CHAIRE ÉPIGÉNÉTIQUE ET MÉMOIRE CELLULAIRE. Année : Reprogrammations développementales, induites et pathologiques
|
|
|
- Alexandra Fitzgerald
- 6 years ago
- Views:
Transcription
1 CHAIRE ÉPIGÉNÉTIQUE ET MÉMOIRE CELLULAIRE Année : Reprogrammations développementales, induites et pathologiques Cours II Etapes de la reprogrammation au cours du développement chez les mammifères 17 mars 2014 Seminaire: Prof. Wolf Reik à 17h30 The Role of DNA Modifications in Epigenetic Reprogramming and Signaling
2 Developmental Programming! TF'x" RNA'' Pol'II' TF'y" RNA'' Pol'II' AAAA' Totipotency Cell division Cell differentiation Morphogenesis AAAA' Pluripotency TF'z" RNA'' Pol'II' AAAA' 1. All cells contain the same genes cell identities depend on which genes are expressed and repressed. 2. These Developmental are established restrictions by transcription imposed factors on the via genome signalling, during cell-cell differentiation communication, are due to positional reversible information epigenetic modifications rather than to permanent genetic changes (Gurdon, 1962)
3 Developmental Programming! TF'x" RNA'' Pol'II' TF'y" RNA'' Pol'II' AAAA' Totipotency Cell division Cell differentiation Morphogenesis AAAA' Pluripotency TF'z" RNA'' Pol'II' AAAA' 1. All cells contain the same genes cell identities depend on which genes are expressed and repressed. 2. These Developmental are established restrictions by transcription imposed factors on the via genome signalling, during cell-cell differentiation communication, are due to positional reversible information epigenetic modifications rather than to permanent genetic changes 3. Changes in gene expression patterns become (Gurdon, heritable 1962) (through mitosis) during development => «Epigenetics»
4 Developmental Programming! Epigenetics: heritable changes in gene function that cannot be explained by changes in DNA sequence. Russo, V.E.A., R.A. Martienssen & A.D. Riggs Eds. (1996) "Epigenetic mechanisms of gene regulation. CSHL Press.
5 Developmental Programming! Epigenetics: heritable changes in gene function that cannot be explained by changes in DNA sequence. Russo, V.E.A., R.A. Martienssen & A.D. Riggs Eds. (1996) "Epigenetic mechanisms of gene regulation. CSHL Press. Eg DNA Methylation an epigenetic modification that can affect gene TF'x" AAAA' expression; RNA'' be propagated over cell Pol'II' division; and lock in the silent state - Important for normal development TF'y" RNA'' Pol'II' AAAA' Cytosine" SAMVCH3' SAM' 5-Methylcytosine" TF'z" RNA'' Pol'II' AAAA' Chromatin histone variants & modifications, and protein complexes such as Polycomb &Trithorax' Heterochroma7n" Inac7ve"gene' Euchroma7n" Ac7ve"gene' Okano M, Bell DW, Haber DA, Li E: DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99: O Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T: The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001, 21: Bledau et al. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development 2014, 141:
6 Chromatin-based Epigenetic Mechanisms Histone Variants and Histone Modifications are: Mediators of chromatin accessibility Platforms for binding proteins Histone modifying enzymes can add or remove these modifications TETs Dnmts De novo: Dnmt3a,3b, 3L Maintenance: Dnmt1 In DNA some cases, methylation histone associated readers and with writers repressed are in complexes, possibly enabling self-perpetuation state of some genes, repeats: and/or spreading in cis Self-templating, stable - but can be removed eg HP1/H3K9me3; PRC2/H3K27me3 etc (actively eg Tet-induced conversion to 5hme; passively during DNA replication)'
7 TF'x" AAAA' expression; RNA'' be propagated over cell Pol'II' Developmental Programming! Epigenetics: heritable changes in gene function that cannot be explained by changes in DNA sequence. Russo, V.E.A., R.A. Martienssen & A.D. Riggs Eds. (1996) "Epigenetic mechanisms of gene regulation. CSHL Press. Eg DNA Methylation an epigenetic modification that can affect gene division; and lock in the silent state TF'y" RNA'' Pol'II' AAAA' Cytosine" SAMVCH3' SAM' 5-Methylcytosine" TF'z" RNA'' Pol'II' AAAA' Chromatin histone variants & modifications, Polycomb &Trithorax complexes' Heterochroma7n" Inac7ve"gene' Euchroma7n" Ac7ve"gene' Epigenetic states are stable but can be reversed: during development, in the germ line, in somatic cells (eg stem cell differentiation), in disease (eg epimutations in cancer), or during nuclear transfer and cloning (reprogramming ): in each case Epigenetic barriers must be overcome Okano M, Bell DW, Haber DA, Li E: DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99: O Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T: The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001, 21: Bledau et al. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development 2014, 141:
8 ! Many types of epigenetic barrier & many ways of overcoming them during development or artificially! Developmental restrictions imposed on the genome during differentiation are due to reversible epigenetic modifications rather than to permanent genetic changes Epigenetic changes allow the maintenance of cell identity but can be overriden by TFs, as well as by active and passive loss
9 Developmental Reprogramming! Somatic Cell Nuclear Transfer (SNCT) Totipotency Cell division Cell differentiation Morphogenesis Pluripotency The success rate of reproductive cloning is very low compared to natural reproduction - due to inappropriate expression of somatic genes, inadequate reactivation of developmental genes Developmental and other epigenetic restrictions errors? imposed (including on the imprinted genome during genes, X differentiation inactivation ) are due to reversible epigenetic modifications rather than to permanent genetic changes => The parental genomes inherited from the gametes are more competent Epigenetic to changes be reprogrammed allow the maintenance to totipotency of cell identity but and can to be support overriden by the TFs, as well as by active and passive loss subsequent changes in cell identity during early embryogenesis?
10 Developmental Reprogramming! Prepare for development (epigenesis) Preserve some epigenetic marks (parental imprints), erase others Zygotic Reprogramming undo gamete programs set up totipotency Dynamic changes in gene expression and epigenetic marks in at least 3 phases of development Germ Line Reprogramming undo somatic program set up germ line program X-chromosome reactivation ICM Reprogramming undo/prevent TE program set up pluripotency X-chromosome reactivation Prepare for the next generation Erase epigenetic history (both programmed and accidental) Prepare for the epiblast (soma and germ line) Adapted from Cantone and Fisher, 2013
11 In the beginning: Two highly specialized cells, the egg and the sperm, fuse to form a totipotent cell, the zygote Protamines+ Primary! Oocyte! Huge maternal store of proteins, mrna (to be translated later) to ensure early development, and to enable reprogramming upon fertilization GV! Oocyte! Meiosis I! Secondary! Oocyte! Spermatogonia! Primary! Spermatocyte! Meiosis I! Secondary! Spermatocyte! Spermatid! 0.15'to'0.2'mm' ' ' ' ' ' ' Fertilization! Meiosis II! ''''''''''''''''3V5'um' Highly packaged genome (protamines, few/ '''''''(0.003' '0.005'mm)' no histones) and a small amount of RNA
12 In the beginning: Two highly specialized cells, the egg and the sperm, fuse to form a totipotent cell, the zygote Accumulation of maternal RNA and Proteins Degradation of maternal RNA and Proteins Primary! Oocyte! GV! Oocyte! Meiosis I! Secondary! Oocyte! Activation of the zygotic (embryonic) genome (ZGA) Spermatogonia! Primary! Spermatocyte! Secondary! Spermatocyte! Spermatid! Fertilization! Meiosis I! Meiosis II! Histone -> Protamine replacement Protamine -> Histone replacement
13 Numerous Maternal Factors Required to Orchestrate Reprogramming and appropriate Activation of the Embryonic Genome A multitude of maternal factors are involved in: erasing gametic epigenomic landscapes, preparing the embryonic genome for appropriate transcription of developmental genes, protecting some regions from reprogramming and ensuring others are silenced
14 Fertilization triggers massive reorganization of the paternal and maternal epigenomes (prior to transcription) Maternal interphase pronucleus Paternal interphase pronucleus The two parental pronuclei remain separate initially Protamine eviction, maternal histone incorporation Adapted Albert and Peters, 2009
15 Fertilization triggers massive reorganization of the paternal and maternal epigenomes (prior to transcription) protamine - histone exchange from Santos & Dean, Reproduction, 2004 Asymmetric Chromatin states
16 Fertilization triggers massive reorganization of the paternal and maternal epigenomes (prior to transcription) H3.3 lys27 methylation has a role in remodeling heterochromatin after fertilization Incorporation of H3.3 into paternal pericentric heterochromatin is important for the initial establishment of pericentromeric heterochromatin through lysine 27. (Akiyama, Suzuki, Matsuda, & Aoki, 2011; Santenard et al., 2010). Mutation of Histone H3.3 lysine K27 to alanine results in a missegregation of chromosomes, developmental arrest and mislocalization of HP1. Same mutation in H3.1 no effect on HP1 localization or development. (Santenard et al., 2010). Adapted Albert and Peters, 2009
17 Fertilization triggers massive reorganization of the paternal and maternal epigenomes (prior to transcription) Maternal histones H3.1, H3.3 TH2A, TH2B Maternal Pool: Proteins + mrna Dnmt1, Polycomb (Eed, Ezh2, YY1) Trithorax (Mll) HP1, Histones Paternal chromatin inheritance: Cairns and Peters groups: 1-15% histones in sperm. How much of this resists reprogramming and what role they might play is still unclear Maternal H3.3 Maternal chromatin inheritance: Some chromatin marks are laid down during oogenesis and are essential for early heterochromatin establishment H4Ac H3K4me3 H3K9Ac H3K9me1 H3K9me2 H3K9me3 H3K4me1 H3K27me1 H4K20me1 protamine - histone exchange H4Ac (++) H3K9Ac H3K9me1 H3K4me1 H3K27me1 H4K20me1 Is zygotic chromatin dynamics actually important for subsequent development? How much is just a consequence of major chromatin remodelling? HIRA van der Heidjden et al, 2005 Maternal Mll2 (TrX) is required for the acquisition and maintenance of H3K4 methylation in the zygote and for normal embyonic gene activation (Andreu-Vieyra et al, 2010) H3K4me3 In the absence of maternal histone variants TH2A/TH2B, paternal genome activation, which accompanies from Santos & Dean, Reproduction, 2004 H3K4me3 and DNA demethylation, is defective. (Shinagawa et al, 2013) NB TH2A/TH2B also enhances OSKM reprogramming during ips!
18 Fertilization triggers massive reorganization of the paternal and maternal epigenomes (prior to transcription) DNA Demethylation (passive) Maternal histones H3.1, H3.3 TH2A, TH2B H4Ac H3K4me3 H3K9Ac H3K9me1 H3K9me2 H3K9me3 H3K4me1 H3K27me1 H4K20me1 Maternal Pool: Proteins + mrna Dnmt1, Polycomb (Eed, Ezh2, YY1) Trithorax (Mll) HP1, Histones Maternal H3.3 protamine - histone exchange DNA demethylation active (ie before DNA replication) & passive H4Ac (++) H3K9Ac H3K9me1 H3K4me1 H3K27me1 H4K20me1 HIRA van der Heidjden et al, 2005 from Santos & Dean, Reproduction, 2004 H3K4me3
19 DNA Methylation in the Mammalian Genome Lineage-specific DNA methylation Participates in maintaining cellular identity' Parent-of-origin specific DNA methylation Transcription-associated DNA methylation of gene bodies Repression of repetitive elements After fertilisation a globally demethylated state is established. Then, a progressively lineage-specific DNA methylome is acquired during pre-implantation development that maintains cellular identity and genomic stability
20 Developmental Dynamics of DNA Methylation Maternal interphase pronucleus (5mC-red) Paternal interphase pronucleus (5hmC-green) W."Reik" A few regions are protected from demethylation: DNA sequences that carry parental imprints Parental imprinting: Paternal or maternal specific gene expression Essential for normal development (Cattanach, Surani, Solter) (see last week) Active demethylation Tet3, BER, Elongator? zygote Passive demethylation (Protection by Stella) morula What about repeats? Transposons, retrotransposons and endogenous retroviruses.
21 Control of Repeat Elements after Fertilization? Retrotransposons, eg endogenous retroviruses (ERVs), present in mammalian genomes must be controlled their expression and mobility/reintegration can be deleterious In the soma and germ line: repression of repeats is via DNA methylation (& pirnas in the latter) In early embryo: global DNA hypomethylation and no pirna machinary mean that repeats can (and some do) become expressed. (Bachvarova, 1988; Efroni et al., 2008; Evsikov et al., 2004; Packer, Manova, & Bachvarova, 1993; Peaston et al., 2004). Might they play a role(s) in early development? (Peaston et al. 2004, Beraldi et al, 2006 and others) DNA meth-independent mechanisms, involving histone modifications, may control repeat silencing and heterochromatin formation during preimplantation development. Fadloun et al «Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA». Peters et al Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell, 107, Fadloun'et'al'
22 Reprogramming in the Zygote Maternal interphase pronucleus (5mC-red) Paternal interphase pronucleus (5hmC-green) W."Reik" Does transient epigenetic asymmetry have a role? Is it simply the result of the different histories of the paternal and maternal (epi)genomes? Does repeat expression have a role, or is it simply a result of incomplete silencing following reprogramming? Adapted Albert and Peters, 2009 Adapted from Cantone and Fisher, 2013 The two parental genomes have different reprogramming requirements This is essential for subsequent development At the same time imprints must be preserved. (Cours IV Mechanisms)
23 Pre-implantation Mouse Development The next steps: The road to extra-embryonic tissue formation and to the embryo proper (pluripotency)
24 Pre-implantation Mouse Development Progressive restriction of cellular plasticity from 4-cell stage (Totipotency lost at this stage in mouse) Early preimplantation mouse development is highly regulative - cleavage stage blastomeres are developmentally plastic and influenced by cell-cell interactions First cell lineage to be specified is the trophectoderm (TE) Beginning at 8-16 cell stage (morula compaction), inside-outside and/or cell polarity changes result in transcription factor (TF) modulation that ultimately translate into cell fate Cdx2 up-regulation in outer cells (via Tead4/Yap1 and Hippo signalling) lead to TE specification Pluripotency markers Oct4, Nanog and Sox2 become progressively up-regulated in inner cells, ICM Oct4 is essential for ICM and its levels distinguish between TE, ICM, PE (no Oct4->TE; hi Oct4-> PE) Second cell lineages to be segregated in the ICM are primitive endoderm (PE) & epiblast (EPI) Through position-dependent and/or Fgf/MAPK signalling determination Adapted from Cockburn and Rossant, JCI, 2010
25 Transcriptional Networks leading to Early Lineage Specification GATA6/Nanog/Oct4 levels and ERK signalling are key for the second Prim Endoderm vs Epiblast decision Positional information triggers signalling and TF modulation - for both lineage specifications, and cell fate progressively becomes locked in epigenetically. Adapted from Albert and Peters, 2009 Pluripotent ground state is transiently present in the pre-epiblast Oct4+/Nanog+/GATA6- cells = ES cells Capable of self-renewal Pluripotent can form teratomas can differentiate into all 3 germ layers Trophectodermal specification is mediated by Tead4, promoting Cdx2 and Eomes expression. TE identity is maintained by Elf5. Elf5 becomes methylated in Epiblast a E4.5 thereby fixing TE/EPI barrier Oct3/4 specifies epiblast fate. Reciprocal inhibition between Cdx2 and Oct3/4 contributes to specification.
26 Pluripotency and the Ground State in ES cells Pluripotency transcriptional network driven by the core transcription factors Oct4, Nanog, Sox2, Klf4 is essential to maintain the undifferentiated state. This network activates genes that are required for ES cell survival and proliferation while repressing target genes that are activated only during differentiation. Transcriptional feed-forward loops, specific mirnas and chromatin states, promote the pluripotency network Oct4'gene'silencing,'degrada6on'of'Nanog,''' mirnamediated'reduc6on'in'oct4,'nanog,' and'sox2'mrna'levels.' NB Ground state is transient in vivo The pluripotency network is rapidly dismantled and only re-established in the germ line
27 Pluripotency and the Ground State in ES cells Pluripotency transcriptional network driven by the core transcription factors Oct4, Nanog and Sox2, is essential to maintain the undifferentiated state. This network activates genes that are required for ES cell survival and proliferation while repressing target genes that are activated only during differentiation. In Primed ESC (not Ground state!): Poised state with «bivalent» domains H3K27me3+H3K4 Transcription factors, Oct4 ESC gene silencing: H3K9me3 DNA methylation, PcG
28 Epigenetic Dynamics during Pre-Implantation Development! Zygotic Reprogramming undo gamete programmes set up totipotency Lessons from X- chromosome inactivation? Germ Line Reprogramming undo somatic program set up germ line program ICM Reprogramming undo/prevent TE program set up pluripotency EpiSC' Cantone & Fisher Nat Struc & Mol Biol revue 2013
29 X-Chromosome Inactivation One of the two X chromosomes must be silenced during early embryogenesis in order for female development to proceed Xa' Xa' X' Xi' Xist"" RNA' Ac6ve'X'chromosome' RNA'Pol'II' Ac' Ac' Ac' Inac6ve'X'chromosome' H3K27' Pc?+ me3' H2A'' Ub' H3K9' me2' Xist"RNA' H3K4' me3' Ac' H3K4' me2' 5Vmethyl'cytosine'' '
30 Developmental Reprogramming in the Inner Cell Mass! Zygotic Reprogramming undo gamete programmes set up totipotency Lessons from X- chromosome inactivation? X inactivation is initially imprinted in the mouse (paternal X only)? Germ Line Reprogramming undo somatic program set up germ line program It then becomes random in the embryo proper (Xm or Xp) EpiSC' Cantone & Fisher Nat Struc & Mol Biol revue 2013
31 X-Chromosome Inactivation during Pre-Implantation Development When is the paternal X silenced? Are cells set aside with an active Xp that will give rise to the embryo proper? Xp (paternal) Morula Xm (maternal) Kay et al, 1994 Huyhn and Lee, 2003 Okamoto et al, 2004 Mak et al, 2004 Patrat et al, cell 2-cell 4-cell 8-cell Morula Blastocyst Zygotic Gene Activation Paternal Xist on Xp active Xist RNA coating of Xp Gene Silencing begins Imprinted inactivation of X P Loss of ACTIVE chromatin marks (Histone H3 and H4 acetylation) Gain of REPRESSIVE marks PcG (PRC2), H3K27me3 H3K9 dimethylation Trophectoderm Xp inactive Takagi and Sasaki, 1975
32 X-Chromosome Inactivation during Pre-Implantation Development When is the paternal X silenced? Are cells set aside with an active Xp that will give rise to the embryo proper? Xp (paternal) Morula Inner cell mass Xp inactive?? Xm (maternal) Kay et al, 1994 Huyhn and Lee, 2003 Okamoto et al, 2004 Mak et al, 2004 Patrat et al, cell 2-cell 4-cell 8-cell Morula Blastocyst Zygotic Gene Activation Paternal Xist on Xp active Xist PRC2' RNA coating of Xp Gene Silencing begins Imprinted inactivation of X P Loss of ACTIVE chromatin marks (Histone H3 and H4 acetylation) All cells have an Xi Gain of REPRESSIVE marks PcG (PRC2), H3K27me3 H3K9 dimethylation Trophectoderm Xp inactive Takagi and Sasaki, 1975
33 The paternal X is reactivated in the ICM (E3.5-E4.5) E3.5 ICM ICM E4.0 ICM ICM TE TE Xist RNA / Nanog / X-linked gene Xist RNA / Nanog / X-linked gene Nanog, Oct4, Sox2 Xist repression Xi reactivation Okamoto et al, 2004 Mak et al, 2004 Navarro et al, 2007 Silva et al, 2009 The inactive Xp (PRC1/2+, mh2a+, but not DNA me) is reprogrammed in the ICM in epiblast cells within 1-2 cell cycles The two Xs are active for 1-2 cell divisions First in vivo evidence of such epigenetic dynamics in ICM (Okamoto et al, 2004; Mak et al, 2004). Random X inactivation then initiates at around E Symptomatic in of embryonic more global lineages reprogramming? Okamoto et al, unpublished Resetting pluripotency following lineage restriction to TE?
34 The Pluripotent State imposes X-chromosome Reactivation Nanog (+Sox2, Oct4?) (Silva et al, Cell 2009) Xist repression + Gene reactivation + Chromatin changes TE PE 3.5 dpc 4.0 dpc 4.5 dpc Epiblast ES cells Two active Xs PE and TE Xp remains inactive
35 The Pluripotency network controls Xist Xist regulation is controlled (partly) by the pluripotency/stem cell factors network: Xist is repressed directly/indirectly by pluripotency factors in mouse ESCs (Navarro et al, 2008, 2010, 2011; Gontan et al, 2012; Minkovsky et al, 2013) Tsix:"Xist'repressor' (Avner,'Brockdorff,'Lee'and'Rougeulle'labs)' Rnf12:"Xist'ac6vator' (Gribnau'lab)'
36 X-inactivation controls the Pluripotency Network Two active Xs block exit from stem cell state and delay differentiation Double dose of unknown X-linked genes inhibits Fgf/MAPK signaling: - reduces DNA methylation levels - prevents down-regulation of stem cell factors X inactivation overrides this block and allows differentiation to proceed in XX cells XCI is controlled by the pluripotency network via Xist repression And it also enables exit from pluripotency in XX cells A developmental checkpoint - to ensure that one X is inactivated before development proceeds? Schulz et al, Cell Stem Cell, 2014
37 What about Human Pre-implantation Development? ZGA ZGA Adapted from Cockburn and Rossant, JCI, 2010 Although stage-specific gene activation seems to be preserved in human and mouse preimplantation development (eg Xue et al, 2013), there seem to be major differences in the timing of events (eg ZGA, implantation), and in the signalling pathways used to modulate TFs and control lineage specification (eg Kuijk et al, 2012) (see Niakan et al, 2012 for review)
38 Constitutive XIST RNA up-regulation but no X inactivation during human pre-implantation development Female No Barr body Male Y No X inactivation for the first 7 days of development! Y ICM shows high XIST expression despite Nanog/Oct4 expression! XIST VERY RNA accumulates different in timing male and and female regulation embryos, to in the TE mouse and ICM cells However genes are expressed from the XIST-associated chromosomes, And no signs of H3K27me3 or a Barr body at day 7 But this is consistent with differences between mescs and hescs: hescs grown in Fgf2/Activin, show fluctuating Xi states Okamoto, Patrat et al. (2011) Evolutionary Diversity of X-chromosome Inactivation in Mammals Nature 472, (=> Claire Rougeulle seminar, March 23rd)
39 Human pre-implantation embryos: Initiation of X inactivation occurs after day 7 First signs of H3K27me3 accumulation on the X chromosome in human day 8 embryos co-cultured on endometrial cells Teklenburg et al, PLoS ONE 2012 XIST becomes monoallelic and X inactivation initiates in human embryos from ~ day 8 onwards around the time of implantation
40 Developmental Reprogramming! Zygotic Reprogramming undo gamete programs set up totipotency Germ Line Reprogramming undo somatic program set up germ line program X-chromosome reactivation ICM Reprogramming undo/prevent TE program set up pluripotency X-chromosome reactivation EpiSC' When is pluripotency lost post-implantation? Adapted from Cantone and Fisher, 2013
41 Pluripotency is rapidly lost in the post-implantation Embryo Levels of pluripotency gene regulatory network activity decline during post-implantation development, reaching a threshold at the onset of somitogenesis (E8.0) where levels become too low to sustain pluripotency. Decreased accessibility of regulatory elements invasion may extinguish pluripotency. Ectopic expression of Oct4 in co-operation with activin/fgf, can revive the pluripotent state initially, but DNA methylation rapidly stabilizes the non-pluripotent state. Osorno et al (2012) Development 139, Hemberger, Dean and Reik, 2009
42 Pluripotency is rapidly lost in the post-implantation Embryo Potential Scenario Active promoters and enhancers have nucleosome-depleted regions (NDRs) that are often occupied by transcription factors and chromatin remodellers. Loss of factor binding during differentiation leads to increased nucleosome occupancy of the regulatory region, providing a substrate for de novo DNA methylation. DNA methylation subsequently provides added stability to the silent state and is likely to be a mechanism for more accurate epigenetic inheritance during cell division. Jones, P. (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond E. Nature Heard, Rev. February Genetics 11 th, ,
43 Developmental Reprogramming! Prepare for development (epigenesis) Preserve some epigenetic marks (parental imprints), erase others Zygotic Reprogramming undo gamete programs set up totipotency Germ Line Reprogramming undo somatic program set up germ line program X-chromosome reactivation ICM Reprogramming undo/prevent TE program set up pluripotency X-chromosome reactivation Prepare for the next generation: Erase epigenetic history (both programmed and accidental) EpiSC' Establish parent-specific information (imprints) Adapted from Cantone and Fisher, 2013 Prepare for the epiblast (soma and germ line)
44 PGC specification: Initiating the genetic program for epigenetic reprogramming Epigenetic Barrier Blimp1/Prdm1 Primed Epiblast E4.5 E5.75 Epigenetic Reprogramming Prdm14 Tcfap2c/Ap2g E7.5 Inner Cell Mass Pluripotent Competence X inactivation DNA methylation Repression of some pluripotency genes Onset of somatic cell differentiation Specification BMP signaling induces Blimp1 (Blimp1 KO -> somatic program) Primordial Germ Cells Repression of somatic program Initiation of epigenetic changes Re-expression of pluripotency genes X-reactivation DNA demethylation [Imprints erased] Epigenetic ground state Somatic cells Courtesy of A. Surani Histone demethylases (Erasure of H3K9me2) Repression of maintenance & de novo DNA methylation Tet1/Tet2 (5mC->5hmC)
45 Reprogramming into the Germ Line! Reprogramming to ensure germ line specific genes are primed and an epigenetic landscape compatible with totipotency to next generation is established. Remodeling is a multistep, coordinated process that requires timely expression of key TFs & appropriate epigenetic modifiers. Kurimoto,'K.,'Yabuta,'Y.,'Ohinata,'Y.,'Shigeta,'M.,'Yamanaka,'K.,'&'Saitou,'M.'(2008).' Complex'genomeVwide'transcrip6on'dynamics'orchestrated'by'Blimp1'for'the'specifica6on'of'the'germ'cell'lineage'in'mice.'Genes'and'Development,'22,' '
46 Reprogramming of PGCs upon entry into the genital ridge E7.5 E8.5 E9.5 E11.5 E12.5 5mC = 12% PGC Specification Epigenetic Migration Reprogramming (Step 1) Reprogramming Loss of H3K9me2 in preparation for How similar or different are the reprogramming DNA Demethylation mechanisms in the later DNA demethylation? X-Reactivation germ line, to those that take place in the inner cell mass/escs Increase in H3K27me3 to compensate for K9 loss eg pluripotency genes? or in the zygote, or during induced pluripotency? Imprints Erasure (Step2) (Cours IV) Courtesy of A. Surani Popp et al, 2010
47 Epigenetic reprogramming in mammals DNA'methyla6on' Imprint'maintenance' Global+demethyla4on+ Ac4ve+and+passive+(paternal)++ Passive+(maternal)+ + Global+ +remethyla4on+ Maintenance'at'some'IAP'elements' 'and'rare'singlevcopy'loci' Global+demethyla4on+ and+imprint+erasure+ in+migra4ng+pgcs+ Imprint'Erasure' Imprint'' Establishment' (male'germ'line)' Global+ +remethyla4on+ Imprint'' Establishment' (female'germ'line)' Zygote' E0.5' E3.5' Blastocyst' Reprogramming+ for+the+epiblast+ E6.5' E8.5'.' Migra6ng'PGCs' E11.5'.'.' E16.5'.'.'.'.'.' Birth' Protamines'(histones) RNAs,'proteins?' Histones+ Maternal+mRNAs,++ Proteins+
48 The Role of DNA Modifications in Epigenetic Reprogramming and Signaling Professor Wolf Reik (Babraham Institute, Cambridge, UK) E. Heard, March 17 th, 2014
Epigenetics DNA methylation. Biosciences 741: Genomics Fall, 2013 Week 13. DNA Methylation
 Epigenetics DNA methylation Biosciences 741: Genomics Fall, 2013 Week 13 DNA Methylation Most methylated cytosines are found in the dinucleotide sequence CG, denoted mcpg. The restriction enzyme HpaII
Epigenetics DNA methylation Biosciences 741: Genomics Fall, 2013 Week 13 DNA Methylation Most methylated cytosines are found in the dinucleotide sequence CG, denoted mcpg. The restriction enzyme HpaII
Lecture 27. Epigenetic regulation of gene expression during development
 Lecture 27 Epigenetic regulation of gene expression during development Development of a multicellular organism is not only determined by the DNA sequence but also epigenetically through DNA methylation
Lecture 27 Epigenetic regulation of gene expression during development Development of a multicellular organism is not only determined by the DNA sequence but also epigenetically through DNA methylation
Histones modifications and variants
 Histones modifications and variants Dr. Institute of Molecular Biology, Johannes Gutenberg University, Mainz www.imb.de Lecture Objectives 1. Chromatin structure and function Chromatin and cell state Nucleosome
Histones modifications and variants Dr. Institute of Molecular Biology, Johannes Gutenberg University, Mainz www.imb.de Lecture Objectives 1. Chromatin structure and function Chromatin and cell state Nucleosome
Research programs involving human early embryos
 Research programs involving human early embryos 1. Understanding normal mammalian embryo development 2. Understanding errors in genetic and epigenetic programs 3. Providing research and therapeutic tools
Research programs involving human early embryos 1. Understanding normal mammalian embryo development 2. Understanding errors in genetic and epigenetic programs 3. Providing research and therapeutic tools
Genetics and Genomics in Medicine Chapter 6 Questions
 Genetics and Genomics in Medicine Chapter 6 Questions Multiple Choice Questions Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the directions
Genetics and Genomics in Medicine Chapter 6 Questions Multiple Choice Questions Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the directions
Stem Cell Epigenetics
 Stem Cell Epigenetics Philippe Collas University of Oslo Institute of Basic Medical Sciences Norwegian Center for Stem Cell Research www.collaslab.com Source of stem cells in the body Somatic ( adult )
Stem Cell Epigenetics Philippe Collas University of Oslo Institute of Basic Medical Sciences Norwegian Center for Stem Cell Research www.collaslab.com Source of stem cells in the body Somatic ( adult )
Strategic delivery: Setting standards Increasing and. Details: Output: Demonstrating efficiency. informing choice.
 Strategic delivery: Setting standards Increasing and informing choice Demonstrating efficiency economy and value Details: Meeting Scientific and Clinical Advances Advisory Committee Agenda item 6 Paper
Strategic delivery: Setting standards Increasing and informing choice Demonstrating efficiency economy and value Details: Meeting Scientific and Clinical Advances Advisory Committee Agenda item 6 Paper
Transcriptional repression of Xi
 Transcriptional repression of Xi Xist Transcription of Xist Xist RNA Spreading of Xist Recruitment of repression factors. Stable repression Translocated Xic cannot efficiently silence autosome regions.
Transcriptional repression of Xi Xist Transcription of Xist Xist RNA Spreading of Xist Recruitment of repression factors. Stable repression Translocated Xic cannot efficiently silence autosome regions.
DNA methylation & demethylation
 DNA methylation & demethylation Lars Schomacher (Group Christof Niehrs) What is Epigenetics? Epigenetics is the study of heritable changes in gene expression (active versus inactive genes) that do not
DNA methylation & demethylation Lars Schomacher (Group Christof Niehrs) What is Epigenetics? Epigenetics is the study of heritable changes in gene expression (active versus inactive genes) that do not
Allelic reprogramming of the histone modification H3K4me3 in early mammalian development
 Allelic reprogramming of the histone modification H3K4me3 in early mammalian development 张戈 Method and material STAR ChIP seq (small-scale TELP-assisted rapid ChIP seq) 200 mouse embryonic stem cells PWK/PhJ
Allelic reprogramming of the histone modification H3K4me3 in early mammalian development 张戈 Method and material STAR ChIP seq (small-scale TELP-assisted rapid ChIP seq) 200 mouse embryonic stem cells PWK/PhJ
Epigenetics. Lyle Armstrong. UJ Taylor & Francis Group. f'ci Garland Science NEW YORK AND LONDON
 ... Epigenetics Lyle Armstrong f'ci Garland Science UJ Taylor & Francis Group NEW YORK AND LONDON Contents CHAPTER 1 INTRODUCTION TO 3.2 CHROMATIN ARCHITECTURE 21 THE STUDY OF EPIGENETICS 1.1 THE CORE
... Epigenetics Lyle Armstrong f'ci Garland Science UJ Taylor & Francis Group NEW YORK AND LONDON Contents CHAPTER 1 INTRODUCTION TO 3.2 CHROMATIN ARCHITECTURE 21 THE STUDY OF EPIGENETICS 1.1 THE CORE
Nuclear reprogramming of sperm and somatic nuclei in eggs and oocytes
 Reprod Med Biol (2013) 12:133 149 DOI 10.1007/s12522-013-0155-z REVIEW ARTICLE Nuclear reprogramming of sperm and somatic nuclei in eggs and oocytes Marta Teperek Kei Miyamoto Received: 27 March 2013 /
Reprod Med Biol (2013) 12:133 149 DOI 10.1007/s12522-013-0155-z REVIEW ARTICLE Nuclear reprogramming of sperm and somatic nuclei in eggs and oocytes Marta Teperek Kei Miyamoto Received: 27 March 2013 /
Today. Genomic Imprinting & X-Inactivation
 Today 1. Quiz (~12 min) 2. Genomic imprinting in mammals 3. X-chromosome inactivation in mammals Note that readings on Dosage Compensation and Genomic Imprinting in Mammals are on our web site. Genomic
Today 1. Quiz (~12 min) 2. Genomic imprinting in mammals 3. X-chromosome inactivation in mammals Note that readings on Dosage Compensation and Genomic Imprinting in Mammals are on our web site. Genomic
Gene Expression DNA RNA. Protein. Metabolites, stress, environment
 Gene Expression DNA RNA Protein Metabolites, stress, environment 1 EPIGENETICS The study of alterations in gene function that cannot be explained by changes in DNA sequence. Epigenetic gene regulatory
Gene Expression DNA RNA Protein Metabolites, stress, environment 1 EPIGENETICS The study of alterations in gene function that cannot be explained by changes in DNA sequence. Epigenetic gene regulatory
Dissecting gene regulation network in human early embryos. at single-cell and single-base resolution
 Dissecting gene regulation network in human early embryos at single-cell and single-base resolution Fuchou Tang BIOPIC, College of Life Sciences Peking University 07/10/2015 Cockburn and Rossant, 2010
Dissecting gene regulation network in human early embryos at single-cell and single-base resolution Fuchou Tang BIOPIC, College of Life Sciences Peking University 07/10/2015 Cockburn and Rossant, 2010
An epigenetic approach to understanding (and predicting?) environmental effects on gene expression
 www.collaslab.com An epigenetic approach to understanding (and predicting?) environmental effects on gene expression Philippe Collas University of Oslo Institute of Basic Medical Sciences Stem Cell Epigenetics
www.collaslab.com An epigenetic approach to understanding (and predicting?) environmental effects on gene expression Philippe Collas University of Oslo Institute of Basic Medical Sciences Stem Cell Epigenetics
Jayanti Tokas 1, Puneet Tokas 2, Shailini Jain 3 and Hariom Yadav 3
 Jayanti Tokas 1, Puneet Tokas 2, Shailini Jain 3 and Hariom Yadav 3 1 Department of Biotechnology, JMIT, Radaur, Haryana, India 2 KITM, Kurukshetra, Haryana, India 3 NIDDK, National Institute of Health,
Jayanti Tokas 1, Puneet Tokas 2, Shailini Jain 3 and Hariom Yadav 3 1 Department of Biotechnology, JMIT, Radaur, Haryana, India 2 KITM, Kurukshetra, Haryana, India 3 NIDDK, National Institute of Health,
DNA METHYLATION IN EARLY MAMMALIAN DEVELOPMENT
 ARISTEA MARIA MAGARAKI DNA METHYLATION IN EARLY MAMMALIAN DEVELOPMENT Supervised by Dr. Petra de Graaf Physiological Chemistry, UMC DNA METHYLATION DURING EARLY MAMMALIAN DEVELOPMENT Master student: Aristea
ARISTEA MARIA MAGARAKI DNA METHYLATION IN EARLY MAMMALIAN DEVELOPMENT Supervised by Dr. Petra de Graaf Physiological Chemistry, UMC DNA METHYLATION DURING EARLY MAMMALIAN DEVELOPMENT Master student: Aristea
Single-cell RNA-Seq profiling of human pre-implantation embryos and embryonic stem cells
 Single-cell RNA-Seq profiling of human pre-implantation embryos and embryonic stem cells Liying Yan,2,5, Mingyu Yang,5, Hongshan Guo, Lu Yang, Jun Wu, Rong Li,2, Ping Liu, Ying Lian, Xiaoying Zheng, Jie
Single-cell RNA-Seq profiling of human pre-implantation embryos and embryonic stem cells Liying Yan,2,5, Mingyu Yang,5, Hongshan Guo, Lu Yang, Jun Wu, Rong Li,2, Ping Liu, Ying Lian, Xiaoying Zheng, Jie
Xist function: bridging chromatin and stem cells
 Review TRENDS in Genetics Vol.23 No.9 Xist function: bridging chromatin and stem cells Anton Wutz Research Institute of Molecular Pathology, Dr. Bohr-Gasse 7, 1030 Vienna, Austria In mammals, dosage compensation
Review TRENDS in Genetics Vol.23 No.9 Xist function: bridging chromatin and stem cells Anton Wutz Research Institute of Molecular Pathology, Dr. Bohr-Gasse 7, 1030 Vienna, Austria In mammals, dosage compensation
Transgenerational epigenetics in the germline cycle of Caenorhabditis elegans
 Transgenerational epigenetics in the germline cycle of Caenorhabditis elegans William G Kelly, Emory University Journal Title: Epigenetics and Chromatin Volume: Volume 7, Number 6 Publisher: BioMed Central
Transgenerational epigenetics in the germline cycle of Caenorhabditis elegans William G Kelly, Emory University Journal Title: Epigenetics and Chromatin Volume: Volume 7, Number 6 Publisher: BioMed Central
Epigenetics Armstrong_Prelims.indd 1 04/11/2013 3:28 pm
 Epigenetics Epigenetics Lyle Armstrong vi Online resources Accessible from www.garlandscience.com, the Student and Instructor Resource Websites provide learning and teaching tools created for Epigenetics.
Epigenetics Epigenetics Lyle Armstrong vi Online resources Accessible from www.garlandscience.com, the Student and Instructor Resource Websites provide learning and teaching tools created for Epigenetics.
Session 2: Biomarkers of epigenetic changes and their applicability to genetic toxicology
 Session 2: Biomarkers of epigenetic changes and their applicability to genetic toxicology Bhaskar Gollapudi, Ph.D The Dow Chemical Company Workshop: Genetic Toxicology: Opportunities to Integrate New Approaches
Session 2: Biomarkers of epigenetic changes and their applicability to genetic toxicology Bhaskar Gollapudi, Ph.D The Dow Chemical Company Workshop: Genetic Toxicology: Opportunities to Integrate New Approaches
Fragile X Syndrome. Genetics, Epigenetics & the Role of Unprogrammed Events in the expression of a Phenotype
 Fragile X Syndrome Genetics, Epigenetics & the Role of Unprogrammed Events in the expression of a Phenotype A loss of function of the FMR-1 gene results in severe learning problems, intellectual disability
Fragile X Syndrome Genetics, Epigenetics & the Role of Unprogrammed Events in the expression of a Phenotype A loss of function of the FMR-1 gene results in severe learning problems, intellectual disability
H3K27me3 may be associated with Oct4 and Sox2 in mouse preimplantation embryos
 H3K27me3 may be associated with Oct4 and Sox2 in mouse preimplantation embryos F.-R. Wu, Y. Zhang, B. Ding, X.-H. Lei, J.-C. Huang, C.-H. Wang, Y. Liu, R. Wang and W.-Y. Li School of Biological Science
H3K27me3 may be associated with Oct4 and Sox2 in mouse preimplantation embryos F.-R. Wu, Y. Zhang, B. Ding, X.-H. Lei, J.-C. Huang, C.-H. Wang, Y. Liu, R. Wang and W.-Y. Li School of Biological Science
Mammalian X-Chromosome Inactivation: An Epigenetics Paradigm
 Mammalian X-Chromosome Inactivation: An Epigenetics Paradigm E. HEARD, * J. CHAUMEIL, O. MASUI, AND I. OKAMOTO CNRS UMR218, Curie Institute, 75248 Paris Cedex 05, France In mammals, dosage compensation
Mammalian X-Chromosome Inactivation: An Epigenetics Paradigm E. HEARD, * J. CHAUMEIL, O. MASUI, AND I. OKAMOTO CNRS UMR218, Curie Institute, 75248 Paris Cedex 05, France In mammals, dosage compensation
STEM CELL GENETICS AND GENOMICS
 STEM CELL GENETICS AND GENOMICS Concise Review: Roles of Polycomb Group Proteins in Development and Disease: A Stem Cell Perspective VINAGOLU K. RAJASEKHAR, a MARTIN BEGEMANN b a Memorial Sloan-Kettering
STEM CELL GENETICS AND GENOMICS Concise Review: Roles of Polycomb Group Proteins in Development and Disease: A Stem Cell Perspective VINAGOLU K. RAJASEKHAR, a MARTIN BEGEMANN b a Memorial Sloan-Kettering
Imprinting. Joyce Ohm Cancer Genetics and Genomics CGP-L2-319 x8821
 Imprinting Joyce Ohm Cancer Genetics and Genomics CGP-L2-319 x8821 Learning Objectives 1. To understand the basic concepts of genomic imprinting Genomic imprinting is an epigenetic phenomenon that causes
Imprinting Joyce Ohm Cancer Genetics and Genomics CGP-L2-319 x8821 Learning Objectives 1. To understand the basic concepts of genomic imprinting Genomic imprinting is an epigenetic phenomenon that causes
Biology Developmental Biology Spring Quarter Midterm 1 Version A
 Biology 411 - Developmental Biology Spring Quarter 2013 Midterm 1 Version A 75 Total Points Open Book Choose 15 out the 20 questions to answer (5 pts each). Only the first 15 questions that are answered
Biology 411 - Developmental Biology Spring Quarter 2013 Midterm 1 Version A 75 Total Points Open Book Choose 15 out the 20 questions to answer (5 pts each). Only the first 15 questions that are answered
Epigenetic Mechanisms
 RCPA Lecture Epigenetic chanisms Jeff Craig Early Life Epigenetics Group, MCRI Dept. of Paediatrics Overview What is epigenetics? Chromatin The epigenetic code What is epigenetics? the interactions of
RCPA Lecture Epigenetic chanisms Jeff Craig Early Life Epigenetics Group, MCRI Dept. of Paediatrics Overview What is epigenetics? Chromatin The epigenetic code What is epigenetics? the interactions of
Epigenetics: The Future of Psychology & Neuroscience. Richard E. Brown Psychology Department Dalhousie University Halifax, NS, B3H 4J1
 Epigenetics: The Future of Psychology & Neuroscience Richard E. Brown Psychology Department Dalhousie University Halifax, NS, B3H 4J1 Nature versus Nurture Despite the belief that the Nature vs. Nurture
Epigenetics: The Future of Psychology & Neuroscience Richard E. Brown Psychology Department Dalhousie University Halifax, NS, B3H 4J1 Nature versus Nurture Despite the belief that the Nature vs. Nurture
Epigenetic Regulation of Health and Disease Nutritional and environmental effects on epigenetic regulation
 Epigenetic Regulation of Health and Disease Nutritional and environmental effects on epigenetic regulation Robert FEIL Director of Research CNRS & University of Montpellier, Montpellier, France. E-mail:
Epigenetic Regulation of Health and Disease Nutritional and environmental effects on epigenetic regulation Robert FEIL Director of Research CNRS & University of Montpellier, Montpellier, France. E-mail:
DNA Methylation and Cancer
 DNA Methylation and Cancer October 25, 2016 Dominic Smiraglia, Ph.D. Department of Cancer Genetics From Alan Wolffe, Science and Medicine, 1999 Vital Statistics Human genome contains 3 billion bp ~ 50,000
DNA Methylation and Cancer October 25, 2016 Dominic Smiraglia, Ph.D. Department of Cancer Genetics From Alan Wolffe, Science and Medicine, 1999 Vital Statistics Human genome contains 3 billion bp ~ 50,000
Epigenetics: A historical overview Dr. Robin Holliday
 Epigenetics 1 Rival hypotheses Epigenisis - The embryo is initially undifferentiated. As development proceeds, increasing levels of complexity emerge giving rise to the larval stage or to the adult organism.
Epigenetics 1 Rival hypotheses Epigenisis - The embryo is initially undifferentiated. As development proceeds, increasing levels of complexity emerge giving rise to the larval stage or to the adult organism.
Introduction: DNA methylation in mammals. DNA+Histones=Chromatin. Long-lasting cellular memory
 Séminaire de présentation des projets de l APR 2010 MIISTÈREDEL ÉCOLOGIE DU DÉVELOPPEMET DURABLE, DU TRASPORT ETDELAMER MethylED: Impact of Endocrine Disruptors on genomic DA methylation patterns in the
Séminaire de présentation des projets de l APR 2010 MIISTÈREDEL ÉCOLOGIE DU DÉVELOPPEMET DURABLE, DU TRASPORT ETDELAMER MethylED: Impact of Endocrine Disruptors on genomic DA methylation patterns in the
Epigenetic Patterns Maintained in Early Caenorhabditis elegans Embryos Can Be Established by Gene Activity in the Parental Germ Cells
 Epigenetic Patterns Maintained in Early Caenorhabditis elegans Embryos Can Be Established by Gene Activity in the Parental Germ Cells Jackelyn K. Arico, Emory University David J Katz, Emory University
Epigenetic Patterns Maintained in Early Caenorhabditis elegans Embryos Can Be Established by Gene Activity in the Parental Germ Cells Jackelyn K. Arico, Emory University David J Katz, Emory University
Genetics and Genomics in Medicine Chapter 6. Questions & Answers
 Genetics and Genomics in Medicine Chapter 6 Multiple Choice Questions Questions & Answers Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the
Genetics and Genomics in Medicine Chapter 6 Multiple Choice Questions Questions & Answers Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the
Lecture 8. Eukaryotic gene regulation: post translational modifications of histones
 Lecture 8 Eukaryotic gene regulation: post translational modifications of histones Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation
Lecture 8 Eukaryotic gene regulation: post translational modifications of histones Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation
Are you the way you are because of the
 EPIGENETICS Are you the way you are because of the It s my fault!! Nurture Genes you inherited from your parents? Nature Experiences during your life? Similar DNA Asthma, Autism, TWINS Bipolar Disorders
EPIGENETICS Are you the way you are because of the It s my fault!! Nurture Genes you inherited from your parents? Nature Experiences during your life? Similar DNA Asthma, Autism, TWINS Bipolar Disorders
Transgenerational Epigenetic Inheritance
 Transgenerational Epigenetic Inheritance TOP ARTICLES SUPPLEMENT CONTENTS EDITORIAL:: Epigenetics: where environment, society and genetics meet Epigenomics Vol. 6 Issue 1 REVIEW: Transgenerational epigenetic
Transgenerational Epigenetic Inheritance TOP ARTICLES SUPPLEMENT CONTENTS EDITORIAL:: Epigenetics: where environment, society and genetics meet Epigenomics Vol. 6 Issue 1 REVIEW: Transgenerational epigenetic
AN INTRODUCTION TO EPIGENETICS DR CHLOE WONG
 AN INTRODUCTION TO EPIGENETICS DR CHLOE WONG MRC SGDP CENTRE, INSTITUTE OF PSYCHIATRY KING S COLLEGE LONDON Oct 2015 Lecture Overview WHY WHAT EPIGENETICS IN PSYCHIARTY Technology-driven genomics research
AN INTRODUCTION TO EPIGENETICS DR CHLOE WONG MRC SGDP CENTRE, INSTITUTE OF PSYCHIATRY KING S COLLEGE LONDON Oct 2015 Lecture Overview WHY WHAT EPIGENETICS IN PSYCHIARTY Technology-driven genomics research
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Embryogenesis begins during oogenesis
 7th Workshop Mammalian Folliculogenesis and Oogenesis ESHRE Campus symposium Stresa, Italy 19 21 April 2012 Acquisition of the oocyte developmental competence Maurizio Zuccotti Embryogenesis begins during
7th Workshop Mammalian Folliculogenesis and Oogenesis ESHRE Campus symposium Stresa, Italy 19 21 April 2012 Acquisition of the oocyte developmental competence Maurizio Zuccotti Embryogenesis begins during
Edinburgh Research Explorer
 Edinburgh Research Explorer Meiosis and retrotransposon silencing during germ cell development in mice Citation for published version: Oellinger, R, Reichmann, J & Adams, IR 2010, 'Meiosis and retrotransposon
Edinburgh Research Explorer Meiosis and retrotransposon silencing during germ cell development in mice Citation for published version: Oellinger, R, Reichmann, J & Adams, IR 2010, 'Meiosis and retrotransposon
Eukaryotic Gene Regulation
 Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Specifying and protecting germ cell fate
 Specifying and protecting germ cell fate Susan Strome 1 and Dustin Updike 2 Abstract Germ cells are the special cells in the body that undergo meiosis to generate gametes and subsequently entire new organisms
Specifying and protecting germ cell fate Susan Strome 1 and Dustin Updike 2 Abstract Germ cells are the special cells in the body that undergo meiosis to generate gametes and subsequently entire new organisms
Joanna Hillman Michael Higgins Lab Oncology for Scientists I 10/29/2015
 Joanna Hillman Michael Higgins Lab Oncology for Scientists I 10/29/2015 ! Define Epigenetics & Genomic Imprinting! Discovery! What is the imprint! Lifecycle of an Imprint DMRs and ICEs! 2 main mechanisms
Joanna Hillman Michael Higgins Lab Oncology for Scientists I 10/29/2015 ! Define Epigenetics & Genomic Imprinting! Discovery! What is the imprint! Lifecycle of an Imprint DMRs and ICEs! 2 main mechanisms
Methylation Dynamics in the Early Mammalian Embryo: Implications of Genome Reprogramming Defects for Development
 CTMI (2006) 310:13 22 c Springer-Verlag Berlin Heidelberg 2006 Methylation Dynamics in the Early Mammalian Embryo: Implications of Genome Reprogramming Defects for Development T. Haaf ( ) Johannes Gutenberg-Universität
CTMI (2006) 310:13 22 c Springer-Verlag Berlin Heidelberg 2006 Methylation Dynamics in the Early Mammalian Embryo: Implications of Genome Reprogramming Defects for Development T. Haaf ( ) Johannes Gutenberg-Universität
Long noncoding RNAs in DNA methylation: new players stepping into the old game
 DOI 10.1186/s13578-016-0109-3 Cell & Bioscience REVIEW Long noncoding RNAs in DNA methylation: new players stepping into the old game Yu Zhao 1, Hao Sun 1,2* and Huating Wang 1,3* Open Access Abstract
DOI 10.1186/s13578-016-0109-3 Cell & Bioscience REVIEW Long noncoding RNAs in DNA methylation: new players stepping into the old game Yu Zhao 1, Hao Sun 1,2* and Huating Wang 1,3* Open Access Abstract
Author(s) Park, Sung-kyu Robin; Yates, John R. the original author and source are
 NAOSITE: Nagasaki University's Ac Title Author(s) The Specification and Global Reprog during Gamete Formation and Early E Samson, Mark; Jow, Margaret M.; Won Colin; Aslanian, Aaron; Saucedo, Is Park, Sung-kyu
NAOSITE: Nagasaki University's Ac Title Author(s) The Specification and Global Reprog during Gamete Formation and Early E Samson, Mark; Jow, Margaret M.; Won Colin; Aslanian, Aaron; Saucedo, Is Park, Sung-kyu
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Epigenetics: Basic Principals and role in health and disease
 Epigenetics: Basic Principals and role in health and disease Cambridge Masterclass Workshop on Epigenetics in GI Health and Disease 3 rd September 2013 Matt Zilbauer Overview Basic principals of Epigenetics
Epigenetics: Basic Principals and role in health and disease Cambridge Masterclass Workshop on Epigenetics in GI Health and Disease 3 rd September 2013 Matt Zilbauer Overview Basic principals of Epigenetics
Repressive Transcription
 Repressive Transcription The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Guenther, M. G., and R. A.
Repressive Transcription The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Guenther, M. G., and R. A.
Epigenetics and inheritance of phenotype variation in livestock
 DOI 10.1186/s13072-016-0081-5 Epigenetics & Chromatin REVIEW Open Access Epigenetics and inheritance of phenotype variation in livestock Kostas A. Triantaphyllopoulos 1*, Ioannis Ikonomopoulos 2 and Andrew
DOI 10.1186/s13072-016-0081-5 Epigenetics & Chromatin REVIEW Open Access Epigenetics and inheritance of phenotype variation in livestock Kostas A. Triantaphyllopoulos 1*, Ioannis Ikonomopoulos 2 and Andrew
Chromatin-Based Regulation of Gene Expression
 Chromatin-Based Regulation of Gene Expression.George J. Quellhorst, Jr., PhD.Associate Director, R&D.Biological Content Development Topics to be Discussed Importance of Chromatin-Based Regulation Mechanism
Chromatin-Based Regulation of Gene Expression.George J. Quellhorst, Jr., PhD.Associate Director, R&D.Biological Content Development Topics to be Discussed Importance of Chromatin-Based Regulation Mechanism
R. Piazza (MD, PhD), Dept. of Medicine and Surgery, University of Milano-Bicocca EPIGENETICS
 R. Piazza (MD, PhD), Dept. of Medicine and Surgery, University of Milano-Bicocca EPIGENETICS EPIGENETICS THE STUDY OF CHANGES IN GENE EXPRESSION THAT ARE POTENTIALLY HERITABLE AND THAT DO NOT ENTAIL A
R. Piazza (MD, PhD), Dept. of Medicine and Surgery, University of Milano-Bicocca EPIGENETICS EPIGENETICS THE STUDY OF CHANGES IN GENE EXPRESSION THAT ARE POTENTIALLY HERITABLE AND THAT DO NOT ENTAIL A
Cluster Dendrogram. dist(cor(na.omit(tss.exprs.chip[, c(1:10, 24, 27, 30, 48:50, dist(cor(na.omit(tss.exprs.chip[, c(1:99, 103, 104, 109, 110,
 A Transcriptome (RNA-seq) Transcriptome (RNA-seq) 3. 2.5 2..5..5...5..5 2. 2.5 3. 2.5 2..5..5...5..5 2. 2.5 Cluster Dendrogram RS_ES3.2 RS_ES3. RS_SHS5.2 RS_SHS5. PS_SHS5.2 PS_SHS5. RS_LJ3 PS_LJ3..4 _SHS5.2
A Transcriptome (RNA-seq) Transcriptome (RNA-seq) 3. 2.5 2..5..5...5..5 2. 2.5 3. 2.5 2..5..5...5..5 2. 2.5 Cluster Dendrogram RS_ES3.2 RS_ES3. RS_SHS5.2 RS_SHS5. PS_SHS5.2 PS_SHS5. RS_LJ3 PS_LJ3..4 _SHS5.2
Epigenetic Inheritance
 (2) The role of Epigenetic Inheritance Lamarck Revisited Lamarck was incorrect in thinking that the inheritance of acquired characters is the main mechanism of evolution (Natural Selection more common)
(2) The role of Epigenetic Inheritance Lamarck Revisited Lamarck was incorrect in thinking that the inheritance of acquired characters is the main mechanism of evolution (Natural Selection more common)
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Selective impairment of methylation maintenance is the major cause of DNA methylation reprogramming in the early embryo
 Arand et al. Epigenetics & Chromatin 2015, 8:1 RESEARCH Open Access Selective impairment of methylation maintenance is the major cause of DNA methylation reprogramming in the early embryo Julia Arand 1,4,
Arand et al. Epigenetics & Chromatin 2015, 8:1 RESEARCH Open Access Selective impairment of methylation maintenance is the major cause of DNA methylation reprogramming in the early embryo Julia Arand 1,4,
Biology 4361 Developmental Biology. October 11, Multiple choice (one point each)
 Biology 4361 Developmental Biology Exam 1 October 11, 2005 Name: ID#: Multiple choice (one point each) 1. Sertoli cells a. surround spermatocytes b. are the structural components of the seminiferous tubules
Biology 4361 Developmental Biology Exam 1 October 11, 2005 Name: ID#: Multiple choice (one point each) 1. Sertoli cells a. surround spermatocytes b. are the structural components of the seminiferous tubules
Oncology 520 Epigene0cs and Cancer. February 16, 2011
 Oncology 520 Epigene0cs and Cancer February 16, 2011 Outline What is epigene0cs? What are examples of epigene0c processes? What is the epigenome and how is it decoded? Are epigene0c mechanisms important
Oncology 520 Epigene0cs and Cancer February 16, 2011 Outline What is epigene0cs? What are examples of epigene0c processes? What is the epigenome and how is it decoded? Are epigene0c mechanisms important
The Pathology of Germ Cell Tumours of the Ovary
 The Pathology of Germ Cell Tumours of the Ovary Professor Mike Wells University of Sheffield Amman, Jordan November 2013 Professor Francisco Paco Nogales I. Primitive germ cell tumors A. Dysgerminoma
The Pathology of Germ Cell Tumours of the Ovary Professor Mike Wells University of Sheffield Amman, Jordan November 2013 Professor Francisco Paco Nogales I. Primitive germ cell tumors A. Dysgerminoma
Epigenetics and Chromatin
 Progress in Molecular and Subcellular Biology 38 Epigenetics and Chromatin Bearbeitet von Philippe Jeanteur 1. Auflage 2008. Taschenbuch. xiii, 266 S. Paperback ISBN 978 3 540 85236 0 Format (B x L): 15,5
Progress in Molecular and Subcellular Biology 38 Epigenetics and Chromatin Bearbeitet von Philippe Jeanteur 1. Auflage 2008. Taschenbuch. xiii, 266 S. Paperback ISBN 978 3 540 85236 0 Format (B x L): 15,5
Recent advances in X-chromosome inactivation Edith Heard
 Recent advances in X-chromosome inactivation Edith Heard X inactivation is the silencing one of the two X chromosomes in XX female mammals. Initiation of this process during early development is controlled
Recent advances in X-chromosome inactivation Edith Heard X inactivation is the silencing one of the two X chromosomes in XX female mammals. Initiation of this process during early development is controlled
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Epigenetics & cancer. Present by : Sanaz Zebardast Under supervision : Dr. Gheibi. 31 December 2016
 Epigenetics & cancer Present by : Sanaz Zebardast Under supervision : Dr. Gheibi 31 December 2016 1 contents Introduction Epigenetic & signaling pathways Epigenetic & integral protein Epigenetic & apoptosis
Epigenetics & cancer Present by : Sanaz Zebardast Under supervision : Dr. Gheibi 31 December 2016 1 contents Introduction Epigenetic & signaling pathways Epigenetic & integral protein Epigenetic & apoptosis
Epigenetic mechanisms and their implications in animal breeding
 Epigenetic mechanisms and their implications in animal breeding Klaus Wimmers, Nares Trakooljul, Michael Oster, Eduard Murani, Siriluck Ponsuksili Leibniz Institute for Farm Animal Biology (FBN), Germany
Epigenetic mechanisms and their implications in animal breeding Klaus Wimmers, Nares Trakooljul, Michael Oster, Eduard Murani, Siriluck Ponsuksili Leibniz Institute for Farm Animal Biology (FBN), Germany
Molecular insights into transgenerational non-genetic inheritance of acquired behaviours
 Molecular insights into transgenerational non-genetic inheritance of acquired behaviours Johannes Bohacek and Isabelle M. Mansuy Abstract Behavioural traits in mammals are influenced by environmental factors,
Molecular insights into transgenerational non-genetic inheritance of acquired behaviours Johannes Bohacek and Isabelle M. Mansuy Abstract Behavioural traits in mammals are influenced by environmental factors,
Biology 4361 Developmental Biology Exam 1 ID#: October 11, 2005
 Biology 4361 Developmental Biology Name: Key Exam 1 ID#: October 11, 2005 Multiple choice (one point each) 1. Primordial germ cells a. are immortal b. produce polar bodies c. are haploid d. are somatic
Biology 4361 Developmental Biology Name: Key Exam 1 ID#: October 11, 2005 Multiple choice (one point each) 1. Primordial germ cells a. are immortal b. produce polar bodies c. are haploid d. are somatic
REVIEWS. X chromosome regulation: diverse patterns in development, tissues and disease
 X chromosome regulation: diverse patterns in development, tissues and disease Xinxian Deng 1, Joel B. Berletch 1, Di K. Nguyen 1 and Christine M. Disteche 1,2 Abstract Genes on the mammalian X chromosome
X chromosome regulation: diverse patterns in development, tissues and disease Xinxian Deng 1, Joel B. Berletch 1, Di K. Nguyen 1 and Christine M. Disteche 1,2 Abstract Genes on the mammalian X chromosome
EPIGENOMICS PROFILING SERVICES
 EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
Methylation reprogramming dynamics and defects in gametogenesis and embryogenesis: implications for reproductive medicine
 Univ.-Prof. Dr. Thomas Haaf Methylation reprogramming dynamics and defects in gametogenesis and embryogenesis: implications for reproductive medicine Epigenetics and DNA methylation Heritable change of
Univ.-Prof. Dr. Thomas Haaf Methylation reprogramming dynamics and defects in gametogenesis and embryogenesis: implications for reproductive medicine Epigenetics and DNA methylation Heritable change of
Chromatin structure and epigenetic regulation of eukaryotic gene expression. Giacomo Cavalli
 Chromatin structure and epigenetic regulation of eukaryotic gene expression Giacomo Cavalli 12.09.2016 10,000 nm DNA compaction in a human nucleus 11 nm 30nm 1bp (0.3nm) compact size DNA length compaction
Chromatin structure and epigenetic regulation of eukaryotic gene expression Giacomo Cavalli 12.09.2016 10,000 nm DNA compaction in a human nucleus 11 nm 30nm 1bp (0.3nm) compact size DNA length compaction
OVERVIEW OF EPIGENETICS
 OVERVIEW OF EIENETICS Date: * Time: 9:00 am - 9:50 am * Room: Berryhill 103 Lecturer: Terry Magnuson 4312 MBRB trm4@med.unc.edu 843-6475 *lease consult the online schedule for this course for the definitive
OVERVIEW OF EIENETICS Date: * Time: 9:00 am - 9:50 am * Room: Berryhill 103 Lecturer: Terry Magnuson 4312 MBRB trm4@med.unc.edu 843-6475 *lease consult the online schedule for this course for the definitive
PRC2 crystal clear. Matthieu Schapira
 PRC2 crystal clear Matthieu Schapira Epigenetic mechanisms control the combination of genes that are switched on and off in any given cell. In turn, this combination, called the transcriptional program,
PRC2 crystal clear Matthieu Schapira Epigenetic mechanisms control the combination of genes that are switched on and off in any given cell. In turn, this combination, called the transcriptional program,
PAGE PROOF. Epigenetic Regulation of the X Chromosomes in C. elegans. Susan Strome 1 and William G. Kelly 2
 15_EPIGEN_p00_strome 6/7/06 2:50 PM Page 1 C H A P T E R 15 Epigenetic Regulation of the X Chromosomes in C. elegans CONTENTS Susan Strome 1 and William G. Kelly 2 1 Department of Biology, Indiana University,
15_EPIGEN_p00_strome 6/7/06 2:50 PM Page 1 C H A P T E R 15 Epigenetic Regulation of the X Chromosomes in C. elegans CONTENTS Susan Strome 1 and William G. Kelly 2 1 Department of Biology, Indiana University,
Epigenetics and Toxicology
 Epigenetics and Toxicology Aline.deconti@fda.hhs.gov Division of Biochemical Toxicology National Center for Toxicology Research U.S.-Food and Drug Administration The views expressed in this presentation
Epigenetics and Toxicology Aline.deconti@fda.hhs.gov Division of Biochemical Toxicology National Center for Toxicology Research U.S.-Food and Drug Administration The views expressed in this presentation
Biology 2C03 Term Test #3
 Biology 2C03 Term Test #3 Instructors: Dr. Kimberley Dej, Ray Procwat Date: Monday March 22, 2010 Time: 10:30 am to 11:20 am Instructions: 1) This midterm test consists of 9 pages. Please ensure that all
Biology 2C03 Term Test #3 Instructors: Dr. Kimberley Dej, Ray Procwat Date: Monday March 22, 2010 Time: 10:30 am to 11:20 am Instructions: 1) This midterm test consists of 9 pages. Please ensure that all
Dynamics of mono, di and tri-methylated histone H3 lysine 4 during male meiotic prophase I. Nuclei were co-stained for H3.1/H3.2. Progressing stages
 Dynamics of mono, di and tri-methylated histone H3 lysine 4 during male meiotic prophase I. Nuclei were co-stained for H3.1/H3.2. Progressing stages of spermatogenesis are shown from left to right. Arrows/dotted
Dynamics of mono, di and tri-methylated histone H3 lysine 4 during male meiotic prophase I. Nuclei were co-stained for H3.1/H3.2. Progressing stages of spermatogenesis are shown from left to right. Arrows/dotted
Name: Xueming Zhao. Professional Title: Professor. Animal embryo biotechnology, mainly including in vitro maturation (IVM), in vitro fertilization
 Name: Xueming Zhao Professional Title: Professor Telephone:86-010-62815892 Fax:86-010-62895971 E-mail: zhaoxueming@caas.cn Website: http://www.iascaas.net.cn/yjspy/dsjj/sssds/dwyzyzypz1/62040.htm Research
Name: Xueming Zhao Professional Title: Professor Telephone:86-010-62815892 Fax:86-010-62895971 E-mail: zhaoxueming@caas.cn Website: http://www.iascaas.net.cn/yjspy/dsjj/sssds/dwyzyzypz1/62040.htm Research
Epigenetic regulation of endogenous genes and developmental processes
 Epigenetic regulation of endogenous genes and developmental processes Epigenetic programming in plants helps control developmental transitions Embryonic to vegetative transition Vegetative to reproductive
Epigenetic regulation of endogenous genes and developmental processes Epigenetic programming in plants helps control developmental transitions Embryonic to vegetative transition Vegetative to reproductive
AGCGGCTGGTGTATGAGG AGCTCGAGCTGCAGGACT AAAGGTGCACTGGTAGCC AGGGCACAAGTGCATGGC CCCGCTGTGCAGAAGTAG
 Supplementary Table S1. Primer sequence information Gene Primer sequence 1 st round PCR 2 nd round PCR Accession# Dnmt3a TCAAGGAGATCATTGATG TGTTCCCACACATGAGCA AGCGGCTGGTGTATGAGG AGCAGATGGTGCAATAGGAC NM_007872.4
Supplementary Table S1. Primer sequence information Gene Primer sequence 1 st round PCR 2 nd round PCR Accession# Dnmt3a TCAAGGAGATCATTGATG TGTTCCCACACATGAGCA AGCGGCTGGTGTATGAGG AGCAGATGGTGCAATAGGAC NM_007872.4
Not IN Our Genes - A Different Kind of Inheritance.! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014
 Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
The Biology and Genetics of Cells and Organisms The Biology of Cancer
 The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
DNA double-strand break repair of parental chromatin in ooplasm and origin of de novo mutations. Peter de Boer
 DNA double-strand break repair of parental chromatin in ooplasm and origin of de novo mutations Peter de Boer Department of Obst.& Gynaecology, Div. Reproductive Medicine Radboud University Nijmegen Medical
DNA double-strand break repair of parental chromatin in ooplasm and origin of de novo mutations Peter de Boer Department of Obst.& Gynaecology, Div. Reproductive Medicine Radboud University Nijmegen Medical
STEM CELL RESEARCH: MEDICAL PROGRESS WITH RESPONSIBILITY
 STEM CELL RESEARCH: MEDICAL PROGRESS WITH RESPONSIBILITY A REPORT FROM THE CHIEF MEDICAL OFFICER S EXPERT GROUP REVIEWING THE POTENTIAL OF DEVELOPMENTS IN STEM CELL RESEARCH AND CELL NUCLEAR REPLACEMENT
STEM CELL RESEARCH: MEDICAL PROGRESS WITH RESPONSIBILITY A REPORT FROM THE CHIEF MEDICAL OFFICER S EXPERT GROUP REVIEWING THE POTENTIAL OF DEVELOPMENTS IN STEM CELL RESEARCH AND CELL NUCLEAR REPLACEMENT
Eukaryotic transcription (III)
 Eukaryotic transcription (III) 1. Chromosome and chromatin structure Chromatin, chromatid, and chromosome chromatin Genomes exist as chromatins before or after cell division (interphase) but as chromatids
Eukaryotic transcription (III) 1. Chromosome and chromatin structure Chromatin, chromatid, and chromosome chromatin Genomes exist as chromatins before or after cell division (interphase) but as chromatids
Testicular stem cells
 Testicular stem cells Dirk G. de Rooij Department of Endocrinology Faculty of Biology, Utrecht University 1. Knowledge on the development of the spermatogenic stem cell lineage 2. Principals of the nature
Testicular stem cells Dirk G. de Rooij Department of Endocrinology Faculty of Biology, Utrecht University 1. Knowledge on the development of the spermatogenic stem cell lineage 2. Principals of the nature
Changes in histone methylation during human oocyte maturation and IVF- or ICSI-derived embryo development
 Changes in histone methylation during human oocyte maturation and IVF- or ICSI-derived embryo development Jie Qiao, M.D., a Yuan Chen, Ph.D., a Li-Ying Yan, Ph.D., a Jie Yan, Ph.D., a Ping Liu, Ph.D.,
Changes in histone methylation during human oocyte maturation and IVF- or ICSI-derived embryo development Jie Qiao, M.D., a Yuan Chen, Ph.D., a Li-Ying Yan, Ph.D., a Jie Yan, Ph.D., a Ping Liu, Ph.D.,
Dedicated to my family
 Dedicated to my family List of Papers This thesis is based on the following papers, which are referred to in the text by their Roman numerals. I Mohammad F*, Pandey RR*, Nagano T, Chakalova L, Mondal
Dedicated to my family List of Papers This thesis is based on the following papers, which are referred to in the text by their Roman numerals. I Mohammad F*, Pandey RR*, Nagano T, Chakalova L, Mondal
Bi-potent Gonads. Sex Determination
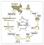 יצירת הגונדות Primordial Germ Cells (PGCs) Somatic cells Genital ridge Bi-potent Gonads Sex Determination Testis and Sperm Ovary and Oocyte Migration of Primordial Germ Cells in the Chick Embryo The
יצירת הגונדות Primordial Germ Cells (PGCs) Somatic cells Genital ridge Bi-potent Gonads Sex Determination Testis and Sperm Ovary and Oocyte Migration of Primordial Germ Cells in the Chick Embryo The
Gametogenesis. To complete this worksheet, select: Module: Continuity Activity: Animations Title: Gametogenesis. Introduction
 Gametogenesis To complete this worksheet, select: Module: Continuity Activity: Animations Title: Gametogenesis Introduction 1. a. Define gametogenesis. b. What cells are gametes? c. What are the two cell
Gametogenesis To complete this worksheet, select: Module: Continuity Activity: Animations Title: Gametogenesis Introduction 1. a. Define gametogenesis. b. What cells are gametes? c. What are the two cell
Animal Development. Lecture 3. Germ Cells and Sex
 Animal Development Lecture 3 Germ Cells and Sex 1 The ovary of sow. The ovary of mare. The ovary of cow. The ovary of ewe. 2 3 The ovary. A generalized vertebrate ovary. (Wilt and Hake, Ch 2, 2004) 4 The
Animal Development Lecture 3 Germ Cells and Sex 1 The ovary of sow. The ovary of mare. The ovary of cow. The ovary of ewe. 2 3 The ovary. A generalized vertebrate ovary. (Wilt and Hake, Ch 2, 2004) 4 The
Review Article Epigenetic Mechanisms of Genomic Imprinting: Common Themes in the Regulation of Imprinted Regions in Mammals, Plants, and Insects
 Genetics Research International Volume 2012, Article ID 585024, 17 pages doi:10.1155/2012/585024 Review Article Epigenetic echanisms of Genomic Imprinting: Common Themes in the Regulation of Imprinted
Genetics Research International Volume 2012, Article ID 585024, 17 pages doi:10.1155/2012/585024 Review Article Epigenetic echanisms of Genomic Imprinting: Common Themes in the Regulation of Imprinted
I) Development: tissue differentiation and timing II) Whole Chromosome Regulation
 Epigenesis: Gene Regulation Epigenesis : Gene Regulation I) Development: tissue differentiation and timing II) Whole Chromosome Regulation (X chromosome inactivation or Lyonization) III) Regulation during
Epigenesis: Gene Regulation Epigenesis : Gene Regulation I) Development: tissue differentiation and timing II) Whole Chromosome Regulation (X chromosome inactivation or Lyonization) III) Regulation during
Mediators and dynamics of DNA methylation
 Mediators and dynamics of DNA methylation Robert Shoemaker, 1 Wei Wang 1 and Kun Zhang 2 As an inherited epigenetic marker occurring mainly on cytosines at CpG dinucleotides, DNA methylation occurs across
Mediators and dynamics of DNA methylation Robert Shoemaker, 1 Wei Wang 1 and Kun Zhang 2 As an inherited epigenetic marker occurring mainly on cytosines at CpG dinucleotides, DNA methylation occurs across
Epigenetic Principles and Mechanisms Underlying Nervous System Function in Health and Disease Mark F. Mehler MD, FAAN
 Epigenetic Principles and Mechanisms Underlying Nervous System Function in Health and Disease Mark F. Mehler MD, FAAN Institute for Brain Disorders and Neural Regeneration F.M. Kirby Program in Neural
Epigenetic Principles and Mechanisms Underlying Nervous System Function in Health and Disease Mark F. Mehler MD, FAAN Institute for Brain Disorders and Neural Regeneration F.M. Kirby Program in Neural
BIOH122 Session 26 Gametogenesis. Introduction. 1. a. Define gametogenesis. b. What cells are gametes?
 BIOH122 Session 26 Gametogenesis Introduction 1. a. Define gametogenesis. b. What cells are gametes? c. What are the two cell division processes that occur during the cell cycle? d. Define the cell cycle.
BIOH122 Session 26 Gametogenesis Introduction 1. a. Define gametogenesis. b. What cells are gametes? c. What are the two cell division processes that occur during the cell cycle? d. Define the cell cycle.
