Lysosomal Delivery of Bioactive Proteins to Living Human Cells via Engineered Exosomes
|
|
|
- Alaina Walker
- 5 years ago
- Views:
Transcription
1 Santa Clara University Scholar Commons Bioengineering Master's Theses Engineering Master's Theses Lysosomal Delivery of Bioactive Proteins to Living Human Cells via Engineered Exosomes Daniel Levy Santa Clara University, Follow this and additional works at: Part of the Biomedical Engineering and Bioengineering Commons Recommended Citation Levy, Daniel, "Lysosomal Delivery of Bioactive Proteins to Living Human Cells via Engineered Exosomes" (2018). Bioengineering Master's Theses This Thesis is brought to you for free and open access by the Engineering Master's Theses at Scholar Commons. It has been accepted for inclusion in Bioengineering Master's Theses by an authorized administrator of Scholar Commons. For more information, please contact
2
3 Lysosomal Delivery of Bioactive Proteins to Living Human Cells via Engineered Exosomes By Daniel Levy Master s Thesis Submitted to the Department of Bioengineering of SANTA CLARA UNIVERSITY in Partial Fulfillment of the Requirements for the degree of Master s of Science in Bioengineering Santa Clara, California 2018 ii
4 ACKNOWLEDGEMENTS I would like to recognize Dr. Biao Lu for his guidance and tutelage for the duration of this project and thesis as my mentor and thesis advisor. I would also like to thank Mai Anh Do, for working on this project alongside me. I also wanted to recognize both Adrian Valones, and Matt Blanco in their support of our research from the Bioengineering Department of Santa Clara University. This project was supported in part by an internal grant from the Department of Engineering at Santa Clara University. iii
5 DISCLOSURE This experiments of this thesis was performed in parallel with Mai Anh Do. Therefore, many figures and data look similar, although they were individually prepared and analyzed. Figures, and figure legends were prepared individually. The experiments for the following figures were performed by: Figure 1 prepared by Dr. Biao Lu. Figures 2, 4, 8 & 9 performed by Mai Anh Do. Figures 3, 5, 6, 7 & 10 performed by Daniel Levy This work was supported by the Department of Engineering at Santa Clara University. iii
6 Lysosomal Delivery of Bioactive Proteins via Engineered Exosomes to Living Human Cells Daniel Levy Department of Bioengineering Santa Clara University 2018 ABSTRACT Exosomes are naturally secreted nanovesicles derived from mammalian cells that are used for intercellular communication in vivo. As a result, they can potentially be used for intracellular delivery of therapeutics for disease treatment. We have developed an exosome pseudotyping approach using vesicular stomatitis virus glycoprotein (VSVG) to produce protein chimeras that optimize production of modified exosomes containing protein therapeutics and facilitate effective delivery to their target cells. To the VSVG transmembrane scaffold, we have fused both fluorescent and luminescent reporters for exosome tracking/visualization and quantification of activity respectively. Through our design, we have shown the biogenesis of VSVG modified exosomes from transfected producer cells through fluorescence imaging and the production of a VSVG-based stable cell line. In addition, we have characterized isolated engineered exosomes and shown that they exhibited the correct size, distribution, and molecular markers, while retaining the bioactivity of their protein cargo. Furthermore, we show that our engineered exosomes and their protein cargo are internalized by multiple cell lines into the endosomal and lysosomal compartments of those cells. Lastly, these modified exosomes can confer their bioactive cargo, either a luminescent reporter or puromycin resistance into these target cells. In summary, this study presents a novel approach to exosome engineering to enhance therapeutic protein loading and delivery, and more importantly, shows the delivery of modified exosomes to intracellular lysosomal compartments. This aspect leads to the assumption that in future studies, these engineered exosomes can be used as a vehicle for delivery of therapeutic proteins for treatment of lysosomal storage diseases. 4
7 TABLE OF CONTENTS INTRODUCTION Current Technologies and Motivation Background Project Goals 7 MATERIALS AND METHODS Design and Construction of VSVG Fusion Proteins Cell Culture Transient Transfection Cell Microscopy Isolation of Exosomes Luciferase Assay ELISA Nanoparticle Tracking Analysis Western Blot Dot Blot Exosome Uptake Assay Stable Cell Line Development HOESCHT Stain Lysosomal Stain Endosomal Stain Cell Counting 10 RESULTS Results Discussion 22 CONCLUSION 23 LIST OF FIGURES 24 REFERENCES 25 5
8 INTRODUCTION 1.1 Current Technologies and Motivation In the past, small-molecule drugs have dominated the pharmacological market around the world. However, as we exhausted the combinations of chemical compounds that can effectively treat disease, it became imperative that more advanced drugs were developed, which lead to the emergence of protein therapeutics [1]. Protein drugs offer numerous advantages over their small-drug counterparts, mostly stemming from their increased specificity to their drug target and thus, reduced off-target effects [2]. Although they offer great promise, there are still several limitations of protein therapeutics, highlighted by our inability to reach intracellular targets via current delivery methods [3, 4]. Included in protein drug limitations are their susceptibility to degradation in vivo and inability to cross the blood-brain barrier in the case of neurological diseases [5]. In an attempt to remedy some of these limitations such as delivery of therapeutics to intracellular targets, many nanoparticle-protein drug conjugates have been investigated such as silica or gold nanoparticles [5, 6]. However, as these nanomaterials are foreign to the body, they can potentially cause a robust immune response, limiting their therapeutic use in vivo [7, 8]. Another limitation of foreign nanomaterials is the lack of degradability that many of these materials hold, leading to concerns about accumulation of foreign substances and potential for toxicity [8]. By themselves, gold or silica nanoparticle-protein drug conjugates also lack the ability to target specific cell types, which is often desired in disease treatment and can lead to higher dosages being utilized to gain a therapeutic response. To remedy these limitations, we present a solution in the use of exosomes as a nano-carrier of therapeutic proteins for use in intracellular drug delivery [9, 10]. 1.2 Background Exosomes are lipid based nanovesicles that shuttle proteins, DNA, RNA, and other lipids between cells [11-13]. They are roughly nanometers in diameter and are formed within the endosomal compartments in mammalian cells where they are known as multiple vesicular bodies (MVB) before being released upon endosomal fusion with the cellular membrane, thus becoming an exosome (Fig 1C) [11, 12, 14, 15]. When the endosome invaginates to form MVBs, cytosolic proteins, DNA, and RNA are enclosed within the MVBs, which then exist within exosomes upon MVB secretion [11]. In mammalian cells, exosomes are secreted by nearly all cell types and as a result, are used in intercellular communication via the transport of proteins, DNA, and RNA [16, 17]. Exosomes deliver their cargo and enter cells via the endocytic pathway using interactions between the exosomal and cellular transmembrane proteins; as exosomes are naturally used as a transport vehicle in cellular communication, it is hypothesized that they can be engineered for use in the delivery of desired therapeutic cargo [18, 19]. Because exosomes are naturally occurring nanovesicles in vivo, they can circumvent many of the limitations of synthetic nanoparticles for protein drug delivery such as a lack of immune response, ability to cross the blood-brain barrier, lack of toxicity, and the ability to attach ligands for targeted cell delivery [5, 9]. As an example, via electroporation techniques, DNA, RNA and small molecule drugs can be loaded into isolated exosomes for increased small-molecule drug delivery or gene therapy techniques [9]. Previously, our group has demonstrated that the naturally occurring exosome surface protein, CD63 could be utilized to shuttle desired proteins into exosomal compartments by creating a chimeric CD63 scaffold-based protein chimera with a reporter protein attached to either 6
9 the N or C-terminus of CD63 [20]. Other naturally occurring exosome surface proteins have also been used for protein loading such as CD81 or CD9 [21]. However, to optimize loading of the protein, and uptake of the modified exosomes by various cell lines, we will be using a pseudotyping approach. Pseudotyping is a method to repackage viruses with new viral envelope proteins [22-24]. This process is typically used to alter and ideally optimize the infectivity of the original virus [25, 26]. One of the most commonly used proteins in pseudotyping is the G glycoprotein of vesicular stomatitis virus (VSVG), due to its high infectivity to a large range of cell types [27]. In addition to this, the VSVG envelope is generated by inward budding of the cell membrane and endosomal compartments, which mimics the mechanism of natural exosome biogenesis [28]. Due to the effective loading of proteins into exosomes and high level of infectivity, rather than using a naturally occurring exosomal protein such as CD63, we will be using VSVG to create desired protein chimeras loaded into, now modified exosomes for therapeutic usage as a proof-of-concept. The proteins that we have attached to the VSVG exosome transmembrane scaffold are either a green or red fluorescent protein, and either Gaussia luciferase or a puromycin resistance protein. The Gaussia luciferase protein, a luminescent reporter allows us to quantify the activity of our engineered exosomes, while the puromycin resistance protein allows us to generate a VSVG based stable cell line for continuous production of modified exosomes which can confer puromycin resistance to target cells upon uptake [29, 30]. The fluorescent proteins allow us to track the biogenesis, and uptake of the engineered exosomes [30]. 1.3 Project Goals Ultimately, the goal of this project is a proof-of-concept that exosomes can be used as an effective delivery vehicle for bioactive proteins in disease treatment. To achieve this, we first needed to show that VSVG could effectively shuttle and anchor our bioactive protein chimeras into extracellularly secreted exosomes. Thus, we needed to track the biogenesis and bioactivity of our modified exosomes within transfected producer cells. Secondly, to achieve our goal, we needed to characterize the isolated exosomes, showing that they had the appropriate size, protein cargo activity, and surface markers that would be expected/necessary. Lastly, we wanted to demonstrate that the VSVG-modified exosomes could be effectively taken up by various mammalian cell lines via the VSVG surface protein, and cellular surface proteins. In addition, the modified exosomes needed to deliver the bioactive protein chimeras to the correct cellular compartments within these target cells. From these three achievements, we believe that will be able to replace our reporter cargo with therapeutic proteins in the future, allowing us to develop an effective delivery system for disease treatment. 7
10 MATERIALS AND METHODS 2.1 Design and Construction of Expression Vectors for VSVG Fusion Proteins Chimeric protein constructs were configured from the 5 à3 end with the following components: a cytomegalovirus promoter, the VSVG signal peptide and full length VSVG, which consists of the extracellular portion, juxtamembrane, proximal, transmembrane, and cytosolic regions of VSVG. Following the full length VSVG, an in-frame insertion of either red fluorescent protein (RFP) or green fluorescent protein (GFP) is attached, followed by either Gaussia luciferase (Gluc) or a puromycin resistance gene (puro) (Fig 1A). At the 3 end, a polyadenylation site was added to finalize the protein construct. The gene expression vectors for the chimeric protein constructs were synthesized by gene synthesis techniques by Genscript (Piscataway Township, NJ, USA). 2.2 Cell Culture Human embryonic kidney cells (HEK 293) were purchased from Alstem (Richmond, CA, USA). Human hepatocellular carcinoma cells (HEPG2), human glioblastoma cells (U87), and mouse adipose tissue fibroblast cells (L929) were purchased from American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco s Modified Eagle s Medium (DMEM) (Gibco; MA, USA), with 10% Fetal Bovine Serum (FBS) (GE Healthcare Life Sciences; Issaquah, WA, USA), and 1% Penicillin Streptomycin (Pen Strep) (Gibco; MA, USA). At ~80% confluence, cells were treated with 0.25% trypsinethylenediamnietraacetic acid (Trypsin-EDTA) (Gibco; MA, USA) for cell dissociation and passaged. All cells were incubated at 37 C and 5% CO Transient Transfection At ~70% confluence, in six-well plates, HEK 293 cells were transfected by a solution containing plasmid DNA (1.3-2 μg/well), PEI transfection reagent (10 μl), and Opti-Mem (200 μl) (Gibco; MA, USA). 2.4 Cell Microscopy Images of live cells were using either fluorescence (Olympus; Waltham, MA, USA) or confocal microscopy (Leica; Buffalo Grove, IL, USA). The software used for image capturing was either cellsens Standard (Olympus; Waltham, MA, USA) or Leica Application Suite X (Leica; Buffalo Grove, IL, USA) for fluorescence and confocal microscopy respectively. Adobe Photoshop CS was used for the overlay, cropping, brightness, and contrast adjustment of images. 2.5 Isolation of Exosomes HEK 293 Cells were cultured to confluence in DMEM, 10% FBS, and 1% Pen Strep in 145 mm tissue culture dishes. They were transiently transfected at ~70% confluence with a solution containing 20 µg plasmid, 100 µl PEI transfection reagent, and 2 ml Opti-Mem. After 24 hours at ~80-90% confluence, cell media was changed to serum free media UltraCulture (Lonza; Portsmouth, NH, USA) for 48 hours. The conditioned media was then collected and centrifuged at 1500g for 10 minutes and filtered through a 0.2 µm filter. Exosomes were then isolated from the collected filtered UltraCulture media using the ExoQuick-TC (System Biosciences; Palo Alto, CA, USA) exosome isolation reagent via the manufacturer s protocol, which entails an incubation at 4 C overnight, before centrifugation at 3000g for 1.5 hours. The supernatant was then removed, and isolated exosomes were re-suspended in PBS (Teknova; Hollister, CA, USA). The re-suspended exosome s protein concentration was measured using NanoDrop Lite (Thermo Fisher Scientific; Fremont, CA, USA). 8
11 2.6 Luciferase Assay Transfected HEK 293 cells were lysed with 500 μl of 1x reporter lysis buffer (Promega; Sunnyvale, CA, USA) in 6 well plates, and the cell lysate was collected μlof either cell lysate or isolated exosomes were added to a 96-well white bottom plate (Corning; Tewsbury, MA, USA). 100 μl of coelenterazine Gaussia Luciferase substrate was added to the wells and luminescence was recorded using an infinite m200pro (Tecan; San Jose, CA, USA) plate reader and icontrol 1.8 software (Tecan; San Jose, CA, USA). 2.7 ELISA A standard curve for CD63 was established using an ExoELISA-ULTRA protein standard (System Biosciences; Palo Alto, CA, USA). 50 μg of isolated sample exosomes were diluted in a volume of 120 μl of Coating Buffer (System Biosciences; Palo Alto, CA, USA). 50 μl of CD63 primary antibody was added to each well at a 1:100 dilution in Blocking Buffer (System Biosciences; Palo Alto, CA, USA) and incubated. 50 μl of secondary antibody at a 1:5000 dilution was then added and incubated. 50 μl of Super-Sensitive TMB ELISA substrate (System Biosciences; Palo Alto, CA, USA) was then added. 50 μl of Stop Buffer was introduced and quantification was performed using an infinite m200pro plate reader and icontrol 1.8 software at 450 nm absorbance. 2.8 Nanoparticle Tracking Analysis (NTA) Exosomes were isolated and resuspended as previously outlined. 150 µl of isolated exosome sample was sent to Particle Characterization Laboratories Inc. (Novato, CA, USA) for NTA analysis per the company s protocol. 2.9 Western Blot Exosomes were isolated as previously outlined. 60 μg of each isolated exosome sample were loaded onto a 4-12% ExpressPlus PAGE gel (GenScript; Piscataway, NJ, USA) with Prestained Protein MW Marker #26612 (ThermoFisher Scientific; Fremont, CA, USA) in their respective wells. Gel Electrophoresis was run using a Mini-PROTEAN Tetra Cell (BioRad; Hercules, CA, USA) and Powerpac power source (BioRad; Hercules, CA, USA). Proteins were transferred to a Amersham Hybond P 0.2 μm PVDF membrane (GE Healthcare Life Sciences; Issaquah, WA, USA) with a semi- dry transfer cell (Bio-Rad). The membrane was then incubated with either primary mouse antibody CD81 mab (Santa Cruz Biotechnology; Santa Cruz, CA, USA) or CD63 (Santa Cruz Biotechnology; Santa Cruz, CA, USA). The membrane was then incubated with anti-mouse HRP-conjugated secondary antibody (ThermoFisher Scientific; Fremont, CA, USA). Blots were visualized with Pierce ECL Western Blotting Substrate (ThermoFisher Scientific; Fremont, CA, USA) on an ImageQuant LAS 500 (GE Healthcare Life Sciences; Issaquah, WA, USA) Dot Blot Exosomes were isolated as previously discussed. 250 μg of isolated exosomes were sent to System Biosciences (Palo Alto, CA, USA) for their ExoCheck service and performed per the company s protocol Exosome Uptake Assay HEK293, U87, HEPG2, and L929 were seeded in 96-well plates. At ~40% confluence, the conditioned media was replaced with 100 μl of UltraCulture serum-free media containing 0.3 μg/μl of isolated exosomes per well. Cells were then imaged at 20x magnification after 6, 24, 48, and 72 hours with either fluorescence or confocal microscopy, as previously outlined Stable Cell Line Development HEK 293 cells were treated with 5 µg/ml puromycin dihydrochloride (ThermoFisher Scientific; Fremont, CA, USA) in DMEM, 10% FBS, and 1% 9
12 Pen Strep, 24 hours post-transfection in a 6-well plate at ~80% confluency. After drug resistant cells grew to confluency, the cells were passaged onto 100 mm plates where they were cultured for 8 weeks. Puromycin resistant cells were viewed under fluorescence microscopy as previously outlined to ensure complete GFP or RFP expression in addition to puromycin resistance HOESCHT Stain Cells were cultured for 24, 48, or 72 hours on glass bottom plates. At the desired time, cell culture medium was removed and replaced with 1:1000 Hoescht (ThermoFisher Scientific; Fremont, CA, USA) in PBS and incubated at 37 C for 10 minutes. The Hoescht solution was removed and cells were washed with PBS before imaging via confocal microscopy as previously described Lysosomal Stain 1 mm LysoTracker Red DND-99 (Invitrogen; Carlsbad, CA, USA) was diluted in cell culture medium to a concentration of 75 nm. Cells were cultured for 48 hours under previously described conditions and culture medium was replaced with the LysoTrackercell media solution and incubated at 37 C for 30 minutes. The LysoTracker solution was then replaced with fresh cell culture medium or PBS before imaging via confocal microscopy Endosomal Stain Cells were cultured for 24 hours on a 500 µl glass bottom plate before 5-7 µl of CellLight Early Endosomes-RFP, BacMam 2.0 (Invitrogen; Carlsbad, CA, USA) or CellLight Late Endosomes-RFP, BacMam 2.0 (Invitrogen; Carlsbad, CA, USA) was added to the cell culture medium. Cells were then cultured for 16 hours at 37 C before imaging via confocal microscopy Cell Counting Cultured cells were trypsinized and collected before being centrifuged at 1500 rpm for 5 min. Cells were then resuspended and mixed with Trypan Blue 0.4% Solution (MP Biomedicals; Solon, Ohio, USA) before being counted on a C-Chip Disposable Hemocytometers (INCYTO; Convington, GA, USA) and Vista Vision Light Microscope (VWR; Visalia, CA, USA). 10
13 RESULTS 3.1 Results 1. Biogenesis of Exosomes Exosomes offer a unique vehicle for intracellular delivery of bioactive cargos to living cells. Being a naturally cell secreted vesicle used in a variety of motifs such as intercellular communication and transport, exosomes can freely travel throughout the body without the threat of an immune response [12]. To harness this property of exosomes, we have developed a chimeric protein via a gene sequence composed of a promoter (CMV), fluorescent reporters GFP or RFP, and either a puromycin resistance gene (Puro) or secretory a Gaussia luciferase (Gluc) reporter all anchored on the exosomal membrane by a vesicular stomatitis viral glycoprotein (VSVG) (Fig 1A, 1B). Figure 1. Exosome pseudotyping design Notes: (A) Design of VSVG constructs. For each construct, VSVG is fused with either a GFP or RFP reporter, in addition to either a Gaussia luciferase (gluc) or Puromycin (Puro) resistance gene. (B) Illustration of the proteinloaded exosomes based on the four gene constructs. The glycoprotein VSVG (black), reporter GFP (green) or RFP (red), and protein cargo gluc (gold) or Puro (purple) are shown. (C) Model illustrating the biogenesis of modified exosomes in mammalian cells. Upon protein production, VSVG migrates to the plasma membrane via its signaling peptide where inward budding generates early endosomes. A second inward budding within the late endosome forms multivesiclular bodies (MVB). Fusion of the late endosome containing MVBs with the plasma membrane results in extracellular release of exosomes. (D) Model illustrating the uptake of modified exosomes in mammalian cells. Modified exosomes are internalized via endocytosis by target cells, resulting in endosome formation, and ultimately either fuse with the lysosomes or cellular membrane for extracellular re-secretion. 11
14 Upon translation of the designed gene sequence, the chimeric protein is to the cell membrane by the VSVG signaling peptide where it is anchored by the VSVG protein itself. Invagination of the cell membrane results in endosome formation with the newly anchored chimeric proteins populating the endosomal membrane [15]. A second invagination of the endosomal compartment results in the formation of multiple vesicular bodies (MVB) which upon secretion, produce exosomes containing the designed chimeric VSVG transmembrane protein (Fig 1C) [21]. By transfecting our gene constructs into HEK293 cells, we tracked our chimeric protein throughout the exosome biogenesis pathway. After the first 6-24 hours, the GFP reporter can be observed at the outer cell membrane before progressing towards the 1 st and 2 nd invaginations after hours and resulting in punctations throughout the cell (Fig 2A-L). These punctations show a stark difference when compared with cytosolic GFP, where a uniform expression is observed, and indicates that reporter GFP expression may be within exosomes (Fig 2M-O). Figure 2. Intracellular tracking of exosome biogenesis Notes: HEK293 cells were transfected with either each of our four VSVG-based plasmid constructs (A-D) or a positive control, EF1-G2F (E). Cells were observed by fluorescence microscopy after 24, 48, and 72 hours. VSVG-reporter modified exosomes aggregate into punctated groups after traveling to the cell membrane and invaginating after 48 hours (A-D) during biogenesis rather than producing a uniform cytosolic expression profile as seen in the positive control at 48 hours (E). 12
15 To confirm that our VSVG based chimeric proteins were indeed located in exosomal compartments within the HEK 293 cells, we co-transfected them with known exosomal marker-reporter constructs CD63 and XPACK (Fig 3). Co-localized expression of reporter proteins was observed within the intracellular exosomes, confirming our initial observations. Figure 3. Colocalization of tracking molecules with exosome markers Notes: HEK293 cells were cotransfected with our VSVG-RFP-gluc (A,C) or VSVG-RFP-puro (B,D) constructs and known exosomal markers: XPACK-GFP (C,D) or CD63-GFP (A,B). After 48 hours incubation, modified exosomes were colocalized into subcellular punctations as demonstrated by the yellow overlap within transfected cells. This confirms that our designed constructs produce protein chimeras that travel to the exosomes within producer cells. Cells were observed by fluorescence microscopy. While the presence of our chimeric protein in exosomes of producer cells was confirmed via imagining, the activity of the bioactive cargos, either secretory Gluc or puromycin resistance within the exosomes were being assayed in parallel. Secretory Gaussia luciferase, attached to our GFP/RFP reporter and VSVG transmembrane anchor produces a luminescent signal and can be measured by a microplate reader. To quantitatively measure luminescence expression, a luciferase assay was run, where we observed a dramatic 70-90x increase in both our GFP and RFP transfected cell lysate over 13
16 untransfected, parental cell lysate (Fig 4A). This indicates that while attached to our VSVG anchoring protein, the bioactivity of the protein cargo is retained within the transfected cells. Separately, the transfected exosome producer cells with puromycin resistance were also assayed to determine cargo activity. By placing the producer cells in cell culture media containing puromycin (3-5 µg/ml) and comparing them visually to untransfected HEK 293 cells, we observed a difference in cell viability and survival (Fig 4B). This experiment further confirms that both our protein cargos remain active when inserted into exosomes via a VSVG transmembrane protein. Figure 4. Engineered tracking markers are biologically active Notes: (A) HEK293 cells were transfected with our VSVG-Gluc constructs, lysed at 48 hours, and combined with the luciferin substrate, where luminescence was recorded via plate reader. Gaussia Luciferase expression in VSVG-gLuc transfected cell lysate compared to mock transfection of parental HEK293 lysate as a negative control show that the protein cargo is biologically active at roughly a 70 and 90-fold increase for VSVG-GFP-Gluc and VSVG-RFP-Gluc respectively. Data is presented as relative light units from three samples (mean ± standard deviation, n =3). (B) At 48 hours post transfection, HEK293 cells previously transfected with VSVG-puro constructs began incubation with 5 µg/ml puromycin media and images were taken at 72 hours post puromycin treatement via fluorescence microscopy. The observed puromycin resistance in VSVG-puro transfected cells shows the biologically active protein cargo was correctly produced and expressed. 14
17 2. Exosome Characterization After observing the biogenesis and activity of our engineered exosomes within producer cells, we isolated our engineered exosomes and further characterized their size distribution, concentration, surface markers, and protein cargo activity. An ELISA assay was run with an antibody against a known exosomal marker, CD63 to determine exosome concentration (exosome number/mg protein) of our isolated engineered exosomes compared with parental HEK 293 exosomes (Fig 5A). The exosome concentrations are similar between the isolated parental HEK 293 exosomes and that of all of our engineered exosome constructs. In addition to the ELISA, Nanoparticle Tracking Analysis (NTA) was performed comparing parental HEK 293 isolated exosomes and our modified exosomes to observe the particle size and distribution of samples. Once again, similar concentrations (exosome number/ml) are observed between the parental exosomes and modified ones (Fig 5B). The NTA also indicates that the particle size is similar between the control exosomes and our engineered nanovesicles ( nm) with a single peak at the correct size being observed. These experiments indicated to us that our engineered exosomes retain their native size and distribution, despite the modifications and presence of a protein cargo. B Particle Size and Distribution 10 Particles/ml 8 Control Exosome C VSVGRFP-Puro ol uc uro Luc -Puro ntr -gl -P -g Co -GFP -GFP -RFP -RFP G SVG SVG SVG V V VS V V Standard (100 nm) D GFP/RFP Confocal Image VSVG-GFP-gLuc VSVG-GFP-Puro VSVG-RFP-Puro Control Control G F P R F P VSVGGFP-gLuc VSVGRFP-gLuc Luciferase Activity x ** x *** x u ol Luc -gl ntr -g FP Co -GFP VG-R VG VS VS Size (100 nm) E Exosome Markers kda Pos GM130 FLOT-1 ICAM VSVG-GFP-gLuc GFP 6 VSVG-RFP-gluc VSVGGFP-Puro VSVG-RFP-Puro Exosomes/Protein (x109 Number/mg) 12 Relative Light Unit Exosome Number VSVG-GFP-Puro A CD63 EpCAM ANXA5 TSG101 Blank CD81 37 CD63 Western Blot CD81 ALIX VSVG-RFP-gLuc c Pos GM130 FLOT-1 ICAM Pos CD81 ALIX VSVG-RFP-puro CD63 EpCAM ANXA5 TSG101 Blank Pos Dot Blot Figure 5 Figure 5. Characterization of exosomes released from producer cells Notes: Exosomes were isolated from serum-free cell culture media of transfected producer cells with Exo-Quick TC (System Biosciences). (A) The total number of exosomes per mg protein of sample was recorded by running an ELISA against a known exosome tetraspanin protein, CD63 antibody. Exosome number per mg protein is similar between all VSVG constructs and the control, unmodified HEK 293 exosomes (~9x109 exosomes/mg protein). (B) Isolated exosomes were analyzed with NTA (Nanosight) indicating similar size distributions between VSVG-based constructs and unmodified HEK 293 exosomes (average size ~ nm). (C) Luciferin was combined with isolated exosomes, and luminescence was recorded via plate reader. Gaussia Luciferase expression in the isolated exosomes shows a biologically active cargo with significantly higher activity levels of a 310 and 910-fold increase for VSVG-GFP-Gluc and VSVG-RFP-Gluc exosomes respectively. Data is presented as relative light units from three samples (mean ± standard deviation, n =3). (D) Isolated modified exosomes were visualized with confocal microscopy. (E) Western blot of isolated modified exosomes was performed with known exosome proteins CD63 and CD81 with sizes ~37 and ~22 kda respectively. A dot blot of isolated modified exosomes from VSVG-RFP-puro and VSVG-RFP-Gluc was performed with known exosome proteins FLOT-1, ICAM, ALIX, CD81, CD63, EpCAM, ANXA5, TSG101 and a negative control protein GM130, showing the presence of correct exosomal molecular markers. 15
18 To detect the presence of the bioactive cargo within our modified exosomes, isolated exosomes were observed via confocal microscopy and compared with isolated parental HEK 293 exosomes. GFP and RFP expression was clearly observed in both the puromycin resistance and Gaussia luciferase modified exosomes but not in parental HEK 293 secreted exosomes (Fig 5D). To quantify the amount of cargo loaded within the isolated exosomes, a luciferase activity assay was run on the isolated modified exosomes containing the Gluc reporter. Luciferase activity was detected at a level of 310, and 910 times in the VSVG- GFP-gLuc and VSVG-RFP-gLuc modified exosomes respectively as compared to parental HEK 293 exosomes (Fig 5C). In conjunction, these two experiments indicated to us that the cargo remained active within the isolated engineered exosomes. Lastly, to further verify that our bioactive cargo was within exosomes, a Dot Blot was performed with known exosomal markers such as CD63, EpCAM, ANXA5, FLOT-1, ICAM, TSG101, ALIX, and CD81, as well as against a negative control marker in GM130. It was observed that both our puromycin resistant and Gaussia luciferase containing exosomes had levels of the known exosomal makers, and not that of the negative control maker, GM130 (Fig 5E). Furthermore, western blots were performed to not only confirm the presence of known exosomal markers CD63 and CD81, but also determine if those markers were the correct protein size, indicating no change from our VSVG based modifications. A molecular weight of kda, and kda was observed for CD63 and CD81 respectively, which is confirmed from literature as the correct size (Fig 5E) [20]. As a whole, these characterization experiments indicate that our protein cargo is active within isolated exosomes, and offer no significant change to particle size, distribution, or expression of native surface proteins. 16
19 3. Exosome Activity in Living Cells After characterization of our isolated engineered exosomes for size distribution, cargo bioactivity, and exosomal markers, we investigated the uptake of the engineered exosomes within various living cell lines in vitro. In our first uptake experiment, isolated exosomes expressing GFP were introduced with serum-free media into a culture of untransfected living cells and observed for 48 hours. It was noted that the modified exosomes containing GFP were successfully taken in by HEK293, HEPG2, U87, and L929 cells after hours incubation time (Fig 6 A-X). The presence of our GFP reporter suggests that not only are the exosomes taken up by various cell types, but that the cargo is biologically active for at least 48 hours post-introduction. HEK293 A GFP B 24 hours 48 hours C E D F 6 hour G U87 6 hour GFP + Phase GFP N 48 hours I K J L U W V X GFP + Phase O Q P R S L929 HepG2 M GFP H 24 hours GFP + Phase GFP T GFP + Phase Figure 6 Figure 6. Uptake of protein-loaded exosomes by mammalian cells Notes: Isolated VSVG-GFP-puro modified exosomes were introduced to HEK293, U87, L929, and HEPG2 cells in culture. After 6, 24 and 48 hours, intracellular localization of the punctated modified exosomes was observed, indicating uptake of the biologically active protein cargo into the target cell. It was hypthoesized that the modified exosomes entered the target cells via endocytosis. Cells were observed via fluorescence microscopy or confocal microscopy. Next, a stable cell line of modified exosome producer cells was established with both our VSVG-GFP-puro and VSVG-RFP-puro gene constructs in media containing either 3 µg/ml or 5 µg/ml puromycin (Fig 7A-H). It has previously been noted by various studies that the cytotoxic nature of VSVG makes it impossible to establish a stable cell line, however, by using a lower concentration of puromycin in cell culture media in conjunction with the inherent robustness of HEK 293 cells, a stable cell line was established [9]. These stable cell lines were used to further ease our research in both the production and uptake of modified exosomes. 17
20 VSVG-GFP-Puro Stable A B VSVG-RFP-Puro Stable E F C VSVG-GFP-Puro D Hoescht G VSVG-RFP-Puro H Hoescht TLD Overlay TLD Overlay Endosomal or Lysosomal Compartment I J K L M VSVG-GFP-Puro Endosome Hoescht N O P Q R TLD Overlay VSVG-GFP-Puro Lysosome Hoescht Figure 7 Figure 7. Intracellular localization of exosome biogenesis in a stable cell line Notes: (A-H) HEK 293 cells were transfected with VSVG-based gene constructs containing a puromycin resistance gene sequence. Cell culture media was replaced with media containing 5 ug/ml purmoycin and surviving cells containing the puromycin resistance gene within their genome were grown for 10 weeks. Cells were observed with confocal microscopy. (I-M) The VSVG-GFP-Puro stable cell line was transfected with an RFP reporter targeting Rab5a, a biomarker associated with early endosomes. The RFP and GFP reporters were observed to be co-localized within similar cellular compartments via confocal microscopy 48 hours after transfection. (N-R) The VSVG-GFP-Puro stable cell line was stained with a red lysosomal dye, and it was observed that the GFP and RFP reporters were colocalized via confocal microscopy 48 hours after plating. TLD Overlay To confirm that the cellular compartment that our engineered chimeric VSVG proteins enter upon biogenesis is initially the endosomes followed by MVBs, our VSVG- GFP-puro stable cell line was transfected with a reporter RFP fused with an early endosomal localizing protein, Rab5a. It was observed that the GFP modified exosomes were co-localized in early endosomal compartments, thus confirming our initial assumption that exosomes are produced from inward budding of endosomes (Fig 7I-M). Separately, our VSVG-GFP-puro stable cell line was chemically stained with a red flurorescent dye localizing within lysosomal compartments and observed via confocal microscopy (Fig 7N- R). It was noted that our modified exosomes within the stable cell line co-localized with a significant fraction of dyed lysosomal compartments. This indicates that a portion of exosome producing endosomes merge into the lysosomal compartment, which is consistent with previous knowledge of endosomal pathways, where the other portion fuses with the cell membrane, secreting its contents [31]. In a separate line of experiments, exosomes from our VSVG-GFP-puro stable cell line were isolated and incubated with our VSVG-RFP-puro for 48 hours. It was observed over this time period that our GFP expressing exosomes not only were taken up by the stable cells, but also that the isolated GFP modified exosomes were co-localized within the exosome producing compartments of the stable producer cells, indicated by yellow 18
21 punctations (Fig 8A-I). To further confirm the previous experiment, our VSVG-GFP-puro and VSVG-RFP-puro stable cell lines were co-cultured for 48 hours. It was hypothesized that modified exosomes from both cell lines would be secreted into the surrounding media, and the GFP/RFP expressing exosomes would be taken up by the opposite stable producer cells. This hypothesis was confirmed by confocal images of the co-culture from hours, which shows a co-localization of GFP and RFP within stable cells, seen as yellow punctations (Fig 8J-R). Exosome Uptake in Stable Cells Exosomes A D G Co-culture J M P VSVG-GFP-Puro B E H VSVG-GFP-Puro K N Q VSVG-RFP-Puro C F I VSVG-RFP-Puro L O R Overlay Overlay 24h 48h 72h 24h 48h 72h Figure 8 Figure 8. Cellular Uptake and Co-localization of Modified Exosomes Notes: (A-I) Modified exosomes isolated from VSVG-GFP-Puro transfected cells were introduced to the VSVG-RFP- Puro stable cell line and observed for 72 hours. Upon uptake of the modified exosomes, it was observed via confocal microscopy that after 48 hours, the reporter GFP and RFP were co-localized within the stable cell line, indicating that they are within the same cellular compartment, which is presumably the endosomes. (J-R) The VSVG-GFP-Puro and VSVG-RFP-Puro stable cell lines were cultured together for 72 hours and observed via confocal microscopy. It was noted that modified exosomes were secreted into the surrounding media and uptaken by the surrounding stable cells. These secreted exosomes were co-localized within the intracellular compartments of the stable cells of the opposite reporter color, further confirming the uptake of modified into the same cellular compartment of stable producer cells. Through the previous lines of experiments, it was demonstrated that modified exosomes were produced in the endosomal compartments, with a population of exosomes within the endosomes fusing with the lysosomal compartments. Furthermore, it was observed that when isolated modified exosomes were introduced to stable exosome producer cells, they were taken up and localized in the same compartments that produce the modified exosomes, presumably the endosomal compartments. Thus, it was hypothesized that the modified exosomes enter the endosomal and lysosomal compartments upon uptake by non-producer, target cells. To confirm this hypothesis, modified GFP expressing exosomes were incubated with parental HEK 293 cells, and subsequently co-transfected 19
22 with a Rab5a based RFP construct targeting the endosomal compartments. It was observed via confocal microscopy that the modified exosomes were localized within the endosomal compartments of non-modified HEK 293 (Fig9A-D) and U87 (Fig9I-L) cells. Exosome uptake in HEK293 A B VSVG-GFP-Puro Endosome - RFP E F G VSVG-GFP-Puro Lysosome - RFP Exosome uptake in U87 I J VSVG-GFP-Puro Endosome - RFP M N VSVG-GFP-Puro Lysosome - RFP D C TLD H TLD K O Overlay Overlay L Hoescht Hoescht P Overlay Overlay Figure 9 Figure 9. Cellular Compartmentalization of Exosomes upon Uptake Notes: (A-D) Isolated VSVG-GFP-Puro exosomes were introduced to parental HEK293 cells in culture for 24 hours before being transfected with an endosomal Rab5a-RFP reporter. After an additional 24 hours, the cells were observed via confocal microscopy and the GFP and RFP reporters were seen to be co-localized within endosomal compartments. (E-H) Again, VSVG-GFP-Puro exosomes were introduced to HEK293 cells in culture for 48 hours before being stained with a red lysosomal dye. The cells were observed via confocal microscopy to have RFP and GFP reporters colocalized within the lysosomal compartments. (I-P) The same procedures were then performed with U87 cells, producing similar observations. Concurrently, modified GFP expressing exosomes were introduced to either nonmodified HEK 293 and U87 cells, however in this case, the cells were stained with a red fluorescent dye localized within lysosomal compartments. Once again, it was observed via confocal microscopy that the modified exosomes were localized within those lysosomal compartments in both HEK 293 (Fig 9E-H) and U87 (Fig 9M-P) cells. Through the previous experiments, we have demonstrated that isolated modified exosomes enter and localize within endosomal and lysosomal compartments upon incubation with various cell lines. However, although GFP and RFP could be observed, we wish to further quantify the levels of bioactivity of the delivered proteins within these cells. To confirm the bioactivity, unmodified HEK 293 cells were incubated with VSVG-GFPGluc modified exosomes for 3-72 hours. At various time points (3, 8, 24, 48 and 72 hours), the HEK 293 cells were washed with PBS to remove modified exosomes not localized within the cells, and subsequently lysed for a luciferase activity assay. Through this timecourse luciferase uptake experiment, it was observed that the unmodified HEK 293 cells incubated with modified exosomes presented increasing luciferase activity for 48 hours, before a slight dip in activity at 72 hours (Fig 10A). This dip in luciferase activity could be due to the degradation of the Gluc protein within the lysosomes. 20
23 A *** *** * * * C B Pre-treatment * 48 h post treatment Control VSVG-GFP-gLuc VSVG-GFP-Puro Figure 10 Figure 10. Bioactivity of Cargo upon Uptake of Modified Exosomes Notes: (A) Isolated modified VSVG-GFP-Gluc exosomes were introduced to parental HEK293 cells and at time points: 3, 8, 24, 48 and 72 hours, the cells were washed with PBS to remove free floating modified exosomes and subsequently lysed. A luciferase assay was performed on the cell lysate with results showing an increase in luminescence over time, likely from uptake of the modified exosome and its cargo. At time points 3, 8, 24, 48, and 72 hours a 1.3, 1.9, 2.8, 4.9, and 4.4-fold increase in bioluminescence of cell lysates was observed respectively. Data is presented as relative light units from three samples (mean ± standard deviation, n =3). (B) Isolated VSVG-GFP-Puro exosomes, or VSVG-GFP-Gluc were added to parental HEK293 cells and incubated for 48 hours. After which, the media was replaced with 3 µg/ml puromycin cell culture media. Cells were observed via fluorescence microscopy for 48 hours post puromycin treatment. It was noted that cells treated with VSVG-GFP-Puro exosomes had a slight resistance to puromycin. (C) After images were taken, the treated cells were collected, stained with Trypan Blue and counted. It was noted that the cells treated with VSVG-GFP-Puro exosomes were ~49% alive as compared to the VSVG-GFP-gLuc negative control which was ~34%. Data is presented as percentage of viable cells from three samples (mean ± standard deviation, n =3). To further confirm that the modified exosomes delivered bioactive proteins to human cells, VSVG-GFP-puro isolated exosomes were incubated with unmodified HEK 293 cells for 48 hours. After 48 hours, the HEK 293 cell culture media was replaced with media containing 3 µg/ml puromycin. The cells were then observed for 24, and 48 hours for loss of cell viability. Through both cell images, and a Trypan blue cell counting method, it was observed that the cells treated with VSVG-GFP-puro exosomes had greater cell viability than HEK 293 cells that were treated with exosomes that did not contain a puromycin resistance protein (~48% vs. ~34%) (Fig 10B, C). Thus, through the previous experiments, it can be deduced that not only do our modified exosomes enter the appropriate cellular compartments, but they also hold the ability to confer their bioactive cargo to target cells upon incubation. 21
24 3.2 Discussion This research has significant implications as it pertains to the development of a novel in vivo drug delivery system. As previously mentioned, engineering exosomes can overcome many limitations of current protein drug delivery systems such as intracellular delivery, in vivo half-life, immunogenicity, and the crossing of the blood-brain barrier [5]. Furthermore, by using VSVG as an exosomal transmembrane anchor, we can optimize both loading and delivery of therapeutic proteins via exosome delivery [32]. Through our research, we have shown that exosomes can be used as an efficient bioactive carrier system to deliver desired protein therapeutics to various cell types. We have demonstrated that we are able to direct biological cargo into exosomes using VSVG as a cellular transmembrane anchor. Upon secretion, these modified exosomes can then be collected, isolated, and purified from their producer cells. We have also shown that while the protein cargo within the modified exosomes is still biologically active, it has little effect on the size distribution and native transmembrane proteins of exosomes. Furthermore, it has been demonstrated that these modified exosomes can be taken up and deliver their biological cargo to various mammalian cell types. We have also shown that not only is the cargo still biologically active upon delivery to target cells, but also enters the endosomal and lysosomal compartments of the target cell. Through the findings of this study, we have multiple avenues of future research to pursue for engineering exosomes as drug delivery vehicles. Because endogenous, non-modified exosomes carry DNA, RNA, and proteins between cells, and we have clearly shown that our modifications do not have an impact on exosome size distribution or molecular markers, it can be extrapolated that we can replace our reporters, GFP/RFP or Gaussia luciferase/puromycin resistance, with a similarly sized therapeutic protein for applications such as disease treatment or enzyme replacement therapies [14, 33, 34]. Also, because we have shown that our engineered exosomes enter the endosomal and lysosomal compartments upon uptake, it would make sense to target a lysosomal storage disease with this future enzyme replacement therapy study. Additionally, rather than using the extracellular domain of VSVG, which is robustly uptaken by a broad spectrum of cell types, we may be able to replace this extracellular region with a targeting ligand, to preferentially direct modified exosomes and thus, its protein cargo to a certain cell type/tissue [35]. Using the example of lysosomal storage disease treatment, an extracellular targeting ligand for macrophages would be physiologically relevant, due to the nature of macrophages in degradation via lysosomal enzymes [36]. In general, with this system, we d theoretically be able to deliver the necessary bioactive therapeutic to the appropriate tissue in vivo. This allows for lower amounts of off-target side effects, which in turn results in a lower necessary dosage required to produce a therapeutic effect [5]. Although we have shown delivery of proteins in this study, it would not be difficult to use our designed constructs to load our engineered exosomes with small molecule drugs, DNA, or RNA, which has far reaching implications in disease treatment of cancer and genetic disease [14, 37]. In future studies, the number of therapeutic proteins within each exosome would need to be determined to estimate the necessary in vivo therapeutic dosage. Furthermore, in our next studies, we could use a therapeutic protein within our modified exosomes for a disease model to determine efficacy of the delivery system and treatment. As previously mentioned, we can also remove the extracellular region of VSVG and replace it with a tissue specific targeting ligand to preferentially deliver our cargo to certain cell types, while also eliminates the potential for an immunogenic response to extracellular VSVG. Ultimately, animal studies would also be optimal to give us a 22
25 better understanding of an exosome-based drug delivery system in vivo. Through these studies, we d prove the utility of exosomes as a nanocarrier of protein therapeutics and open the door for delivery of other biologically active therapeutics using this system. 23
26 CONCLUSION We have demonstrated that VSVG has the ability to shuttle and anchor bioactive proteins into the secreted exosomal membranes, where, engineered exosomes can be collected and used as a delivery vehicle of desired therapeutic cargo to the endosomal and lysosomal cellular compartments of targeted cells. Our dual reporter system of GFP/RFP and Gluc/puro designed constructs was present in our modified exosomes, allowing us to track their movement visually and perform quantitative analysis. Our modified exosomes overcome many limitations of current protein drug delivery methods such as in vivo half-life and immunogenicity to name a few. Because exosomes are used naturally in cellular communication pathways, they offer unique properties in vivo that can be levied in drug delivery such as tumor cell penetration, and the ability to cross the blood-brain barrier. Engineered exosomes offer an exciting avenue for drug delivery research but needs to be studied in depth before therapeutic engineered exosomes can find its way into the clinical setting. 24
Exo-Glow TM Exosome Labeling Kits
 Exo-Glow TM Exosome Labeling Kits Cat# EXOR100A-1 Cat# EXOG200A-1 Cat# EXOC300A-1 User Manual Store kit at -20 o C on receipt Version 8 3/10/2017 A limited-use label license covers this product. By use
Exo-Glow TM Exosome Labeling Kits Cat# EXOR100A-1 Cat# EXOG200A-1 Cat# EXOC300A-1 User Manual Store kit at -20 o C on receipt Version 8 3/10/2017 A limited-use label license covers this product. By use
EXOTESTTM. ELISA assay for exosome capture, quantification and characterization from cell culture supernatants and biological fluids
 DATA SHEET EXOTESTTM ELISA assay for exosome capture, quantification and characterization from cell culture supernatants and biological fluids INTRODUCTION Exosomes are small endosome-derived lipid nanoparticles
DATA SHEET EXOTESTTM ELISA assay for exosome capture, quantification and characterization from cell culture supernatants and biological fluids INTRODUCTION Exosomes are small endosome-derived lipid nanoparticles
Long term stability of isolated human serum derived exosomes
 Long term stability of isolated human serum derived exosomes Candice de Boer (PhD student) Regenerative Medicine Laboratory Supervisor: Associate Professor Neil Davies Exosomes First discovered during
Long term stability of isolated human serum derived exosomes Candice de Boer (PhD student) Regenerative Medicine Laboratory Supervisor: Associate Professor Neil Davies Exosomes First discovered during
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
Production of Exosomes in a Hollow Fiber Bioreactor
 Production of Exosomes in a Hollow Fiber Bioreactor John J S Cadwell, President and CEO, FiberCell Systems Inc INTRODUCTION Exosomes are small lipid membrane vesicles (80-120 nm) of endocytic origin generated
Production of Exosomes in a Hollow Fiber Bioreactor John J S Cadwell, President and CEO, FiberCell Systems Inc INTRODUCTION Exosomes are small lipid membrane vesicles (80-120 nm) of endocytic origin generated
Exo Check Exosome Antibody Arrays
 Exo Check Exosome Antibody Arrays Cat # EXORAY200A 4, Cat #EXORAY210A 8 User Manual Please see individual components for storage conditions Version 1 5/23/2018 A limited use label license covers this product.
Exo Check Exosome Antibody Arrays Cat # EXORAY200A 4, Cat #EXORAY210A 8 User Manual Please see individual components for storage conditions Version 1 5/23/2018 A limited use label license covers this product.
MagCapture Exosome Isolation Kit PS Q&A
 MagCapture Exosome Isolation Kit PS Q&A Specifications and performance P.1 Comparison of the conventional method P.2 Operation methods and composition P.4 Amount of starting sample P.5 Analysis after exosomes
MagCapture Exosome Isolation Kit PS Q&A Specifications and performance P.1 Comparison of the conventional method P.2 Operation methods and composition P.4 Amount of starting sample P.5 Analysis after exosomes
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Exosome ELISA Complete Kits
 Exosome ELISA Complete Kits EXOEL-CD9A-1, EXOEL-CD63A-1, EXOEL-CD81A-1 User Manual See PAC for Storage Conditions for Individual Components Version 12 4/17/2017 A limited-use label license covers this
Exosome ELISA Complete Kits EXOEL-CD9A-1, EXOEL-CD63A-1, EXOEL-CD81A-1 User Manual See PAC for Storage Conditions for Individual Components Version 12 4/17/2017 A limited-use label license covers this
Instructions. Fuse-It-mRNA easy. Shipping and Storage. Overview. Kit Contents. Specifications. Note: Important Guidelines
 Membrane fusion is a highly efficient method for transfecting various molecules and particles into mammalian cells, even into sensitive and primary cells. The Fuse-It reagents are cargo-specific liposomal
Membrane fusion is a highly efficient method for transfecting various molecules and particles into mammalian cells, even into sensitive and primary cells. The Fuse-It reagents are cargo-specific liposomal
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Exosome ELISA Complete Kits
 Exosome ELISA Complete Kits EXOEL-CD9A-1, EXOEL-CD63A-1, EXOEL-CD81A-1 User Manual See PAC for Storage Conditions for Individual Components Version 12 4/17/2017 A limited-use label license covers this
Exosome ELISA Complete Kits EXOEL-CD9A-1, EXOEL-CD63A-1, EXOEL-CD81A-1 User Manual See PAC for Storage Conditions for Individual Components Version 12 4/17/2017 A limited-use label license covers this
BMDCs were generated in vitro from bone marrow cells cultured in 10 % RPMI supplemented
 Supplemental Materials Figure S1. Cultured BMDCs express CD11c BMDCs were generated in vitro from bone marrow cells cultured in 10 % RPMI supplemented with 15 ng/ml GM-CSF. Media was changed and fresh
Supplemental Materials Figure S1. Cultured BMDCs express CD11c BMDCs were generated in vitro from bone marrow cells cultured in 10 % RPMI supplemented with 15 ng/ml GM-CSF. Media was changed and fresh
EXOCET Exosome Quantitation Assay
 EXOCET Exosome Quantitation Assay EXOCET96A-1 User Manual Check Package Contents for Storage Temperatures Version 3 5/30/2017 A limited-use label license covers this product. By use of this product, you
EXOCET Exosome Quantitation Assay EXOCET96A-1 User Manual Check Package Contents for Storage Temperatures Version 3 5/30/2017 A limited-use label license covers this product. By use of this product, you
Exo-Check Exosome Antibody Array (Neuro) Standard Kit
 Exo-Check Exosome Antibody Array (Neuro) Standard Kit Cat # EXORAY500A-4, EXORAY500A-8 User Manual Please see individual components for storage conditions Version 1 8/21/2018 A limited-use label license
Exo-Check Exosome Antibody Array (Neuro) Standard Kit Cat # EXORAY500A-4, EXORAY500A-8 User Manual Please see individual components for storage conditions Version 1 8/21/2018 A limited-use label license
Optimization of the Fuse-It-mRNA Protocol for L929 Cells in the µ-plate 24 Well
 Optimization of the Fuse-It-mRNA Protocol for L929 Cells in the µ-plate 24 Well 1. General Information... 1 2. Background... 1 3. Material and Equipment Required... 2 4. Experimental Procedure and Results...
Optimization of the Fuse-It-mRNA Protocol for L929 Cells in the µ-plate 24 Well 1. General Information... 1 2. Background... 1 3. Material and Equipment Required... 2 4. Experimental Procedure and Results...
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates
 HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
Pre-made Reporter Lentivirus for JAK-STAT Signaling Pathway
 Pre-made Reporter for JAK-STAT Signaling Pathway Cat# Product Name Amounts LVP937-P or: LVP937-P-PBS ISRE-GFP (Puro) LVP938-P or: LVP938-P-PBS ISRE-RFP (Puro) LVP939-P or: LVP939-P-PBS ISRE-Luc (Puro)
Pre-made Reporter for JAK-STAT Signaling Pathway Cat# Product Name Amounts LVP937-P or: LVP937-P-PBS ISRE-GFP (Puro) LVP938-P or: LVP938-P-PBS ISRE-RFP (Puro) LVP939-P or: LVP939-P-PBS ISRE-Luc (Puro)
Influenza A H1N1 HA ELISA Pair Set
 Influenza A H1N1 HA ELISA Pair Set for H1N1 ( A/Puerto Rico/8/1934 ) HA Catalog Number : SEK11684 To achieve the best assay results, this manual must be read carefully before using this product and the
Influenza A H1N1 HA ELISA Pair Set for H1N1 ( A/Puerto Rico/8/1934 ) HA Catalog Number : SEK11684 To achieve the best assay results, this manual must be read carefully before using this product and the
Analyses of Intravesicular Exosomal Proteins Using a Nano-Plasmonic System
 Supporting Information Analyses of Intravesicular Exosomal Proteins Using a Nano-Plasmonic System Jongmin Park 1, Hyungsoon Im 1.2, Seonki Hong 1, Cesar M. Castro 1,3, Ralph Weissleder 1,4, Hakho Lee 1,2
Supporting Information Analyses of Intravesicular Exosomal Proteins Using a Nano-Plasmonic System Jongmin Park 1, Hyungsoon Im 1.2, Seonki Hong 1, Cesar M. Castro 1,3, Ralph Weissleder 1,4, Hakho Lee 1,2
Ready-to-use Lentiviral Particles for intracelular labeling
 Ready-to-use Lentiviral Particles for intracelular labeling (LocLight TM Living cell imaging lentivirus for sub-cellular localization) LocLight TM cell organelle labeling lentivirus are provided as 200ul/per
Ready-to-use Lentiviral Particles for intracelular labeling (LocLight TM Living cell imaging lentivirus for sub-cellular localization) LocLight TM cell organelle labeling lentivirus are provided as 200ul/per
ab CytoPainter Golgi/ER Staining Kit
 ab139485 CytoPainter Golgi/ER Staining Kit Instructions for Use Designed to detect Golgi bodies and endoplasmic reticulum by microscopy This product is for research use only and is not intended for diagnostic
ab139485 CytoPainter Golgi/ER Staining Kit Instructions for Use Designed to detect Golgi bodies and endoplasmic reticulum by microscopy This product is for research use only and is not intended for diagnostic
B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small cell lung cancer
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
Pre-made Reporter Lentivirus for MAPK/ERK Signal Pathway
 Pre-made Reporter for MAPK/ERK Signal Pathway Cat# Product Name Amounts LVP957-P or: LVP957-P-PBS SRE-GFP (Puro) LVP958-P or: LVP958-P-PBS SRE-RFP (Puro) LVP959-P or: LVP959-P-PBS SRE-Luc (Puro) LVP960-P
Pre-made Reporter for MAPK/ERK Signal Pathway Cat# Product Name Amounts LVP957-P or: LVP957-P-PBS SRE-GFP (Puro) LVP958-P or: LVP958-P-PBS SRE-RFP (Puro) LVP959-P or: LVP959-P-PBS SRE-Luc (Puro) LVP960-P
Protocol for Gene Transfection & Western Blotting
 The schedule and the manual of basic techniques for cell culture Advanced Protocol for Gene Transfection & Western Blotting Schedule Day 1 26/07/2008 Transfection Day 3 28/07/2008 Cell lysis Immunoprecipitation
The schedule and the manual of basic techniques for cell culture Advanced Protocol for Gene Transfection & Western Blotting Schedule Day 1 26/07/2008 Transfection Day 3 28/07/2008 Cell lysis Immunoprecipitation
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Pre-made Lentiviral Particles for Fluorescent Proteins
 Pre-made Lentiviral Particles for Fluorescent Proteins Catalog# Product Name Amounts Fluorescent proteins expressed under sucmv promoter: LVP001 LVP001-PBS LVP002 LVP002-PBS LVP011 LVP011-PBS LVP012 LVP012-PBS
Pre-made Lentiviral Particles for Fluorescent Proteins Catalog# Product Name Amounts Fluorescent proteins expressed under sucmv promoter: LVP001 LVP001-PBS LVP002 LVP002-PBS LVP011 LVP011-PBS LVP012 LVP012-PBS
Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
 Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Catalog Number KA1538 48 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use...
Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Catalog Number KA1538 48 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use...
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening
 Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation
 SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplementary Information
 Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
EPIGENTEK. EpiQuik Global Acetyl Histone H3K27 Quantification Kit (Colorimetric) Base Catalog # P-4059 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Global Acetyl Histone H3K27 Quantification Kit (Colorimetric) Base Catalog # P-4059 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Acetyl Histone H3K27 Quantification Kit (Colorimetric)
EpiQuik Global Acetyl Histone H3K27 Quantification Kit (Colorimetric) Base Catalog # P-4059 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Acetyl Histone H3K27 Quantification Kit (Colorimetric)
EPIGENTEK. EpiQuik Global Histone H4 Acetylation Assay Kit. Base Catalog # P-4009 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Global Histone H4 Acetylation Assay Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Histone H4 Acetylation Assay Kit is suitable for specifically measuring global
EpiQuik Global Histone H4 Acetylation Assay Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Histone H4 Acetylation Assay Kit is suitable for specifically measuring global
genome edited transient transfection, CMV promoter
 Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
04_polarity. The formation of synaptic vesicles
 Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
XCF TM COMPLETE Exosome and cfdna Isolation Kit (for Serum & Plasma)
 XCF TM COMPLETE Exosome and cfdna Isolation Kit (for Serum & Plasma) Cat# XCF100A-1 User Manual Store kit components at +4ºC and +25ºC Version 1 2/2/2017 A limited-use label license covers this product.
XCF TM COMPLETE Exosome and cfdna Isolation Kit (for Serum & Plasma) Cat# XCF100A-1 User Manual Store kit components at +4ºC and +25ºC Version 1 2/2/2017 A limited-use label license covers this product.
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION
 Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
Islet viability assay and Glucose Stimulated Insulin Secretion assay RT-PCR and Western Blot
 Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Global Histone H3 Acetylation Assay Kit
 Global Histone H3 Acetylation Assay Kit Catalog Number KA0633 96 assays Version: 06 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Global Histone H3 Acetylation Assay Kit Catalog Number KA0633 96 assays Version: 06 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
20X Buffer (Tube1) 96-well microplate (12 strips) 1
 PROTOCOL MitoProfile Rapid Microplate Assay Kit for PDH Activity and Quantity (Combines Kit MSP18 & MSP19) 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MSP20 Rev.1 DESCRIPTION MitoProfile Rapid Microplate
PROTOCOL MitoProfile Rapid Microplate Assay Kit for PDH Activity and Quantity (Combines Kit MSP18 & MSP19) 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MSP20 Rev.1 DESCRIPTION MitoProfile Rapid Microplate
Pre-made Reporter Lentivirus for NF-κB Signal Pathway
 Pre-made Reporter for NF-κB Signal Pathway Cat# Product Name Amounts LVP965-P or: LVP965-P-PBS NFKB-GFP (Puro) LVP966-P or: LVP966-P-PBS NFKB-RFP (Puro) LVP967-P or: LVP967-P-PBS NFKB-Luc (Puro) LVP968-P
Pre-made Reporter for NF-κB Signal Pathway Cat# Product Name Amounts LVP965-P or: LVP965-P-PBS NFKB-GFP (Puro) LVP966-P or: LVP966-P-PBS NFKB-RFP (Puro) LVP967-P or: LVP967-P-PBS NFKB-Luc (Puro) LVP968-P
Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit
 PROTOCOL Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit DESCRIPTION Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit Sufficient materials
PROTOCOL Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit DESCRIPTION Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit Sufficient materials
Fluorescence Microscopy
 Fluorescence Microscopy Imaging Organelles Mitochondria Lysosomes Nuclei Endoplasmic Reticulum Plasma Membrane F-Actin AAT Bioquest Introduction: Organelle-Selective Stains Organelles are tiny, specialized
Fluorescence Microscopy Imaging Organelles Mitochondria Lysosomes Nuclei Endoplasmic Reticulum Plasma Membrane F-Actin AAT Bioquest Introduction: Organelle-Selective Stains Organelles are tiny, specialized
EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
 EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Total Histone H3 Acetylation Detection Fast Kit (Colorimetric)
Instructions. Fuse-It-Color. Overview. Specifications
 Membrane fusion is a novel and highly superior method for incorporating various molecules and particles into mammalian cells. Cargo-specific liposomal carriers are able to attach and rapidly fuse with
Membrane fusion is a novel and highly superior method for incorporating various molecules and particles into mammalian cells. Cargo-specific liposomal carriers are able to attach and rapidly fuse with
Mesenchymal stem cells release exosomes that transfer mirnas to endothelial cells and promote angiogenesis
 Mesenchymal stem cells release exosomes that transfer mirnas to endothelial cells and promote angiogenesis Tanja Wagner 1 Introduction 2 Mesenchymal stem cells (MSCs) non-haematopoietic, multipotent stem
Mesenchymal stem cells release exosomes that transfer mirnas to endothelial cells and promote angiogenesis Tanja Wagner 1 Introduction 2 Mesenchymal stem cells (MSCs) non-haematopoietic, multipotent stem
Protein MultiColor Stable, Low Range
 Product Name: DynaMarker Protein MultiColor Stable, Low Range Code No: DM670L Lot No: ******* Size: 200 μl x 3 (DM670 x 3) (120 mini-gel lanes) Storage: 4 C Stability: 12 months at 4 C Storage Buffer:
Product Name: DynaMarker Protein MultiColor Stable, Low Range Code No: DM670L Lot No: ******* Size: 200 μl x 3 (DM670 x 3) (120 mini-gel lanes) Storage: 4 C Stability: 12 months at 4 C Storage Buffer:
The Schedule and the Manual of Basic Techniques for Cell Culture
 The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
Purification and Fluorescent Labeling of Exosomes Asuka Nanbo 1*, Eri Kawanishi 2, Ryuji Yoshida 2 and Hironori Yoshiyama 3
 Purification and Fluorescent Labeling of Exosomes Asuka Nanbo 1*, Eri Kawanishi 2, Ryuji Yoshida 2 and Hironori Yoshiyama 3 1 Graduate School of Medicine, Hokkaido University, Sapporo, Japan; 2 Graduate
Purification and Fluorescent Labeling of Exosomes Asuka Nanbo 1*, Eri Kawanishi 2, Ryuji Yoshida 2 and Hironori Yoshiyama 3 1 Graduate School of Medicine, Hokkaido University, Sapporo, Japan; 2 Graduate
Large Scale Infection for Pooled Screens of shrna libraries
 Last modified 01/11/09 Large Scale Infection for Pooled Screens of shrna libraries Biao Luo, Glenn Cowley, Michael Okamoto, Tanaz Sharifnia This protocol can be further optimized if cells being used are
Last modified 01/11/09 Large Scale Infection for Pooled Screens of shrna libraries Biao Luo, Glenn Cowley, Michael Okamoto, Tanaz Sharifnia This protocol can be further optimized if cells being used are
Essential Medium, containing 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin. Huvec were cultured in
 Supplemental data Methods Cell culture media formulations A-431 and U-87 MG cells were maintained in Dulbecco s Modified Eagle s Medium. FaDu cells were cultured in Eagle's Minimum Essential Medium, containing
Supplemental data Methods Cell culture media formulations A-431 and U-87 MG cells were maintained in Dulbecco s Modified Eagle s Medium. FaDu cells were cultured in Eagle's Minimum Essential Medium, containing
Service and Collaboration
 Summary Section 7 Introduction 68 Exosome and Microvesicle Isolation 69 EV protein quantification, sceening and profiling 69 Nanoparticle Tracking Analysis (NTA) 70 EV Nucleic Acid isolation 70 Nucleic
Summary Section 7 Introduction 68 Exosome and Microvesicle Isolation 69 EV protein quantification, sceening and profiling 69 Nanoparticle Tracking Analysis (NTA) 70 EV Nucleic Acid isolation 70 Nucleic
NF-κB p65 (Phospho-Thr254)
 Assay Biotechnology Company www.assaybiotech.com Tel: 1-877-883-7988 Fax: 1-877-610-9758 NF-κB p65 (Phospho-Thr254) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02015 Please read the provided manual
Assay Biotechnology Company www.assaybiotech.com Tel: 1-877-883-7988 Fax: 1-877-610-9758 NF-κB p65 (Phospho-Thr254) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02015 Please read the provided manual
Human Immunodeficiency Virus type 1 (HIV-1) gp120 / Glycoprotein 120 ELISA Pair Set
 Human Immunodeficiency Virus type 1 (HIV-1) gp120 / Glycoprotein 120 ELISA Pair Set Catalog Number : SEK11233 To achieve the best assay results, this manual must be read carefully before using this product
Human Immunodeficiency Virus type 1 (HIV-1) gp120 / Glycoprotein 120 ELISA Pair Set Catalog Number : SEK11233 To achieve the best assay results, this manual must be read carefully before using this product
Molecular Trafficking
 SCBM 251 Molecular Trafficking Assoc. Prof. Rutaiwan Tohtong Department of Biochemistry Faculty of Science rutaiwan.toh@mahidol.ac.th Lecture outline 1. What is molecular trafficking? Why is it important?
SCBM 251 Molecular Trafficking Assoc. Prof. Rutaiwan Tohtong Department of Biochemistry Faculty of Science rutaiwan.toh@mahidol.ac.th Lecture outline 1. What is molecular trafficking? Why is it important?
Supporting Information
 Supporting Information The Effects of Spacer Length and Composition on Aptamer-Mediated Cell-Specific Targeting with Nanoscale PEGylated Liposomal Doxorubicin Hang Xing +, [a] Ji Li +, [a] Weidong Xu,
Supporting Information The Effects of Spacer Length and Composition on Aptamer-Mediated Cell-Specific Targeting with Nanoscale PEGylated Liposomal Doxorubicin Hang Xing +, [a] Ji Li +, [a] Weidong Xu,
A Microfluidic ExoSearch Chip for Multiplexed Exosome Detection Towards Bloodbased Ovarian Cancer Diagnosis
 Electronic Supplementary Material (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2015 Supplementary Information A Microfluidic ExoSearch Chip for Multiplexed Exosome Detection
Electronic Supplementary Material (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2015 Supplementary Information A Microfluidic ExoSearch Chip for Multiplexed Exosome Detection
Exosomes Immunocapture and Isolation Tools: Immunobeads
 Product Insert Exosomes Immunocapture and Isolation Tools: Immunobeads HBM-BOLF-##/##-# HBM-BOLC-##/##-# HBM-BTLF-##/##-# Overall Exosome capture from Human Biological fluids Overall Exosome capture from
Product Insert Exosomes Immunocapture and Isolation Tools: Immunobeads HBM-BOLF-##/##-# HBM-BOLC-##/##-# HBM-BTLF-##/##-# Overall Exosome capture from Human Biological fluids Overall Exosome capture from
Chapter 3. Expression of α5-megfp in Mouse Cortical Neurons. on the β subunit. Signal sequences in the M3-M4 loop of β nachrs bind protein factors to
 22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
VIROMER ONE. standardized transfection reagent
 VIROMER ONE standardized transfection reagent 1 Doing transfection is like making coffee. Same procedure, somewhat different result each time you do it. Then came Nespresso. An easy, comfortable and clean
VIROMER ONE standardized transfection reagent 1 Doing transfection is like making coffee. Same procedure, somewhat different result each time you do it. Then came Nespresso. An easy, comfortable and clean
Human LDL Receptor / LDLR ELISA Pair Set
 Human LDL Receptor / LDLR ELISA Pair Set Catalog Number : SEK10231 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized in
Human LDL Receptor / LDLR ELISA Pair Set Catalog Number : SEK10231 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized in
PROTOCOL: OPTIMIZATION OF LENTIVIRAL TRANSDUCTION USING SPINFECTION
 Last Modified: April 2018 Last Review: October 2018 PROTOCOL: OPTIMIZATION OF LENTIVIRAL TRANSDUCTION USING SPINFECTION Table of Contents 1. Brief Description 1 2. Materials and Reagents.1 3. Optimization
Last Modified: April 2018 Last Review: October 2018 PROTOCOL: OPTIMIZATION OF LENTIVIRAL TRANSDUCTION USING SPINFECTION Table of Contents 1. Brief Description 1 2. Materials and Reagents.1 3. Optimization
Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
Gladstone Institutes, University of California (UCSF), San Francisco, USA
 Fluorescence-linked Antigen Quantification (FLAQ) Assay for Fast Quantification of HIV-1 p24 Gag Marianne Gesner, Mekhala Maiti, Robert Grant and Marielle Cavrois * Gladstone Institutes, University of
Fluorescence-linked Antigen Quantification (FLAQ) Assay for Fast Quantification of HIV-1 p24 Gag Marianne Gesner, Mekhala Maiti, Robert Grant and Marielle Cavrois * Gladstone Institutes, University of
Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival
 Supplementary Information for Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival Tatsuro Kawamura 1, Makoto Kawatani 1, Makoto Muroi, Yasumitsu Kondoh,
Supplementary Information for Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival Tatsuro Kawamura 1, Makoto Kawatani 1, Makoto Muroi, Yasumitsu Kondoh,
Pre-made Lentiviral Particles for intracelular labeling: (LocLight TM Living cell imaging lentivirus for sub-cellular localization)
 Pre-made Lentiviral Particles for intracelular labeling: (LocLight TM Living cell imaging lentivirus for sub-cellular localization) LocLight TM cell organelle labeling lentivirus is provided as 200ul/per
Pre-made Lentiviral Particles for intracelular labeling: (LocLight TM Living cell imaging lentivirus for sub-cellular localization) LocLight TM cell organelle labeling lentivirus is provided as 200ul/per
EPIGENTEK. EpiQuik Global Histone H3 Acetylation Assay Kit. Base Catalog # P-4008 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Global Histone H3 Acetylation Assay Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Histone H3 Acetylation Assay Kit is suitable for specifically measuring global
EpiQuik Global Histone H3 Acetylation Assay Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Histone H3 Acetylation Assay Kit is suitable for specifically measuring global
Chapter 6. Antigen Presentation to T lymphocytes
 Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
PRODUCT INFORMATION & MANUAL
 PRODUCT INFORMATION & MANUAL 0.4 micron for Overall Exosome Isolation (Cell Media) NBP2-49826 For research use only. Not for diagnostic or therapeutic procedures. www.novusbio.com - P: 303.730.1950 - P:
PRODUCT INFORMATION & MANUAL 0.4 micron for Overall Exosome Isolation (Cell Media) NBP2-49826 For research use only. Not for diagnostic or therapeutic procedures. www.novusbio.com - P: 303.730.1950 - P:
6. In figure 3e, the catalase is actually catalase mrna, and should be labeled appropriately.
 Reviewers' comments: Reviewer #1 (Remarks to the Author): Summary: The authors engineered an exosome-producing genetic system (EXOtic) that, when transiently transfected into mammalian cells, is capable
Reviewers' comments: Reviewer #1 (Remarks to the Author): Summary: The authors engineered an exosome-producing genetic system (EXOtic) that, when transiently transfected into mammalian cells, is capable
Human SH-SY5Y neuroblastoma cells (A.T.C.C., Manassas, VA) were cultured in DMEM, F-12
 SUPPLEMENTARY METHODS Cell cultures Human SH-SY5Y neuroblastoma cells (A.T.C.C., Manassas, VA) were cultured in DMEM, F-12 Ham with 25 mm HEPES and NaHCO 3 (1:1) and supplemented with 10% (v/v) FBS, 1.0
SUPPLEMENTARY METHODS Cell cultures Human SH-SY5Y neuroblastoma cells (A.T.C.C., Manassas, VA) were cultured in DMEM, F-12 Ham with 25 mm HEPES and NaHCO 3 (1:1) and supplemented with 10% (v/v) FBS, 1.0
Electron micrograph of phosphotungstanic acid-stained exosomes derived from murine
 1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
Lumino Firefly Luciferase Assay
 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Lumino Firefly Luciferase Assay (Cat. # 786 1267, 786 1268) think proteins! think G-Biosciences
G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Lumino Firefly Luciferase Assay (Cat. # 786 1267, 786 1268) think proteins! think G-Biosciences
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): The submitted manuscript by Heegon Kim reports on a cooperative targeting system in which synthetic and biological nano-components participate
Reviewers' comments: Reviewer #1 (Remarks to the Author): The submitted manuscript by Heegon Kim reports on a cooperative targeting system in which synthetic and biological nano-components participate
Cellular compartments
 Cellular compartments 1. Cellular compartments and their function 2. Evolution of cellular compartments 3. How to make a 3D model of cellular compartment 4. Cell organelles in the fluorescent microscope
Cellular compartments 1. Cellular compartments and their function 2. Evolution of cellular compartments 3. How to make a 3D model of cellular compartment 4. Cell organelles in the fluorescent microscope
Constitutive Reporter Lentiviral Vectors Expressing Fluorescent Proteins
 Constitutive Reporter Lentiviral Vectors Expressing Fluorescent Proteins www.vectalys.com/products/ Constitutive Reporter Lentiviral Vectors Catalog Number referring to this User Manual: 0008VCT; 0009VCT;
Constitutive Reporter Lentiviral Vectors Expressing Fluorescent Proteins www.vectalys.com/products/ Constitutive Reporter Lentiviral Vectors Catalog Number referring to this User Manual: 0008VCT; 0009VCT;
LDL Uptake Flow Cytometry Assay Kit
 LDL Uptake Flow Cytometry Assay Kit Item No. 601470 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
LDL Uptake Flow Cytometry Assay Kit Item No. 601470 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Supplementary Material
 Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Extracellular Vesicle Quantification Kits
 Summary Section 3 Introduction 34 ExoTEST TM Ready to Use ELISA quantification kit 35 Exo-FACS Ready to Use kit 38 Ordering information Exosome research is a relatively novel and exciting field of biomedical
Summary Section 3 Introduction 34 ExoTEST TM Ready to Use ELISA quantification kit 35 Exo-FACS Ready to Use kit 38 Ordering information Exosome research is a relatively novel and exciting field of biomedical
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human
 Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Influenza B Hemagglutinin / HA ELISA Pair Set
 Influenza B Hemagglutinin / HA ELISA Pair Set Catalog Number : SEK11053 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized
Influenza B Hemagglutinin / HA ELISA Pair Set Catalog Number : SEK11053 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
EPIGENTEK. EpiQuik HDAC2 Assay Kit (Colorimetric) Base Catalog # P-4006 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik HDAC2 Assay Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik HDAC2 Assay Kit is very suitable for measuring HDAC2 levels from various fresh tissues and
EpiQuik HDAC2 Assay Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik HDAC2 Assay Kit is very suitable for measuring HDAC2 levels from various fresh tissues and
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
INSTRUCTIONS Pierce Primary Cardiomyocyte Isolation Kit
 INSTRUCTIONS Pierce Primary Cardiomyocyte Isolation Kit 88281 Number Description 88281 Pierce Primary Cardiomyocyte Isolation Kit, contains sufficient reagents to isolate cardiomyocytes from 50 neonatal
INSTRUCTIONS Pierce Primary Cardiomyocyte Isolation Kit 88281 Number Description 88281 Pierce Primary Cardiomyocyte Isolation Kit, contains sufficient reagents to isolate cardiomyocytes from 50 neonatal
Product Manual. Human LDLR ELISA Kit. Catalog Number. FOR RESEARCH USE ONLY Not for use in diagnostic procedures
 Product Manual Human LDLR ELISA Kit Catalog Number STA-386 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cholesterol is an essential component of cellular membranes,
Product Manual Human LDLR ELISA Kit Catalog Number STA-386 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cholesterol is an essential component of cellular membranes,
Impact of hyper-o-glcnacylation on apoptosis and NF-κB activity SUPPLEMENTARY METHODS
 SUPPLEMENTARY METHODS 3D culture and cell proliferation- MiaPaCa-2 cell culture in 3D was performed as described previously (1). Briefly, 8-well glass chamber slides were evenly coated with 50 µl/well
SUPPLEMENTARY METHODS 3D culture and cell proliferation- MiaPaCa-2 cell culture in 3D was performed as described previously (1). Briefly, 8-well glass chamber slides were evenly coated with 50 µl/well
Human HBcAb IgM ELISA kit
 Human HBcAb IgM ELISA kit Catalog number: NR-R10163 (96 wells) The kit is designed to qualitatively detect HBcAb IgM in human serum or plasma. FOR RESEARCH USE ONLY. NOT FOR DIAGNOSTIC OR THERAPEUTIC PURPOSES
Human HBcAb IgM ELISA kit Catalog number: NR-R10163 (96 wells) The kit is designed to qualitatively detect HBcAb IgM in human serum or plasma. FOR RESEARCH USE ONLY. NOT FOR DIAGNOSTIC OR THERAPEUTIC PURPOSES
Supplemental Data Figure S1 Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells.
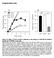 Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Validation & Assay Performance Summary
 Validation & Assay Performance Summary LanthaScreen IGF-1R GripTite Cells Cat. no. K1834 Modification Detected: Phosphorylation of Multiple Tyr Residues on IGF-1R LanthaScreen Cellular Assay Validation
Validation & Assay Performance Summary LanthaScreen IGF-1R GripTite Cells Cat. no. K1834 Modification Detected: Phosphorylation of Multiple Tyr Residues on IGF-1R LanthaScreen Cellular Assay Validation
Oxford Expression Technologies Ltd
 Oxford Expression Technologies Ltd Founded in 2007 as a spin out from Oxford Brookes University and Natural Environment Research Council Technology based on the insect baculovirus expression vectors (BEVs)
Oxford Expression Technologies Ltd Founded in 2007 as a spin out from Oxford Brookes University and Natural Environment Research Council Technology based on the insect baculovirus expression vectors (BEVs)
