Interpreting Diverse Genomic Data Using Gene Sets
|
|
|
- David Spencer
- 5 years ago
- Views:
Transcription
1 Interpreting Diverse Genomic Data Using Gene Sets giovanni FHCRC February 2012
2 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red: Copy number alterations; Blue: Point mutations.
3 Why gene-set analysis? Improvements in interpretability of experimental results. Detection of subtle correlated changes in sets. Detection of set-level biological signals. Integration of diverse data sources.
4 The birthplaces of gene set analysis: I Tavazoie etal Nature Genetics 2000 Hypergeometric p-value.
5 The birthplaces of gene set analysis: II Mirnics et al Neuron 2000 Molecular Characterization of Schizophrenia Viewed by Microarray Analysis of Gene Expression in Prefrontal Cortex.
6 A Formalism for Two-Stage Gene Set Analysis Binary response vector Y (phenotype, class label, case-control...) one for each of N samples G N matrix X of genetic information on samples G S binary membership matrix M Stage I Testing of differences between groups for each gene. Compute for each gene g a score s g (X,Y ), capturing the relationship between the genomic measurements and a phenotype of interest. Stage II Testing of differences in scores between sets. Take the scores computed in Stage I as data, and look for association between the scores and the columns of M.
7 An Early Gene Set Analysis Chowers etal Human Molecular Genetics, 2003 Distribution of standard deviations for expression ratios of all genes of known function on the array (solid line), photoreceptor genes (dashed line), and genes involved in cell proliferation (dotted line).
8 Gene Set Enrichment Analysis Mootha et al Nature Genetics, 2003; Subramanian PNAS 2005
9 Caveats I: Biology SET QUALITY SET OVERLAP TISSUE SPECIFICITY PATHWAY TOPOLOGY
10 Caveats II: Statistics GENES ARE NOT INDEPENDENT WHAT IS THE NULL HYPOTHESIS? BIG SET BIAS
11 Outline, References and Acknowledgments MANY TECHNOLOGIES S. Tyekucheva, L. Marchionni, R. Karchin and G. Parmigiani Integrating diverse genomic data using gene sets. Genome Biol., 12: R105, ATOMS S.M. Boca, H. Corrada Bravo, B. Caffo, J.T. Leek and G. Parmigiani. A decision-theory approach to interpretable set analysis for high-dimensional data. JHU Biostat Working Paper 211, PATIENTS S.M. Boca, K.W. Kinzler, V.E. Velculescu, B. Vogelstein and G. Parmigiani. Patient oriented gene-set analysis for cancer mutation data. Genome Biol., 11: R112, 2010.
12 Multiple data types Phenotype (Pa,ents) Set C Gene s Gene s Gene s Gene s Set B Set Z Genes Set A
13 Gene-centric approaches for multiple data types Binary response vector Y (phenotype, class label, case-control...) G N matrix X of genetic information on samples G S binary membership matrix M Stage I Stage II s g (X 1,...,X D,Y ) t s (s,m s ) Integration sg(x 1 d,y )... sg D (X d,y ) t s (s 1...s D,M s ) Meta-analysis sg(x 1 d,y )... sg D (X d,y ) t s (s 1,M s )... t s (s D,M s ) Visualization
14 Clustering Sets to Compare Experiments
15 Integrative more powerful than Meta-analytic A B Expression 1 Expression 2 CNV 1 CNV 2 LR Avg. p value Min. p value True positive rate Expression 1 Expression 2 CNV 1 CNV 2 LR Avg. p value Min. p value Altered fraction False positive rate Independent Sets ROC for classification of spiked-in sets
16 Integrative more robust than Meta-analytic A B Expression 1 Expression 2 CNV 1 CNV 2 LR Avg. p value Min. p value True positive rate Expression 1 Expression 2 CNV 1 CNV 2 LR Avg. p value Min. p value Altered fraction False positive rate Chromosomal Segments ROC for classification of spiked-in sets
17 Integrative discovers novel sets (a) Synthetic sets Canonical pathways Fraction of exclusively discovered sets Expression 1 Expression 2 CN 1 CN 2 Fraction of exclusively discovered sets Expression 1 Expression 2 CN 1 CN Number of top sets Number of top sets (b) Synthetic sets Canonical pathways Fraction of exclusively discovered sets INT Avg. p value Min. p value Fraction of exclusively discovered sets INT Avg. p value Min. p value Number of top sets Number of top sets
18 GBM Color key Value Color key Value Color key 2 2 Value E2 C2 C2 C2 C1 WNT pathway C1 E2 Stress pathway C1 E2 Glycolysis pathway E1 E1 E1 PGK1 ENO1 TPI1 GPI ALDOB PFKL PKLR HK1 MAP2K4 TRADD MAP2K3 TANK MAP2K6 JUN IKBKG TNFRSF1A MAP3K14 CRADD CASP2 NFKB1 TRAF2 RIPK1 ATF1 CHUK LTA MAP4K2 NFKBIA MAPK14 IKBKB TNF MAPK8 APC WIF1 GSK3B FZD1 CSNK1A1 NLK CTNNB1 HDAC1 MAP3K7 CCND1 CSNK2A1 TLE1 CTBP1 CSNK1D PPARD FRAT1 BTRC AXIN1 MAP3K7IP1 MYC CREBBP
19 Enrichment of 3 intersecting pathways for ER+ BC Wnt pathway 28.7 % (261) 29.4% (225) 14.0% (23) 42.0% (175) Cell cycle 38.0% (235) 27.1% (7) 61.2% (6) 29.0% (31) Ubiquitin mediated proteolysis 26.1% (231) 24.5% (187) X% (Y): X% out of Y genes are estimated to have densities from the alternative distribution.
20 Decision Theoretic Angle Divide genes into atoms based on sets. Truth is the list of alternatives. We search for estimators among the unions of atoms. The estimators are based on the loss function: (1 w) # of FD +w # of MD. The posterior expected loss is: (1 w) EFD + w EMD.
21 Atomic False Discovery Rate We define the atomic false discovery rate for atom A as: AFDR(A) = FD(A)/n A. Theorem (Boca et al., 2010) Atom A is included in the Bayes estimator if and only if the atomic FDR is thresholded by w: ÂFDR(A) w. 1 ÂFDR estimates the fraction of alternatives in an atom.
22 Atomic FDR measures enrichment 1 EFDR p value enrichment fraction enrichment fraction
23 Altered Pathways in Glioblastoma Parsons 2008
24 Gene-centric vs Patient Centric Scores log10(q values) obtained by counting altered samples log10(q values) obtained by combining gene scores
25 Outline, References and Acknowledgments MANY TECHNOLOGIES S. Tyekucheva, L. Marchionni, R. Karchin and G. Parmigiani Integrating diverse genomic data using gene sets. Genome Biol., 12: R105, ATOMS S.M. Boca, H. Corrada Bravo, B. Caffo, J.T. Leek and G. Parmigiani. A decision-theory approach to interpretable set analysis for high-dimensional data. JHU Biostat Working Paper 211, PATIENTS S.M. Boca, K.W. Kinzler, V.E. Velculescu, B. Vogelstein and G. Parmigiani. Patient oriented gene-set analysis for cancer mutation data. Genome Biol., 11: R112, 2010.
Interpreting Diverse Genomic Data Using Gene Sets
 Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Interpreting Diverse Genomic Data Using Gene Sets giovanni parmigiani@dfci.harvard.edu FHCRC February 2012 Gene Sets Leary 2008 PNAS Alterations in the combined FGF, EGFR, ERBB2 and PIK3 pathways. Red:
Introduction to Gene Sets Analysis
 Introduction to Svitlana Tyekucheva Dana-Farber Cancer Institute May 15, 2012 Introduction Various measurements: gene expression, copy number variation, methylation status, mutation profile, etc. Main
Introduction to Svitlana Tyekucheva Dana-Farber Cancer Institute May 15, 2012 Introduction Various measurements: gene expression, copy number variation, methylation status, mutation profile, etc. Main
Package CancerMutationAnalysis
 Type Package Package CancerMutationAnalysis Title Cancer mutation analysis Version 1.2.1 Author Giovanni Parmigiani, Simina M. Boca March 25, 2013 Maintainer Simina M. Boca Imports
Type Package Package CancerMutationAnalysis Title Cancer mutation analysis Version 1.2.1 Author Giovanni Parmigiani, Simina M. Boca March 25, 2013 Maintainer Simina M. Boca Imports
* Kyoto Encyclopedia of Genes and Genomes.
 Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al.
 Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al. Holger Höfling Gad Getz Robert Tibshirani June 26, 2007 1 Introduction Identifying genes that are involved
Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al. Holger Höfling Gad Getz Robert Tibshirani June 26, 2007 1 Introduction Identifying genes that are involved
CancerMutationAnalysis package
 CancerMutationAnalysis package Giovanni Parmigiani Dana-Farber Cancer Institute and Harvard School of Public Health email: gp@jimmy.harvard.edu, Simina M. Boca Georgetown University Medical Center email:
CancerMutationAnalysis package Giovanni Parmigiani Dana-Farber Cancer Institute and Harvard School of Public Health email: gp@jimmy.harvard.edu, Simina M. Boca Georgetown University Medical Center email:
Supplementary figures legends
 Supplementary Information to Nijmegen Breakage Syndrome fibroblasts and ipscs: cellular models for uncovering diseaseassociated signaling pathways and establishing a screening platform for anti-oxidants
Supplementary Information to Nijmegen Breakage Syndrome fibroblasts and ipscs: cellular models for uncovering diseaseassociated signaling pathways and establishing a screening platform for anti-oxidants
Integrated Analysis of Copy Number and Gene Expression
 Integrated Analysis of Copy Number and Gene Expression Nexus Copy Number provides user-friendly interface and functionalities to integrate copy number analysis with gene expression results for the purpose
Integrated Analysis of Copy Number and Gene Expression Nexus Copy Number provides user-friendly interface and functionalities to integrate copy number analysis with gene expression results for the purpose
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Expanded View Figures
 Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
Comparison of Gene Set Analysis with Various Score Transformations to Test the Significance of Sets of Genes
 Comparison of Gene Set Analysis with Various Score Transformations to Test the Significance of Sets of Genes Ivan Arreola and Dr. David Han Department of Management of Science and Statistics, University
Comparison of Gene Set Analysis with Various Score Transformations to Test the Significance of Sets of Genes Ivan Arreola and Dr. David Han Department of Management of Science and Statistics, University
SUPPLEMENTARY FIGURES: Supplementary Figure 1
 SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
Module 3: Pathway and Drug Development
 Module 3: Pathway and Drug Development Table of Contents 1.1 Getting Started... 6 1.2 Identifying a Dasatinib sensitive cancer signature... 7 1.2.1 Identifying and validating a Dasatinib Signature... 7
Module 3: Pathway and Drug Development Table of Contents 1.1 Getting Started... 6 1.2 Identifying a Dasatinib sensitive cancer signature... 7 1.2.1 Identifying and validating a Dasatinib Signature... 7
Statistical Analysis of Single Nucleotide Polymorphism Microarrays in Cancer Studies
 Statistical Analysis of Single Nucleotide Polymorphism Microarrays in Cancer Studies Stanford Biostatistics Workshop Pierre Neuvial with Henrik Bengtsson and Terry Speed Department of Statistics, UC Berkeley
Statistical Analysis of Single Nucleotide Polymorphism Microarrays in Cancer Studies Stanford Biostatistics Workshop Pierre Neuvial with Henrik Bengtsson and Terry Speed Department of Statistics, UC Berkeley
Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors
 Wang et al LEGENDS TO SUPPLEMENTARY INFORMATION Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors A. Induced expression of estrogen receptor α (ERα) in AME vs PDA
Wang et al LEGENDS TO SUPPLEMENTARY INFORMATION Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors A. Induced expression of estrogen receptor α (ERα) in AME vs PDA
Nature Medicine: doi: /nm.3967
 Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Nature Neuroscience: doi: /nn Supplementary Figure 1. Behavioral training.
 Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Characteriza*on of Soma*c Muta*ons in Cancer Genomes
 Characteriza*on of Soma*c Muta*ons in Cancer Genomes Ben Raphael Department of Computer Science Center for Computa*onal Molecular Biology Soma*c Muta*ons and Cancer Clonal Theory (Nowell 1976) Passenger
Characteriza*on of Soma*c Muta*ons in Cancer Genomes Ben Raphael Department of Computer Science Center for Computa*onal Molecular Biology Soma*c Muta*ons and Cancer Clonal Theory (Nowell 1976) Passenger
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
Supplementary figures
 Supplementary figures Supplementary figure 1: Pathway enrichment for each comparison. The first column shows enrichment with differentially expressed genes between the diet groups at t = 0, the second
Supplementary figures Supplementary figure 1: Pathway enrichment for each comparison. The first column shows enrichment with differentially expressed genes between the diet groups at t = 0, the second
LTA Analysis of HapMap Genotype Data
 LTA Analysis of HapMap Genotype Data Introduction. This supplement to Global variation in copy number in the human genome, by Redon et al., describes the details of the LTA analysis used to screen HapMap
LTA Analysis of HapMap Genotype Data Introduction. This supplement to Global variation in copy number in the human genome, by Redon et al., describes the details of the LTA analysis used to screen HapMap
Expert-guided Visual Exploration (EVE) for patient stratification. Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland
 Expert-guided Visual Exploration (EVE) for patient stratification Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland Oncoscape.sttrcancer.org Paul Lisa Ken Jenny Desert Eric The challenge Given - patient clinical
Expert-guided Visual Exploration (EVE) for patient stratification Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland Oncoscape.sttrcancer.org Paul Lisa Ken Jenny Desert Eric The challenge Given - patient clinical
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/7/303/303ra139/dc1 Supplementary Materials for Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular
www.sciencetranslationalmedicine.org/cgi/content/full/7/303/303ra139/dc1 Supplementary Materials for Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular
False Discovery Rates and Copy Number Variation. Bradley Efron and Nancy Zhang Stanford University
 False Discovery Rates and Copy Number Variation Bradley Efron and Nancy Zhang Stanford University Three Statistical Centuries 19th (Quetelet) Huge data sets, simple questions 20th (Fisher, Neyman, Hotelling,...
False Discovery Rates and Copy Number Variation Bradley Efron and Nancy Zhang Stanford University Three Statistical Centuries 19th (Quetelet) Huge data sets, simple questions 20th (Fisher, Neyman, Hotelling,...
CPM (x 10-3 ) Tregs +Teffs. Tregs alone ICOS CLTA-4
 A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
Figure 1: Effects of cisplatin on survival of lung cancer cells.
 Figure 1 Figure 1: Effects of cisplatin on survival of lung cancer cells. To determine the IC 50 concentration of cisplatin, cells were treated with various concentrations of cisplatin and cell survival
Figure 1 Figure 1: Effects of cisplatin on survival of lung cancer cells. To determine the IC 50 concentration of cisplatin, cells were treated with various concentrations of cisplatin and cell survival
Development of Carcinoma Pathways
 The Construction of Genetic Pathway to Colorectal Cancer Moriah Wright, MD Clinical Fellow in Colorectal Surgery Creighton University School of Medicine Management of Colon and Diseases February 23, 2019
The Construction of Genetic Pathway to Colorectal Cancer Moriah Wright, MD Clinical Fellow in Colorectal Surgery Creighton University School of Medicine Management of Colon and Diseases February 23, 2019
Figure S2. Distribution of acgh probes on all ten chromosomes of the RIL M0022
 96 APPENDIX B. Supporting Information for chapter 4 "changes in genome content generated via segregation of non-allelic homologs" Figure S1. Potential de novo CNV probes and sizes of apparently de novo
96 APPENDIX B. Supporting Information for chapter 4 "changes in genome content generated via segregation of non-allelic homologs" Figure S1. Potential de novo CNV probes and sizes of apparently de novo
Supplementary Material
 Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Risk-prediction modelling in cancer with multiple genomic data sets: a Bayesian variable selection approach
 Risk-prediction modelling in cancer with multiple genomic data sets: a Bayesian variable selection approach Manuela Zucknick Division of Biostatistics, German Cancer Research Center Biometry Workshop,
Risk-prediction modelling in cancer with multiple genomic data sets: a Bayesian variable selection approach Manuela Zucknick Division of Biostatistics, German Cancer Research Center Biometry Workshop,
Error Detection based on neural signals
 Error Detection based on neural signals Nir Even- Chen and Igor Berman, Electrical Engineering, Stanford Introduction Brain computer interface (BCI) is a direct communication pathway between the brain
Error Detection based on neural signals Nir Even- Chen and Igor Berman, Electrical Engineering, Stanford Introduction Brain computer interface (BCI) is a direct communication pathway between the brain
Case Study - Informatics
 bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
bd@jubilantbiosys.com Case Study - Informatics www.jubilantbiosys.com Validating as a Prognostic marker and Therapeutic Target for Multiple Cancers Introduction Human genome projects and high throughput
Multiplexed Cancer Pathway Analysis
 NanoString Technologies, Inc. Multiplexed Cancer Pathway Analysis for Gene Expression Lucas Dennis, Patrick Danaher, Rich Boykin, Joseph Beechem NanoString Technologies, Inc., Seattle WA 98109 v1.0 MARCH
NanoString Technologies, Inc. Multiplexed Cancer Pathway Analysis for Gene Expression Lucas Dennis, Patrick Danaher, Rich Boykin, Joseph Beechem NanoString Technologies, Inc., Seattle WA 98109 v1.0 MARCH
Identifying Causal Genes and Dysregulated Pathways in Complex Diseases
 Identifying Causal Genes and Dysregulated Pathways in Complex Diseases Yoo-Ah Kim, Stefan Wuchty, Teresa M. Przytycka* National Center for Biotechnology Information, National Library of Medicine, National
Identifying Causal Genes and Dysregulated Pathways in Complex Diseases Yoo-Ah Kim, Stefan Wuchty, Teresa M. Przytycka* National Center for Biotechnology Information, National Library of Medicine, National
Cancer. The fundamental defect is. unregulated cell division. Properties of Cancerous Cells. Causes of Cancer. Altered growth and proliferation
 Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Supplementary Figure 1. Copy Number Alterations TP53 Mutation Type. C-class TP53 WT. TP53 mut. Nature Genetics: doi: /ng.
 Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
An improved hybrid of SVM and SCAD for pathway analysis
 www.bioinformation.net Hypothesis Volume 7(4) An improved hybrid of SVM and SCAD for pathway analysis Muhammad Faiz Misman*, Mohd Saberi Mohamad, Safaai Deris, Afnizanfaizal Abdullah, Siti Zaiton Mohd
www.bioinformation.net Hypothesis Volume 7(4) An improved hybrid of SVM and SCAD for pathway analysis Muhammad Faiz Misman*, Mohd Saberi Mohamad, Safaai Deris, Afnizanfaizal Abdullah, Siti Zaiton Mohd
S1 Appendix: Figs A G and Table A. b Normal Generalized Fraction 0.075
 Aiello & Alter (216) PLoS One vol. 11 no. 1 e164546 S1 Appendix A-1 S1 Appendix: Figs A G and Table A a Tumor Generalized Fraction b Normal Generalized Fraction.25.5.75.25.5.75 1 53 4 59 2 58 8 57 3 48
Aiello & Alter (216) PLoS One vol. 11 no. 1 e164546 S1 Appendix A-1 S1 Appendix: Figs A G and Table A a Tumor Generalized Fraction b Normal Generalized Fraction.25.5.75.25.5.75 1 53 4 59 2 58 8 57 3 48
Outlier Analysis. Lijun Zhang
 Outlier Analysis Lijun Zhang zlj@nju.edu.cn http://cs.nju.edu.cn/zlj Outline Introduction Extreme Value Analysis Probabilistic Models Clustering for Outlier Detection Distance-Based Outlier Detection Density-Based
Outlier Analysis Lijun Zhang zlj@nju.edu.cn http://cs.nju.edu.cn/zlj Outline Introduction Extreme Value Analysis Probabilistic Models Clustering for Outlier Detection Distance-Based Outlier Detection Density-Based
Meta-analysis of gene coexpression networks in the post-mortem prefrontal cortex of patients with schizophrenia and unaffected controls
 Mistry et al. BMC Neuroscience 2013, 14:105 RESEARCH ARTICLE Open Access Meta-analysis of gene coexpression networks in the post-mortem prefrontal cortex of patients with schizophrenia and unaffected controls
Mistry et al. BMC Neuroscience 2013, 14:105 RESEARCH ARTICLE Open Access Meta-analysis of gene coexpression networks in the post-mortem prefrontal cortex of patients with schizophrenia and unaffected controls
Nature Genetics: doi: /ng Supplementary Figure 1. Rates of different mutation types in CRC.
 Supplementary Figure 1 Rates of different mutation types in CRC. (a) Stratification by mutation type indicates that C>T mutations occur at a significantly greater rate than other types. (b) As for the
Supplementary Figure 1 Rates of different mutation types in CRC. (a) Stratification by mutation type indicates that C>T mutations occur at a significantly greater rate than other types. (b) As for the
Search settings MaxQuant
 Search settings MaxQuant Briefly, we used MaxQuant version 1.5.0.0 with the following settings. As variable modifications we allowed Acetyl (Protein N-terminus), methionine oxidation and glutamine to pyroglutamate
Search settings MaxQuant Briefly, we used MaxQuant version 1.5.0.0 with the following settings. As variable modifications we allowed Acetyl (Protein N-terminus), methionine oxidation and glutamine to pyroglutamate
Biostatistical modelling in genomics for clinical cancer studies
 This work was supported by Entente Cordiale Cancer Research Bursaries Biostatistical modelling in genomics for clinical cancer studies Philippe Broët JE 2492 Faculté de Médecine Paris-Sud In collaboration
This work was supported by Entente Cordiale Cancer Research Bursaries Biostatistical modelling in genomics for clinical cancer studies Philippe Broët JE 2492 Faculté de Médecine Paris-Sud In collaboration
Nature Methods: doi: /nmeth.3115
 Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
November 9, Johns Hopkins School of Medicine, Baltimore, MD,
 Fast detection of de-novo copy number variants from case-parent SNP arrays identifies a deletion on chromosome 7p14.1 associated with non-syndromic isolated cleft lip/palate Samuel G. Younkin 1, Robert
Fast detection of de-novo copy number variants from case-parent SNP arrays identifies a deletion on chromosome 7p14.1 associated with non-syndromic isolated cleft lip/palate Samuel G. Younkin 1, Robert
Susceptibility Prediction in Familial Colon Cancer
 Susceptibility Prediction in Familial Colon Cancer Giovanni Parmigiani gp@jhu.edu Cancer Risk Prediction Models: A Workshop on Development, Evaluation, and Application NCI, May 2004 CROSS-PLATFORM COMPARISON
Susceptibility Prediction in Familial Colon Cancer Giovanni Parmigiani gp@jhu.edu Cancer Risk Prediction Models: A Workshop on Development, Evaluation, and Application NCI, May 2004 CROSS-PLATFORM COMPARISON
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice
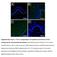 Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Nature Genetics: doi: /ng Supplementary Figure 1. Mutational signatures in BCC compared to melanoma.
 Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
FONS Nové sekvenační technologie vklinickédiagnostice?
 FONS 2010 Nové sekvenační technologie vklinickédiagnostice? Sekvenování amplikonů Sequence capture Celogenomové sekvenování FONS 2010 Sekvenování amplikonů Amplicon sequencing - amplicon sequencing enables
FONS 2010 Nové sekvenační technologie vklinickédiagnostice? Sekvenování amplikonů Sequence capture Celogenomové sekvenování FONS 2010 Sekvenování amplikonů Amplicon sequencing - amplicon sequencing enables
CoINcIDE: A framework for discovery of patient subtypes across multiple datasets
 Planey and Gevaert Genome Medicine (2016) 8:27 DOI 10.1186/s13073-016-0281-4 METHOD CoINcIDE: A framework for discovery of patient subtypes across multiple datasets Catherine R. Planey and Olivier Gevaert
Planey and Gevaert Genome Medicine (2016) 8:27 DOI 10.1186/s13073-016-0281-4 METHOD CoINcIDE: A framework for discovery of patient subtypes across multiple datasets Catherine R. Planey and Olivier Gevaert
Fluxion Biosciences and Swift Biosciences Somatic variant detection from liquid biopsy samples using targeted NGS
 APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
APPLICATION NOTE Fluxion Biosciences and Swift Biosciences OVERVIEW This application note describes a robust method for detecting somatic mutations from liquid biopsy samples by combining circulating tumor
Inferring Biological Meaning from Cap Analysis Gene Expression Data
 Inferring Biological Meaning from Cap Analysis Gene Expression Data HRYSOULA PAPADAKIS 1. Introduction This project is inspired by the recent development of the Cap analysis gene expression (CAGE) method,
Inferring Biological Meaning from Cap Analysis Gene Expression Data HRYSOULA PAPADAKIS 1. Introduction This project is inspired by the recent development of the Cap analysis gene expression (CAGE) method,
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
Aspects of Statistical Modelling & Data Analysis in Gene Expression Genomics. Mike West Duke University
 Aspects of Statistical Modelling & Data Analysis in Gene Expression Genomics Mike West Duke University Papers, software, many links: www.isds.duke.edu/~mw ABS04 web site: Lecture slides, stats notes, papers,
Aspects of Statistical Modelling & Data Analysis in Gene Expression Genomics Mike West Duke University Papers, software, many links: www.isds.duke.edu/~mw ABS04 web site: Lecture slides, stats notes, papers,
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Illustration of the working of network-based SVM to confidently predict a new (and now confirmed) ASD gene. Gene CTNND2 s brain network neighborhood that enabled its prediction by
Supplementary Figure 1 Illustration of the working of network-based SVM to confidently predict a new (and now confirmed) ASD gene. Gene CTNND2 s brain network neighborhood that enabled its prediction by
Cancer. The fundamental defect is. unregulated cell division. Properties of Cancerous Cells. Causes of Cancer. Altered growth and proliferation
 Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Cancer The fundamental defect is unregulated cell division. Properties of Cancerous Cells Altered growth and proliferation Loss of growth factor dependence Loss of contact inhibition Immortalization Alterated
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Gene Ontology 2 Function/Pathway Enrichment. Biol4559 Thurs, April 12, 2018 Bill Pearson Pinn 6-057
 Gene Ontology 2 Function/Pathway Enrichment Biol4559 Thurs, April 12, 2018 Bill Pearson wrp@virginia.edu 4-2818 Pinn 6-057 Function/Pathway enrichment analysis do sets (subsets) of differentially expressed
Gene Ontology 2 Function/Pathway Enrichment Biol4559 Thurs, April 12, 2018 Bill Pearson wrp@virginia.edu 4-2818 Pinn 6-057 Function/Pathway enrichment analysis do sets (subsets) of differentially expressed
Introduction to Computational Neuroscience
 Introduction to Computational Neuroscience Lecture 5: Data analysis II Lesson Title 1 Introduction 2 Structure and Function of the NS 3 Windows to the Brain 4 Data analysis 5 Data analysis II 6 Single
Introduction to Computational Neuroscience Lecture 5: Data analysis II Lesson Title 1 Introduction 2 Structure and Function of the NS 3 Windows to the Brain 4 Data analysis 5 Data analysis II 6 Single
Cancer outlier differential gene expression detection
 Biostatistics (2007), 8, 3, pp. 566 575 doi:10.1093/biostatistics/kxl029 Advance Access publication on October 4, 2006 Cancer outlier differential gene expression detection BAOLIN WU Division of Biostatistics,
Biostatistics (2007), 8, 3, pp. 566 575 doi:10.1093/biostatistics/kxl029 Advance Access publication on October 4, 2006 Cancer outlier differential gene expression detection BAOLIN WU Division of Biostatistics,
cn.mops - Mixture of Poissons for CNV detection in NGS data Günter Klambauer Institute of Bioinformatics, Johannes Kepler University Linz
 Software Manual Institute of Bioinformatics, Johannes Kepler University Linz cn.mops - Mixture of Poissons for CNV detection in NGS data Günter Klambauer Institute of Bioinformatics, Johannes Kepler University
Software Manual Institute of Bioinformatics, Johannes Kepler University Linz cn.mops - Mixture of Poissons for CNV detection in NGS data Günter Klambauer Institute of Bioinformatics, Johannes Kepler University
Nature Methods: doi: /nmeth Supplementary Figure 1. Activity in turtle dorsal cortex is sparse.
 Supplementary Figure 1 Activity in turtle dorsal cortex is sparse. a. Probability distribution of firing rates across the population (notice log scale) in our data. The range of firing rates is wide but
Supplementary Figure 1 Activity in turtle dorsal cortex is sparse. a. Probability distribution of firing rates across the population (notice log scale) in our data. The range of firing rates is wide but
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD
 Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Introduction to LOH and Allele Specific Copy Number User Forum
 Introduction to LOH and Allele Specific Copy Number User Forum Jonathan Gerstenhaber Introduction to LOH and ASCN User Forum Contents 1. Loss of heterozygosity Analysis procedure Types of baselines 2.
Introduction to LOH and Allele Specific Copy Number User Forum Jonathan Gerstenhaber Introduction to LOH and ASCN User Forum Contents 1. Loss of heterozygosity Analysis procedure Types of baselines 2.
Whole-genome detection of disease-associated deletions or excess homozygosity in a case control study of rheumatoid arthritis
 HMG Advance Access published December 21, 2012 Human Molecular Genetics, 2012 1 13 doi:10.1093/hmg/dds512 Whole-genome detection of disease-associated deletions or excess homozygosity in a case control
HMG Advance Access published December 21, 2012 Human Molecular Genetics, 2012 1 13 doi:10.1093/hmg/dds512 Whole-genome detection of disease-associated deletions or excess homozygosity in a case control
Biomarker adaptive designs in clinical trials
 Review Article Biomarker adaptive designs in clinical trials James J. Chen 1, Tzu-Pin Lu 1,2, Dung-Tsa Chen 3, Sue-Jane Wang 4 1 Division of Bioinformatics and Biostatistics, National Center for Toxicological
Review Article Biomarker adaptive designs in clinical trials James J. Chen 1, Tzu-Pin Lu 1,2, Dung-Tsa Chen 3, Sue-Jane Wang 4 1 Division of Bioinformatics and Biostatistics, National Center for Toxicological
Model-Based Detection of Spiculated Lesions in Mammograms
 Medical Image Analysis (1998) volume 3, number?, pp 1 23 c Oxford University Press Model-Based Detection of Spiculated Lesions in Mammograms R. Zwiggelaar 1, T.C. Parr 1, J.E. Schumm 1, I.W. Hutt 1, C.J.
Medical Image Analysis (1998) volume 3, number?, pp 1 23 c Oxford University Press Model-Based Detection of Spiculated Lesions in Mammograms R. Zwiggelaar 1, T.C. Parr 1, J.E. Schumm 1, I.W. Hutt 1, C.J.
Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes.
 Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Integrative analysis of survival-associated gene sets in breast cancer
 Varn et al. BMC Medical Genomics (2015) 8:11 DOI 10.1186/s12920-015-0086-0 RESEARCH ARTICLE Open Access Integrative analysis of survival-associated gene sets in breast cancer Frederick S Varn 1, Matthew
Varn et al. BMC Medical Genomics (2015) 8:11 DOI 10.1186/s12920-015-0086-0 RESEARCH ARTICLE Open Access Integrative analysis of survival-associated gene sets in breast cancer Frederick S Varn 1, Matthew
Review: Genome assembly Reads
 Assembly validation Review: Genome assembly Reads Contigs Scaffolds Chromosome Review: Mate pair data Overlap-Layout-Consensus AMOS project: A Modular Open Source assembler Importing data to an AMOS bank
Assembly validation Review: Genome assembly Reads Contigs Scaffolds Chromosome Review: Mate pair data Overlap-Layout-Consensus AMOS project: A Modular Open Source assembler Importing data to an AMOS bank
Identification of regions with common copy-number variations using SNP array
 Identification of regions with common copy-number variations using SNP array Agus Salim Epidemiology and Public Health National University of Singapore Copy Number Variation (CNV) Copy number alteration
Identification of regions with common copy-number variations using SNP array Agus Salim Epidemiology and Public Health National University of Singapore Copy Number Variation (CNV) Copy number alteration
ARTICLE RESEARCH. Macmillan Publishers Limited. All rights reserved
 Extended Data Figure 6 Annotation of drivers based on clinical characteristics and co-occurrence patterns. a, Putative drivers affecting greater than 10 patients were assessed for enrichment in IGHV mutated
Extended Data Figure 6 Annotation of drivers based on clinical characteristics and co-occurrence patterns. a, Putative drivers affecting greater than 10 patients were assessed for enrichment in IGHV mutated
Numerous hypothesis tests were performed in this study. To reduce the false positive due to
 Two alternative data-splitting Numerous hypothesis tests were performed in this study. To reduce the false positive due to multiple testing, we are not only seeking the results with extremely small p values
Two alternative data-splitting Numerous hypothesis tests were performed in this study. To reduce the false positive due to multiple testing, we are not only seeking the results with extremely small p values
T-Cell Network Example for GeneNet (August 2015) or later
 T-Cell Network Example for GeneNet 1.2.13 (August 2015) or later This note reproduces the T-Cell network example from R. Opgen-Rhein and K. Strimmer. 2006a. Using regularized dynamic correlation to infer
T-Cell Network Example for GeneNet 1.2.13 (August 2015) or later This note reproduces the T-Cell network example from R. Opgen-Rhein and K. Strimmer. 2006a. Using regularized dynamic correlation to infer
Bioimaging and Functional Genomics
 Bioimaging and Functional Genomics Elisa Ficarra, EPF Lausanne Giovanni De Micheli, EPF Lausanne Sungroh Yoon, Stanford University Luca Benini, University of Bologna Enrico Macii,, Politecnico di Torino
Bioimaging and Functional Genomics Elisa Ficarra, EPF Lausanne Giovanni De Micheli, EPF Lausanne Sungroh Yoon, Stanford University Luca Benini, University of Bologna Enrico Macii,, Politecnico di Torino
Supplementary Information. Supplementary Figures
 Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Supplementary Material. Part I: Sample Information. Part II: Pathway Information
 Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Supplementary Material Part I: Sample Information Three NPC cell lines, CNE1, CNE2, and HK1 were treated with CYC202. Gene expression of 380 selected genes were collected at 0, 2, 4, 6, 12 and 24 hours
Lecture 20. Disease Genetics
 Lecture 20. Disease Genetics Michael Schatz April 12 2018 JHU 600.749: Applied Comparative Genomics Part 1: Pre-genome Era Sickle Cell Anaemia Sickle-cell anaemia (SCA) is an abnormality in the oxygen-carrying
Lecture 20. Disease Genetics Michael Schatz April 12 2018 JHU 600.749: Applied Comparative Genomics Part 1: Pre-genome Era Sickle Cell Anaemia Sickle-cell anaemia (SCA) is an abnormality in the oxygen-carrying
Nature Getetics: doi: /ng.3471
 Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Terapias personalizadas en cancer
 Terapias personalizadas en cancer Atanasio Pandiella Cancer Research Center Salamanca, Spain 20 Aniversario Farmacia Univali, Itajaì Genomic variability of tumors Personalised therapies in cancer To find
Terapias personalizadas en cancer Atanasio Pandiella Cancer Research Center Salamanca, Spain 20 Aniversario Farmacia Univali, Itajaì Genomic variability of tumors Personalised therapies in cancer To find
Clustered mutations of oncogenes and tumor suppressors.
 Supplementary Figure 1 Clustered mutations of oncogenes and tumor suppressors. For each oncogene (red dots) and tumor suppressor (blue dots), the number of mutations found in an intramolecular cluster
Supplementary Figure 1 Clustered mutations of oncogenes and tumor suppressors. For each oncogene (red dots) and tumor suppressor (blue dots), the number of mutations found in an intramolecular cluster
Screening for novel oncology biomarker panels using both DNA and protein microarrays. John Anson, PhD VP Biomarker Discovery
 Screening for novel oncology biomarker panels using both DNA and protein microarrays John Anson, PhD VP Biomarker Discovery Outline of presentation Introduction to OGT and our approach to biomarker studies
Screening for novel oncology biomarker panels using both DNA and protein microarrays John Anson, PhD VP Biomarker Discovery Outline of presentation Introduction to OGT and our approach to biomarker studies
TADA: Analyzing De Novo, Transmission and Case-Control Sequencing Data
 TADA: Analyzing De Novo, Transmission and Case-Control Sequencing Data Each person inherits mutations from parents, some of which may predispose the person to certain diseases. Meanwhile, new mutations
TADA: Analyzing De Novo, Transmission and Case-Control Sequencing Data Each person inherits mutations from parents, some of which may predispose the person to certain diseases. Meanwhile, new mutations
Frequency(%) KRAS G12 KRAS G13 KRAS A146 KRAS Q61 KRAS K117N PIK3CA H1047 PIK3CA E545 PIK3CA E542K PIK3CA Q546. EGFR exon19 NFS-indel EGFR L858R
 Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Next-Gen Analytics in Digital Pathology
 Next-Gen Analytics in Digital Pathology Cliff Hoyt, CTO Cambridge Research & Instrumentation April 29, 2010 Seeing life in a new light 1 Digital Pathology Today Acquisition, storage, dissemination, remote
Next-Gen Analytics in Digital Pathology Cliff Hoyt, CTO Cambridge Research & Instrumentation April 29, 2010 Seeing life in a new light 1 Digital Pathology Today Acquisition, storage, dissemination, remote
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits
 AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
MicroRNA expression profiling and functional analysis in prostate cancer. Marco Folini s.c. Ricerca Traslazionale DOSL
 MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
Feature Vector Denoising with Prior Network Structures. (with Y. Fan, L. Raphael) NESS 2015, University of Connecticut
 Feature Vector Denoising with Prior Network Structures (with Y. Fan, L. Raphael) NESS 2015, University of Connecticut Summary: I. General idea: denoising functions on Euclidean space ---> denoising in
Feature Vector Denoising with Prior Network Structures (with Y. Fan, L. Raphael) NESS 2015, University of Connecticut Summary: I. General idea: denoising functions on Euclidean space ---> denoising in
Bayesian Prediction Tree Models
 Bayesian Prediction Tree Models Statistical Prediction Tree Modelling for Clinico-Genomics Clinical gene expression data - expression signatures, profiling Tree models for predictive sub-typing Combining
Bayesian Prediction Tree Models Statistical Prediction Tree Modelling for Clinico-Genomics Clinical gene expression data - expression signatures, profiling Tree models for predictive sub-typing Combining
Introduction. We can make a prediction about Y i based on X i by setting a threshold value T, and predicting Y i = 1 when X i > T.
 Diagnostic Tests 1 Introduction Suppose we have a quantitative measurement X i on experimental or observed units i = 1,..., n, and a characteristic Y i = 0 or Y i = 1 (e.g. case/control status). The measurement
Diagnostic Tests 1 Introduction Suppose we have a quantitative measurement X i on experimental or observed units i = 1,..., n, and a characteristic Y i = 0 or Y i = 1 (e.g. case/control status). The measurement
Cost effective, computer-aided analytical performance evaluation of chromosomal microarrays for clinical laboratories
 University of Iowa Iowa Research Online Theses and Dissertations Summer 2012 Cost effective, computer-aided analytical performance evaluation of chromosomal microarrays for clinical laboratories Corey
University of Iowa Iowa Research Online Theses and Dissertations Summer 2012 Cost effective, computer-aided analytical performance evaluation of chromosomal microarrays for clinical laboratories Corey
Reward prediction based on stimulus categorization in. primate lateral prefrontal cortex
 Reward prediction based on stimulus categorization in primate lateral prefrontal cortex Xiaochuan Pan, Kosuke Sawa, Ichiro Tsuda, Minoro Tsukada, Masamichi Sakagami Supplementary Information This PDF file
Reward prediction based on stimulus categorization in primate lateral prefrontal cortex Xiaochuan Pan, Kosuke Sawa, Ichiro Tsuda, Minoro Tsukada, Masamichi Sakagami Supplementary Information This PDF file
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines.
 Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
Transform genomic data into real-life results
 CLINICAL SUMMARY Transform genomic data into real-life results Biomarker testing and targeted therapies can drive improved outcomes in clinical practice New FDA-Approved Broad Companion Diagnostic for
CLINICAL SUMMARY Transform genomic data into real-life results Biomarker testing and targeted therapies can drive improved outcomes in clinical practice New FDA-Approved Broad Companion Diagnostic for
Molecular Subtyping of Endometrial Cancer: A ProMisE ing Change
 Molecular Subtyping of Endometrial Cancer: A ProMisE ing Change Charles Matthew Quick, M.D. Associate Professor of Pathology Director of Gynecologic Pathology University of Arkansas for Medical Sciences
Molecular Subtyping of Endometrial Cancer: A ProMisE ing Change Charles Matthew Quick, M.D. Associate Professor of Pathology Director of Gynecologic Pathology University of Arkansas for Medical Sciences
Working Memory (Goal Maintenance and Interference Control) Edward E. Smith Columbia University
 Working Memory (Goal Maintenance and Interference Control) Edward E. Smith Columbia University Outline Goal Maintenance Interference resolution: distraction, proactive interference, and directed forgetting
Working Memory (Goal Maintenance and Interference Control) Edward E. Smith Columbia University Outline Goal Maintenance Interference resolution: distraction, proactive interference, and directed forgetting
Clinical evaluation of microarray data
 Clinical evaluation of microarray data David Amor 19 th June 2011 Single base change Microarrays 3-4Mb What is a microarray? Up to 10 6 bits of Information!! Highly multiplexed FISH hybridisations. Microarray
Clinical evaluation of microarray data David Amor 19 th June 2011 Single base change Microarrays 3-4Mb What is a microarray? Up to 10 6 bits of Information!! Highly multiplexed FISH hybridisations. Microarray
