SUPPLEMENTAL DATA SUPPLEMENTAL FIGURES
|
|
|
- Blake Spencer
- 5 years ago
- Views:
Transcription
1 SUPPLEMENTAL DATA SUPPLEMENTAL FIGURES Supplemental Figure 1. Major terpenes present in axenic (sterile) cultures of Marchantia polymorpha harvested after 0, 3, 6, 12 months of growth. 1
2 Supplemental Figure 2. Measurement of terpene synthase-like gene expression based on the Fragments per kilobase of exon per million fragments mapped (FPKM). We relied on the methods of Yeo et al. (2013) to validate the use of map reads to gain a relative appreciation for transcripts levels in M. polymorpha, which were corroborated by RT-PCR measurements (see Supplemental Figure 5). Yeo, Y.S. et al. (2013) Functional identification of valerena-1,10-diene synthase, a terpene synthase catalyzing a unique chemical cascade in the biosynthesis of biologically active sesquiterpenes in Valeriana officinalis. J. Biol. Chem. 288:
3 Supplemental Figure 3. Relative expression of the terpene synthase-like genes (MpMTPSL) from axenic as well as non-axenic M. polymorpha tissue grown for 0 (A), 3 (B), 6 (C) and 12 (D) months. Qualitative PCR was performed for terpene synthase like genes using 250 ng of cdna templates from axenic as well as non-axenic cultures of M. polymorpha as described in detail in the Materials and Methods. 3
4 Supplemental Figure 4. Web logo based consensus sequence pattern for Marchantia terpene synthase like genes and Aspartate-rich substrate binding motif description using clustalw alignment. Aspartate-rich substrate binding motif was calculated using WebLogo based on an alignment of the 9 Marchantia terpene synthase like genes (MpMTPSL1-9) (A). The bigger the letter, the more conserved the amino acid site. The alignment was produced from commercially available Mac-vector plugins for Clustal W using MpMTPSL1-9 along with the bacterial pentalene synthase gene (GenBank Accession AAA19131) (B). The conserved metal binding motifs, the DDXXD motif (1) and the NDXXSXXXE motif (2) are highlighted in red. 4
5 Supplemental Figure 5. Clustal W alignment of the diterpene synthase-like genes present in M. polymorpha (MpDTPS 1-4) centered on Class I (DXDD) and Class II (DDXXD) divalent metal binding motifs. Alignment was produced from commercially available Clustal W plugins for MacVector using MpDTPS 1-4 along with Physcomitrella patens ent-kaurene synthase gene (PpCPS/KS) (GenBank Accession AB ). The conserved aspartate-rich motifs for Class-I and Class-II are highlight by red blocks. Supplemental Figure 6. Purification of recombinant MpMTPSL terpene synthases. Coomassie Blue-stained SDS-PAGE gel, showing recombinant MpMTPSL-2, 3, 4, 5, 6 and 7 expressed in E. coli BL21-DE3 and after Co 2+ -affinity purification. Separation of 2.5 µg of each protein was performed on 10% discontinuous SDS-polyacrylamide gels. 5
6 Supplemental Figure 7A. Enzyme kinetic determinations for MpMTPSL2, 3, 4, 5, 6, 7. Enzyme assays (50 μl) were set up with purified MpMTPSL1-7 at 100 nm and the indicated concentration of 3 H-NPP, 3 H-GPP, and 3 H-FPP. Assays were incubated for 5 min at 37 C and stopped by addition of 50 μl stop buffer. The reactions were then extracted with 200 μl of hexane and radioactivity determined in aliquots by scintillation spectrometry. The data was analyzed using the Prism Graphpad 6.0. Data represents mean of triplicate assays. 6
7 Supplemental Figure 7B. Enzyme kinetic determinations for MpMTPSL4 and MpMTPSL9 for total (non-scrubbed) and all hydrocarbon (scrubbed) reaction products. Enzyme assays (50 μl) were set up with purified MpMTPSL4 or MpMTPSL9 at 100 nm and the indicated concentration of 3 H-FPP. Assays were incubated for 5 min at 37 C and stopped by addition of 50 μl stop buffer. The reactions were then extracted with 200 μl of hexane, aliquots which were subject to silica-scrub or not prior to scintillation counting. The data was analyzed using the Prism Graphpad 6.0. Data represents mean of triplicate assays. 7
8 Supplemental Figure 8. GC chromatograms of terpene reaction product(s) generated by MpMTPSL3 in vitro and in vivo. GC chromatograms of the in vitro products formed by MpMTPSL3 (~100 nm) incubated with 100 µm FPP (A) and the in vivo products generated by E. coli (B) or yeast (C) cultures expressing the MpMTPS3 gene. GC chromatogram of extractable terpenes from M. polymorpha (D) and annotated for the overlapping products (dashed black line) present in multiple samples. 8
9 Supplemental Figure 9. Gas chromatogram of terpene reaction product(s) generated by MpMTPSL5 in vitro and in vivo. GC chromatograms of the in vitro products formed by MpMTPSL5 (~100 nm) incubated with 100 µm FPP (A) and the in vivo products generated by E. coli (B) or yeast (C) cultures expressing the MpMTPSL5 gene. GC chromatogram of extractable terpenes from M. polymorpha (D) and annotated for the overlapping products (dashed black line) present in multiple samples. 9
10 Supplemental Figure 10. Gas chromatogram of terpenes generated by MpMTPSL7 in vitro and in vivo. GC chromatograms of the in vitro products formed by MpMTPSL7 (~100 nm) incubated with 100 µm FPP (A) and the in vivo products generated by E. coli (B) or yeast (C) cultures expressing the MpMTPSL7 gene. GC chromatogram of extractable terpenes from M. polymorpha (D) and annotated for the overlapping products (dashed black line) present in multiple samples. 10
11 Supplemental Figure 11. Gas chromatogram of terpenes generated by MpMTPSL9 in vitro and in vivo. GC chromatograms of the in vitro products formed by MpMTPSL9 (~100 nm) incubated with 100 µm FPP (A) and the in vivo products generated by E. coli (B) and GC chromatogram of extractable terpenes from M. polymorpha (C) and annotated for the overlapping products (dashed black line) present in multiple samples. 11
12 Supplemental Figure 12. GC chromatogram of terpene reaction product(s) generated in vitro by MpMTPSL6. GC chromatogram of the in vitro products formed by MpMTPSL6 (~100 nm) incubated with 100 µm GPP (A) in comparison to a chromatogram for ocimene (B) where the two peaks in chromatogram represents cis-β-ocimene (1) and trans-β-ocimene (2). 12
13 Supplemental Figure 13. GC chromatograms of terpene reaction product(s) generated by MpMTPSL2 in vitro using NPP as substrate. GC chromatograms of the in vitro products formed by MpMTPSL2 (100 nm) incubated with 100 µm NPP (A) in comparison to the D-limonene in M. polymorpha (0 month axenic culture, prior to developmental accumulation of most sesquiterpenes), and an authentic D-limonene standard. 13
14 Supplemental Figure 14. Mass spectra of selected compounds produced by MpMTPSL4 as shown in Figure 6 and for (-) alpha-gurjunene standard. 14
15 Supplemental Figure 15. GC-MS analysis of diterpenes products generated by coexpression of MpDTPS4, GGPP synthase plus An2 or OsCPS2 or AgAS D621A, which are copalyl diphosphate synthases (CPS) that produce ent, syn and normal CPP from GGPP, respectively. MpDTPS4 reacts with ent-cpp to form ent-kaurene when co-expressed with An2 from maize, while it remains largely unreactive with syn and normal CPP as indicated by unreacted syn and normal copalol, respectively when co-expressed with OsCPS2 or AgAS D
16 1 H -1 H COSY HMBC NOESY H OH 17 H OH H H Supplemental Figure 16. Numbering and selected 1 H- 1 H COSY, HMBC and NOESY correlations for atiseranol. 16
17 Supplemental Figure 17. Phylogenetic relationships of MpDTPS1, 3 and 4 to other plant mono-functional CPSs and KSs and a bifunctional KS from Physcomitrella as inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The Marchantia diterpene synthases (MpDTPS) phylogenetic tree was constructed based on the amino acid alignment presented in Supplemental Data Set 3. These terpene synthase sequences were downloaded from BLASTP search analysis tool at NCBI (Altschul et. al. 1997). Sequences for mono-functional CPSs and KSs and a bi-functional KS from Physcomitrella were selected based on sequence similarities. The multiple 17
18 sequence alignment was performed with the amino acid sequences of the 107 selected genes (including M. polymorpha diterpene synthase genes) using a commercially available MacVector program with default parameters (Rastogi, 2000) (Supplemental Data Set 3), and the phylogenetic tree was built using MEGA 6.0 (Tamura et al., 2013). The parameters used were Poisson model as substitution model with uniform rates and complete deletion for gaps or missing data. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Branches corresponding to partitions reproduced in less than 50% of the bootstrap replicates were collapsed via the program algorithm, and the tree was visualized with FigTree version ( Altschul, S.F. Madden, T.L. Schäffer, A.A. Zhang, J. Zhang, Z. Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: Rastogi, P.A. (2000). MacVector. Integrated sequence analysis for the Macintosh. Methods Mol Biol. 132: Saitou, N. and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: Supplemental Figure 18. Intron-exon organization of MpDTPS1 to 4 in comparison to a typical mono-functional diterpene synthase (CPS) found in Arabidopsis (AT4g02780) (Sun and Kamiya, 1994). The data is based on in silico analysis of M. polymorpha genomic sequences available in the NCBI SRA database in comparison to the assembled transcriptome of M. polymorpha. The SRA data was downloaded and assembled in the CLC work bench ver. 4.7 as discussed earlier in case of our transcriptome assembly. Sun, T.P. and Kamiya, Y. (1994) The Arabidopsis GA1 locus encodes the cyclase entkaurene synthetase A of gibberellin biosynthesis. Plant Cell 6:
19 Supplemental Figure 19. Phylogenetic analysis of the terpene synthase-like and diterpene synthase-like proteins from M. polymorpha in relationship to bacterial, fungal and plant terpene synthase proteins (see Supplemental Data Sets 2 and 4). Maximum likelihood phylogenetic tree analyses were performed with 59 amino acid sequences (see Supplementary Data Set 4) aligned across 285 positions. The terpene synthase sequences used include some sequences obtained from another liverwort (Pellia endiviifolia) transcriptome (Alaba et al., 2015), which have not been functionally 19
20 verified, plus validated terpene synthases from select bacteria and fungi, and examples across the evolutionary spectrum of plants based on the sequence similarity network (Figure 9). The maximum likelihood tree was generated using PhyML program interface in Seaview 4.0 (Gouy et al. 2010). Selection of best fit model was based on results provided by the Prottest server (Abascal et al 2005). The parameters used to generate the consensus phylogenetic tree were BIONJ as the starting tree (Gascuel 1997), using the LG substitution model with the four rate of substitution categories, estimated gamma distribution parameter and 1000 bootstrap repetitions. Bootstrap values are shown on each branches as percentage of replicates associated with end-point sequence clusters. The annotation of genes isolated from different organisms is shaded by different colors. Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21: Alaba, S., et al. (2015). The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants. New Phytol. 206: Gascuel, O. (1997). BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14: Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:
21 Supplemental Figure 20. Evolutionary relationships of Marchantia terpenes synthase proteins to other terpene synthase proteins presented in Supplemental Data Sets 2 and 4. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The optimal tree with the sum of branch length = is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (5000 replicates) are shown next to the branches (Felsenstein 1985). The 21
22 evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 5). The analysis involved 59 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 285 positions in the final Supplemental Data Set 4. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016). Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: Kumar S., Stecher G., and Tamura K. (2015). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: Saitou N. and Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: Zuckerkandl E. and Pauling L. (1965). Evolutionary divergence and convergence in proteins. Edited in Evolving Genes and Proteins by V. Bryson and H.J. Vogel, pp Academic Press, New York. 22
23 Supplemental Figure 21. Developmental time course for the amounts of the major MpMTPSL4 products found in M. polymorpha. Major MpMTPSL4 products measured during the development of M. polymorpha. ( )-α- Gurjunene is the major hydrocarbon produced by MpMTPSL4 (A). The time courses for the changes in the abundance of two suspected sesquiterpene alcohols found in M. polymoprha and produced by MpMTPSL4 are also shown: (relative retention time (RRT) to dodecane standard) (B) and (RRT to dodecane standard) (C). 23
24 SUPPLEMENTAL TABLES Supplemental Table 1. TBLASTN search of Marchantia transcriptome database (42,617 contigs) using archetypical mono- and sesqui-terpene synthases. Hits to M. polymorpha genes are noted in bold. Query Hits in M. polymorpha transcriptomic database E-value Score Gaps (%) Betacubebene synthase [Magnolia grandiflora] gb ACC ir ed)contig26612denovoassembly (MpDTPS3) ir ed)contig9617denovoassembly (MpDTPS4) ir ed)contig7218denovoassembly (MpDTPS1) ir ed)contig27100denovoassembly (contig shows similarity to diterpene synthase from Abies balsamea) ir ed)contig6926denovoassembly (MpDTPS2) 8.35E E E E E AtTPS11 (Thujopsene synthase or Alphabarbatene synthase [Arabidopsis thaliana] gb Q4KSH9.2 ired)contig27100denovoassembly(conti g shows similarity to diterpene synthase from Abies balsamea) ired)contig7218denovoassembly (MpDTPS1) ired)contig9617denovoassembly (MpDTPS4) 5.84E E E
25 ired)contig26612denovoassembly (MpDTPS3) ired)contig6926denovoassembly (MpDTPS2) 4.38E E Santalene and bergamotene synthase [Solanum habrochaites] gb ACJ ired)contig26612denovoassembly (MpDTPS3) ired)contig6926denovoassembly (MpDTPS2) ired)contig7218denovoassembly (MpDTPS1) ired)contig9617denovoassembly (MpDTPS4) ired)contig27745denovoassembly (contig similar to copalyl diphosphate synthase [Taiwania cryptomerioides] gb AFE ) ired)contig27100denovoassembly (contig similar to levopimaradiene/abietadiene synthase [Picea abies] gb AAS ) 7.48E E E E E E S-limonene synthase [Mentha spicata] gb AAC ired)contig26612denovoassembly (MpDTPS3) ired)contig9617denovoassembly (MpDTPS4) 1.58E E
26 ired)contig7218denovoassembly (MpDTPS1) ired)contig27100denovoassembly (contig similar to levopimaradiene/abietadiene synthase [Picea abies] gb AAS ) ired)contig6926denovoassembly (MpDTPS2) ired)contig6926denovoassembly (MpDTPS2) 3.20E E E E (+)- germacrene D synthase [Solidago canadensis] gb AAR EAS_TOBA C 5-epiaristolochene ired)contig26612denovoassembly (MpDTPS3) ired)contig9617denovoassembly (MpDTPS4) ired)contig27100denovoassembly (contig similar to levopimaradiene/abietadiene synthase [Picea abies] gb AAS ) ired)contig7218denovoassembly (MpDTPS1) ired)contig6926denovoassembly (MpDTPS2) ired)contig26612denovoassembly (MpDTPS3) 4.11E E E E E E
27 synthase sp Q40577 ired)contig9617denovoassembly (MpDTPS4) ired)contig27100denovoassembly (contig similar to levopimaradiene/abietadiene synthase [Picea abies] gb AAS ) ired)contig7218denovoassembly (MpDTPS1) ired)contig6926denovoassembly (MpDTPS2) 6.33E E E E VTSS1_HYO MU Vetispiradiene synthase 1 (HVS1) sp Q ired)contig26612denovoassembly ired)contig9617denovoassembly (MpDTPS4) ired)contig7218denovoassembly (MpDTPS1) ired)contig27100denovoassembly (contig similar to levopimaradiene/abietadiene synthase [Picea abies] gb AAS ) ired)contig6926denovoassembly (MpDTPS2) 1.21E E E E E
28 Supplemental Table 2. Pfam domain search of the M. polymorpha assembled contigs with PF The assembled contigs for M. polymorpha were translated using the six frame translation module within Geneious (Geneious Pro v5.5 created by Biomatters; available from The six frame translated sequences were then screened for the Pfam PF01397 domain using HMMER3.0 ( No. Contig identified PFAM Domain Used HMM cut off E-value Top hit BLATSX (NCBI) E-value Gene Cloned MHA_AA_NoIndex_L 006_R1_001_pf_(pair ed)_contig_26612_de _Novo_Assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pair ed)_contig_7218_de_ Novo_Assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pair ed)_contig_9617_de_ Novo_Assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pair ed)_contig_6926_de_ Novo_Assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pair ed)_contig_27100_de _Novo_Assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pair ed)_contig_27745_de _Novo_Assembly PF PF PF PF PF PF BAJ entkaurene synthase [Jungermannia subulata] Length: 886 BAJ entkaurene synthase [Jungermannia subulata] Length: 886 ADB (-)- ent-kaurene synthase [Picea sitchensis] Length: 757 BAJ entkaurene synthase [Jungermannia subulata] Length: 886 AEL diterpene synthase TPS2, partial [Abies balsamea] Length=852 BAJ entkaurene synthase [Jungermannia subulata] Length= MpDTPS3 5E-133 MpDTPS1 8E-116 MpDTPS4 0 MpDTPS2 3.00E E-74 Partial (5' missing) Partial (5' and 3'missing) 28
29 Supplemental Table 3. Pfam domain search of the M. polymorpha assembled contigs with PF The assembled contigs for M. polymorpha were translated using the six frame translation module within Geneious (Geneious Pro v5.5 created by Biomatters; available from The six frame translated sequences were then screened for the Pfam PF03936 domain using HMMER3.0 ( No Contig identified Pfam domain used HMM cut off E-value MHA_AA_NoIndex_L 006_R1_001_pf_(pai PF red)_contig_9617_de _Novo_Assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pai PF red)_contig_26612_d e_novo_assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pai PF red)_contig_7218_de _Novo_Assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pai PF red)_contig_27100_d e_novo_assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pai PF red)_contig_6926_de _Novo_Assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pai PF red)_contig_27182_d e_novo_assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pai PF red)_contig_9911_de _Novo_Assembly Top hit BLATSX (NCBI) ADB (-)- ent-kaurene synthase [Picea sitchensis] Length: 757 BAJ entkaurene synthase [Jungermannia subulata] Length: 886 BAJ entkaurene synthase [Jungermannia subulata] Length: 886 AEL diterpene synthase TPS2, partial [Abies balsamea]= length 852 BAJ entkaurene synthase [Jungermannia subulata] Length: 886 WP_ hypothetical protein [Streptomyces sp. AA0539] Length: E-value Gene status 8E-116 MpDTPS4 0 MpDTPS3 5E-133 MpDTPS1 3.00E- 92 Partial (5' missing) 0 MpDTPS2 2.00E- 19 MpMTPSL1 351 WP_ Terpene synthase, 8.00E-11 MpMTPSL2 metal-binding protein [Plesiocystis 29
30 pacifica] Length: MHA_AA_NoIndex_L 006_R1_001_pf_(pai PF red)_contig_9134_de _Novo_Assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pai PF03936 red)_contig_29158_d e_novo_assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pai PF03936 red)_contig_14438_d e_novo_assembly MHA_AA_NoIndex_L 006_R1_001_pf_(pai PF red)_contig_27591_d e_novo_assembly YP_ terpene synthase metal-binding domain-containing protein [Nocardia brasiliensis ATCC ] Length: 755 gb ACU Terpene synthase metal-binding domain protein [Chitinophaga pinensisdsm 2588]Length=321 gb EGO putative terpene cyclase [Serpula lacrymans var. lacrymans S7.9]Length=342 Geosmin synthase [Streptomyces sp. LaPpAH-95] WP_ L ength: E E E E- 13 MpMTPSL4 MpMTPSL5 MpMTPSL8 MpMTPSL3 30
31 Supplemental Table 4. Pfam domain search of the M. polymorpha assembled contigs with PF The assembled contigs for M. polymorpha were translated using the six frame translation module within Geneious (Geneious Pro v5.5 created by Biomatters; available from The six frame translated sequences were then screened for the Pfam PF06330 domain using HMMER3.0 ( No Contig identified Pfam domain used HMM cut off E-value MHA_AA_NoIndex_ L006_R1_001_pf_(p PF aired)_contig_13913 _De_Novo_Assembl y MHA_AA_NoIndex_ L006_R1_001_pf_(p aired)_contig_17864 PF _De_Novo_Assembl y MHA_AA_NoIndex_ L006_R1_001_pf_(p PF aired)_contig_9134_ De_Novo_Assembly MHA_AA_NoIndex_ L006_R1_001_pf_(p aired)_contig_27182 PF _De_Novo_Assembl y MHA_AA_NoIndex_ L006_R1_001_pf_(p aired)_contig_27591 PF _De_Novo_Assembl y MHA_AA_NoIndex_ L006_R1_001_pf_(p PF aired)_contig_9911_ De_Novo_Assembly Top hit BLATSX (NCBI) trichodiene synthase [Fusarium sambucinum] ACZ Length: 247 XP_ predicted protein [Postia placenta Mad-698-R] Length: 305 YP_ terpene synthase metal-binding domain-containing protein [Nocardia brasiliensis ATCC ] Length: 755 WP_ hypothetical protein [Streptomyces sp. AA0539] Length: 351 Geosmin synthase [Streptomyces sp. LaPpAH-95] WP_ Le ngth: 737 WP_ Terpene synthase, metal-binding protein [Plesiocystis pacifica] Length: 355 E-value 7.00E- 15 XP_ E E E E- 11 Putative gene name assigned MpMTPSL7 MpMTPSL6 MpMTPSL4 MpMTPSL1 MpMTPSL3 MpMTPSL2 31
32 7 8 MHA_AA_NoIndex_ L006_R1_001_pf_(p aired)_contig_29158 PF06330 _De_Novo_Assembl y 10 MHA_AA_NoIndex_ L006_R1_001_pf_(p aired)_contig_14438 PF _De_Novo_Assembl y gb ACU Terpene synthase metal-binding domain protein [Chitinophaga pinensisdsm 2588]Length=321 gb EGO putative terpene cyclase [Serpula lacrymans var. lacrymans S7.9]Length= E E- 06 MpMTPSL5 MpMTPSL8 9 MHA_AA_NoIndex_ L006_R1_001_pf_(p PF aired)_contig_6926_ De_Novo_Assembly BAJ entkaurene synthase [Jungermannia subulata] Length: MpDTPS2 32
33 Supplemental Table 5. Summary of Pfam domain searches of the M. polymorpha transcriptome (45,309 contigs, assembled from the NCBI SRA database SRP according to Sharma et al. 2013) for PF01397, PF03936, and PF06330 motifs. Contig number Top hit BLATSX (NCBI) E-value Assigned gene name WP_ hypothetical protein [Streptomyces sp. AA0539] 9798 WP_ Terpene synthase, metal-binding protein [Plesiocystis pacifica] 2E-19 MpMTPSL1 8E-11 MpMTPSL2 4252, 4254, Geosmin synthase [Streptomyces sp. LaPpAH-95] WP_ E-13 MpMTPSL3 2193, YP_ terpene synthase metal-binding domaincontaining protein [Nocardia brasiliensis ATCC ] 9297, gb ACU Terpene synthase metal-binding domain protein [Chitinophaga pinensis DSM 2588] XP_ predicted protein [Postia placenta Mad-698- R] 8770, trichodiene synthase [Fusarium sambucinum] ACZ gb EGO putative terpene cyclase [Serpula lacrymans var. lacrymans S7.9] Select seq gb ACU Terpene synthase metal-binding domain protein [Chitinophaga pinensis DSM 2588] ACU E-16 MpMTPSL4 1E-10 MpMTPSL5 7E-11 MpMTPSL6 7E-15 MpMTPSL MpMTPSL8 7E-14 Partial 33
34 18987 Trichodiene synthase [Penicillium expansum] KGO Similar to Trichodiene synthase; acc. no. Q6A1B7 [Pyronema omphalodes CBS ] CCX E-10 MpMTPSL Partial 34
35 Supplemental Table 6. References sequences used for similarity and identity comparisons. Symbol Gene Accession References SmMTPSL1 SmMTPSL17 SmMTPSL26 SmMTPSL22 SmTPS9 SmTPS10 PhyTPS TRI5_FUSSP TASY_TAXBR TEAS PrAS ScGS MpMTPSL1 MpMTPSL2 MpMTPSL3 MpMTPSL4 MpMTPSL5 hypothetical protein SELMODRAFT_ [Selaginella moellendorffii] hypothetical protein SELMODRAFT_ [Selaginella moellendorffii] hypothetical protein SELMODRAFT_ [Selaginella moellendorffii] hypothetical protein SELMODRAFT_ [Selaginella moellendorffii] kaurene synthase [Selaginella moellendorffii] terpene synthase, partial [Selaginella moellendorffii] ent-kaurene synthase [Physcomitrella patens] sesquiterpene cyclase gene from the trichotheceneproducing fungus Fusarium sporotrichioides taxadiene synthase [Taxus brevifolia] 5-epi-aristolochene synthase XP_ Li et al XP_ Li et al XP_ Li et al XP_ Li et al XP_ Li et al AFR34003 Li et al BAF61135 Hayashi et al AAN05035 Hohn and Beremand 1989 AAC Wildung and Croteau, 1996 Q40577 Facchini and Chappell 1992 Aristolochene synthase Q Proctor and Hohn, 1993 cyclase [Streptomyces NP_ Hsiao and Kirby, coelicolor A3(2)] 2008 Marchantia Terpene KU This study synthase like gene Marchantia Terpene KU This study synthase like gene Marchantia Terpene KU This study synthase like gene Marchantia Terpene KU This study synthase like gene Marchantia Terpene KU This study synthase like gene 35
36 MpMTPSL6 MpMTPSL7 MpMTPSL8 MpMTPSL9 MpDTPS1 MpDTPS3 MpDTPS4 4S-LS Marchantia Terpene synthase like gene Marchantia Terpene synthase like gene Marchantia Terpene synthase like gene Marchantia Terpene synthase like gene Marchantia Diterpene synthase like gene Marchantia Diterpene synthase like gene Marchantia Diterpene synthase like gene 4S-limonene synthase [Mentha spicata] KU This study KU This study KU This study KU This study KU This study KU This study KU This study AAC37366 Colby et al 1993 Li, G. et al. (2012). Nonseed plant Selaginella moellendorffii has both seed plant and microbial types of terpene synthases. Proc. Natl. Acad. Sci. 109: Hayashi, K.I., Kawaide, H., Notomi, M., Sakigi, Y., Matsuo, A., and Nozaki, H. (2006). Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 580: Hohn, T.M. and Beremand, P.D. (1989). Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene 79: Wildung, M.R. and Croteau, R. (1996). A cdna clone for taxadiene synthase, the diterpene cyclase that catalyzes the committed step of taxol biosynthesis. J. Biol. Chem. 271: Facchini, P.J. and Chappell, J. (1992). Gene family for an elicitor-induced sesquiterpene cyclase in tobacco. Proc. Natl. Acad. Sci. U. S. A. 89: Proctor, R.H. and Hohns, T.M. (1993). Aristolochene Synthase isolation, characterization, and bacterial expression of a sesquiterpenoid biosynthetic gene (Aril) from Penicillium roqueforti. J. Biol. Chem. 268: Hsiao, N.-H. and Kirby, R. (2008). Comparative genomics of Streptomyces avermitilis, Streptomyces cattleya, Streptomyces maritimus and Kitasatospora aureofaciens using a Streptomyces coelicolor microarray system. Antonie Van Leeuwenhoek 93:
37 Colby, S.M., Alonso, W.R., Katahira, E.J., McGarvey, D.J., and Croteau, R. (1993). 4Slimonene synthase from the oil glands of spearmint (Mentha spicata). cdna isolation, characterization, and bacterial expression of the catalytically active monoterpene cyclase. J. Biol. Chem. 268: Supplemental Table 7. Kinetic constants for the terpene synthase-like enzymes from M. polymorpha. Kinetic analysis was performed using purified His-tagged MpMTPSL proteins with preferred substrates from Table 1 (ND-No activity detected). Enzyme Preferred substrate KM (µm) kcat (s -1 ) kcat/km (s -1 mm -1 ) MpMTPSL1 ND MpMTPSL2 NPP ± ± MpMTPSL3 FPP 4.71 ± ± MpMTPSL4 FPP ± ± MpMTPSL5 FPP 8.93 ± ± MpMTPSL6 NPP 1.42 ± ± MpMTPSL7 FPP ± ± MpMTPSL8 ND MpMTPSL9 FPP ± ±
38 Supplemental Table 8. 1 H-NMR for compound 7 (from Figure 6) in CDCl3. The structure of peak 7 was determined based on 1 H, 13 C, 1 H- 1 H-gCOSY, and 1 H- 13 C- HSQC experiments and on comparison with 1 H and 13 C data of other known aromadendrene alcohols. The 1 H-NMR demonstrated 2 singlet methyl groups at δh 1.25 and δh 1.04 and two methyl doublets at δh 0.86 and δh 0.91 (d, J= 6.5 Hz.) corresponding to H3-14 and H3-15. The 13 C-NMR revealed an absence of olefinic carbons in the δc ppm range, and revealed the presence of a quarternary alcohol at δc Because (+)-ledol, (+)-globulol, and (-)-viridiflorol all exhibit a C-10 secondary alcohol at δc 75.0 ppm, the downfield shift of the δc 83.4 alcohol indicates that it is likely at the C-1 or C-5 bridgehead carbon (Figure 7). The 1 H- 1 H-gCOSY revealed all of the expected couplings for a gurjunene skeleton, except that the H-6 cyclopropane ring proton was present as a doublet in cis configuration with H-7 (H-6= d, J=10 Hz, H-7= td, J=10, 6, 1). In the other gurjunene alcohols, H-6 appears as a triplet coupling with H-5 and H-7 (Kaplan et al., 2000). This suggests that the position of the quarternary alcohol to be at the C-5 bridgehead carbon Position δh H (1H, m, complex) H (1H, m, complex) H-3eq (1H, m, complex) H-3ax (1H, m, complex) H (1H, m, complex) H H (1H, d, J= 10) H (1H, ddd, J= 10, 9, 6) H-8eq (1H, m, complex) H-8ax (1H, m, complex) H-9eq (1H, m, complex) H-9ax (1H, m, complex) H (1H, m, complex) H H (3H, s) H (3H, s) H (1H, d, J= 6) H (1H, d, J= 6). 38
39 Supplemental Table C-NMR data for compound 7 (from Figure 6), (+)-globulol, and (+)-ledol in CDCl3. δ C position 7 (+)-globulol a ledol b C C C C C C C C C C C C C C C a) Toyota, M., Tanaka, M., and Asakawa, Y. (1999). A revision of the 13C NMR spectral assignment of globulol. Spectroscopy 14: b) Kaplan, M. a, Pugialli, H.R.L., Lopes, D., Gottlieb, H.E., Auxiliadora, M., and Kaplan, C. (2000). The stereochemistry of ledol from Renealmia chrysotrycha: an NMR study. Phytochemistry 55: recorded on a Varian JNMR 400 MHz spectrometer at 100 MHz. 39
40 Supplemental Table H and 13 C NMR data for atiseranol. NMR experiments were conducted on a Bruker Avance 700 spectrometer equipped with a 5-mm HCN cryogenic probe. Structural analysis was performed using one-dimensional 1 H, and two-dimensional DQF-COSY, HSQC, HMQC, HMBC, and NOESY experiment spectra acquired at 700 MHz, and one-dimensional 13 C spectrum (174 MHz) using standard experiments from the Bruker TopSpin version 1.4 software. An analysis of the DQF-COSY and HSQC spectra led to the unambiguous assignment of the protons and corresponding carbon signals. Correlations from the HMBC spectra were used to build the planar structure, and the stereochemistry was determined by the NOESY experiment. In the NOESY spectrum, the correlations of H3-17/H-13a, H3-17/H-14b, H3-20/H-13b and H3-20/H-14a indicated that 16-OH possessed a β-configuration. Chemical shifts were referenced using known chloroform ( 13 C 77.23, 1 H 7.24 ppm) signals offset from TMS, and compared to those previously reported. Position MPld40 δ H δ C 1 a 1.45 (1H, m) 39.9 b 0.73 (1H, m) 2 a 1.48 (1H, m) 18.8 b 1.28 (1H, m) 3 a 1.29 (1H, m) 42.8 b 1.05 (1H, m) (1H, m) a 1.36 (1H, m) 19.3 b 1.20 (1H, m) (1H, m) (1H, m) (1H, m) (1H, m) (1H, m) (1H, m) a 1.56 (1H, m) 24.7 b 1.38 (1H, m) 14 a 1.73 (1H, m) 27.9 b 0.70 (1H, m) (1H, m) (1H, m) (3H, s) (3H, s) (3H, s) (3H, s)
41 Supplemental Table 11. Primers used for cloning full-length MpMTPSL genes. Gene Primer Sequences (5 3 ) MpMTPSL1 MpMTPSL2 MpMTPSL2 MpMTPSL-1_F1 ATGTCTGCGAGGGACAATGG MpMTPSL-1_R1 TTAGATGATTCTGTGGTCTT MpMTPSL-2_F1 ATGGCGGCAAAAGCTCACAGGAGTTT MpMTPSL-2_R1 TTATCTCAAGTAGTGGTCCT SK299 ATGATCGGACTGGTGAGCAGTATTTATG MpMTPSL-2_R1 TTATCTCAAGTAGTGGTCCT MpMTPSL3 MpMTPSL4 MpMTPSL5 MpMTPSL-3F MpMTPSL-3R MpMTPSL-4F MpMTPSL-4R SK202F K199R ATGGCTTCACAACACTCGGCATCTACC CTACATTTTCAGATTTTCTTCTATC ATGGCACCAACTTTAGACTCGG CTAGACACTGTTGATGTGTTTTAC ATGGCCCCAAGTTTAGACTCGGATTCTAC GTAGAACAAATCATCAAGCGTCAGTAA MpMTPSL6 MpMTPSL7 MpMTPSL-6_F1 ATGAAGCCCATCATGGTGAGCTCTG MpMTPSL-6_R1 TTAAACGAAATCACCTTGGAACCAATCGTC MpMTPSL-7_F1 ATGTCGAGCATGGCGAGCTGTG MpMTPSL-7_R1 TTAAATGAGATTACCTTGGAACCAGCCG MpMTPSL8 MpMTPSL9 SK261F SK262R SK417F SK418R ATGGGCCCAAGTTTAAACTCG CTACCTTTTTAGATTTCCTGTGTCTG CATGCCATGGATGACGAAGACGCTTCCGGCT CCCAAGCTTCTACACACAGTCACCCGCGAACC 41
42 Supplemental Table 12. Primers used for cloning full length MpDTPS genes. Gene Primer Sequences (5 3 ) MpDTPS1 MpDTPS2 MpDTPS3 MpDTPS4 SK005F GGGAATTCCATATGATGCTAGCCGTCGATGAACCGAC SK006R CCCAAGCTTCTACTCGCTGGGGAAGCGTTTG SK007F GGGAATTCCATATGATGGCGAGCTCGACTGCC SK008R GGAATTCTCAAGAGAGCACGGGTTCG SK009F GGAATTCCATATGATGGCATTCTCGTTAGCAGG SK010R ATAAGAATGCGGCCGCTCAGGCCACAGGCTCGAAGAGTAG SK341F AGCCATGATTTCGAATGATGAGG SK343R CTAGGCCTGTTCACTTTCGATGG 42
43 Supplemental Table 13. Primers used for DNA sequencing. Gene Primer Sequence (5 3 ) SK187R SK161F SK162F GGAAACCGAAAATGTCATTATGCCAACC GTGACAGATCTCTTCGTCAAAGCTC ACTTGTGGCTCGAGTACTGTGAAAG MpMTPSL1 SK163F SK164F MpMTPSL-1_sF2 ATCTCGGCTCTTACGTCTCGAATAC CAACAAGGTTCTCATGTGGTTCTTC GATGTTGATAAATCACGAGG MpMTPSL-1_sF3 MpMTPSL-1_sR2 MpMTPSL-1_sR3 SK167F SK168F CAGAACTACTTACTGGGATGT TTGTTCCATGCTTGGGCGCC GCCATTGTCGAGCTTTGACG ATCAATGAGTTCGAGACAAGAGTGG TCACAGGAGTTTCTCCAAGCTTATG MpMTPSL2 SK169F SK170F MpMTPSL-2_sF2 TTAGATTTGCAGCCTTTAGCATCAG TCTGATAGCCGAGTTCAATGAAAAG GAGTTCAATGAAAAGGCCCA MpMTPSL-2_sR2 MpMTPSL-2_sR3 SK228F ACCACAAGATAACCTTCTGC TGATACTCTGCGGATAATCT TCCTGAGCTTATGGGCTATGTGTTC MpMTPSL3 SK229F SK230R SK282F SK283R GTGGAATGACCCTGAGAACAAGAG CTCTTCAGCTGCCTACGAAACTGTC ACAAGGACAGTTTCGTAGGC CAATGTCTGTGTACTCGGGAG SK194R TTGGGGTAACGGTGCTGGAAATGTG 43
44 44 SK195R GTAGATGCCGAGTGTTGTGAAGCCA MpMTPSL4 SK196R TGCCCATAGAACTCAACCAGTCAAAAG SK197R GCATTCGGTAGTCTTGGGGTTCAGTC SK221R CTGGGCGATTCTAGTCGTATACTC SK222F CTCCGATTTCACATATTCCCAACAC SK223F GAGTATACGACTAGAATCGCCCAG SK240R CGAAGGGTGACGTAATCAGCCACATTC SK286F ATTTCAGAGCCCCATACATCC SK287R ATGCCCATAGAACTCAACCAG MpMTPSL5 SK284F TTGGTTCTCTTGTGGACTGAC SK285R AGCCTCAATCTCCATCAACG SK185R GAAAACAAGAAACGGCATCACACCTCC SK186R CTTGTTCCGAGAGACAGCCCGTTATGT SK198F CCTGTTGAGTGTATATCTAGACAGGCTG SK199R TTACTGACGCTTGATGATTTGTTCTAC SK202F ATGGCCCCAAGTTTAGACTCGGATTCTA C MpMTPSL6 SK121F CATTCTTTTCATCGTCACTGCG SK122R TCGTTGATGTGGCTTGAGG SK295F GAACATCACCGCTTTGGAAG MpMTPSL7 SK038F ATGTCGAGCATGGCGAGCTGTGGTGC SK039R CTAAATGAGATTACCTTGGAACCAGC SK119F GTCAGTTAGAGCACGAGTACAG SK120R GAAATAGAGGAAGGTGAGGCG
45 SK180R SK076F SK200F SK201R SK211F SK212R SK251F CGAGTCGGATGTAGTTTCTTGTAC GATTTAGAAATGCCTGCATGGACTATC CCTGTTGAGTGTATATCTAGACAAGCTG CTACCTTTTTAGATTTCCTGTGTCTG ATGGCGGCCAAATTCTCTAAGCTTATTG CTACCTTTTTAGATTTCCTGTGTCTG CCTGACAACGAGCGATTACTGGAA MpMTPSL8 SK252F SK253F SK254F SK261F CTGATTGCATCAGAGTTCGACGAC TTCATGAATACGACTGCCAATCTTAC AGACTGGATTCCTGGAACGCACGAG ATGGGCCCAAGTTTAAACTCG SK262R SK187R SK188R SK288R SK289R CTACCTTTTTAGATTTCCTGTGTCTG GGAAACCGAAAATGTCATTATGCCAACC GATGTCAGTACAAGCGGCCAGCATATTC GGTCGCTCTCCGACTGCCTTAG CTCGTTGTCAGGGTCATTCCAC MpMTPSL9 SK419F SK420F SK421R SK422R GATGCCTGATGGTGGAGC TACAACAAGATATATCCACTGATCCC CGTCCTCGAACTCCTTGTAA CACGTGAGTGATAATGTTTCTCG 45
46 Supplemental Table 14. Primers used for cloning MpMTPSL genes into bacterial (E. coli) protein expression vectors and their restriction site. Gene Primer Sequence (5 3 ) Restriction Site Vector MpMTPSL1 MpMTPSL2 MpMTPSL3 MpMTPSL4 MpMTPSL5 MpMTPSL6 MpMTPSL7 MpMTPSL8 MpMTPSL9 SK159F SK160R SK227F SK292R SK171F SK172R SK173F SK174R SK207F SK208R SK273F SK275R SK077F SK078R SK259F SK260R SK423F SK424R CATATGATGTCTGCGAGGGACAATG GGGCTATC AAGCTTTTTGATGATTCTGTGGTCTT CGTTGTATC TATACCATGGGCATGGCGGCAAAAG CTCACAG CGCGGATCCCTGTCTCAAGTAGTGG TCCTCTTTTTGGTATC CATATGATGGCTTCACAACACTCGG CATCTACCG AAGCTTCTTCATTTTCAGATTTTCTT CTATCTCAG GAGCTCATGGCACCAACTTTAGACT CGGATTCTAC AAGCTTCTTGACACTGTTGATGTGTT TTAC CGGGATCCATGGCCCCAAGTTTAGA CTCG ATAAGAATGCGGCCGCTTACTGACG CTTGATGATTTG TCTAGA GAAGGAGAATGAAGCCCATC CCCAAGCTTGTAAACGAAATCACCT TG GGAATTCCATATGTCGAGCATGGCG AGCTGTGGTGC GGAATTCCTAAATGAGATTACCTTG GAACCAGC CGCGGATCCATGGGCCCAAGTTTAA ACTC ACGCGTCGACCTACCTTTTTAGATTT CC GGGAATTCCATATGACGAAGACGCT TCCGGCT CCGGAATTCCTACACACAGTCACCC GCGAACC NdeI HindIII NcoI BamHI NdeI HindIII SacI HindIII BamHI NotI XbaI HindIII NdeI EcoRI BamHI SalI NdeI EcoRI pet28a pet28a pet28a pet28b pet28a pet28a pet28a pet28b pet28a 46
47 Supplemental Table 15. Primers used for cloning MpMTPSL genes into specific restriction sites within yeast expression vectors. Gene Primer Sequences (5 3 ) Restriction Site Vector MpMTPSL1 MpMTPSL3 MpMTPSL4 MpMTPSL5 MpMTPSL6 MpMTPSL7 MpMTPSL9 mmtps1notif mmtps1speir mmtps3notif mmtps3speir mmtps4speif mmtps4sacir mmtps5notif mmtps5speir mmtps6notif mmtps6speir mmtps7sacif mmtps7pacir SK445F SK446R GGGGCGGCCGCAAAACAATGT CTGCGAGGGACAATGGGGCT GACTAGTTTAGATGATTCTGTG GTTTCGTTG GGGGCGGCCGCAAAACAATGG CTTCACAACACTCGGCATC GACTAGTCTACATTTTCAGATT TTCTTCTATC GACTAGTAAAACAATGGCACCA ACTTTAGACTCGG GGGGAGCTCCTAGACACTGTT GATGTGTTTTAC GGGGCGGCCGCAAAACAATGC AGGAGATTCTTTACTTCC GACTAGTTTAAACGAAATCACC TTGGAACC GGGGCGGCCGCAAAACAATGA AGCCCATCATGGTGAGCTC GACTAGTTTAAACGAAATCACC TTGGAACC GGGGAGCTCAAAACAATGGCG AGCTGTGGTGCCGGGAAC CCTTAATTAATTACGCCAGCTA TTTAGGTGACAC GGGGCGGCCGCAAAACAATGA CGAAGACGCTTCCG GACTAGTTCTACACACAGTCAC CCGCGAACC NotI SpeI NotI SpeI SpeI SacI NotI SpeI NotI SpeI SacI PacI NotI SpeI X-HIS X-HIS X-HIS X-HIS X-HIS X-HIS X-HIS 47
48 Supplemental Table 16. Primers used for qualitative RT-PCR. Gene Primer Sequence (5 3 ) MpMTPSL1 MpMTPSL2 MpMTPSL3 MpMTPSL4 MpMTPSL5 MpMTPSL6 MpMTPSL7 MpMTPSL8 SK309F SK310R SK302F SK303R SK282F SK283R SK286F SK287R SK284F SK285R SK121F SK122R SK119F SK120R SK311F SK312R CCTTTGTTCAATTGTGTCTCCG ACAAGTCCCTGAAAGCTGATG CTTGAAATCTGGAGCGACATG AGAGCAGAACCACAAGATAACC ACAAGGACAGTTTCGTAGGC CAATGTCTGTGTACTCGGGAG ATTTCAGAGCCCCATACATCC ATGCCCATAGAACTCAACCAG TTGGTTCTCTTGTGGACTGAC AGCCTCAATCTCCATCAACG CATTCTTTTCATCGTCACTGCG TCGTTGATGTGGCTTGAGG GTCAGTTAGAGCACGAGTACAG GAAATAGAGGAAGGTGAGGCG GAAATAGAGGAAGGTGAGGCG TGCCCAAAGAAACGATCCAG MpMTPSL9 SK419F GATGCCTGATGGTGGAGC 48
49 Actin GAPDH SK421R SK101F SK102R SK280F SK281R CGTCCTCGAACTCCTTGTAA ATGAAGATTCTGACCGAGCG GAAGTCCAGGGCAATGTAGG GTCATTCAGAGTACCCACCG TCCCTTCATTTCGCCTTCAG MpDTPS1 SK314F SK313R CTACTTCTCAACTCTGTCGTGC GGACACATACTTCAGCTCATCG MpDTPS2 SK316F SK315R TGGAGGTTGCAGGAATTACAG CTTGGTCAGTCTCTTCGTGTG MpDTPS3 SK318F SK317R CTCGCTCTGCCCTACAAAG TCCCAGTCCAAAATCTCGTG MpDTPS4 SK320F SK321R CAGCAAAATACAAGGGCGTG AAACTAACCCAGCTCGTGTC 49
Supplementary data Characterization of terpene synthase gene family in Artemisia annua L
 POJ 7(5):373-381 (2014) ISSN:1836-3644 Supplementary data Characterization of terpene synthase gene family in Artemisia annua L Weizhang Jia, Ying Wang *, Xuesong Yu, Ling Zhang, Shunhui Liu, Zebo Huang
POJ 7(5):373-381 (2014) ISSN:1836-3644 Supplementary data Characterization of terpene synthase gene family in Artemisia annua L Weizhang Jia, Ying Wang *, Xuesong Yu, Ling Zhang, Shunhui Liu, Zebo Huang
The BLAST search on NCBI ( and GISAID
 Supplemental materials and methods The BLAST search on NCBI (http:// www.ncbi.nlm.nih.gov) and GISAID (http://www.platform.gisaid.org) showed that hemagglutinin (HA) gene of North American H5N1, H5N2 and
Supplemental materials and methods The BLAST search on NCBI (http:// www.ncbi.nlm.nih.gov) and GISAID (http://www.platform.gisaid.org) showed that hemagglutinin (HA) gene of North American H5N1, H5N2 and
Supplementary Information
 Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
sesquiterpenes monoterpenes mg gfw -1 fruit skin peel seed whole fruit
 mg gfw -1 14 12 1 8 6 4 2 sesquiterpenes monoterpenes fruit skin peel seed whole fruit Supplementary Figure S1. Terpene amount in fruit skin, seeds, and whole fruits of Japanese pepper. Monoterpenes (β-phellandrene,
mg gfw -1 14 12 1 8 6 4 2 sesquiterpenes monoterpenes fruit skin peel seed whole fruit Supplementary Figure S1. Terpene amount in fruit skin, seeds, and whole fruits of Japanese pepper. Monoterpenes (β-phellandrene,
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the
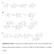 Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Rajesh Kannangai Phone: ; Fax: ; *Corresponding author
 Amino acid sequence divergence of Tat protein (exon1) of subtype B and C HIV-1 strains: Does it have implications for vaccine development? Abraham Joseph Kandathil 1, Rajesh Kannangai 1, *, Oriapadickal
Amino acid sequence divergence of Tat protein (exon1) of subtype B and C HIV-1 strains: Does it have implications for vaccine development? Abraham Joseph Kandathil 1, Rajesh Kannangai 1, *, Oriapadickal
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted. residues are colored red. Numbers indicate the position of each residue.
 Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
Characterizing fatty acids with advanced multinuclear NMR methods
 Characterizing fatty acids with advanced multinuclear NMR methods Fatty acids consist of long carbon chains ending with a carboxylic acid on one side and a methyl group on the other. Most naturally occurring
Characterizing fatty acids with advanced multinuclear NMR methods Fatty acids consist of long carbon chains ending with a carboxylic acid on one side and a methyl group on the other. Most naturally occurring
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Name: Due on Wensday, December 7th Bioinformatics Take Home Exam #9 Pick one most correct answer, unless stated otherwise!
 Name: Due on Wensday, December 7th Bioinformatics Take Home Exam #9 Pick one most correct answer, unless stated otherwise! 1. What process brought 2 divergent chlorophylls into the ancestor of the cyanobacteria,
Name: Due on Wensday, December 7th Bioinformatics Take Home Exam #9 Pick one most correct answer, unless stated otherwise! 1. What process brought 2 divergent chlorophylls into the ancestor of the cyanobacteria,
* H W-M 1,3-alkyl shift * * *
 FPP squalene synthase allylic cation 1' PP 2' 3' FPP electrophilic addition results tertiary cation PP loss of proton with formation of cyclopropane ring PP presqualene PP loss of diphosphate results in
FPP squalene synthase allylic cation 1' PP 2' 3' FPP electrophilic addition results tertiary cation PP loss of proton with formation of cyclopropane ring PP presqualene PP loss of diphosphate results in
Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66
 SUPPORTING INFORMATION belonging to the manuscript: Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66 by Changsheng Wu 1, 2, Gilles P. van Wezel 1, *, and Young Hae
SUPPORTING INFORMATION belonging to the manuscript: Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66 by Changsheng Wu 1, 2, Gilles P. van Wezel 1, *, and Young Hae
Principles of phylogenetic analysis
 Principles of phylogenetic analysis Arne Holst-Jensen, NVI, Norway. Fusarium course, Ås, Norway, June 22 nd 2008 Distance based methods Compare C OTUs and characters X A + D = Pairwise: A and B; X characters
Principles of phylogenetic analysis Arne Holst-Jensen, NVI, Norway. Fusarium course, Ås, Norway, June 22 nd 2008 Distance based methods Compare C OTUs and characters X A + D = Pairwise: A and B; X characters
Supporting Information
 Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
Electronic Supporting Information for. Mycobacterium tuberculosis H37Rv 3377c encodes the diterpene cyclase for producing the halimane skeleton
 Electronic Supporting Information for Mycobacterium tuberculosis 37Rv 3377c encodes the diterpene cyclase for producing the halimane skeleton Chiaki Nakano, a Tomoo Okamura, a Tsutomu Sato, a Tohru Dairi
Electronic Supporting Information for Mycobacterium tuberculosis 37Rv 3377c encodes the diterpene cyclase for producing the halimane skeleton Chiaki Nakano, a Tomoo Okamura, a Tsutomu Sato, a Tohru Dairi
Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study.
 Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study. In this study, one fluorophosphonate (FP, 1), three nitrophenol phosphonate probes
Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study. In this study, one fluorophosphonate (FP, 1), three nitrophenol phosphonate probes
2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein
![2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein](/thumbs/86/94743397.jpg) Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
TWO NEW ELLAGIC ACID GLYCOSIDES FROM LEAVES OF DIPLOPANAX STACHYANTHUS
 Journal of Asian Natural Products Research, December 2004, Vol. 6 (4), pp. 271 276 TWO NEW ELLAGIC ACID GLYCOSIDES FROM LEAVES OF DIPLOPANAX STACHYANTHUS XIAO-HONG YAN and YUE-WEI GUO* State Key Laboratory
Journal of Asian Natural Products Research, December 2004, Vol. 6 (4), pp. 271 276 TWO NEW ELLAGIC ACID GLYCOSIDES FROM LEAVES OF DIPLOPANAX STACHYANTHUS XIAO-HONG YAN and YUE-WEI GUO* State Key Laboratory
Cytochalasins from an Australian marine sediment-derived Phomopsis sp. (CMB-M0042F): Acid-mediated intra-molecular cycloadditions enhance
 SUPPORTING INFORMATION Cytochalasins from an Australian marine sediment-derived Phomopsis sp. (CMB-M42F): Acid-mediated intra-molecular cycloadditions enhance chemical diversity Zhuo Shang, Ritesh Raju,
SUPPORTING INFORMATION Cytochalasins from an Australian marine sediment-derived Phomopsis sp. (CMB-M42F): Acid-mediated intra-molecular cycloadditions enhance chemical diversity Zhuo Shang, Ritesh Raju,
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
Phylogenomics. Antonis Rokas Department of Biological Sciences Vanderbilt University.
 Phylogenomics Antonis Rokas Department of Biological Sciences Vanderbilt University http://as.vanderbilt.edu/rokaslab High-Throughput DNA Sequencing Technologies 454 / Roche 450 bp 1.5 Gbp / day Illumina
Phylogenomics Antonis Rokas Department of Biological Sciences Vanderbilt University http://as.vanderbilt.edu/rokaslab High-Throughput DNA Sequencing Technologies 454 / Roche 450 bp 1.5 Gbp / day Illumina
Supplementary Figure 1. HGL-DTG levels in uninduced and herbivory induced leaves. Total HGL-
 Supplementary Figure 1. HGL-DTG levels in uninduced and herbivory induced leaves. Total HGL- DTG concentrations [F 1,4 = 36.88, P 0.0037; significant differences (threshold: P 0.05) between means (± SE)
Supplementary Figure 1. HGL-DTG levels in uninduced and herbivory induced leaves. Total HGL- DTG concentrations [F 1,4 = 36.88, P 0.0037; significant differences (threshold: P 0.05) between means (± SE)
Cloning and Expression of a Bacterial CGTase and Impacts on Phytoremediation. Sarah J. MacDonald Assistant Professor Missouri Valley College
 Cloning and Expression of a Bacterial CGTase and Impacts on Phytoremediation Sarah J. MacDonald Assistant Professor Missouri Valley College Phytoremediation of Organic Compounds Phytodegradation: Plants
Cloning and Expression of a Bacterial CGTase and Impacts on Phytoremediation Sarah J. MacDonald Assistant Professor Missouri Valley College Phytoremediation of Organic Compounds Phytodegradation: Plants
ITS accuracy at GenBank. Conrad Schoch Barbara Robbertse
 ITS accuracy at GenBank Conrad Schoch Barbara Robbertse Improving accuracy Barcode tag in GenBank Barcode submission tool Standards RefSeq Targeted Loci Well validated sequences already in GenBank Bacteria
ITS accuracy at GenBank Conrad Schoch Barbara Robbertse Improving accuracy Barcode tag in GenBank Barcode submission tool Standards RefSeq Targeted Loci Well validated sequences already in GenBank Bacteria
proteins Diterpene cyclases and the nature of the isoprene fold
 proteins STRUCTURE O FUNCTION O BIOINFORMATICS Diterpene cyclases and the nature of the isoprene fold Rong Cao, 1 Yonghui Zhang, 2 Francis M. Mann, 3 Cancan Huang, 1 Dushyant Mukkamala, 1 Michael P. Hudock,
proteins STRUCTURE O FUNCTION O BIOINFORMATICS Diterpene cyclases and the nature of the isoprene fold Rong Cao, 1 Yonghui Zhang, 2 Francis M. Mann, 3 Cancan Huang, 1 Dushyant Mukkamala, 1 Michael P. Hudock,
RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays
 Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Applied Microbiology and Biotechnology. Supplementary Material
 Applied Microbiology and Biotechnology Supplementary Material Biochemical characterization of a GH70 protein from Lactobacillus kunkeei DSM 12361 with two catalytic domains involving branching sucrase
Applied Microbiology and Biotechnology Supplementary Material Biochemical characterization of a GH70 protein from Lactobacillus kunkeei DSM 12361 with two catalytic domains involving branching sucrase
Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and Gerard D. Wright.
 Supplementary Data for TetX is a Flavin-Dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and
Supplementary Data for TetX is a Flavin-Dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol
 Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
Hands-On Ten The BRCA1 Gene and Protein
 Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
A New Cadinane Sesquiterpene from the Marine Brown Alga Dictyopteris divaricata
 Molecules 2009, 4, 2273-2277; doi:0.3390/molecules4062273 Article OPEN ACCESS molecules ISSN 420-3049 www.mdpi.com/journal/molecules A New Cadinane Sesquiterpene from the Marine Brown Alga Dictyopteris
Molecules 2009, 4, 2273-2277; doi:0.3390/molecules4062273 Article OPEN ACCESS molecules ISSN 420-3049 www.mdpi.com/journal/molecules A New Cadinane Sesquiterpene from the Marine Brown Alga Dictyopteris
Supporting information (protein purification, kinetic characterization, product isolation, and characterization by NMR and mass spectrometry):
 Supporting Information Mechanistic studies of a novel C-S lyase in ergothioneine biosynthesis: the involvement of a sulfenic acid intermediate Heng Song, 1 Wen Hu, 1,2 Nathchar Naowarojna, 1 Ampon Sae
Supporting Information Mechanistic studies of a novel C-S lyase in ergothioneine biosynthesis: the involvement of a sulfenic acid intermediate Heng Song, 1 Wen Hu, 1,2 Nathchar Naowarojna, 1 Ampon Sae
THE JOURNAL OF ANTIBIOTICS. Polyketomycin, a New Antibiotic from Streptomyces sp. MK277-AF1. II. Structure Determination
 THE JOURNAL OF ANTIBIOTICS Polyketomycin, a New Antibiotic from Streptomyces sp. MK277-AF1 II. Structure Determination ISAO MOMOSE, WEI CHEN, HIKARU NAKAMURA, HIROSHI NAGANAWA, HIRONOBU IINUMA and TOMIO
THE JOURNAL OF ANTIBIOTICS Polyketomycin, a New Antibiotic from Streptomyces sp. MK277-AF1 II. Structure Determination ISAO MOMOSE, WEI CHEN, HIKARU NAKAMURA, HIROSHI NAGANAWA, HIRONOBU IINUMA and TOMIO
A Novel Labda-7,13E-dien-15-ol-Producing Bifunctional Diterpene Synthase from Selaginella moellendorffii
 Biochemistry, Biophysics and Molecular Biology Publications Biochemistry, Biophysics and Molecular Biology 9-5-2011 A Novel Labda-7,13E-dien-15-ol-Producing Bifunctional Diterpene Synthase from Selaginella
Biochemistry, Biophysics and Molecular Biology Publications Biochemistry, Biophysics and Molecular Biology 9-5-2011 A Novel Labda-7,13E-dien-15-ol-Producing Bifunctional Diterpene Synthase from Selaginella
Luminescent platforms for monitoring changes in the solubility of amylin and huntingtin in living cells
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Project PRACE 1IP, WP7.4
 Project PRACE 1IP, WP7.4 Plamenka Borovska, Veska Gancheva Computer Systems Department Technical University of Sofia The Team is consists of 5 members: 2 Professors; 1 Assist. Professor; 2 Researchers;
Project PRACE 1IP, WP7.4 Plamenka Borovska, Veska Gancheva Computer Systems Department Technical University of Sofia The Team is consists of 5 members: 2 Professors; 1 Assist. Professor; 2 Researchers;
Supplementary Material (ESI) for Chemical Communications This journal is (c) The Royal Society of Chemistry 2008
 Experimental Details Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich Chemical Company and were used as received. 2-DOS and neamine were kindly provided by Dr. F. Huang. Paromamine
Experimental Details Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich Chemical Company and were used as received. 2-DOS and neamine were kindly provided by Dr. F. Huang. Paromamine
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Simple copper/tempo catalyzed aerobic dehydrogenation. of benzylic amines and anilines
 Simple copper/tempo catalyzed aerobic dehydrogenation of benzylic amines and anilines Zhenzhong Hu and Francesca M. Kerton,* Department of Chemistry, Memorial University of Newfoundland, St. John s, NL,
Simple copper/tempo catalyzed aerobic dehydrogenation of benzylic amines and anilines Zhenzhong Hu and Francesca M. Kerton,* Department of Chemistry, Memorial University of Newfoundland, St. John s, NL,
Biosynthesis of Isoprenoids
 Biosynthesis of Isoprenoids David Wang s Natural Products Class Terpene Mankind has used terpenes that are extracted from plants for many different purposes as fragrances and flavors, as pharmaceutical
Biosynthesis of Isoprenoids David Wang s Natural Products Class Terpene Mankind has used terpenes that are extracted from plants for many different purposes as fragrances and flavors, as pharmaceutical
MODULE 4: SPLICING. Removal of introns from messenger RNA by splicing
 Last update: 05/10/2017 MODULE 4: SPLICING Lesson Plan: Title MEG LAAKSO Removal of introns from messenger RNA by splicing Objectives Identify splice donor and acceptor sites that are best supported by
Last update: 05/10/2017 MODULE 4: SPLICING Lesson Plan: Title MEG LAAKSO Removal of introns from messenger RNA by splicing Objectives Identify splice donor and acceptor sites that are best supported by
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Non-Conjugated Double Bonds
 1 H-NMR Spectroscopy of Fatty Acids and Their Derivatives Non-Conjugated Double Bonds The introduction of one double bond gives rise to several peaks in the NMR spectrum compared to the saturated chains
1 H-NMR Spectroscopy of Fatty Acids and Their Derivatives Non-Conjugated Double Bonds The introduction of one double bond gives rise to several peaks in the NMR spectrum compared to the saturated chains
Supporting Information
 Supporting Information Wiley-VCH 2008 69451 Weinheim, Germany Supporting Information Enantioselective Cu-catalyzed 1,4-Addition of Various Grignard Reagents to Cyclohexenone using Taddol-derived Phosphine-Phosphite
Supporting Information Wiley-VCH 2008 69451 Weinheim, Germany Supporting Information Enantioselective Cu-catalyzed 1,4-Addition of Various Grignard Reagents to Cyclohexenone using Taddol-derived Phosphine-Phosphite
Bioactivity Based Molecular Networking for the Discovery of Drug Lead in Natural Product Bioassay-Guided Fractionation
 Bioactivity Based Molecular Networking for the Discovery of Drug Lead in Natural Product Bioassay-Guided Fractionation Louis-Félix Nothias,,, Mélissa Nothias-Esposito,, # Ricardo da Silva,, Mingxun Wang,,,
Bioactivity Based Molecular Networking for the Discovery of Drug Lead in Natural Product Bioassay-Guided Fractionation Louis-Félix Nothias,,, Mélissa Nothias-Esposito,, # Ricardo da Silva,, Mingxun Wang,,,
Masatoshi Shibuya,Takahisa Sato, Masaki Tomizawa, and Yoshiharu Iwabuchi* Department of Organic Chemistry, Graduate School of Pharmaceutical Sciences,
 Oxoammonium ion/naclo 2 : An Expedient, Catalytic System for One-pot Oxidation of Primary Alcohols to Carboxylic Acid with Broad Substrate Applicability Masatoshi Shibuya,Takahisa Sato, Masaki Tomizawa,
Oxoammonium ion/naclo 2 : An Expedient, Catalytic System for One-pot Oxidation of Primary Alcohols to Carboxylic Acid with Broad Substrate Applicability Masatoshi Shibuya,Takahisa Sato, Masaki Tomizawa,
Parsing a multifunctional biosynthetic gene cluster from rice: Biochemical characterization of CYP71Z6 & 7
 Biochemistry, Biophysics and Molecular Biology Publications Biochemistry, Biophysics and Molecular Biology 11-4-2011 Parsing a multifunctional biosynthetic gene cluster from rice: Biochemical characterization
Biochemistry, Biophysics and Molecular Biology Publications Biochemistry, Biophysics and Molecular Biology 11-4-2011 Parsing a multifunctional biosynthetic gene cluster from rice: Biochemical characterization
Supplemental Data. Landgraf et al. (2014). Plant Cell /tpc
 C ATGTCAAGGATAGTAGCGGAAAATATGTTACAAGGGGGAGAAAATGTACA ATTTTATGATCAAAGAGTACAACAAGCCATGGAGATGTCACAAGCCAGCG CGTACTCTTCACCCACCCTAGGCCAAATGCTAAAGCGCGTGGGAGACGTG AGAAAAGAAGTCACCGGCGACGAAACTCCGGTGCACCGGATTCTCGATAT
C ATGTCAAGGATAGTAGCGGAAAATATGTTACAAGGGGGAGAAAATGTACA ATTTTATGATCAAAGAGTACAACAAGCCATGGAGATGTCACAAGCCAGCG CGTACTCTTCACCCACCCTAGGCCAAATGCTAAAGCGCGTGGGAGACGTG AGAAAAGAAGTCACCGGCGACGAAACTCCGGTGCACCGGATTCTCGATAT
Supporting Information for:
 Supporting Information for: Methylerythritol Cyclodiphosphate (MEcPP) in Deoxyxylulose Phosphate Pathway: Synthesis from an Epoxide and Mechanisms Youli Xiao, a Rodney L. Nyland II, b Caren L. Freel Meyers
Supporting Information for: Methylerythritol Cyclodiphosphate (MEcPP) in Deoxyxylulose Phosphate Pathway: Synthesis from an Epoxide and Mechanisms Youli Xiao, a Rodney L. Nyland II, b Caren L. Freel Meyers
Overview of the Expressway Cell-Free Expression Systems. Expressway Mini Cell-Free Expression System
 Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Supporting information
 Electronic Supplementary Material (ESI) for Catalysis Science & Technology. This journal is The Royal Society of Chemistry 214 Striking difference between alkane and olefin metathesis by the well-defined
Electronic Supplementary Material (ESI) for Catalysis Science & Technology. This journal is The Royal Society of Chemistry 214 Striking difference between alkane and olefin metathesis by the well-defined
Nitro-Grela-type complexes containing iodides. robust and selective catalysts for olefin metathesis
 Supporting Information for Nitro-Grela-type complexes containing iodides robust and selective catalysts for olefin metathesis under challenging conditions. Andrzej Tracz, 1,2 Mateusz Matczak, 1 Katarzyna
Supporting Information for Nitro-Grela-type complexes containing iodides robust and selective catalysts for olefin metathesis under challenging conditions. Andrzej Tracz, 1,2 Mateusz Matczak, 1 Katarzyna
Supplemental Information
 Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Supporting Materials. Experimental Section. internal standard TMS (0 ppm). The peak patterns are indicated as follows: s, singlet; d,
 CuBr-Catalyzed Efficient Alkynylation of sp 3 C-H Bonds Adjacent to a itrogen Atom Zhiping Li and Chao-Jun Li* Department of Chemistry, McGill University, 801 Sherbrooke St. West, Montreal, Quebec H3A
CuBr-Catalyzed Efficient Alkynylation of sp 3 C-H Bonds Adjacent to a itrogen Atom Zhiping Li and Chao-Jun Li* Department of Chemistry, McGill University, 801 Sherbrooke St. West, Montreal, Quebec H3A
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
SpliceDB: database of canonical and non-canonical mammalian splice sites
 2001 Oxford University Press Nucleic Acids Research, 2001, Vol. 29, No. 1 255 259 SpliceDB: database of canonical and non-canonical mammalian splice sites M.Burset,I.A.Seledtsov 1 and V. V. Solovyev* The
2001 Oxford University Press Nucleic Acids Research, 2001, Vol. 29, No. 1 255 259 SpliceDB: database of canonical and non-canonical mammalian splice sites M.Burset,I.A.Seledtsov 1 and V. V. Solovyev* The
Cloning and Expression of a Subfamily 1.4 Lipase from Bacillus licheniformis IBRL-CHS2
 Tropical Life Sciences Research, 27(Supp. 1), 145 150, 2016 Cloning and Expression of a Subfamily 1.4 Lipase from Bacillus licheniformis IBRL-CHS2 1 Nidyaletchmy Subba Reddy, 1 Rashidah Abdul Rahim *,
Tropical Life Sciences Research, 27(Supp. 1), 145 150, 2016 Cloning and Expression of a Subfamily 1.4 Lipase from Bacillus licheniformis IBRL-CHS2 1 Nidyaletchmy Subba Reddy, 1 Rashidah Abdul Rahim *,
Identification of mirnas in Eucalyptus globulus Plant by Computational Methods
 International Journal of Pharmaceutical Science Invention ISSN (Online): 2319 6718, ISSN (Print): 2319 670X Volume 2 Issue 5 May 2013 PP.70-74 Identification of mirnas in Eucalyptus globulus Plant by Computational
International Journal of Pharmaceutical Science Invention ISSN (Online): 2319 6718, ISSN (Print): 2319 670X Volume 2 Issue 5 May 2013 PP.70-74 Identification of mirnas in Eucalyptus globulus Plant by Computational
Bioinformatics Laboratory Exercise
 Bioinformatics Laboratory Exercise Biology is in the midst of the genomics revolution, the application of robotic technology to generate huge amounts of molecular biology data. Genomics has led to an explosion
Bioinformatics Laboratory Exercise Biology is in the midst of the genomics revolution, the application of robotic technology to generate huge amounts of molecular biology data. Genomics has led to an explosion
Manja Henze, Dorothee Merker and Lothar Elling. 1. Characteristics of the Recombinant β-glycosidase from Pyrococcus
 S1 of S17 Supplementary Materials: Microwave-Assisted Synthesis of Glycoconjugates by Transgalactosylation with Recombinant Thermostable β-glycosidase from Pyrococcus Manja Henze, Dorothee Merker and Lothar
S1 of S17 Supplementary Materials: Microwave-Assisted Synthesis of Glycoconjugates by Transgalactosylation with Recombinant Thermostable β-glycosidase from Pyrococcus Manja Henze, Dorothee Merker and Lothar
Supporting Information
 Supporting Information Identification, characterization and molecular adaptation of class I redox systems for the production of multi-functionalized diterpenoids Christian Görner a,, Patrick Schrepfer
Supporting Information Identification, characterization and molecular adaptation of class I redox systems for the production of multi-functionalized diterpenoids Christian Görner a,, Patrick Schrepfer
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Comparing Amino Acid Sequences Abstract
 Teacher Guide Comparing Amino Acid Sequences Abstract In this hands-on activity, students work in small groups to compare amino acid sequences for a particular protein from a to the same protein from 5
Teacher Guide Comparing Amino Acid Sequences Abstract In this hands-on activity, students work in small groups to compare amino acid sequences for a particular protein from a to the same protein from 5
Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum
 FEBS Letters 583 (2009) 475 480 journal homepage: www.febsletters.org Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum Dana Morrone
FEBS Letters 583 (2009) 475 480 journal homepage: www.febsletters.org Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum Dana Morrone
CONSTRUCTION OF PHYLOGENETIC TREE USING NEIGHBOR JOINING ALGORITHMS TO IDENTIFY THE HOST AND THE SPREADING OF SARS EPIDEMIC
 CONSTRUCTION OF PHYLOGENETIC TREE USING NEIGHBOR JOINING ALGORITHMS TO IDENTIFY THE HOST AND THE SPREADING OF SARS EPIDEMIC 1 MOHAMMAD ISA IRAWAN, 2 SITI AMIROCH 1 Institut Teknologi Sepuluh Nopember (ITS)
CONSTRUCTION OF PHYLOGENETIC TREE USING NEIGHBOR JOINING ALGORITHMS TO IDENTIFY THE HOST AND THE SPREADING OF SARS EPIDEMIC 1 MOHAMMAD ISA IRAWAN, 2 SITI AMIROCH 1 Institut Teknologi Sepuluh Nopember (ITS)
Dana Morrone Iowa State University. Matthew L. Hillwig Iowa State University. Matthew E. Mead Iowa State University. Luke Lowry Iowa State University
 Biochemistry, Biophysics and Molecular Biology Publications Biochemistry, Biophysics and Molecular Biology 5-1-2011 Evident and latent plasticity across the rice diterpene synthase family with potential
Biochemistry, Biophysics and Molecular Biology Publications Biochemistry, Biophysics and Molecular Biology 5-1-2011 Evident and latent plasticity across the rice diterpene synthase family with potential
Acaulins A and B, Trimeric Macrodiolides from Acaulium. Table of Contents
 Supporting Information Acaulins A and B, Trimeric Macrodiolides from Acaulium sp. H-JQSF Ting Ting Wang, Ying Jie Wei, Hui Ming Ge, Rui Hua Jiao, Ren Xiang Tan* * Corresponding author. E-mail: rxtan@nju.edu.cn
Supporting Information Acaulins A and B, Trimeric Macrodiolides from Acaulium sp. H-JQSF Ting Ting Wang, Ying Jie Wei, Hui Ming Ge, Rui Hua Jiao, Ren Xiang Tan* * Corresponding author. E-mail: rxtan@nju.edu.cn
A putative MYB35 ortholog is a candidate for the sex-determining genes in Asparagus
 Supplementary figures for: A putative MYB35 ortholog is a candidate for the sex-determining genes in Asparagus officinalis Daisuke Tsugama, Kohei Matsuyama, Mayui Ide, Masato Hayashi, Kaien Fujino, and
Supplementary figures for: A putative MYB35 ortholog is a candidate for the sex-determining genes in Asparagus officinalis Daisuke Tsugama, Kohei Matsuyama, Mayui Ide, Masato Hayashi, Kaien Fujino, and
Electronic Supplementary Information
 Electronic Supplementary Information ~ Experimental Procedures and Spectral/Analytical Data ~ Use of Dimethyl Carbonate as a Solvent Greatly Enhances the Biaryl Coupling of Aryl Iodides and Organoboron
Electronic Supplementary Information ~ Experimental Procedures and Spectral/Analytical Data ~ Use of Dimethyl Carbonate as a Solvent Greatly Enhances the Biaryl Coupling of Aryl Iodides and Organoboron
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
CHAPTER 6 METABOLIC PATHWAY RECONSTRUCTION
 66 CHAPTER 6 METABOLIC PATHWAY RECONSTRUCTION 6.1 GENERAL A metabolic pathway is a series of biochemical reactions that occur within a cell and these chemical reactions are catalyzed by certain enzymes.
66 CHAPTER 6 METABOLIC PATHWAY RECONSTRUCTION 6.1 GENERAL A metabolic pathway is a series of biochemical reactions that occur within a cell and these chemical reactions are catalyzed by certain enzymes.
Mechanisms of alternative splicing regulation
 Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Supplemental Information For: The genetics of splicing in neuroblastoma
 Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
Identification and characterization of multiple splice variants of Cdc2-like kinase 4 (Clk4)
 Identification and characterization of multiple splice variants of Cdc2-like kinase 4 (Clk4) Vahagn Stepanyan Department of Biological Sciences, Fordham University Abstract: Alternative splicing is an
Identification and characterization of multiple splice variants of Cdc2-like kinase 4 (Clk4) Vahagn Stepanyan Department of Biological Sciences, Fordham University Abstract: Alternative splicing is an
Topic 6 Structure Determination Revision Notes
 1) Introduction Topic 6 Structure Determination Revision Notes Mass spectrometry, infrared spectroscopy and NMR spectroscopy can be used to determine the structure of unknown compounds 2) Mass spectrometry
1) Introduction Topic 6 Structure Determination Revision Notes Mass spectrometry, infrared spectroscopy and NMR spectroscopy can be used to determine the structure of unknown compounds 2) Mass spectrometry
SUPPLEMENTARY INFORMATION
 ARTICLE NUMBER: 16198 DOI: 10.1038/NMICROBIOL.2016.198 Genome reduction in an abundant and ubiquitous soil bacterium, Candidatus Udaeobacter copiosus Tess E Brewer 1, 2, Kim M Handley 3, Paul Carini 1,
ARTICLE NUMBER: 16198 DOI: 10.1038/NMICROBIOL.2016.198 Genome reduction in an abundant and ubiquitous soil bacterium, Candidatus Udaeobacter copiosus Tess E Brewer 1, 2, Kim M Handley 3, Paul Carini 1,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
SMPD 287 Spring 2015 Bioinformatics in Medical Product Development. Final Examination
 Final Examination You have a choice between A, B, or C. Please email your solutions, as a pdf attachment, by May 13, 2015. In the subject of the email, please use the following format: firstname_lastname_x
Final Examination You have a choice between A, B, or C. Please email your solutions, as a pdf attachment, by May 13, 2015. In the subject of the email, please use the following format: firstname_lastname_x
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Hao D. H., Ma W. G., Sheng Y. L., Zhang J. B., Jin Y. F., Yang H. Q., Li Z. G., Wang S. S., GONG Ming*
 Comparison of transcriptomes and gene expression profiles of two chilling- and drought-tolerant and intolerant Nicotiana tabacum varieties under low temperature and drought stress Hao D. H., Ma W. G.,
Comparison of transcriptomes and gene expression profiles of two chilling- and drought-tolerant and intolerant Nicotiana tabacum varieties under low temperature and drought stress Hao D. H., Ma W. G.,
Evolution of hepatitis C virus in blood donors and their respective recipients
 Journal of General Virology (2003), 84, 441 446 DOI 10.1099/vir.0.18642-0 Short Communication Correspondence Jean-François Cantaloube jfc-ets-ap@gulliver.fr Evolution of hepatitis C virus in blood donors
Journal of General Virology (2003), 84, 441 446 DOI 10.1099/vir.0.18642-0 Short Communication Correspondence Jean-François Cantaloube jfc-ets-ap@gulliver.fr Evolution of hepatitis C virus in blood donors
Supplementary Figure 1. ESI/MS/MS analyses of native and de-acetylated S2A Supplementary Figure 2. Partial 1D 1H NMR spectrum of S2A
 Supplementary Figure 1. ESI/MS/MS analyses of native and de-acetylated S2A. Panel A, Positive ESI mass spectra of native (N) and de-acetylated (DA) non-active S2A were obtained, and demonstrated a shift
Supplementary Figure 1. ESI/MS/MS analyses of native and de-acetylated S2A. Panel A, Positive ESI mass spectra of native (N) and de-acetylated (DA) non-active S2A were obtained, and demonstrated a shift
Exploring HIV Evolution: An Opportunity for Research Sam Donovan and Anton E. Weisstein
 Microbes Count! 137 Video IV: Reading the Code of Life Human Immunodeficiency Virus (HIV), like other retroviruses, has a much higher mutation rate than is typically found in organisms that do not go through
Microbes Count! 137 Video IV: Reading the Code of Life Human Immunodeficiency Virus (HIV), like other retroviruses, has a much higher mutation rate than is typically found in organisms that do not go through
Preparation of Stable Aziridinium Ions and Their Ring Openings
 Supplementary Information Preparation of Stable Aziridinium Ions and Their Ring Openings Yongeun Kim a Hyun-Joon Ha*, a Sae Young Yun b and Won Koo Lee,*,b a Department of Chemistry and Protein Research
Supplementary Information Preparation of Stable Aziridinium Ions and Their Ring Openings Yongeun Kim a Hyun-Joon Ha*, a Sae Young Yun b and Won Koo Lee,*,b a Department of Chemistry and Protein Research
Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus
 Supplemental Information Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus Torben Hofeditz, 1 Claudia Eva-Maria Unsin 2, Jutta
Supplemental Information Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus Torben Hofeditz, 1 Claudia Eva-Maria Unsin 2, Jutta
SUPPLEMENTARY INFORMATION. Divergent TLR7/9 signaling and type I interferon production distinguish
 SUPPLEMENTARY INFOATION Divergent TLR7/9 signaling and type I interferon production distinguish pathogenic and non-pathogenic AIDS-virus infections Judith N. Mandl, Ashley P. Barry, Thomas H. Vanderford,
SUPPLEMENTARY INFOATION Divergent TLR7/9 signaling and type I interferon production distinguish pathogenic and non-pathogenic AIDS-virus infections Judith N. Mandl, Ashley P. Barry, Thomas H. Vanderford,
Graphene Quantum Dots-Band-Aids Used for Wound Disinfection
 Supporting information Graphene Quantum Dots-Band-Aids Used for Wound Disinfection Hanjun Sun, Nan Gao, Kai Dong, Jinsong Ren, and Xiaogang Qu* Laboratory of Chemical Biology, Division of Biological Inorganic
Supporting information Graphene Quantum Dots-Band-Aids Used for Wound Disinfection Hanjun Sun, Nan Gao, Kai Dong, Jinsong Ren, and Xiaogang Qu* Laboratory of Chemical Biology, Division of Biological Inorganic
Ambient temperature regulated flowering time
 Ambient temperature regulated flowering time Applications of RNAseq RNA- seq course: The power of RNA-seq June 7 th, 2013; Richard Immink Overview Introduction: Biological research question/hypothesis
Ambient temperature regulated flowering time Applications of RNAseq RNA- seq course: The power of RNA-seq June 7 th, 2013; Richard Immink Overview Introduction: Biological research question/hypothesis
Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana
 Mol Genet Genomics (2002) 267: 730 745 DOI 10.1007/s00438-002-0709-y ORIGINAL PAPER S. Aubourg Æ A. Lecharny Æ J. Bohlmann Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis
Mol Genet Genomics (2002) 267: 730 745 DOI 10.1007/s00438-002-0709-y ORIGINAL PAPER S. Aubourg Æ A. Lecharny Æ J. Bohlmann Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis
In vitro conversion of chanoclavine-i aldehyde to the stereoisomers festuclavine and pyroclavine was controlled by the second reduction step
 1 Electronic Supporting Information to: In vitro conversion of chanoclavinei aldehyde to the stereoisomers festuclavine and pyroclavine was controlled by the second reduction step Marco Matuschek a, Christiane
1 Electronic Supporting Information to: In vitro conversion of chanoclavinei aldehyde to the stereoisomers festuclavine and pyroclavine was controlled by the second reduction step Marco Matuschek a, Christiane
Going Nowhere Fast: Lentivirus genetic sequence evolution does not correlate with phenotypic evolution.
 Going Nowhere Fast: Lentivirus genetic sequence evolution does not correlate with phenotypic evolution. Brian T. Foley, PhD btf@lanl.gov HIV Genetic Sequences, Immunology, Drug Resistance and Vaccine Trials
Going Nowhere Fast: Lentivirus genetic sequence evolution does not correlate with phenotypic evolution. Brian T. Foley, PhD btf@lanl.gov HIV Genetic Sequences, Immunology, Drug Resistance and Vaccine Trials
Supplementary Information
 Supplementary Information J. Braz. Chem. Soc., Vol. 24, No. 12, S1-S21, 2013. Printed in Brazil - 2013 Sociedade Brasileira de Química 0103-5053 $6.00+0.00 SI Rui He, a,b,c Bochu Wang,*,b Toshiyuki Wakimoto,
Supplementary Information J. Braz. Chem. Soc., Vol. 24, No. 12, S1-S21, 2013. Printed in Brazil - 2013 Sociedade Brasileira de Química 0103-5053 $6.00+0.00 SI Rui He, a,b,c Bochu Wang,*,b Toshiyuki Wakimoto,
Labelling Studies on the Biosynthesis of Terpenes in Fusarium fujikuroi. Institut für Organische Chemie, Hagenring 30, Braunschweig
 Supplementary Information: Labelling Studies on the Biosynthesis of Terpenes in Fusarium fujikuroi Christian A. Citron, a Nelson L. Brock, a Bettina Tudzynski, b Jeroen S. Dickschat* a a Institut für Organische
Supplementary Information: Labelling Studies on the Biosynthesis of Terpenes in Fusarium fujikuroi Christian A. Citron, a Nelson L. Brock, a Bettina Tudzynski, b Jeroen S. Dickschat* a a Institut für Organische
FINAL ANNOTATION REPORT: Drosophila virilis Fosmid 11 (48P14) Robert Carrasquillo Bio 4342
 FINAL ANNOTATION REPORT: Drosophila virilis Fosmid 11 (48P14) Robert Carrasquillo Bio 4342 2006 TABLE OF CONTENTS I. Overview... 3 II. Genes... 4 III. Clustal Analysis... 15 IV. Repeat Analysis... 17 V.
FINAL ANNOTATION REPORT: Drosophila virilis Fosmid 11 (48P14) Robert Carrasquillo Bio 4342 2006 TABLE OF CONTENTS I. Overview... 3 II. Genes... 4 III. Clustal Analysis... 15 IV. Repeat Analysis... 17 V.
User Guide. Protein Clpper. Statistical scoring of protease cleavage sites. 1. Introduction Protein Clpper Analysis Procedure...
 User Guide Protein Clpper Statistical scoring of protease cleavage sites Content 1. Introduction... 2 2. Protein Clpper Analysis Procedure... 3 3. Input and Output Files... 9 4. Contact Information...
User Guide Protein Clpper Statistical scoring of protease cleavage sites Content 1. Introduction... 2 2. Protein Clpper Analysis Procedure... 3 3. Input and Output Files... 9 4. Contact Information...
Divergent Construction of Pyrazoles via Michael Addition of N-Aryl Hydrazones to 1,2-Diaza-1,3-dienes
 Divergent Construction of Pyrazoles via Michael Addition of N-Aryl Hydrazones to 1,2-Diaza-1,3-dienes Serena Mantenuto, Fabio Mantellini, Gianfranco Favi,* and Orazio A. Attanasi Department of Biomolecular
Divergent Construction of Pyrazoles via Michael Addition of N-Aryl Hydrazones to 1,2-Diaza-1,3-dienes Serena Mantenuto, Fabio Mantellini, Gianfranco Favi,* and Orazio A. Attanasi Department of Biomolecular
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq
 Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
by Micah Johnson, Michael Turner, Elena Poiata, Stephanie Vadasz, Nathan Yardley, and Maria Moreno
 MCDB 201L: Molecular Biology Laboratory Professor: Maria Moreno By submitting this essay, I attest that it is my own work, completed in accordance with University regulations. Micah Johnson Abstract Cloning
MCDB 201L: Molecular Biology Laboratory Professor: Maria Moreno By submitting this essay, I attest that it is my own work, completed in accordance with University regulations. Micah Johnson Abstract Cloning
Supporting Information
 Supporting Information Unconventional Passerini Reaction towards α-aminoxyamides Ajay L. Chandgude, Alexander Dömling* Department of Drug Design, University of Groningen, Antonius Deusinglaan 1, 9713 AV
Supporting Information Unconventional Passerini Reaction towards α-aminoxyamides Ajay L. Chandgude, Alexander Dömling* Department of Drug Design, University of Groningen, Antonius Deusinglaan 1, 9713 AV
Fluorescent probes for detecting monoamine oxidase activity and cell imaging
 Fluorescent probes for detecting monoamine oxidase activity and cell imaging Xuefeng Li, Huatang Zhang, Yusheng Xie, Yi Hu, Hongyan Sun *, Qing Zhu * Supporting Information Table of Contents 1. General
Fluorescent probes for detecting monoamine oxidase activity and cell imaging Xuefeng Li, Huatang Zhang, Yusheng Xie, Yi Hu, Hongyan Sun *, Qing Zhu * Supporting Information Table of Contents 1. General
