., Mader, J., Rocha, S. & Komander, D. Cezanne (OTUD7B) regulates HIF-α homeostasis in a proteasome-independent manner. EMBO Rep. 5, (204). 69. Qin, Y. et al. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog. 0, e (204). 70. Komander, D. et al. Molecular discrimination of structurally equivalent Lys -linked and linear polys. EMBO Rep. 0, (2009). 7. Rahighi, S. et al. Specific recognition of linear s by NEMO is important for NF-κB activation. Cell 36, (2009). 72. Damgaard, R. B. et al. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol. Cell 46, (202). 73. Inn, K. S. et al. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol. Cell 4, (20). 74. Gatti, M. et al. RNF68 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 0, (205). 75. Liu, J. et al. Rhbdd3 controls autoimmunity by suppressing the production of IL-6 by dendritic cells via K27-linked ubiquitination of the regulator NEMO. Nat. Immunol. 5, (204). 76. Johnson, E. S., Ma, P. C., Ota, I. M. & Varshavsky, A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270, (995). 77. Yuan, W. C. et al. K33-linked polyubiquitination of coronin 7 by Cul3-KLHL20 ubiquitin E3 ligase regulates protein trafficking. Mol Cell 54, (204). 78. Ordureau, A. et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and synthesis. Mol. Cell 56, (204). 79. Sarraf, S. A. et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, (203). 80. Hospenthal, M. K., Freund, S. M. & Komander, D. Assembly, analysis and architecture of atypical s. Nat. Struct. Mol. Biol. 20, (203). 8. Rutz, S. et al. Deubiquitinase DUBA is a post-translational brake on interleukin-7 production in T cells. Nature 58, (205). 82. Okamoto, K. et al. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 494, (203). 83. Gudjonsson, T. et al. TRIP2 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 50, (202). 84. Wang, M., Cheng, D., Peng, J. & Pickart, C. M. Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J. 25, (2006). 85. Michel, M. A. et al. Assembly and specific recognition of k29- and k33-linked polyubiquitin. Mol. Cell 58, (205). 86. Jin, L. et al. iquitin-dependent regulation of COPII coat size and function. Nature 2, (202). 87. Werner, A. et al. Cell-fate determination by ubiquitin-dependent regulation of translation. Nature 525, (205). 88. Licchesi, J. D. et al. An ankyrin-repeat ubiquitin-binding domain determines TRABID s specificity for atypical s. Nat. Struct. Mol. Biol. 9, 62 7 (202). 89. Virdee, S., Ye, Y., Nguyen, D. P., Komander, D. & Chin, J. W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 6, (200). 90. Kristariyanto, Y. A. et al. K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin. Mol. Cell 58, (205). 9. Yuan, L. et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat. Cell Biol. 7, 69 8 (205). 92. Ernst, R., Mueller, B., Ploegh, H. L. & Schlieker, C. The otubain YOD is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol. Cell 36, (2009). 93. Bingol, B. et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 50, (204). 94. Durcan, T. M. et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 33, (204). 95. Boname, J. M. et al. Efficient internalization of MHC I requires lysine- and lysine- mixed linkage polys. Traffic, (200). 96. Koegl, M. et al. A novel ubiquitination factor, E4, is involved in multi assembly. Cell 96, (999). 97. Tsuchiya, H., Tanaka, K. & Saeki, Y. The parallel reaction monitoring method contributes to a highly sensitive poly quantification. Biochem. Biophys. Res. Commun. 436, (203). NATURE CELL BIOLOGY VOLUME 8 NUMBER 6 JUNE
8 98. Saeki, Y., Tayama, Y., Toh-e, A. & Yokosawa, H. Definitive evidence for Ufd2-catalyzed elongation of the through Lys linkage. Biochem. Biophys. Res. Commun. 320, (2004). 99. Richly, H. et al. A series of ubiquitin binding factors connects CDC/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 20, (2005). 00. Hwang, C. S., Shemorry, A., Auerbach, D. & Varshavsky, A. The N-end rule pathway is mediated by a complex of the RING-type r and HECT-type Ufd4 ubiquitin ligases. Nat. Cell Biol. 2, (200). 0. Brown, N. G. et al. RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex. Proc. Natl Acad. Sci. USA 2, (205). 02. Yu, H., King, R. W., Peters, J. M. & Kirschner, M. W. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr. Biol. 6, (996). 03. Williamson, A. et al. Regulation of initiation to control the timing of substrate degradation. Mol. Cell 42, (20). 04. Wickliffe, K. E., Lorenz, S., Wemmer, D. E., Kuriyan, J. & Rape, M. The mechanism of linkage-specific elongation by a single-subunit e2. Cell 44, (20). 05. Grice, G. L. et al. The proteasome distinguishes between heterotypic and homotypic lysine--linked polys. Cell Rep. 2, (205). 06. Kim, N. C. et al. VCP is essential for mitochondrial quality control by PINK/Parkin and this function is impaired by VCP mutations. Neuron 78, (203). 07. Locke, M., Toth, J. I. & Petroski, M. D. Lys- and Lys-linked s interact with p97 during endoplasmic-reticulum-associated degradation. Biochem. J. 459, (204). 08. Zhang, Z. et al. Ter94 ATPase complex targets k-linked ubiquitinated ci to proteasomes for partial degradation. Dev. Cell 25, (203). 09. Kim, H. T. et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitinprotein ligases (E3s) synthesize nondegradable forked s containing all possible isopeptide linkages. J. Biol. Chem. 282, (2007). 0. Cao, Z., Xiong, J., Takeuchi, M., Kurama, T. & Goeddel, D. V. TRAF6 is a signal transducer for interleukin-. Nature 383, (996).. Emmerich, C. H. et al. Activation of the canonical IKK complex by K/M-linked hybrid s. Proc. Natl Acad. Sci. USA 0, (203). 2. Wertz, I. E. et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature 528, (205). 3. Prudden, J. et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, (2007). 4. Uzunova, K. et al. iquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282, (2007). 5. Xie, Y. et al. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 282, (2007). 6. Branigan, E., Plechanovova, A., Jaffray, E. G., Naismith, J. H. & Hay, R. T. Structural basis for the RING-catalyzed synthesis of K-linked s. Nat. Struct. Mol. Biol. 22, (205). 7. Wertz, I. E. et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430, (2004). 8. Tatham, M. H. et al. RNF4 is a poly-sumo-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 0, (2008). 9. Lallemand-Breitenbach, V. et al. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 0, (2008). 20. Gibbs-Seymour, I. et al. iquitin-sumo circuitry controls activated fanconi anemia ID complex dosage in response to DNA damage. Mol. Cell 57, (205). 2. Yin, Y. et al. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26, (202). 22. Galanty, Y., Belotserkovskaya, R., Coates, J. & Jackson, S. P. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26, (202). 23. Nakasone, M. A., Livnat-Levanon, N., Glickman, M. H., Cohen, R. E. & Fushman, D. Mixed-linkage s send mixed messages. Structure 2, (203). 24. Peng, J. et al. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2, (2003). 25. Rikova, K. et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 3, (2007). 26. Swaney, D. L. et al. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 0, (203). 27. Swaney, D. L., Rodriguez-Mias, R. A. & Villen, J. Phosphorylation of ubiquitin at Ser65 affects its polymerization, targets, and proteome-wide turnover. EMBO Rep. 6, 3 44 (205). 28. Yamano, K. & Youle, R. J. PINK is degraded through the N-end rule pathway. Autophagy 9, (203). 29. Jin, S. M. et al. Mitochondrial membrane potential regulates PINK import and proteolytic destabilization by PARL. J. Cell Biol. 9, (200). 30. Narendra, D. P. et al. PINK is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e (200). 3. Kazlauskaite, A. et al. Parkin is activated by PINK-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, (204). 32. Kane, L. A. et al. PINK phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, (204). 33. Koyano, F. et al. iquitin is phosphorylated by PINK to activate parkin. Nature 50, (204). 34. Wauer, T. et al. iquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 34, (205). 35. Heo, J. M., Ordureau, A., Paulo, J. A., Rinehart, J. & Harper, J. W. The PINK-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK activation to promote mitophagy. Mol. Cell 60, 7 20 (205). 36. Lazarou, M. et al. The ubiquitin kinase PINK recruits autophagy receptors to induce mitophagy. Nature 524, (205). 37. Herhaus, L. & Dikic, I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 6, (205). 38. Wauer, T., Simicek, M., Schubert, A. & Komander, D. Mechanism of phospho-ubiquitininduced PARKIN activation. Nature 524, (205). 39. Ohtake, F. et al. iquitin acetylation inhibits poly elongation. EMBO Rep. 6, (205). 40. Cui, J. et al. Glutamine deamidation and dysfunction of ubiquitin/nedd8 induced by a bacterial effector family. Science 329, (200). 4. Psakhye, I. & Jentsch, S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 5, (202). 42. Matyskiela, M. E., Lander, G. C. & Martin, A. Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 20, (203). 43. Saeki, Y. et al. Lysine -linked poly may serve as a targeting signal for the 26S proteasome. EMBO J. 28, (2009). 44. Shin, J. H. et al. PARIS (ZNF746) repression of PGC-α contributes to neurodegeneration in Parkinson s disease. Cell 44, (20). 45. Dimova, N. V. et al. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B. Nat. Cell Biol. 4, (202). 46. Shabek, N. et al. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol. Cell, (202). 47. Suzuki, T. & Varshavsky, A. Degradation signals in the lysine-asparagine sequence space. EMBO J. 8, (999).. Crosas, B. et al. iquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 27, (2006). 49. Newton, K. et al. iquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 34, (2008). 50. Valkevich, E. M., Sanchez, N. A., Ge, Y. & Strieter, E. R. Middle-down mass spectrometry enables characterization of branched s. Biochemistry 53, (204). 586 NATURE CELL BIOLOGY VOLUME 8 NUMBER 6 JUNE 206
Ubiquitin modifications
 REVIEW Ubiquitin modifications Cell Research (2016) :1-24. www.nature.com/cr npg Kirby N Swatek 1, David Komander 1 1 Medical Research Council Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge,
REVIEW Ubiquitin modifications Cell Research (2016) :1-24. www.nature.com/cr npg Kirby N Swatek 1, David Komander 1 1 Medical Research Council Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge,
Molecular Graphics Perspective of Protein Structure and Function
 Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Protein methylation CH 3
 Protein methylation CH 3 methionine S-adenosylmethionine (SAM or adomet) Methyl group of the methionine is activated by the + charge of the adjacent sulfur atom. SAM S-adenosylhomocysteine homocysteine
Protein methylation CH 3 methionine S-adenosylmethionine (SAM or adomet) Methyl group of the methionine is activated by the + charge of the adjacent sulfur atom. SAM S-adenosylhomocysteine homocysteine
SUPPLEMENTARY INFORMATION
 Figure S1. Silver staining and immunoblotting of the purified TAK1 kinase complex. The TAK1 kinase complex was purified through tandem affinity methods (Protein A and FLAG), and aliquots of the purified
Figure S1. Silver staining and immunoblotting of the purified TAK1 kinase complex. The TAK1 kinase complex was purified through tandem affinity methods (Protein A and FLAG), and aliquots of the purified
Study of different types of ubiquitination
 Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
three forms: c-cbl, CBLb and CBL-3) MDM2, MDMX and SCF Fbxo11
 Supplementary information S1 (table) Reported Neddylation Substrates Target protein Biological function of target Effect of neddylation E2 E3 Regulation of neddylation Cullins RING ubiquitin E3 ligases
Supplementary information S1 (table) Reported Neddylation Substrates Target protein Biological function of target Effect of neddylation E2 E3 Regulation of neddylation Cullins RING ubiquitin E3 ligases
1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled
 Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Proteasomes. When Death Comes a Knock n. Warren Gallagher Chem412, Spring 2001
 Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Summary of Endomembrane-system
 Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
K11-linked ubiquitin chains as novel regulators of cell division
 Review K11-linked ubiquitin chains as novel regulators of cell division Katherine E. Wickliffe, Adam Williamson, Hermann-Josef Meyer, Aileen Kelly and Michael Rape Department of Molecular and Cell Biology,
Review K11-linked ubiquitin chains as novel regulators of cell division Katherine E. Wickliffe, Adam Williamson, Hermann-Josef Meyer, Aileen Kelly and Michael Rape Department of Molecular and Cell Biology,
Signal Transduction Cascades
 Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
The ubiquitin-proteasome system. in bone biology. University of Nottingham. ECTS PhD Training July 5 th Dr Rob Layfield
 The ubiquitin-proteasome system in bone biology Dr Rob Layfield University of Nottingham ECTS PhD Training July 5 th 2009 Images reproduced from: http://www.bostonbiochem.com/overview.php?prod=ubchains
The ubiquitin-proteasome system in bone biology Dr Rob Layfield University of Nottingham ECTS PhD Training July 5 th 2009 Images reproduced from: http://www.bostonbiochem.com/overview.php?prod=ubchains
Removal of Shelterin Reveals the Telomere End-Protection Problem
 Removal of Shelterin Reveals the Telomere End-Protection Problem DSB Double-Strand Breaks causate da radiazioni stress ossidativo farmaci DSB e CROMATINA Higher-order chromatin packaging is a barrier to
Removal of Shelterin Reveals the Telomere End-Protection Problem DSB Double-Strand Breaks causate da radiazioni stress ossidativo farmaci DSB e CROMATINA Higher-order chromatin packaging is a barrier to
REGULATION OF ENZYME ACTIVITY. Medical Biochemistry, Lecture 25
 REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
Homework Hanson section MCB Course, Fall 2014
 Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
The Nobel Prize in Chemistry 2004
 The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
Post-translational modifications of proteins in gene regulation under hypoxic conditions
 203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
Lecture 10. G1/S Regulation and Cell Cycle Checkpoints. G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint
 Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Complexity DNA. Genome RNA. Transcriptome. Protein. Proteome. Metabolites. Metabolome
 DNA Genome Complexity RNA Transcriptome Systems Biology Linking all the components of a cell in a quantitative and temporal manner Protein Proteome Metabolites Metabolome Where are the functional elements?
DNA Genome Complexity RNA Transcriptome Systems Biology Linking all the components of a cell in a quantitative and temporal manner Protein Proteome Metabolites Metabolome Where are the functional elements?
Cell Cycle, Mitosis, and Microtubules. LS1A Final Exam Review Friday 1/12/07. Processes occurring during cell cycle
 Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Biol403 MAP kinase signalling
 Biol403 MAP kinase signalling The mitogen activated protein kinase (MAPK) pathway is a signalling cascade activated by a diverse range of effectors. The cascade regulates many cellular activities including
Biol403 MAP kinase signalling The mitogen activated protein kinase (MAPK) pathway is a signalling cascade activated by a diverse range of effectors. The cascade regulates many cellular activities including
Insulin mrna to Protein Kit
 Insulin mrna to Protein Kit A 3DMD Paper BioInformatics and Mini-Toober Folding Activity Student Handout www.3dmoleculardesigns.com Insulin mrna to Protein Kit Contents Becoming Familiar with the Data...
Insulin mrna to Protein Kit A 3DMD Paper BioInformatics and Mini-Toober Folding Activity Student Handout www.3dmoleculardesigns.com Insulin mrna to Protein Kit Contents Becoming Familiar with the Data...
BCHM3972 Human Molecular Cell Biology (Advanced) 2013 Course University of Sydney
 BCHM3972 Human Molecular Cell Biology (Advanced) 2013 Course University of Sydney Page 2: Immune Mechanisms & Molecular Biology of Host Defence (Prof Campbell) Page 45: Infection and Implications for Cell
BCHM3972 Human Molecular Cell Biology (Advanced) 2013 Course University of Sydney Page 2: Immune Mechanisms & Molecular Biology of Host Defence (Prof Campbell) Page 45: Infection and Implications for Cell
Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013
 Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
The K48-K63 Branched Ubiquitin Chain Regulates NF-kB Signaling
 Article The K48-K63 Branched Ubiquitin Chain Regulates NF-kB Signaling Graphical Abstract Authors Fumiaki Ohtake, Yasushi Saeki, Satoshi Ishido, Jun Kanno, Keiji Tanaka Correspondence otake-fm@igakuken.or.jp
Article The K48-K63 Branched Ubiquitin Chain Regulates NF-kB Signaling Graphical Abstract Authors Fumiaki Ohtake, Yasushi Saeki, Satoshi Ishido, Jun Kanno, Keiji Tanaka Correspondence otake-fm@igakuken.or.jp
Critical Review. Protein Monoubiquitination and Polyubiquitination Generate Structural Diversity to Control Distinct Biological Processes
 IUBMB Life, 64(2): 136 142, February 2012 Critical Review Protein Monoubiquitination and Polyubiquitination Generate Structural Diversity to Control Distinct Biological Processes Martin Sadowski 1, Randy
IUBMB Life, 64(2): 136 142, February 2012 Critical Review Protein Monoubiquitination and Polyubiquitination Generate Structural Diversity to Control Distinct Biological Processes Martin Sadowski 1, Randy
October 26, Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Biochemistry 2 Recita0on Amino Acid Metabolism
 Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Antigen Presentation to T lymphocytes
 Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antibodies and T cell receptors
Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antibodies and T cell receptors
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cellular functions of protein degradation
 Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Ch. 18 Regulation of Gene Expression
 Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Cbl ubiquitin ligase: Lord of the RINGs
 Cbl ubiquitin ligase: Lord of the RINGs Not just quite interesting - really interesting! A cell must be able to degrade proteins when their activity is no longer required. Many eukaryotic proteins are
Cbl ubiquitin ligase: Lord of the RINGs Not just quite interesting - really interesting! A cell must be able to degrade proteins when their activity is no longer required. Many eukaryotic proteins are
Chapt 15: Molecular Genetics of Cell Cycle and Cancer
 Chapt 15: Molecular Genetics of Cell Cycle and Cancer Student Learning Outcomes: Describe the cell cycle: steps taken by a cell to duplicate itself = cell division; Interphase (G1, S and G2), Mitosis.
Chapt 15: Molecular Genetics of Cell Cycle and Cancer Student Learning Outcomes: Describe the cell cycle: steps taken by a cell to duplicate itself = cell division; Interphase (G1, S and G2), Mitosis.
Eukaryotic Gene Regulation
 Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Effects of Second Messengers
 Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
HIV-1 genome organization
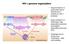 HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
Lecture 14 - The cell cycle and cell death
 02.17.10 Lecture 14 - The cell cycle and cell death The cell cycle: cells duplicate their contents and divide The cell cycle may be divided into 4 phases The cell cycle triggers essential processes (DNA
02.17.10 Lecture 14 - The cell cycle and cell death The cell cycle: cells duplicate their contents and divide The cell cycle may be divided into 4 phases The cell cycle triggers essential processes (DNA
Chemistry 107 Exam 4 Study Guide
 Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
RNA Processing in Eukaryotes *
 OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
Chapter 6. Antigen Presentation to T lymphocytes
 Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Zool 3200: Cell Biology Exam 4 Part I 2/3/15
 Name: Key Trask Zool 3200: Cell Biology Exam 4 Part I 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Name: Key Trask Zool 3200: Cell Biology Exam 4 Part I 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Histones modifications and variants
 Histones modifications and variants Dr. Institute of Molecular Biology, Johannes Gutenberg University, Mainz www.imb.de Lecture Objectives 1. Chromatin structure and function Chromatin and cell state Nucleosome
Histones modifications and variants Dr. Institute of Molecular Biology, Johannes Gutenberg University, Mainz www.imb.de Lecture Objectives 1. Chromatin structure and function Chromatin and cell state Nucleosome
General information. Cell mediated immunity. 455 LSA, Tuesday 11 to noon. Anytime after class.
 General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
Mechanisms of alternative splicing regulation
 Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting
 Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Intrinsic cellular defenses against virus infection
 Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Regulators of Cell Cycle Progression
 Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Practice Exam 2 MCBII
 1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
B F. Location of MHC class I pockets termed B and F that bind P2 and P9 amino acid side chains of the peptide
 Different MHC alleles confer different functional properties on the adaptive immune system by specifying molecules that have different peptide binding abilities Location of MHC class I pockets termed B
Different MHC alleles confer different functional properties on the adaptive immune system by specifying molecules that have different peptide binding abilities Location of MHC class I pockets termed B
Lecture 15. Signal Transduction Pathways - Introduction
 Lecture 15 Signal Transduction Pathways - Introduction So far.. Regulation of mrna synthesis Regulation of rrna synthesis Regulation of trna & 5S rrna synthesis Regulation of gene expression by signals
Lecture 15 Signal Transduction Pathways - Introduction So far.. Regulation of mrna synthesis Regulation of rrna synthesis Regulation of trna & 5S rrna synthesis Regulation of gene expression by signals
Lecture 2: Virology. I. Background
 Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
Chapter 11: Enzyme Catalysis
 Chapter 11: Enzyme Catalysis Matching A) high B) deprotonated C) protonated D) least resistance E) motion F) rate-determining G) leaving group H) short peptides I) amino acid J) low K) coenzymes L) concerted
Chapter 11: Enzyme Catalysis Matching A) high B) deprotonated C) protonated D) least resistance E) motion F) rate-determining G) leaving group H) short peptides I) amino acid J) low K) coenzymes L) concerted
基醫所. The Cell Cycle. Chi-Wu Chiang, Ph.D. IMM, NCKU
 基醫所 The Cell Cycle Chi-Wu Chiang, Ph.D. IMM, NCKU 1 1 Introduction to cell cycle and cell cycle checkpoints 2 2 Cell cycle A cell reproduces by performing an orderly sequence of events in which it duplicates
基醫所 The Cell Cycle Chi-Wu Chiang, Ph.D. IMM, NCKU 1 1 Introduction to cell cycle and cell cycle checkpoints 2 2 Cell cycle A cell reproduces by performing an orderly sequence of events in which it duplicates
Intracellular Compartments and Protein Sorting
 Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004
 Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
The recognition of ubiquitinated proteins by the proteasome
 Cell. Mol. Life Sci. (2016) 73:3497 3506 DOI 10.1007/s00018-016-2255-5 Cellular and Molecular Life Sciences REVIEW The recognition of ubiquitinated proteins by the proteasome Guinevere L. Grice 1 James
Cell. Mol. Life Sci. (2016) 73:3497 3506 DOI 10.1007/s00018-016-2255-5 Cellular and Molecular Life Sciences REVIEW The recognition of ubiquitinated proteins by the proteasome Guinevere L. Grice 1 James
Propagation of the Signal
 OpenStax-CNX module: m44452 1 Propagation of the Signal OpenStax College This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section,
OpenStax-CNX module: m44452 1 Propagation of the Signal OpenStax College This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section,
Mechanism of Ubiquitin-Chain Formation by the Human Anaphase-Promoting Complex
 Mechanism of Ubiquitin-Chain Formation by the Human Anaphase-Promoting Complex Lingyan Jin, 1,2 Adam Williamson, 1,2 Sudeep Banerjee, 1 Isabelle Philipp, 1 and Michael Rape 1, * 1 Department of Molecular
Mechanism of Ubiquitin-Chain Formation by the Human Anaphase-Promoting Complex Lingyan Jin, 1,2 Adam Williamson, 1,2 Sudeep Banerjee, 1 Isabelle Philipp, 1 and Michael Rape 1, * 1 Department of Molecular
2015 AP Biology Unit #4 Quiz 1 Cell Communication, Cancer and The Cell Cycle Week of November
 Name: Class: Date: 2015 AP Biology Unit #4 Quiz 1 Cell Communication, Cancer and The Cell Cycle Week of 16-20 November Multiple Choice Identify the choice that best completes the statement or answers the
Name: Class: Date: 2015 AP Biology Unit #4 Quiz 1 Cell Communication, Cancer and The Cell Cycle Week of 16-20 November Multiple Choice Identify the choice that best completes the statement or answers the
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
Cancer. Throughout the life of an individual, but particularly during development, every cell constantly faces decisions.
 Cancer Throughout the life of an individual, but particularly during development, every cell constantly faces decisions. Should it divide? Yes No--> Should it differentiate? Yes No-->Should it die? Yes-->Apoptosis
Cancer Throughout the life of an individual, but particularly during development, every cell constantly faces decisions. Should it divide? Yes No--> Should it differentiate? Yes No-->Should it die? Yes-->Apoptosis
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Newly Recognized Components of the Innate Immune System
 Newly Recognized Components of the Innate Immune System NOD Proteins: Intracellular Peptidoglycan Sensors NOD-1 NOD-2 Nod Protein LRR; Ligand Recognition CARD RICK I-κB p50 p65 NF-κB Polymorphisms in Nod-2
Newly Recognized Components of the Innate Immune System NOD Proteins: Intracellular Peptidoglycan Sensors NOD-1 NOD-2 Nod Protein LRR; Ligand Recognition CARD RICK I-κB p50 p65 NF-κB Polymorphisms in Nod-2
Key Concept B F. How do peptides get loaded onto the proper kind of MHC molecule?
 Location of MHC class I pockets termed B and F that bind P and P9 amino acid side chains of the peptide Different MHC alleles confer different functional properties on the adaptive immune system by specifying
Location of MHC class I pockets termed B and F that bind P and P9 amino acid side chains of the peptide Different MHC alleles confer different functional properties on the adaptive immune system by specifying
RECAP (1)! In eukaryotes, large primary transcripts are processed to smaller, mature mrnas.! What was first evidence for this precursorproduct
 RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
RECAP (1) In eukaryotes, large primary transcripts are processed to smaller, mature mrnas. What was first evidence for this precursorproduct relationship? DNA Observation: Nuclear RNA pool consists of
p53 and Apoptosis: Master Guardian and Executioner Part 2
 p53 and Apoptosis: Master Guardian and Executioner Part 2 p14arf in human cells is a antagonist of Mdm2. The expression of ARF causes a rapid increase in p53 levels, so what would you suggest?.. The enemy
p53 and Apoptosis: Master Guardian and Executioner Part 2 p14arf in human cells is a antagonist of Mdm2. The expression of ARF causes a rapid increase in p53 levels, so what would you suggest?.. The enemy
Signaling. Dr. Sujata Persad Katz Group Centre for Pharmacy & Health research
 Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
ubiquitinylation succinylation butyrylation hydroxylation crotonylation
 Supplementary Information S1 (table) Overview of histone core-modifications histone, residue/modification H1 H2A methylation acetylation phosphorylation formylation oxidation crotonylation hydroxylation
Supplementary Information S1 (table) Overview of histone core-modifications histone, residue/modification H1 H2A methylation acetylation phosphorylation formylation oxidation crotonylation hydroxylation
Enzyme-coupled Receptors. Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors
 Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Einführung in die Genetik
 Einführung in die Genetik Prof. Dr. Kay Schneitz (EBio Pflanzen) http://plantdev.bio.wzw.tum.de schneitz@wzw.tum.de Prof. Dr. Claus Schwechheimer (PlaSysBiol) http://wzw.tum.de/sysbiol claus.schwechheimer@wzw.tum.de
Einführung in die Genetik Prof. Dr. Kay Schneitz (EBio Pflanzen) http://plantdev.bio.wzw.tum.de schneitz@wzw.tum.de Prof. Dr. Claus Schwechheimer (PlaSysBiol) http://wzw.tum.de/sysbiol claus.schwechheimer@wzw.tum.de
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature13418 Supplementary Results: USP30 opposes autophagic flux In HEK-293 cells, USP30 overexpression increased basal LC3-II levels, dependent on enzymatic activity,
SUPPLEMENTARY INFORMATION doi:10.1038/nature13418 Supplementary Results: USP30 opposes autophagic flux In HEK-293 cells, USP30 overexpression increased basal LC3-II levels, dependent on enzymatic activity,
Chapter 10. Regulatory Strategy
 Chapter 10 Regulatory Strategy Regulation of enzymatic activity: 1. Allosteric Control. Allosteric proteins have a regulatory site(s) and multiple functional sites Activity of proteins is regulated by
Chapter 10 Regulatory Strategy Regulation of enzymatic activity: 1. Allosteric Control. Allosteric proteins have a regulatory site(s) and multiple functional sites Activity of proteins is regulated by
KEY CONCEPT QUESTIONS IN SIGNAL TRANSDUCTION
 Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER
 REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER RNA Splicing Lecture 3, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Splice
REGULATED SPLICING AND THE UNSOLVED MYSTERY OF SPLICEOSOME MUTATIONS IN CANCER RNA Splicing Lecture 3, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Splice
Cell Communication. Chapter 11. Biology Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 11 Cell Communication PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp
Chapter 11 Cell Communication PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp
Life Science 1A Final Exam. January 19, 2006
 ame: TF: Section Time Life Science 1A Final Exam January 19, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use non-erasable
ame: TF: Section Time Life Science 1A Final Exam January 19, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use non-erasable
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
Section 6. Junaid Malek, M.D.
 Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Prof. R. V. Skibbens. Cell Cycle, Cell Division and Cancer (Part 2)
 Prof. R. V. Skibbens November 22, 2010 BIOS 10: BioScience in the 21 st Century Cell Cycle, Cell Division and Cancer (Part 2) Directionality - clocks go in only one direction G1 doesn t have replication-inducing
Prof. R. V. Skibbens November 22, 2010 BIOS 10: BioScience in the 21 st Century Cell Cycle, Cell Division and Cancer (Part 2) Directionality - clocks go in only one direction G1 doesn t have replication-inducing
Chapter 13: Vesicular Traffic
 Chapter 13: Vesicular Traffic Know the terminology: ER, Golgi, vesicle, clathrin, COP-I, COP-II, BiP, glycosylation, KDEL, microtubule, SNAREs, dynamin, mannose-6-phosphate, M6P receptor, endocytosis,
Chapter 13: Vesicular Traffic Know the terminology: ER, Golgi, vesicle, clathrin, COP-I, COP-II, BiP, glycosylation, KDEL, microtubule, SNAREs, dynamin, mannose-6-phosphate, M6P receptor, endocytosis,
Genetic information flows from mrna to protein through the process of translation
 Genetic information flows from mrn to protein through the process of translation TYPES OF RN (RIBONUCLEIC CID) RN s job - protein synthesis (assembly of amino acids into proteins) Three main types: 1.
Genetic information flows from mrn to protein through the process of translation TYPES OF RN (RIBONUCLEIC CID) RN s job - protein synthesis (assembly of amino acids into proteins) Three main types: 1.
PROTEIN TRAFFICKING. Dr. SARRAY Sameh, Ph.D
 PROTEIN TRAFFICKING Dr. SARRAY Sameh, Ph.D Overview Proteins are synthesized either on free ribosomes or on ribosomes bound to endoplasmic reticulum (RER). The synthesis of nuclear, mitochondrial and peroxisomal
PROTEIN TRAFFICKING Dr. SARRAY Sameh, Ph.D Overview Proteins are synthesized either on free ribosomes or on ribosomes bound to endoplasmic reticulum (RER). The synthesis of nuclear, mitochondrial and peroxisomal
Mitosis and the Cell Cycle
 Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
2013 John Wiley & Sons, Inc. All rights reserved. PROTEIN SORTING. Lecture 10 BIOL 266/ Biology Department Concordia University. Dr. S.
 PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
Chapter 31. Completing the Protein Life Cycle: Folding, Processing and Degradation. Biochemistry by Reginald Garrett and Charles Grisham
 Chapter 31 Completing the Protein Life Cycle: Folding, Processing and Degradation Biochemistry by Reginald Garrett and Charles Grisham Essential Question How are newly synthesized polypeptide chains transformed
Chapter 31 Completing the Protein Life Cycle: Folding, Processing and Degradation Biochemistry by Reginald Garrett and Charles Grisham Essential Question How are newly synthesized polypeptide chains transformed
PRC2 crystal clear. Matthieu Schapira
 PRC2 crystal clear Matthieu Schapira Epigenetic mechanisms control the combination of genes that are switched on and off in any given cell. In turn, this combination, called the transcriptional program,
PRC2 crystal clear Matthieu Schapira Epigenetic mechanisms control the combination of genes that are switched on and off in any given cell. In turn, this combination, called the transcriptional program,
Chapter 9. Cellular Signaling
 Chapter 9 Cellular Signaling Cellular Messaging Page 215 Cells can signal to each other and interpret the signals they receive from other cells and the environment Signals are most often chemicals The
Chapter 9 Cellular Signaling Cellular Messaging Page 215 Cells can signal to each other and interpret the signals they receive from other cells and the environment Signals are most often chemicals The
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
C) You find that the Raf kinase is not constitutively active. What was necessary in the previous assay to show any Raf kinase activity?
 PROBLEM SET 3 1. You have obtained immortalized liver cells from a patient who died of Wilson s disease, an inherited disorder of copper metabolism marked by neuronal degeneration and hepatic cirrhosis.
PROBLEM SET 3 1. You have obtained immortalized liver cells from a patient who died of Wilson s disease, an inherited disorder of copper metabolism marked by neuronal degeneration and hepatic cirrhosis.
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001
 BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
BIOLOGY. Cell Communication CAMPBELL. Reece Urry Cain Wasserman Minorsky Jackson. Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick
 CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 11 Cell Communication Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick Cellular Messaging Cells can signal to
CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 11 Cell Communication Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick Cellular Messaging Cells can signal to
The T cell receptor for MHC-associated peptide antigens
 1 The T cell receptor for MHC-associated peptide antigens T lymphocytes have a dual specificity: they recognize polymporphic residues of self MHC molecules, and they also recognize residues of peptide
1 The T cell receptor for MHC-associated peptide antigens T lymphocytes have a dual specificity: they recognize polymporphic residues of self MHC molecules, and they also recognize residues of peptide
BIO 5099: Molecular Biology for Computer Scientists (et al)
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
