USA. Supplemental Information
|
|
|
- Jody Shelton
- 5 years ago
- Views:
Transcription
1 Cloning and sequencing of the kedarcidin biosynthetic gene cluster from Streptoalloteichus sp. ATCC revealing new insights into biosynthesis of the enediyne family of antitumor antibiotics Jeremy R. Lohman, a Sheng-Xiong Huang, a Geoffrey P. Horsman, b Paul E. Dilfer, a Tingting Huang, a Yihua Chen, b Evelyn Wendt-Pienkowski, b and Ben Shen *,a,b,c,d a Department of Chemistry, The Scripps Research Institute, Jupiter, Florida 33458, USA b Division of Pharmaceutical Sciences, School of Pharmacy, University of Wisconsin-Madison, Madison, Wisconsin, 53705, USA c Department of Molecular Therapeutics, The Scripps Research Institute, Jupiter, Florida 33458, USA d Natural Products Library Initiative at The Scripps Research Institute, The Scripps Research Institute, Jupiter, Florida 33458, USA * The Scripps Research Institute, 130 Scripps Way, #3A1, Jupiter, Florida 33458, USA; Tel: (561) ; Fax: (561) ; shenb@scripps.edu Supplemental Information Fig. S1. The amino acid sequence of KedA in comparison with other known apoproteins Fig. S2. The original and revised structures of the KED chromophore Fig. S3. Enediyne natural products whose structures have been determined Fig. S4. HPLC and MS analysis of the KED chromophore Fig. S5. SDS-PAGE analysis of purified KedF Fig. S6. Comparative analysis of the KED, C-1027, and MDP gene clusters supporting the proposed pathway for (R)-2-aza-3-chloro- -tyrosine in KED biosynthesis Fig. S7. Comparative analysis of the KED, NCS, and MDP gene cluster supporting the proposed pathway for 3-hydroxy-7,8-dimethoxy-6-isopropoxy-2-naphthoic acid in KED biosynthesis References S2 S3 S4 S5 S6 S7 S8 S9 S1
2 Fig S1. The amino acid sequence of the KED apoprotein deduced from keda and comparison with the KED apoprotein determined by Edman degradation 1 as well as other known apoproteins. 2 KedA, deduced from keda (the keda gene has also been recently deposited to the GeneBank under accession no. ADG ); KedA*, determined by Edman degradation 1 ; CagA (accession no. D10827), C-1027; AxnA (D11457), the enediyne structure is not known (AxnA is identical to CagA except for the D122A variation, which could be resulted from sequence error); NcsA (D10996), neocarzinostatin; McmA (D90006), auromomycin (whose structure has not been determined). EFE72430 (from Streptomyces ghanaensis ATCC 14672), ACZ98704 (from Streptosporangium roseum), EDX26137 (from Streptomyces sp. Mg1), and ADJ47157 (from Amycolatopsis mediterranei, strain U-32) are predicted apoproteins from putative enediyne biosynthetic gene clusters unveiled during genome sequencing efforts. Neither their enediyne chromoprotein production nor the enediyne chromophore structures have been established. The leader peptides are shown in italic and the first amino acid of the mature apoproteins are shown in red. ACZ98704 was predicted to have two cut sites. In addition to the predicted N-terminus of Ala-Ser-Ala-Ala-Val, two variants of the KedA apoprotein with a Ser-Ala-Ala-Val or an Ala-Ala-Val terminus were isolated. 1 S2
3 Fig. S2. The original (1, proposed in ,5 ) and revised structures (2, revised in and 3, revised in 2007, 7 is considered to be the final correct structure) of kedarcidin (KED) chromophore. S3
4 Fig. S3. Enediyne natural products whose structures 8 have been determined: (A) 9-membered enediynes C-1027, neocarzinostatin (NCS), maduropetin (MDP), kedarcidin (KED), N1999A2, sporolides A and B, cyanosporasides A and B, and fijiolides A and B. C-1027, NCS, MDP, and KED were isolated as chromproteins. N1999A2 was isolated as the enedyne chromophore alone. The sporolides, cyanosporasides, and fijiolides were isolated as aromatized products, whose enediyne forms were hypothetical; no apoprotein has been associated with these compounds. (B) 10-membered enediynes, all of which were isolated as discrete small molecules. S4
5 Fig S4. HPLC and HRESIMS analysis of the KED chromophore confirming the enediyne form and supporting the aromatized forms. 9,10 (I) HPLC chromatograms showing freshly prepared EtOAc extract of the KED chromophore from a KED chromoprotein sample (top panel) and the same EtOAc extract kept at room temperature overnight (bottom panel). Enediyne form ( ); aromatized form ( ). (II) Structures of the enediyne and aromatized forms of the KED chromophore supported by HRESIMS analysis. HRESIMS spectra of (III) the KED chromophore enediyne form with [M + H] + ion at m/z and (IV) the KED chromophore aromatized form with [M + H] + ion at m/z S5
6 Fig S5. SDS-PAGE (5-15%) analysis of purified KedF used in characterizing KedF as an epoxide hydrolase: lane 1, MW standard and lane 2, purified KedF. The predicted molecular weight of the recombinant KedF is 44.8 kda. S6
7 Fig S6. Comparative analysis of the KED, C-1027, and MDP biosynthetic gene clusters supporting the proposed pathway for (R)-2-aza-3-chloro- -tyrosine in KED biosynthesis: (A) genes encoding enzymes with high sequence homology among the three clusters for the L-tyrosine-derived moieties in C-1027 and MDP biosynthesis and for (R)-2-aza-3-chloro- -tyrosine in KED biosynthesis and (B) their proposed pathways. S7
8 Fig. S7. Comparative analysis of the KED, NCS, and MDP biosynthetic gene clusters supporting the proposed pathway for 3-hydroxy-7,8-dimethoxy-6-isopropoxy-2-naphthoic acid in KED biosynthesis: (A) genes encoding enzymes with high sequence homology among the three clusters for aromatic acid moieties in KED, NCS and MDP biosynthesis, (B) domain organization of the three iterative type I PKSs MdpB, NcsB, and KedU38 (KS, ketosynthase; AT, acyl transferase; DH, dehydratase; KR, ketoreductase; ACP, acyl carrier protein) and their proposed pathways for the biosynthesis of the nascent aromatic acids, and (C) tailoring steps for converting the nascent aromatic acids to fully modified aromatic acid moieties and their activation and incorporation in MDP, NCS, and KED biosynthesis. S8
9 References 1. S. J. Hofstead, J. A. Matson, A. R. Malacko and H. Marquardt, J. Antibiot. 1992, 45, J. S. Thorson, B. Shen, R. E. Whitwam, W. Liu and Y. Li, Bioorg. Chem. 1999, 27, V. T. T. Hang, T. S. Kim, T.-J. Oh and J. K. Sohng, Biotechnol. Bioproc. Eng. 2011, 16, J. E. Leet, D. R. Schroeder, S. J. Hofstead, J. Golik, K. L. Colson, S. Huang, S. E. Klohr, T. W. Doyle and J. A. Matson, J. Am. Chem. Soc. 1992, 114, J. E. Leet, D. R. Schroeder, D. R. Langley, K. L. Colson, S. Huang, S. E. Klohr, M. S. Lee, J. Golik, S. J. Hofstead, T. W. Doyle and J. A. Matson, J. Am. Chem. Soc. 1993, 115, S. Kawata, S. Ashizawa and M. Hirama, J. Am. Chem. Soc. 1997, 119, F. Ren, P. C. Hogan, A. J. Anderson and A. G. Myers, J. Am. Chem. Soc. 2007, 129, S. G. Van Lanen and B. Shen, Curr. Topics Med. Chem. 2008, 8, A. G. Myers, A. R. Hurd and P. C. Hogan, J. Am. Chem. Soc. 2002, 124, K. Ogawa, Y. Koyama, I. Ohashi, I. Sato and M Hirama, Angew. Chem. Int. Ed. 2009, 48, S9
The Chemistry of Nine-Membered. Enediyne Natural Products
 The Chemistry of ine-mbered Enediyne atural Products Zhang Wang MacMillan Group eting April 17, 2013 atural Products Sharing a Unique Structure C cyanosporaside A [] R 2 R sporolide A C [] [] R 1 R 1 =,
The Chemistry of ine-mbered Enediyne atural Products Zhang Wang MacMillan Group eting April 17, 2013 atural Products Sharing a Unique Structure C cyanosporaside A [] R 2 R sporolide A C [] [] R 1 R 1 =,
Published on Web 10/06/2007
 Published on Web 10/06/2007 Characterization of the Maduropeptin Biosynthetic Gene Cluster from Actinomadura madurae ATCC 39144 Supporting a Unifying Paradigm for Enediyne Biosynthesis Steven G. Van Lanen,
Published on Web 10/06/2007 Characterization of the Maduropeptin Biosynthetic Gene Cluster from Actinomadura madurae ATCC 39144 Supporting a Unifying Paradigm for Enediyne Biosynthesis Steven G. Van Lanen,
Enediyne Natural Products: Biosynthesis and Prospect Towards Engineering Novel Antitumor Agents
 Current Medicinal Chemistry, 2003, 10, 2317-2325 2317 Enediyne atural Products: Biosynthesis and Prospect Towards Engineering ovel Antitumor Agents Ben Shen 1,2*, Wen Liu 1 and Koichi onaka 1 1 Division
Current Medicinal Chemistry, 2003, 10, 2317-2325 2317 Enediyne atural Products: Biosynthesis and Prospect Towards Engineering ovel Antitumor Agents Ben Shen 1,2*, Wen Liu 1 and Koichi onaka 1 1 Division
Antitumor Antibiotics: Bleomycin, Enediynes, and Mitomycin
 Chem. Rev. 2005, 105, 739 758 739 Antitumor Antibiotics: Bleomycin, Enediynes, and Mitomycin Ute Galm, Martin H. Hager, Steven G. Van Lanen, Jianhua Ju, Jon S. Thorson,*, and Ben Shen*,, Division of Pharmaceutical
Chem. Rev. 2005, 105, 739 758 739 Antitumor Antibiotics: Bleomycin, Enediynes, and Mitomycin Ute Galm, Martin H. Hager, Steven G. Van Lanen, Jianhua Ju, Jon S. Thorson,*, and Ben Shen*,, Division of Pharmaceutical
Investigations of Pactamycin Biosynthesis Andrew Osborn
 Investigations of Pactamycin Biosynthesis Andrew Osborn Research Mentors: Dr. Taifo Mahmud, Dr. Kerry McPhail A Bioresource Research Thesis Seminar Presentation Streptomyces pactum produces the antitumor
Investigations of Pactamycin Biosynthesis Andrew Osborn Research Mentors: Dr. Taifo Mahmud, Dr. Kerry McPhail A Bioresource Research Thesis Seminar Presentation Streptomyces pactum produces the antitumor
Lomaiviticin Biosynthesis Employs a New Strategy for Starter Unit Generation
 Lomaiviticin Biosynthesis Employs a New Strategy for Starter Unit Generation The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters
Lomaiviticin Biosynthesis Employs a New Strategy for Starter Unit Generation The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters
Neocarzinostatin Complex: Chromophore Analogs, Binding Structure and Stabilization Mechanism
 Neocarzinostatin Complex: Chromophore Analogs, Binding Structure and Stabilization Mechanism Masahiro Hirama Department of Chemistry, Faculty of Science, Tohoku University, Abstract: Design, synthesis,
Neocarzinostatin Complex: Chromophore Analogs, Binding Structure and Stabilization Mechanism Masahiro Hirama Department of Chemistry, Faculty of Science, Tohoku University, Abstract: Design, synthesis,
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the
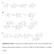 Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Critical Review Iterative Type I Polyketide Synthases Involved in Enediyne Natural Product Biosynthesis
 Critical Review Iterative Type I Polyketide Synthases Involved in Enediyne Natural Product Biosynthesis Xiaolei Chen 1* Rui Ji 2* Xin Jiang 3 Rongqiang Yang 2 Fuli Liu 4 Ying Xin 5 1 Department of Chemistry,
Critical Review Iterative Type I Polyketide Synthases Involved in Enediyne Natural Product Biosynthesis Xiaolei Chen 1* Rui Ji 2* Xin Jiang 3 Rongqiang Yang 2 Fuli Liu 4 Ying Xin 5 1 Department of Chemistry,
Two Types of Inulin Fructotransferases
 Materials 2011, 4, 1543-1547; doi:10.3390/ma4091543 Short Note OPEN ACCESS materials ISSN 1996-1944 www.mdpi.com/journal/materials Two Types of Inulin Fructotransferases Kazutomo Haraguchi National Food
Materials 2011, 4, 1543-1547; doi:10.3390/ma4091543 Short Note OPEN ACCESS materials ISSN 1996-1944 www.mdpi.com/journal/materials Two Types of Inulin Fructotransferases Kazutomo Haraguchi National Food
This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth 2 points.
 MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
Supplementary Information
 Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
The Neocarzinostatin Biosynthetic Gene Cluster from Streptomyces carzinostaticus ATCC Involving Two Iterative Type I Polyketide Synthases
 Chemistry & Biology, Vol. 12, 293 302, March, 2005, 2005 Elsevier Ltd All rights reserved. DOI 10.1016/j.chembiol.2004.12.013 The Neocarzinostatin Biosynthetic Gene Cluster from Streptomyces carzinostaticus
Chemistry & Biology, Vol. 12, 293 302, March, 2005, 2005 Elsevier Ltd All rights reserved. DOI 10.1016/j.chembiol.2004.12.013 The Neocarzinostatin Biosynthetic Gene Cluster from Streptomyces carzinostaticus
Supplementary Information
 Supplementary Information Assembly and Clustering of Natural Antibiotics Guides Target Identification Chad W. Johnston 1,2, Michael A. Skinnider 1,2, Chris A. Dejong 1,2, Philip N. Rees 1,2, Gregory M.
Supplementary Information Assembly and Clustering of Natural Antibiotics Guides Target Identification Chad W. Johnston 1,2, Michael A. Skinnider 1,2, Chris A. Dejong 1,2, Philip N. Rees 1,2, Gregory M.
Nucleophilic Addition to a p-benzyne Derived from an Enediyne: A New Mechanism for Halide Incorporation into Biomolecules
 Nucleophilic Addition to a p-benzyne Derived from an Enediyne: A New Mechanism for Halide Incorporation into Biomolecules Perrin, C. L.; Rodgers, B. L.; O Connor, J. M. J. Am. Chem. Soc. 2007, ASAP John
Nucleophilic Addition to a p-benzyne Derived from an Enediyne: A New Mechanism for Halide Incorporation into Biomolecules Perrin, C. L.; Rodgers, B. L.; O Connor, J. M. J. Am. Chem. Soc. 2007, ASAP John
Module Contact: Dr Paul McDermott, PHA Copyright of the University of East Anglia Version 1
 UIVERSITY F EAST AGLIA School of Pharmacy Main Series UG Examination 2012-13 LIFE SCIECES CHEMISTRY PHA-1ECY Time allowed: 2 hours You must answer FUR (4) of the SIX (6) questions. Use a SEPARATE answer
UIVERSITY F EAST AGLIA School of Pharmacy Main Series UG Examination 2012-13 LIFE SCIECES CHEMISTRY PHA-1ECY Time allowed: 2 hours You must answer FUR (4) of the SIX (6) questions. Use a SEPARATE answer
THE JOURNAL OF ANTIBIOTICS. Polyketomycin, a New Antibiotic from Streptomyces sp. MK277-AF1. II. Structure Determination
 THE JOURNAL OF ANTIBIOTICS Polyketomycin, a New Antibiotic from Streptomyces sp. MK277-AF1 II. Structure Determination ISAO MOMOSE, WEI CHEN, HIKARU NAKAMURA, HIROSHI NAGANAWA, HIRONOBU IINUMA and TOMIO
THE JOURNAL OF ANTIBIOTICS Polyketomycin, a New Antibiotic from Streptomyces sp. MK277-AF1 II. Structure Determination ISAO MOMOSE, WEI CHEN, HIKARU NAKAMURA, HIROSHI NAGANAWA, HIRONOBU IINUMA and TOMIO
SUPPORTING INFORMATION
 SUPPORTING INFORMATION Reduction of Aromatic and Heterocyclic Aromatic N-Hydroxylamines by Human Cytochrome P450 2S1 Wang, K., and Guengerich, F. P. (2013) Chem. Res. Toxicol. 26, 000-000 Figure S1. HPLC
SUPPORTING INFORMATION Reduction of Aromatic and Heterocyclic Aromatic N-Hydroxylamines by Human Cytochrome P450 2S1 Wang, K., and Guengerich, F. P. (2013) Chem. Res. Toxicol. 26, 000-000 Figure S1. HPLC
Human fatty acid synthase: Assembling recombinant halves of the fatty acid synthase subunit protein reconstitutes enzyme activity
 Proc. Natl. Acad. Sci. USA Vol. 94, pp. 12326 12330, November 1997 Biochemistry Human fatty acid synthase: Assembling recombinant halves of the fatty acid synthase subunit protein reconstitutes enzyme
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 12326 12330, November 1997 Biochemistry Human fatty acid synthase: Assembling recombinant halves of the fatty acid synthase subunit protein reconstitutes enzyme
Supporting Information
 Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
Glycolysis - Plasmodium
 Apicomplexan Biochemistry Basics Toxoplasma Good cell biology model Genome sequencing not completed Virtual pathways Cryptosporidium The strange one Genome sequence completed Virtual pathways Plasmodium
Apicomplexan Biochemistry Basics Toxoplasma Good cell biology model Genome sequencing not completed Virtual pathways Cryptosporidium The strange one Genome sequence completed Virtual pathways Plasmodium
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted. residues are colored red. Numbers indicate the position of each residue.
 Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
Lecture 3. Tandem MS & Protein Sequencing
 Lecture 3 Tandem MS & Protein Sequencing Nancy Allbritton, M.D., Ph.D. Department of Physiology & Biophysics 824-9137 (office) nlallbri@uci.edu Office- Rm D349 Medical Science D Bldg. Tandem MS Steps:
Lecture 3 Tandem MS & Protein Sequencing Nancy Allbritton, M.D., Ph.D. Department of Physiology & Biophysics 824-9137 (office) nlallbri@uci.edu Office- Rm D349 Medical Science D Bldg. Tandem MS Steps:
AA s are the building blocks of proteins
 Chamras Chemistry 106 Lecture otes Chapter 24: Amino Acids, Peptides, and Proteins General Formula: () n (') α-amino Acids: (n = 1) Example: Amino Acids and Proteins: Glycine Alanine Valine AA s are the
Chamras Chemistry 106 Lecture otes Chapter 24: Amino Acids, Peptides, and Proteins General Formula: () n (') α-amino Acids: (n = 1) Example: Amino Acids and Proteins: Glycine Alanine Valine AA s are the
Beta Amyloid Peptides
 Beta Amyloid Peptides The Most Comprehensive Collection for Alzheimer s Disease Academic Services Pharmaceutical Services Beta Amyloid Peptides The Most Comprehensive Collection for Alzheimer s Disease
Beta Amyloid Peptides The Most Comprehensive Collection for Alzheimer s Disease Academic Services Pharmaceutical Services Beta Amyloid Peptides The Most Comprehensive Collection for Alzheimer s Disease
sequence-specific manner
 Proc. Natl. Acad. Sci. USA Vol. 90, pp. 2822-2826, April 1993 Biochemistry Kedarcidin chromophore: An enediyne that cleaves DNA in a sequence-specific manner N. ZEIN*t, K. L. COLSONt, J. E. LEET*, D. R.
Proc. Natl. Acad. Sci. USA Vol. 90, pp. 2822-2826, April 1993 Biochemistry Kedarcidin chromophore: An enediyne that cleaves DNA in a sequence-specific manner N. ZEIN*t, K. L. COLSONt, J. E. LEET*, D. R.
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Department of Chemistry, University of California, Davis, One Shields Avenue, Davis, CA 95616, USA
 Journal of Industrial Microbiology & Biotechnology (2001) 27, 378 385 D 2001 Nature Publishing Group 1367-5435/01 $17.00 www.nature.com/jim The biosynthetic gene cluster for the anticancer drug bleomycin
Journal of Industrial Microbiology & Biotechnology (2001) 27, 378 385 D 2001 Nature Publishing Group 1367-5435/01 $17.00 www.nature.com/jim The biosynthetic gene cluster for the anticancer drug bleomycin
COMMUNICATION Acyl chain elongation drives ketosynthase substrate selectivity in trans-acyl transferase polyketide synthases
 Acyl chain elongation drives ketosynthase substrate selectivity in trans-acyl transferase polyketide synthases Matthew Jenner [a], José Pedro Afonso [a], Hannah R. Bailey [a], Sarah Frank [b], Annette
Acyl chain elongation drives ketosynthase substrate selectivity in trans-acyl transferase polyketide synthases Matthew Jenner [a], José Pedro Afonso [a], Hannah R. Bailey [a], Sarah Frank [b], Annette
Supporting Information
 Supporting Information A new series of cytotoxic pyrazoline derivatives as potential anticancer agents induces cell cycle arrest and apoptosis Hong Wang 1,, Jinhong Zheng 1,, Weijie Xu 1, Cheng Chen 1,
Supporting Information A new series of cytotoxic pyrazoline derivatives as potential anticancer agents induces cell cycle arrest and apoptosis Hong Wang 1,, Jinhong Zheng 1,, Weijie Xu 1, Cheng Chen 1,
Continuation of Antibiotics
 Continuation of Antibiotics CLASSIFICATIN F ANTIBITICS According to their mechanism of action Antibiotics as disturber with the biosynthesis of protein These antibiotics all target the bacterial ribosome
Continuation of Antibiotics CLASSIFICATIN F ANTIBITICS According to their mechanism of action Antibiotics as disturber with the biosynthesis of protein These antibiotics all target the bacterial ribosome
The biosynthetic gene cluster for the anticancer
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Liangcheng Du Publications Published Research - Department of Chemistry December 2001 The biosynthetic gene cluster for
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Liangcheng Du Publications Published Research - Department of Chemistry December 2001 The biosynthetic gene cluster for
Web: https://www.maynoothuniversity.ie/biology 1
 Running head: Integrated systems in Aspergillus fumigatus. Interplay between Gliotoxin Resistance, Secretion and the Methyl/Methionine Cycle in Aspergillus fumigatus. Rebecca A. Owens 1, Grainne O Keeffe
Running head: Integrated systems in Aspergillus fumigatus. Interplay between Gliotoxin Resistance, Secretion and the Methyl/Methionine Cycle in Aspergillus fumigatus. Rebecca A. Owens 1, Grainne O Keeffe
Supplementary Information
 Supplementary Information Precursors of trnas are stabilized by methylguanosine cap structures Takayuki Ohira and Tsutomu Suzuki Department of Chemistry and Biotechnology, Graduate School of Engineering,
Supplementary Information Precursors of trnas are stabilized by methylguanosine cap structures Takayuki Ohira and Tsutomu Suzuki Department of Chemistry and Biotechnology, Graduate School of Engineering,
Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis
 Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/1/e1500678/dc1 Supplementary Materials for Chemical synthesis of erythropoietin glycoforms for insights into the relationship between glycosylation pattern and
advances.sciencemag.org/cgi/content/full/2/1/e1500678/dc1 Supplementary Materials for Chemical synthesis of erythropoietin glycoforms for insights into the relationship between glycosylation pattern and
Finding the Sweet Spot- Mechanism Guided Design of Glycosidase Inhibitors. Jahnabi Roy CHEM 575 Seminar 11/01/12
 Finding the Sweet Spot- Mechanism Guided Design of Glycosidase Inhibitors Jahnabi Roy CHEM 575 Seminar 11/01/12 Glycans and Glycosyl Hydrolases http://cellbiology.med.unsw.edu.au/units/science/lecture0803.htm
Finding the Sweet Spot- Mechanism Guided Design of Glycosidase Inhibitors Jahnabi Roy CHEM 575 Seminar 11/01/12 Glycans and Glycosyl Hydrolases http://cellbiology.med.unsw.edu.au/units/science/lecture0803.htm
Metal Species for Amide Hydrolysis. Literature Seminar, Kiyomichi SHINODA (M1)
 Metal Species for Amide ydrolysis Literature Seminar, 20130511 Kiyomichi SIDA (M1) Table of Contents 1 Introduction 2 Residue- or Sequence-Selective ydrolysis of Amides 3 Protein-Selective ydrolysis of
Metal Species for Amide ydrolysis Literature Seminar, 20130511 Kiyomichi SIDA (M1) Table of Contents 1 Introduction 2 Residue- or Sequence-Selective ydrolysis of Amides 3 Protein-Selective ydrolysis of
BCH Graduate Survey of Biochemistry
 BCH 5045 Graduate Survey of Biochemistry Instructor: Charles Guy Producer: Ron Thomas Director: Glen Graham Lecture 7 Slide sets available at: http://hort.ifas.ufl.edu/teach/guyweb/bch5045/index.html David
BCH 5045 Graduate Survey of Biochemistry Instructor: Charles Guy Producer: Ron Thomas Director: Glen Graham Lecture 7 Slide sets available at: http://hort.ifas.ufl.edu/teach/guyweb/bch5045/index.html David
Level and activity of D-amino acids in mouse brain tissue and blood
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 SUPPLEMENTARY INFORMATION Level and activity of D-amino acids in mouse brain tissue and blood Choyce A. Weatherly 1, Siqi Du 1, Curran Parpia 1, Polan T. Santos 2, Adam
1 2 3 4 5 6 7 8 9 10 11 12 13 14 SUPPLEMENTARY INFORMATION Level and activity of D-amino acids in mouse brain tissue and blood Choyce A. Weatherly 1, Siqi Du 1, Curran Parpia 1, Polan T. Santos 2, Adam
Lecture 11 - Biosynthesis of Amino Acids
 Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
Automated synthesis of backbone protected peptides
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Electronic Supplementary Information Automated synthesis of backbone protected peptides Abu-Baker
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Electronic Supplementary Information Automated synthesis of backbone protected peptides Abu-Baker
Chapter 3: Amino Acids and Peptides
 Chapter 3: Amino Acids and Peptides BINF 6101/8101, Spring 2018 Outline 1. Overall amino acid structure 2. Amino acid stereochemistry 3. Amino acid sidechain structure & classification 4. Non-standard
Chapter 3: Amino Acids and Peptides BINF 6101/8101, Spring 2018 Outline 1. Overall amino acid structure 2. Amino acid stereochemistry 3. Amino acid sidechain structure & classification 4. Non-standard
Purification of Glucagon3 Interleukin-2 Fusion Protein Derived from E. coli
 Purification of Glucagon3 Interleukin-2 Fusion Protein Derived from E. coli Hye Soon Won Dept. of Chem. Eng. Chungnam National University INTRODUCTION Human interleukin-2(hil-2) - known as T Cell Growth
Purification of Glucagon3 Interleukin-2 Fusion Protein Derived from E. coli Hye Soon Won Dept. of Chem. Eng. Chungnam National University INTRODUCTION Human interleukin-2(hil-2) - known as T Cell Growth
Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis
 Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis Hongcheng Liu*, Gomathinayagam Ponniah, Alyssa Neil, Rekha Patel, Bruce Andrien Protein Characterization,
Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis Hongcheng Liu*, Gomathinayagam Ponniah, Alyssa Neil, Rekha Patel, Bruce Andrien Protein Characterization,
Supplementary Information. Amino group carrier protein-mediated secondary metabolite biosynthesis in Streptomyces
 Supplementary Information Amino group carrier protein-mediated secondary metabolite biosynthesis in Streptomyces Fumihito Hasebe 1, Kenichi Matsuda 1, Taro Shiraishi 1, Yushi Futamura 2, Takeshi Nakano
Supplementary Information Amino group carrier protein-mediated secondary metabolite biosynthesis in Streptomyces Fumihito Hasebe 1, Kenichi Matsuda 1, Taro Shiraishi 1, Yushi Futamura 2, Takeshi Nakano
Overproduction of ristomycin A by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17
 Supplemental Material for verproduction of ristomycin A by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17 Marius Spohn, a orbert Kirchner, b,c Andreas Kulik, a, Angelika Jochim,
Supplemental Material for verproduction of ristomycin A by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17 Marius Spohn, a orbert Kirchner, b,c Andreas Kulik, a, Angelika Jochim,
MITOCW watch?v=ddt1kusdoog
 MITOCW watch?v=ddt1kusdoog The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
MITOCW watch?v=ddt1kusdoog The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
Aflatoxin biosynthesis and regulation
 Aflatoxin biosynthesis and regulation Kenneth Ehrlich Southern Regional Research Center New rleans Introduction AF/ST biosynthesis involves expression of at least 25 genes Genes are organized in a cluster
Aflatoxin biosynthesis and regulation Kenneth Ehrlich Southern Regional Research Center New rleans Introduction AF/ST biosynthesis involves expression of at least 25 genes Genes are organized in a cluster
Chemical Biology, Option II Mechanism Based Proteomic Tagging Case History CH1
 Proteome Wide Screening of Serine Protease Activity Proc Natl Acad Sci 1999, 97, 14694; Proteomics 2001, 1, 1067; Proc Natl Acad Sci 2002, 99, 10335; Biochemistry 2001, 40, 4005; J. Am. Chem. Soc., 2005,
Proteome Wide Screening of Serine Protease Activity Proc Natl Acad Sci 1999, 97, 14694; Proteomics 2001, 1, 1067; Proc Natl Acad Sci 2002, 99, 10335; Biochemistry 2001, 40, 4005; J. Am. Chem. Soc., 2005,
,^ Ben Shen, and Steven D. Bruner*,
 9590 Biochemistry 2009, 48, 9590 9598 DOI: 10.1021/bi901257q Molecular Basis of Substrate Promiscuity for the SAM-Dependent O-Methyltransferase NcsB1, Involved in the Biosynthesis of the Enediyne Antitumor
9590 Biochemistry 2009, 48, 9590 9598 DOI: 10.1021/bi901257q Molecular Basis of Substrate Promiscuity for the SAM-Dependent O-Methyltransferase NcsB1, Involved in the Biosynthesis of the Enediyne Antitumor
Supporting Information
 Supporting Information Cyclic Peptidyl Inhibitors against Human Peptidyl-Prolyl Isomerase Pin1 Tao Liu, Yu Liu, Hung-Ying Kao, *,, and Dehua Pei Table of Contents: Table S1. Structures of amino acid building
Supporting Information Cyclic Peptidyl Inhibitors against Human Peptidyl-Prolyl Isomerase Pin1 Tao Liu, Yu Liu, Hung-Ying Kao, *,, and Dehua Pei Table of Contents: Table S1. Structures of amino acid building
VOL. 56 NO. 2, FEB THE JOURNAL OF ANTIBIOTICS pp YM and YM , Novel Thiopeptide Antibiotics Produced
 VOL. 56 NO. 2, FEB. 2003 THE JOURNAL OF ANTIBIOTICS pp. 129-134 YM-266183 and YM-266184, Novel Thiopeptide Antibiotics Produced by Bacillus cereus Isolated from a Marine Sponge II. Structure Elucidation
VOL. 56 NO. 2, FEB. 2003 THE JOURNAL OF ANTIBIOTICS pp. 129-134 YM-266183 and YM-266184, Novel Thiopeptide Antibiotics Produced by Bacillus cereus Isolated from a Marine Sponge II. Structure Elucidation
Supporting Information for:
 Supporting Information for: Methylerythritol Cyclodiphosphate (MEcPP) in Deoxyxylulose Phosphate Pathway: Synthesis from an Epoxide and Mechanisms Youli Xiao, a Rodney L. Nyland II, b Caren L. Freel Meyers
Supporting Information for: Methylerythritol Cyclodiphosphate (MEcPP) in Deoxyxylulose Phosphate Pathway: Synthesis from an Epoxide and Mechanisms Youli Xiao, a Rodney L. Nyland II, b Caren L. Freel Meyers
Improve Protein Analysis with the New, Mass Spectrometry- Compatible ProteasMAX Surfactant
 Improve Protein Analysis with the New, Mass Spectrometry- Compatible Surfactant ABSTRACT Incomplete solubilization and digestion and poor peptide recovery are frequent limitations in protein sample preparation
Improve Protein Analysis with the New, Mass Spectrometry- Compatible Surfactant ABSTRACT Incomplete solubilization and digestion and poor peptide recovery are frequent limitations in protein sample preparation
ESI and MALDI Mass Spectrometry. S. Sankararaman Department of Chemistry Indian Institute of Technology Madras Chennai
 ESI and MALDI Mass Spectrometry S. Sankararaman Department of Chemistry Indian Institute of Technology Madras Chennai 600036 sanka@iitm.ac.in MODULE 22 ESI-Q MASS SPECTROMETRY - examples Mol. Wt = 1140
ESI and MALDI Mass Spectrometry S. Sankararaman Department of Chemistry Indian Institute of Technology Madras Chennai 600036 sanka@iitm.ac.in MODULE 22 ESI-Q MASS SPECTROMETRY - examples Mol. Wt = 1140
Modeling holo-acp:dh and holo-acp:kr complexes of modular polyketide synthases: a docking and molecular dynamics study
 Anand and Mohanty BMC Structural Biology 2012, 12:10 RESEARCH ARTICLE Open Access Modeling holo-acp:dh and holo-acp:kr complexes of modular polyketide synthases: a docking and molecular dynamics study
Anand and Mohanty BMC Structural Biology 2012, 12:10 RESEARCH ARTICLE Open Access Modeling holo-acp:dh and holo-acp:kr complexes of modular polyketide synthases: a docking and molecular dynamics study
MS/MS Scan Modes. Eötvös University, Budapest April 16, MS/MS Scan Modes. Árpád Somogyi. Product Ion Scan Select. Scan. Precursor Ion Scan Scan
 MS/MS Modes Árpád Somogyi Eötvös University, Budapest April 16, 2012 MS/MS Modes Product Ion Precursor Ion Neutral Loss Δ ed Reaction Monitoring (SRM) 1 modes in a triple quadrupole (QqQ) (one quadrupole
MS/MS Modes Árpád Somogyi Eötvös University, Budapest April 16, 2012 MS/MS Modes Product Ion Precursor Ion Neutral Loss Δ ed Reaction Monitoring (SRM) 1 modes in a triple quadrupole (QqQ) (one quadrupole
Defining HbA1c: the indispensable decision to approach measurement standardization
 Defining HbA1c: the indispensable decision to approach measurement standardization Jan-Olof Jeppsson IFCC WG for HbA1c standardization Clinical Chemistry, Malmö University Hospital Sweden 6 November, 2007
Defining HbA1c: the indispensable decision to approach measurement standardization Jan-Olof Jeppsson IFCC WG for HbA1c standardization Clinical Chemistry, Malmö University Hospital Sweden 6 November, 2007
PREDICTING THE EXPRESSION AND SOLUBILITY OF MEMBRANE PROTEINS
 PREDICTING THE EXPRESSION AND SOLUBILITY OF MEMBRANE PROTEINS Mark E. Dumont, Michael A. White, Kathy Clark, Elizabeth J. Grayhack, and Eric. M. Phizicky Unknowns in Membrane Protein Expression, and Solubilization
PREDICTING THE EXPRESSION AND SOLUBILITY OF MEMBRANE PROTEINS Mark E. Dumont, Michael A. White, Kathy Clark, Elizabeth J. Grayhack, and Eric. M. Phizicky Unknowns in Membrane Protein Expression, and Solubilization
1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled
 Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
AMINO ACIDS STRUCTURE, CLASSIFICATION, PROPERTIES. PRIMARY STRUCTURE OF PROTEINS
 AMINO ACIDS STRUCTURE, CLASSIFICATION, PROPERTIES. PRIMARY STRUCTURE OF PROTEINS Elena Rivneac PhD, Associate Professor Department of Biochemistry and Clinical Biochemistry State University of Medicine
AMINO ACIDS STRUCTURE, CLASSIFICATION, PROPERTIES. PRIMARY STRUCTURE OF PROTEINS Elena Rivneac PhD, Associate Professor Department of Biochemistry and Clinical Biochemistry State University of Medicine
Three Dimensional Mapping and Imaging of Neuropeptides and Lipids in Crustacean Brain
 Three Dimensional Mapping and Imaging of Neuropeptides and Lipids in Crustacean Brain Using the 4800 MALDI TOF/TOF Analyzer Ruibing Chen and Lingjun Li School of Pharmacy and Department of Chemistry, University
Three Dimensional Mapping and Imaging of Neuropeptides and Lipids in Crustacean Brain Using the 4800 MALDI TOF/TOF Analyzer Ruibing Chen and Lingjun Li School of Pharmacy and Department of Chemistry, University
SUPPORTING INFORMATION FOR
 SUPPORTING INFORMATION FOR Cytotoxic bagremycins from Mangrove-derived Streptomyces sp. Q22 Lei Chen, Weiyun Chai, Wenling Wang, Tengfei Song, Xiao-Yuan Lian,*,, Zhizhen Zhang,*, *Corresponding Authors.
SUPPORTING INFORMATION FOR Cytotoxic bagremycins from Mangrove-derived Streptomyces sp. Q22 Lei Chen, Weiyun Chai, Wenling Wang, Tengfei Song, Xiao-Yuan Lian,*,, Zhizhen Zhang,*, *Corresponding Authors.
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Chemical constituents from Agrimonia pilosa Ledeb. and their chemotaxonomic significance Wei-jie Liu, Xue-qian Hou, Hao Chen, Jing-yu Liang*, Jian-bo Sun** Department of Natural
SUPPLEMENTARY MATERIAL Chemical constituents from Agrimonia pilosa Ledeb. and their chemotaxonomic significance Wei-jie Liu, Xue-qian Hou, Hao Chen, Jing-yu Liang*, Jian-bo Sun** Department of Natural
Preparation, isolation and characterization of N α -Fmoc-peptide isocyanates: Solution synthesis of oligo-α-peptidyl ureas
 SUPPORTING INFORMATION Preparation, isolation and characterization of N α -Fmoc-peptide isocyanates: Solution synthesis of oligo-α-peptidyl ureas Vommina V. Suresh Babu*, Basanagoud S. Patil, and Rao Venkataramanarao
SUPPORTING INFORMATION Preparation, isolation and characterization of N α -Fmoc-peptide isocyanates: Solution synthesis of oligo-α-peptidyl ureas Vommina V. Suresh Babu*, Basanagoud S. Patil, and Rao Venkataramanarao
Amino acids-incorporated nanoflowers with an
 Amino acids-incorporated nanoflowers with an intrinsic peroxidase-like activity Zhuo-Fu Wu 1,2,+, Zhi Wang 1,+, Ye Zhang 3, Ya-Li Ma 3, Cheng-Yan He 4, Heng Li 1, Lei Chen 1, Qi-Sheng Huo 3, Lei Wang 1,*
Amino acids-incorporated nanoflowers with an intrinsic peroxidase-like activity Zhuo-Fu Wu 1,2,+, Zhi Wang 1,+, Ye Zhang 3, Ya-Li Ma 3, Cheng-Yan He 4, Heng Li 1, Lei Chen 1, Qi-Sheng Huo 3, Lei Wang 1,*
Mammalian-type Glycosylation l in LEXSY
 Mammalian-type Glycosylation l in LEXSY Case Study: Recombinant hu Erythropoietin Jena Bioscience GmbH Loebstedter Str. 80 07749 Jena, Germany Tel.: +49-3641-628-5000 Fax: +49-3641-628-5100 628 e-mail:
Mammalian-type Glycosylation l in LEXSY Case Study: Recombinant hu Erythropoietin Jena Bioscience GmbH Loebstedter Str. 80 07749 Jena, Germany Tel.: +49-3641-628-5000 Fax: +49-3641-628-5100 628 e-mail:
2. Which of the following amino acids is most likely to be found on the outer surface of a properly folded protein?
 Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Background: Polyketide synthases (PKSs) are bacterial multienzyme systems. Results: The polyketide products of reconstituted act and tcm PKSs were
 Research Paper 607 Heterologous expression, purification, reconstitution and kinetic analysis of an extended type polyketide synthase Robert JX Zawadal and Chaitan Khosla1y213 Background: Polyketide synthases
Research Paper 607 Heterologous expression, purification, reconstitution and kinetic analysis of an extended type polyketide synthase Robert JX Zawadal and Chaitan Khosla1y213 Background: Polyketide synthases
Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study.
 Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study. In this study, one fluorophosphonate (FP, 1), three nitrophenol phosphonate probes
Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study. In this study, one fluorophosphonate (FP, 1), three nitrophenol phosphonate probes
Life Sciences 1A Midterm Exam 2. November 13, 2006
 Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Applying Molecular Networking for the Detection of
 Supporting Information Applying Molecular Networking for the Detection of Natural Sources and Analogues of the Selective Gq Protein Inhibitor FR900359 Raphael Reher, Markus Kuschak, Nina Heycke, Suvi Annala,
Supporting Information Applying Molecular Networking for the Detection of Natural Sources and Analogues of the Selective Gq Protein Inhibitor FR900359 Raphael Reher, Markus Kuschak, Nina Heycke, Suvi Annala,
2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry
 Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
Regulation of P450. b. competitive-inhibitor but not substrate. c. non-competitive inhibition. 2. Covalent binding to heme or protein
 10. Inhibition of P450 Regulation of P450 a. competitive-2 chemicals metabolized by same isoform b. competitive-inhibitor but not substrate c. non-competitive inhibition 11. Induction of P450 1. Non-covalent
10. Inhibition of P450 Regulation of P450 a. competitive-2 chemicals metabolized by same isoform b. competitive-inhibitor but not substrate c. non-competitive inhibition 11. Induction of P450 1. Non-covalent
Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels. Supplementary Information
 Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels Craig T. Armstrong, Philip E. Mason, J. L. Ross Anderson and Christopher E. Dempsey
Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels Craig T. Armstrong, Philip E. Mason, J. L. Ross Anderson and Christopher E. Dempsey
Author(s) Sugihara, Shinji; Orikasa, Yoshitak. Citation Biotechnology Letters, 30(3): 411-4
 An EntD-like phosphopantetheinyl tr Title Photobacterium profundum SS9 comple Moritella marina strain MP-1 involv docosahexaenoic acid Author(s) Sugihara, Shinji; Orikasa, Yoshitak Citation Biotechnology
An EntD-like phosphopantetheinyl tr Title Photobacterium profundum SS9 comple Moritella marina strain MP-1 involv docosahexaenoic acid Author(s) Sugihara, Shinji; Orikasa, Yoshitak Citation Biotechnology
Molecular Biology and Etiology of FOV in Cotton
 Molecular Biology and Etiology of FOV in Cotton Jinggao Liu Alois A. Bell Jaemin Cho Robert D. Stipanovic Lorraine Puckhaber Jose Quintana Southern Plains Agricultural Research Center Agricultural Research
Molecular Biology and Etiology of FOV in Cotton Jinggao Liu Alois A. Bell Jaemin Cho Robert D. Stipanovic Lorraine Puckhaber Jose Quintana Southern Plains Agricultural Research Center Agricultural Research
CS612 - Algorithms in Bioinformatics
 Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
Supplementary Results
 upplementary Results upplementary Table 1. Primers used in this study. upplementary Figure 1. PCP3-TE assay in the absence of ul. upplementary Figure 2. Multiple alignment of the sulfazecin TE, highlighting
upplementary Results upplementary Table 1. Primers used in this study. upplementary Figure 1. PCP3-TE assay in the absence of ul. upplementary Figure 2. Multiple alignment of the sulfazecin TE, highlighting
Post-translational Modifications to Human Bile Acid CoA:Amino Acid N- acyltransferase
 Post-translational Modifications to uman Bile Acid oa:amino Acid - acyltransferase Erin Shonsey UAB Graduate Student September 12, 2006 onjugation of Bile Acids SoA + + 3 2 2 S 3 - hbat oa-s 2 2 S - 3
Post-translational Modifications to uman Bile Acid oa:amino Acid - acyltransferase Erin Shonsey UAB Graduate Student September 12, 2006 onjugation of Bile Acids SoA + + 3 2 2 S 3 - hbat oa-s 2 2 S - 3
LC-MS/MS for the quantification of Peptide biomarker and mixture of closely related Protein in formulation
 EUROPEAN BIOANALYSIS FORUM Barcelona, November 14-16, 2012 LC-MS/MS for the quantification of Peptide biomarker and mixture of closely related Protein in formulation Luc-Alain SAVOY CONTENT Part I: SGS
EUROPEAN BIOANALYSIS FORUM Barcelona, November 14-16, 2012 LC-MS/MS for the quantification of Peptide biomarker and mixture of closely related Protein in formulation Luc-Alain SAVOY CONTENT Part I: SGS
BIOL 158: BIOLOGICAL CHEMISTRY II
 BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
Chapter 3 Substrate specificity of iterative PKS: Role of KS domains
 Chapter 3 Substrate specificity of iterative PKS: Role of KS domains 3.1 INTRODUCTION Type I iterative polyketides are characterized by a single set of catalytic active sites which are used iteratively
Chapter 3 Substrate specificity of iterative PKS: Role of KS domains 3.1 INTRODUCTION Type I iterative polyketides are characterized by a single set of catalytic active sites which are used iteratively
Identification, characterization of a flavin-containing monooxygenase MoA and its function in a
 1 Applied Microbiology and Biotechnology 2 Supplementary Material 3 4 Identification, characterization of a flavin-containing monooxygenase MoA and its function in a specific sophorolipid molecule metabolism
1 Applied Microbiology and Biotechnology 2 Supplementary Material 3 4 Identification, characterization of a flavin-containing monooxygenase MoA and its function in a specific sophorolipid molecule metabolism
Microbial Metabolism Quorum Sensing and Biofilms
 1 Microbial Metabolism Quorum Sensing and Biofilms Ching-Tsan Huang ( 黃慶璨 ) Office: Agronomy Hall, Room 111 Tel: (02) 33664454 E-mail: cthuang@ntu.edu.tw 2 Discovery of Cell-to-Cell Communication Stage
1 Microbial Metabolism Quorum Sensing and Biofilms Ching-Tsan Huang ( 黃慶璨 ) Office: Agronomy Hall, Room 111 Tel: (02) 33664454 E-mail: cthuang@ntu.edu.tw 2 Discovery of Cell-to-Cell Communication Stage
Mass Spectrometry. Mass spectrometer MALDI-TOF ESI/MS/MS. Basic components. Ionization source Mass analyzer Detector
 Mass Spectrometry MALDI-TOF ESI/MS/MS Mass spectrometer Basic components Ionization source Mass analyzer Detector 1 Principles of Mass Spectrometry Proteins are separated by mass to charge ratio (limit
Mass Spectrometry MALDI-TOF ESI/MS/MS Mass spectrometer Basic components Ionization source Mass analyzer Detector 1 Principles of Mass Spectrometry Proteins are separated by mass to charge ratio (limit
Proteins are sometimes only produced in one cell type or cell compartment (brain has 15,000 expressed proteins, gut has 2,000).
 Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Comparison of Biochemical Properties of the Original. and Newly Identified Oleate Hydratases from
 Comparison of Biochemical Properties of the Original and Newly Identified Oleate Hydratases from Stenotrophomonas maltophilia Woo-Ri Kang, a Min-Ju Seo, a Kyung-Chul Shin, a Jin-Byung Park, b Deok-Kun
Comparison of Biochemical Properties of the Original and Newly Identified Oleate Hydratases from Stenotrophomonas maltophilia Woo-Ri Kang, a Min-Ju Seo, a Kyung-Chul Shin, a Jin-Byung Park, b Deok-Kun
Ectopic expression of the minimal whie polyketide synthase generates a library of aromatic polyketides of diverse sizes and shapes
 Proc. Natl. Acad. Sci. USA Vol. 96, pp. 3622 3627, March 1999 Biochemistry Ectopic expression of the minimal whie polyketide synthase generates a library of aromatic polyketides of diverse sizes and shapes
Proc. Natl. Acad. Sci. USA Vol. 96, pp. 3622 3627, March 1999 Biochemistry Ectopic expression of the minimal whie polyketide synthase generates a library of aromatic polyketides of diverse sizes and shapes
Insulin Resistance. Biol 405 Molecular Medicine
 Insulin Resistance Biol 405 Molecular Medicine Insulin resistance: a subnormal biological response to insulin. Defects of either insulin secretion or insulin action can cause diabetes mellitus. Insulin-dependent
Insulin Resistance Biol 405 Molecular Medicine Insulin resistance: a subnormal biological response to insulin. Defects of either insulin secretion or insulin action can cause diabetes mellitus. Insulin-dependent
VOL. 55 NO. 10, OCT THE JOURNAL OF ANTIBIOTICS pp
 VOL. 55 NO. 10, OCT. 2002 THE JOURNAL OF ANTIBIOTICS pp. 919-923 Detection of the Mevalonate Pathway in Streptomyces Species Using the 3-Hydroxy-3- methylglutaryl Coenzyme A Reductase Gene those with only
VOL. 55 NO. 10, OCT. 2002 THE JOURNAL OF ANTIBIOTICS pp. 919-923 Detection of the Mevalonate Pathway in Streptomyces Species Using the 3-Hydroxy-3- methylglutaryl Coenzyme A Reductase Gene those with only
Microwave heating in peptide side chain modification via sulfhydryl reaction
 Microwave heating in peptide side chain modification via sulfhydryl reaction E. Calce and S. De Luca* Institute of Biostructures and Bioimaging, National Research Council, 80134 Naples, Italy stefania.deluca@cnr.it
Microwave heating in peptide side chain modification via sulfhydryl reaction E. Calce and S. De Luca* Institute of Biostructures and Bioimaging, National Research Council, 80134 Naples, Italy stefania.deluca@cnr.it
Steps Towards Deciphering the Post-Polyketide Synthase Tailoring Steps in the Phoslactomycin Biosynthesis Pathway
 Portland State University PDXScholar Dissertations and Theses Dissertations and Theses Spring 7-23-2015 Steps Towards Deciphering the Post-Polyketide Synthase Tailoring Steps in the Phoslactomycin Biosynthesis
Portland State University PDXScholar Dissertations and Theses Dissertations and Theses Spring 7-23-2015 Steps Towards Deciphering the Post-Polyketide Synthase Tailoring Steps in the Phoslactomycin Biosynthesis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Problem-solving Test: The Mechanism of Protein Synthesis
 Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
Interlocking of primary and secondary metabolism in antibiotic-producing actinomycetes
 Interlocking of primary and secondary metabolism in antibiotic-producing actinomycetes E. Stegmann 1, B. adatsch 1, C. Kittel 1,. Puk 1, R. Menges 1, R. Süßmuth 2, W. Wohlleben 1 1 Institute of Microbiology,
Interlocking of primary and secondary metabolism in antibiotic-producing actinomycetes E. Stegmann 1, B. adatsch 1, C. Kittel 1,. Puk 1, R. Menges 1, R. Süßmuth 2, W. Wohlleben 1 1 Institute of Microbiology,
New Insights into Peptidoglycan Biosynthesis. Louis B. Rice Warren Alpert Medical School of Brown University Providence, RI
 New Insights into Peptidoglycan Biosynthesis Louis B. Rice Warren Alpert Medical School of Brown University Providence, RI Outline of Presentation Review role of penicillin-binding proteins in cell wall
New Insights into Peptidoglycan Biosynthesis Louis B. Rice Warren Alpert Medical School of Brown University Providence, RI Outline of Presentation Review role of penicillin-binding proteins in cell wall
Chapter 4. Erythronolide B and the Erythromycins
 124 Chapter 4 Erythronolide B and the Erythromycins Isolation and tructure Isolated in 1952 from the soil bacteria actinomycetes, 1 the erythromycins are a distinguished family of natural products by virtue
124 Chapter 4 Erythronolide B and the Erythromycins Isolation and tructure Isolated in 1952 from the soil bacteria actinomycetes, 1 the erythromycins are a distinguished family of natural products by virtue
Graphene Quantum Dots-Band-Aids Used for Wound Disinfection
 Supporting information Graphene Quantum Dots-Band-Aids Used for Wound Disinfection Hanjun Sun, Nan Gao, Kai Dong, Jinsong Ren, and Xiaogang Qu* Laboratory of Chemical Biology, Division of Biological Inorganic
Supporting information Graphene Quantum Dots-Band-Aids Used for Wound Disinfection Hanjun Sun, Nan Gao, Kai Dong, Jinsong Ren, and Xiaogang Qu* Laboratory of Chemical Biology, Division of Biological Inorganic
