|
|
|
- Posy Robbins
- 5 years ago
- Views:
Transcription
1
2
3
4
5
6 Supplementary Table 1. Annotated candidate SNV and indels in melanoma tumors. Patient C1 Patient C2 Patient C3 Patient C4 Patient C5 Patient C6 Patient C7 Patient C10 Chr Position Reference Allele Sample Allele Variation Type Gene Region Gene Symbol Transcript ID Protein Variant Pre Prog Pre* Prog Pre Prog Pre Prog Pre Prog Pre Prog Pre Prog Pre Prog PolyPhen- 2 Conservation Translation SIFT Function Function phylop p- Impact Prediction Prediction value Variant Findings dbsnp ID COSMIC ID RD AF RD AF RD AF RD AF RD AF RD AF RD AF RD AF RD AF RD AF RD AF RD AF RD AF RD AF RD AF RD AF G A SNV Exonic; ncrnacasp9 NM_ p.S270F missense Damaging Probably Dam 1.68E A C SNV Exonic; ncrnacasp9 NM_ p.L34R missense Damaging Probably Dam 2.70E G A SNV Exonic STMN1 NM_ p.P152S missense Tolerated Benign G A SNV Exonic SNRNP40; ZCCNM_ p.S20N missense C T SNV Exonic TIE1 NM_ p.S431F missense Damaging Probably Dam 4.55E C T SNV Exonic TIE1 NM_ p.A1060V missense Damaging Possibly Dama 1.42E G A SNV Exonic PRKAA2 NM_ p.E168K missense Damaging Probably Dam 1.57E G A SNV Exonic LPAR3 NM_ p.R105C missense Tolerated Probably Dam 6.86E C T SNV Exonic COL24A1 NM_ p.R1538Q missense Tolerated Possibly Dama 3.98E C T SNV Splice Site COL24A1 NM_ E G A SNV Exonic COL11A1 NM_ p.P1320S missense Damaging Probably Dam 2.37E G T SNV Exonic NRAS NM_ p.Q61K missense Damaging Possibly Dama 1.19E ; ; 580; C T SNV Exonic NRAS NM_ p.G12D missense Damaging Benign 8.17E ; G A SNV Exonic NGF NM_ p.R83C missense Damaging Benign 1.27E G A SNV Exonic ITGA10 NM_ p.D384N missense Damaging Probably Dam 5.36E GGGGGTGGGCGGGGG Deletion Exonic EFNA3 NM_ p.V71_G74del in- frame C T SNV Exonic INSRR NM_ p.R1157H missense Damaging Probably Dam 2.53E C A SNV Exonic INSRR NM_ p.G380C missense Damaging Probably Dam 3.03E G A SNV Exonic NTRK1 NM_ p.E193K missense Damaging Benign C T SNV Exonic TNN NM_ p.P31L missense Tolerated Benign G A SNV Exonic TNN NM_ p.G870E missense Damaging Probably Dam 1.86E C T SNV Exonic TNN NM_ p.P981S missense Tolerated Benign G A SNV Exonic TNN NM_ p.G1106R missense Damaging Probably Dam 1.67E G A SNV Exonic TNR NM_ p.P1342S missense Damaging Probably Dam 1.16E G A SNV Exonic TNR NM_ p.S1220F missense Damaging Possibly Dama 6.93E C T SNV Exonic TNR NM_ p.D743N missense Tolerated Benign 9.57E C T SNV Exonic CACNA1E NM_ p.S776F missense Damaging Probably Dam 9.14E C T SNV Exonic CACNA1E NM_ p.H1317Y missense Activating Benign 1.18E G A SNV Exonic CACNA1E NM_ p.G1373E missense Damaging Probably Dam 7.38E C A SNV Exonic CACNA1E NM_ p.H1446N missense Tolerated Benign 8.73E C T SNV Exonic CACNA1E NM_ p.S2007L missense Tolerated Benign 4.33E C T SNV Exonic LAMC1 NM_ p.R1087C missense Damaging Benign 1.34E G A SNV Exonic LAMC2 NM_ p.G998E missense Damaging Probably Dam 2.52E G A SNV Exonic CACNA1S NM_ p.S1854F missense Damaging Benign C T SNV Exonic CACNA1S NM_ p.G1724E missense Tolerated Benign A C SNV Exonic CACNA1S NM_ p.V1449G missense Damaging Possibly Dama 1.12E TGTCGTCGTCGTGTCGTCGTCGInsertion Exonic PPP2R5A NM_ p.S2_S3insP in- frame G A SNV Exonic PPP2R5A NM_ p.E69K missense Tolerated Possibly Dama 9.82E C T SNV Exonic AKT3 NM_ p.E17K missense Damaging Probably Dam 2.47E ; ; G A SNV Exonic SOS1 NM_ p.R826* stop gain C T SNV Exonic IL1R1 NM_ p.P85S missense Damaging Probably Dam 2.25E C A SNV Exonic IL1R1 NM_ p.Q125K missense Tolerated Benign G A SNV Exonic G6PC2 NM_ p.D139N missense Tolerated Benign 3.91E G A SNV Exonic G6PC2 NM_ p.G186S missense Damaging Probably Dam 1.11E A G SNV Exonic ITGAV NM_ p.E763G missense Tolerated Benign 4.43E G A SNV Exonic COL3A1 NM_ p.G483R missense Damaging Possibly Dama 4.20E C T SNV Exonic COL3A1 NM_ p.P830L missense Tolerated Benign 5.60E G A SNV Exonic COL3A1 NM_ p.E1096K missense Tolerated Probably Dam 1.12E C A SNV Exonic COL3A1 NM_ p.L1265M missense Damaging Probably Dam 7.28E G A SNV Exonic COL5A2 NM_ p.R1060W missense Damaging Probably Damaging C T SNV Exonic COL5A2 NM_ p.G135E missense Damaging Probably Dam 1.68E G A SNV Exonic IDH1 NM_ p.R132C missense Damaging Benign 1.61E ; C T SNV Exonic FN1 NM_ p.G966R missense Tolerated Benign 5.12E G T SNV Exonic IRS1 NM_ p.S1213R missense Tolerated Benign G A SNV Exonic COL4A3 NM_ p.G366E missense Damaging Probably Dam 4.01E G A SNV Exonic COL4A3 NM_ p.G449E missense Damaging Probably Dam 4.93E G GA Insertion Exonic RAF1 NM_ p.Q601fs* frameshift 6.19E C T SNV Exonic TGFBR2 NM_ p.L386F missense Damaging Probably Dam 4.10E G A SNV Exonic LAMB2 NM_ p.R1626W missense Tolerated Benign G A SNV Exonic CACNA1D NM_ p.V236I missense Damaging Possibly Dama 1.05E T C SNV Exonic CACNA1D NM_ p.C494R missense Tolerated Benign 6.38E G A SNV Exonic; ncrnacacna2d3 NR_ p.a754t missense Tolerated Benign 3.32E C T SNV Exonic; ncrnacacna2d3 NR_ p.p786s missense Tolerated Benign 3.93E C T SNV Exonic; ncrnacacna2d3 NR_ p.p786l missense Damaging Possibly Dama 3.93E C T SNV Exonic FLNB NM_ p.T1193I missense Damaging Probably Dam 5.19E G A SNV Exonic COL6A6 NM_ p.E555K missense Tolerated Benign 8.55E C T SNV Exonic MECOM NM_ p.E104K missense Tolerated Probably Dam 1.77E C T SNV Exonic MECOM NM_ p.E101K missense Tolerated Benign C T SNV Exonic PIK3CA NM_ p.R230* stop gain G A SNV Exonic FGF12 NM_ p.S76L missense Damaging Possibly Dama 2.81E G GA Insertion Exonic PAK2 NM_ p.E146Gfs* unknown 6.25E G A SNV Exonic PDGFRA NM_ p.E34K missense Tolerated Benign 1.47E C T SNV Exonic PDGFRA NM_ p.S942F missense Damaging Possibly Dama 3.18E T C SNV Exonic KDR NM_ p.M1285V missense Tolerated Benign T G SNV Exonic KDR NM_ p.I256L missense Tolerated Benign GAGCAGC GAGCAGCAGCInsertion Exonic FGF5 NM_ p.Q224dup in- frame G A SNV Exonic IBSP NM_ p.G159E missense Activating Benign G A SNV Exonic FGF2; NUDT6 NM_ p.P5S; p.p174s missense Damaging Probably Dam 4.12E G A SNV Exonic; ncrnapdgfc NM_ p.R253C missense Damaging Probably Dam 3.63E A C SNV Exonic RAPGEF2 NM_ p.T1119P missense Tolerated Benign 5.22E C T SNV Exonic RAPGEF2 NM_ p.T1366M missense Tolerated Benign 1.77E C T SNV Exonic SPOCK3 NM_ p.G366E missense Damaging Probably Dam 1.98E CTCATCAT CTCAT Deletion Exonic VEGFC NM_ p.M419del unknown G A SNV Exonic CASP3 NM_ p.R64W missense Damaging Probably Dam 1.47E T C SNV Exonic ADAMTS12 NM_ p.Y1305C missense Tolerated Probably Dam 5.48E G A SNV Exonic PRLR NM_ p.P500L missense Tolerated Benign C T SNV Exonic IL7R NM_ p.L421F missense Tolerated Benign G A SNV Exonic FYB NM_ p.P137L missense Activating Benign G A SNV Exonic GHR NM_ p.E519K missense Tolerated Possibly Dama 1.46E A T SNV Exonic ITGA1 NM_ p.N780I missense Damaging Benign 6.79E G A SNV Exonic ITGA2 NM_ p.G370R missense Tolerated Possibly Dama 6.12E
7 G A SNV Exonic ITGA2 NM_ p.G370E missense Tolerated Possibly Dama 2.62E TCAACAACAACTCAACAACAACDeletion Exonic MAP3K1 NM_ p.T949del in- frame 1.11E G A SNV Exonic THBS4 NM_ p.D739N missense Damaging Probably Dam 8.22E G A SNV Exonic THBS4 NM_ p.D869N missense Damaging Probably Dam 4.51E G A SNV Exonic RASGRF2 NM_ p.W225* stop gain 8.65E A C SNV Exonic RASGRF2 NM_ p.K311Q missense Tolerated Benign 7.46E T A SNV Splice Site TGFBI NM_ E G A SNV Exonic; ncrnacsf1r NM_ p.P347L missense Tolerated Possibly Damaging C T SNV Exonic PDGFRB NM_ p.E141K missense Tolerated Benign C T SNV Exonic COL11A2 NM_ p.G1063E missense Damaging Probably Dam 1.87E G A SNV Exonic LAMA4 NM_ p.R1073* stop gain 6.86E G A SNV Exonic MAP3K5 NM_ p.R1370* stop gain 7.78E C T SNV Exonic TAB2 NM_ p.S588F missense Damaging Probably Dam 2.98E C T SNV Exonic RAC1 NM_ p.P29S missense Damaging Possibly Dama 5.32E ; C T SNV Exonic RAC1 NM_ p.P29L missense Damaging Possibly Dama 5.32E G A SNV Exonic EGFR NM_ p.E967K missense Damaging Probably Dam 1.24E G A SNV Exonic NCF1 NM_ p.S99G missense C T SNV Exonic GNGT1 NM_ p.S34F missense Damaging Probably Dam 2.99E G A SNV Exonic COL1A2 NM_ p.G538D missense Damaging Probably Dam 5.51E C T SNV Exonic GNB2 NM_ p.S97F missense Damaging Possibly Dama 1.57E C T SNV Exonic RELN NM_ p.R3458Q missense Damaging Benign 5.15E G A SNV Exonic RELN NM_ p.P2620L missense Damaging Probably Dam 1.41E A T SNV Exonic LAMB1 NM_ p.V1451D missense Damaging Benign 2.47E C A SNV Exonic LAMB4 NM_ p.G1195V missense Tolerated Benign 5.55E C G SNV Exonic MET NM_ p.F931L missense Tolerated Benign A G SNV Exonic MET NM_ p.N1288S missense Damaging Possibly Dama 8.47E C T SNV Exonic FLNC NM_ p.L50F missense Damaging Probably Dam 4.63E G A SNV Exonic FLNC NM_ p.V978M missense Tolerated Possibly Dama 6.10E CTGGTGGT CTGGT Deletion Exonic CREB3L2 NM_ p.T100del in- frame A T SNV Exonic BRAF NM_ p.V600E missense Damaging Probably Dam 7.87E ; 476; AC TT SNV Exonic BRAF NM_ p.V600K missense Damaging Possibly Dama 6.62E ; C T SNV Exonic NOS3 NM_ p.S260F missense Damaging Probably Dam 3.03E G A SNV Exonic NOS3 NM_ p.E901K missense Tolerated Possibly Dama 4.40E CACAAC CAC Deletion Exonic PPP2CB NM_ p.C269del in- frame 4.07E G A SNV Exonic EIF4EBP1 NM_ p.R73K missense Tolerated Benign G A SNV Exonic NSMAF NM_ p.S484F missense Damaging Possibly Dama 4.15E C T SNV Exonic SNX31 NM_ p.E132K missense Tolerated Benign C T SNV Exonic ANGPT1 NM_ p.D77N missense Damaging Benign 1.13E G A SNV Exonic PTK2 NM_ p.P469S missense Tolerated Benign 1.54E C T SNV Exonic IFNA17 NM_ p.E106K missense Damaging Benign AT ATT Insertion Exonic IFNA17 NM_ p.H57Qfs* frameshift G A SNV Exonic CDKN2A (p16i NM_ p.P114L synonymous; mdamaging Probably Dam 1.99E ; 12476; ; 53077; G A SNV Exonic CDKN2A (p16i NM_ p.P81L synonymous; mdamaging Probably Dam 2.01E ; C T SNV Exonic CDKN2A (p14anm_ p.v51m missense Damaging Probably Dam 2.34E ; T C SNV Exonic PRKACG NM_ p.Y123C missense Damaging Probably Damaging C T SNV Exonic NTRK2 NM_ p.R691C missense Damaging Probably Dam 2.95E G A SNV Exonic SYK NM_ p.G514R missense Damaging Probably Dam 1.34E TAAAA TAAAAAA Insertion Exonic TNC NM_ p.I1505fs* frameshift G A SNV Exonic TLR4 NM_ p.G111E missense Damaging Probably Dam 1.21E C T SNV Exonic TLR4 NM_ p.H256Y missense Activating Benign G A SNV Exonic TLR4 NM_ p.E445K missense Tolerated Benign G A SNV Exonic PPP6C NM_ p.P168S missense Damaging Probably Dam 1.14E C T SNV Exonic TSC1 NM_ p.V127I missense Tolerated Benign G A SNV Exonic COL5A1 NM_ p.G54R missense Damaging Probably Dam 2.05E G A SNV Exonic COL5A1 NM_ p.D854N missense Damaging Probably Dam 8.95E G A SNV Exonic CACNA1B NM_ p.G633R missense Damaging Probably Dam 2.49E C T SNV Exonic CACNA1B NM_ p.P2143S missense Tolerated C T SNV Exonic IL2RA NM_ p.G155R missense Damaging Probably Dam 4.13E T A SNV Exonic ITGA8 NM_ p.I921L missense Tolerated Benign C T SNV Exonic ITGA8 NM_ p.A149T missense Damaging Probably Dam 4.45E G A SNV Splice Site CACNB2 NM_ E CATTATT CATT Deletion Exonic PTEN NM_ p.I33del in- frame 4.66E ; 4972; CAAAAA CAAAAAA Insertion Exonic PTEN NM_ p.V166fs* frameshift 3.00E C T SNV Exonic PTEN NM_ p.P169S missense Damaging Probably Dam 1.64E C T SNV Exonic NFKB2 NM_ p.P430S missense Tolerated Benign C T SNV Exonic; ncrnafgfr2 NM_ p.R626Q missense Damaging Probably Dam 7.80E C T SNV Exonic; ncrnafgfr2 NM_ p.G489E missense Damaging Probably Dam 5.98E T C SNV Exonic RRAS2 NM_ p.Y105C missense Tolerated Possibly Damaging C T SNV Exonic PTPN5 NM_ p.D534N missense Damaging Probably Dam 9.98E C T SNV 5'UTR; Exonic BDNF NM_ p.R13K missense Tolerated Benign 2.40E G A SNV Exonic TRAF6 NM_ p.P183L missense Tolerated Benign AGG AGGG Insertion Exonic CREB3L1 NM_ p.D509fs frameshift 3.08E G A SNV Exonic TNKS1BP1 NM_ p.P1386S missense Tolerated Benign G A SNV Splice Site PPP2R5B NM_ E C T SNV Exonic CACNA2D4 NM_ p.W685* stop gain 1.50E C T SNV Exonic CACNA2D4 NM_ p.R522Q missense Tolerated Benign 2.28E ; G A SNV Exonic CACNA2D4 NM_ p.P413L missense Tolerated Benign G A SNV Exonic CACNA1C NM_ p.G1053E missense Damaging Probably Dam 2.87E G C SNV Exonic CACNA1C NM_ p.E1392Q missense Activating Benign 2.51E C T SNV Exonic FGF6 NM_ p.E119K missense Damaging Probably Dam 7.05E G A SNV Exonic GNB3 NM_ p.R251H missense Tolerated Possibly Dama 3.95E C T SNV Exonic GYS2 NM_ p.G338R missense Damaging Probably Dam 1.14E C T SNV Exonic EIF4B NM_ p.S585F missense Damaging Benign 7.59E A C SNV Exonic MDM2 NM_ p.Q414H missense Tolerated Probably Damaging G A SNV Exonic PTPRR NM_ p.S342F missense Damaging Probably Dam 3.08E G A SNV Exonic KITLG NM_ p.Q243* stop gain 3.82E C T SNV Exonic RPL6 NM_ p.D120N missense Damaging Possibly Dama 3.96E G A SNV Exonic TAOK3 NM_ p.R306* stop gain 1.30E C T SNV Exonic FLT1 NM_ p.E1091K missense Damaging Probably Dam 6.81E G A SNV Exonic FLT1 NM_ p.A80V missense Tolerated Benign T A SNV Exonic RB1 NM_ p.I227N missense Damaging Probably Dam 4.52E G A SNV Splice Site RB1 NM_ E C T SNV Exonic RB1 NM_ p.S463L missense Damaging Probably Dam 4.54E G A SNV Splice Site RB1 NM_ E C T SNV Exonic RB1 NM_ p.R656W missense Tolerated Benign 6.08E G A SNV Exonic COL4A1 NM_ p.P991S missense Tolerated Probably Dam 8.69E G A SNV Exonic COL4A1 NM_ p.S967F missense Tolerated Benign G A SNV Exonic COL4A1 NM_ p.S628F missense Tolerated Benign
8 G A SNV Exonic COL4A2 NM_ p.G720S missense Damaging Probably Dam 2.14E C T SNV Exonic COL4A2 NM_ p.S829F missense Tolerated Probably Damaging T TA Insertion Exonic PCK2 NM_ p.I237Rfs* frameshift 1.99E G C SNV Exonic SOS2 NM_ p.Q178E missense Damaging Benign 7.91E C T SNV Exonic; ncrnatcl1a NM_ p.D105N missense Damaging Probably Dam 6.68E C T SNV Exonic RASGRP1 NM_ p.E319K missense Damaging Possibly Dama 8.36E A T SNV Exonic CASC5 NM_ p.T601S missense Tolerated Probably Damaging G T SNV Exonic CASC5 NM_ p.E1800* stop gain 1.74E G T SNV Splice Site TYRO3 NM_ E G A SNV Exonic GNB5 NM_ p.A263V missense Tolerated Possibly Dama 2.05E C A SNV Exonic MAP2K1 NM_ p.P124Q missense Damaging Probably Dam 1.19E C T SNV Exonic ITGA11 NM_ p.W390* stop gain 5.61E G A SNV Exonic NTHL1 NM_ p.R21W missense Damaging Benign C T SNV Exonic PRKCB NM_ p.P638S missense Tolerated Benign 1.49E C T SNV Exonic CD19 NM_ p.P376S missense Tolerated Benign G A SNV Exonic TAOK2 NM_ p.V959M missense Damaging Possibly Dama 6.01E G A SNV Exonic RBL2 NM_ p.E478K missense Tolerated Benign 2.08E C T SNV Exonic SDR42E1 NM_ p.R92Q missense Tolerated Benign A C SNV Exonic TP53 NM_ p.V218G missense Damaging Probably Dam 2.74E ; T A SNV Exonic VTN NM_ p.R324S missense Tolerated Benign C T SNV Exonic ERBB2 NM_ p.P617S missense Tolerated Benign G T SNV Exonic ITGA2B NM_ p.A788D missense Damaging Probably Dam 8.91E G A SNV Exonic ITGA2B NM_ p.Q642* stop gain 6.50E C T SNV Exonic MAP3K14 NM_ p.G248R missense Benign G A SNV Splice Site ITGB3 NM_ E C T SNV Exonic NGFR NM_ p.S24F missense Tolerated Benign C T SNV Exonic NGFR NM_ p.P370L missense Damaging Benign C T SNV Exonic ITGA3 NM_ p.P354L missense Tolerated Benign C T SNV Exonic COL1A1 NM_ p.G233E missense Damaging Probably Dam 6.93E C T SNV Exonic COL1A1 NM_ p.R220Q missense Tolerated Probably Dam 7.41E C T SNV Exonic CACNA1G NM_ p.S111F missense Damaging Possibly Dama 2.14E G A SNV Exonic TUBD1; RPS6KNM_ p.D37N missense Tolerated Benign 3.61E C T SNV Exonic CSH2 NM_ p.D197N missense Tolerated Benign A G SNV Exonic GH2 NM_ p.W241R synonymous; mactivating Benign G A SNV Exonic CACNG4 NM_ p.R171Q missense Damaging Possibly Damaging A T SNV Exonic MAP2K6 NM_ p.I56L missense Tolerated Probably Dam 1.77E G A SNV Exonic ITGB4 NM_ p.M1447I missense Tolerated Benign 3.30E G A SNV Exonic LAMA3 NM_ p.R706Q missense Damaging Possibly Dama 1.60E C T SNV Exonic MKNK2 NM_ p.D32N missense Tolerated Benign C G SNV Exonic MAP2K2 NM_ p.C125S missense Damaging Probably Dam 2.15E G A SNV Exonic CREB3L3 NM_ p.G397R missense Tolerated Benign C T SNV Exonic CACNA1A NM_ p.M1587I missense Damaging Benign 3.24E C T SNV Exonic PKN1 NM_ p.R543W missense Tolerated Possibly Damaging C T SNV Exonic MAP1S NM_ p.P760L missense Damaging Benign C G SNV Exonic COMP NM_ p.G645R missense Damaging Probably Dam 3.51E G A SNV Exonic AXL NM_ p.E818K missense Damaging Probably Dam 1.32E G A SNV Exonic COL6A2 NM_ p.V158M missense Damaging Probably Dam 7.83E C T SNV Exonic RAC2 NM_ p.G10E missense Damaging Possibly Dama 1.11E C T SNV Exonic PLA2G6 NM_ p.A664T missense Damaging Benign 2.30E G A SNV Exonic MAPK12 NM_ p.P282S missense Tolerated Benign 3.87E- 03 X C T SNV Exonic VCX3B (includenm_ p.v170m missense Tolerated Benign X C T SNV Exonic VCX3B (includenm_ p.v160m missense Tolerated Benign X C T SNV Exonic VCX3B (includenm_ p.v120m missense Tolerated Benign X C T SNV Exonic VCX3B (includenm_ p.a17t missense Activating Benign X G T SNV Exonic VCX3B (includenm_ p.t15k missense Tolerated Benign X C G SNV Exonic VCX3B (includenm_ p.a70g missense Activating Benign X G A SNV Exonic VCX3B (includenm_ p.e162k missense Damaging Possibly Dama 1.56E- 03 X A G SNV Exonic VCX3B (includenm_ p.m180v missense Tolerated Benign X A G SNV Exonic VCX3B (includenm_ p.d30g missense Tolerated Benign X C T SNV Exonic CACNA1F NM_ p.D892N missense Damaging Possibly Dama 1.84E- 04 X TTCCTCCTCCTCTTCCTCCTCCTCInsertion Exonic CACNA1F NM_ p.E825dup in- frame X G A SNV Exonic MED12 NM_ p.G1441R missense Damaging Probably Dam 8.83E- 06 X G A SNV Exonic COL4A6 NM_ p.P1378L missense Tolerated Possibly Dama 1.69E- 03 X C T SNV Exonic COL4A6 NM_ p.D1289N missense Tolerated Benign 3.01E X G A SNV Exonic COL4A6 NM_ p.P659S missense Tolerated Benign X C T SNV Exonic COL4A6 NM_ p.G569D missense Tolerated Probably Dam 3.52E- 04 X C T SNV Exonic COL4A6 NM_ p.G458S missense Damaging Probably Dam 4.72E- 04 X G A SNV Exonic COL4A5 NM_ p.G126R missense Damaging Probably Dam 2.48E- 06 X G A SNV Exonic COL4A5 NM_ p.G1669R missense Damaging 1.00E- 06 X C T SNV Exonic MTCP1 NM_ p.E75K missense Tolerated Benign RD, Read Depth; AF, Allele Frequency; Chr, Chromosome *The EDT tumor was used as the Pre for Patient C2
9 Supplementary Table 2. Amplification and Sequencing Primers Forward Reverse BRAF GGCTCTCGGTTATAAGATGGC ACAGGAAACGCACCATATCC MEK1 CGTTACCCGGGTCCAAAATG CTTTGTCACAGGTGAAATGC CATGGATGGAGGTTCTCTGG AGGGCTTGACATCTCTGTGC MEK2 CTCCCGGCCCGCCCCCTATG GTGGAGGCGCCAGCCTGTCC GTCAGCATCGCGGTTCTCC TCACCCCGAAGTCACACAG MEK1/2 TGCAGAAGAAGCTGGAGGAGC N-RAS AGCTTGAGGTTCTTGCTGGT TCAGGACCAGGGTGTCAGTG AKT1 AGCGCCAGCCTGAGAGGA TCTCCATCCCTCCAAGCTAT GACAGGTGGAAGAACAGCT
10 Supplementary Table 3. Exome sequencing data summary Patient Tumor % coverage of Mean read depth target regions of target regions (>10x) C1 Pre Prog C2 Pre* Prog C3 Pre Prog C4 Pre Prog C5 Pre Prog C6 Pre Prog C7 Pre Prog C10 Pre Prog *The EDT tumor was used as the Pre for Patient C2
UNIVERSITY OF TORINO DEPARTMENT OF ONCOLOGY. Giorgio V. Scagliotti University of Torino Dipartment of Oncology
 Giorgio V. Scagliotti University of Torino Dipartment of Oncology giorgio.scagliotti@unito.it Molecular landscape of MM not fully characterized to allow personalized treatment Recurrent genetic alterations
Giorgio V. Scagliotti University of Torino Dipartment of Oncology giorgio.scagliotti@unito.it Molecular landscape of MM not fully characterized to allow personalized treatment Recurrent genetic alterations
Frequency(%) KRAS G12 KRAS G13 KRAS A146 KRAS Q61 KRAS K117N PIK3CA H1047 PIK3CA E545 PIK3CA E542K PIK3CA Q546. EGFR exon19 NFS-indel EGFR L858R
 Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
B16F1 B16F10 Supplemental Figure S1
 B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies
 Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Systematic investigation of cancer-associated somatic point mutations in SNP databases HyunChul Jung 1,2, Thomas Bleazard 3, Jongkeun Lee 1 and Dongwan Hong 1 1. Cancer Genomics
SUPPLEMENTARY INFORMATION Systematic investigation of cancer-associated somatic point mutations in SNP databases HyunChul Jung 1,2, Thomas Bleazard 3, Jongkeun Lee 1 and Dongwan Hong 1 1. Cancer Genomics
Jennifer Hauenstein Oncology Cytogenetics Emory University Hospital Atlanta, GA
 Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Illumina s Cancer Research Portfolio and Dedicated Workflows
 Illumina s Cancer Research Portfolio and Dedicated Workflows Michael Sohn Clinical Sales Specialist Spain&Italy 2017 2017 Illumina, Inc. All rights reserved. Illumina, 24sure, BaseSpace, BeadArray, BlueFish,
Illumina s Cancer Research Portfolio and Dedicated Workflows Michael Sohn Clinical Sales Specialist Spain&Italy 2017 2017 Illumina, Inc. All rights reserved. Illumina, 24sure, BaseSpace, BeadArray, BlueFish,
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study. February 2018
 Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Insights from Sequencing the Melanoma Exome
 Insights from Sequencing the Melanoma Exome Michael Krauthammer, MD PhD, December 2 2015 Yale University School Yof Medicine 1 2012 Exome Screens and Results Exome Sequencing of 108 sun-exposed melanomas
Insights from Sequencing the Melanoma Exome Michael Krauthammer, MD PhD, December 2 2015 Yale University School Yof Medicine 1 2012 Exome Screens and Results Exome Sequencing of 108 sun-exposed melanomas
Genomic Medicine: What every pathologist needs to know
 Genomic Medicine: What every pathologist needs to know Stephen P. Ethier, Ph.D. Professor, Department of Pathology and Laboratory Medicine, MUSC Director, MUSC Center for Genomic Medicine Genomics and
Genomic Medicine: What every pathologist needs to know Stephen P. Ethier, Ph.D. Professor, Department of Pathology and Laboratory Medicine, MUSC Director, MUSC Center for Genomic Medicine Genomics and
Accel-Amplicon Panels
 Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
SureSelect Cancer All-In-One Custom and Catalog NGS Assays
 SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
IntelliGENSM. Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community.
 IntelliGENSM Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community. NGS TRANSFORMS GENOMIC TESTING Background Cancers may emerge as a result of somatically
IntelliGENSM Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community. NGS TRANSFORMS GENOMIC TESTING Background Cancers may emerge as a result of somatically
The Promise of Epilepsy Genetics A Personal & Scientific Perspective December 3, 2012
 The Promise of Epilepsy Genetics A Personal & Scientific Perspective December 3, 2012 Tracy Dixon-Salazar, Ph.D. University of California, San Diego American Epilepsy Society Annual Meeting 1 Disclosure
The Promise of Epilepsy Genetics A Personal & Scientific Perspective December 3, 2012 Tracy Dixon-Salazar, Ph.D. University of California, San Diego American Epilepsy Society Annual Meeting 1 Disclosure
Supplementary Information
 Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making
 Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making November 20, 2014 Capturing Value in Next Generation Sequencing Symposium Douglas Johnson MD, MSCI Vanderbilt-Ingram
Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making November 20, 2014 Capturing Value in Next Generation Sequencing Symposium Douglas Johnson MD, MSCI Vanderbilt-Ingram
The PI3K/AKT Signaling Pathway: associations of mirnas with dysregulated gene expression in colorectal cancer
 1 RESEARCH ARTICLE The PI3K/AKT Signaling Pathway: associations of mirnas with dysregulated gene expression in colorectal cancer Martha L. Slattery 1, Lila E. Mullany 1, Lori C. Sakoda 2, Roger K. Wolff
1 RESEARCH ARTICLE The PI3K/AKT Signaling Pathway: associations of mirnas with dysregulated gene expression in colorectal cancer Martha L. Slattery 1, Lila E. Mullany 1, Lori C. Sakoda 2, Roger K. Wolff
6/12/2018. Disclosures. Clinical Genomics The CLIA Lab Perspective. Outline. COH HopeSeq Heme Panels
 Clinical Genomics The CLIA Lab Perspective Disclosures Raju K. Pillai, M.D. Hematopathologist / Molecular Pathologist Director, Pathology Bioinformatics City of Hope National Medical Center, Duarte, CA
Clinical Genomics The CLIA Lab Perspective Disclosures Raju K. Pillai, M.D. Hematopathologist / Molecular Pathologist Director, Pathology Bioinformatics City of Hope National Medical Center, Duarte, CA
Supplementary Table S1: Primers for DNA sequencing (A), quantification of. relative mrna expression (B), and copy number analysis (C)
 Supplementary Table S1: Primers for DNA sequencing (A), quantification of relative mrna expression (B), and copy number analysis (C) A. DNA sequencing (1-4) Genes Primers Sequence EGFR exon 18 EGFR E18F
Supplementary Table S1: Primers for DNA sequencing (A), quantification of relative mrna expression (B), and copy number analysis (C) A. DNA sequencing (1-4) Genes Primers Sequence EGFR exon 18 EGFR E18F
Fluxion Biosciences and Swift Biosciences Somatic variant detection from liquid biopsy samples using targeted NGS
 APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
Characterisation of structural variation in breast. cancer genomes using paired-end sequencing on. the Illumina Genome Analyser
 Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
5 th July 2016 ACGS Dr Michelle Wood Laboratory Genetics, Cardiff
 5 th July 2016 ACGS Dr Michelle Wood Laboratory Genetics, Cardiff National molecular screening of patients with lung cancer for a national trial of multiple novel agents. 2000 NSCLC patients/year (late
5 th July 2016 ACGS Dr Michelle Wood Laboratory Genetics, Cardiff National molecular screening of patients with lung cancer for a national trial of multiple novel agents. 2000 NSCLC patients/year (late
Next Generation Sequencing: What Will it Mean for the Pathologist?
 Next Generation Sequencing: What Will it Mean for the Pathologist? Christopher Corless, MD, PhD Professor of Pathology, Oregon Health & Science University Chief Medical Officer, Knight Diagnostic Laboratories
Next Generation Sequencing: What Will it Mean for the Pathologist? Christopher Corless, MD, PhD Professor of Pathology, Oregon Health & Science University Chief Medical Officer, Knight Diagnostic Laboratories
EXAMPLE. - Potentially responsive to PI3K/mTOR and MEK combination therapy or mtor/mek and PKC combination therapy. ratio (%)
 Dr Kate Goodhealth Goodhealth Medical Clinic 123 Address Road SUBURBTOWN NSW 2000 Melanie Citizen Referring Doctor Your ref Address Dr John Medico 123 Main Street, SUBURBTOWN NSW 2000 Phone 02 9999 9999
Dr Kate Goodhealth Goodhealth Medical Clinic 123 Address Road SUBURBTOWN NSW 2000 Melanie Citizen Referring Doctor Your ref Address Dr John Medico 123 Main Street, SUBURBTOWN NSW 2000 Phone 02 9999 9999
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester
 Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
Supplementary Figure 1. Copy Number Alterations TP53 Mutation Type. C-class TP53 WT. TP53 mut. Nature Genetics: doi: /ng.
 Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
COSMIC - Catalogue of Somatic Mutations in Cancer
 COSMIC - Catalogue of Somatic Mutations in Cancer http://cancer.sanger.ac.uk/cosmic https://academic.oup.com/nar/articl e-lookup/doi/10.1093/nar/gkw1121 Data In Large-scale systematic screens Detailed
COSMIC - Catalogue of Somatic Mutations in Cancer http://cancer.sanger.ac.uk/cosmic https://academic.oup.com/nar/articl e-lookup/doi/10.1093/nar/gkw1121 Data In Large-scale systematic screens Detailed
Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD
 Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD Neuropathology Fellow Division of Neuropathology Center for Personalized Diagnosis (CPD) Glial
Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD Neuropathology Fellow Division of Neuropathology Center for Personalized Diagnosis (CPD) Glial
Shashikant Kulkarni, M.S (Medicine)., Ph.D., FACMG Associate Professor of Pathology & Immunology Associate Professor of Pediatrics and Genetics
 Shashikant Kulkarni, M.S (Medicine)., Ph.D., FACMG Associate Professor of Pathology & Immunology Associate Professor of Pediatrics and Genetics Director of Cytogenomics and Molecular Pathology Evidence-based
Shashikant Kulkarni, M.S (Medicine)., Ph.D., FACMG Associate Professor of Pathology & Immunology Associate Professor of Pediatrics and Genetics Director of Cytogenomics and Molecular Pathology Evidence-based
Next generation sequencing on FFPE tumor tissues for lung and colorectal carcinoma diagnostics and research Sakari Knuutila
 Next generation sequencing on FFPE tumor tissues for lung and colorectal carcinoma diagnostics and research Sakari Knuutila Faculty of Medicine / Haartman Institute / Department of Pathology / Sakari Knuutila
Next generation sequencing on FFPE tumor tissues for lung and colorectal carcinoma diagnostics and research Sakari Knuutila Faculty of Medicine / Haartman Institute / Department of Pathology / Sakari Knuutila
Clinical Grade Genomic Profiling: The Time Has Come
 Clinical Grade Genomic Profiling: The Time Has Come Gary Palmer, MD, JD, MBA, MPH Senior Vice President, Medical Affairs Foundation Medicine, Inc. Oct. 22, 2013 1 Why We Are Here A Shared Vision At Foundation
Clinical Grade Genomic Profiling: The Time Has Come Gary Palmer, MD, JD, MBA, MPH Senior Vice President, Medical Affairs Foundation Medicine, Inc. Oct. 22, 2013 1 Why We Are Here A Shared Vision At Foundation
The Role of Next Generation Sequencing in Solid Tumor Mutation Testing
 The Role of Next Generation Sequencing in Solid Tumor Mutation Testing Allie H. Grossmann MD PhD Department of Pathology, University of Utah Division of Anatomic Pathology & Oncology, ARUP Laboratories
The Role of Next Generation Sequencing in Solid Tumor Mutation Testing Allie H. Grossmann MD PhD Department of Pathology, University of Utah Division of Anatomic Pathology & Oncology, ARUP Laboratories
Exons from 137 genes were targeted for hybrid selection and massively parallel sequencing.
 SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
Reporting TP53 gene analysis results in CLL
 Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
BWA alignment to reference transcriptome and genome. Convert transcriptome mappings back to genome space
 Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
An Integrative Pharmacogenomic Approach Identifies Two-drug. Combination Therapies for Personalized Cancer Medicine
 An Integrative Pharmacogemic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine Yin Liu 1,2,3, Teng Fei 4,5, Xiaoqi Zheng 6, Myles Brown 4,5, Peng Zhang 7*, X. Shirley
An Integrative Pharmacogemic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine Yin Liu 1,2,3, Teng Fei 4,5, Xiaoqi Zheng 6, Myles Brown 4,5, Peng Zhang 7*, X. Shirley
CITATION FILE CONTENT/FORMAT
 CITATION For any resultant publications using please cite: Matthew A. Field, Vicky Cho, T. Daniel Andrews, and Chris C. Goodnow (2015). "Reliably detecting clinically important variants requires both combined
CITATION For any resultant publications using please cite: Matthew A. Field, Vicky Cho, T. Daniel Andrews, and Chris C. Goodnow (2015). "Reliably detecting clinically important variants requires both combined
The Center for PERSONALIZED DIAGNOSTICS
 The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Next generation histopathological diagnosis for precision medicine in solid cancers
 Next generation histopathological diagnosis for precision medicine in solid cancers from genomics to clinical application Aldo Scarpa ARC-NET Applied Research on Cancer Department of Pathology and Diagnostics
Next generation histopathological diagnosis for precision medicine in solid cancers from genomics to clinical application Aldo Scarpa ARC-NET Applied Research on Cancer Department of Pathology and Diagnostics
Dr Yvonne Wallis Consultant Clinical Scientist West Midlands Regional Genetics Laboratory
 Dr Yvonne Wallis Consultant Clinical Scientist West Midlands Regional Genetics Laboratory Personalised Therapy/Precision Medicine Selection of a therapeutic drug based on the presence or absence of a specific
Dr Yvonne Wallis Consultant Clinical Scientist West Midlands Regional Genetics Laboratory Personalised Therapy/Precision Medicine Selection of a therapeutic drug based on the presence or absence of a specific
APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST PROSPECTIVE
 AMP COMPANION MEETING SYMPOSIUM AT USCAP 2015 NEXT-GENERATION OF PATHOLOGY: ROLE OF PATHOLOGIST IN NGS-BASED PERSONALIZED MEDICINE APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST
AMP COMPANION MEETING SYMPOSIUM AT USCAP 2015 NEXT-GENERATION OF PATHOLOGY: ROLE OF PATHOLOGIST IN NGS-BASED PERSONALIZED MEDICINE APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST
Next generation diagnostics Bringing high-throughput sequencing into clinical application
 Next generation diagnostics Bringing high-throughput sequencing into clinical application Leonardo A. Meza-Zepeda, PhD Translational Genomics Group Institute for Cancer Research Leonardo.Meza-Zepeda@rr-research.no
Next generation diagnostics Bringing high-throughput sequencing into clinical application Leonardo A. Meza-Zepeda, PhD Translational Genomics Group Institute for Cancer Research Leonardo.Meza-Zepeda@rr-research.no
Variant Classification. Author: Mike Thiesen, Golden Helix, Inc.
 Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Supplementary Material. Part I: Sample Information. Part II: Pathway Information
 Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Frampton et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Supplementary Figure 1A Base substitution detection
SUPPLEMENTARY INFORMATION Frampton et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Supplementary Figure 1A Base substitution detection
MET skipping mutation, EGFR
 New NSCLC biomarkers in clinical research: detection of MET skipping mutation, EGFR T790M, and other important biomarkers Fernando López-Ríos Laboratorio de Dianas Terapéuticas Hospital Universitario HM
New NSCLC biomarkers in clinical research: detection of MET skipping mutation, EGFR T790M, and other important biomarkers Fernando López-Ríos Laboratorio de Dianas Terapéuticas Hospital Universitario HM
Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ
 Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ G E NETIC T E CHNOLOGIST MEDICAL G E NETICS, CARDIFF To Cover Background to the project Choice of panel Validation process Genes on panel, Protocol
Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ G E NETIC T E CHNOLOGIST MEDICAL G E NETICS, CARDIFF To Cover Background to the project Choice of panel Validation process Genes on panel, Protocol
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:.38/nature83 Supplementary figure An Overview of Experimental Flow Hypothesis: The tumor microenvironment has a significant impact on cancer cell chemoresistance. Screen Coculture
SUPPLEMENTARY INFORMATION doi:.38/nature83 Supplementary figure An Overview of Experimental Flow Hypothesis: The tumor microenvironment has a significant impact on cancer cell chemoresistance. Screen Coculture
CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE
 CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE is_clinical dbsnp MITO GENE chr1 13273 G C heterozygous - - -. - DDX11L1 chr1 949654 A G Homozygous 52 - - rs8997 - ISG15 chr1 1021346 A G heterozygous
CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE is_clinical dbsnp MITO GENE chr1 13273 G C heterozygous - - -. - DDX11L1 chr1 949654 A G Homozygous 52 - - rs8997 - ISG15 chr1 1021346 A G heterozygous
Secuenciación masiva: papel en la toma de decisiones
 Secuenciación masiva: papel en la toma de decisiones Cancer is a Genetic Disease Development of cancer is driven by the acquisition of somatic genetic alterations: Nonsynonymous point mutations: missense.
Secuenciación masiva: papel en la toma de decisiones Cancer is a Genetic Disease Development of cancer is driven by the acquisition of somatic genetic alterations: Nonsynonymous point mutations: missense.
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each
 Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Oncomine Focus assay panel and Oncomine Knowledgebase Reporter.
 Oncomine Focus assay panel and Oncomine Knowledgebase Reporter. How it can help to identify relevant alteration and early phase trials. Dr Isabelle SOUBEYRAN Dr Emmanuel KHALIFA Molecular Pathology Unit
Oncomine Focus assay panel and Oncomine Knowledgebase Reporter. How it can help to identify relevant alteration and early phase trials. Dr Isabelle SOUBEYRAN Dr Emmanuel KHALIFA Molecular Pathology Unit
Personalised cancer care Information for Medical Specialists. A new way to unlock treatment options for your patients
 Personalised cancer care Information for Medical Specialists A new way to unlock treatment options for your patients Contents Optimised for clinical benefit 4 Development history 4 Full FIND IT panel vs
Personalised cancer care Information for Medical Specialists A new way to unlock treatment options for your patients Contents Optimised for clinical benefit 4 Development history 4 Full FIND IT panel vs
Nature Genetics: doi: /ng Supplementary Figure 1. Depths and coverages in whole-exome and targeted deep sequencing data.
 Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Supplementary Figure 1 Depths and coverages in whole-exome and targeted deep sequencing data. Depth (top) and coverage (bottom) of whole-exome sequencing for 38 independent JPN cases (mean depth = 130)
Developments in small cell lung cancer G. Giaccone, MD PhD Chief, Medical Oncology Branch and Affiliates National Cancer Institute Bethesda MD USA
 Developments in small cell lung cancer G. Giaccone, MD PhD Chief, Medical Oncology Branch and Affiliates National Cancer Institute Bethesda MD USA Geneva April 20, 2012 Neuroendocrine tumors of lung Typical
Developments in small cell lung cancer G. Giaccone, MD PhD Chief, Medical Oncology Branch and Affiliates National Cancer Institute Bethesda MD USA Geneva April 20, 2012 Neuroendocrine tumors of lung Typical
Diagnostica Molecolare!
 Diagnostica Molecolare! Aldo Scarpa Unità Diagnostica Molecolare Azienda Ospedaliera Universitaria Integrata di Verona e ARC-NET Centro di Ricerca Applicata sul Cancro PDTA CARCINOMA POLMONARE - IL PAZIENTE
Diagnostica Molecolare! Aldo Scarpa Unità Diagnostica Molecolare Azienda Ospedaliera Universitaria Integrata di Verona e ARC-NET Centro di Ricerca Applicata sul Cancro PDTA CARCINOMA POLMONARE - IL PAZIENTE
Supplementary Figure 1. Estimation of tumour content
 Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes
 Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes KM Kortüm 1,8 *, EK Mai 3,4 *, NH Hanafiah 3 *, CX Shi 1, YX Zhu 1, L Bruins 1, S Barrio 1,
Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes KM Kortüm 1,8 *, EK Mai 3,4 *, NH Hanafiah 3 *, CX Shi 1, YX Zhu 1, L Bruins 1, S Barrio 1,
MSI positive MSI negative
 Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
The mutations that drive cancer. Paul Edwards. Department of Pathology and Cancer Research UK Cambridge Institute, University of Cambridge
 The mutations that drive cancer Paul Edwards Department of Pathology and Cancer Research UK Cambridge Institute, University of Cambridge Previously on Cancer... hereditary predisposition Normal Cell Slightly
The mutations that drive cancer Paul Edwards Department of Pathology and Cancer Research UK Cambridge Institute, University of Cambridge Previously on Cancer... hereditary predisposition Normal Cell Slightly
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Identification of genomic alterations in cervical cancer biopsies by exome sequencing
 Chapter- 4 Identification of genomic alterations in cervical cancer biopsies by exome sequencing 105 4.1 INTRODUCTION Athough HPV has been identified as the prime etiological factor for cervical cancer,
Chapter- 4 Identification of genomic alterations in cervical cancer biopsies by exome sequencing 105 4.1 INTRODUCTION Athough HPV has been identified as the prime etiological factor for cervical cancer,
Webinar Series. Characterizing cancers from liquid biopsies and FFPE samples: The rise of targeted sequencing panels. Participating experts
 Characterizing cancers from liquid biopsies and FFPE samples: The rise of targeted sequencing panels April 13, 2016 Webinar Series Brought to you by the Science/ AAAS Custom Publishing Office Participating
Characterizing cancers from liquid biopsies and FFPE samples: The rise of targeted sequencing panels April 13, 2016 Webinar Series Brought to you by the Science/ AAAS Custom Publishing Office Participating
What is the status of the technologies of "precision medicine?
 Session 2: What is the status of the technologies of "precision medicine? Gideon Blumenthal, MD, Clinical Team Leader, Thoracic and Head/Neck Oncology, Center for Drug Evaluation and Research (CDER), U.S.
Session 2: What is the status of the technologies of "precision medicine? Gideon Blumenthal, MD, Clinical Team Leader, Thoracic and Head/Neck Oncology, Center for Drug Evaluation and Research (CDER), U.S.
The linking of specific cancer genetic alterations to molecular targeted therapies is driving a new era of personalised medicine
 The linking of specific cancer genetic alterations to molecular targeted therapies is driving a new era of personalised medicine Oncologica addresses this new era of precision medicine by exploiting state
The linking of specific cancer genetic alterations to molecular targeted therapies is driving a new era of personalised medicine Oncologica addresses this new era of precision medicine by exploiting state
Sample Report. Result: POSITIVE Mutations Detected: NRAS-G13V, TP53-R282W, TP53-G245D, TP53-R175H, BRAF-V600M, MSI-High
 Sample Report PATIENT INFORMATION Name: DOB: Age: Sex: Address: Doe, John 09/04/1969 49 Male South Plainfield, NJ, 07080 SAMPLE INFORMATION Date Collected: Date Received: Date of Report: lab ID: Testing
Sample Report PATIENT INFORMATION Name: DOB: Age: Sex: Address: Doe, John 09/04/1969 49 Male South Plainfield, NJ, 07080 SAMPLE INFORMATION Date Collected: Date Received: Date of Report: lab ID: Testing
MEDICAL GENOMICS LABORATORY. Non-NF1 RASopathy panel by Next-Gen Sequencing and Deletion/Duplication Analysis of SPRED1 (NNP-NG)
 Non-NF1 RASopathy panel by Next-Gen Sequencing and Deletion/Duplication Analysis of SPRED1 (NNP-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with
Non-NF1 RASopathy panel by Next-Gen Sequencing and Deletion/Duplication Analysis of SPRED1 (NNP-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with
TP53 mutational profile in CLL : A retrospective study of the FILO group.
 TP53 mutational profile in CLL : A retrospective study of the FILO group. Fanny Baran-Marszak Hopital Avicenne Bobigny France 2nd ERIC workshop on TP53 analysis in CLL, Stresa 2017 TP53 abnormalities :
TP53 mutational profile in CLL : A retrospective study of the FILO group. Fanny Baran-Marszak Hopital Avicenne Bobigny France 2nd ERIC workshop on TP53 analysis in CLL, Stresa 2017 TP53 abnormalities :
SUPPLEMENTAL MATERIAL. Characterization and discovery of novel mirnas and mornas in JAK2V617F. mutated SET2 cells
 SUPPLEMENTAL MATERIAL Characterization and discovery of novel mirnas and mornas in JAK2V617F mutated SET2 cells Stefania Bortoluzzi 1*, Andrea Bisognin 1*, et al. 1 Supplemental methods Short RNA library
SUPPLEMENTAL MATERIAL Characterization and discovery of novel mirnas and mornas in JAK2V617F mutated SET2 cells Stefania Bortoluzzi 1*, Andrea Bisognin 1*, et al. 1 Supplemental methods Short RNA library
Targeted Molecular Diagnostics for Targeted Therapies in Hematological Disorders
 Targeted Molecular Diagnostics for Targeted Therapies in Hematological Disorders Richard D. Press, MD, PhD Dept of Pathology Knight Cancer Institute Knight Diagnostic Labs Oregon Health & Science University
Targeted Molecular Diagnostics for Targeted Therapies in Hematological Disorders Richard D. Press, MD, PhD Dept of Pathology Knight Cancer Institute Knight Diagnostic Labs Oregon Health & Science University
Click to edit Master /tle style
 Click to edit Master /tle style Tel: (314) 747-7337 Toll Free: (866) 450-7697 Fax: (314) 747-7336 Email: gps@wustl.edu Website: gps.wustl.edu GENETIC TESTING IN CANCER Ka/nka Vigh-Conrad, PhD Genomics
Click to edit Master /tle style Tel: (314) 747-7337 Toll Free: (866) 450-7697 Fax: (314) 747-7336 Email: gps@wustl.edu Website: gps.wustl.edu GENETIC TESTING IN CANCER Ka/nka Vigh-Conrad, PhD Genomics
Assessing the Economics of Genomic Medicine. Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center
 Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Looking Beyond the Standard-of- Care : The Clinical Trial Option
 1 Looking Beyond the Standard-of- Care : The Clinical Trial Option Terry Mamounas, M.D., M.P.H., F.A.C.S. Medical Director, Comprehensive Breast Program UF Health Cancer Center at Orlando Health Professor
1 Looking Beyond the Standard-of- Care : The Clinical Trial Option Terry Mamounas, M.D., M.P.H., F.A.C.S. Medical Director, Comprehensive Breast Program UF Health Cancer Center at Orlando Health Professor
Vertical Magnetic Separation of Circulating Tumor Cells and Somatic Genomic-Alteration Analysis in Lung Cancer Patients
 Vertical Magnetic Separation of Circulating Cells and Somatic Genomic-Alteration Analysis in Lung Cancer Patients Chang Eun Yoo 1,2#, Jong-Myeon Park 3#, Hui-Sung Moon 1,2, Je-Gun Joung 2, Dae-Soon Son
Vertical Magnetic Separation of Circulating Cells and Somatic Genomic-Alteration Analysis in Lung Cancer Patients Chang Eun Yoo 1,2#, Jong-Myeon Park 3#, Hui-Sung Moon 1,2, Je-Gun Joung 2, Dae-Soon Son
IMPLEMENTING NEXT GENERATION SEQUENCING IN A PATHOLOGY LABORATORY
 Madrid, Spain IMPLEMENTING NEXT GENERATION SEQUENCING IN A PATHOLOGY LABORATORY Dr. JL Rodríguez Peralto NGS Ion Torrent Oncomine Focus Assay - Implementation experience for EGFR mutation detection
Madrid, Spain IMPLEMENTING NEXT GENERATION SEQUENCING IN A PATHOLOGY LABORATORY Dr. JL Rodríguez Peralto NGS Ion Torrent Oncomine Focus Assay - Implementation experience for EGFR mutation detection
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature13898 Supplementary Information Table 1 Kras mutation status of carcinogen-induced mouse lung adenomas Tumour Treatment Strain Grade Genotype Kras status (WES)* Kras status (Sanger) 32T1
doi:10.1038/nature13898 Supplementary Information Table 1 Kras mutation status of carcinogen-induced mouse lung adenomas Tumour Treatment Strain Grade Genotype Kras status (WES)* Kras status (Sanger) 32T1
Supplementary Figure 1: Func8onal Network Analysis of Kinases Significantly Modulated by MERS CoV Infec8on and Conserved Across All Time Points
 A. B. 8 4 Supplementary Figure : Func8onal Network Analysis of Kinases Significantly Modulated by MERS CoV Infec8on and Conserved Across All Time Points Examined. A) Venn diagram analysis of kinases significantly
A. B. 8 4 Supplementary Figure : Func8onal Network Analysis of Kinases Significantly Modulated by MERS CoV Infec8on and Conserved Across All Time Points Examined. A) Venn diagram analysis of kinases significantly
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
p.arg119gly p.arg119his p.ala179thr c.540+1g>a c.617_633+6del Prediction basis structure
 a Missense ATG p.arg119gly p.arg119his p.ala179thr p.ala189val p.gly206trp p.gly206arg p.arg251his p.ala257thr TGA 5 UTR 1 2 3 4 5 6 7 3 UTR Splice Site, Frameshift b Mutation p.gly206trp p.gly4fsx50 c.138+1g>a
a Missense ATG p.arg119gly p.arg119his p.ala179thr p.ala189val p.gly206trp p.gly206arg p.arg251his p.ala257thr TGA 5 UTR 1 2 3 4 5 6 7 3 UTR Splice Site, Frameshift b Mutation p.gly206trp p.gly4fsx50 c.138+1g>a
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research
 Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Nature Genetics: doi: /ng Supplementary Figure 1. Country distribution of GME samples and designation of geographical subregions.
 Supplementary Figure 1 Country distribution of GME samples and designation of geographical subregions. GME samples collected across 20 countries and territories from the GME. Pie size corresponds to the
Supplementary Figure 1 Country distribution of GME samples and designation of geographical subregions. GME samples collected across 20 countries and territories from the GME. Pie size corresponds to the
Nature Genetics: doi: /ng Supplementary Figure 1. TCGA data set on HNSCCs reanalyzed in this study.
 Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
Development of Circulating Tumor DNA
 Development of Circulating Tumor DNA Title of presentation Arial Bold 30pt in White Biomarkers Secondary title 22pt using Arial Next in White Generation Sequencing Brian Dougherty PhD, MBA Translational
Development of Circulating Tumor DNA Title of presentation Arial Bold 30pt in White Biomarkers Secondary title 22pt using Arial Next in White Generation Sequencing Brian Dougherty PhD, MBA Translational
MEDICAL POLICY Genetic Testing for Breast and Ovarian Cancers
 POLICY: PG0067 ORIGINAL EFFECTIVE: 07/30/02 LAST REVIEW: 01/25/18 MEDICAL POLICY Genetic Testing for Breast and Ovarian Cancers GUIDELINES This policy does not certify benefits or authorization of benefits,
POLICY: PG0067 ORIGINAL EFFECTIVE: 07/30/02 LAST REVIEW: 01/25/18 MEDICAL POLICY Genetic Testing for Breast and Ovarian Cancers GUIDELINES This policy does not certify benefits or authorization of benefits,
Supplementary Table 1. Information on the 174 single nucleotide variants identified by whole-exome sequencing
 Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Genetic heterogeneity and transcriptional plasticity drive resistance during immuno- and targeted therapy
 Genetic heterogeneity and transcriptional plasticity drive resistance during immuno- and targeted therapy Reinhard Dummer, Daniel Widmer, Daniela Mihic, Phil Cheng, Lars French, Verena Paulitzschke, Mitch
Genetic heterogeneity and transcriptional plasticity drive resistance during immuno- and targeted therapy Reinhard Dummer, Daniel Widmer, Daniela Mihic, Phil Cheng, Lars French, Verena Paulitzschke, Mitch
Figure S4. 15 Mets Whole Exome. 5 Primary Tumors Cancer Panel and WES. Next Generation Sequencing
 Figure S4 Next Generation Sequencing 15 Mets Whole Exome 5 Primary Tumors Cancer Panel and WES Get coverage of all variant loci for all three Mets Variant Filtering Sequence Alignments Index and align
Figure S4 Next Generation Sequencing 15 Mets Whole Exome 5 Primary Tumors Cancer Panel and WES Get coverage of all variant loci for all three Mets Variant Filtering Sequence Alignments Index and align
Next generation sequencing analysis - A UK perspective. Nicholas Lea
 Next generation sequencing analysis - A UK perspective Nicholas Lea King s HMDC LMH is part of an integrated pathology service at King s Haematological Malignancy Diagnostic Centre (HMDC) HMDC serves population
Next generation sequencing analysis - A UK perspective Nicholas Lea King s HMDC LMH is part of an integrated pathology service at King s Haematological Malignancy Diagnostic Centre (HMDC) HMDC serves population
Nature Neuroscience: doi: /nn Supplementary Figure 1. Missense damaging predictions as a function of allele frequency
 Supplementary Figure 1 Missense damaging predictions as a function of allele frequency Percentage of missense variants classified as damaging by eight different classifiers and a classifier consisting
Supplementary Figure 1 Missense damaging predictions as a function of allele frequency Percentage of missense variants classified as damaging by eight different classifiers and a classifier consisting
Supplementary Figure 1
 Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases.
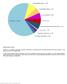 Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Concurrent Practical Session ACMG Classification
 Variant Effect Prediction Training Course 6-8 November 2017 Prague, Czech Republic Concurrent Practical Session ACMG Classification Andreas Laner / Anna Benet-Pagès 1 Content 1. Background... 3 2. Aim
Variant Effect Prediction Training Course 6-8 November 2017 Prague, Czech Republic Concurrent Practical Session ACMG Classification Andreas Laner / Anna Benet-Pagès 1 Content 1. Background... 3 2. Aim
MutationTaster & RegulationSpotter
 MutationTaster & RegulationSpotter Pathogenicity Prediction of Sequence Variants: Past, Present and Future Dr. rer. nat. Jana Marie Schwarz Klinik für Pädiatrie m. S. Neurologie Exzellenzcluster NeuroCure
MutationTaster & RegulationSpotter Pathogenicity Prediction of Sequence Variants: Past, Present and Future Dr. rer. nat. Jana Marie Schwarz Klinik für Pädiatrie m. S. Neurologie Exzellenzcluster NeuroCure
Select analysis on the next pages. Sample request and sending address see last page. Institut für Pathologie und Molekularpathologie
 Diagnostic Tumor Genome Analysis Schmelzbergstrasse 12 8091 Zürich Tel.: (+41) 044 255 3929 Fax.: (+41) 044 255 4416 Client (address, telephone number): ngs.pathologie@usz.ch www.pathologie.usz.ch Sample-Nr:
Diagnostic Tumor Genome Analysis Schmelzbergstrasse 12 8091 Zürich Tel.: (+41) 044 255 3929 Fax.: (+41) 044 255 4416 Client (address, telephone number): ngs.pathologie@usz.ch www.pathologie.usz.ch Sample-Nr:
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma. Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute
 Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Enabling Personalized
 Molecular Enabling Personalized Diagnostics Medicine- Targeted Sequencing: NGS-based solutions Silvia Dorn Roel Reinders- Andreas Diplas Friday, 19.06.2015 Company Overview Founded in April 2011 Development
Molecular Enabling Personalized Diagnostics Medicine- Targeted Sequencing: NGS-based solutions Silvia Dorn Roel Reinders- Andreas Diplas Friday, 19.06.2015 Company Overview Founded in April 2011 Development
Whole Exome Sequenced Characteristics
 Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
Published in: PLoS Genetics. DOI: /journal.pgen Link to publication
 Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. Dayeh, Tasnim; Volkov, Petr; Salö,
Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. Dayeh, Tasnim; Volkov, Petr; Salö,
