Nature Genetics: doi: /ng Supplementary Figure 1. TCGA data set on HNSCCs reanalyzed in this study.
|
|
|
- Solomon Thomas
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each.
2 Supplementary Figure 2 The methylation-defined H3 K36 subgroup consists of samples carrying damaging NSD1 or H3 K36M alterations. (a) Our analysis reveals that the H3K36 methylation cluster (Fig. 1) is comprised of 61 samples. Ten samples carry H3K36M alterations, while 51 samples have NSD1 alterations, with one carrying both. One sample had no available genomic/transcriptome data. (b) H3K36M alteraions are observed in a wide array of histone H3 genes, supporting the dominant nature of this K-to-M substitution as reported 32. (c) Samples in the H3K36 cluster are enriched for nonsense (in red) NSD1 mutations. Missense mutations (in green) are often observed near or at the functional SET domain.
3 Supplementary Figure 3 IGV screenshots of complex genomic events in NSD1 using RNA-seq data. (a) A focal deletion of a set of 10 exons in sample BA (b) Near absent expression of NSD1 in sample CN (c) Aberrant NSD1 splicing, possibly due to a translocation in sample D (d) A Focal deletion of a set of 14 exons in sample UF-A7JH
4 Supplementary Figure 4 Heat map showing hierarchical clustering of HNSCC samples based on DNA methylation and genetic alterations in other genes associated with HNSSC. (Top) Previous 4-class, expression-based classification (Atypical (black), Classical (red), Mesenchymal (green), Basal(blue)). (Middle) Other genes involved in the H3K36 methylation pathways are infrequently mutated in HNSCC and do not vary within HPV- HNSCC subgroups. (Bottom) Only TP53, CASP8, NSD1 and H3K36M vary significantly among HPV- HNSCC subgroups. For color coding of methylation groups and anatomical locations, please see Figure 1.
5 Supplementary Figure 5 Mean sample methylation levels of HNSCC. Mean sample methylation levels of HNSCC subgroups showing that NSD1 and H3K36M alterations are similarly associated with a DNA hypomethylated phenotype compared to the other four methylation clusters identified in HNSCCs.
6 Supplementary Figure 6 Global DNA methylation, NSD1 expression level and the types of mutations in the H3 K36M cluster. (a,b) Samples with NSD1 mutations outside the H3K36M cluster (grey bars) show higher global DNA methylation levels (a) and higher NSD1 gene expression levels (b) compared to samples in the H3K36 cluster (blue bars). (c) Samples in the H3K36 cluster frequently carry NSD1 truncating (nonsense and frameshift) mutations compared to NSD1 mutated samples in the other HNSCC subgroups.
7 Supplementary Figure 7 Mean somatic mutations per sample of HNSCC subgroups. Mean somatic mutations per sample of HNSCC subgroups showing that NSD1 mutant samples have higher somatic mutation count than H3K36M samples or other HNSCC subgroups.
8 Supplementary Figure 8 NSD1 but not H3 K36M mutant samples have smoking-induced somatic mutation signatures. (a) In NSD1 samples, similar to lung adenocarcinoma (LUAD) and lung squamous cell carcinomas (LUSC), a large proportion of somatic mutations is accounted by the smoking-associated signature S4 (red). In comparison, other HNSCC samples (left) have much less smoking-induced somatic mutations. (b) Unsupervised clustering of mutation frequencies groups NSD1, but not H3K36M, with lung cancers, suggesting the mutational spectrum in NSD1 samples is associated with smoking. (c) A five-signature model identifies UV exposure (S1), Temozolomide (S2) and smoking (S4) as major mutation generation processes.
9 Supplementary Figure 9 Kaplan Meier curve for HNSCC patients, grouped by DNA-methylation-defined molecular subgroups.
10 Supplementary Figure 10 Anatomical distribution of HNSCC samples relative to K36M and NSD1 alterations. Anatomical distribution of HNSCC samples showing that H3K36M samples are exclusively found in the oral cavity whereas the majority of NSD1 mutated samples are found in the larynx and oral cavity.
11 Supplementary Figure 11 NSD1 and NSD2 expression in the H3K36 cluster and other HNSCC subgroups (a) Samples in the H3K36 cluster show lower NSD1 expression levels. (b) NSD2 levels are uniform among al HNSCC subgroups.
12 Supplementary Figure 12 RNA-expression clustering of HNSCC samples. Similar to our findings using DNA methylation, we observe a group of samples enriched in NSD1/H3K36M alterations (H3K36 cluster) and another enriched for HPV+ HNSCC. For color coding of methylation groups and anatomical locations, please see Figure 1.
13 Supplementary Figure 13 Gene ontology enrichment analysis on H3K36-specific differentially expressed genes. Differential expression analysis performed on H3K36 samples versus all other HNSCCs tumors, controlling for anatomical location.
14 Supplementary Figure 14 Western blots for H3 K36M and NSD1 in HNSSC cell lines. Full length immunoblots including molecular weights that were used in Figure 4.
15 Supplementary Table 1. NSD1 mutations in samples included in the H3K36 cluster. Hugo Symbol NSD1, Chromosome 5. Sample ID Protein Mutation Start End Position Reference Variant Allele Mutation Change Type Position Allele Allele Frequency Assessor TCGA-BA A-01D p.r788x Nonsense C T TCGA-CV A-01D p.h1872fs Frame_Shift_Del AT TCGA-CV A-31D p.q1153x Nonsense C T NA TCGA-BB A-11D p.e990x Nonsense G T TCGA-BA A-11D p.s707x Nonsense C G TCGA-CV A-41D p.g1678w Missense G T Medium TCGA-CV A-11D p.e1534x Nonsense G T 0.75 TCGA-CR A-11D p.g1524fs Frame_Shift_Del G TCGA-CR A-11D p.p2567t Missense C A 0.1 Neutral TCGA-CR A-11D p.q1989x Nonsense C T TCGA-CR A-11D p.t723s Missense A T Neutral TCGA-CR A-11D p.e1979x Nonsense G T TCGA-CV A-11D p.s1086fs Frame_Shift_Del AGTAAAG TCGA-CR A-11D p.s1528fs Frameshift Insertion A TCGA-CV A-11D p.s1359x Nonsense C A TCGA-CV A-11D p.a2009t Missense G A High TCGA-CR A-01D p.y1997c Missense A G High TCGA-CN A-01D p.g1928x Nonsense G T TCGA-CN A-01D p.g959x Nonsense G T TCGA-CN A-01D p.s589fs Frame_Shift_Del TT TCGA-CN A-01D p.k1433x Nonsense A T TCGA-F A-11D p.m1094fs Frame_Shift_Del G TCGA-CV A-11D p.c1619s Missense G C Medium TCGA-CV A-11D p.c792x Nonsense C A TCGA-CV A-11D p.r1473x Nonsense C T TCGA-CV-A461-01A-41D-A25Y-08 p.r1757fs Frame_Shift_Del G TCGA-CV-A461-01A-41D-A25Y-08 p.s981fs frameshift insertion G TCGA-BA-A4IF-01A-11D-A25Y-08 p.p1665l Missense C T Medium TCGA-CN-A497-01A-11D-A24D-08 p.e1391x Nonsense G T TCGA-CN-A497-01A-11D-A24D-08 p.w2032x Nonsense G A TCGA-CN-A63W-01A-11D-A30E-08 p.w1160x Nonsense G A Note Sample also has NSD1 frame shift deletion and nonsense mutation Sample also has NSD1 frame shift deletion and nonsense mutation Sample also has NSD1 nonsense mutation TCGA-CN-A641-01A-11D-A30E-08 p.c1897f Missense G T High Sample also has NSD1 frame shift insertion TCGA-CN-A641-01A-11D-A30E-08 p.i1946fs frameshift insertion T TCGA-CN-A641-01A-11D-A30E-08 p.r1948i Missense CG AT Medium Sample also has NSD1 frame shift insertion TCGA-CN-A63T-01A-11D-A28R-08 Splice G A 0.2 TCGA-CN-A63T-01A-11D-A28R-08 p.e1970a Missense A C High Sample also has NSD1 splicing mutation TCGA-BB-A5HZ-01A-21D-A28R-08 p.g1095fs Frame_Shift_Del GCCACTTAA TCGA-BB-A5HZ-01A-21D-A28R-08 p.i1291fs Frameshift Insertion T 0.2 TCGA-P3-A5QF-01A-11D-A28R-08 p.r2005q Missense G A Medium TCGA-KU-A66S-01A-21D-A30E-08 p.h1616y Missense C T Medium Sample also has NSD1 frame shift deletion TCGA-KU-A66S-01A-21D-A30E-08 p.r1952fs Frame_Shift_Del G TCGA-CN-A63U-01A-11D-A30E-08 p.e1501x Nonsense G T TCGA-CQ A-21D-A30E-08 p.q679x Nonsense C T TCGA-BA-A6DL-01A-21D-A30E-08 p.r1072x Nonsense C T TCGA-BA-A6DL-01A-21D-A30E-08 p.r1200fs Frame_Shift_Del GGGATGAG TCGA-H7-A6C5-01A-11D-A30E-08 p.1664_1665nonframe_shift_del TCC TCGA-D6-A6EO-01A-11D-A31L-08 p.e2028fs Frame_Shift_Del A TCGA-D6-A74Q-01A-11D-A34J-08 p.p753fs Frame_Shift_Del A TCGA-CN-A6V3-01A-12D-A34J-08 p.c1710y Missense G A High TCGA-CN-A6V3-01A-12D-A34J-08 p.w1769c Missense G T 0.45 Medium TCGA-UF-A71D-01A-12D-A34J-08 p.t922fs Frame_Shift_Del C TCGA-BA-A6DE-01A-22D-A31L-08 p.e1575x Nonsense G T TCGA-CV-A6JU-01A-11D-A31L-08 p.y1941fs Frameshift Insertion T TCGA-QK-A6VB-01A-12D-A34J-08 p.y1834fs Frameshift Insertion A TCGA-P3-A6T8-01A-11D-A34J-08 p.s2664fs Frame_Shift_Del T TCGA-UF-A719-01A-12D-A34J-08 p.s1528y Missense C A 0.7 Low Sample also has H3K36M mutation TCGA-UF-A7JF-01A-11D-A34J-08 p.e1853x Nonsense G T TCGA-QK-A8Z8-01A-11D-A p.h1872fs Frame_Shift_Del AT
16 Supplementary Table 2. Complex genomic events in H3K36 samples with no NSD1 or H3K36M mutations by Whole Exome Sequencing. LOH=Loss Of Heterozygosity; NA=Not Available Sample Result GISTIC NSD1 GISTIC NSD1 Gene Value Gene Threshold CV-7091 LOH CN-4731 Focal Deletion of 8 exons D Splicing defect CN-6988 LOH BA-4074 No expression NA NA CV-7428 Not sequenced CV-7435 No expression UF-A7JH Focal deletion exons
17 Supplementary Table 3. NSD1 mutations in samples excluded from the H3K36 cluster. Hugo Symbol NSD1, Chromosome 5. Sample ID Protein Start End Reference Variant Allele Mutation Mutation Type Change Position Position Allele Allele Frequency Assessor TCGA-CN A-02D p.c1710s Missense G C High TCGA-HD A-11D p.e1202fs Frame_Shift_Del G - NA TCGA-DQ A-11D p.p803fs Frame_Shift_Del C - NA TCGA-CR A-11D p.d1489n Missense G A Neutral TCGA-CR A-11D p.l2054r Missense T G NA High TCGA-CV A-21D p.e2467q Missense G C Low TCGA-CV A-21D p.r1984x Nonsense C T TCGA-CR A-11D p.r1700x Nonsense C T TCGA-CV A-11D p.d1992h Missense G C Neutral TCGA-CV A-11D p.d2002h Missense G C Medium TCGA-CN A-01D p.s744x Nonsense C G TCGA-CV A-11D p.i2113m Missense C G Low TCGA-CV A-11D p.d325v Missense A T Medium TCGA-CV A-21D p.s96c Missense C G Neutral TCGA-CV A-11D p.p434l Missense C T Low TCGA-CV A-11D p.p434s Missense C T Low TCGA-D A-11D p.v366l Missense G T Low TCGA-CV-A468-01A-11D-A25Y-08 p.r1320x Nonsense C T TCGA-CV-A45Z-01A-21D-A25D-08 p.k601x Nonsense A T TCGA-F7-A623-01A-11D-A28R-08 p.e1520k Missense G A Low TCGA-UF-A71B-01A-12D-A34J-08 p.g566e Missense G A Neutral TCGA-TN-A7HL-01A-11D-A34J-08 p.r1634q Missense G A Medium TCGA-P3-A6T6-01A-11D-A34J-08 p.e1516x Nonsense G T TCGA-QK-AA3J-01A-11D-A p.r788x Nonsense C T
18 Supplementary Table 4. H3K36M Mutations in TCGA HNSCCs Hugo Protein Start End Reference Variant Allele Sample ID Mutation Type Symbol Change Chromosome Position Position Allele Allele Frequency H3F3B TCGA-CQ A-11D p.k37m* nonsynonymous SNV T A H3F3B TCGA-QK-A8Z7-01A-11D-A p.k37m nonsynonymous SNV T A HIST1H3C TCGA-CN A-01D p.k37m nonsynonymous SNV A T HIST1H3C TCGA-CQ A-11D p.k37m nonsynonymous SNV A T HIST1H3C TCGA-CQ-A4C9-01A-11D-A25D-08 p.k37m nonsynonymous SNV A T HIST1H3C TCGA-MT-A67D-01A-31D-A30E-08 p.k37m nonsynonymous SNV A T HIST1H3E TCGA-CV A-11D p.k37m nonsynonymous SNV A T HIST1H3G TCGA-UF-A7JD-01A-11D-A34J-08 p.k37m nonsynonymous SNV T A HIST1H3I TCGA-CV A-11D p.k37m nonsynonymous SNV T A HIST1H3I TCGA-UF-A719-01A-12D-A34J-08 p.k37m nonsynonymous SNV T A HIST2H3D TCGA-CX A-11D p.k37m nonsynonymous SNV T A *K37M is the universal nomenclature when describing amino acid changes in genomic data. We note that histone proteins have the first methionine removed from the final product thus shifting the numbering by one amino acid. In the context of protein sequences, this mutation will generally be referred to as K36M.
19 Supplementary Table 6. Meta analysis of previous HNSCC sequencing studies reveals other NSD1 and H3K36M mutations. Table adapted from Riaz, Morris, Lee, & Chan. Unraveling the molecular genetics of head and neck cancer through genome-wide approaches. Genes Dis 1; 75-86, 2014 Sequencing Study (# Samples Sequenced) Stransky (74) NSD1 p.c1710* (Larynx, Smoker, HPV-), p.c2124y (Larynx, Smoker, HPV-), p.p1726h (Larynx, Smoker, HPV-), p.r1233fs (Larynx, Smoker, HPV-), p.r1984q (Hypopharynx, Smoker, HPV-), p.y1834fs (Larynx, Smoker, HPV-), splicing (Oral Cavity, Smoker, HPV-) H3K36M HIST1H3H (Oral Cavity, Smoker, HPV-) Agrawal (32) 0 0 Pickering (40) p.r2017q (Tongue, Smoker, HPV-), p.w2032r (FOM, Smoker, HPV-), 0 p.r1984x (FOM, Smoker, HPV-) India ICGC (50) p.q517* (Gingivo-buccal oral, NA, NA), p.s958l (Gingivo-buccal oral, NA, NA), p.r1811* (Gingivo-buccal oral, NA, NA), p.c1911f (Gingivo-buccal oral, NA, NA) 0
Whole Exome Sequenced Characteristics
 Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
BWA alignment to reference transcriptome and genome. Convert transcriptome mappings back to genome space
 Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Supplementary Table 2. Identified causative mutations and/or mutation candidates.
 Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Nature Genetics: doi: /ng Supplementary Figure 1. Somatic coding mutations identified by WES/WGS for 83 ATL cases.
 Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Nature Immunology: doi: /ni Supplementary Figure 1. RNA-Seq analysis of CD8 + TILs and N-TILs.
 Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
 Supplementary Figures Figure S1. Patient cohorts and study design. To define and interrogate the genetic alterations underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
Supplementary Figures Figure S1. Patient cohorts and study design. To define and interrogate the genetic alterations underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
Supplementary Figure 1: Classification scheme for non-synonymous and nonsense germline MC1R variants. The common variants with previously established
 Supplementary Figure 1: Classification scheme for nonsynonymous and nonsense germline MC1R variants. The common variants with previously established classifications 1 3 are shown. The effect of novel missense
Supplementary Figure 1: Classification scheme for nonsynonymous and nonsense germline MC1R variants. The common variants with previously established classifications 1 3 are shown. The effect of novel missense
Supplementary Figure 1
 Supplementary Figure 1 A B HEC1A PTEN +/+ HEC1A PTEN -/- 16 HEC1A PTEN -/- 22 PTEN RAD51 % Cells with RAD51 foci 40 -IR +IR 30 20 10 * *! TUBULIN 0 HEC1A PTEN+/+ HEC1A PTEN-/- 16 HEC1A PTEN-/- 22 C D 1.0
Supplementary Figure 1 A B HEC1A PTEN +/+ HEC1A PTEN -/- 16 HEC1A PTEN -/- 22 PTEN RAD51 % Cells with RAD51 foci 40 -IR +IR 30 20 10 * *! TUBULIN 0 HEC1A PTEN+/+ HEC1A PTEN-/- 16 HEC1A PTEN-/- 22 C D 1.0
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis
 The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
6.3 DNA Mutations. SBI4U Ms. Ho-Lau
 6.3 DNA Mutations SBI4U Ms. Ho-Lau DNA Mutations Gene expression can be affected by errors that occur during DNA replication. Some errors are repaired, but others can become mutations (changes in the nucleotide
6.3 DNA Mutations SBI4U Ms. Ho-Lau DNA Mutations Gene expression can be affected by errors that occur during DNA replication. Some errors are repaired, but others can become mutations (changes in the nucleotide
Nature Genetics: doi: /ng Supplementary Figure 1. Clinical timeline for the discovery WES cases.
 Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
UW359 Ovary 3c 3 Serous Recurrent 68 BRCA1 816delGT BRCA1 del exon 1-2. UW417 Ovary 3c 3 Serous Primary 38 BRCA1 1675delA
 Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
Nature Medicine: doi: /nm.3967
 Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplemental Figure 1. Genes showing ectopic H3K9 dimethylation in this study are DNA hypermethylated in Lister et al. study.
 mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
Supplementary Table 1. PIK3CA mutation in colorectal cancer
 Liao X et al. PIK3CA Mutation in Colorectal Cancer. Page 1 Supplementary Table 1. PIK3CA mutation in colorectal cancer Exon Domain Nucleotide change* Amino acid change* cases 9 Helical c.1621t>a p.e541t
Liao X et al. PIK3CA Mutation in Colorectal Cancer. Page 1 Supplementary Table 1. PIK3CA mutation in colorectal cancer Exon Domain Nucleotide change* Amino acid change* cases 9 Helical c.1621t>a p.e541t
Importance of minor TP53 mutated clones in the clinic
 Importance of minor TP53 mutated clones in the clinic Davide Rossi, M.D., Ph.D. Hematology IOSI - Oncology Institute of Southern Switzerland IOR - Institute of Oncology Reserach Bellinzona - Switzerland
Importance of minor TP53 mutated clones in the clinic Davide Rossi, M.D., Ph.D. Hematology IOSI - Oncology Institute of Southern Switzerland IOR - Institute of Oncology Reserach Bellinzona - Switzerland
Tumor suppressor genes D R. S H O S S E I N I - A S L
 Tumor suppressor genes 1 D R. S H O S S E I N I - A S L What is a Tumor Suppressor Gene? 2 A tumor suppressor gene is a type of cancer gene that is created by loss-of function mutations. In contrast to
Tumor suppressor genes 1 D R. S H O S S E I N I - A S L What is a Tumor Suppressor Gene? 2 A tumor suppressor gene is a type of cancer gene that is created by loss-of function mutations. In contrast to
Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations
 Genomic sequencing of meningiomas identifies oncogenic SMO and AKT mutations Priscilla K. Brastianos*; Peleg M. Horowitz*; Sandro Santagata; Robert T. Jones; Aaron McKenna; Gad Getz; Keith L. Ligon; Emanuele
Genomic sequencing of meningiomas identifies oncogenic SMO and AKT mutations Priscilla K. Brastianos*; Peleg M. Horowitz*; Sandro Santagata; Robert T. Jones; Aaron McKenna; Gad Getz; Keith L. Ligon; Emanuele
Nature Getetics: doi: /ng.3471
 Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Biomarker development in the era of precision medicine. Bei Li, Interdisciplinary Technical Journal Club
 Biomarker development in the era of precision medicine Bei Li, 23.08.2016 Interdisciplinary Technical Journal Club The top ten highest-grossing drugs in the United States help between 1 in 25 and 1 in
Biomarker development in the era of precision medicine Bei Li, 23.08.2016 Interdisciplinary Technical Journal Club The top ten highest-grossing drugs in the United States help between 1 in 25 and 1 in
CDH1 truncating alterations were detected in all six plasmacytoid-variant bladder tumors analyzed by whole-exome sequencing.
 Supplementary Figure 1 CDH1 truncating alterations were detected in all six plasmacytoid-variant bladder tumors analyzed by whole-exome sequencing. Whole-exome sequencing of six plasmacytoid-variant bladder
Supplementary Figure 1 CDH1 truncating alterations were detected in all six plasmacytoid-variant bladder tumors analyzed by whole-exome sequencing. Whole-exome sequencing of six plasmacytoid-variant bladder
Supplementary Information
 Supplementary Information JAK1 truncating mutations in gynecologic cancer define new role of cancerassociated protein tyrosine kinase aberrations Yuan Ren, Yonghong Zhang, Richard Z. Liu, David A. Fenstermacher,
Supplementary Information JAK1 truncating mutations in gynecologic cancer define new role of cancerassociated protein tyrosine kinase aberrations Yuan Ren, Yonghong Zhang, Richard Z. Liu, David A. Fenstermacher,
SUPPLEMENTARY INFORMATION. Intron retention is a widespread mechanism of tumor suppressor inactivation.
 SUPPLEMENTARY INFORMATION Intron retention is a widespread mechanism of tumor suppressor inactivation. Hyunchul Jung 1,2,3, Donghoon Lee 1,4, Jongkeun Lee 1,5, Donghyun Park 2,6, Yeon Jeong Kim 2,6, Woong-Yang
SUPPLEMENTARY INFORMATION Intron retention is a widespread mechanism of tumor suppressor inactivation. Hyunchul Jung 1,2,3, Donghoon Lee 1,4, Jongkeun Lee 1,5, Donghyun Park 2,6, Yeon Jeong Kim 2,6, Woong-Yang
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/7/283/283ra54/dc1 Supplementary Materials for Clonal status of actionable driver events and the timing of mutational processes in cancer evolution
www.sciencetranslationalmedicine.org/cgi/content/full/7/283/283ra54/dc1 Supplementary Materials for Clonal status of actionable driver events and the timing of mutational processes in cancer evolution
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Computer Science, Biology, and Biomedical Informatics (CoSBBI) Outline. Molecular Biology of Cancer AND. Goals/Expectations. David Boone 7/1/2015
 Goals/Expectations Computer Science, Biology, and Biomedical (CoSBBI) We want to excite you about the world of computer science, biology, and biomedical informatics. Experience what it is like to be a
Goals/Expectations Computer Science, Biology, and Biomedical (CoSBBI) We want to excite you about the world of computer science, biology, and biomedical informatics. Experience what it is like to be a
Section Chapter 14. Go to Section:
 Section 12-3 Chapter 14 Go to Section: Content Objectives Write these Down! I will be able to identify: The origin of genetic differences among organisms. The possible kinds of different mutations. The
Section 12-3 Chapter 14 Go to Section: Content Objectives Write these Down! I will be able to identify: The origin of genetic differences among organisms. The possible kinds of different mutations. The
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first
 Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Nature Genetics: doi: /ng Supplementary Figure 1. Mutational signatures in BCC compared to melanoma.
 Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
Variant Classification. Author: Mike Thiesen, Golden Helix, Inc.
 Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Reporting TP53 gene analysis results in CLL
 Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
MSI positive MSI negative
 Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS
 MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS How do different alleles arise? ( allele : form of a gene; specific base sequence at a site on DNA) Mutations: heritable changes in genes Mutations occur in DNA
MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS How do different alleles arise? ( allele : form of a gene; specific base sequence at a site on DNA) Mutations: heritable changes in genes Mutations occur in DNA
Nature Genetics: doi: /ng Supplementary Figure 1. Rates of different mutation types in CRC.
 Supplementary Figure 1 Rates of different mutation types in CRC. (a) Stratification by mutation type indicates that C>T mutations occur at a significantly greater rate than other types. (b) As for the
Supplementary Figure 1 Rates of different mutation types in CRC. (a) Stratification by mutation type indicates that C>T mutations occur at a significantly greater rate than other types. (b) As for the
gliomas. Fetal brain expected who each low-
 Supplementary Figure S1. Grade-specificity aberrant expression of HOXA genes in gliomas. (A) Representative RT-PCR analyses of HOXA gene expression in human astrocytomas. Exemplified glioma samples include
Supplementary Figure S1. Grade-specificity aberrant expression of HOXA genes in gliomas. (A) Representative RT-PCR analyses of HOXA gene expression in human astrocytomas. Exemplified glioma samples include
Supplementary Figure 1
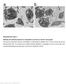 Supplementary Figure 1 Platelets from affected patients are comparable to controls on electron micrograph. Thin-section transmission electron micrographs of representative platelets from a normal control
Supplementary Figure 1 Platelets from affected patients are comparable to controls on electron micrograph. Thin-section transmission electron micrographs of representative platelets from a normal control
WHEN DO MUTATIONS OCCUR?
 WHEN DO MUTATIONS OCCUR? While most DNA replicates with fairly high accuracy, mistakes do happen. DNA polymerase sometimes inserts the wrong nucleotide or too many or too few nucleotides into a sequence.
WHEN DO MUTATIONS OCCUR? While most DNA replicates with fairly high accuracy, mistakes do happen. DNA polymerase sometimes inserts the wrong nucleotide or too many or too few nucleotides into a sequence.
Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well
 Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well plate at cell densities ranging from 25-225 cells in
Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well plate at cell densities ranging from 25-225 cells in
Genetic alterations of histone lysine methyltransferases and their significance in breast cancer
 Genetic alterations of histone lysine methyltransferases and their significance in breast cancer Supplementary Materials and Methods Phylogenetic tree of the HMT superfamily The phylogeny outlined in the
Genetic alterations of histone lysine methyltransferases and their significance in breast cancer Supplementary Materials and Methods Phylogenetic tree of the HMT superfamily The phylogeny outlined in the
Session 4 Rebecca Poulos
 The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 20
The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 20
Bio 111 Study Guide Chapter 17 From Gene to Protein
 Bio 111 Study Guide Chapter 17 From Gene to Protein BEFORE CLASS: Reading: Read the introduction on p. 333, skip the beginning of Concept 17.1 from p. 334 to the bottom of the first column on p. 336, and
Bio 111 Study Guide Chapter 17 From Gene to Protein BEFORE CLASS: Reading: Read the introduction on p. 333, skip the beginning of Concept 17.1 from p. 334 to the bottom of the first column on p. 336, and
Nature Genetics: doi: /ng.2995
 Supplementary Figure 1 Kaplan-Meier survival curves of patients with brainstem tumors. (a) Comparison of patients with PPM1D mutation versus wild-type PPM1D. (b) Comparison of patients with PPM1D mutation
Supplementary Figure 1 Kaplan-Meier survival curves of patients with brainstem tumors. (a) Comparison of patients with PPM1D mutation versus wild-type PPM1D. (b) Comparison of patients with PPM1D mutation
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies
 Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each
 Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Genomic Methods in Cancer Epigenetic Dysregulation
 Genomic Methods in Cancer Epigenetic Dysregulation Clara, Lyon 2018 Jacek Majewski, Associate Professor Department of Human Genetics, McGill University Montreal, Canada A few words about my lab Genomics
Genomic Methods in Cancer Epigenetic Dysregulation Clara, Lyon 2018 Jacek Majewski, Associate Professor Department of Human Genetics, McGill University Montreal, Canada A few words about my lab Genomics
Supplementary Figure 1. Copy Number Alterations TP53 Mutation Type. C-class TP53 WT. TP53 mut. Nature Genetics: doi: /ng.
 Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
 Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Nature Genetics: doi: /ng Supplementary Figure 1. Country distribution of GME samples and designation of geographical subregions.
 Supplementary Figure 1 Country distribution of GME samples and designation of geographical subregions. GME samples collected across 20 countries and territories from the GME. Pie size corresponds to the
Supplementary Figure 1 Country distribution of GME samples and designation of geographical subregions. GME samples collected across 20 countries and territories from the GME. Pie size corresponds to the
Supplementary Figure 1 Overall study design
 Supplementary Figure 1 Overall study design We obtained 19 tumour specimens from 14 men with prostate cancer who harboured pathogenic germline BRCA2-mutations. Germline DNA was available for five patients
Supplementary Figure 1 Overall study design We obtained 19 tumour specimens from 14 men with prostate cancer who harboured pathogenic germline BRCA2-mutations. Germline DNA was available for five patients
Nature Medicine: doi: /nm.4439
 Figure S1. Overview of the variant calling and verification process. This figure expands on Fig. 1c with details of verified variants identification in 547 additional validation samples. Somatic variants
Figure S1. Overview of the variant calling and verification process. This figure expands on Fig. 1c with details of verified variants identification in 547 additional validation samples. Somatic variants
Exploring TCGA Pan-Cancer Data at the UCSC Cancer Genomics Browser
 Exploring TCGA Pan-Cancer Data at the UCSC Cancer Genomics Browser Melissa S. Cline 1*, Brian Craft 1, Teresa Swatloski 1, Mary Goldman 1, Singer Ma 1, David Haussler 1, Jingchun Zhu 1 1 Center for Biomolecular
Exploring TCGA Pan-Cancer Data at the UCSC Cancer Genomics Browser Melissa S. Cline 1*, Brian Craft 1, Teresa Swatloski 1, Mary Goldman 1, Singer Ma 1, David Haussler 1, Jingchun Zhu 1 1 Center for Biomolecular
NGS in tissue and liquid biopsy
 NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
Expanded View Figures
 EMO Molecular Medicine Proteomic map of squamous cell carcinomas Hanibal ohnenberger et al Expanded View Figures Figure EV1. Technical reproducibility. Pearson s correlation analysis of normalised SILC
EMO Molecular Medicine Proteomic map of squamous cell carcinomas Hanibal ohnenberger et al Expanded View Figures Figure EV1. Technical reproducibility. Pearson s correlation analysis of normalised SILC
Session 4 Rebecca Poulos
 The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 28
The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 28
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
MEDICAL GENOMICS LABORATORY. Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG)
 Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Hands-On Ten The BRCA1 Gene and Protein
 Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples
 A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples Sona Pekova, MD., PhD. Chambon Ltd., Laboratory for molecular diagnostics, Prague, Czech
A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples Sona Pekova, MD., PhD. Chambon Ltd., Laboratory for molecular diagnostics, Prague, Czech
Smoking, human papillomavirus infection, and p53 mutation as risk factors in oropharyngeal cancer: a case-control study
 RESEARCH FUND FOR THE CONTROL OF INFECTIOUS DISEASES Smoking, human papillomavirus infection, and p53 as risk factors in oropharyngeal cancer: a case-control study PKS Chan *, JSY Chor, AC Vlantis, TL
RESEARCH FUND FOR THE CONTROL OF INFECTIOUS DISEASES Smoking, human papillomavirus infection, and p53 as risk factors in oropharyngeal cancer: a case-control study PKS Chan *, JSY Chor, AC Vlantis, TL
Supplementary Figure 1. Estimation of tumour content
 Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
DNA-seq Bioinformatics Analysis: Copy Number Variation
 DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
Expanded View Figures
 Molecular Systems iology Tumor CNs reflect metabolic selection Nicholas Graham et al Expanded View Figures Human primary tumors CN CN characterization by unsupervised PC Human Signature Human Signature
Molecular Systems iology Tumor CNs reflect metabolic selection Nicholas Graham et al Expanded View Figures Human primary tumors CN CN characterization by unsupervised PC Human Signature Human Signature
Epigenetics. Jenny van Dongen Vrije Universiteit (VU) Amsterdam Boulder, Friday march 10, 2017
 Epigenetics Jenny van Dongen Vrije Universiteit (VU) Amsterdam j.van.dongen@vu.nl Boulder, Friday march 10, 2017 Epigenetics Epigenetics= The study of molecular mechanisms that influence the activity of
Epigenetics Jenny van Dongen Vrije Universiteit (VU) Amsterdam j.van.dongen@vu.nl Boulder, Friday march 10, 2017 Epigenetics Epigenetics= The study of molecular mechanisms that influence the activity of
JULY 21, Genetics 101: SCN1A. Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology
 JULY 21, 2018 Genetics 101: SCN1A Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology Disclosures: I have no financial interests or relationships to disclose. Objectives 1. Review genetic
JULY 21, 2018 Genetics 101: SCN1A Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology Disclosures: I have no financial interests or relationships to disclose. Objectives 1. Review genetic
MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS. (Start your clickers)
 MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS (Start your clickers) How do mutations arise? And how do they affect a cell and its organism? Mutations: heritable changes in genes Mutations occur in DNA But
MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS (Start your clickers) How do mutations arise? And how do they affect a cell and its organism? Mutations: heritable changes in genes Mutations occur in DNA But
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Chapter 12-4 DNA Mutations Notes
 Chapter 12-4 DNA Mutations Notes I. Mutations Introduction A. Definition: Changes in the DNA sequence that affect genetic information B. Mutagen= physical or chemical agent that interacts with DNA to cause
Chapter 12-4 DNA Mutations Notes I. Mutations Introduction A. Definition: Changes in the DNA sequence that affect genetic information B. Mutagen= physical or chemical agent that interacts with DNA to cause
Nature Genetics: doi: /ng Supplementary Figure 1. Alternative splicing events in the 5K panel.
 Supplementary Figure 1 Alternative splicing events in the 5K panel. The majority of cryptic splicing occurred by creation of an AG or GT (Type I). While some other mutations increased the usage of a nearby
Supplementary Figure 1 Alternative splicing events in the 5K panel. The majority of cryptic splicing occurred by creation of an AG or GT (Type I). While some other mutations increased the usage of a nearby
Computational Systems Biology: Biology X
 Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
CANCER GENETICS PROVIDER SURVEY
 Dear Participant, Previously you agreed to participate in an evaluation of an education program we developed for primary care providers on the topic of cancer genetics. This is an IRB-approved, CDCfunded
Dear Participant, Previously you agreed to participate in an evaluation of an education program we developed for primary care providers on the topic of cancer genetics. This is an IRB-approved, CDCfunded
Supplementary Figure 1. Experimental paradigm. A combination of genome and exome sequencing coupled with array-comparative genome hybridization was
 Supplementary Figure 1. Experimental paradigm. A combination of genome and exome sequencing coupled with array-comparative genome hybridization was performed on a total of 85 SS patients. Data filtration
Supplementary Figure 1. Experimental paradigm. A combination of genome and exome sequencing coupled with array-comparative genome hybridization was performed on a total of 85 SS patients. Data filtration
SUPPLEMENTARY FIGURES: Supplementary Figure 1
 SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma.
 Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Dynamic reprogramming of DNA methylation in SETD2- deregulated renal cell carcinoma
 /, Vol. 7, No. 2 Dynamic reprogramming of DNA methylation in SETD2- deregulated renal cell carcinoma Rochelle L. Tiedemann 1, Ryan A. Hlady 2, Paul D. Hanavan 3, Douglas F. Lake 3, Raoul Tibes 4,5, Jeong-Heon
/, Vol. 7, No. 2 Dynamic reprogramming of DNA methylation in SETD2- deregulated renal cell carcinoma Rochelle L. Tiedemann 1, Ryan A. Hlady 2, Paul D. Hanavan 3, Douglas F. Lake 3, Raoul Tibes 4,5, Jeong-Heon
GENE EXPRESSION. Amoeba Sisters video 3pk9YVo. Individuality & Mutations
 Amoeba Sisters video https://www.youtube.com/watch?v=giez 3pk9YVo GENE EXPRESSION Individuality & Mutations Complete video handout http://www.amoebasisters.com/uploads/ 2/1/9/0/21902384/video_recap_of_muta
Amoeba Sisters video https://www.youtube.com/watch?v=giez 3pk9YVo GENE EXPRESSION Individuality & Mutations Complete video handout http://www.amoebasisters.com/uploads/ 2/1/9/0/21902384/video_recap_of_muta
Cancer Informatics Lecture
 Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
COMPUTATIONAL OPTIMISATION OF TARGETED DNA SEQUENCING FOR CANCER DETECTION
 COMPUTATIONAL OPTIMISATION OF TARGETED DNA SEQUENCING FOR CANCER DETECTION Pierre Martinez, Nicholas McGranahan, Nicolai Juul Birkbak, Marco Gerlinger, Charles Swanton* SUPPLEMENTARY INFORMATION SUPPLEMENTARY
COMPUTATIONAL OPTIMISATION OF TARGETED DNA SEQUENCING FOR CANCER DETECTION Pierre Martinez, Nicholas McGranahan, Nicolai Juul Birkbak, Marco Gerlinger, Charles Swanton* SUPPLEMENTARY INFORMATION SUPPLEMENTARY
DNA Analysis in Glycogen storage disease
 DNA Analysis in Glycogen storage disease Nick Beauchamp PhD Sheffield, Sheffield Children s NHS Foundation Trust 14th October 2010 Glycogen Synthase Type 0 Glycogen Synthesis and Breakdown Type IV UDPGlucose
DNA Analysis in Glycogen storage disease Nick Beauchamp PhD Sheffield, Sheffield Children s NHS Foundation Trust 14th October 2010 Glycogen Synthase Type 0 Glycogen Synthesis and Breakdown Type IV UDPGlucose
Supplementary Figure 1. Spitzoid Melanoma with PPFIBP1-MET fusion. (a) Histopathology (4x) shows a domed papule with melanocytes extending into the
 Supplementary Figure 1. Spitzoid Melanoma with PPFIBP1-MET fusion. (a) Histopathology (4x) shows a domed papule with melanocytes extending into the deep dermis. (b) The melanocytes demonstrate abundant
Supplementary Figure 1. Spitzoid Melanoma with PPFIBP1-MET fusion. (a) Histopathology (4x) shows a domed papule with melanocytes extending into the deep dermis. (b) The melanocytes demonstrate abundant
Functional analysis of DNA variants
 Functional analysis of DNA variants GS011143, Introduction to Bioinformatics The University of Texas GSBS program, Fall 2012 Ken Chen, Ph.D. Department of Bioinformatics and Computational Biology UT MD
Functional analysis of DNA variants GS011143, Introduction to Bioinformatics The University of Texas GSBS program, Fall 2012 Ken Chen, Ph.D. Department of Bioinformatics and Computational Biology UT MD
MMB (MGPG) Non traditional Inheritance Epigenetics. A.Turco
 MMB (MGPG) 2017 Non traditional Inheritance Epigenetics A.Turco NON TRADITIONAL INHERITANCE EXCEPTIONS TO MENDELISM - Genetic linkage (2 loci close to each other) - Complex or Multifactorial Disease (MFD)
MMB (MGPG) 2017 Non traditional Inheritance Epigenetics A.Turco NON TRADITIONAL INHERITANCE EXCEPTIONS TO MENDELISM - Genetic linkage (2 loci close to each other) - Complex or Multifactorial Disease (MFD)
User s Manual Version 1.0
 User s Manual Version 1.0 #639 Longmian Avenue, Jiangning District, Nanjing,211198,P.R.China. http://tcoa.cpu.edu.cn/ Contact us at xiaosheng.wang@cpu.edu.cn for technical issue and questions Catalogue
User s Manual Version 1.0 #639 Longmian Avenue, Jiangning District, Nanjing,211198,P.R.China. http://tcoa.cpu.edu.cn/ Contact us at xiaosheng.wang@cpu.edu.cn for technical issue and questions Catalogue
Molecular Pathology of Ovarian Carcinoma with Morphological Correlation
 Molecular athology of Ovarian Carcinoma with Morphological Correlation Kathleen R. Cho, M.D. Comprehensive Cancer Center and Departments of athology and Internal Medicine University of Michigan Medical
Molecular athology of Ovarian Carcinoma with Morphological Correlation Kathleen R. Cho, M.D. Comprehensive Cancer Center and Departments of athology and Internal Medicine University of Michigan Medical
Chapter 11 Gene Expression
 Chapter 11 Gene Expression 11-1 Control of Gene Expression Gene Expression- the activation of a gene to form a protein -a gene is on or expressed when it is transcribed. -cells do not always need to produce
Chapter 11 Gene Expression 11-1 Control of Gene Expression Gene Expression- the activation of a gene to form a protein -a gene is on or expressed when it is transcribed. -cells do not always need to produce
Nature Genetics: doi: /ng Supplementary Figure 1. Phenotypic characterization of MES- and ADRN-type cells.
 Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Identifying Mutations Responsible for Rare Disorders Using New Technologies
 Identifying Mutations Responsible for Rare Disorders Using New Technologies Jacek Majewski, Department of Human Genetics, McGill University, Montreal, QC Canada Mendelian Diseases Clear mode of inheritance
Identifying Mutations Responsible for Rare Disorders Using New Technologies Jacek Majewski, Department of Human Genetics, McGill University, Montreal, QC Canada Mendelian Diseases Clear mode of inheritance
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Clinical Spectrum and Genetic Mechanism of GLUT1-DS. Yasushi ITO (Tokyo Women s Medical University, Japan)
 Clinical Spectrum and Genetic Mechanism of GLUT1-DS Yasushi ITO (Tokyo Women s Medical University, Japan) Glucose transporter type 1 (GLUT1) deficiency syndrome Mutation in the SLC2A1 / GLUT1 gene Deficiency
Clinical Spectrum and Genetic Mechanism of GLUT1-DS Yasushi ITO (Tokyo Women s Medical University, Japan) Glucose transporter type 1 (GLUT1) deficiency syndrome Mutation in the SLC2A1 / GLUT1 gene Deficiency
Supplementary Figure 1
 Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.
 Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Exomes and Beyond Addressing the Genome with OneSeq. Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA
 Exomes and Beyond Addressing the Genome with OneSeq Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA Technologies to Detect Various Types of Mutations RESOLUTION Targeted
Exomes and Beyond Addressing the Genome with OneSeq Madhuri Hegde, PhD, FACMG Emory University Emory Genetics Laboratory Atlanta, GA Technologies to Detect Various Types of Mutations RESOLUTION Targeted
Central Dogma. Central Dogma. Translation (mrna -> protein)
 Central Dogma Central Dogma Translation (mrna -> protein) mrna code for amino acids 1. Codons as Triplet code 2. Redundancy 3. Open reading frames 4. Start and stop codons 5. Mistakes in translation 6.
Central Dogma Central Dogma Translation (mrna -> protein) mrna code for amino acids 1. Codons as Triplet code 2. Redundancy 3. Open reading frames 4. Start and stop codons 5. Mistakes in translation 6.
Integration of Cancer Genome into GECCO- Genetics and Epidemiology of Colorectal Cancer Consortium
 Integration of Cancer Genome into GECCO- Genetics and Epidemiology of Colorectal Cancer Consortium Ulrike Peters Fred Hutchinson Cancer Research Center University of Washington U01-CA137088-05, PI: Peters
Integration of Cancer Genome into GECCO- Genetics and Epidemiology of Colorectal Cancer Consortium Ulrike Peters Fred Hutchinson Cancer Research Center University of Washington U01-CA137088-05, PI: Peters
Introduction to LOH and Allele Specific Copy Number User Forum
 Introduction to LOH and Allele Specific Copy Number User Forum Jonathan Gerstenhaber Introduction to LOH and ASCN User Forum Contents 1. Loss of heterozygosity Analysis procedure Types of baselines 2.
Introduction to LOH and Allele Specific Copy Number User Forum Jonathan Gerstenhaber Introduction to LOH and ASCN User Forum Contents 1. Loss of heterozygosity Analysis procedure Types of baselines 2.
Identification of genomic alterations in cervical cancer biopsies by exome sequencing
 Chapter- 4 Identification of genomic alterations in cervical cancer biopsies by exome sequencing 105 4.1 INTRODUCTION Athough HPV has been identified as the prime etiological factor for cervical cancer,
Chapter- 4 Identification of genomic alterations in cervical cancer biopsies by exome sequencing 105 4.1 INTRODUCTION Athough HPV has been identified as the prime etiological factor for cervical cancer,
