Molecular and genetic characterization of peroxisome biogenesis disorders Ebberink, M.S.
|
|
|
- Jemimah Hopkins
- 5 years ago
- Views:
Transcription
1 UvA-DARE (Digital Academic Repository) Molecular and genetic characterization of peroxisome biogenesis disorders Ebberink, M.S. Link to publication Citation for published version (APA): Ebberink, M. S. (010). Molecular and genetic characterization of peroxisome biogenesis disorders General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: or a letter to: Library of the University of Amsterdam, Secretariat, Singel 45, 101 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam ( Download date: 6 Dec 018
2 Chapter Genetic Classification and Mutational Spectrum of More Than 600 Patients with a Zellweger Syndrome Spectrum Disorder Merel S. Ebberink, Petra A.W. Mooijer, Jeannette Gootjes, Janet Koster, Ronald J.A. Wanders, Hans R. Waterham Laboratory Genetic Metabolic Diseases, Academic Medical Centre at the University of Amsterdam, Amsterdam, the Netherlands. Submitted for publication
3 Chapter Abstract Introduction: The autosomal recessive Zellweger syndrome spectrum (ZSS) disorders comprise a main subgroup of the peroxisome biogenesis disorders. The ZSS disorders can be caused by mutations in any of 1 different currently identified PEX genes resulting in severe, often lethal, multi-systemic disorders. Objective: To get insight into the spectrum of PEX gene defects among ZSS disorders and to investigate if additional human PEX genes are required for functional peroxisome biogenesis, we assigned over 600 ZSS fibroblast cell lines to different genetic complementation groups. Methods: Fibroblast cell lines were subjected to a complementation assay involving fusion by means of polyethylene glycol or a PEX cdna transfection assay specifically developed for this purpose. For the majority of the cell lines we subsequently determined the underlying mutations by sequence analysis of the implicated PEX genes. Results and Discussion: The PEX cdna transfection assay allows for the rapid identification of PEX genes defective in ZSS patients. The assignment of over 600 fibroblast cell lines to different genetic complementation groups provides the most comprehensive and representative overview of the frequency distribution of the different PEX gene defects. We did not identify any novel genetic complementation group, suggesting that all PEX gene defects resulting in peroxisome deficiency are currently known. 3
4 Genetic Classification and Mutational Spectrum of ZSS Introduction Peroxisome biogenesis disorders (MIM ) include the Zellweger syndrome spectrum (ZSS) disorders and Rhizomelic Chondrodysplasia Punctata type I (Weller et al., 003). The latter is caused by mutations in the PEX7 gene, encoding a cytosolic receptor involved in the import of a small subset of peroxisomal matrix proteins (Braverman et al., 00; Motley et al., 00). The ZSS disorders include the Zellweger syndrome (ZS, MIM 14100), neonatal adrenoleukodystrophy (NALD, MIM 0370) and infantile Refsum disease (IRD, MIM 66510), which represent a spectrum of disease severity with ZS being the most, and IRD the least severe disorder. ZSS disorders are autosomal recessive disorders and can be caused by a defect in any of at least 1 different PEX genes (Steinberg et al., 006). These PEX genes encode proteins called peroxins that are involved in various stages of peroxisomal protein import and/or the biogenesis of peroxisomes. Severe defects in one of these PEX genes result in a complete absence of functional peroxisomes as observed in the Zellweger Syndrome phenotype. The NALD or IRD phenotypes are often associated with residual peroxisomal functions and/or partial functional peroxisomes (i.e. peroxisomal mosaicism; Wanders and Waterham, 006; Weller et al., 003)). Many human PEX genes have been identified on the basis of sequence similarity with yeast PEX genes identified by functional complementation of peroxisomedeficient yeast mutants. At present more than 30 different yeast peroxins have been identified that are involved in different peroxisomal processes, including the formation of peroxisomal membranes, peroxisomal growth, fission and proliferation, and import of matrix proteins (Platta and Erdmann, 007). Of these yeast peroxins, 19 have been implicated in peroxisome protein import and biogenesis, whereas in humans, so far only 13 peroxins have been identified that are required for these processes. To study whether human peroxisome biogenesis requires the action of additional peroxins, we set out to assign primary skin fibroblasts from patients diagnosed with a ZSS disorder to different genetic complementation groups. Following the complementation analysis, we performed mutation analysis of the respective PEX genes in most of the cell lines. We here report an overview of our genetic complementation studies in more than 600 patient cell lines with a Zellweger syndrome spectrum disorder. In addition, we provide an overview of all mutations we identified in these patient cell lines, completed with previously reported mutations in the respective PEX genes. Methods Cell lines For this study, we used 613 independent primary skin fibroblast cell lines from non-related patients that had been sent to our laboratory since 1985 for diagnostic evaluation and which were diagnosed with a ZSS disorder. The diagnosis is based on metabolite analysis in plasma, which ideally involves analysis of very long chain fatty acids (VLCFAs), bile acid intermediates, phytanic acid, pristanic acid, pipecolic acid and plasmalogens, and/or detailed studies in fibroblasts, including 33
5 Chapter VLCFA analysis, C6:0 and pristanic acid β-oxidation, phytanic acid α-oxidation and dihydroxyacetonephosphate acyltransferase activity analysis, and the absence of peroxisomes assessed by catalase immunofluorescence (IF) microscopy (Wanders and Waterham, 005). The cell lines were cultured in DMEM medium (Gibco, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Bio-Whittaker), 100 U/ ml penicillin, 100 µg/ml streptomycin and 5 mm Hepes buffer with L-glutamine in a humidified atmosphere of 5% CO and at 37 C. Genetic complementation assays To assign cell lines to genetic complementation groups, we used a previously described polyethylene glycol (PEG)- mediated cell fusion assay (Brul et al., 1988) and a PEX cdna transfection assay, specifically developed for this purpose. In the latter assay, ZSS cell lines are co-transfected separately with the different PEX cdnas subcloned in the pcdna3 expression vector and an egfp-skl vector (Waterham et al., 007). The egfp-skl vector expresses egfp tagged with the peroxisomal targeting signal -SKL, which allows immediate visual detection of peroxisomes when present. The transfections were performed with the NHDF Nucleofector Kit (Amaxa, Cologne, Germany) and the Nucleofector Device (Amaxa, Cologne, Germany) using the standard program U-3, which in our hands results in high transfection efficiencies ranging from 70-80% for primary skin fibroblasts. We used the following transfection order of PEX expression vectors based on the estimated frequency of the different PEX gene defects: 1) PEX1, PEX6 and PEX1; ) PEX, PEX10 and PEX6; 3) PEX5, PEX13 and PEX14. Transfection with PEX3, PEX16 and PEX19 was performed when the patient fibroblasts show no peroxisomal membrane remnants based on immunofluorescence microscopy with antibodies against ALDP, a peroxisomal membrane protein. Cells were examined by means of fluorescence microscopy 48 to 7 hours after transfection to determine the subcellular localization of the peroxisomal reporter protein egfp-skl. Mutation analysis Mutation analysis was performed by either sequencing all exons plus flanking intronic sequences of the PEX genes amplified by PCR from genomic DNA or by sequencing PEX cdnas prepared from total RNA fractions. Genomic DNA was isolated from skin fibroblasts using the NucleoSpin Tissue genomic DNA purification kit (Macherey-nagel, Germany, Düren). Total RNA was isolated from skin fibroblasts using Trizol (Invitrogen, Carlsbad, CA) extraction, after which cdna was prepared using a first strand cdna synthesis kit for RT-PCR (Roche, Mannheim, Germany). All forward and reverse primers (sequences available on request) were tagged with a -1M13 (5- TGTAAAACGACGGCCAGT-3 ) sequence or M13rev (5 -CAGGAAACAGCTATGACC-3 ) sequence, respectively. PCR fragments were sequenced in two directions using -1M13 and M13rev primers by means of BigDye Terminator v1.1 Cycle Sequencing Kits (Applied Biosystems, Foster City, CA, USA) and analyzed on an Applied Biosystems 377A automated DNA sequencer, following the manufacturer s protocol (Applied Biosystems, Foster City, CA, USA). Mutation analysis of PEX1 (GenBank accession number NM_000466) was performed by sequencing the coding region of the PEX1 cdna and/ or exons amplified from gdna and by RFLP analysis to identify the frequently occurring p.g843d mutation. Mutations 34
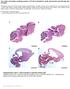
6 Genetic Classification and Mutational Spectrum of ZSS in the PEX gene were identified by sequencing exon 4 from gdna (NM_000318). The PEX3 (NM_003630) gene was analyzed by sequencing of the coding region of the PEX3 cdna (NM_003630). Mutation analysis of PEX5L (X84899), PEX10 (NM_00617), PEX1 (NM_00086), PEX13 (NM_00618), PEX14 (NM_004565), PEX16 (NM_004813), PEX19 (NM_00857) and PEX6 (NM_01799) were performed by sequencing of all exons plus flanking intronic sequences amplified from gdna. Sequences were compared to the indicated reference sequences with nucleotide numbering starting at the first adenine of the translation initiation codon ATG. Results and Discussion Genetic complementation Initially, we used a previously reported cell fusion complementation assay (Brul et al., 1988). Because this assay is rather laborious, we developed a more rapid and Figure 1. Complementation testing by PEG fusion and by PEX cdna transfection. Principle of the PEG fusion method. (A), a tester cell line with a known PEX gene defect and a patient cell line are fused by PEG treatment resulting in multi-nucleated cells. Cells are examined by catalase immunofluorescence microscopy to determine complementation (B) or non complementation (C). Principle of the PEX cdna transfection assay. The pcdna3-pex expression vectors are co-transfected separately but together with egfp-skl into patient cells (D). Cells are examined by direct fluorescent microscopy to determine complementation (E) or non complementation (F). 35
7 Chapter direct complementation assay taking advantage of the known identities of the 1 human PEX genes that can cause a ZSS disorder (for principle see Figure 1). The PEX cdna transfection assay worked very well for cell lines displaying complete peroxisome deficiencies, and for cell lines displaying different degrees of peroxisomal mosaicism that are sensitive to culturing at 40ºC, which exacerbates the defect in peroxisome biogenesis (Gootjes et al., 004d). We assigned 613 different ZSS cell lines to genetic complementation groups (Table 1). The results give a representative overview of the spectrum of PEX gene defects among ZSS disorders with PEX1 (MIM60136), PEX6 (MIM601498) and PEX1 (MIM601758) defects being the most common. Although we received ZSS cell lines from different parts of the world, they predominantly (>90%) are of Caucasian origin and thus the spectrum of PEX gene defects primarily relates to the Caucasian race. Table 1. Frequency distribution of PEX gene defects in ZSS. Classification of the 613 different patient cell lines was done by either genetic complementation (cdna transfection of PEG fusion) or by sequence analysis. PEX gene Genetic complementation (number of cell lines) Frequency (%) PEX PEX 3 4 PEX3 3 <1 PEX5 13 PEX PEX PEX PEX PEX14 3 <1 PEX PEX19 4 <1 PEX6 1 3 Somewhat unexpected, all the analyzed ZSS cell lines could be assigned to one of the known genetic complementation groups and no novel complementation group was identified. In a recent study, where 90 cell lines were assigned to genetic complementation groups using a similar approach, also no novel complementation groups were identified (Krause et al., 009). This suggests that, in contrast to yeast where 19 different PEX genes have been implicated, peroxisome protein import and biogenesis in humans would only require 1 PEX genes. However, it should be emphasized that to be suitable for genetic complementation testing, cell lines need to display a complete peroxisome deficiency in the majority of cells. It therefore remains possible that defects in other putative human PEX genes cannot be identified by this approach, because they don t result in peroxisome deficiency or perhaps lead to different clinical presentations, as recently exemplified by a very distinct peroxisomal and mitochondrial fission defect due to a dominant negative mutation in the DLP1 36
8 Genetic Classification and Mutational Spectrum of ZSS gene (Waterham et al., 007). In addition, defects in certain PEX genes may also be without consequences due to gene redundancy. For example, the yeast PEX4 gene, which is associated with complete peroxisome deficiency in yeast, appears to have 3 human ortholoques (Grou et al., 008). Considering that these ortholoques are encoded by different genes and each can promote the ubiquitination of PEX5 (MIM600414) on its own, it seems unlikely that a defect in one of these proteins will cause a peroxisomal disorder (Grou et al., 008). Mutation analysis After the assignment of patient cell lines to genetic complementation groups, we performed mutation analysis of the respective PEX genes in the majority of the cell lines. We here report the mutations we identified in the different PEX genes. The reported mutations are assumed to be pathogenic for the following reasons: 1) They are the only sequence variants identified in the coding regions or flanking intronic sequences of the different PEX genes, which were demonstrated to be defective by the fact that peroxisome biogenesis in the cell line is restored after transfection with the respective PEX cdna (i.e. functional assay). ) The sequence variants were not listed in the SNP database or encountered in more than 100 control chromosomes. It should be noted that in the vast majority of cell lines we identified either two heterozygous or one homozygous putative pathogenic mutation. In few cell lines we only found one heterozygous mutation after sequencing gdna, which in most cases appeared homozygously expressed when analyzing the respective PEX cdna, indicating that the second, unidentified mutation affects gene transcription or mrna stability. PEX1 is the most affected gene among patients with a ZSS disorder. It encodes a protein belonging to the AAA ATPase protein family (ATPases Associated with various cellular Activities) and contains two ATP-binding folds. Table lists all mutations identified in the cell lines we analyzed, completed with mutations reported in literature. Among these mutations a few common mutations have been identified (Maxwell et al., 005). The most common mutation in PEX1 is a missense mutation in the second ATP-binding domain (c.58g>a) leading to p.g843d. This mutation reduces the binding between PEX1 and PEX6 and is known for its temperature sensitivity, meaning that when fibroblasts of patients homozygous for G843D are cultured at 30 C, they regain the ability to import catalase and other peroxisomal matrix proteins, as well as the various peroxisomal metabolic functions (Imamura et al., 1998a; Maxwell et al., 005). Patients homozygous for the G843D mutation usually present with the milder IRD phenotype. The second most common mutation in PEX1 is c.079_098inst, which results in a frame shift predicted to cause a truncated PEX1 protein (Collins and Gould, 1999), although Northern blot analysis showed that this mutation results in low steady state PEX1 mrna levels (Maxwell et al., 005). Patients homozygous for this mutation are affected with the severe ZS phenotype. The c. 916delA mutation is the third most common mutation in PEX1; this mutation leads to a frame shift. When combined with previously reported PEX1 mutations, a total of 81 different mutations has now been identified in the PEX1 gene, of which 3 have not been reported previously. 37
9 Chapter Table. Mutations in the PEX1 gene. Number of patients Mutations identified in this study Homozygouzygous Hetero- Exon Nucleotide Amino acid Reference 1 c.t>c disruption startcodon 3 this study 1 c.3g>a disruption startcodon 1 this study 1 c.5g>a p.wx 1 this study 1 c.56_57deltg p.v19gfsx48 1 this study c.130-a>t - 1 this study i c.73+1g>a - 1 this study 3 c.74g>c p.v9l (a) 3 c.343_344insct p.d115afsx18 1 this study 4 c.434_448delinsgcaa p.v145_ 1 (b) Q150delinsGfsX4 5 c.547c>t p.r183x 1 (c) 5 c.569c>a p.s190x 4 this study 5 c.616c>t p.q06x 1 this study 5 c.781c>t p.q61x (d) 5 c delaa p.q61rfsx8 3 (c) 5 c.788_789delca p.t63fsx5 1 (b) 5 c.904delg p.a30qfsx3 (a,e) 5 c.911_91delct p.s304fsx3 1 (m) 5 c.1007t>c p.v336a 1 this study 5 c.1108_1109insa p.i370nfsx 1 (a) 5 c.1193delt p.i398tfsx7 1 this study i5 c.139+1g>t - 1 (c) 6 c.1340t>c p.l447s 1 this study 7 c.1411c>t p.q471x 1 this study 8 c.1501_150delct p.l501x 1 this study 8 c.1509_1510dupta p.v504x 1 this study 8 c.1579a>g p.t56a 1 this study 9 c.1663t>c p.s555p 1 this study i9 c g>t - 1 (b) 10 c.1706a>c p.h569p 1 this study 10 c.1713_1716deltcac p.h571fsx10 1 this study 10 c.1716_1717delca p.h57qfsx19 (c) 10 c.1777g>a p.g593r 1 (b) 11 c.184dela p.e615kfsx30 1 this study 11 c.1865_1866inscagtgtgga - (f) 11 c.1886_1887delgt p.c69x 1 this study 11 c.1891delg p.a631lfsx14 1 this study 11 c.1897c>t p.r633x (d) 1 r.1900_070del p.g634_h690del (g) 1 c.1957t>a p.w653r 1 this study 1 c.1960_1961dupcagtgtgga p.t651_w653dup 1 (f,h,i,j) 1 c.1976t>a p.v659d 1 this study 1 c.1991t>c p.l664p (d) 1 c.008c>a p.l670m 1 (b) 1 c.034_035delca p.h678qfsx15 1 (k) 1 c.071+1g>t - 5 (b) 38
10 Genetic Classification and Mutational Spectrum of ZSS Table. (continue) Mutation Number of patient identified in this study Exon Nucleotide Amino Acids Homozygouzygous Hetro- Reference 13 c.083_085delatg p.m659del (m) 13 c delgataa p.m695ifsx45 (k) 13 c.097_098inst p.i700fsx (a,b,f,k,l) 13 c.4c>t p.q74x this study 13 c.7_416del p.e743nfsxfs (f) 14 c.364g>a Deletion exon 14 1 this study 14 c.368c>t p.r790x 1 (e) 14 c.383c>t p.r795x 1 (l) 14 c.387t>c p.l796p 1 (m) 14 c.391_39deltc p.r798sfsx35 (a) 14 c.39c>g p.r798g 1 (e) 15 c.58g>a p.g843d 4 6 (b,e,f,k,l) 15 c.537_545delatgaagtta p.h846_ (k) instcatggt R849delinsLfsX bpinsex15 - (j) 16 c.614c>t p.r87x (f) 16 c.633_635 deltgt p.l879del 1 1 this study 16 c.636t>c p.l879s 3 this study 16 c.654c>g p.t885r 1 thid study 17 c.730dela p.l910fsx (b) 17 c.760dela p.a91lfsx40 1 (this study) 18 c.814_818delctttg p.f938lfsx (f) 18 c.845c>t p.r949w 1 this study 18 c.846g>a p.r949q (b) 18 c.916dela p.g973afsx (e,f,k) 18 c.96+1g>a - 1 (k) 18 c.96+_96+3inst - (l) 18 c.96+t>c - (b) 19 c.97-15t_308- Delition exon 19 and this study 34AdelinsATAGTATAGA & c _ exon 0 19 c.966t>c p.i989t (b) 19 c.99c>t p.r998x (e) 19 c.993g>a p.r998q (a) 19 c.30_304delcct p.p1008del (k) 0 c.3038g>a p.r1013h (m) 0 c.3180_3181inst p.g1061wfsx16 (l) 0 c.307+1g>c - (l) 1 c.387c>g p.s1096x (m) 1 - p.y116x (f) 3 c delagtc p.q131hfsx3 1 (c, m) 4 c.3850t>c p.x184qnextx9 1 (b) (a) Maxwell et al., 005 (f) Preuss et al., 00 (k) Steinberg et al., 004 (b) Walter et al., 001 (g) Tamura et al., 1998 (l) Collins and Gould, 1999 (c) Yik et al., 009 (h) Gartner et al., 1999 (m) Rosewich et al, 005 (d) Tamura et al., 001 (i) Portsteffen et al., 1997 (e) Maxwell et al., 00 (j) Reuber et al.,
11 Chapter The PEX (MIM ) gene was the first gene reported to be mutated in ZSS patients. PEX is an integral membrane protein with two transmembrane domains and a zinc-binding motif, probably involved in the poly-ubiquitination of the peroxisomal import receptor PEX5 (Platta et al., 009). The PEX gene contains four exons, but the entire coding sequence is included in exon 4. One mutation in the PEX gene (c.355c>t) was found to be associated with temperature sensitivity: cells cultured at 30 C show partial catalase import (Imamura et al., 1998b). The patient fibroblasts homozygous for the c.669g>a mutation displayed a mosaic peroxisomal pattern, characterized by import competency of catalase in 30% of the cells (Gootjes et al., 003; Gootjes et al., 004c). In this study, six novel mutations have been identified, making a total of 18 different mutations currently identified in the PEX gene (Table 3). Table 3. Mutations in the PEX gene. Mutations Number of patients identified in this study Exon Nucleotide Amino acid Homozygous Heterozygous Reference 4 c.115c>t p.q39x (b) 4 c.163 G>A p.e55k (a, c) 4 c.73delt p.n9tfsx (a) 4 c.79_83delgagat p.r94sfsx5 1 (d) 4 c.86c>t p.q96x 1 this study 4 c.35inst p.c109lfsx14 1 this study 4 c.339_345delcaggtgg p.r114x 1 1 this study 4 c.355c>t p.r119x 4 1 (a, c, e) 4 c.373c>t p.r15x (a, c, f) 4 c.550delc p.r184vfsx8 3 (g) 4 c.64delg p.k15sfsx 1 (g) 4 c.669g>a p.w3x (d) 4 c.739t>c p.c47r 1 (d) 4 c.78a>g p.h61r 1 this study 4 c.791g>a p.c64y 1 this study 4 c.834_838deltactt p.f78lfsx3 1 (b) 4 c.866insa p.s89fsx36 1 this study (a) Steinberg et al., 004 (d) Gootjes et al., 004a (g) Shimozawa et al., 000b (b) Krause et al., 006 (e) Shimozawa et al., 1993 (c) Imamura et al., 1998b (f) Shimozawa et al., 1998 PEX3 (MIM603164) is an integral membrane protein with two putative membranespanning domains. The exact function of PEX3 is unclear, but it is assumed that PEX3 plays a role in insertion of peroxisomal membrane proteins into the peroxisomal membrane based on the absence of peroxisomal ghosts in cell lines with a defect in PEX3 (Ghaedi et al., 000; Muntau et al., 000). In total, six different mutations in PEX3 were identified of which four have been reported previously (Table 4). 40
12 Genetic Classification and Mutational Spectrum of ZSS Table 4. Mutations in the PEX3 gene. Mutation Number of patients identified in this study Exon Nucleotide Amino acid Homozygous Heterozygous Reference c.157c>t p.r53x (a) 4 c.38_331delataa p.i110vfsx3 1 this study 5 r.334_393del p.f11_v131del 1 this study 7 c.543_544inst p.v18cfsx (a, b, c) 10 c.856c>t p.r86x 1 (d) 11 c.94-8t>g p.s314rfsx3 (b, e) (a) South et al., 000 (c) Shimozawa et al., 000a (e) Ghaedi et al., 000 (b) Muntau et al., 000 (d) Dursun et al., 009 PEX5 is a protein with seven di-aromatic pentapeptide repeats (WxxxF/Y) essential for docking to the peroxisomal membrane and for binding to either PEX13 (MIM601789) or PEX14 (MIM601791, (Saidowsky et al., 001; Weller et al., 003) and with seven tetrapeptide repeats (TPRs) to mediate the interaction with the PTS1 sequence. In humans, two functional protein variants of PEX5 are produced as a result of alternative splicing of the PEX5 mrna. The longest variant, PEX5L, contains an additional 111bp encoding 37 amino acids, due to alternative splicing of exon 7 (Dodt et al., 001). The shorter protein, PEX5S, has been reported to be exclusively involved in peroxisomal PTS1 protein import, whereas PEX5L mediates both PTS1 and PTS protein import. Recently, we published 11 mutations in the PEX5 gene (Ebberink et al., 009). These mutations combined with previously reported mutations are listed in table 5. Table 5. Mutations in the PEX5 gene. Mutations Exon Nucleotide Amino acid Number of patients identified in this study Homozygous Heterozygous (a) Ebberink et al., 009 (b) Dodt et al., 1995 (c ) Shimozawa et al., 1999b Reference 5 c.548_55-55delins38inv p.d150gfsx33 1 (a) 7 c.86c>t p.r76x 1 (a) 10 c.1090c>t p.q364x 1 (a) 11 [c.144a>g]+[c.548- p.n415s 1 (a) 549dupATCG; c.604g>c] i1 c g>a p.r394s, r.395_465del 1 (a) 1 c.158c>t p.r40x (a) 1 c.179c>t p.r47x (b) i14 c.1561-a>c r.1561_1718del, 1 (a) r.1561_1584del 14 c.1578t>g pn56k (a, b) 14 c.1669c>t p.r557w 1 (a) 15 c.1799c>g p.s600w 1 (a, c) 41
13 Chapter The PEX6 gene consists of 17 exons and mutations in the PEX6 gene were found to be scattered throughout the whole gene. PEX6 is also a member of the AAA ATPase family and interacts with PEX1. PEX6 is required for import of peroxisomal matrix proteins and/or vesicle fusion. At present, 77 different mutations in PEX6 gene have been identified, a complete overview of which has been reported very recently and thus will not been shown here (Ebberink et al., 010a). PEX10 (MIM60859) is an integral membrane protein and contains two transmembrane domains and a C-terminal zinc-binding motif, like PEX and PEX1. Two mrna splice forms of PEX10 have been identified, resulting from the use of a different splice acceptor site at the 3 end of intron 3. The longer form accounts for 10% of the PEX10 mrna in the cell and appears to be slightly less functional (Warren et al., 000). Combined with previously reported PEX10 mutations, 6 different mutations have been identified in the PEX10 gene of which nine have not been reported previously (Table 6). Table 6. Mutations in the PEX10 gene. Mutations Number of patients identified in this study Exon Nucleotide Amino acid Homozygous Heterozygous Reference 1 c.t>c disruption startcodon 1 (a) 1 c.3g>a disruption startcodon 1 this study 1 c.4delg p.apfsx9 (b) 1 c.13_8del, ins0bp p.a5pfsx46 (c) 3 c.03c>a p.t68n 1 this study 3 c.08g>c p.g70r 1 this study 3 c.11g>a p.e71k 1 this study 3 c.33a>g p.q78r 1 this study 3 c.337delc p.l113wfsx40 (d) 3 c.35c>t p.q118x 1 this study 3 c.373c>t p.r15x (e) i3 c.600+1g>a - 1 (e) 4 c.697delg p.a13pfsx131 1 this study 4 c.704_705insa p.l36afsx10 (b, c) 4 c.646_647insa p.s16kfsx13 1 this study 4 c.730c>t p.r44x 1 (b, f) 5 c.814_815delct p.l7vfsx (g) 5 c.830t>c p.l77p (d) 5 c.835g>t p.e79x 1 (b) 5 c.868c>g p.h90d (b) 5 c.870c>g p.h90q (e) 5 c.881g>a p.w94x (f) 5 c.89g>t p.c301f (b) 6 c.919t>c p.c307r 1 this study 6 c.93g>a p.r311q 1 (a) (a) Regal et al., 010 (d) Steinberg et al., 009 (g) Okumoto et al., 1998a (b) Steinberg et al., 004 (e) Warren et al., 1998 (c) Warren et al., 000 (f) Krause et al., 006 4
14 Genetic Classification and Mutational Spectrum of ZSS Mutations in the PEX1 gene are the third common cause of a ZSS disorder: 9% of our patient group has a mutation in PEX1. PEX1 is one of the RING finger proteins; it contains two transmembrane domains and a C-terminal zinc-binding motif. All 34 identified PEX1 mutations are listed in table 7, including 8 that have not been reported before. The ps30f mutation (c.959c>t) is the most common PEX1 mutation and is located in the zinc-binding domain. Patients homozygous Table 7. Mutations in the PEX1 gene. Mutations Number of patients identified in this study Exon Nucleotide Amino acid Homozygous Heterozygous Reference 1 c.16dupg p.a6gfsx11 1 this study 1 c.6_7delca p.t9sfsx7 (a) 1 c.10a>t p.r34s 1 (b) 1 c.16+1g>t (a) c.0_04delctt p.l68del (a) c.0t>g p.y74d 1 this study c.60_61insaa p.y87x (c) c.68_71delaaga p.k90efsx3 (a) c.73a>t p.r91s 1 (d) c.308_309inst p.l103ffsx3 1 (d) c.378_380delttc p.l16del 1 this study c.445_454deltcttcccgct p.s149gfsx15 1 this study c.460c>t p.r154x 1 this study c.478_479delca p.y156x 1 this study c.531_533delaca p.q178del (e) c.533_535delaac p.q178del 1 1 (c) c.538c>t p.r180x (f, g) c.541_54inst p.y181lfsx37 (c) c.604c>t p.r0x 3 (d) c.65c>t p.q09x (d) i c.681-a>c - (c) 3 c.684_687deltagt p.srfsx3 (a) 3 c.691a>t p.k31x (h) 3 c.733_734dupgcct p.l45cfsx19 1 (c, f) 3 c.744_745inst p.t49yfsx14 1 (a) 3 c.775c>t p.q59x 1 this study 3 c.875_876delct p.s9x 1 (g) 3 c.887_888deltc p.l96fs3007x 1 1 (c, d, f) 3 c.887delt p.l96pfsx5 (c) 3 c.888_889delct p.l97tfsx1 1 3 (a, c) 3 c.90g>a p.c307y 1 this study 3 c.949c>t p.l317f 1 (d) 3 c.959c>t p.s30f 13 (c) 3 c.1047_1049delaca p.q349del (c) (a) Chang and Gould, 1998 (d) Gootjes et al., 004c (g) Okumoto et al., 1998b (b) Zeharia et al., 007 (e) Yik et al., 009 (h) Okumoto and Fujiki, 1997 (c) Steinberg et al., 004 (f) Chang et al.,
15 Chapter for this mutation presented an atypical biochemical phenotype in skin fibroblasts: the peroxisomal parameters are mostly normal to slightly abnormal. The mutation causes a temperature-sensitive phenotype: when cultured at 30 C, all cells contain catalase import competent peroxisomes, when cultured at 37 C only, 70% of the cells contain catalase positive peroxisomes and when cultured at 40 C none of the cells contain catalase positive peroxisomes (Gootjes et al., 004a). Table 8. Mutations in the PEX13 gene. Mutations Number of patients identified in this study Exon Nucleotide Amino acid Homozygous Heterozygous Reference i1 c.9+t>g - 1 this study c.107_10del p.g36dfsx61 (a) c.439_441delatg p.m147del 1 this study c.676c>t p.r6x this study c.70g>a p.w34x 1 (b) 4 c.937t>g p.w313g (c) 4 c.970g>t p.g34x (b) 4 c.977t>c p.i36t 1 (b, d) 4 c.980c>g p.p37r 1 this study 1-4 c.1_ (a) 1 Deletion of 147,308 bp that included the PEX13 gene and 70,094 bp upstream and 45,69 bp downstream of the PEX13 gene (a) Al-Dirbashi et al., 009 (c) Krause et al., 006 (b) Shimozawa et al., 1999a (d) Liu et al., 1999 PEX13 is a peroxisomal integral membrane protein with two putative membrane domains. The C-terminus contains an SH3 domain, which binds to pentapeptide repeats -4 of PEX5. The N-terminal part of yeast PEX13 interacts with PEX7. Table 8 lists ten different PEX13 mutations of which six have been reported before. The I36T mutation causes a temperature-sensitive biochemical phenotype in fibroblasts: peroxisomes still import residual levels of PTS1 and PTS proteins at 37 C but not at elevated temperature. (Liu et al., 1999; Shimozawa et al., 1999b). Table 9. Mutations in the PEX14 gene. Mutaties Exon Nucleotide Amino acid Number of patients identified in this study Homozygous Heterozygous Reference 3 c.85-?_170+?del p.[ile9_lys56del;gly57glyfsx] (a) 7 c.553c>t p.gln185ter (b) 7 c.585+1g>t r.585+1_37ins 1 this study (a) Huybrechts et al., 008 (b) Shimozawa et al.,
16 Genetic Classification and Mutational Spectrum of ZSS The PEX14 gene encodes a peroxisomal membrane protein that was initially identified as a peroxisomal docking factor for the PTS1 receptor PEX5 via binding to the amino-terminal WxxxF/Y motifs of PEX5. More recently, it was proposed that PEX14 is also the site from which PEX5 leaves the peroxisomal compartment (Azevedo and Schliebs, 006). A mutation in PEX14 is the least common cause of ZZS disorder. Three different mutations have now been identified in the PEX14 gene of which 1 has not been reported previously (Table 9). The PEX16 (MIM603360) gene encodes an integral peroxisomal membrane protein involved in peroxisomal membrane assembly. Two different groups of PEX16- defective patients have been reported; patients with a severe clinical presentation of whom the fibroblasts displayed a defect in import of peroxisomal matrix and membrane proteins, resulting in a total absence of peroxisomal remnants (Honsho et al., 1998; Shimozawa et al., 00) and, very recently, several patients with a relatively mild clinical phenotype of the fibroblasts showed enlarged, import-competent peroxisomes (Ebberink et al, 010b). In total eight different mutations have been identified in the PEX16 gene of which one has not been reported previously (Table 10). Table 10. Mutations in the PEX16 gene. Mutations Number of patients identified in this study Exon Nucleotide Amino acid Homozygous Heterozygous Reference i5 c.460+ insg - 1 this study 6 c.56c>t p.r176x (a, b) 8 c.753_755deltgt p.v5del 1 (c) 9 c.865c>a p.p89t 1 (c) i11 c _ (c) i10 c.95+t>c R98fsX38 (d) 11 c.984delg p.i330sfsx7 1 (c) 11 c.99a>g p.y331c 1 (c) (a) Honsho et al., 1998 (c) Ebberink et al., 010b (d) Shimozawa et al., 00 (b) South and Gould, 1999 The PEX19 (MIM60079) gene encodes a mainly cytosolic protein that binds many peroxisomal membrane proteins, suggesting that PEX19 functions as a peroxisomal receptor for PMP (Sacksteder et al., 000). The patients with a defect in PEX19 presented with a severe ZS phenotype and the fibroblasts displayed the absence of peroxisomal ghosts (Matsuzono et al., 1999). Table 11 lists three different PEX19 mutations of which two have not been reported before. PEX6 (MIM608666) is a membrane protein with one putative membrane-spanning domain. Human PEX6 is the ortholoque of yeast PEX15. Some of the reported mutations were found to lead to temperature-sensitive biochemical phenotype (Matsumoto et al., 003a; Matsumoto et al., 003b). Combined with previously reported PEX6 mutations, different mutations have now been identified in the PEX6 gene of which three have not been reported previously (Table 1). 45
17 Chapter Table 11. Mutations in the PEX19 gene. Mutations Number of patients identified in this study Exon Nucleotide Amino acid Homozygous Heterozygous Reference 3 c.30dela p.k30fsx13 1 this study 6 c.739c>t p.q57x 1 this study 6 c.763_764insa p.m55nfsx4 1 (a) (a) Matsuzono et al., 1999 Table 1. Mutations in the PEX6 gene. Mutations Number of patients identified in this study Exon Nucleotide Amino acid Homozygous Heterozygous Reference c.t>c disruption startcodon (a, b) c.34_35insc p.l1pfsx103 (a) c.37_38delag p.r13gfsx164 (c) c.73_79delgtgcgcg p.v5rfsx55 1 this study c.131t>c p.l44p 1 (b) c.134t>c p.l45p (a, b) c.185g>a p.w6x 1 this study c.19_16del p.s64rfsx10 (c) i c.30+1g>t p.t77fsx139 (b) 3 c.54_55inst p.l85lfsx93 (a, b) 3 c.65g>a p.g89r (a) 3 c.9c>t p.r98w 10 (a, b, c) 3 c.96g>a p.w99x (c) 3 c.315g>a p.w105x 1 this study 3 c.350c>t p.p117l (b) 3 c.353c>g p.p118r (c) i3 c.371+t>c - (c) 4 c.46_548dup1bpinst p.a143_v18dup (b) +G183V 4 c.574c>t p.r19x 1 (c) i4 c.667+t>c - (c) 5 r.668_814 p.g3_p71del (a) 6 c.86delc p.r88afsx366 (b) (a) Matsumoto et al., 003b (b) Weller et al., 005 (c) Steinberg et al., 004 Conclusions In summary, we developed an efficient genetic complementation assay, which allows for rapid identification of the defective PEX gene in fibroblast cell lines of ZSS patients. This assay has been included in our diagnostic work up of patients diagnosed with a ZSS disorder, followed by sequence analysis of the defective PEX gene. After analysis of 613 independent cell lines, we did not detect cell lines with a novel defective PEX gene suggesting that only mutations in one of the currently known PEX genes leads to complete peroxisome deficiency and a Zellweger Syndrome-like presentation. 46
18 Genetic Classification and Mutational Spectrum of ZSS Acknowledgements We thank to Dr. S.J. Gould for the gift of PEX pcdna3 plasmids, J. Haasjes, W. Oostheim, M, de Gijsel and R.A. Jibodh for their assistance in different parts of this study and Dr. S. Ferdinandusse for critical reading of the manuscript. This study was supported by a grant from the Prinses Beatrix Fonds (MAR 03_016) and by the FP6 European Union Project peroxisomes (LSHG-CT ). We gratefully acknowledge all doctors and medical specialists who referred patient material to our laboratory. References Al-Dirbashi OY, Shaheen R, Al-Sayed M, Al-Dosari M, Makhseed N, Abu SL, Santa T, Meyer BF, Shimozawa N, Alkuraya FS Zellweger syndrome caused by PEX13 deficiency: report of two novel mutations. Am J Med Genet A 149A: Azevedo JE, Schliebs W Pex14p, more than just a docking protein. Biochim Biophys Acta 1763: Braverman N, Chen L, Lin P, Obie C, Steel G, Douglas P, Chakraborty PK, Clarke JT, Boneh A, Moser A, Moser H, Valle D. 00. Mutation analysis of PEX7 in 60 probands with rhizomelic chondrodysplasia punctata and functional correlations of genotype with phenotype. Hum Mutat 0: Brul S, Westerveld A, Strijland A, Wanders RJ, Schram AW, Heymans HS, Schutgens RB, van den Bosch H, Tager JM Genetic heterogeneity in the cerebrohepatorenal (Zellweger) syndrome and other inherited disorders with a generalized impairment of peroxisomal functions. A study using complementation analysis. J Clin Invest 81: Chang CC, Gould SJ Phenotype-genotype relationships in complementation group 3 of the peroxisome-biogenesis disorders. Am J Hum Genet 63: Chang CC, Warren DS, Sacksteder KA, Gould SJ PEX1 interacts with PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J Cell Biol 147: Collins CS, Gould SJ Identification of a common PEX1 mutation in Zellweger syndrome. Hum Mutat 14: Dodt G, Braverman N, Wong C, Moser A, Moser HW, Watkins P, Valle D, Gould SJ Mutations in the PTS1 receptor gene, PXR1, define complementation group of the peroxisome biogenesis disorders. Nat Genet 9: Dodt G, Warren D, Becker E, Rehling P, Gould SJ Domain mapping of human PEX5 reveals functional and structural similarities to Saccharomyces cerevisiae Pex18p and Pex1p. J Biol Chem 76: Dursun A, Gucer S, Ebberink MS, Yigit S, Wanders RJ, Waterham HR Zellweger syndrome with unusual findings: non-immune hydrops fetalis, dermal erythropoiesis and hypoplastic toe nails. J Inherit Metab Dis. Ebberink MS, Koster J, Wanders RJ, Waterham HR. 010a. Spectrum of PEX6 mutations in Zellweger syndrome spectrum patients. Hum Mutat 31:E Ebberink MS, Mooyer PA, Koster J, Dekker CJ, Eyskens FJ, Dionisi-Vici C, Clayton PT, Barth PG, Wanders RJ, Waterham HR Genotype-phenotype correlation in PEX5-deficient peroxisome biogenesis defective cell lines. Hum Mutat 30: Ebberink MS, Csanyi B, Chong WK, Denis S, Sharp P, Mooijer PAW, Dekker CJM, Spooner C, Ngu LH, De Sousa C, Wanders RJA, Fietz MJ, Clayton PT, Waterham HR, Ferdinandusse S. 010b. Identification of an unusual variant peroxisome biogenesis disorder caused by mutations in the PEX16 gene. J Med Genet. (accepted) Gartner J, Preuss N, Brosius U, Biermanns M Mutations in PEX1 in peroxisome biogenesis disorders: G843D and a mild clinical phenotype. J Inherit Metab Dis : Ghaedi K, Honsho M, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y PEX3 is the causal gene responsible for peroxisome membrane assembly-defective Zellweger syndrome of complementation group G. Am J Hum Genet 67: Gootjes J, Mandel H, Mooijer PA, Roels F, Waterham HR, Wanders RJ Resolution of the molecular 47
19 Chapter defect in a patient with peroxisomal mosaicism in the liver. Adv Exp Med Biol 544: Gootjes J, Schmohl F, Waterham HR, Wanders RJ. 004a. Novel mutations in the PEX1 gene of patients with a peroxisome biogenesis disorder. Eur J Hum Genet 1: Gootjes J, Skovby F, Christensen E, Wanders RJ, Ferdinandusse S. 004b. Reinvestigation of trihydroxycholestanoic acidemia reveals a peroxisome biogenesis disorder. Neurology 6: Gootjes J, Elpeleg O, Eyskens F, Mandel H, Mitanchez D, Shimozawa N, Suzuki Y, Waterham HR, Wanders RJ. 004c. Novel mutations in the PEX gene of four unrelated patients with a peroxisome biogenesis disorder. Pediatr Res 55: Gootjes J, Schmohl F, Mooijer PA, Dekker C, Mandel H, Topcu M, Huemer M, Von Schutz M, Marquardt T, Smeitink JA, Waterham HR, Wanders RJ. 004d. Identification of the molecular defect in patients with peroxisomal mosaicism using a novel method involving culturing of cells at 40 degrees C: implications for other inborn errors of metabolism. Hum Mutat 4: Grou CP, Carvalho AF, Pinto MP, Wiese S, Piechura H, Meyer HE, Warscheid B, Sa-Miranda C, Azevedo JE Members of the ED (UbcH5) Family Mediate the Ubiquitination of the Conserved Cysteine of Pex5p, the Peroxisomal Import Receptor. J Biol Chem 83: Honsho M, Tamura S, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y Mutation in PEX16 is causal in the peroxisome-deficient Zellweger syndrome of complementation group D. Am J Hum Genet 63: Huybrechts SJ, Van Veldhoven PP, Hoffman I, Zeevaert R, de Vos R, Demaerel P, Brams M, Jaeken J, Fransen M, Cassiman D Identification of a novel PEX14 mutation in Zellweger syndrome. J Med Genet 45: Imamura A, Tamura S, Shimozawa N, Suzuki Y, Zhang Z, Tsukamoto T, Orii T, Kondo N, Osumi T, Fujiki Y. 1998a. Temperature-sensitive mutation in PEX1 moderates the phenotypes of peroxisome deficiency disorders. Hum Mol Genet 7: Imamura A, Tsukamoto T, Shimozawa N, Suzuki Y, Zhang Z, Imanaka T, Fujiki Y, Orii T, Kondo N, Osumi T. 1998b. Temperature-sensitive phenotypes of peroxisome-assembly processes represent the milder forms of human peroxisome-biogenesis disorders. Am J Hum Genet 6: Krause C, Rosewich H, Thanos M, Gartner J Identification of novel mutations in PEX, PEX6, PEX10, PEX1, and PEX13 in Zellweger spectrum patients. Hum Mutat 7:1157. Krause C, Rosewich H, Gartner J Rational diagnostic strategy for Zellweger syndrome spectrum patients. Eur J Hum Genet. Liu Y, Bjorkman J, Urquhart A, Wanders RJ, Crane DI, Gould SJ PEX13 is mutated in complementation group 13 of the peroxisome-biogenesis disorders. Am J Hum Genet 65: Matsumoto N, Tamura S, Fujiki Y. 003a. The pathogenic peroxin Pex6p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. Nat Cell Biol 5: Matsumoto N, Tamura S, Furuki S, Miyata N, Moser A, Shimozawa N, Moser HW, Suzuki Y, Kondo N, Fujiki Y. 003b. Mutations in novel peroxin gene PEX6 that cause peroxisome-biogenesis disorders of complementation group 8 provide a genotype-phenotype correlation. Am J Hum Genet 73: Matsuzono Y, Kinoshita N, Tamura S, Shimozawa N, Hamasaki M, Ghaedi K, Wanders RJ, Suzuki Y, Kondo N, Fujiki Y Human PEX19: cdna cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc Natl Acad Sci U S A 96: Maxwell MA, Allen T, Solly PB, Svingen T, Paton BC, Crane DI. 00. Novel PEX1 mutations and genotype-phenotype correlations in Australasian peroxisome biogenesis disorder patients. Hum Mutat 0: Maxwell MA, Leane PB, Paton BC, Crane DI Novel PEX1 coding mutations and 5 UTR regulatory polymorphisms. Hum Mutat 6:79. Motley AM, Brites P, Gerez L, Hogenhout E, Haasjes J, Benne R, Tabak HF, Wanders RJ, Waterham HR. 00. Mutational spectrum in the PEX7 gene and functional analysis of mutant alleles in 78 patients with rhizomelic chondrodysplasia punctata type 1. Am J Hum Genet 70: Muntau AC, Mayerhofer PU, Paton BC, Kammerer S, Roscher AA Defective peroxisome membrane synthesis due to mutations in human PEX3 causes Zellweger syndrome, complementation group G. Am J Hum Genet 67: Okumoto K, Fujiki Y PEX1 encodes an integral membrane protein of peroxisomes. Nat Genet 17:
20 Genetic Classification and Mutational Spectrum of ZSS Okumoto K, Itoh R, Shimozawa N, Suzuki Y, Tamura S, Kondo N, Fujiki Y. 1998a. Mutations in PEX10 is the cause of Zellweger peroxisome deficiency syndrome of complementation group B. Hum Mol Genet 7: Okumoto K, Shimozawa N, Kawai A, Tamura S, Tsukamoto T, Osumi T, Moser H, Wanders RJ, Suzuki Y, Kondo N, Fujiki Y. 1998b. PEX1, the pathogenic gene of group III Zellweger syndrome: cdna cloning by functional complementation on a CHO cell mutant, patient analysis, and characterization of PEX1p. Mol Cell Biol 18: Platta HW, Erdmann R The peroxisomal protein import machinery. FEBS Lett 581: Platta HW, El MF, Baumer BE, Schlee D, Girzalsky W, Erdmann R Pex and pex1 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol 9: Portsteffen H, Beyer A, Becker E, Epplen C, Pawlak A, Kunau WH, Dodt G Human PEX1 is mutated in complementation group 1 of the peroxisome biogenesis disorders. Nat Genet 17: Preuss N, Brosius U, Biermanns M, Muntau AC, Conzelmann E, Gartner J. 00. PEX1 mutations in complementation group 1 of Zellweger spectrum patients correlate with severity of disease. Pediatr Res 51: Régal L, Ebberink MS, Goemans N, Wanders RJA, De Meirleir L, Jaeken J, Schrooten M, Van Coster R, Waterham HR Mutations in PEX10 are a cause of autosomal recessive ataxia. Ann Neurol. (accepted) Reuber BE, Germain-Lee E, Collins CS, Morrell JC, Ameritunga R, Moser HW, Valle D, Gould SJ Mutations in PEX1 are the most common cause of peroxisome biogenesis disorders. Nat Genet 17: Rosewich H, Ohlenbusch A, Gärtner J Genetic and clinical aspects of Zellweger spectrum patients with PEX1 mutations. J Med Genet. 005 Sep;4(9):e58. Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol 148: Saidowsky J, Dodt G, Kirchberg K, Wegner A, Nastainczyk W, Kunau WH, Schliebs W The diaromatic pentapeptide repeats of the human peroxisome import receptor PEX5 are separate high affinity binding sites for the peroxisomal membrane protein PEX14. J Biol Chem 76: Shimozawa N, Suzuki Y, Orii T, Moser A, Moser HW, Wanders RJ Standardization of complementation grouping of peroxisome-deficient disorders and the second Zellweger patient with peroxisomal assembly factor-1 (PAF-1) defect. Am J Hum Genet 5: Shimozawa N, Suzuki Y, Tomatsu S, Nakamura H, Kono T, Takada H, Tsukamoto T, Fujiki Y, Orii T, Kondo N A novel mutation, R15X in peroxisome assembly factor-1 responsible for Zellweger syndrome. Hum Mutat Suppl 1:S134-S136. Shimozawa N, Zhang Z, Suzuki Y, Imamura A, Tsukamoto T, Osumi T, Fujiki Y, Orii T, Barth PG, Wanders RJ, Kondo N. 1999a. Functional heterogeneity of C-terminal peroxisome targeting signal 1 in PEX5- defective patients. Biochem Biophys Res Commun 6: Shimozawa N, Suzuki Y, Zhang Z, Imamura A, Toyama R, Mukai S, Fujiki Y, Tsukamoto T, Osumi T, Orii T, Wanders RJ, Kondo N. 1999b. Nonsense and temperature-sensitive mutations in PEX13 are the cause of complementation group H of peroxisome biogenesis disorders. Hum Mol Genet 8: Shimozawa N, Suzuki Y, Zhang Z, Imamura A, Ghaedi K, Fujiki Y, Kondo N. 000a. Identification of PEX3 as the gene mutated in a Zellweger syndrome patient lacking peroxisomal remnant structures. Hum Mol Genet 9: Shimozawa N, Zhang Z, Imamura A, Suzuki Y, Fujiki Y, Tsukamoto T, Osumi T, Aubourg P, Wanders RJ, Kondo N. 000b. Molecular mechanism of detectable catalase-containing particles, peroxisomes, in fibroblasts from a PEX-defective patient. Biochem Biophys Res Commun 68: Shimozawa N, Nagase T, Takemoto Y, Suzuki Y, Fujiki Y, Wanders RJ, Kondo N. 00. A novel aberrant splicing mutation of the PEX16 gene in two patients with Zellweger syndrome. Biochem Biophys Res Commun 9: Shimozawa N, Tsukamoto T, Nagase T, Takemoto Y, Koyama N, Suzuki Y, Komori M, Osumi T, Jeannette G, Wanders RJ, Kondo N Identification of a new complementation group of the peroxisome biogenesis disorders and PEX14 as the mutated gene. Hum Mutat 3: South ST, Gould SJ Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol 144: South ST, Sacksteder KA, Li X, Liu Y, Gould SJ Inhibitors of COPI and COPII do not block PEX3-49
21 Chapter mediated peroxisome synthesis. J Cell Biol 149: Steinberg S, Chen L, Wei L, Moser A, Moser H, Cutting G, Braverman N The PEX Gene Screen: molecular diagnosis of peroxisome biogenesis disorders in the Zellweger syndrome spectrum. Mol Genet Metab 83:5-63. Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW Peroxisome biogenesis disorders. Biochim Biophys Acta 1763: Steinberg SJ, Snowden A, Braverman NE, Chen L, Watkins PA, Clayton PT, Setchell KD, Heubi JE, Raymond GV, Moser AB, Moser HW A PEX10 defect in a patient with no detectable defect in peroxisome assembly or metabolism in cultured fibroblasts. J Inherit Metab Dis 3: Tamura S, Okumoto K, Toyama R, Shimozawa N, Tsukamoto T, Suzuki Y, Osumi T, Kondo N, Fujiki Y Human PEX1 cloned by functional complementation on a CHO cell mutant is responsible for peroxisome-deficient Zellweger syndrome of complementation group I. Proc Natl Acad Sci U S A 95: Tamura S, Matsumoto N, Imamura A, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y Phenotypegenotype relationships in peroxisome biogenesis disorders of PEX1-defective complementation group 1 are defined by Pex1p-Pex6p interaction. Biochem J 357: Walter C, Gootjes J, Mooijer PA, Portsteffen H, Klein C, Waterham HR, Barth PG, Epplen JT, Kunau WH, Wanders RJ, Dodt G Disorders of peroxisome biogenesis due to mutations in PEX1: phenotypes and PEX1 protein levels. Am J Hum Genet 69: Wanders RJ, Waterham HR Peroxisomal disorders I: biochemistry and genetics of peroxisome biogenesis disorders. Clin Genet 67: Wanders RJ, Waterham HR Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem 75: Warren DS, Morrell JC, Moser HW, Valle D, Gould SJ Identification of PEX10, the gene defective in complementation group 7 of the peroxisome-biogenesis disorders. Am J Hum Genet 63: Warren DS, Wolfe BD, Gould SJ Phenotype-genotype relationships in PEX10-deficient peroxisome biogenesis disorder patients. Hum Mutat 15: Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med 356: Weller S, Gould SJ, Valle D Peroxisome biogenesis disorders. Annu Rev Genomics Hum Genet 4: Weller S, Cajigas I, Morrell J, Obie C, Steel G, Gould SJ, Valle D Alternative splicing suggests extended function of PEX6 in peroxisome biogenesis. Am J Hum Genet 76: Yik WY, Steinberg SJ, Moser AB, Moser HW, Hacia JG Identification of novel mutations and sequence variation in the Zellweger syndrome spectrum of peroxisome biogenesis disorders. Hum Mutat 30:E467-E480. Zeharia A, Ebberink MS, Wanders RJ, Waterham HR, Gutman A, Nissenkorn A, Korman SH A novel PEX1 mutation identified as the cause of a peroxisomal biogenesis disorder with mild clinical phenotype, mild biochemical abnormalities in fibroblasts and a mosaic catalase immunofluorescence pattern, even at 40 degrees C. J Hum Genet 5:
P eroxisomes are ubiquitous components of eukaryotic. Genetic and clinical aspects of Zellweger spectrum patients ONLINE MUTATION REPORT
 1of6 ONLINE MUTATION REPORT Genetic and clinical aspects of Zellweger spectrum patients with PEX1 mutations H Rosewich, A Ohlenbusch, J Gärtner... J Med Genet 2005;42:e58 (http:///cgi/content/full/42/9/e58).
1of6 ONLINE MUTATION REPORT Genetic and clinical aspects of Zellweger spectrum patients with PEX1 mutations H Rosewich, A Ohlenbusch, J Gärtner... J Med Genet 2005;42:e58 (http:///cgi/content/full/42/9/e58).
Citation for published version (APA): Klouwer, F. C. C. (2018). Zellweger spectrum disorders: From bench to bedside
 UvA-DARE (Digital Academic Repository) Zellweger spectrum disorders Klouwer, F.C.C. Link to publication Citation for published version (APA): Klouwer, F. C. C. (2018). Zellweger spectrum disorders: From
UvA-DARE (Digital Academic Repository) Zellweger spectrum disorders Klouwer, F.C.C. Link to publication Citation for published version (APA): Klouwer, F. C. C. (2018). Zellweger spectrum disorders: From
The Role of Organic Acids in the Diagnosis of Peroxisomal Biogenesis Disorders
 The Role of Organic Acids in the Diagnosis of Peroxisomal Biogenesis Disorders Catherine Dibden Northern General Hospital Sheffield Children s Hospital Peroxisomes Small sub-cellular organelles Present
The Role of Organic Acids in the Diagnosis of Peroxisomal Biogenesis Disorders Catherine Dibden Northern General Hospital Sheffield Children s Hospital Peroxisomes Small sub-cellular organelles Present
MRI as diagnostic tool in early-onset peroxisomal disorders
 MRI as diagnostic tool in early-onset peroxisomal disorders M.S. van der Knaap, MD, PhD E. Wassmer, MD N.I. Wolf, MD, PhD P. Ferreira, MD M. Topçu, MD R.J.A. Wanders, PhD H.R. Waterham, PhD S. Ferdinandusse,
MRI as diagnostic tool in early-onset peroxisomal disorders M.S. van der Knaap, MD, PhD E. Wassmer, MD N.I. Wolf, MD, PhD P. Ferreira, MD M. Topçu, MD R.J.A. Wanders, PhD H.R. Waterham, PhD S. Ferdinandusse,
Identification of an unusual variant peroxisome biogenesis disorder caused by mutations in the PEX16 gene
 Identification of an unusual variant peroxisome biogenesis disorder caused by mutations in the PEX16 gene Merel Ebberink, Barbara Csanyi, Simone Denis, Peter Sharp, Petra Mooijer, Conny Dekker, Peter Clayton,
Identification of an unusual variant peroxisome biogenesis disorder caused by mutations in the PEX16 gene Merel Ebberink, Barbara Csanyi, Simone Denis, Peter Sharp, Petra Mooijer, Conny Dekker, Peter Clayton,
Metabolic control of peroxisome abundance
 Journal of Cell Science 112, 1579-1590 (1999) Printed in Great Britain The Company of Biologists Limited 1999 JCS0105 1579 Metabolic control of peroxisome abundance Chia-Che Chang 1, Sarah South 1, Dan
Journal of Cell Science 112, 1579-1590 (1999) Printed in Great Britain The Company of Biologists Limited 1999 JCS0105 1579 Metabolic control of peroxisome abundance Chia-Che Chang 1, Sarah South 1, Dan
Arginine improves peroxisome functioning in cells from patients with a mild peroxisome biogenesis disorder
 Berendse et al. Orphanet Journal of Rare Diseases 2013, 8:138 RESEARCH Open Access Arginine improves peroxisome functioning in cells from patients with a mild peroxisome biogenesis disorder Kevin Berendse
Berendse et al. Orphanet Journal of Rare Diseases 2013, 8:138 RESEARCH Open Access Arginine improves peroxisome functioning in cells from patients with a mild peroxisome biogenesis disorder Kevin Berendse
Molecular and genetic characterization of peroxisome biogenesis disorders Ebberink, M.S.
 UvA-DARE (Digital Academic Repository) Molecular and genetic characterization of peroxisome biogenesis disorders Ebberink, M.S. Link to publication Citation for published version (APA): Ebberink, M. S.
UvA-DARE (Digital Academic Repository) Molecular and genetic characterization of peroxisome biogenesis disorders Ebberink, M.S. Link to publication Citation for published version (APA): Ebberink, M. S.
Supplementary Figure 1
 Supplementary Figure 1 A B HEC1A PTEN +/+ HEC1A PTEN -/- 16 HEC1A PTEN -/- 22 PTEN RAD51 % Cells with RAD51 foci 40 -IR +IR 30 20 10 * *! TUBULIN 0 HEC1A PTEN+/+ HEC1A PTEN-/- 16 HEC1A PTEN-/- 22 C D 1.0
Supplementary Figure 1 A B HEC1A PTEN +/+ HEC1A PTEN -/- 16 HEC1A PTEN -/- 22 PTEN RAD51 % Cells with RAD51 foci 40 -IR +IR 30 20 10 * *! TUBULIN 0 HEC1A PTEN+/+ HEC1A PTEN-/- 16 HEC1A PTEN-/- 22 C D 1.0
Supplementary Table 2. Identified causative mutations and/or mutation candidates.
 Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Supplementary Table 2. Identified causative mutations and/or mutation candidates. Nonsense mutations base change aa change Average depth Result of next generation in 432 patient Hereditary form of the
Metabolomics to study functional consequences in peroxisomal disorders Herzog, K.
 UvA-DARE (Digital Academic Repository) Metabolomics to study functional consequences in peroxisomal disorders Herzog, K. Link to publication Citation for published version (APA): Herzog, K. (2017). Metabolomics
UvA-DARE (Digital Academic Repository) Metabolomics to study functional consequences in peroxisomal disorders Herzog, K. Link to publication Citation for published version (APA): Herzog, K. (2017). Metabolomics
UW359 Ovary 3c 3 Serous Recurrent 68 BRCA1 816delGT BRCA1 del exon 1-2. UW417 Ovary 3c 3 Serous Primary 38 BRCA1 1675delA
 Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
Supplementary Table 1. Cases with deleterious germline mutations, somatic HR mutations, and somatic PTEN mutations. ID Site Stage Grade Histology Tumor Age Germline mutation(s) a Somatic HR mutation(s)
UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy. Link to publication
 UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy Link to publication Citation for published version (APA): Franken, R. (2016). Marfan syndrome: Getting
UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy Link to publication Citation for published version (APA): Franken, R. (2016). Marfan syndrome: Getting
UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy. Link to publication
 UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy Link to publication Citation for published version (APA): Franken, R. (2016). Marfan syndrome: Getting
UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy Link to publication Citation for published version (APA): Franken, R. (2016). Marfan syndrome: Getting
Defining the peroxisome
 0 2 4 6 8 0 2 4 6 8 20 Defining the peroxisome PEROXISOMAL DISEASES Ronald JA Wanders Laboratory for Genetic Metabolic Disorders Department of Pediatrics & Clinical Chemistry Academic Medical Center University
0 2 4 6 8 0 2 4 6 8 20 Defining the peroxisome PEROXISOMAL DISEASES Ronald JA Wanders Laboratory for Genetic Metabolic Disorders Department of Pediatrics & Clinical Chemistry Academic Medical Center University
Identification of three distinct peroxisomal protein import defects in patients with peroxisome biogenesis disorders
 Journal of Cell Science 108, 1817-1829 (1995) Printed in Great Britain The Company of Biologists Limited 1995 1817 Identification of three distinct peroxisomal protein import defects in patients with peroxisome
Journal of Cell Science 108, 1817-1829 (1995) Printed in Great Britain The Company of Biologists Limited 1995 1817 Identification of three distinct peroxisomal protein import defects in patients with peroxisome
Hereditary disorders of peroxisomal metabolism.
 Hereditary disorders of peroxisomal metabolism http://www.cytochemistry.net/cell-biology/perox.jpg Peroxisomes single membrane organelles from less than 100 to more than 1000 per eucaryotic cell more than
Hereditary disorders of peroxisomal metabolism http://www.cytochemistry.net/cell-biology/perox.jpg Peroxisomes single membrane organelles from less than 100 to more than 1000 per eucaryotic cell more than
A novel HPLC-based method to diagnose peroxisomal D-bifunctional protein enoyl-coa hydratase deficiency
 methods A novel HPLC-based method to diagnose peroxisomal D-bifunctional protein enoyl-coa hydratase deficiency Jolein Gloerich,* Simone Denis,* Elisabeth G. van Grunsven,* Georges Dacremont, Ronald J.
methods A novel HPLC-based method to diagnose peroxisomal D-bifunctional protein enoyl-coa hydratase deficiency Jolein Gloerich,* Simone Denis,* Elisabeth G. van Grunsven,* Georges Dacremont, Ronald J.
UvA-DARE (Digital Academic Repository) Improving aspects of palliative care for children Jagt, C.T. Link to publication
 UvA-DARE (Digital Academic Repository) Improving aspects of palliative care for children Jagt, C.T. Link to publication Citation for published version (APA): Jagt, C. T. (2017). Improving aspects of palliative
UvA-DARE (Digital Academic Repository) Improving aspects of palliative care for children Jagt, C.T. Link to publication Citation for published version (APA): Jagt, C. T. (2017). Improving aspects of palliative
UNCORRECTED PROOF ARTICLE IN PRESS
 BBAMCR-15512; No. of pages: 16; 4C: 6, 9, 11 + model Biochimica et Biophysica Acta xx (2006) xxx xxx www.elsevier.com/locate/bbamcr 1 Review 2 Peroxisome biogenesis disorders 3 Steven J. Steinberg a,b,,
BBAMCR-15512; No. of pages: 16; 4C: 6, 9, 11 + model Biochimica et Biophysica Acta xx (2006) xxx xxx www.elsevier.com/locate/bbamcr 1 Review 2 Peroxisome biogenesis disorders 3 Steven J. Steinberg a,b,,
Citation for published version (APA): Von Eije, K. J. (2009). RNAi based gene therapy for HIV-1, from bench to bedside
 UvA-DARE (Digital Academic Repository) RNAi based gene therapy for HIV-1, from bench to bedside Von Eije, K.J. Link to publication Citation for published version (APA): Von Eije, K. J. (2009). RNAi based
UvA-DARE (Digital Academic Repository) RNAi based gene therapy for HIV-1, from bench to bedside Von Eije, K.J. Link to publication Citation for published version (APA): Von Eije, K. J. (2009). RNAi based
Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM
 Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
Wanders. Ether lipid biosynthesis: alkyldihydroxyacetonephosphate synthase protein deficiency leads to reduced dihydroxyacetonephosphate
 Ether lipid biosynthesis: alkyl-dihydroxyacetonephosphate synthase protein deficiency leads to reduced dihydroxyacetonephosphate acyltransferase activities E. C. J. M. de Vet, 1, * L. Ijlst, W. Oostheim,
Ether lipid biosynthesis: alkyl-dihydroxyacetonephosphate synthase protein deficiency leads to reduced dihydroxyacetonephosphate acyltransferase activities E. C. J. M. de Vet, 1, * L. Ijlst, W. Oostheim,
ALD database, diagnostic dilemmas and the need for translational metabolism
 ALD database, diagnostic dilemmas and the need for translational metabolism Stephan Kemp, Ph.D. Associate Professor & AMC Principal Investigator Genetic Metabolic Diseases & Pediatrics Academic Medical
ALD database, diagnostic dilemmas and the need for translational metabolism Stephan Kemp, Ph.D. Associate Professor & AMC Principal Investigator Genetic Metabolic Diseases & Pediatrics Academic Medical
Complementation analysis in patients with the clinical phenotype of a generalised peroxisomal disorder
 J Med Genet 1996;33:295-299 Division of Medical and Molecular Genetics, United Medical and Dental Schools of Guy's and St Thomas's Hospitals, 7th/8th floors, Guy's Hospital Tower, London Bridge, London
J Med Genet 1996;33:295-299 Division of Medical and Molecular Genetics, United Medical and Dental Schools of Guy's and St Thomas's Hospitals, 7th/8th floors, Guy's Hospital Tower, London Bridge, London
Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson s Disease Guides Genetic Diagnosis
 Ivyspring International Publisher 638 Theranostics Research Paper 2016; 6(5): 638-649. doi: 10.7150/thno.14596 Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson
Ivyspring International Publisher 638 Theranostics Research Paper 2016; 6(5): 638-649. doi: 10.7150/thno.14596 Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson
Hereditary disorders of peroxisomal metabolism.
 Hereditary disorders of peroxisomal metabolism http://www.cytochemistry.net/cell-biology/perox.jpg Peroxisomes single membrane organelles from less than 100 to more than 1000 per eukaryotic cell more than
Hereditary disorders of peroxisomal metabolism http://www.cytochemistry.net/cell-biology/perox.jpg Peroxisomes single membrane organelles from less than 100 to more than 1000 per eukaryotic cell more than
Whole Exome Sequenced Characteristics
 Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
Supplementary Tables Supplementary Table 1: Patient characteristics of 45 whole exome sequenced HNSCC tumors Whole Exome Sequenced Characteristics Tumors (n=45) Age, years Median (range) 61.0 (19-90) Sex,
Lactase, sucrase-isomaltase, and carbamoyl phosphate synthase I expression in human intestine van Beers, E.H.
 UvA-DARE (Digital Academic Repository) Lactase, sucrase-isomaltase, and carbamoyl phosphate synthase I expression in human intestine van Beers, E.H. Link to publication Citation for published version (APA):
UvA-DARE (Digital Academic Repository) Lactase, sucrase-isomaltase, and carbamoyl phosphate synthase I expression in human intestine van Beers, E.H. Link to publication Citation for published version (APA):
Characterizing scaphoid nonunion deformity using 2-D and 3-D imaging techniques ten Berg, P.W.L.
 UvA-DARE (Digital Academic Repository) Characterizing scaphoid nonunion deformity using 2-D and 3-D imaging techniques ten Berg, P.W.L. Link to publication Citation for published version (APA): ten Berg,
UvA-DARE (Digital Academic Repository) Characterizing scaphoid nonunion deformity using 2-D and 3-D imaging techniques ten Berg, P.W.L. Link to publication Citation for published version (APA): ten Berg,
variant led to a premature stop codon p.k316* which resulted in nonsense-mediated mrna decay. Although the exact function of the C19L1 is still
 157 Neurological disorders primarily affect and impair the functioning of the brain and/or neurological system. Structural, electrical or metabolic abnormalities in the brain or neurological system can
157 Neurological disorders primarily affect and impair the functioning of the brain and/or neurological system. Structural, electrical or metabolic abnormalities in the brain or neurological system can
Bacterial meningitis in adults: Host and pathogen factors, treatment and outcome Heckenberg, S.G.B.
 UvA-DARE (Digital Academic Repository) Bacterial meningitis in adults: Host and pathogen factors, treatment and outcome Heckenberg, S.G.B. Link to publication Citation for published version (APA): Heckenberg,
UvA-DARE (Digital Academic Repository) Bacterial meningitis in adults: Host and pathogen factors, treatment and outcome Heckenberg, S.G.B. Link to publication Citation for published version (APA): Heckenberg,
Summary of Results CARRIER STATUS DNA INSIGHT. Result: Carrier, Heterozygote. Protected Health Information. Propionic Acidemia
 Test Results Reviewed & Approved by: Laboratory Director, Nilesh Dharajiya,.D. SAPLE REPORT CARRIER STATUS DNA INSIGHT PERSONAL DETAILS DOB Jan 1, 19XX ETHNICITY Asian ORDERING HEALTHCARE PROFESSIONAL
Test Results Reviewed & Approved by: Laboratory Director, Nilesh Dharajiya,.D. SAPLE REPORT CARRIER STATUS DNA INSIGHT PERSONAL DETAILS DOB Jan 1, 19XX ETHNICITY Asian ORDERING HEALTHCARE PROFESSIONAL
Citation for published version (APA): van Munster, B. C. (2009). Pathophysiological studies in delirium : a focus on genetics.
 UvA-DARE (Digital Academic Repository) Pathophysiological studies in delirium : a focus on genetics van Munster, B.C. Link to publication Citation for published version (APA): van Munster, B. C. (2009).
UvA-DARE (Digital Academic Repository) Pathophysiological studies in delirium : a focus on genetics van Munster, B.C. Link to publication Citation for published version (APA): van Munster, B. C. (2009).
UvA-DARE (Digital Academic Repository) Genetic basis of hypertrophic cardiomyopathy Bos, J.M. Link to publication
 UvA-DARE (Digital Academic Repository) Genetic basis of hypertrophic cardiomyopathy Bos, J.M. Link to publication Citation for published version (APA): Bos, J. M. (2010). Genetic basis of hypertrophic
UvA-DARE (Digital Academic Repository) Genetic basis of hypertrophic cardiomyopathy Bos, J.M. Link to publication Citation for published version (APA): Bos, J. M. (2010). Genetic basis of hypertrophic
CHAPTER IV RESULTS Microcephaly General description
 47 CHAPTER IV RESULTS 4.1. Microcephaly 4.1.1. General description This study found that from a previous study of 527 individuals with MR, 48 (23 female and 25 male) unrelated individuals were identified
47 CHAPTER IV RESULTS 4.1. Microcephaly 4.1.1. General description This study found that from a previous study of 527 individuals with MR, 48 (23 female and 25 male) unrelated individuals were identified
Supplementary appendix
 Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Wells JM, Farris RF, Gosdin TA, et al. Pulmonary
Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Wells JM, Farris RF, Gosdin TA, et al. Pulmonary
Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future?
 Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future? Tony Marinakii Purine Research Laboratory Biochemical Sciences Whole exome sequencing No doubt
Whole exome sequencing as a first line test: Is there even a role for metabolic biochemists in the future? Tony Marinakii Purine Research Laboratory Biochemical Sciences Whole exome sequencing No doubt
The importomer A peroxisomal membrane complex involved in protein translocation into the peroxisome matrix
 Biochimica et Biophysica Acta 1763 (2006) 1613 1619 www.elsevier.com/locate/bbamcr Review The importomer A peroxisomal membrane complex involved in protein translocation into the peroxisome matrix Naganand
Biochimica et Biophysica Acta 1763 (2006) 1613 1619 www.elsevier.com/locate/bbamcr Review The importomer A peroxisomal membrane complex involved in protein translocation into the peroxisome matrix Naganand
THE ROLE OF CFTR MUTATIONS IN CAUSING CYSTIC FIBROSIS (CF)
 THE ROLE OF CFTR MUTATIONS IN CAUSING CYSTIC FIBROSIS (CF) Vertex Pharmaceuticals Incorporated, 50 Northern Avenue, Boston, MA 02210. Vertex and the Vertex triangle logo are registered trademarks of Vertex
THE ROLE OF CFTR MUTATIONS IN CAUSING CYSTIC FIBROSIS (CF) Vertex Pharmaceuticals Incorporated, 50 Northern Avenue, Boston, MA 02210. Vertex and the Vertex triangle logo are registered trademarks of Vertex
Beta Thalassemia Case Study Introduction to Bioinformatics
 Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
UvA-DARE (Digital Academic Repository) The artificial pancreas Kropff, J. Link to publication
 UvA-DARE (Digital Academic Repository) The artificial pancreas Kropff, J. Link to publication Citation for published version (APA): Kropff, J. (2017). The artificial pancreas: From logic to life General
UvA-DARE (Digital Academic Repository) The artificial pancreas Kropff, J. Link to publication Citation for published version (APA): Kropff, J. (2017). The artificial pancreas: From logic to life General
SALSA MLPA KIT P060-B2 SMA
 SALSA MLPA KIT P6-B2 SMA Lot 111, 511: As compared to the previous version B1 (lot 11), the 88 and 96 nt DNA Denaturation control fragments have been replaced (QDX2). Please note that, in contrast to the
SALSA MLPA KIT P6-B2 SMA Lot 111, 511: As compared to the previous version B1 (lot 11), the 88 and 96 nt DNA Denaturation control fragments have been replaced (QDX2). Please note that, in contrast to the
Gezinskenmerken: De constructie van de Vragenlijst Gezinskenmerken (VGK) Klijn, W.J.L.
 UvA-DARE (Digital Academic Repository) Gezinskenmerken: De constructie van de Vragenlijst Gezinskenmerken (VGK) Klijn, W.J.L. Link to publication Citation for published version (APA): Klijn, W. J. L. (2013).
UvA-DARE (Digital Academic Repository) Gezinskenmerken: De constructie van de Vragenlijst Gezinskenmerken (VGK) Klijn, W.J.L. Link to publication Citation for published version (APA): Klijn, W. J. L. (2013).
JULY 21, Genetics 101: SCN1A. Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology
 JULY 21, 2018 Genetics 101: SCN1A Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology Disclosures: I have no financial interests or relationships to disclose. Objectives 1. Review genetic
JULY 21, 2018 Genetics 101: SCN1A Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology Disclosures: I have no financial interests or relationships to disclose. Objectives 1. Review genetic
Supplementary Document
 Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Nature Genetics: doi: /ng Supplementary Figure 1. TCGA data set on HNSCCs reanalyzed in this study.
 Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
Supplementary Figure 1 TCGA data set on HNSCCs reanalyzed in this study. Summary of the TCGA dataset on HNSCCs re-analyzed in this study and the respective numbers of samples available within each. Supplementary
BIOCHEMICAL AND MOLECULAR HETEROGENEITY IN CARNITINE PALMITOYLTRANSFERASE II DEFICIENCY
 BIOCHEMICAL AND MOLECULAR HETEROGENEITY IN CARNITINE PALMITOYLTRANSFERASE II DEFICIENCY Carmen Sousa, Helena Fonseca, Hugo Rocha, Ana Marcão, Laura Vilarinho, Luísa Diogo, Sílvia Sequeira, Cristina Costa,
BIOCHEMICAL AND MOLECULAR HETEROGENEITY IN CARNITINE PALMITOYLTRANSFERASE II DEFICIENCY Carmen Sousa, Helena Fonseca, Hugo Rocha, Ana Marcão, Laura Vilarinho, Luísa Diogo, Sílvia Sequeira, Cristina Costa,
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
MRC-Holland MLPA. Description version 19;
 SALSA MLPA probemix P6-B2 SMA Lot B2-712, B2-312, B2-111, B2-511: As compared to the previous version B1 (lot B1-11), the 88 and 96 nt DNA Denaturation control fragments have been replaced (QDX2). SPINAL
SALSA MLPA probemix P6-B2 SMA Lot B2-712, B2-312, B2-111, B2-511: As compared to the previous version B1 (lot B1-11), the 88 and 96 nt DNA Denaturation control fragments have been replaced (QDX2). SPINAL
Citation for published version (APA): Parigger, E. M. (2012). Language and executive functioning in children with ADHD Den Bosch: Boxpress
 UvA-DARE (Digital Academic Repository) Language and executive functioning in children with ADHD Parigger, E.M. Link to publication Citation for published version (APA): Parigger, E. M. (2012). Language
UvA-DARE (Digital Academic Repository) Language and executive functioning in children with ADHD Parigger, E.M. Link to publication Citation for published version (APA): Parigger, E. M. (2012). Language
MRC-Holland MLPA. Description version 29; 31 July 2015
 SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0114. As compared to the previous B2 version (lot 0813 and 0912), 11 target probes are replaced or added, and 10 new reference probes are included. P082
SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0114. As compared to the previous B2 version (lot 0813 and 0912), 11 target probes are replaced or added, and 10 new reference probes are included. P082
Peroxisomal Disorders
 Peroxisomal Disorders George Gray Birmingham Childrens Hospital Peroxisomal Disorders Peroxisomes are large single membrane bound organelles that are present in the cytoplasm of all cells. They are formed
Peroxisomal Disorders George Gray Birmingham Childrens Hospital Peroxisomal Disorders Peroxisomes are large single membrane bound organelles that are present in the cytoplasm of all cells. They are formed
Clinimetrics, clinical profile and prognosis in early Parkinson s disease Post, B.
 UvA-DARE (Digital Academic Repository) Clinimetrics, clinical profile and prognosis in early Parkinson s disease Post, B. Link to publication Citation for published version (APA): Post, B. (2009). Clinimetrics,
UvA-DARE (Digital Academic Repository) Clinimetrics, clinical profile and prognosis in early Parkinson s disease Post, B. Link to publication Citation for published version (APA): Post, B. (2009). Clinimetrics,
Peroxisomal disorders I: biochemistry and genetics of peroxisome biogenesis disorders
 Clin Genet 2004: 67: 107 133 Copyright # Blackwell Munksgaard 2004 Printed in Singapore. All rights reserved CLINICAL GENETICS doi: 10.1111/j.1399-0004.2004.00329.x Mini Review Peroxisomal disorders I:
Clin Genet 2004: 67: 107 133 Copyright # Blackwell Munksgaard 2004 Printed in Singapore. All rights reserved CLINICAL GENETICS doi: 10.1111/j.1399-0004.2004.00329.x Mini Review Peroxisomal disorders I:
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
MRC-Holland MLPA. Description version 30; 06 June 2017
 SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0517, C1-0114. As compared to the previous B2 version (lot B2-0813, B2-0912), 11 target probes are replaced or added, and 10 new reference probes are
SALSA MLPA probemix P081-C1/P082-C1 NF1 P081 Lot C1-0517, C1-0114. As compared to the previous B2 version (lot B2-0813, B2-0912), 11 target probes are replaced or added, and 10 new reference probes are
The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation
 The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation Anita Becker-Heck#, Irene Zohn#, Noriko Okabe#, Andrew Pollock#, Kari Baker Lenhart,
The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation Anita Becker-Heck#, Irene Zohn#, Noriko Okabe#, Andrew Pollock#, Kari Baker Lenhart,
Intron p Hybrid with p.leu245val p. Gly336Cys. p.leu62phe p. Ala137Val p. Asn152Thr hybrid with p.leu245val p.
 RHD negative, i.e. null Phenotype Allele name Nucleotide change Exon Predicted amino acid change D RHD*01N.01 RHD deletion 1-10 p.0 Allele name detail Comments D RHD*08N.01 RHD*Pseudogene RHD* Ψ 37- bp
RHD negative, i.e. null Phenotype Allele name Nucleotide change Exon Predicted amino acid change D RHD*01N.01 RHD deletion 1-10 p.0 Allele name detail Comments D RHD*08N.01 RHD*Pseudogene RHD* Ψ 37- bp
Identification of a novel in-frame de novo mutation in SPTAN1 in intellectual disability and pontocerebellar atrophy
 Hamdan et al., Identification of a novel in-frame de novo mutation in SPTAN1 in intellectual disability and pontocerebellar atrophy Supplementary Information Gene screening and bioinformatics PCR primers
Hamdan et al., Identification of a novel in-frame de novo mutation in SPTAN1 in intellectual disability and pontocerebellar atrophy Supplementary Information Gene screening and bioinformatics PCR primers
Mutational and phenotypical spectrum of phenylalanine hydroxylase deficiency in Denmark
 Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
Supplementary Online Content
 Supplementary Online Content Ku CA, Hull S, Arno G, et al. Detailed clinical phenotype and molecular genetic findings in CLN3-associated isolated retinal degeneration. JAMA Ophthalmol. Published online
Supplementary Online Content Ku CA, Hull S, Arno G, et al. Detailed clinical phenotype and molecular genetic findings in CLN3-associated isolated retinal degeneration. JAMA Ophthalmol. Published online
Citation for published version (APA): Wijkerslooth de Weerdesteyn, T. R. (2013). Population screening for colorectal cancer by colonoscopy
 UvA-DARE (Digital Academic Repository) Population screening for colorectal cancer by colonoscopy de Wijkerslooth, T.R. Link to publication Citation for published version (APA): Wijkerslooth de Weerdesteyn,
UvA-DARE (Digital Academic Repository) Population screening for colorectal cancer by colonoscopy de Wijkerslooth, T.R. Link to publication Citation for published version (APA): Wijkerslooth de Weerdesteyn,
Acute lung injury in children : from viral infection and mechanical ventilation to inflammation and apoptosis Bern, R.A.
 UvA-DARE (Digital Academic Repository) Acute lung injury in children : from viral infection and mechanical ventilation to inflammation and apoptosis Bern, R.A. Link to publication Citation for published
UvA-DARE (Digital Academic Repository) Acute lung injury in children : from viral infection and mechanical ventilation to inflammation and apoptosis Bern, R.A. Link to publication Citation for published
Tobacco control policies and socio-economic inequalities in smoking cessation Bosdriesz, J.R.
 UvA-DARE (Digital Academic Repository) Tobacco control policies and socio-economic inequalities in smoking cessation Bosdriesz, J.R. Link to publication Citation for published version (APA): Bosdriesz,
UvA-DARE (Digital Academic Repository) Tobacco control policies and socio-economic inequalities in smoking cessation Bosdriesz, J.R. Link to publication Citation for published version (APA): Bosdriesz,
Thyroid disease and haemostasis: a relationship with clinical implications? Squizzato, A.
 UvA-DARE (Digital Academic Repository) Thyroid disease and haemostasis: a relationship with clinical implications? Squizzato, A. Link to publication Citation for published version (APA): Squizzato, A.
UvA-DARE (Digital Academic Repository) Thyroid disease and haemostasis: a relationship with clinical implications? Squizzato, A. Link to publication Citation for published version (APA): Squizzato, A.
Nature Genetics: doi: /ng Supplementary Figure 1. Alternative splicing events in the 5K panel.
 Supplementary Figure 1 Alternative splicing events in the 5K panel. The majority of cryptic splicing occurred by creation of an AG or GT (Type I). While some other mutations increased the usage of a nearby
Supplementary Figure 1 Alternative splicing events in the 5K panel. The majority of cryptic splicing occurred by creation of an AG or GT (Type I). While some other mutations increased the usage of a nearby
X-LINKED ADRENOLEUKODISTROPHY: PROFILES OF VERY LONG CHAIN FATTY ACIDS IN PLASMA AND FIBROBLASTS IN EIGTH SERBIAN PATIENTS
 Indian Journal of Clinical Biochemistry, 2007 / 22 (2) 118-122 X-LINKED ADRENOLEUKODISTROPHY: PROFILES OF VERY LONG CHAIN FATTY ACIDS IN PLASMA AND FIBROBLASTS IN EIGTH SERBIAN PATIENTS Sanja Grkovic,
Indian Journal of Clinical Biochemistry, 2007 / 22 (2) 118-122 X-LINKED ADRENOLEUKODISTROPHY: PROFILES OF VERY LONG CHAIN FATTY ACIDS IN PLASMA AND FIBROBLASTS IN EIGTH SERBIAN PATIENTS Sanja Grkovic,
UvA-DARE (Digital Academic Repository) Vascular factors in dementia and apathy Eurelings, Lisa. Link to publication
 UvA-DARE (Digital Academic Repository) Vascular factors in dementia and apathy Eurelings, Lisa Link to publication Citation for published version (APA): Eurelings, L. S. M. (2016). Vascular factors in
UvA-DARE (Digital Academic Repository) Vascular factors in dementia and apathy Eurelings, Lisa Link to publication Citation for published version (APA): Eurelings, L. S. M. (2016). Vascular factors in
Studies on inflammatory bowel disease and functional gastrointestinal disorders in children and adults Hoekman, D.R.
 UvA-DARE (Digital Academic Repository) Studies on inflammatory bowel disease and functional gastrointestinal disorders in children and adults Hoekman, D.R. Link to publication Citation for published version
UvA-DARE (Digital Academic Repository) Studies on inflammatory bowel disease and functional gastrointestinal disorders in children and adults Hoekman, D.R. Link to publication Citation for published version
Citation for published version (APA): Sivapalaratnam, S. (2012). The molecular basis of early onset cardiovascular disease
 UvA-DARE (Digital Academic Repository) The molecular basis of early onset cardiovascular disease Sivapalaratnam, S. Link to publication Citation for published version (APA): Sivapalaratnam, S. (2012).
UvA-DARE (Digital Academic Repository) The molecular basis of early onset cardiovascular disease Sivapalaratnam, S. Link to publication Citation for published version (APA): Sivapalaratnam, S. (2012).
UvA-DARE (Digital Academic Repository) An electronic nose in respiratory disease Dragonieri, S. Link to publication
 UvA-DARE (Digital Academic Repository) An electronic nose in respiratory disease Dragonieri, S. Link to publication Citation for published version (APA): Dragonieri, S. (2012). An electronic nose in respiratory
UvA-DARE (Digital Academic Repository) An electronic nose in respiratory disease Dragonieri, S. Link to publication Citation for published version (APA): Dragonieri, S. (2012). An electronic nose in respiratory
UvA-DARE (Digital Academic Repository) Toothbrushing efficacy Rosema, N.A.M. Link to publication
 UvA-DARE (Digital Academic Repository) Toothbrushing efficacy Rosema, N.A.M. Link to publication Citation for published version (APA): Rosema, N. A. M. (2015). Toothbrushing efficacy. General rights It
UvA-DARE (Digital Academic Repository) Toothbrushing efficacy Rosema, N.A.M. Link to publication Citation for published version (APA): Rosema, N. A. M. (2015). Toothbrushing efficacy. General rights It
MEDICAL GENOMICS LABORATORY. Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG)
 Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Building blocks for return to work after sick leave due to depression de Vries, Gabe
 UvA-DARE (Digital Academic Repository) Building blocks for return to work after sick leave due to depression de Vries, Gabe Link to publication Citation for published version (APA): de Vries, G. (2016).
UvA-DARE (Digital Academic Repository) Building blocks for return to work after sick leave due to depression de Vries, Gabe Link to publication Citation for published version (APA): de Vries, G. (2016).
Bile acid abnormalities in peroxisomal disorders
 Bile acid abnormalities in peroxisomal disorders Sacha Ferdinandusse Lab. Genetic Metabolic Diseases Academic Medical Center Amsterdam Peroxisomes play an important role in bile acid biosynthesis Bile
Bile acid abnormalities in peroxisomal disorders Sacha Ferdinandusse Lab. Genetic Metabolic Diseases Academic Medical Center Amsterdam Peroxisomes play an important role in bile acid biosynthesis Bile
Infantile Refsum Disease: Influence of Dietary Treatment on Plasma Phytanic Acid Levels
 JIMD Reports DOI 10.1007/8904_2015_487 CASE REPORT Infantile Refsum Disease: Influence of Dietary Treatment on Plasma Phytanic Acid Levels Maria João Nabais Sá Júlio C. Rocha Manuela F. Almeida Carla Carmona
JIMD Reports DOI 10.1007/8904_2015_487 CASE REPORT Infantile Refsum Disease: Influence of Dietary Treatment on Plasma Phytanic Acid Levels Maria João Nabais Sá Júlio C. Rocha Manuela F. Almeida Carla Carmona
Beta Thalassemia Sami Khuri Department of Computer Science San José State University Spring 2015
 Bioinformatics in Medical Product Development SMPD 287 Three Beta Thalassemia Sami Khuri Department of Computer Science San José State University Hemoglobin Outline Anatomy of a gene Hemoglobinopathies
Bioinformatics in Medical Product Development SMPD 287 Three Beta Thalassemia Sami Khuri Department of Computer Science San José State University Hemoglobin Outline Anatomy of a gene Hemoglobinopathies
Advances in Abdominal Aortic Aneurysm Care - Towards personalized, centralized and endovascular care van Beek, S.C.
 UvA-DARE (Digital Academic Repository) Advances in Abdominal Aortic Aneurysm Care - Towards personalized, centralized and endovascular care van Beek, S.C. Link to publication Citation for published version
UvA-DARE (Digital Academic Repository) Advances in Abdominal Aortic Aneurysm Care - Towards personalized, centralized and endovascular care van Beek, S.C. Link to publication Citation for published version
Names for ABO (ISBT 001) Blood Group Alleles
 Names for ABO (ISBT 001) blood group alleles v1.1 1102 Names for ABO (ISBT 001) Blood Group Alleles General description: The ABO system was discovered as in 1900 and is considered the first and clinically
Names for ABO (ISBT 001) blood group alleles v1.1 1102 Names for ABO (ISBT 001) Blood Group Alleles General description: The ABO system was discovered as in 1900 and is considered the first and clinically
Names for H (ISBT 018) Blood Group Alleles
 Names for H (ISBT 018) Blood Group Alleles General description: The H blood group system consists of one antigen, H, that is carried on glycolipids and glycoproteins on the RBC membrane, where it is synthesised
Names for H (ISBT 018) Blood Group Alleles General description: The H blood group system consists of one antigen, H, that is carried on glycolipids and glycoproteins on the RBC membrane, where it is synthesised
IMPORT OF PEROXISOMAL MATRIX
 Annu. Rev. Biochem. 2000. 69:399 418 Copyright c 2000 by Annual Reviews. All rights reserved IMPORT OF PEROXISOMAL MATRIX AND MEMBRANE PROTEINS S. Subramani, Antonius Koller, and William B. Snyder Department
Annu. Rev. Biochem. 2000. 69:399 418 Copyright c 2000 by Annual Reviews. All rights reserved IMPORT OF PEROXISOMAL MATRIX AND MEMBRANE PROTEINS S. Subramani, Antonius Koller, and William B. Snyder Department
Human Adrenoleukodystrophy Protein and Related Peroxisomal ABC Transporters Interact with the Peroxisomal Assembly Protein PEX19p
 Biochemical and Biophysical Research Communications 271, 144 150 (2000) doi:10.1006/bbrc.2000.2572, available online at http://www.idealibrary.com on Human Adrenoleukodystrophy Protein and Related Peroxisomal
Biochemical and Biophysical Research Communications 271, 144 150 (2000) doi:10.1006/bbrc.2000.2572, available online at http://www.idealibrary.com on Human Adrenoleukodystrophy Protein and Related Peroxisomal
MRC-Holland MLPA. Description version 29;
 SALSA MLPA KIT P003-B1 MLH1/MSH2 Lot 1209, 0109. As compared to the previous lots 0307 and 1006, one MLH1 probe (exon 19) and four MSH2 probes have been replaced. In addition, one extra MSH2 exon 1 probe,
SALSA MLPA KIT P003-B1 MLH1/MSH2 Lot 1209, 0109. As compared to the previous lots 0307 and 1006, one MLH1 probe (exon 19) and four MSH2 probes have been replaced. In addition, one extra MSH2 exon 1 probe,
AMORE (Ablative surgery, MOulage technique brachytherapy and REconstruction) for childhood head and neck rhabdomyosarcoma Buwalda, J.
 UvA-DARE (Digital Academic Repository) AMORE (Ablative surgery, MOulage technique brachytherapy and REconstruction) for childhood head and neck rhabdomyosarcoma Buwalda, J. Link to publication Citation
UvA-DARE (Digital Academic Repository) AMORE (Ablative surgery, MOulage technique brachytherapy and REconstruction) for childhood head and neck rhabdomyosarcoma Buwalda, J. Link to publication Citation
Iron and vitamin D deficiency in children living in Western-Europe Akkermans, M.D.
 UvA-DARE (Digital Academic Repository) Iron and vitamin D deficiency in children living in Western-Europe Akkermans, M.D. Link to publication Citation for published version (APA): Akkermans, M. D. (2017).
UvA-DARE (Digital Academic Repository) Iron and vitamin D deficiency in children living in Western-Europe Akkermans, M.D. Link to publication Citation for published version (APA): Akkermans, M. D. (2017).
Prediction of toxicity in concurrent chemoradiation for non-small cell lung cancer Uijterlinde, W.I.
 UvA-DARE (Digital Academic Repository) Prediction of toxicity in concurrent chemoradiation for non-small cell lung cancer Uijterlinde, W.I. Link to publication Citation for published version (APA): Uijterlinde,
UvA-DARE (Digital Academic Repository) Prediction of toxicity in concurrent chemoradiation for non-small cell lung cancer Uijterlinde, W.I. Link to publication Citation for published version (APA): Uijterlinde,
HMG Advance Access published May 27, 2013
 HMG Advance Access published May 27, 2013 1 Functional Analysis of PEX13 Mutation in Zellweger Syndrome Spectrum Patient Reveals Novel Homooligomerization of PEX13 and its Role in Human Peroxisome Biogenesis
HMG Advance Access published May 27, 2013 1 Functional Analysis of PEX13 Mutation in Zellweger Syndrome Spectrum Patient Reveals Novel Homooligomerization of PEX13 and its Role in Human Peroxisome Biogenesis
Molecular Characterization of the NF2 Gene in Korean Patients with Neurofibromatosis Type 2: A Report of Four Novel Mutations
 Korean J Lab Med 2010;30:190-4 DOI 10.3343/kjlm.2010.30.2.190 Original Article Diagnostic Genetics Molecular Characterization of the NF2 Gene in Korean Patients with Neurofibromatosis Type 2: A Report
Korean J Lab Med 2010;30:190-4 DOI 10.3343/kjlm.2010.30.2.190 Original Article Diagnostic Genetics Molecular Characterization of the NF2 Gene in Korean Patients with Neurofibromatosis Type 2: A Report
UvA-DARE (Digital Academic Repository) Genetic variation in Helicobacter pylori Pan, Z. Link to publication
 UvA-DARE (Digital Academic Repository) Genetic variation in Helicobacter pylori Pan, Z. Link to publication Citation for published version (APA): Pan, Z. (1999). Genetic variation in Helicobacter pylori
UvA-DARE (Digital Academic Repository) Genetic variation in Helicobacter pylori Pan, Z. Link to publication Citation for published version (APA): Pan, Z. (1999). Genetic variation in Helicobacter pylori
Diagnostic research in perspective: examples of retrieval, synthesis and analysis Bachmann, L.M.
 UvA-DARE (Digital Academic Repository) Diagnostic research in perspective: examples of retrieval, synthesis and analysis Bachmann, L.M. Link to publication Citation for published version (APA): Bachmann,
UvA-DARE (Digital Academic Repository) Diagnostic research in perspective: examples of retrieval, synthesis and analysis Bachmann, L.M. Link to publication Citation for published version (APA): Bachmann,
Mutational Spectrum of FAM83H: The C-Terminal Portion is Required for Tooth Enamel Calcification
 HUMAN MUTATION Mutation in Brief #1014, 29:E95-E99, (2008) Online MUTATION IN BRIEF Mutational Spectrum of FAM83H: The C-Terminal Portion is Required for Tooth Enamel Calcification Sook-Kyung Lee 1, Jan
HUMAN MUTATION Mutation in Brief #1014, 29:E95-E99, (2008) Online MUTATION IN BRIEF Mutational Spectrum of FAM83H: The C-Terminal Portion is Required for Tooth Enamel Calcification Sook-Kyung Lee 1, Jan
MRC-Holland MLPA. Description version 18; 09 September 2015
 SALSA MLPA probemix P090-A4 BRCA2 Lot A4-0715, A4-0714, A4-0314, A4-0813, A4-0712: Compared to lot A3-0710, the 88 and 96 nt control fragments have been replaced (QDX2). This product is identical to the
SALSA MLPA probemix P090-A4 BRCA2 Lot A4-0715, A4-0714, A4-0314, A4-0813, A4-0712: Compared to lot A3-0710, the 88 and 96 nt control fragments have been replaced (QDX2). This product is identical to the
Enzyme replacement therapy in Fabry disease, towards individualized treatment Arends, M.
 UvA-DARE (Digital Academic Repository) Enzyme replacement therapy in Fabry disease, towards individualized treatment Arends, M. Link to publication Citation for published version (APA): Arends, M. (2017).
UvA-DARE (Digital Academic Repository) Enzyme replacement therapy in Fabry disease, towards individualized treatment Arends, M. Link to publication Citation for published version (APA): Arends, M. (2017).
Long Term Follow-Up Clinical Guidelines for X-linked Adrenoleukodystrophy
 Long Term Follow-Up Clinical Guidelines for X-linked Adrenoleukodystrophy Gerald Raymond, M.D. Department of Pediatrics and Neurology Penn State Medical Center Hershey, PA June 20, 2018 Disclosure Information
Long Term Follow-Up Clinical Guidelines for X-linked Adrenoleukodystrophy Gerald Raymond, M.D. Department of Pediatrics and Neurology Penn State Medical Center Hershey, PA June 20, 2018 Disclosure Information
July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label)
 July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label) p AHLJ090 AKT1_33765 AKT1 33765 p.e17k c.49g>a C T 2 AHBKFRM BRAF_473
July 2015 Assay ID Assay Name Gene COSMIC ID Amino acid change Nucleotide change Wild type allele (VIC label) Mutant allele (FAM label) p AHLJ090 AKT1_33765 AKT1 33765 p.e17k c.49g>a C T 2 AHBKFRM BRAF_473
MRC-Holland MLPA. Description version 07; 26 November 2015
 SALSA MLPA probemix P266-B1 CLCNKB Lot B1-0415, B1-0911. As compared to version A1 (lot A1-0908), one target probe for CLCNKB (exon 11) has been replaced. In addition, one reference probe has been replaced
SALSA MLPA probemix P266-B1 CLCNKB Lot B1-0415, B1-0911. As compared to version A1 (lot A1-0908), one target probe for CLCNKB (exon 11) has been replaced. In addition, one reference probe has been replaced
The role of media entertainment in children s and adolescents ADHD-related behaviors: A reason for concern? Nikkelen, S.W.C.
 UvA-DARE (Digital Academic Repository) The role of media entertainment in children s and adolescents ADHD-related behaviors: A reason for concern? Nikkelen, S.W.C. Link to publication Citation for published
UvA-DARE (Digital Academic Repository) The role of media entertainment in children s and adolescents ADHD-related behaviors: A reason for concern? Nikkelen, S.W.C. Link to publication Citation for published
Bio 111 Study Guide Chapter 17 From Gene to Protein
 Bio 111 Study Guide Chapter 17 From Gene to Protein BEFORE CLASS: Reading: Read the introduction on p. 333, skip the beginning of Concept 17.1 from p. 334 to the bottom of the first column on p. 336, and
Bio 111 Study Guide Chapter 17 From Gene to Protein BEFORE CLASS: Reading: Read the introduction on p. 333, skip the beginning of Concept 17.1 from p. 334 to the bottom of the first column on p. 336, and
