Nature Genetics: doi: /ng Supplementary Figure 1
|
|
|
- Amberly Hunter
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1 Multiple samples from five patients (P4, P8, P14, P15 and P17) with Barrett s esophagus and adjacent EAC show that the poor overlap is not a result of sampling bias. Bar graphs showing the number of SNVs for samples of Barrett s esophagus (BE) and cancer (C) that are either present in all samples (teal), present in all BE samples and absent in all cancer samples (pink), or present in all cancer samples and absent in all BE samples (gray) for each particular patient. Purple bars show the number of remaining variants not accounted for in the other groups (for example, present in a proportion of samples).
2 Supplementary Figure 2 EAC samples display multiple copy number aberrations compared to paired Barrett s samples. (a) Copy number plots showing two examples of Barrett s esophagus genomes that are majority copy number 2, compared to the matched EAC samples, which contain multiple gains and losses. (b) Similar plots but for patients where copy number events can be seen in the Barrett s esophagus sample as well as in the EAC sample (but are not significant enough to make a call). (c) Examples of copy number events in the Barrett s esophagus sample that are not seen in the matching EAC sample. The top track shows the depth ratio in a 20-kb window, and the bottom track shows the B-allele ratio.
3 Supplementary Figure 3 Principal-component analysis of mutational context. The counts of each different mutational context in each of the three sets of SNVs (common, Barrett s esophagus (BE) unique, EAC unique) were used as input for a principal-component analysis. There is some separation between the common SNVs and the unique SNVs in the second component, but the differences between the groups are not striking (6.9% variance in the second component). (b) Plot showing the weights of the 96 different mutational contexts. Four mutational contexts C(A>C)C, G(C>G)G and C(A>T)C (all at the top right-hand side of the plot) as well as T(C>G)A (bottom right) seem to be the main cause for the separation in the second component.
4 Supplementary Figure 4 Copy number plot for one of the Barrett s esophagus samples with low-grade dysplasia. Copy number plot for one of the Barrett s esophagus samples with low-grade dysplasia that was sent for whole-genome sequencing. Large deletions on multiple chromosomes, including chromosomes 5, 11, 13, 18 and 21, can be seen.
5 Supplementary Table 1: Patient demographics and tumor characteristics for the esophageal adenocarcinoma patient cohort Patient characteristics n = 23 Sex, n (%) Male 19 (82.6) Female 4 (17.4) Age, years (%) < Median Range Tumor stage, n (%) IA IB IIA IIB IIIA IIIB IIIC IV Missing information Differentiation, n (%) Well/Moderate Moderate Moderate/poor Poor Barrett s esophagus pathology No dysplasia Indefinite for dysplasia Low grade dysplasia High grade dysplasia Maximum Length of Barrett s esophagus (cm) Median Range Length of tumor (mm) Median Range 3 (13.0) 13 (56.5) 7 (30.4) (13.0) 4 (17.4) 6 (26.1) 0 (0.0) 12 (52.2) 5 (21.7) 4 (17.4) 17 (73.9) 1 (4.3) 3 (13.0)
6 Supplementary Table 2: Esophageal adenocarcinoma patient information. Patient Centre Gender Age* TNM7 Stage Germline tissue BE maximal length (cm) Tumor length (mm) 1 Addenbrookes Hospital, Cambridge M 62 T2N0M0 IB Blood NA 40 2 St Thomas Hospital, London F 55 T2N1M0 IIB Blood Addenbrookes Hospital, Cambridge M 61 T3N2M1 IV Blood NA 80 4 Royal Surrey County Hospital, Guildford M 58 T3N1M0 IIIA Blood Gloucestershire Royal Hospital, Gloucester M 64 T1N0M0 IA Blood NA 22 6 Gloucestershire Royal Hospital, Gloucester M 73 T3N1Mx NA Blood NA 31 7 Addenbrookes Hospital, Cambridge M 75 T3N0M0 IIA Blood NA 20 8 Salford Royal Infirmary, Manchester M 65 T2N0M0 IB Blood Addenbrookes Hospital, Cambridge M 67 T1N0M0 IA Blood Addenbrookes Hospital, Cambridge F 79 T2N0M0 IB Blood Addenbrookes Hospital, Cambridge M 67 T4N2M0 IIIC Normal esophagus Addenbrookes Hospital, Cambridge M 65 T2N1M0 IIB Blood NA Royal Surrey County Hospital, Guildford F 61 T2N0M0 IB Blood 15 NA 14 St Thomas Hospital, London M 68 T3N1M0 IIIA Blood Royal Surrey County Hospital, Guildford M 75 T3N3M1 IV Blood Addenbrookes Hospital, Cambridge M 65 T3N1M0 IIIA Blood NA Royal Surrey County Hospital, Guildford M 82 T3N1M0 IIIA Blood St Thomas Hospital, London M 61 T3N1M0 IIIA Blood Addenbrookes Hospital, Cambridge M 75 T3N0M0 IIA Blood NA Edinburgh Royal Infirmary, Edinburgh F 79 T3N1M0 IIIA Blood NA NA 21 Royal Surrey County Hospital, Guildford M 61 T1N0M0 IA Blood Royal Surrey County Hospital, Guildford M 50 T3N3M0 IIIC Blood Addenbrookes Hospital, Cambridge M 64 TxNxMx NA Blood 2 NA *Age at time of diagnosis. NA = not available.
7 Supplementary Table 3: Paired Barrett s esophagus, esophageal adenocarcinoma (EAC) and normal sample sequencing statistics. mapq = mapping quality, HiQ = high quality bases. The error rate HiQ gives the percentage of high quality bases that have mismatches to the reference genome. The sample purity for the EAC and Barrett s esophagus samples are also given. Patient Sample type purity-filtered pairs % aligned pairs median insert size % mapq=0 error rate HiQ genome coverage mapq>3 Callable genome totalnumbersnps Sample purity 1 EAC 1,244,932, Normal 590,520, Barrett's 1,195,274, EAC 1,482,353, Normal 557,666, Barrett's 931,330, EAC 1,009,393, Normal 1,161,403, Barrett's 986,130, EAC 1,229,915, Normal 702,258, Barrett's 921,904, EAC 980,497, Normal 659,466, Barrett's 880,868, EAC 1,115,303, Normal 748,774, Barrett's 953,373, EAC 1,249,130, Normal 620,715,
8 7 Barrett's 1,199,786, EAC 1,357,442, Normal 762,452, Barrett's 946,965, EAC 1,092,099, Normal 577,280, Barrett's 1,038,034, EAC 950,588, Normal 807,792, Barrett's 935,820, EAC 995,657, Normal 639,793, Barrett's 1,135,954, EAC 1,068,844, Normal 559,984, Barrett's 955,544, EAC 1,338,170, Normal 715,607, Barrett's 916,018, EAC 1,024,602, Normal 752,798, Barrett's 1,015,964, EAC 1,428,413, Normal 791,855, Barrett's 1,130,452, EAC 1,066,283, Normal 770,574, Barrett's 944,468, EAC 1,039,100, Normal 716,508,
9 17 Barrett's 1,168,839, EAC 1,258,875, Normal 696,954, Barrett's 1,011,091, EAC 1,295,238, Normal 623,088, Barrett's 1,195,290, EAC 1,299,881, Normal 622,692, Barrett's 1,039,910, EAC 1,070,994, Normal 642,977, Barrett's 1,034,752, EAC 1,027,607, Normal 510,088, Barrett's 913,440, EAC 1,363,277, Normal 579,686, Barrett's 1,408,290,
10 Supplementary Table 4: Information linking the EGA identifier found in EGAD to the patients presented in this manuscript. EGA Identifier Sample type Patient LP Barrett's 1 LP Tumor 1 LP DNA_H01 Barrett's 2 LP DNA_C03 Barrett's 2 LP DNA_D03 Barrett's 2 LP DNA_H01 Tumor 2 LP DNA_D01 Barrett's 3 LP DNA_D01 Tumor 3 LP DNA_E01 Barrett's 4 LP DNA_E02 Barrett's 4 LP DNA_F02 Barrett's 4 LP DNA_D01 Tumor 4 LP DNA_G02 Tumor 4 LP DNA_C01 Barrett's 5 LP DNA_A02 Tumor 5 LP DNA_C01 Barrett's 6 LP DNA_C01 Tumor 6 LP Barrett's 7 LP Tumor 7 LP DNA_G01 Barrett's 8 LP DNA_H02 Barrett's 8 LP DNA_G01 Tumor 8 LP DNA_A03 Tumor 8 LP DNA_B03 Tumor 8 LP DNA_A01 Barrett's 9
11 LP DNA_A01 Tumor 9 LP DNA_A01 Barrett's 10 LP DNA_A01 Tumor 10 LP DNA_A01 Barrett's 11 LP DNA_A03 Tumor 11 LP DNA_A01 Barrett's 12 LP DNA_A01 Tumor 12 LP DNA_F01 Barrett's 13 LP DNA_F01 Tumor 13 LP DNA_E01 Barrett's 14 LP DNA_A01 Barrett's 14 LP DNA_E01 Tumor 14 LP DNA_C01 Tumor 14 LP DNA_D01 Tumor 14 LP DNA_A01 Barrett's 15 LP DNA_E01 Barrett's 15 LP DNA_F01 Barrett's 15 LP DNA_C01 Tumor 15 LP DNA_G01 Tumor 15 LP DNA_H01 Tumor 15 LP DNA_A02 Barrett's 16 LP DNA_F03 Tumor 16 LP DNA_D01 Barrett's 17 LP DNA_A02 Barrett's 17 LP DNA_B02 Barrett's 17 LP DNA_C02 Barrett's 17 LP DNA_D02 Barrett's 17 LP DNA_B02 Tumor 17 LP DNA_F01 Barrett's 18
12 LP DNA_F01 Tumor 18 LP DNA_A01 Barrett's 19 LP DNA_A01 Tumor 19 LP DNA_A01 Barrett's 20 LP DNA_A01 Tumor 20 LP DNA_B01 Barrett's 21 LP DNA_B01 Tumor 21 LP DNA_B01 Barrett's 22 LP DNA_E01 Tumor 22 LP Barrett's 23 LP Tumor 23 barretts_1c Barrett's with gastric metaplasia AHM1051 barretts_1a Barrett's with intestinal metaplasia AHM1051 barretts_1d Barrett's with low grade dysplasia AHM1051 barretts_1e Early cancer AHM1051 SS CSS.casava18 FFPE Barrett's with dysplasia AHM1051 SS CSS.casava18 FFPE Barrett's with gastric metaplasia AHM1051 SS CSS.casava18 FFPE Barrett's with high grade dysplasia AHM1051 SS CSS.casava18 FFPE Barrett's with intestinal metaplasia AHM1051 SS CSS.casava18 FFPE Barrett's with intestinal metaplasia AHM1051 SS CSS.casava18 FFPE Barrett's with low grade dysplasia AHM1051 SS CSS.casava18 FFPE germline AHM1051 barretts_1b germline AHM1051
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Nature Biotechnology: doi: /nbt.1904
 Supplementary Information Comparison between assembly-based SV calls and array CGH results Genome-wide array assessment of copy number changes, such as array comparative genomic hybridization (acgh), is
Supplementary Information Comparison between assembly-based SV calls and array CGH results Genome-wide array assessment of copy number changes, such as array comparative genomic hybridization (acgh), is
Nature Genetics: doi: /ng Supplementary Figure 1. Rates of different mutation types in CRC.
 Supplementary Figure 1 Rates of different mutation types in CRC. (a) Stratification by mutation type indicates that C>T mutations occur at a significantly greater rate than other types. (b) As for the
Supplementary Figure 1 Rates of different mutation types in CRC. (a) Stratification by mutation type indicates that C>T mutations occur at a significantly greater rate than other types. (b) As for the
The 8 th Edition of TNM for Lung Cancer: The IASLC Proposals.
 13 th Cambridge Chest Meeting 2015. The 8 th Edition of TNM for Lung Cancer: The IASLC Proposals. Peter Goldstraw, Honorary Consultant Thoracic Surgeon, Royal Brompton Hospital, Emeritus Professor of Thoracic
13 th Cambridge Chest Meeting 2015. The 8 th Edition of TNM for Lung Cancer: The IASLC Proposals. Peter Goldstraw, Honorary Consultant Thoracic Surgeon, Royal Brompton Hospital, Emeritus Professor of Thoracic
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits
 AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage
 An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage Aaron M Newman1,2,7, Scott V Bratman1,3,7, Jacqueline To3, Jacob F Wynne3, Neville C W Eclov3, Leslie A Modlin3,
An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage Aaron M Newman1,2,7, Scott V Bratman1,3,7, Jacqueline To3, Jacob F Wynne3, Neville C W Eclov3, Leslie A Modlin3,
Supplementary Figure 1. Estimation of tumour content
 Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
DNA-seq Bioinformatics Analysis: Copy Number Variation
 DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/7/283/283ra54/dc1 Supplementary Materials for Clonal status of actionable driver events and the timing of mutational processes in cancer evolution
www.sciencetranslationalmedicine.org/cgi/content/full/7/283/283ra54/dc1 Supplementary Materials for Clonal status of actionable driver events and the timing of mutational processes in cancer evolution
BWA alignment to reference transcriptome and genome. Convert transcriptome mappings back to genome space
 Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Supplementary Information
 Supplementary Information - chimeric fusion transcript in human gastric cancer promotes tumorigenesis through activation of PI3K/AKT signaling Sun Mi Yun, Kwiyeom Yoon, Sunghoon Lee, Eunjeong Kim, Seong-Ho
Supplementary Information - chimeric fusion transcript in human gastric cancer promotes tumorigenesis through activation of PI3K/AKT signaling Sun Mi Yun, Kwiyeom Yoon, Sunghoon Lee, Eunjeong Kim, Seong-Ho
Nature Methods: doi: /nmeth.3115
 Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Gastric Cancer Histopathology Reporting Proforma
 Gastric Cancer Histopathology Reporting Proforma Mandatory questions (i.e. protocol standards) are in bold (e.g. S1.01). S1.01 Identification Family name Given name(s) Date of birth Sex Male Female Intersex/indeterminate
Gastric Cancer Histopathology Reporting Proforma Mandatory questions (i.e. protocol standards) are in bold (e.g. S1.01). S1.01 Identification Family name Given name(s) Date of birth Sex Male Female Intersex/indeterminate
Quality ID #249 (NQF 1854): Barrett s Esophagus National Quality Strategy Domain: Effective Clinical Care
 Quality ID #249 (NQF 1854): Barrett s Esophagus National Quality Strategy Domain: Effective Clinical Care 2018 OPTIONS FOR INDIVIDUAL MEASURES: REGISTRY ONLY MEASURE TYPE: Process DESCRIPTION: Percentage
Quality ID #249 (NQF 1854): Barrett s Esophagus National Quality Strategy Domain: Effective Clinical Care 2018 OPTIONS FOR INDIVIDUAL MEASURES: REGISTRY ONLY MEASURE TYPE: Process DESCRIPTION: Percentage
Quality ID #249 (NQF 1854): Barrett s Esophagus National Quality Strategy Domain: Effective Clinical Care
 Quality ID #249 (NQF 1854): Barrett s Esophagus National Quality Strategy Domain: Effective Clinical Care 2018 OPTIONS FOR INDIVIDUAL MEASURES: CLAIMS ONLY MEASURE TYPE: Process DESCRIPTION: Percentage
Quality ID #249 (NQF 1854): Barrett s Esophagus National Quality Strategy Domain: Effective Clinical Care 2018 OPTIONS FOR INDIVIDUAL MEASURES: CLAIMS ONLY MEASURE TYPE: Process DESCRIPTION: Percentage
Colorspace & Matching
 Colorspace & Matching Outline Color space and 2-base-encoding Quality Values and filtering Mapping algorithm and considerations Estimate accuracy Coverage 2 2008 Applied Biosystems Color Space Properties
Colorspace & Matching Outline Color space and 2-base-encoding Quality Values and filtering Mapping algorithm and considerations Estimate accuracy Coverage 2 2008 Applied Biosystems Color Space Properties
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first
 Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Abstract. Optimization strategy of Copy Number Variant calling using Multiplicom solutions APPLICATION NOTE. Introduction
 Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Supplementary Information. Supplementary Figures
 Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer
 Supplementary Information Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer Lars A. Forsberg, Chiara Rasi, Niklas Malmqvist, Hanna Davies, Saichand
Supplementary Information Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer Lars A. Forsberg, Chiara Rasi, Niklas Malmqvist, Hanna Davies, Saichand
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Supplementary Methods
 Supplementary Methods Short Read Preprocessing Reads are preprocessed differently according to how they will be used: detection of the variant in the tumor, discovery of an artifact in the normal or for
Supplementary Methods Short Read Preprocessing Reads are preprocessed differently according to how they will be used: detection of the variant in the tumor, discovery of an artifact in the normal or for
Guidelines for the assessment of mismatch repair (MMR) status in Colorectal Cancer
 Guidelines for the assessment of mismatch repair (MMR) status in Colorectal Cancer Start date: May 2015 Review date: April 2018 1 Background Mismatch repair (MMR) deficiency is seen in approximately 15%
Guidelines for the assessment of mismatch repair (MMR) status in Colorectal Cancer Start date: May 2015 Review date: April 2018 1 Background Mismatch repair (MMR) deficiency is seen in approximately 15%
Supplementary Tables. Supplementary Figures
 Supplementary Files for Zehir, Benayed et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients Supplementary Tables Supplementary Table 1: Sample
Supplementary Files for Zehir, Benayed et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients Supplementary Tables Supplementary Table 1: Sample
Nature Medicine: doi: /nm.3967
 Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
AVENIO ctdna Analysis Kits The complete NGS liquid biopsy solution EMPOWER YOUR LAB
 Analysis Kits The complete NGS liquid biopsy solution EMPOWER YOUR LAB Analysis Kits Next-generation performance in liquid biopsies 2 Accelerating clinical research From liquid biopsy to next-generation
Analysis Kits The complete NGS liquid biopsy solution EMPOWER YOUR LAB Analysis Kits Next-generation performance in liquid biopsies 2 Accelerating clinical research From liquid biopsy to next-generation
2019 COLLECTION TYPE: MIPS CLINICAL QUALITY MEASURES (CQMS) MEASURE TYPE: Process
 Quality ID #249 (NQF 1854): Barrett s Esophagus National Quality Strategy Domain: Effective Clinical Care Meaningful Measure Area: Transfer of Health Information and Interoperability 2019 COLLECTION TYPE:
Quality ID #249 (NQF 1854): Barrett s Esophagus National Quality Strategy Domain: Effective Clinical Care Meaningful Measure Area: Transfer of Health Information and Interoperability 2019 COLLECTION TYPE:
Frequency(%) KRAS G12 KRAS G13 KRAS A146 KRAS Q61 KRAS K117N PIK3CA H1047 PIK3CA E545 PIK3CA E542K PIK3CA Q546. EGFR exon19 NFS-indel EGFR L858R
 Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
CDH1 truncating alterations were detected in all six plasmacytoid-variant bladder tumors analyzed by whole-exome sequencing.
 Supplementary Figure 1 CDH1 truncating alterations were detected in all six plasmacytoid-variant bladder tumors analyzed by whole-exome sequencing. Whole-exome sequencing of six plasmacytoid-variant bladder
Supplementary Figure 1 CDH1 truncating alterations were detected in all six plasmacytoid-variant bladder tumors analyzed by whole-exome sequencing. Whole-exome sequencing of six plasmacytoid-variant bladder
Supplementary note: Comparison of deletion variants identified in this study and four earlier studies
 Supplementary note: Comparison of deletion variants identified in this study and four earlier studies Here we compare the results of this study to potentially overlapping results from four earlier studies
Supplementary note: Comparison of deletion variants identified in this study and four earlier studies Here we compare the results of this study to potentially overlapping results from four earlier studies
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols.
 Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Single-strand DNA library preparation improves sequencing of formalin-fixed and paraffin-embedded (FFPE) cancer DNA
 www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Single-strand DNA library preparation improves sequencing of formalin-fixed and paraffin-embedded (FFPE) DNA Supplementary Materials
www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Single-strand DNA library preparation improves sequencing of formalin-fixed and paraffin-embedded (FFPE) DNA Supplementary Materials
AD (Leave blank) TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients
 AD (Leave blank) Award Number: W81XWH-12-1-0444 TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients PRINCIPAL INVESTIGATOR: Mark A. Watson, MD PhD CONTRACTING ORGANIZATION:
AD (Leave blank) Award Number: W81XWH-12-1-0444 TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients PRINCIPAL INVESTIGATOR: Mark A. Watson, MD PhD CONTRACTING ORGANIZATION:
CITATION FILE CONTENT/FORMAT
 CITATION For any resultant publications using please cite: Matthew A. Field, Vicky Cho, T. Daniel Andrews, and Chris C. Goodnow (2015). "Reliably detecting clinically important variants requires both combined
CITATION For any resultant publications using please cite: Matthew A. Field, Vicky Cho, T. Daniel Andrews, and Chris C. Goodnow (2015). "Reliably detecting clinically important variants requires both combined
MSI positive MSI negative
 Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
MEDICAL GENOMICS LABORATORY. Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG)
 Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Dr Yvonne Wallis Consultant Clinical Scientist West Midlands Regional Genetics Laboratory
 Dr Yvonne Wallis Consultant Clinical Scientist West Midlands Regional Genetics Laboratory Personalised Therapy/Precision Medicine Selection of a therapeutic drug based on the presence or absence of a specific
Dr Yvonne Wallis Consultant Clinical Scientist West Midlands Regional Genetics Laboratory Personalised Therapy/Precision Medicine Selection of a therapeutic drug based on the presence or absence of a specific
NPQR Quality Payment Program (QPP) Measures 21_18247_LS.
 NPQR Quality Payment Program (QPP) Measures 21_18247_LS MEASURE ID: QPP 99 MEASURE TITLE: Breast Cancer Resection Pathology Reporting pt Category (Primary Tumor) and pn Category (Regional Lymph Nodes)
NPQR Quality Payment Program (QPP) Measures 21_18247_LS MEASURE ID: QPP 99 MEASURE TITLE: Breast Cancer Resection Pathology Reporting pt Category (Primary Tumor) and pn Category (Regional Lymph Nodes)
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical
 Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Tumours of the Oesophagus & Gastro-Oesophageal Junction Histopathology Reporting Proforma
 Tumours of the Oesophagus & Gastro-Oesophageal Junction Histopathology Reporting Proforma Mandatory questions (i.e. protocol standards) are in bold (e.g. S1.01). S1.01 Identification Family name Given
Tumours of the Oesophagus & Gastro-Oesophageal Junction Histopathology Reporting Proforma Mandatory questions (i.e. protocol standards) are in bold (e.g. S1.01). S1.01 Identification Family name Given
Disclosure. Summary. Circulating DNA and NGS technology 3/27/2017. Disclosure of Relevant Financial Relationships. JS Reis-Filho, MD, PhD, FRCPath
 Circulating DNA and NGS technology JS Reis-Filho, MD, PhD, FRCPath Director of Experimental Pathology, Department of Pathology Affiliate Member, Human Oncology and Pathogenesis Program Disclosure of Relevant
Circulating DNA and NGS technology JS Reis-Filho, MD, PhD, FRCPath Director of Experimental Pathology, Department of Pathology Affiliate Member, Human Oncology and Pathogenesis Program Disclosure of Relevant
Supplementary Figures
 Supplementary Figures Supplementary Fig 1. Comparison of sub-samples on the first two principal components of genetic variation. TheBritishsampleisplottedwithredpoints.The sub-samples of the diverse sample
Supplementary Figures Supplementary Fig 1. Comparison of sub-samples on the first two principal components of genetic variation. TheBritishsampleisplottedwithredpoints.The sub-samples of the diverse sample
Nature Genetics: doi: /ng Supplementary Figure 1. PCA for ancestry in SNV data.
 Supplementary Figure 1 PCA for ancestry in SNV data. (a) EIGENSTRAT principal-component analysis (PCA) of SNV genotype data on all samples. (b) PCA of only proband SNV genotype data. (c) PCA of SNV genotype
Supplementary Figure 1 PCA for ancestry in SNV data. (a) EIGENSTRAT principal-component analysis (PCA) of SNV genotype data on all samples. (b) PCA of only proband SNV genotype data. (c) PCA of SNV genotype
Evaluation of BRCA1/2 and homologous recombination defects in ovarian cancer and impact on clinical outcomes
 Evaluation of BRCA1/2 and homologous recombination defects in ovarian cancer and impact on clinical outcomes Melinda S. Yates, PhD Department of Gynecologic Oncology & Reproductive Medicine University
Evaluation of BRCA1/2 and homologous recombination defects in ovarian cancer and impact on clinical outcomes Melinda S. Yates, PhD Department of Gynecologic Oncology & Reproductive Medicine University
Using the 7 th edition American Joint Commission on Cancer (AJCC) Cancer Staging Manual to Determine Esophageal Cancer Staging in SEER-Medicare Data
 Paper PH10 Using the 7 th edition American Joint Commission on Cancer (AJCC) Cancer Staging Manual to Determine Esophageal Cancer Staging in SEER-Medicare Data Johnita L. Byrd, Emory University School
Paper PH10 Using the 7 th edition American Joint Commission on Cancer (AJCC) Cancer Staging Manual to Determine Esophageal Cancer Staging in SEER-Medicare Data Johnita L. Byrd, Emory University School
Mutation Profile to Predict Tumor Stage in Lung Adenocarcinoma
 Mutation Profile to Predict Tumor Stage in Lung Adenocarcinoma 1 st Calvin Kuo Mechanical Engineering Department Stanford University Stanford, USA calvink@stanford.edu Abstract Lung adenocarcinoma is among
Mutation Profile to Predict Tumor Stage in Lung Adenocarcinoma 1 st Calvin Kuo Mechanical Engineering Department Stanford University Stanford, USA calvink@stanford.edu Abstract Lung adenocarcinoma is among
plasma MATCH Andrew Wardley,
 in partnership with plasma MATCH A multiple parallel cohort, open-label, multi-centre phase IIa clinical trial of circulating tumour DNA screening to direct targeted therapies in patients with advanced
in partnership with plasma MATCH A multiple parallel cohort, open-label, multi-centre phase IIa clinical trial of circulating tumour DNA screening to direct targeted therapies in patients with advanced
Performance Characteristics BRCA MASTR Plus Dx
 Performance Characteristics BRCA MASTR Plus Dx with drmid Dx for Illumina NGS systems Manufacturer Multiplicom N.V. Galileïlaan 18 2845 Niel Belgium Table of Contents 1. Workflow... 4 2. Performance Characteristics
Performance Characteristics BRCA MASTR Plus Dx with drmid Dx for Illumina NGS systems Manufacturer Multiplicom N.V. Galileïlaan 18 2845 Niel Belgium Table of Contents 1. Workflow... 4 2. Performance Characteristics
Lung Cancer Genetics: Common Mutations and How to Treat Them David J. Kwiatkowski, MD, PhD. Mount Carrigain 2/4/17
 Lung Cancer Genetics: Common Mutations and How to Treat Them David J. Kwiatkowski, MD, PhD Mount Carrigain 2/4/17 Histology Adenocarcinoma: Mixed subtype, acinar, papillary, solid, micropapillary, lepidic
Lung Cancer Genetics: Common Mutations and How to Treat Them David J. Kwiatkowski, MD, PhD Mount Carrigain 2/4/17 Histology Adenocarcinoma: Mixed subtype, acinar, papillary, solid, micropapillary, lepidic
Detection of aneuploidy in a single cell using the Ion ReproSeq PGS View Kit
 APPLICATION NOTE Ion PGM System Detection of aneuploidy in a single cell using the Ion ReproSeq PGS View Kit Key findings The Ion PGM System, in concert with the Ion ReproSeq PGS View Kit and Ion Reporter
APPLICATION NOTE Ion PGM System Detection of aneuploidy in a single cell using the Ion ReproSeq PGS View Kit Key findings The Ion PGM System, in concert with the Ion ReproSeq PGS View Kit and Ion Reporter
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma. Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute
 Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
 Supplementary Figures Figure S1. Patient cohorts and study design. To define and interrogate the genetic alterations underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
Supplementary Figures Figure S1. Patient cohorts and study design. To define and interrogate the genetic alterations underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
Risk group criteria for tailoring adjuvant treatment in patients with endometrial cancer : a validation study of the GOG criteria
 Risk group criteria for tailoring adjuvant treatment in patients with endometrial cancer : a validation study of the GOG criteria Suk-Joon Chang, MD, Hee-Sug Ryu MD Gynecologic Cancer Center Department
Risk group criteria for tailoring adjuvant treatment in patients with endometrial cancer : a validation study of the GOG criteria Suk-Joon Chang, MD, Hee-Sug Ryu MD Gynecologic Cancer Center Department
Variant Classification. Author: Mike Thiesen, Golden Helix, Inc.
 Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Nature Neuroscience: doi: /nn Supplementary Figure 1. Missense damaging predictions as a function of allele frequency
 Supplementary Figure 1 Missense damaging predictions as a function of allele frequency Percentage of missense variants classified as damaging by eight different classifiers and a classifier consisting
Supplementary Figure 1 Missense damaging predictions as a function of allele frequency Percentage of missense variants classified as damaging by eight different classifiers and a classifier consisting
Gastric (Stomach) Cancer
 Gastric (Stomach) Cancer Gastric cancer is a disease in which malignant (cancer) cells form in the lining of the stomach. The stomach is a J-shaped organ in the upper abdomen. It is part of the digestive
Gastric (Stomach) Cancer Gastric cancer is a disease in which malignant (cancer) cells form in the lining of the stomach. The stomach is a J-shaped organ in the upper abdomen. It is part of the digestive
Nature Genetics: doi: /ng Supplementary Figure 1. Country distribution of GME samples and designation of geographical subregions.
 Supplementary Figure 1 Country distribution of GME samples and designation of geographical subregions. GME samples collected across 20 countries and territories from the GME. Pie size corresponds to the
Supplementary Figure 1 Country distribution of GME samples and designation of geographical subregions. GME samples collected across 20 countries and territories from the GME. Pie size corresponds to the
Dr Rick Tearle Senior Applications Specialist, EMEA Complete Genomics Complete Genomics, Inc.
 Dr Rick Tearle Senior Applications Specialist, EMEA Complete Genomics Topics Overview of Data Processing Pipeline Overview of Data Files 2 DNA Nano-Ball (DNB) Read Structure Genome : acgtacatgcattcacacatgcttagctatctctcgccag
Dr Rick Tearle Senior Applications Specialist, EMEA Complete Genomics Topics Overview of Data Processing Pipeline Overview of Data Files 2 DNA Nano-Ball (DNB) Read Structure Genome : acgtacatgcattcacacatgcttagctatctctcgccag
LTA Analysis of HapMap Genotype Data
 LTA Analysis of HapMap Genotype Data Introduction. This supplement to Global variation in copy number in the human genome, by Redon et al., describes the details of the LTA analysis used to screen HapMap
LTA Analysis of HapMap Genotype Data Introduction. This supplement to Global variation in copy number in the human genome, by Redon et al., describes the details of the LTA analysis used to screen HapMap
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
stomach p63/krt5 CDH17 CEACAM5 CEACAM7 ANXA13 ANXA10 CLDN18 VSIG1 TFF2 VIL1 GDA
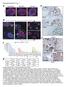 Supplementary Fig. 1 a esophagus Barrett s e FABP1 Ecad stomach TFF3 b p63/krt5 CDH17 REG4 CDH17 f c signal intensity p63/krt5 EsoSC 5000 4000 3000 2000 CDH17 1500 1000 Barrett s KRT14 KRT6A KRT13 KRT5
Supplementary Fig. 1 a esophagus Barrett s e FABP1 Ecad stomach TFF3 b p63/krt5 CDH17 REG4 CDH17 f c signal intensity p63/krt5 EsoSC 5000 4000 3000 2000 CDH17 1500 1000 Barrett s KRT14 KRT6A KRT13 KRT5
Characterisation of structural variation in breast. cancer genomes using paired-end sequencing on. the Illumina Genome Analyser
 Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
MEDICAL GENOMICS LABORATORY. Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG)
 Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with receipt) Saliva (OGR-575 DNA Genotek;
Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with receipt) Saliva (OGR-575 DNA Genotek;
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10495 WWW.NATURE.COM/NATURE 1 2 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 3 4 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 5 6 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 7 8 WWW.NATURE.COM/NATURE
doi:10.1038/nature10495 WWW.NATURE.COM/NATURE 1 2 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 3 4 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 5 6 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 7 8 WWW.NATURE.COM/NATURE
Chromatin marks identify critical cell-types for fine-mapping complex trait variants
 Chromatin marks identify critical cell-types for fine-mapping complex trait variants Gosia Trynka 1-4 *, Cynthia Sandor 1-4 *, Buhm Han 1-4, Han Xu 5, Barbara E Stranger 1,4#, X Shirley Liu 5, and Soumya
Chromatin marks identify critical cell-types for fine-mapping complex trait variants Gosia Trynka 1-4 *, Cynthia Sandor 1-4 *, Buhm Han 1-4, Han Xu 5, Barbara E Stranger 1,4#, X Shirley Liu 5, and Soumya
Supplementary Note. Nature Genetics: doi: /ng.2928
 Supplementary Note Loss of heterozygosity analysis (LOH). We used VCFtools v0.1.11 to extract only singlenucleotide variants with minimum depth of 15X and minimum mapping quality of 20 to create a ped
Supplementary Note Loss of heterozygosity analysis (LOH). We used VCFtools v0.1.11 to extract only singlenucleotide variants with minimum depth of 15X and minimum mapping quality of 20 to create a ped
Treatment of oligometastatic NSCLC
 Treatment of oligometastatic NSCLC Jarosław Kużdżał Department of Thoracic Surgery Jagiellonian University Collegium Medicum, John Paul II Hospital, Cracow New idea? 14 NSCLC patients with solitary extrathoracic
Treatment of oligometastatic NSCLC Jarosław Kużdżał Department of Thoracic Surgery Jagiellonian University Collegium Medicum, John Paul II Hospital, Cracow New idea? 14 NSCLC patients with solitary extrathoracic
of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed.
 Supplementary Note The potential association and implications of HBV integration at known and putative cancer genes of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed. Human telomerase
Supplementary Note The potential association and implications of HBV integration at known and putative cancer genes of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed. Human telomerase
MY LUNG CANCER CARE PLAN
 MY LUNG CANCER CARE PLAN MANAGE DIAGNOSIS TRACK TREATMENTS & APPOINTMENTS MONITOR SYMPTOMS FREE TO BREATHE SUPPORT LINE (844) 835-4325 A FREE resource for lung cancer patients & caregivers We are Free
MY LUNG CANCER CARE PLAN MANAGE DIAGNOSIS TRACK TREATMENTS & APPOINTMENTS MONITOR SYMPTOMS FREE TO BREATHE SUPPORT LINE (844) 835-4325 A FREE resource for lung cancer patients & caregivers We are Free
COLORECTAL PATHWAY GROUP, MANCHESTER CANCER. Guidelines for the assessment of mismatch. Colorectal Cancer
 COLORECTAL PATHWAY GROUP, MANCHESTER CANCER Guidelines for the assessment of mismatch repair (MMR) status in Colorectal Cancer January 2015 1 Background Mismatch repair (MMR) deficiency is seen in approximately
COLORECTAL PATHWAY GROUP, MANCHESTER CANCER Guidelines for the assessment of mismatch repair (MMR) status in Colorectal Cancer January 2015 1 Background Mismatch repair (MMR) deficiency is seen in approximately
Structural Variation and Medical Genomics
 Structural Variation and Medical Genomics Andrew King Department of Biomedical Informatics July 8, 2014 You already know about small scale genetic mutations Single nucleotide polymorphism (SNPs) Deletions,
Structural Variation and Medical Genomics Andrew King Department of Biomedical Informatics July 8, 2014 You already know about small scale genetic mutations Single nucleotide polymorphism (SNPs) Deletions,
University Mainz. Early Gastric Cancer. Ralf Kiesslich. Johannes Gutenberg University Mainz, Germany. Early Gastric Cancer 15.6.
 Ralf Kiesslich Johannes Gutenberg University Mainz, Germany DIAGNOSIS Unmask lesions - Chromoendoscopy -NBI Red flag technology - Autofluorescence Surface and detail analysis - Magnifying endoscopy - High
Ralf Kiesslich Johannes Gutenberg University Mainz, Germany DIAGNOSIS Unmask lesions - Chromoendoscopy -NBI Red flag technology - Autofluorescence Surface and detail analysis - Magnifying endoscopy - High
Histopathology of Endoscopic Resection Specimens from Barrett's Esophagus
 Histopathology of Endoscopic Resection Specimens from Barrett's Esophagus Br J Surg 38 oct. 1950 Definition of Barrett's esophagus A change in the esophageal epithelium of any length that can be recognized
Histopathology of Endoscopic Resection Specimens from Barrett's Esophagus Br J Surg 38 oct. 1950 Definition of Barrett's esophagus A change in the esophageal epithelium of any length that can be recognized
The 8th Edition of the TNM Classification for Lung Cancer Background, Innovations and Implications for Clinical Practice
 The 8th Edition of the TNM Classification for Lung Cancer Background, Innovations and Implications for Clinical Practice University of Torino Lecture 28th June 2017 Torino, Italy Ramón Rami-Porta Thoracic
The 8th Edition of the TNM Classification for Lung Cancer Background, Innovations and Implications for Clinical Practice University of Torino Lecture 28th June 2017 Torino, Italy Ramón Rami-Porta Thoracic
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research
 Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Supplementary Figure 1. A - MSH6 p.val282thrfs*10: Endome. Endome. Colon
 Supplementary Figure 1 - MSH6 p.val282hrfs*10: 1780 1785 C C C C C 1 B - MSH6 p.phe1088leufs*5: 1820 1810 Rectal Rectal Gastric C C C C C C Rectal C 2 C - MSH6 p.rg1172lysfs*5: 1745 1740 Ovarian 3 D -
Supplementary Figure 1 - MSH6 p.val282hrfs*10: 1780 1785 C C C C C 1 B - MSH6 p.phe1088leufs*5: 1820 1810 Rectal Rectal Gastric C C C C C C Rectal C 2 C - MSH6 p.rg1172lysfs*5: 1745 1740 Ovarian 3 D -
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
The 7th Edition of TNM in Lung Cancer.
 10th European Conference Perspectives in Lung Cancer. Brussels, March 2009. The 7th Edition of TNM in Lung Cancer. Peter Goldstraw, Consultant Thoracic Surgeon, Royal Brompton Hospital, Professor of Thoracic
10th European Conference Perspectives in Lung Cancer. Brussels, March 2009. The 7th Edition of TNM in Lung Cancer. Peter Goldstraw, Consultant Thoracic Surgeon, Royal Brompton Hospital, Professor of Thoracic
Breast and ovarian cancer in Serbia: the importance of mutation detection in hereditary predisposition genes using NGS
 Breast and ovarian cancer in Serbia: the importance of mutation detection in hereditary predisposition genes using NGS dr sc. Ana Krivokuća Laboratory for molecular genetics Institute for Oncology and
Breast and ovarian cancer in Serbia: the importance of mutation detection in hereditary predisposition genes using NGS dr sc. Ana Krivokuća Laboratory for molecular genetics Institute for Oncology and
Assessing Laboratory Performance for Next Generation Sequencing Based Detection of Germline Variants through Proficiency Testing
 Assessing Laboratory Performance for Next Generation Sequencing Based Detection of Germline Variants through Proficiency Testing Karl V. Voelkerding, MD Professor of Pathology University of Utah Medical
Assessing Laboratory Performance for Next Generation Sequencing Based Detection of Germline Variants through Proficiency Testing Karl V. Voelkerding, MD Professor of Pathology University of Utah Medical
November 9, Johns Hopkins School of Medicine, Baltimore, MD,
 Fast detection of de-novo copy number variants from case-parent SNP arrays identifies a deletion on chromosome 7p14.1 associated with non-syndromic isolated cleft lip/palate Samuel G. Younkin 1, Robert
Fast detection of de-novo copy number variants from case-parent SNP arrays identifies a deletion on chromosome 7p14.1 associated with non-syndromic isolated cleft lip/palate Samuel G. Younkin 1, Robert
Nature Genetics: doi: /ng Supplementary Figure 1. Mutational signatures in BCC compared to melanoma.
 Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
The 100,000 Genomes Project Harnessing the power of genomics for NHS rare disease and cancer patients
 The 100,000 Genomes Project Harnessing the power of genomics for NHS rare disease and cancer patients Dr Richard Scott, Clinical Lead for Rare Disease Dr Nirupa Murugaesu, Clinical Lead for Cancer Four
The 100,000 Genomes Project Harnessing the power of genomics for NHS rare disease and cancer patients Dr Richard Scott, Clinical Lead for Rare Disease Dr Nirupa Murugaesu, Clinical Lead for Cancer Four
Large-scale identity-by-descent mapping discovers rare haplotypes of large effect. Suyash Shringarpure 23andMe, Inc. ASHG 2017
 Large-scale identity-by-descent mapping discovers rare haplotypes of large effect Suyash Shringarpure 23andMe, Inc. ASHG 2017 1 Why care about rare variants of large effect? Months from randomization 2
Large-scale identity-by-descent mapping discovers rare haplotypes of large effect Suyash Shringarpure 23andMe, Inc. ASHG 2017 1 Why care about rare variants of large effect? Months from randomization 2
Paris classification (2003) 삼성의료원내과이준행
 Paris classification (2003) 삼성의료원내과이준행 JGCA classification - Japanese Gastric Cancer Association - Type 0 superficial polypoid, flat/depressed, or excavated tumors Type 1 polypoid carcinomas, usually attached
Paris classification (2003) 삼성의료원내과이준행 JGCA classification - Japanese Gastric Cancer Association - Type 0 superficial polypoid, flat/depressed, or excavated tumors Type 1 polypoid carcinomas, usually attached
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing)
 Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
No mutations were identified.
 Hereditary High Cholesterol Test ORDERING PHYSICIAN PRIMARY CONTACT SPECIMEN Report date: Aug 1, 2017 Dr. Jenny Jones Sample Medical Group 123 Main St. Sample, CA Kelly Peters Sample Medical Group 123
Hereditary High Cholesterol Test ORDERING PHYSICIAN PRIMARY CONTACT SPECIMEN Report date: Aug 1, 2017 Dr. Jenny Jones Sample Medical Group 123 Main St. Sample, CA Kelly Peters Sample Medical Group 123
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
COLORECTAL PATHWAY GROUP, MANCHESTER CANCER. Guidelines for the assessment of mismatch. Colorectal Cancer
 COLORECTAL PATHWAY GROUP, MANCHESTER CANCER Guidelines for the assessment of mismatch repair (MMR) status in Colorectal Cancer March 2017 1 Background Mismatch repair (MMR) deficiency is seen in approximately
COLORECTAL PATHWAY GROUP, MANCHESTER CANCER Guidelines for the assessment of mismatch repair (MMR) status in Colorectal Cancer March 2017 1 Background Mismatch repair (MMR) deficiency is seen in approximately
Genetic alterations of histone lysine methyltransferases and their significance in breast cancer
 Genetic alterations of histone lysine methyltransferases and their significance in breast cancer Supplementary Materials and Methods Phylogenetic tree of the HMT superfamily The phylogeny outlined in the
Genetic alterations of histone lysine methyltransferases and their significance in breast cancer Supplementary Materials and Methods Phylogenetic tree of the HMT superfamily The phylogeny outlined in the
Integration of Cancer Genome into GECCO- Genetics and Epidemiology of Colorectal Cancer Consortium
 Integration of Cancer Genome into GECCO- Genetics and Epidemiology of Colorectal Cancer Consortium Ulrike Peters Fred Hutchinson Cancer Research Center University of Washington U01-CA137088-05, PI: Peters
Integration of Cancer Genome into GECCO- Genetics and Epidemiology of Colorectal Cancer Consortium Ulrike Peters Fred Hutchinson Cancer Research Center University of Washington U01-CA137088-05, PI: Peters
Nature Neuroscience: doi: /nn Supplementary Figure 1. Behavioral training.
 Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
ACE ImmunoID Biomarker Discovery Solutions ACE ImmunoID Platform for Tumor Immunogenomics
 ACE ImmunoID Biomarker Discovery Solutions ACE ImmunoID Platform for Tumor Immunogenomics Precision Genomics for Immuno-Oncology Personalis, Inc. ACE ImmunoID When one biomarker doesn t tell the whole
ACE ImmunoID Biomarker Discovery Solutions ACE ImmunoID Platform for Tumor Immunogenomics Precision Genomics for Immuno-Oncology Personalis, Inc. ACE ImmunoID When one biomarker doesn t tell the whole
Prognosis of esophageal squamous cell carcinoma patients with preoperative radiotherapy: Comparison of different cancer staging systems
 Thoracic Cancer ISSN 1759-7706 ORIGINAL ARTICLE Prognosis of esophageal squamous cell carcinoma patients with preoperative radiotherapy: Comparison of different cancer staging systems Qifeng Wang 1 *,
Thoracic Cancer ISSN 1759-7706 ORIGINAL ARTICLE Prognosis of esophageal squamous cell carcinoma patients with preoperative radiotherapy: Comparison of different cancer staging systems Qifeng Wang 1 *,
Surgical Problems in Proximal GI Cancer Management Cardia Tumours Question #1: What are cardia tumours?
 Surgical Problems in Proximal GI Cancer Management Cardia Tumours Question #1: What are cardia tumours? Question #2: How are cardia tumours managed? Michael F. Humer December 3, 2005 Vancouver, BC Case
Surgical Problems in Proximal GI Cancer Management Cardia Tumours Question #1: What are cardia tumours? Question #2: How are cardia tumours managed? Michael F. Humer December 3, 2005 Vancouver, BC Case
LUNG CANCER PATHOLOGY: UPDATE ON NEUROENDOCRINE LUNG TUMORS
 LUNG CANCER PATHOLOGY: UPDATE ON NEUROENDOCRINE LUNG TUMORS William D. Travis, M.D. Attending Thoracic Pathologist Memorial Sloan Kettering Cancer Center New York, NY PULMONARY NE TUMORS CLASSIFICATION
LUNG CANCER PATHOLOGY: UPDATE ON NEUROENDOCRINE LUNG TUMORS William D. Travis, M.D. Attending Thoracic Pathologist Memorial Sloan Kettering Cancer Center New York, NY PULMONARY NE TUMORS CLASSIFICATION
Nature Genetics: doi: /ng Supplementary Figure 1. Somatic coding mutations identified by WES/WGS for 83 ATL cases.
 Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
