Supplementary Data Figure S1. A) PKI-587 suppression of p-akt in A498 and (+/- 10
|
|
|
- Abner Lambert
- 5 years ago
- Views:
Transcription
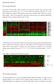
1 Supplementary Data Figure S1. A) PKI-587 suppression of p-akt in A498 and (+/- 10 μm Verapamil), and B) PKI-587 suppression of p-akt in BT474, & U87MG [μm] A) p-akt T308 +/-Verapamil Akt [μm] A Akt p-akt T308 +/-Verapamil B) BT [μm] p-akt T308 Akt U87MG [μm] p-akt T308 Akt
2 Supplementary Data Figure S2. In MDA-MB-361: A) PKI-587 Induction of cparp at 1 h; and B) PKI-587 vs mtor inhibitor (MTI-178) effect on cparp, pakt-t308, pakt-s473, and total Akt. A) [μm] 0.25 h cparp < pakt T308 1 h cparp <pakt T308 cparp 2 h <pakt T308
3 B)
4 Supplementary Data Figure S3. PKI-587 Anti-tumor Effect in the MDA-MB-361 Xenograft Model. A) Dose response (1, 5, 9 regimen), B) MDA-MB-361 tumor regression caused by PKI-587 at 20 mg/kg (1, 5, 9) or Taxol 60 mg/kg (IP once). A)
5 B)
6 Supplementary Data Figure S4. PKI-587 Efficacy in the BT474 Breast Xenograft Model. A) PKI-587 antitumor effect at 5, and B) 10 mg/kg (1, 5, 9 regimen). A) B)
7 Supplementary Figure S5. Activation of Caspase 3/7 in HCT116 and H1975 cells in vitro by PKI-587 alone, or in combination. A. B. C.
8 Supplementary Data Figure S6. PKI-587 Efficacy in the H1975 (NSCLC) Xenograft Model. A) PKI-587 antitumor effect against H1975 tumors (~ 400 mm 3 ) at 10 mg/kg (dx5, twice); or B) against H1975 tumors (~ 1000 mm 3 ) at 10 mg/kg (dx5, twice) & 25 mg/kg (once weekly, twice). C) Panel B to day 7 with error bars. A) B) C)
9 Supplementary Data Figure S7. PKI-587 (20 mg/kg; 1, 5, 9 regimen) Efficacy in the H1975 (NSCLC) Orthotopic Model.
10 Supplementary Data Figure S8. PKI-587 Dose response in the U87MG Glioma Xenograft Model.
11 Supplementary Figure S9. HKI-272 effect on pmapk & pakt; PKI-587 effect on pmapk - in H1975 cells. A) HKI-272 effect on pakt-t308 and pmapk in H1975 (NSCLC) tumor cells, B) PKI-587 effect on pmapk in H1975 (NSCLC) tumor cells. A) B)
12 Supplementary Figure S10. Durable in vivo effect of PKI-587 (25 mg/kg) on PI3K/Akt/mTOR effectors in the MDA-MB-361 xenograft model.
13 Supplementary Figure Legends: for Antitumor Efficacy of PKI-587, a Highly Potent Dual PI3K/mTOR Kinase Inhibitor. Supplementary Data Figure S1. A) Suppression of p-akt T308 in and A498 (renal) cells, after 4 h exposure to PKI-587 in vitro. In A498 and p-akt T308 was also assessed in the presence of 10 μm verapamil (+). Total Akt is also shown. B) Suppression of p-akt T308 in BT474 (breast), and U87MG (glioma) cells after 4 h exposure to PKI-587 in vitro. Total Akt is shown. Supplementary Data Figure S2. A) Cleaved PARP induction in MDA-MB-361 cells after 0.25, 1, and 2 h exposure to PKI-587 in vitro; B) PKI-587 vs mtor inhibitor (MTI- 178) effect on cparp, pakt-t308, pakt-s473, and total Akt in MDA-MB-361 (4 h exposure). Supplementary Data Figure S3. A) MED determination from the MDA-MB-361 breast xenograft model with PKI-587 given at 1.56, 3.125, and 6.25 mg/kg; on a day 1, 5, 9 regimen to day14; B) MDA-MB-361 tumor regression caused by PKI-587 at 20 mg/kg (1, 5, 9) or Taxol 60 mg/kg (IP once). Supplementary Data Figure S4. A) PKI-587 efficacy in the BT-474 (breast; Her2+, PIK3CA [K111N]) xenograft model at 5 mg/kg (, left), and B) at 10 mg/kg (, right) in intermittent (1, 5, 9, etc) dosing format. PKI-587 was administered to day 21 in both studies. Supplementary Data Figure S5. A) Caspase 3/7 activation by PKI-587 in HCT116; or PKI-587 plus camptothecin (Cpt) (at 320 nm) at 24 h. B) Caspase 3/7 activation by
14 PKI-587 in HCT116; or PKI-587 plus PD (at 100 nm) at 24 h. C) Caspase 3/7 activation by PKI-587 in H1975, alone and with HKI-272 (at 250 nm) at 24 h. Supplementary Data Figure S6. PKI-587 efficacy in the H1975 (NSCLC; EFGR L858R/T790M) xenograft model at A) 10 mg/kg; regimen: daily for 5 days (dx5), 2 day dosing holiday, then a second dx5 round against 400 mm 3 tumors (); B) PKI-587 at 10 mg/kg, daily for 5 days (dx5), 7 day dosing holiday, then a second dx5 round (), or at 25 mg/kg (, right) once weekly (days 1, 7, and 14) - against large (1000 mm 3 ) tumors; C) Panel B graph to day 7 with error bars. Supplementary Data Figure S7. PKI-587 at 20 mg/kg in the H1975 (EGFR; L858R/T790M) orthotopic lung cancer model (day 1, 5, 9 regimen). Solid black line indicates treated mice, dashed line indicates control group. The last (end) dose in the treated group is denoted by the symbol, ^, at day 65. Supplementary Data Figure S8. PKI-587 efficacy in the U87MG (PTEN negative) xenograft model, administered in dose response format at 12.5, 6.25, 3.12, and 1.56 mg/kg daily for 5 days (dx5). PKI-587 at 6.25 ( ) and 12.5 mg/kg () had a cytostatic effect on tumors. Supplementary Data Figure S9. HKI-272 effect on pmapk & pakt; PKI-587 effect on pmapk - in H1975 cells. A) HKI-272 effect on pakt-t308 and pmapk in H1975 (NSCLC) tumor cells; B) PKI-587 effect on pmapk in H1975 (NSCLC) tumor cells. Supplementary Data Figure S10. Durable in vivo effect of PKI-587 (25 mg/kg) on PI3K/Akt/mTOR effectors in the MDA-MB-361 xenograft model.
15 . Supplementary Table S1. PKI-587 IC50s from 43 human tumor cell lines (Caliper). Cell Line Origin Mutation(s) IC50 mm BT-549 Breast PTEN, RB, TP HS578T Breast HRAS, TP53, CDKN2A MDA-MB-231 Breast KRAS, BRAF, TP53,CDKN2A BT549 Breast PTEN, RB, TP MCF7 Breast PIK3CA, CDKN2A SF-539 CNS PTEN, RB, TP SNB-19 CNS PTEN, TP53, CDKN2A SNB-75 CNS TP SF-268 CNS EGFR, TP53, CDKN2A U251 CNS PTEN, TP53, CDKN2A Colo205 Colon BRAF, TP53, APC HCC2998 Colon KRAS, RB, TP53, APC HCT116 Colon KRAS, PIK3CA SW620 Colon KRAS, TP53, APC KM-12 Colon PTEN, TP53, BRCA HOP92 Lung TP53, CDKN2A NCI-H23 Lung KRAS, STK11, TP NCI-226 Lung CDKN2A EKVX Lung TP NCI-H460 Lung KRAS, PIK3CA, STK A549 Lung KRAS, CDKN2A, STK HOP62 Lung KRAS, TP53, CDKN2A SK-MEL28 Melanoma BRAF, EGFR, TP UACC-257 Melanoma BRAF LOX-IMVI Melanoma BRAF, CDKN2A 0.01 MB-435 Melanoma BRAF, TP53, CDKN2A SK-MEL5 Melanoma BRAF, STK11, CDKN2A Malme3M Melanoma BRAF, CDKN2A UACC-62 Melanoma BRAF, PTEN, CDKN2A M14 Melanoma BRAF, TP SK-MEL2 Melanoma NRAS, TP OVCAR-4 Ovarian TP OVCAR-5 Ovarian KRAS, CDKN2A IGROV-1 Ovarian PTEN, TP53, BRCA SK-OV3 Ovarian PIK3CA, TP53, CDKN2A, APC OV-CAR-3 Ovarian TP NCI-ADR-RES Ovarian ERBB2, TP OVCAR-8 Ovarian ERBB2, TP PC3 Prostate PTEN, TP SN-12C Renal TP Caki-1 Renal CDKN2A RXF-393 Renal PTEN, TP53, CDKN2A A498 Renal VHL, CDKN2A 0.493
16 Supplementary Data Table S2. ATP competitive profile of PKI-587. PKI-587: IC50 mm at various ATP concentrations ATP [mm] Observed IC50 [mm] ATP [mm] Ratio^ at ATP [mm] (^IC50/IC50 at 25 mm) Theoretical* increase in IC50 if ATP competitive *Cheng and Prussoff (1973) Biochemical Pharmacol. 22:3099
17 Supplementary Table S3. PKI-587 data from 236 human kinase panel (Invitrogen). PKI-587 was tested at 1 M. Kinase Tested % Inh. Kinase Tested % Inh. Kinase Tested % Inh. Kinase Tested % Inh. Kinase Tested % Inh. Kinase Tested % Inh. ABL1 0 CSF1R (FMS) 5 FGR 0 MAP4K2 (GCK) 1 PASK 7 ROS1-3 ABL1 E255K -5 CSK 3 FLT1 (VEGFR1) 3 MAP4K4 (HGK) 12 PDGFRA (PDGFR alpha) 3 RPS6KA1 (RSK1) 2 ABL1 G250E -1 CSNK1A1 (CK1 alpha 1) 11 FLT3 7 MAP4K5 (KHS1) 9 PDGFRA D842V 7 RPS6KA2 (RSK3) -1 ABL1 T315I 0 CSNK1D (CK1 delta) -4 FLT3 D835Y 3 MAPK1 (ERK2) 3 PDGFRA T674I 13 RPS6KA3 (RSK2) -3 ABL1 Y253F 1 CSNK1E (CK1 epsilon) 14 FLT4 (VEGFR3) 2 MAPK10 (JNK3) -4 PDGFRA V561D 4 RPS6KA4 (MSK2) 12 ABL2 (Arg) -4 CSNK1G1 (CK1 gamma 1) 0 FRAP1 (mtor) 101 MAPK11 (p38 beta) 9 PDGFRB (PDGFR beta) 4 RPS6KA5 (MSK1) 10 ACVR1B (ALK4) -7 CSNK1G2 (CK1 gamma 2) 4 FRK (PTK5) 3 MAPK12 (p38 gamma) 3 PDK1-4 RPS6KA6 (RSK4) 0 ADRBK1 (GRK2) 4 CSNK1G3 (CK1 gamma 3) 0 FYN -1 MAPK13 (p38 delta) -4 PHKG1 9 RPS6KB1 (p70s6k) 7 ADRBK2 (GRK3) 3 CSNK2A1 (CK2 alpha 1) 5 GRK4-5 MAPK14 (p38 alpha) -6 PHKG2 9 SGK (SGK1) -1 AKT1 (PKB alpha) 0 CSNK2A2 (CK2 alpha 2) 9 GRK5-11 MAPK3 (ERK1) 0 PIM1 4 SGK2 3 AKT2 (PKB beta) -2 DAPK3 (ZIPK) 1 GRK6 0 MAPK8 (JNK1) -7 PIM2 7 SGKL (SGK3) 2 AKT3 (PKB gamma) -2 DCAMKL2 (DCK2) -3 GRK7-16 MAPK9 (JNK2) -8 PKN1 (PRK1) 5 SRC 1 ALK 2 DYRK1A -1 GSK3A (GSK3 alpha) -1 MAPKAPK2-3 PLK1 6 SRC N1 1 AMPK A1/B1/G1-5 DYRK1B -2 GSK3B (GSK3 beta) 2 MAPKAPK3-7 PLK2 2 SRMS (Srm) -9 AMPK A2/B1/G1 1 DYRK3 0 HCK 4 MAPKAPK5 (PRAK) -1 PLK3-8 SRPK1-4 AURKA (Aurora A) 6 DYRK4-1 HIPK1 (Myak) -1 MARK1 (MARK) 2 PRKACA (PKA) 4 SRPK2 4 AURKB (Aurora B) 0 EEF2K 2 HIPK4-1 MARK2 3 PRKCA (PKC alpha) 0 STK22B (TSSK2) -2 AURKC (Aurora C) -2 EGFR (ErbB1) 0 IGF1R -3 MATK (HYL) -4 PRKCB1 (PKC beta I) 3 STK22D (TSSK1) -1 AXL 5 EGFR (ErbB1) L858R 3 IKBKB (IKK beta) 4 MELK 5 PRKCB2 (PKC beta II) 10 STK23 (MSSK1) -1 BLK 9 EGFR (ErbB1) L861Q 5 IKBKE (IKK epsilon) 6 MERTK (cmer) -3 PRKCD (PKC delta) 6 STK24 (MST3) 19 BMX 2 EGFR (ErbB1) T790M 3 INSR -3 MET (cmet) 16 PRKCE (PKC epsilon) 6 STK25 (YSK1) 12 BRAF 58 EGFR (ErbB1) T790M L858R -4 INSRR (IRR) -1 MET M1250T -2 PRKCG (PKC gamma) 10 STK3 (MST2) 8 BRAF V600E 75 EPHA1-2 IRAK4 4 MINK1 15 PRKCH (PKC eta) 2 STK4 (MST1) 8 BRSK1 (SAD1) 2 EPHA2-11 ITK 1 MST1R (RON) 5 PRKCI (PKC iota) -2 SYK 3 BTK 0 EPHA3-1 JAK1-3 MST4 16 PRKCN (PKD3) -3 TAOK2 (TAO1) 24 CAMK1D (CaMKI delta) 0 EPHA4 1 JAK2-3 MUSK 15 PRKCQ (PKC theta) 1 TBK1 5 CAMK2A (CaMKII alpha) 7 EPHA5-2 JAK2 JH1 JH2 5 MYLK2 (skmlck) -1 PRKCZ (PKC zeta) 4 TEK (Tie2) 4 CAMK2B (CaMKII beta) 10 EPHA8-3 JAK2 JH1 JH2 V617F 6 NEK1 1 PRKD1 (PKC mu) 2 TYK2 5 CAMK2D (CaMKII delta) 4 EPHB1 3 JAK3-7 NEK2 7 PRKD2 (PKD2) -5 TYRO3 (RSE) 1 CAMK4 (CaMKIV) 4 EPHB2 0 KDR (VEGFR2) -1 NEK4 1 PRKG1 3 YES1-2 CDC42 BPA (MRCKA) 5 EPHB3 0 KIT 9 NEK6 2 PRKG2 (PKG2) 6 ZAP70-8 CDC42 BPB (MRCKB) 2 EPHB4-4 KIT T670I 4 NEK7 1 PRKX 6 CDK1/cyclin B 0 ERBB2 (HER2) 6 LCK 8 NEK9 5 PTK2 (FAK) -2 CDK2/cyclin A 6 ERBB4 (HER4) -4 LTK (TYK1) 0 NTRK1 (TRKA) -5 PTK2B (FAK2) -9 CDK5/p25 5 FER 1 LYN A 7 NTRK2 (TRKB) 0 PTK6 (Brk) -12 CDK5/p35 6 FES (FPS) 5 LYN B 6 NTRK3 (TRKC) -1 RAF1 (craf) Y340D Y341D 29 CHEK1 (CHK1) 0 FGFR1 1 MAP2K1 (MEK1) 10 PAK2 (PAK65) 0 RET 1 CHEK2 (CHK2) 6 FGFR2 3 MAP2K2 (MEK2) 8 PAK3 14 RET V804L -1 CLK1 0 FGFR3 16 MAP2K6 (MKK6) -1 PAK4 3 RET Y791F 2 CLK2-3 FGFR3 K650E 0 MAP3K8 (COT) 17 PAK6 11 ROCK1 5 CLK3-6 FGFR4 1 MAP3K9 (MLK1) -9 PAK7 (KIAA1264) 7 ROCK2 0
18 Supplementary Data Table S4. Effect of Verapamil on PKI-587 IC50 values in A498, 786-0, DLD1, and H1299. Growth Inhibition IC50 mm Cell Line PKI-587 PKI-587 & Verapamil [10 mm] s A DLD H
19 Supplementary DataTable S5. PK/Safety for PKI-587. PK 3 mg/kg (Single dose, nude mouse) Plasma 25 mg/kg (Single dose, nude mouse) Plasma Time (h) ng/ml s Time (h) ng/ml s T 1/2 (h) 4.9 T 1/2 (h) 14.4 Clp (ml/min/kg) 17 Clp (ml/min/kg) 7 Vss (L/kg) 2.4 Vss (L/kg) 7.2 AUC (0-infin) (h/ng/ml) 2863 AUC (0-infin) (h/ng/ml) Safety (3 mg/kg) Clearance (CLp): nude mouse (13 ml/min/kg), monkey (3.9 ml/min/kg), dog (7 ml/min/kg), and rat (17 ml/min/kg) Vd ss (volume of distibution): Moderate/high in nude mice (3 L/kg), rat (9 L/kg), dog (4 L/kg), monkey (2.4 L/kg) Apparent t 1/2 : 5.2 h (nude mice), 11.5 h (rat), 16 h (dog), 18.5 h (monkey) Intrinsic Clearance in Hepatocytes: Low in rat, dog, human; moderate in monkey Exposure Ratios (3-cycle [1,5,9 regimen] studies, MTD/MED) } Rat: ER = 2.1 } Dog: ER = 7.2 Metabolism, etc Major Human Metabolic Pathways: No major metabolites; profile similar across species CYP Inhibition: Drug/Drug Interaction Not Likely pgp Interaction: Not an Inhibitor Primary Drug Metabolism/PK issues: No Issues Genotoxicity: Screening Ames - Negative
Supplementary information. Supplementary Table 1. Kinase Type Format N Average (IC 50 )
 Supplementary information Supplementary Table 1 Kinase Type Format N Average (IC 50 ) SD ALK Y Lantha 6 > 10 n.d. AURORA_A S/T Caliper 6 > 10 n.d. AXL Y Lantha 3 > 10 n.d. BTK Y Caliper 6 > 10 n.d. cabl
Supplementary information Supplementary Table 1 Kinase Type Format N Average (IC 50 ) SD ALK Y Lantha 6 > 10 n.d. AURORA_A S/T Caliper 6 > 10 n.d. AXL Y Lantha 3 > 10 n.d. BTK Y Caliper 6 > 10 n.d. cabl
Supplemental text file, including:
 Supplemental text file, including: Supplemental Table S1: Kinase profile for XL147 Supplemental Table S2: Effects of XL147 on PI3K Pathway Signaling in MCF7 and PC-3 Cells Supplemental Table S3: XL147
Supplemental text file, including: Supplemental Table S1: Kinase profile for XL147 Supplemental Table S2: Effects of XL147 on PI3K Pathway Signaling in MCF7 and PC-3 Cells Supplemental Table S3: XL147
SUPPLEMENTAL DATA. Supplemental Experimental Procedures
 SUPPLEMENTAL DATA An ATP-competitive mtor inhibitor reveals rapamycin-resistant functions of mtorc1 Carson C. Thoreen, Seong A. Kang, Jae Won Chang, Qingsong Liu, Jianming Zhang, Yi Gao, Laurie J. Reichling,
SUPPLEMENTAL DATA An ATP-competitive mtor inhibitor reveals rapamycin-resistant functions of mtorc1 Carson C. Thoreen, Seong A. Kang, Jae Won Chang, Qingsong Liu, Jianming Zhang, Yi Gao, Laurie J. Reichling,
Supporting Information. Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series
 Supporting Information Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1 Lewis Gazzard, a,* Karen Williams, g Huifen Chen, a Lorraine Axford,
Supporting Information Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1 Lewis Gazzard, a,* Karen Williams, g Huifen Chen, a Lorraine Axford,
Brigatinib overcomes ALK resistance mechanisms preclinically Zhang et al Supplementary information
 Supplementary information The potent ALK inhibitor brigatinib (AP26113) overcomes mechanisms of resistance to first- and second-generation ALK inhibitors in preclinical models Sen Zhang, Rana Anjum, Rachel
Supplementary information The potent ALK inhibitor brigatinib (AP26113) overcomes mechanisms of resistance to first- and second-generation ALK inhibitors in preclinical models Sen Zhang, Rana Anjum, Rachel
2. Appendix Tables legend General Legend applicable for Table S1 to S4 (Page 10)
 Appendix Data The hvps- signalling module counteracts inhibition of the PIK-Akt pathway to maintain mtorc activity and tumour growth Ruzica Bago, Eeva Sommer, Pau Castel, Claire Crafter, Fiona P. Bailey,
Appendix Data The hvps- signalling module counteracts inhibition of the PIK-Akt pathway to maintain mtorc activity and tumour growth Ruzica Bago, Eeva Sommer, Pau Castel, Claire Crafter, Fiona P. Bailey,
Overcoming Resistance to HER2 Inhibitors Through State-Specific Kinase Binding
 Supplementary Information Overcoming Resistance to Inhibitors Through State-Specific Kinase Binding Chris J. Novotny 1, Sirkku Pollari 2, Jin H. Park 3, Mark A. Lemmon 3,4, Weijun Shen 2*, Kevan M. Shokat
Supplementary Information Overcoming Resistance to Inhibitors Through State-Specific Kinase Binding Chris J. Novotny 1, Sirkku Pollari 2, Jin H. Park 3, Mark A. Lemmon 3,4, Weijun Shen 2*, Kevan M. Shokat
Supplementary Table 1. In vitro side effect profiling study for LDN/OSU
 Supplementary Table 1. In vitro side effect profiling study for LDN/OSU-0212320. Receptor Percent Inhibition 10 µm Neurotransmitter Related Adenosine, Non-selective 7.29% Adrenergic, Alpha 1, Non-selective
Supplementary Table 1. In vitro side effect profiling study for LDN/OSU-0212320. Receptor Percent Inhibition 10 µm Neurotransmitter Related Adenosine, Non-selective 7.29% Adrenergic, Alpha 1, Non-selective
2,4,6-Triaminopyrimidine as a Novel Hinge Binder in a Series of PI3Kδ Selective Inhibitors
 SUPPORTING INFORMATION 2,4,6-Triaminopyrimidine as a Novel Hinge Binder in a Series of PI3Kδ Selective Inhibitors Leena Patel,* Jayaraman Chandrasekhar, Jerry Evarts, Aaron C. Haran, Carmen Ip, Joshua
SUPPORTING INFORMATION 2,4,6-Triaminopyrimidine as a Novel Hinge Binder in a Series of PI3Kδ Selective Inhibitors Leena Patel,* Jayaraman Chandrasekhar, Jerry Evarts, Aaron C. Haran, Carmen Ip, Joshua
Supplementary Materials for
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 217 Supplementary Materials for Chemical genomic analysis of GPR35 signaling Heidi (Haibei)
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 217 Supplementary Materials for Chemical genomic analysis of GPR35 signaling Heidi (Haibei)
Mechanisms of resistance to JAK inhibitors. L. Knoops
 Mechanisms of resistance to JAK inhibitors L. Knoops 1 : Resistance to tyrosine kinase inhibition in cancer are related in sequence and structure. The main diagram illustrates the similarity between the
Mechanisms of resistance to JAK inhibitors L. Knoops 1 : Resistance to tyrosine kinase inhibition in cancer are related in sequence and structure. The main diagram illustrates the similarity between the
Staurosporine Tethered Peptide Ligands for camp-dependent Protein Kinase A (PKA): Optimization and Selectivity Profiling
 Staurosporine Tethered Peptide Ligands for camp-dependent Protein Kinase A (PKA): Optimization and Selectivity Profiling Carolyn D. Shomin, Scott C. Meyer and Indraneel Ghosh* Supplementary Information
Staurosporine Tethered Peptide Ligands for camp-dependent Protein Kinase A (PKA): Optimization and Selectivity Profiling Carolyn D. Shomin, Scott C. Meyer and Indraneel Ghosh* Supplementary Information
Supplementary information
 Supplementary information Pyk2 activates the NLRP3 inflammasome by directly phosphorylating ASC and contributes to inflammasome-dependent peritonitis I-Che Chung 1, Chun-Nan OuYang 1, Sheng-Ning Yuan 1,
Supplementary information Pyk2 activates the NLRP3 inflammasome by directly phosphorylating ASC and contributes to inflammasome-dependent peritonitis I-Che Chung 1, Chun-Nan OuYang 1, Sheng-Ning Yuan 1,
Supplemental Table S1: Inhibition of HDAC class I and class II family by CUDC-101 (IC50 in nm)
 Supplemental Table S1: Inhibition of HDAC class I and class II family by CUDC-101 (IC50 in nm) Class I Class II HDAC1 HDAC2 HDAC3 HDAC8 HDAC4 HDAC5 HDAC6 HDAC7 HDAC9 HDAC10 4.5 12.6 9.1 79.8 13.2 11.4
Supplemental Table S1: Inhibition of HDAC class I and class II family by CUDC-101 (IC50 in nm) Class I Class II HDAC1 HDAC2 HDAC3 HDAC8 HDAC4 HDAC5 HDAC6 HDAC7 HDAC9 HDAC10 4.5 12.6 9.1 79.8 13.2 11.4
Loss of viability Z score threshold. Gene not essential
 Gene essential Z score gene A BT20 BT474 CAL120 CAL51 CAMA1 HCC1143 HCC202 JIMT1 MCF7 MDAM134 MDAMB157 MDAMB231 MDAMB453 MDAMB468 SKBR3 SUM149 SUM44 T47D VP229 Loss of viability Z score threshold BT20
Gene essential Z score gene A BT20 BT474 CAL120 CAL51 CAMA1 HCC1143 HCC202 JIMT1 MCF7 MDAM134 MDAMB157 MDAMB231 MDAMB453 MDAMB468 SKBR3 SUM149 SUM44 T47D VP229 Loss of viability Z score threshold BT20
Content Prioritization And Content Entry and Quality Control Process
 Content Prioritization And Content Entry and Quality Control Process The process of data capture begins with the definition of the content module or sub-module to be built (see figure 1). Broadly we define
Content Prioritization And Content Entry and Quality Control Process The process of data capture begins with the definition of the content module or sub-module to be built (see figure 1). Broadly we define
Supplemental Materials
 Irreversible inhibition of BTK kinase by a novel highly selective inhibitor CHMFL-BTK- 11 suppresses inflammatory response in rheumatoid arthritis model Hong Wu 1,2,7, Qiong Huang 3,7, Ziping Qi 1,7, Yongfei
Irreversible inhibition of BTK kinase by a novel highly selective inhibitor CHMFL-BTK- 11 suppresses inflammatory response in rheumatoid arthritis model Hong Wu 1,2,7, Qiong Huang 3,7, Ziping Qi 1,7, Yongfei
Supporting Information
 Supporting Information Bommi-Reddy et al. 10.1073/pnas.0806574105 Fig. S1. Reintroducing wild-type pvhl in VHL / cells does not affect cell growth in vitro.(a) Immunoblot analysis of 786-O and RCC4 VHL
Supporting Information Bommi-Reddy et al. 10.1073/pnas.0806574105 Fig. S1. Reintroducing wild-type pvhl in VHL / cells does not affect cell growth in vitro.(a) Immunoblot analysis of 786-O and RCC4 VHL
Identification of 4-phenylquinolin-2(1H)-one as a specific allosteric inhibitor of Akt
 Supporting Information Identification of 4-phenylquinolin-2(1H)-one as a specific allosteric inhibitor of Akt Bill X. Huang 1, Kenny Newcomer 1, Karl Kevala 1, Elena Barnaeva 2, Wei Zheng 2, Xin Hu 2,
Supporting Information Identification of 4-phenylquinolin-2(1H)-one as a specific allosteric inhibitor of Akt Bill X. Huang 1, Kenny Newcomer 1, Karl Kevala 1, Elena Barnaeva 2, Wei Zheng 2, Xin Hu 2,
SUPPLEMENTAL TABLES. Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance
 SUPPLEMENTAL TABLES Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance Ping Chen 1, Nathan V. Lee 2, Wenyue Hu 3, Meirong Xu 2, Rose Ann Ferre 1, Hieu Lam 2, Simon Bergqvist 2,
SUPPLEMENTAL TABLES Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance Ping Chen 1, Nathan V. Lee 2, Wenyue Hu 3, Meirong Xu 2, Rose Ann Ferre 1, Hieu Lam 2, Simon Bergqvist 2,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:.38/nature83 Supplementary figure An Overview of Experimental Flow Hypothesis: The tumor microenvironment has a significant impact on cancer cell chemoresistance. Screen Coculture
SUPPLEMENTARY INFORMATION doi:.38/nature83 Supplementary figure An Overview of Experimental Flow Hypothesis: The tumor microenvironment has a significant impact on cancer cell chemoresistance. Screen Coculture
50 nmoles substrate. 8,000 Test Points based on 100 nm in reaction
 Product Insert IMAP Substrates About the IMAP Substrates To facilitate the customization of the IMAP assay to fit your specific needs, Molecular Devices offers a wide range of validated substrates and
Product Insert IMAP Substrates About the IMAP Substrates To facilitate the customization of the IMAP assay to fit your specific needs, Molecular Devices offers a wide range of validated substrates and
Antitumor Activity of CUDC-305, a Novel Oral HSP90 Inhibitor, in Solid and Hematological Tumor Xenograft Models
 Antitumor Activity of CUDC-5, a Novel Oral HSP Inhibitor, in Solid and Hematological Tumor Xenograft Models Rudi Bao, MD/PhD April 1, 2 AACR 1th Annual Meeting 2 Experimental and Molecular Therapeutics
Antitumor Activity of CUDC-5, a Novel Oral HSP Inhibitor, in Solid and Hematological Tumor Xenograft Models Rudi Bao, MD/PhD April 1, 2 AACR 1th Annual Meeting 2 Experimental and Molecular Therapeutics
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study. February 2018
 Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Targeted Agent and Profiling Utilization Registry (TAPUR ) Study February 2018 Precision Medicine Therapies designed to target the molecular alteration that aids cancer development 30 TARGET gene alterations
Targeting S-Adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A
 Targeting S-Adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A Supplementary information Casey L. Quinlan* 1, Stephen E. Kaiser* 2, Ben Bolaños 2, Dawn Nowlin 1, Rita Grantner 1,
Targeting S-Adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A Supplementary information Casey L. Quinlan* 1, Stephen E. Kaiser* 2, Ben Bolaños 2, Dawn Nowlin 1, Rita Grantner 1,
Phosphorylation Site Company Cat #
 Supplemental Table 1. Antibodies used for RPPA analysis. Label Protein Phosphorylation Site Company Cat # Used on MDA_CLSS Used on MDA_Pilot 4EBP1 4EBP1 Cell Signaling 9452 No 4EBP1.pS65 4EBP1 S65 Cell
Supplemental Table 1. Antibodies used for RPPA analysis. Label Protein Phosphorylation Site Company Cat # Used on MDA_CLSS Used on MDA_Pilot 4EBP1 4EBP1 Cell Signaling 9452 No 4EBP1.pS65 4EBP1 S65 Cell
IntelliGENSM. Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community.
 IntelliGENSM Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community. NGS TRANSFORMS GENOMIC TESTING Background Cancers may emerge as a result of somatically
IntelliGENSM Integrated Oncology is making next generation sequencing faster and more accessible to the oncology community. NGS TRANSFORMS GENOMIC TESTING Background Cancers may emerge as a result of somatically
Supporting Information
 Supporting Information Discovery of Highly Isoform Selective rally Bioavailable Phosphoinositide-3-Kinase (PI3K)- Inhibitors. ils Pemberton,,* Mickael Mogemark,,* Susanne Arlbrandt, Peter Bold, Rhona J
Supporting Information Discovery of Highly Isoform Selective rally Bioavailable Phosphoinositide-3-Kinase (PI3K)- Inhibitors. ils Pemberton,,* Mickael Mogemark,,* Susanne Arlbrandt, Peter Bold, Rhona J
Discovery of Orally Efficacious Phosphoinositide-3-Kinase delta Inhibitors with Improved
 SUPPORTIG IFORMATIO Discovery of Orally Efficacious Phosphoinositide-3-Kinase delta Inhibitors with Improved Metabolic Stability Leena Patel,* Jayaraman Chandrasekhar, Jerry Evarts, Kristen Forseth, Aaron
SUPPORTIG IFORMATIO Discovery of Orally Efficacious Phosphoinositide-3-Kinase delta Inhibitors with Improved Metabolic Stability Leena Patel,* Jayaraman Chandrasekhar, Jerry Evarts, Kristen Forseth, Aaron
Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making
 Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making November 20, 2014 Capturing Value in Next Generation Sequencing Symposium Douglas Johnson MD, MSCI Vanderbilt-Ingram
Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making November 20, 2014 Capturing Value in Next Generation Sequencing Symposium Douglas Johnson MD, MSCI Vanderbilt-Ingram
Supplementary Information. Targeting BRK-positive Breast Cancer with Small
 Supplementary Information Targeting BRK-positive Breast Cancer with Small Molecule Kinase Inhibitors Jie Jiang 1#, Fu Gui 1#, Zhixiang He 1#, Li Li 1, Yunzhan Li 1, Shunying Li 2, Xinrui Wu 1, Zhou Deng
Supplementary Information Targeting BRK-positive Breast Cancer with Small Molecule Kinase Inhibitors Jie Jiang 1#, Fu Gui 1#, Zhixiang He 1#, Li Li 1, Yunzhan Li 1, Shunying Li 2, Xinrui Wu 1, Zhou Deng
SUPPLEMENTARY INFORMATION
 Supplementary Information S3 TAM- family small molecule kinase inhibitors in development Compound Indication(s) Target Profile Develop Primary Target MERTK TYRO3 Other targets ment Phase Refs Cabozantinib
Supplementary Information S3 TAM- family small molecule kinase inhibitors in development Compound Indication(s) Target Profile Develop Primary Target MERTK TYRO3 Other targets ment Phase Refs Cabozantinib
Next generation sequencing on FFPE tumor tissues for lung and colorectal carcinoma diagnostics and research Sakari Knuutila
 Next generation sequencing on FFPE tumor tissues for lung and colorectal carcinoma diagnostics and research Sakari Knuutila Faculty of Medicine / Haartman Institute / Department of Pathology / Sakari Knuutila
Next generation sequencing on FFPE tumor tissues for lung and colorectal carcinoma diagnostics and research Sakari Knuutila Faculty of Medicine / Haartman Institute / Department of Pathology / Sakari Knuutila
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/322/ra38/dc1 Supplementary Materials for Dynamic Reprogramming of Signaling Upon Met Inhibition Reveals a Mechanism of Drug Resistance in Gastric Cancer Andrea
www.sciencesignaling.org/cgi/content/full/7/322/ra38/dc1 Supplementary Materials for Dynamic Reprogramming of Signaling Upon Met Inhibition Reveals a Mechanism of Drug Resistance in Gastric Cancer Andrea
SureSelect Cancer All-In-One Custom and Catalog NGS Assays
 SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
Supplementary Material. Part I: Sample Information. Part II: Pathway Information
 Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Supplementary Data Supplementary Table S1. IC 50 values of ipatasertib on cell viability in cell lines with
 Supplementary Data Supplementary Table S1. IC 50 values of ipatasertib on cell viability in cell lines with or without alterations in PTEN or PIK3CA. Cell Line Tissue Type Ipatasertib IC 50 (μmol/l) a
Supplementary Data Supplementary Table S1. IC 50 values of ipatasertib on cell viability in cell lines with or without alterations in PTEN or PIK3CA. Cell Line Tissue Type Ipatasertib IC 50 (μmol/l) a
ALM301: Allosteric Isoform selective Akt inhibitor
 ALM301: Allosteric Isoform selective Akt inhibitor Background Akt1/2 selective inhibitors ALM301 Back-up compounds Akt2 selective inhibitors Approaches to Akt Inhibition Both ATP competitive and allosteric
ALM301: Allosteric Isoform selective Akt inhibitor Background Akt1/2 selective inhibitors ALM301 Back-up compounds Akt2 selective inhibitors Approaches to Akt Inhibition Both ATP competitive and allosteric
Understanding Signaling Pathways by Modifying Sensitivity to PLX4720 in B-RAF V600E Melanoma
 Understanding Signaling Pathways by Modifying Sensitivity to PLX4720 in B-RAF V600E Melanoma Muska Hassan NCI-ICBP Summer Fellow Broad Institute of MIT and Harvard: Cancer Program Mentor: Cory Johannessen,
Understanding Signaling Pathways by Modifying Sensitivity to PLX4720 in B-RAF V600E Melanoma Muska Hassan NCI-ICBP Summer Fellow Broad Institute of MIT and Harvard: Cancer Program Mentor: Cory Johannessen,
SUPPLEMENTARY INFORMATION
 Meta-Analysis Supplementary Fig.. Schematic of experimental design. Large scale shrna screens for proliferation/viability were performed in 9 cell lines using a library of 52 shrnas targeting 957 genes
Meta-Analysis Supplementary Fig.. Schematic of experimental design. Large scale shrna screens for proliferation/viability were performed in 9 cell lines using a library of 52 shrnas targeting 957 genes
AACR 101st Annual Meeting 2010, Washington D.C. Experimental and Molecular Therapeutics Section 29; Abstract #3855
 Investigation of the Growth Inhibitory Activity of the MEK Inhibitor ARRY-162 in Combination with Everolimus in a Variety of KRas and PI3K Pathway Mutant Cancers Brian Tunquist, Tyler Risom, Debbie Anderson,
Investigation of the Growth Inhibitory Activity of the MEK Inhibitor ARRY-162 in Combination with Everolimus in a Variety of KRas and PI3K Pathway Mutant Cancers Brian Tunquist, Tyler Risom, Debbie Anderson,
Out-Patient Billing CPT Codes
 Out-Patient Billing CPT Codes Updated Date: August 3, 08 Client Billed Molecular Tests HPV DNA Tissue Testing 8764 No Medicare Billed - Molecular Tests NeoARRAY NeoARRAY SNP/Cytogenetic No 89 NeoLAB NeoLAB
Out-Patient Billing CPT Codes Updated Date: August 3, 08 Client Billed Molecular Tests HPV DNA Tissue Testing 8764 No Medicare Billed - Molecular Tests NeoARRAY NeoARRAY SNP/Cytogenetic No 89 NeoLAB NeoLAB
RXDX-101 & RXDX-102. Justin Gainor, MD February 20 th, 2014
 RXDX-101 & RXDX-102 Justin Gainor, MD February 20 th, 2014 Background Chromosomal fusions are important oncogenic drivers in NSCLC - ALK Rearrangements (4-6%) - ROS1 Rearrangements (1-2%) - RET Rearrangements
RXDX-101 & RXDX-102 Justin Gainor, MD February 20 th, 2014 Background Chromosomal fusions are important oncogenic drivers in NSCLC - ALK Rearrangements (4-6%) - ROS1 Rearrangements (1-2%) - RET Rearrangements
Genomic Medicine: What every pathologist needs to know
 Genomic Medicine: What every pathologist needs to know Stephen P. Ethier, Ph.D. Professor, Department of Pathology and Laboratory Medicine, MUSC Director, MUSC Center for Genomic Medicine Genomics and
Genomic Medicine: What every pathologist needs to know Stephen P. Ethier, Ph.D. Professor, Department of Pathology and Laboratory Medicine, MUSC Director, MUSC Center for Genomic Medicine Genomics and
Frequency(%) KRAS G12 KRAS G13 KRAS A146 KRAS Q61 KRAS K117N PIK3CA H1047 PIK3CA E545 PIK3CA E542K PIK3CA Q546. EGFR exon19 NFS-indel EGFR L858R
 Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
What is the status of the technologies of "precision medicine?
 Session 2: What is the status of the technologies of "precision medicine? Gideon Blumenthal, MD, Clinical Team Leader, Thoracic and Head/Neck Oncology, Center for Drug Evaluation and Research (CDER), U.S.
Session 2: What is the status of the technologies of "precision medicine? Gideon Blumenthal, MD, Clinical Team Leader, Thoracic and Head/Neck Oncology, Center for Drug Evaluation and Research (CDER), U.S.
NanoBRET: A Quantitative Technique to Measure Kinase Target Engagement in Live Cells Matthew Robers
 anobret: A Quantitative Technique to Measure Kinase Target Engagement in Live Cells Matthew Robers Sr. Research Scientist & Group Leader Premise: Targets may behave differently in cells than in isolation
anobret: A Quantitative Technique to Measure Kinase Target Engagement in Live Cells Matthew Robers Sr. Research Scientist & Group Leader Premise: Targets may behave differently in cells than in isolation
PLX7486 Background Information October Candidate for CRUK Combinations Alliance
 PLX7486 Background Information October 2015 Candidate for CRUK Combinations Alliance 1 Oct 2015 Plexxikon s Development Pipeline Compound Target Cancer Indication Stage of Development Pre- IND Ph1 Ph2
PLX7486 Background Information October 2015 Candidate for CRUK Combinations Alliance 1 Oct 2015 Plexxikon s Development Pipeline Compound Target Cancer Indication Stage of Development Pre- IND Ph1 Ph2
Supplemental Table 1. Biochemical and Cellular Potency and Selectivity of PF
 Supplemental Table 1. Biochemical and Cellular Potency and Selectivity of PF- 02341066 Assay IC 50 nm Selectivity Ratio d Biochemical Activity In Vitro c-met/hgfr enzyme (Ki, nm) a 4 NA Cellular Activity
Supplemental Table 1. Biochemical and Cellular Potency and Selectivity of PF- 02341066 Assay IC 50 nm Selectivity Ratio d Biochemical Activity In Vitro c-met/hgfr enzyme (Ki, nm) a 4 NA Cellular Activity
LavaCell. Fluorescent Cell Stain. (version A) Catalog No
 LavaCell Fluorescent Cell Stain (version A) Catalog No. 15004 Active Motif North America 1914 Palomar Oaks Way, Suite 150 Carlsbad, California 92008, USA Toll free: 877 222 9543 Telephone: 760 431 1263
LavaCell Fluorescent Cell Stain (version A) Catalog No. 15004 Active Motif North America 1914 Palomar Oaks Way, Suite 150 Carlsbad, California 92008, USA Toll free: 877 222 9543 Telephone: 760 431 1263
Investigations of helminth kinomes - from understanding parasite biology to drug discovery. Andreas J. Stroehlein
 Investigations of helminth kinomes - from understanding parasite biology to drug discovery Andreas J. Stroehlein BSc Bioinformatics The Pan-Proteome of the Bacterial Pathogens Causing Bovine Respiratory
Investigations of helminth kinomes - from understanding parasite biology to drug discovery Andreas J. Stroehlein BSc Bioinformatics The Pan-Proteome of the Bacterial Pathogens Causing Bovine Respiratory
Clinical Grade Genomic Profiling: The Time Has Come
 Clinical Grade Genomic Profiling: The Time Has Come Gary Palmer, MD, JD, MBA, MPH Senior Vice President, Medical Affairs Foundation Medicine, Inc. Oct. 22, 2013 1 Why We Are Here A Shared Vision At Foundation
Clinical Grade Genomic Profiling: The Time Has Come Gary Palmer, MD, JD, MBA, MPH Senior Vice President, Medical Affairs Foundation Medicine, Inc. Oct. 22, 2013 1 Why We Are Here A Shared Vision At Foundation
For Simultaneously Detecting the Relative Level of Tyrosine Phosphorylation of Human Receptor Tyrosine Kinases (RTKs).
 RayBio Phosphorylation Antibody Array I For Simultaneously Detecting the Relative Level of Tyrosine Phosphorylation of Human Receptor Tyrosine Kinases (RTKs). User Manual (Revised Jul 25, 2007) (Cat# AAH-PRTK-1-2;
RayBio Phosphorylation Antibody Array I For Simultaneously Detecting the Relative Level of Tyrosine Phosphorylation of Human Receptor Tyrosine Kinases (RTKs). User Manual (Revised Jul 25, 2007) (Cat# AAH-PRTK-1-2;
Tratamiento neoadyuvante: Enfermedad residual como marcador de resistencia Carlos L. Arteaga, MD Vanderbilt-Ingram Cancer Center Vanderbilt
 Tratamiento neoadyuvante: Enfermedad residual como marcador de resistencia Carlos L. Arteaga, MD Vanderbilt-Ingram Cancer Center Vanderbilt University Neoadjuvant (preoperative) therapy Surgery Systemic
Tratamiento neoadyuvante: Enfermedad residual como marcador de resistencia Carlos L. Arteaga, MD Vanderbilt-Ingram Cancer Center Vanderbilt University Neoadjuvant (preoperative) therapy Surgery Systemic
Jennifer Hauenstein Oncology Cytogenetics Emory University Hospital Atlanta, GA
 Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Response and resistance to BRAF inhibitors in melanoma
 Response and resistance to BRAF inhibitors in melanoma Keith T. Flaherty, M.D. Massachusetts General Hospital Cancer Center Disclosures Roche/Genentech: consultant GlaxoSmithKline: consultant BRAF mutations
Response and resistance to BRAF inhibitors in melanoma Keith T. Flaherty, M.D. Massachusetts General Hospital Cancer Center Disclosures Roche/Genentech: consultant GlaxoSmithKline: consultant BRAF mutations
Kinases 1. Kinase Inhibitors 13. Kinase Inhibitor Sets 21. Kinase Substrates 22. Kinase Activators 24. Phosphatases 26. Phosphatase Inhibitors 26
 Table of contents Kinases 1 Kinase Inhibitors 13 Kinase Inhibitor Sets 21 Kinase Substrates 22 Kinase Activators 24 Phosphatases 26 Phosphatase Inhibitors 26 Cytokines 27 Antibodies 28 Research Sets 28
Table of contents Kinases 1 Kinase Inhibitors 13 Kinase Inhibitor Sets 21 Kinase Substrates 22 Kinase Activators 24 Phosphatases 26 Phosphatase Inhibitors 26 Cytokines 27 Antibodies 28 Research Sets 28
Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD
 Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD Neuropathology Fellow Division of Neuropathology Center for Personalized Diagnosis (CPD) Glial
Clinically Useful Next Generation Sequencing and Molecular Testing in Gliomas MacLean P. Nasrallah, MD PhD Neuropathology Fellow Division of Neuropathology Center for Personalized Diagnosis (CPD) Glial
Fluxion Biosciences and Swift Biosciences Somatic variant detection from liquid biopsy samples using targeted NGS
 APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
NMS Drug Discovery Platform. NMS-ROL partnership opportunities. Antonella Isacchi Director, Biotechnology Dept. Kinase Platform Coordinator
 MS Drug Discovery Platform MS-RL partnership opportunities Antonella Isacchi Director, Biotechnology Dept. Kinase Platform Coordinator MS-RL Meeting 28 January 2011 utline of the presentation Introduction
MS Drug Discovery Platform MS-RL partnership opportunities Antonella Isacchi Director, Biotechnology Dept. Kinase Platform Coordinator MS-RL Meeting 28 January 2011 utline of the presentation Introduction
How to compute a semantic similarity threshold. Charles Bettembourg, Christian Diot, Olivier Dameron
 How to compute a semantic similarity threshold Charles Bettembourg, Christian Diot, Olivier Dameron Abstract The analysis of gene annotations related to Gene Ontology plays an important role in the interpretation
How to compute a semantic similarity threshold Charles Bettembourg, Christian Diot, Olivier Dameron Abstract The analysis of gene annotations related to Gene Ontology plays an important role in the interpretation
EXAMPLE. - Potentially responsive to PI3K/mTOR and MEK combination therapy or mtor/mek and PKC combination therapy. ratio (%)
 Dr Kate Goodhealth Goodhealth Medical Clinic 123 Address Road SUBURBTOWN NSW 2000 Melanie Citizen Referring Doctor Your ref Address Dr John Medico 123 Main Street, SUBURBTOWN NSW 2000 Phone 02 9999 9999
Dr Kate Goodhealth Goodhealth Medical Clinic 123 Address Road SUBURBTOWN NSW 2000 Melanie Citizen Referring Doctor Your ref Address Dr John Medico 123 Main Street, SUBURBTOWN NSW 2000 Phone 02 9999 9999
VEGFR. 1
 VEGFR VEGFRs (vascular endothelial growth factor receptors) are tyrosine kinase receptors responsible for binding with VEGF to initiate signal cascades that stimulate angiogenesis among other effects.
VEGFR VEGFRs (vascular endothelial growth factor receptors) are tyrosine kinase receptors responsible for binding with VEGF to initiate signal cascades that stimulate angiogenesis among other effects.
Development of Rational Drug Combinations for Oncology Indications - An Industry Perspective
 Development of Rational Drug Combinations for Oncology Indications An Industry Perspective Stuart Lutzker, MDPhD Vice President Oncology Early Development Genentech Inc 1 Conventional Oncology Drug Development
Development of Rational Drug Combinations for Oncology Indications An Industry Perspective Stuart Lutzker, MDPhD Vice President Oncology Early Development Genentech Inc 1 Conventional Oncology Drug Development
The Center for PERSONALIZED DIAGNOSTICS
 The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
Predictive biomarker profiling of > 1,900 sarcomas: Identification of potential novel treatment modalities
 Predictive biomarker profiling of > 1,900 sarcomas: Identification of potential novel treatment modalities Sujana Movva 1, Wenhsiang Wen 2, Wangjuh Chen 2, Sherri Z. Millis 2, Margaret von Mehren 1, Zoran
Predictive biomarker profiling of > 1,900 sarcomas: Identification of potential novel treatment modalities Sujana Movva 1, Wenhsiang Wen 2, Wangjuh Chen 2, Sherri Z. Millis 2, Margaret von Mehren 1, Zoran
SUPPORTING INFORMATION
 SUPPORTING INFORMATION Discovery and Preclinical Characterization of 6-Chloro-5-[4-(1- hydroxycyclobutyl)phenyl]-1h-indole-3-carboxylic Acid (PF-06409577), a Direct Activator of Adenosine Monophosphate-activated
SUPPORTING INFORMATION Discovery and Preclinical Characterization of 6-Chloro-5-[4-(1- hydroxycyclobutyl)phenyl]-1h-indole-3-carboxylic Acid (PF-06409577), a Direct Activator of Adenosine Monophosphate-activated
Gene Symbol Accession Alias/Prev Symbol Official Full Name AAK1 NM_ KIAA1048, DKFZp686K16132 AP2 associated kinase 1 (AAK1) AATK
 Gene Symbol Accession Alias/Prev Symbol Official Full Name AAK1 NM_014911.2 KIAA1048, DKFZp686K16132 AP2 associated kinase 1 (AAK1) AATK NM_001080395.2 AATYK, AATYK1, KIAA0641, LMR1, LMTK1, p35bp apoptosis-associated
Gene Symbol Accession Alias/Prev Symbol Official Full Name AAK1 NM_014911.2 KIAA1048, DKFZp686K16132 AP2 associated kinase 1 (AAK1) AATK NM_001080395.2 AATYK, AATYK1, KIAA0641, LMR1, LMTK1, p35bp apoptosis-associated
DMSO GNF7156 GNF4877 DA/ U Ed/ nilusni M o u s e b e t a c e l l p r o li f e r a t i o n GNF4877 G N F G N F Ki6 e 3
 GNF4877 Vehicle % E d u + In s + c e lls Insulin/ Ki67 Insulin/EdU/DAPI a b DMSO GNF7156 GNF4877 c d M o u s e b e ta c e ll p r o life r a tio n e GNF4877 4 3 G N F 7 1 5 6 C h iro n 9 9 2 1 2 Insulin/Ki67
GNF4877 Vehicle % E d u + In s + c e lls Insulin/ Ki67 Insulin/EdU/DAPI a b DMSO GNF7156 GNF4877 c d M o u s e b e ta c e ll p r o life r a tio n e GNF4877 4 3 G N F 7 1 5 6 C h iro n 9 9 2 1 2 Insulin/Ki67
Illumina s Cancer Research Portfolio and Dedicated Workflows
 Illumina s Cancer Research Portfolio and Dedicated Workflows Michael Sohn Clinical Sales Specialist Spain&Italy 2017 2017 Illumina, Inc. All rights reserved. Illumina, 24sure, BaseSpace, BeadArray, BlueFish,
Illumina s Cancer Research Portfolio and Dedicated Workflows Michael Sohn Clinical Sales Specialist Spain&Italy 2017 2017 Illumina, Inc. All rights reserved. Illumina, 24sure, BaseSpace, BeadArray, BlueFish,
Personalised cancer care Information for Medical Specialists. A new way to unlock treatment options for your patients
 Personalised cancer care Information for Medical Specialists A new way to unlock treatment options for your patients Contents Optimised for clinical benefit 4 Development history 4 Full FIND IT panel vs
Personalised cancer care Information for Medical Specialists A new way to unlock treatment options for your patients Contents Optimised for clinical benefit 4 Development history 4 Full FIND IT panel vs
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): The authors have presented data demonstrating activation of AKT as a common resistance mechanism in EGFR mutation positive, EGFR TKI resistant
Reviewers' comments: Reviewer #1 (Remarks to the Author): The authors have presented data demonstrating activation of AKT as a common resistance mechanism in EGFR mutation positive, EGFR TKI resistant
Accel-Amplicon Panels
 Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST PROSPECTIVE
 AMP COMPANION MEETING SYMPOSIUM AT USCAP 2015 NEXT-GENERATION OF PATHOLOGY: ROLE OF PATHOLOGIST IN NGS-BASED PERSONALIZED MEDICINE APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST
AMP COMPANION MEETING SYMPOSIUM AT USCAP 2015 NEXT-GENERATION OF PATHOLOGY: ROLE OF PATHOLOGIST IN NGS-BASED PERSONALIZED MEDICINE APPLICATIONS OF NEXT GENERATION SEQUENCING IN SOLID TUMORS - PATHOLOGIST
Targeting the Hsp90-Cdc37-client protein interaction to disrupt Hsp90 chaperone machinery
 Li et al. Journal of Hematology & Oncology (2018) 11:59 https://doi.org/10.1186/s13045-018-0602-8 REVIEW Targeting the Hsp90-Cdc37-client protein interaction to disrupt Hsp90 chaperone machinery Ting Li
Li et al. Journal of Hematology & Oncology (2018) 11:59 https://doi.org/10.1186/s13045-018-0602-8 REVIEW Targeting the Hsp90-Cdc37-client protein interaction to disrupt Hsp90 chaperone machinery Ting Li
August 17, Dear Valued Client:
 August 7, 08 Re: CMS Announces 6-Month Period of Enforcement Discretion for Laboratory Date of Service Exception Policy Under the Medicare Clinical Laboratory Fee Schedule (the 4 Day Rule ) Dear Valued
August 7, 08 Re: CMS Announces 6-Month Period of Enforcement Discretion for Laboratory Date of Service Exception Policy Under the Medicare Clinical Laboratory Fee Schedule (the 4 Day Rule ) Dear Valued
Yong Wu, Ph.D. Division of Cancer Research and Training (DCRT) Charles R. Drew University of Medicine & Science
 Yong Wu, Ph.D. Division of Cancer Research and Training (DCRT) Charles R. Drew University of Medicine & Science Jay Vadgama, Ph.D Chief, Division of Cancer Research and Training Background One in 8 women
Yong Wu, Ph.D. Division of Cancer Research and Training (DCRT) Charles R. Drew University of Medicine & Science Jay Vadgama, Ph.D Chief, Division of Cancer Research and Training Background One in 8 women
Sample Metrics. Allele Frequency (%) Read Depth Ploidy. Gene CDS Effect Protein Effect. LN Metastasis Tumor Purity Computational Pathology 80% 60%
 Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
ENMD-2076, an Oral Aurora A and Angiogenesis Kinase Inhibitor
 ENMD-2076, an Oral Aurora A and Angiogenesis Kinase Inhibitor Dr. Mark R. Bray VP Research, EntreMed, Inc. AACR Annual Meeting New Drugs on the Horizon 2 April 2008 1 Disclosure Information: AACR New Drugs
ENMD-2076, an Oral Aurora A and Angiogenesis Kinase Inhibitor Dr. Mark R. Bray VP Research, EntreMed, Inc. AACR Annual Meeting New Drugs on the Horizon 2 April 2008 1 Disclosure Information: AACR New Drugs
I. Diagnosis of the cancer type in CUP
 Latest Research: USA I. Diagnosis of the cancer type in CUP II. Outcomes of site-specific therapy of the cancer type in CUP a. Prospective clinical trial b. Retrospective clinical trials 1 Latest Research:
Latest Research: USA I. Diagnosis of the cancer type in CUP II. Outcomes of site-specific therapy of the cancer type in CUP a. Prospective clinical trial b. Retrospective clinical trials 1 Latest Research:
Exons from 137 genes were targeted for hybrid selection and massively parallel sequencing.
 SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
SUPPLEMENTARY TABLES Supplementary Table 1. Targeted Genes ABL1 ABL2 AKT1 AKT2 AKT3 ALK APC ATM AURKA BCL2 BRAF BRCA1 BRCA2 CCND1 CCNE1 CDC73 CDH1 CDK4 CDK6 CDK8 CDKN1A CDKN2A CEBPA CHEK1 CHEK2 CREBBP
Mass spectrometric genotyping (OncoMap) Sensitivity of genotyping Laser capture microdissection Quantifying Langerhans cells in LCH samples
 Mass spectrometric genotyping (OncoMap) DNA was quantified using picogreen analysis, then subjected to whole genome amplification and used for further analysis only if DNA fingerprinting demonstrated non-biased
Mass spectrometric genotyping (OncoMap) DNA was quantified using picogreen analysis, then subjected to whole genome amplification and used for further analysis only if DNA fingerprinting demonstrated non-biased
Oncomine Focus assay panel and Oncomine Knowledgebase Reporter.
 Oncomine Focus assay panel and Oncomine Knowledgebase Reporter. How it can help to identify relevant alteration and early phase trials. Dr Isabelle SOUBEYRAN Dr Emmanuel KHALIFA Molecular Pathology Unit
Oncomine Focus assay panel and Oncomine Knowledgebase Reporter. How it can help to identify relevant alteration and early phase trials. Dr Isabelle SOUBEYRAN Dr Emmanuel KHALIFA Molecular Pathology Unit
Diagnostic test Suggested website label Description Hospitals available
 Diagnostic test Suggested website label Description Hospitals available Abbott Molecular Inc, PATHVYSION HER-2 DNA Probe Kit (FISH) PathVysion kit A diagnostic tool used to determine whether a particular
Diagnostic test Suggested website label Description Hospitals available Abbott Molecular Inc, PATHVYSION HER-2 DNA Probe Kit (FISH) PathVysion kit A diagnostic tool used to determine whether a particular
ras Multikinase Inhibitor Multikinase Inhibitor 0.1
 a ras ** * ** * ** ** ** ** un in et m lu Se ib SL G 32 W 7 So 50 ra 74 fe ni b LY W 294 or 0 tm 02 R an ap n a in Ev my er cin ol im BE us Z2 3 En P 5 za I13 st 0 au rin D as SP ati 60 nib C 012 is 5
a ras ** * ** * ** ** ** ** un in et m lu Se ib SL G 32 W 7 So 50 ra 74 fe ni b LY W 294 or 0 tm 02 R an ap n a in Ev my er cin ol im BE us Z2 3 En P 5 za I13 st 0 au rin D as SP ati 60 nib C 012 is 5
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Executive Summary. Reproduction prohibited v
 Kinases are a large family of proteins that have now become firmly established as a major class of drug targets for the pharmaceutical industry. The sequencing of the Human Genome has led to the identification
Kinases are a large family of proteins that have now become firmly established as a major class of drug targets for the pharmaceutical industry. The sequencing of the Human Genome has led to the identification
Assessing the Economics of Genomic Medicine. Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center
 Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Osamu Tetsu, MD, PhD Associate Professor Department of Otolaryngology-Head and Neck Surgery School of Medicine, University of California, San
 Osamu Tetsu, MD, PhD Associate Professor Department of Otolaryngology-Head and Neck Surgery School of Medicine, University of California, San Francisco Lung Cancer Classification Pathological Classification
Osamu Tetsu, MD, PhD Associate Professor Department of Otolaryngology-Head and Neck Surgery School of Medicine, University of California, San Francisco Lung Cancer Classification Pathological Classification
Dox. R26-rtTA Tyr-CreERT2. any ink/arf, no rtta (n=8) ink/arf +/+ (n=5) Day 0 Day 11 Day 18 Day 28
 A 4OHT Dox hraf iip tumors inras ddh 2 O -RT Ink/Arf / Pten l/ l R26-lsl-rtTA Tyr-reERT2 TetO-hRAF V6E Ink/Arf / Pten / R26-rtTA Tyr-reERT2 TetO-hRAF V6E Ink/Arf / Pten / R26-rtTA Tyr-reERT2 TetO-hRAF
A 4OHT Dox hraf iip tumors inras ddh 2 O -RT Ink/Arf / Pten l/ l R26-lsl-rtTA Tyr-reERT2 TetO-hRAF V6E Ink/Arf / Pten / R26-rtTA Tyr-reERT2 TetO-hRAF V6E Ink/Arf / Pten / R26-rtTA Tyr-reERT2 TetO-hRAF
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester
 Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
Dr David Guttery Senior PDRA Dept. of Cancer Studies and CRUK Leicester Centre University of Leicester dsg6@le.ac.uk CFDNA/CTDNA Circulating-free AS A LIQUID DNA BIOPSY (cfdna) Tumour Biopsy Liquid Biopsy
Discovery of a Highly Potent and Selective Indenoindolone Type 1 pan-flt3 Inhibitor
 Supporting Information Discovery of a Highly Potent and Selective Indenoindolone Type 1 pan-flt3 Inhibitor John M. Hatcher a,b,e, Ellen Weisberg c,e, Taebo Sim d, Richard M. Stone c, Suiyang Liu c, James
Supporting Information Discovery of a Highly Potent and Selective Indenoindolone Type 1 pan-flt3 Inhibitor John M. Hatcher a,b,e, Ellen Weisberg c,e, Taebo Sim d, Richard M. Stone c, Suiyang Liu c, James
blood mononuclear cells were stained with AAD, annexin, CD3, CD4, CD25, LAP and specific
 Supplementary figure legends Figure 1: Selective expression of regulatory markers on CD4 + LAP + T cells. Human peripheral blood mononuclear cells were stained with AAD, annexin, CD3, CD4, CD25, LAP and
Supplementary figure legends Figure 1: Selective expression of regulatory markers on CD4 + LAP + T cells. Human peripheral blood mononuclear cells were stained with AAD, annexin, CD3, CD4, CD25, LAP and
1.Basis of resistance 2.Mechanisms of resistance 3.How to overcome resistance. 13/10/2017 Sara Redaelli
 Dott.ssa Sara Redaelli 13/10/2017 1.Basis of resistance 2.Mechanisms of resistance 3.How to overcome resistance Tumor Heterogeneity: Oncogenic Drivers in NSCLC The Promise of Genotype-Directed Therapy
Dott.ssa Sara Redaelli 13/10/2017 1.Basis of resistance 2.Mechanisms of resistance 3.How to overcome resistance Tumor Heterogeneity: Oncogenic Drivers in NSCLC The Promise of Genotype-Directed Therapy
Kinome Profiling: The Potential in ER-Negative Patients. Charles M. Perou, Ph.D. Departments of Genetics and Pathology
 Kinome Profiling: The Potential in ER-Negative Patients Charles M. Perou, Ph.D. Departments of Genetics and Pathology Lineberger Comprehensive Cancer Center University of North Carolina Chapel Hill, North
Kinome Profiling: The Potential in ER-Negative Patients Charles M. Perou, Ph.D. Departments of Genetics and Pathology Lineberger Comprehensive Cancer Center University of North Carolina Chapel Hill, North
Combinations of targeted therapies in oncology an industry view
 Combinations of targeted therapies in oncology an industry view Dr Susan Galbraith SVP Head of Oncology imed AstraZeneca WIN 2014 Symposium 23-24 June Paris France Disclosures I am a full time employee
Combinations of targeted therapies in oncology an industry view Dr Susan Galbraith SVP Head of Oncology imed AstraZeneca WIN 2014 Symposium 23-24 June Paris France Disclosures I am a full time employee
Signaling Through Immune System Receptors (Ch. 7)
 Signaling Through Immune System Receptors (Ch. 7) 1. General principles of signal transduction and propagation. 2. Antigen receptor signaling and lymphocyte activation. 3. Other receptors and signaling
Signaling Through Immune System Receptors (Ch. 7) 1. General principles of signal transduction and propagation. 2. Antigen receptor signaling and lymphocyte activation. 3. Other receptors and signaling
Secuenciación masiva: papel en la toma de decisiones
 Secuenciación masiva: papel en la toma de decisiones Cancer is a Genetic Disease Development of cancer is driven by the acquisition of somatic genetic alterations: Nonsynonymous point mutations: missense.
Secuenciación masiva: papel en la toma de decisiones Cancer is a Genetic Disease Development of cancer is driven by the acquisition of somatic genetic alterations: Nonsynonymous point mutations: missense.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Frampton et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Supplementary Figure 1A Base substitution detection
SUPPLEMENTARY INFORMATION Frampton et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Supplementary Figure 1A Base substitution detection
