p.arg119gly p.arg119his p.ala179thr c.540+1g>a c.617_633+6del Prediction basis structure
|
|
|
- Norma Byrd
- 5 years ago
- Views:
Transcription
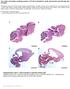
1 a Missense ATG p.arg119gly p.arg119his p.ala179thr p.ala189val p.gly206trp p.gly206arg p.arg251his p.ala257thr TGA 5 UTR UTR Splice Site, Frameshift b Mutation p.gly206trp p.gly4fsx50 c.138+1g>a Prediction probably damaging Prediction basis structure c.540+1g>a c.617_633+6del c.633+1g>c Substitution effect 1.1.2: structural effect, buried site, overpacking Overpacking at buried site c.797g>a; p.arg266gln c.797+1g>a c.797+2_797+5del Prophyler MAPP P- value p.arg266gln benign alignment N/A p.arg119gly probably damaging alignment N/A 9.923*E-4 p.arg119his probably damaging alignment N/A p.ala189val benign alignment N/A 0,2072 p.ala179thr probably damaging alignment N/A 3.392*E-5 p.gly206arg p.arg251his p.ala257thr probably damaging probably damaging probably damaging structure structure structure 1.1.2: structural effect, buried site, overpacking Overpacking at buried site 1.1.1: structural effect, buried site, hydrophobicity Hydrophobicity change at buried site 1.1.1: structural effect, buried site, hydrophobicity Hydrophobicity change at buried site 5.365*E Supplementary Figure 1 Localization and effect of PYCR1 mutations. ( a) Genomic localization. Note that mutations accumulate in exons 4-6 of PYCR1. Red= position of amino acids essential for NAD(P)H binding. Orange= 100% conserved residues. Yellow= >80% conserved residues. White= spice site and frameshift mutations. ( b) Assessment of effect of missense mutations. The computational tools PolyPhen ( and Prophyler ( were used to estimate the effects of the PYCR1 missense mutations on protein function. The first column shows the mutation, columns 2-4 show the PolyPhen predictions, and column 5 shows the Prophyler MAPP P value. Low P-values are strong predictions for deleteriousness, high P-values mean the variant is unlikely to be deleterious. Note that mutation p.arg266gln, which is predicted to be benign by both PolyPhen and Prophyler on the basis of protein characteristics, is predicted to affect splicing by altering the invariable donor splice site at the 3' end of exon 6. Splicing is not judged by these programs. The mutation p.ala189val was also judged to be benign by both programs. In this case, the other allele carries the Arg119His mutation which is predicted to be deleterious. As the p.ala189val change was not present in more than 100 control samples, it likely results in reduced residual activity. However, we cannot rule out a rare variant.
2 a fold expression change fold expression change Fold expression change b ,4 1,2 1,0 0,8 0,6 Liver Kidney Heart Skin Brain Eye spinal nerve Bone Osteoblast Osteoclast Liver Kidney Heart Skin Brain Eye spinal nerve Bone Osteoblast mosteoclast Liver Kidney Heart Skin Brain Eye spinal nerve Bone Osteoblast Osteoclast Liver Kidney Heart Skin Brain Eye Pycr1 Pycr2 Pycrl P5cs spinal nerve Bone Osteoblast Osteoclast 0,4 0,2 0,0 Ctr 1 Ctr 2 HJ DI MJ Ctr 1 Ctr 2 HJ DI MJ Ctr 1 Ctr 2 HJ DI MJ PYCR1 PYCR2 P5CS Supplementary Figure 2 Expression of genes involved in the proline metabolic pathway determined by quantitative PCR. ( a) Expression of genes involved in proline metabolism in tissues from 4 day old mice normalized to expression levels in the heart determined by quantitative PCR. Note extremely high Pycr1 expression levels in osteoblasts and skin. Pycr2 and Pycrl are also most highly expressed in osteoblasts, but otherwise show uniform expression, as does P5cs. ( b) Gene expression changes in fibroblasts from individuals with PYCR1 mutations and controls. While the Gly206Trp mutation in patient TP does not affect PYCR1 expression levels the mutations in DI and SJ clearly reduce mrna levels due to nonsense-mediated decay. Expression of PYCR2 and P5CS is not affected.
3 a [ mol/l] Ctr (n=8) Pat (n=13) Ctr (n=8) Pat (n=13) Proline Ornithine b [ mol/l] Ctr (n=4) Pat (n=4) Proline c % cells BrdU positive 60% 50% 40% 30% 20% 10% 0% Ctr (n=3) Pat (n=3) Ctr (n=3) Pat (n=3) Ctr (n=3) Pat (n=3) 0mMPro 0,2mMPro 5mMPro Supplementary Figure 3 Loss of PYCR1 does not cause hypoprolinemia or a cell proliferation defect. ( a) Proline and ornithine levels in individuals with PYCR1 mutations (Pat) compared to controls (Ctr). Note non-significant (p=0,051) reduction of proline concentrations in affected individuals. ( b) Proline levels in fibroblast lysates from controls and from individuals with PYCR1 mutation. No significant differenced were detectable. (c) Proliferation rates of fibroblasts from individuals with PYCR1 mutations (Pat) and controls (Ctr) grown in medium with different proline concentrations. No significant differences were observed, but mutant cells showed slightly higher proliferation. Representative result of two independent experiments. Ctr = control; Pat = patient. Errors are given as s.d..
4 a b c background DCDHF only d # Cells DCDHF + H 2 O Ctrl (n=2) Pat (n=4) Ctrl (n=2) Pat (n=4) DCDHF Intrinsic ROS level ROS level after treatment with 1mM H2O2 Supplementary Figure 4 Mitochondrial function in fibroblasts and lymphoblastoid cell lines. (a) Respiratory rates (oxygen consumption) were measured in 7 x 106 cultured, digitonin-permeabilized fibroblasts from patients and normal controls. No significant differences were found in respiratory chain function. (b) Respiratory rates (oxygen consumption) were measured in 2 x 107 cultured,digitonin-permeabilized EBV-immortalized lymphoblastoid cell lines (LCL) from patients, heterozygous parents and normal controls. No significant differences were found in respiratory chain function between the different genotypes. ( c) ROS detection by DCDHF. Representative traces showing fluorescence intensity levels without DCDHF, whith DCDHF and with DCDHF and H2O 2. ( c) Quantita- tive comparison of mean DCDHF fluorescence intensity in controls and patients with and without H O. Representative results of one experiment are given. 2 2
5 a b c Control d Pycr1 Mo e Pycr2 Mo f g Pycr1+2 Mo h Pycr1+2 Mo + PYCR1 i Control j Pycr1+2 Mo Pycr1+2 Mo + PYCR1 k l m Control Pycr1+2 Mo Pycr1+2 Mo + PYCR1 n o p Supplementary Figure 5 Morpholino (MO)-mediated knockdown of Pycr1 and Pycr2 in zebrafish leads to malformation, epidermal atrophy and increased apoptosis. ( a-h) Control, Pycr1 morphant, Pycr2 morphant, Pycr1+2 morphant and Pycr1+2 morphant zebrafish injected with 20pg human PYCR1 mrna at stage 24h. ( f-h) Control, Pycr 1+2 morphant and Pycr1+2 morphant zebrafish injected with 20 pg human PYCR1 mrna at stage 48 hours. Note reduced length of morphant fishes, abnormally bowed tail and loosening of the epidermis around the yolk sac. The severity of the phenotype increases when Pycr1 and Pycr2 are both knocked down. Only subtle changes are seen after rescue of the morphant phenotype with 20pg of human PYCR1 mrna. ( i,j) Dermal atrophy in Pycr1+2 morphants. In 48 hours-old control fish () i adouble-layered epidermis surrounds the yolk sac. In Pycr1+2 morphants ( j) this epidermis is atrophic. Goldner trichrome staining. ( k-m) Acridine orange staining of apoptotic cells at stage 24h. Note significant increased number of apoptotic cells in morphants and almost complete reversion of the phenotype by co-injection of the human PYCR1 mrna. ( n-p) Acridine staining of apoptotic cells in the tail, higher magnification.
6
7 Supplementary Table 1 Results of high throughput sequencing Strategy flowchart 5 lanes of Illumina sequence 47,880,719 raw reads Bfast uniquely aligned reads 19,271,149 (40%) Total mismatches to reference in chr17: ,611,834 Mismatches to reference >= 2x coverage, % 71,495 Mismatches to reference >= 2x coverage, homozygous (80-100%) 3,083 Mismatches to reference in dbsnp 2,531 Mismatches to reference not in dbsnp 552 Mismatches causing a coding change in UCSC genes 11 Intersection of genes with mutations in other samples
8 Supplementary Table 1 Results of high throughput sequencing List of mismatches on chr17 pos Chromosome Mismatch position Mismatch description and Coverage chr C >T(10:10:100%[F:7:70% R:3:30%]) chr G >A(15:15:100%[F:13:86.7% R:2:13.3%]) chr C >T(8:8:100%[F:7:87.5% R:1:12.5%]) chr G >A(2:2:100%[F:0:0% R:2:100%]) chr C >T(2:2:100%[F:0:0% R:2:100%]) chr C >A(2:2:100%[F:2:100% R:0:0%]) chr C >T(7:7:100%[F:6:85.7% R:1:14.3%]) chr T >C(2:2:100%[F:2:100% R:0:0%]) chr T >G(2:2:100%[F:2:100% R:0:0%]) chr T >C(3:3:100%[F:0:0% R:3:100%]) chr T >G(16:16:100%[F:3:18.8% R:13:81.2%]) chr G >A(5:5:100%[F:4:80% R:1:20%]) chr C >T(2:2:100%[F:2:100% R:0:0%]) chr A >T(11:11:100%[F:8:72.7% R:3:27.3%]) chr G >A(4:4:100%[F:0:0% R:4:100%]) chr A >C(2:2:100%[F:0:0% R:2:100%]) chr G >A(2:2:100%[F:2:100% R:0:0%]) chr T >C(12:12:100%[F:7:58.3% R:5:41.7%]) chr C >G(2:2:100%[F:2:100% R:0:0%]) chr G >C(2:2:100%[F:0:0% R:2:100%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr DEL:C > (5:5:100%[F:1 R:4]) chr T >C(3:3:100%[F:3:100% R:0:0%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr G >C(25:25:100%[F:11:44% R:14:56%]) chr C >T(9:9:100%[F:4:44.4% R:5:55.6%]) chr G >A(3:3:100%[F:3:100% R:0:0%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr T >C(2:2:100%[F:0:0% R:2:100%]) chr T >C(2:2:100%[F:0:0% R:2:100%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr A >G(7:7:100%[F:5:71.4% R:2:28.6%]) chr C >T(20:20:100%[F:10:50% R:10:50%]) chr C >G(3:3:100%[F:1:33.3% R:2:66.7%]) chr C >G(2:2:100%[F:1:50% R:1:50%]) chr T >G(43:43:100%[F:18:41.9% R:25:58.1%]) chr T >C(6:6:100%[F:1:16.7% R:5:83.3%]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr T >C(2:2:100%[F:1:50% R:1:50%]) chr C >G(2:2:100%[F:2:100% R:0:0%]) chr G >A(11:11:100%[F:5:45.5% R:6:54.5%]) chr C >G(2:2:100%[F:1:50% R:1:50%]) chr C >T(55:55:100%[F:4:7.27% R:51:92.7%]) chr C >A(11:11:100%[F:3:27.3% R:8:72.7%]) chr C >G(3:3:100%[F:0:0% R:3:100%]) chr C >T(16:16:100%[F:11:68.8% R:5:31.2%])
9 chr G >A(11:11:100%[F:6:54.5% R:5:45.5%]) chr A >G(2:2:100%[F:0:0% R:2:100%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr A >G(4:4:100%[F:1:25% R:3:75%]) chr DEL:T > (8:8:100%[F:3 R:5]) chr C >T(12:12:100%[F:9:75% R:3:25%]) chr T >G(6:6:100%[F:2:33.3% R:4:66.7%]) chr G >A(39:39:100%[F:22:56.4% R:17:43.6%]) chr T >C(8:8:100%[F:0:0% R:8:100%]) chr DEL:GCAC > (3:3:100%[F:3 R:0]) chr A >G(4:4:100%[F:4:100% R:0:0%]) chr DEL:T > (2:2:100%[F:0 R:2]) chr T >G(2:2:100%[F:1:50% R:1:50%]) chr G >A(2:2:100%[F:2:100% R:0:0%]) chr G >A(9:9:100%[F:8:88.9% R:1:11.1%]) chr A >C(2:2:100%[F:1:50% R:1:50%]) chr G >A(2:2:100%[F:1:50% R:1:50%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr T >C(2:2:100%[F:2:100% R:0:0%]) chr A >C(2:2:100%[F:0:0% R:2:100%]) chr T >G(2:2:100%[F:1:50% R:1:50%]) chr C >G(17:17:100%[F:0:0% R:17:100%]) chr C >A(2:2:100%[F:0:0% R:2:100%]) chr C >T(15:15:100%[F:9:60% R:6:40%]) chr T >A(3:3:100%[F:0:0% R:3:100%]) chr T >A(2:2:100%[F:0:0% R:2:100%]) chr A >C(2:2:100%[F:0:0% R:2:100%]) chr T >A(4:4:100%[F:4:100% R:0:0%]) chr G >C(4:4:100%[F:3:75% R:1:25%]) chr C >G(19:19:100%[F:11:57.9% R:8:42.1%]) chr G >T(3:3:100%[F:3:100% R:0:0%]) chr INS: >T(2:2:100%[F:1 R:0]) chr A >G(18:18:100%[F:8:44.4% R:10:55.6%]) chr C >A(4:4:100%[F:4:100% R:0:0%]) chr T >C(2:2:100%[F:0:0% R:2:100%]) chr C >T(4:4:100%[F:3:75% R:1:25%]) chr C >G(2:2:100%[F:2:100% R:0:0%]) chr C >T(2:2:100%[F:2:100% R:0:0%]) chr INS: >C(2:2:100%[F:1 R:0]) chr T >A(8:8:100%[F:1:12.5% R:7:87.5%]) chr A >T(2:2:100%[F:2:100% R:0:0%]) chr T >C(22:22:100%[F:12:54.5% R:10:45.5%]) chr G >A(2:2:100%[F:2:100% R:0:0%]) chr INS: >A(3:3:100%[F:3 R:0]) chr C >G(2:2:100%[F:0:0% R:2:100%]) chr DEL:A > (2:2:100%[F:2 R:0]) chr DEL:AC > (3:3:100%[F:3 R:0]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr C >T(16:16:100%[F:8:50% R:8:50%]) chr G >A(9:9:100%[F:5:55.6% R:4:44.4%])
10 chr C >A(2:2:100%[F:0:0% R:2:100%]) chr C >T(22:22:100%[F:12:54.5% R:10:45.5%]) chr A >G(55:55:100%[F:23:41.8% R:32:58.2%]) chr C >A(2:2:100%[F:2:100% R:0:0%]) chr C >G(2:2:100%[F:0:0% R:2:100%]) chr C >A(12:12:100%[F:9:75% R:3:25%]) chr G >A(16:16:100%[F:13:81.2% R:3:18.8%]) chr T >G(4:4:100%[F:1:25% R:3:75%]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr T >G(2:2:100%[F:2:100% R:0:0%]) chr T >C(3:3:100%[F:0:0% R:3:100%]) chr G >A(2:2:100%[F:2:100% R:0:0%]) chr T >A(2:2:100%[F:0:0% R:2:100%]) chr C >G(19:19:100%[F:17:89.5% R:2:10.5%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr C >T(2:2:100%[F:0:0% R:2:100%]) chr C >T(3:3:100%[F:3:100% R:0:0%]) chr A >G(3:3:100%[F:0:0% R:3:100%]) chr G >A(19:19:100%[F:14:73.7% R:5:26.3%]) chr DEL:T > (2:2:100%[F:0 R:2]) chr C >T(13:13:100%[F:8:61.5% R:5:38.5%]) chr C >A(2:2:100%[F:2:100% R:0:0%]) chr G >A(5:5:100%[F:1:20% R:4:80%]) chr G >C(3:3:100%[F:1:33.3% R:2:66.7%]) chr INS: >C(6:6:100%[F:3 R:0]) chr A >G(15:15:100%[F:3:20% R:12:80%]) chr G >A(6:6:100%[F:1:16.7% R:5:83.3%]) chr A >T(2:2:100%[F:1:50% R:1:50%]) chr G >C(15:15:100%[F:6:40% R:9:60%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr T >C(28:28:100%[F:24:85.7% R:4:14.3%]) chr A >C(25:25:100%[F:13:52% R:12:48%]) chr G >A(11:11:100%[F:7:63.6% R:4:36.4%]) chr A >G(25:25:100%[F:21:84% R:4:16%]) chr C >G(26:26:100%[F:12:46.2% R:14:53.8%]) chr A >C(4:4:100%[F:2:50% R:2:50%]) chr DEL:CTT > (2:2:100%[F:1 R:1]) chr G >T(2:2:100%[F:1:50% R:1:50%]) chr G >A(36:36:100%[F:20:55.6% R:16:44.4%]) chr G >T(8:8:100%[F:5:62.5% R:3:37.5%]) chr C >A(6:6:100%[F:2:33.3% R:4:66.7%]) chr G >C(10:10:100%[F:0:0% R:10:100%]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr DEL:G > (2:2:100%[F:2 R:0]) chr T >G(8:8:100%[F:6:75% R:2:25%]) chr T >C(2:2:100%[F:0:0% R:2:100%]) chr G >A(22:22:100%[F:16:72.7% R:6:27.3%]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr G >A(20:20:100%[F:15:75% R:5:25%]) chr G >A(3:3:100%[F:0:0% R:3:100%])
11 chr A >G(2:2:100%[F:0:0% R:2:100%]) chr G >A(2:2:100%[F:2:100% R:0:0%]) chr A >G(14:14:100%[F:8:57.1% R:6:42.9%]) chr G >A(12:12:100%[F:8:66.7% R:4:33.3%]) chr G >T(14:14:100%[F:1:7.14% R:13:92.9%]) chr T >C(8:8:100%[F:5:62.5% R:3:37.5%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr A >G(2:2:100%[F:1:50% R:1:50%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr C >T(5:5:100%[F:3:60% R:2:40%]) chr T >G(3:3:100%[F:1:33.3% R:2:66.7%]) chr T >A(8:8:100%[F:0:0% R:8:100%]) chr T >G(5:5:100%[F:5:100% R:0:0%]) chr C >T(11:11:100%[F:8:72.7% R:3:27.3%]) chr A >T(2:2:100%[F:2:100% R:0:0%]) chr G >T(2:2:100%[F:0:0% R:2:100%]) chr G >T(2:2:100%[F:0:0% R:2:100%]) chr G >A(3:3:100%[F:3:100% R:0:0%]) chr C >T(13:13:100%[F:9:69.2% R:4:30.8%]) chr A >G(2:2:100%[F:0:0% R:2:100%]) chr A >G(25:25:100%[F:18:72% R:7:28%]) chr T >C(12:12:100%[F:6:50% R:6:50%]) chr G >A(3:3:100%[F:1:33.3% R:2:66.7%]) chr C >G(2:2:100%[F:0:0% R:2:100%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr G >T(2:2:100%[F:2:100% R:0:0%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr C >T(4:4:100%[F:2:50% R:2:50%]) chr C >T(9:9:100%[F:5:55.6% R:4:44.4%]) chr A >G(4:4:100%[F:1:25% R:3:75%]) chr T >A(10:10:100%[F:9:90% R:1:10%]) chr G >A(2:2:100%[F:0:0% R:2:100%]) chr C >T(19:19:100%[F:17:89.5% R:2:10.5%]) chr C >G(2:2:100%[F:2:100% R:0:0%]) chr T >C(23:23:100%[F:11:47.8% R:12:52.2%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr A >C(6:6:100%[F:6:100% R:0:0%]) chr T >C(3:3:100%[F:2:66.7% R:1:33.3%]) chr T >C(4:4:100%[F:3:75% R:1:25%]) chr C >T(2:2:100%[F:2:100% R:0:0%]) chr T >C(2:2:100%[F:1:50% R:1:50%]) chr C >A(4:4:100%[F:4:100% R:0:0%]) chr G >A(10:10:100%[F:6:60% R:4:40%]) chr G >A(11:11:100%[F:7:63.6% R:4:36.4%]) chr A >G(20:20:100%[F:12:60% R:8:40%]) chr C >T(14:14:100%[F:9:64.3% R:5:35.7%]) chr INS: >C(2:2:100%[F:2 R:0]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr T >C(2:2:100%[F:2:100% R:0:0%]) chr A >T(2:2:100%[F:2:100% R:0:0%])
12 chr A >T(2:2:100%[F:2:100% R:0:0%]) chr G >A(6:6:100%[F:4:66.7% R:2:33.3%]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr T >G(2:2:100%[F:2:100% R:0:0%]) chr T >G(17:17:100%[F:11:64.7% R:6:35.3%]) chr G >T(2:2:100%[F:2:100% R:0:0%]) chr DEL:T > (2:2:100%[F:0 R:2]) chr A >G(2:2:100%[F:1:50% R:1:50%]) chr G >A(2:2:100%[F:1:50% R:1:50%]) chr A >G(28:28:100%[F:17:60.7% R:11:39.3%]) chr T >C(2:2:100%[F:0:0% R:2:100%]) chr C >T(21:21:100%[F:11:52.4% R:10:47.6%]) chr T >G(9:9:100%[F:6:66.7% R:3:33.3%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr T >C(17:17:100%[F:14:82.4% R:3:17.6%]) chr G >A(3:3:100%[F:3:100% R:0:0%]) chr C >T(8:8:100%[F:5:62.5% R:3:37.5%]) chr DEL:A > (2:2:100%[F:1 R:1]) chr T >C(8:8:100%[F:4:50% R:4:50%]) chr C >A(2:2:100%[F:1:50% R:1:50%]) chr T >C(8:8:100%[F:4:50% R:4:50%]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr A >G(2:2:100%[F:1:50% R:1:50%]) chr DEL:A > (2:2:100%[F:2 R:0]) chr INS: >A(17:17:100%[F:9 R:0]) chr C >G(2:2:100%[F:0:0% R:2:100%]) chr A >T(2:2:100%[F:0:0% R:2:100%]) chr T >G(2:2:100%[F:2:100% R:0:0%]) chr G >A(4:4:100%[F:2:50% R:2:50%]) chr T >C(2:2:100%[F:2:100% R:0:0%]) chr A >G(15:15:100%[F:7:46.7% R:8:53.3%]) chr A >C(2:2:100%[F:1:50% R:1:50%]) chr C >G(2:2:100%[F:0:0% R:2:100%]) chr C >T(2:2:100%[F:2:100% R:0:0%]) chr INS: >T(23:23:100%[F:7 R:0]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr C >A(2:2:100%[F:0:0% R:2:100%]) chr INS: >T(5:5:100%[F:5 R:0]) chr C >T(7:7:100%[F:4:57.1% R:3:42.9%]) chr A >G(2:2:100%[F:1:50% R:1:50%]) chr G >A(11:11:100%[F:5:45.5% R:6:54.5%]) chr G >A(13:13:100%[F:9:69.2% R:4:30.8%]) chr INS: >G(4:4:100%[F:1 R:0]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr T >G(28:28:100%[F:12:42.9% R:16:57.1%]) chr C >G(9:9:100%[F:2:22.2% R:7:77.8%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr T >C(2:2:100%[F:0:0% R:2:100%]) chr C >T(2:2:100%[F:1:50% R:1:50%]) chr C >G(13:13:100%[F:5:38.5% R:8:61.5%])
13 chr T >C(3:3:100%[F:2:66.7% R:1:33.3%]) chr T >C(34:34:100%[F:20:58.8% R:14:41.2%]) chr G >A(4:4:100%[F:4:100% R:0:0%]) chr INS: >G(2:2:100%[F:2 R:0]) chr C >T(14:14:100%[F:9:64.3% R:5:35.7%]) chr C >G(2:2:100%[F:1:50% R:1:50%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr DEL:G > (2:2:100%[F:0 R:2]) chr G >T(8:8:100%[F:8:100% R:0:0%]) chr A >T(3:3:100%[F:3:100% R:0:0%]) chr C >T(24:24:100%[F:13:54.2% R:11:45.8%]) chr DEL:A > (2:2:100%[F:1 R:1]) chr C >T(7:7:100%[F:6:85.7% R:1:14.3%]) chr C >A(57:57:100%[F:14:24.6% R:43:75.4%]) chr T >A(20:20:100%[F:14:70% R:6:30%]) chr C >T(2:2:100%[F:2:100% R:0:0%]) chr C >A(3:3:100%[F:3:100% R:0:0%]) chr T >C(11:11:100%[F:6:54.5% R:5:45.5%]) chr C >A(5:5:100%[F:5:100% R:0:0%]) chr G >A(9:9:100%[F:6:66.7% R:3:33.3%]) chr G >A(4:4:100%[F:0:0% R:4:100%]) chr T >G(2:2:100%[F:1:50% R:1:50%]) chr T >C(2:2:100%[F:0:0% R:2:100%]) chr C >T(2:2:100%[F:1:50% R:1:50%]) chr C >T(4:4:100%[F:0:0% R:4:100%]) chr C >G(2:2:100%[F:0:0% R:2:100%]) chr T >G(3:3:100%[F:2:66.7% R:1:33.3%]) chr A >G(2:2:100%[F:1:50% R:1:50%]) chr G >A(8:8:100%[F:3:37.5% R:5:62.5%]) chr C >T(21:21:100%[F:13:61.9% R:8:38.1%]) chr G >C(24:24:100%[F:8:33.3% R:16:66.7%]) chr T >C(3:3:100%[F:3:100% R:0:0%]) chr C >T(18:18:100%[F:4:22.2% R:14:77.8%]) chr DEL:CTTCTG > (2:2:100%[F:2 R:0]) chr A >T(6:6:100%[F:3:50% R:3:50%]) chr G >C(21:21:100%[F:11:52.4% R:10:47.6%]) chr C >A(3:3:100%[F:3:100% R:0:0%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr G >C(10:10:100%[F:2:20% R:8:80%]) chr T >C(2:2:100%[F:1:50% R:1:50%]) chr T >C(8:8:100%[F:1:12.5% R:7:87.5%]) chr C >A(2:2:100%[F:1:50% R:1:50%]) chr T >C(2:2:100%[F:2:100% R:0:0%]) chr T >G(2:2:100%[F:2:100% R:0:0%]) chr INS: >G(21:21:100%[F:20 R:0]) chr A >G(16:16:100%[F:13:81.2% R:3:18.8%]) chr G >C(27:27:100%[F:20:74.1% R:7:25.9%]) chr A >G(7:7:100%[F:3:42.9% R:4:57.1%]) chr C >G(3:3:100%[F:0:0% R:3:100%]) chr A >G(18:18:100%[F:16:88.9% R:2:11.1%])
14 chr T >C(6:6:100%[F:0:0% R:6:100%]) chr G >A(6:6:100%[F:4:66.7% R:2:33.3%]) chr A >G(12:12:100%[F:10:83.3% R:2:16.7%]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr G >C(11:11:100%[F:5:45.5% R:6:54.5%]) chr G >A(19:19:100%[F:6:31.6% R:13:68.4%]) chr C >T(7:7:100%[F:3:42.9% R:4:57.1%]) chr G >T(2:2:100%[F:2:100% R:0:0%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr G >T(2:2:100%[F:1:50% R:1:50%]) chr C >T(13:13:100%[F:8:61.5% R:5:38.5%]) chr T >C(14:14:100%[F:9:64.3% R:5:35.7%]) chr INS: >A(4:4:100%[F:4 R:0]) chr C >G(2:2:100%[F:1:50% R:1:50%]) chr T >A(3:3:100%[F:0:0% R:3:100%]) chr T >C(42:42:100%[F:37:88.1% R:5:11.9%]) chr DEL:TG > (5:5:100%[F:4 R:1]) chr G >T(2:2:100%[F:2:100% R:0:0%]) chr C >G(20:20:100%[F:11:55% R:9:45%]) chr A >T(2:2:100%[F:2:100% R:0:0%]) chr G >T(3:3:100%[F:2:66.7% R:1:33.3%]) chr A >C(2:2:100%[F:0:0% R:2:100%]) chr A >T(5:5:100%[F:4:80% R:1:20%]) chr A >G(23:23:100%[F:5:21.7% R:18:78.3%]) chr G >A(5:5:100%[F:4:80% R:1:20%]) chr A >G(2:2:100%[F:0:0% R:2:100%]) chr C >G(2:2:100%[F:2:100% R:0:0%]) chr C >T(14:14:100%[F:11:78.6% R:3:21.4%]) chr C >T(6:6:100%[F:3:50% R:3:50%]) chr G >A(53:53:100%[F:47:88.7% R:6:11.3%]) chr A >G(34:34:100%[F:16:47.1% R:18:52.9%]) chr A >G(3:3:100%[F:2:66.7% R:1:33.3%]) chr T >G(2:2:100%[F:1:50% R:1:50%]) chr G >T(6:6:100%[F:6:100% R:0:0%]) chr G >A(8:8:100%[F:4:50% R:4:50%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr C >T(2:2:100%[F:2:100% R:0:0%]) chr C >T(9:9:100%[F:5:55.6% R:4:44.4%]) chr C >G(3:3:100%[F:3:100% R:0:0%]) chr T >C(2:2:100%[F:0:0% R:2:100%]) chr C >A(2:2:100%[F:0:0% R:2:100%]) chr G >A(8:8:100%[F:4:50% R:4:50%]) chr DEL:C > (4:4:100%[F:3 R:1]) chr G >A(2:2:100%[F:0:0% R:2:100%]) chr G >A(17:17:100%[F:6:35.3% R:11:64.7%]) chr T >C(2:2:100%[F:0:0% R:2:100%]) chr T >G(2:2:100%[F:0:0% R:2:100%]) chr G >T(2:2:100%[F:0:0% R:2:100%]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr T >A(17:17:100%[F:7:41.2% R:10:58.8%])
15 chr A >C(2:2:100%[F:2:100% R:0:0%]) chr A >C(2:2:100%[F:1:50% R:1:50%]) chr T >C(3:3:100%[F:1:33.3% R:2:66.7%]) chr A >G(18:18:100%[F:6:33.3% R:12:66.7%]) chr C >T(12:12:100%[F:9:75% R:3:25%]) chr C >A(23:23:100%[F:12:52.2% R:11:47.8%]) chr G >A(42:42:100%[F:31:73.8% R:11:26.2%]) chr A >G(13:13:100%[F:6:46.2% R:7:53.8%]) chr T >C(2:2:100%[F:1:50% R:1:50%]) chr G >T(2:2:100%[F:2:100% R:0:0%]) chr A >T(12:12:100%[F:10:83.3% R:2:16.7%]) chr A >T(2:2:100%[F:0:0% R:2:100%]) chr C >T(4:4:100%[F:3:75% R:1:25%]) chr G >T(31:31:100%[F:12:38.7% R:19:61.3%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr T >G(2:2:100%[F:2:100% R:0:0%]) chr C >G(8:8:100%[F:1:12.5% R:7:87.5%]) chr A >C(3:3:100%[F:0:0% R:3:100%]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr C >T(2:2:100%[F:2:100% R:0:0%]) chr C >G(6:6:100%[F:5:83.3% R:1:16.7%]) chr DEL:CAG > (3:3:100%[F:1 R:2]) chr C >T(2:2:100%[F:2:100% R:0:0%]) chr T >G(2:2:100%[F:1:50% R:1:50%]) chr INS: >C(7:7:100%[F:0 R:0]) chr G >C(21:21:100%[F:9:42.9% R:12:57.1%]) chr T >A(12:12:100%[F:3:25% R:9:75%]) chr T >C(5:5:100%[F:1:20% R:4:80%]) chr C >T(7:7:100%[F:3:42.9% R:4:57.1%]) chr G >A(19:19:100%[F:11:57.9% R:8:42.1%]) chr G >T(2:2:100%[F:2:100% R:0:0%]) chr INS: >A(2:2:100%[F:2 R:0]) chr G >A(6:6:100%[F:1:16.7% R:5:83.3%]) chr T >A(3:3:100%[F:0:0% R:3:100%]) chr C >G(2:2:100%[F:1:50% R:1:50%]) chr G >A(4:4:100%[F:2:50% R:2:50%]) chr G >A(15:15:100%[F:5:33.3% R:10:66.7%]) chr G >C(26:26:100%[F:9:34.6% R:17:65.4%]) chr G >A(42:42:100%[F:28:66.7% R:14:33.3%]) chr A >C(10:10:100%[F:9:90% R:1:10%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr G >A(41:41:100%[F:25:61% R:16:39%]) chr C >T(38:38:100%[F:26:68.4% R:12:31.6%]) chr C >T(8:8:100%[F:5:62.5% R:3:37.5%]) chr C >T(15:15:100%[F:7:46.7% R:8:53.3%]) chr A >G(7:7:100%[F:7:100% R:0:0%]) chr C >T(19:19:100%[F:9:47.4% R:10:52.6%]) chr C >G(3:3:100%[F:3:100% R:0:0%]) chr T >G(3:3:100%[F:0:0% R:3:100%]) chr T >G(2:2:100%[F:0:0% R:2:100%])
16 chr T >C(3:3:100%[F:3:100% R:0:0%]) chr T >C(12:12:100%[F:6:50% R:6:50%]) chr INS: >C(2:2:100%[F:2 R:0]) chr C >T(9:9:100%[F:7:77.8% R:2:22.2%]) chr C >T(25:25:100%[F:14:56% R:11:44%]) chr A >T(2:2:100%[F:2:100% R:0:0%]) chr G >C(2:2:100%[F:0:0% R:2:100%]) chr C >T(3:3:100%[F:0:0% R:3:100%]) chr C >A(4:4:100%[F:0:0% R:4:100%]) chr G >T(45:45:100%[F:24:53.3% R:21:46.7%]) chr T >C(19:19:100%[F:15:78.9% R:4:21.1%]) chr A >G(9:9:100%[F:8:88.9% R:1:11.1%]) chr G >C(2:2:100%[F:1:50% R:1:50%]) chr G >A(25:25:100%[F:8:32% R:17:68%]) chr G >A(11:11:100%[F:11:100% R:0:0%]) chr G >A(31:31:100%[F:25:80.6% R:6:19.4%]) chr C >T(3:3:100%[F:0:0% R:3:100%]) chr A >C(2:2:100%[F:0:0% R:2:100%]) chr C >G(2:2:100%[F:2:100% R:0:0%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr T >G(2:2:100%[F:2:100% R:0:0%]) chr G >A(8:8:100%[F:5:62.5% R:3:37.5%]) chr A >C(2:2:100%[F:1:50% R:1:50%]) chr G >A(2:2:100%[F:0:0% R:2:100%]) chr G >C(24:24:100%[F:16:66.7% R:8:33.3%]) chr G >C(6:6:100%[F:3:50% R:3:50%]) chr A >T(12:12:100%[F:8:66.7% R:4:33.3%]) chr T >C(3:3:100%[F:0:0% R:3:100%]) chr A >G(8:8:100%[F:6:75% R:2:25%]) chr C >A(2:2:100%[F:0:0% R:2:100%]) chr T >A(2:2:100%[F:2:100% R:0:0%]) chr G >T(2:2:100%[F:2:100% R:0:0%]) chr C >T(14:14:100%[F:11:78.6% R:3:21.4%]) chr A >C(3:3:100%[F:2:66.7% R:1:33.3%]) chr DEL:A > (29:29:100%[F:22 R:7]) chr C >A(9:9:100%[F:2:22.2% R:7:77.8%]) chr A >G(12:12:100%[F:6:50% R:6:50%]) chr T >A(2:2:100%[F:0:0% R:2:100%]) chr A >C(2:2:100%[F:0:0% R:2:100%]) chr G >C(27:27:100%[F:16:59.3% R:11:40.7%]) chr T >C(2:2:100%[F:2:100% R:0:0%]) chr G >T(9:9:100%[F:2:22.2% R:7:77.8%]) chr A >G(4:4:100%[F:2:50% R:2:50%]) chr G >T(2:2:100%[F:2:100% R:0:0%]) chr T >C(26:26:100%[F:17:65.4% R:9:34.6%]) chr DEL:TT > (2:2:100%[F:2 R:0]) chr G >A(3:3:100%[F:3:100% R:0:0%]) chr A >G(3:3:100%[F:3:100% R:0:0%]) chr C >A(2:2:100%[F:0:0% R:2:100%]) chr C >T(5:5:100%[F:3:60% R:2:40%])
17 chr T >C(43:43:100%[F:28:65.1% R:15:34.9%]) chr DEL:C > (2:2:100%[F:2 R:0]) chr C >G(23:23:100%[F:15:65.2% R:8:34.8%]) chr C >G(2:2:100%[F:2:100% R:0:0%]) chr C >T(2:2:100%[F:0:0% R:2:100%]) chr C >T(12:12:100%[F:7:58.3% R:5:41.7%]) chr C >T(25:25:100%[F:19:76% R:6:24%]) chr T >G(18:18:100%[F:15:83.3% R:3:16.7%]) chr G >A(2:2:100%[F:0:0% R:2:100%]) chr G >A(2:2:100%[F:2:100% R:0:0%]) chr C >G(90:90:100%[F:8:8.89% R:82:91.1%]) chr G >A(3:3:100%[F:2:66.7% R:1:33.3%]) chr T >G(2:2:100%[F:2:100% R:0:0%]) chr G >A(2:2:100%[F:2:100% R:0:0%]) chr G >C(2:2:100%[F:0:0% R:2:100%]) chr C >T(17:17:100%[F:9:52.9% R:8:47.1%]) chr C >T(30:30:100%[F:19:63.3% R:11:36.7%]) chr A >G(24:24:100%[F:16:66.7% R:8:33.3%]) chr A >C(3:3:100%[F:1:33.3% R:2:66.7%]) chr T >G(2:2:100%[F:2:100% R:0:0%]) chr C >T(20:20:100%[F:10:50% R:10:50%]) chr C >T(12:12:100%[F:7:58.3% R:5:41.7%]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr A >G(5:5:100%[F:1:20% R:4:80%]) chr G >A(12:12:100%[F:8:66.7% R:4:33.3%]) chr G >A(34:34:100%[F:14:41.2% R:20:58.8%]) chr INS: >C(21:21:100%[F:8 R:0]) chr C >T(30:30:100%[F:14:46.7% R:16:53.3%]) chr T >A(2:2:100%[F:2:100% R:0:0%]) chr T >A(6:6:100%[F:6:100% R:0:0%]) chr T >G(2:2:100%[F:2:100% R:0:0%]) chr G >A(20:20:100%[F:10:50% R:10:50%]) chr T >C(2:2:100%[F:0:0% R:2:100%]) chr T >C(8:8:100%[F:7:87.5% R:1:12.5%]) chr G >C(2:2:100%[F:1:50% R:1:50%]) chr G >A(10:10:100%[F:5:50% R:5:50%]) chr C >T(16:16:100%[F:8:50% R:8:50%]) chr G >T(3:3:100%[F:3:100% R:0:0%]) chr A >G(16:16:100%[F:6:37.5% R:10:62.5%]) chr T >C(6:6:100%[F:6:100% R:0:0%]) chr A >C(3:3:100%[F:3:100% R:0:0%]) chr C >T(19:19:100%[F:16:84.2% R:3:15.8%]) chr C >A(2:2:100%[F:2:100% R:0:0%]) chr A >C(2:2:100%[F:0:0% R:2:100%]) chr C >T(7:7:100%[F:5:71.4% R:2:28.6%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr T >C(10:10:100%[F:1:10% R:9:90%]) chr G >A(10:10:100%[F:5:50% R:5:50%]) chr A >C(2:2:100%[F:1:50% R:1:50%]) chr C >G(2:2:100%[F:2:100% R:0:0%])
18 chr A >G(9:9:100%[F:7:77.8% R:2:22.2%]) chr C >T(8:8:100%[F:5:62.5% R:3:37.5%]) chr T >C(7:7:100%[F:4:57.1% R:3:42.9%]) chr G >C(2:2:100%[F:0:0% R:2:100%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr G >C(20:20:100%[F:12:60% R:8:40%]) chr A >C(7:7:100%[F:0:0% R:7:100%]) chr A >G(16:16:100%[F:10:62.5% R:6:37.5%]) chr C >T(20:20:100%[F:13:65% R:7:35%]) chr T >C(12:12:100%[F:4:33.3% R:8:66.7%]) chr G >A(6:6:100%[F:3:50% R:3:50%]) chr DEL:AGTG > (5:5:100%[F:2 R:3]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr C >A(2:2:100%[F:0:0% R:2:100%]) chr A >G(2:2:100%[F:0:0% R:2:100%]) chr A >C(2:2:100%[F:1:50% R:1:50%]) chr G >T(4:4:100%[F:2:50% R:2:50%]) chr INS: >TA(2:2:100%[F:2 R:0]) chr G >A(4:4:100%[F:2:50% R:2:50%]) chr A >G(2:2:100%[F:2:100% R:0:0%]) chr C >G(2:2:100%[F:1:50% R:1:50%]) chr C >T(5:5:100%[F:4:80% R:1:20%]) chr C >T(20:20:100%[F:18:90% R:2:10%]) chr C >T(14:14:100%[F:3:21.4% R:11:78.6%]) chr G >A(2:2:100%[F:2:100% R:0:0%]) chr C >T(8:8:100%[F:2:25% R:6:75%]) chr G >A(6:6:100%[F:2:33.3% R:4:66.7%]) chr G >A(13:13:100%[F:11:84.6% R:2:15.4%]) chr C >T(7:7:100%[F:5:71.4% R:2:28.6%]) chr C >T(2:2:100%[F:1:50% R:1:50%]) chr A >C(7:7:100%[F:6:85.7% R:1:14.3%]) chr G >T(2:2:100%[F:2:100% R:0:0%]) chr T >G(2:2:100%[F:1:50% R:1:50%]) chr C >T(6:6:100%[F:0:0% R:6:100%]) chr C >T(19:19:100%[F:10:52.6% R:9:47.4%]) chr INS: >G(4:4:100%[F:1 R:0]) chr C >T(3:3:100%[F:2:66.7% R:1:33.3%]) chr T >G(2:2:100%[F:1:50% R:1:50%]) chr T >C(10:10:100%[F:5:50% R:5:50%]) chr A >C(2:2:100%[F:2:100% R:0:0%]) chr C >T(2:2:100%[F:0:0% R:2:100%]) chr A >G(19:19:100%[F:9:47.4% R:10:52.6%]) chr C >T(4:4:100%[F:0:0% R:4:100%]) chr T >C(4:4:100%[F:4:100% R:0:0%]) chr T >C(3:3:100%[F:0:0% R:3:100%]) chr C >A(2:2:100%[F:1:50% R:1:50%]) chr A >C(5:5:100%[F:4:80% R:1:20%]) chr C >A(7:7:100%[F:4:57.1% R:3:42.9%]) chr G >A(10:10:100%[F:6:60% R:4:40%]) chr G >A(9:9:100%[F:3:33.3% R:6:66.7%])
19 chr C >G(2:2:100%[F:0:0% R:2:100%]) chr C >T(7:7:100%[F:2:28.6% R:5:71.4%]) chr C >T(18:18:100%[F:16:88.9% R:2:11.1%]) chr T >C(4:4:100%[F:2:50% R:2:50%]) chr T >C(2:2:100%[F:1:50% R:1:50%]) chr T >A(5:5:100%[F:1:20% R:4:80%])
20 Supplementary Table 1 Results of high throughput sequencing List of novel non synonymous sequence changes Known_Gene Known_Gene_URL Coding_Region Strand Mutation Mutation_Type Mutation_Position Codon_Position Codon_Change AA_Change ST6GALNAC2 bin/hggene?hgg_gene=uc002jsg.2 chr17: A VAR chr17: CAT >AAT M >I RNF213 bin/hggene?hgg_gene=uc002jyh.1 chr17: G VAR chr17: ATC >GTC I >V PYCR1 bin/hggene?hgg_gene=uc002kcr.1 chr17: T VAR chr17: GCG >GTG R >H UTS2R bin/hggene?hgg_gene=uc002ker.1 chr17: G VAR chr17: TCC >GCC S >A SGSH bin/hggene?hgg_gene=uc002jxz.2 chr17: G VAR chr17: CTC >CTG E >Q C17orf90 bin/hggene?hgg_gene=uc002kbb.2 chr17: G VAR chr17: GGT >GGG T >P SLC16A3 bin/hggene?hgg_gene=uc002kea.1 chr17: G VAR chr17: GCC >GGC A >G
21 Supplementary Table 2 Primer and morpholino sequences Supplementary Table 2.1 Genomic DNA primer sequences Exon Primer Sequence 1 right GAACCTCCACCCCTCCAG left CTCCCCATTCCCAGGAAG 2 right AGCCAAAGCTAGCCATGAAG left CAGGTCCCAAGAGCAATCAG 3-4 right CAGCATTCTCTGTGCCATTC left TTCTCCTCCTTCCCTTCTGG 5-6 right GGTGGAAGAGGACCTGATTG left ATCTGCTGAGTGCGTGAATG 7 right TGGTGATGCTGAGCTGATTC left TTCTCACACGGGAAGGAGAG Product Size [bp] Supplementary Table 2.2 Human cdna primer sequences used for RT-PCR Gene Primer Sequence Product Size [bp] GAPDH PYCR1 PYCR2 P5CS right right right right TGCACCACCAACTGCTTAGC ATGAGCGTGGGCTTCATC TCGGCTCACAAGATAATAGCC GGCTGATGGCCTTGTATGAG left left left left GGCATGGACTGTGGTCATGAG GGGAGCTAGCCATTATCTTGTG TTCACCGTCTCCTTGTTGC TCATGGAAATCCAAATTGGTC Supplementary Table 2.3 Mouse cdna primer sequences used for RT-PCR Gene Primer Sequence Product Size [bp] Gapdh Pycr1 Pycr2 Pycrl P5cs right right right right right GGGAAGCCCATCACCATCTT AGACATGGACCAAGCTACGG GTCGGCTCACAAGATAATAGCC GTCTCAGTGGCAGTGGTGTG GCATTCTTCACCTCCTGACC left left left left left CGGCCTCACCCCATTTG TCACAGCCAGGAAGAGCAC TCCTTATTGCTTCGGGTCAG CAGGGTCTGAGCAGCAATG CGGCAGAGATCCTCAACTTC Supplementary Table 2.4 Morpholino sequences Species Morpholino sequence Gene Xenopus laevis 5 -AGCTCCTATGAAGCCCACACTCATG-3 Pycr1 Danio rerio 5 -CAGCTCCGATAAATCCCACACTCAT-3 Pycr1 Danio rerio 5 -CCGCTCCAATGAAGCCCACACTCAT-3 Pycr2
Supplementary Document
 Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Beta Thalassemia Case Study Introduction to Bioinformatics
 Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
CHAPTER IV RESULTS Microcephaly General description
 47 CHAPTER IV RESULTS 4.1. Microcephaly 4.1.1. General description This study found that from a previous study of 527 individuals with MR, 48 (23 female and 25 male) unrelated individuals were identified
47 CHAPTER IV RESULTS 4.1. Microcephaly 4.1.1. General description This study found that from a previous study of 527 individuals with MR, 48 (23 female and 25 male) unrelated individuals were identified
CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE
 CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE is_clinical dbsnp MITO GENE chr1 13273 G C heterozygous - - -. - DDX11L1 chr1 949654 A G Homozygous 52 - - rs8997 - ISG15 chr1 1021346 A G heterozygous
CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE is_clinical dbsnp MITO GENE chr1 13273 G C heterozygous - - -. - DDX11L1 chr1 949654 A G Homozygous 52 - - rs8997 - ISG15 chr1 1021346 A G heterozygous
Genetic Testing and Analysis. (858) MRN: Specimen: Saliva Received: 07/26/2016 GENETIC ANALYSIS REPORT
 GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
Beta Thalassemia Sami Khuri Department of Computer Science San José State University Spring 2015
 Bioinformatics in Medical Product Development SMPD 287 Three Beta Thalassemia Sami Khuri Department of Computer Science San José State University Hemoglobin Outline Anatomy of a gene Hemoglobinopathies
Bioinformatics in Medical Product Development SMPD 287 Three Beta Thalassemia Sami Khuri Department of Computer Science San José State University Hemoglobin Outline Anatomy of a gene Hemoglobinopathies
Concurrent Practical Session ACMG Classification
 Variant Effect Prediction Training Course 6-8 November 2017 Prague, Czech Republic Concurrent Practical Session ACMG Classification Andreas Laner / Anna Benet-Pagès 1 Content 1. Background... 3 2. Aim
Variant Effect Prediction Training Course 6-8 November 2017 Prague, Czech Republic Concurrent Practical Session ACMG Classification Andreas Laner / Anna Benet-Pagès 1 Content 1. Background... 3 2. Aim
Supplemental Data. Shin et al. Plant Cell. (2012) /tpc YFP N
 MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
variant led to a premature stop codon p.k316* which resulted in nonsense-mediated mrna decay. Although the exact function of the C19L1 is still
 157 Neurological disorders primarily affect and impair the functioning of the brain and/or neurological system. Structural, electrical or metabolic abnormalities in the brain or neurological system can
157 Neurological disorders primarily affect and impair the functioning of the brain and/or neurological system. Structural, electrical or metabolic abnormalities in the brain or neurological system can
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most
 Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
SUPPLEMENTARY INFORMATION
 Supplementary Information included with Nature MS 2008-02-01484B by Colantonio et al., entitled The dynein regulatory complex is required for ciliary motility and otolith biogenesis in the inner ear. This
Supplementary Information included with Nature MS 2008-02-01484B by Colantonio et al., entitled The dynein regulatory complex is required for ciliary motility and otolith biogenesis in the inner ear. This
Abbreviations: P- paraffin-embedded section; C, cryosection; Bio-SA, biotin-streptavidin-conjugated fluorescein amplification.
 Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at
 Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
Supplementary Figure 1
 Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation
 The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation Anita Becker-Heck#, Irene Zohn#, Noriko Okabe#, Andrew Pollock#, Kari Baker Lenhart,
The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation Anita Becker-Heck#, Irene Zohn#, Noriko Okabe#, Andrew Pollock#, Kari Baker Lenhart,
SUPPLEMENTAL FIGURE 1
 SUPPLEMENTL FIGURE 1 C Supplemental Figure 1. pproach for removal of snorns from Rpl13a gene. () Wild type Rpl13a exonintron structure is shown, with exo in black and intronic snorns in red rectangles.
SUPPLEMENTL FIGURE 1 C Supplemental Figure 1. pproach for removal of snorns from Rpl13a gene. () Wild type Rpl13a exonintron structure is shown, with exo in black and intronic snorns in red rectangles.
Figure S1. Analysis of genomic and cdna sequences of the targeted regions in WT-KI and
 Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
WDR62 is associated with the spindle pole and mutated in human microcephaly
 WDR62 is associated with the spindle pole and mutated in human microcephaly Adeline K. Nicholas, Maryam Khurshid, Julie Désir, Ofélia P. Carvalho, James J. Cox, Gemma Thornton, Rizwana Kausar, Muhammad
WDR62 is associated with the spindle pole and mutated in human microcephaly Adeline K. Nicholas, Maryam Khurshid, Julie Désir, Ofélia P. Carvalho, James J. Cox, Gemma Thornton, Rizwana Kausar, Muhammad
CD31 5'-AGA GAC GGT CTT GTC GCA GT-3' 5 ' -TAC TGG GCT TCG AGA GCA GT-3'
 Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
Supplementary Figure 1
 Supplementary Figure 1 3 3 3 1 1 Bregma -1.6mm 3 : Bregma Ref) Http://www.mbl.org/atlas165/atlas165_start.html Bregma -.18mm Supplementary Figure 1 Schematic representation of the utilized brain slice
Supplementary Figure 1 3 3 3 1 1 Bregma -1.6mm 3 : Bregma Ref) Http://www.mbl.org/atlas165/atlas165_start.html Bregma -.18mm Supplementary Figure 1 Schematic representation of the utilized brain slice
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
The Complexity of Simple Genetics
 The Complexity of Simple Genetics? The ciliopathies: a journey into variable penetrance and expressivity Bardet-Biedl Syndrome Allelism at a single locus is insufficient to explain phenotypic variability
The Complexity of Simple Genetics? The ciliopathies: a journey into variable penetrance and expressivity Bardet-Biedl Syndrome Allelism at a single locus is insufficient to explain phenotypic variability
Integration Solutions
 Integration Solutions (1) a) With no active glycosyltransferase of either type, an ii individual would not be able to add any sugars to the O form of the lipopolysaccharide. Thus, the only lipopolysaccharide
Integration Solutions (1) a) With no active glycosyltransferase of either type, an ii individual would not be able to add any sugars to the O form of the lipopolysaccharide. Thus, the only lipopolysaccharide
BIOLOGY 621 Identification of the Snorks
 Name: Date: Block: BIOLOGY 621 Identification of the Snorks INTRODUCTION: In this simulation activity, you will examine the DNA sequence of a fictitious organism - the Snork. Snorks were discovered on
Name: Date: Block: BIOLOGY 621 Identification of the Snorks INTRODUCTION: In this simulation activity, you will examine the DNA sequence of a fictitious organism - the Snork. Snorks were discovered on
Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards
 Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards incubated in 100 % ethanol overnight at 4 C and embedded in
Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards incubated in 100 % ethanol overnight at 4 C and embedded in
Reporting TP53 gene analysis results in CLL
 Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Identification of a novel in-frame de novo mutation in SPTAN1 in intellectual disability and pontocerebellar atrophy
 Hamdan et al., Identification of a novel in-frame de novo mutation in SPTAN1 in intellectual disability and pontocerebellar atrophy Supplementary Information Gene screening and bioinformatics PCR primers
Hamdan et al., Identification of a novel in-frame de novo mutation in SPTAN1 in intellectual disability and pontocerebellar atrophy Supplementary Information Gene screening and bioinformatics PCR primers
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05883 SUPPLEMENTARY INFORMATION Supplemental Figure 1 Prostaglandin agonists and antagonists alter runx1/cmyb expression. a-e, Embryos were exposed to (b) PGE2 and (c) PGI2 (20μM) and
doi: 10.1038/nature05883 SUPPLEMENTARY INFORMATION Supplemental Figure 1 Prostaglandin agonists and antagonists alter runx1/cmyb expression. a-e, Embryos were exposed to (b) PGE2 and (c) PGI2 (20μM) and
Names for ABO (ISBT 001) Blood Group Alleles
 Names for ABO (ISBT 001) blood group alleles v1.1 1102 Names for ABO (ISBT 001) Blood Group Alleles General description: The ABO system was discovered as in 1900 and is considered the first and clinically
Names for ABO (ISBT 001) blood group alleles v1.1 1102 Names for ABO (ISBT 001) Blood Group Alleles General description: The ABO system was discovered as in 1900 and is considered the first and clinically
Variant Classification. Author: Mike Thiesen, Golden Helix, Inc.
 Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
SUPPLEMENTARY DATA. Supplementary Table 1. Primer sequences for qrt-pcr
 Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
SUPPLEMENTARY INFORMATION. Rare independent mutations in renal salt handling genes contribute to blood pressure variation
 SUPPLEMENTARY INFORMATION Rare independent mutations in renal salt handling genes contribute to blood pressure variation Weizhen Ji, Jia Nee Foo, Brian J. O Roak, Hongyu Zhao, Martin G. Larson, David B.
SUPPLEMENTARY INFORMATION Rare independent mutations in renal salt handling genes contribute to blood pressure variation Weizhen Ji, Jia Nee Foo, Brian J. O Roak, Hongyu Zhao, Martin G. Larson, David B.
Genotype-Phenotype in Egyptian Patients with Nephropathic Cystinosis. (December 2012 report)
 Genotype-Phenotype in Egyptian Patients with Nephropathic Cystinosis (December 2012 report) This is the first study of the genotype of Nephropathic Cystinosis (NC) patients in Egypt and the region of North
Genotype-Phenotype in Egyptian Patients with Nephropathic Cystinosis (December 2012 report) This is the first study of the genotype of Nephropathic Cystinosis (NC) patients in Egypt and the region of North
MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS
 MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS How do different alleles arise? ( allele : form of a gene; specific base sequence at a site on DNA) Mutations: heritable changes in genes Mutations occur in DNA
MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS How do different alleles arise? ( allele : form of a gene; specific base sequence at a site on DNA) Mutations: heritable changes in genes Mutations occur in DNA
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
*To whom correspondence should be addressed. This PDF file includes:
 www.sciencemag.org/cgi/content/full/science.1212182/dc1 Supporting Online Material for Partial Retraction to Detection of an Infectious Retrovirus, XMRV, in Blood Cells of Patients with Chronic Fatigue
www.sciencemag.org/cgi/content/full/science.1212182/dc1 Supporting Online Material for Partial Retraction to Detection of an Infectious Retrovirus, XMRV, in Blood Cells of Patients with Chronic Fatigue
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM.
 Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
BIOL2005 WORKSHEET 2008
 BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
Table S1. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9. Assay (s) Target Name Sequence (5 3 ) Comments
 SUPPLEMENTAL INFORMATION 2 3 Table S. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9 genes. Assay (s) Target Name Sequence (5 3 ) Comments CDC M InfA Forward (NS), CDC M
SUPPLEMENTAL INFORMATION 2 3 Table S. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9 genes. Assay (s) Target Name Sequence (5 3 ) Comments CDC M InfA Forward (NS), CDC M
MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS. (Start your clickers)
 MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS (Start your clickers) How do mutations arise? And how do they affect a cell and its organism? Mutations: heritable changes in genes Mutations occur in DNA But
MUTATIONS, MUTAGENESIS, AND CARCINOGENESIS (Start your clickers) How do mutations arise? And how do they affect a cell and its organism? Mutations: heritable changes in genes Mutations occur in DNA But
Journal of Cell Science Supplementary information. Arl8b +/- Arl8b -/- Inset B. electron density. genotype
 J. Cell Sci. : doi:.4/jcs.59: Supplementary information E9. A Arl8b /- Arl8b -/- Arl8b Arl8b non-specific band Gapdh Tbp E7.5 HE Inset B D Control al am hf C E Arl8b -/- al am hf E8.5 F low middle high
J. Cell Sci. : doi:.4/jcs.59: Supplementary information E9. A Arl8b /- Arl8b -/- Arl8b Arl8b non-specific band Gapdh Tbp E7.5 HE Inset B D Control al am hf C E Arl8b -/- al am hf E8.5 F low middle high
Supplementary Materials
 Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Supplementary Figure 1 a
 Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary Materials and Methods
 DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
MEDICAL GENOMICS LABORATORY. Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG)
 Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma.
 Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
EGFR shrna A: CCGGCGCAAGTGTAAGAAGTGCGAACTCGAGTTCGCACTTCTTACACTTGCG TTTTTG. EGFR shrna B: CCGGAGAATGTGGAATACCTAAGGCTCGAGCCTTAGGTATTCCACATTCTCTT TTTG
 Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Nature Genetics: doi: /ng Supplementary Figure 1. Brain magnetic resonance imaging in patient 9 at age 3.6 years.
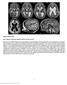 Supplementary Figure 1 Brain magnetic resonance imaging in patient 9 at age 3.6 years. (a d) Axial T2-weighted images show a marked degree of ventriculomegaly. Note the diencephalic mesencephalic junction
Supplementary Figure 1 Brain magnetic resonance imaging in patient 9 at age 3.6 years. (a d) Axial T2-weighted images show a marked degree of ventriculomegaly. Note the diencephalic mesencephalic junction
THE UMD TP53 MUTATION DATABASE UPDATES AND BENEFITS. Pr. Thierry Soussi
 THE UMD TP53 MUTATION DATABASE UPDATES AND BENEFITS Pr. Thierry Soussi thierry.soussi@ki.se thierry.soussi@upmc.fr TP53: 33 YEARS AND COUNTING STRUCTURE FUNCTION RELATIONSHIP OF WILD AND MUTANT TP53 1984
THE UMD TP53 MUTATION DATABASE UPDATES AND BENEFITS Pr. Thierry Soussi thierry.soussi@ki.se thierry.soussi@upmc.fr TP53: 33 YEARS AND COUNTING STRUCTURE FUNCTION RELATIONSHIP OF WILD AND MUTANT TP53 1984
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research
 Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Supplementary Table 2. Conserved regulatory elements in the promoters of CD36.
 Supplementary Table 1. RT-qPCR primers for CD3, PPARg and CEBP. Assay Forward Primer Reverse Primer 1A CAT TTG TGG CCT TGT GCT CTT TGA TGA GTC ACA GAA AGA ATC AAT TC 1B AGG AAA TGA ACT GAT GAG TCA CAG
Supplementary Table 1. RT-qPCR primers for CD3, PPARg and CEBP. Assay Forward Primer Reverse Primer 1A CAT TTG TGG CCT TGT GCT CTT TGA TGA GTC ACA GAA AGA ATC AAT TC 1B AGG AAA TGA ACT GAT GAG TCA CAG
Identifying Mutations Responsible for Rare Disorders Using New Technologies
 Identifying Mutations Responsible for Rare Disorders Using New Technologies Jacek Majewski, Department of Human Genetics, McGill University, Montreal, QC Canada Mendelian Diseases Clear mode of inheritance
Identifying Mutations Responsible for Rare Disorders Using New Technologies Jacek Majewski, Department of Human Genetics, McGill University, Montreal, QC Canada Mendelian Diseases Clear mode of inheritance
Supplementary Figure 1
 A B D Relative TAp73 mrna p73 Supplementary Figure 1 25 2 15 1 5 p63 _-tub. MDA-468 HCC1143 HCC38 SUM149 MDA-468 HCC1143 HCC38 SUM149 HCC-1937 MDA-MB-468 ΔNp63_ TAp73_ TAp73β E C Relative ΔNp63 mrna TAp73
A B D Relative TAp73 mrna p73 Supplementary Figure 1 25 2 15 1 5 p63 _-tub. MDA-468 HCC1143 HCC38 SUM149 MDA-468 HCC1143 HCC38 SUM149 HCC-1937 MDA-MB-468 ΔNp63_ TAp73_ TAp73β E C Relative ΔNp63 mrna TAp73
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Ch 7 Mutation. A heritable change in DNA Random Source of genetic variation in a species may be advantageous, deleterious, neutral
 Ch 7 Mutation A heritable change in DNA Random Source of genetic variation in a species may be advantageous, deleterious, neutral Mutation (+ sexual reproduction) + natural selection = evolution Types
Ch 7 Mutation A heritable change in DNA Random Source of genetic variation in a species may be advantageous, deleterious, neutral Mutation (+ sexual reproduction) + natural selection = evolution Types
Supplemental Information. Cancer-Associated Fibroblasts Neutralize. the Anti-tumor Effect of CSF1 Receptor Blockade
 Cancer Cell, Volume 32 Supplemental Information Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors Vinit Kumar, Laxminarasimha
Cancer Cell, Volume 32 Supplemental Information Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors Vinit Kumar, Laxminarasimha
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the
 Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
What do you think of when you here the word genome?
 What do you think of when you here the word genome? What do you think of when you here the word genome? Personal Genomics Outline Review of pre-lab work Genomics and Medicine Case Overview & Assignment
What do you think of when you here the word genome? What do you think of when you here the word genome? Personal Genomics Outline Review of pre-lab work Genomics and Medicine Case Overview & Assignment
Detection of 549 new HLA alleles in potential stem cell donors from the United States, Poland and Germany
 HLA ISSN 2059-2302 BRIEF COMMUNICATION Detection of 549 new HLA alleles in potential stem cell donors from the United States, Poland and Germany C. J. Hernández-Frederick 1, N. Cereb 2,A.S.Giani 1, J.
HLA ISSN 2059-2302 BRIEF COMMUNICATION Detection of 549 new HLA alleles in potential stem cell donors from the United States, Poland and Germany C. J. Hernández-Frederick 1, N. Cereb 2,A.S.Giani 1, J.
Supplemental Information For: The genetics of splicing in neuroblastoma
 Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
McAlpine PERK-GSK3 regulates foam cell formation. Supplemental Material. Supplementary Table I. Sequences of real time PCR primers.
 Mclpine PERK-GSK3 regulates foam cell formation Supplemental Material Supplementary Table I. Sequences of real time PCR primers. Primer Name Primer Sequences (5-3 ) Product Size (bp) GRP78 (human) Fwd:
Mclpine PERK-GSK3 regulates foam cell formation Supplemental Material Supplementary Table I. Sequences of real time PCR primers. Primer Name Primer Sequences (5-3 ) Product Size (bp) GRP78 (human) Fwd:
Molecular Characterization of the NF2 Gene in Korean Patients with Neurofibromatosis Type 2: A Report of Four Novel Mutations
 Korean J Lab Med 2010;30:190-4 DOI 10.3343/kjlm.2010.30.2.190 Original Article Diagnostic Genetics Molecular Characterization of the NF2 Gene in Korean Patients with Neurofibromatosis Type 2: A Report
Korean J Lab Med 2010;30:190-4 DOI 10.3343/kjlm.2010.30.2.190 Original Article Diagnostic Genetics Molecular Characterization of the NF2 Gene in Korean Patients with Neurofibromatosis Type 2: A Report
Bioinformatics Laboratory Exercise
 Bioinformatics Laboratory Exercise Biology is in the midst of the genomics revolution, the application of robotic technology to generate huge amounts of molecular biology data. Genomics has led to an explosion
Bioinformatics Laboratory Exercise Biology is in the midst of the genomics revolution, the application of robotic technology to generate huge amounts of molecular biology data. Genomics has led to an explosion
CITATION FILE CONTENT/FORMAT
 CITATION For any resultant publications using please cite: Matthew A. Field, Vicky Cho, T. Daniel Andrews, and Chris C. Goodnow (2015). "Reliably detecting clinically important variants requires both combined
CITATION For any resultant publications using please cite: Matthew A. Field, Vicky Cho, T. Daniel Andrews, and Chris C. Goodnow (2015). "Reliably detecting clinically important variants requires both combined
BWA alignment to reference transcriptome and genome. Convert transcriptome mappings back to genome space
 Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
MSI positive MSI negative
 Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
Characterizing intra-host influenza virus populations to predict emergence
 Characterizing intra-host influenza virus populations to predict emergence June 12, 2012 Forum on Microbial Threats Washington, DC Elodie Ghedin Center for Vaccine Research Dept. Computational & Systems
Characterizing intra-host influenza virus populations to predict emergence June 12, 2012 Forum on Microbial Threats Washington, DC Elodie Ghedin Center for Vaccine Research Dept. Computational & Systems
Description of Supplementary Files. File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables
 Description of Supplementary Files File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables Supplementary Figure 1: (A), HCT116 IDH1-WT and IDH1-R132H cells were
Description of Supplementary Files File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables Supplementary Figure 1: (A), HCT116 IDH1-WT and IDH1-R132H cells were
Genome 371, Autumn 2018 Quiz Section 9: Genetics of Cancer Worksheet
 Genome 371, Autumn 2018 Quiz Section 9: Genetics of Cancer Worksheet All cancer is due to genetic mutations. However, in cancer that clusters in families (familial cancer) at least one of these mutations
Genome 371, Autumn 2018 Quiz Section 9: Genetics of Cancer Worksheet All cancer is due to genetic mutations. However, in cancer that clusters in families (familial cancer) at least one of these mutations
AP VP DLP H&E. p-akt DLP
 A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP
 Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP Frequency Domain Mammals a 3 S/P b Mammals a 3 S/P b
Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP Frequency Domain Mammals a 3 S/P b Mammals a 3 S/P b
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle.
 Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplemental Information. Th17 Lymphocytes Induce Neuronal. Cell Death in a Human ipsc-based. Model of Parkinson's Disease
 Cell Stem Cell, Volume 23 Supplemental Information Th17 Lymphocytes Induce Neuronal Cell Death in a Human ipsc-based Model of Parkinson's Disease Annika Sommer, Franz Maxreiter, Florian Krach, Tanja Fadler,
Cell Stem Cell, Volume 23 Supplemental Information Th17 Lymphocytes Induce Neuronal Cell Death in a Human ipsc-based Model of Parkinson's Disease Annika Sommer, Franz Maxreiter, Florian Krach, Tanja Fadler,
Mutation Screening and Association Studies of the Human UCP 3 Gene in Normoglycemic and NIDDM Morbidly Obese Patients
 Mutation Screening and Association Studies of the Human UCP 3 Gene in Normoglycemic and NIDDM Morbidly Obese Patients Shuichi OTABE, Karine CLEMENT, Séverine DUBOIS, Frederic LEPRETRE, Veronique PELLOUX,
Mutation Screening and Association Studies of the Human UCP 3 Gene in Normoglycemic and NIDDM Morbidly Obese Patients Shuichi OTABE, Karine CLEMENT, Séverine DUBOIS, Frederic LEPRETRE, Veronique PELLOUX,
Pathophysiology of inherited kidney disorders: lessons from the zebrafish
 Pathophysiology of inherited kidney disorders: lessons from the zebrafish Francisco J. Arjona Radboud Institute for Molecular Life Sciences Radboud university medical center Nijmegen, The Netherlands www.physiomics.eu
Pathophysiology of inherited kidney disorders: lessons from the zebrafish Francisco J. Arjona Radboud Institute for Molecular Life Sciences Radboud university medical center Nijmegen, The Netherlands www.physiomics.eu
Supplemental Methods Supplemental Table 1. Supplemental Figure 1. Supplemental Figure 2. Supplemental Figure 3. Supplemental Figure 4.
 Supplemental Methods TGF-B1 ELISA Supernatants were collected from AT2 cells cultured for 1, 2, 3, or 4 days and frozen at -80 degrees C until use in the ELISA. A commercially available mouse TGF-B1 Duo
Supplemental Methods TGF-B1 ELISA Supernatants were collected from AT2 cells cultured for 1, 2, 3, or 4 days and frozen at -80 degrees C until use in the ELISA. A commercially available mouse TGF-B1 Duo
Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
 Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION
 DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION Gentzon Hall 1,2#, Brandon Lane 1,3#, Megan Chryst-Ladd 1,3, Guanghong Wu 1,3, Jen-Jar Lin 4, XueJun
DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION Gentzon Hall 1,2#, Brandon Lane 1,3#, Megan Chryst-Ladd 1,3, Guanghong Wu 1,3, Jen-Jar Lin 4, XueJun
The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep
 SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Yatsenko AN, Georgiadis AP, Röpke A, et al. X-linked TEX11
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Yatsenko AN, Georgiadis AP, Röpke A, et al. X-linked TEX11
The Human Major Histocompatibility Complex
 The Human Major Histocompatibility Complex 1 Location and Organization of the HLA Complex on Chromosome 6 NEJM 343(10):702-9 2 Inheritance of the HLA Complex Haplotype Inheritance (Family Study) 3 Structure
The Human Major Histocompatibility Complex 1 Location and Organization of the HLA Complex on Chromosome 6 NEJM 343(10):702-9 2 Inheritance of the HLA Complex Haplotype Inheritance (Family Study) 3 Structure
Insulin Resistance. Biol 405 Molecular Medicine
 Insulin Resistance Biol 405 Molecular Medicine Insulin resistance: a subnormal biological response to insulin. Defects of either insulin secretion or insulin action can cause diabetes mellitus. Insulin-dependent
Insulin Resistance Biol 405 Molecular Medicine Insulin resistance: a subnormal biological response to insulin. Defects of either insulin secretion or insulin action can cause diabetes mellitus. Insulin-dependent
Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM
 Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep
 Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep Tom Walsh, PhD Division of Medical Genetics University of Washington Next generation sequencing Sanger sequencing gold
Using the Bravo Liquid-Handling System for Next Generation Sequencing Sample Prep Tom Walsh, PhD Division of Medical Genetics University of Washington Next generation sequencing Sanger sequencing gold
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Lats1/2 deleted ihbs and ihps showed decreased transcripts of hepatocyte related genes (a and b) Western blots (a) and recombination PCR (b) of control and
Supplementary Figure 1 Supplementary Figure 1. Lats1/2 deleted ihbs and ihps showed decreased transcripts of hepatocyte related genes (a and b) Western blots (a) and recombination PCR (b) of control and
Table S1. Primers used to quantitatively amplify the human mirnas precursors and indicated genes
 Table S1. Primers used to quantitatively amplify the human mirnas precursors and indicated genes Forward primer (5 3 ) Rervese primer (5 3 ) U6 CTCGCTTCGGCAGCACA AACGCTTCACGAATTTGCGT 5S TACGGCCATACCACCCTGAA
Table S1. Primers used to quantitatively amplify the human mirnas precursors and indicated genes Forward primer (5 3 ) Rervese primer (5 3 ) U6 CTCGCTTCGGCAGCACA AACGCTTCACGAATTTGCGT 5S TACGGCCATACCACCCTGAA
6/12/2018. Disclosures. Clinical Genomics The CLIA Lab Perspective. Outline. COH HopeSeq Heme Panels
 Clinical Genomics The CLIA Lab Perspective Disclosures Raju K. Pillai, M.D. Hematopathologist / Molecular Pathologist Director, Pathology Bioinformatics City of Hope National Medical Center, Duarte, CA
Clinical Genomics The CLIA Lab Perspective Disclosures Raju K. Pillai, M.D. Hematopathologist / Molecular Pathologist Director, Pathology Bioinformatics City of Hope National Medical Center, Duarte, CA
Characterisation of structural variation in breast. cancer genomes using paired-end sequencing on. the Illumina Genome Analyser
 Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
UW359 Ovary 3c 3 Serous Recurrent 68 BRCA1 816delGT BRCA1 del exon 1-2. UW417 Ovary 3c 3 Serous Primary 38 BRCA1 1675delA
 Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
without LOI phenotype by breeding female wild-type C57BL/6J and male H19 +/.
 Sakatani et al. 1 Supporting Online Material Materials and methods Mice and genotyping: H19 mutant mice with C57BL/6J background carrying a deletion in the structural H19 gene (3 kb) and 10 kb of 5 flanking
Sakatani et al. 1 Supporting Online Material Materials and methods Mice and genotyping: H19 mutant mice with C57BL/6J background carrying a deletion in the structural H19 gene (3 kb) and 10 kb of 5 flanking
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
A guide to understanding variant classification
 White paper A guide to understanding variant classification In a diagnostic setting, variant classification forms the basis for clinical judgment, making proper classification of variants critical to your
White paper A guide to understanding variant classification In a diagnostic setting, variant classification forms the basis for clinical judgment, making proper classification of variants critical to your
Culture Density (OD600) 0.1. Culture Density (OD600) Culture Density (OD600) Culture Density (OD600) Culture Density (OD600)
 A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
The Effect of Epstein-Barr Virus Latent Membrane Protein 2 Expression on the Kinetics of Early B Cell Infection
 The Effect of Epstein-Barr Virus Latent Membrane Protein 2 Expression on the Kinetics of Early B Cell Infection Laura R. Wasil, Monica J. Tomaszewski, Aki Hoji, David T. Rowe* Department of Infectious
The Effect of Epstein-Barr Virus Latent Membrane Protein 2 Expression on the Kinetics of Early B Cell Infection Laura R. Wasil, Monica J. Tomaszewski, Aki Hoji, David T. Rowe* Department of Infectious
