Running title: Human adipocyte metabolism at the genome-scale
|
|
|
- Abel Patrick
- 5 years ago
- Views:
Transcription
1 Integration of clinical data with genome-scale metabolic model of the human adipocyte Running title: Human adipocyte metabolism at the genome-scale Adil Mardinoglu 1, Rasmus Agren 1, Caroline Kampf 2, Anna Asplund 2, Intawat Nookaew 1, Peter Jacobson 3, Andrew J. Walley 4, Philippe Froguel 4,5, Lena M. Carlsson 3, Mathias Uhlen 6 and Jens Nielsen 1, * 1 Department of Chemical and Biological Engineering, Chalmers University of Technology, Gothenburg, Sweden 2 Department of Immunology, Genetics and Pathology, Science for Life Laboratory, Uppsala University, Uppsala, Sweden 3 Department of Molecular and Clinical Medicine and Center for Cardiovascular and Metabolic Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden 4 Department of Genomics of Common Diseases, School of Public Health, Imperial College London, Hammersmith Hospital, London, UK 5 CNRS 8090-Institute of Biology, Pasteur Institute, Lille, France 6 Department of Proteomics, School of Biotechnology, AlbaNova University Center, Royal Institute of Technology (KTH), Stockholm, Sweden * Corresponding author nielsenj@chalmers.se Tel: Fax:
2 Table of Contents Supplementary Text... 3 Adipocyte genome-scale metabolic model coverage on functions... 3 Analysis of transcriptome data for subcutaneous adipose tissue and integrated analysis... 5 Integration of clinical data into the genome-scale metabolic model for adipocytes... 6 Supplementary Tables... 7 Table S Table S Table S Table S Table S Table S Table S Supplementary Figures Figure S Figure S Figure S Figure S Figure S Figure S Figure S Figure S Supplementary Datasets Supplementary References
3 Supplementary Text Adipocyte genome-scale metabolic model coverage on functions The comprehensive iadipocytes1809 covers major known metabolism related pathways as well as extensive knowledge of lipid metabolism in human adipocytes. Fat absorption and transport to adipocytes: The process of fat digestion is complex and involves multi coordination of lingual, gastric, intestinal, biliary and pancreatic functions. The short and medium-chain FAs resulting from the digestion process are absorbed in the gut and then transported to the liver, via the portal vein, where it is oxidized. Other yields of the digestion process including long-chain FAs, monoacylglycerol (MAG), lysophospholipids and cholesterols are combined with bile salts and absorbed by the intestine wall. The longchain FAs are converted to TAGs as well as cholesterol and lysophospholipids are converted to their esters in the intestine and recently formed TAGs, phospholipids (PLs) and esters are combined into de novo synthesized apolipoproteins to form chylomicrons. Produced chylomicrons in enterocytes are transported to the bloodstream via the lymph vessels and they are to be used as FA source by the adipose and other soft tissues. Chylomicrons in the bloodstream are hydrolyzed to MAG, FAs, and glycerol by lipoprotein lipase (LPL) to be stored as TAGs in adipocytes or to be used as an energy source. Although small amount of FAs are synthesized by de novo FA synthesis inside the cell, most of the FAs deposited in adipocytes are taken up from two potential sources: non-esterified FAs (NEFAs) and lipoproteins (Table S5) including chylomicrons, very-low-density lipoprotein (VLDL), low-density lipoproteins (LDL) and high-density lipoproteins (HDL) through LPL (Duncan et al., 2007; Guilherme et al., 2008; Large et al., 2004). There is a clear preference for uptake of FAs from lipoproteins compared to plasma NEFAs whereas chylomicrons are a better source of FAs in the post-prandial state and VLDLs are a better source of FAs in post-absorptive state (Bickerton et al., 2007; Large et al., 2004). After the removal of the FAs in chylomicrons by adipocytes, remnants of chylomicrons are cleared from the blood by the liver LDL receptors and their related proteins. The liver catabolizes chylomicrons remnants, forms VLDL by combining resynthesized TAGs with small amounts of cholesterol and PLs and releases them into the blood to be used as FAs sources for adipocytes in the post-absorptive state. FAs in VLDL are taken up through LPL hydrolysis by adipocytes and remnants of VLDL are cleared by the liver to be transformed into intermediate-density lipoproteins (IDL) and finally into LDL. Adipose tissue stores more than 25% of the total body cholesterol and de novo synthesized cholesterol and cholesterol taken up via lipoproteins are integrated into the plasma membrane and lipid droplets (LD) structure. In adipocytes, de novo cholesterol synthesis is limited and the greater part of the cholesterol in the cell is taken up from lipoproteins through LPL hydrolysis. LDL and HDL are the potential carriers for cholesterol and CEs from plasma to adipocytes where CEs are hydrolyzed to free cholesterols and re-esterified. In the reconstructed GEM for adipocytes, 59 different common long and very long chain FAs (Table S4) in human plasma can be taken up as NEFAs and lipoproteins (Figure S5, S6 and 3
4 S7). Cholesterol and its 59 different CEs can also be taken up from LDLs and HDLs. In the post-prandial state, FAs and CEs can be incorporated into LD structures and in the postabsorptive state LDs can be broken down to FAs and CEs. De novo synthesis of fatty acids: Even though factors such as background diet, physical activity, genetics and hormones can influence de novo FAs synthesis, synthetic processes are quite limited in adipocytes. The main sources of the glycerol-3-phosphate for FA synthesis are glucose that is taken up from blood by the insulin-regulated GLUT-4 transporter, lactate/pyruvate that can be converted to glycerol-3-phosphate via glyceroneogenesis, and from catabolism of amino acids. The synthetic process starts with the breakdown of the excess dietary carbohydrates to acetyl-coa that is a precursor for biosynthesis of palmitic acid (C:16:0). Palmitic acid can be further elongated to longer chain saturated FAs by adding acetyl groups, through the action of FA elongation systems in the cytoplasm. In mammalian systems, desaturation of de novo synthesized saturated FAs stops with the formation of the n- 9 series monounsaturated fatty acids (MUFA) performed by 9 desaturase. The products of de novo synthesis are esterified with glycerol to form TAGs and the TAGs are stored in LDs. A healthy subject has the capacity of synthesizing about 20% percent of the FAs in newly formed TAGs via de novo synthesis and this knowledge is used in the formation of LDs in the model (Strawford et al., 2004). In the GEM, with the uptake of the essential FAs linoleate and linolenate, 57 different long and very long chain FAs can be synthesized and can be incorporated into the formation of LDs. Metabolism of linoleate and linolenate: linoleate (LA), unsaturated omega-6 fatty acid, and linolenate (ALA), polyunsaturated omega-3 fatty acid cannot be synthesized by mammalian systems since mammals cannot introduce double bonds between Δ10 and the methyl terminal end. LA and ALA are essential FAs for the synthesis of arachidonate (AA) and eicosapentaenoate (EPA), that serve as precursors for long-chain polyunsaturated fatty acids (LCPUFAs) and eicosanoids and they must be obtained from the diet. After the uptake of LA and ALA, different steps are involved as follows: double bond is inserted at the Δ6 position of LA and ALA by the action of Δ6 desaturase, two carbon chains are elongated, another double bond is inserted at the Δ5 position by Δ5-desaturase to form AA (20:4n-6) and EPA (20:5n- 3), two carbon chain is elongated to form 22:4n-6 and 22:5n-3, and further two carbon chains are elongated to produce 24:4n-6 and 24:5n-3, respectively. Subsequently, 24:4n-6 and 24:5n- 3 are desaturated by Δ6 desaturase to yield 24:5n-6 and 24:6n-3 that are shortened two carbon chain by the beta oxidation in the peroxisome to form docosapentaenoic acid (DPA) and docosahexaenoic acid (DHA). The LA and ALA pathways are independent of each other and there are no crossover reactions between them. There is, however, a competition between the two pathways as the reactions in both pathways are catalyzed by the same enzymes (Figure S6). In the GEM, the eicosanoids such as prostaglandins (PG), prostacyclins (PGI), thromboxanes (TX), leukotrienes (LT), hydroperoxytetraenoic acids (HPETE), hydroxyeicosatetraenoic acids (HETE) and lipoxins are formed via long-chain omega-6 and omega-3 C20 PUFA. This 4
5 biological function is performed by two different enzymes such as Cyclooxygenase (COX) enzyme that converts the C20 FAs to prostanoids including PG, PGI and TX and lipooxygenase (LOX) enzyme that converts the C20 FAs to HPETE and further to LT, HETE and lipoxins. There are also two isoforms of COX enzymes including COX 1 that controls the physiological roles of eicosanoids, and COX 2 which is activated by some biological processes such as inflammation. DHA-derived signaling molecules docosanoids have also been formed via LA and ALA pathways by COX-2 and 5-LOX enzymes. Docosanoids are also termed as D-series resolvins and protectins (neuroprotectins D1) that are both neuroprotective and anti-inflammatory. In addition, both the eicosanoids and the docosanoids are involved in different biological processes such as inflammation, immune response and cell growth and proliferation. Omega-3 and Omega-6 FAs regulates the enzymes that perform the lipid metabolism (i.e. inhibit FA synthesis in adipose tissue and contain transcription of the leptin gene that regulates appetite, body weight and adiposity) whereas omega-3 FAs have a number of other anti-inflammatory effects via cytokines family of proteins. Oxidation of fatty acids: During starvation, the TAGs in which the body stores energy provide FAs as a direct energy source for other tissues and a substrate for production of ketone bodies. In the model, -oxidation of LCFA occurs in the mitochondria and -oxidation of VLCFA starts in the peroxisome and continues either in the peroxisome or the mitochondria. Transport of FAs (acyl-coa) into the mitochondria requires the four step carnitine shuttle whereas transport to the peroxisome does not. In the model, phytanic acid that is taken up from the blood cannot undergo -oxidation due to its β-methyl branch. Therefore, it is broken down into pristanic acid by removal of a single carbon from the carboxyl end via -oxidation that takes place in the peroxisome. Other Major Metabolic Pathways: Adipose tissue takes up glucose in the post-prandial and post-absorptive state (McQuaid et al., 2011) and most of the glucose taken up by adipocytes enters glycolysis, which is an important pathway for TAGs synthesis by providing glycerol-3- phosphate necessary for FAs esterification. Some part of the glucose may also enter the tricarboxylic acid cycle (TCA cycle) for energy production. Analysis of transcriptome data for subcutaneous adipose tissue and integrated analysis Gene expression in subcutaneous adipose tissue (SAT) of subjects involved in Swedish Obese Subjects (SOS) Sip Pair Study (Table S1) was analyzed. Gene expression levels of overweight and obese groups are compared with the lean group and the Limma package was used to identify significant genes that were over- or under expressed (adjusted p-values cutoff level < 0.001). Linear models were fitted to the data using linear and robust regression separately before applying an empirical Bayes shrinkage method. Correction for multiple testing was performed using Storey s FDR procedure on the p-values of the shrunk test statistics to generate q-values. The q-values, generated for each probe set in each group were calculated using the Reporter algorithm (Oliveira et al., 2008; Patil and Nielsen, 2005) in order to summarize the transcriptional changes in terms of more general features such as biological process Gene Ontology (BP:GO) terms (Berardini et al., 2010) and KEGG pathways (Kanehisa et al., 2012). Figures S1 and S2 show the output for KEGG pathways for male and female subjects, respectively (cutoff p <1e-04). Figures S3 and S4 show the output for BP:GO 5
6 for male and female subjects, respectively (cutoff p <1e-04). During the mapping of the probe-sets to the KEGG pathways and BP:GO terms, the manufacturer mapping file was used. Integration of clinical data into the genome-scale metabolic model for adipocytes In order to reconstruct a predictive and functional model for adipocytes in white adipose tissue (WAT), it is necessary to incorporate some relevant experimental measurements (i.e. Fatty Acid (FA) composition of plasma and WAT). Mitrou et. al. (Mitrou et al., 2009) investigated insulin action on glucose disposal in SAT and muscle tissue after the consumption of a mixed meal in different healthy subject groups. The study consisted of 30 obese non-diabetic women (age 34±1 year, body mass index (BMI) 47±1 kg/m²) and 10 lean women (age 39±4 year, BMI 23±1 kg/m²). The glucose and insulin levels were measured at the veins draining the abdominal SAT and forearm muscles and in the radial artery for 360 min. Fasting TAGs was increased in obese subjects (1.56±0.2 mmol/l) versus lean subjects (0.93±0.1 mmol/l) and HDL-cholesterol was decreased in obese subjects (0.95±0.05 mmol/l) versus lean subjects (1.2±0.07mmol/l). Total cholesterol and LDL-cholesterol are also increased in obese subjects (5.3±0.4 mmol/l and 3.7±0.4 mmol/l) versus lean subjects (4.4±0.2 mmol/l and 2.8±0.2 mmol/l). In post-prandial state, glucose uptake in SAT in obese subjects is less than lean subjects (0.45±0.1 versus 1.1±0.17 µmol/min per 100 ml tissue). However, the average glucose uptake rate in obese subjects (0.275±0.04 mmol/min) is more than lean subjects (0.12±0.02 mmol/min) when SAT glucose uptake was multiplied by tissue mass and expressed as per total fat mass. In obese subjects, plasma insulin level is increased comparing to lean subjects and the glucose uptake rate in SAT is not altered in both postprandial and post-absorptive states (approximately 0.45 µmol/min per 100 ml tissues). Bickerton et. al. (Bickerton et al., 2007) probe the components of fat metabolism in postprandial and post-absorptive states by using a combination of stable isotope labeling and arteriovenous difference measurements. The uptake of the FAs derived from chylomicron- TAGs, VLDL-TAGs and circulating NEFAs are quantified in adipose tissue and skeletal muscle. In post-prandial state, the adipose tissue takes up FAs from both dietary (chylomicron) and VLDL-TAGs with greater fractional extraction of the chylomicron-tags. Significant amount of plasma NEFAs that are minor in comparison to chylomicron-tags FAs are also taken up in the post-prandial state but not in the post-absorptive state. In the late post-absorptive state (6 hours after meal), adipose tissue NEFA uptake rate of the LPLderived fatty acids (~320 nmol/min per 100 g tissues) and plasma NEFA (~-180 nmol/min per 100 g tissues) is reported and used in our study. 6
7 Supplementary Tables Table S1. Clinical data for lean and obese subjects obtained from Swedish Obese Subjects (SOS) Sib Pair Study (Walley et al., 2012). In the SOS Sib Pair Study, human subcutaneous adipose tissue (SAT) samples are obtained from subjects in order to understand the molecular mechanism of the obesity since SAT displays obesity-related changes in gene expression (Wellen and Hotamisligil, 2003). In the SOS Sib Pair Study, in post-absorptive state, the measurements of anthropometry, fat mass (FM), fat-free mass (FFM), blood pressure (BP), fasting glucose, total cholesterol, triacylglycerols, high-density lipoprotein cholesterol (HDLcholesterol), low-density lipoprotein cholesterol (LDL-cholesterol), serum insulin, serum C peptide (C-peptide), and highly sensitive Creactive protein (hs-crp) were performed. Values are given as means ± SD unless stated otherwise. Obesity-discordant Sib-pair study, a sub-study of the Swedish Obese Subjects study FEMALE MALE Variable/Group Lean Overweight Obese Lean Overweight Obese BMI Range (kg/m2) 18.5< BMI< BMI<30 30 BMI 18.5< BMI< BMI<30 30 BMI Sample size (n) Age 36.5 ± ± ± ± ± ± 9.4 BMI (kg/m2) 22.0 ± ± ± ± ± ± 5.1 Waist 76.0 ± ± ± ± ± ± 11.1 WHR 0.79 ± ± ± ± ± ± 0.06 FFM (kg) 48.6 ± ± ± ± ± ± 7.2 FM (kg) 14.9 ± ± ± ± ± ± 8.2 Systolic BP (mmhg) ± ± ± ± ± ± 14.6 Diastolic BP (mmhg) 64.7 ± ± ± ± ± ± 9.5 Insulin (mu/l) 5.07 ± ± ± ± ± ± 8.98 HOMA-IR 1.04 ± ± ± ± ± ± 2.03 C-peptide (mmol/l) 0.46 ± ± ± ± ± ± 0.44 Hs-CRP (mg/l) 1.90 ± ± ± ± ± ± 3.40 Glucose (mmol/l) 4.60 ± ± ± ± ± ± 0.55 Triacylglycerol (mmol/l) 0.69 ± ± ± ± ± ± 0.52 Total Cholesterol (mmol/l) 4.13 ± ± ± ± ± ± 0.69 LDL-cholesterol (mmol/l) 2.37 ± ± ± ± ± ± 0.50 HDL-cholesterol (mmol/l) 1.45 ± ± ± ± ± ±
8 Table S2. Versions of the databases used in the reconstruction of the Human Metabolic Reaction (HMR) database and genome-scale metabolic model for adipocytes, iadipocytes1809. Database Export format Version Downloaded from Recon 1 Flat file, SBML Jan 31, EHMN Flat file June 6, HepatoNet1 Flat file March 1, Reactome Flat file, SBML October 17, HumanCyc Flat file, SBML KEGG Flat file, KGML 48 ftp://ftp.genome.jp/pub/kegg/ HPA Flat file 10 Uniprot Flat file January 9, HMDB Flat file LipidMap Flat file November 9, Table S3. Content of genome-scale metabolic model for adipocytes, iadipocytes1809, Human Metabolic Reaction (HMR) database and previously published literature-based models. GEMs Reactions Metabolites Genes iadipocytes (2497)* 1809 Human Metabolic Reaction (HMR) database (3160)* 3668 ihuman Recon EHMN HepatoNet iab * unique metabolites in the model. 8
9 Table S4. List of the fatty acids used in the genome-scale metabolic model for adipocytes, iadipocytes1809. ID LM_ID SYSTEMATIC_NAME OUR_MODEL_NAME SYNONYMS 1 LMFA Dodecanoic acid lauric acid Lauric acid (C12:0) 2 LMFA Tridecanoic acid tridecylic acid Tridecylic acid(c13:0) 3 LMFA Tetradecanoic acid myristic acid Myristic acid (C14:0) 4 LMFA E-tetradecenoic acid (9E)-tetradecenoic acid Myristoleic acid (C14:1,n-5) 5 LMFA Z-tetradecenoic acid (7Z)-tetradecenoic acid Tetradecenoic acid (C14:1,n-7) 6 LMFA Z-tetradecenoic acid physeteric acid Physeteric acid (C14:1,n-9) 7 LMFA Pentadecanoic acid pentadecylic acid Pentadecanoic acid (C15:0) 8 LMFA Hexadecanoic acid palmitate Palmitic acid (C16:0) 9 LMFA Z-hexadecenoic acid palmitolate Hexadecenoic acid (C16:1,n-7) 10 LMFA Z-hexadecenoic acid 7-palmitoleic acid Hexadecenoic acid (C16:1,n-9) 11 LMFA Heptadecanoic acid margaric acid Margaric acid (C17:0) 12 LMFA Z-heptadecenoic acid (10Z)-heptadecenoic acid Heptadecenoic acid (C17:1,n-7) 13 LMFA Z-heptadecenoic acid 9-heptadecylenic acid Heptadecenoic acid (C17:1,n-8) 14 LMFA Octadecanoic acid stearate Stearic acid (C18:0) 15 LMFA Z-octadecenoic acid (13Z)-octadecenoic acid Octadecenoic acid (C18:1,n-5) 16 LMFA Z-octadecenoic acid cis-vaccenic acid Octadecenoic acid (C18:1,n-7) 17 LMFA Z-octadecenoic acid oleate Oleic acid (C18:1,n-9) 18 LMFA E-octadecenoic acid elaidate Elaidic acid (C18:1,n-9) 19 LMFA Z-octadecenoic acid (7Z)-octadecenoic acid Octadecenoic acid (C18:1,n-11) 20 LMFA Z,9Z-octadecadienoic acid (6Z,9Z)-octadecadienoic acid Isolinoleic acid (C18:2,n-9) 21 LMFA Nonadecanoic acid nonadecylic acid Nonadecylic acid(c19:0) 22 LMFA Eicosanoic acid eicosanoate Eicosanoic acid (C20:0) 23 LMFA Z-eicosenoic acid (13Z)-eicosenoic acid Eicosenoic acid (C20:1,n-7) 24 LMFA Z-eicosenoic acid cis-gondoic acid Eicosenoic acid (C20:1,n-9) 25 LMFA Z-eicosenoic acid 9-eicosenoic acid Eicosenoic acid (C20:1,n-11) 26 LMFA Z,11Z-eicosadienoic acid 8,11-eicosadienoic acid Eicosadienoic acid (C20:2,n-9) 27 LMFA ,8,11-eicosatrienoic acid mead acid Mead acid (C20:3,n-9) 28 LMFA Heneicosanoic acid henicosanoic acid Henicosanoic acid(c21:0) 29 LMFA Docosanoic acid behenic acid Behenic acid (C22:0) 9
10 30 LMFA Z-docosenoic acid cis-erucic acid Docosenoic acid (C22:1,n-9) 31 LMFA Z-docosenoic acid cis-cetoleic acid Docosenoic acid (C22:1,n-11) 32 LMFA Tricosanoic acid tricosanoic acid Tricosanoic-acid (C23:0) 33 LMFA Tetracosanoic acid lignocerate Lignoceric acid (C24:0) 34 LMFA Z-tetracosenoic acid nervonic acid Nervonic acid (C24:1,n-9) 35 LMFA Hexacosanoic acid cerotic acid Cerotic acid (C26:0) 36 LMFA Z-hexacosenoic acid ximenic acid Ximenic acid (C26:1,n-9) 37 LMFA Z,12Z,15Z-octadecatrienoic acid linolenate Linolenic acid (C18:3,n-3)* 38 LMFA Z,9Z,12Z,15Z-octadecatetraenoic acid stearidonic acid Stearidonic-acid (C18:4,n-3) 39 LMFA Z,11Z,14Z,17Z-eicosatetraenoic acid omega-3-arachidonic acid Eicosatetraenoic acid (C20:4,n-3) 40 LMFA Z,8Z,11Z,14Z,17Z-eicosapentaenoic acid EPA Timnodonic acid (C20:5,n-3) 41 LMFA Z,10Z,13Z,16Z,19Z-docosapentaenoic acid DPA Clupanodonic acid (C22:5,n-3) 42 LMFA Z,12Z,15Z,18Z,21Z-tetracosapentaenoic acid (9Z,12Z,15Z,18Z,21Z)-TPA Tetracosapentaenoic acid (C24:5,n-3) 43 LMFA Z,9Z,12Z,15Z,18Z,21Z-tetracosahexaenoic acid (6Z,9Z,12Z,15Z,18Z,21Z)-THA Tetracosahexaenoic acid (C24:6,n-3) 44 LMFA Z,7Z,10Z,13Z,16Z,19Z-docosahexaenoic acid DHA Docosahexaenoic acid (C22:6, n-3) 45 LMFA Z,14Z,17Z-eicosatrienoic acid (11Z,14Z,17Z)-eicosatrienoic acid Eicosatrienoic acid (C20:3,n-3) 46 LMFA Z,16Z,19Z-docosatrienoic acid 13,16,19-docosatrienoic acid Docosatrienoic-acid(C22:3,n-3) 47 N/A 10Z,13Z,16Z,19Z-docosatetraenoic acid 10,13,16,19-docosatetraenoic acid Docosatetraenoic acid C(22:4,n-3) 48 N/A 12Z,15Z,18Z,21Z-tetracosatetraenoic acid 12,15,18,21-tetracosatetraenoic acid Tetracosatetraenoic acid C(24:4,n-3) 49 LMFA Z,12Z-octadecadienoic acid linoleate Linoleic acid (C18:2, n-6)* 50 LMFA Z,9Z,12Z-octadecatrienoic acid gamma-linolenate Gamolenic acid (C18:3,n-6) 51 LMFA Z,11Z,14Z-eicosatrienoic acid dihomo-gamma-linolenate Eicosatrienoic acid (C20:3,n-6) 52 LMFA Z,8Z,11Z,14Z-eicosatetraenoic acid arachidonate Arachidonic acid (C20:4,n-6) 53 LMFA Z,10Z,13Z,16Z-docosatetraenoic acid adrenic acid Adrenic acid (C22:4,n-6) 54 LMFA Z,12Z,15Z,18Z-tetracosatetraenoic acid (9Z,12Z,15Z,18Z)-TTA Tetracosatetraenoic acid (C24:4,n-6) 55 LMFA Z,9Z,12Z,15Z,18Z-tetracosapentaenoic acid (6Z,9Z,12Z,15Z,18Z)-TPA Tetracosapentaenoic acid (C24:5,n-6) 56 LMFA Z,7Z,10Z,13Z,16Z-docosapentaenoic acid (4Z,7Z,10Z,13Z,16Z)-DPA Docosapentaenoic acid (C22:5,n-6) 57 LMFA Z,14Z-eicosadienoic acid (11Z,14Z)-eicosadienoic acid Eicosadienoic acid (C20:2,n-6) 58 LMFA Z,16Z-docosadienoic acid (13Z,16Z)-docosadienoic acid Docosadienoic acid (C22:2,n-6) 59 LMFA Z,13Z,16Z-docosatriynoic acid 10,13,16-docosatriynoic acid Docosatriynoic acid (C22:3,n-6) 10
11 Table S5. Lipid composition of plasma lipoproteins that are fatty acid and cholesterol source for adipocytes (Caballero, 2009; Mills et al., 1984). Lipoproteins Composition (wt%) Size (nm) Origin Cholesterol CE TAGs Phospholipids Protein Chylomicrons Intestine VLDL Liver IDL VLDL LDL VLDL & IDL HDL Liver & Intestine Table S6. Lipid composition of lipid droplets (LDs) in adipocytes (Bartz et al., 2007). LDs are rich in triacylglycerols (TAGs), cholesterol esters (CEs) and an unknown neutral lipid (~19%) that migrated between CEs and TAGs, ether neutral lipid monoalk(en)yl diacylglycerol (MADAG). LDs also contain phospholipids including phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), ether-linked phosphatidylcholine (epc), ether-linked phosphatidylethanolamine (epe), lyso phosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE) and small amount of phosphatidylserine (PS) or sphingomyelin (SM). Relative amount of lipid (%) Phospholipids (%) TAGs 44.0 CE 34.0 MADAG 19.0 NEFAs 0.5 Cholesterol 0.5 Phospholipids 2.0 PC PE PI PS SM epc epe LPC LPE
12 Table S7. Random sampling algorithm results for obese subjects compared to lean subjects. Reactions change both in flux and expression in the same direction. DW: Down regulated. Reaction RXNID Male Female EQUATION SUBSYSTEM DW: HMR_0005 HMR_ acylglycerol-chylomicron pool[c] + H2O[c] => fatty acid- Acylglycerides metabolism LD-TG1 pool[c] + glycerol[c] DW: HMR_0002 HMR_ H2O[s] + TAG-chylomicron pool[s] => 1,2-diacylglycerolchylomicron Acylglycerides metabolism pool[s] + fatty acid-chylomicron pool[s] DW: HMR_0003 HMR_ ,2-diacylglycerol-chylomicron pool[s] + H2O[s] => 1- Acylglycerides metabolism acylglycerol-chylomicron pool[s] + fatty acid-chylomicron pool[s] DW: HMR_0010 HMR_ acylglycerol-VLDL pool[c] + H2O[c] => fatty acid-ld-tg1 Artificial reactions pool[c] + glycerol[c] DW: HMR_3275 HMR_ FAD[m] + linoleoyl-coa[m] => FADH2[m] + trans,cis,cis- 2,9,12-octadecatrienoyl-CoA[m] DW: HMR_3278 HMR_ (3S)-3-hydroxylinoleoyl-CoA[m] + NAD+[m] => 3- oxolinoleoyl-coa[m] + H+[m] + NADH[m] DW: HMR_3279 HMR_ oxolinoleoyl-CoA[m] + CoA[m] => acetyl-coa[m] + cis,cis-palmito-7,10-dienoyl-coa[m] DW: HMR_3280 HMR_ FAD[m] + cis,cis-palmito-7,10-dienoyl-coa[m] => FADH2[m] + trans,cis,cis-2,7,10-hexadecatrienoyl-coa[m] DW: HMR_3277 HMR_ H2O[m] + trans,cis,cis-2,9,12-octadecatrienoyl-coa[m] => (3S)-3-hydroxylinoleoyl-CoA[m] DW: HMR_3288 HMR_ cis,cis-3,6-dodecadienoyl-coa[m] => trans,cis-lauro-2,6- dienoyl-coa[m] DW: HMR_3282 HMR_ (3S)-3-hydroxy-cis,cis-palmito-7,10-dienoyl-CoA[m] + NAD+[m] => 3-oxo-cis,cis-7,10-hexadecadienoyl-CoA[m] + H+[m] + NADH[m] DW: HMR_3286 HMR_ (S)-hydroxy-(5Z,8Z)-tetradecadienoyl-CoA[m] + NAD+[m] => 3-oxo-cis,cis-5,8-tetradecadienoyl-CoA[m] + H+[m] + NADH[m] DW: HMR_3292 HMR_ (3S)-3-hydroxydodec-cis-6-enoyl-CoA[m] + NAD+[m] => 3- oxolaur-6-cis-enoyl-coa[m] + H+[m] + NADH[m] DW: HMR_3298 HMR_ (2E)-decenoyl-CoA[m] <=> trans-3-decenoyl-coa[m] DW: HMR_3293 HMR_ oxolaur-6-cis-enoyl-CoA[m] + CoA[m] => 4-cis-decenoyl- CoA[m] + acetyl-coa[m] 12
13 DW: HMR_3283 HMR_ oxo-cis,cis-7,10-hexadecadienoyl-CoA[m] + CoA[m] => acetyl-coa[m] + cis,cis-myristo-5,8-dienoyl-coa[m] DW: HMR_3287 HMR_ oxo-cis,cis-5,8-tetradecadienoyl-CoA[m] + CoA[m] => acetyl-coa[m] + cis,cis-3,6-dodecadienoyl-coa[m] DW: HMR_3284 HMR_ FAD[m] + cis,cis-myristo-5,8-dienoyl-coa[m] => FADH2[m] + trans-2-cis,cis-5,8-tetradecatrienoyl-coa[m] DW: HMR_3296 HMR_ trans-4-cis-decadienoyl-CoA[m] + H+[m] + NADH[m] => NAD+[m] + trans-3-decenoyl-coa[m] DW: HMR_3281 HMR_ H2O[m] + trans,cis,cis-2,7,10-hexadecatrienoyl-coa[m] => (3S)-3-hydroxy-cis,cis-palmito-7,10-dienoyl-CoA[m] DW: HMR_3285 HMR_ H2O[m] + trans-2-cis,cis-5,8-tetradecatrienoyl-coa[m] => 3(S)-hydroxy-(5Z,8Z)-tetradecadienoyl-CoA[m] DW: HMR_3290 HMR_ H2O[m] + trans,cis-lauro-2,6-dienoyl-coa[m] => (3S)-3- hydroxydodec-cis-6-enoyl-coa[m] DW: HMR_3429 HMR_ (7Z,10Z,13Z,16Z,19Z)-docosapentaenoyl-CoA [m] + 10 CoA[m] + 5 FAD[m] + 10 H2O[m] + 10 NAD+[m] => 5 FADH2[m] + 10 H+[m] + 10 NADH[m] + 11 acetyl-coa[m] DW: HMR_3430 HMR_ (4Z,7Z,10Z,13Z,16Z,19Z)-docosahexaenoyl-CoA[m] + 10 CoA[m] + 4 FAD[m] + 10 H2O[m] + 10 NAD+[m] => 4 FADH2[m] + 10 H+[m] + 10 NADH[m] + 11 acetyl-coa[m] DW: HMR_3426 HMR_ CoA[m] + 5 FAD[m] + 8 H2O[m] + 8 NAD+[m] + linolenoyl-coa[m] => 5 FADH2[m] + 8 H+[m] + 8 NADH[m] + 9 acetyl-coa[m] DW: HMR_3151 HMR_ (S)-hydroxyoctanoyl-CoA[m] + NAD+[m] => 3-oxooctanoyl- CoA[m] + H+[m] + NADH[m] DW: HMR_3158 HMR_ (S)-hydroxyhexanoyl-CoA[m] + NAD+[m] => 3-oxohexanoyl- CoA[m] + H+[m] + NADH[m] DW: HMR_3150 HMR_ (2E)-octenoyl-CoA[m] + H2O[m] => (S)-hydroxyoctanoyl- CoA[m] DW: HMR_3157 HMR_ (2E)-hexenoyl-CoA[m] + H2O[m] => (S)-hydroxyhexanoyl- CoA[m] DW: HMR_3149 HMR_ FAD[m] + octanoyl-coa[m] => (2E)-octenoyl-CoA[m] + FADH2[m] DW: HMR_3153 HMR_ oxooctanoyl-CoA[m] + CoA[m] => acetyl-coa[m] + hexanoyl-coa[m] DW: HMR_3156 HMR_ FAD[m] + hexanoyl-coa[m] => (2E)-hexenoyl-CoA[m] + 13
14 FADH2[m] DW: HMR_3144 HMR_ (S)-hydroxydecanoyl-CoA[m] + NAD+[m] => 3-oxodecanoyl- CoA[m] + H+[m] + NADH[m] DW: HMR_3143 HMR_ (2E)-decenoyl-CoA[m] + H2O[m] => (S)-hydroxydecanoyl- CoA[m] DW: HMR_3146 HMR_ oxodecanoyl-CoA[m] + CoA[m] => acetyl-coa[m] + octanoyl-coa[m] DW: HMR_3137 HMR_ (S)-3-hydroxydodecanoyl-CoA[m] + NAD+[m] => 3- oxododecanoyl-coa[m] + H+[m] + NADH[m] DW: HMR_3142 HMR_ FAD[m] + decanoyl-coa[m] => (2E)-decenoyl-CoA[m] + FADH2[m] DW: HMR_3139 HMR_ oxododecanoyl-CoA[m] + CoA[m] => acetyl-coa[m] + decanoyl-coa[m] DW: HMR_3136 HMR_ (2E)-dodecenoyl-CoA[m] + H2O[m] => (S)-3- hydroxydodecanoyl-coa[m] DW: HMR_3123 HMR_ (S)-3-hydroxyhexadecanoyl-CoA[m] + NAD+[m] => 3- oxopalmitoyl-coa[m] + H+[m] + NADH[m] DW: HMR_3125 HMR_ oxopalmitoyl-CoA[m] + CoA[m] => acetyl-coa[m] + myristoyl-coa[m] DW: HMR_3122 HMR_ (2E)-hexadecenoyl-CoA[m] + H2O[m] => (S)-3- hydroxyhexadecanoyl-coa[m] DW: HMR_3121 HMR_ FAD[m] + palmitoyl-coa[m] => (2E)-hexadecenoyl-CoA[m] + FADH2[m] DW: HMR_3130 HMR_ (S)-3-hydroxytetradecanoyl-CoA[m] + NAD+[m] => 3- oxotetradecanoyl-coa[m] + H+[m] + NADH[m] DW: HMR_3132 HMR_ oxotetradecanoyl-CoA[m] + CoA[m] => acetyl-coa[m] + lauroyl-coa[m] DW: HMR_3128 HMR_ FAD[m] + myristoyl-coa[m] => (2E)-tetradecenoyl-CoA[m] + FADH2[m] DW: HMR_3129 HMR_ (2E)-tetradecenoyl-CoA[m] + H2O[m] => (S)-3- hydroxytetradecanoyl-coa[m] DW: HMR_3421 HMR_ (7Z,10Z,13Z,16Z)-docosatetraenoyl-CoA[m] + 10 CoA[m] + 6 FAD[m] + 10 H2O[m] + 10 NAD+[m] => 6 FADH2[m] + 10 H+[m] + 10 NADH[m] + 11 acetyl-coa[m] DW: HMR_3228 HMR_ (S)-3-hydroxyoleyleoyl-CoA[m] + NAD+[m] => 3-oxooleoyl- CoA[m] + H+[m] + NADH[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) 14
15 DW: HMR_3229 HMR_ oxooleoyl-CoA[m] + CoA[m] => 7-hexadecenoyl-CoA[m] + acetyl-coa[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_3227 HMR_ H2O[m] + trans,cis-octadeca-2,9-dienoyl-coa[m] => (S)-3- hydroxyoleyleoyl-coa[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_3226 HMR_ FAD[m] + oleoyl-coa[m] => FADH2[m] + trans,cisoctadeca-2,9-dienoyl-coa[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_3232 HMR_ (S)-3-hydroxy-7-hexadecenoyl-CoA[m] + NAD+[m] => 3- oxo-7-hexadecenoyl-coa[m] + H+[m] + NADH[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_3233 HMR_ oxo-7-hexadecenoyl-CoA[m] + CoA[m] => 5-tetradecenoyl- CoA[m] + acetyl-coa[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_3230 HMR_ hexadecenoyl-CoA[m] + FAD[m] => FADH2[m] + trans,cis-hexadeca-2,7-dienoyl-coa[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_3231 HMR_ H2O[m] + trans,cis-hexadeca-2,7-dienoyl-coa[m] => (S)-3- hydroxy-7-hexadecenoyl-coa[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_3239 HMR_ (2E)-dodecenoyl-CoA[m] <=> (3Z)-dodecenoyl-CoA[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_3236 HMR_ NAD+[m] + cis-(3s)-hydroxytetradec-5-enoyl-coa[m] => 3- oxomyrist-5-enoyl-coa[m] + H+[m] + NADH[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_3237 HMR_ oxomyrist-5-enoyl-CoA[m] + CoA[m] => (3Z)-dodecenoyl- CoA[m] + acetyl-coa[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_3234 HMR_ tetradecenoyl-CoA[m] + FAD[m] => FADH2[m] + trans,cismyristo-2,5-dienoyl-coa[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_3235 HMR_ H2O[m] + trans,cis-myristo-2,5-dienoyl-coa[m] => cis-(3s)- hydroxytetradec-5-enoyl-coa[m] Beta oxidation of unsaturated fatty acids (n-9) (mitochondrial) DW: HMR_4137 HMR_ CoA[m] + NAD+[m] + pyruvate[m] => CO2[m] + H+[m] + Glycolysis / Gluconeogenesis NADH[m] + acetyl-coa[m] DW: HMR_3212 HMR_ FAD[m] + propanoyl-coa[m] => FADH2[m] + acrylyl- Propanoate metabolism CoA[m] DW: HMR_4735 HMR_ H2O[m] + acrylyl-coa[m] => 3-hydroxypropionyl-CoA[m] beta-alanine metabolism DW: HMR_4741 HMR_ hydroxypropionyl-CoA[m] + H2O[m] => CoA[m] + hydracrylate[m] DW: HMR_2726 HMR_ (7Z,10Z,13Z,16Z,19Z)-docosapentaenoyl-CoA [c] + L- carnitine[c] <=> (7Z,10Z,13Z,16Z,19Z)- docosapentaenoylcarnitine[c] + CoA[c] DW: HMR_2730 HMR_ (4Z,7Z,10Z,13Z,16Z,19Z)-docosahexaenoyl-CoA[c] + L- carnitine[c] <=> CoA[c] + docosahexaenoylcarnitine[c] Propanoate metabolism Carnitine shuttle (cytosolic) Carnitine shuttle (cytosolic) 15
16 DW: HMR_2742 HMR_ CoA[c] + linoleic-carnitine[c] <=> L-carnitine[c] + linoleoyl- Carnitine shuttle (cytosolic) CoA[c] DW: HMR_2727 HMR_ (7Z,10Z,13Z,16Z,19Z)-docosapentaenoylcarnitine[c] + L- Carnitine shuttle (mitochondrial) carnitine[m] <=> (7Z,10Z,13Z,16Z,19Z)- docosapentaenoylcarnitine[m] + L-carnitine[c] DW: HMR_2731 HMR_ L-carnitine[c] + docosahexaenoylcarnitine[m] <=> L- Carnitine shuttle (mitochondrial) carnitine[m] + docosahexaenoylcarnitine[c] DW: HMR_2744 HMR_ L-carnitine[c] + linoleic-carnitine[m] <=> L-carnitine[m] + Carnitine shuttle (mitochondrial) linoleic-carnitine[c] DW: HMR_2722 HMR_ (5Z,8Z,11Z,14Z,17Z)-eicosapentaenoylcarnitine[c] + L- Carnitine shuttle (mitochondrial) carnitine[m] <=> (5Z,8Z,11Z,14Z,17Z)- eicosapentaenoylcarnitine[m] + L-carnitine[c] DW: HMR_2712 HMR_ L-carnitine[c] + octadecatrienoylcarnitine[m] <=> L- Carnitine shuttle (mitochondrial) carnitine[m] + octadecatrienoylcarnitine[c] DW: HMR_2729 HMR_ (7Z,10Z,13Z,16Z,19Z)-docosapentaenoyl-CoA [m] + L- Carnitine shuttle (mitochondrial) carnitine[m] <=> (7Z,10Z,13Z,16Z,19Z)- docosapentaenoylcarnitine[m] + CoA[m] DW: HMR_2732 HMR_ (4Z,7Z,10Z,13Z,16Z,19Z)-docosahexaenoyl-CoA[m] + L- Carnitine shuttle (mitochondrial) carnitine[m] <=> CoA[m] + docosahexaenoylcarnitine[m] DW: HMR_2745 HMR_ CoA[m] + linoleic-carnitine[m] <=> L-carnitine[m] + linoleoyl-coa[m] Carnitine shuttle (mitochondrial) DW: HMR_4295 HMR_ ATP[m] + H+[m] + HCO3-[m] + acetyl-coa[m] => ADP[m] + Pi[m] + malonyl-coa[m] Fatty acid biosynthesis (evenchain) DW: HMR_4655 HMR_ H+[c] + NADPH[c] + folate[c] <=> NADP+[c] + Folate metabolism dihydrofolate[c] DW: HMR_0534 HMR_ linoleoyl-coa[c] + sn-glycerol-3-phosphate[c] => 1- Glycerolipid metabolism acylglycerol-3p-lin[c] + CoA[c] DW: HMR_0453 HMR_ H+[c] + NADPH[c] + glyceraldehyde[c] <=> NADP+[c] + glycerol[c] Glycerolipid metabolism DW: HMR_6409 HMR_ H+[m] + NADH[m] + lipoamide[m] <=> NAD+[m] + dihydrolipoamide[m] Glycine, serine and threonine metabolism DW: HMR_1263 HMR_ ,11-dihydro-LTB4-CoA[c] + AMP[c] + PPi[c] <=> 10,11- Leukotriene metabolism dihydro-ltb4[c] + ATP[c] + CoA[c] DW: HMR_1264 HMR_ ,11-dihydro-LTB4-CoA[m] + AMP[m] + PPi[m] <=> 10,11- dihydro-ltb4[m] + ATP[m] + CoA[m] Leukotriene metabolism DW: HMR_6914 HMR_ H+[m] + O2[m] + 4 ferrocytochrome C[m] => 4 H+[c] + 2 Oxidative phosphorylation 16
17 H2O[m] + 4 ferricytochrome C[m] DW: HMR_6918 HMR_ H+[m] + 2 ferricytochrome C[m] + ubiquinol[m] => 4 H+[c] Oxidative phosphorylation + 2 ferrocytochrome C[m] + ubiquinone[m] DW: HMR_6921 HMR_ H+[m] + NADH[m] + ubiquinone[m] => 4 H+[c] + Oxidative phosphorylation NAD+[m] + ubiquinol[m] DW: HMR_6912 HMR_ H2O[m] + PPi[m] => 2 Pi[m] Oxidative phosphorylation DW: HMR_4004 HMR_ ADP[m] <=> AMP[m] + ATP[m] Purine metabolism DW: HMR_4002 HMR_ ADP[c] <=> AMP[c] + ATP[c] Purine metabolism DW: HMR_0007 HMR_ H2O[s] + TAG-VLDL pool[s] => 1,2-diacylglycerol-VLDL Transport, extracellular pool[s] + fatty acid-vldl pool[s] DW: HMR_0008 HMR_ ,2-diacylglycerol-VLDL pool[s] + H2O[s] => 1-acylglycerol- Transport, extracellular VLDL pool[s] + fatty acid-vldl pool[s] DW: HMR_4885 HMR_ H2O[c] <=> H2O[s] Transport, extracellular DW: HMR_8741 HMR_ fumarate[c] + sulfite[m] <=> fumarate[m] + sulfite[c] Transport, mitochondrial DW: HMR_5043 HMR_ H+[c] + Pi[c] <=> H+[m] + Pi[m] Transport, mitochondrial DW: HMR_4888 HMR_ H2O[c] <=> H2O[m] Transport, mitochondrial DW: HMR_6330 HMR_ AKG[c] + Pi[m] <=> AKG[m] + Pi[c] Transport, mitochondrial DW: HMR_4145 HMR_ CoA[m] + citrate[m] <=> H2O[m] + OAA[m] + acetyl- CoA[m] DW: HMR_4209 HMR_ AKG[m] + thiamin-pp[m] => 3-carboxy-1-hydroxypropyl- ThPP[m] + CO2[m] DW: HMR_6413 HMR_ carboxy-1-hydroxypropyl-ThPP[m] + lipoamide[m] => S- succinyldihydrolipoamide[m] + thiamin-pp[m] DW: HMR_6414 HMR_ CoA[m] + S-succinyldihydrolipoamide[m] <=> dihydrolipoamide[m] + succinyl-coa[m] Tricarboxylic acid cycle and glyoxylate/dicarboxylate metabolism Tricarboxylic acid cycle and glyoxylate/dicarboxylate metabolism Tricarboxylic acid cycle and glyoxylate/dicarboxylate metabolism Tricarboxylic acid cycle and glyoxylate/dicarboxylate metabolism DW: HMR_4456 HMR_ citrate[m] <=> isocitrate[m] Tricarboxylic acid cycle and glyoxylate/dicarboxylate metabolism DW: HMR_4408 HMR_ H2O[c] + fumarate[c] <=> malate[c] Tricarboxylic acid cycle and glyoxylate/dicarboxylate 17
18 DW: HMR_3958 HMR_ NADP+[m] + isocitrate[m] => AKG[m] + CO2[m] + H+[m] + NADPH[m] DW: HMR_4652 HMR_ fumarate[m] + ubiquinol[m] <=> succinate[m] + ubiquinone[m] metabolism Tricarboxylic acid cycle and glyoxylate/dicarboxylate metabolism Tricarboxylic acid cycle and glyoxylate/dicarboxylate metabolism DW: HMR_8743 HMR_ FADH2[m] + fumarate[m] <=> FAD[m] + succinate[m] Tricarboxylic acid cycle and glyoxylate/dicarboxylate metabolism DW: HMR_4147 HMR_ CoA[m] + GTP[m] + succinate[m] <=> GDP[m] + Pi[m] + succinyl-coa[m] Tricarboxylic acid cycle and glyoxylate/dicarboxylate metabolism DW: HMR_3160 HMR_ oxohexanoyl-CoA[m] + CoA[m] => acetyl-coa[m] + butyryl-coa[m] DW: HMR_3163 HMR_ FAD[m] + butyryl-coa[m] => FADH2[m] + crotonyl-coa[m] Butanoate metabolism DW: HMR_3164 HMR_ H2O[m] + crotonyl-coa[m] => (S)-3-hydroxybutyryl-CoA[m] Tryptophan metabolism DW: HMR_3166 HMR_ (S)-3-hydroxybutyryl-CoA[m] + NAD+[m] => H+[m] + Tryptophan metabolism NADH[m] + acetoacetyl-coa[m] DW: HMR_3215 HMR_ (R)-methylmalonyl-CoA[m] => succinyl-coa[m] Valine, leucine, and isoleucine metabolism DW: HMR_3213 HMR_ methylmalonyl-coa[m] => (R)-methylmalonyl-CoA[m] Valine, leucine, and isoleucine metabolism 18
19 Obese Normal (Up) Obese Normal (Down) Overweight Normal (Up) Overweight Normal (Down) Supplementary Figures Antigen processing and presentation Leukocyte transendothelial mig ration Protein processing in endoplasmic reticulum Rheumatoid a rthritis Asthma Arrhythmogenic right ventricular cardiomyopathy (ARVC) Systemic lupus erythematosus Viral myocarditis Regulation of actin cytos keleton Pathogenic Escherichia coli infection Fc gamma R mediated phagocytosis Focal adhesion Toxoplasmosis Vibrio cholerae infection Hypertrophic cardiomyopathy (HCM) D Glutamine and D glutamate metabolism p53 signaling path way Osteoclast differentiation Bacterial invasion of epithelial cells Complement and coagulation cascades Shigellosis Dilated cardio myopathy Leishmaniasis Cell adhesion molecules (CAMs) Chemokine signaling path way B cell receptor signaling path way Mucin type O Glycan biosynthesis Amoebiasis Protein export ECM receptor interaction Adherens junction Intestinal immune network for IgA production Other glycan degradation Drug metabolism other enzymes Endocytosis Proximal tubule bicarbonate reclamation Nitrogen metabolism Oxidative phosphorylation Parkinsons disease Aldosterone regulated sodium reabso rption Peroxisome Insulin signaling path way Adipocytokine signaling path way Cyanoamino acid metabolism Arginine and proline metabolism Phenylalanine metabolism Huntingtons disease Vitamin digestion and abso rption Neuroactive ligand receptor inte raction Fatty acid biosynthesis Glycine, serine and threonine metabolism Lysine degradation Tryptophan metabolism Butanoate metabolism Pyruvate metabolism Fatty acid elongation Citrate cycle (TCA cycle) Glyoxylate and dicarboxylate metabolism Lysosome Staphylococcus aureus infection Phagosome Metabolic path ways Fatty acid metabolism Valine, leucine and isoleucine deg radation Propanoate metabolism Figure S1. Enrichment in KEGG pathways between obese, overweight and normal (lean) male subjects, as calculated by the Reporter Features algorithm. Red and blue represent enrichment in up-regulated and down-regulated genes respectively. Deep red/blue represents 1e
20 Obese Normal (Up) Obese Normal (Down) Overweight Normal (Up) Overweight Normal (Down) Systemic lupus erythematosus Leukocyte transendothelial mig ration Cell adhesion molecules (CAMs) Focal adhesion Protein processing in endoplasmic reticulum Pathogenic Escherichia coli infection Regulation of actin cytos keleton Fc gamma R mediated phagocytosis Viral myocarditis Lysosome Staphylococcus aureus infection Antigen processing and presentation Phagosome Shigellosis Arrhythmogenic right ventricular cardiomyopathy (ARVC) ECM receptor interaction Osteoclast differentiation Chemokine signaling path way Natural killer cell mediated cytot oxicity Dilated cardio myopathy Rheumatoid a rthritis Leishmaniasis Intestinal immune network for IgA production Allograft rejection Autoimmune thyroid disease Graft versus host disease Type I diabetes mellitus Toxoplasmosis Asthma B cell receptor signaling path way Vibrio cholerae infection Bacterial invasion of epithelial cells Other glycan degradation Proteasome Fc epsilon RI signaling path way p53 signaling path way Glycosaminoglycan biosynthesis chondroitin sul fate Mucin type O Glycan biosynthesis Toll like receptor signaling path way Protein digestion and abso rption Amoebiasis Tight junction Hematopoietic cell lineage Amino sugar and nucleotide sugar metabolism Hypertrophic cardiomyopathy (HCM) D Glutamine and D glutamate metabolism Complement and coagulation cascades Sphingolipid metabolism Endocytosis Neurotrophin signaling path way Proximal tubule bicarbonate reclamation Sulfur relay system Phenylalanine metabolism Tyrosine metabolism Glycolysis Gluconeogenesis Nitrogen metabolism Valine, leucine and isoleucine biosynthesis mtor signaling pathway One carbon pool by folate Vitamin digestion and abso rption Lysine biosynthesis Parkinsons disease Drug metabolism cytochrome P Cyanoamino acid metabolism Insulin signaling path way Glycine, serine and threonine metabolism Aldosterone regulated sodium reabso rption Adipocytokine signaling path way Tryptophan metabolism Lysine degradation Glyoxylate and dicarboxylate metabolism Metabolic path ways Pyruvate metabolism Butanoate metabolism Fatty acid elongation Citrate cycle (TCA cycle) Valine, leucine and isoleucine degradation Propanoate metabolism Ribosome Fatty acid metabolism Fatty acid biosynthesis Figure S2. Enrichment in KEGG pathways between obese, overweight and normal (lean) female subjects, as calculated by the Reporter Features algorithm. Red and blue represent enrichment in up-regulated and down-regulated genes respectively. Deep red/blue represents 1e
21 Obese Normal (Up) Obese Normal (Down) Overweight Normal (Up) Overweight Normal (Down) GO: tolerance induction to lipopolysaccharide GO: negative regulation of osteoclast proliferation GO: negative regulation of toll like receptor 4 signaling pathway GO: B 1 B cell homeostasis GO: regulation of vascular wound healing GO: negative regulation of CD40 signaling pathway GO: positive regulation of stress activated protein kinase signaling cascade GO: positive regulation of glial cell proliferation GO: positive regulation of chemokine production GO: negative regulation of neural precursor cell proliferation GO: positive regulation of collagen biosynthetic process GO: regulation of respiratory gaseous exchange by neurological system process GO: positive regulation of tyrosine phosphorylation of STAT protein GO: cellular iron ion homeostasis GO: post translational protein modification GO: protein N linked glycosylation via asparagine GO: Tie receptor signaling pathway GO: regulation of cellular senescence GO: cellular response to heat GO: glycerol ether metabolic process GO: negative regulation of endothelial cell migration GO: response to virus GO: protein O linked glycosylation GO: negative regulation of tumor necrosis factor production GO: chemotaxis GO: negative regulation of canonical Wnt receptor signaling pathway GO: superoxide anion generation GO: defense response to virus GO: regulation of innate immune response GO: cellular response to heparin GO: enzyme linked receptor protein signaling pathway GO: prostate gland epithelium morphogenesis GO: motor axon guidance GO: negative regulation of odontogenesis of dentine containing tooth GO: B cell receptor signaling pathway GO: response to wounding GO: negative regulation of toll like receptor 2 signaling pathway GO: negative regulation of blood vessel endothelial cell migration GO: cell redox homeostasis GO: negative regulation of cartilage development GO: negative regulation of angiogenesis GO: response to drug GO: neurotransmitter secretion GO: glutamate secretion GO: ossification GO: positive regulation of Fc receptor mediated stimulatory signaling pathway GO: cell morphogenesis involved in differentiation GO: cell matrix adhesion GO: positive regulation of mast cell degranulation GO: amino acid transport GO: beta selection GO: positive regulation of interleukin 3 biosynthetic process GO: serotonin secretion GO: axon extension involved in axon guidance GO: regulation of cellular amino acid metabolic process GO: establishment of protein localization GO: positive regulation of muscle hyperplasia GO: collagen fibril organization GO: endodermal digestive tract morphogenesis GO: leukocyte differentiation GO: antigen processing and presentation of peptide antigen via MHC class I GO: negative chemotaxis GO: response to organic cyclic compound GO: positive regulation of protein phosphorylation GO: response to estradiol stimulus GO: glutamine catabolic process GO: regulation of cell shape GO: small GTPase mediated signal transduction GO: positive regulation of cell activation GO: negative regulation of chemokine mediated signaling pathway GO: apoptosis involved in luteolysis GO: negative regulation of leukocyte chemotaxis GO: muscle contraction GO: regulation of osteoblast proliferation GO: osteoclast proliferation GO: actin cytoskeleton organization GO: interferon gamma mediated signaling pathway GO: cytokine mediated signaling pathway GO: leukotriene biosynthetic process GO: innate immune response GO: peripheral nervous system axon regeneration GO: antigen processing and presentation of peptide or polysaccharide antigen via MHC class II GO: negative regulation of protein localization at cell surface GO: axon guidance GO: cytoplasmic sequestering of protein GO: negative regulation of DNA damage response, signal transduction by p53 class mediator GO: N terminal protein myristoylation GO: nucleobase containing compound interconversion GO: de novo posttranslational protein folding GO: T cell costimulation GO: regulation of MAPKKK cascade GO: positive regulation of cell proliferation GO: protein lipoylation GO: response to carbohydrate stimulus GO: anti apoptosis GO: cell junction assembly GO: adherens junction organization GO: response to peptide hormone stimulus GO: protein localization at cell surface GO: regulation of chondrocyte differentiation GO: actin crosslink formation GO: cortical actin cytoskeleton organization GO: cardiac cell differentiation GO: positive regulation of Wnt receptor signaling pathway, planar cell polarity pathway GO: positive regulation of cyclin dependent protein kinase activity GO: extracellular matrix organization GO: antigen processing and presentation of exogenous peptide antigen via MHC class I GO: positive regulation of type IIa hypersensitivity GO: spindle assembly involved in mitosis GO: negative regulation of protein processing GO: response to misfolded protein GO: negative regulation of activin receptor signaling pathway GO: response to vitamin E GO: negative regulation of apoptosis GO: negative regulation of cellular component movement GO: response to ethanol GO: regulation of bone mineralization GO: proteolysis GO: positive regulation of protein ubiquitination involved in ubiquitin dependent protein catabolic process GO: positive regulation of urine volume GO: heart development GO: catecholamine metabolic process GO: cell migration involved in sprouting angiogenesis GO: mitochondrial DNA replication GO: negative regulation of cellular response to growth factor stimulus GO: negative regulation of lamellipodium assembly GO: negative regulation of retinal ganglion cell axon guidance GO: induction of negative chemotaxis GO: negative regulation of mononuclear cell migration GO: chemorepulsion involved in postnatal olfactory bulb interneuron migration GO: corticospinal neuron axon guidance through spinal cord GO: negative regulation of small GTPase mediated signal transduction GO: negative regulation of neutrophil chemotaxis GO: negative regulation of smooth muscle cell chemotaxis GO: leukocyte migration GO: response to cortisol stimulus GO: actin filament bundle assembly GO: cell adhesion GO: Roundabout signaling pathway GO: induction of synaptic plasticity by chemical substance GO: complement activation GO: positive regulation of ERK1 and ERK2 cascade GO: regulation of apoptosis GO: pinocytosis GO: inflammatory response GO: signal transduction GO: response to calcium ion GO: platelet activation GO: response to axon injury GO: negative regulation of vascular permeability GO: positive regulation of vascular permeability GO: Mo molybdopterin cofactor biosynthetic process GO: modulation by virus of host transcription GO: positive regulation of axon extension involved in regeneration GO: protein homotetramerization GO: glutamine biosynthetic process GO: energy reserve metabolic process GO: response to insulin stimulus GO: cellular amino acid biosynthetic process GO: cellular response to insulin stimulus GO: long chain fatty acyl CoA biosynthetic process GO: positive regulation of fatty acid beta oxidation GO: negative regulation of insulin secretion GO: cellular nitrogen compound metabolic process GO: pyruvate metabolic process GO: positive regulation of glutamate secretion GO: regulation of acetyl CoA biosynthetic process from pyruvate GO: regulation of gluconeogenesis GO: glucocorticoid metabolic process GO: negative regulation of receptor biosynthetic process GO: primitive erythrocyte differentiation GO: basophil chemotaxis GO: negative regulation of sequestering of triglyceride GO: fatty acid beta oxidation using acyl CoA dehydrogenase GO: thymine metabolic process GO: valine catabolic process GO: beta alanine catabolic process GO: lipid metabolic process GO: negative regulation of appetite GO: positive regulation of protein autophosphorylation GO: negative regulation of protein dephosphorylation GO: mammary gland alveolus development GO: fibroblast growth factor receptor signaling pathway GO: prostate gland growth GO: negative regulation of astrocyte differentiation GO: behavior GO: mesoderm development GO: positive regulation of glycogen biosynthetic process GO: negative regulation of ATPase activity GO: ethanol oxidation GO: negative regulation of sodium ion transport GO: regulation of cell death GO: camera type eye morphogenesis GO: synaptic transmission, glycinergic GO: brown fat cell differentiation GO: positive regulation of protein kinase C signaling cascade GO: neuron fate specification GO: L lysine catabolic process GO: regulation of fatty acid metabolic process GO: hydrogen sulfide biosynthetic process GO: fructose 2,6 bisphosphate metabolic process GO: carnitine metabolic process, CoA linked GO: thiamine containing compound metabolic process GO: neurotransmitter uptake GO: folic acid metabolic process GO: multicellular organismal protein metabolic process GO: phosphatidic acid biosynthetic process GO: cellular biogenic amine metabolic process GO: fatty acid transport GO: fatty acid biosynthetic process GO: triglyceride biosynthetic process GO: cochlea development GO: negative regulation of cell death GO: auditory receptor cell differentiation GO: response to selenium ion GO: L serine catabolic process GO: water homeostasis GO: negative regulation of blood pressure GO: cellular response to prostaglandin stimulus GO: regulation of chromosome organization GO: succinyl CoA metabolic process GO: phenylethylamine metabolic process GO: oxaloacetate metabolic process GO: fatty acid alpha oxidation GO: negative regulation of NFAT protein import into nucleus GO: mitochondrial electron transport, ubiquinol to cytochrome c GO: chromatin mediated maintenance of transcription GO: induction of positive chemotaxis GO: regulation of vasodilation GO: vascular transport GO: negative regulation of macrophage derived foam cell differentiation GO: malate metabolic process GO: muscle atrophy GO: respiratory electron transport chain GO: respiratory system process GO: L alanine metabolic process GO: regulation of renal sodium excretion GO: positive regulation of phospholipase A2 activity GO: thymine catabolic process GO: negative regulation of glycolysis GO: biosynthetic process GO: response to mercury ion GO: regulation of blood vessel size by renin angiotensin GO: renin angiotensin regulation of aldosterone production GO: positive regulation of NAD(P)H oxidase activity GO: cellular response to magnesium ion GO: hydrogen peroxide mediated programmed cell death GO: positive regulation of mast cell chemotaxis GO: tricarboxylic acid cycle GO: positive regulation of fatty acid oxidation GO: valine metabolic process GO: mechanoreceptor differentiation GO: regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter GO: regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter GO: regulation of glycolysis by positive regulation of transcription from RNA polymerase II promoter GO: cellular component movement GO: blood coagulation GO: immune response GO: platelet degranulation GO: fatty acid beta oxidation GO: fatty acid metabolic process GO: water soluble vitamin metabolic process GO: branched chain family amino acid catabolic process GO: acetyl CoA metabolic process GO: cellular lipid metabolic process GO: vitamin metabolic process GO: response to corticosterone stimulus GO: regulation of glucocorticoid biosynthetic process GO: glucocorticoid mediated signaling pathway GO: blood coagulation, extrinsic pathway GO: activation of Rap GTPase activity GO: peptide cross linking GO: protein folding GO: response to amine stimulus GO: positive regulation of smooth muscle cell proliferation GO: epithelial cell differentiation involved in prostate gland development GO: cell cell junction organization GO: negative regulation of bone resorption GO: developmental maturation GO: complement activation, classical pathway GO: neutrophil chemotaxis GO: molybdopterin cofactor biosynthetic process GO: cell maturation GO: centromere complex assembly GO: transcription from RNA polymerase II promoter GO: embryonic forelimb morphogenesis GO: positive regulation of intracellular protein kinase cascade GO: mammary gland duct morphogenesis GO: positive regulation of cellular metabolic process GO: carnitine biosynthetic process Figure S3. Enrichment in biological process Gene Ontology (BP:GO) terms between obese, overweight and normal (lean) male subjects, as calculated by the Reporter Features algorithm. Red and blue represent enrichment in up-regulated and down-regulated genes respectively. Deep red/blue represents 1e
22 Obese Normal (Up) Obese Normal (Down) Overweight Normal (Up) Overweight Normal (Down) GO: negative regulation of neutrophil chemotaxis GO: induction of negative chemotaxis GO: negative regulation of small GTPase mediated signal transduction GO: negative regulation of smooth muscle cell chemotaxis GO: negative regulation of mononuclear cell migration GO: corticospinal neuron axon guidance through spinal cord GO: negative regulation of retinal ganglion cell axon guidance GO: negative regulation of cellular response to growth factor stimulus GO: negative regulation of lamellipodium assembly GO: B cell receptor signaling pathway GO: proteolysis GO: activation of innate immune response GO: proteolysis involved in cellular protein catabolic process GO: induction of synaptic plasticity by chemical substance GO: peripheral nervous system axon regeneration GO: interferon beta production GO: developmental maturation GO: response to cortisol stimulus GO: negative regulation of leukocyte chemotaxis GO: regulation of MAPKKK cascade GO: protein N linked glycosylation via asparagine GO: apoptosis involved in luteolysis GO: positive regulation of stress activated protein kinase signaling cascade GO: actin cytoskeleton organization GO: negative regulation of ubiquitin protein ligase activity involved in mitotic cell cycle GO: response to manganese ion GO: positive regulation of muscle hyperplasia GO: positive regulation of Fc receptor mediated stimulatory signaling pathway GO: protein polymerization GO: neutrophil chemotaxis GO: nucleobase containing compound interconversion GO: post translational protein modification GO: DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest GO: positive regulation of interleukin 3 biosynthetic process GO: beta selection GO: defense response GO: serotonin secretion GO: anaphase promoting complex dependent proteasomal ubiquitin dependent protein catabolic process GO: positive regulation of ubiquitin protein ligase activity involved in mitotic cell cycle GO: protein polyubiquitination GO: regulation of ubiquitin protein ligase activity involved in mitotic cell cycle GO: cellular response to hormone stimulus GO: transmembrane transport GO: peptide cross linking GO: cellular protein catabolic process GO: cellular response to heat GO: sphingolipid catabolic process GO: positive regulation of mast cell degranulation GO: copper ion transport GO: cell proliferation GO: negative regulation of DNA damage response, signal transduction by p53 class mediator GO: chemorepulsion involved in postnatal olfactory bulb interneuron migration GO: transport GO: ceramide metabolic process GO: positive regulation of peptidyl tyrosine phosphorylation GO: positive regulation of protein phosphorylation GO: ossification GO: antigen processing and presentation GO: anti apoptosis GO: cell cycle checkpoint GO: PMA inducible membrane protein ectodomain proteolysis GO: glutamate uptake involved in synaptic transmission GO: response to magnesium ion GO: prostanoid metabolic process GO: cellular response to tumor necrosis factor GO: positive regulation of granulocyte macrophage colony stimulating factor biosynthetic process GO: negative regulation of microtubule depolymerization GO: age dependent response to oxidative stress GO: bile acid and bile salt transport GO: G2 M transition checkpoint GO: negative regulation of transferase activity GO: response to amine stimulus GO: attachment of GPI anchor to protein GO: M phase specific microtubule process GO: response to gravity GO: regulation of vascular wound healing GO: epithelial cell differentiation involved in mammary gland alveolus development GO: positive regulation of cell activation GO: tolerance induction to lipopolysaccharide GO: positive regulation of chemokine production GO: positive regulation of alpha beta T cell differentiation GO: neuromuscular process controlling posture GO: positive regulation of cellular component movement GO: S phase of mitotic cell cycle GO: negative regulation of endothelial cell migration GO: ruffle organization GO: cell cell junction organization GO: adherens junction organization GO: protein O linked glycosylation GO: positive regulation of type IIa hypersensitivity GO: response to cytokine stimulus GO: lysosome organization GO: antigen processing and presentation of exogenous peptide antigen via MHC class I GO: response to carbohydrate stimulus GO: positive regulation of glial cell proliferation GO: negative regulation of chemokine mediated signaling pathway GO: response to insecticide GO: M G1 transition of mitotic cell cycle GO: regulation of release of sequestered calcium ion into cytosol GO: vesicle transport along microtubule GO: release of cytochrome c from mitochondria GO: positive regulation of protein binding GO: regulation of DNA recombination GO: negative regulation of protein binding GO: G1 S transition of mitotic cell cycle GO: sarcomere organization GO: calcium ion dependent exocytosis GO: osteoclast differentiation GO: cytoplasmic sequestering of protein GO: chondrocyte development GO: acute phase response GO: positive regulation of tyrosine phosphorylation of STAT protein GO: cellular response to heparin GO: enzyme linked receptor protein signaling pathway GO: secretion by cell GO: amino acid transport GO: positive regulation of peptidyl serine phosphorylation GO: negative regulation of interleukin 12 production GO: cell cell signaling GO: response to vitamin A GO: carbohydrate metabolic process GO: glycolipid catabolic process GO: oligosaccharide catabolic process GO: positive regulation of transforming growth factor beta1 production GO: cellular response to lipopolysaccharide GO: positive regulation of angiogenesis GO: axon extension involved in axon guidance GO: superoxide anion generation GO: positive regulation of macrophage activation GO: cellular response to interleukin 1 GO: positive regulation of vesicle fusion GO: ganglioside catabolic process GO: glycosaminoglycan biosynthetic process GO: musculoskeletal movement GO: superoxide metabolic process GO: response to ethanol GO: response to oxidative stress GO: positive regulation of smooth muscle cell proliferation GO: positive regulation of ERK1 and ERK2 cascade GO: peptidyl proline modification GO: catecholamine metabolic process GO: osteoclast proliferation GO: lipid storage GO: positive regulation of I kappab kinase NF kappab cascade GO: regulation of osteoblast proliferation GO: response to virus GO: vesicle mediated transport GO: skeletal system development GO: positive regulation of secretion GO: regulation of epithelial cell migration GO: protein transport GO: negative regulation of actin filament polymerization GO: cortical actin cytoskeleton organization GO: positive regulation of receptor internalization GO: response to unfolded protein GO: glutamate secretion GO: cell cell adhesion GO: membrane protein ectodomain proteolysis GO: muscle contraction GO: positive regulation of endothelial cell migration GO: protein localization at cell surface GO: response to stress GO: neurotransmitter secretion GO: cellular membrane organization GO: blood coagulation, extrinsic pathway GO: antigen processing and presentation of endogenous peptide antigen via MHC class I GO: post Golgi vesicle mediated transport GO: interleukin 27 mediated signaling pathway GO: regulation of interferon gamma mediated signaling pathway GO: leukocyte cell cell adhesion GO: protein catabolic process GO: actin filament capping GO: epidermal growth factor receptor signaling pathway GO: GTP catabolic process GO: protein deglycosylation GO: negative chemotaxis GO: glutamine catabolic process GO: protein O linked glycosylation via serine GO: cell matrix adhesion GO: T cell receptor signaling pathway GO: protein O linked glycosylation via threonine GO: regulation of immune response GO: T cell costimulation GO: interspecies interaction between organisms GO: type I interferon mediated signaling pathway GO: innate immune response GO: leukocyte migration GO: de novo posttranslational protein folding GO: cell junction assembly GO: complement activation GO: antigen processing and presentation of peptide or polysaccharide antigen via MHC class II GO: complement activation, classical pathway GO: Roundabout signaling pathway GO: positive regulation of Wnt receptor signaling pathway, planar cell polarity pathway GO: cell morphogenesis involved in differentiation GO: integrin mediated signaling pathway GO: cell redox homeostasis GO: response to calcium ion GO: pinocytosis GO: response to drug GO: collagen fibril organization GO: chemotaxis GO: regulation of cell shape GO: response to peptide hormone stimulus GO: leukotriene biosynthetic process GO: small GTPase mediated signal transduction GO: response to wounding GO: response to organic cyclic compound GO: cellular response to extracellular stimulus GO: protein folding GO: glycerol ether metabolic process GO: apoptosis GO: response to axon injury GO: regulation of cellular amino acid metabolic process GO: regulation of apoptosis GO: platelet activation GO: signal transduction GO: negative regulation of apoptosis GO: actin filament bundle assembly GO: positive regulation of cell proliferation GO: negative regulation of vascular permeability GO: positive regulation of protein ubiquitination involved in ubiquitin dependent protein catabolic process GO: electron transport chain GO: antigen processing and presentation of peptide antigen via MHC class I GO: cellular nitrogen compound metabolic process GO: axon guidance GO: cell adhesion GO: interferon gamma mediated signaling pathway GO: cytokine mediated signaling pathway GO: cellular component movement GO: blood coagulation GO: inflammatory response GO: platelet degranulation GO: cellular protein metabolic process GO: immune response GO: endocrine pancreas development GO: lipid metabolic process GO: carnitine biosynthetic process GO: viral infectious cycle GO: ethanol oxidation GO: negative regulation of sequestering of triglyceride GO: positive regulation of vascular permeability GO: Mo molybdopterin cofactor biosynthetic process GO: cellular lipid metabolic process GO: protein homotetramerization GO: translation GO: gene expression GO: translational elongation GO: viral transcription GO: translational termination GO: translational initiation GO: branched chain family amino acid catabolic process GO: positive regulation of cellular metabolic process GO: acetyl CoA metabolic process GO: fatty acid metabolic process GO: water soluble vitamin metabolic process GO: fatty acid beta oxidation GO: positive regulation of fatty acid oxidation GO: vitamin metabolic process GO: L lysine catabolic process GO: hydrogen sulfide biosynthetic process GO: succinyl CoA metabolic process GO: valine metabolic process GO: regulation of glycolysis by positive regulation of transcription from RNA polymerase II promoter GO: regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter GO: phospholipid biosynthetic process GO: regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter GO: regulation of acetyl CoA biosynthetic process from pyruvate GO: molybdopterin cofactor biosynthetic process GO: phosphatidic acid biosynthetic process GO: positive regulation of fatty acid beta oxidation GO: modulation by virus of host transcription GO: positive regulation of axon extension involved in regeneration GO: response to insulin stimulus GO: negative regulation of receptor biosynthetic process GO: ribosomal large subunit biogenesis GO: glutamine biosynthetic process GO: beta alanine catabolic process GO: thymine metabolic process GO: response to corticosterone stimulus GO: valine catabolic process GO: cellular response to insulin stimulus GO: regulation of gluconeogenesis GO: glucocorticoid mediated signaling pathway GO: water homeostasis GO: positive regulation of glycogen biosynthetic process GO: regulation of glucocorticoid biosynthetic process GO: cardiac muscle tissue development GO: transcription from RNA polymerase II promoter GO: prostate gland growth GO: negative regulation of protein refolding GO: thiamine containing compound metabolic process GO: negative regulation of kinase activity GO: muscle atrophy GO: positive regulation of cerebellar granule cell precursor proliferation GO: canonical Wnt receptor signaling pathway involved in positive regulation of wound healing GO: respiratory system process GO: canonical Wnt receptor signaling pathway involved in positive regulation of endothelial cell migration GO: canonical Wnt receptor signaling pathway involved in positive regulation of cell cell adhesion GO: mitochondrial DNA replication GO: response to selenium ion GO: mrna metabolic process GO: RNA metabolic process GO: mammary gland alveolus development GO: viral reproduction GO: blood vessel remodeling GO: negative regulation of cell death GO: bone mineralization involved in bone maturation GO: positive regulation of calcineurin NFAT signaling pathway GO: positive regulation of protein import into nucleus, translocation GO: chondroitin sulfate proteoglycan biosynthetic process GO: glucose metabolic process GO: induction of apoptosis GO: muscle hypertrophy GO: negative regulation of protein dephosphorylation GO: regulation of transcription, DNA dependent GO: glycolate metabolic process GO: satellite cell maintenance involved in skeletal muscle regeneration GO: myotube cell development GO: regulation of translational initiation GO: NAD metabolic process GO: hydrogen peroxide catabolic process GO: neurotransmitter catabolic process GO: mitochondrion transport along microtubule GO: positive regulation of glucose import GO: NAD biosynthetic process GO: purine base biosynthetic process GO: tricarboxylic acid cycle GO: aerobic respiration GO: glucocorticoid metabolic process GO: regulation of transcription from RNA polymerase II promoter by nuclear hormone receptor GO: transcription, DNA dependent GO: negative regulation of translational initiation GO: fatty acid beta oxidation using acyl CoA dehydrogenase GO: triglyceride biosynthetic process GO: cellular response to magnesium ion GO: neurotransmitter uptake GO: activation of mitotic anaphase promoting complex activity GO: regulation of B cell apoptosis GO: negative regulation of cyclin dependent protein kinase activity involved in G1 S GO: regulation of myeloid cell apoptosis GO: respiratory electron transport chain GO: negative regulation of macrophage derived foam cell differentiation GO: hydrogen peroxide mediated programmed cell death GO: biosynthetic process GO: ovarian follicle rupture GO: peptidyl histidine phosphorylation GO: basophil chemotaxis GO: positive regulation of mast cell chemotaxis GO: primitive erythrocyte differentiation GO: lysine catabolic process GO: cell migration involved in sprouting angiogenesis GO: negative regulation of protein phosphorylation GO: negative regulation of cell proliferation GO: transmembrane receptor protein tyrosine kinase signaling pathway GO: heart development GO: response to glucose stimulus GO: negative regulation of catalytic activity GO: cellular amino acid biosynthetic process GO: fatty acid transport GO: NADPH oxidation GO: negative regulation of signal transduction GO: negative regulation of interleukin 8 production GO: negative regulation of appetite GO: cochlea development GO: mechanoreceptor differentiation GO: steroid hormone mediated signaling pathway GO: pyruvate metabolic process GO: regulation of fatty acid metabolic process GO: regulation of cell death GO: L serine catabolic process GO: brown fat cell differentiation GO: negative regulation of cholesterol storage GO: regulation of insulin receptor signaling pathway GO: detection of stimulus GO: folic acid metabolic process GO: heart valve morphogenesis GO: positive regulation of insulin receptor signaling pathway GO: negative regulation of focal adhesion assembly GO: protein stabilization GO: negative regulation of molecular function GO: aging GO: negative regulation of insulin secretion GO: reflex GO: CD8 positive, alpha beta T cell differentiation GO: purine nucleotide biosynthetic process GO: regulation of cyclin dependent protein kinase activity GO: alpha beta T cell differentiation GO: one carbon metabolic process GO: righting reflex GO: mammary gland duct morphogenesis GO: pyridine nucleotide biosynthetic process GO: triglyceride catabolic process GO: cellular response to prostaglandin stimulus GO: negative regulation of somatostatin secretion GO: positive regulation of phosphatidylinositol 3 kinase cascade GO: negative regulation of interferon gamma mediated signaling pathway GO: protein localization to centrosome GO: response to reactive oxygen species GO: neuron maturation GO: response to mercury ion GO: rrna transport GO: induction of apoptosis by oxidative stress GO: cellular sodium ion homeostasis GO: positive regulation of protein complex disassembly GO: protein pyridoxal 5 phosphate linkage via peptidyl N6 pyridoxal phosphate L lysine GO: cysteine biosynthetic process GO: cellular response to cobalt ion GO: bronchus morphogenesis GO: negative regulation of mitochondrial fusion GO: negative regulation of membrane potential GO: negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning GO: lipoprotein metabolic process GO: negative regulation of glycolysis GO: regulation of adenylate cyclase activity GO: negative regulation of transcription, DNA dependent GO: insulin receptor signaling pathway GO: fibroblast growth factor receptor signaling pathway GO: carnitine shuttle GO: positive regulation of mesenchymal cell proliferation GO: energy reserve metabolic process GO: ribosome biogenesis GO: myoblast proliferation GO: glycogen catabolic process GO: response to molecule of fungal origin GO: embryonic camera type eye development GO: positive chemotaxis GO: positive regulation of glycolysis GO: negative regulation of transforming growth factor beta receptor signaling pathway GO: glycine biosynthetic process from serine GO: chromatin modification GO: prostate gland stromal morphogenesis GO: centrosome cycle GO: regulation of protein metabolic process GO: regulation of fatty acid oxidation GO: negative regulation of B cell apoptosis GO: glucocorticoid receptor signaling pathway GO: positive regulation of NAD(P)H oxidase activity GO: homotypic cell cell adhesion GO: regulation of blood vessel size by renin angiotensin GO: farnesyl diphosphate metabolic process GO: low density lipoprotein particle remodeling GO: renin angiotensin regulation of aldosterone production GO: cell cell junction assembly GO: prostate gland epithelium morphogenesis GO: negative regulation of transporter activity GO: protein dephosphorylation GO: positive regulation of glucose metabolic process GO: peptidyl tyrosine dephosphorylation GO: regulation of establishment or maintenance of cell polarity GO: intracellular receptor mediated signaling pathway GO: negative regulation of G1 S transition of mitotic cell cycle GO: protein tetramerization GO: fatty acyl CoA biosynthetic process GO: negative regulation of ATPase activity GO: positive regulation of phospholipase A2 activity GO: organ development GO: auditory receptor cell differentiation GO: regulation of vasodilation GO: cellular biogenic amine metabolic process GO: positive regulation of cell fate specification GO: asymmetric cell division GO: regulation of endodeoxyribonuclease activity Figure S4. Enrichment in biological process Gene Ontology (BP:GO) terms between obese, overweight and normal (lean) female subjects, as calculated by the Reporter Features algorithm. Red and blue represent enrichment in up-regulated and down-regulated genes respectively. Deep red/blue represents 1e
23 Figure S5. Illustration of omega 9 and other fatty acids metabolism in the genome-scale metabolic model for adipocytes, iadipocytes
24 Figure S6. Illustration of linoleate (LA), unsaturated omega-6 fatty acid and linolenate (ALA), polyunsaturated omega-3 fatty acid acids metabolism in the genome-scale metabolic model for adipocytes, iadipocytes1809. LA and ALA are essential fatty acids and should be taken with the diet. 24
25 Figure S7. Illustration of odd chain fatty acids metabolism in the genome-scale metabolic model for adipocytes, iadipocytes
26 Figure S8. Reporter Metabolites obtained in the same way as in Figure 5, but using the previously published GEM, iab586. Metabolites with p-values lower than 6e-3 are shown, rather than the top 20 metabolites, since there were only a few metabolites around which the genes were significantly up regulated. 26
LIPID METABOLISM. Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI
 LIPID METABOLISM Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI Lipid metabolism is concerned mainly with fatty acids cholesterol Source of fatty acids from dietary fat de novo
LIPID METABOLISM Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI Lipid metabolism is concerned mainly with fatty acids cholesterol Source of fatty acids from dietary fat de novo
Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler
 Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler DR CALUX Boys Girls Database Systemic lupus erythematosus 4.4 0.0021 6.7
Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler DR CALUX Boys Girls Database Systemic lupus erythematosus 4.4 0.0021 6.7
Supplementary Figure 1.
 Supplementary Figure 1. Increased expression of cell cycle pathway genes in insulin + Glut2 low cells of STZ-induced diabetic islets. A) random blood glucose measuers of STZ and vehicle treated MIP-GFP
Supplementary Figure 1. Increased expression of cell cycle pathway genes in insulin + Glut2 low cells of STZ-induced diabetic islets. A) random blood glucose measuers of STZ and vehicle treated MIP-GFP
BCM 221 LECTURES OJEMEKELE O.
 BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
Identification of Tissue-Specific Protein-Coding and Noncoding. Transcripts across 14 Human Tissues Using RNA-seq
 Identification of Tissue-Specific Protein-Coding and Noncoding Transcripts across 14 Human Tissues Using RNA-seq Jinhang Zhu 1, Geng Chen 1, Sibo Zhu 1,2, Suqing Li 3, Zhuo Wen 3, Bin Li 1, Yuanting Zheng
Identification of Tissue-Specific Protein-Coding and Noncoding Transcripts across 14 Human Tissues Using RNA-seq Jinhang Zhu 1, Geng Chen 1, Sibo Zhu 1,2, Suqing Li 3, Zhuo Wen 3, Bin Li 1, Yuanting Zheng
23.1 Lipid Metabolism in Animals. Chapter 23. Micelles Lipid Metabolism in. Animals. Overview of Digestion Lipid Metabolism in
 Denniston Topping Caret Copyright! The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Chapter 23 Fatty Acid Metabolism Triglycerides (Tgl) are emulsified into fat droplets
Denniston Topping Caret Copyright! The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Chapter 23 Fatty Acid Metabolism Triglycerides (Tgl) are emulsified into fat droplets
Systems biology approaches and pathway tools for investigating cardiovascular disease
 Systems biology approaches and pathway tools for investigating cardiovascular disease Craig E. Wheelock 1,2*, Åsa M. Wheelock 2,3,4, Shuichi Kawashima 5, Diego Diez 2, Minoru Kanehisa 2,5, Marjan van Erk
Systems biology approaches and pathway tools for investigating cardiovascular disease Craig E. Wheelock 1,2*, Åsa M. Wheelock 2,3,4, Shuichi Kawashima 5, Diego Diez 2, Minoru Kanehisa 2,5, Marjan van Erk
Published on Second Faculty of Medicine, Charles University (http://www.lf2.cuni.cz )
 Published on Second Faculty of Medicine, Charles University (http://www.lf2.cuni.cz ) Biochemistry Submitted by Marie Havlová on 8. February 2012-0:00 Syllabus of Biochemistry Mechanisms of enzyme catalysis.
Published on Second Faculty of Medicine, Charles University (http://www.lf2.cuni.cz ) Biochemistry Submitted by Marie Havlová on 8. February 2012-0:00 Syllabus of Biochemistry Mechanisms of enzyme catalysis.
LIPID METABOLISM
 LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
BIOL2171 ANU TCA CYCLE
 TCA CYCLE IMPORTANCE: Oxidation of 2C Acetyl Co-A 2CO 2 + 3NADH + FADH 2 (8e-s donated to O 2 in the ETC) + GTP (energy) + Heat OVERVIEW: Occurs In the mitochondrion matrix. 1. the acetyl portion of acetyl-coa
TCA CYCLE IMPORTANCE: Oxidation of 2C Acetyl Co-A 2CO 2 + 3NADH + FADH 2 (8e-s donated to O 2 in the ETC) + GTP (energy) + Heat OVERVIEW: Occurs In the mitochondrion matrix. 1. the acetyl portion of acetyl-coa
ANSC/NUTR 618 Lipids & Lipid Metabolism
 I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
Integrative Metabolism: Significance
 Integrative Metabolism: Significance Energy Containing Nutrients Carbohydrates Fats Proteins Catabolism Energy Depleted End Products H 2 O NH 3 ADP + Pi NAD + NADP + FAD + Pi NADH+H + NADPH+H + FADH2 Cell
Integrative Metabolism: Significance Energy Containing Nutrients Carbohydrates Fats Proteins Catabolism Energy Depleted End Products H 2 O NH 3 ADP + Pi NAD + NADP + FAD + Pi NADH+H + NADPH+H + FADH2 Cell
CHM333 LECTURE 34: 11/30 12/2/09 FALL 2009 Professor Christine Hrycyna
 Lipid Metabolism β-oxidation FA Acetyl-CoA Triacylglycerols (TAGs) and glycogen are the two major forms of stored energy in vertebrates Glycogen can supply ATP for muscle contraction for less than an hour
Lipid Metabolism β-oxidation FA Acetyl-CoA Triacylglycerols (TAGs) and glycogen are the two major forms of stored energy in vertebrates Glycogen can supply ATP for muscle contraction for less than an hour
Biological role of lipids
 Lipids Lipids Organic compounds present in living organisms, insoluble in water but able to be extracted by organic solvents such as: chloroform, acetone, benzene. Extraction = the action of taking out
Lipids Lipids Organic compounds present in living organisms, insoluble in water but able to be extracted by organic solvents such as: chloroform, acetone, benzene. Extraction = the action of taking out
Lipids digestion and absorption, Biochemistry II
 Lipids digestion and absorption, blood plasma lipids, lipoproteins Biochemistry II Lecture 1 2008 (J.S.) Triacylglycerols (as well as free fatty acids and both free and esterified cholesterol) are very
Lipids digestion and absorption, blood plasma lipids, lipoproteins Biochemistry II Lecture 1 2008 (J.S.) Triacylglycerols (as well as free fatty acids and both free and esterified cholesterol) are very
Introduction to the Study of Lipids
 Introduction to the Study of Lipids Factors to Consider in the Study of Biomolecules What are the features of the basic building blocks? (ex: monosaccharides, alcohols, fatty acids, amino acids) 1) General
Introduction to the Study of Lipids Factors to Consider in the Study of Biomolecules What are the features of the basic building blocks? (ex: monosaccharides, alcohols, fatty acids, amino acids) 1) General
Erythrocyte Essential Fatty Acids
 Biolab reference: PHWN\ABCD\A12 Referred by: Dr.. Patient: Mr Sample Patient Date of birth: 16/01/969 Your reference: Date: 04/01/2012 Sex: Male Sample date: 03/01/2012 Erythrocyte Essential Fatty Acids
Biolab reference: PHWN\ABCD\A12 Referred by: Dr.. Patient: Mr Sample Patient Date of birth: 16/01/969 Your reference: Date: 04/01/2012 Sex: Male Sample date: 03/01/2012 Erythrocyte Essential Fatty Acids
OVERVIEW M ET AB OL IS M OF FR EE FA TT Y AC ID S
 LIPOLYSIS LIPOLYSIS OVERVIEW CATABOLISM OF FREE FATTY ACIDS Nonesterified fatty acids Source:- (a) breakdown of TAG in adipose tissue (b) action of Lipoprotein lipase on plasma TAG Combined with Albumin
LIPOLYSIS LIPOLYSIS OVERVIEW CATABOLISM OF FREE FATTY ACIDS Nonesterified fatty acids Source:- (a) breakdown of TAG in adipose tissue (b) action of Lipoprotein lipase on plasma TAG Combined with Albumin
6. How Are Fatty Acids Produced? 7. How Are Acylglycerols and Compound Lipids Produced? 8. How Is Cholesterol Produced?
 Lipid Metabolism Learning bjectives 1 How Are Lipids Involved in the Generationand Storage of Energy? 2 How Are Lipids Catabolized? 3 What Is the Energy Yield from the xidation of Fatty Acids? 4 How Are
Lipid Metabolism Learning bjectives 1 How Are Lipids Involved in the Generationand Storage of Energy? 2 How Are Lipids Catabolized? 3 What Is the Energy Yield from the xidation of Fatty Acids? 4 How Are
Integration Of Metabolism
 Integration Of Metabolism Metabolism Consist of Highly Interconnected Pathways The basic strategy of catabolic metabolism is to form ATP, NADPH, and building blocks for biosyntheses. 1. ATP is the universal
Integration Of Metabolism Metabolism Consist of Highly Interconnected Pathways The basic strategy of catabolic metabolism is to form ATP, NADPH, and building blocks for biosyntheses. 1. ATP is the universal
Lipid Metabolism. Catabolism Overview
 Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
Bio 366: Biological Chemistry II Test #1, 100 points (7 pages)
 Bio 366: Biological Chemistry II Test #1, 100 points (7 pages) READ THIS: Take a numbered test and sit in the seat with that number on it. Remove the numbered sticker from the desk, and stick it on the
Bio 366: Biological Chemistry II Test #1, 100 points (7 pages) READ THIS: Take a numbered test and sit in the seat with that number on it. Remove the numbered sticker from the desk, and stick it on the
Roles of Lipids. principal form of stored energy major constituents of cell membranes vitamins messengers intra and extracellular
 Roles of Lipids principal form of stored energy major constituents of cell membranes vitamins messengers intra and extracellular = Oxidation of fatty acids Central energy-yielding pathway in animals. O
Roles of Lipids principal form of stored energy major constituents of cell membranes vitamins messengers intra and extracellular = Oxidation of fatty acids Central energy-yielding pathway in animals. O
Integration of clinical data with a genome-scale metabolic model of the human adipocyte
 Molecular Systems Biology 9; Article number 649; doi:10.1038/msb.2013.5 Citation: Molecular Systems Biology 9:649 www.molecularsystemsbiology.com Integration of clinical data with a genome-scale metabolic
Molecular Systems Biology 9; Article number 649; doi:10.1038/msb.2013.5 Citation: Molecular Systems Biology 9:649 www.molecularsystemsbiology.com Integration of clinical data with a genome-scale metabolic
Plasma lipoproteins & atherosclerosis by. Prof.Dr. Maha M. Sallam
 Biochemistry Department Plasma lipoproteins & atherosclerosis by Prof.Dr. Maha M. Sallam 1 1. Recognize structures,types and role of lipoproteins in blood (Chylomicrons, VLDL, LDL and HDL). 2. Explain
Biochemistry Department Plasma lipoproteins & atherosclerosis by Prof.Dr. Maha M. Sallam 1 1. Recognize structures,types and role of lipoproteins in blood (Chylomicrons, VLDL, LDL and HDL). 2. Explain
Chapter 16 - Lipid Metabolism
 Chapter 16 - Lipid Metabolism Fatty acids have four major physiologic roles in the cell: Building blocks of phospholipids and glycolipids Added onto proteins to create lipoproteins, which targets them
Chapter 16 - Lipid Metabolism Fatty acids have four major physiologic roles in the cell: Building blocks of phospholipids and glycolipids Added onto proteins to create lipoproteins, which targets them
Oxidation of Long Chain Fatty Acids
 Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
Nafith Abu Tarboush DDS, MSc, PhD
 Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Lipids (cholesterol, cholesterol esters, phospholipids & triacylglycerols) combined with proteins (apolipoprotein) in
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Lipids (cholesterol, cholesterol esters, phospholipids & triacylglycerols) combined with proteins (apolipoprotein) in
BBSG 501 Section 4 Metabolic Fuels, Energy and Order Fall 2003 Semester
 BBSG 501 Section 4 Metabolic Fuels, Energy and Order Fall 2003 Semester Section Director: Dave Ford, Ph.D. Office: MS 141: ext. 8129: e-mail: fordda@slu.edu Lecturers: Michael Moxley, Ph.D. Office: MS
BBSG 501 Section 4 Metabolic Fuels, Energy and Order Fall 2003 Semester Section Director: Dave Ford, Ph.D. Office: MS 141: ext. 8129: e-mail: fordda@slu.edu Lecturers: Michael Moxley, Ph.D. Office: MS
number Done by Corrected by Doctor Faisal Al-Khatibe
 number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
Fatty Acid and Triacylglycerol Metabolism 1
 Fatty Acid and Triacylglycerol Metabolism 1 Mobilization of stored fats and oxidation of fatty acids Lippincott s Chapter 16 What is the first lecture about What is triacylglycerol Fatty acids structure
Fatty Acid and Triacylglycerol Metabolism 1 Mobilization of stored fats and oxidation of fatty acids Lippincott s Chapter 16 What is the first lecture about What is triacylglycerol Fatty acids structure
Fats and Lipids (Ans570)
 Fats and Lipids (Ans570) Outlines Fats and Lipids Structure, nomenclature Phospholipids, Sterols, and Lipid Derivatives Lipid Oxidation Roles of fat in food processing and dietary fat Lipid and fat analysis:
Fats and Lipids (Ans570) Outlines Fats and Lipids Structure, nomenclature Phospholipids, Sterols, and Lipid Derivatives Lipid Oxidation Roles of fat in food processing and dietary fat Lipid and fat analysis:
By: Dr Hadi Mozafari 1
 Biological lipids are a chemically diverse group of compounds, the common and defining feature of which is their insolubility in water. By: Dr Hadi Mozafari 1 Fats and oils are the principal stored forms
Biological lipids are a chemically diverse group of compounds, the common and defining feature of which is their insolubility in water. By: Dr Hadi Mozafari 1 Fats and oils are the principal stored forms
CHY2026: General Biochemistry. Lipid Metabolism
 CHY2026: General Biochemistry Lipid Metabolism Lipid Digestion Lipid Metabolism Fats (triglycerides) are high metabolic energy molecules Fats yield 9.3 kcal of energy (carbohydrates and proteins 4.1 kcal)
CHY2026: General Biochemistry Lipid Metabolism Lipid Digestion Lipid Metabolism Fats (triglycerides) are high metabolic energy molecules Fats yield 9.3 kcal of energy (carbohydrates and proteins 4.1 kcal)
Summary of fatty acid synthesis
 Lipid Metabolism, part 2 1 Summary of fatty acid synthesis 8 acetyl CoA + 14 NADPH + 14 H+ + 7 ATP palmitic acid (16:0) + 8 CoA + 14 NADP + + 7 ADP + 7 Pi + 7 H20 1. The major suppliers of NADPH for fatty
Lipid Metabolism, part 2 1 Summary of fatty acid synthesis 8 acetyl CoA + 14 NADPH + 14 H+ + 7 ATP palmitic acid (16:0) + 8 CoA + 14 NADP + + 7 ADP + 7 Pi + 7 H20 1. The major suppliers of NADPH for fatty
Lecture 5: Cell Metabolism. Biology 219 Dr. Adam Ross
 Lecture 5: Cell Metabolism Biology 219 Dr. Adam Ross Cellular Respiration Set of reactions that take place during the conversion of nutrients into ATP Intricate regulatory relationship between several
Lecture 5: Cell Metabolism Biology 219 Dr. Adam Ross Cellular Respiration Set of reactions that take place during the conversion of nutrients into ATP Intricate regulatory relationship between several
Lecture 36. Key Concepts. Overview of lipid metabolism. Reactions of fatty acid oxidation. Energy yield from fatty acid oxidation
 Lecture 36 Lipid Metabolism 1 Fatty Acid Oxidation Ketone Bodies Key Concepts Overview of lipid metabolism Reactions of fatty acid oxidation Energy yield from fatty acid oxidation Formation of ketone bodies
Lecture 36 Lipid Metabolism 1 Fatty Acid Oxidation Ketone Bodies Key Concepts Overview of lipid metabolism Reactions of fatty acid oxidation Energy yield from fatty acid oxidation Formation of ketone bodies
Lipid metabolism. Degradation and biosynthesis of fatty acids Ketone bodies
 Lipid metabolism Degradation and biosynthesis of fatty acids Ketone bodies Fatty acids (FA) primary fuel molecules in the fat category main use is for long-term energy storage high level of energy storage:
Lipid metabolism Degradation and biosynthesis of fatty acids Ketone bodies Fatty acids (FA) primary fuel molecules in the fat category main use is for long-term energy storage high level of energy storage:
Intermediary metabolism. Eva Samcová
 Intermediary metabolism Eva Samcová Metabolic roles of tissues Four major tissues play a dominant role in fuel metabolism : liver, adipose, muscle, and brain. These tissues do not function in isolation.
Intermediary metabolism Eva Samcová Metabolic roles of tissues Four major tissues play a dominant role in fuel metabolism : liver, adipose, muscle, and brain. These tissues do not function in isolation.
REACTOME: Nonsense Mediated Decay (NMD) REACTOME:72764 Eukaryotic Translation Termination. REACTOME:72737 Cap dependent Translation Initiation
 A REACTOME:975957 Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) REACTOME:975956 Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) REACTOME:927802
A REACTOME:975957 Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) REACTOME:975956 Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) REACTOME:927802
Lipid and Amino Acid Metabolism
 CHEM 3331 Fundamentals of Biochemistry Chapter 14 Lipid and Amino Acid Metabolism Organic and Biochemistry for Today Spencer L. Seager / Michael R. Slabaugh Mr. Kevin A. Boudreaux Angelo State University
CHEM 3331 Fundamentals of Biochemistry Chapter 14 Lipid and Amino Acid Metabolism Organic and Biochemistry for Today Spencer L. Seager / Michael R. Slabaugh Mr. Kevin A. Boudreaux Angelo State University
Biosynthesis of Fatty Acids. By Dr.QUTAIBA A. QASIM
 Biosynthesis of Fatty Acids By Dr.QUTAIBA A. QASIM Fatty Acids Definition Fatty acids are comprised of hydrocarbon chains terminating with carboxylic acid groups. Fatty acids and their associated derivatives
Biosynthesis of Fatty Acids By Dr.QUTAIBA A. QASIM Fatty Acids Definition Fatty acids are comprised of hydrocarbon chains terminating with carboxylic acid groups. Fatty acids and their associated derivatives
Chapter 26 Biochemistry 5th edition. phospholipids. Sphingolipids. Cholesterol. db=books&itool=toolbar
 http://www.ncbi.nlm.nih.gov/sites/entrez? db=books&itool=toolbar 1 The surface of a soap bubble is a bilayer formed by detergent molecules 2 Chapter 26 Biochemistry 5th edition phospholipids Sphingolipids
http://www.ncbi.nlm.nih.gov/sites/entrez? db=books&itool=toolbar 1 The surface of a soap bubble is a bilayer formed by detergent molecules 2 Chapter 26 Biochemistry 5th edition phospholipids Sphingolipids
MPS Advanced Plant Biochemistry Course
 MPS 587 - Advanced Plant Biochemistry Course Lipids section guest lecture: Philip Bates Post-doc Browse lab Office: 453 Clark Hall Email: phil_bates@wsu.edu Lecture 9 Lipids I 1. What is a lipid? 2. Fatty
MPS 587 - Advanced Plant Biochemistry Course Lipids section guest lecture: Philip Bates Post-doc Browse lab Office: 453 Clark Hall Email: phil_bates@wsu.edu Lecture 9 Lipids I 1. What is a lipid? 2. Fatty
BIOSYNTHESIS OF FATTY ACIDS. doc. Ing. Zenóbia Chavková, CSc.
 BIOSYNTHESIS OF FATTY ACIDS doc. Ing. Zenóbia Chavková, CSc. The pathway for the of FAs is not the reversal of the oxidation pathway Both pathways are separated within different cellular compartments In
BIOSYNTHESIS OF FATTY ACIDS doc. Ing. Zenóbia Chavková, CSc. The pathway for the of FAs is not the reversal of the oxidation pathway Both pathways are separated within different cellular compartments In
Multiple choice: Circle the best answer on this exam. There are 12 multiple choice questions, each question is worth 3 points.
 CHEM 4420 Exam 4 Spring 2015 Dr. Stone Page 1 of 6 Name Use complete sentences when requested. There are 120 possible points on this exam. Therefore there are 20 bonus points. Multiple choice: Circle the
CHEM 4420 Exam 4 Spring 2015 Dr. Stone Page 1 of 6 Name Use complete sentences when requested. There are 120 possible points on this exam. Therefore there are 20 bonus points. Multiple choice: Circle the
Organic molecules highly hydrophobic and water insoluble.
 UNIT 5. LIPIDS OUTLINE 5.1. Introduction. 5.2. Fatty acids. 5.3. Eicosanoids. 5.4. Triacylglycerols = Triglycerides. 5.5. Waxes. 5.6. Membrane lipids: glycerophospholipids and sphingolipids. 5.7. Isoprenoids
UNIT 5. LIPIDS OUTLINE 5.1. Introduction. 5.2. Fatty acids. 5.3. Eicosanoids. 5.4. Triacylglycerols = Triglycerides. 5.5. Waxes. 5.6. Membrane lipids: glycerophospholipids and sphingolipids. 5.7. Isoprenoids
Synthesis and degradation of fatty acids Martina Srbová
 Synthesis and degradation of fatty acids Martina Srbová martina.srbova@lfmotol.cuni.cz Fatty acids (FA) mostly an even number of carbon atoms and linear chain in esterified form as component of lipids
Synthesis and degradation of fatty acids Martina Srbová martina.srbova@lfmotol.cuni.cz Fatty acids (FA) mostly an even number of carbon atoms and linear chain in esterified form as component of lipids
Biosynthesis of Triacylglycerides (TG) in liver. Mobilization of stored fat and oxidation of fatty acids
 Biosynthesis of Triacylglycerides (TG) in liver Mobilization of stored fat and oxidation of fatty acids Activation of hormone sensitive lipase This enzyme is activated when phosphorylated (3,5 cyclic AMPdependent
Biosynthesis of Triacylglycerides (TG) in liver Mobilization of stored fat and oxidation of fatty acids Activation of hormone sensitive lipase This enzyme is activated when phosphorylated (3,5 cyclic AMPdependent
AMINO ACIDS NON-ESSENTIAL ESSENTIAL
 Edith Frederika Introduction A major component of food is PROTEIN The protein ingested as part of our diet are not the same protein required by the body Only 40 to 50 gr of protein is required by a normal
Edith Frederika Introduction A major component of food is PROTEIN The protein ingested as part of our diet are not the same protein required by the body Only 40 to 50 gr of protein is required by a normal
CHAPTER 28 LIPIDS SOLUTIONS TO REVIEW QUESTIONS
 28 09/16/2013 17:44:40 Page 415 APTER 28 LIPIDS SLUTINS T REVIEW QUESTINS 1. The lipids, which are dissimilar substances, are arbitrarily classified as a group on the basis of their solubility in fat solvents
28 09/16/2013 17:44:40 Page 415 APTER 28 LIPIDS SLUTINS T REVIEW QUESTINS 1. The lipids, which are dissimilar substances, are arbitrarily classified as a group on the basis of their solubility in fat solvents
Integration Of Metabolism
 Integration Of Metabolism Metabolism Consist of Highly Interconnected Pathways The basic strategy of catabolic metabolism is to form ATP, NADPH, and building blocks for biosyntheses. 1. ATP is the universal
Integration Of Metabolism Metabolism Consist of Highly Interconnected Pathways The basic strategy of catabolic metabolism is to form ATP, NADPH, and building blocks for biosyntheses. 1. ATP is the universal
MILK BIOSYNTHESIS PART 3: FAT
 MILK BIOSYNTHESIS PART 3: FAT KEY ENZYMES (FROM ALL BIOSYNTHESIS LECTURES) FDPase = fructose diphosphatase Citrate lyase Isocitrate dehydrogenase Fatty acid synthetase Acetyl CoA carboxylase Fatty acyl
MILK BIOSYNTHESIS PART 3: FAT KEY ENZYMES (FROM ALL BIOSYNTHESIS LECTURES) FDPase = fructose diphosphatase Citrate lyase Isocitrate dehydrogenase Fatty acid synthetase Acetyl CoA carboxylase Fatty acyl
Chapter 8. Functions of Lipids. Structural Nature of Lipids. BCH 4053 Spring 2001 Chapter 8 Lecture Notes. Slide 1. Slide 2.
 BCH 4053 Spring 2001 Chapter 8 Lecture Notes 1 Chapter 8 Lipids 2 Functions of Lipids Energy Storage Thermal Insulation Structural Components of Membranes Protective Coatings of Plants and Insects Hormonal
BCH 4053 Spring 2001 Chapter 8 Lecture Notes 1 Chapter 8 Lipids 2 Functions of Lipids Energy Storage Thermal Insulation Structural Components of Membranes Protective Coatings of Plants and Insects Hormonal
Fatty acids synthesis
 Fatty acids synthesis The synthesis start from Acetyl COA the first step requires ATP + reducing power NADPH! even though the oxidation and synthesis are different pathways but from chemical part of view
Fatty acids synthesis The synthesis start from Acetyl COA the first step requires ATP + reducing power NADPH! even though the oxidation and synthesis are different pathways but from chemical part of view
Cholesterol and its transport. Alice Skoumalová
 Cholesterol and its transport Alice Skoumalová 27 carbons Cholesterol - structure Cholesterol importance A stabilizing component of cell membranes A precursor of bile salts A precursor of steroid hormones
Cholesterol and its transport Alice Skoumalová 27 carbons Cholesterol - structure Cholesterol importance A stabilizing component of cell membranes A precursor of bile salts A precursor of steroid hormones
OBJECTIVE. Lipids are largely hydrocarbon derivatives and thus represent
 Paper 4. Biomolecules and their interactions Module 20: Saturated and unsaturated fatty acids, Nomenclature of fatty acids and Essential and non-essential fatty acids OBJECTIVE The main aim of this module
Paper 4. Biomolecules and their interactions Module 20: Saturated and unsaturated fatty acids, Nomenclature of fatty acids and Essential and non-essential fatty acids OBJECTIVE The main aim of this module
E.coli Core Model: Metabolic Core
 1 E.coli Core Model: Metabolic Core 2 LEARNING OBJECTIVES Each student should be able to: Describe the glycolysis pathway in the core model. Describe the TCA cycle in the core model. Explain gluconeogenesis.
1 E.coli Core Model: Metabolic Core 2 LEARNING OBJECTIVES Each student should be able to: Describe the glycolysis pathway in the core model. Describe the TCA cycle in the core model. Explain gluconeogenesis.
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM. Fatty Acid Elongation and Desaturation
 ANSC/NUTR 618 LIPIDS & LIPID METABOLISM I. Fatty acid elongation A. General 1. At least 60% of fatty acids in triacylglycerols are C18. 2. Free palmitic acid (16:0) synthesized in cytoplasm is elongated
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM I. Fatty acid elongation A. General 1. At least 60% of fatty acids in triacylglycerols are C18. 2. Free palmitic acid (16:0) synthesized in cytoplasm is elongated
Lipid Metabolism. Remember fats?? Triacylglycerols - major form of energy storage in animals
 Remember fats?? Triacylglycerols - major form of energy storage in animals Your energy reserves: ~0.5% carbs (glycogen + glucose) ~15% protein (muscle, last resort) ~85% fat Why use fat for energy? 1 gram
Remember fats?? Triacylglycerols - major form of energy storage in animals Your energy reserves: ~0.5% carbs (glycogen + glucose) ~15% protein (muscle, last resort) ~85% fat Why use fat for energy? 1 gram
Lehninger 5 th ed. Chapter 17
 Lehninger 5 th ed. Chapter 17 December 26, 2010 Prof. Shimon Schuldiner Email: Shimon.Schuldiner@huji.ac.il Phone: 6585992 CHAPTER 17 Fatty Acid Catabolism Key topics: How fats are digested in animals
Lehninger 5 th ed. Chapter 17 December 26, 2010 Prof. Shimon Schuldiner Email: Shimon.Schuldiner@huji.ac.il Phone: 6585992 CHAPTER 17 Fatty Acid Catabolism Key topics: How fats are digested in animals
Comparison of Nutrients, Nutrient Ratios and Other Food Components in NDSR and the ASA24
 Comparison of Nutrients, Nutrient Ratios and Other Food Components in and the Category Primary Energy Sources Alcohol Total Fat Total Protein Energy (kilocalories) Total Carbohydrate Energy (kilojoules)
Comparison of Nutrients, Nutrient Ratios and Other Food Components in and the Category Primary Energy Sources Alcohol Total Fat Total Protein Energy (kilocalories) Total Carbohydrate Energy (kilojoules)
Objectives By the end of lecture the student should:
 Objectives By the end of lecture the student should: Illustrate α oxidation of fatty acids. Understand ω oxidation of fatty acids. List sources and fates of active acetate. Discuss eicosanoids. 2- α Oxidation
Objectives By the end of lecture the student should: Illustrate α oxidation of fatty acids. Understand ω oxidation of fatty acids. List sources and fates of active acetate. Discuss eicosanoids. 2- α Oxidation
Principles of Anatomy and Physiology
 Principles of Anatomy and Physiology 14 th Edition CHAPTER 25 Metabolism and Nutrition Metabolic Reactions Metabolism refers to all of the chemical reactions taking place in the body. Reactions that break
Principles of Anatomy and Physiology 14 th Edition CHAPTER 25 Metabolism and Nutrition Metabolic Reactions Metabolism refers to all of the chemical reactions taking place in the body. Reactions that break
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 28 Fatty Acid Synthesis 2013 W. H. Freeman and Company Chapter 28 Outline 1. The first stage of fatty acid synthesis is transfer
Chemistry 1120 Exam 4 Study Guide
 Chemistry 1120 Exam 4 Study Guide Chapter 12 12.1 Identify and differentiate between macronutrients (lipids, amino acids and saccharides) and micronutrients (vitamins and minerals). Master Tutor Section
Chemistry 1120 Exam 4 Study Guide Chapter 12 12.1 Identify and differentiate between macronutrients (lipids, amino acids and saccharides) and micronutrients (vitamins and minerals). Master Tutor Section
CHAPTER 28 LIPIDS SOLUTIONS TO REVIEW QUESTIONS
 HAPTER 28 LIPIDS SLUTINS T REVIEW QUESTINS 1. The lipids, which are dissimilar substances, are arbitrarily classified as a group on the basis of their solubility in fat solvents and their insolubility
HAPTER 28 LIPIDS SLUTINS T REVIEW QUESTINS 1. The lipids, which are dissimilar substances, are arbitrarily classified as a group on the basis of their solubility in fat solvents and their insolubility
Lipid Metabolism * OpenStax
 OpenStax-CNX module: m46462 1 Lipid Metabolism * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you will be able
OpenStax-CNX module: m46462 1 Lipid Metabolism * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you will be able
Chapter 22, Fatty Acid Metabolism CH 3 (CH 2 ) 14 CO 2 R C C O2 CH 2 OH O R. Lipase + 3 H 2 O
 hapter 22, Fatty Acid Metabolism Pages: 603-613 I. Introduction - Fatty acids have 4 major physiological roles: - omponents of phospholipids and glycolipids (membranes) - Attachment to Proteins targets
hapter 22, Fatty Acid Metabolism Pages: 603-613 I. Introduction - Fatty acids have 4 major physiological roles: - omponents of phospholipids and glycolipids (membranes) - Attachment to Proteins targets
Biochemistry Sheet 27 Fatty Acid Synthesis Dr. Faisal Khatib
 Page1 بسم رلاهللا On Thursday, we discussed the synthesis of fatty acids and its regulation. We also went on to talk about the synthesis of Triacylglycerol (TAG). Last time, we started talking about the
Page1 بسم رلاهللا On Thursday, we discussed the synthesis of fatty acids and its regulation. We also went on to talk about the synthesis of Triacylglycerol (TAG). Last time, we started talking about the
Lecture: 26 OXIDATION OF FATTY ACIDS
 Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Optimising. membranes
 Optimising Neuronal membranes Lipids are classified as 1. Simple lipids oils and fats 2. Complex lipids a) Phospholipids b) Glycosphingolipids containing a fatty acid, sphingosine and a CHO c) Lipoproteins
Optimising Neuronal membranes Lipids are classified as 1. Simple lipids oils and fats 2. Complex lipids a) Phospholipids b) Glycosphingolipids containing a fatty acid, sphingosine and a CHO c) Lipoproteins
number Done by Corrected by Doctor Nayef Karadsheh
 number 13 Done by Asma Karameh Corrected by Saad hayek Doctor Nayef Karadsheh Gluconeogenesis This lecture covers gluconeogenesis with aspects of: 1) Introduction to glucose distribution through tissues.
number 13 Done by Asma Karameh Corrected by Saad hayek Doctor Nayef Karadsheh Gluconeogenesis This lecture covers gluconeogenesis with aspects of: 1) Introduction to glucose distribution through tissues.
UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DIVISION OF BASIC MEDICAL SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY
 1 UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DIVISION OF BASIC MEDICAL SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY GLUCOSE HOMEOSTASIS An Overview WHAT IS HOMEOSTASIS? Homeostasis
1 UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DIVISION OF BASIC MEDICAL SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY GLUCOSE HOMEOSTASIS An Overview WHAT IS HOMEOSTASIS? Homeostasis
Voet Biochemistry 3e John Wiley & Sons, Inc.
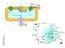 * * Voet Biochemistry 3e Lipid Metabolism Part I: (Chap. 25, sec.1-3) Glucose C 6 H 12 O 6 + 6 O 2 6 CO 2 + 6 H 2 O G o = -2823 kj/mol Fats (palmitic acid) C 16 H 32 O 2 + 23 O 2 16 CO 2 + 16 H 2 O G o
* * Voet Biochemistry 3e Lipid Metabolism Part I: (Chap. 25, sec.1-3) Glucose C 6 H 12 O 6 + 6 O 2 6 CO 2 + 6 H 2 O G o = -2823 kj/mol Fats (palmitic acid) C 16 H 32 O 2 + 23 O 2 16 CO 2 + 16 H 2 O G o
Metabolism of acylglycerols and sphingolipids. Martina Srbová
 Metabolism of acylglycerols and sphingolipids Martina Srbová Types of glycerolipids and sphingolipids 1. Triacylglycerols function as energy reserves adipose tissue (storage of triacylglycerol), lipoproteins
Metabolism of acylglycerols and sphingolipids Martina Srbová Types of glycerolipids and sphingolipids 1. Triacylglycerols function as energy reserves adipose tissue (storage of triacylglycerol), lipoproteins
Leen Alsahele. Razan Al-zoubi ... Faisal
 25 Leen Alsahele Razan Al-zoubi... Faisal last time we started talking about regulation of fatty acid synthesis and degradation *regulation of fatty acid synthesis by: 1- regulation of acetyl CoA carboxylase
25 Leen Alsahele Razan Al-zoubi... Faisal last time we started talking about regulation of fatty acid synthesis and degradation *regulation of fatty acid synthesis by: 1- regulation of acetyl CoA carboxylase
Formalin-fixed paraffin-embedded sections of liver from a recipient mouse sacrificed after two rounds
 Supplementary figure legends Supplementary Figure 1 Fah + hepatocytes in a Fah -/- mouse transplanted with sorted cells. Formalin-fixed paraffin-embedded sections of liver from a recipient mouse sacrificed
Supplementary figure legends Supplementary Figure 1 Fah + hepatocytes in a Fah -/- mouse transplanted with sorted cells. Formalin-fixed paraffin-embedded sections of liver from a recipient mouse sacrificed
Metabolomic Data Analysis with MetaboAnalystR
 Metabolomic Data Analysis with MetaboAnalystR Name: guest623839008349489450 February 7, 2018 1 Background Understanding the functional importance of metabolites in untargeted metabolomics is limited due
Metabolomic Data Analysis with MetaboAnalystR Name: guest623839008349489450 February 7, 2018 1 Background Understanding the functional importance of metabolites in untargeted metabolomics is limited due
Energy storage in cells
 Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
Fatty acids and phospholipids
 PYS 4xx Intro 2 1 PYS 4xx Intro 2 - Molecular building blocks We now describe in more detail the nomenclature and composition of several classes of compounds of relevance to the cell, including: membrane
PYS 4xx Intro 2 1 PYS 4xx Intro 2 - Molecular building blocks We now describe in more detail the nomenclature and composition of several classes of compounds of relevance to the cell, including: membrane
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM Lipoprotein Metabolism
 ANSC/NUTR 618 LIPIDS & LIPID METABOLISM Lipoprotein Metabolism I. Chylomicrons (exogenous pathway) A. 83% triacylglycerol, 2% protein, 8% cholesterol plus cholesterol esters, 7% phospholipid (esp. phosphatidylcholine)
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM Lipoprotein Metabolism I. Chylomicrons (exogenous pathway) A. 83% triacylglycerol, 2% protein, 8% cholesterol plus cholesterol esters, 7% phospholipid (esp. phosphatidylcholine)
BCH 4054 Spring 2001 Chapter 24 Lecture Notes
 BCH 4054 Spring 2001 Chapter 24 Lecture Notes 1 Chapter 24 Fatty Acid Catabolism 2 Fatty Acids as Energy Source Triglycerides yield 37 kj/g dry weight Protein 17 kj/g Glycogen 16 kj/g (even less wet weight)
BCH 4054 Spring 2001 Chapter 24 Lecture Notes 1 Chapter 24 Fatty Acid Catabolism 2 Fatty Acids as Energy Source Triglycerides yield 37 kj/g dry weight Protein 17 kj/g Glycogen 16 kj/g (even less wet weight)
Fatty acid breakdown
 Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 27 Fatty Acid Degradation Dietary Lipid (Triacylglycerol) Metabolism - In the small intestine, fat particles are coated with bile
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 27 Fatty Acid Degradation Dietary Lipid (Triacylglycerol) Metabolism - In the small intestine, fat particles are coated with bile
Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins
 Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins - Cholesterol: It is a sterol which is found in all eukaryotic cells and contains an oxygen (as a hydroxyl group OH) on Carbon number
Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins - Cholesterol: It is a sterol which is found in all eukaryotic cells and contains an oxygen (as a hydroxyl group OH) on Carbon number
The art of tracing dietary fat in humans. Leanne Hodson
 The art of tracing dietary fat in humans Leanne Hodson Dietary fat Other lipoproteins: IDL, LDL, HDL Hodson and Fielding linical Lipidology (2010) Relationship between blood & dietary fatty acids Typically:
The art of tracing dietary fat in humans Leanne Hodson Dietary fat Other lipoproteins: IDL, LDL, HDL Hodson and Fielding linical Lipidology (2010) Relationship between blood & dietary fatty acids Typically:
SUPPLEMENTARY DATA Supplementary Figure 1. Body weight and fat mass of AdicerKO mice.
 SUPPLEMENTARY DATA Supplementary Figure 1. Body weight and fat mass of AdicerKO mice. Twelve week old mice were subjected to ad libitum (AL) or dietary restriction (DR) regimens for three months. (A) Body
SUPPLEMENTARY DATA Supplementary Figure 1. Body weight and fat mass of AdicerKO mice. Twelve week old mice were subjected to ad libitum (AL) or dietary restriction (DR) regimens for three months. (A) Body
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM. Triacylglycerol and Fatty Acid Metabolism
 ANSC/NUTR 618 LIPIDS & LIPID METABOLISM II. Triacylglycerol synthesis A. Overall pathway Glycerol-3-phosphate + 3 Fatty acyl-coa à Triacylglycerol + 3 CoASH B. Enzymes 1. Acyl-CoA synthase 2. Glycerol-phosphate
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM II. Triacylglycerol synthesis A. Overall pathway Glycerol-3-phosphate + 3 Fatty acyl-coa à Triacylglycerol + 3 CoASH B. Enzymes 1. Acyl-CoA synthase 2. Glycerol-phosphate
Review Session 1. Control Systems and Homeostasis. Figure 1.8 A simple control system. Biol 219 Review Sessiono 1 Fall 2016
 Control Systems and Homeostasis Review Session 1 Regulated variables are kept within normal range by control mechanisms Keeps near set point, or optimum value Control systems local and reflex Input signal
Control Systems and Homeostasis Review Session 1 Regulated variables are kept within normal range by control mechanisms Keeps near set point, or optimum value Control systems local and reflex Input signal
INTEGRATIVE MEDICINE BLOOD - EDTA Result Range Units
 D Collected : 00-00-0000 INTEGRATIVE MEDICINE BLOOD - EDTA Result Range Units RED CELL FATTY ACID PROFILE Red Cell Fatty Acid Summary Saturated Fats, Total 36.99 19.30-39.40 Monounsaturated Fats, Total
D Collected : 00-00-0000 INTEGRATIVE MEDICINE BLOOD - EDTA Result Range Units RED CELL FATTY ACID PROFILE Red Cell Fatty Acid Summary Saturated Fats, Total 36.99 19.30-39.40 Monounsaturated Fats, Total
BIOLOGY 103 Spring 2001 MIDTERM LAB SECTION
 BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
: Overview of EFA metabolism
 Figure 1 gives an overview of the metabolic fate of the EFAs when consumed in the diet. The n-6 and n-3 PUFAs when consumed in the form of dietary triglyceride from various food sources undergoes digestion
Figure 1 gives an overview of the metabolic fate of the EFAs when consumed in the diet. The n-6 and n-3 PUFAs when consumed in the form of dietary triglyceride from various food sources undergoes digestion
2-more complex molecules (fatty acyl esters) as triacylglycerols.
 ** Fatty acids exist in two forms:- 1-free fatty acids (unesterified) 2-more complex molecules (fatty acyl esters) as triacylglycerols. ** most tissues might use fatty acids as source of energy during
** Fatty acids exist in two forms:- 1-free fatty acids (unesterified) 2-more complex molecules (fatty acyl esters) as triacylglycerols. ** most tissues might use fatty acids as source of energy during
Fatty Acid Degradation. Catabolism Overview. TAG and FA 11/11/2015. Chapter 27, Stryer Short Course. Lipids as a fuel source diet Beta oxidation
 Fatty Acid Degradation Chapter 27, Stryer Short Course Catabolism verview Lipids as a fuel source diet Beta oxidation saturated Unsaturated dd chain Ketone bodies as fuel Physiology High energy More reduced
Fatty Acid Degradation Chapter 27, Stryer Short Course Catabolism verview Lipids as a fuel source diet Beta oxidation saturated Unsaturated dd chain Ketone bodies as fuel Physiology High energy More reduced
Supplementary Information
 Supplementary Information Serum and synovial fluid lipidomic profiles predict obesity-associated osteoarthritis, synovitis, and wound repair Chia-Lung Wu 1,2, Kelly A. Kimmerling 1,2, Dianne Little 3,
Supplementary Information Serum and synovial fluid lipidomic profiles predict obesity-associated osteoarthritis, synovitis, and wound repair Chia-Lung Wu 1,2, Kelly A. Kimmerling 1,2, Dianne Little 3,
Synthesis of Fatty Acids and Triacylglycerol
 Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
Synthesis of Fatty Acids and Triacylglycerol Lippincott s Chapter 16 Fatty Acid Synthesis Mainly in the Liver Requires Carbon Source: Acetyl CoA Reducing Power: NADPH 8 CH 3 COO C 15 H 33 COO Energy Input:
Pathways of changes in hypermethylation in F1 offspring exposed to intrauterine hyperglycemia
 Supplementary Table 1. Pathways of changes in hypermethylation in F1 offspring exposed to intrauterine hyperglycemia Pathway Enrichment Score (-log10[p value]) p-value genes Glycosaminoglycan degradation
Supplementary Table 1. Pathways of changes in hypermethylation in F1 offspring exposed to intrauterine hyperglycemia Pathway Enrichment Score (-log10[p value]) p-value genes Glycosaminoglycan degradation
Dr. Nafith Abu Tarboush
 4 Dr. Nafith Abu Tarboush June 24 th 2013 Ahmad Moayd 1 Definition and general properties refer to slide no. 2 Lipids: macromolecules made from Alcohol and Fatty acid bonded by ester linkage. Amphipathic
4 Dr. Nafith Abu Tarboush June 24 th 2013 Ahmad Moayd 1 Definition and general properties refer to slide no. 2 Lipids: macromolecules made from Alcohol and Fatty acid bonded by ester linkage. Amphipathic
