Supplementary Figure 1. HGL-DTG levels in uninduced and herbivory induced leaves. Total HGL-
|
|
|
- Oliver McKinney
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1. HGL-DTG levels in uninduced and herbivory induced leaves. Total HGL- DTG concentrations [F 1,4 = 36.88, P ; significant differences (threshold: P 0.05) between means (± SE) determined by Fisher s LSD test (one-way ANOVA); n= 3] and proportions of malonylated and non-malonylated forms in uninduced and M. sexta herbivory-induced N. attenuata leaves. Together with the data in Fig. 1b, which shows proportions of various HGL-DTGs in uninduced and induced leaves, this data shows the quantitative increase contributed by increases in Lyc4, Nic1 and Nic2 contents in herbivory-induced leaves.
2 Supplementary Figure 2: Structure and key HMBC correlations ( 1 H 13 C) of 3-O-[αrhamnopyranosyl-(1 4)-β-glucopyranosyl]-17-hydroxygeranyllinalool (RGHGL). The colors of the arrows correspond to the colors of the cross signals in the spectra shown in Supplementary Fig. 3-7.
3 Supplementary Figure 3: Partial HMBC spectrum (500 MHz, MeOH-d 4 ) of RGHGL
4 Supplementary Figure 4: Partial HMBC spectrum (500 MHz, MeOH-d 4 ) of RGHGL
5 Supplementary Figure 5: Partial HMBC spectrum (500 MHz, MeOH-d 4 ) of RGHGL
6 Supplementary Figure 6: Partial HSQC spectrum (500 MHz, MeOH-d 4 ) of RGHGL
7 Supplementary Figure 7: Partial HMBC spectrum (500 MHz, MeOH-d 4 ) of RGHGL
8 Supplementary Figure 8. U(H)PLC/ ESI-QTOF-MS based analysis of Lyc4, RGHGL and rebaudioside A. Standard curves of (a) Lyc4 (b) RGHGL and (c) rebaudioside A (internal standard) revealed linear responses of U(H)PLC/ESI-QTOF MS to them (n= 3 for each concentration of a compound); limit of detection of rebaudioside A was 0.3 ng. Standard curves of (d) Lyc4 and (e) RGHGL (f) rebaudioside A revealed linear responses of HPLC-ELS1A to them (n= 3 for each concentration of a compound); rebaudioside A was used as an external standard for the relative quantification of these compounds by HPLC-ELS1A and its limit of detection was 15 ng. Plots showing linearity in extraction efficiency of (g) Lyc4 and (h) RGHGL (i) rebaudioside A, from standard addition experiments with frass; for both the compounds, extraction efficiency was >90% when 2, 4, 6, 8, 16 and 32 µg of each compound was spiked to 100 mg frass (n= 3 for each spiking concentration) before extraction and analyzed by HPLC-ELS1A.
9 Supplementary Figure 9. M. sexta possesses three BGs; MsBG2 and MsBG3 transcripts are not upregulated upon Lyc4 ingestion. (a) Phylogeny of M. sexta glycoside hydrolases (only ORFs) deciphered using Clustal-W (thousand bootstrapping trials; only the bootstrap values >50 displayed) showing that M. sexta transcriptome contains three BGs; MsBG2 and MsBG3 are 60.2% and 51.5% similar to MsBG1, respectively. BG2 transcripts (relative to ubiquitin) in midguts of fourth-instar larvae feeding on (b) Lyc4-containing EV and Lyc4-deplete irggpps plants (n= 6) and (c) artificial diet containing water (control), 6 mm Lyc4 or RGHGL (n= 6). BG3 transcripts (relative to ubiquitin) in midguts of fourth-instar larvae feeding on (d) Lyc4-containing EV and Lyc4-deplete irggpps plants (n= 6) and (e) artificial diet containing water (control), 6 mm Lyc4 or RGHGL (n= 6).
10 Supplementary Figure 10. Characterization of transgenic irbg1 N. attenuata lines and BG1- silenced M. sexta larvae. (a) Southern hybridization over the HindIII digested genomic DNA, showing the presence of a single copy of transgene fragment inserted in two independently generated transgenic irbg1 N. attenuata lines ( and 379-8); wild type control shows the absence of transgene insertion. 1kb DNA ladder was used as a size marker. (b) Northern hybridization showing the presence of BG1 small RNA in irbg1 (375-10) and irbg1 (379-8) N. attenuata leaves and in the midguts of fourth-instar larvae feeding on the respective stable transgenic lines; RNA from EV leaves and midgut of larvae those were feeding on EV leaves were used as negative controls, respectively. (c) BG1 transcripts (relative to ubiquitin) in midguts of fourth-instar larvae feeding on the second independently transformed irbg1 line (379-8) [F 1,10 = 940.5, P ; significant differences (threshold: P 0.05) between means (± SE) determined by Fisher s LSD test (one-way ANOVA); n= 6], showing equal silencing as that achieved by feeding on irbg1 (375-10) (shown in Fig. 3E). (d) Flowering EV and independently transformed irbg1 plants ( and 379-8) plants showing that the transformation of PMRi construct did not affect plant morphology. (e) Concentrations of various secondary metabolites in leaves of EV and independently transformed irbg1 lines ( and 379-8) (n= 5); since morphologies and the secondary metabolite concentrations and MsBG1 silencing efficiencies of and did not differ, was randomly selected for the further experimentation. Transcripts (relative to ubiquitin) of (f) MsBG2 (60.2% similar to MsBG1) and (g) MsBG3 (51.5% similar to MsBG1) in the midguts of fourth-instar EV- and irbg1-feeding larvae showing that the off-target silencing of MsBG2 and MsBG3 had not occurred while silencing MsBG1 (n= 6).
11 Supplementary Figure 11. Characterization of BG1-silenced M. sexta larvae. Concentration of (a) Lyc4 [midgut: F 1,11 = 2.96, P 0.05; hindgut: F 1,11 = 6.41, P 0.02; hemolymph: F 1,8 = 14.9, P 0.005; fat body+ skin: F 1,11 = 5.926, P 0.03; significant differences (threshold: P 0.05) between means (± SE) determined by Fisher s LSD test (one-way ANOVA, separately conducted for each tissue);] and (b) RGHGL [ midgut: F 1,11 = 13.7, P 0.005; significant differences (threshold: P 0.05) between means (± SE) determined by Fisher s LSD test (one-way ANOVA, separately conducted for each tissue);] in foregut, midgut, hindgut, hemolymph, Malpighian tubules and skin with fat body of fourth-instar larvae feeding on EV and irbg1 (375-10) N. attenuata plants. For Lyc4 as well as RGHGL concentrations in foregut, midgut, hindgut, hemolymph, Malpighian tubules and skin with fat body of EV-fed larvae, n= 7, 7, 7, 6, 5 and 7, respectively and for Lyc4 concentration in foregut, midgut, hindgut, hemolymph, Malpighian tubules and skin with fat body of irbg1-fed larvae, n= 7, 6, 6, 4, 5 and 6, respectively. (c) Lyc4 and RGHGL are not degraded in frass over the 24 h period of the excretion efficiency determination assays. Fresh frass was spiked with each metabolite to attain the final concentration of 0.5%; the spiked frass was extracted and analyzed after zero and 24 h of incubation to quantify the recovered metabolite (n= 3). (d) Stages of molting impairment leading to mortality in M. sexta larvae feeding on irbg1 plants.
12 Supplementary Figure 12. BG1 transcripts and Lyc4 and RGHGL concentrations in larvae feeding on Lyc4- and RGHGL-supplemented leaves. (a) BG1 transcripts (relative to ubiquitin) in midguts [F 7,40 = 9253, P ; significant differences (threshold: P 0.05) between means (± SE) determined by Games Howell test (Welch s ANOVA); n= 6] and (b) Lyc4 [F 7,24 = 75.39, P ; significant differences (threshold: P 0.05) between means (± SE) determined by Games Howell test (Welch s ANOVA); n= 4] and (c) RGHGL [F 7,24 = 50.77, P ; significant differences (threshold: P 0.05) between means (± SE) determined by Games Howell test (Welch s ANOVA); n= 4] concentrations in the bodies of M. sexta larvae after 8d feeding on water coated EV, irbg1, irggpps and B G leaves, Lyc4 coated (final concentration 6 mm) irggpps and B G leaves and RGHGL coated (final concentration 6 mm) irggpps and B G leaves. (d) Lyc4 and RGHGL coated on irggpps leaf are not degraded over the 24 h period. Each detached leaf was coated with Lyc4 or RGHGL to attain the final concentration of 6 mm and was extracted and analyzed after zero and 24 h of incubation to quantify the recovered metabolite (n= 5).
13 Supplementary Figure 13. Spiders do not choose between EV- irbg1- and irggpps-fed larvae. Spider s prey capture and killing (%) in choice assays (1 h) on second-instar M. sexta larvae feeding on (a) EV or irbg1, (b) EV or irggpps and (c) irbg1 and irggpps plants (n= 30 in all assays). (d) Spider s prey capture and killing (%) [significant differences (P 0.05) determined by Fisher s exact test of frequencies; n= 30] and (e) prey ingestion (% of total killed) [significant differences (P 0.05) determined by Fisher s exact test of frequencies; n= 30] in no-choice assays (1 h) on second-instar water-, Lyc4- or RGHGL-coated (final concentration 6 mm for both Lyc4 and RGHGL) M. sexta larvae feeding on AD. Spider s prey capture and killing (%) in choice assays (1 h) on AD-feeding second-instar M. sexta larvae coated with (f) water or Lyc4 (final concentration 6 mm) [significant difference (P 0.05) determined by Fisher s exact test of frequencies; n= 21], (g) water or RGHGL (final concentration 6 mm) (n= 21) and (h) Lyc4 (final concentration 6 mm) or RGHGL (final concentration 6 mm) [significant difference (P 0.05) determined by Fisher s exact test of frequencies; n= 21]. Please note that prey capture and killing percentages using AD-fed larvae are higher than those obtained using irggpps-fed larvae (Fig. 6 d-f and h-i) because AD does not contain nicotine, which larvae ingest from irggpps plants and exhale to deter spiders. (i) Lyc4 and RGHGL topically coated to the larval body are not degraded over the period of choice or no-choice assays (1 h). Each larva was coated with either Lyc4 or RGHGL to attain the final concentration of 6 mm; it was washed after zero and 1 h of incubation and the wash was analyzed to quantify the recovered metabolite (n= 3).
14 Supplementary Table 1. 1 H NMR (500 MHz) and 13 C NMR (125 MHz) chemical shifts for RGHGL in MeOH-d 4 Position δ H, multiplet, J (Hz) δ C Position δ H, multiplet, J (Hz) δ C 1a 5.23, dd, 17.8, 1.2 Glucose b 5.20, dd, 11.1, 1.2 1' 4.36, d, , dd, 17.8, ' 3.20, dd, 8.0, ' 3.42, dd, 9.2, , m ' 3.52, dd, 9.4, , m ' 3.24, m , t, * 6'a 3.76, dd, 12.0, 'b 3.64, dd, 12.0, , m 41.2** Rhamnose , m " 4.85, d, , t, * 2" 3.83, dd, 3.3, " 3.63, dd, 9.6, , m 40.9** 4" 3.40, dd, 9.6, , m " 3.96, dq, 9.4, , t, " 1.26, d, , s , s , s , s , s 23.3 *, **: May be interchanged
15 Supplementary Table 2. M. sexta gene primers used in various experiments Primer pair No Gene Primer sequences (5'-3') Use 1 MsBG1.1 2 MsBG1.2 3 MsBG1.3 4 MsBG2 5 MsBG3 For- CTCGCTTGTTATGGCGGGT Rev- GCGGCAGTGGCTGCAC For- TCGTCCTTCTCGCTTGTTATG Rev- GCTGCACCAAACAGAAATCC For- TCGAGTCCTAGCTCTCTCATCA Rev- GGACAAAACGTGTCACATGGTA For- CCACGGTCACAAGTTAAGGC Rev- CTTTTGCCGTCCTCATTCCAC For- GGATCTGCCGCAAAGACTG Rev- TCACTCTATCGCCGAAGTTCTC Amplification of 301bp fragment to clone in PMRi vector Transcript quantification and testing the silencing efficiency of M. sexta BG1 Generation of probe for northern hybridization Transcript quantification and testing the co-silencing efficiency of M. sexta BG2 Transcript quantification and testing the co-silencing efficiency of M. sexta BG3
16 Supplementary Table 3. APHIS notification numbers for importing transgenic N. attenuata seeds and releasing plants at the field station, Utah, USA. Line Import # Year Release# EV n r irggpps n r (NaGGPPS NCBI accession no. EF382626) irbg m r (MsBG1 NCBI accession no. FK816842)
S3. U(H)PLC/ESI-QTOF-MS
 Supporting Information Corrected February 16, 2016 Supporting information Materials and Methods Stem-feeding nicotine to N. attenuata leaves Waldbauer assays for nicotine budgeting Kinetics of nicotine
Supporting Information Corrected February 16, 2016 Supporting information Materials and Methods Stem-feeding nicotine to N. attenuata leaves Waldbauer assays for nicotine budgeting Kinetics of nicotine
METABOSCAPE A METABOLITE PROFILING PIPELINE DRIVEN BY AUTOMATIC COMPOUND IDENTIFICATION
 METABOSCAPE A METABOLITE PROFILING PIPELINE DRIVEN BY AUTOMATIC COMPOUND IDENTIFICATION OR W TO LINK HRAM QTOF PLANT METABOLOMICS DATA TO BIOLOGY Aiko Barsch, Bruker Daltonics, Bremen, Germany 1 Outline
METABOSCAPE A METABOLITE PROFILING PIPELINE DRIVEN BY AUTOMATIC COMPOUND IDENTIFICATION OR W TO LINK HRAM QTOF PLANT METABOLOMICS DATA TO BIOLOGY Aiko Barsch, Bruker Daltonics, Bremen, Germany 1 Outline
Supplementary Information
 Supplementary Information New Highly Oxygenated Germacranolides from Carpesium divaricatum and their Cytotoxic Activity Tao Zhang, 1 Jin-Guang Si, 1, 2 Qiu-Bo Zhang, 1 Gang Ding, 1 and Zhong-Mei Zou 1*
Supplementary Information New Highly Oxygenated Germacranolides from Carpesium divaricatum and their Cytotoxic Activity Tao Zhang, 1 Jin-Guang Si, 1, 2 Qiu-Bo Zhang, 1 Gang Ding, 1 and Zhong-Mei Zou 1*
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the
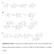 Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
BIOL 208: Principles of Ecology and Evolution Lab Week 5, 6, &7. Bioenergetics of Caterpillars
 Background BIOL 08: Principles of Ecology and Evolution Lab Week 5,, &7 The tobacco hornworm larva (Manduca sexta) eats the leaves of a wide range of solanaceous plants (tomato, tobacco). It passes through
Background BIOL 08: Principles of Ecology and Evolution Lab Week 5,, &7 The tobacco hornworm larva (Manduca sexta) eats the leaves of a wide range of solanaceous plants (tomato, tobacco). It passes through
Marine Streptomyces sp. derived antimycin analogues. suppress HeLa cells via depletion HPV E6/E7 mediated by
 Marine Streptomyces sp. derived antimycin analogues suppress HeLa cells via depletion HPV E6/E7 mediated by ROS-dependent ubiquitin proteasome system Weiyi Zhang 1, +, Qian Che 1, 2, +, Hongsheng Tan 1,
Marine Streptomyces sp. derived antimycin analogues suppress HeLa cells via depletion HPV E6/E7 mediated by ROS-dependent ubiquitin proteasome system Weiyi Zhang 1, +, Qian Che 1, 2, +, Hongsheng Tan 1,
ncounter TM Analysis System
 ncounter TM Analysis System Molecules That Count TM www.nanostring.com Agenda NanoString Technologies History Introduction to the ncounter Analysis System CodeSet Design and Assay Principals System Performance
ncounter TM Analysis System Molecules That Count TM www.nanostring.com Agenda NanoString Technologies History Introduction to the ncounter Analysis System CodeSet Design and Assay Principals System Performance
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted. residues are colored red. Numbers indicate the position of each residue.
 Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
CHAPTER 4 RESULTS. showed that all three replicates had similar growth trends (Figure 4.1) (p<0.05; p=0.0000)
 CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein
![2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked. amino-modification products by acrolein](/thumbs/86/94743397.jpg) Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
Supplementary Information 2,6,9-Triazabicyclo[3.3.1]nonanes as overlooked amino-modification products by acrolein Ayumi Tsutsui and Katsunori Tanaka* Biofunctional Synthetic Chemistry Laboratory, RIKEN
control kda ATGL ATGLi HSL 82 GAPDH * ** *** WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi iwat gwat ibat
 body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
Supplementary Figures
 Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
SUPPLEMENTARY MATERIAL
 SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Supplementary Information
 Supplementary Information The fate of W chromosomes in hybrids between wild silkmoths, Samia cynthia ssp.: no role in sex determination and reproduction Atsuo Yoshido, Frantisek Marec, Ken Sahara Supplementary
Supplementary Information The fate of W chromosomes in hybrids between wild silkmoths, Samia cynthia ssp.: no role in sex determination and reproduction Atsuo Yoshido, Frantisek Marec, Ken Sahara Supplementary
Supporting Information
 Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
Supplementary Information
 Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Summary and conclusions
 Summary and conclusions 133 Part 1: Toxicity effects of Cry toxins upon hemocoelic delivery to A. janata larvae The first investigation was aimed to find out whether other modes of delivery of Cry toxins
Summary and conclusions 133 Part 1: Toxicity effects of Cry toxins upon hemocoelic delivery to A. janata larvae The first investigation was aimed to find out whether other modes of delivery of Cry toxins
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66
 SUPPORTING INFORMATION belonging to the manuscript: Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66 by Changsheng Wu 1, 2, Gilles P. van Wezel 1, *, and Young Hae
SUPPORTING INFORMATION belonging to the manuscript: Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66 by Changsheng Wu 1, 2, Gilles P. van Wezel 1, *, and Young Hae
Delineation of the Role of Glycosylation in the Cytotoxic Properties of Quercetin using Novel Assays in Living Vertebrates
 Delineation of the Role of Glycosylation in the Cytotoxic Properties of Quercetin using Novel Assays in Living Vertebrates Si-Hwan Park,, Hyun Jung Kim,, Soon-Ho Yim, Ah-Ra Kim, Nisha Tyagi, Haihong Shen,
Delineation of the Role of Glycosylation in the Cytotoxic Properties of Quercetin using Novel Assays in Living Vertebrates Si-Hwan Park,, Hyun Jung Kim,, Soon-Ho Yim, Ah-Ra Kim, Nisha Tyagi, Haihong Shen,
Supplementary Figures
 Supplementary Figures 9 10 11 Supplementary Figure 1. Old plants are more resistant to insect herbivores than young plants. (a) Image of young (1-day-old, 1D) and old (-day-old, D) plants of Arabidopsis
Supplementary Figures 9 10 11 Supplementary Figure 1. Old plants are more resistant to insect herbivores than young plants. (a) Image of young (1-day-old, 1D) and old (-day-old, D) plants of Arabidopsis
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus
 Supplemental Information Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus Torben Hofeditz, 1 Claudia Eva-Maria Unsin 2, Jutta
Supplemental Information Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus Torben Hofeditz, 1 Claudia Eva-Maria Unsin 2, Jutta
Ambient temperature regulated flowering time
 Ambient temperature regulated flowering time Applications of RNAseq RNA- seq course: The power of RNA-seq June 7 th, 2013; Richard Immink Overview Introduction: Biological research question/hypothesis
Ambient temperature regulated flowering time Applications of RNAseq RNA- seq course: The power of RNA-seq June 7 th, 2013; Richard Immink Overview Introduction: Biological research question/hypothesis
Inquiry Master 6.1A Cabbage White Butterfly Inquiry, Care, Preparation, and Maintenance Calendar
 Inquiry Master 6.1A Cabbage White Butterfly Inquiry, Care, Preparation, and Maintenance Calendar This calendar lists the approximate number of days after laying the eggs on the soil of the radish plants
Inquiry Master 6.1A Cabbage White Butterfly Inquiry, Care, Preparation, and Maintenance Calendar This calendar lists the approximate number of days after laying the eggs on the soil of the radish plants
Behavioral generalization
 Supplementary Figure 1 Behavioral generalization. a. Behavioral generalization curves in four Individual sessions. Shown is the conditioned response (CR, mean ± SEM), as a function of absolute (main) or
Supplementary Figure 1 Behavioral generalization. a. Behavioral generalization curves in four Individual sessions. Shown is the conditioned response (CR, mean ± SEM), as a function of absolute (main) or
In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described
 Supplemental Methods In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described previously 1 2 using 32P-labeled 211 bp fragment from 3 HS1. Footprinting reaction mixes contained
Supplemental Methods In vitro DNase I foot printing. In vitro DNase I footprinting was performed as described previously 1 2 using 32P-labeled 211 bp fragment from 3 HS1. Footprinting reaction mixes contained
Phenomena first observed in petunia
 Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Supporting information
 Supporting information Rhabdopeptide/Xenortide-like Peptides from Xenorhabdus innexi with Terminal Amines Showing Potent Anti-protozoal Activity Lei Zhao,, Marcel Kaiser,, Helge B. Bode *,, Molekulare
Supporting information Rhabdopeptide/Xenortide-like Peptides from Xenorhabdus innexi with Terminal Amines Showing Potent Anti-protozoal Activity Lei Zhao,, Marcel Kaiser,, Helge B. Bode *,, Molekulare
SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila
 SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila Ayako Kohyama-Koganeya, 1,2 Takuji Nabetani, 1 Masayuki Miura, 2,3 Yoshio Hirabayashi
SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila Ayako Kohyama-Koganeya, 1,2 Takuji Nabetani, 1 Masayuki Miura, 2,3 Yoshio Hirabayashi
Genome-editing via Oviductal Nucleic Acids Delivery (GONAD) system: a novel microinjection-independent genome engineering method in mice
 Supplementary Information Genome-editing via Oviductal Nucleic Acids Delivery (GONAD) system: a novel microinjection-independent genome engineering method in mice Gou Takahashi, Channabasavaiah B Gurumurthy,
Supplementary Information Genome-editing via Oviductal Nucleic Acids Delivery (GONAD) system: a novel microinjection-independent genome engineering method in mice Gou Takahashi, Channabasavaiah B Gurumurthy,
Supplemental Materials and Methods Plasmids and viruses Quantitative Reverse Transcription PCR Generation of molecular standard for quantitative PCR
 Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced to express
 Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
 MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
Supplementary Figure 1. ESI/MS/MS analyses of native and de-acetylated S2A Supplementary Figure 2. Partial 1D 1H NMR spectrum of S2A
 Supplementary Figure 1. ESI/MS/MS analyses of native and de-acetylated S2A. Panel A, Positive ESI mass spectra of native (N) and de-acetylated (DA) non-active S2A were obtained, and demonstrated a shift
Supplementary Figure 1. ESI/MS/MS analyses of native and de-acetylated S2A. Panel A, Positive ESI mass spectra of native (N) and de-acetylated (DA) non-active S2A were obtained, and demonstrated a shift
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Histogram showing hybridization signals for chicken (left) and quail (right) genomic DNA analyzed by Chicken GeneChip (n=3). www.nature.com/nature 1 Supplementary Figure 2. Independent
Supplementary Figure 1. Histogram showing hybridization signals for chicken (left) and quail (right) genomic DNA analyzed by Chicken GeneChip (n=3). www.nature.com/nature 1 Supplementary Figure 2. Independent
Nature Biotechnology: doi: /nbt.1904
 Supplementary Information Comparison between assembly-based SV calls and array CGH results Genome-wide array assessment of copy number changes, such as array comparative genomic hybridization (acgh), is
Supplementary Information Comparison between assembly-based SV calls and array CGH results Genome-wide array assessment of copy number changes, such as array comparative genomic hybridization (acgh), is
Ohio Vegetable & Small Fruit Research & Development Program 2007 Report on Research
 Ohio Vegetable & Small Fruit Research & Development Program 2007 Report on Research Project Title: New Corn Earworm Management for Fresh Market Sweet Corn Principal Investigator(s): Jim Jasinski, Celeste
Ohio Vegetable & Small Fruit Research & Development Program 2007 Report on Research Project Title: New Corn Earworm Management for Fresh Market Sweet Corn Principal Investigator(s): Jim Jasinski, Celeste
Lab Tuesday: Virus Diseases
 Lab Tuesday: Virus Diseases Quiz for Bacterial Pathogens lab (pp 67-73) and Biocontrol of Crown Gall (p. 113-117), Observation of Viral Movement in Plants (p. 119), and Intro section for Viruses (pp. 75-77).
Lab Tuesday: Virus Diseases Quiz for Bacterial Pathogens lab (pp 67-73) and Biocontrol of Crown Gall (p. 113-117), Observation of Viral Movement in Plants (p. 119), and Intro section for Viruses (pp. 75-77).
Inhibition of Cdk5 Promotes β-cell Differentiation from Ductal Progenitors
 Inhibition of Cdk5 Promotes β-cell Differentiation from Ductal Progenitors Ka-Cheuk Liu, Gunter Leuckx, Daisuke Sakano, Philip A. Seymour, Charlotte L. Mattsson, Linn Rautio, Willem Staels, Yannick Verdonck,
Inhibition of Cdk5 Promotes β-cell Differentiation from Ductal Progenitors Ka-Cheuk Liu, Gunter Leuckx, Daisuke Sakano, Philip A. Seymour, Charlotte L. Mattsson, Linn Rautio, Willem Staels, Yannick Verdonck,
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis
 APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
Primer DNA sequence (5 to 3 )
 Supplemental Table 1 Oligonucleotide primers used in the study Primer DNA sequence (5 to 3 ) Plasmodium falciparum 18s forward Plasmodium falciparum 18s reverse Plasmodium falciparum MSP1 forward Plasmodium
Supplemental Table 1 Oligonucleotide primers used in the study Primer DNA sequence (5 to 3 ) Plasmodium falciparum 18s forward Plasmodium falciparum 18s reverse Plasmodium falciparum MSP1 forward Plasmodium
Characterizing fatty acids with advanced multinuclear NMR methods
 Characterizing fatty acids with advanced multinuclear NMR methods Fatty acids consist of long carbon chains ending with a carboxylic acid on one side and a methyl group on the other. Most naturally occurring
Characterizing fatty acids with advanced multinuclear NMR methods Fatty acids consist of long carbon chains ending with a carboxylic acid on one side and a methyl group on the other. Most naturally occurring
Feeding Measurements
 Feeding Measurements Measures of Feeding Rates (or specific mortality rate) Clearance Rate Concept Clearance Rate (F) = Volume cleared of prey pred -1 time -1 predator prey A consumer may have different
Feeding Measurements Measures of Feeding Rates (or specific mortality rate) Clearance Rate Concept Clearance Rate (F) = Volume cleared of prey pred -1 time -1 predator prey A consumer may have different
Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis
 Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
sirna count per 50 kb small RNAs matching the direct strand Repeat length (bp) per 50 kb repeats in the chromosome
 Qi et al. 26-3-2564C Qi et al., Figure S1 sirna count per 5 kb small RNAs matching the direct strand sirna count per 5 kb small RNAs matching the complementary strand Repeat length (bp) per 5 kb repeats
Qi et al. 26-3-2564C Qi et al., Figure S1 sirna count per 5 kb small RNAs matching the direct strand sirna count per 5 kb small RNAs matching the complementary strand Repeat length (bp) per 5 kb repeats
Supporting Information
 Supporting Information Harries et al. 1.173/pnas.9923916 A Fig. S1. Disruption of microfilaments within epidermal cells after treatment with 5 M Lat. Images of N. benthamiana cells are from plants expressing
Supporting Information Harries et al. 1.173/pnas.9923916 A Fig. S1. Disruption of microfilaments within epidermal cells after treatment with 5 M Lat. Images of N. benthamiana cells are from plants expressing
Potential use of High iron and low phytate GM rice and their Bio-safety Assessment
 Potential use of High iron and low phytate GM rice and their Bio-safety Assessment Dr. Karabi Datta University of Calcutta, India Background High iron rice and iron bioavailability Micronutrient deficiency
Potential use of High iron and low phytate GM rice and their Bio-safety Assessment Dr. Karabi Datta University of Calcutta, India Background High iron rice and iron bioavailability Micronutrient deficiency
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
chapter 1 - fig. 2 Mechanism of transcriptional control by ppar agonists.
 chapter 1 - fig. 1 The -omics subdisciplines. chapter 1 - fig. 2 Mechanism of transcriptional control by ppar agonists. 201 figures chapter 1 chapter 2 - fig. 1 Schematic overview of the different steps
chapter 1 - fig. 1 The -omics subdisciplines. chapter 1 - fig. 2 Mechanism of transcriptional control by ppar agonists. 201 figures chapter 1 chapter 2 - fig. 1 Schematic overview of the different steps
Supplementary Figure 1
 Supplementary Figure 1 5 microns C7 B6 unclassified H19 C7 signal H19 guide signal H19 B6 signal C7 SNP spots H19 RNA spots B6 SNP spots colocalization H19 RNA classification Supplementary Figure 1. Allele-specific
Supplementary Figure 1 5 microns C7 B6 unclassified H19 C7 signal H19 guide signal H19 B6 signal C7 SNP spots H19 RNA spots B6 SNP spots colocalization H19 RNA classification Supplementary Figure 1. Allele-specific
Plant Cell Biology; Identification and manipulation of plant quality traits
 Plant Cell Biology; Identification and manipulation of plant quality traits Phil Morris, Mark Robbins, Joe Gallagher and Ana Winters Mechanisms of protein protection in forages 30 Determining the constraints
Plant Cell Biology; Identification and manipulation of plant quality traits Phil Morris, Mark Robbins, Joe Gallagher and Ana Winters Mechanisms of protein protection in forages 30 Determining the constraints
Suppl Video: Tumor cells (green) and monocytes (white) are seeded on a confluent endothelial
 Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Department of Molecular Ecology, Max Planck Institute for Chemical Ecology, Jena, Germany b
 The Plant Cell, Vol. 22: 273 292, January 2010, www.plantcell.org ã 2010 American Society of Plant Biologists Jasmonate and pphsystemin Regulate Key Malonylation Steps in the Biosynthesis of 17-Hydroxygeranyllinalool
The Plant Cell, Vol. 22: 273 292, January 2010, www.plantcell.org ã 2010 American Society of Plant Biologists Jasmonate and pphsystemin Regulate Key Malonylation Steps in the Biosynthesis of 17-Hydroxygeranyllinalool
Not IN Our Genes - A Different Kind of Inheritance.! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014
 Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
Phosphate buffered saline (PBS) for washing the cells TE buffer (nuclease-free) ph 7.5 for use with the PrimePCR Reverse Transcription Control Assay
 Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Supplemental Data. Deinlein et al. Plant Cell. (2012) /tpc
 µm Zn 2+ 15 µm Zn 2+ Growth (% of control) empty vector NS1 NS2 NS3 NS4 S. pombe zhfδ Supplemental Figure 1. Functional characterization of. halleri NS genes in Zn 2+ hypersensitive S. pombe Δzhf mutant
µm Zn 2+ 15 µm Zn 2+ Growth (% of control) empty vector NS1 NS2 NS3 NS4 S. pombe zhfδ Supplemental Figure 1. Functional characterization of. halleri NS genes in Zn 2+ hypersensitive S. pombe Δzhf mutant
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Abstract. Optimization strategy of Copy Number Variant calling using Multiplicom solutions APPLICATION NOTE. Introduction
 Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Supplementary Information
 Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Lab Tuesday: Virus Diseases
 Lab Tuesday: Virus Diseases Quiz for Bacterial Pathogens lab (pp 69-75) and Biocontrol of Crown Gall (p. 115-119), Observation of Viral Movement in Plants (p. 121), and Intro section for Viruses (pp. 77-79).
Lab Tuesday: Virus Diseases Quiz for Bacterial Pathogens lab (pp 69-75) and Biocontrol of Crown Gall (p. 115-119), Observation of Viral Movement in Plants (p. 121), and Intro section for Viruses (pp. 77-79).
Male 30. Female. Body weight (g) Age (weeks) Age (weeks) Atg7 f/f Atg7 ΔCD11c
 ody weight (g) ody weight (g) 34 3 Male 3 27 Female 26 24 22 18 7 9 11 13 15 17 19 21 23 21 18 15 7 9 11 13 15 17 19 21 23 Age (weeks) Age (weeks) Supplementary Figure 1. Lean phenotypes in mice regardless
ody weight (g) ody weight (g) 34 3 Male 3 27 Female 26 24 22 18 7 9 11 13 15 17 19 21 23 21 18 15 7 9 11 13 15 17 19 21 23 Age (weeks) Age (weeks) Supplementary Figure 1. Lean phenotypes in mice regardless
Supplementary Information
 Supplementary Information Precursors of trnas are stabilized by methylguanosine cap structures Takayuki Ohira and Tsutomu Suzuki Department of Chemistry and Biotechnology, Graduate School of Engineering,
Supplementary Information Precursors of trnas are stabilized by methylguanosine cap structures Takayuki Ohira and Tsutomu Suzuki Department of Chemistry and Biotechnology, Graduate School of Engineering,
FONS Nové sekvenační technologie vklinickédiagnostice?
 FONS 2010 Nové sekvenační technologie vklinickédiagnostice? Sekvenování amplikonů Sequence capture Celogenomové sekvenování FONS 2010 Sekvenování amplikonů Amplicon sequencing - amplicon sequencing enables
FONS 2010 Nové sekvenační technologie vklinickédiagnostice? Sekvenování amplikonů Sequence capture Celogenomové sekvenování FONS 2010 Sekvenování amplikonů Amplicon sequencing - amplicon sequencing enables
Cloning and Expression of a Bacterial CGTase and Impacts on Phytoremediation. Sarah J. MacDonald Assistant Professor Missouri Valley College
 Cloning and Expression of a Bacterial CGTase and Impacts on Phytoremediation Sarah J. MacDonald Assistant Professor Missouri Valley College Phytoremediation of Organic Compounds Phytodegradation: Plants
Cloning and Expression of a Bacterial CGTase and Impacts on Phytoremediation Sarah J. MacDonald Assistant Professor Missouri Valley College Phytoremediation of Organic Compounds Phytodegradation: Plants
Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples
 DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
SUPPORTING INFORMATION
 SUPPORTING INFORMATION Phosphine-Mediated Disulfide Metathesis in Aqueous Media Rémi Caraballo, Morakot Sakulsombat, and Olof Ramström* KTH - Royal Institute of Technology, Department of Chemistry Teknikringen
SUPPORTING INFORMATION Phosphine-Mediated Disulfide Metathesis in Aqueous Media Rémi Caraballo, Morakot Sakulsombat, and Olof Ramström* KTH - Royal Institute of Technology, Department of Chemistry Teknikringen
Supporting information
 Supporting information Figure legends Supplementary Table 1. Specific product ions obtained from fragmentation of lithium adducts in the positive ion mode comparing the different positional isomers of
Supporting information Figure legends Supplementary Table 1. Specific product ions obtained from fragmentation of lithium adducts in the positive ion mode comparing the different positional isomers of
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Cryopreservation alters CD62L expression by CD4 T cells. Freshly isolated (left) or cryopreserved PBMCs (right) were stained with the mix of antibodies described
Supplementary Figure 1 Supplementary Figure 1: Cryopreservation alters CD62L expression by CD4 T cells. Freshly isolated (left) or cryopreserved PBMCs (right) were stained with the mix of antibodies described
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the
 Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
Supplementary Information
 Supplementary Information J. Braz. Chem. Soc., Vol. 24, No. 12, S1-S21, 2013. Printed in Brazil - 2013 Sociedade Brasileira de Química 0103-5053 $6.00+0.00 SI Rui He, a,b,c Bochu Wang,*,b Toshiyuki Wakimoto,
Supplementary Information J. Braz. Chem. Soc., Vol. 24, No. 12, S1-S21, 2013. Printed in Brazil - 2013 Sociedade Brasileira de Química 0103-5053 $6.00+0.00 SI Rui He, a,b,c Bochu Wang,*,b Toshiyuki Wakimoto,
genomics for systems biology / ISB2020 RNA sequencing (RNA-seq)
 RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
Overview of the Expressway Cell-Free Expression Systems. Expressway Mini Cell-Free Expression System
 Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Supplementary Figures
 J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
Supporting Information
 Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice
 Supplementary Methods: Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice and gently meshed in DMEM containing 10% FBS to prepare for single cell suspensions. CD4 + CD25
Supplementary Methods: Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice and gently meshed in DMEM containing 10% FBS to prepare for single cell suspensions. CD4 + CD25
Chapter 5. Viral infections (I)
 Chapter 5. Viral infections (I) 1. Properties of virus - Virus: derived from Latin and means poison or stench (foul odor) - Definition an infectious, potentially pathogenic nucleoprotein entity which reproduces
Chapter 5. Viral infections (I) 1. Properties of virus - Virus: derived from Latin and means poison or stench (foul odor) - Definition an infectious, potentially pathogenic nucleoprotein entity which reproduces
II. The effect of strain and IGF-1 gene transfer using SMGT techniques on some semen characteristics:-
 The experimental work of this study was carried out in poultry farm of the Department of Animal Production, Faculty of Agriculture, Benha University, Egypt with cooperation of Genetic Engineering and Biotechnology
The experimental work of this study was carried out in poultry farm of the Department of Animal Production, Faculty of Agriculture, Benha University, Egypt with cooperation of Genetic Engineering and Biotechnology
Soluble ADAM33 initiates airway remodeling to promote susceptibility for. Elizabeth R. Davies, Joanne F.C. Kelly, Peter H. Howarth, David I Wilson,
 Revised Suppl. Data: Soluble ADAM33 1 Soluble ADAM33 initiates airway remodeling to promote susceptibility for allergic asthma in early life Elizabeth R. Davies, Joanne F.C. Kelly, Peter H. Howarth, David
Revised Suppl. Data: Soluble ADAM33 1 Soluble ADAM33 initiates airway remodeling to promote susceptibility for allergic asthma in early life Elizabeth R. Davies, Joanne F.C. Kelly, Peter H. Howarth, David
Supplementary Online Content
 Supplementary Online Content Fumagalli D, Venet D, Ignatiadis M, et al. RNA Sequencing to predict response to neoadjuvant anti-her2 therapy: a secondary analysis of the NeoALTTO randomized clinical trial.
Supplementary Online Content Fumagalli D, Venet D, Ignatiadis M, et al. RNA Sequencing to predict response to neoadjuvant anti-her2 therapy: a secondary analysis of the NeoALTTO randomized clinical trial.
Scheme S1. Synthesis of glycose-amino ligand.
 Scheme S1. Synthesis of glycose-amino ligand. 5-Chloro-1-pentyl-2,3,4,6-tetra-O-acetyl-ß-D-glucopyranoside S2 To a solution of penta-o-acetyl-ß-d-glucopyranoside S1 (3.0 g, 7.69 mmol) and 5-chloropentan-1-ol
Scheme S1. Synthesis of glycose-amino ligand. 5-Chloro-1-pentyl-2,3,4,6-tetra-O-acetyl-ß-D-glucopyranoside S2 To a solution of penta-o-acetyl-ß-d-glucopyranoside S1 (3.0 g, 7.69 mmol) and 5-chloropentan-1-ol
Comparison of ESI-MS Spectra in MassBank Database
 BMEI 2008 Comparison of ESI-MS Spectra in MassBank Database Hisayuki Horai 1,2, Masanori Arita 1,2,3,4, Takaaki Nishioka 1,2 1 IAB, Keio Univ., 2 JST-BIRD, 3 Univ. of Tokyo, 4 RIKEN PSC 1 Table of Contents
BMEI 2008 Comparison of ESI-MS Spectra in MassBank Database Hisayuki Horai 1,2, Masanori Arita 1,2,3,4, Takaaki Nishioka 1,2 1 IAB, Keio Univ., 2 JST-BIRD, 3 Univ. of Tokyo, 4 RIKEN PSC 1 Table of Contents
ExpressArt FFPE Clear RNAready kit
 Features and Example Results General problems with FFPE samples Formalin-fixation of tissues results in severe RNA fragmentation, as well as in RNA RNA, RNA-DNA and RNA protein cross-linking, which impairs
Features and Example Results General problems with FFPE samples Formalin-fixation of tissues results in severe RNA fragmentation, as well as in RNA RNA, RNA-DNA and RNA protein cross-linking, which impairs
Cytochalasins from an Australian marine sediment-derived Phomopsis sp. (CMB-M0042F): Acid-mediated intra-molecular cycloadditions enhance
 SUPPORTING INFORMATION Cytochalasins from an Australian marine sediment-derived Phomopsis sp. (CMB-M42F): Acid-mediated intra-molecular cycloadditions enhance chemical diversity Zhuo Shang, Ritesh Raju,
SUPPORTING INFORMATION Cytochalasins from an Australian marine sediment-derived Phomopsis sp. (CMB-M42F): Acid-mediated intra-molecular cycloadditions enhance chemical diversity Zhuo Shang, Ritesh Raju,
Supporting Information
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Supporting Information Enzyme-activatable Probe with a Self-immolative Linker for Rapid and Sensitive
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Supporting Information Enzyme-activatable Probe with a Self-immolative Linker for Rapid and Sensitive
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol
 Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
ddm1a (PFG_3A-51065) ATG ddm1b (PFG_2B-60109) ATG osdrm2 (PFG_3A-04110) osdrm2 osdrm2 osdrm2
 Relative expression.6.5.4.3.2.1 OsDDM1a OsDDM1b OsDRM2 TG TG TG ddm1a (PFG_3-5165) P1 P3 ddm1b (PFG_2-619) P2 531bp 5233bp P1 P4 P3 P2 F1 R1 F1 TG TG TG F1 R1 C ddm1a -/- -/- ddm1b -/- +/- +/- -/- D (PFG_3-411)
Relative expression.6.5.4.3.2.1 OsDDM1a OsDDM1b OsDRM2 TG TG TG ddm1a (PFG_3-5165) P1 P3 ddm1b (PFG_2-619) P2 531bp 5233bp P1 P4 P3 P2 F1 R1 F1 TG TG TG F1 R1 C ddm1a -/- -/- ddm1b -/- +/- +/- -/- D (PFG_3-411)
Probe. Hind III Q,!?R'!! /0!!!!D1"?R'! vector. Homologous recombination
 Supple-Zhang Page 1 Wild-type locus Targeting construct Targeted allele Exon Exon3 Exon Probe P1 P P3 FRT FRT loxp loxp neo vector amh I Homologous recombination neo P1 P P3 FLPe recombination Q,!?R'!!
Supple-Zhang Page 1 Wild-type locus Targeting construct Targeted allele Exon Exon3 Exon Probe P1 P P3 FRT FRT loxp loxp neo vector amh I Homologous recombination neo P1 P P3 FLPe recombination Q,!?R'!!
Protein SD Units (P-value) Cluster order
 SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
Fluorescent probes for detecting monoamine oxidase activity and cell imaging
 Fluorescent probes for detecting monoamine oxidase activity and cell imaging Xuefeng Li, Huatang Zhang, Yusheng Xie, Yi Hu, Hongyan Sun *, Qing Zhu * Supporting Information Table of Contents 1. General
Fluorescent probes for detecting monoamine oxidase activity and cell imaging Xuefeng Li, Huatang Zhang, Yusheng Xie, Yi Hu, Hongyan Sun *, Qing Zhu * Supporting Information Table of Contents 1. General
